Significance
In dividing cells, long-lived proteins are continuously diluted by being partitioned into newly formed daughter cells. Conversely, short-lived proteins are cleared from a cell primarily by proteolysis rather than cell division. Thus, when a cell stops dividing, there is a natural tendency for long-lived proteins to accumulate relative to short-lived proteins. This effect is disruptive to cells and leads to the accumulation of aged and damaged proteins over time. Here, we analyzed the degradation of thousands of proteins in dividing and nondividing (quiescent) skin cells. Our results demonstrate that quiescent cells avoid the accumulation of long-lived proteins by enhancing their degradation through pathways involving the lysosome. This mechanism may be important for promotion of protein homeostasis in aged organisms.
Keywords: protein degradation, quiescence, protein homeostasis, lysosome, quantitative proteomics
Abstract
In dividing cells, cytoplasmic dilution is the dominant route of clearance for long-lived proteins whose inherent degradation is slower than the cellular growth rate. Thus, as cells transition from a dividing to a nondividing state, there is a propensity for long-lived proteins to become stabilized relative to short-lived proteins, leading to alterations in the abundance distribution of the proteome. However, it is not known if cells mount a compensatory response to counter this potentially deleterious proteostatic disruption. We used a proteomic approach to demonstrate that fibroblasts selectively increase degradation rates of long-lived proteins as they transition from a proliferating to a quiescent state. The selective degradation of long-lived proteins occurs by the concurrent activation of lysosomal biogenesis and up-regulation of macroautophagy. Through this mechanism, quiescent cells avoid the accumulation of aged long-lived proteins that would otherwise result from the absence of cytoplasmic dilution by cell division.
In dividing cells, proteins can be cleared by two general mechanisms: dilution by cytokinesis and proteolytic degradation (1, 2). Long-lived proteins with half-lives longer than the cellular growth rate are primarily removed by cell division. Conversely, short-lived proteins with half-lives shorter than the cellular growth rate are predominantly cleared by the cell’s degradation machinery (3, 4) (Fig. 1A). This dichotomy can potentially have a significant effect on the abundance distribution of the proteome as cells alternate between dividing and nondividing states. In the absence of compensatory mechanisms, cell division arrest would be expected to preferentially stabilize long-lived proteins and increase their expression levels relative to short-lived proteins. Such an effect has been observed in transformed cell lines where growth-arresting anticancer drugs lead to the selective accumulation of long-lived proteins (1). Indeed, it has been argued that cell death resulting from the proteomic imbalance induced by growth-arresting drugs provides a potential therapeutic strategy against rapidly growing tumors (1).
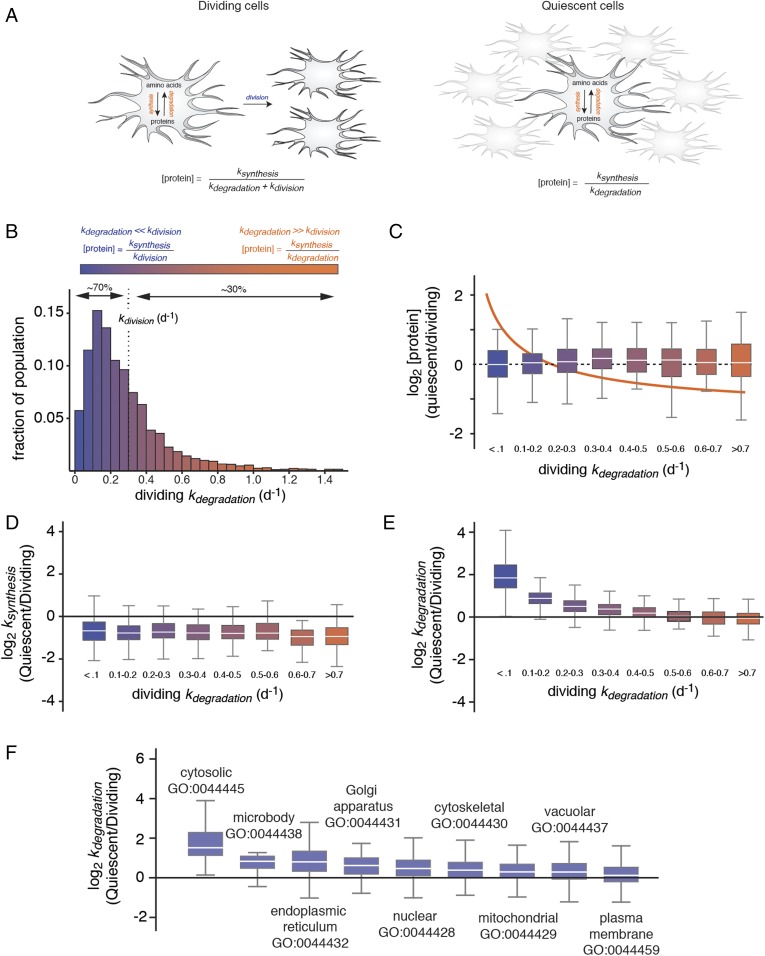
Fig. 1.
Selective degradation of long-lived proteins mitigates their accumulation in quiescent cells. (A) Schematic outlines the relationship between [protein], ksynthesis, kdegradation, and kdivision in dividing and quiescent cells. (B) The measured distribution of kdegradation in dividing cells indicating the percentage of the proteome with kdegradation values greater or less than kdivision. These two classes of proteins are referred to as “short-lived” and “long-lived,” and their clearance is dominated by degradation and cell division, respectively. (C) Relationship between relative protein expression levels in quiescent and dividing cells to kdegradation in dividing cells. Box plots indicate the distribution of log2 [protein] ratios within different ranges of kdegradation in dividing cells. The box indicates the interquartile range (IQR), and the line indicates the median. Far outliers (>1.5*IQR) were excluded. The color scale refers to distribution shown in B. The orange line indicates the theoretical relationship between log2 [protein] ratios and kdegradation in the absence of changes in kdegradation and ksynthesis between dividing and quiescent cells (see kinetic model). (D) Relationship between relative protein synthesis rates in quiescent and dividing cells to kdegradation in dividing cells. (E) Relationship between relative protein degradation rates in quiescent and dividing cells to kdegradation in dividing cells. (F) Box plots indicate the distribution of log2 ratio of kdegradation measurements between quiescent and dividing cells for proteins mapped to different gene ontology (GO) component accessions.
However, the problem of proteomic imbalance may not be limited to transformed cells exposed to growth-arresting drugs. Many untransformed cell types naturally alternate between a state of proliferation and a state of reversible cell cycle arrest known as quiescence (5). Quiescence-induced stabilization of long-lived proteins may therefore represent a general proteostatic disruption that impacts many different cell types. Whether such an imbalance occurs as cells naturally transition from a proliferating to a quiescent state or whether they mount a compensatory response to counter this proteostatic disruption is not known.
To gain insight into the nature and mechanism of global changes in proteome distribution and dynamics under quiescence, we utilized a proteomics approach to investigate dermal fibroblasts as they transition from a dividing to a contact-inhibited state. In vivo, dermal fibroblasts are primarily maintained in a quiescent state and enter a proliferative state as part of the wound healing response (6). Proliferating fibroblasts can reenter the quiescent state upon contact with neighboring cells (5). Contact-inhibited fibroblasts remain metabolically active and carry out the synthesis and secretion of extracellular matrix proteins such as collagen fibers that form the basement membrane of connective tissues (7, 8). The failure of fibroblasts to achieve quiescence upon completion of wound healing results in excessive scarring and fibrotic disease (7).
Our results indicate that, upon entering quiescence, fibroblasts enhance rates of protein degradation for much of their proteome, and that this effect is most pronounced for long-lived proteins. The enhanced protein degradative flux is achieved through at least two concurrent mechanisms: increased biogenesis of lysosomal compartments and activation of macroautophagy. Our quantitative analysis indicates that enhancements of protein degradation rates plays an important role in diminishing protein accumulation and maintaining protein homeostasis as fibroblasts transition from a dividing state to a quiescent state. We suggest that enhancement of protein degradation represents a universal cellular response to quiescence designed to diminish the accumulation of aged proteins that would otherwise result from the absence of cytoplasmic dilution by cell division.
Results and Discussion
Selective Degradation of Long-lived Proteins in Quiescent Fibroblasts.
We first used time-resolved analysis of fractional isotopic labeling to measure protein degradation rate constants (kdegradation) on a proteome-wide scale for dividing fibroblasts (Fig. S1). The approach used mass spectrometry and dynamic Stable Isotope Labeling with Amino acids in Cell culture (SILAC) to detect and quantify the incorporation kinetics of isotopically labeled amino acids in proteins over time (9–11). The kdegradation values were analyzed for 3,861 protein groups (Dataset S1). The distribution of 2,857 kdegradation measurements that passed the quality control thresholds (see Materials and Methods) is shown in Fig. 1B in relation to the growth rate (kdivision). We observed that ∼30% and 70% of the fibroblast proteome is being degraded at rates that exceed or lag the cell division rate, respectively (Fig. 1B).
Given that a significant fraction of the proteome is being degraded at rates that are far slower than the rate of cell division, it would be expected that, in the absence of a compensatory mechanism, the induction of quiescence should result in a major shift in the abundance distribution of the proteome caused by the selective accumulation of long-lived proteins with respect to short-lived proteins (Fig. 1C, line). To determine if such a proteomic imbalance occurs, we globally measured relative changes in protein levels between dividing and contact-inhibited cells using a canonical steady-state SILAC approach (Fig. 1C, Fig. S2, and Dataset S2). Strikingly, the results indicated that there is no overall enrichment of long-lived proteins with respect to short-lived proteins in quiescent cells.
The above results suggested that fibroblasts may be mounting a selective response to reduce the expression levels of long-lived protein upon reaching quiescence. Such a response could potentially occur through a reduction in rates of synthesis or an enhancement in degradation rates of long-lived proteins. To distinguish between these two possibilities, we concurrently measured proteome-wide changes in steady-state protein levels and protein degradation rates in dividing and quiescent cells (Figs. S2 and S3 and Datasets S1 and S2). Since steady-state protein levels are established by the combined effects of protein synthesis and degradation (see formal kinetic model in Materials and Methods), these data enabled us to globally calculate the ratio of protein synthesis rates between quiescent and dividing cells under steady-state conditions (Fig. 1D and Fig. S4). The data indicate that the rates of protein synthesis are significantly decreased in quiescent cells. This result was not entirely surprising given that the down-regulation of protein synthesis is a well-described feature of quiescent cells and has been shown to occur through the inhibition of transcription, ribosome biogenesis, and translational initiation (12–14). The reduction in protein synthesis was verified by analyzing the accumulation of nascent proteins by monitoring the incorporation of the noncanonical amino acid azidohomoalanine (AHA) (15) (Fig. S4). However, the decrease in synthesis rates is not correlated to protein stability and long-lived and short-lived proteins are equally impacted by quiescence (Fig. 1D). More unexpectedly, our results also indicated that, upon entering quiescence, fibroblasts enhance rates of protein degradation, and that this effect is most pronounced for long-lived proteins (Fig. 1E and Fig. S5). Indeed, for the most stable subset of the proteome (kdegradation < 0.1 d−1), the degradation rate constant is reduced by a median factor of four. This effect provides a putative explanation for the lack of enrichment of long-lived proteins in quiescent cells. Increases in degradation rates were observed across proteins mapped to diverse gene ontologies. However, among major localization categories, cytosolic proteins were most dramatically impacted (Fig. 1F).
Up-Regulation of Macroautophagy in Quiescent Cells.
We next set out to identify the mechanism of selective degradation of long-lived proteins in quiescent cells. In eukaryotic cells, the predominant protein degradation pathways with broad selectivity are the ubiquitin proteasome system (UPS) and macroautophagy (16, 17). Under basal conditions, the former is thought to be the primary degradation pathway for short-lived proteins whereas the latter is thought to be responsible for much of the constitutive turnover of long-lived proteins (16, 18, 19). Previous studies reported that macroautophagy is activated in quiescent transformed mouse embryonic fibroblasts and quiescent muscle stem cells (20, 21), although the impact of this effect on proteome degradation kinetics was not investigated. We therefore considered whether up-regulation of macroautophagy may be partly responsible for the selective degradation of long-lived proteins in quiescent human dermal fibroblasts.
We first verified that macroautophagy is activated in quiescent dermal fibroblasts (Fig. 2A). The level of SQSTM1/p62 protein, a receptor and substrate of macroautophagy, is reduced in quiescent cells. The quiescence-induced reduction in SQSTM1/p62 levels is not observed at the mRNA level as determined by RNA-seq (see Up-Regulation of Lysosomal Biogenesis in Quiescent Cells). Thus, the observed posttranscriptional reduction in p62 protein levels is consistent with enhanced autophagic degradation (22–25). Additionally, the level of lipidated LC3 (LC3-II), a marker of autophagosomes, is significantly increased in quiescent cells (22, 26).
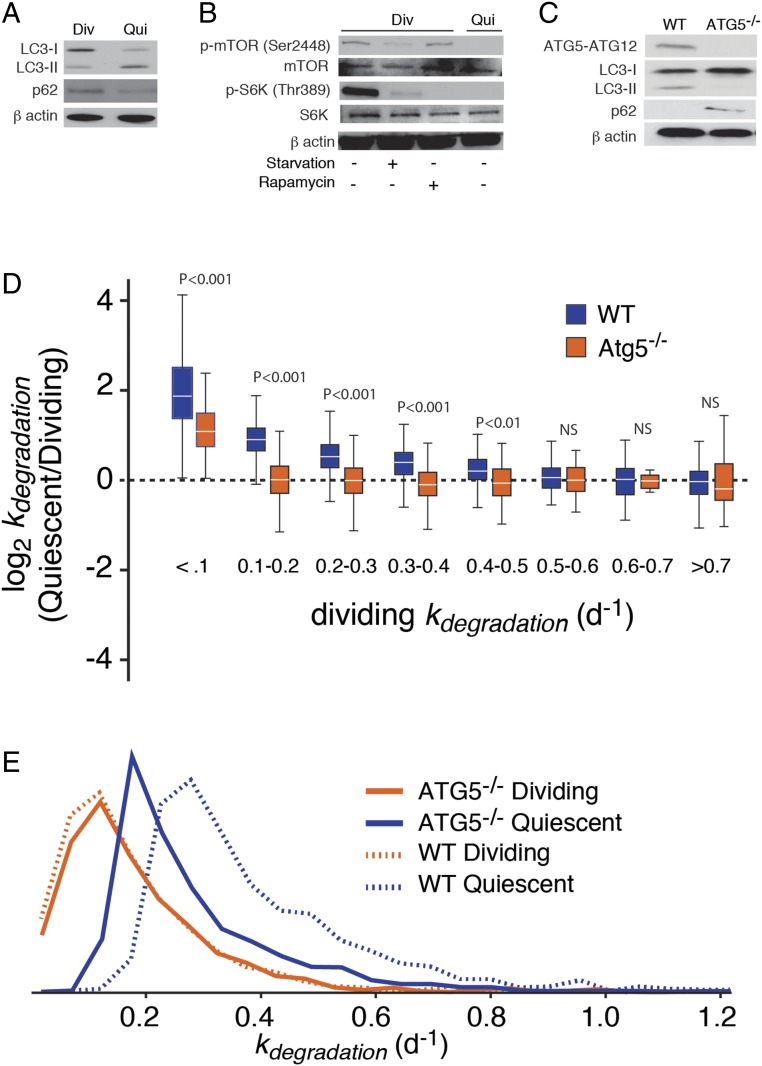
Fig. 2.
Up-regulation of macroautophagy in quiescent cells contributes to the degradation of long-lived proteins in quiescent cells. (A) Western blots showing the accumulation of LC3-II and depletion of p62 in wild-type quiescent cells. (B) Western blots showing the reduced levels of Ser-2448 phosphorylation of mTOR and phosphorylated S6 kinase in quiescent cells. (C) Western blots indicating the absence of ATG5 protein (normally expressed in the cell complexed to ATG12), depletion of LC3-II, and accumulation of p62 in ATG5−/− cells. (D) Relationship between relative protein degradation rates in quiescent and dividing ATG5−/− cells to kdegradation, in comparison with wild-type cells. Comparisons were conducted by two-sided Mann−Whitney U test. NS (not significant) indicates a P value greater than 0.05. (E) Distribution of kdegradation measurements for wild-type and ATG5−/− cells. The data indicate that the enhancement of kdegradation in quiescent cells is significantly diminished in ATG5−/− cells.
In mammalian cells, the activation of mammalian target of rapamycin (mTOR) pathway has been associated with the elevation of protein synthesis, inhibition of autophagy, and promotion of cell growth (18, 27, 28). We therefore considered whether quiescence-induced inhibition of mTOR lies upstream of autophagic activation and repression of protein synthesis observed in contact-inhibited fibroblasts. A connection between TOR repression and quiescence was observed in a few previous studies (29). For example, treatment of yeast with rapamycin has been shown to result in cell cycle arrest in a G0/G1 state (30). In another study, cell cycle reentry of quiescent Drosophila neuroblasts was shown to be dependent on TOR and its activation by the phosphatidylinositol-3-kinase-AKT (PI3K) pathway (31). Phosphorylation of mTOR at Ser-2448 is a biomarker of the activation-state PI3K pathway and its downstream up-regulation of mTOR activity (32, 33). Here, we observed that the level of Ser-2448 phosphorylation, but not total mTOR, is greatly reduced in quiescent fibroblasts (Fig. 2B). The repression of the TOR pathway in quiescent cells was confirmed by the dramatic reduction in the level of phosphorylated (but not total) S6 kinase, its canonical substrate (28). This effect was mirrored in cells where TOR was inhibited by amino acid starvation or addition of rapamycin. The results are consistent with the idea that the activation of autophagy and repression of protein synthesis in quiescent fibroblasts are linked to the repression of the TOR pathway. However, clarifying the detailed mechanisms that link contact inhibition to TOR inhibition, autophagic activation and translational repression will require further study.
Next, we quantified the contribution of activated macroautophagy to the enhancement of protein degradation rates in quiescent cells. Proteomic analyses were carried out in dividing and quiescent fibroblasts lacking the ATG5 gene known to be required for canonical macroautophagy (17). The CRISPR-mediated creation and characterization of human ATG5−/− dermal fibroblasts has been described in a previous study (9), and the inhibition of macroautophagy in this mutant was confirmed in this study (Fig. 2C). Strikingly, we observed that the quiescence-induced enhancement of degradation rates observed in wild-type cells was greatly diminished, but not entirely eliminated, in ATG5−/− cells (Fig. 2 D and E and Dataset S1). This observation is consistent with previously reported proteostatic phenotypes of ATG5−/− fibroblasts (9). For example, we observed that quiescent ATG5−/− fibroblasts are significantly larger and contain more total protein mass per cell in comparison with wild-type cells and selectively accumulate long-lived proteins (Fig. S6).
Up-Regulation of Lysosomal Biogenesis in Quiescent Cells.
The analysis of ATG5−/− cells supported the idea that macroautophagy significantly contributes to the selective clearance of long-lived proteins in quiescent cells. We next considered whether the activation of macroautophagy in quiescent cells (evidenced by markers of autophagosomes) occurs concurrently with expansion of lysosomal compartments (the location of autophagic degradation). We searched for evidence for up-regulation of lysosomal biogenesis by analyzing global changes in the transcriptome by RNA-seq (Fig. 3, Fig. S7, and Dataset S3) and steady-state protein levels by SILAC (Fig. S2 and Dataset S2). Previous transcriptomic studies had investigated fibroblasts quiescence using microarray-based technologies, and our RNA-seq results were largely consistent with the most comprehensive of these studies (Fig. S7). Gene ontology analysis indicated that many of the observed changes in mRNA and protein levels were consistent with the known role of quiescent fibroblasts as producers of protein components of the extracellular matrix (e.g., extracellular proteins and proteins associated with the secretory pathway were up-regulated) (Figs. S2 and S7 and Dataset S4). Other observed changes in expression levels were less expected. For example, we observed a dramatic increase in levels of MHC class I proteins in quiescent cells (see Catalytic Activity of the Proteasome in Quiescent Cells). Importantly, in support of our hypothesis, we also observed a significant up-regulation of resident lysosomal proteins in quiescent cells (Fig. 3A and Fig. S7).
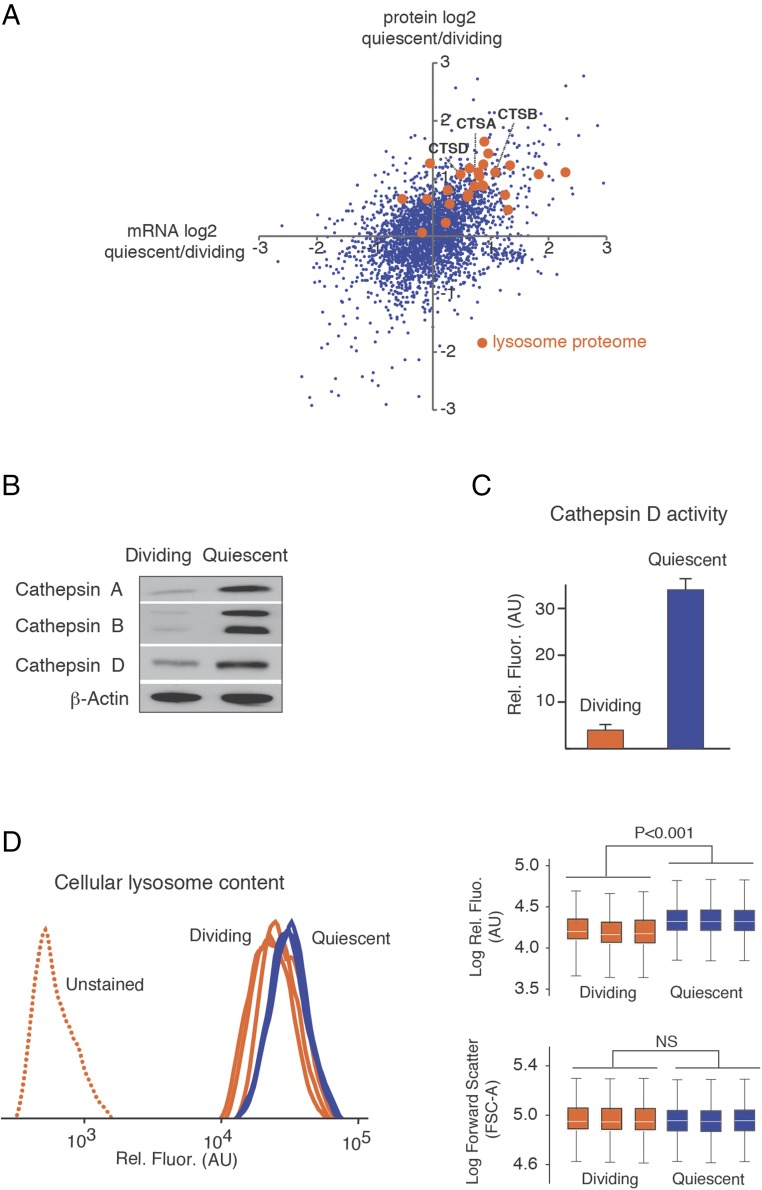
Fig. 3.
Up-regulation of lysosomal biogenesis in quiescent cells. (A) The scatter plot shows the differential expression of lysosomal genes at protein and mRNA levels as determined by SILAC and RNA-Seq, respectively. Orange dots highlight the lysosomal proteome (51). Three cathepsin genes, analyzed in B, are highlighted. (B) Western blots indicating the up-regulation of cathepsins A, B, and D in quiescent cells. (C) Increased cathepsin D activity in quiescent cell extracts based on a cell-free assay. (D) Increased accumulation of lysosomes in quiescent cells measured by flow cytometry of cells stained with LysoGreen. The box plots indicate the complete range (whiskers), interquartile range (box), and median (white line) of the measurements for replicate experiments. The forward scatter data indicate that cell size was not altered by quiescence. Comparisons were conducted by two-sided Mann−Whitney U test. NS (not significant) indicates a P value greater than 0.05.
Transcriptionally up-regulated lysosomal proteins include the hydrolases cathepsins A, B, and D (Fig. 3A). The up-regulation of cathepsins was validated by Western blots (Fig. 3 A and B). Using a cell-free assay, we also demonstrated that the total activity of one representative cathepsin (cathepsin D) is significantly increased in extracts obtained from quiescent cells (Fig. 3C). Increased cathepsin D activity during quiescence was previously observed in 3T3 fibroblasts (34). Although cathepsins are primarily localized to lysosomes where they catalyze the basal degradation of cellular proteins, they can also be released into the cytoplasm under deleterious conditions where they can trigger apoptotic cell death. However, our quiescent cells do not have other hallmarks of apoptotic cells (e.g., caspase 3 activation), and the up-regulation of cathepsins appears to be a cellular response to quiescence rather than a marker of cell death (Fig. S7). Using fluorescence-activated cell sorting (FACS) analysis of fibroblasts stained with a lysosome-specific dye, we detected a significant expansion of lysosomal compartments in quiescent cells in comparison with dividing cells (Fig. 3D). Thus, the up-regulation of lysosomal proteins in quiescent cells appears to coincide with an overall increase in lysosomal volume.
Catalytic Activity of the Proteasome in Quiescent Cells.
We next considered whether quiescence additionally influences the activity of the UPS. We observed that, in cell-free assays, the chymotrypsin-like activity of the proteasome against a synthetic peptide substrate is significantly increased in extracts obtained from quiescent cells in comparison with dividing cells (Fig. 4A). This result was somewhat unexpected given that, as a group, the expression levels of proteasomal subunits are largely unaffected by quiescence (Fig. 4B). Two of the exceptions are the proteasome components PA28α and PS28β (encoded by PSME1 and PSME2 genes) that comprise the regulatory 11S cap of the immunoproteasome (35). The expression levels of PA28α/β are elevated in quiescent cells at mRNA and protein levels as validated by qPCR and Western blots (Fig. 4 C and D).
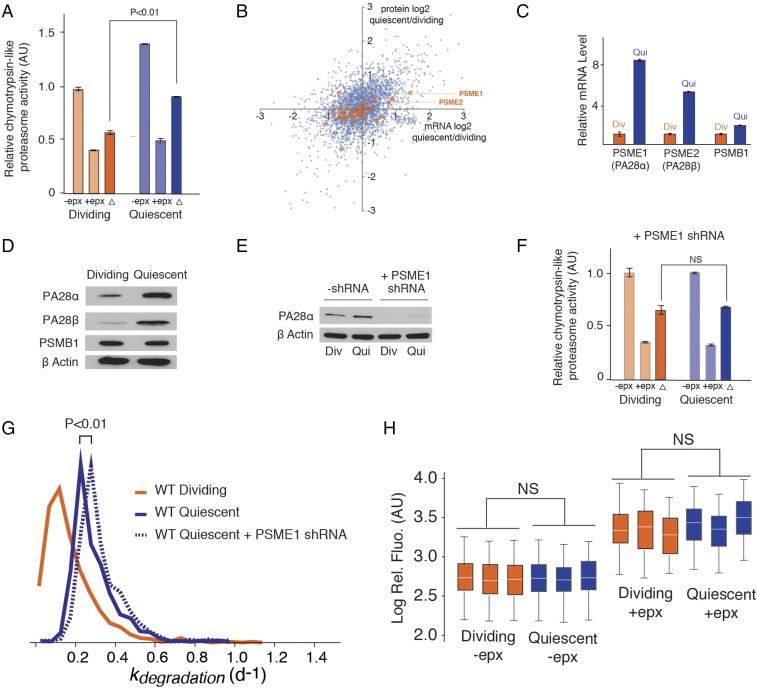
Fig. 4.
Proteasome activity does not contribute to the enhancement of degradation rates in quiescent cells. (A) Proteasome activity is enhanced in quiescent cells based on in vitro chymotrypsin-like activity assays; −epx (epoxomicin) indicates total chymotrypsin-like activity, +epx indicates chymotrypsin-like activity from nonproteasome sources, and the difference (∆) is a measurement of chymotrypsin-like activity from the proteasome. (B) Changes in the mRNA and protein expression levels of proteasome subunits determined by RNA-Seq and SILAC. The data indicate the up-regulation of 11S subunits PSME1 (PA28α) and PSME2 (PA28β) and were validated by (C) qPCR and (D) Western blots. The 20S core subunit PSMB1 is included as a control. (E) Knockdown of PSME1 by shRNA. (F) Knockdown of PSME1 eliminates the enhancement of proteasome activity in quiescent cells based on chymotrypsin-like activity assays. (G) Knockdown of PSME1 does not influence protein degradation rates (kdegradation) in quiescent cells as determined by dynamic SILAC experiments. (H) Expression level of a fluorescent proteasome substrate (pZsProSensor1) is unaffected by quiescence. Fibroblasts were transfected with pZsProSensor1 expression vector in dividing or contact-inhibited quiescent states, with or without subsequent addition of epoxomicin. The fluorescence distribution of cells was analyzed by FACS. Box plots were generated as described in Fig. 3D. Comparisons were conducted by two-sided Mann−Whitney U test. NS (not significant) indicates a P value greater than 0.05.
Substrate access to the core 20S proteasome is activated by regulatory protein complexes that bind to the ends of the core catalytic chamber (36). The best-described proteasome activator is the 19S regulatory complex (also known as PA700) that is known to stimulate the degradation of polyubiquitinated substrates by an ATP-dependent mechanism (37, 38). PA28α/β comprises an alternative regulatory complex, 11S (also known as PA28) (37). The 11S is thought to contribute to peptide generation and presentation on MHC class I molecules for recognition by cytotoxic T lymphocytes (39). The association of PA28α/β to acquired immunity is supported by its high expression levels in immune tissues and induction by IFNγ (40). PA28α/β has been shown to enhance the activity of 20S proteasomes, in its canonical and immunoproteasome configurations, against model peptide substrates (41). However, PA28α/β does not appear to recognize polyubiquitinated proteins nor hydrolyze ATP, and therefore its potential contribution to enhancing degradation rates of cellular proteins is uncertain (42). Instead, PA28α/β appears to play an auxiliary role in the MHC class I pathway by transferring peptides from the proteasome to the MHC loading complex (43, 44), lengthening the peptide products to an optimal length for MHC loading (43), or augmenting class I presentation by altering the cleavage site within a polypeptide (45, 46).
Based on these previous findings, it remained unclear whether the up-regulation of PA28α/β could be a contributing factor to decreased proteome half-lives observed in quiescent cells. To address this question, we knocked down the expression level of PA28α using an shRNA construct targeted against the PSME1 gene (Fig. 4E). The knockdown of PA28α eliminated the quiescence-induced elevation of proteasome activity against the target peptide substrate in vitro (Fig. 4F). We next conducted a global comparison of protein degradation rates between wild-type and PA28α knockdown cells under quiescent conditions (Fig. 4G and Dataset S1). Our global data indicate that the knockdown of PA28α had relatively little impact on mitigating the enhanced protein degradation rates in quiescent cells. Indeed, it appears that cellular protein degradation rates increase slightly in that absence of PA28α. Thus, although the up-regulation of PA28α/β in quiescent cells results in enhanced degradation of synthetic peptide substrates in vitro, this altered activity is not indicative of a PA28α/β-induced increase in protein turnover kinetics in vivo. Consistent with these results, we observed that the level of a heterologously expressed fluorescent reporter proteasome substrate [pZsProSensor-1 encoding the degradation domain of mouse ornithine decarboxylase protein fused to the COOH terminus of ZsGreen (47)] does not change as cells transition from a dividing to a quiescent state (Fig. 4H). Together, the results suggest that escalation of macroautophagy and lysosomal biogenesis, and not UPS activation, are the dominant factors contributing to increased protein degradation in quiescent cells (Fig. 5A). Instead, the up-regulation of PA28α/β and MHC1 proteins (see Up-Regulation of Lysosomal Biogenesis in Quiescent Cells), may be indicative of the up-regulation of the class I pathway and a higher basal level of antigen presentation in quiescent cells.
Fig. 5.
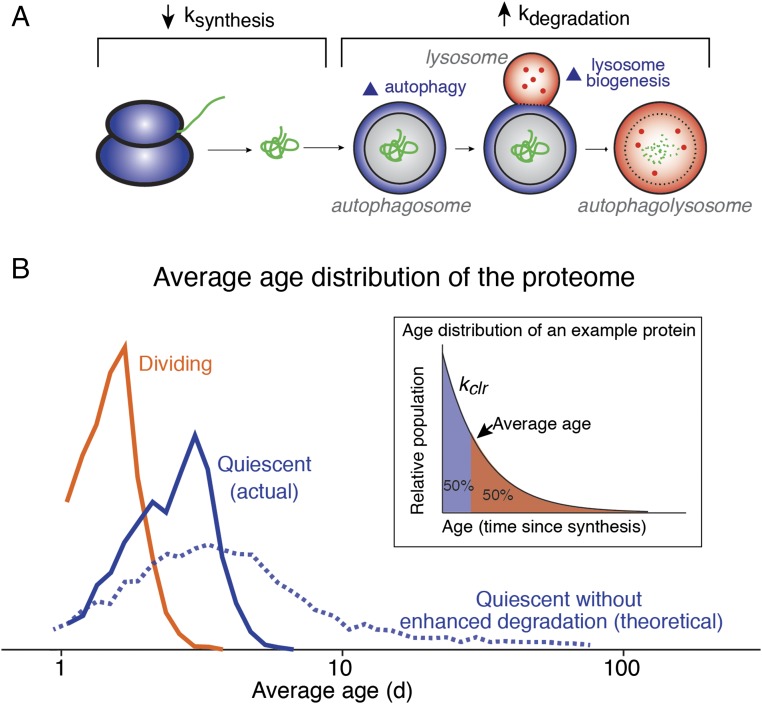
Regulation of protein dynamics in quiescent cells and its ramification for protein homeostasis. (A) In quiescent fibroblasts, protein synthesis rates are reduced and protein degradation rates are increased. The latter effect is caused by concurrent up-regulation of lysosome biogenesis and macroautophagy and is limited to long-lived proteins. (B) The distribution of the average age of protein populations in quiescent and dividing cells. The average age of a given protein population can be experimentally determined by its clearance rate (Inset). In the theoretical scenario where the quiescence-induced up-regulation of protein degradation is lacking, the age distribution of the proteome would be expected to significantly increase.
Conclusions
In dividing cells, proteins may be cleared either by cytoplasmic dilution or degradation. Whereas degradation is the predominant route of clearance for short-lived proteins, long-lived proteins (whose half-lives are significantly longer than the doubling time of the cell) are predominantly cleared from a cell through the process of proliferation (1, 48). Thus, as cells transition from a dividing to a nondividing state, the clearance kinetics of long-lived proteins decline, whereas short-lived proteins remain unaffected. If no compensatory response is initiated, this phenomenon would be expected to result in a significant global increase in cellular concentrations of long-lived proteins relative to short-lived proteins as dividing cells transition to quiescence. However, we have shown here that there is no significant proteome-wide correlation between protein half-lives and quiescence-induced changes in steady-state protein levels.
Our results provide an explanation for this apparent paradox. Activation of autophagy-mediated lysosomal degradation increases rates of degradation for long-lived proteins, partially offsetting the reduction in their clearance rates. Conversely, short-lived proteins, known to be primarily degraded at a faster rate by the UPS, are largely impervious to the up-regulation of lysosomal degradation in quiescent cells. Thus, the preferential degradation of stable proteins by quiescent-induced autophagy can be explained without invoking a specific receptor-mediated mechanism comparable to canonical forms of “selective autophagy.” Rather, this apparent selectivity arises from the fact that basal nonselective autophagy is the rate-determining form of clearance only for long-lived proteins that are not robust targets of more-rapid forms of degradation such as the UPS.
It is important to note that the impairment of macroautophagy does not completely abolish the quiescence-induced enhancement of degradation rates. Thus, it is possible that alternative degradation pathways may also contribute to the degradation of long-lived proteins in nonproliferating cells. For example, although the UPS pathway does not appear to be globally impacted by quiescence, we cannot rule out the possibility that contact inhibition modulates the selectivity of UPS, leading to degradation of specific substrates in quiescent cells.
In addition to preventing global changes in the abundance distribution of the proteome, the selective degradation of stable proteins may play a second important role in maintaining long-term homeostasis in quiescent cells. The total clearance rate of a protein, established by the sum contribution of cell division and protein degradation, dictates the age distribution of its molecular population at steady state (Fig. 5B). Hence, as the rate of clearance for a given protein decreases, the average age (elapsed time after synthesis) of its molecular population increases. Accordingly, if no compensatory mechanism is initiated, the cessation of cell division in quiescent cells would be expected to result in a significant proteome-wide increase in aged proteins. Fig. 5B depicts the quantitation of this effect based on the global measurements of degradation kinetics and the cellular proliferation rates in this study. A dramatic increase in the average age of cellular proteins could potentially lead to increased accumulation of damaged and aggregated proteins in quiescent cells over time. The activation of lysosomal degradation significantly mitigates this effect and halts the accumulation of aged, and potentially damaged, proteins in nonproliferating quiescent cells. Thus, the up-regulation of protein degradation by lysosomal degradation in quiescent cells may represent an adaptive response for the promotion of protein homeostasis in postmitotic tissues in aged organisms. Whether this proteostatic response is shared among diverse quiescent cell types and tissues remains to be determined.
Materials and Methods
Cell Culture.
Cell culture Human primary fibroblasts were maintained in Eagle’s Minimum Essential Medium (ATCC) supplemented with 15% FBS (Invitrogen), 100 U/mL penicillin, 100 U/mL streptomycin at 37 °C with 5% CO2. Human embryonic kidney cell line 293FT (Life Technologies) was maintained in DMEM (Life Technologies) supplemented with 10% FBS (Invitrogen), 100 U/mL penicillin, 100 U/mL streptomycin at 37 °C with 5% CO2.
Stable Isotope Labeling.
The media utilized for isotopic labeling was Eagle’s minimum essential medium (ATCC) supplemented with 15% dialyzed FBS (Thermo Scientific), 100 U/mL penicillin, and 100 U/mL streptomycin. Cells were gradually adapted to this media by replacing normal FBS with dialyzed FBS within four passages. Cells were then plated at a density of 500,000 cells per 10-cm plate.
One day after plating, the dividing cultures were switched to MEM labeling media for SILAC (Thermo Scientific) supplemented with l-arginine:HCl (13C6, 99%) and l-lysine:2HCl (13C6, 99%; Cambridge Isotope Laboratories) at concentrations of 0.1264 g/L and 0.087 g/L and 15% dialyzed FBS (Thermo Scientific). Cells were collected after 0, 1, 2, and 3 d of labeling and washed with PBS, and cell pellets were frozen before further analysis. Eight days after plating, the confluent quiescent cultures were switched to MEM labeling media for SILAC (Thermo Scientific) supplemented with l-arginine:HCl (13C6, 99%) and l-lysine:2HCl (13C6, 99%; Cambridge Isotope Laboratories) at concentrations of 0.1264 g/L and 0.087 g/L and 15% dialyzed FBS (Thermo Scientific). Cells were collected after 0, 2, 4, and 6 d of labeling and washed with PBS, and cell pellets were frozen before further analysis.
Cell Cycle Analysis by Flow Cytometry.
Wild-type cells were cultured as described in Cell Culture, cells were collected, and 1 × 106 cells were centrifuged at 300 × g for 5 min and washed once with 1× PBS. While mixing and resuspending cells, 1 mL of ice-cold 70% ethanol was slowly added to fix cells. Then 200 μL of fixed cells were transferred to a new tube and centrifuged at 300 × g for 5 min and washed once with PBS; 200 μL of Muse Cell Cycle reagent from Muse Cell Cycle Kit (Millipore) was used for each tube. Cells were incubated for 30 min at room temperature in the dark. Muse Cell Analyzer (Millipore) was used to analyze the DNA content of each sample.
Cell Size Measurement by Flow Cytometry.
Wild-type and ATG5−/− cells were cultured as described in Cell Culture, cells were collected, and 1 × 106 cells were centrifuged at 300 × g for 5 min and washed once with 1× PBS. While mixing and resuspending cells, 1 mL of ice-cold 70% ethanol was slowly added to fix cells. Then 200 μL of fixed cells were transferred to a new tube and centrifuged at 300 × g for 5 min and washed once with PBS. Cells were analyzed on a LSR II instrument (BD) with forward scatter (FSC) at 400 V and side scatter (SSC) at 200 V.
Analysis of mTOR Pathway.
Human primary fibroblasts were seeded in 500,000 cells per plate and grown for 2 d and for 8 d to obtain dividing and quiescent cultures, respectively. For starvation treatment, dividing cells were washed using PBS twice, and then media was switched to EBSS media (Sigma) for 2 h. For rapamycin treatment, 10 nM rapamycin was added to cells for 24 h. After treatment, dividing and quiescent cultures were collected, and cell pellets were stored in −80 °C for Western blots.
Western Blots.
Cells were lysed with ice-cold lysis buffer (10 mM Tris⋅HCl pH 8.0, 0.15 M NaCl, 0.5% Nonidet P-40, 0.48% SDS). Cell lysates were centrifuged at 14,500 × g for 10 min. For Western blot analysis, 30 µg was separated by electrophoresis in 10% polyacrylamide gels and transferred to polyvinylidene difluoride membranes using Trans-Blot SD Semi-Dry Electrophoretic Transfer Cell (Biorad). After 1 h incubation at room temperature in Tris-buffered saline Tween (TBST)/5% nonfat milk, the membrane was incubated with the indicated antibodies at 4 °C overnight. The membranes were then washed with TBST/0.1% Tween 20, and the corresponding secondary antibodies were applied to the membranes for 1 h at room temperature. The membranes were then washed with TBST/0.1% Tween 20, and the detection of signal was done with an enhanced chemiluminescence detection kit (Pierce). The primary antibodies and the corresponding dilutions utilized for the Western blots were p27 Kip1 (D69C12) XP Rabbit mAb: 1:1,000 (Cell Signaling Technology); Anti beta-Actin Antibody: 1:2000 (Abcam); Anti-p62 (SQSTM1) Antibody: 1:1,000 (MBL International); Anti-LC3 Antibody: 1:1,000 (MBL International); Phospho-mTOR (Ser2448) Antibody: 1:1,000 (Cell Signaling Technology); mTOR(ab2732): 1:2,000 (Abcam); 20S Proteasome α2 Antibody (B-4): 1:200 (Santa Cruz Biotechnology); PA28α Antibody: 1:1,000 (Cell Signaling Technology); PA28β Antibody: 1:200 (Santa Cruz Biotechnology); PA28γ Antibody (47): sc-136025 (Santa Cruz Biotechnology); Cathepsin A (A-19): 1:200 (Santa Cruz Technology), Cathepsin B (H-5): 1:500 (Santa Cruz Technology), Cathepsin D (c-20): 1:200 (Santa Cruz Technology); Phospho-p70 S6 Kinase (Thr389)(108D2): 1:1,000 (Cell Signaling Biotechnology); S6 Kinase(E175): 1:1,000 (Abcam); and Phospho-S6 Ribosomal Protein (Ser235/236) (D57.2.2E): 1:1,000 (Cell Signaling Biotechnology). In instances where multiple proteins were analyzed from the same set of extracts (e.g., dividing versus quiescent), each extract was loaded on multiple lanes of the same SDS/PAGE gel and, following transfer onto the membrane, was cut into multiple “strips.” Each strip, containing a replicate of the set of extracts, was probed with a different primary antibody against a specific target protein and, subsequently, with the corresponding secondary antibody.
l-AHA Labeling.
Human primary fibroblasts were seeded in 500,000 cells per plate and grown for 2 d and for 8 d for dividing cultures and quiescent cultures, respectively. Before AHA labeling, media was replaced with Roswell Park Memorial Institute (RPMI), no methionine (Thermo Scientific) media supplemented with 15% FBS, 100 U/mL penicillin, and 100 U/mL streptomycin, and cultures were incubated in 37 °C for 30 min to deplete methionine from the media. Then, media was replaced by RPMI, no methionine media supplemented with 15% FBS, 100 U/mL penicillin, 100 U/mL streptomycin, and 4 mM l-AHA; 100 μg/mL cycloheximide was used to inhibit protein synthesis as negative control. After 1 h incubation with AHA, cells were collected and lysed; 50 μL of 1 mg/mL lysate was used for click reaction using Click Chemistry Protein Reaction Buffer Kit (Click Chemistry Tools); 20 μL of click reaction product was used for Western blot and probed by Horseradish peroxidase streptavidin (Thermo Fisher Scientific).
Mass Spectrometry Sample Preparation.
Lysis buffer was composed of 8 M urea, 75 mM NaCl, 50 mM Tris, pH 8.5; 50 μL of lysis buffer was used for one million cells. Cell lysis was conducted by adding lysis buffer to pelleted cells, followed by vortexing and sonication using a QSonica sonicator. Sonication cycles consisted of 5 × 60 s sonications with 1-min incubations on ice between each cycle. Samples were then centrifuged for 10 min at 15,000 × g, and the supernatant was collected. Protein concentration was determined by bicinchoninic acid (BCA) assays; 100 μg of each extract was removed, and brought up to 100 μL in 100 mM ammonium bicarbonate. This dilution lowered the urea concentration to less than 1.5 M such that trypsinization efficiency was not inhibited. Then 2 μg of trypsin (Thermo Scientific) was added to each extract, and the samples were allowed to incubate overnight at 37 °C. Disulfide bonds were reduced with the addition of Bond-Breaker TCEP Solution (Thermo Scientific) to a final concentration of 5 mM with incubation at 55 °C for 1 h. Reduced cysteines were alkylated by the addition of iodoacetamide at a final concentration of 10 mM by incubating in the dark at room temperature for 30 min. Subsequently, an additional 2 μg of trypsin was added to the samples and incubated for 3 h at 37 °C. Formic acid was added to a final concentration of 1% to stop trypsin activity. Samples were dried down in a Centrivap concentrator (Labconco). Digested samples were reconstituted in 300 μL of 0.1% TFA and were fractionated using a Pierce High pH Reversed-Phase Peptide Fractionation Kit (Thermo Scientific). Briefly, the columns were prepared by adding 300 μL of acetonitrile and centrifuging at 5,000 × g for 2 min (twice). The same was then done using 0.1% TFA. Samples were then loaded onto the column and centrifuged at 3,000 × g for 2 min, then washed with HPLC-grade water (JT Baker). Eight fractions were then collected, using increasing amounts of acetonitrile in 0.1% triethylamine; the percentages of acetonitrile were 10%, 12.5%, 15%, 17.5%, 20%, 22.5%, 25%, and 50%. Fractions were dried down in a Centrivap concentrator (Labconco), resuspended in 50 μL of 0.1% TFA, and proceeded to mass spectrometry analysis.
LC-MS/MS Analysis.
Six microliters of each high-pH fraction was analyzed on a Q Exactive Plus mass spectrometer (Thermo Scientific) coupled with an Easy nLC-1000 pump (Thermo Scientific). Columns were hand-pulled using 100 μm of fused silica, which was packed with 30 cm of 1.8 μm, 120 Angstrom C18 beads (Sepax). Mobile phase A was 0.1% formic acid in water, and mobile phase B was 0.1% formic acid in acetonitrile. Peptides were separated using a gradient of 8 to 30% B over 145 min, 30 to 50% B over 10 min, and 50 to 70% B over 4 min, holding at 70% B for 5 min. The gradient returned to 0% B in 4 min, and held there for 10 min to prepare for the next injection. The flow rate throughout the run was held at 300 μL/min. A data-dependent top 10 MS2 method was used for peptide detection and fragmentation. For each cycle, one full scan from m/z 400 to 1,700 was acquired in the Orbitrap at a resolution of 70,000 at m/z = 200, with an automatic gain control (AGC) target of 3e6, and a maximum injection time of 50 ms. After each full scan, the top 10 most intense ions were selected for fragmentation in the HCD cell, followed by detection in the Orbitrap at a resolution of 35,000 at m/z = 200. The AGC target was set to 1e5, with a maximum injection time of 150 ms, an isolation width of 0.5 Da, and a normalized collision energy of 35. A dynamic exclusion window of 25 s was used, while excluding unassigned, +1, and greater than +5 charge states. The polysiloxane 445.12003 m/z was used for lock mass.
Peptides were identified from MS/MS spectra by searching against the Homo sapiens Swiss-Prot database using the integrated Andromeda search engine with the software MaxQuant (49). SILAC peptide and protein quantification was performed with MaxQuant using default parameters. For each peptide, the heavy/light (H/L) ratio was determined by a regression model fitted to all isotopic peaks within all scans that the peptide eluted in. H/L ratio for each protein was determined as the median of all of the peptides assigned to the protein (50). For a given protein, H/(H+L) ratio was calculated based on the H/L ratio from MaxQuant outputs. All H/(H+L) ratios at all time points were combined to obtain an aggregated plot for the kinetics of labeling. The aggregated plots were fitted to a single exponential function by least-square fitting to obtain the first-order clearance rate constant (kclr) for each protein. For dividing cells, the experimentally measured rate of cellular proliferation (log2/doubling time) was subtracted from the measured kclr to determine the first-order rate constant for degradation (kdeg). For dividing cells, the proliferation rate was experimentally determined to be ∼0.30 d−1. For quiescent cells, kclr measurements were assumed to equal kdeg. Refer to the kinetic model described below (see Kinetic Model) for further details.
RNA-Seq.
Total RNA was isolated using the RNeasy Plus Kit (Qiagen) per manufacturer’s recommendations. RNA concentration was determined with the NanopDrop 1000 spectrophotometer (NanoDrop), and RNA quality was assessed with the Agilent Bioanalyzer (Agilent). The TruSeq RNA Sample Preparation Kit V2 (Illumina) was used for next-generation sequencing library construction per manufacturer’s protocols. Briefly, mRNA was purified from 100 ng of total RNA with oligo-dT magnetic beads and fragmented. First-strand cDNA synthesis was performed with random hexamer priming followed by second-strand cDNA synthesis. End repair and 3′ adenylation was then performed on the double-stranded cDNA. Illumina adaptors were ligated to both ends of the cDNA, purified by gel electrophoresis, and amplified with PCR primers specific to the adaptor sequences to generate amplicons of ∼200 bp to 500 bp in size. The amplified libraries were hybridized to the Illumina single-end flow cell and amplified using the cBot (Illumina) at a concentration of 8 pM per lane. Single end reads of 100 nt were generated for each sample and aligned to the organism specific reference genome.
shRNA Knockdown.
PSME1 knockdown assay was performed using shRNA obtained from GE Dharmacon. Inhibition of expression of PSME1 was achieved by infection with viruses made from 293T cells using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s protocol. Forty-eight hours after infection, 0.5 μg/mL puromycin was used for selection for 5 d. Puromycin-resistant cells were selected for further analysis.
Lysosomal Content Detection by Flow Cytometry.
Cells were plated at a density of 500,000 cells per 10-cm plate. Three replicates for dividing culture and quiescent culture were prepared. CytoPainter Lysosomal Staining Kit (Abam) was used for staining. Cells were collected, and 1 × 106 were centrifuged at 300 × g for 5 min and resuspended in 500 μL of culturing media. Dye-working solution was made by diluting 20 μL of LysoGreen Indicator in 10 mL of Live Cell Staining Buffer; 500 μL of dye-working solution was added to each sample. Cells were incubated in a 37 °C, 5% CO2 incubator for 30 min. Cells were analyzed by BD LSR II (BD), using Blue F filter.
Cathepsin D Activity Assay.
Cathepsin D activity was measured by using Cathepsin D Activity Assay Kit (Abcam). Cells were collected, and 350 μL of lysis buffer was used to lyse 1 million cells; 5 μL of lysate was incubated in substrate/buffer solution for 75 min at 37 °C. Enzyme activity was measured by monitoring release of the fluorescent cleavage product, MCA. Activity measurements from parallel reactions containing 0.7 µM protease inhibitor pepstatin A were subtracted from the activity measurements obtained without the inhibitor. Fluorescence was quantified by Infinite M100 (Tecan) at Ex/Em = 328/460 nm.
Proteasome Activity Assays.
In vitro proteasome activity was measured by using 20S Proteasome activity assay kit (Millipore); 75 μg of total protein in 50 μL of cell lysate was incubated in assay buffer containing 25 nM Hepes, pH 7.4, 0.05 mL of EDTA, 0.05% mM Nonidet P-40, 0.001% SDS, and 25 μg of the proteasome substrate, LLVY-AMC, for 90 min at 37 °C. Enzyme activity was measured by monitoring release of the fluorescent LLVYAMC cleavage product, 7-amino-4-methylcoumarin. Activity measurements from parallel reactions containing 10 μM proteasome inhibitor epoxomicin were subtracted from the activity measurements obtained without the inhibitor. For in vivo analysis of proteasome activity, the proteasome sensor plasmid pZsProSensor-1 (Clontech Laboratories) was transfected into dividing and quiescent cells using TransIT-X2 transfection reagent (Mirus Bio). The pZsProSensor-1 plasmid encodes a destabilized fluorescent protein consisting of amino acids 422 to 461 of mouse ornithine decarboxylase fused to the COOH terminus of ZsGreen1. This fusion protein is rapidly degraded by the proteasome in a ubiquitin-independent manner. For half of the transfected cultures, 10 μM proteasome inhibitor epoxomicin was added to the media 40 h after transfection. Flow cytometry analysis was conducted as described in Cell Cycle Analysis by Flow Cytometry.
Kinetic Model.
The kinetic model applied in this study is based on the following assumptions:
-
i)
Protein synthesis is a zero order process with respect to protein concentration.
-
ii)
Protein degradation occurs at a constant fractional rate that is uniform for the entire protein pool. Thus, protein degradation can be modeled as a first-order process with respect to protein concentration.
-
iii)
The total protein concentration of each cell does not change during the experimental time course, and the system is at steady state.
Based on these assumptions, we can devise the following rate equations for the accumulation of labeled proteins:
| [1] |
where
| [2] |
Here, ksyn is the zero-order rate constant for protein synthesis, kdeg is the first-order rate constant for protein degradation, kdiv is the first-order rate constant for cell division, and kclr is the total clearance rate of the protein.
We solve for [labeled protein](t) using the constraint [labeled protein](0) = 0.
| [3] |
where
| [4] |
Since our protein labeling measurements are fractional (i.e., internally normalized with respect to total steady-state protein levels), the observed fractional labeling is derived as
| [5] |
In quiescent cells, where the rate of cell division is zero, Eqs. 4 and 5 simplify to
| [6] |
| [7] |
The ratio of synthesis rates of quiescent cells to synthesis rates of proliferating cells as a function of ratios of steady-state protein levels and degradation rate constants can be derived by dividing Eq. 6 by Eq. 4,
| [8] |
In the theoretical scenario where kdeg is identical between quiescent and proliferating cells (theoretical plot in Fig. 5B), then
| [9] |
| [10] |
| [11] |
and hence
| [12] |
In this theoretical scenario, protein clearance rates in quiescent cells can be determined by measuring the difference between experimentally determined rates of protein clearance and cell division in proliferating cells.
Data Access and Accession Numbers.
All data for the dynamic time-resolved isotopic labeling experiments and steady-state SILAC experiments are available at the ProteomeXchange Consortium: accession no. PXD004937. For RNA-Seq experiments, raw sequence reads and differential expression results are available in the Gene Expression Omnibus: accession no. GSE86867. The following link can be used to view the record GSE86867: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=szepqemapdwdvad&acc=GSE86867.
Institutional Approval.
Required approval of experiments was provided by University of Rochester Research Subjects Review Board.
Supplementary Material
Acknowledgments
We thank Dragony Fu and his laboratory for helpful discussions, and Vera Gorbunova, Andrei Seluanov, and their laboratories for providing cell lines and advice. We also thank John Ashton and Jason Myers at the University of Rochester Genomics Resource Center for sequencing analyses. This work was supported by National Institutes of Health Grant R35 GM119502 and National Science Foundation Grant MCB-1350165 CAREER.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The proteomic data reported in this paper have been deposited in the ProteomeXchange Consortium database (accession no. PXD004937). The RNA-seq data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE86867).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1710238114/-/DCSupplemental.
References
- 1.Eden E, et al. Proteome half-life dynamics in living human cells. Science. 2011;331:764–768. doi: 10.1126/science.1199784. [DOI] [PubMed] [Google Scholar]
- 2.Claydon AJ, Beynon R. Proteome dynamics: Revisiting turnover with a global perspective. Mol Cell Proteomics. 2012;11:1551–1565. doi: 10.1074/mcp.O112.022186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 4.Pratt JM, et al. Dynamics of protein turnover, a missing dimension in proteomics. Mol Cell Proteomics. 2002;1:579–591. doi: 10.1074/mcp.m200046-mcp200. [DOI] [PubMed] [Google Scholar]
- 5.Coller HA, Sang L, Roberts JM. A new description of cellular quiescence. PLoS Biol. 2006;4:e83. doi: 10.1371/journal.pbio.0040083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin P. Wound healing–Aiming for perfect skin regeneration. Science. 1997;276:75–81. doi: 10.1126/science.276.5309.75. [DOI] [PubMed] [Google Scholar]
- 7.Desmoulière A, Chaponnier C, Gabbiani G. Tissue repair, contraction, and the myofibroblast. Wound Repair Regen. 2005;13:7–12. doi: 10.1111/j.1067-1927.2005.130102.x. [DOI] [PubMed] [Google Scholar]
- 8.Lemons JMS, et al. Quiescent fibroblasts exhibit high metabolic activity. PLoS Biol. 2010;8:e1000514. doi: 10.1371/journal.pbio.1000514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang T, Shen S, Qu J, Ghaemmaghami S. Global analysis of cellular protein flux quantifies the selectivity of basal autophagy. Cell Reports. 2016;14:2426–2439. doi: 10.1016/j.celrep.2016.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang T, et al. Kinetics of precursor labeling in stable isotope labeling in cell cultures (SILAC) experiments. Anal Chem. 2014;86:11334–11341. doi: 10.1021/ac503067a. [DOI] [PubMed] [Google Scholar]
- 11.Mann M. Functional and quantitative proteomics using SILAC. Nat Rev Mol Cell Biol. 2006;7:952–958. doi: 10.1038/nrm2067. [DOI] [PubMed] [Google Scholar]
- 12.Braun EL, Fuge EK, Padilla PA, Werner-Washburne M. A stationary-phase gene in Saccharomyces cerevisiae is a member of a novel, highly conserved gene family. J Bacteriol. 1996;178:6865–6872. doi: 10.1128/jb.178.23.6865-6872.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuge EK, Braun EL, Werner-Washburne M. Protein synthesis in long-term stationary-phase cultures of Saccharomyces cerevisiae. J Bacteriol. 1994;176:5802–5813. doi: 10.1128/jb.176.18.5802-5813.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levine EM, Becker Y, Boone CW, Eagle H. Contact inhibition, macromolecular synthesis, and polyribosomes in cultured human diploid fibroblasts. Proc Natl Acad Sci USA. 1965;53:350–356. doi: 10.1073/pnas.53.2.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dieterich DC, Link AJ, Graumann J, Tirrell DA, Schuman EM. Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT) Proc Natl Acad Sci USA. 2006;103:9482–9487. doi: 10.1073/pnas.0601637103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ciechanover A. Proteolysis: From the lysosome to ubiquitin and the proteasome. Nat Rev Mol Cell Biol. 2005;6:79–87. doi: 10.1038/nrm1552. [DOI] [PubMed] [Google Scholar]
- 17.Mizushima N. Autophagy: Process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 18.Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhattacharyya S, Yu H, Mim C, Matouschek A. Regulated protein turnover: Snapshots of the proteasome in action. Nat Rev Mol Cell Biol. 2014;15:122–133. doi: 10.1038/nrm3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.García-Prat L, et al. Autophagy maintains stemness by preventing senescence. Nature. 2016;529:37–42. doi: 10.1038/nature16187. [DOI] [PubMed] [Google Scholar]
- 21.Valentin M, Yang E. Autophagy is activated, but is not required for the G0 function of BCL-2 or BCL-xL. Cell Cycle. 2008;7:2762–2768. doi: 10.4161/cc.7.17.6595. [DOI] [PubMed] [Google Scholar]
- 22.Klionsky DJ, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition) Autophagy. 2016;12:1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Komatsu M, et al. Essential role for autophagy protein Atg7 in the maintenance of axonal homeostasis and the prevention of axonal degeneration. Proc Natl Acad Sci USA. 2007;104:14489–14494. doi: 10.1073/pnas.0701311104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang QJ, et al. Induction of autophagy in axonal dystrophy and degeneration. J Neurosci. 2006;26:8057–8068. doi: 10.1523/JNEUROSCI.2261-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nezis IP, et al. Ref(2)P, the Drosophila melanogaster homologue of mammalian p62, is required for the formation of protein aggregates in adult brain. J Cell Biol. 2008;180:1065–1071. doi: 10.1083/jcb.200711108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Castino R, Fiorentino I, Cagnin M, Giovia A, Isidoro C. Chelation of lysosomal iron protects dopaminergic SH-SY5Y neuroblastoma cells from hydrogen peroxide toxicity by precluding autophagy and Akt dephosphorylation. Toxicol Sci. 2011;123:523–541. doi: 10.1093/toxsci/kfr179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kamada Y, et al. Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol Cell Biol. 2010;30:1049–1058. doi: 10.1128/MCB.01344-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 29.Valcourt JR, et al. Staying alive: Metabolic adaptations to quiescence. Cell Cycle. 2012;11:1680–1696. doi: 10.4161/cc.19879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barbet NC, et al. TOR controls translation initiation and early G1 progression in yeast. Mol Biol Cell. 1996;7:25–42. doi: 10.1091/mbc.7.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sousa-Nunes R, Yee LL, Gould AP. Fat cells reactivate quiescent neuroblasts via TOR and glial insulin relays in Drosophila. Nature. 2011;471:508–512. doi: 10.1038/nature09867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chiang GG, Abraham RT. Phosphorylation of mammalian target of rapamycin (mTOR) at Ser-2448 is mediated by p70S6 kinase. J Biol Chem. 2005;280:25485–25490. doi: 10.1074/jbc.M501707200. [DOI] [PubMed] [Google Scholar]
- 33.Choe G, et al. Analysis of the phosphatidylinositol 3′-kinase signaling pathway in glioblastoma patients in vivo. Cancer Res. 2003;63:2742–2746. [PubMed] [Google Scholar]
- 34.Lockwood TD, Shier WT. Regulation of acid proteases during growth, quiescence and starvation in normal and transformed cells. Nature. 1977;267:252–254. doi: 10.1038/267252a0. [DOI] [PubMed] [Google Scholar]
- 35.Ferrington DA, Gregerson DS. Immunoproteasomes: Structure, function, and antigen presentation. Prog Mol Biol Transl Sci. 2012;109:75–112. doi: 10.1016/B978-0-12-397863-9.00003-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanaka K. The proteasome: Overview of structure and functions. Proc Jpn Acad Ser B Phys Biol Sci. 2009;85:12–36. doi: 10.2183/pjab.85.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Benaroudj N, Zwickl P, Seemüller E, Baumeister W, Goldberg AL. ATP hydrolysis by the proteasome regulatory complex PAN serves multiple functions in protein degradation. Mol Cell. 2003;11:69–78. doi: 10.1016/s1097-2765(02)00775-x. [DOI] [PubMed] [Google Scholar]
- 38.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rechsteiner M, Realini C, Ustrell V. The proteasome activator 11 S REG (PA28) and class I antigen presentation. Biochem J. 2000;345:1–15. [PMC free article] [PubMed] [Google Scholar]
- 40.Tanahashi N, et al. Molecular properties of the proteasome activator PA28 family proteins and γ-interferon regulation. Genes Cells. 1997;2:195–211. doi: 10.1046/j.1365-2443.1997.d01-308.x. [DOI] [PubMed] [Google Scholar]
- 41.Ma CP, Slaughter CA, DeMartino GN. Identification, purification, and characterization of a protein activator (PA28) of the 20 S proteasome (macropain) J Biol Chem. 1992;267:10515–10523. [PubMed] [Google Scholar]
- 42.Dubiel W, Pratt G, Ferrell K, Rechsteiner M. Purification of an 11 S regulator of the multicatalytic protease. J Biol Chem. 1992;267:22369–22377. [PubMed] [Google Scholar]
- 43.Whitby FG, et al. Structural basis for the activation of 20S proteasomes by 11S regulators. Nature. 2000;408:115–120. doi: 10.1038/35040607. [DOI] [PubMed] [Google Scholar]
- 44.Yamano T, et al. Two distinct pathways mediated by PA28 and hsp90 in major histocompatibility complex class I antigen processing. J Exp Med. 2002;196:185–196. doi: 10.1084/jem.20011922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li J, et al. Lysine 188 substitutions convert the pattern of proteasome activation by REGgamma to that of REGs alpha and beta. EMBO J. 2001;20:3359–3369. doi: 10.1093/emboj/20.13.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dick TP, et al. Coordinated dual cleavages induced by the proteasome regulator PA28 lead to dominant MHC ligands. Cell. 1996;86:253–262. doi: 10.1016/s0092-8674(00)80097-5. [DOI] [PubMed] [Google Scholar]
- 47.Li X, Coffino P. Degradation of ornithine decarboxylase: Exposure of the C-terminal target by a polyamine-inducible inhibitory protein. Mol Cell Biol. 1993;13:2377–2383. doi: 10.1128/mcb.13.4.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Claydon AJ, Thom MD, Hurst JL, Beynon RJ. Protein turnover: Measurement of proteome dynamics by whole animal metabolic labelling with stable isotope labelled amino acids. Proteomics. 2012;12:1194–1206. doi: 10.1002/pmic.201100556. [DOI] [PubMed] [Google Scholar]
- 49.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotech. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 50.de Godoy LM, et al. Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature. 2008;455:1251–1254. doi: 10.1038/nature07341. [DOI] [PubMed] [Google Scholar]
- 51.Schröder BA, Wrocklage C, Hasilik A, Saftig P. The proteome of lysosomes. Proteomics. 2010;10:4053–4076. doi: 10.1002/pmic.201000196. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.