Significance
Dysregulation of signaling via heterotrimeric G proteins leads to pathogenesis. Thus, developing an efficient armamentarium to study G protein regulation is crucial for understanding the molecular basis of disease. The classical view of G protein activation as an exclusive function of G protein-coupled receptors has been challenged by the discovery of nonreceptor G protein activators. Dysregulation of a family of such nonreceptor activators has been linked to human disorders like cancer or birth defects, but the underlying mechanisms remain poorly understood due to the lack of experimental tools. Here, we use protein engineering to rationally design a genetically encoded inhibitor of these G protein activators and demonstrate its usefulness to block aberrant signaling in cancer cells and abrogate developmental malformations in animal embryos.
Keywords: GPCR, GEF, Girdin, DAPLE, integrin
Abstract
Activation of heterotrimeric G proteins by cytoplasmic nonreceptor proteins is an alternative to the classical mechanism via G protein-coupled receptors (GPCRs). A subset of nonreceptor G protein activators is characterized by a conserved sequence named the Gα-binding and activating (GBA) motif, which confers guanine nucleotide exchange factor (GEF) activity in vitro and promotes G protein-dependent signaling in cells. GBA proteins have important roles in physiology and disease but remain greatly understudied. This is due, in part, to the lack of efficient tools that specifically disrupt GBA motif function in the context of the large multifunctional proteins in which they are embedded. This hindrance to the study of alternative mechanisms of G protein activation contrasts with the wealth of convenient chemical and genetic tools to manipulate GPCR-dependent activation. Here, we describe the rational design and implementation of a genetically encoded protein that specifically inhibits GBA motifs: GBA inhibitor (GBAi). GBAi was engineered by introducing modifications in Gαi that preclude coupling to every known major binding partner [GPCRs, Gβγ, effectors, guanine nucleotide dissociation inhibitors (GDIs), GTPase-activating proteins (GAPs), or the chaperone/GEF Ric-8A], while favoring high-affinity binding to all known GBA motifs. We demonstrate that GBAi does not interfere with canonical GPCR-G protein signaling but blocks GBA-dependent signaling in cancer cells. Furthermore, by implementing GBAi in vivo, we show that GBA-dependent signaling modulates phenotypes during Xenopus laevis embryonic development. In summary, GBAi is a selective, efficient, and convenient tool to dissect the biological processes controlled by a GPCR-independent mechanism of G protein activation mediated by cytoplasmic factors.
The core mechanism of heterotrimeric G protein regulation consists of activation by G protein-coupled receptors (GPCRs) via nucleotide exchange (1). GPCRs are guanine nucleotide exchange factors (GEFs) that promote the exchange of GDP for GTP on Gα subunits, which, in turn, leads to the dissociation of Gαβγ heterotrimers. Subsequently, both Gα-GTP and “free” Gβγ subunits modulate the activity of downstream effectors. However, there is a complex network of accessory proteins that modulate G protein signaling beyond the action of GPCRs. This includes guanine nucleotide dissociation inhibitors (GDIs) (2, 3), which bind Gα and lock it in an inactive GDP-bound state, and GTPase-activating proteins (GAPs) (4), which accelerate the rate of GTP hydrolysis by Gα. Ric-8 proteins are also G protein binding partners that facilitate Gα folding and stability while also possessing GEF activity in vitro (5). More recently, we and others have characterized a group of nonreceptor proteins that possess GEF activity for Gαi proteins in vitro and promote G protein signaling in cells (6–9) as determined by measurements that reflect either the formation of GTP-bound Gαi (e.g., cAMP reduction, Gα-GTP specific antibodies) (6, 10–12) or free Gβγ [e.g., bioluminescence resonance energy transfer (BRET)/FRET biosensors or PI3K-Akt signaling] (6, 7, 11, 13, 14). This family of nonreceptor GEFs is characterized by containing the Gα-binding and activating (GBA) motif (6–9), a 30- to 35-residue-long sequence that is necessary and sufficient for Gαi binding and activation. The GBA motif has been found in proteins from worms to humans (15), suggesting that G protein activation by GBA motif-containing proteins is an ancient mechanism of signaling regulation that appeared at least 300 Mya.
The GBA motif has been identified in four mammalian proteins to date: Gα-interacting vesicle-associated protein (GIV; also known as Girdin), Dishevelled-associating protein with high frequency of leucines (DAPLE), Calnuc, and NUCB2 (6, 7, 9). Although there is evidence that the GBA motif of Calnuc and NUCB2 binds and activates Gαi in vitro (9), the biological consequences of this have not yet been elucidated. For example, Calnuc binds to Gαi on the surface of the Golgi apparatus (16) and modulates intracellular trafficking (17), but it is unclear whether one is a consequence of the other or whether the function of the GBA motif plays a role. On the other hand, the biological functions of the GBA motifs of GIV and DAPLE have been characterized more extensively. Both GIV and DAPLE regulate signal transduction via G protein activation downstream of membrane receptors that are not necessarily GPCRs, like receptor tyrosine kinases or integrins (6, 8, 18). As a consequence, they impact a variety of cellular processes (6, 8, 18), like cell motility, proliferation, or autophagy, and their dysregulation is associated with human diseases, such as cancer, liver fibrosis, nephrotic syndrome, and insulin resistance (8, 19). However, it is unclear if other functions previously described for GIV and DAPLE are GBA-dependent. For example, it is not known if the role of GIV in memory and angiogenesis (20–22), or the possible role of DAPLE in embryonic development (23–25), is mediated via the GBA motif.
The development of tools that modulate or interfere with different steps of G protein signaling is closely intertwined with advances in the molecular understanding of this signaling mechanism and the development of novel therapeutics. This is well exemplified by the fact that >25% of US Food and Drug Administration-approved drugs are direct modulators of GPCRs. More recently, there has also been success in the identification of small-molecule inhibitors that target Gα or Gβγ subunits directly (26), or even some of their modulators, like regulator of G protein signaling (RGS) proteins (GAPs) (27) or GoLoco motif proteins (GDIs) (28). In addition to small molecules, an important approach to study G protein signaling has relied on the use of toxins, like pertussis, cholera, or Pasteurella multocida toxins (29). Pertussis toxin has been particularly useful to define and study G protein signaling mechanisms. This toxin ADP ribosylates α-subunits of the Gi/o family and specifically precludes their coupling to GPCRs. For over three decades, sensitivity to pertussis toxin has been used as an operational definition to mark molecular mechanisms or biological processes that are mediated by GPCR-Gi/o signaling. Another approach has been the use of genetically encoded tools, like the C-terminal domain of GRK2 (30). This domain, originally termed “beta-adrenergic receptor kinase 1 carboxy terminus (βARKct),” binds with high affinity to free Gβγ subunits and occludes the surface normally used by the G protein to bind and activate its effectors. Analogous to the generic use of pertussis toxin for GPCR-Gi/o coupling, βARKct has been widely used for decades to define and study Gβγ-dependent events. Other peptides or protein domains that preclude Gα or Gβγ binding to effectors have also been described and validated in applications ranging from in vitro biochemistry to animal models (26, 31, 32).
In contrast to this wealth of tools to manipulate other components of the G protein regulatory machinery, advances in the study of GBA-mediated G protein regulation have been dampened by the lack of experimental tools. There is no small molecule or biological agent that specifically inhibits this signaling mechanism. Because the GBA motif is embedded in multifunctional proteins, assessing the role of its G protein regulatory function in different biological processes has been limited to multistep genetic manipulations. To date, the most convincing approach for this has been to genetically blunt the expression of the endogenous GBA protein and replace it with a version in which the G protein regulatory function has been disabled by mutagenesis. This is not only cumbersome and labor-intensive but has numerous technical limitations, like the difficulty in manipulating and delivering (e.g., via viral particles) large genes (e.g., ∼6 kb for GIV and DAPLE) efficiently, or limited control over varying levels of expression of the proteins of interest. In addition to these limitations that hamper the clear interpretation of results or preclude experimental design in some settings (like whole animals), this approach does not account for possible compensatory effects mediated by other GBA proteins that are not targeted simultaneously in the same system. To overcome some of these limitations and expand the repertoire of tools to investigate the unknown functions of GBA proteins, we set out to develop a genetically encoded synthetic protein that functions as a generic inhibitor of GBA motifs. All GBA motifs described to date are located within structurally disordered regions well separated from other functional domains of the proteins in which they are embedded (33), so we reasoned that they could be specifically blocked if they bound to a protein that precluded their interaction with the target G protein. We chose to achieve this goal by rationally modifying Gαi itself instead of another synthetic scaffold for several reasons. By definition, Gαi binds generically to all GBA motifs, and the structural basis for this interaction has been recently characterized in detail by NMR spectroscopy, computational modeling, and biochemistry (33). We leveraged this information, along with a wealth of prior structural and biochemical information on other G protein regulatory interactions, to engineer modifications in Gαi that favor GBA association and preclude its binding to other interacting partners. Here, we show a comprehensive biochemical characterization of the resulting “GBA inhibitor” (GBAi) and its validation in two different biological systems (i.e., cancer cells in culture, vertebrate embryos). Our results establish GBAi as a selective and efficient tool to dissect the biological processes controlled by a GPCR-independent mechanism of G protein activation mediated by cytoplasmic factors.
Results and Discussion
Rational Design of GBAi.
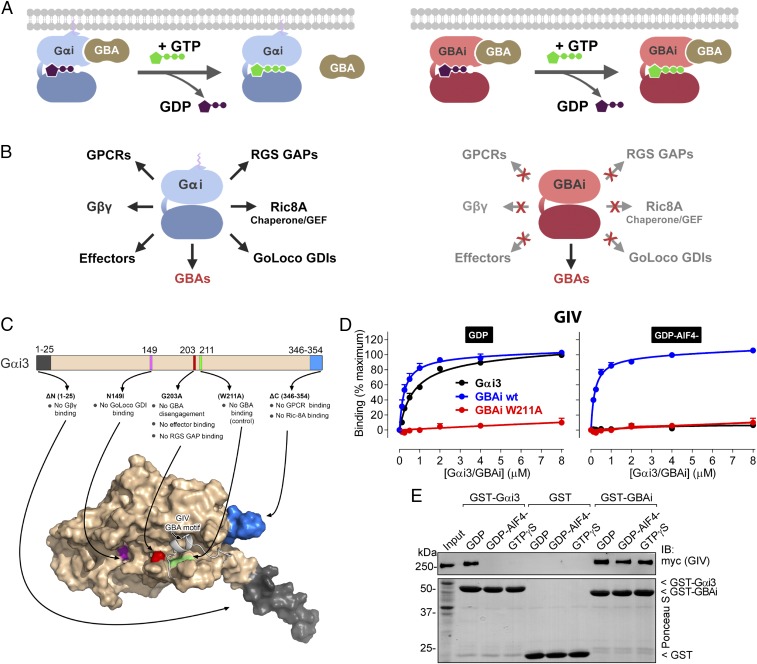
We envisioned the design of a genetically encoded protein to inhibit GBA proteins (GBAi) based on Gαi3 as the starting template. We sought to engineer two main properties to achieve efficient and specific inhibition of GBA motifs. One was that GBAi remains bound to GBA motifs even after exchange of GDP for GTP (Fig. 1A), which is known to cause disengagement of GBA motifs from Gαi proteins (6, 8, 9, 15). This property would favor the constitutive association with GBA motifs in cells. The second property was that GBAi does not interact with Gαi binding partners that are not GBA proteins (Fig. 1B). This would make the action of GBAi specific for GBA motifs. To achieve these goals, we introduced a number of modifications in Gαi3 based on prior knowledge from the literature (Fig. 1C).
Fig. 1.
Rational design of GBAi, a specific inhibitor of GBA motifs. (A, Left) GBA motifs bind to Gαi in its inactive conformation (bound to GDP, purple), but not in its active conformation (bound to GTP, green). (A, Right) Desired property of Gαi-derived GBAi is that its binding to GBA motifs is not reversed upon nucleotide exchange of GDP for GTP, thereby resulting in constitutive high-affinity binding to GBA motifs in cells. (B) Desired property of Gαi-derived GBAi is a lack of binding to any other Gαi binding partner (GPCRs, Gβγ, effectors, RGS GAPs, GoLoco GDIs, or the chaperone/GEF Ric-8A; Left), thereby resulting in specific binding and inhibition of only GBA motifs (Right). (C, Top) Schematic of the modifications engineered into Gαi3 to generate GBAi. The G203A mutation disrupts a conformational change of the SwII region that normally occurs upon GTP binding and is required to bind effectors and RGS GAPs and, at the same time, precludes GBA binding. Additional mutations/deletions were introduced to prevent binding of Gβγ (ΔN 1–25), GPCRs (ΔC 346–354), GoLoco GDIs (N149I), and Ric-8A (ΔC 346–354). The W211A mutation is known to inhibit binding of Gαi3 to all GBA motifs described to date (i.e., from GIV, DAPLE, Calnuc, NUCB2) and is used as a negative control throughout this study. (C, Bottom) Localization of each modification is shown in a previously described model of Gαi3 bound to the GBA motif of GIV (33). (D) GBAi binds to the GBA motif of GIV with the same affinity as Gαi3, and GBAi-GIV binding is not affected by the activation mimetic GDP-AlF4−. Binding of a fluorescently labeled GIV peptide corresponding to its GBA motif (residues 1,671–1,701) to the indicated concentrations of purified His-tagged Gαi3 (black), GBAi wt (blue), or GBAi W211A (red) was determined by FP in the presence of GDP (Left) or GDP-AlF4− (Right). Data were normalized to maximal binding and fitted to a one-site binding model (solid lines). Results from three independent experiments are expressed as mean ± SEM. (E) GBAi binds the same to full-length GIV in the presence of GDP, GDP-AlF4−, or GTPγS, whereas Gαi3 binds GIV only in the presence of GDP. Lysates of HEK293T cells expressing full-length, myc-tagged GIV were incubated with GST, GST-Gαi3, or GST-GBAi immobilized on glutathione-agarose beads in the presence of GDP, GDP-AlF4−, or GTPγS (as indicated). Resin-bound proteins were eluted, separated by SDS/PAGE, and analyzed by Ponceau S-staining and immunoblotting (IB) as indicated. Input = 10% of the amount of lysate used in each pulldown. One experiment of two with very similar results is shown.
Mutation of Gly203 to Ala (G203A) is known to permit nucleotide binding and hydrolysis with properties similar to those of Gαi wild-type (wt) at physiological concentrations of Mg2+ (∼1 mM), but disrupts a change in conformation of the Switch II region (SwII) that occurs in the wt protein upon GTP binding (34, 35). This change in conformation in Gαi wt leads to its disengagement from G protein binding partners that bind to inactive, GDP-bound Gα (e.g., Gβγ motifs, GBA motifs) and promotes binding to effectors of Gα-GTP (6, 8, 9, 36, 37). Based on this, we reasoned that introducing the G203A mutation would preclude GBA disengagement from GBAi even upon loading of GTP. At the same time, this mutation would also preclude binding to effectors and RGS GAPs, which bind to GTP-bound active conformations of Gα by making direct contacts with the SwII (37–39). To disrupt GBAi binding to GoLoco GDIs, another major group of Gα binding partners, we mutated Asn149 to Ile (N149I), which is known to disrupt Gαi binding to all GoLoco motifs without affecting other properties of the G protein (40). We deleted the last nine C-terminal residues (ΔC) with the dual purpose of disrupting binding to GPCRs (41–43) and the chaperone/GEF Ric-8A (44). The first 25 N-terminal residues (ΔN) were also deleted to preclude binding to Gβγ subunits based on the facts that the N terminus is one of the major Gβγ contacts in the heterotrimeric structure and that previous reports have shown that deletion of the N terminus of Gαi3 (45) or other Gα subunits (46–48) is sufficient to abolish Gβγ binding. Neither ΔC nor ΔN has significant effects on Gα nucleotide binding or hydrolysis (33, 49). G203A, N149I, ΔC, or ΔN is known or predicted to have no effect on GBA motif binding based on structural insights and/or experimental data (33) (Fig. 1C).
GPCRs, Gβγ, effectors, RGS GAPs, GoLoco GDIs, and Ric-8A are, to our knowledge, all of the Gαi binding partners for which a biological function has been clearly established. Thus, the set of mutations described above is expected to preserve binding to GBA motifs without interfering with the function of other Gαi binding partners. To further rule out possible off-target effects of GBAi, we designed a mutated version in which Trp211 is replaced by Ala (W211A) to be used as a negative control in all of our experiments (Fig. 1C). This mutation has been shown to disrupt binding of Gαi3 to every GBA motif described to date (6, 7, 9), but it has no significant effect on G protein structural integrity or the ability to bind and hydrolyze nucleotides (50). Thus, disruption of GBAi-mediated effects by the W211A mutation will increase confidence in its specificity for inhibiting GBA motifs. We next proceeded to validate that the properties predicted from the effect of individual mutations described above hold true when present simultaneously in GBAi.
GBAi Is Properly Folded and Binds Nucleotides.
First, we assessed the structural integrity of purified GBAi. For this, we monitored its thermal denaturation by differential scanning fluorimetry assays carried out in the presence or absence of an excess of nucleotides (Fig. S1). This assay is equivalent to that recently established by Sun et al. (51) to assess the overall stability and nucleotide binding ability of Gαi1 mutants. The GBAi melting temperature (Tm) in this assay was comparable to that of wt Gαi3 in the absence of added nucleotides (∼40 °C). Addition of an excess amount of GDP (250 μM) increased the Tm of both GBAi and Gαi3 only moderately (∼ +4 °C), whereas addition of the same concentration of the nonhydrolyzable GTP analog guanosine 5′-O-[γ-thio]triphosphate (GTPγS) had a more marked effect (∼ +20 °C; Fig. S1). The larger extent of the stabilization upon GTPγS binding compared with GDP binding is consistent with previous observations with Gαi1 (51). In contrast, addition of GTPγS to Gαi3 S47R, a recently characterized GTP-binding deficient mutant (52), did not recapitulate the marked thermal stabilization observed for GBAi or Gαi3 (Fig. S1). Taken together, these findings indicate that GBAi is folded properly and retains the ability to bind nucleotides.
GBAi Binds to GBA Motifs and Does Not Dissociate upon GTP Binding.
To start characterizing its biochemical properties, we compared the binding of GBAi to GBA motifs with that of Gαi3 and examined the effect of GTP mimetics on the binding. For this, GBAi was purified from bacteria and binding to a peptide corresponding to the GBA motif of GIV (residues 1,671–1,701) was measured using a fluorescence polarization (FP) assay. GIV is the prototypical protein with a GBA motif, and we have previously shown that peptides corresponding to the GBA motif fully recapitulate the binding properties of the protein (33, 53). GBAi binds to the GIV-derived peptide with an affinity similar to that of Gαi3 in the presence of GDP (Fig. 1D and Table S1). The equilibrium dissociation constants (Kds) were ∼0.5 μM, which is consistent with previously reported values using different methods (6, 13). As expected, binding to Gαi3 was almost abolished in the presence of GDP-AlF4− (which mimics the GTP-bound transition state) (39) (Fig. 1D). In contrast, GBAi binding affinity was the same in the presence of GDP or GDP-AlF4−. The GBAi W211A mutant did not bind GIV in the presence of either GDP or GDP-AlF4−, demonstrating that it is a bona fide negative control for GBA binding. Equivalent results were obtained when Gαi3 and GBAi preloaded with the nonhydrolyzable GTP analog GTPγS (instead of GDP-AlF4−) were compared with GDP in the same FP assay (Fig. S2). We also confirmed that binding of GBAi to full-length GIV, instead of to the GBA motif peptide used in FP assays, is not disrupted by GDP-AlF4− or GTPγS in pulldown assays (Fig. 1E). These results indicate that GBAi binds to GIV with an affinity comparable to that of Gαi3-GDP and that, contrary to what occurs with Gαi3, GBAi binding is not disrupted upon GTP binding.
Next, we investigated the properties of the interaction of GBAi with other GBA motifs. For this, we carried out FP experiments as above with peptides derived from the three other mammalian proteins with a GBA motif described to date (i.e., DAPLE, Calnuc, NUCB2) (Fig. S3A). The results were analogous to those obtained with GIV, indicating that GBAi binds to each one of the GBA motifs with the same affinity as Gαi3-GDP and that the binding is not affected by GTP mimetics but can be abolished by the W211A mutation (Fig. S3 B and C and Table S1). Similar observations were made when the interactions of full-length DAPLE, Calnuc, and NUCB2 were tested using a protein–protein binding assay different from FP (i.e., GST pulldowns) (Fig. S3D), which further validates our conclusions on the properties of GBAi binding. Finally, we also tested if the binding mode of GBA motifs was conserved between GBAi and Gαi3. Previous structural and biochemical studies have shed light onto how GBA motifs engage G proteins. A critical feature is that a set of hydrophobic residues in the GBA motif is used to make contacts with a groove formed by the SwII and α3 helix of Gαi3 (6–9, 15, 54). Mutation of a Phe conserved across all GBA motifs (Fig. S3A) to Ala (F → A) has been previously shown to markedly reduce their binding to Gαi3 (6–9, 15). We found that the same occurs with GBAi (Fig. S4), supporting that GBA motifs bind to GBAi and Gαi3 in a similar manner. Taken together, these findings show that GBAi binds efficiently to all known GBA motifs, and does not disengage from the GBA motifs upon GTP binding, and that the W211A mutation disrupts its binding to GBA motifs.
GBAi Does Not Bind to Gβγ.
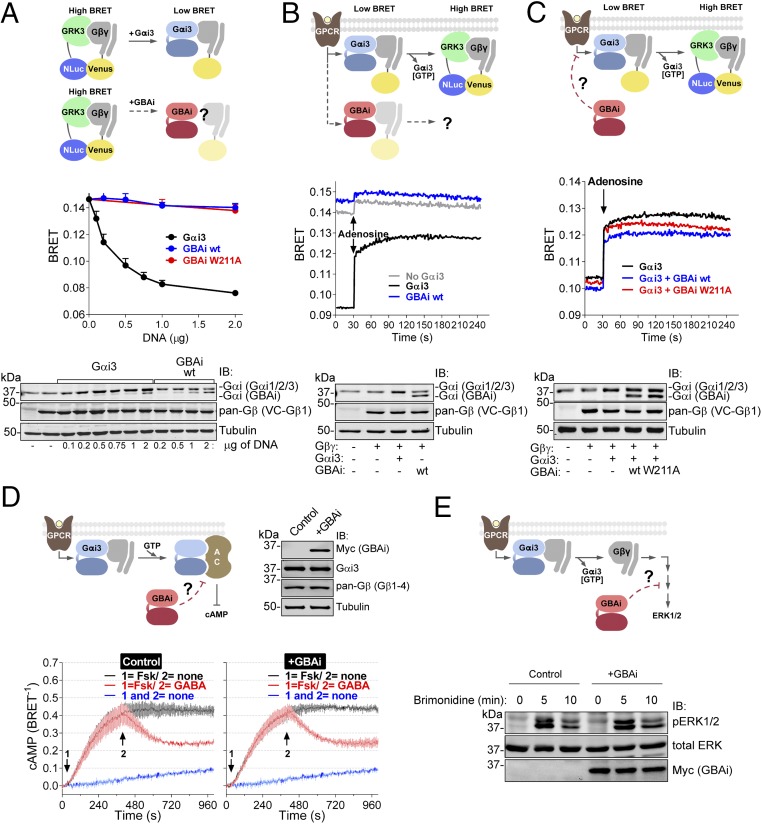
Next, we investigated if GBAi fulfills its second desired property that was part of our original design (Fig. 1 A and B); that is, it does not interact with Gαi binding partners that are not GBA proteins. We focused our initial efforts on Gβγ because it binds constitutively to Gα subunits with high affinity and is the predominant binding partner in cells. An initial concern was that, according to one report (55), Gβγ subunits can interact with Gαt (α-transducin) even if the N terminus is deleted. However, these observations were based on assays carried out in vitro using supraphysiological concentrations of G proteins (55) and are in discrepancy with several other reports showing that deletion of the N terminus of Gαi3 or other Gα subunits disrupts Gβγ binding under more physiological conditions (45–48). To directly compare binding of GBAi and Gαi3 to Gβγ, we used a previously described (56, 57) BRET assay in HEK293T cells. In this assay, BRET results from the interaction between free Gβγ and its effector GRK3 (Fig. 2A), which can be inhibited by Gα binding to Gβγ due to competition for an overlapping interaction region with GRK3. As expected, expression of Gβγ alone in the absence of exogenous Gα resulted in high levels of BRET that were quenched upon expression of increasing amounts of Gαi3 in a dose-dependent manner (Fig. 2A). In contrast, expression of comparable amounts of GBAi did not reduce the BRET signal, indicating that GBAi does not associate with Gβγ in cells. To further validate this conclusion, we carried out immunoprecipitation experiments in MCF-7 cells (Fig. S5). Equal amounts of epitope-tagged Gαi3 and GBAi (wt or W211A) were coexpressed with exogenous Gβγ and immunoprecipitated. Exogenous and endogenous Gβγ was present in Gαi3 immunoprecipitates, whereas Gβγ was not detected in the GBAi immunoprecipitates (Fig. S5). These results indicate that GBAi does not bind to Gβγ in cells.
Fig. 2.
GBAi does not interfere with GPCR-mediated activation of G proteins. (A) GBAi does not associate with Gβγ in mammalian cells as determined by BRET. (Top) Schematic of the BRET assay used to monitor the association of Gαi3 or GBAi with Gβγ. In the absence of Gαi3, Venus-tagged Gβγ (V-Gβγ, BRET acceptor) associates with mas-GRK3ct-NLuc (GRK3, BRET donor) inducing a high BRET signal. Expression of Gαi3 competitively displaces V-Gβγ from mas-GRK3ct-NLuc and decreases BRET. (Middle) HEK293T cells were transfected with the indicated amounts of plasmids encoding for Gαi3, GBAi wt, and GBAi W211A, and equal amounts of the BRET donor (mas-GRK3ct-NLuc) and acceptor (V-Gβγ). BRET was measured under resting (unstimulated) conditions and is presented as mean ± SEM of four independent experiments. (Bottom) Protein expression of Gαi3, GBAi wt, and V-Gβγ was assessed by immunoblotting (IB) with the indicated antibodies. The upper band detected by the Gαi antibody corresponds to exogenous Gαi3 plus endogenous Gαi1, Gαi2, and Gαi3, whereas the lower band corresponds to GBAi. (B) GBAi does not couple to GPCRs in mammalian cells as determined by BRET. (Top) Schematic depicting the BRET assay used to determine the coupling of Gαi3 or GBAi to GPCRs. Under resting conditions, V-Gβγ associates with Gαi3 and BRET is low. Upon GPCR stimulation, the Gαi3:V-Gβγ heterotrimer dissociates and V-Gβγ interacts with mas-GRK3ct-NLuc, leading to an increase in BRET. (Middle) HEK293T cells were transfected with plasmids encoding for Gαi3 (0.5 μg) or GBAi wt (2 μg), along with plasmids for the BRET donor and acceptor (mas-GRK3ct-NLuc and V-Gβγ), as well as for the adenosine 1 receptor. BRET was measured every second. After 30 s of measurement under resting conditions, cells were stimulated with adenosine (10 μM). One representative experiment of three is shown. (Bottom) Protein expression of Gαi3, GBAi wt, and V-Gβγ was assessed by IB with the indicated antibodies. (C) GBAi does not interfere with GPCR-mediated activation of Gαi3 as determined by BRET. (Top) Schematic depicting the BRET experiment used to assess the possible interference of GBAi with Gαi3 coupling to GPCRs. The experimental design is as in B except that GBAi is coexpressed, along with Gαi3 and the rest of the BRET assay components, to test if it could impair the G protein-dependent BRET increase observed upon GPCR stimulation. (Middle) Experiments were carried out exactly as in B except that GBAi (wt or W211A, 2 μg) and Gαi3 (0.5 μg) were expressed simultaneously in the same cells. One representative experiment of three is shown. (Bottom) Protein expression of Gαi3, GBAi, and V-Gβγ was assessed by IB with the indicated antibodies. (D) GBAi does not interfere with GPCR-mediated regulation of cAMP by Gαi. (Top Left) Schematic depicting the experiment used to assess the possible interference of GBAi with GPCR-mediated regulation of cAMP levels. (Top Right) Protein expression of GBAi, Gαi3, and Gβγ was assessed by IB with the indicated antibodies. (Bottom) HEK293T cells were transfected with plasmids encoding for Nluc-EPAC-VV (0.05 μg) and GABABRs (0.2 μg) in the presence (+GBAi) or absence (control) of GBAi wt (2 μg). BRET was measured every 4 s. Forskolin (Fsk, 1 μM) and GABA (1 μM) were added (sequentially) at the indicated times. The blue trace corresponds to unstimulated control cells and is duplicated in both panels as a visual reference of the baseline. Results from three independent experiments are expressed as mean ± SEM. (E) GBAi does not interfere with GPCR-mediated regulation of ERK1/2 by Gβγ. (Top) Schematic depicting the experiment used to assess the possible interference of GBAi with GPCR-mediated regulation of ERK1/2. (Bottom) HEK293T cells were transfected with plasmids encoding for α2A-AR (0.2 μg) in the presence (+GBAi) or absence (control) of GBAi wt (2 μg). Cells were serum-starved overnight and then stimulated with brimonidine (5 μM) for the indicated times. Cell lysates were immunoblotted with the indicated antibodies. One representative experiment of three is shown.
GBAi Does Not Interfere with GPCR-Mediated G Protein Signaling.
The C-terminal tails of Gα subunits and their association with Gβγ subunits are obligatory requirements for coupling to GPCRs (41–43). To confirm that GBAi, which lacks these two features, does not couple to GPCRs, we used the BRET-based assay described above. In cells expressing Gαi3 and the GPCR adenosine 1 receptor (in addition to the BRET donor and acceptor), adenosine led to a rapid increase in BRET, which reflects the dissociation of Gαi3:Gβγ heterotrimers upon activation (Fig. 2B). On the contrary, in cells expressing GBAi instead of Gαi3, no significant BRET increase was observed in response to adenosine (Fig. 2B). In agreement with Fig. 2A, the basal BRET signal before adenosine stimulation was already as high as in cells not expressing either GBAi or Gαi3, which reflects the lack of association of GBAi with Gβγ (Fig. 2B) and also explains the insensitivity to GPCR stimulation. To further validate that GBAi does not interfere with GPCR-mediated signaling, we tested the effect of expressing GBAi on the activation of Gαi3:Gβγ heterotrimers using the same BRET-based assay (Fig. 2C). We found that the BRET response caused by the dissociation of Gαi3:Gβγ heterotrimers upon adenosine stimulation was not affected by coexpression of GBAi (wt or W211A) with Gαi3. Together, these observations show that GBAi does not couple to GPCRs and does not interfere with GPCR-mediated activation of G proteins.
To further validate that GBAi does not interfere with GPCR-mediated signaling, we investigated its effect on downstream G protein-dependent readouts. First, we monitored cAMP levels in HEK293T cells using a previously described BRET-based biosensor (“nanoluc-exchange protein directly activated by cAMP-VenusVenus,” Nluc-EPAC-VV) (56) in which BRET efficiency is inversely proportional to cAMP levels. Upon GPCR stimulation, GTP-bound Gαi subunits dampen cAMP production by directly inhibiting the activity of adenylyl cyclases. As expected, addition of the adenylyl cyclase activator forskolin elevated the intracellular levels of cAMP, which were efficiently decreased upon subsequent stimulation of the Gi-coupled GPCR GABAB receptor (GABABR) (Fig. 2D). Expression of GBAi did not alter either forskolin-stimulated cAMP levels or the inhibitory response after GABABR stimulation (Fig. 2D). The same lack of effect by GBAi was observed when a different Gi-coupled GPCR was used [α2A-adrenergic receptor (α2A-AR)] or when forskolin-mediated cAMP elevation was inhibited by prestimulation of the GABABR (Fig. S6). Next, we investigated if GBAi interfered with ERK1/2 activation upon stimulation of the α2A-AR, a Gi signaling response that has been previously shown to be mediated by Gβγ rather that Gα (58, 59). We found that ERK1/2 activation (as determined by its phosphorylation) in response to α2A-AR stimulation was not altered upon GBAi expression (Fig. 2E). Together, these results indicate that GBAi does not interfere with GPCR-mediated regulation of G protein signaling.
GBAi Does Not Interact with Gαi Effectors and the Gαi Regulators RGS GAPs, GoLoco GDIs, and Ric-8A.
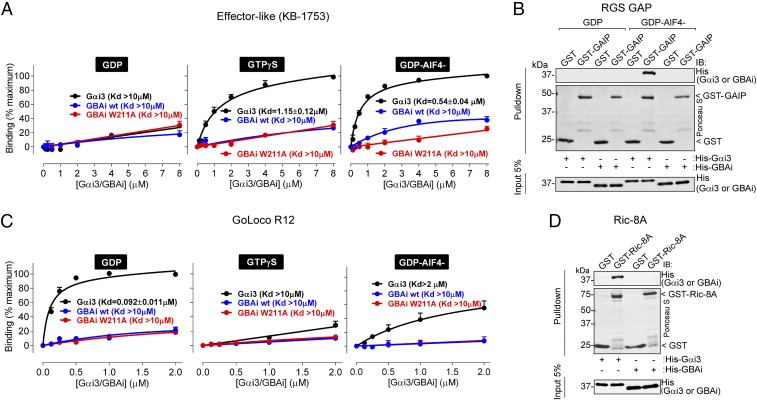
In addition to GPCRs and Gβγ subunits, Gαi proteins bind to effectors and various regulators. An effector of Gα subunits can be defined as a protein whose function is modulated upon specific binding of the G protein in its GTP-bound active conformation. Our results in Fig. 1D and Figs. S2 and S3 suggest that GBAi cannot adopt an active conformation because it does not disengage from GBA motifs in the presence of GTP mimetics, presumably due to the properties associated with the G203A mutation. This is further supported by the fact that GBAi does not interfere with Gαi regulation of its effector adenylyl cyclase in cells (Fig. 2D). Next, we tested directly if GBAi binds to KB-1753, a synthetic peptide that binds to Gαi subunits like an effector (37). KB-1753 is the only effector-like molecule that has been crystalized in complex with Gαi to date, and its biochemical properties have been extensively characterized (37). Using a fluorescently labeled KB-1753 peptide in FP assays, we found that, as expected, it bound to Gαi3 preferentially when preincubated with GTPγS or GDP-AlF4− versus GDP, whereas no binding to GBAi was detected in any of the three conditions (Fig. 3A). The same lack of GBAi binding was observed when binding was assessed in pulldown assays (Fig. S7A). Similar experiments were carried out with GST-fused GAIP (also known as RGS19), a representative member of the RGS GAP family. RGS GAPs also bind to GTP-loaded active conformations of Gαi proteins, with a preference for the transition state mimicked by GDP-AlF4− (39). As for KB-1753, GAIP bound to Gαi3, but not GBAi, in the GDP-AlF4− (Fig. 3B) or GTPγS (Fig. S7B) condition. We also confirmed the lack of GBAi binding to GAIP in cell-based assays by carrying out coimmunoprecipitations after expression of the proteins of interest in HEK293T cells (Fig. S8A). Taken together with the results shown in Fig. 1 and Figs. S2 and S3, these findings indicate that GBAi cannot adopt an active conformation capable of engaging effectors or other regulators that bind to Gα-GTP like RGS proteins.
Fig. 3.
GBAi does not bind to Gαi effectors or the Gαi regulators RGS GAPs, GoLoco GDIs, and Ric-8A. (A) GBAi does not bind to the effector-like peptide KB-1753. Binding of a fluorescently labeled KB-1753 peptide to the indicated concentrations of purified His-tagged Gαi3 (black), GBAi wt (blue), or GBAi W211A (red) was determined by FP in the presence of GDP, GTPγS, or GDP-AlF4− as indicated. Data were normalized to maximal binding and fitted to a one-site binding model (solid lines) to calculate the indicated Kds. KB-1753 binds Gαi3 with high affinity in the presence of GDP-AlF4− or GTPγS and low affinity in the presence of GDP, whereas GBAi does not bind in any of the three conditions. Results from three independent experiments are expressed as mean ± SEM. (B) GBAi does not bind to the RGS GAP protein GAIP. Purified GST or GST-GAIP was immobilized on glutathione-agarose beads and incubated with purified His-Gαi3 or His-GBAi in the presence of GDP or GDP-AlF4− as indicated. Resin-bound proteins were eluted, separated by SDS/PAGE, and analyzed by Ponceau S-staining and immunoblotting (IB) as indicated. GST-GAIP binds His-Gαi3 in the presence of GDP-AlF4−, but not GDP, whereas it does not bind to GBAi either in the presence of GDP or GDP-AlF4−. One representative experiment of three is shown. (C) GBAi does not bind to the GoLoco GDI motif of RGS12 (GoLoco R12). Binding of a fluorescently labeled peptide corresponding to GoLoco R12 (RGS12 residues 1,185–1,221) to the indicated concentrations of purified His-tagged Gαi3 (black), GBAi wt (blue), or GBAi W211A (red) was determined by FP in the presence of GDP (Left), GTPγS (Center), or GDP-AlF4− (Right). Data were normalized to maximal binding and fitted to a one-site binding model (solid lines) to calculate the indicated Kds. GoLoco R12 binds Gαi3 with high affinity in the presence of GDP and low affinity in the presence of GTPγS or GDP-AlF4−, whereas GBAi binding is almost or completely absent under the same conditions. Results from three independent experiments are expressed as mean ± SEM. (D) GBAi does not bind to the chaperone/GEF protein Ric-8A. Experiments were carried out exactly as in B except that GST–Ric-8A was used instead of GST–KB-1753 and all conditions were tested in the presence of GDP. GST–Ric-8A binds to His-Gαi3 but not to His-GBAi. One representative experiment of three is shown.
Next, we investigated if GBAi binds to GoLoco GDIs, another major family of Gαi regulators (3). Much like proteins with a GBA motif, GoLoco GDIs bind to GDP-bound Gαi with a marked preference over GTP-bound Gαi (2, 3). Because the N149I mutation in Gαi has been previously shown to abolish binding to all GoLoco motif-containing proteins (40), we reasoned that GBAi would not bind to this class of G protein regulators. For this, we compared the binding of Gαi3 and GBAi to the GoLoco motif of RGS12 (GoLoco R12) as a representative example using an FP assay analogous to that used in Fig. 1D. Consistent with previous reports (60), GoLoco R12 bound to Gαi3-GDP with high affinity (Kd below 100 nM) (Fig. 3C). In contrast GBAi binding was nearly absent and even weaker than the binding of GoLoco R12 to active (GDP-AlF4−) Gαi3 (Fig. 3C). We also found that GBAi expressed in mammalian cells does not bind to a truncated form of RGS12 containing its GoLoco motif but lacking its RGS domain (Fig. S8B).
Finally, we tested if GBAi interacts with the nonreceptor GEF/chaperone Ric-8A and found that it does not interact with either when using purified proteins in vitro (Fig. 3D) or by coimmunoprecipitation in mammalian cells (Fig. S8C). This is not surprising because it has been previously reported that a deletion of the last nine residues of Gαi, like the one present in GBAi (Fig. 1C), completely abolishes Ric-8A binding (44).
GBAi Inhibits GIV-Mediated Enhancement of PI3K-Akt Signaling in Response to Integrin Stimulation.
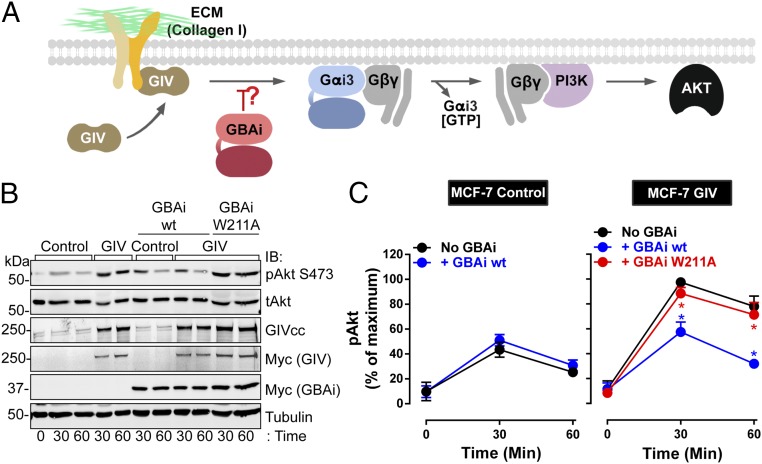
The results presented so far demonstrate that GBAi displays all of the desired properties described in Fig. 1 to behave as an efficient and specific inhibitor of GBA proteins. To provide proof-of-principle evidence that GBAi can indeed function as an inhibitor of the biological function of GBA motifs, we tested it in a well-characterized system. We have recently described that GIV promotes integrin signaling in cancer cells by activating G proteins via its GBA motif (18, 61). Others have arrived at a similar conclusion independently (62). The mechanism by which GIV promotes integrin signaling was elucidated using loss-of-function and gain-of-function genetic approaches, along with pharmacological manipulations, and was corroborated in multiple cell lines (18, 61). Thus, this is a robust system for testing GBAi. In brief, GIV is recruited to active integrins, leading to G protein activation via its GBA motif and subsequent enhancement of Gβγ-PI3K-Akt signaling (Fig. 4A). To interrogate this mechanism with GBAi, we used MCF-7 cells expressing a control plasmid or a plasmid encoding for GIV (Fig. 4 B and C). MCF-7 cells are poorly invasive breast cancer cells that naturally express very low levels of GIV (18, 61). Upon exogenous expression of GIV, MCF-7 cells gain proinvasive traits (7, 18, 61), including enhanced integrin-Akt signaling. Consistent with our previous observations (18, 61), GIV expression in MCF-7 cells did not affect Akt activation (as determined by the levels of Akt phosphorylated at S473) in unstimulated cells (time 0 in the figures) (Fig. S9), but potentiated it in response to collagen I stimulation compared with control cells (Fig. 4 B and C, compare black lines/symbols in Fig. 4C). Importantly, expression of GBAi wt inhibited the enhancement of Akt activation in GIV-expressing cells (compare blue with black lines/symbols in Fig. 4C, Right) to the levels of activation observed in control cells (not expressing exogenous GIV and in the absence of GBAi) (Fig. 4 B and C, black lines/symbols in Fig. 4C, Left). On the other hand, GBAi wt had no effect on the activation of Akt in control cells not expressing exogenous GIV (compare blue and black lines/symbols in Fig. 4C, Left), suggesting that GBAi specifically inhibits the GIV-dependent response. Moreover, the inhibition mediated by GBAi wt was not reproduced by the GBA binding-deficient mutant W211A (Fig. 4 B and C, compare red with black lines/symbols in Fig. 4C, Left), which indicates that the action of GBAi is due to inhibition of GIV’s GBA motif and not to some other spurious effect. These results demonstrate that GBAi functions as a specific inhibitor of GBA-mediated signaling of GIV in cells.
Fig. 4.
GBAi inhibits GIV-mediated potentiation of PI3K-Akt signaling upon integrin stimulation. (A) Schematic of a previously described mechanism (18) by which GIV potentiates PI3K-Akt signaling in response to integrin stimulation via its GBA motif. Stimulation of cells with extracellular matrix (ECM) proteins (e.g., collagen I) triggers the recruitment of GIV to the intracellular tail of integrins, which, in turn, leads to GBA-dependent G protein signaling. PI3K-Akt activation is achieved via free Gβγ subunits released from Gi heterotrimers upon GIV GBA action. (B and C) GBAi wt, but not GBAi W211A, inhibits GIV-mediated potentiation of Akt activation upon collagen I stimulation of MCF-7 cells. MCF-7 cells stably expressing a vector control or full-length GIV and transfected with myc-GBAi wt (blue) or myc-GBAi W211A (red), as indicated, were lifted from culture dishes; kept in suspension for 1 h in serum-free media (time 0); and stimulated by plating on collagen I-coated culture dishes for 30 and 60 min. One representative immunoblot result from four independent experiments is shown in A, and the results for the quantification of Akt activation [as determined by levels of phosphorylated Akt (pAkt)] expressed as mean ± SEM are shown in C. *P < 0.05, using the Student’s t test (blue, compared with no GBAi; red, compared with GBAi wt). tAkt, total Akt.
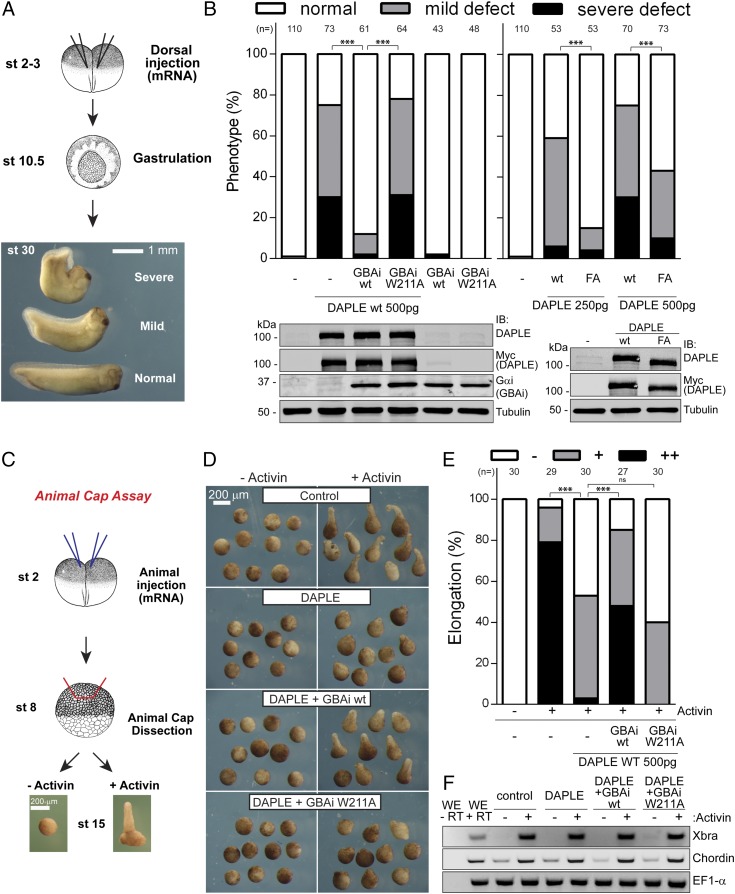
GBAi Inhibits DAPLE-Mediated Gastrulation and Convergent Extension Defects in Xenopus laevis Embryos.
The inhibition of GIV-mediated enhancement of integrin signaling in cancer cells is an important validation benchmark for GBAi. Next, we simultaneously interrogated if GBAi possesses two important features: (i) usefulness in revealing uncharacterized biological activities of GBA motifs and (ii) effectiveness in blocking GBA-mediated processes in whole organisms in vivo. For this, we investigated the role of the GBA motif of DAPLE in embryogenesis using Xenopus laevis as a model of vertebrate development.
Noncanonical Wnt signaling is crucial for the orchestration of cell movements during gastrulation, and Dishevelled is a core component of this pathway (63). DAPLE has been previously shown to play a role in embryonic development, and it is believed to do so, at least in part, by modulating noncanonical Wnt signaling via direct interaction with Dishevelled (23, 24). On the other hand, we have recently shown that that the GBA motif of DAPLE is also required for efficient noncanonical Wnt signaling in isolated cells (6). However, it is unknown if the GBA motif is important for the effects of DAPLE on embryo development. Consistent with previous reports (23, 24), we found that dorsal injection of DAPLE mRNA (encoding residues 1,217–2,028, containing both the GBA and Dishevelled binding motifs) into frog embryos caused gastrulation defects with high frequency (Fig. 5 A and B). Coexpression of GBAi wt with DAPLE almost completely blunted the gastrulation defects induced by DAPLE, while the same amount of GBAi wt alone had no effect (Fig. 5 A and B). On the other hand, coexpression of GBAi W211A with DAPLE did not diminish the frequency of gastrulation defects (Fig. 5 A and B). These results indicate that DAPLE induces gastrulation defects via its GBA motif. To further validate this using a GBAi-independent approach, we carried out analogous experiments in frog embryos to compare the effect of DAPLE wt and a validated GEF-deficient mutant of its GBA motif [i.e., F1675A (FA) (6)] on gastrulation. We found that the FA mutation dramatically reduced the frequency of gastrulation defects induced by DAPLE (Fig. 5 A and B), validating that this phenotype is caused by the DAPLE GBA motif. The incomplete reduction of the phenotype with the FA mutant is not surprising because it has been previously shown that the FA mutation does not completely abolish GBA function in other experimental settings (6, 15, 54). However, this suggests that GBAi can be more efficient than the FA mutation in inhibiting the function of GBA motifs. Previous reports have shown that, much like the FA mutation, deletion of the Dishevelled binding motif of DAPLE also impairs its role in embryonic development in the same experimental system as described here (23, 24). Thus, it is conceivable that both G protein and Dishevelled binding by DAPLE work coordinately in this context.
Fig. 5.
GBAi inhibits DAPLE-mediated convergent extension defects in X. laevis embryos. (A and B) GBAi wt or the F1675A mutation in DAPLE inhibits developmental defects induced by DAPLE. (A) Schematic depicting the assay to assess DAPLE-induced developmental defects. mRNAs are injected equatorially in both dorsal blastomeres of two- to four-cell embryos [stage (st) 2–3], and phenotypes are assessed at st 30. Representative phenotypes are shown: normal or dorsally bent embryos (mild or severe). (B, Left) DAPLE (500 pg), GBAi wt (1 ng), or GBAi W211A (1 ng) mRNAs were injected as indicated, and the frequency of phenotypes is assessed at st 30. The total number of embryos analyzed from three independent experiments for each group is indicated on the top of the graph. (Bottom) Protein expression of DAPLE and GBAi was assessed by immunoblotting (IB) with the indicated antibodies. (B, Right) DAPLE wt or DAPLE FA mRNAs (250 pg or 500 pg) were injected, and phenotypes were analyzed as in the left graph. (Bottom) Protein expression of DAPLE wt and DAPLE FA (250 pg of mRNA) was assessed by IB with the indicated antibodies. (C–F) GBAi wt, but not GBAi W211A, blocks DAPLE-mediated inhibition of convergent extension movements. (C) DAPLE (500 pg) and GBAi (wt or W211A, 1 ng) were coinjected into the animal hemisphere of both blastomeres of two-cell embryos. Animal caps were dissected at st 8, treated (or not treated) with activin to induce elongation, and analyzed for elongation at st 15. Representative pictures of activin-treated versus untreated caps injected with the indicated mRNAs (D) and the frequency of different elongation phenotypes [E; −, none, +, mild, or ++, strong elongation] are shown. The total number of embryos analyzed from three independent experiments for each group is indicated on the top of the graph. (F) RT-PCR of activin-mediated gene induction from caps at st 10.5 injected with the indicated mRNAs and treated or not treated with activin. Whole embryos (WE) with (+RT) or without (−RT) the reverse transcriptase reaction are shown on the left lanes as positive and negative controls, respectively. ***P < 0.001, χ2 test.
We performed additional experiments with GBAi to substantiate that DAPLE causes gastrulation defects via its GBA motif by impinging on a process that relies heavily on noncanonical Wnt signaling. Gastrulation defects, such as those we observed in Fig. 5A, frequently arise from alterations in a set of cell movements commonly referred to as convergent extension (64). Noncanonical Wnt signaling is an obligatory requirement for convergent extension movements (64). Experimentally, convergent extension can be assessed with an assay that involves the treatment of embryo explants with activin (Fig. 5C). Briefly, stimulation of animal caps excised at stage 8 with activin induces the expression of mesodermal differentiation genes, which, in turn, causes the release of noncanonical Wnt ligands that drive convergent extension cell movements and subsequent explant elongation (65). Expression of DAPLE by mRNA microinjection led to a marked attenuation of activin-induced animal cap elongation (Fig. 5 D and E). The effect of DAPLE was efficiently reverted by GBAi wt, but not by the GBA-binding deficient W211A mutant (Fig. 5 D and E), indicating that the GBA motif of DAPLE is required for the observed attenuation of animal cap elongation. We ruled out that the observed effects of DAPLE were due to disruption of activin-stimulated mesodermal differentiation by performing RT-PCR assays (Fig. 5F). We found that activin stimulation led to mesodermal differentiation in all experimental conditions, as determined by induction of the mesodermal markers Xbra and Chordin (Fig. 5F). Taken together, these findings indicate that the underlying cause of the gastrulation phenotypes caused by DAPLE via its GBA motif is a defect in convergent extension movements.
In summary, we have described here the rational design, validation, and implementation of a synthetic protein that can be used as a tool to investigate the functions of GBA motifs in vitro and in vivo. The development of tools to interdict specific steps of G protein signaling has proven extremely valuable in the past to advance our knowledge of this core mechanism of cell communication (26, 29). There is a parallelism between GBAi and one of the tools that has historically contributed to major advances in the field of G protein signaling (i.e., pertussis toxin). While pertussis toxin uncouples Gαi subunits from their canonical GEFs (i.e., GPCRs), GBAi uncouples Gαi subunits from a family of atypical GEFs that are not membrane receptors. In the same way that pertussis toxin serves as a tool to generically assess the role of GPCR-Gi coupling in a myriad of biological contexts, we hope that GBAi will serve a similar purpose for investigating the consequences of GBA-Gi coupling.
Materials and Methods
Kds were determined by FP measurements, and protein–protein binding was assessed by GST pulldown or immunoprecipitation assays as previously described (6, 7, 9, 53, 66), with minor modifications. Collagen I stimulation experiments were carried out exactly as in the study by Leyme et al. (18), and BRET-based measurement was carried out as described previously (52, 56, 57, 67), with minor modifications. For X. laevis experiments, fertilized eggs were microinjected by in vitro-transcribed mRNA and analyzed as indicated in the main text. Statistical significance between various conditions was assessed by the Student’s t test or χ2 test. A full description of materials and methods used in this study is provided in Supporting Information.
Supplementary Material
Acknowledgments
We are indebted to Nevin Lambert (Augusta University) for providing critical reagents and for extensive discussions to set up and optimize the BRET assays. We also thank Kirill Martemyanov (Scripps Research Institute), Pradipta Ghosh and Marilyn Farquhar (University of California, San Diego), John Sondek (University of North Carolina at Chapel Hill), Joe Blumer and Steve Lanier (Medical University of South Carolina), Paul Slessinger (Mount Sinai School of Medicine), and Steve Sprang (University of Montana) for providing reagents. This work was supported by NIH Grants R01GM112631 and R01GM108733, American Cancer Society Grants RSG-13-362-01-TBE and IRG-72-001-36, and the Karin Grunebaum Cancer Research Foundation (to M.G.-M.) and NIH Grant R01GM098367 (to I.D.). V.D. is a recipient of a postdoctoral fellowship from the Hartwell Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. S.R.S. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1707992114/-/DCSupplemental.
References
- 1.Gilman AG. G proteins: Transducers of receptor-generated signals. Annu Rev Biochem. 1987;56:615–649. doi: 10.1146/annurev.bi.56.070187.003151. [DOI] [PubMed] [Google Scholar]
- 2.Sato M, Blumer JB, Simon V, Lanier SM. Accessory proteins for G proteins: Partners in signaling. Annu Rev Pharmacol Toxicol. 2006;46:151–187. doi: 10.1146/annurev.pharmtox.46.120604.141115. [DOI] [PubMed] [Google Scholar]
- 3.Willard FS, Kimple RJ, Siderovski DP. Return of the GDI: The GoLoco motif in cell division. Annu Rev Biochem. 2004;73:925–951. doi: 10.1146/annurev.biochem.73.011303.073756. [DOI] [PubMed] [Google Scholar]
- 4.Ross EM, Wilkie TM. GTPase-activating proteins for heterotrimeric G proteins: Regulators of G protein signaling (RGS) and RGS-like proteins. Annu Rev Biochem. 2000;69:795–827. doi: 10.1146/annurev.biochem.69.1.795. [DOI] [PubMed] [Google Scholar]
- 5.Tall GG. Ric-8 regulation of heterotrimeric G proteins. J Recept Signal Transduct Res. 2013;33:139–143. doi: 10.3109/10799893.2013.763828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aznar N, et al. Daple is a novel non-receptor GEF required for trimeric G protein activation in Wnt signaling. Elife. 2015;4:e07091. doi: 10.7554/eLife.07091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia-Marcos M, Ghosh P, Farquhar MG. GIV is a nonreceptor GEF for G alpha i with a unique motif that regulates Akt signaling. Proc Natl Acad Sci USA. 2009;106:3178–3183. doi: 10.1073/pnas.0900294106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia-Marcos M, Ghosh P, Farquhar MG. GIV/Girdin transmits signals from multiple receptors by triggering trimeric G protein activation. J Biol Chem. 2015;290:6697–6704. doi: 10.1074/jbc.R114.613414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia-Marcos M, Kietrsunthorn PS, Wang H, Ghosh P, Farquhar MG. G protein binding sites on Calnuc (nucleobindin 1) and NUCB2 (nucleobindin 2) define a new class of G(alpha)i-regulatory motifs. J Biol Chem. 2011;286:28138–28149. doi: 10.1074/jbc.M110.204099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin C, et al. Structural basis for activation of trimeric Gi proteins by multiple growth factor receptors via GIV/Girdin. Mol Biol Cell. 2014;25:3654–3671. doi: 10.1091/mbc.E14-05-0978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Midde KK, et al. Multimodular biosensors reveal a novel platform for activation of G proteins by growth factor receptors. Proc Natl Acad Sci USA. 2015;112:E937–E946. doi: 10.1073/pnas.1420140112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lopez-Sanchez I, et al. GIV/Girdin is a central hub for profibrogenic signalling networks during liver fibrosis. Nat Commun. 2014;5:4451. doi: 10.1038/ncomms5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma GS, et al. Therapeutic effects of cell-permeant peptides that activate G proteins downstream of growth factors. Proc Natl Acad Sci USA. 2015;112:E2602–E2610. doi: 10.1073/pnas.1505543112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parag-Sharma K, et al. Membrane recruitment of the non-receptor protein GIV/Girdin is sufficient for activating heterotrimeric G proteins. J Biol Chem. 2016;291:27098–27111. doi: 10.1074/jbc.M116.764431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coleman BD, et al. Evolutionary conservation of a GPCR-independent mechanism of trimeric G protein activation. Mol Biol Evol. 2016;33:820–837. doi: 10.1093/molbev/msv336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weiss TS, et al. Galpha i3 binding to calnuc on Golgi membranes in living cells monitored by fluorescence resonance energy transfer of green fluorescent protein fusion proteins. Proc Natl Acad Sci USA. 2001;98:14961–14966. doi: 10.1073/pnas.261572098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin P, Fischer T, Lavoie C, Huang H, Farquhar MG. Calnuc plays a role in dynamic distribution of Galphai but not Gbeta subunits and modulates ACTH secretion in AtT-20 neuroendocrine secretory cells. Mol Neurodegener. 2009;4:15. doi: 10.1186/1750-1326-4-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leyme A, Marivin A, Perez-Gutierrez L, Nguyen LT, Garcia-Marcos M. Integrins activate trimeric G proteins via the nonreceptor protein GIV/Girdin. J Cell Biol. 2015;210:1165–1184. doi: 10.1083/jcb.201506041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghosh P. Heterotrimeric G proteins as emerging targets for network based therapy in cancer: End of a long futile campaign striking heads of a Hydra. Aging (Albany NY) 2015;7:469–474. doi: 10.18632/aging.100781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kitamura T, et al. Regulation of VEGF-mediated angiogenesis by the Akt/PKB substrate Girdin. Nat Cell Biol. 2008;10:329–337. doi: 10.1038/ncb1695. [DOI] [PubMed] [Google Scholar]
- 21.Enomoto A, et al. Roles of disrupted-in-schizophrenia 1-interacting protein girdin in postnatal development of the dentate gyrus. Neuron. 2009;63:774–787. doi: 10.1016/j.neuron.2009.08.015. [DOI] [PubMed] [Google Scholar]
- 22.Nakai T, et al. Girdin phosphorylation is crucial for synaptic plasticity and memory: A potential role in the interaction of BDNF/TrkB/Akt signaling with NMDA receptor. J Neurosci. 2014;34:14995–15008. doi: 10.1523/JNEUROSCI.2228-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oshita A, et al. Identification and characterization of a novel Dvl-binding protein that suppresses Wnt signalling pathway. Genes Cells. 2003;8:1005–1017. doi: 10.1111/j.1365-2443.2003.00692.x. [DOI] [PubMed] [Google Scholar]
- 24.Kobayashi H, et al. Novel Daple-like protein positively regulates both the Wnt/beta-catenin pathway and the Wnt/JNK pathway in Xenopus. Mech Dev. 2005;122:1138–1153. doi: 10.1016/j.mod.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 25.Ekici AB, et al. Disturbed Wnt signalling due to a mutation in CCDC88C causes an autosomal recessive non-syndromic hydrocephalus with medial diverticulum. Mol Syndromol. 2010;1:99–112. doi: 10.1159/000319859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smrcka AV. Molecular targeting of Gα and Gβγ subunits: A potential approach for cancer therapeutics. Trends Pharmacol Sci. 2013;34:290–298. doi: 10.1016/j.tips.2013.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sjögren B, Blazer LL, Neubig RR. Regulators of G protein signaling proteins as targets for drug discovery. Prog Mol Biol Transl Sci. 2010;91:81–119. doi: 10.1016/S1877-1173(10)91004-1. [DOI] [PubMed] [Google Scholar]
- 28.Kimple AJ, et al. A high throughput fluorescence polarization assay for inhibitors of the GoLoco motif/G-alpha interaction. Comb Chem High Throughput Screen. 2008;11:396–409. doi: 10.2174/138620708784534770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Milligan G, Kostenis E. Heterotrimeric G-proteins: A short history. Br J Pharmacol. 2006;147(Suppl 1):S46–S55. doi: 10.1038/sj.bjp.0706405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koch WJ, Hawes BE, Inglese J, Luttrell LM, Lefkowitz RJ. Cellular expression of the carboxyl terminus of a G protein-coupled receptor kinase attenuates G beta gamma-mediated signaling. J Biol Chem. 1994;269:6193–6197. [PubMed] [Google Scholar]
- 31.Schumacher SM, et al. A peptide of the RGS domain of GRK2 binds and inhibits Gα(q) to suppress pathological cardiac hypertrophy and dysfunction. Sci Signal. 2016;9:ra30. doi: 10.1126/scisignal.aae0549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Charpentier TH, et al. Potent and selective peptide-based inhibition of the G protein Gαq. J Biol Chem. 2016;291:25608–25616. doi: 10.1074/jbc.M116.740407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Opakua AI, et al. Molecular mechanism of Gαi activation by non-GPCR proteins with a Gα-binding and activating motif. Nat Commun. 2017;8:15163. doi: 10.1038/ncomms15163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raw AS, Coleman DE, Gilman AG, Sprang SR. Structural and biochemical characterization of the GTPgammaS-, GDP.Pi-, and GDP-bound forms of a GTPase-deficient Gly42 → Val mutant of Gialpha1. Biochemistry. 1997;36:15660–15669. doi: 10.1021/bi971912p. [DOI] [PubMed] [Google Scholar]
- 35.Berghuis AM, Lee E, Raw AS, Gilman AG, Sprang SR. Structure of the GDP-Pi complex of Gly203 → Ala gialpha1: A mimic of the ternary product complex of galpha-catalyzed GTP hydrolysis. Structure. 1996;4:1277–1290. doi: 10.1016/s0969-2126(96)00136-0. [DOI] [PubMed] [Google Scholar]
- 36.Wall MA, Posner BA, Sprang SR. Structural basis of activity and subunit recognition in G protein heterotrimers. Structure. 1998;6:1169–1183. doi: 10.1016/s0969-2126(98)00117-8. [DOI] [PubMed] [Google Scholar]
- 37.Johnston CA, et al. Minimal determinants for binding activated G alpha from the structure of a G alpha(i1)-peptide dimer. Biochemistry. 2006;45:11390–11400. doi: 10.1021/bi0613832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Waldo GL, et al. Kinetic scaffolding mediated by a phospholipase C-beta and Gq signaling complex. Science. 2010;330:974–980. doi: 10.1126/science.1193438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tesmer JJ, Berman DM, Gilman AG, Sprang SR. Structure of RGS4 bound to AlF4–activated G(i alpha1): Stabilization of the transition state for GTP hydrolysis. Cell. 1997;89:251–261. doi: 10.1016/s0092-8674(00)80204-4. [DOI] [PubMed] [Google Scholar]
- 40.Willard FS, et al. A point mutation to Galphai selectively blocks GoLoco motif binding: Direct evidence for Galpha.GoLoco complexes in mitotic spindle dynamics. J Biol Chem. 2008;283:36698–36710. doi: 10.1074/jbc.M804936200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oldham WM, Van Eps N, Preininger AM, Hubbell WL, Hamm HE. Mechanism of the receptor-catalyzed activation of heterotrimeric G proteins. Nat Struct Mol Biol. 2006;13:772–777. doi: 10.1038/nsmb1129. [DOI] [PubMed] [Google Scholar]
- 42.Rasmussen SG, et al. Crystal structure of the β2 adrenergic receptor-Gs protein complex. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Flock T, et al. Universal allosteric mechanism for Gα activation by GPCRs. Nature. 2015;524:173–179. doi: 10.1038/nature14663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thomas CJ, et al. The nucleotide exchange factor Ric-8A is a chaperone for the conformationally dynamic nucleotide-free state of Gαi1. PLoS One. 2011;6:e23197. doi: 10.1371/journal.pone.0023197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Graf R, Mattera R, Codina J, Estes MK, Birnbaumer L. A truncated recombinant alpha subunit of Gi3 with a reduced affinity for beta gamma dimers and altered guanosine 5′-3-O-(thio)triphosphate binding. J Biol Chem. 1992;267:24307–24314. [PubMed] [Google Scholar]
- 46.Denker BM, Neer EJ, Schmidt CJ. Mutagenesis of the amino terminus of the alpha subunit of the G protein Go. In vitro characterization of alpha o beta gamma interactions. J Biol Chem. 1992;267:6272–6277. [PubMed] [Google Scholar]
- 47.Journot L, Pantaloni C, Bockaert J, Audigier Y. Deletion within the amino-terminal region of Gs alpha impairs its ability to interact with beta gamma subunits and to activate adenylate cyclase. J Biol Chem. 1991;266:9009–9015. [PubMed] [Google Scholar]
- 48.Neer EJ, Pulsifer L, Wolf LG. The amino terminus of G protein alpha subunits is required for interaction with beta gamma. J Biol Chem. 1988;263:8996–9000. [PubMed] [Google Scholar]
- 49.Nanoff C, et al. The carboxyl terminus of the Galpha-subunit is the latch for triggered activation of heterotrimeric G proteins. Mol Pharmacol. 2006;69:397–405. doi: 10.1124/mol.105.016725. [DOI] [PubMed] [Google Scholar]
- 50.Thomas CJ, et al. Uncoupling conformational change from GTP hydrolysis in a heterotrimeric G protein alpha-subunit. Proc Natl Acad Sci USA. 2004;101:7560–7565. doi: 10.1073/pnas.0304091101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun D, et al. Probing Gαi1 protein activation at single-amino acid resolution. Nat Struct Mol Biol. 2015;22:686–694. doi: 10.1038/nsmb.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marivin A, et al. Dominant-negative Gα subunits are a mechanism of dysregulated heterotrimeric G protein signaling in human disease. Sci Signal. 2016;9:ra37. doi: 10.1126/scisignal.aad2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.DiGiacomo V, et al. The Gαi-GIV binding interface is a druggable protein-protein interaction. Sci Rep. 2017;7:8575. doi: 10.1038/s41598-017-08829-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Garcia-Marcos M, et al. Functional characterization of the guanine nucleotide exchange factor (GEF) motif of GIV protein reveals a threshold effect in signaling. Proc Natl Acad Sci USA. 2012;109:1961–1966. doi: 10.1073/pnas.1120538109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Herrmann R, Heck M, Henklein P, Hofmann KP, Ernst OP. Signal transfer from GPCRs to G proteins: Role of the G alpha N-terminal region in rhodopsin-transducin coupling. J Biol Chem. 2006;281:30234–30241. doi: 10.1074/jbc.M600797200. [DOI] [PubMed] [Google Scholar]
- 56.Masuho I, et al. Distinct profiles of functional discrimination among G proteins determine the actions of G protein-coupled receptors. Sci Signal. 2015;8:ra123. doi: 10.1126/scisignal.aab4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hollins B, Kuravi S, Digby GJ, Lambert NA. The c-terminus of GRK3 indicates rapid dissociation of G protein heterotrimers. Cell Signal. 2009;21:1015–1021. doi: 10.1016/j.cellsig.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koch WJ, Hawes BE, Allen LF, Lefkowitz RJ. Direct evidence that Gi-coupled receptor stimulation of mitogen-activated protein kinase is mediated by G beta gamma activation of p21ras. Proc Natl Acad Sci USA. 1994;91:12706–12710. doi: 10.1073/pnas.91.26.12706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hawes BE, van Biesen T, Koch WJ, Luttrell LM, Lefkowitz RJ. Distinct pathways of Gi- and Gq-mediated mitogen-activated protein kinase activation. J Biol Chem. 1995;270:17148–17153. doi: 10.1074/jbc.270.29.17148. [DOI] [PubMed] [Google Scholar]
- 60.Kimple RJ, et al. RGS12 and RGS14 GoLoco motifs are G alpha(i) interaction sites with guanine nucleotide dissociation inhibitor activity. J Biol Chem. 2001;276:29275–29281. doi: 10.1074/jbc.M103208200. [DOI] [PubMed] [Google Scholar]
- 61.Leyme A, Marivin A, Garcia-Marcos M. GIV/Girdin creates a positive feedback loop that potentiates outside-in integrin signaling in cancer cells. J Biol Chem. 2016;291:8269–8282. doi: 10.1074/jbc.M115.691550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lopez-Sanchez I, et al. Focal adhesions are foci for tyrosine-based signal transduction via GIV/Girdin and G proteins. Mol Biol Cell. 2015;26:4313–4324. doi: 10.1091/mbc.E15-07-0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sokol SY. Analysis of Dishevelled signalling pathways during Xenopus development. Curr Biol. 1996;6:1456–1467. doi: 10.1016/s0960-9822(96)00750-6. [DOI] [PubMed] [Google Scholar]
- 64.Wallingford JB, Fraser SE, Harland RM. Convergent extension: The molecular control of polarized cell movement during embryonic development. Dev Cell. 2002;2:695–706. doi: 10.1016/s1534-5807(02)00197-1. [DOI] [PubMed] [Google Scholar]
- 65.Tada M, Smith JC. Xwnt11 is a target of Xenopus Brachyury: Regulation of gastrulation movements via Dishevelled, but not through the canonical Wnt pathway. Development. 2000;127:2227–2238. doi: 10.1242/dev.127.10.2227. [DOI] [PubMed] [Google Scholar]
- 66.Maziarz M, Garcia-Marcos M. Fluorescence polarization assays to measure interactions between Gα subunits of heterotrimeric G proteins and regulatory motifs. Methods Cell Biol. 2017;142:133–143. doi: 10.1016/bs.mcb.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Maziarz M, Garcia-Marcos M. Rapid kinetic BRET measurements to monitor G protein activation by GPCR and non-GPCR proteins. Methods Cell Biol. 2017;142:145–157. doi: 10.1016/bs.mcb.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Le-Niculescu H, Niesman I, Fischer T, DeVries L, Farquhar MG. Identification and characterization of GIV, a novel Galpha i/s-interacting protein found on COPI, endoplasmic reticulum-Golgi transport vesicles. J Biol Chem. 2005;280:22012–22020. doi: 10.1074/jbc.M501833200. [DOI] [PubMed] [Google Scholar]
- 69.De Vries L, et al. RGS-GAIP, a GTPase-activating protein for Galphai heterotrimeric G proteins, is located on clathrin-coated vesicles. Mol Biol Cell. 1998;9:1123–1134. doi: 10.1091/mbc.9.5.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Garcia-Marcos M, Ghosh P, Ear J, Farquhar MG. A structural determinant that renders G alpha(i) sensitive to activation by GIV/girdin is required to promote cell migration. J Biol Chem. 2010;285:12765–12777. doi: 10.1074/jbc.M109.045161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thomas CJ, Tall GG, Adhikari A, Sprang SR. Ric-8A catalyzes guanine nucleotide exchange on G alphai1 bound to the GPR/GoLoco exchange inhibitor AGS3. J Biol Chem. 2008;283:23150–23160. doi: 10.1074/jbc.M802422200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fischer T, De Vries L, Meerloo T, Farquhar MG. Promotion of G alpha i3 subunit down-regulation by GIPN, a putative E3 ubiquitin ligase that interacts with RGS-GAIP. Proc Natl Acad Sci USA. 2003;100:8270–8275. doi: 10.1073/pnas.1432965100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lin P, Yao Y, Hofmeister R, Tsien RY, Farquhar MG. Overexpression of CALNUC (nucleobindin) increases agonist and thapsigargin releasable Ca2+ storage in the Golgi. J Cell Biol. 1999;145:279–289. doi: 10.1083/jcb.145.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stols L, et al. A new vector for high-throughput, ligation-independent cloning encoding a tobacco etch virus protease cleavage site. Protein Expr Purif. 2002;25:8–15. doi: 10.1006/prep.2001.1603. [DOI] [PubMed] [Google Scholar]
- 75.Cabrita LD, Dai W, Bottomley SP. A family of E. coli expression vectors for laboratory scale and high throughput soluble protein production. BMC Biotechnol. 2006;6:12. doi: 10.1186/1472-6750-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Park TJ, Gray RS, Sato A, Habas R, Wallingford JB. Subcellular localization and signaling properties of dishevelled in developing vertebrate embryos. Curr Biol. 2005;15:1039–1044. doi: 10.1016/j.cub.2005.04.062. [DOI] [PubMed] [Google Scholar]
- 77.National Research Council . Guide for the Care and Use of Laboratory Animals. 8th Ed National Academies Press; Washington, DC: 2011. [Google Scholar]
- 78.Hubrecht-Laboratorium (Embryologisch Instituut) Nieuwkoop PD, Faber J. Normal Table of Xenopus laevis (Daudin); A Systematical and Chronological Survey of the Development from the Fertilized Egg Till the End of Metamorphosis. North-Holland; Amsterdam: 1956. [Google Scholar]
- 79.Habas R, Kato Y, He X. Wnt/Frizzled activation of Rho regulates vertebrate gastrulation and requires a novel Formin homology protein Daam1. Cell. 2001;107:843–854. doi: 10.1016/s0092-8674(01)00614-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.