In this issue of JEM, Krejciova et al. report that astrocytes derived from human iPSCs can replicate human CJD prions. These observations provide a new, potentially very valuable model for studying human prions in cellula and for identifying antiprion compounds that might serve as clinical candidates. Furthermore, they add to the evidence that astrocytes may not be just innocent bystanders in prion diseases.
Abstract
In this issue of JEM, Krejciova et al. (https://doi.org/10.1084/jem.20161547) report that astrocytes derived from human iPSCs can replicate human CJD prions. These observations provide a new, potentially very valuable model for studying human prions in cellula and for identifying antiprion compounds that might serve as clinical candidates. Furthermore, they add to the evidence that astrocytes may not be just innocent bystanders in prion diseases.

Insight from Adriano Aguzzi and Yingjun Liu
Prions are the infectious agents that cause Creutzfeldt–Jakob disease (CJD) and other fatal neurodegenerative disorders affecting both humans and animals. Prions consist of PrPSc, a pathological isoform of the cellular prion protein PrPC that is expressed in neurons, astrocytes, and oligodendrocytes of the adult brain. Nucleation and self-sustained propagation of PrPSc is the major event driving the progression of prion diseases (Aguzzi et al., 2008). Although prion diseases are rare, they are invariably lethal, and no disease-modifying therapies exist. This sad situation is compounded by a dearth of validated therapeutic targets, which stems from our incomplete understanding of the biochemical and cellular networks involved in the pathogenesis (Aguzzi et al., 2017). Because of the gaps in our understanding of the mechanisms underlying prion replication, propagation, and neurotoxicity, prions continue to represent a significant threat to public health. Now, a study by a coalition of Scottish laboratories (see Krejciova et al. in this issue) has provided new tools and interesting insights into the aforementioned issues.
The rationale for studying prion diseases goes beyond the goal of providing therapeutic options because prions yield a robust and flexible model for studying general pathways of neurodegeneration in the context of protein aggregation disease. You see, when laboratory mice are infected with prions, they develop a disease that has all the characteristics of naturally occurring mammalian prion diseases, including the typical histological triad of neuronal loss, spongiosis, and reactive gliosis. This is not true for animal models of other neurodegeneration such as Alzheimer’s, Parkinson’s, and other diseases. APP transgenic mice, for example, develop amyloid plaques consisting of Aβ aggregates, yet do not reproduce many other aspects of Alzheimer’s disease. Hence, one may argue that the mechanism active in neurodegeneration (such as microglia and astrocyte activation) may be best studied in prion infection models and then tested in other protein aggregation diseases.
For all their advantages, however, animal models of prion disease do not allow full dissection of the causal chain of events underlying prion replication and toxicity; in vitro models are essential to reach these goals. Although certain neuroblastoma cell lines were found to replicate prions (Enari et al., 2001; Klöhn et al., 2003), none of these lines appear to experience overt neurotoxicity as a consequence (Li et al., 2010).
Against this backdrop, prion-infectible human cell lines would be desirable for advancing the understanding of human prion diseases and for developing therapeutic interventions. However, such experimental systems have been surprisingly elusive: none of the most popular cell lines allows for replication of human CJD prions, which may substantially differ from other prion strains. An early report that SH-SY5Y neuroblastoma cells can be infected with human prions seems dubious because these cells express only trace amounts of PrPC (unpublished observations). These limitations have hampered the mechanistic study of human prion diseases and may have also hindered the discovery of human therapies because many antiprion compounds are only effective in specific animal species (Aguzzi and Polymenidou, 2004; Bolognesi and Legname, 2015; Aguzzi et al., 2017).
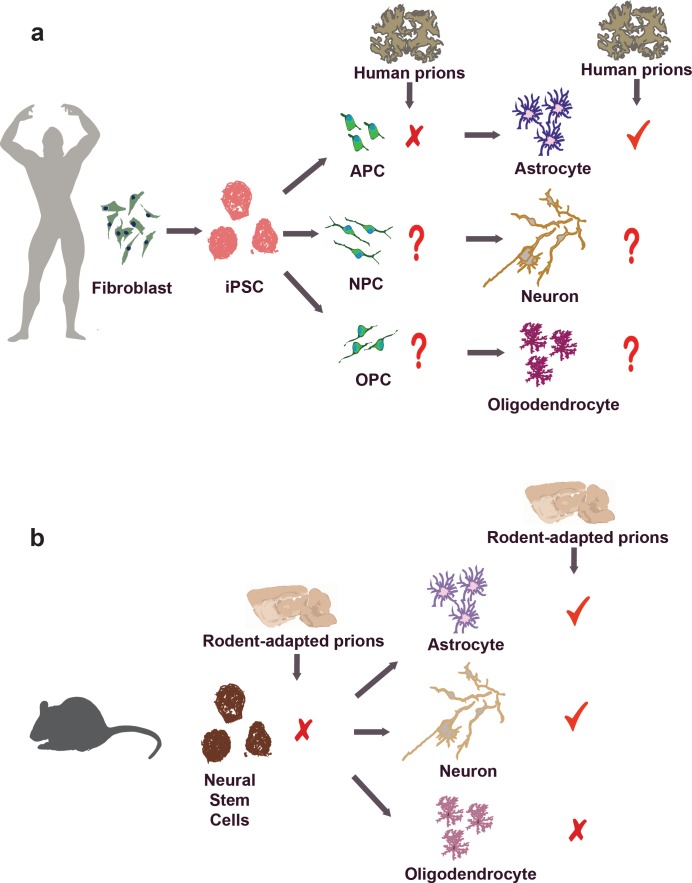
Krejciova et al. (2017) have found that human CJD brain extracts can elicit prion replication in astrocytes that were grown out of human induced pluripotent stem cells (iPSCs; see figure). A common Met/Val polymorphism at codon 129 of the PRNP gene controls susceptibility of humans to prion infections, with homozygous individuals (Met/Met and Val/Val) being overrepresented in collectives of CJD patients. Moreover, variant CJD, which is thought to represent the transmission of bovine spongiform encephalopathy (“mad cow disease”) to humans, has almost exclusively affected homozygous Met/Met individuals. This finding was replicated in the iPSC-derived astrocytes. In view of the potentially unlimited supply of genetically homogeneous iPSC-derived astrocytes, this model might become useful for screening chemical libraries for compounds that inhibit human prion replication.
(a) Human iPSC-derived astrocyte supports the replication of CJD prions. In contrast to differentiated astrocytes, however, human iPSC-derived APCs failed to replicate human prions despite expression of PrPC. It is still unclear whether human iPSC-derived neurons, oligodendrocytes, and their precursors (NPC and OPC, respectively) are capable of supporting human prion replication. (b) Rodent studies have showed that differentiated neurons and astrocytes, but neither undifferentiated neurospheres nor oligodendrocytes, support prion replication.
The new findings also provide additional evidence that astrocytes might contribute to the pathogenesis of prion diseases. Previous studies had suggested that PrPSc deposits in astrocytes during prion infection both in human and in rodents (Diedrich et al., 1991; Kovács et al., 2005). Also, we and others had reported that PrnpZH1/ZH1 mice (Büeler et al., 1993) expression a PrPC transgene driven by a glial fibrillary acidic protein (GFAP) promoter exhibited prion replication in vivo (Raeber et al., 1997) with PrPSc accumulating in astrocytes (Jeffrey et al., 2004). Although these studies suggested that astrocytes may directly contribute to prion replication and propagation, we deemed them somewhat dubious (including those to which we had contributed) because the GFAP promoter fragment used for the generation of these mice was later found to be ectopically active in certain neuronal populations (Marino et al., 2000; Zhuo et al., 2001; Casper and McCarthy, 2006), thereby sowing doubts whether prion replication occurred truly in astrocytes. The unambiguous replication of CJD prions in human iPSC-derived astrocytes goes a long way toward vindicating the Raeber publication, yet the astrocytes used by Krejciova et al. (2017), like those from any other iPSC-based systems, may be contaminated with small numbers of neuronal cells. Complete, reliable elimination of all contaminating neurons would be desirable but may not be technically attainable.
Krejciova’s findings may provide a tool to address a long-standing enigma: why is it that some cell types support prion replication whereas others do not, despite high levels of PrPC expression? Perhaps the assembly of infectious prions requires a molecular nanomachine (the “prion replicase”) that is only present in certain cell types. Alternatively, replication-incompetent cells may clear prions more efficiently than others. In some in vitro systems, prion clearance may be as simple as prion dilution: if cells divide faster than they can replicate prions, the latter will over time become inevitably diluted. In contrast with the differentiated astrocytes, human iPSC-derived astrocyte precursor cells (APCs) failed to support the replication of human prions despite expression of PrPC. This is in line with studies using rodents showing that differentiated neurons and astrocytes, but neither undifferentiated neurospheres nor oligodendrocytes, support prion replication (Cronier et al., 2004; Prinz et al., 2004; Herva et al., 2010). The quest for the elusive prion replicase could be crucially aided by genome-wide siRNA and CRISPR screens of replication-competent cell lines.
Although astrocytes are indispensable to the integrity of the central nervous system, the Barres laboratory has recently shown that microglia-activated astrocytes can significantly contribute to neurotoxicity (Liddelow et al., 2017). These findings raise the question of whether prions modulate the polarization of astrocytes toward a protective or a toxic phenotype and whether such modulation is cell autonomous or requires the intervention of microglia.
Prions are bona fide infectious agents, and prion titers can be measured with methods similar to those used in diagnostic virology. Do the iPSC-derived astrocytes indeed replicate prion infectivity, or do they simply store exogenous infectivity without multiplying it? The presence of prions is often equated with the appearance of proteinase K–resistant PrP, yet prion infectivity can exist in the absence of protein resistance (Leske et al., 2017) and vice versa (Giles et al., 2017). The Scottish authors have addressed this issue by testing whether prion-infected astrocytes would transmit the infection to a second generation of naive astrocytes. This is an important test, which should always be performed before claiming that any protein aggregate (be it composed of Aβ, synuclein, tau, or else) is a “prion.” Ideally, the multiplication of the agent should be proved by rigorous titration experiment of input and output prions, e.g., by limiting dilution.
A more specialized question relates to the maintenance of “strainness.” Prions come in the form of many different strains, each one of which displays heritable, specific incubation times and target areas in the brain. The physicochemical underpinnings of strainness are likely specified by the conformation of PrPSc aggregates (Sigurdson et al., 2007), which may be switched by passage through specific cell types. It will be interesting to assess whether astrocytes impose specific constraints onto the conformation of replicated prions. Finally, it is conceivable that astrocytes and neurons potentiate the toxicity of prions by amplifying prions produced by each other in a ping-pong mechanism.
In summary, the availability of a new cellular model of prion infection extends the toolbox available for prion science and enables a range of potentially important experiments. Because studies of human genetics have thus far failed to identify any risk factors for prion diseases other than the PRNP gene itself, cell lines may contribute to filling that gap—in conjunction with modern technologies such as bar-coded CRISPR libraries and next-gen sequencing. The brief report of Krejciova et al. (2017) may therefore contribute to the basic understanding of human prion replication and to accelerating drug development for human prion diseases.
References
- Aguzzi A., et al. Annu. Rev. Pathol. 2008 doi: 10.1146/annurev.pathmechdis.3.121806.154326. [DOI] [PubMed] [Google Scholar]
- Aguzzi A., et al. Annu. Rev. Pharmacol. Toxicol. 2017 doi: 10.1146/annurev-pharmtox-010617-052745. [DOI] [Google Scholar]
- Aguzzi A., and Polymenidou M. Cell. 2004 doi: 10.1016/S0092-8674(03)01031-6. [DOI] [PubMed] [Google Scholar]
- Bolognesi M.L., and Legname G. Expert Opin. Drug Discov. 2015 doi: 10.1517/17460441.2015.1016498. [DOI] [PubMed] [Google Scholar]
- Büeler H., et al. Cell. 1993 doi: 10.1016/0092-8674(93)90360-3. [DOI] [Google Scholar]
- Casper K.B., and McCarthy K.D. Mol. Cell. Neurosci. 2006 doi: 10.1016/j.mcn.2005.12.006. [DOI] [PubMed] [Google Scholar]
- Cronier S., et al. Proc. Natl. Acad. Sci. USA. 2004 doi: 10.1073/pnas.0402725101. [DOI] [Google Scholar]
- Diedrich J.F., et al. Proc. Natl. Acad. Sci. USA. 1991 doi: 10.1073/pnas.88.2.375. [DOI] [Google Scholar]
- Enari M., et al. Proc. Natl. Acad. Sci. USA. 2001 doi: 10.1073/pnas.151242598. [DOI] [Google Scholar]
- Giles K., et al. Cold Spring Harb. Perspect. Biol. 2017 doi: 10.1101/cshperspect.a023499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herva M.E., et al. J. Neurosci. Methods. 2010 doi: 10.1016/j.jneumeth.2010.02.022. [DOI] [PubMed] [Google Scholar]
- Jeffrey M., et al. Ann. Neurol. 2004 doi: 10.1002/ana.20093. [DOI] [PubMed] [Google Scholar]
- Klöhn P.C., et al. Proc. Natl. Acad. Sci. USA. 2003 doi: 10.1073/pnas.1834432100. [DOI] [Google Scholar]
- Kovács G.G., et al. Am. J. Pathol. 2005 doi: 10.1016/S0002-9440(10)62252-3. [DOI] [Google Scholar]
- Krejciova Z., et al. J. Exp. Med. 2017 doi: 10.1084/jem.20161547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leske H., et al. PLoS One. 2017 doi: 10.1371/journal.pone.0170503. [DOI] [Google Scholar]
- Li J., et al. Science. 2010 doi: 10.1126/science.1183218. [DOI] [Google Scholar]
- Liddelow, et al. Nature. 2017 doi: 10.1038/nature21029. [DOI] [Google Scholar]
- Marino S., et al. 2000. Genes Dev. 14:994–1004. [PMC free article] [PubMed] [Google Scholar]
- Prinz M., et al. J. Neurosci. 2004 doi: 10.1523/JNEUROSCI.0122-04.2004. [DOI] [Google Scholar]
- Raeber, et al. EMBO J. 1997 doi: 10.1093/emboj/16.20.6057. [DOI] [Google Scholar]
- Sigurdson, et al. Nat. Methods. 2007 doi: 10.1038/nmeth1131. [DOI] [PubMed] [Google Scholar]
- Zhuo L., et al. Genesis. 2001 doi: 10.1002/gene.10008. [DOI] [Google Scholar]