Significance
To protect biodiversity for the long term, nature reserves and other protected areas need to represent a broad range of different genetic types. However, genetic data are expensive and time-consuming to obtain. Here we show that freely available environmental and geographic variables can be used as effective surrogates for genetic data in conservation planning. This means that conservation planners can, with some confidence, design protected area systems to represent intraspecific genetic diversity without investing in expensive programs to obtain and analyze genetic data.
Keywords: conservation, biodiversity, AFLP, genetic diversity, alpine plants
Abstract
Protected areas buffer species from anthropogenic threats and provide places for the processes that generate and maintain biodiversity to continue. However, genetic variation, the raw material for evolution, is difficult to capture in conservation planning, not least because genetic data require considerable resources to obtain and analyze. Here we show that freely available environmental and geographic distance variables can be highly effective surrogates in conservation planning for representing adaptive and neutral intraspecific genetic variation. We obtained occurrence and genetic data from the IntraBioDiv project for 27 plant species collected over the European Alps using a gridded sampling scheme. For each species, we identified loci that were potentially under selection using outlier loci methods, and mapped their main gradients of adaptive and neutral genetic variation across the grid cells. We then used the cells as planning units to prioritize protected area acquisitions. First, we verified that the spatial patterns of environmental and geographic variation were correlated, respectively, with adaptive and neutral genetic variation. Second, we showed that these surrogates can predict the proportion of genetic variation secured in randomly generated solutions. Finally, we discovered that solutions based only on surrogate information secured substantial amounts of adaptive and neutral genetic variation. Our work paves the way for widespread integration of surrogates for genetic variation into conservation planning.
Protected areas spearhead conservation efforts (1). They buffer species from anthropogenic impacts, providing places for them to persist. Since the resources available for conservation are limited, protected areas need to be sited in places that fulfill conservation objectives for minimal cost (2). To achieve this, conservation planning exercises often generate plans for entire networks of protected areas (prioritizations) to preserve species and broad-scale biodiversity processes (3). After determining which species to protect [e.g., using evolutionary distinctiveness (4)], conservation planners need to ensure that prioritizations represent the intraspecific genetic variation found within species to secure their long-term persistence (5–7). As a consequence, there has been increasing interest in designing prioritizations that fulfill this objective (8–12).
Although the strength of natural selection is a continuous force, genetic variation is often classified as adaptive or neutral (reviewed in ref. 13). Adaptive genetic variation is associated with loci that significantly affect fitness. Typically, such “adaptive” loci are detected due to anomalous patterns among individuals (e.g., refs. 14 and 15), although phenotypic–genetic associations are more likely to identify functionally important genomic regions for individual species (e.g., ref. 16). By representing the full range of adaptive variation, including adaptations of which we are currently unaware, protected areas can enhance a species’ capacity to persist in a range of different conditions (5, 17). In contrast, neutral genetic variation is associated with loci that do not significantly affect fitness but instead reflect the evolutionary history of different populations. By representing the full range of neutral variation, protected areas can safeguard against the adverse effects of low genetic diversity (18). Thus, optimally sited protected area networks would represent patterns of both adaptive and neutral genetic variation (6).
Recently, the use of surrogate data has been proposed to generate prioritizations that capture intraspecific genetic variation (10, 19, 20)—without needing to use genetic data directly. Since adaptation is ultimately driven by selection pressures, environmental variables have been proposed as surrogates for adaptive genetic variation (e.g., refs. 3 and 19). Specifically, by conserving individuals in a broad range of environmental conditions, we might expect to capture individuals with a diverse set of local adaptations, and, overall, capture a large proportion of the adaptive genetic variation present in the species. However, the effectiveness of this approach remains unverified. Neutral genetic variation arises from a reduction in gene flow between populations. Over the last few years, the field of phylogeography has predominantly focused on describing neutral genetic diversity, and so the landscape factors that affect neutral variation are relatively well understood (reviewed in ref. 21). Potential surrogates for neutral genetic variation have been based on variables that predict the level of connectivity between different areas. For instance, according to the isolation by distance model (22), populations located farther apart are predicted to experience less gene flow and, in turn, share less genetic material at neutral loci. Based on this idea, spreading conservation priorities evenly across the geographic distribution of an island network has been found to capture more neutral genetic variation (20). However, this remains untested in spatially contiguous systems where connectivity is complicated by many additional factors [e.g., land use (23)]—the typical situation in most conservation planning exercises.
Here we determine whether environmental and geographic surrogates capture adaptive and neutral genetic variation, respectively, in the context of conservation planning. We use distribution and genomic data for 27 alpine plant species in the European Alps that were obtained by the IntraBioDiv project (24). The data were collected following a gridded sampling scheme which we adopted as planning units for this study. We show that, for most species, the spatial patterns of adaptive and neutral genetic variation correlated with environmental [obtained from WorldClim (25)] and geographic variables. We also show that, for most species, the level of association is strong enough to be operational for conservation planning. Finally, we demonstrate that using these surrogates results in prioritizations that secure a substantial proportion of intraspecific genetic variation.
Results
We detected putatively adaptive genetic variation in 10 of the 27 plant species. We used two outlier detection methods—that solely used genetic data and did not use environmental data—to identify loci under selection. These methods returned reasonably consistent results (mean 87.92% loci per species assigned the same classification ±8.75 SD; Dataset S1). Of the species that were associated with adaptive genetic variation, only a small proportion of loci were classified as being adaptive (mean 3.01% loci per species ±1.95 SD). After identifying the loci showing strong signals of selection, we used nonmetric multidimensional scaling (NMDS) analyses to identify the main gradients of adaptive (if detected) and neutral variation for each species. Generally, only a small number of continuous dimensions were needed to sufficiently describe their patterns of adaptive ( for all species; SI Appendix, Table S1) and neutral genetic variation ( for all species; SI Appendix, Table S1). The resulting ordinations were used to construct an adaptive (if detected) and neutral genetic space for each species. The spatial distribution of these genetic spaces (SI Appendix, Figs. S1–S27) generally showed substantial spatial autocorrelation (e.g., Cerastium uniflorum and Dryas octopetala; SI Appendix, Figs. S6 and S8), suggesting that the proposed surrogate variables have the potential to be effective for conservation planning.
We verified that the spatial patterns in environmental variation correlated with the broad-scale patterns in adaptive genetic variation for each species. We also verified that there was a correlation between neutral genetic variation and variation in geographic position. For each species, we constructed dissimilarity matrices expressing differences between the planning units where individuals were detected based on the units’ (i) environmental characteristics and (ii) geographic position, and also the (iii) adaptive (if adaptive loci detected) and (iv) neutral genetic characteristics of the individuals found inside them. The spatial patterns of environmental variation were significantly correlated with the patterns of adaptive genetic variation for 8 of the 10 species associated with adaptive variation (P < 0.05; mean 0.09 marginal ± 0.06 SD for significant models; SI Appendix, Table S2). Similarly, geographic distance between planning units was also correlated significantly with the spatial patterns of neutral genetic variation among planning units for 26 of the 27 species (P < 0.05; mean 0.21 marginal ± 0.16 SD for significant models; SI Appendix, Table S2). Thus, for most species, planning units that contained different environmental conditions contained individuals with different adaptive genetic characteristics, and planning units that were farther apart tended to contain individuals with different neutral genetic characteristics. After verifying these correlations, our next step was to determine whether they were strong enough to be useful for conservation planning.
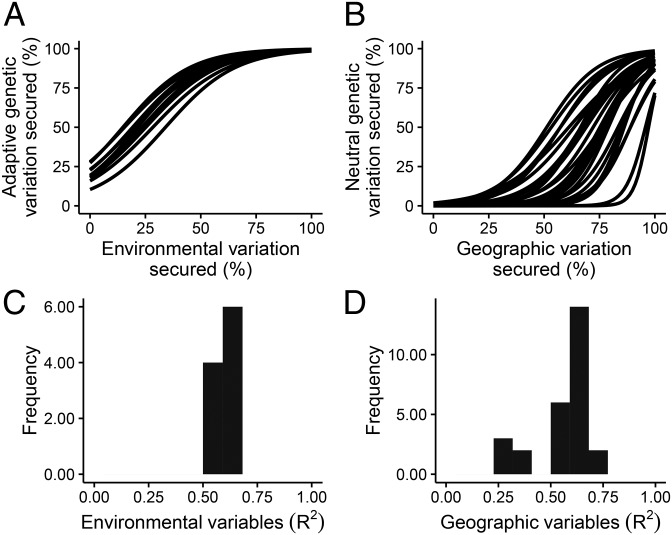
For each species, we generated a suite of 10,000 random prioritizations, and calculated the proportion of environmental variation and adaptive (if present) genetic variation that each prioritization captured. We then repeated this process, and calculated the proportion of variation in geographic position and neutral genetic variation they sampled. The environmental and geographic variables were moderately effective predictors for the genetic variation represented by the randomly generated prioritizations (Fig. 1 and SI Appendix, Figs. S28 and S29). The relationship between the proportion of genetic and surrogate variation secured in the prioritizations varied among the species (species × proportion of environmental variation interaction term: ; species × proportion of geographic variation interaction term: ; < 0.001). Post hoc analyses showed that these relationships were positive for all species (environmental: minimum Z = 40.71, maximum ; geographic: minimum Z = 23.23, maximum ; SI Appendix, Table S3). After establishing that the proportion of surrogate variation secured in a prioritization also predicted the proportion of genetic variation it secured, our final step was to determine whether prioritizations generated using targets to represent variation in the surrogate variables were more effective at representing intraspecific genetic variation.
Fig. 1.
The proportion of (A) adaptive (10 species) and (B) neutral (27 species) genetic variation represented in a suite of randomly generated prioritizations for different species as a function of the surrogate variation they also secured (lines). The distribution of values (C and D) for the 37 models shown in A and B.
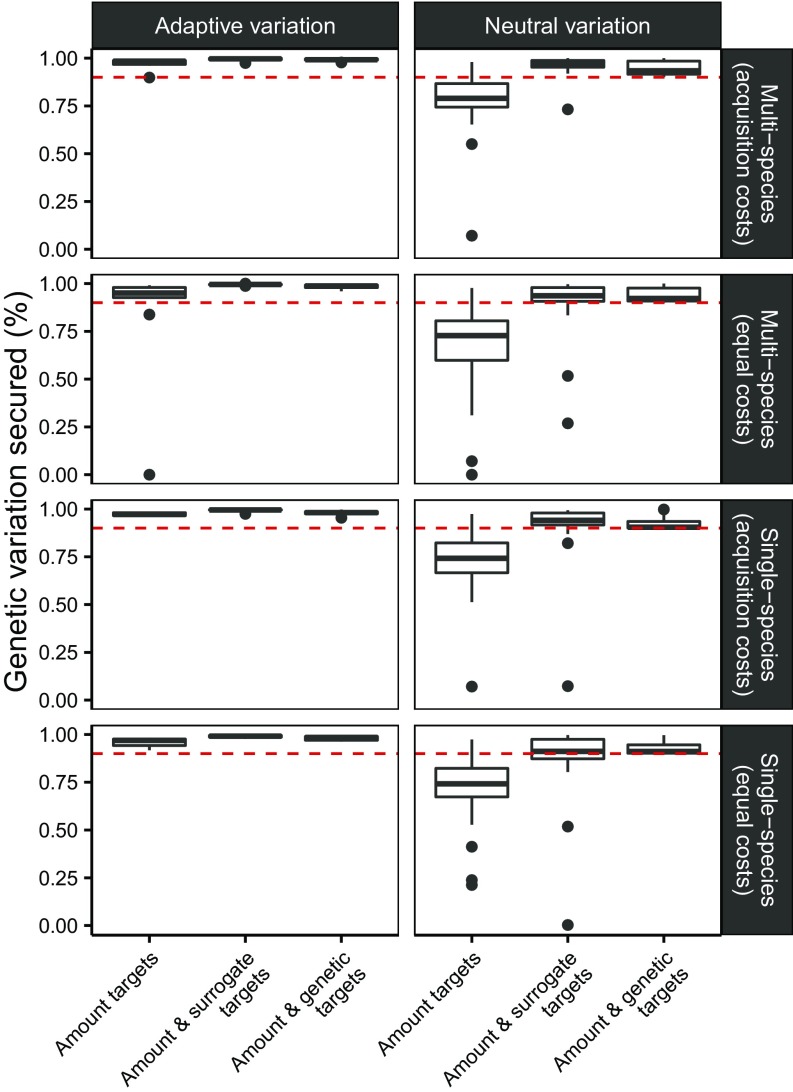
To determine whether environmental and geographic targets could improve the effectiveness of prioritizations in representing genetic variation, we generated prioritizations using (i) “amount targets” reflecting the traditional approach of representing a certain proportion of each species’ geographic distribution, (ii) “amount and surrogate targets” where we targeted representation of the environmental and geographic surrogates as well as a certain proportion of each species’ geographic distribution, and (iii) “amount and genetic targets” where we targeted representation of the directly measured genetic variation as well as a certain proportion of each species’ geographic distribution. To determine whether these patterns were robust, for each of the three combinations of the targets, we generated prioritizations under four scenarios: (i) single species with equal costs, (ii) single species with acquisition costs, (iii) multispecies with equal costs, and (iv) multispecies with acquisition costs (Fig. 2 and SI Appendix, Fig. S30).
Fig. 2.
The proportion of adaptive and neutral genetic variation secured for each species in solutions based on different targets and scenarios. Box plots show the median, and the 25th and 75th percentiles. Whiskers show the data at 1.5 times the interquantile range. Points show data outside the whiskers. Red lines show the target proportion of genetic variation.
The proportion of genetic variation secured in a given prioritization was not found to depend on any interaction between the planning scenario, targets used to generate the prioritization, and the type of genetic variation measured (; ). The proportion of genetic variation secured in a prioritization did depend, however, on the targets used to generate the prioritization (; ). Most notably, prioritizations based on the surrogate variables represented significantly more genetic variation than amount-based prioritizations (93.25% ± 13.81 SD overall genetic variation secured versus 78.39% ± 23.54 SD; ; ), and were not distinguishable from prioritizations based on measured genetic data (94.68% ± 3.69 SD; ; ). These results indicate that environmental and geographic surrogates in this case far outperform traditional amount-based conservation planning, and perform almost as well as a conservation plan based directly on genetic data.
Overall, the prioritizations tended to secure a greater proportion of adaptive genetic variation than neutral genetic variation (adaptive: mean 95.9% ± 8.23 secured across all prioritizations; neutral: mean 72.08% ± 24.06; ; ). Thus, regardless of the targets used to generate a prioritization, or the planning context under which the prioritization was generated, prioritizations tend to secure more adaptive than neutral genetic variation.
The average proportion of genetic variation secured by prioritizations varied under different scenarios (; ). Specifically, prioritizations generated under single-species scenario with equal costs (mean 78.42% genetic variation secured ±23.54) secured less genetic variation than those under the single species with acquisition costs (mean 88.74% genetic variation secured ±15.54; ; ) or the multispecies with acquisition cost scenarios (mean 91.91% genetic variation secured ±12.31; ; ). These results suggest the proportion of genetic variation secured in a prioritization may be sensitive to acquisition cost.
Discussion
We have shown that broad-scale environmental and geographic variables can be effective as surrogates for adaptive and neutral genetic variation in conservation planning. Our study investigates this using field measurements of genetic variation for a suite of plant species across an ecoregion (26). For most species, the proportion of surrogate variation captured in a prioritization predicted the proportion of genetic variation that was also captured in the prioritization. Moreover, for most species, prioritizations generated using geographic targets secured a greater proportion of neutral genetic variation than traditional conservation planning methods. These environmental and geographic surrogates were based on freely available datasets and, with the exception of a few places with poor climatic data (25), could be applied to any study region across the world.
Our results demonstrate that environmental and geographic variables are effective surrogates for most species considered here (Fig. 2). Despite this, geographic distance was a surprisingly poor surrogate for neutral genetic variation for a few species (e.g., Gentiana nivalis = 0.3; Gypsophila repens = 0.25; Luzula alpinopilosa = 0.28). One explanation for this result is that geographic distance will be a poor surrogate for neutral genetic variation where spatial genetic structure is complicated by additional factors (10, 23, 27). For example, some species can maintain relatively high connectivity between distant populations [e.g., wind dispersing plants (24)] and, in some places, gene flow can be disrupted by landscape features [e.g., anthropogenically modified land (23)]. Overall, such species were the exception in our analysis, and both surrogates performed moderately well for most species.
Our results show that environmental and geographic surrogates can be used to capture genetic variation in prioritizations. The next question is, what percentage of surrogate variation should planners target to preserve genetic variation? To secure at least 90% of the species’ neutral genetic variation, prioritizations needed to sample 94.45% ± 6.64 SD of the variation in geographic position among the planning units occupied by each species. Additionally, to secure at least 90% of each species’ adaptive genetic variation, prioritizations needed to sample 57.01% ± 6.67 SD of the environmental variation in planning units occupied by each of the species. For all species, however, it was possible to generate solutions that secured a large proportion of the surrogate variation (>90%) and only a small proportion of genetic variation (<20%; SI Appendix, Figs. S28 and S29). Planners may avoid such outcomes by using both amount- and surrogate-based targets. Depending on the study area and the species of conservation interest, higher targets may be needed to increase the likelihood that prioritizations will secure a large proportion of the intraspecific genetic variation for all of the species in the planning exercise.
Our results suggest that conservation planners can secure a representative sample of intraspecific adaptive genetic variation using conventional reserve selection methods without needing surrogate or genetic data (Fig. 2). This finding was evident in both the single-species and multispecies prioritizations. One explanation for the single-species prioritizations is that, because the spatial patterns of adaptive variation tended to cluster into a few main groups (e.g., Carex sempervirens and C. uniflorum; SI Appendix, Figs. S5 and S6), a random selection of planning units across a species’ geographic distribution would have a fairly high chance of capturing individuals that belonged to several of the main genetic groups. This mechanism also explains why adaptive variation accumulated much more quickly than neutral genetic variation in the randomly generated prioritizations (Fig. 1). On the other hand, one potential explanation for the multispecies prioritization is that, because it was generated using a comprehensive set of species—each with their own habitat preferences—the solution was forced to secure each species in a range of different suitable habitats and, in turn, capture a representative sample of the species’ adaptive genetic variation. If future studies on other taxa and biomes verify that conventional reserve selection methods do indeed conserve intraspecific adaptive genetic variation, this finding could have large implications for conservation planning.
There are several limitations associated with our analysis. Firstly, the size of the planning units we used (∼20 × 22.5 km) is larger than is typically used in regional conservation planning. We used this resolution because the genetic data were collected at this scale. While we could have interpolated the genetic data to a finer resolution, this would have introduced additional spatial autocorrelation and biased our analysis. Secondly, we used geographic distances as surrogates for neutral genetic variation. Although distances that incorporate data on dispersal cost may perform better (e.g., topography), such distances often require species-specific scaling (e.g., ref. 23) and so cannot easily be used in multispecies planning exercises. Thirdly, we used amplified fragment length polymorphism data (AFLP) (28) to describe genetic variation. While next-generation sequencing provides higher-resolution genetic information (13), we know of no suitable multispecies genomic dataset, and our methodology would still have used only the main gradients of the genetic variation to generate prioritizations in a feasible period. Furthermore, even with modern population genomic approaches, a survey can, at best, hope to identify markers that are linked to functionally adaptive variants. Fourthly, to our knowledge, none of the species we investigated here are at serious risk of extinction (29, 30). Since anthropogenic processes can alter patterns of species’ genetic variation (e.g., ref. 23), future work should aim to establish whether these surrogates are effective for imperiled species. Finally, conventional conservation planning exercises are sensitive to the quality and completeness of the underlying distribution data (31), and it is likely that the effectiveness of these surrogates is also sensitive to data quality. For instance, prioritizations generated using distribution data that omitted populations in specific habitats may fail to represent certain adaptations. Recent advances in distribution modeling have provided a wealth of data for conservation planners (32), and so they should endeavor to obtain high-quality data where possible. Nevertheless, our findings pave the way for the widespread integration of evolutionary processes at the intraspecific level into reserve selection.
Genetic data provide a broad range of insights into a species’ persistence and are fundamental to managing certain kinds of conservation problems (7). However, conservation interventions often need to be implemented urgently before the highest-quality data are available (33). We found broad-scale environmental and geographic variables to be effective surrogates for representing adaptive and neutral genetic variation for most species in our study system. We call for further studies to examine these surrogates in other taxa and biomes. In cases where genetic data are not available, careful use of such surrogates in conservation planning could vastly improve the chances of long-term biodiversity persistence for relatively little additional cost.
Materials and Methods
Study System.
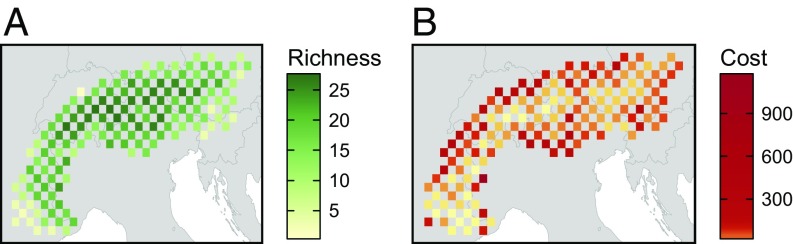
We used data for 27 alpine plant species in the European Alps collected by the IntraBioDiv project (ref. 24 and Fig. 3A). This dataset has been used to explore patterns of adaptive (e.g., ref. 34) and neutral genetic variation (e.g., ref. 26), and the potential for species richness as a surrogate for genetic diversity (35). Data were collected using a longitude by latitude grid (∼20 × 22.5 km; SI Appendix, Fig. S31). Project surveyors visited every second grid cell, and, for each species, they collected samples from three individuals if any individuals were found. They genotyped samples using AFLPs, and constructed matrices denoting the presence of polymorphisms at loci for each species (mean 130.7 ± 54.9 SD markers genotyped per species; for more information, see ref. 26). Thus, the dataset contains information describing the genomic properties of individuals for each species over a geographic grid. We used these data because they comprised comparable genetic information for a range of species with different evolutionary and life histories collected using a standardized sampling scheme. To permit replication and validation of this study, all of our data, code, and results are stored in an online repository (www.github.com/jeffreyhanson/genetic-surrogates) and are also available under the digital object identifier (DOI) 10.5281/zenodo.843625. All spatial and statistical analyses were conducted in R (version 3.3.2) (36).
Fig. 3.
Map of the study area showing (A) the pattern of species richness and (B) the distribution of acquisition cost depicted using a quantile-based color ramp. Squares denote planning units.
Landscape Data.
We adopted the sampling grid used to collect data as planning units to develop prioritizations. Of the total 388 cells in the grid, we used the 149 cells that contained samples for subsequent analysis. We calculated the total human population density inside each planning unit [1 km2 resolution from the Global Rural-Urban Mapping Project (37)] and used this to represent acquisition cost (Fig. 3B).
We created environmental and geographic surrogate variables for each species (SI Appendix, Fig. S32). To describe the geographic location of each planning unit (SI Appendix, Fig. S33), we projected the grid into an equidistant coordinate system (Europe Equidistant Conic; ESRI:102031), calculated the centroid of each grid cell, and extracted their 2D coordinates. To describe the environmental characteristics of each planning unit (SI Appendix, Fig. S34), we obtained 19 climatic layers [ resolution; obtained from WorldClim (25)], projected them and the planning units into an equal-area coordinate system (Europe Lambert Conformal Conic; ESRI:102014), and computed planning unit averages for each climatic variable. To reduce dimensionality, for each species, we subjected the climatic values associated with the planning units they were found in to a principal components analysis (PCA). We used the first three principal components to characterize climatic variation found across the species’ geographic distributions inside the study area (mean 90.26% variation described ±0.99 SD of the total climatic variation; SI Appendix, Table S4). Thus, we constructed a 2D geographic space as a potential surrogate for neutral genetic variation, and a 3D environmental space as a potential surrogate for adaptive genetic variation for each species. In these spaces, each planning unit was associated with a single point. Planning units that contained environmental conditions that were more comparable, or were located in places with higher spatial proximity, were associated with points that were closer together in these environmental or geographic spaces.
Adaptive and Neutral Genetic Data.
To investigate the effectiveness of our surrogates, we first needed to identify which of the sampled loci were adaptive (SI Appendix, Fig. S35). Following recommended practices, we used two outlier detection methods to achieve this [an individual- and a population-level method (38)]. The basic premise underpinning such methods is that neutral loci are expected to exhibit a certain level of variation, and loci that deviate from this expectation are likely to be under selection (13). The advantage of these methods—in contrast with environmental association analyses—is that they do not use environmental data, which would have introduced an element of circularity into our analysis. Loci identified by both outlier detection methods were treated as adaptive, and the remainder were treated as neutral. To minimize false positives, we omitted loci from both methods where the global frequency of the minor allele was less than 10%, and treated them as neutral.
The first outlier detection method involved fitting multinominal-Dirichlet models implemented in BayeScan (version 2.1) (14). We adopted a similar methodology to Bothwell et al. (34) and applied it to each of the species separately. Following their methodology, we initially grouped conspecifics into genetic lineages to further minimize false positives (SI Appendix, Figs. S36–S62), by fitting admixture and correlated alleles models implemented in Structure [version 2.3.4 (39); 20 replicates per species; 5,000 admixture burn-in iterations; 300,000 burn-in iterations; 400,000 total iterations] using the number of lineages previously determined by Alvarez et al. (26) and combining replicate runs using ClumPP [version 1.1.2 (40); greedy algorithm based on the statistic; 1,000 iterations]. We then ran BayeScan for each species using these lineages (1:1 prior odds; four replicates per species; 20 pilot runs; 100,000 burn-in iterations; 110,000 total iterations thinned by 10 iterations) using a suitable false discovery rate [ (41)]. We omitted individuals if their population membership was uncertain (maximum membership probability ).
The second outlier detection method involved fitting PCAs to identify outlier loci [implemented in the pcadapt R package (42)]. To enable comparisons between the two outlier detection methods, we used the same individuals in this analysis as in the BayeScan analysis. For each of the 27 species, we first imputed missing data by replacing missing values with the average frequency among conspecifics, then ran a PCA over the loci matrix, and extracted the minimum number of components needed to secure the overarching population level variation among the loci (10% of the variation in loci). We then computed q values using Mahalanobis distances, and used the same false discovery rate used in the BayeScan analysis to identify loci under selection [using the qvalue R package (43)].
After classifying loci as adaptive or neutral, we mapped the main gradients of the adaptive (if detected) and neutral genetic variation for each species. We discarded the population groupings, and partitioned species’ adaptive and neutral loci into separate matrices. We applied NMDS [implemented in the vegan R package (44)] using Gower distances [via the cluster R package (45)] to derive continuous variables that described the main gradients of adaptive and neutral genetic variation separately for each species (SI Appendix, Table S1). To ensure that the ordinations described a sufficient amount of the genetic variation, we ran successive scaling analyses with increasing dimensionality until a sufficient stress value was obtained (maximum stress value ≤ 0.25; 100 random starts for each analysis). Since each grid cell had multiple samples per species, we averaged the ordinated values for conspecifics in the same grid cell.
Thus, we constructed an adaptive (if detected) and neutral genetic space for each species. For a given species, each planning unit in which it occurred was associated with a multidimensional point in the species’ adaptive genetic space (if adaptive genetic variation was detected) and another multidimensional point in the species’ neutral genetic space. Planning units that were closer together in these genetic spaces were occupied by individuals with more comparable AFLP data. By spreading out conservation effort across these genetic spaces, and, in turn, selecting planning units occupied by individuals with increasingly different polymorphisms, prioritizations can secure more genetic variation.
Prioritization Method.
We used the raptr R package to generate solutions (12). This toolkit can identify the cheapest set of planning units required to preserve both a target proportion of the species’ geographic range (using amount-based targets) and a target proportion of intraspecific variation (using space-based targets). We used the environmental and geographic surrogate spaces, and the adaptive (if detected) and neutral genetic spaces, to generate and evaluate solutions. Solutions associated with negative values—because they secured very little genetic variation—were replaced with zeros to facilitate statistical analysis. We solved all reserve selection problems to within 10% of optimality (using Gurobi; version 7.0.2).
Computational and Statistical Analyses.
Our first aim was to determine whether the environmental and geographic variables correlate with the spatial patterns of adaptive and neutral genetic variation. To achieve this, for each species, we created dissimilarity matrices using Euclidean distances and the data in the surrogate and genetic spaces. These matrices showed differences between the planning units occupied by the species in terms of the units’ (i) geographic position, (ii) environmental characteristics, and (iii) adaptive (if detected) and (iv) neutral genetic characteristics of the individuals inside them.
We fitted maximum likelihood population effects (MLPE) models using maximum likelihood to investigate correlations between the dissimilarity matrices [using the lme4 R package (46)]. These models use random effects to accommodate the structure of dissimilarity matrices. For each species, we fitted an MLPE model to the species’ dissimilarity matrices based on geographic position and neutral genetic variation. If adaptive loci were detected, we also fitted an MLPE model to the species’ dissimilarity matrices describing environmental and adaptive genetic variation. All data variables were z-transformed to improve convergence. To test whether the surrogates explained the genetic variation, we compared each model to its null model using a test, and applied Bonferroni corrections.
Our second aim was to determine whether the environmental and geographic variables were effective surrogates for adaptive and neutral genetic variation. For each species, we generated 20,000 prioritizations by randomly selecting different combinations of planning units that the species occupied. For half of these random prioritizations, we calculated the proportion of geographic variation and neutral genetic variation they secured, and, for the other half, we calculated the proportion of environmental variation and adaptive genetic variation they secured. These calculations were performed using the raptr R package.
We fitted two full generalized linear models with logit link functions. The first model was fit to the proportion of adaptive genetic variation secured in a prioritization using the proportion of environmental variation also secured in the prioritization. The second model was fit to the proportion of neutral genetic variation secured in a prioritization using the proportion of geographic variation also secured in the prioritization. Additionally, both models contained a variable indicating the species for which the prioritizations were generated, and an interaction term. They were subjected to backward stepwise term deletion routines to assess term significance. Post hoc analyses were conducted to assess trends for each species using Bonferroni corrections [using the multcomp R package (47)]. To assess the performance of the surrogates, we refit these models separately for each species and computed the Cragg and Uhler’s pseudo value [using the pscl R package (48)].
Our third aim was to determine whether surrogate-based targets improved representation of genetic variation in prioritizations. As previously mentioned, we generated prioritizations using different combinations of targets and conservation planning scenarios. We used 20% amount-based targets for each species in all prioritizations to secure an adequate proportion of the species’ distributions. Based on the results from our previous analysis, we used 97.5% surrogate-based targets and 90% genetic-based targets. We generated a single solution for each target/scenario combination, except for the “single-species (equal costs) with amount-based targets” combination, for which we generated 1,000 replicates because it had many optimal solutions. We computed the proportion of adaptive (if present) and neutral genetic variation secured for each species in the prioritizations.
We fitted generalized linear mixed-effects models with logit link functions to evaluate the prioritizations (using the lme4 R package). We fitted a full model to the proportion of genetic variation secured in a given prioritization. This model contained categorical variables indicating the targets (amount only, amount and surrogate targets, or amount and genetic targets), the planning scenario (single species, multispecies, or multispecies with cost), the type of genetic variation measured (adaptive or neutral), and all interactions between them. Data for same species were accommodated using a random intercept term. The full model was subjected to a backward stepwise term deletion routine to assess significance. A post hoc analysis was conducted using Tukey contrasts with Bonferroni corrections (using the multcomp R package).
Supplementary Material
Acknowledgments
J.O.H. was supported by an Australian Government Research Training Program Scholarship. R.A.F. has an Australian Research Council Future Fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The code, data, and results reported in this paper have been deposited in GitHub (10.5281/zenodo.843625).
See Commentary on page 12638.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1711009114/-/DCSupplemental.
References
- 1.Watson JE, Dudley N, Segan DB, Hockings M. The performance and potential of protected areas. Nature. 2014;515:67–72. doi: 10.1038/nature13947. [DOI] [PubMed] [Google Scholar]
- 2.Margules CR, Pressey RL. Systematic conservation planning. Nature. 2000;405:243–253. doi: 10.1038/35012251. [DOI] [PubMed] [Google Scholar]
- 3.Cowling RM, Pressey RL. Rapid plant diversification: Planning for an evolutionary future. Proc Natl Acad Sci USA. 2001;98:5452–5457. doi: 10.1073/pnas.101093498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Isaac NJ, Turvey ST, Collen B, Waterman C, Baillie JE. Mammals on the EDGE: Conservation priorities based on threat and phylogeny. PLoS One. 2007;2:e296. doi: 10.1371/journal.pone.0000296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crandall KA, Bininda-Emonds ORP, Mace GM, Wayne RK. Considering evolutionary processes in conservation biology. Trends Ecol Evol. 2000;15:290–295. doi: 10.1016/s0169-5347(00)01876-0. [DOI] [PubMed] [Google Scholar]
- 6.Moritz C. Strategies to protect biological diversity and the evolutionary processes that sustain it. Syst Biol. 2002;51:238–254. doi: 10.1080/10635150252899752. [DOI] [PubMed] [Google Scholar]
- 7.Hendry AP, et al. Evolutionary biology in biodiversity science, conservation, and policy: A call to action. Evolution. 2010;64:1517–1528. doi: 10.1111/j.1558-5646.2010.00947.x. [DOI] [PubMed] [Google Scholar]
- 8.Diniz JAF, Telles M. Optimization procedures for establishing reserve networks for biodiversity conservation taking into account population genetic structure. Genet Mol Biol. 2006;29:207–214. [Google Scholar]
- 9.Carvalho SB, et al. Spatial conservation prioritization of biodiversity spanning the evolutionary continuum. Nat Ecol Evol. 2017;1:0151. doi: 10.1038/s41559-017-0151. [DOI] [PubMed] [Google Scholar]
- 10.Potts AJ, Hedderson TA, Cowling RM. Testing large-scale conservation corridors designed for patterns and processes: Comparative phylogeography of three tree species. Diversity Distrib. 2013;19:1418–1428. [Google Scholar]
- 11.Nielsen ES, Beger M, Henriques R, Selkoe KA, von der Heyden S. Multispecies genetic objectives in spatial conservation planning. Conserv Biol. 2017;31:872–882. doi: 10.1111/cobi.12875. [DOI] [PubMed] [Google Scholar]
- 12.Hanson JO, Rhodes JR, Possingham HP, Fuller RA. raptr: Representative and adequate prioritization toolkit in R. Methods Ecol Evol. 2017 doi: 10.1111/2041-210X.12862. [DOI] [Google Scholar]
- 13.Schoville SD, et al. Adaptive genetic variation on the landscape: Methods and cases. Annu Rev Ecol Evol Syst. 2012;43:23–43. [Google Scholar]
- 14.Foll M, Gaggiotti O. A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: A Bayesian perspective. Genetics. 2008;180:977–993. doi: 10.1534/genetics.108.092221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duforet-Frebourg N, Bazin E, Blum MG. Genome scans for detecting footprints of local adaptation using a Bayesian factor model. Mol Biol Evol. 2014;31:2483–2495. doi: 10.1093/molbev/msu182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steiner CC, Weber JN, Hoekstra HE. Adaptive variation in beach mice produced by two interacting pigmentation genes. PLoS Biol. 2007;5:1–10. doi: 10.1371/journal.pbio.0050219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sgro CM, Lowe AJ, Hoffmann AA. Building evolutionary resilience for conserving biodiversity under climate change. Evol Appl. 2011;4:326–337. doi: 10.1111/j.1752-4571.2010.00157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moritz C. Defining evolutionarily significant units for conservation. Trends Ecol Evol. 1994;9:373–375. doi: 10.1016/0169-5347(94)90057-4. [DOI] [PubMed] [Google Scholar]
- 19.Carvalho SB, Brito JC, Crespo EJ, Possingham HP. Incorporating evolutionary processes into conservation planning using species distribution data: A case study with the western Mediterranean herpetofauna. Diversity Distrib. 2011;17:408–421. [Google Scholar]
- 20.Ponce-Reyes R, Clegg SM, Carvalho SB, McDonald-Madden E, Possingham HP. Geographical surrogates of genetic variation for selecting island populations for conservation. Divers Distrib. 2014;20:640–651. [Google Scholar]
- 21.Avise JC. Phylogeography: Retrospect and prospect. J Biogeogr. 2009;36:3–15. [Google Scholar]
- 22.Wright S. Isolation by distance. Genetics. 1943;28:114–138. doi: 10.1093/genetics/28.2.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dudaniec RY, et al. Dealing with uncertainty in landscape genetic resistance models: A case of three co-occurring marsupials. Mol Ecol. 2016;25:470–486. doi: 10.1111/mec.13482. [DOI] [PubMed] [Google Scholar]
- 24.Meirmans P, Goudet J, Gaggiotti O. IntraBioDiv Consortium Ecology and life history affect different aspects of the population structure of 27 high-alpine plants. Mol Ecol. 2011;20:3144–3155. doi: 10.1111/j.1365-294X.2011.05164.x. [DOI] [PubMed] [Google Scholar]
- 25.Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. Very high resolution interpolated climate surfaces for global land areas. Int J Climatol. 2005;25:1965–1978. [Google Scholar]
- 26.Alvarez N, et al. History or ecology? Substrate type as a major driver of spatial genetic structure in alpine plants. Ecol Lett. 2009;12:632–640. doi: 10.1111/j.1461-0248.2009.01312.x. [DOI] [PubMed] [Google Scholar]
- 27.Fortuna MA, Albaladejo RG, Fernández L, Aparicio A, Bascompte J. Networks of spatial genetic variation across species. Proc Natl Acad Sci USA. 2009;106:19044–19049. doi: 10.1073/pnas.0907704106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vos P, et al. AFLP: A new technique for DNA fingerprinting. Nucleic Acids Res. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bornand C, et al. 2016 Rote Liste Gefässpflanzen. Gefährdete Arten der Schweiz. German. Available at https://www.infoflora.ch/en/assets/content/documents/roteliste_pflanzen_d_20160908.pdf. Accessed August 10, 2017.
- 30.International Union for Conservation of Nature 2017 The IUCN Red List of Threatened Species. Version 2017-1. Available at http://www.iucnredlist.org. Accessed August 10, 2017.
- 31.Wilson KA, Westphal MI, Possingham HP, Elith J. Sensitivity of conservation planning to different approaches to using predicted species distribution data. Biol Conserv. 2005;122:99–112. [Google Scholar]
- 32.Guisan A, et al. Predicting species distributions for conservation decisions. Ecol Lett. 2013;16:1424–1435. doi: 10.1111/ele.12189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ceballos G, Ehrlich PR, Dirzo R. Biological annihilation via the ongoing sixth mass extinction signaled by vertebrate population losses and declines. Proc Natl Acad Sci USA. 2017;114:E6089–E6096. doi: 10.1073/pnas.1704949114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bothwell H, et al. Identifying genetic signatures of selection in a non-model species, alpine gentian (Gentiana nivalis L.), using a landscape genetic approach. Conserv Genet. 2013;14:467–481. [Google Scholar]
- 35.Taberlet P, et al. Genetic diversity in widespread species is not congruent with species richness in alpine plant communities. Ecol Lett. 2012;15:1439–1448. doi: 10.1111/ele.12004. [DOI] [PubMed] [Google Scholar]
- 36.R Core Team 2014 R: A Language and Environment for Statistical Computing, Version 3.3.2. Available at http://www.R-project.org/. Accessed November 1, 2016.
- 37.CIESEN, Columbia University; International Food Policy Research Institute; The World Bank; Centro Internacional de Agricultura Tropical 2011 Global Rural-Urban Mapping Project, Version 1 (GRUMP v1): Urban Extents Grid. Available at dx.doi.org/10.7927/H4GH9FVG. Accessed April 13, 2016.
- 38.de Villemereuil P, Frichot É, Bazin É, François O, Gaggiotti OE. Genome scan methods against more complex models: When and how much should we trust them? Mol Ecol. 2014;23:2006–2019. doi: 10.1111/mec.12705. [DOI] [PubMed] [Google Scholar]
- 39.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: Dominant markers and null alleles. Mol Ecol Notes. 2007;7:574–578. doi: 10.1111/j.1471-8286.2007.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jakobsson M, Rosenberg NA. CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;23:1801–1806. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- 41.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Series B Methodol. 1995;57:289–300. [Google Scholar]
- 42.Luu K, Blum MG, Duforet-Frebourg N. 2016 Pcadapt: Fast Principal Component Analysis for Outlier Detection. R Package Version 3.0.2. Available at https://CRAN.R-_project.org/package=pcadapt. Accessed April 14, 2017.
- 43.Storey JD, Bass AJ, Dabney A, Robinson D. 2015 qvalue: Q-Value Estimation for False Discovery Rate Control. R Package Version 2.6.0. Available at github.com/jdstorey/qvalue. Accessed April 14, 2017.
- 44.Oksanen J, et al. 2015 Vegan: Community ecology package. R package version 2.4-1. Available at CRAN.R-project.org/package=vegan. Accessed April 14, 2017.
- 45.Maechler M, Rousseeuw P, Struyf A, Hubert M, Hornik K. 2015 Cluster: Cluster Analysis Basics and Extensions. R Package Version 2.0.5. Available at CRAN.R-_project.org/package=cluster. Accessed April 14, 2017.
- 46.Bates D, Mächler M, Bolker B, Walker S. Fitting linear mixed-effects models using lme4. J Stat Software. 2015;67:1–48. [Google Scholar]
- 47.Hothorn T, Bretz F, Westfall P. Simultaneous inference in general parametric models. Biometrical J. 2008;50:346–363. doi: 10.1002/bimj.200810425. [DOI] [PubMed] [Google Scholar]
- 48.Jackman S. 2015 pscl: Classes and Methods for R Developed in the Political Science Computational Laboratory, Stanford University. R Package Version 1.4.9. Available at pscl.stanford.edu. Accessed April 14, 2017.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.