Abstract
AURKB (Aurora Kinase B) is a serine/threonine kinase better known for its role at the mitotic kinetochore during chromosome segregation. Here, we demonstrate that AURKB localizes to the telomeres in mouse embryonic stem cells, where it interacts with the essential telomere protein TERF1. Loss of AURKB function affects TERF1 telomere binding and results in aberrant telomere structure. In vitro kinase experiments successfully identified Serine 404 on TERF1 as a putative AURKB target site. Importantly, in vivo overexpression of S404-TERF1 mutants results in fragile telomere formation. These findings demonstrate that AURKB is an important regulator of telomere structural integrity.
INTRODUCTION
Telomeres are specialized structures located at the ends of linear chromosomes. In mammalian cells, they comprise of many kilobases of tandem ‘TTAGGG’ repeats. Telomere length maintenance can directly impact the replicative capacity of cells. Incomplete replication of the 5′ end of telomeres by canonical DNA polymerases results in the shortening of telomere length with every cell division. When telomeres become critically short, they activate checkpoints leading to growth arrest or cell death. Thus, a balance of telomere attrition and elongation is necessary for continual proliferation of cells. Cells capable of indefinite proliferation, including pluripotent embryonic stem cells (ESCs), have very long telomeres and express telomerase, a reverse transcriptase that adds telomere DNA de novo to directly lengthen telomere DNA (1–4). Only ESCs with long telomeres exhibit authentic developmental pluripotency, while telomerase-deficient ESCs with short telomeres show reduced capacities for proliferation and stable differentiation (5,6), illustrating the importance of telomere integrity for maintaining stem cell pluripotency.
There is increasing evidence that the telomere chromatin state of pluripotent cells is distinct from that of somatic cells (2,3,7). ESCs and induced pluripotent stem cells (iPSCs) show a lower density of heterochromatic marks including histone H3 Lysine 9 trimethylation and H4 Lysine 20 trimethylation at the telomeres (2,7). Furthermore, we and others have shown that histone variant H3.3, chromatin remodeler ATRX, transcription factors TBX3, ZSCAN4, TCSTV1/3 and PML bodies are important for telomere function in ESCs (7–15) These studies suggest that telomere chromatin maintenance in pluripotent cells requires a unique subset of regulators.
In mammalian cells, telomere DNA is bound by the hexameric protein complex called Shelterin, which consists of TERF1, TERF2, RAP1, TIN2, POT1 and TPP1. The Shelterin complex suppresses an aberrant DNA damage response, and controls telomere length and DNA replication (16,17). TERF1, which binds the double-stranded TTAGGG repeats, is essential for regulating telomere lengthening and DNA replication. Gene knockout of TERF1 results in early embryonic lethality in mice (18), while conditional deletion in adult tissue compartments leads to exhausted adult stem cell reserves and premature tissue degeneration (19,20). At the cellular level, loss of TERF1 results in telomeric DNA fragility and damage, and formation of aberrant telomeric DNA bridges during mitosis (18,20–27), while a high level of TERF1 leads to aberrant telomere DNA shortening and dysfunction (22,23,25,26). These studies suggest the importance of a tight regulation of TERF1 level at the telomeres. Recent studies have shown that TERF1 is expressed at a very high level in pluripotent cells, and TERF1 upregulation is essential for the generation of iPSCs and establishment of ESCs from mouse early blastocysts (3,28). Although it remains unclear how TERF1 contributes to the pluripotent state, it is evident that TERF1 level at the telomeres is differentially regulated in various cell types. Indeed, a number of studies have shown that TERF1 binding at the telomeres can be regulated by post-translational modifications, including phosphorylation by a multitude of kinases such as Ataxia telangiectasia mutated (ATM) kinase, Cyclin-dependent kinase 1 (CDK1) and Polo-like kinase 1 (PLK1) (29).
Aurora Kinase B (AURKB) is a serine/threonine kinase that is better known for its role in controlling chromosome segregation during mitosis. Through its highly regulated spatio-temporal association and phosphorylation of an array of interacting proteins (for example, CENP-A, INCENP and MCAK) at the centromere/kinetochore, AURKB controls mitotic spindle assembly/checkpoint, chromosome alignment and cytokinesis (30). Loss of AURKB results in defective mitotic progression and chromosome mis-segregation, leading to genetic instability (31–34). Recent studies have also linked AURKB to proteins unrelated to chromosome segregation including factors regulating cellular pluripotency, suggesting that AURKB plays other important cellular functions (35–38).
Here, we show that AURKB localizes to telomeres in pluripotent mouse ESCs, but not in their differentiated counterparts, in a cell-cycle regulated manner. Inhibition of AURKB activity results in increased binding of TERF1 at the telomeres and the formation of multiple telomeric signals (MTS), an indication of telomere fragility. Considering TERF1 binding at the telomeres is affected by its phosphorylation status (39–44), we investigated the interaction of AURKB with TERF1. We show by in vitro assays that AURKB phosphorylates TERF1 at Serine 404 (S404), a conserved amino acid residue found within the DNA binding domain of TERF1. In addition, the in vivo expression of the phospho-null S404A-TERF1 mutant results in increased binding at the telomeres and MTS formation, recapitulating the phenotype of AURKB-depleted cells. In contrast, the phospho-mimic S404E-TERF1 mutant is impaired in telomere DNA binding, suggesting that phosphorylation of TERF1-S404 functions to release TERF1 from telomere DNA. Moreover, S404E-TERF1 mutant acts in a dominant negative fashion by stripping endogenous TERF1 from the telomeres and its long-term expression results in aberrant telomere lengthening. Together, we show for the first time that AURKB is important for telomere maintenance in mouse ESCs, where it phosphorylates TERF1 and regulates its binding at the telomeres. Our study provides important insights into the functions of AURKB and TERF1 in telomere maintenance.
MATERIALS AND METHODS
Cell cultures
Cell lines including human HT1080 (fibrosarcoma), super-telomerase HT1080 (45), Alternative lengthening of telomere (ALT) positive SKLU1 (lung adenocarcinoma), SV40 transformed fibroblast GM847 and mouse NIH3T3 (transformed fibroblasts) cell lines were cultured in DMEM with 10% FCS. Mouse ES129.1 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 12% heat-inactivated FCS and 103 units/mL of leukemic inhibiting factor (LIF) and 0.1 mM β-mercaptoethanol.
Antibodies
Primary antibodies used were as follows: rabbit polyclonal antisera against mouse TERF1 (27); mouse monoclonal antisera against AURKB (BD Transduction Laboratories, #611082); mouse monoclonal antisera against GFP (Roche, #11814460001), rabbit polyclonal antisera against phosphorylated H3.3 serine 31 (Active Motif, #39637), mouse monoclonal antisera against TERF2 (Santa Cruz, #sc-47693) and rat monoclonal antisera against hemagglutinin (HA) tag (Roche, #11867423001). Secondary antisera used were Alexa Fluor® 488/594 or horseradish peroxidase conjugated donkey/goat anti-mouse or anti-rabbit (Molecular Probes, Invitrogen).
Immunofluorescence analyses and telomere fluorescence in situ hybridization (TEL-FISH)
Cells were treated with 100 ng/ml Colcemid (mitotic spindle poison; Gibco) for 1 h at 37°C to enrich for mitotic cells. For immunofluorescence of cytospun preparations, cells were harvested and resuspended in a hypotonic solution of 0.075M KCl before cells were cytospun onto glass slides. Chromosome preparations were incubated in KCM buffer (120 mM KCl, 20 mM NaCl, 10 mM Tris–HCl at pH 7.2, 0.5 mM ethylenediaminetetraacetic acid (EDTA) 0.1% [v/v] Triton X-100, and protease inhibitor) and processed for immunofluorescence as previously described (46). Slides were extracted in KCM buffer containing 0.5% Triton X-100, blocked in KCM buffer containing 1% BSA (bovine serum albumin, Roche) and incubated with the relevant primary and secondary antibodies for 1 h at 37°C. After each round of antibody incubation, slides were washed thrice in KCM buffer. Slides were then fixed in 4% formaldehyde/KCM and mounted in mounting medium (Vetashield).
For immunofluorescence of pre-fixed cells, cells were fixed in 1% formaldehyde/phosphate-buffered saline (PBS) before being cytopsun onto glass slides. Samples were extracted with 0.5% Triton X-100/PBS, blocked in 1% BSA and incubated with the relevant primary and secondary antibodies at 37°C. After each round of antibody incubation, slides were washed thrice in 0.2% Tween-20/PBS. Slides were fixed in 4% formaldehyde/PBS and mounted in mounting medium (Vetashield).
For FISH analyses, cells in hypotonic solution were fixed in ice-cold methanol/acetic acid (3:1 ratio). Cells were dropped onto slides and allowed to dry overnight. Slides were rehydrated in PBS, treated with 0.5 μg/ml RNaseA, fixed in 4% formaldehyde/PBS, followed by serial ethanol dehydration. Chromosome preparations were briefly heat-denatured and hybridized with telomere C-strand AlexaFluor® 488-conjugated telomere PNA (peptide nucleic acid) probe; or in conjunction with CENP-B box Cy5-conjugated centromere PNA probe (PNA Bio Inc.) in hybridization solution (10 mM Tris–HCl pH 7.2, 70% deionized formamide/0.5% Blocking Reagent (Roche)) at 80°C. After 1–2 h of incubation, slides were sequentially rinsed in FISH buffer I (10 mM Tris–HCl pH 7.2, 70% formamide, 0.1% BSA) and FISH buffer II (0.1M Tris–HCl pH 7.5, 0.15M NaCl, 0.08% Tween-20) before being fixed in serial ethanol dehydration.
For telomere CO-FISH (chromosome orientation fluorescence in situ hybridization) analyses, cells were treated for 12 h with 10 μM BrdU:BrdC (3:1), followed by Colcemid for 1 h. Cells were then harvested and fixed as described above. Prior to hybridization with labeled telomere probes, slides were treated with RNase A (0.2 μg/ml) for 10 min, stained with Hoechst 33258 (0.5 μg/ml in 2×SSC) and UV irradiated. The BrdU/C labeled DNA strand was digested with Exonuclease III and fixed with serial ethanol dehydration. Slides were then hybridized with G-strand AlexaFluor® 594-conjugated telomere PNA probe (PNA Bio Inc.) for 2 h, briefly washed with in FISH Buffer I, and hybridized with C-strand AlexaFluor® 488-conjugated telomere PNA probe for a further 2 h. Slides were then washed and fixed as with normal FISH protocol.
All images were collected using a Zeiss Imager M2 fluorescence microscope linked to an AxioCam MRm CCD camera system and processed using the Zen software 2011 (Carl Zeiss Microscopy).
Custom Python scripts and modules were used to quantify the amount of TERF1 present at telomeres (https://github.com/jni/trf1-quantification). The script used the scikit-image library (47) to set levels of threshold for images, quantify properties of each TERF1 spot and perform NumPy normalization (48). Briefly, discrete and circular TERF1 foci at the telomere ends of mitotic chromosomes were set at a threshold level using Otsu’s method (49), giving a discrete and connected region of pixels for each foci. For each discrete TERF1 foci, the mean, maximum and total arbitrary fluorescence intensity (AFU) units of TERF1 (red) and DAPI (blue) channels were measured, respectively. To compare the amount of binding per unit of DNA, the summary statistic was presented as the mean intensity ratio of TERF1 to DAPI. To examine the relative abundance of Telomere (TEL-FISH) and Centromere FISH signals, we set a threshold level for each channel independently using the Otsu method as described above, then computed the mean AFU for points above the threshold level (https://github.com/jni/mean-intensities).
Comparisons between TEL-FISH/MTS formation datasets and TERF1 mean intensity fluorescence datasets were performed using an unpaired, two-tailed Student's t test. For all tests, α was assumed to be 0.05.
siRNA depletion experiments and real-time PCR analyses
siRNA oligonucleotides specific for mouse AURKB (set #1: SMARTpool: ON-TARGETplus siRNA, Dharmacon; set #2: Ambion s74513), TERF1 and TERF2 (SMARTpool: ON-TARGETplus siRNA, Dharmacon) were transfected into mouse ESCs using the Lipofectamine 2000 Transfection Reagent (Thermo Scientific). As controls, medium GC-content scramble siRNA oligonucleotides (Thermo Scientific) were included in the experiments. For 72 and 96 h siRNA knockdown experiments, cells were re-transfected with siRNAs after 48 h to maintain continuous depletion of gene expression.
RNA was prepared according to the manufacturer’s protocol (Roche) after 48–96 h of transfection and treated with DNAse using the Promega RQ1 reagent (Promega). cDNA was synthesized using the cDNA Reverse Transcriptase kit (Life Technologies). The expression levels of target genes were quantitated with the FastStart DNA Green Sybr using the LightCycler (Roche). As an internal control, primers specific for Actin was used in real-time polymerase chain reaction (PCR) analysis. The comparative cycle threshold (CT) method was used for data analyses and relative fold difference was expressed as 2−ΔΔCT. The primers used in real-time PCR analysis include:
AURKB: 5′GATCCCAGAACAAGCAGCCT3′ and 5′TCGATTTCGATCTCTCGGCG3′;
ACTIN: 5′TCCCTGGAGAAGAGCTACGA3′ and 5′AGCACTGTGTTGGCGTACAG3′;
TERF1: 5′ACAGCGCCGAGGCTATTATT3′ and 5′GTGTAATACGCTCATCAACT3′;
TERF2: 5′AAGTGGAACAGCCCTAACGG3′ and 5′TTCACCTGGTGCCTGAACTT 3′;
TBX3: 5′GGTAAGGCAGACCCCGAAAT3′ and 5′GCTTGGGAAGGCCAAAGTAAAT3′;
NANOG: 5′TTGCTTACAAGGGTCTGCTACT3′ and 5′ACTGGTAGAAGAATCAGGGCT3′.
Construction and expression of GST-TERF1 fragments and in vitro binding assay
pGEX-N- and C-terminal GST-TERF1 constructs were generated by cloning N- (amino acids 1–251) and C-terminal fragments of TERF1 (amino acids 240–421) into pGEX-2T plasmid using BamH1 and EcoR1 sites. These plasmids were transformed into BL21 bacterial cells and induced (using 1 mM Isopropyl-β-D-thiogalactoside (IPTG)) for expression for 6–8 h. Bacterial cells were lysed in ice-cold TNE buffer (10 mM Tris–HCl pH 7.8, 1% (v/v) NP40, 150 mM NaCl, 1mM EDTA, 1% (v/v) Triton X-100 and protease inhibitors), and GST-fusion proteins were purified using Gluthathione Sepharose beads (GE Healthcare Life Sciences), before being incubated with ESC lysates for 4 h at 4°C. Beads were washed several times in ice cold RIPA buffer (150 mM NaCl, 50 mM Tris–HCl at pH 7.5, 0.25% sodium deoxycholate, 0.1% NP40, 0.1% SDS, 1 mM NaF, 1 mM sodium orthovanadate and protease inhibitor), boiled in sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer prior to SDS-PAGE and western blotting using appropriate antibodies.
Construction of GFP-WT and mutant (S404A and S404E) TERF1 DNA constructs
gBLOCK DNA fragments corresponding to the N- and C-terminal TERF1 were purchased from IDT technologies. These fragments carry either wild-type (WT) or mutant (S404A and S404E) TERF1 cDNA sequences. They were cloned into pEGFP-C1 (Gibco) plasmid at NcoI and EcoR1 sites using the Gibson Assembly Kit (New England Biolabs). These constructs were used for the transient expression of WT and mutant GFP-TERF1. gBLOCK DNA fragments of WT and mutant (S404A and S404E) TERF1 fragments were also cloned into pHL-EF1a-SphcCas9-iP-A (Addgene) at NcoI and EcoR1 sites. These constructs were used for the generation of stable cell lines expressing WT and mutant GFP-TERF1. To generate expression construct carrying HA tagged TERF1, gBLOCK DNA fragments from IDT technologies were cloned into pHL-EF1a-SphcCas9-iP-A at NcoI and EcoR1 sites. Transfection reactions using respective constructs were carried out using the Lipofectamine 2000 system (Invitrogen) according to the manufacturer's instructions over 24 h prior to immunofluorescence and DNA-FISH analyses.
In vitro kinase assay and identification of the phosphorylation site by MS-based proteomics
One microgram of the peptide ‘NWAKILSHYKFNNRTSVMLKDRWRTMKRLK’ (Chinapeptides) was incubated with 0.5 μg of recombinant human AURKB protein (Abcam) in the presence of Magnesium/ATP cocktail (Merck Millipore) for 1 h at 37°C. The in vitro kinase assay products were digested in solution with 10 ng Lys-C at 37°C overnight. Digested peptides were desalted and analysed by data dependent acquisition LC-MS/MS. Raw files were converted to the mzML format and searched with the search engine COMET (50) against a decoy human protein database (Swissprot) including common contaminants. Methionine oxidation and phosphorylation on serine, threonine and tyrosine were set as differential modifications. Peptide-spectra-matches were statistically validated using the trans-proteomic pipeline (51). Peptides found to be phosphorylated were further investigated by targeted MS-based proteomics using parallel-reaction monitoring (PRM) or multiple-reaction monitoring (MRM) in order to refine the phosphorylation site localization. PRM and MRM measurements were then analyzed and visualized in Skyline (52).
Chromatin immunoprecipitation/quantitative PCR (ChIP/qPCR) analyses
Cells were harvested and crosslinked with 1% formaldehyde in PBS for 10 min at room temperature. Excess formaldehyde was quenched with glycine at a final concentration of 0.25 M. Cells were washed with PBS, pelleted and lysed in cold cell lysis buffer (10 mM Tris pH 8, 10 mM NaCl, 0.2% NP40 and protease inhibitors). Nuclei were centrifugated, resuspended in 50 mM Tris pH 8, 10 mM EDTA and 1% sodium dodecyl sulphate (SDS), and sonicated with a Bioruptor (Diagenode) to obtain chromatin fragments of 500 bp or less. The chromatin suspension was diluted in dilution buffer (20 mM Tris pH 8, 2 mM EDTA, 150 mM NaCl, 1% Triton X-100 and 0.01% SDS and protease inhibitors) and pre-cleared with Protein A sepharose beads at 4°C. Pre-cleared chromatin was immunoprecipitated with antibody-bound beads at 4°C overnight. For each chromatin immunoprecipitation (ChIP) reaction, 2 to 5 μg of antibody and 20 μl of Protein A Sepharose or magnetic beads (50% slurry) were used. The immunoprecipitated material was washed and eluted in 100 mM NaHCO3 and 1% SDS. The eluted material was treated with 100μg/mL RNaseA and 200 μg/ml Proteinase K and reverse-crosslinked at 65°C overnight. DNA was phenol/chloroform extracted and precipitated using tRNA and glycogen as carriers. Purified ChIP DNA was used as template for qPCR using the primers corresponding to telomeric repeats and GAPDH (as a negative control). ChIP qPCR values were expressed as relative to input, and normalized further against H3 ChIP.
DNA primers for ChIP include:
Telomere DNA: 5′GGTTTTTGAGGGTGAGGGTGAGGGTGAGGGTGAGGGT3′ and 5′TCCCGACTATCCCTATCCCTATCCCTATCCCTATCCCTA3′, and
GAPDH: 5′AGAGAGGGAGGAGGGGAAATG3′ and 5′AACAGGGAGGAGCAGAGAGCAC3′
RESULTS
AURKB localizes to the telomeres of mitotic and interphase mouse ESCs
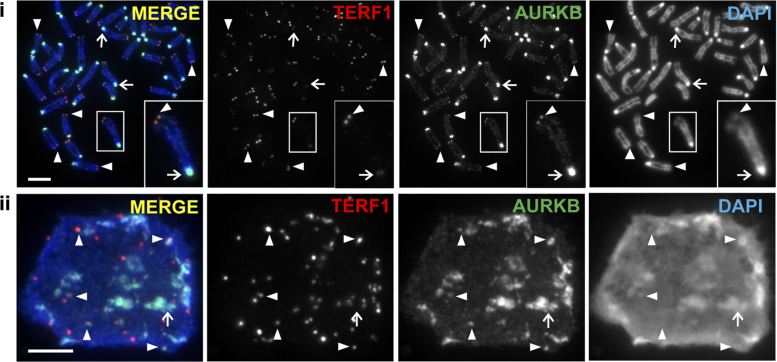
Telomere maintenance is important for stem cell pluripotency (5,6,28,53), whilst recent studies have shown a role for Aurora kinases in regulating ESC pluripotency (54,55). In this study, we investigated the potential role of Aurora kinases in regulating telomere function in pluripotent ES cells. Immunofluorescence analyses were performed on mouse ES129.1 cells using antibodies against AURKB and TERF1 (a telomere marker) (Figure 1). Consistent with previous reports, AURKB localized to the centromeric regions of mitotic chromosomes (32). However, it also localized to the telomeres on mitotic chromosomes as indicated by colocalization with TERF1 staining (Figure 1i). In a proportion of interphase nuclei, AURKB could also be seen localized to a subset of telomeres (Figure 1ii). A similar observation was made in ESCs expressing GFP-tagged AURKB (Supplementary Figure S1A), and in a mitotic population without treatment with microtubule spindle toxin (Supplementary Figure S1B).
Figure 1.
AURKB localizes at the telomeres in mouse ESCs. Localization of AURKB at telomeres in mitotic (i) and interphase (ii) mouse ES129.1 cells, as shown by colocalization with TERF1 (indicated by arrowheads). AURKB also localized to the pericentric regions on mitotic chromosomes (i) and in interphase cells (ii), as seen by its enrichment at the DAPI-dense regions (indicated by arrows). Scalebars represent 5 μm.
To determine if localization of AURKB at telomeres was cell-cycle-dependent, mouse ESCs were synchronized at the G1/S border using thymidine block treatment, released and harvested at 2-h intervals as they progressed into S phase and mitosis. We previously showed that mouse ESCs (i.e. ES129.1 cells) progressed through S phase by 2–6 h, and telomere DNA replication occurs predominantly at late S phase (4–6 h post-release) (7). AURKB presence at the telomeres peaked at 4–6 h post-release, with 60–76% of cells showing six or more AURKB/TERF1 co-localizing foci (Supplementary Figure S2). The data indicate that the presence of AURKB at the telomeres is most prominent during telomere replication occurring at late S phase and persists throughout mitosis (7).
Telomeric localization of AURKB is specific to ESCs and is lost without TERF1.
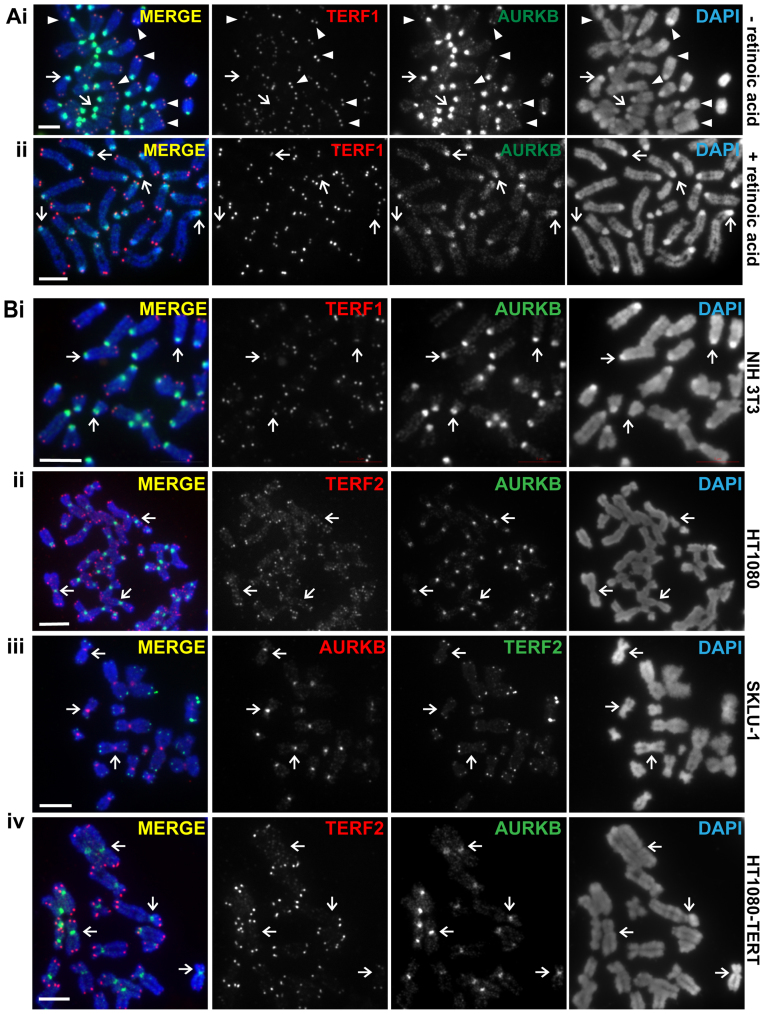
Telomere chromatin undergoes dynamic remodeling during cellular differentiation (7,11,15). To examine whether AURKB telomere localization was affected by the induction of cellular differentiation, we withdrew LIF from ESC culture and added retinoid acid (RA) (Figure 2A) (56). We had previously shown that LIF withdrawal and RA treatment led to a loss of ES cellular pluripotency, as shown by loss of OCT4 expression (7). Differentiated cells also showed reduced levels of TBX3 and NANOG, but no significant reduction of AURKB or TERF1 expression level was detected (Supplementary Figure S3A). In control cells, 93% of mitotic spreads showed clear co-localization of AURKB with TERF1 at the telomeres (Figure 2Ai and Supplementary Figure S3B). After 8 days of treatment, only 26% of cells showed AURKB/TERF1 co-localization (Figure 2Aii and Supplementary Figure S3B). Our findings indicate that AURKB is lost from the telomeres as cells differentiate. Consistent with this, AURKB was not detected at the telomeres of a range of non-pluripotent cell types, including mouse NIH3T3 and human HT1080 (Figure 2B). It was also not detected at telomeres in telomerase-null, Alternative Lengthening Mechanism (ALT) positive SKLU1 and GM847 cancer cells (not shown) (57) and telomerase-overexpressing HT1080 cells (45) (Figure 2B), suggesting that AURKB localization at telomeres is uncoupled from telomere length. We also examined the importance of Shelterin, i.e. TERF1 and TERF2 in the recruitment of AURKB at the telomeres. Mouse ESCs were subjected to siRNA depletion of TERF1 and TERF2, respectively (Supplementary Figure S4A). Compared to cells subjected to scramble control and TERF2-specific siRNA depletion, depletion of TERF1 led to loss of AURKB at the telomeres but not at the pericentric regions (Supplementary Figure S4A).
Figure 2.
AURKB localization at telomere is linked to stem cell pluripotency. (A) AURKB localizes to the telomeres of mitotic mouse ES129.1 cells (arrowheads in (i)), but is lost in ES129.1 cells subjected to retinoic acid treatment differentiation (ii). Note that AURKB localization at pericentric heterochromatin is not lost in differentiated cells (arrows in Ai-ii). (B) AURKB localizes to the pericentric heterochromatin (arrows) but not to the telomeres of somatic, non-ESCs including mouse NIH3T3 (i) and human HT1080 (ii), telomerase-negative SKLU1 ALT cancer (iii) and telomerase overexpressing HT1080 (iv) cells. In mouse cells, TERF1 was used as a telomere marker. In human cells, TERF2 antibody was used as the telomere marker as the TERF1 antibody did not work in human cell types. Scalebars represent 5μm.
Loss of AURKB in mouse ESCs results in aberrant multiple telomeric signals (MTS).
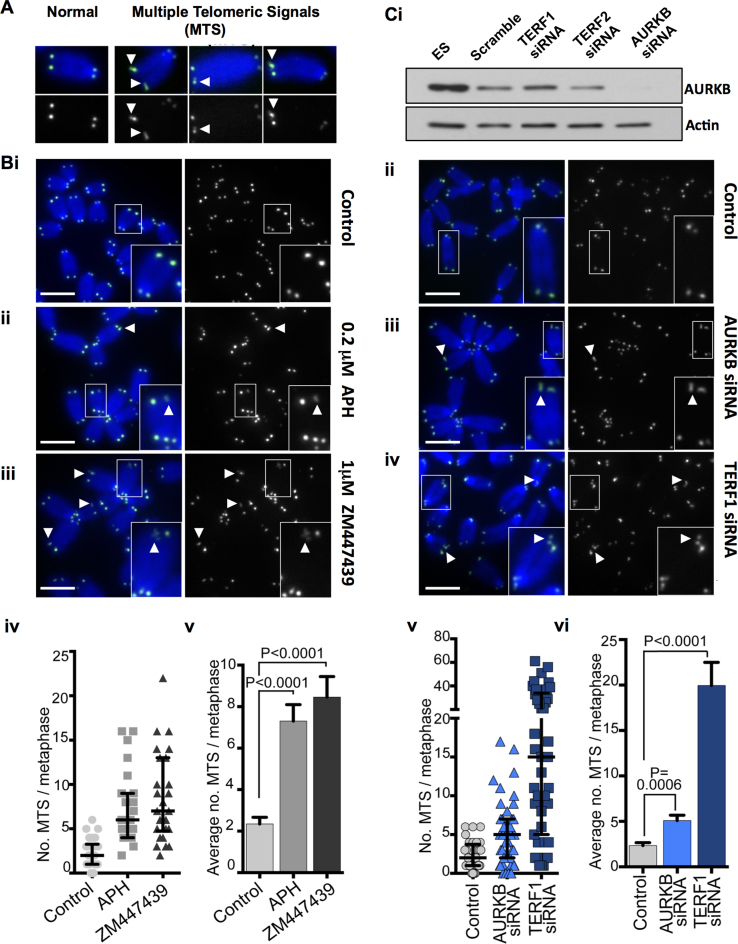
To determine the role of AURKB in controlling telomere function, AURKB activity was inhibited by treatment with 1 μM ZM447439, an AURKB inhibitor (31) without causing deleterious cell-cycle changes in mouse ESCs (55). Consistent with inhibition of AURKB activity, a reduced level of H3 Serine 10 phosphorylation was observed (Supplementary Figure S4B). After 24 h of ZM447439 treatment, the most notable telomere phenotype was the aberrant structure at metaphase telomeres. Normal telomere fluorescence in situ hybridization (TEL-FISH) signals at individual chromatid ends are represented by single signals of nearly equal intensity between sister chromatid ends (examples are shown in Figure 3A). After AURKB inhibition, there was a significant 4-fold increase in the incidence of MTS formation on the chromatid ends, from an average frequency of 2.3 MTS/metaphase in untreated cells to 8.5 MTS/metaphase in ZM447439 treated mouse ESCs (Figure 3B). ZM447439 treatment in mouse embryonic fibroblasts did not result in a significant increase in MTS formation (Supplementary Figure S5A), suggesting that AURKB function in telomere integrity is specific to ESCs. MTS has previously been associated with TERF1 depletion and conditions known to cause replication fork collapse, such as aphidicolin (APH) treatment (16) (which was used as a positive control, Figure 3B).
Figure 3.
Loss of AURKB activity in ESCs results in the formation of MTS. (A) Examples of MTS (obtained with APH treatment) shown. (B) Representative metaphase images of untreated control mouse ES129.1 cells (i) and those treated with either 0.2 µM APH (ii) or 1 µM AURKB inhibitor ZM447439 (iii) for 24 h. TEL-FISH analyses indicated that 24 h of 1 µM ZM447439 treatment resulted in an increase in MTS formation from an average of 2.3 of MTS/metaphase in untreated control cells to 8.5 MTS/metaphase in ZM447439 treated cells (P < 0.0001; N = 1000 chromosomes from three biological replicates), compared to an average of 7.3 MTS/metaphase in cells treated with 0.2 µM APH (P < 0.0001; N = 1000 chromosomes from three biological replicates) (iv and v). (C) Western blot analyses of AURKB and actin in ES129.1 cells subjected to scramble control siRNA and siRNA depletion of TERF1, TERF2 and AURKB, respectively (i). Representative images of metaphase ES129.1 cells subjected to scramble control siRNA (ii; negative control), 72 h of AURKB (iii) and TERF1 siRNA depletion (iv), respectively. About 72 h of AURKB depletion resulted in aberrant MTS formation, increasing from an average of 2.3 MTS/metaphase in cells subjected to scramble control siRNA depletion to 5.1 MTS/metaphase in AURKB-depleted cells (P = 0.0006, N = 1200 chromosomes from three biological replicates) (v and vi). As a comparison, 72 h of TERF1 siRNA depletion caused an average of 19.95 MTS/metaphase (P < 0.0001; Cv and vi). Magnified images of the boxed chromosomes in B and C are shown in the inset, with examples of MTS indicated by the arrowheads. Each point in scatterplots (Biv and Cv) represents of the number of MTS in a single metaphase spread, with error bars showing Q1, Q2 and Q3 values. P-values are indicated in column graphs (Biv and Cv). Scalebars represent 5 μm.
To further examine the role of AURKB at the telomeres of mouse ESCs, we analysed the effect of siRNA depletion of AURKB (set #1) in mouse ESCs (Figure 3C and Supplementary Figure S5B). In agreement with our findings using AURKB inhibitor (Figure 3B), siRNA depletion of AURKB also resulted in MTS formation, with an increase in the average of MTS/metaphase from 2.3 to 5.1 (Figure 3C). AURKB siRNA depletion using an independent AURKB siRNA oligonucleotide (set #2) led to a similar increase of MTS formation (Supplementary Figure S5C). As a control, we also depleted TERF1, which led to a high level of MTS formation (19.95 MTS/metaphase spread; Figure 3C and Supplementary Figure S5B) (19,24). Combined, our data suggest that AURKB is required for the maintenance of telomere integrity in mouse ESCs.
AURKB is a novel TERF1 kinase: AURKB binds and phosphorylates TERF1 at S404 in vitro
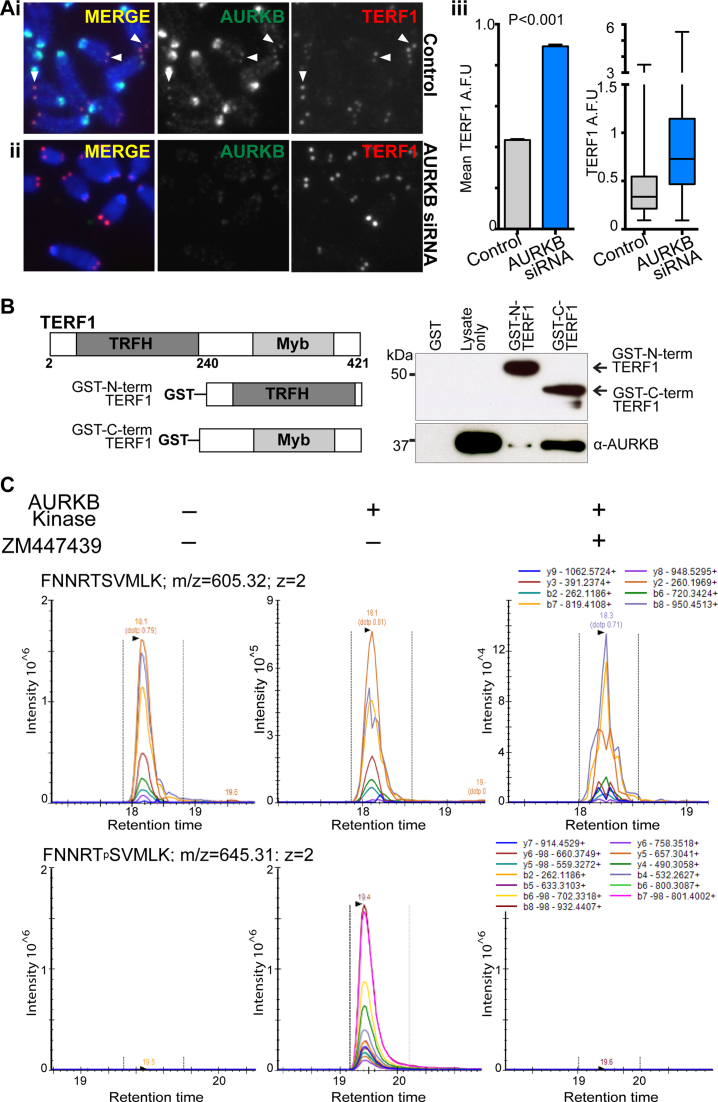
We hypothesized that the increase in MTS formation in ESCs depleted of AURKB activity could be associated with compromised control of TERF1 function at the telomeres. To investigate this, fluorescence intensity of TERF1 staining on mitotic chromosomes was measured in both control and AURKB-depleted cells (set #1) (Figure 4A). The loss of AURKB function led to a significant increase in TERF1 binding at the telomeres (66.7%; Figure 4A). This increase in TERF1 binding upon AURKB inhibition was also detected by ChIP experiments using GFP-TERF1 expressing cells (Supplementary Figure S6A). However, unlike the impact on TERF1 binding, AURKB inhibition did not affect histone H3 occupancy, or the level of TERF2 at the telomeres (Supplementary Figure S6A and B). Combined, our data suggest that TERF1 level at the telomeres is negatively regulated by the presence of AURKB, and illustrate the specific effect of AURKB activity on TERF1 binding at the telomeres.
Figure 4.
AURKB interacts with and phosphorylates TERF1 in mouse ESCs. (A) Seventy-two hours siRNA depletion of AURKB results in increased TERF1 binding at the telomeres in ES129.1 cells. Representative images of control (i) and AURKB-depleted cells (ii) stained for AURKB and TERF1 are shown. Loss of AURKB function led to a 66.7% increase in mean fluorescence staining intensity in TERF1 binding at the telomeres (P < 0.0001, >4000 TERF1 foci from three biological replicates). Error bars represent standard error of the mean, with box and whiskers plot showing Q1, Q2 and Q3 values (iii). P-values are indicated in column graphs. Scalebars represent 5 μm. (B) Schematic on the left shows the GST-fusion proteins of N- and C-terminal TERF1 used in the in vitro binding assay. GST-fusion TERF1 fragments were immobilized on glutathione agarose beads and incubated with ES129.1 cell lysates. Western blot analyses showed a strong interaction between AURKB and C-terminal of TERF1, but not with N-terminal TERF1 or GST-only peptides. (C) Targeted MS-based proteomics on a 30 amino acid synthetic peptide spanning N389-K418 of TERF1 incubated with recombinant AURKB in the presence or absence of an AURBK inhibitor in an in vitro kinase assay and digested with Lys-C. Analysis by multiple-reaction monitoring (MRM). Only 10 amino acid sequence N399-K408 (spanning S404) is shown for clarity. Arrows indicate Lys-C cleavage sites. The theoretical b- and y-ion fragment series after higher-energy collision dissociation (HCD) are indicated above and below the peptide sequence. MRM traces of the two relevant peptides FNNRTSVMLK and FNNRTpSVMLK across the three different conditions. The mass-to-charge ratio (m/z), the charge (z) and the monitored transitions are shown for each peptide (see Supplementary Figures S7 and 8 for further details).
To further investigate the relationship between AURKB and TERF1, we determined if AURKB interacted with TERF1. Reciprocal co-immunoprecipitation experiments in GFP-TERF1 expressing mouse ESCs showed that GFP-TERF1 co-purified with AURKB, indicating that TERF1 and AURKB directly interact in vivo in mouse ESCs (Supplementary Figure S6C). Furthermore, two GST-TERF1 fusion proteins carrying the N- and C-terminal fragments of TERF1 were generated. The GST-N-terminal TERF1 (amino acids 2–251) encompassed the TRFH domain that controls TERF1 dimerization and protein–protein interactions, whereas the GST-C-terminal TERF1 (amino acids 240–421) spanned the DNA-binding Myb-like domain (58) (Figure 4B). These GST fusion proteins were purified and immobilized onto glutathione-agarose beads prior to incubation with mouse ESC lysates. As shown in Figure 4B, AURKB co-purified with GST-C-terminal TERF1 protein, but not with the negative control GST-only protein. Only a low level of AURKB was found bound to the GST-N-terminal-TERF1, indicating that AURKB interacts predominantly with the C-terminal fragment of TERF1 (Figure 4B).
Next, we used GPS2.1, a phosphorylation prediction tool (59) to identify potential AURKB sites on the C-terminal fragment of TERF1. One of the highest ranked putative AURKB sites, S404, was found located within the highly conserved Myb-like DNA binding domain (58). To determine whether AURKB could phosphorylate S404 on TERF1, mass spectrometric analyses were undertaken on a 30 amino acid synthetic TERF1 peptide spanning N389 to K418 following incubation with recombinant AURKB in the presence or absence of an AURKB inhibitor (Figure 4C; Supplementary Figures S7 and 8). The results demonstrated that S404 was readily phosphorylated in presence of AURKB (Figure 4Civ). Phosphorylation of the adjacent residue T403 (not identified as a potential AURKB site) was also detected but at a lower intensity (Supplementary Figure S8). Upon addition of the AURKB inhibitor, phosphorylation of both S404 and T403 was significantly reduced, suggesting that AURKB selectively phosphorylates these residues (Supplementary Figure S7).
S404-TERF1 phospho-mutants show aberrant TERF1 binding in vivo and MTS formation
In order to determine the functional consequences of TERF1 T403 and S404 phosphorylation, GFP-tagged WT-TERF1, and the phospho-null mutants (T403A-TERF1 and S404A-TERF1), were transiently expressed in mouse ESCs. GFP-T403A-TERF1 was extremely poorly expressed (Supplementary Figure S9A) and thus, binding at the telomere was not detectable (not shown). Both WT-TERF1 and S404A-TERF1 showed robust protein expression and clear telomeric binding (Supplementary Figure S9C and D). Given expression of T403A-TERF1 could only be partially rescued by treatment with the proteasome inhibitor MG132, suggesting its instability (Supplementary Figure S9A), we chose to focus on the importance of S404-TERF1 in subsequent analyses. To further characterize the importance of S404 phosphorylation, we also generated the phospho-mimetic mutant GFP-S404E-TERF1. Despite showing a robust protein expression (Supplementary Figure S9B), GFP-TERF1-S404E binding was not detected at the telomeres in both interphase and mitotic cells (Supplementary Figure S9C and D). A diffuse GFP-staining was observed throughout the nucleoplasm in S404E-TERF1 expressing cells, instead of the distinct focal pattern at the telomeres found in WT-TERF1 expressing cells (Supplementary Figure S9C and D). The reduced telomeric binding of TERF1-S404E suggests that S404 phosphorylation may function to release TERF1 from the telomeres.
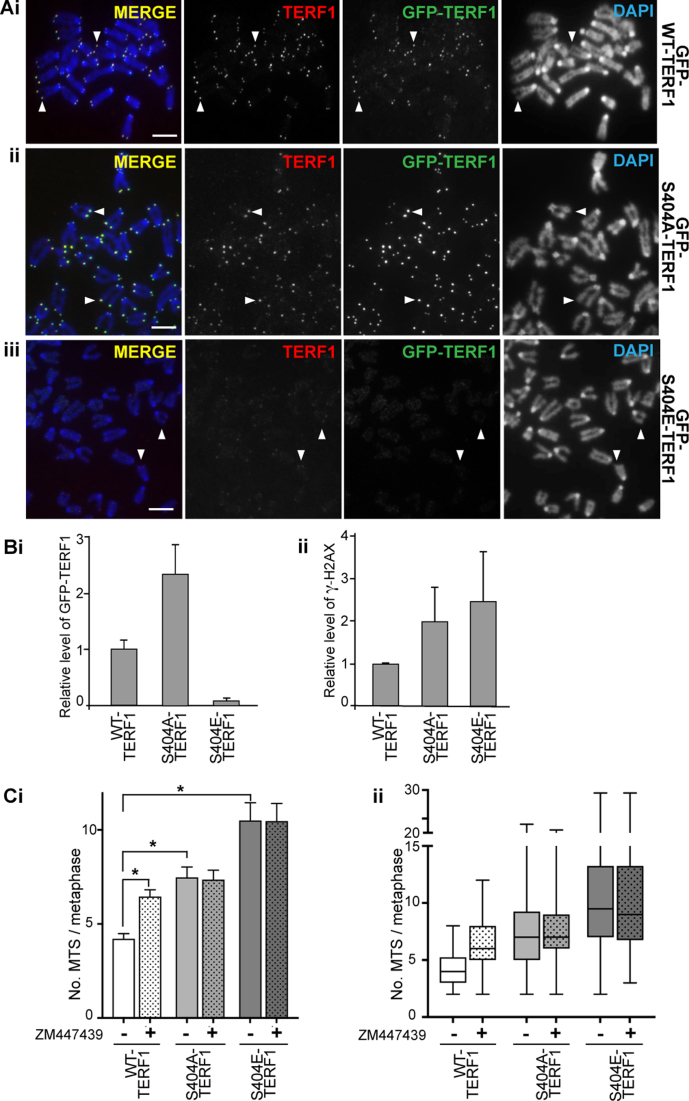
To study the long-term impact of S404 mutation on TERF1 binding and telomere function, we generated cells stably expressing GFP-TERF1 mutants (Figure 5 and Supplementary Figure S10). Similar to the transient expression experiments (Supplementary Figure S9), S404A-TERF1 was not affected in its ability to bind telomeres, whereas S404E-TERF1 failed to bind to the telomeres (Figure 5A). ChIP experiments were performed on stable GFP-TERF1 expression clones to assess the impact of S404 mutation on TERF1 binding. We had previously established and validated a ChIP/telomeric PCR assay to assess chromatin properties at telomeres in mouse ESCs (7,60). Compared to GFP-WT-TERF1 expressing cells, GFP-S404A-TERF1 cells showed a moderate increase in telomere binding, while GFP-S404E-TERF1 cells were greatly reduced in the level of telomere binding (Figure 5Bi and Supplementary Figure S10B). We also performed immunofluorescence analyses in cells depleted of endogenous TERF1 and expressing HA-tagged siRNA-resistant WT and S404 mutant TERF1. Consistent with our data with GFP-TERF1 cells, both HA-WT-TERF1 and HA-S404A-TERF1 bound to telomeres, but HA-S404E-TERF1 failed to bind (Supplementary Figure S11).
Figure 5.
Phosphorylation of S404-TERF1 controls TERF1 binding to the telomere. (A) Mouse ESCs stably expressing GFP-tagged WT-TERF1 (i), phospho-null TERF1-S404A (ii) and phospho-mimic TERF1-S404E (iii), respectively. Cytospun metaphase chromosomes showed clear binding of GFP-WT-TERF1 (i) and GFP-S404A-TERF1 (ii) at the telomeres (arrowheads). However, GFP-S404E-TERF1 binding could not be detected at the telomeres (arrowheads in iii). In GFP-S404E-TERF1 cells, staining of endogenous TERF1 was also greatly reduced (iii). (B) ChIP/qPCR experiments in GFP-tagged WT-TERF1, S404A-TERF1 and S404E-TERF1 cells. When compared to that of GFP-WT-TERF1, GFP-S404A-TERF1 binding at telomere DNA was increased by 2.3-fold while GFP-S404E-TERF1 showed a greatly reduced binding (∼7% of GFP-WT-TERF1 binding) (i). ChIP/qPCR analyses showing increases in γ-H2AX at telomeres in GFP-S404A and S404E-TERF1 stable cell lines (ii). (C) MTS formation was assessed in GFP-WT-TERF1 cells with and without 24 h of ZM447439 treatment. Both GFP-S404A-TERF1 and GFP-S404E-TERF1 cell lines showed increased MTS formation; increased from 4.2 MTS/metaphase in GFP-WT-TERF1 cells to 7.5 and 10.5 MTS/metaphase in GFP-S404A-TERF1 and GFP-S404E-TERF1 cells, respectively. ZM447439 treatment resulted in increased MTS formation, from 4.2 to 6.5 MTS/metaphase in GFP-WT-TERF1 cells. However, ZM447439 treatment did not result in further increase in MTS formation in GFP-S404A-TERF1 and GFP-S404E-TERF1 cells, respectively. The error bars in column graphs (B and Ci) represent standard error of the mean. Asterisk in (Ci) indicates P-values < 0.001; N > 1300 chromosomes from over 30 mitotic spreads per condition from three biological replicates. Box and whiskers plot shown in (C) represent Q1, Q2 and Q3, with the maximum and minimum values plotted. Scalebars represent 5 μm.
TEL-FISH analysis was performed to assess if the S404-TERF1 mutant cells showed compromised telomere integrity. Both GFP-S404A and S404E-TERF1 mutant cells showed increased MTS formation when compared to GFP-WT-TERF1 cells. Importantly, ZM447439 treatment did not result in further MTS formation in GFP-S404-TERF1 mutant cells, but resulted in a significant increase in MTS formation in GFP-WT-TERF1 cells (Figure 5C), suggesting that S404-TERF1 was the primary target of AURKB at the telomeres. In addition, ChIP/qPCR and immunofluorescence analyses detected increased levels of γ-H2AX, a DNA damage mark, at the telomeres in both S404A and S404E-TERF1 cells when compared to that in WT-TERF1 expressing cells (Figure 5Bii and Supplementary Figure S12). These findings are consistent with previous reports that MTS formation is accompanied by the presence of DNA damage marker γ-H2AX (20,24).
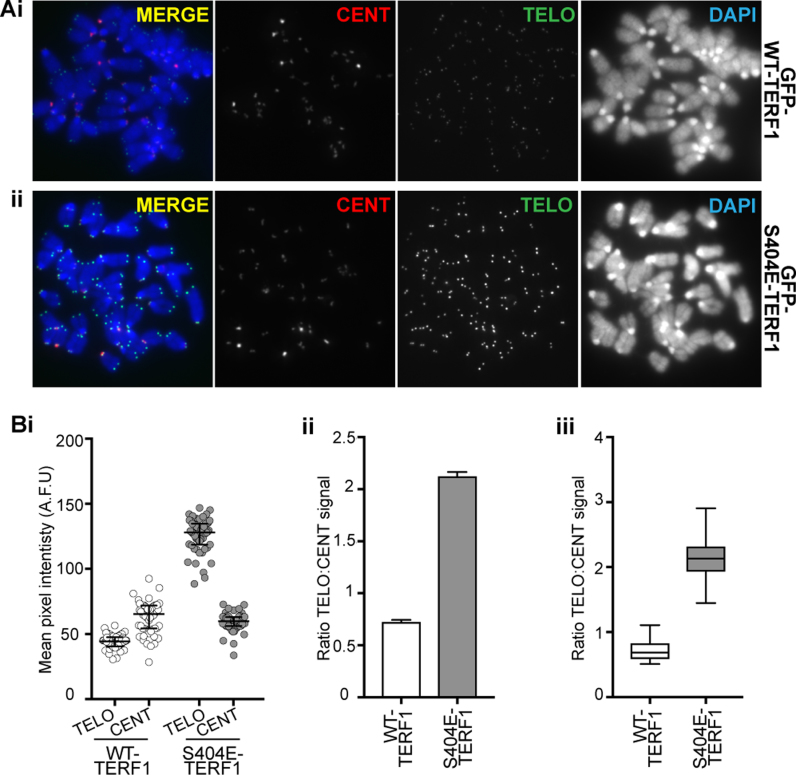
Compared to GFP-WT-TERF1 cells, GFP-S404A-TERF1 cells showed increased MTS formation, from an average frequency of 4.2 MTS/metaphase in GFP-WT-TERF1 cells to 7.5 MTS/metaphase (Figure 5C), suggesting that loss of S404-TERF1 phosphorylation is associated with MTS formation, consistent with the increase in MTS formation seen in cells depleted of AURKB function (Figure 3B and C). However, GFP-S404E-TERF1 cells showed an even greater increase in MTS formation, to 10.5 MTS/metaphase, despite minimal binding of GFP-S404E-TERF1 at the telomeres (Figure 5Ci). Immunofluorescence analyses showed that not only there was no detectable GFP-TERF1-S404E binding at the telomeres, but there was also a decrease in the level of endogenous TERF1 binding at the telomeres (Figure 5Aiii). This suggests that long-term expression of phospho-mimic S404E mutant could be acting in a dominant negative fashion, depleting telomeres of TERF1 binding, resulting in aberrant telomere phenotype as indicated by the MTS formation. Another prediction from this observation is that long-term expression of GFP-S404E-TERF1 (and loss of TERF1 binding) would result in increased telomere length (22). Indeed, TEL-FISH signals in GFP-S404E-TERF1 increased in intensity when compared of GFP-WT-TERF1 cells after 3 months in culture (Figure 6A). When normalized against the centromere-FISH signal, the overall TEL-FISH signal in GFP-S404E-TERF1 cells was increased by ∼2.9 times than that in GFP-WT-TERF1 cells (Figure 6B). The increase in TEL-FISH intensity suggests that long-term dominant-negative effect of S404E-TERF1 results in telomere lengthening in these cells. Together our data suggest that AURKB phosphorylates S404 of TERF1 to regulate its binding at the telomeres of mouse ESCs.
Figure 6.
Long-term expression of S404E-TERF1 results in telomere lengthening. (A) Representative images of GFP-WT-TERF1 (i) and GFP-S404E-TERF1 cells (ii). Telomere length in these cells was analysed using a TEL-FISH quantitative protocol using DNA probes specific to telomere (green) and centromere (red) sequences. TEL-FISH signals in the GFP-S404E GFP-S404A-TERF1 and GFP-S404E-TERF1 cells were increased in intensity when compared to those in GFP-WT-TERF1 cells. (B) Fluorescence quantitation showed that telomeres in GFP-TERF1-S404E were longer than those in GFP-WT-TERF1 cells. (i) The scatter plot shows the mean pixel intensities of TEL-FISH centromere satellite and signals for all metaphase spreads, respectively. The centromere FISH signals between the WT and mutant GFP-TERF1 cell lines were similar in levels. TEL-FISH signals were normalized against centromere-FISH signals, as shown as column graphs (ii) and box and whiskers plots (iii). TEL-FISH signals in GFP-S404E-TERF1 cells were increased by almost three times than those in GFP-WT-TERF1 cells, indicating an increase in telomere length. The error bars in (Bi) represent standard error of the mean. (P = 0.0001, > 50 spreads from three biological replicates were assessed per cell line). Box and whiskers plot shown in (Bii) represent Q1, Q2 and Q3, with the maximum and minimum values plotted.
DISCUSSION
AURKB is well known to be important for chromosome segregation through its roles at the centromere and mitotic spindle during mitosis (61); however, a recent study showed that AURKB could also be found at the pericentric and subtelomeric regions in mouse ESCs (55). In this study, we have demonstrated for the first time, the specific localization of AURKB at the telomeres of mouse ESCs, which is lost as cells differentiate (Figures 1 and 2). AURKB localization at telomeres is not found in a range of other cell types, including those with long telomeres, and telomerase overexpression (Figure 2). These findings suggest that AURKB telomeric localization is uncoupled from telomerase status or telomere length. Additionally, the loss of AURKB at the telomeres of ESCs undergoing differentiation is not a consequence of telomere shortening, but more likely due to the dynamic differentiation-dependent remodeling of ESC telomere chromatin (7). We have also shown that loss of AURKB, either through drug-mediated inhibition or siRNA depletion, results in MTS formation and increased TERF1 binding at telomeres (Figures 3 and 4). Our data indicate that AURKB can phosphorylate the integral telomere DNA-binding Shelterin protein TERF1 at S404 (within the DNA-binding domain) in vitro. Although S404-TERF1 phosphorylation has not been detected in vivo, and this is a limitation of the current study; our in vivo S404-TERF1 mutation studies strongly suggests that S404-TERF1 phosphorylation negatively regulates TERF1 binding to telomeric DNA (Figure 5). Furthermore, we have demonstrated that loss of TERF1-S404 phosphorylation (by expression of S404A-TERF1 mutant) leads to increased MTS formation, recapitulating the telomere phenotype seen in AURKB-depleted mouse ESCs (Figures 5 and 6). Interestingly, a long-term expression of S404E-TERF1 leads to a greater increase in MTS formation, accompanied by increased telomere length. This is likely caused by a dominant negative impact of S404E mutation on the endogenous TERF1 function. Consistently, we have also observed an increase in DNA damage at telomeres in both S404A and S404E-TERF1 mutant cell lines (Figure 6). Together, our study shows the role of AURKB as a novel TERF1 kinase and it is required for maintaining telomere integrity in mouse ESCs.
TERF1 binding at telomeric DNA is dynamic (62), and controlled by post-translational modifications. TERF1 phosphorylation can be driven by a slew of kinases including ATM, CDK1, PLK and Aurora kinase A (40–42,63). In addition, these kinases target many different residues within TERF1 for varied functions. For example, in human cells, T344-TERF1 is phosphorylated by CDK1 to prime TERF1 phosphorylation by PLK1 to stimulate TERF1 binding to telomeric DNA (63). CDK1 also targets T371-TERF1 for phosphorylation but this keeps TERF1 from binding to telomeres (41). In this study, we identified a novel phosphorylation target of AURKB in S404-TERF1, which is located within the highly conserved DNA-binding domain. To our knowledge, this is the first time phosphorylation of S404 (or the equivalent residue on human TERF1) has been reported. AURKB can potentially target multiple residues within the same substrate (64–66). Although we cannot exclude that AURKB may also target other residues within TERF1, we have provided clear evidence that phosphorylation of this single S404 residue is sufficient to release it from telomere chromatin. It is also important to note that AURKB inhibition in S404-TERF1 mutant cells did not result in a further increase in MTS formation. This strongly suggests that the increase in MTS formation in AURKB-inhibited cells is linked to the loss of S404-TERF1 phosphorylation in mouse ESCs (Figure 5C).
Furthermore, considering AURKB-depleted cells show increased telomeric TERF1 binding and S404E-TERF1 is impaired in telomere binding, we propose that AURKB phosphorylates S404 to release TERF1 from telomere DNA. Through the regulation of S404 phosphorylation, AURKB controls TERF1 binding to ensure proper telomere maintenance in ESCs. These data align with previous findings that TERF1 levels must be strictly regulated for correct telomere function, as both loss (20,24) and overexpression (19,22,23,25,67) of TERF1 have detrimental effects on cells. Considering TERF1 is expressed at very high levels in pluripotent cells (3,28), the control of TERF1 binding though AURKB phosphorylation may represent an important additional layer of regulation in mouse ESCs. The presence of such a rapid and reversible mechanism to regulate TERF1 binding is useful, given that TERF1 is involved in a number of processes including telomere length control and telomere DNA replication throughout the cell cycle (19,22–24).
In conclusion, our study has identified AURKB as a novel TERF1 kinase and S404 phosphorylation of TERF1 regulates its binding and telomere integrity in mouse ESCs. Considering that AURKB was recently identified as a factor associated with pluripotency (54), and has also been implicated in the control of telomerase activity (55), our study provides important mechanistic insights into how AURKB could contribute to the biology of pluripotent stem cells.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Samuel Rodgers for his help with protein immunoprecipitation analysis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Health and Medical Research Council of Australia; Monash Postgraduate Discovery Scholarship (to B.V.); Australia Research Council (ARC) Future Fellowship Award (to L.H.W.). Funding for open access charge: ARC.
Conflict of interest statement. None declared.
REFERENCES
- 1. Lee H.W., Blasco M.A., Gottlieb G.J., Horner J.W. 2nd, Greider C.W., DePinho R.A.. Essential role of mouse telomerase in highly proliferative organs. Nature. 1998; 392:569–574. [DOI] [PubMed] [Google Scholar]
- 2. Marion R.M., Strati K., Li H., Tejera A., Schoeftner S., Ortega S., Serrano M., Blasco M.A.. Telomeres acquire embryonic stem cell characteristics in induced pluripotent stem cells. Cell Stem Cell. 2009; 4:141–154. [DOI] [PubMed] [Google Scholar]
- 3. Varela E., Schneider R.P., Ortega S., Blasco M.A.. Different telomere-length dynamics at the inner cell mass versus established embryonic stem (ES) cells. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:15207–15212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liu L., Bailey S.M., Okuka M., Munoz P., Li C., Zhou L., Wu C., Czerwiec E., Sandler L., Seyfang A. et al. . Telomere lengthening early in development. Nat. Cell Biol. 2007; 9:1436–1441. [DOI] [PubMed] [Google Scholar]
- 5. Huang J., Wang F., Okuka M., Liu N., Ji G., Ye X., Zuo B., Li M., Liang P., Ge W.W. et al. . Association of telomere length with authentic pluripotency of ES/iPS cells. Cell Res. 2011; 21:779–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pucci F., Gardano L., Harrington L.. Short telomeres in ESCs lead to unstable differentiation. Cell Stem Cell. 2013; 12:479–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wong L.H., Ren H., Williams E., McGhie J., Ahn S., Sim M., Tam A., Earle E., Anderson M.A., Mann J. et al. . Histone H3.3 incorporation provides a unique and functionally essential telomeric chromatin in embryonic stem cells. Genome Res. 2009; 19:404–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang Q., Dan J., Wang H., Guo R., Mao J., Fu H., Wei X., Liu L.. Tcstv1 and Tcstv3 elongate telomeres of mouse ES cells. Sci. Rep. 2016; 6:19852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zimmermann M., Kibe T., Kabir S., de Lange T.. TRF1 negotiates TTAGGG repeat-associated replication problems by recruiting the BLM helicase and the TPP1/POT1 repressor of ATR signaling. Genes Dev. 2014; 28:2477–2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Delbarre E., Ivanauskiene K., Kuntziger T., Collas P.. DAXX-dependent supply of soluble (H3.3-H4) dimers to PML bodies pending deposition into chromatin. Genome Res. 2013; 23:440–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wong L.H., McGhie J.D., Sim M., Anderson M.A., Ahn S., Hannan R.D., George A.J., Morgan K.A., Mann J.R., Choo K.H.. ATRX interacts with H3.3 in maintaining telomere structural integrity in pluripotent embryonic stem cells. Genome Res. 2010; 20:351–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ivanauskiene K., Delbarre E., McGhie J.D., Kuntziger T., Wong L.H., Collas P.. The PML-associated protein DEK regulates the balance of H3.3 loading on chromatin and is important for telomere integrity. Genome Res. 2014; 24:1584–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Voon H.P., Wong L.H.. New players in heterochromatin silencing: histone variant H3.3 and the ATRX/DAXX chaperone. Nucleic Acids Res. 2016; 44:1496–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Voon H.P., C P., W L.H.. Compromised telomeric heterochromatin promotes ALTernative lengthening of telomeres. Trends Cancer. 2016; 2:114–116. [DOI] [PubMed] [Google Scholar]
- 15. Chang F.T., McGhie J.D., Chan F.L., Tang M.C., Anderson M.A., Mann J.R., Andy Choo K.H., Wong L.H.. PML bodies provide an important platform for the maintenance of telomeric chromatin integrity in embryonic stem cells. Nucleic Acids Res. 2013; 41:4447–4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sfeir A., de Lange T.. Removal of shelterin reveals the telomere end-protection problem. Science. 2012; 336:593–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005; 19:2100–2110. [DOI] [PubMed] [Google Scholar]
- 18. Karlseder J., Kachatrian L., Takai H., Mercer K., Hingorani S., Jacks T., de Lange T.. Targeted deletion reveals an essential function for the telomere length regulator Trf1. Mol. Cell. Biol. 2003; 23:6533–6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Munoz P., Blanco R., de Carcer G., Schoeftner S., Benetti R., Flores J.M., Malumbres M., Blasco M.A.. TRF1 controls telomere length and mitotic fidelity in epithelial homeostasis. Mol. Cell. Biol. 2009; 29:1608–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Martinez P., Thanasoula M., Munoz P., Liao C., Tejera A., McNees C., Flores J.M., Fernandez-Capetillo O., Tarsounas M., Blasco M.A.. Increased telomere fragility and fusions resulting from TRF1 deficiency lead to degenerative pathologies and increased cancer in mice. Genes Dev. 2009; 23:2060–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ye J.Z., de Lange T.. TIN2 is a tankyrase 1 PARP modulator in the TRF1 telomere length control complex. Nat. Genet. 2004; 36:618–623. [DOI] [PubMed] [Google Scholar]
- 22. van Steensel B., de Lange T.. Control of telomere length by the human telomeric protein TRF1. Nature. 1997; 385:740–743. [DOI] [PubMed] [Google Scholar]
- 23. Smogorzewska A., van Steensel B., Bianchi A., Oelmann S., Schaefer M.R., Schnapp G., de Lange T.. Control of human telomere length by TRF1 and TRF2. Mol. Cell. Biol. 2000; 20:1659–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sfeir A., Kosiyatrakul S.T., Hockemeyer D., MacRae S.L., Karlseder J., Schildkraut C.L., de Lange T.. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009; 138:90–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ohki R., Ishikawa F.. Telomere-bound TRF1 and TRF2 stall the replication fork at telomeric repeats. Nucleic Acids Res. 2004; 32:1627–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lisaingo K., Uringa E.J., Lansdorp P.M.. Resolution of telomere associations by TRF1 cleavage in mouse embryonic stem cells. Mol. Biol. Cell. 2014; 25:1958–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Iwano T., Tachibana M., Reth M., Shinkai Y.. Importance of TRF1 for functional telomere structure. J. Biol. Chem. 2004; 279:1442–1448. [DOI] [PubMed] [Google Scholar]
- 28. Schneider R.P., Garrobo I., Foronda M., Palacios J.A., Marion R.M., Flores I., Ortega S., Blasco M.A.. TRF1 is a stem cell marker and is essential for the generation of induced pluripotent stem cells. Nat. Commun. 2013; 4:1946. [DOI] [PubMed] [Google Scholar]
- 29. Walker J.R., Zhu X.D.. Post-translational modifications of TRF1 and TRF2 and their roles in telomere maintenance. Mech. Ageing Dev. 2012; 133:421–434. [DOI] [PubMed] [Google Scholar]
- 30. Carmena M., Wheelock M., Funabiki H., Earnshaw W.C.. The chromosomal passenger complex (CPC): from easy rider to the godfather of mitosis. Nat. Rev. Mol. Cell Biol. 2012; 13:789–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ditchfield C., Johnson V.L., Tighe A., Ellston R., Haworth C., Johnson T., Mortlock A., Keen N., Taylor S.S.. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J. Cell Biol. 2003; 161:267–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Honda R., Korner R., Nigg E.A.. Exploring the functional interactions between Aurora B, INCENP, and survivin in mitosis. Mol. Biol. Cell. 2003; 14:3325–3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Terada Y., Tatsuka M., Suzuki F., Yasuda Y., Fujita S., Otsu M.. AIM-1: a mammalian midbody-associated protein required for cytokinesis. EMBO J. 1998; 17:667–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fernandez-Miranda G., Trakala M., Martin J., Escobar B., Gonzalez A., Ghyselinck N.B., Ortega S., Canamero M., Perez de Castro I., Malumbres M.. Genetic disruption of aurora B uncovers an essential role for aurora C during early mammalian development. Development. 2011; 138:2661–2672. [DOI] [PubMed] [Google Scholar]
- 35. van der Waal M.S., Hengeveld R.C., van der Horst A., Lens S.M.. Cell division control by the chromosomal passenger complex. Exp. Cell Res. 2012; 318:1407–1420. [DOI] [PubMed] [Google Scholar]
- 36. Gully C.P., Velazquez-Torres G., Shin J.H., Fuentes-Mattei E., Wang E., Carlock C., Chen J., Rothenberg D., Adams H.P., Choi H.H. et al. . Aurora B kinase phosphorylates and instigates degradation of p53. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:E1513–E1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wu L., Ma C.A., Zhao Y., Jain A.. Aurora B interacts with NIR-p53, leading to p53 phosphorylation in its DNA-binding domain and subsequent functional suppression. J. Biol. Chem. 2011; 286:2236–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yang C., Tang X., Guo X., Niikura Y., Kitagawa K., Cui K., Wong S.T., Fu L., Xu B.. Aurora-B mediated ATM serine 1403 phosphorylation is required for mitotic ATM activation and the spindle checkpoint. Mol. Cell. 2011; 44:597–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chen Y.C., Teng S.C., Wu K.J.. Phosphorylation of telomeric repeat binding factor 1 (TRF1) by Akt causes telomere shortening. Cancer Investig. 2009; 27:24–28. [DOI] [PubMed] [Google Scholar]
- 40. McKerlie M., Lin S., Zhu X.D.. ATM regulates proteasome-dependent subnuclear localization of TRF1, which is important for telomere maintenance. Nucleic Acids Res. 2012; 40:3975–3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. McKerlie M., Zhu X.D.. Cyclin B-dependent kinase 1 regulates human TRF1 to modulate the resolution of sister telomeres. Nat. Commun. 2011; 2:371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ohishi T., Hirota T., Tsuruo T., Seimiya H.. TRF1 mediates mitotic abnormalities induced by Aurora-A overexpression. Cancer Res. 2010; 70:2041–2052. [DOI] [PubMed] [Google Scholar]
- 43. Tong A.S., Stern J.L., Sfeir A., Kartawinata M., de Lange T., Zhu X.D., Bryan T.M.. ATM and ATR signaling regulate the recruitment of human telomerase to telomeres. Cell Rep. 2015; 13:1633–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wu Y., Xiao S., Zhu X.D.. MRE11-RAD50-NBS1 and ATM function as co-mediators of TRF1 in telomere length control. Nat. Struct. Mol. Biol. 2007; 14:832–840. [DOI] [PubMed] [Google Scholar]
- 45. Cristofari G., Lingner J.. Telomere length homeostasis requires that telomerase levels are limiting. EMBO J. 2006; 25:565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wong L.H., Saffery R., Anderson M.A., Earle E., Quach J.M., Stafford A.J., Fowler K.J., Choo K.H.. Analysis of mitotic and expression properties of human neocentromere-based transchromosomes in mice. J. Biol. Chem. 2005; 280:3954–3962. [DOI] [PubMed] [Google Scholar]
- 47. van der Walt S., Schonberger J.L., Nunez-Iglesias J., Boulogne F., Warner J.D., Yager N., Gouillart E., Yu T., Contributors S.I.. scikit-image: image processing in Python. Peerj. 2014; 2:e453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. van der Walt S., Colbert S.C., Varoquaux G.. The NumPy array: a structure for efficient numerical computation. Comput. Sci. Eng. 2011; 13:22–30. [Google Scholar]
- 49. Otsu N. Threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern. 1979; 9:62–66. [Google Scholar]
- 50. Eng J.K., Jahan T.A., Hoopmann M.R.. Comet: an open-source MS/MS sequence database search tool. Proteomics. 2013; 13:22–24. [DOI] [PubMed] [Google Scholar]
- 51. Keller A., Nesvizhskii A.I., Kolker E., Aebersold R.. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 2002; 74:5383–5392. [DOI] [PubMed] [Google Scholar]
- 52. MacLean B., Tomazela D.M., Shulman N., Chambers M., Finney G.L., Frewen B., Kern R., Tabb D.L., Liebler D.C., MacCoss M.J.. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010; 26:966–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang F., Yin Y., Ye X., Liu K., Zhu H., Wang L., Chiourea M., Okuka M., Ji G., Dan J. et al. . Molecular insights into the heterogeneity of telomere reprogramming in induced pluripotent stem cells. Cell Res. 2012; 22:757–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lee D.F., Su J., Ang Y.S., Carvajal-Vergara X., Mulero-Navarro S., Pereira C.F., Gingold J., Wang H.L., Zhao R., Sevilla A. et al. . Regulation of embryonic and induced pluripotency by aurora kinase-p53 signaling. Cell Stem Cell. 2012; 11:179–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mallm J.P., Rippe K.. Aurora Kinase B regulates telomerase activity via a centromeric RNA in stem cells. Cell Rep. 2015; 11:1667–1678. [DOI] [PubMed] [Google Scholar]
- 56. Strickland S., Mahdavi V.. The induction of differentiation in teratocarcinoma stem cells by retinoic acid. Cell. 1978; 15:393–403. [DOI] [PubMed] [Google Scholar]
- 57. Bryan T.M., Englezou A., Gupta J., Bacchetti S., Reddel R.R.. Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J. 1995; 14:4240–4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bianchi A., Smith S., Chong L., Elias P., de Lange T.. TRF1 is a dimer and bends telomeric DNA. EMBO J. 1997; 16:1785–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Xue Y., Liu Z., Cao J., Ma Q., Gao X., Wang Q., Jin C., Zhou Y., Wen L., Ren J.. GPS 2.1: enhanced prediction of kinase-specific phosphorylation sites with an algorithm of motif length selection. Protein Eng. Des. Sel. 2011; 24:255–260. [DOI] [PubMed] [Google Scholar]
- 60. Udugama M., FT M.C., Chan F.L., Tang M.C., Pickett H.A., JD R.M., Mayne L., Collas P., Mann J.R., Wong L.H.. Histone variant H3.3 provides the heterochromatic H3 lysine 9 tri-methylation mark at telomeres. Nucleic Acids Res. 2015; 43:10227–10237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Carmena M., Ruchaud S., Earnshaw W.C.. Making the Auroras glow: regulation of Aurora A and B kinase function by interacting proteins. Curr. Opin. Cell Biol. 2009; 21:796–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Mattern K.A., Swiggers S.J., Nigg A.L., Lowenberg B., Houtsmuller A.B., Zijlmans J.M.. Dynamics of protein binding to telomeres in living cells: implications for telomere structure and function. Mol. Cell. Biol. 2004; 24:5587–5594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wu Z.Q., Yang X., Weber G., Liu X.. Plk1 phosphorylation of TRF1 is essential for its binding to telomeres. J. Biol. Chem. 2008; 283:25503–25513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Liokatis S., Stutzer A., Elsasser S.J., Theillet F.X., Klingberg R., van Rossum B., Schwarzer D., Allis C.D., Fischle W., Selenko P.. Phosphorylation of histone H3 Ser10 establishes a hierarchy for subsequent intramolecular modification events. Nat. Struct. Mol. Biol. 2012; 19:819–823. [DOI] [PubMed] [Google Scholar]
- 65. Hirota T., Lipp J.J., Toh B.H., Peters J.M.. Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin. Nature. 2005; 438:1176–1180. [DOI] [PubMed] [Google Scholar]
- 66. Goto H., Yasui Y., Nigg E.A., Inagaki M.. Aurora-B phosphorylates Histone H3 at serine28 with regard to the mitotic chromosome condensation. Genes Cells. 2002; 7:11–17. [DOI] [PubMed] [Google Scholar]
- 67. Leman A.R., Dheekollu J., Deng Z., Lee S.W., Das M.M., Lieberman P.M., Noguchi E.. Timeless preserves telomere length by promoting efficient DNA replication through human telomeres. Cell Cycle. 2012; 11:2337–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.