Abstract
MicroRNAs form an essential class of post-transcriptional gene regulator of eukaryotic species, and play critical parts in development and disease and stress responses. MicroRNAs may originate from various genomic loci, have structural characteristics, and appear in canonical or modified forms, making them subtle to detect and analyze. We present miRvial, a robust computational method and companion software package that supports parameter adjustment and visual inspection of candidate microRNAs. Extensive results comparing miRvial and six existing microRNA finding methods on six model organisms, Mus musculus, Drosophila melanogaste, Arabidopsis thaliana, Oryza sativa, Physcomitrella patens and Chlamydomonas reinhardtii, demonstrated the utility and rigor of miRvial in detecting novel microRNAs and characterizing features of microRNAs. Experimental validation of several novel microRNAs in C. reinhardtii that were predicted by miRvial but missed by the other methods illustrated the superior performance of miRvial over the existing methods. miRvial is open source and available at https://github.com/SystemsBiologyOfJianghanUniversity/miRvial.
INTRODUCTION
MicroRNAs (miRNAs) form a class of short, noncoding RNAs that play critical regulatory roles in animals and plants (1). They are generated from long primary RNA transcripts with hairpin-shaped fold-back structures. Most of miRNA precursors are processed by the canonical miRNA biogenesis pathway involving cleavage activities of the RNase type III enzymes Drosha and Dicer in animals or DICER-LIKE enzymes in plants. The ∼22-nt long mature miRNAs exert their functions after being loaded into RNA-induced silencing complexes (1).
Several computational methods have been developed for miRNA identification (2–7), most of which take advantage of small-RNA profiling data from next-generation sequencing (NGS) (8–10). The combination of NGS profiling, computational analysis and experimental validation has produced a collection of genuine miRNAs in many organisms and deepened our understanding of the diversity and conservation of miRNAs. The results also made it feasible to characterize miRNAs in gene regulation and elucidate their broad functions in development, stress response and complex diseases (1,11).
Despite much success, it remains challenging to identify miRNAs with high sensitivity and specificity. One difficulty lies in the diversity of miRNAs and heterogeneity of their biogenesis. Besides genomic loci hosting exclusively miRNAs, miRNAs may arise from sites where other noncoding RNAs reside (12,13). Noncanonical miRNAs have miRNA-hairpin structures but bypass canonical miRNA processing steps. For example, their production requires Dicer but not Drosha nor Dgcr8 (13). Secondly, miRNA structures may vary considerably. miRNA hairpins may differ drastically in size and structure within the same organism or across species. For instance, miRNA hairpins are typically shorter in animals than in plants. The number of bulges along the stem of a hairpin structure can also vary to a large extent between animal and plant miRNAs. Taken together, the diversity of miRNAs makes it challenging to accurately classify miRNAs only based on computational features. The third problem is that many existing methods are tailored to miRNAs in specific model species, e.g. Mus musculus (mouse) or Drosophila melanogaste (fruitfly) in animals, or Arabidopsis thaliana or Oryza sativa (rice) in plants. However, as a class of gene expression regulators in eukaryotic organisms, little effort has been made to reliably and comprehensively identify canonical and noncanonical miRNAs in diverse organisms. Moreover, some of the existing methods have parameters hard coded or parameters difficult to tune, making them ineffective for finding novel miRNAs; the drawback is exacerbated particularly in finding and analyzing miRNAs in less-studied species.
Based upon our extensive experience in identifying and analyzing miRNAs in human (14,15), plants (16,17) and viruses (18,19), we aim at a robust method and software tool for identification and analysis of miRNAs in various organisms. In this paper, we describe the essential steps required for identifying miRNAs from deep sequencing data with respect to underlying miRNA biogenesis. We implemented these features and steps in miRvial (miRNAs via integrative analysis), a robust computational method and software package for miRNA identification and analysis. Comparing to the existing methods, miRvial supports parameter adjustment and visual inspection of candidate miRNAs so that it is versatile for accurate identification of miRNAs in various animal and plant species. We also present experimental results comparing miRvial and six popular existing methods on mouse, fruitfly, Arabidopsis, rice, moss and algae to demonstrate the utility and power of miRvial in identifying novel microRNA candidates and characterizing features and expression patterns of miRNA detected.
MATERIALS AND METHODS
Data collection
We utilized in this study sequencing-based small-RNA profiling data from six species, Mus musculus (mouse), Drosophila melanogaster (fluitfly), Arabidopsis thaliana, Oryza sativa (rice), Physcomitrella patens (moss) and Chlamydomonas reinhardtii (algae). Detailed information of the datasets can be found in Supplemental Table S1. The data for the first four species were from NCBI databases. The data on mouse (GSE20384) were collected from three mouse tissues—brain, ovary and testes—and embryonic day 7.5 (E7.5), day 9.5 (E9.5) and day 12.5 (E12.5), as well as whole newborn mice. The data on fluitfly (GSE12840) were collected from modENCODE project under the conditions of late embryo, larval, pupal and adult head. The data on moss (GSE5103) were collected under three conditions, i.e. 7-day wild-type P. patens in protonemata; 14-day-old wild-type in protonemata and young Gametophores and ∼60-day old wild-type mature in gametophores and sporophytes. The data on Arabidopsis (GSM632205–GSM632209) were collected from seedlings of 4-week old wildtype and transgenic plants grown under 22°C or 30°C. The data on rice (GSE32973) were collected from seedling, root, shoot and panicle of O. sativa japonica.
Sequencing based small-RNA profiling
We collected the data for small-RNA species in algae C. reinhardtii strain CC503 cw92 mt+, obtained from the Chlamy Center (http://www.chlamy.org). C. reinhardtii cells were grown in 2-amino-2-(hydroxymethyl)-1,3-propanediol (TRIS)-acetate-phosphate (TAP) medium (Harris, 1989) at 22–26°C under a 12 h:12 h light/dark cycle. Four-day-old cells were collected, and total RNA was isolated with Trizol reagent (Invitrogen). Total RNA of 1 ug was used for the standard small RNA libraries construction. Small-RNA libraries were prepared according to the manufacturers’ protocols using the NEBNext® Small RNA Library Prep Set for SOLiD™ (NEW ENGLAND BioLabs). Briefly, 3′ adapters were ligated to small RNAs and then RT primers were introduced to seal the 3′ end, 5′ adapters ligated. A reverse transcription reaction was performed thereafter, and the resulting cDNA was amplified using the barcoded primers for each sample with SOLiD5500 RNA-Seq BC01-BC96 (Life Technologies). The PCR products were resolved on the 6% PAGE gels and the fragments from 110 to 130 bp were selected. The yield and the size distribution of the amplified cDNA were assessed using the Aglient High sensitivity DNA Kit on the 2100 Bioanalyzer Instruments (Agilent Technologies). The barcoded libraries were mixed in a pool at the same concentration. Pemplate bead preparation, emulsion PCR, deposition and sequence were performed according to the standard protocol.
Major steps of miRvial
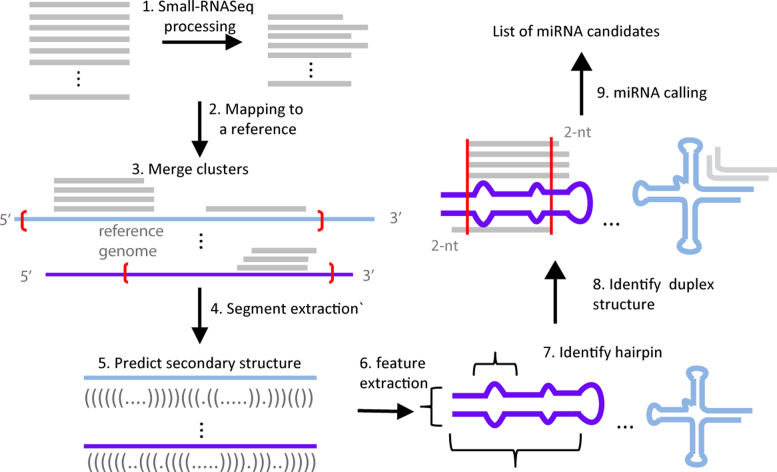
miRvial has several steps (Figure 1). Raw sequencing reads were first trimmed to remove 3′-end adapter sequences by an in-house method that recursively searches for the longest substring of the adaptor appearing within a sequence read. If a raw read did not have a substring of the adaptor longer than 6-nt, it was considered to carry no adaptor and discarded. Other adopter trimming methods, e.g. fastqx-tool (http://hannonlab.cshl.edu/fastx_toolkit/), may be used in this step. The high-quality and adaptor removed reads, called qualified reads hereafter, were aligned to the corresponding reference genome by Bowtie (version 0.12.7 release) (20) with parameters ‘-k 10 –m 30’. Reads that can be mapped to >30 genomic loci were discarded, while up to 10 valid alignments will be reported as multiple-mapped reads.
Figure 1.
Major steps and flow chart of miRvial. It shows the steps on how raw sequencing reads are initially processed, including 3′ adapter trimming (step 1); the remaining reads are aligned to a reference genome in step 2, where loci with sufficient reads are merged (red brackets in step 3) and extracted for secondary structure analysis (steps 4 and 5). miRvial identifies miRNA-like hairpins using a representation of three features based on secondary structures (steps 6 and 7). It searches for miRNA duplexes with the characteristic ∼2-nt 3′-overhangs using the alignment of sequencing reads (step 8). Application of the steps lead to a list of candidate miRNAs in the reference genome.
Adjacent reads (gapLen in the software manual) were merged to register the initial loci of candidate miRNAs. The parameters for different species can be adjusted, e.g. values of gapLen and mincopy (50 and 10, respectively) were used for mouse miRNAs. The loci containing sufficient numbers of reads (mincopy in the software manual) were then subject to RNA secondary structure analysis. Folding structures of the (merged) loci were obtained by a RNA-fold program such as RNAfold in the ViennaRNA package (21) or Mfold (22). miRvial could parse and analyze multiple secondary structures. For example, RNAsubopt can generate suboptimal secondary structures within a user-defined range. miRvial considers all of the structures from the folding program and search for miRNAs with alternative suboptimal secondary structures. The length of the genomic segment surrounding a genomic locus to be folded was determined by taking into consideration the average length of a miRNA precursor and the sequencing reads around the locus. Since the average precursor length varies across species, a parameter ‘ext_Len’ was introduced to the miRvial software to accommodate this length variation. The first 5′-end position covered by a sequencing read was extended by ‘ext_Len’ nt upstream and the last 3′-end position covered by a sequencing read was extended by ‘ext_Len’ nt downstream to form the genomic segment to be folded. The default value for ‘ext_Len’ is 150-nt for animal species and 300-nt for plant species, and can be adjusted if needed.
The values of three features of a secondary structure of a potential hairpin, i.e., the stem length, hairpin length, and size of the maximal bulge, were computed. The stem length is the number of base pairs of a stretch of sufficiently long consecutive ‘matched’ base pairs of the folding structure allowing a few unpaired bases (e.g., at least 17 base pairs with a maximum 5 unpaired bases were used to detect mouse miRNAs). The hairpin length is the sum of its stem and loop. The size of the maximal bulge is the number of unpaired bases along a stem (Supplemental Figure S1). A hairpin structure is detected to exist in a secondary structure if these values satisfy the criteria for the three parameters, i.e., ‘base_pair_num’, ‘hairpin_len’ and ‘buldge_gap’, for the species considered, e.g., these parameters for miRNAs in mouse were set to 17, 120 and 8, respectively. miRvial evaluates the entire secondary structure of a locus to find stems. These parameters can be adjusted as shown in Figure 2. An illustrative example is shown in Supplemental Figure S1.
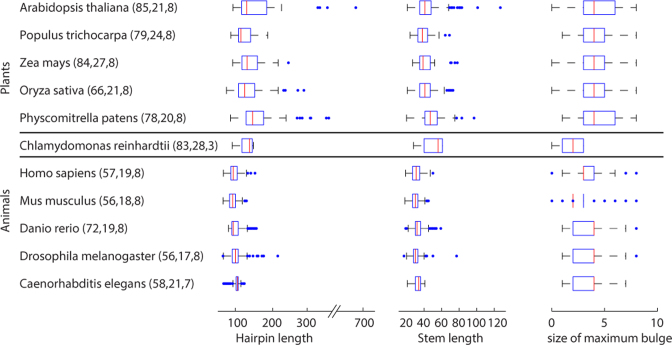
Figure 2.
Variation of miRNA hairpins measured in three structural features. The first column lists model species. The hairpin length and the size of maximum bulge are in the unit of the number of nucleotides. The stem length is in the unit of the number of base pairs. The central mark of a box plot represents the median value in an organism, while the edges of the box indicate the 25th and 75th percentiles. The plotted whiskers correspond to approximately 99% coverage of data points, and outliers are plotted as individual points outside the whiskers. Values in parenthesis represent the values for the three parameters set up in miRvial. miRNA information was retrieved from miRBase version 21.
Finally, the following four miRNA-annotation criteria were adopted to detect candidate miRNAs: (i) occurrence of miRNA reads on the arms of predicted hairpin structures; (ii) presence of no less than a user-determined miRNA reads of the highest frequency on predicted hairpins (in the current study, we chose 10 minimal reads); (iii) presence of possible miRNA* sequencing reads unless searching for noncanonical miRNAs and (iv) presence of ∼2-nt 3′ overhangs on miRNA/miRNA* duplexes. Since the parameters and filtering criteria of miRvial are species-specific, for convenience we provided parameters of model species in the current package. These files, named ‘param_$species.txt’, can be modified for running miRVial.
The candidate miRNAs could be further reviewed via a graphic display, i.e. all small RNA reads were aligned to their corresponding hairpin sequence and visually inspected. The read with the highest read count was preferentially selected as the mature miRNA sequence. The secondary structure for each hairpin could also be visualized to verify the miRNA* sequence.
The existing methods compared
We systematically compared miRvial with six existing miRNA prediction methods. Three of them focus on animal species – miRDeep2 (3), miRTRAP (2) and MIReNA (4) – and the other three are tailored to plans – miRDeep-P (5), miRPlant (6) and miRA (7). If the six tools have parameters similar to the ones in miRvial, we used the same values as in miRvial. Running scripts and parameters can be found in Supplemental Files S1 (available at https://github.com/SystemsBiologyOfJianghanUniversity/miRvial). We also surveyed the features of a total of 11 existing miRNA identification tools (Supplemental Table S2). With respect to these existing methods, miRvial offers unique features of examining suboptimal RNA secondary structures, flexible parameter adjustment, and visualizing miRNA precursor folding structures, which in combination greatly improve performance.
Validation of novel miRNAs in algae
To validate the results of novel miRNAs in C. reinhardtii predicted by miRvial, five novel miRNAs in C. reinhardtii strain CC-503 were further analyzed by Reverse Transcription-Polymerase Chain Reaction (RT-PCR). The 72 h cells cultured at 25°C in TAP medium were collected. Small RNAs were extracted using miRcute miRNA Isolation Kit (TIANGEN BIOTECH), and first-strand cDNA synthesis was performed by miRcute miRNA First-Strand cDNA Synthesis Kit (TIANGEN BIOTECH). A synthetic exogenous reference miRNA Std1 (5′- GCTATATGCAAGTCCGGCCATAC-3′) was introduced as a positive control. Forward Primers for the PCR amplifications were listed in Supplemental Table S3 and the reverse primer was provided by the first-strand cDNA synthesis kit. PCR products were resolved on 4% agarose gels.
RESULTS
miRvial was designed to follow the essential steps of the canonical miRNA biogenesis pathway and further enhanced by additional steps to make it robust (Figure 1). Briefly, raw sequencing reads are processed to remove 3′ adapter for sequencing, and high-quality or qualified reads are then mapped to the reference genome and/or expressed sequence tags. The genomic loci with a sufficient number of mapped reads are processed to merge neighboring loci if they are close to one another. The (merged) loci are extended on both directions and the corresponding genomic sequences are then extracted. The length of the extension is adjusted to reflect possible variances in miRNA hairpin lengths in different species. RNA secondary structures are predicted in silico and, as an option, visually inspected to identify candidate miRNA hairpins, followed by the detection of RNA-RNA duplex structures formed by sequencing reads as a prominent feature for miRNAs. In the following, we discuss these major steps in detail.
Initial data processing steps are important
The initial steps for preprocessing of sequencing data, including adaptor removing and quality control, and alignment of reads play an important role in miRNA identification (Figure 1 and Methods). NGS is error prone, and raw sequencing reads from NGS need to be processed to remove erroneous reads, i.e. those that carry no 3′ sequencing adaptors or have low sequencing quality. The adaptor-trimmed, high-quality reads are referred to as qualified reads or simply reads in the rest of the discussion. The way of aligning qualified reads to the reference genome or ESTs also affects miRNA identification. For example, it is important to allow a read to be aligned to multiple loci because paralogous miRNAs from multiple genomic loci are prevalent in animals and plants, and are categorized as individual members of a miRNA family. For instance, murine let-7 derives from 13 distinct loci in the mouse genome and rice MIR169 has 20 individual members. Moreover, read abundance is another factor to consider. Loci with singleton reads or few accumulative reads may be discarded since insufficient reads reduce the accuracy of prediction. RNAs with low expression may simply reflect transcriptional noises, by-products of RNA processing or RNA degradation products (23).
Fold-back hairpins are characteristic, structural signatures of miRNAs
Secondary structures are fundamental features of RNA molecules that can be exploited to distinguish one type of RNA from another. For example, the characteristic hairpin shaped structure of a miRNA differs from the typical cloverleaf structure of a tRNA, making it possible to distinguish miRNAs from many other noncoding RNAs. Recent NGS-based small-RNA profiling assays have made it possible to probe secondary structures in vivo (24) and in vitro (25). However, as RNA secondary structures are computationally predicted, biological relevant structures are sometimes suboptimal. Moreover, transcripts may indeed fold into alternative secondary structures by nature, exemplified by mammalian miR-1983 derived from an alternatively folded tRNA transcript (13,26). A list of computationally suboptimal structures can thus be generated using Mfold (22) or RNAfold (21), which led to an increased sensitivity as shown in later experiments.
miRNA hairpins may vary drastically in size and structure within the same organism or across species. For example, miRNA hairpins are typically shorter in animals than in plants (Figure 2), indicating that hairpin lengths need to be taken into account in miRNA identification. Other factors such as the length of a stem and the number of bulges along the stem of a hairpin structure can also vary to a large extent between animal and plant miRNAs (Figure 2).
We thus introduced in miRvial a new representation of a hairpin structure with three features, namely the length of the hairpin, the length of the stem and the size of the maximum bulge on the stem (see Methods). The three features capture the intrinsic properties of a hairpin, and thus were used to distinguish miRNAs from other RNA molecules. Furthermore, in order to accommodate the diversity of miRNAs and variation of miRNA precursors and hairpin structures, these three features can be adjusted in the miRvial software pipeline. For example, following the statistics in Figure 2, a stem of a murine miRNA hairpin is at least 18 base pairs, a bulge on the stem has no more than eight unpaired bases, and the total hairpin length is at least 56-nt. A structure is considered to contain a hairpin as long as the features satisfy the parameters for an organism. An illustrative example is in Supplemental Figure S1, where the three features are used to distinguish a miRNA hairpin from other noncoding RNA structures.
The RNA/RNA duplex is another characteristic feature for most miRNA genes. A miRNA duplex consists of two annealed RNA strands with a ∼2-nt 3′ overhang, which reflects a key characteristic of cleavage activities of endonuclease Dicers in miRNA biogenesis. The next step is thus to identify whether a RNA/RNA duplex is present on a secondary structure using the aligned reads (see Materials and Methods). If such a duplex is present, the locus is then considered as a candidate miRNA with a high confidence.
Increase miRNA detection sensitivity by considering atypical miRNAs
miRvial identifies noncanonical miRNAs as miRNAs with no sequencing reads for miRNA*. As a result, it can find atypical miRNAs without miRNA/miRNA* duplexes and may also include canonical miRNAs whose miRNAs* have not been detected due insufficient sequencing depth. Although the presence of a duplex structure can increase the likelihood of a genuine miRNA, noncanonical miRNAs may not obey this rule. As an extreme example, the precursor of noncanonical miRNAs miR451 is directly loaded into AGO2 after Drosha processing, thus bypassing Dicer processing. As a result, the dominant mature miR-451, which is 23-nt in length and is derived from the 5p-arm, spans across the hairpin loop, while a few miRNAs are derived from the 3p-arm of miR-451. Thus, miR-451 precursor has an atypical structure with no canonical RNA duplex on its hairpin (27,28). More seriously, predicted secondary RNA structures may not be the actual structures, making the alignment of mature miRNAs not form canonical duplexes (Supplemental Figure S2). Furthermore, mature miRNA species on one arm of a hairpin may not be detected due to insufficient profiling depth of sequencing (10) or a high degradation rate of miRNAs in the cell.
miRvial deals with these issues by deviating from the stringent criterion on the presence of a duplex to produce a list of candidate miRNAs and their genomic loci. For noncanonical miRNAs, information of genomic loci where sequencing reads are mapped to may facilitate identification of noncanonical miRNAs. For example, when reads are aligned to both ends of a short intron, they may represent a miRtron, where the intronic region constitutes a construct of a miRNA precursor (29) (Supplemental Figure S3). The user may provide intron sequences as input to let miRvial search for noncanonical miRNAs. miRvial also provides an option for visualization of candidate miRNAs with sequences surrounding their loci.
Seeing is believing—graphic display of candidate miRNAs
The modularized scripts of the miRvial software can be called consecutively to carry out miRNA discovery and analysis. Moreover, miRvial offers scripts to convert text outputs to a user friendly visual output to display alternative RNA secondary structures of candidate miRNA precursors with information of sequencing data and to facilitate visual inspection and selection of genuine miRNAs (Figure 3). This graphic interface makes miRvial easy to use, flexible and robust comparing to the existing methods, to be discussed next.
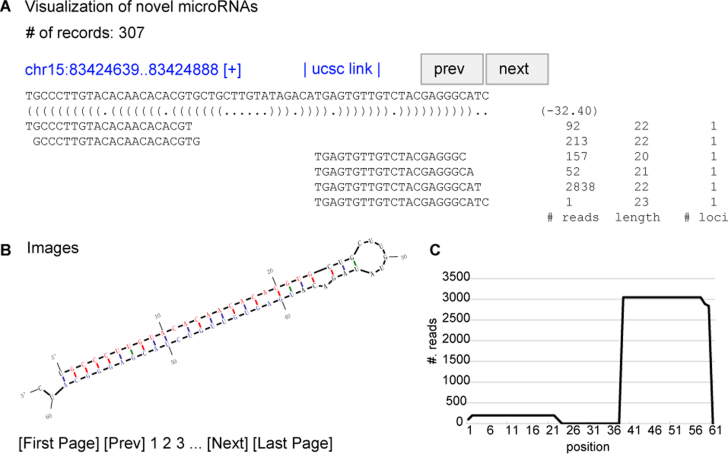
Figure 3.
A schematic graphic output from miRvial. miRvial provides graphic output to assist visual inspection and selection of genuine miRNAs. Shown is an example of a miRNA detected by miRvial. (A) The upper panel gives the number of candidate miRNAs miRvial reported; one in this case shown. The panel shows the unique genomic locus of the miRNA, along with the predicted RNA secondary structure represented in parentheses, and the folding energy in a negative value. The lines below show the aligned sequencing reads, followed by the number of reads (# reads), the length of the unique read (length), and the number of mappable loci of the unique read (#. loci). (B) The lower-left plot shows the hairpin structure, where putative mature miRNAs are highlighted in color (blue from 5p-arm and red from 3p-arm). (C) The lower-right plot shows the distribution of reads in the predicted precursor sequence.
miRvial outperforms six existing methods
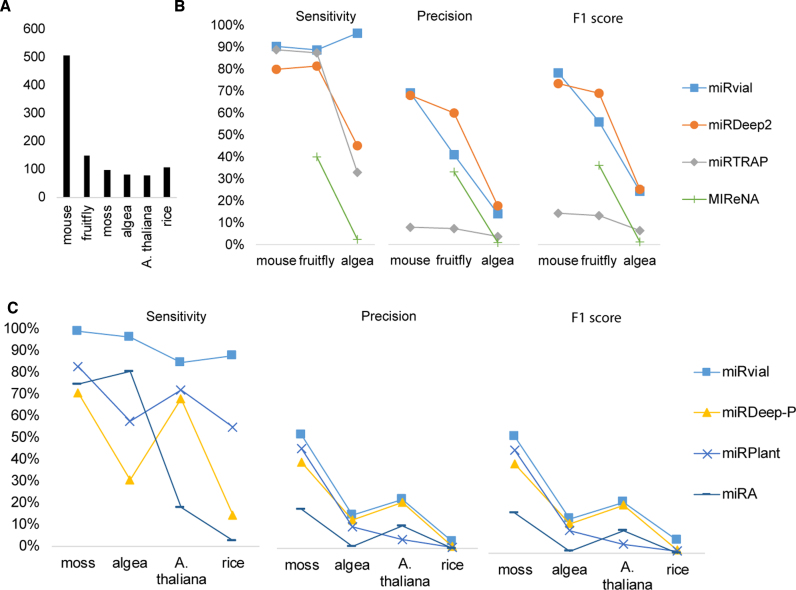
We compared miRvial with six existing methods on six organisms, i.e. M. musculus (mouse), D. melanogaster (fruitfly), A. thaliana, O. sativa (rice), P. patens (moss) and C. reinhardtii (algae). The small RNA-seq data for the first four species were retrieved from public sources (Supplemental Table S1) and that for C. reinhardtii was generated specifically for the current study so that experimental validation of some novel miRNAs could be carried out (to be discussed below). Take mouse as an example; a total of 60 million small RNAs were sequenced from three mouse tissues—brain, ovary, and testes, as well as whole newborn and embryonic day 7.5 (E7.5), E9.5 and E12.5. Much effort has been devoted to annotating and validating murine miRNAs, leading to 506 well-curated canonical and noncanonical miRNAs (10). For algae miRNAs, we used 82 reannotated miRNAs (miRBase version 21) and newly identified miRNAs as true positives. We also used 150, 78, 106 and 98 highly confident miRNAs of fruitfly, Arabidopsis, rice and moss, respectively, from miRBase (version 21) as benchmarks (i.e. true positives) to evaluate the performance of miRNA finding methods compared (Figure 4A).
Figure 4.
Comparison of the methods for miRNA prediction. (A) The known miRNAs in mouse, fruitfly, moss, algae and A. thaliana, as annotated in miRBase or previous studies (see the main text), are used as true positives for comparison. (B) The performance of miRvial, miRDeep2, miRTRAP and MIReNA on three animal species. (C) The performance of miRvial, miRDeep-P, miRPlant and miRA on plant organisms. Sensitivity is true positives divided by the number of known miRNA. Precision is true positives divided by the number of predicted positive miRNAs. F1 score is 2 × precision × sensitivity divided by (precision + sensitivity), which is the harmonic mean of precision and sensitivity.
We used sensitivity, precision and F1 score to quantify the results from the methods compared. We did not use other quality measures, such as specificity or accuracy, because true negatives in the data that we tested were not known. Sensitivity, which has also been commonly referred to as recall, is the ratio of the number of true positive predictions to the number of actual positives in the data. Precision is the ratio of the number of true positive predictions to the total number of predicted positives. F1 score is the harmonic mean of precision and sensitivity.
miRvial has the highest sensitivity in predicting miRNAs in both animals and plants. For example, miRvial has sensitivities of 90.32% and 88.67% in predicting mouse and fruitfly miRNAs, respectively. In comparison, miRDeep2 and miRTRAP have slightly lower sensitivities than miRvial in mouse and fruitfly miRNAs, but fell short to a large extent in algae miRNAs (96% of miRvial versus 45% of miRDeep2 and 33% of miRTRAP, Figure 4B). miReNA is the most lavish among four methods compared in predicting candidate miRNAs in animal species (Figure 4A and B). As a result, miReNA may miss many true miRNAs, resulting in a very low sensitivity on all animal species (Figure 4B). Moreover, miRvial was able to detect 39 of the 45 currently known noncanonical miRNAs in mouse (10), showing a high sensitivity on noncanonical miRNAs.
miRvial also has the highest sensitivity in predicting miRNAs in plant species, especially on rice (87.7%), moss (99.0% sensitivity) and algae (96.3%) (Figure 4C). In comparison, the other three prediction methods have lower sensitivities, exemplified by miRA, which reported too many candidate miRNAs and resulted to a low precision and possibly a high false positive rate; its highest precision is only 16.7 % on moss and its precision is as low as 0.9% on algae (Figure 4C). Surprisingly, even though it predicted many more miRNA candidates than miRvial, e.g. 7250 versus 566 on algae, miRA has a lower sensitivity than miRvial, e.g. 80.4% versus 96.3% on algae, making it noncompetitive among the three methods. Overall, miRvial achieved the highest sensitivity among the methods on all five species, where the other methods fell behind (Figure 4).
Additionally, miRvial also has the highest precisions and F1 scores on four out of six tests. For example, miRvial has 69% precision and 78% F1 score on mouse (Figure 4B), compared favorably with miRDeep2 (68% precision and 73% F1 score) and miRTRAP (8% precision and 14% F1 score). Note that miRvial has higher sensitivities on fruitfly (88.67%) and algae (96.34%), whereas miRDeep2 has lower sensitivities (81.33% in fruitfly and 45.12% in algae). Nevertheless, miRvial was slightly outperformed by miRDeep2 on fruitfly and algae (Figure 4B). For example, miRvial has a slight lower precision than miRDeep2 (14.0% versus 17.5%) on algae, as the latter reported substantially fewer candidate miRNAs, i.e. 211 versus 566 (Figure 4B). Despite its importance in evolution, C. reinhardtii has not been well studied, so that it may have more genuine miRNAs that remain to be identified; five of these novel miRNAs are experimentally validated in the current study, as discussed below. Therefore, the actual precision of miRvial on algae should be higher than what is reported in Figure 4.
Several miRNAs, including some canonical ones, were missed by the other methods, including murine miR-106 and miR-223, which may be functionally important as it is highly expressed in neutrophils (10), reflected by sufficient sequencing reads. Moreover, recent studies reported miRNAs discovered from the fungus Neurospora crassa (30,31), which have atypical RNA structure, i.e. long hairpins (∼300nt) and RNA-RNA duplexes with 5′ (instead of 3′) overhangs. Such miRNAs do not follow the community-accepted miRNA criteria and they were not detected by miRvial and all the other methods that we compared under that stringent criteria.
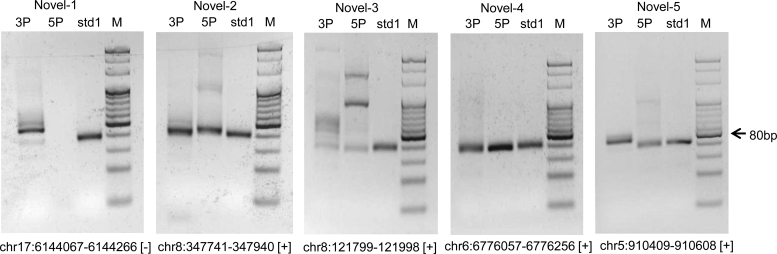
This indicates that the criteria used by these methods, including a RNA/RNA duplex to be supported by some sequencing reads, may be too stringent to facilitate identification of these miRNAs. Some of the criteria can be relaxed to increase the sensitivity of these methods. For example, to increase miRvial's sensitivity, we relaxed the criterion of presence of a duplex so that miRvial reported a set of candidate loci comprising 503 of the 506 (99.4% sensitivity) known murine miRNAs (Supplemental Table S4). Functionally important miRNAs and noncanonical miRNAs, such as miR-223 and miR-451, were rescued by this revision, increasing the effectiveness of miRvial. Additionally, miRvial also identified 142 of the 150 known (95%) miRNAs in D. melanogaster, a 6% increase from the results under the stringent criteria. Finally, miRvial can detect 15 of 25 fungus candidates with less stringent criteria, achieving a sensitivity of 60.0% and a precision of 40.5% (Supplemental Table S4). One caveat is that miRvial allows relaxation of parameters (primarily the presence of sequenced miRNA*) to increase the sensitivity, but doing so may decrease the precision (Supplemental Table S4), indicating the importance of including the duplex rule for finding bona fide miRNAs. To further assess the power of miRvial, we next studied miRNAs in C. reinhardtii using the three methods. We first profiled small-RNA species in C. reinhardtii using deep sequencing (see Methods). We then predicted and prioritized novel miRNAs from all of the three methods based on the small-RNA profiling data, and selected five novel candidate miRNAs which were identified by miRvial but missed by the other methods since miRvial reported more candidate miRNAs than miRDeep2 and miRTRAP is not competitive as discussed above. The five novel miRNAs were experimentally validated by RT-PCR. Reverse transcription fragments for all these five miRNAs were detected (about 80bp with the miRNAs plus the adaptors, Figure 5). In addition, four out of five miRNAs have visible products from both 5p and 3p arms of their hairpin structures, indicating their endogenous expression in the strain of a wild background C. reinhardtii (CC503). This result indicated that the five predicted novel miRNAs were expressed in algae and our miRvial system is effective in identifying new miRNAs.
Figure 5.
RT-PCR validation of five novel miRNAs in Chlamydomonas reinhardtii. The appearances of miRNAs on the 3p- and 5p-arms of five novel miRNA candidates are tested by RT-PCR. The PCR products on 4% agarose gels are marked as below. 3P and 5P: 3p-arms and 5p-arms of miRNAs, respectively; std1: a synthetic exogenous reference gene as a positive control; M: 20 bp DNA marker; the arrow points to the location of 80 bp on the marker; the genomic loci of the miRNAs are listed below the figures.
DISCUSSION
Diversity of microRNAs
It is difficult to have a single definition to succinctly encompass all types of miRNAs, as diverse miRNA biogenesis pathways are prevalent in plants and animals (26,32). Recent evidences show miRNAs can be derived from short hairpins that bypass Drosha processing and enter Dicer machinery directly, such as precursors of mmu-mir-320 (13), snoRNA-derived miRNAs in human (12) and herpesvirus-encoded miRNAs in mouse (18,19,33) and monkey (34). miRNAs are also evolutionarily related to other endogenous small RNAs, particularly siRNAs.
Large structural variations of miRNAs also made it challenging to accurately predict miRNAs even with the data from sequencing-based small-RNA expression profiling. For example, miRNA hairpins are typically longer in plants than in animals, as shown in Figure 2. Mature miRNAs on the 5p- and 3p-arms on miRNA duplexes in plants may thus be widely separated (35). Furthermore, many long miRNA precursors in plants may host multiple, distinct miRNA duplexes and produce genuine miRNAs (16). For example, the length of miR319 precursor in model plant Arabidopsis is ∼170-nt long and can accommodate 3 distinct miRNA duplexes and miRNAs (16). Long hairpin structures are excellent substrates for both miRNA and siRNA processing, giving rise to both miRNAs and siRNAs (32,36). While multiple phased miRNAs are processed by DCL1 on long hairpin substrates, other Dicer-like enzymes (e.g. DCL2, DCL3 and DCL4 in Arabidopsis) are able to process long hairpins as well, giving rise to endo-siRNAs of various sizes (37). Long hairpins occasionally appear in animal genomes. In Drosophila melanogaster, a few miRNAs, e.g. dme-mir-997, may originate from a long hairpin. The precursor of murine miRtron miR-3102 is relatively long (104-nt) and encodes two consecutive miRNAs arranged next to each other (10). On the other hand, long hairpin RNAs may also express to give rise to siRNAs in mammalian cells, e.g. embryonic stem cells and ovaries (38), in which antiviral interferon response is not active for eliminating extensive double-stranded RNAs.
Some poorly annealed hairpins seem to also fit into miRNA biogenesis pathways. For example, some miRtrons with internal loops of 4- or 5-nt are shown to be processed into miRNAs by experimental assays (39). Computational prediction of secondary structures also indicates that mmu-miR-106b and mmu-miR-3070a may carry more unpaired bases than others (as shown in miRBase, version 21), suggesting that Dicer can tolerate a variety of substrates with loosely paired structures (39). Robust computational approaches thus are needed to take into account such atypical structures when identifying hairpin structures. Supported by a visualization tool, miRvial represents a secondary structure with three features and identifies a hairpin structure as long as the features satisfy species-specific criteria that cover a broad range of structures.
Integration of data from multiple sources
Sequencing-based small-RNA expression profiling under multiple conditions, computational analysis and experimental validation assays can be integrated to facilitate accurate identification of miRNAs. Particularly, data from mutants of miRNA microprocessor complexes can significantly enhance identification of genuine miRNAs (13,40). This can help identify miRNAs without extensive analysis of secondary structures when in combination of data from normal conditions, thus providing an orthogonal means to the conventional approaches that heavily depend on structure analysis. Furthermore, it adds confidence to miRNA annotation if the targets of a candidate miRNA are determined (40). To this end, effective assays, such as PAR-clip (41) in animals and Degradome in plants (42), have been devised to detect interactions between a miRNA and its targets. A rich resource in StarBase can be used to identify targets of new candidate miRNAs in some model organisms (43).
Methods without sequencing-based profiling data
A few methods have been developed based only on computational prediction of miRNAs without harnessing sequencing data. In addition to RNA secondary structures, these methods depend upon conservation of miRNAs, since conservation through evolution is a strong filter for genuine and potentially functional genetic units including miRNAs. The most successful methods for computational miRNA finding rely upon conservation of miRNA candidates across related species (44,45). In particular, conserved hairpins that diverge more quickly in their terminal loops relative to the hairpin stems are more likely to be genuine miRNAs (45,46).
The idea of using sequence features in a classifier has also been explored in miRNA prediction. In particularly, sequence features were used in a support vector machines (SVM) based classifier for identifying miRtrons in flies (39). As a complementary approach, these classification-based methods can be used to rank candidates of miRNAs.
DATA AVAILABILITY
The sequencing data of small RNA species in Chlamydomonas reinhardtii that we generated for and analyzed in the current study have been deposited into NCBI Sequence Read Archive (SRA) under accession number of SRP091654.
Supplementary Material
ACKNOWLEDGEMENTS
Author contributions: W.Z. perceived the research and designed the overall study. J.X. and W.Z. designed the experiments and miRvial software and wrote the paper. J.X. and K.Z. developed the miRvial software. J.Z. and H.P. prepared samples of algae and profiled the expression of small RNA species using RNA-seq. T.L. experimentally validated miRNAs in algae. J.X., L.L. and Z.F. compared six miRNA identification tools on five organisms.
Footnotes
Present address: Jing Xia, Yale Stem Cell Center and Department of Cell Biology, Yale University, New Haven, CT 06519, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Talent Development Program of Wuhan; Municipal Government of Wuhan, Hubei, China [2014070504020241]; Jianghan University, Wuhan, China; United States National Institutes of Health [R01GM100364]; United States National Science Foundation [DBI-0743797]. Funding for open access charge: Municipal Government of Wuhan [2014070504020241] and United States National Institutes of Health [R01GM100364].
Conflict of interest statement. None declared.
REFERENCES
- 1. Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009; 136:215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hendrix D., Levine M., Shi W.. miRTRAP, a computational method for the systematic identification of miRNAs from high throughput sequencing data. Genome Biol. 2010; 11:R39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Friedländer M.R., Mackowiak S.D., Li N., Chen W., Rajewsky N.. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012; 40:37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mathelier A., Carbone A.. MIReNA: finding microRNAs with high accuracy and no learning at genome scale and from deep sequencing data. Bioinformatics. 2010; 26:2226–2234. [DOI] [PubMed] [Google Scholar]
- 5. Yang X., Li L.. miRDeep-P: a computational tool for analyzing the microRNA transcriptome in plants. Bioinformatics. 2011; 27:2614–2615. [DOI] [PubMed] [Google Scholar]
- 6. An J., Lai J., Sajjanhar A., Lehman M.L., Nelson C.C.. miRPlant: an integrated tool for identification of plant miRNA from RNA sequencing data. BMC Bioinformatics. 2014; 15:1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Evers M., Huttner M., Dueck A., Meister G., Engelmann J.C.. miRA: adaptable novel miRNA identification in plants using small RNA sequencing data. BMC Bioinformatics. 2015; 16:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Berezikov E., Robine N., Samsonova A., Westholm J.O., Naqvi A., Hung J., Okamura K., Dai Q., Bortolamiol-becet D., Martin R. et al. . Deep annotation of Drosophila melanogaster microRNAs yields insights into their processing, modification, and emergence. Genome Res. 2011; 21:203–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Morin R.D., Connor M.D.O., Griffith M., Kuchenbauer F., Delaney A., Prabhu A. liisa, Zhao Y., Mcdonald H., Zeng T., Hirst M. et al. . Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008; 18:610–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chiang H.R., Schoenfeld L.W., Ruby J.G., Auyeung V.C., Spies N., Baek D., Johnston W.K., Russ C., Luo S., Babiarz J.E. et al. . Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev. 2010; 24:992–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Carthew R.W., Sontheimer E.J.. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009; 136:642–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ender C., Krek A., Friedländer M.R., Beitzinger M., Weinmann L., Chen W., Pfeffer S., Rajewsky N., Meister G.. A human snoRNA with microRNA-like functions. Mol. Cell. 2008; 32:519–528. [DOI] [PubMed] [Google Scholar]
- 13. Babiarz J.E., Ruby J.G., Wang Y., Bartel D.P., Blelloch R.. Mouse ES cells express endogenous shRNAs, siRNAs, and other microprocessor-independent, dicer-dependent small RNAs. Genes Dev. 2008; 22:2773–2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xia J., Joyce C.E., Bowcock A.M., Zhang W.. Noncanonical microRNAs and endogenous siRNAs in normal and psoriatic human skin. Hum. Mol. Genet. 2013; 22:737–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Joyce C.E., Zhou X., Xia J., Ryan C., Thrash B., Menter A., Zhang W., Bowcock A.M.. Deep sequencing of small RNAs from human skin reveals major alterations in the psoriasis miRNAome. Hum. Mol. Genet. 2011; 20:4025–4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang W., Gao S., Zhou X., Xia J., Chellappan P., Zhou X., Zhang X., Jin H.. Multiple distinct small RNAs originate from the same microRNA precursors. Genome Biol. 2010; 11:R81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zeng C., Wang W., Zheng Y., Chen X., Bo W., Song S., Zhang W., Peng M.. Conservation and divergence of microRNAs and their functions in Euphorbiaceous plants. Nucleic Acids Res. 2010; 38:981–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xia J., Zhang W.. Noncanonical MicroRNAs and Endogenous siRNAs in Lytic Infection of Murine Gammaherpesvirus. PLoS One. 2012; 7:e47863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Reese T.A., Xia J., Johnson L.S., Zhou X., Zhang W., Virgin H.W.. Identification of novel microRNA-like molecules generated from herpesvirus and host tRNA transcripts. J. Virol. 2010; 84:10344–10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hofacker I.L. Vienna RNA secondary structure server. Nucleic Acids Res. 2003; 31:3429–3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003; 31:3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Struhl K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat. Struct. Mol. Biol. 2007; 14:103–105. [DOI] [PubMed] [Google Scholar]
- 24. Ding Y., Tang Y., Kwok C.K., Zhang Y., Bevilacqua P.C., Assmann S.M.. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature. 2014; 505:696–700. [DOI] [PubMed] [Google Scholar]
- 25. Kertesz M., Wan Y., Mazor E., Rinn J.L., Nutter R.C., Chang H.Y., Segal E.. Genome-wide measurement of RNA secondary structure in yeast. Nature. 2010; 467:103–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yang J.S., Lai E.C.. Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants. Mol. Cell. 2011; 43:892–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yang J.-S., Maurin T., Robine N., Rasmussen K.D., Jeffrey K.L., Chandwani R., Papapetrou E.P., Sadelain M., O’Carroll D. et al. . Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:15163–15168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cifuentes D., Xue H., Taylor D.W., Patnode H., Mishima Y., Cheloufi S., Ma E., Mane S., Hannon G.J., Lawson N. et al. . A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010; 328:1694–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ruby J.G., Jan C.H., Bartel D.P.. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007; 448:83–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yang Q., Li L., Xue Z., Ye Q., Zhang L., Li S., Liu Y.. Transcription of the major Neurospora crassa microRNA–like small RNAs relies on RNA polymerase III. PLOS Genet. 2013; 9:e1003227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lee H.-C., Li L., Gu W., Xue Z., Crosthwaite S.K., Pertsemlidis A., Lewis Z.A., Freitag M., Selker E.U., Mello C.C. et al. . Diverse pathways generate microRNA-like RNAs and dicer-independent small interfering RNAs in fungi. Mol. Cell. 2010; 38:803–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Axtell M.J., Westholm J.O., Lai E.C.. Vive la différence: biogenesis and evolution of microRNAs in plants and animals. Genome Biol. 2011; 12:221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bogerd H.P., Karnowski H.W., Cai X., Shin J., Pohlers M., Cullen B.R.. A mammalian herpesvirus uses noncanonical expression and processing mechanisms to generate viral MicroRNAs. Mol. Cell. 2010; 37:135–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cazalla D., Xie M., Steitz J.A.. A primate herpesvirus uses the integrator complex to generate viral MicroRNAs. Mol. Cell. 2015; 43:982–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cuperus J.T., Fahlgren N., Carrington J.C.. Evolution and functional diversification of MIRNA genes. Plant Cell. 2011; 23:431–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chellappan P., Xia J., Zhou X., Gao S., Zhang X., Coutino G., Vazquez F., Zhang W., Jin H.. siRNAs from miRNA sites mediate DNA methylation of target genes. Nucleic Acids Res. 2010; 38:6883–6894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rajagopalan R., Vaucheret H., Trejo J., Bartel D.P.. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006; 20:3407–3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Okamura K., Lai E.C.. Endogenous small interfering RNAs in animals. Nat. Rev. Mol. Cell Biol. 2008; 9:673–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chung W.-J., Agius P., Westholm J.O., Chen M., Okamura K., Robine N., Leslie C.S., Lai E.C.. Computational and experimental identification of mirtrons in Drosophila melanogaster and Caenorhabditis elegans. Genome Res. 2011; 21:286–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Meyers B.C., Axtell M.J., Bartel B., Bartel D.P., Baulcombe D., Bowman J.L., Cao X., Carrington J.C., Chen X., Green P.J. et al. . Criteria for annotation of plant MicroRNAs. Plant Cell. 2008; 20:3186–3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hafner M., Landthaler M., Burger L., Khorshid M., Hausser J., Berninger P., Rothballer A., Ascano M., Jungkamp A.-C., Munschauer M. et al. . Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010; 141:129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Addo-Quaye C., Eshoo T.W., Bartel D.P., Axtell M.J.. Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr. Biol. 2008; 18:758–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yang J.-H., Li J.-H., Shao P., Zhou H., Chen Y.-Q., Qu L.-H.. starBase: a database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Res. 2011; 39:D202–D209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Grad Y., Aach J., Hayes G.D., Reinhart B.J., Church G.M., Ruvkun G., Kim J.. Computational and experimental identification of C. elegans microRNAs. Mol. Cell. 2016; 11:1253–1263. [DOI] [PubMed] [Google Scholar]
- 45. Lai E., Tomancak P., Williams R., Rubin G.. Computational identification of Drosophila microRNA genes. Genome Biol. 2003; 4:R42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Berezikov E., Guryev V., van de Belt J., Wienholds E., Plasterk R.H.A., Cuppen E.. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2016; 120:21–24. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data of small RNA species in Chlamydomonas reinhardtii that we generated for and analyzed in the current study have been deposited into NCBI Sequence Read Archive (SRA) under accession number of SRP091654.