Significance
Breeding protocols in biotechnology focus on fast short-term responses by selecting extreme phenotypes from maximally diverse pools of heritable variants. However, when interactions among alleles are unpredictable, optimizing long-term responses may require intermediate stages with low or modest improvement. By subjecting mutant libraries of an antibiotic-degrading enzyme of different sizes to selection with a new antibiotic, we find support for this prediction: despite slower initial improvement due to limited genetic variation, the final alleles from two small libraries reached the highest antibiotic-resistance levels overall due to a broader exploration of the adaptive landscape. Our results present a cautionary tale for “greedy” adaptation protocols in biotechnology and call for quantitative data on mutation supplies in the clinical evolution of antibiotic resistance.
Keywords: adaptation, mutation supply, antibiotic resistance, fitness landscape, experimental evolution
Abstract
Populations with large mutation supplies adapt via the “greedy” substitution of the fittest genotype available, leading to fast and repeatable short-term responses. At longer time scales, smaller mutation supplies may in theory lead to larger improvements when distant high-fitness genotypes more readily evolve from lower-fitness intermediates. Here we test for long-term adaptive benefits from small mutation supplies using in vitro evolution of an antibiotic-degrading enzyme in the presence of a novel antibiotic. Consistent with predictions, large mutant libraries cause rapid initial adaptation via the substitution of cohorts of mutations, but show later deceleration and convergence. Smaller libraries show on average smaller initial, but also more variable, improvements, with two lines yielding alleles with exceptionally high resistance levels. These two alleles share three mutations with the large-library alleles, which are known from previous work, but also have unique mutations. Replay evolution experiments and analyses of the adaptive landscape of the enzyme suggest that the benefit resulted from a combination of avoiding mutational cohorts leading to local peaks and chance. Our results demonstrate adaptive benefits from limited mutation supplies on a rugged fitness landscape, which has implications for artificial selection protocols in biotechnology and argues for a better understanding of mutation supplies in clinical settings.
The supply of mutations is a fundamental determinant of the dynamics and repeatability of adaptation, particularly in asexual populations (1, 2). Populations with large mutation supplies, such as large populations or populations with a high mutation rate, are expected to show “greedy” adaptation via the substitution of the fittest genotypes available, i.e., those that are most likely to survive clonal interference (3). By benefitting high-fitness genotypes, populations with large mutation supplies thus enhance both the short-term rate and repeatability of adaptation (1, 3–5). Similarly, methods used in agriculture and biotechnology for breeding animals, plants and enzymes with desired properties are often greedy and aimed at maximizing short-term responses by selecting extreme phenotypes from maximally diverse pools of heritable variants (6).
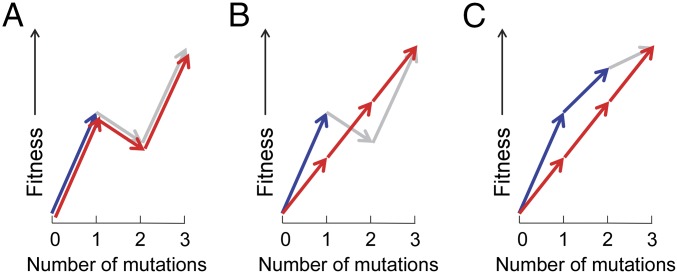
However, at longer time scales when adaptation involves the sequential substitution of multiple mutations, the relationship between a population’s mutation supply and adaptation is complicated by epistasis. For instance, sign epistasis may introduce local fitness maxima, where populations with large mutation supplies are repeatedly “trapped,” whereas populations with smaller mutation supplies may either escape via the substitution of a deleterious mutation followed by adaptation (7) (Fig. 1A) or avoid these traps by substituting smaller-benefit mutations (4, 5, 8, 9) (Fig. 1B). Very large mutation supplies could lead to increased rates of long-term adaptation again, by allowing the escape from local peaks via the substitution of mutational cohorts (1, 5, 10). Magnitude epistasis may yield an additional, more subtle, benefit for populations with small mutation supplies by introducing variation in the flatness of pathways leading to the same peak (Fig. 1C). Greedy pathways, with large initial and smaller later improvements, are expected to take more time to arrive at the maximum than flatter pathways with more consistent improvements per step, due to stronger within-pathway clonal interference (11).
Fig. 1.
Possible adaptive benefits from small mutation supplies in the presence of epistasis. In each panel, blue arrows indicate greedy mutational pathways expected for populations with large mutation supplies; red arrows show pathways expected for smaller mutation supplies; and gray arrows show unrealized mutations within the time period given (e.g., of an evolution experiment). Note that although arrows indicate individual mutations here, the mechanisms remain valid when arrows each reflect cohorts of mutations. (A) Sign epistasis causes multiple fitness peaks, where escape to the higher peak requires the substitution of a deleterious mutation, which is more likely for small populations (7). (B) Sign epistasis causes multiple fitness peaks, where populations with large mutation supplies end at a local maximum, whereas smaller-benefit mutations, given a chance in the absence of clonal interference, avoid this trap and reach the global maximum (4, 5). (C) Magnitude epistasis causes greedy pathways to arrive later at the global maximum than pathways with similar fitness improvements per step realized under small mutation supplies due to reduced within-pathway clonal interference (11).
Small mutation supplies may thus provide long-term adaptive benefits despite slower short-term improvement whenever long-term adaptation is contingent on earlier genotypes of submaximal fitness. The size of the benefit will depend also on the actual topography of the fitness landscape (4, 5, 8, 9). For example, when the largest-benefit mutations available lead toward a local fitness maximum, populations with small mutation supplies may generally show greater long-term improvements (4). However, when path steepness and peak height are not correlated, populations with small mutation supplies may only occasionally benefit from their greater adaptive heterogeneity. Beyond the notion that fitness landscapes are often complex, little is known about actual topographies (12–18), and very few studies showed the mediating effect from mutation supply. One empirical study reported a long-term adaptive benefit of small populations in a rich nutrient environment (4). In that study, some small populations of the bacterium Escherichia coli reached higher fitness than any population of 50-fold larger size. Because these populations showed deviating fitness trajectories with slow early improvements, the fitness landscape was inferred to be rugged, but no genetic information was provided. More than 80 y ago, Wright (7) predicted a related but distinct adaptive benefit for structured populations, where mutation supplies are not different, but local demes may cross fitness valleys via the substitution of deleterious mutations followed by population-wide adaptation. Support for a structured-population benefit was found in a recent study with E. coli (19), whereas a similar study with Saccharomyces cerevisiae found no support (20).
Here, we test for adaptive benefits from small mutation supplies during the in vitro evolution of the antibiotic-degrading enzyme TEM-1 β-lactamase in the presence of cefotaxime. TEM-1 is the ancestral allele of a family of more than 175 β-lactamases, each with increased activity against diverse synthetic β-lactams (21). Although currently other β-lactamases, such as the CTX-M family, are more prevalent in clinical bacterial isolates, TEM-1 has become a major model in experimental evolution. Not only could clinical patterns of molecular evolution be recapitulated in the laboratory (22, 23), TEM-1 has also been used to address basic evolutionary questions. For example, recent work with TEM-1 revealed molecular details that determine its evolvability (14, 24–28), and showed that its fitness landscape is rugged (16, 27, 29, 30). This causes TEM alleles selected from large mutation supplies to often follow the same trajectory involving three common mutations (16). In the present study, we ask whether reducing the supply of mutations will release populations from adaptive constraints caused by clonal interference and open up new trajectories, possibly including ones leading to higher resistance.
Results
Evolution Experiment.
To test for possible adaptive benefits from small mutation supplies, we compared adaptation of large and small libraries of mutants of TEM-1 β-lactamase under maximal selection for increased resistance to the novel substrate cefotaxime (CTX; Fig. S1). Four large and four small libraries of TEM-alleles created by error-prone PCR were subjected to selection in a gradient of CTX concentrations by inoculating bottles with growth medium and twofold increasing CTX concentrations with ∼10 times the library size. Our aim was to contrast selection using large libraries (LL; ∼105–106 transformants), known to often lead to the substitution of three common mutations [E104K, M182T, and G238S (16)], with selection using the smallest possible libraries that allow adaptation (SL). Because the minimum population size required for adaptation was a priori unknown, SL lines were represented by three library sizes each, containing 1%, 0.1%, and 0.01% of the transformants present in LL lines. After 48 h, a single random bacterial clone was selected from the highest CTX concentration allowing bacterial growth and its β-lactamase gene was sequenced, because sequencing of multiple clones suggested that these conditions lead to the (near) fixation of a single-best genotype (16). For LL lines, the selected clone was directly used as template for a new round of evolution; for SL lines, we continued with the clone from the smallest of the three selected SL populations that had substituted at least one nonsynonymous mutation. As a result, the population size of SL lines varied during the experiment (Fig. S2). Four rounds of evolution under LL and SL conditions were followed by two rounds using LL conditions to allow the approach of a nearby optimum. Colony counts on selective media indicated that geometric mean library sizes of LL and SL lines during the first four rounds were 5.6 × 105 and 3.2 × 102, respectively.
Two SL Alleles Evolve High CTX Resistance.
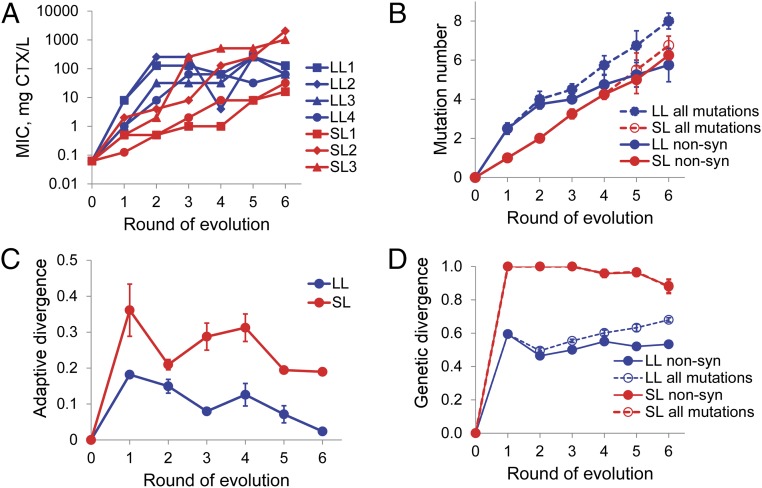
Both LL and SL lines evolved large increases in CTX resistance, with improvements in the minimal inhibitory concentration (MIC) of ∼250–32,000-fold (16–2,048 mg CTX/L relative to 0.0625 mg CTX/L for TEM-1; Fig. 2A). Although LL and SL lines reach on average similar final improvements (10.25 MIC doublings for LL versus 11.5 for SL lines, U = 8, P > 0.10), they also show clear differences in phenotypic dynamics: LL lines rapidly increase resistance in the first two rounds and then level off, whereas SL lines improve more slowly initially, but continue at a steady pace. The decline in resistance of LL2 between rounds 3 and 4 resulted from the isolation of a minority genotype with a deleterious mutation (R259S), because sequencing 20 further clones from this population identified the selected mutation in only one case and the clone reverted this mutation in the next round causing the recovery of MIC (Figs. 2A and 3B) Most notably, SL2 and SL3 evolved 8- to 32-fold higher resistance than any of the LL lines, despite their much smaller size.
Fig. 2.
Dynamics and repeatability of phenotypic and genomic evolution. (A) Changes in CTX resistance of the four LL (blue) and four SL (red) lines. Shown are the median of three replicate MIC assays per line and round. (B) Average number of mutations in LL (blue) and SL (red) lines for nonsynonymous mutations (solid lines) and all mutations (dashed lines). (C) Relative phenotypic divergence, measured as the average pairwise difference in MIC-step improvement relative to TEM-1, divided by the sum of MIC-step improvements of both lines. (D) Relative genetic divergence, considering only nonsynonymous or all mutations, measured as the pairwise Hamming distance divided by the sum of both Hamming distances relative to TEM-1. Error bars are SEM based on variation among the four replicates.
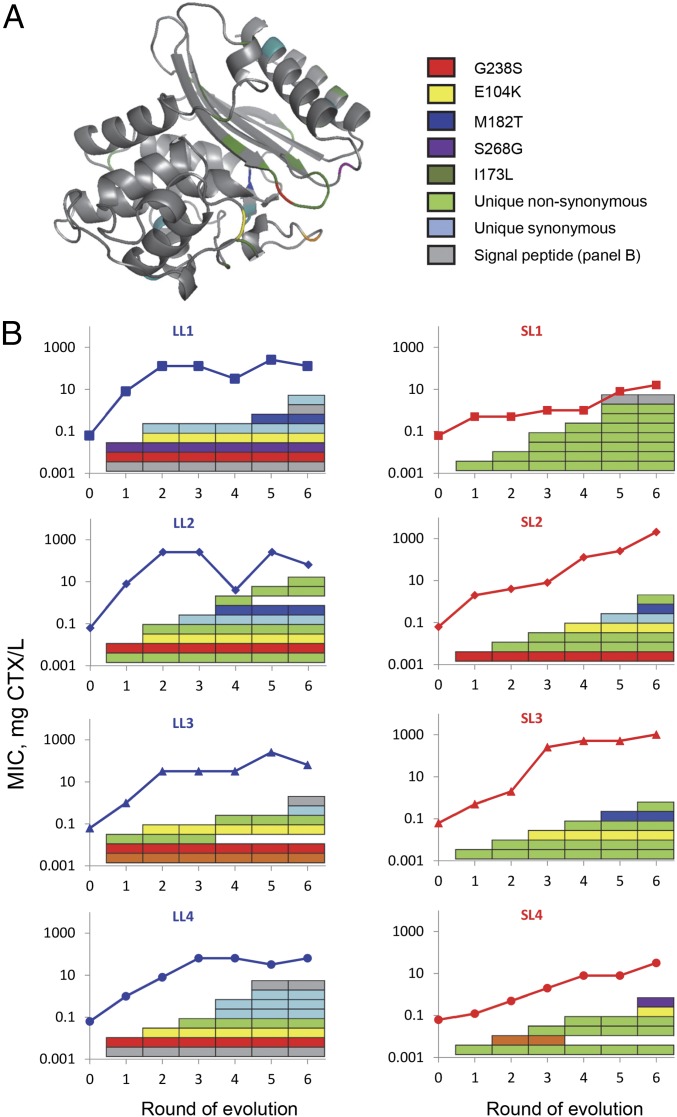
Fig. 3.
Mutational pathways to increased CTX resistance. (A) Folded β-lactamase enzyme with positions of shared and unique mutations. (B) MIC (lines) and mutational pathways (colored rectangles) for the LL (Left, blue) and SL lines (Right, red). Also mutations in the signal peptide are shown (in gray), including identity of shared mutations; see Fig. S2 for identity of all mutations.
LL and SL lines substituted similar total numbers of mutations (Fig. 2B; on average 7.25 and 6.5 per LL and SL line, respectively, of which 5.0 and 5.75 are nonsynonymous; U > 3, P > 0.10 in both cases), and high-MIC lines SL2 and SL3 were no exception (six nonsynonymous mutations each and seven and six mutations in total, respectively). Note that these numbers exclude one reversion (LL2) and two substitutions replacing previous mutations at the same site (LL3 and SL4), so that the total number of mutational events is slightly higher than the number of mutations present in the final alleles. LL lines substituted relatively more synonymous mutations than SL lines (Fig. 2B; Fisher’s P = 0.027), which partly reflects our selection criterion that SL lines should show at least one nonsynonymous mutation. Total mutation number does not correlate with resistance (Spearman rank correlation, all mutations: r = −0.358, n = 8, P > 0.1; nonsynonymous mutations only: r = −0.069, n = 8, P > 0.1).
As observed in previous work (16, 27), LL lines adapted by using, among other mutations, three common mutations in strict order: G238S (round 1) and E104K (round 2), followed by M182T in two of the four lines (Fig. 3 and Fig. S2). SL lines used more diverse mutations, and substituted each a different mutation in round 1: E240G, G238S, G238A, and R164H. These mutations are all known to have large effects on CTX resistance (26) and competitive fitness in the presence of CTX (28). Interestingly, high-MIC lines SL2 and SL3 also substituted these common-pathway mutations (except that SL3 replaces glycine at position 238 by alanine instead of serine) in the same order, suggesting a shared molecular basis for their adaptation.
Relationship Between MIC and Competitive Fitness.
To verify that MIC was a good proxy for fitness under the selective conditions of our experiment, we performed bulk competitions with the eight final genotypes under conditions similar to those of the evolution experiment, i.e., at maximum CTX concentrations and initial cell densities similar as for the LL lines. Illumina sequencing of population DNA at five time points and ∼500,000-fold coverage was used to measure changes in allele frequencies (Fig. S3A). Competitive fitness of each allele relative to the other seven shows a positive correlation with resistance (r = 0.806, n = 8, P = 0.0157; Fig. S3B), confirming that MIC changes are associated with fitness increases.
SL and LL lines differed not only in the size of their mutation supply, but also in initial cell density during selection in the first four rounds, because inocula of ∼10 times the library size were used. This lower initial cell density of SL lines allowed for more generations of selection, which could have affected the efficiency of selection in SL versus LL lines. To test this, we compared the outcome of selection of samples from the same mutant library under three conditions with threefold replication, always at maximum CTX concentration: 100% library inoculated at 10 times the library size (LL size/LL density), 1% library sample inoculated at 10 times the library size (SL size/SL density), and 1% library sample inoculated at 1,000 times the library size (SL size/LL density). However, initial cell density had no effect on the type (or even frequency) of selected mutants, whereas library size did (Fig. S4). Thus, at least initially, differences in mutation supply, not differences in cell density, affected the outcome of selection.
Examining the Cause of the Small Mutation-Supply Benefit.
To understand the high resistance evolved by SL2 and SL3, we performed several further analyses. We first examined the general pattern of phenotypic and genetic changes for LL and SL lines. Consistent with expectations that small mutation supplies allow a broader exploration of the fitness landscape, SL lines show a greater divergence than LL lines, both at the level of resistance (Fig. 2C) and genotype (Fig. 2D). In addition, whereas LL lines show rapid initial improvement followed by stasis, SL lines initially adapt more slowly but also more steadily, partly reflecting our requirement that they substitute at least one nonsynonymous mutation in the first four rounds (Fig. 2A; see Fig. S5 and Table S1 for a formal statistical test).
We then looked in more detail at the evolutionary trajectories of high-MIC lines SL2 and SL3. The fact that common large-benefit mutations E104K, M182T, and G238S (or G238A) are used by SL2 and SL3, as well as by two LL lines (LL1 and LL2), indicates that the additional mutations in these lines play a crucial role. A comparison of the MIC estimates of the final alleles of SL2 (2,048 mg CTX/L) and SL3 (1,024 mg CTX/L) with those of LL1 (128 mg CTX/L) and LL2 (64 mg CTX/L) and threefold mutant E104K/M182T/G238S (256 mg CTX/L) (16, 26) indicates that the additional mutations in SL2 and SL3 are required to explain the high MIC of these alleles. The comparison also suggests two possible scenarios for the additional mutations in LL1 and LL2: (i) they are deleterious hitchhikers or (ii) they are beneficial (at least those selected in the first rounds), but cause later adaptive constraints. We can reject the first possibility: the MIC of LL1 and LL2 is at least fourfold higher than that of SL2 and SL3 after the first round, and all additional nonsynonymous mutations in LL1 and LL2 are either known to be beneficial (28) or have been observed in selection experiments using CTX (23). We therefore hypothesize that the early greedy substitution of additional mutations together with common mutations G238S and E104K constrains the further adaptation of the LL lines.
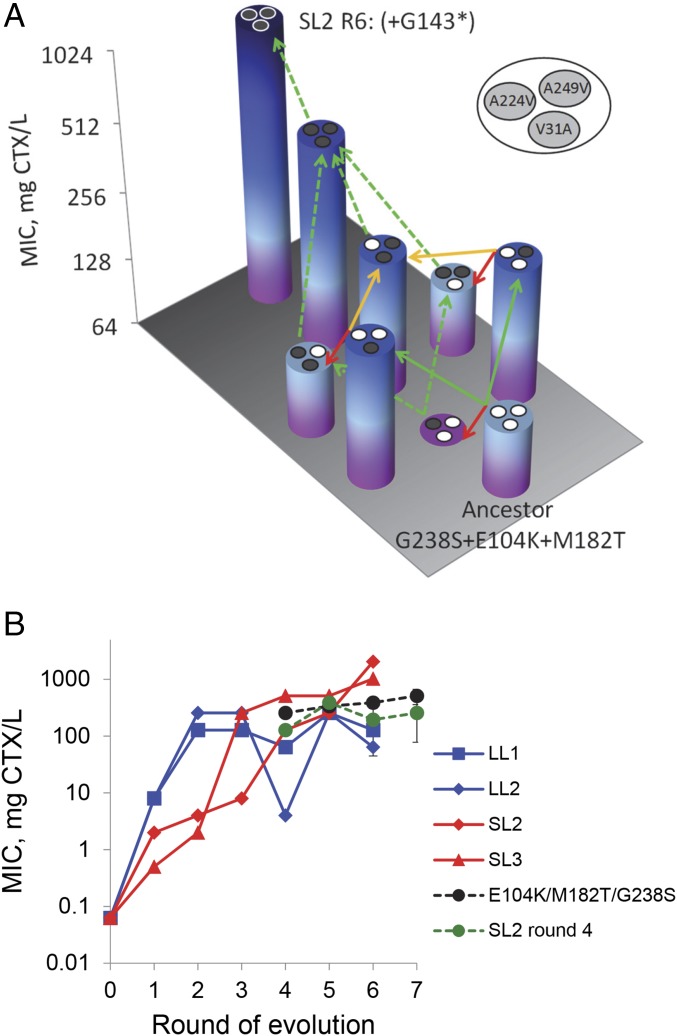
To understand how the additional mutations in the LL lines constrain adaptation, we performed a few further analyses. Because mutations G238S, E104K, and M182T were selected in the LL lines, as well as in previous studies (16, 24, 27, 30, 31), we first asked whether allele SL2 is selectively accessible from the genotype containing these three mutations, using two independent analyses. We first constructed and analyzed the resistance landscape connecting threefold mutant E104K/M182T/G238S and the genotype containing the six nonsynonymous mutations of final allele SL2 (neglecting synonymous mutation G143* obtained in round 5). Although the MIC estimates were slightly lower than previous estimates (for threefold mutant and SL2) and suggest that G143* is beneficial, they also show that sign epistasis constrains SL2’s trajectory: each pathway from threefold mutant to final allele SL2 contains at least one step without MIC improvement (Fig. 4A). Second, we evolved the threefold mutant for three rounds and fivefold replication under LL conditions. Although they reach higher resistance than LL1 and LL2, none of the replicate lines reached MIC values (Fig. 4B) or used mutations similar to SL2 (Fig. S6). Together, these analyses suggest that allele SL2 is selectively inaccessible from common threefold mutant E104K/M182T/G238S due to sign epistasis, which makes not only the type of mutations important but also their order.
Fig. 4.
Understanding the adaptive benefit of SL2. (A) Resistance landscape showing all possible pathways between threefold mutant E104K/M182T/G238S and the sixfold mutant containing all nonsynonymous mutations of final allele SL2; final allele SL2 is also shown. Green arrows indicate an increase in MIC along a mutational trajectory to the sixfold mutant, whereas red arrows indicate a decrease in MIC and yellow arrows indicate neutral steps. Dashed green arrows indicate MIC increases in trajectories that are not accessible due to negative or neutral steps. (B) Comparison of CTX resistance (MIC) of the two high-MIC small-mutation supply lines (SL2 and SL3; red) with the two LL lines containing shared common mutations E104K/M182T/G238S (LL1 and LL2; blue), together with the average resistance of the five replicate lines evolved for three rounds under LL conditions with threefold mutant E104K/M182T/G238S (black) and allele SL2 from round 4 (green; Fig. S6). Error bars are SEM based on MIC variation among the five replicates.
To test whether the selection of final allele SL2 was contingent upon mutation type and order, we then asked if allele SL2 from round 4 (when the MIC is still comparable to that of other lines) is at a unique position in the fitness landscape from where high-resistance genotypes are readily accessible. To address this, we evolved allele SL2 from round 4 for three rounds with five replicates under LL conditions. Surprisingly, none of the replicate lines evolved final allele SL2 or alleles with similar resistance levels (Fig. 4B and Fig. S6). Two lines gained common mutation M182T, but no line selected A249V or G143*. A comparison of the MIC of SL2 after rounds 4 and 6 suggests that, in addition to M182T, either A249V or G143* or both were required to reach the final high MIC of SL2 (Fig. 4B). We therefore conclude that the selection of final allele SL2 also crucially depended on the chance occurrence of specific mutations, even at this later stage.
Discussion
By subjecting replicate libraries of mutant TEM-1 β-lactamase alleles of varying size to selection with a novel antibiotic for multiple rounds, we found two small-library lines to evolve alleles with higher resistance levels than any of the large-library lines. The observed benefit was associated with greater divergence and slower initial improvements relative to populations with large libraries (Fig. 2 and Fig. S5). The parallel pattern of rapid initial improvement followed by stasis observed for the large-library lines suggested that the early greedy selection of high-fitness genotypes containing multiple mutations caused later adaptive constraints. Replay evolution experiments and analyses of the fitness landscape of one high-resistance allele (Fig. 4) confirmed that sign epistasis among the resistance mutations likely caused adaptive constraints, consistent with previous analyses of the fitness landscape of this enzyme (16, 27, 29, 30). However, our inability to obtain alleles with resistance levels comparable to SL2 and SL3 in a replay experiment with allele SL2 from round 4 (Fig. 4B and Fig. S6) also indicates a crucial contribution of chance during the occurrence and selection of mutations.
In theory, epistasis may provide a small mutation-supply benefit in three distinct ways: (i) by introducing local maxima that are escaped more effectively by small populations (7) (Fig. 1A); (ii) by introducing local maxima that are avoided via nongreedy pathways allowed by small mutation supplies (4, 5, 8, 9); or (iii) by causing variation in within-pathway clonal interference and allowing faster adaptation via nongreedy pathways (11) (Fig. 1C). As explanation for our results, the second mechanism seems most important. The first mechanism was unimportant, because selection was strong and the MIC trajectories of SL2 and SL3 increased at each step (Fig. 3B), rejecting a significant role for deleterious mutations. Instead and consistent with previous work (16, 27, 29, 30), sign epistasis likely constrained adaptation in our experiment (Fig. 4), providing support for the second mechanism. However, the third mechanism may also have contributed to the SL benefit, particularly because we fixed a single genotype each round (see next paragraph): the shared mutations of LL and high-MIC SL lines suggest that they ascend the same adaptive peak, and the dynamics of resistance and genetic changes (Fig. 2) are consistent with predictions from this mechanism (Fig. 1C).
Besides mutation supply, initial cell density during selection differed for SL and LL lines during the first four rounds of evolution. By starting selection with populations of roughly 10 times the size of the library sample, we equalized the probability of mutants present in LL and SL libraries to survive drift at their initial low frequency (32), but also increased the duration of selection for SL libraries. This difference could have affected our results in two possible ways. First, it may have increased the probability of fixation of small-benefit mutations under SL relative to LL conditions. This effect may have been particularly strong because we selected a single mutant during each round, which amplified differences in final allele frequency. Second, the protection of lower-resistant mutants by high-resistant alleles via reduction of the environmental antibiotic concentration (33, 34) may have been less important under SL than LL conditions, because lower-resistant mutants would need to survive more generations in initially high-antibiotic concentrations under SL conditions. Despite these potential differences in selective conditions in SL and LL lines, selection with a mutant library showed no effect from initial cell density on the outcome of selection during the first round (Fig. S4). However, we cannot rule out that weaker selection in the LL lines may have affected the outcome in later rounds, when selective benefits of mutations were smaller and we sometimes failed to select additional mutations (Fig. S2).
How can we understand the high resistance of alleles SL2 and SL3 at a molecular level? Similar to other enzymes adapting to a new substrate (35–37), TEM-1’s adaptation to CTX results from a combination of mutations that increase enzyme activity and those that compensate the destabilizing pleiotropic effects of activity-enhancing mutations and restore enzyme abundance (38, 39). For example, SL2 and SL3 combine mutations G238S/A, E104K, and S268T, known to increase TEM’s activity on CTX, with mutations M182T, V31A, A224V, T265M, and D35E, which are known to stabilize the folded enzyme (14, 23, 26, 40). Because the LL lines also combine activity-enhancing (e.g., G238S, E104K) with stabilizing mutations (e.g., H153R, R275Q/L) (14, 40), the adaptive benefits of SL2 and SL3 seem to result from finding combinations of mutations that better optimize these two enzyme properties, yielding a high dose of active enzyme at a minimum cost per cell (37). The picture that emerges is that pleiotropy between enzyme activity and stability causes few accessible mutational trajectories to high CTX resistance, which is more efficiently tracked by testing mutations one-by-one than via the greedy selection of multiple mutations at once.
Finally, what are the broader implications of our findings? They highlight in the first place the fundamental importance of the interplay between fitness landscape topography and population dynamics in determining the tempo and mode of evolution (12, 17). Similar small-population adaptive benefits as observed here for TEM may be expected under strong epistatic constraints, such as for the adaptation of proteins encoded by single genes (12, 41) or already well-adapted genotypes (42). Second, our findings are a cautionary tale for breeding protocols used in biotechnology (6, 43), which often aim at maximizing the genetic variation subjected to selection, or use iterative approaches where the best replacement in each step is identified and used in further rounds (44). Such protocols lead to rapid improvements, but may also close the door to pathways initiated by smaller improvements, or even neutral or deleterious changes, that eventually lead to better enzymes. Particularly when selective constraints are known (45), it may be worth using more prudent protocols to minimize these constraints—for instance, by screening for mutants showing limited improvement (44) or subdividing mutants into smaller selection pools (46).
Third, are there any clinical implications, given that our results are based on a notorious antibiotic-resistance enzyme? Population bottlenecks occur during the colonization of host organisms by commensal and pathogenic bacteria (47). It is tempting to speculate that these bottlenecks could contribute to ameliorating selective constraints imposed by a rugged fitness landscape, including for antibiotic resistance. However, we think that similar small-population benefits as observed for TEM are less likely during the clinical evolution of antibiotic resistance, due to the much higher number of, and weaker epistasis among, adaptive mutations occurring throughout the genome (12, 41). Other differences that prevent the direct translation of our findings to the clinic are lower mutation supplies, selection with different antibiotics, and the regular horizontal transfer of β-lactamases in clinical settings. Consistent with this notion, although all 29 mutations we found were known from previous evolution experiments, only 12 occur in clinical isolates (23), which contain, on average, fewer mutations (2.6 vs. 5.4 in our alleles). Therefore, predicting the evolutionary trajectories to antibiotic resistance in clinical settings remains a daunting task for which quantitative assessment of key factors, including mutation supplies, is urgently needed (48).
Almost a century ago, Wright (7) foresaw the “perils of greed” for large unstructured populations adapting on a rugged fitness landscape on theoretical grounds. Our results add to a growing body of recent empirical support for this view (4, 18, 19, 43, 46) by showing adaptive benefits for an antibiotic resistance enzyme from small mutation supplies.
Materials and Methods
Strains and Plasmids.
E. coli strain DH5αE (Invitrogen) was used as a host for all plasmids. Plasmid pACSE3 (22) was used as the vector for cloning and expressing TEM alleles. This ∼5.1-kb plasmid contains a tetracycline-resistance marker and replicates at ∼10 copies per cell.
In Vitro Evolution.
Detailed methods for the in vitro mutagenesis, cloning, expression, selection of mutant TEM alleles, and MIC assays are provided as Supporting Information. Briefly, the GeneMorph II Mutagenesis Kit (Stratagene) is used to introduce random mutations in the coding region of TEM-1 (on average ∼1.6 mutations per kilobase). Digested amplicons were then cloned into vector pACSE3 and transformed into competent DH5αE cells. We started the experiment by running four independent error-prone PCR reactions, using the conditions described above. After digestion, ligation, and transformation, the cells were allowed to recover in 2 mL SOC (super optimal broth with catabolite repression) medium for 90 min. After recovery, we added 1.6 mL of the SOC to 500 mL LB + Tet (creating a LL), and we used the remaining 400 µL SOC with transformants to create the 100×, 1,000×, and 10,000× diluted libraries (using steps of 10-fold dilution in 3.6 mL LB; the SLs). Library size was estimated by plating samples on LB–tetracycline (Tet). Immediately after dilution, each library was also added to 500 mL LB + Tet and grown overnight at 37 °C for library expansion and subsequent selection. After this initial round of evolution (where the small libraries were actually fractions of the large library of each line), we continued the experiment using error-prone PCR reactions (on the selected clone from the previous round as template) for the SL and LL lines. Round 5 and 6 deviated from the first four rounds in that for the SL lines LL libraries were also used to allow all lines to approach a peak nearby. For selection, a series of bottles containing 50 mL Mueller Hinton medium (Merck) was inoculated with CTX (stock solution in 0.1 M NaPO4, pH 7.0; Sigma) concentrations in twofold increments, as well as isopropyl-β-d-thiogalactopyranoside (IPTG) at 50 μM. After 48 h of selection, a single random clone was isolated from the highest CTX concentration with bacterial growth, and its plasmid isolated and used for the next round of evolution.
Bulk Competitions and Statistical Methods.
Supplementary Material
Acknowledgments
We thank Martijn Schenk, Thomas van Dijk, Edouard Severing, Joachim Krug, and Dan Tawfik for useful comments. This work was supported by European Community Seventh Framework Programme (FP7/2007-2013) Grant 225167 “e-FLUX” and by Deutsche Forschungsgemeinschaft within SFB 680 “Molecular Basis of Evolutionary Innovations.”
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1712999114/-/DCSupplemental.
References
- 1.Szendro IG, Franke J, de Visser JAGM, Krug J. Predictability of evolution depends nonmonotonically on population size. Proc Natl Acad Sci USA. 2013;110:571–576. doi: 10.1073/pnas.1213613110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vogwill T, Phillips RL, Gifford DR, MacLean RC. Divergent evolution peaks under intermediate population bottlenecks during bacterial experimental evolution. Proc R Soc B. 2016;283:20160749. doi: 10.1098/rspb.2016.0749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gerrish PJ, Lenski RE. The fate of competing beneficial mutations in an asexual population. Genetica. 1998;102-103:127–144. [PubMed] [Google Scholar]
- 4.Rozen DE, Habets MGJL, Handel A, de Visser JAGM. Heterogeneous adaptive trajectories of small populations on complex fitness landscapes. PLoS One. 2008;3:e1715. doi: 10.1371/journal.pone.0001715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ochs IE, Desai MM. The competition between simple and complex evolutionary trajectories in asexual populations. BMC Evol Biol. 2015;15:55. doi: 10.1186/s12862-015-0334-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Packer MS, Liu DR. Methods for the directed evolution of proteins. Nat Rev Genet. 2015;16:379–394. doi: 10.1038/nrg3927. [DOI] [PubMed] [Google Scholar]
- 7.Wright S. The roles of mutation, inbreeding, crossbreeding and selection in evolution. Proceedings of the Sixth International Congress of Genetics. 1932;1:356–366. [Google Scholar]
- 8.Handel A, Rozen DE. The impact of population size on the evolution of asexual microbes on smooth versus rugged fitness landscapes. BMC Evol Biol. 2009;9:236. doi: 10.1186/1471-2148-9-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jain K, Krug J, Park S-C. Evolutionary advantage of small populations on complex fitness landscapes. Evolution. 2011;65:1945–1955. doi: 10.1111/j.1558-5646.2011.01280.x. [DOI] [PubMed] [Google Scholar]
- 10.Weinreich DM, Chao L. Rapid evolutionary escape by large populations from local fitness peaks is likely in nature. Evolution. 2005;59:1175–1182. [PubMed] [Google Scholar]
- 11.Ogbunugafor CB, Eppstein MJ. Competition along trajectories governs adaptation rates towards antimicrobial resistance. Nat Ecol Evol. 2016;1:7. doi: 10.1038/s41559-016-0007. [DOI] [PubMed] [Google Scholar]
- 12.de Visser JAGM, Krug J. Empirical fitness landscapes and the predictability of evolution. Nat Rev Genet. 2014;15:480–490. doi: 10.1038/nrg3744. [DOI] [PubMed] [Google Scholar]
- 13.Weinreich DM, Lan Y, Wylie CS, Heckendorn RB. Should evolutionary geneticists worry about higher-order epistasis? Curr Opin Genet Dev. 2013;23:700–707. doi: 10.1016/j.gde.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bershtein S, Goldin K, Tawfik DS. Intense neutral drifts yield robust and evolvable consensus proteins. J Mol Biol. 2008;379:1029–1044. doi: 10.1016/j.jmb.2008.04.024. [DOI] [PubMed] [Google Scholar]
- 15.Blount ZD, Borland CZ, Lenski RE. Historical contingency and the evolution of a key innovation in an experimental population of Escherichia coli. Proc Natl Acad Sci USA. 2008;105:7899–7906. doi: 10.1073/pnas.0803151105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salverda MLM, et al. Initial mutations direct alternative pathways of protein evolution. PLoS Genet. 2011;7:e1001321. doi: 10.1371/journal.pgen.1001321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tenaillon O, et al. Tempo and mode of genome evolution in a 50,000-generation experiment. Nature. 2016;536:165–170. doi: 10.1038/nature18959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woods RJ, et al. Second-order selection for evolvability in a large Escherichia coli population. Science. 2011;331:1433–1436. doi: 10.1126/science.1198914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nahum JR, et al. A tortoise-hare pattern seen in adapting structured and unstructured populations suggests a rugged fitness landscape in bacteria. Proc Natl Acad Sci USA. 2015;112:7530–7535. doi: 10.1073/pnas.1410631112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kryazhimskiy S, Rice DP, Desai MM. Population subdivision and adaptation in asexual populations of Saccharomyces cerevisiae. Evolution. 2012;66:1931–1941. doi: 10.1111/j.1558-5646.2011.01569.x. [DOI] [PubMed] [Google Scholar]
- 21.Bush K. Evolution of β-lactamases: Past, present, and future. In: Dougherty TJ, Pucci MJ, editors. Antibiotic Discovery and Development. Springer; Boston: 2012. pp. 427–453. [Google Scholar]
- 22.Barlow M, Hall BG. Predicting evolutionary potential: In vitro evolution accurately reproduces natural evolution of the tem beta-lactamase. Genetics. 2002;160:823–832. doi: 10.1093/genetics/160.3.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salverda MLM, De Visser JAGM, Barlow M. Natural evolution of TEM-1 β-lactamase: Experimental reconstruction and clinical relevance. FEMS Microbiol Rev. 2010;34:1015–1036. doi: 10.1111/j.1574-6976.2010.00222.x. [DOI] [PubMed] [Google Scholar]
- 24.Bratulic S, Toll-Riera M, Wagner A. Mistranslation can enhance fitness through purging of deleterious mutations. Nat Commun. 2017;8:15410. doi: 10.1038/ncomms15410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deng Z, et al. Deep sequencing of systematic combinatorial libraries reveals β-lactamase sequence constraints at high resolution. J Mol Biol. 2012;424:150–167. doi: 10.1016/j.jmb.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schenk MF, Szendro IG, Krug J, de Visser JAGM. Quantifying the adaptive potential of an antibiotic resistance enzyme. PLoS Genet. 2012;8:e1002783. doi: 10.1371/journal.pgen.1002783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schenk MF, et al. Role of pleiotropy during adaptation of TEM-1 β-lactamase to two novel antibiotics. Evol Appl. 2015;8:248–260. doi: 10.1111/eva.12200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stiffler MA, Hekstra DR, Ranganathan R. Evolvability as a function of purifying selection in TEM-1 β-lactamase. Cell. 2015;160:882–892. doi: 10.1016/j.cell.2015.01.035. [DOI] [PubMed] [Google Scholar]
- 29.Schenk MF, Szendro IG, Salverda MLM, Krug J, de Visser JAGM. Patterns of epistasis between beneficial mutations in an antibiotic resistance gene. Mol Biol Evol. 2013;30:1779–1787. doi: 10.1093/molbev/mst096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weinreich DM, Delaney NF, Depristo MA, Hartl DL. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science. 2006;312:111–114. doi: 10.1126/science.1123539. [DOI] [PubMed] [Google Scholar]
- 31.Hall BG. Predicting evolution by in vitro evolution requires determining evolutionary pathways. Antimicrob Agents Chemother. 2002;46:3035–3038. doi: 10.1128/AAC.46.9.3035-3038.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kimura M. On the probability of fixation of mutant genes in a population. Genetics. 1962;47:713–719. doi: 10.1093/genetics/47.6.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nicoloff H, Andersson DI. Indirect resistance to several classes of antibiotics in cocultures with resistant bacteria expressing antibiotic-modifying or -degrading enzymes. J Antimicrob Chemother. 2016;71:100–110. doi: 10.1093/jac/dkv312. [DOI] [PubMed] [Google Scholar]
- 34.Yurtsev EA, Chao HX, Datta MS, Artemova T, Gore J. Bacterial cheating drives the population dynamics of cooperative antibiotic resistance plasmids. Mol Syst Biol. 2013;9:683. doi: 10.1038/msb.2013.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DePristo MA, Weinreich DM, Hartl DL. Missense meanderings in sequence space: A biophysical view of protein evolution. Nat Rev Genet. 2005;6:678–687. doi: 10.1038/nrg1672. [DOI] [PubMed] [Google Scholar]
- 36.Mehta SC, Rice K, Palzkill T. Natural variants of the KPC-2 carbapenemase have evolved increased catalytic efficiency for ceftazidime hydrolysis at the cost of enzyme stability. PLoS Pathog. 2015;11:e1004949. doi: 10.1371/journal.ppat.1004949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soskine M, Tawfik DS. Mutational effects and the evolution of new protein functions. Nat Rev Genet. 2010;11:572–582. doi: 10.1038/nrg2808. [DOI] [PubMed] [Google Scholar]
- 38.Knies JL, Cai F, Weinreich DM. Enzyme efficiency but not thermostability drives cefotaxime resistance evolution in TEM-1 β-lactamase. Mol Biol Evol. 2017;34:1040–1054. doi: 10.1093/molbev/msx053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang X, Minasov G, Shoichet BK. Evolution of an antibiotic resistance enzyme constrained by stability and activity trade-offs. J Mol Biol. 2002;320:85–95. doi: 10.1016/S0022-2836(02)00400-X. [DOI] [PubMed] [Google Scholar]
- 40.Kather I, Jakob RP, Dobbek H, Schmid FX. Increased folding stability of TEM-1 β-lactamase by in vitro selection. J Mol Biol. 2008;383:238–251. doi: 10.1016/j.jmb.2008.07.082. [DOI] [PubMed] [Google Scholar]
- 41.Watson RA, Weinreich DM, Wakeley J. Genome structure and the benefit of sex. Evolution. 2011;65:523–536. doi: 10.1111/j.1558-5646.2010.01144.x. [DOI] [PubMed] [Google Scholar]
- 42.Hwang S, Park S-C, Krug J. Genotypic complexity of Fisher’s geometric model. Genetics. 2017;206:1049–1079. doi: 10.1534/genetics.116.199497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaltenbach M, Tokuriki N. Dynamics and constraints of enzyme evolution. J Exp Zool B Mol Dev Evol. 2014;322:468–487. doi: 10.1002/jez.b.22562. [DOI] [PubMed] [Google Scholar]
- 44.Goldsmith M, Tawfik DS. Enzyme engineering by targeted libraries. Methods Enzymol. 2013;523:257–283. doi: 10.1016/B978-0-12-394292-0.00012-6. [DOI] [PubMed] [Google Scholar]
- 45.Miton CM, Tokuriki N. How mutational epistasis impairs predictability in protein evolution and design. Protein Sci. 2016;25:1260–1272. doi: 10.1002/pro.2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dickinson BC, Leconte AM, Allen B, Esvelt KM, Liu DR. Experimental interrogation of the path dependence and stochasticity of protein evolution using phage-assisted continuous evolution. Proc Natl Acad Sci USA. 2013;110:9007–9012. doi: 10.1073/pnas.1220670110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moxon ER, Murphy PA. Haemophilus influenzae bacteremia and meningitis resulting from survival of a single organism. Proc Natl Acad Sci USA. 1978;75:1534–1536. doi: 10.1073/pnas.75.3.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hughes D, Andersson DI. Evolutionary trajectories to antibiotic resistance. Annu Rev Microbiol. 2017;71:579–596. doi: 10.1146/annurev-micro-090816-093813. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.