Matsuzawa-Ishimoto et al. show that autophagy gene ATG16L1, which is associated with inflammatory diseases of the gastrointestinal tract, is essential for preventing necroptotic cell death and loss of Paneth cells in the intestinal epithelium.
Abstract
A variant of the autophagy gene ATG16L1 is associated with Crohn’s disease, an inflammatory bowel disease (IBD), and poor survival in allogeneic hematopoietic stem cell transplant recipients. We demonstrate that ATG16L1 in the intestinal epithelium is essential for preventing loss of Paneth cells and exaggerated cell death in animal models of virally triggered IBD and allogeneic hematopoietic stem cell transplantation. Intestinal organoids lacking ATG16L1 reproduced this loss in Paneth cells and displayed TNFα-mediated necroptosis, a form of programmed necrosis. This cytoprotective function of ATG16L1 was associated with the role of autophagy in promoting mitochondrial homeostasis. Finally, therapeutic blockade of necroptosis through TNFα or RIPK1 inhibition ameliorated disease in the virally triggered IBD model. These findings indicate that, in contrast to tumor cells in which autophagy promotes caspase-independent cell death, ATG16L1 maintains the intestinal barrier by inhibiting necroptosis in the epithelium.
Introduction
Autophagy involves sequestration of cytosolic material into double-membrane vesicles termed autophagosomes, which subsequently fuse with the lysosome, leading to the degradation and recycling of the contents. A role for autophagy in the mucosal barrier is suggested by the genetic association between ATG16L1 and small intestinal Crohn’s disease, a major form of inflammatory bowel disease (IBD; Wlodarska et al., 2015; Cadwell, 2016). ATG16L1 is part of a complex that lipidates the ubiquitin-like molecule LC3 to promote autophagosome formation and function. The risk allele of ATG16L1 associated with IBD susceptibility (ATG16L1T300A) introduces a caspase-cleavage site that destabilizes the protein product and reduces autophagy in the presence of TNFα (Lassen et al., 2014; Murthy et al., 2014). How decreased autophagy relates to the intestinal barrier dysfunction and inflammation characteristic of IBD is under intense investigation.
One mechanism by which autophagy supports viability and counters inflammation is through the removal of cytotoxic material such as depolarized mitochondria that produce ROS (Mariño et al., 2014; Cadwell, 2016). Consistent with this prosurvival function, induction of autophagy by cytosolic HMGB1 in the intestinal epithelium is associated with protection from apoptosis (Zhu et al., 2015). However, autophagy is essential for salivary gland tissue degradation during Drosophila melanogaster development (Berry and Baehrecke, 2007) and mediates cell death in stressed neurons in Caenorhabditis elegans and mammals (Samara et al., 2008; Liu et al., 2013). In transformed cells, autophagy can facilitate a form of regulated necrosis termed necroptosis that is triggered by TNFα or other inflammatory signals (Chen et al., 2011). The autophagosome serves as a scaffold for the necroptosis signaling complex upon deletion of Map3k7, a tumor suppressor gene commonly mutated in prostate cancer (Goodall et al., 2016). Necroptosis occurs when receptor-interacting serine-threonine kinase 1 (RIPK1) and RIPK3 interact and activate mixed-lineage kinase domain–like protein (MLKL; Zhang et al., 2009; Sun et al., 2012). MLKL has been suggested to execute necroptosis through regulation of mitochondrial fission, but this mechanism has been contested (Pasparakis and Vandenabeele, 2015). Adding to this confusion, mitochondrial degradation through autophagy (mitophagy) can promote or prevent necroptosis in animal models of chronic obstructive pulmonary disease and ischemia/reperfusion injury, respectively (Mizumura et al., 2014; Lu et al., 2016). Therefore, the intersection among autophagy, mitochondrial homeostasis, and necrotic cell death is complex and potentially cell type dependent.
We previously demonstrated that murine norovirus (MNV) infection of mice with a germline gene-trap mutation in Atg16L1 (Atg16L1HM mice; HM, hypomorph) induces morphological and functional defects in Paneth cells (Cadwell et al., 2008, 2010), antimicrobial epithelial cells in the small intestinal crypt (Vaishnava et al., 2008; Adolph et al., 2013). This observation in MNV-infected Atg16L1HM mice led us to identify similar Paneth cell defects in resection specimens from Crohn’s disease patients homozygous for the ATG16L1T300A risk allele (Cadwell et al., 2008). Also, MNV-infected Atg16L1HM mice display additional pathologies when treated with dextran sodium sulfate (DSS), such as blunted villi in the small intestine (Cadwell et al., 2010). The persistent strain of MNV that induces these intestinal abnormalities in Atg16L1HM mice does not typically induce disease in immunocompetent mice. In fact, we recently showed that MNV infection promotes intestinal development and protects against injury in antibiotic-treated WT C57BL/6 (B6) mice from DSS (Kernbauer et al., 2014). An outstanding question is why an otherwise beneficial enteric virus induces disease pathologies when autophagy is reduced.
In addition to this virally triggered model of IBD, Atg16L1HM mice are susceptible to graft-versus-host disease (GVHD) after allogeneic hematopoietic stem cell transplantation (allo-HSCT), a procedure used to treat malignant and nonmalignant blood disorders (Hubbard-Lucey et al., 2014). Notably, the same ATG16L1T300A allele linked to IBD is associated with poor survival after allo-HSCT in humans (Holler et al., 2010; Hubbard-Lucey et al., 2014). TNFα blockade ameliorates disease in both the virally triggered IBD and the GVHD models in Atg16L1HM mice (Cadwell et al., 2010; Hubbard-Lucey et al., 2014). Thus, it is possible that preventing TNFα-induced pathology is a conserved function of autophagy in these two disease conditions. To understand the role of ATG16L1 and the autophagy machinery in dampening inflammation at the mucosal barrier, we investigated the mechanisms by which ATG16L1 plays a cytoprotective function in the intestinal epithelium in both the virally triggered IBD and GVHD models.
Results
ATG16L1 in the epithelium protects against exacerbated intestinal injury after MNV infection
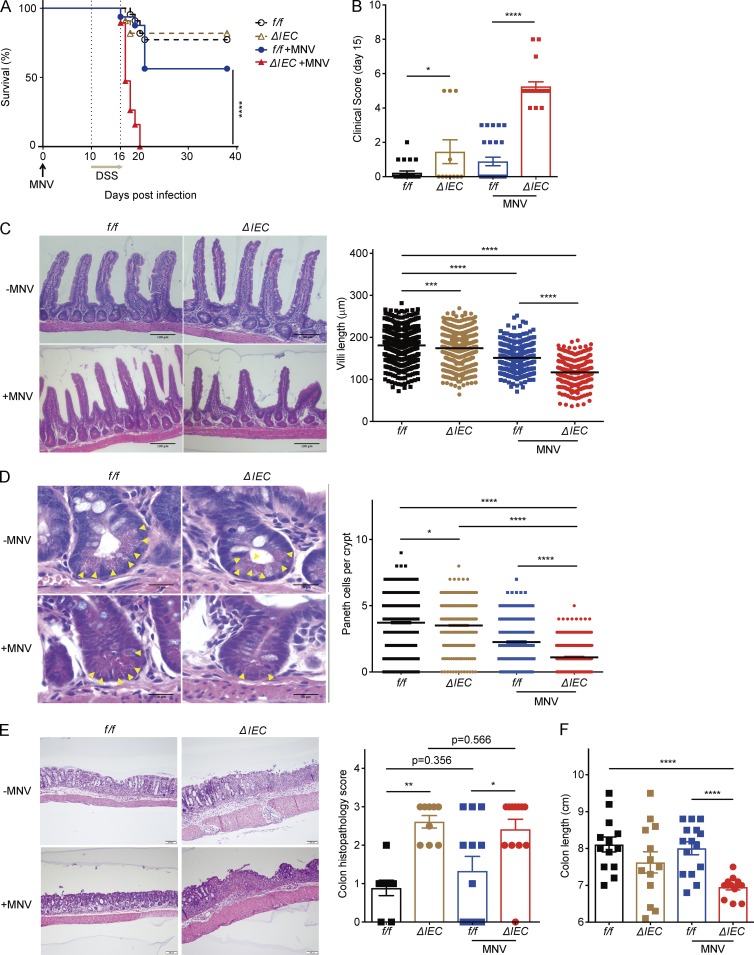
Given that MNV behaves similarly to symbiotic bacteria in WT B6 mice (Kernbauer et al., 2014), the virally triggered IBD model may reveal mechanisms involved in tolerating the presence of microbes in the gut. However, the cell type-specific function of ATG16L1 in this model has not been investigated. Autophagy in intestinal epithelial cells (IECs) is critical for protection against Salmonella enterica Typhimurium and secretion of antimicrobial molecules and mucin (Adolph et al., 2013; Benjamin et al., 2013; Conway et al., 2013; Patel et al., 2013). Thus, we examined susceptibility of Atg16L1f/f;villin-Cre mice in which Atg16L1 is deleted in IECs (Atg16L1ΔIEC) to DSS treatment in the presence or absence of MNV. Atg16L1ΔIEC mice receiving MNV+DSS displayed higher lethality and clinical disease score compared with similarly treated littermate Atg16L1f/f mice and uninfected groups (Fig. 1, A and B). MNV-infected Atg16L1ΔIEC mice, but not uninfected mice, displayed blunted villi in the small intestine and a decrease in Paneth cells (Fig. 1, C and D). In contrast, there was no significant difference in goblet cells even in the presence of MNV infection (Fig. S1 A). Atg16L1ΔIEC mice displayed more severe colon histopathology compared with Atg16L1f/f mice regardless of MNV infection; however, shortening of colon length was particularly striking in Atg16L1ΔIEC mice infected with MNV (Fig. 1, E and F). Additionally, analyses of a panel of cytokines indicated that TNFα was increased in sera of Atg16L1ΔIEC mice in an MNV-dependent manner (Fig. S1 B). MNV burden was similar in Atg16L1ΔIEC and Atg16L1f/f mice (Fig. S1 C). Therefore, these results raise the possibility that deletion of Atg16L1 sensitizes IECs to the inflammatory response to the virus.
Figure 1.
ATG16L1 in the epithelium protects against intestinal injury after MNV infection. (A and B) Survival (A) and clinical disease score (B) of Atg16L1f/f (f/f) and Atg16L1ΔIEC (ΔIEC) mice treated with 5% DSS ± MNV infection. n = 22 (f/f), 11 (ΔIEC), 16 (f/f + MNV), and 19 (ΔIEC + MNV). (C and D) Representative H&E images of small intestine harvested from mice treated as in A and quantification of villi length (C) and Paneth cells (yellow arrowheads; D). At least 50 villi and crypts were quantified per mouse. n = 9 (f/f), 9 (ΔIEC), 6 (f/f + MNV), and 6 (ΔIEC + MNV). (E and F) Representative H&E images of colon and quantification of colon histopathology (E) and colon length (F) from mice treated as in A. n = 13 (f/f), 13 (ΔIEC), 15 (f/f + MNV), and 13 (ΔIEC + MNV). (C–E) Bars: 100 µm (C and E); 20 µm (D). Data points represent individual mice in B, E, and F, individual villi in C, and individual crypts in D. Bars represent mean ± SEM, and at least two independent experiments were performed. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by Mantel-Cox in A and one-way ANOVA and Tukey’s test in B–F.
Deletion of Atg16L1 in the intestinal epithelium worsens GVHD
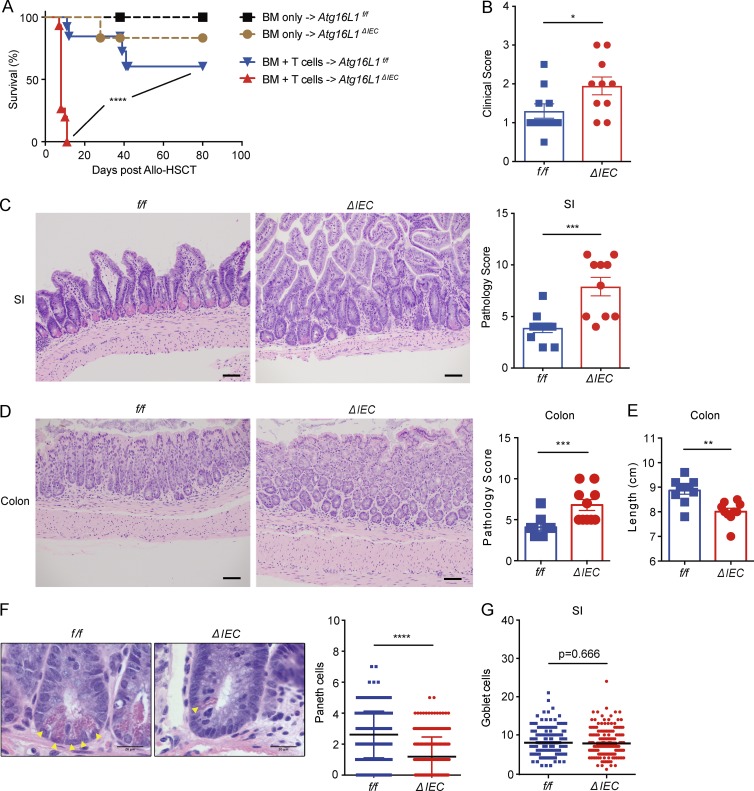
Next, we wished to validate this role of ATG16L1 in IECs in a second model of intestinal inflammation. GVHD is often accompanied by a compromised intestinal barrier (Hill and Ferrara, 2000). To elicit GVHD, we used an animal model of allo-HSCT similar to our previous study (Hubbard-Lucey et al., 2014) in which bone marrow (BM) cells with or without T cells from B10.BR donor mice were transplanted into lethally irradiated Atg16L1ΔIEC mice, which are on the B6 background. Compared with Atg16L1f/f recipients, Atg16L1ΔIEC mice receiving BM and 2 × 106 T cells displayed increased mortality and clinical GVHD score (Fig. 2, A and B). Mice that received BM without T cells did not display this severe lethality (Fig. 2 A), supporting the T cell dependence of the model. Because of the rapid course of the disease, we sacrificed the recipients on day 4 after allo-HSCT for all of our further analyses. We found that Atg16L1ΔIEC mice displayed exacerbated small intestinal and colonic histopathology and a decrease in colon length (Fig. 2, C–E). Atg16L1ΔIEC mice showed more lymphocytic infiltration in the crypts, apoptotic glandular epithelial cells, and crypt regeneration compared with Atg16L1f/f recipients in the small intestine (Fig. 2 C). In addition, the number of Paneth cells, but not goblet cells, in Atg16L1ΔIEC mice was significantly decreased compared with Atg16L1f/f recipients (Fig. 2, F and G). The number of CD4+ and CD8+ T cells, dendritic cells, B220+ B cells, and CD11b+ granulocytes in the lamina propria of the small intestine and colon were similar in Atg16Lf/f and Atg16L1ΔIEC recipients (Fig. S1, D–K). TNFα was readily detectable in the sera of both recipients, and IFN-γ was significantly increased in Atg16L1ΔIEC mice (Fig. S1 L). Collectively, these results indicate that Atg16L1 deletion in IECs was sufficient to confer increased susceptibility in models of IBD and allo-HSCT, and the rapid lethality observed in Atg16L1ΔIEC allo-HSCT recipients does not correlate with obvious differences in the magnitude of cell-mediated immune mediators that are typically associated with GVHD. Instead, these results raise the possibility that ATG16L1 deficiency enhances epithelial tissue damage.
Figure 2.
Deletion of Atg16L1 in the intestinal epithelium of allo-HSCT recipients worsens GVHD. (A) Survival of lethally irradiated Atg16L1f/f (f/f) and Atg16L1ΔIEC (ΔIEC) mice transplanted with 5 × 106 T cell–depleted BM cells with or without 2 × 106 splenic T cells from donor B10.BR mice. n = 5 (f/f, BM only), 6 (ΔIEC, BM only), 13 (f/f, BM + T cells), and 15 (ΔIEC, BM + T cells). (B–E) Mice receiving BM and T cells from B10.BR mice as in A were sacrificed 4 d after allo-HSCT and analyzed for macroscopic signs of GVHD (B; see Materials and methods). Representative images of H&E-stained sections and pathology score of small intestine (C) and colon (D) and quantification of colon length (E). n = 10 mice per group. (F) Representative higher magnification H&E images from C and quantification of Paneth cells (yellow arrowheads). n = 5 mice per group. At least 50 crypts were quantified per mouse. (G) Quantification of goblet cells from sections in C stained with periodic acid–Schiff (PAS)/Alcian blue of the small intestine. At least 50 villi were quantified per mouse. n = 5 mice per group. (C, D, and F) Bars: 200 µm (C and D); 20 µm (F). Data points present individual mice in B–E, individual crypts in F, and individual villi in G. Bars represent mean ± SEM, and at least two independent experiments were performed. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by Mantel-Cox in A and Mann-Whitney U test in B–G.
ATG16L1 protects against epithelial cell death
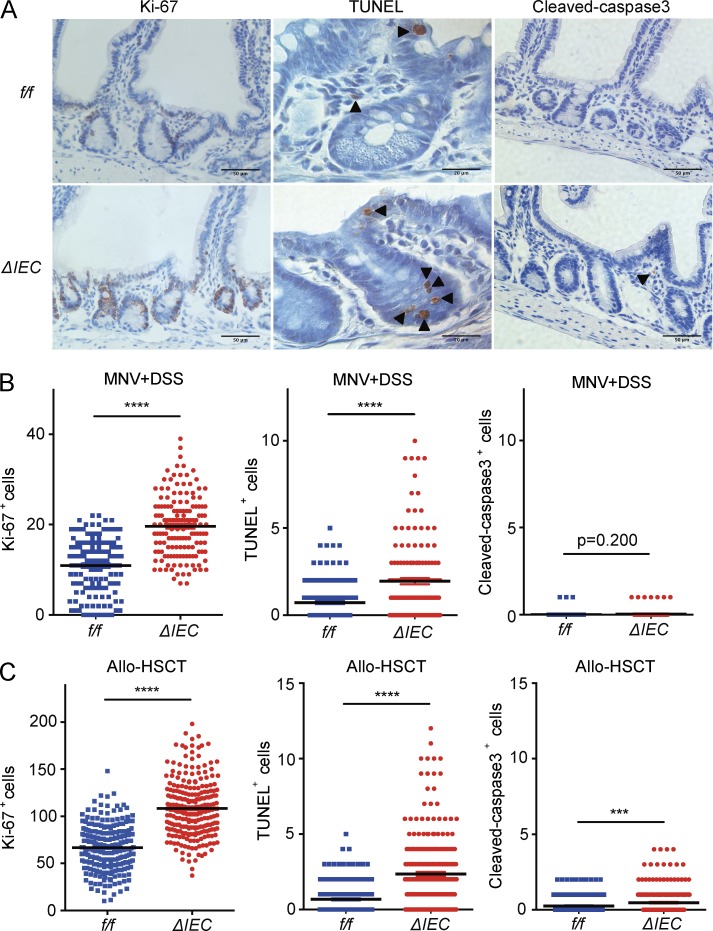
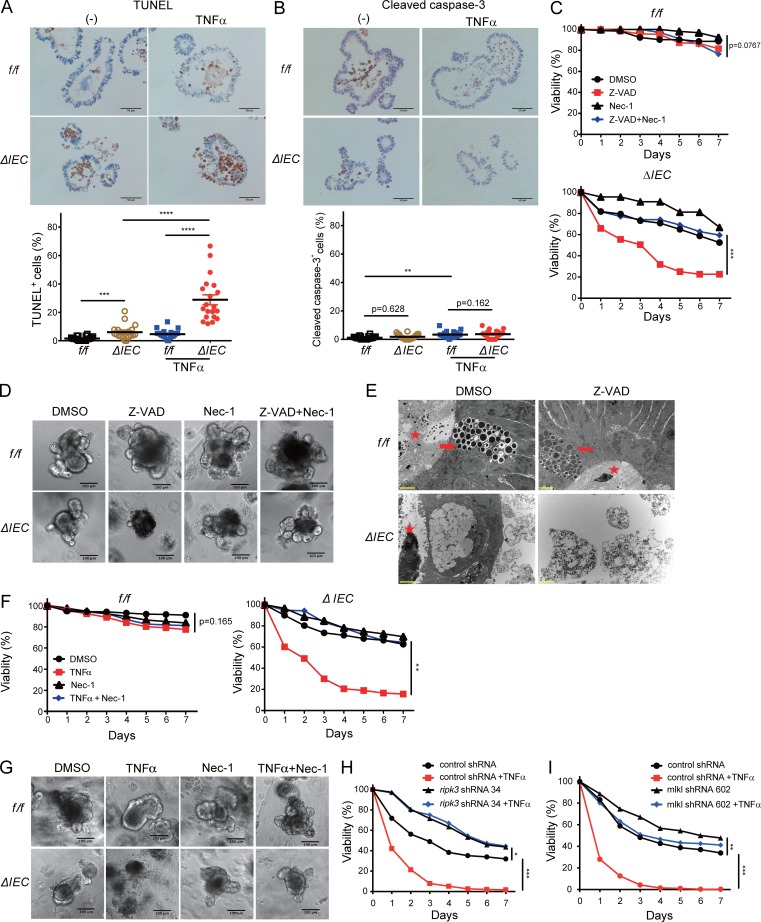
We next examined markers of epithelial turnover in intestinal tissue harvested from the above mice. In the MNV+DSS model, there was a striking increase in IECs positive for the proliferation marker Ki-67 in Atg16L1ΔIEC compared with Atg16Lf/f mice (Fig. 3, A and B). Additionally, a large number of IECs in Atg16L1ΔIEC mice were positive for terminal deoxynucleotidyl transferase–mediated dUTP nick-end labeling (TUNEL; Fig. 3, A and B), indicative of cell death. TUNEL staining was striking in the crypt-base of the small intestine, where Paneth cells are located (Ramanan and Cadwell, 2016). In contrast, we did not observe a similar degree of staining with the apoptosis marker cleaved caspase-3 (CC3; Fig. 3, A and B). There was also an increase in IECs positive for Ki-67 and TUNEL in Atg16L1ΔIEC mice compared with Atg16Lf/f mice in the allo-HSCT model (Fig. 3 C). The number of CC3+ cells were modest and did not overlap with the TUNEL staining, which was again enriched in the crypt-base (Fig. 3 C). Thus, ATG16L1 has a critical role in IEC homeostasis in both the virally triggered IBD model and the GVHD model. The enrichment of TUNEL+ cells in the absence of a similar increase in CC3 staining in the Atg16L1ΔIEC intestines raises the possibility that ATG16L1 prevents a nonapoptotic form of cell death (Gold et al., 1994; Grasl-Kraupp et al., 1995; Imagawa et al., 2016).
Figure 3.
ATG16L1 protects against cell death in the small intestinal epithelium. (A and B) Representative images (A) and quantification of Ki-67, TUNEL, and CC3 staining (B) of small intestine harvested from MNV-infected Atg16L1f/f (f/f) and Atg16L1ΔIEC (ΔIEC) mice on day 15 after MNV infection. Arrowheads indicate IECs positive for the indicated markers. Bars, 20 µm for TUNEL and 50 µm for Ki-67 and CC3 staining. At least 50 crypt–villus units were quantified per mouse from three mice per group. (C) Ki-67-, TUNEL-, and CC3-positive IECs were quantified as in B in small intestinal sections from Atg16L1f/f and Atg16L1ΔIEC mice on day 4 after allo-HSCT. At least 50 crypt–villus units were quantified per mouse from five mice per group. Data points in B and C represent individual crypts. Bars represent mean ± SEM, and at least two independent experiments were performed. ***, P < 0.001; ****, P < 0.0001 by unpaired t test.
ATG16L1 is necessary for viability of intestinal organoids
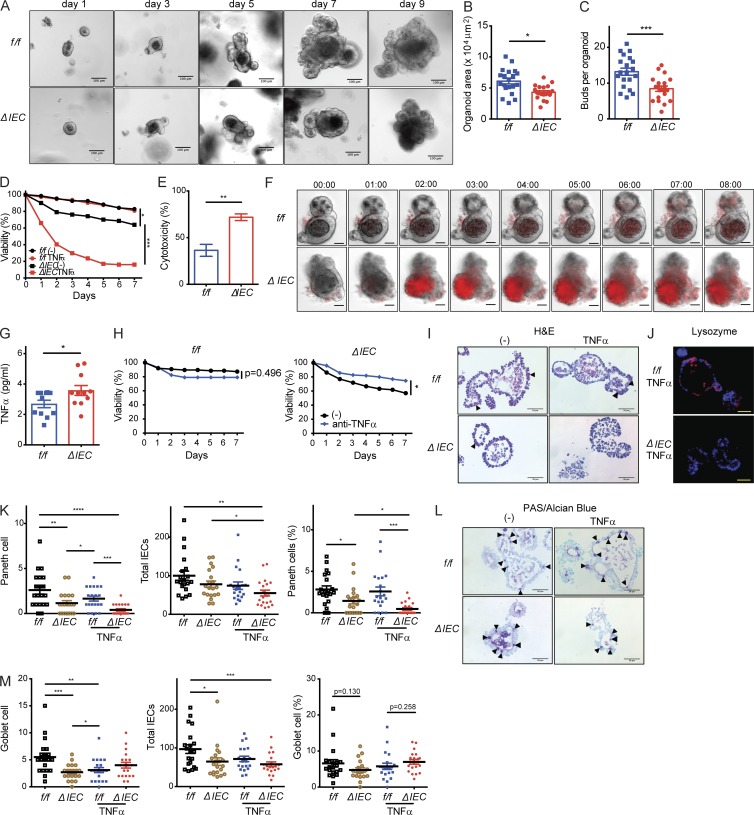
To examine the mechanism by which ATG16L1 maintains IECs, we derived organoids from small intestinal crypts harvested from Atg16L1f/f and Atg16L1ΔIEC mice. A previous study reported that Atg16L1 mutant Paneth cells have reduced capacity to promote organoid formation when cocultured with Lgr5+ stem cells (Lassen et al., 2014). Consistent with this finding, we found that organoids generated from Atg16L1ΔIEC mice were smaller and contained fewer buds compared with those generated from Atg16L1f/f mice (Fig. 4, A–C). Although the number of crypts isolated from each genotype were similar, and ATG16L1 deficiency had little effect on the expression of the stem cell marker Lgr5 (Fig. S2, A and B), we observed that up to 40% of the Atg16L1ΔIEC organoids lost viability over time on the basis of their collapsed structure and complete absence of buds (Fig. 4 D). Propidium iodide (PI) uptake and lactate dehydrogenase (LDH) release assays confirmed that an increased amount of cell death occurs in Atg16L1ΔIEC organoids (Fig. 4 E and Fig. S2 C). Loss in viability of ATG16L1-deficient organoids, but not those derived from control mice, was significantly exacerbated when cultured in the presence of TNFα (Fig. 4 D). Live imaging analyses showed that Atg16L1ΔIEC organoids rapidly take up PI in the media upon exposure to TNFα (Fig. 4 F and Video 1). These findings indicate that ATG16L1 is necessary for survival of intestinal organoids and resistance to TNFα.
Figure 4.
ATG16L1 is necessary for the survival of intestinal organoids and Paneth cells. (A–C) Representative images of small intestinal organoids derived from Atg16L1f/f (f/f) and Atg16L1ΔIEC (ΔIEC) mice on the indicated days of culture (A). Area (B) and bud number (C) were quantified on day 7. At least 20 organoids were quantified from three mice each. (D) Viability of organoids on the basis of morphology (see Fig. S2 C) ± 20 ng/ml TNFα. n ≥ 10 mice each. (E) LDH cytotoxicity of organoids on day 2 quantified by LDH release assay. n = 3 mice each. (F) Time-series images corresponding to Video 1 in which organoids were treated with 20 ng/ml TNFα at time 0 in the presence of PI (red). Digits represent hours:minutes. Representative of three independent repeats. (G) Quantification of TNFα in culture supernatants from D on day 3. n = 11 per group. (H) Viability of organoids ± 20 µg/ml anti-TNFα antibody. n = 3 mice each. (I–K) Representative H&E images (I) and immunofluorescence images (J) of lysozyme (red) and DAPI (blue) of organoids on day 5. TNFα was added on day 4. Triangles indicate Paneth cells in I. (K) Total number of Paneth cells (left), total number of IECs (middle), and frequency of Paneth cells normalized to total IECs (right) were quantified from images in I. At least 20 organoids were quantified from three mice each. (L and M) Representative images of goblet cells (triangles) in periodic acid–Schiff (PAS)/Alcian blue staining of organoids from I (L) and quantification of total number of goblet cells (left), total number of IECs (middle), and frequency of Paneth cells normalized to total IECs (right; M). At least 20 organoids were quantified from three mice each. (A, F, I, J, and L) Bars: 100 µm (A); 50 µm (F, I, J, and L). Data points in B, C, G, K, and M represent individual organoids, and data points in D, E, and H are mean of three technical replicates. Bars represent mean ± SEM, and at least two independent experiments were performed. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by unpaired t test in B, C, D, E, G, and H and one-way ANOVA and Tukey’s test in K and M.
It is possible that the reason why some Atg16L1ΔIEC organoids die in the absence of exogenous TNFα administration is that low levels of TNFα are produced by the organoids. Indeed, TNFα was detectable in the supernatant of Atg16L1ΔIEC organoids in the absence of exogenous TNFα administration (Fig. 4 G). Anti-TNFα antibodies improved viability of Atg16L1ΔIEC organoids from 60% to 80% and had no effect on Atg16L1f/f organoids (Fig. 4 H). Thus, Atg16L1ΔIEC organoids spontaneously produce factors, including TNFα, that mediate cell death. In sharp contrast to these findings with organoids derived from the small intestine, the viability of Atg16L1f/f and Atg16L1ΔIEC organoids derived from the colon was similar, even upon addition of exogenous TNFα (Fig. S2, D and E).
The observation that Atg16L1ΔIEC colonic organoids remain viable in the presence of TNFα may be related to the lack of Paneth cells in the colon. Thus, we examined the effect of Atg16L1 deletion on Paneth cells in small intestinal organoids. Paneth cells were quantified by light microscopy in day 5 cultures, a time point at which a sufficient number of intact Atg16L1ΔIEC organoids can be analyzed. We found that the absolute number of Paneth cells, as well as the proportion of Paneth cells normalized to total epithelial cells, were significantly reduced in Atg16L1ΔIEC organoids compared with Atg16L1f/f organoids (Fig. 4, I and K). The addition of TNFα the day before analysis reduced Paneth cell numbers further in Atg16L1ΔIEC organoids (Fig. 4, I and K). Immunofluorescence analysis of the Paneth cell granule protein lysozyme confirmed that TNFα treatment led to an almost complete loss of these cells in Atg16L1ΔIEC organoids (Fig. 4 J). In contrast, the proportion of goblet cells normalized to total cells was similar between the genotypes (Fig. 4, L and M). We noted that ATG16L1 deficiency decreased the absolute number of goblet cells, but this finding is likely a reflection of the smaller size of Atg16L1ΔIEC organoids (Fig. 4, B and C). Also, TNFα treatment increased rather than decreased goblet cell numbers in Atg16L1ΔIEC organoids. Thus, Paneth cells in Atg16L1ΔIEC organoids are preferentially sensitive to TNFα-induced depletion.
ATG16L1 protects small intestinal organoids from necroptosis
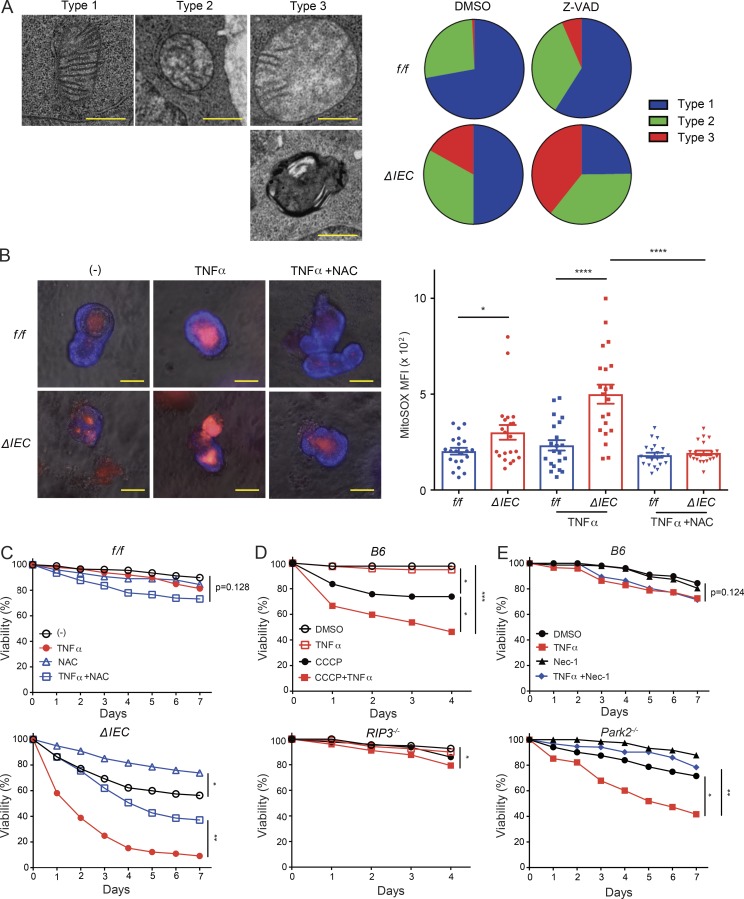
Similar to the findings in vivo, Atg16L1ΔIEC organoids treated with TNFα displayed a high proportion of TUNEL+ cells relative to CC3+ cells; TUNEL and CC3 staining were both low in TNFα-treated Atg16L1f/f organoids (Fig. 5, A and B). Rather than improve viability, the apoptosis (pan-caspase) inhibitor Z-VAD-FMK exacerbated cell death in Atg16L1ΔIEC organoids (Fig. 5, C and D). Cells displaying morphology consistent with necrotic cell death, such as discontinuous plasma membrane and swelling of organelles, were detected in Atg16L1ΔIEC organoids treated with Z-VAD-FMK by transmission electron microscopy (TEM) analyses (Fig. 5 E and Fig. S3 A). These results suggest that ATG16L1 deficiency increases susceptibility to nonapoptotic cell death.
Figure 5.
ATG16L1 inhibits necroptosis in intestinal organoids. (A and B) Representative images and quantification of TUNEL+ (A) and CC3+ (B) cells per organoid on day 5 normalized to total IEC. TNFα was added on day 4. At least 20 organoids were quantified from three mice each. (C and D) Quantification of viability (C) and representative images of organoids (D) from Atg16L1f/f and Atg16L1ΔIEC mice on day 5 ± Z-VAD-FMK (Z-VAD) and Nec-1. (E) Representative TEM images of organoids on day 3. Z-VAD was added on day 0. Stars indicate lumen side of organoid, and arrows indicate Paneth cell. (F and G) Quantification of viability (F) and representative images of organoids (G) from Atg16L1f/f and Atg16L1ΔIEC mice on day 5 ± TNFα and Nec-1. (H and I) Viability of Atg16L1ΔIEC organoids transduced with lentiviruses encoding shRNAs targeting Ripk3 (H) and Mlkl (I) or a nonspecific control, ±TNFα. (A, B, D, E, and G) Bars: 50 µm (A and B); 100 µm (D and G); 2 µm (E). Data points in A and B represent individual organoids, and data points in C, F, H, and I are mean of three technical replicates. Bars represent mean ± SEM, and at least two independent experiments were performed. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by one-way ANOVA and Tukey’s test in A and B and unpaired t test in C, F, H, and I.
In contrast to pyroptosis mediated by caspase-1, Z-VAD-FMK promotes necroptosis by preventing caspase-8 from functioning as a negative regulator of RIPK1/3 signaling (Hitomi et al., 2008; Kaiser et al., 2011; Oberst et al., 2011). We found that decreased viability of Atg16L1ΔIEC organoids treated with Z-VAD-FMK was reversed by the RIPK1 inhibitor Necrostatin-1 (Nec-1; Degterev et al., 2005; Fig. 5 C). TNFα-induced cell death in Atg16L1ΔIEC organoids was also rescued by Nec-1 (Fig. 5, F and G). We next examined the effect of Ripk3 and Mlkl knockdown (KD) using two different hairpins per target gene introduced by lentiviral transduction. Wnt-3a was added to the culture media to facilitate the lentiviral transduction procedure, which we confirmed did not alter the effect of TNFα on Atg16L1ΔIEC organoids (Fig. S3 B). Atg16L1ΔIEC organoids transduced with the Ripk3 or Mlkl shRNAs, but not control shRNA, displayed improved survival in the presence of TNFα (Fig. 5, H and I; and Fig. S3, C–F). These results indicate that TNFα induces necroptosis in Atg16L1ΔIEC organoids.
ATG16L1 protects intestinal organoids from necroptosis by maintaining mitochondrial homeostasis
Autophagy has been shown to mediate the degradation of caspase-8 to inhibit TRAIL-mediated apoptosis in a colorectal cancer cell line (Hou et al., 2010). However, we did not detect a convincing difference in RIPK1, RIPK3, MLKL, or caspase-8 levels when comparing Atg16L1f/f and Atg16L1ΔIEC organoids (Fig. S4 A). Instead, we detected an increase in phosphorylated-RIPK3 (p-RIP3) and p-MLKL in Atg16L1ΔIEC organoids after TNFα treatment (Fig. S4 A). These results are consistent with a model in which ATG16L1 prevents enhanced signaling rather than degradation of the RIPK3/MLKL complex. The limited availability of protein that can be harvested from organoids precluded biochemical analyses of additional posttranslational modifications or protein interactions, and thus, we pursued other potential mechanisms that can explain the role of ATG16L in epithelial viability.
Removal of damaged mitochondria is a well-established function of autophagy (Randow and Youle, 2014). Therefore, we examined mitochondria in the ultrastructural images from Fig. 5 E. In these analyses, we did not discriminate between different cell types because there was an insufficient number of Paneth cells in Atg16L1ΔIEC for quantification. Atg16L1ΔIEC epithelial cells contained a high proportion of morphologically aberrant mitochondria that are swollen and missing cristae, which was exacerbated by the addition of Z-VAD-FMK (Fig. 6 A). Atg16L1ΔIEC organoids also contained a higher degree of staining with the fluorescent mitochondrial superoxide indicator MitoSOX in their epithelium, especially after treatment with TNFα (Fig. 6 B). To determine the contribution of elevated ROS to loss in viability, we treated organoids with the antioxidant N-acetyl-l-cysteine (NAC). NAC reduced MitoSOX staining and improved survival of TNFα-treated Atg16L1ΔIEC organoids and even improved survival of unstimulated Atg16L1ΔIEC organoids (Fig. 6, B and C). These results suggest that mitochondrial ROS accumulation contributes to the impaired survival of Atg16L1ΔIEC organoids.
Figure 6.
ATG16L1 protects intestinal organoids from necroptosis by maintaining mitochondrial homeostasis. (A) Representative TEM images of mitochondria classified into type 1 (cristae are maintained), type 2 (mildly swollen and a moderate decrease in the number of visible cristae), and type 3 (severely swollen and >70% of cristae are missing, or highly dysmorphic and electron dense). At least 20 cells in each organoid were analyzed per group from three mice each. (B) Representative fluorescent images of organoids on day 3 treated ± TNFα and NAC and stained with MitoSOX Red. Quantification of MitoSOX mean fluorescence intensity (MFI) in the epithelium shown. Staining in the lumen was excluded. At least 20 organoids were quantified from three mice each. (C–E) Viability of organoids from Atg16L1f/f and Atg16L1ΔIEC mice treated ± TNFα and NAC (C), B6 and Rip3−/− mice treated ± TNFα and CCCP (D), and B6 and Park2−/− mice treated ± TNFα and Nec-1 (E). n = 3 mice each. (A and B) Bars: 0.5 µm (A); 50 µm (B). Data points in B represent individual organoids, and data points in C–E are mean of three technical replicates. Bars represent mean ± SEM, and at least two independent experiments were performed. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by one-way ANOVA and Tukey’s test in B and unpaired t test in C–E.
Next, we examined whether disruption of mitochondria is sufficient to confer sensitivity to necroptosis. WT B6 organoids treated with the mitochondrial uncoupler carbonyl cyanide m-chlorophenylhydrazone (CCCP) displayed impaired survival that was exacerbated by TNFα (Fig. 6 D). In contrast, organoids derived from Ripk3−/− mice displayed a modest decrease in viability only after 4 d of adding CCCP and TNFα in the culture (Fig. 6 D). We next examined the effect of Parkin (Park2) deletion on organoid survival. Parkin is an E3 ligase that ubiquitinates mitochondrial outer membrane proteins to mediate selective degradation through autophagy (Randow and Youle, 2014). Park2−/− organoids were susceptible to TNFα-induced death, which was rescued by Nec-1 addition (Fig. 6 E). Additionally, knocking down another autophagy gene, Atg7, also led to impaired survival in the presence of TNFα, which was rescued by Nec-1 (Fig. S4, B and C). These data indicate that genetic or chemical disruption of autophagy and mitochondrial homeostasis can confer susceptibility to necroptosis in IECs.
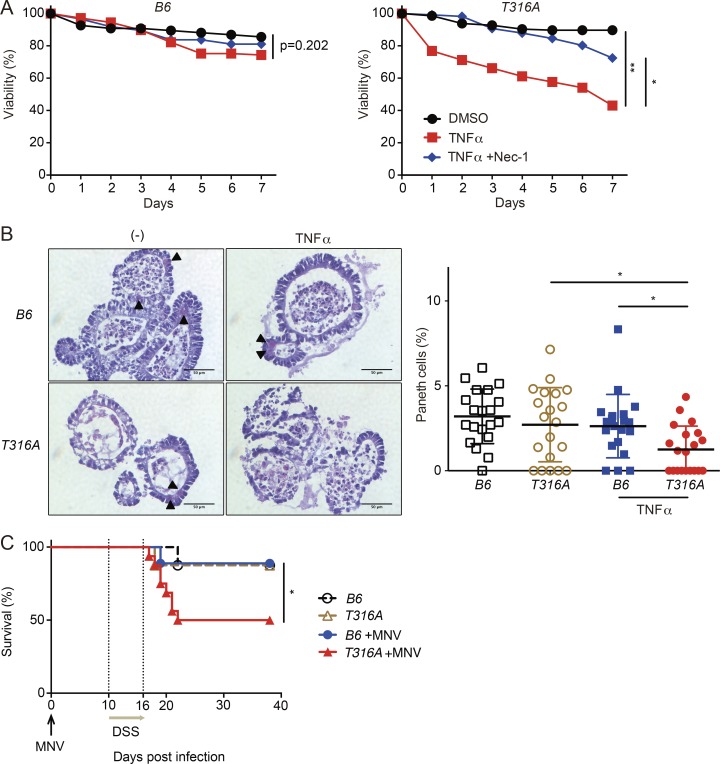
To determine whether the disease variant of ATG16L1 confers sensitivity to cell death, we examined organoids generated from Atg16L1T316A knock-in mice harboring the murine equivalent of the human risk allele (Murthy et al., 2014). Atg16L1T316A organoids treated with TNFα displayed a loss in viability that was prevented by Nec-1 (Fig. 7 A). In addition, TNFα treatment caused a decrease in Paneth cells in Atg16L1T316A organoids that was not observed in the WT B6 controls (Fig. 7 B). Moreover, Atg16L1T316A mice were susceptible to lethality after MNV+DSS treatment (Fig. 7 C). These data support the relevance of our model to genetic susceptibility underlying Crohn’s disease.
Figure 7.
The ATG16L1 risk variant is associated with loss of intestinal organoid viability and virally triggered disease. (A and B) Viability of organoids from B6 and Atg16L1T316A (T316A) mice treated ± TNFα and Nec-1. n = 3 mice each (A). Representative H&E images and quantification of Paneth cells per organoid on day 7 normalized to total IECs (B). TNFα was added on day 0. Arrowheads indicate Paneth cells. At least 20 organoids were analyzed from three mice each. Bars, 50 µm. (C) Survival of B6 and Atg16L1T316A (T316A) mice treated with 5% DSS ± MNV infection. n = 8 (f/f), 8 (T316A), 9 (f/f + MNV), and 16 (T316A + MNV). Data points in A are mean of three technical replicates, and data points in B represent individual organoids. Bars represent mean ± SEM, and at least two independent experiments were performed. *, P < 0.05; **, P < 0.01 by unpaired t test in A, one-way ANOVA and Tukey’s test in B, and Mantel-Cox in C.
TNFα and RIPK1 inhibition ameliorates disease in ATG16L1-deficient mice
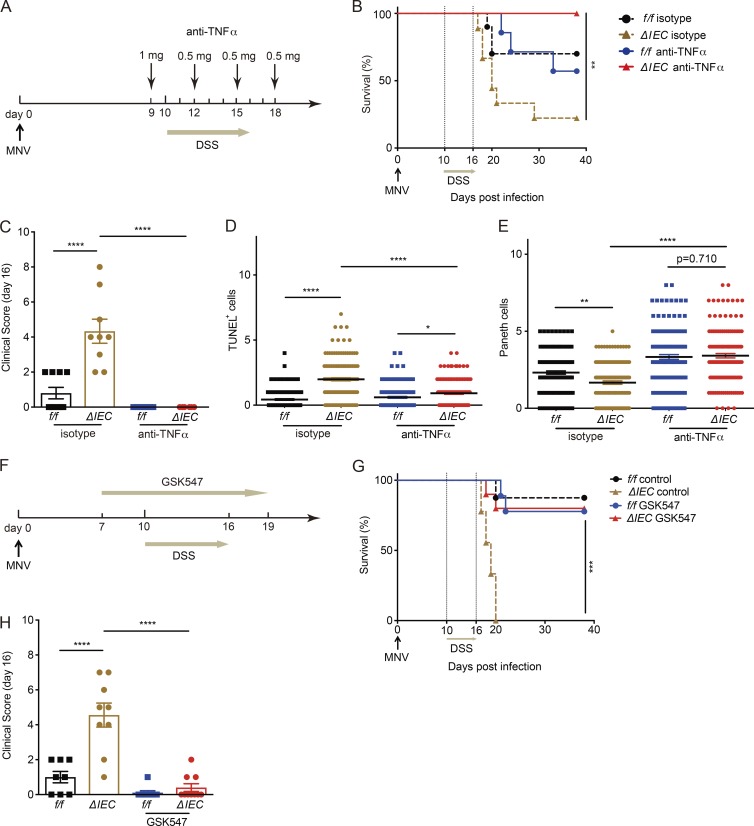
We examined whether anti-TNFα antibody or RIPK1 inhibition prevent cell death in vivo and ameliorate disease. Consistent with our hypothesis, administration of TNFα-blocking antibodies led to 100% survival of Atg16L1ΔIEC mice receiving MNV+DSS, eliminated all signs of morbidity, reduced cell death, and restored Paneth cell numbers (Fig. 8, A–E). This finding likely reflects a function of RIPK1 signaling downstream of TNFα because administration of the RIPK1 inhibitor Nec-1s (Takahashi et al., 2012) reduced TUNEL+ cells in the crypts of Atg16L1ΔIEC mice and restored Paneth cell numbers to levels similar to Atg16L1f/f mice (Fig. S5, A and B). To determine whether RIPK1 represents a target for therapeutic intervention, we tested a newly generated necroptosis inhibitor with improved specificity for RIPK1, GSK547. We confirmed the efficacy of GSK547 in blocking necroptosis in vitro (Fig. S5 C) and found that it rescues survival of Atg16L1ΔIEC mice treated with MNV+DSS (Fig. 8, F–H). These data indicate that ATG16L1 has a critical cytoprotective function in vitro and in vivo in the presence of TNFα.
Figure 8.
TNFα and RIPK1 inhibition ameliorates disease in ATG16L1-deficient mice. (A) Schematic representation of anti-TNFα treatment in MNV+DSS model. (B and C) Survival of Atg16L1f/f (f/f) and Atg16L1ΔIEC (ΔIEC) mice treated with anti-TNFα or isotype control antibodies were monitored for survival (B) and clinical disease score (C) on day 16. n = 7 (f/f, isotype), 9 (ΔIEC, isotype), 7 (f/f, anti-TNF), and 10 (ΔIEC, anti-TNF). (D and E) Number of TUNEL+ (D) and Paneth (E) cells per crypt on day 15 in mice treated ± anti-TNFα. At least 50 crypts were quantified from three mice per group. (F) Schematic representation of GSK547 treatment in MNV+DSS model. (G and H) Atg16L1f/f and Atg16L1ΔIEC mice treated with either GSK547 or control chow were monitored for survival (G) and clinical disease score on day 16 (H). n = 8 (f/f, control), 9 (ΔIEC, control), 9 (f/f, GSK547), and 10 (ΔIEC, GSK547). Data points in C and H represent individual mice, and data points in D and E represent individual crypts. Bars represent mean ± SEM, and at least two independent experiments were performed. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by Mantel-Cox in B and G and one-way ANOVA and Tukey’s test in C–E and H.
Discussion
We found that Atg16L1 deletion in IECs is sufficient to exacerbate disease in models of IBD and GVHD and is associated with a defect in epithelial turnover marked by a reduction in Paneth cell numbers. In the IBD model, disease was dependent on MNV, likely reflecting the as yet poorly understood immune response to this intestinal virus, which includes production of TNFα and other cytokines (McCartney et al., 2008; Kim et al., 2011; Fang et al., 2013). The dependence of MNV is a critical aspect of our findings given our previous observation that MNV is an otherwise beneficial virus (Kernbauer et al., 2014). How animal viruses affect host physiology beyond their roles as pathogens remains obscure, and our knowledge of the long-term impact of intestinal viruses is far behind that of symbiotic bacteria (Cadwell, 2015). Our results support the paradigm that context, such as host genotype, determine whether a given infectious agent has a beneficial or adverse effect on the host.
The cytoprotective function of ATG16L1 was reproduced remarkably well in intestinal organoids, allowing us to gain insight into the underlying cell biological basis of our observations. Several findings in this in vitro model indicate that ATG16L1 is blocking necroptosis rather than apoptosis or pyroptosis: death of ATG16L1-deficient organoids is exacerbated by Z-VAD-FMK, loss of viability occurs in the absence of caspase-3 cleavage, and survival is rescued by blocking RIPK1, RIPK3, or MLKL. Consistent with these observations, blocking TNFα or RIPK1 in vivo in MNV+DSS-treated ATG16L1-deficient mice reduces epithelial cell death, restores Paneth cell numbers, and ameliorates disease. Additional mechanistic experiments are required to characterize the in vivo cell death modality and the specific role of Paneth cells. Other groups have shown that depletion of Paneth cells can occur in a manner dependent on RIPK3, and that Crohn’s disease patients display RIPK3+ Paneth cells in the small intestine (Günther et al., 2011; Simmons et al., 2016). Our observation that organoids and mice harboring the Atg16L1 risk variant reproduce observations made with complete Atg16L1 deficiency provides a strong rationale for pursuing future in vivo experiments that link our findings to these observations in the literature and offers an opportunity to identify pathogenesis events that are specific to this genotype as a way to segregate patients.
The link between necroptosis in ATG16L1-deficient organoids and mitochondrial ROS is supported by a recent study showing that Pink1 deleted fibroblasts, which are deficient in mitophagy, display increased ROS and susceptibility to Z-VAD-FMK–induced cell death (Lu et al., 2016). However, mitochondria are dispensable for necroptosis in T cells treated with Z-VAD-FMK and TNFα (Tait et al., 2013). A critical difference between our system and many of the other experimental models is that we can induce necroptosis with TNFα in the absence of Z-VAD-FMK; other studies investigating necroptosis typically use caspase-8 inhibition to shunt the cell death pathway away from apoptosis. It remains possible that when caspase-8 is intact, mitochondrial ROS contribute to necroptosis by altering signaling. ROS and TNFα signaling are known to have a complex bidirectional relationship, and ROS can alter activation of molecules without affecting their total levels (Blaser et al., 2016). This potential mechanism would be consistent with findings demonstrating that mitochondria do not function downstream of MLKL during necroptosis (Wang et al., 2012; Murphy et al., 2013; Remijsen et al., 2014). Also, it is unlikely that TNFα functions alone in vivo. Interferons (both type I and IFN-γ) are notable because they are associated with viral infections, display significant crosstalk with autophagy, and trigger necroptosis (Pasparakis and Vandenabeele, 2015; Cadwell, 2016). Animal models of GVHD indicate that interferons can either promote or ameliorate inflammation (Blazar et al., 2012; Fischer et al., 2017). We suggest that organoid cultures can help deconvolute the intersection between these key inflammatory cytokines and ROS activity in the epithelium.
Our findings also contrast with studies demonstrating a role for autophagy in promoting necroptosis in cancer cells. In prostate cells in which autophagy promotes RIPK1-MLKL interaction and necroptosis, deletion of the tumor suppressor Map3k7 is necessary for sensitization to necroptosis (Goodall et al., 2016), suggesting that autophagy may have a distinct role in cell viability during tumorigenesis. To our knowledge, we are the first to investigate this pathway in a primary 3D culture model, which may explain why we found that autophagy has an inhibitory effect on necroptosis.
Although we found that Atg7 KD or Park2 deletion renders organoids susceptible to TNFα-induced necroptosis, it remains possible that ATG16L1 has functions outside of mitophagy that contribute to cell death. Recent studies have provided compelling evidence that intestinal inflammation occurs when IRE1α mediates unresolved ER stress downstream of ATG16L1 deletion in Paneth cells (Adolph et al., 2013; Diamanti et al., 2017; Tschurtschenthaler et al., 2017). The findings presented here indicate that disruption in mitochondrial homeostasis in Paneth cells is another deleterious consequence of ATG16L1 deficiency, which likely synergizes with ER stress. ER stress has been implicated in necroptosis (Saveljeva et al., 2015). Also, with the emergence of unconventional forms of autophagy (i.e., autophagy-related pathways), it will be important to consider functions of ATG16L1 that are independent of known roles of autophagy. Elucidating the detailed molecular intersection between ATG16L1 and necroptosis in IECs will be an important future direction.
Our study establishes an intimate relationship between the ATG16L1 risk allele, necrotic cell death, and Paneth cell dysfunction, all of which have been independently linked to IBD. Additionally, our findings indicate that this three-way relationship may apply to GVHD as well. Improvements in the specificity of necroptosis inhibitors will allow us to test the efficacy of therapeutically targeting this pathway in these disease models and may ultimately lead to new treatment options in patients.
Materials and methods
Mice
Age- and gender-matched 6–12-wk-old mice on the C57BL/6J (B6) background were used. Atg16L1f/f;villinCre (Atg16L1ΔIEC) and littermate control Atg16L1f/f mice were generated for experiments by crossing Cre-positive and Cre-negative mice provided by S. Virgin (Washington University School of Medicine, St. Louis, MO). B6, B10.BR, and Park2−/− mice were purchased from The Jackson Laboratory and bred onsite to generate animals for experimentation. RIP3−/− mice were provided by G. Miller (NYU School of Medicine, New York, NY). Atg16L1T316A mice were provided by M. van Lookeren Campaigne (Genentech). All animal studies were performed according to approved protocols by the New York University School of Medicine and Memorial Sloan Kettering Cancer Center Institutional Animal Care and Use Committees.
Virally triggered disease model
MNV CR6 concentrated stock was prepared as described (Cadwell et al., 2010; Kernbauer et al., 2014). Mice were infected orally by pipette with 3 × 106 PFUs resuspended in 25 µl PBS. Plaque assay was used to quantify viral burden in 2 cm of ileum or colon as previously described (Kernbauer et al., 2014). On day 10 postinfection, mice were administered 5% DSS (TdB Consultancy) in their drinking water for 6 d, which was replaced by regular drinking water for the remainder of the experiment. Mice were injected i.p. with 1 mg of either anti-TNFα (XT3.1) or IgG1 isotype control (HRPN) antibodies (Bio X Cell) 1 d before DSS treatment and received subsequent injections of 0.5 mg every 3 d. Mice were fed with chow containing 0.833 g/kg GSK547 from day 7 to day 19 or injected i.p. with Nec-1s (4 mg/kg; BioVision) or 7% DMSO (Fisher BioReagents) every day from day 9 to day 14. Clinical disease score was quantified on the basis of five parameters, including diarrhea (0–2), hunched posture (0–2), fur ruffling (0 or 1), mobility (0–2), and blood stool (0–1), in which eight was the maximum score for the pathology.
Hematopoietic stem cell transplantation
Allo-HSCTs were performed as previously described (Hubbard-Lucey et al., 2014) with 1,100-cGy split-dosed irradiation of recipients transplanted with 5 × 106 BM cells after T cell depletion with anti-Thy-1.2 and low-TOX-M rabbit complement (Cedarlane Laboratories). Donor T cells were prepared by harvesting splenocytes and enriching T cells by Miltenyi MACS purification of CD5 (routinely >90% purity). Recipient mice were monitored daily for survival, and clinical GVHD was quantified at the time of sacrificing mice for five clinical parameters (weight loss, hunched posture, activity, fur ruffling, and skin lesions) on a scale from 0 to 2. A clinical GVHD score was generated by the summation of the five criteria as previously described (Cooke et al., 1998).
Intestinal organoids
Small intestinal crypts were isolated and cultured as described previously (Sato et al., 2009). In brief, crypts of proximal small intestine were counted and embedded in 30 µl of Matrigel (Corning) at 10,000 crypts/ml and cultured in DMEM/F-12 (Thermo Fisher Scientific) in the presence of 100 IU penicillin and 100 µg/ml streptomycin (Corning), 125 µg/ml gentamicin (Gibco), 2 mM l-glutamine (Corning), 20 ng/ml mEGF (PeproTech), 100 ng/ml mNoggin (R&D Systems), and 500 ng/ml mR-Spondin 1 (R&D Systems), hereafter referred as ENR medium. Surface area of organoids was quantified with ImageJ (National Institutes of Health). Colonic crypts were isolated and cultured as described previously (Sato et al., 2011; Yui et al., 2012). Crypts of colon were counted and embedded in Matrigel and cultured in the ENR medium plus 50 ng/ml Wnt-3a, 1× B-27, and N-2 supplement (Thermo Fisher Scientific). For viability assays, crypts were embedded in 10 µl Matrigel at 5,000 crypts/ml and cultured in 96-well culture plate in triplicate with or without 50 µM Z-VAD-FMK (ApexBio), 20 µM Nec-1 (Sigma-Aldrich), 20 ng/ml mTNFα (PeproTech), 20 µg/ml anti-TNFα antibody (R&D Systems), 100 µM NAC (Sigma-Aldrich), and 400 nM GSK547 (GlaxoSmithKline). For CCCP treatment, organoids were prepared from small intesitnal crypts in a manner similar to other experiments except on day 5 were mechanically dissociated into crypt domains by pipetting and cultured with or without 5 µM CCCP (Sigma-Aldrich) and 20 ng/ml mTNFα. Percentage viable organoids was determined by daily quantification of the number of intact organoids. Opaque organoids with condensed structures or those that have lost adherence were excluded. Dead organoids were marked by staining with 100 µg/ml PI (Sigma-Aldrich) and 100 µg/ml Hoechst 33342 (Invitrogen). LDH release was measured by harvesting supernatant from organoid cultures on day 2 and using CytoTox-ONE Homogeneous Membrane Integrity Assay (Promega) according to the manufacturer’s protocol. Mitochondrial ROS were measured by staining day 3 organoids with MitoSOX Red for live-cell imaging (Thermo Fisher Scientific) according to the manufacturer’s protocol. The mean fluorescence intensity in the epithelium of at least 20 organoids were analyzed using ImageJ software (National Institutes of Health).
Histology and immunohistochemistry
Quantification of all microscopy data were performed blind. Intestinal sections were prepared and stained with H&E and periodic acid–Schiff/Alcian blue as previously described (Cadwell et al., 2010; Kernbauer et al., 2014). Villus height was quantified for at least 50 villi per mouse in ileal sections that were cut perpendicular to the villus-crypt axis as defined by the presence of a visible crypt lumen from the orifice to the base of the crypt, thereby minimizing tissue orientation-based artifacts (Cadwell et al., 2010). Colon histopathology was scored by Y.D. by grading semiquantitatively as 0 (no change) to 4 (most severe) for the following inflammatory lesions: severity of chronic inflammation, crypt abscess, and granulomatous inflammation; and for the following epithelial lesions: hyperplasia, mucin depletion, ulceration, and crypt loss (Leach et al., 1996). Intestinal GVHD was scored by C.L. as previously described (Hubbard-Lucey et al., 2014). For immunohistochemistry, formalin fixed paraffin-embedded sections were deparaffinized online and antigen retrieved in Ventana Cell Conditioner 2 (Citrate). Endogenous peroxidase activity was blocked with hydrogen peroxide. Unconjugated polyclonal rabbit anti-mouse CC3 (asp-175; Cell Signaling Technology) and unconjugated rabbit anti-mouse Ki67 (SP7; Lab Vision) were used, and both Ki-67 and CC3 were detected with anti-rabbit, HRP-conjugated multimer and visualized with 3,3-diaminobenzidene and enhanced with copper sulfate. TUNEL staining was performed using the In situ Cell Death Detection POD kit (Roche Diagnostics) on the Discovery XT according to the manufacturer’s protocol. Appropriate positive and negative controls were run in parallel to study sections. At least 50 crypts per mouse were observed to count CC3+, Ki-67+ cells, and TUNEL+ cells. For intestinal organoids, frozen sections were prepared by fixing with 4% paraformaldehyde (Electro Microscopy Sciences) and cryoprotecting with 20% sucrose (Sigma-Aldrich). Fixed organoids were mixed with NEG-50 (Thermo Fisher Scientific) and frozen in cryomold with 2-methylbutane (Themo Fisher Scientific) cooled by dry ice. 5-µm sections were made by Cryostat (Micron HM350; Thermo Fisher Scientific). The sections were air-dried overnight at room temperature and then stained with anti–Lysozyme C (C-19; Santa Cruz) and followed by the secondary Alexa Fluor 594 (Invitrogen) antibody. At least 20 organoids were observed per sample to count TUNEL+, CC3+, Paneth, and goblet cells in organoids. Histology and immunohistochemistry samples were analyzed using Zeiss Axioplan with Spot camera. Bright-field/fluorescent images and video imaging of live organoids were performed using a Zeiss AxioObserver.Z1 with EC Plan-Neofluar 10×/0.30 lens, FL Filter Set 43 HE Cy3, and Axiocam 503 Mono operated with Zen Blue software. For live video imaging, 20 ng/ml mTNFα and 100 µg/ml PI were added before imaging, and time-lapse images of multiple fields were collected at 5-min intervals over 19 h. Images were processed and quantified using ImageJ.
Immunoblotting
Small intestinal organoids at day 3 were stimulated with 20 ng/ml mTNFα for 2 h, then organoids were incubated in lysis buffer (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% Triton X-100, 10% glycerol, and 1× Halt protease and phosphatase inhibitor cocktail; Thermo Fisher Scientific) on ice for 20 min and centrifuged at 14,000 g for 20 min. Samples were resolved on Bolt 4%–12% Bis-Tris Plus Gels (Invitrogen) and transferred to polyvinylidene fluoride membranes. The following antibodies were used for immunoblotting studies: anti–β-actin (AC-15; Sigma-Aldrich), anti–Phospho-MLKL (Ser345) and anti-MLKL (Cell Signaling), anti-RIP3 (phospho S232; Abcam), anti-RIP3 (AHP1797; AbD Serotec), anti–caspase-8 (1G12; Enzo Life Sciences), anti-RIP (D94C12; Cell Signaling Technology), and anti-Atg16L (Sigma-Aldrich). Secondary antibodies (mouse anti–rabbit [211-032-171], goat anti–mouse [115-035-174], and goat anti–rat [112-035-175], respectively) were purchased from Jackson Laboratories, and IRDye 680RD goat anti–rabbit IgG (925-68071) and IRDye 800CW goat anti–mouse IgG (925-32210) were purchased from LI-COR.
Flow cytometry and cytokine analyses
Antibodies were obtained from BD Biosciences PharMingen (CD8 and CD11c), and BioLegend (CD4, CD11b, B220, and I-AE). Cells were stained for 20 min at 4°C in PBS with 0.5% BSA (PBS/BSA) after Fc block, washed, and resuspended in DAPI (Sigma-Aldrich) in PBS/BSA to exclude dead cells. Flow cytometry was performed on an LSR II (BD Biosciences) and analyzed with FlowJo (Tree Star Software). For cytokine quantification, blood was collected into microcentrifuge tubes, allowed to clot, and centrifuged, and the serum was collected. ProcartaPlex Multiplex Immunoassay was conducted per the manufacturer’s instructions (Affymetrix). Results were acquired with a Luminex 200 instrument and analyzed with xPONENT software (Luminex Corporation).
Lentivirus infection and gene KD
RIPK3 shRNA (TRCN0000022536 and TRCN0000022534), MLKL shRNA (TRCN0000022602 and TRCN0000022599), ATG7 shRNA (TRCN0000375444), and nontarget shRNA control (SHC016) constructs were purchased from Sigma-Aldrich. Each lentiviral construct along with lentiviral packaging mix (pLP1, pLP2, and VSVG; Sigma-Aldrich) was cotransfected into 293FT cells using Lipofectamine 3000 (Thermo Fisher Scientific) according to the manufacturer’s protocol, then the supernatant containing lentivirus was harvested and concentrated using Lenti-X concentrator (Clontech). The transduction procedure was adapted from a previously described method (Koo et al., 2011). In brief, organoids from Atg16L1ΔIEC mice were cultured in antibiotic-free ENR medium for 5 d, and at day 3 the medium was changed to ENR plus 50 ng/ml Wnt-3a (R&D Systems) and 10 mM nicotinamide (Sigma-Aldrich). The organoids were mechanically dissociated into single cells by pipetting, and the fragments were incubated with TrypLE Express (Gibco) for 5 min at 37°C and centrifuged at 1000 g for 5 min. The cell clusters were combined with viral suspension containing the ENR growth factors and 50 ng/ml Wnt-3a, 10 mM nicotinamide, 10 µM Y-27632 dihydrochloride (Sigma-Aldrich), and 8 µg/ml Polybrene (Millipore). The cells were transferred into 48-well culture plate, and the plate was centrifuged at 600 g at 32°C for 60 min. Finally, the cells were embedded in 30 µl of Matrigel and cultured in 24-well plate in antibiotic-free ENR medium containing Wnt-3a, nicotinamide, and Y-27632 dihydrochloride. 2 d after infection the medium was changed to ENR plus Wnt-3a with or without mTNFα.
Transmission electron microscopy
Intestinal organoids at day 3 were fixed with 2.5% glutaraldehyde, 2% paraformaldehyde in 0.1 M sodium cacodylate buffer, pH 7.2, for 2 h and postfixed with 1% osmium tetroxide for 1.5 h at room temperature, then processed in a standard manner and embedded in EMbed 812 (Electron Microscopy Sciences). Semithin sections were cut at 1 mm and stained with 1% toluidine blue to find the structure of interests. Then ultrathin sections (60 nm) were cut, mounted on 200-mesh copper grids, and stained with uranyl acetate and lead citrate. Stained grids were examined under Philips CM-12 electron microscope and photographed with a Gatan (4k × 2.7k) digital camera. To quantify mitochondria, at least 20 epithelial cells in each organoid were observed, and mitochondria were classified as type 1 (mitochondria whose cristae are maintained), type 2 (mildly swollen and a moderate decrease in the number of visible cristae), and type 3 (severely swollen and >70% of cristae are missing, or highly dysmorphic electron dense mitochondria).
Quantitative RT-PCR
Total RNA was extracted from organoids at day 2 after lentiviral transduction or at day 3 after crypt dissemination for Lgr5 with TRIzol Reagent (Invitrogen) and cDNA was synthesized using ProtoScript First Strand cDNA Synthesis kit (NEB) according to the manufacturer’s protocol. Quantitative PCR was performed on a Roche 480 II LightCycler. Gene expression was normalized to Gapdh using the following primers: Ripk3, 5′-AGGCTTCTAAAGCGAGTGATGT-3′ and 5′-TGAAGTCTTGTCTACCAACTCAGC-3′; Atg7, 5′-CAGTTTCCAGTCCGTTGAAGTCCT-3′ and 5′-GGGTCCATACATCCACTGAGGTTC-3′; Lgr5, 5′-CCTACTCGAAGACTTACCCAG-3′ and 5′-GCATTGGGGTGAATGATAGCA-3′; and Gapdh, 5′-TGGCCTTCCGTGTTCCTAC-3′ and 5′-GAGTTGCTGTTGAAGTCGCA-3′.
Statistical analysis
GraphPad Prism version 7 was used for statistical analysis. Differences between two groups were assessed using two-tailed unpaired Student’s t test when data were distributed normally; otherwise Mann-Whitney U test was used. An ANOVA with Tukey’s multiple-comparisons test was used to evaluate experiments involving multiple groups. Survival was analyzed using the Mantel-Cox log-rank test.
Online supplemental material
Fig. S1 shows additional analyses of Atg16L1ΔIEC mice subjected to MNV+DSS or allo-HSCT and is related to Figs. 1 and 2. Fig. S2 shows the effect of ATG16L1 deletion on small intestinal stem cells and colonic organoids and representative pictures of PI staining and is related to Fig. 4. Fig. S3 shows additional analyses of the effect of Z-VAD and TNFα on Atg16L1ΔIEC organoids and the efficacy of Ripk3 and Mlkl KD and is related to Fig. 5. Fig. S4 shows Western blotting analyses of necroptosis signaling molecules in organoids and quantification of cell death in ATG7-deficient organoids and is related to Fig. 6. Fig. S5 shows quantification of cell death and Paneth cells in mice treated with Nec-1s and in vitro efficacy of GSK547 and is related to Fig. 8. Video 1 shows live imaging analysis of organoids treated with TNFα and is related to Fig. 4.
Supplementary Material
Acknowledgments
We would like to thank Hiromichi Shimizu (University of California, San Francisco), Nobukatsu Horita (University of Michigan), Deepshika Ramanan (NYU School of Medicine), Eugene Rudensky (NYU School of Medicine), the NYUSoM Microcopy Core (NIH RR023704), the NYUSoM Histopathology Core and Immunohistochemistry Core, the NYUSoM Flow Cytometry and Cell Sorting Center (NIH 5P30CA016087-33), and the Laboratory of Comparative Pathology and the Molecular Cytology Core and the Small-Animal Imaging Core Facility (NIH P30 CA008748) for advice and technical support. Also, we would like to thank Menno van Lookeren Campaigne (Genentech) for providing Atg16L1T316A mice.
This work was supported by National Institutes of Health grants R01 HL123340 (K. Cadwell, M.R.M. van den Brink), R01 DK093668 (K. Cadwell), R01 DK103788 (K. Cadwell), R01 AI121244 (K. Cadwell), R01 HL069929 (M.R.M. van den Brink), R01 AI100288 (M.R.M. van den Brink), R01 AI080455 (M.R.M. van den Brink), and R01 AI101406 (M.R.M. van den Brink); a Faculty Scholar grant from the Howard Hughes Medical Institute, the Stony Wold-Herbert Fund, and philanthropy from Bernard Levine (K. Cadwell); the Radiation Effects Research Foundation–National Institute of Allergy and Infectious Diseases, Experimental Therapeutics Center of Memorial Sloan Kettering Cancer Center funded by W. H. Goodwin and A. Goodwin, the Lymphoma Foundation, Alex’s Lemonade Stand Foundation, the Geoffrey Beene Cancer Research Center at Memorial Sloan Kettering Cancer Center, and the Susan and Peter Solomon Divisional Genomics Program (M.R.M. van den Brink); the Uehara Memorial Foundation and the Mochida Memorial Foundation for Medical and Pharmaceutical Research (Y. Matsuzawa-Ishimoto); and the American Society for Blood and Marrow Transplantation (Y. Shono). K. Cadwell is a Burroughs Wellcome Fund Investigator in the Pathogenesis of Infectious Diseases. This research was also supported by the Parker Institute for Cancer Immunotherapy at Memorial Sloan Kettering Cancer Center.
The authors declare no competing financial interests.
Author contributions: Y. Matsuzawa-Ishimoto, Y. Shono, M.R.M. van den Brink, and K. Cadwell formulated the original hypothesis, designed the study, and analyzed the results. Y. Matsuzawa-Ishimoto performed the MNV+DSS and organoid experiments, and Y. Shono performed the allo-HSCT experiments. L.E. Gomez and V.M. Hubbard-Lucey contributed to MNV+DSS and allo-HSCT experiments. J. Neil contributed to virus quantification analysis. M.Z. Dewan provided technical assistance in preparing frozen sections of intestinal organoids. M. Cammer provided technical assistance in microscopy analysis. S.R. Lieberman and A. Lazrak provided assistance in allo-HSCT experiments. J.M. Marinis, A. Beal, P.A. Harris, and J. Bertin contributed to experiments involving GSK547. C. Liu and Y. Ding performed histopathology analyses. Y. Matsuzawa-Ishimoto, Y. Shono, M.R.M. van den Brink, and K. Cadwell wrote the manuscript, and all authors commented on the manuscript, data, and conclusions.
Footnotes
Abbreviations used:
- allo-HSCT
- allogeneic hematopoietic stem cell transplantation
- CCCP
- carbonyl cyanide m-chlorophenylhydrazone
- DSS
- dextran sodium sulfate
- GVHD
- graft-versus-host disease
- HM
- hypomorph
- IBD
- inflammatory bowel disease
- IEC
- intestinal epithelial cell
- KD
- knockdown
- LDH
- lactate dehydrogenase
- MLKL
- mixed-lineage kinase domain–like protein
- MNV
- murine norovirus
- NAC
- N-acetyl-l-cysteine
- PI
- propidium iodide
- TEM
- transmission electron microscopy
- TUNEL
- terminal deoxynucleotidyl transferase–mediated dUTP nick-end labeling
References
- Adolph T.E., Tomczak M.F., Niederreiter L., Ko H.J., Böck J., Martinez-Naves E., Glickman J.N., Tschurtschenthaler M., Hartwig J., Hosomi S., et al. . 2013. Paneth cells as a site of origin for intestinal inflammation. Nature. 503:272–276. 10.1038/nature12599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin J.L., Sumpter R. Jr., Levine B., and Hooper L.V.. 2013. Intestinal epithelial autophagy is essential for host defense against invasive bacteria. Cell Host Microbe. 13:723–734. 10.1016/j.chom.2013.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry D.L., and Baehrecke E.H.. 2007. Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell. 131:1137–1148. 10.1016/j.cell.2007.10.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaser H., Dostert C., Mak T.W., and Brenner D.. 2016. TNF and ROS crosstalk in inflammation. Trends Cell Biol. 26:249–261. 10.1016/j.tcb.2015.12.002 [DOI] [PubMed] [Google Scholar]
- Blazar B.R., Murphy W.J., and Abedi M.. 2012. Advances in graft-versus-host disease biology and therapy. Nat. Rev. Immunol. 12:443–458. 10.1038/nri3212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell K. 2015. The virome in host health and disease. Immunity. 42:805–813. 10.1016/j.immuni.2015.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell K. 2016. Crosstalk between autophagy and inflammatory signalling pathways: balancing defence and homeostasis. Nat. Rev. Immunol. 16:661–675. 10.1038/nri.2016.100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell K., Liu J.Y., Brown S.L., Miyoshi H., Loh J., Lennerz J.K., Kishi C., Kc W., Carrero J.A., Hunt S., et al. . 2008. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 456:259–263. 10.1038/nature07416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell K., Patel K.K., Maloney N.S., Liu T.C., Ng A.C., Storer C.E., Head R.D., Xavier R., Stappenbeck T.S., and Virgin H.W.. 2010. Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. Cell. 141:1135–1145. 10.1016/j.cell.2010.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S.Y., Chiu L.Y., Maa M.C., Wang J.S., Chien C.L., and Lin W.W.. 2011. zVAD-induced autophagic cell death requires c-Src-dependent ERK and JNK activation and reactive oxygen species generation. Autophagy. 7:217–228. 10.4161/auto.7.2.14212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway K.L., Kuballa P., Song J.H., Patel K.K., Castoreno A.B., Yilmaz O.H., Jijon H.B., Zhang M., Aldrich L.N., Villablanca E.J., et al. . 2013. Atg16l1 is required for autophagy in intestinal epithelial cells and protection of mice from Salmonella infection. Gastroenterology. 145:1347–1357. 10.1053/j.gastro.2013.08.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke K.R., Krenger W., Hill G., Martin T.R., Kobzik L., Brewer J., Simmons R., Crawford J.M., van den Brink M.R., and Ferrara J.L.. 1998. Host reactive donor T cells are associated with lung injury after experimental allogeneic bone marrow transplantation. Blood. 92:2571–2580. [PubMed] [Google Scholar]
- Degterev A., Huang Z., Boyce M., Li Y., Jagtap P., Mizushima N., Cuny G.D., Mitchison T.J., Moskowitz M.A., and Yuan J.. 2005. Chemical inhibitor of nonapoptotic cell death with therapeutic potential for ischemic brain injury. Nat. Chem. Biol. 1:112–119. 10.1038/nchembio711 [DOI] [PubMed] [Google Scholar]
- Diamanti M.A., Gupta J., Bennecke M., De Oliveira T., Ramakrishnan M., Braczynski A.K., Richter B., Beli P., Hu Y., Saleh M., et al. . 2017. IKKα controls ATG16L1 degradation to prevent ER stress during inflammation. J. Exp. Med. 214:423–437. 10.1084/jem.20161867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang H., Tan M., Xia M., Wang L., and Jiang X.. 2013. Norovirus P particle efficiently elicits innate, humoral and cellular immunity. PLoS One. 8:e63269 10.1371/journal.pone.0063269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer J.C., Bscheider M., Eisenkolb G., Lin C.C., Wintges A., Otten V., Lindemans C.A., Heidegger S., Rudelius M., Monette S., et al. . 2017. RIG-I/MAVS and STING signaling promote gut integrity during irradiation- and immune-mediated tissue injury. Sci. Transl. Med. 9:9 10.1126/scitranslmed.aag2513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold R., Schmied M., Giegerich G., Breitschopf H., Hartung H.P., Toyka K.V., and Lassmann H.. 1994. Differentiation between cellular apoptosis and necrosis by the combined use of in situ tailing and nick translation techniques. Lab. Invest. 71:219–225. [PubMed] [Google Scholar]
- Goodall M.L., Fitzwalter B.E., Zahedi S., Wu M., Rodriguez D., Mulcahy-Levy J.M., Green D.R., Morgan M., Cramer S.D., and Thorburn A.. 2016. The autophagy machinery controls cell death switching between apoptosis and necroptosis. Dev. Cell. 37:337–349. 10.1016/j.devcel.2016.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasl-Kraupp B., Ruttkay-Nedecky B., Koudelka H., Bukowska K., Bursch W., and Schulte-Hermann R.. 1995. In situ detection of fragmented DNA (TUNEL assay) fails to discriminate among apoptosis, necrosis, and autolytic cell death: a cautionary note. Hepatology. 21:1465–1468. [DOI] [PubMed] [Google Scholar]
- Günther C., Martini E., Wittkopf N., Amann K., Weigmann B., Neumann H., Waldner M.J., Hedrick S.M., Tenzer S., Neurath M.F., and Becker C.. 2011. Caspase-8 regulates TNF-α-induced epithelial necroptosis and terminal ileitis. Nature. 477:335–339. 10.1038/nature10400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill G.R., and Ferrara J.L.. 2000. The primacy of the gastrointestinal tract as a target organ of acute graft-versus-host disease: rationale for the use of cytokine shields in allogeneic bone marrow transplantation. Blood. 95:2754–2759. [PubMed] [Google Scholar]
- Hitomi J., Christofferson D.E., Ng A., Yao J., Degterev A., Xavier R.J., and Yuan J.. 2008. Identification of a molecular signaling network that regulates a cellular necrotic cell death pathway. Cell. 135:1311–1323. 10.1016/j.cell.2008.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holler E., Landfried K., Meier J., Hausmann M., and Rogler G.. 2010. The role of bacteria and pattern recognition receptors in GVHD. Int. J. Inflamm. 2010:814326 10.4061/2010/814326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou W., Han J., Lu C., Goldstein L.A., and Rabinowich H.. 2010. Autophagic degradation of active caspase-8: a crosstalk mechanism between autophagy and apoptosis. Autophagy. 6:891–900. 10.4161/auto.6.7.13038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard-Lucey V.M., Shono Y., Maurer K., West M.L., Singer N.V., Ziegler C.G., Lezcano C., Motta A.C., Schmid K., Levi S.M., et al. . 2014. Autophagy gene Atg16L1 prevents lethal T cell alloreactivity mediated by dendritic cells. Immunity. 41:579–591. 10.1016/j.immuni.2014.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imagawa Y., Saitoh T., and Tsujimoto Y.. 2016. Vital staining for cell death identifies Atg9a-dependent necrosis in developmental bone formation in mouse. Nat. Commun. 7:13391 10.1038/ncomms13391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser W.J., Upton J.W., Long A.B., Livingston-Rosanoff D., Daley-Bauer L.P., Hakem R., Caspary T., and Mocarski E.S.. 2011. RIP3 mediates the embryonic lethality of caspase-8-deficient mice. Nature. 471:368–372. 10.1038/nature09857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kernbauer E., Ding Y., and Cadwell K.. 2014. An enteric virus can replace the beneficial function of commensal bacteria. Nature. 516:94–98. 10.1038/nature13960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.G., Park J.H., Reimer T., Baker D.P., Kawai T., Kumar H., Akira S., Wobus C., and Núñez G.. 2011. Viral infection augments Nod1/2 signaling to potentiate lethality associated with secondary bacterial infections. Cell Host Microbe. 9:496–507. 10.1016/j.chom.2011.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo B.K., Stange D.E., Sato T., Karthaus W., Farin H.F., Huch M., van Es J.H., and Clevers H.. 2011. Controlled gene expression in primary Lgr5 organoid cultures. Nat. Methods. 9:81–83. 10.1038/nmeth.1802 [DOI] [PubMed] [Google Scholar]
- Lassen K.G., Kuballa P., Conway K.L., Patel K.K., Becker C.E., Peloquin J.M., Villablanca E.J., Norman J.M., Liu T.C., Heath R.J., et al. . 2014. Atg16L1 T300A variant decreases selective autophagy resulting in altered cytokine signaling and decreased antibacterial defense. Proc. Natl. Acad. Sci. USA. 111:7741–7746. 10.1073/pnas.1407001111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach M.W., Bean A.G., Mauze S., Coffman R.L., and Powrie F.. 1996. Inflammatory bowel disease in C.B-17 scid mice reconstituted with the CD45RBhigh subset of CD4+ T cells. Am. J. Pathol. 148:1503–1515. [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Shoji-Kawata S., Sumpter R.M. Jr., Wei Y., Ginet V., Zhang L., Posner B., Tran K.A., Green D.R., Xavier R.J., et al. . 2013. Autosis is a Na+,K+-ATPase-regulated form of cell death triggered by autophagy-inducing peptides, starvation, and hypoxia-ischemia. Proc. Natl. Acad. Sci. USA. 110:20364–20371. 10.1073/pnas.1319661110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W., Sun J., Yoon J.S., Zhang Y., Zheng L., Murphy E., Mattson M.P., and Lenardo M.J.. 2016. Mitochondrial protein PGAM5 regulates mitophagic protection against cell necroptosis. PLoS One. 11:e0147792 10.1371/journal.pone.0147792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariño G., Niso-Santano M., Baehrecke E.H., and Kroemer G.. 2014. Self-consumption: the interplay of autophagy and apoptosis. Nat. Rev. Mol. Cell Biol. 15:81–94. 10.1038/nrm3735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney S.A., Thackray L.B., Gitlin L., Gilfillan S., Virgin H.W., and Colonna M.. 2008. MDA-5 recognition of a murine norovirus. PLoS Pathog. 4:e1000108 10.1371/journal.ppat.1000108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizumura K., Cloonan S.M., Nakahira K., Bhashyam A.R., Cervo M., Kitada T., Glass K., Owen C.A., Mahmood A., Washko G.R., et al. . 2014. Mitophagy-dependent necroptosis contributes to the pathogenesis of COPD. J. Clin. Invest. 124:3987–4003. 10.1172/JCI74985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy J.M., Czabotar P.E., Hildebrand J.M., Lucet I.S., Zhang J.G., Alvarez-Diaz S., Lewis R., Lalaoui N., Metcalf D., Webb A.I., et al. . 2013. The pseudokinase MLKL mediates necroptosis via a molecular switch mechanism. Immunity. 39:443–453. 10.1016/j.immuni.2013.06.018 [DOI] [PubMed] [Google Scholar]
- Murthy A., Li Y., Peng I., Reichelt M., Katakam A.K., Noubade R., Roose-Girma M., DeVoss J., Diehl L., Graham R.R., and van Lookeren Campagne M.. 2014. A Crohn’s disease variant in Atg16l1 enhances its degradation by caspase 3. Nature. 506:456–462. 10.1038/nature13044 [DOI] [PubMed] [Google Scholar]
- Oberst A., Dillon C.P., Weinlich R., McCormick L.L., Fitzgerald P., Pop C., Hakem R., Salvesen G.S., and Green D.R.. 2011. Catalytic activity of the caspase-8-FLIP(L) complex inhibits RIPK3-dependent necrosis. Nature. 471:363–367. 10.1038/nature09852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasparakis M., and Vandenabeele P.. 2015. Necroptosis and its role in inflammation. Nature. 517:311–320. 10.1038/nature14191 [DOI] [PubMed] [Google Scholar]
- Patel K.K., Miyoshi H., Beatty W.L., Head R.D., Malvin N.P., Cadwell K., Guan J.L., Saitoh T., Akira S., Seglen P.O., et al. . 2013. Autophagy proteins control goblet cell function by potentiating reactive oxygen species production. EMBO J. 32:3130–3144. 10.1038/emboj.2013.233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanan D., and Cadwell K.. 2016. Intrinsic defense mechanisms of the intestinal epithelium. Cell Host Microbe. 19:434–441. 10.1016/j.chom.2016.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randow F., and Youle R.J.. 2014. Self and nonself: how autophagy targets mitochondria and bacteria. Cell Host Microbe. 15:403–411. 10.1016/j.chom.2014.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remijsen Q., Goossens V., Grootjans S., Van den Haute C., Vanlangenakker N., Dondelinger Y., Roelandt R., Bruggeman I., Goncalves A., Bertrand M.J., et al. . 2014. Depletion of RIPK3 or MLKL blocks TNF-driven necroptosis and switches towards a delayed RIPK1 kinase-dependent apoptosis. Cell Death Dis. 5:e1004 10.1038/cddis.2013.531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samara C., Syntichaki P., and Tavernarakis N.. 2008. Autophagy is required for necrotic cell death in Caenorhabditis elegans. Cell Death Differ. 15:105–112. 10.1038/sj.cdd.4402231 [DOI] [PubMed] [Google Scholar]
- Sato T., Vries R.G., Snippert H.J., van de Wetering M., Barker N., Stange D.E., van Es J.H., Abo A., Kujala P., Peters P.J., and Clevers H.. 2009. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 459:262–265. 10.1038/nature07935 [DOI] [PubMed] [Google Scholar]
- Sato T., Stange D.E., Ferrante M., Vries R.G., Van Es J.H., Van den Brink S., Van Houdt W.J., Pronk A., Van Gorp J., Siersema P.D., and Clevers H.. 2011. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 141:1762–1772. 10.1053/j.gastro.2011.07.050 [DOI] [PubMed] [Google Scholar]
- Saveljeva S., Mc Laughlin S.L., Vandenabeele P., Samali A., and Bertrand M.J.. 2015. Endoplasmic reticulum stress induces ligand-independent TNFR1-mediated necroptosis in L929 cells. Cell Death Dis. 6:e1587 10.1038/cddis.2014.548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons A.N., Kajino-Sakamoto R., and Ninomiya-Tsuji J.. 2016. TAK1 regulates Paneth cell integrity partly through blocking necroptosis. Cell Death Dis. 7:e2196 10.1038/cddis.2016.98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Wang H., Wang Z., He S., Chen S., Liao D., Wang L., Yan J., Liu W., Lei X., and Wang X.. 2012. Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell. 148:213–227. 10.1016/j.cell.2011.11.031 [DOI] [PubMed] [Google Scholar]
- Tait S.W., Oberst A., Quarato G., Milasta S., Haller M., Wang R., Karvela M., Ichim G., Yatim N., Albert M.L., et al. . 2013. Widespread mitochondrial depletion via mitophagy does not compromise necroptosis. Cell Reports. 5:878–885. 10.1016/j.celrep.2013.10.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N., Duprez L., Grootjans S., Cauwels A., Nerinckx W., DuHadaway J.B., Goossens V., Roelandt R., Van Hauwermeiren F., Libert C., et al. . 2012. Necrostatin-1 analogues: critical issues on the specificity, activity and in vivo use in experimental disease models. Cell Death Dis. 3:e437 10.1038/cddis.2012.176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschurtschenthaler M., Adolph T.E., Ashcroft J.W., Niederreiter L., Bharti R., Saveljeva S., Bhattacharyya J., Flak M.B., Shih D.Q., Fuhler G.M., et al. . 2017. Defective ATG16L1-mediated removal of IRE1α drives Crohn’s disease-like ileitis. J. Exp. Med. 214:401–422. 10.1084/jem.20160791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaishnava S., Behrendt C.L., Ismail A.S., Eckmann L., and Hooper L.V.. 2008. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc. Natl. Acad. Sci. USA. 105:20858–20863. 10.1073/pnas.0808723105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Jiang H., Chen S., Du F., and Wang X.. 2012. The mitochondrial phosphatase PGAM5 functions at the convergence point of multiple necrotic death pathways. Cell. 148:228–243. 10.1016/j.cell.2011.11.030 [DOI] [PubMed] [Google Scholar]
- Wlodarska M., Kostic A.D., and Xavier R.J.. 2015. An integrative view of microbiome-host interactions in inflammatory bowel diseases. Cell Host Microbe. 17:577–591. 10.1016/j.chom.2015.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yui S., Nakamura T., Sato T., Nemoto Y., Mizutani T., Zheng X., Ichinose S., Nagaishi T., Okamoto R., Tsuchiya K., et al. . 2012. Functional engraftment of colon epithelium expanded in vitro from a single adult Lgr5+ stem cell. Nat. Med. 18:618–623. 10.1038/nm.2695 [DOI] [PubMed] [Google Scholar]
- Zhang D.W., Shao J., Lin J., Zhang N., Lu B.J., Lin S.C., Dong M.Q., and Han J.. 2009. RIP3, an energy metabolism regulator that switches TNF-induced cell death from apoptosis to necrosis. Science. 325:332–336. 10.1126/science.1172308 [DOI] [PubMed] [Google Scholar]
- Zhu X., Messer J.S., Wang Y., Lin F., Cham C.M., Chang J., Billiar T.R., Lotze M.T., Boone D.L., and Chang E.B.. 2015. Cytosolic HMGB1 controls the cellular autophagy/apoptosis checkpoint during inflammation. J. Clin. Invest. 125:1098–1110. 10.1172/JCI76344 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.