Significance
The contribution of SNARE-mediated membrane fusion to the different stages along the autophagic process is not well understood. In this work we demonstrate that inhibition of SNARE priming leads to impairment of the autophagic flux manifested by the accumulation of mature autophagosomes that do not fuse with the lysosomes. Our results suggest that SNARE priming is essential for the fusion of the autophagosome with the lysosome but not for autophagosome formation.
Keywords: autophagy, SNARE priming, NSF, αSNAP, autophagosome biogenesis
Abstract
Autophagy, a unique intracellular membrane-trafficking pathway, is initiated by the formation of an isolation membrane (phagophore) that engulfs cytoplasmic constituents, leading to generation of the autophagosome, a double-membrane vesicle, which is targeted to the lysosome. The outer autophagosomal membrane consequently fuses with the lysosomal membrane. Multiple membrane-fusion events mediated by SNARE molecules have been postulated to promote autophagy. αSNAP, the adaptor molecule for the SNARE-priming enzyme N-ethylmaleimide-sensitive factor (NSF) is known to be crucial for intracellular membrane fusion processes, but its role in autophagy remains unclear. Here we demonstrated that knockdown of αSNAP leads to inhibition of autophagy, manifested by an accumulation of sealed autophagosomes located in close proximity to lysosomes but not fused with them. Under these conditions, moreover, association of both Atg9 and the autophagy-related SNARE protein syntaxin17 with the autophagosome remained unaffected. Finally, our results suggested that under starvation conditions, the levels of αSNAP, although low, are nevertheless sufficient to partially promote the SNARE priming required for autophagy. Taken together, these findings indicate that while autophagosomal–lysosomal membrane fusion is sensitive to inhibition of SNARE priming, the initial stages of autophagosome biogenesis and autophagosome expansion remain resistant to its loss.
Most intracellular membrane fusion events are mediated by soluble NSF attachment protein receptor (SNARE) molecules. A set of three Q/t-SNAREs on the target membrane and one R/v-SNARE on the trafficking vesicle create a SNARE–pin complex, bringing the membrane and the vesicle into close proximity and thus enabling their fusion (1, 2). Once the two have fused, the individual SNARE molecules are released from their complex, in an ATP-dependent manner, by the enzyme N-ethylmaleimide sensitive factor (NSF) and its adaptor soluble NSF-attachment protein alpha (αSNAP) (3). These two proteins are universal components of cell trafficking and fusion, and deletion of either of them is lethal to the cell (4). Recruitment of the ATPase NSF to the SNARE complex is fully dependent on the presence of αSNAP. Three αSNAP molecules direct the homohexameric NSF to a SNARE protein complex (5), stimulating ATP hydrolysis and thereby inducing the conformational changes in the SNARE molecules that result in their “priming” and enable them to promote subsequent events (6). Besides αSNAP, there are two additional SNAP isoforms: βSNAP, which promotes membrane fusion in the brain (7), and γSNAP, recently shown to stimulate SNARE disassembly specifically in endosomes (8). Changes in intracellular αSNAP levels have also been implicated in various pathological conditions such as neurodegenerative diseases, diabetes, and cancer (reviewed in ref. 9).
Autophagy is a catabolic process that targets proteins and organelles for degradation in the lysosome. It is initiated by the formation of an isolation membrane (also termed phagophore) that elongates and engulfs cytoplasmatic constituents to form the autophagosome, a double-membrane vesicle, which is then targeted to the lysosome. These sequential stages of autophagosome biogenesis demand significant membrane remodeling and membrane fusion processes. There is no evidence however for retrograde transport from this vesicle back to the original membrane, suggesting that SNARE priming might not be essential for autophagosome biogenesis.
The role of SNARE molecules in the autophagic process in yeast and in mammalian cells was recently reported. SNARE complexes that promote fusion of vesicles with the lysosome in yeast have been identified in both the cytoplasm-to-vacuole (Cvt) pathway and starvation-induced autophagy (10), as well as in phagophore elongation (11). In mammalian cells and in Drosophila, SNARE molecules were found essential for autophagosomal–lysosomal membrane fusion (12, 13). The role of SNARE priming in autophagosome biogenesis, however, remains unclear. It was originally reported that Sec18p and Sec17p, NSF, and SNAP yeast orthologs are essential for autophagosomal–vacuolar membrane fusion but not for autophagosome formation (14). Nair et al. (15), however, have recently indicated that autophagosome formation in yeast may also be affected by mutations in these genes. In the mammalian system, it was suggested that SNARE molecules are essential for autophagosome biogenesis; yet the role of NSF and αSNAP in this system was not directly addressed (16, 17).
In the present study, we used an siRNA approach to examine the role of SNARE priming in autophagy. We showed that hampering of SNARE priming by knockdown of αSNAP leads to the inhibition of autophagy accompanied by the accumulation of LC3-and p62-labeled autophagosomes in the vicinity of lysosomes. Moreover, our results suggested that the translocation of syntaxin17 (an autophagy-related SNARE protein) from the mitochondria to the autophagosomes is not affected by the reduction in αSNAP levels, nor does it affect the association of Atg9 vesicles with the autophagic membrane. Importantly, starved cells were relatively more resistant to αSNAP knockdown, possibly because of heightened association of NSF molecules with the membrane at locations where it promotes SNARE priming. In summary, our results imply that SNARE priming is essential for the production of autophagosomes capable of fusion with the lysosomes, but are fairly dispensable to the membrane supply needed for autophagosomal formation.
Results
SNARE Priming Is Essential for Autophagic Flux.
SNARE priming, predominantly mediated by NSF and αSNAP, is essential for all intracellular membrane fusion events. Its role in autophagy, however, remains unclear. In an attempt to elucidate its role, we studied the effect of αSNAP knockdown on the autophagic activity of HeLa cells. Autophagic flux was monitored in the presence or absence of the lysosomal inhibitor bafilomycin A (BafA) using both Western blot and immunofluorescence analysis. αSNAP depletion led to a decrease in autophagic flux, as indicated by the accumulation of LC3II (the lipidated form of LC3, the mammalian ortholog of the yeast Atg8) and of p62, a well-characterized autophagic cargo (Fig. 1 A and B). Moreover, unlike the perinuclear distribution of the labeled vesicles in control cells, αSNAP knockdown led to the dispersion of these labeled vesicles throughout the cytoplasm (Fig. 1B). Similar results were obtained when GATE-16, another mAtg8 protein family member, was used to monitor autophagy (Fig. S1 A and B). Of note, knockdown of NSF affected the autophagic flux yet to a lesser extent compared with the control siRNA (Fig. S1 E and F). In addition, the expression of an NSF dominant negative mutant (NSF E329Q) resulted in the inhibition of the autophagic flux (Fig. S1 C and D). Autophagic activity of GFP-LC3–expressing cells was also monitored by ImageStream analysis as an alternative approach to determine the effect on autophagy upon αSNAP knockdown (18). By measuring bright detail intensity (BDI), which combines both texture and intensity features (19), we show that the autophagic flux is inhibited upon αSNAP knockdown (Fig. 1C and Fig. S1G). To rule out the possibility of an indirect effect of αSNAP knockdown on autophagy, cells depleted of αSNAP were transfected with an siRNA-resistant αSNAP fused to its N terminus with a FLAG tag. This treatment resulted in the full recovery of autophagy in cells expressing the siRNA-resistant FLAG-αSNAP (full line in Fig. 1D) compared with the recovery of neighboring nontransfected cells (dashed line in Fig. 1D). Restoration of the autophagic flux by the siRNA-resistant FLAG-αSNAP was also evident when cell lysates were subjected to Western blot analysis (Fig. 1E).
Fig. 1.
SNARE priming is essential for autophagic flux. (A, Left) HeLa cells were transfected with nontargeting (control) siRNA (non tgt) or αSNAP siRNA (siαSNAP) using DharmaFECT 1 transfection reagent. After 48 h, the cells were incubated for 4 h in the absence or presence of 0.1 µM BafA, lysed with RIPA extraction buffer, and then analyzed by Western blotting. (A, Right) At least four independent trials were quantified, using Image Studio Lite software normalized to the nontargeting control sample. Values are means ± SE. (B, Left) HeLa cells were treated as in A, fixed with 100% methanol, immunostained with anti-LC3 and anti-p62 antibodies, and analyzed by an Olympus confocal microscope. (Scale bar, 20 µm.) Large magnification of stained cells is presented in the Right column. (B, Right) At least 80 control cells (not treated with BafA) were quantified, using Imaris ×64 8.2.0 software, Bitplane. ***P < 0.0001. Values are means ± SE. (C) HeLa cells stably expressing GFP-LC3 were treated as in A, then harvested by trypsin, fixed with 100% methanol, and stained with DAPI. Cells were then subjected to ImageStream analysis, as described in Materials and Methods. (Left) Four independent trials were analyzed and quantified by IDEAS v. 6.2 software. Values are means ± SE. (Right) Representative BDI histogram of ImageStream analysis. (D) HeLa cells were transfected as in A, and after 24 h were transfected with siRNA-resistant FLAG-tagged αSNAP using jetPEI transfection reagent for a further 24 h. The cells were then incubated for 4 h in the absence or presence of 0.1 µM BafA, fixed with 100% methanol, stained with anti-p62 and anti-FLAG antibodies, and analyzed by an Olympus confocal microscope. (Scale bar, 20 µm.) (E) Cells were treated as in D, lysed with RIPA extraction buffer, and then analyzed by Western blotting.
Autophagosome Formation Does Not Require SNARE Priming.
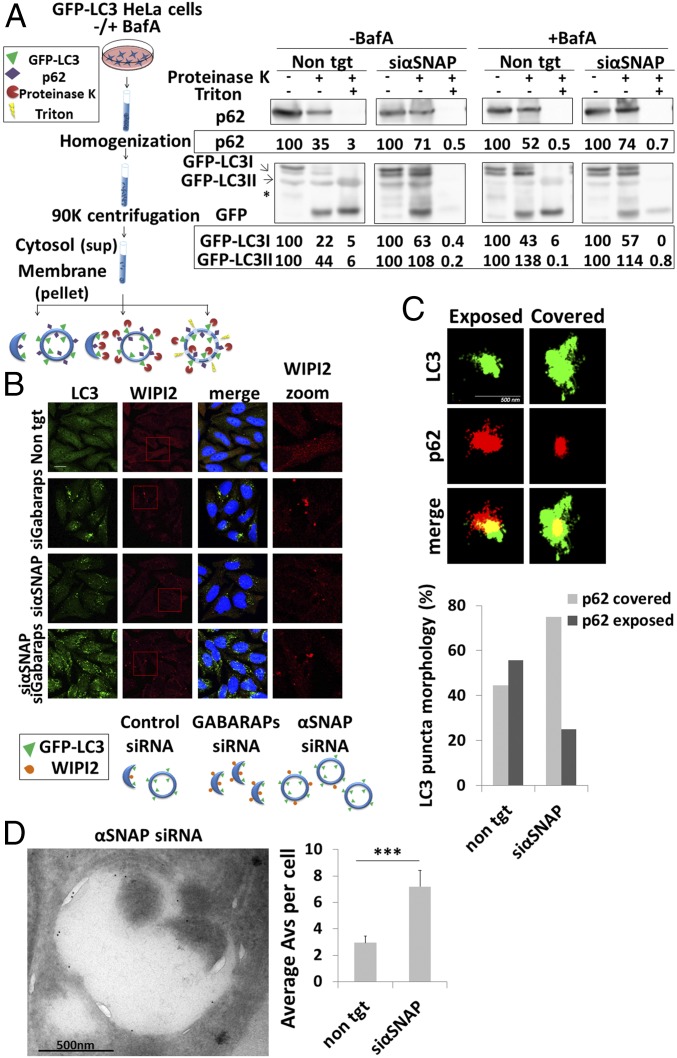
To determine whether the autophagy-related structures accumulated in response to αSNAP knockdown are open phagophores or sealed autophagosomes, we isolated membranes from GFP-LC3–expressing cells and subjected them to treatment with proteinase K in the presence or absence of the detergent Triton X-100 (Fig. 2A, Left column). GFP-LC3 would be expected to be protected from protease treatment only if located within sealed autophagosomes. Triton X-100 was used in these experiments to dissolve the membranes exposing the protected proteins to the protease. Notably, protease-induced cleavage of the exposed GFP-LC3 is known to result in LC3 degradation and accumulation of free GFP molecules (20). As shown in Fig. 2A (Right column), αSNAP knockdown led to the accumulation of protease-protected LC3 and p62, indicative of the accumulation of sealed autophagosomes. In cells transfected with nontargeting siRNA, protected LC3 and p62 were detectable only in relatively low amounts, consistent with the notion that under steady-state conditions the numbers of autophagosomes present are small. As a control for this system, cells were incubated in the presence of BafA (known to accumulate mature autophagosomes and autolysosomes), with resulting accumulation of protected LC3II and p62 in both control and αSNAP-knockdown cells (Fig. 2A). Taken together, these results suggest that SNARE priming is essential principally for late stages of autophagy but remain dispensable for the formation of the double-membrane autophagosome.
Fig. 2.
Autophagosome formation does not require SNARE priming. (A, Left) Depiction of the protection assay scheme described in Materials and Methods. (A, Right) HeLa cells stably expressing GFP-LC3 were transfected with nontargeting (control) siRNA (non tgt) or αSNAP siRNA (siαSNAP) using DharmaFECT 1 transfection reagent. After 48 h, the cells were incubated for 4 h in the absence or presence of 0.1 µM BafA and were then homogenized, centrifuged, and subjected to Proteinase K and Triton X-100 treatment as described in Materials and Methods. Protein levels were quantified using Image Studio Lite software. (B, Top immunofluorescent analysis) HeLa cells were transfected with nontargeting (control) siRNA (non tgt), αSNAP siRNA (siαSNAP), the siRNA of three GABARAP family members (siGABARAPs), or both siαSNAP and siGABARAPs, using DharmaFECT 1 transfection reagent. After 48 h, the cells were fixed with 100% methanol, immunostained with anti-LC3 and anti-WIPI2 antibodies, and analyzed by an Olympus confocal microscope. (Scale bar, 20 µm.) Large magnification of WIPI staining is presented in the Right column. (B, Bottom scheme) Depiction of the autophagy-related bodies detected under the different treatments. (C, Top images) HeLa cells were transected as in A. After 48 h the cells were fixed with 100% methanol, immunostained with anti-LC3 and anti-p62 antibodies, and subjected to Dual Color 3D STORM imaging as described in Materials and Methods. (C, Bottom graph) Ratio quantification of LC3- and p62-puncta morphologies; n = 25. (D, Left) HeLa cells stably expressing GFP-LC3 were transfected as in A. Cryosections of fixed cells were immunolabeled with anti-GFP antibodies and analyzed by TEM as described in Materials and Methods. (D, Right) Analysis of the GFP-labeled structures by ImageJ software; n > 40, ***P < 0.004. Values are means ± SE.
Additional evidence for the possibility that the vesicles, which accumulate upon αSNAP depletion, are mature autophagosomes was obtained by the monitoring of WIPI2 (an early phagophore marker that disassociates from the phagophore upon autophagosome formation) (21). As shown in Fig. 2B and Fig. S2A, knockdown of all GABARAP family members, led to large accumulation of LC3-labeled vesicles that contain WIPI2 (22). In contrast, the LC3-labeled vesicles that accumulated in response to αSNAP knockdown were only partially labeled with WIPI2, indicating that most of the vesicles, which accumulated under those conditions, represent a later stage of autophagosome formation (Fig. 2B and Fig. S2A). Furthermore, knockdown of αSNAP together with GABARAP knockdown led to the accumulation of a LC3 and WIPI2 positive structure, supporting the notion that αSNAP acts downstream of autophagosome formation (Fig. 2B and Fig. S2A).
Next, the LC3- and p62-labeled puncta were analyzed using superresolution microscopy. p62 puncta, which were fully covered by LC3, indicating sealed autophagosomes, were more prevalent upon knockdown of αSNAP compared with structures in which the p62 signal was exposed (Fig. 2C and Movies S1 and S2). As expected in the control cells, equal populations of the two structure types were depicted.
To further determine the nature of the membrane structures that accumulate upon αSNAP depletion, cells stably expressing GFP-LC3 were analyzed by correlative light immunoelectron microscopy (Fig. 2D and Fig. S2B). Evidently, knockdown of αSNAP resulted in the accumulation of membrane vesicles labeled with GFP-LC3. LC3 was found on both the inner (luminal) and the outer (cytosolic) sides of these membranes, indicating that these vesicles are indeed sealed double membrane autophagosomes. Unlike in control cells, in cells knocked down for αSNAP the GFP-LC3–labeled membranal structures that we detected were closed, and no significant differences were found in average autophagosomal sizes between the control and αSNAP knockdown cells (Fig. S2 B and C). Furthermore, a more detailed cellular morphology obtained by transmission electron microscopy, indicated that αSNAP knockdown led to accumulation of double membrane autophagosomes engulfing cytoplasmic material (Fig. S2C).
αSNAP Is Essential for Priming of Autophagy-Related SNARE Complexes.
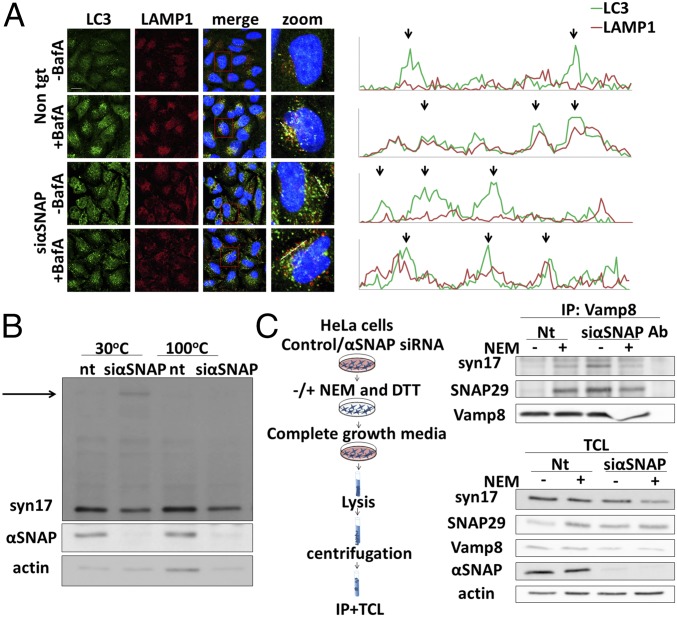
Different approaches were used to analyze the ability of mature autophagosomes that accumulate upon αSNAP depletion to fuse with the lysosome. First, we used immunofluorescence analysis to monitor, in the absence of αSNAP, the accumulation of LC3-labeled (Fig. 3A) or GFP-GATE16–labeled (Fig. S1A) autophagosomes that were colocalized with anti–LAMP1-labeled lysosomes. These autophagosomes were indeed detectable only in the vicinity of the lysosome, indicating that membrane fusion between these organelles was impaired. Notably, αSNAP siRNA did not affect lysosomal acidification, excluding the possibility of an indirect effect on lysosomal activity (Fig. S3A).
Fig. 3.
αSNAP is essential for priming autophagy-related SNARE complexes. (A, Left) HeLa cells were transfected with nontargeting (control) siRNA (non tgt) or αSNAP siRNA (siαSNAP) using DharmaFECT 1 transfection reagent. After 48 h, the cells were fixed with 100% methanol, immunostained with anti-LC3 and anti-LAMP1 antibodies, and then analyzed by an Olympus confocal microscope. (Scale bar, 20 µm.) Large magnification of stained cells is presented in the Right column. (A, Right) Intensity profiles of the marked lines in the large magnification column were obtained by Olympus FluoView FV 1000 analysis software. (B) HeLa cells were transfected and treated as in A, lysed with RIPA extraction buffer, heated to the indicated temperatures, and analyzed by Western blotting. nt, nontargeting (control) siRNA. (C) HeLa cells were transfected as in A, treated with or without NEM and DTT, lysed with SNARE lysis buffer, and immunoprecipitated with Vamp8 antibodies. (Left) Immunoprecipitation scheme as detailed in Materials and Methods. (Right Top) IP with Vamp8 antibodies. (Bottom) TCL, total cell lysates; Ab, antibodies with no lysate incubation; IP, immunoprecipitation. Further details are provided in Materials and Methods.
Next, we examined the formation of the SNARE complexes that mediate membrane fusion between autophagosomes and the lysosome. For that purpose, we took advantage of the fact that SNARE complexes are often resistant to SDS (23). As depicted in Fig. 3B, when samples obtained from cells treated with αSNAP siRNA were separated on SDS/PAGE, syntaxin17 was found to migrate as a large SDS-resistant complex that dissociates upon boiling. By means of an immunoprecipitation approach, we then examined the effect of αSNAP knockdown on an autophagy-related SNARE complex consisting of syntaxin17, SNAP29, and VAMP8 (12). Using anti-VAMP8 antibodies, we found that this SNARE complex is immunoprecipitated only upon αSNAP knockdown or when the general alkylator N-ethylmaleimide (NEM) was used to block NSF activity (24) (Fig. 3C). When we used anti-syntaxin17 antibodies to immunoprecipitate the complex, only SNAP29 coimmunoprecipitated (Fig. S3B). Importantly an accumulation of the Vamp4–syntaxin6–syntaxin16–Vti1 complex, involved in the endocytic pathway was also evident upon αSNAP knockdown (Fig. S3C). Furthermore, other intracellular trafficking pathways such as ligand-induced endocytosis of EGFR as well as endoplasmic reticulum (ER)-to-Golgi trafficking of ManII were inhibited upon αSNAP knockdown, indicating a general effect on membrane fusion processes under these conditions (Fig. S4). Taken together, these results indicated that αSNAP is predominantly needed for priming of the SNARE complex required for fusion of the autophagosomal and lysosomal membranes while autophagosome formation is less sensitive to its absence.
Translocation of Syntaxin17 and Atg9 to the Autophagosome Is SNARE-Priming Independent.
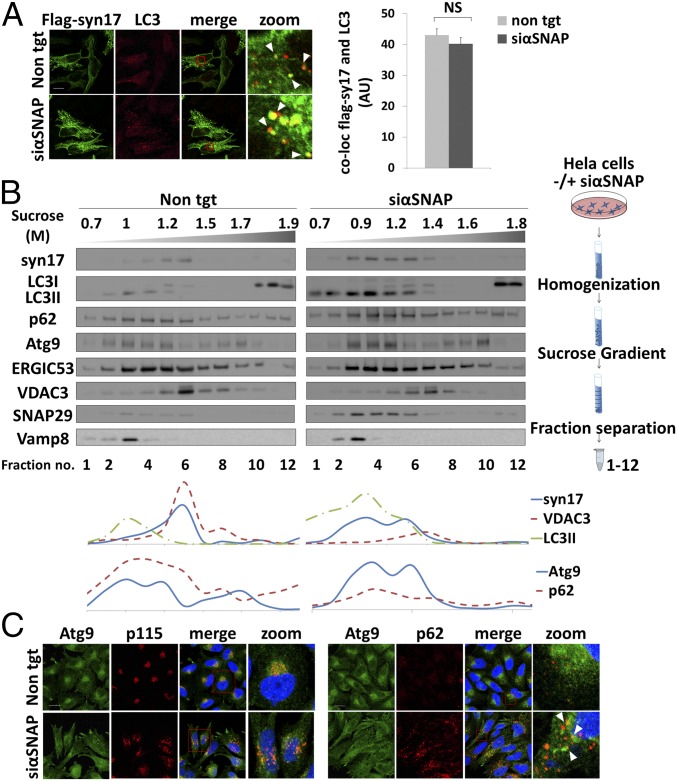
Syntaxin17 reportedly translocates from the mitochondria to mature autophagosomes (25). We therefore examined whether such translocation is dependent on αSNAP. Confocal microscopy analysis revealed that αSNAP knockdown does not affect the translocation of syntaxin17 to autophagosomes (Fig. 4A). Subcellular fractionation by sucrose gradient flotation assay showed that in control cells, syntaxin17 cofractionates mainly with the mitochondrial marker VDAC3 (Fig. 4B, fraction 6). A small amount, however, cofractionated with the autophagosomal markers LC3 and p62, consistently with the small numbers of autophagosomes present under these conditions (Fig. 4B, fractions 3 and 4). Since αSNAP knockdown leads to accumulation of autophagosomes, we observed—as expected—a marked increase in syntaxin17 cofractionated with the autophagosomal markers (Fig. 4B), suggesting that its translocation to autophagosomes is independent of αSNAP. Notably, knockdown of αSNAP did not affect the migration of VAMP8 and SNAP29 on the sucrose gradient. Altogether, these findings suggested that recruitment of syntaxin17 to the autophagosomal membrane is αSNAP independent.
Fig. 4.
Translocation of syntaxin17 and Atg9 to the autophagosome is SNARE-priming independent. (A, Left) HeLa cells were transfected with nontargeting (control) siRNA (non tgt) or αSNAP siRNA (siαSNAP) using DharmaFect 1 transfection reagent. After 24 h, the cells were transfected with FLAG-tagged syntaxin17 using jetPEI transfection reagent for an additional 24 h. Cells were then fixed with 100% methanol, stained with anti-LC3 and anti-FLAG antibodies, and analyzed by an Olympus confocal microscope. (Scale bar, 20 µm.) Right column shows large magnification of stained cells. (Right) At least three independent trials were quantified, using Imaris ×64 8.2.0 software, Bitplane. NS, not significant. Values are means ± SE. (B, Top Left) HeLa cells were transfected as in A. After 48 h, the cells were homogenized, floated upon a sucrose gradient as described in Materials and Methods, and quantified by Image Studio Lite software. (Top Right) Flotation assay scheme as detailed in Materials and Methods. (Bottom) Protein levels in the fractions were quantified using Image Studio Lite software. (C) HeLa cells were transfected as in A, fixed with 4% paraformaldehyde, immunostained with anti-Atg9, anti-p115, and anti-p62 antibodies, and analyzed by the Olympus confocal microscope. (Scale bar, 20 µm.) Large magnification of stained cells is presented in the Right column of each panel.
Atg9, the only identified transmembrane protein needed for the formation of complete autophagosomes, is synthesized in the ER, and shuttles between the Golgi and endosomes (26, 27). We used subcellular fractionation and immunofluorescence microscopy to examine the effect of αSNAP knockdown on the subcellular localization of Atg9. Under control conditions, Atg9 cofractionated with ERGIC-53, a marker of the ER–Golgi intermediate compartment (Fig. 4B, fraction 5), and with the autophagosomal markers p62 and LC3 (Fig. 4B, fractions 3 and 4). Similar results were obtained by confocal microscopy analysis (Fig. 4C, Right column). Upon αSNAP knockdown, the ability of Atg9 to reach autophagosomes was preserved (Fig. 4B), although it was dispersed together with the Golgi marker p115, a peripheral Golgi-associated protein, within the cell, probably because of Golgi dispersion upon αSNAP inhibition (Fig. 4C, Left column). These results suggested that the contribution of Atg9 to autophagosome biogenesis does not depend on SNARE priming. This finding is in agreement with a report indicating that Atg9 vesicles are found in the vicinity of autophagic membranes but do not fuse with them (26).
A Low Level of αSNAP Is Sufficient to Partially Promote Autophagy Under Starvation Conditions.
We next examined the effect of αSNAP depletion on autophagy induced by starvation. As shown in Fig. 5, αSNAP knockdown led to inhibition of the autophagic flux also in starved cells (Fig. 5 A and B and Fig. S1 C–F). Apparently, the effect of αSNAP depletion on starvation-induced autophagy was lower than that observed under control conditions. Statistical analysis showed significant increase in autophagosome number but not size in cells following αSNAP siRNA transfection (Fig. 5B and Figs. S2C and S5A). To determine whether the remaining αSNAP molecules are sufficient to target NSF to membranes enabling SNARE priming under starvation, we separated cells by fractionating them into high-speed supernatant (HSS) (cytosolic fraction) and high-speed pellet (HSP) (membrane fraction). We found that both αSNAP and NSF accumulate in the pellets of nontarget siRNA-transfected cells, and that upon starvation the levels of αSNAP and NSF detected in the pellet fraction are higher than under fed conditions (Fig. 5C). Upon αSNAP depletion, however, whereas NSF was found both in the supernatant and in the pellet, residual amounts of αSNAP were detected only in the pellet. This finding was consistent with the role of αSNAP as an adaptor protein that limits the ability of NSF to bind membranes. In addition, upon starvation, the levels of the SNARE molecules syntaxin17 and SNAP29 in the pellet in αSNAP knockdown cells were decreased, pointing to SNARE complex disassembly (Fig. 5C). Moreover, both in the absence and in the presence of NEM, the amount of SNAP29 that coimmunoprecipitated with syntaxin17 was lower in αSNAP-depleted starved cells than in cells under fed conditions (Fig. S5B). Our results thus suggested that under starvation the low levels of αSNAP are capable of binding NSF molecules to partially promote the SNARE priming needed for autophagy.
Fig. 5.
A low level of αSNAP is sufficient to partially promote autophagy under starvation conditions. (A, Left) HeLa cells were transfected with nontargeting (control) siRNA (non tgt) or αSNAP siRNA using DharmaFECT 1 transfection reagent. After 48 h, the cells were incubated for 4 h in normal growth medium or EBSS in the absence or presence of 0.1 µM BafA, lysed with RIPA extraction buffer, and then analyzed by Western blotting. (A, Right) At least four independent trials were quantified, using Image Studio Lite software normalized to the nontargeting control sample. Values are means ± SE. Starv, starvation conditions. (B, Left) HeLa cells were treated as in A, fixed with 100% methanol, immunostained with anti-LC3 antibodies, analyzed by a Leica confocal microscope using the same threshold for all samples and deconvolved using Huygens software. Z-stack projection was done using ImageJ software. (Scale bar, 10 µm.) Large magnification of stained cells is presented at Right. (Middle) Quantification of LC3 puncta using LAS X software, Leica Microsystems. n > 1,000, ***P < 0.0001. Values are means ± SE. (Right) HeLa cells stably expressing GFP-LC3 were treated as in A, then harvested by trypsin, fixed with 100% methanol, and stained with DAPI. Cells were then subjected to ImageStream analysis, as described in Materials and Methods. Four independent trials were analyzed and quantified by IDEAS v. 6.2 software. (C) HeLa cells were transfected as in A, homogenized, and fractionated as described in Materials and Methods. Total cell homogenates are presented on the Right.
Discussion
Little is known about the participation of SNARE priming in the autophagic process, and specifically in autophagosome biogenesis. In this study, by knocking down αSNAP—a protein that serves as an essential adaptor for almost all processes involving intracellular membrane fusion—we showed that although autophagosomes are still formed in its absence, their ability to fuse with the lysosomes is compromised. This resulted in inhibition of the autophagic flux manifested by the accumulation of sealed autophagosomes marked by LC3, p62, and syntaxin17. We further demonstrated that αSNAP is not needed for the association of syntaxin17 and Atg9 with the autophagosomal membrane. Taken together, our results imply that SNARE disassembly is essential for fusion of the autophagosome with the lysosome, but not for phagophore initiation or membrane supply during autophagosome biogenesis. Notably, however, our results did not exclude the need for SNARE-dependent membrane fusion for the initial steps of autophagosome biogenesis.
SNARE priming is driven by NSF, an enzyme for which αSNAP acts as a structural adaptor. Both activities are essential for cell survival. Here we used the siRNA approach to study the role of SNARE priming in autophagy and chose to focus on the adaptor molecule αSNAP, as knockdown of the enzyme NSF resulted in a weaker effect on the autophagic flux most likely due to residual NSF enzymatic activity. It is important to note that αSNAP knockdown affects, equally and without bias, not only a specific SNARE molecule but all SNARE priming events, thus preventing unbalanced changes in other intracellular trafficking processes. Unexpectedly, our results indicated that while knockdown of αSNAP suppresses the autophagic flux, it does not inhibit the formation of autophagosomes supporting the notion that autophagosome biogenesis is less sensitive to the loss of SNARE priming.
Syntaxin17 was recently identified as an essential SNARE molecule for autophagy. Initially it was reported to be incorporated into mature autophagosomes to promote fusion between the autophagosomal and the lysosomal membranes (12, 13), whereas a more recent study suggested that it might take part in the formation of the isolation membrane (28). Interestingly, syntaxin17 was also found in mitochondria, where it promotes mitochondrial division independently of its SNARE domain as well as ER calcium homeostasis and shifts to its autophagic role upon starvation (25). Our present results showed that syntaxin17 is located in the mitochondria under control conditions, but in the absence of αSNAP it accumulates on autophagosomes, indicating that its delivery to these organelles does not require SNARE priming. It therefore seems that its accumulation on autophagosomes simply reflects their inability to mature and fuse with the lysosome. Notably, we show that αSNAP knockdown in our system is accompanied by an accumulation of other, autophagy unrelated, SNARE complexes as well as inhibition of general trafficking processes, indicating that multiple SNARE-dependent trafficking processes were affected.
The participation of Atg9 in autophagosome biogenesis has been extensively studied both in yeast and in mammalian cells (26, 27). In mammals it was found to be localized in transient proximity to phagophores but not incorporated into them (26), and to be recycled to endosomal compartments from the Golgi, where it seems to be at its most profuse (29). Our data showed that the contribution of Atg9 to autophagosome biogenesis is independent of SNARE priming. While the localization of Atg9 in the cell was affected when SNARE priming was inhibited (probably owing to dispersion of the Golgi) its localization to autophagosomes remained unaffected, consistently with the notion that Atg9-associated vesicles do not fuse with autophagosomes (26, 30).
Starvation is a well-established inducer of autophagy, known to accelerate the biogenesis of autophagosomes and their consequent lysosomal degradation. Nevertheless we found that αSNAP knockdown in starved cells leads to a partial inhibition of the autophagic flux compared with cells grown under complete medium. In an attempt to provide evidence to explain this phenomenon, we provide data suggesting that upon starvation, higher levels of NSF are targeted to membranes, where they can sufficiently promote the SNARE priming needed for autophagy. We thus speculate that under starvation αSNAP and NSF are targeted to SNARE complexes specifically involved in autophagosomal–lysosomal membrane fusion. The mechanism responsible for the regulation of this process will be the subject of future studies.
Taken together, our findings indicated that the membrane fusion of autophagosomes with the lysosome is sensitive to SNARE priming inhibition, while autophagosome formation is resistant to its loss. A plausible scenario is that low NSF and αSNAP levels allow priming of SNARE complexes required for autophagosome formation but not of SNAREs that act in autophagosome–lysosome fusion; alternatively, our data may indicate that the process is sufficiently supported by nascent, unpaired SNARE molecules.
Materials and Methods
See Supporting Information for additional methods.
Cell Culture and Transfection.
HeLa cells were grown on αMEM medium supplemented with 10% FCS at 37 °C in 5% CO2. For siRNA silencing, subconfluent HeLa cells were transfected with different siRNA pools or oligos using DharmaFect 1 transfection reagent (Dharmacon) according to the manufacturer’s instructions. Experiments were performed 48 h after transfection. Plasmid transfection for protein overexpression was conducted using jetPEI transfection reagent (Polyplus transfection) according to the manufacturer’s instructions. Starvation conditions were achieved by washing cells three times with PBS and incubating them in Earle’s balanced salt solution (EBSS) medium at 37 °C for 4 h. Lysosomal degradation was inhibited by incubating cells in the presence of 100 nM BafA (LC Laboratories). Total cell extracts were produced using RIPA extraction buffer (0.1 M NaCl, 5 mM EDTA, 0.1 M Na2HPO4/NaH2PO4 pH 7.5, 1% Triton, 0.5% DOC, 0.1% SDS) with a mixture of protease inhibitors (Calbiochem). Protein levels were measured by Image Studio Lite software. Levels of protein degradation were calculated by dividing the protein levels of BafA-treated samples by those of untreated samples.
Fluorescence Microscopy.
Cells were plated on sterile coverslips (13-mm diameter) and cultured under the conditions indicated. Cells were fixed with 4% paraformaldehyde in PBS for 10 min and permeabilized with 0.1% Triton X-100 for 8 min. When anti-LC3 antibodies were used, cells were fixed and permeabilized with cold methanol for 7 min at −20 °C. Cells were blocked by incubation with 10% FCS in PBS for 30 min at room temperature, followed by incubation for 1 h with the primary antibody. Cells were then incubated with the secondary antibody for 30 min. Confocal images were obtained with an FV1000 Olympus laser-scanning confocal microscope equipped with a UPLS APO 60× N.A. 1.35 oil immersion lens or with a Leica TCS SP8 laser-scanning confocal microscope equipped with a HCPL APO CS2 63×/1.40 oil immersion lens as indicated. Images were analyzed by FluoView FV1000 software (Olympus) or Huygens and LASX software, respectively. Threshold for autophagosome quantifications in all samples was 0.031 μm3. Live cell imaging was obtained with a DeltaVision microscope (Applied Precision). Colocalization values between the different markers were analyzed from at least two individual experiments by Imaris ×64 8.2.0 software, Bitplane.
Supplementary Material
Acknowledgments
We thank Vladimir Kiss for his assistance in confocal microscopy. This study was supported in part by the Israeli Science Foundation (Grant 1247/15), the Legacy Heritage Fund (Grant 1935/16), and the Minerva Foundation, with funding from the Federal German Ministry for Education and Research.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. S.A.T. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1705572114/-/DCSupplemental.
References
- 1.Jahn R, Scheller RH. SNAREs: Engines for membrane fusion. Nat Rev Mol Cell Biol. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- 2.Südhof TC, Rothman JE. Membrane fusion: Grappling with SNARE and SM proteins. Science. 2009;323:474–477. doi: 10.1126/science.1161748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Söllner T, et al. SNAP receptors implicated in vesicle targeting and fusion. Nature. 1993;362:318–324. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- 4.Chae TH, Kim S, Marz KE, Hanson PI, Walsh CA. The hyh mutation uncovers roles for alpha Snap in apical protein localization and control of neural cell fate. Nat Genet. 2004;36:264–270. doi: 10.1038/ng1302. [DOI] [PubMed] [Google Scholar]
- 5.Burgoyne RD, Morgan A. Secretory granule exocytosis. Physiol Rev. 2003;83:581–632. doi: 10.1152/physrev.00031.2002. [DOI] [PubMed] [Google Scholar]
- 6.Morgan A, Dimaline R, Burgoyne RD. The ATPase activity of N-ethylmaleimide-sensitive fusion protein (NSF) is regulated by soluble NSF attachment proteins. J Biol Chem. 1994;269:29347–29350. [PubMed] [Google Scholar]
- 7.Whiteheart SW, et al. SNAP family of NSF attachment proteins includes a brain-specific isoform. Nature. 1993;362:353–355. doi: 10.1038/362353a0. [DOI] [PubMed] [Google Scholar]
- 8.Inoue H, et al. γ-SNAP stimulates disassembly of endosomal SNARE complexes and regulates endocytic trafficking pathways. J Cell Sci. 2015;128:2781–2794. doi: 10.1242/jcs.158634. [DOI] [PubMed] [Google Scholar]
- 9.Andreeva AV, Kutuzov MA, Voyno-Yasenetskaya TA. A ubiquitous membrane fusion protein alpha SNAP: A potential therapeutic target for cancer, diabetes and neurological disorders? Expert Opin Ther Targets. 2006;10:723–733. doi: 10.1517/14728222.10.5.723. [DOI] [PubMed] [Google Scholar]
- 10.Tooze SA, Abada A, Elazar Z. Endocytosis and autophagy: Exploitation or cooperation? Cold Spring Harb Perspect Biol. 2014;6:a018358. doi: 10.1101/cshperspect.a018358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mari M, et al. An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J Cell Biol. 2010;190:1005–1022. doi: 10.1083/jcb.200912089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Itakura E, Kishi-Itakura C, Mizushima N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 2012;151:1256–1269. doi: 10.1016/j.cell.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 13.Takáts S, et al. Autophagosomal Syntaxin17-dependent lysosomal degradation maintains neuronal function in Drosophila. J Cell Biol. 2013;201:531–539. doi: 10.1083/jcb.201211160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishihara N, et al. Autophagosome requires specific early Sec proteins for its formation and NSF/SNARE for vacuolar fusion. Mol Biol Cell. 2001;12:3690–3702. doi: 10.1091/mbc.12.11.3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nair U, et al. SNARE proteins are required for macroautophagy. Cell. 2011;146:290–302. doi: 10.1016/j.cell.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moreau K, Ravikumar B, Renna M, Puri C, Rubinsztein DC. Autophagosome precursor maturation requires homotypic fusion. Cell. 2011;146:303–317. doi: 10.1016/j.cell.2011.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Naydenov NG, Harris G, Morales V, Ivanov AI. Loss of a membrane trafficking protein αSNAP induces non-canonical autophagy in human epithelia. Cell Cycle. 2012;11:4613–4625. doi: 10.4161/cc.22885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shvets E, Fass E, Elazar Z. Utilizing flow cytometry to monitor autophagy in living mammalian cells. Autophagy. 2008;4:621–628. doi: 10.4161/auto.5939. [DOI] [PubMed] [Google Scholar]
- 19.Demishtein A, Porat Z, Elazar Z, Shvets E. Applications of flow cytometry for measurement of autophagy. Methods. 2015;75:87–95. doi: 10.1016/j.ymeth.2014.12.020. [DOI] [PubMed] [Google Scholar]
- 20.Nair U, Thumm M, Klionsky DJ, Krick R. GFP-Atg8 protease protection as a tool to monitor autophagosome biogenesis. Autophagy. 2011;7:1546–1550. doi: 10.4161/auto.7.12.18424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Polson HE, et al. Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy. 2010;6:506–522. doi: 10.4161/auto.6.4.11863. [DOI] [PubMed] [Google Scholar]
- 22.Weidberg H, et al. LC3 and GATE-16 N termini mediate membrane fusion processes required for autophagosome biogenesis. Dev Cell. 2011;20:444–454. doi: 10.1016/j.devcel.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 23.Hayashi T, Yamasaki S, Nauenburg S, Binz T, Niemann H. Disassembly of the reconstituted synaptic vesicle membrane fusion complex in vitro. EMBO J. 1995;14:2317–2325. doi: 10.1002/j.1460-2075.1995.tb07226.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glick BS, Rothman JE. Possible role for fatty acyl-coenzyme A in intracellular protein transport. Nature. 1987;326:309–312. doi: 10.1038/326309a0. [DOI] [PubMed] [Google Scholar]
- 25.Arasaki K, et al. A role for the ancient SNARE syntaxin 17 in regulating mitochondrial division. Dev Cell. 2015;32:304–317. doi: 10.1016/j.devcel.2014.12.011. [DOI] [PubMed] [Google Scholar]
- 26.Orsi A, et al. Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, are required for autophagy. Mol Biol Cell. 2012;23:1860–1873. doi: 10.1091/mbc.E11-09-0746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamamoto H, et al. Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J Cell Biol. 2012;198:219–233. doi: 10.1083/jcb.201202061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hamasaki M, et al. Autophagosomes form at ER-mitochondria contact sites. Nature. 2013;495:389–393. doi: 10.1038/nature11910. [DOI] [PubMed] [Google Scholar]
- 29.Lamb CA, et al. TBC1D14 regulates autophagy via the TRAPP complex and ATG9 traffic. EMBO J. 2016;35:281–301. doi: 10.15252/embj.201592695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karanasios E, et al. Autophagy initiation by ULK complex assembly on ER tubulovesicular regions marked by ATG9 vesicles. Nat Commun. 2016;7:12420. doi: 10.1038/ncomms12420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boncompain G, et al. Synchronization of secretory protein traffic in populations of cells. Nat Methods. 2012;9:493–498. doi: 10.1038/nmeth.1928. [DOI] [PubMed] [Google Scholar]
- 32.Laufman O, Hong W, Lev S. The COG complex interacts directly with Syntaxin 6 and positively regulates endosome-to-TGN retrograde transport. J Cell Biol. 2011;194:459–472. doi: 10.1083/jcb.201102045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tokuyasu KT. Application of cryoultramicrotomy to immunocytochemistry. J Microsc. 1986;143:139–149. doi: 10.1111/j.1365-2818.1986.tb02772.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.