Significance
The Plasmodium falciparum circumsporozoite protein (CSP) has been studied for decades as a potential immunogen, but little structural information is available on how antibodies recognize the immunodominant NANP repeats within CSP. The most advanced vaccine candidate is RTS,S, which includes multiple NANP repeats. Here, we analyzed two functional antibodies from an RTS,S trial and determined the number of repeats that interact with the antibody Fab fragments using isothermal titration calorimetry and X-ray crystallography. Using negative-stain electron microscopy, we also established how the antibody binds to the NANP repeat region in a recombinant CSP construct. The structural features outlined here provide a rationale for structure-based immunogen design to improve upon the efficacy of the current RTS,S vaccine.
Keywords: malaria, circumsporozoite protein, antibodies, X-ray crystallography, EM
Abstract
Acquired resistance against antimalarial drugs has further increased the need for an effective malaria vaccine. The current leading candidate, RTS,S, is a recombinant circumsporozoite protein (CSP)-based vaccine against Plasmodium falciparum that contains 19 NANP repeats followed by a thrombospondin repeat domain. Although RTS,S has undergone extensive clinical testing and has progressed through phase III clinical trials, continued efforts are underway to enhance its efficacy and duration of protection. Here, we determined that two monoclonal antibodies (mAbs 311 and 317), isolated from a recent controlled human malaria infection trial exploring a delayed fractional dose, inhibit parasite development in vivo by at least 97%. Crystal structures of antibody fragments (Fabs) 311 and 317 with an (NPNA)3 peptide illustrate their different binding modes. Notwithstanding, one and three of the three NPNA repeats adopt similar well-defined type I β-turns with Fab311 and Fab317, respectively. Furthermore, to explore antibody binding in the context of P. falciparum CSP, we used negative-stain electron microscopy on a recombinant shortened CSP (rsCSP) construct saturated with Fabs. Both complexes display a compact rsCSP with multiple Fabs bound, with the rsCSP–Fab311 complex forming a highly organized helical structure. Together, these structural insights may aid in the design of a next-generation malaria vaccine.
Malaria remains one of the world’s most important public health challenges with an estimated 212 million cases and 429,000 deaths in 2015 (1). Although these numbers have been declining over the past 15 years, further progress is being thwarted by the appearance of drug-resistant parasite strains and insecticide-resistant mosquitoes (2). To promote malaria elimination and eradication campaigns, the WHO has prioritized development of a malaria vaccine (2). The most advanced vaccine at present is RTS,S, a Plasmodium falciparum circumsporozoite protein (CSP)-based vaccine. In a phase III clinical trial, RTS,S reduced the incidence of clinical malaria by ∼51% (over the first 14 mo) in children who were 5–17 mo old when they received the first of three doses (3). Protection was highest immediately after vaccination, waned over time, and was enhanced by a fourth dose 18 mo after dose 3 (4). Over 48 mo (median), vaccine efficacy was 28% following three doses and 36% after four doses (5). Thus, this vaccine represents a major advance for the malaria field, particularly with recent reports of drug-resistant P. falciparum in South East Asia (6). An important objective is to improve and extend the efficacy of antimalarial vaccines by exploiting knowledge of how protective antibodies recognize CSP and how vaccine-induced immunity impacts the parasite life cycle.
RTS,S targets the pre-erythrocytic stage of the P. falciparum life cycle, in which sporozoites are introduced into humans from the mosquito and then migrate to the liver. The sporozoite is coated with CSP, which is required for sporozoite development in infected mosquitos and for adhesion and invasion of hepatocytes in humans (7–9). CSP is comprised of an immunogenic central repeat region flanked by two conserved regions, the N-terminal domain and the C-terminal α-thrombospondin repeat (αTSR) domain with a glycosylphosphatidylinositol (GPI) anchor for attachment to the sporozoite membrane (10–12). The repeat region of the P. falciparum 3D7 strain CSP consists of four NVDP and 38 NANP repeats (13), although the exact number can differ per strain (14, 15), and is predicted to be structurally disordered (16). The first three NVDP repeats are interspersed between the first three NANP repeats, while a fourth is located after the first 20 NANP repeats.
CSP-based vaccine efforts, including the RTS,S vaccine, have focused primarily on these immunogenic repeats. RTS,S contains 19 NANP repeats and the C-terminal αTSR domain without the GPI anchor that are fused to hepatitis B viral surface protein, such that virus-like particles are formed (17, 18). When combined with the adjuvant AS01, very robust immune responses against the repeat region can be obtained shortly following vaccination with serum antibody concentrations of over 100 μg/mL (19). Clinical trials employing the human challenge model indicate that protection may be improved by modulating the dose and administration schedule (20, 21). In a recent phase IIa RTS,S/AS01B controlled human malaria infection (CHMI) trial, the standard administration regimen of three monthly vaccinations resulted in 63% protection, while vaccination with a delayed fractional dose showed 87% protection (21). Over 100 antibodies to the NANP repeats and 20 against the C-terminal region of CSP have been identified by sequencing plasmablast mRNAs derived from both the standard and the delayed-fractional dose arms and from protected and nonprotected individuals. In all groups within the trial, two related IGHV3 heavy-chain variable regions (HV3-30 and HV3-33) predominate in the antibody sequences.
Here, we sought to understand the binding and specificity of mAbs derived from a protected individual within the fractionated dose administration arm of the CHMI trial. Our goal was to understand the nature and specificity of RTS,S-elicited Abs at the molecular level to guide structure-aided vaccine design. We selected two individual NANP-specific human mAbs, 311 and 317 [HV3-33 and HV3-30, with different IGLV1 light chains (LV1-40 and KV1-5, respectively)] for structural study. Both antibodies inhibited infection in vivo using transgenic Plasmodium berghei in which its CSP protein was replaced with P. falciparum CSP. X-ray structures were determined for the NANP repeats in complex with antibody fragments (Fabs) derived from mAbs 311 and 317, and negative-stain electron microscopy (nsEM) evaluated how the NANP repeats were recognized in the context of CSP. Despite the intrinsic disorder of the NANP repeat region, we show that these Fabs recognize distinct secondary structures within the repeat region that likely are in equilibrium with its generally unfolded state in solution or that possibly represent different states on CSP. Furthermore, a supramolecular structure is formed when the antibodies are bound to a recombinant form of CSP. These important structural features provide a platform to redesign the NANP repeat region in the current RTS,S vaccine to improve the efficacy and duration of the immune response.
Results
In Vivo Evaluation of Protection.
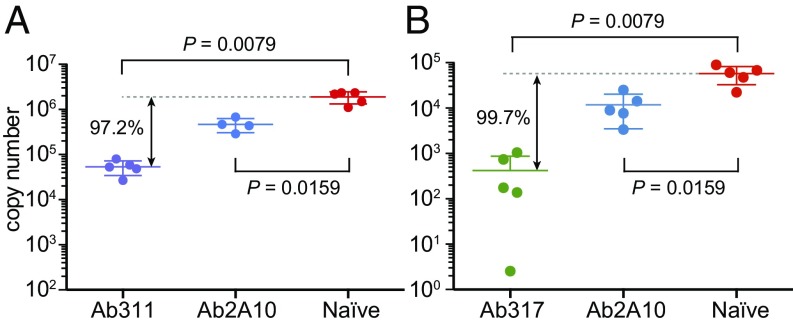
Mice were administered mAbs 311 and 317 at a dose of 100 and 300 μg, respectively, and then were challenged 5–10 min later with P. berghei that was engineered to express full-length CSP from the P. falciparum 3D7 strain (22). The parasite liver load was assessed in the mice at 40 h postchallenge by qPCR for P. berghei-specific 18S rRNA (23). The mAbs 311 and 317 inhibited parasite development by at least 97% (97.2% and 99.7%, respectively) (Fig. 1), compared with only 75–82% inhibition for a previously reported anti-NANP antibody, 2A10 (300 μg dose) (24). Thus, mAbs 311 and 317 represent functional antibodies that are associated with strong protection.
Fig. 1.
Antibody inhibition of malaria infection in mice. Parasite liver load 40 h post challenge with a chimeric P. berghei strain as assessed by qPCR for P. berghei-specific 18S rRNA after administration of Ab311 (100 μg) (A) and Ab317 (300 μg) (B). Significant protection is observed compared with naive mice, with 97.2% and 99.7% inhibition of parasite development for Ab311 and Ab317, respectively, while a previously reported antibody, 2A10 (300 μg dose) (24) showed only 75–82% inhibition of the parasite liver load. The P values were determined using the Mann–Whitney U test. Only four data points are available for 2A10 in A because one mouse died.
Epitope Mapping for Fab Fragments of 317 and 311 mAbs.
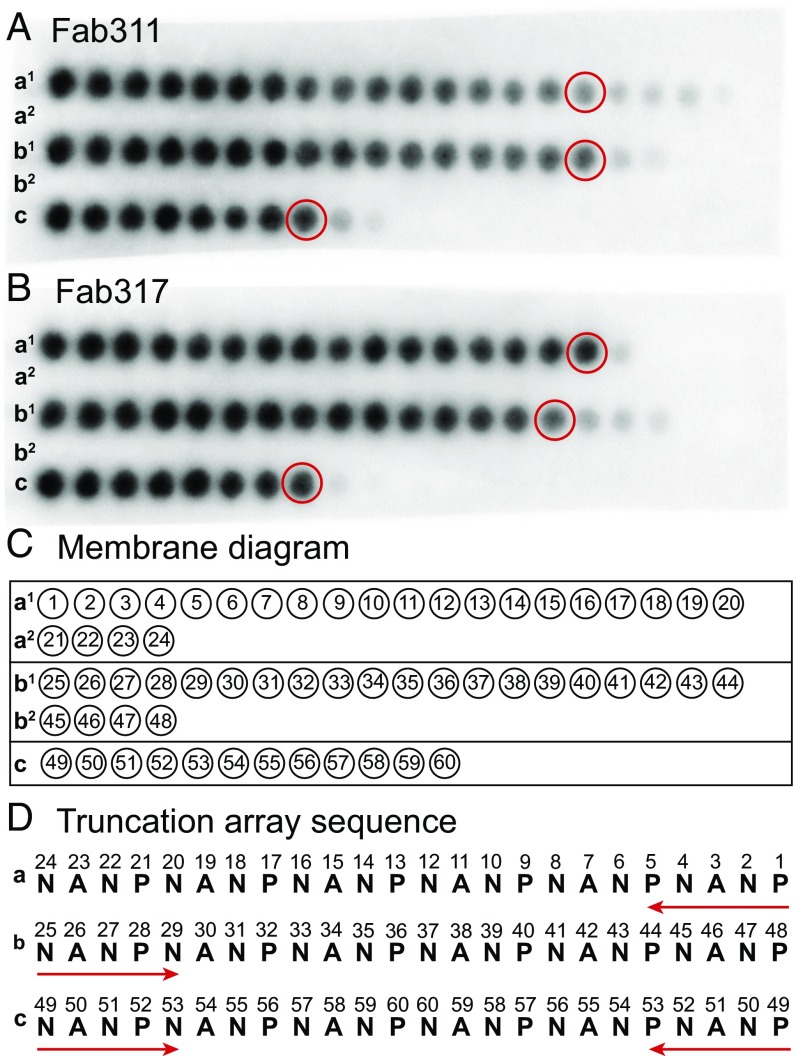
To determine the minimal NANP repeat sequence for binding, a customized peptide array (PepSpot) was designed that contains a series of truncated peptides derived from a 24-mer (NANP)6 peptide, which were spot-synthesized onto a cellulose membrane. Fab311 and Fab317 bind strongly to 10 or more amino acids regardless of the truncation approach, suggesting that a minimum epitope for strong binding consists of around 2.5 NANP repeats (Fig. 2). Additional weak spots for Fab311 indicate it can still bind shorter peptides, down to 1.5 NANP repeats (Fig. 2), whereas Fab317 requires a minimum of two NANP repeats. We therefore selected a 12-mer Ac-NPNANPNANPNA-NH2 peptide, (NPNA)3, for structural and binding studies.
Fig. 2.
Epitope mapping using truncation peptide arrays. (A and B) The PepSpot membrane is shown for Fab311 (A) and Fab317 (B) and consists of five rows of the spotted peptides (a1, a2, b1, b2, and c). Dark spots indicate strong Fab binding. (C and D) Schematic of the location of the peptide spots on the membrane. The numbers within the circles refer to the numbers in the truncation array sequence (D). Rows a1 and a2 correspond to a truncation array starting from the C terminus of the (NANP)6 peptide, rows b1 and b2 are truncations from the N terminus, and row c represents truncations from both the N terminus and C terminus simultaneously. The peptides that appear to have the minimal number of repeats for strong Fab binding are circled in red in A and B.
Peptide Affinity Measurements.
The (NPNA)3 peptides bind strongly to the antibodies with Kds of 305 ± 60 nM for Fab311 and 78 ± 16 nM for Fab317 (Table 1 and Fig. S1). To determine the minimal epitope for optimal binding, binding affinities for an (NPNA)2 peptide were measured. The Kd for (NPNA)2 binding to Fab311 (296 ± 19 nM) is almost identical to that of the (NPNA)3 peptide, whereas the Kd for (NPNA)2 binding to Fab317 increased 2.2-fold (173 ± 8 nM). Although NVDP repeats were not present in the RTS,S vaccine, we decided to test whether such a sequence could be accommodated by the antibodies, because they are present in the wild-type CSP. Fab311 has approximately fivefold lower affinity for peptides Ac-NPNVDPNANPNV-NH2 and Ac-DPNANPNVDPNA-NH2 (Kds of 1.79 ± 0.19 μM and 1.37 ± 0.39 μM, respectively), whereas Fab317 bound to these same peptides with substantially lower (Kd of 12.09 ± 2.49 μM) and fourfold higher (Kd of 0.45 ± 0.07 μM) affinity, respectively (Table 1 and Fig. S2).
Table 1.
Dissociation constants for NANP/NVDP repeat-containing peptides obtained from ITC affinity measurements
| Binding parameter | (NPNA)3 | (NPNA)2 | NPNVDPNANPNV | DPNANPNVDPNA |
| Fab311 | ||||
| No. of sites | 0.74 ± 0.02 | 1.50 ± 0.08 | 1.24 ± 0.02 | 1.20 ± 0.03 |
| Kd, μM | 0.305 ± 0.060 | 0.296 ± 0.019 | 1.787 ± 0.189 | 1.368 ± 0.390 |
| ∆H, cal/mol | −29,823 ± 261 | −16,363 ± 261 | −18,657 ± 378 | −20,200 ± 1,078 |
| ∆S, cal−1⋅mol−1⋅degree−1 | −70 ± 1 | −25 ± 1 | −36 ± 2 | −41 ± 4 |
| Fab317 | ||||
| No. of sites | 1.27 ± 0.02 | 1.41 ± 0.05 | 1.72 ± 0.13* | 1.64 ± 0.05 |
| Kd, μM | 0.078 ± 0.016 | 0.173 ± 0.008 | 12.091 ± 2.488* | 0.448 ± 0.068 |
| ∆H, cal/mol | −15,700 ± 225 | −12,090 ± 221 | −9,657 ± 426* | −9,387 ± 152 |
| ∆S, cal−1⋅mol−1⋅degree−1 | −20 ± 1 | −10 ± 1 | −10 ± 2* | −2 ± 1 |
All values are the average and SD of triplicate experiments except for those with an asterisk (*), which are from duplicate experiments.
Crystal Structures of Fab311 and Fab317 in Complex with the (NPNA)3 Peptide.
Fab311 and Fab317 crystals with the (NPNA)3 peptide diffracted to 2.1 Å and 2.4 Å, respectively. Fab311–peptide crystallized in space group P212121 with a single complex in the crystal asymmetric unit (asu), whereas Fab317–peptide crystallized in space group P21 with two complexes in the asu. The Fabs were numbered with the Kabat system, and the (NPNA)3 peptides were numbered from 2 to 13, with the N-terminal acetyl group and C-terminal NH2 group being assigned as residues 1 and 14, respectively.
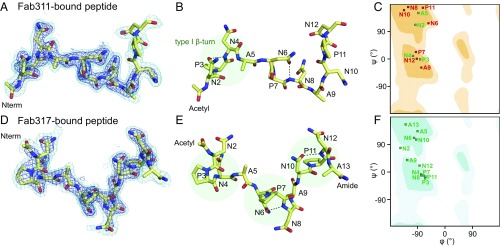
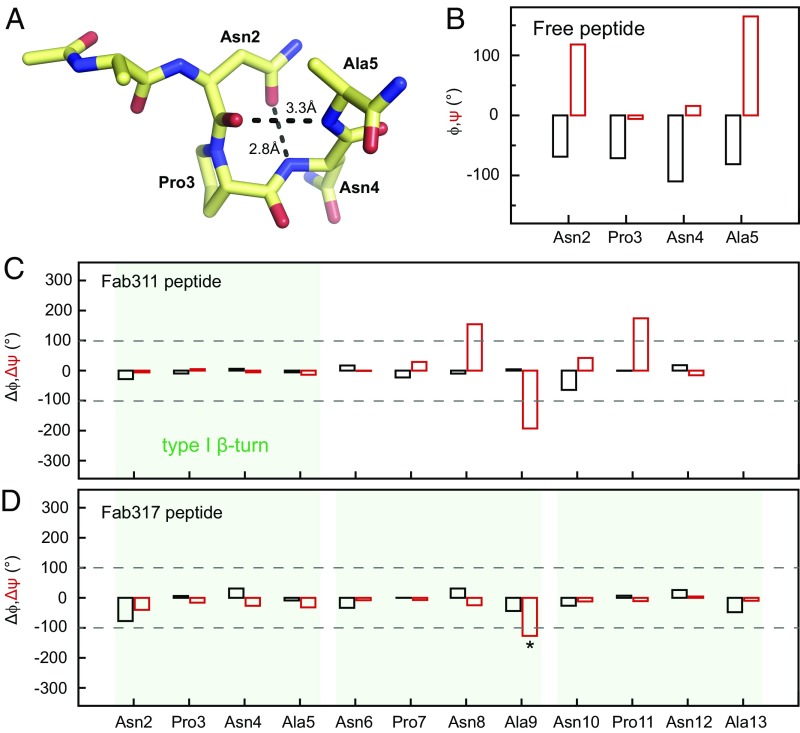
The electron density for the peptides in both complexes is generally well defined. The (NPNA)3 peptide forms largely extended structures in both antibody-binding sites with kinks in the peptide that arise from defined secondary structures (Fig. 3). Dihedral angle analysis shows that type I β-turns (residues i to i + 3) are found for the NPNA repeats, which are stabilized by hydrogen bonding of the first Asn side chain (residue i) to the backbone amide of the next Asn (i + 2) (Fig. 4). The N-terminal NPNA repeats of both peptides adopt type I β-turns. While this cadence of type I β-turns continues in the Fab317-bound peptide, the Fab311-bound peptide maintains a more extended structure for the second and third NPNA repeats, both of which deviate from canonical type I β-turns (Fig. 4 B and E). Inspection of 2Fo-Fc maps contoured at 2.0σ and 0.8σ (Fig. 4 A and D) reveals that the electron density for the peptide bound to Fab311 is weaker after Ala9 and absent for the C-terminal Ala13. In contrast, the (NPNA)3 peptide bound to Fab317 is well defined for all peptide residues except Ala13.
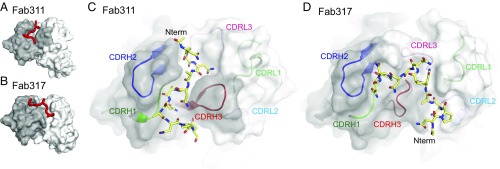
Fig. 3.
Crystal structures of (NPNA)3 peptides in complex with Fab311 and Fab317. (A and B) Surface representation of the variable domains of Fab311 (A) and Fab317 (B) with the (NPNA)3 peptide represented by a red tube. The heavy- and light-chain variable domains are colored dark and light gray, respectively. (C and D) Paratope representation for Fab311 (C) and Fab317 (D) with a transparent dark gray surface for the heavy chain and a transparent light gray surface for the light chain. The underlying CDR loops are shown in cartoon representation and are colored green (H1), blue (H2), red (H3), light green (L1), light blue (L2), and pink (L3). The (NPNA)3 peptide is shown in stick representation (yellow carbons). The N terminus of each peptide is indicated (Nterm).
Fig. 4.
Structural analysis of antibody-bound peptides. (A and D) 2Fo-Fc electron density maps contoured at 2.0σ (blue) and 0.8σ (cyan) for peptide bound to Fab311 (A) and Fab317 (D). The peptide is shown in stick representation (yellow carbons). (B and E) Type I β-turns are highlighted by transparent green circles for peptide bound to Fab311 (B) and Fab317 (E). Intrapeptide hydrogen bonds that emulate a pseudo 310 turn between the first Asn sidechain and amide backbone of the third residue in the turn are shown as black dashed lines. (C and F) Ramachandran plots for the dihedral angles of Fab311-bound peptide (C) and Fab317-bound peptide (F). Residues that have typical dihedral angles indicative of canonical NPNA type I β-turns are colored green; otherwise they are colored red. The β-sheet region is in the dark shaded region of the plot in the upper left quadrant, and the α-helical region is in the central region on the left around ψ of −30°. The Fab311-bound and Fab317-bound peptides have one and three canonical type I β-turns, respectively.
The peptide interaction with the complementarity-determining region (CDR) loops is very different in each structure, however. In Fab311, the peptide resides in a groove that runs parallel to the heavy and light chains. The peptide N-terminal region is located between CDR H2 and CDR L3, and the peptide then threads between CDR H2, CDR H3, and CDR H1. The Fab311 CDR H3 (12 residues) helps form the groove in which the peptide resides (Fig. 3 A and C). The heavy chain contributes 80.2% of the buried surface area (BSA) (574 Å2 on the Fab and 520 Å2 on the peptide) from CDR H1 (17.4%), H2 (30.5%), and H3 (32.3%), while the light chain contributes only 19.3%, mainly CDR L3 (17.3%). The remaining 0.5% is contributed by a heavy-chain framework residue. In Fab317, the peptide binds in a more perpendicular mode compared with Fab311 and occupies a relatively shallow binding pocket that initially starts between the light and heavy chains and then bends sharply and traverses from the light to the heavy chain (Fig. 3 B and D). The heavy chain contributes 58.2% of the BSA from CDR H1 (5.4%), H2 (21.7%), and H3 (31.1%), while the light chain contributes 39.3% from CDR L1 (10.9%), L2 (7.7%), and L3 (20.7%), with the remaining 2.5% from a light-chain framework residue. The BSAs on Fab317 and (NPNA)3 peptide are 533 Å2 and 580 Å2, respectively. Since CDR H3 and CDR L3 contribute ∼30% and ∼20%, respectively, of the BSA for both Fab311 and Fab317, differences in the binding mode arise mainly from different interactions with light- and heavy-chain CDR1 and CDR2.
Hydrogen-bonding networks and amino acid BSAs highlight additional differences. Most of the hydrogen bonds between Fab311 and peptide are made with the C-terminal half of the peptide (Table S2), although its N-terminal region (Pro3, Asn4, and Ala5) engages Fab311 through a hydrogen-bonding network that includes four interfacial water molecules (Fig. S3). Furthermore, Pro3 and Pro7 of the (NPNA)3 peptide are buried in the Fab311 paratope groove with BSAs of 76 Å2 and 75 Å2, respectively, and engage in CH/π interactions with Phe58H and Trp52H (Fig. S3). The peptide-binding site on Fab317 does not contain any observable interfacial water molecules. Hydrogen-bonding partners with Fab317 are evenly distributed throughout the peptide (Asn4, Asn6, Ala9, Pro11, and Asn12) (Table S2). In contrast, Pro3 and Pro7 have small BSAs of 22 Å2 and 10 Å2, respectively. Pro11 has a larger BSA of 78 Å2, compared with only 7 Å2 in the Fab311–peptide complex.
nsEM of CSP with Protective Antibodies.
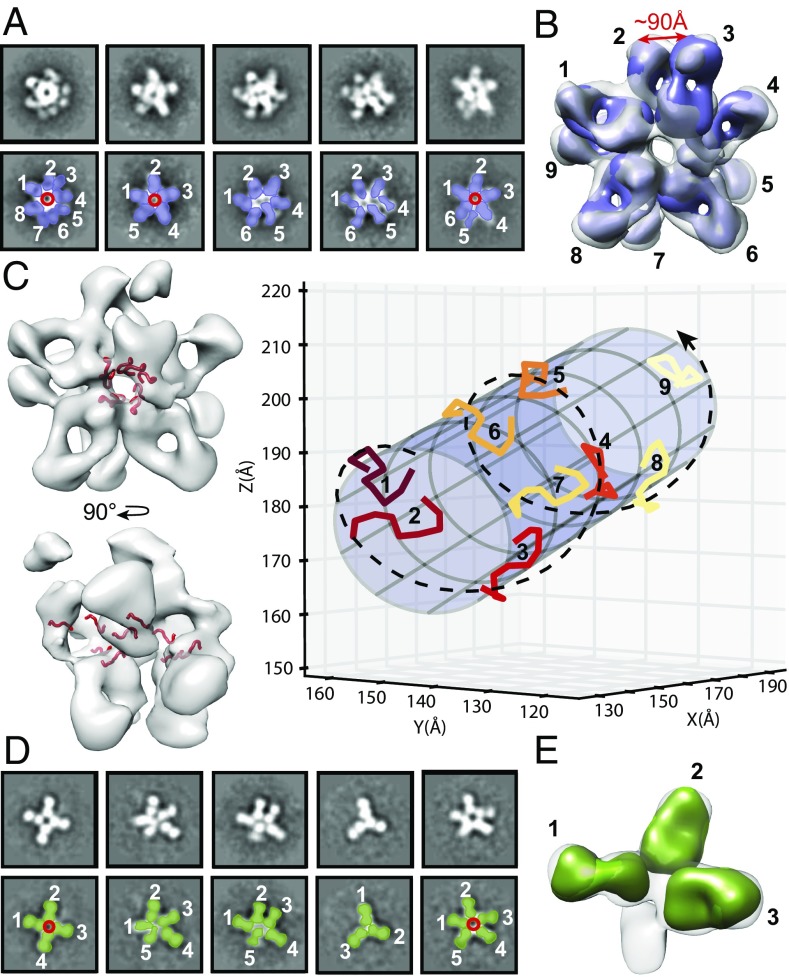
Single-particle nsEM was then utilized to visualize the stoichiometry and overall molecular organization of the CSP–Fab complexes using an engineered recombinant version of CSP with a reduced number of NANP/NVDP repeats [hereafter, “recombinant shortened CSP” (rsCSP): 19/3 repeats instead of 38/4 for the P. falciparum 3D7 strain]. Visual inspection of the 2D class averages for the Fab311–rsCSP complex indicate a binding stoichiometry of five or more per rsCSP molecule (Fig. 5A), while in Fab317–rsCSP up to five Fabs are bound (Fig. 5D and Fig. S4). Further 3D classification and refinement of the Fab311–rsCSP complex data using C1 symmetry revealed up to nine Fabs bound per rsCSP molecule (Fig. 5B). However, at lower threshold values of the negative-stain map, additional Fab densities became visible (threshold values of 0.274, 0.223, and 0.179 reveal five, eight, and nine Fabs per complex, respectively), perhaps as a result of substoichiometric complexes that were averaged with the rest of the data. Docking the Fab311–(NPNA)3 crystal structures into the EM density map shows that the peptides and, by extension, rsCSP adopt an extended, left-handed, helical conformation with a fitted radius of ∼15 Å when bound to this antibody (Fig. 5C and Fig. S4C). The Ala9 and Asn2 Cα atoms in the crystal structure of two adjacent (NPNA)3 peptides are, on average, 13 ± 2 Å apart when fitted into the EM reconstruction, and about five peptides complete one turn around the helical axis. In total, we observe two full turns that comprise a distance of 97 Å between the N termini of the first and last peptides in the helical structure (Fig. 5 B and C). Notably, there is little or no density in the map that would correspond to the N- or C-terminal domains of the rsCSP, which may be the result of intrinsic disorder or flexibility relative to the ordered NANP–Fab–bound repeats. In contrast, the Fab317–rsCSP complex had fewer Fabs bound, and therefore a smaller portion of the structure was resolved. While 2D classification demonstrates up to five Fabs bound, the 3D refinement converged to three Fabs bound per rsCSP, which may be attributable to various stoichiometries, as seen in the class averages (Fig. 5 D and E). Similar to Fab311–rsCSP, the Fabs in the Fab317–rsCSP complex are in very close proximity. The differences in overall shape between the two complexes indicate some plasticity of rsCSP. We do not know whether its conformation is influenced by Fab binding, as the unliganded CSP structure is not known.
Fig. 5.
nsEM for rsCSP bound to Fab311 and Fab317. (A and D) Five selected representative class averages for the rsCSP–Fab311 (A) and rsCSP–Fab317 (D) complexes, false colored to show the location of rsCSP (red), Fab311 (purple), and Fab317 (green). Fabs are labeled by number in white. (B) The refined 3D model confirmed the presence of multiple densities for Fab311, for which a total count of nine Fab311s could be observed at the low threshold level. The distance between the heavy-chain C termini of Fab311 nos. 2 and 3 was measured at 90 Å, which could be accommodated in an IgG. (C) The (NPNA)3 peptide (red) from the Fab311 crystal structure was docked into the 3D model (Left) and revealed a helical shape looking into the central hole of the rsCSP complex and along its length (Right). The docked peptides in the nsEM map for the rsCSP–Fab311 complex were fitted to a cylinder with a radius of 15.2 Å. The peptides are colored using a color progression, and the helical organization is shown by the dashed spiral line. (D and E) Reaching convergence for the rsCSP–Fab317 complex (E) was difficult due to the various stoichiometries seen in the 2D class averages (D). The unmodeled blob may be a remnant of a fourth Fab fragment that is present in some of the complexes.
Discussion
The Minimal Epitope for Binding Consists of Two or Three NPNA Repeats.
The buried surface areas for the (NPNA)3 peptides bound to Fab311 and Fab317 indicate that all residues are engaged in Fab contacts except for Ala13 with Fab311 (Fig. S5). However, the main epitope of the Fab311-bound peptide is likely smaller than three NPNA repeats, as the last four residues have much weaker electron density and are likely more disordered (Fig. 4A). PepSpot epitope and isothermal titration calorimetry (ITC) affinity measurements (Fig. 2 and Table 1) confirm the hypothesis that the minimum epitope for optimal binding is two and three NPNA units for Fab311 and Fab317, respectively. These results are consistent with a previous study in which (NANP)3 peptides were able to efficiently inhibit binding of anti-NANP mAbs to P. falciparum sporozoites (25).
NPNA Repeats Can Adopt Type I β-Turns in Solution and in Complex with Antibody.
Previous studies on the NANP repeats focused on the unbound state using NMR spectroscopy (26) and X-ray crystallography (27) in which the NPNA units (residues i to i + 3) within NANP peptides of different lengths were in a dynamic equilibrium between a disordered state and an ordered type I β-turn. The highly populated type I β-turn is stabilized by a hydrogen bond between the side-chain of Asn (residue i) and the backbone amide of the next Asn (residue i + 2). Our crystal structures reveal that the bound peptides also adopt NPNA type I β-turns (Fig. 4). Specifically, peptides bound to Fab311 and Fab317 both have their N-terminal NPNA repeat in a canonical type I β-turn. The Fab317-bound peptide continues to display type I β-turns in a NPNA-like cadence, whereas the additional NPNA repeats in the Fab311-bound peptide have dihedral angles that deviate from a canonical type I β-turn (Fig. 4 C and F). Comparison of the dihedral angles for each NPNA repeat in our peptide to the ANPNA crystal structure (27) establishes that the type I β-turns in our Fab-bound peptides are nearly identical in conformation (Fig. 6). The peptide structures bound to Fabs also agree with NMR solution experiments that the CSP repeat region follows a cadence of NPNA repeats with each repeat having a high probability of forming a type I β-turn (26). Although the central NPNA repeat in the peptide bound to Fab311 does not form a canonical type I β-turn, the Asn6 side chain still hydrogen bonds to the Asn8 backbone amide (Fig. 4B). Interestingly, the relative orientation of this central NPNA repeat to the heavy-chain variable CDR loops is similar to that of the third NPNA repeat in the Fab317-bound peptide, which is expected since their heavy-chain germline genes are closely related (Fig. S6). Furthermore, a high Pearson correlation coefficient between the BSAs of the backbone and side-chain atoms for the second NPNA repeat of the Fab311 (NPNA)3 peptide with the third NPNA unit of the Fab317 (NPNA)3 peptide indicates a striking similarity in their binding mode (Fig. S7). These results are corroborated by similar hydrogen-bond networks for these residues, although the donor/acceptor residues on the Fab fragment are different (Table S2). Thus, elicitation of these antibodies may have originated by recognition of a single type I β-turn from which the antibodies independently evolved to increase the number of contacts and thus their affinity for the peptide repeats.
Fig. 6.
Comparison of dihedral angles shows similarities between the bound and free peptides. (A) X-ray structure of the free ANPNA peptide shows a type I β-turn in which the Asn2 (residue i) OD1 also hydrogen bonds to the backbone amide of Asn4 (i + 2) (27). (B) Plot of the dihedral angles for the NPNA unit in the ANPNA X-ray structure; ϕ and ψ are shown in black and red, respectively. (C and D) Plots of the dihedral angle differences between each of the NPNA units for the peptide bound to Fab311 (C) or Fab317 (D) and the NPNA unit of the free peptide; Δϕ and Δψ are shown in black and red, respectively. Type I β-turns are highlighted by transparent green boxes. The dihedral angle differences are relatively small within each NPNA type I β-turn, except for Ala9 Δψ in the Fab317 peptide (asterisk). This deviation from the NPNA type I β-turn in solution reflects a change in direction at the end of the NPNA repeat rather than a disruption of the canonical type I β-turn.
The CSP Antigen Adopts a Collapsed Conformation in Its Antibody-Bound Form.
To date, the handful of studies that have attempted to elucidate the structure of full-length CSP, suggest that CSP exists in two conformations: open and collapsed (28, 29). CSP is speculated to be present in the collapsed form on the sporozoite surface during migration to the liver and in the open form when it fully extends to engage the host cell receptor for entry (28). However, determining the structure of full-length CSP has been challenging, which is not surprising since ∼75% of the CSP sequence, including the entire central NANP repeat region and part of the N-terminal domain, is predicted to be disordered (16). Here, we were able to obtain high-quality nsEM images for rsCSP saturated with Fab311 or Fab317. Despite the proposed intrinsic disorder of free CSP, both complexes display a collapsed state of the antigen with closely apposed Fabs observed in the EM projection. The rsCSP–Fab311 complex, in particular, adopts a large supramolecular structure that appears to form a spiral with the Fabs radiating tangentially from the surface of the CSP repeats (Fig. 5 B and C). Many questions remain as to whether this same structure is formed on the sporozoite surface or whether antibodies help induce this particular conformation. The C termini of the heavy chains of Fab311 are 90 Å apart (Fig. 5B), suggesting that an individual IgG is capable of binding to two adjacent epitopes within a single rsCSP antigen. Alternatively, given the avidity effects of having two antigen-binding modules (Fabs) per antibody, antibodies may be able to crosslink CSP molecules on the sporozoite surface. Notwithstanding, transition between CSP topologies may be important for the infectious lifecycle of malaria, and therefore the antibodies described here and others may be useful probes to further understand its mechanism of action.
Epitope Identification and Binding Stoichiometry.
The nsEM reconstructions indicate that up to nine copies of Fab311 are present on rsCSP, compared with only five for Fab317. From our epitope-mapping experiments, the crystal structure of the Fab311–peptide complex, and ITC affinity measurements, we established that the minimum Fab311 epitope consists of two NPNA repeats. Since the total number of NANP repeats in rsCSP is 19 with three interspersed NVDP repeats, that leaves a stretch of 16 NANP repeats or eight epitopes available for binding, which is slightly less than the nine Fabs that we observe in the nsEM map. There are two possible explanations for these observations. First, Fab311 can also bind with reasonable affinity to at least one of the NVDP repeats on CSP even though these sequences are not present on the RTS,S vaccine. If these NVDP repeats were fully available for antibody binding, the number of potential epitopes could increase to 11 [(19 NANP + 3 NVDP)/2]. A second possibility is that Fab311 can bind with reasonable affinity to single NANP repeats that are flanked by NVDP repeats. To further investigate the binding differences observed in the nsEM reconstructions between the antibodies, we performed ITC experiments to measure the binding of Fab311 and Fab317 to peptides that contained both NANP and NVDP repeats. Affinity measurements confirm that Fab311 has a reasonable affinity for the NVDP region, with Kds of 1.37 and 1.79 μM for the Ac-DPNANPNVDPNA-NH2 and Ac-NPNVDPNANPNV-NH2 peptides, respectively, compared with 0.45 and 12.09 μM for Fab317. Although Fab317 is able to bind the peptide tightly when the NVDP repeat is downstream from an NANP repeat, binding is abrogated when the order is reversed. Hence, the different binding stoichiometry for Fab311 and Fab317 can more likely be rationalized by the preferred minimum Fab317 epitope consisting of about three NPNA repeats and probably no binding to the NVDP repeat region. Thus, approximately five [(19 NANP – 3 NANP)/3] binding sites are available, which is exactly what we observe with nsEM.
Implications for Vaccine Efforts and Injectable Biologics.
Our crystal structures show both similarities and differences in the way the same peptide is recognized by the two antibodies. The antibodies recognize the NPNA repeats as well-defined β-turns, modified turns, or as more extended structures. Additionally, nsEM reconstructions show that the NANP repeat region can be further stabilized by antibody interactions in the context of the rsCSP. These findings provide exciting opportunities for structure-assisted immunogen design. Since NMR data suggest that these type I β-turn conformations represent a major population of the NPNA repeats in solution, and since we observe NPNA type I β-turns in peptides bound to highly protective antibodies, stabilizing these secondary structure elements in next-generation CSP immunogens may elicit an improved antibody response. Indeed, attempts to stabilize type I β-turns in the NANP repeat go back to 1990 with covalent hydrogen-bond mimics (30). Five years later, the NPNA type I β-turn conformation was stabilized in solution by replacing proline with (S)-α-methylproline (31). Both approaches elicited polyclonal antisera in rabbits against the designed/synthetized peptides that recognized intact sporozoites. We propose that rational NANP-based immunogen design may start at the level of stabilizing the NPNA type I β-turns and gradually build in more complexity to mimic the superstructure of the repeat units that we observe by nsEM.
Finally, efforts are underway to determine if mAbs themselves can be used as interventions to prevent malaria parasite infection. To accomplish this goal, the mAbs will need to be optimized to achieve protection at the lowest possible serum concentration. The structures presented here provide a good starting point for further mAb engineering and for identifying mAbs with improved binding characteristics. As a single sporozoite is covered with hundreds of CSP molecules, which are continually shed (32), it is beneficial to use as little antibody as possible per sporozoite to target all sporozoites that enter the body. In this study, Fab317 would appear to be the more promising of the two antibodies in that regard. Further studies will assess whether other epitopes are available for both immunogen design and for antibody parsimony.
Materials and Methods
Detailed material and methods are provided in SI Materials and Methods.
Evaluation of the Protective Activity of mAbs.
mAbs 311 and 317 were administered to 7- to 8-wk-old C57BL/6 mice by i.v. injection before challenge with P. berghei chimeric sporozoites expressing P. falciparum CSP (3D7 strain) (22). Antibody inhibition of parasite development was analyzed by comparing the parasite liver burdens of experimental and naïve control mice. C57Bl/6 mice were housed in the animal facility of the Johns Hopkins Bloomberg School of Public Health. All animal procedures were approved by the Animal Care and Use Committee at Johns Hopkins University, protocol no. MO16H35.
Protein Production.
Fab311 and Fab317 were expressed in FreeStyle 293F cells (Invitrogen) and purified using a lambda or kappa affinity column (GE Healthcare) followed by cation exchange chromatography (monoS). rsCSP was expressed in Escherichia coli (SHUFFLE cells; New England Biolabs) and purified as described (33). The rsCSP construct has three NVDP repeats interspersed with three NANP repeats followed by a continuous region of 16 NANP repeats.
PepSpot Analysis.
A customized (NANP)6 peptide truncation array was purchased from JPT Peptide Technologies GmbH. Binding of Fab311 and Fab317 was visualized using HRP-conjugated anti-human Fab secondary antibodies.
ITC.
ITC affinity measurements were performed using a MicroCal Auto-iTC200 instrument (GE Healthcare).
X-Ray Crystallography.
Fab–(NPNA)3 peptide complexes were made by adding the peptide to the Fab in a 5:1 molar ratio. Crystals were obtained by sitting-drop vapor diffusion. X-ray diffraction data were collected at the Stanford Synchrotron Radiation Lightsource (SSRL) BL12-2 and the Advanced Photon Source (APS) BL23ID-B.
nsEM.
rsCSP was saturated with Fabs and purified by size-exclusion chromatography. Complexes were imaged on a FEI Tecnai Spirit T12 electron microscope equipped with a Tietz TVIPS CMOS camera.
Supplementary Material
Acknowledgments
We thank Erik Jongert and Constantinos Kurt Wibmer for critical reading of the manuscript and Robyn L. Stanfield for technical expertise. This work was funded by PATH’s Malaria Vaccine Initiative under a collaborative agreement with The Scripps Research Institute. The work reported here is dependent on a more comprehensive analysis of antibodies elicited in a CHMI study conducted by a partnership of Walter Reed Army Institute of Research, GlaxoSmithKline Biologicals SA, Atreca Inc., and PATH’s Malaria Vaccine Initiative. The CHMI trial is registered with ClinicalTrials.gov, no. NCT01857869. The SSRL, SLAC National Accelerator Laboratory, is supported by the US Department of Energy (DOE), Office of Science, Office of Basic Energy Sciences under Contract DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research and by NIH National Institute of General Medical Sciences (NIGMS), including Grant P41GM103393. GM/CA @ APS is funded in whole or in part by National Cancer Institute Grant ACB-12002 and NIGMS Grant AGM-12006. This research used the resources of the APS, a US DOE Office of Science User Facility operated by Argonne National Laboratory under Contract DE-AC02-06CH11357. The Eiger 16M detector was funded by NIH Office of Research Infrastructure Programs, High-End Instrumentation Grant 1S10OD012289-01A1. The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the DOE, NIGMS, or NIH.
Footnotes
Conflict of interest statement: W.V. and D.E. are employees of and own equity in Atreca, Inc.
Data deposition: The X-ray structure factors and coordinates have been deposited in the Protein Data Bank [PDB ID codes 6AXK for Fab311-(NPNA)3 and 6AXL for Fab317-(NPNA)3]. The EM reconstructions and maps are deposited in the Electron Microscopy Data Bank [accession codes EMD-7068 (rsCSP + Fab311) and EMD-7069 (rsCSP + Fab317)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1715812114/-/DCSupplemental.
References
- 1.World Health Organization . World Malaria Report 2016. WHO; Geneva: 2016. [Google Scholar]
- 2.WHO Malaria Vaccine Funders Group . Malaria Vaccine Technology Roadmap. WHO; Geneva: 2013. [Google Scholar]
- 3.Agnandji ST, et al. RTS,S Clinical Trials Partnership First results of phase 3 trial of RTS,S/AS01 malaria vaccine in African children. N Engl J Med. 2011;365:1863–1875. doi: 10.1056/NEJMoa1102287. [DOI] [PubMed] [Google Scholar]
- 4.RTS,S Clinical Trials Partnership Efficacy and safety of the RTS,S/AS01 malaria vaccine during 18 months after vaccination: A phase 3 randomized, controlled trial in children and young infants at 11 African sites. PLoS Med. 2014;11:e1001685. doi: 10.1371/journal.pmed.1001685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.RTS,S Clinical Trials Partnership Efficacy and safety of RTS,S/AS01 malaria vaccine with or without a booster dose in infants and children in Africa: Final results of a phase 3, individually randomised, controlled trial. Lancet. 2015;386:31–45. doi: 10.1016/S0140-6736(15)60721-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Imwong M, Hien TT, Thuy-Nhien NT, Dondorp AM, White NJ. Spread of a single multidrug resistant malaria parasite lineage (PfPailin) to Vietnam. Lancet Infect Dis. 2017;17:1022–1023. doi: 10.1016/S1473-3099(17)30524-8. [DOI] [PubMed] [Google Scholar]
- 7.Rich KA, George FW, 4th, Law JL, Martin WJ. Cell-adhesive motif in region II of malarial circumsporozoite protein. Science. 1990;249:1574–1577. doi: 10.1126/science.2120774. [DOI] [PubMed] [Google Scholar]
- 8.Ancsin JB, Kisilevsky R. A binding site for highly sulfated heparan sulfate is identified in the N terminus of the circumsporozoite protein: Significance for malarial sporozoite attachment to hepatocytes. J Biol Chem. 2004;279:21824–21832. doi: 10.1074/jbc.M401979200. [DOI] [PubMed] [Google Scholar]
- 9.Coppi A, et al. Heparan sulfate proteoglycans provide a signal to Plasmodium sporozoites to stop migrating and productively invade host cells. Cell Host Microbe. 2007;2:316–327. doi: 10.1016/j.chom.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nussenzweig V, Nussenzweig RS. Circumsporozoite proteins of malaria parasites. Bull Mem Acad R Med Belg. 1989;144:493–504. [PubMed] [Google Scholar]
- 11.Plassmeyer ML, et al. Structure of the Plasmodium falciparum circumsporozoite protein, a leading malaria vaccine candidate. J Biol Chem. 2009;284:26951–26963. doi: 10.1074/jbc.M109.013706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doud MB, et al. Unexpected fold in the circumsporozoite protein target of malaria vaccines. Proc Natl Acad Sci USA. 2012;109:7817–7822. doi: 10.1073/pnas.1205737109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bowman S, et al. The complete nucleotide sequence of chromosome 3 of Plasmodium falciparum. Nature. 1999;400:532–538. doi: 10.1038/22964. [DOI] [PubMed] [Google Scholar]
- 14.Lockyer MJ, Schwarz RT. Strain variation in the circumsporozoite protein gene of Plasmodium falciparum. Mol Biochem Parasitol. 1987;22:101–108. doi: 10.1016/0166-6851(87)90073-9. [DOI] [PubMed] [Google Scholar]
- 15.Bowman NM, et al. Comparative population structure of Plasmodium falciparum circumsporozoite protein NANP repeat lengths in Lilongwe, Malawi. Sci Rep. 2013;3:1990. doi: 10.1038/srep01990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guy AJ, et al. Insights into the immunological properties of intrinsically disordered malaria proteins using proteome scale predictions. PLoS One. 2015;10:e0141729. doi: 10.1371/journal.pone.0141729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Wilde M, Cohen J. 2001. US Patent 6,169,171.
- 18.Cohen J, Nussenzweig V, Nussenzweig R, Vekemans J, Leach A. From the circumsporozoite protein to the RTS, S/AS candidate vaccine. Hum Vaccin. 2010;6:90–96. doi: 10.4161/hv.6.1.9677. [DOI] [PubMed] [Google Scholar]
- 19.Kester KE, et al. RTS,S Vaccine Evaluation Group Randomized, double-blind, phase 2a trial of falciparum malaria vaccines RTS,S/AS01B and RTS,S/AS02A in malaria-naive adults: Safety, efficacy, and immunologic associates of protection. J Infect Dis. 2009;200:337–346. doi: 10.1086/600120. [DOI] [PubMed] [Google Scholar]
- 20.Stoute JA, et al. A preliminary evaluation of a recombinant circumsporozoite protein vaccine against Plasmodium falciparum malaria. RTS,S Malaria Vaccine Evaluation Group. N Engl J Med. 1997;336:86–91. doi: 10.1056/NEJM199701093360202. [DOI] [PubMed] [Google Scholar]
- 21.Regules JA, et al. Fractional third and fourth dose of RTS,S/AS01 malaria candidate vaccine: A phase 2a controlled human malaria parasite infection and immunogenicity study. J Infect Dis. 2016;214:762–771. doi: 10.1093/infdis/jiw237. [DOI] [PubMed] [Google Scholar]
- 22.Espinosa DA, et al. Robust antibody and CD8(+) T-cell responses induced by P. falciparum CSP adsorbed to cationic liposomal adjuvant CAF09 confer sterilizing immunity against experimental rodent malaria infection. NPJ Vaccines. 2017;2:10. doi: 10.1038/s41541-017-0011-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bruña-Romero O, et al. Detection of malaria liver-stages in mice infected through the bite of a single Anopheles mosquito using a highly sensitive real-time PCR. Int J Parasitol. 2001;31:1499–1502. doi: 10.1016/s0020-7519(01)00265-x. [DOI] [PubMed] [Google Scholar]
- 24.Zavala F, Cochrane AH, Nardin EH, Nussenzweig RS, Nussenzweig V. Circumsporozoite proteins of malaria parasites contain a single immunodominant region with two or more identical epitopes. J Exp Med. 1983;157:1947–1957. doi: 10.1084/jem.157.6.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zavala F, et al. Rationale for development of a synthetic vaccine against Plasmodium falciparum malaria. Science. 1985;228:1436–1440. doi: 10.1126/science.2409595. [DOI] [PubMed] [Google Scholar]
- 26.Dyson HJ, Satterthwait AC, Lerner RA, Wright PE. Conformational preferences of synthetic peptides derived from the immunodominant site of the circumsporozoite protein of Plasmodium falciparum by 1H NMR. Biochemistry. 1990;29:7828–7837. doi: 10.1021/bi00486a008. [DOI] [PubMed] [Google Scholar]
- 27.Ghasparian A, Moehle K, Linden A, Robinson JA. Crystal structure of an NPNA-repeat motif from the circumsporozoite protein of the malaria parasite Plasmodium falciparum. Chem Commun (Camb) 2006:174–176. doi: 10.1039/b510812h. [DOI] [PubMed] [Google Scholar]
- 28.Herrera R, et al. Reversible conformational change in the Plasmodium falciparum circumsporozoite protein masks its adhesion domains. Infect Immun. 2015;83:3771–3780. doi: 10.1128/IAI.02676-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patra AP, Sharma S, Ainavarapu SR. Force spectroscopy of the Plasmodium falciparum vaccine candidate circumsporozoite protein suggests a mechanically pliable repeat region. J Biol Chem. 2017;292:2110–2119. doi: 10.1074/jbc.M116.754796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Satterthwait AC, et al. The conformational restriction of synthetic vaccines for malaria. Bull World Health Organ. 1990;68:17–25. [PMC free article] [PubMed] [Google Scholar]
- 31.Bisang C, et al. Stabilization of type-I β-turn conformations in peptides containing the NPNA-repeat motif of the Plasmodium falciparum circumsporozoite protein by substituting proline for (S)-α-methylproline. J Am Chem Soc. 1995;117:7904–7915. [Google Scholar]
- 32.Stewart MJ, Vanderberg JP. Malaria sporozoites leave behind trails of circumsporozoite protein during gliding motility. J Protozool. 1988;35:389–393. doi: 10.1111/j.1550-7408.1988.tb04115.x. [DOI] [PubMed] [Google Scholar]
- 33.Schwenk R, et al. IgG2 antibodies against a clinical grade Plasmodium falciparum CSP vaccine antigen associate with protection against transgenic sporozoite challenge in mice. PLoS One. 2014;9:e111020. doi: 10.1371/journal.pone.0111020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bruña-Romero O, et al. Detection of malaria liver-stages in mice infected through the bite of a single Anopheles mosquito using a highly sensitive real-time PCR. Int J Parasitol. 2001;31:1499–1502. doi: 10.1016/s0020-7519(01)00265-x. [DOI] [PubMed] [Google Scholar]
- 35.Anthis NJ, Clore GM. Sequence-specific determination of protein and peptide concentrations by absorbance at 205 nm. Protein Sci. 2013;22:851–858. doi: 10.1002/pro.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 37.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Biasini M, et al. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42:W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bordoli L, et al. Protein structure homology modeling using SWISS-MODEL workspace. Nat Protoc. 2009;4:1–13. doi: 10.1038/nprot.2008.197. [DOI] [PubMed] [Google Scholar]
- 40.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 41.Lepore R, Olimpieri PP, Messih MA, Tramontano A. PIGSPro: Prediction of immunoGlobulin structures v2. Nucleic Acids Res. 2017;45:W17–W23. doi: 10.1093/nar/gkx334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Connolly ML. The molecular surface package. J Mol Graph. 1993;11:139–141. doi: 10.1016/0263-7855(93)87010-3. [DOI] [PubMed] [Google Scholar]
- 45.Gelin BR, Karplus M. Side-chain torsional potentials: Effect of dipeptide, protein, and solvent environment. Biochemistry. 1979;18:1256–1268. doi: 10.1021/bi00574a022. [DOI] [PubMed] [Google Scholar]
- 46.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 47.McDonald IK, Thornton JM. Satisfying hydrogen bonding potential in proteins. J Mol Biol. 1994;238:777–793. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
- 48.Lovell SC, et al. Structure validation by Cα geometry: ϕ, ψ and Cβ deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 49.Suloway C, et al. Automated molecular microscopy: The new Leginon system. J Struct Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 50.Lander GC, et al. Appion: An integrated, database-driven pipeline to facilitate EM image processing. J Struct Biol. 2009;166:95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Voss NR, Yoshioka CK, Radermacher M, Potter CS, Carragher B. DoG Picker and TiltPicker: Software tools to facilitate particle selection in single particle electron microscopy. J Struct Biol. 2009;166:205–213. doi: 10.1016/j.jsb.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ogura T, Iwasaki K, Sato C. Topology representing network enables highly accurate classification of protein images taken by cryo electron-microscope without masking. J Struct Biol. 2003;143:185–200. doi: 10.1016/j.jsb.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 53.Scheres SH. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA. cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 55.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.