Summary
Caloric restriction, leanness and decreased activity of insulin/insulin-like growth factor 1 (IGF-1) receptor signaling are associated with increased longevity in a wide range of organisms from Caenorhabditis elegans to humans. Fat-specific insulin receptor knock-out (FIRKO) mice represent an interesting dichotomy, with leanness and increased lifespan, despite normal or increased food intake. To determine the mechanisms by which a lack of insulin signaling in adipose tissue might exert this effect, we performed physiological and gene expression studies in FIRKO and control mice as they aged. At the whole body level, FIRKO mice demonstrated an increase in basal metabolic rate and respiratory exchange ratio. Analysis of gene expression in white adipose tissue (WAT) of FIRKO mice from 6 to 36 months of age revealed persistently high expression of the nuclear-encoded mitochondrial genes involved in glycolysis, tricarboxylic acid cycle, β-oxidation and oxidative phosphorylation as compared to expression of the same genes in WAT from controls that showed a tendency to decline in expression with age. These changes in gene expression were correlated with increased cytochrome c and cytochrome c oxidase subunit IV at the protein level, increased citrate synthase activity, increased expression of peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α) and PGC-1β, and an increase in mitochondrial DNA in WAT of FIRKO mice. Together, these data suggest that maintenance of mitochondrial activity and metabolic rates in adipose tissue may be important contributors to the increased lifespan of the FIRKO mouse.
Keywords: calorie restriction, diabetes, insulin resistance, mitochondria, obesity, PGC-1α
Introduction
While many factors have been suggested to contribute to longevity, studies over the last several years have revealed particularly important roles for the activity of the insulin/insulin-like growth factor 1 (IGF-1) signaling systems in lifespan of diverse organisms, ranging from yeast to rodents (Katic & Kahn, 2005; van Heemst et al., 2005). Thus, Drosophila melanogaster and Caenorhabditis elegans in which insulin/IGF receptor signaling have been disrupted (Dorman et al., 1995; Clancy et al., 2001; Kenyon, 2005), as well as mice with heterozygous knock-out of the IGF-1 receptor (Holzenberger et al., 2003) and mice over-expressing the Klotho gene (Kurosu et al., 2005), which suppresses insulin/IGF-1 signaling, all have increased longevity. Caloric restriction, leanness and small body size have also been long associated with increased lifespan in many species, including rodents and humans (Longo & Finch, 2003; McCulloch & Gems, 2003; Rogina & Helfand, 2004). While the precise connection between these pathways and longevity remains unknown, it is clear that the aging process is regulated by some of the same pathways that affect growth, development and metabolism, and the impact of these pathways on longevity is an evolutionary conserved process.
Recently, we developed a mouse model that incorporates some, but not all of these above features, which allowed us to address the question of whether leanness, independent of caloric restriction, could be important in determination of longevity. Fat-specific insulin receptor knock-out (FIRKO) mice, in which there is a tissue-specific inactivation of insulin receptor signaling in fat only, have lower fat mass and lower body weight than control littermates, and this occurs despite eating at least equivalent amounts of food (Bluher et al., 2002). FIRKO mice are also protected against age-related and hyperphagia-induced obesity, as well as obesity-related glucose intolerance and other metabolic abnormalities (Bluher et al., 2002). Besides these physiological changes, both male and female FIRKO mice have an ~134-day increase in mean lifespan (18%), with parallel increases in median and maximum lifespans (Bluher et al., 2003). In contrast, survival up to 24 months of age was not different in vascular endothelial cell insulin receptor knock-out (VENIRKO) mice as compared to control (Vicent et al., 2003). These results indicate that adipose tissue itself may produce, store or clear some factor or factors, or that insulin resistance in fat can alter metabolism in some manner, which affects the overall life expectancy of the animal.
There are two theories about how leanness and energy metabolism may be associated with longevity (Harman, 1956; Brand, 2000; Speakman et al., 2004). Both consider mitochondrial activity and free-radical production, but involve completely contrary predictions. The ‘rate of living-free-radical theory’ suggests a negative association between longevity and mitochondrial metabolism (Harman, 1956), while the ‘uncoupling to survive’ hypothesis suggests that the correlation with mitochondrial activity (uncoupling/metabolism) is positive (Brand, 2000; Speakman et al., 2004). Both theories are supported by experimental data, making the issue of the role of metabolism in longevity controversial. In the present study, we have assessed mitochondrial activity, energy metabolism and gene expression in FIRKO mice during aging to define the role of these pathways in longevity and determined how isolated insulin resistance in adipose tissue might affect lifespan.
Results
Increased metabolic rate in FIRKO mice
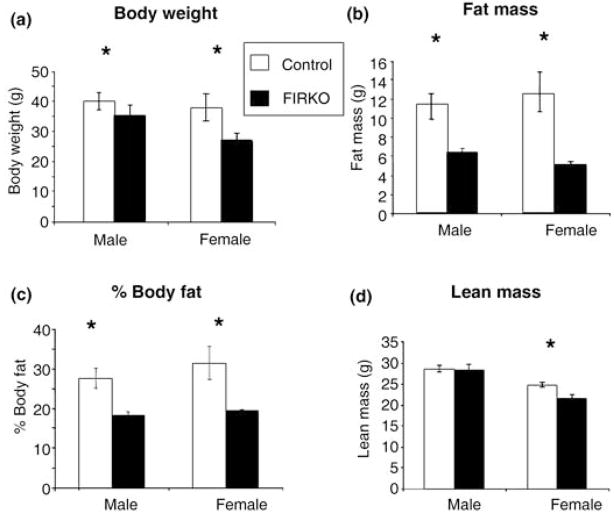
As we have previously shown, FIRKO mice have lower body weight than controls throughout life (Bluher et al., 2002) (Fig. 1a). Dual energy X-ray absorptiometry (DEXA) scans performed in 10-month-old mice demonstrated that in both males and females this difference in body weight was primarily due to a decrease in fat mass as compared to controls (Fig. 1b,c). In addition, female FIRKO mice had a small decrease in lean body mass (Fig. 1d).
Fig. 1.
Body weight and body composition by dual energy X-ray absorptiometry (DEXA) scan of control and fat-specific insulin receptor knock-out (FIRKO) mice. Male and female FIRKO mice had consistently and significantly lower body weights than control mice (P < 0.05). DEXA scan analysis was performed at 10 months of age and decreased fat mass and percentage of body fat in both genders (P < 0.01), as well as decreased lean mass in female FIRKO mice (P < 0.01). Statistical analysis was done using a Student’s t-test (*P < 0.05), and results are expressed as average ± SEM.
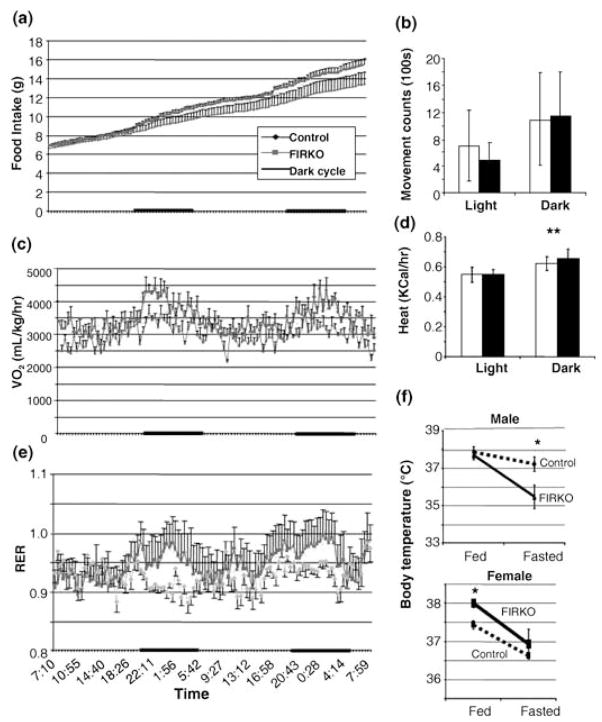
Assessment of cumulative food intake in 10-month-old FIRKO mice using a comprehensive lab animal monitoring system (CLAMS) apparatus revealed that this decrease in body weight and fat mass occurred despite the fact that FIRKO mice had significantly higher (10.9% ± 4.7) food intake than controls (Fig. 2a). In addition, FIRKO mice showed the same (during dark periods) or even lower (during light periods) physical activity throughout the 48-h measurement period (Fig. 2b). Thus, FIRKO mice are lean, while eating more and being less active. In addition, although FIRKO and control mice had similar heat production during light periods, FIRKO mice had higher heat production compared to controls during dark periods, that is, periods of feeding [0.658 ± 0.059 kcal h−1 (FIRKO) vs. 0.620 ± 0.046 kcal h−1 (controls)] (Fig. 2d). In the fed state, female FIRKO mice had higher body temperature than controls (Fig. 2f). This was not observed in the male FIRKO mice, but the latter did exhibit a higher drop in body temperature in the fasted state than control mice.
Fig. 2.
Metabolic parameters in fat-specific insulin receptor knock-out (FIRKO) mice assessed using a CLAMS apparatus. Ten- to 11-month-old male and female mice were acclimated to the apparatus for 2 days, then studied for two additional days. (a) Cumulative food intake was increased (10.9% on average) in FIRKO mice compared to controls. (b) Physical activity measured by light beam breaks was 46% lower (P < 0.005) in FIRKO mice during a light period, but did not differ during the dark period. (c) Oxygen consumption of FIRKO mice expressed per kg body weight was increased 14.5% ± 11.7 and 20.6% ± 10.0 during the light and a dark cycle, respectively (P < 0.001). (d) Heat production was slightly, but significantly higher in FIRKO compared to control mice during the dark period (6.8%, P < 0.005). (e) Respiratory exchange ratio (RER), a measure of total body metabolism, was significantly higher (3.9%, P < 0.001) during the dark period in FIRKO compared to control mice. (f) Basal body temperature was measured in both of male and female mice in the fed and fasted state as described in the Experimental procedures. In (a–e), analysis was done by comparing an average of all light vs. an average of all dark periods for all metabolic parameters. Statistical analysis was done using a Student’s t-test to compare light vs. dark period in each group of animals as well as light vs. light and dark vs. dark periods in different groups of animals, and results are expressed as average ± SEM.
To understand better the mechanism of their sustained lower body weight, oxygen consumption and respiratory exchange ratio (RER) were measured. Oxygen consumption was increased on average by 14.5% (3195 ± 204 mL kg−1 h−1 vs. 2789 ± 261 mL kg−1 h−1) and 20.6% (3812 ± 334 mL kg−1 h−1 vs. 3173 ± 243 mL kg−1 h−1) during light and dark cycles, respectively, in FIRKO mice compared to controls (Fig. 2c). Likewise, the RER was increased by 3.9% in FIRKO mice [0.960 ± 0.021 (FIRKO) vs. 0.924 ± 0.019 (controls), P < 0.001] during the dark period (Fig. 2e). The result for oxygen consumption during a dark cycle was similarly increased when expressed per animal (data not shown). Thus, the lack of insulin signaling in fat not only reduces lipid storage in these depots, but also increases whole body energy metabolism and keeps FIRKO mice lean despite normal food intake.
Analysis of gene expression in white adipose tissue from FIRKO
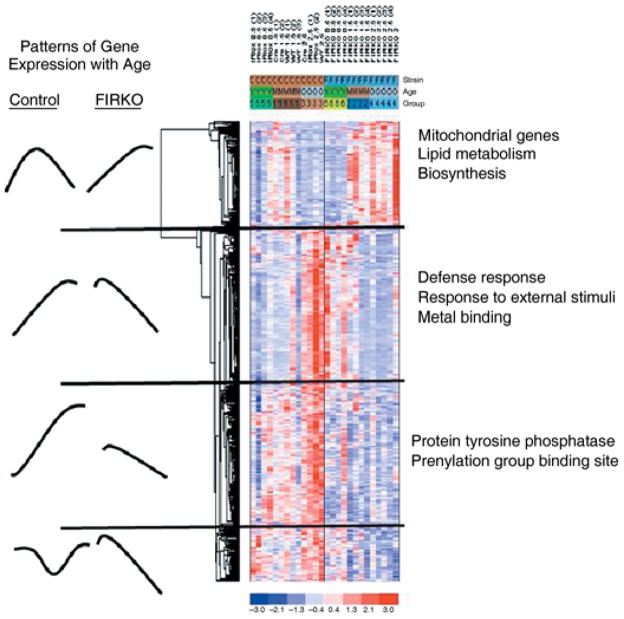
To understand better the observed changes in metabolism of FIRKO mice, and to define the contribution of lack of insulin signaling in white adipose tissue (WAT) on gene expression, we performed a microarray analysis of WAT isolated from young (6 months old), middle-aged (1.5 years old) and old (2.5–3 years old) FIRKO and control mice. This analysis was done using Affymetrix mouse 430A chips (Santa Clara, CA, USA), containing 22 690 probe sets, representing approximately 14 000 genes. Analysis of variance (ANOVA) was performed using a level of significance of P < 0.01 on genes whose expression was significantly changed in fat of 2.5- to 3-year-old FIRKO mice compared to control mice, and those genes whose expression was altered were further subjected to both MappFinder and clustering analysis to determine patterns of expression with age.
MappFinder (www.Grenmapp.org) analysis revealed that the most significantly and the most coordinately changed pathways in WAT of old mice were those that involved mitochondrial electron transport chain and ion transport (Table 1). Clustering analysis confirmed the significant involvement of these pathways and further defined four distinct patterns of expression (Fig. 3). Group I was characterized by genes that increased in middle-aged mice and decreased in old control mice, but progressively increased in adipose of FIRKO mice with age. This group consisted primarily of nuclear-encoded mitochondrial genes involved in oxidative phosphorylation, lipid metabolism and biosynthesis. Group II consisted of genes that increased with age in control mice and decreased with age in FIRKO mice. This group included genes involved in defense mechanisms, response to external stimuli and metal binding. Group III consisted of genes coding for protein tyrosine phosphatases and protein prenylation and had a similar pattern as the Group II, except that expression of genes in the middle-aged control group of mice was higher, and their expression in FIRKO young mice was lower than in Group II. Group IV genes exhibited a pattern with high expression in controls (with a tendency to decreased expression in middle-aged mice), as compared to lower and progressively declining expression in FIRKO mice with age. This group was functionally heterogeneous, and included some genes involved in transcription [e.g. mortality factor 4 like 2 (MORF-related gene X), telomerase binding protein 23 (Tbp23), proliferating cell nuclear antigen (PCNA), histone H1c and H2A histone family member Z], and genes involved in metabolism [e.g. glucose-regulated protein 8, UDP-Gal:βGlcNAc β-1,4-galactosyltransferase, polypeptide 1 and acyl-CoA synthetase long-chain family member 4 (Acsl4) and tyrosine 3-monoxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide (Ywhaz), integrin V α and interferon gamma receptor 2]. The complete data sets are available on the Diabetes Genome Anatomy Program website (www.diabetesgenome.org).
Table 1.
MappFinder analysis of the most significantly and most coordinately changed pathways in old fat-specific insulin receptor knock-out (FIRKO) compared to old control mice
| GO name | Number changed | Number measured | Number in GO* | Changed (%) | % on chip | Z score | Permuted P | Adjusted P |
|---|---|---|---|---|---|---|---|---|
| Cation transporter activity | 45 | 140 | 175 | 80 | 32 | 5.375 | < 0.001 | 0.077 |
| Sodium ion transporter activity | 9 | 13 | 20 | 65 | 69 | 5.303 | < 0.001 | 0.079 |
| NADH dehydrogenase (ubiquinone) activity | 9 | 13 | 24 | 54 | 69 | 5.303 | < 0.001 | 0.079 |
| Primary active transporter activity | 49 | 163 | 192 | 85 | 80 | 5.071 | < 0.001 | 0.208 |
| Mitochondrial electron transport chain | 15 | 32 | 44 | 73 | 47 | 4.848 | < 0.001 | 0.451 |
| Metal ion transporter activity | 16 | 36 | 46 | 78 | 44 | 4.742 | < 0.001 | 0.458 |
| Hydrogen ion transporter activity | 29 | 87 | 108 | 81 | 33 | 4.532 | < 0.001 | 0.773 |
| Monovalent inorganic cation transporter activity | 29 | 87 | 110 | 79 | 33 | 4.532 | < 0.001 | 0.773 |
| Mitochondrion | 104 | 452 | 582 | 78 | 23 | 4.358 | < 0.001 | 0.82 |
| Ion transporter activity | 48 | 176 | 220 | 80 | 27 | 4.247 | < 0.001 | 0.842 |
| Oxidoreductase act/on NADH or NADPH/quinone or similar | 9 | 17 | 24 | 71 | 53 | 4.219 | < 0.001 | 0.849 |
| Mitochondrial inner membrane | 30 | 97 | 122 | 80 | 31 | 4.134 | < 0.001 | 0.907 |
| NADH dehydrogenase activity | 7 | 12 | 13 | 92 | 58 | 4.057 | 0.002 | 0.93 |
GD: Gene Ontology Database
Fig. 3.
Analysis of microarray data obtained from RNA isolated from white adipose tissue from young, middle-aged and old mice. Tissues were isolated from mice at 6 months, 1.5 years and 2.5–3 years of age, RNA was extracted and level of gene expression assessed using Affymetrix murine 450A arrays. The CEL files generated by the Affymetrix Microarray Suite version 5.0 (MAS 5.0; Affymetrix) were analyzed using dChip version 1.3 software (http://www.dchip.org). Subset of genes (probesets) that were shown to be differentially expressed in old animals were additionally analyzed by analysis of variance (ANOVA) test with P < 0.01. Comparison of gene expression in control and FIRKO animals during aging showed four different patterns of expression as described in Results.
Increased expression of genes whose products are involved in glycolysis, tricarboxylic acid cycle, mitochondrial electron transport and β-oxidation in old FIRKO mice
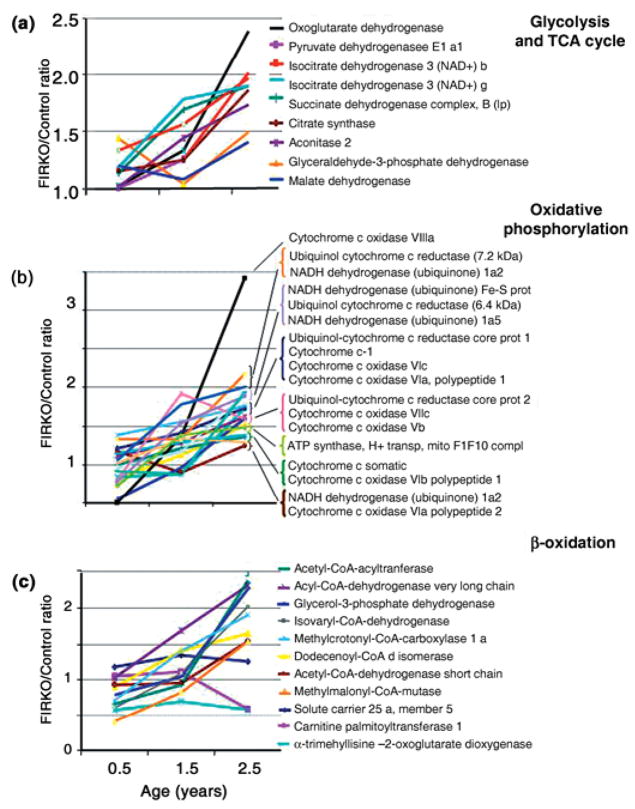
The most significantly and coordinately changed group of genes in adipose of old FIRKO mice as compared to controls, as defined by MappFinder analysis, was the group of nuclear-encoded mitochondrial genes. The pattern of this change is most apparent when one expresses the data from the three different age groups as the ratio of expression at each age. Thus, the ratio of expression between FIRKO and control mice for many of the genes whose products are involved in regulation of glycolysis and tricarboxylic acid (TCA) cycle, fatty acid β-oxidation and oxidative phosphorylation increased progressively with age (Fig. 4a–c).
Fig. 4.
Expression of genes in fat-specific insulin receptor knock-out (FIRKO) and control mice as a function of age. The relative fold expression in FIRKO vs. control mice obtained by microarray analysis was calculated for those genes whose protein products are involved in glycolysis, the tricarboxylic acid (TCA) cycle, oxidative phosphorylation and β-oxidation.
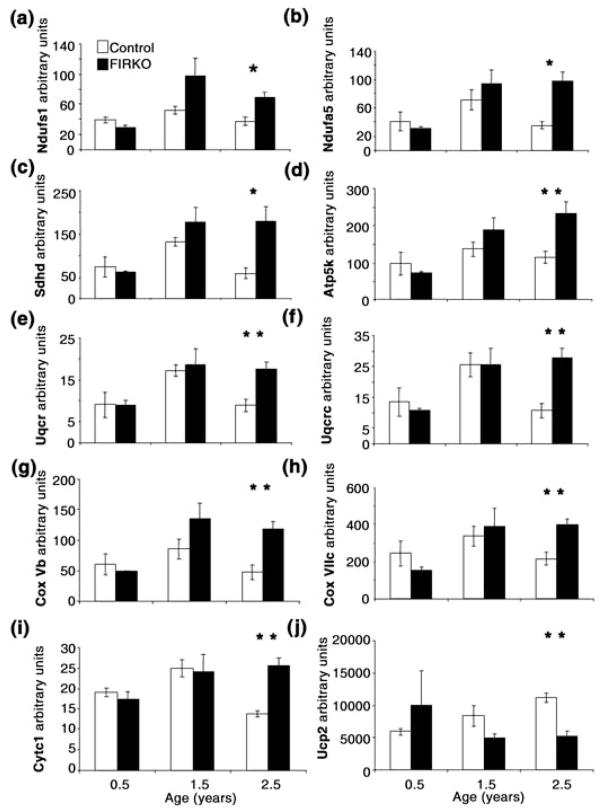
To understand the contributors to the increase in ratio, the expression of individual genes during aging was analyzed. In general, for these genes, mRNA expression in WAT of the two genotypes was similar at 6 months of age, increased in both control and FIRKO groups in 1.5-year-old animals (but usually to a greater extent in FIRKO than in control), then decreased significantly in 2.5- to 3-year-old control animals, while remaining either the same or further increasing in old FIRKO animals. The Affymetrix results were confirmed using real-time quantitative reverse transcriptase–polymerase chain reaction (qRT-PCR) analysis for many of these genes (Fig. 5a–i).
Fig. 5.
Quantitative reverse transcriptase–polymerase chain reaction (RT-PCR) analysis of genes encoding mitochondrial proteins involved in oxidative phosphorylation (electron transport) and oxidative stress response. (a,b) Two subunits [NADH dehydrogenase (ub) Fe-S protein 1 (Ndufs1) and NADH dehydrogenase (ubiquinone) 1 α subcomplex, 5 (Nudfa5)] of mitochondrial Complex I. (c) Succinate dehydrogenase (Sdhd) (Complex II). (d) ATP synthase, H+ transporting, mitochondrial F1F0 complex, subunit e (Atp5k), subunit of Complex V. (e,f) Two subunits [ubiquinol–cytochrome c reductase (6.4 kDa) subunit (Uqcr) and ubiquinol–cytochrome c reductase core protein 2 (Uqcrc2)] of Complex III. (g,h) Two subunits [cytochrome c oxidase, subunit Vb (COX 5b) and cytochrome c oxidase, subunit VIIc (COX 7c)] of mitochondrial Complex IV. (i) Expression of cytochrome c1 (Cyc1). (j) Uncoupling protein 2 (Ucp2). Statistical analysis was performed using a Student’s t-test (*P < 0.05, **P < 0.01) for the comparison of different genotypes in each age group, and results are expressed as average ± SEM. All genes, except Nudfs1, Atp5k and Ucp2, had also a significantly decreased expression in old compared to middle-aged control mice. In addition, all oxidative phosphorylation genes had significantly increased expression in old FIRKO compared to young FIRKO mice (P ≤ 0.01).
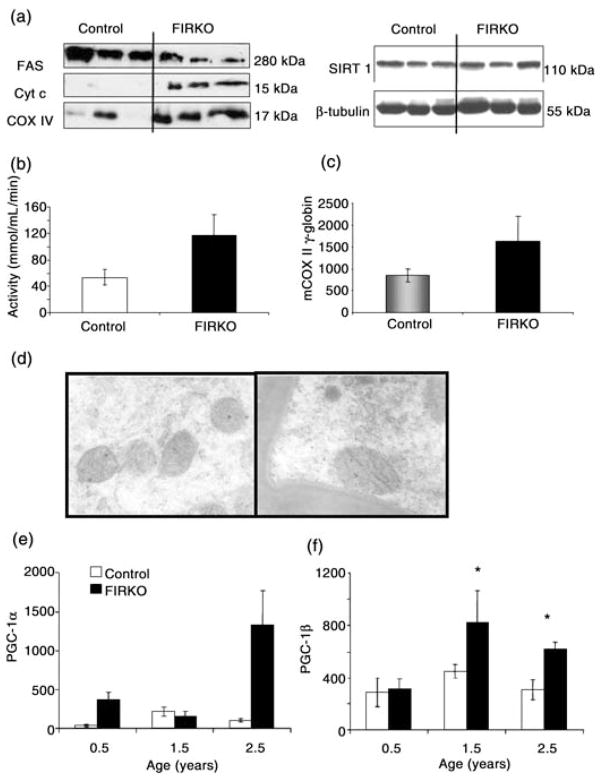
For several genes, we confirmed these changes in expression at the protein level by Western blot analysis of fat from 2-year-old mice. Thus, Western blot analysis showed increased protein levels of cytochrome c oxidase subunit IV (COX IV), as well as increased levels of cytochrome c protein, in WAT isolated from FIRKO mice compared to control, which was in agreement with the microarray data (Fig. 6a). In addition, in line with microarray and protein data, assay of citrate synthase enzymatic activity showed a twofold increase in WAT isolated from 1.5- to 2-year-old FIRKO mice compared to controls (Fig. 6b), although this did not reach statistical significance due to low number (n = 3) of samples per group available from old animals. In contrast, there was no change in the level of expression of Sirt1, a protein deacetylase whose expression has been linked to longevity in lower species (Lin et al., 2000; Tissenbaum & Guarente, 2001; Bitterman et al., 2002; Anderson et al., 2003a), or in the level of β-tubulin in fat of FIRKO mice. On the other hand, expression of the mitochondrial sirtuin, SIRT3, was increased 1.6-fold (P = 0.05) in old FIRKO mice compared to old control mice. These results suggest increased cellular metabolism and respiration in WAT of old FIRKO mice as compared to controls, and this is consistent with the fact that old FIRKO mice have a higher metabolic rate than controls.
Fig. 6.
Conformation of microarray analysis and biological assays. (a) Western blot analysis of white adipose tissue isolated from 1.5–2-year-old mice for cytochrome c (Cyt c) and cytochrome c oxidase, subunit IV (COX IV). Note that SIRT1 expression was not different in fat-specific insulin receptor knock-out (FIRKO) compared to control mice. (b) Citrate synthase assay was performed as described in Experimental procedures and showed an almost threefold increased activity in FIRKO compared to control mice (P = 0.055). (c) Ratio of mitochondrial (COX II) vs. nuclear (β-globin) DNA obtained by quantitative reverse transcriptase–polymerase chain reaction (qRT-PCR) showed significantly increased (P = 0.02) mDNA copy number per cell in FIRKO compared to control mice. (d) Electron micrographs of adipose tissue of 2-year-old control and FIRKO mice (original magnification ×99750). (e,f) RT-PCR analysis showed almost significantly increased expression of peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α) (P = 0.07) and a significant increase of expression of PGC-1β in old FIRKO compared to control mice (P = 0.017). Statistical analysis was done using a Student’s t-test (*P < 0.05) for the comparison of different genotypes in each age group, and results are expressed as average ± SEM.
Mitochondrial morphology and mitochondrial DNA analysis in WAT from FIRKO and control mice
Increased expression of mitochondrial genes and proteins can reflect either higher number of mitochondria per cell or higher activity of each individual mitochondrion. qRT-PCR of mCOX II and β-globin DNA showed a significantly increased ratio of mitochondrial to nuclear DNA content in the fat of FIRKO mice compared to controls (Fig. 6c). Preliminary analysis of electron microscopic images of adipose tissue from control and FIRKO mice at 10 months of age showed a variability of mitochondrial size in both strains, but demonstrated a modest increase in mitochondrial size in the fat of FIRKO as compared to control mice (Fig. 6d). Although microarray analysis did not detect a difference in expression of peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α) and PGC-1β in FIRKO mice compared to control, expression of both PGC-1α and PGC-1β was higher in adipose tissue of old FIRKO mice compared to controls when analyzed by qRT-PCR (Fig. 6e,f), although for PGC-1α this did not reach statistical significance, probably due to the generally low expression of this gene in WAT. However, taken together, these data suggest that the increased mitochondrial activity in WAT of FIRKO mice compared to old controls is primarily due to an increased number or mass of mitochondria.
Discussion
Aging is accompanied by a progressive loss of physiological functions that increases the probability of death. The exact molecular mechanisms that result in aging, however, are still not fully understood. Studies over the last several years have revealed particularly important roles of the insulin/IGF-1 signaling systems, caloric intake and nutrition, body fat content, pathways involved in oxidative stress and control of protein acetylation via the sirtuin family of protein deacetylases (Martin et al., 1996; Honda & Honda, 1999; Guarente & Kenyon, 2000; Clancy et al., 2001; Cohen et al., 2004; Katic & Kahn, 2005).
Dysregulation of energy homeostasis is also clearly related to aging in several ways. First, alterations in insulin/IGF-1 signaling produce alterations in energy homeostasis. Second, it is well known that leanness is associated with increased longevity, and moderate caloric restriction in all species from C. elegans to humans is associated with increase in lifespan (Kagawa, 1978; Cefalu et al., 1997; Vanfleteren & Braeckman, 1999; Weyer et al., 2000; Rogina et al., 2002; Walford et al., 2002; Anderson et al., 2003b; Longo & Finch, 2003; Mattison et al., 2003). Conversely, aging and age-related diseases, such as type 2 diabetes (Martin et al., 1992), coronary heart disease (Facchini et al., 1992, 2001; Laws & Reaven, 1993), hypertension (Facchini et al., 2001) and neurodegenerative diseases (Schubert et al., 2004), are associated with obesity, alterations in body fat distribution and insulin resistance (Enzi et al., 1986; Fraze et al., 1987; Kohrt et al., 1993). While body weight is a function of both energy intake and energy expenditure and energy intake is related to lifespan, the role of energy expenditure in longevity is less clear. In fact, in lower species, there are data to suggest that both decreased (Wisniewski et al., 1999) and increased (Greenberg & Boozer, 2000; Lin et al., 2002; Barros et al., 2004) energy expenditure may be associated with longevity.
Adipose tissue plays a central role in the regulation of energy balance, participating in both energy storage and energy expenditure (Spiegelman & Flier, 2001). Studies in several genetically modified mice models have linked decreased fat mass with longevity, including C/EBPβ knock-in (β/β) (Chiu et al., 2004), translational inhibitor 4E-BP1 (Eif4ebp1−/−) knock-out (Tsukiyama-Kohara et al., 2001), and c-Cbl knock-out (Molero et al., 2004). FIRKO mice have a disruption of the insulin signaling system in adipose tissue only and are resistant to age- and hyperphagia-induced obesity and obesity-related whole body insulin resistance (Bluher et al., 2002). In addition, they have an 18% increase in median, mean and maximal lifespan (Bluher et al., 2003). FIRKO mice therefore provide a unique opportunity to determine the role of insulin action on adipose tissue and energy expenditure in longevity.
We find that aging FIRKO mice have reduced body fat mass (when expressed as either percentage of body weight or absolute mass) and that this occurs despite normal to increased food intake as compared to control mice. In fact, when caloric intake is expressed as per gram body weight (gbw), the difference between FIRKO mice (0.44 cal gbw−1) and controls (0.34 cal gbw−1) is quite striking (i.e. about 30%) and this difference in body weight is maintained throughout life. The current study indicates that this discrepancy between increased food intake and lower body weight in FIRKO mice can be explained by an increase in whole body metabolic rate. Thus, FIRKO mice have increased oxygen consumption (VO2) and RER, and this difference is greatest during the dark phase that corresponds to the feeding time for rodents.
As noted above, while there is an association between metabolic rates in lower organisms and longevity, the exact nature of this relationship in higher organisms has been unclear. In Saccharomyces cerevisiae, increased metabolic rate has been shown to relate to increased lifespan (Anderson et al., 2003b), whereas lower metabolic rates have been shown in long-lived C. elegans (Dillin et al., 2002; Shi et al., 2005). Results in D. melanogaster have shown no change in metabolic rate by either calorie restriction or the lack of insulin/IGF-1 signaling, both known to extend lifespan of the flies (Hulbert et al., 2004), and results obtained from rodents are controversial (Speakman et al., 2004; Selman et al., 2005). This has led to two opposing hypotheses: one that states that decreased metabolic rate is required for an extended lifespan (‘rate of living’ theory) (Harman, 1956) and another that relates increased metabolism with the prolonged lifespan (‘uncoupling to survive’ theory) (Brand, 2000; Speakman et al., 2004). Our finding that FIRKO mice have higher metabolic rates than controls supports the latter hypothesis.
Although the metabolic rate is higher in FIRKO mice than controls, their voluntary activity is unchanged and body temperature in the fed state is higher only in the female FIRKO mice. Male FIRKO mice do exhibit a higher drop in body temperature in the fasted state than control mice, suggesting that they might be more dependent on dietary-induced thermogenesis. As FIRKO mice lack insulin receptors in all fat depots, including brown adipose tissue, and have smaller brown and white fat mass, it is possible that the change in brown fat, rather than white fat, accounts for the change in thermoregulation.
Mitochondria are the main source of energy for cells, and their size and number per cell differ depending on cellular energetic needs. Mitochondria also have a very important role in thermogenesis, oxidative stress, survival and apoptosis (Duchen, 2004). Many studies have shown declining mitochondrial activity in aging and age-related diseases, which is consistent with the notion that maintenance of mitochondrial activity could result in prolonged life. For example, a decline of mitochondrial activity during normal aging has been shown in multiple tissues (Kwong & Sohal, 2000), including muscle (Kerner et al., 2001), heart (Lesnefsky et al., 2001) and brain (Ojaimi et al., 1999; Haripriya et al., 2004; Manczak et al., 2005; Ross, 2005), and decreased expression of nuclear-encoded mitochondrial genes and mitochondrial activity has been observed in human muscle related to insulin resistance and type 2 diabetes mellitus (Patti et al., 2003), as well as aging (Petersen et al., 2003; Lyons et al., 2006). On the other hand, long-term calorie-restricted mice as well as c/EBPβ knock-in and Eif4ebp1−/− mice show increased mitochondrial activity in WAT (Tsukiyama-Kohara et al., 2001; Chiu et al., 2004), and in the latter two cases, this is associated with the higher metabolism of mice.
In this study, we focused on differences in gene expression in WAT of old FIRKO and control mice, because MappFinder analysis showed that the expression of genes in the mitochondrial pathway in old groups of mice was most significantly changed (Table 1). While most differentially expressed genes between young groups were those involved in defense response and response to external stimuli, our study demonstrates an increase in expression of genes whose products are associated with mitochondrial activity, including those involved in glycolysis, β-oxidation, the TCA cycle and oxidative phosphorylation, in WAT from aging FIRKO vs. control mice. These differences were not found in young animals and are mainly a result of a decline in expression of these genes in normal aging control mice, while their expression remains high or even increases further in old FIRKO.
Cytochrome c and COX IV were among genes whose expression was significantly increased in WAT of old FIRKO compared to old control mice both at the RNA and protein levels. Citrate synthase expression and activity were also increased in adipose tissue of old FIRKO mice. Citrate synthase is the initial enzyme of the TCA cycle and catalyzes the reaction between acetyl coenzyme A and oxaloacetic acid to form citric acid. This enzyme is an exclusive marker of the mitochondrial matrix and its activity was increased about twofold in adipose tissue from old FIRKO mice compared to controls.
Higher mitochondrial activity can reflect either a higher number/mass of mitochondria per cell or the same number of mitochondria with higher activity per mitochondrion, or a combination of both. Electron micrographs and assessment of the ratio between a mitochondrial and nuclear DNA content suggest that the increased mitochondrial activity in FIRKO mice may be due to an increase in the number or mass of mitochondria. Further studies of this point will be required. Increased expression of the transcriptional coactivators PGC-1α and PGC-1β in WAT of FIRKO mice is consistent with this result, and over-expression of PGC-1α in adipose tissue has been shown to increase mitochondrial content and energy expenditure by this tissue (Tiraby et al., 2003). Altered levels of the SIRT family of nicotinamide adenine dinucleotide (NAD)-dependent protein deacetylases have been associated with longevity, and SIRT1 has been shown to interact with PGC-1α in control of gene expression (Nemoto et al., 2005). However, SIRT1 expression was not different at the mRNA or protein level between FIRKO and control WAT. On the other hand, SIRT3 expression was increased in adipose tissue of old FIRKO mice as compared to controls. SIRT3 is present in mitochondria (Onyango et al., 2002), and caloric restriction and cold exposure have also been reported to elevate SIRT3 expression in brown adipose tissue and regulate thermogenesis through PGC-1α and UCP1 (Shi et al., 2005). SIRT3 has also been shown to be reduced in muscle of mice with insulin-deficient diabetes, indicating a link between insulin action and this sirtuin (Yechoor et al., 2004).
One potential caveat of this study is that the fat samples were taken from mice matched by age, but by definition, these mice were at somewhat different parts of their maximal life expectancy. Thus, at 30 months of age, control mice are at 86% of their maximum lifespan, while at the same age, the FIRKO mice are only at 76% of their maximum lifespan. As mice age, occurrence of age-related diseases might also affect gene expression and metabolism in fat. Therefore, we cannot be sure that if we had been able to measure the expression of mitochondrial genes in WAT in equivalently older FIRKO mice at 86% or more of their maximal lifespan that mitochondrial gene expression would not decrease in a manner similar to that observed in control mice. However, the results shown in Fig. 3 demonstrate that almost all genes analyzed show even higher expression in the WAT of 30-month-old FIRKO mice than in 18 -month-old FIRKO mice, indicating that, at least up to that point in the lifecycle of these mice, there is no drop-off in gene expression of these mitochondrial genes. Furthermore, even if FIRKO mice do eventually demonstrate a similar decrease in expression as they approach the limit of their lifespan, one could consider this drop-off in expression of mitochondrial genes as a true ‘marker’ of aging. Also, it is possible that the decrease in the expression of mitochondrial genes in control mice WAT is at least in part a consequence of changes in insulin sensitivity with aging, as normal mice become more insulin resistant with age, whereas FIKRO mice remain lean and insulin sensitive as they age (Bluher et al., 2002).
While both FIRKO mice and calorie-restricted animals have prolonged lifespan, some of the mechanisms appear to be similar, whereas others are different in these two models. Thus, both FIRKO mice and calorie-restricted rodents have increased whole body insulin sensitivity (Sugimoto & Erikson, 1985; Barzilai et al., 1998; Bluher et al., 2002), but at the level of fat, WAT of calorie-restricted mice is more insulin sensitive than adipose from controls (Higami et al., 2004), whereas adipose tissue from FIRKO mice is completely insulin resistant due to the lack of insulin receptors (Bluher et al., 2002). Whether the effect of tissue-specific insulin resistance to increase mitochondrial activity is specific to adipose tissue is unclear. Expression of mitochondrial genes is decreased in muscle of humans with insulin resistance due to type 2 diabetes (Patti et al., 2003), and mitochondrial activity in muscle as measured by levels of ATP is decreased in elderly humans (Petersen et al., 2003). On the other hand, mitochondrial gene expression is not changed in muscle or liver of mice with tissue-specific insulin receptor knockouts (MIRKO and LIRKO) (Yechoor et al., 2004), although the latter animals have not been studied for gene expression beyond 6 months of age. Microarray data from both calorie-restricted and FIRKO mice (available at www.diabetesgenome.org), as well as biological data from FIRKO adipocytes in vitro (Bluher et al., 2003) suggest increased lipolysis, which could contribute to the smaller adipose tissue mass in both models compared to controls throughout life. These results, together with the results obtained from other adipose tissue-specific knock-out mice with decreased adipocyte sizes and increased respiration, also suggest that increased metabolism and adenosine triphosphate (ATP) production in WAT of old FIRKO mice could be a consequence of decreased size of adipocytes rather than the lack of insulin receptor.
One might imagine that the increased mitochondrial activity in FIRKO mice is associated with increased production of reactive oxygen species and increased oxidative stress in the cells (Harman, 1956). However, our microarray data reveal lower levels of expression of Ucp2 and Hif1α in FIRKO fat than in controls, suggesting decreased oxidative stress in adipose tissue of FIRKO mice (Fig. 5j and data at www.diabetesgenome.org). In addition, it has been shown that higher respiratory activity decreases release of mitochondrial reactive oxygen species and extends longevity in yeast (Barros et al., 2004). Also, it may be that PGC-1α, whose expression is increased in FIRKO adipose tissue, is not only stimulating mitochondrial biogenesis, but also regulating genes that protect cells from oxidative stress by reducing reactive oxygen species accumulation, increasing mitochondrial membrane potential and reducing apoptosis (Valle et al., 2005). It is worth noting that FIRKO mice have altered adipokine secretion with higher leptin levels and higher adiponectin expression in WAT (Bluher et al., 2002). To what extent these might contribute to the beneficial metabolic effects or changes in mitochondrial activity in other organs, such as muscle, liver, brain, intestine or pancreas, remains to be determined.
Finally, it is known that a genetic background can influence both physiology and lifespan of mice. As both control and FIRKO mice were on mixed background, we were careful to use littermates as controls for this gene expression study. Thus, we do not think that the observed differences could be due to strain differences. In addition, there are no data about difference in lifespan between 129sv and C57Bl6/J mouse strains.
In summary, our data indicate that FIRKO mice have increased metabolism and increased expression of genes and proteins involved in mitochondrial oxidative metabolism in the WAT compared to controls as they age. They also have lower expression of genes involved in oxidative stress, which together with their decreased fat mass and higher energy expenditure may support their increase in lifespan. These results show a close relationship between aging and energy metabolism at molecular level and confirm a central role of adipose tissue in longevity. In addition, the increased longevity in FIRKO mice and their protection from metabolic diseases, such as diabetes and obesity, suggests that increasing the metabolic rate of WAT as drugs might provide a target for pharmacological treatment to increase lifespan and improve metabolic homeostasis.
Experimental procedures
Animals
All animal experiments were conducted in accordance with mandated standards of humane care approved by the institutional animal care and use committees of Joslin Diabetes Center and Beth Israel Deaconess Medical Center. All mice were housed in a mouse facility on a 12-h light/dark cycle and had ad libitum access to standard chow (Purina Autoclavable Laboratory Mouse Breeder Chow, St. Louis, MO, USA) and water. Male control (Lox/Lox) and FIRKO mice, both on mixed 129sv/C57Bl6/J background, were killed at the age of 6, 18 and 30–36 months by inhalation of CO2. The epididymal WAT was immediately isolated, frozen in liquid nitrogen and stored at −80 °C until use.
Body composition and metabolic analysis
Ten-month-old male ad libitum-fed control mice and FIRKO mice were used for metabolic analysis. Analysis of fat and lean body mass was conducted using DEXA (Lunar PIXImus2 densitometer; GE Medical Systems, Madison, WI, USA) in mice anesthetized with Avertin (tribromoethanol:tert-amyl alcohol, 0.015 mL g−1 i.p.). For metabolic analysis, six 10-month-old male mice of both genotypes (FIRKO and controls) were housed individually, given ad libitum access to food and evaluated for ambulatory activity using an OPTO-M3 sensor system (Comprehensive Laboratory Animal Monitoring System, CLAMS; Columbus Instruments, Columbus, OH, USA). A score of ambulatory activity was determined by consecutive photobeam breaks occurring in adjacent beams. Beam breaks were counted for 60 s consecutively for each 12 cages, cycling five times per hour during two full light/dark cycles.
Indirect calorimetry was measured on the same mice using an open-circuit Oxymax system (Columbus Instruments, OH, USA). Mice were individually housed in Plexiglas cages through which air of known O2 concentration was passed at a constant flow rate. After a 48-h acclimation period, exhaust air was sampled 60 s in every 12 min in each cage consecutively for 48 h in the fed state for the determination of O2 and CO2. Core body temperature was also measured in 10-month-old male and female mice (control and FIRKO) fed ad libitum with normal or high fat chow, or after 10-h light phase fasting. A rodent thermocouple probe attached to Thermalert THS (Physitemp, Braintree Scientific, Braintree, MA, USA) equipped with a rectal probe was inserted 1 cm into the rectum and read after stabilization.
RNA preparation and microarray analysis
Total RNA was extracted from WAT from young (four IRlox and four FIRKO mice), middle-aged [five control (two Cre/lox and three WT) and four FIRKO mice] and old [four control (one WT, one Cre/lox and two IRlox) and six FIRKO (four 2.5 years old and two 3 years old)] mice using Qiagen minicolumns (Valencia, CA, USA). The purified total RNA was used to synthesize double-stranded cDNA (dsDNA) using Superscript Choice System (Invitrogen) with an oligo-dT primer containing T7 RNA polymerase promoter. After purification of dscDNA, in vitro transcription (IVT) was performed by using the BioArray High Yield RNA transcript labeling kit (Enzo, Farmingdale, NY, USA). Subsequently, biotin-labeled cRNA was purified and fragmented. Fragmented cRNAs from each mouse was then individually hybridized to 27 murine 450A arrays (Affymetrix) at 45 °C for 16 h. After hybridization, the gene chips were automatically washed, stained with streptavidin–phycoerythrin using a fluidics system and scanned with a Hewlett Packard GeneArray Scanner (Affymetrix).
Statistical analysis of microarray data
The 26 CEL files (because of the insufficient quality, a sample isolated from one 3-year-old FIRKO mouse was omitted from further analysis) generated by the Affymetrix Microarray Suite version 5.0 (MAS 5.0; Affymetrix) were analyzed using dChip v1.3 software (http://www.dchip.org). Expression data were normalized at perfect match and mismatch probe level using the invariant set normalization method, and then model-based expression values were calculated (Li & Wong, 2001). The default implementation of dChip used the perfect match/mismatch intensity matrix. MOE430A gene information files were used to provide annotation information to probe sets, such as accession number, LocusLink ID, gene name, gene ontology terms, protein domain terms, pathway terms, chromosome terms and gene description.
For comparison of global gene expression profiles between two sample sets (e.g. FIRKO old vs. FIRKO young groups), the group means and standard errors were computed by pooling arrays considering measurement accuracy, where samples in the same group are regarded as ‘replicates’ (Li & Wong, 2001). The following filtering criteria were applied when comparing the two groups: a minimum of a 1.2-fold change between the group means; a minimum of 100 for absolute difference between the two group means; and a P-value < 0.05. We also used the 90% confidence boundary as the lower confidence boundary of fold changes for filtering. The t-statistic was computed as (mean 1 − mean 2)/√[SE (mean 1)2 + SE(mean 2)2], and its P-value is computed based on the t-distribution.
To find genes correlated with strain and age, we also performed supervised ANOVA using these variables as ‘factor’. Median false discovery rate (FDR) for all comparisons was computed by permutation analysis using dChip and was found to be less than 5%. For the data in this manuscript, ANOVA was performed using an even more stringent level of significance of P < 0.01. To identify novel sample clusters and their associated ‘signature genes’, we performed hierarchical clustering using genes obtained by samples comparison and ANOVA (Eisen et al., 1998).
qRT-PCR
The 26 samples isolated from young, middle-aged and old control (13 samples) and FIRKO (13 samples) mice, with four to five samples in each group, were used for qRT-PCR. qRT-PCR was performed with the Sybr Green kit (Applied Biosystems, Foster City, CA, USA) using an ABI PRISM 7000 Sequence Detection System (Applied Biosciences) according to the instructions provided by the manufacturer. TBP was used as an internal control based on our results showing that its expression was not affected by knock-out of insulin receptor. The primers were designed using Primer Express version 3.0 (Applied Biosystems, Foster City, CA, USA). The sequences of primers were as follows: (i) NADH dehydrogenase (ubiquinone) 1a, 5 (Complex I) (5′-GCG GAG CCA GAT GT T AAA AA-3′ and 5′-CCA TCC ACC ATC TGA CAC TG-3′), (ii) NADH dehydrogenase (ubiquinone) Fe-S protein 1 (Complex 1) (5′-TTG GGA ACA ACA GGA AGA GG-3′ and 5′-T TC CCA CTG CAT CCA TTA CA-3′), (iii) succinate dehydrogenase, complex B (Ip) (Complex II) (5′-CTG GTG GAA CGG AGA CAA GT-3′ and 5′-GT T AAG CCA ATG CTC GCT TC-3′), (iv) ubiquinol-cytochrome c reductase core protein 2 (Complex III) (5′-TCC AAC AAC TTG GGA ACC TC-3′ and 5′-GGT GCT GTG GTG ACA TTG AG-3′), (v) ubiquinol-cytochrome c reductase (6.4 kDa) (Complex III) (5′-GGG GTG ACC CTG AGT AT T GA-3′ and 5′-ATG TAA GGC ACC CAG TCC AG-3′), (vi), cytochrome c oxidase VIIc (Complex IV) (5′-AGA ACT TCC AGC AGC GAC AT-3′ and 5′-TAA AGA AAG GTG CGG CAA AC-3′), (vii) cytochrome c oxidase Vb (Complex IV) (5′-CAG AAG GGA CTG GAC CCA TA-3′ and 5′-ATA ACA CAG GGG CTC AGT GG-3′), (viii) ATP synthase, H+ transport, mitochondrial F1F10 complex, subunit e (Complex V) (5′-CGG T TC AGG TCT CTC CAC TC-3′ and 5′-TGA CGC CTC ACT TGA GAA TG-3′), (ix) cytochrome c, somatic (5′-GCC CGG AAC GAA TTA AAA AT-3′ and 5′-CCA GGT GAT GCC TTT GTT CT-3′), (x) cytochrome c1 (5′-GCT ACC CAT GGT CTC ATC GT-3′ and 5′-CAT CAT CAT TAG GGC CAT CC-3′), (xi) TBP (5′-ACG CT T CAC CAA TGA CTC CTA TG-3′ and 5′-TGA CTG CAG CAA ATC GCT TGG-3′), (xiii) PGCa (5′-GTC AAC AGC AAA AGC CAC AA-3′ and 5′-TCT GGG GTC AGA GGA AGA GA-3′), (xiv) PGCb (5′-CTT GCT TTT CCC AGA TGA GG-3′ and 5′-CCC TGT CCG TGA GGA ACG-3′), (xv) pyruvate dehydrogenase 1 (5′-GTC ACC ACA GTG CTC ACC AG-3′ and 5′-TGA AGC CAT GTG CTC GAT AG-3′).
For calculating mitochondrial vs. nuclear DNA content in the cells, RT-PCR was performed using 75 ng DNA isolated from three control and three FIRKO mice (all 2 years old), as a template and primers for intronic region of β-globin (5′-ATC CAG GT T ACA AGG CAG CT-3′ and 5′-GGG AAA CAT AGA CAG GGG-3′) and for mitochondrial COX II gene (5′-CCA TCC CAG GCC GAC TAA-3′ and 5′-AAT T TC AGA GCA T TG GCC ATA GA-3′).
Western blotting
Epididymal adipose tissue was isolated from three FIRKO and three control (Lox/Lox) 2-year-old mice, immediately transferred in liquid nitrogen and stored at −80 °C until further processing. The tissue was homogenized in a lysis buffer [Buffer A (25 mM Tris–HCl (pH = 7.4), 2 mM Na3VO4, 10 mM NaF, 10 mM Na4P2O7, 1 mM EGTA, 1 mM EDTA and 10 nM okadaic acid) pH = 7.4, 10% glycerol, 1%NP40 and protease inhibitors]. After sonication, the lysate was centrifuged in Eppendorf centrifuge at 12 000 g for 15 min, supernatants were transferred to new tubes and used for total cell lysate preparation by addition of loading buffer (sample loading buffer, Sigma). Equal amounts of proteins were loaded on sodium dodecyl sulphate (SDS)-polyacrylamide gels and electrophorized for about 2 h at a constant of 95 V. Proteins were transferred to nitrocellulose membranes (80 min and constant 200 mA), at 4 °C, blocked in Blocking Reagent (Roche, Indianapolis, IN, USA), incubated for 2 h at room temperature with primary antibodies (anti-COX IV and anti-FAS, Abcam, Cambridge, MA, USA; anti-cytochrome c, Bimol, USA; anti-Sirt1, Upstate Biotechnology, Lake Placid, NY, USA; and anti-β-tubulin, Sigma), washed thoroughly with Tris-buffered saline (TBS)/0.05%Tween-20 for 60 min and incubated with an appropriate secondary antibody (anti-rabbit, anti-mouse) and anti-actin horseradish peroxidase (HRP) conjugated. After 60 min, membranes were thoroughly washed and the signal was quantitated using chemiluminescence (BM Chemiluminescence Blotting Substrate, Roche, Mannheim, Germany).
Citrate synthase activity
Citrate synthase activity was measured using a citrate synthase assay kit (Sigma) according to the manufacturer’s instructions. In brief, frozen WAT from three FIRKO and three IRlox mice, all 2 years old, was homogenized in CelLytic™ buffer (Sigma-Aldrich, St. Louis, MO, USA), centrifuged and supernatant was used to measure citrate synthase activity. The assay was performed in 96-well plates, and the activity (μmol mL−1 min−1) was measured after the addition of oxaloacetic acid (OAA) in a Micro-plate Scanning Spectrophotometer (Bio-Tek Instruments, Inc., Winooski, VT, USA).
Electron microscopy
Adipose tissue was harvested from the 2.5-year-old animals, cut into small pieces and submersed immediately into a fixative (2% paraformaldehyde, 2% gluteraldehyde, in a 0.1-M phosphate buffer, pH 7.4) where it remained for several hours at room temperature before being stored at 4 °C. After washing in a 0.1-M phosphate buffer, the tissue was postfixed for 2 h with a 2% osmium tetroxide solution in a Millonig’s buffer, pH 7.4, washed again and stored at 4 °C. Ascending gradations of alcohol (50–100%) were used for dehydration, after which propylene oxide was used as the clearing agent. The tissue samples were in infiltration solution (75% propylene oxide, 25% Araldite 502 epoxy resin, and 50% propylene oxide, 50% Araldite 502 epoxy resin) for several hours and then placed into a vacuum desiccator overnight, after which they were embedded in fresh Araldite 502 epoxy resin using BEEM capsules. The resin was then cured in 60 °C for 48 h. The tissue blocks were trimmed and 1 μ sections were obtained using LKB Nova ultramicrotome. Ultrathin sections (60–80 nm) were cut from selected sample blocks using a Diatome Diamond Knife (LKB Nova ultramicrotome), mounted on 75 mesh copper grids and stained with uranyl acetate and lead citrate before being observed on the Philips 301 transmission electron microscope.
Acknowledgments
This work has been supported by National Institutes of Health (NIH) grant DK31036 and DK60837, the Diabetes Genome Anatomy Project. We would like to thank Laureen Mazzola and Lin Pei for a technical help and help with animals, and Dr. Jenny Gunton for very useful advice with statistical analysis.
References
- Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Sinclair DA. Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae. Nature. 2003a;423:181–185. doi: 10.1038/nature01578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson RM, Latorre-Esteves M, Neves AR, Lavu S, Medvedik O, Taylor C, Howitz KT, Santos H, Sinclair DA. Yeast life-span extension by calorie restriction is independent of NAD fluctuation. Science. 2003b;302:2124–2126. doi: 10.1126/science.1088697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros MH, Bandy B, Tahara EB, Kowaltowski AJ. Higher respiratory activity decreases mitochondrial reactive oxygen release and increases life span in Saccharomyces cerevisiae. J Biol Chem. 2004;279:49883–49888. doi: 10.1074/jbc.M408918200. [DOI] [PubMed] [Google Scholar]
- Barzilai N, Banerjee S, Hawkins M, Chen W, Rossetti L. Caloric restriction reverses hepatic insulin resistance in aging rats by decreasing visceral fat. J Clin Invest. 1998;101:1353–1361. doi: 10.1172/JCI485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J Biol Chem. 2002;277:45099–45107. doi: 10.1074/jbc.M205670200. [DOI] [PubMed] [Google Scholar]
- Bluher M, Kahn BB, Kahn CR. Extended longevity in mice lacking the insulin receptor in adipose tissue. Science. 2003;299:572–574. doi: 10.1126/science.1078223. [DOI] [PubMed] [Google Scholar]
- Bluher M, Michael MD, Peroni OD, Ueki K, Carter N, Kahn BB, Kahn CR. Adipose tissue selective insulin receptor knockout protects against obesity and obesity-related glucose intolerance. Dev Cell. 2002;3:25–38. doi: 10.1016/s1534-5807(02)00199-5. [DOI] [PubMed] [Google Scholar]
- Brand MD. Uncoupling to survive? The role of mitochondrial inefficiency in ageing. Exp Gerontol. 2000;35:811–820. doi: 10.1016/s0531-5565(00)00135-2. [DOI] [PubMed] [Google Scholar]
- Cefalu WT, Wagner JD, Wang ZQ, Bell-Farrow AD, Collins J, Haskell D, Bechtold R, Morgan T. A study of caloric restriction and cardiovascular aging in cynomolgus monkeys (Macaca fascicularis): a potential model for aging research. J Gerontol A Biol Sci Med Sci. 1997;52:B10–B19. doi: 10.1093/gerona/52a.1.b10. [DOI] [PubMed] [Google Scholar]
- Chiu CH, Lin WD, Huang SY, Lee YH. Effect of a C/EBP gene replacement on mitochondrial biogenesis in fat cells. Genes Dev. 2004;18:1970–1975. doi: 10.1101/gad.1213104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy DJ, Gems D, Harshman LG, Oldham S, Stocker H, Hafen E, Leevers SJ, Partridge L. Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science. 2001;292:104–106. doi: 10.1126/science.1057991. [DOI] [PubMed] [Google Scholar]
- Cohen HY, Miller C, Bitterman KJ, Wall NR, Hekking B, Kessler B, Howitz KT, Gorospe M, De Cabo R, Sinclair DA. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305:390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- Dillin A, Hsu AL, Arantes-Oliveira N, Lehrer-Graiwer J, Hsin H, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Rates of behavior and aging specified by mitochondrial function during development. Science. 2002;298:2398–2401. doi: 10.1126/science.1077780. [DOI] [PubMed] [Google Scholar]
- Dorman JB, Albinder B, Shroyer T, Kenyon C. The age-1 and daf-2 genes function in a common pathway to control the lifespan of Caenorhabditis elegans. Genetics. 1995;141:1399–1406. doi: 10.1093/genetics/141.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchen MR. Roles of mitochondria in health and disease. Diabetes. 2004;53(Suppl 1):S96–S102. doi: 10.2337/diabetes.53.2007.s96. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enzi G, Gasparo M, Biondetti PR, Fiore D, Semisa M, Zurlo F. Subcutaneous and visceral fat distribution according to sex, age, and overweight, evaluated by computed tomography. Am J Clin Nutr. 1986;44:739–746. doi: 10.1093/ajcn/44.6.739. [DOI] [PubMed] [Google Scholar]
- Facchini F, Hollenbeck CB, Chen YN, Chen YD, Reaven GM. Demonstration of a relationship between white blood cell count, insulin resistance, and several risk factors for coronary heart disease in women. J Intern Med. 1992;232:267–272. doi: 10.1111/j.1365-2796.1992.tb00582.x. [DOI] [PubMed] [Google Scholar]
- Facchini FS, Hua N, Abbasi F, Reaven GM. Insulin resistance as a predictor of age-related diseases. J Clin Endocrinol Metab. 2001;86:3574–3578. doi: 10.1210/jcem.86.8.7763. [DOI] [PubMed] [Google Scholar]
- Fraze E, Chiou YA, Chen YD, Reaven GM. Age-related changes in postprandial plasma glucose, insulin, and free fatty acid concentrations in nondiabetic individuals. J Am Geriatr Soc. 1987;35:224–228. doi: 10.1111/j.1532-5415.1987.tb02313.x. [DOI] [PubMed] [Google Scholar]
- Greenberg JA, Boozer CN. Metabolic mass, metabolic rate, caloric restriction, and aging in male Fischer 344 rats. Mech Ageing Dev. 2000;113:37–48. doi: 10.1016/s0047-6374(99)00094-9. [DOI] [PubMed] [Google Scholar]
- Guarente L, Kenyon C. Genetic pathways that regulate ageing in model organisms. Nature. 2000;408:255–262. doi: 10.1038/35041700. [DOI] [PubMed] [Google Scholar]
- Haripriya D, Devi MA, Kokilavani V, Sangeetha P, Panneerselvam C. Age-dependent alterations in mitochondrial enzymes in cortex, striatum and hippocampus of rat brain–potential role of L-carnitine. Biogerontology. 2004;5:355–364. doi: 10.1007/s10522-004-2575-y. [DOI] [PubMed] [Google Scholar]
- Harman D. Aging: a theory based on free radical and radiation chemistry. J Gerontol. 1956;11:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- van Heemst D, Beekman M, Mooijaart SP, Heijmans BT, Brandt BW, Zwaan BJ, Slagboom PE, Westendorp RG. Reduced insulin/IGF-1 signalling and human longevity. Aging Cell. 2005;4:79–85. doi: 10.1111/j.1474-9728.2005.00148.x. [DOI] [PubMed] [Google Scholar]
- Higami Y, Pugh TD, Page GP, Allison DB, Prolla TA, Weindruch R. Adipose tissue energy metabolism: altered gene expression profile of mice subjected to long-term caloric restriction. FASEB J. 2004;18:415–417. doi: 10.1096/fj.03-0678fje. [DOI] [PubMed] [Google Scholar]
- Holzenberger M, Dupont J, Ducos B, Leneuve P, Geloen A, Even PC, Cervera P, Le Bouc Y. IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature. 2003;421:182–187. doi: 10.1038/nature01298. [DOI] [PubMed] [Google Scholar]
- Honda Y, Honda S. The daf-2 gene network for longevity regulates oxidative stress resistance and Mn-superoxide dismutase gene expression in Caenorhabditis elegans. FASEB J. 1999;13:1385–1393. [PubMed] [Google Scholar]
- Hulbert AJ, Clancy DJ, Mair W, Braeckman BP, Gems D, Partridge L. Metabolic rate is not reduced by dietary-restriction or by lowered insulin/IGF-1 signalling and is not correlated with individual lifespan in Drosophila melanogaster. Exp Gerontol. 2004;39:1137–1143. doi: 10.1016/j.exger.2004.04.006. [DOI] [PubMed] [Google Scholar]
- Kagawa Y. Impact of Westernization on the nutrition of Japanese: changes in physique, cancer, longevity and centenarians. Prev Med. 1978;7:205–217. doi: 10.1016/0091-7435(78)90246-3. [DOI] [PubMed] [Google Scholar]
- Katic M, Kahn CR. The role of insulin and IGF-1 signaling in longevity. Cell Mol Life Sci. 2005;62:320–343. doi: 10.1007/s00018-004-4297-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C. The plasticity of aging: insights from long-lived mutants. Cell. 2005;120:449–460. doi: 10.1016/j.cell.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Kerner J, Turkaly PJ, Minkler PE, Hoppel CL. Aging skeletal muscle mitochondria in the rat: decreased uncoupling protein-3 content. Am J Physiol Endocrinol Metab. 2001;281:E1054–E1062. doi: 10.1152/ajpendo.2001.281.5.E1054. [DOI] [PubMed] [Google Scholar]
- Kohrt WM, Kirwan JP, Staten MA, Bourey RE, King DS, Holloszy JO. Insulin resistance in aging is related to abdominal obesity. Diabetes. 1993;42:273–281. [PubMed] [Google Scholar]
- Kurosu H, Yamamoto M, Clark JD, Pastor JV, Nandi A, Gurnani P, McGuinness OP, Chikuda H, Yamaguchi M, Kawaguchi H, Shimomura I, Takayama Y, Herz J, Kahn CR, Rosenblatt KP, Kuro-o M. Suppression of aging in mice by the hormone Klotho. Science. 2005;309:1829–1833. doi: 10.1126/science.1112766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong LK, Sohal RS. Age-related changes in activities of mitochondrial electron transport complexes in various tissues of the mouse. Arch Biochem Biophys. 2000;373:16–22. doi: 10.1006/abbi.1999.1495. [DOI] [PubMed] [Google Scholar]
- Laws A, Reaven GM. Insulin resistance and risk factors for coronary heart disease. In: Ferrannini E, editor. Clinical Endocrinology and Metabolism: Insulin Resistance and Disease. London: Baillière Tindall; 1993. pp. 1063–1078. [DOI] [PubMed] [Google Scholar]
- Lesnefsky EJ, Gudz TI, Moghaddas S, Migita CT, Ikeda-Saito M, Turkaly PJ, Hoppel CL. Aging decreases electron transport complex III activity in heart interfibrillar mitochondria by alteration of the cytochrome c binding site. J Mol Cell Cardiol. 2001;33:37–47. doi: 10.1006/jmcc.2000.1273. [DOI] [PubMed] [Google Scholar]
- Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- Lin SJ, Kaeberlein M, Andalis AA, Sturtz LA, Defossez PA, Culotta VC, Fink GR, Guarente L. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature. 2002;418:344–348. doi: 10.1038/nature00829. [DOI] [PubMed] [Google Scholar]
- Longo VD, Finch CE. Evolutionary medicine: from dwarf model systems to healthy centenarians? Science. 2003;299:1342–1346. doi: 10.1126/science.1077991. [DOI] [PubMed] [Google Scholar]
- Lyons CN, Mathieu-Costello O, Moyes CD. Regulation of skeletal muscle mitochondrial content during aging. J Gerontol A Biol Sci Med Sci. 2006;61:3–13. doi: 10.1093/gerona/61.1.3. [DOI] [PubMed] [Google Scholar]
- Manczak M, Jung Y, Park BS, Partovi D, Reddy PH. Time-course of mitochondrial gene expressions in mice brains: implications for mitochondrial dysfunction, oxidative damage, and cytochrome c in aging. J Neurochem. 2005;92:494–504. doi: 10.1111/j.1471-4159.2004.02884.x. [DOI] [PubMed] [Google Scholar]
- Martin GM, Austad SN, Johnson TE. Genetic analysis of ageing: role of oxidative damage and environmental stresses. Nat Genet. 1996;13:25–34. doi: 10.1038/ng0596-25. [DOI] [PubMed] [Google Scholar]
- Martin BC, Warram JH, Krolewski AS, Bergman RN, Soeldner JS, Kahn CR. Role of glucose and insulin resistance in development of type 2 diabetes mellitus: results of a 25-year follow-up study. Lancet. 1992;340:925–929. doi: 10.1016/0140-6736(92)92814-v. [DOI] [PubMed] [Google Scholar]
- Mattison JA, Lane MA, Roth GS, Ingram DK. Calorie restriction in rhesus monkeys. Exp Gerontol. 2003;38:35–46. doi: 10.1016/s0531-5565(02)00146-8. [DOI] [PubMed] [Google Scholar]
- McCulloch D, Gems D. Body size, insulin/IGF signaling and aging in the nematode Caenorhabditis elegans. Exp Gerontol. 2003;38:129–136. doi: 10.1016/s0531-5565(02)00147-x. [DOI] [PubMed] [Google Scholar]
- Molero JC, Jensen TE, Withers PC, Couzens M, Herzog H, Thien CB, Langdon WY, Walder K, Murphy MA, Bowtell DD, James DE, Cooney GJ. c-Cbl-deficient mice have reduced adiposity, higher energy expenditure, and improved peripheral insulin action. J Clin Invest. 2004;114:1326–1333. doi: 10.1172/JCI21480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto S, Fergusson MM, Finkel T. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1α. J Biol Chem. 2005;280:16456–16460. doi: 10.1074/jbc.M501485200. [DOI] [PubMed] [Google Scholar]
- Ojaimi J, Masters CL, Opeskin K, McKelvie P, Byrne E. Mitochondrial respiratory chain activity in the human brain as a function of age. Mech Ageing Dev. 1999;111:39–47. doi: 10.1016/s0047-6374(99)00071-8. [DOI] [PubMed] [Google Scholar]
- Onyango P, Celic I, McCaffery JM, Boeke JD, Feinberg AP. SIRT3, a human SIR2 homologue, is an NAD-dependent deacetylase localized to mitochondria. Proc Natl Acad Sci USA. 2002;99:13653–13658. doi: 10.1073/pnas.222538099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patti ME, Butte AJ, Crunkhorn S, Cusi K, Berria R, Kashyap S, Miyazaki Y, Kohane I, Costello M, Saccone R, Landaker EJ, Goldfine AB, Mun E, DeFronzo R, Finlayson J, Kahn CR, Mandarino LJ. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: potential role of PGC1 and NRF1. Proc Natl Acad Sci USA. 2003;100:8466–8471. doi: 10.1073/pnas.1032913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen KF, Befroy D, Dufour S, Dziura J, Ariyan C, Rothman DL, DiPietro L, Cline GW, Shulman GI. Mitochondrial dysfunction in the elderly: possible role in insulin resistance. Science. 2003;300:1140–1142. doi: 10.1126/science.1082889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci USA. 2004;101:15998–16003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogina B, Helfand SL, Frankel S. Longevity regulation by Drosophila Rpd3 deacetylase and caloric restriction. Science. 2002;298:1745. doi: 10.1126/science.1078986. [DOI] [PubMed] [Google Scholar]
- Ross CM. Folate, mitochondria, ROS, and the aging brain. Am J Med. 2005;118:1174–1175. doi: 10.1016/j.amjmed.2005.03.033. [DOI] [PubMed] [Google Scholar]
- Schubert M, Gautam D, Surjo D, Ueki K, Baudler S, Schubert D, Kondo T, Alber J, Galldiks N, Kustermann E, Arndt S, Jacobs AH, Krone W, Kahn CR, Bruning JC. Role for neuronal insulin resistance in neurodegenerative diseases. Proc Natl Acad Sci USA. 2004;101:3100–3105. doi: 10.1073/pnas.0308724101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selman C, Phillips T, Staib JL, Duncan JS, Leeuwenburgh C, Speakman JR. Energy expenditure of calorically restricted rats is higher than predicted from their altered body composition. Mech Ageing Dev. 2005;126:783–793. doi: 10.1016/j.mad.2005.02.004. [DOI] [PubMed] [Google Scholar]
- Shi T, Wang F, Stieren E, Tong Q. SIRT3, a mitochondrial sirtuin deacetylase, regulates mitochondrial function and thermogenesis in brown adipocytes. J Biol Chem. 2005;280:13560–13567. doi: 10.1074/jbc.M414670200. [DOI] [PubMed] [Google Scholar]
- Speakman JR, Talbot DA, Selman C, Snart S, McLaren JS, Redman P, Krol E, Jackson DM, Johnson MS, Brand MD. Uncoupled and surviving: individual mice with high metabolism have greater mitochondrial uncoupling and live longer. Aging Cell. 2004;3:87–95. doi: 10.1111/j.1474-9728.2004.00097.x. [DOI] [PubMed] [Google Scholar]
- Spiegelman BM, Flier JS. Obesity and the regulation of energy balance. Cell. 2001;104:531–543. doi: 10.1016/s0092-8674(01)00240-9. [DOI] [PubMed] [Google Scholar]
- Sugimoto Y, Erikson RL. Phosphatidylinositol kinase activities in normal and Rous sarcoma virus-transformed cells. Mol Cell Biol. 1985;5:3194–3198. doi: 10.1128/mcb.5.11.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiraby C, Tavernier G, Lefort C, Larrouy D, Bouillaud F, Ricquier D, Langin D. Acquirement of brown fat cell features by human white adipocytes. J Biol Chem. 2003;278:33370–33376. doi: 10.1074/jbc.M305235200. [DOI] [PubMed] [Google Scholar]
- Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- Tsukiyama-Kohara K, Poulin F, Kohara M, DeMaria CT, Cheng A, Wu Z, Gingras AC, Katsume A, Elchebly M, Spiegelman BM, Harper ME, Tremblay ML, Sonenberg N. Adipose tissue reduction in mice lacking the translational inhibitor 4E-BP1. Nat Med. 2001;7:1128–1132. doi: 10.1038/nm1001-1128. [DOI] [PubMed] [Google Scholar]
- Valle I, Alvarez-Barrientos A, Arza E, Lamas S, Monsalve M. PGC-1α regulates the mitochondrial antioxidant defense system in vascular endothelial cells. Cardiovasc Res. 2005;66:562–573. doi: 10.1016/j.cardiores.2005.01.026. [DOI] [PubMed] [Google Scholar]
- Vanfleteren JR, Braeckman BP. Mechanisms of life span determination in Caenorhabditis elegans. Neurobiol Aging. 1999;20:487–502. doi: 10.1016/s0197-4580(99)00087-1. [DOI] [PubMed] [Google Scholar]
- Vicent D, Ilany J, Kondo T, Naruse K, Fisher SJ, Kisanuke Y, Bursell S, Yanagisawa M, King GL, Kahn CR. The role of endothelial signalling in the regulation of vascular tone and insulin resistance. J Clin Invest. 2003;111:1373–1380. doi: 10.1172/JCI15211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walford RL, Mock D, Verdery R, MacCallum T. Calorie restriction in biosphere 2: alterations in physiologic, hematologic, hormonal, and biochemical parameters in humans restricted for a 2-year period. J Gerontol A Biol Sci Med Sci. 2002;57:B211–B224. doi: 10.1093/gerona/57.6.b211. [DOI] [PubMed] [Google Scholar]
- Weyer C, Walford RL, Harper IT, Milner M, MacCallum T, Tataranni PA, Ravussin E. Energy metabolism after 2 y of energy restriction: the biosphere 2 experiment. Am J Clin Nutr. 2000;72:946–953. doi: 10.1093/ajcn/72.4.946. [DOI] [PubMed] [Google Scholar]
- Wisniewski D, Strife A, Swendeman S, Erdjument-Bromage H, Geromanos S, Kavanaugh WM, Tempst P, Clarkson B. A novel SH2-containing phosphatidylinositol 3–5-trisphosphate 5-phosphatase (SHIP2) is constitutively tyrosine phosphorylated and associated with src homologous and collagen gene (SHC) in chronic myelogenous leukemia progenitor cells. Blood. 1999;93:2707–2720. [PubMed] [Google Scholar]
- Yechoor VK, Patti ME, Ueki K, Laustsen PG, Saccone R, Rauniyar R, Kahn CR. Distinct pathways of insulin-regulated versus diabetes-regulated gene expression: an in vivo analysis in MIRKO mice. Proc Natl Acad Sci USA. 2004;101:16525–16530. doi: 10.1073/pnas.0407574101. [DOI] [PMC free article] [PubMed] [Google Scholar]