Significance
Fur is a key regulator of bacterial iron homeostasis and regulates the derepression of iron acquisition systems when iron is limited. However, it is unclear whether Fur-regulated genes are derepressed coordinately or in a sequential manner. Here, we describe a graded response for the Bacillus subtilis Fur regulon as the intracellular iron pool is gradually depleted. Three distinct sets of genes are sequentially expressed, and the induction order can be explained, at least in part, by differences in Fur affinity to its target operator sites. These results provide insights into the distinct roles of Fur target genes and contribute to our understanding of bacterial metalloregulatory systems.
Keywords: Fur regulon, iron limitation, graded response, iron-sparing response
Abstract
Bacterial cells modulate transcription in response to changes in iron availability. The ferric uptake regulator (Fur) senses intracellular iron availability and plays a central role in maintaining iron homeostasis in Bacillus subtilis. Here we utilized FrvA, a high-affinity Fe2+ efflux transporter from Listeria monocytogenes, as an inducible genetic tool to deplete intracellular iron. We then characterized the responses of the Fur, FsrA, and PerR regulons as cells transition from iron sufficiency to deficiency. Our results indicate that the Fur regulon is derepressed in three distinct waves. First, uptake systems for elemental iron (efeUOB), ferric citrate (fecCDEF), and petrobactin (fpbNOPQ) are induced to prevent iron deficiency. Second, B. subtilis synthesizes its own siderophore bacillibactin (dhbACEBF) and turns on bacillibactin (feuABC) and hydroxamate siderophore (fhuBCGD) uptake systems to scavenge iron from the environment and flavodoxins (ykuNOP) to replace ferredoxins. Third, as iron levels decline further, an “iron-sparing” response (fsrA, fbpAB, and fbpC) is induced to block the translation of abundant iron-utilizing proteins and thereby permit the most essential iron-dependent enzymes access to the limited iron pools. ChIP experiments demonstrate that in vivo occupancy of Fur correlates with derepression of each operon, and the graded response observed here results, at least in part, from higher-affinity binding of Fur to the “late”-induced genes.
Iron is indispensable for cell growth and survival for most bacteria. In many natural environments, particularly under neutral pH and aerobic conditions, iron limitation is a big challenge for bacteria due to the extremely low solubility and bioavailability of the Fe3+ ion. However, iron can also be toxic to cells when present in excess due to its participation in Fenton chemistry, which generates reactive oxygen radicals resulting in macromolecular damage and cell death (1, 2). Therefore, bacteria require regulatory systems to maintain iron homeostasis. The ferric uptake regulator (Fur) senses intracellular iron levels and plays a critical role in bacterial iron homeostasis by virtue of its ability to regulate transcription of genes involved in iron uptake, storage, and efflux (3, 4).
The Fur regulon has been well characterized in Bacillus subtilis and includes more than 50 genes, many of which are implicated in iron uptake, endogenous siderophore (bacillibactin) synthesis and uptake, and xenosiderophore uptake (siderophore made by other organisms) (5). Under iron-sufficient conditions, Fur binds to Fe2+ in the cytosol, and the resultant holo-Fur binds to its target DNA operators and functions as a repressor; under iron-limited conditions, apo-Fur dissociates from DNA, and the repression by Fur is relieved. Derepression of the Fur regulon leads to expression of numerous iron uptake systems to scavenge iron and adapt to iron deficiency. These include transporters for the import of elemental iron (EfeUOB), ferric citrate (YfmCDEF, renamed as FecCDEF to reflect their physiological role in ferric citrate import), hydroxamate siderophores (ferrioxamine and ferrichrome) (FhuBCGD-YxeB), and petrobactin (YclNOPQ, renamed as FpbNOPQ to reflect their role in ferric petrobactin import) (6).
In addition, B. subtilis synthesizes its own siderophore, bacillibactin (BB) (DhbACEBF), and a cognate uptake system (FeuABC-YusV), both of which are under Fur regulation (5, 7, 8). A phosphopantetheinyl transferase (encoded by sfp) is also required for BB biosynthesis (9, 10). Most laboratory strains of B. subtilis [those derived from the legacy strain B. subtilis 168 (11)] carry an sfp null mutation (sfp0) (9, 10) and can only produce the BB precursor 2,3-dihydroxybenzoic acid (DHBA) and its glycine conjugate (DHBG) [collectively assigned as DHB(G)]. The DHB(G) compounds bind to Fe3+ with modest affinity, while BB binds to Fe3+ with very high affinity (7, 12). BB uptake shares the same pathway as enterobactin (FeuABC-YusV) (8) and is under two levels of regulation: iron-dependent repression by Fur and substrate-specific activation by Btr (bacillibactin transport regulator), an AraC family transcriptional activator (13, 14). In addition to its prototypic role as an Fe2+-sensing repressor, emerging evidence indicates that in some cases, apo-Fur (without its coregulator Fe2+-bound) also binds DNA, and both forms of Fur may also function as transcriptional activators (15–17).
To adapt to iron limitation, bacteria also reconfigure their proteome to reduce synthesis of abundant, iron-utilizing proteins that are not essential for growth (an “iron-sparing” response), and they express alternative, iron-independent versions of some proteins. Together, these responses enable the cell to prioritize utilization of iron (13, 18, 19). In B. subtilis, Fur regulates FsrA, a noncoding Fur-regulated small RNA, which mediates the iron-sparing response with the help of three putative RNA chaperones (FbpABC) (13, 19). FsrA and its coregulators block the translation of low-priority iron-utilizing enzymes such as aconitase and succinate dehydrogenase and enable high-priority iron-dependent proteins to utilize the limited iron (13, 19). In addition, cells may substitute iron-independent flavodoxin for ferrodoxin, an abundant iron–sulfur protein involved in electron transfer reactions. Expression of flavodoxins (YkuNOP) is under regulation of Fur (5, 20).
Another transcriptional regulator that plays an important role in B. subtilis iron homeostasis is PerR, which modulates the adaptive response to peroxide stress, including peroxide detoxification, heme biosynthesis, and iron storage genes (21–26). PerR also regulates pfeT, a P1B4-type ATPase recently characterized as an Fe2+ efflux transporter (27). Expression of pfeT is induced under two conditions, oxidative stress and excess iron (27). A homolog of PfeT in Listeria monocytogenes, FrvA, has been identified as a high-affinity Fe2+ exporter, and its expression imposes severe iron limitation that leads to derepression of the Fur and PerR regulons in B. subtilis (28).
In this study, we utilized L. monocytogenes FrvA as an inducible genetic tool to deplete intracellular iron. By monitoring the transcription of Fur-regulated genes as cells transition from iron sufficiency to deficiency we determined that the Fur regulon is derepressed in three distinguishable waves. The first wave comprises uptake systems for elemental iron (efeUOB), ferric citrate (fecCDEF), and petrobactin (fpbNOPQ); in the second wave, B. subtilis synthesizes its own siderophore bacillibactin (dhbACEBF) and turns on bacillibactin uptake (feuABC) along with hydroxamate siderophore uptake (fhuBCGD) and flavodoxins (ykuNOP); last, as iron levels decrease further, the iron-sparing response (fsrA, fbpAB, and fbpC) is induced to prioritize iron utilization. Furthermore, the stepwise induction of Fur-regulated genes correlates with operator occupancy in vivo and can be explained, at least in part, by differences in protein–DNA-binding affinity.
Results
Expression of FrvA Imposes Iron Deprivation and Inhibits Cell Growth of B. subtilis.
Due to the high activity and redundancy of iron import systems, iron starvation can be difficult to achieve under laboratory conditions without the use of chelating agents such as dipyridyl. Here, we explored the use of the Fe2+ efflux transporter FrvA as a genetic tool to study iron limitation responses in B. subtilis. The resulting gradual depletion of intracellular iron levels may more closely mimic the natural situation wherein intracellular iron is depleted by growth.
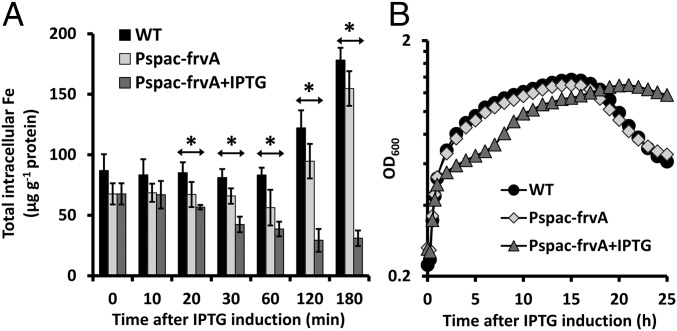
To validate its use as an iron depletion system, we monitored the effects of FrvA expression on intracellular metal levels using inductively coupled plasma mass spectrometry (ICP-MS). The uninduced cells harboring an isopropyl β-d-1-thiogalactopyranoside (IPTG)-inducible ectopic copy of frvA (WT Pspac-frvA) displayed a slightly reduced level of intracellular iron compared with WT, presumably due to the leaky expression from the Pspac promoter. When FrvA was induced by IPTG, intracellular iron levels declined by twofold after 30 min of treatment (Fig. 1A). By contrast, intracellular iron levels in both WT and uninduced cells increased significantly (about twofold) after 2 h (Fig. 1A), probably due to high iron demand as the growing cells enter into late logarithmic phase (transition phase) growth, which is characterized by increased respiratory activity (29). The intracellular level of other metals such as Mn and Co was not significantly changed during the course of the experiment (SI Appendix, Fig. S1).
Fig. 1.
Induction of L. monocytogenes FrvA imposes iron deprivation and inhibits cell growth of B. subtilis. (A) Levels of intracellular metals (Fe, Mn, and Co) were monitored for 3 h after 1 mM IPTG addition by inductively coupled plasma mass spectrometry (ICP-MS). The total concentration of ions was expressed as µg ion per gram of protein (mean ± SD; n = 3). Significant differences between WT and induced Pspac-frvA cells are determined by two-tailed t test as indicated: *P < 0.01. (B) Representative growth curves of the same strains as A in LB medium amended with 10 µM FeSO4. For IPTG-treated cells, 1 mM IPTG was added to cell culture when OD600 reaches ∼0.25. Experiments were performed at least three times with three biological replicates.
To evaluate the effects of FrvA expression on growth, we grew cells to early exponential phase (OD600 ∼ 0.25) in LB medium amended with 10 µM of FeSO4, induced expression of FrvA with 1 mM of IPTG, and monitored growth. The uninduced cells grew as well as WT in the first 3 h and displayed very minor growth inhibition afterward, presumably due to leaky expression of FrvA from Pspac (28) (Fig. 1B). In contrast, when FrvA was induced, cell growth was inhibited after ∼2 h (Fig. 1B). Thus, there is a substantial period of time (between ∼30–90 min after induction) where iron levels are significantly reduced (more than twofold reduction of total intracellular iron) with little if any apparent growth inhibition. Therefore, we chose this period of time to explore the graded response of the Fur regulon.
Fur-Regulated Genes Are Derepressed in a Stepwise Fashion.
To provide a global overview of the transcriptional changes as cells transition from iron sufficiency to iron deficiency we analyzed changes in RNA levels at 20, 30, 60, and 120 min after FrvA induction using oligonucleotide-based microarrays (SI Appendix, Fig. S2). In this analysis, most Fur-regulated operons were expressed at a low level before induction, as expected for cells grown in LB supplemented with 10 µM iron. In a direct comparison between cells carrying the Pspac-frvA construct grown with and without IPTG induction, the majority of the Fur regulon was derepressed (at least transiently) between 20 and 60 min after induction. In many cases, the maximal fold induction was comparable to that noted in a comparison of the fur mutant to WT (in strains lacking Pspac-frvA). In general, it appeared that some genes were induced at earlier time points, while induction of others was only evident at late time points, suggesting that the Fur regulon might be derepressed in a gradual manner in response to iron availability. Consistent with our prior study (28), induction of the PerR regulon in response to iron limitation was also apparent at late time points (3 h) and also in cultures with IPTG added at the time of subculture (steady state) (SI Appendix, Fig. S3A). This derepression is noted for peroxide detoxification enzymes (katA, ahpC, and ahpF) and a miniferritin that may function in iron storage (mrgA). These results were further confirmed by quantitative PCR (qPCR) (SI Appendix, Fig. S3B).
We were surprised to note that the two iron uptake systems most important for growth in LB medium (30), elemental iron uptake [EfeUOB; formerly, YwbLMN (31)] and ferric citrate import [FecCDEF; formerly, YfmCDEF (7)] were not highly induced during the course of this experiment (SI Appendix, Fig. S2). We hypothesized that these operons might be already partially derepressed in the uninduced cells due to mild iron deprivation resulting from leaky expression of FrvA. To test this idea, we used qPCR to monitor the expression of selected Fur-regulated operons in LB with 10 µM iron. We included the uptake systems for elemental iron (efeUOB), ferric citrate (fecCDEF), hydroxamate siderophores (fhuBCGD), and petrobactin (fpbNOPQ), as well as flavodoxins (ykuNOP), BB biosynthesis (dhbACEBF), BB uptake (feuABC), and iron-sparing response (fsrA, fbpAB, and fbpC) operons. Relative derepression of the first gene in each operon was analyzed by comparing gene expression in the induced cells as a percentage of derepression as observed in a fur mutant versus WT. Among all of the genes tested, three genes, fpbN, efeU, and fecC, were strongly derepressed (30–55% derepression) even at time 0 (SI Appendix, Fig. S4), presumably due to leaky expression of FrvA.
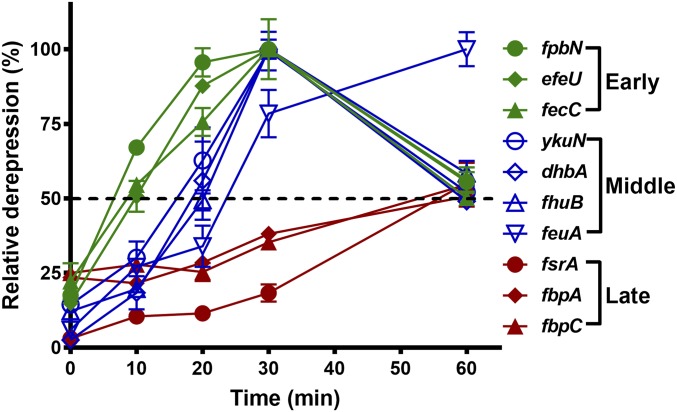
We hypothesized that a higher concentration of iron in the medium might enable a more complete repression of the Fur regulon. We therefore checked repression in LB medium amended with 25, 50, and 100 µM iron and determined that 25 µM was sufficient to fully repress Fur-regulated genes. We therefore repeated our qPCR analysis using LB medium amended with 25 µM. For this analysis we used 50% derepression as the threshold to define the order of gene induction. Three waves of derepression were revealed with 50% derepression at ∼10, 20, and 60 min (Fig. 2). As judged by monitoring of the first gene in each operon, we can resolve early (fpbN, efeU, and fecC), middle (fhuB, ykuN, dhbA, and feuA), and late (fsrA, fbpA, and fbpC) genes (Fig. 2). These results are consistent with the first set of experiments using 10 µM FeSO4 with the exception of feuA being assigned as a middle gene (Fig. 2 and SI Appendix, Fig. S4). The late induction of the FsrA/FbpABC iron-sparing response is expected to lead to translational down-regulation of a large number of target genes, which, as shown previously, is reflected as a reduction of steady-state mRNA levels (13). Indeed, we also observed a decrease in the mRNA levels of many FsrA-regulated genes at later time points (1 and 2 h) (SI Appendix, Fig. S5).
Fig. 2.
Fur-regulated genes are induced in a stepwise manner as a function of iron depletion. Cells were grown at 37 °C in LB medium overnight and subcultured (1:100 ratio) into fresh LB medium amended with 25 µM FeSO4. At OD600 ∼0.25, 1 mM IPTG was added to induce expression of FrvA. Quantitative PCR was conducted using a specific primer set for each gene (SI Appendix, Table S2). The housekeeping gene 23S rRNA was used as an internal control. Relative expression of the first gene in each operon at different times was analyzed by comparing gene expression in the IPTG-induced cells versus that in wild-type cells. The percentage of derepression was normalized based on the full derepression observed in a fur null mutant versus WT cells, which was set as 100% derepression for each gene tested.
Stepwise Induction of Fur-Regulated Genes Correlates with Operator Occupancy.
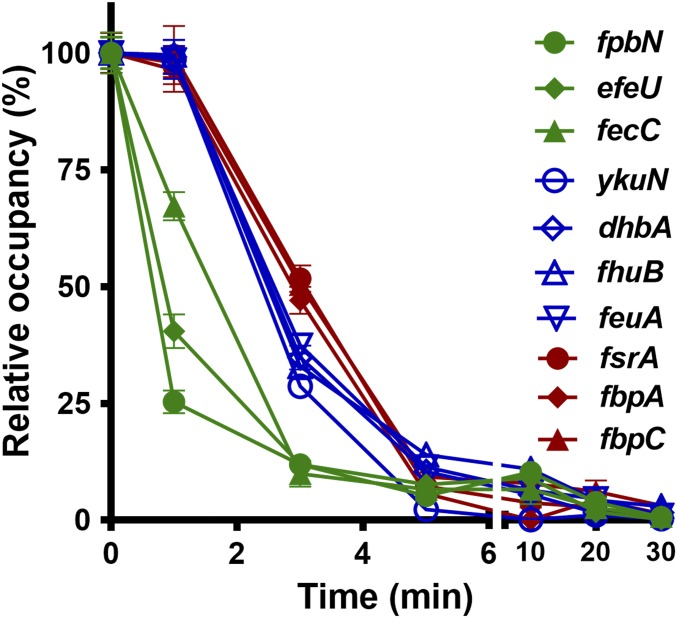
The molecular mechanism for the stepwise derepression of Fur-regulated genes may simply be that Fur operator occupancy is sequentially affected as a function of iron depletion. To test this idea, we constructed a fully functional chromosomal FLAG-tagged Fur in both WT and Pspac-frvA (SI Appendix, Fig. S6). Using chromatin immunoprecipitation (ChIP) coupled with qPCR, we found that occupancy of Fur on the operator sites tested declined rapidly upon iron depletion. Fur occupancy decreased significantly at the sites of three early genes (fpbN, efeU, and fecC) by 1 min; the differences of Fur occupancy among the three sets of genes became evident by 3 min, with only ∼10% occupancy for early genes (fpbN, efeU, and fecC), ∼35% occupancy for middle genes (fhuB, ykuN, dhbA, and feuA), and ∼55% occupancy for late genes (fsrA, fbpA, and fbpC) (Fig. 3). More importantly, the three distinct patterns revealed by ChIP (Fig. 3) correspond to the same three sets of early, middle, and late genes determined by qPCR (Fig. 2). The timescale of Fur dissociation from its operators (Fig. 3) appears to be considerably faster than the resulting accumulation of the corresponding mRNAs (Fig. 2). However, the ChIP procedure likely underestimates the time required for loss of operator binding since the times noted do not account for the time required to harvest cells by centrifugation before protein–DNA cross-linking. Moreover, the mRNA measurements reflect the cumulative effect of Fur dissociation, RNAP recruitment, and transcription, leading to an increase in mRNA levels. Despite the differences in timescale, these results support the general notion that Fur dissociates from early gene operator sites more quickly as intracellular iron levels decrease.
Fig. 3.
Operator occupancy correlates with stepwise derepression of the Fur regulon. Fur occupancy on different promoter sites was evaluated by chromatin immunoprecipitation (ChIP) using anti-FLAG antibodies. Coimmunoprecipitated DNA was quantified by qPCR using specific primer sets to the promoter regions of the target genes as listed in SI Appendix, Table S2. DNA enrichment was calculated based on the input DNA (1% of total DNA used for each ChIP experiment). Fur occupancy at different sites was set as 100% at time-point 0. Dates are presented as the relative percentage (%) of occupancy at different time points after 1 mM IPTG induction (mean ± SD; n = 3). No significant DNA enrichment was observed for gyrA that is used as a nonspecific negative control.
Stepwise Induction of Fur-Regulated Genes Is Correlated with Protein–DNA-Binding Affinity.
As the FrvA efflux pump gradually depletes iron from the cytosol the concentration of metallated Fur protein is predicted to decrease. We therefore hypothesized that the earliest induced operons in the Fur regulon would correspond to those with operator sites that bind holo-Fur weakly and, conversely, that the late-induced genes would bind holo-Fur with higher affinity. We determined the biochemical binding affinity in vitro using electrophoretic mobility shift assay (EMSA). The equilibrium dissociation constant (Kd) for Fur binding to different target operators provides an estimate of the concentration of active protein required for half-maximal repression. In support of our hypothesis, the stepwise induction of Fur regulon correlates, at least in part, with protein–DNA-binding affinity (Table 1 and SI Appendix, Figs. S7 and S8). For example, Fur binds to the operators of early genes with relatively low affinity (Kd value of ∼3 and 5.6 nM for fecC and fpbN, respectively), to those of middle gene with higher affinity (0.8 nM for fhuB, 0.9 nM for feuA, and 2.1 nM for dhbA), and with highest affinity to those for late genes (0.5–0.6 nM for all three late genes, fsrA, fbpA, and fbpC) (Table 1 and SI Appendix, Figs. S7 and S8). However, there are some exceptions. For instance, the induction of efeU, which binds Fur with high affinity (a Kd value of 1.5 nM), occurs as early as that of fpbN (a Kd value of 5.6 nM). Fur binds to some early and middle genes with an affinity difference of three- to fourfold (Table 1 and SI Appendix, Figs. S7 and S8), suggesting that other factors may also affect the regulation of some Fur target genes. Indeed, previous studies have revealed that multiple transcription factors such as ResD, NsrR, and Fur interact with one another and coregulate transcription of ykuN and other Fur-regulated genes during anaerobic fermentation and nitrate respiration (32, 33). It is also known that the bacillibactin uptake system (FeuABC) is under regulation of both Fur and Btr (13, 14) (see below).
Table 1.
Binding affinity of B. subtilis Fur to its target promoters
| Order of induction | Promoter | Kd*(nM) |
| Early | fpbN | 5.6 ± 0.5 |
| efeU | 1.5 ± 0.3 | |
| fecC | 3 ± 0.5 | |
| Middle | ykuN | 2.1 ± 0.4 |
| dhbA | 2.1 ± 0.5 | |
| fhuB | 0.9 ± 0.2 | |
| feuA | 0.8 ± 0.2 | |
| Late | fsrA | 0.6 ± 0.2 |
| fbpA | 0.6 ± 0.1 | |
| fbpC | 0.5 ± 0.1 |
Kd values were calculated using GraphPad Prism 5 (mean ± SD, n = 3) (SI Appendix, Figs. S7 and S8).
Stepwise Induction of Fur Regulon in Response to Iron Limitation in Strains Expressing Bacillibactin.
In most environmental isolates of B. subtilis, the mature siderophore BB is synthesized and exported by YmfD, a major facilitator superfamily transporter (34). The ferric–siderophore complex (Fe3+-BB) is then imported by the FeuABC-YusV system and hydrolyzed by the BesA esterase to release iron (8). Once imported, BB binds to the Btr transcription activator and stimulates transcription of the feuABC operon. BB binds to Fe3+ with remarkably high affinity, comparable to the chemically similar enterobactin produced by enteric bacteria (7, 12), and is therefore an effective iron-scavenging agent. Indeed, sfp+ strains have higher growth rate and yield in minimal medium amended with either 25 µM (SI Appendix, Fig. S9A) or 1 mM iron (SI Appendix, Fig. S9B) compared with sfp0 strains. Many laboratory strains (based on B. subtilis 168) are sfp0 and therefore only produce the BB precursors DHB(G), which bind Fe3+ with only modest affinity (lower than citrate) and may not function as an effective siderophore in natural settings (7, 35). Moreover, DHB(G) is ineffective in activating Btr, although Btr does have significant stimulatory activity even in the absence of siderophore (14).
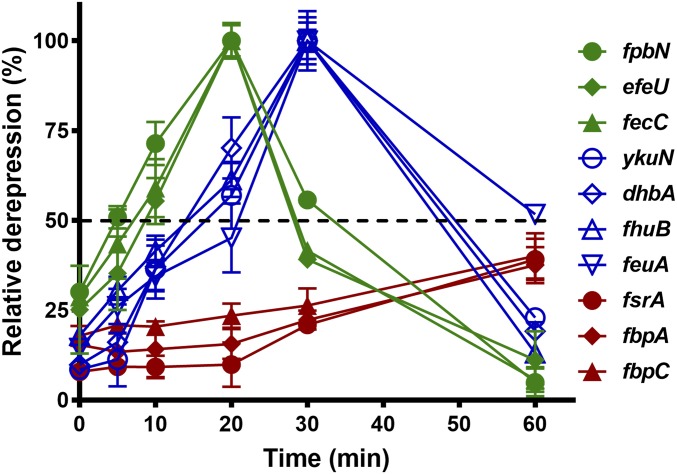
To further explore the graded response of the Fur regulon in strains producing BB, we monitored the induction of Fur target genes in sfp+ strains harboring an IPTG-inducible copy of frvA (sfp+ Pspac-frvA) as cells transition from iron sufficiency (LB + 25 µM iron) to deficiency. Consistent with the studies above (Fig. 2), Fur-regulated genes were derepressed in a stepwise manner with early (ypbN, efeU, and fecC), middle (fhuB, ykuN, dhbA, and feuA), and late (fsrA, fbpA, and fbpC) genes (Fig. 4). However, a few changes were notable: induction of all early and middle genes was shifted a little earlier, while induction of the late genes (fsrA, fbpA, and fbpC) was shifted later (Fig. 4). This suggests that efficient iron acquisition through Fe3+-BB delays proteomic remodeling via the iron-sparing response.
Fig. 4.
Fur-regulated genes are induced in a similar stepwise manner as a function of iron depletion in strains producing bacillibactin. Overnight cell culture was inoculated with 1:100 ratio into fresh LB medium amended with 25 µM FeSO4. After OD600 reaches ∼0.25, 1 mM IPTG was added to induce expression of FrvA. Total RNA was extracted and subjected to cDNA synthesis followed by qPCR. Relative expression of each gene at different times was monitored by comparing gene expression in the IPTG-induced cells versus that in sfp+ WT cells. The percentage of derepression was evaluated based on the full derepression observed in sfp+ fur mutant versus sfp+ WT, which was set as 100% derepression.
Inactivation of Bacillibactin Uptake Affects Induction of the Fur-Regulated Genes.
Expression of feuA is induced ∼20-fold upon iron depletion in a sfp0 strain (SI Appendix, Fig. S10A). Previous studies revealed that BB, but not the precursor DHBA, increases binding of Btr to the feuA promoter and stimulates transcription (13, 14). Indeed, the expression of feuA was derepressed up to 68-fold in a sfp+ strain (SI Appendix, Fig. S10C), indicating a more than threefold activation by BB. In the absence of Btr, feuA mRNA levels were reduced >30-fold, although Fur repression of feuA is still relieved by iron depletion (SI Appendix, Fig. S10 B and D). This effect can also be observed in disk-diffusion assays (SI Appendix, Fig. S11A). BB and DHB(G) compounds bind to Fe3+ and form complexes of purple color apparent around the zone of inhibition resulting from overexpression of FrvA (SI Appendix, Fig. S11A). As expected, inactivation of bacillibactin uptake by deletion of btr leads to clearly visible accumulation of DHB(G) or BB in the medium (SI Appendix, Fig. S11A). This result was confirmed by quantitation of the secreted Fe3+-DHB(G) or Fe3+-BB complex (SI Appendix, Fig. S11 B and C).
Although deletion of btr does not change the order of derepression of Fur target genes (SI Appendix, Fig. S10 E and F), most genes are induced earlier than observed in experiments done in WT cells (Figs. 2 and 4). In sfp0 strains, 50% derepression shifts from 10, 20, 60 min to 5, 14, or 40 min for early, middle, or late genes, respectively (SI Appendix, Fig. S10E). In sfp+ strains, 50% derepression shifts from 8, 15, 70 min to 3, 8, or 35 min for early, middle, or late genes, respectively (SI Appendix, Fig. S10F). These results suggest that biosynthesis and uptake of BB is critical for maintaining iron homeostasis upon iron limitation. These results also confirm that Btr is required for significant expression of the feuA operon, and in the absence of Btr the production of iron chelators (BB or its precursors) that cannot be efficiently imported back into the cell results in a more rapid onset of iron starvation (SI Appendix, Fig. S10 E and F).
Iron-Sparing Response Induced by Iron Depletion Leads to an Increase in Intracellular Labile Iron.
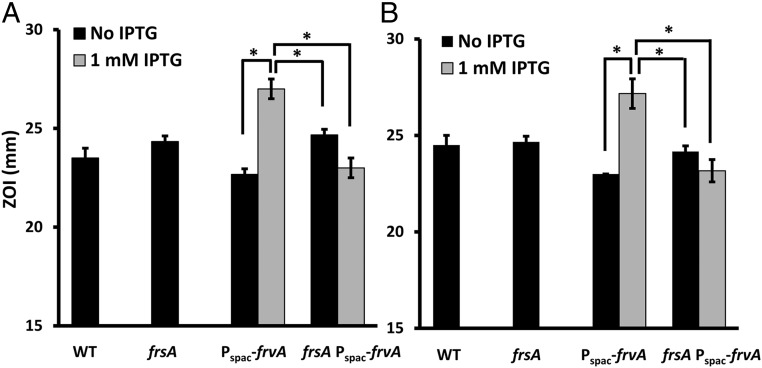
As shown above, when cells experience severe iron limitation the iron-sparing response is engaged to modulate cellular proteomes and prioritize the use of limited iron (Figs. 2 and Fig. 4 and SI Appendix, Figs. S4, S5, and S10). Since the translation of many abundant, low-priority iron-utilizing proteins is blocked by FsrA and its coregulators, we anticipated that intracellular labile iron pools might actually increase under these conditions, despite the fact that total intracellular iron is depleted (Fig. 1A). Indeed, in Escherichia coli expression of the small RNA RyhB, analogous to FsrA in B. subtilis, leads to an increase in the intracellular iron pool (36, 37). To test this notion, we monitored the intracellular labile pool by testing sensitivity to streptonigrin (SN), an antibiotic that responds to intracellular labile iron levels (38). Indeed, when FrvA is expressed, it depletes total intracellular iron but the intracellular labile iron level increases, as judged by the higher sensitivity to SN in both LB plates without or with supplementing exogenous iron (Fig. 5). In the absence of the FsrA-mediated iron-sparing response, this increase in intracellular labile iron no longer occurs (Fig. 5), suggesting that the iron-sparing response is chiefly, if not solely, responsible for this increase.
Fig. 5.
The iron-sparing response induced by iron depletion leads to an increase in intracellular labile iron. Sensitivity to streptonigrin (SN) was monitored using a disk-diffusion assay in LB agar plates containing either 0 (A) or 100 μM FeSO4 (B). Strains tested in both panels are wild-type (WT, CU1065), fsrA null mutant, Pspac-frvA, and fsrA Pspac-frvA. The data are expressed as the diameter (mean ± SD; n = 3) of the inhibition zone (mm). Significant differences determined by two-tailed t test are indicated: *P < 0.05.
Discussion
Iron homeostasis relies on the ability of bacterial cells to express iron acquisition systems including siderophores and their uptake systems. In B. subtilis, iron limitation activates expression of uptake systems for elemental iron, ferric citrate, and several xenosiderophores, as well as for biosynthesis and uptake of the endogenous siderophore bacillibactin. Our results show that as intracellular iron levels decline Fur-regulated genes are induced in a stepwise manner with three distinguishable waves. The first response to iron limitation is the induction of uptake systems for petrobactin (FpbNOPQ), elemental iron (EfeUOB), and ferric citrate (FecCDEF). Petrobactin (PB), a photoreactive catecholate siderophore, is synthesized by marine bacteria (39) and some members of the Bacillus cereus group (40–43), including Bacillus anthracis (41). PB-mediated iron acquisition may play an important physiological role in cell survival within some ecological niches. For example, in B. anthracis PB biosynthesis is required for survival in macrophages and virulence in mice (44, 45). PB, elemental iron, and ferric citrate likely represent major iron sources generally available in the environment (46, 47), consistent with the early induction of their uptake systems.
As intracellular iron levels decline further, more efficient siderophore-mediated acquisition systems are needed to support growth. The second wave includes production of BB (DhbACEBF) and expression of several uptake systems: FeuABC for both BB and enterobactin (8), and FhuBCGD for hydroxamate siderophores (ferrichrome and ferrioxamine) (5). Both enterobactin and BB exhibit remarkably high affinity to iron, 10−34.3 or 10−33.1 M, respectively (7); while hydroxamate siderophores have lower affinity to iron ranging from 10−25.2 to 10−27.5 M (7). In addition, as part of a common strategy against nutrient limitation, cells replace 4Fe-4S ferrodoxins with alternative iron-independent flavodoxins to help alleviate iron demand (48). B. subtilis contains two flavodoxins, YkuN and YkuP, which are encoded by the ykuNOP operon. They have been shown to function as electron donors for fatty acid desaturation equivalently as ferrodoxin (49). More generally, flavodoxins have been used as a biological marker for iron limitation in ecological settings (50), indicating this adaptive response is widespread.
As iron bioavailability declines further, the iron-sparing response is induced to remodel the proteome and conserve iron. The iron-sparing response has been well studied in several microorganisms. In E. coli, it is mediated by a small RNA RyhB, which with the assistance of an RNA chaperone blocks the translation of nonessential iron-utilizing proteins (18, 51). In B. subtilis, this response is mediated by a small RNA FsrA and its three coregulators FbpA, FbpB and FbpC. FsrA binds target mRNAs through RNA-mRNA base-pairing and blocks translation of its targets to reduce iron demand (13, 19).
Collectively our results demonstrate that as cells transition from iron sufficiency to deficiency, B. subtilis expresses (i) iron uptake systems to prevent deficiency, (ii) alternative noniron proteins to replace iron enzymes and reduce iron demand and high-affinity siderophore-mediated import systems to scavenge iron, and (iii) a small RNA FsrA and its partner proteins to prioritize iron utilization. Nevertheless, the graded response of Fur regulon to iron limitation may extend further. A recent study using genome footprinting coupled with high-throughput sequencing (GeF-Seq) revealed that under anaerobic conditions B. subtilis Fur binds to many DNA sites in vivo in addition to the previously established Fur regulon (33). E. coli Fur has also been shown to play a regulatory role beyond iron metabolism, including DNA synthesis and biofilm formation (15). Moreover, a graded response of Fur regulon to iron starvation may also be present in other bacteria. Indeed, a graded response was reported in Crocosphaera watsonii, a unicellular diazotrophic marine cyanobacterium (52), however the underlying mechanism was not characterized.
In natural settings, metal limitation may occur gradually as intracellular metal levels fall. As a result, it is unlikely that transcription regulators turn on their regulon all at once, but rather as a graded response. One remarkable example is the derepression of Zur regulon in response to zinc limitation in Streptomyces coelicolor (53) and B. subtilis (54). As cells transition from zinc sufficiency to deficiency, Zur-regulated genes are also induced in a stepwise manner. In B. subtilis it has been demonstrated that as an initial response to zinc deprivation alternative ribosomal proteins are derepressed to mobilize zinc from intracellular zinc pools. In the second wave, high-affinity uptake systems are induced to import zinc. Finally, as zinc levels decline further, late genes are induced to replace Zn-dependent proteins needed to ensure ribosome assembly and folate synthesis (54). Together with the results reported here, this example illustrates how bacterial cells prioritize their responses to metal limitation.
Materials and Methods
Materials and methods are described in the SI Appendix, including growth conditions, quantification of intracellular metal ion by ICP-MS, growth curve assay, Western blot, disk-diffusion assay, siderophore quantification, ChIP-qPCR, and EMSA.
Supplementary Material
Acknowledgments
We thank members of the J.D.H. laboratory for helpful comments. This work was supported by a grant from the National Institutes of Health (to J.D.H.) (R35GM122461).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1713008114/-/DCSupplemental.
References
- 1.Imlay JA. Pathways of oxidative damage. Annu Rev Microbiol. 2003;57:395–418. doi: 10.1146/annurev.micro.57.030502.090938. [DOI] [PubMed] [Google Scholar]
- 2.Park S, You X, Imlay JA. Substantial DNA damage from submicromolar intracellular hydrogen peroxide detected in Hpx- mutants of Escherichia coli. Proc Natl Acad Sci USA. 2005;102:9317–9322. doi: 10.1073/pnas.0502051102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Helmann JD. Specificity of metal sensing: Iron and manganese homeostasis in Bacillus subtilis. J Biol Chem. 2014;289:28112–28120. doi: 10.1074/jbc.R114.587071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fleischhacker AS, Kiley PJ. Iron-containing transcription factors and their roles as sensors. Curr Opin Chem Biol. 2011;15:335–341. doi: 10.1016/j.cbpa.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baichoo N, Wang T, Ye R, Helmann JD. Global analysis of the Bacillus subtilis Fur regulon and the iron starvation stimulon. Mol Microbiol. 2002;45:1613–1629. doi: 10.1046/j.1365-2958.2002.03113.x. [DOI] [PubMed] [Google Scholar]
- 6.Zawadzka AM, et al. Characterization of a Bacillus subtilis transporter for petrobactin, an anthrax stealth siderophore. Proc Natl Acad Sci USA. 2009;106:21854–21859. doi: 10.1073/pnas.0904793106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ollinger J, Song KB, Antelmann H, Hecker M, Helmann JD. Role of the Fur regulon in iron transport in Bacillus subtilis. J Bacteriol. 2006;188:3664–3673. doi: 10.1128/JB.188.10.3664-3673.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miethke M, et al. Ferri-bacillibactin uptake and hydrolysis in Bacillus subtilis. Mol Microbiol. 2006;61:1413–1427. doi: 10.1111/j.1365-2958.2006.05321.x. [DOI] [PubMed] [Google Scholar]
- 9.May JJ, Wendrich TM, Marahiel MA. The dhb operon of Bacillus subtilis encodes the biosynthetic template for the catecholic siderophore 2,3-dihydroxybenzoate-glycine-threonine trimeric ester bacillibactin. J Biol Chem. 2001;276:7209–7217. doi: 10.1074/jbc.M009140200. [DOI] [PubMed] [Google Scholar]
- 10.Quadri LE, et al. Characterization of Sfp, a Bacillus subtilis phosphopantetheinyl transferase for peptidyl carrier protein domains in peptide synthetases. Biochemistry. 1998;37:1585–1595. doi: 10.1021/bi9719861. [DOI] [PubMed] [Google Scholar]
- 11.Zeigler DR, et al. The origins of 168, W23, and other Bacillus subtilis legacy strains. J Bacteriol. 2008;190:6983–6995. doi: 10.1128/JB.00722-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dertz EA, Xu J, Stintzi A, Raymond KN. Bacillibactin-mediated iron transport in Bacillus subtilis. J Am Chem Soc. 2006;128:22–23. doi: 10.1021/ja055898c. [DOI] [PubMed] [Google Scholar]
- 13.Smaldone GT, et al. A global investigation of the Bacillus subtilis iron-sparing response identifies major changes in metabolism. J Bacteriol. 2012;194:2594–2605. doi: 10.1128/JB.05990-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaballa A, Helmann JD. Substrate induction of siderophore transport in Bacillus subtilis mediated by a novel one-component regulator. Mol Microbiol. 2007;66:164–173. doi: 10.1111/j.1365-2958.2007.05905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seo SW, et al. Deciphering Fur transcriptional regulatory network highlights its complex role beyond iron metabolism in Escherichia coli. Nat Commun. 2014;5:4910. doi: 10.1038/ncomms5910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delany I, Rappuoli R, Scarlato V. Fur functions as an activator and as a repressor of putative virulence genes in Neisseria meningitidis. Mol Microbiol. 2004;52:1081–1090. doi: 10.1111/j.1365-2958.2004.04030.x. [DOI] [PubMed] [Google Scholar]
- 17.Yu C, Genco CA. Fur-mediated activation of gene transcription in the human pathogen Neisseria gonorrhoeae. J Bacteriol. 2012;194:1730–1742. doi: 10.1128/JB.06176-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Massé E, Vanderpool CK, Gottesman S. Effect of RyhB small RNA on global iron use in Escherichia coli. J Bacteriol. 2005;187:6962–6971. doi: 10.1128/JB.187.20.6962-6971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaballa A, et al. The Bacillus subtilis iron-sparing response is mediated by a Fur-regulated small RNA and three small, basic proteins. Proc Natl Acad Sci USA. 2008;105:11927–11932. doi: 10.1073/pnas.0711752105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lawson RJ, von Wachenfeldt C, Haq I, Perkins J, Munro AW. Expression and characterization of the two flavodoxin proteins of Bacillus subtilis, YkuN and YkuP: Biophysical properties and interactions with cytochrome P450 BioI. Biochemistry. 2004;43:12390–12409. doi: 10.1021/bi049131t. [DOI] [PubMed] [Google Scholar]
- 21.Lee JW, Helmann JD. The PerR transcription factor senses H2O2 by metal-catalysed histidine oxidation. Nature. 2006;440:363–367. doi: 10.1038/nature04537. [DOI] [PubMed] [Google Scholar]
- 22.Faulkner MJ, Helmann JD. Peroxide stress elicits adaptive changes in bacterial metal ion homeostasis. Antioxid Redox Signal. 2011;15:175–189. doi: 10.1089/ars.2010.3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zuber P. Management of oxidative stress in Bacillus. Annu Rev Microbiol. 2009;63:575–597. doi: 10.1146/annurev.micro.091208.073241. [DOI] [PubMed] [Google Scholar]
- 24.Chen L, Helmann JD. Bacillus subtilis MrgA is a Dps(PexB) homologue: Evidence for metalloregulation of an oxidative-stress gene. Mol Microbiol. 1995;18:295–300. doi: 10.1111/j.1365-2958.1995.mmi_18020295.x. [DOI] [PubMed] [Google Scholar]
- 25.Herbig AF, Helmann JD. Roles of metal ions and hydrogen peroxide in modulating the interaction of the Bacillus subtilis PerR peroxide regulon repressor with operator DNA. Mol Microbiol. 2001;41:849–859. doi: 10.1046/j.1365-2958.2001.02543.x. [DOI] [PubMed] [Google Scholar]
- 26.Fuangthong M, Herbig AF, Bsat N, Helmann JD. Regulation of the Bacillus subtilis fur and perR genes by PerR: Not all members of the PerR regulon are peroxide inducible. J Bacteriol. 2002;184:3276–3286. doi: 10.1128/JB.184.12.3276-3286.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guan G, et al. PfeT, a P1B4 -type ATPase, effluxes ferrous iron and protects Bacillus subtilis against iron intoxication. Mol Microbiol. 2015;98:787–803. doi: 10.1111/mmi.13158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pi H, Patel SJ, Argüello JM, Helmann JD. The Listeria monocytogenes Fur-regulated virulence protein FrvA is an Fe(II) efflux P1B4 -type ATPase. Mol Microbiol. 2016;100:1066–1079. doi: 10.1111/mmi.13368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mueller JP, Taber HW. Structure and expression of the cytochrome aa3 regulatory gene ctaA of Bacillus subtilis. J Bacteriol. 1989;171:4979–4986. doi: 10.1128/jb.171.9.4979-4986.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Faulkner MJ, Ma Z, Fuangthong M, Helmann JD. Derepression of the Bacillus subtilis PerR peroxide stress response leads to iron deficiency. J Bacteriol. 2012;194:1226–1235. doi: 10.1128/JB.06566-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miethke M, Monteferrante CG, Marahiel MA, van Dijl JM. The Bacillus subtilis EfeUOB transporter is essential for high-affinity acquisition of ferrous and ferric iron. Biochim Biophys Acta. 2013;1833:2267–2278. doi: 10.1016/j.bbamcr.2013.05.027. [DOI] [PubMed] [Google Scholar]
- 32.Henares B, et al. The ResD response regulator, through functional interaction with NsrR and fur, plays three distinct roles in Bacillus subtilis transcriptional control. J Bacteriol. 2014;196:493–503. doi: 10.1128/JB.01166-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chumsakul O, et al. Genome-wide analysis of ResD, NsrR, and Fur binding in Bacillus subtilis during anaerobic fermentative growth by in vivo footprinting. J Bacteriol. 2017;199:e00086–e17. doi: 10.1128/JB.00086-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miethke M, Schmidt S, Marahiel MA. The major facilitator superfamily-type transporter YmfE and the multidrug-efflux activator Mta mediate bacillibactin secretion in Bacillus subtilis. J Bacteriol. 2008;190:5143–5152. doi: 10.1128/JB.00464-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chipperfield JR, Ratledge C. Salicylic acid is not a bacterial siderophore: A theoretical study. Biometals. 2000;13:165–168. doi: 10.1023/a:1009227206890. [DOI] [PubMed] [Google Scholar]
- 36.Jacques JF, et al. RyhB small RNA modulates the free intracellular iron pool and is essential for normal growth during iron limitation in Escherichia coli. Mol Microbiol. 2006;62:1181–1190. doi: 10.1111/j.1365-2958.2006.05439.x. [DOI] [PubMed] [Google Scholar]
- 37.Gerstle K, Klätschke K, Hahn U, Piganeau N. The small RNA RybA regulates key-genes in the biosynthesis of aromatic amino acids under peroxide stress in E. coli. RNA Biol. 2012;9:458–468. doi: 10.4161/rna.19065. [DOI] [PubMed] [Google Scholar]
- 38.Yeowell HN, White JR. Iron requirement in the bactericidal mechanism of streptonigrin. Antimicrob Agents Chemother. 1982;22:961–968. doi: 10.1128/aac.22.6.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barbeau K, Zhang G, Live DH, Butler A. Petrobactin, a photoreactive siderophore produced by the oil-degrading marine bacterium Marinobacter hydrocarbonoclasticus. J Am Chem Soc. 2002;124:378–379. doi: 10.1021/ja0119088. [DOI] [PubMed] [Google Scholar]
- 40.Abergel RJ, Zawadzka AM, Raymond KN. Petrobactin-mediated iron transport in pathogenic bacteria: Coordination chemistry of an unusual 3,4-catecholate/citrate siderophore. J Am Chem Soc. 2008;130:2124–2125. doi: 10.1021/ja077202g. [DOI] [PubMed] [Google Scholar]
- 41.Koppisch AT, et al. Petrobactin is the primary siderophore synthesized by Bacillus anthracis str. Sterne under conditions of iron starvation. Biometals. 2005;18:577–585. doi: 10.1007/s10534-005-1782-6. [DOI] [PubMed] [Google Scholar]
- 42.Koppisch AT, et al. Petrobactin is produced by both pathogenic and non-pathogenic isolates of the Bacillus cereus group of bacteria. Biometals. 2008;21:581–589. doi: 10.1007/s10534-008-9144-9. [DOI] [PubMed] [Google Scholar]
- 43.Wilson MK, Abergel RJ, Raymond KN, Arceneaux JE, Byers BR. Siderophores of Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis. Biochem Biophys Res Commun. 2006;348:320–325. doi: 10.1016/j.bbrc.2006.07.055. [DOI] [PubMed] [Google Scholar]
- 44.Cendrowski S, MacArthur W, Hanna P. Bacillus anthracis requires siderophore biosynthesis for growth in macrophages and mouse virulence. Mol Microbiol. 2004;51:407–417. doi: 10.1046/j.1365-2958.2003.03861.x. [DOI] [PubMed] [Google Scholar]
- 45.Abergel RJ, et al. Anthrax pathogen evades the mammalian immune system through stealth siderophore production. Proc Natl Acad Sci USA. 2006;103:18499–18503. doi: 10.1073/pnas.0607055103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Frawley ER, et al. Iron and citrate export by a major facilitator superfamily pump regulates metabolism and stress resistance in Salmonella Typhimurium. Proc Natl Acad Sci USA. 2013;110:12054–12059. doi: 10.1073/pnas.1218274110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pi H, Helmann JD. Ferrous iron efflux systems in bacteria. Metallomics. 2017;9:840–851. doi: 10.1039/c7mt00112f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Merchant SS, Helmann JD. Elemental economy: Microbial strategies for optimizing growth in the face of nutrient limitation. Adv Microb Physiol. 2012;60:91–210. doi: 10.1016/B978-0-12-398264-3.00002-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chazarreta-Cifre L, Martiarena L, de Mendoza D, Altabe SG. Role of ferredoxin and flavodoxins in Bacillus subtilis fatty acid desaturation. J Bacteriol. 2011;193:4043–4048. doi: 10.1128/JB.05103-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Erdner DL, Anderson DM. Ferredoxin and flavodoxin as biochemical indicators of iron limitation during open‐ocean iron enrichment. Limnol Oceanogr. 1999;44:1609–1615. [Google Scholar]
- 51.Massé E, Salvail H, Desnoyers G, Arguin M. Small RNAs controlling iron metabolism. Curr Opin Microbiol. 2007;10:140–145. doi: 10.1016/j.mib.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 52.Jacq V, Ridame C, L’Helguen S, Kaczmar F, Saliot A. Response of the unicellular diazotrophic cyanobacterium Crocosphaera watsonii to iron limitation. PLoS One. 2014;9:e86749. doi: 10.1371/journal.pone.0086749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shin JH, et al. Graded expression of zinc-responsive genes through two regulatory zinc-binding sites in Zur. Proc Natl Acad Sci USA. 2011;108:5045–5050. doi: 10.1073/pnas.1017744108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shin JH, Helmann JD. Molecular logic of the Zur-regulated zinc deprivation response in Bacillus subtilis. Nat Commun. 2016;7:12612. doi: 10.1038/ncomms12612. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.