Rogers et al. (1) compare the sharing of derived alleles among the genomes of Africans, non-Africans, a Neandertal, and a Denisovan to infer the demographic history of archaic humans. They estimate that the effective population size (Ne) of Neandertals was ∼15,000 individuals and that Neandertals and Denisovans separated from each other shortly after their ancestor separated from modern humans (∼300 generations). These estimates are at odds with previous results (2) that suggested a smaller Ne for Neandertals [ranging from 1,000–5,000 individuals according to pairwise sequentially Markovian coalescent (PSMC) estimates of Ne over time] and a longer common branch for the Neandertal–Denisovan ancestor (5,000–10,000 generations). We note that the difference in assumed mutation rate between the two publications cannot account for the differences in estimates.
A short Neandertal–Denisovan common branch would predict that the divergence between Neandertals and Denisovans is similar to the divergence between Africans and Denisovans. Coalescent simulations show that the Rogers et al. (1) model predicts a ratio of Neandertal–Denisovan to African–Denisovan divergence of 0.85. This is larger than the observed ratio (0.76 using the Altai Neandertal, Denisovan, and an African Mbuti genome). In contrast, the previous model [F(A|B) + PSMC] predicts 0.68–0.79. The high ratio produced by the Rogers et al. (1) model indicates an underestimate of the Neandertal–Denisova common lineage length.
The Rogers et al. (1) model predicts a large Ne for Neandertals that is at odds with the low heterozygosity in the Altai Neandertal. To explain this inconsistency, the authors suggest that Neandertals lived in a highly substructured population. However, further information is available in the form of published low-coverage Neandertal genomes (2, 3). These genomes showed a smaller divergence from the Altai Neandertal genome (2.5–3% of the divergence between humans and the human–chimpanzee ancestor) than the expected divergence of two random Neandertal lineages under the model proposed by Rogers et al. (1) (∼4%). For comparison, two individuals sampled from a population with the demographic history of the highly inbred Altai Neandertal (inferred via PSMC) would yield an estimate of 1.1%.
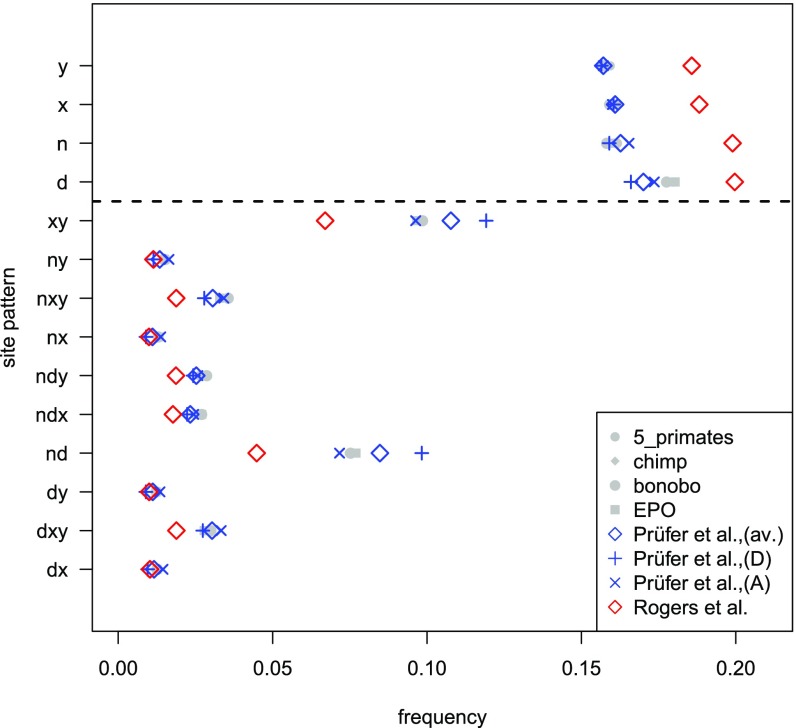
Rogers et al. (1) used the relative frequency of patterns of shared derived changes but ignored derived changes that are specific to one of the archaic or modern humans. While their model produces a marginally better fit for the frequencies of shared derived patterns, we find that the previous model fits the data substantially better when considering all derived changes (Fig. 1). This indicates that Rogers et al. (1) fit statistics that are not sufficiently informative to infer demography and split times.
Fig. 1.
Frequencies of 14 site patterns among the genomes of the Altai Neandertal (n), the Denisovan (d), a Yoruban (x), and a French (y) from ref. 2. Derived alleles in the data (gray) were determined using chimpanzee or bonobo as outgroup, or using the Enredo–Pecan–Ortheus (EPO) inferred chimpanzee ancestor or an inferred ancestor according to five primate outgroup genomes. Coalescent simulations using the estimates from Prüfer et al. (2) are shown in blue [(D), split estimates F(Yoruban|Denisova) and F(Altai|Denisova); (A), split estimates F(Yoruban|Altai) and F(Denisova|Altai); (av.), average of archaic and Yoruban split estimates] and Rogers et al. (1) in red.
We conclude that the Rogers et al. (1) model does not capture some of the basic features of available data. A relatively small effective population size for Neandertals and a longer shared branch for Neandertals and Denisova remain better supported.
Supplementary Material
Acknowledgments
We thank Svante Pääbo, Nick Patterson, and David Reich for helpful comments.
Footnotes
The authors declare no conflict of interest.
Data deposition: Data and simulation scripts are available at https://bioinf.eva.mpg.de/comment_rogers.
References
- 1.Rogers AR, Bohlender RJ, Huff CD. Early history of Neanderthals and Denisovans. Proc Natl Acad Sci USA. 2017;114:9859–9863. doi: 10.1073/pnas.1706426114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prüfer K, et al. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature. 2014;505:43–49. doi: 10.1038/nature12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Green RE, et al. A draft sequence of the Neandertal genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]