Significance
Respiratory complex I, a redox-coupled proton pumping enzyme, is central to aerobic metabolism in mammalian mitochondria and implicated in many neuromuscular disorders. One of its substrates, ubiquinone-10, binds in an unusually long and narrow channel, which is at the intersection of the enzyme’s electron and proton transfer modules and a hotspot for disease-causing mutations. Here, we use a minimal, self-assembled respiratory chain to study complex I catalyzing with ubiquinones of different isoprenoid chain lengths. We show that the channel enhances the affinity of long-chain quinones, assists in their transfer along the channel, and organizes them for product release. Finally, we discuss how efficient binding and dissociation processes may help to link redox catalysis to proton pumping for energy conversion.
Keywords: bioenergetics, coenzyme Q10, electron transport chain, mitochondria, NADH:ubiquinone oxidoreductase
Abstract
Respiratory complex I (NADH:ubiquinone oxidoreductase), one of the largest membrane-bound enzymes in mammalian cells, powers ATP synthesis by using the energy from electron transfer from NADH to ubiquinone-10 to drive protons across the energy-transducing mitochondrial inner membrane. Ubiquinone-10 is extremely hydrophobic, but in complex I the binding site for its redox-active quinone headgroup is ∼20 Å above the membrane surface. Structural data suggest it accesses the site by a narrow channel, long enough to accommodate almost all of its ∼50-Å isoprenoid chain. However, how ubiquinone/ubiquinol exchange occurs on catalytically relevant timescales, and whether binding/dissociation events are involved in coupling electron transfer to proton translocation, are unknown. Here, we use proteoliposomes containing complex I, together with a quinol oxidase, to determine the kinetics of complex I catalysis with ubiquinones of varying isoprenoid chain length, from 1 to 10 units. We interpret our results using structural data, which show the hydrophobic channel is interrupted by a highly charged region at isoprenoids 4–7. We demonstrate that ubiquinol-10 dissociation is not rate determining and deduce that ubiquinone-10 has both the highest binding affinity and the fastest binding rate. We propose that the charged region and chain directionality assist product dissociation, and that isoprenoid stepping ensures short transit times. These properties of the channel do not benefit the exhange of short-chain quinones, for which product dissociation may become rate limiting. Thus, we discuss how the long channel does not hinder catalysis under physiological conditions and the possible roles of ubiquinone/ubiquinol binding/dissociation in energy conversion.
Respiratory complex I (NADH:ubiquinone oxidoreductase) (1) is a major entry point to the electron transport chain of oxidative phosphorylation in mammalian mitochondria. It catalyzes NADH oxidation coupled to ubiquinone reduction and captures the free energy produced to transport protons (2, 3) across the mitochondrial inner membrane, supporting ATP synthesis and transport processes. Because complex I is essential for regenerating NAD+ to sustain the tricarboxylic acid cycle and fatty acid oxidation, and an important contributor to cellular reactive oxygen species production (4), mutations in its subunits and assembly factors cause a wide range of inherited neuromuscular and metabolic diseases (5).
Mammalian complex I is a large (1 MDa) membrane-bound enzyme of 45 subunits. Due to advances in single-particle electron cryomicroscopy (cryoEM), knowledge of its structure (6–8) has surged forward recently, and descriptions of the 14 core subunits, in mammalian (6–8), fungal (9) and bacterial (10) enzymes, have laid a new foundation for mechanistic studies. NADH is oxidized by a flavin near the top of the hydrophilic domain of the L-shaped complex. Then, electrons are transferred along a chain of seven iron–sulfur (FeS) clusters to ubiquinone, bound at the interface of the hydrophilic and membrane domains. The membrane domain contains four antiporter-like units, considered to each transport one proton per cycle. They are connected by elements indicative of ion-transport activities, including π-bulges, loops in transmembrane helices (TMHs), and a series of buried charged residues, which also connects them to the quinone-binding region. Although molecular simulations have suggested how conformational, protonation, and hydration changes could propagate through the membrane domain to drive proton transfer events (11), how the energy from quinone reduction is captured and transferred to proton translocation is currently unknown. Reactions that may initiate the proton-transfer cascade include movement of a conserved aspartate upon quinone reduction (12), double reduction of Q to Q2− or its subsequent protonation to QH2 (13), and/or quinone binding/quinol dissociation. It has also been suggested that a permanently bound quinone shuttles between two positions in the channel, requiring an additional site for the exchangeable substrate to bind (14).
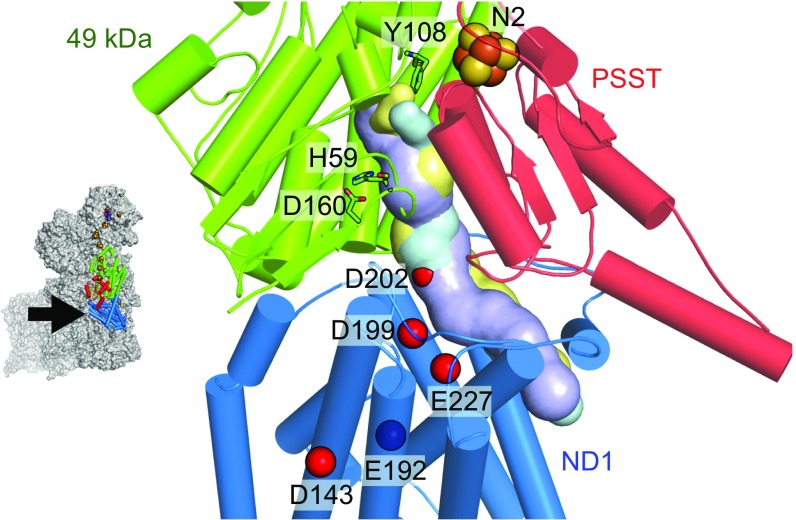
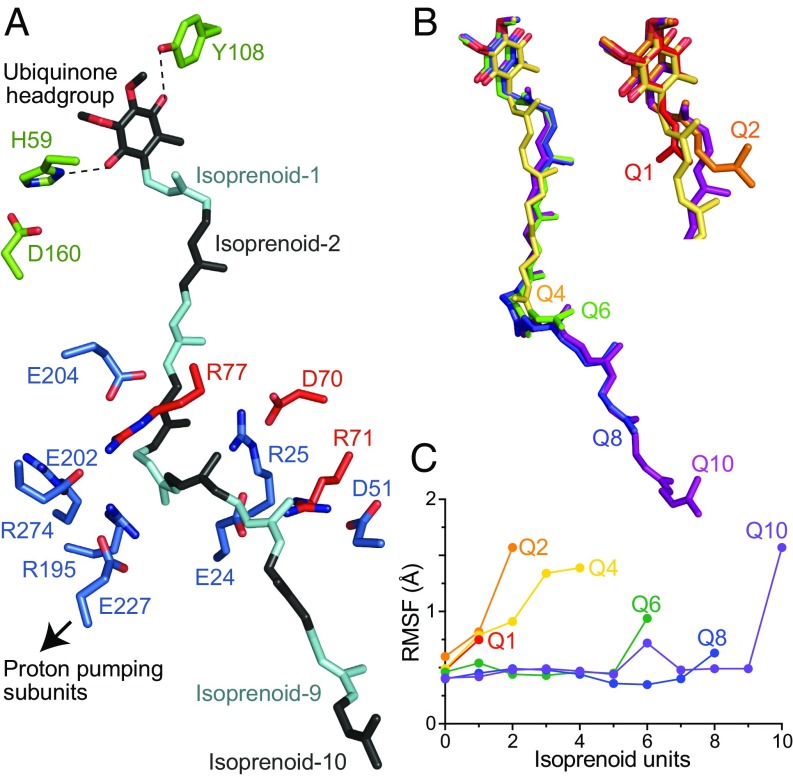
Here, we focus on ubiquinone binding and reduction by mammalian complex I. Strikingly, the binding site for the redox-active ubiquinone headgroup is ∼20 Å above the membrane interface and thought to be accessed by a long, narrow channel (Fig. 1) that has been identified in all of the structures described so far, but not yet confirmed experimentally. The headgroup binds in a cleft between the 49 kDa and PSST subunits (we use the nomenclature for the bovine enzyme throughout) and both mutational studies in Yarrowia lipolytica (15) and Escherichia coli (16), and densities observed in structural data from Thermus thermophilus complex I (10), indicate that it forms hydrogen bonds with H5949 kDa and Y10849 kDa, placing it within 12 Å of the terminal FeS cluster, N2. At the base of the cleft, the predicted channel meets subunit ND1, and runs along its interface with the 49-kDa and PSST subunits before exiting into the membrane; in total, it is long enough to accommodate most of the ∼50-Å-long isoprenoid tail of ubiquinone-10. Many structurally diverse inhibitors are thought to bind in the channel (17) and it is a hotspot for both pathophysiological mutations (ref. 5 and https://www.mitomap.org) and site-directed variants that affect catalysis (18, 19).
Fig. 1.
The proposed ubiquinone-binding channel in mammalian complex I. The 49-kDa, PSST, and ND1 subunits from bovine complex I (5LC5.pdb) (6) are in cartoon, with the surfaces of predicted quinone-binding cavities in the aligned structures of the bovine (6) (cyan), porcine (7) (yellow), and T. thermophilus (10) (purple) enzymes. The quinone headgroup is considered to hydrogen bond to Y108 and H59, and H59 to D160. The carboxylate groups of acidic residues connecting the quinone-binding region to the proton-pumping subunits are shown by red spheres. Cavities were generated using the Caver 3.0 PyMOL plugin (33) with a 1.4-Å probe.
Here, we have used proteoliposomes (PLs) to determine the kinetics of complex I catalysis with a series of ubiquinone substrates of varying isoprenoid tail length, from ubiquinone-1 (Q1) to ubiquinone-10 (Q10). Previous attempts to investigate the effects of quinone tail length used native membranes supplemented with exogenous quinones, following removal of the endogenous Q10 by lyophilization and pentane extraction (20–22). However, these studies were compromised because (i) the quinone exchange procedures were detrimental to catalysis (the specific activities were 10-fold lower than observed here); and (ii) effects on complex I activity were obscured by the catalysis of other enzymes: Lenaz et al. (22) reconstituted Q1 to Q10 into lyophilized pentane-extracted mitochondria and measured O2 consumption, which requires complexes III and IV, whereas Estornell et al. (20) similarly reconstituted Q10 and measured cytochrome c reduction, which requires complex III. More recently, Fato et al. (21) assayed lyophilized pentane-extracted bovine mitochondria reconstituted with Q3, Q5, and Q10 with an NADH-Q1 reductase assay. In contrast, the PLs used here (23) contain highly active bovine complex I (CI, to oxidize NADH and reduce ubiquinone to ubiquinol), the cyanide-insensitive nonprotonmotive alternative oxidase from Trypanosoma brucei brucei (AOX, to reoxidize ubiquinol to ubiquinone and reduce O2) and varying concentrations of different ubiquinones. The system and its kinetic properties can be defined precisely through measurements of the protein, phospholipid, and ubiquinone contents and have been developed to ensure complex I is rate determining. We compare the kinetic parameters of different ubiquinones with the structural and physicochemical properties of the quinone-binding channel to reveal determinants of catalysis.
Results
Ubiquinone Reduction by Complex I Is Rate Limiting for Catalysis.
The CI-AOX PLs described here catalyze NADH:O2 oxidoreduction (the NADH:O2 reaction) by redox cycling ubiquinone/ubiquinol: complex I reduces ubiquinone to ubiquinol and AOX reoxidizes it. To use the NADH:O2 reaction to investigate ubiquinone reduction by complex I it must be rate limiting. Thus, the NADH (200 µM) and O2 (200–250 µM) concentrations used are substantially higher than the KM values of 79 ± 8 µM for complex I (measured with the NADH:O2 reaction) and 10–20 µM for AOX (24). Furthermore, the kcat value for NADH oxidation by the flavin in bovine complex I is >5,000 s−1 (25), more than 10 times faster than the maximum rate of NADH:ubiquinone oxidoreduction, so NADH oxidation does not limit catalysis.
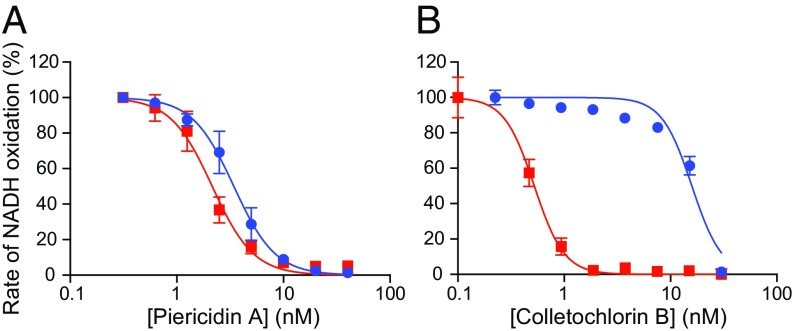
The complex I and AOX used here display similar turnover rates in solution assays with ubiquinone-1, typically 250–300 s−1 and 200–250 s−1, respectively. Thus, to increase the rate of ubiquinol oxidation, PLs were created starting from a molar ratio of 1:25 CI:AOX. The ratio then effectively doubles to 1:46 ± 1.1 (mean value ± SD for all samples) because NADH can only access complex I oriented with its active site outwards (77 ± 10% of the total), whereas AOX substrates can access it in both orientations, and because AOX incorporates into PLs more efficiently than complex I (80 ± 15% and 59 ± 11%, respectively). This high ratio provides a strong expectation for complex I being rate limiting, and thus for the ubiquinone/ubiquinol pool being predominantly oxidized during catalysis (23). To test this expectation, titrations on the NADH:O2 reaction by Q10-containing PLs with a high (1:51) or low (1:1.5) CI:AOX ratio (defined for outward facing complex I) were performed with complex I (piericidin A)- and AOX (colletochlorin B) (26)-specific inhibitors. For both ratios, piericidin A inhibition builds rapidly at low concentrations (Fig. 2A) and the IC50 values are similar (2.0 ± 1.1 and 3.4 ± 1.1 nM, or 8.7 and 8.0 piericidin A per oriented complex I, respectively). In contrast, the high ratio PLs were unresponsive to low colletochlorin B concentrations (Fig. 2B) and displayed an IC50 value 30 times greater (15.9 ± 0.03 and 0.52 ± 0.03 nM, respectively, or 0.7 and 1.4 colletochlorin B per AOX). Thus, in the high ratio (typical) preparation, most of the AOX can be inhibited with little effect on catalysis, confirming that AOX is not rate limiting. High ratio PLs containing Q8, Q6, and Q4, or tested with Q2, all gave colletochlorin B IC50 values of 17.6 ± 1.3 nM, matching that for Q10. Only for Q1 was the IC50 value for colletochlorin B different; its much higher value of ∼5 µM suggests colletochlorin B is a poor inhibitor of ubiquinol-1 oxidation by AOX, or that Q1 is a poor complex I substrate.
Fig. 2.
Complex I is rate limiting for catalysis in CI-AOX PLs. Normalized rates of the NADH:O2 reaction by PLs containing high (1:51, blue) and low (1:1.5, red) ratios of oriented-CI:AOX are shown. Both preparations contained ∼10 mM Q10. (A) Inhibition by piericidin A. (B) Inhibition by colletochlorin B.
Measurement of KM and kcat Values.
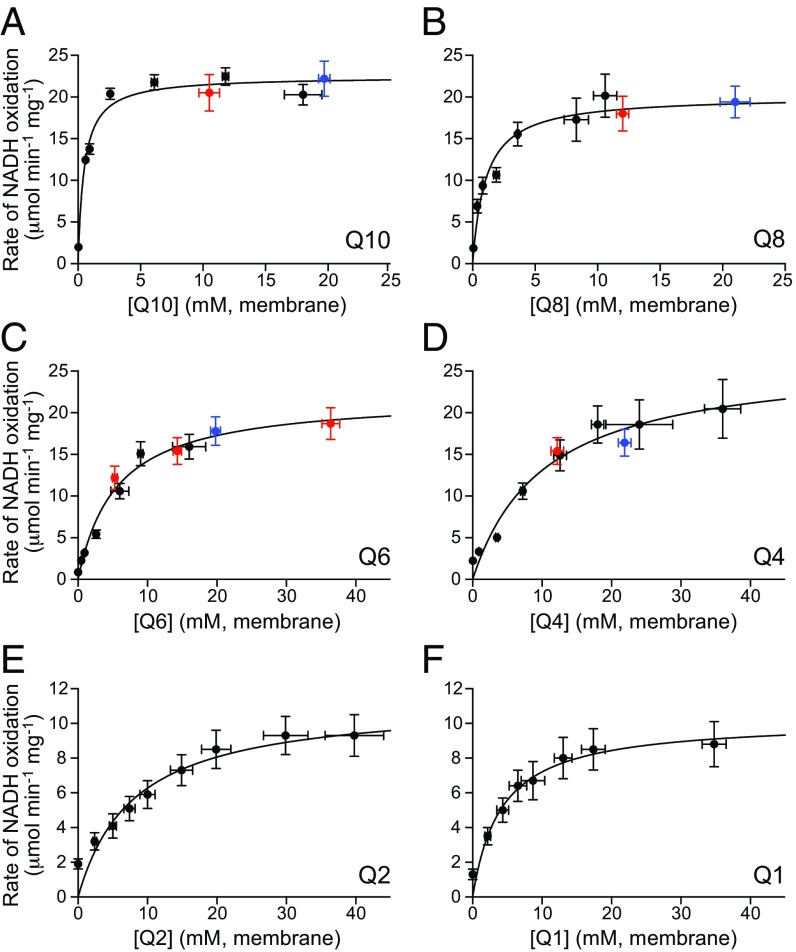
Fig. 3 shows Michaelis–Menten curves for complex I catalyzing with Q10, Q8, Q6, Q4, Q2, and Q1. The highly hydrophobic Q10, Q8, Q6, and Q4 molecules [calculated logP values (cyclohexane/water) 19.4, 15.7, 12, and 8.3, respectively (https://www.ncbi.nlm.nih.gov/pccompound)] were incorporated into the PL membranes during reconstitution so each point is from a different preparation. The samples for each curve were prepared together in batches, then their quinone, phospholipid, complex I, and AOX concentrations were determined. Quinone concentrations in the membrane were calculated by assuming that 1 mg of phospholipid occupies ∼1 µL (27), so 1 nmol of quinone per milligram phospholipid is equivalent to 1 mM. The more hydrophilic Q2 and Q1 molecules were added from ethanolic stock solutions to quinone-free PLs and considered to partition between the aqueous and membrane phases. Their membrane concentrations were calculated from the phase volumes and membrane/water logP values, 4.0 for Q2 and 2.9 for Q1 (28). Micelle formation was not considered because the critical micelle concentration of Q2 of 14.0 µM (29) is above the maximum 10-µM concentration added, and it was confirmed that all of the Q2 added could be reduced by complex I. Addition of protonophore uncouplers, such as gramicidin, did not increase the rate so they were not included. Finally, to account for variations in enzyme activity between the quinone-specific batch preparations, two further batches containing samples from each different quinone were prepared (see red and blue points in Fig. 3); the Q1 and Q2 datasets are from quinone-free PLs included in these batches. The Q10 KM dataset was used as the reference and scaling parameters for the other datasets derived by simultaneous nonlinear least squares fitting; the scaling parameters were applied uniformly and so affect only Vmax, not KM.
Fig. 3.
Michaelis–Menten curves for reduction of Q1–Q10 by complex I. (A) Q10, (B) Q8, (C) Q6, (D) Q4, (E) Q2, and (F) Q1. The black points are from sets of PLs with different quinone concentrations; the datasets for Q1–Q8 were adjusted to the Q10 dataset using the red and blue datasets and scaling factors of 1.44 (Q8), 1.35 (Q6), 1.86 (Q4), 1.57 (red points), and 1.60 (blue points). Each value is the mean of at least three replicates ± the SD propagated from each underlying measurement.
Dependence of KM and Vmax on Isoprenoid Chain Length.
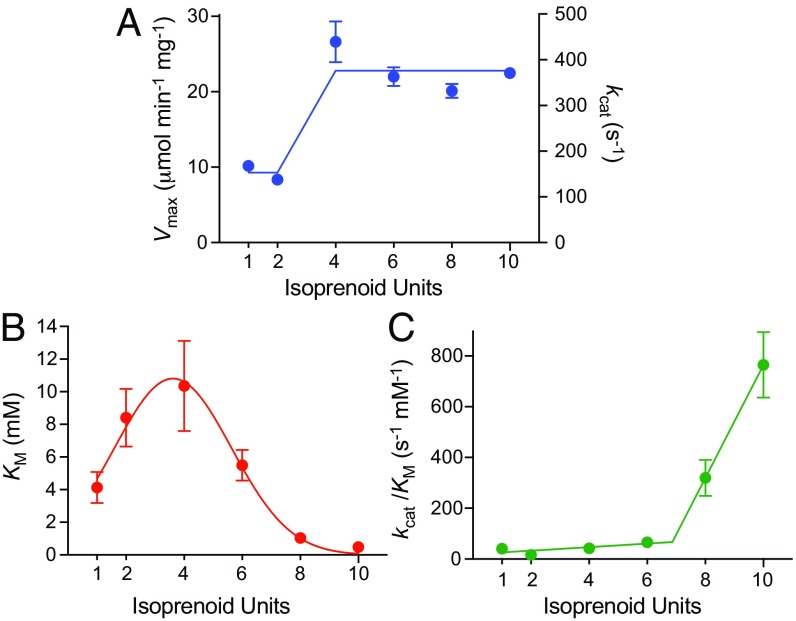
The KM and Vmax values for each ubiquinone (from Fig. 3) are summarized in Fig. 4. Fig. 4A shows that Vmax (or kcat) is biphasic and averages to 23 ± 2 µmol⋅min−1⋅mg−1 (380 s−1) for Q10, Q8, Q6, and Q4, and 9.3 ± 0.9 µmol⋅min−1⋅mg−1 (150 s−1) for Q2 and Q1. Previous studies, using pentane-extracted mitochondria, also observed the highest NADH:O2 activities from Q7 to Q10, with Q1 to Q4 supporting only ∼30% of the Q10 value (21, 22). Fig. 4B shows that KM displays a bell-shaped curve with the highest value at Q4. We note that the Q10 KM value reported here of 0.48 mM is lower than that of 3.9 mM we measured previously (23) due to (i) improvements to the AOX preparation that have increased its specific activity two- to threefold, (ii) improvements to our quinone quantification protocol, and (iii) the omission of alamethicin. Alamethicin was used previously (23) to open pores in the membrane to allow NADH to access all of the complex I, but we have now found that 11.25 µg⋅mL−1 increases the apparent Q10 KM value from 0.5 ± 0.1–1.3 ± 0.4 mM. Alamethicin has been reported to sequester membrane-bound fatty acids (30) so it may sequester quinones also, and to induce structural changes in the membrane (31). Finally, Fig. 4C shows that the catalytic efficiency or pseudo second order rate constant, kcat/KM, increases markedly for Q8 and Q10. Previously, kcat/KM was reported to exhibit a bell-shaped dependence on substrate hydrophobicity (32) but the earlier study used only quinones with alkyl chains up to 11 carbons long, whereas the same trend is not replicated here using polyisoprenoid ubiquinones with partition coefficients that vary much more.
Fig. 4.
Dependence of the Michaelis–Menten parameters on isoprenoid chain length. (A) Vmax and kcat values shown with average values of 9.3 ± 0.9 µmol⋅min−1⋅mg−1 (150 ± 15 s−1) for Q1–Q2 and 23 ± 2 µmol⋅min−1⋅mg−1 (380 ± 39 s−1) for Q4–Q10. (B) KM values; the curve is only to guide the eye. (C) kcat/KM values with two linear fits (gradients 7.0 and 220 mM−1⋅s−1⋅isoprenoid unit−1). Values are from the data in Fig. 3 and also given in Table S1.
Analysis of Structural Data and Docking of Q10.
To investigate how our data correlate with structural features in complex I, we used the Caver software (33) to detect and compare the proposed quinone-binding sites in available structures. Fig. S1 shows that the bovine (6), porcine (7), T. thermophilus (10), and Y. lipolytica (9) structures describe a common channel. The same channel is truncated in the ovine structure (8) by the 49-kDa subunit β1–β2 loop, and similarly constricted in the Y. lipolytica structure (9). Variations in the channels may arise from their medium resolutions and/or different enzyme states; both the ovine and Y. lipolytica enzymes were proposed to be in the “deactive” state (8, 9) that is unable to reduce quinone, whereas the cryoEM dataset for the bovine structure (6) was classified into three states and only the structure allocated to the “active” state was used here. The bovine, porcine, and T. thermophilus channels are overlaid in Fig. 1, highlighting their similarity.
To better define how quinones bind in the channel, we modeled Q10 into the channel identified in the bovine complex [Protein Data Bank (PDB) ID: 5LC5] (6) and relaxed the system by atomistic molecular dynamics (MD) simulations (Fig. 5A). Subsequently, by truncating the relaxed Q10 model, we performed further MD simulations for Q8, Q6, Q4, Q2, and Q1 (Fig. 5B). Analyses of the Q10-bound structure showed that its channel closely matches the original channel (Fig. S1) and, in all cases, the modeled protein structures around the bound quinones remain in similar conformations except that, for Q1–Q6 and the empty site, the sidechain of F224ND1 moves into the position of isoprenoid-7 (Fig. S2A). Notably, the root-mean-square fluctuations (RMSFs) of the bound quinone variants suggest that the long isoprenoid chains of Q8 and Q6, which overlay closely on that of Q10 (Fig. 5B), are spatially tightly constrained by the channel, whereas the shorter chains of Q4, Q2, and Q1 have much greater conformational freedom (Fig. 5C).
Fig. 5.
Modeled structures of bovine complex I with Q1–Q10 bound. (A) The modeled Q10 molecule with its isoprenoids in black and cyan, alternately, alongside the sidechains of charged residues within 5 Å (red, PSST; blue, ND1). Green: 49-kDa subunit residues hydrogen bonded to the ubiquinone headgroup. (B) The overlaid modeled structures for Q10, Q8, Q6, and Q4, and Inset for Q10, Q4, Q2, and Q1. (C) RMSFs for each quinone species from MD simulations, colored as in B.
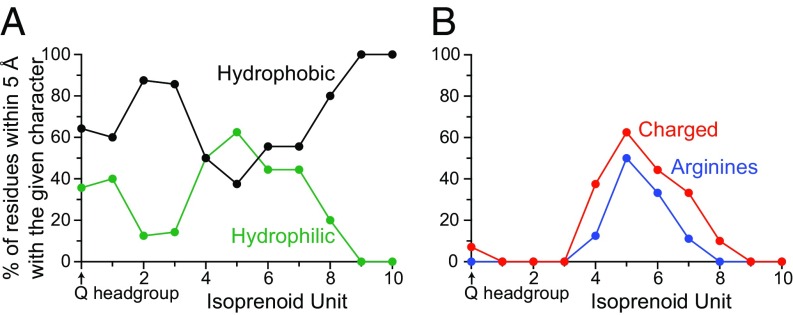
Strikingly, Fig. 6 shows how the properties of residues close to the modeled Q10 vary along it (Fig. S3 presents equivalent data for Q8–Q1). The environment of the headgroup plus isoprenoids 1–3, in the cleft between the PSST and 49-kDa subunits, is primarily hydrophobic and uncharged. Conversely, that of isoprenoids 4–7 contains many charged residues. In particular, a group of highly conserved arginines (Fig. 5A) form a channel elbow that produces a ∼100° kink in the modeled Q10 between isoprenoids 4 and 5. Notably, the hydrophobic faces of the Arg guanidiniums form π-stacking interactions with the isoprenoids, while their edges form polar and electrostatic interactions with a set of conserved Glu/Asp residues (Fig. 5A), consistent with the known behavior of guanidium groups (34). In the modeled structures, interactions between the charged residues separate them into two groups (Fig. S2B), with the group involving residues on ND1 TMH1 showing less positional variation than that involving primarily residues on the ND1 TMH5–6 loop (Fig. S2C), noted previously for its flexibility (6). Finally, for isoprenoids 8 and 9 the expected hydrophobic nature of the ubiquinone-binding channel is reestablished.
Fig. 6.
The protein environment of the modeled bound Q10 molecule in the structure of bovine CI. (A) Percentage of residues within 5 Å of each isoprenoid that are hydrophobic (A, F, I, L, M, P, V, W, and Y) or hydrophilic (C, D, E, G, H, K, N, Q, R, S, and T). (B) Percentage of residues within 5 Å of each isoprenoid that are canonically charged (D, E, H, K, and R) and that are arginines.
Discussion
Entry and Exit of Quinone/Quinol to and from the Channel.
In the classical Michaelis–Menten model, kcat encapsulates all of the steps that follow formation of the enzyme–substrate complex, so the fact that kcat is constant for Q4–Q10 (Fig. 4A) suggests that product dissociation, which is expected to be chain-length dependent, is not rate limiting. Alternative rate-limiting steps include the electron–proton transfers for quinone reduction, the coupled processes leading to proton translocation, and all of the reactions required to complete the catalytic cycle.
For Q2 and Q1, kcat (150 s−1) is substantially lower than for Q4–Q10 (380 s−1) (Fig. 4A) so the rate-limiting step has either slowed or changed identity. Both possibilities indicate that a chain-length dependent step, such as product dissociation, has slowed down. Furthermore, the pseudo second-order rate constant kcat/KM [where KM = (koff + kcat)/kon] provides a guide to the rate constant for substrate binding (kon) since it approximates to it when kcat > koff and reflects how fast the rate increases with increasing concentration, in substrate-limiting conditions. kcat/KM increases substantially with isoprenoid tail length (Fig. 4C). Both observations are consistent with rates of binding and dissociation (i.e., channel transit rates) increasing with isoprenoid chain length, and being limiting only for Q2 and Q1. We propose three explanations. First, the short isoprenoid tails of Q1, Q2, and Q4 do not overlay their respective Q10 isoprenoids (Fig. 5B) so they are conformationally mobile within the site (Fig. 5C): their dissociation may be hindered by lack of directionality, due to lack of a guiding anchor extending down the channel. The tail of Q10 extends into the membrane and may guide the entire dissociation process. Second, for the shorter quinones, an additional molecule(s) may enter the channel behind the substrate, impeding its dissociation. Third, the channel entrance is at a similar depth below the membrane interface to the favored position for the ubiquinone-10 headgroup (35), whereas shorter tail quinones may distribute differently in the membrane, affecting their binding rates.
Finally, while the simple Michaelis–Menten mechanism provides a convenient framework for interpreting our data, it does not account for the unusual nature of the ubiquinone-binding channel in complex I. Transfer along the whole channel is unlikely to occur over a single transition state but over a complex energy surface that may form local minima and transient binding sites. Longer chain species may move faster along the channel because their stepwise movements incur minimal changes. For example, moving a Q10 molecule by one step requires only the residues (and water molecules) around the headgroup and the leading isoprenoid to reorganize (at all other positions one isoprenoid simply replaces another), whereas moving a short chain quinone requires the region behind it to reorganize also. The energy surface for Q10 transit along the channel may therefore be flatter. In this way, the unusually long substrate-binding channel in complex I need not exert a rate-limiting effect on catalysis.
Effect of Isoprenoid Chain Length on Binding Affinity.
In classical Michaelis–Menten kinetics, KM is expressed as (koff + kcat)/kon so, for Q4–Q10, where kcat is constant (Fig. 4A) but KM decreases with increasing chain length (Fig. 4B), koff decreases and/or kon increases. Both possibilities suggest that Kd (= koff/kon) decreases, such that Q4 binds most weakly and Q6, Q8, and Q10 progressively more tightly. The comparison does not extend to Q2 and Q1 because kcat is different, but as their kcat values are similar, the same considerations suggest Q1 binds more tightly than Q2. Thus, although a robust comparison between Q4 and Q2 is currently not possible, we may infer that the curve describing Kd qualitatively resembles that of KM.
The relative free-energy changes that result from moving each quinone species from a low dielectric (membrane) environment into its complex I binding site were evaluated using continuum electrostatics calculations (36), revealing a linear dependence on isoprenoid number (Fig. S4). The trend is consistent with the decreasing Kd proposed for Q4–Q10, but not with the decrease proposed for Q4–Q1. Intriguingly, the discrepancy can be explained by considering that koff for Q4, Q2, and Q1 may be decreased, as discussed above, by their lacking a guiding chain for dissociation and by additional substrate molecules blocking their exit. By decreasing koff, these factors “cage” short chain species in the binding site, independently of their intrinsic affinity for it, and decrease Kd. Notably, many tight-binding hydrophobic complex I inhibitors match the dimensions of Q1–Q4 and are considered to occupy the same binding site; density attributed to piericidin A, which resembles Q3, was observed to overlay density attributed to decylubiquinone in crystallographic maps from T. thermophilus complex I (10). Thus, the potent inhibition of these molecules may also partly result from caging effects.
The concept of a highly charged channel for the hydrophobic isoprenoid chain, as observed for isoprenoids 4–7 (Figs. 5 and 6), is intrinsically challenging. Charged cavities in proteins tend to fill with water molecules (37), and indeed, in MD simulations on the structure of T. thermophilus complex I, waters accumulate in the channel, especially in the charged region (11). If waters are present it may be that (i) the channel has become hydrated artificially during the extended handling required for structural work, or (ii) during every catalytic cycle the quinone displaces waters, making an entropic contribution to offset the enthalpy loss of breaking charged and polar interactions. In fact, an unfavorable binding enthalpy from the charged region may mitigate the increasing affinity due to binding an increasing number of hydrophobic units within a hydrophobic channel and be important for efficient product release. Higher resolution structural data are required for further understanding of this intriguing structural feature.
Finally, increased contributions to the binding affinity from the second, nonpolar region at isoprenoids 8 and 9 cause a marked increase in kcat/KM (Fig. 4C). Because kcat is constant from Q4 to Q10, kcat/KM is dominated by the reciprocal of KM and the apparent discontinuity in Fig. 4C results simply from a point of inflection in the KM curve.
How Is Proton Translocation Coupled to Quinone Reduction?
The identity of the coupling point, at which the redox reaction initiates proton translocation, is currently the most important unknown feature of the complex I mechanism. Molecular dynamics simulations of the movement of D16049 kDa away from H5949 kDa, as a result of proton transfer from H59 to the nascent quinol, have been used to illustrate one possibility (12). Alternatively, could conformational changes triggered by quinone/quinol moving along the channel, particularly through the charged section, trigger proton translocation? If so, short chain quinones must activate the mechanism as effectively as long chain quinones, since they elicit the same proton-pumping stoichiometry (2, 3). As Fig. 5A shows, the charged residues surrounding the channel are, like D160, connected to the center of the membrane domain by a chain of acidic residues (6–10, 12), suggesting how quinone/quinol binding/dissociation may be communicated to the proton translocation machinery. To elucidate, challenge, and add to proposals for the mechanistic coupling point in complex I require higher resolution structures set in different states, alongside mutational, functional, and computational studies to provide complementary strategies and perspectives on tackling this difficult problem.
Experimental Methods
Preparation of AOX.
The construct described previously for overexpression of AOX from T. brucei brucei (38) was modified by removing the N-terminal mitochondrial targeting peptide (residues 1–24) and replacing the 6xHis tag with a Twin-Strep tag (IBA GmbH) (39). The modified AOX was overexpressed in E. coli strain FN102 (38) in a 60-L fermenter as described previously (23, 38), except by using 100 µg mL−1 ampicillin instead of carbenicillin. The culture was incubated at 30 °C, 60% O2 saturation until OD600 ∼0.6, then expression induced with 25 µM isopropyl β-d-1-thiogalactopyranoside for 12 h. Cells (200–300 g) were collected by centrifugation, then membranes were prepared immediately (23) and resuspended to ∼30 mg protein mL−1 in 50 mM Tris⋅HCl (pH 8.0 at 4 °C) for storage at −80 °C.
All of the following steps were at 4 °C: 20 mL of membranes were solubilized at 6 mg protein mL−1 in 25 mM Tris⋅HCl (pH 8.0), 200 mM MgSO4, 1.4% (wt/vol) n-octyl-glucopyranoside (Anagrade, Anatrace) and 20% (vol/vol) glycerol for 1 h then centrifuged (165,000 × g, 30 min). The supernatant was loaded onto a ∼8-mL column of Strep-Tactin Superflow high capacity resin (IBA GmbH) preequilibrated in strep buffer: 20 mM Tris⋅HCl (pH 8.0 at 4 °C), 50 mM MgSO4, 160 mM NaCl and 20% (vol/vol) glycerol (39). AOX was eluted in strep buffer supplemented with 2.5 mM desthiobiotin (Sigma-Aldrich) and 0.042% n-dodecyl-β-d-maltopyranoside (DDM) (Anagrade, Anatrace) (39). Pooled AOX-containing fractions were concentrated 10-fold, then dialyzed for 6 h against 2 L of strep buffer plus 0.042% DDM (23). Typical preparations yielded 3–5 mg AOX and were ≥95% pure by SDS/PAGE.
Preparation of Complex I.
Mitochondrial membranes were prepared from Bos taurus (bovine) heart (40) then complex I was prepared as described previously (23), with minor modifications. Membranes were solubilized at 5 mg mL−1 protein in 1% DDM. The Q-Sepharose buffers consisted of 20 mM Tris⋅HCl (pH 7.55 at 4 °C), 2 mM EDTA, 10% (vol/vol) ethylene glycol, 0.2% (wt/vol) DDM, 0.02% asolectin (total soy lipid extract, Avanti Polar Lipids), and 0.02% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS). Size exclusion chromatography was conducted on a Superose 6 Increase 10/300 column (GE Healthcare) in 20 mM Tris⋅HCl (pH 7.55 at 4 °C), 200 mM NaCl, 0.05% DDM, and 10% (vol/vol) glycerol. Samples were flash frozen for storage at ∼20 mg⋅mL−1 in 30% (vol/vol) glycerol.
Preparation of CI-AOX PLs.
Chloroform solutions (25 mg mL−1) of bovine heart phosphatidylcholine (PC), phosphatidylethanolamine (PE) and cardiolipin (CL) were from Avanti Polar Lipids. Chloroform stock solutions of Q10 (Sigma Aldrich), Q8 (Avanti Polar Lipids), Q6 (Avanti Polar Lipids) and Q4 (Santa Cruz Biotechnology) were created at 2–4 mM. PLs were prepared as described previously (23), starting from mixtures of 8 mg PC, 1 mg PE, 1 mg CL and quinone (as required).
Characterization of PLs.
To quanitify the quinone present, 90 µL of ethanol (HPLC grade Chromasolv; Sigma-Aldrich) was added to 10 µL of PL solution, sonicated (1 min) and centrifuged (16,300 × g, 10 min). A total of 50 µL of supernatant was injected onto a Nucleosil 100–5C18 (Hichrom) column, and run at 30 °C at 800 µL⋅min−1 on an Agilent 1100 series HPLC in 70% ethanol, 30% methanol, 0.07% HClO4, and 50 mM NaClO4 for Q10 and Q8, or in 100% methanol, 0.07% HClO4 and 50 mM NaClO4 for Q6 and Q4 (retention times 9, 6, 10, and 5 min, respectively). Concentrations were determined by comparison with known standards. Total phospholipid contents were determined as detailed previously (23). Total protein contents were quantified by the amido black assay, which is insensitive to detergents and high phospholipid concentrations (41). The NADH:APAD+ oxidoreduction assay was used to determine the total complex I content and orientation together with 15 µg⋅mL−1 alamethicin to allow NADH into the PL lumen (23). The AOX content was taken as the difference between the total protein and complex I contents.
Catalytic Activity Assays.
Activity assays were at 32 °C in 10 mM Tris-SO4 (pH 7.5 at 32 °C), 50 mM KCl, 200 µM NADH with ∼1.5 mg protein mL−1 PLs (23). NADH oxidation was monitored at 340–380 nm (ε = 4.81 mM−1⋅cm−1) on a Molecular Devices Spectramax 384 Plus platereader. Q2 and Q1 (Sigma-Aldrich) were added in ethanol. Inhibitor-insensitive rates were determined in 0.5 µM piericidin A as 0.30 ± 0.03 µmol NADH min−1⋅mg CI−1 for all quinones except Q1 (which exhibited a concentration-dependent inhibitor-insensitive rate) and have been subtracted from the data reported.
Molecular Modeling of Ubiquinone-Bound Structures.
Ubiquinone-10 was docked into a channel identified by the HOLE software (42) in the structure of bovine complex I [PDB ID: 5LC5 (6)] with missing amino acid sidechains modeled. Then, 20 ns of classical MD simulation (integration time step 2 fs, 310 K) were performed using NAMD2 (43) with the CHARMM36 force field (44), and density functional theory (DFT)-derived parameters for all cofactors (12). Backbone atoms of the 49 kDa, ND1, and PSST subunits were harmonically restrained (force constant 2 kcal⋅mol−1⋅Å−2), with positional constraints for all other subunits. The A57–V61 and L106–S110 loops of the 49-kDa subunit that carry H59 and Y108 were not restrained. For the Q1–Q8-bound structures, the Q10 molecule was truncated and each variant relaxed for a further 10 ns. Free energies for quinone solvation into complex I were estimated by the molecular mechanics–generalized Born surface area (MM-GBSA) method (36), using 50 snapshots extracted from 10-ns MD simulations.
Supplementary Material
Acknowledgments
We thank C. Humphreys & Sons Abattoir (Chelmsford) for providing bovine hearts and Sotiria Tavoulari for help with the amido black assay. Computing time was provided by the SuperMuc at The Leibniz Rechenzentrum (Grant pr48de). This work was supported by the Medical Research Council (Grant U105663141 to J.H.) and by the German Research Foundation (V.R.I.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The modeled structures with Q1–Q10 bound and with the empty site are available from the University of Cambridge data repository (https://doi.org/10.17863/CAM.13786).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1714074114/-/DCSupplemental.
References
- 1.Hirst J. Mitochondrial complex I. Annu Rev Biochem. 2013;82:551–575. doi: 10.1146/annurev-biochem-070511-103700. [DOI] [PubMed] [Google Scholar]
- 2.Jones AJY, Blaza JN, Varghese F, Hirst J. Respiratory complex I in Bos taurus and Paracoccus denitrificans pumps four protons across the membrane for every NADH oxidized. J Biol Chem. 2017;292:4987–4995. doi: 10.1074/jbc.M116.771899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Galkin AS, Grivennikova VG, Vinogradov AD. →H+/2e- stoichiometry in NADH-quinone reductase reactions catalyzed by bovine heart submitochondrial particles. FEBS Lett. 1999;451:157–161. doi: 10.1016/s0014-5793(99)00575-x. [DOI] [PubMed] [Google Scholar]
- 4.Murphy MP. How mitochondria produce reactive oxygen species. Biochem J. 2009;417:1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fassone E, Rahman S. Complex I deficiency: Clinical features, biochemistry and molecular genetics. J Med Genet. 2012;49:578–590. doi: 10.1136/jmedgenet-2012-101159. [DOI] [PubMed] [Google Scholar]
- 6.Zhu J, Vinothkumar KR, Hirst J. Structure of mammalian respiratory complex I. Nature. 2016;536:354–358. doi: 10.1038/nature19095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu M, Gu J, Guo R, Huang Y, Yang M. Structure of mammalian respiratory supercomplex I1III2IV1. Cell. 2016;167:1598–1609. doi: 10.1016/j.cell.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 8.Fiedorczuk K, et al. Atomic structure of the entire mammalian mitochondrial complex I. Nature. 2016;538:406–410. doi: 10.1038/nature19794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zickermann V, et al. Structural biology. Mechanistic insight from the crystal structure of mitochondrial complex I. Science. 2015;347:44–49. doi: 10.1126/science.1259859. [DOI] [PubMed] [Google Scholar]
- 10.Baradaran R, Berrisford JM, Minhas GS, Sazanov LA. Crystal structure of the entire respiratory complex I. Nature. 2013;494:443–448. doi: 10.1038/nature11871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Di Luca A, Gamiz-Hernandez AP, Kaila VRI. Symmetry-related proton transfer pathways in respiratory complex I. Proc Natl Acad Sci USA. 2017;114:E6314–E6321. doi: 10.1073/pnas.1706278114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma V, et al. Redox-induced activation of the proton pump in the respiratory complex I. Proc Natl Acad Sci USA. 2015;112:11571–11576. doi: 10.1073/pnas.1503761112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Efremov RG, Sazanov LA. The coupling mechanism of respiratory complex I–A structural and evolutionary perspective. Biochim Biophys Acta. 2012;1817:1785–1795. doi: 10.1016/j.bbabio.2012.02.015. [DOI] [PubMed] [Google Scholar]
- 14.Wikström M, Sharma V, Kaila VRI, Hosler JP, Hummer G. New perspectives on proton pumping in cellular respiration. Chem Rev. 2015;115:2196–2221. doi: 10.1021/cr500448t. [DOI] [PubMed] [Google Scholar]
- 15.Tocilescu MA, et al. The role of a conserved tyrosine in the 49-kDa subunit of complex I for ubiquinone binding and reduction. Biochim Biophys Acta. 2010;1797:625–632. doi: 10.1016/j.bbabio.2010.01.029. [DOI] [PubMed] [Google Scholar]
- 16.Sinha PK, et al. Conserved amino acid residues of the NuoD segment important for structure and function of Escherichia coli NDH-1 (complex I) Biochemistry. 2015;54:753–764. doi: 10.1021/bi501403t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murai M, Miyoshi H. Current topics on inhibitors of respiratory complex I. Biochim Biophys Acta. 2016;1857:884–891. doi: 10.1016/j.bbabio.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 18.Angerer H, et al. Tracing the tail of ubiquinone in mitochondrial complex I. Biochim Biophys Acta. 2012;1817:1776–1784. doi: 10.1016/j.bbabio.2012.03.021. [DOI] [PubMed] [Google Scholar]
- 19.Sinha PK, et al. Critical roles of subunit NuoH (ND1) in the assembly of peripheral subunits with the membrane domain of Escherichia coli NDH-1. J Biol Chem. 2009;284:9814–9823. doi: 10.1074/jbc.M809468200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Estornell E, et al. Saturation kinetics of coenzyme Q in NADH and succinate oxidation in beef heart mitochondria. FEBS Lett. 1992;311:107–109. doi: 10.1016/0014-5793(92)81378-y. [DOI] [PubMed] [Google Scholar]
- 21.Fato R, et al. Steady-state kinetics of the reduction of coenzyme Q analogs by complex I (NADH:ubiquinone oxidoreductase) in bovine heart mitochondria and submitochondrial particles. Biochemistry. 1996;35:2705–2716. doi: 10.1021/bi9516034. [DOI] [PubMed] [Google Scholar]
- 22.Lenaz G, Castelli A, Littarru GP, Bertoli E, Folkers K. Specificity of lipids and coenzyme Q in mitochondrial NADH and succin-oxidase of beef heart and S. cerevisiae. Arch Biochem Biophys. 1971;142:407–416. doi: 10.1016/0003-9861(71)90504-2. [DOI] [PubMed] [Google Scholar]
- 23.Jones AJY, et al. A self-assembled respiratory chain that catalyzes NADH oxidation by ubiquinone-10 cycling between complex I and the alternative oxidase. Angew Chem Int Ed Engl. 2016;55:728–731. doi: 10.1002/anie.201507332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Young L, et al. Probing the ubiquinol-binding site of recombinant Sauromatum guttatum alternative oxidase expressed in E. coli membranes through site-directed mutagenesis. Biochim Biophys Acta. 2014;1837:1219–1225. doi: 10.1016/j.bbabio.2014.01.027. [DOI] [PubMed] [Google Scholar]
- 25.Birrell JA, Yakovlev G, Hirst J. Reactions of the flavin mononucleotide in complex I: A combined mechanism describes NADH oxidation coupled to the reduction of APAD+, ferricyanide, or molecular oxygen. Biochemistry. 2009;48:12005–12013. doi: 10.1021/bi901706w. [DOI] [PubMed] [Google Scholar]
- 26.Shiba T, et al. Structure of the trypanosome cyanide-insensitive alternative oxidase. Proc Natl Acad Sci USA. 2013;110:4580–4585. doi: 10.1073/pnas.1218386110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagle JF, Tristram-Nagle S. Structure of lipid bilayers. Biochim Biophys Acta. 2000;1469:159–195. doi: 10.1016/s0304-4157(00)00016-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fato R, Battino M, Degli Esposti M, Parenti Castelli G, Lenaz G. Determination of partition and lateral diffusion coefficients of ubiquinones by fluorescence quenching of n-(9-anthroyloxy)stearic acids in phospholipid vesicles and mitochondrial membranes. Biochemistry. 1986;25:3378–3390. doi: 10.1021/bi00359a043. [DOI] [PubMed] [Google Scholar]
- 29.Battino M, Fahmy T, Lenaz G. Determination of the critical micelle concentration of short-chain ubiquinones in model systems. Biochim Biophys Acta. 1986;851:377–384. [Google Scholar]
- 30.Afanasyeva EF, Syryamina VN, Dzuba SA. Alamethicin can capture lipid-like molecules in the membrane. J Chem Phys. 2017;146:011103. doi: 10.1063/1.4973703. [DOI] [PubMed] [Google Scholar]
- 31.Wang KF, Nagarajan R, Camesano TA. Antimicrobial peptide alamethicin insertion into lipid bilayer: A QCM-D exploration. Colloids Surf B Biointerfaces. 2014;116:472–481. doi: 10.1016/j.colsurfb.2014.01.036. [DOI] [PubMed] [Google Scholar]
- 32.Degli Esposti M, et al. The specificity of mitochondrial complex I for ubiquinones. Biochem J. 1996;313:327–334. doi: 10.1042/bj3130327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chovancova E, et al. CAVER 3.0: A tool for the analysis of transport pathways in dynamic protein structures. PLOS Comput Biol. 2012;8:e1002708. doi: 10.1371/journal.pcbi.1002708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Armstrong CT, Mason PE, Anderson JLR, Dempsey CE. Arginine side chain interactions and the role of arginine as a gating charge carrier in voltage sensitive ion channels. Sci Rep. 2016;6:21759. doi: 10.1038/srep21759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Galassi VV, Arantes GM. Partition, orientation and mobility of ubiquinones in a lipid bilayer. Biochim Biophys Acta. 2015;1847:1560–1573. doi: 10.1016/j.bbabio.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 36.Genheden S, Ryde U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin Drug Discov. 2015;10:449–461. doi: 10.1517/17460441.2015.1032936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang L, Hermans J. Hydrophilicity of cavities in proteins. Proteins. 1996;24:433–438. doi: 10.1002/(SICI)1097-0134(199604)24:4<433::AID-PROT3>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 38.Nihei C, et al. Purification of active recombinant trypanosome alternative oxidase. FEBS Lett. 2003;538:35–40. doi: 10.1016/s0014-5793(03)00120-0. [DOI] [PubMed] [Google Scholar]
- 39.Schmidt TGM, et al. Development of the Twin-Strep-tag® and its application for purification of recombinant proteins from cell culture supernatants. Protein Expr Purif. 2013;92:54–61. doi: 10.1016/j.pep.2013.08.021. [DOI] [PubMed] [Google Scholar]
- 40.Blaza JN, Serreli R, Jones AJY, Mohammed K, Hirst J. Kinetic evidence against partitioning of the ubiquinone pool and the catalytic relevance of respiratory-chain supercomplexes. Proc Natl Acad Sci USA. 2014;111:15735–15740. doi: 10.1073/pnas.1413855111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaplan RS, Pedersen PL. Determination of microgram quantities of protein in the presence of milligram levels of lipid with amido black 10B. Anal Biochem. 1985;150:97–104. doi: 10.1016/0003-2697(85)90445-2. [DOI] [PubMed] [Google Scholar]
- 42.Smart OS, Neduvelil JG, Wang X, Wallace BA, Sansom MSP. HOLE: A program for the analysis of the pore dimensions of ion channel structural models. J Mol Graph. 1996;14:354–360, 376. doi: 10.1016/s0263-7855(97)00009-x. [DOI] [PubMed] [Google Scholar]
- 43.Phillips JC, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Best RB, et al. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ(1) and χ(2) dihedral angles. J Chem Theory Comput. 2012;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.