Significance
Clinical treatment of Helicobacter pylori using combination therapy is greatly challenged by the undesired killing of commensal bacteria and progressive development of drug resistance. To address these issues, we developed pH-sensitive, helix–coil conformation transitionable, antimicrobial polypeptides as a single therapeutic agent to selectively kill H. pylori under acidic condition in the stomach with minimal toxicity to commensal bacteria and diminished drug resistance. Through the control of the secondary structure transition, the polypeptides showed unappreciable toxicity to commensal bacteria and tissues at physiological pH when they adopted random coiled conformation, while the restoration of helical structure in the acidic stomach allowed the polypeptide to regain membrane disruptive capability to effectively and selectively kill H. pylori, including drug-resistant strains.
Keywords: antimicrobial peptide, α-helix, conformational transition, H. pylori, pH sensitiveness
Abstract
Current clinical treatment of Helicobacter pylori infection, the main etiological factor in the development of gastritis, gastric ulcers, and gastric carcinoma, requires a combination of at least two antibiotics and one proton pump inhibitor. However, such triple therapy suffers from progressively decreased therapeutic efficacy due to the drug resistance and undesired killing of the commensal bacteria due to poor selectivity. Here, we report the development of antimicrobial polypeptide-based monotherapy, which can specifically kill H. pylori under acidic pH in the stomach while inducing minimal toxicity to commensal bacteria under physiological pH. Specifically, we designed a class of pH-sensitive, helix–coil conformation transitionable antimicrobial polypeptides (HCT-AMPs) (PGA)m-r-(PHLG-MHH)n, bearing randomly distributed negatively charged glutamic acid and positively charged poly(γ-6-N-(methyldihexylammonium)hexyl-l-glutamate) (PHLG-MHH) residues. The HCT-AMPs showed unappreciable toxicity at physiological pH when they adopted random coiled conformation. Under acidic condition in the stomach, they transformed to the helical structure and exhibited potent antibacterial activity against H. pylori, including clinically isolated drug-resistant strains. After oral gavage, the HCT-AMPs afforded comparable H. pylori killing efficacy to the triple-therapy approach while inducing minimal toxicity against normal tissues and commensal bacteria, in comparison with the remarkable killing of commensal bacteria by 65% and 86% in the ileal contents and feces, respectively, following triple therapy. This strategy renders an effective approach to specifically target and kill H. pylori in the stomach while not harming the commensal bacteria/normal tissues.
It is widely accepted that Helicobacter pylori, with 50% of the population worldwide infected, is the main etiological factor for the development of gastritis, gastric ulcers, and gastric carcinoma (1–4). To eradicate H. pylori, triple therapy is recommended as first-line therapy in the clinical setting, which involves the use of a combination of two antibiotics for optimal bacterial killing and a proton pump inhibitor (PPI) for increasing the gastric pH to enhance the antimicrobial activity and stability of antibiotics in the gastric fluid (5–8). However, the combination therapy is often associated with various side effects (9, 10). In particular, it leads to undesired elimination of commensal bacteria which are closely related to various physiological and metabolic processes (11–13), the development of the immune system (13–15), and a range of diseases, such as inflammatory bowel disease (16, 17), colon cancer (18, 19), Parkinson’s disease (20), obesity (21–23), diabetes (24), atherosclerosis (25, 26), and allergy (27, 28). For example, gut microbiota play an important role in colorectal carcinogenesis, and colorectal cancer patients show significantly reduced microbial diversity in feces (18, 19). In addition, the efficacy of triple therapy is hurdled by the constant increment of drug resistance, and the resistance to any of the three drugs will make the treatment end with failure (29–31). The antimicrobial activity of antibiotics could also be insufficient to eradicate bacteria for the patients that are acid hypersecretors or extensive metabolizers, wherein PPI fails to increase the gastric pH (32, 33). To address these critical issues, anti-H. pylori therapy should be designed to feature selective killing of H. pylori with potentially diminished resistance.
Antimicrobial peptides (AMPs) have recently emerged as promising antimicrobial candidates, which are capable of disrupting bacterial membrane structure to combat multidrug-resistant microbes (34–39). It is shown that AMPs can kill H. pylori in vivo as a single agent (40–42). However, these antimicrobial agents often suffer from high cytotoxicity (e.g., hemolysis), poor proteolytic stability, and low selectivity (37, 43). We recently developed a class of radially amphiphilic (RA) antimicrobial polypeptides with a hydrophobic helical core and a charged exterior shell, affording potent antimicrobial activity that are associated with their helical structure (44, 45). While these RA polypeptides afford several advantages over conventional AMPs, such as simplicity in design and stability against proteases (44), they lack the capability of differentiating pathogenic bacteria from commensal bacteria, which will cause nonspecific killing of commensal bacteria when applied for anti-H. pylori therapy.
Considering that H. pylori thrive in the stomach under unique acidic condition (with a mean gastric pH of ∼2 in human) while commensal bacteria reside in the intestine with relatively neutral pH (6–8) (46–48), we herein developed a class of pH-sensitive, helix–coil conformation transitionable antimicrobial polypeptides (HCT-AMPs) (PGA)m-r-(PHLG-MHH)n, with both anionic groups (glutamic acid) and cationic groups (tertiary amine) in the polypeptide side chains (Fig. 1A). The HCT-AMPs display distorted helix at physiological pH due to the intramolecular electrostatic interactions between the anionic carboxylate and cationic amine groups, ultimately leading to minimal toxicity to intestinal commensal bacteria, while under the acidic condition in the stomach, the HCT-AMPs transform to the helical conformation due to the protonation of the carboxylate groups and the depletion of the side-chain electrostatic interaction, which result in potent antimicrobial efficacy against H. pylori in the stomach. These HCT-AMPs showed comparable H. pylori killing efficacy as clinically used triple therapy in a mouse model, with inhibited toxicity against normal tissues and commensal bacteria. Triple therapy killed 65% and 86% of commensal bacteria in the ileal contents and feces, respectively.
Fig. 1.
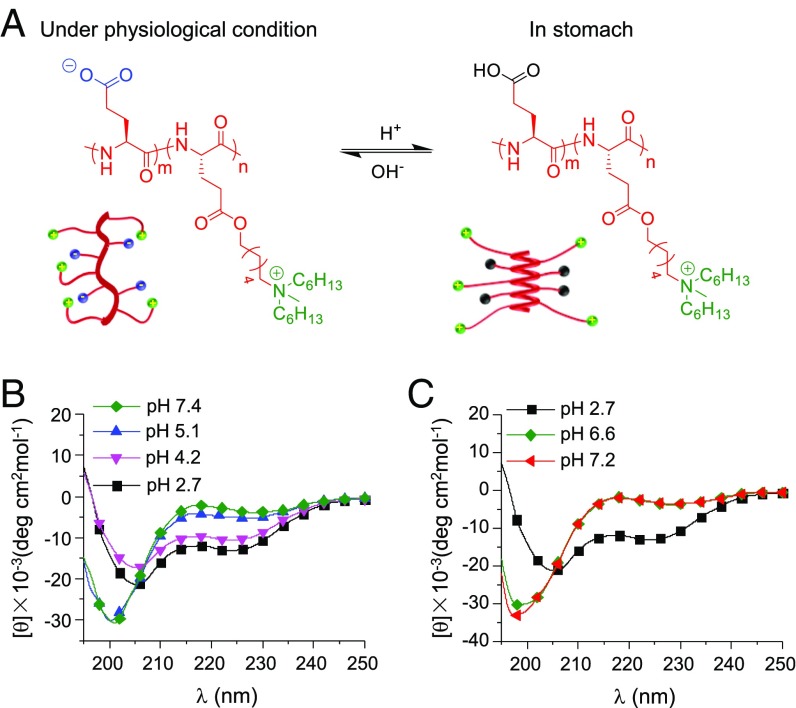
HCT-AMPs display pH-sensitive helix–coil transition. (A) Schematic illustration of the pH-responsive conformation transition of HCT-AMP (PGA)18-r-(PHLG-MHH)20. It adopts random coiled conformation at physiological pH to impart low toxicity while transforming to helical conformation under acidic condition in the stomach to induce potent antimicrobial activity against H. pylori. The green balls represent cationic amine groups, the blue balls represent anionic group (-COO), and the black balls represent neutral groups (-COOH). CD spectra of PL2 (4.4 μM) at various pH values adjusted from 7.4 to 2.7 with 1 M HCl (B) and from 2.7 to 7.2 with 1 M NaOH (C).
Results
HCT-AMPs Display pH-Sensitive Helix–Coil Transition.
Random copolypeptides PL2, (PGA)18-r-(PHLG-MHH)20, with anionic glutamic acid (Glu) and cationic poly(γ-6-N-(methyldihexylammonium)hexyl-l-glutamate) (PHLG-MHH) residues (Fig. 1A) were developed via the ring-opening polymerization of l-γ-(6-chlorohexyl)-Glu–based N-carboxyanhydrides (NCA) and l-tert-butyl-Glu-NCA (tBLG-NCA) (i), followed by amination (ii) and trifluoroacetic acid-assisted deesterification (iii) (44, 49) (SI Appendix, Scheme S2). pH-independent helical polypeptide and nonhelical polypeptides [prepared with D,l-γ-(6-chlorohexyl)-Glu-NCA) and DL-tert-butyl-Glu-NCA as monomers] were synthesized as control polypeptides, named as PL1 = (PHLG-MHH)20, PDL2 = (PDLGA)20-r-(PHDLG-MHH)25 (SI Appendix, Table S1 and Scheme S2). The structure of the polypeptides was confirmed by 1H-NMR spectra (SI Appendix, Figs. S1 and S2).
The secondary structure of the polypeptides at different pH value was then investigated by circular dichroism (CD). As shown in Fig. 1B, the secondary structure of PL2 was related to the charge status of PGA (pKa ∼ 4.5) (SI Appendix, Fig. S3). Particularly, at pH ≥ 5.1, when the carboxyl groups exhibited negative charge, PL2 adopted random coiled conformation due to the intermolecular electrostatic interaction between the negatively charged carboxyl groups and the positively charged amine groups of PHLG-MHH. In contrast, at pH ≤ 4.2 when the carboxyl groups were protonated, PL2 restored typical helical conformation as evidenced by the double minima at 208 and 222 nm, which was attributed to the depletion of side-chain charge interactions. Distortion of helical conformation was noted when the pH was adjusted back to neutral (Fig. 1C). In consistence with such findings, PL1 containing only the cationic PHLG-MHH segment displayed stable helical conformation independent of pH change (SI Appendix, Fig. S4A). PDL2, a racemic analog of PL2, also demonstrated pH-independent nonhelical conformation.
HCT-AMPs Selectively Kill H. pylori in Vitro.
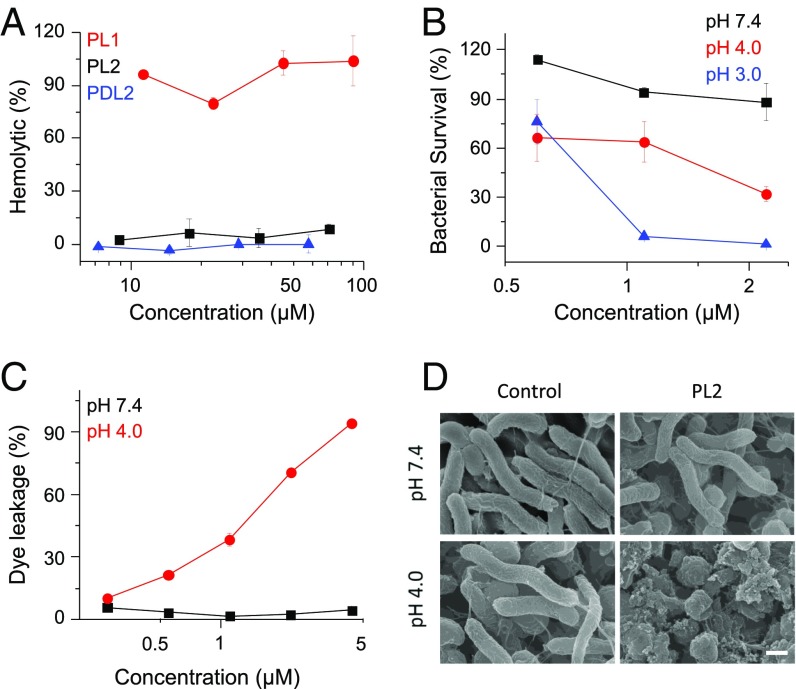
With respect to its pH-responsive secondary structure, the HCT-AMP is expected to show low toxicity at neutral pH while showing high antibacterial activity against H. pylori in the acidic gastric environment. To test this hypothesis, we first evaluated the hemolytic activity of the polypeptides at pH 7.4. PL2 and PDL2, affording random coiled structure at pH 7.4, which showed no hemolytic activity at a high concentration up to 70 μM, while PL1 with stable helical structure caused remarkable hemolysis at 10 μM, which further substantiated the helical conformation-dependent membrane toxicity against erythrocytes (Fig. 2A). Additionally, PL2 showed low antibacterial activity against Escherichia coli DH5α and MG1655 at pH 7.4, affording the minimal inhibitory concentration (MIC) higher than 70 μM. Such results suggest that PL2 would not kill commensal bacteria in the intestine with relatively neutral pH. We then determined the bactericidal activity of HCT-AMPs against H. pylori SS1 strain under various pH conditions. Upon incubation of SS1 with PL2 at pH 7.4 for 1 h, no bactericidal activity was noted. However, notable bacterial killing was achieved by PL2 at pH 4.0, and further decreased pH value correlated to higher bactericidal activity (Fig. 2B). As a comparison, the nonhelical PDL2 exhibited unappreciable bactericidal activity at both pH 7.4 and 3.0 and the helical PL1 killed H. pylori SS1 at both conditions (SI Appendix, Fig. S4 B and C). These results collectively indicate that the restoration of helical structure is essential for PL2 to selectively kill H. pylori under the gastric acidic condition, and the loss of helical conformation under intestinal neutral condition may contribute to the low toxicity against commensal bacteria.
Fig. 2.
HCT-AMPs selectively kill H. pylori under acidic condition in vitro. (A) The hemolytic activity of polypeptides at pH 7.4. PL1, PL2, and PDL2, dissolved in PBS (pH 7.4) at various concentrations, was incubated with fresh rabbit blood for 1 h. Hemoglobin release was measured by UV absorbance at 576 nm using a microplate reader. (B) The survival rate of H. pylori SS1 after incubation with PL2 for 1 h at various pHs. PL2, dissolved in the Tris⋅HCl buffer at various pHs (pH 7.4, 4.0, 3.0), was incubated with SS1 at corresponding pHs in Brucella broth (BB) medium supplied with fresh urea (10 mM), 10% FBS, and vancomycin (5 µg/mL). The bacterial count was determined by counting colony-forming units (cfu) of alive bacteria with agar plating. Bacteria incubated with Tris⋅HCl buffer only at corresponding pH were served as 100% survival. (C) Extent of ANTS/DPX efflux in negatively charged liposomes after treatment with PL2 at various concentrations at pH 7.4 or pH 4.0. (D) SEM images of SS1 after treatment with Tris⋅HCl buffer or PL2 at pH 7.4 or pH 4.0. SS1 bacterial cells were incubated with or without PL2 (2.2 µM) at pH 7.4 or 4.0 for 30 min. (Scale bar, 0.5 μm.)
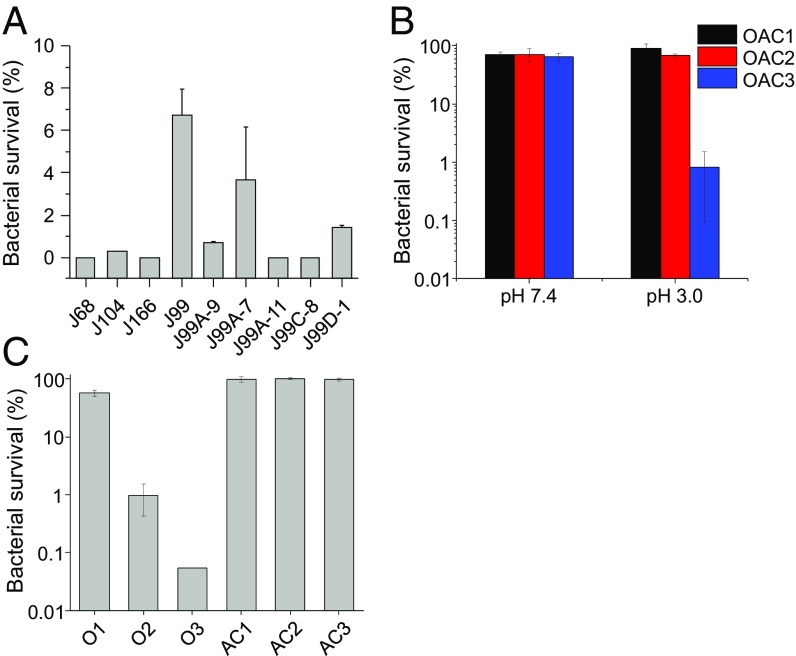
A combination of omeprazole, amoxicillin, and clarithromycin (OAC) is used for the treatment of H. pylori infection in clinic (8). We further explored the bactericidal activity of these antibiotics under the same condition to allow direct comparison with PL2. Although the MIC of amoxicillin and clarithromycin against SS1 was as low as 0.13 and 0.07 µM, respectively, amoxicillin and clarithromycin only killed ∼40% and ∼60% of the bacteria at pH 7.4 after 1 h incubation at high concentrations of 273.7 and 133.7 µM, respectively, indicating the slow function of antibiotics (SI Appendix, Fig. S5A). Moreover, these two antibiotics showed no bactericidal activity against SS1 at pH 3.0, rationalizing why PPI is demanded to raise the pH of the stomach to potentiate the antibiotic-mediated treatment of H. pylori infection in vivo. A combination of OAC effectively killed SS1 in 1 h at both pH 7.4 and 3.0 (SI Appendix, Fig. S5B). The antibacterial activity of OAC at pH 3.0 was attributed to the bacterial killing effect of omeprazole, while a combination of amoxicillin and clarithromycin (AC) showed no antibacterial activity at pH 3.0 (SI Appendix, Fig. S5C). It should be noted that omeprazole performs differently in vitro and in vivo. Omeprazole is used for the increase of the gastric pH to enhance the antimicrobial activity and stability of antibiotics in the gastric fluid in vivo (5–8). It suppresses stomach acid secretion via specific inhibition of the H+/K+-ATPase system found at the secretory surface of gastric parietal cells. Because of such unique mechanism, omeprazole cannot increase the pH value of the acidic medium during the bacterial killing study in vitro. Instead, omeprazole readily converts to the active sulfenamide form and causes a substantial decrease in survival of H. pylori under acidic condition in vitro (50).
The bactericidal mechanism of HCT-AMPs was further explored by the vesicle leakage assessment as well as bacterial morphology observation. PL2 was labeled with Cy5 (Cy5-PL2), and its binding with bacteria membranes was observed using fluorescence microscopy. More Cy5-PL2 bound to SS1 bacterial cells at pH 4.0 than at pH 7.4 (SI Appendix, Fig. S6), which was attributed to the formation of cationcally helical structure at acidic pH that promoted binding of polypeptides to phospholipid bilayers. The membrane activity of the polypeptides was studied at both pH 7.4 and 4.0 by assessing dye leakage from anionic liposomes, a commonly used model to simulate the phosphatidylethanolamine-rich bacterial cell membrane (44). After incubation, PL2 induced great dye leakage from the liposome at pH 4.0 while inducing minimal dye leakage at pH 7.4 (Fig. 2C). These results demonstrate that the acid-triggered helix formation of HCT-AMPs allows them to directly disrupt the bacteria membranes, a mechanism that most AMPs utilize to kill bacteria. Comparatively, control polypeptide PL1 induced notable dye leakage at both pH 7.4 and 4.0, while PDL2 induced unappreciable dye leakage under both conditions, which well correlated to the helical and nonhelical conformation of PL1 and PDL2, respectively (SI Appendix, Fig. S4D). In support of the acid-activated membrane disruption, we further observed dramatic damage of the bacterial membranes by scanning electron microscopy (SEM) after incubation with PL2 at pH 4.0 (Fig. 2D). At pH 7.4, PL2 minimally affected the bacterial morphology, consistent with its minimal membrane activity at the nonhelical state. Control polypeptide PL1 showed membrane disruption toward SS1 at both pH 7.4 and 4.0, while PDL2 did not affect the morphology of bacteria under both conditions (SI Appendix, Fig. S4E). Taken together, these results indicate that HCT-AMPs are able to selectively kill bacteria under acidic condition via acid-triggered helix formation and helix-assisted bacterial membrane disruption.
The emergence of drug-resistant H. pylori is the main reason for clinical treatment failure (29, 30, 33). The resistance to clarithromycin, metronidazole, tetracycline, fluoroquinolones, as well as rifamycin, has become a serious issue (29, 30, 33). As such, we further tested the antibacterial activity of HCT-AMPs against clinically isolated strains, including clarithromycin-resistant J99A-7, J99A-11, J99C-8, and J99D-1 (51, 52) (SI Appendix, Fig. S5 D and E). PL2 effectively killed over 90% of these bacterial strains at pH 3.0 after 1 h incubation at a concentration of 4.4 µM (Fig. 3A), which further substantiated its potency in overcoming the drug resistance toward effective anti-H. pylori therapy. OAC could not effectively kill drug-resistant bacteria J99A-11 and J99D-1 at pH 7.4. It instead killed bacteria at pH 3.0 in a concentration-dependent manner (Fig. 3B and SI Appendix, Fig. S5F), which was also attributed to the bacterial killing effect of omeprazole (Fig. 3C and SI Appendix, Fig. S5G).
Fig. 3.
HCT-AMPs effectively kill clinically isolated drug-resistant H. pylori. (A) The bactericidal activity of PL2 against clinically isolated H. pylori strains. PL2, dissolved in the Tris⋅HCl buffer (pH 3.0), was incubated with clinically isolated strains in BB medium supplied with fresh urea, FBS, and vancomycin (pH 3.0) for 1 h. The final concentration of PL2 was 4.4 µM. (B) The antibacterial activity of triple therapy against J99A-11 at pH 7.4 or 3.0. Concentrations of OAC in OAC1 are 20.0, 6.8, and 1.9 µM, respectively; in OAC2 are 40.0, 13.6, and 3.8 µM, respectively; in OAC3 are 80.0, 27.2, and 7.6 µM, respectively. (C) The antibacterial activity of omeprazole (O) and a combination of amoxicillin and clarithromycin (AC) against J99A-11 at pH 3.0. O1, O2, and O3 represents the concentration of omeprazole at 20.0, 40.0, and 80.0 µM, respectively. AC1, AC2, and AC3 represents the concentration of AC at 6.8 and 1.9; 13.6 and 3.8 µM; and 27.2 and 7.6 µM, respectively.
The application of AMPs often hurdles by the short durations of antimicrobial activity due to their rapid digestion by endogenous proteases (37). The RA polypeptide, with densely packed hydrophobic side chains forming a hydrophobic cortex to protect the polypeptide backbone amide bonds, was shown to be more stable against proteolysis compared with typical AMPs (44). We herein also tested the proteolytic stability of HCT-AMPs against pepsin, the main digestive protease in the stomach. After incubation with pepsin at pH 4.0 for up to 24 h, HPLC analysis showed that PL2 was resistant to pepsin-mediated degradation (SI Appendix, Fig. S7).
HCT-AMPs Selectively Kill H. pylori in Vivo.
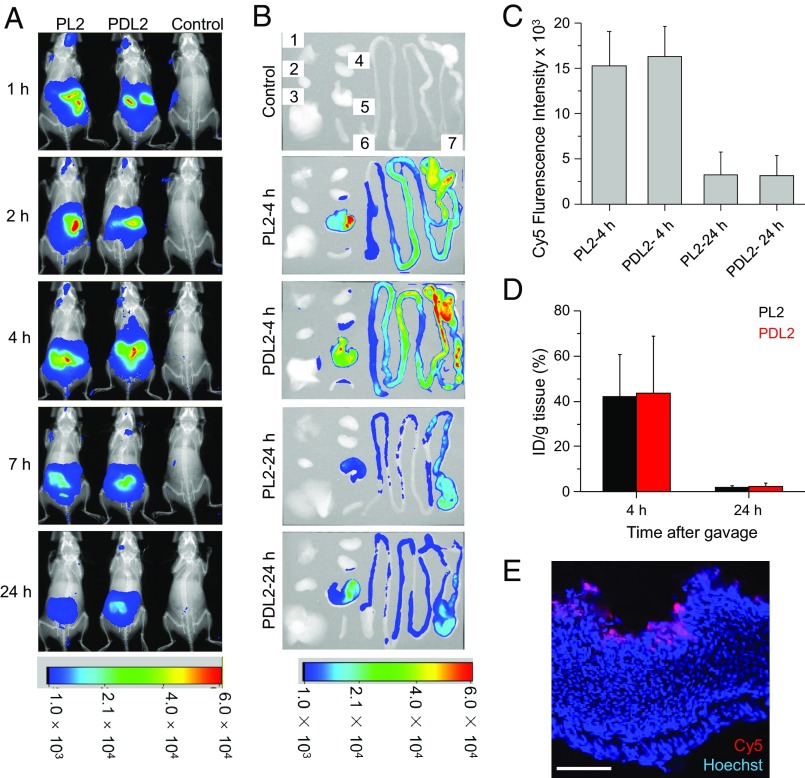
We then evaluated the therapeutic efficacy of HCT-AMPs against H. pylori SS1 in vivo. We first studied the biodistribution of HCT-AMPs after oral gavage of Cy5-labeled polypeptides. PL2 and PDL2 showed similar biodistribution profiles, with the majority of the polypeptides retained in the stomach and intestines within 4 h and gradually excreted within 24 h (Fig. 4A). By collecting major organs 4 h post-oral gavage, we further observed strong fluorescence in the gastric tissue (Fig. 4 B and C). Such observation was further supported by the quantification of the gastric retention of PL2, which reached ∼40% ID/g 4 h postgavage and notably decreased to ∼6% 24 h postgavage (Fig. 4D). It should be mentioned that 4 h is a relatively long time compared with the reported gastric emptying time of mice (3). As such, these results indicate that HCT-AMPs could be effectively retained in the stomach against gastric emptying, likely due to the electrostatic interactions between the anionic mucus and the polypeptides that possess positive charges under acidic condition. We further studied the penetration of Cy5-PL2 into gastric mucosa by observing the cryosections of mouse stomach collected 4 h after oral gavage. The confocal image revealed a thin layer of Cy5-PL2 on the luminal side of the gastric mucosa, confirming the diffusion of PL2 toward the gastric epithelium and its retention in the mucus layer (Fig. 4E).
Fig. 4.
In vivo distribution of Cy5-labeled HCT-AMPs following oral gavage. (A) Representative whole-body fluorescence imaging of C57BL/6J mice treated with Cy5-PL2 and Cy5-PDL2 (2.6 μmol/kg). Cy5-PL2, Cy5-PDL2, and PBS were administrated by oral gavage. Mice were then imaged with the Bruker Xtreme In-Vivo Fluorescence Imaging System at 1, 2, 4, 7, and 24 h postinjection (p.i.). (B) Representative ex vivo fluorescence imaging of major organs (1, 2, 3, 4, 5, 6, and 7 represents lung, heart, liver, kidney, stomach, spleen, and intestines, respectively) from C57BL/6J mice treated with Cy5-PL2 and Cy5-PDL2. Major organs were harvested and imaged ex vivo 4 h or 24 h p.i. of Cy5-PL2 and Cy5-PDL2. (C) Ex vivo fluorescence intensity of stomach harvested from mice receiving the treatment in B. Ex vivo images were quantified by measuring fluorescence intensity at selected region of interest. All values were expressed as means ± SD (n = 3). (D) Retention of Cy5-PL2 and Cy5-PDL2 in mouse stomach 4 and 24 h after oral gavage. Stomach samples were harvested and homogenized, and the lysates were used to determine the amount of Cy5 retained in the tissues with a fluorescence spectrometer. (E) Confocal image of mouse stomach following treatment with Cy5-PL2. (Scale bar, 100 μm.)
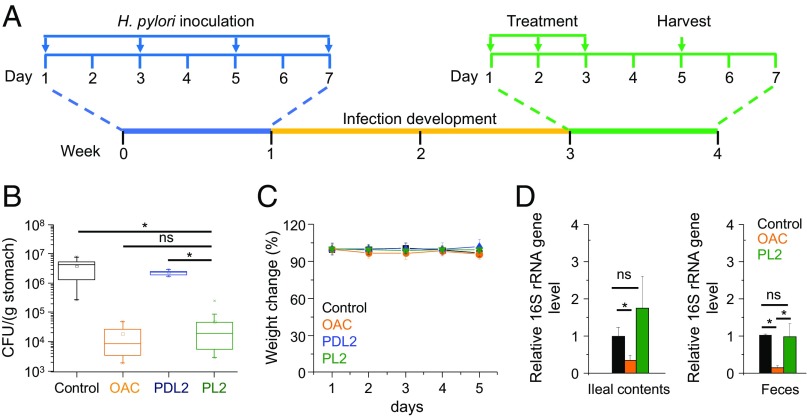
We then infected mice with H. pylori SS1 (1 × 108 cfu per animal) by oral gavage every other day for four times (Fig. 5A). Two weeks after inoculation, infected mice were divided into four groups and treated with PBS (control), triple-therapy OAC, PDL2, or PL2. PL2 showed similar bacteria killing ability as the OAC treatment, with a decrease of bacterial burden by ∼100× compared with the control group. PDL2, although displaying similar biodistribution profiles as PL2, showed no significant decrease of bacterial burden (Fig. 5B). These results indicate that the formation of membrane-active helical structure is essential for killing H. pylori. Because the pH in the mouse stomach (∼3.0 when fed and ∼4.0 when fasted) is higher than that in the human stomach (53), it is expected that the HCT-AMPs may show even higher H. pylori killing efficiency in the human stomach.
Fig. 5.
Anti-H. pylori efficacy of HCT-AMPs in vivo. (A) The study protocol of H. pylori inoculation, infection development, and treatments in C57BL/6J mice. Each mouse was administered with SS1 (OD600 = 2, 0.2 mL) intragastrically through oral gavage every other day for four times (on days 1, 3, 5, and 7, respectively), and the infection was allowed to develop for 2 wk. Mice were then treated with control (PBS), triple therapy (OAC, omeprazole 400 µmol/kg, amoxicillin 68 µmol/kg, and clarithromycin 19.1 μmol/kg), PDL2, and PL2 (2.6 μmol/kg) once daily for a consecutive 3 d. (B) Bacterial burden in the stomach of H. pylori-infected mice treated with PBS, triple-control therapy, PDL2, and PL2 (n ≥ 6). (C) Bodyweight change of mice following treatment of various formulations as in B. (D) The killing effect of PL2 against commensal bacteria determined by measuring the bacterial load in the feces and ileal contents of mice after a daily gavage of PL2, control (5% DMSO), and OAC for three consecutive days. The bacterial load was determined by quantitative real-time PCR. The 16S rRNA gene level was normalized to the tissue weight (n ≥ 6). All of the data are represented as average ± SD and analyzed by Student’s t test (*P ≤ 0.05). “ns” represents no significant difference (P > 0.05).
The toxicity of PL2 was further explored. No obvious change of animal body weight was noticed following treatment with PL2 as described above, indicating the low toxicity of PL2 in vivo (Fig. 5C). To analyze the toxicity of PL2 toward the stomach, we performed H&E-staining assay, the terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay, and detected caspase 3/8 activity of mouse stomach after treatments. No significant inflammation of stomach or injury of mucosa layer was observed after gavage of PL2 according to the H&E-staining assay (SI Appendix, Fig. S8 A–C). PL2 administration did not induce apoptosis in stomach cells, as measured by the TUNEL assay and the activities of caspase 3 and 8 (SI Appendix, Fig. S8 D–G). The plasma levels of alanine aminotransferase, aspartate aminotransferase, creatinine, urea nitrogen, sodium ion, and potassium ion showed no significant change after the PL2 treatment (SI Appendix, Table S2), verifying the lack of significant acute damage toward the liver and kidney as well as the balance of electrolytes in the blood. The low toxicity of PL2 was further confirmed by H&E staining of intestines, liver, and kidney, with barely detectable histological abnormality (SI Appendix, Fig. S9). More importantly, PL2 did not cause undesired killing of the commensal bacteria in the ileal contents and feces of mice, while OAC treatment killed commensal bacteria in the ileal contents and feces by 65% and 86%, respectively (Fig. 5D). These results collectively indicate the low side effect of HCT-AMPs toward normal cells or commensal bacteria.
Discussion
In this study, we designed a class of pH-responsive HCT-AMPs as a single bactericidal agent that can specifically target H. pylori under the acid condition in the stomach. Random polypeptide copolymers (PGA)m-r-(PHLG-MHH)n were developed with randomly distributed negatively charged Glu residues and positively charged PHLG-MHH residues. The conformation and membrane activity of the polypeptides depend on the charge state of Glu residues. At physiological pH, the polypeptides adopt random coiled conformation with low toxicity, while in the stomach under acidic condition they are converted to the helical conformation with potent membrane disruption capability to effectively kill H. pylori. With such design, HCT-AMPs showed minimal toxicity against normal tissues/commensal bacteria but in vivo H. pylori killing efficacy comparable to the triple therapy, with remarkably improved selectivity of anti-H. pylori therapy.
Commensal bacteria have gained increased attention due to their important functions during the physiological and metabolic processes as well as the development of the immune system (11, 13, 14). Undesired alteration of the microbiome can disturb the symbiotic relationship between resident microorganisms and the digestive tract, and thus induce the occurrence as well as progression of various diseases, including but not limited to inflammatory bowel disease (16, 17), colon cancer (18, 19), Parkinson’s disease (20), obesity (21–23), diabetes (24), atherosclerosis (25, 26), and allergy (27, 28). Recent research has also shown that the elimination of commensal bacteria significantly alleviates the efficacy of anticancer therapeutics, including CTLA-4 and PD-L1 blockade-mediated cancer immunotherapy (54–56). Therefore, development of therapeutics that can selectively kill H. pylori without harming commensal bacteria is highly attractive and important. The triple therapy, used in clinical H. pylori treatment, killed 65% and 86% of the commensal bacteria in the ileal contents and feces, respectively, while the pH-sensitive HCT-AMPs showed unappreciable toxicity against commensal bacteria (Fig. 5D). This strategy therefore represents an ideal and promising approach to target and selectively kill H. pylori in the stomach without provoking damage to commensal bacteria.
Apart from the undesired toxicity, the triple therapy also suffers from the progressive increase of drug resistance that undermines its therapeutic efficacy against H. pylori-induced gastric diseases (29–31, 33, 57). High resistance rates are noted for clinically used antibiotics, such as 60–70% for metronidazole, 20–38% for clarithromycin, and 30–38% for levofloxacin (57). In the current study, HCT-AMPs kill bacteria mainly by disrupting the membrane integrity (Fig. 2D), a highly destructive mechanism for bacterial killing with low susceptibility for resistance development (34–37, 58, 59). As such, we demonstrated that HCT-AMPs could effectively kill clinically isolated drug-resistant bacterial strains (Fig. 3A). The clinical success of triple therapy also faces great hurdles by the resistance to PPI, because in many patients, PPI cannot effectively increase their gastric pH, thus leading to low antimicrobial activity of antibiotics in the gastric fluid (32, 33). The HCT-AMPs developed herein kill H. pylori under acidic condition as a single agent, and they exhibited increased anti-H. pylori efficacy with the decrease of pH (Fig. 2B), effectively bypassing the problem of PPI resistance. Based on these understandings, it is expected that the HCT-AMPs would outperform the classical triple therapy toward anti-H. pylori therapy.
In conclusion, we developed a class of pH-sensitive HCT-AMPs as a single therapeutic agent to target and selectively treat H. pylori infection in the stomach. The HCT-AMPs can specifically kill H. pylori, including drug-resistant strains, under acidic conditions, and showed minimal toxicity against normal tissues/commensal bacteria. The pH-sensitive HCT-AMPs greatly outperform the triple therapy that suffers from undesired toxicity as well as drug resistance. This strategy therefore provides a safe and effective approach to overcome the critical challenges in the treatment of H. pylori-induced gastric diseases, and would render promising utilities toward anti-H. pylori therapy.
Materials and Methods
Animals.
Female C57BL/6J mice were purchased from The Jackson Labs (Bar Harbor, ME) for the efficacy studies and biodistribution studies. Feed and water were available ad Librium. Artificial light was provided in a 12 h/12 h cycle. The animal protocol was reviewed and approved by the Illinois Institutional Animal Care and Use Committee (IACUC) of University of Illinois at Urbana–Champaign. For the toxicity studies, male ICR mice (6–8 weeks, 18–22 g) were obtained from Shanghai Slaccas Experimental Animal Co., Ltd. (Shanghai, China) and were housed in a specific-pathogen–free room. The animal protocols were approved by the Institutional Animal Care and Use Committee, Soochow University.
PL2 was synthesized via the reaction of PCHLG20-r-PtBLG18 with N-methyldihexylamine followed by removal of the tert-butyl group by trifluoracetic acid. Details describing preparation and characterization of HCT-AMPs, in vitro and in vivo antibacterial assays, and hemolytic assay can be found in SI Appendix.
Supplementary Material
Acknowledgments
J. Cheng acknowledges partial support from the NSF (CHE 1508710 and 1308485) for the design and synthesis of polypeptides. L.-F.C. and J. Cheng acknowledge support from the NIH (R21AI117080) for antibacterial peptides against H. pylori. L.Y. acknowledges support from the National Natural Science Foundation of China (51403145, 51573123, and 51722305), the Ministry of Science and Technology of China (2016YFA0201200), and Priority Academic Program Development of Jiangsu Higher Education Institutions. R.M.P. acknowledges support from the NIH (R01 DK 58587, R01 CA 77955, P01 CA 116087, and P30 DK 058404).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1710408114/-/DCSupplemental.
References
- 1.Malfertheiner P. Infection: Bismuth improves PPI-based triple therapy for H. pylori eradication. Nat Rev Gastroenterol Hepatol. 2010;7:538–539. doi: 10.1038/nrgastro.2010.131. [DOI] [PubMed] [Google Scholar]
- 2.Kodaman N, et al. Human and Helicobacter pylori coevolution shapes the risk of gastric disease. Proc Natl Acad Sci USA. 2014;111:1455–1460. doi: 10.1073/pnas.1318093111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thamphiwatana S, Gao W, Obonyo M, Zhang L. In vivo treatment of Helicobacter pylori infection with liposomal linolenic acid reduces colonization and ameliorates inflammation. Proc Natl Acad Sci USA. 2014;111:17600–17605. doi: 10.1073/pnas.1418230111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farrell P. Pathogenesis: Infections causing gastric cancer. Nat Microbiol. 2016;1:16038. doi: 10.1038/nmicrobiol.2016.38. [DOI] [PubMed] [Google Scholar]
- 5.Brown D. Antibiotic resistance breakers: Can repurposed drugs fill the antibiotic discovery void? Nat Rev Drug Discov. 2015;14:821–832. doi: 10.1038/nrd4675. [DOI] [PubMed] [Google Scholar]
- 6.Fock KM, Graham DY, Malfertheiner P. Helicobacter pylori research: Historical insights and future directions. Nat Rev Gastroenterol Hepatol. 2013;10:495–500. doi: 10.1038/nrgastro.2013.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rimbara E, Fischbach LA, Graham DY. Optimal therapy for Helicobacter pylori infections. Nat Rev Gastroenterol Hepatol. 2011;8:79–88. doi: 10.1038/nrgastro.2010.210. [DOI] [PubMed] [Google Scholar]
- 8.Tonge KA, et al. Eradication of Helicobacter-pylori using clarithromycin, omeprazole and amoxicillin for one week. Gastroenterology. 1995;108:A242. [Google Scholar]
- 9.Bell GD, et al. Experience with ‘triple’ anti-Helicobacter pylori eradication therapy: Side effects and the importance of testing the pre-treatment bacterial isolate for metronidazole resistance. Aliment Pharmacol Ther. 1992;6:427–435. doi: 10.1111/j.1365-2036.1992.tb00556.x. [DOI] [PubMed] [Google Scholar]
- 10.Tefera S, et al. Bismuth-based combination therapy for Helicobacter pylori-associated peptic ulcer disease (metronidazole for eradication, ranitidine for pain) Am J Gastroenterol. 1996;91:935–941. [PubMed] [Google Scholar]
- 11.Littman DR, Pamer EG. Role of the commensal microbiota in normal and pathogenic host immune responses. Cell Host Microbe. 2011;10:311–323. doi: 10.1016/j.chom.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brestoff JR, Artis D. Commensal bacteria at the interface of host metabolism and the immune system. Nat Immunol. 2013;14:676–684. doi: 10.1038/ni.2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hepworth MR, et al. Innate lymphoid cells regulate CD4+ T-cell responses to intestinal commensal bacteria. Nature. 2013;498:113–117. doi: 10.1038/nature12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manichanh C, Borruel N, Casellas F, Guarner F. The gut microbiota in IBD. Nat Rev Gastroenterol Hepatol. 2012;9:599–608. doi: 10.1038/nrgastro.2012.152. [DOI] [PubMed] [Google Scholar]
- 17.Ray K. IBD. Understanding gut microbiota in new-onset Crohn’s disease. Nat Rev Gastroenterol Hepatol. 2014;11:268. doi: 10.1038/nrgastro.2014.45. [DOI] [PubMed] [Google Scholar]
- 18.Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol. 2014;12:661–672. doi: 10.1038/nrmicro3344. [DOI] [PubMed] [Google Scholar]
- 19.Tjalsma H, Boleij A, Marchesi JR, Dutilh BE. A bacterial driver-passenger model for colorectal cancer: Beyond the usual suspects. Nat Rev Microbiol. 2012;10:575–582. doi: 10.1038/nrmicro2819. [DOI] [PubMed] [Google Scholar]
- 20.Sampson TR, et al. Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson’s disease. Cell. 2016;167:1469–1480.e12. doi: 10.1016/j.cell.2016.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perry RJ, et al. Acetate mediates a microbiome-brain-β-cell axis to promote metabolic syndrome. Nature. 2016;534:213–217. doi: 10.1038/nature18309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buffington SA, et al. Microbial reconstitution reverses maternal diet-induced social and synaptic deficits in offspring. Cell. 2016;165:1762–1775. doi: 10.1016/j.cell.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turnbaugh PJ, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 24.Hill JH, Franzosa EA, Huttenhower C, Guillemin K. A conserved bacterial protein induces pancreatic beta cell expansion during zebrafish development. Elife. 2016;5:e20145. doi: 10.7554/eLife.20145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jonsson AL, Bäckhed F. Role of gut microbiota in atherosclerosis. Nat Rev Cardiol. 2017;14:79–87. doi: 10.1038/nrcardio.2016.183. [DOI] [PubMed] [Google Scholar]
- 26.Chistiakov DA, Bobryshev YV, Kozarov E, Sobenin IA, Orekhov AN. Role of gut microbiota in the modulation of atherosclerosis-associated immune response. Front Microbiol. 2015;6:671. doi: 10.3389/fmicb.2015.00671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tlaskalová-Hogenová H, et al. Commensal bacteria (normal microflora), mucosal immunity and chronic inflammatory and autoimmune diseases. Immunol Lett. 2004;93:97–108. doi: 10.1016/j.imlet.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 28.Kamada N, Seo SU, Chen GY, Núñez G. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol. 2013;13:321–335. doi: 10.1038/nri3430. [DOI] [PubMed] [Google Scholar]
- 29.Wu WM, Yang YS, Sun G. Recent insights into antibiotic resistance in Helicobacter pylori eradication. Gastroenterol Res Pract. 2012;2012:723183. doi: 10.1155/2012/723183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gerrits MM, van Vliet AH, Kuipers EJ, Kusters JG. Helicobacter pylori and antimicrobial resistance: Molecular mechanisms and clinical implications. Lancet Infect Dis. 2006;6:699–709. doi: 10.1016/S1473-3099(06)70627-2. [DOI] [PubMed] [Google Scholar]
- 31.Graham DY, Shiotani A. New concepts of resistance in the treatment of Helicobacter pylori infections. Nat Clin Pract Gastroenterol Hepatol. 2008;5:321–331. doi: 10.1038/ncpgasthep1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McColl KEL, el-Omar E, Gillen D. Interactions between H. pylori infection, gastric acid secretion and anti-secretory therapy. Br Med Bull. 1998;54:121–138. doi: 10.1093/oxfordjournals.bmb.a011663. [DOI] [PubMed] [Google Scholar]
- 33.Mégraud F, Lamouliatte H. Review article: The treatment of refractory Helicobacter pylori infection. Aliment Pharmacol Ther. 2003;17:1333–1343. doi: 10.1046/j.1365-2036.2003.01592.x. [DOI] [PubMed] [Google Scholar]
- 34.Hancock REW, Sahl HG. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat Biotechnol. 2006;24:1551–1557. doi: 10.1038/nbt1267. [DOI] [PubMed] [Google Scholar]
- 35.Radzishevsky IS, et al. Improved antimicrobial peptides based on acyl-lysine oligomers. Nat Biotechnol. 2007;25:657–659. doi: 10.1038/nbt1309. [DOI] [PubMed] [Google Scholar]
- 36.Nederberg F, et al. Biodegradable nanostructures with selective lysis of microbial membranes. Nat Chem. 2011;3:409–414. doi: 10.1038/nchem.1012. [DOI] [PubMed] [Google Scholar]
- 37.Fjell CD, Hiss JA, Hancock REW, Schneider G. Designing antimicrobial peptides: Form follows function. Nat Rev Drug Discov. 2011;11:37–51. doi: 10.1038/nrd3591. [DOI] [PubMed] [Google Scholar]
- 38.Porter EA, Wang X, Lee HS, Weisblum B, Gellman SH. Non-haemolytic beta-amino-acid oligomers. Nature. 2000;404:565. doi: 10.1038/35007145. [DOI] [PubMed] [Google Scholar]
- 39.Tew GN, et al. De novo design of biomimetic antimicrobial polymers. Proc Natl Acad Sci USA. 2002;99:5110–5114. doi: 10.1073/pnas.082046199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Makobongo MO, Gancz H, Carpenter BM, McDaniel DP, Merrell DS. The oligo-acyl lysyl antimicrobial peptide C12K-2β12 exhibits a dual mechanism of action and demonstrates strong in vivo efficacy against Helicobacter pylori. Antimicrob Agents Chemother. 2012;56:378–390. doi: 10.1128/AAC.00689-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu L, Fang Y, Huang Q, Wu J. A rigidity-enhanced antimicrobial activity: A case for linear cationic α-helical peptide HP(2-20) and its four analogues. PLoS One. 2011;6:e16441. doi: 10.1371/journal.pone.0016441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Makobongo MO, Gilbreath JJ, Merrell DS. Nontraditional therapies to treat Helicobacter pylori infection. J Microbiol. 2014;52:259–272. doi: 10.1007/s12275-014-3603-5. [DOI] [PubMed] [Google Scholar]
- 43.Engler AC, et al. Emerging trends in macromolecular antimicrobials to fight multi-drug-resistant infections. Nano Today. 2012;7:201–222. [Google Scholar]
- 44.Xiong M, et al. Helical antimicrobial polypeptides with radial amphiphilicity. Proc Natl Acad Sci USA. 2015;112:13155–13160. doi: 10.1073/pnas.1507893112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xiong M, et al. Bacteria-assisted activation of antimicrobial polypeptides by a random-coil to helix transition. Angew Chem Int Ed Engl. 2017;56:10826–10829. doi: 10.1002/anie.201706071. [DOI] [PubMed] [Google Scholar]
- 46.Press AG, et al. Gastrointestinal pH profiles in patients with inflammatory bowel disease. Aliment Pharmacol Ther. 1998;12:673–678. doi: 10.1046/j.1365-2036.1998.00358.x. [DOI] [PubMed] [Google Scholar]
- 47.Dressman JB, et al. Upper gastrointestinal (GI) pH in young, healthy men and women. Pharm Res. 1990;7:756–761. doi: 10.1023/a:1015827908309. [DOI] [PubMed] [Google Scholar]
- 48.Dressman JB. Comparison of canine and human gastrointestinal physiology. Pharm Res. 1986;3:123–131. doi: 10.1023/A:1016353705970. [DOI] [PubMed] [Google Scholar]
- 49.Lu H, Cheng J. Hexamethyldisilazane-mediated controlled polymerization of alpha-amino acid N-carboxyanhydrides. J Am Chem Soc. 2007;129:14114–14115. doi: 10.1021/ja074961q. [DOI] [PubMed] [Google Scholar]
- 50.Mcgowan CC, Cover TL, Blaser MJ. The proton pump inhibitor omeprazole inhibits acid survival of Helicobacter-pylori by a urease-independent mechanism. Gastroenterology. 1994;107:1572–1578. doi: 10.1016/0016-5085(94)90121-x. [DOI] [PubMed] [Google Scholar]
- 51.Peek RM, Jr, et al. Helicobacter pylori strain-specific genotypes and modulation of the gastric epithelial cell cycle. Cancer Res. 1999;59:6124–6131. [PubMed] [Google Scholar]
- 52.Israel DA, et al. Helicobacter pylori genetic diversity within the gastric niche of a single human host. Proc Natl Acad Sci USA. 2001;98:14625–14630. doi: 10.1073/pnas.251551698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McConnell EL, Basit AW, Murdan S. Measurements of rat and mouse gastrointestinal pH, fluid and lymphoid tissue, and implications for in-vivo experiments. J Pharm Pharmacol. 2008;60:63–70. doi: 10.1211/jpp.60.1.0008. [DOI] [PubMed] [Google Scholar]
- 54.Vétizou M, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350:1079–1084. doi: 10.1126/science.aad1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sivan A, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350:1084–1089. doi: 10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Iida N, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 2013;342:967–970. doi: 10.1126/science.1240527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang M. High antibiotic resistance rate: A difficult issue for Helicobacter pylori eradication treatment. World J Gastroenterol. 2015;21:13432–13437. doi: 10.3748/wjg.v21.i48.13432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ong ZY, Wiradharma N, Yang YY. Strategies employed in the design and optimization of synthetic antimicrobial peptide amphiphiles with enhanced therapeutic potentials. Adv Drug Deliv Rev. 2014;78:28–45. doi: 10.1016/j.addr.2014.10.013. [DOI] [PubMed] [Google Scholar]
- 59.Fox JL. Antimicrobial peptides stage a comeback. Nat Biotechnol. 2013;31:379–382. doi: 10.1038/nbt.2572. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.