Abstract
The aminoglycoside resistance conferred by an N1-methylation of A1408 in 16S rRNA by a novel plasmid-mediated methyltransferase NpmA can be a future health threat. In the present study, we have determined crystal structures of the bacterial ribosomal decoding A site with an A1408m1A antibiotic-resistance mutation both in the presence and absence of aminoglycosides. G418 and paromomycin both possessing a 6′-OH group specifically bind to the mutant A site and disturb its function as a molecular switch in the decoding process. On the other hand, binding of gentamicin with a 6′-NH3+ group to the mutant A site could not be observed in the present crystal structure. These observations agree with the minimum inhibitory concentration of aminoglycosides against Escherichia coli. In addition, one of our crystal structures suggests a possible conformational change of A1408 during the N1-methylation reaction by NpmA. The structural information obtained explains how bacteria acquire resistance against aminoglycosides along with a minimum of fitness cost by the N1-methylation of A1408 and provides novel information for designing the next-generation aminoglycoside.
INTRODUCTION
Aminoglycosides such as neomycin and gentamicin are a class of antibiotics that cause mRNA miscoding (1,2), inhibit translocation (3,4) and inhibit ribosome recycling (5), and have been widely used in treatment of serious bacterial infectious diseases. They are produced by Streptomyces (-mycin) and Micromonospora (-micin) species (6). These aminoglycoside producers have several mechanisms to prevent self-toxicity by nature; (i) modifying the target site, (ii) modifying aminoglycoside itself, (iii) decreasing intracellular drug concentration (7,8). As the WHO (World Health Organization) recently warned about global threat of antibiotic resistance (9), pathogenic bacteria acquired these antibiotic-resistance mechanisms from the antibiotic producers due to the excessive, inappropriate and often unnecessary use of antibiotics.
Aminoglycosides except streptomycin commonly target the ribosomal A-site molecular switch in 16S rRNA composed of highly conserved 15 nt residues (Figure 1) (10). During the decoding process, the molecular switch changes its conformation between ‘off’ (with tucked-in A1492 and/or A1493) and ‘on’ (with bulged-out A1492 and A1493) states, and discriminates a single cognate-tRNA from several near-cognate tRNAs by recognitions between the bulged-out A1492/A1493 residues and the first two base pairs in the tRNA–mRNA complex through A-minor interactions (11–13). The binding specificity of aminoglycosides to the bacterial A-site switch is mainly conferred by its ring I, which makes a pseudo pair with the universally conserved A1408 in the bacterial A site through two hydrogen bonds (Supplementary Figure S1) (12–26). The interaction forces the A site to adopt the ‘on’ state even when near-cognate tRNAs are delivered to the A site, thereby reducing the accuracy of the decoding process resulting to cell death. Therefore, it is reasonable that the aminoglycoside producers possess chromosomally encoded 16S rRNA methyltransferase (e.g. KamA, KamB, CmnU and Kmr) as a self-defense system that methylates the N1 position of A1408 to disturb the pseudo-pair formation (27). However, in 2007, Wachino et al. first isolated an aminoglycoside-resistant Escherichia coli strain possessing a plasmid-encoded A1408-N1-methyltransferase NpmA from Japanese clinical settings (28). Since the npmA gene is encoded in a plasmid, rapid spread of antibiotic-resistant bacteria by its horizontal transfer between pathogens can be a future threat. Recently, structural insights into the methylation of A1408 by A1408-N1-methyltransferases have been revealed by crystal structures of NpmA and KamB themselves and NpmA in complex with the substrate 30S ribosome (29–31). These structural information are useful for designing A1408-N1-methyltransferase inhibitors. In the present study, in order to obtain a structural basis for the intrinsic and acquired antibiotic resistance conferred by the N1-methylation of A1408, we have determined crystal structures of the m1A1408-modified A site both in the presence and absence of aminoglycosides.
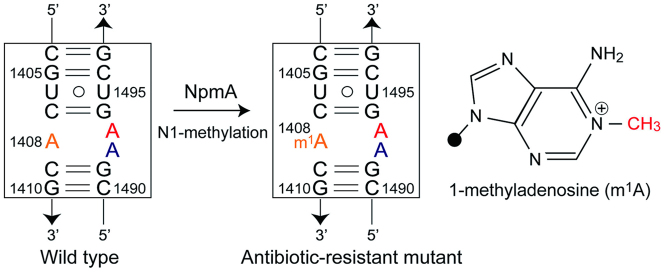
Figure 1.
Secondary structure of the bacterial ribosomal A site with the antibiotic-resistant A1408m1A modification and chemical structure of 1-methyladenosine (m1A). The rRNA residues are numbered according to the numbering used in Escherichia coli 16S rRNA. The A/m1A1408, A1492 and A1493 residues are colored in orange, blue and red, respectively.
MATERIALS AND METHODS
Sample preparations and crystallizations
An RNA duplex designed to fold as a double helix containing two bacterial A sites with the A1408m1A antibiotic-resistant modification was used in the present study (Supplementary Figure S2). Such RNA duplexes have been extensively used as successful models in a series of crystallographic studies of the bacterial (14–26), protozoal (32–34) and human (35–38) ribosomal RNA A sites, since their crystal packing interactions mimic a molecular crowding environment around the A site in the ribosome (see examples below) (39). A chemically synthesized RNA oligomer (Gene Design Inc., Japan) was purified by 20% polyacrylamide gel electrophoresis under a denaturing condition containing 3.2 M urea and then desalted by C18 reversed-phase chromatography. Prior to crystallization, RNA solutions (1 mM RNA, 50 mM sodium cacodylate (pH = 7.0)) with and without 2 mM aminoglycoside (Figure 2; G418, Paromomycin or Gentamicin) were prepared. Crystallizations were performed at 20°C by the hanging-drop vapor diffusion method by mixing 1 μl of RNA solution and 1 μl of crystallization solution containing 50 mM sodium cacodylate (pH 7.0), 0–10 mM spermine tetrahydrochloride, 1–10% (v/v) 2-methyl-2,4-pentanediol and 10–750 mM cation chlorides. The crystallization droplets were equilibrated against 250 μl of a reservoir solution containing 40% 2-methyl-2,4-pentanediol. Four types of crystals were obtained both in the absence and presence of aminoglycosides G418, Paromomycin and Gentamicin (A1408m1A, A1408m1A-G418, A1408m1A-Paromomycin and A1408m1A-Gentamicin crystals, hereafter). Detailed crystallization conditions are summarized in Supplementary Table S1.
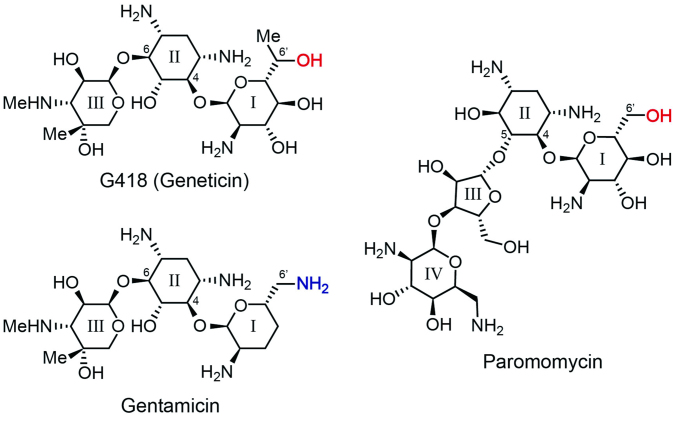
Figure 2.
Chemical structures of G418 (geneticin), gentamicin and paromomycin.
Data collections and structure determinations
X-ray datasets of the four crystals were collected at 100K with synchrotron radiation at the structural biology beamlines, NW12A, BL-17A and BL-5A, of the Photon Factory (Tsukuba, Japan). Each dataset was processed with the program CrystalClear (Rigaku Americas Corp., TX, USA). The obtained intensity data were further converted to structure-factor amplitudes using the program TRUNCATE from the CCP4 suite (40). The statistics of data collections and the crystal data are summarized in Table 1.
Table 1. Crystal data, statistics of data collections and structure refinements.
| Crystal code | A1408m1A | A1408m1A-G418 | A1408m1A-Paromomycin | A1408m1A-Gentamicin |
|---|---|---|---|---|
| PDB-ID | 4WCP | 4WCQ | 4WCR | 4WCS |
| Crystal data | ||||
| Space group | C2 | P21212 | P21212 | P21212 |
| Unit cell (Å, °) | a = 159.7 | a = 33.4 | a = 31.6 | a = 32.6 |
| b = 46.5 | b = 89.8 | b = 90.8 | b = 92.0 | |
| c = 38.8 | c = 46.8 | c = 46.8 | c = 47.7 | |
| β = 96.4 | ||||
| Z a | 1.5 | 1 | 1 | 1 |
| Data collection | ||||
| Beamline | NW12A of PF | BL-17A of PF | BL-17A of PF | BL-5A of PF |
| Wavelength (Å) | 1.0 | 0.98 | 0.98 | 1.0 |
| Resolution (Å) | 38.5-2.4 | 44.9-2.1 | 32.6-3.5 | 42.3-3.1 |
| of the outer shell (Å) | 2.5-2.4 | 2.2-2.1 | 3.6-3.5 | 3.2-3.1 |
| Unique reflections | 11198 | 8419 | 1848 | 2851 |
| Completeness (%) | 99.4 | 96.6 | 97.2 | 99.3 |
| in the outer shell (%) | 99.5 | 98.4 | 100.0 | 100.0 |
| R merge b (%) | 4.2 | 5.9 | 14.3 | 6.8 |
| in the outer shell (%) | 33.0 | 36.5 | 48.7 | 28.4 |
| Redundancy | 7.0 | 6.4 | 5.8 | 6.3 |
| in the outer shell | 6.9 | 6.7 | 6.2 | 6.7 |
| I/σ(I) | 22.4 | 15.4 | 8.2 | 13.0 |
| in the outer shell | 4.7 | 4.3 | 2.2 | 2.7 |
| Structure refinement | ||||
| Resolution range (Å) | 38.5-2.4 | 44.9-2.1 | 30.0-3.5 | 40.0-3.1 |
| Used reflections | 11197 | 8418 | 1847 | 2847 |
| R-factorc (%) | 17.7 | 20.7 | 20.5 | 25.5 |
| R free d (%) | 21.6 | 25.1 | 22.8 | 28.4 |
| Average B-factor | ||||
| of RNA | 48.1 | 39.1 | 137.8 | 107.5 |
| of ligand | 32.6 | 132.2 | ||
| of water | 51.6 | 42.9 | 78.7 | 126.0 |
| No. of atoms | ||||
| of RNA | 1461 | 920 | 937 | 920 |
| of ligand | - | 68 | 42 | - |
| of water | 137 | 142 | 8 | 19 |
| R.m.s.d. bond length (Å) | 0.005 | 0.021 | 0.058 | 0.007 |
| R.m.s.d. bond angles (°) | 1.0 | 1.6 | 1.0 | 1.1 |
aNumber of RNA duplex in the asymmetric unit.
b R merge = 100 × Σhklj|Ihklj – <Ihklj>| / Σhklj<Ihklj>.
c R-factor = 100 × Σ||Fo| – |Fc|| / Σ|Fo|, where |Fo| and |Fc| are optimally scaled observed and calculated structure factor amplitudes, respectively.
dCalculated using a random set containing 10% of observations.
The initial phases of the four crystals were derived with the molecular replacement method by using the program AutoMR from the Phenix suite (41,42). The molecular structures were constructed and manipulated with the programs Coot (43,44). The atomic parameters of each structure were refined using the program phenix.refine from the Phenix suite (41,45) through a combination of simulated-annealing, crystallographic conjugate gradient minimization refinements and B-factor refinements. The statistics of structure refinements are summarized in Table 1. Molecular drawings were made using the program PyMOL (46). The atomic coordinates and experimental data of the A1408m1A, A1408m1A-G418, A1408m1A-Paromomycin and A1408m1A-Gentamicin crystals have been deposited in the Protein Data Bank (PDB) with the ID codes 4WCP, 4WCQ, 4WCR and 4WCS, respectively (Supplementary Figure S2).
RESULTS
Aminoglycoside binding to the m1A1408-modified A site
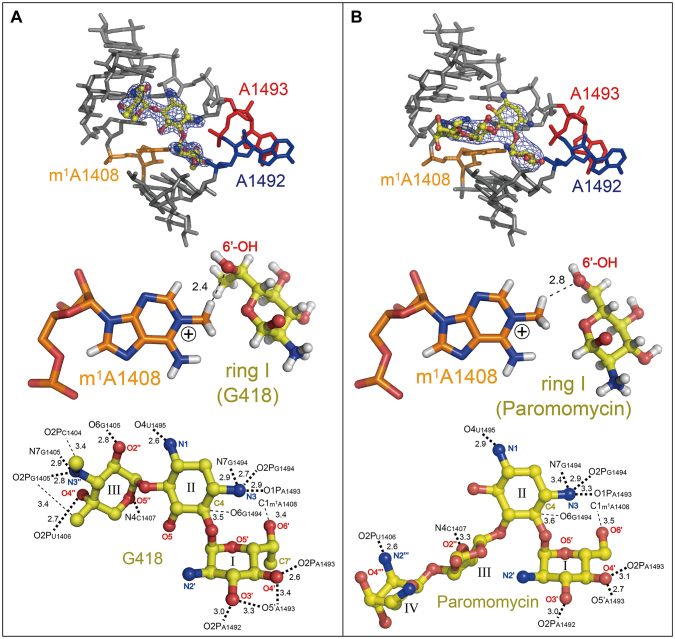
In the A1408m1A-G418 and A1408m1A-Paromomycin crystals, aminoglycosides G418 and paromomycin both possessing a 6′-OH group specifically bind to the m1A1408-modified A site and force it to adopt the ‘on’ state conformation with fully bulged-out A1492 and A1493 (Figure 3, top). It is very important to note that these bulged-out adenines recognize two consecutive Watson–Crick G = C base pairs in a symmetry-related molecule through the A-minor interactions (Supplementary Figure S3). This crystal packing interaction perfectly mimics the recognition between the A site and the tRNA–mRNA complex occurring in the ribosome (12,13,39). Ring I of G418 and paromomycin stack on top of G1491. A short distance between the CH3 group attached to N1 of m1A1408 and the 6′-OH group of ring I suggests that a C-H…O interaction may exist between them and supports formation of a pseudo pair between m1A1408 and ring I (Figure 3, middle). Another possibility is that 6′-OH of ring I simply accommodates in the binding pocket without any steric/electrostatic hindrance with m1A1408. On the opposite side of ring I, the O3′ and O4′ atoms make hydrogen bonds to the phosphate oxygen atoms of A1492 and A1493 (Figure 3, bottom). In addition, ring II, which is also referred to as 2-DOS (2-deoxystreptamine) ring and important for the specific recognition of the A site, makes hydrogen bonds from its N3 atom to the phosphate oxygen atoms of A1493 and A1494 (Figure 3, bottom), so that A1492 and A1493 are fixed in the bulged-out conformation. G418 that belongs to the 4,6-disubstituted subclass makes 17 direct contacts (13 hydrogen bonds and 4 C-H…O interactions) to the mutant A site (Figure 3A, bottom). On the other hand, paromomycin that belongs to the 4,5-disubstituted subclass makes 11 direct contacts (9 hydrogen bonds and 2 C-H…O interactions) (Figure 3B, bottom).
Figure 3.
Binding of aminoglycosides, G418 (A) and paromomycin (B), to the ‘on’ states of the m1A1408-modified A site. Overviews (top), pseudo pairs between m1A1408 and ring I (middle) and detailed interactions of aminoglycosides to the mutant A site (bottom) are shown. The m1A1408, A1492 and A1493 residues are colored in orange, blue and red, respectively. In the top figures, local Fo-Fc omit maps calculated after removing aminoglycosides are drawn in dark blue at 3 σ contour level. In the middle figures, hydrogen atoms colored in white are added for a better understanding. Hydrogen bonds and C-H…O interactions are shown as bold and thin dashed lines with distances in Å, respectively.
In the A1408m1A-Gentamicin crystal obtained in the presence of gentamicin with a 6′-NH3+ group, binding of the aminoglycoside to the mutant A site could not be observed (Supplementary Figure S2). We cannot exclude a possibility that gentamicin did not bind only in the present crystal form due to, for example, crystal packing effect or degrade of gentamicin. However, it can be assumed by a molecular modelling that ring I with the 6′-NH3+ group is less likely to form a pseudo pair with m1A1408, because the NH3+ group may repel the CH3 group attached to the positively-charged N1 atom (Figure 4).
Figure 4.
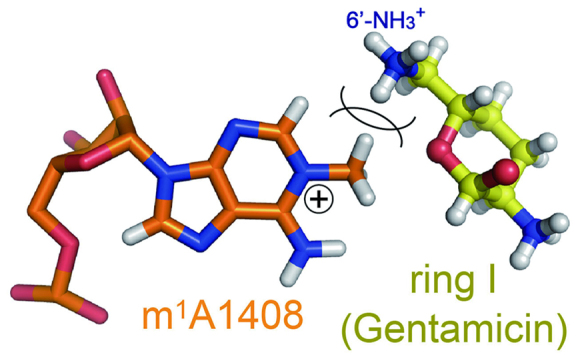
Molecular modelling of an impossible pseudo pair between m1A1408 and ring I of gentamicin. The 6′-NH3+ group repels the CH3 group attached to the positively-charged N1 atom of m1A1408.
DISCUSSION
Minimum inhibitory concentration
The present crystal structures shown above agree with the minimum inhibitory concentrations (MICs) of aminoglycosides against E. coli (Supplementary Table S2) (28). Escherichia coli CSH-2 strain without the npmA gene is susceptible to both paromomycin (MICCSH-2 = 0.5 μg/ml) and gentamicin (MICCSH-2 ≤ 0.06 μg/ml). On the other hand, the clinically isolated antibiotic-resistant E. coli ARS3 strain and E. coli CSH-2 strain possessing a conjugative plasmid carrying the npmA gene (CSH-2 (pARS3)) exhibit high-level resistance to gentamicin with the 6′-NH3+ group (MICARS3 > 256 μg/ml; MICCSH-2 (pARS3) = 125 μg/ml). These antibiotic-resistant strains exhibit low to moderate level resistance to paromomycin with the 6′-OH group (MICARS3 = 64 μg/ml; MICCSH-2 (pARS3) = 4 μg/ml) probably due to attenuation of pseudo-pair stability by the N1-methylation of A1408 (Supplementary Figure S1), but it can also be said that paromomycin is moderately active against the mutant strains. Although MICs of G418 against the antibiotic-resistant strains are not reported, it is expected by our crystal structure that the compound with the 6′-OH group should be active against resistant bacteria with N1-methyltransferase.
Effects of the N1-methylation of A1408 on translation efficiency and fitness of the host
From a total of four crystals obtained in this study, nine m1A1408-modified A site structures were observed (Supplementary Figure S2). These structures, as well as accumulated wild-type A site structures obtained from crystal structures of the 30S and 70S ribosomes (12,13) and the A-site RNA model (14–26,38), allow us to speculate and compare the motions of the modified and wild-type A site molecular switches. In the case of the wild-type, the A site molecular switch takes relatively stable ‘off’ states, in which one of the two adenines form a cis Watson–Crick A○A base pair with A1408 through N1…H-N6 (38,47) (Figure 5A and Supplementary Figure S4). On the other hand, in the case of the m1A1408-modified A site, the A site switch cannot take any stable ‘off’ state, since the CH3 group attached to N1 of m1A1408 inhibits base-pair formation with A1492 or A1493 (Figure 5B). Since the modified A site takes the ‘on’ state with two bulged-out adenines, which is identical to that of the wild-type A site, the m1A1408-modified A site maintains its function as the molecular switch. These observations suggest that the ON/OFF switching might favorably occur in the m1A1408-modified A site compared to the wild-type. Recent study revealed that there is a simple linear trade-off between speed and accuracy of decoding (48). Therefore, it can be expected that the energetically ‘softer’ mutant A-site molecular switch may display lower accuracy of translation compared to the wild-type one.
Figure 5.
Local structures of the ‘off’ states of the wild-type (A; PDB-ID = 3BNL (33)) and m1A1408-modified (B) A sites. A dashed line indicates a hydrogen bond. The CH3 group attached to N1 of m1A1408 is shown as an orange sphere.
Recently, effect of m1A1408 modification on translation efficiency and fitness of the host have carefully been analyzed by Grillot-Courvalin and co-workers (49). According to their analyses, several important insights have been elucidated; (i) expression of NpmA interferes with endogenous methylation at the neighboring C1407 residue (m5C1407 modification) by blocking action of a housekeeping methyltransferase RsmF. (ii) Escherichia coli strains with and without the npmA gene had the same growth rate, but in vitro competition experiments between them revealed a small disadvantage of the former cell (loss of 2.7% per generation). On the other hand, (iii) there was no reduction in growth rate of E. coli without the housekeeping rsmF gene relative to the wild-type E. coli, and no fitness decrease in competition experiments performed between E. coli strains with and without the rsmF gene (relative fitness of 1). In addition, dual luciferase assays showed that (iv) a decrease in the +1 reading frame maintenance (16%) and in UGA readthrough (20%) was observed in the presence of NpmA, and a decrease in the UGA readthrough (24%) was also observed in the absence of RsmF. These results are summed up that the exogenous N1-methylation at A1408 is a part of the causes of the slight fitness cost and the decreasing of translation accuracy. The present comparative analysis of the motions of the A-site molecular switches with and without m1A1408 modification suggests at least that the N1-methylation of A1408 affects fitness cost and translation accuracy only at a minimum level but provides a good level of resistance against aminoglycosides.
A possible conformational change of A1408
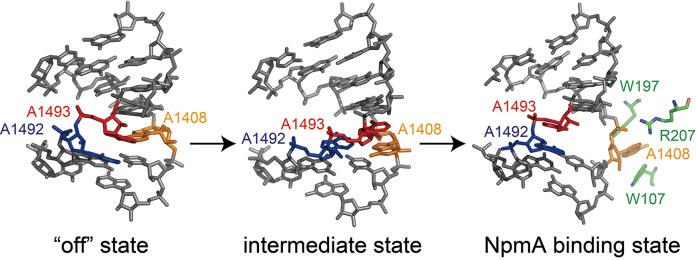
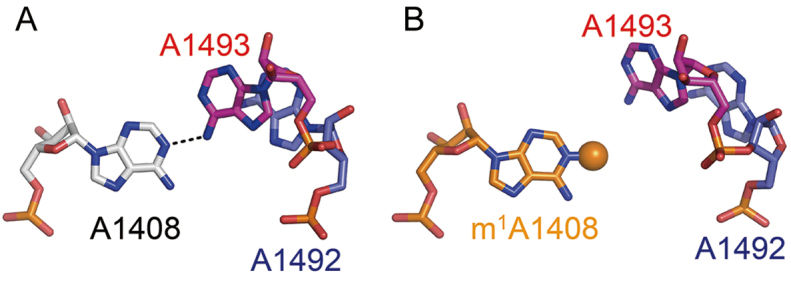
Based on the crystal structure of NpmA in complex with the substrate 30S ribosome, Dunkle et al. proposed how NpmA binds to the 30S ribosome and changes its conformation for inducing conformational changes of the ribosomal RNA that allows A1408 to enter the active site of NpmA to be methylated (31). The A1408 residue is located opposite the conformationally dynamic A1492 and A1493 residues. Thus, A1408 is likely to be easily bulged out, and then might be captured and stabilized by NpmA residues R207 and W107/W197, respectively. However, despite the accumulated structural information of the bacterial ribosome, structure of the A site with bulged-out A1408 has not been reported so far. Recently, we have successfully captured a partially bulged-out conformation of A1408 in our previously reported crystal structure of the bacterial A site in complex with a synthetic aminoglycoside (Figure 6, middle) (26). The structure may be one of the intermediate states of the A1408 flipping. The partially bulged-out A1408 could easily be captured by R207, and then be taken into the catalytic core of NpmA composed of W107 and W197 (Figure 6).
Figure 6.
A possible conformational change of A1408. In the ‘off’ state (left; PDB ID = 3BNL (38)), A1408 (orange) stays inside the A-site helix and forms a base pair with A1493 (red) through a hydrogen bond. In a possible intermediate state (middle; PDB ID = 4PDQ (26)), A1492 (blue) and A1493 (red) are bulged in and A1408 (orange) is partially bulged out. In the NpmA binding state (right; PDB ID = 4OX9 (31)), A1408 is fully bulged out and stabilized by NpmA residues R207, W107 and W197 (green).
CONCLUSION
In the present study, we have shown how bacteria acquire high-level resistance against aminoglycosides with the 6′-NH3+ group along with a minimum of fitness cost by the N1-methylation of A1408 in 16S rRNA. In addition, we have confirmed at the atomic level that aminoglycosides with the 6′-OH group are still effective against antibiotic-resistant bacteria possessing the A1408-N1-methyltransferase. These observations indicate that a hydrogen-bond acceptor at the 6′ position of ring I is an essential element for designing the next-generation aminoglycosides (33,50,51). Taken together with a result of our previous crystallographic study on the A1408G spontaneous mutation at chromosomal level found in clinical isolates (32), structural bases for the antibiotic resistance conferred by mutations at A1408 in 16S rRNA are now completely elucidated. Obtained structural information is of great value to overcome a part of the issue of antibiotic resistance.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the Photon Factory for provision of synchrotron radiation facilities (No. 2012G519 and 2014G532) and acknowledge the staff of the BL-5A, BL-17A and NW12A beamlines.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACCESSION NUMBERS
PDB IDs: 4WCP, 4WCQ, 4WCR and 4WCS.
FUNDING
Ministry of Education, Culture, Sports, Science and Technology, Japan (MEXT) Grant-in-Aid for Young Scientists (B) [23790054, 26860025]; MEXT Grant-in-Aid for Scientific Research (C) [17K08248]; Kurata Memorial Hitachi Science and Technology Foundation, Kurata Grant [2013-20] (in part); Ichiro Kanehara Foundation [15KI192] (in part); Japan Science Society, Sasakawa Scientific Research Grant [28-325] (to H.K.); SUNBOR Scholarship (to H.K.). Funding for open access charge: MEXT, Grant-in-Aid for Scientific Research (C) [17K08248].
Conflict of interest statement. None declared.
REFERENCES
- 1. Davies J., Gorini L., Davies B.D.. Misreading of RNA codewords induced by aminoglycoside antibiotics. Mol. Pharmacol. 1965; 1:93–106. [PubMed] [Google Scholar]
- 2. Davies J., Jones D.S., Khorana H.G.. A further study of misreading of codons induced by streptomycin and neomycin using ribopolynucleotides containing two nucleotides in alternating sequence as templates. J. Mol. Biol. 1966; 18:48–57. [DOI] [PubMed] [Google Scholar]
- 3. Cabañas M.J., Vázquez D., Modolell J.. Inhibition of ribosomal translocation by aminoglycoside antibiotics. Biochem. Biophys. Res. Commun. 1987; 83:991–997. [DOI] [PubMed] [Google Scholar]
- 4. Misumi M., Nishimura T., Komai T., Tanaka N.. Interaction of kanamycin and related antibiotics with the large subunit of ribosomes and the inhibition of translocation. Biochem. Biophys. Res. Commun. 1978; 84:358–365. [DOI] [PubMed] [Google Scholar]
- 5. Hirokawa G., Kiel M.C., Muto A., Selmer M., Raj V.S., Liljas A., Igarashi K., Kaji H., Kaji A.. Post-termination complex disassembly by ribosome recycling factor, a functional tRNA mimic. EMBO J. 2002; 21:2272–2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pipersberg W., Aboshanab K.M., Schmid-Beiβner H., Wehmeier U.F.. Arya DP. The biochemistry and genetics of aminoglycoside producers. Aminoglycoside Antibiotics, from Chemical Ciology to Drug Discovery. 2007; Hoboken: Wiley-Interscience; 15–118. [Google Scholar]
- 7. Shakya T., Wright G.D.. Arya DP. Mechanisms of aminoglycoside amtibiotic resistance. Aminoglycoside Antibiotics, from Chemical Biology to Drug Discovery. 2007; Hoboken: Wiley-Interscience; 119–140. [Google Scholar]
- 8. Cundiffe E. How antibiotic-producing organisms avoid suicide. Annu. Rev. Microbiol. 1989; 43:207–233. [DOI] [PubMed] [Google Scholar]
- 9. World Health Organization (WHO) WHO report on antibiotic resistance: Antimicrobial resistance: global report on surveillance 2014. 2014; Geneva. [Google Scholar]
- 10. Kondo J., Westhof E.. Gualerzi C, Brandi L, Fabbretti A, Pon C. Aminoglycoside antibiotics: structural decoding of inhibitors targeting the ribosomal decoding A site. Antibiotics: Targets, Mechanisms and Resistance. 2013; Weinheim: Wiley-VCH; 453–470. [Google Scholar]
- 11. Kondo J., Westhof E.. Arya DP. Structural comparisons between prokaryotic and eukaryotic ribosomal decoding A sites free and complexed with aminoglycosides. Aminoglycoside Antibiotics, from Chemical Biology to Drug Discovery. 2007; Hoboken: Wiley-Interscience; 209–223. [Google Scholar]
- 12. Ogle J.M., Brodersen D.E., Clemons W.M. Jr, Tarry M.J., Carter A.P., Ramakrishnan V.. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001; 292:897–902. [DOI] [PubMed] [Google Scholar]
- 13. Selmer M., Dunham C.M., Murphy F.V. 4th, Weixlbaumer A., Petry S., Kelley A.C., Weir J.R., Ramakrishnan V.. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006; 313:1935–1942. [DOI] [PubMed] [Google Scholar]
- 14. Vicens Q., Westhof E.. Crystal structure of paromomycin docked into the eubacterial ribosomal decoding A site. Structure. 2001; 9:647–658. [DOI] [PubMed] [Google Scholar]
- 15. Vicens Q., Westhof E.. Crystal structure of a complex between the aminoglycoside tobramycin and an oligonucleotide containing the ribosomal decoding a site. Chem. Biol. 2002; 9:747–755. [DOI] [PubMed] [Google Scholar]
- 16. Vicens Q., Westhof E.. Crystal structure of geneticin bound to a bacterial 16S ribosomal RNA A site oligonucleotide. J. Mol. Biol. 2003; 326:1175–1188. [DOI] [PubMed] [Google Scholar]
- 17. François B., Szychowski J., Adhikari S.S., Pachamuthu K., Swayze E.E., Griffey R.H., Migawa M.T., Westhof E., Hanessian S.. Antibacterial aminoglycosides with a modified mode of binding to the ribosomal-RNA decoding site. Angew. Chem. Int. Ed. Engl. 2004; 43:6735–6738. [DOI] [PubMed] [Google Scholar]
- 18. François B., Russell R.J.M., Murray J.B., Aboul-ela F., Masquida B., Vicens Q., Westhof E.. Crystal structures of complexes between aminoglycosides and decoding A site oligonucleotides: role of the number of rings and positive charges in the specific binding leading to miscoding. Nucleic Acids Res. 2005; 33:5677–5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Han Q., Zhao Q., Fish S., Simonsen K.B., Vourloumis D., Froelich J.M., Wall D., Hermann T.. Molecular recognition by glycoside pseudo base pairs and triples in an apramycin-RNA complex. Angew. Chem. Int. Ed. Engl. 2005; 44:2694–2700. [DOI] [PubMed] [Google Scholar]
- 20. Kondo J., François B., Russell R.J., Murray J.B., Westhof E.. Crystal structure of the bacterial ribosomal decoding site complexed with amikacin containing the γ-amino-α-hydroxybutyryl (haba) group. Biochimie. 2006; 88:1027–1031. [DOI] [PubMed] [Google Scholar]
- 21. Kondo J., Pachamuthu K., François B., Szychowski J., Hanessian S., Westhof E.. Crystal structure of the bacterial decoding site complex with a synthetic doubly functionalized paromomycin derivative: a new specific binding mode to and A-minor motif enhances in vitro antibacterial activity. ChemMedChem. 2007; 2:1631–1638. [DOI] [PubMed] [Google Scholar]
- 22. Hanessian S., Pachamuthu K., Szychowski J., Giguére A., Swayze E., Migawa M.T., François B., Kondo J., Westhof E.. Structure-based design, synthesis and A-site rRNA co-crystal complexes of novel amphiphilic aminoglycoside antibiotics with new binding modes: A synergistic hydrophobic effect against resistant bacteria. Bioorg. Med. Chem. Lett. 20:7097–7101. [DOI] [PubMed] [Google Scholar]
- 23. Szychowski J., Kondo J., Zahr O., Auclair K., Westhof E., Hanessian S., Keillor J.W.. Inhibition of aminoglycoside-deactivation enzymes APH(3′)-IIIa and AAC(6′)-Ii by amphiphilic paromomycin O2’’-ether analogues. ChemMedChem. 2011; 6:1961–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kondo J., Koganei M., Kasahara T.. Crystal structure and specific binding mode of sisomicin to the bacterial ribosomal decoding site. ACS Med. Chem. Lett. 2012; 3:741–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Maianti J.P., Kanazawa H., Dozzo P., Matias R.D., Feeney L.A., Armstrong E.S., Hildebrandt D.J., Kane T.R., Gliedt M.J., Goldblum A.A. et al. . Toxicity modulation, resistance enzyme evasion, and A-site X-ray structure of broad-spectrum antibacterial neomycin analogs. ACS Chem. Biol. 2014; 9:2067–2073. [DOI] [PubMed] [Google Scholar]
- 26. Hanessian S., Saavedra O.M., Vilchis-Reyes M.A., Maianti J.P., Kanazawa H., Dozzo P., Matias R.D., Serio A., Kondo J.. Synthesis, broad spectrum antibacterial activity, and X-ray co-crystal structure of the decoding bacterial ribosomal A-site with 4′-deoxy-4′-fluoro neomycin analogs. Chem. Sci. 2014; 5:4621–4632. [Google Scholar]
- 27. Wachino J., Arakawa Y.. Exogenously acquired 16S rRNA methyltransferases found in aminoglycoside-resistant pathogenic Gram-negative bacteria: an update. Drug Resist. Updat. 2012; 15:133–148. [DOI] [PubMed] [Google Scholar]
- 28. Wachino J., Shibayama K., Kurokawa H., Kimura K., Yamane K., Suzuki S., Shibata N., Ike Y., Arakawa Y.. Novel plasmid-mediated 16S rRNA m1A1408 methyltransferase, npmA, found in a clinically isolated Esherichia coli strain resistant to structurally diverse aminoglycoside. Antimicrob. Agents Chemother. 2007; 51:4401–4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Macmaster R., Zelinskaya N., Savic M., Rankin C.R., Conn G.L.. Structural insights into the function of aminoglycoside-resistance A1408 16S rRNA methyltransferases from antibiotic-producing and human pathogenic bacteria. Nucleic Acids Res. 2010; 38:7791–7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Husain N., Obranić S., Koscinski L., Seetharaman J., Babić F., Bujnicki J.M., Maravić-Vlahoviček G., Sivaraman J.. Structural basis for the methylation of A1408 in 16S rRNA by a panaminoglycoside resistance methyltransferase NpmA from a clinical isolate and analysis of the NpmA interactions with the 30S ribosomal subunit. Nucleic Acids Res. 2011; 39:1903–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dunkle J.A., Vinal K., Desai P.M., Zelinskaya N., Savic M., West D.M., Conn G.L., Dunham C.M.. Molecular recognition and modification of the 30S ribosome by the aminoglycoside-resistance methyltransferase NpmA. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:6275–6280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kondo J. A structural basis for the antibiotic resistance conferred by an A1408G mutation in 16S rRNA and for the antiprotozoal activity of aminoglycosides. Angew. Chem. Int. Ed. Engl. 2012; 51:465–468. [DOI] [PubMed] [Google Scholar]
- 33. Kondo J., Koganei M., Maianti J.P., Ly V.L., Hanessian S.. Crystal structures of a bioactive 6‘-hydroxy variant of sisomicin bound to the bacterial and protozoal ribosomal decoding sites. ChemMedChem. 2013; 8:733–739. [DOI] [PubMed] [Google Scholar]
- 34. Shalev M., Kondo J., Kopelyanskiy D., Jaffe C., Adir L.N., Baasov T.. Identification of the molecular attributes required for aminoglycoside activity against Leishmania. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:13333–13338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kondo J., Urzhumtsev A., Westhof E.. Two conformational states in the crystal structure of the Homo sapiens cytoplasmic ribosomal decoding A site. Nucleic Acids Res. 2006; 34:676–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kondo J., François B., Urzhumtsev A., Westhof E.. Crystal structure of the Homo sapiens cytoplasmic ribosomal decoding site complexed with apramycin. Angew. Chem. Int. Ed. Engl. 2006; 45:3310–3314. [DOI] [PubMed] [Google Scholar]
- 37. Kondo J., Hainrichson M., Nudelman I., Shallom-Shezifi D., Barbieri C.M., Pilch D.S., Westhof E., Baasov T.. Differential selectivity of natural and synthetic aminoglycosides towards the eukaryotic and prokaryotic decoding A sites. ChemBioChem. 2007; 8:1700–1709. [DOI] [PubMed] [Google Scholar]
- 38. Kondo J., Westhof E.. The bacterial and mitochondrial ribosomal A-site molecular switches possess different conformational substates. Nucleic Acids Res. 2008; 36:2654–2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kondo J., Sauter C., Masquida B.. Hartmann RK, Binderein A, Schön A, Westhof E. RNA crystallization. Handbook of RNA Biochemistry: Second, Completely Revised and Enlarged Edition. 2014; Weinheim: Wiley-VCH; 481–498. [Google Scholar]
- 40. Collaborative Computational Project, Number 4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr. 1994; D50:760–763. [DOI] [PubMed] [Google Scholar]
- 41. Adams P.D., Afonine P.D., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.-W., Kapral G.J., Grosse-Kunstleve R.W. et al. . PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. 2010; D66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J.. Phaser crystallographic software. J. Appl. Cryst. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta Crystallogr. 2004; D60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 44. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta Crystallogr. 2010; D66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Afonine P.V., Grosse-Kunstleve R.W., Echols N., Headd J.J., Moriarty N.W., Mustyakimov M., Terwilliger T.C., Urzhumtsev A., Zwart P.H., Adams P.D.. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. 2012; D68:352–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. DeLano W.L. The PyMOL Molecular Graphics System. 2008; Palo Alto: DeLano Scientific LLC. [Google Scholar]
- 47. Shandrick S., Zhao Q., Han Q., Ayida B.K., Takahashi M., Winters G.C., Simonsen K.B., Vourloumis D., Hermann T.. Monitoring molecular recognition of the ribosomal decoding site. Angew. Chem. Int. Ed. Engl. 2004; 43:3177–3182. [DOI] [PubMed] [Google Scholar]
- 48. Johansson M., Zhang J., Ehrenberg M.. Genetic code translation displays a linear trade-off between efficiency and accuracy of tRNA selection. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:131–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lioy V.S., Goussard S., Guerinrau V., Yoon E-J., Courvalin P., Galimand M., Grillot-Courvalin C.. Aminoglycoside resistance 16S rRNA methyltransferases block endogenous methylation, affect translation efficiency and fitness of the host. RNA. 2014; 20:382–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hainrichson M., Nudelman I., Baasov T.. Designer aminoglycosides: the race to develop improced antibiotics and compuonds for the treatment of human genetic diseases. Org. Biomol. Chem. 2008; 6:227–239. [DOI] [PubMed] [Google Scholar]
- 51. Duscha S., Boukari H., Shcherbakov D., Salian S., Silva S., Kendall A., Kato T., Akbergenov R., Perez-Fernandez D., Bernet B. et al. . Identification and ecaluation of improced 4‘-O-(alkyl) 4,5-disubstituted 2-deoxystraptamines as next-generation aminoglycoside antibiotics. Mbio. 2014; 5, doi:10.1128/mBio.01827-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.