Abstract
Osteopontin (OPN) plays an important role in cancer progression, however its prognostic significance and its downstream factors are largely elusive. In this study, we have shown that expression of OPN was significantly higher in bladder cancer specimens with higher T-stage or tumor grades. In addition, a high level of OPN was significantly associated with poorer survival in two independent bladder cancer patient cohorts totaling 389 bladder cancer patients with available survival data. We further identified Matrix metallopeptidase 9 (MMP9) and S100 calcium-binding protein A8 (S100A8) were both downstream factors for OPN in bladder cancer specimens and bladder cancer cell lines. Expression of OPN was significantly positively associated with that of MMP9 and S100A8, while overexpression of OPN resulted in upregulation of MMP9 and S100A8, and knockdown of OPN showed consistent downregulation of MMP9 and S100A8 expression levels. Importantly, expression levels of both MMP9 and S100A8 were significantly associated with higher T-stage, higher tumor grade and a shorter survival time in the bladder cancer patients. Interestingly, OPN expression only predicted survival in MMP9-high, but not MMP9-low subgroups, and in S100A8-low but not S100A8-high subgroups. Our results suggest that OPN, MMP9 and S100A8 all play a significant role in bladder cancer progression and are potential prognostic markers and therapeutic targets in bladder cancer. The mechanistic link between these three genes and bladder cancer progression warrants further investigation.
Keywords: Osteopontin, MMP9, S100A8, bladder, prognosis.
Introduction
Bladder cancer is a major cause of cancer-related death worldwide, with about 380,000 new cases and 150,000 deaths per year 1. More than 90% of bladder cancers are transitional cell carcinoma (TCC), and around 5% and 2% remaining cases are squamous cell carcinoma and adenocarcinoma, respectively 2. TCC can be further divided into muscle invasive or non-muscle invasive; with muscle invasive bladder cancer has a poorer prognosis and a 10-year survival rate of only 50% 2.
Osteopontin (OPN) is a phosphorylated sialoprotein in human encoded by highly conserved gene SPP1. OPN is expressed approximately as 34-kDa protein, and its full-length form is of 60-kDa 3 and secreted by various cells and plays an important role in cellular processes related to cancer development 4. It has specific splice variants: OPN-a, OPN-b and OPN-c 5. OPN-c has been known to be a strong prognosis marker in breast, ovarian, lung and colorectal cancer cells 5 and found expressed in bladder cancer cells 6. Alteration in numerous malignant cells is reported to induce higher OPN expression compared to normal cells 7, 8. The higher the OPN expression, more drastic effects are observed in cell adhesion, cell proliferation, migration and extracellular matrix (ECM) invasion rates through interacting with αvβ3 integrin that is present in a variety of cells. OPN exists as a soluble cytokinase and a constituent of ECM, which ECM has a distinctive composition - the type IV collagen 9, 10. OPN is shown to promote chemoresistance 11, stem cell properties / phenotype 12, autophagy 13, angiogenesis 14, cell migration 15, cell invasion 16, 17, metastasis via epithelial-mesenchymal transition 18, 19 and immunosuppression in metastatic niche 20 in different cancers. It is also a prognostic marker in human cancers 21 including breast cancer 22, 23, colorectal cancer 24, lung cancer 25, 26, head and neck squamous cell carcinoma 27, gastric cancer 28, 29 and prostate cancer 30. We have also previously shown that OPN promotes breast cancer progression 23, and have identified its upstream 31, 32 and downstream targets 33. Nonetheless, little is known regarding the role of OPN in bladder cancer.
Previously, Ang et al. demonstrated that OPN plasma levels were elevated in higher stage of bladder transitional cell carcinoma 34. Zhao also showed that high OPN plasma level was related to the more aggressive phenotype of bladder cancer 35. Interestingly, plasma OPN level has been used to predict the disease stage and reoccurrence incidents showing similar results with higher level of plasma OPN found to be significantly associated with both higher T-stage and lymph node invasion 36. Further study also revealed that increased level of OPN mRNA was associated with both muscle invasiveness and advanced disease stage, however, with a limited number of specimens (n = 77), no statistically significant survival difference was found between OPN-high and OPN-low groups of patients 37. Study of OPN in vitro also found that knockdown of OPN in human bladder cancer cells reduced its proliferation and migration 38. These studies have demonstrated the importance of OPN in bladder cancer progression. Nevertheless, the downstream targets of OPN and the mechanisms of OPN-mediated bladder cancer progression are largely unknown, and the prognostic significance of mRNA expression of OPN in bladder cancer remains a question. In this study, we aimed to investigate the expression level of OPN and its prognostic value and identified potential downstream targets of OPN in bladder cancer.
Results
OPN overexpression in bladder cancer specimens
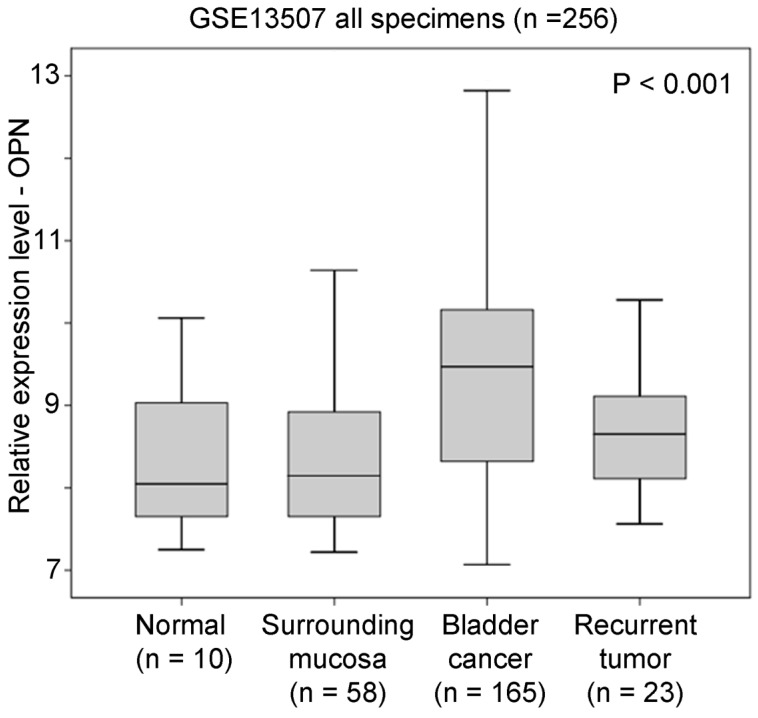
In GSE13507 datasets, there were four different types of specimens, including normal, surrounding mucosa, bladder cancer, and recurrent tumor. An ANOVA analysis showed that OPN expression was significantly higher in bladder cancer compared to other specimens (p < 0.001, Figure 1). By post-hoc analysis, bladder cancer had a significantly higher level of OPN expression compared to surrounding mucosa (Games-Howell test, p < 0.001), suggesting that OPN may play an important role in early development of bladder cancer.
Figure 1.
The association between OPN expression and bladder cancer. A box plot showing the expression level of OPN in different bladder specimens.
The association between OPN expression and clinicopathological parameters in bladder cancer
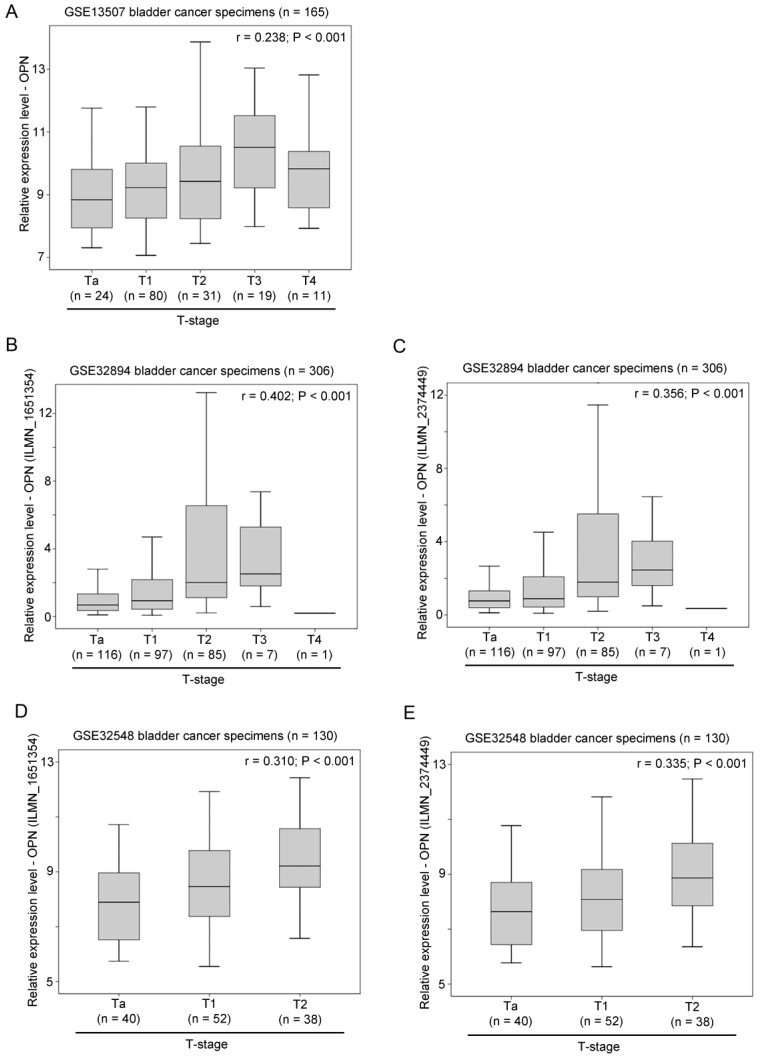
The association between OPN expression and T-stage was tested in three independent bladder cancer cohorts. In GSE13507, OPN expression was significantly positively correlated with T-stage (r = 0.238, p < 0.001; Figure 2A). Similar results were obtained in GSE32894 and GSE32548. In these two datasets, there were two probes for OPN and these two probes (ILMN_1651354 and ILMN_2374449) were tested independently. As shown in Figure 2B and 2C, OPN expression, as dictated by ILMN_1651354 and ILMN_2374449, was significantly positively correlated with T-stage, respectively, in GSE32894 (r = 0.402, p < 0.001 and r = 0.356, p < 0.001, respectively). As shown in Figure 2D and 2E, OPN expression was significantly positively correlated with T-stage in GSE32548 for the probe IDs of ILMN_1651354 (r = 0.310, p < 0.001) and ILMN_2374449 (r = 0.335, p < 0.001).
Figure 2.
The association between OPN expression and T-stage. (A) A box plot showing the expression level of OPN in bladder cancer with different stages in patient cohort GSE13507. Box plots showing the expression level of OPN based on the probe (B) ILMN_1651354 and (C) ILMN_2374449 in bladder cancer with different stages in bladder in patient cohort GSE32894. Box plots showing the expression level of OPN based on the probe (D) ILMN_1651354 and (E) ILMN_2374449 in bladder cancer with different stages in in bladder in patient cohort GSE32548.
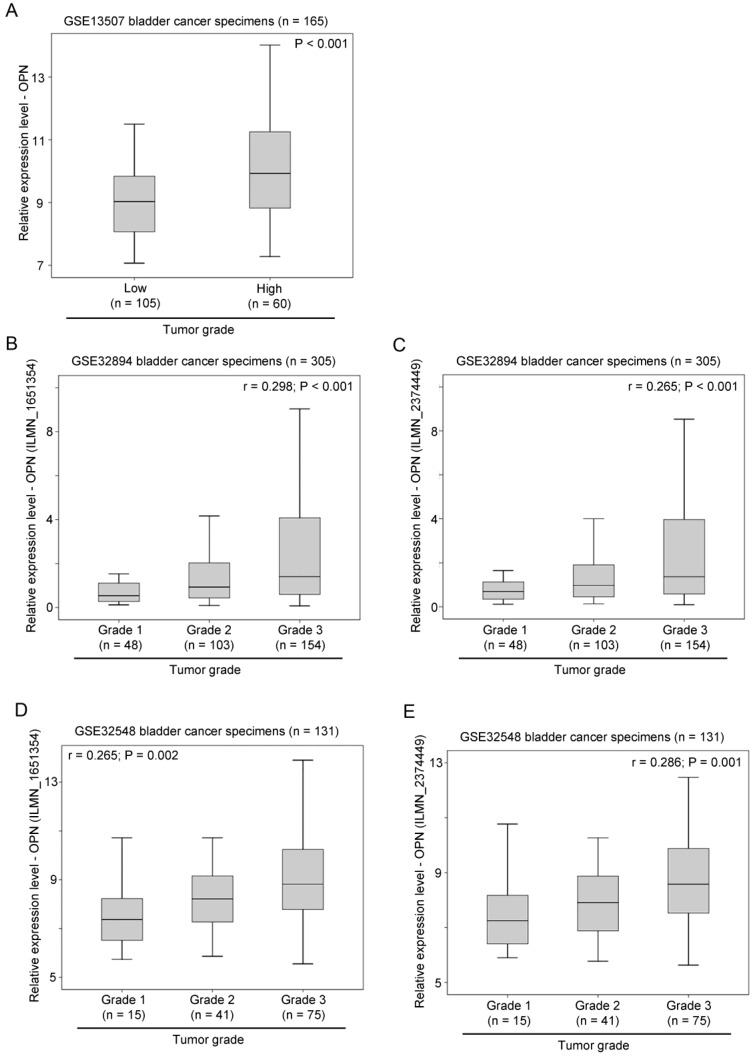
The association between OPN expression and tumor grading was also tested in three independent bladder cancer cohorts. In GSE13507, OPN expression was significantly higher in tumors with a high histological grade compared to those with a low histological grade (p < 0.001; Figure 3A). Similarly, in GSE32894, OPN expression was significantly positively correlated with tumor grading for probe ILMN_1651354 (r = 0.298, p < 0.001; Figure 3B) and probe ILMN_2374449 (r = 0.265, p < 0.001; Figure 3C). In another cohort, GSE32548, OPN expression was also significantly positively correlated with tumor grading for probe ILMN_1651354 (r = 0.265, p = 0.002; Figure 3D) and (r = 0.286, p = 0.001; Figure 3E).
Figure 3.
The association between OPN expression and tumor grade. (A) A box plot showing the expression level of OPN in bladder cancer with different tumor grades in patient cohort GSE13507. Box plots showing the expression level of OPN based on the probe (B) ILMN_1651354 and (C) ILMN_2374449 in bladder cancer with different tumor grades in bladder in patient cohort GSE32894. Box plots showing the expression level of OPN based on the probe (D) ILMN_1651354 and (E) ILMN_2374449 in bladder cancer with different tumor grades in in bladder in patient cohort GSE32548.
The association between OPN expression and survival in bladder cancer patients
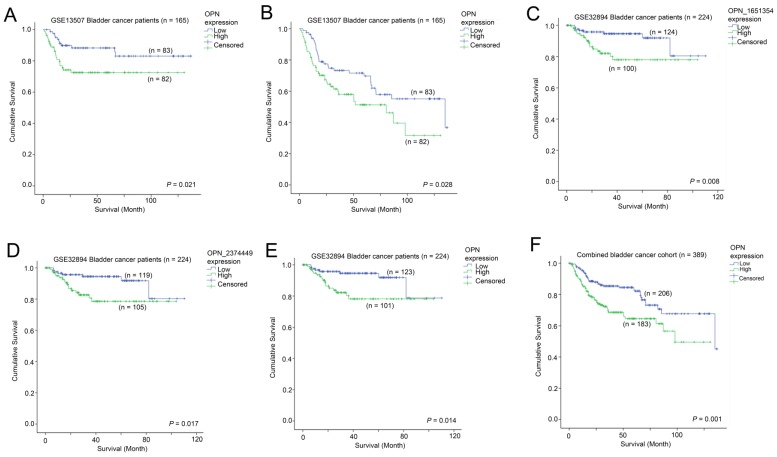
The association between OPN expression and survival was tested in two independent bladder cancer cohorts in which survival data was available. In GSE13507, a high-level expression of OPN was significantly associated with a shorter disease specific survival time (p = 0.021; Figure 4A), while it was also significantly associated with a shorter overall survival time (p = 0.028; Figure 4B). Similarly, in GSE32894, a high-level expression of OPN was significantly associated with a shorter overall survival time for probe ILMN_1651354 (p = 0.008; Figure 4C), probe ILMN_2374449 (p = 0.017; Figure 4D) and the average of the two probes (p = 0.014; Figure 4E). When the data of the two cohorts were combined, patients whose tumors expressed OPN at a high-level had a mean survival time of 85.5 months (95% CI = 75.4 - 95.7 months) and those patients whose tumors expressed OPN at a low level had a mean survival time of 107.1 (95% CI = 98.7 - 115.5). A high-level expression of OPN was significantly associated with a shorter overall survival time in these 389 bladder cancer patients (p = 0.001; Figure 4F).
Figure 4.
The association between OPN expression and survival. Kaplan-Meier analysis showing survival curves for patients whose tumor expressed a high-level (Green) and a low level (Blue) of OPN in bladder cancer patients for (A) cancer specific survival and (B) overall survival in GSE13507. Kaplan-Meier analysis showing survival curves for patients whose tumor expressed a high-level (Green) and a low level (Blue) of OPN in bladder cancer patients for overall survival based on (C) probe ILMN_1651354, (D) ILMN_2374449 and (E) combined OPN average in GSE32894. (F) Kaplan-Meier analysis showing survival curves for the combined bladder cancer patient cohort.
Identification of OPN co-expressing genes in bladder cancer
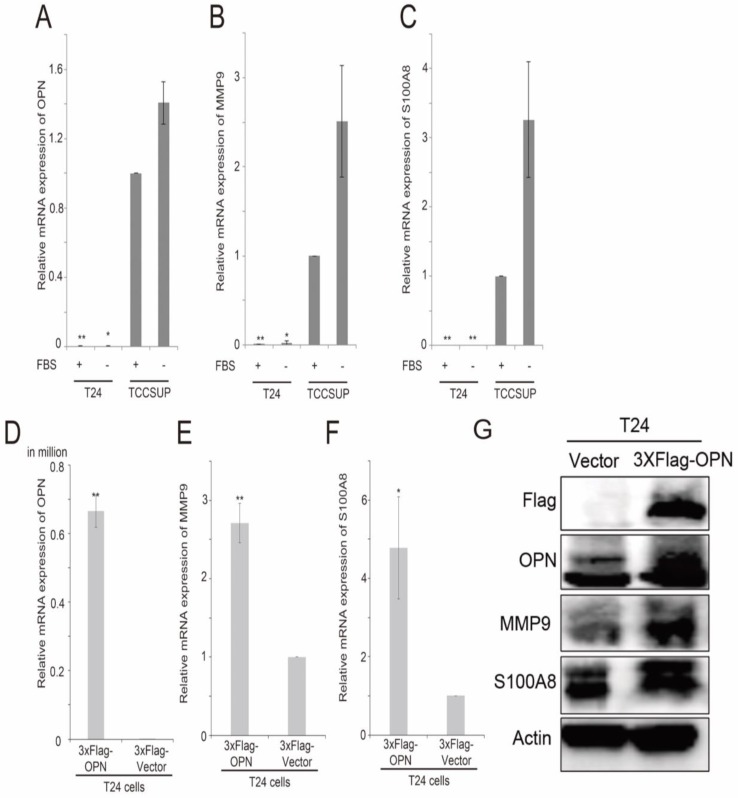
By using 2-sample Welch's T-test, several genes were found to be consistently and significantly differentially expressed in the two bladder cancer datasets, GSE13507 and GSE32894, and have been shown to have a link in published articles. Two metastasis-associated genes, S100A8 and MMP9, three cluster of differentiation genes, CD14, CD80 and CD163, and two oxygen pathway genes, MOX1 and NOX4, were found to be significantly higher in OPN-high tumors compared to OPN-low tumors. The correlation between the expression levels of these genes was analyzed by Spearman's rank test, and we found that the expression levels of these genes were significantly correlated with that of OPN (Table 1). The expression levels of S100A8 and OPN have both been shown to be increased in stroma in prostate cancer with loss of JunB expression 39. Knockdown of OPN in colon cancer suppressed the expression of MMP9 40. Since OPN is a metastasis-associated gene, the correlation between OPN and MMP9, and between OPN and S100A8 were investigated in vitro. We tested the expression levels of OPN, MMP9 and S100A8 in two high grade bladder cancer cell lines, T24 and TCCSUP cells. Interestingly, TCCSUP cells expressed OPN (Figure 5A), MMP9 (Figure 5B) and S100A8 (Figure 5C) at a significantly higher level compared to T24 cells regardless of the availability of serum. The gene expression level of OPN was observed highest in serum-free and growth media of TCCSUP cells (grade IV), which was about 85.9-fold change, and lesser than one-fold for T24 cells. Since T24 cells expressed a low level of OPN, MMP9 and S100A8, this cell line was used to further study the association between these three genes via OPN overexpression. This is accomplished using a flag epitope tagged to the amplified OPN gene. Overexpression of OPN in T24 cells resulted in an increased mRNA expression of OPN (Figure 5D), MMP9 (Figure 5E) and S100A8 (Figure 5F). The results were confirmed in gene expression level with a fold change of 4.8 and 2.7 for S100A8 and MMP9 respectively, and with western blot analysis. As shown in Figure 5G, overexpression of OPN resulted in an increase in OPN, MMP9 and S100A8 protein levels in T24 cells. The protein levels of S100A8 and MMP9 corresponded to the elevated OPN expression, and the intensity of S100A8 band was increased significantly, while MMP9 band demonstrated obvious clear band compared to the vector control. Additionally, we performed an siRNA transient transfection to knockdown the OPN in TCCSUP cell line and a decrease in MMP9 and S100A8 protein expression levels were observed respectively (Figure S1). These collective results clearly propose that MMP9 and S100A8 are the downstream targets of OPN-mediated metastasis.
Table 1.
GSE13507 Bladder cancer dataset (n = 165); Correlation of CD133 and other gene expressions
| OPN | S100A8 | MMP9 | CD14 | CD80 | CD163 | HMOX1 | NOX4 | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Spearman's rho test | OPN | Correlation Coefficient | 1.000 | 0.394 | 0.475 | 0.440 | 0.366 | 0.550 | 0.432 | 0.278 |
| P value | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | |||
| S100A8 | Correlation Coefficient | 0.394 | 1.000 | 0.454 | 0.419 | 0.213 | 0.413 | 0.477 | 0.316 | |
| P value | <0.001 | <0.001 | <0.001 | 0.006 | <0.001 | <0.001 | <0.001 | |||
| MMP9 | Correlation Coefficient | 0.475 | 0.454 | 1.000 | 0.633 | 0.491 | 0.630 | 0.537 | 0.236 | |
| P value | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | 0.002 | |||
| CD14 | Correlation Coefficient | 0.440 | 0.419 | 0.633 | 1.000 | 0.480 | 0.823 | 0.486 | 0.198 | |
| P value | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | 0.011 | |||
| CD80 | Correlation Coefficient | 0.366 | 0.213 | 0.491 | 0.480 | 1.000 | 0.463 | 0.351 | 0.161 | |
| P value | <0.001 | 0.006 | <0.001 | <0.001 | <0.001 | <0.001 | 0.039 | |||
| CD163 | Correlation Coefficient | 0.550 | 0.413 | 0.630 | 0.823 | 0.463 | 1.000 | 0.596 | 0.134 | |
| P value | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | 0.085 | |||
| HMOX1 | Correlation Coefficient | 0.432 | 0.477 | 0.537 | 0.486 | 0.351 | 0.596 | 1.000 | 0.056 | |
| P value | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | 0.477 | |||
| NOX4 | Correlation Coefficient | 0.278 | 0.316 | 0.236 | 0.198 | 0.161 | 0.134 | 0.056 | 1.000 | |
| P value. | <0.001 | <0.001 | 0.002 | 0.011 | 0.039 | 0.085 | 0.477 | |||
Figure 5.
The analysis of expression levels of OPN, MMP9 and S100A8 in bladder cancer cell lines. Histograms showing qPCR analysis for (A) OPN, (B) MMP9 and (C) S100A8 in T24 and TCCSUP bladder cancer cell lines. Histograms showing qPCR analysis for (D) OPN, (E) MMP9 and (F) S100A8 in T24 cells overexpressing OPN and vector control and (G) western blot analysis for protein levels of OPN, MMP9, S100A8 and Actin in T24 cells overexpressing OPN (3xF-OPN) and vector control (3xFlag). n=9, **p<0.05 vs. control and is statistically analyzed with T-test.
The associations between expression of MMP9 and S100A8, and clinicopathological parameters in bladder cancer
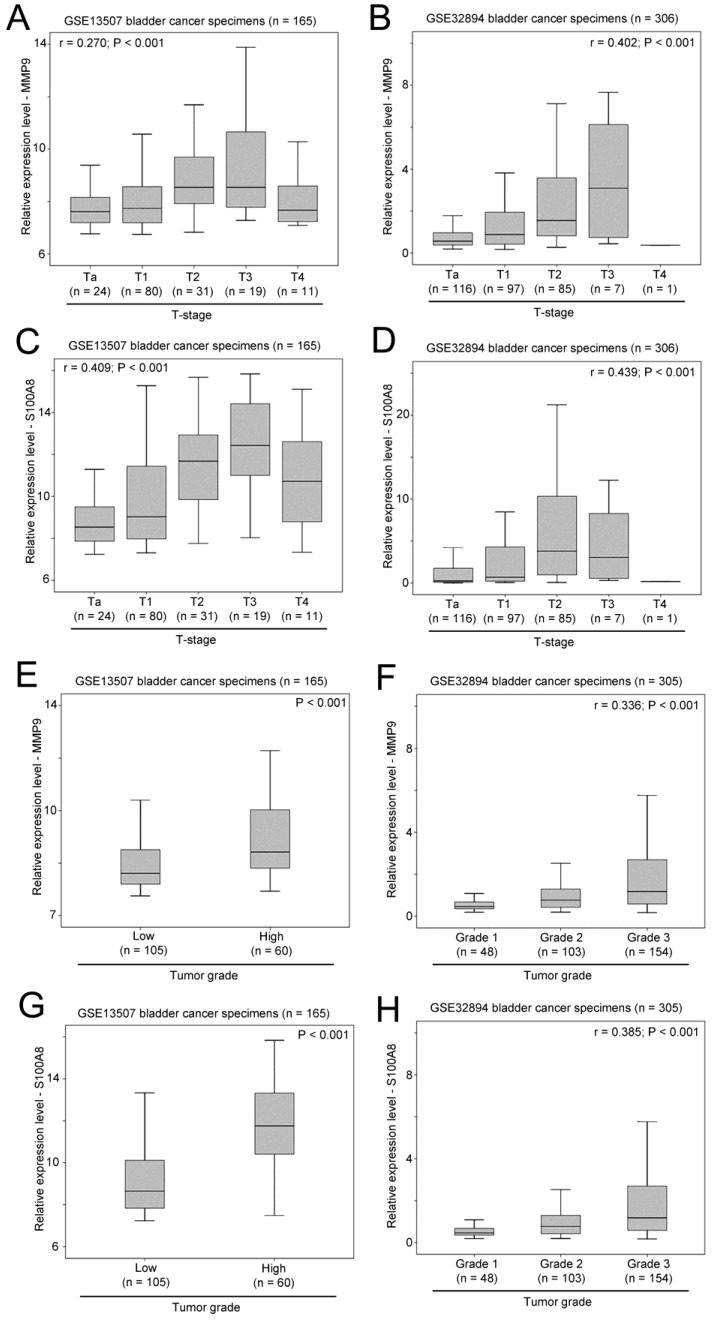
As shown in Figure 6A and 6B, a significant positive correlation was observed between MMP9 expression and T-stage in two independent bladder cancer cohorts. Similarly, the expression levels of S100A8 was also significantly positively correlated with T-stage in these two bladder patient cohorts (Figure 6C and 6D). In addition, increased levels of expression of MMP9 (Figure 6E and 6F) and S100A8 (Figure 6G and 6H) were observed in bladder cancer specimens with higher tumor grade in both cohorts. Our results suggest that both the expression levels of MMP9 and S100A8 are associated with a more aggressive phenotypes of bladder cancer.
Figure 6.
The association between expression levels of MMP9 and S100A8, and clinicopathological parameters. Box plots showing the expression levels of MMP9 in bladder cancer specimens with different T-stage in patient cohort (A) GSE13507 and (B) GSE32894. Box plots showing the expression levels of S100A8 in bladder cancer specimens with different T-stage in patient cohort (C) GSE13507 and (D) GSE32894. Box plots showing the expression levels of MMP9 in bladder cancer specimens with different tumor grades in patient cohort (E) GSE13507 and (F) GSE32894. Box plots showing the expression levels of S100A8 in bladder cancer specimens with different tumor grades in patient cohort (G) GSE13507 and (H) GSE32894.
The associations between expression of MMP9 and S100A8, and survival in bladder cancer
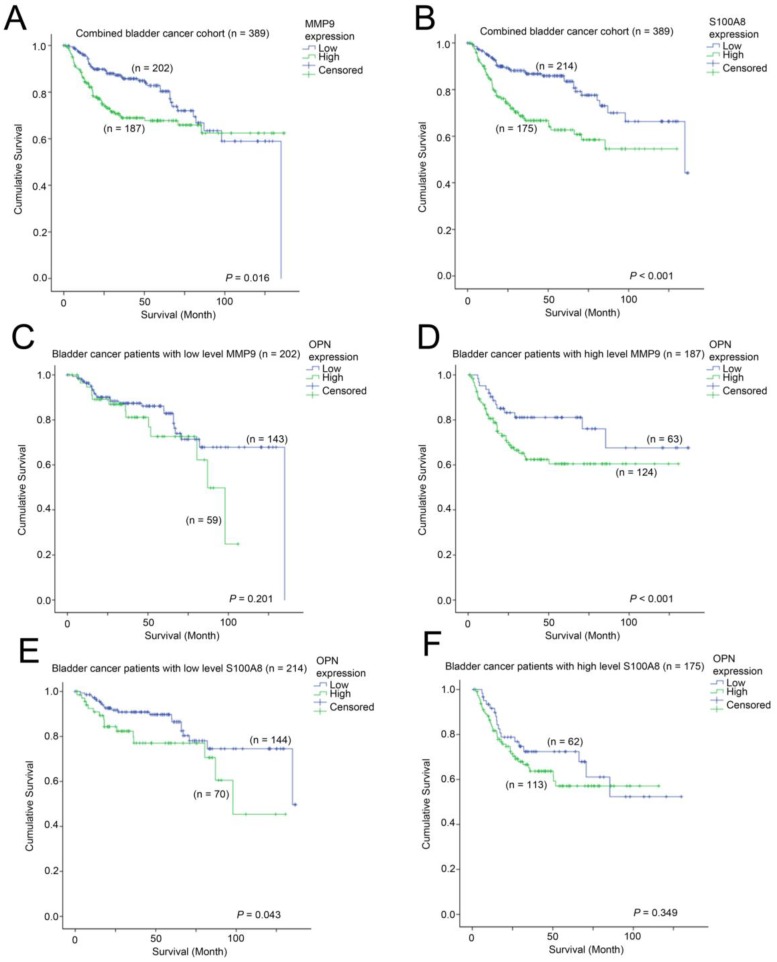
We further investigated whether MMP9 and S100A8 are prognostic indicators in bladder cancer. In patients, whose bladder cancer had a low level of expression of MMP9 had a mean survival time of 102 months (95% CI = 93 - 111 months) compared to those patients whose bladder cancer had a mean survival time of 95 months (95% CI = 86 - 105 months). As shown in Figure 7A, a high-level of MMP9 was significantly associated with a shorter survival time in bladder cancer patients (p = 0.016). Similarly, patients whose bladder cancer expressed a high-level of S100A8 had a mean survival time of 84 months (75 - 94 months), which was significantly shorter compared to those patients whose bladder cancer expressed a low level of S100A8, with a mean survival time of 108 months (95% CI = 100 - 117 months, p < 0.001; Figure 7B). We also investigated how these two genes interacted with OPN-survival association in these two bladder cancer patient cohorts. While the survival time was not significantly different between patients whose tumor expressed a low level of OPN vs. those with a high-level of OPN in the MMP9-low patient subgroup (Figure 7C), while in the MMP9-high subgroup, patients whose bladder cancer had a high-level of OPN expression had a significantly shorter survival time compared to those patients whose cancer had a low level of OPN-expression (p = 0.037; Figure 7D). In contrast, OPN was significantly associated with survival only in bladder cancer patients whose cancer had a low level expression of S100A8 (p = 0.043; Figure 7E), but not in those with a high-level expression of S100A8 (Figure 7F).
Figure 7.
The association between MMP9, S100A8 and OPN with survival. Kaplan-Meier analysis showing survival curves for patients expressing different levels of (A) MMP9 and (B) S100A8 in the combined bladder cancer patient cohort. Kaplan-Meier analysis showing survival curves for patients expressing different levels of OPN in bladder cancer patients whose tumor expressed (C) a low level of MMP9, (D) a high-level of MMP9, (E) a low-evel of S100A8 and (F) a high-level of S100A8.
Discussion
OPN has been shown to play an important role in disease progression in multiple types of cancer 5, 6, 21. The upregulated OPN level is shown to correlate with malignant grade and metastasis 5, 6, 41. One reason could be due to the OPN production by lymphocytes and macrophages although they are not expressed in circulating monocytes. These white blood cells are reported by Sodek and colleagues (2000) to induce OPN concentration in diseased tissues 42. Furthermore, those compiled GEO datasets have revealed and supported co-expressing genes upregulated along with higher OPN expression in advanced bladder cancer. Subsequently, we decided to further study MMP9 and S100A8 to establish the correlation between OPN and these co-expressed genes. MMP9 has been reported to be co-expressed with OPN 43, 44, while S100A8 with OPN in several publications 39, 45. Our results showed that the upregulation of MMP9 and S100A8 gene and protein levels correspond to the OPN overexpression and downregulation of these genes/proteins with knockdown of OPN. Matrix metalloproteinase, (MMP) family is an important ECM-degrading proteinase 7, 46, 47 and one of metalloproteinase activities is to facilitate cell migration 48. ECM, however, consists a barrier that prevents migration. Type IV collagen, a distinctive component of ECM and is degraded by the enzyme encoded by MMP9 gene because it consists of a collagen binding domain which is located between the catalytic region and C-terminal hemopexin domain 46. Therefore, breaking down the collagen barrier is necessary for migration with ECM degradation process. MMP claims to selectively degrade certain components of ECM and subsequently releases growth factors that exist within the ECM 46. Stawarski et al. reported that intercellular intergrin signaling pathway is activated when the ECM protein is cleaved 49. OPN molecule can be cleaved by several MMPs, including MMP9 resulting in an N-terminal section consists of the RGD sequence 50. The highly conserved RGD domain of OPN interacts with several integrins, such as αvβ3, and this interaction (OPN/αvβ3) mediates cell adhesion and migration 42. Lenga et al. found that OPN-/- fibroblasts have low attachment to collagen gels and impair the formation of super mature focal adhesion which cause weak adhesions to ECM 51. This depicts the importance of OPN in affecting and changing the cancer phenotypic characteristics.
MMP9 has been reported to be involved in the pathogenesis of bladder cancer and other cancers when it is upregulated 47, 52. Both pro-MMP9 and its active form have a PEX-domain that either docks or/and form heterodimeratization with the surface receptor, CD44 in the OPN molecule of T24 cells. This docking is predicted to eventually lead to cell survival via Lyn kinase activation and heterodimerization binding that causes subsequent phosphorlysation of kinase effectors, effect upon EGFR activation. MMP9 active form supports cell survival and migration, thus this suggests why MMP9 and OPN expression levels are higher in advanced invasive cancer cells, e.g., TCCSUP compared to T24 46. Therefore, upregulation of OPN and its co-expressing gene, MMP9 play an important role in cancer cell migratory and adhesive properties.
In bladder cancer, overexpression of OPN is correlated with cancer-specific survival of upper urinary tract urothelial carcinoma with limited number specimens 53. In the present study, we found that a high-level expression of OPN mRNA was significantly associated with T-stage and tumor grade, and most importantly, it was consistently associated with shorter disease-specific and overall survival in two independent bladder cancer patient cohorts. This finding is consistent with the study published in 2017 by Hussain et al 41 which revealed that OPN was significantly overexpressed in invasive bladder cancer compared to healthy tissue. Importantly, we identified two metastasis-related genes, MMP9 and S100A8, that could potentially be the downstream targets of OPN promoting bladder cancer progression. These two genes were upregulated by OPN overexpression in bladder cancer cell lines, and a high-level expression of these two genes, individually, was also associated with higher T-stage, tumor grade, and poorer survival. Meanwhile, we investigated the siRNA knockdown of OPN gene in TCCSUP cell line, we found that the downregulation of OPN reduced the expression of MMP9 and S100A8. Wu et al. reported that knockdown of OPN has inhibited the MMP9 expression 40 and Lim et al. observed that MMP9 is downregulated in S100A8 knockdown cancer cells 54. Our results support and further extend previous publication studies. Our results suggest that OPN could be an important prognostic marker and a therapeutic target in bladder cancer, and that MMP9 and S100A8 could be potential downstream targets for OPN-mediated metastasis.
In general, the MMP9 expression level is notably lower than OPN expression in the cell basal level because MMP9 is secreted in its latent form and generated upon activation. The OPN/αvβ3 signaling is known to control CD44 surface expression, MMP9 stimulation, and migration in osteoclast 42, 55. We hypothesized that OPN/αvβ3 signaling would cause similar effect in T24 cells. Phillip et al. reported that OPN does stimulate tumor growth, cell migration and invasion; it also further activates the matrix metalloproteinases 7. OPN/αvβ3 signaling can potentiate MMP production by activating the downstream kinase effector, the FAK in addition to the ERK pathway. This pathway not only promotes Fos expression via phosphorylation, which further stimulates MMPs assembly, but also directly activate pro-MMP2 42. MMP9 (82 kDa) is secreted in its proenzyme form, pro-MMP9 (92 kDa).
The relationship between OPN and MMP9 has been demonstrated in aortic mesenchymal cells, in which OPN treatment of vascular smooth muscle cells enhanced pro-MMP9 activity 43, and MMP9 was upregulated by OPN in cardiac and skeletal muscle resulting in cardiac abnormalities 56. In gastric cancer, OPN also promotes metastasis via MMP9 activation 57 while in colon cancer cells, knockdown of OPN resulted in reduced expression of MMP9 44. MMP9 activity has been shown to be higher in invasive bladder cancer compared to normal control 58. Knockdown of MMP9 by microRNAs in bladder cancer cells also resulted in decreased cell invasion 59. However, the prognostic significance of MMP9 has not yet been fully elucidated. In the present study, we have shown that increased expression of MMP9 was correlated with higher T-stage, tumor grade and a shorter survival in bladder cancer. Our results suggest that MMP9 may be an important prognostic marker for bladder cancer. Interestingly, since OPN-survival association was only observed in MMP9-high patient subgroup, OPN-mediated bladder cancer progression may be dependent on the presence of high-level of MMP9.
Some of the intercellular functions that are affected when some S100 proteins undergo conformational changes are regulation of calcium homeostasis, cell growth and migration, cell cycle and phosphorylation 60. Any alterations during cell development could promote potential oncogenesis progression. Little is known about the relationship between OPN and S100A8. A highly metastatic colon cancer cell line expressed elevated levels of OPN, MMP9 and S100A8 45, while in a prostate cancer model, the tumor stroma was altered with increased expression of OPN and S100A8 39. S100A8 was demonstrated to have higher expression in bladder cancer compared to normal bladder tissue 61, and S100A8 alone was shown to be a prognostic marker 62 while an S100A8 gene signature was shown to be a strong predictor of disease progression for bladder cancer 63. In line with these results, our results showed that a high-level expression of S100A8 was associated with higher T-stage and tumor grade, and a shorter survival time.
From our results, we have verified that the OPN overexpression lead to an increase in MMP9 and S100A8 expression, while the knockdown of OPN showed converse effect in MMP9 and S100A8 expression levels. Based on these results, we evidently proposed that MMP9 and S100A8 are downstream of OPN. The expression levels of these three genes were positively correlated in human bladder cancer specimens, the mechanism of OPN-mediated upregulation of MMP9 and S100A8 was not investigated in the present study. Although we found that OPN, MMP9 and S100A8 are all prognostic markers for bladder cancer patients, we have not fully elucidated how the interactions of these three genes promote bladder cancer progression in this study. The establishment of mechanistic insight on the interactions of these three genes with bladder cancer progression will lead to a better understanding of how OPN promotes bladder cancer progression. Nonetheless, our results have demonstrated that OPN may regulate MMP9 and S100A8 to promote bladder cancer progression, and the expression levels of OPN may be a prognostic marker and therapeutic target for human bladder cancer.
In conclusion, taken together, OPN, MMP9 and S100A8 all play significant roles in bladder cancer progression and are potential prognostic markers and therapeutic targets in bladder cancer.
Materials & Methods
Cell lines and reagents
Two poorly differentiated bladder cancer cell lines T24 and TCCSUP were purchased from ATCC. These cell lines were grown in cell culture media (T24 in McCoy's 5a medium modified, Gibco: 16600082; TCCSUP in Minimum essential medium Gibco: 11095080 and 1 mM sodium pyruvate) added with 10% fetal bovine serum, 1% penicillin/streptomycin.
RNA purification and cDNA preparation
Gene expressions were induced in all bladder cell lines by culturing in 10% serum and another batch following a 24-h duration of serum starvation prior RNA purification (RNeasy Mini Kit, 74106), followed by converting 1 μg of samples to synthesized cDNA using a thermal cycler (C1000 Touch, Bio Rad) (High Capacity cDNA Reverse Transcription Kit, 4368813, Applied Biosystems).
qPCR validation
The amounts of three target genes such as OPN, MMP9 and S100A8 cDNA present in the bladder cell lines were quantified by quantitative PCR using TaqMan probes using GAPDH as an endogenous control. Equivalent amount of cDNA was subjected to quantitative PCR detection. The final concentration of the 1X master mix (Taqman Universal PCR Master Mix, #4304437) followed by respective probes (SPP1: Hs00959010_m1; MMP9: Hs00234579_m1; S100A8: Hs00374264_g1; GAPDH: Hs02758991_g1; Life Technologies). The parameters were set using the Bio-Rad CFX Manager where FAM scan mode detection with 40 cycles were selected. Data normalization was accomplished using the GAPDH and normalized values were subjected to a 2-ΔΔCt formula to quantify the fold change. Individual CT and intrasample variable values, the mean ± SD were calculated from the generated results.
Total protein quantification and western blot analysis
The cell line samples were homogenized with ice-cold RIPA lysis buffer that was added with protease inhibitor (Complete EDTA-free, #10634200, Roche) and phosphatase inhibitor (PhosSTOP, #04906837001, Roche) and centrifuged at 20,000 x g for 30 minutes at 4°C. Supernatants were collected and kept in -80°C. Bradford method was used to quantify the protein concentration required for western blot sample loading (Pierce BCA Protein Assay Kit, #23225). All samples were dissolved in 1X LDS sample buffer and 1X reducing agent (Life Technologies) and heated for 5 minutes at 95°C. An equal concentration of protein was electrophoresed and separated in NuPage 4-12% Bis-Tris gel (Invitrogen, #NP0322) and transferred to a 0.2 um nitrocellulose membrane (#IB301032) using iBlot (Invitrogen). Blocking was carried out in 5% bovine serum albumin, 0.1% Tween 20 in PBS for non-specific binding for an hour at room temperature. The membranes were incubated overnight at 4°C with mouse monoclonal primary antibodies: OPN, LFMb-14 (1:200, sc-73631, Santa Cruz biotechnology); anti-flag (1:2000, F1804, Sigma), MMP9 2c3, (1:500, sc-21733, Santa Cruz Biotechnology), Anti-HS100A8 (1:500, MAB4570, R&D Systems,) and normalized with actin, C4 (1:5000, sc-47778, Santa Cruz Biotechnology). They were washed and incubated for 60 minutes at room temperature with secondary antibody, goat anti-mouse (1:5000, HRP, sc-2055, Santa Cruz Biotechnology). The membrane was visualized by chemiluminescent HRP substrate reagent (1:1) (Immobilon Western, WBKLS0500, Millipore) using ChemiDoc MP Imaging System (Bio Rad).
Molecular cloning and gene transient transfection
OPN-c gene of pcDNA3-OPN-V5 was a gift from Steven Johnson (Addgene plasmid # 11617). It was amplified by Phusion HF DNA polymerase (M0530L) and purified by the QIAquick PCR Purification Kit (#28106) and QIA gel Extraction kit (#28706). The PCR product was cut with NEB CutSmart and ligated overnight at 16ºC to p3XFLAG-CMV™-7.1 expression vector (E7533 SIGMA). Transformation was performed with competent E. coli cells (One Shot Top10, #C404003, Thermofisher). The overexpression gene transient transfection was carried out with Lipofectamine 3000 reagent (Invitrogen, CA, USA), while transient knockdown using Mission® esiRNA (EHU018231 SIGMA) with Lipofectamine 2000 following the manufacturer's protocol. The cell was seeded to be 60-70% confluent prior transfection. The ratio of plasmid DNA/Lipofectamine 3000 was selected as 2.5 μg / 3.75 μl for the OPN overexpression transfection and siRNA/Lipofectamine 2000 for transient knockdown was used at 1200 ng / 4 μl process (5× 105 cells in 3 ml complete culture medium per well of a 6-well plate). The cell was post-transfected for 48 hours at 37°C, then the transfection efficacy was determined by qPCR quantification and western blot analysis.
GEO Dataset computational analysis -- Extraction of clinical and microarray gene expression data from bladder cancer patient datasets
Two bladder cancer patient datasets, GSE13507 (n = 256) 64 and GSE32894 (n = 308) 65 GSE32548 (n = 131) 66 were identified from Gene Expression Omnibus database based on the following search criteria: 1.) bladder or urothelial cancer, 2.) available information on clinicopathological parameters, 3.) available information on gene expressions and 4.) with sample size more than 100. The GEO website has standardized URLs for its individual datasets, e.g. the overall summary information about the microarray dataset GSE13507 can be accessed at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE13507. These two datasets were publicly available. For each GEO data series, links are provided at the bottom of the page to the Series Matrix File(s), which contain the expression values for each gene (probeset) and each microarray. The URLs to the Series Matrix File(s) are also standardized. For GSE13507, the URL was http://ftp.ncbi.nlm.nih.gov/pub/geo/DATA/SeriesMatrix/GSE13507. The files in gzip format were then unzipped to the tab-delimited text format, which contain detailed information for statistical analysis. R scripting was used to extract the expression values of a small number of genes (probesets) of interest and the clinical data from the data matrixes downloaded from GEO.
Correlations of gene expression levels and clinical data
The associations between expression levels of genes were analyzed by Spearman's rank test. Expression levels were further divided into high and low levels using the median expression level as the cut-off point for Kaplan-Meier survival analysis. Results were compared by log-rank test. These statistical analyses were performed using SPSS 19.0 and a p-value of < 0.05 was considered as significant.
Identification of OPN co-expressing genes
Patients were stratified into two groups based on the expression levels of OPN as described above. The gene expression patterns of patients in OPN-low subgroup and those in the OPN-high subgroup (whose survival was significantly poorer) were compared. Probesets that were differentially expressed between these two subgroups were identified by 2-sample Welch's T-test. This test was used to avoid the type I error due to unequal variances of the values of probesets between subgroups. Briefly, a Welch's t test was applied to each probeset corresponding to a certain gene in the data matrix using our own Java application MyStats. P values and the differential expression in fold changes for all the probesets were generated as tab-delimited worksheets of Excel for further analysis.
Identification of therapeutic targets for bladder cancer patients overexpressing OPN
Patients who had their tumors expressing a high-level of OPN were stratified into two groups based on their survival status (alive or deceased). Differential expressions of different probesets between patients in the OPN high-alive subgroup and those in the OPN high-deceased subgroup were identified as described above.
Statistical analysis
Statistical analysis between gene was performed using the R-Script and Levene's Test for homogeneity and followed by T-test in primary qPCR experiment. Other data are expressed as means ± SD. Mean between groups were compared using a T-test and difference between multiple group were analyzed by 1-way ANOVA in Prism 6.0. In all analysis, P < 0.05 was considered statistically significant.
Supplementary Material
Supplementary figure S1.
Acknowledgments
This study was supported by the University of Macau Multi-Year Research Grant (MYRG2015-00065-FHS) to HFK.
Authors' contributions
JPCW and RW carried out the experiments, while LP and OT performed the statistical analysis. SDZ, QW, HFY, ME and HFK analyzed and interpreted the GEO DataSets. JPCW, RW, OT drafted the manuscript. HFY and HFK participated in the design of the study and revised the manuscript. All authors read and approved the final manuscript.
Abbreviations
- OPN
Osteopontin
- MMP9
Matrix metallopeptidase 9
- S100A8
S100 calcium-binding protein A8
- TCC
Transitional cell carcinoma
- ECM
extracellular matrix
- FAK
Focal adhesion kinase.
References
- 1.Knowles MA, Hurst CD. Molecular biology of bladder cancer: new insights into pathogenesis and clinical diversity. Nat Rev Cancer. 2015;15:25–41. doi: 10.1038/nrc3817. [DOI] [PubMed] [Google Scholar]
- 2.Li HT, Duymich CE, Weisenberger DJ, Liang G. Genetic and Epigenetic Alterations in Bladder Cancer. Int Neurourol J. 2016;20:S84–94. doi: 10.5213/inj.1632752.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sodek J, Ganss B, McKee MD. Osteopontin. Crit Rev Oral Biol Med. 2000;11:279–303. doi: 10.1177/10454411000110030101. [DOI] [PubMed] [Google Scholar]
- 4.Johnston NI, Gunasekharan VK, Ravindranath A, O'Connell C, Johnston PG, El-Tanani MK. Osteopontin as a target for cancer therapy. Front Biosci. 2008;13:4361–72. doi: 10.2741/3009. [DOI] [PubMed] [Google Scholar]
- 5.Ran W, Wong JPC, Kwok HF. Osteopontin - a promising biomarker for cancer therapy. J Cancer. 2017;8:2173–83. doi: 10.7150/jca.20480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mirza M, Shaughnessy E, Weber GF, Hurley JK, Vanpatten KA, Pestano G. Grading, Staging and prognosing Cancer using Osteopontin-C. Google Patents; 2008. [Google Scholar]
- 7.Philip S, Bulbule A, Kundu GC. Osteopontin stimulates tumor growth and activation of promatrix metalloproteinase-2 through nuclear factor-κB-mediated induction of membrane type 1 matrix metalloproteinase in murine melanoma cells. Journal of Biological Chemistry. 2001;276:44926–35. doi: 10.1074/jbc.M103334200. [DOI] [PubMed] [Google Scholar]
- 8.Wang D, Yamamoto S, Hijiya N, Benveniste EN, Gladson CL. Transcriptional regulation of the human osteopontin promoter: functional analysis and DNA-protein interactions. Oncogene; 2000. p. 19. [DOI] [PubMed] [Google Scholar]
- 9.Lu P, Weaver VM, Werb Z. The extracellular matrix: a dynamic niche in cancer progression. The Journal of cell biology. 2012;196:395–406. doi: 10.1083/jcb.201102147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lund SA, Giachelli CM, Scatena M. The role of osteopontin in inflammatory processes. Journal of cell communication and signaling. 2009;3:311–22. doi: 10.1007/s12079-009-0068-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gu T, Ohashi R, Cui R, Tajima K, Yoshioka M, Iwakami S. et al. Osteopontin is involved in the development of acquired chemo-resistance of cisplatin in small cell lung cancer. Lung Cancer. 2009;66:176–83. doi: 10.1016/j.lungcan.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 12.Pietras A, Katz AM, Ekstrom EJ, Wee B, Halliday JJ, Pitter KL. et al. Osteopontin-CD44 signaling in the glioma perivascular niche enhances cancer stem cell phenotypes and promotes aggressive tumor growth. Cell Stem Cell. 2014;14:357–69. doi: 10.1016/j.stem.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu G, Fan X, Tang M, Chen R, Wang H, Jia R. et al. Osteopontin induces autophagy to promote chemo-resistance in human hepatocellular carcinoma cells. Cancer Lett. 2016;383:171–82. doi: 10.1016/j.canlet.2016.09.033. [DOI] [PubMed] [Google Scholar]
- 14.Raja R, Kale S, Thorat D, Soundararajan G, Lohite K, Mane A. et al. Hypoxia-driven osteopontin contributes to breast tumor growth through modulation of HIF1alpha-mediated VEGF-dependent angiogenesis. Oncogene. 2014;33:2053–64. doi: 10.1038/onc.2013.171. [DOI] [PubMed] [Google Scholar]
- 15.Cheng HC, Liu YP, Shan YS, Huang CY, Lin FC, Lin LC. et al. Loss of RUNX3 increases osteopontin expression and promotes cell migration in gastric cancer. Carcinogenesis. 2013;34:2452–9. doi: 10.1093/carcin/bgt218. [DOI] [PubMed] [Google Scholar]
- 16.Lin J, Myers AL, Wang Z, Nancarrow DJ, Ferrer-Torres D, Handlogten A. et al. Osteopontin (OPN/SPP1) isoforms collectively enhance tumor cell invasion and dissemination in esophageal adenocarcinoma. Oncotarget. 2015;6:22239–57. doi: 10.18632/oncotarget.4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yin M, Soikkeli J, Jahkola T, Virolainen S, Saksela O, Holtta E. Osteopontin promotes the invasive growth of melanoma cells by activating integrin alphavbeta3 and down-regulating tetraspanin CD9. Am J Pathol. 2014;184:842–58. doi: 10.1016/j.ajpath.2013.11.020. [DOI] [PubMed] [Google Scholar]
- 18.Jia R, Liang Y, Chen R, Liu G, Wang H, Tang M. et al. Osteopontin facilitates tumor metastasis by regulating epithelial-mesenchymal plasticity. Cell Death Dis. 2016;7:e2564. doi: 10.1038/cddis.2016.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dong Q, Zhu X, Dai C, Zhang X, Gao X, Wei J. et al. Osteopontin promotes epithelial-mesenchymal transition of hepatocellular carcinoma through regulating vimentin. Oncotarget. 2016;7:12997–3012. doi: 10.18632/oncotarget.7016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sangaletti S, Tripodo C, Sandri S, Torselli I, Vitali C, Ratti C. et al. Osteopontin shapes immunosuppression in the metastatic niche. Cancer Res. 2014;74:4706–19. doi: 10.1158/0008-5472.CAN-13-3334. [DOI] [PubMed] [Google Scholar]
- 21.Weber GF, Lett GS, Haubein NC. Osteopontin is a marker for cancer aggressiveness and patient survival. Br J Cancer. 2010;103:861–9. doi: 10.1038/sj.bjc.6605834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bramwell V, Doig G, Tuck A, Wilson S, Tonkin K, Vandenberg T. et al. Osteopontin (OPN): A novel tumor marker of potential utility in metastatic breast cancer. J Clin Oncol. 2004;22:575. [Google Scholar]
- 23.Rudland PS, Platt-Higgins A, El-Tanani M, De Silva Rudland S, Barraclough R, Winstanley JH. et al. Prognostic significance of the metastasis-associated protein osteopontin in human breast cancer. Cancer Res. 2002;62:3417–27. [PubMed] [Google Scholar]
- 24.Zhao M, Liang F, Zhang B, Yan W, Zhang J. The impact of osteopontin on prognosis and clinicopathology of colorectal cancer patients: a systematic meta-analysis. Sci Rep. 2015;5:12713. doi: 10.1038/srep12713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takenaka M, Hanagiri T, Shinohara S, Yasuda M, Chikaishi Y, Oka S. et al. Serum level of osteopontin as a prognostic factor in patients who underwent surgical resection for non-small-cell lung cancer. Clin Lung Cancer. 2013;14:288–94. doi: 10.1016/j.cllc.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 26.Mack PC, Redman MW, Chansky K, Williamson SK, Farneth NC, Lara PN Jr. et al. Lower osteopontin plasma levels are associated with superior outcomes in advanced non-small-cell lung cancer patients receiving platinum-based chemotherapy: SWOG Study S0003. J Clin Oncol. 2008;26:4771–6. doi: 10.1200/JCO.2008.17.0662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lim AM, Rischin D, Cao H, Fisher R, McArthur GA, Peters LJ. et al. Prognostic significance of plasma osteopontin in patients with locally advanced head and neck squamous cell carcinoma (HNSCC) treated with chemoradiation on TROG 02.02 phase III trial. J Clin Oncol. 2011;29:5506. doi: 10.1158/1078-0432.CCR-11-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yazici O, Akbulut H, Akgun N, Utkan G, Yalcin B, Icli F. The role of serum osteopontin (OPN) as a prognostic and predictive marker in patients with advanced gastric cancer. J Clin Oncol. 2011;29:e14627. [Google Scholar]
- 29.Gu X, Gao XS, Ma M, Qin S, Qi X, Li X, Prognostic significance of osteopontin expression in gastric cancer: a meta-analysis. Oncotarget; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prager AJ, Peng CR, Lita E, McNally D, Kaushal A, Sproull M. et al. Urinary osteopontin as a marker for localized and metastatic prostate cancer. J Clin Oncol. 2011;29:e15147. [Google Scholar]
- 31.El-Tanani M, Platt-Higgins A, Rudland PS, Campbell FC. Ets gene PEA3 cooperates with beta-catenin-Lef-1 and c-Jun in regulation of osteopontin transcription. J Biol Chem. 2004;279:20794–806. doi: 10.1074/jbc.M311131200. [DOI] [PubMed] [Google Scholar]
- 32.Ravindranath A, Yuen HF, Chan KK, Grills C, Fennell DA, Lappin TR. et al. Wnt-beta-catenin-Tcf-4 signalling-modulated invasiveness is dependent on osteopontin expression in breast cancer. Br J Cancer. 2011;105:542–51. doi: 10.1038/bjc.2011.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kurisetty VV, Johnston PG, Johnston N, Erwin P, Crowe P, Fernig DG. et al. RAN GTPase is an effector of the invasive/metastatic phenotype induced by osteopontin. Oncogene. 2008;27:7139–49. doi: 10.1038/onc.2008.325. [DOI] [PubMed] [Google Scholar]
- 34.Ang C, Chambers AF, Tuck AB, Winquist E, Izawa JI. Plasma osteopontin levels are predictive of disease stage in patients with transitional cell carcinoma of the bladder. BJU Int. 2005;96:803–5. doi: 10.1111/j.1464-410X.2005.05716.x. [DOI] [PubMed] [Google Scholar]
- 35.Zhao L, Wang Y, Qu N, Huang C, Chen L. Significance of plasma osteopontin levels in patients with bladder urothelial carcinomas. Mol Diagn Ther. 2012;16:311–6. doi: 10.1007/s40291-012-0005-4. [DOI] [PubMed] [Google Scholar]
- 36.Park MG, Oh MM, Yoon JH, Park JY, Park HS, Moon DG. et al. The value of plasma osteopontin levels as a predictive factor of disease stage and recurrence in patients with bladder urothelial carcinoma: a prospective study. Kaohsiung J Med Sci. 2012;28:526–30. doi: 10.1016/j.kjms.2012.04.014. [DOI] [PubMed] [Google Scholar]
- 37.Zaravinos A, Volanis D, Lambrou GI, Delakas D, Spandidos DA. Role of the angiogenic components, VEGFA, FGF2, OPN and RHOC, in urothelial cell carcinoma of the urinary bladder. Oncol Rep. 2012;28:1159–66. doi: 10.3892/or.2012.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu ST, Guo C, Ding X, Fan WJ, Zhang FH, Xu WL. et al. Role of osteopontin in the regulation of human bladder cancer proliferation and migration in T24 cells. Mol Med Rep. 2015;11:3701–7. doi: 10.3892/mmr.2015.3202. [DOI] [PubMed] [Google Scholar]
- 39.Thomsen M, Bakiri L, Hasenfuss S, Wu H, Morente M, Wagner E. Loss of JUNB/AP-1 promotes invasive prostate cancer. Cell Death & Differentiation. 2015;22:574–82. doi: 10.1038/cdd.2014.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu X-L, Lin K-J, Bai A-P, Wang W-X, Meng X-K, Su X-L. et al. Osteopontin knockdown suppresses the growth and angiogenesis of colon cancer cells. World journal of gastroenterology: WJG. 2014;20:10440. doi: 10.3748/wjg.v20.i30.10440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hussain SA, Palmer DH, Syn W-K, Sacco JJ, Greensmith R, Elmetwali T. et al. Gene expression profiling in bladder cancer identifies potential therapeutic targets. International Journal of Oncology. 2017;50:1147–59. doi: 10.3892/ijo.2017.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sodek J, Ganss B, McKee M. Osteopontin. Critical Reviews in Oral Biology & Medicine. 2000;11:279–303. doi: 10.1177/10454411000110030101. [DOI] [PubMed] [Google Scholar]
- 43.Lai CF, Seshadri V, Huang K, Shao JS, Cai J, Vattikuti R. et al. An osteopontin-NADPH oxidase signaling cascade promotes pro-matrix metalloproteinase 9 activation in aortic mesenchymal cells. Circ Res. 2006;98:1479–89. doi: 10.1161/01.RES.0000227550.00426.60. [DOI] [PubMed] [Google Scholar]
- 44.Wu XL, Lin KJ, Bai AP, Wang WX, Meng XK, Su XL. et al. Osteopontin knockdown suppresses the growth and angiogenesis of colon cancer cells. World J Gastroenterol. 2014;20:10440–8. doi: 10.3748/wjg.v20.i30.10440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y, Davis C, Ryan J, Janney C, Pena MM. Development and characterization of a reliable mouse model of colorectal cancer metastasis to the liver. Clin Exp Metastasis. 2013;30:903–18. doi: 10.1007/s10585-013-9591-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bauvois B. New facets of matrix metalloproteinases MMP-2 and MMP-9 as cell surface transducers: outside-in signaling and relationship to tumor progression. Biochimica et Biophysica Acta (BBA)-Reviews on Cancer. 2012;1825:29–36. doi: 10.1016/j.bbcan.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 47.Zeng FC, Cen S, Tang ZY, Kang XL. Elevated matrix metalloproteinase-9 expression may contribute to the pathogenesis of bladder cancer. Oncology letters. 2016;11:2213–22. doi: 10.3892/ol.2016.4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vu TH, Werb Z. Matrix metalloproteinases: effectors of development and normal physiology. Genes & development. 2000;14:2123–33. doi: 10.1101/gad.815400. [DOI] [PubMed] [Google Scholar]
- 49.Stawarski M, Stefaniuk M, Wlodarczyk J. Matrix metalloproteinase-9 involvement in the structural plasticity of dendritic spines. Frontiers in neuroanatomy; 2014. p. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lindsey ML, Zouein FA, Tian Y, Padmanabhan Iyer R, de Castro Brás LE. Osteopontin is proteolytically processed by matrix metalloproteinase 9 1. Canadian journal of physiology and pharmacology. 2015;93:879–86. doi: 10.1139/cjpp-2015-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lenga Y, Koh A, Perera AS, McCulloch CA, Sodek J, Zohar R. Osteopontin expression is required for myofibroblast differentiation. Circulation research. 2008;102:319–27. doi: 10.1161/CIRCRESAHA.107.160408. [DOI] [PubMed] [Google Scholar]
- 52.Annaházi A, Ábrahám S, Farkas K, Rosztóczy A, Inczefi O, Földesi I. et al. A pilot study on faecal MMP-9: a new noninvasive diagnostic marker of colorectal cancer. British journal of cancer. 2016;114:787–92. doi: 10.1038/bjc.2016.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ke HL, Chang LL, Yang SF, Lin HH, Li CC, Wu DC. et al. Osteopontin overexpression predicts poor prognosis of upper urinary tract urothelial carcinoma. Urol Oncol. 2011;29:703–9. doi: 10.1016/j.urolonc.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 54.Lim SY, Yuzhalin AE, Gordon-Weeks AN, Muschel RJ. Tumor-infiltrating monocytes/macrophages promote tumor invasion and migration by upregulating S100A8 and S100A9 expression in cancer cells. Oncogene; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Desai B. Identifying the Key Players in Osteopontin/alpha(v)beta(3)-mediated Migration and Invasion of Metastatic Prostate Cancer Cells. University of Maryland, Baltimore; 2008. [Google Scholar]
- 56.Dahiya S, Givvimani S, Bhatnagar S, Qipshidze N, Tyagi SC, Kumar A. Osteopontin-stimulated expression of matrix metalloproteinase-9 causes cardiomyopathy in the mdx model of Duchenne muscular dystrophy. J Immunol. 2011;187:2723–31. doi: 10.4049/jimmunol.1101342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Song G, Ouyang G, Mao Y, Ming Y, Bao S, Hu T. Osteopontin promotes gastric cancer metastasis by augmenting cell survival and invasion through Akt-mediated HIF-1alpha up-regulation and MMP9 activation. J Cell Mol Med. 2009;13:1706–18. doi: 10.1111/j.1582-4934.2008.00540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nutt JE, Durkan GC, Mellon JK, Lunec J. Matrix metalloproteinases (MMPs) in bladder cancer: the induction of MMP9 by epidermal growth factor and its detection in urine. BJU Int. 2003;91:99–104. doi: 10.1046/j.1464-410x.2003.04020.x. [DOI] [PubMed] [Google Scholar]
- 59.Wu WB, Wang W, Du YH, Li H, Xia SJ, Liu HT. MicroRNA-3713 regulates bladder cell invasion via MMP9. Sci Rep. 2016;6:32374. doi: 10.1038/srep32374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen H, Xu C, Qing'e Jin ZL. S100 protein family in human cancer. American journal of cancer research. 2014;4:89. [PMC free article] [PubMed] [Google Scholar]
- 61.Tolson JP, Flad T, Gnau V, Dihazi H, Hennenlotter J, Beck A. et al. Differential detection of S100A8 in transitional cell carcinoma of the bladder by pair wise tissue proteomic and immunohistochemical analysis. Proteomics. 2006;6:697–708. doi: 10.1002/pmic.200500033. [DOI] [PubMed] [Google Scholar]
- 62.Ha YS, Kim MJ, Yoon HY, Kang HW, Kim YJ, Yun SJ. et al. mRNA Expression of S100A8 as a Prognostic Marker for Progression of Non-Muscle-Invasive Bladder Cancer. Korean J Urol. 2010;51:15–20. doi: 10.4111/kju.2010.51.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim SK, Kim EJ, Leem SH, Ha YS, Kim YJ, Kim WJ. Identification of S100A8-correlated genes for prediction of disease progression in non-muscle invasive bladder cancer. BMC Cancer. 2010;10:21. doi: 10.1186/1471-2407-10-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee J-S, Leem S-H, Lee S-Y, Kim S-C, Park E-S, Kim S-B. et al. Expression signature of E2F1 and its associated genes predict superficial to invasive progression of bladder tumors. Journal of clinical oncology. 2010;28:2660–7. doi: 10.1200/JCO.2009.25.0977. [DOI] [PubMed] [Google Scholar]
- 65.Sjödahl G, Lauss M, Lövgren K, Chebil G, Gudjonsson S, Veerla S. et al. A molecular taxonomy for urothelial carcinoma. Clinical Cancer Research. 2012;18:3377–86. doi: 10.1158/1078-0432.CCR-12-0077-T. [DOI] [PubMed] [Google Scholar]
- 66.Lindgren D, Sjödahl G, Lauss M, Staaf J, Chebil G, Lövgren K. et al. Integrated genomic and gene expression profiling identifies two major genomic circuits in urothelial carcinoma. PloS one. 2012;7:e38863. doi: 10.1371/journal.pone.0038863. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figure S1.