Abstract
Chromosome conformation is an important feature of metazoan gene regulation1,2; however, enhancer–promoter contact remodeling during cellular differentiation remains poorly understood3. To address this, genome-wide promoter capture Hi-C (CHi-C)1,4 was performed during epidermal differentiation5. Two classes of enhancer–promoter contacts associated with differentiation-induced genes were identified. The first class (‘gained’) increased in contact strength during differentiation in concert with enhancer acquisition of the H3K27ac activation mark. The second class (‘stable’) were pre-established in undifferentiated cells, with enhancers constitutively marked by H3K27ac. The stable class was associated with the canonical conformation regulator cohesin, whereas the gained class was not, implying distinct mechanisms of contact formation and regulation. Analysis of stable enhancers identified a new, essential role for a constitutively expressed, lineage-restricted ETS-family transcription factor, EHF, in epidermal differentiation. Furthermore, neither class of contacts was observed in pluripotent cells, suggesting that lineage-specific chromatin structure is established in tissue progenitor cells and is further remodeled in terminal differentiation.
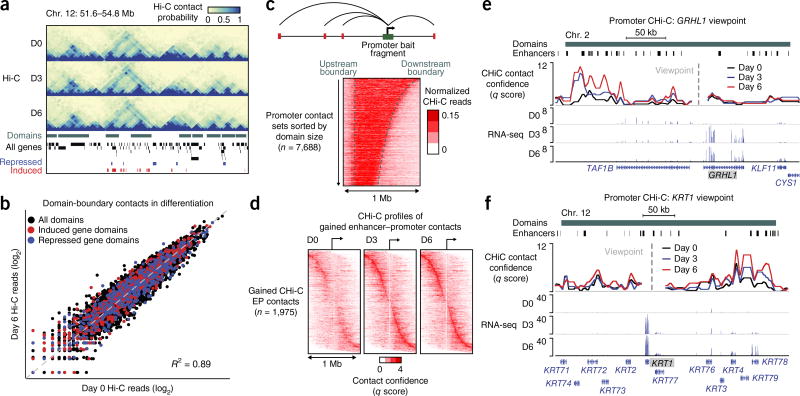
Chromatin architecture dynamics in the terminal differentiation of somatic tissues are not well understood6–10. Hi-C was therefore applied to progenitor and differentiating primary human epidermal keratinocytes, identifying interaction domains separated by boundaries exhibiting stable positions from undifferentiated progenitor-containing cell populations (day 0) to early (day 3) and late (day 6) calcium-induced differentiation in vitro (Fig. 1a). A global set of contacts was defined that are anchored at the boundaries of previously defined contact domains11, and Hi-C read counts were then compared for these contacts during different stages of differentiation (Fig. 1b and Supplementary Table 1). Domain-boundary contacts were stable during differentiation (Fig. 1b and Supplementary Fig. 1a). RNA-seq performed across the differentiation time course (Supplementary Table 2) demonstrated that >95% of induced and repressed genes resided in domains with stable boundaries (Supplementary Fig. 1b), suggesting that regulatory dynamics occur via intradomain contacts.
Figure 1.
Remodeling of the enhancer–promoter contact landscape in terminal differentiation. (a) Genomic locus of keratin-family genes. From top to bottom: Hi-C interaction matrices, self-interacting domains (green), and genes induced (red) or repressed (blue) on day 3 or day 6 of epidermal cell differentiation identified by RNA-seq. (b) Scatterplot of Hi-C read counts supporting contacts between domain boundaries. Contacts enclosing domains containing differentially expressed genes are highlighted. (c) Heat map of CHi-C read count profiles observed for bait HindIII fragments residing within a domain. Each row represents the contact signal from a single promoter, promoters are aligned by distance to the upstream domain boundary, and rows are sorted in order of domain size. (d) Heat maps of CHi-C q scores (determined by CHiCAGO) for 1,975 enhancer contacts gained with target promoters during epidermal cell differentiation. Contacts are sorted in order of enhancer–promoter (EP) distance. (edgeR, FDR < 0.1; fold change > 2). (e,f) Genomic loci for the GRHL1 (e) and KRT1 (f) genes with promoter CHi-C signal. Enhancers correspond to promoter-distal H3K27ac ChIP–seq peaks.
To explore this possibility, genome-wide promoter CHi-C was performed during differentiation. Histone H3 lysine 27 acetylation (H3K27ac) ChIP–seq data provided distal H3K27ac peaks used to identify putative enhancers in contact with promoters12,13. Distal H3K27ac peaks drove enhancer reporter activity and exhibited expected patterns for other histone marks relative to promoters (Supplementary Fig. 1c,d). CHi-C identified 207,663 enhancer–promoter contacts and 89,752 promoter–promoter contacts throughout differentiation. Both classes of contacts were largely restricted to single domains (Fig. 1c and Supplementary Fig. 1e). Notably, 3,575 enhancer–promoter contacts had increased CHi-C signal as differentiation progressed, with 1,975 exhibiting a >2-fold change (Fig. 1d and Supplementary Table 3). A connection between gained contacts and induction of differentiation-related genes was exemplified by enhancer–promoter interactions involving the key differentiation-associated genes GRHL1 and KRT1 (refs. 14,15) (Fig. 1e,f and Supplementary Fig. 2a,b).
3,207 enhancer–promoter contacts with decreased signal in differentiation were also observed, of which 1,481 exhibited a >2-fold change (Supplementary Fig. 2c and Supplementary Table 3). 735 genes induced or repressed in differentiation were engaged in a dynamic contact, consistent with contact network rewiring within domains during differentiation. In contrast to contacts at domain boundaries, promoter-associated contacts with increased strength exhibited a global bias for association with the promoters of differentiation-induced genes, and a similar bias was observed for contacts with reduced signal and the promoters of repressed genes (Supplementary Fig. 2d). Gene ontology (GO) term analysis of induced genes associated with gained contacts showed enrichment for epidermal differentiation terms, whereas repressed genes associated with lost contacts were enriched for GO terms related to the progenitor state, such as proliferation (Supplementary Fig. 2e).
In addition to dynamic enhancer–promoter contacts, the promoters of differentiation-induced genes also engaged in stable, pre-established enhancer–promoter contacts. 65% (502 genes) were engaged exclusively in stable enhancer–promoter contacts, as compared to 6% (43 genes) that were engaged exclusively in gained contacts (Supplementary Fig. 3a,b) and 29% (222 genes) that were engaged in both gained and stable contacts. The former group echoes recent findings in Drosophila melanogaster embryogenesis, where the vast majority of enhancer–promoter contacts are pre-established before gene induction8, and with findings at Hox loci in mouse development, where tissue-invariant structural contacts form a universal architecture that guides tissue-specific enhancer–promoter interactions16,17. Differentiation genes within each category of enhancer–promoter contacts had similar GO terms related to epidermal differentiation (Supplementary Fig. 3c). The GO term enrichment for the gained-only set was limited owing to its small size; however, these genes were induced at similar levels to the other two sets and included genes of the known differentiation-associated kallikrein, late cornified envelope, and MAF families (Supplementary Fig. 3d and Supplementary Table 4). Interestingly, genes involved in both gained and stable contacts (GS genes) exhibited more lineage-specific expression than stable-contact-only genes (Supplementary Fig. 3e).
The relationship between enhancer chromatin state, enhancer–promoter contacts, and gene expression was next examined. Enhancers that were dynamically marked (gained or lost) for H3K27ac during differentiation were first identified (Supplementary Table 5). Gene induction was highly correlated with the number of H3K27ac-gained enhancers in contact with genes; similarly, gene repression was correlated with the number of H3K27ac-lost enhancers in contact with genes in progenitors (Fig. 2a). This result is consistent with observations from the hair follicle, within which dramatic H3K27ac dynamics accompany stem cell differentiation18. Interestingly, distal regions that gained contacts with repressed genes were enriched for a bivalent enhancer state as determined by chromHMM in progenitor cells, suggesting a negative regulatory role (Supplementary Fig. 4a). Overall, these results support a model in which multiple classes of enhancers with distinct H3K27ac dynamics interact with a gene, not only to provide regulatory robustness19 but also to increase the magnitude of gene induction.
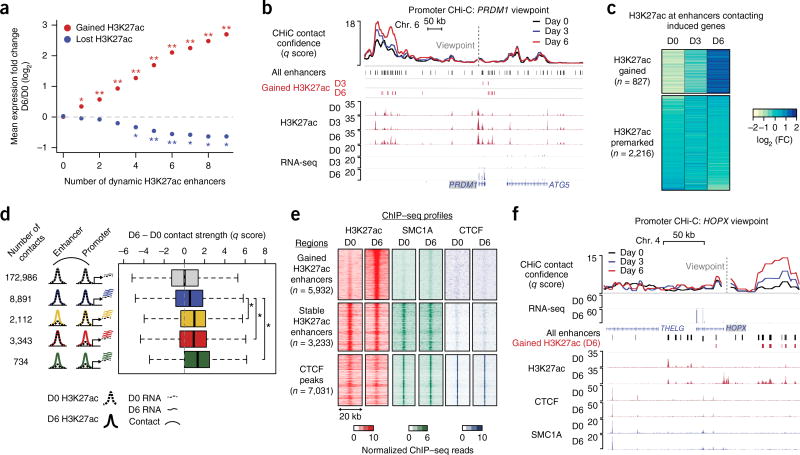
Figure 2.
Enhancer activation and contact dynamics are linked independently of cohesin. (a) Mean change in expression for genes in CHi-C contacts with enhancers displaying gained or lost H3K27ac status (empirical FDR, *, FDR < 0.05; **, FDR < 0.01). (b) Genomic locus of the PRDM1 gene, including CHi-C q score and H3K27ac ChIP–seq signal tracks. Gained enhancers correspond to H3K27ac peaks with significant gain in H3K27ac signal on day 3 or day 6 versus day 0 (edgeR, FDR < 0.05; fold change > 2). (c) Heat map of the fold change in H3K27ac ChIP–seq read count. Regions are separated into the set with gained H3K27ac signal on day 6 versus day 0 (edgeR, FDR < 0.05; fold change > 2) and the set with premarked H3K27ac on day 6 versus day 0 (edgeR, FDR > 0.7). (d) Box-and-whisker plots of the difference in contact q score between day 6 and day 0. Contact sets are defined by H3K27ac dynamics at the promoter locus (bait HindIII fragment) or enhancer. Dashed lines denote day 0 for either H3K27ac signal or mRNA expression of differentiation-related genes; solid lines denote corresponding signals for day 6. Each box represents the median and interquartile range; whiskers extend to 1.5 times the interquartile range (empirical FDR, *, FDR < 0.01). (e) Heat maps of H3K27ac, SMC1A, and CTCF ChIP–seq signals at gained enhancers, stable enhancers, and CTCF peaks (stable enhancers and CTCF peaks were restricted to chromosome 1 to approximately match the number of gained enhancer peaks). (f) HOPX locus, including CHi-C q score and ChIP–seq tracks for H3K27ac, CTCF, and SMC1A.
An example of such a mixed-class contact landscape was found at the PRDM1 gene, which contacts both gained and stable H3K27ac peaks (Fig. 2b and Supplementary Fig. 4b). Extending this analysis to all 3,043 enhancers in contact with differentiation-induced genes, 827 significantly gained H3K27ac during differentiation, whereas 2,216 showed stable levels of H3K27ac (Fig. 2c). Promoters contacting differentiation-related genes showed a similar pattern (Supplementary Fig. 4c). Analysis of pluripotent cells and distant somatic tissues showed that stable enhancers and, to a greater degree, H3K27ac-gained enhancers were specifically marked by H3K27ac in keratinocytes (Supplementary Fig. 5a–d). Analysis of promoter CHi-C data from human embryonic stem cells (hESCs) showed that both gained and pre-established enhancer–promoter contacts associated with differentiation genes showed significant reduction in signal in hESCs relative to kerationcytes, unlike the tissue-invariant contacts described in the mouse Hox loci16,17 (Supplementary Fig. 5e–g). Moreover, clear hESC-specific contact signal was detected at the SOX21 gene that has hESC-specific expression (Supplementary Fig. 5h). These results suggest that a set of somatic enhancer–promoter contacts is established at a developmental stage after pluripotency but before induction of terminal differentiation genes.
The relationship between enhancer or promoter chromatin state activation, as measured by H3K27ac, and contact dynamics was next examined. Notably, gain of H3K27ac at an enhancer or promoter during differentiation was associated with significant increases in contact strength (Fig. 2d and Supplementary Fig. 6a). In contrast, constitutively H3K27ac-marked enhancers and promoters showed generally stable contact strength.
The conformation-associated factors CTCF and cohesin influence diverse classes of contacts in many settings11,7,20. To understand the potential roles of these factors in the regulatory landscape of epidermal differentiation, their genomic distribution was examined. Analysis of CTCF ChIP–seq peaks found widespread colocalization of CTCF and the cohesin subunit SMC1A (Fig. 2e, bottom). These sites were enriched at domain boundaries, as expected (Supplementary Fig. 6b)2,21. Cohesin, but not CTCF, was associated with constitutively H3K27ac-marked enhancers, as has been observed in other systems7,22 (Fig. 2e). Unexpectedly, minimal cohesin binding occurred at enhancers that gained H3K27ac during differentiation (Fig. 2e and Supplementary Fig. 6c). The absence of cohesin was exemplified at the HOPX locus, which exhibited dramatically increased contacts between the HOPX promoter and distal enhancers that gained H3K27ac during differentiation (Fig. 2f and Supplementary Fig. 6d). Few of the 77,155 enhancer and promoter regions exhibited significant cohesin dynamics relative to day 0 (3 and 4 regions gained and 12 and 1 regions lost on days 3 and 6, respectively, at false discovery rate (FDR) < 0.1, no minimum fold change; Supplementary Fig. 6e,f). Therefore, progenitor-established enhancer–promoter contacts are associated with premarked H3K27ac and constitutive cohesin binding at enhancers, whereas enhancer–promoter contacts acquired in differentiation are associated with enhancers that gain H3K27ac and lack cohesin.
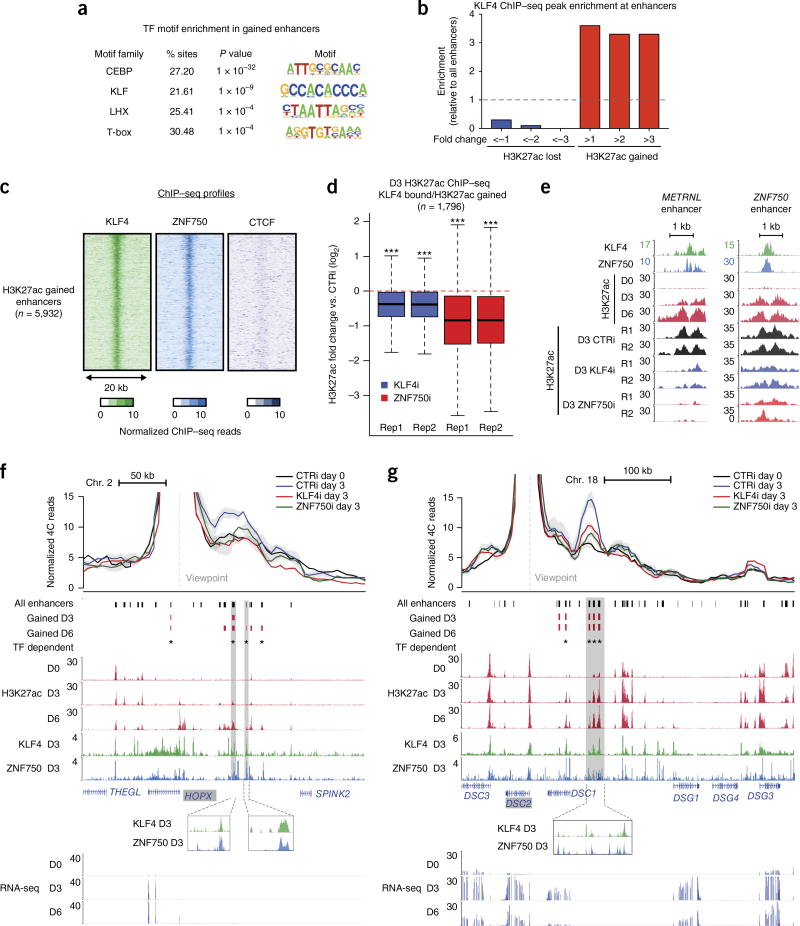
Given the absence of cohesin at H3K27ac-gained enhancers, these regions were analyzed for transcription factor motifs corresponding to factors potentially regulating contacts and H3K27ac dynamics. C/EBP- and KLF-family motifs were highly enriched, consistent with the findings that C/EBPα, C/EBPβ, and the differentiation-induced transcription factor KLF4 are essential for epidermal differentiation23–25 (Fig. 3a). KLF4 binding was enriched at enhancers that acquired H3K27ac and depleted at enhancers that lost H3K27ac during differentiation (Fig. 3b). Moreover, ZNF750—another essential, differentiation-specific transcription factor that physically interacts with KLF4 to drive differentiation24,26—colocalized with KLF4 at H3K27ac-gained enhancers (Fig. 3c). KLF4- and ZNF750-bound enhancers also frequently overlapped super-enhancers (Supplementary Fig. 7a).
Figure 3.
Induced transcription factors couple regulation of chromatin activation and conformation. (a) Enrichment of transcription factor (TF) motifs identified by HOMER in enhancers gained on day 6 versus enhancers lost on day 6. Motifs are ranked by −log10 (P value). (b) Enrichment of day 3 KLF4 ChIP–seq peaks in H3K27ac peak subsets determined by fold change in H3K27ac dynamics on day 3 versus day 0. (c) Heat maps of KLF4, ZNF750, and CTCF ChIP–seq signal on day 3 at enhancers with gained H3K27ac signal. (d) Box-and-whisker plots representing relative H3K27ac ChIP–seq signal in control versus transcription factor–knockdown conditions, for two biological replicates of each knockdown. Regions analyzed were H3K27ac-gained enhancers bound by KLF4. Keratinocytes were treated with siRNAs targeting KLF4 (KLF4i), ZNF750 (ZNF750i), or a scrambled control (CTRi) and placed in differentiation conditions for 3 d. Each box represents the median and interquartile range; whiskers extend to 1.5 times the interquartile range (empirical FDR, ***, FDR < 10 × 10−3). (e) Representative genomic loci depicting the effect of KLF4 or ZNF750 depletion at enhancers. Both loci exhibit significant loss of H3K27ac upon KLF4 or ZNF750 depletion (edgeR, FDR < 0.05). (f) UMI-4C profile of interactions anchored by the HOPX promoter in control and KLF4- or ZNF750-knockdown conditions. Error bands represent s.e.m. between replicates. Vertical shaded boxes represent KLF4- or ZNF750-binding sites coinciding with gained H3K27ac. Gained H3K27ac peaks in normal differentiation that exhibit H3K27ac signal loss in KLF4 or ZNF750 knockdown (edgeR, FDR < 0.05) are flagged as transcription factor dependent. (g) As in f, with the UMI-4C profile anchored by the DSC2 promoter.
These findings raised the possibility that KLF4 and ZNF750 contribute to both enhancer activation and enhancer–promoter contact formation. To address this possibility, H3K27ac ChIP–seq was performed in differentiating cells treated with small interfering RNAs (siRNAs) targeting either KLF4 or ZNF70 (Supplementary Fig. 7b). Depletion of either factor impaired acquisition of H3K27ac at regions bound by these factors that gained H3K27ac in differentiation (Fig. 3d,e), confirming that both are required for enhancer commissioning. Comparison of genome-wide H3K27ac signal in transcription factor–depleted cells, normally differentiating cells, and cells of other lineages indicated that transcription factor depletion altered epidermal differentiation but did not alter epidermal identity (Supplementary Fig. 7c).
To assess whether KLF4 and ZNF750 also influence enhancer–promoter contacts, contact strength in transcription factor–depleted cells was measured by targeted chromosome conformation capture with unique molecular identifiers (UMI-4C)27. Promoter-centric contact profiles for HOPX, DSC2, and PRDM1, three differentiation-related genes regulated by KLF4 and ZNF750 (Supplementary Fig. 7d), were chosen for analysis. Depleting either KLF4 or ZNF750 decreased the strength of differentiation-specific contacts between H3K27ac-gained enhancers bound by these factors and target promoters, whereas other enhancers retained contact (Fig. 3f,g and Supplementary Fig. 7e). However, the binding of KLF4 and ZNF750 to many other regions suggests that these factors do not specify contact and chromatin state dynamics at all enhancers with which they associate. Indeed, a dynamic contact between the KLF4- and ZNF750-bound promoters of DSC2 and DSC3 was not regulated by either transcription factor. Therefore, the differentiation-induced transcription factors KLF4 and ZNF750 are required for both acquisition of H3K27ac and establishment of enhancer–promoter contacts at a subset of enhancers targeting differentiation-associated genes.
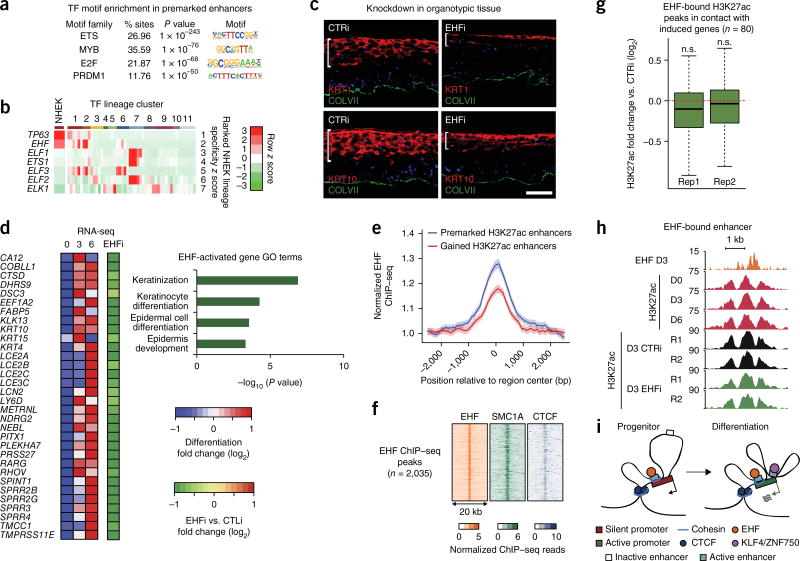
Pre-established enhancers have been linked to gene regulation in other tissues such as the intestine and blood8,28,29, so enhancers in stable contact with differentiation-induced genes were analyzed for transcription factor motifs that might correspond to factors regulating this process. The most common enriched motifs corresponded to the ETS family30 (Fig. 4a and Supplementary Fig. 8a). The EHF ETS-family transcription factor showed the most lineage-specific expression31 in stratified epithelia (Fig. 4b and Supplementary Fig. 8b), similar to the master epithelial regulator TP63 (encoding p63)32. EHF expression was largely stable during differentiation (Supplementary Fig. 8c–e). EHF depletion in organotypic human epidermal tissue, which retains the architecture and differentiation of intact epidermis33, impaired expression of key differentiation-related genes (Fig. 4c and Supplementary Fig. 9a,b). RNA-seq analysis of EHF-depleted epidermal tissue demonstrated that EHF loss altered expression of 400 genes (Supplementary Fig. 9c and Supplementary Table 6). EHF-dependent genes included keratin, SPRR, and LCE genes and were associated with GO terms that included keratinocyte differentiation and epidermis development (Fig. 4d), whereas EHF-repressed genes were enriched for functions associated with other lineages (Supplementary Fig. 9c). EHF is thus required both for induction of differentiation-related genes and for repression of ectopic gene expression.
Figure 4.
The transcription factor EHF associates with premarked H3K27ac elements and is essential for epidermal differentiation. (a) Top enriched transcription factor motifs identified by HOMER in premarked H3K27ac elements in contact with the promoters of differentiation-induced genes. (b) Heat map representing z scores of transcription factor expression values determined by RNA-seq from the Roadmap Epigenomics Project. Each cell corresponds to the average expression z score across the cell type groups identified through hierarchical clustering of all genes. Rows are sorted by the z score in the cluster containing epidermal keratinocytes, and the z scores for epidermal keratinocytes are shown separately in the leftmost column. Cluster IDs are as follows: 1, epithelial; 2, cancer cell line; 3, simple epithelial; 4, extramedullary hematopoietic; 5, muscle; 6, gastrointestinal/hepatic; 7, immune; 8, neural; 9, mesenchymal; 10, embryonic; 11, other. (c) Immunofluorescence microscopy of organotypic epidermis treated with siRNAs targeting EHF (EHFi) or a scrambled control. The white bar highlights the height of the region marked by the differentiation-specific proteins KRT1 and KRT10. Scale bar, 50 µm. (d) Heat map representing mRNA expression of differentiation-induced genes in EHF-depleted organotypic epidermis relative to normal control. GO terms were derived for EHF-dependent genes. (e) EHF ChIP–seq metaplot signal at H3K27-premarked (n = 5,932) and H3K27ac-gained (n = 3,233) putative enhancers. Error bands represent 98% boostrapped confidence intervals. (f) Heat maps of day 3 EHF, SMC1A, and CTCF ChIP–seq profiles at all EHF ChIP–seq peaks. (g) Box-and-whisker plots representing relative H3K27ac ChIP–seq signal at day 3 of differentiation in control versus EHF-knockdown conditions. Each distribution represents a biological replicate. Regions analyzed are H3K27ac peaks bound by EHF and in contact with the promoters of differentiation-induced genes. Each box represents the median and interquartile range; whiskers extend to 1.5 times the interquartile range (empirical FDR, n.s., FDR > 0.05). (h) Representative genomic locus demonstrating the effect of EHF depletion at EHF-bound enhancers. (i) Working model. Induction of differentiation-related genes involves two types of enhancer–promoter interactions that occur within CTCF- and cohesin-bound domains: (i) H3K27ac premarked enhancers, bound by cohesin and by constitutively expressed transcription factors, such as EHF, in stable contact with differentiation-related genes and (ii) enhancers that bind inducible transcription factors, such as KLF4 and ZNF750, to gain H3K27ac marks and increase contact with differentiation-related genes.
Consistent with the constitutive expression of EHF, EHF binding was detected at stably H3K27ac-marked, ETS-motif-containing enhancers that contacted EHF-dependent differentiation-related genes (Supplementary Fig. 9d). EHF binding at these sites remained largely stable as cells transitioned from the progenitor state to differentiation. EHF ChIP–seq analysis in differentiating cells identified loci highly enriched for the ETS motif (Supplementary Fig. 9e) and demonstrated enriched binding at constitutively H3K27ac-marked enhancers over those that dynamically gained H3K27ac, consistent with the motif analysis (Fig. 4e). Additionally, these EHF-bound, H3K27ac-premarked enhancers generally engaged in stable contacts with differentiation-associated genes (Supplementary Fig. 9f). Consistent with the observation that constitutive enhancers are enriched for cohesin, EHF-bound regions exhibited widespread co-occupancy with SMC1A (Fig. 4f).
To determine whether EHF directly maintains enhancer–promoter contacts and chromatin state at enhancers with which it is associated, H3K27ac ChIP–seq and UMI-4C were performed in EHF-depleted differentiating cells. EHF-bound enhancers in contact with the promoters of differentiation-induced genes maintained H3K27ac levels in the absence of EHF, while enhancer–promoter contacts involving EHF-bound enhancers at the EHF-regulated DSG3 locus were also unaffected (Fig. 4g,h and Supplementary Fig. 9g,h). These data suggest that EHF regulates gene expression in a manner distinct from KLF4 and ZNF750, namely by using a chromatin state and contact scaffold maintained by other factors, such as DNMT3A and DNMT3B34, to prime gene induction.
Here we used chromosome conformation analysis to identify two classes of enhancer–promoter contacts in epidermal differentiation. The first gained enhancer–promoter contact strength during differentiation in concert with enhancer acquisition of H3K27ac. Unlike previously studied enhancers, these dynamic enhancers exhibited minimal binding to cohesin, instead relying on differentiation-induced transcription factors, such as KLF4 and ZNF750, to regulate the contacts and underlying enhancer activation. The second class of enhancer–promoter contacts, in contrast, was pre-established in progenitor cells but not in embryonic stem cells and connected constitutively H3K27ac-marked enhancers with differentiation-induced promoters. These enhancers were constitutively bound by cohesin as well as stably expressed, essential transcription factors, such as EHF. These findings inform a model (Fig. 4i) in which two enhancer–promoter contact classes with distinct regulatory mechanisms cooperate to induce the expression of differentiation-related genes within stable domains bounded by CTCF and cohesin. More broadly, these results suggest that progenitor cells partially pre-establish a regulatory apparatus that is fully engaged in terminal differentiation.
URLs. HOMER, http://biowhat.ucsd.edu/homer/index.html; Roadmap Epigenomics Project online data portal, http://egg2.wustl.edu/roadmap/data/byFileType/chromhmmSegmentations/ChmmModels/coreMarks/jointModel/final/; GWAS Catalog from EMBL, http://www.ebi.ac.uk/gwas/docs/file-downloads.
ONLINE METHODS
Cell culture
Primary human keratinocytes were isolated from fresh, surgically discarded neonatal foreskin. Keratinocytes were grown in 1:1 KCSFM and Medium 154 (Life Technologies). Keratinocytes were induced to differentiate by addition of 1.2 mM calcium for 3 or 6 d at full confluence. For regenerated organotypic epidermal tissue, 1 million siRNA-treated keratinocytes were seeded onto devitalized human dermis for 4 d and then harvested for RNA or protein.
Hi-C library preparation
Hi-C library generation was carried out in biological duplicate samples at days 0, 3, and 6 as described previously with the following modifications1. After fixation in 2% formaldehyde for 10 min, 20 to 30 million cells were Dounce homogenized in 10 ml of ice-cold lysis buffer ten times on ice with a tight pestle, incubated on ice for 15 min, and then Dounce homogenized a further ten times. After overnight digestion with HindIII at 37 °C, DNA ends were labeled with biotin-14-dATP (Life Technologies) in a Klenow end-filling reaction and ligated in nuclei overnight. After phenol-chloroform purification, the DNA concentration was measured using Quant-iT PicoGreen (Life Technologies), and 40 µg of DNA was sheared to an average size of 400 bp, using the manufacturer’s instructions (Covaris). The sheared DNA was end repaired, adenine tailed, and double size selected using AMPure XP beads to isolate DNA ranging from 250 to 550 bp in size. Ligation fragments marked by biotin were immobilized using MyOne Streptavidin C1 DynaBeads (Invitrogen) and ligated to paired-end adaptors (Illumina). The immobilized Hi-C libraries were amplified using PE PCR 1.0 and PE PCR 2.0 primers (Illumina) with 6–9 PCR amplification cycles.
Biotinylated RNA bait library design
Biotinylated 120-mer RNA baits were designed to target both ends of HindIII restriction fragments overlapping the Ensembl promoters of protein-coding, noncoding, antisense, snRNA, miRNA, and snoRNA transcripts. A target sequence was valid if its GC content ranged between 25 and 65% and the sequence contained no more than two consecutive Ns and was within 330 bp of the HindIII restriction fragment terminus.
Promoter capture Hi-C
Capture Hi-C of promoters was carried out in biological duplicate samples at days 0, 3, and 6 with SureSelect target enrichment, using the custom-designed biotinylated RNA bait library and custom paired-end blockers according to the manufacturer’s instructions (Agilent Technologies). After library enrichment, a post-capture PCR amplification step was carried out using PE PCR 1.0 and PE PCR 2.0 primers with 4–6 PCR amplification cycles. Hi-C and CHi-C libraries were sequenced on the Illumina HiSeq 2000 platform.
UMI-4C library preparation and analysis
4C libraries were generated as previously described24 with the following modifications. Contacts were generated in intact nuclei following the in situ Hi-C protocol11 to minimize spurious ligations. After ligation, nuclei were spun down and resuspended in 95 µl of PK Buffer (10 mM Tris pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.5% SDS) and 5 µl of 20 mg/ml proteinase K (Thermo Fisher, AM2548). Protein was degraded at 55 °C for 45 min, and cross-links were reversed at 68 °C for at least 2 h. 5 µl of 5 mg/ml RNase A (Affymetrix, 70194Z) was then added, and samples were incubated at 37 °C for 30 min. DNA was ethanol precipitated, and the pellet was resuspended in 200 µl of 10 mM Tris pH 8.0. DNA was quantified with a Nanodrop, and 5–10 µg of DNA was taken into sonication and subsequent UMI-4C library construction. We generated two biological replicate UMI-4C libraries for each experiment and then performed two PCRs per replicate for each anchor locus (resulting in a total of four libraries for each locus per experiment). HiC-Pro35 was used to align paired-end reads, assign reads to MboI restriction fragments, filter for spurious contacts, and count reads supporting contact between 5-kb bins. Bins corresponding to anchor loci (primer-binding sites) were used to plot profiles for each sample. Profiles were scaled by the total read pairs associated with the anchor bin in each sample, divided by 1,000, to account for variable read depth. Profiles were plotted in R and smoothed with the rollmean function of the zoo package.
Chromatin immunoprecipitation
For ChIP followed by qPCR, 2–5 million primary keratinocytes were used as starting material; for ChIP–seq, 10–30 million cells were used. ChIP was performed essentially as described24. Pulldown was performed with 10 µg of ChIP–seq-validated antibody against H3K27ac (Abcam, ab4729), CTCF (Millipore, 07-729), SMC1A (Bethyl, A300-055A), or EHF (Santa Cruz Biotechnology, sc-166653). Staph A cells were used for pulldown. DNA was purified using Qiagen QIAquick PCR Purification columns and subjected to qPCR (primer sequences in Supplementary Table 7). For qPCR, the percentage of input signal was calculated, and error bars represent s.e.m. calculated using GraphPad Prism. ChIP–seq libraries were prepared with the NEBNext ChIP–seq library prep kit (NEB) using AMPure beads (Agencourt) for purification.
Immunoblotting
For immunoblot analysis of EHF, total protein was extracted in RIPA buffer with Complete Mini, EDTA-free Protease inhibitor cocktail tablets (Roche), and 30 µg of total protein was resolved by 4–12% SDS–PAGE and transferred to a nitrocellulose membrane. The membrane was incubated in primary antibody (Santa Cruz Biotechnology, sc-166653) overnight at 4 °C at a concentration of 1:500 and in secondary antibody (LI-COR) for 1 h at room temperature. The LI-COR Odyssey Clx instrument was used for protein detection, and expression of actin was detected to confirm equal loading.
Immunofluorescence microscopy
7-µm sections of organotypic epidermal tissue were fixed with 4% formaldehyde for 15 min followed by blocking in PBS with 2.5% normal goat serum, 0.3% Triton X-100, and 2% BSA for 30 min. Primary antibodies against keratin-1 (Covance, PRB-149P; 1:2,000 dilution), keratin-10 (Neomarkers, MS611P; 1:350 dilution), collagen VII (Calbiochem, 234192; 1:200 dilution), and collagen VII (Chemicon, MAB2500; 1:200 dilution) were added overnight at 4 °C, and secondary antibodies were added for 1 h at room temperature. Quantification of immunofluorescence signal was performed with ImageJ, and Student’s t tests were used to compare biological samples.
qRT–PCR gene expression analysis
The RNeasy plus kit (Qiagen) was used to extract total RNA. Approximately 1 µg was used as input to the iSCRIPT cDNA synthesis kit (Bio-Rad). cDNA and primers were mixed with Maxima SYBR Green Mastermix (Fermentas), and the Roche 480 LightCycler instrument was used for qPCR.
Enhancer reporter assays
Reporter constructs were generated by cloning regions of interest into the lentiviral pGreenFire vector (System Biosciences). Firefly luciferase activity was measured using the Dual-Luciferase Reporter Assay System from Promega. Luminescence was normalized to proviral integrants as previously described26.
siRNA-mediated knockdown
For siRNA-mediated gene transfer, 1 × 106 primary human keratinocytes were electroporated with 1 nmol of siRNA oligonucleotide, using the human keratinocyte Nucleofector kit (Lonza). Dharmacon ON-TARGET siRNA sequences were as follows: ZNF750 (5′-CCACCAGAGTTTCCACATA-3′), KLF4 (5′-TGACCAGGCACTACCGTAA-3′), EHF siRNA A (5′-GGAAGGAGGTGGTGTAATGTT-3′), and EHF siRNA B (5′-GACGAGAACTATTTATATG-3′). EHF siRNA B was used for RNA-seq, H3K27ac ChIP–seq, and UMI-4C experiments.
RNA-seq library preparation, data processing, and dynamic expression calls
RNA-seq libraries were prepared with TruSeq RNA Library Prep Kit v2 (Illumina). For quantitative comparison of transcriptional changes in differentiation, paired-end RNA-seq reads were obtained from biological duplicates at days 0, 3, and 6, with a read depth of 6 × 107 read pairs per sample per time point. For RNA-seq with knockdown in organotypic tissue, three technical replicates were obtained for both EHF and CTR knockdown, with a read depth of 3 × 107 read pairs per sample. Reads were aligned to the GENCODE v19 transcriptome in hg19 using STAR aligner version 2.4.1d with default settings. RSEM version 1.2.21 was then used to calculate expected read counts per gene that were input to edgeR version 3.4.2 for differential expression analysis. Analysis of the read count distribution indicated that a threshold of five reads per gene generally separated expressed from unexpressed genes, so all genes with fewer than five reads were excluded from edgeR analysis. Enriched GO terms for RNA-seq differentially expressed gene sets were identified using DAVID36. Signal tracks were generated using previously published data33 by first using BEDTools genomeCoveragebed to produce bedGraph files scaled to 1 million reads per data set. Then, the UCSC Genome Browser utility bedGraphToBigWig was used with default parameters to generate bigwig files.
Motif enrichment analysis
The HOMER tools function findMotifsGenome was used with default parameters and a region size of 500 bp to compare sets of genomic regions for differential motif enrichment. When comparing sets of H3K27ac peaks, we identified sets of narrow open chromatin sites within these peaks by identifying peaks of ATAC–seq summits. ATAC–seq data were generated in a previous study37. The results of the known motif enrichments are presented.
Hi-C and CHi-C data processing, heat maps, interaction calls, and visualization
1.36 × 109 total Hi-C reads and 1.09 × 109 total CHi-C reads were processed using the HiCUP pipeline38, which aligns reads, filters artifact fragments (such as circularized reads and re-ligations), and removes duplicates. To quantitatively score CHi-C interaction strength, we used the CHiCAGO pipeline39, which normalizes reads for sequencing depth and fragment visibility bias and provides a statistical assessment of contact strength by comparing observed reads supporting a particular interaction to a global distance-dependent background model. Interactions between baits with FDR < 0.01 were considered to be promoter–promoter interactions. Interactions between baits and non-bait fragments with FDR < 0.01 were considered to be enhancer–promoter interactions if the non-bait fragment overlapped an H3K27ac peak at any day of the time course. The edgeR package40 version 3.4.2 was used to call significantly increased or decreased interactions on the basis of read counts supporting interactions. A previous study described bias based on contact distance associated with the identification of differential interactions from Hi-C-like data41. We extended this insight to our analysis by splitting CHi-C contacts into two distance regimes of greater or less than 150 kb. Differential interaction calls from each set were thresholded by FDR and fold change and were combined to form the final set of dynamic promoter–promoter or enhancer–promoter interactions. We also noticed that many CHi-C contacts were supported by relatively low read counts to allow for confident differential signal identification40. For this reason, we excluded from edgeR analysis CHi-C contacts that were not supported by at least 15 reads in at least one replicate. For visualization of CHi-C read counts associated with an individual promoter, counts corresponding to contact between individual HindIII fragments and the bait fragment were scaled by the total number of reads emanating from the bait fragment divided by 1,000. This normalization accounted for variable read depth and efficiency of bait capture across samples. Profiles were plotted in R and smoothed with the rollmean function of the zoo package. In all Genome Browser examples of CHi-C signal, the enhancer track corresponds to H3K27ac peaks that do not overlap CHi-C promoter bait HindIII fragments. For comparisons to hESCs, CHi-C data were acquired from the Open Science Framework (accession SDBG4; ref. 42). Global comparisons between keratinocytes and hESCs were performed on quantile-normalized read counts based on the union set of contacts called in hESCs and keratinocytes.
ChIP–seq data processing, heat map generation, and edgeR analysis
KLF4 and ZNF750 ChIP–seq data were generated in a previous study33 (GEO accession GSE57702). H3K27ac, CTCF, SMC1A, and EHF ChIP–seq analyses were performed in undifferentiated and differentiating primary human keratinocytes, with an average range of 20–25 × 106 reads per independent ChIP–seq experiment. H3K27ac and SMC1A ChIP–seq analyses were performed in biological duplicates. ChIP–seq reads were mapped to the hg19 genome with Bowtie2 using default parameters. Aligned reads were filtered for minimum MAPQ of 30, and duplicates were removed using SAMtools. Signal tracks were generated by first using the BEDTools genomeCoverageBed tool to produce bedGraph files scaled to 10 million reads per data set. Then, the UCSC Genome Browser utility bedGraphToBigWig was used with default parameters to generate bigwig files. For H3K27ac, SMC1A, and CTCF signal tracks, we performed linear scaling to normalize for ChIP efficiency. To achieve this, we adjusted the genomeCoverageBed scaling factors by the median read count value observed in a union set of peak calls for each ChIP target from all days. All ChIP–seq signal tracks are displayed with a minimum y axis of 1. Peaks were called using MACS2 with default parameters except for KLF4 and EHF, for which the confidence threshold was adjusted to account for spurious peak calling identified by visual inspection. P values of 1 × 10−8 and 1 × 10−4 were used for KLF4 and EHF peak calls, respectively. Heat maps of ChIP–seq signal profiles were generated with the HOMER (http://biowhat.ucsd.edu/homer/index.html) tool annotatePeaks with the following parameters: -ghist 50, -size 10000. ChIP–seq peaks exhibiting differential H3K27ac or SMC1A signal across the time course were identified using edgeR similarly to above. We noticed that two of the six SMC1A ChIP–seq data sets had relatively low signal-to-background ratios as determined by calculating the fraction of reads in peaks (FRiP)43. Two replicates with a FRiP score below 2% were excluded from edgeR analysis. MA plots representing SMC1A ChIP signal magnitude and dynamics at enhancers (Supplementary Fig. 2d,e) were generated using the smoothScatter function in R.
Calculation of enrichment of gene sets associated with dynamic contact sets
To determine whether sets of dynamic enhancer–promoter contacts were preferentially associated with dynamically expressed genes, we first identified the proportion of all enhancer–promoter contacts in which the contact bait region overlapped the transcription start site (TSS) of a gene in the gene set. We compared the overlap proportion of the dynamic contact sets to the total contact set and report that ratio as the overlap enrichment of the dynamic set. This process was carried out for contact and gene sets identified using edgeR as described above.
Calculation of empirical false discovery rate for dynamics of gene expression based on contacting enhancer status
For Figure 2a, we determined an empirical FDR to identify a gene set with a mean RNA-seq fold change of equal or greater magnitude than what was observed in the experiment. We sampled all genes 1,000 times using a number of samples equal to the size of the test set.
Gene association with dynamic and stable contacts
For Supplementary Figure 3, induced genes were associated with either gained or stable CHi-C contacts at the last time point of differentiation. The stable contact set was defined as having an edgeR FDR >0.7 for dynamic contact signal and a minimum of 15 reads for at least one time point. The dynamic contact set was identified by applying a threshold of FDR < 0.1 and a minimum of 15 reads for at least one time point, as described above. Genes were then classified for promoters associating with gained, stable, or both types of contact.
Roadmap Epigenomics Project chromHMM analysis
The core 15-state chromHMM model for normal human epidermal keratinocytes generated by the Roadmap Epigenomics Project was acquired from the online data portal at http://egg2.wustl.edu/roadmap/data/byFileType/chromhmmSegmentations/ChmmModels/coreMarks/jointModel/final/. ChromHMM v1.1 was used to assess enrichment of genomic regions across each state with the OverlapEnrichment function.
Calculation of empirical false discovery rate for dynamics of contact sets
We calculated empirical FDRs through sampling for the comparisons of contact strength dynamics between pairs of contact sets (Fig. 2d and Supplementary Figs. 4b and 8e). In each case, we considered the stable contact set as a reference and used 1,000 repeated samplings of equal number as the test sample from that set to determine how often a distribution with a mean as or more divergent could be drawn.
Calculation of KLF4 ChIP–seq peak enrichment at dynamic H3K27ac regions
To determine whether KLF4 preferentially binds at dynamic H3K27ac regions, we first calculated the proportion of all H3K27ac peak regions that overlapped a KLF4 peak. We then calculated the proportion of dynamic H3K27ac regions (at various thresholds) that overlapped KLF4 peaks and report the ratio of proportions between the dynamic set of H3K27ac peaks and the total set as the enrichment.
Super-enhancer identification and enrichment analysis
Super-enhancers were identified at each time point using the ROSE (https://bitbucket.org/young_computation/rose) tool with H3K27ac ChIP–seq peaks as input. Default stitching distance and a TSS exclusion distance of 2 kb were used. Enrichment of transcription factor–bound or dynamic contact-associated enhancers was calculated by determining the proportion of all enhancers or specific subsets overlapping super-enhancers. The ratio of these two proportions is reported as the enrichment of the enhancer subset overlap with super-enhancers.
Cell type specificity of ETS transcription factor family member expression
We downloaded gene-level expression read counts for all paired-end, poly(A)-selected RNA-seq data from the Roadmap Epigenomics Project. Expression levels were quantile normalized, and we calculated the Pearson correlation for all pairs of samples. The resulting correlation matrix was clustered using the heatmap.2 function of the gplots package in R, and lineage-associated clusters were manually annotated. We calculated the z score of expression for each gene across the collection of cell types.
Statistics
Analysis-specific statistics were applied as described in each subsection. Sets of significantly differential elements (genes, genomic regions, or contacts) were identified with edgeR. When sampling a subset of elements (genes, genomic intervals, or contacts) from a larger set to compare distributions, empirical FDRs were calculated by randomly sampling sets of equal size to the test set. The mean value of the test set was then compared to the collection of random samples to determine how frequently a random sample exhibited at least as extreme a mean value. For other comparisons of distributions, the two-sided Kolmogorov–Smirnov test was employed in R, and absolute P values are reported. The minimum P value calculated for this test in R is 2.2 × 10−16. For comparisons of immunofluorescence intensity from organotypic tissue, Student’s t test was used.
A Life Sciences Reporting Summary for this publication is available.
Code availability
Custom scripts described in the Online Methods will be made available upon request.
Data availability
All sequencing data are available through the Gene Expression Omnibus (GEO) via accession GSE84662. Source data are provided for Figures 1–4 and Supplementary Figures 1–8.
Supplementary Material
Acknowledgments
We thank J. Wysocka, A. Oro, R. Flynn, O. Wapinski, and A. Brunet for presubmission review. This work was supported by the US Veterans Affairs Office of Research and Development and NIH/NIAMS grant AR45192 to P.A.K. and by NIH grant U01 HG007919 to M.S., H.Y.C., W.J.G., A.K., and P.A.K. This work was supported in part by funding from the Biotechnology and Biological Sciences Research Council (BB/J004480/1) and the European Community’s Seventh Framework Programme (MODHEP consortium 259743) to P.F.
Footnotes
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
B.C.B., A.J.R., and M.F.-M. designed and executed experiments, analyzed data, and wrote the manuscript. I.H., M.R.M., D.S.K., L.D.B., Z.Z., M. Spivakov, and V.L.-P. executed experiments, analyzed data, and contributed to design of experimentation. J.C., M. Spivakov, and S.W.W. developed and executed the CHiCAGO analysis pipeline. W.J.G., H.Y.C., M. Snyder, A.K., P.F., and H.Y.C. contributed to experimental design and analysis. P.A.K. designed experiments, analyzed data, and wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Mifsud B, et al. Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat. Genet. 2015;47:598–606. doi: 10.1038/ng.3286. [DOI] [PubMed] [Google Scholar]
- 2.Dixon JR, Gorkin DU, Ren B. Chromatin domains: the unit of chromosome organization. Mol. Cell. 2016;62:668–680. doi: 10.1016/j.molcel.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dixon JR, et al. Chromatin architecture reorganization during stem cell differentiation. Nature. 2015;518:331–336. doi: 10.1038/nature14222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schoenfelder S, et al. The pluripotent regulatory circuitry connecting promoters to their long-range interacting elements. Genome Res. 2015;25:582–597. doi: 10.1101/gr.185272.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lopez-Pajares V, et al. A lncRNA–MAF:MAFB transcription factor network regulates epidermal differentiation. Dev. Cell. 2015;32:693–706. doi: 10.1016/j.devcel.2015.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kagey MH, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ji X, et al. 3D chromosome regulatory landscape of human pluripotent cells. Cell Stem Cell. 2016;18:262–275. doi: 10.1016/j.stem.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghavi-Helm Y, et al. Enhancer loops appear stable during development and are associated with paused polymerase. Nature. 2014;512:96–100. doi: 10.1038/nature13417. [DOI] [PubMed] [Google Scholar]
- 9.Jin F, et al. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503:290–294. doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Phillips-Cremins JE, et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell. 2013;153:1281–1295. doi: 10.1016/j.cell.2013.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rao SSP, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Creyghton MP, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rada-Iglesias A, et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boglev Y, et al. The unique and cooperative roles of the Grainy head-like transcription factors in epidermal development reflect unexpected target gene specificity. Dev. Biol. 2011;349:512–522. doi: 10.1016/j.ydbio.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 15.Fuchs E, Green H. The expression of keratin genes in epidermis and cultured epidermal cells. Cell. 1978;15:887–897. doi: 10.1016/0092-8674(78)90273-8. [DOI] [PubMed] [Google Scholar]
- 16.Montavon T, et al. A regulatory archipelago controls Hox genes transcription in digits. Cell. 2011;147:1132–1145. doi: 10.1016/j.cell.2011.10.023. [DOI] [PubMed] [Google Scholar]
- 17.Lonfat N, Montavon T, Darbellay F, Gitto S, Duboule D. Convergent evolution of complex regulatory landscapes and pleiotropy at Hox loci. Science. 2014;346:1004–1006. doi: 10.1126/science.1257493. [DOI] [PubMed] [Google Scholar]
- 18.Adam RC, et al. Pioneer factors govern super-enhancer dynamics in stem cell plasticity and lineage choice. Nature. 2015;521:366–370. doi: 10.1038/nature14289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spitz F, Furlong EEM. Transcription factors: from enhancer binding to developmental control. Nat. Rev. Genet. 2012;13:613–626. doi: 10.1038/nrg3207. [DOI] [PubMed] [Google Scholar]
- 20.Li L, et al. Widespread rearrangement of 3D chromatin organization underlies Polycomb-mediated stress-induced silencing. Mol. Cell. 2015;58:216–231. doi: 10.1016/j.molcel.2015.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dowen JM, et al. Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell. 2014;159:374–387. doi: 10.1016/j.cell.2014.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Segre JA, Bauer C, Fuchs E. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat. Genet. 1999;22:356–360. doi: 10.1038/11926. [DOI] [PubMed] [Google Scholar]
- 24.Boxer LD, Barajas B, Tao S, Zhang J, Khavari PA. ZNF750 interacts with KLF4 and RCOR1, KDM1A, and CTBP1/2 chromatin regulators to repress epidermal progenitor genes and induce differentiation genes. Genes Dev. 2014;28:2013–2026. doi: 10.1101/gad.246579.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lopez RG, et al. C/EBPα and β couple interfollicular keratinocyte proliferation arrest to commitment and terminal differentiation. Nat. Cell Biol. 2009;11:1181–1190. doi: 10.1038/ncb1960. [DOI] [PubMed] [Google Scholar]
- 26.Sen GL, et al. ZNF750 is a p63 target gene that induces KLF4 to drive terminal epidermal differentiation. Dev. Cell. 2012;22:669–677. doi: 10.1016/j.devcel.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwartzman O, et al. UMI-4C for quantitative and targeted chromosomal contact profiling. Nat. Methods. 2016;13:685–691. doi: 10.1038/nmeth.3922. [DOI] [PubMed] [Google Scholar]
- 28.Samstein RM, et al. Foxp3 exploits a pre-existent enhancer landscape for regulatory T cell lineage specification. Cell. 2012;151:153–166. doi: 10.1016/j.cell.2012.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim T-H, et al. Broadly permissive intestinal chromatin underlies lateral inhibition and cell plasticity. Nature. 2014;506:511–515. doi: 10.1038/nature12903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fossum SL, et al. Ets homologous factor regulates pathways controlling response to injury in airway epithelial cells. Nucleic Acids Res. 2014;42:13588–13598. doi: 10.1093/nar/gku1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kundaje A, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Truong AB, Kretz M, Ridky TW, Kimmel R, Khavari PA. p63 regulates proliferation and differentiation of developmentally mature keratinocytes. Genes Dev. 2006;20:3185–3197. doi: 10.1101/gad.1463206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kretz M, et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493:231–235. doi: 10.1038/nature11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rinaldi L, et al. Dnmt3a and Dnmt3b associate with enhancers to regulate human epidermal stem cell homeostasis. Cell Stem Cell. 2016;19:491–501. doi: 10.1016/j.stem.2016.06.020. [DOI] [PubMed] [Google Scholar]
- 35.Servant N, et al. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol. 2015;16:259. doi: 10.1186/s13059-015-0831-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 37.Bao X, et al. A novel ATAC–seq approach reveals lineage-specific reinforcement of the open chromatin landscape via cooperation between BAF and p63. Genome Biol. 2015;16:284. doi: 10.1186/s13059-015-0840-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wingett S, et al. HiCUP: pipeline for mapping and processing Hi-C data. F1000Res. 2015;4:1310. doi: 10.12688/f1000research.7334.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cairns J, et al. CHiCAGO: robust detection of DNA looping interactions in Capture Hi-C data. Genome Biol. 2016;17:127. doi: 10.1186/s13059-016-0992-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klein FA, et al. FourCSeq: analysis of 4C sequencing data. Bioinformatics. 2015;31:3085–3091. doi: 10.1093/bioinformatics/btv335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Freire-Pritchett P, et al. Global reorganisation of cis-regulatory units upon lineage commitment of human embryonic stem cells. eLife. 2017;6:e21926. doi: 10.7554/eLife.21926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Landt SG, et al. ChIP–seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012;22:1813–1831. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data are available through the Gene Expression Omnibus (GEO) via accession GSE84662. Source data are provided for Figures 1–4 and Supplementary Figures 1–8.