Rujano et al. report mutations in ATP6AP2 leading to liver disease, immunodeficiency, and psychomotor impairment. ATP6AP2 deficiency impairs the assembly and function of the V-ATPase proton pump, causing defects in protein glycosylation and autophagy.
Abstract
The biogenesis of the multi-subunit vacuolar-type H+-ATPase (V-ATPase) is initiated in the endoplasmic reticulum with the assembly of the proton pore V0, which is controlled by a group of assembly factors. Here, we identify two hemizygous missense mutations in the extracellular domain of the accessory V-ATPase subunit ATP6AP2 (also known as the [pro]renin receptor) responsible for a glycosylation disorder with liver disease, immunodeficiency, cutis laxa, and psychomotor impairment. We show that ATP6AP2 deficiency in the mouse liver caused hypoglycosylation of serum proteins and autophagy defects. The introduction of one of the missense mutations into Drosophila led to reduced survival and altered lipid metabolism. We further demonstrate that in the liver-like fat body, the autophagic dysregulation was associated with defects in lysosomal acidification and mammalian target of rapamycin (mTOR) signaling. Finally, both ATP6AP2 mutations impaired protein stability and the interaction with ATP6AP1, a member of the V0 assembly complex. Collectively, our data suggest that the missense mutations in ATP6AP2 lead to impaired V-ATPase assembly and subsequent defects in glycosylation and autophagy.
Introduction
The multi-subunit vacuolar-type H+-ATPase (V-ATPase) acidifies intracellular organelles, thereby controlling several events in the secretory and endocytic pathway, such as proteolytic processing, protein degradation, autophagy, and glycosylation. Structurally, it consists of a proton pore (V0 sector) and ATP hydrolysis domain (V1 sector), each composed of several core subunits (Forgac, 2007). In addition, there are two accessory subunits named ATP6AP1 and ATP6AP2.
ATP6AP1 has recently been shown to be the orthologue of the yeast V-ATPase assembly factor Voa1 (Jansen et al., 2016a). Voa1 cooperates with four other chaperones (Vma21, Vma12, Vma22, and Pkr1) to assemble the V0 sector in the ER (Ryan et al., 2008). The newly assembled proton pore is then transported to the Golgi, where the preassembled V1 sector is added from the cytoplasm to constitute a functional holoenzyme (Forgac, 2007). Mutations in ATP6AP1 lead to a congenital disorder of glycosylation (CDG), which is a rapidly growing disease group caused by the deficiency in components of the glycosylation machineries and, more generally, in factors important for ER and Golgi homeostasis (Freeze et al., 2014; Scott et al., 2014; Hennet and Cabalzar, 2015). ATP6AP1 deficiency manifests with hypogammaglobulinemia as well as liver and cognitive abnormalities (Jansen et al., 2016a). Similar clinical phenotypes occur in patients with mutations in either one of the two other putative orthologues of yeast assembly factors TMEM199 (Vma12) and CCDC115 (Vma22; Jansen et al., 2016b,c), suggesting that V0 misassembly is the common pathogenic process in these syndromes.
The other accessory subunit ATP6AP2 has so far not been associated with CDG. In contrast, exon-skipping mutations in ATP6AP2 have been associated with cognitive disorders with Parkinsonism, spasticity, epilepsy, and intellectual disability (Ramser et al., 2005; Korvatska et al., 2013). Functionally, the protein has been suggested to act as a (pro)renin receptor (Nguyen et al., 2002) and in several signaling pathways (Buechling et al., 2010; Cruciat et al., 2010; Hermle et al., 2010, 2013; Schafer et al., 2015). The involvement of ATP6AP2 in V-ATPase function has been demonstrated in various cell culture and animal studies (Kinouchi et al., 2010, 2013; Riediger et al., 2011; Hermle et al., 2013; Trepiccione et al., 2016). In mice, the deletion of Atp6ap2 in cardiomyocytes, hepatocytes, or podocytes results in a significant decrease of V0 subunits and autophagy defects (Kinouchi et al., 2010, 2013; Kissing et al., 2017). Yet how ATP6AP2 mechanistically contributes to V-ATPase function remains to be determined.
Here, we identify missense mutations in ATP6AP2 that cause a metabolic disorder with phenotypic similarity to the deficiency of other V0 assembly factors. Using complementary in vitro and in vivo approaches, we show that ATP6AP2 interacts with members of the ER-based V0 assembly complex. The interaction is impaired by the identified missense mutations in ATP6AP2, causing reduced V-ATPase activity, defective autophagy, and ultimately, compromised cellular and organismal homeostasis. Our results suggest that ATP6AP2 has a crucial role in V-ATPase assembly, both in invertebrates and vertebrates.
Results
Mutations in X-linked ATP6AP2 cause immunodeficiency, liver disease, psychomotor impairment, and cutis laxa
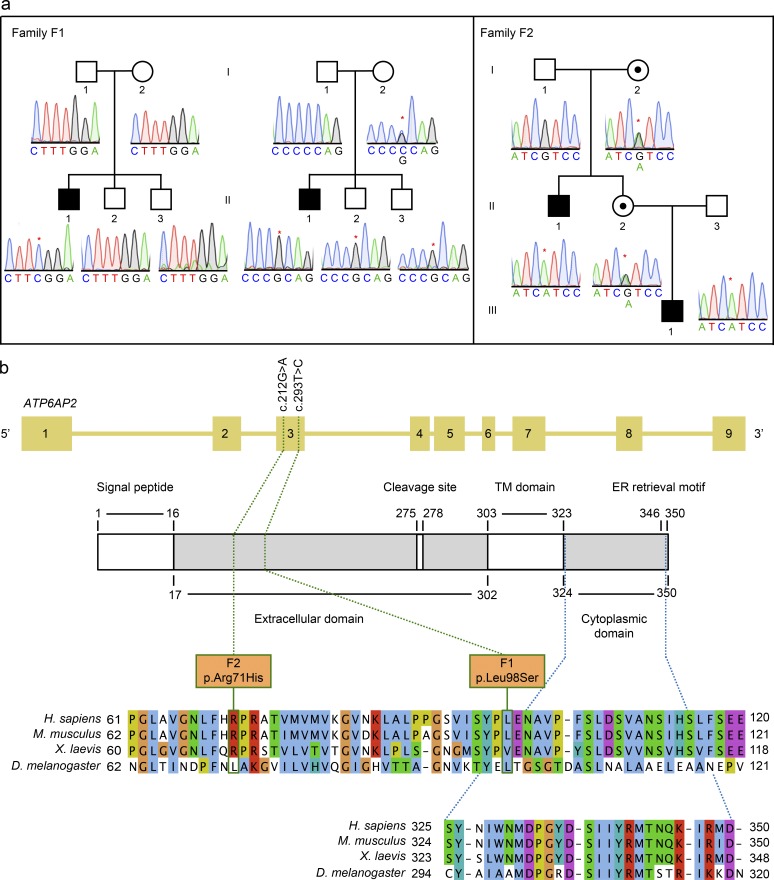
Two hemizygous mutations in ATP6AP2 were identified by whole exome sequencing in three male individuals from two unrelated families included in cohorts of unsolved CDG cases. A Portuguese boy (P1) was shown to carry a hemizygous missense mutation c.293T>C (p.L98S) in exon 3 (Fig. 1, a and b). The mutation was absent in the parents and the two brothers. In contrast, the neighboring polymorphism c.268C>G was transmitted by the heterozygous mother to all sons. Together, this suggested that p.L98S, which is evolutionary conserved between vertebrates and invertebrates, is a de novo mutation. Another hemizygous missense mutation, c.212G>A (p.R71H), was found in the same exon in two individuals (P2 and P3) of a German family (Fig. 1, a and b). Both mutations were absent from more than 60,000 control individuals in the ExAC server (http://exac.broadinstitute.org/).
Figure 1.
ATP6AP2 mutations cause a novel glycosylation disorder. (a) Pedigrees and sequence profiles of ATP6AP2 mutations in families F1 and F2. Whole exome sequencing identified two mutations in three patients from two different families. Partial chromatograms show X-linked segregation for all patients. Family F1 shows on the left side the segregation of the de novo missense pathogenic mutation (c.293T>C) identified in patient P1 (F1-II.1), and on the right side the segregation of the maternally inherited SNP (c.268C>G) to patient P1 and healthy siblings (F1-II.2 and F1-II.3). Pedigree for family F2 shows the segregation of the maternally inherited missense mutation (c.212G>A) to patient P2 (F2-II.1) and a healthy female sibling (F2-II.2). Patient P3 (F2-III.1) inherited the same mutation as P2 from healthy individual F2-II.2. Black-filled squares indicate affected males, whereas black dots in white-filled circles indicate healthy carrier females. The asterisk indicates the respective nucleotide change. (b) Schematic representation of the intron–exon structure of ATP6AP2 and the encoded protein. Numbers from 1–9 represent the different exons of the ATP6AP2 gene. The domain structure of the protein is schematized underneath, and the numbers delimiting the different domains refer to amino acids positions. In white are depicted the signal peptide, the cleavage site, the transmembrane (TM) domain, and C-terminal ER retrieval motif. The green dotted lines indicate the positions of the missense mutations within the families, both at the nucleotide and the protein level (green boxes). The blue dotted lines indicate the position of the cytoplasmic domain. The conservation of the affected amino acids is illustrated by the sequence alignments of the region containing the missense mutations (upper) and the cytoplasmic tail (lower) from Homo sapiens ATP6AP2 in Mus musculus, Xenopus laevis, and D. melanogaster, respectively. Although p.L98S is conserved in Drosophila, p.R71H is not. The ER retrieval motif is KxRxx in vertebrates and KKxx in Drosophila.
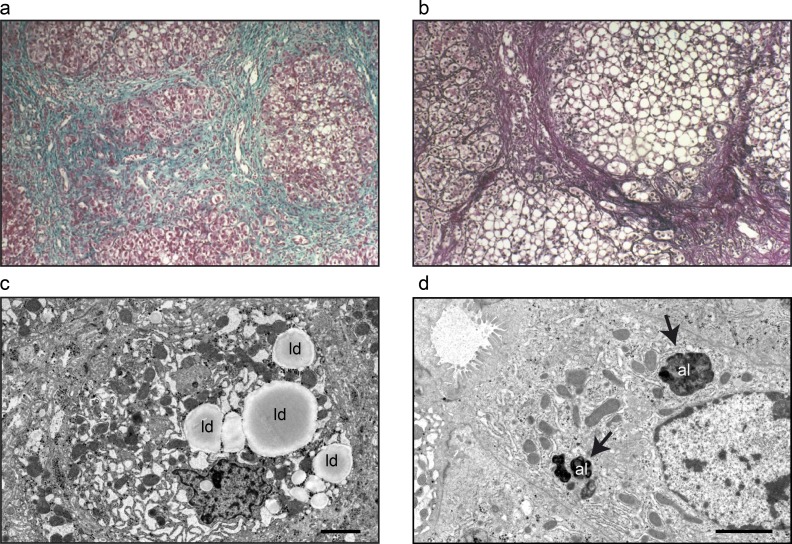
The predominant symptoms of the patients are hepatopathy and immunodeficiency. For P1, ascites, collateral vascularization, and hepatosplenomegaly and increased liver transaminases aspartate transaminase (AST) and alanine transaminase (ALT) led to a liver biopsy at 16 mo of age showing micronodular cirrhosis and steatosis (Table 1 and Fig. 2, a and b). Recurrent infections initiated immunological analysis that showed low levels of Igs, poor polysaccharide antibody response, decreased level of CD4+, and increased levels of CD8+ lymphocytes (Table 1). Treatment with weekly subcutaneous Ig led to decreased infection frequency. P2 suffered from recurrent pulmonary and upper respiratory tract infections throughout infancy and childhood. Plasmaelectrophoresis revealed hypogammaglobulinemia, and further investigation led to the diagnosis of IgG subclass deficiency with reduced IgG1 and IgG3. IgG substitution was consequently initiated, under which the patient’s general health status improved. Repeated ultrasounds showed hepatosplenomegaly with inhomogeneous and hyperechogenic structure. Serum cholesterol and the transaminase enzymes AST and ALT were mildly elevated (Table 1). P3 developed severe liver failure at 5 mo of age. Liver biopsy revealed lipid accumulation and enlarged vacuolar structures within hepatocytes (Fig. 2, c and d). Similar to the other two patients, he has repeatedly suffered from infections, but his immunological status has so far been normal. Additional clinical features for all patients were cutis laxa (P1, P2, and P3), dysmorphic features (P1 and P2), mild intellectual disability (P1 and P2), and mild ataxia (P2; Table 1).
Table 1. Clinical data of patients with ATP6AP2 mutations.
| Parameter | Patient 1 | Patient 2 | Patient 3 |
|---|---|---|---|
| Sex | Male | Male | Male |
| Current age | 17 yr | 21 yr | 10 mo |
| Consanguinity | None | None | None |
| Ethnicity | Caucasian | Caucasian | Caucasian |
| Mutation cDNA | c.293C>T | c.212G>A | c.212G>A |
| Mutation protein | p.Leu98Ser (L98S) | p. Arg71His (R71H) | p. Arg71His (R71H) |
| Glycosylation defect | IEF: tetrasialotransferrin: 34.2% (norm: 48.5–65.3%); trisialotransferrin: 28.9% (norm: 5.5–15.1%); disialotransferrin: 15% (norm: 2.0–6.1%); monosialotransferrin: 3.3% (norm: 0.0–3.7%); asialotransferrin: 1.5% (norm: 0%) | HPLC: tetrasialotransferrin 55.17% (norm: 85.7–94.0%); trisialotransferrin 33.11% (norm: 1.16–6.36%); disialotransferrin 8.92% (norm: 0.38–1.82%); monosialotransferrin 1.68% (norm: 0%) | HPLC: tetrasialotransferrin 35.7% (norm: 85.7–94.0%); trisialotransferrin 36.4% (norm: 1.16–6.36%); disialotransferrin 18.7% (norm: 0.38–1.82%); monosialotransferrin 9.0% (norm: 0%) |
| Onset of symptoms | 5 mo | Directly after birth | Directly after birth |
| Liver | Prolonged neonatal jaundice and persistent hepatosplenomegaly with recurrent episodes of hypoalbuminemia and ascites | No hepatomegaly in childhood, but liver parenchyma of an inhomogeneous, hyperechoic structure. | Prolonged neonatal jaundice and hepatosplenomegaly. Cholestasis. Irregular liver parenchyma with portosystemic shunts and ascites. Hypoalbuminemia, hyperammonemia. Coagulopathy. Listed for liver transplantation. |
| Today: hepatomegaly, mild elevation of liver enzymes | |||
| Liver biopsy | Micronodular hepatic cirrhosis with moderate macrovesicular steatosis (biopsy at 8 mo), elevated copper | Not available | Diffuse micronodular hepatic cirrhosis |
| Elevated copper | |||
| Neonatal icterus | + | - | + |
| Splenomegaly | + | + | + |
| Infections | Recurrent severe infections (e.g., sepsis, peritonitis) | Recurring pulmonary and upper respiratory tract infections throughout infancy and childhood | Recurring upper respiratory tract infections, positive blood cultures with pneumococcus at one occasion |
| Neurological symptoms | Mild cognitive impairment | Ataxic gait, mild cognitive impairment | Normal |
| Cutis laxa | Mild to moderate | Pronounced, improvement over time | Pronounced |
| Hypogammaglo-bulinemia | + | + | - |
| IgG | 2.31 g/L (reference: 5.6–13.8 g/L) | Reduced IgG1 and IgG3 | - |
| IgM | <0.17 g/L | 18 mg/dl (reference: 40–230 mg/dl) | - |
| IgA | <0.08 g/L | <15 mg/dl (reference: 70–400 mg/dl) | - |
| Increased transaminases | At 17 yr, mild elevation of transaminases (AST 61 U/l [reference: 15–46 UI/l], ALT 51 U/l [reference: 10–40 U/l]) | Ranging from mild elevation at age 9 yr: AST 92 U/l (reference: 8–60 U/l), ALT 59 U/l (reference: <44 U/l) to pronounced hepatic affection (AST 135 U/l) | Within the first year: AST 100–160 U/l, (reference: <71 U/l) |
| Serum copper | Not available | At 1 yr: 1.5 µmol/l (reference: 10–30 µmol/l) | Normal serum copper at the age of 4 mo: 89 µg/dl (reference: 65–165 µg/dl) |
| Normalized over the following months | |||
| Serum lipids | At age 17 yr (liver normal): | At age 20 yr: | At age 5 mo: |
| Cholesterol: 172 (reference: 200 mg/dl) | Cholesterol: 199 (reference: 200 mg/dl) | Cholesterol: 254 (reference: 81–147 mg/dl) | |
| LDL-cholesterol: 90 (reference: 50–130 md/dl) | LDL-cholesterol: 149 (reference: 50–130 md/dl) | ||
| Other laboratory findings | Low factor V and VI | Low factor XI (58% [reference: >70%]) and free protein S (57.5% [reference: 60–140%]) | Low factors II (27.7% [reference: 60–120%]), V (35.9% [reference: 55–130%]), VII (21.9% [reference: 47–130%]), IX (17.27% [reference: 36–136%]), and XI (23.9% [reference: 49–134%]) |
| Factor V: 30% (reference: 65–148%), factor VII: 24% (reference: 58–115%) | |||
| Other clinical findings | Mild dysmorphic features | Low-set ears, micrognathia, a flat and wide-set chest, laterally facing nipples, and hypospadia |
IEF, isoelectric focusing.
Figure 2.
Liver histology and fibroblasts of patients. Immunohistochemistry and transmission electron microscopy of liver tissue from patient 1 (P1) and patient 3 (P3). (a and b) Liver biopsy from P1 displays micronodular cirrhosis (a; Masson’s trichrome staining, 100×) and macro- and microvacuolar steatosis (b; Gordon and Sweet's staining, 200×). (c and d) Electron micrographs from single hepatocytes in liver biopsies from P3 with accumulations of autolysosomes (al; arrows) and lipid droplets (ld). Bars: (c) 5 µm; (d) 2 µm.
Collectively, missense mutations in ATP6AP2 cause a new syndrome, predominantly affecting the liver, immune system, central nervous system, and skin connective tissue.
ATP6AP2 deficiency causes hypoglycosylation of serum proteins in patients and mice
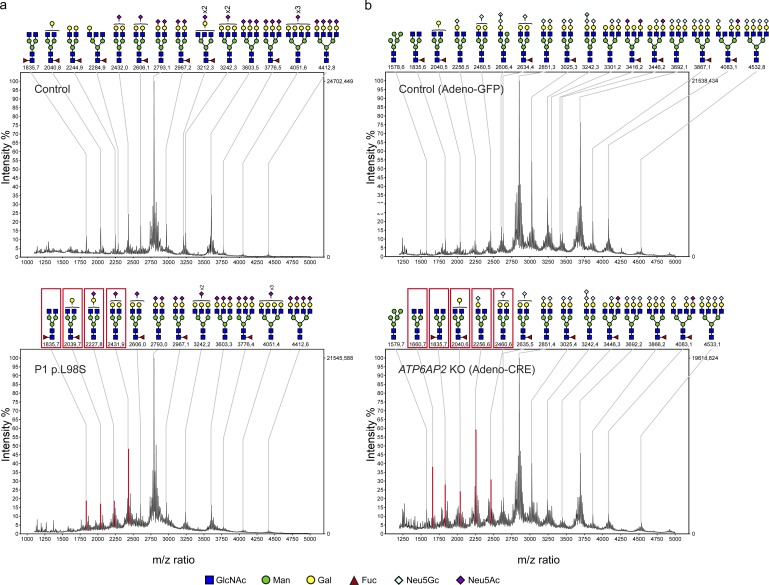
Analysis of protein N-glycosylation showed that all patient sera had abnormal transferrin glycosylation profiles with decreased tetrasialotransferrin and increased tri-, di-, and monosialotransferrin (Fig. S1, a and b; and not depicted). HPLC analysis confirmed that only half of the transferrin molecules (53.3% in patient 2 and 50.28% in P3) were present as the correctly glycosylated tetrasialo-transferrin (Fig. S1, a and b). N-linked glycans on total plasma glycoproteins were analyzed using matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry (MS), which showed a relative increase in the undersialylated glycans in patient 1 (Fig. 3 a).
Figure 3.
Hypoglycosylation of serum proteins in patient and mouse samples. (a) N-linked glycans of serum from a healthy individual (top) and from P1 with the L98S mutation in ATP6AP2 (bottom) were analyzed by MALDI-TOF MS. (b) Representative MALDI-TOF MS profiles of adeno-GFP- (n = 7) and adeno-CRE- (n = 5) injected mice (complete data can be found in Fig. S4). Red boxes and peaks in the lower panels of a and b indicate the increase of biantennary glycans (compared with most extensively branched N-glycans in the control), which also contain less undersialylated structures.
So far, ATP6AP2 has not been implicated in any glycosylation disorder. Moreover, analysis of protein glycosylation in a patient carrying one of the previously described exon-skipping variants in ATP6AP2 (Korvatska et al., 2013) showed a normal transferrin glycosylation profile and only a slight undersialylation of total serum proteins (Fig. S2, a and b). Therefore, we tested whether ATP6AP2 deficiency was sufficient to cause serum glycosylation abnormalities. Mice carrying a floxed allele of ATP6AP2 (Riediger et al., 2011) were injected with adeno-CRE (or adeno-GFP as a control) to induce an acute reduction of ATP6AP2 in the liver (Fig. S3 a). Serum analysis showed elevation of the liver enzymes AST and ALT and high levels of cholesterol in the Cre-injected mice, suggesting liver damage and metabolic abnormalities (Fig. S3, b and c). MALDI–TOF MS showed an increase of unsialylated and mono-sialylated N-glycans and a decrease in tri-sialylated N-glycans compared with adeno-GFP–injected mice (Fig. 3 b and Fig. S3 d). Thus, reduced ATP6AP2 expression in the liver is sufficient to recapitulate the glycosylation defects of the patients.
Disease-associated mutations target ATP6AP2 for degradation
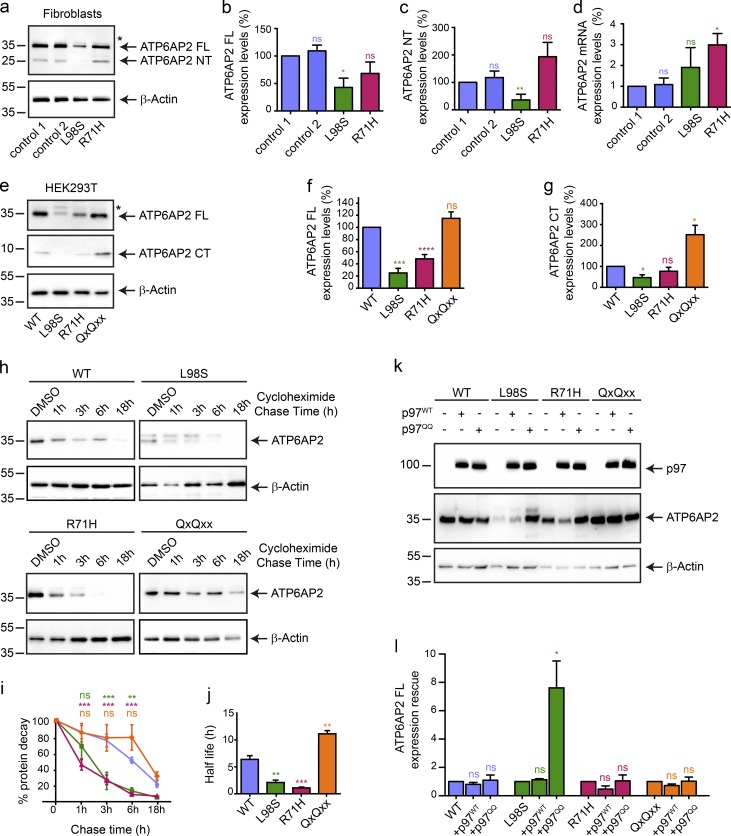
ATP6AP2 is a type I transmembrane protein that undergoes two proteolytic cleavages in the Golgi to generate a luminal or extracellular N-terminal fragment (NT) and a C-terminal fragment (CT) harboring a transmembrane domain and a short cytoplasmic tail (Fig. 1 b; Cousin et al., 2009; Nakagawa et al., 2017). The cytoplasmic tail contains an RxKxx ER retrieval motif that mediates retrograde transport to the ER (Nilsson et al., 1989; Schefe et al., 2006; Sihn et al., 2013). To investigate the effect of the missense mutations on expression and cleavage of ATP6AP2, we performed Western blotting in fibroblasts derived from patients P1 (ATP6AP2L98S) and P3 (ATP6AP2R71H). Steady-state levels of full-length (FL) ATP6AP2 were significantly reduced in ATP6AP2L98S but not ATP6AP2R71H fibroblasts, in comparison to ATP6AP2WT from two different healthy controls (Fig. 4, a and b). In contrast, mRNA levels of ATP6AP2R71H but not of ATP6AP2L98S were elevated (Fig. 4 d). Moreover, upon transient overexpression of HA-tagged ATP6AP2L98S and ATP6AP2R71H in HEK293T cells, the levels of both mutant proteins were decreased in comparison to the WT (Fig. 4, e and f). To understand whether the reduced levels of the overexpressed mutants were a result of decreased stability, we performed a cycloheximide chase assay in HEK293T cells. Whereas ATP6AP2WT was still present after 18 h of cycloheximide pretreatment, ATP6AP2L98S and ATP6AP2R71H were barely detectable after 3 h, indicating that both mutant proteins have a shorter half-life and are targeted for degradation (Fig. 4, h–j).
Figure 4.
ATP6AP2 mutants p.L98S and p.R71H are degraded via ERAD. (a) Endogenous levels of ATP6AP2 FL and NT in control and patient fibroblasts using anti-ATP6AP2-NT. The asterisk * indicates glycosylated form of ATP6AP2L98S. (b and c) Expression levels of ATP6AP2 FL and NT relative to those of β-actin were calculated. All data are mean ± SEM of three independent experiments. ns, not significant; *, P < 0.05; **, P < 0.01. The statistical significance was determined by an unpaired Student’s t test. (d) ATP6AP2 mRNA quantification in control and patient fibroblasts. All data are mean ± SEM of three independent experiments. ns, not significant; *, P < 0.05. The statistical significance was determined by an unpaired Student’s t test. (e) Steady-state levels of overexpressed ATP6AP2WT-HA, ATP6AP2L98S-HA, ATP6AP2R71H-HA, and ATP6AP2QxQxx-HA at 48 h after transfection in HEK293T cells. Anti-HA reveals FL and CT. The asterisk indicates glycosylated form of ATP6AP2L98S. (f and g) Expression levels of ATP6AP2 FL and CT relative to those of β-actin were calculated. All data are mean ± SEM of five independent experiments. ns, not significant; *, P < 0.05; ***, P < 0.001; ****, P < 0.0001. The statistical significance was determined by an unpaired Student’s t test. (h–j) Turnover of ATP6AP2 WT, L98S, R71H, and QxQxx was determined by cycloheximide (100 µM) chase experiments. The chase started 24 h after transfection and was allowed for the indicated time. The percentage of the protein decay was graphically reported (i), and the difference of the in cellulo half-life between the WT, L98S, R71H, and QxQxx was analyzed in j. Blots are representative of four individual experiments. All data are mean ± SEM of four independent experiments. ns, not significant; **, P < 0.01; ***, P < 0.001. The statistical significance was determined by a regular two-way ANOVA followed by a Bonferroni multiple comparisons test in i and an unpaired Student’s t test in j. (k) HEK293T cells were transiently cotransfected with the CT-tagged construct ATP6AP2 WT, L98S, R71H, and QxQxx and p97-WT-His or p97-QQ-His. Samples were analyzed with anti-HA and anti-His antibodies. (l) Expression levels of ATP6AP2 FL in the presence of p97WT or p97QQ relative to those of β-actin were calculated. All data are mean ± SEM of three independent experiments. ns, not significant; *, P < 0.05. The statistical significance was determined by an unpaired Student’s t test. Molecular mass is indicated in kilodaltons.
Additionally, endogenous and overexpressed ATP6AP2L98S showed reduced levels of the cleavage products NT and CT (Fig. 4, a, c, e, and g), which suggested a defect in the export out of the ER. Therefore, we asked whether degradation of this mutant involved the ER-associated degradation (ERAD) pathway, by which misfolded transmembrane proteins are targeted for proteasomal degradation via retrotranslocation into the cytoplasm. To inhibit ERAD, we coexpressed WT and dominant-negative versions of p97 (p97WT and p97QQ, respectively), a crucial component of the retrotranslocation machinery (Ye et al., 2001). Whereas p97QQ but not p97WT caused a significant accumulation of ATP6AP2L98S, ATP6AP2WT and ATP6AP2R71H steady-state levels were unchanged (Fig. 4, k and l). Importantly, a version of ATP6AP2 with mutations in the C-terminal ER retrieval motif (ATP6AP2QxQxx) not only was unaffected by both p97 constructs (Fig. 4, k and l) but also showed more cleavage (Fig. 4, e and g) and the longest half-life (Fig. 4, h–j). These findings were confirmed by localization studies of the different constructs in HeLa cells (Fig. S4, a and b). Whereas ATP6AP2WT and ATP6AP2R71H showed both ER and Golgi localization, ATP6AP2L98S localized primarily to the ER. ATP6AP2QxQxx showed an exclusive Golgi localization. Altogether, these results suggest that although the reduced retrograde transport of ATP6AP2QxQxx causes more protein stability, the increased ER localization of ATP6AP2L98S promotes its targeting for ERAD. For ATP6AP2R71H, which showed higher endogenous steady-state levels and more Golgi localization than ATP6AP2L98S, the precise degradation pathway remains unclear.
ATP6AP2L98S is posttranslationally modified by N-glycan addition
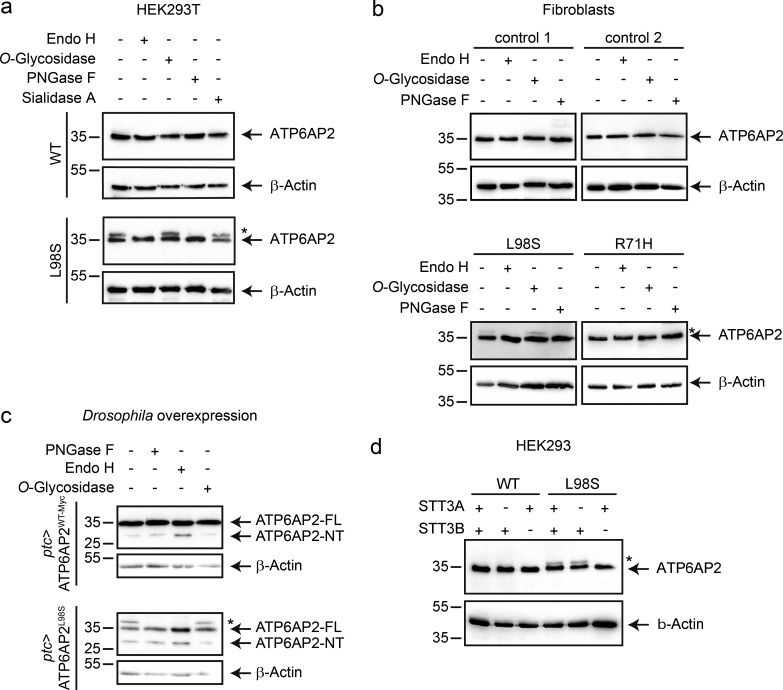
Apart from lower expression levels, overexpressed ATP6AP2L98S in HEK293T cells and endogenous ATP6AP2L98S in patient fibroblasts also exhibited an additional slower migrating band in immunoblots (Fig. 4, a and e; and Fig. 5, a and b). As the majority of proteins synthesized in the ER undergo glycosylation, we tested whether this band was the result of glycan addition. For this, we treated protein lysates with different deglycosylation enzymes. Whereas O-glycosidase and sialidase A treatment did not affect the mobility of the upper band, addition of the N-glycosidases endoglycosidase H (Endo H) and peptide-N-glycosidase F (PNGase F) completely eliminated it (Fig. 5, a and b), suggesting that a fraction of ATP6AP2L98S is N-glycosylated and localized in the ER.
Figure 5.
The mutant ATP6AP2L98S migrates as a double band as a result of posttranslational N-glycosylation. (a) HEK293T cells were transiently transfected with C-terminally HA-tagged constructs of WT and L98S. Protein extracts were immunoblotted with anti-HA antibody after incubation with (+) and without (-) Endo H, O-glycosidase, PNGase F, or sialidase A. Asterisk indicates slower migrating band for ATP6AP2L98S. (b) The expression of endogenous ATP6AP2 was analyzed in two different control fibroblasts from healthy donors and fibroblasts from P1 (p.L98S) and P3 (p.R71H). Cell lysates were separated by SDS-PAGE after incubation with (+) or without (-) Endo H, O-glycosidase, or PNGase F, and the proteins were revealed with anti-ATP6AP2-NT antibody. (c) ATP6AP2WT and ATP6AP2L98S were overexpressed with patched(ptc)-GAL4 in flies, and lysates were analyzed by immunoblot with anti-ATP6AP2-NT antibody after incubation with (+) and without (-) Endo H, O-glycosidase, or PNGase F. (d) Parental HEK293, STT3A KO, and STT3B KO cells were transiently transfected with ATP6AP2WT or ATP6AP2L98S. Cell lysates were analyzed by immunoblotting and revealed with anti-HA antibody. β-Actin served as a loading control. Data in a–d are representative of three, three, two, and five experiments, respectively. Molecular mass is indicated in kilodaltons.
N-glycosylation in the ER typically occurs cotranslationally through oligosaccharyltransferases complexes harboring the STT3A subunit (Cherepanova et al., 2016). However, when glycosylation sites are skipped or when proteins are misfolded, N-glycans can also be added posttranslationally by STT3B-containing oligosaccharyltransferase complexes (Sato et al., 2012; Cherepanova et al., 2016). To investigate whether the N-glycan addition in ATP6AP2L98S occurred co- or posttranslationally, we expressed ATP6AP2L98S in STT3A and STT3B KO cells (Cherepanova and Gilmore, 2016). We found that the slower migrating band ATP6AP2L98S was present in STT3A but not in STT3B KO cells (Fig. 5 d), suggesting that ATP6AP2L98S is subject to posttranslational N-glycosylation in the ER. Given that this mutant protein is targeted for ERAD, the results suggest that the N-glycan is added as a result of misfolding.
The p.L98S mutation causes developmental defects and decreased protein stability in Drosophila
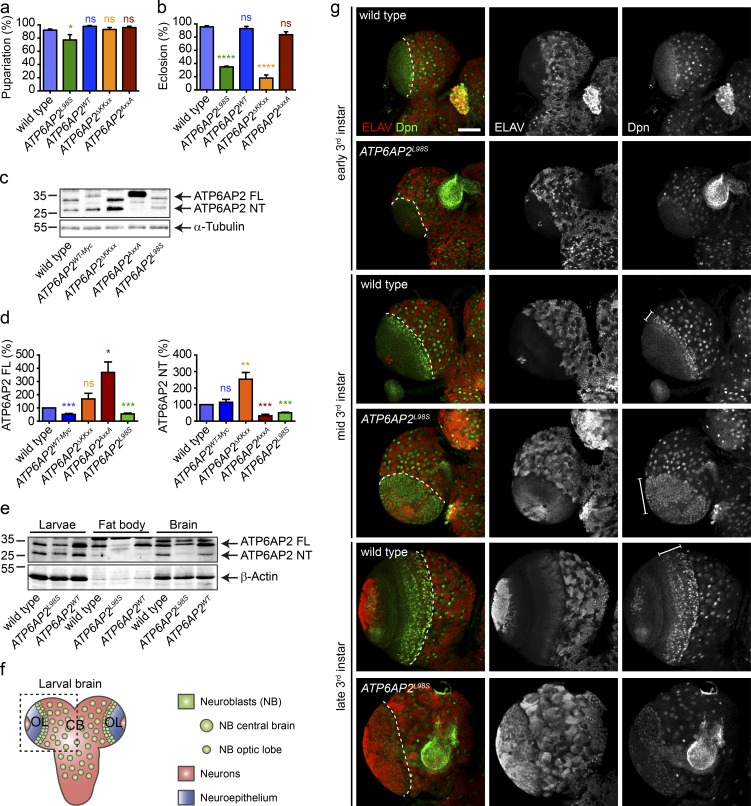
To better understand the consequences of the p.L98S mutation on animal development and homeostasis, we turned to Drosophila, where the leucine residue at position 98 is conserved (Fig. 1 b). Null alleles of the Drosophila orthologue of ATP6AP2 (also known as VhaM8.9 or VhaPRR) are early lethal (Hermle et al., 2013). Thus, we generated an ATP6AP2 transgene carrying the p.L98S mutation and used a genetic rescue approach in which we expressed ATP6AP2L98S mutant under the ATP6AP2 promoter and in the background of the ATP6AP2Δ1 null allele (hereafter referred to as ATP6AP2L98S; see Table S1 for precise genotypes; Hermle et al., 2013). In parallel, we analyzed versions of WT ATP6AP2 with and without a C-terminal Myc tag (ATP6AP2WT-Myc and ATP6AP2WT, respectively), a deletion of the ER retrieval motif (ATP6AP2ΔKKxx), and a mutant of the furin cleavage site (ATP6AP2AxxA) to assess the importance of ATP6AP2 proteolytic cleavage in the function of ATP6AP2. Expression of ATP6AP2WT and ATP6AP2AxxA led to full restoration of viability into adulthood (Fig. 6, a and b). In contrast, ATP6AP2L98S mildly impaired larval-to-pupal transition (Fig. 6 a), and both ATP6AP2L98S and ATP6AP2ΔKKxx significantly reduced the percentage of adults eclosing from the pupal case (Fig. 6 b). These adult survivors demonstrated poor mobility, decreased or absent climbing capabilities, blistered wings, and reduced head size and died 3–4 d after eclosion (not depicted).
Figure 6.
L98S mutation causes decreased viability and premature differentiation of neural progenitors in Drosophila. (a) Percentage of pupariation of the different ATP6AP2 mutants used in this study. (b) Percentage of eclosion of adult flies. All data are mean ± SEM of three to five independent experiments (50–100 animals per genotype per experiment). *, P < 0.05; ****, P < 0.0001. Significance was determined by one-way ANOVA followed by a Bonferroni multiple comparisons test. (c) Western blot of ATP6AP2 in whole larvae extracts from WT and rescued flies showing expression levels and cleavage pattern of endogenous ATP6AP2 and the different rescue transgenes. α-Tubulin is used as loading control. (d) Relative levels of FL (left) and NT (right) ATP6AP2 mutants compared with endogenous ATP6AP2. All data are mean ± SEM of seven independent experiments (5 animals per genotype per experiment). ns, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001. The statistical significance was determined by a paired Student’s t test. (e) Western blot of ATP6AP2 in whole larvae, fat body, and brain extracts of WT, ATP6AP2L98S, and ATP6AP2WT rescue flies. β-Actin is used as loading control. Blot is representative of four independent experiments (5, 15, and 50 animals per genotype per experiment [larval, fat body, and brain extracts, respectively]). (f) Schematic representation of the larval Drosophila brain. CB, central brain; OL, optic lobe. (g) Representative micrographs (out of three independent experiments; 5–10 brains per genotype per experiment) for WT and ATP6AP2L98S brain lobes from early (top) mid (middle) and late (bottom) third instar (third) larvae stained with the pan-neuronal marker ELAV (red in the merged panels) and the neuroblasts marker Deadpan (Dpn, green in the merged panels). Bar, 50 µm. Dashed lines in the merged panel separate the optic lobe (left) from the central brain (right). The region occupied by optic lobe neuroblasts is indicated with lines in mid and late third instar larvae. Molecular mass is indicated in kilodaltons.
Next, we analyzed ATP6AP2 protein expression and processing by Western blotting in whole larval extracts. Compared with endogenous ATP6AP2, ATP6AP2WT-Myc showed reduced levels of FL protein and similar levels of cleaved NT. In ATP6AP2ΔKKxx larvae, FL and NT levels were higher compared with endogenous control (Fig. 6, c and d), indicating enhanced cleavage and stability of the protein as found for ATP6AP2QxQxx overexpression in HEK293T cells. As expected, proteolytic cleavage was suppressed in ATP6AP2AxxA with a concomitant increase of the FL protein. Importantly, in ATP6AP2L98S larvae, the levels of both FL and NT were reduced compared with WT controls in whole larval lysates (Fig. 6, c and d), confirming the decreased stability observed for the mutant proteins in patient fibroblasts and HEK293T cells. Although glycosylated forms of ATP6AP2L98S were not seen when expressed under its own promoter (Fig. 6, c and e), ATP6AP2L98S was also N-glycosylated in Drosophila upon overexpression with the UAS-Gal4 system (Fig. 5 c). Collectively, the p.L98S mutation in Drosophila impairs ATP6AP2 protein stability and is sufficient to cause developmental defects.
Flies with the p.L98S mutation present brain developmental defects and impaired lipid homeostasis
To understand the cause of the compromised viability of the ATP6AP2L98S flies, we turned to two tissues affected in the patients: the central nervous system and the fat body, a liver-like tissue. We first examined whether differences in expression levels and processing could be observed for ATP6AP2 in these tissues. We found that FL endogenous ATP6AP2 and FL transgenic ATP6AP2WT were similarly expressed in whole larval extracts, fat body, and larval brain (Fig. 6 e). In contrast, FL ATP6AP2L98S expression was highly reduced, especially in fat body tissue. Furthermore, minimal cleavage of ATP6AP2 was observed in fat body for both endogenous and transgenic constructs, which could indicate a higher need for the FL form of ATP6AP2 in the fat body (Fig. 6 e).
To assess the consequences of the p.L98S mutation in central nervous system development, we examined the developing brain in ATP6AP2L98S third instar larvae. Using immunolabeling, we found that the p.L98S mutation strongly affected the optic lobes, which are one of the major structures of the Drosophila brain (Fig. 6, f and g). The optic lobes develop from neuroepithelial cells, which first increase in number through symmetric divisions and are later converted into optic lobe neuroblasts (Yasugi et al., 2008; Egger et al., 2010). These neuroblasts divide asymmetrically to self-renew and to generate neurons. We observed that in ATP6AP2L98S brains there is an expansion of optic lobe neuroblasts, marked by the expression of Deadpan (Dpn; Fig. 6 g). Whereas in WT mid third instar larval brains, only three to four cell rows of optic lobe neuroblasts could be observed, in ATP6AP2L98S mutant brains, most of the optic lobe was occupied by Dpn-positive cells, indicating a premature conversion to neuroblasts possibly at the expense of neuroepithelial cells. Consistent with a premature and increased neurogenesis, we found an increase in cells positive for the pan-neuronal marker ELAV in the optic lobes and the central brain of mid and late third instar ATP6AP2L98S larvae (Fig. 6 g). These observations suggest that ATP6AP2 plays an important role in Drosophila neural development, which is altered by the patient’s missense mutation.
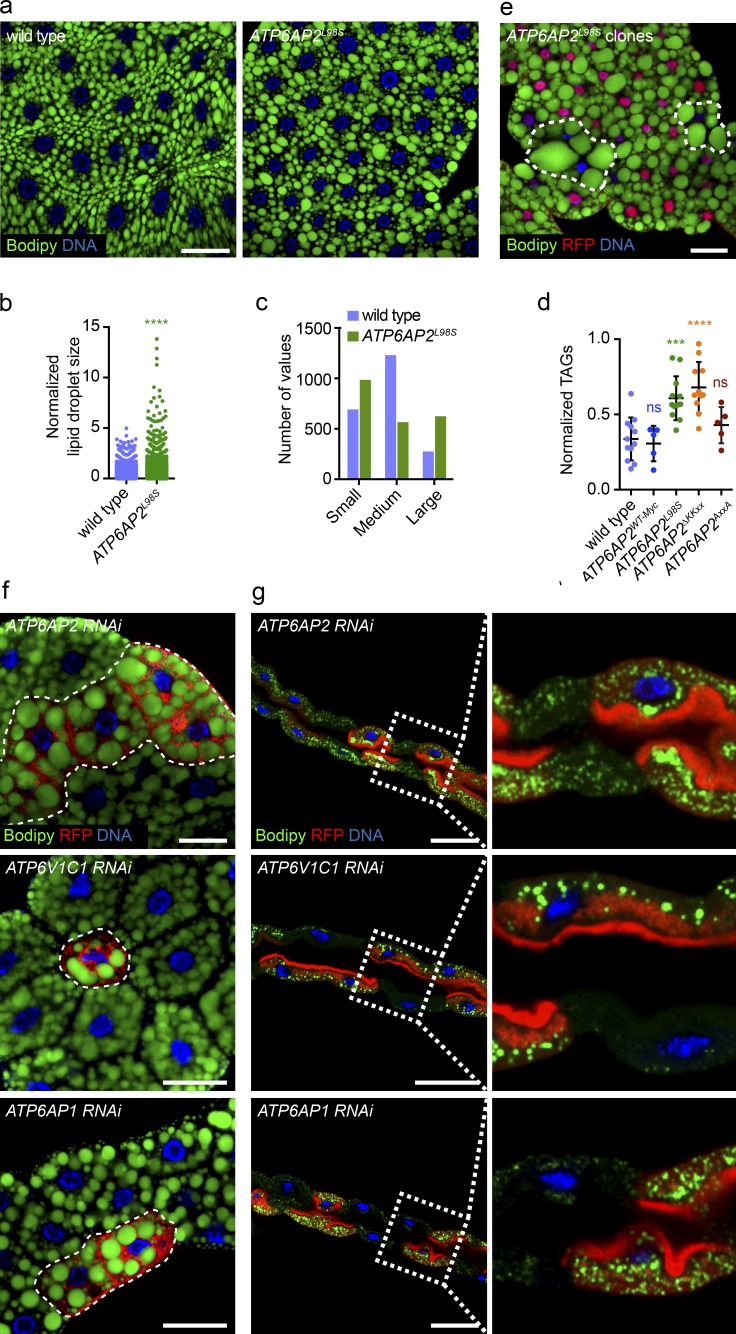
The larval fat body is featured by a high amount of lipid droplets, which are storage organelles for triglycerides (TAGs) and cholesteryl esters (Ugur et al., 2016). Compared with WT, we found an increase in the size of lipid droplets in ATP6AP2L98S mutant fat body cells (Fig. 7, a and b) as well as an increase in the number of small and large lipid droplets with a concomitant decrease of medium-sized droplets (Fig. 7 c). The increase in lipid droplets was accompanied by an increase in the levels of total TAGs in ATP6AP2L98S mutant larvae (Fig. 7 d). An increase in TAGs was also observed in ATP6AP2ΔKKxx but not in ATP6AP2AxxA or ATP6AP2WT-Myc animals. Furthermore, in clones expressing ATP6AP2L98S in the null background, we found a strong increase in lipid droplet size compared with the surrounding ATP6AP2WT-Myc tissue (Fig. 7 e, and see Table S1 for precise genotype). A similar phenotype was seen in clonal populations of cells in which ATP6AP2 was down-regulated using RNAi (Fig. 7 f), suggesting that ATP6AP2L98S is a loss-of-function mutation. Furthermore, RNAi-mediated knockdown of ATP6AP2 also provoked lipid droplet accumulation in other tissues, such as Malpighian tubules (Fig. 7 g) and pupal wings (see Fig. 9 h).
Figure 7.
L98S mutation in Drosophila ATP6AP2 causes impaired lipid metabolism. (a) Bodipy (green) staining of wandering third instar larval fat bodies of WT and ATP6AP2L98S rescue. (b) Quantification of lipid droplet size in WT and ATP6AP2L98S third instar larval fat bodies. Each dot represents one lipid droplet. Data are from four independent experiments (5–10 animals per genotype per experiment) in which a total of 28 and 34 fat bodies were analyzed for WT and ATP6AP2L98S, respectively. ****, P < 0.0001. Significance was determined by a Kolmogorov-Smirnov test. (c) Binning of data from (b) showing the distribution of lipid droplets by size category (small, medium, large). (d) Total TAG levels assayed enzymatically in WT, ATP6AP2WT-Myc, ATP6AP2L98S, ATP6AP2ΔKKxx, and ATP6AP2AxxA wandering third instar larvae. Lines represent the mean ± SD of 5–10 independent experiments (5 animals per genotype per experiment). ns, not significant; ***, P = 0.0001; ****, P < 0.0001. Significance was determined by one-way ANOVA followed by a Bonferroni multiple comparisons test. (e) Analysis of lipid droplets in clonal populations of fat body cells expressing ATP6AP2L98S (RFP-negative) surrounded by ATP6AP2WT-Myc cells (RFP-positive). Lipid droplets stained with Bodipy (green). DNA stained with Hoechst (blue). (f) Analysis of lipid droplets (green) in clonal populations of fat body cells and expressing RNAi against ATP6AP2, ATP6V1C1, and ATP6AP1 (RFP-positive) surrounded by WT cells (RFP-negative). (g) Analysis of lipid droplets (green) in clonal populations of Malpighian tubule cells expressing RNAi against ATP6AP2, ATP6V1C1, and ATP6AP1 (RFP-positive) surrounded by WT cells (RFP-negative). Right panels are magnifications of the insets demarked in left panels. (a and e–g) Bars, 50 µm. Micrographs of clonal analyses are representative of at least three independent experiments (10–15 animals per genotype per experiment).
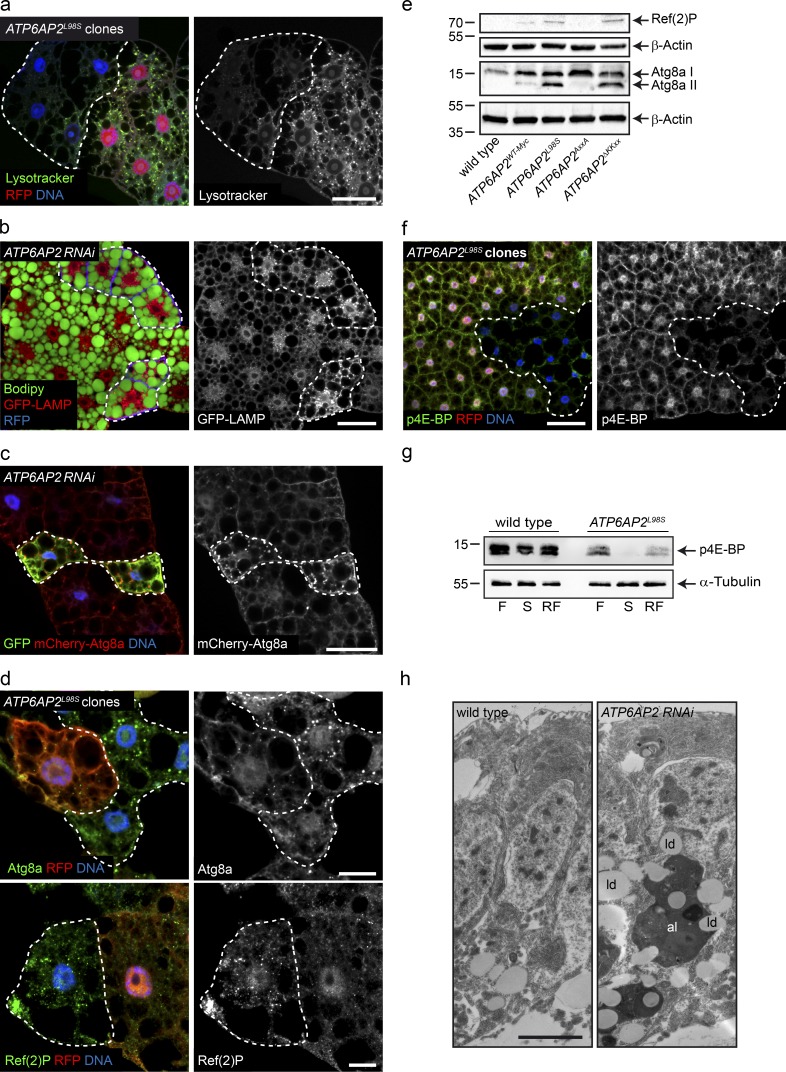
Figure 9.
ATP6AP2 knockdown and ATP6AP2L98S mutation lead to lipid accumulation as a result of autophagic defects. (a) Lysotracker labeling (green in merged panel) of ATP6AP2L98S clonal populations (RFP-negative) of fat body cells surrounded by ATP6AP2WT-Myc cells (RFP-positive). Bar, 50 µm. (b) GFP-LAMP accumulation (red in merged panel) and increased lipid droplet size in ATP6AP2 knockdown cells (RFP-positive, blue in merged panel). Lipid droplets stained with Bodipy (green in merged panel). Bar, 50 µm. (c) Atg8a-mCherry accumulation (red in merged panel) in ATP6AP2 knockdown cells (GFP-positive, green in merged panel). Bar, 50 µm. (d) Endogenous Atg8a (upper) and Ref(2)P (lower) staining (green in merged panel) in ATP6AP2L98S clonal populations (RFP-negative) of fat body cells surrounded by ATP6AP2WT-Myc cells (RFP-positive). Bars, 20 µm. (e) Western blot of Ref(2)P and Atg8a in whole larvae extracts of WT and ATP6AP2WT-Myc, ATP6AP2L98S, ATP6AP2AxxA, and ATP6AP2ΔKKxx flies. β-Actin is used as loading control. Blot is representative of five independent experiments (five animals per genotype per experiment). (f) p-4E-BP staining of ATP6AP2L98S clonal populations (RFP-negative) of fat body cells surrounded by ATP6AP2WT-Myc cells (RFP-positive). Bar, 50 µm. (g) Western blot of p4E-BP in fat body extracts of WT and ATP6AP2L98S third instar larvae in fed (F), starved (S), and refed (RF) conditions. α-Tubulin is used as loading control. Blot is representative of four independent experiments (20–25 animals per genotype per experiment). (h) EM micrographs of pupal wings in which ATP6AP2 has been down-regulated in the dorsal compartment using RNAi driven by apterous-Gal4. As opposed to WT cells from the ventral epithelium (left), knockdown cells (right) show accumulation of lipid droplets (ld) and other organelles in large autolysosomes (al). Bar, 200 nm. Micrographs of clonal analyses are representative of at least three independent experiments (10–15 animals per genotype per experiment). Molecular mass is indicated in kilodaltons.
Finally, we tested whether the down-regulation of other V-ATPase–associated genes could cause the same phenotype. Indeed, we found increased lipid droplet size in fat body and Malpighian tubules upon knockdown of ATP6V1C1 and ATP6AP1 (also known as Vha44 and VhaAC45, respectively; Fig. 7, f and g), suggesting that lipid accumulation in ATP6AP2L98S cells is a general V-ATPase–dependent phenotype.
ATP6AP2 forms a complex with other V-ATPase assembly factors
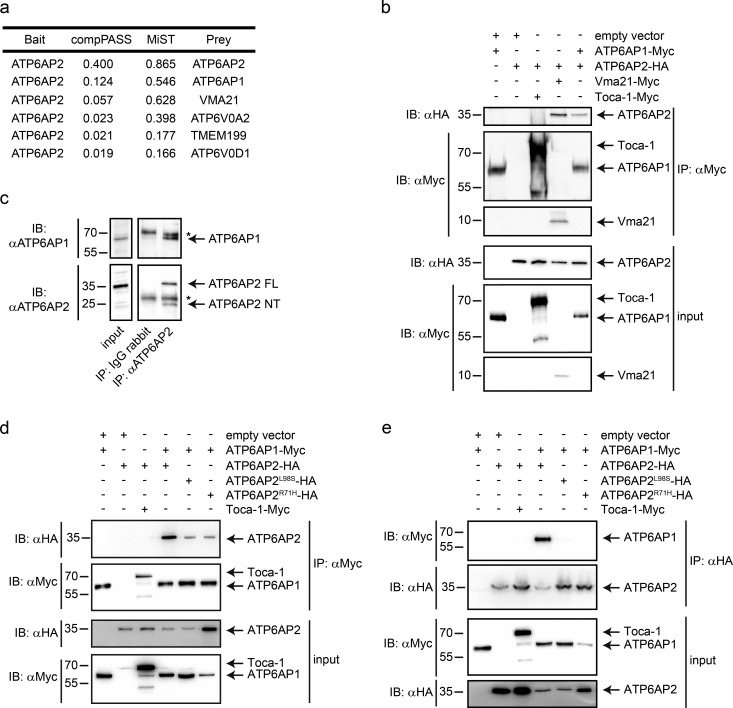
Given the phenotypic overlap between the ATP6AP2L98S mutant and ATP6V1C1 and ATP6AP1 knockdown in flies, we analyzed whether ATP6AP2 could interact with V-ATPase subunits and assembly factors and, if yes, whether the missense mutations could affect these interactions. We performed affinity purification coupled to MS (AP-MS) in HEK293T cells transiently overexpressing ATP6AP2WT. Among the top-ranking interactors, we found assembly factors for the V0 sector of the V-ATPase (ATP6AP1, VMA21, and TMEM199) and V0 subunits themselves (ATP6V0A2 and ATP6V0D1; Fig. 8 a), three of which have been implicated in CDG (ATP6AP1, TMEM199, and ATP6V0A2; Kornak et al., 2008; Jansen et al., 2016a,c). For the top candidates ATP6AP1 and VMA21, the interaction was confirmed by coimmunoprecipitation (coIP) of overexpressed tagged proteins in HEK293T cells (Fig. 8 b), and for ATP6AP1 also by endogenous coIP (Fig. 8 c). Moreover, in a yeast two-hybrid (Y2H) screen on a mouse cDNA library, we found ATP6AP1 as a binding partner of ATP6AP2. Mapping experiments with different truncated constructs suggested that the interaction requires the N-terminal luminal domains of both proteins, which in the case of ATP6AP2 harbors both amino acids mutated in our patients. Accordingly, both mutations impaired the interaction with ATP6AP1 in Y2H (Fig. S5) and in reciprocal coIP experiments (Fig. 8, d and e). In contrast, a construct with a deletion of exon 4 mimicking the effect of the exon-skipping mutation in P4 bound with similar strength to ATP6AP1 as ATP6AP2WT (Fig. S2 c). Together, these results suggest that ATP6AP2 forms a complex with other V-ATPase assembly factors, and that this interaction is impaired by the CDG-associated missense mutations.
Figure 8.
ATP6AP2 interacts with V-ATPase assembly factors. (a) Table showing the list of top-ranking interactors of ATP6AP2 based on two different score systems (CompPASS and MiST; Jäger et al., 2011). (b) CoIPs in HEK293T cells using the indicated constructs. Proteins were immunoprecipitated with anti-Myc antibody, and cell lysates were subjected to immunoblotting with anti-HA and anti-Myc antibodies. (c) Endogenous immunoprecipitation of ATP6AP2 in HEK293T cells using anti-ATP6AP2 (NT) antibody or control IgG. Immunoprecipitates were analyzed with anti-ATP6AP2 and anti-ATP6AP1. Lysate inputs of both proteins are on the left. The asterisks indicate heavy and light chains of the antibodies. (d) CoIPs in HEK293T cells using the indicated constructs. Proteins were immunoprecipitated with anti-Myc antibody, and cell lysates were subjected to immunoblotting with anti-HA and anti-Myc antibodies. (e) Reciprocal coIP experiment using anti-HA antibodies for immunoprecipitation. Toca-1-Myc is a negative control. Data are representative of eight (b), three (c), five (d), and two (e) independent experiments. Molecular mass is indicated in kilodaltons.
ATP6AP2 mutations leads to defects in autophagy
To address the functional consequences of the decreased interactions with V-ATPase assembly factors, we analyzed V-ATPase activity in ATP6AP2L98S fat body cells by staining with the acidotropic dye Lysotracker. ATP6AP2L98S mutant clones showed a reduction in Lysotracker-positive organelles compared with the WT surrounding tissue, indicating reduced acidity (Fig. 9 a). In accordance with the importance of V-ATPase–mediated acidification for autophagic degradation (Mauvezin et al., 2015), we found a correlation between lipid droplet size increase in knockdown cells and impaired lysosomal activity as determined using the GFP-LAMP reporter that accumulates when lysosome function is impaired (Fig. 9 b; Pulipparacharuvil et al., 2005). Moreover, we observed an accumulation of the autophagosomal marker Atg8a-mCherry (LC3 in mammals) in fat body cells expressing ATP6AP2 RNAi (Fig. 9 c) as well as an increase of endogenous Atg8a in ATP6AP2L98S clones in fat body (Fig. 9 d).
To further characterize the autophagy defect, we analyzed the processing of Atg8a by Western blotting. Atg8a can be visualized as one or two forms: a nonprocessed cytosolic form (Atg8a-I) and a faster migrating membrane-associated lipidated form (Atg8a-II), which is the active form involved in autophagosome formation. We detected an increase in Atg8a-II in ATP6AP2L98S larval lysates compared with WT and ATP6AP2WT-Myc (Fig. 9 e). To distinguish between increased autophagosome formation or decreased autophagic degradation (or a combination of both), we tested the levels of Ref(2)p (p62 in mammals). Consistent with decreased autophagic degradation, Ref(2)p was increased in ATP6AP2L98S clones (Fig. 9 d) and ATP6AP2L98S whole larval lysates (Fig. 9 e). Importantly, this is in agreement with the increased LC3-II, p62, and LAMP1/2 levels in mouse liver tissue acutely depleted of ATP6AP2 (Fig. S3 a; Kissing et al., 2017). Finally, we found an increase of Atg8a–II and Ref(2)p in ATP6AP2ΔKKxx, but not in ATP6AP2AxxA larval lysates (Fig. 9 e), suggesting that ER retrieval but not cleavage is essential for ATP6AP2 function in autophagy.
The V-ATPase has also been involved in the activation of mammalian target of rapamycin (mTOR) signaling (Zoncu et al., 2011), which conversely triggers autophagy when inhibited (Noda and Ohsumi, 1998). Thus, to analyze whether the ATP6AP2L98S mutant could alter mTOR activity, we tested the phosphorylation status of 4E-BP, an important downstream effector of mTOR. Fat body clones expressing ATP6AP2L98S showed reduced levels of phospho-4E-BP (p4E-BP) compared with the surrounding ATP6AP2WT-Myc tissue (Fig. 9 f). Likewise, p4E-BP was reduced in Western blotting of fed, starved, and refed ATP6AP2L98 whole larvae compared with control animals (Fig. 9 g), suggesting that stimulation of autophagy cause by mTOR inactivation in cells where lysosomal degradation is defective could contribute to the pathology. These findings were supported by transmission electron microscopy studies performed in pupal wings, in which knockdown cells showed a strong accumulation of lipid droplets and other organelles in large electron-dense vacuoles (Fig. 9 h). These vacuoles most likely represent autolysosomes with reduced degradative capacity and are reminiscent of the vacuoles found in the liver biopsy of P3 (Fig. 2 d).
Collectively, these results suggest that defects in V-ATPase activity, autophagy, and mTOR signaling are key to the disease caused by ATP6AP2 missense mutations, particularly with regard to the steatohepatitis.
Discussion
Genetic diseases caused by defective V-ATPase function are clinically diverse. Mutations in the core subunits ATP6V1B1 and ATP6V0A4 manifest in cells specialized in proton secretion such as renal intercalated cells and osteoclasts where these subunits are highly expressed (Forgac, 2007). In contrast, mutations in ubiquitously expressed core subunits and V0 assembly factors give rise to more systemic disorders (Kornak et al., 2008; Kortüm et al., 2015; Jansen et al., 2016a,b,c; Van Damme et al., 2017). The clinical similarity of ATP6AP2 deficiency with mutations in other assembly factors, most notably ATP6AP1, strongly argues for a role of ATP6AP2 in the assembly of the V0 sector of the V-ATPase. In support of this hypothesis, we show that ATP6AP2 not only interacts with V0 assembly factors but also requires retrograde delivery to the ER for function as has previously been shown for several yeast assembly factors (Malkus et al., 2004; Ryan et al., 2008; Jansen et al., 2016a). Moreover, the identified missense mutations impair the interaction with ATP6AP1 and can cause reduced proton pump activity.
The acute depletion of ATP6AP2 in the mouse liver was sufficient to recapitulate the glycosylation and autophagy defects of the patients. Mechanistically, both direct and indirect effects on the glycosylation machinery are possible in the absence of V-ATPase activity. Whereas some glycosylation enzymes like sialidases require low pH for optimal activity (al-Awqati et al., 1992), defects in V-ATPase assembly or function could also cause secondary defects in ER and Golgi homeostasis that are responsible for the glycosylation defects. Thus, it remains to be determined how ATP6AP2 deficiency affects glycosylation and how far the glycosylation defect contributes to the clinical symptoms.
The observed defect in autophagic flux can, on the other hand, more easily be linked to V-ATPase dysfunction and to the clinical phenotype. As V-ATPase–mediated acidification is generally indispensable for the final degradation step in autophagy, it can be argued that a ubiquitous small decrease in V-ATPase activity primarily manifests in organs with a high need for autophagy. Whereas plasma cells of the immune system depend on high rates of autophagy to counteract ER expansion when Ig synthesis is high (Pengo et al., 2013), dysfunctional lipophagy can cause steatosis of the liver (Singh et al., 2009; Baiceanu et al., 2016). The latter process was recapitulated in the Drosophila mutant, where fat body cells showed lipid accumulation along with reduced organellar acidity and decreased lysosomal degradation. Aggravating the pathology, the decreased mTORC1 activation, observed in these cells, may permit an inappropriate induction of autophagosome formation, autophagosome maturation, and fusion with lysosomes despite the inability to execute degradation in the final step of autophagy (Ramachandran et al., 2013; Mauvezin et al., 2015). Such a block in autophagy flux after its induction is supported by the observed lipidation of LC3/Atg8a and p62/Ref(2)P accumulation and may explain the enlarged autolysosomes seen in the liver biopsies and in the epithelial cells of the fly wings.
Whether the intellectual disability and mild ataxia in patients with ATP6AP2 missense mutations are also a consequence of reduced autophagy awaits further testing. By removing dysfunctional organelles and macromolecules, autophagy is beyond dispute an important survival mechanism in postmitotic neurons (Hara et al., 2006; Mariño et al., 2011). Accordingly, the neuron-specific down-regulation of ATP6AP2 in Drosophila and mouse has been shown to lead to autophagy defects and cognitive impairment (Dubos et al., 2015). However, the premature neuronal differentiation phenotype in the developing ATP6AP2L98S fly brains argues for additional defects in signaling pathways involved in neural development. One candidate pathway is Notch signaling that depends on proper V-ATPase function (Yan et al., 2009; Vaccari et al., 2010) and is required to maintain the pool of neural progenitors in the developing fly brain (Bowman et al., 2008; Egger et al., 2010; Wang et al., 2011). Indeed, Notch phenotypes were previously found in ATP6AP2 mutant wing clones (Hermle et al., 2013), and more importantly, reduction of Notch signaling because of inhibition of the V-ATPase has been shown to deplete neural stem cells in mice by promoting their differentiation (Lange et al., 2011).
Interestingly, late-onset cerebral manifestations such as Parkinsonism and epilepsy are the main problems in patients with exon-skipping mutations in ATP6AP2 (Ramser et al., 2005; Korvatska et al., 2013). However, these patients do not exhibit any obvious liver, immune, or skin defects. Furthermore, we found almost normal glycosylation in one of these patients as well as no effect on the interaction with ATP6AP1 upon deletion of exon 4 (Fig. S2, a–c). These results suggest that the clinical manifestation of ATP6AP2 deficiency depends on the severity of the mutations. Indeed, whereas the exon-skipping mutations have only partial penetrance reducing the levels of full-size ATP6AP2 transcripts by <50% (Korvatska et al., 2013), the missense mutations described here lead to impaired protein stability and the inability to fully rescue the embryonic lethality of the Drosophila null mutation. In particular, we provide evidence that ATP6AP2L98S is misfolded in the ER and targeted for ERAD. Thus, it is very likely that missense mutations have a stronger impact on overall ATP6AP2 function than the exon-skipping mutations.
In summary, we identify three individuals with a new glycosylation and autophagy disorder caused by different point mutations in the luminal domain of ATP6AP2. Our functional studies using cell culture, Drosophila, and mouse models suggest that the missense mutations reduce the interaction with V0 assembly factors and, consequently, V-ATPase activity. Interestingly, the lack of the ER retrieval motif caused more cleavage and led to reduced survival, fat accumulation, and autophagy defects in Drosophila, suggesting that retrograde Golgi-to-ER transport prevents cleavage in the Golgi and is required for proper ATP6AP2 function. Moreover, we demonstrate that proteolytic processing of ATP6AP2 is tissue-dependent (Fig. 6 e), similar to what has been shown before for ATP6AP1 (Jansen et al., 2016a). Our results thus provide a better understanding of ATP6AP2 functions and pave the way for additional studies on how ATP6AP2 processing and trafficking controls V-ATPase assembly.
Materials and methods
Ethics statement
Research on patients’ cells was prospectively reviewed and approved by the Ethical Committee of the University Hospitals of Leuven and Münster.
Exome sequencing
For the p.L98S case, mutation identification was achieved by whole exome sequencing. Genomic DNA was sheared by sonication, platform-specific adaptors were ligated, and the resulting fragments were size selected. The library was captured using the SeqCap EZ Human Exome Library v3.0 (Roche NimbleGen), and paired end (2 × 101 bp) sequenced on a HiSeq2000 (Illumina). Reads were aligned to the human reference genome (hg19) using BWA (v0.6.2), and duplicate reads were removed using Picard MarkDuplicates (v1.78). Local realignment around indels, base quality score recalibration, and variant calling were performed using GATK (v2.4.9) RealignerTargetCreator, IndelRealigner, BaseRecalibrator, and UnifiedGenotyper. Variants were annotated using Annovar (v11-02-2013). Synonymous variants were excluded, whereas variants with a frequency <5% in the 1000 Genomes project, ESP, GoNL, and our in-house database were further considered. Subsequent prioritization was then applied based on recessive inheritance, conservation, and pathogenicity prediction scores.
For the p.R71H cases, whole exome analysis of the patients DNA was done on an Illumina HiSeq2500. Subsequent prioritization in both cases was then applied based on recessive inheritance, conservation, and pathogenicity prediction scores.
Sanger sequencing
Total RNA was isolated from the primary fibroblasts of the patient and control cells using the RNeasy kit (QIAGEN). 2 µg of purified total RNA was subjected to RT with the First-Strand cDNA synthesis kit (GE Healthcare) following the manufacturer’s instructions. For PCRs, cDNAs obtained after RT were diluted 1:5.
Genomic DNA was extracted from white blood cells from the patient and controls using the DNeasy Blood & Tissue kit (QIAGEN) according to the manufacturer’s protocol. Primers were designed (Primer Blast) to amplify the different exons of the ATP6AP2 gene (available from GenBank under accession no. NM_005765.2), including at least 50 bp of the flanking intronic regions. Sequences of the primers for exon 3 are as follows: ATP6AP2_E03_F: 5′-ACCGCTTTGTGCTTTTCAAT-3′; ATP6AP2_E03_R: 5′-TCACCCACATTGCCTGAATA-3′.
DNA of the patients and family members of family 2 was isolated from EDTA blood using the DNeasy Blood & Tissue kit (QIAGEN) according to the manufacturer’s protocol. Primer sequences used for Sanger sequencing of ATP6AP2 (exon 3) were as follows: ATP6AP2 (exon 3) forward: 5′-TTGTCAGTGTCATCTCAGAACCCC-3′; ATP6AP2 (exon 3) reverse: 5′-TGGCTCTCTTTTTCAAGCCTCTGG-3′. Primers were designed with Primer Blast (Ye et al., 2012).
Glycosylation studies
Capillary zone electrophoresis and isoelectric focusing of serum transferrin were performed as previously described (Zühlsdorf et al., 2015). Glycosylation of serum transferrin was additionally assessed via HPLC using the CDT in serum kit by Chromesystems according to the manufacturer’s protocol.
For MALDI-TOF MS analysis, aliquots of 20 µl of mouse sera were mixed with lysis buffer (50 mM phosphate buffer, pH 7.9, containing 0.5% SDS and 1% β-mercaptoethanol) and incubated at 100°C for 20 min. After cooling and the addition of phosphate buffer and 1% Nonidet P-40, three units of PNGase F (Roche Diagnostics) were added, and samples were incubated for 24 h at 37°C. Released glycans were purified on a graphitized carbon column (Alltech). Glycans were permethylated as previously described (Faid et al., 2007) and purified on an SPE C18 column (Alltech). Permethylated glycans were then dissolved in methanol/water (1:1 vol/vol) and mixed with an equal volume of dihydroxyaminobenzoic acid matrix (Aldrich; 20 mg/ml in methanol/water 1:1 vol/vol). Mass spectra were acquired using an Applied Biosystems/MDS Sciex 4800 MALDI-TOF/TOF analyzer at fixed laser intensity for 1000 shots/spectrum. A total of 5000 shots were accumulated in reflectron positive ion mode MS for each sample.
Mouse experiments
Generation of the ATP6AP2fl/fl mutant mouse line was described previously (Riediger et al., 2011). For in vivo transduction, 109 of adenoviral infectious particles were diluted in 0.9% NaCl and administered retro-orbitally in a total volume of 100 µl per mouse. Mice were sacrificed 10 d after injection. Adenovirus coding for GFP was used as a control in all experiments. GFP and GFP-Cre adenoviral vectors were described previously (Nemazanyy et al., 2013). Animals were fed a standard chow diet (Teklad global protein diet; 20% protein, 75% carbohydrate, 5% fat). All mice were sacrificed at 2–4 pm in a random-fed state. All animal studies were approved by the Direction Départementale des Services Vétérinaires, Préfecture de Police, Paris, France. Liver transaminases ALT and AST and total cholesterol levels in serum were determined with a multiparametric automate Olympus AU 400.
Plasmids
The cDNAs of Vma21, Atp6ap1, and Atp6ap2 were generated by gene synthesis (Invitrogen GeneArt Gene Synthesis service) and subcloned using HindIII or NcoI and BamHI sites into pFrog-Myc/HA (a pcDNA3-derivative), resulting in C-terminal tagged versions of the respective proteins as previously described (Blanz et al., 2015). A version of ATP6AP2 lacking exon 4 (101-132aa) was subcloned from the pCS2 vector (Korvatska et al., 2013) into pFrog-Myc. For the AP-MS approach, ATP6AP2 cDNA was subcloned into pDNA4/TO with a triple FLAG-tag. All constructs were verified by sequencing. To obtain all the mutants of ATP6AP2 with single and double amino acid substitutions (ATP6AP2-L98S-HA, ATP6AP2-R71H-HA, and ATP6AP2-QxQxx-HA), the construct pFrog-ATP6AP2-HA was used as a template, and site-direct mutagenesis reactions were performed with the QuickChange II Site-Directed Mutagenesis kit (Agilent) according to the manufacturer’s instruction using the following oligos: -ATP6AP2-L98S-HA: forward: 5′-CCCTTCGGAGAATGCAGTTCCTTTTAGTCTAGACAGC-3′, reverse: 5′-CTCCGAAGGGTAGGAGATGACGCTGCCTGC-3′; -ATP6AP2-R71H-HA-forward: 5′-TTCCACATGCCACGGGCTACCATTATGGTGATGG-3′, reverse: 5′-CCCGTGGATGGTGGAATAGGTTACCCACGGCAAGCCC-3′; -ATP6AP2-QxQxx-HA- (sequential mutagenesis): forward: 5′-ATCAGCAGATTCGAATAGATGGATCCTACCCA-3′, reverse: 5′-AATCTGCTGATTTGTCATCCTATAGATGATGC-3′; forward: 5′-GATTCAAATAGATGGATCCTACCCATACGACG-3′, reverse: 5′-CTATTTGAATCTGCTGATTTGTCATCCTATAG-3′.
pcDNA3.1-p97-WT-His and pcDNA3.1-p97-QQ-His were kindly provided by Y. Yihong (National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD).
RNA extraction, RT, and quantitative real-time PCR
Total RNA was isolated from fibroblasts using a QIAGEN RNA extraction kit. cDNA was prepared using reverse transcription iScript cDNA Synthesis kit (Bio-Rad). Relative expression levels of ATP6AP2 were determined using the Power SYBR Green PCR Master Mix (ThermoFisher) and the following primers: ATP6AP2 exon 2 forward, 5′-CTGCATTGTCCATGGGCTTC-3′, and ATP6AP2 exon 3 reverse, 5′-AACAGGTTACCCACTGCGAG-3′. Expression levels were normalized to HPRT.
Cell culture, Western blotting, and immunoprecipitation
Primary fibroblasts from patients and controls were grown from a skin biopsy and cultured in Nutrient Mixture F-12 DMEM/F-12 (ThermoFisher) supplemented with 10% FBS at 37°C under 5% CO2. Research on patients’ cells was prospectively reviewed and approved by the Ethical Committees of the University Hospital of Leuven and Münster. HEK293T cells were routinely grown at 37°C in DMEM (ThermoFisher) containing 10% FBS. HEK293 STT3A KO, STT3B KO, and the parental HEK293 cells were cultured as described previously (Cherepanova and Gilmore, 2016). Cells were transfected by using Lipofectamine 2000 (Invitrogen) in Opti-MEM (ThermoFisher) according to the manufacturers’ instructions.
The expression of ATP6AP2 forms was analyzed by Western blotting 48 h after transfection. For this, cells were harvested in 50 µl of lysis buffer (1% Triton X-100 in PBS) supplemented with protease inhibitor cocktail (Roche). After 30-min incubation in lysis buffer at 4°C, the lysates were then cleared by centrifugation for 10 min at 13,200 rpm at 4°C. The protein concentration was determined using a Pierce BCA Protein Assay kit (Thermo Scientific). For each experiment, aliquots of the lysate (20 µg total protein per aliquot) were denatured using 1× SDS sample buffer for 25 min at 55°C and analyzed by SDS-PAGE. For immunoblotting, proteins were transferred to nitrocellulose filters by using the iBlot2 Dry Blotting system (ThermoFisher) and then blocked for 1 h in blocking buffer (5% nonfat dry milk, 0.2% Tween-20 in PBS). The membranes were incubated overnight with the specific primaries antibodies, followed by HPR-conjugated secondary antibodies. To assess relative protein levels, band intensity was measured with the ImageJ software.
For the coIP (anti-Myc), HEK293T cells were transfected with 1 µg of ATP6AP2WT-HA, 1 µg of ATP6AP2L98S-HA, and 1 µg of ATP6AP2R71H-HA. For the reciprocal coIP (anti-HA), cells were transfected with 1 µg of ATP6AP2WT-HA, 2 µg of ATP6AP2L98S-HA, and 2 µg of ATP6AP2R71H-HA. Cells were harvested in 100 µl of EBC lysis buffer (120 mM NaCl, 50 mM Tris [pH 8.0], 5 mM EDTA, 50 mM Hepes, and 0,5% NP-40) supplemented with protease inhibitor. After 1 h incubation in lysis buffer at 4°C, the lysates were then centrifuged for 10 min at 13,200 rpm at 4°C, and the protein concentration was determined as before. For each sample, 2 mg of protein was incubated overnight with 1 µg of mouse mAb anti-Myc at 4°C. Next, 50 µl of Dynabeads Protein G (Thermo Scientific) was added 1 h at 4°C and washed according to the manufacturer’s instructions. The immunoprecipitated proteins were eluted by addition of 30 µl of 4× SDS sample buffer, followed by 5 min incubation at 95°C. Initial lysates and immunoprecipitated proteins were analyzed by SDS-PAGE and immunoblotted with specific antibodies. For the endogenous immunoprecipitation, 2 mg of lysates were incubated overnight with either rabbit Ig (as control) or with 2 µg of rabbit polyclonal antibody anti-ATP6AP2 at 4°C. 100 µl of Dynabeads Protein G was added, and the proteins were eluted and analyzed by Western blotting as described in the previous paragraph.
The following antibodies were used: rabbit polyclonal anti-Myc antibody (sc-789) and mouse monoclonal anti-Myc antibody (sc-40) both from Santa Cruz Biotechnology, rat monoclonal anti-HA antibody (11867423001; Roche), rabbit polyclonal anti-ATP6AP1 antibody (ab176609; Abcam), and rabbit polyclonal anti-ATP6AP2 antibody (HPA003156), mouse monoclonal anti–β-actin antibody (A1978), and mouse monoclonal antihistidine antibody (H1029), all from Sigma-Aldrich. Secondary antibodies used were antirat, antirabbit, and antimouse Horseradish Peroxidase Conjugated (Thermo Scientific).
Indirect immunofluorescence studies
HeLa cells were grown on glass coverslips in DMEM (ThermoFisher) supplemented with 10% FBS. Cells were transfected with Lipofectamine 2000 (Invitrogen) in Opti-MEM (ThermoFisher) according to the manufacturer’s instructions. The localization of ATP6AP2 construct was analyzed 48 h after transfection. For this, cells were fixed with 4% formaldehyde in PBS for 30 min at room temperature. Formaldehyde was quenched with 0.1 M glycine in PBS for 5 min on ice, and cells were made permeable with 0.1% Triton X-100 in PBS 10 min at room temperature. Cells were blocked for 30 min at room temperature in BSA 3% and labeled with the appropriate primary antibodies overnight at 4°C in 0.1% PBS–Tween-20 supplemented with 3% BSA. After washing, cells were incubated 1 h at room temperature with secondary antibodies (dilution 1:200) and Hoechst (0.5 µg ml−1) in PBS. Coverslips were mounted in Moviol. Primary antibodies used were rat anti-HA (11867423001; Roche), mouse anti-Calnexin (ALX-804-014-R100; Enzo Life Science), and rabbit anti-Giantin (Antibody platform Institut Curie). Secondary antibodies used were fluorescent conjugated Alexa Fluor 488 and Alexa Fluor 555 (Invitrogen Molecular Probes).
Analysis of protein stability and glycosidase inhibitor treatments
For the cycloheximide chase assay, 30, 42, 45, and 46 h after transfection, culture medium was changed for the chase medium (DMEM supplemented with 20 mM Hepes), and cycloheximide (Sigma-Aldrich) was added to reach a final concentration of 100 µM. Vehicle controls were treated with DMSO for 18 h.
For the deglycosylation digestion, 20 µg of cell lysates were treated according to the manufacturer's instructions and incubated for 1 h at 37°C with 500 U of Endo H, 500 U of PNGase F, 40,000 U of O-glycosidase (New England BioLabs), or 0.005 U Sialidase A (Prozyme).
Proteomics
Transfection of HEK293 cells and AP-MS were performed as previously described (Jäger et al., 2011). In brief, pools of HEK293 cells transiently expressing ATP6AP2-3×FLAG were used to precipitate ATP6AP2 and associated protein complexes using affinity gels followed by MS analysis. Biological triplicates of ATP6AP2-3xFLAG were performed, and only those proteins that were detected in all spectral counts are shown. Data were analyzed with two AP-MS scoring algorithms, MiST (Jäger et al., 2011) and CompPASS (Sowa et al., 2009).
Y2H analysis
The Y2H screen was performed by Hybrigenics Services. The coding sequence of the human ATP6AP2 wt NT17-275 was PCR-amplified and cloned into pB27 as a N-terminal fusion to bait fragment (LexA-ATP6AP2 wt NT17-275). The constructs were checked by sequencing and used as a bait to screen a random-primed mouse kidney cDNA library constructed into pP6. pB27 and pP6 derive from the original pBTM116 (Vojtek and Hollenberg, 1995) and pGADGH (Bartel et al., 1993) plasmids, respectively. ULTImate Y2H screening was performed against the mouse kidney cDNA library using ATP6AP2 wt NT17-275 as a bait. The prey fragments cloned in frame with the Gal4 activation domain into plasmid pP6, derived from the original pGADGH (Bartel et al., 1993), were checked by sequencing.
For 1-by-1 screening, the prey fragment for the human ATP6AP1 (ATP6AP1-NT30-254, PCR amplified from the coding sequence of the human ATP6AP1-NT30-254), human ATP6AP2L98SNT17-275 (PCR amplified), and human ATP6AP2R71HNT17-275 (fragment synthesized) were cloned in frame with the LexA DNA binding domain into pB27 (LexA-ATP6AP1-NT30-254 and LexA-ATP6AP2L98SNT17-275). The prey fragments for the human ATP6AP2 wt NT17-275, the human ATP6AP2L98SNT17-275, and the ATP6AP2R71HNT17-275 were cloned in frame with the Gal4 AD into plasmid pP7 (AD-ATP6AP2 wt NT17-275, AD-ATP6AP2L98SNT17-275, and AD-ATP6AP2R71HNT17-275). pP7 derives from the original pGADGH (Bartel et al., 1993). The pP7 prey plasmid used in the control assay is derived from the pP6 plasmid. All constructs were checked by sequencing the entire inserts.
Fly strains and husbandry
Drosophila melanogaster stocks were raised on standard cornmeal food at 25°C. The following stocks were used: w1118 (used as WT control), FRT82B, ATP6AP2Δ1, ATP6AP2>ATP6AP2WT-Myc, ATP6AP2>ATP6AP2WT, and ATP6AP2>ATP6AP2AxxA, which have all been described in (Hermle et al., 2013), ATP6AP2>ATP6AP2L98S, ATP6AP2>ATP6AP2ΔKKxx, and UAS>ATP6AP2L98S were generated in this study (see next paragraph), tub-GFP–LAMP (gift from H. Krämer, UT Southwestern Medical Center, Dallas, TX), hs-Flp; tub-FRT-Gal80-FRT-Gal4, UAS–mCD8–mRFP (gift from F. Bosveld, Institut Curie, Paris, France), hs-Flp; UAS-Dcr2; R4-mCherry-Atg8, Actin-FRT-CD2-FRT-Gal4,UAS-GFPnls/TM6B (gift from G. Juhasz, Eotvos Lorand University, Budapest, Hungary), UAS-ATP6AP2RNAi, GD5830, UAS-ATP6V1C1RNAi, KK101527, UAS-ATP6AP1RNAi, GD48017 (all from VDRC Stock Center), FRT42D, ubi-mRFPnls (Bloomington Stock Center), hs-Flp;; Tub-FRT-Gal80-FRT-Gal4, UAS-mCD8-RFP (gift from F. Bosveld), and Gal4 drivers ptc-Gal4 and ap-Gal4 (Bloomington Stock Center). For the precise genotype used in each figure, see also Table S1.
ATP6AP2>ATP6AP2L98S-, ATP6AP2>ATP6AP2ΔKKxx-, and UAS>ATP6AP2L98S-expressing transgenic flies have been created by site-directed mutagenesis using the QuickChange II Site directed Mutagenesis kit (Agilent) according to the manufacturer’s instructions. For ATP6AP2>ATP6AP2L98S and ATP6AP2>ATP6AP2ΔKKxx, mutagenesis was performed on the pattB plasmid containing the WT extended gene region of ATP6AP2 (Hermle et al., 2013) using the following oligos: -ATP6AP2L98S-: forward: 5′-CGTTCCGGAGCCTGTCGACTCGTAGGTCTTGACG-3′, reverse: 5′-CGTTCCGGAGCCTGTCGACTCGTAGGTCTTGACG-3′; -ATP6AP2ΔKKxx-: forward: 5′-CCTTAGTTGTCCTTCTAGATGCGGGTAGAGGTC-3′, reverse: 5′-GACCTCTACCCGCATCTAGAAGGACAACTAAGG-3′.
For UAS>ATP6AP2L98S, mutagenesis was done on the pUASg-HAattB plasmid containing the coding sequence of ATP6AP2 optimised by GeneArt (Invitrogen) with a final stop codon that impedes the expression of the HA tag, using the following primers: forward: 5′-CGTGAAGACCTACGAGTCGACCGGCAGCGGCACC-3′, reverse: 5′-GGTGCCGCTGCCGGTCGACTCGTAGGTCTTCACG-3′.
All constructs were injected into flies with the attP landing site at 86FB by Bestgene.
Clones were generated using FLP/FRT or flip-out techniques (Xu and Rubin, 1993). Clones in fat bodies were induced in embryos by 2 h heat shock at 37°C (except for clones generated with the hs-Flp, UAS-Dcr2;Actin>CD2>Gal4;UAS-GFP,R4-mCherry-Atg8a/TM6B stock, which form spontaneous clones without heat shock) and analyzed 4 d later in wandering third instar larvae.
Percentage of pupariation and eclosion
50–100 third instar larvae of each genotype were collected 90 ± 4 h after egg laying on standard food plates supplemented with dried yeast (2–3 h egg collections) and reared on tubes containing standard food at 25°C. The number of larvae that underwent pupariation was scored twice a day. Pupae were then left to eclose, and the number of surviving adult flies was scored. Five independent experiments were performed with genotypes WT, ATP6AP2WT-Myc, ATP6AP2WT, and ATP6AP2L98S. ATP6AP2ΔKKxx and ATP6AP2AxxA were scored in three independent experiments.
Western blotting of Drosophila extracts
For whole larval extracts, five wandering third instar larvae per genotype were collected and washed once in PBS, ethanol 70%, and water to remove food, surface bacteria, and other contaminants. Larvae were transferred to an Eppendorf tube containing 100 µl of lysis buffer (50 mM Tris-base, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100) supplemented with protease and phosphatase inhibitors cocktails (Roche) and homogenized three times with a pellet pestle for 10–15 s on ice. For fat body and brain extracts, for preparation of lysates from third instar fat bodies and larval brains, whole fat bodies from 15 larvae per genotype and 50 brains per genotype were dissected in PBS, transferred to Eppendorf tubes containing 50 µl of lysis buffer, incubated for 15 min on ice, and lysed by sonication. Lysates were cleared by centrifugation at maximum speed for 10 min, and the supernatants were transferred to clean Eppendorf tubes. Protein concentration was determined using the Pierce BCA Protein Assay kit (Thermo Scientific) according to the manufacturer’s instructions. Samples (20 µg) were analyzed by SDS-PAGE followed by immunoblotting. The following antibodies were used: guinea pig anti-ATP6AP2 (Hermle et al., 2013), mouse anti–β-actin antibody (Sigma-Aldrich, A1978), mouse anti–α-tubulin (Sigma-Aldrich, T6199), rabbit anti-Ref(2)P (gift from T.E. Rusten), rabbit anti–p4E-BP (Cell Signaling, 2855) and rabbit anti-Atg8a (gift from G. Juhasz). Secondary antibodies used were anti–guinea pig, anti–rabbit, and anti–mouse Horseradish Peroxidase Conjugated (Thermo Scientific).
Staging, starvation, and refeeding of larvae
0-2 h egg collections were done at 25°C using egg collection chambers on 60-mm petri dishes containing standard food supplemented with dry yeast. 72 h after larval hatching, 50 larvae (mid third instar) were transferred to vials containing a filter paper soaked in PBS and kept at 25°C for 6 h (starvation). Subsequently, half of the starved larvae were transferred to dishes containing yeast paste and kept at 25°C for 30 min (refeeding). Fat bodies from fed, starved, and refed larvae were dissected and processed as before for Western blotting analyses.
Immunohistochemistry
Brains and fat bodies from third instar larvae were dissected in PBS, fixed for 20 min (30 min in the case of p4E-BP staining) in 4% paraformaldehyde in PBS (+0.1% Triton X-100 for brains), washed three times in PBS-T (PBS + 0.3% Triton X-100 for brains and PBS + 0.1% Triton X-100 for fat bodies), and incubated overnight at 4°C with primary antibodies diluted in PBS-T. After washing, tissues were incubated overnight at 4°C for brains and 2 h at room temperature for fat bodies with secondary antibodies (dilution 1:200) and Hoechst (0.5 µg ml−1) diluted in PBS-T. Tissues were washed twice in PBS-T followed by a wash in PBS. Tissues were mounted in mounting medium (1.25% n-propyl gallate, 75% glycerol, 25% H2O). Primary antibodies used were rabbit anti-Dpn (1:200, gift from R. Basto), rabbit anti-GFP (1:500, Antibody Platform Institut Curie), rat anti-RFP (1:200, 5F8, ChromoTek), rabbit anti-mCherry (1:100, Clontech), rat anti-ELAV (1:100, 7E8A10, Developmental Studies Hybridoma Bank), guinea-pig anti-ATP6AP2 (Hermle et al., 2013), rabbit anti-Ref(2)P (gift from T.E. Rusten), rabbit anti-Atg8a (gift from G. Juhasz) and rabbit anti–p4E-BP (2855, Cell Signaling). Secondary antibodies used were fluorescent conjugated Alexa Fluor 488, Alexa Fluor 555, Alexa Fluor 633, and Alexa Fluor 647 (Invitrogen Molecular Probes).
Bodipy and LysoTracker staining
For Bodipy labeling, dissected fat bodies were fixed for 30 min in 4% paraformaldehyde in PBS, washed in PBS, and incubated with BODIPY 493/503 (2.5 µg ml−1, Molecular Probes) and Hoechst (0.5 µg ml−1) diluted in PBS for 30 min. Tissues were mounted as described in the Immunohistochemistry section.
For Lysotracker labeling, dissected fat bodies were incubated with LysoTracker Green DND-26 (100 nM, Molecular Probes) and Hoechst (0.5 µg ml−1) diluted in PBS for 3 min, washed in PBS, and fixed in 4% paraformaldehyde in PBS for 10 min. Fat bodies were washed three times in PBS and mounted as before.
Tissue imaging
All images were acquired on a Leica TCS SP8 equipped with a 405-nm laser line and a White Light Laser with a 63x/1.4 DIC Lambda blue PLAN APOCHROMATE objective. Images were processed with Fiji (Schindelin et al., 2012) and Adobe Photoshop.
TAG assay
Five wandering third instar larvae per genotype were collected and washed once in PBS, ethanol 70%, and water to remove food, surface bacteria, and other contaminants. Larvae were transferred to an Eppendorf tube containing 200 µl of PBS-T (PBS+0.05% Triton X-100 supplemented with protease inhibitors, Roche) and homogenized three times with a pellet pestle for 10–15 s on ice. Homogenates were subsequently sonicated. 10 µl of homogenized sample was collected for protein determination, and the rest of the homogenate was incubated at 70°C for 10 min. 20 µl of the sample and also standards were added to tubes containing 20 µl PBS-T + PI (baseline) or 20 µl TAG reagent (Sigma T2449) and incubated for 30 min at 37°C. Samples were centrifuged at maximum speed for 3 min, and 7.5 µl of each sample supernatant was transferred to clear-bottom 96-well plate containing 22.5 µl of PBS-T. 100 µl of free glycerol reagent (Sigma F6428) was added, and the mix was incubated at 37°C for 5 min. Absorbance was measured in a spectrophotometer at 560 nm. TAG concentration for each sample was determined by subtracting the absorbance for the free glycerol in the untreated samples (baseline) from the total glycerol concentration in samples that were incubated in triglyceride reagent. As a final step, samples were normalized to the corresponding protein concentrations.
Electron microscopy
Drosophila prepupal wings (5.5 h APF) were dissected and directly fixed with 4% paraformaldehyde and 1% glutaraldehyde in 0.1 M PB overnight at 4°C. After fixation, wings were washed in PB, treated with 2% OsO4, stained with uranyl acetate, dehydrated, and embedded in epoxy resin (Durcupan ACM, Fluka, Sigma-Aldrich). 40 nm ultrathin cross sections in the proximal-distal axis were cut using an ultramicrotome (Leica UC6) and analyzed with a Zeiss LEO 906 transmission electron microscope operated at 80 kV and equipped with a 2K CCD camera (Tröndle).
Online supplemental material
Fig. S1 shows the glycosylation diagnostics for patients P2 and P3. Fig. S2 shows the glycosylation diagnostics of the patient with the exon-skipping mutation (P4) and the normal interaction of ATP6AP2 Δexon4 with ATP6AP1. Fig. S3 shows the characterization of ATP6AP2 cKO mice. Fig. S4 shows the subcellular localization of the different ATP6AP2 constructs in HeLa cells. Fig. S5 shows the interaction between ATP6AP2 and ATP6AP1 using the Y2H method. Table S1 shows the Drosophila genotypes used in this study.
Supplementary Material
Acknowledgments
We thank Luisa Diogo and Emilia Faria for clinical data on the patient with the p.L98S mutation. We are grateful to Gabor Juhasz, Aurelio Teleman, Frank Perez, Renata Basto, and Tor Erik Rusten and the Developmental Studies Hybridoma Bank for providing antibodies. We thank Gabor Juhasz, Helmut Krämer, Floris Bosveld, the Bloomington stock center and Vienna Drosophila RNAi Center for providing flies. We also thank Yihong Ye for providing the p97 plasmids, Olga Korvatska for the ATP6AP2 Δ4 plasmid, Reid Gilmore for the STT3 KO cells, and Geneviève Nguyen/Michael Bader for the ATP6AP2 KO mice. We are grateful to Eva Gleixner for preparing fly samples for the electron microscopy experiments. We thank the Imagine Microscopy platform for assistance with microscopy. We are grateful to Jaak Jaeken, Vincent Cantagrel, Floris Bosveld, Massimo D’Agostino, Fred Bernard, Tom Stevens, and Ari Helenius for critically reading the manuscript.
This work has been supported by the ATIP-Avenir program, the Fondation Bettencourt-Schueller (Liliane Bettencourt Chair of Developmental Biology), state funding by the Agence Nationale de la Recherche under the Investissements d’avenir program (grant no. ANR-10-IAHU-01), and a NEPHROFLY grant (no. ANR-14-ACHN-0013) to M. Simons. This research was also supported by an Agence Nationale de la Recherche grant (no. SOLV-CDG) to F. Foulquier and a NUTRISENSPIK grant (no. ANR-16-CE14-0029) to G. Panasyuk. G. Matthijs received support from the European Union’s Horizon 2020 research and innovation program under the ERA-NET Cofund action (grant no. 643578); the research was funded by the Research Foundation (FWO, Flanders; EURO-CDG-2 project). F. Folquier, G. Matthijs, and R. Péanne belong to LIA GLYCOLAB4CDG (International Associated Laboratory), funded by CNRS (France) and FWO. R. Péanne is a postdoctoral researcher (Pegasus Marie Curie Fellow) of the FWO. T.D. Bird and W.H. Raskind receive support from the National Institutes of Health (grant no. R01NS069719) and the Department of Veterans Affairs. This work is supported by National Funds through the Fundação para a Ciência e a Tecnologia (Portuguese national funding agency for science, research and technology) in the frameworks of the UID/Multi/00215/2013 project–Unit for Multidisciplinary Research in Biomedicine–UMIB/ICBAS/UP.
The authors declare no competing financial interests.
Author contributions: T. Marquardt, G. Matthijs, E.M. Maier, S. Nobre, P. Garcia, D. Quelhas, P. Freisinger, T.D. Bird, W.H. Raskind, and J.H. Park recruited patients and gathered detailed clinical information and data for the study. J. Reunert, T. Marquardt, R. Péanne, D. Rymen, E. Souche, and G. Matthijs identified mutations in the human ATP6AP2 gene. S. Duvet, F. Foulquier, G. Matthijs, R. Péanne, D. Rymen, J. Reunert, J.H. Park, T. Marquardt, and Y. Wada performed glycosylation studies. G. Panasyuk designed, performed, and analyzed the mouse experiments. M. Cannata Serio designed, performed, and analyzed the experiments in patient fibroblasts and HEK293 cells. S. Jäger, N.J. Krogan, and M. Schwake. performed proteomics. M.A. Rujano and V. Hauser designed, performed, and analyzed the Drosophila experiments. M.C. Guida generated Drosophila tools. O. Kretz performed TEM on Drosophila samples. M. Simons, T. Marquardt, and G. Matthijs conceived of and supervised the project and analyzed the data. M. Simons wrote the paper with help from M. A. Rujano and M. Cannata Serio. All authors critically reviewed the paper.
Footnotes
Abbreviations used:
- AD
- activation domain
- ALT
- alanine transaminase
- AP-MS
- affinity purification coupled to MS
- AST
- aspartate transaminase
- CDG
- congenital disorder of glycosylation
- coIP
- coimmunoprecipitation
- CT
- C-terminal fragment
- Dpn
- Deadpan
- Endo H
- endoglycosidase H
- ERAD
- ER-associated degradation
- FL
- full-length
- MALDI-TOF
- matrix-assisted laser desorption/ionization time-of-flight
- MS
- mass spectrometry
- mTOR
- mammalian target of rapamycin
- NT
- N-terminal fragment
- p4E-BP
- phospho-4E-BP
- PNGase F
- peptide-N-glycosidase F
- TAG
- triglyceride
- V-ATPase
- vacuolar-type H+-ATPase
- Y2H
- yeast two-hybrid
References
- al-Awqati Q., Barasch J., and Landry D.. 1992. Chloride channels of intracellular organelles and their potential role in cystic fibrosis. J. Exp. Biol. 172:245–266. [DOI] [PubMed] [Google Scholar]
- Baiceanu A., Mesdom P., Lagouge M., and Foufelle F.. 2016. Endoplasmic reticulum proteostasis in hepatic steatosis. Nat. Rev. Endocrinol. 12:710–722. 10.1038/nrendo.2016.124 [DOI] [PubMed] [Google Scholar]
- Bartel P., Chien C.T., Sternglanz R., and Fields S.. 1993. Elimination of false positives that arise in using the two-hybrid system. Biotechniques. 14:920–924. [PubMed] [Google Scholar]
- Blanz J., Zunke F., Markmann S., Damme M., Braulke T., Saftig P., and Schwake M.. 2015. Mannose 6-phosphate-independent Lysosomal Sorting of LIMP-2. Traffic. 16:1127–1136. 10.1111/tra.12313 [DOI] [PubMed] [Google Scholar]
- Bowman S.K., Rolland V., Betschinger J., Kinsey K.A., Emery G., and Knoblich J.A.. 2008. The tumor suppressors Brat and Numb regulate transit-amplifying neuroblast lineages in Drosophila. Dev. Cell. 14:535–546. 10.1016/j.devcel.2008.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buechling T., Bartscherer K., Ohkawara B., Chaudhary V., Spirohn K., Niehrs C., and Boutros M.. 2010. Wnt/Frizzled signaling requires dPRR, the Drosophila homolog of the prorenin receptor. Curr. Biol. 20:1263–1268. 10.1016/j.cub.2010.05.028 [DOI] [PubMed] [Google Scholar]
- Cherepanova N.A., and Gilmore R.. 2016. Mammalian cells lacking either the cotranslational or posttranslocational oligosaccharyltransferase complex display substrate-dependent defects in asparagine linked glycosylation. Sci. Rep. 6:20946 10.1038/srep20946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanova N., Shrimal S., and Gilmore R.. 2016. N-linked glycosylation and homeostasis of the endoplasmic reticulum. Curr. Opin. Cell Biol. 41:57–65. 10.1016/j.ceb.2016.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousin C., Bracquart D., Contrepas A., Corvol P., Muller L., and Nguyen G.. 2009. Soluble form of the (pro)renin receptor generated by intracellular cleavage by furin is secreted in plasma. Hypertension. 53:1077–1082. 10.1161/HYPERTENSIONAHA.108.127258 [DOI] [PubMed] [Google Scholar]
- Cruciat C.M., Ohkawara B., Acebron S.P., Karaulanov E., Reinhard C., Ingelfinger D., Boutros M., and Niehrs C.. 2010. Requirement of prorenin receptor and vacuolar H+-ATPase-mediated acidification for Wnt signaling. Science. 327:459–463. 10.1126/science.1179802 [DOI] [PubMed] [Google Scholar]
- Dubos A., Castells-Nobau A., Meziane H., Oortveld M.A., Houbaert X., Iacono G., Martin C., Mittelhaeuser C., Lalanne V., Kramer J.M., et al. 2015. Conditional depletion of intellectual disability and Parkinsonism candidate gene ATP6AP2 in fly and mouse induces cognitive impairment and neurodegeneration. Hum. Mol. Genet. 24:6736–6755. 10.1093/hmg/ddv380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger B., Gold K.S., and Brand A.H.. 2010. Notch regulates the switch from symmetric to asymmetric neural stem cell division in the Drosophila optic lobe. Development. 137:2981–2987. 10.1242/dev.051250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faid V., Chirat F., Seta N., Foulquier F., and Morelle W.. 2007. A rapid mass spectrometric strategy for the characterization of N- and O-glycan chains in the diagnosis of defects in glycan biosynthesis. Proteomics. 7:1800–1813. 10.1002/pmic.200600977 [DOI] [PubMed] [Google Scholar]
- Forgac M. 2007. Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat. Rev. Mol. Cell Biol. 8:917–929. 10.1038/nrm2272 [DOI] [PubMed] [Google Scholar]
- Freeze H.H., Chong J.X., Bamshad M.J., and Ng B.G.. 2014. Solving glycosylation disorders: fundamental approaches reveal complicated pathways. Am. J. Hum. Genet. 94:161–175. 10.1016/j.ajhg.2013.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T., Nakamura K., Matsui M., Yamamoto A., Nakahara Y., Suzuki-Migishima R., Yokoyama M., Mishima K., Saito I., Okano H., and Mizushima N.. 2006. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 441:885–889. 10.1038/nature04724 [DOI] [PubMed] [Google Scholar]
- Hennet T., and Cabalzar J.. 2015. Congenital disorders of glycosylation: a concise chart of glycocalyx dysfunction. Trends Biochem. Sci. 40:377–384. 10.1016/j.tibs.2015.03.002 [DOI] [PubMed] [Google Scholar]
- Hermle T., Saltukoglu D., Grünewald J., Walz G., and Simons M.. 2010. Regulation of Frizzled-dependent planar polarity signaling by a V-ATPase subunit. Curr. Biol. 20:1269–1276. 10.1016/j.cub.2010.05.057 [DOI] [PubMed] [Google Scholar]
- Hermle T., Guida M.C., Beck S., Helmstädter S., and Simons M.. 2013. Drosophila ATP6AP2/VhaPRR functions both as a novel planar cell polarity core protein and a regulator of endosomal trafficking. EMBO J. 32:245–259. 10.1038/emboj.2012.323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäger S., Cimermancic P., Gulbahce N., Johnson J.R., McGovern K.E., Clarke S.C., Shales M., Mercenne G., Pache L., Li K., et al. 2011. Global landscape of HIV-human protein complexes. Nature. 481:365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen E.J., Timal S., Ryan M., Ashikov A., van Scherpenzeel M., Graham L.A., Mandel H., Hoischen A., Iancu T.C., Raymond K., et al. 2016a ATP6AP1 deficiency causes an immunodeficiency with hepatopathy, cognitive impairment and abnormal protein glycosylation. Nat. Commun. 7:11600 10.1038/ncomms11600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen J.C., Cirak S., van Scherpenzeel M., Timal S., Reunert J., Rust S., Pérez B., Vicogne D., Krawitz P., Wada Y., et al. 2016b CCDC115 Deficiency Causes a Disorder of Golgi Homeostasis with Abnormal Protein Glycosylation. Am. J. Hum. Genet. 98:310–321. 10.1016/j.ajhg.2015.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen J.C., Timal S., van Scherpenzeel M., Michelakakis H., Vicogne D., Ashikov A., Moraitou M., Hoischen A., Huijben K., Steenbergen G., et al. 2016c TMEM199 Deficiency Is a Disorder of Golgi Homeostasis Characterized by Elevated Aminotransferases, Alkaline Phosphatase, and Cholesterol and Abnormal Glycosylation. Am. J. Hum. Genet. 98:322–330. 10.1016/j.ajhg.2015.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinouchi K., Ichihara A., Sano M., Sun-Wada G.H., Wada Y., Kurauchi-Mito A., Bokuda K., Narita T., Oshima Y., Sakoda M., et al. 2010. The (pro)renin receptor/ATP6AP2 is essential for vacuolar H+-ATPase assembly in murine cardiomyocytes. Circ. Res. 107:30–34. 10.1161/CIRCRESAHA.110.224667 [DOI] [PubMed] [Google Scholar]
- Kinouchi K., Ichihara A., Sano M., Sun-Wada G.H., Wada Y., Ochi H., Fukuda T., Bokuda K., Kurosawa H., Yoshida N., et al. 2013. The role of individual domains and the significance of shedding of ATP6AP2/(pro)renin receptor in vacuolar H(+)-ATPase biogenesis. PLoS One. 8:e78603 10.1371/journal.pone.0078603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissing S., Rudnik S., Damme M., Lullmann-Rauch R., Ichihara A., Kornak U., Eskelinen E.L., Jabs S., Heeren J., De Brabander J.K., et al. 2017. Disruption of the vacuolar-type H+-ATPase complex in liver causes MTORC1-independent accumulation of autophagic vacuoles and lysosomes. Autophagy. 13:670–685. 10.1080/15548627.2017.1280216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornak U., Reynders E., Dimopoulou A., van Reeuwijk J., Fischer B., Rajab A., Budde B., Nürnberg P., Foulquier F., Lefeber D., et al. ARCL Debré-type Study Group . 2008. Impaired glycosylation and cutis laxa caused by mutations in the vesicular H+-ATPase subunit ATP6V0A2. Nat. Genet. 40:32–34. 10.1038/ng.2007.45 [DOI] [PubMed] [Google Scholar]
- Kortüm F., Caputo V., Bauer C.K., Stella L., Ciolfi A., Alawi M., Bocchinfuso G., Flex E., Paolacci S., Dentici M.L., et al. 2015. Mutations in KCNH1 and ATP6V1B2 cause Zimmermann-Laband syndrome. Nat. Genet. 47:661–667. 10.1038/ng.3282 [DOI] [PubMed] [Google Scholar]
- Korvatska O., Strand N.S., Berndt J.D., Strovas T., Chen D.H., Leverenz J.B., Kiianitsa K., Mata I.F., Karakoc E., Greenup J.L., et al. 2013. Altered splicing of ATP6AP2 causes X-linked parkinsonism with spasticity (XPDS). Hum. Mol. Genet. 22:3259–3268. 10.1093/hmg/ddt180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange C., Prenninger S., Knuckles P., Taylor V., Levin M., and Calegari F.. 2011. The H(+) vacuolar ATPase maintains neural stem cells in the developing mouse cortex. Stem Cells Dev. 20:843–850. 10.1089/scd.2010.0484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkus P., Graham L.A., Stevens T.H., and Schekman R.. 2004. Role of Vma21p in assembly and transport of the yeast vacuolar ATPase. Mol. Biol. Cell. 15:5075–5091. 10.1091/mbc.E04-06-0514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariño G., Madeo F., and Kroemer G.. 2011. Autophagy for tissue homeostasis and neuroprotection. Curr. Opin. Cell Biol. 23:198–206. 10.1016/j.ceb.2010.10.001 [DOI] [PubMed] [Google Scholar]
- Mauvezin C., Nagy P., Juhász G., and Neufeld T.P.. 2015. Autophagosome-lysosome fusion is independent of V-ATPase-mediated acidification. Nat. Commun. 6:7007 10.1038/ncomms8007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T., Suzuki-Nakagawa C., Watanabe A., Asami E., Matsumoto M., Nakano M., Ebihara A., Uddin M.N., and Suzuki F.. 2017. Site-1 protease is required for the generation of soluble (pro)renin receptor. J. Biochem. 161:369–379. 10.1093/jb/mvw080 [DOI] [PubMed] [Google Scholar]
- Nemazanyy I., Blaauw B., Paolini C., Caillaud C., Protasi F., Mueller A., Proikas-Cezanne T., Russell R.C., Guan K.L., Nishino I., et al. 2013. Defects of Vps15 in skeletal muscles lead to autophagic vacuolar myopathy and lysosomal disease. EMBO Mol. Med. 5:870–890. 10.1002/emmm.201202057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen G., Delarue F., Burcklé C., Bouzhir L., Giller T., and Sraer J.D.. 2002. Pivotal role of the renin/prorenin receptor in angiotensin II production and cellular responses to renin. J. Clin. Invest. 109:1417–1427. 10.1172/JCI0214276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson T., Jackson M., and Peterson P.A.. 1989. Short cytoplasmic sequences serve as retention signals for transmembrane proteins in the endoplasmic reticulum. Cell. 58:707–718. 10.1016/0092-8674(89)90105-0 [DOI] [PubMed] [Google Scholar]
- Noda T., and Ohsumi Y.. 1998. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J. Biol. Chem. 273:3963–3966. 10.1074/jbc.273.7.3963 [DOI] [PubMed] [Google Scholar]
- Pengo N., Scolari M., Oliva L., Milan E., Mainoldi F., Raimondi A., Fagioli C., Merlini A., Mariani E., Pasqualetto E., et al. 2013. Plasma cells require autophagy for sustainable immunoglobulin production. Nat. Immunol. 14:298–305. 10.1038/ni.2524 [DOI] [PubMed] [Google Scholar]
- Pulipparacharuvil S., Akbar M.A., Ray S., Sevrioukov E.A., Haberman A.S., Rohrer J., and Krämer H.. 2005. Drosophila Vps16A is required for trafficking to lysosomes and biogenesis of pigment granules. J. Cell Sci. 118:3663–3673. 10.1242/jcs.02502 [DOI] [PubMed] [Google Scholar]
- Ramachandran N., Munteanu I., Wang P., Ruggieri A., Rilstone J.J., Israelian N., Naranian T., Paroutis P., Guo R., Ren Z.P., et al. 2013. VMA21 deficiency prevents vacuolar ATPase assembly and causes autophagic vacuolar myopathy. Acta Neuropathol. 125:439–457. 10.1007/s00401-012-1073-6 [DOI] [PubMed] [Google Scholar]
- Ramser J., Abidi F.E., Burckle C.A., Lenski C., Toriello H., Wen G., Lubs H.A., Engert S., Stevenson R.E., Meindl A., et al. 2005. A unique exonic splice enhancer mutation in a family with X-linked mental retardation and epilepsy points to a novel role of the renin receptor. Hum. Mol. Genet. 14:1019–1027. 10.1093/hmg/ddi094 [DOI] [PubMed] [Google Scholar]
- Riediger F., Quack I., Qadri F., Hartleben B., Park J.K., Potthoff S.A., Sohn D., Sihn G., Rousselle A., Fokuhl V., et al. 2011. Prorenin receptor is essential for podocyte autophagy and survival. J. Am. Soc. Nephrol. 22:2193–2202. 10.1681/ASN.2011020200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan M., Graham L.A., and Stevens T.H.. 2008. Voa1p functions in V-ATPase assembly in the yeast endoplasmic reticulum. Mol. Biol. Cell. 19:5131–5142. 10.1091/mbc.E08-06-0629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Sako Y., Sho M., Momohara M., Suico M.A., Shuto T., Nishitoh H., Okiyoneda T., Kokame K., Kaneko M., et al. 2012. STT3B-dependent posttranslational N-glycosylation as a surveillance system for secretory protein. Mol. Cell. 47:99–110. 10.1016/j.molcel.2012.04.015 [DOI] [PubMed] [Google Scholar]
- Schafer S.T., Han J., Pena M., von Bohlen Und Halbach O., Peters J., and Gage F.H.. 2015. The Wnt adaptor protein ATP6AP2 regulates multiple stages of adult hippocampal neurogenesis. J. Neurosci. 35:4983–4998. 10.1523/JNEUROSCI.4130-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schefe J.H., Menk M., Reinemund J., Effertz K., Hobbs R.M., Pandolfi P.P., Ruiz P., Unger T., and Funke-Kaiser H.. 2006. A novel signal transduction cascade involving direct physical interaction of the renin/prorenin receptor with the transcription factor promyelocytic zinc finger protein. Circ. Res. 99:1355–1366. 10.1161/01.RES.0000251700.00994.0d [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott K., Gadomski T., Kozicz T., and Morava E.. 2014. Congenital disorders of glycosylation: new defects and still counting. J. Inherit. Metab. Dis. 37:609–617. 10.1007/s10545-014-9720-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sihn G., Burckle C., Rousselle A., Reimer T., and Bader M.. 2013. (Pro)renin receptor: subcellular localizations and functions. Front. Biosci. (Elite Ed.). 5:500–508. 10.2741/E631 [DOI] [PubMed] [Google Scholar]
- Singh R., Kaushik S., Wang Y., Xiang Y., Novak I., Komatsu M., Tanaka K., Cuervo A.M., and Czaja M.J.. 2009. Autophagy regulates lipid metabolism. Nature. 458:1131–1135. 10.1038/nature07976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowa M.E., Bennett E.J., Gygi S.P., and Harper J.W.. 2009. Defining the human deubiquitinating enzyme interaction landscape. Cell. 138:389–403. 10.1016/j.cell.2009.04.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trepiccione F., Gerber S.D., Grahammer F., López-Cayuqueo K.I., Baudrie V., Păunescu T.G., Capen D.E., Picard N., Alexander R.T., Huber T.B., et al. 2016. Renal Atp6ap2/(Pro)renin Receptor Is Required for Normal Vacuolar H+-ATPase Function but Not for the Renin-Angiotensin System. J. Am. Soc. Nephrol. 27:3320–3330. 10.1681/ASN.2015080915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugur B., Chen K., and Bellen H.J.. 2016. Drosophila tools and assays for the study of human diseases. Dis. Model. Mech. 9:235–244. 10.1242/dmm.023762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaccari T., Duchi S., Cortese K., Tacchetti C., and Bilder D.. 2010. The vacuolar ATPase is required for physiological as well as pathological activation of the Notch receptor. Development. 137:1825–1832. 10.1242/dev.045484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme T., Gardeitchik T., Mohamed M., Guerrero-Castillo S., Freisinger P., Guillemyn B., Kariminejad A., Dalloyaux D., van Kraaij S., Lefeber D.J., et al. 2017. Mutations in ATP6V1E1 or ATP6V1A Cause Autosomal-Recessive Cutis Laxa. Am. J. Hum. Genet. 100:216–227. 10.1016/j.ajhg.2016.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vojtek A.B., and Hollenberg S.M.. 1995. Ras-Raf interaction: two-hybrid analysis. Methods Enzymol. 255:331–342. 10.1016/S0076-6879(95)55036-4 [DOI] [PubMed] [Google Scholar]
- Wang W., Liu W., Wang Y., Zhou L., Tang X., and Luo H.. 2011. Notch signaling regulates neuroepithelial stem cell maintenance and neuroblast formation in Drosophila optic lobe development. Dev. Biol. 350:414–428. 10.1016/j.ydbio.2010.12.002 [DOI] [PubMed] [Google Scholar]
- Xu T., and Rubin G.M.. 1993. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 117:1223–1237. [DOI] [PubMed] [Google Scholar]
- Yan Y., Denef N., and Schüpbach T.. 2009. The vacuolar proton pump, V-ATPase, is required for notch signaling and endosomal trafficking in Drosophila. Dev. Cell. 17:387–402. 10.1016/j.devcel.2009.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasugi T., Umetsu D., Murakami S., Sato M., and Tabata T.. 2008. Drosophila optic lobe neuroblasts triggered by a wave of proneural gene expression that is negatively regulated by JAK/STAT. Development. 135:1471–1480. 10.1242/dev.019117 [DOI] [PubMed] [Google Scholar]
- Ye J., Coulouris G., Zaretskaya I., Cutcutache I., Rozen S., and Madden T.L.. 2012. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 13:134 10.1186/1471-2105-13-134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Meyer H.H., and Rapoport T.A.. 2001. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature. 414:652–656. 10.1038/414652a [DOI] [PubMed] [Google Scholar]
- Zoncu R., Bar-Peled L., Efeyan A., Wang S., Sancak Y., and Sabatini D.M.. 2011. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science. 334:678–683. 10.1126/science.1207056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zühlsdorf A., Park J.H., Wada Y., Rust S., Reunert J., DuChesne I., Grüneberg M., and Marquardt T.. 2015. Transferrin variants: pitfalls in the diagnostics of Congenital disorders of glycosylation. Clin. Biochem. 48:11–13. 10.1016/j.clinbiochem.2014.09.022 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.