Abstract
Neuregulin-1β (NRG-1β) is critical for cardiac development and repair, and recombinant forms are currently being assessed as possible therapeutics for systolic heart failure. We previously demonstrated that recombinant NRG-1β reduces cardiac fibrosis in an animal model of cardiac remodeling and heart failure, suggesting that there may be direct effects on cardiac fibroblasts. Here we show that NRG-1β receptors (ErbB2, ErbB3, and ErbB4) are expressed in normal human cardiac ventricular (NHCV) fibroblast cell lines. Treatment of NHCV fibroblasts with recombinant NRG-1β induced activation of the AKT pathway, which was phosphoinositide 3-kinase (PI3K)-dependent. Moreover, the NRG-1β-induced PI3K/AKT signaling in these cells required phosphorylation of both ErbB2 and ErbB3 receptors at tyrosine (Tyr)1248 and Tyr1289 respectively. RNASeq analysis of NRG-1β-treated cardiac fibroblasts obtained from three different individuals revealed a global gene expression signature consistent with cell growth and survival. We confirmed enhanced cellular proliferation and viability in NHCV fibroblasts in response to NRG-1β, which was abrogated by PI3K, ErbB2, and ErbB3 inhibitors. NRG-1β also induced production and secretion of cytokines (interleukin-1α and interferon-γ) and pro-reparative factors (angiopoietin-2, brain-derived neurotrophic factor, and crypto-1), suggesting a role in cardiac repair through the activation of paracrine signaling.
Keywords: Neuregulin, Cardiac fibroblast, ErbB3, Survival, RNASeq
1. Introduction
Cardiac muscle is highly susceptible to injury caused by ischemia, inflammation, pressure overload and/or volume overload. Cardiac damage incurred during injurious processes, such as myocardial infarction (MI), results in the initiation of the wound healing processes in response to cardiomyocyte death leading to replacement of normal cardiac tissue with a stiff, collagenous scar. While this fibrotic scar is necessary to the integrity of the heart, its extreme rigidity and lack of functioning cardiomyocytes perturb the heart's natural mechanical properties, accelerating pathogenic remodeling and promoting development of heart failure. The conversion of fibroblasts to collagen-producing myofibroblasts (myoFbs) is a hallmark of cardiac tissue remodeling subsequent to injury, but their persistence after this initial healing phase contributes to adverse extracellular matrix (ECM) remodeling and collagen deposition that interferes with normal heart functions [1].
NRG-1β (neuregulin-1β) is an epidermal growth factor family member, which is absolutely required for embryonic heart development [2–5]. More recently, it has become evident that NRG-1β also plays a role in cardiac repair in the adult heart [4,6] and may contribute to the beneficial effects of exercise on cardiac and skeletal muscles [7,8]. The main source of NRG-1β in the heart is the cardiac microvascular endothelium [6,9]. Endothelial cell-derived NRG-1β is activated by proteolysis, after which it induces both autocrine and paracrine signals within the myocardium [10–14] and possibly distant signaling, as circulating levels are associated with ischemic and coronary artery disease [15,16].
NRG-1β transduces signaling in cardiac myocytes and endothelial cells via erythroblastic leukemia viral oncogene homolog tyrosine kinase receptors (ErbB2, ErbB3, and/or ErbB4). NRG-1β-bound ErbB4 forms a homo-dimer or hetero-dimerizes with one of the other ErbBs, leading to activation of the intracellular tyrosine kinase domain and signal transduction. ErbB2 contains a functional tyrosine kinase domain but lacks a NRG binding site. ErbB3, on the other hand is kinase dead, but when bound by NRG-1β can dimerize with another ErbB receptor (typically ErbB2) and thereby relay the signal. Upon ligand binding, ErbB receptors are activated via autophosphorylation, which results in one of several downstream signaling cascades, dependent upon cell type and condition [17–19]. Known NRG-inducible cardiac signaling pathways include phosphoinositide 3-kinase (PI3K)/AKT, Ras/ERK, and Src/FAK [17–19]. In cardiomyocytes, for example, NRG-1β influences sarcomeric organization, induces proliferation, and increases survival [10,20–23]. In embryonic stem cells, NRG-1β promotes differentiation into cardiomyocytes [24–26]. NRG-1β can also influence blood pressure via the rostral ventrolateral medulla and modulate circulating immune cell functions [6].
Our laboratory previously reported improved cardiac function in both rat and swine models of myocardial infarction (MI) in animals treated with the glial growth factor 2 (GGF2) [27] isoform of NRG-1β, which contains a unique kringle domain in addition to the Ig-like and EGF-like domains that are common to other isoforms [19]. These studies led to the currently ongoing clinical trial examining the safety and tolerability of GGF2 in human subjects with systolic heart failure. We also demonstrated previously ErbB receptor expression and signaling in primary rodent cardiac stromal cells [27], as well as reduced fibrosis and MyoFb percentages in remote-from-infarct cardiac tissues of post-MI swine [27]. These preliminary experiments suggested NRG-1β may regulate cardiac fibroblast signaling and activity.
In this study, we investigated the effects of NRG-1β treatment on primary human cardiac ventricular fibroblasts and herein describe the presence of functional ErbB signaling in human fibroblasts and involvement of NRG/ErbB in fibroblast-mediated signaling, a significant finding given their importance in cardiac remodeling.
2. Materials and methods
2.1. Cell culture and reagents
Primary human cardiac ventricular fibroblasts (NHCF-V, cc-2904) were purchased from Lonza Walkersville Inc. (Walkersville, MD), expanded per manufacturer's protocol, and stored in 1 mL aliquots in Recovery Cell Culture Freezing Medium (Gibco/Thermo Fisher Scientific, Waltham, MA) in liquid nitrogen. All media, additives, and sub-culturing reagents were purchased from Lonza. Cells were routinely cultured in FGM-3 BulletKit (cc-cc4526), which includes Fibroblast Basal Medium (cc-3301) supplemented with FGM-3 SingleQuots (cc-4525). FGM-3 SingleQuots include recombinant human insulin (cc-4021), gentamycin sulfate amphotericin-B (cc-4081), and recombinant human fibroblasts growth factor-B (cc-4065). Expansion/splitting was accomplished by rinsing cells in HEPES Buffered Saline Solution (cc-5024) followed by detachment with Trypsin/EDTA Solution (cc-5012), inactivation of trypsin with Trypsin Neutralization Solution (TNS, cc-5002) and centrifugation at 1000g for 5 min. Unless otherwise stated, cells were serum-starved for 24 h followed by treatments in Fibroblast Basal Medium supplemented with gentamycin only (50 µg/mL, 17–528Z, Lonza). A total of five different NHCF-V cell lines (cc-2904, Lonza) were used in this study. Donor characteristics are provided in Table 1.
Table 1.
General characteristics of the human cardiac ventricular fibroblast donors.
| Tissue acq. no. | Age | Race | Sex | Experiments |
|---|---|---|---|---|
| 20731 | 42 Y | Caucasian | Male | Preliminary optimization experiments |
| 25981 | 45 Y | Caucasian | Male | Optimization, Western blotting, qPCR, preliminary cytokine profiling |
| 27277 | 32 Y | Caucasian | Female | Western blotting, qPCR, RNASeq, flow, cytokine arrays |
| 27741 | 52 Y | Caucasian | Male | Western blotting, qPCR, RNASeq, flow, proliferation and viability, cytokine arrays |
| 27321 | 31 Y | Caucasian | Female | Western blotting, qPCR, RNASeq, flow, proliferation and viability, cytokine arrays |
2.2. RNA sequencing (RNASeq)
Total RNA was isolated using the RNEasy Midi kit (Qiagen, Valencia, CA) per manufacturer's instruction. Quality control, mRNA enrichment and cDNA library preparation were performed by the Vanderbilt Technologies and Genomics (VANTAGE) Core at Vanderbilt. RNA integrity was assessed using the Bioanalyzer 2100 (Agilent). The Illumina Truseq RNA sample prep kit was employed for targeted analysis of polyadenylated transcripts. Paired-end sequencing was conducted on the Illumina HiSeq 2500. The resulting FASTQ data files for each sample were transferred to the Basespace Sequence Hub and aligned to the hg19 human reference genome assembly using the STAR aligner (Dobin et al., Bioinformatics 2012) with the following parameters: (1) pseudoautosomal regions masking, (2) mapping quality score of at least 37 (3) and paired end reads mapped to same chromosome with expected orientations.
Resulting BAM files were imported into Partek Genomics Suite 6.6 software, followed by quantification, normalization, and differential gene expression. Reads were quantified based on the RefSeq transcriptome, and normalized reads were represented as RPKM (Reads Per Kilobase of transcript per Million mapped reads).
2.3. Quantitative polymerase chain reaction (qPCR)
Total RNA was converted to cDNA with iScript (Bio-Rad). Pre-validated primers for human ErbB2, ErbB3, and ErbB4 receptors were purchased from Qiagen. Relative gene expression for each receptor was assessed using iTaq Universal SYBR Green Supermix (Bio-Rad) in a Bio-Rad CFX instrument, according to manufacturer's protocol. Briefly, ~200 ng of cDNA was mixed with 2× supermix mix, RNase-free water and 1 µM of primers, for a total reaction volume of 10 µL. A typical protocol included for polymerase activation at 95 °C for 30 s, and 40 cycles as follows: denaturation (5 s at 95 °C), annealing/extension (30 s at 60 °C), followed by melt-curve analysis. The comparative threshold method used to calculate fold-differences in Excel. Pre-validated human GAPDH primers served as internal controls to normalize target gene expression across different samples. Levels of each ErbB receptor was analyzed at P0, P1, and P2 (where P0 = pre-passaged cells from Lonza before subsequent passage in our laboratory). The experiment was performed for three cell lines acquired from different individuals. At least three technical replicates were also included to ensure reproducibility.
2.4. Western blot analysis
Cells were lysed with RIPA buffer (Millipore), supplemented with protease and phosphatase inhibitors (Roche) and mechanical disrupted using 29½ gauge needles (Becton Dickinson). Lysates were then centrifuged at 14,000 RPM for 15 min followed by measurement of proteins in the resulting supernatants by Bradford assay (Bio-Rad). Proteins (30 µg/lane) were mixed with 4× loading buffer (Bio-Rad), loaded into 4–20% TGX polyacrylamide gels (Bio-Rad) and subjected to gel electrophoresis. Proteins were then transferred to PVDF membranes (0.45 µm pore size, Millipore) at 100 V for 1 h at 4 °C. Membranes were rinsed in tris-buffered saline (TBS) and then blocked in 5% blocking buffer in TBS with 0.1% Tween-20 (TBST) for 1 h. After washing in TBST, membranes were incubated with primary antibody diluted in 5% bovine serum albumin (BSA, Sigma) in TBST overnight at 4 °C. After additional washing, membranes were probed with horseradish peroxidase (HRP)-linked anti-rabbit antibodies (Cell Signaling Technologies) in 5% blocking buffer for 1 h at room temperature, washed again, incubated for 5 min with chemiluminescent reagent, and exposed to X-ray film. Supersignal West Dura Extended Duration Substrate (Thermo Scientific) was used to detect phosphorylated ErbB2. All other proteins were visualized using Clarity Western ECL Substrate (Bio-Rad).
2.5. Immunoblot pathway arrays
The Human Phospho-kinase Proteome Profiler Array (ARY003B, R&D Systems, Minneapolis, MN) was used to detect phosphorylation of multiple kinases simultaneously, per manufacturer's directions. This panel includes 43 kinase phosphorylation sites, 2 related total proteins, 3 reference controls, and 1 negative control spotted in duplicate onto two separate membranes. Briefly, arrays were blocked with Array Buffer 1 for 1 h at room temperature, followed by incubation with ~200 µg of protein lysates (per membrane) overnight at 4 °C. All remaining steps, including washes, hybridization with primary and enzyme-linked secondary antibodies, and chemi-luminescence substrate exposure steps were accomplished the following day using manufacturer-provided instructions and reagents. Blots were visualized using X-ray film with exposures ranging from 10 s to 10 min, followed by dark-room development and densitometry analysis using the reference spots for normalization. Negative control spots produced no detectable signal for any of the samples, even after 10 min of exposure time.
2.6. Flow cytometry
Cells were detached using StemPro Accutase (Gibco), counted and treated with Human TruStain FcX™ (Biolegend, San Diego, CA) to prevent non-specific binding. Cell surface-expressed ErbB receptors were assayed using phycoerythrin (PE)-conjugated anti-human ErbB2/Her2 (Fab1129P) and IgG2B isotype-matched control (IC0041P), anti-hErbB3/Her3 (Fab3481P) and IgG1 control (IC002P), or anti-hErbB4/Her4 (Fab11311P) and IgG2A (IC003P) isotype control as follows: 1 × 106 cells were incubated with 5 µL of each antibody in 100 µL of FACS buffer (0.5% BSA and 2 mM EDTA in PBS) for 30 min at 4 °C. The cells were then washed 3 times with FACs buffer and immediately analyzed on a FACSCanto flow cytometer with DIVA software (Becton Dickinson, Franklin Lakes, NJ). Dead cells were eliminated from the analysis using 7-AAD (BD Phermingen). For each experiment, we gated on single live cells and used isotype controls for each fluorophore to establish the gates. Data analysis was done using FlowJo software (Tree Star, Inc.).
2.7. Cell proliferation
BrdU Assay Kit #6813 (Cell Signaling) was used to detect BrdU incorporation, per manufacturer's instructions, as an assessment of cell proliferation. Briefly, ~10,000 cells/well were seeded into a 96-well plate and allowed to attach overnight. Cells were subsequently washed with HEPES and then exposed to various concentrations of NRG-1β with or without inhibitors for 24 h. Cells were then incubated with 1× BrdU for an additional 24 h, followed by fixation/denaturation (100 µL/well for 30 min at RT) and incubation with 1× detection antibody for 1 h. After washing, cells were incubated with HRP-linked secondary antibody for 30 min, washed again and then incubated with TMB substrate for 30 min before application of STOP solution. Absorbance at 450 nm was read using an Epoch microplate spectrophotometer (BioTek Instruments), followed by background subtraction and statistical analysis.
2.8. Cell survival
To induce stress-associated death, cells were exposed to various concentrations of hydrogen peroxide (H2O2) in media containing gentamycin and 2% serum. Cell viability was assessed using CellTiter Blue (Promega), per manufacturer's instructions. Fluorescence (560EX/590EM) was subsequently measured using a GloMax 96 Microplate Luminometer (Promega).
2.9. Cytokines
The Proteome Profiler Human XL Cytokine Array Kit (ARY022, R&D Systems, Minneapolis, MN) was used to detect secreted cytokines, per manufacturer's directions. This panel includes 102 human cytokines, 3 reference controls, and 1 negative control spotted in duplicate onto a single membrane. NHCV fibroblasts obtained from 4 different individuals were untreated or treated with 30 ng/mL of NRG-1β for 24 h, and 500 µL of cell culture supernatant was run on each array.
2.10. Statistics
All data are expressed as means ± SEM. Statistical comparisons made between two variables were performed using the Student's t-test (for pairwise comparisons). Comparisons between more than two variables were performed using one-way ANOVA with a Tukey's post hoc test. p-Values of <0.05 were considered statistically significant. For RNASeq, Partek was used to perform pairwise comparisons of average group values and one-way ANOVA for the four groups. Only transcripts that resulted in a fold-change of at least 1.5 and p value of <0.05 were considered significantly altered.
3. Results
3.1. Human ventricular cardiac fibroblasts express ErbB receptors
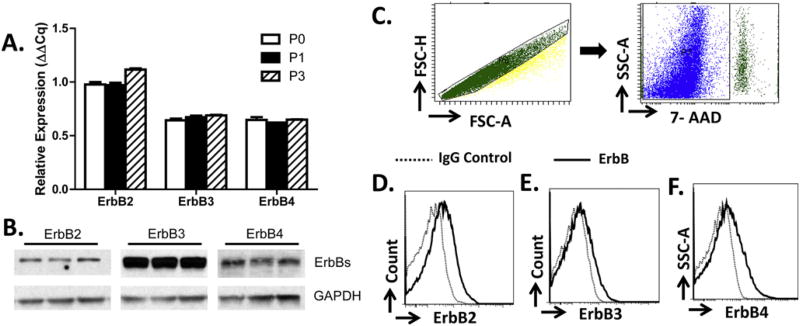
We previously reported expression of neuregulin-1β (NRG-1β) receptors (ErbB2, ErbB3, and ErbB4) in rodent primary cardiac stromal cells [27]. To determine if normal human cardiac ventricular (NHCV) fibroblasts also express all three ErbB receptors, we employed three different approaches; first, we performed quantitative PCR (qPCR) on cells after three different passages. As shown in Fig. 1A, all three ErbB receptors are expressed by NHCV fibroblasts, and this expression was stable across passages. Second, we performed Western blot analysis and similarly found protein expression of ErbB2, ErbB3, and ErbB4 receptors (Fig. 1B). Third, we performed flow cytometry using a gating strategy that allowed us to select live single cells (Fig. 1C). We found that compared to the isotype controls, there was a marked increase in cell surface expression of ErbB2 (Fig. 1D), ErbB3 (Fig. 1E) and ErbB4 (Fig. 1F). These results demonstrate that human fibroblasts express all three ErbB receptors on the cell surface at both a pre- and a post-transcriptional level.
Fig. 1.
NHCV fibroblasts express ErbB Receptors. A, relative mRNA levels of ErbB2, ErbB3, and ErbB4 receptors in normal human ventricular cardiac (NHCV) fibroblasts after three passages is shown. B, Western blot analysis shows protein expression of ErbB2, ErbB3, and ErbB4 receptors in unstimulated NHCF cells. Anti-rabbit GAPDH served as a loading control. C, gating strategy to identify single live cells. Representative flow cytometry histograms showing isotype controls and surface expression of ErbB2 (D), ErbB3 (E), and ErbB4 (F) (n = 3 in 3 independent experiments).
3.2. NRG-1β induces pro-survival gene expression in NHCV fibroblasts
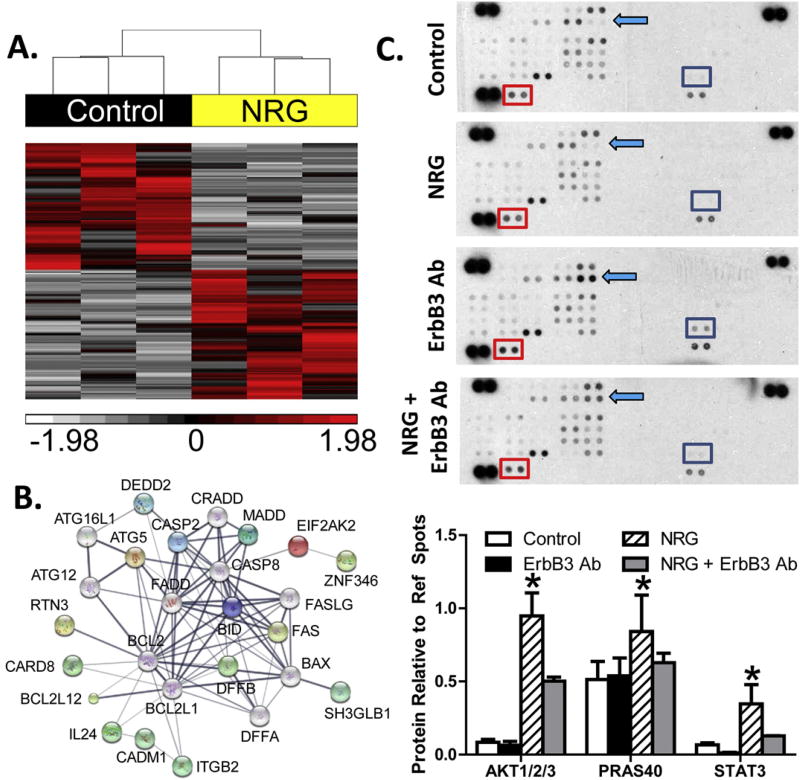
To determine the functional significance of ErbB receptor expression on human fibroblasts, NHCV cells from three different individuals were treated with NRG-1β for 6 h and RNA sequencing performed. Post-alignment analysis identified 776 transcripts as significantly altered between untreated and NRG-1β-treated fibroblasts, which upon hierarchical clustering distinguished between controls and treatments (Fig. 2A). As shown in Table 2, further analysis of these altered genes revealed enriched functions consistent with cellular proliferation and survival, including Ras signaling, regulation of apoptosis and cell death, and signaling pathways that are classically associated with cancer. Additionally, network analysis confirmed that proteins encoded by altered genes were functionally associated. Apoptotic genes that were found to be interrelated with one another via overlapping signaling pathways based on STRING Network analysis are shown in Fig. 2B. As expected, ErbB signaling was identified as an enriched signaling pathway in NRG-1β-treated fibroblasts (Table 2).
Fig. 2.
RNA sequencing and protein array results for neuregulin-treated fibroblasts. A, hierarchical clustering of RNA transcript RPKM (Reads Per Kilobase per Million mapped reads) for differential genes between untreated and NRG-1β-treated NHCV fibroblasts. The cluster was produced using Partek Genomics Suite with average linkage and Euclidian distance measures. Bright red, white, and black indicate the highest, lowest, and median normalized reads, respectively. Vertical dendrograms represent the individual donor fibroblasts, of which there are three replicates for each group. B, differentially expressed genes associated with cell death formed a functional protein interaction network when analyzed using STRING software code and libraries, which are freely available online (string-db.org). C, representative immunoblots show differentially expressed phosphorylated AKT pathway proteins in NHCV fibroblasts that were untreated or treated for 30 min with NRG-1β, with and without pre-treatment with MAB3481, an ErbB3 blocking antibody. Each protein is represented on the array in duplicate, and each array includes three positive references (top left, top right, and bottom left) as well as negative reference spots (bottom right). Colored squares indicate significantly differential proteins. D, graphical representation of differentially expressed proteins identified using immunoblot arrays. Data includes densitometry analysis results for four replicate experiments (n = 4, *p < 0.05 for NRG-1β-treated cells, when compared to the various control treatments).
Table 2.
Enriched functions of human fibroblasts treated with NRG-1β.
| Ontology/pathway | p value | Up-regulated | Down-regulated |
|---|---|---|---|
| Protein localization | 0.001342 | BID, STRADA, AP3S1, MTX2, PEX5, SRGN, SRP19, SRP54, STAM, SYTL3, SYTL4, SDCBP, STX10, TLK1 | RAP3IP, RPAIN, SH3GLB1, SNAPIN, AURKB, CLTA, GOSR1, SNX1, SNX2, VTI1A, ZFYVE16 |
| Ras protein signal transduction | 0.00176 | RASGRP3, ARHGEF3, RHOA, SDCBP | RASSF1, GNB1, APOE, RGL2, RREB1 |
| Apoptosis (ontology) | 0.009091 | ATG5, BID, DFFB, FAS, FAIM, NLRP12, RASSF5, ARHGEF3, CARD8, DEDD2, RTN3, RNF130, SRGN, TRIM69, ZNF346 | ADAMTSL4, BCL2L13, MADD, MCF2L, SH3GLB1, BLCAP, CIB1, CASP2, CADM1, EIF2AK2, ITGB2, IL24, PIGT, PUF60, ZDHHC16 |
| Cell death | 0.019755 | L1CAM, RASSF5, ATXN10 + 15 genes above | 15 genes listed above |
| Pathways in cancer | 0.007181 | BID, FAS, MAX, RASSF5, SMAD2, PTCH1, RHOA, TPM3, VHL, WNT2B | RASSF1, EGF, IKBKG, MSH6, PAX8, PIK3R3, RARB |
| ErbB signaling pathway | 0.019615 | PAK6, RPS6KB1, ABL2 | CAMK2G, EGF, NRG2, PIK3R3 |
| Apoptosis (pathway) | 0.056577 | BID, DFFB, FAS, IRAK4 | IKBKG, PIK3R3, PRKACB |
We next performed a screening assay to identify early phosphorylation events in NRG-1β-treated NHCV fibroblasts, using duplicate spotted immunoblot protein arrays to compare untreated versus NRG-1β-treated NHCV fibroblasts and detected significantly higher levels of phosphorylated AKT. Interestingly, PRAS40 and STAT3 were also phosphorylated in response to NRG-1β treatment (Fig. 2C). Pre-treatment with an ErbB3 blocking antibody (MAB3481) markedly reduced the levels of all three of these key phospho-signaling molecules (Fig. 2C). These results were reproducible and significant across three replicate experiments (Fig. 2D).
3.3. NRG-1β activates the phosphoinositide 3-kinase (PI3K)/AKT pathway in NHCV fibroblasts
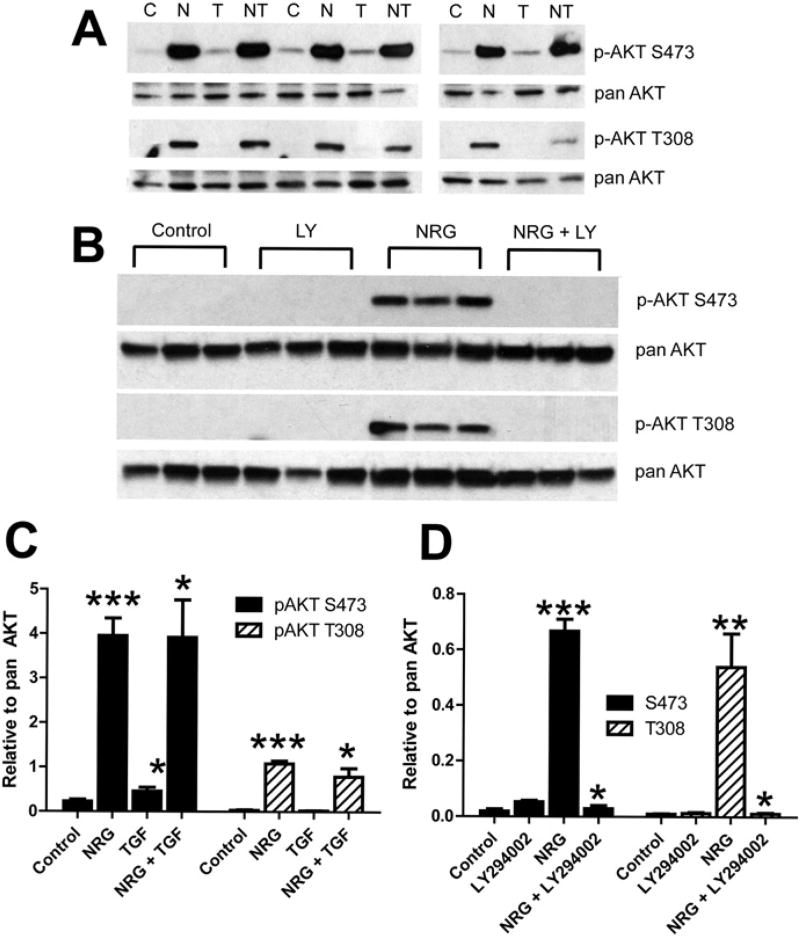
To examine the effect of NRG-1β on the PI3K/AKT pathway, NHCV fibroblasts were treated with recombinant NRG-1β for 30 min and the phosphorylation of two key amino acids of AKT assessed by Western blot analysis. As shown, NRG-1β treatment consistently induced phosphorylation of AKT at both serine (S)473 and threonine (T)308 (Fig. 3A and C). The NRG-1β-induced phosphorylation was consistently observed and statistically significant at both phosphorylation sites (Fig. 3A). On the other hand, these alterations were neither observed in NHCV cells exposed to the pro-fibrotic factor TGF-β, nor did TGF-β change the levels of phosphorylated AKT at S473 or T308 when the cells were treated with both NRG-1β and TGF-β (Fig. 3A). Pre-treatment of NHCV cells with the PI3K inhibitor LY40092 abolished phosphorylation of AKT at both S473 and T308 (Fig. 3B), indicating that NRG-1β activation of AKT in these cells is PI3K-dependent.
Fig. 3.
Neuregulin-1β treatment induces AKT activation in human cardiac fibroblasts. A, Western blot analysis of protein lysates of NHCV fibroblasts treated for 30 min with 30 ng/mL of NRG-1β. Bands shown are 60 kDa and were detected using antibodies that recognize phosphorylated AKT (pAKT) at serine position 473 (S473) or threonine position 308 (T308). Blots re-probed with pan AKT served as protein loading controls. Experimental conditions included untreated control cells, denoted by a “C”, and cells treated with NRG-1β (N), 1 ng/mL TGF-β (T), or both NRG-1β and TGF-1β (NT). Representative blots are shown. B, Western blots of protein lysates of NHCV fibroblasts probed with pAKT at S473 or T308 and re-probed with pan AKT. Treatments shown (in triplicate) include untreated control cells, and cells treated with the PI3K inhibitor LY294002 (LY), 30 ng/mL of NRG-1β (NRG), or both LY and NRG. C and D, Graphical representations of densitometry analysis results for the differentially expressed proteins shown in A and B, respectively (n = 3, *p < 0.05, **p < 0.01, and ***p < 0.001, respectively for NRG-1β-treated cells, when compared to appropriate control treatments).
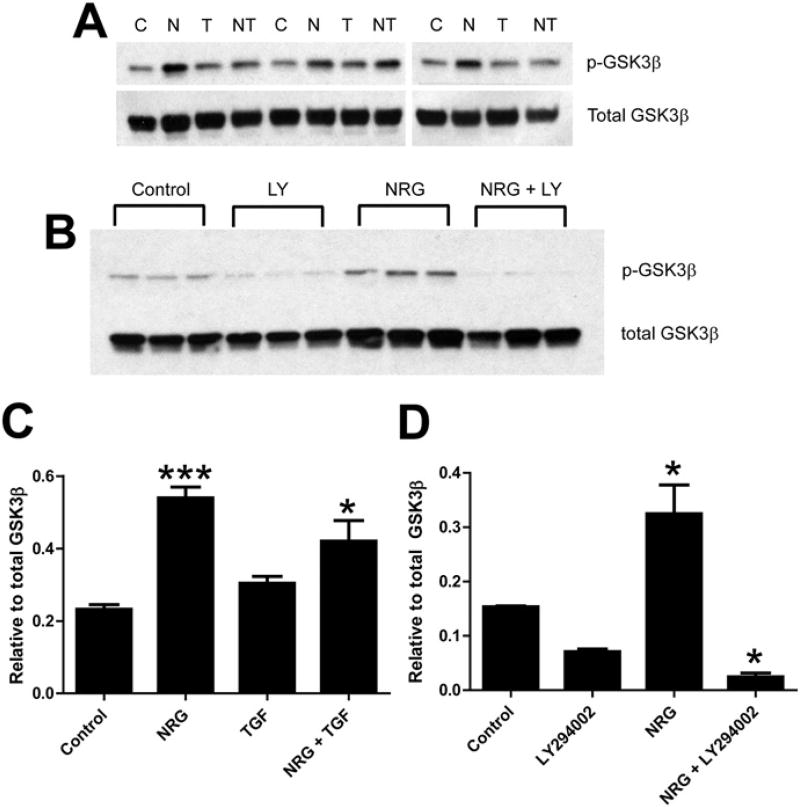
GSK-3β, a known downstream target of AKT kinase activity, was likewise phosphorylated in NHCV cells upon stimulation with exogenous NRG-1β (Fig. 4A and C). As was observed for AKT, TGF-β did not significantly affect the phosphorylation of GSK-3β when given alone or in conjunction with NRG-1β. Also, GSK-3β phosphorylation was significantly reduced in cells exposed to LY49002 (Fig. 4B and D), as was observed for AKT phosphorylation. Considered together, these results strongly suggest that NRG-1β activates the PI3K-AKT-GSK-3β signaling axis in human cardiac fibroblasts.
Fig. 4.
Neuregulin-1β treatment induces phosphorylation of GSK-3β in human cardiac fibroblasts. A, Western blot analysis of protein lysates of NHCV fibroblasts treated for 30 min with 30 ng/mL of NRG-1β. Bands shown are 46 kDa and were detected using anti-rabbit GSK-3β and after re-probing with pan GSK-3β. Experimental conditions included untreated control cells, denoted by a “C”, and cells treated with NRG-1β (N), 1 ng/mL TGF-β (T), or both NRG-1β and TGF-1β (NT). Representative blots are shown. B, Western blots of protein lysates of NHCV fibroblasts probed first with pGSK-3β and then with pan GSK-3β (as a loading control). Treatments shown (in triplicate) include untreated control cells, and cells treated with the PI3K inhibitor LY294002 (LY), 30 ng/mL of NRG-1β (NRG), or both LY and NRG. C and D, Graphical representations of densitometry analysis results for the differentially expressed proteins shown in A and B, respectively (*p < 0.05 and ***p < 0.001, respectively for NRG-1β-treated cells, when compared to appropriate control treatments).
3.4. NRG-1β-induces ErbB3-dependent activation of ErbB2 in NHCV fibroblasts
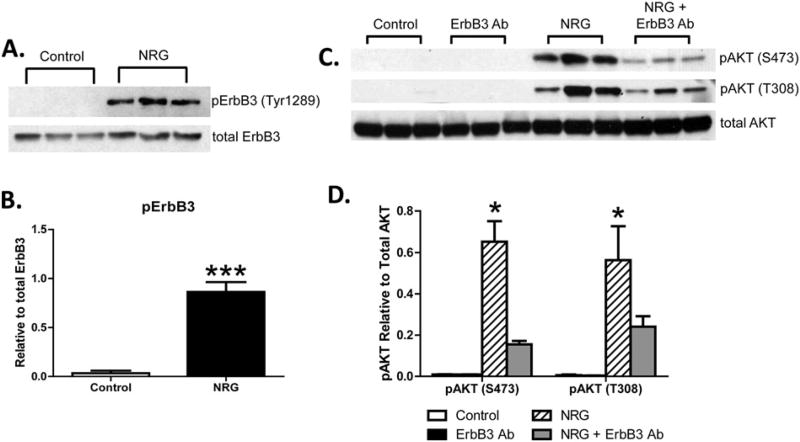
We next measured ErbB receptor activation and found that ErbB3 receptor was phosphorylated at a key PI3K site (Tyr1289) in human fibroblasts treated with NRG-1β (Fig. 5A). Phospho-ErbB3 was not detectable in unstimulated NHCV cells (Fig. 5A), and NRG-1β-mediated induction was significant and reproducible by 20 min post-treatment (Fig. 5B). Inhibition of ErbB3 with MAB3481 suppressed AKT phosphorylation at S473 and T308 (Fig. 5C, D).
Fig. 5.
Neuregulin-1β treatment induces phosphorylation of ErbB3 receptor in human cardiac fibroblasts. A, Western blot analysis of protein lysates of NHCV fibroblasts treated for 30 min with 30 ng/mL of NRG-1β. Bands shown are 185 kDa and were detected using an anti-rabbit antibodies that recognize phosphorylated ErbB3 (pErbB) at tyrosine position 1289 (Tyr1289) and pan ErbB3. B, Graphical representation shows densitometry analysis result for blot shown in A. C, Western blots of protein lysates of NHCV fibroblasts probed with pAKT at S473 or T308 and re-probed with pan AKT. Treatments shown (in triplicate) include untreated control cells, and cells treated with an ErbB3 blocking antibody (MAB3481), 30 ng/mL of NRG-1β (NRG), or both ErbB3Ab and NRG. Graphical representations of densitometry analysis results for the differentially expressed proteins are shown in C (n=3, *p < 0.05, ***p < 0.001 for NRG-1β-treated cells compared to untreated fibroblasts).
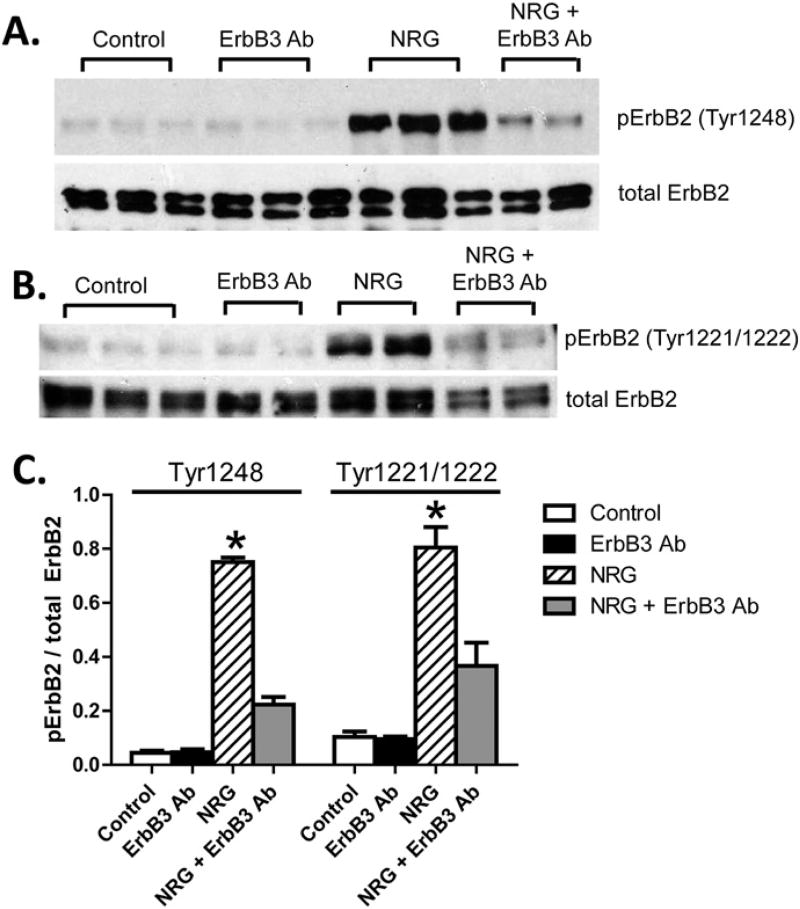
We next examined ErbB2 activation, because NRG-1β/ErbB3 signaling requires dimerization with one of the other ErbB receptors, most typically ErbB2. ErbB2 receptor phosphorylation at Tyr1248 was detectable by Western blot analysis in NHCV fibroblasts treated for 20 min with NRG-1β (Fig. 6A). ErbB2 phosphorylation at Tyr1221/1222 was also present in response to NRG-1β (Fig. 6B). These phosphorylation events were dependent upon activation of ErbB3, as demonstrated by significant suppression of phospho-ErbB2 Tyr1248 and Tyr1221/1222 levels by the ErbB3 inhibitor MAB3481 (Fig. 6).
Fig. 6.
Neuregulin-1β treatment induces phosphorylation of ErbB2 receptor in human cardiac fibroblasts. A, Western blot analysis of protein lysates of NHCV fibroblasts treated for 30 min with 30 ng/mL of NRG-1β. Bands shown are 185 kDa and were detected using an anti-rabbit antibodies that recognize phosphorylated ErbB2 (pErbB2) at tyrosine position 1289 (Tyr1248) and pan ErbB2. A representative blot is shown. B, Western blot analysis of protein lysates of NHCV fibroblasts treated for 30 min with 30 ng/mL of NRG-1β. Bands shown are 185 kDa and were detected using anti-rabbit antibodies that recognize phosphorylated ErbB2 (pErbB2) at tyrosine positions 1221 and 1222 (Tyr1221/1222), and pan ErbB2. C, Graphical representation showing densitometry analysis result for blots shown in A and B. Asterisks represent significance (n= 3, *p < 0.05 for NRG-1β-treated cells compared to controls).
3.5. NRG-1β enhances proliferation and survival of human cardiac fibroblasts
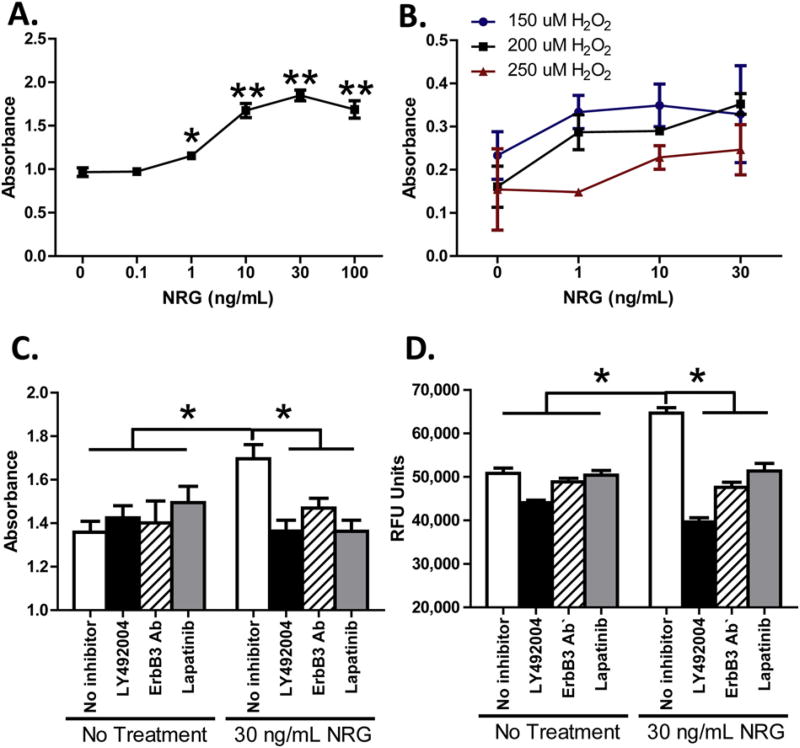
We next measured BrdU incorporation in NHCV fibroblasts and found a dose-dependent increase in cellular proliferation in response to NRG-1 β treatment (Fig. 7A). Similarly, NRG-1β increased human cardiac fibroblast survival in the presence of hydrogen peroxide (H2O2) (Fig. 7B). Blockade of PI3K (LY294002), ErbB3 (MAB3481), or ErbB2 (Lapatinib) suppressed NRG-1β-induced cell proliferation (Fig. 7A and C), as well as viability of fibroblasts exposed to 200 µM H2O2 (Fig. 7B and D).
Fig. 7.
NRG enhances proliferation and survival of human cardiac fibroblasts. A, graphical representation of BrdU incorporation in NHCV fibroblasts treated with increasing dosages of NRG (1–100 ng/mL), as shown on the abscissa. Cells were treated for 24 h, then treated with BrdU, incubated an additional 24 h, and the absorbance recorded as shown on the ordinate. B, graphical representation of cell viability in NHCV fibroblasts exposed to hydrogen peroxide (H2O2) and treated with NRG for 24 h. Absorbance at 590/600 nm is shown on the ordinate and concentration of NRG is displayed on the abscissa. Three concentrations of H2O2 are shown, as represented by blue (150 µm), black (200 µm), and red (250 µm) lines. C, results of cell proliferation assays for untreated and NRG-treated NHCV fibroblasts with and without pre-treatment with inhibitors for PI3K (LY294002, ErbB3 (MAB3481), or ErbB2 (Lapatinib). Treatments and BrdU incorporation were performed as described in A. D, results of viability assays for untreated and NRG-treated NHCV fibroblasts with and without pre-treatment with inhibitors for PI3K (LY294002), ErbB3 (MAB3481), or ErbB2 (Lapatinib). Untreated and treated cells were challenged with 200 µm for 24 h to induce cell death, as described in B (*p < 0.05 for NRG-1β-treated cells compared to controls).
3.6. NRG-1β induces secretion of cytokines from human cardiac fibroblasts
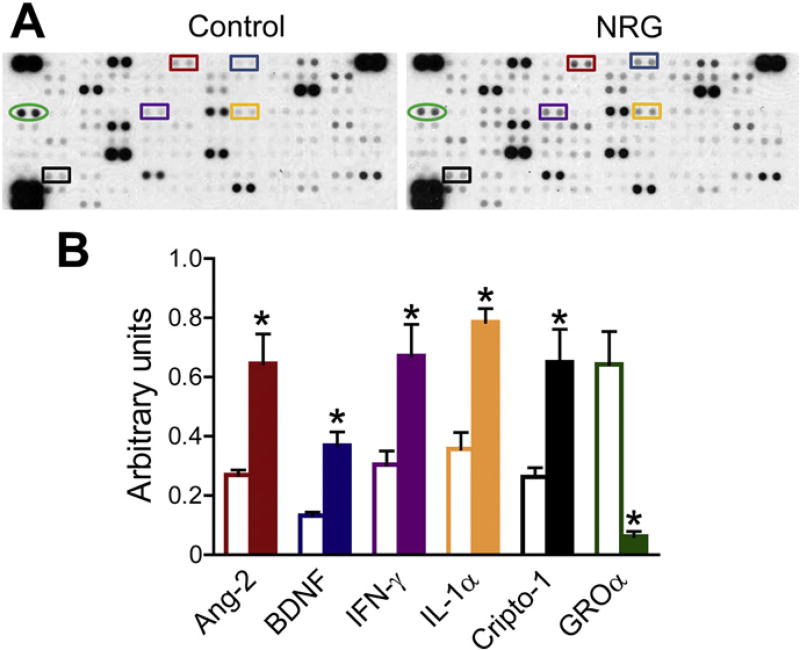
In the post-MI setting, proliferating fibroblasts are important for angiogenesis and other inflammatory-mediated reparative processes. We therefore screened the secretome in NHCV fibroblasts to identify NRG-1β inducible cytokines. In supernatants from NHCV cells treated for 24 h, we found induction of several cytokines, which were reproducibly secreted in response to NRG-1β treatment in three different fibroblast populations (Fig. 8A). Reproducible and statistically significant cytokines induced by NRG-1β treatment included angiopoietin-2 (Ang-2), brain-derived neurotropic protein (BDNF), interferon-γ (IFN-γ), interleukin-1α (IL-1α), and Cryptic family 1 (Cripto-1) (Fig. 8B). Growth-related α protein (GROα) production and secretion, on the other hand, was inhibited by NRG-1β treatment in NHCV fibroblasts (Fig. 8).
Fig. 8.
Cytokines secreted from neuregulin-treated fibroblasts. A, representative immunoblots show differentially expressed cytokines in the supernatants collected from NHCV fibroblasts that were untreated or treated for 24 h with NRG-1β. Each cytokine is represented on the array in duplicate, and each array includes three positive references (top left, top right, and bottom left) as well as negative reference spots (bottom right). Colored squares indicate significantly differential cytokines. B, graphical representation of differentially expressed cytokines identified using cytokine profile arrays. Data includes densitometry analysis results for three replicate experiments conducted using NHCV fibroblast cell populations established from three separate individuals (n = 3, *p < 0.05 for NRG-1β-treated cells, when compared to the various control treatments). Open and solid bars represent untreated control cells and NRG-1β-treated cells, respectively. Bar colors correspond to colors of squares shown in A.
4. Discussion
The present studies describe a new pathway of NRG-1β/ErbB receptor signaling in cardiac fibroblasts (Fig. 9). We demonstrate for the first time that NRG-1β induces phosphorylation of ErbB3 and ErbB2 receptors, resulting in activation of signaling in association with proliferative, pro-survival, and expression of cytokines with known roles in multiple processes, including recruitment of phagocytic immune cells, angiogenesis and activation of stem cells (Fig. 9). These findings have interesting and important implications for understanding the function of NRG-1β in the heart, including understanding how ErbB-targeted therapies for cancer may alter cardiovascular function. As importantly, these findings have implications for the development of recombinant NRGs for the treatment of cardiovascular and other diseases.
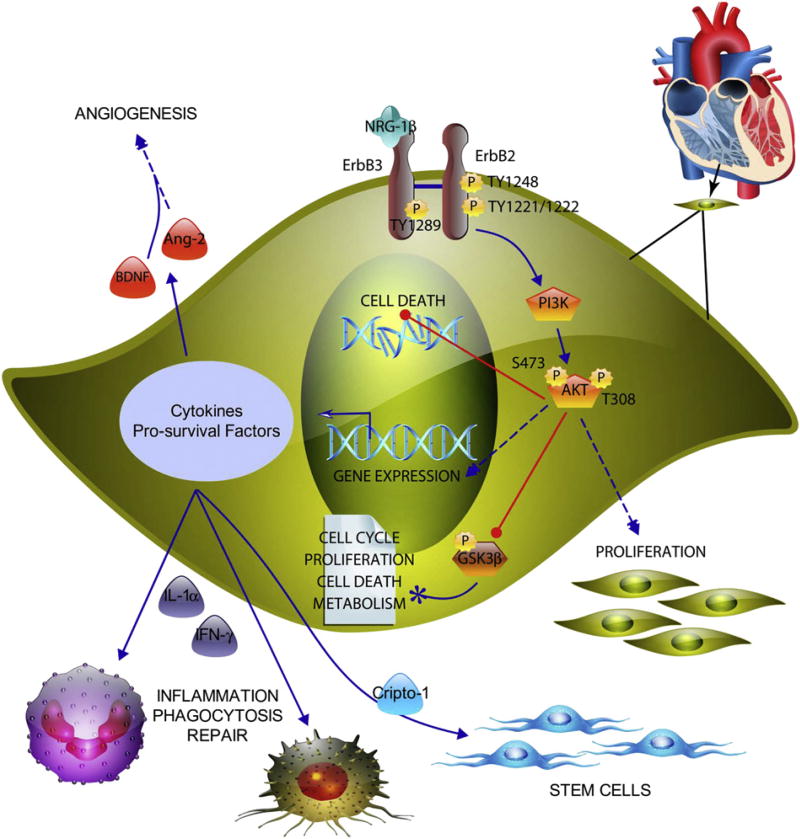
Fig. 9.
Model illustrating the intracellular signaling pathway leading to cell proliferation and survival in response to NRG-1β treatment. NRG-1β induces phosphorylation of the kinase-dead ErbB3 receptor, which leads to recruitment and phosphorylation of the kinase active ErbB2 receptor. This leads to activation of phosphoinositide-3 kinase (PI3K) with subsequent phosphorylation of protein kinase B (AKT) at serine (S)473 and threonine (T)308. AKT phosphorylates and inactivates GSK-3β, which participates in multiple signaling pathways, per the literature (paper icon).
NRG-1β/ErbB receptor signaling in cardiac cells has been well established, although the major focus to date has been on cardiomyocytes and endothelial cells. Our finding that ErbB receptor signaling occurs in human cardiac ventricular fibroblasts builds upon our previous work demonstrating ErbB receptor expression in rat and mouse cardiac stromal cells [27], as well as that by other investigators showing that ErbB receptor expression in fibroblasts from other tissues [28], most recently in skin fibroblasts [29,30].
It appears that all three ErbB receptors are expressed in normal NHCV fibroblasts and are functionally active. The main route of signaling activated by recombinant NRG-1β under the condition studied appears to be formation of ErbB2/ErbB3 heterodimers, since inhibition of either of these receptors significantly reduced downstream signaling and functions. It is likely, however, that ErbB4 also contributes to NRG-1β-mediated signaling in fibroblasts, because the ErbB3 blocking antibody reduced but did not eliminate ErbB2 phosphorylation. Whether ErbB4 activity in these cells occurs in conjunction with ErbB3-mediated signaling or is a compensatory consequence of ErbB3 inhibition will require further study. NRG-1β/ErbB signaling might lead to feedback regulation of ErbB receptor expression in cardiac fibroblasts via endocytic downregulation or via ubiquitination and proteolysis, as suggested by previous studies of ErbB receptors [31]. It is also possible that cardiac fibroblast ErbB3 or ErbB4 receptor is activated by other ligands, such as betacellulin, epiregulin, heparin-binding EGF-like growth factor, or other forms of NRG (e.g., NRG-1α or NRG-2).
Increased proliferation and survival is a hallmark of NRG-1β-mediated signaling in a variety of cell types, but NRG-1β-stimulated production of cytokines in cardiac fibroblasts is a novel finding. Angiopoietin (Ang)-2 protein levels in supernatants from normal human cardiac ventricular fibroblasts obtained from three different individuals were significantly increased in response to NRG-1β administration (Fig. 8). Ang-2 is a tightly-regulated vascular growth factor important for postnatal angiogenesis [32,33]. In response to endothelial damage, dysfunction or ischemia, Ang-2 is released from activated endothelial cells and acts coordinately with vascular endothelial growth factor (VEGF) to destabilize the vasculature and promote angiogenesis [34–36]. During inflammation or angiogenesis, Ang-2 acts as a partial antagonist of the Ang-1-Tie2 signaling system, which in essence “inhibits endothelial cell quiescence”.
BDNF, which was also induced by NRG-1β treatment, is a circulating neurotrophic cytokine that coordinates the regulation of energy homeostasis, including glucose metabolism in peripheral tissues, cardiovascular functions, and circadian rhythm [37]. BDNF has been well-characterized as a growth and pro-survival factor in a variety of cell types, including neurons [38], skeletal muscle stem cells, [39] and more recently cardiac progenitor cells [40]. BDNF also promotes angiogenesis, improves left ventricular function under ischemic conditions [41,42] and protects against adverse cardiac remodeling after myocardial infarction [43,44]. Plasma levels of BDNF were found to be significantly decreased in patients with heart failure, and circulating levels of BDNF levels were associated with cardiac disease severity [45].
Of note, we found that NRG-1β increases production of IFN-γ and IL-1α, which are potent immunomodulatory cytokines with pleiotropic effects, dependent upon cell type and context [46]. For example, IFN-γ is pro-inflammatory but can also induce autophagy and may thereby contribute to cardiomyocyte survival [47,48]. Similarly, IL-1α is triggered as part of the immune response after myocardial infarction, which initiates the phagocytic clearance of necrotic/apoptotic cells and debris and thus promotes cardiac repair [49].
We also found that NRG-1β mediates increased production of Cripto-1, a multifunctional embryonic protein that controls stem cell biology [50], including maintenance of postnatal stem cells (e.g., skeletal muscle satellite cells), epithelial-to-mesenchymal-transition and tumorigenesis [51–53]. Cripto-1 is important for early heart morphogenesis [54], and is required for inducible differentiation of embryonic stem cells to a cardiac lineage [55,56]. Mice in which the Cripto-1 gene has been disrupted die in utero due to the inability of precardiac mesoderm to differentiate into functional cardiomyocytes [57]. Notably, Cripto-1 contains an EGF-like domain, which although incapable of direct binding to ErbB receptors, can indirectly enhance ErbB4 phosphorylation [58].
In contrast to the aforementioned cytokines, we found that NRG-1β induced downregulation of GROα in human cardiac fibroblasts. GROα (also called CXCL1), which is a ligand for CXC receptor 2 (CXCR2), a growth-related oncogene and chemoattractant for neutrophils [59,60], basophils [59], monocytes [61], and T cells [62]. GROα can induce the proliferation of endothelial cells in culture and angiogenesis in vivo [63,64]. Excessive GROα/CXCR2 signaling, however, can lead to endothelial dysfunction and vascular lesions. GROα has been implicated in thrombin-induced angiogenesis [65], atherosclerosis [66], and aortic aneurisms [67] and may contribute to coronary artery disease [64].
To our knowledge, this is the first study to demonstrate an important role of NRG-1β in regulating cardiac fibroblast signaling via the ErbB receptors leading to increased cell proliferation and survival. Since a known function of cardiac fibroblasts is to maintain the integrity of the cardiac extracellular matrix, the observed effects of NRG-1β on these cells suggest a role for ErbB signaling in the anti-fibrotic effects that have been observed in animal models of heart failure treated with recombinant NRG [27,68]. The modulatory effects of NRG/ErbB signaling on reactive oxygen species (ROS)-induced fibroblast survival warrant further exploration, given the established role of ROS in activation of a pro-fibrotic phenotype in cardiac fibroblasts [69]. Further exploration of these effects are important to understand in the context of developing recombinant NRGs engineered to selectively activate ErbB4 homodimers [70], which may or may not be sufficient to alter fibroblast biology.
Although the cardiac fibroblasts used were of human origin, one significant limitation of this study is an inability to extrapolate our results to the in vivo cardiac environment. How NRG-1β signaling might regulate fibroblasts during post-MI remodeling is likely complex, and additional studies using co-cultures of fibroblasts with other cardiac cells, animal models and human fibroblasts obtained from diseased heart explants are clearly warranted. Possible mechanisms of action include NRG-1β-mediated reduction in injury-primed fibroblast activation and matrix deposition, alterations of fibroblast-cardiomyocyte interactions, and/or paracrine modulation of inflammatory cell activities through fibroblast production and secretion of immune cell regulatory factors.
In summary, we demonstrate mitogenic and pro-survival activities for NRG-1β in human cardiac ventricular fibroblasts, which to our knowledge have never been previously reported. These data suggest a mechanism of action for NRG-1β-mediated fibroblast signaling via ErbB2 and ErbB3 dimerization leading to the activation of the PI3K/AKT pathway and secretion of cytokines (Fig. 9). Considered together, these data suggest that NRG-1β may contribute to cardiac repair through activation of paracrine signaling in fibroblasts (Fig. 9).
Acknowledgments
This work was supported by the National Institutes of Health [K01HL130497, 2T32HL007411-36, K01HL121045, U01 HL100398]. The Vanderbilt VANTAGE Core provided technical assistance for this work. VANTAGE is supported in part by CTSA Grant (5UL1 RR024975-03), the Vanderbilt Ingram Cancer Center (P30 CA68485), the Vanderbilt Vision Center (P30 EY08126), and NIH/NCRR (G20 RR030956).
Downstream consequences of these NRG/ErbB3-induced signaling events include transcriptional alterations, enhanced cellular proliferation, and a decrease in H2O2-mediated cell death. Also shown are the downstream effects of NRG-1β signaling, including secretion of cytokines (IL-1α, IFN-γ, Ang-2, BDNF, and Cripto-1) that are known to play important roles in multiple processes, including recruitment of phagocytic immune cells, angiogenesis and activation of stem cells. The illustration was produced using ePath 3D software (Protein Lounge, San Diego, CA).
Glossary
- NRG
neuregulin
- NHCV
normal human cardiac ventricular
- Tyr
tyrosine
- MI
myocardial infarction
- myoFb
myofibroblast
- ECM
extracellular matrix
- ErbB
erythroblastic leukemia viral oncogene homolog tyrosine kinase receptor
- PI3K
phosphoinositide 3-kinase
- GGF2
glial growth factor 2
- RNASeq
RNA sequencing
- qPCR
quantitative polymerase chain reaction
- PE
phycoerythrin
- Ang-2
angiopoietin-2
- BDNF
brain-derived neurotropic protein
- IFN-γ
interferon-γ
- IL-1α
interleukin-1α
- Cripto-1
cryptic family 1
- GROα
growth-related α
- VEGF
vascular endothelial growth factor
- CXCR2
CXC receptor 2
Footnotes
Disclosures
None.
References
- 1.Fan D, Takawale A, Lee J, Kassiri Z. Cardiac fibroblasts, fibrosis and extracellular matrix remodeling in heart disease. Fibrogenesis Tissue Repair. 2012;5:15. doi: 10.1186/1755-1536-5-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meyer D, Birchmeier C. Multiple essential functions of neuregulin in development. Nature. 1995;378:386–390. doi: 10.1038/378386a0. [DOI] [PubMed] [Google Scholar]
- 3.Kramer R, Bucay N, Kane DJ, Martin LE, Tarpley JE, Theill LE. Neuregulins with an Ig-like domain are essential for mouse myocardial and neuronal development. Proc. Natl. Acad. Sci. U. S. A. 1996;93:4833–4838. doi: 10.1073/pnas.93.10.4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Odiete O, Hill MF, Sawyer DB. Neuregulin in cardiovascular development and disease. Circ. Res. 2012;111:1376–1385. doi: 10.1161/CIRCRESAHA.112.267286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fuller SJ, Sivarajah K, Sugden PH. ErbB receptors, their ligands, and the consequences of their activation and inhibition in the myocardium. J. Mol. Cell. Cardiol. 2008;44:831–854. doi: 10.1016/j.yjmcc.2008.02.278. [DOI] [PubMed] [Google Scholar]
- 6.Galindo CL, Ryzhov S, Sawyer DB. Neuregulin as a heart failure therapy and mediator of reverse remodeling. Curr. Heart Fail. Rep. 2014;11:40–49. doi: 10.1007/s11897-013-0176-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai MX, Shi XC, Chen T, Tan ZN, Lin QQ, Du SJ, et al. Exercise training activates neuregulin 1/ErbB signaling and promotes cardiac repair in a rat myocardial infarction model. Life Sci. 2016;149:1–9. doi: 10.1016/j.lfs.2016.02.055. [DOI] [PubMed] [Google Scholar]
- 8.Ennequin G, Boisseau N, Caillaud K, Chavanelle V, Gerbaix M, Metz L, et al. Exercise training and return to a well-balanced diet activate the neuregulin 1/ErbB pathway in skeletal muscle of obese rats. J. Physiol. 2015;593:2665–2677. doi: 10.1113/JP270026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rupert CE, Coulombe KL. The roles of neuregulin-1 in cardiac development, homeostasis, and disease. Biomark. Insights. 2015;10:1–9. doi: 10.4137/BMI.S20061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuramochi Y, Cote GM, Guo X, Lebrasseur NK, Cui L, Liao R, et al. Cardiac endothelial cells regulate reactive oxygen species-induced cardiomyocyte apoptosis through neuregulin-1beta/erbB4 signaling. J. Biol. Chem. 2004;279:51141–51147. doi: 10.1074/jbc.M408662200. [DOI] [PubMed] [Google Scholar]
- 11.Noireaud J, Andriantsitohaina R. Recent insights in the paracrine modulation of cardiomyocyte contractility by cardiac endothelial cells. Biomed. Res. Int. 2014;2014:923805. doi: 10.1155/2014/923805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lemmens K, Segers VF, Demolder M, De Keulenaer GW. Role of neuregulin-1/ErbB2 signaling in endothelium-cardiomyocyte cross-talk. J. Biol. Chem. 2006;281:19469–19477. doi: 10.1074/jbc.M600399200. [DOI] [PubMed] [Google Scholar]
- 13.Kalinowski A, Plowes NJ, Huang Q, Berdejo-Izquierdo C, Russell RR, Russell KS. Metalloproteinase-dependent cleavage of neuregulin and autocrine stimulation of vascular endothelial cells. FASEB J. 2010;24:2567–2575. doi: 10.1096/fj.08-129072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iivanainen E, Paatero I, Heikkinen SM, Junttila TT, Cao R, Klint P, et al. Intra- and extracellular signaling by endothelial neuregulin-1. Exp. Cell Res. 2007;313:2896–2909. doi: 10.1016/j.yexcr.2007.03.042. [DOI] [PubMed] [Google Scholar]
- 15.Ky B, Kimmel SE, Safa RN, Putt ME, Sweitzer NK, Fang JC, et al. Neuregulin-1 beta is associated with disease severity and adverse outcomes in chronic heart failure. Circulation. 2009;120:310–317. doi: 10.1161/CIRCULATIONAHA.109.856310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geisberg CA, Wang G, Safa RN, Smith HM, Anderson B, Peng XY, et al. Circulating neuregulin-1beta levels vary according to the angiographic severity of coronary artery disease and ischemia. Coron. Artery Dis. 2011;22:577–582. doi: 10.1097/MCA.0b013e32834d3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Falls DL. Neuregulins: functions, forms, and signaling strategies. Exp. Cell Res. 2003;284:14–30. doi: 10.1016/s0014-4827(02)00102-7. [DOI] [PubMed] [Google Scholar]
- 18.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat. Rev. Mol. Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 19.Carraway KL, 3rd, Burden SJ. Neuregulins and their receptors. Curr. Opin. Neurobiol. 1995;5:606–612. doi: 10.1016/0959-4388(95)80065-4. [DOI] [PubMed] [Google Scholar]
- 20.Fukazawa R, Miller TA, Kuramochi Y, Frantz S, Kim YD, Marchionni MA, et al. Neuregulin-1 protects ventricular myocytes from anthracycline-induced apoptosis via erbB4-dependent activation of PI3-kinase/Akt. J. Mol. Cell. Cardiol. 2003;35:1473–1479. doi: 10.1016/j.yjmcc.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 21.Kuramochi Y, Guo X, Sawyer DB. Neuregulin activates erbB2-dependent src/FAK signaling and cytoskeletal remodeling in isolated adult rat cardiac myocytes. J. Mol. Cell. Cardiol. 2006;41:228–235. doi: 10.1016/j.yjmcc.2006.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuramochi Y, Lim CC, Guo X, Colucci WS, Liao R, Sawyer DB. Myocyte contractile activity modulates norepinephrine cytotoxicity and survival effects of neuregulin-1beta. Am. J. Physiol. Cell Physiol. 2004;286:C222–C229. doi: 10.1152/ajpcell.00312.2003. [DOI] [PubMed] [Google Scholar]
- 23.Zhao YY, Sawyer DR, Baliga RR, Opel DJ, Han X, Marchionni MA, et al. Neuregulins promote survival and growth of cardiac myocytes. Persistence of ErbB2 and ErbB4 expression in neonatal and adult ventricular myocytes. J. Biol. Chem. 1998;273:10261–10269. doi: 10.1074/jbc.273.17.10261. [DOI] [PubMed] [Google Scholar]
- 24.Kim HS, Cho JW, Hidaka K, Morisaki T. Activation of MEK-ERK by heregulin-beta1 promotes the development of cardiomyocytes derived from ES cells. Biochem. Biophys. Res. Commun. 2007;361:732–738. doi: 10.1016/j.bbrc.2007.07.045. [DOI] [PubMed] [Google Scholar]
- 25.Wang Z, Xu G, Wu Y, Guan Y, Cui L, Lei X, et al. Neuregulin-1 enhances differentiation of cardiomyocytes from embryonic stem cells. Med. Biol. Eng. Comput. 2009;47:41–48. doi: 10.1007/s11517-008-0383-2. [DOI] [PubMed] [Google Scholar]
- 26.Hao J, Galindo CL, Tran TL, Sawyer DB. Neuregulin-1beta induces embryonic stem cell cardiomyogenesis via ErbB3/ErbB2 receptors. Biochem. J. 2014;458:335–341. doi: 10.1042/BJ20130818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galindo CL, Kasasbeh E, Murphy A, Ryzhov S, Lenihan S, Ahmad FA, et al. Anti-remodeling and anti-fibrotic effects of the neuregulin-1beta glial growth factor 2 in a large animal model of heart failure. J. Am. Heart Assoc. 2014;3:e000773. doi: 10.1161/JAHA.113.000773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andrianifahanana M, Wilkes MC, Repellin CE, Edens M, Kottom TJ, Rahimi RA, et al. ERBB receptor activation is required for profibrotic responses to transforming growth factor beta. Cancer Res. 2010;70:7421–7430. doi: 10.1158/0008-5472.CAN-10-0232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim JS, Choi IG, Lee BC, Park JB, Kim JH, Jeong JH, et al. Neuregulin induces CTGF expression in hypertrophic scarring fibroblasts. Mol. Cell. Biochem. 2012;365:181–189. doi: 10.1007/s11010-012-1258-2. [DOI] [PubMed] [Google Scholar]
- 30.Jumper N, Hodgkinson T, Paus R, Bayat A. A role for neuregulin-1 in promoting keloid fibroblast migration via ErbB2-mediated signaling. Acta Derm. Venereol. 2016 doi: 10.2340/00015555-2587. [ePub ahead of print] [DOI] [PubMed] [Google Scholar]
- 31.Roepstorff K, Grovdal L, Grandal M, Lerdrup M, van Deurs B. Endocytic downregulation of ErbB receptors: mechanisms and relevance in cancer. Histochem. Cell Biol. 2008;129:563–578. doi: 10.1007/s00418-008-0401-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gale NW, Thurston G, Hackett SF, Renard R, Wang Q, McClain J, et al. Angiopoietin-2 is required for postnatal angiogenesis and lymphatic patterning, and only the latter role is rescued by Angiopoietin-1. Dev. Cell. 2002;3:411–423. doi: 10.1016/s1534-5807(02)00217-4. [DOI] [PubMed] [Google Scholar]
- 33.Maisonpierre PC, Suri C, Jones PF, Bartunkova S, Wiegand SJ, Radziejewski C, et al. Angiopoietin-2, a natural antagonist for Tie2 that disrupts in vivo angiogenesis. Science. 1997;277:55–60. doi: 10.1126/science.277.5322.55. [DOI] [PubMed] [Google Scholar]
- 34.Fiedler U, Scharpfenecker M, Koidl S, Hegen A, Grunow V, Schmidt JM, et al. The Tie-2 ligand angiopoietin-2 is stored in and rapidly released upon stimulation from endothelial cell Weibel-Palade bodies. Blood. 2004;103:4150–4156. doi: 10.1182/blood-2003-10-3685. [DOI] [PubMed] [Google Scholar]
- 35.Augustin HG, Koh GY, Thurston G, Alitalo K. Control of vascular morphogenesis and homeostasis through the angiopoietin-Tie system. Nat. Rev. Mol. Cell Biol. 2009;10:165–177. doi: 10.1038/nrm2639. [DOI] [PubMed] [Google Scholar]
- 36.Lobov IB, Brooks PC, Lang RA. Angiopoietin-2 displays VEGF-dependent modulation of capillary structure and endothelial cell survival in vivo. Proc. Natl. Acad. Sci. U. S. A. 2002;99:11205–11210. doi: 10.1073/pnas.172161899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marosi K, Mattson MP. BDNF mediates adaptive brain and body responses to energetic challenges. Trends Endocrinol. Metab. 2014;25:89–98. doi: 10.1016/j.tem.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Binder DK, Scharfman HE. Brain-derived neurotrophic factor. Growth Factors. 2004;22:123–131. doi: 10.1080/08977190410001723308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clow C, Jasmin BJ. Brain-derived neurotrophic factor regulates satellite cell differentiation and skeltal muscle regeneration. Mol. Biol. Cell. 2010;21:2182–2190. doi: 10.1091/mbc.E10-02-0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Samal R, Ameling S, Dhople V, Sappa PK, Wenzel K, Volker U, et al. Brain derived neurotrophic factor contributes to the cardiogenic potential of adult resident progenitor cells in failing murine heart. PLoS One. 2015;10:e0120360. doi: 10.1371/journal.pone.0120360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kermani P, Hempstead B. Brain-derived neurotrophic factor: a newly described mediator of angiogenesis. Trends Cardiovasc. Med. 2007;17:140–143. doi: 10.1016/j.tcm.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Y, Sun L, Huan Y, Zhao H, Deng J. Application of bFGF and BDNF to improve angiogenesis and cardiac function. J. Surg. Res. 2006;136:85–91. doi: 10.1016/j.jss.2006.04.034. [DOI] [PubMed] [Google Scholar]
- 43.Okada S, Yokoyama M, Toko H, Tateno K, Moriya J, Shimizu I, et al. Brain-derived neurotrophic factor protects against cardiac dysfunction after myocardial infarction via a central nervous system-mediated pathway. Arterioscler. Thromb. Vasc. Biol. 2012;32:1902–1909. doi: 10.1161/ATVBAHA.112.248930. [DOI] [PubMed] [Google Scholar]
- 44.Hang P, Zhao J, Cai B, Tian S, Huang W, Guo J, et al. Brain-derived neurotrophic factor regulates TRPC3/6 channels and protects against myocardial infarction in rodents. Int. J. Biol. Sci. 2015;11:536–545. doi: 10.7150/ijbs.10754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takashio S, Sugiyama S, Yamamuro M, Takahama H, Hayashi T, Sugano Y, et al. Significance of low plasma levels of brain-derived neurotrophic factor in patients with heart failure. Am. J. Cardiol. 2015;116:243–249. doi: 10.1016/j.amjcard.2015.04.018. [DOI] [PubMed] [Google Scholar]
- 46.Nathan C. Points of control in inflammation. Nature. 2002;420:846–852. doi: 10.1038/nature01320. [DOI] [PubMed] [Google Scholar]
- 47.Garcia AG, Wilson RM, Heo J, Murthy NR, Baid S, Ouchi N, et al. Interferon-gamma ablation exacerbates myocardial hypertrophy in diastolic heart failure. Am. J. Physiol. Heart Circ. Physiol. 2012;303:H587–H596. doi: 10.1152/ajpheart.00298.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paulus GL, Xavier RJ. Autophagy and checkpoints for intracellular pathogen defense. Curr. Opin. Gastroenterol. 2015;31:14–23. doi: 10.1097/MOG.0000000000000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frangogiannis NG. Inflammation in cardiac injury, repair and regeneration. Curr. Opin. Cardiol. 2015;30:240–245. doi: 10.1097/HCO.0000000000000158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bianco C, Rangel MC, Castro NP, Nagaoka T, Rollman K, Gonzales M, et al. Role of Cripto-1 in stem cell maintenance and malignant progression. Am. J. Pathol. 2010;177:532–540. doi: 10.2353/ajpath.2010.100102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guardiola O, Lafuste P, Brunelli S, Iaconis S, Touvier T, Mourikis P, et al. Cripto regulates skeletal muscle regeneration and modulates satellite cell determination by antagonizing myostatin. Proc. Natl. Acad. Sci. U. S. A. 2012;109:E3231–E3240. doi: 10.1073/pnas.1204017109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bianco C, Strizzi L, Ebert A, Chang C, Rehman A, Normanno N, et al. Role of human cripto-1 in tumor angiogenesis. J. Natl. Cancer Inst. 2005;97:132–141. doi: 10.1093/jnci/dji011. [DOI] [PubMed] [Google Scholar]
- 53.Watanabe K, Meyer MJ, Strizzi L, Lee JM, Gonzales M, Bianco C, et al. Cripto-1 is a cell surface marker for a tumorigenic, undifferentiated subpopulation in human embryonal carcinoma cells. Stem Cells. 2010;28:1303–1314. doi: 10.1002/stem.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dono R, Scalera L, Pacifico F, Acampora D, Persico MG, Simeone A. The murine cripto gene: expression during mesoderm induction and early heart morphogenesis. Development. 1993;118:1157–1168. doi: 10.1242/dev.118.4.1157. [DOI] [PubMed] [Google Scholar]
- 55.Xu C, Liguori G, Adamson ED, Persico MG. Specific arrest of cardiogenesis in cultured embryonic stem cells lacking Cripto-1. Dev. Biol. 1998;196:237–247. doi: 10.1006/dbio.1998.8862. [DOI] [PubMed] [Google Scholar]
- 56.Bianco C, Cotten C, Lonardo E, Strizzi L, Baraty C, Mancino M, et al. Cripto-1 is required for hypoxia to induce cardiac differentiation of mouse embryonic stem cells. Am. J. Pathol. 2009;175:2146–2158. doi: 10.2353/ajpath.2009.090218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu C, Liguori G, Persico MG, Adamson ED. Abrogation of the Cripto gene in mouse leads to failure of postgastrulation morphogenesis and lack of differentiation of cardiomyocytes. Development. 1999;126:483–494. doi: 10.1242/dev.126.3.483. [DOI] [PubMed] [Google Scholar]
- 58.Bianco C, Kannan S, De Santis M, Seno M, Tang CK, Martinez-Lacaci I, et al. Cripto-1 indirectly stimulates the tyrosine phosphorylation of erb B-4 through a novel receptor. J. Biol. Chem. 1999;274:8624–8629. doi: 10.1074/jbc.274.13.8624. [DOI] [PubMed] [Google Scholar]
- 59.Geiser T, Dewald B, Ehrengruber MU, Clark-Lewis I, Baggiolini M. The interleukin-8-related chemotactic cytokines GRO alpha, GRO beta, and GRO gamma activate human neutrophil and basophil leukocytes. J. Biol. Chem. 1993;268:15419–15424. [PubMed] [Google Scholar]
- 60.Bozic CR, Kolakowski LF, Jr, Gerard NP, Garcia-Rodriguez C, von Uexkull-Guldenband C, Conklyn MJ, et al. Expression and biologic characterization of the murine chemokine KC. J. Immunol. 1995;154:6048–6057. [PubMed] [Google Scholar]
- 61.Schwartz D, Andalibi A, Chaverri-Almada L, Berliner JA, Kirchgessner T, Fang ZT, et al. Role of the GRO family of chemokines inmonocyte adhesion to MM-LDL-stimulated endothelium. J. Clin. Invest. 1994;94:1968–1973. doi: 10.1172/JCI117548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jinquan T, Frydenberg J, Mukaida N, Bonde J, Larsen CG, Matsushima K, et al. Recombinant human growth-regulated oncogene-alpha induces T lymphocyte chemotaxis. A process regulated via IL-8 receptors by IFN-gamma, TNF-alpha, IL-4, IL-10, and IL-13. J. Immunol. 1995;155:5359–5368. [PubMed] [Google Scholar]
- 63.Bechara C, Chai H, Lin PH, Yao Q, Chen C. Growth related oncogene-alpha (GRO-alpha): roles in atherosclerosis, angiogenesis and other inflammatory conditions. Med. Sci. Monit. 2007;13:RA87–RA90. [PubMed] [Google Scholar]
- 64.Bechara C, Wang X, Chai H, Lin PH, Yao Q, Chen C. Growth-related oncogene-alpha induces endothelial dysfunction through oxidative stress and downregulation of eNOS in porcine coronary arteries. Am. J. Physiol. Heart Circ. Physiol. 2007;293:H3088–H3095. doi: 10.1152/ajpheart.00473.2007. [DOI] [PubMed] [Google Scholar]
- 65.Caunt M, Hu L, Tang T, Brooks PC, Ibrahim S, Karpatkin S. Growth-regulated oncogene is pivotal in thrombin-induced angiogenesis. Cancer Res. 2006;66:4125–4132. doi: 10.1158/0008-5472.CAN-05-2570. [DOI] [PubMed] [Google Scholar]
- 66.Boisvert WA, Santiago R, Curtiss LK, Terkeltaub RA. A leukocyte homologue of the IL-8 receptor CXCR-2 mediates the accumulation of macrophages in atherosclerotic lesions of LDL receptor-deficient mice. J. Clin. Invest. 1998;101:353–363. doi: 10.1172/JCI1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Middleton RK, Lloyd GM, Bown MJ, Cooper NJ, London NJ, Sayers RD. The pro-inflammatory and chemotactic cytokine microenvironment of the abdominal aortic aneurysm wall: a protein array study. J. Vasc. Surg. 2007;45:574–580. doi: 10.1016/j.jvs.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 68.Hill MF, Patel AV, Murphy A, Smith HM, Galindo CL, Pentassuglia L, et al. Intravenous glial growth factor 2 (GGF2) isoform of neuregulin-1beta improves left ventricular function, gene and protein expression in rats after myocardial infarction. PLoS One. 2013;8:e55741. doi: 10.1371/journal.pone.0055741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Richter K, Kietzmann T. Reactive oxygen species and fibrosis: further evidence of a significant liaison. Cell Tissue Res. 2016;365:591–605. doi: 10.1007/s00441-016-2445-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jay SM, Murthy AC, Hawkins JF, Wortzel JR, Steinhauser ML, Alvarez LM, et al. An engineered bivalent neuregulin protects against doxorubicin-induced cardiotoxicity with reduced proneoplastic potential. Circulation. 2013;128:152–161. doi: 10.1161/CIRCULATIONAHA.113.002203. [DOI] [PMC free article] [PubMed] [Google Scholar]