Significance
Cytochrome oxidase is a widespread respiratory enzyme that conserves energy released when oxygen is reduced by pumping protons across the membrane in which it is located. Here, we use atomistic simulations of the whole bovine enzyme to investigate properties of the H channel, a structure that has been proposed to provide the pathway for pumped protons in mammalian forms of the enzyme. These studies show that although parts of the structure could function in this manner, a gap persists. This gap could be bridged only if a buried histidine becomes protonated. Based on these simulations, we propose that the H channel acts as a dielectric well, modulating effects of buried charge changes.
Keywords: cell respiration, electron transfer, proton pumping, dielectric well, protein hydration
Abstract
Proton pumping A-type cytochrome c oxidase (CcO) terminates the respiratory chains of mitochondria and many bacteria. Three possible proton transfer pathways (D, K, and H channels) have been identified based on structural, functional, and mutational data. Whereas the D channel provides the route for all pumped protons in bacterial A-type CcOs, studies of bovine mitochondrial CcO have led to suggestions that its H channel instead provides this route. Here, we have studied H-channel function by performing atomistic molecular dynamics simulations on the entire, as well as core, structure of bovine CcO in a lipid-solvent environment. The majority of residues in the H channel do not undergo large conformational fluctuations. Its upper and middle regions have adequate hydration and H-bonding residues to form potential proton-conducting channels, and Asp51 exhibits conformational fluctuations that have been observed crystallographically. In contrast, throughout the simulations, we do not observe transient water networks that could support proton transfer from the N phase toward heme a via neutral His413, regardless of a labile H bond between Ser382 and the hydroxyethylfarnesyl group of heme a. In fact, the region around His413 only became sufficiently hydrated when His413 was fixed in its protonated imidazolium state, but its calculated pKa is too low for this to provide the means to create a proton transfer pathway. Our simulations show that the electric dipole moment of residues around heme a changes with the redox state, hence suggesting that the H channel could play a more general role as a dielectric well.
Cytochrome c oxidase (CcO) is a respiratory energy-transducing enzyme. It catalyzes electron transfer from cytochrome c to molecular oxygen, conserving the released energy as a charge and proton gradient across the membrane in which it is located (1). Mammalian mitochondrial CcOs are composed of at least 13 different polypeptides (2). They are members of a diverse “superfamily” of homologous proteins (3–5), falling within the “A1” subgroup, which includes many bacterial CcOs and quinol oxidases. All A1-type CcOs share a remarkably similar catalytic core formed by three subunits (I, II, and III), which house the redox-active metal centers CuA; heme a; and a binuclear center (BNC) composed of heme a3, CuB, and a catalytic tyrosine. Structures of several A1-type bacterial CcOs (6, 7) and of one mitochondrial form (bovine) (8) have been solved at atomic resolution (Fig. 1). The electron transfer pathway and many details of the oxygen reduction chemistry have been resolved (9–11). However, although the basic principles of redox-coupled proton pumping are well established, the specific atomic details remain in dispute, particularly the roles of three possible proton transfer pathways (D, K, and H channels) that have been identified in the A-type CcOs based on crystal structure data (9–11).
Fig. 1.
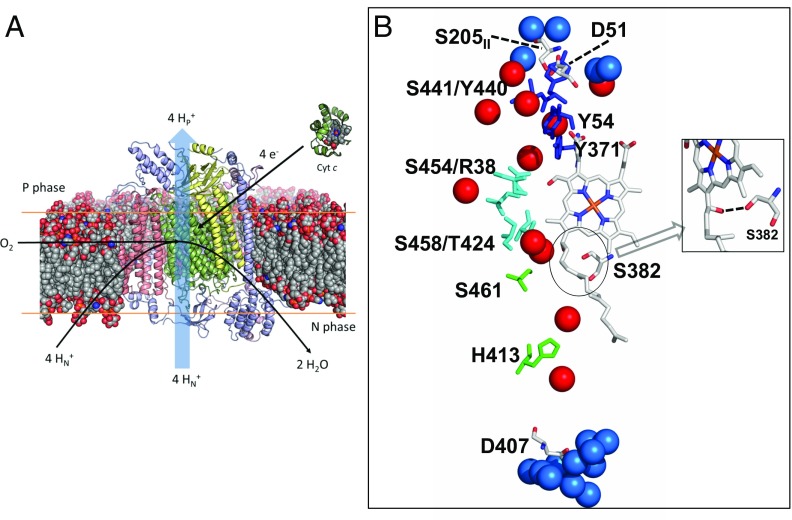
Structure of bovine CcO and key features of the H channel. (A) Thirteen-subunit bovine CcO is shown in its approximate position embedded in a lipid bilayer. Subunits I, II, and III are shown in green, yellow, and pink, respectively, with additional subunits shown in blue. (B) Details of the H channel. In the main text, residues that are components of the putative H channel are divided into top (Y54, Y371, Y440, and S441; shown in blue), middle (R38, T424, S454, and S458; shown in cyan), and lower (H413 and S461; shown in green). Other residues discussed in the main text are D51 at the P-phase interface, its H-bonded partner S205 of subunit II, D407 at the N phase, and S382 of helix X (details are provided in main text). (Inset) Reoriented view to illustrate the H bond between S382 and the hydroxyethylfarnesyl chain of heme a. Water molecules within the H channel are shown as red spheres. Additional waters within 5 Å of D51 or S205II at the P phase, or within 5 Å of D407 at the N phase, are shown as blue spheres. Protein, heme cofactor, and water molecules were drawn using coordinates of oxidized bovine CcO (PDB ID code 1V54) (31). All amino acid numbering corresponds to the sequences of CcO from Bos taurus, unless otherwise stated.
In bacterial CcOs, structural data, together with a wide range of biophysical and spectroscopic studies, strongly support a model for proton/electron coupling that involves only the D and K channels in internal proton transfers (9, 12, 13). The K channel delivers the first two substrate protons into the BNC. The D channel provides part of the route for the remaining two substrate protons and all four translocated protons. Experimental support for these proton-conducting functions has come particularly from studies of mutant forms of bacterial CcOs (14). However, some significant issues remain. For example, in all CcO structures determined by X-ray crystallography, there is no connection for proton transfer from Glu242, the residue at the end of the D pathway, either to the BNC or to the likely exit route of the translocated protons. There is also the question of how the D channel could act as a conduit for both substrate and translocated protons. Unless gated precisely, the protons would always be driven into the BNC for exergonic water production, hence “short-circuiting” the coupling process, rather than being driven across the membrane against a protonmotive force. One possible solution to these points has come from classical molecular dynamics (MD) simulations suggesting that water molecules dynamically reorganize at different stages of the catalytic process to form a transient H-bonded link either to a proton trap above the hemes or to the BNC (15–18). Recent multiscale reactive MD simulations performed on the Rhodobacter sphaeroides CcO structure have provided free-energy profiles and predicted rates of internal proton transfer from the top of the D channel to both the proton trap and the BNC for wild-type (19) and decoupling mutants (20). Interestingly, internal proton transfer rates calculated from these simulations are found to be in good agreement with the kinetic data from electrometric experiments (21). Direct experimental evidence for functional water molecule reorganization has come from FTIR studies of bovine and bacterial CcOs (22), although their location(s) within the channels remain(s) unclear.
In contrast, the third hydrophilic H channel has been suggested to conduct translocated protons in bovine mitochondrial CcO, based upon structural and functional studies (8, 10). This H channel (Fig. 1B) is separate from the BNC and the D and K channels. The lower part consists of a domain leading from mitochondrial matrix toward the hydroxyethylfarnesyl and formyl groups of heme a. It is proposed to open and become hydrated after the BNC has become reduced, creating a proton pathway for entry of four protons into a proton-collecting array close to a bound Mg2+ ion “above” the BNC (23). Subsequent binding of oxygen closes this channel, and one proton is proposed to be released into the P phase with each reduction/oxidation of heme a via an amide bond gate between Tyr440 and Ser441 (24) and an H-bonded network to Asp51 at the P-phase surface (10). Support has come from structural perturbations induced by redox/ligand state changes in the water channel (25–28), the Asp51 residue (29), and the proposed proton-collecting site around the bound Mg2+ (23). These observations, together with effects of H-channel mutations on coupling efficiencies in a chimeric bovine/human CcO construct (30, 31), have led to the proposal that these structures provide the route for translocated protons in mammalian mitochondrial CcOs. This contrasts with bacterial CcOs (32), where the H channel is less complete, particularly lacking Asp51 and the YS residues, whose amide linkage is proposed to form a gate. Furthermore, mutations introduced into the bacterial H channel are without effect (32), although some similar redox-induced crystallographic structural changes are observed in a serine residue that influences the opening of the bovine water channel (Ser382 in bovine enzyme) (33–35). It also contrasts with yeast (Saccharomyces cerevisiae) mitochondrial CcO: This has a clear H channel (36) (although also lacking Asp51 and the proposed YS gate), but mutations within it do not affect proton translocation, whereas D-channel mutations induce effects similar to those induced in bacterial CcOs (37, 38). An alternative role of the H channel has been suggested in bacterial and yeast CcOs as a “dielectric channel or well,” providing groups that can cooperatively reorient their dipoles/charges in response to redox changes, and hence modulate function (13).
Direct experimental testing of the functions of the H channel in mammalian forms of CcO is challenging, particularly because of the difficulties in introducing suitable mutations into the mitochondrially encoded subunit I and in direct kinetic observation of specific water and proton movements. Considerable advances have been made in MD simulations of large protein structures embedded in aqueous media and/or membrane bilayers, including large coupled electron transfer complexes (39, 40). Such simulations, in conjunction with quantum mechanical (QM) calculations of metal center structures (41), have been successfully applied to regions of CcOs to predict oxygen reduction mechanism (42), water structure and protonic gating of the D channel in the vicinity of Glu242 (15, 18, 19, 43, 44), and behavior of the K channel (45, 46). Here, we apply these methods to predict the dynamic properties of the H channel in an atomic model of membrane-embedded bovine CcO (Models and Methods and Tables 1–3) with a focus on its possible role as a proton channel and/or a dielectric well.
Table 1.
C1 model systems and their simulation lengths
| Model system | CuA | Heme a | BNC | His413 | Asp51 | Glu242 | Tyr244 | Lys319 | Asp364 | No. of replica and simulation lengths, ns |
| C1-I | OX | OX | OX* | D (0) | P (0) | P (0) | D (−1) | D (0) | P (0) | 1 × 300 |
| 3 × 40† | ||||||||||
| C1-II | OX | OX | OX* | D (0) | P (0) | D (−1) | D (−1) | D (0) | P (0) | 200 |
| C1-III | OX | OX | OX* | D (0) | P (0) | P (0) | D (−1) | P (+1) | P (0) | 200 |
| C1-IV | OX | OX | OX‡ | D (0) | P (0) | P (0) | P (0) | D (0) | P (0) | 200 |
| C1-V | OX | OX | OX§ | D (0) | P (0) | P (0) | D (−1) | D (0) | P (0) | 200 |
| C1-VI | OX | OX | OX* | P (+1) | P (0) | P (0) | D (−1) | D (0) | P (0) | 150 |
Protonation state changes of key amino acid residues (D, deprotonated, P, protonated; net charge in parentheses) compared with the original C1-I simulation are highlighted in bold. The protonation pattern within the BNC in C1-IV and C1-V was varied from that in C1-I as defined in the footnotes. OX, oxidized.
BNC structure of Fe[III]-OH−....H2O-Cu[II] TyrO−.
With hydrated H-channel snapshots taken from C1-VI.
BNC structure of Fe[III]-OH−....H2O-Cu[II] TyrOH.
BNC structure of Fe[III]-H2O....H2O-Cu[II] TyrO−.
Table 3.
C3 model systems and their simulation lengths
| Model system | CuA | Heme a | BNC* | His413 | Asp51 | Glu242 | Tyr244 | Lys319 | Asp364 | No. of replica and simulation lengths, ns |
| C3-I (FR)† | RED | RED | RED | D (0) | D (−1) | P (0) | P (0) | P (+1) | P (0) | 1 × 996 |
| 1 × 952 | ||||||||||
| C3-II (FR)‡ | RED | RED | RED | D (0) | D (−1) | P (0) | P (0) | P (+1) | P (0) | 5 × 30 |
| C3-III (FR)§ | RED | RED | RED | D (0) | D (−1) | P (0) | P (0) | P (+1) | P (0) | 5 × 15 |
| C3-IV (FR)¶ | RED | RED | RED | D (0) | D (−1) | P (0) | P (0) | P (+1) | P (0) | 5 × 15 |
RED, reduced.
Reduced BNC structure of Fe[II]……..Cu[I] TyrOH.
BNC reduced with CuA/heme a reduced.
Simulations started with a fully hydrated lower part of the H channel.
Simulations started with a H3O+ modeled below His413.
Simulations started with a H3O+ modeled above His413.
Results
H-Channel Dynamics and Hydration.
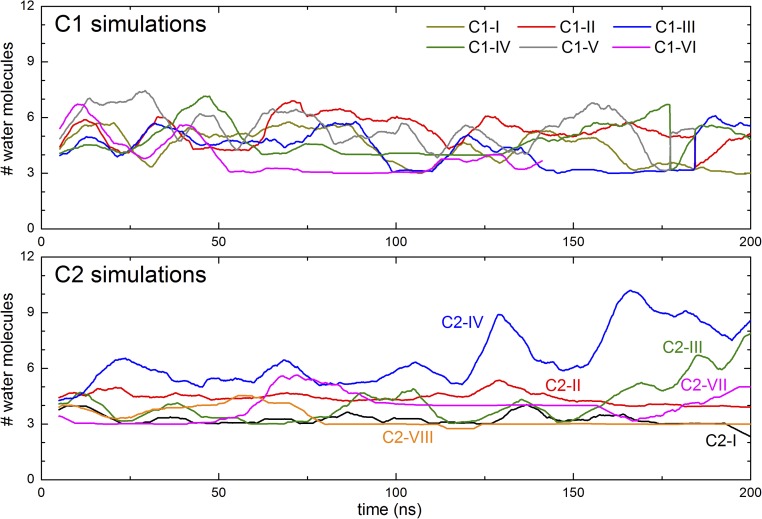
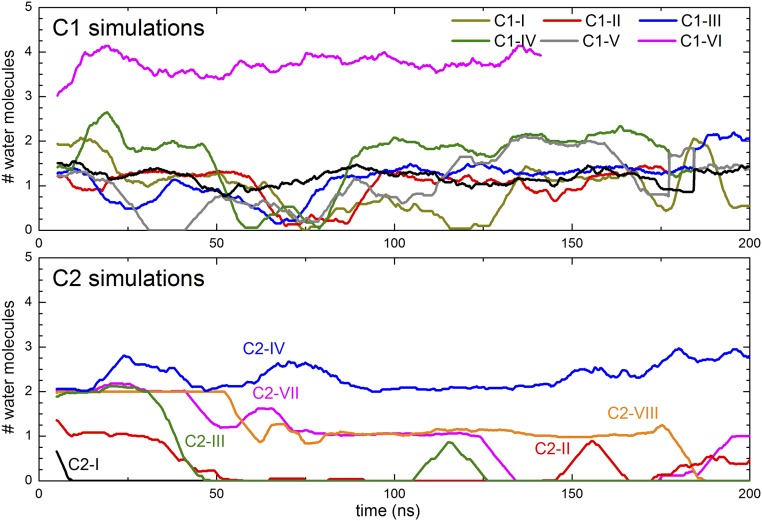
We first analyzed the dynamics and number of water molecules in the H-channel region. Average numbers of water molecules were calculated over entire simulation trajectories by counting water oxygens within 5 Å of key H-channel residues (subunit I residues His413, Ser461, Thr424, Ser454, Ser458, Arg38, Tyr440, Tyr441, Tyr371, and Tyr54; Fig. 1). These results show that almost all water molecules within the entire H-channel region exchange with the solvent during the simulation time scales but that, despite that fact, their number does not change significantly throughout the simulations (Table 4). For all C1 simulations, the average number of water molecules ranges from 20 to 22, with the sole exception of C1-VI (in which His413 was modeled in its cationic form; Table 1). For the majority of C2 simulations, the number of water molecules in the H channel is found to fluctuate between 20 and 32, with the exception of system C2-IV, in which His413 was again modeled in its cationic form. To get further insights into these dynamics, we performed additional analyses by dividing the H channel into three regions: an upper region between the top of heme a and Asp51 that includes residues 54, 371, 440, and 441; a central region adjacent to heme a that includes residues 38, 424, 454, and 458; and a lower region that includes residues 461 and 413, which spans Asp407 in the N phase and the central region (Fig. 1B).
Table 4.
Average number of water molecules in the H channel from C1 and C2 systems
| Simulation system | Average no. of water molecules | Simulation system | Average no. of water molecules |
| C1-I | 22 ± 4 | C2-I (replica 1/2) | 32 ± 6/27 ± 4 |
| C1-II | 20 ± 2 | C2-II (replica 1/2) | 20 ± 3/19 ± 3 |
| C1-III | 20 ± 2 | C2-III | 27 ± 5 |
| C1-IV | 22 ± 2 | C2-IV | 42 ± 9 |
| C1-V | 20 ± 2 | C2-V | 32 ± 6 |
| C1-VI | 25 ± 3 | C2-VI | 22 ± 3 |
| C2-VII (replica 1/2) | 31 ± 7/33 ± 4 | ||
| C2-VIII (replica 1/2) | 28 ± 5/22 ± 4 |
Upper Region of the H Channel.
We first considered the upper region of the H channel containing polar amino acid residues Tyr54, Tyr371, Tyr440, and Ser441. Structural data show that this region is relatively hydrophilic and contains the majority of the crystallographically defined H-channel water molecules [nine in fully oxidized and 11 in fully reduced (FR) bovine CcO crystal structures (Protein Data Bank [PDB] ID codes 1V54 and 1V55, respectively) (31)]. Our simulations reveal additional hydration in this region (SI Appendix, Tables S1 and S4). Crystallographic (29) and FTIR (47, 48) data indicate that Asp51 is relatively buried and protonated in fully oxidized enzyme, with its side chain forming hydrogen bonds with Ser441 and Ser205 of subunits I and II, respectively (10). Accordingly, since all C1 simulations were performed with fully oxidized enzyme, all had Asp51 protonated. The hydrogen bond with the side chain of Ser205 was found to be stable throughout these simulations (Table 5), in agreement with the structural and FTIR data, except for C1-I, where the H bond was found to be broken, leading to a larger average Asp51-Ser205 distance and recruitment of several additional waters into this region (SI Appendix, Table S1).
Table 5.
Average distance between D51 and the S205 (subunit II)
| Simulation system | Distance, Å | Simulation system | Distance, Å |
| C1-I | 4.6 ± 1.2 | C2-I (replica 1/2) | 9.6 ± 3.2/6.0 ± 1.8 |
| C1-II | 3.5 ± 0.2 | C2-II (replica 1/2) | 4.4 ± 1.1/5.3 ± 1.7 |
| C1-III | 3.6 ± 0.2 | C2-III | 5.2 ± 1.6 |
| C1-IV | 3.6 ± 0.2 | C2-IV | 9.1 ± 3.5 |
| C1-V | 3.5 ± 0.2 | C2-V | 4.9 ± 1.3 |
| C1-VI | 3.5 ± 0.1 | C2-VI | 6.1 ± 1.6 |
| C2-VII (replica 1/2) | 7.4 ± 1.8/9.1 ± 2.3 | ||
| C2-VIII (replica 1/2) | 6.6 ± 2.0/3.9 ± 0.8 |
Distance is calculated between the Cγ atom of Asp51 and Oγ atom of Ser205.
FTIR data indicate that reduction of heme a and/or CuA leads to Asp51 deprotonation (47, 49). This is supported by the structure of the FR enzyme, which has been modeled with deprotonated Asp51 that loses its bonding to Ser205 and becomes more hydrated (10). For the majority of C2 simulations, performed on the experimentally characterized P-type catalytic states and various redox states of heme a/CuA, we modeled Asp51 in its deprotonated state (except for C2-VI; Table 2). In these cases, it was observed that the H bond between Ser205 and deprotonated Asp51 is broken (Table 5) and Asp51 orients toward the solvent. This change in orientation of Asp51 is correlated with a large influx of water into the upper H-channel region (Table 4), as also observed in the C1-I simulation described above, in which the Asp51-Ser205 bond was again broken. Overall, the simulations are consistent with structural data showing that protonated Asp51 is relatively buried and stabilized by H bonding with Ser205 and that deprotonation leads to bond loss and hydration. This behavior of Asp51 and the high hydration of this region are in accord with the earlier suggestion (10) that there is a dynamic H-bond interconnectivity in the region that could facilitate proton transfer. However, the reorientation of Asp51 and its associated hydration increase do not induce any major conformational changes in the middle or lower parts of the H channel between heme a and the N phase, where viable proton conduction pathways are less evident (discussed below).
Table 2.
C2 model systems and their simulation lengths
| Model system | CuA | Heme a | BNC* | His413 | Asp51 | Glu242 | Tyr244 | Lys319 | Asp364 | No. of replica and simulation lengths, ns |
| C2-I | RED | OX | PM | D (0) | D (−1) | P (0) | D (0) | P (+1) | P (0) | 2 × 200 |
| C2-II | OX | RED | PM | D (0) | D (−1) | P (0) | D (0) | P (+1) | P (0) | 2 × 200 |
| C2-III | OX | OX | PR | D (0) | D (−1) | P (0) | D (−1) | P (+1) | P (0) | 1 × 200 |
| C2-IV | OX | RED | PM | P (+1) | D (−1) | P (0) | D (0) | P (+1) | P (0) | 1 × 200 |
| C2-V | RED | OX | PM | P (+1) | D (−1) | P (0) | D (0) | P (+1) | P (0) | 1 × 100 |
| C2-VI | RED | OX | PM | D (0) | P (0) | P (0) | D (0) | P (+1) | P (0) | 1 × 100 |
| C2-VII (MV)† | OX | OX | RED | D (0) | D (−1) | P (0) | P (0) | P (+1) | P (0) | 2 × 200 |
| C2-VIII (FR)‡ | RED | RED | RED | D (0) | D (−1) | P (0) | P (0) | P (+1) | P (0) | 2 × 200 |
Protonation state changes of key amino acid residues (D, deprotonated; P, protonated; net charge in parentheses) compared with C2-I/II/III simulations are highlighted in bold. Configurations of the BNC in different states are defined in the footnotes. MV, mixed valence; OX, oxidized; RED, reduced.
BNC had chemical structures of: PM, Fe[IV] = O2−....HO−-Cu[II] TyrO•; PR, Fe[IV] = O2−....HO−-Cu[II] TyrO−; RED, Fe[II]……..Cu[I] TyrOH.
BNC reduced with CuA/heme a oxidized.
BNC reduced with CuA/heme a reduced.
It should be noted that the higher water occupancy in some C2 simulations (Table 4) results primarily from influx of water molecules into the hydrophilic upper region of the H channel in response to the loss of hydrogen bonding between deprotonated Asp51 and Ser205. The data in SI Appendix, Tables S1–S6 using C1 and C2 systems further illustrate that the additional hydration associated with loss of the Asp51-Ser205 hydrogen bonding is primarily confined to the upper region, whereas variations in hydration in the middle and lower parts of H channel are much smaller.
Middle Region of the H Channel.
The middle section of the H channel, adjacent to heme a, potentially links Ser461 to the upper H-bonded and hydrated network. It contains polar amino acid residues and four buried water molecules in both oxidized (PDB ID code 1V54) and reduced (PDB ID code 1V55) crystal structures (Fig. 1). In all simulations, residues Arg38, Thr424, Ser454, and Ser458 remain stable in their crystallographic positions (SI Appendix, Fig. S1). The hydration level persists during the simulations and, due to the rigid structure of this region, the average water occupancy does not change much during the simulations in any of the states studied (SI Appendix, Tables S2 and S5). The water molecules do exchange with solvent waters, and their number does fluctuate in some states (SI Appendix, Tables S2 and S5; also Fig. 2), although the crystallographically defined water sites remain fully occupied (Fig. 3). Analysis of simulation trajectories reveals that the bound water molecules are expelled by waters that transiently appear in the vicinity during the simulations and that the exchanges occur on a nanosecond time scale. Overall, the simulation data indicate that sufficient waters could appear in this region to create connectivity from S461 up to the upper H-bonded network. However, the only protonatable residue is Arg38, which is unlikely to deprotonate easily, and most or all of any proton pathway would have to be provided by waters in that region.
Fig. 2.
Time dependencies of numbers of water molecules adjacent to heme a. Only water molecules within 4 Å of the N and O atoms of the side chains of residues 461, 382, 458, 424, 38, 454, 371, 451, 54, and 428 are included. The plot shown is a running average of 100 simulation snapshots taken at 100-ps time intervals.
Fig. 3.
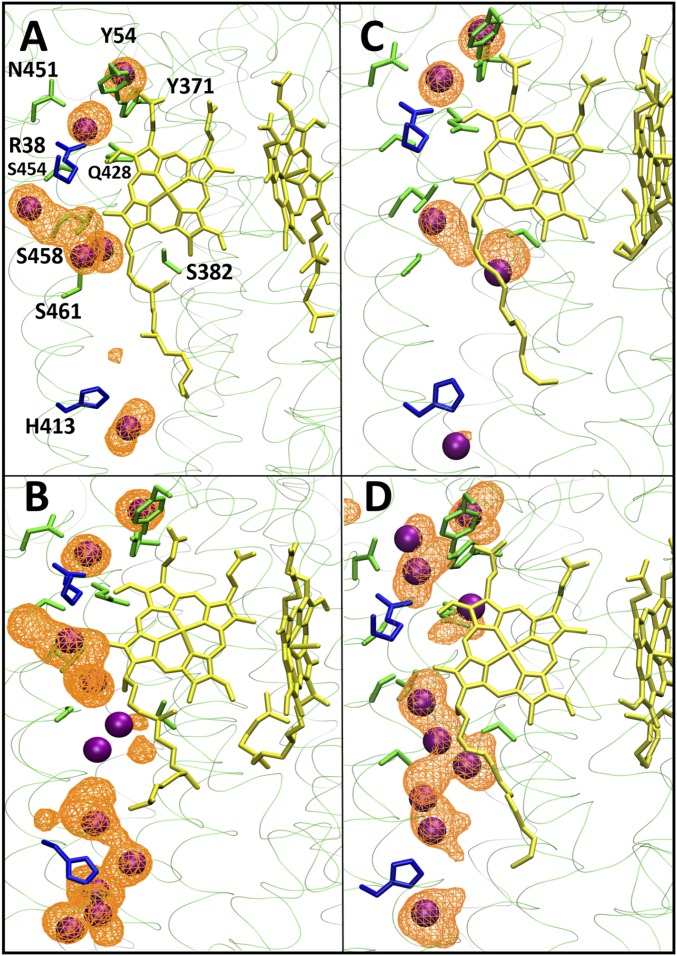
Occupancy of water molecules in the neighborhood of His413 and heme a (yellow) from simulations C1-I (A), C1-VI (B), C2-II (C), and C2-IV (D). Occupancy is displayed as an isosurface with an isovalue of 0.08 (A and B) or 0.10 (C and D). Instantaneous positions of water molecules are shown as purple spheres, polar amino acid residues are shown in green, and basic residues are shown in blue. Hydrogens are not shown for clarity.
Lower Region of the H Channel.
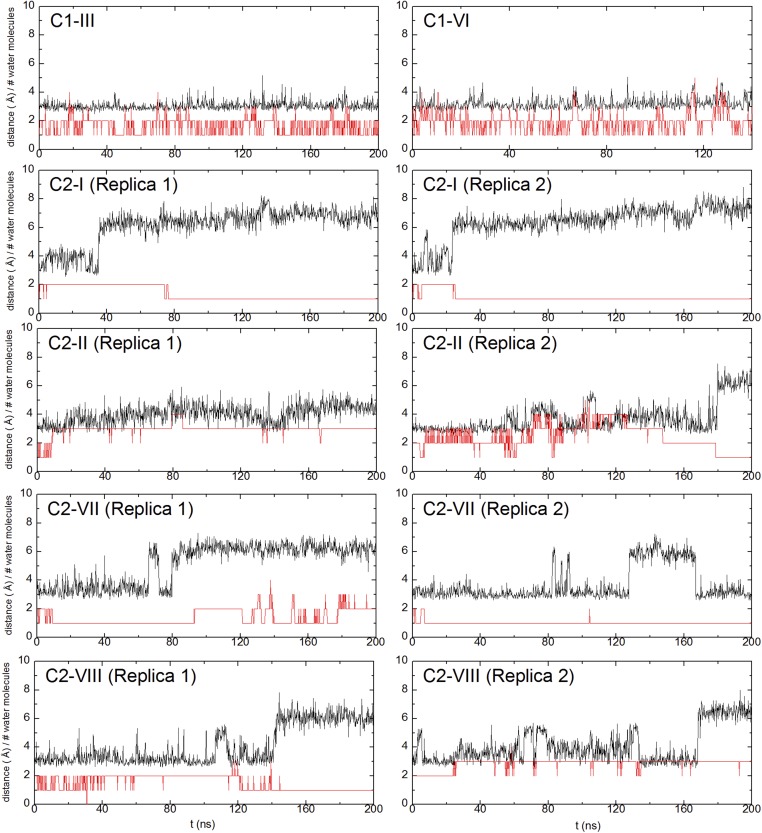
The lower section links Asp407 in the N phase to the middle region via residues His413 and Ser461 (Fig. 1). His413, a partly conserved residue for which the H channel is named, is located in a predominantly hydrophobic pocket with two crystallographically defined water molecules above and below (Fig. 1 and SI Appendix, Fig. S2). However, in the crystal structure of fully oxidized enzyme, there is a connectivity gap from there to Ser461, Ser458, and the two water molecules above them. The side chain of nearby Ser382 (Fig. 1) hydrogen-bonds to the −OH of the hydroxyethylfarnesyl chain of heme a. In the FR structure, however, this H bond is broken and Ser382 moves such that a cavity is formed that has been proposed to allow water to enter, and hence to allow proton transfer from the N phase via His413 into the middle region (23, 26). Indeed, in C1 simulations carried out with the oxidized form of CcO, the hydrogen bond between the −OH groups of Ser382 and the hydroxyethylfarnesyl chain of heme a persists, as do the two closest waters that are above Ser461/Ser458 (Fig. 4). For C2 simulations, this hydrogen bond tends to dissociate in both P-type catalytic intermediate states or with the BNC reduced (Fig. 4). Hence, although this dissociation is consistent with the structure of the FR state, the data show that dissociation of the H bond is not specifically correlated with any significant additional hydration of the cavity between His413 and Ser461. In all simulations that failed to increase hydration in this region, the common feature instead was that all had His413 in its neutral state; in all of these, the low hydration persists and the occupancy of the two water sites above and below His413 does not fluctuate much (snapshots in Fig. 3 A and C), including in those cases where the BNC metals are reduced (C2-VII and C2-VIII). To further test this behavior, two ∼1-μs MD simulations were performed in the FR state with the C3 system (Table 3). Again, hydration remained minimal in the regions immediately above and below His413 in these long-time-scale simulations, with only 1.6 ± 1.1 and 1.3 ± 0.8 water molecules within 5 Å of His413 and Ser461 in two simulation replicas. These conclusions are also supported by data in Fig. 5 and SI Appendix, Tables S3 and S6, which show that this low water occupancy persists throughout the simulation times in all redox states studied for both C1 and C2 systems, provided that the His413 is neutral. This suggests that there is a permanent barrier to proton transfer in this region, regardless of the Ser382 conformation, in contrast to proposals based on structural and FTIR data. These findings agree with observations in R. sphaeroides CcO (33), where the same type of structural change of the equivalent serine has been observed. In this enzyme, the H channel is not operative, and recent kinetic data on wild-type and a Ser425Ala mutant show no major differences in their reaction kinetics or coupling efficiencies (50).
Fig. 4.
Time dependencies of Ser382 conformation and occupancy of water molecules. Ser382 conformation is represented by the H bond between the side-chain O atom of Ser382 and the O atom of the hydroxyethylfarnesyl group of heme a (black trace; also Fig. 1B). The numbers of water molecules within 6 Å of the latter are also plotted (red trace).
Fig. 5.
Time dependencies of occupancy of water molecules in the neighborhood of His413. The plot shown is a running average of 100 simulation snapshots taken at 100-ps time intervals. Only water molecules whose oxygen atoms are within 4 Å of the nitrogen atoms of His413 side chain are considered.
A low pKa of His413 is in accord with a location that is surrounded by hydrophobic residues (SI Appendix, Fig. S2). However, if the BNC is oxidized, heme a does display a weak pH dependency of its redox midpoint potential (Em) over a wide pH range (51, 52), consistent with weak redox Bohr effects on one or more groups. It was shown that this is linked to a small proton uptake from the inner (51) or both (53) sides of the membrane upon reduction of heme a. Given its location, one of these groups might conceivably be His413, which, if it became protonated, might influence the surrounding region and its hydration level. To explore this further, we first calculated the pKa of His413 by performing continuum electrostatic calculations on simulation snapshots in different redox states of the enzyme. We observed that the reduction of heme a causes an increase in the pKa of His413 of about 1.5 pKa units, but with values still too low (pKa < 5) to result in significant protonation. This viewpoint is also supported by two independent computational approaches (Models and Methods).
Nevertheless, we simulated the effect of protonating His413 (total charge of +1) on the occupancy and dynamics of water molecules in the lower part of the H channel (Tables 1 and 2). In both C1 and C2 simulations, forced protonation of His413 does indeed induce more water molecules to appear in its vicinity, providing connectivity between the N side of the membrane and His413, and between His413 and Ser461 (Figs. 3 and 5 and SI Appendix, Tables S3 and S6), in sharp contrast to the case of neutral His413, in which no such water wires form (Fig. 3, compare A and C with B and D). However, recalculation of its pKa using snapshots from simulations with protonated His413 again indicated predicted pKas (<5) too low to support any significant His413 protonation with a concomitant increase in hydration. Hence, these data tend to rule out the opening of the channel by this effect.
As an additional test of the stability of hydration in the lower region of the H channel, eight short MD simulations (three of setup C1-I and five of C3-II) were performed starting with a fully hydrated H channel and a neutral His413. In all of these, water occupancy in the lower part of the H channel rapidly declined within the first 5–20 ns (SI Appendix, Figs. S3 and S4), strongly supporting the notion that hydration above and below neutral His413 is not stable. Based on C1-I simulations, we also reconstructed the free-energy profile associated with wetting of the lower part of the H channel (SI Appendix, Fig. S5) using the dynamic weighted histogram analysis (54). These results further confirm that when neutral, His413 destabilizes hydration in the lower H channel by at least 2–3 kcal/mol.
Given the importance of the charge and position of a proton in hydration and in proton translocation through this pathway, we did separate classical and quantum mechanics/molecular mechanics (QM/MM) MD simulations in which a proton was treated explicitly (Models and Methods). First, data from multiple independent classical simulations show that an H3O+ ion is unstable in the H channel, particularly in the lower segment between His413 and Asp407. In all five independent simulations, the H3O+ ion exited the region within first 15 ns of simulation time to the bulk aqueous phase at the N side of the membrane. A similar scenario was observed when QM/MM MD simulations were performed by simulating an explicit proton on water molecules connecting His413 to Asp407 (SI Appendix, Fig. S2). Only when the proton was modeled on the water H-bonded to His413 did it form doubly protonated His413, whereas in various other cases, the proton rapidly diffused toward Asp407 (already during initial energy minimization; also Movie S1). Interestingly, in a relatively longer 15-ps QM/MM MD simulation of protonated Asp407, the proton never escaped the latter residue. In fact, the water chain connecting protonated Asp407 to His413 disassembled (Movie S2) in agreement with classical simulations.
Overall, the data from multiscale simulations suggest that proton uptake from the N side of the membrane via His413 is strongly unfavorable because of kinetic (absence of water wire) as well as thermodynamic (low pKa of His413) reasons.
Redox-Coupled Changes.
Based on the simulation data presented above, the lower half of the H channel is unlikely to provide a pathway for coupled proton translocation. However, it may be expected that redox change of heme a is accompanied by some structural or hydration rearrangements in its environment induced by the net charge change (13). Comparisons of simulation data of states in which heme a is oxidized or reduced do not show any major differences in the dynamics of residues or hydration in the region next to heme a. In addition, as shown above, a redox Bohr protonation of His413 appears to be very unlikely. To explore more subtle possible redox-coupled changes that may occur in this region, we calculated the electric dipole moment of the side chains of selected amino acid residues close to heme a (residues Tyr54, Tyr371, Gln428, Arg38, Ser454, Ser458, Thr424, Ser382, and Ser461). The data from multiple independent simulations in different redox states show that reduction of heme a (compare C2-I vs. C2-II and C2-VII vs. C2-VIII) is coupled to reduction in the magnitude of the electric dipole moment of nearby residues (Table 6 and SI Appendix, Fig. S6), consistent with the net charge change in the iron/heme system from +1 (oxidized) to 0 (reduced). Although polarization effects, such as charge fluctuations, are not accounted for in the simulations reported here, both the C1 and C2 systems give roughly the same magnitude for the oxidized state, and the subtle difference in the electric dipole moment in different redox states is consistently observed in the C2 simulations. Such changes are to be expected when a net charge change occurs on a buried group and are consistent with a general function of part or all of the H-channel structure as a “dielectric well,” (13) where small concerted changes in polarizable groups occur to counteract the buried charge change.
Table 6.
Average electric dipole moment of side chains of amino acid residues next to heme a
| Simulation system | Average electric dipole moment, D |
| C1 simulations | From 20.1 ± 2.1–21.6 ± 1.9 |
| C2-I (replica 1/2) | 21.5 ± 1.8/21.8 ± 1.7 |
| C2-II (replica 1/2) | 16.8 ± 2.2/18.3 ± 2.0 |
| C2-VII (replica 1/2) | 20.6 ± 2.5/21.2 ± 1.4 |
| C2-VIII (replica 1/2) | 18.6 ± 2.2/17.4 ± 2.5 |
Average is made over the entire simulation trajectory and comprises residues 54, 371, 428, 38, 454, 458, 424, 382, and 461 of subunit I.
Discussion
We have performed fully atomistic classical MD simulations on the entire structure of CcO and on various protonation and redox configurations on a smaller three-subunit model, with a focus on the possible role of the H channel in redox-driven proton pumping. Data from multiple independent simulations on both systems show consistent behavior in the main elements of the H channel, much of which is consistent with crystallographic data. Overall, there are no large-scale conformational changes, apart from those involving Asp51 and Ser382, that have been described in structural work. Here, for clarity of discussion, we have divided the H channel into three functional regions.
The upper region connects the top of heme a with Asp51 at the P-phase border and includes residues Tyr54, Tyr371, Tyr440, and Ser441. Crystallographic data show that this region is hydrated, and simulations indicate additional hydration, especially if Asp51 becomes deprotonated, a state in which it tends to move out of its relatively buried location (29). FTIR data (47, 49) have suggested that deprotonation is linked to heme a/CuA reduction, although this is surprising since it is the opposite of a classical redox-linked Bohr effect (55). It is also inconsistent with the −20- to −30-mV pH dependency of heme a redox potential when the BNC is oxidized (51, 52) and with even weaker pH dependency when the BNC is reduced and CO-ligated (56). This anomaly is yet to be explained. Nevertheless, both the structural data and the simulations performed here suggest that ample hydrated pathways are available in this part of the structure to form possible proton relay pathways. Hence, it seems feasible that this region could indeed form a potentially gated exit pathway to the P phase for protons accumulated in the proton trap region.
The middle region of the H channel adjacent to heme a includes residues Ser458, Arg38, Ser454, Gln428, Tyr371, and Tyr54. As shown here, these residues stay more or less stable throughout the simulations. The few waters observed in crystal structures remain, and their number even increases somewhat in the simulations. Hence, although it is a relatively rigid region and with Arg38 as the only protonatable residue (but with a high pKa), sufficient H bonding between residues and waters could be envisaged to provide a proton-conducting pathway.
However, proton connectivity between this region and the N phase at Asp407 is more problematic. This lower region runs from Ser461 to Asp407 via His413. Only two water molecules are observed in the crystal structures immediately above and below His413, which are insufficient to form a proton-conducting pathway. In oxidized CcO, Ser382 makes an H bond to the −OH group of the hydroxyethylfarnesyl chain of heme a. However, in FR CcO, this bond breaks and Ser382 moves into helix X (10). This has been proposed to form a cavity that hydrates this region sufficiently to enable proton transfer. In the simulations, Ser382 does undergo a conformational change that is in agreement with the structural data. However, its dissociation from the hydroxyethylfarnesyl group does not induce the postulated bridging of the gap between His413 and Ser461 with additional waters. Indeed, the same type of structural change of the equivalent serine has been observed in R. sphaeroides CcO, where an H channel is not operative (33), and in which recent kinetic data on wild-type and a Ser425Ala mutant show no major differences in their reaction kinetics or coupling efficiencies (50). It is not conserved in the structurally and biochemically well-characterized B-type oxidase from Thermus thermophilus, the quinol oxidase from Escherichia coli, and various C-type oxidases (57). Hence, its conformational change may be more associated with properties or stability of the heme a rather than having a channel-opening function.
Our simulations show that this lack of water-based connectivity in the region around His413 persists as long as the latter is neutral, which is its calculated protonation state. Only when simulations are performed with protonated His413 does rapid hydration and transient water wire formation occur in the lower part of the H channel. However, the pKa of this His is likely to be too low to be able to form its imidazolium state, even when heme a becomes reduced. First, its environment is rather nonpolar (SI Appendix, Fig. S2). Second, continuum electrostatic calculations indicate a very low pKa, even when heme a is reduced. Third, the weak Em pH dependency of heme a (51, 52) shows that no single residue has a strongly linked redox Bohr protonation. Therefore, since the neutral protonation state is very likely the physiologically relevant state, we conclude that the absence of sufficient water density in the lower part of the channel remains the major concern in models in which the structure functions in conducting pumped protons across the structure. It might also be noted that His413 is not well conserved in eukaryotic CcOs, being absent, for example, from yeast and many insect and worm species, highlighting the notion that if the H channel is indeed operative as a proton channel, this function is unlikely to be widespread in eukaryotes. Indeed, direct tests with mutants of S. cerevisiae CcO have shown that it does not function in this way in this enzyme (11, 37, 38).
Overall, the data presented here are unable to support the viewpoint that the H channel is a proton-pumping route in bovine CcO unless the unlikely protonation of His413 can be demonstrated. An alternative (or perhaps in bovine CcO, an additional) role for the H channel in all A-type CcOs as a dielectric well of the channel was suggested (13). In such a role, the function of the more polar residues is to adjust their dipoles in response to the change of charge on the buried heme when it changes redox state, since charge neutralization is not achieved with a redox Bohr protonation. Some evidence that this is occurring to some extent comes here from the calculated overall electric dipole moment of the residues surrounding heme a. Their polarity increases on oxidation of heme a, consistent with the net +1 charge of this state. Whether the polarity of these groups could also be altered by allosteric effects of supernumerary subunits or by ligand binding, hence providing a means for external factors to modulate internal kinetic properties, remains to be tested experimentally.
Models and Methods
Model System Construction.
Three different model systems (C1, C2, and C3) were constructed from the oxidized bovine CcO crystal structure (PDB ID code 1V54) (31) to perform classical MD simulations. In model system C1, the entire 13-subunit monomeric CcO was immersed in a lipid bilayer formed by 50% phosphatidylcholine (12:0/12:0), 30% phosphatidylethanolamine (12:0/12:0), and 20% phosphatidylinositol (14:0/14:0), which was then solvated with transferable intermolecular potential 3 (TIP3) water molecules, and with K+ and Cl− ions to account for 100 mM ionic strength. The entire membrane-protein-solvent system was constructed using CHARMM-GUI tools (58, 59) and consisted of ∼390,000 atoms. A second smaller model system (C2) comprising ∼280,000 atoms was also constructed to study different intermediates of the catalytic cycle of CcO and to test the consistency of results in comparison to the large model system, C1. C2 consisted of the three core catalytic subunits (I, II, and III) immersed in a hybrid lipid bilayer formed by dianionic cardiolipin (18:2/18:2/18:2/18:2), phosphatidylcholine (18:2/18:2), and phosphatidylethanolamine (18:2/18:2) in a ratio of 1:3.38:3.05, which mimics the composition of the inner mitochondrial membrane (also ref. 17). Water (TIP3) was added as a solvent, and 100 mM ionic strength was established with Na+ and Cl− ions. The third model system (C3, comprising ca. 130,000 atoms) was also constructed to study long-time-scale behavior of hydration in the lower part of the H channel. For this, a two-subunit enzyme was immersed in a hybrid lipid bilayer (same as C2), and two independent simulations of up to 1 μs each, as well as multiple short simulations of 15–30 ns, were performed in the FR state of the enzyme (Table 3).
Fully atomistic classical MD simulations were performed in different redox/protonation states, as described in Tables 1 (C1), 2 (C2), and 3 (C3). All internal water molecules present in the PDB ID code 1V54 atomic structure were retained. Standard protonation states at pH 7 were used for all titratable residues except those listed in Tables 1–3. All simulations were performed using NAMD (60) software, together with the CHARMM force field for protein, lipids, water, and ions (61, 62). Parameters for a classical H3O+ ion were taken from an earlier study of Sagnella and Voth (63). The parameters for metal centers were obtained from an earlier study by Johansson et al. (64). MD simulations were carried out at physiological temperature and pressure using 1- or 2-fs time steps for systems C2 and C1/C3, respectively. The particle mesh Ewald method (65), as implemented in NAMD, was used to treat long-range electrostatics. The simulation trajectory data were saved every 10 or 100 ps, and Visual Molecular Dynamics software was used for trajectory visualization and analyses (66).
The C1 system in state I (oxidized; Table 1) was equilibrated for 100 ns, and the resulting structure was used to initiate C1 II–VI simulations. In these further states, the protonation states of functionally important residues and/or the protonation pattern within the BNC was altered. The total simulation time for all C1 systems was ca. 1.3 μs. In the smaller C2 model systems, both redox [oxidized (OX); reduced (RED)] states of metals and protonation states of key amino acids were varied, with particular focus on the influence on the H-channel structure of electron transfer from CuA to heme a (C2-II versus C2-I) and from heme a to a BNC intermediate (C2-II versus C2-III), and of reduction of heme a/CuA (C2-VII versus C2-VIII). The total simulation time of all C2 systems was ca. 2.2 μs, and for all C3 systems, it was ∼2.3 μs.
Continuum electrostatic calculations were also performed on snapshots obtained from MD simulations of system C2 to estimate pKas. Up to 100 simulation snapshots were chosen to calculate the pKas in different redox/protonation states of the enzyme. The MEAD program (67) was used to calculate intrinsic pKa and site–site interaction energies, and KARLSBERG software (68) was used to estimate the final pKa of the residue by a full-titration calculation of all titratable sites in a three-subunit enzyme using reference pKa values for Arg, Asp, Glu, His, Lys, and Tyr, as well as propionate groups of 12.0, 4.5, 4.6, 6.2, 10.4, 9.7, and 4.8, respectively. To further validate the obtained pKa values, we also performed pKa estimation using Propka (69, 70) and Alchemical (71) free-energy simulations, as implemented in NAMD, using a small model system containing only subunit I and the pKa of H78 of lysozyme as a reference value. Alchemical simulations consisted of two independent trajectories with 21 windows each for both the protein and the reference (overall, 20 ns). Regardless of the method used, we consistently obtained low pKa values for His413 (pKa < 5).
Six QM/MM MD simulations were performed using the Turbomole and Amber packages (72–74). The QM region consisted of His413, Asp407, and water molecules around these residues (Movies S1 and S2). Link atoms (hydrogens) were introduced between Cα and Cβ atoms of His413 as well as Asp407. An extra proton was modeled on His413, on Asp407, or next to the water molecules in the QM region (ca. 8.5, 6.2, 2.9, and 2.3 Å from Asp407), followed by a 1,000-step conjugate gradient geometry optimization and MD at 310 K (Langevin thermostat) for 5–15 ps using a 1-fs time step. The Tao, Perdew, Staroverov, Scuseria (TPSS) density functional (75) and def2-SVP split valence polarization basis set (76), along with resolution of identity approximation (77) and density function theory DFT-D3 dispersion correction (78), were used.
System Stability.
Analysis of simulation trajectories revealed that C1 large model systems stabilized with an rmsd plateaued in the range of 2–3 Å (SI Appendix, Fig. S7). For C2 systems, the rmsd stabilized at 3–4 Å (SI Appendix, Fig. S8). These data suggest that simulations had converged sufficiently to be able to observe any rapid dynamics of water molecules in the H channel.
Simulations on model system C1 showed no large-scale conformational changes in the residues involved in the H channel induced by variations in the possible ground state of the oxidized BNC or by changes in the protonation states of the selected amino acid residues. When final resulting structures of C1 I–VI simulations were compared for the positions of key H-channel residues (Arg38, Asp51, Tyr54, Tyr371, His413, Thr424, Tyr440, Tyr441, Ser454, Ser458, and Ser461), the rmsd of all nonhydrogen atoms was smaller than 0.95 Å, and their positions were remarkably similar to those in the crystal structure. A similar scenario was observed when final snapshots of C2 simulations were compared. The rmsd for all C2 simulations was ≤0.9 Å, except for system C2-II (rmsd of 1.02 Å). Despite the differences in system sizes and lipid compositions of model systems C1 and C2, the structures obtained from simulations are remarkably similar (SI Appendix, Fig. S1).
Supplementary Material
Acknowledgments
V.S. thanks Prof. Mårten Wikström for many helpful discussions. V.S. received research funding from the Academy of Finland and Magnus Ehrnrooth Foundation. P.G.J. received funding from the Spanish Ministry of Economy and Competitiveness (Grant MINECO/FEDER-CTQ2015-65033-P) and the Juan de la Cierva Fellowship (IJCI-2014-20615). P.R.R. was funded by the Biotechnology and Biological Sciences Research Council (Grants BB/K001094/1 and BB/L020165/1). E.R. was supported by Engineering and Physical Sciences Research Council (EPSRC) Grant EP/N020669/1 and Biotechnology and Biological Sciences Research Council (BBSRC) Grant BB/N007700/1. The computational resources for this project were generously provided by the Center for Scientific Computing, Finland, and, in part, by the Tampere Center for Scientific Computing, Finland. We acknowledge computer time on the Advanced Research Computing High End Resource (ARCHER) via the UK High-End Computing Consortium for Biomolecular Simulation, supported by the EPSRC (Grant EP/L000253/1) and the computational resources of the NIH Biowulf cluster. P.G.J. received computing time allocation from Centro de computación científica at the Universidad Autónoma de Madrid.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. P.B. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1708628114/-/DCSupplemental.
References
- 1.Wikström MKF. Proton pump coupled to cytochrome c oxidase in mitochondria. Nature. 1977;266:271–273. doi: 10.1038/266271a0. [DOI] [PubMed] [Google Scholar]
- 2.Kadenbach B, Hüttemann M. The subunit composition and function of mammalian cytochrome c oxidase. Mitochondrion. 2015;24:64–76. doi: 10.1016/j.mito.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 3.García-Horsman JA, Barquera B, Rumbley J, Ma J, Gennis RB. The superfamily of heme-copper respiratory oxidases. J Bacteriol. 1994;176:5587–5600. doi: 10.1128/jb.176.18.5587-5600.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sousa FL, et al. The superfamily of heme-copper oxygen reductases: Types and evolutionary considerations. Biochim Biophys Acta. 2012;1817:629–637. doi: 10.1016/j.bbabio.2011.09.020. [DOI] [PubMed] [Google Scholar]
- 5.Hemp J, Gennis RB. In: Results and Problems in Cell Differentiation. Schäfer G, Penefsky HS, editors. Springer; Berlin: 2008. pp. 1–31. [PubMed] [Google Scholar]
- 6.Iwata S, Ostermeier C, Ludwig B, Michel H. Structure at 2.8 A resolution of cytochrome c oxidase from Paracoccus denitrificans. Nature. 1995;376:660–669. doi: 10.1038/376660a0. [DOI] [PubMed] [Google Scholar]
- 7.Svensson-Ek M, et al. The X-ray crystal structures of wild-type and EQ(I-286) mutant cytochrome c oxidases from Rhodobacter sphaeroides. J Mol Biol. 2002;321:329–339. doi: 10.1016/s0022-2836(02)00619-8. [DOI] [PubMed] [Google Scholar]
- 8.Tsukihara T, et al. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A. Science. 1996;272:1136–1144. doi: 10.1126/science.272.5265.1136. [DOI] [PubMed] [Google Scholar]
- 9.Wikström M, Sharma V, Kaila VRI, Hosler JP, Hummer G. New perspectives on proton pumping in cellular respiration. Chem Rev. 2015;115:2196–2221. doi: 10.1021/cr500448t. [DOI] [PubMed] [Google Scholar]
- 10.Yoshikawa S, Shimada A. Reaction mechanism of cytochrome c oxidase. Chem Rev. 2015;115:1936–1989. doi: 10.1021/cr500266a. [DOI] [PubMed] [Google Scholar]
- 11.Rich PR. Mitochondrial cytochrome c oxidase: Catalysis, coupling and controversies. Biochem Soc Trans. 2017;45:813–829. doi: 10.1042/BST20160139. [DOI] [PubMed] [Google Scholar]
- 12.Brzezinski P, Gennis RB. Cytochrome c oxidase: Exciting progress and remaining mysteries. J Bioenerg Biomembr. 2008;40:521–531. doi: 10.1007/s10863-008-9181-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rich PR, Maréchal A. Functions of the hydrophilic channels in protonmotive cytochrome c oxidase. J R Soc Interface. 2013;10:20130183. doi: 10.1098/rsif.2013.0183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fetter JR, et al. Possible proton relay pathways in cytochrome c oxidase. Proc Natl Acad Sci USA. 1995;92:1604–1608. doi: 10.1073/pnas.92.5.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim YC, Wikström M, Hummer G. Kinetic gating of the proton pump in cytochrome c oxidase. Proc Natl Acad Sci USA. 2009;106:13707–13712. doi: 10.1073/pnas.0903938106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wikström M, et al. Gating of proton and water transfer in the respiratory enzyme cytochrome c oxidase. Proc Natl Acad Sci USA. 2005;102:10478–10481. doi: 10.1073/pnas.0502873102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharma V, Enkavi G, Vattulainen I, Róg T, Wikström M. Proton-coupled electron transfer and the role of water molecules in proton pumping by cytochrome c oxidase. Proc Natl Acad Sci USA. 2015;112:2040–2045. doi: 10.1073/pnas.1409543112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goyal P, Yang S, Cui Q. Microscopic basis for kinetic gating in cytochrome c oxidase: Insights from QM/MM analysis. Chem Sci (Camb) 2015;6:826–841. doi: 10.1039/c4sc01674b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liang R, Swanson JMJ, Peng Y, Wikström M, Voth GA. Multiscale simulations reveal key features of the proton-pumping mechanism in cytochrome c oxidase. Proc Natl Acad Sci USA. 2016;113:7420–7425. doi: 10.1073/pnas.1601982113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang R, Swanson JMJ, Wikström M, Voth GA. Understanding the essential proton-pumping kinetic gates and decoupling mutations in cytochrome c oxidase. Proc Natl Acad Sci USA. 2017;114:5924–5929. doi: 10.1073/pnas.1703654114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Belevich I, Verkhovsky MI, Wikström M. Proton-coupled electron transfer drives the proton pump of cytochrome c oxidase. Nature. 2006;440:829–832. doi: 10.1038/nature04619. [DOI] [PubMed] [Google Scholar]
- 22.Maréchal A, Rich PR. Water molecule reorganization in cytochrome c oxidase revealed by FTIR spectroscopy. Proc Natl Acad Sci USA. 2011;108:8634–8638. doi: 10.1073/pnas.1019419108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yano N, et al. The Mg2+-containing water cluster of mammalian cytochrome c oxidase collects four pumping proton equivalents in each catalytic cycle. J Biol Chem. 2016;291:23882–23894. doi: 10.1074/jbc.M115.711770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamiya K, Boero M, Tateno M, Shiraishi K, Oshiyama A. Possible mechanism of proton transfer through peptide groups in the H-pathway of the bovine cytochrome c oxidase. J Am Chem Soc. 2007;129:9663–9673. doi: 10.1021/ja070464y. [DOI] [PubMed] [Google Scholar]
- 25.Yoshikawa S, Muramoto K, Shinzawa-Itoh K, Mochizuki M. Structural studies on bovine heart cytochrome c oxidase. Biochim Biophys Acta. 2012;1817:579–589. doi: 10.1016/j.bbabio.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 26.Kubo M, et al. Effective pumping proton collection facilitated by a copper site (CuB) of bovine heart cytochrome c oxidase, revealed by a newly developed time-resolved infrared system. J Biol Chem. 2013;288:30259–30269. doi: 10.1074/jbc.M113.473983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shimada A, et al. A nanosecond time-resolved XFEL analysis of structural changes associated with CO release from cytochrome c oxidase. Sci Adv. 2017;3:e1603042. doi: 10.1126/sciadv.1603042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishigami I, et al. Crystal structure of CO-bound cytochrome c oxidase determined by serial femtosecond X-ray crystallography at room temperature. Proc Natl Acad Sci USA. 2017;114:8011–8016. doi: 10.1073/pnas.1705628114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoshikawa S, et al. Redox-coupled crystal structural changes in bovine heart cytochrome c oxidase. Science. 1998;280:1723–1729. doi: 10.1126/science.280.5370.1723. [DOI] [PubMed] [Google Scholar]
- 30.Shimokata K, et al. The proton pumping pathway of bovine heart cytochrome c oxidase. Proc Natl Acad Sci USA. 2007;104:4200–4205. doi: 10.1073/pnas.0611627104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsukihara T, et al. The low-spin heme of cytochrome c oxidase as the driving element of the proton-pumping process. Proc Natl Acad Sci USA. 2003;100:15304–15309. doi: 10.1073/pnas.2635097100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee H-M, et al. Mutations in the putative H-channel in the cytochrome c oxidase from Rhodobacter sphaeroides show that this channel is not important for proton conduction but reveal modulation of the properties of heme a. Biochemistry. 2000;39:2989–2996. doi: 10.1021/bi9924821. [DOI] [PubMed] [Google Scholar]
- 33.Qin L, et al. Redox-dependent conformational changes in cytochrome C oxidase suggest a gating mechanism for proton uptake. Biochemistry. 2009;48:5121–5130. doi: 10.1021/bi9001387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koepke J, et al. High resolution crystal structure of Paracoccus denitrificans cytochrome c oxidase: New insights into the active site and the proton transfer pathways. Biochim Biophys Acta. 2009;1787:635–645. doi: 10.1016/j.bbabio.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 35.Liu J, Hiser C, Ferguson-Miller S. Role of conformational change and K-path ligands in controlling cytochrome c oxidase activity. Biochem Soc Trans. 2017;45:1087–1095. doi: 10.1042/BST20160138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maréchal A, Meunier B, Lee D, Orengo C, Rich PR. Yeast cytochrome c oxidase: A model system to study mitochondrial forms of the haem-copper oxidase superfamily. Biochim Biophys Acta. 2012;1817:620–628. doi: 10.1016/j.bbabio.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dodia RJ. 2014. Structure-function relationship of mitochondrial cytochrome c oxidase: Redox centres, proton pathways and isozymes. PhD thesis (University College London, London)
- 38.Maréchal A, Haraux F, Meunier B, Rich PR. Determination of H+/e ratios in mitochondrial yeast cytochrome c oxidase. Biochim Biophys Acta Bioenerg. 2014;1837:e100. [Google Scholar]
- 39.Postila PA, et al. Atomistic determinants of co-enzyme Q reduction at the Qi-site of the cytochrome bc1 complex. Sci Rep. 2016;6:33607. doi: 10.1038/srep33607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharma V, et al. Redox-induced activation of the proton pump in the respiratory complex I. Proc Natl Acad Sci USA. 2015;112:11571–11576. doi: 10.1073/pnas.1503761112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sharma V, Karlin KD, Wikström M. Computational study of the activated O(H) state in the catalytic mechanism of cytochrome c oxidase. Proc Natl Acad Sci USA. 2013;110:16844–16849. doi: 10.1073/pnas.1220379110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blomberg MRA, Siegbahn PEM. The mechanism for proton pumping in cytochrome c oxidase from an electrostatic and quantum chemical perspective. Biochim Biophys Acta. 2012;1817:495–505. doi: 10.1016/j.bbabio.2011.09.014. [DOI] [PubMed] [Google Scholar]
- 43.Woelke AL, et al. Exploring the possible role of Glu286 in CcO by electrostatic energy computations combined with molecular dynamics. J Phys Chem B. 2013;117:12432–12441. doi: 10.1021/jp407250d. [DOI] [PubMed] [Google Scholar]
- 44.Kaila VRI, Verkhovsky MI, Hummer G, Wikström M. Glutamic acid 242 is a valve in the proton pump of cytochrome c oxidase. Proc Natl Acad Sci USA. 2008;105:6255–6259. doi: 10.1073/pnas.0800770105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sharma V, Wikström M. The role of the K-channel and the active-site tyrosine in the catalytic mechanism of cytochrome c oxidase. Biochim Biophys Acta. 2016;1857:1111–1115. doi: 10.1016/j.bbabio.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 46.Woelke AL, Galstyan G, Knapp E-W. Lysine 362 in cytochrome c oxidase regulates opening of the K-channel via changes in pKA and conformation. Biochim Biophys Acta. 2014;1837:1998–2003. doi: 10.1016/j.bbabio.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 47.Okuno D, Iwase T, Shinzawa-Itoh K, Yoshikawa S, Kitagawa T. FTIR detection of protonation/deprotonation of key carboxyl side chains caused by redox change of the Cu(A)-heme a moiety and ligand dissociation from the heme a3-Cu(B) center of bovine heart cytochrome c oxidase. J Am Chem Soc. 2003;125:7209–7218. doi: 10.1021/ja021302z. [DOI] [PubMed] [Google Scholar]
- 48.Maréchal A, Meunier B, Rich PR. Assignment of the CO-sensitive carboxyl group in mitochondrial forms of cytochrome c oxidase using yeast mutants. Biochim Biophys Acta. 2012;1817:1921–1924. doi: 10.1016/j.bbabio.2012.03.036. [DOI] [PubMed] [Google Scholar]
- 49.Dodia R, Maréchal A, Bettini S, Iwaki M, Rich PR. IR signatures of the metal centres of bovine cytochrome c oxidase: Assignments and redox-linkage. Biochem Soc Trans. 2013;41:1242–1248. doi: 10.1042/BST20130087. [DOI] [PubMed] [Google Scholar]
- 50.Vilhjálmsdóttir J, Johansson A-L, Brzezinski P. Structural changes and proton transfer in cytochrome c oxidase. Sci Rep. 2015;5:12047. doi: 10.1038/srep12047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Artzatbanov VY, Konstantinov AA, Skulachev VP. Involvement of intramitochondrial protons in redox reactions of cytochrome alpha. FEBS Lett. 1978;87:180–185. doi: 10.1016/0014-5793(78)80327-5. [DOI] [PubMed] [Google Scholar]
- 52.Moody AJ, Rich PR. The effect of pH on redox titrations of haem a in cyanide-liganded cytochrome-c oxidase: Experimental and modelling studies. Biochim Biophys Acta. 1990;1015:205–215. doi: 10.1016/0005-2728(90)90022-v. [DOI] [PubMed] [Google Scholar]
- 53.Mitchell R. 1991. The nature and significance of the pH dependence of electron equilibration in the cytochrome c–cytochrome c oxidase system. PhD thesis (King’s College London, London)
- 54.Rosta E, Hummer G. Free energies from dynamic weighted histogram analysis using unbiased Markov state model. J Chem Theory Comput. 2015;11:276–285. doi: 10.1021/ct500719p. [DOI] [PubMed] [Google Scholar]
- 55.Papa S, Guerrieri F, Izzo G. Redox Bohr-effects in the cytochrome system of mitochondria. FEBS Lett. 1979;105:213–216. doi: 10.1016/0014-5793(79)80614-6. [DOI] [PubMed] [Google Scholar]
- 56.Ellis WR, Jr, Wang H, Blair DF, Gray HB, Chan SI. Spectroelectrochemical study of the cytochrome a site in carbon monoxide inhibited cytochrome c oxidase. Biochemistry. 1986;25:161–167. doi: 10.1021/bi00349a023. [DOI] [PubMed] [Google Scholar]
- 57.Sharma V, Puustinen A, Wikström M, Laakkonen L. Sequence analysis of the cbb3 oxidases and an atomic model for the Rhodobacter sphaeroides enzyme. Biochemistry. 2006;45:5754–5765. doi: 10.1021/bi060169a. [DOI] [PubMed] [Google Scholar]
- 58.Jo S, Kim T, Iyer VG, Im W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J Comput Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 59.Lee J, et al. CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J Chem Theory Comput. 2016;12:405–413. doi: 10.1021/acs.jctc.5b00935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Phillips JC, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.MacKerell AD, Jr, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 62.Klauda JB, et al. Update of the CHARMM all-atom additive force field for lipids: Validation on six lipid types. J Phys Chem B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sagnella DE, Voth GA. Structure and dynamics of hydronium in the ion channel gramicidin A. Biophys J. 1996;70:2043–2051. doi: 10.1016/S0006-3495(96)79773-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Johansson MP, Kaila VR, Laakkonen L. Charge parameterization of the metal centers in cytochrome c oxidase. J Comput Chem. 2008;29:753–767. doi: 10.1002/jcc.20835. [DOI] [PubMed] [Google Scholar]
- 65.Darden T, York D, Pederson L. Particle mesh Ewald: An N.log(N) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 66.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 67.Bashford D, Gerwert K. Electrostatic calculations of the pKa values of ionizable groups in bacteriorhodopsin. J Mol Biol. 1992;224:473–486. doi: 10.1016/0022-2836(92)91009-e. [DOI] [PubMed] [Google Scholar]
- 68.Rabenstein B, Knapp EW. Calculated pH-dependent population of CO-myoglobin conformers. Biophys J. 2001;80:1141–1150. doi: 10.1016/S0006-3495(01)76091-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Søndergaard CR, Olsson MHM, Rostkowski M, Jensen JH. Improved treatment of ligands and coupling effects in empirical calculation and rationalization of pKa values. J Chem Theory Comput. 2011;7:2284–2295. doi: 10.1021/ct200133y. [DOI] [PubMed] [Google Scholar]
- 70.Olsson MHM, Søndergaard CR, Rostkowski M, Jensen JH. PROPKA3: Consistent treatment of internal and surface residues in empirical pKa predictions. J Chem Theory Comput. 2011;7:525–537. doi: 10.1021/ct100578z. [DOI] [PubMed] [Google Scholar]
- 71.Dixit SB, Chipot C. Can absolute free energies of association be estimated from molecular mechanical simulations? The biotin-streptavidin system revisited. J Phys Chem A. 2001;105:9795–9799. [Google Scholar]
- 72.Ahlrichs R, Bär M, Häser M, Horn H, Kölmel C. Electronic structure calculations on workstation computers: The program system Turbomole. Chem Phys Lett. 1989;162:165–169. [Google Scholar]
- 73.Case DA, et al. 2017. AMBER 2017 (University of California, San Francisco)
- 74. TURBOMOLE (2016) TURBOMOLE: Program Package for ab initio Electronic Structure Calculations (TURBOMOLE GmbH, Karlsruhe, Germany), Version 7.1.
- 75.Tao J, Perdew JP, Staroverov VN, Scuseria GE. Climbing the density functional ladder: Nonempirical meta-generalized gradient approximation designed for molecules and solids. Phys Rev Lett. 2003;91:146401. doi: 10.1103/PhysRevLett.91.146401. [DOI] [PubMed] [Google Scholar]
- 76.Weigend F, Ahlrichs R. Balanced basis sets of split valence, triple zeta valence and quadruple zeta valence quality for H to Rn: Design and assessment of accuracy. Phys Chem Chem Phys. 2005;7:3297–3305. doi: 10.1039/b508541a. [DOI] [PubMed] [Google Scholar]
- 77.Eichkorn K, Treutler O, Öhm H, Häser M, Ahlrichs R. Auxiliary basis sets to approximate Coulomb potentials. Chem Phys Lett. 1995;242:652–660. [Google Scholar]
- 78.Grimme S, Ehrlich S, Goerigk L. Effect of the damping function in dispersion corrected density functional theory. J Comput Chem. 2011;32:1456–1465. doi: 10.1002/jcc.21759. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.