Significance
RNAi is an RNA-induced gene-silencing pathway that is shared among various organisms. Better understanding of RNAi is urgently needed to improve our knowledge of RNA-mediated gene regulation and to advance the field of functional genomics and its application to gene therapy. We counted with high precision the number of transcripts in each cell’s nucleus and cytoplasm as a function of silencing time to investigate the role of small RNA secondary structures such as loop length and stem mismatches. We screened various structural features of small RNAs and discovered a distinct role of each structural element that contributes to gene-silencing kinetics. We provide a helpful guideline for designing small RNAs for more efficient gene silencing.
Keywords: RNA interference, single-molecule fluorescence in situ hybridization, premiRNA, secondary structure, Ago loading
Abstract
PremicroRNAs (premiRNAs) possess secondary structures consisting of a loop and a stem with multiple mismatches. Despite the well-characterized RNAi pathway, how the structural features of premiRNA contribute to dicing and subsequent gene-silencing efficiency remains unclear. Using single-molecule FISH, we demonstrate that cytoplasmic mRNA, but not nuclear mRNA, is reduced during RNAi. The dicing rate and silencing efficiency both increase in a correlated manner as a function of the loop length. In contrast, mismatches in the stem drastically diminish the silencing efficiency without impacting the dicing rate. We show that this decoupling effect is not due to the loading to the RNA-induced silencing complex, RNA uptake, or cellular dicing. We postulate that the stem mismatches perturb the handover of the cleaved miRNAs from Dicer to Argonaute, leading to poor strand selection. Our results imply that the stem structures prevalent in cellular miRNAs have suboptimal silencing efficiency.
RNAi is an important biological pathway in which small RNA molecules bearing a sequence complementary to target mRNAs lead to the down-regulation of corresponding gene expression (1). It is conserved in a wide variety of organisms including animals, plants, and fungi and is predicted to influence more than 60% of human protein-coding genes (2). Due to the ease of applying small RNAs to deplete a specific gene of interest, RNAi has revolutionized the study of functional genomics and has paved the way for gene therapy (3–5).
RNAi is a multistep process that initiates from the biogenesis of miRNA or siRNA from precursor RNAs and results in the inhibition of a target gene by an RNA-induced silencing complex (RISC) (6–8). First, a precursor RNA (premiRNA or presiRNA) is trimmed down to 21- to 23-bp dsRNA (miRNA or siRNA) by a ribonuclease, Dicer (9–12). Second, Dicer hands over the cleaved miRNA or siRNA to Argonaute (Ago). Third, a guide strand, which bears complementarity to the target mRNA, is selected over a passenger strand via cleavage or unwinding of the passenger strand. Fourth, the guide strand is incorporated into an active RISC composed of Dicer, TAR RNA-binding protein (TRBP), and Ago (13–15). Fifth, the active RISC bearing a guide strand searches for a target mRNA by guide strand:mRNA complementarity (16, 17). Last, when the target is found, gene expression from the mRNA is down-regulated by either mRNA degradation or translational repression (18).
RNAs that induce gene silencing display two distinct structures, represented by premiRNA and presiRNA. PremiRNA is an endogenous, genome-encoded sequence transcribed by RNAP II in the nucleus, processed by Drosha/DGCR8, and exported from the nucleus to the cytoplasm (6). PresiRNA is typically exogenously applied to cells by various delivery methods. The majority of cellular premiRNAs possess a stem–loop structure in which the stem bears multiple mismatches, whereas presiRNAs are typically designed to be fully duplexed without mismatch. The effect of loop size on the dicing kinetics was reported by previously Tomari and coworkers (19) and Zeng and coworkers (20), but its relationship to the silencing efficiency has not been tested. While the effect of mismatches between the guide strand and target mRNA has been extensively studied (21–24), the effect of mismatch between guide and passenger strand has not been systematically examined.
We sought to study how different structural features found in premiRNAs and presiRNAs modulate overall gene-silencing efficiency by measuring two key steps in the RNAi pathway: (i) dicing kinetics to measure how quickly premiRNA or presiRNA is cleaved by Dicer, and (ii) silencing rate to quantitate how quickly cellular mRNAs are degraded as a result of silencing. We applied structural variants of silencing RNAs possessing different loop sizes or stem mismatches to learn about their role in altering dicing kinetics and mRNA silencing efficiency.
We further used single-molecule FISH (smFISH) (25, 26) to quantify cellular mRNAs with high accuracy. The smFISH technique enables the visualization of individual mRNAs as diffraction-limited spots in single cells without losing the spatial information of the mRNAs (27). In fact, mRNA imaging by smFISH enables spatial separation between nuclear mRNA and cytoplasmic mRNA in individual cells. Based on such spatial resolution, we demonstrate that cytoplasmic mRNAs are selectively lost during RNAi while nuclear mRNAs are maintained without change. We show that the overall RNAi efficiency depends heavily on the structure of the silencing RNA. RNA with a longer loop induces faster dicing and enhanced target gene silencing, indicating a correlation between dicing and silencing. For RNAs with no stem mismatches, dicing is the rate-determining step of RNAi. In contrast, for RNAs with mismatches in the stem, the dicing rate is not affected at all, but silencing efficiency is drastically diminished, strongly indicating a decoupling between dicing and silencing. We demonstrate that such decoupling is not due to differences in RNA uptake, guide strand selection, or mRNA target search, but likely is due to the step in which Dicer hands over the cleaved RNA substrate to Ago. Our study reveals how a longer loop size promotes dicing and silencing while mismatches on the RNA stem significantly diminish gene silencing without affecting dicing.
Results
Quantification of RNAi-Induced mRNA Reduction Using smFISH.
We employed smFISH to study RNAi with at a single-cell resolution. We chose a target gene for silencing to meet the following criteria. First, the number of transcripts per cell should not vary too much from cell to cell, since a homogeneous expression profile is critical for distinguishing RNAi-dependent changes from other stochastic RNA-level fluctuations caused by transcription activity and/or mRNA decay. Second, the number of mRNAs per cell should be sufficiently high so that mRNA reduction by RNAi can be tracked reliably. Third, the abundance or concentration of target gene should not exceed the spatially distinguishable density of the smFISH signal. Housekeeping genes are good candidates because they are constitutively expressed in all cells and are likely to exhibit low cell-to-cell variability in the numbers of mRNAs. Based on these criteria, we selected lamin A, a housekeeping gene, as our target gene, which displays 300–400 mRNAs per HeLa cell with low cell-to-cell variability (28).
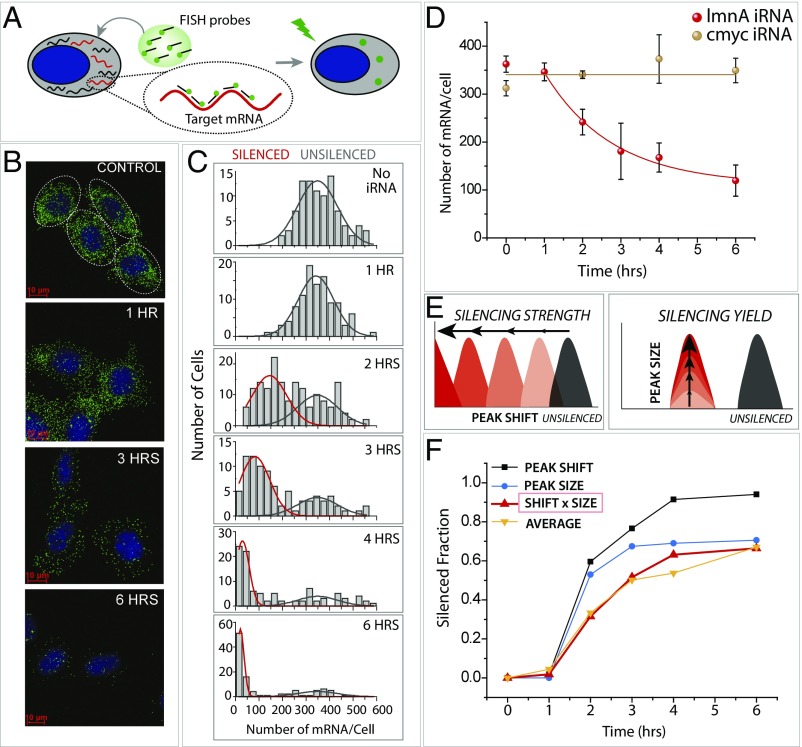
The smFISH probes were prepared by chemically synthesizing 32 DNA oligonucleotides (each 20 nt long), all possessing a complementary sequence to lamin A mRNA. They were each labeled with Cy3 dye (Methods and Fig. 1A). HeLa cells treated under different silencing conditions were fixed and permeabilized, and the FISH probes were hybridized to visualize individual lamin A mRNAs. The cells were also stained with DAPI to visualize the nucleus (Methods and Fig. 1A). The smFISH image in the control sample (no silencing) displays DAPI staining of the nucleus in blue and distinct green fluorescent spots representing individual lamin A transcripts, ∼350 mRNAs per cell (Fig. 1B, Top). We initiated silencing in HeLa cells by transfecting short dsRNAs designed to induce RNAi to silence lamin A. (For simplicity, we henceforth refer to the various types of small dsRNAs used for gene silencing in this study as “iRNA” for “interfering RNA.”) Ten minutes after transfection with iRNAs (U5 iRNA was used here; see Tables S1 and S2), the cell culture medium was refreshed to wash out excess iRNA. Cells were collected at 0, 1, 2, 3, 4, and 6 h after transfection and were prepared for smFISH imaging.
Fig. 1.
Quantitation of RNA silencing in single cells using smFISH. (A) smFISH is performed by applying 32 fluorescently labeled oligonucleotides bearing a sequence complementary to the target mRNA (lamin A) to fixed and permeabilized cells. It enables visualizing individual mRNA molecules in single cells. (B) smFISH images of individual cells in control cells (no iRNA) and 1, 3, and 6 h after iRNA treatment. (C) Quantification of smFISH data plotted as a function of hours after silencing. (D) Target-specific and nonspecific iRNA treatment leads to LMNA mRNA reduction only in case of lmnA iRNA. (E) Two types of parameters that characterize efficiency of RNAi are horizontal peak shift and vertical peak size reduction. (F) Peak shift, peak size, peak shift × peak size, and the average (average number of mRNAs tracked over time of silencing was converted to a silenced fraction) per cell plotted over time of RNA silencing.
Imaging involves taking 25–35 z-sections of every 300 nm across the entire cell thickness to capture all of the mRNAs in cells. Each green signal above a threshold intensity was counted as one single mRNA, and the number of mRNAs in individual cells was used to build a histogram at each time point. As expected from silencing, the number of mRNAs diminished over time (Fig. 1B). The histograms show primarily two peaks, with one representing the unsilenced population of cells (Fig. 1C, black outline fit) and the other peak corresponding to silenced cells (Fig. 1C, red outline fit).
To test the specificity of gene silencing, we treated the cells with an iRNA designed against the c-Myc gene for silencing and applied the same smFISH probes prepared for lamin A transcripts. The average number of lamin A mRNA over time after silencing revealed that only the iRNA against lamin A (red circles in Fig. 1D), but not the iRNA against c-Myc (brown circles Fig. 1D), reduced the number of lamin A mRNAs, confirming gene-specific silencing by the iRNA treatment. We obtained a similar silencing result in a different human cell line, A549, albeit with a slightly slower silencing kinetics (Fig. S1A). Interestingly, our result suggests that there is a finite time delay of at least 1 h after the iRNA is introduced to cells until mRNA reduction is observed (Fig. 1 C and D). This is not due to the process of RNA uptake, because we wash out the excess RNAs at 10 min after transfection, suggesting that RNA uptake is a rapid process that occurs within the first 10 min of transfection. Rather, the delay likely represents the time required for the iRNA to be released from endosomes that encapsulate the iRNA at the time of transfection, as well as the process of RNAi that precedes mRNA cleavage (Fig. 1 C and D).
The histogram analysis of single mRNA counts allows us to quantify the silencing effect in two ways. The first is the peak shift, which measures how much the center of the peak position moves from a high to a low number of mRNA, reflecting silencing strength. The second is the peak size, i.e., the fraction of cells in the silenced population, indicating silencing yield (Fig. 1E). Together, both the peak shift and peak size report on the efficiency of gene silencing. Therefore, we analyzed both parameters for all the data obtained for this study, as shown in the silencing histogram in Fig. 1C and the analysis of that histogram in Fig. 1F. The silenced cell population fitted with red curve shifted to the left while the unsilenced population reduced over time without a peak shift (Fig. 1C). Our analysis shows that while the peak shift approaches 90%, the peak size reaches ∼70%, suggesting that ∼30% of cells do not undergo efficient gene silencing up to 6 h after inducing RNAi at the tested condition. To test if the unsilenced population is the result of inefficient RNA uptake, we quantified the cellular RNA uptake by using fluorescently labeled iRNA. We observed 98% efficiency (Fig. S1B), suggesting highly efficient RNA uptake by nearly all cells. We also questioned if the inefficient silencing in 30% of cells is due to the iRNA concentration used for silencing. To address this question, we transfected cells with 1 nM iRNA, which is 30-fold lower than that used in Fig. 1, and found that 1 nM iRNA induced a higher unsilenced population (Fig. S1C). However, the unsilenced population was reduced significantly at 24 h regardless of iRNA concentration, suggesting that the unsilenced cell population comes from slower gene silencing and not from failed RNA uptake (Fig. S1C). Our analysis scheme has the advantage of reporting on the peak size, which provides a proxy for the homogeneity of RNAi activation or kinetics, and the peak shift, which represents silencing strength among various iRNA structures.
Nuclear mRNA Remains Unchanged While Cytoplasmic mRNA Undergoes Silencing.
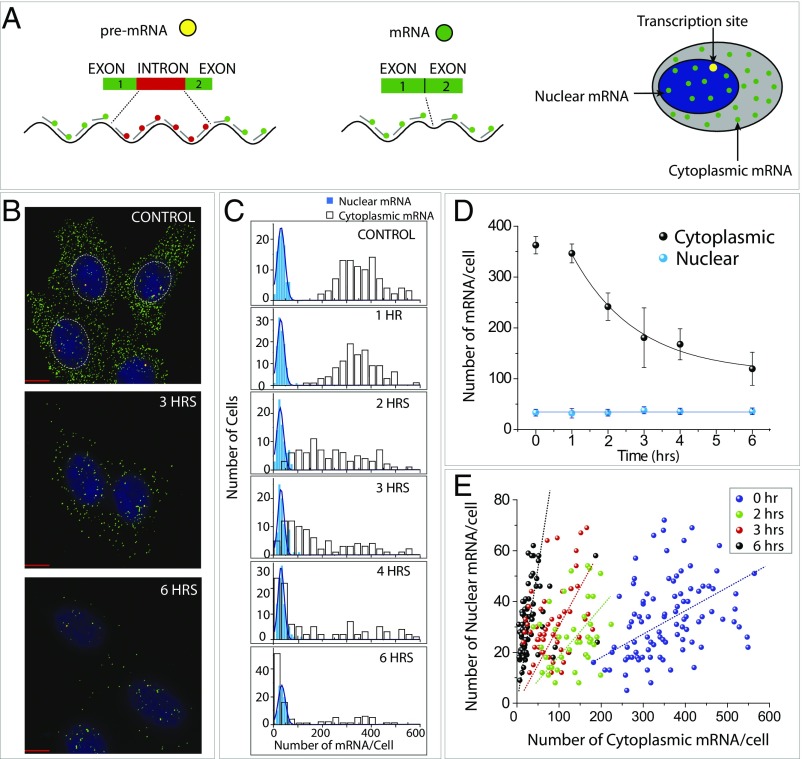
The smFISH method enables not only the quantification of mRNA but also the localization of individual transcripts in cells with high spatial resolution. Based on the clear separation between nucleus and cytoplasm enabled by DAPI staining, we counted the nuclear and cytoplasmic pools of mRNA as a function of silencing time. In the nucleus, there are two types of smFISH signals: mature mRNA and the pre-mRNA, which has not been spliced, likely located at the site of active transcription. To distinguish between these two types of mRNAs, we performed dual-color smFISH in which the exon and intron were color-coded differently. The FISH probes for the exon and intron were labeled with Cy3 (green) and Cy5 (red), respectively. In this scheme, pre-mRNA appeared as yellow (overlap of green and red) and the mature mRNA as green fluorescent spots (Fig. 2A). The representative dual-color smFISH image displays both yellow and green spots in the nucleus (Fig. 2B; the periphery of the DAPI-stained nucleus is marked with a white dashed circle).
Fig. 2.
RNAi reduces cytoplasmic mRNA without changing nuclear mRNA. (A) Visualization of nuclear mRNA by using dual-color smFISH including green-labeled probes designed against the exon and the red probes against the intron. Yellow spots are produced by the overlap of green and red represent active transcription sites, whereas green spots are nuclear mRNA. (B) Representative dual-color smFISH image displaying the exon and intron of lamin A mRNA taken at 0, 3, and 6 h after iRNA treatment. (C) The histogram shows nuclear (blue bars) and cytoplasmic (white bars) mRNAs as a function of silencing time. (D) Count of nuclear (blue) vs. cytoplasmic mRNA (black) per cell. (E) The number of nuclear mRNA per cell plotted against number of cytoplasmic mRNA per cell for 1, 2, 3, and 6 h postsilencing.
We plotted the number of the nuclear mRNA (light blue bars) vs. cytoplasmic mRNA (white bars) in individual cells over the time of silencing (Fig. 2C). This revealed that the nuclear mRNA, excluding the pre-mRNA, was maintained at a similar number, in contrast to the rapidly reducing numbers of cytoplasmic mRNA induced by RNAi (Fig. 2 C and D). The average number of nuclear lamin A mRNA is about 40, which is approximately one tenth of the cytoplasmic mRNA in unsilenced cells (Fig. 2D). An independent plot of nuclear mRNA vs. cytoplasmic mRNA in individual cells over time of silencing shows the spread of nuclear mRNA between 10 and 70 with most cells exhibiting 20–50 mRNAs per nucleus (Fig. 2E). This plot revealed a positive correlation between the number of cytoplasmic and nuclear mRNA regardless of RNAi condition (Fig. 2E). The slope of 0.09 for unsilenced cells (0 h) suggests the ratio of nuclear:cytoplasmic RNA is ∼1:10 (Fig. S2, Left). When this balance is perturbed by RNAi, the ratio changes approximately to 1:5 (2 h), 1:3 (4 h), and 1:1 (6 h).
Next, we tested the protein level of lamin A over time after silencing using immunofluorescence. The reduction kinetics of the lamin A protein revealed an ∼36-h delay compared with mRNA silencing (Fig. S2, Right). Such delay may scale with the doubling time of HeLa cells and the turnover rate of lamin A protein, based on the previous work which reported the characteristic cell cycle for HeLa cell as about 20 h and the median half-life of the 4,100 proteins measured in a nondividing HeLa cell as 36 h (29). Taken together, our results suggest that the number of the cytoplasmic mRNA, rather than the number of nuclear mRNA or the protein level, is the best proxy for evaluating the silencing efficiency. Therefore, we focus on cytoplasmic mRNA quantification as a primary means of evaluating the role of iRNA structure in RNAi efficiency.
Dicing Kinetics Correlates with Gene-Silencing Efficiency.
The RNAi pathway entails sequential steps of distinct biochemical reactions (Fig. 3A). First, the dsRNA (premiRNA or presiRNA) undergoes cleavage by Dicer, assisted by TRBP. The cleaved dsRNA is handed over from Dicer to Ago. The strands of the dsRNA are separated, and the guide strand (displayed as red line in Fig. 3A) is selected and used for mRNA target search. Upon finding the target, the mRNA is degraded or held from translation in the context of P-body, both resulting in translational repression. Along the RNAi pathway, a dsRNA morphs into three different forms starting with the initial dsRNA with or without a loop (Dicer substrate), followed by a shortened dsRNA after dicing (Dicer product), and subsequently to two types of ssRNA after strand selection, guide and passenger strand (Fig. 3A). siRNAs or miRNAs that are short enough to be loaded to Ago can bypass the dicing step (30). We sought to examine if the dicing kinetics and RNAi efficiency are related by testing series of Dicer substrates that require dicing before silencing. For this section, we designed series of dsRNAs that will yield the same Dicer product, so that the steps that follow dicing will be identical (Table S3). This strategy ensures that the difference seen in silencing can be attributed to the dicing step.
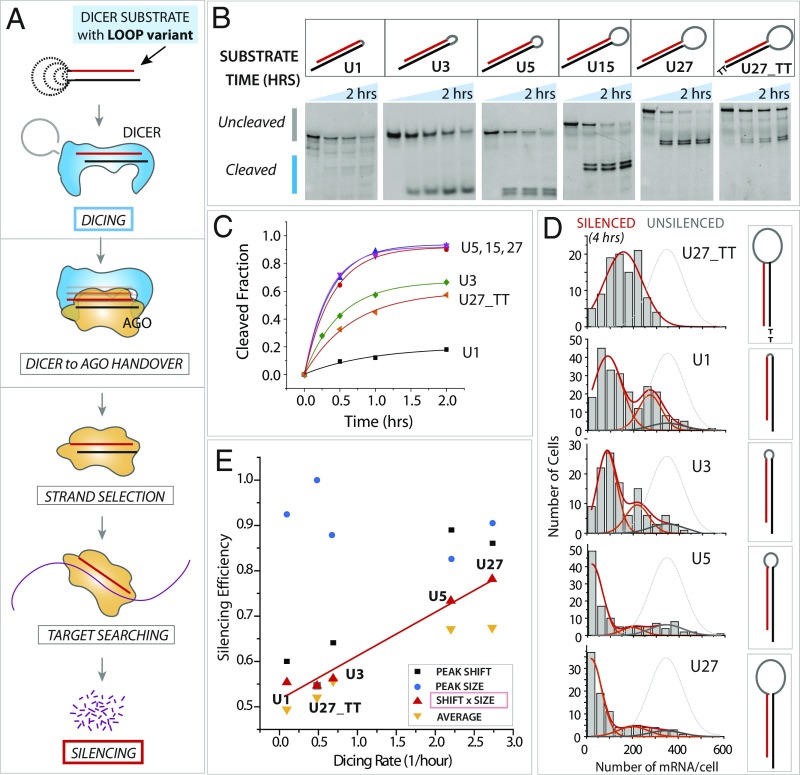
Fig. 3.
Dicing kinetics is correlated with gene-silencing efficiency. (A) Schematic of the RNAi pathway, which involves dicing, Dicer-to-Ago handover, strand selection, target searching, and mRNA silencing. (B) Dicing results on loop length variants of U1, U5, U15, U27, and U27 with TT overhang. (C) Quantification of dicing rate. (D) mRNA histogram of loop length variants after 4 h of silencing. (E) Silencing efficiency plotted against dicing rate for loop length variants. The peak size (blue symbols) is similar for all RNAs, but the peak shift (black symbols) and shift × size (red symbols) show a linear correlation between the dicing rate and silencing efficiency. See Fig. 1 E and F for fitting parameters.
First, we investigated the effect of loop length on RNAi efficiency. The loop is the main feature that differentiates premiRNA (with a loop) from presiRNA (without a loop). Evidence indicates that having a loop (premiRNA) accelerates dicing kinetics and that a larger loop size further enhances the cleavage rate by Dicer (19, 20, 31). What is unclear is whether that relationship is also reflected in mRNA silencing, i.e., if a longer loop length, which induces a faster dicing rate, results in more efficient gene silencing. For example, if postdicing steps include major rate-limiting steps, the difference in the dicing rate among loop variants would be masked. In other words, the correlation between the dicing rate and RNAi efficiency can be expected only if the rate-limiting step is the dicing step. We prepared series of hairpin-structured dsRNAs with different sizes of loop (1, 3, 5, 15, or 27 nt) referred to as “U1,” “U3,” “U5,” “U15,” and “U27,” respectively, since the loop sequence was composed of polyuracil to avoid unintended secondary structures within the loop. Each substrate was subjected to a dicing assay in which fluorescently labeled RNA substrates were incubated with purified human Dicer and the cleaved products were resolved on denaturing PAGE and visualized by a fluorescence gel imager (Methods and Fig. 3B). Consistent with the previous reports (20, 32), the dicing rate was faster for dsRNA with a longer loop size (Fig. 3C). We note that the multiple bands seen in U1 are likely caused by a short loop that distorts the helical structure of the stem. This pattern disappears from U3, suggesting U3 and the longer loops than U3 allow proper formation of the RNA stem.
In parallel, the same set of substrates was applied to cells to silence the lamin A gene. The silencing efficiency was quantified by counting the number of mRNAs (smFISH spots) after 4 h of RNA transfection in HeLa cells (Fig. 3D), because it is difficult to observe differences in silencing at longer time point when most cells already have undergone gene silencing regardless of different silencing kinetics (Figs. 1 and 2). The smFISH data analyzed from individual cells and compiled to histograms clearly show that silencing is more efficient with longer looped substrates (5–27 nt), as evident from the higher degree of peak shift observed for U5 and U27 (Fig. 3D). The histogram was dissected into a peak size and peak shift analysis, as before (Fig. 1 E and F). Silencing efficiency (peak shift × peak size) plotted against the dicing rate for the loop-length variants shows that dicing kinetics and silencing efficiency are correlated, suggesting that the dicing step is likely the rate-limiting step for these substrates (Fig. 3E). The dicing rate and silencing efficiency measured for U27_TT (which is identical to U27 except for two deoxy-thymidines at the 3′ overhang) yielded slow dicing and low silencing efficiency, hence the data located along the linear fit (Fig. 3E). This is likely due to the TTs reducing the affinity of RNA to Dicer, consistent with a previous report (33).
Interestingly, for all histograms, only the peak shift, but not the peak size, correlated with the dicing rate (Fig. 3E). The constant level of peak size (light blue circles) shows that the fraction of cells undergoing active silencing is similar, independent of loop size or a DNA tail of iRNA. In contrast, the different levels of peak shift (black squares) and the product of size and shift (red triangles) indicate that silencing efficiency varied substantially depending on the loop size (Fig. 3E). The averaged silencing efficiency, which was independently calculated by taking the average number of mRNAs in all cells, also correlated with the dicing rate, further supporting the correlation between the dicing kinetics and the gene-silencing efficiency (Fig. 3E). Taking these findings together, we show that the small loop size in the range of 1–3 nt leads to slow dicing, giving rise to inefficient silencing, whereas longer loops of 5 nt or above induce fast dicing and efficient silencing. Such correlation was also observed when the loop sequence composition was changed from polyuracil to mixed bases taken from prelet7 miRNA (Fig. S3), supporting the dicing step as the rate-limiting step.
Stem Mismatch Breaks the Correlation Between Dicing and Silencing.
Next, we studied the effect of mismatches in the stem, which are prevalent in most miRNAs but not in siRNAs. Previous studies reported on the effect of mismatches in Ago loading and unwinding of the siRNA substrates (34, 35). However, how mismatches in the stem influence RNAi efficiency remains elusive. For this study, we prepared a series of RNA substrates with the same loop length (U27) and one or multiple mismatches along the duplex stem. In all RNAs, we kept the same guide strand so that the subsequent downstream processes, including mRNA target search, pairing with the target mRNA, and gene silencing, will not differ among different stem variants. Therefore, the stem variants were created by introducing single and multiple mismatches exclusively on the passenger strand (Fig. 4A). The positions of the multiple mismatches were adopted from a well-known miRNA structure, prelet7 (36).
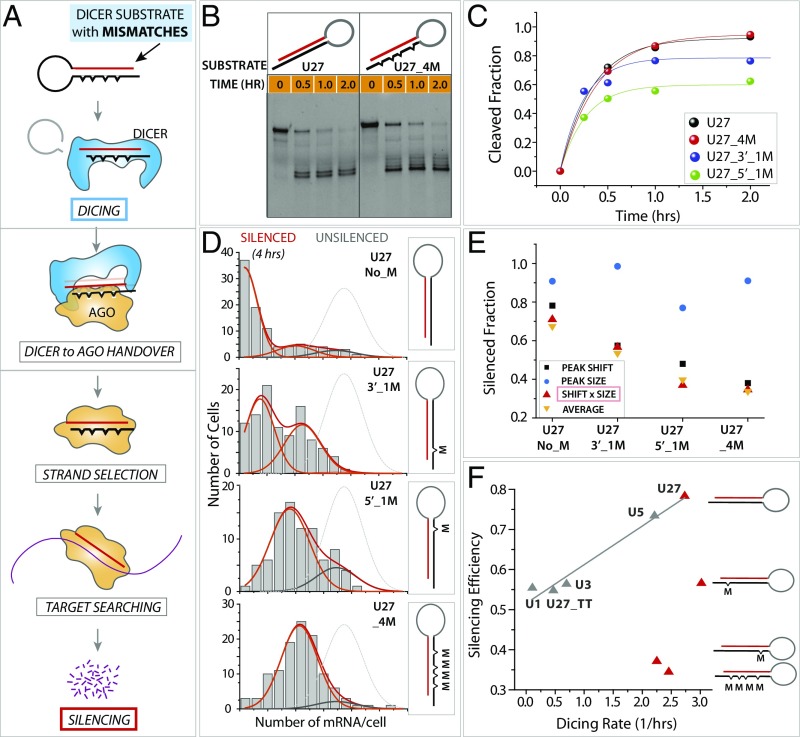
Fig. 4.
Stem mismatch breaks the correlation between dicing and silencing. (A) Schematic of the RNAi pathway. Small RNAs with varying numbers of stem mismatches are applied to dicing and silencing measurement. (B) Dicing result of U27 with no mismatch and U27_4M with four mismatches in the stem. (C) Quantification of dicing kinetics. (D) mRNA histogram of silencing induced by different stem-mismatched RNA substrates. (E) Analysis of the histogram in D for the four stem-mismatch variants. (F) Silencing efficiency plotted against dicing rate for stem-mismatch variants.
When we performed the dicing assay using stem structure-mismatch variants with no to four mismatches along the stem, we obtained similar dicing kinetics for all dsRNAs possessing no mismatch to four mismatches along the stem, strongly suggesting that the mismatches on the stem do not hamper the RNA cleavage activity by Dicer (Fig. 4 B and C). In contrast, the RNA silencing induced by the same set of stem-mismatch variants revealed a dramatic difference in silencing efficiency. Remarkably, the constructs with mismatches displayed drastically diminished silencing efficiency, as seen by inefficient mRNA reduction at 4 h after iRNA transfection, compared with the construct with no mismatch (Fig. 4D). Dissection of the smFISH data by peak size and peak shift shows that while the peak size remains unchanged, peak shift shows a substantially reduced silencing. Again, the peak shift reflects the silencing strength while peak size serves to confirm that RNAi is equivalently active in all conditions (Fig. 4E). In summary, the mismatches along the dsRNA stem significantly diminish the RNAi efficiency without influencing the dicing kinetics and thereby break the correlation between the dicing rate and the RNAi efficiency (Fig. 4F).
To test if this decoupling effect arises from a gene-specific case, we tested dicing and silencing of an unrelated gene, poly(A)-binding protein C (PABPC). In the same manner, we prepared iRNAs containing unstructured or structured stems designed against PABPC mRNA and applied them to dicing and silencing platforms. We observed that the dicing rate was similar, but the silencing efficiency was greatly diminished by the stem mismatches, strongly supporting the stem mismatch-dependent decoupling of dicing and silencing (Fig. S4). Next, we tested if the observed decoupling effect of dicing and silencing may result from in vitro dicing rates generated from using purified Dicer. To test the cellular dicing rate, we used HeLa cell lysates and fluorescently labeled iRNA substrates with and without stem mismatches and probed for the cleaved vs. uncleaved RNA. The result shows that the cellular dicing rate is comparable in the two substrates, ruling out the possibility that the low silencing by stem-mismatched RNA is due to a decreased cellular dicing rate (Fig. S5A). We also checked if the effect might result from less-efficient iRNA uptake caused by the stem mismatches. To check this, we transfected cells with eight different fluorescently labeled iRNAs. Then, each iRNA was isolated from the cell lysate and quantified by gel electrophoresis. When the iRNA uptake was plotted against RNA silencing efficiency, we did not see any correlation, indicating that the RNA uptake is not responsible for the different silencing efficiency of iRNAs with different structures (Fig. S5). Taken together, our findings suggest that the low silencing efficiency induced by stem mismatches is not due to a gene-specific effect, discrepancy between in vitro and cellular dicing, or RNA uptake but results from postdicing steps of RNAi. The observation that RNAi efficiency is greatly diminished by the stem mismatches is intriguing, given that most cellular miRNAs harbor mismatches along the stem (37).
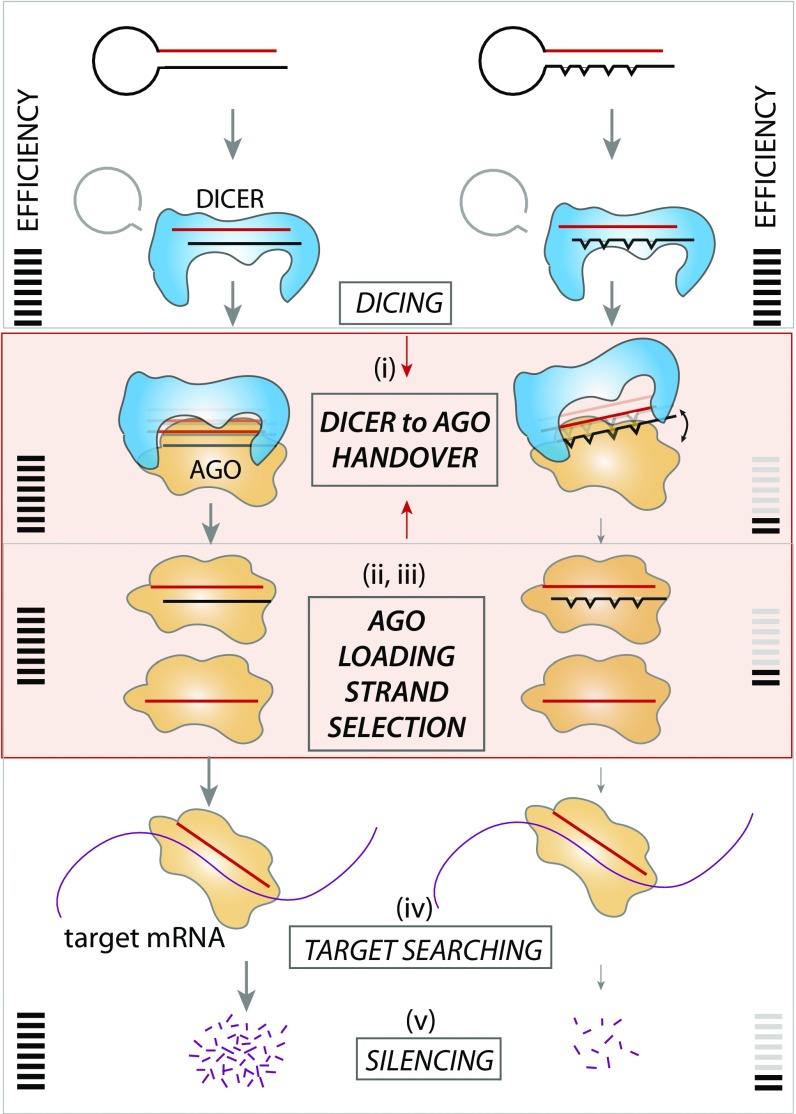
Stem Mismatch Impedes Dicer-to-Ago Handover and Subsequent Ago Loading.
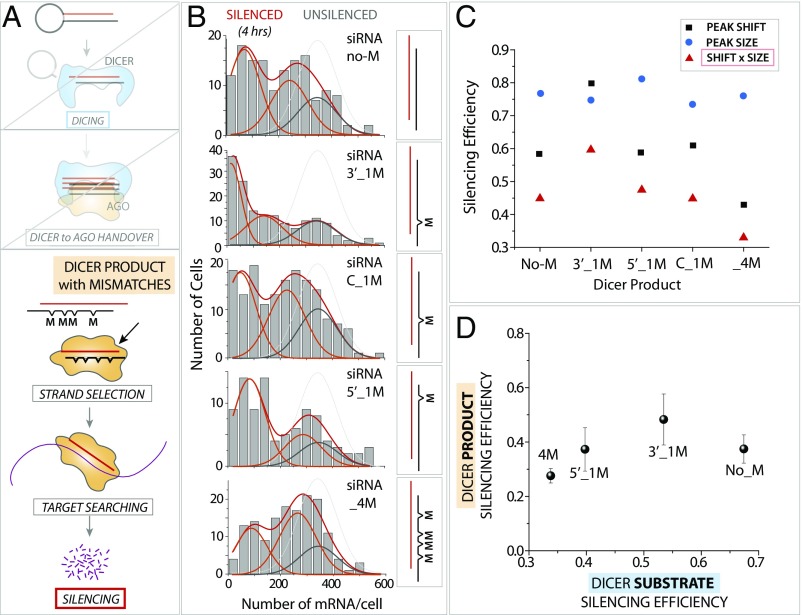
Which postdicing step may be responsible for the poor silencing induced by stem mismatches? The possible rate-limiting steps in the postdicing process include (i) the process by which Dicer hands over cleaved RNA product to Ago; (ii) the unwinding of the guide and passenger strands; (iii) strand selection; (iv) target search; and (v) mRNA degradation (see Fig. 6). We can rule out steps of iv and v (target search and mRNA degradation), because all iRNA substrates used in the current study have an identical guide strand, which should produce the same outcome for target search and mRNA degradation. To address if step ii, unwinding of guide and passenger strand, or iii, strand selection, is responsible for the decoupling effect, we prepared iRNAs that represent the precleaved Dicer product that can be directly loaded to Ago for further downstream processing. The precleaved RNA enables bypassing both the dicing reaction and Dicer-to-Ago handover steps, thereby probing only the steps starting after the Ago loading (Fig. 5A). Therefore, this scheme allows one to distinguish between pre-Ago loading and post-Ago loading. If the post-Ago loading is the major rate-limiting step responsible for the reduced silencing efficiency induced by stem mismatches, we would expect to observe the same degree of difference in Dicer products as in Dicer substrates. We prepared a series of Dicer products that are devoid of loop structure and contain mismatches identical to the Dicer substrates used above (Fig. 4). As before, we quantified the RNAi efficiency after 4 h of RNA transfection using smFISH.
Fig. 6.
Summary: Mismatches in the stem of dsRNA do not interfere with dicing but drastically diminish silencing efficiency. Our result reveals that this is likely due to perturbed Dicer-to-Ago handover in which mismatches in the stem disrupt the A-form helical structure of dsRNA, making it difficult to fit into Ago pocket.
Fig. 5.
Stem mismatch does not impact downstream of AGO. (A) Schematic of the RNAi pathway. A Dicer product bearing the same mismatch variation as the Dicer substrate is applied to the silencing assay. (B) mRNA histogram for Dicer product with varying mismatches. (C) Silencing efficiency induced by the Dicer product with the stem mismatches. (D) Silencing efficiency of the Dicer product with varying mismatches plotted against the silencing efficiency of the Dicer substrate with the same mismatches.
Overall, the silencing efficiency for the Dicer products was lower than that for the Dicer substrates that require dicing (Fig. 5B). Interestingly, a 3′ single mismatch produced more efficient silencing than the rest of the substrates, including the one with no mismatch (Fig. 5 B and C). The analysis of peak size and shift demonstrated again that the peak shift, not peak size, reflected the difference in gene silencing by differently structured iRNAs (Fig. 5C). To compare the stem-mismatch effect in Dicer product vs. Dicer substrates, we plotted the two sets of data against each other (Fig. 5D). As seen, while stem mismatches produced substantial differences in Dicer substrate, the same mismatches in Dicer product did not exhibit much variation in silencing efficiency, as indicated by a narrow distribution in the y axis (Fig. 5D). This result indicates that the stem mismatches do not significantly alter the steps downstream of Ago loading.
To further analyze the effect of mismatch in lowering silencing efficiency, we tested if and how Ago loading outcome is modulated by mismatches in the RNA stem. We prepared doubly labeled iRNAs (guide strand labeled with Cy3 and passenger strand with Cy5) with or without mismatches, applied them to HeLa cell lysate, and performed an EMSA (Fig. S6A). The result reveals several important ways in which silencing efficiency is modulated by stem mismatches. First, the overall Ago loading is lower in the case of four mismatches (Fig. S6B, total in navy blue), which is likely due to inefficient handover of diced RNA from Dicer to Ago2. Second, the presence of mismatches interferes with the guide strand loading to Ago2 (Fig. S6B, guide in green). Third, the mismatches promote the nonproductive passenger strand loading to Ago2 (Fig. S6B, passenger in red). All three effects together contribute to the reduction of silencing efficiency. This result also implies that the mismatches on the RNA stem may lead to compromised strand-selection bias, leading to increased formation of passenger–Ago complexes. Taken together, these results show that, while stem mismatches do not interfere with RNA uptake, dicing, and post-Ago loading, they impede Ago-loading efficiency and strand-selection accuracy. Therefore, we conclude that the stem mismatches likely hinder steps i–iii, the process by which Dicer hands over cleaved RNA product to Ago and the concomitant selection of the guide strand (Fig. 6).
Discussion
In this study, we took advantage of the smFISH technique, which enables single mRNA counting in single cells. Even though the smFISH cannot report on the translational repression induced by RNAi, it enables precise quantification of RNAi at the mRNA level. We uncovered features of the RNAi pathway that cannot be addressed by other preexisting mRNA detection methods. Conventional methods for mRNA detection require pulling mRNA from a batch of cells to generate enough signal for quantification, averaging out the mRNA level over a large cell population. The use of smFISH to study RNAi provides several unique advantages over the conventional mRNA detection methods: (i) highly accurate and direct mRNA quantification without the need for amplification; (ii) the ability to count single mRNA molecules in single cells; (iii) image-based analysis providing localization of individual mRNAs, enabling quantitation of nuclear mRNA and cytoplasmic mRNA separately; and (iv) (related to ii and iii) capturing cell-to-cell variation in mRNA copy number. Interestingly, we observed cell-to-cell heterogeneity in response to RNAi represented by the distinct unsilenced and silenced cell population (Fig. 1C).
The independent detection of cytoplasmic and nuclear mRNA revealed that RNAi reduces only cytoplasmic mRNA without affecting nuclear mRNA. It has been reported that RNAi happens mainly in the cytoplasm, not in the nucleus, because Ago is primarily present in cytoplasm (38–40). On the other hand, several reports suggested the possibility of RNAi activity in the nucleus (41). Indeed, the nuclear RNAi pathway plays an important role in the centromere function of Schizosaccharomyces pombe and in the sexual reproduction of ciliate Tetrahymena. In addition, some compartments of RNAi exist in the nucleus of NRDE-3 in Caenorhabditis elegans and in Piwi proteins in Drosophila and mouse. Our results demonstrate that only the cytoplasmic mRNA is reduced significantly by RNAi, while the nuclear mRNA is maintained at a constant level in HeLa cells, suggesting that RNAi mainly impacts cytoplasmic mRNA in mammalian systems. Such results point toward a regulatory mechanism that sustains a steady level of nuclear mRNA concentration regardless of the changes in the cytoplasmic mRNA. One possibility is that the rate of transcription and the rate of mRNA export from the nucleus to the cytoplasm are kept in balance to maintain a constant mRNA concentration in the nucleus. In addition, we observed a positive linear correlation between the number of mRNAs in the nucleus and in the cytoplasm regardless of RNAi (Fig. 2E). This may reflect the abundance of mRNA that scales with the cellular volume to maintain a constant mRNA concentration in individual cells (28). For example, smaller cells may have lower numbers of both nuclear and cytoplasmic mRNA, and vice versa. Furthermore, this reflects another layer of regulation, which may involve an active communication between the nucleus and cytoplasm to maintain a homeostasis of mRNA numbers across both compartments. When such balance is perturbed by RNAi, the nuclear mRNA is sustained, but the cytoplasmic mRNA is selectively lost.
It is critical to understand how the structure of iRNA affects the RNAi pathway, since such knowledge can be applied directly to enhance RNAi efficiency, especially for biomedical purposes. We chose to investigate major structural features present in majority of premiRNAs. First, we examined the effect of loop size, since all premiRNAs possess a loop that requires cleavage by Dicer. Our results indicate that a bigger loop size induces faster dicing and more efficient silencing. The loop-size variants displayed a highly linear correlation between dicing rate and silencing efficiency, implying that dicing is a rate-limiting step that determines silencing outcome. Second, we modulated the stem structure by introducing one or multiple mismatches on the passenger strand. Most cellular miRNAs contain mismatches in the central region as well as in the seed and/or 3′-mid regions of the stem (37). In our measurement, all stem variants showed the similar dicing kinetics, consistent with the previous report (20), but showed significantly reduced silencing efficiency. RNA composition can influence Dicer-mediated cleavage and corresponding silencing efficiency (42), and thermal stability of the stem may contribute to Dicer cleavage (43). Nevertheless, the mismatches we generated on the stem of dsRNA did not alter the cleavage preference by Dicer, suggesting that dicing is not a rate-determining step for silencing efficiency for the stem variants.
Previous studies on mismatches between a guide strand and target mRNA demonstrated that the complementarity of the seed region is critical for target search and that mismatches in the center inhibit the target cleavage (44–46). However, the impact of mismatches between guide and passenger strand on the RNAi pathway has not been carefully examined. Therefore, our study is significant in unveiling the functional role of miRNA structure. Based on our results, we postulate that the mismatched stem structure interferes with the process in which Dicer hands over the cleaved RNA product to Ago. This may be explained by the structure of human Ago2, which organizes dsRNA into an A-form helix by extensive interactions with the RNA backbone (47, 48). Such structural constraints embedded in Ago2 are well suited for accommodating a dsRNA in A-form helical shape. Therefore, dsRNA with mismatches that distort and disrupt the A-form helical structure will not fit into the RNA-binding pocket of Ago2. Such steric hindrance may contribute to inefficient silencing. Based on our results shown in Fig. 5, such an effect may not be distinguished when siRNA (Dicer product) engages with Ago, since this loading process appears to be inefficient in general. In contrast, Dicer may hand over the cleaved product to Ago for the iRNA without mismatches proficiently but may do so less proficiently for the substrates with mismatches.
Our finding that the stem mismatches diminish the efficiency of gene silencing (Fig. 4 D–F) is not fully consistent with previous reports which showed that central mismatches promote Ago loading and that seed or 3′-mid mismatches facilitate unwinding (34, 35). Our result showing higher RNAi efficiency for the Dicer product with a single 3′ seed mismatch than for the Dicer product with no mismatch agrees with the previous studies. However, these results cannot be compared directly because of differences in the RNA constructs used in this study and the previous studies, which have not extended their findings on dicing and unwinding to mRNA silencing in cells.
In addition to loop size and stem mismatches, we investigated the effect of DNA substitution at the 3′ overhang. The DNA substitution of a 3′ overhang is expected to protect the 3′ end structure from processive 3′-exonuclease in serum, which can lead to more efficient RNAi (49). In addition to DNA substitution of a 3′ overhang, various chemical modifications at the 3′ end of RNA have been developed to protect against exonuclease digest and thereby improve RNAi efficiency (50). In our measurement, DNA substitution increased the RNAi efficiency for Dicer product, which agrees with the protective role of the DNA overhang, but decreased the efficiency for Dicer substrate (Fig. 3 and Fig. S6). This difference may be explained by the following. Dicer product is loaded onto Ago, which binds DNA and RNA overhang indiscriminately (51). In contrast, Dicer substrate requires loading onto Dicer, which displays lower affinity toward the DNA overhang (20). Therefore, despite the protective cap provided by the DNA overhang, it is favorable only in the case of Dicer product. This suggests that the chemical modifications to protect the 3′overhang may not necessarily guarantee a higher efficiency of RNA silencing.
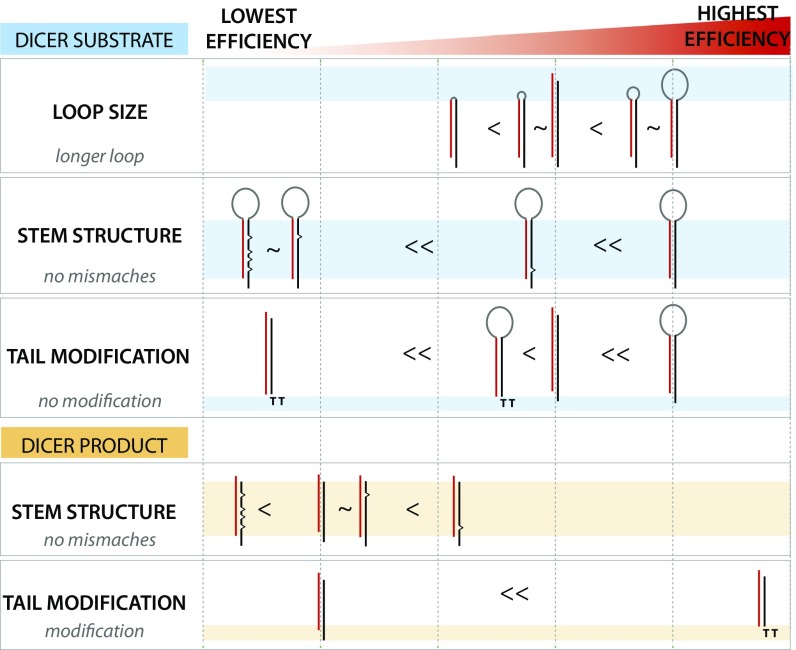
We summarize how the dsRNA structures such as loop size, stem mismatches, and 3′ overhang affect RNAi efficiency in Fig. 7. Dicer substrates (Top, blue stripe) containing no stem mismatches show more potent gene-silencing efficiency than Dicer product (Bottom, orange stripe) in general, which is consistent with the previous report (52). However, this trend does not hold when the stem mismatches or DNA overhang are incorporated. The stem mismatches strongly inhibit the RNAi efficiency of Dicer substrate, and the DNA substitution of the 3′ overhang in the Dicer product showed substantially enhanced RNAi efficiency compared with the one in the Dicer substrate. This information provides a guideline for designing a synthetic dsRNA for efficient gene silencing. Taken together, our study defines the role of two primary structural components of premiRNA—the loop and the stem. While loop size correlates well with both dicing and silencing, stem mismatch greatly interferes with silencing efficiency without compromising the dicing rate. Furthermore, this striking finding implies that the majority of cellular miRNAs, which harbor extensive stem structures, are likely built for less-than-optimal silencing efficiency. Our results also suggest that disease-linked mutations that occur in miRNA coding genes may act by altering miRNA’s stem structure, thereby modulating the gene-silencing efficiency.
Fig. 7.
Silencing efficiency of structured RNAs. RNA substrates tested in this study are ordered from the lowest to highest efficiency of gene silencing from left to right. (Upper) Dicer substrate. (Lower) Dicer product. Each panel is subdivided into structural variants.
Methods
Preparation of iRNA and smFISH Probes.
All the iRNAs were chemically synthesized and purified using RNase-free HPLC by Integrated DNA Technologies, and all of the smFISH probes were designed and synthesized with 5′ amine modification by LGC Biosearch Technologies. The sequences of iRNAs and smFISH probes are described in Tables S1–S3. We labeled 0.5–2 nmol of the smFISH probes or iRNA with fluorophores by incubating them with 0.2 or 0.1 mg of Cy3 or Cy5 (GE Healthcare) in 50 μL of 100 mM NaHCO3 buffer, pH 8.5, overnight and removed the excess dye by two rounds of ethanol precipitation. For ethanol precipitation, we mixed the labeling mixture with 2.5 volumes of ice-cold 100% ethanol and 1/10 volume of 5 M NaCl, incubated them at −20 °C for 30 min, and then centrifuged them down at the maximum speed (>14,000 × g) for 30 min. The precipitated pellets were recovered by removing the supernatant and were rinsed using ice-cold 70% ethanol. After ethanol precipitation, the pellets of iRNAs or smFISH probes were dried in the air at room temperature and were dissolved in T50 buffer [10 mM Tris (pH 7.5) and 50 mM NaCl]. The single-stranded iRNAs were annealed to generate duplex or stem–loop–structured iRNAs in T50 buffer by heating them at 90 °C for 2–5 min and cooling them slowly to room temperature for duplex structures or quickly on ice for stem–loop structures.
Cell Culture and Transfection.
Human HeLa cells were cultured at 37 °C in (DMEM; Thermo Fisher) supplemented with 10% FBS (Corning Cellgro), 1% penicillin-streptomycin 100× (Corning Cellgro), 2 mM l-glutamate (Thermo Fisher), and 1 mM sodium pyruvate (Thermo Fisher). Human A549 cells were cultured at 37 °C in RPMI medium 1640 (Thermo Fisher) supplemented with 10% FBS (Corning Cellgro) and 1% penicillin-streptomycin 100×. The cells were plated on two-well culture chambers (Lab-Tek) 1 d before transfection of iRNA that induces gene silencing for the smFISH experiment. Synthetic iRNAs (3 or 30 nM) were transfected in cells using RNAiMax reagent (Invitrogen).
The use of mammalian cells was approved by Johns Hopkins Homewood Institutional Review Board.
Quantification of RNA Uptake by RNA Extraction from Cell Lysate.
To quantify RNA uptake by transfection, we seeded HeLa cells in a 60-mm culture dish and transfected them with 3 nM of Cy3-labeled iRNA for 4 h using RNAiMax reagent. We rinsed out the Cy3-labeled iRNAs that had not been taken up. Then we extracted total RNAs from cells using the mirVana miRNA Isolation Kit (Thermo Fisher Scientific) and performed a denaturing PAGE at 200 V for 35–40 min. We imaged the gel and measured Cy3 intensity to quantify RNA uptake using Amersham Imager 600 (GE Healthcare).
Dicing Assay Using Denaturing PAGE.
We mixed 20 nM of Cy3-labeled dsRNA and 25.6 μM of purified human Dicer in dicing buffer [20 mM Tris (pH 7.5), 25 mM NaCl, 1 mM DTT and 3 mM MgCl2] and incubated the mixture for 0, 15, 30, 60, or 120 min at 37 °C. Then we stopped the dicing reaction by adding 10% of glycerol, heating it for 2 min at 90 °C, and cooling it quickly on the ice and performed a 15% denaturing polyacrylamide/urea gel electrophoresis (Bio Rad) at 200 V for 35–45 min. We took the gel image using an Amersham Imager 600 for quantification. For the cellular dicing assay, we mixed 20 nM of doubly labeled dsRNA with cell lysates instead of the purified human Dicer.
Sample Preparation for smFISH.
We fixed cells using 4% paraformaldehyde for 10 min at room temperature and permeabilized them using either 70% ethanol for 1 h or 0.5% Triton-X for 10 min. We incubated the permeabilized cells with the mixture of hybridization buffer [10 (wt/vol) % dextran sulfate, 10% formamide and 2× SSC] and the FISH probe set at a final concentration of 2.5 nM overnight. The hybridized cells were incubated twice in wash buffer (10% formamide and 2× SSC) for 30 min at 37 °C and were stored in PBS for imaging. The cells were stained with 5 μg/mL of DAPI for 10 min at room temperature before imaging.
Fluorescence Imaging of smFISH and Immunostaining and Single mRNA Counting Using FISH-QUANT.
We used a Zeiss ApoTome microscope and a Nikon Ni-E microscope for smFISH imaging and a Zeiss LSM 600 confocal microscope for immunofluorescence imaging. To obtain the 3D image of entire cells, we performed multiple z-stack images at a 300-nm or 500-nm resolution. We counted the number of single mRNAs per cell using FISH-QUANT software (53). The boundary of nucleus was defined based on the DAPI staining.
Data Analysis of mRNA Histogram.
We fit the mRNA population by multiple Gaussian peaks. First, we assigned the unsilenced peak (gray curve in Figs. 3D, 4D, and 5B) using the same mean value and peak width from the cytoplasmic mRNA population of cells that were not treated with any iRNAs. Second, we provided the subfitting for silenced peaks (orange curves in Figs. 3D, 4D, and 5B). This shows that silencing is not homogeneous, i.e., there are populations of efficiently silenced (left orange peaks in Figs. 3D, 4D, and 5B), poorly silenced (middle orange peaks in Figs. 3D, 4D, and 5B), and yet-to-be-silenced (gray peaks in Figs. 3D, 4D, and 5B) cells. We calculated the peak shift by averaging the peak shifts from the unsilenced peak for every silenced population, and peak size from the sum of the silenced population divided by the total population.
Supplementary Material
Acknowledgments
We thank Dr. Jaya Sarkar for careful proofreading of the manuscript and members of the S.M. laboratory and the Taekjip Ha laboratory for helpful discussions. This work was supported by American Cancer Society Grant RSG-12-066-01-DMC, NIH Grants 1DP2 GM105453 and R01 GM115631, and the National Science Foundation Physics Frontiers Center Program (0822613) through the Center for the Physics of Living Cells (H.R.K., A.G., and S.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1710298114/-/DCSupplemental.
References
- 1.Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bobbin ML, Rossi JJ. RNA interference (RNAi)-based therapeutics: Delivering on the promise? Annu Rev Pharmacol Toxicol. 2016;56:103–122. doi: 10.1146/annurev-pharmtox-010715-103633. [DOI] [PubMed] [Google Scholar]
- 4.Dorsett Y, Tuschl T. siRNAs: Applications in functional genomics and potential as therapeutics. Nat Rev Drug Discov. 2004;3:318–329. doi: 10.1038/nrd1345. [DOI] [PubMed] [Google Scholar]
- 5.Scherr M, Eder M. RNAi in functional genomics. Curr Opin Mol Ther. 2004;6:129–135. [PubMed] [Google Scholar]
- 6.Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- 7.Wilson RC, Doudna JA. Molecular mechanisms of RNA interference. Annu Rev Biophys. 2013;42:217–239. doi: 10.1146/annurev-biophys-083012-130404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Macrae IJ, et al. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- 10.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 11.Lee YS, et al. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- 12.Hutvágner G, et al. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 13.Leuschner PJ, Ameres SL, Kueng S, Martinez J. Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep. 2006;7:314–320. doi: 10.1038/sj.embor.7400637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 15.Rand TA, Petersen S, Du F, Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621–629. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 16.Meister G, et al. Human argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 17.Liu J, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 18.Iwakawa HO, Tomari Y. The functions of MicroRNAs: mRNA decay and translational repression. Trends Cell Biol. 2015;25:651–665. doi: 10.1016/j.tcb.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 19.Tsutsumi A, Kawamata T, Izumi N, Seitz H, Tomari Y. Recognition of the pre-miRNA structure by Drosophila Dicer-1. Nat Struct Mol Biol. 2011;18:1153–1158. doi: 10.1038/nsmb.2125. [DOI] [PubMed] [Google Scholar]
- 20.Feng Y, Zhang X, Graves P, Zeng Y. A comprehensive analysis of precursor microRNA cleavage by human Dicer. RNA. 2012;18:2083–2092. doi: 10.1261/rna.033688.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jackson AL, et al. Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA. 2006;12:1179–1187. doi: 10.1261/rna.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ameres SL, Martinez J, Schroeder R. Molecular basis for target RNA recognition and cleavage by human RISC. Cell. 2007;130:101–112. doi: 10.1016/j.cell.2007.04.037. [DOI] [PubMed] [Google Scholar]
- 23.Pusch O, et al. Nucleotide sequence homology requirements of HIV-1-specific short hairpin RNA. Nucleic Acids Res. 2003;31:6444–6449. doi: 10.1093/nar/gkg876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haley B, Zamore PD. Kinetic analysis of the RNAi enzyme complex. Nat Struct Mol Biol. 2004;11:599–606. doi: 10.1038/nsmb780. [DOI] [PubMed] [Google Scholar]
- 25.Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods. 2008;5:877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Femino AM, Fay FS, Fogarty K, Singer RH. Visualization of single RNA transcripts in situ. Science. 1998;280:585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- 27.Crosetto N, Bienko M, van Oudenaarden A. Spatially resolved transcriptomics and beyond. Nat Rev Genet. 2015;16:57–66. doi: 10.1038/nrg3832. [DOI] [PubMed] [Google Scholar]
- 28.Padovan-Merhar O, et al. Single mammalian cells compensate for differences in cellular volume and DNA copy number through independent global transcriptional mechanisms. Mol Cell. 2015;58:339–352. doi: 10.1016/j.molcel.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cambridge SB, et al. Systems-wide proteomic analysis in mammalian cells reveals conserved, functional protein turnover. J Proteome Res. 2011;10:5275–5284. doi: 10.1021/pr101183k. [DOI] [PubMed] [Google Scholar]
- 30.Filipowicz W. RNAi: The nuts and bolts of the RISC machine. Cell. 2005;122:17–20. doi: 10.1016/j.cell.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 31.Chakravarthy S, Sternberg SH, Kellenberger CA, Doudna JA. Substrate-specific kinetics of Dicer-catalyzed RNA processing. J Mol Biol. 2010;404:392–402. doi: 10.1016/j.jmb.2010.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang X, Zeng Y. The terminal loop region controls microRNA processing by Drosha and Dicer. Nucleic Acids Res. 2010;38:7689–7697. doi: 10.1093/nar/gkq645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou J, et al. Deep sequencing analyses of DsiRNAs reveal the influence of 3′ terminal overhangs on dicing polarity, strand selectivity, and RNA editing of siRNAs. Mol Ther Nucleic Acids. 2012;1:e17. doi: 10.1038/mtna.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kawamata T, Tomari Y. Native gel analysis for RISC assembly. Methods Mol Biol. 2011;725:91–105. doi: 10.1007/978-1-61779-046-1_7. [DOI] [PubMed] [Google Scholar]
- 35.Yoda M, et al. ATP-dependent human RISC assembly pathways. Nat Struct Mol Biol. 2010;17:17–23. doi: 10.1038/nsmb.1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pasquinelli AE, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 37.Kawamata T, Seitz H, Tomari Y. Structural determinants of miRNAs for RISC loading and slicer-independent unwinding. Nat Struct Mol Biol. 2009;16:953–960. doi: 10.1038/nsmb.1630. [DOI] [PubMed] [Google Scholar]
- 38.Leung AK, Calabrese JM, Sharp PA. Quantitative analysis of argonaute protein reveals microRNA-dependent localization to stress granules. Proc Natl Acad Sci USA. 2006;103:18125–18130. doi: 10.1073/pnas.0608845103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee YS, et al. Silencing by small RNAs is linked to endosomal trafficking. Nat Cell Biol. 2009;11:1150–1156. doi: 10.1038/ncb1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gibbings DJ, Ciaudo C, Erhardt M, Voinnet O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat Cell Biol. 2009;11:1143–1149. doi: 10.1038/ncb1929. [DOI] [PubMed] [Google Scholar]
- 41.Castel SE, Martienssen RA. RNA interference in the nucleus: Roles for small RNAs in transcription, epigenetics and beyond. Nat Rev Genet. 2013;14:100–112. doi: 10.1038/nrg3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 43.Ahmed F, Kaundal R, Raghava GP. PHDcleav: A SVM based method for predicting human Dicer cleavage sites using sequence and secondary structure of miRNA precursors. BMC Bioinformatics. 2013;14(Suppl 14):S9. doi: 10.1186/1471-2105-14-S14-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salomon WE, Jolly SM, Moore MJ, Zamore PD, Serebrov V. Single-molecule imaging reveals that argonaute reshapes the binding properties of its nucleic acid guides. Cell. 2015;162:84–95. doi: 10.1016/j.cell.2015.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y, Sheng G, Juranek S, Tuschl T, Patel DJ. Structure of the guide-strand-containing argonaute silencing complex. Nature. 2008;456:209–213. doi: 10.1038/nature07315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Y, et al. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature. 2008;456:921–926. doi: 10.1038/nature07666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schirle NT, MacRae IJ. The crystal structure of human argonaute2. Science. 2012;336:1037–1040. doi: 10.1126/science.1221551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schirle NT, Sheu-Gruttadauria J, MacRae IJ. Structural basis for microRNA targeting. Science. 2014;346:608–613. doi: 10.1126/science.1258040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chiu YL, Rana TM. siRNA function in RNAi: A chemical modification analysis. RNA. 2003;9:1034–1048. doi: 10.1261/rna.5103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bramsen JB, et al. A large-scale chemical modification screen identifies design rules to generate siRNAs with high activity, high stability and low toxicity. Nucleic Acids Res. 2009;37:2867–2881. doi: 10.1093/nar/gkp106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lingel A, Simon B, Izaurralde E, Sattler M. Nucleic acid 3′-end recognition by the argonaute2 PAZ domain. Nat Struct Mol Biol. 2004;11:576–577. doi: 10.1038/nsmb777. [DOI] [PubMed] [Google Scholar]
- 52.Kim DH, et al. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat Biotechnol. 2005;23:222–226. doi: 10.1038/nbt1051. [DOI] [PubMed] [Google Scholar]
- 53.Mueller F, et al. FISH-quant: Automatic counting of transcripts in 3D FISH images. Nat Methods. 2013;10:277–278. doi: 10.1038/nmeth.2406. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.