Abstract
RNA interference defends against RNA viruses and retro-elements within an organism's genome. It is triggered by duplex siRNAs, of which one strand is selected to confer sequence-specificity to the RNA induced silencing complex (RISC). In Drosophila, Dicer-2 (Dcr-2) and the double-stranded RNA binding domain (dsRBD) protein R2D2 form the RISC loading complex (RLC) and select one strand of exogenous siRNAs according to the relative thermodynamic stability of base-pairing at either end. Through genome editing we demonstrate that Loqs-PD, the Drosophila homolog of human TAR RNA binding protein (TRBP) and a paralog of R2D2, forms an alternative RLC with Dcr-2 that is required for strand choice of endogenous siRNAs in S2 cells. Two canonical dsRBDs in Loqs-PD bind to siRNAs with enhanced affinity compared to miRNA/miRNA* duplexes. Structural analysis, NMR and biophysical experiments indicate that the Loqs-PD dsRBDs can slide along the RNA duplex to the ends of the siRNA. A moderate but notable binding preference for the thermodynamically more stable siRNA end by Loqs-PD alone is greatly amplified in complex with Dcr-2 to initiate strand discrimination by asymmetry sensing in the RLC.
INTRODUCTION
RNA interference (RNAi) is a sequence-specific RNA degradation process that serves to defend against invading RNA viruses (1–3) and retro-transposons (4–7). RNAi is triggered by double-stranded RNA (dsRNA), which is processed into 21 nt siRNA duplexes with 2 nt 3′-overhangs by the RNase III family enzyme Dicer. In Drosophila melanogaster, Dcr-2 is dedicated to the processing of dsRNA into siRNAs, whereas Dcr-1 operates in the related microRNA (miRNA) pathway. A common theme in the biogenesis of both miRNAs and siRNAs is the pairing of an RNaseIII-family nuclease with a small partner protein that contains two or more instances of the dsRNA binding domain (dsRBD) (8–14). While virus-derived sequences are called siRNAs, the transposon-targeting sequences are usually referred to as endo-siRNAs to point out their endogenous origin.
The dsRBD has a conserved fold with αßßßα topology (15,16) that mediates binding to double-stranded RNA molecules as well as protein–protein interactions (17). Structural studies of RNA-bound domains revealed mainly contacts to the ribose-phosphate backbone of an A-form RNA helix without sequence-specificity (18–20). In certain cases, particular deviations from a perfect dsRNA helix can be specifically recognized by a dsRBD (21). Despite the fact that dsRBDs often occur in multiple instances within the same protein or within a protein complex, only few studies have examined their interplay at the structural level (20–24) or analyzed the roles of tandem dsRBD for RNA binding (18,22,25,26). It is thus important to characterize the molecular function and biological activity of tandem dsRBD domains using structural and biophysical approaches combined with functional analysis.
Although the structure of an siRNA duplex is pseudo-symmetric, one strand is preferentially incorporated into RISC. Strand preference depends on the relative stability of base-pairing at either end of the siRNA (referred to as thermodynamic asymmetry sensing), such that the less stably base-paired 5′-end preferentially enters the Argonaute protein. Several mechanisms for strand selection can operate, such as 5′-nucleotide preference (e.g. 5′-U) of the Ago-protein sensing of unpaired nucleotides or phosphorylation state (for synthetic siRNAs) or the formation of specific protein complexes that aid during Ago-loading (27–31). In Drosophila, loading of Ago2 with siRNAs depends on the RISC loading complex (RLC) composed of Dcr-2 and the dsRBD protein R2D2 (32–35). The RLC but not Dcr-2 alone binds asymmetrically to duplex siRNAs. Therefore, it has been proposed that R2D2 serves as the sensor for asymmetry detection (32). In humans, it appears that the TRBP/Dicer complex can bind siRNAs asymmetrically (36,37) but this may not be required during small RNA loading (34,38,39). Biochemical studies have demonstrated partial redundancy between the paralogs Loqs-PD and R2D2 (40,41) and deep sequencing experiments have revealed that some Ago2-loaded siRNAs are present even in r2d2 null mutant animals (42). However, the specific function of Loqs-PD during RISC loading has not been clarified.
Here, we have performed deep sequencing of small RNAs after genome editing in cultured S2-cells to demonstrate that R2D2 can be replaced by its paralog Loqs-PD for loading of endogenous and exogenous siRNAs but not for loading of Ago2-tropic miRNAs. NMR-derived solution structures show that the Loqs dsRBD domains adopt canonical dsRBD folds. Single and tandem dsRBDs can bind independently to RNA with moderate preference for siRNA over pre-miRNA or miRNA/miRNA* substrates. Site-specific cross-linking is most efficient at the siRNA ends and demonstrates that the complex of Dcr-2 with Loqs-PD possesses strand discrimination activity analogous to Dcr-2/R2D2. Notably, NMR data indicate that the dsRBDs slide along the double-stranded RNA helix and do not mediate sequence-specific contacts to the RNA. The tandem dsRBDs of Loqs have a weak but notable preference to bind the more stably base-paired end of an siRNA duplex. This moderate preference is strongly enhanced in the Dcr-2/Loqs-PD complex, indicating their cooperation during the strand selection process. Identification of Loqs-PD as the alternative RISC loading complex subunit has thus allowed us to examine the role of both individual Drosophila RLC components during the initiation of guide strand selection. Our results indicate that the dsRBD protein serves as siRNA asymmetry sensor for the alternative RLC, and demonstrate that the combined action of Dcr-2 with Loqs-PD is essential for faithful strand discrimination.
MATERIALS AND METHODS
Protein expression, purification
For expression of full length Loqs-PD (residues 1–359), truncated Loqs-PDΔN (residues 129–359), Loqs-PDΔNC (residues 129–322) and the two dsRNA-binding domains dsRBD1 (residues 129–211) and dsRBD2 (residues 245–322) a modified pET-M11 vector with an N-terminal His6-ZZ-tag and a following tobacco etch virus (TEV) protease cleavage site was used. All plasmids were verified by sequencing, transformed into Escherichia coli BL21 (DE3) cells for protein expression and cultivated in LB or minimal M9T medium containing either or both of 2g/l [U-13C]-glucose and 1g/l 15N ammonium chloride. See supplementary methods for details on the purification procedure. His6-tagged Dcr-2 was expressed in Sf21 cells using the BAC-to-BAC Baculovirus Expression System (Invitrogen), purified via a HisTrap HP column (GE Healthcare) (8) followed by a HiPrep 16/60 Sephacryl S200 gel filtration column (GE Healthcare).
NMR spectroscopy
Proteins and protein–RNA complexes were studied in sodium phosphate buffer pH 6.5 and 7.2. 5% D2O was added for locking. Protein samples were measured at 298 K and imino NOESY spectra of RNA were recorded at 278 or 298 K. Spectra were recorded on Bruker AVIII 500, AVIII 600, AVIII 750, AVIII 800 and AVIII 900 NMR spectrometers with cryogenic (TCI) triple resonance gradient probes, or standard TXI probes for AVIII 750. Data were processed using Topspin 3.0 and NMRPipe/Draw (43) and analyzed with Sparky 3 (T.D. Goddard and D.G. Kneller, SPARKY 3, University of California, San Francisco, USA). Backbone assignments were derived from HNCA, HNCACB, HNCACO, HNCO and HNCOCACB experiments, side chain assignments were obtained from HCCH-TOCSY, 15N- and 13C-edited NOESY-HSQC experiments (44). NMR titration experiments were performed with individual dsRBD1 or dsRBD2 domains and 21-nt endo-siRNA with 2 nt 3′ overhangs (ban-siRNA), (5′-UCAGCUUUCAAAAUGAUCUCACU-3′, 5′-UGAGAUCAUUUUGAAAGCUGAUU-3′) at protein concentrations of 100 μM with protein:RNA ratios as follows: 1:0, 1:0.25, 1:0.5, 1:0.75, 1:1, 1:1.25. For Loqs-PDΔNC protein concentrations of 200 μM and molar ratios of 1:0, 1:0.12, 1:0.47, 1:0.7, 1:0.9, 1:1.1 were used. See Supplementary Information for further details.
Binding assays
Please refer to the Supplementary Information for details on RNA sample preparation. 10 nM FITC-labeled RNA oligos were incubated in binding buffer (10 mM Hepes, pH 7.4, 100 mM KAc, 2 mM MgAc and 5 mM DTT) with increasing amounts of recombinant protein for 30 min at RT in a black, non-binding 96-well plate (Greiner bio-one). Anisotropy values were recorded in an Infinite M1000 plate reader (Tecan). For EMSA experiments, binding reactions were mixed 4:1 with binding buffer supplemented with 50% glycerol and resolved on native 4% PAA gels for 20–24 min at 200V. A Typhoon 9400 variable mode imager (GE Healthcare) was used to image the fluorescence signal. Quantification of EMSA bands was performed with Multigauge V3.0 software (FujiFilm), and binding curves were fitted with the Hill equation in Origin 8.1.
Crosslinking experiments
The substrate oligonucleotides were radioactively labeled using polynucleotide kinase (Fermentas) and γ32P-ATP; the non-labeled strands were phosphorylated with non-radioactive ATP. 10nM radioactively labeled, thiouridine containing siRNA duplexes were incubated with 1 μM dsRBD1, 1 μM dsRBD2, 500 nM LoqsPDΔNC, 500 nM LoqsPD or 250 nM Dcr-2/LoqsPD complex for 30 min at room temperature. Crosslinking was induced with 3 × 500 mJ/cm2 of 365 nm wavelength light, and the samples were resolved on a 10 or 15% PAA gel for 1 h at 170 V. The radioactive signals were read out using a phosphorimager screen on a Typhoon imager and quantified using Multigauge V3.0 software.
Cell culture and deep sequencing experiments
Drosophila S2-cells were grown and transfected in Schneider's medium as described before (45). For genome editing, a cas9 expression plasmid was co-transfected with a PCR-generated sgRNA expression cassette. One week later, the cells were diluted, clones were picked, amplified and the induced mutations were characterized. Further details and oligonucleotide sequences are provided in the supplementary section. The cells were then treated with 300 ng/ml of dsRNA targeting Renilla luciferase, 100 ng/ml of dsRNA targeting firefly luciferase, 30 ng/ml of dsRNA targeting GFP and 10 ng/ml of dsRNA targeting DsRed. Total RNA was isolated 3 days later and processed for comparative Illumina sequencing with and without a β-elimination step as previously described (46). To compare the different sequencing datasets, a normalization to the sum of all miRNA-matching reads was performed. Further details on data analysis are given in the supplementary section.
RESULTS
R2D2 is dispensable for loading of siRNAs into Ago2 in Drosophila S2-cells
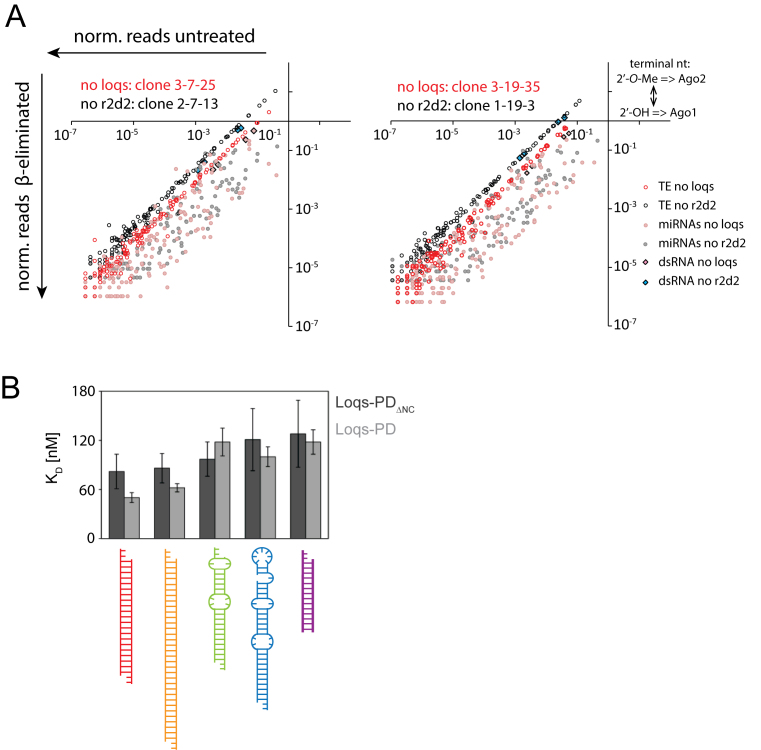
R2D2 was originally identified via co-purification with Dcr-2. Genetic analysis revealed that it is required for the loading, but not the production, of siRNAs in Drosophila embryos (8). Subsequent studies indicated that the PD isoform of Loqs, an R2D2 paralog involved in the endo-siRNA pathway, can also interact with Dcr-2 (47–49). Based on data obtained with mutant flies, a sequential model was proposed where Loqs-PD interacts with Dcr-2 during the dicing reaction while R2D2 is an obligatory partner during the loading reaction of all siRNAs (40,48,50,51). In S2 cells, however, R2D2 was dispensable for the function of endogenous siRNAs but required for full silencing activity of experimentally introduced dsRNA (52,53), thus arguing for the existence of an alternative RLC for endo-siRNAs (Supplementary Figure S1A).
To clarify whether this discrepancy was due to technical differences (depletion of factors via RNAi in cell culture vs. mutant alleles in vivo) we generated r2d2 as well as loqs mutant cell lines via CRISPR-cas9 mediated genome editing. Absence of the proteins was verified by sequencing of the mutated loci (data not shown) and western blotting (Supplementary Figure S1B and data not shown). We analyzed the small RNA content via deep sequencing, exploiting the fact that the 2′-O-methyl modified, Ago2-loaded siRNAs are resistant to oxidation by periodate (54,55) to determine their loading state. A comparison of the predominantly Ago1-loaded miRNAs (sensitive to oxidation and β-elimination) with Ago2-loaded transposon-targeting endo-siRNAs allows to assess the functionality of RISC loading complex. Strikingly, we found that both endogenous and exogenous siRNAs can be loaded normally in the complete absence of r2d2. When loqs was inactivated, however, the endogenous siRNAs were partially sensitive to β-elimination (Figure 1A). This effect was consistently observed in three independent experiments involving two distinct loqs and r2d2 mutant cell clones (Supplementary Figure S2). Based on the axis intercept of trendlines through the TE-matching endo-siRNA data points, we estimate that the amount of Ago2-loaded siRNAs was 3.5 ± 1.2 fold lower in loqs mutant cells (average ± SD, three biological replicates, Supplementary Figure S2A–C). The same was true for siRNAs derived from exogenously added dsRNA but not for Ago2-tropic miRNAs such as miR-276-5p or miR-277-3p (Supplementary Figure S2D). R2D2 is nonetheless required for full efficiency of experimental RNAi in S2-cells (52), indicating that the two paralogs can function redundantly in siRNA loading. The remaining transposon-targeting siRNAs in either mutant showed no difference in the average ΔΔG (around 1 kcal/mol in all cases, range was from 0.90 to 1.05 kcal/mol). Furthermore, the 5′-nucleotide bias was essentially the same (∼38% U, ∼29%A, ∼13%G and ∼20%C) for all genotypes. We conclude that the remaining transposon-targeting endo-siRNAs are bona fide Ago2 loaded, consistent with our 2′-O-methylation state analysis.
Figure 1.
The PD isoform of Loqs is of central importance for siRNA biogenesis in S2-cells. (A) Small RNA sequencing libraries were created from genomically engineered S2 cell lines lacking either r2d2 (lines 1-19-3 and 2-7-13, black or grey) or loqs (lines 3-19-35 and 3-7-25, red or pink). For both cell lines, libraries were generated directly from total RNA and after oxidation/beta-elimination to assess Ago2 loading state. Small RNAs were summed up for each transposon consensus sequence (open circles, transposon sequences can be several kb long) or miRNA (filled circles). Transposon-targeting endo-siRNAs and siRNAs derived from exogenously added siRNAs remained preferentially Ago2-loaded ( = separated from miRNAs) in the absence of r2d2 whereas the absence of loqs reduced their association with Ago2. The loqs mutation leads to a mild reduction in mature miRNA and siRNA levels because of its role during the nucleolytic processing steps (Dcr-1/Loqs-PB and Dcr-2/Loqs-PD complexes). These effects affect miRNAs and siRNAs comparably, though (see also Supplementary Figure S2) and are thus compensated in the miRNA-normalized data shown here. (B) Binding affinities for recombinant Loqs-PDΔNC (dark gray) and full-length Loqs-PD (light gray) determined by fluorescence anisotropy measurements for different RNA substrates; see Table 1 for the binding affinities and Supplementary Figure S3A for titration curves.
While loqs is required to silence a transgene-derived endo-siRNA reporter, r2d2 is required for silencing by the preferentially Ago2-loaded miR-277 (Supplementary Figure S1C). Thus, S2-cells apparently have two partially reduntant RISC loading complexes: Dcr-2/R2D2 and Dcr-2/Loqs-PD.
Loqs dsRBDs bind dsRNA independently
It has been difficult to study the biochemical and structural properties of the dsRBD-containing RLC component in the past because recombinant production of R2D2 is challenging in the absence of Dcr-2. Fortunately, we could express Loqs-PD as well as its individual dsRNA binding domains. Fluorescence anisotropy measurements were performed to determine the binding affinity of the Loqs dsRBDs to various RNA substrates (Figure 1B, Supplementary Figure S3A). The sequence of our model substrates is derived from the Drosophila miRNA bantam, which was modified to mimic an siRNA duplex (21-nt siRNA duplex with 2 nt 3′ overhangs, 23 nt strand length, which will be termed ban-siRNA in the following) and a longer dsRNA (35 nt strand length). The individual dsRBDs bind their substrates with comparable affinities (Supplementary Figure S3B and Tables 1 and 4). There is no pronounced difference in binding affinity between the different RNA constructs, which represent both the substrates and the products of dicing reactions (see supplement for further information).
Table 1. Binding affinities determined by fluorescence anisotropy [nM].
| RNA | Loqs dsRBD1 | Loqs dsRBD2 | LoqsΔNC | Loqs-PD |
|---|---|---|---|---|
| siRNA | 228 ± 27 | 278 ± 24 | 82 ± 21 | 50 ± 6 |
| dsRNA | 191 ± 29 | 229 ± 48 | 86 ± 18 | 62 ± 5 |
| miRNA | 233 ± 17 | 241 ± 75 | 97 ± 21 | 118 ± 17 |
| pre-miRNA | 221 ± 43 | 218 ± 61 | 121 ± 38 | 100 ± 12 |
| 14 ntRNA | 365 ± 62 | 334 ± 8 | 128 ± 41 | 118 ± 15 |
Table 4. Hill coefficients obtained from fitting the fluorescence anisotropy data.
| RNA | Loqs dsRBD1 | Loqs dsRBD2 | LoqsΔNC | Loqs-PD |
|---|---|---|---|---|
| siRNA | 7.9 ± 3.0 | 3.4 ± 2.1 | 3.3 ± 0.9 | 1.3 ± 0.2 |
| dsRNA | 3.9 ± 1.0 | 3.0 ± 1.2 | 3.2 ± 0.8 | 2.5 ± 0.2 |
| miRNA | 5.1 ± 1.6 | 3.8 ± 1.6 | 5.1 ± 2.9 | 3.3 ± 0.8 |
| pre-miRNA | 4.5 ± 0.8 | 2.6 ± 0.7 | 4.6 ± 0.9 | 2.4 ± 0.7 |
| 14 ntRNA | 7.7 ± 2.4 | 2.0 ±1.7 | 3.9 ± 1.0 | 2.9 ± 0.7 |
The RNA binding region of Loqs proteins comprises the tandem dsRBDs, which are connected by a 45-residue linker (LoqsΔNC). When comparing the binding of single and tandem dsRBDs only a moderate increase in binding affinity is observed. The gain in affinity is ∼3-fold for the construct containing only the two domains connected via the linker and ∼5-fold for full-length Loqs-PD (Table 1). The increase in affinity for the two dsRBDs together is clearly less than expected for additive binding, which should have led to a sub-picomolar affinity based on our measurements for the single dsRBDs (56). We thus conclude that RNA binding does not involve concomitant engagement of the two dsRBDs, which rather appear to bind independently to the RNA.
Loqs dsRBDs adopt a dsRBD fold with a canonical RNA binding interface
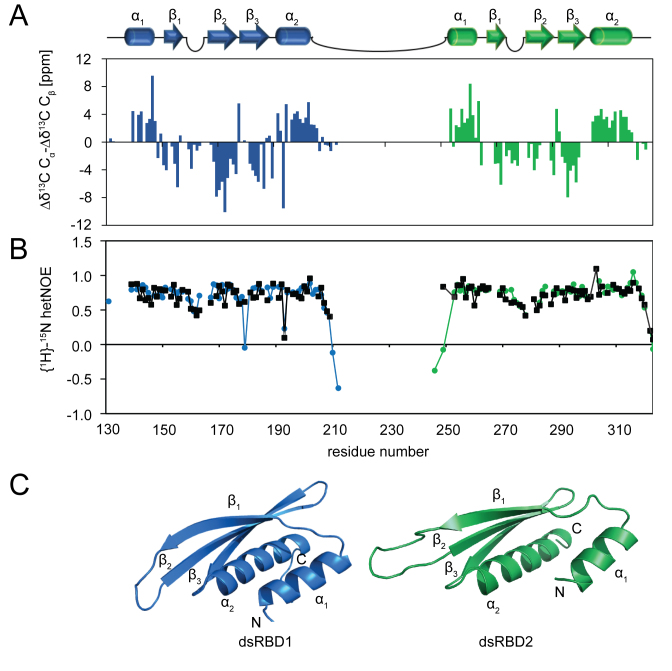
To analyze molecular details of the LoqsPD-dsRNA interaction we determined the structures of the first and second dsRBD, which are common to all Loqs isoforms (Supplementary Figure S1A). NMR 1H,15N-correlation spectra of the single domains superimpose very well with those of Loqs-PDΔNC (Supplementary Figure S4A). This demonstrates that the individual dsRBDs do not strongly interact in solution in the absence of RNA, and is consistent with NMR 15N relaxation data (Supplementary Figure S4B). {1H}–15N heteronuclear NOE data (Figure 2B) indicate that the two domains are connected by a flexible linker (residues 206–251) that allows the domains to tumble independently in solution. The structures of the two dsRBDs were determined using ROSETTA and the NOE-based RASREC-ROSETTA protocol (57). Structural statistics are shown in Table 2. Both dsRBDs exhibit the canonical αβββα-topology (15,16) with two helices packed from one side onto a triple-stranded β-sheet connected by short flexible loops (Figure 2A and C).
Figure 2.
NMR analysis and structures of Loqs dsRBD1 and dsRBD2. (A) NMR 13C secondary chemical shifts (Δδ13Cα-Δδ13Cβ) of the single domains (dsRBD1 in blue, dsRBD2 in green) indicate that both Loqs dsRBDs exhibit the conserved αβββα topology. Secondary structure elements are depicted as cartoons on top. (B) Low {1H}–15N heteronuclear NOE data show that the linker connecting dsRBD1 and dsRBD2 is highly flexible in solution. Specific loops that contribute to RNA binding in the two dsRBDs also exhibit some flexibility. Data for single domains are shown in black, data for the tandem construct are colored in blue (dsRBD1) and green (dsRBD2). (C) The NMR-derived structures show that the Loqs dsRBDs adopt a canonical dsRBD fold. Ensembles of the 10 lowest energy structures are shown in Supplementary Figure S4C.
Table 2. Structural statistics for the NMR structures of dsRBD1 and dsRBD2.
| dsRBD1 (aa 137-205) | dsRBD2 (aa 215–318) | |
|---|---|---|
| Rosetta structural statistics | ||
| NOEs | ||
| With unique assignment | 887 | |
| With intra-residue assignment | 559 | |
| With sequential assignment |i – j| = 1 | 180 | |
| With medium-range assignment 1< |i – j| < 5 | 52 | |
| With long-range assignment |i – j| > 5 | 96 | |
| Eliminated due to distance violations | 89 | |
| Eliminated due to network, minimal peak volume and maximal assignments | 81 | |
| Coordinate precision r.m.s.d. [Å] a | ||
| Backbone | 2.12 | 1.04 |
| Heavy atom | 8.46 | 5.79 |
| Ramachandran statistics [%] b | ||
| Favored regions | 99.3 | 96.8 |
| Allowed regions | 0.6 | 3.1 |
| Outlier regions | 0.1 | 0.2 |
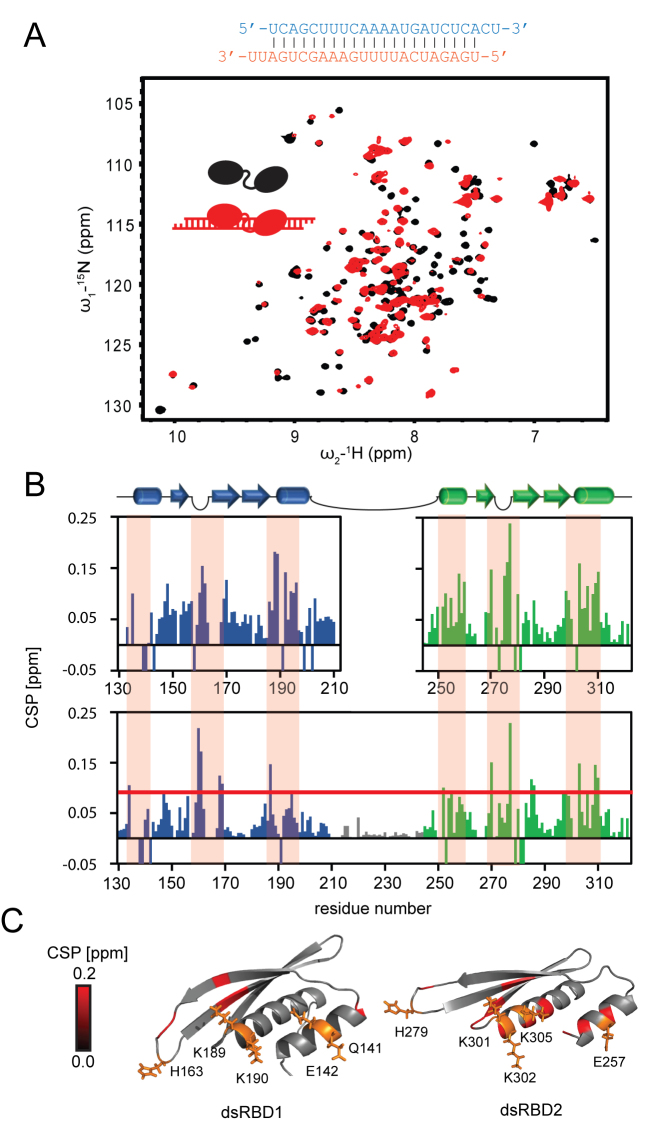
To map the RNA binding interface on the dsRBDs we performed NMR titrations with unlabeled RNA to 15N-labeled single dsRBD1 and dsRBD2 domains as well as the tandem domain construct Loqs-PDΔNC. Significant changes in NMR chemical shifts are observed upon addition of a ban-siRNA (Figure 3A and B). Mapping of the NMR chemical shift perturbations (CSPs) onto the structures shows that three regions are involved in dsRNA binding: the two helices (α1, α2 and α3, α4 in dsRBD1 and dsRBD2, respectively), and the loops connecting strands β2–β3 and β5–β6, respectively, in each domain (Figure 3C). Similar CSPs are observed upon titration of single domains or the tandem domain (Figure 3B), confirming that both domains interact individually with the RNA with the canonical binding surface. Importantly, the NMR titrations demonstrate that the linker connecting dsRBD1 and dsRBD2 remains flexible and does not mediate contacts to the RNA.
Figure 3.
Loqs dsRBD1 and 2 bind siRNA independently and via conserved binding mode. (A) 1H,15N correlation NMR spectra of Loqs-PDΔNC free (black) and bound to ban-siRNA (red), the RNA sequence is shown above. (B) Upon titration of ban-siRNA the amide signals in the tandem dsRBD protein exhibit significant chemical shift perturbations (CSP), while the linker (gray) is not affected. The single domains titrated with the same RNA (graphs on top) show a similar pattern of CSP demonstrating that both domains bind RNA independently. Negative bars indicate broadened lines and non-traceable signals. Residues with CSP above the red line (mean ± SD) are highlighted in C. The orange shaded boxes indicate main RNA binding regions. (C) The CSP induced by RNA binding are plotted on the domain structures and reveal three binding regions: The first helix (α1, α3), the loop (L1, L2) and the N terminal part of the second helix (α2, α4) are involved in RNA binding. Loqs thus binds via the typical dsRBD interface. Side chains of typical conserved RNA binding residues are shown as orange sticks. We confirmed that the Loqs dsRBD1 and dsRBD2 mediate canonical interactions with the dsRNA by similarity searches using DALI (77). Both Loqs dsRBDs superimpose well with dsRBD-dsRNA complex structures from the respective top hits (Supplementary Figure S4D, Supplementary Table S1).
Loqs-PD slides along the siRNA duplex with preferential binding at the siRNA termini
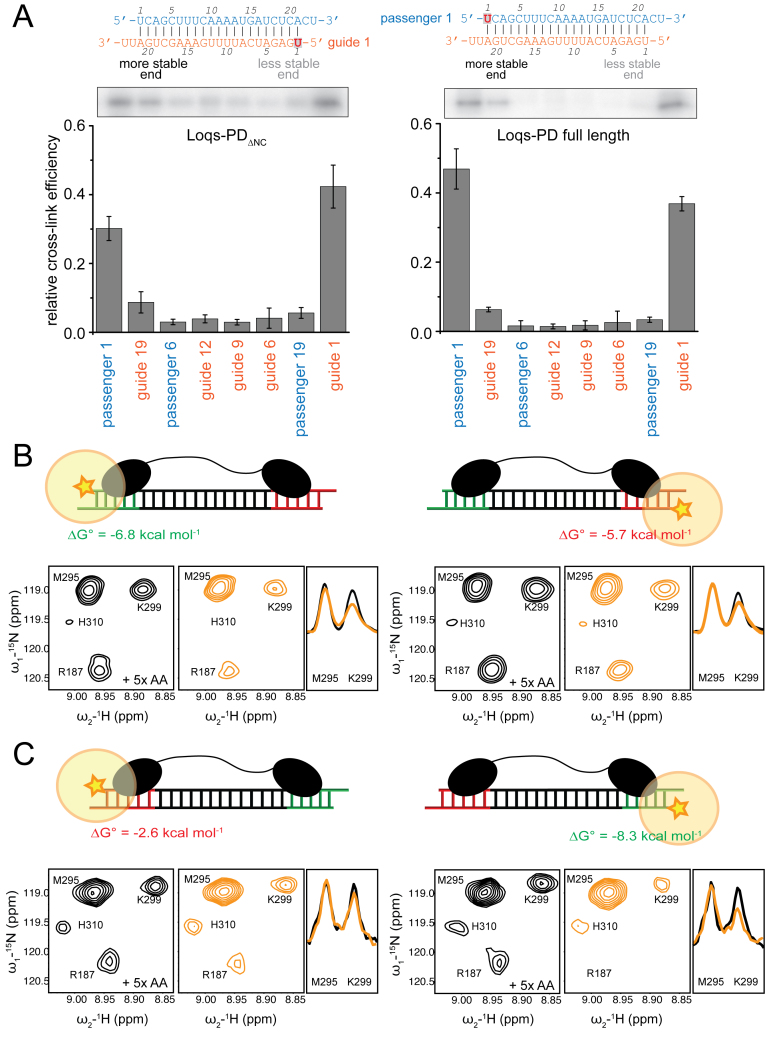
Based on our analysis of S2-cell RNA (Figure 1A) we expected that the Dcr-2/Loqs-PD complex can perform a similar function as the Dcr-2/R2D2 complex, which interprets the difference in base-pairing stability at either end of the siRNA (32). To test this hypothesis, we employed 4-thio-uridine labeled synthetic RNA substrates to allow site-specific protein–RNA cross-links. We created a series of substrates that report protein binding along the entire siRNA molecule (Supplementary Figure S5A). For both Loqs-PDΔNC and full-length Loqs-PD, we found that the cross-linking efficiency is higher at either end of the RNA duplex than in the center; this was true even when the corresponding mismatch-containing miRNA/miRNA* duplex was used (data not shown). In this assay, a clear preference for the more stably base-paired end of the duplex was not detected at any of the interrogated positions (Figure 4A), perhaps due to the fact that the thermodynamic asymmetry of this model duplex is moderate as A–U versus G–C base pair differences occur only at the fifth position. The same cross-linking profile was observed for the individual dsRBDs (Supplementary Figure S5B).
Figure 4.
Loqs-PD preferentially resides at the termini of a duplex siRNA. (A) Site-specific cross-linking experiments were carried out with site-specifically 4-thio-uridine labeled RNAs (see Supplementary Figure S5 for further details). The RNAs were radioactively labeled, incubated with recombinant protein, cross-linked with 365 nm UV light and separated by gel electrophoresis. Both full-length Loqs-PD and Loqs-PDΔNC preferentially react with labeled uridine-residues at either end of the dsRNA. The bar graphs represent a quantitative analysis of three independent experiments (mean ± SD). (B) Intermolecular paramagnetic relaxation enhancements (PREs (Supplementary Figure S6A) observed in 1H,15N HSQC NMR spectra with 15N-labeled Loqs-PD bound to an RNA duplex with a TEMPO-spin label (orange star in the schematic pictures) attached to either the more stable (left) or the less stable (right) 5′ end. Zoomed views of NMR spectra in the diamagnetic and paramagnetic state, are shown in black and orange, respectively. A stronger reduction in signal intensities for the paramagnetic state observed on the left indicates a preferential binding to the more stable end of the siRNA. For the M295 and K299 amide 1D traces taken from the 2D spectra are shown on the right to demonstrate the PRE-induced line-broadening. (C) Intermolecular PREs observed for a highly asymmetric RNA. Note, that the thermodynamic stability of this RNA duplex is inverted compared to (B). The significantly stronger PRE effects observed relate to the larger thermodynamic asymmetry of the duplex. The RNA sequence and experimental PRE data are shown in Supplementary Figure S6B.
The current model for the Dcr-2/R2D2 RLC postulates that R2D2 initiates strand selection by binding to the more stably base-paired end, although the molecular mechanism for this is unknown. To explore whether the Loqs-PD subunit of the alternative RLC may have an intrinsic preference for the more stably base-paired siRNA we analyzed intermolecular NMR paramagnetic relaxation enhancements (PREs). PREs were measured by the attachment of a paramagnetic TEMPO spin label separately to either 5′ end of the RNA duplex (58,59). The spin label causes distance-dependent line-broadening of NMR signals within a range of up to ≈20 Å distance and the resulting PREs thus report on spatial proximity between protein and the RNA-attached spin label. PRE measurements are more sensitive to detect weak differences (60,61) than the biochemical approaches using site-specific cross-linking. PREs were measured by comparing 1H,15N correlation NMR experiments of the Loqs-PDΔNC protein bound to ban-siRNA with terminally attached TEMPO spin labels in the paramagnetic and diamagnetic state (58). Moderate intermolecular PREs are observed for several protein residues (R187, K299, H310) when the spin label is attached to the more stably base-paired end of the RNA (G1 in Figure 4B, Supplementary Figure S6A), indicating a stronger interaction with this region of the RNA duplex. In contrast, only very weak PRE effects are detectable in similar regions when a spin label is attached to the less stably base-paired end (G24 in Figure 4B, Supplementary Figure S6A). To further support this conclusion, we designed an siRNA duplex with a very pronounced thermodynamic asymmetry (Figure 4C, Supplementary Figure S6B). Compared to the ban-siRNA this highly asymmetric duplex shows substantially increased PRE effects to the more stable end of the RNA duplex, thus supporting the notion that the dsRBDs sense the thermodynamically more stable regions of a RNA helix. These data demonstrate that Loqs-PD is not bound with comparably affinity to the two termini of the RNA duplex but has a small, but notable intrinsic preference to bind to the more stably base-paired end.
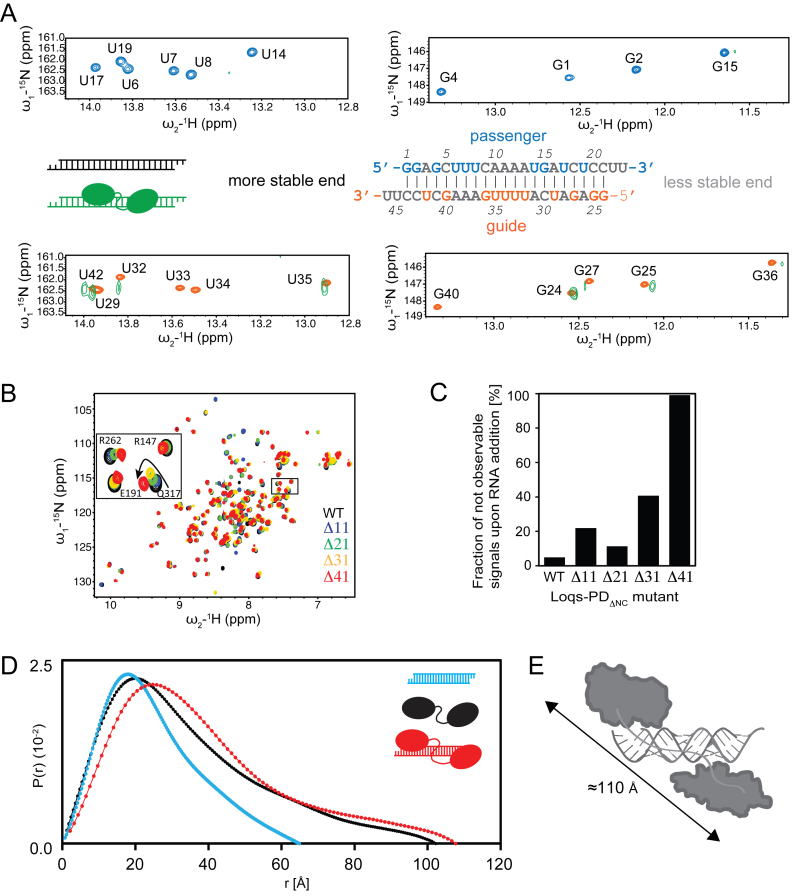
We compared imino NOESY and 1H,15N imino correlation spectra of the free and protein-bound RNA to map the Loqs-PD binding on to the siRNA duplex. Upon addition of unlabeled Loqs-PDΔNC protein all NMR signals from the observed isotope-labeled RNA show severe line broadening (Figure 5A). Imino NMR signals of several guanine and uracil bases could still be traced, while all uracil imino signals in the U-rich central region (U33, U34) are broadened beyond detection (Figure 5A). This likely is an indication of sliding of the protein along the double-helical RNA duplex, where varying chemical environments in the different dynamically exchanging binding modes cause line-broadening. The NMR chemical shift perturbations thus demonstrate that Loqs exhibits a highly dynamic binding to the dsRNA that involves sliding along the RNA duplex.
Figure 5.
Loqs slides along RNA and the linker is needed for proper domain arrangement. (A) Zoomed view of 1H,15N NMR correlation spectra of the imino region of the ban-siRNA in the free (blue and orange, depending on which RNA strand is isotope-labeled and thus NMR observable) and protein-bound (green) form. Imino signals in the less stable 5′ region (i.e. residues 15–23 in the passenger and 24–36 in the guide strand) experience less line-broadening (reflecting weaker interactions with the protein) than those in the more stable end. The line-broadening observed indicates sliding of the protein along the RNA. Note, that only bases (G, U) with imino protons are detectable, while A and C bases are not detectable in this experiment and are therefore shown in light gray in the secondary structure of the RNA. (B) Superposition of 1H,15N HSQC NMR spectra of wildtype Loqs-PDΔNC free (black) and linker mutants as indicated by different colors. The zoomed view shows that shortening of the linker leads to chemical shift changes along a linear trajectory, while a complete deletion of the linker causes strong chemical shift changes in a different direction (red), indicating significant structural perturbation or rigidification of the domain arrangements in the tandem dsRBDs upon linker deletion. (C) The fraction of unobservable NMR signals in 1H,15N NMR correlation spectra in the presence of RNA for tandem dsRBD proteins, wildtype and with different linker deletions is shown. (D) SAXS-derived pairwise distance distribution P(r) for siRNA (blue), Loqs-PDΔNC(black) and the 1:1 complex (red).
Dynamic siRNA recognition by the Loqs-PD tandem dsRBDs
To explore whether the linker plays a role for the dynamic siRNA recognition, we prepared Loqs-PDΔNC mutants with varying linker lengths (Δ11, Δ21, Δ31, Δ41) connecting the two dsRBDs (Supplementary Figure S7A). Potential interactions of the dsRBDs where analyzed by comparing NMR chemical shifts in 1H,15N correlation spectra (Figure 5B, Supplementary Figure S7B). Analysis of the linker deletion tandem domains in the absence of RNA shows only negligible changes in the chemical shifts of the individual domains. However, an almost complete deletion of the linker (Δ41) leads to more pronounced effects on the NMR spectra indicating that the relative domain arrangement is affected, presumably by steric strain. A significant compaction with decreasing linker length is also indicated by SAXS analysis (Supplementary Figure S7D). When bound to siRNA, the protein–RNA binding interface is not much affected by shortening of the linker as similar chemical shift perturbations are observed upon RNA binding independent of the linker length. Nevertheless, a gradual shortening of the linker leads to increased line broadening, which suggests more pronounced sliding of the protein on the dsRNA ligand (Figure 5B and C, Supplementary Figure S7B and C). Most likely, the protein with a shortened linker cannot adopt a domain arrangement required for optimal RNA contacts of both domains and switches thus to a dynamic binding mode where sometimes only one of the dsRBDs may be bound to the RNA. Consistent with this fluorescence anisotropy experiments show that the binding affinity of the Δ41 linker deletion with our dsRNA model substrates exhibits a two-fold reduction compared to the wild type linker, and is thus comparable with a single dsRBD (Table 1; Supplementary Figure S7F).
To characterize the overall shape of the protein–RNA complex we compared SAXS experiments for the free tandem dsRBD protein Loqs-PDΔNC, the free ban-siRNA and a complex of the two (Figure 5D; Table 3). The SAXS data for the unbound Loqs-PDΔNC indicate a maximum dimension of ≈100 Å (Figure 5D). This distance is consistent with the presence of an unstructured linker, where the two domains are flexibly connected. The dynamic arrangement of the two dsRBDs in the free protein is further confirmed by SAXS data for the Loqs linker mutants (Supplementary Figure S7D; Table 3), where the maximum distance decreases proportional with shortening of the linker.
Table 3. SAXS data of Loqs-PDΔNC, linker mutants and protein–RNA complexes.
| Sample | R g [Å] | Dmax [Å] | Porod [Å3] |
|---|---|---|---|
| Loqs-PDΔNC 2.5 mg/ml | 26.94 ± 0.46 | 91.9 | 34156.2 |
| Loqs-PDΔNCΔ21 2.4 mg/ml | 22.65 ± 1.03 | 78.7 | 26153.6 |
| Loqs-PDΔNCΔ41 9.3 mg/ml | 21.20 ± 0.20 | 72.6 | 25538.6 |
| Loqs-PDΔNC 2.0 mg/ml + ban-siRNA | 32.69 ± 0.48 | 114.4 | 56691.6 |
| Loqs-PDΔNCΔ21 1.8 mg/ml + ban-siRNA | 24.17 ± 0.40 | 84.29 | 38595.9 |
| Loqs-PDΔNCΔ31 1.3 mg/ml + ban-siRNA | 21.61 ± 0.30 | 75.34 | 23990.9 |
SAXS data of the protein–RNA complex indicate a maximum distance of ≈115 Å. This is consistent with the preferential binding of the two dsRBDs to the termini of the siRNA duplex, for which a pairwise distance of ≈110 Å is expected (Figure 5E). Interestingly, upon truncation of the linker connecting the tandem dsRBDs in the protein–RNA complexes the dimension of the complex as indicated by SAXS decreases accordingly (Supplementary Figure S7E). This is consistent with a dynamic binding and sliding of the two dsRBDs on the dsRNA helix, where shortening of the linker will restrict the spatial separation of the two domains. The combined analysis of the NMR and SAXS data shows that both dsRBDs exhibit a dynamic binding mode that involves sliding along the dsRNA helix, and preferentially reside at the siRNA termini.
We also analyzed whether the linker is important for the interaction of Loqs-PD with Dcr-2. For this, we transfected a myc-tagged version of Loqs-PD Δ41 into an S2-cell clone harboring a genomically integrated Flag2-tag at the C-terminus of Dcr-2 (62). Then, we immunoprecipitated the endogenous Dcr-2 protein and confirmed by Western blot that myc-Loqs-PDΔ41 was still associated with Dcr-2 (Supplementary Figure S7G). In summary, we conclude that the inter-domain linker enables simultaneous tandem-dsRBD binding of Loqs-PD. Interaction with Dcr-2, on the other hand, is independent of the linker.
Loqs-PD and Dcr-2 together can interpret the thermodynamic asymmetry of an siRNA
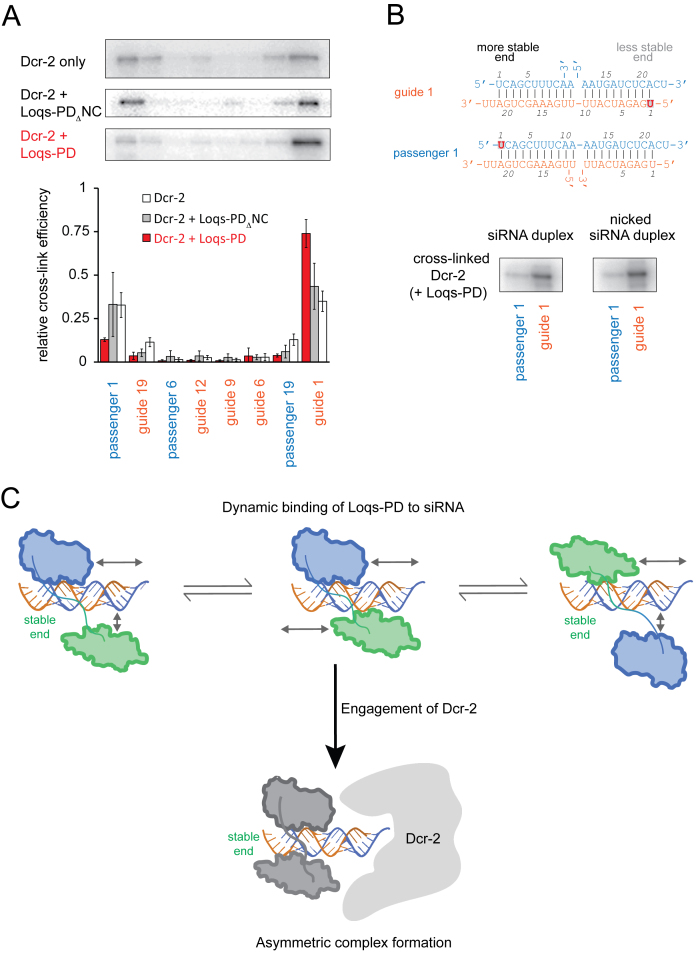
The NMR data indicate only a very small binding preference towards the thermodynamically more stable end of the siRNA. We therefore investigated the role of Dcr-2 for guide strand selection. First, we used recombinant Dcr-2 and cross-linked it to our siRNA model substrates. Unlike previous publications (32,33) we were able to observe crosslinking of Dcr-2 alone to our oligonucleotide substrates. Like Loqs-PD, Dcr-2 showed more efficient crosslinking to the ends of the siRNA duplex and we did not detect a preference for the less stably base paired end of our moderately asymmetric siRNA (Figure 6A). In contrast, upon addition of a 1.5–2-fold excess of full-length Loqs-PD over Dcr-2 to the binding reaction, asymmetric binding of Dcr-2 with a preference for the less stably base-paired end was induced (Figure 6A). This capacity does not reflect competitive displacement from the more stably base-paired end as it requires protein–protein interaction between Dcr-2 and Loqs-PD: The double-domain construct Loqs-PDΔNC that lacks the C-terminal 37 amino acids involved in the association with Dcr-2 (47,52) was unable to induce preferential binding of Dcr-2 to the less stably base paired end of the siRNA (Figure 6A). Thus, the alternative RISC loading complex has properties that differ from each of its subunits alone.
Figure 6.
Dcr-2 and Loqs-PD can interpret the thermodynamic siRNA asymmetry. (A) Site-specific cross-linking of Dcr-2 to 4-thio-uridine containing, radioactively labeled siRNAs revealed preferential association of Dcr-2 with the ends of a ban-siRNA; however, only the complex of full-length Loqs-PD with Dcr-2 can interpret the differential base-pairing stability at either end. The autoradiographs show cross-linked Dcr-2 protein, the bars below are a quantification of three independent experiments (mean ± SD). (B) To test whether the asymmetry sensing is communicated via distortions along the dsRNA helix, we created nicked siRNA duplexes (depicted on top) and cross-linked the Dcr-2 / Loqs-PD complex to either end. We could not detect any difference between the normal and the nicked siRNA duplex. (C) The dsRBD protein moiety of RLC (either Loqs-PD or R2D2) contributes to strand selection by identifying the more stably base-paired end. This may be facilitated by a prolonged sampling period, during which the dsRBD protein steps along the siRNA until Dcr-2 also engages in binding and fixes asymmetric complex formation.
Why can the hetero-duplex of Dcr-2 and a dsRBD protein sense the differential base pairing stability at the siRNA ends so efficiently? Since the two proteins must engage in a protein–protein interaction for this purpose, it may be that RLC induces bending or torsion of the RNA double helix, and that this is more easily accommodated with Dcr-2 bound to the less stably base-paired end. A similar communication via double-stranded nucleic acids has recently been described for proteins bound to DNA (63,64). An siRNA with a nick between positions 11 and 12 of the passenger strand should reduce the transmission of bending or torsion from one siRNA end to the other. Yet, this molecule was bound with identical affinity as the normal siRNA by the Loqs dsRBDs (Supplementary Figure S5C) and asymmetric binding of the Dcr2–LoqsPD complex to the nicked siRNA duplex was retained (Figure 6B). Thus, propagation of RNA conformational changes along the RNA duplex is not essential for guide strand selection.
DISCUSSION
The siRNA duplexes used to trigger RNA interference are structurally symmetric but one of the two strands can become preferentially incorporated into RISC. This functional asymmetry is determined by the relative base-pairing stability at either end of the siRNA duplex (27,28). Incorporation of this concept into siRNA design rules is now essential for every knock-down experiment, yet we do not fully understand the molecular mechanisms behind it. Functional characterization of the Drosophila Dcr-2/R2D2 RISC loading complex (35,65) provided the first protein entity capable of interpreting the differential base pairing stability at the ends of an siRNA (32). Since Dcr-2 alone cannot fulfill this task, R2D2 was proposed to confer asymmetric binding of RLC on the siRNA (32). However, the individual contribution of R2D2 has been difficult to assess because it is degraded in the absence of Dcr-2 and recombinant production is challenging. Here, we employed genome engineering to demonstrate that the PD-isoform of Loqs, a homolog of human TRBP, forms an alternative RLC (aRLC) with Dcr-2 that is required to load transposon-targeting endo-siRNAs into Ago2 in cultured cells. We could recombinantly express Loqs-PD, enabling us to examine its function with structural and biochemical approaches. Our results indicate that the two double-stranded RNA binding domains associate independently of each other with RNA. This is possible as the tandem dsRBDs are connected via a long, unstructured linker region that uncouples their behavior in solution. Analysis of imino NMR signals in the RNA indicates a highly dynamic binding mode, which may be required to search and scan the RNA double helix. Truncation or deletion of the flexible linker perturbs the tandem dsRBD binding mode. Site-specific crosslinking confirms that the two proteins form an alternative RISC loading complex, which is able to interpret the asymmetry of the base-pairing stability at the termini of an siRNA. Importantly, our NMR data indicate that Loqs-PD has a moderate but notable intrinsic ability to recognize the more stably base-paired end of an siRNA duplex. This likely reflects that a more stable RNA helical confirmation provides an optimal binding surface for dsRBD binding. Our results therefore indicate a molecular mechanism for initiation of the strand selection process via asymmetry sensing by the dsRBD subunit of the RLC. Nevertheless, this preference is greatly enhanced in the protein complex with Dcr-2.
Does the alternative RLC exist only in S2-cells?
Previous reports that define RLC for loading of Ago2 in Drosophila relied thus far exclusively on genetic and biochemical experiments with r2d2 mutant flies. Yet, a genome-wide RNAi screen with three different reporter constructs in cell culture recovered neither loqs nor r2d2 (53). Our genome editing and deep sequencing experiments in this manuscript clearly demonstrate that a redundant RLC activity linked to Loqs exists in S2-cells. This is consistent with data from a genome-wide screen of DNA-damage induced siRNA generation where only loqs, but not r2d2, was recovered as a candidate (66). Is the alternative RLC that we describe in S2-cells a particular idiosyncrasy of this cell line? Transposon-targeting endo-siRNAs were slightly sensitive to oxidation and beta-elimination (an indication of reduced 2′-O-methyl modification and thus reduced Ago2 loading) in loqs-PD mutants and some Ago2-loaded siRNAs remain even in r2d2 null mutant flies (42,52), suggesting that alternative RLC also acts in flies at a low level. Possibly, the action of alternative RLC is influenced by the relative expression levels of Loqs-PD and R2D2; in this case, the importance of RLC and alternative RLC (aRLC) may vary between tissues and cell lines. An important question is whether the two forms of RLC, which clearly co-exist in S2 cells (Supplementary Figure S1C), display specificity for certain small RNA substrates. Both complexes can load endogenous and exogenous siRNAs into Ago2, indicating that the contribution of each RLC may be dictated by their relative abundance. Unfortunately, we were unable to derive double-mutant clones to test the extent of redundancy between Loqs-PD and R2D2 directly. Previously, we had found that exogenous siRNAs were partially dependent on R2D2 and that isoform-specific knock-down of Loqs-PD only increased the efficiency of experimental RNAi (52). Our knock-out cells do not recapitulate this difference. One cause for this may be the fact that we deleted all isoforms of Loqs, while the knock-downs were specifically targeting Loqs-PD. It is conceivable that in the absence of co-factors for Dcr-1 (i.e. Loqs-PB or Loqs-PA), some R2D2 is being sequestered. Alternatively, the growth time required in our genome editing procedure may select for cells that have undergone compensatory changes which are absent in the knock-down approach. In any case, however, the fact that exogenous RNAi is not completely abrogated upon loss of R2D2 is consistent in both approaches and clearly indicates that a redundant activity must exist.
RNA binding by the tandem dsRBDs of Loqs-PD
The assembly of multiple ligand binding sites on a single protein can lead to improved binding due to avidity effects and/or even cooperativity. This is exploited in both natural (e.g. oxygen transport by hemoglobin or bivalent binding by vertebrate antibodies) and synthetic (e.g. multimerization of epitope tags on proteins or transcription factor binding sites in promoters) biomolecular interactions. In multi-domain RNA binding proteins, various modes of RNA binding have been observed ranging from independent to preformed domain-domain arrangements (67–72). A flexible linker connecting two RBDs may thereby functionally uncouple the RNA binding contributions of individual domains (56). Here, we observed only a moderate increase in binding affinity when comparing the individual versus tandem Loqs-PD dsRBDs. Similar observations were made for the human homolog TRBP and several other, multiple dsRBD containing proteins (26). According to our deletion analysis, a minimal linker length is required in order for the two dsRBDs to functionally interact. Presumably, this is to accommodate the restraints imposed by the rigid, helical structure of dsRNA. Single molecule studies suggested that longer linkers, e.g. the 61 amino acid linker in TRBP, favor a binding mode that allows the protein to ‘slide’ along dsRNA whereas shorter linkers, e.g. the 25 amino acids in Staufen1, lead to a rather stationary binding (26). The task of Loqs-PD in aRLC is to provide a structure-specific recognition of short siRNA helices in a reversible manner to allow subsequent hand-over to the effector protein Ago2. Single-molecule experiments have revealed that the Loqs-PB isoform and its human homolog TRBP can slide along immobilized dsRNA (73). Our NMR data support these observations and show that binding of Loqs to dsRNA is dynamic in solution and involves sliding along the dsRNA helix. The native 45-residue interdomain linker length may thus constitute an optimal compromise for sufficiently strong, yet mobile and reversible interaction with the duplex RNA.
Discrimination of the siRNA guide strand by dsRBD proteins
One of the most intriguing aspects of siRNA biogenesis is the identification of the guide strand based on the differential base pairing stability at either end (27,28). In the case of Drosophila, the RISC loading complex (RLC) composed of Dcr-2 and the dsRBD cofactor R2D2 can interpret the thermodynamic asymmetry (32). While it is a matter of debate whether loading of mammalian RISC with siRNAs requires the Dicer/TRBP complex for strand discrimination (34,38,39,74,75), it is clear that during human RISC assembly thermodynamic asymmetry is considered independent of other features, such as structural imperfections or terminal nucleotide identity (27). Noland and colleagues demonstrated that the human Dicer/TRBP complex or a fusion of human Dicer with the tandem dsRBDs from the spermatid perinuclear RNA binding protein (SPNR) can bind asymmetrically to siRNA duplexes. Further studies suggested TRBP to sense thermodynamic asymmetry on its own (36). Here, we could demonstrate that the complex of Drosophila Dcr-2 and Loqs-PD can also serve as a sensor for siRNA asymmetry and represents an alternative RLC. As for the human complex (33) efficient sensing depends on a protein–protein interaction between Dcr-2 and Loqs-PD. Evolution has thus produced distinct approaches for loading Drosophila Ago2. Nevertheless, the presence of a dsRBD partner protein for Dcr-2 is an essential and conserved requirement. Due to imperfect base-pairing in the precursors of microRNAs, their loading into Ago proteins may potentially be distinct from siRNA loading. Intriguingly, though, the dsRBD domains present in Loqs-PD are also present in the Loqs-PB isoform that partners with Dcr-1 for miRNA biogenesis.
Cross-linking of R2D2 to siRNA ends is influenced by the phosphorylation state of the 5′-end (32), indicating that recognition of siRNA ends may be possible for dsRBD proteins. A likely scenario is thus initial binding at a random position, followed by sliding (Figure 6C). Considering that an individual dsRBD covers a stretch of ∼12 bp on duplex RNA, this ‘sliding’ along an siRNA may rather represent a sequential engagement of either of the two dsRBDs and could also be envisaged as an oscillation between alternative binding states. Our NMR PRE data indicate that Loqs-PD has a weak preference to bind the more stably base-paired end, which is expected to be more rigid and represent a more canonical double-stranded RNA conformation. However, this preference appears too weak to explain the observed loading bias. Thus, an amplification mechanism exists upon formation of RLC together with Dcr-2. The NMR PRE data did not indicate a preference for a particular dsRBD in Loqs-PD to bind to the more stable base-paired end, arguing that the flexible linker indeed connects two functionally equivalent domains.
Taken together, we propose that after initial binding, a dynamic ‘scanning’ mode of RNA–protein association is established. In this mode, RLC slides or oscillates along the RNA duplex towards either end with the help of its tandem dsRBD subunit in a state that is not yet loading competent. Transition from the scanning mode to loading-competent RLC probably involves conformational changes in Dcr-2, perhaps equivalent to the state transitions described by Sinha and colleagues for the dicing reaction (76). Binding of the siRNA in the thermodynamically ‘correct’ orientation may prolong the RLC residence time, favor conformational transitions and thereby amplify the strand discrimination capability. In addition, a potential dimerization of aRLC as described for the TAF11/Dcr-2/R2D2 complex (37) may contribute to the selectivity. Finally, we cannot exclude that the Dcr-2 protein on its own also displays a weak preference for the less stably paired end, which was - like the binding preference of Loqs-PD - not detectable by our cross-link approach. Dynamic, solution-based assays may reveal if a weak preference also exists for Dcr-2 and, importantly, reveal how the strand discrimination ability is amplified during the RLC conformational transitions.
DATA AVAILABILITY
NMR chemical shifts, structural restraints and the atomic coordinate of dsRBD1 and dsRBD2 are deposited in the BMRB (accession 341244,34123) and the PDB (accession 5NPG, 5NPA), respectively. The sequencing data has been deposited at the European Nucleotide Archive (ENA) with the accession number PRJEB20483.
Supplementary Material
ACKNOWLEDGEMENTS
We thank NMR time at the Bavarian NMR Center and SAXS measurements at the facility of the SFB1035 at Department Chemie, Technische Universität München.
Author Contributions: J.N.T. performed cloning, protein expression and RNA transcription and purification, NMR experiments and data analysis. S.F. performed binding assays and cell experiments. T.K. analyzed NMR spectra. R.S. performed SAXS measurements and data analysis. A.G. performed SLS measurements. C.W., M. J. and C.K. synthesized spin labelled RNA. R.B. and S.K. performed cell experiments and binding assays. O.L. and C.H. performed structure calculations. K.F. and M.S. designed the study. J.N.T., S.F., K.F. and M.S. wrote the manuscript. All authors discussed the results and commented on the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
FUNDING
Deutsche Forschungsgemeinschaft [SFB1035, GRK1721 to M.S., FO360/2, SFB646 to K.F.]. Funding for open access charge: Internal Funds.
Conflict of interest statement. None declared.
REFERENCES
- 1. van Rij R.P., Saleh M.C., Berry B., Foo C., Houk A., Antoniewski C., Andino R.. The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster. Genes Dev. 2006; 20:2985–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang X.H., Aliyari R., Li W.X., Li H.W., Kim K., Carthew R., Atkinson P., Ding S.W.. RNA interference directs innate immunity against viruses in adult Drosophila. Science. 2006; 312:452–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. van Mierlo J.T., van Cleef K.W., van Rij R.P.. Defense and counterdefense in the RNAi-based antiviral immune system in insects. Methods Mol. Biol. 2011; 721:3–22. [DOI] [PubMed] [Google Scholar]
- 4. Chung W.J., Okamura K., Martin R., Lai E.C.. Endogenous RNA interference provides a somatic defense against Drosophila transposons. Curr. Biol. 2008; 18:795–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Czech B., Malone C.D., Zhou R., Stark A., Schlingeheyde C., Dus M., Perrimon N., Kellis M., Wohlschlegel J.A., Sachidanandam R. et al. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008; 453:798–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ghildiyal M., Seitz H., Horwich M.D., Li C., Du T., Lee S., Xu J., Kittler E.L., Zapp M.L., Weng Z. et al. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science. 2008; 320:1077–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Okamura K., Chung W.J., Ruby J.G., Guo H., Bartel D.P., Lai E.C.. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature. 2008; 453:803–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu Q., Rand T.A., Kalidas S., Du F., Kim H.E., Smith D.P., Wang X.. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science. 2003; 301:1921–1925. [DOI] [PubMed] [Google Scholar]
- 9. Denli A.M., Tops B.B., Plasterk R.H., Ketting R.F., Hannon G.J.. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004; 432:231–235. [DOI] [PubMed] [Google Scholar]
- 10. Han J., Lee Y., Yeom K.H., Kim Y.K., Jin H., Kim V.N.. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004; 18:3016–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Landthaler M., Yalcin A., Tuschl T.. The human DiGeorge syndrome critical region gene 8 and Its D. melanogaster homolog are required for miRNA biogenesis. Curr. Biol.: CB. 2004; 14:2162–2167. [DOI] [PubMed] [Google Scholar]
- 12. Chendrimada T.P., Gregory R.I., Kumaraswamy E., Norman J., Cooch N., Nishikura K., Shiekhattar R.. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005; 436:740–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Forstemann K., Tomari Y., Du T., Vagin V.V., Denli A.M., Bratu D.P., Klattenhoff C., Theurkauf W.E., Zamore P.D.. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 2005; 3:e236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jiang F., Ye X., Liu X., Fincher L., McKearin D., Liu Q.. Dicer-1 and R3D1-L catalyze microRNA maturation in Drosophila. Genes Dev. 2005; 19:1674–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bycroft M., Grunert S., Murzin A.G., Proctor M., St Johnston D.. NMR solution structure of a dsRNA binding domain from Drosophila staufen protein reveals homology to the N-terminal domain of ribosomal protein S5. EMBO J. 1995; 14:3563–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kharrat A., Macias M.J., Gibson T.J., Nilges M., Pastore A.. Structure of the dsRNA binding domain of E. coli RNase III. EMBO J. 1995; 14:3572–3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Doyle M., Jantsch M.F.. New and old roles of the double-stranded RNA-binding domain. J. Struct. Biol. 2002; 140:147–153. [DOI] [PubMed] [Google Scholar]
- 18. Yamashita S., Nagata T., Kawazoe M., Takemoto C., Kigawa T., Guntert P., Kobayashi N., Terada T., Shirouzu M., Wakiyama M. et al. Structures of the first and second double-stranded RNA-binding domains of human TAR RNA-binding protein. Protein Sci. 2011; 20:118–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ryter J.M., Schultz S.C.. Molecular basis of double-stranded RNA-protein interactions: structure of a dsRNA-binding domain complexed with dsRNA. EMBO J. 1998; 17:7505–7513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huang Y., Ji L., Huang Q., Vassylyev D.G., Chen X., Ma J.B.. Structural insights into mechanisms of the small RNA methyltransferase HEN1. Nature. 2009; 461:823–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stefl R., Oberstrass F.C., Hood J.L., Jourdan M., Zimmermann M., Skrisovska L., Maris C., Peng L., Hofr C., Emeson R.B. et al. The solution structure of the ADAR2 dsRBM-RNA complex reveals a sequence-specific readout of the minor groove. Cell. 2010; 143:225–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nanduri S., Carpick B.W., Yang Y., Williams B.R., Qin J.. Structure of the double-stranded RNA-binding domain of the protein kinase PKR reveals the molecular basis of its dsRNA-mediated activation. EMBO J. 1998; 17:5458–5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sohn S.Y., Bae W.J., Kim J.J., Yeom K.H., Kim V.N., Cho Y.. Crystal structure of human DGCR8 core. Nat. Struct. Mol. Biol. 2007; 14:847–853. [DOI] [PubMed] [Google Scholar]
- 24. Jayachandran U., Grey H., Cook A.G.. Nuclear factor 90 uses an ADAR2-like binding mode to recognize specific bases in dsRNA. Nucleic Acids Res. 2016; 44:1924–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tian B., Mathews M.B.. Functional characterization of and cooperation between the double-stranded RNA-binding motifs of the protein kinase PKR. J. Biol. Chem. 2001; 276:9936–9944. [DOI] [PubMed] [Google Scholar]
- 26. Wang X., Vukovic L., Koh H.R., Schulten K., Myong S.. Dynamic profiling of double-stranded RNA binding proteins. Nucleic Acids Res. 2015; 43:7566–7576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Khvorova A., Reynolds A., Jayasena S.D.. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003; 115:209–216. [DOI] [PubMed] [Google Scholar]
- 28. Schwarz D.S., Hutvagner G., Du T., Xu Z., Aronin N., Zamore P.D.. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003; 115:199–208. [DOI] [PubMed] [Google Scholar]
- 29. Mi S., Cai T., Hu Y., Chen Y., Hodges E., Ni F., Wu L., Li S., Zhou H., Long C. et al. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell. 2008; 133:116–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Takeda A., Iwasaki S., Watanabe T., Utsumi M., Watanabe Y.. The mechanism selecting the guide strand from small RNA duplexes is different among argonaute proteins. Plant Cell Physiol. 2008; 49:493–500. [DOI] [PubMed] [Google Scholar]
- 31. Montgomery T.A., Howell M.D., Cuperus J.T., Li D., Hansen J.E., Alexander A.L., Chapman E.J., Fahlgren N., Allen E., Carrington J.C.. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell. 2008; 133:128–141. [DOI] [PubMed] [Google Scholar]
- 32. Tomari Y., Matranga C., Haley B., Martinez N., Zamore P.D.. A protein sensor for siRNA asymmetry. Science. 2004; 306:1377–1380. [DOI] [PubMed] [Google Scholar]
- 33. Noland C.L., Ma E., Doudna J.A.. siRNA repositioning for guide strand selection by human Dicer complexes. Mol. Cell. 2011; 43:110–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Noland C.L., Doudna J.A.. Multiple sensors ensure guide strand selection in human RNAi pathways. RNA. 2013; 19:639–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pham J.W., Pellino J.L., Lee Y.S., Carthew R.W., Sontheimer E.J.. A Dicer-2-dependent 80s complex cleaves targeted mRNAs during RNAi in Drosophila. Cell. 2004; 117:83–94. [DOI] [PubMed] [Google Scholar]
- 36. Gredell J.A., Dittmer M.J., Wu M., Chan C., Walton S.P.. Recognition of siRNA asymmetry by TAR RNA binding protein. Biochemistry. 2010; 49:3148–3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liang C., Wang Y., Murota Y., Liu X., Smith D., Siomi M.C., Liu Q.. TAF11 Assembles the RISC Loading Complex to Enhance RNAi Efficiency. Mol. Cell. 2015; 59:807–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kim Y., Yeo J., Lee J.H., Cho J., Seo D., Kim J.S., Kim V.N.. Deletion of human tarbp2 reveals cellular microRNA targets and cell-cycle function of TRBP. Cell Rep. 2014; 9:1061–1074. [DOI] [PubMed] [Google Scholar]
- 39. Betancur J.G., Tomari Y.. Dicer is dispensable for asymmetric RISC loading in mammals. RNA. 2012; 18:24–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Marques J.T., Kim K., Wu P.H., Alleyne T.M., Jafari N., Carthew R.W.. Loqs and R2D2 act sequentially in the siRNA pathway in Drosophila. Nat. Struct. Mol. Biol. 2010; 17:24–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nishida K.M., Miyoshi K., Ogino A., Miyoshi T., Siomi H., Siomi M.C.. Roles of R2D2, a cytoplasmic D2 body component, in the endogenous siRNA pathway in Drosophila. Mol. Cell. 2013; 49:680–691. [DOI] [PubMed] [Google Scholar]
- 42. Mirkovic-Hosle M., Forstemann K.. Transposon defense by endo-siRNAs, piRNAs and somatic pilRNAs in Drosophila: contributions of Loqs-PD and R2D2. PLoS One. 2014; 9:e84994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., Bax A.. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995; 6:277–293. [DOI] [PubMed] [Google Scholar]
- 44. Sattler M., Schleucher J., Griesinger C.. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. J. Biomol. NMR. 1999; 6:277–293. [Google Scholar]
- 45. Shah C., Forstemann K.. Monitoring miRNA-mediated silencing in Drosophila melanogaster S2-cells. Biochim. Biophys. Acta. 2008; 1779:766–772. [DOI] [PubMed] [Google Scholar]
- 46. Schmidts I., Bottcher R., Mirkovic-Hosle M., Forstemann K.. Homology directed repair is unaffected by the absence of siRNAs in Drosophila melanogaster. Nucleic Acids Res. 2016; 44:8261–8271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hartig J.V., Esslinger S., Bottcher R., Saito K., Forstemann K.. Endo-siRNAs depend on a new isoform of loquacious and target artificially introduced, high-copy sequences. EMBO J. 2009; 28:2932–2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Miyoshi K., Miyoshi T., Hartig J.V., Siomi H., Siomi M.C.. Molecular mechanisms that funnel RNA precursors into endogenous small-interfering RNA and microRNA biogenesis pathways in Drosophila. RNA. 2010; 16:506–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhou R., Czech B., Brennecke J., Sachidanandam R., Wohlschlegel J.A., Perrimon N., Hannon G.J.. Processing of Drosophila endo-siRNAs depends on a specific Loquacious isoform. RNA. 2009; 15:1886–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cenik E.S., Fukunaga R., Lu G., Dutcher R., Wang Y., Tanaka Hall T.M., Zamore P.D.. Phosphate and R2D2 restrict the substrate specificity of Dicer-2, an ATP-driven ribonuclease. Mol. Cell. 2011; 42:172–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Okamura K., Robine N., Liu Y., Liu Q., Lai E.C.. R2D2 organizes small regulatory RNA pathways in Drosophila. Mol. Cell. Biol. 2010; 31:884–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hartig J.V., Forstemann K.. Loqs-PD and R2D2 define independent pathways for RISC generation in Drosophila. Nucleic Acids Res. 2011; 39:3836–3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhou R., Hotta I., Denli A.M., Hong P., Perrimon N., Hannon G.J.. Comparative analysis of argonaute-dependent small RNA pathways in Drosophila. Mol. Cell. 2008; 32:592–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Vagin V.V., Sigova A., Li C., Seitz H., Gvozdev V., Zamore P.D.. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006; 313:320–324. [DOI] [PubMed] [Google Scholar]
- 55. Elmer K., Helfer S., Mirkovic-Hosle M., Forstemann K.. Analysis of endo-siRNAs in Drosophila. Methods Mol. Biol. 2014; 1173:33–49. [DOI] [PubMed] [Google Scholar]
- 56. Shamoo Y., Abdul-Manan N., Williams K.R.. Multiple RNA binding domains (RBDs) just don’t add up. Nucleic Acids Res. 1995; 23:725–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Neumuller R.A., Richter C., Fischer A., Novatchkova M., Neumuller K.G., Knoblich J.A.. Genome-wide analysis of self-renewal in Drosophila neural stem cells by transgenic RNAi. Cell Stem Cell. 2011; 8:580–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wunderlich C.H., Huber R.G., Spitzer R., Liedl K.R., Kloiber K., Kreutz C.. A novel paramagnetic relaxation enhancement tag for nucleic acids: a tool to study structure and dynamics of RNA. ACS Chem. Biol. 2013; 8:2697–2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wunderlich C.H., Juen M.A., LeBlanc R.M., Longhini A.P., Dayie T.K., Kreutz C.. Stable isotope-labeled RNA phosphoramidites to facilitate dynamics by NMR. Methods Enzymol. 2015; 565:461–494. [DOI] [PubMed] [Google Scholar]
- 60. Gobl C., Madl T., Simon B., Sattler M.. NMR approaches for structural analysis of multidomain proteins and complexes in solution. Prog. Nucl. Magn. Reson. Spectrosc. 2014; 80:26–63. [DOI] [PubMed] [Google Scholar]
- 61. Clore G.M., Iwahara J.. Theory, practice, and applications of paramagnetic relaxation enhancement for the characterization of transient low-population states of biological macromolecules and their complexes. Chem. Rev. 2009; 109:4108–4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bottcher R., Hollmann M., Merk K., Nitschko V., Obermaier C., Philippou-Massier J., Wieland I., Gaul U., Forstemann K.. Efficient chromosomal gene modification with CRISPR/cas9 and PCR-based homologous recombination donors in cultured Drosophila cells. Nucleic Acids Res. 2014; 42:e89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wang Y.M., Tegenfeldt J.O., Reisner W., Riehn R., Guan X.J., Guo L., Golding I., Cox E.C., Sturm J., Austin R.H.. Single-molecule studies of repressor-DNA interactions show long-range interactions. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:9796–9801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kim S., Brostromer E., Xing D., Jin J., Chong S., Ge H., Wang S., Gu C., Yang L., Gao Y.Q. et al. Probing allostery through DNA. Science. 2013; 339:816–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tomari Y., Du T., Haley B., Schwarz D.S., Bennett R., Cook H.A., Koppetsch B.S., Theurkauf W.E., Zamore P.D.. RISC assembly defects in the Drosophila RNAi mutant armitage. Cell. 2004; 116:831–841. [DOI] [PubMed] [Google Scholar]
- 66. Merk K., Breinig M., Bottcher R., Krebs S., Blum H., Boutros M., Forstemann K.. Splicing stimulates siRNA formation at Drosophila DNA double-strand breaks. PLoS Genet. 2017; 13:e1006861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Rohrl C., Meisslitzer-Ruppitsch C., Bittman R., Li Z., Pabst G., Prassl R., Strobl W., Neumuller J., Ellinger A., Pavelka M. et al. Combined light and electron microscopy using diaminobenzidine photooxidation to monitor trafficking of lipids derived from lipoprotein particles. Curr. Pharm. Biotechnol. 2012; 13:331–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Leitinger G., Masich S., Neumuller J., Pabst M.A., Pavelka M., Rind F.C., Shupliakov O., Simmons P.J., Kolb D.. Structural organization of the presynaptic density at identified synapses in the locust central nervous system. J. Comp. Neurol. 2012; 520:384–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Esslinger S.M., Schwalb B., Helfer S., Michalik K.M., Witte H., Maier K.C., Martin D., Michalke B., Tresch A., Cramer P. et al. Drosophila miR-277 controls branched-chain amino acid catabolism and affects lifespan. RNA Biol. 2013; 10:1042–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Giuliani F., Giuliani G., Bauer R., Rabouille C.. Innexin 3, a new gene required for dorsal closure in Drosophila embryo. PLoS One. 2013; 8:e69212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Giuliani G., Giuliani F., Volk T., Rabouille C.. The Drosophila RNA-binding protein HOW controls the stability of dgrasp mRNA in the follicular epithelium. Nucleic Acids Res. 2014; 42:1970–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Glisovic T., Bachorik J.L., Yong J., Dreyfuss G.. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008; 582:1977–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Koh H.R., Kidwell M.A., Ragunathan K., Doudna J.A., Myong S.. ATP-independent diffusion of double-stranded RNA binding proteins. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:151–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Murchison E.P., Partridge J.F., Tam O.H., Cheloufi S., Hannon G.J.. Characterization of Dicer-deficient murine embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:12135–12140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kawamata T., Tomari Y.. Making RISC. Trends Biochem. Sci. 2010; 35:368–376. [DOI] [PubMed] [Google Scholar]
- 76. Sinha N.K., Trettin K.D., Aruscavage P.J., Bass B.L.. Drosophila dicer-2 cleavage is mediated by helicase- and dsRNA termini-dependent states that are modulated by Loquacious-PD. Mol. Cell. 2015; 58:406–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Markstein M., Dettorre S., Cho J., Neumuller R.A., Craig-Muller S., Perrimon N.. Systematic screen of chemotherapeutics in Drosophila stem cell tumors. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:4530–4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Yan D., Neumuller R.A., Buckner M., Ayers K., Li H., Hu Y., Yang-Zhou D., Pan L., Wang X., Kelley C. et al. A regulatory network of Drosophila germline stem cell self-renewal. Dev. Cell. 2014; 28:459–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
NMR chemical shifts, structural restraints and the atomic coordinate of dsRBD1 and dsRBD2 are deposited in the BMRB (accession 341244,34123) and the PDB (accession 5NPG, 5NPA), respectively. The sequencing data has been deposited at the European Nucleotide Archive (ENA) with the accession number PRJEB20483.