Significance
The recent advances in cancer immunotherapy motivated us to investigate the clonal structure of the T cell receptor repertoire in breast tumors, normal breast, and blood in the same individuals. We found quantitatively distinct clonal structures in all three tissues, which enabled us to predict whether tissue is normal or tumor solely by comparing the repertoire of the tissue with blood. T cell receptor sequences shared between patients’ tumors are rare and, in general, do not appear to be specific to the cancer.
Keywords: T cell receptor, breast cancer, repertoire sequencing
Abstract
Tumor-infiltrating T cells play an important role in many cancers, and can improve prognosis and yield therapeutic targets. We characterized T cells infiltrating both breast cancer tumors and the surrounding normal breast tissue to identify T cells specific to each, as well as their abundance in peripheral blood. Using immune profiling of the T cell beta-chain repertoire in 16 patients with early-stage breast cancer, we show that the clonal structure of the tumor is significantly different from adjacent breast tissue, with the tumor containing ∼2.5-fold greater density of T cells and higher clonality compared with normal breast. The clonal structure of T cells in blood and normal breast is more similar than between blood and tumor, and could be used to distinguish tumor from normal breast tissue in 14 of 16 patients. Many T cell sequences overlap between tissue and blood from the same patient, including ∼50% of T cells between tumor and normal breast. Both tumor and normal breast contain high-abundance “enriched” sequences that are absent or of low abundance in the other tissue. Many of these T cells are either not detected or detected with very low frequency in the blood, suggesting the existence of separate compartments of T cells in both tumor and normal breast. Enriched T cell sequences are typically unique to each patient, but a subset is shared between many different patients. We show that many of these are commonly generated sequences, and thus unlikely to play an important role in the tumor microenvironment.
The immune system is thought to play an integral role throughout the life cycle of many cancers, including preventing initiation, suppressing development, and influencing treatment and patient outcomes (1, 2). Genomic alterations in tumors create immunogenic targets that can be recognized as nonself and eliminated by cytotoxic CD8+ T cells (3). At the same time, this process imposes a selective pressure on the tumor to evade this surveillance (4, 5), sometimes hijacking immune mechanisms that can then be targets for immunotherapy (6, 7). Breast cancer is less immunogenic than other cancers (8), but tumor-infiltrating lymphocytes (TILs) have been observed in all subtypes and shown to have prognostic value in human epidermal growth factor receptor 2 (HER2)-positive breast cancer and triple-negative (estrogen receptor-negative, progesterone receptor-negative, and HER2 receptor-negative) breast cancer (recently reviewed in refs. 9–12).
High-throughput DNA sequencing of the recombined V(D)J region of the T cell receptor beta-chain (TCRB) has become a standard technique for quantifying the distribution of millions of T cells in a biological sample (13–15). One cell in 100,000 is reliably detected (16), improving clinical monitoring of pathogenic immune cells in various blood cancers (17). Large (>600 individuals) public databases of TCRB data have been generated and used to infer individual major histocompatibility complex (MHC) alleles and cytomegalovirus (CMV) exposure (18). Even though recent developments in single-cell methods can identify both alpha- and beta-chain sequences (19–21), the peptide-MHC target of each T cell is generally not known. Regardless, characterizing the T cell receptor repertoire over a range of conditions can provide insight into the subset of T cell sequences that may be relevant in a variety of clinical applications (22).
Exploratory studies of the T cell repertoire in tumors from several cancers have found differing repertoires between colorectal tumors and adjacent mucosal tissue (23), intratumoral heterogeneity in renal (24) and esophageal (25) carcinomas, spatial homogeneity in ovarian cancer (26), two subgroups of T cell repertoires in pancreatic cancer (27), increased clonality of CD4+ T cells in non-small cell lung cancer (NSCLC) compared with CD19+ B cell and CD8+ T cell compartments (28), and stereotyped shifts in the repertoire after cryoablation and immunotherapy in breast cancer (29). Large-scale genomic studies extracting T cell sequences from bulk RNA-sequencing data from The Cancer Genome Atlas (TCGA) observed a strong correlation between T cell diversity and tumor mutation load in a variety of cancer types (30). Exome data from TCGA were used to derive an immune DNA signature for breast cancer that correlated with clinical outcomes (31). Single-cell sequencing has also been applied to immune cells infiltrating breast cancer. In one study (20), a subset of CD8+ T cells with matching alpha- and beta-chains was detected in breast tumors and sentinel lymph nodes from multiple patients.
In this study we use TCRB sequencing to determine the T cell repertoire in matched samples of peripheral blood, tumor, and adjacent normal breast tissue from 16 patients with early-stage breast cancer. We characterize T cell repertoires from these tissues and show that the clonal structure of the tumor is different from normal breast and blood, and can be used, in principle, to distinguish tumor from normal breast. We identify and describe a subset of “enriched” TCRB sequences with high abundance in each tumor and absent or low abundance in normal breast. Lastly, we characterize a subset of complementarity-determining region 3 (CDR3) sequences that are shared between different patients and argue that most of these are likely to be common sequences that are unlikely to play an important role in the tumor microenvironment.
Results
Study Population.
Of 16 invasive breast cancers, 12 tumors were estrogen receptor-positive (ER+), progesterone receptor-positive (PR+), and HER2-negative (HER2−). Among these 12 tumors, eight were invasive ductal carcinomas and four were invasive lobular carcinomas, grades 1–2 with low proliferation rates (11 of 12 had Ki67 < 15%) and 1 to 3 cm in size (10 of 12 were less than 3 cm in diameter). The remaining four tumors had various receptor statuses, including an 8-cm triple-negative (ER−/PR−/HER2−) high-grade tumor with a high proliferation rate (Ki67 of 50–70%) and an ER+/PR+/HER2+ invasive mucinous carcinoma. Two patients received neoadjuvant chemotherapy before surgery, and one patient had hepatitis C infection (Table 1).
Table 1.
Clinical information for each patient
| Patient identification | Age, y | Type | Size, cm | Grade | Ki67, % | ER/PR/HER2 |
| BR01 | 66 | IDC | 1.2 | 2 | <5 | +/+/− |
| BR07 | 71 | IDC | 1.2 | 1 | 1–5 | +/+/− |
| BR13 | 58 | IDC | 1.0 | 2 | 10–15 | +/+/− |
| BR15 | 72 | IDC | 2.0 | 2 | 5–10 | +/+/− |
| BR16 | 56 | IDC | 1.8 | 2 | 5–10 | +/+/− |
| BR17* | 66 | IDC | 2.1 | 2 | 10–15 | +/+/− |
| BR18 | 49 | IDC | 2.0 | 1–2 | 21 | +/+/− |
| BR21 | 45 | IDC | 2.5 | 1 | 1–5 | +/+/− |
| BR19 | 68 | ILC | 2.4 | 1 | 5–10 | +/+/− |
| BR20 | 67 | ILC | 4.7 | 1 | 10 | +/+/− |
| BR14 | 61 | ILC | 1.9 | 1 | 5–10 | +/+/− |
| BR26† | 43 | ILC | 3.2 | 1 | 5–15 | +/+/− |
| BR22 | 36 | IMC | 2.0 | 2 | <5 | +/+/+ |
| BR05 | 54 | IDC | 2.5 | 2 | 10–15 | +/−/− |
| BR25 | 57 | IDC | 2.2 | 3 | 20–30 | −/−/+ |
| BR24† | 55 | IDC | 8.0 | 3 | 50–70 | −/−/− |
IDC, invasive ductal carcinoma; ILC, invasive lobular carcinoma; IMC, invasive mucinous carcinoma.
Indicates patient with hepatitis C infection.
Indicates patients receiving therapy before tumor removal.
Breast Tumors Contain a Larger Proportion of T Cells and a More Clonal Repertoire than Normal Breast Tissue.
V(D)J rearrangements within the TCRB locus are PCR-amplified with V- and J-gene–specific primers, and the sequence of the CDR3 was determined using high-throughput sequencing of genomic DNA isolated from peripheral blood, tumor, and normal breast tissue (Fig. 1A). The absolute abundance of input template molecules for each nucleic acid sequence, which is an estimate of the number of input T cells, is determined from synthetic spike-in control sequences (details are provided in Materials and Methods). The abundance of individual T cell clonotypes is determined by combining templates from molecules with the same TCRB V-gene family, J-gene segment, and productive (i.e., in-frame) CDR3 amino acid sequence. A total of 2,651,842 templates across 1,097,674 unique clonotypes were detected, with individual abundances ranging from one to 28,508 templates per clonotype (details are provided in SI Appendix, Table S1).
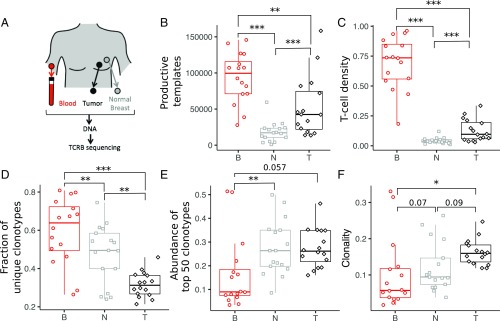
Fig. 1.
T cell abundance and repertoire diversity in blood, tumor, and adjacent normal breast tissue. (A) Matched trio of samples was obtained from blood PBMCs, tumor, and normal breast tissue for each patient. Genomic DNA was isolated from each sample, and the TCRB region was PCR-amplified before Illumina sequencing. The set of unique clonotypes, defined as TCRB sequences with the same V family, J gene, and CDR3 amino acid sequence, and the number of input template molecules for each clonotype comprise the TCRB repertoire in each sample. The number of T cells in each tissue is estimated from the abundance of in-frame TCRB template molecules (B), and the number density is determined as the fraction of templates relative to the number of input cells (C) (SI Appendix, Table S1). Repertoire diversity for each tissue is estimated from the fraction of templates corresponding to unique clonotypes (D), the cumulative abundance of the top 50 clonotypes in each sample (E), and the clonality (F). Additional details are provided in Materials and Methods. Unless otherwise indicated, *P < 0.05; **P < 0.005; ***P < 0.0005. B, blood; N, normal breast; T, tumor.
The size of the TCRB repertoire is different for each tissue. The number of productive templates represents the number of T cells in each sample, and is greatest in blood (median of 99,500 per sample), followed by tumor (median of 42,500 per sample) and normal breast tissue (median of 17,200 per sample; Fig. 1B). Normalizing the number of productive templates by the number of input cells (SI Appendix, Table S2) results in an estimate of the overall T cell density in each tissue. Tumors contain ∼2.5-fold higher density of infiltrating T cells than adjacent normal breast tissue (median of 9.6% and 3.8%, respectively; P < 0.0005; Fig. 1C), with both containing a lower density of T cells than found in circulating peripheral blood mononuclear cells (PBMCs; median of ∼74%).
The structure of the repertoires also differs between the three tissues. The number of unique clonotypes is approximately twofold larger in tumor (median of ∼12,500) compared with normal breast (median of ∼7,000; P < 0.01), with fourfold more detected in blood (median of ∼46,000) compared with tumor (SI Appendix, Fig. S1A and Table S1). The corresponding fraction of T cells with unique sequences, however, is lower in tumor (median of 0.31) than in either blood (median of 0.64; P < 0.0005) or normal breast (median of 0.49; P < 0.005; Fig. 1D). The cumulative abundance within the top 50 clonotypes is similar between tumor and normal breast (median of ∼27%), with both having approximately threefold higher abundance than blood (median of ∼9%; Fig. 1E). The clonality in the tumor (median of 0.16) is higher than in blood (median of 0.10; P < 0.05) and normal breast tissue (median of 0.12; P < 0.09; Fig. 1F). Both metrics indicate that the repertoire of T cells in the tumor is less diverse than in normal breast tissue. Recent tools to estimate total repertoire diversity (32) indicate that the differences we observe in unique clonotypes and diversity (SI Appendix, Fig. S1 B and C, respectively) are robust to the sampling depth used here. Neither T cell density nor clonality discriminates between ductal (n = 8) and lobular (n = 4) ER+/PR+/HER2− breast tumors (SI Appendix, Fig. S2). The small study size prevents further tumor subtype analysis.
Abundant Clonotypes Are Often Detected in Multiple Tissues from the Same Patient.
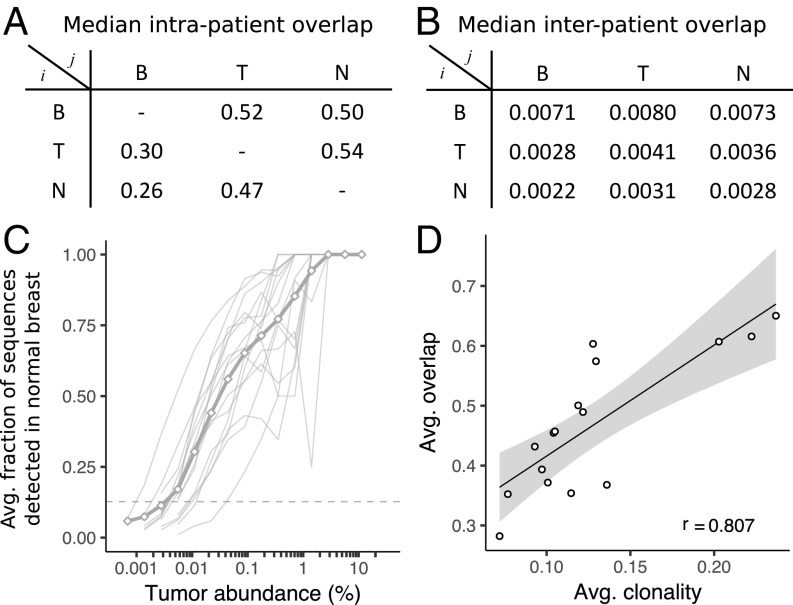
The overlap between two repertoires is defined here as the fraction of T cells in one sample with at least one identical clonotype in the other sample, and is calculated over all pairs of samples (SI Appendix, Fig. S3). Averaging over the intrapatient tissue combinations shows that approximately half of the templates in tumor and normal breast overlap with each other and with blood, whereas 25–30% of the templates in blood overlap with either tissue (Fig. 2A). Approximately 100-fold less overlap is observed between tissues from different patients (Fig. 2B). Clonotypes with large abundance in one tissue are more frequently detected in other tissues from the same patient (Fig. 2C and SI Appendix, Fig. S4). The degree of this overlap is highly variable from patient to patient, but the trends are similar within the different tissue compartments of the same patient (SI Appendix, Fig. S5). The average overlap across each patient’s tissues is strongly correlated with the average clonality (Fig. 2D), indicating that the observed patient-to-patient variability is likely a property of the underlying repertoire. This is consistent with higher clonality repertoires containing more abundant clonotypes, which have a greater probability of being detected in multiple tissues. Sequences detected in all three tissues are also biased toward high-abundance clonotypes, with the fraction of templates detected in all three tissues nearly 30% compared with less than 10% between any two tissues (SI Appendix, Fig. S6).
Fig. 2.
Overlap of TCRB templates between tissues and patients. The overlap fraction between two samples is defined as the fraction of templates in tissue j (columns) with sequences that are also detected in tissue i (rows), with the median value reported for within (A) and between (B) patient tissue combinations. (C) On average (thick gray line) and individually (thin gray lines), sequences with high abundance in the tumor are more likely to be detected in normal breast. Similar trends hold for other tissue combinations (SI Appendix, Fig. S4). (D) Overlap between samples observed for each patient is strongly correlated (linear regression with 95% confidence interval and Pearson correlation coefficient, r) with the clonality of the patient’s repertoire. Avg., Average; B, blood; N, normal breast; T, tumor.
Tumor, Normal Breast, Blood, and Contralateral Breast Clonotype Abundances in a Single Patient.
Despite the significant fraction of overlapping T cells between tissues within the same patient, some highly abundant clonotypes in one tissue are either not detected or detected at very low levels in the other tissues. Scatter plots of the abundance of each TCRB sequence between tumor and normal breast for all patients (SI Appendix, Fig. S9) indicate that such clonotypes exist over a range of abundances, with varying amounts detected in blood from each patient (SI Appendix, Figs. S10 and S11).
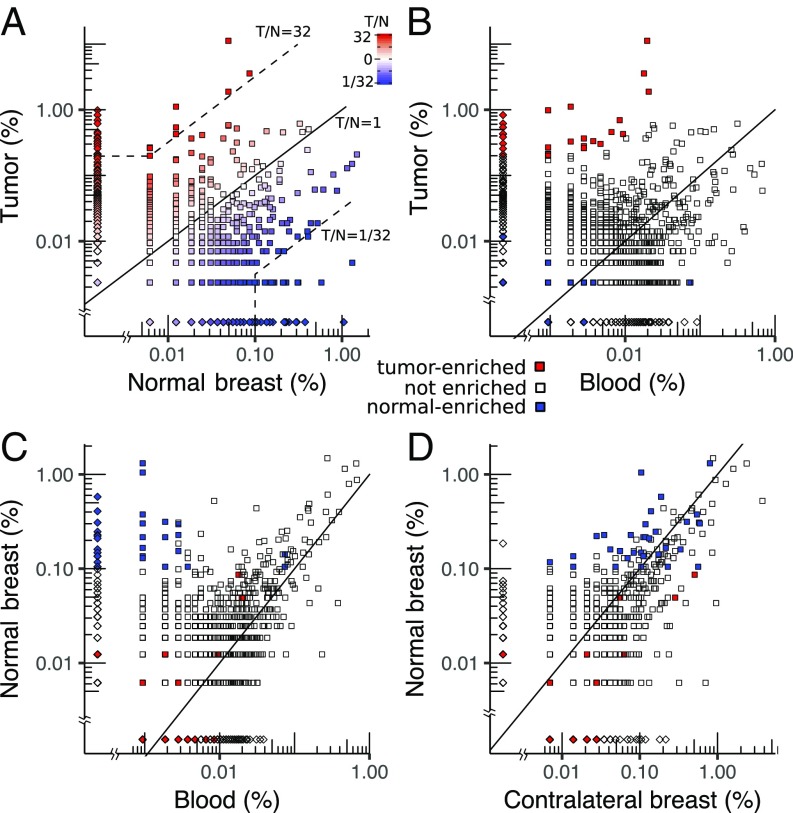
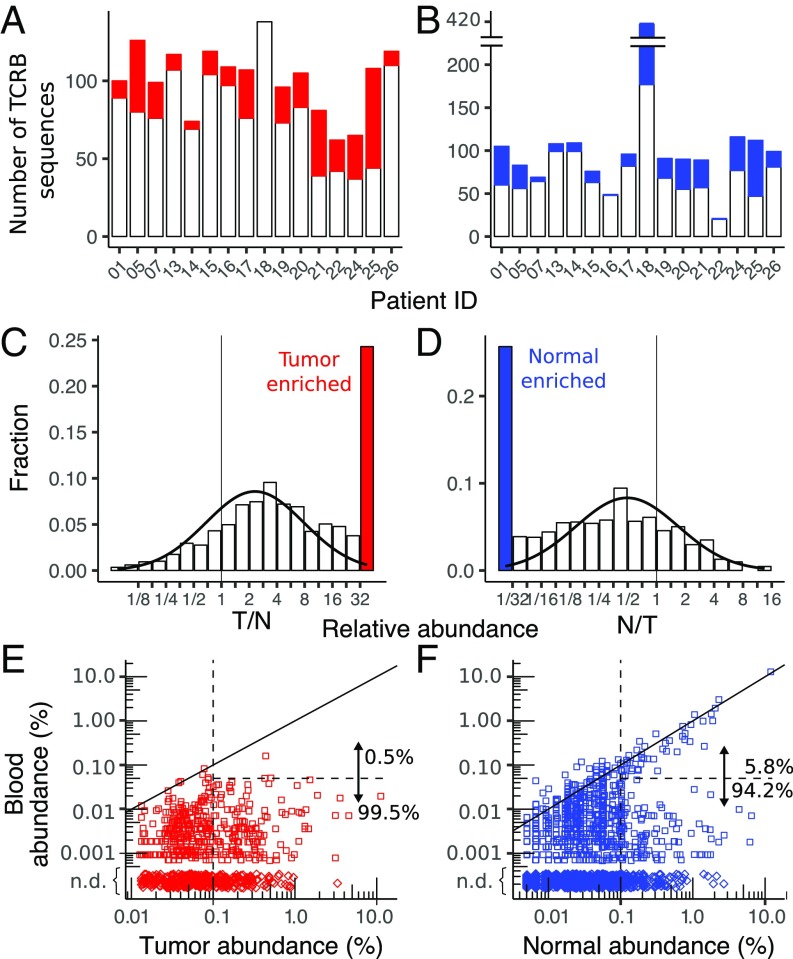
In patient BR21, for example, 1,802 sequences (comprising ∼45% of the templates) overlap between tumor and normal breast, and span a broad range of abundances from 0.002 to 11% (Fig. 3A). One hundred fifty-four clonotypes are present with abundance greater than 0.1% in either tissue, including 32 in tumor (9.5% of templates) that are missing from normal breast and 16 in normal breast (4% of templates) that are missing from tumor. To characterize this subset of “tumor-enriched” sequences, we define them to include sequences that have abundance greater than 0.1% in tumor and at least 32-fold greater abundance relative to normal breast (details are provided in Materials and Methods). In the tumor, approximately half (42 of 81) of the abundant sequences (29% of templates) are enriched, whereas in normal breast, approximately one-third (32 of 89) of abundant sequences (9% of templates) are enriched. Unlike BR21, where the top eight clonotypes in the tumor are all enriched, most patients typically have several clonotypes with high abundance in both tumor and normal breast tissue (SI Appendix, Fig. S9), suggesting that high-abundance clonotypes in the tumor are not specific to the tumor.
Fig. 3.
Scatter plots of clonotype abundance for patient BR21. (A) Tumor (T) vs. normal breast (N). (B) Tumor vs. blood. (C) Normal breast vs. blood. (D) Normal breast vs. contralateral normal breast. Each point represents a clonotype detected in one (diamonds) or both (squares) samples. Clonotypes with equal abundance in both samples lie along the diagonal (solid black line), whereas clones enriched in the tumor relative to normal breast or normal relative to tumor are defined as relative abundance greater than 32 and absolute abundance greater than 0.1% (dashed lines). Note that many sequences, especially at low abundance, contain the same number of templates in both samples but are only represented by a single point (the distribution of clonotype abundances is shown in SI Appendix, Fig. S7).
At the sequencing depth used in this study, ∼60% of all TCRB templates in tumor and normal breast have clonotypes that are detected in blood (∼24% of the templates in blood), including a similar fraction of tumor-enriched (24 of 42; Fig. 3B) and normal-enriched (17 of 32; Fig. 3C) clonotypes. Most tumor-enriched clonotypes correspond to low-abundance clonotypes in blood (median is less than 0.001%), which, together, total only 0.15–0.17% of templates.
BR21 is the only patient with a second normal sample from the contralateral breast (Fig. 3D). The top clonotypes in the two normal tissues are very similar, with 109 of 113 sequences (∼18% of templates) detected in both tissues. Unlike the case in blood, where the shared clonotypes have very low abundance, the shared clonotypes in the contralateral breast are highly correlated with those in normal breast tissue (Pearson correlation = 0.60; SI Appendix, Fig. S8A). All of the normal-enriched sequences are also detected in the contralateral breast.
Pearson correlation coefficients of the abundance from the top 100 clonotypes between each pair of tissues (SI Appendix, Figs. S10 and S11) indicate that normal breast, blood, and contralateral breast are more correlated with each other (0.45–0.6) than with the tumor (−0.13 to −0.03; SI Appendix, Fig. S8A). This trend is consistent across many of the patients, with larger correlations between normal breast and blood (median correlation of 0.40) compared with normal breast and tumor (median correlation of 0.08) or blood and tumor (median correlation of 0.01; SI Appendix, Fig. S8B).
Classifying Tumor and Normal Breast from the T Cell Repertoire.
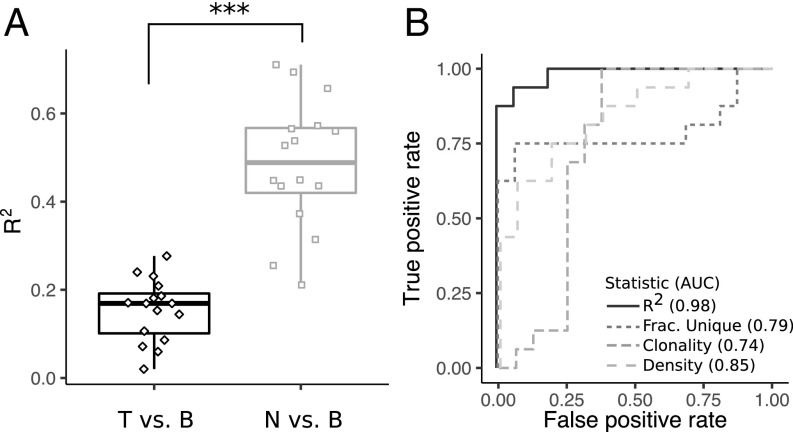
The differences in clonal structure between tumor and normal breast (Fig. 1) suggest that the T cell repertoire could be used to identify a hypothetical biopsy of uncertain tissue as either normal breast or tumor. The larger correlation between normal breast and blood compared with tumor and blood (SI Appendix, Fig. S8B) indicates that including a matched blood sample along with a tissue biopsy would further improve the performance of such an assay. We find that the fraction of the variance in the data captured by a linear model fit to the abundance of the top 100 clonotypes in blood with the corresponding abundance in either tissue [i.e., the coefficient of determination (R2) of the fit lines in SI Appendix, Figs. S10 and S11] distinguishes tumor (median of 0.17) from normal breast (median of 0.49; P < 3.5e5; Fig. 4A). Receiver operator curves comparing performance of the R2 metric with T cell density, clonality, and unique T cell fraction show that all four have reasonable performance but that the R2 metric has a peak sensitivity and specificity of ∼0.94 and a larger area under the curve (AUC) of 0.98 compared with the others (AUCs of 0.85, 0.74, and 0.79, respectively; Fig. 4B).
Fig. 4.
Distinguishing between TCRB repertoires from tumor and normal breast. (A) R2 resulting from fitting the tumor vs. blood abundance and normal breast vs. blood abundance (fit lines are shown in SI Appendix, Figs. S10 and S11). ***P < 0.0005. (B) Receiver operator curves and corresponding AUC values for metrics that distinguish repertoires between tumor and normal breast. B, blood; Frac., fraction; N, normal breast; T, tumor.
Subsets of Sequences Are Highly Enriched in Tumors and in Normal Breast Tissues.
The number of tumor-enriched sequences varies across patients from five to 64 (median of 22) sequences per sample, corresponding to 0.9–29.4% (median of 6.7%) of templates in each tumor (Fig. 5A). While the exact value of the threshold used to define enriched is somewhat arbitrary, the distribution of relative abundances indicates that the proportion of clonotypes with tumor/normal breast greater than 32 exceeds the amount expected from a log-normal distribution fit to the binned data (Fig. 5C). There also exists a similar distribution of enriched sequences in normal breast tissue where the number of normal breast-enriched sequences per sample varies between 1 and 65 (median of 18), with template abundances ranging from 0.9 to 29% (median of 6.7%; Fig. 5B). The overall number of normal breast-enriched sequences also exceeds what is expected from a log-normal fit (Fig. 3D). In contrast, the number of enriched sequences with abundance less than 0.1% in tumor and normal breast is relatively low and does not exceed the amount predicted from a log-normal fit to the distribution (SI Appendix, Fig. S12).
Fig. 5.
Enriched sequences in tumor and normal breast. The total number of sequences with abundance greater than 0.1% in each patient, including the number that are enriched in the tumor relative to normal breast (A) and in the normal breast relative to tumor (B), is shown The relatively large number of enriched sequences in normal breast from BR18 is likely due to the low-input amount of normal breast DNA in this sample (SI Appendix, Table S1), which can be seen in the small number of low-abundance (<0.1%) sequences (SI Appendix, Fig. S12). The distribution of enriched sequences across all patients for sequences in the tumor (C) and normal breast (D, not including the outlier BR18), including a curve fit to a log normal distribution (solid black line), is shown. N, normal breast; T, tumor. Scatter plots compare the enriched clonotypes in tumor (E) and normal breast (F) with their abundance in peripheral blood. Both high- and low-abundance (0.1%, vertical dashed line) clonotypes are shown, and the fractions of sequences that are highly correlated with blood (solid diagonal line) and with abundance greater than 0.05% (horizontal dashed line) are enumerated. Clonotypes detected in tumor or normal breast but not detected (n.d.) in blood (♢) are depicted with arbitrary tissue abundance.
Across all patients, ∼40% of tumor-enriched and 32% of normal breast-enriched clonotypes were detected in peripheral blood. For tumor-enriched sequences, there is no correlation between the abundances in tumor and blood, with only two of 370 tumor-enriched sequences detected above 0.05% abundance in blood (Fig. 3E). In normal breast, however, 44 of 761 (6%) enriched sequences exceed 0.05% abundance in blood and are highly correlated with the abundance in normal tissue (Fig. 3F). Thus, normal breast-enriched sequences seem to have two subsets, one with abundances that are correlated with those in blood and the other where they are not. Tumor-enriched clonotypes only contain the subset of sequences that are uncorrelated with the abundance in blood.
Shared Sequences Between Patients Are Consistent with Common Sequences.
The subset of TCRB CDR3 sequences shared across multiple samples may contain clonotypes that recognize a common shared antigen, such as a breast cancer epitope, or sequences that occur naturally across multiple people. We show below that commonly shared CDR3 sequences have similar properties in the three tissue compartments and, unlike tumor-enriched sequences, are consistent with commonly occurring CDR3 sequences in a population of healthy donors.
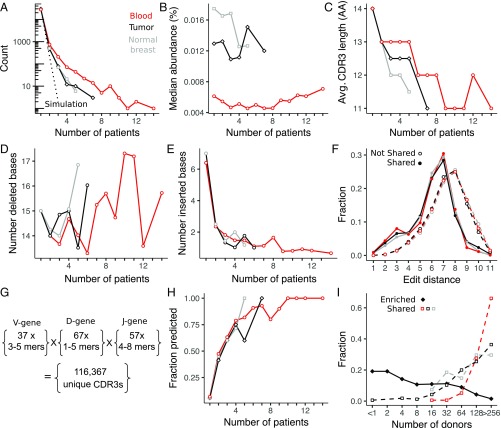
The fraction of clonotypes with shared CDR3 sequences between two patients across the different tissue combinations is well described (within ∼20%) by a standard capture-recapture model with a single-fit parameter representing the total number of productive sequences in the population (M = 2 × 106; SI Appendix, Fig. S13). To compare the degree of sharing between different tissues, we compare the overlap among the top ∼2,000 most abundant CDR3 sequences in each tissue (Materials and Methods). Using the inferred population size M, we simulated the expected fraction of sequences detected in multiple patients and show that the amount of sharing exceeds what is expected from random sampling (Fig. 6A). Approximately 2% of abundant tumor and normal breast sequences and 4% of those in blood are shared across two or more patients. In particular, the tail of the distribution is larger than expected, with 102, 13, and 6 highly shared sequences detected in more than five patients from blood, tumor, and normal breast, respectively. The capture-recapture model ignores clonotype abundance within each patient, but this assumption is consistent with the minimal bias observed in the abundance of highly shared CDR3 sequences compared with unshared sequences (Fig. 6B).
Fig. 6.
Interpatient T cell sharing of clonotypes between tissues. (A) Fraction of blood, tumor, and normal breast tissue sequences found in one (not shared) or more (shared) patients compared with the expected fraction by random sampling (dashed lines). (B) Median abundance of clonotypes detected in multiple patients is not sensitive to the degree of sharing. Compared with clonotypes that are not shared across multiple patients, shared clonotypes have a shorter CDR3 amino acid (AA) length (C), which can be attributed to a similar number of deleted (D) but fewer inserted (E), nucleotide bases in the junctional regions between V-D and D-J genes. Avg., Average. (F) Diversity of CDR3 sequences is estimated from the distribution of edit distances (length = 13 aa), which is shifted to lower values for shared sequences. Number and length of germline V-, D-, and J-gene amino acid sequence fragments used in a recombination model of low-diversity CDR3 sequences (G) and the fraction of CDR3 sequences detected by the model (H) are shown (details are provided in Materials and Methods). (I) Results from a database query of 585 healthy donors (18) indicate that shared CDR3 sequences are also likely to be shared in a population of healthy donors, whereas enriched CDR3 sequences are much less likely to be shared in the general population.
Several metrics indicate that highly shared sequences are less diverse than private sequences. First, average CDR3 length is shorter for shared sequences, decreasing from 14.5 aa in nonshared sequences to ∼11 aa in the most highly shared sequences (Fig. 6C). The number of nucleic acid bases deleted from the CDR3 region of shared sequences is relatively unchanged (Fig. 6D), but the number of inserted bases in the CDR3 region decreases sharply from approximately six nucleic acids in nonshared sequences to approximately one nucleic acid in shared sequences (Fig. 6E). Lastly, the edit distance between all pairs of randomly selected nonshared sequences with a length of 13 aa has a relatively smooth distribution that is peaked at eight amino acid bases. In contrast, the edit distance distribution between shared CDR3 sequences is shifted toward shorter lengths, with a shoulder at approximately three amino acid bases indicating a less diverse subset (Fig. 6F).
We used a simple model to compute low-diversity recombinations from all germline amino acid contributions from the V-, D-, and J-gene segments to the CDR3 region. The artificial CDR3s include amino acid deletions, but no insertions, and result in 116,367 unique sequences (Fig. 6G, further details provided in Materials and Methods). Eighty percent of these are not detected in our study, mostly due to nonuniform V- and J-gene usage in actual repertoires; 10% were detected in nonshared clonotypes, with the remaining 10% distributed across all shared CDR3s. After subsampling, the fraction of shared CDR3s predicted by the model increases rapidly with the degree of sharing, with 100% of the most frequently observed clonotypes in each tissue predicted by the model (Fig. 6H).
Lastly, shared CDR3 sequences in tumor, blood, and normal breast are almost all relatively common in a database of T cell repertoires from 585 healthy donors (18) (Fig. 6I). In contrast, tumor-enriched sequences are much less frequently observed in the database, with ∼50% of CDR3s detected in less than five donors. Both tumor-enriched and shared CDR3 sequences are similarly represented in male and female donors (SI Appendix, Fig. S15), which is expected for the commonly shared sequences but not for enriched sequences, suggesting that the most highly enriched T cells in the tumor may not be recognizing antigens specific to female breast cancer.
Discussion
In this work, we performed bulk T cell repertoire sequencing from peripheral blood, adjacent normal breast, and tumor from 16 prospectively collected patients with breast cancer, most of which were early-stage, treatment-naive, ER+/PR+/HER2− subtype tumors. Advantages of bulk repertoire sequencing include its high sensitivity for detecting T cell clonotypes (16), which may be missed by traditional staining (29), and the simplicity of genomic DNA input, which is not subjected to the variability of single-cell isolation techniques (33) required in cytometry. The disadvantage is that some important cellular details are missed, such as the heavy- and light-chain pairing and the expression of functional markers, such as CD4, CD8, FOXP3, CTLA4, and PDL1 (19, 34), unless cells have been previously sorted (15, 35). Our goal was to characterize the differences in repertoires between adjacent normal breast tissue and tumor to determine if there was evidence for subsets of T cells that might play a role in the tumor microenvironment (36).
Recent guidelines have been established for evaluating TILs in breast cancer via standard hematoxylin and eosin (H&E) staining (37), and clinical studies have shown that outcomes are correlated with the number of infiltrating T cells, especially for triple-negative and HER2+ subtypes (38). Quantifying T cells via sequencing is highly reproducible and correlates well with H&E staining but with higher sensitivity (29). Immunohistochemistry indicates that T cells in healthy breast tissue are mostly located in lobules, with CD8+ T cells ∼10-fold more abundant than CD4+ T cells (39). Immune infiltrates isolated from tumor and normal breast contain similar amounts of CD8+ T cells (∼20% of T cells), whereas CD4+ T cells in tumor comprise ∼40% of T cells compared with ∼20% in normal breast tissue (40). We show that repertoires from PBMCs, tumor, and normal breast are distinct, with large differences in the number and density of T cells detected in each tissue compartment. The tumor repertoire is less diverse than in normal breast, as indicated by the lower fraction of unique T cells and increased clonality in the tumor (Fig. 1 E and F). These results are consistent with a similar study comparing the effects of cryoablation and immunotherapy on TCRB repertoires in patients with early-stage breast cancer (29).
These repertoire metrics have predictive value and could be used to classify unknown tissue samples taken from tumor or normal breast. The performance in this small cohort varies from an AUC of 0.74 for clonality to an AUC of 0.85 for T cell density. Since repertoires from normal breast and blood are more similar than those from tumor and blood, a better metric can be determined by comparing the tissue and blood repertoires. Using the R2 goodness of fit between each tissue with blood, the AUC increases to 0.98 with only two samples not properly classified.
Extensive overlap between tissues within the same patient is expected due to normal blood perfusion. On average ∼50% of T cells in either tissue sample were found to overlap with the other tissue or blood samples from the same patient. This fraction varied from patient to patient and was correlated with the average clonality of the repertoires in each patient, which is consistent with the more clonal repertoires containing larger clonotypes that are more frequently sampled in different tissues.
In addition to many frequently overlapping clonotypes, most patient tumors contained a subset of clonotypes (median of 22) that are highly enriched in the tumor relative to normal breast. In a few patients (BR05, BR21, and BR22 in SI Appendix, Fig. S9), these clonotypes are clearly dominant in the tumor; however, in most patients, high-abundance clonotypes in the tumor are also highly abundant in the normal breast, suggesting that they are less likely to play an important role in the tumor microenvironment. Methods have been developed to identify the subset of clonotypes that change abundance in serial blood samples following an external perturbation (41). Analogous subsets are not as clear between breast tumors and normal tissue, but there is a population of highly enriched clonotypes in each tissue type across the cohort of patients (Fig. 5 C and D). We focused our analysis of enriched clonotypes on those with at least 0.1% abundance due to limited sequencing depth and multiple reports that tumor-reactive T cells identified in other cancers typically exceed 0.1%. For example, in melanoma and early-stage NCSLC, ∼3% (42) and 0.2–1.5% (43) of infiltrating CD8+ T cells are tumor-reactive, with sorting on PD1+ in melanoma further enriching the population to 1–10% (44). CD4+ T cells may also be tumor-reactive (45, 46), with 0.13–0.3% of expanded CD4+ T cells being tumor-reactive in melanoma (47).
Enriched clonotypes in normal breast tissue show two subsets in peripheral blood: one that is highly correlated with blood and the other distributed over a range of low (or zero) abundance in blood. We interpret the first subset as sequences from blood that perfuse normal tissue, whereas the second subset may represent a compartment of tissue-resident T cells in normal breast (48). The abundance distribution of tumor-enriched clonotypes in blood resembles this second subset seen in normal breast and does not contain clonotypes that are highly correlated with blood. While some of the tumor-enriched T cells may play a direct role in the tumor microenvironment, many T cells in this population may instead represent a compartment of tissue-specific resident T cells that are distinct from blood and performing standard immune surveillance (49).
Public T cells expressing the same TCR sequence but generated in different individuals have been identified for numerous infectious diseases, autoimmune disorders, and some cancers (50). In cancer, this process is thought to require T cells that recognize epitopes from mutated proteins that are presented by similar major MHC molecules (3), and thus occur more often in cancers with larger numbers of mutations (8). We detect a small fraction of TCRB CDR3s across multiple tumors, but our analysis suggests most of these are low-diversity sequences that are unlikely to be tumor-specific. First, CDR3 sequences shared across tumors follow similar trends as those shared in blood and normal breast, with a similar amount detected in normal breast as in tumor. Shared clonotypes were not strongly biased by abundance and contained shorter CDR3 lengths with many fewer inserted nucleotide bases into the junction regions, as reported in earlier studies (15, 51) and recently reviewed (52). A simple model generating all CDR3 recombinations from germline amino acid sequences with minimal insertions was able to predict many of the shared sequences, including all of the most highly shared sequences. These findings are consistent with more detailed physical models of the nucleotide sequence recombination (53) that show commonly occurring sequences have a higher likelihood of being generated.
Shared and tumor-enriched sequences are distinct, with only two CDR3s detected in both sets. We also queried a large public database of T cell repertoires in blood from 585 healthy volunteers (18). Enriched sequences were detected in many fewer healthy donors than shared sequences, supporting the hypothesis that enriched sequences may play a specialized role in a subset of patients, whereas most shared sequences are common in the population. Interestingly, tumor-enriched sequences were equally prevalent in male and female volunteers, suggesting that they may not be specific to breast cancer.
Conclusions
By comparing bulk T cell repertoires from tumor and normal breast tissue in a cohort of patients with early-stage breast cancer, we were able to determine the clonal structure of infiltrating lymphocytes. These clonal structures were quite distinct between tumor and normal breast, and we were able to distinguish tissues as normal or tumor solely on the basis of comparing T cell repertoire with blood. We identified a subset of T cells in each patient that were highly abundant in the tumor compared with normal breast; this population may represent a clinically relevant subset for future investigation. In many patients, however, these clonotypes were dominated by more abundant clonotypes, which were also highly abundant in normal breast, thus highlighting the large amount of background T cells in the tumor that complicate isolating and identifying tumor-specific T cells.
Materials and Methods
Sample Collection.
Informed consent was obtained from patients with invasive breast cancers greater than 1 cm under Stanford Institutional Review Board-approved Research Protocol 5630. Whole blood (5–10 mL) was collected in EDTA tubes before surgery and processed within 2 h. After tumor resection, tumor tissue was visually identified and a portion was excised and placed in prechilled RNAlater. Samples of adjacent normal breast tissue were obtained from sites 2–4 cm away from the tumor, rinsed with PBS, and placed in a separate tube of prechilled RNAlater. For one patient undergoing simultaneous double mastectomy, normal breast tissue was also obtained from the contralateral breast.
Sample Processing.
Plasma was removed after separation by centrifugation for 10 min at 1,600 × g. The remaining blood cells were resuspended in PBS, and mononuclear cells were isolated via Ficoll-Paque centrifugation with Sepmate-50 tubes (Stem Cell Technologies). Cells were cryopreserved in 90% FBS and 10% DMSO, divided into aliquots containing 2 to 4 million cells, and stored in liquid nitrogen until use. Cells were thawed at room temperature, diluted with PBS, pelleted at 350 RCF, and resuspended in RLTplus buffer (Qiagen) with 1% beta-mercaptoethanol (Sigma) and 0.05% Reagent DX anti-foaming agent (no. 19088; Qiagen). Lysate was passed through a QIAshredder (Qiagen), and DNA and RNA were extracted using an AllPrep DNA/RNA Mini kit (Qiagen) following the manufacturer’s instructions.
DNA Isolation.
Tumor and normal breast tissue samples were divided into pieces, incubated in RNAlater overnight, and transferred to −20 °C the following day. For the tumor, 20–50 mg of tissue was diced with a fresh razor blade and homogenized in RLTplus buffer containing 1% beta-mercaptoethanol and 0.5% reagent DX antifoaming agent using a Tissuelyzer II (Qiagen) with oscillation frequency set to 30/s for 3–9 min. Homogenized lysate was centrifuged at 20,000 RCF for 3 min, and the supernatant was removed and passed through a QIAshredder before proceeding with the Allprep Mini kit for DNA and RNA isolation. An on-column digestion with 20 μL of proteinase-K (Qiagen) dissolved in 60 μL of buffer AW1 was implemented before washing the DNA column with buffer AW2. DNA yield from tumors was variable, but, typically, 20–50 mg of tumor was required for >3 μg of DNA. Normal breast tissue was processed similarly except 100–300 mg of tissue was often required to obtain sufficient (>3 μg) DNA due to high lipid content and low density of cells. In particular, care was taken to avoid the lipid layer after the 20,000 × g centrifugation step. DNA was quantified with fluorometric quantitation (Qubit High Sensitivity DNA kit) and UV spectroscopy (Nanodrop 1000) (SI Appendix, Table S2). To minimize any contamination, samples were processed at separate times and all work was performed in a PCR cabinet (AirClean).
Immunosequencing.
TCRB sequencing was performed on genomic DNA purified from blood, tumor, and normal breast tissue using the ImmunoSeq Intro kit (Adaptive Biotechnologies) following the manufacturer’s instructions. Briefly, two genomic DNA replicates per sample, each containing either 0.5 μg (blood) or 1.5 μg (tumor, normal breast; details are provided in SI Appendix, Table S1) are independently amplified using 2× Multiplex PCR Master mix (31 cycles; no. 206151; Qiagen) with proprietary primers and spike-in control sequences for absolute quantification (13, 54). The product is cleaned up with AMPure XP magnetic beads, and amplified again with eight additional cycles of PCR to attach Illumina sequencing adapters containing sample-specific barcodes. An 87-bp fragment that includes the CDR3 region and flanking V and J genes is sequenced using single-ended 150-bp reads of seven samples (including two replicates for each sample) multiplexed onto a single lane of an Illumina MiSeq sequencer with version 3.0 chemistry. The raw sequencing data are uploaded to Adaptive Biotechnologies and processed using the company’s proprietary pipeline. Results for each sample are reported to the user after merging data from both replicates and include absolute quantification of template molecules for all nucleic acid sequences detected; CDR3 amino acid sequence for in-frame molecules; V-, D-, and J-gene segment identification; the most likely number of bases deleted from each gene segment contributing to the CDR3; and the most likely number of bases inserted into either junction. The average read coverage of each sample was ∼10-fold (details are provided in SI Appendix, Table S2).
Data Analysis.
Processed sequence data for each sample were downloaded from Adaptive Biotechnologies and analyzed with custom scripts in bash, awk, and R. Unless otherwise noted, clonotypes are defined here as productive recombinations containing a V-gene family, CDR3 amino acid sequence, and J-gene segment. Sequence abundance is calculated as the number of templates for that sequence divided by the total number of templates in the sample. After normalization via the spike-in control sequence, the assay is reported to be quantitative to one T cell in 20,000 (54). The T cell density is determined by dividing the total number of templates by the total number of input cells estimated from the input amount of DNA. Clonality represents the distribution of clone sizes and is defined as 1 minus the normalized Shannon entropy of the TCRB abundances (i.e., the Pielou evenness; SI Appendix, Table S1). Using this definition, clonality ranges from zero, where all T cells are evenly distributed across all clonotypes, to unity, where all T cells are represented by a single clonotype. P values between tissues are calculated by a paired Wilcoxon rank sum test in R unless otherwise noted, with *, **, and *** indicating P values less than 0.05, 0.005, and 0.0005, respectively. Estimates of diversity in unique clonotypes and clonality were performed using Recon analysis with default parameters (32).
Overlapping Templates.
The fraction of templates in patient k, tissue j that are shared with patient k′, tissue i is defined as the sum of the abundances of all sequences in sample (j,k) that were also detected at least once in (i,k′). As a result, and the fraction of overlapping sequences between tissues with large differences in total numbers of sequences can be represented (SI Appendix, Fig. S3). Intrapatient and interpatient averages are determined by finding the median over tissue pairs (i,j) when k = k′ and k ≠ k′, respectively (Fig. 2 A and B).
Capture-Recapture Model.
The number of overlapping sequences between tissues (i,j) is modeled as
where and are the unique sequences observed in each tissue, and M represents the total number of unique sequences in the study population and is determined by minimizing the ratio, g, of modeled and measured counts over all pairs of tissues between patients (k ≠ k′):
Results and a detailed comparison of data to the model are provided in SI Appendix, Fig. S13.
Tumor-Normal Classifier.
Receiver operator curves and the AUC were computed with the ROCR package in R using the density of T cells, fraction of unique clonotypes, and clonality (Fig. 1) as metrics to distinguish repertoires from tumor and normal tissue. A classifier based on the R2 value comparing the top 100 clones in blood with tumor and normal breast was also evaluated. Linear regression was performed between the abundances of the top 100 clonotypes in blood and the corresponding abundances in tumor, and the fraction of variance captured by the fit (R2) was tabulated for each sample (SI Appendix, Fig. S10). The same process was repeated between the top 100 clonotypes in blood with normal breast (SI Appendix, Fig. S11).
Enriched Sequences.
Sequences between pairs of tissues are plotted on log–log plots, where sequences with zero counts in one of the samples are plotted at 1/4 of the lowest abundance in that sample. Tumor sequence n is enriched if it has abundance greater than 0.1% and relative abundance in the normal breast exceeding 32 (i.e., Tn/Nn ≥ 32), including clonotypes not detected in normal breast (i.e., Nn = 0). Similarly, enriched normal sequences are defined as sequences with greater than 0.1% abundance in the normal breast and a ratio with the tumor exceeding 32 (i.e., Nn/Tn ≥ 32).
Interpatient Sequence Sharing.
The number of patients with at least one template detected for each CDR3 sequence is independently tallied for the three tissue compartments, and the sequences are binned according to the number of shared patients (SI Appendix, Fig. S14). To more easily compare sharing between samples and tissues, this process is repeated for the top 1,945 CDR3s from each patient (only BR18N has fewer sequences). The expected amount of sharing (dashed lines in Fig. 6A and SI Appendix, Fig. S14) is determined by random sampling sequences from a population of size M = 2 × 106 according to the sampling depth in each patient and tallying the number of common clonotypes across patients. The median abundance, CDR3 length, and number of inserted/deleted nucleotide bases in the junction are computed for each bin. For sequences where the D gene could not be resolved, the number of deleted bases is conservatively estimated at 10 deleted nucleotides, which is the maximum number of deleted D-gene bases reported. The edit distance between two CDR3 sequences was calculated as the number of amino acid base changes required to transform one CDR3 sequence into the other. The distribution of edit distances between all pairs of CDR3 sequences with a length of 13 aa from shared sequences (detected in more than one patient) was compared with nonshared sequences (only detected in one patient).
CDR3 Recombination Model.
Commonly shared low-diversity sequences are generated from the international ImMunoGeneTics information system (IMGT) germline amino acid sequences for the human V, D, and J genes. Amino acid sequences contributing to the CDR3 region are extracted from the 5′-end of each V gene starting with the conserved cysteine (or nearest equivalent in some V genes), the 3′-end of each J gene ending in the conserved phenylalanine, and the full D gene, including all three reading frames. All combinations of these sequences are then used to generate a list of low-diversity CDR3 sequences. To reflect our observation that shared CDR3s are shorter with many deletions and few insertions, all possible amino acid truncations of the D gene and one to two amino acid truncations from the 3′-end of the J gene are also included. No inserted amino acids are included. In general, nucleic acid sequences are ignored except when either end of the D gene or the 3′-end of the J gene contains two nucleic acids, thus strongly biasing the amino acid usage at this position. These additional amino acids are not strictly germline, but are also included. A detailed list of sequences from each gene is provided in SI Appendix, Table S3.
Database of Healthy Donors.
To query the database of 585 healthy bone marrow donors (18), a list of the top 400 shared and 858 enriched CDR3 sequences was compiled. Shared sequences include those CDR3s that are detected in at least two tumor samples, two normal breast samples, or four blood samples. These sequences were provided to Adaptive Biotechnologies, where a query on their internal database was performed, and the subset of ImmunoSeq data from each patient’s matching clonotypes was provided to us. For each TCRB sequence, the number of patients, the relative abundances, and the corresponding nucleic acid sequences were tallied for enriched/shared sequences and male/female donors.
Data Availability.
All immunosequencing data underlying this study can be analyzed and freely downloaded from the Adaptive Biotechnologies immuneACCESS site at https://clients.adaptivebiotech.com/pub/beausang-2017-pnas.
Supplementary Material
Acknowledgments
We thank Yasemin Sucu for help in processing samples, Dr. David Hamm and his team at Adaptive Biotechnologies for assistance in the database query, and Florian Rubelt for helpful discussions. This study received funding from the Howard Hughes Medical Institute, Mattias Westman, the John and Marva Warnock Research Fund, and the Debra and Andrew Rachleff Research Fund.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the immuneACCESS database, https://clients.adaptivebiotech.com/pub/beausang-2017-pnas.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1713863114/-/DCSupplemental.
References
- 1.Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD. Cancer immunoediting: From immunosurveillance to tumor escape. Nat Immunol. 2002;3:991–998. doi: 10.1038/ni1102-991. [DOI] [PubMed] [Google Scholar]
- 2.Fridman WH, Pagès F, Sautès-Fridman C, Galon J. The immune contexture in human tumours: Impact on clinical outcome. Nat Rev Cancer. 2012;12:298–306. doi: 10.1038/nrc3245. [DOI] [PubMed] [Google Scholar]
- 3.Heemskerk B, Kvistborg P, Schumacher TNM. The cancer antigenome. EMBO J. 2013;32:194–203. doi: 10.1038/emboj.2012.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blankenstein T, Coulie PG, Gilboa E, Jaffee EM. The determinants of tumour immunogenicity. Nat Rev Cancer. 2012;12:307–313. doi: 10.1038/nrc3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen DS, Mellman I. Oncology meets immunology: The cancer-immunity cycle. Immunity. 2013;39:1–10. doi: 10.1016/j.immuni.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 6.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coulie PG, Van den Eynde BJ, van der Bruggen P, Boon T. Tumour antigens recognized by T lymphocytes: At the core of cancer immunotherapy. Nat Rev Cancer. 2014;14:135–146. doi: 10.1038/nrc3670. [DOI] [PubMed] [Google Scholar]
- 8.Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science. 2015;348:69–74. doi: 10.1126/science.aaa4971. [DOI] [PubMed] [Google Scholar]
- 9.Dushyanthen S, et al. Relevance of tumor-infiltrating lymphocytes in breast cancer. BMC Med. 2015;13:202. doi: 10.1186/s12916-015-0431-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwa M, Adams S. Prognostic and predictive value of tumor-infiltrating lymphocytes in breast cancer. Curr Breast Cancer Rep. 2016;8:1–13. [Google Scholar]
- 11.Miyan M, Schmidt-Mende J, Kiessling R, Poschke I, de Boniface J. Differential tumor infiltration by T-cells characterizes intrinsic molecular subtypes in breast cancer. J Transl Med. 2016;14:227. doi: 10.1186/s12967-016-0983-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Savas P, et al. Clinical relevance of host immunity in breast cancer: From TILs to the clinic. Nat Rev Clin Oncol. 2016;13:228–241. doi: 10.1038/nrclinonc.2015.215. [DOI] [PubMed] [Google Scholar]
- 13.Robins HS, et al. Comprehensive assessment of T-cell receptor β-chain diversity in alphabeta T cells. Blood. 2009;114:4099–4107. doi: 10.1182/blood-2009-04-217604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Freeman JD, Warren RL, Webb JR, Nelson BH, Holt RA. Profiling the T-cell receptor beta-chain repertoire by massively parallel sequencing. Genome Res. 2009;19:1817–1824. doi: 10.1101/gr.092924.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robins HS, et al. Overlap and effective size of the human CD8+ T cell receptor repertoire. Sci Transl Med. 2010;2:47ra64. doi: 10.1126/scitranslmed.3001442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robins H, et al. Ultra-sensitive detection of rare T cell clones. J Immunol Methods. 2012;375:14–19. doi: 10.1016/j.jim.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu D, et al. High-throughput sequencing detects minimal residual disease in acute T lymphoblastic leukemia. Sci Transl Med. 2012;4:134ra63. doi: 10.1126/scitranslmed.3003656. [DOI] [PubMed] [Google Scholar]
- 18.Emerson RO, et al. Immunosequencing identifies signatures of cytomegalovirus exposure history and HLA-mediated effects on the T cell repertoire. Nat Genet. 2017;49:659–665. doi: 10.1038/ng.3822. [DOI] [PubMed] [Google Scholar]
- 19.Han A, Glanville J, Hansmann L, Davis MM. Linking T-cell receptor sequence to functional phenotype at the single-cell level. Nat Biotechnol. 2014;32:684–692. doi: 10.1038/nbt.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Munson DJ, et al. Identification of shared TCR sequences from T cells in human breast cancer using emulsion RT-PCR. Proc Natl Acad Sci USA. 2016;113:8272–8277. doi: 10.1073/pnas.1606994113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Howie B, et al. High-throughput pairing of T cell receptor α and β sequences. Sci Transl Med. 2015;7:301ra131. doi: 10.1126/scitranslmed.aac5624. [DOI] [PubMed] [Google Scholar]
- 22.Kirsch I, Vignali M, Robins H. T-cell receptor profiling in cancer. Mol Oncol. 2015;9:2063–2070. doi: 10.1016/j.molonc.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sherwood AM, et al. Tumor-infiltrating lymphocytes in colorectal tumors display a diversity of T cell receptor sequences that differ from the T cells in adjacent mucosal tissue. Cancer Immunol Immunother. 2013;62:1453–1461. doi: 10.1007/s00262-013-1446-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerlinger M, et al. Ultra-deep T cell receptor sequencing reveals the complexity and intratumour heterogeneity of T cell clones in renal cell carcinomas. J Pathol. 2013;231:424–432. doi: 10.1002/path.4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Z, et al. T cell receptor β-chain repertoire analysis reveals intratumour heterogeneity of tumour-infiltrating lymphocytes in oesophageal squamous cell carcinoma. J Pathol. 2016;239:450–458. doi: 10.1002/path.4742. [DOI] [PubMed] [Google Scholar]
- 26.Emerson RO, et al. High-throughput sequencing of T-cell receptors reveals a homogeneous repertoire of tumour-infiltrating lymphocytes in ovarian cancer. J Pathol. 2013;231:433–440. doi: 10.1002/path.4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bai X, et al. Characteristics of tumor infiltrating lymphocyte and circulating lymphocyte repertoires in pancreatic cancer by the sequencing of T cell receptors. Sci Rep. 2015;5:13664. doi: 10.1038/srep13664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu W, et al. A high density of tertiary lymphoid structure B cells in lung tumors is associated with increased CD4+ T cell receptor repertoire clonality. Oncoimmunology. 2015;4:e1051922. doi: 10.1080/2162402X.2015.1051922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Page DB, et al. Deep sequencing of T-cell receptor DNA as a biomarker of clonally expanded TILs in breast cancer after immunotherapy. Cancer Immunol Res. 2016;4:835–844. doi: 10.1158/2326-6066.CIR-16-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li B, et al. Landscape of tumor-infiltrating T cell repertoire of human cancers. Nat Genet. 2016;48:725–732. doi: 10.1038/ng.3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levy E, et al. Immune DNA signature of T-cell infiltration in breast tumor exomes. Sci Rep. 2016;6:30064. doi: 10.1038/srep30064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaplinsky J, Arnaout R. Robust estimates of overall immune-repertoire diversity from high-throughput measurements on samples. Nat Commun. 2016;7:11881. doi: 10.1038/ncomms11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Steinert EM, et al. Quantifying memory CD8 T cells reveals regionalization of immunosurveillance. Cell. 2015;161:737–749. doi: 10.1016/j.cell.2015.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chung W, et al. Single-cell RNA-seq enables comprehensive tumour and immune cell profiling in primary breast cancer. Nat Commun. 2017;8:15081. doi: 10.1038/ncomms15081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qi Q, et al. Diversity and clonal selection in the human T-cell repertoire. Proc Natl Acad Sci USA. 2014;111:13139–13144. doi: 10.1073/pnas.1409155111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gajewski TF, Schreiber H, Fu Y-X. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol. 2013;14:1014–1022. doi: 10.1038/ni.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salgado R, et al. International TILs Working Group 2014 The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: Recommendations by an International TILs Working Group 2014. Ann Oncol. 2015;26:259–271. doi: 10.1093/annonc/mdu450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Denkert C, et al. Tumor-infiltrating lymphocytes and response to neoadjuvant chemotherapy with or without carboplatin in human epidermal growth factor receptor 2-positive and triple-negative primary breast cancers. J Clin Oncol. 2015;33:983–991. doi: 10.1200/JCO.2014.58.1967. [DOI] [PubMed] [Google Scholar]
- 39.Degnim AC, et al. Immune cell quantitation in normal breast tissue lobules with and without lobulitis. Breast Cancer Res Treat. 2014;144:539–549. doi: 10.1007/s10549-014-2896-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ruffell B, et al. Leukocyte composition of human breast cancer. Proc Natl Acad Sci USA. 2012;109:2796–2801. doi: 10.1073/pnas.1104303108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DeWitt WS, et al. Dynamics of the cytotoxic T cell response to a model of acute viral infection. J Virol. 2015;89:4517–4526. doi: 10.1128/JVI.03474-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Rooij N, et al. Tumor exome analysis reveals neoantigen-specific T-cell reactivity in an ipilimumab-responsive melanoma. J Clin Oncol. 2013;31:e439–e442. doi: 10.1200/JCO.2012.47.7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McGranahan N, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016;351:1463–1469. doi: 10.1126/science.aaf1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pasetto A, et al. Tumor- and neoantigen-reactive T-cell receptors can be identified based on their frequency in fresh tumor. Cancer Immunol Res. 2016;4:734–743. doi: 10.1158/2326-6066.CIR-16-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kreiter S, et al. Mutant MHC class II epitopes drive therapeutic immune responses to cancer. Nature. 2015;520:692–696. doi: 10.1038/nature14426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tran E, et al. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science. 2014;344:641–645. doi: 10.1126/science.1251102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Linnemann C, et al. High-throughput epitope discovery reveals frequent recognition of neo-antigens by CD4+ T cells in human melanoma. Nat Med. 2015;21:81–85. doi: 10.1038/nm.3773. [DOI] [PubMed] [Google Scholar]
- 48.Schenkel JM, Masopust D. Tissue-resident memory T cells. Immunity. 2014;41:886–897. doi: 10.1016/j.immuni.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Park CO, Kupper TS. The emerging role of resident memory T cells in protective immunity and inflammatory disease. Nat Med. 2015;21:688–697. doi: 10.1038/nm.3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li H, Ye C, Ji G, Han J. Determinants of public T cell responses. Cell Res. 2012;22:33–42. doi: 10.1038/cr.2012.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Venturi V, Price DA, Douek DC, Davenport MP. The molecular basis for public T-cell responses? Nat Rev Immunol. 2008;8:231–238. doi: 10.1038/nri2260. [DOI] [PubMed] [Google Scholar]
- 52.Callan CG, Jr, Mora T, Walczak AM. Repertoire sequencing and the statistical ensemble approach to adaptive immunity. Curr Opin Syst Biol. 2017;1:44–47. [Google Scholar]
- 53.Murugan A, Mora T, Walczak AM, Callan CG., Jr Statistical inference of the generation probability of T-cell receptors from sequence repertoires. Proc Natl Acad Sci USA. 2012;109:16161–16166. doi: 10.1073/pnas.1212755109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carlson CS, et al. Using synthetic templates to design an unbiased multiplex PCR assay. Nat Commun. 2013;4:2680. doi: 10.1038/ncomms3680. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All immunosequencing data underlying this study can be analyzed and freely downloaded from the Adaptive Biotechnologies immuneACCESS site at https://clients.adaptivebiotech.com/pub/beausang-2017-pnas.