Abstract
In Escherichia coli, the level of the ATP–DnaA initiator is increased temporarily at the time of replication initiation. The replication origin, oriC, contains a duplex-unwinding element (DUE) flanking a DnaA-oligomerization region (DOR), which includes twelve DnaA-binding sites (DnaA boxes) and the DNA-bending protein IHF-binding site (IBS). Although complexes of IHF and ATP–DnaA assembly on the DOR unwind the DUE, the configuration of the crucial nucleoprotein complexes remains elusive. To resolve this, we analyzed individual DnaA protomers in the complex and here demonstrate that the DUE–DnaA-box-R1–IBS–DnaA-box-R5M region is essential for DUE unwinding. R5M-bound ATP–DnaA predominantly promotes ATP–DnaA assembly on the DUE-proximal DOR, and R1-bound DnaA has a supporting role. This mechanism might support timely assembly of ATP–DnaA on oriC. DnaA protomers bound to R1 and R5M directly bind to the unwound DUE strand, which is crucial in replication initiation. Data from in vivo experiments support these results. We propose that the DnaA assembly on the IHF-bent DOR directly binds to the unwound DUE strand, and timely formation of this ternary complex regulates replication initiation. Structural features of oriC support the idea that these mechanisms for DUE unwinding are fundamentally conserved in various bacterial species including pathogens.
INTRODUCTION
The initiation of bacterial DNA replication requires unwinding of the double helix at a replication origin sequence, which proceeds via the regulated construction of high-order nucleoprotein complexes. In Escherichia coli, chromosomal replication initiation is promoted by binding of the DNA-bending protein IHF and multiple molecules of ATP-bound DnaA protein (ATP–DnaA) to specific elements of the oriC sequence (Figure 1A) (1–5). This interaction results in formation of an initiation complex, which promotes unwinding of an AT-rich region of oriC and loading of DnaB helicases onto the resulting single-stranded DNA (ssDNA), enabling the formation of sister replisomes, and bidirectional chromosomal replication (6,7).
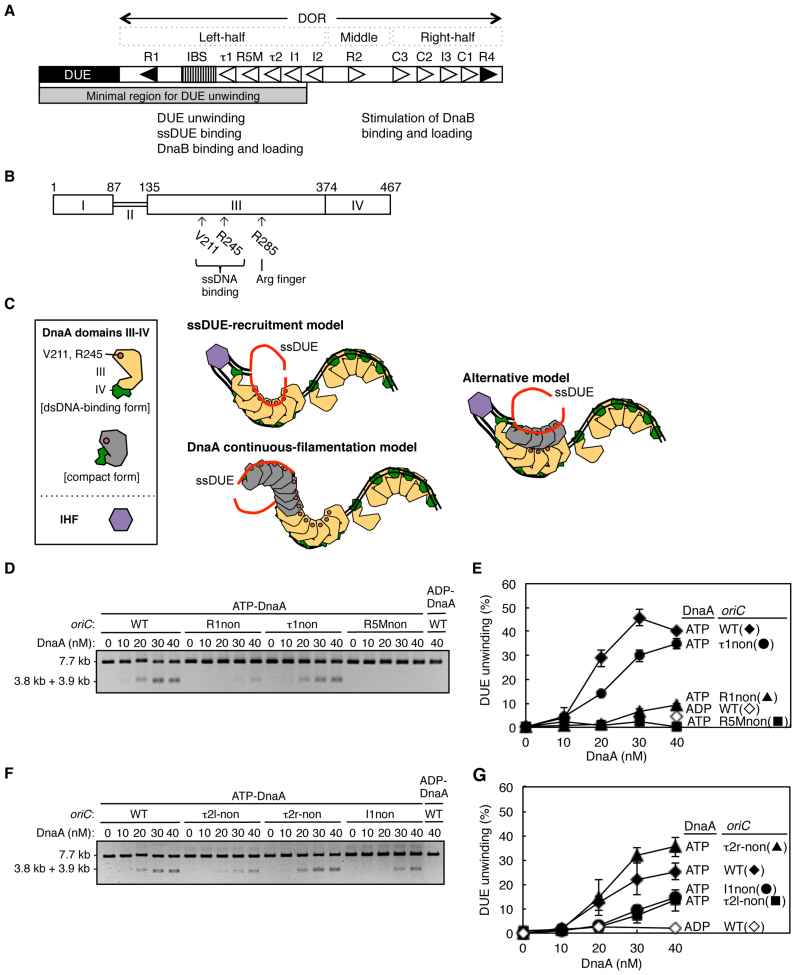
Figure 1.
Determination of the key DnaA boxes required for oriC DUE unwinding. (A) Overall structure of oriC. The duplex-unwinding element (DUE; closed bar), DnaA-oligomerization region (DOR; open bar), and IHF-binding site (IBS; striped pattern box) are indicated. High-affinity DnaA boxes (R1 and R4) are indicated by closed triangles, and moderate-affinity (R2) and low-affinity sites (R5M, τ1–2, I1–3 and C1–3) are indicated by open triangles. The functions of left half and middle-right half of the DOR are indicated. The minimal region for DUE unwinding determined in this study is indicated by a gray bar (also see text). (B) DnaA protein domain structure. Domains of E. coli DnaA are indicated by open boxes with relevant amino acid numbering. The residues investigated in this study and their functions are indicated. (C) Proposed models of DUE-unwinding-complex structure. For simplicity, only DnaA domains III and IV are shown. DnaA domain III bound to ssDNA (and not dsDNA) is indicated by light-gray shading, and IHF by a purple hexagon. IHF binding bends DNA to >160°. ssDUE strands are shown in red. (D–G) DUE-unwinding assay. ATP–DnaA or ADP–DnaA, along with IHF (32 nM), were incubated for 3 min at 38°C with oriC DNA (1.32 nM) containing DnaA-box mutations, followed by incubation for 200 s at 38°C with P1 nuclease (4 U). Resultant DNAs were analyzed by gel electrophoresis shown in (D) and (F). The percentages of DNA digested by P1 nuclease out of the total input DNA are shown as ‘DUE unwinding (%)’ in (E) and (G). Two independent experiments were carried out, and both data, mean values, and representative gel images in a black-white inverted mode are shown.
The cellular level of ATP–DnaA fluctuates during the cell cycle, peaking at the time of replication initiation (8). The ATP that is bound to DnaA is hydrolyzed soon after initiation, yielding inactive ADP–DnaA, and substantially decreasing the level of ATP–DnaA (2,9). Before the next round of initiation occurs in a timely manner, ADP–DnaA must be converted back to ATP–DnaA by a regulated nucleotide-exchange reaction (10,11). Also, transcription of the dnaA gene is regulated to increase before replication initiation and to decease after it (12–14). De novo synthesized DnaA molecules would bind ATP which is much more abundant in cells than ADP. Thus, timely ATP–DnaA function at oriC responding to the temporal increase in the ATP–DnaA level is important.
The DnaA protein consists of four functional domains (Figure 1B) (4,15). Domain I interacts site-specifically with several proteins, including DnaB and DiaA, which stimulates ATP–DnaA assembly (16–19). However, domains I-II are basically dispensable for specific unwinding of oriC (19). Domain II is a flexible linker (16,20). Domain III contains AAA+ (ATPase Associated with various cellular Activities) motifs that are involved in tight ATP/ADP binding, ATP hydrolysis, and DnaA–DnaA interactions (4,5,15,21–24). Like typical AAA+ proteins, DnaA AAA+ domain can construct a homo-oligomer with helical structure (21). The AAA+ arginine-finger motif Arg285 has an important role in enabling the multimeric ATP–DnaA interaction with oriC (23). This motif, in one DnaA protomer, recognizes ATP bound to an adjacent DnaA protomer, promoting structural modification of the DnaA oligomers, which enables formation of an initiation-competent complex at oriC (21,23). In addition, AID motifs (Arg227 and Leu290) are required for construction of unwinding-competent DnaA complexes and are suggested to be exposed on the domain III surface, participating in domain III-domain III interactions (24). Also, AAA+ Box VII motif Arg281 is required for stable formation of DnaA oligomers on oriC (22). ATP–DnaA molecules cooperatively bind to a DNA region bearing DnaA binding sites with specific intervening spaces (25,26). H/B motifs (Val211 and Arg245) in domain III sustain oriC unwinding by directly binding to the T-rich strand of the unwound duplex-unwinding element (ssDUE) (Supplementary Figure S1A) (27,28). Domain IV binds to a 9 bp sequence (DnaA box) via a helix-turn-helix motif (29).
oriC consists of the DUE and the DnaA-oligomerization region (DOR) (Figure 1A). The DUE includes three AT-rich 13-mer repeats, and the DOR has 12 DnaA boxes (R1/2/4/5M, I1/2/3, τ1/2, and C1/2/3) and a single IHF-binding site (IBS) (1,23,26,30,31). The R1 and R4 DnaA boxes, located at the ends of the DOR, perfectly match the consensus sequence (TTATNCACA), and bind ATP–DnaA or ADP–DnaA with high affinity (23,30,31). The R2 box has moderate affinity, and the other boxes have low affinities for DnaA. ATP–DnaA assembly on the low-affinity DnaA boxes is stimulated by cooperative binding via interaction between the arginine-finger motif of a protomer and ATP tightly bound to the next protomer (21,23,25,26,28,30,31).
The DOR can be divided to three functional regions (the left half, middle, and right half) on which DnaA molecules form structurally distinct subcomplexes (24,26,32,33) (Figure 1A). The left-half and right-half DOR includes a set of DnaA boxes (R1-τ1-R5M-τ2-I1-I2 and C3-C2-I3-C1-R4, respectively) and the middle region includes a single DnaA box (R2). The left-half DOR–DnaA subcomplex unwinds the DUE with the aid of IHF-dependent DNA bending; the middle and right-half DOR–DnaA subcomplexes are totally dispensable for DUE unwinding (24,34). The left-half and right-half subcomplexes are thought to independently interact with a DnaB helicase for bidirectional loading (24,33,34). Those DnaA subcomplexes form basically independently and AID motifs stimulate formation of the left-half DOR–DnaA subcomplex rather than that of the right-half subcomplex (24,33). DnaA bound to the R1 and R4 boxes might act as assembly cores in the left-half and right-half subcomplexes (26). In addition, although the τ2 box in the left-half DOR has a degenerated consensus sequence and thus its orientation cannot be determined only by its sequence, it is recently suggested to be the same as that with the R1 box (33). This is consistent with cooperative binding modes of DnaA occurring on a region bearing closely spaced DnaA boxes in the same orientation (26).
For mechanism of DUE unwinding, the ssDUE-recruitment model has been proposed (32,34). In this model, ssDUE is recruited to the left-half DnaA subcomplex via IHF-induced DNA bending, and directly binds to H/B motifs of DnaA molecules assembled on the DOR (Figure 1C). A speculative alternative model suggests that ssDUE-bound DnaA molecules bind to the left-half DnaA subcomplex through DnaA–DnaA interactions (1,34). Another model proposes continuous filamentation, with DnaA forming a filament from the DOR to ssDUE (5,27,35). In this model, ATP–DnaA bound to the R1 box interacts with other ATP–DnaA molecules, resulting in formation of extended oligomers of ATP–DnaA bound to ssDUE. In the alternative two models, H/B motifs are required for DnaA molecules bound to ssDUE, but not for those bound to the DOR, and in the third model, continuous binding of DnaA molecules occurs from the DOR to the ssDUE. The ssDUE-recruitment model is the only one that proposes that a DnaA oligomer complexed with the DOR simultaneously and directly binds ssDUE. This proposal has not yet been proved, although binding of DnaA to ssDUE in the absence of the DOR is known to be inefficient compared with binding in the presence of the DOR (28). Consistently, our recent study based on structures of DnaA-DNA complexes supported the hypothesis that a single DnaA molecule can simultaneously bind ssDUE and dsDNA by H/B motifs of domain III and domain IV (33).
To clarify the mechanism of DUE unwinding, we investigated the specific roles of DnaA protomers in the formation of the left-half DnaA subcomplex. Our results show that binding of DnaA protomers to R1 and R5M boxes is essential for DUE unwinding, whereas other DnaA boxes are stimulatory, or even dispensable. Notably, R5M-bound DnaA acted as a core for DnaA assembly on the left-half DOR, whereas R1-bound DnaA was supportive. The ssDUE-binding activity of DnaA protomers bound to R1 and R5M was essential to DUE unwinding. Results of in vivo analyses were basically consistent with the in vitro data. These results are important for understanding the regulation of ATP–DnaA assembly on oriC and the structure of the functional complex for DUE unwinding, and those support the ssDUE-recruitment model. In addition, overall similarity in sequences of the origin regions suggests that these molecular bases for DNA unwinding are evolutionarily conserved of many eubacterial species including important pathogens.
MATERIALS AND METHODS
Proteins, strains, DNA and plasmids
DnaA and its derivatives used in this study were prepared as described previously (28,32). His-DnaA domain III-IV was overexpressed and purified by the methods similar to wild-type DnaA, as described previously (36,37). Strains, DNA, and plasmids used are described in Supplementary Information.
Buffers
Buffer P contained 60 mM Hepes–KOH (pH 7.6), 0.1 mM zinc acetate, 8 mM magnesium acetate, 30% [v/v] glycerol, and 0.32 mg/ml bovine serum albumin (BSA). Buffer N contained 50 mM Hepes–KOH (pH 7.6), 2.5 mM magnesium acetate, 0.3 mM EDTA, 7 mM dithiothreitol (DTT), 0.007% [v/v] Triton X-100 and 20% [v/v] glycerol. Buffer F contained 25 mM Hepes–KOH (pH 7.6), 5 mM calcium acetate, 2.8 mM magnesium acetate, 45 mM ammonium sulfate, 4 mM DTT, 10% [v/v] glycerol, 0.2% [v/v] Triton X-100, 0.5 mg/ml BSA, 7 μg/ml poly (dA-dT) and 7 μg/ml poly (dI-dC). Buffer G contained 20 mM Hepes–KOH (pH7.6), 1 mM EDTA, 4 mM DTT, 5 mM magnesium acetate and 10% [v/v] glycerol.
DUE-unwinding assay
The assay was essentially performed as described previously (17,32,33). Briefly, EcoDnaA, ChiDnaA, and its derivatives were preincubated for 15 min at 4°C with 3 μM ATP or ADP, respectively. The resulting ATP– or ADP–DnaA was incubated at 38°C for 3 min in 20 μl buffer P containing a derivative of 1.32 nM M13oriCMS9, 32 nM IHF and 100 mM KCl, followed by further incubation with 4 U P1 nuclease (Wako) for 200 s. After reactions were stopped by addition of 0.5% SDS, one-third portions of each purified DNA sample was digested with EcoRI, which yielded 3.8 and 3.9 kb fragments for DUE-unwound M13oriCMS9 (33). The resultant DNA samples were analyzed by 1% agarose-gel electrophoresis and Gelstar (Lonza) staining.
EMSA of DnaA complex formation
The assay was essentially performed as described previously (24,32). Briefly, 35 nM DNA fragments were incubated at 30°C for 10 min in 10 μl buffer G containing ATP-EcoDnaA or ADP-EcoDnaA, 0.25 mg/ml BSA, and 200 ng λ phage DNA as a competitor, followed by 2% agarose-gel electrophoresis at room temperature, Gelstar staining, and densitometric scanning. In addition, reaction mixtures described above were analyzed by 4% polyacrylamide-gel electrophoresis in Tris-borate buffer at 4°C and Gelstar staining so that DnaA binding to the R1 box could be specifically detected.
DNase I-footprint assay
The assay was essentially performed as described previously (23,28), with minor modifications in the length of oriC fragment and the concentration of competitor DNA for detection of IHF binding. DnaA was incubated for 10 min at 30°C in 10 μl buffer F containing 2.4 nM end-labeled oriC fragment (346 bp), 7 μg/ml poly (dA-dT), 7 μg/ml poly (dI-dC) and 3 mM ATP or ADP in the presence of absence of IHF, followed by further incubation with 2.5 mU DNase I (NEB) for 4 min. Purified DNA was analyzed by 5% sequencing-gel electrophoresis and a BAS-2500 image analyzer (Fuji).
EMSA of ssDUE recruitment
The assay was essentially performed as described previously (32,34). Briefly, wild-type or mutant EcoDnaA and/or ChiDnaA, preincubated with 3 μM ATP or ADP, were incubated with 16 nM 32P-labeled ssDUE derivatives (ssDUE M28, ssDUE M28rev, ssDUE-dsR1WT, ssDUE-dsR1non or ssDUE-dsR1Tma) at 30°C for 10 min in 5 μl buffer G containing 35 nM DOR derivatives, 80 mM KCl, 0.25 mg/ml BSA, 2 mM ATP, and 25 ng λ phage DNA, followed by 4% polyacrylamide-gel electrophoresis at 4°C.
Flowcytometry analysis
The analysis was essentially performed as described previously (11,38). Briefly, cells were exponentially grown until the absorbance of the culture (A660) reached 0.1. Portions of the cultures were immediately chilled in 70% ethanol and used for analysis of cell mass (volume) using a Multisizer 3 Coulter counter (Beckman Coulter). Incubation of remaining portions of the cultures was continued for 4 h in the presence of 300 μg/ml rifampicin and 10 μg/ml cephalexin for run-out replication of the chromosomal DNA, followed by DNA staining with SYTOX Green (Life Technologies) and analysis of the cellular DNA content with a FACS Calibur flow cytometer (BD Biosciences).
RESULTS
DnaA boxes R1 and R5M are the most important for DUE unwinding
We previously showed that the oriC region containing the DUE and the left-half DOR exhibits full activity in DUE unwinding (Figure 1A) (24,34). To determine the essential region for DUE unwinding in more detail, we first performed a DUE unwinding assay with a series of truncated oriC constructs, in a reaction buffer with a physiological ionic concentration (100 mM KCl) (Supplementary Figure S1B). In this assay, unwound DUE was specifically digested by P1 nuclease. The minimal left-half DOR that sustained activity in DUE unwinding contained a region from the R1 box to the I1 box (Figure 1A and Supplementary Figure S1B and C).
To understand the contribution of each DnaA box to DUE-unwinding activity, similar assays were performed with oriC-containing plasmids bearing substitution mutations in individual DnaA boxes R1, τ1, R5M, τ2 and I1. Consistent with previous results (25,34), mutation of the R1 box excluding specific affinity for DnaA (R1non) (31) severely impaired DUE unwinding, although a slight residual activity remained (Figure 1D and E). Mutation of the R5M box (R5Mnon) also severely inactivated DUE unwinding (Figure 1D and E). In addition, mutation of the I1 box moderately impaired DUE unwinding (Figure 1F and G). By contrast, DUE unwinding was only slightly inhibited by mutation of τ1 (τ1non) (Figure 1D and E).
Due to degenerated consensus sequence, the DnaA-binding sequence of the τ2 box has two possible orientations, designated τ2l (leftward) and τ2r (rightward) (Supplementary Figure S1D) (23,33); although we recently analyzed these sequences using specific mutations (33), in this study other mutations which specifically alter the sequence with either orientation (i.e., τ2l-non and τ2r-non) were analyzed separately to have additional evidence. Mutation of the τ2l-non, but not the τ2r-non, moderately impaired DUE unwinding (Figure 1F and G).
These results indicate that the R5M box and the R1 box have important roles in DUE unwinding, that the τ2 and the I1 boxes contribute to this process but are not essential and that the τ1 box is dispensable under these conditions. The τ2 sequence seems to function in the leftward orientation (Figure 1A), which is consistent with head-to-tail cooperative binding of DnaA and with the result of our recent study using other τ2 mutations (33). These results are fundamentally consistent with those of a previous study of the minimum oriC region required for DUE unwinding, which was conducted with a lower salt concentration (34), although the supportive role for the τ2 and the I1 was shown in the present assay. In general, higher salt concentrations more severely inhibit non-specific or weak binding between proteins and between protein and nucleic acids. Under the previous low-salt conditions, non-specific interaction of DnaA with sequences substituted at the τ2 and I1 boxes might be allowed (34).
DnaA box R5M (but not R1) is a prerequisite for ATP–DnaA assembly on the left-half DOR
To investigate the specific roles of DnaA boxes R5M and R1 in DUE unwinding, we first analyzed their roles in DnaA assembly by electrophoretic-mobility-shift assay (EMSA). As we showed previously (24), the left-half DOR fragment and DnaA formed a high-order complex in an ATP binding-dependent manner (Figure 2A and B). The left-half DOR with the R1non mutation was only slightly less effective at promoting complex formation than the wild-type fragment. By contrast, the fragment with the R5Mnon mutation was severely impaired in complex formation, with a level of band shifting as low as that of the wild-type fragment in the presence of ADP–DnaA (Figure 2A and B). Decreases in complex formation on these mutant fragments are basically consistent with previous studies which analyzed R1 and R5M mutants separately (25,26). Also, when complex formation with those DORs was similarly analyzed using polyacrylamide gel, specific binding of DnaA to the R1 site was detected and overall results were consistent with the above data (Supplementary Figure S2A). Higher-order complexes constructed with the R1non fragment migrated at a slightly faster rate than those with the wilt-type fragment, which is consistent with the idea that, even in the absence of DnaA binding to R1, DnaA complexes are constructed in the R5M-I2 region by cooperative binding of DnaA molecules, resulting in a complex with considerable stability (Supplementary Figure S2A) (also see Discussion). Slight differences in complex formation efficiency might be caused by differences in experimental conditions (e.g. buffer) between agarose and polyacrylamide gel electrophoresis.
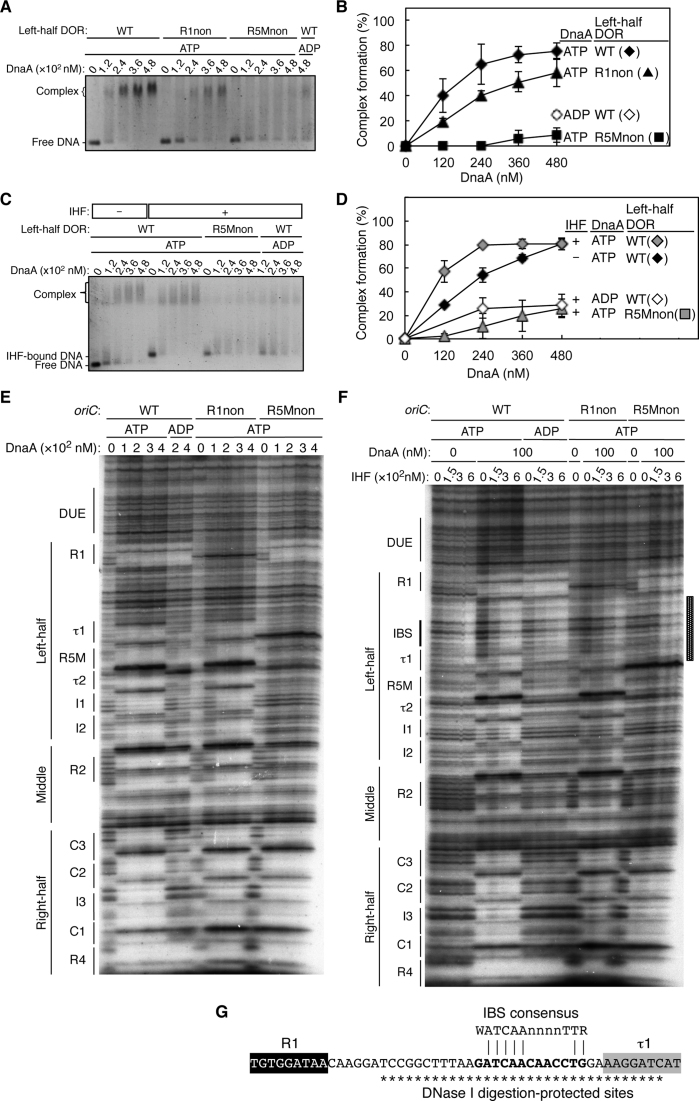
Figure 2.
DnaA bound to R5M, but not R1, is essential for DnaA assembly on the left-half DOR. (A–D) EMSA with the left-half DOR bearing R1non or R5Mnon mutations. Each DNA fragment (35 nM) was incubated with the indicated amount of DnaA in the absence (A) or presence (C) of 72 nM IHF, then analyzed by 2% agarose-gel electrophoresis. The amount of DNA complexed with DnaA relative to the input DNA is shown as ‘Complex formation (%)’ in (B) and (D). Two independent experiments were carried out, and both data, mean values, and representative gel images in a black-white inverted mode are shown. (E–G) DNase I-footprint assay with oriC fragments (346 bp) containing R1non or R5Mnon mutations. Indicated amounts of ATP–DnaA or ADP–DnaA were incubated for 10 min at 30°C with 2.4 nM 32P-end-labeled oriC and 3 mM ATP or ADP in the absence (E) or presence (F) of 150–600 nM IHF, followed by DNase I digestion and gel-electrophoresis analysis. Positions of DNA motifs are indicated. The sequence of the region protected by IHF binding is shown in (G). The IBS consensus sequence and DNase I digestion-protected sites are indicated by bold lettering and asterisks, respectively.
Consistent with the results of the DUE unwinding assay (Figure 1D–G), complex formation with the fragments bearing τ2l-non or I1non mutations was moderately inhibited, whereas the τ1non and τ2r-non mutations did not affect complex formation (Supplementary Figure S2B and C).
To determine the extent of protein–DNA interactions, we performed DNase I-footprint analysis on oriC sequences with R1non or R5Mnon mutations (Figure 2E). As we showed previously (23), ATP–DnaA bound to all the low-affinity sites on the wild-type oriC, whereas ADP–DnaA bound predominantly to the R1 and R4 boxes and also to the R2 box. Footprint patterns of ATP–DnaA in the presence of the R1non mutation were essentially the same as those on the wild-type oriC, except for the absence of protection at the R1non site itself and a slight reduction in the levels of protection of the DnaA boxes in the left-half DOR at the lowest DnaA concentration (100 nM). This reduction is basically consistent with previous studies using DMS footprint experiments (25,39): severe inhibition in the DnaA assembly in the previous study might be caused by a lower input DnaA/oriC ratio. The ratio was at most about 13 in the previous studies (25,39) but it was 42 at the point of 100 nM DnaA in the present study (Figure 2E).
However, in the presence of the R5Mnon mutation, ATP–DnaA binding to the τ1–I2 region was severely inhibited, even though protection of the R1 box from DNase I digestion persisted. By contrast, with the exception of R2, binding of ATP–DnaA to DnaA boxes on the right-half DOR was not substantially inhibited. These results are consistent with a previous study (25) as well as with our EMSA results (Figure 2A and B; Supplementary Figure S2A), and with our previous finding that DnaA subcomplexes formed on each half-DOR independently (24).
Taken together, these results suggest that the R5M-bound DnaA, rather than the R1-bound DnaA, has a pivotal role in formation of the left-half DnaA subcomplex. This proposed mechanism is consistent with the location and orientation of the sites in oriC, and with evidence from a previous study indicating that effective cooperative binding of DnaA molecules depends on a short distance (2–5 bp) between DnaA-binding sites (26). Our observation of moderate inhibition of DnaA binding at the R2 box in the absence of R5M-DnaA binding (i.e. in the case of R5Mnon) is consistent with the possibility that DnaA molecules bound at R2 and I2 may weakly interact with each other via domain I–domain I interactions (34,40).
IHF stimulates ATP–DnaA assembly in an R5M box-dependent manner
IHF stimulates DnaA-dependent DUE unwinding of the supercoiled form of oriC, and even of linearized oriC (34,41). To further investigate the role of R5M in DnaA assembly, we performed EMSA and DNase I-footprint assays in the presence of IHF. In the EMSA, addition of IHF stimulated ATP–DnaA assembly on the wild-type oriC (Figure 2C and D), consistent with previous results (42,43). However, with an oriC substrate that had an R5Mnon mutation, DnaA assembly was inhibited even in the presence of IHF.
In the DNase I-footprint assay, IHF protected a 33 bp region covering the IBS and 7 bp of the 9 bp constituting the τ1 box (Figure 2F and G). IHF binding of such a long region is consistent with previous data, and would be caused by a bent DNA structure enclosing the bound IHF, as IHF bends a 35 bp DNA substrate by >160° (11,44). This result corresponds with the observed dispensability of τ1 in DUE unwinding (Figure 1D and E), and with the previous suggestion that DnaA might not bind to the τ1 box of superhelical oriC (26). If the binding of IHF physically inhibits DnaA binding to τ1, it does so without apparent adverse effects on DUE unwinding. Also, apparent Kd of IHF for the IHF-binding consensus sequence and that of DnaA for DNA containing the R2 box under the same experimental conditions are reported to be 37 and 213 nM respectively (45). In addition, the cellular amount of IHF is reported to be about 2.9-to-7.5-fold higher than that of DnaA in exponentially growing cells (46). Based on these data, it would be unlikely that DnaA binding to τ1 results in inhibition of IHF binding to oriC.
In the presence of ATP–DnaA, IHF stimulated protection of the R5M–I2 region of wild-type as previously shown (Figure 2F) (42,43). Notably, IHF stimulated protection of the R5M–I2 region of the R1non fragment, but not the R5Mnon fragment (Figure 2F). In addition, ATP–DnaA binding to R2 was inhibited on the R5Mnon fragment even in the presence of IHF. By contrast, IHF did not affect ATP–DnaA binding to the right-half DOR R4–C3 region. Previously, protection of the I3 box also is reported to be stimulated by IHF in the presence of DnaA (42), but under our present conditions using a higher DnaA concentration and a linear DNA fragment, this site was efficiently protected even without IHF, consistent with our previous data (23,24,28). These results suggest that R5M-bound ATP–DnaA stimulates cooperative ATP–DnaA binding to the τ2–I2 region, and even to the R2 box, even in the presence of IHF.
DUE unwinding depends on ssDNA binding of the R1- and R5M-bound DnaA
To determine requirements for ssDNA binding of the R1- and R5M-bound DnaA in DUE unwinding, we first analyzed a chimeric DnaA (ChiDnaA) consisting of E. coli DnaA (EcoDnaA) domains I–III and Thermotoga maritima DnaA (TmaDnaA) domain IV, which specifically binds to its cognate DnaA-binding sequence (TmaDnaA box), but not to EcoDnaA boxes, with at least moderate affinity (Figure 3A and Supplementary Figure S3C and D) (32,33,47). The DUE of oriC bearing a TmaDnaA box at the R1 site (R1Tma) (Figure 3B) was previously shown to be efficiently unwound in the presence of ATP-EcoDnaA and ATP-ChiDnaA (or even ADP-ChiDnaA) (32). This unwinding was dependent on the Arg285 of ChiDnaA, suggesting that the Arg finger of the R1-bound DnaA orients inwards, interacting with the flanking ATP–DnaA of the left-half DOR subcomplex (Figure 3C) (32,33). In the previous study (32), the relative position of TmaDnaA box in the R1 box region is optimized for replicational initiation (Figure 3B). Based on this, in the present study we introduced TmaDnaA box sequence at the R5M box similarly to the R1 box (Figure 3B).
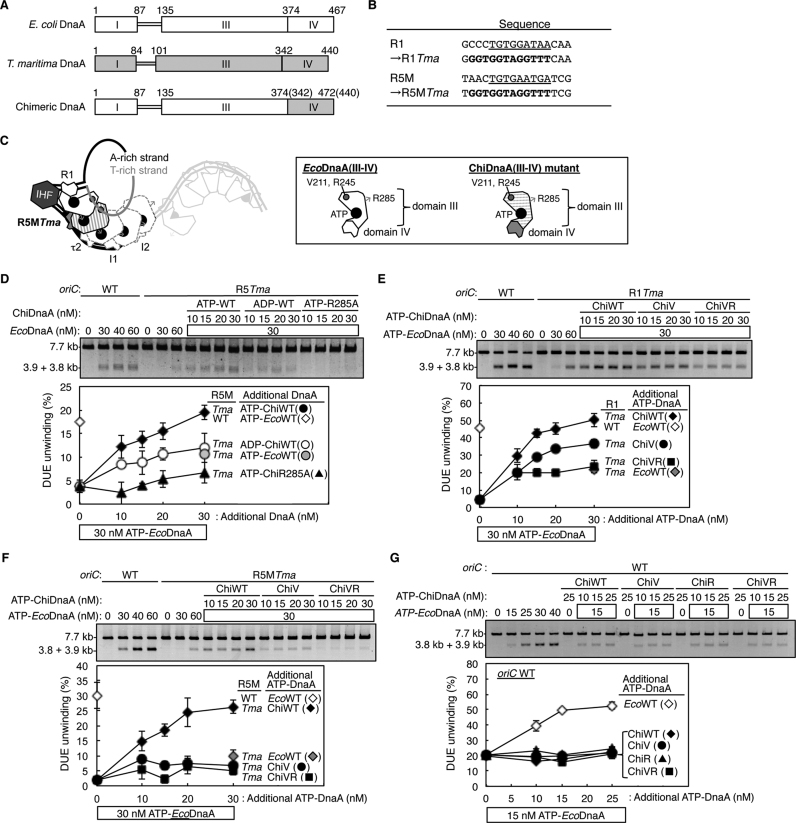
Figure 3.
H/B motifs of DnaA bound to R1 and R5M are important for DUE unwinding. (A) Structures of E. coli DnaA (EcoDnaA), T. maritima DnaA (TmaDnaA), and chimeric DnaA (ChiDnaA). Domains of EcoDnaA and TmaDnaA are indicated by open and gray boxes, respectively. (B) Sequences of R1 and R5M boxes substituted with TmaDnaA box. (C) A schematic of DUE unwinding assay using ChiDnaA, EcoDnaA, and R5MTma oriC plasmid. ChiDnaA mutant (indicated by striped domain III and gray domain IV) was suggested to bind specifically to R5MTma site. The other DnaA boxes were occupied by EcoDnaAs (indicated by open shapes of domain III and IV). (D) DUE-unwinding assays with an R5MTma oriC plasmid. An oriC plasmid (1.32 nM) with the R5MTma substitution was incubated for 3 min at 38°C with wild-type ATP-EcoDnaA (EcoWT) and indicated amounts of ATP/ADP-chimeric DnaA (ChiWT) or its R285A mutant (ChiR285A), followed by incubation with P1 nuclease. Resultant DNAs were analyzed by gel electrophoresis. The percentages of DNA digested by P1 nuclease relative to the total input DNA are shown as ‘DUE unwinding (%)’. (E–G) DUE-unwinding assay with ssDUE-binding-variant ChiDnaA. oriC plasmid with (E and F) or without (G) the substitution of R1Tma (E) or R5MTma (F) were incubated as described above, except for the use of ATP-ChiDnaA V211A (ChiV) and ATP-ChiDnaA V211A R245A (ChiVR). For panels D–G, two independent experiments were carried out, and both data, mean values and representative gel images in a black–white inverted mode are shown.
DUE unwinding of oriC bearing a TmaDnaA box at the R5M site (R5MTma) occurred in the presence of 30 nM ATP-EcoDnaA in manner dependent on ATP-ChiDnaA, but not ATP-ChiDnaA R285A (Figure 3D). The unwinding activity with ATP-ChiDnaA reached a maximal level similar to that sustained by the wild-type oriC with ATP-EcoDnaA. DUE unwinding was moderately inhibited with ADP-ChiDnaA in the presence of EcoDnaA (Figure 3D). The moderate activity of ADP-ChiDnaA could be caused by elevated affinity for the R5M site, compared to that of EcoDnaA for the original R5M box (Supplementary Figure S3C and D). This might result in moderate stimulation of DnaA assembly in the R5M-flanking region, causing moderate activity in DUE unwinding. Based on our previous study (32), the Arg finger of R5M-bound DnaA, but not the bound nucleotide, would be crucial for interaction with the flanking τ2-binding DnaA, thereby stimulating DnaA assembly in the R5M-I2 region. The present data of ChiDnaA R285A showing severe inhibition of DUE unwinding of oriC bearing R5Tma is consistent with this idea.
Unwinding of DUE in oriC bearing a TmaDnaA box at the R1 site (R1Tma) was fully sustained when both of ATP-EcoDnaA and ATP-ChiDnaA were sufficient (Figure 3E). These results support the idea that the structure of the complex containing EcoDnaA and R5M-bound ChiDnaA, as well as that containing R1-bound ChiDnaA (32), is fundamentally similar to that of the native oriC with EcoDnaA. The roles for ATP and the Arg finger in the R5M-bound DnaA suggest that R5M-DnaA interacts with DnaA molecules on both sides (that is, at R1 and τ2l, because this assay included IHF, which inhibits DnaA–τ1 binding), and that these interactions stimulate formation of functional initiation complexes. This idea is consistent also with the crucial role for R5M-DnaA in DnaA assembly on the left-half DOR (Figure 2).
Next, the introduction of mutations in the ssDUE-binding H/B motif of ChiDnaA (ChiV211A and ChiV211A R245A) was assessed. If direct ssDUE binding by DnaA protomers bound to R1 and R5M is essential for DUE unwinding (Figure 1C), binding of these ChiDnaA variants to the R1Tma and R5MTma sites should impair DUE unwinding, even with EcoDnaA bound to the other DnaA boxes. Affinities of the purified ChiDnaA variants for nucleotide and TmaDnaA box were similar to those of the ‘wild-type’ ChiDnaA (Supplementary Figure S3A–D). Notably, with the R1Tma oriC, ChiV211A and ChiV211A R245A proteins were impaired in DUE unwinding with a slight residual activity, even in the presence of EcoDnaA (Figure 3E). Moreover, with the R5MTma oriC, both ChiV211A and ChiV211A R245A proteins had very little DUE-unwinding activity (Figure 3F). ChiDnaA itself did not stimulate DUE unwinding of wild-type oriC, demonstrating the binding specificity of ChiDnaA for TmaDnaA box sequences (Figure 3G). In addition, if DnaA box-unbound DnaA molecules bind to R1- and R5M-bound DnaAs and those additional DnaAs bind ssDUE (Alternative model in Figure 1C), addition of ChiDnaA should stimulate unwinding of wild-type oriC in the presence of EcoDnaA. However, addition of ChiDnaA did not stimulate initiation (Figure 3G); similar results are shown also in our previous study (32). Taken together, at least under the conditions used here, these results support the idea that direct binding of ssDUE by DnaA protomers bound to R1 and R5M is important for DUE unwinding, and that ssDUE binding by DnaA bound to R5M is more effective than that by DnaA bound to R1.
Direct role for R5M-bound DnaA in ssDUE binding
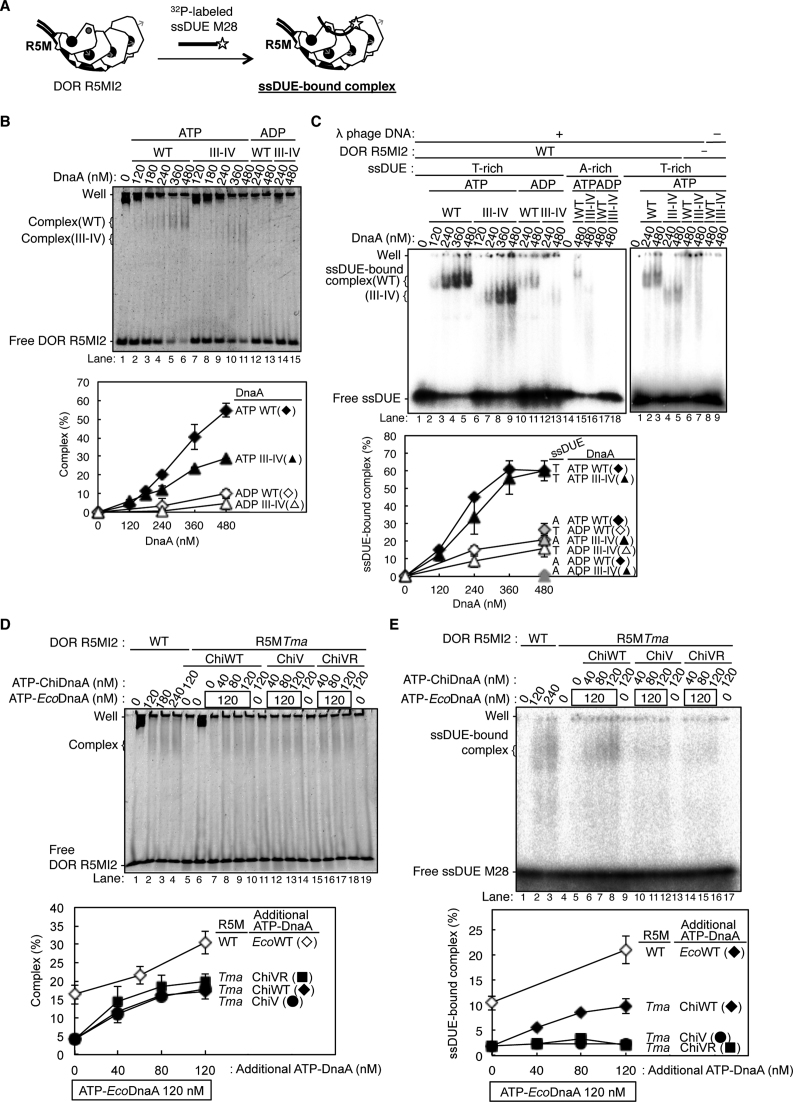
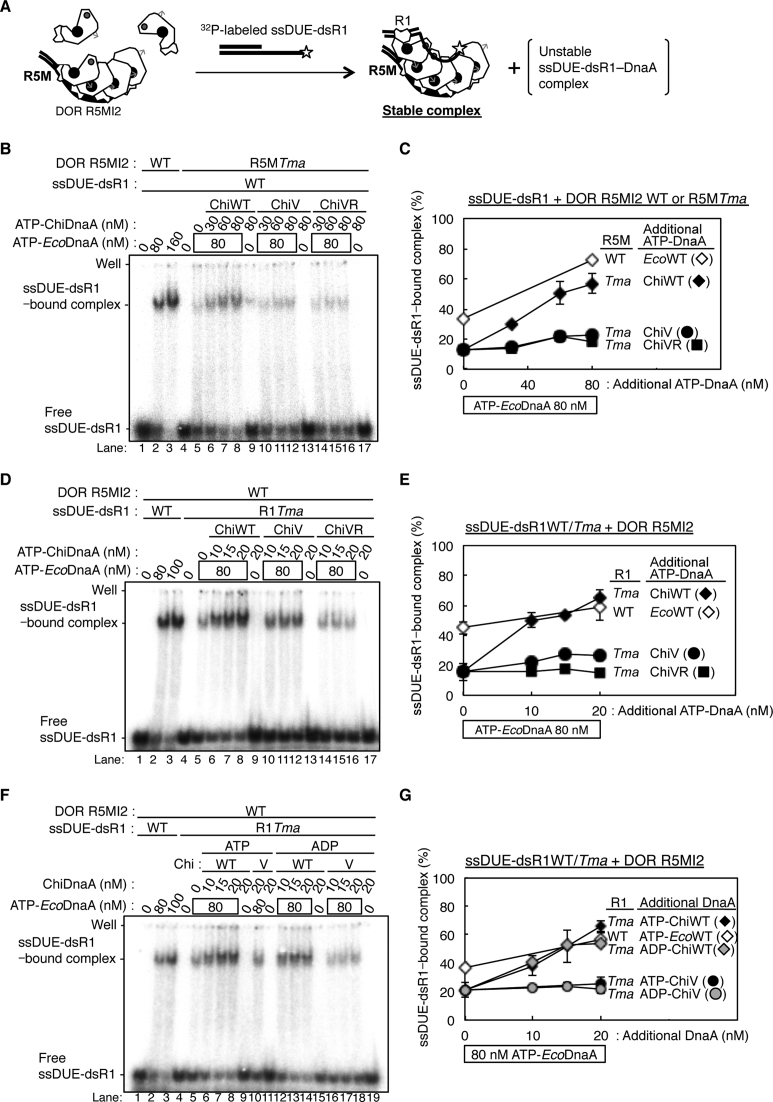
ATP–DnaA multimers constructed on DOR specifically bind to the T-rich (not the A-rich) strand of the ssDUE (24,28,32,34). Because the crucial role for R5M-bound DnaA in DUE unwinding was shown (Figure 3), we first elucidate the ssDUE binding of R5M-bound DnaA by EMSA with a combination of two DNA elements: a 32P-labeled ssDUE (the T-rich strand 28-mer, ssDUE M28, or the A-rich strand 28-mer, ssDUE M28rev) and a truncated left-half DOR spanning the R5M box to the I2 box (DOR R5MI2). ATP–DnaA should form an oligomer on the DOR, which binds the ssDUE M28, resulting in a ternary complex (Figure 4A) (28,34). It is demonstrated that whereas ssDUE binding of DOR-free DnaA is inefficient, DnaA complex formation with DOR drastically enhances the ssDUE binding activity (28,34). This is reasonably explained by the idea that functional DnaA oligomers are constructed on DOR and the resultant DnaA oligomer binds ssDUE using multiple (at least two) binding sites in ssDUE and the H/B motif residues of the DnaA oligomer, which effectively stabilizes ssDUE binding (28,34). In the present study, deletion of the R1– τ1 region from the left-half DOR excludes possible ssDUE binding by the R1-bound DnaA, thereby being capable of detecting R5M-bound DnaA-specific activity in the absence of the τ1 box (Figures 1A and 4A).
Figure 4.
Roles of DnaA bound to R5M in ssDUE binding to the DnaA–oriC complex. (A) A schematic of EMSA of ssDUE recruitment. ATP–DnaA was incubated with the DOR R5MI2 fragment and 32P-end-labeled ssDUE M28, followed by electrophoretic-mobility-shift assay. For simplicity, only DnaA domains III–IV are shown. ssDUE M28 is shown with thick black lines, 32P with a star, and the DOR R5MI2 DNA with thin black lines. ATP and Arg finger are also shown as in Figure 3C. (B and C) EMSA using the DOR R5MI2 and EcoDnaA or its truncated form bearing domain III-IV. Indicated amounts of the ATP or ADP form of DnaA were incubated for 10 min at 30°C with (+) or without (–) the DOR R5MI2 (35 nM) and λ phage DNA in the absence (B) and presence(C) of ssDUE M28 (T-rich strand) and ssDUE M28rev (A-rich strand), followed by 4% polyacrylamide-gel electrophoresis. Two independent experiments were carried out, and representative gel images are shown. The amounts of DOR R5MI2 complexed with DnaA, or ssDUE M28 and ssDUE M28rev bound to the DnaA–DOR R5MI2 complexes were quantified as ‘Complex (%)’ in (B) and ‘ssDUE-bound complex (%)’ in (C), and both data and mean values are shown. λ phage DNA included as a competitor remained near the gel wells, which is not included in Complex (%) and ssDUE-bound complex (%). (D and E) EMSA using DOR R5MI2 with R5MTma substitution (DOR R5MI2-R5MTma), EcoDnaA, ChiDnaA(ChiWT) or its derivatives (ChiV, ChiVR) in the absence (D) and presence(E) of ssDUE M28 as described above. Two independent experiments were carried out, representative gel images are shown, and both data, mean values and detected complexes (%) are also shown as described above. For panels B and D, representative gel images are shown in a black-white inverted mode.
First, we analzyed DnaA complex formation on DOR R5MI2 bearing four low-affinity sites (i.e., R5M, τ2, I1 and I2). Previously DnaA self-oligomerization was analyzed using glutaraldehyde, which indicates that without DNA, DnaA monomers and homodimers are mainly detected (48). We used this method with or without DOR R5MI2. As shown in Supplementary Figure S4A, without the DNA (or with ssDUE), monomers were predominant and homodimers were detected moderately (Supplementary Figure S4A, lanes 10–11), consistent with previous results. In contrast, higher oligomers including up to pentamers were detected in the presence of R5MI2 (Supplementary Figure S4A, lanes 3–7). Given that homodimers are moderately detected even without the DNA, these data are consistent with the idea that at most four DnaA molecules can directly bind to the DNA, constructing homo-oligomers. Notably, formation of these higher oligomers was evidently stimulated by ATP (but not ADP) binding of DnaA (Supplementary Figure S4A, lanes 7 versus 9). Thus, these data support out idea that ATP–DnaA tetramers are constructed on R5MI2 depending on a cooperative manner.
Furthermore, to examine the idea that construction of DnaA complex on R5MI2 would depend on cooperative binding more strongly than that on DNA bearing high affinity sites, we performed EMSA using R5MI2 and R5MI2(R14) which carried four high-affinity sites (R1 box sequences) instead of the four low-affinity sites. As shown in Supplementary Figure S4B, whereas the intermediate binding complexes were detected for R5MI2(R14), those were only little for R5MI2 (Supplementary Figure S4B, lanes 1–6 versus 9–14). Also, DnaA complex formation on R5MI2 was dependent on ATP–DnaA (but not ADP–DnaA) (Supplementary Figure S4B, lanes 7–8 versus 15–16), consistent with the cross-linking experiments. These data are consistent with the idea that formation of ATP–DnaA oligomers on R5MI2 highly depends on cooperative binding and binding intermediates are very unstable.
In addition, we performed EMSA using R5MI2 with or without an individual substitution of the low affinity sites with the nonsense sequence excluding specific affinity for DnaA. As shown in Supplementary Figure S4C, unlike the R5MI2, the derivative bearing R5Mnon substantially inactive in constructing high-order ATP–DnaA complexes, consistent with the data of DNase I footprint (Figure 2). The derivative bearing τ2l-non, I1non or I2non had only moderate or slight activities in high-order ATP–DnaA complex formation. There results suggest that at least three low-affinity sites are required for stable construction of the DnaA complexes and DnaA binding to the R5M site has an essential role to sustain the stability of the complexes.
Next, we performed EMSA experiments using R5MI2 and ssDUE. When ATP-EcoDnaA was co-incubated, complex formation with DOR R5MI2 was detected (Figure 4B), and significant band shift of ssDUE M28 also was detected in a manner dependent on ATP-EcoDnaA and DOR R5MI2, indicating formation of a ternary complex composed of ATP-EcoDnaA molecules, ssDUE, and DOR R5MI2 (Figure 4C and Supplementary Figure S4A). When 175 fmol (35 nM) of DOR R5MI2 and 2.4 pmol (480 nM) of ATP-EcoDnaA were co-incubated, the resultant DnaA complexes included 102 fmol of DOR R5MI2 (about 60% of the input) and bound 48 fmol of ssDUE (∼60% of the input ssDUE) (Figure 4B, lane 6 and 4C, lane 5). Formation of moderately smeared bands of DOR R5MI2-DnaA complexes in the gel might be caused by partial degradation of the complexes during electrophoresis. Slight background at gel wells was seen in a competitor λ phage DNA-dependent, but DOR-independent manner, which could be caused by non-specific aggregations due to a propensity of DnaA (Figure 4C, right panel, lanes 2–3, 6 versus 8). These are consistent with our previous results (28,34).
Previous studies using P1 nuclease assay demonstrate that DnaA domains I-II are dispensable for DUE unwinding (19). Even when truncated DnaA protein bearing only domains III-IV was used for EMSA of ssDUE recruitment, ssDUE bound to complexes of DOR R5MI2 and ATP-EcoDnaA domains III-IV molecules at a level comparable to the case of the full-length ATP–DnaA (Figure 4B and C). Specificities for ATP–DnaA and the T-rich strand of ssDUE also were sustained (Figure 4B and C). These results also are consistent with the idea that ssDUE directly binds to DnaA domain III of DnaA complexed with DOR in a process of DUE unwinding.
When a DOR R5MI2 derivative bearing TmaDnaA box at the R5M site (R5MI2-R5MTma) was used, formation of EcoDnaA complexes on the DOR derivative were severely inhibited (Figure 4D, lane 7 and 4E, lane 5). This is consistent with the crucial roles for R5M-DnaA in ATP–DnaA assembly on the left-half DOR and DUE unwinding (Figures 2 and 3). Consistently, higher-order complexes with ssDUE were reconstructed by addition of ATP-ChiDnaA (Figure 4D, lanes 8–10; Figure 4E, lanes 6–9). Complex formation with DOR R5MI2-R5MTma, ATP-ChiDnaA and 120 nM ATP-EcoDnaA reached a level similar to that of complex formation with the wild-type DOR R5MI2 and 120 nM ATP-EcoDnaA. However, when ChiDnaA V211A or V211A R245A mutant was used, binding of ssDUE to the DOR-DnaA complexes was severely inhibited (Figure 4E, lanes 10–17) although the mutant ChiDnaA proteins as well as the ‘wild-type’ ChiDnaA were complexed with DOR R5MI2-R5MTma even in the presence of ATP-EcoDnaA (Figure 4D, lanes 12–19). These results support the idea that direct binding of the R5M-bound DnaA to ssDUE is crucial for the ternary complex formation.
Direct role for R1-bound DnaA in ssDUE binding
To analyze the role for R1-DnaA using similar EMSA, we used the 3’-end labeled 28-mer ssDUE conjugated to double-stranded (ds) DNA bearing the R1 box (ssDUE-dsR1) (Figure 5A). In these experiments, DnaA was preincubated with DOR R5MI2, followed by further incubation with ssDUE-dsR1. As we previously showed (32,34), DnaA bound to the R1 box of ssDUE-dsR1 stimulates formation of ternary complexes including left-half DOR without the R1 box. Briefly, ssDUE-dsR1 DNA has about 2-fold higher activity in binding to the R1-deleted DOR–-DnaA complexes than ssDUE (34). In addition, the Arg finger (but not ATP) of the R1-bound DnaA is essential for the stimulation of ssDUE-dsR1 binding to the DOR-DnaA complexes, consistent with the idea that the R1-bound DnaA interacts with ATP–DnaA bound to the edge (i.e. R5M site) of the R5MI2 DNA (32) (Figure 5A).
Figure 5.
Roles of DnaA bound to R1 and R5M in ssDUE recruitment into the DnaA–oriC complex. (A) A schematic of EMSA of ssDUE recruitment using 32P-end-labeled ssDUE-conjugated dsDNA bearing DnaA box R1 (ssDUE-dsR1). ATP–DnaA was preincubated with DOR R5MI2, followed by co-incubation with ssDUE-dsR1. Possible complexes are illustrated as in Figure 3C. See text for details. (B and C) EMSA of ssDUE recruitment with DOR R5MI2 and DOR R5MI2-R5MTma. Indicated amounts of the ATP form of EcoDnaA and ChiDnaA (ChiWT) or its derivatives (ChiV, ChiVR) were pre-incubated for 5 min on ice with the DOR R5MI2 or DOR R5MI2-R5MTma (35 nM), followed by further incubation for 10 min at 30°C with ssDUE-dsR1 (16 nM). A representative gel image is shown. The amounts of ssDUE-dsR1 bound to the DnaA–DOR R5MI2 complexes were quantified as ‘ssDUE-dsR1−bound complex (%)’. Two independent experiments were carried out, and both data and mean values are shown in (C). λ phage DNA included as a competitor remained near the gel wells, which is not included in ssDUE-dsR1−bound complex (%). (D–G) EMSA of ssDUE recruitment with ssDUE-dsR1 bearing the R1Tma substitution. Experiments were performed as described above, except for ssDUE-dsR1 (WT) and ssDUE-dsR1Tma (R1Tma). The ATP-form (D and E) or ADP-form (F and G) ChiDnaA wild-type and its derivatives were analyzed. Two independent experiments were carried out, and quantified as described above, and both data, mean values and ssDUE-dsR1−bound complex (%) are shown in (E) and (G), respectively.
When ATP-EcoDnaA was co-incubated, a shifted band of ssDUE-dsR1 was predominantly detected in a manner dependent on DOR R5MI2, indicating formation of a stable ternary complex (Figure 5B, D and F, lanes 1–3 and Supplementary Figure S4D, lanes 1–5). Consistent results were obtained when truncated DnaA protein consisting of only domains III-IV was used (Supplementary Figure S4E). In the absence of DOR R5MI2, ssDUE-dsR1 signals smeared, indicating that only unstable irregular complexes were formed with ATP-EcoDnaA (Supplementary Figure S4D, lanes 3–13). These are consistent with our previous results (32,34) and the data shown in Figure 4B and C.
When DOR R5MI2-R5MTma was used in this assay, formation of ternary complexes with ssDUE-dsR1 and ATP-EcoDnaA was stimulated in a manner dependent on ATP-ChiDnaA, but not ATP-ChiDnaA V211A or ATP-ChiDnaA V211A R245A (Figure 5B and C). Only faint shifted bands were detected with ATP-EcoDnaA in the absence of ChiDnaA, which could be caused by interaction of ssDUE-dsR1–ATP-EcoDnaA unstable complexes with EcoDnaA unstably bound to the τ2-I1-I2 boxes. Basically similar unstable complexes could be formed in the presence of ATP-EcoDnaA and the mutant ChiDnaAs. These results are consistent with the results of Figure 4 and thus support the important role for the R5M-bound DnaA in ssDUE binding.
When ssDUE-dsR1 derivative bearing TmaDnaA box at the R1 site (ssDUE-dsR1Tma) was incubated with DOR R5MI2, EcoDnaA and ChiDnaA, ternary complexes were formed at a level similar to that with ssDUE-dsR1 and EcoDnaA, which was dependent on the Val211 and Arg245 residues of ChiDnaA (Figure 5D and E). These data consistent with the idea that the R1-bound DnaA binds ssDUE simultaneously using the H/B motif residues. In the absence of ChiDnaA, a ternary complex was detected at a moderate level (Figure 5D, lanes 1–3 versus lane 5), which would be caused by ssDUE binding to R5MI2-DnaA complexes, consistent with our present and previous results (Figure 4B) (32). Consistent results were further obtained in similar experiments using a left-half DOR lacking R1 (left-half DORΔR1) instead of DOR R5MI2 (Supplementary Figure S5).
Also, even when ADP-ChiDnaA was preincubated with ssDUE-dsR1Tma, ternary complex formation was stimulated as seen for ATP-ChiDnaA (Figure 5F and G). These results indicate that the R1-bound DnaA protomer does not require ATP binding during this process, which is consistent with our previous results that the ATP and ADP forms of the R1-bound DnaA are similarly active in DUE unwinding (32).
These results indicated that direct binding of the R1-bound DnaA to ssDUE was important for ternary complex formation of the left-half DOR–DnaA–ssDUE, even though the R1 box was located in cis to the ssDUE in this experiment (Figure 5A). As the R1-bound DnaA interacts with both the R1 box and ssDUE of ssDUE-dsR1 simultaneously, it might cause a structural change, such as DNA bending, thereby stimulating interaction of the ssDUE region with DnaA bound to the DOR (Figure 5A). Taken together, these results are consistent with the idea that R1-bound DnaA interacts with R5M-bound DnaA, and that both of these DnaA protomers directly bind to ssDUE, resulting in stable ternary complex formation.
Chromosomal mutation of R5M, rather than R1, severely inhibits replication initiation
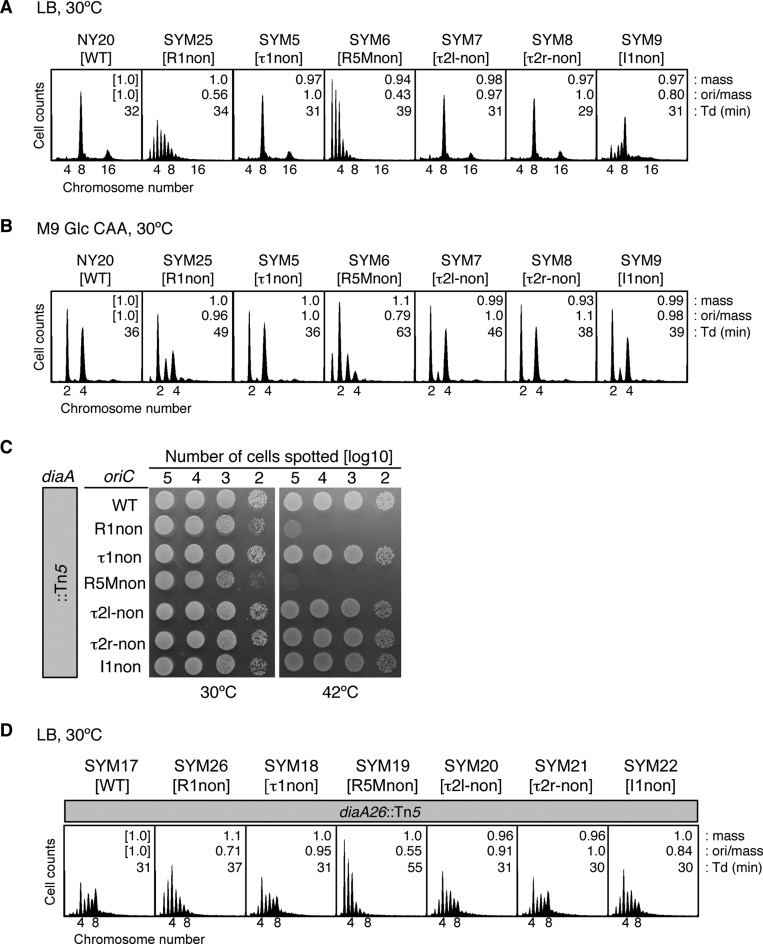
The in vivo roles of individual DnaA protomers in the minimum region for DUE unwinding were analyzed by construction of E. coli strains with R1non (SYM25), τ1non (SYM5), R5Mnon (SYM6), τ2l-non (SYM7), τ2r-non (SYM8) and I1non (SYM9) mutations in the chromosomal oriC sequence. Inhibitions of initiation in cells bearing the mutant R1 or R5M are previously reported (25,49). Doubling times at 30°C of SYM25, SYM5, SYM7, SYM8 and SYM9 cells were similar to those of wild-type (NY20) cells, whereas those of R5Mnon cells (SYM6) were longer (Figure 6A and B). Cell-cycle parameters were determined by flow cytometry, as in previous studies (11,17,18,32,38). The initiation of replication and cell division of growing cells were inhibited by rifampicin and cephalexin, and the cells were further incubated to enable run-out replication of the chromosomes. The resultant copy number of chromosomes per cell (determined by flow cytometry) corresponds to the number of copies of oriC at the time of drug addition (50). The ori/mass ratio was deduced to indicate the relative level of replication initiation activity (11,38).
Figure 6.
Analysis of chromosomal oriC mutants. (A and B) Flow-cytometry analysis of a series of chromosomal oriC mutants bearing DnaA-box nonsense sequences. Relevant oriC genotypes of strains are shown as brackets. Cells were exponentially grown at 30°C in LB medium (A) or M9 Glc CAA medium (B), followed by further incubation for run-out replication. DNA contents were quantified with a flow cytometer, and are indicated with the equivalent chromosome numbers on the x axes. Cell sizes (mass) at the time of drug addition were quantified using a Coulter counter, and the mass and ori/mass ratio relative to those of NY20 cells and the doubling time (Td) of cells are indicated at the top right of each panel. (C) Spot test of diaA-null cells with oriC mutations. Serial dilutions of the full-growth cultures (∼109 cells/ml) at 30°C were spotted onto LB-agar plates and incubated for 9 h at 42°C or for 13 h at 30°C. (D) Flow-cytometry analysis of diaA-null cells with oriC mutations. Experiments were performed as described for panel A.
When grown at 30°C in LB medium, wild-type cells had a major peak of eight chromosomes and a minor peak of 16 chromosomes (Figure 6A). By contrast, in SYM25 cells (R1non), the predominant peak was for four chromosomes, with a minor peak of eight chromosomes, and several peaks corresponding to abnormal numbers (three, five, six, and seven chromosomes), indicating that replication initiation was inhibited. Notably, replication initiation in SYM6 cells (R5Mnon) was further inhibited, with a predominant peak of two chromosomes, and other peaks corresponding to three, four, and five chromosomes. SYM9 cells (I1non) had moderate inhibition of initiation; although the predominant peak corresponded to eight chromosomes, the peak for four chromosomes rather than that for 16 chromosomes was evident. SYM5 cells (τ1non) and SYM8 cells (τ2r-non) had peak patterns similar to those of wild-type cells, consistent with the in vitro results (Figure 1D–G).
The mutant strains had a similar relationship to the wild type when grown in M9 medium containing 0.2% glucose and 0.2% casamino acids (M9 Glc CAA) medium (Figure 6B). However, whereas, in LB, SYM7 cells (τ2l-non) had a peak pattern similar to that of wild-type cells, in M9 Glc CAA, the pattern suggested a slight inhibition in initiation, similar to that of SYM9 cells (I1non). The results with R1non, and R5Mnon mutations are basically consistent with those of previous reports (25,49), and suggest that R5M-bound DnaA has a greater role in initiation than R1-bound DnaA, which is consistent with the in vitro results (Figures 1D and E and 2).
The τ1non, τ2l-non, τ2r-non and R5M-non mutations changed the GATC sequence, the Dam methylation site, but initiation regulation was not changed by τ1non and τ2r-non, indicating that individual methylation at the τ1 and τ2 sites is not important for initiation regulation. Also, a recent report shows by detailed mutant analysis that the methylation of GATC sequence in R5M is not required for initiation regulation (51), consistent with the idea that initiation inhibition by R5Mnon mutation is independent of methylation of this site.
Importance of R1 and R5M in the absence of DiaA
The DiaA protein binds to DnaA and stimulates DnaA assembly on oriC, thereby stimulating DUE unwinding and replication initiation (17,37). DnaA-box mutations were combined with wild-type and mutant diaA. Whereas wild-type diaA cells with either wild-type or substituted oriC DnaA boxes grew at both 30°C and 42°C (Supplementary Figure S6A), growth of diaA-disrupted SYM26 (R1non) and SYM19 (R5Mnon), but not cells with other DnaA-box mutations, was inhibited slightly at 30°C and severely at 42°C (Figure 6C and Supplementary Figure S6A).
Flow cytometry on cells grown at 30°C in LB medium demonstrated that diaA-disrupted, wild-type oriC cells (SYM17) were inhibited for replication initiation (with a main peak of four chromosomes), and also had asynchronous initiations with abnormal numbers of chromosomes (Figure 6D, cf. Figure 6A), as we previously reported (17,37). Notably, the peak of eight chromosomes was reduced in diaA-disrupted, R1non cells (SYM26) relative to SYM17. Replication initiation was further affected in diaA-disrupted, R5Mnon cells (SYM19), in which the eight-chromosome peak was absent and the peaks from one to three chromosomes were enhanced, relative to SYM17. Cells with I1non (SYM22) had moderate inhibition, indicated by a decrease in the eight-chromosome peak compared with that of SYM17 cells. Cells with τ2l-non (SYM20) had a similar pattern of peaks to SYM22, consistent with the result of the DUE-unwinding assay (Figure 1F and G). As the patterns of peaks of wild-type diaA cells with τ2l-non and I1non mutations differed (Figure 6A), DiaA might support DnaA binding to the τ2 site rather than the I1 site. diaA-disrupted cells with τ1non (SYM18) or τ2r-non (SYM21) had peak patterns similar to those of SYM17 cells. These results suggest that inhibition of replication initiation in the presence of DnaA box mutations is largely independent of DiaA, thus supporting the importance of R1 and R5M even in the absence of DiaA.
ssDUE binding of R1- and R5M-DnaA is important in chromosome replication
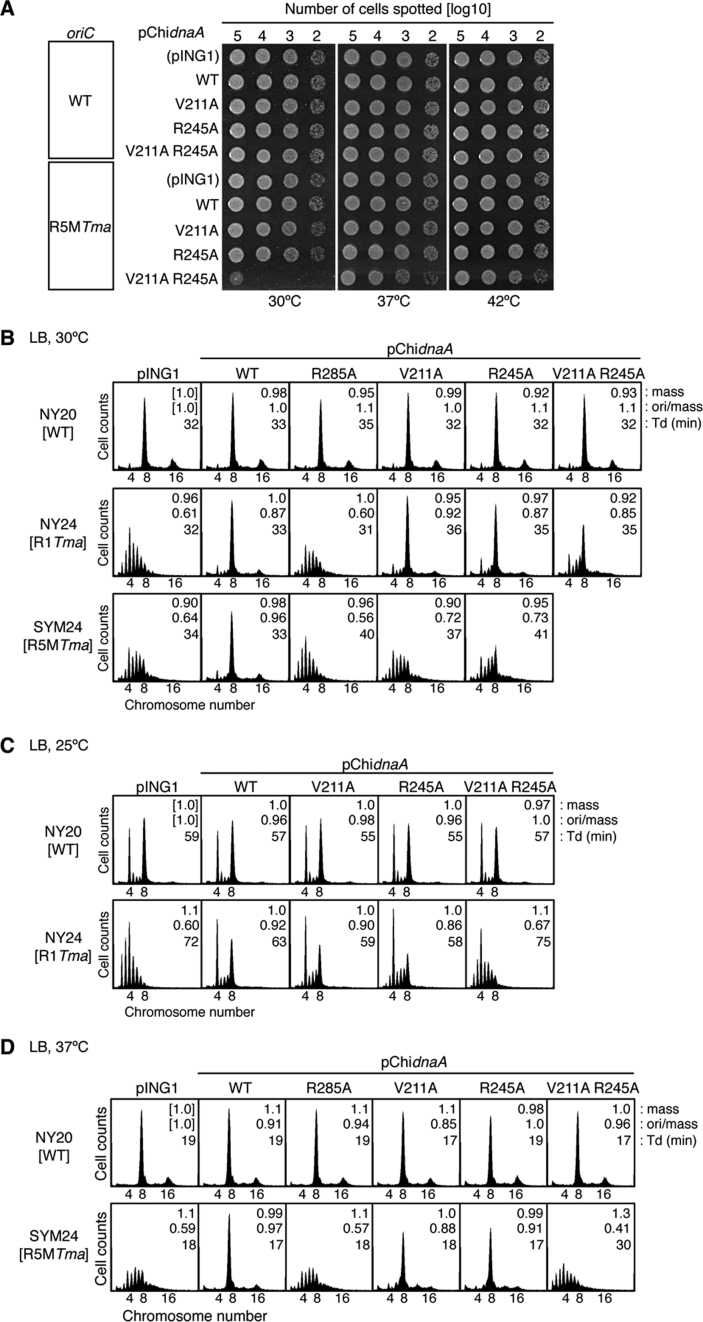
To investigate the roles of Val211 and Arg245 of DnaA bound to R1 and R5M in the initiation of chromosome replication, we analyzed cells with chromosomal oriC substitution mutations R1Tma (NY24) and R5MTma (SYM24) containing pING1-derived plasmids (pChidnaA) encoding ChiDnaA (pChiWT), ChiV211A, ChiR245A, or ChiV211A R245A. pING1 is a pBR322 derivative bearing the araB promoter and araC repressor gene (52). The substitution R1Tma, but not R5MTma, preserved the GATC sequences. ‘Leaky’ expression of the ChiDnaA genes (without arabinose induction) resulted in a similar level, which was moderate, compared to that from the endogenous chromosomal dnaA gene (Supplementary Figure S6B and C). Wild-type dnaA cells bearing pChiWT sustain normal growth rates and replication cycle regulation, as indicated in our previous study (32) and in the present study (Figure 7). Similarly, wild-type dnaA cells bearing pChidnaA V211A R245A sustained normal growth rates and replication cycle regulation (Figure 7). These suggest that expression of those chimeric DnaAs in cells do not substantially inhibit cell growth and chromosomal replication. At 30°C, NY24 (R1Tma) and SYM24 (R5MTma) cells bearing pING1 grew at a similar rate (32 and 34 min, respectively) to wild-type cells (NY20) with pING1 (32 min).
Figure 7.
Roles of DnaA bound to R1 and R5M in vivo. (A) Spot test of cells with the chromosomal R5MTma mutation (or with wild-type oriC) and pECTM plasmid (pING1) containing the wild-type (WT) chimeric dnaA gene (pChidnaA) or its derivatives (pChidnaA V211A, R245A and V211A R245A). Serial dilutions of the full-growth cultures (∼109 cells/ml) at 42°C were spotted onto LB-agar plates and incubated for 9 h at 42°C or for 13 h at 30°C. (B–D) Flow-cytometry analysis. Cells with the wild-type (WT) chromosomal oriC or with R1Tma or R5MTma mutations along with pChidnaA (WT) or its parent vector (pING1) or derivatives were grown at 30°C (B), 25°C (C) or 37°C (D) in LB medium, followed by further incubation for run-out replication. DNA contents were quantified with a flow cytometer, and are indicated with the equivalent chromosome numbers on the x axes. Cell sizes (mass) at the time of drug addition were quantified with a Coulter counter, and the mass and ori/mass ratio relative to those of NY20 cells with pING1 and the doubling time (Td) of cells are indicated at the top right of each panel.
Formation of colonies at 30, 37 and 42°C was similar in NY24 (R1Tma) and NY20 (wild-type) cells with pChiWT, pChiV211A, pChiR245A or pChiV211A R245A plasmids (Supplementary Figure S6D). Similar results were obtained in SYM24 (R5MTma) cells, except for those containing pChiV211A R245A (Figure 7A), which had severely reduced colony formation at 30°C, supporting the idea that ssDUE-binding by DnaA bound to R5M is important for replication initiation in vivo.
Flow cytometry showed similar numbers of chromosomes in NY20 (wild-type) cells with different plasmids, grown at 30°C, with the eight-chromosome peak predominant (Figure 7B). NY24 (R1Tma) cells with pING1 had a similar peak pattern to that of SYM25 (R1non) cells (Figure 6A). NY24 cells with pChiWT, but not pChiR285A, were similar to NY20 with pING1, as previously described (32), indicating that ChiDnaA bound to the R1Tma site for replication initiation. Although NY24 cells with pChiV211A or pChiR245A had peak patterns similar to those of NY24–pChiWT, those with pChiV211A R245A had a smaller eight-chromosome peak and an increase in abnormal peaks of five to seven chromosomes. When these sets of cells were grown at 25°C, the initiation inhibition in NY24–pChiV211A R245A cells was more evident (Figure 7C, cf. Figure 7B). These results demonstrate the importance of ssDUE binding by DnaA bound to R1 in vivo.
In SYM24 (R5MTma)–pING1 cells at 30°C, the four-chromosome peak was predominant and asynchronous initiations occurred (Figure 7B), meaning inhibition of initiation, largely similar to in SYM6 (R5Mnon) cells (Figure 6A). The peak patterns of SYM24–pChiWT cells were similar to those of NY20–pChiWT cells, demonstrating the functioning of cellular ChiDnaA at the oriC R5MTma site. Also, this data mean that the GATC sequence change at this site is not important in initiation regulation. Notably, replication initiation was severely inhibited in SYM24 cells with pChiR285A, pChiV211A or pChiR245A, corroborating our in vitro results (Figures 2–5). Growth of these cells at 37°C (Figure 7D) showed severe inhibition of replication initiation in SYM24–pChiR285A and SYM24–pChiV211A R245A cells, but only slight inhibition in SYM24–pChiV211A and SYM24–pChiR245A. At higher temperatures, DUE unwinding might be stimulated by elevated thermal motion, which could explain why growth of SYM24–pChiV211A R245A cells was severely inhibited at 30°C, but only slightly at 37 and 42°C (Figure 7A). This proposed mechanism could also explain the different requirements for the V211 and R245 residues in DnaA bound to R1 for growth at 25°C and 30°C (Figure 7B and C).
In addition, inhibition of initiation in SYM6 (R5Mnon) cell is severer than that in SYM24 (R5MTma) (Figures 6 and 7). Whereas R5Mnon sequence completely excludes bases identical to the 9-mer DnaA box consensus, 12-mer TmaDnaA box sequence has three bases which are identical to the 9-mer DnaA box consensus (Figure 3B) (47). Although this partial coincidence does not result in substantial affinity in vitro (32), it might cause a minimum affinity for EcoDnaA in vivo, only partially stimulating initiation. Such difference in initiation was not seen for R1 (SYM26 and NY24). Thus, this partial activity would be significant only in the low-affinity site such as R5M.
DISCUSSION
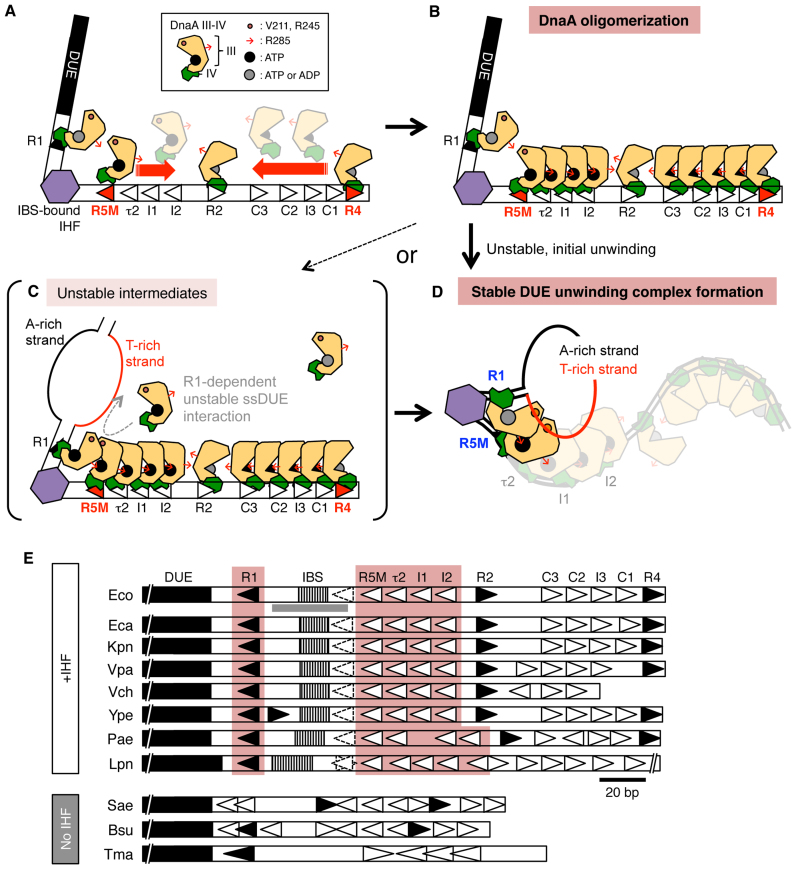
To uncover important details of the molecular dynamics of the replication initiation complex in E. coli, we investigated the roles of individual DnaA protomers in vitro and in vivo. Our results demonstrated that DnaA bound to the R5M box, rather than the R1 box, has a predominant role in ATP–DnaA assembly on the left-half DOR (Figure 2), and that DnaA bound to R1 or R5M binds ssDUE in vitro, which is important for DUE unwinding (Figures 3–5). The data from in vivo and in vitro experiments were consistent, and indicated the importance of the ssDUE-binding activity of the DnaA bound to R5M in replication initiation (Figures 6 and 7). Use of the R5M box, rather than R1, as the key for DnaA assembly on the left-half DOR could enable timely replication initiation. Contrary to the R1 box, which is occupied with DnaA throughout the cell cycle (1), binding of ATP–DnaA to the R5M box should occur when the cellular ATP–DnaA level increases, just before initiation, enabling recruitment of ATP–DnaA (but not ADP–DnaA) to the τ2–I1 region. Also, our results support the ssDUE-recruitment model of initiation at oriC.
The results of EMSA and DNase I-footprint experiments suggested that R5M-bound ATP–DnaA acts as the assembly core of DnaA in the left-half DOR (Figure 8A and B), and this function of the R5M-DnaA does not depend on the R1-bound DnaA, even in the presence of IHF (Figure 2C, D and F). This result is consistent with the known requirement for a short intervening space (2–5 bp) between two DnaA boxes for stimulation of cooperative DnaA binding (Figure 8A) (26). In the presence of IHF, R1-bound DnaA slightly stimulated DnaA binding to R5M, which in turn stimulated DnaA binding to the I1–I2 region. IHF-dependent sharp DNA bending might promote direct interaction between the R1-bound and R5M-bound DnaA molecules, stimulating overall DnaA assembly (Figure 8A). The roles for ATP binding and the Arg finger of the R5M-bound DnaA for DUE unwinding are consistent with this mechanism (Figure 3D); the ATP of the R5M-bound DnaA would interact with the R1-bound DnaA Arg finger (32). These mechanisms could form part of a switch for timely replication initiation coordinated with the increase of the ATP–DnaA level during the cell cycle.
Figure 8.
Model of DUE-unwinding-complex formation. (A–D) Model for oriC-complex dynamics. The R5M-bound and R4-bound DnaAs have a central role in ATP–DnaA assembly on each half of the DOR (A and B). Both ATP–DnaA and ADP–DnaA can bind to the R1 and R4 boxes for initiation of DNA replication (32). The Arg fingers of each DnaA molecule (indicated by small red arrows) are expected to be oriented inward within the DOR, stimulating ATP–DnaA complex formation in opposite directions, except for the R2-bound DnaA (33). In the DUE-unwinding process, binding of IHF causes DNA bending, enabling interaction between DnaA molecules bound to R1 and R5M boxes. The DUE is unwound, and the resultant T-rich strand of ssDUE (red line) is recruited into the DnaA protomers and binds to DnaA bound to R1 and R5M, resulting in stable DUE unwinding that can initiate replication (D). If the R1-bound DnaA is in the ATP form, unstable intermediates might be formed by binding of free ATP–DnaA to ssDUE via interaction with the R1-bound ATP–DnaA (C), As domains I-II are basically dispensable for DUE unwinding, those domains are omitted. (E) oriC sequences of representative bacteria. The DUE, IBS, and IHF binding region shown in Figure 2F and G are indicated by black, striped, and gray bars, respectively. DnaA boxes with complete match to the consensus sequence and with moderate similarity are indicated by closed and open triangles, respectively. DnaA box τ1 and its corresponding sites are indicated by dotted lines. DnaA boxes with left-ward orientation in possible left-half DORs are highlighted. Abbreviations: Eco, Escherichia coli K-12; Eca, Erwinia carotovora atroseptica SCRI1043; Kpn, Klebsiella pneumonia 342; Vpa, Vibrio parahaemolyticus RIMD 2210633 chromosome I; Vch, Vibrio cholerae O1 biovar eltor str. N16961 chromosome I; Ype, Yersinia pestis CO92; Pae, Pseudomonas aeruginosa PAO1; Lpn, Legionella pneumophila str. Paris; Sae, Staphylococcus aureus RF122; Bsu, Bacillus subtilis QB928; Tma, Thermotoga maritima. See Supplementary Table S1 for sequences.
Even in the absence of R1-bound DnaA, ATP–DnaA molecules construct complexes on the left-half DOR bearing only low affinity DnaA boxes (Figure 2 and Supplementary Figure S4A-C). We infer that even weak binding of multiple DnaA molecules to these sites can effectively concordantly enhance cooperative binding, resulting in enhanced complex formation. Similar cooperative binding effects using multiple weak interactions are known as the linkage effect (53). Briefly, even if a single DnaA molecule can not stably bind to a single low-affinity site, binding of multiple DnaA molecules to DNA bearing multiple low-affinity sites can be enhanced in a cooperative manner by DnaA domain III-domain III interactions, resulting in complex formation with much higher affinity for DNA as a complex.
Also, for in vivo residual activities in initiation of cells bearing mutant R1 box or R5M box, we suggest a possibility that DnaA bound to R2 box stimulates construction of a partial DnaA subcomplex on the left-half DOR, probably through DnaA domain I-domain I interaction and that the resultant partial DnaA complexes sustain a partial activity in DUE unwinding and DnaB helicase loading. Our previous data of deletion analysis of DOR suggest the stimulatory role for R2-bound DnaA in DnaA subcomplex formation on the left-half DOR (34); consistent results are also reported (25).
Our results in vitro showed that the ssDUE-binding activities of DnaA bound to R1 and to R5M are important for DUE unwinding, and for binding of DOR–DnaA complexes to the T-rich strand of the DUE (Figures 3–5), which was supported by in vivo analyses (Figure 7). Consistent with the in vitro data (Figures 4 and 5), even in vivo, severe inhibition of initiation was seen in the R5MTma cells with ChiDnaA V211A R245A, but not with ChiWT (Figure 7). Moreover, wild-type oriC cells sustained basically normal initiation regulation in the presence of ChiDnaA V211A R245A (Figure 7). The results support the ssDUE-recruitment model rather than the continuous-filamentation model or the alternative model (Figure 1C and 8D; Supplementary Figure S7). The predominant roles for R1-bound and R5M-bound DnaA in ssDUE binding are consistent with our previous finding that two T-rich regions within ssDUE are required for ssDUE binding to DOR–DnaA complexes (28), and also with the co-crystal structure of ssDNA and an oligomeric domain III of the DnaA ortholog from a hyperthermophilic bacterium (27). The role of R1-bound DnaA in ssDUE binding is also consistent with the strict requirement for the spacing between the DUE and the R1 box (34,54); even a 1 bp insertion or a 2 bp deletion in this space diminishes the DUE-unwinding activity. In addition, DnaA bound to τ2 and I1 was probably supportive for DUE unwinding (Figures 1 and 2 and Supplementary Figure S1 and S2), and might bind ssDUE in concert with R1-bound and R5M-bound DnaA. These supportive roles might underlie the residual levels of initiation in cells with DnaA V211A and R245A variants bound to the R1 or R5M boxes. These structural and functional features are all well consistent with the near-atomic model of the fundamental structure of the initiation complex (33): particularly, in the model the H/B-motifs of R1- and R5M-bound DnaA are exposed on the surface and close to the left edge of DOR (Figure 8D; Supplementary Figure S7).
Previously published evidence that supports the ssDUE-recruitment model includes the activity of ADP–DnaA bound to R1 in initiation in vitro (32). ADP–DnaA at the R1 box is unlikely to enable the formation of DnaA multimers toward the DUE region, as required in the continuous-filamentation model (Figure 1C). Further evidence comes from the results of footprint experiments, which demonstrate that ATP–DnaA does not form multimers covering the region from the R1 box to the DUE (Figure 2E and F) (17,23,24,28). In addition, high concentrations of DnaA are required for ssDNA binding in the absence of a DnaA-bound DOR (28). Finally, IHF binding to the DOR stimulates DUE unwinding (34). All this evidence is consistent with the ssDUE-recruitment model. However, we do not completely exclude other possibilities; for example, given that DOR-free ATP–DnaA molecules form unstable complexes with ssDUE-dsR1, free ATP–DnaA molecules might bind ssDUE to form unstable intermediate complexes via interaction with the R1-bound DnaA when the R1-bound DnaA is the ATP form (Figure 8C). Also, if ssDUE recruitment mechanism does not function by some physiological or genetic alterations, DnaA could bind ssDUE by an alternative mechanism to rescue initiation of replication.
The continuous filamentation model suggests that DnaA domain III, which binds to ssDUE, is located near domain IV via rotation using a flexible loop between the two domains, inhibiting DnaA box binding by domain IV (Figure 1C) (21,27,35). A previous study (35) showed that addition of 13-mer dsDNA bearing R1 box inhibited oligomerization of DnaA on ssDUE (which was detected by glutaraldehyde cross-linking), yielding the R1 box-bound DnaA monomers which do not stably bind ssDNA. Although these data are interpreted to support the continuous filamentation model (27), there is a reasonable alternative interpretation that only when DnaA molecules construct an oligomer bound to DOR (but not monomers bound to separated DNA fragments bearing a single DnaA box), stable ssDUE binding is sustained. This is consistent with the ssDUE recruitment model. We previously demonstrated that ssDUE binding of DnaA is not stimulated by addition of DNA bearing only R1 site whereas it is effectively stimulated by DNA bearing multiple DnaA boxes (34). In addition, we demonstrated also that two T-rich regions in the ssDUE are essential in binding to DnaA oligomers constructed on DOR (28). In the present study, we showed that two DnaA molecules bound the R1 and R5M boxes are crucial in ssDUE binding of the left-half DOR-DnaA complexes. All these data are consistent with the ssDUE recruitment model.
In addition, in the previous paper (35), E. coli DnaA S421A, S421D, S421K and S421Y, which bear a mutation in domain IV, are reported to inhibit DUE unwinding. Although affinity of these proteins for a single DnaA box R1 was indicated to be sustained, these analyses lacked controls such as non-specific sequence. More importantly, specific affinities of these mutant DnaAs for low-affinity DnaA boxes were not analyzed. In the crystal structure, S421 residue directly interacts with a backbone phosphate within DnaA box DNA (29), and thus there is a possibility that the direct S421-DNA interaction might be more effective for binding to low-affinity sites than for binding to high-affinity sites. Based on this possibility, we suggest an interpretation that DnaA binding specificity to DnaA box or DnaA oligomerization on DOR including the low-affinity sites is reduced in these S421 mutants, resulting in inhibition in DUE unwinding. Aquifex aeolicus (aa) counterparts of E. coli DnaA S421A and S421Y, aaDnaA S350A and S350Y sustained the affinity for ssDNA at a level similar to the wild-type control (35), consistent with this idea. Partial decrease in affinity for ssDNA of aaDnaA S350D and S350K (35) could be an indirect consequence of the mutants-specific altered interaction between domains III and IV, as Asp (D) and Lys (K) can produce electrostatic interactions. In the crystal structure of aaDnaA domains III-IV complexed with ssDNA, aaDnaA S350 is located near domain III of the flanking DnaA molecule bound to ssDNA and is apart from ssDNA (27), which suggests a possibility that these mutants affect DnaA-DnaA interaction.
The replication origin oriV of the RK2 plasmid has iterons to which its cognate initiator protein TrfA binds, and a DUE with four AT-rich 13-mer repeats that resemble the DUE 13-mers of E. coli oriC (55). Notably, TrfA complexes assembled on the oriV iterons bind to the bottom strand of the 13-mer repeats in a sequence-dependent manner, resulting in a tripartite nucleoprotein complex (55,56), which is fundamentally similar to the T-rich ssDUE–DnaA–DOR tripartite complex. In the oriC region of E. coli-proximal species, sequences corresponding to the DUE, IBS, the consensus DnaA boxes R1 and R4, and the semi-consensus DnaA boxes including even τ1 are highly conserved (Figure 8E and Supplementary Table S1). The DUE-unwinding mechanism involving ssDUE recruitment could be broadly conserved in many bacteria. In addition, in E. coli, because IHF binds to the oriC IBS only temporally before initiation (9), τ1-bound DnaA could assist in stimulating DnaA assembly while IHF is not yet bound to the IBS. Also, in cells defective in IHF function, τ1-bound DnaA could participate in rescuing initiation under specific conditions. Those possibilities are based on the role for IHF in stimulation of cooperative DnaA binding in vitro (30) and should be explored in future. Inclusion of IHF in EMSA for ssDUE recruitment reduced binding specificities of ChiDnaA, inhibiting this analysis (data not shown). In the evolutionarily distant bacteria Staphylococcus aureus, Bacillus subtilis, and T. maritima which lack genes encoding IHF (Figure 8E and Supplementary Table S1), HU protein (a structural and functional homolog of IHF, which preferentially binds to AT-rich sequences) is conserved (57). Although T. maritima oriC is unwound depending of TmaDnaA and E. coli HU in vitro (47), Initiation mechanisms in these species remain to be further analyzed. Different mechanisms might be used in different bacterial species, and even in E. coli in specific genetic and environmental conditions.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the Research Support Center, Graduate School of Medical Sciences, Kyushu University for DNA sequencing.
Footnotes
Present address: Yasunori Noguchi, DNA Replication Group, Institute of Clinical Sciences, Imperial College London, London W12 0NN, UK.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
MEXT/JSPS KAKENHI [26291004, 16H00775, 17H03656, 15K18479, 16J02075]; JSPS pre-doctoral fellowship (to Y.S.). Funding for open access charge: JSPS/MEXT.
Conflict of interest statement. None declared.
REFERENCES
- 1. Leonard A.C., Grimwade J.E.. The orisome: structure and function. Front. Microbiol. 2015; 6:545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Katayama T., Ozaki S., Keyamura K., Fujimitsu K.. Regulation of the replication cycle: conserved and diverse regulatory systems for DnaA and oriC. Nat. Rev. Microbiol. 2010; 8:163–170. [DOI] [PubMed] [Google Scholar]
- 3. Wolański M., Donczew R., Zawilak-Pawlik A., Zakrzewska-Czerwińska J.. oriC-encoded instructions for the initiation of bacterial chromosome replication. Front. Microbiol. 2015; 5:735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kaguni J.M. Replication initiation at the Escherichia coli chromosomal origin. Curr. Opin. Chem. Biol. 2011; 15:606–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Costa A., Hood I.V., Berger J.M.. Mechanisms for initiating cellular DNA replication. Annu. Rev. Biochem. 2013; 82:25–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. O’Donnell M., Langston L., Stillman B.. Principles and concepts of DNA replication in bacteria, archaea, and eukarya. Cold Spring Harb. Perspect. Biol. 2013; 5:a010108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chodavarapu S., Kaguni J.M.. Replication initiation in bacteria. Enzymes. 2016; 39:1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kurokawa K., Nishida S., Emoto A., Sekimizu K., Katayama T.. Replication cycle-coordinated change of the adenine nucleotide-bound forms of DnaA protein in Escherichia coli. EMBO J. 1999; 18:6642–6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kasho K., Katayama T.. DnaA binding locus datA promotes DnaA-ATP hydrolysis to enable cell cycle-coordinated replication initiation. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:936–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fujimitsu K., Senriuchi T., Katayama T.. Specific genomic sequences of E. coli promote replicational initiation by directly reactivating ADP–DnaA. Genes Dev. 2009; 23:1221–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kasho K., Fujimitsu K., Matoba T., Oshima T., Katayama T.. Timely binding of IHF and Fis to DARS2 regulates ATP–DnaA production and replication initiation. Nucleic Acids Res. 2014; 42:13134–13149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Campbell J.L., Kleckner N.. E. coli oriC and the dnaA gene promoter are sequestered from dam methyltransferase following the passage of the chromosomal replication fork. Cell. 1990; 62:967–979. [DOI] [PubMed] [Google Scholar]
- 13. Theisen P.W., Grimwade J.E., Leonard A.C., Bogan J.A., Helmstetter C.E.. Correlation of gene transcription with the time of initiation of chromosome replication in Escherichia coli. Mol. Microbiol. 1993; 10:575–584. [DOI] [PubMed] [Google Scholar]
- 14. Gon S., Camara J.E., Klungsøyr H.K., Crooke E., Skarstad K., Beckwith J.. A novel regulatory mechanism couples deoxyribonucleotide synthesis and DNA replication in Escherichia coli. EMBO J. 2006; 25:1137–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ozaki S., Katayama T.. DnaA structure, function, and dynamics in the initiation at the chromosomal origin. Plasmid. 2009; 62:71–82. [DOI] [PubMed] [Google Scholar]
- 16. Abe Y., Jo T., Matsuda Y., Matsunaga C., Katayama T., Ueda T.. Structure and function of DnaA N-terminal domains: Specific sites and mechanisms in inter-DnaA interaction and in DnaB helicase loading on oriC. J. Biol. Chem. 2007; 282:17816–17827. [DOI] [PubMed] [Google Scholar]
- 17. Keyamura K., Fujikawa N., Ishida T., Ozaki S., Su’etsugu M., Fujimitsu K., Kagawa W., Yokoyama S., Kurumizaka H., Katayama T.. The interaction of DiaA and DnaA regulates the replication cycle in E. coli by directly promoting ATP DnaA-specific initiation complexes. Genes Dev. 2007; 21:2083–2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Keyamura K., Abe Y., Higashi M., Ueda T., Katayama T.. DiaA dynamics are coupled with changes in initial origin complexes leading to helicase loading. J. Biol. Chem. 2009; 284:25038–25050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sutton M.D., Carr K.M., Vicente M., Kaguni J.M.. Escherichia coli DnaA protein. the N-terminal domain and loading of DnaB helicase at the E. coli chromosomal origin. J. Biol. Chem. 1998; 273:34255–34262. [DOI] [PubMed] [Google Scholar]
- 20. Nozaki S., Ogawa T.. Determination of the minimum domain II size of Escherichia coli DnaA protein essential for cell viability. Microbiology. 2008; 154:3379–3384. [DOI] [PubMed] [Google Scholar]
- 21. Erzberger J.P., Mott M.L., Berger J.M.. Structural basis for ATP-dependent DnaA assembly and replication-origin remodeling. Nat. Struct. Mol. Biol. 2006; 13:676–683. [DOI] [PubMed] [Google Scholar]
- 22. Felczak M.M., Kaguni J.M.. The box VII motif of Escherichia coli DnaA protein is required for DnaA oligomerization at the E. coli replication origin. J. Biol. Chem. 2004; 279:51156–51162. [DOI] [PubMed] [Google Scholar]
- 23. Kawakami H., Keyamura K., Katayama T.. Formation of an ATP–DnaA-specific initiation complex requires DnaA arginine 285, a conserved motif in the AAA+ protein family. J. Biol. Chem. 2005; 280:27420–27430. [DOI] [PubMed] [Google Scholar]
- 24. Ozaki S., Noguchi Y., Hayashi Y., Miyazaki E., Katayama T.. Differentiation of the DnaA-oriC subcomplex for DNA unwinding in a replication initiation complex. J. Biol. Chem. 2012; 287:37458–37471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kaur G., Vora M.P., Czerwonka C.A., Rozgaja T.A., Grimwade J.E., Leonard A.C.. Building the bacterial orisome: high-affinity DnaA recognition plays a role in setting the conformation of oriC DNA. Mol. Microbiol. 2014; 91:1148–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rozgaja T.A., Grimwade J.E., Iqbal M., Czerwonka C., Vora M., Leonard A.C.. Two oppositely oriented arrays of low-affinity recognition sites in oriC guide progressive binding of DnaA during Escherichia coli pre-RC assembly. Mol. Microbiol. 2011; 82:475–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Duderstadt K.E., Chuang K., Berger J.M.. DNA stretching by bacterial initiators promotes replication origin opening. Nature. 2011; 478:209–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ozaki S., Kawakami H., Nakamura K., Fujikawa N., Kagawa W., Park S.-Y., Yokoyama S., Kurumizaka H., Katayama T.. A common mechanism for the ATP–DnaA-dependent formation of open complexes at the replication origin. J. Biol. Chem. 2008; 283:8351–8362. [DOI] [PubMed] [Google Scholar]
- 29. Fujikawa N., Kurumizaka H., Nureki O., Terada T., Shirouzu M., Katayama T., Yokoyama S.. Structural basis of replication origin recognition by the DnaA protein. Nucleic Acids Res. 2003; 31:2077–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McGarry K.C., Ryan V.T., Grimwade J.E., Leonard A.C.. Two discriminatory binding sites in the Escherichia coli replication origin are required for DNA strand opening by initiator DnaA-ATP. Proc. Natl. Acad. Sci. U. S. A. 2004; 101:2811–2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schaper S., Messer W.. Interaction of the initiator protein DnaA of Escherichia coli with its DNA target. J. Biol. Chem. 1995; 270:17622–17626. [DOI] [PubMed] [Google Scholar]
- 32. Noguchi Y., Sakiyama Y., Kawakami H., Katayama T.. The Arg fingers of key DnaA protomers are oriented inward of the replication origin oriC and stimulate DnaA subcomplexes in the initiation complex. J. Biol. Chem. 2015; 290:20295–20312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shimizu M., Noguchi Y., Sakiyama Y., Kawakami H., Katayama T., Takada S.. Near-atomic structural model for bacterial DNA replication initiation complex and its functional insights. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:E8021–E8030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ozaki S., Katayama T.. Highly organized DnaA-oriC complexes recruit the single-stranded DNA for replication initiation. Nucleic Acids Res. 2012; 40:1648–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Duderstadt K.E., Mott M.L., Crisona N.J., Chuang K., Yang H., Berger J.M.. Origin remodeling and opening in bacteria rely on distinct assembly states of the DnaA initiator. J. Biol. Chem. 2010; 285:28229–28239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Su’etsugu M., Shimuta T., Ishida T., Kawakami H., Katayama T.. Protein associations in DnaA-ATP hydrolysis mediated by the Hda-replicase clamp complex. J. Biol. Chem. 2005; 280:6528–6536. [DOI] [PubMed] [Google Scholar]
- 37. Ishida T., Akimitsu N., Kashioka T., Hatano M., Kubota T., Ogata Y., Sekimizu K., Katayama T.. DiaA, a novel DnaA-binding protein, ensures the timely initiation of Escherichia coli chromosome replication. J. Biol. Chem. 2004; 279:45546–45555. [DOI] [PubMed] [Google Scholar]
- 38. Kasho K., Tanaka H., Sakai R., Katayama T.. Cooperative DnaA binding to the negatively supercoiled datA locus stimulates DnaA-ATP hydrolysis. J. Biol. Chem. 2017; 292:1251–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Miller D.T., Grimwade J.E., Betteridge T., Rozgaja T., Torgue J.J.-C., Leonard A.C.. Bacterial origin recognition complexes direct assembly of higher-order DnaA oligomeric structures. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:18479–18484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Felczak M.M., Simmons L.A., Kaguni J.M.. An essential tryptophan of Escherichia coli DnaA protein functions in oligomerization at the E. coli replication origin. J. Biol. Chem. 2005; 280:24627–24633. [DOI] [PubMed] [Google Scholar]
- 41. Hwang D.S., Kornberg A.. Opening of the replication origin of Escherichia coli by DnaA protein with protein HU or IHF. J. Biol. Chem. 1992; 267:23083–23086. [PubMed] [Google Scholar]
- 42. Grimwade J.E., Ryan V.T., Leonard A.C.. IHF redistributes bound initiator protein, DnaA, on supercoiled oriC of Escherichia coli. Mol. Microbiol. 2000; 35:835–844. [DOI] [PubMed] [Google Scholar]
- 43. Ryan V.T., Grimwade J.E., Nievera C.J., Leonard A.C.. IHF and HU stimulate assembly of pre-replication complexes at Escherichia coli oriC by two different mechanisms. Mol. Microbiol. 2002; 46:113–124. [DOI] [PubMed] [Google Scholar]
- 44. Rice P.A., Yang S., Mizuuchi K., Nash H.A.. Crystal structure of an IHF-DNA complex: a protein-induced DNA U-turn. Cell. 1996; 87:1295–1306. [DOI] [PubMed] [Google Scholar]
- 45. Azam T.A., Ishihama A.. Twelve species of the nucleoid-associated protein from Escherichia coli. Sequence recognition specificity and DNA binding affinity. J. Biol. Chem. 1999; 274:33105–33113. [DOI] [PubMed] [Google Scholar]
- 46. Ali Azam T., Iwata A., Nishimura A., Ueda S., Ishihama A.. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J. Bacteriol. 1999; 181:6361–6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ozaki S., Fujimitsu K., Kurumizaka H., Katayama T.. The DnaA homolog of the hyperthermophilic eubacterium Thermotoga maritima forms an open complex with a minimal 149-bp origin region in an ATP-dependent manner. Genes Cells. 2006; 11:425–438. [DOI] [PubMed] [Google Scholar]
- 48. Simmons L.A., Felczak M., Kaguni J.M.. DnaA protein of Escherichia coli: Oligomerization at the E. coli chromosomal origin is required for initiation and involves specific N-terminal amino acids. Mol. Microbiol. 2003; 49:849–858. [DOI] [PubMed] [Google Scholar]
- 49. Weigel C., Messer W., Preiss S., Welzeck M., Morigen, Boye E.. The sequence requirements for a functional Escherichia coli replication origin are different for the chromosome and a minichromosome. Mol. Microbiol. 2001; 40:498–507. [DOI] [PubMed] [Google Scholar]
- 50. Skarstad K., Bernander R., Boye E.. Analysis of DNA replication in vivo by flow cytometry. Meth. Enzymol. 1995; 262:604–613. [DOI] [PubMed] [Google Scholar]
- 51. Jha J.K., Chattoraj D.K.. Inactivation of individual SeqA binding sites of the E. coli origin reveals robustness of replication initiation synchrony. PLoS ONE. 2016; 11:e0166722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Katayama T. The mutant DnaAcos protein which overinitiates replication of the Escherichia coli chromosome is inert to negative regulation for initiation. J. Biol. Chem. 1994; 269:22075–22079. [PubMed] [Google Scholar]
- 53. Stauffer M.E., Chazin W.J.. Structural mechanisms of DNA replication, repair, and recombination. J. Biol. Chem. 2004; 279:30915–30918. [DOI] [PubMed] [Google Scholar]
- 54. Hsu J., Bramhill D., Thompson C.M.. Open complex formation by DnaA initiation protein at the Escherichia coli chromosomal origin requires the 13-mers precisely spaced relative to the 9-mers. Mol. Microbiol. 1994; 11:903–911. [DOI] [PubMed] [Google Scholar]
- 55. Wegrzyn K., Fuentes-Perez M.E., Bury K., Rajewska M., Moreno-Herrero F., Konieczny I.. Sequence-specific interactions of rep proteins with ssDNA in the AT-rich region of the plasmid replication origin. Nucleic Acids Res. 2014; 42:7807–7818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wawrzycka A., Gross M., Wasaznik A., Konieczny I.. Plasmid replication initiator interactions with origin 13-mers and polymerase subunits contribute to strand-specific replisome assembly. Proc. Natl. Acad. Sci. U. S. A. 2015; 112:E4188–E4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Swinger K.K., Rice P.A.. IHF and HU: flexible architects of bent DNA. Curr. Opin. Struct. Biol. 2004; 14:28–35. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.