Abstract
Histone variants and their chaperones are key regulators of eukaryotic transcription, and are critical for normal development. The histone variant H3.3 has been shown to play important roles in pluripotency and differentiation, and although its genome-wide patterns have been investigated, little is known about the role of its dynamic turnover in transcriptional regulation. To elucidate the role of H3.3 dynamics in embryonic stem cell (ESC) biology, we generated mouse ESC lines carrying a single copy of a doxycycline (Dox)-inducible HA-tagged version of H3.3 and monitored the rate of H3.3 incorporation by ChIP-seq at varying time points following Dox induction, before and after RA-induced differentiation. Comparing H3.3 turnover profiles in ESCs and RA-treated cells, we identified a hyperdynamic H3.3-containing nucleosome at the −1 position in promoters of genes expressed in ESCs. This dynamic nucleosome is restricted and shifted downstream into the +1 position following differentiation. We suggest that histone turnover dynamics provides an additional mechanism involved in expression regulation, and that a hyperdynamic −1 nucleosome marks promoters in ESCs. Our data provide evidence for regional regulation of H3.3 turnover in ESC promoters, and calls for testing, in high resolution, the dynamic behavior of additional histone variants and other structural chromatin proteins.
INTRODUCTION
Chromatin, comprised of DNA and its packaging proteins, is a central component in regulating embryonic stem cell (ESC) identity and plasticity (1). The basic component of chromatin structure is the nucleosome, which is made up of DNA wrapped around two copies each of the histone proteins H2A, H2B, H3 and H4. Most histones are stably associated with DNA, and new incorporation occurs mostly during DNA replication, although a smaller histone fraction, including, notably, histone variants, exhibits dynamic exchange with the soluble pool of nucleoplasmic histones (2). In ESCs this dynamic interaction is enhanced (3,4), resulting in a hyperdynamic chromatin state in ESCs, which is believed to be functionally important for the maintenance of pluripotency (3,5–9).
In recent years, accumulating evidence suggest important roles for histone variants in shaping the epigenetic landscape of ESCs and in regulating pluripotency and early differentiation events (10,11). In particular, the histone variant H3.3 was shown to play a key role in maintaining ESC pluripotency by regulating gene expression programs important for lineage specification (12–14). H3.3 is ubiquitously expressed and its deposition in chromatin is replication independent and mediated by two chaperone systems, HIRA and DAXX (15). H3.3 turnover is linked to a variety of key aspects of chromatin biology (16,17), and is involved in both gene expression activation (18) and the silencing of repetitive elements (19). Distinct mechanisms of histone deposition and eviction (i.e. turnover) pertain to the dynamics of H3.3 at different chromatin regions (20). In mouse fibroblasts fast turnover was found on promoters and enhancers of active genes, correlating, as expected, with the presence of active histone marks (21). In post mitotic neurons, rapid H3.3 turnover was shown to be essential for normal brain development, and in both neurons and glia, H3.3 histone turnover patterns are critical in mediating neuronal activity-dependent gene expression, synaptic connectivity and cognition (22). In ESCs, the turnover patterns seem to be more complex and high turnover rates are not restricted to active genes. Instead, dynamic exchange of H3.3 was also found around promoters of silent genes as well as repressed repetitive elements (13,18,19). However, at a global scale, H3.3 was the only chromatin protein tested, which did not show the characteristic hyperdynamic binding of chromatin proteins in ESCs (3). Finally, HIRA and H3.3 are both required for the establishment of the suppressive H3K27me3 mark at promoters of developmentally regulated genes in ESCs (23). These somewhat confounding results, prompted us to study the genome-wide incorporation of H3.3 in ESCs and during early ESC differentiation, examine its dynamic exchange patterns, and test its potential involvement in regulating gene expression and/or silencing during differentiation.
MATERIALS AND METHODS
Cell culture
Cells were cultured on gelatinized tissue culture plates in ESC media (Dulbecco modified Eagle medium supplemented with 10% defined fetal bovine serum, 100 U/ml penicillin, 100 mg/ml streptomycin, 2 mmol/l L-glutamine, 5 mg/ml MEM non-essential amino acids, 0.12 mmol/l β-mercaptoethanol and 1000 U/ml leukemia inhibitory factor) supplemented with the 2i inhibitors cocktail (mitogen activated protein kinase (MAPK)/extracellular-signal-regulated kinase (ERK) kinase (MEK) inhibitor PD0325901 and the glycogen synthase kinase 3 (GSK3) inhibitor CHIR99021). This media was shown to maintain mES cell self-renewal without the addition of exogenous factors (24). Hygromycin b was also added as selection against loss of the H3.3-HA construct for several passages. For retinoic acid (RA) induced differentiation, cells were grown on gelatin-coated dishes for 4 days in ESC media without LIF and 2i supplemented with 1 μM RA. All cells were cultured at 37°C in 5% CO2.
Immunofluorescence
Cells, grown on plastic or cover-slips, were fixed in 4% paraformaldehyde (15 min, room temperature), washed three times (phosphate-buffered saline (PBS), 5 min, room temperature), permeabilized (0.5% Triton X-100 in PBS, 5 min, room temperature) and incubated with the primary antibodies diluted 1:20–1:400 (1 h, room temperature or 4°C overnight) in 10% serum in PBS. Cells were then washed three times (PBS, 5 min, room temperature), incubated with secondary antibodies diluted 1:1000 (1 h, room temperature) in 10% serum in PBS, washed again (PBS, 5 min, room temperature), DAPI stained (5 min, room temperature), washed (PBS, 5 min, room temperature) and mounted on a microscope slide with anti-fade (Dako, Glostrup, Denmark). Cells grown on plastic were kept in PBS until imaged. Detection was carried out using the relevant secondary antibodies conjugated to Alexa488 (A21202, A21206, A11055) or Alexa568 (A1003, A10042, A11057, Molecular Probes).
Protein extraction and western blots
For whole cell extraction: trypsinized cells were washed once with cold PBS, centrifuged (500 g, 4°C, 5 min) and resuspended in ice-cold hypotonic Lysis Buffer (20 mM Tris–Cl pH 7.5, 0.2 ethylenediaminetetraacetic acid (EDTA) mM, 0.5 mM 1,4-Dithiothreitol (DTT), protease inhibitors cocktail 1:100 [Sigma]). Cells were incubated for 10 min on ice. A similar volume of High Salt Buffer (20 mM Tris–Cl pH 7.5, 0.2 EDTA mM, 0.5 mM DTT, 1 M NaCl) was added and cells were incubated (10 min on ice). Cells were then centrifuged (20 000 g, 4°C, 30 min) and the supernatants were collected for further analysis. Protein concentrations were determined using the Bradford assay. Western blot detection was performed using LAS-3000 (FUJI FILM). Quantification was performed using ImageJ. For chromatin protein extraction: fractionation of chromatin bound proteins and nucleoplasmic proteins was done as previously described (25). Protein extraction was performed on KH2 ESCs treated with Dox for different times. The chromatin bound fraction was separated on 4–20% gradient Bis-Tris SDS gels (BioRad), blotted, and incubated with the primary antibodies. Detection for western blot was with anti-mouse anti-rabbit or anti-goat antibodies conjugated to HRP (115-035-062, 111-035-144, 705-035-147 Jackson ImmunoResearch, respectively).
Chromatin immunoprecipitation (ChIP)
Chromatin immunoprecipitation (ChIP) was performed as described previously (26). In short, cells from different time points after dox addition, with and without 4 days retinoic acid (4dRA) treatment, were collected and fixed. We normalized the cell number by the WB HA signal, since populations from different time points express different amounts of H3.3-HA. IP was done using Magna ChIP™ kit (Millipore) and DNA was purified using QIAquick PCR purification kit (Qiagen). ChIP Grade HA tag antibody was used (ab9110 from Abcam). Libraries from two input fractions and six bound fractions (ES or 4dRA cells incubated with dox for 1, 4 and 8 h) were prepared as previously described (27) and sequenced at the Technion Genome Center by Illumina HiSeq 2500 machine. Two biological replicas for each fraction were prepared and sequenced. More details can be found in the Supplementary data.
RNA extraction, library preparation and sequencing
RNA was prepared using the RNesay kit (Qiagen) according to the manufacturer's instructions, library preparation was done with QuantSeq 3′ mRNA-Seq Library Prep Kit by Lexogen. Four RNA libraries were prepared: from ESCs and 4dRA cells without or with 4H Dox addition. Libraries were subjected to single-end sequencing using Illumina Next-Seq 500 platform.
Bioinformatics and statistical analysis
Genomic mapping was done using Bowtie (28) using the mouse genome version mm9. Mapping considered only unique hits with up to three mismatches per read. Following mapping, each read was extended to a length of 500 bp and read coverage was calculated in 50 bp bins. This was done to account for limited read coverage in H3.3 histone ChIP-seq. Similar analysis with 150 bp read extension was too noisy, while longer extension led to less specific estimation of turnover rates. Genome-wide read coverage profiles were then normalized to a total of 10M mapped reads per experiment. These were saved as supplementary bigwig files. TSS/metagene plots were created using the deepTools package (v1.5.12) (29).
Turnover rate estimation
Turnover dynamics were estimated by taking the log2 of the ratio between the 1 and 8 h H3.3_HA ChIP-seq coverage (after normalizing each experiment to 10M coverage). While this ratio is not identical to a full physical model of genomic nucleosome integration, following an external switching signal (30), it allows us to estimate the overall dynamics by which each genomic locus integrates newly generated H3.3 nucleosomes. At the first time points (e.g. 1 h) only very dynamic loci show integration of H3.3 nucleosomes, resulting with strong ChIP-seq signal for these few positions. By the later time points, additional genomic loci (with slower turnover dynamics) show H3.3 integration. Due to the relatively higher number of the loci, their overall ChIP enrichment is lower. Thus the ratio between early (1 h) and late (8 h) time points offer a direct approximation of turnover dynamics. We take the logarithm of this ratio (in base 2) to normalize-out some general trends and preprocessing normalizations (e.g. due to different sequencing depth for each experiments), and to allow more accurate visualizations (e.g. x2 and x1/2 changes showing a similar magnitude of +1 and −1 in log2 scale). Changes in turnover dynamics, between mESCs before and after 4 days of RA induction were calculating by comparing the log2 ratios at the two cell populations, namely ESC log2(1 /8 h HA enrichment) minus RA log2(1/8 h HA enrichment).
RNA-seq analysis
To group genes based on RNA-seq expression levels, genes were divided to seven groups based on the difference in gene expression in ES and following 4 days of RA induction (both, following 4H Dox). Specifically, we estimated the expression level of each gene (FPKM) using cufflinks (31), and then annotated every gene as one of three non-differential groups (<2-fold change between ES and RA conditions): (i) low-average expression <1 FPKM; (ii) mid-average expression between 1 and 100 FPKM; (iii) high-average expression larger than 100 FPKM; or as one of four differential groups: (iv) down5: RA expression is at least 5-fold smaller than ES expression level; (v) down2: RA expression is 2- to 5-fold smaller than ES; (vi) up5: RA expression is at least 5-fold higher than ES expression level; and (vii) up2: RA expression is at 2- to 5-fold higher than ES expression.
RESULTS
H3.3 turnover dynamics can be measured using time-ChIP
To test the dynamic incorporation and exchange of the histone variant H3.3 in pluripotent ESCs and during early differentiation, we engineered mouse KH2 ESCs (32) to express a single copy of H3.3-HA located downstream of the Type I Collagen (Col1A1) locus under the control of Doxycyclin (Dox), thus providing a genetically encoded pulse-chase method (18,21,33). We monitored the rate of incorporation of the HA-tagged H3.3 at different time points following Dox induction, similar to the ‘time-ChIP’ method previously reported (34).
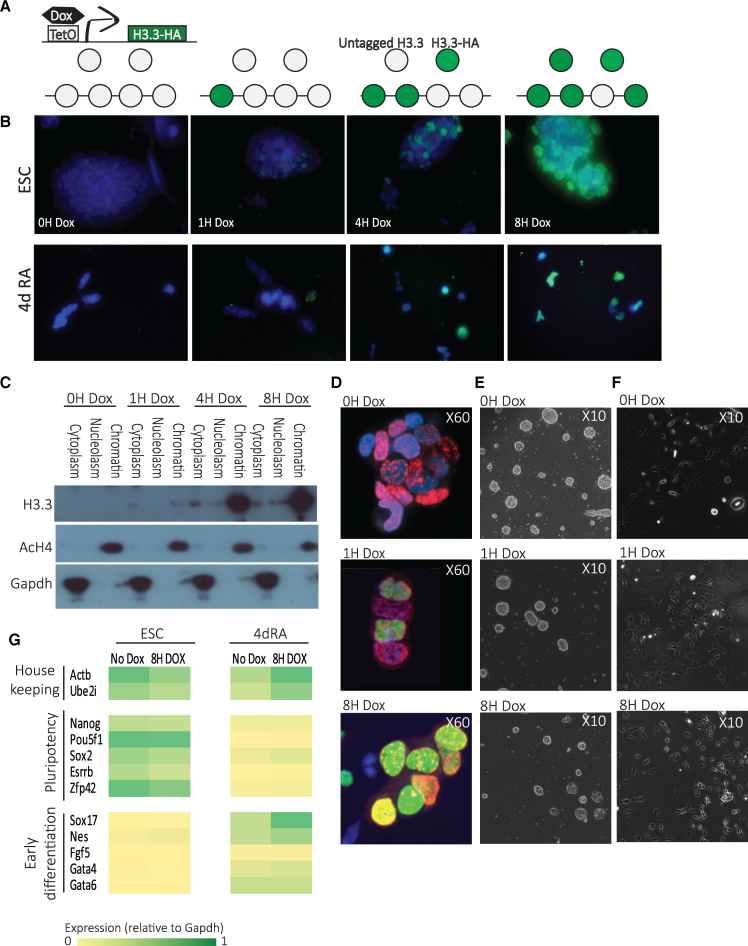
Upon Dox addition, a pool of tagged H3.3 immediately begins to accumulate, reaching saturation within 8 h (Figure 1A). To verify the inducibility and test the kinetics of the system, Dox was added to the media at different times and H3.3-HA expression levels were measured by immunofluorescence (IF, Figure 1B) and western blots (WB, Figure 1C and Supplementary Figure S1A). IF demonstrated proper chromatin localization of H3.3-HA in ESCs (Figure 1B, top) and 4-day RA-treated cells (Figure 1B, bottom), and both IF and WB demonstrated proper induction of H3.3-HA expression. To further confirm appropriate chromatin incorporation of the H3.3-HA protein, we divided the cells into three cellular fractions (cytoplasm, nucleoplasm, chromatin-bound) before performing WBs, and quantified the WB signal strength using ImageJ, with GAPDH and acetylated H4 antibodies used as controls (Supplementary Figure S1B). Once again, H3.3-HA was found exclusively in the chromatin-bound fraction (Figure 1C and Supplementary Figure S1B). Cell counts and IdU staining demonstrated normal cell cycle kinetics and proliferation rates (Figure 1D and Supplementary Figure S1C), and no change was observed in morphology of ESC or RA treated cells following Dox induction (Figure 1E and F). Growth characteristics and expression of pluripotency markers before and after RA induced differentiation with and without Dox induction were as expected (Figure 1G; Supplementary Figure S1D and E). IF on RA-treated cells demonstrated that H3.3-HA induction follows the same pattern as in ESCs. These cells displayed normal differentiated phenotypes following 8 h Dox induction (Figure 1E). H3.3-HA expression and localization to chromatin was detected as early as 1 h post induction and showed maximal levels at 8 h post-induction (Figure 1B and C).
Figure 1.
Genome-wide measurements of H3.3 dynamics. (A) A schematic diagram showing components of the doxycycline (Dox) histone H3.3-HA induction system in KH2 ESC and the use of this system for assaying H3.3 turnover dynamics. (B) HA-tagged H3.3 expression in ESC (upper panel) and in 4d RA cells (lower panel) was induced by Dox addition at different times and immunolabeled with anti-HA antibodies. DAPI staining overlay is shown in blue. (C) Time course western blot following fractionation of ESC cellular extracts to three fractions showing protein levels of transgenic HA-H3.3 (lower panel) compared to acetylated H4 (upper panel, chromatin bound) and GAPDH (middle panel, cytoplasmatic) expression at time points from 0 to 8 h (0–8 h) after Dox addition. (D) HA (green) and Bromodeoxyuridine (BrdU) (red) immunostaining of KH2 ESCs treated with IdU for 2 h and Dox for 0, 1 and 8 h. Nuclei were stained with DAPI and analyzed by confocal microscopy. (E) Representative bright-field images of KH2 ESCs after 0, 1 and 8 h with Dox show no significant difference in colonies morphology. (F) Representative bright-field images of 4dRA cells after 0, 1 and 8 h with Dox show no significant difference in cell morphology. (G) Expression levels of pluripotency, selected lineage marker and housekeeping genes in KH2 ESCs and 4dRA cells, with and without Dox addition. Normalized to Gapdh expression.
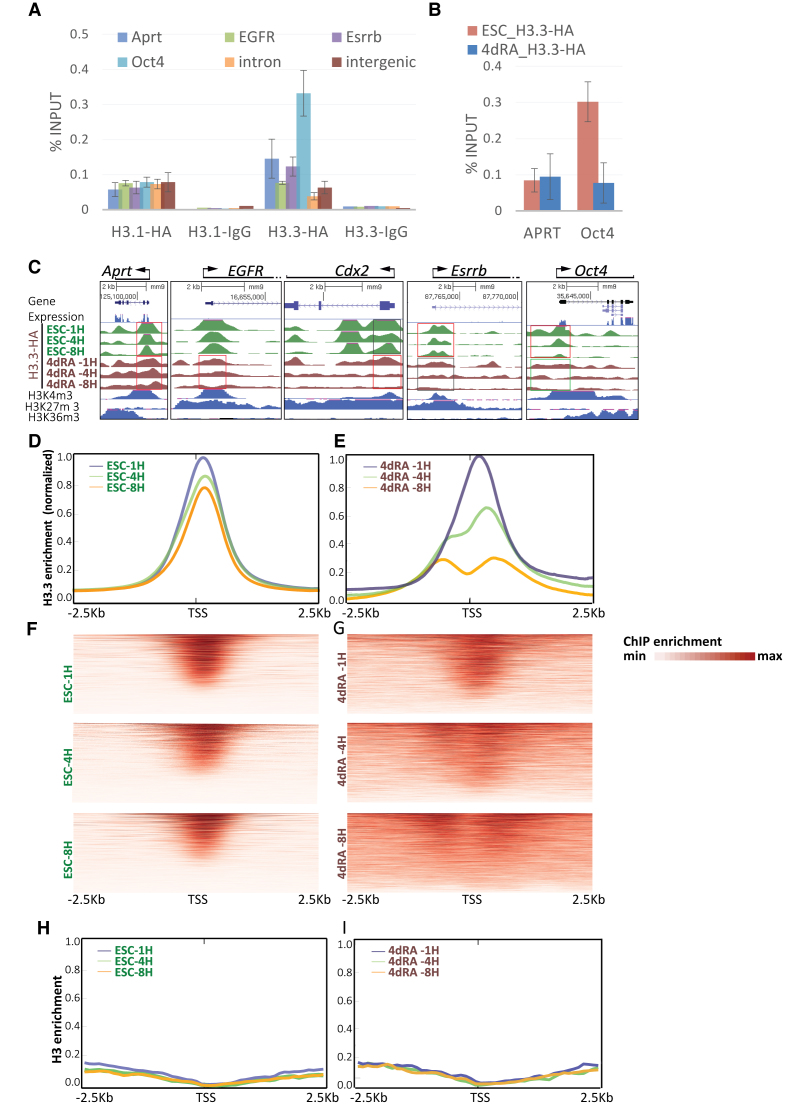
To further confirm the validity of HA ChIP, we tested the enrichment and depletion of H3.3-HA using anti-HA antibodies on previously validated H3.3-positive and H3.3-negative promoters (13) following 4 h of Dox induction, and found an excellent agreement (Figure 2A and Supplementary Figure S1F). An equivalent KH2 cell line expressing the canonical histone variant H3.1-HA did not show preference to any gene or genomic region. IgG antibody was used as a negative control.
Figure 2.
HA ChIP-seq shows faster dynamics for H3.3 in RA treated cells. (A) ChIP-PCR for H3.3-HA, H3.1-HA and IgG on KH2 ESCs incubated with Dox for 4H, using primers for the indicated genes. Aprt is a house keeping gene, EGFR is expressed in differentiated cells, Essrb and Oct4 are expressed in ESCs, and intron and intergenic regions were used as a negative controls. Enrichment was calculated relative to input. (B) ChIP-PCR for H3.3-HA on KH2 ESCs and 4dRA treated cells incubated with Dox for 4H, using primers for Aprt and Oct4. (C) UCSC screenshots of the indicated genes of H3.3-HA ChIP-seq data on KH2 ESCs induced with Dox for 1, 4 and 8 h (1, 4, 8 h—green density plots) and on 4dRA cells induced with Dox (brown density plots). Boxes show regions of rapid (red), moderate (gray) or slow (green) H3.3 replacement. mRNA tracks are shown in blue. H3K4me3, H3K27me3 and H3K36me3 tracks are shown in blue. (D) Metagene averaged plot representation of ESC HA ChIP-seq enrichment profiles around the TSS for H3.3-HA from the three time points following Dox induction. (E) Metagene averaged ChIP-seq enrichment profiles for 4dRA induced cells around the TSS for H3.3-HA from the three time points following Dox induction. (F) TSS-aligned heatmaps of ESC H3.3 ChIP-seq for three time points datasets. (G) Same, for 4dRA H3.3 ChIP-seq datasets. (H) Metagene averaged plot representation of ChIP-seq enrichment profiles in ESCs around the TSS for canonical H3 in the three indicated time points following Dox induction. (I) Same as ‘H’ for 4dRA cells.
H3.3 turnover is elevated around TSSs
We next wished to examine the genome-wide incorporation kinetics of H3.3 in both ESCs and during differentiation. We performed ChIP followed by high-throughput sequencing (ChIP-seq) for HA at 1, 4 and 8 h following Dox induction, on both ESCs and in ESCs treated for 4 days with retinoic acid (4dRA), as well as RNA-seq, for both conditions. We sequenced two input fractions and six bound fractions in two biological replicates. The ChIP-seq results, aligned to UCSC mm9, are in agreement with previously published observations (13,23,33) and with re-analyzed published data of H3.3 time-ChIP in ESCs (14,34) or MEFs (21) (Supplementary Figure S2A). As expected, the data show a strong correlation between H3.3 enrichment and gene expression (Figure 2B and C, low Aprt enrichment in both ESC and 4dRA, high Oct4 enrichment in ESC only) as well as H3K4me3 binding (Figure 2C, all expressed genes). As previously reported, non-expressed ‘bivalent’ genes (i.e. promoter enriched for both active H3K4me3 and suppressive H3K27me3) are also enriched for H3.3 (e.g. EGFR, Figure 2C) while pluripotency specific genes (e.g. Esrrb, Oct4, Figure 2C and Supplementary Figure S1F) are depleted for H3.3 following differentiation (14,21,34).
Metagene analysis around the transcription start site (TSS) region (±2.5 Kb, Figure 2D) showed minor global reduction in H3.3 enrichment over the 8 h time-point in ESCs. The 4dRA cells, however, showed a global and pronounced decrease in enrichment over time in the TSS (Figure 2E). When comparing the different time-points, we observed that while ESCs show a rapid and very specific uptake of H3.3 in the TSS region in all time points (Figure 2D and F), in the RA-treated cells H3.3 incorporation gradually becomes wide-spread upstream and downstream of the TSS and lower in the TSS itself (Figure 2E and G). Similar analysis of canonical H3 showed low and uniform enrichment at all time points in both ESCs and 4dRA cells (Figure 2H and I). Since the difference between the 8 and the 1 h time point reflects the incorporation rate, these results demonstrate that H3.3 turnover is faster and more narrowly centered around the TSS in the RA-treated cells, while in ESCs H3.3 is exchanged over a wider region around the TSS. In short, H3.3 turnover rates are, on average, globally elevated around the TSS (Figure 2D–G) and differentiated cells show faster turnover rate than ESCs.
Clustering H3.3 turnover correlates with transcription and function
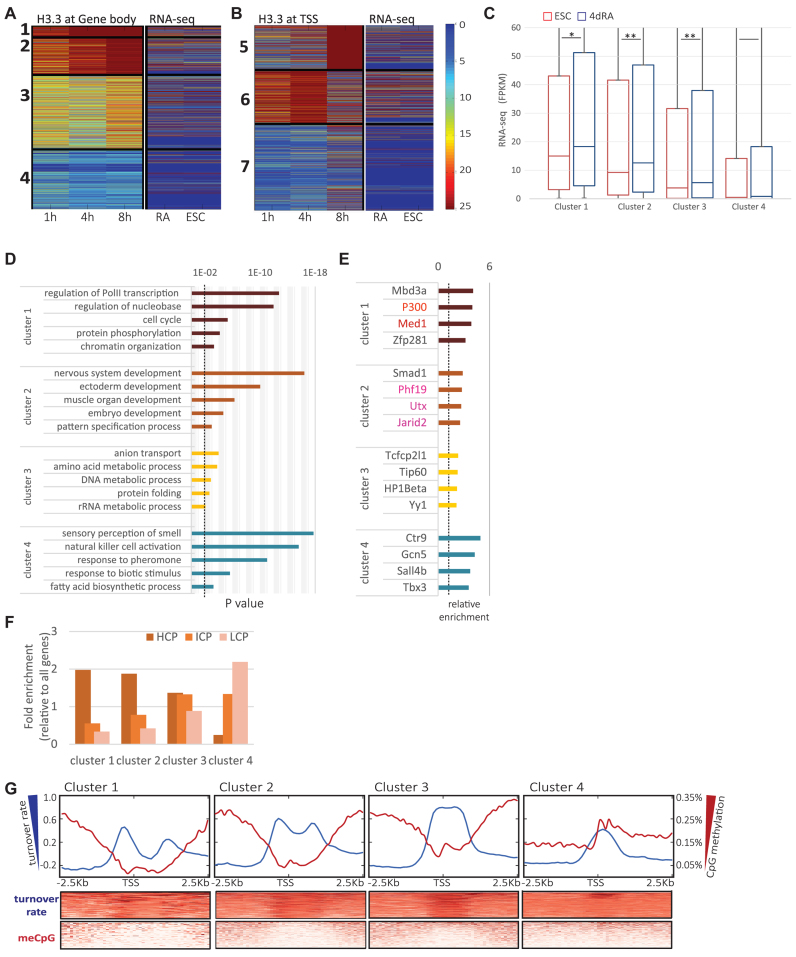
Next, we wished to test whether H3.3 turnover dynamics separates genes into functional groups. To this end, we clustered the turnover data using k-means based on average H3.3 ChIP-seq signal along the gene body (GB, Figure 3A, k = 4) or at the TSS (Figure 3B, k = 3). The TSS clusters were clustered into high turnover (Cluster 6), low turnover (Cluster 5) and low H3.3 enrichment (Cluster 7), which is the largest cluster. To reveal gene expression patterns in each group, we compared with our RNA-seq data and validated the expected correlation: the fast TSS turnover cluster contain the highly expressed genes while Cluster 7 contains mostly silenced genes (Figure 3B). To ensure that the Dox treatment itself did not alter the transcriptional profile of our cells, we first compared RNA-seq profiles with and without Dox induction, and found no significant influence on transcription (+Dox, Supplementary Figure S2B and C, right panel). In contrast to the TSS turnover pattern, inside the gene body the turnover rate is lower and the incorporation is more gradual. Interestingly, the correlation between the turnover in the TSS and in the gene body seem to be very weak or nonexistent (Supplementary Figure S2B and C). The four gene body clusters are correlated with descending transcription levels (Figure 3A and C). Cluster 1, which is the smallest cluster, shows high enrichment scores in all three time-points and contains highly expressed genes. Cluster 2 contains genes in which H3.3 is more gradually accumulated over time compared with Cluster 1, but shows a significantly higher turnover rates than Cluster 3, which includes genes with intermediate levels of incorporation and expression, and especially Cluster 4, which contains mostly silent genes with very little H3.3 incorporation.
Figure 3.
Clustering of H3.3 dynamics correlates with expression. (A) Heatmap showing H3.3-HA ChIP-seq signal (normalized read coverage, averaged over gene body) in ESCs (mean of two biological replicates) 1, 4 and 8 h following Dox induction (left). Data were clustered using standard K-means clustering algorithm (with K = 4) after removal of genes with missing values. Shown is the optimal clustering from 20 randomized initializations per dataset. K values were chosen from a range of tested values. RNA-seq data (FPKM values) from ESC and 4dRA cells are shown on the right. Color scaling for both ChIP-seq and RNA-seq as in (B). (B) Heatmap showing maximal H3.3-HA ChIP-seq signal (normalized read coverage, maximal value at 1 Kb windows surrounding TSSs). Data were clustered as before (with K = 3). The right panel shows the RNA-seq results of ESCs and 4dRA cells, 4 h after Dox addition. RNA-seq results (FPKM) of ESCs without Dox addition show similar patterns (Supplementary Figure S2B and C). (C) Box plot of expression levels (FPKM) in ESCs and 4dRA of gene body clusters. Change between ESCs and RA expression values is significant in clusters 1–3 (one asterisk denote P < 0.05, two asterisks denote P < 0.01; Kolmogorov–Smirnov test). Although Clusters 1 and 2 appear quite similar, they are highly statistically significantly different (P < 3 × 10−9, Kolmogorov–Smirnov test). Cluster 3 is significantly different from Clusters 1 (P < 2 × 10−77) and Cluster 2 (P < 1 × 10−105). Cluster 4 differs significantly from all the rest (P < 10−100 in all cases). Similar significant differences were calculated for the 4dRA cells. (D) GO analysis for the gene body clusters shown in panel A using the PANTHERDB online tool (http://www.pantherdb.org/). The five most enriched GO terms (functional classification) statistical significant over-representation (P < 0.01, dotted line) are shown for each class. Note that transcriptional regulation functions are enriched in Cluster 1, while developmental genes are over-represented in Cluster 2. (E) Epigenomic features analysis of each cluster, using the BindDB online tool (http://bind-db.huji.ac.il/). The 4 most enriched features in each cluster are presented (P < 0.01; hypergeometric test). Note the enrichment of super enhancer related factors in Cluster 1 (red) and the enrichment of Polycomb related factors in Cluster 2 (pink). (F) Enrichment scores of %CpG in ESCs on promoters of the four gene-body clusters shown in A: High (HCP, brown), intermediate (ICP, orange) and low (LCP, pink). (G) CpG methylation levels in ESCs versus the turnover rate (log2 ratio of H3.3 ChIP-seq signal in 1 versus 8 h) around the TSS, grouped by clusters. The three clusters which contain expressed genes (Clusters 1–3) show negative correlations between CpG methylation and H3.3 turnover dynamics.
To further characterize gene-body turnover clusters, we applied gene ontology (GO) analysis to each cluster separately using the PANTHER web tool (35) (Figure 3D). Cluster 1, which is characterized by the highest H3.3 enrichment scores within the gene body, was enriched with GO terms largely associated with chromatin and transcription (Figure 3D, top) and Cluster 2 shows enrichment of GO terms associated with development. We further performed in silico ‘reverse ChIP’ analysis for the different clusters using available ChIP-seq datasets we previously collected in the form of BindDB (36), and identified several transcription factors (TFs) that are selectively and significantly enriched within each of the four clusters (Figure 3E). Genes in Cluster 1 were enriched for features of super-enhancers including p300 and Med1 while genes in Cluster 2 were enriched with PRC2 components including Phf19, Utx and Jarid2. To determine whether the clusters are associated with different sets of promoters based on CpG content, we classified the promoters into groups of High-, Intermediate- and Low-CpG content within the four clusters (Figure 3F). Previous studies showed that the majority of genes with high CpG content promoters (HCP genes) in both ESCs and differentiated cells are marked by histone H3K4me3 and RNAPII occupancy, regardless of whether the gene is active or repressed (37–39). Analysis of the DNA methylation status (meCpG) around the TSS of the clusters revealed, in Clusters 1–3, a negative correlation between H3.3 turnover rate (log2 ratio between the 1 and 8 h time-points at promoter regions, see below) and CpG methylation (Figure 3G). Cluster 4 contains low/non-expressed genes and their TSS region is methylated, as expected. However, the H3.3 turnover rate is still higher than the background levels. It is interesting to note that in the absence of the H3.3 chaperone HIRA, normal CpG methylation pattern was lost during mouse oogenesis (40). Thus, H3.3 turnover might play an important role in establishing a proper methylation pattern during normal development. The enrichment of the super-enhancer signature in Cluster 1 and the PRC2 signature in Cluster 2, prompted us to further focus on genes adjacent to super-enhancers in ESCs, which are highly expressed (41), and bivalent genes, which are marked by both H3K4me3 and H3K27me3 (39,42–44).
H3.3 turnover reflects transcriptional states in ESCs
These differences in H3.3 dynamics between ESCs and 4dRA prompted us to examine the nature of these changes, particularly in relation to gene expression. To this end, we performed functional gene classification by grouping the genes according to their expression levels in ESCs and 4dRA (Figure 4A). We estimated turnover rates for each gene (TSS and gene body) by taking the log2 ratio between the 1 and 8 h time-points at promoter regions, as previously performed (33), and compared the general turnover rate of the genome in both ESCs and 4dRA (Figure 4B). To test whether H3.3 turnover rates reflect transcription rates, we analyzed the ‘high’ (genes highly expressed in both ESCs and 4dRA) and ‘low’ (genes not expressed in either condition) expression groups (Figure 4A) separately. Interestingly, comparing H3.3 turnover rates in ESCs and RA-treated cells, we found that the wider turnover peak in ESCs becomes more concentrated around the TSS in 4dRA (Figure 4B). While the ‘low’ expression group showed a small difference in H3.3 turnover between ESCs and 4dRA, the ‘high’ expression group showed a wider exchange region around the TSS in the undifferentiated ESCs (Figure 4C). In contrast to TSS turnover dynamics, in which the highly expressed genes showed, as expected, the fastest H3.3 TSS turnover rates (bold lines) and the lowly expressed genes showed the slowest turnover rates (thin lines). However, in the gene body itself and upstream to the TSS region, we observed opposite trends: the highly expressed genes, which are also relatively enriched with H3.3, showed a slower H3.3 turnover rate than the lowly-expressed and the silent genes (Figure 3C and Supplementary Figure S2D). These results demonstrate that H3.3 is enriched in highly expressed genes but while the H3.3 turnover rate around the TSS is mainly a function of expression, H3.3 turnover rate upstream to the TSS and inside the gene body displays a more complex pattern.
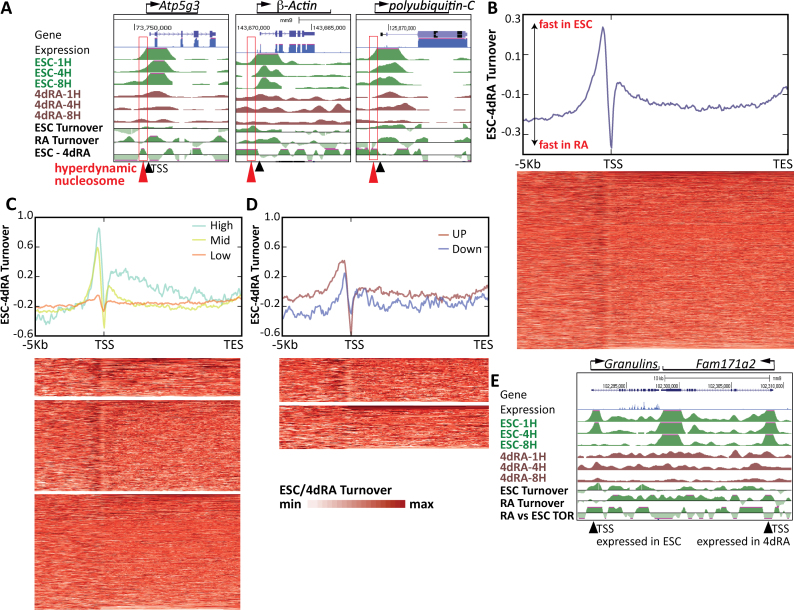
Figure 4.
H3.3 turnover rate and distribution changes during differentiation. (A) Comparison of RNA-seq data in ESCs (X-axis) and after 4 days RA treatment (Y-axis). Dots are colored according to RA/ESC expression level changes. (B) Metagene profile of average turnover rate of all genes in ESCs (red) and following 4 days RA treatment (4dRA, blue) on a linear scale from TSS to TES. Inset shows a narrower window of 10 Kb in linear scale around the TSS. Aligned heatmaps are shown below. Turnover rate was measured using log2 of promoter HA data for 1 h divided by 8 h. Hence, high values signify rapid turnover rate and low levels imply slow turnover rate. (C) Metagene profile of average turnover rate of the highly expressed genes (thick lines) and lowly expressed genes (thin lines) in ESCs (red) and following 4 days RA treatment (4dRA, blue). Aligned heatmaps are shown below. (D) Metagene profile of average turnover rate around the TSS of the highly expressed genes (thick lines) and lowly expressed genes (thin lines) in ESCs (red) and following 4 days RA treatment (4dRA, blue). Aligned heatmaps are shown below. (E) Metagene profile of average turnover rate around the TSS of the upregulated genes following 4 days RA treatment (‘Up’—thick lines) and downregulated genes (thin lines) in ESCs (red) and following 4 days RA treatment (4dRA, blue). Aligned heatmaps are shown below. (F) Metagene profile of average turnover rate around the TSS of the H3K4me3/H3K27me3 bivalent genes (‘K4+K27+’—thick lines) and H3K4me3 only genes (‘K4+’—thin lines) in ESCs (red) and following 4 days RA treatment (4dRA, blue). Aligned heatmaps are shown below. (G) Average H3.3 turnover rate in a 10 Kb window around conventional ESC enhancers (pink) and super-enhancers (blue). (H) Average H3.3 turnover rate in a 20 Kb window around ESC enhancers marked with H3K122ac and H3K27ac (pink) and those marked only with H3K122ac (blue).
Analyzing the 5 Kb region surrounding the TSS, the ‘high’ group once again shows a distinct turnover profile in ESCs, with a slightly elevated turnover upstream to the TSS (Figure 4D). The ‘Mid’ group shows similar although less distinct patterns (Supplementary Figure S2C). To look for turnover patterns characteristic of changes in expression, we focused on turnover rates in the ‘Up’ (genes upregulated in 4dRA samples) and the ‘Down’ (genes downregulated in 4dRA samples) groups in ESC and in 4dRA. While the H3.3 turnover profiles in the ‘Up’ and ‘Down’ groups are indistinct in the 4dRA samples, in ESCs, the ‘Up’ group shows a dip around the TSS, with higher turnover rates both upstream and downstream (Figure 4E). ESCs are generally relatively enriched with H3K4me3/H3K27me3 bivalent genes, some of which are resolved upon differentiation (42–45). PRC1-positive bivalent domains appear functionally distinct as they retain H3K27me3 more efficiently upon differentiation, show stringent conservation of the chromatin state and associate more strongly with developmental genes (46). We first compared the H3.3 turnover rates in both TSSs and gene bodies of bivalent genes, to that of genes marked only with H3K4me3 or H3K27me3 (Supplementary Figure S3A). H3K4me3-marked genes showed a higher H3.3 turnover rate in their TSS but a lower turnover rate in their gene body compared with the H3K27me3-marked genes. Bivalent genes showed a mixed pattern, where their TSS H3.3 turnover was as high as the H3K4me3-marked genes, while the gene-body turnover was as high as the H3K27me3-marked genes. This might reflect the poised state of a bivalent gene, in which the TSS is ready to transcribe while the gene body maintains a closed chromatin state with a higher turnover rate, as observed in silent genes. Zooming in into the 5 Kb region around the TSS (Supplementary Figure S3B), a finer picture is revealed: the peak around the TSS in H3K4me3-only genes is mostly upstream to the TSS, whereas in bivalent genes it is more enriched downstream to the TSS. Interestingly, when we grouped the genes according to H3K4me3 (‘K4’) and H3K27me3 (‘K27’) enrichment, we observed that while the ‘K4’ group turnover pattern resembles that of the ‘high’ group, the ‘K4/K27’ group resembles the ‘Up’ group. This suggests that in ESCs, H3.3 turnover corresponds to the genes’ expression and epigenetic profile, while in differentiated cells the turnover patterns are more uniform and do not reflect the genes’ activity (Figure 4F). When we analyzed the association of each of these groups of genes (‘K4-only’, ‘K27-only’ and bivalent) with the different clusters, we observed that while Cluster 1 is enriched with ‘K4-only’ genes, Cluster 2 is more enriched with bivalent genes, hinting that the difference in gene body H3.3 dynamics represents a functional difference between these gene groups (Supplementary Figure S3C). Moreover, in the bivalent gene group, ‘PRC1+PRC2’-positive bivalent domains appear functionally distinct from ‘PRC2 only’ as they retain H3K27me3 more efficiently upon differentiation, show stringent conservation of the chromatin state and associate more strongly with developmental genes (46). While the PRC2-only genes show no difference between the clusters, Cluster 2 is specifically and significantly enriched with the PRC1+2 labeled genes (Supplementary Figure S3D), indicating a link between a specific H3.3 turnover pattern and this important group of developmental genes. Zooming in into the turnover rate of the bivalent gene group, we find that the PRC1+2 bound genes show faster H3.3 turnover in the TSS (Supplementary Figure S3E) but not in the gene body when compared to their PRC2-only counterparts (Supplementary Figure S3F). Thus, H3.3 turnover seems to be involved in the fine-tuning of bivalent gene expression.
H3.3 is highly dynamic in a particular set of enhancers
H3.3 was recently shown to display particularly fast turnover rates in super-enhancer regions, where also the most significant changes were detected following differentiation (34). We therefore tested H3.3 turnover dynamics on enhancers and super-enhancers in our system. Our RNA-seq data confirmed that super-enhancer associated genes (as listed in Whyte et al. (40)) are expressed at higher levels than enhancer-associated genes in both ESCs and RA-induced cells (Supplementary Figure S4A). Correspondingly, H3.3 turnover rates were slower in the gene body of super-enhancer-related genes compared with enhancer-related genes (Supplementary Figure S4B). This change was specific to gene body ESCs and was not observed in the TSS, or in the gene bodies of RA-treated cells. Cluster 1 showed the most dramatic enrichment of super-enhancer proximal genes (Supplementary Figure S4C), concomitant to the high enrichment of Med1 and p300 binding on Cluster 1 genes (Figure 3E). We observed high H3.3 enrichment as well as high turnover rates at the epicenter of the super-enhancers (Figure 4G). Epicenters are short (<2 kb) active subdomains within the super-enhancers, which are particularly enriched for TF binding sites and allow for cooperative binding. Conventional enhancers also show a peak at the center, but since they are in general much shorter, the peak is, respectively, narrower (Figure 4G). However, when we zoom out to the full 10 Kb sequences and the surrounding genomic region, we see high H3.3 enrichment but slower turnover inside the super-enhancers than the average surrounding genomic regions (Supplementary Figure S4D). These changes in nucleosome density are not global, since ChIP for canonical H3 did not show any significant differences between the enhancer center and its surroundings (Supplementary Figure S4E).
More recently, a new class of active functional enhancers, marked by H3K122ac, but which lack H3K27ac, was suggested to regulate a subset of developmental genes in mouse ESCs (47). These enhancers are also enriched for H3K64ac, H3K27me3 and H2A.Z, as well as for H3K4me1, DNAseI hypersensitivity, and p300. Comparing these newly identified enhancers (H3K122ac+/H3K27ac-) to ‘conventional’ enhancers (H3K122ac+/H3K27ac+) we found a selective high turnover peak of H3.3 at the H3K122ac+/H3K27ac- but not the H3K122ac+/H3K27ac+ enhancers (Figure 4H). Importantly, both groups are highly enriched for H3.3. This suggests that H3.3 turnover rate is not only important around genes but also inside gene regulatory regions, and in particular, its dynamic turnover is highly selective for poised enhancers.
To test another group of non-genic potential regulatory regions, we also analyzed H3.3 enrichment and turnover in the topological-associated domains (TADs) boundaries, which were reported to play a role in gene expression regulation in ESCs (48). TAD boundaries are conserved in evolution, and enriched for housekeeping genes and active chromatin modifications. We found enrichment of H3.3 on TAD boundaries, but lower turnover rate than their genomic surrounding (Supplementary Figure S4F). This finding is in agreement with previous observation in human differentiated cells (49), and likely reflects the special chromatin context of TADs boundaries.
A hyperdynamic H3.3 nucleosome marks promoters in ESCs
Finally, we concentrated on H3.3 turnover dynamics in the promoter regions of ESCs and 4dRA. Analyzing the distributions of the turnover peaks in ESCs and 4dRA, we noticed that the wider ESC peak corresponds roughly to a genomic size of a single nucleosome. This suggested that ESCs may contain a hyperdynamic nucleosome immediately upstream to the TSS, which is not present or less frequent in the 4dRA cells. To test this, we zoomed in on several of the ‘high’ category genes and, remarkably, found evidence for such a dynamic nucleosome just upstream to the TSS, not present in the 4dRA cells (Figure 5A, red arrows). To test whether such a H3.3 hyperdynamic ‘−1’ nucleosome is a general feature of ESCs, we calculated the H3.3 turnover rate in ESCs versus RA-treated cells (i.e. log2(1/8 h HA enrichment) in ESC - log2(1/8 h HA enrichment) in 4dRA). This analysis revealed an exceptionally strong peak (i.e. high H3.3 turnover rate) immediately upstream to the TSS, in ESCs relatively to 4dRA (Figure 5B). This high turnover is mostly significant in highly expressed and poised genes (Figure 5C and D). While the deposition map itself (Figure 2D) shows that most of the H3.3 deposition in ESCs occurs around and downstream of the TSS, the resolution is such that the deposition area covers 2 kb (roughly, from 1 kb upstream to 1 kb downstream of the TSS, slightly shifted downstream, Figure 2D). By analyzing turnover rates rather than H3.3 deposition per se (Figure 4B), we realized that H3.3 turnover peaks almost exactly at the TSS itself, with RA-treated cells showing a defined peak at the TSS and ESCs showing a wider turnover region spanning roughly a nucleosome-size region upstream to the TSS (Figure 4D). To strengthen our approach, we performed similar turnover rate calculations using previously published datasets (14,21,34) and also repeated the H3.3 time-ChIP experiments. Reassuringly, we find that the turnover rates in the differentiated cells (MEFs in this case) (Supplementary Figure S5A and B) show a more defined peak above the TSS (similar to the situation in our RA cells), while in the ESCs the distribution is wider upstream, reflecting the ‘hyperdynamic’ phenomenon that we identified.
Figure 5.
ESCs contain a hyperdynamic nucleosome upstream to the TSS. (A) USCS screenshot maps of the indicated highly expressed housekeeping genes H3.3-HA turnover tracks in ESCs and in 4dRA cells as well as turnover rate change. RNA-seq tracks are shown in blue. The TSS is marked with a black triangle; the −1 hyperdynamic nucleosome is marked with a red triangle. (B) Turnover rate change (ESC log2(1/8 h HA enrichment)-(RA log2(1/8 h HA enrichment) analysis reveals a hyperdynamic −1 nucleosome in ESCs. Aligned heatmaps are shown below. (C) Turnover rate change between ESCs and 4dRA in highly-expressed (light blue), mid-range (green) and lowly expressed (red) genes. Aligned heatmaps are shown below. (D) Turnover rate change between ESCs and 4dRA in up-regulated (red) and down-regulated (blue) genes following differentiation. Aligned heatmaps are shown below. (E) UCSC screenshots of adjacent down-regulated (Grn, left side) and up-regulated (Fam171a2, right) genes turnover rate in ESC and 4dRA cells. ESC RNA-seq track is shown in blue. TSSs are indicated with black triangles.
This analysis also revealed a negative peak just downstream to the hyperdynamic −1 nucleosome, suggesting that +1 nucleosome is more dynamic in the 4dRA cells. These data show that although the overall genomic turnover rate of H3.3 increases in differentiated cells, a specific −1 nucleosome is hyperdynamic in pluripotent ESCs and restricts upon differentiation, either shifting downstream, or otherwise replaced by a +1 nucleosome following differentiation. To further examine the association between the newly identified hyperdynamic −1 nucleosome and expression, we tested the ESC/4dRA turnover profiles in our expression clusters (Supplementary Figure S5C). We first confirmed that Clusters 1 and 2 retain mostly expressed genes (Supplementary Figure S5D), while Cluster 4 inhabits the low/no expression genes in both ESCs and 4dRA cells. Genes that were either down- or up-regulated during differentiation did not show any cluster preference (Supplementary Figure S5E). Genes differentially expressed between ESCs and 4dRA overall show a similar trend although the −1 nucleosome in the up-regulated genes is more dynamic than the +1 nucleosome in the down-regulated genes (Figure 5D and E), suggesting that a hyperdynamic nucleosome marks both expressed and poised genes in ESCs. Taken together, our data reveal a hyperdynamic nucleosome at the −1 position in ESCs, providing a high-resolution window into the dynamics of the otherwise stably associated core histone variant H3.3 (50).
DISCUSSION
Using a Dox-inducible single copy integration of H3.3-HA in mouse ESCs, we were able to follow its genome-wide incorporation dynamics. Our data show that H3.3 turnover provides an additional layer of information, which may be used to reveal classes of co-regulated gene families (51).
Specifically, we found that super-enhancers (41) contain an H3.3-enriched nucleosome at its epi-center, in agreement with a recent parallel study (34). In addition, we identified an even stronger enrichment for H3.3 inside a more recently identified class of enhancers marked with H3K122ac (47), in contrast to ‘regular’ enhancers where no such H3.3-enriched nucleosome is present. Since H3K122 is located on the lateral surface of the histone octamer, the acetylation of this residue was suggested to function by directly altering nucleosomal stability and mobility (52,53). The finding of histone H3 globular domain acetylation (K122ac) in a family of enhancers (47), suggests that opening of local chromatin structure might be an important facet of enhancer function. H3.3 high turnover at those sites support this idea and suggest that hyperdynamic H3.3 is a basic characteristic of many active enhancers. Correlating H3.3 dynamics with enhancer datasets may thus assist in further identifying and classifying specific enhancer families.
Our most intriguing observation revealed here is the presence of a hyperdynamic H3.3 nucleosome in ESCs, which is restricted and either shifted downstream or replaced by a different +1 nucleosome upon differentiation. Interestingly, similar results were reported recently following chemical mapping of nucleosome positioning, in which high nucleosome occupancy is observed in the −1 position in mouse ESCs (54). Previously, we and others showed that chromatin proteins are associated more loosely in pluripotent compared with differentiated cells (3,4,7,55–58), supporting a ‘hyperdynamic’ chromatin state in pluripotency (59). We hypothesized that this open chromatin reflects the promiscuous transcription program we observed in mouse ESCs (60). However, it was later argued that this global transcription is a result of growth conditions which include serum, and when grown in the presence of GSK3 and MAPK/ERK inhibitors (known as ‘2i’) ESCs assume a ‘naïve’ state and no longer display spurious transcription (61). Regardless, in our original studies, H3.3 was the only chromatin-related protein tested, which did not show increased global turnover dynamics in ESCs (50), and hence it posed a particularly interesting case for genome-wide incorporation studies. To ensure no serum-related artifacts, we cultured our cells in the current study in ground state 2i conditions. Remarkably, while we observed a slight overall increase in H3.3 incorporation dynamics following differentiation, when we compared the meta-gene turnover profiles between ESCs and RA-treated cells we found a single hyperdynamic nucleosome in ESCs at the −1 position, immediately upstream to the TSS. This is reminiscent of the promoter-associated peaks of MacroH2A2, which were previously shown to possess relatively rapid exchange dynamics in ESCs (33). It would be interesting to test whether H3.3 and MacroH2A2 reside within the same dynamic nucleosome. Our previous studies, which involved photobleaching-related methods (i.e. FRAP, FLIP), could not have identified a single hyperdynamic nucleosome because such photobleaching-based methods provide a global average and do not possess the resolution to identify such regional behavior. It would now be interesting to test the genome-wide turnover dynamics of additional chromatin-related proteins, as was done for H3.3 and MacroH2A2 (13,20,21,33,34), including core histones. The latter would pose a bigger challenge due to changes in cell cycle dynamics following differentiation. One of the major advantages of using a replication-independent histone variant for this study is that it is not S-phase dependent and hence should not be majorly affected by changes in cell cycle kinetics. By contrast, since core histones are incorporated during S-phase, changes in cell cycle, especially in the fraction of cells that are in S-phase during sampling, may affect the analysis.
While previous reports compared ESCs to either MEFs or in vivo derived NPCs, this study is the first to directly compare undifferentiated ‘ground state’ ESCs with differentiated ESCs. Another novelty of our work is our focus on H3.3 dynamics within the gene body rather than the TSS. We show that while the TSS dynamics is directly correlated with transcription, as shown before us, the dynamics of gene body H3.3 is correlated with other regulatory properties of the genes. Overall, our results are in agreement with previous studies addressing H3.3 turnover in ESCs, but a direct comparison is problematic since other reports used a system of H3.3 dissociation, while we analyzed H3.3 incorporation. Nonetheless, comparing H3.3 turnover in ESCs versus MEFs (21), and employing similar statistical analyses to the ones we performed here, confirmed the presence of the hyperdynamic H3.3 nucleosome we identified (Supplementary Figure S5A).
To conclude, by monitoring H3.3 incorporation dynamics over time before and following ESC differentiation, we identify a hyperdynamic H3.3-containing nucleosome at the −1 position, marking promoters of expressed genes in ESCs. Following differentiation, this hyperdynamic nucleosome is restricted and shifted downstream (or otherwise replaced by a different ‘+1’ nucleosome) into the reading frame of the corresponding gene. This may explain earlier observations by Felsenfeld and colleagues showing that nucleosome depleted regions are in fact enriched with H3.3 (18), suggesting that dynamic turnover may be more important in regulating transcription than the presence/absence of a histone or a particular histone modification. Our data thus provide evidence for regional regulation of H3.3 turnover in ESC promoters, and calls for testing, in high resolution, the dynamic behavior of additional histone variants and other structural chromatin proteins.
DATA AVAILABILITY
The sequencing data from the study have been submitted to the NCBI Gene Expression Omnibus (GEO), accession number GSE88745.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Malka Nissim-Rafinia for technical assistance.
Author contributions: conceived and designed the experiments: S.S., S.M. and E.M. Performed the experiments: S.S., B.K., J.D.A. and D.M.S. Analyzed the data: T.K. and S.S. Wrote the paper: S.S. and E.M.
Footnotes
Present addresses:
Sharon Schlesinger, Department of animal science, The Robert H. Smith Faculty of Agriculture, Food, and Environment, The Hebrew University of Jerusalem, Rehovot 7610001, Israel.
Jose David Aguilera, Department of animal science, The Robert H. Smith Faculty of Agriculture, Food, and Environment, The Hebrew University of Jerusalem, Rehovot 7610001, Israel.
Divya Mundackal Sivaraman, Cell Conversion Technology Unit, RIKEN Center for Life Science Technologies, Yokohama 2300045, Japan.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Israel Science Foundation (ISF) [1140/17 to E.M., 913/15 to T.K., 761/17 to S.S.]; Marie-Curie Career Integration Grant [618327 to T.K.]; European Research Council (to E.M.); Israeli Center of Excellence (I-CORE) for Gene Regulation in Complex Human Disease [41/11 to T.K.]; Israeli Center of Excellence (I-CORE) for Chromatin and RNA in Gene Regulation [1796/12 to T.K.]; Arthur Gutterman Chair for Stem Cell Research (to E.M.). Funding for open access charge: ISF [1140/17].
Conflict of interest statement. None declared.
REFERENCES
- 1. Probst A.V., Almouzni G.. Heterochromatin establishment in the context of genome-wide epigenetic reprogramming. Trends Genet. 2011; 27:177–185. [DOI] [PubMed] [Google Scholar]
- 2. Ahmad K., Henikoff S.. Epigenetic consequences of nucleosome dynamics. Cell. 2002; 111:281–284. [DOI] [PubMed] [Google Scholar]
- 3. Meshorer E., Misteli T.. Chromatin in pluripotent embryonic stem cells and differentiation. Nat. Rev. Mol. Cell Biol. 2006; 7:540–546. [DOI] [PubMed] [Google Scholar]
- 4. Melcer S., Meshorer E.. Chromatin plasticity in pluripotent cells. Essays Biochem. 2010; 48:245–262. [DOI] [PubMed] [Google Scholar]
- 5. Mattout A., Meshorer E.. Chromatin plasticity and genome organization in pluripotent embryonic stem cells. Curr. Opin. Cell Biol. 2010; 22:334–341. [DOI] [PubMed] [Google Scholar]
- 6. Gaspar-Maia A., Alajem A., Meshorer E., Ramalho-Santos M.. Open chromatin in pluripotency and reprogramming. Nat. Rev. Mol. Cell Biol. 2011; 12:36–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Boskovic A., Eid A., Pontabry J., Ishiuchi T., Spiegelhalter C., Raghu Ram E.V., Meshorer E., Torres-Padilla M.E.. Higher chromatin mobility supports totipotency and precedes pluripotency in vivo. Genes Dev. 2014; 28:1042–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen T., Dent S.Y.. Chromatin modifiers and remodellers: regulators of cellular differentiation. Nat. Rev. Genet. 2014; 15:93–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dang-Nguyen T.Q., Torres-Padilla M.E.. How cells build totipotency and pluripotency: nuclear, chromatin and transcriptional architecture. Curr. Opin. Cell Biol. 2015; 34:9–15. [DOI] [PubMed] [Google Scholar]
- 10. Skene P.J., Henikoff S.. Histone variants in pluripotency and disease. Development. 2013; 140:2513–2524. [DOI] [PubMed] [Google Scholar]
- 11. Turinetto V., Giachino C.. Multiple facets of histone variant H2AX: a DNA double-strand-break marker with several biological functions. Nucleic Acids Res. 2015; 43:2489–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wong L.H., Ren H., Williams E., McGhie J., Ahn S., Sim M., Tam A., Earle E., Anderson M.A., Mann J. et al. Histone H3.3 incorporation provides a unique and functionally essential telomeric chromatin in embryonic stem cells. Genome Res. 2009; 19:404–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Goldberg A.D., Banaszynski L.A., Noh K.-M.M., Lewis P.W., Elsaesser S.J., Stadler S., Dewell S., Law M., Guo X., Li X. et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010; 140:678–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ha M., Kraushaar D.C., Zhao K.. Genome-wide analysis of H3.3 dissociation reveals high nucleosome turnover at distal regulatory regions of embryonic stem cells. Epigenet. Chromatin. 2014; 7:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Szenker E., Lacoste N., Almouzni G.. A developmental requirement for HIRA-dependent H3.3 deposition revealed at gastrulation in Xenopus. Cell Rep. 2012; 1:730–740. [DOI] [PubMed] [Google Scholar]
- 16. Ahmad K., Henikoff S.. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell. 2002; 9:1191–1200. [DOI] [PubMed] [Google Scholar]
- 17. Ray-Gallet D., Quivy J.P., Scamps C., Martini E.M., Lipinski M., Almouzni G.. HIRA is critical for a nucleosome assembly pathway independent of DNA synthesis. Mol. Cell. 2002; 9:1091–1100. [DOI] [PubMed] [Google Scholar]
- 18. Jin C., Zang C., Wei G., Cui K., Peng W., Zhao K., Felsenfeld G.. H3.3/H2A.Z double variant-containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nat. Genet. 2009; 41:941–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Elsasser S.J., Noh K.M., Diaz N., Allis C.D., Banaszynski L.A.. Histone H3.3 is required for endogenous retroviral element silencing in embryonic stem cells. Nature. 2015; 522:240–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mito Y., Henikoff J.G., Henikoff S.. Genome-scale profiling of histone H3.3 replacement patterns. Nat. Genet. 2005; 37:1090–1097. [DOI] [PubMed] [Google Scholar]
- 21. Kraushaar D.C., Jin W., Maunakea A., Abraham B., Ha M., Zhao K.. Genome-wide incorporation dynamics reveal distinct categories of turnover for the histone variant H3.3. Genome Biol. 2013; 14:R121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Maze I., Wenderski W., Noh K.M., Bagot R.C., Tzavaras N., Purushothaman I., Elsasser S.J., Guo Y., Ionete C., Hurd Y.L. et al. Critical role of histone turnover in neuronal transcription and plasticity. Neuron. 2015; 87:77–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Banaszynski L.A., Wen D., Dewell S., Whitcomb S.J., Lin M., Diaz N., Elsasser S.J., Chapgier A., Goldberg A.D., Canaani E. et al. Hira-dependent histone H3.3 deposition facilitates PRC2 recruitment at developmental loci in ES cells. Cell. 2013; 155:107–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Silva J., Barrandon O., Nichols J., Kawaguchi J., Theunissen T.W., Smith A.. Promotion of reprogramming to ground state pluripotency by signal inhibition. PLoS Biol. 2008; 6:e253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mendez J., Stillman B.. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 2000; 20:8602–8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schlesinger S., Goff S.P.. Silencing of proviruses in embryonic cells: efficiency, stability and chromatin modifications. EMBO Rep. 2013; 14:73–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Blecher-Gonen R., Barnett-Itzhaki Z., Jaitin D., Amann-Zalcenstein D., Lara-Astiaso D., Amit I.. High-throughput chromatin immunoprecipitation for genome-wide mapping of in vivo protein-DNA interactions and epigenomic states. Nat. Protoc. 2013; 8:539–554. [DOI] [PubMed] [Google Scholar]
- 28. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ramirez F., Ryan D.P., Gruning B., Bhardwaj V., Kilpert F., Richter A.S., Heyne S., Dundar F., Manke T.. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016; 44:W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dion M.F., Kaplan T., Kim M., Buratowski S., Friedman N., Rando O.J.. Dynamics of replication-independent histone turnover in budding yeast. Science. 2007; 315:1405–1408. [DOI] [PubMed] [Google Scholar]
- 31. Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L.. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010; 28:511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Beard C., Hochedlinger K., Plath K., Wutz A., Jaenisch R.. Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis. 2006; 44:23–28. [DOI] [PubMed] [Google Scholar]
- 33. Yildirim O., Hung J.H., Cedeno R.J., Weng Z., Lengner C.J., Rando O.J.. A system for genome-wide histone variant dynamics in ES cells reveals dynamic MacroH2A2 replacement at promoters. PLoS Genet. 2014; 10:e1004515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Deaton A.M., Gomez-Rodriguez M., Mieczkowski J., Tolstorukov M.Y., Kundu S., Sadreyev R.I., Jansen L.E., Kingston R.E.. Enhancer regions show high histone H3.3 turnover that changes during differentiation. Elife. 2016; 5:e15316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mi H., Poudel S., Muruganujan A., Casagrande J.T., Thomas P.D.. PANTHER version 10: expanded protein families and functions, and analysis tools. Nucleic Acids Res. 2016; 44:D336–D342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Livyatan I., Aaronson Y., Gokhman D., Ashkenazi R., Meshorer E.. BindDB: an integrated database and webtool platform for “Reverse-ChIP” epigenomic analysis. Cell Stem Cell. 2015; 17:647–648. [DOI] [PubMed] [Google Scholar]
- 37. Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K.. High-resolution profiling of histone methylations in the human genome. Cell. 2007; 129:823–837. [DOI] [PubMed] [Google Scholar]
- 38. Guenther M.G., Levine S.S., Boyer L.A., Jaenisch R., Young R.A.. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007; 130:77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mikkelsen T.S., Ku M., Jaffe D.B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.-K.K., Koche R.P. et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007; 448:553–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nashun B., Hill P.W., Smallwood S.A., Dharmalingam G., Amouroux R., Clark S.J., Sharma V., Ndjetehe E., Pelczar P., Festenstein R.J. et al. Continuous histone replacement by Hira is essential for normal transcriptional regulation and de novo DNA methylation during mouse oogenesis. Mol. Cell. 2015; 60:611–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Whyte W.A., Orlando D.A., Hnisz D., Abraham B.J., Lin C.Y., Kagey M.H., Rahl P.B., Lee T.I., Young R.A.. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013; 153:307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bernstein B.E., Mikkelsen T.S., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K. et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006; 125:315–326. [DOI] [PubMed] [Google Scholar]
- 43. Azuara V., Perry P., Sauer S., Spivakov M., Jorgensen H.F., John R.M., Gouti M., Casanova M., Warnes G., Merkenschlager M. et al. Chromatin signatures of pluripotent cell lines. Nat. Cell Biol. 2006; 8:532–538. [DOI] [PubMed] [Google Scholar]
- 44. Harikumar A., Meshorer E.. Chromatin remodeling and bivalent histone modifications in embryonic stem cells. EMBO Rep. 2015; 16:1609–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shema E., Jones D., Shoresh N., Donohue L., Ram O., Bernstein B.E.. Single-molecule decoding of combinatorially modified nucleosomes. Science. 2016; 352:717–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ku M., Koche R.P., Rheinbay E., Mendenhall E.M., Endoh M., Mikkelsen T.S., Presser A., Nusbaum C., Xie X., Chi A.S. et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008; 4:e1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pradeepa M.M., Grimes G.R., Kumar Y., Olley G., Taylor G.C., Schneider R., Bickmore W.A.. Histone H3 globular domain acetylation identifies a new class of enhancers. Nat. Genet. 2016; 48:681–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Dixon J.R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J.S., Ren B.. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012; 485:376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dunleavy E.M., Almouzni G., Karpen G.H.. H3.3 is deposited at centromeres in S phase as a placeholder for newly assembled CENP-A in G (1) phase. Nucleus. 2011; 2:146–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Meshorer E., Yellajoshula D., George E., Scambler P.J., Brown D.T., Misteli T.. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev. Cell. 2006; 10:105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Aaronson Y., Livyatan I., Gokhman D., Meshorer E.. Systematic identification of gene family regulators in mouse and human embryonic stem cells. Nucleic Acids Res. 2016; 44:4080–4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tropberger P., Schneider R.. Scratching the (lateral) surface of chromatin regulation by histone modifications. Nat. Struct. Mol. Biol. 2013; 20:657–661. [DOI] [PubMed] [Google Scholar]
- 53. Tropberger P., Pott S., Keller C., Kamieniarz-Gdula K., Caron M., Richter F., Li G., Mittler G., Liu E.T., Buhler M. et al. Regulation of transcription through acetylation of H3K122 on the lateral surface of the histone octamer. Cell. 2013; 152:859–872. [DOI] [PubMed] [Google Scholar]
- 54. Voong L.N., Xi L., Sebeson A.C., Xiong B., Wang J.P., Wang X.. Insights into nucleosome organization in mouse embryonic stem cells through chemical mapping. Cell. 2016; 167:1555–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bhattacharya D., Talwar S., Mazumder A., Shivashankar G.V.. Spatio-temporal plasticity in chromatin organization in mouse cell differentiation and during Drosophila embryogenesis. Biophys. J. 2009; 96:3832–3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Melcer S., Hezroni H., Rand E., Nissim-Rafinia M., Skoultchi A., Stewart C.L., Bustin M., Meshorer E.. Histone modifications and lamin A regulate chromatin protein dynamics in early embryonic stem cell differentiation. Nat. Commun. 2012; 3:910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Manukyan M., Singh P.B.. Epigenome rejuvenation: HP1beta mobility as a measure of pluripotent and senescent chromatin ground states. Sci. Rep. 2014; 4:4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sustackova G., Legartova S., Kozubek S., Stixova L., Pachernik J., Bartova E.. Differentiation-independent fluctuation of pluripotency-related transcription factors and other epigenetic markers in embryonic stem cell colonies. Stem Cells Dev. 2012; 21:710–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gaspar-Maia A., Alajem A., Polesso F., Sridharan R., Mason M.J., Heidersbach A., Ramalho-Santos J., McManus M.T., Plath K., Meshorer E. et al. Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature. 2009; 460:863–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Efroni S., Duttagupta R., Cheng J., Dehghani H., Hoeppner D.J., Dash C., Bazett-Jones D.P., Le Grice S., McKay R.D., Buetow K.H. et al. Global transcription in pluripotent embryonic stem cells. Cell Stem Cell. 2008; 2:437–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Marks H., Kalkan T., Menafra R., Denissov S., Jones K., Hofemeister H., Nichols J., Kranz A., Stewart A.F., Smith A. et al. The transcriptional and epigenomic foundations of ground state pluripotency. Cell. 2012; 149:590–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data from the study have been submitted to the NCBI Gene Expression Omnibus (GEO), accession number GSE88745.