Abstract
We report here a robust, tunable, and reversible transcription control system for endogenous genes. The REMOTE-control system (Reversible Manipulation of Transcription at Endogenous loci) employs enhanced lac repression and tet activation systems. With this approach, we show in mouse embryonic stem cells that endogenous Dnmt1 gene transcription could be up- or downregulated in a tunable, inducible, and reversible manner across nearly two orders of magnitude. Transcriptional repression of Dnmt1 by REMOTE-control was potent enough to cause embryonic lethality in mice, reminiscent of a genetic knockout of Dnmt1 and could substantially suppress intestinal polyp formation when applied to an ApcMin model. Binding by the enhanced lac repressor was sufficiently tight to allow strong attenuation of transcriptional elongation, even at operators located many kilobases downstream of the transcription start site and to produce invariably tight repression of all of the strong viral/mammalian promoters tested. Our approach of targeting tet transcriptional activators to the endogenous Dnmt1 promoter resulted in robust upregulation of this highly expressed housekeeping gene. Our system provides exquisite control of the level, timing, and cell-type specificity of endogenous gene expression, and the potency and versatility of the system will enable high resolution in vivo functional analyses.
INTRODUCTION
Genetic manipulation of mammals has been integral to the interrogation of gene function in a native biological context (1). Loss- or gain-of-function approaches by genetic knockout or transgenic technologies have long served as a direct, powerful means to reveal gene function. However, these dichotomous (on/off) and non-temporal approaches are not well-suited for understanding the full functional spectrum of a gene as the functional manifestation of a gene depends critically on spatio-temporal context and its distinct expression levels therein. Therefore, an ability to control the timing, location, and level of a gene expression would be ideal for precise functional characterization. Conditional knockout technologies have enabled spatio-temporal regulation, but limitations associated with their dichotomous nature, such as a cell lethality, continue to pose challenges and do not allow for studying the phenotypic consequences of variable levels of gene expression. Conditional knockdown approaches using tet-regulated shRNA or miRNA have helped to fill this void, but their successful implementation is not universal (2–4). Off-target effects remain an intrinsic problem for RNAi (5) and are difficult to control in vivo. CRISPR/Cas-mediated technologies have provided a new, versatile means of controlling endogenous gene expression and will greatly improve our study of gene function (6,7), although the efficacy of in vivo transcriptional control using CRISPR/Cas systems is unclear.
In principle, modification of endogenous gene promoters with binding sequences for transcriptional repressors or activators with regulatory ligands would allow for tunable, inducible and reversible gene expression control. Indeed, a few reports have described such exquisite control of endogenous gene expression by prokaryotic binary systems (8–11). However, the insufficient potency of the existing binary systems and the technical challenge associated with the modification of endogenous genes by conventional gene targeting have hampered their broad application. Recently, the latter challenge has been substantially mitigated by the introduction of CRISPR/Cas-mediated genome editing technologies, with which endogenous gene modification has now become a routine task (12–16).
Here, we provide (i) an engineered binary system sufficiently potent for endogenous gene repression, (ii) a first proof of principle for the inducible transcriptional upregulation of an endogenous gene from its cognate promoter in mice and (iii) a standardized and efficient application strategy.
We have engineered an enhanced lacI repression system that is substantially stronger than the wild-type lac system (>100 times) and produces consistently tight repression of all of the widely used, strong viral/mammalian promoters we tested. We also show that tight repression can be achieved even through inhibition of transcription elongation from operators located in an intron far downstream of transcription start sites, providing a simple strategy for repression without affecting the endogenous promoter. Moreover, we were able to upregulate expression 6.5-fold from an already highly expressed gene, Dnmt1, by targeting tet activators to its endogenous promoter. The availability of efficient regulatory ligands for both the lac repressor (IPTG) and the tet activator (doxycycline) makes the REMOTE-control system tunable and reversible (Figure 1A), providing exquisite versatility for high resolution in vivo functional analyses. The system could be expanded to other organisms and binary systems.
Figure 1.
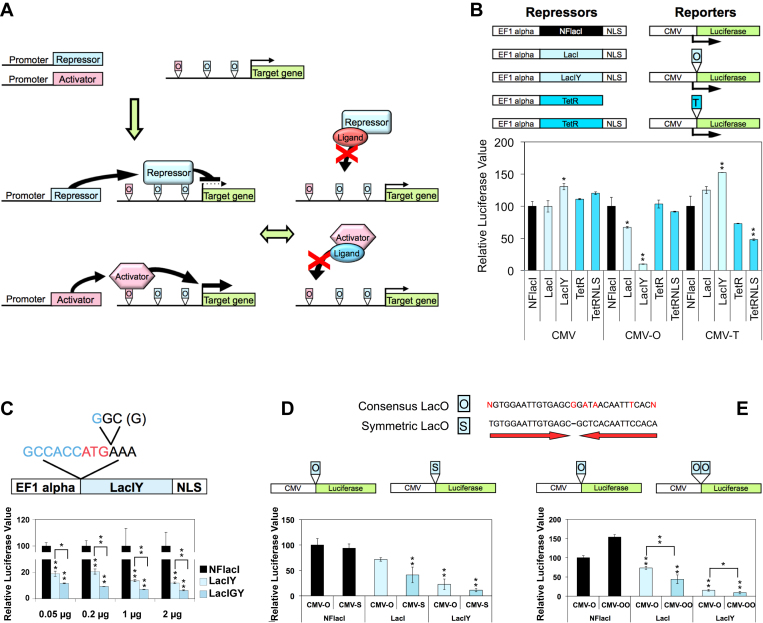
In Vitro Optimization of the REMOTE-control System. (A) Schematic view of the REMOTE-control system. The transcription of a target gene can be up- or downregulated upon the binding of activators or repressors to the corresponding operators. Regulatory ligands of the repressor and transactivator reverse the transcriptional regulations. (B) Repressor and reporter constructs used in the transient reporter assay: non-functional lacI (NFlacI); wild-type lacI (lacI); lacI with a tight-binding mutation (lacIY); wild-type tet repressor (TetR); tet repressor with a nuclear localization signal (tetRNLS); lac operator (O); and tet operator (T). CMV-O and CMV-T refer to the reporter containing a lacO or a tetO, respectively. Reporters (50 ng/well in 96-well plate) and corresponding repressors were co-transfected into NIH/3T3 cells in a 1:1 molar ratio and assayed 24 h after transfection (one-way ANOVA). See also Supplementary Figure S1A–C. (C) Top: lacIY with a stabilizing amino acid insertion; the Kozak sequence is indicated in blue. Bottom: Reporter assays for lac repressors containing the stabilizing amino acid. Experiments were performed in 24-well plates. Reporter plasmid (2 μg/well) was co-transfected with four different amounts of repressor plasmids as indicated. Luciferase activity was measured 48 h after transfection (between two groups: t-tests, between more than two groups: one-way ANOVA). (D) Sequence of the consensus (O) and symmetric lac operators (S). The reporter (50 ng/well in 96-well plate) harboring either the consensus or symmetric operator was co-transfected with repressor as indicated (between two groups: t-tests, between more than two groups: one-way ANOVA). (E) The reporter containing either one or two operators (50 ng/well in 96-well plate) was co-transfected with repressor as indicated (between two groups: t-tests, between more than two groups: one-way ANOVA). Luciferase data are presented as percent relative to the reference value set to 100%. Data are represented as mean ± SEM (n = 3). *P ≤ 0.05, **P ≤ 0.01.
MATERIALS AND METHODS
Luciferase reporter assay
NIH/3T3 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (tetracycline-free, Hyclone) and 1% penicillin/streptomycin. Lipofectamine 2000 (Thermo Fisher Scientific, Waltham, MA, USA) was used for transfection. The Dual Luciferase Reporter Assay Kit (Promega, Madison, WI, USA) was used to determine luciferase expression. The ratio of firefly to renilla luminescence intensity was calculated and used as a measure for promoter activity.
Gene-targeting
The Dnmt1LO and LGT alleles were generated using a conventional gene-targeting procedure. ES cells were maintained in HEPES-buffered (20 mM [pH 7.3]) DMEM supplemented with 15% fetal calf serum (GE Heathcare Life Sciences, Pittsburgh, PA, USA), 0.1 mM nonessential amino acids (Thermo Fisher Scientific, Waltham, MA, USA), 0.1 mM β-mercaptoethanol (Sigma-Aldrich, St. Louis, MO, USA), and penicillin–streptomycin (Thermo Fisher Scientific, Waltham, MA). ES cells were grown on feeder layers of gamma-irradiated embryonic fibroblast cells and supplemented with leukemia inhibitory factor (LIF) (Thermo Fisher Scientific, Waltham, MA) at 106 U/ml. ES cells were electroporated with 20 μg of linearized DNA in PBS at a set voltage 230 V and a capacitance of 500 μF with a Bio-Rad GenePulser II. Antibiotic selection was initiated the following day and continued for 8–11 days before picking. G418 was applied to the culture medium at a concentration of 300 μg/ml. PCR screening was performed using EXL DNA polymerase (Agilent Technologies, Santa Clara, CA, USA) with following primers; 5′-TGCATCCTAGGCCTATTAATATTC-3′ (forward) and 5′-GAAGTGACTGTCACACAAGCACA-3′ (reverse). Cre recombinase was introduced by electroporation as described above. Ganciclovir (510 ng/ml) selection was performed to screen for the TkNeo removed ES cells. PCR screening was performed with following primers; 5′-GGTTCAGAAGGGCCCTCCTT-3′ (forward) and 5′-CCAGGCAGAGTCTCTTTGATAG-3′ (reverse).
RT-PCR assays
Total RNA was prepared using TRIzol (Thermo Fisher Scientific, Waltham, MA). cDNAs were made using Superscript III reverse transcriptase (Thermo Fisher Scientific, Waltham, MA), as per the manufacturer's instructions, and amplified by PCR with Taq polymerase. See Supplementary Materials and Methods for additional details.
Gene expression array
Eight RNA samples were analyzed using the Illumina MouseRef-8 v2.0 Expression BeadChip array (Illumina) at the University of Southern California Epigenome Center. The quality of the RNA samples was assessed using spectrophotometry and capillary electrophoresis. All samples were quantified on an ND-8000 (ThermoScientific). The samples were then run on Experion RNA StdSens Analysis chip (Bio-rad) to ensure that the RNA was of sufficient quality, free from DNA contamination and not overly degraded. All samples that passed the initial QC checks were processed using TotalPrep RNA Amplification Kit (Ambion). First, 500 ng of RNA from each sample was converted using reverse transcription to single-stranded cDNA. Next, the second cDNA strand was synthesized, and the resulting double-stranded cDNA was purified. This cDNA was then converted to biotin-labeled cRNA through an overnight in vitro transcription step. The cRNA was then purified using the provided filter columns and was quantified by spectrophotometry using the ND-8000. The cRNA samples were brought to a concentration of 150 ng/μl. cRNA (750 ng) was mixed with a hybridization buffer and added to a MouseRef-8 v2.0 Expression BeadChip (Illumina). The samples were allowed to incubate overnight on the chips and were then stained and scanned using an Illumina HD Beadarray scanner. Distribution of densities and signal intensities were highly similar between all samples. Data underwent log2 transformation and quantile normalization using Partek Genomics Suite 6.5 software (Partek). Fold changes were calculated from the average signal intensity from the normalized Data. Data from gene expression array experiments have been deposited in GEO (accession number: GSE28386).
Statistics
All quantitative data were collected from experiments performed in at least triplicate, and expressed as mean ± standard Error. All experiments, except the ones in Figure 5C, D, and Supplementary Figure S7D, were analyzed in Prism as follows: differences between two groups were assayed by two-tailed unpaired t-tests and differences between more than two groups were assayed by one-way ANOVA, where Dunnett's t-tests were used to determine which groups (if any) differed significantly from the control. The remaining experiments were assayed in R 3.2.0 via ANOVA, but instead of Dunnett's t-tests, false discovery rate corrected Welch's t-tests were used to determine which groups differed significantly from the control; this method was chosen to appropriately account for the strong heteroskedasticity present in these data. Regardless of software, significant differences were considered when P < 0.05; *P ≤ 0.05 and **P ≤ 0.01.
Figure 5.
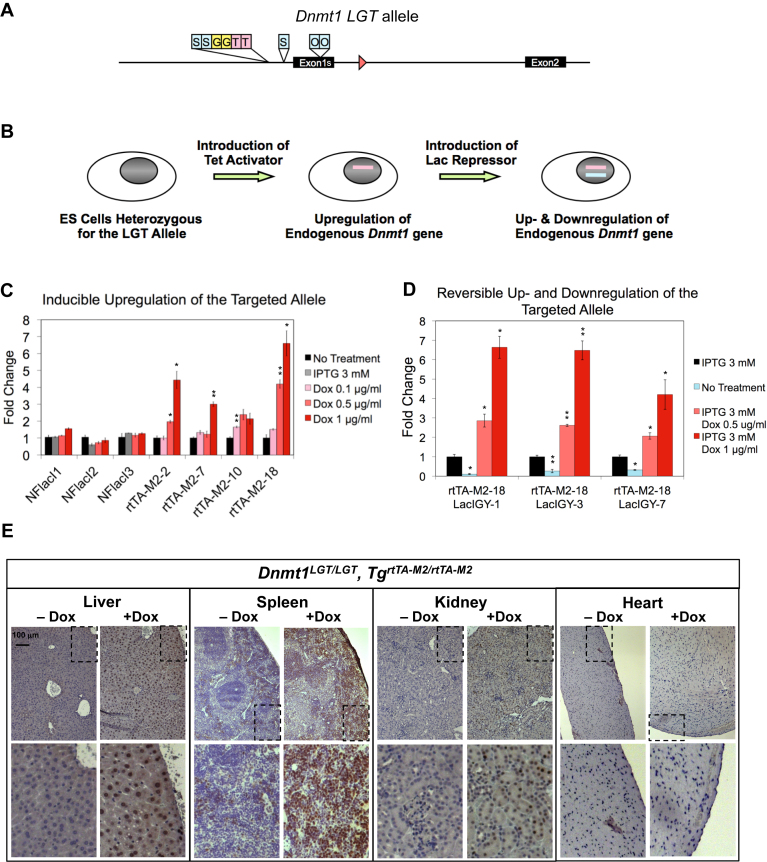
Up- and downregulation of endogenous Dnmt1 expression. (A) Gene-targeting and screening were performed as described in Figure 3. (B) NFlacI or rtTA-M2 was stably introduced into the targeted ES cells through electroporation followed by G418 selection. lacIGY was stably introduced into the rtTA-M2-expressing ES cells via electroporation followed by puromycin selection. Pink and blue lines indicate the integrated rtTA-M2 and lacIGY transgenes, respectively. (C) Dnmt1LGT expression in the presence of IPTG or different doses of doxycycline. ES cell clones were subjected to real-time RT-PCR assays using a reaction specific for the targeted allele. (D) Up- and downregulation of the Dnmt1LGT allele by lacIGY and rtTA-M2. Pcna was used for normalization in all experiments. (E) Immunostaining results of Dnmt1 protein from various tissues with and without doxycycline treatment. Mice were treated with a doxycycline diet for one month. Data are represented as mean ± SEM (n = 3). *P ≤ 0.05, **P ≤ 0.01 (Welch's t-tests, see Methods for additional details).
RESULTS
In vitro optimization of the REMOTE-control system
The first step in developing an effective transcriptional control system capable of both activation and repression is to evaluate which system is best suited for each task. We focused our efforts on the lac and tet systems as two of the best characterized and most widely used reversible binary transcriptional regulatory systems. To test their repressive capabilities, we constructed reporters with a single-copy lac operator (lacO) or tet operator (tetO) placed between the CMV promoter and the luciferase gene. All repressor constructs were engineered to have identical plasmid backbones and promoters to ensure a fair comparison of their repressive capacity per gene copy. Given differences in translation efficiency and protein stability (see below), that measure is more appropriate for transgenic applications than a comparison of repressor proteins on an equimolar basis (Figure 1B).
We performed the comparison with five different repressors: a lac repressor with codon usage optimized for mammalian cells, producing a wild-type repressor protein (17) (referred to as lacI); a lac repressor with a tight-binding mutation (lacIY); a non-functional lac repressor control (NFlacI); a wild-type tet repressor (TetR); and a tet repressor with nuclear localization signal (TetRNLS). We generated lacIY by adopting a tight-binding mutation identified in E. coli that increases the repressor-to-operator binding affinity ∼100-fold (18). We engineered tetRNLS by inserting the nuclear localization signal used for the lac repressors into the tet repressor.
The lacO or tetO insertion alone had little effect on the promoter in the absence of repressors (Supplementary Figure S1A). The reporter and repressor plasmids were transiently introduced into NIH/3T3 cells at a 1:1 plasmid molar ratio. Among the repressors, the tight-binding mutant lacIY showed the strongest repression of the CMV promoter (a 90% reduction of luminescence relative to NFlacI), while other repressors showed modest repression (Figure 1B). To exclude possible biases, including the fact that lac and tet repressors were tested on two different reporters separately, we also used reporters containing both operators in tandem in both sequential configurations, and with repressors introduced at various concentrations (up to an 8-fold molar excess) (Supplementary Figure S1B and C). For this comparison, we used lacIY and tetRNLS as the best-performing repressors for each system. We found that lacIY repressed the CMV promoter substantially better under all experimental conditions (Supplementary Figure S1B and C). Based on the level of repression and the less stringent constraints on the position of its operator, we chose the lac operator/repressor system as the repression module for the REMOTE-control system.
A lysine residue at the N-terminus of a protein is a primary destabilizing amino acid in mammals (19). We therefore inserted a stabilizing amino acid into the lacIY repressor to mask its N-terminal lysine. We chose glycine, since it is one of the four stabilizing amino acids in mice (19) and because its optimal codon in mammals (GGC) adds a critical component of the Kozak consensus eukaryotic translation initiation sequence (guanine at +4) (20) when placed immediately adjacent to the initiation codon (Figure 1C). This glycine insertion in lacIY, giving lacIGY, resulted in improved repression at four different repressor-to-reporter ratios (Figure 1C).
The consensus lac operator is an imperfect palindromic 29-bp sequence with just four mispairs (Figure 1D). A 30-bp perfectly symmetric lac operator sequence interacts with the lac repressor 8–10 times more strongly than the consensus sequence (21). We used this modified lac operator to further improve operator/repressor binding affinity. The reporter having the symmetric operator sequence was repressed more strongly by both lacI and lacIY than the reporter having a consensus operator (Figure 1D). Finally, we were able to further improve repression by about 50% by simply doubling the number of operator sequences (Figure 1E).
Modification of the Dnmt1 promoter
Homozygous knockout of the Dnmt1 gene generally results in cell death, interfering with our ability to use conditional knockouts to study Dnmt1 deficiency in vivo (22). Moreover, Dnmt1 is a highly expressed housekeeping gene, and robust overexpression of the full-length protein has been difficult to achieve in cell culture (23) and has resulted in embryonic lethality in vivo (24). Thus, this gene was a good candidate for testing our REMOTE-control system.
We first defined the minimally effective Dnmt1 somatic promoter in the mouse (25). A 281-bp fragment (pDL5) spanning the transcription start site (–182 to +99) conferred full expression potential in NIH/3T3 cells as previously reported (26) (Data not shown). We next selected candidate operator insertion sites within this 281-bp fragment containing the Dnmt1 promoter that avoided disrupting evolutionarily conserved sequences or putative transcription factor binding sites obtained from a screen for consensus binding sites and from functional studies (Figure 2A) (26,27). We also included a candidate insertion site in the 5′ UTR (+99), as we had observed efficient repression from an operator located downstream of transcription start site of the CMV promoter (Figure 1B, Supplementary Figure S1B and C). We devised a PCR-based cloning method using a shared set of just three restriction enzymes for the promoter deletions and operator insertions, thus streamlining the construction of plasmids with multiple operator insertions at any desired location (Supplementary Figure S2A). This method is in principle applicable to any promoter.
Figure 2.
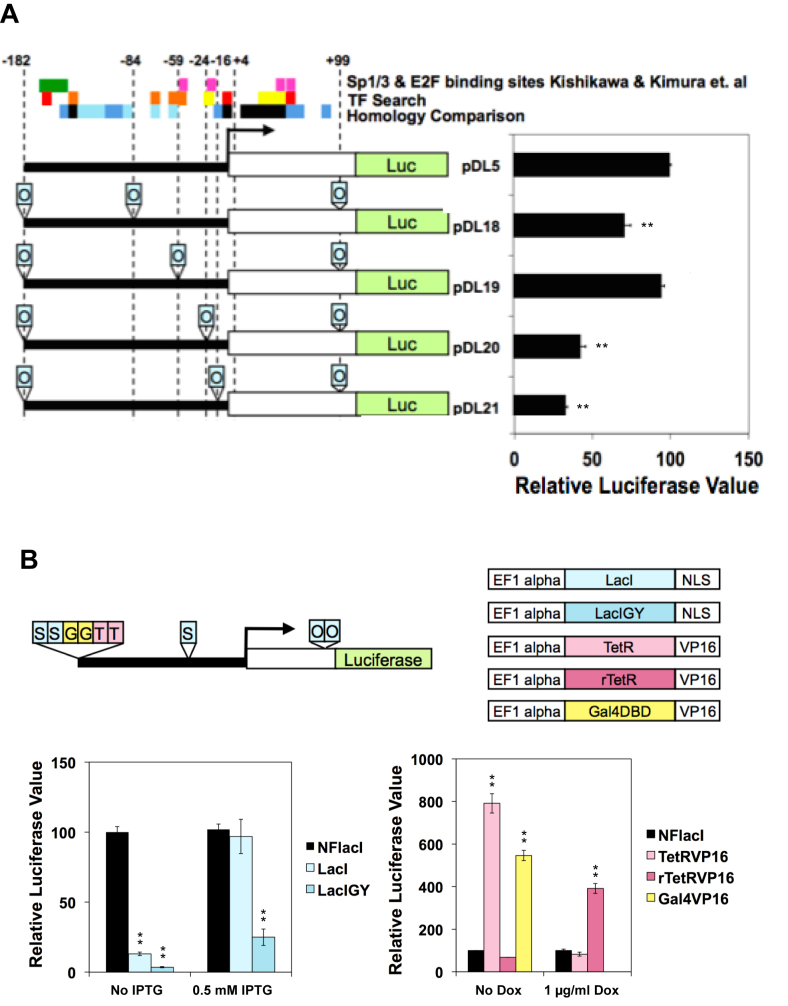
Applying the System to the Dnmt1 Promoter. (A) Reporter assay with the Dnmt1 promoters modified with lac operator (O) sequences in the absence of the lac repressor. The 281-bp sequence immediately upstream of the somatic transcription start site was subjected to a transcription factor binding site search (TFSEARCH: http://www.cbrc.jp/research/db/TFSEARCH.html) and to homology comparison with rat and human DNMT1 promoter sequences. Putative transcription binding sites are color coded from red to light yellow according to their scores (100–96, red; 95–91, orange; 90–85, yellow). Regions with high homology are shown as black. Green and pink rectangles represent Sp1/3 and E2F binding sites, respectively (one-way ANOVA). (B) The reporter containing the modified Dnmt1 promoter and repressors were transfected in the presence or absence of IPTG (left panel). See Supplementary Figure S2C for higher amounts of IPTG treatment. The reporter and transactivators were transfected in the presence or absence of doxycycline (right panel) (one-way ANOVA). Luciferase data are presented as percent relative to the reference value set to 100%. Data are represented as mean ± SEM (n = 3). *P ≤ 0.05, **P ≤ 0.01.
We inserted lacO sequences at the selected sites and assessed their effects on promoter efficacy in the absence of repressor. Insertions at –182, –59 and +99 (pDL19) resulted in a promoter strength comparable to that of the unmodified promoter (Figure 2A). We then tested the symmetric operator and double-operator sequences at –182, –59 and +99. Location –59 could tolerate only a single operator (data not shown), but location +99 could accommodate a double consensus operator without a noticeable effect on promoter efficacy (data not shown). However, the double symmetric operator severely suppressed luciferase activity when located immediately downstream of the transcription start site of any promoter construct in the absence of the lac repressor (data not shown). This is likely attributable to the formation of strong hairpin structures in the 5′ UTR of the mature mRNA, since we did not observe this effect of double symmetric operators when they were in an intron (Figure 6A, Supplementary Figure S7A). In summary, we inserted a double symmetric operator at –182, a single symmetric operator at –59 and a double consensus operator at +99. We further modified the Dnmt1 promoter with tet operators and gal4 DNA binding sequences (DBS) at –182 to achieve upregulation using the tetVP16 (tTA) and gal4VP16 transactivators.
Figure 6.
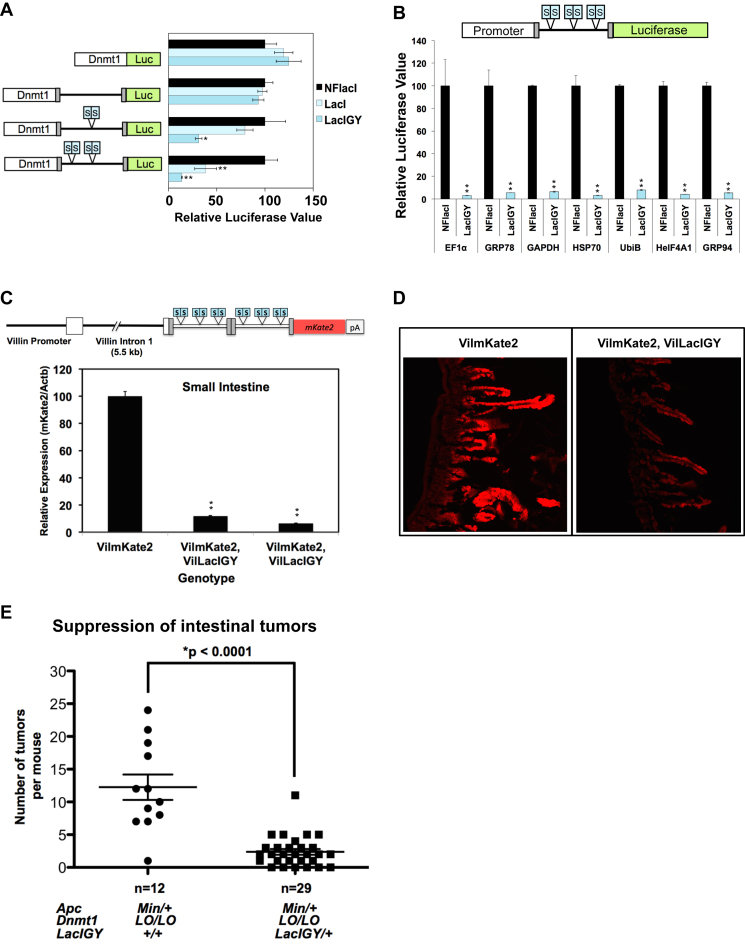
Attenuating transcription elongation by lac repressor and applying the repression system to other promoters. (A) Attenuation of transcription elongation. The rabbit β-globin intron (black line flanked by gray boxes) was modified with symmetric operators under the control of the Dnmt1 promoter (one-way-ANOVA). (B) Attenuation of transcription elongation of various promoters (t-tests). Two symmetric lac operators were inserted at three locations within a rabbit β-globin intron. Luciferase data are presented as percent relative to the reference value set to 100%. (C) The VilmKate2 transgenic mouse line. The rabbit β-globin intron was modified with symmetric operators under the control of the Villin promoter. mKate2 expression in the small intestine from mice with or without lacIGY (one-way ANOVA). (D) Confocal mKate2 images of small intestine with and without lacIGY expression. (E) Tumor multiplicity in the ApcMin mice analyzed for tumor multiplicity at six months of age (Mann–Whitney U test). Data are represented as mean ± SEM (n = 3). *P ≤ 0.05, **P ≤ 0.01
Up- and downregulation of the Dnmt1 promoter by the REMOTE-control system in vitro
We first tested whether the lac repressor could repress the modified Dnmt1 promoter by introducing the lacI or lacIGY plasmids into NIH/3T3 cells together with the reporter plasmid containing the modified Dnmt1 promoter. Both the wild-type and modified lac repressors successfully repressed transcription from the Dnmt1 promoter with operator insertions (Figure 2B). Almost complete repression (3.7% residual expression) was achieved with lacIGY. The repression by lacI was completely reversed by 0.5 mM IPTG (isopropyl β-d-1-thiogalactopyranoside), while that treatment restored only 25% of unregulated expression for lacIGY. We also tested whether repression by lacIGY could be completely reversed by molar concentrations of IPTG from 0.5 mM to 10 mM. Even the best restoration, at 3 mM IPTG, gave only 40% of unregulated expression (Supplementary Figure S2B). However, subsequent experiments in stably transfected single-copy cell lines and in mice showed almost complete derepression of lacIGY by IPTG (Figure 4D and E, Supplementary Figure S6B). This suggests that the incomplete reversion in transient transfection experiments might be due to the high reporter plasmid copy number and large amounts of lacIGY in transiently transfected cells.
Figure 4.
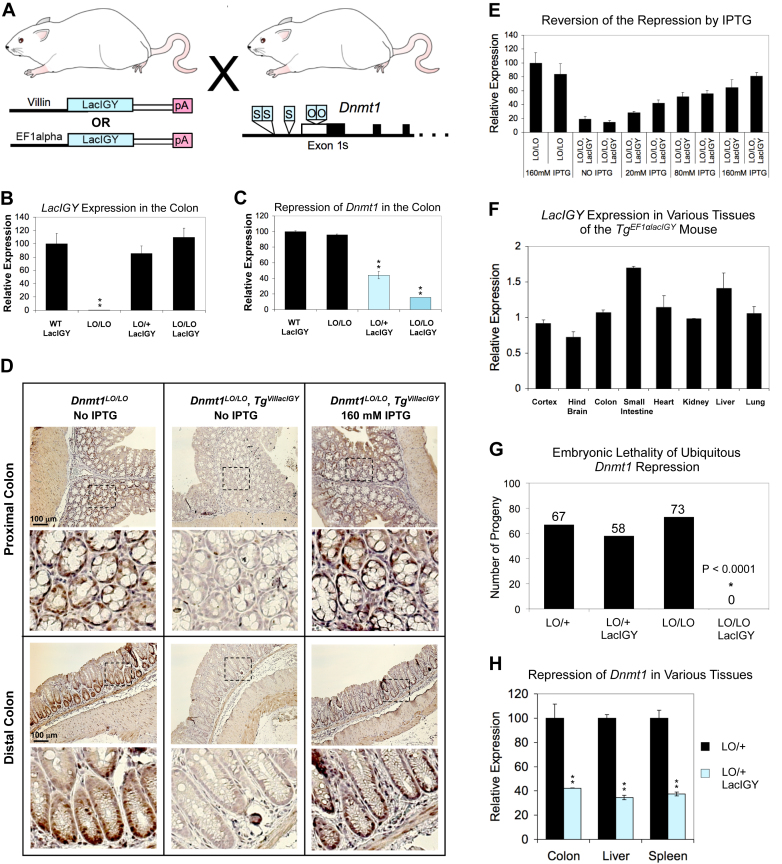
In vivo downregulation of Dnmt1. (A) The lacIGY transgenic (left) and Dnmt1LO (right) mice. The promoters driving lacIGY expression are indicated before the transgene. White thin boxes indicate the rabbit β-globin intron. (B) LacIGY expression from the colon (one-way-ANOVA). (C) Dnmt1 expression in the colon from mice with or without the targeted allele (Dnmt1LO) and lacIGY, using a real-time RT-PCR reaction designed to detect both the wild-type and targeted Dnmt1 alleles (one-way ANOVA). (D) Immunostaining of Dnmt1 protein from the colon with and without IPTG treatment. For three weeks, mice were given IPTG-containing water kept in a light-protected bottle. (E) Real-time RT-PCR analysis showing the in vivo reversion of the repression from IPTG treatment. (F) lacIGY expression normalized by Xist in tissues from the EFlacIGY transgenic line. (G) Genotyping results from the Dnmt1LO/LO × Dnmt1LO/+/TgEFlacIGY cross, demonstrating the embryonic lethal phenotype of Dnmt1LO/LO, TgEFlacIGYmice. Genotyping was performed at birth (Chi-square test). (H) Dnmt1 expression from the targeted allele (Dnmt1LO) in tissues of Dnmt1LO/+/TgEFlacIGY mice (t-tests). Data are represented as mean ± SEM (n = 3). *P ≤ 0.05, **P ≤ 0.01
To achieve upregulation of the Dnmt1 promoter, we introduced the tetVP16 or gal4VP16 transactivator plasmids into NIH/3T3 cells. In transient reporter assays, we observed 5-fold and 8-fold inductions for gal4VP16 and tetVP16, respectively, and the 8-fold induction by tetVP16 was completely abolished by doxycycline treatment, an antagonist of tet (Figure 2B). We also tested a reverse version of tetVP16, rtetVP16 (rtTA), which binds to the operator when doxycycline is bound to achieve inducible upregulation. With 1 μg/ml of doxycycline, rtetVP16 increased the expression by 4-fold, and no induction was observed in the absence of doxycycline (Figure 2B). With these three modifications together, we could achieve repression of the Dnmt1 promoter down to 0.03-fold and upregulation up to 8-fold. We chose the tet system over the gal4 system for our upregulation approach, owing to its reversibility, but in principle, the gal4 system could be used to add additional controls to the Dnmt1 expression regulation by recruitment of other transcription regulators, such as histone modifying enzymes or transcription factors.
Applying the REMOTE-control system to the endogenous Dnmt1 promoter and testing the repression system in ES cells
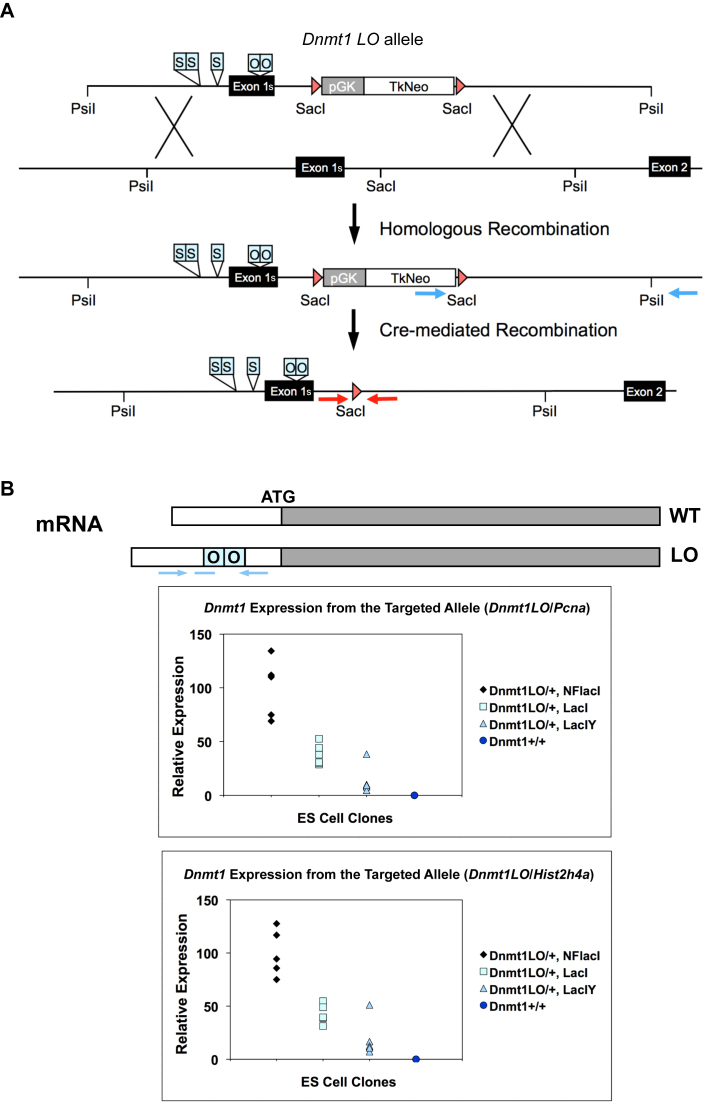
We identified three locations at which to insert lac operators without affecting the activity of the minimally effective Dnmt1 promoter (–182 to +99). We used homologous recombination in mouse embryonic stem cells to introduce lac operators at the endogenous Dnmt1 promoter to generate the modified Dnmt1 allele designated Dnmt1LO (Figure 3A). We stably introduced lac repressor constructs into the targeted ES cells to test repression efficacy in the context of a native chromatin configuration prior to testing the system in mice. We developed a real-time RT-PCR reaction specific for the Dnmt1LO allele to analyze the level of repression of the targeted allele (Figure 3B). We performed expression analysis with RNA samples from several independent clones of each of the ES cell lines. We observed ∼60% transcriptional repression with wild-type lacI, but in excess of 90% repression in five out of six ES cell clones expressing lacIY (Figure 3B). The lacI and lacIY clones showed comparable levels of expression (Supplementary Figure S3A and B), so we infer that the difference in repression efficacy is likely due to binding affinity differences between the two repressor proteins.
Figure 3.
Modifying the endogenous Dnmt1 promoter and repression of the endogenous Dnmt1 by lac repressors in ES cells. (A) Gene-targeting was performed using a replacement-type vector containing the Dnmt1 promoter modified with lac operator sequences as depicted. Blue arrows indicate the primers used in PCR screening after G418 selection. The selectable marker was removed from the targeted ES cells using Cre recombinase. PCR screening was performed across the single loxP generated as a result of the recombination that is unique to the targeted allele using primers shown as red arrows after ganciclovir selection. (B) NFlacI, lacI, and lacIY were stably introduced into the targeted ES cells through electroporation followed by hygromycin selection. Independent clones from each ES cell line were subjected to the real-time RT-PCR assays using a reaction specific for the targeted allele as indicated by blue arrows (primers) and line (probe). Dnmt1LO expression was normalized against Pcna (left panel) or against histone H4 (Hist2h4a) (Right panel). Expression levels of lacI and lacIGY are shown in Supplementary Figure S3.
In vivo downregulation of Dnmt1 expression by the Lac system
We introduced the Dnmt1LO modification into the mouse germline and generated Dnmt1LO/LO homozygous mice (Figure 4A). We did not observe any difference in Dnmt1 expression in the colon between Dnmt1+/+ and Dnmt1LO/LO mice in the absence of lac repressor (Figure 4C), confirming that our lacO modification had not disrupted normal promoter function. We next developed tissue-specific and ubiquitous transgenic mouse lines for the lacIGY (Figure 4A). We generated intestine-specific lines using the Villin promoter, which restricts the expression of a transgene to the intestine, colon and kidney proximal tubules (28) (Figure 4A and B). Among the three transgenic lines we developed, line 3 showed a robust, intestine-specific expression of lacIGY. We crossed this transgenic line with the Dnmt1LO mice and generated Dnmt1LO/LO/TgVillacIGY mice to test the in vivo efficacy of transcriptional repression. Dnmt1LO/LO/TgVillacIGY mice reduced Dnmt1 expression to 15% of the levels observed in Dnmt1LO/LO mice without the repressor transgene (Figure 4C). These results show for the first time that the lac repressor can inhibit transcription of an endogenous gene at its cognate locus in mice. We confirmed the reduction in Dnmt1 expression at the protein level by immunostaining. Nuclear staining of Dnmt1 protein disappeared almost completely from colonic crypts of Dnmt1LO/LO/TgVillacIGYmice (Figure 4D).
We next investigated whether repression by the lac repressor could be reversed by IPTG treatment. Previous studies showed that IPTG can be efficiently applied to various mouse tissues via oral administration or intravenous injection and can effectively inhibit lac repression of exogenous transgenes in mice (17). We added IPTG to the drinking water for 3 weeks at concentrations of 20, 80 or 160 mM. Repression was reversed in a dose-dependent manner (Figure 4E). In contrast to the partial reversion in the transient transfection experiments (Figure 2B, Supplementary Figure S2B), expression in vivo was restored to 90% of unregulated expression at 160 mM IPTG (Figure 4E). Extended IPTG treatment for up to 7 months did not result in any notable deleterious effects. In addition, IPTG withdrawal resumed the tight repression of Dnmt1 in 2–3 days (Supplementary Figure S4A).
We also tested the repression system in a broader spectrum of tissues using a ubiquitously expressing lacIGY line, with expression driven by the human elongation factor 1 alpha (EF1α) promoter (Figure 4A). One of the transgenic lines (line 11) exhibited high lacIGY expression across all the organs analyzed (Figure 4F). We observed that ubiquitous downregulation of Dnmt1 by the lac system resulted in embryonic lethality (Chi-square test, P < 0.0001, Figure 4G). A genetically modified Dnmt1 hypomorphic line having 10% of the expression of wild-type mice is viable (29). From this, it can be surmised that the level of residual Dnmt1 expression in Dnmt1LO/LO/TgEFlacIGY is likely <10% in at least some essential tissues, impairing proper development. Expression analysis from mice heterozygous for the Dnmt1LO targeted allele showed that in the presence of lacIGY, transcription was reduced to approximately half-normal levels, consistent with almost complete repression of the Dnmt1LO allele, with the residual transcription from the remaining wild-type allele (Figure 4H).
DNA methylation and transcriptome analyses showed both global and gene-specific DNA methylation and gene expression changes upon the inhibition of Dnmt1 (Supplementary Figure S5A-G, Tables S1–S3). This provides further support for the potency of our enhanced repression system, since trace amounts of Dnmt1 are sufficient to maintain substantial levels of genomic DNA methylation in cell lines and mice (30,31). As a surrogate for estimation of global methylation, we analyzed mouse repetitive element B1, B2 and IAP (Supplementary Figure S5A and B).
Inducible and reversible Up- and downregulation of endogenous Dnmt1
It has been particularly challenging to achieve strong overexpression of full-length Dnmt1 in vitro and in vivo (23,24). A 2.5-fold overexpression of Dnmt1 from a Dnmt1 BAC clone was found to result in embryonic lethality (24). No transgenic lines of Dnmt1 have been reported, despite numerous attempts. Here, we report a new approach to achieve upregulation of Dnmt1 by targeting transcriptional transactivators to the endogenous Dnmt1 promoter. We combined two copies each of tet operators and of gal4 DNA binding sequences with the lac operator insertions described above to create a Dnmt1LGT allele with more flexible control, i.e. allowing either upregulation or repression of transcription depending on which transacting protein is present (Figure 5A).
To test whether endogenous Dnmt1 expression can be upregulated from the Dnmt1LGT allele, we stably introduced an advanced version of the inducible tet activator (rtTA-M2) into the targeted ES cells (Figure 5B). In the absence of doxycycline, Dnmt1 expression from the rtTA-M2-expressing cells was comparable to that from control ES cells, but upon treatment with doxycycline, Dnmt1 expression was increased up to 6.5-fold (Figure 5C and D, Supplementary Figure S6A and B). We confirmed that the Dnmt1LGT allele could also be downregulated 70–90% by introduction of the lacIGY repressor (Figure 5D). Thus, we were able to achieve either up- or downregulation of an endogenous allele in the same cells, using doxycycline to control activator binding and IPTG to reverse repressor binding.
We next introduced the Dnmt1LGT modification into the mouse germline and generated Dnmt1LGT/LGT homozygous mice. As we found from the colon, the modifications have no deleterious effect on the activity the endogenous Dnmt1 promoter (Figure 4C, Supplementary Figure S6C and D). We crossed them with the available ROSA26-rtTA-M2 transgenic line to test in vivo upregulation of Dnmt1 (32). We observed a strong upregulation of Dnmt1 from the liver, spleen, and kidney (Figure 5E). This provides the first experimental evidence that endogenous gene expression in mice can be upregulated from its cognate promoter by targeted recruitment of a transcriptional activator. We found no detectable upregulation in the heart, which could be explained by the proliferation-dependent expression pattern of Dnmt1 and the sparse population of proliferation-competent cells in the heart (33) (Figure 5E). Thus, the stimulatory effects of rtTA-M2 on transcription can enhance expression of a transcriptionally competent gene, but may not be sufficient to turn on a transcriptionally silent gene. This inability may be overcome by increasing the number of activator binding sites and the expression level of activator.
Transcriptional repression by attenuation of transcription elongation
Attenuation of transcription elongation by the lac repressor has been controversial. While successful blockage of transcription elongation has been reported (34), elongation factor SII enables RNA polymerase II to proceed through DNA-bound lac repressors at high efficiency (35). We hypothesized that the enhanced binding affinity of our modified lac system might be strong enough to overcome the effects of elongation factor SII. We tested this by inserting operators into the rabbit β-globin intron under the control of the Dnmt1 promoter.
We observed moderate repression at intronic operators with the wild-type lac and tet repressors (Figure 6A, Supplementary Figure S7A), but the enhanced lac repressor indeed resulted in strong transcriptional repression (Figure 6A–D, Supplementary Figure S7A and B). We found that attenuation of transcription at downstream operators could be readily achieved with other strong promoters, including CMV, Ubc and even EF1α, one of the strongest mammalian promoters (Supplementary Figure S7B). We found no correlation between the level of residual expression and the strength of the promoters, suggesting that the repression capacity of our enhanced lacI system exceeds the transcriptional strength of all of the robust promoters we tested (Figure 6B, Supplementary Figure S7B and C).
In order to determine the in vivo efficacy of transcriptional attenuation by the REMOTE-control system, we generated a transgenic line with a reporter gene containing the lacO inserted rabbit β-globin intron and crossed with the TgVillacIGY mice (Figure 6C). We observed more than 90% repression of the reporter in the small intestine (Figure 6C and D). This successful repression by inhibiting transcription elongation underscores the strength of the lacIGY binding, and further demonstrates the utility of our system. Furthermore, this method enables rapid application of the REMOTE-control system by simplifying the placement of operator sequences without disrupting normal expression in the absence of repressor, as introns contain far fewer binding sites for transcription factors than do promoters. This approach could also be advantageous for controlling genes with multiple promoters.
Suppression of intestinal polyp formation through repression of Dnmt1 by the REMOTE-control system
Mouse models having lower Dnmt1 expression have significantly fewer intestinal tumors induced by genetic mutations in the Mlh1 or Apc gene (29,36,37). These studies show that Dnmt1 plays an essential role for tumor initiation in these mouse models. Using our REMOTE-control system with the ApcMin model, we found that intestine-specific reduction of Dnmt1 expression down to 15% resulted in substantial suppression of intestinal polyp formation (Figure 6E). Unlike previous studies using Dnmt1 hypomorphic mouse models where Dnmt1 expression is reduced uniformly across all cell types, our system achieves intestinal epithelial cell-specific Dnmt1 repression. This allows us to attribute the observed strong intestinal antineoplastic effect of Dnmt1 repression specifically to epithelial cell-autonomous effects. This result not only demonstrates the in vivo functionality and potency of the system, but also provides new experimental opportunities that the system has to offer for investigating the role of DNA methylation beyond tumorigenesis. For example, since the system allows a reversible control, it is now possible to ask whether Dnmt1 also plays a role in the maintenance of the established tumors. The mouse model can be maintained upon IPTG treatment to allow tumors to grow under normal Dnmt1 expression, and IPTG can be withdrawn to repress Dnmt1 expression after tumors are formed. The induction kinetics of IPTG (2–3 days) and of Doxycycline (6–10 days) are sufficiently responsive for such tumor initiation and maintenance studies (Supplementary Figure S4A) (2,38–40). This question is particularly relevant in light of the increasing interest in the use of epigenetic therapy in human malignancies.
DISCUSSION
Significant effort has been expended to achieve more refined control of gene expression to recapitulate pathological processes or to experimentally intervene in biological pathways with precision. Advances in molecular genetics enable temporo-spatial dissection of gene function by conditionally activating or inactivating target genes in living organisms. Site-specific binary recombination technologies, such as the Cre/loxP system, have been widely used to achieve conditional inactivation. The Cre/loxP system now allows for inducible activation through conditional excision of a stop element in the target gene (41). However, in these recombination-based approaches, the ability to control gene function is lost once deletion has taken place. Further, the dichotomous nature of the system does not allow assessment of phenotypic effects across a continuous spectrum of gene expression levels.
The REMOTE-control system offers reversible and tunable expression control that can be applied and withdrawn multiple times in a living organism, allowing us to test the reversibility of a phenotype and to investigate gene function at different expression levels. The potency and multi-functionality of the REMOTE-control system makes it an attractive alternative to conventional and conditional knockout approaches.
Control of endogenous gene expression has also been achieved by several fusion transcriptional activators or repressors, such as KRAB-fused or VP64-fused ZNF, TALE or dCas9 proteins (6,42,43). Of these, the dCas9 system offers the greatest flexibility and potential, allowing the targeting of any genomic loci by introducing RNA sequences complementary to the regions of interest. However, the recruitment of powerful eukaryotic repressors, such as the KRAB and its interacting protein KAP1, results in permanent silencing of target genes by inducing promoter DNA methylation and heterochromatin, which can spread over several tens of kilobases, (44,45). In contrast, the REMOTE-control system achieves repression by steric hindrance (46). Indeed, we were able to reverse the repression caused by the lacIGY repressor after several months of suppression in mice (see Figure 4D and E). Notably, we were also able to fully restore expression from the targeted allele after over two months of repression in heterozygote ES cells (Dnmt1LGT/+) (Supplementary Figure S6B), which maintain global DNA methylation levels comparable to those of wild-type ES cells (see Supplementary Figure S5A and B) (47). This ability to temporarily affect endogenous gene expression should provide greater precision for investigating gene function in vivo.
In this study, our enhanced lac system was able to overcome the transcriptional strength of all the robust promoters tested, suggesting promising general applicability and reliability. A limitation of the REMOTE-control is the potential challenge associated with the insertion of the effector binding sites without affecting target gene expression. Prokaryotic repressors achieve repression by steric hindrance with the transcription machinery (48). This short-range mode of action imposes constraints on the design of the target promoter configuration, and increases the risk of affecting transcription in the absence of repressor binding while our enhanced lac repressor successfully repressed both CMV and Dnmt1 promoters from operators located outside the critical promoter regions. Our intron-based alternative repression approach substantially mitigates this risk and thus improves the general applicability of the REMOTE-control system. The enhanced lac system was also able to tightly repress transcription at operators located several kilobases downstream of the transcription start site (Figure 6C and D). Importantly, this approach may be less susceptible to variation among different genes, tissues, and organisms because the repression is likely achieved through physical hindrance between two components, the Pol II transcription elongation machinery and the lac repressor–operator complexes (Figure 6B–D, Supplementary Figure S7B and C) (46). The large evolutionary distance to the originating species of the regulatory components and the complexity of the operator sequences provides for a high degree of target specificity in the control of gene expression in mammals (49). The availability of regulatory ligands (IPTG for the lac repressor and doxycycline for the tet activator) provides temporal and dose-dependent expression control. Furthermore, libraries of repressors and activators can be established and applied to any endogenous gene with predictable spatial and/or temporal regulation. For example, existing tet transactivator transgenic mouse lines can be readily applied to achieve upregulation of the endogenous Dnmt1 gene in most mouse tissues, which will be a useful resource because the gene plays important roles in many biological and pathological processes, including cancer.
Gene upregulation by the REMOTE-control system has several advantages over other inducible transgenic approaches. It eliminates the necessity of generating multiple transgenic lines to rule out position effects of the transgene insertion, as the upregulation is achieved at the endogenous locus. In addition, our approach is well-suited for genes with a robust basal expression because it enhances expression from an already strong endogenous promoter, whereas other transgenic approaches induce expression from minimal viral promoters. In this study, we were able to increase the activity of the robust Dnmt1 promoter up to 6.5-fold (see Figure 5C–E). We were even able to upregulate the EF1α promoter, one of the strongest known promoters, 3.5-fold (see Supplementary Figure S7D), which would be difficult to achieve with minimal viral promoters. Finally, our approach retains elements of natural regulation, including innate cis-regulatory elements, which may confer tissue specificity, cell-cycle control, and splicing variants, which would be difficult to achieve with transgenic approaches. On the other hand, our approach might have a limitation in inducing expression from promoters with low transcription potential, such as ones in strong heterochromatic regions as indicated by our results from mouse heart (Figure 5E). It remains to be tested whether such promoters can also be upregulated with higher levels of activator and more binding sites.
The potency and multi-utility of our system provide the capability to control the level, timing, and location of endogenous gene expression and will enable in vivo functional analyses at much higher resolutions. With the recent advent of CRISPR/Cas-mediated gene-targeting technology, genetic manipulation of endogenous loci has been readily accomplished (13–16). This technical advance will greatly facilitate the insertion of regulatory sequences for the REMOTE-control system and will enable facile application of the technology.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Heidi Scrable for providing the mammalian lacI (Mayo Clinic, Rochester, MN), Dr Daniel Louvard (Institut Curie, Paris, France) for the Villin promoter, Dr Laurie Jackson-Grusby (Childrens Hospital, Boston, MA) for her contributions to the early development of this project and for the mouse Dnmt1 antibody. We thank Dr Nancy Wu and Dr Robert Maxson for help in the generation of transgenic and knockout mice. We also thank Dr Marconett and Dr Hinoue for their help in the analysis of expression array data. We thank David Nadziejka for carefully editing the manuscript. We also thank Dr Mary Winn and Zachary Madaj for their help in the statistical analysis of the data. We are also grateful to the members of the Laird laboratory for helpful discussions and assistance.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [CA075090, DA030325, CA157918 to P.W.L.]. Funding for open access charge: National Institutes of Health [CA157918].
Conflict of interest statement. None declared.
REFERENCES
- 1. Nagy A., Perrimon N., Sandmeyer S., Plasterk R.. Tailoring the genome: the power of genetic approaches. Nat. Genet. 2003; 33(Suppl):276–284. [DOI] [PubMed] [Google Scholar]
- 2. Premsrirut P.K., Dow L.E., Kim S.Y., Camiolo M., Malone C.D., Miething C., Scuoppo C., Zuber J., Dickins R.A., Kogan S.C. et al. . A rapid and scalable system for studying gene function in mice using conditional RNA interference. Cell. 2011; 145:145–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kelly A., Hurlstone A.F.. The use of RNAi technologies for gene knockdown in zebrafish. Brief. Funct. Genomics. 2011; 10:189–196. [DOI] [PubMed] [Google Scholar]
- 4. Chen C.M., Chiu S.L., Shen W., Cline H.T.. Co-expression of Argonaute2 enhances short hairpin RNA-induced RNA interference in Xenopus CNS neurons in vivo. Front. Neurosci. 2009; 3:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Qiu S., Adema C.M., Lane T.. A computational study of off-target effects of RNA interference. Nucleic Acids Res. 2005; 33:1834–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gilbert L.A., Horlbeck M.A., Adamson B., Villalta J.E., Chen Y., Whitehead E.H., Guimaraes C., Panning B., Ploegh H.L., Bassik M.C. et al. . Genome-scale CRISPR-mediated control of gene repression and activation. Cell. 2014; 159:647–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Konermann S., Brigham M.D., Trevino A.E., Joung J., Abudayyeh O.O., Barcena C., Hsu P.D., Habib N., Gootenberg J.S., Nishimasu H. et al. . Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015; 517:583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Itzhaki J.E., Gilbert C.S., Porter A.C.. Construction by gene targeting in human cells of a ‘conditional' CDC2 mutant that rereplicates its DNA. Nat. Genet. 1997; 15:258–265. [DOI] [PubMed] [Google Scholar]
- 9. Tanaka K.F., Ahmari S.E., Leonardo E.D., Richardson-Jones J.W., Budreck E.C., Scheiffele P., Sugio S., Inamura N., Ikenaka K., Hen R.. Flexible accelerated STOP tetracycline operator-knockin (FAST): a versatile and efficient new gene modulating system. Biol. Psychiatry. 2010; 67:770–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sun R., Zhao K., Shen R., Cai L., Yang X., Kuang Y., Mao J., Huang F., Wang Z., Fei J.. Inducible and reversible regulation of endogenous gene in mouse. Nucleic Acids Res. 2012; 40:e166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Richardson-Jones J.W., Craige C.P., Guiard B.P., Stephen A., Metzger K.L., Kung H.F., Gardier A.M., Dranovsky A., David D.J., Beck S.G. et al. . 5-HT1A autoreceptor levels determine vulnerability to stress and response to antidepressants. Neuron. 2010; 65:40–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hsu P.D., Lander E.S., Zhang F.. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014; 157:1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chu V.T., Weber T., Graf R., Sommermann T., Petsch K., Sack U., Volchkov P., Rajewsky K., Kuhn R.. Efficient generation of Rosa26 knock-in mice using CRISPR/Cas9 in C57BL/6 zygotes. BMC Biotechnol. 2016; 16:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bishop K.A., Harrington A., Kouranova E., Weinstein E.J., Rosen C.J., Cui X., Liaw L.. CRISPR/Cas9-mediated insertion of loxP sites in the mouse Dock7 gene provides an effective alternative to use of targeted embryonic stem cells. G3 (Bethesda). 2016; 6:2051–2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Aida T., Chiyo K., Usami T., Ishikubo H., Imahashi R., Wada Y., Tanaka K.F., Sakuma T., Yamamoto T., Tanaka K.. Cloning-free CRISPR/Cas system facilitates functional cassette knock-in in mice. Genome Biol. 2015; 16:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yang H., Wang H., Shivalila C.S., Cheng A.W., Shi L., Jaenisch R.. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013; 154:1370–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cronin C.A., Gluba W., Scrable H.. The lac operator-repressor system is functional in the mouse. Genes Dev. 2001; 15:1506–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schmitz A., Coulondre C., Miller J.H.. Genetic studies of the lac repressor. V. Repressors which bind operator more tightly generated by suppression and reversion of nonsense mutations. J. Mol. Biol. 1978; 123:431–454. [DOI] [PubMed] [Google Scholar]
- 19. Varshavsky A. The N-end rule: functions, mysteries, uses. Proc. Natl. Acad. Sci. U.S.A. 1996; 93:12142–12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kozak M. Recognition of AUG and alternative initiator codons is augmented by G in position +4 but is not generally affected by the nucleotides in positions +5 and +6. EMBO J. 1997; 16:2482–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sadler J.R., Sasmor H., Betz J.L.. A perfectly symmetric lac operator binds the lac repressor very tightly. Proc. Natl. Acad. Sci. U.S.A. 1983; 80:6785–6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jackson-Grusby L., Beard C., Possemato R., Tudor M., Fambrough D., Csankovszki G., Dausman J., Lee P., Wilson C., Lander E. et al. . Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat. Genet. 2001; 27:31–39. [DOI] [PubMed] [Google Scholar]
- 23. Choi S.H., Heo K., Byun H.M., An W., Lu W., Yang A.S.. Identification of preferential target sites for human DNA methyltransferases. Nucleic Acids Res. 2011; 39:104–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Biniszkiewicz D., Gribnau J., Ramsahoye B., Gaudet F., Eggan K., Humpherys D., Mastrangelo M.A., Jun Z., Walter J., Jaenisch R.. Dnmt1 overexpression causes genomic hypermethylation, loss of imprinting, and embryonic lethality. Mol. Cell. Biol. 2002; 22:2124–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mertineit C., Yoder J.A., Taketo T., Laird D.W., Trasler J.M., Bestor T.H.. Sex-specific exons control DNA methyltransferase in mammalian germ cells. Development. 1998; 125:889–897. [DOI] [PubMed] [Google Scholar]
- 26. Kishikawa S., Murata T., Kimura H., Shiota K., Yokoyama K.K.. Regulation of transcription of the Dnmt1 gene by Sp1 and Sp3 zinc finger proteins. Eur. J. Biochem. 2002; 269:2961–2970. [DOI] [PubMed] [Google Scholar]
- 27. Kimura H., Nakamura T., Ogawa T., Tanaka S., Shiota K.. Transcription of mouse DNA methyltransferase 1 (Dnmt1) is regulated by both E2F-Rb-HDAC-dependent and -independent pathways. Nucleic Acids Res. 2003; 31:3101–3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Robine S., Jaisser F., Louvard D.. Epithelial cell growth and differentiation. IV. Controlled spatiotemporal expression of transgenes: new tools to study normal and pathological states. Am. J. Physiol. 1997; 273:G759–762. [DOI] [PubMed] [Google Scholar]
- 29. Gaudet F., Hodgson J.G., Eden A., Jackson-Grusby L., Dausman J., Gray J.W., Leonhardt H., Jaenisch R.. Induction of tumors in mice by genomic hypomethylation. Science. 2003; 300:489–492. [DOI] [PubMed] [Google Scholar]
- 30. Egger G., Jeong S., Escobar S.G., Cortez C.C., Li T.W., Saito Y., Yoo C.B., Jones P.A., Liang G.. Identification of DNMT1 (DNA methyltransferase 1) hypomorphs in somatic knockouts suggests an essential role for DNMT1 in cell survival. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:14080–14085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rhee I., Jair K.W., Yen R.W., Lengauer C., Herman J.G., Kinzler K.W., Vogelstein B., Baylin S.B., Schuebel K.E.. CpG methylation is maintained in human cancer cells lacking DNMT1. Nature. 2000; 404:1003–1007. [DOI] [PubMed] [Google Scholar]
- 32. Linhart H.G., Lin H., Yamada Y., Moran E., Steine E.J., Gokhale S., Lo G., Cantu E., Ehrich M., He T. et al. . Dnmt3b promotes tumorigenesis in vivo by gene-specific de novo methylation and transcriptional silencing. Genes Dev. 2007; 21:3110–3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mollova M., Bersell K., Walsh S., Savla J., Das L.T., Park S.Y., Silberstein L.E., Dos Remedios C.G., Graham D., Colan S. et al. . Cardiomyocyte proliferation contributes to heart growth in young humans. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:1446–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Deuschle U., Hipskind R.A., Bujard H.. RNA polymerase II transcription blocked by Escherichia coli lac repressor. Science. 1990; 248:480–483. [DOI] [PubMed] [Google Scholar]
- 35. Reines D., Mote J. Jr. Elongation factor SII-dependent transcription by RNA polymerase II through a sequence-specific DNA-binding protein. Proc. Natl. Acad. Sci. U.S.A. 1993; 90:1917–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Laird P.W., Jackson-Grusby L., Fazeli A., Dickinson S.L., Jung W.E., Li E., Weinberg R.A., Jaenisch R.. Suppression of intestinal neoplasia by DNA hypomethylation. Cell. 1995; 81:197–205. [DOI] [PubMed] [Google Scholar]
- 37. Trinh B.N., Long T.I., Nickel A.E., Shibata D., Laird P.W.. DNA methyltransferase deficiency modifies cancer susceptibility in mice lacking DNA mismatch repair. Mol. Cell. Biol. 2002; 22:2906–2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stevenson M., Carlisle R., Davies B., Preece C., Hammett M., Liu W.L., Fisher K.D., Ryan A., Scrable H., Seymour L.W.. Development of a positive-readout mouse model of siRNA pharmacodynamics. Mol. Ther. Nucleic Acids. 2013; 2:e133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Grespi F., Ottina E., Yannoutsos N., Geley S., Villunger A.. Generation and evaluation of an IPTG-regulated version of Vav-gene promoter for mouse transgenesis. PLoS One. 2011; 6:e18051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Seibler J., Kleinridders A., Kuter-Luks B., Niehaves S., Bruning J.C., Schwenk F.. Reversible gene knockdown in mice using a tight, inducible shRNA expression system. Nucleic Acids Res. 2007; 35:e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ventura A., Kirsch D.G., McLaughlin M.E., Tuveson D.A., Grimm J., Lintault L., Newman J., Reczek E.E., Weissleder R., Jacks T.. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007; 445:661–665. [DOI] [PubMed] [Google Scholar]
- 42. de Groote M.L., Verschure P.J., Rots M.G.. Epigenetic Editing: targeted rewriting of epigenetic marks to modulate expression of selected target genes. Nucleic Acids Res. 2012; 40:10596–10613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Konermann S., Brigham M.D., Trevino A.E., Joung J., Abudayyeh O.O., Barcena C., Hsu P.D., Habib N., Gootenberg J.S., Nishimasu H. et al. . Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2014; 517:583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Peng H., Ivanov A.V., Oh H.J., Lau Y.F., Rauscher F.J. 3rd. Epigenetic gene silencing by the SRY protein is mediated by a KRAB-O protein that recruits the KAP1 co-repressor machinery. J. Biol. Chem. 2009; 284:35670–35680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Groner A.C., Meylan S., Ciuffi A., Zangger N., Ambrosini G., Denervaud N., Bucher P., Trono D.. KRAB-zinc finger proteins and KAP1 can mediate long-range transcriptional repression through heterochromatin spreading. PLoS Genet. 2010; 6:e1000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ptashne M. Principles of a switch. Nat. Chem. Biol. 2011; 7:484–487. [DOI] [PubMed] [Google Scholar]
- 47. Chan M.F., van Amerongen R., Nijjar T., Cuppen E., Jones P.A., Laird P.W.. Reduced rates of gene loss, gene silencing, and gene mutation in Dnmt1-deficient embryonic stem cells. Mol. Cell. Biol. 2001; 21:7587–7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ptashne M., Gilbert W.. Genetic repressors. Sci. Am. 1970; 222:36–44. [DOI] [PubMed] [Google Scholar]
- 49. Labow M.A., Baim S.B., Shenk T., Levine A.J.. Conversion of the lac repressor into an allosterically regulated transcriptional activator for mammalian cells. Mol. Cell. Biol. 1990; 10:3343–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.