Significance
Diflavin-linked disulfide oxidoreductases (DDORs) are structurally related to the low-molecular-weight type NADP-dependent thioredoxin reductases, although they do not share a common function. The biochemical and structural characterization of DDORs has revealed a previously unreported mechanism for the transfer of reducing equivalents in flavoenzymes. The present work illustrates the extent to which nature has experimented with flavins as enzyme cofactors in the evolution of redox reactions.
Keywords: flavoprotein, transfer of reducing equivalents, redox active disulfide, Rossmann fold, sulfhydryl
Abstract
Flavoproteins participate in a wide variety of physiologically relevant processes that typically involve redox reactions. Within this protein superfamily, there exists a group that is able to transfer reducing equivalents from FAD to a redox-active disulfide bridge, which further reduces disulfide bridges in target proteins to regulate their structure and function. We have identified a previously undescribed type of flavin enzyme that is exclusive to oxygenic photosynthetic prokaryotes and that is based on the primary sequence that had been assigned as an NADPH-dependent thioredoxin reductase (NTR). However, our experimental data show that the protein does not transfer reducing equivalents from flavins to disulfides as in NTRs but functions in the opposite direction. High-resolution structures of the protein from Gloeobacter violaceus and Synechocystis sp. PCC6803 obtained by X-ray crystallography showed two juxtaposed FAD molecules per monomer in redox communication with an active disulfide bridge in a variant of the fold adopted by NTRs. We have tentatively named the flavoprotein “DDOR” (diflavin-linked disulfide oxidoreductase) and propose that its activity is linked to a thiol-based transfer of reducing equivalents in bacterial membranes. These findings expand the structural and mechanistic repertoire of flavoenzymes with oxidoreductase activity and pave the way to explore new protein engineering approaches aimed at designing redox-active proteins for diverse biotechnological applications.
The capacity for the controlled oxidation of sulfhydryl groups in proteins and metabolites to regulate cellular processes and physiological states has been exploited by evolution in all kingdoms of living organisms (1–5). Redox elements involved in these modifications include cofactors in metallo- and flavoproteins participating in the transfer of reducing equivalents. In most types of cells, the thioredoxin (Trx) and glutaredoxin (Grx) systems are largely responsible for the regulation of the oxidation state of thiols in response to metabolic need (6). The Trx system is composed of Trx, a small protein with a redox-active CxxC motif, and a thioredoxin reductase (TR) to reduce Trx. In the Grx system, Grxs—structurally similar to Trx but physicochemically different—are primarily reduced nonenzymatically by glutathione (GSH), itself reduced with NADPH by glutathione reductase. Typically, Trxs participate in dithiol/disulfide exchange reactions that result in a change in the structural and functional properties of target proteins, whereas Grxs are mainly involved in glutathionylation and Fe–S cluster assembly (7–9).
Thiol-based redox regulation assumes a primary role in oxygenic photosynthesis due to the production of molecular oxygen and a change in the redox environment during the diurnal cycle (10, 11). Oxyphotosynthetic organisms have evolved a sophisticated network of redox signaling and modulation to regulate cell processes in response to these changes. In cyanobacteria and plastids, the Trx system plays a prominent role in diurnal and oxidative regulation. For this purpose, organisms use two main types of TRs, ferredoxin (Fdx):Trx reductase (FTR) and NADP-thioredoxin reductase (NTR), that function coordinately in the regulation of processes in response to changing environmental conditions (12). FTR is a monomeric metalloenzyme composed of a 4Fe–4S center and a redox-active CxxC motif that catalyzes the transfer of reducing equivalents from reduced Fdx to oxidized Trx (13). In this way, an electronic signal is converted to a thiol signal. NTR is a homodimeric flavoenzyme containing a noncovalently bound FAD and a redox-active CxxC motif that reduces Trx at the expense of NADPH (14). Chloroplasts and some cyanobacteria contain a special form of NTR, called “NTRC,” in which the enzyme is fused to a functional Trx partner in a single polypeptide chain (15); NTRC plays a central role in antioxidant metabolism (16). The FTR and NTR systems are interesting examples of how structurally and functionally distinct classes of cofactor-dependent enzymes have evolved to regulate Trx-linked processes in cyanobacteria and plastids.
Recent work has increased the number of members of the flavoenzyme family active in Trx reduction in oxyphotosynthetic organisms, including an enzyme called “DTR” (for “deeply-rooted thioredoxin reductase”) that reduces Trxs in a pyridine nucleotide-independent manner. DTR is present in some cyanobacteria and marine algae (17), but the physiological electron donor has not been identified.
With the aim of further characterizing the different members of the TR superfamily in cyanobacteria, we have studied the gene products of Gloeobacter violaceus gll2934 and Synechocystis sp. PCC 6803 slr0600. Although these proteins are annotated as NTRs in the databases, the enzymes exhibit unexpected properties that preclude a function as Trx reductases, and their physiological role remains elusive. Among other properties, we have found that each monomer of the dimeric protein contains two juxtaposed flavins, one of which is in redox communication with a disulfide in a CxxC motif. We have thus provisionally named the protein “DDOR” (for “diflavin-linked disulfide oxidoreductase”). It appears that DDOR has diversified its function across members of the NTR-related protein family by evolving specific structural motifs; in oxyphotosynthetic organisms, two flavin cofactors fit together in a juxtaposed position in the enzyme. To our knowledge, these properties represent a previously unrecognized mechanism for the transfer of reducing equivalents that expands the structural and mechanistic repertoire of flavoenzymes with oxidoreductase activity.
Results
DDOR Lacks the Archetypical Pyridine Nucleotide-Binding Site.
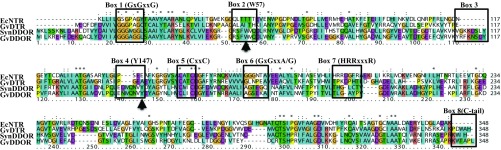
A sequence comparison (Fig. 1) and phylogenetic analyses (Fig. S1) of annotated TRs in cyanobacteria clearly distinguish three groups of NTR-related enzymes: the archetypal NTR (and the related NTRC), the recently characterized NADP-independent DTR (17), and a third uncharacterized group herein named DDOR or diflavin-linked disulfide oxidoreductase (see below).
Fig. 1.
Protein sequence alignment of EcNTR, GvDTR, SynDDOR, and GvDDOR. Boxes 1 and 5 include the conserved motif for FAD binding (GxGxxG) and the redox-active Cys (CxxC), respectively. Motifs for NAD(P)H binding in pyridine nucleotide-dependent TRs are depicted in boxes 6 (GxGxxA/G) and 7 (HRRxxxR). Regions specific for the DDOR family are shown in boxes 2–4 and include two conserved aromatic residues (W57 and Y147; positions are indicated with arrows) for π–π stacking interactions with the isoalloxazine group of a flavin prosthetic group (Fig. 3B) and the insertion of seven amino acids in a region essential for Trx binding in NTRs. C-terminal extensions in DTR and DDOR are indicated in box 8. Residue conservation colors are shown according to default ClustalX parameters (39).
The primary sequence of DDOR proteins shares with NTRs an FAD-binding site (GxGxxG) and a conserved CxxC motif, a hallmark of redox-active proteins (Fig. 1, boxes 1 and 5); however, they lack the common GxGxxA/G and HRRxxxR motifs necessary for pyridine nucleotide binding (Fig. 1, boxes 6 and 7). In addition to altered pyridine nucleotide-binding sites, variations in DDOR sequences comprised a C-terminal extension with a conserved aromatic residue (Fig. 1, box 8); other insertions detected in the multiple sequence protein alignment include modifications located in a region that participates in Trx binding in NTRs (18, 19) and that can be used as a signature sequence for this protein family (Fig. 1, boxes 2–4).
The distribution of DDOR together with NTR/NTRC and DTR in selected cyanobacterial species is shown in Table 1. There is not a clear pattern of distribution in cyanobacteria. Some organisms with DDOR do not possess archetypal NTR/NTRC but do possess DTR, as is the case of G. violaceus; others, such as Synechocystis sp. PC6803, have DDOR but not NTR/NTRC or DTR (Table 1). This observation opens an intriguing scenario whereby Trx reduction by a flavoenzyme is independent of NADPH in these organisms. To provide a complete view of the diversity of the NTR-related protein superfamily in cyanobacteria, we characterized two members of the DDOR protein group: the gene product of Gloeobacter gll2934 and the sole NTR-annotated sequence in the model cyanobacterium Synechocystis sp. PCC 6803 (slr0600).
Table 1.
TR (FTR, NTR/NTRC, and DTR) and DDOR composition of selected cyanobacteria
| Species | FTR | NTR | NTRC | DTR | DDOR |
| Acaryochloris marina MBIC11017 | + | — | + | + | + |
| Anabaena variabilis ATCC 29413 | + | — | + | — | + |
| Gloeobacter violaceus PCC 7421* | — | — | — | + | + |
| Leptolyngbya sp. PCC 7376 | + | — | — | + | + |
| Nostoc sp. PCC 7120 | + | + | + | — | + |
| Synechococcus sp. JA-3-3Ab* | + | — | + | — | + |
| Synechocystis sp. PCC 6803 | + | — | — | — | + |
| Thermosynechococcus elongatus BP-1 | + | — | + | — | — |
| Trichodesmium erythraeum IMS101 | + | — | — | + | — |
| Prochlorococcus marinus SS120 | — | — | + | + | — |
| Prochlorococcus marinus MIT9313 | — | — | — | + | — |
Members of early-branching lineages of bacteria.
NADPH Is Not the Direct Donor of Reducing Equivalents to DDOR Flavoproteins.
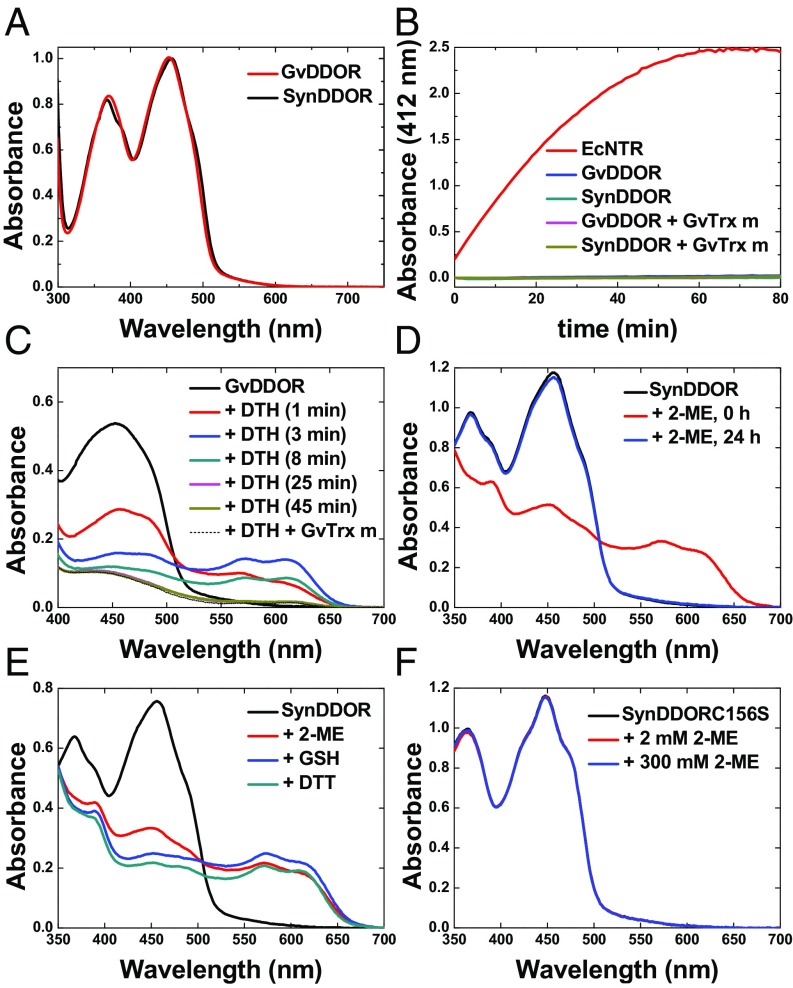
In light of the sequence divergence in the pyridine nucleotide-binding motifs, we produced Gloeobacter and Synechocystis DDOR proteins (GvDDOR and SynDDOR, respectively) for their functional and structural characterization. The proteins exhibited spectral features typical of flavin-containing enzymes (20), with absorption maxima at 367–370 and 452–454 nm (Fig. 2A). However, the lack of a distinct shoulder at 470 nm indicates that DDORs have a flavin in a more polar environment than the flavins of NTR, as shown by crystallography (see below) (20). In contrast to typical NTRs (21), GvDDOR and SynDDOR failed to catalyze the reduction of 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB) in either the presence or absence of Trx using NADPH as source of reducing equivalents (Fig. 2B). Following an approach previously applied to DTR (17), a microcalorimetric assay was employed to measure the affinity of the DDOR enzyme for pyridine nucleotides. The isothermal titration calorimetry (ITC)-binding isotherms obtained from the titration of DDOR with 3-acetylpyridine adenine dinucleotide phosphate (AADP), a nonreducible analog of NADPH (18), failed to result in detectable binding (Fig. S2A). We therefore concluded that DDORs are pyridine nucleotide-independent flavoenzymes.
Fig. 2.
Analysis of DDOR activity. (A) UV-visible absorption spectra of GvDDOR and SynDDOR proteins in buffer (20 mM potassium phosphate, 100 mM KCl, pH 7.6). (B) Activity of EcNTR, GvDDOR, and SynDDOR (250 nM) reducing 5 mM DTNB with 150 nM NADPH as a source of reducing equivalents measured as absorbance changes at 412 nm. Experiments were also performed in the presence of GvTrx-m. (C) Reduction of GvDDOR with DTH in buffer [100 mM potassium phosphate (pH 6.8), 100 mM KCl, and 2 mM EDTA] under anaerobic conditions. The time after addition of DTH is indicated in parentheses. The spectrum obtained after the addition of homologous Trx-m to the reduced system is displayed as a dashed black line. (D) Spectral changes of GvDDOR during incubation with 10 mM 2-ME at 5 min and 24 h. (E) Spectral changes after incubation of GvDDOR with 10 mM dithiol (DTT) and monothiol (GSH and 2-ME) substrates. (F) Incubation of the mutant protein SynDDORC156S with 2-ME.
DDORs Do Not Reduce Trx.
The reduction of the flavin component of DDOR was tested with sodium dithionite (DTH), a well-known nonphysiological reductant of the flavin moiety, by monitoring changes in the visible flavin spectrum upon reductant addition under anaerobic conditions. We observed that DTH was able to slowly reduce the GvDDOR enzyme, at first resulting in a decrease of the absorption band at 453 nm and the appearance of bands at 567 nm and 608 nm that were displaced to 573 nm and 611 nm with time (Fig. 2C). The comparatively rapid changes are attributable to a neutral (blue) flavin semiquinone (SQ) species; the slower changes are due to the further reduction of the SQ (22). Reoxidation of the enzyme was not observed after the addition of m-type Trx from Gloeobacter (GvTrx-m) under our conditions (Fig. 2C). This behavior contrasted with Escherichia coli NTR (EcNTR) and GvDTR, whose flavins were reduced by DTH without the accumulation of SQ intermediates in a reaction quickly reversed by Trx addition (17, 20). These results provided evidence that the GvDDOR flavoprotein is not a TR.
The DDOR Flavin Cofactor Is Reduced by Small Thiols.
During the process of protein purification it was observed that GvDDOR and SynDDOR proteins showed a yellow color, typical of oxidized flavin, that changed to a brownish color when low concentrations of 2-mercaptoethanol (2-ME) were added to the buffer; this effect was reversed after a few hours in air (Fig. 2D). The UV-visible spectra clearly showed a reduction of the flavin cofactor of DDOR primarily to the blue SQ with 2-ME (Fig. 2D); interestingly, the peak at 390 nm suggests the formation of some sulfur adduct to the flavin at the C4a position (23). The three thiol reductants 2-ME, GSH, and DTT formed different amounts of C4a adduct (Fig. 2E); DTT, having the lowest redox potential, resulted in the least adduct, and 2-ME, with the highest redox potential, resulted in the most adduct. These results indicate that the two-electron transfer from the exogenous thiols to FAD is mediated by a redox-active disulfide. To confirm this possibility, site-directed mutagenesis was performed to convert the C-terminal cysteine of the CxxC motif to serine in the SynDDOR enzyme (SynDDORC156S). Residues of this redox motif were chosen as the reactive sulfhydryl candidates, since no other Cys residues are conserved in the sequence in members of this protein family (Fig. 1). The chemical state of the isoalloxazine moiety of the flavin cofactor in the SynDDORC156S mutated protein proved to be unaffected by thiol compounds (Fig. 2F), indicating that the CxxC active site is in direct two-electron redox communication with the flavin.
Crystal Structures of DDORs Show Two FAD Cofactors Per Monomer.
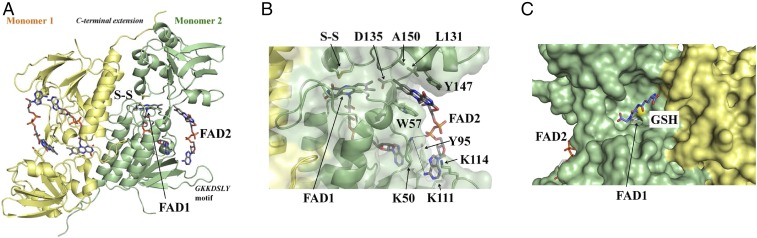
To further characterize DDOR, we crystallized and obtained high-resolution structures of the enzyme using X-ray crystallography. The structures of the SynDDOR and GvDDOR proteins were determined at 2.0- and 2.2-Å resolution, respectively (Table S1). Due to the high structural similarity between the two proteins (Fig. S3), we refer to SynDDOR in the next sections, unless otherwise stated. Similar to NTRs, the DDORs are homodimers (Fig. 3A) that displayed essentially identical structures. Each displayed two Rossmann fold domains per monomer (24) analogous to the FAD- and NADP-binding domains in NTRs (Fig. S4). The redox-active CxxC motif of the pseudoNADP-binding domain in SynDDOR forms a disulfide bridge (Fig. 3A). The relative orientation of the two domains in DDORs bring the C-terminal Cys of the CxxC redox motif close to the re face of the FAD isoalloxazine ring (FAD1), adjacent to the flavin C4a atom. This conformation is equivalent to the flavin-oxidizing (FO) conformation described in NTRs (Fig. 4A and Fig. S4B) (25).
Fig. 3.
Crystal structure of DDOR. (A) Ribbon drawing of SynDDOR homodimers. Each monomer is represented in yellow or green, respectively. Flavins and the redox-active Cys amino acids that were found to form a disulfide bridge in the crystal are depicted in stick representation. Unique features of DDOR structure include a C-terminal extension in monomers that contribute to the dimer interface and a GKKDSLY motif (which corresponds to the SynDDOR protein sequence as shown in Fig. 1, box 1) located between two β-strands in the FAD-binding domain that formed an extension loop. (B) Stabilization of FAD2 by protein residues, shown in stick representation, in monomer 2. Two aromatic residues (W57 and Y147) that stack over FAD2 and form π-stackings are represented. Salt bridges are shown as dashed lines. The disulfide and the two FAD cofactors are in stick models. Protein monomers are represented as ribbons and molecular surfaces. (C) Structural evidence of GSH bound to DDOR determined from crystals of GvDDOR soaked in GSH followed by cryo-trapping. Flavin cofactors are shown in stick form in light gray. Protein monomers are represented as surfaces, and redox-active Cys is shown as yellow spacefill.
Fig. 4.
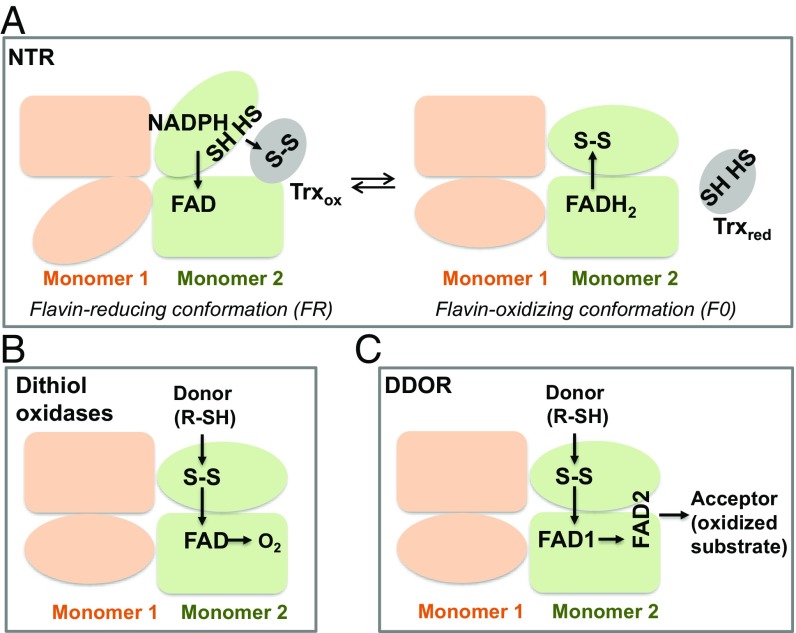
Schematic representation of the structures and functions of (A) NTR, (B) dithiol oxidases, and (C) DDOR. The three enzymes are homodimers; each monomer is represented in orange or green. For simplicity, the working model for only one monomer is shown in each case. (A) In NTRs, the catalytic mechanism involves a major conformational change within one domain in a monomer in which reducing equivalents are transferred first from NADPH to FAD (FR conformation) and then from reduced FAD to the redox-active cysteine residues in NTR (FO conformation). Subsequently, reducing equivalents are transferred to Trx. (B) Dithiol oxidases with an NTR-fold catalyze the formation of disulfide bridges in small molecules via a redox-active CxxC motif and a FAD cofactor that uses oxygen as final electron acceptor. (C) DDOR uses a previously unreported strategy of juxtaposed redox centers: two FADs and one redox-active disulfide within a monomer. The reaction mechanism would involve an initial attack by a sulfhydryl group of the substrate on the redox-active disulfide in DDOR. The reducing equivalents would be transferred to the vicinal FAD1 that would reduce FAD2, which in turn would transfer the reducing equivalents to an external acceptor substrate.
Coordination of the FAD1 cofactor in the FAD-binding domain of the DDORs superimposes well to the binding of oxidized FAD in NTRs. In comparing the DDOR and NTR structures, a distinguishing feature that emerged was an insert of amino acids (the GKKDSLY motif in the Synechocystis protein sequence) (Fig. 1, box 3) in the FAD binding-domain of DDOR. These amino acids form an extended loop in the putative Trx-binding region of NTRs (Fig. S4A) (18). Further, a C-terminal extension in DDORs not present in NTRs established tight contacts with the pseudoNADP-binding domain of the opposite monomer (Figs. S4A and S5A). This interaction results in a different orientation of the pseudoNADP-binding domain in DDOR compared with NTR that perturbs the region of the external crevice, impeding the binding of the pyridine cofactor (Figs. S4 and S6).
Perhaps the most striking feature in DDOR was the appearance of a clear, interpretable electron density corresponding to a second FAD cofactor (FAD2)—not predictable from sequence—trapped in a monomer at the interface of the two domains (Fig. 3A and Fig. S7A). The binding of FAD2 is stabilized by the side-chains of conserved amino acids from both domains of a monomer (Fig. 3B and Fig. S5B); FAD2 is not directly involved in crystal-packing contacts in any of the four crystal structures obtained in this study, indicating that it is not a crystallographic artifact. The presence of FAD2 in DDOR is not an artifact of protein manipulation, since the protein was subjected to gel filtration and dialysis before crystallization. Moreover, the results were reproduced with a protein sample not reconstituted with exogenous FAD (SynDDOR in Table S1). Of particular note, the aromatic isoalloxazine of FAD2 was sandwiched between the indole ring of a tryptophan and the phenolic ring of a tyrosine that lock it into place (Fig. 3B). The two key aromatic residues for FAD2 binding occur in all sequences of the DDOR protein family (Fig. 1, boxes 2 and 4) but not in NTRs or related sequences, suggesting a functional role for the binding site. The distances between the isoalloxazine ring and the aromatic rings are ∼3.2 and 3.4 Å, respectively, resulting in π–π stacking interactions that likely contribute to the stability of FAD2 binding. In our structure, the C7 and C8 methyl groups of the flavin isoalloxazine rings of FAD1 and FAD2 are juxtaposed (less than 4 Å apart) and make an angle of about 150°, forming an almost continuous structure that suggests a direct interflavin redox transfer that is independent of amino acids in the protein (Fig. 3B). To our knowledge, such an arrangement of two flavin cofactors within the same polypeptide chain has not been previously observed in this protein fold and is unique to DDOR.
On a final note, the presence of the C-terminal extension together with the simultaneous binding of FAD2 by amino acids of the two Rossmann fold domains within a monomer indicates that the conformational flexibility of the enzyme is severely restricted. This rigidity precludes the rotation of the pseudoNADP-binding domain with respect to the FAD-binding domain, which would be instrumental in exposing the reduced CxxC redox-active site for the interaction observed with oxidized Trx in NTRs (Fig. 4A).
DDORs Displayed Structural but Not Functional Similarities with Thiol Oxidase Flavoproteins.
A search for structural homologs using the DALI server (26) retrieved proteins with the Protein Data Bank (PDB) ID codes 4fk1, 4ntd, and 4jnA as the most similar to SynDDOR (Z-scores of 33.7, 32.6, and 32.5 and rmsds of 2.8, 3.6, and 2.6 Å, respectively); comparisons restricted to the pseudoNADP-binding domain resulted in even better structural alignments (DALI Z-scores of 20.5, 17.0, and 16.4 and rmsds of 1.2, 1.9, and 1.6 Å, respectively). The PDB code 4fk1 corresponds to the TR2 protein of Bacillus anthracis with unknown function (27); a structural comparison using DALI server (26) showed that 4fk1 is a homolog to 4ntd. Proteins with 4ntd and 4jnA PDB ID codes relate to HlmI and DepH proteins, respectively, two NAD(P)H-independent flavin dithiol oxidases with an NTR-fold that catalyze the formation of disulfide bridges in small molecules via a redox-active CxxC motif and a FAD cofactor. Both use oxygen as final electron acceptor (Fig. 4B) (28–30). Hints of the molecular basis of the substrate binding were obtained for DepH cocrystallized with a substrate analog (28); in another study, the HlmI structure contained a GSH molecule in the substrate pocket (30). Analysis of the SynDDOR structure revealed a cavity in the vicinity of the CxxC motif similar to the substrate-binding pocket observed in DepH bound to a substrate analog (dm-FK228). Therefore, we undertook an approach in which SynDDOR and GvDDOR crystals were incubated with reducing thiols GSH, DTT, and 2-ME. Soaking the yellow DDOR crystals resulted in a rapid bleaching of the color, suggestive of FAD reduction. GSH-reduced GvDDOR protein crystals were stable and suitable for X-ray diffraction data collection, and the structure was refined against data to 2-Å resolution (GvDDOR:GSH complex in Table S1). The structure of GvDDOR:GSH complex was essentially the same as the GvDDOR structure (rmsd of 0.47 Å), except for the appearance of a poorly defined density blob near the C-terminal residue of the CxxC motif that was interpreted as a GSH molecule (Fig. 3C and Fig. S7B).
Together, our structural studies point to a strong similarity between DDOR and dithiol oxidases. However, in contrast to DepH, HlmI, and other dithiol oxidases (30), DDORs are diflavin proteins. As discussed above, stable blue neutral SQ intermediates and C4a adducts in DDOR were formed after treatment of the purified protein with different thiol compounds in a reaction mediated by the CxxC motif. Because flavin reduction was reversed after a few hours of exposure to air (Fig. 2D), we conducted an assay to determine if oxygen could serve as an electron acceptor for DDOR (31). The addition of either DTT or GSH to DDOR did not cause a detectable increase of oxygen consumption relative to control samples (Fig. S2B), suggesting that DDOR reacts poorly with oxygen.
DDOR Is a Peripheral Membrane Protein.
As a first step toward determining the function of DDOR, we explored its intracellular localization in two Synechocystis strains that expressed the protein fused to a triple-HA epitope tag. Extracts prepared from cells expressing the tagged construct were fractionated to separate soluble and membrane proteins. Using immunoblotting to monitor, we found that SynDDOR was localized in the membrane fraction. However, the protein could be solubilized by mild detergent treatment at high ionic strength, consistent with an extrinsic membrane association (fraction II in Fig. S2C). We concluded that SynDDOR is a peripheral membrane protein, suggesting an activity that relates to a membrane-associated process.
Discussion
Recently, a number of NTR-related flavoproteins have been found to reduce Trx in a pyridine nucleotide-independent manner in both bacteria and archaea (17, 32, 33). The findings support the view that cells have developed multiple ways to modulate Trx-linked processes during evolution. Further, a group of thiol/disulfide-linked flavoproteins catalyzing a diverse set of chemical reactions has been identified that shows the same fold as NTRs but is independent of Trx (28–30, 34). The group includes enzymes that use molecular oxygen for the oxidation of dithiols to disulfides in natural products with antibiotic, antitumor, and virulence factor activities (30, 34). Dithiol oxidases have a modified substrate-binding pocket for pyridine nucleotides and do not require a conformational shift for the transfer of reducing equivalents to the substrate during the catalytic cycle, in contrast to NTRs (18). Thus, although the catalytic roles of the redox cofactors (CxxC motif and FAD) are well conserved in NTRs and dithiol oxidases, the enzymes have evolved modified essential structural elements and divergent functions. The present work expands the group of NTR-related flavoproteins with thiol-dependent activity to include DDOR, a previously unrecognized diflavin disulfide oxidoreductase family.
By sequence comparisons we have identified DDOR in a large, ecologically and physiologically diverse group of oxygenic phototrophic bacteria, including free marine and freshwater cyanobacteria. The DDOR-encoding gene is present in the genome of the primordial cyanobacteria Gloeobacter, which lack thylakoids, suggesting that the gene was present in the ancestor of cyanobacteria. The retention of the gene by a diverse group of cyanobacteria highlights its functional significance in these organisms. Surprisingly, DDOR function has been related to Grxs in Synechocystis, although contradictory results were reported and further studies are necessary (35, 36). Here, slr0600 gene-disruption experiments have shown that the absence of the enzyme resulted in cells with unusual sensitivity to oxidative stress and high light (37). It seems likely, therefore, that DDOR plays a role in controlling cellular redox status. A full understanding of DDOR function awaits the identification of its redox partner(s).
Unexpectedly, three redox groups were found per DDOR monomer, including a CxxC active site in redox communication with two FAD molecules (FAD1 and FAD2). Remarkably, the isoalloxazine ring of FAD2 was found at a distance and relative orientation favorable to interflavin redox transfer with FAD1 in the same monomer. This finding is consistent with the results showing communication between exogenous thiols and oxidized flavins via the disulfide bridge and equilibrium of redox states between the two flavin cofactors in the DDOR enzyme. Moreover, FAD2 was found in a geometry that could provide the direct transfer of reducing equivalents from the aromatic system to the acceptor. Although flavin reduction was slowly reversed in air in vitro, molecular oxygen was a poor acceptor of electrons.
Collectively, these data allowed us to formulate a working model for DDOR activity (Fig. 4C). The reaction mechanism would involve the initial attack by a sulfhydryl group of the substrate to the redox-active disulfide adjacent to the re-face of FAD1 to form a mixed disulfide and a thiolate; the newly formed thiolate forms an adduct with the vicinal FAD1. The reduction of FAD1 is complete when a second sulfhydryl group attacks the mixed disulfide bond and the enzymatic redox-active cysteines are oxidized, reforming the disulfide bond at the active site of DDOR. Subsequently, FAD1 would reduce FAD2, which, in turn, would transfer reducing equivalents to an external acceptor substrate. Our model assumes that, during the catalytic cycle, FAD2 would be in permanent or transient contact with the acceptor redox partner in a reaction that would not involve major domain reorganization in DDOR. The ample diversity of potential physiological donors and acceptors makes it difficult to assign a particular candidate based on present evidence. Physiologically, the donor of reducing equivalents could be a small thiol substrate that is properly accommodated in the cavity near the CxxC motif at the donor-binding site. Possible donors could include a redox protein (or peptide) or a small molecule (such as GSH, reduced Cys, or a metabolite). Our results are consistent with GSH or a surrogate serving as the physiological donor. However, further experiments are required to corroborate this possibility. On the other hand, the crystal structures show a solvent-exposed acceptor-binding site where a variety of one or two electron-carrying molecules (such as quinones) or cofactors in proteins (for example, heme, Fe–S clusters, or flavins) could serve as efficient acceptors of the reducing equivalents.
Fractionation experiments have indicated that the DDOR enzyme is associated with the bacterial membrane where processes such as photosynthesis, respiration, and Fe–S cluster assembly take place. It thus seems possible that DDOR activity is linked to membrane-bound cofactors or redox proteins that are part of a multicomponent system. Whether flavins in DDOR would mediate transfer of redox equivalents to or from the membrane and connect the oxidation of sulfhydryls to the reduction of a membranous substrate such as a quinone or a metallocenter remains to be investigated.
In summary, we have solved high-resolution structures of a type of flavin oxidoreductase, DDOR, that uses an unprecedented strategy of juxtaposed redox centers—two FADs and one redox-active disulfide in a single polypeptide chain. Our understanding of the mechanism by which DDOR functions is incomplete, and identification of the physiological substrates for this family of enzymes represents an important issue that deserves further investigation. The present work attests to the richness of the structural and functional diversity of flavin-dependent NTR-related proteins and further illustrates the extent to which evolution has experimented with flavins as enzyme cofactors in the evolution of redox reactions. Finally, our results pave the way to explore new protein-engineering approaches in the design of redox-active proteins for a spectrum of biotechnological applications.
Materials and Methods
gll2934 (Gloeobacter violaceus) and slr0600 (Synechocystis sp. PCC6803) ORFs were inserted into the pET28a expression vector (Novagen). Recombinant proteins were produced in the Rosetta(DE3) E. coli cell strain and were purified from the soluble fraction using Ni2+ HiPrep (GE Healthcare) and gel-filtration HiPrep 16/60 Sephacryl S300 (GE Healthcare) chromatography. Enzymatic assays were performed essentially as described in ref. 17. Protein crystals were grown at room temperature by the vapor-diffusion method. Diffraction data were collected using synchrotron radiation at the i03 (Diamond Light Source) and XALOC (38) (ALBA Synchrotron) beamlines. Synechocystis sp. PCC 6803 cells were transformed with a plasmid that contained the slr0600 gene fused to a HA tag. Cells were grown photoautotrophically. Additional information on materials and methods can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Charles H. Williams of the University of Michigan for a sample of AADP and editing expertise; Drs. Luis Rubio and Emilio Jiménez for help and guidance with the DTH experiments; Elena Andrés Galván for excellent technical help; and the beamline staff at the Diamond Light Source and ALBA Synchrotron for their assistance in data collection. This work was supported by Spanish Ministerio de Economía, Industria y Competitividad Grants BFU2016-80343-P and BIO2016-75634-P. R.M.B. is supported by a Ramón y Cajal contract from the Spanish Ministerio de Economía, Industria y Competitividad. L.L.-M. is supported by a postdoctoral contract from Universidad de Sevilla. The research leading to these results received funding from the European Community’s Seventh Framework Program (FP7/2007–2013) under BioStruct-X Grant Agreement 7687.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors reported in this paper have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 5JRI, 5K0A, 5ODE, and 5N0J).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1713698114/-/DCSupplemental.
References
- 1.Green J, Paget MS. Bacterial redox sensors. Nat Rev Microbiol. 2004;2:954–966. doi: 10.1038/nrmicro1022. [DOI] [PubMed] [Google Scholar]
- 2.Klomsiri C, Karplus PA, Poole LB. Cysteine-based redox switches in enzymes. Antioxid Redox Signal. 2011;14:1065–1077. doi: 10.1089/ars.2010.3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dalle-Donne I, Rossi R, Colombo G, Giustarini D, Milzani A. Protein S-glutathionylation: A regulatory device from bacteria to humans. Trends Biochem Sci. 2009;34:85–96. doi: 10.1016/j.tibs.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 4.Brandes N, Schmitt S, Jakob U. Thiol-based redox switches in eukaryotic proteins. Antioxid Redox Signal. 2009;11:997–1014. doi: 10.1089/ars.2008.2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schafer FQ, Buettner GR. Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radic Biol Med. 2001;30:1191–1212. doi: 10.1016/s0891-5849(01)00480-4. [DOI] [PubMed] [Google Scholar]
- 6.Holmgren A. Thioredoxin and glutaredoxin systems. J Biol Chem. 1989;264:13963–13966. [PubMed] [Google Scholar]
- 7.Grek CL, Zhang J, Manevich Y, Townsend DM, Tew KD. Causes and consequences of cysteine S-glutathionylation. J Biol Chem. 2013;288:26497–26504. doi: 10.1074/jbc.R113.461368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rouhier N, Couturier J, Johnson MK, Jacquot J-P. Glutaredoxins: Roles in iron homeostasis. Trends Biochem Sci. 2010;35:43–52. doi: 10.1016/j.tibs.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee S, Kim SM, Lee RT. Thioredoxin and thioredoxin target proteins: From molecular mechanisms to functional significance. Antioxid Redox Signal. 2013;18:1165–1207. doi: 10.1089/ars.2011.4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balsera M, Uberegui E, Schürmann P, Buchanan BB. Evolutionary development of redox regulation in chloroplasts. Antioxid Redox Signal. 2014;21:1327–1355. doi: 10.1089/ars.2013.5817. [DOI] [PubMed] [Google Scholar]
- 11.Schürmann P, Buchanan BB. The ferredoxin/thioredoxin system of oxygenic photosynthesis. Antioxid Redox Signal. 2008;10:1235–1274. doi: 10.1089/ars.2007.1931. [DOI] [PubMed] [Google Scholar]
- 12.Jacquot J-P, Eklund H, Rouhier N, Schürmann P. Structural and evolutionary aspects of thioredoxin reductases in photosynthetic organisms. Trends Plant Sci. 2009;14:336–343. doi: 10.1016/j.tplants.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 13.Dai S, et al. Structural snapshots along the reaction pathway of ferredoxin-thioredoxin reductase. Nature. 2007;448:92–96. doi: 10.1038/nature05937. [DOI] [PubMed] [Google Scholar]
- 14.Waksman G, Krishna TSR, Williams CH, Jr, Kuriyan J. Crystal structure of Escherichia coli thioredoxin reductase refined at 2 A resolution. Implications for a large conformational change during catalysis. J Mol Biol. 1994;236:800–816. [PubMed] [Google Scholar]
- 15.Serrato AJ, Pérez-Ruiz JM, Spínola MC, Cejudo FJ. A novel NADPH thioredoxin reductase, localized in the chloroplast, which deficiency causes hypersensitivity to abiotic stress in Arabidopsis thaliana. J Biol Chem. 2004;279:43821–43827. doi: 10.1074/jbc.M404696200. [DOI] [PubMed] [Google Scholar]
- 16.Pérez-Ruiz JM, et al. Rice NTRC is a high-efficiency redox system for chloroplast protection against oxidative damage. Plant Cell. 2006;18:2356–2368. doi: 10.1105/tpc.106.041541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buey RM, et al. A new member of the thioredoxin reductase family from early oxygenic photosynthetic organisms. Mol Plant. 2017;10:212–215. doi: 10.1016/j.molp.2016.06.019. [DOI] [PubMed] [Google Scholar]
- 18.Lennon BW, Williams CH, Jr, Ludwig ML. Twists in catalysis: Alternating conformations of Escherichia coli thioredoxin reductase. Science. 2000;289:1190–1194. doi: 10.1126/science.289.5482.1190. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Z, et al. Crystal structure of Saccharomyces cerevisiae cytoplasmic thioredoxin reductase Trr1 reveals the structural basis for species-specific recognition of thioredoxin. Biochim Biophys Acta. 2009;1794:124–128. doi: 10.1016/j.bbapap.2008.09.011. [DOI] [PubMed] [Google Scholar]
- 20.Prongay AJ, Williams CH., Jr Oxidation-reduction properties of Escherichia coli thioredoxin reductase altered at each active site cysteine residue. J Biol Chem. 1992;267:25181–25188. [PubMed] [Google Scholar]
- 21.Holmgren A, Bjornstedt M. Thioredoxin and thioredoxin reductase. Methods Enzymol. 1995;252:199–208. doi: 10.1016/0076-6879(95)52023-6. [DOI] [PubMed] [Google Scholar]
- 22.Zanetti G, Williams CH, Jr, Massey V. Influence of photoirradiation on the oxidation-reduction state of thioredoxin reductase. J Biol Chem. 1968;243:4013–4019. [PubMed] [Google Scholar]
- 23.Thorpe C, Williams CH., Jr Spectral evidence for a flavin adduct in a monoalkylated derivative of pig heart lipoamide dehydrogenase. J Biol Chem. 1976;251:7726–7728. [PubMed] [Google Scholar]
- 24.Kuriyan J, et al. Convergent evolution of similar function in two structurally divergent enzymes. Nature. 1991;352:172–174. doi: 10.1038/352172a0. [DOI] [PubMed] [Google Scholar]
- 25.Mulrooney SB, Williams CH., Jr Evidence for two conformational states of thioredoxin reductase from Escherichia coli: Use of intrinsic and extrinsic quenchers of flavin fluorescence as probes to observe domain rotation. Protein Sci. 1997;6:2188–2195. doi: 10.1002/pro.5560061013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holm L, Sander C. The FSSP database: Fold classification based on structure-structure alignment of proteins. Nucleic Acids Res. 1996;24:206–209. doi: 10.1093/nar/24.1.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gustafsson TN, Sahlin M, Lu J, Sjöberg B-M, Holmgren A. Bacillus anthracis thioredoxin systems, characterization and role as electron donors for ribonucleotide reductase. J Biol Chem. 2012;287:39686–39697. doi: 10.1074/jbc.M112.413427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li J, Wang C, Zhang Z-M, Cheng Y-Q, Zhou J. The structural basis of an NADP+-independent dithiol oxidase in FK228 biosynthesis. Sci Rep. 2014;4:4145. doi: 10.1038/srep04145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li B, Walsh CT. Streptomyces clavuligerus HlmI is an intramolecular disulfide-forming dithiol oxidase in holomycin biosynthesis. Biochemistry. 2011;50:4615–4622. doi: 10.1021/bi200321c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scharf DH, et al. Flavoenzyme-catalyzed formation of disulfide bonds in natural products. Angew Chem Int Ed Engl. 2014;53:2221–2224. doi: 10.1002/anie.201309302. [DOI] [PubMed] [Google Scholar]
- 31.Mattevi A. To be or not to be an oxidase: Challenging the oxygen reactivity of flavoenzymes. Trends Biochem Sci. 2006;31:276–283. doi: 10.1016/j.tibs.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 32.Hernandez HH, Jaquez OA, Hamill MJ, Elliott SJ, Drennan CL. Thioredoxin reductase from Thermoplasma acidophilum: A new twist on redox regulation. Biochemistry. 2008;47:9728–9737. doi: 10.1021/bi8006753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Susanti D, Loganathan U, Mukhopadhyay B. A novel F420-dependent thioredoxin reductase gated by low potential FAD: A tool for redox regulation in an anaerobe. J Biol Chem. 2016;291:23084–23100. doi: 10.1074/jbc.M116.750208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang C, Wesener SR, Zhang H, Cheng Y-Q. An FAD-dependent pyridine nucleotide-disulfide oxidoreductase is involved in disulfide bond formation in FK228 anticancer depsipeptide. Chem Biol. 2009;16:585–593. doi: 10.1016/j.chembiol.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 35.Li M, et al. Identification of novel targets of cyanobacterial glutaredoxin. Arch Biochem Biophys. 2007;458:220–228. doi: 10.1016/j.abb.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 36.Marteyn B, Domain F, Legrain P, Chauvat F, Cassier-Chauvat C. The thioredoxin reductase-glutaredoxins-ferredoxin crossroad pathway for selenate tolerance in Synechocystis PCC6803. Mol Microbiol. 2009;71:520–532. doi: 10.1111/j.1365-2958.2008.06550.x. [DOI] [PubMed] [Google Scholar]
- 37.Hishiya S, et al. Binary reducing equivalent pathways using NADPH-thioredoxin reductase and ferredoxin-thioredoxin reductase in the cyanobacterium Synechocystis sp. strain PCC 6803. Plant Cell Physiol. 2008;49:11–18. doi: 10.1093/pcp/pcm158. [DOI] [PubMed] [Google Scholar]
- 38.Juanhuix J, et al. Developments in optics and performance at BL13-XALOC, the macromolecular crystallography beamline at the ALBA synchrotron. J Synchrotron Radiat. 2014;21:679–689. doi: 10.1107/S160057751400825X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li J, et al. Site-directed mutagenesis by combination of homologous recombination and DpnI digestion of the plasmid template in Escherichia coli. Anal Biochem. 2008;373:389–391. doi: 10.1016/j.ab.2007.10.034. [DOI] [PubMed] [Google Scholar]
- 41.Budisa N, et al. High-level biosynthetic substitution of methionine in proteins by its analogs 2-aminohexanoic acid, selenomethionine, telluromethionine and ethionine in Escherichia coli. Eur J Biochem. 1995;230:788–796. doi: 10.1111/j.1432-1033.1995.tb20622.x. [DOI] [PubMed] [Google Scholar]
- 42.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 43.Sheldrick GM. Experimental phasing with SHELXC/D/E: Combining chain tracing with density modification. Acta Crystallogr D Biol Crystallogr. 2010;66:479–485. doi: 10.1107/S0907444909038360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pape T, Schneider TR. HKL2MAP: A graphical user interface for macromolecular phasing with SHELX programs. J Appl Cryst. 2004;37:843–844. [Google Scholar]
- 45.Vonrhein C, et al. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr D Biol Crystallogr. 2011;67:293–302. doi: 10.1107/S0907444911007773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tickle IJ, et al. STARANIS. Global Phasing Ltd.; Cambridge, UK: 2016. [Google Scholar]
- 47.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 51.Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 53.Rippka R, Deruelles J, Waterbury JB, Herman M, Stanier RY. Generic assigment, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol. 1979;111:1–61. [Google Scholar]
- 54.Ferino F, Chauvat F. A promoter-probe vector-host system for the cyanobacterium, Synechocystis PCC6803. Gene. 1989;84:257–266. doi: 10.1016/0378-1119(89)90499-x. [DOI] [PubMed] [Google Scholar]
- 55.Omata T, Ohmori M, Arai N, Ogawa T. Genetically engineered mutant of the cyanobacterium Synechococcus PCC 7942 defective in nitrate transport. Proc Natl Acad Sci USA. 1989;86:6612–6616. doi: 10.1073/pnas.86.17.6612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mérida A, Leurentop L, Candau P, Florencio FJ. Purification and properties of glutamine synthetases from the cyanobacteria Synechocystis sp. strain PCC 6803 and Calothrix sp. strain PCC 7601. J Bacteriol. 1990;172:4732–4735. doi: 10.1128/jb.172.8.4732-4735.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Akif M, Suhre K, Verma C, Mande SC. Conformational flexibility of Mycobacterium tuberculosis thioredoxin reductase: crystal structure and normal-mode analysis. Acta Crystallogr D Biol Crystallogr. 2005;61:1603–1611. doi: 10.1107/S0907444905030519. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.