Abstract
Mitochondrial polycistronic transcripts are extensively processed to give rise to functional mRNAs, rRNAs and tRNAs; starting with the release of tRNA elements through 5′-processing by RNase P (MRPP1/2/3-complex) and 3′-processing by RNase Z (ELAC2). Here, we show using in vitro experiments that MRPP1/2 is not only a component of the mitochondrial RNase P but that it retains the tRNA product from the 5′-processing step and significantly enhances the efficiency of ELAC2-catalyzed 3′-processing for 17 of the 22 tRNAs encoded in the human mitochondrial genome. Furthermore, MRPP1/2 retains the tRNA product after ELAC2 processing and presents the nascent tRNA to the mitochondrial CCA-adding enzyme. Thus, in addition to being an essential component of the RNase P reaction, MRPP1/2 serves as a processing platform for several down-stream tRNA maturation steps in human mitochondria. These findings are of fundamental importance for our molecular understanding of disease-related mutations in MRPP1/2, ELAC2 and mitochondrial tRNA genes.
INTRODUCTION
Mitochondria, powerhouses of the eukaryotic cell, have their own genome and gene expression system. Functional mitochondrial gene expression is crucial for the biogenesis of the respiratory chain, and thereby for cellular metabolism. Defects in mitochondrial gene expression are a direct cause for debilitating mitochondrial diseases, but also imply age-related diseases and ageing (1,2).
The human mitochondrial genome is 16.6 kB long, circular and encodes 37 genes including 13 mRNAs, 22 tRNAs and 2 rRNAs. The genome resides in nucleoids, which are discrete, compact structures about 100 nm in diameter (3,4). The genome is transcribed bi-directionally to produce long polycistronic heavy- and light-strand precursor RNA transcripts. These transcripts must be further processed to release the mature mitochondrial RNAs essential for organellar translation, and hence for respiratory chain biogenesis (5–7). The large majority of mRNA and rRNA elements are flanked by one or more tRNA elements. In what is known as the tRNA punctuation model (6), excision of the tRNA elements releases all other elements and is a prerequisite for RNA maturation. This extensive processing is presumably co-transcriptional, and is mainly performed in distinct mitochondrial RNA granules located close to the nucleoid (8–10). Mitochondrial RNA maturation is a complex process which involves a battery of nucleases and a more detailed molecular understanding is desired to link aberrant RNA processing to mitochondrial disease (11).
The 5′- and 3′-processing of the mitochondrial tRNA elements are performed by the mitochondrial RNase P and RNase Z endonucleases, respectively, both of which are located within or close to the mitochondrial RNA granules (10). After maturation of the tRNA ends, the 3′-terminal cytosine-cytosine-adenine (3′-CCA) triplet is added by the mitochondrial CCA-adding enzyme, completing the three core processing steps required for nearly all mitochondrial tRNAs. The human mitochondrial RNase P is composed of three protein subunits: mitochondrial RNase P proteins 1, 2 and 3 (MRPP1, MRPP2 and MRPP3, respectively) (12). MRPP1 and MRPP2 form a strong protein complex (MRPP1/2) (12,13), which is not only involved in 5′-processing of the tRNA elements by RNase P but is also responsible for the N1-methylation of adenosine and guanosine at position 9 (m1A9 and m1G9, respectively) of human mitochondrial tRNAs (13–15). MRPP2 is a multifunctional protein involved in other cellular processes unrelated to mitochondrial RNase P activity such as amino acid catabolism and lipid metabolism (16,17). Interestingly, the 5′-processing and N1-methylation processes of MRPP1/2 are uncoupled (13). The endonuclease activity of mitochondrial RNase P is mediated by MRPP3, but is strictly dependent on the presence of MRPP1/2 (12,18,19).
RNase Z mediates tRNA 3′-processing, which involves removing extra 3′ nucleotides immediately after the discriminator base from tRNA precursors. The RNase Z proteins belong to the metallo-β-lactamase superfamily ELAC1/2 (20–22). Mammals encode multiple forms of RNase Z with ELAC1 located in the nucleus and in the cytosol (23–25), while ELAC2 resides in the nucleus and in mitochondria (24–26).
The tRNA genes present in the mitochondrial genome do not encode for the 3′-terminal CCA triplet; hence, the next essential step in tRNA maturation is the post-transcriptional addition of the conserved 3′-CCA sequence required for aminoacylation. The reaction is catalyzed by the CCA-adding enzyme, which uses cytidine triphosphate and adenosine triphosphate as substrates to catalyze a sequence-specific nucleotide polymerization without a requirement for a nucleic acid template (27,28).
It has been shown that the three core processing steps of human mitochondrial tRNA maturation—5′-cleavage, 3′-cleavage and the 3′-CCA addition—follow an ordered process (1,24,29–31) without evidence for a direct link between the different steps. However, during earlier work on the crystal structure of the nuclease domain of mitochondrial RNase P (19), we noted that RNase P was not able to perform multiple-turnover reactions. Here, we have further investigated this issue, and observed that, although MRPP3 performs multiple-turnover reactions, the MRPP1/2 complex is only capable of single-turnover catalysis in the RNase P catalyzed tRNA 5′-processing. Furthermore, we demonstrate that MRPP1/2 is not only responsible for mitochondrial RNase P-based 5′-leader removal, but also that it remains attached to the tRNA substrate to enable further downstream processing steps including 3′-tail removal by ELAC2 and 3′-CCA addition by the CCA-adding enzyme. Specifically, for the large majority of human mitochondrial tRNAs, the MRPP1/2 complex significantly enhances ELAC2-catalyzed tRNA 3′-processing. The MRPP1/2 complex thereby takes an unexpected central role in the core processing steps required for human mitochondrial tRNA maturation.
MATERIALS AND METHODS
Plasmids
The plasmids for the expression of human mitochondrial RNase P protein 1 (MRPP1; UniProt: Q7L0Y3; residues 40–403; also called tRNA methyltransferase 10C homolog, TRMT10C), human mitochondrial RNase P protein 2 (MRPP2; UniProt: Q99714; residues 1–261; also called short chain dehydrogenase/reductase 5C member 1, SDR5C1; or 17β-hydroxysteroid dehydrogenase type 10, HSD10) and human mitochondrial RNase P protein 3 (MRPP3; UniProt: O15091; residues 45–583; also called proteinaceous RNase P, PRORP) were as previously reported (19). The gene encoding human mitochondrial zinc phosphodiesterase ELAC protein 2 (ELAC2; UniProt: Q9BQ52; residues 31–826) was synthesized (DNA2.0) and cloned into the pJExpress 411 vector, which adds an N-terminal His6-tag and a TEV cleavage site to the N-terminus of the protein. The gene encoding human mitochondrial CCA-adding enzyme (UniProt: Q96Q11; residues 42–434; also called mitochondrial CCA tRNA nucleotidyltransferase 1, TRNT1) was synthesized (Gen9) and cloned into the pNIC28-Bsa4 expression vector (GenBank: EF198106), which adds an N-terminal His6-tag and a TEV cleavage site to the N-terminus of the protein. Correct cloning was confirmed by DNA sequencing (Source Biosciences).
Protein expression and purification
All recombinant proteins were produced in Escherichia coli strain KRX (Promega) using isopropyl β-D-1-thiogalactopyranoside and rhamnose for induction of protein expression. The MRPP1/2 complex was produced essentially as previously described (19) and stored in 20 mM Tris pH 7.6, 200 mM NaCl, 10%(v/v) glycerol and 2 mM Tris(2-carboxyethyl)phosphine (TCEP). MRPP3 was purified as previously reported (19) and stored in 20 mM Na-HEPES pH 7.5, 300 mM NaCl, 10%(v/v) glycerol and 0.5 mM TCEP. ELAC2 was purified using nickel-affinity, Q anion exchange and size exclusion chromatography in a final buffer of 50 mM Tris pH 8.0, 150 mM NaCl, 10%(v/v) glycerol and 5 mM TCEP. The N-terminal His6-tag was removed by TEV protease treatment. The CCA-adding enzyme was purified using nickel-affinity, heparin affinity and size exclusion chromatography in a final buffer of 20 mM Na-HEPES pH 7.6, 150 mM NaCl, 5%(v/v) glycerol and 2 mM TCEP. All proteins were concentrated by ultrafiltration and then flash cooled in liquid nitrogen and stored at −80°C.
Preparation of tRNAs
Template DNA for Homo sapiens mitochondrial pre-tRNA substrates were synthesized (Integrated DNA Technologies). The tRNA constructs were either directly produced by run-off transcription or were joined 5′ of the GlmS ribozyme. See Supplementary Tables S1–3 for details of the construct design. The template DNA was amplified by polymerase chain reaction using Phusion® High-Fidelity DNA Polymerase (New England BioLabs) or Phire II DNA Polymerase (Thermo Fisher Scientific). In vitro run-off transcription was performed with T7 RNA polymerase at 37°C for 1 to 3 h in reaction mixtures ranging from 50 μl to 5 ml and consisting of 30 mM Na-HEPES pH 8.0, 27 mM MgCl2, 4 mM NTP mix, 10 mM TCEP, 2 mM spermidine and 0.01% Triton X-100. For transcription of tRNA constructs joined to the GlmS ribozyme, 2 mM glucosamine-6-phosphate was added to the reaction mixture to initiate the cleavage. The tRNA constructs co-transcribed with the GlmS ribozyme were applied over a Mono Q column and eluted with a linear gradient of 20 mM K-HEPES pH 7.6, 1 M KCl and 10 mM MgCl2. The final tRNA samples were buffer exchanged to 20 mM K-HEPES pH 7.6, 100 mM KCl and 10 mM MgCl2 and concentrated by ultrafiltration. The remaining tRNA constructs were purified after in vitro transcription using a Quick-RNA Miniprep kit (Zymo Research) and stored in nuclease free water. All tRNA samples were flash cooled in liquid nitrogen and stored at −80°C.
RNase P and RNase Z cleavage assay
The tRNA 5′- and 3′-cleavage activity was assayed in a cleavage buffer containing 20 mM K-HEPES pH 7.6, 130–160 mM KCl, 2 mM MgCl2, 2 mM TCEP and 0.1 mg/ml bovine serum albumin (BSA), unless otherwise stated. The reaction mixes included 200 nM pre-tRNA, 800 nM MRPP1/2, 50 nM MRPP3 and/or 50 nM ELAC2, or various combinations of the protein components. Reactions containing MRPP1/2 were further supplemented with 5 μM S-adenosyl methionine (SAM). The reaction mixes were pre-incubated at 30°C for 10 min, and the reaction was initiated by addition of MRPP3 and/or ELAC2 with subsequent incubation at 30°C for 20 min for pre-tRNATyr samples and 60 min for pre-tRNAHis samples, unless otherwise stated in the figure legends. For the pre-mature forms tRNATyr(38,17), tRNAHis(25,S) and tRNAHis(0,S), 1 U/μl RiboLock RNase Inhibitor (Thermo Fisher Scientific) was added to the reaction mix to minimize unspecific background cleavage. This did not change the overall outcome of the experiment. For the RNase P and RNase Z dilution experiments, the concentrations of the proteins are given in the figure legends. For the salt dependent reactions, the KCl concentration was adjusted accordingly. The reaction products were separated by 7M urea, 10% polyacrylamide-gel electrophoresis (PAGE) gels in 1×Tris/Borate/EDTA (TBE) running buffer. After electrophoresis, the reaction products were visualized on a Geldoc system (Bio-Rad) by staining with SYBR Green II (Invitrogen).
Assays of CCA-adding activity
The CCA tRNA nucleotidyltransferase assays were carried out in assay buffer containing 20 mM K-HEPES pH 7.6, 30 or 150 mM KCl, 5 mM MgCl2, 0.5 mM NTPs, 2 mM TCEP and 0.1 mg/ml BSA using 200 nM pre-tRNA samples, 800 nM MRPP1/2 (if applicable) and 50 nM CCA-adding enzyme. The reaction mixes were pre-incubated at 30°C for 10 min, and the reaction was initiated by the addition of CCA-adding enzyme. The reactions were incubated for 15 and 60 min at 30°C, and the reaction products were separated by 7M urea 10% PAGE in 1× TBE running buffer and analysed using SYBR Green II (Invitrogen) staining.
Gel filtration chromatography
For reconstitution, MRPP1/2 and tRNA were mixed in a 1:2 molar ratio in the presence of equimolar amounts of SAM and nicotinamide adenine dinucleotide (NADH) for 10 min at 4°C, and then the samples were applied to a Superdex S200 10/300 GL column equilibrated in 20 mM K-HEPES pH 7.6, 130 mM KCl, 5%(v/v) glycerol, 5 mM MgCl2 and 2 mM TCEP. When enzymatic reactions were combined with gel filtration chromatography experiments, the corresponding MRPP1/2:tRNA complexes were first reconstituted and purified either on a pre-equilibrated Superdex S200 10/300 GL column or on a HiPrep 16/60 Sephacryl S300 HR column as required. The reconstituted MRPP1/2:tRNA complexes were incubated for 60 min at 30°C with MRPP3 in a 2:1 molar ratio, with ELAC2 in a 3:1 molar ratio, or with CCA-adding enzyme in a 2:1 molar ratio (supplemented with a 2-fold amount of NTP mix). The reaction mixes were resolved on a Superdex S200 10/300 GL column pre-equilibrated in the above given buffer with detection of the absorption at 280 nm. Fractions of interest were analyzed by 7M urea 10% PAGE in 1× TBE running buffer with subsequent SYBR Green II (Invitrogen) staining, and by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) using 4–12% Bis-Tris gels (Novex or Bolt by Invitrogen) in 1× 4-Morpholineethanesulfonicacid (MES) running buffer. All columns were from GE Healthcare Life Sciences.
Determination of tRNA-binding constants
The binding affinity of the MRPP1/2 complex to mitochondrial tRNATyr processing intermediates Y(5,6), Y(0,6), Y(0,0) and Y(0,CCA) were investigated through electrophoretic mobility shift assays. For every tRNA intermediate, a series of 20 μl reaction mixtures were prepared, each containing 250 nM tRNA and varying amounts of MRPP1/2 complex (0–800 nM) in the presence of 5 μM SAM in cleavage buffer (see above). The reaction mixes (in triplicates) were incubated at 30°C for 10 min before separation on 4% agarose EX E-gels (Invitrogen) at 4°C. The gels were imaged using a Geldoc XT system (Bio-Rad) and the band intensities were calculated using ImageLab 4.1 (BioRad). Kd values were determined using non-linear regression analysis in GraphPad Prism 7 (GraphPad).
RESULTS
tRNA remains bound to MRPP1/2 after RNase P cleavage
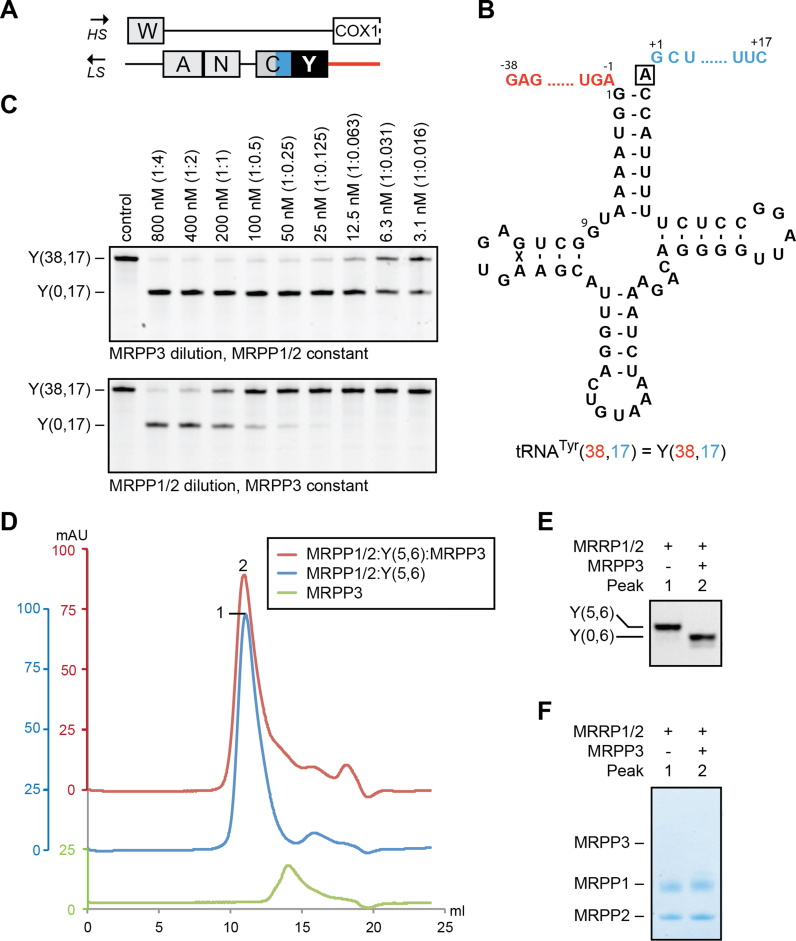
To characterize the effects of the relative enzyme concentrations on human mitochondrial RNase P-cleavage efficiency, we initially used pre-tRNATyr(38,17) as a substrate (where (X,Y) indicates the number of additional nucleotides of the 5′-leader and 3′-tail, respectively). The pre-tRNATyr is located at the 5′-end of the cluster of tRNA genes Tyr-Cys-Asn-Ala located on the light-strand (Figure 1A and B). We varied the amounts of MRPP1/2 complex or MRPP3 in separate runs. The results showed that 5′-processing of pre-tRNATyr(38,17) by mitochondrial RNase P exhibited different dependencies on MRPP1/2 and MRPP3 concentrations. MRPP3 displayed full activity at sub-stoichiometric ratios of protein:tRNA indicative of multiple-turnover reactions (Figure 1C, top panel). In contrast, when there was less MRPP1/2 complex than tRNA substrate, there was a quick decline in activity (Figure 1C, bottom panel). Based on this observation, we hypothesized that MRPP1/2 remains attached to the 5′-processed tRNA. To test this, we reconstituted MRPP1/2 with pre-tRNATyr(5,6). This resulted in the formation of a complex that was stable during gel filtration (Peak 1 in Figure 1D–F). When MRPP3 was added to the pre-formed MRPP1/2:pre-tRNATyr(5,6) complex, the 5′-leader of the pre-tRNA was removed, while the 5′-processed pre-tRNATyr(0,6) remained attached to MRPP1/2 (Peak 2 in Figure 1D–F).
Figure 1.
Role of MRPP1/2 in RNase P function on tRNATyr. (A) Schematic representation of the light-strand (LS) tRNA cluster Tyr-Cys-Asn-Ala and its complementary heavy-strand (HS). (B) Secondary structure of human pre-tRNATyr(38,17) (Y(38,17)) having 38 nt as 5′-leader and 17 nt as 3′-trailer. The discriminator base is boxed. (C) RNase P dilution series on 200 nM tRNATyr(38,17). Top: MRPP3 dilution (from 800 to 3.1 nM) and MRPP1/2 constant (800 nM). Bottom: MRPP1/2 dilution (from 800 to 3.1 nM) and MRPP3 constant (50 nM). The molar ratio of tRNA to protein is given in brackets. The reactions were incubated for 1 h. (D–F) Gel filtration chromatograms of MRPP1/2:pre-tRNATyr(5,6) prior and after MRPP3 treatment as well as of MRPP3 alone (D). Peaks of interest are numbered and the peaks tRNA and protein content are visualized by urea PAGE (E) and SDS-PAGE (F), respectively.
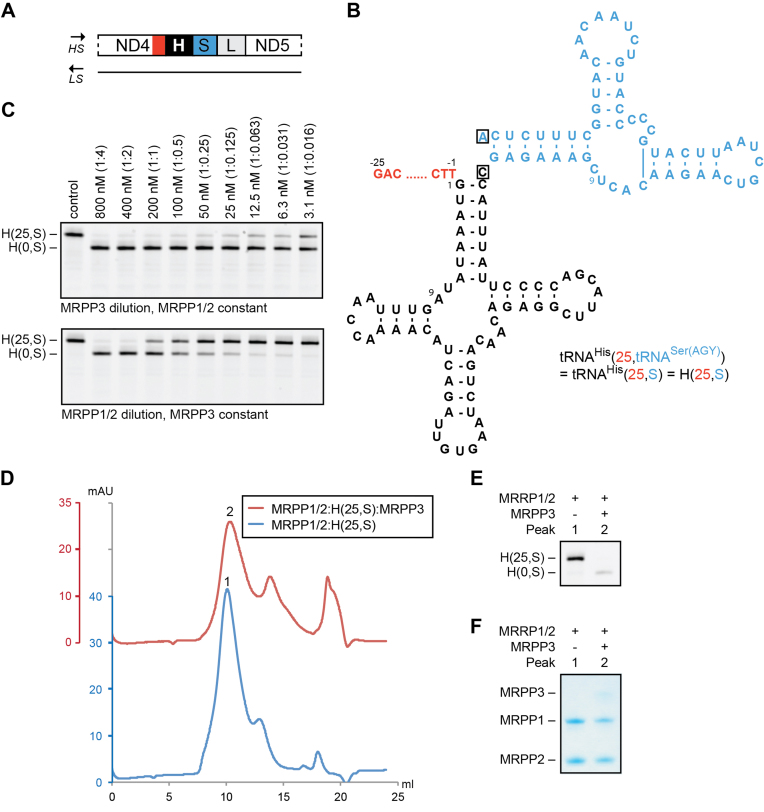
We then continued with a more complex mitochondrial tRNA substrate from the heavy-strand: pre-tRNAHis(25,S); where S corresponds to the complete 59 nucleotide-long tRNASer(AGY) (Figure 2A and B). tRNAHis is located at the 5′-end of a cluster of tRNAs—His-Ser(AGY)-Leu(CUN)—located between the ND4 and ND5 coding sequences. For tRNAHis(25,S), we observed the same general pattern as with pre-tRNATyr. Specifically, there was a strong concentration dependency on the MRPP1/2 complex, while MRPP3 appeared to perform multiple-turnover reactions in the mitochondrial RNase P cleavage reaction (Figure 2C). Interestingly, RNase P did not process the His-Ser(AGY) junction, shown by the fact that the incubation of pre-tRNAHis(25,S) with RNase P gave only pre-tRNAHis(0,S) and not pre-tRNAHis(0,0) and pre-tRNASer(0,0) as products. Gel filtration experiments confirmed that MRPP1/2 was able to form a strong complex with pre-tRNAHis(25,S) (Peak 1 in Figure 2D–F). Moreover, after the RNase P cleavage reaction, the product pre-tRNAHis(0,S) remained bound to MRPP1/2 (Peak 2 in Figure 2D–F).
Figure 2.
Role of MRPP1/2 in RNase P function on tRNAHis. (A) Schematic representation of the heavy-strand (HS) tRNA cluster His-Ser(AGY)-Leu(CUN). The 5′- and 3′-region of tRNAHis (in black) are highlighted in red and blue, respectively. (B) Secondary structure of pre-tRNAHis(25,tRNASer(AGY)) (H(25,S)) having 25 nt as 5′-leader (in red) and the complete tRNASer(AGY) as 3′-trailer (in blue). The discriminator bases are boxed. (C) RNase P dilution series on 200 nM tRNAHis(25,S). Top: MRPP3 dilution (from 800 to 3.1 nM) and MRPP1/2 constant (800 nM). Bottom: MRPP1/2 dilution (from 800 to 3.1 nM) and MRPP3 constant (50 nM). The molar ratio of tRNA to protein is shown in parentheses. The reactions were incubated for 1 h. (D–F) Gel filtration chromatograms of MRPP1/2:pre-tRNAHis(25,S) prior and after MRPP3 treatment (D). Peaks of interest are numbered, and tRNA and protein content of the peaks are visualized by urea PAGE (E) and SDS-PAGE (F), respectively.
MRPP1/2 enhances the activity of ELAC2 for the majority of mitochondrial tRNAs
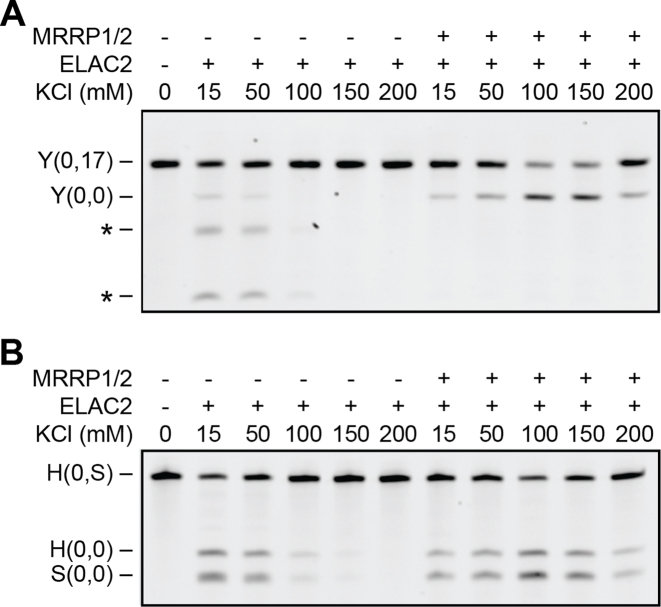
Since MRPP1/2 remained bound to the 5′-processed pre-tRNA generated by mitochondrial RNase P cleavage in both our experiments reported here, we asked whether MRPP1/2 also played a role in the ensuing tRNA 3′-tail removal catalyzed by ELAC2. To this end, we assayed ELAC2 activity on pre-tRNATyr(0,17) both in the absence and presence of MRPP1/2. We found that ELAC2 alone was only active under non-physiological conditions with low ionic strength characterized by a low cleavage rate and a significant amount of mis-cleavage (Figure 3A). In contrast, when MRPP1/2 was present, the tRNA 3′-tail was efficiently cleaved off under all experimental conditions tested, with maximal cleavage efficiency under physiological conditions (100–150 mM KCl) (Figure 3A). The mis-cleavage of pre-tRNATyr(0,17) observed for ELAC2 under low-ionic strength conditions was completely abolished in the presence of MRPP1/2. For the heavy-strand substrate, pre-tRNAHis(0,S), we observed a similar pattern, whereby ELAC2 alone cleaved only under non-physiological conditions at very low ionic strength (Figure 3B). As in the case of pre-tRNATyr, in the presence of MRPP1/2 we observed cleavage of pre-tRNAHis(0,S) under all experimental conditions with maximum activity in the physiological salt range.
Figure 3.
MRPP1/2 supports ELAC2 based 3′-tail removal. (A and B) ELAC2 activity in the absence and presence of 800 nM MRPP1/2 at different salt concentrations (15–200 mM KCl) on pre-tRNATyr(0,17) (Y(0,17)) (A) and pre-tRNAHis(0,S) (H(0,S)) (B). Mis-cleavage on Y(0,17) is indicated by asterisks. The reaction mixes of H(0,S) were incubated for 1 h.
To test if the MRPP1/2 complex played a general role in ELAC2 3′-tRNA processing, we analyzed the effect of MRPP1/2 on ELAC2 activity for all 22 pre-tRNA substrates present in human mitochondria. The results demonstrated that for the large majority of human mitochondrial tRNAs, 3′-tail removal catalyzed by ELAC2 was enhanced in the presence of MRPP1/2 (Table 1 and Supplementary Figure S1). Specifically, once the MRPP1/2 complex was present, the ELAC2-cleavage efficiency was strongly enhanced for 12 tRNA substrates, and moderately enhanced for five tRNA substrates. For the heavy-strand substrate, tRNALeu(UUR), as well as for the light-strand substrates, tRNASer(UCN), tRNAAsn and tRNAGln, there were no significant differences with and without added MRPP1/2 complex. Interestingly, ELAC2 activity on tRNASer(AGY)(0,tRNALeu(CUN)+15) was drastically inhibited in the presence of MRPP1/2. To confirm that it was the MRPP1/2 complex, and not any of the individual proteins, that supported the ELAC2-catalyzed removal of the 3′-tail, we also tested the activity of ELAC2 in the presence of MRPP1, MRPP2 and MRPP1/2 complex. The results confirmed that only the full MRPP1/2 complex supported the ELAC2-cleavage reaction (Supplementary Figure S3).
Table 1. Level of support by MRPP1/2 on ELAC2 mediated 3′-tail removal.
| heavy-strand: | light-strand: | ||
|---|---|---|---|
| tRNA | Effect of MRPP1/2 | tRNA | Effect of MRPP1/2 |
| Phe(0,15) | + | Pro(0,15) | ++ |
| Val(0,15) | ++ | Glu(0,15) | ++ |
| LeuUUR(0,15) | +/- | SerUCN(0,15) | +/- |
| Ile(0,15) | + | Tyr(0,17) | ++ |
| Met(0,15) | + | Cys(0,15) | ++ |
| Trp(0,15) | ++ | Asn(0,15) | +/- |
| Asp(0,15) | ++ | Ala(0,15) | ++ |
| Lys(0,15) | ++ | Gln(0,15) | +/- |
| Gly(0,15) | + | ||
| Arg(0,15) | ++ | ||
| His(0,S)a | ++ | ||
| SerAGY(0,L1+15)b | - - | ||
| LeuCUN(0,15) | + | ||
| Thr(0,15) | ++ |
++strong enhancement of ELAC2 efficiency in presence of MRPP1/2.
+moderate enhancement of ELAC2 efficiency in presence of MRPP1/2.
+/-no significant difference in ELAC2 efficiency in the absence or presence of MRPP1/2.
- -strong decrease of ELAC2 efficiency in presence of MRPP1/2.
aS, SerAGY
bL1, LeuCUN
See also Supplementary Figure S1.
tRNA remains bound to MRPP1/2 after ELAC2 cleavage
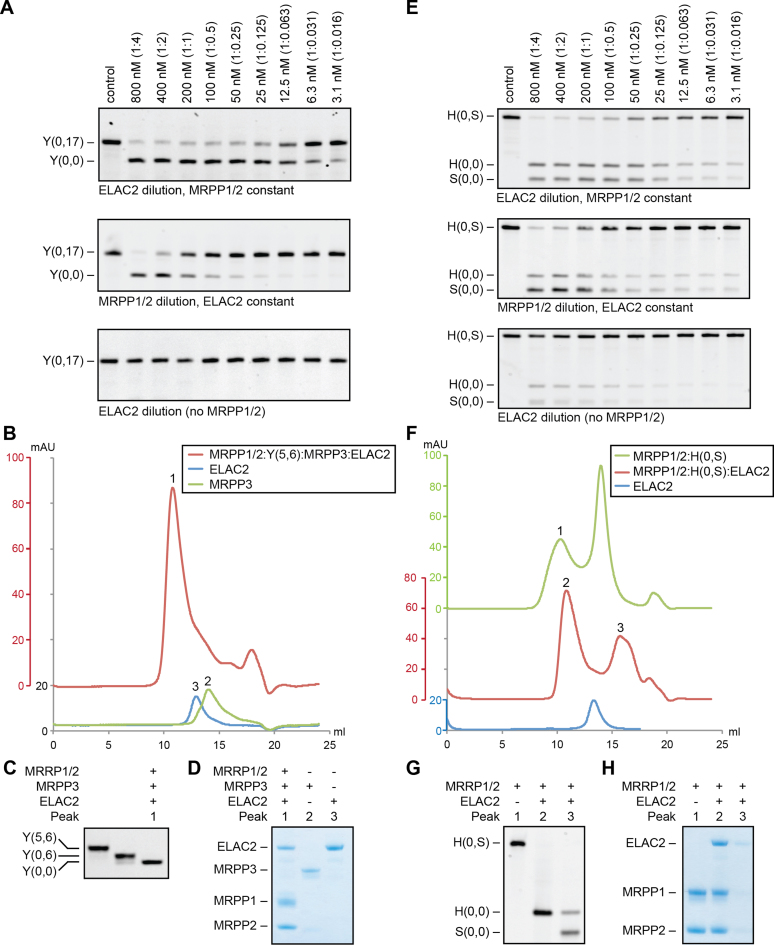
To investigate the interplay between MRPP1/2 and ELAC2 in more detail, we varied the relative concentrations of the components. Specifically, we varied the concentrations of ELAC2 and MRPP1/2 under physiological conditions using pre-tRNATyr(0,17) as a substrate. We found that ELAC2 took part in multiple-turnover reactions, whereas MRPP1/2 only performed a single-turnover reaction in the RNase Z catalyzed tRNA 3′-processing (Figure 4A). Further analysis using gel filtration revealed that the incubation of a preformed MRPP1/2:pre-tRNATyr(5,6) complex with MRPP3 and ELAC2 resulted in the formation of tRNATyr(0,0), which remained attached to MRPP1/2 (Peak 1 in Figure 4B–D). Low amounts of ELAC2 also remained bound to the resulting MRPP1/2:pre-tRNATyr(0,0) complex (Figure 4D and Supplementary Figure S4), which suggests a weak interaction between ELAC2 and MRPP1/2:pre-tRNATyr(0,0) even after 3′-tail removal.
Figure 4.
Role of MRPP1/2 in RNase Z function on tRNATyr and tRNAHis. (A) RNase Z dilution experiments on pre-tRNATyr(0,17) (Y(0,17)). Top: ELAC2 dilution series (800–3.1 nM) in the presence of 800 nM MRPP1/2. Middle: MRPP1/2 dilution series (800–3.1 nM) in the presence of 50 nM ELAC2. Bottom: ELAC2 dilution series (800–3.1 nM) in the absence of MRPP1/2. The reactions were incubated for 1 h. The molar ratio of tRNA to protein is shown in parentheses. (B–D) Gel filtration chromatograms of MRPP1/2:pre-tRNATyr(5,6) after treatment with MRPP3 and ELAC2, as well as of MRPP3 and ELAC2 alone. Peaks of interest are numbered and tRNA and protein content of the peaks is visualized by urea PAGE (C) and SDS-PAGE (D), respectively. For comparison, also pre-tRNATyr(5,6) and pre-tRNATyr(0,6) from Figure 1E are shown in (C). (E) RNase Z dilution experiments on pre-tRNAHis(0,S) (H(0,S)). Reaction condition are as given in (A). (F–H) Gel filtration chromatograms of MRPP1/2:pre-tRNAHis(0,S) prior to and after treatment with ELAC2. Peaks of interest are numbered, and tRNA and protein content of the peaks is visualized by urea PAGE (G) and SDS-PAGE (H), respectively.
A similar pattern was observed for pre-tRNAHis(0,S), where RNase Z processing of the His-Ser(AGY) junction released tRNAHis(0,0) and tRNASer(AGY)(0,0). Here, the ELAC2 activity declined below a 1:1 molar ratio of MRPP1/2 to pre-tRNAHis(0,S) (Figure 4E). ELAC2 was also able to process pre-tRNAHis(0,S) in the absence of MRPP1/2, but with a 125-fold reduction in cleavage efficiency. Moreover, we treated a preformed MRPP1/2:pre-tRNAHis(0,S) complex with ELAC2, and followed this by gel filtration analysis. The resulting chromatogram showed two main peaks corresponding to MRPP1/2:pre-tRNAHis(0,0) and free tRNASer(AGY)(0,0) (Peaks 2 and 3 in Figure 4F–H). Peak 3, which contains free tRNASer(AGY)(0,0), also contained low amounts of pre-tRNAHis(0,0), presumably originating from carry-over from Peak 2. Since the tRNAs studied above were from tRNA clusters we tested if the 3′-processing of a single-gene tRNA follows the same pattern. To this end, we varied the concentrations of ELAC2 and MRPP1/2 under physiological conditions using pre-tRNAArg(0,15) from the heavy strand as a substrate. Once again, we found that ELAC2 took part in multiple-turnover reactions, whereas MRPP1/2 only performed a single-turnover reaction in the RNase Z catalyzed tRNA 3′-processing and that MRPP1/2 very significantly enhanced ELAC2 catalyzed tRNA 3′-processing (Supplementary Figure S2).
The order of tRNA processing is unchanged even in the presence of MRPP1/2
We had now shown for tRNATyr(0,17) and tRNAHis(0,S) that MRPP1/2 was needed for both 5′- and 3′-tRNA maturation. However, the mitochondrial tRNA processing order was established in vivo or using mitochondrial extracts (29–31) and not individually purified proteins in reconstituted complexes as in the work presented here. To test whether the RNA-processing order was altered by the presence of MRPP1/2 during the 5′- and 3′-processing steps, we used pre-tRNATyr(38,17) and pre-tRNAHis(25,S) as substrates in cleavage reactions with mitochondrial RNase P and RNase Z. The results showed that the in vitro maturation of tRNA continued to follow the canonical order (Figure 5A and B). Specifically, RNase P first had to remove the 5′-leader from the pre-tRNA before ELAC2 was able to remove the 3′-tail. This held true even in the presence of the MRPP1/2 complex and during prolonged incubation (90 min). However, for tRNASer(AGY)(0,tRNALeu(CUN)+15), we found that ELAC2, in the presence of MRPP1/2, removed the 15 nucleotide long 3′ tail from tRNALeu(CUN) even in presence of the 5′ uncleaved tRNASer(AGY) (Supplementary Figure S1D).
Figure 5.
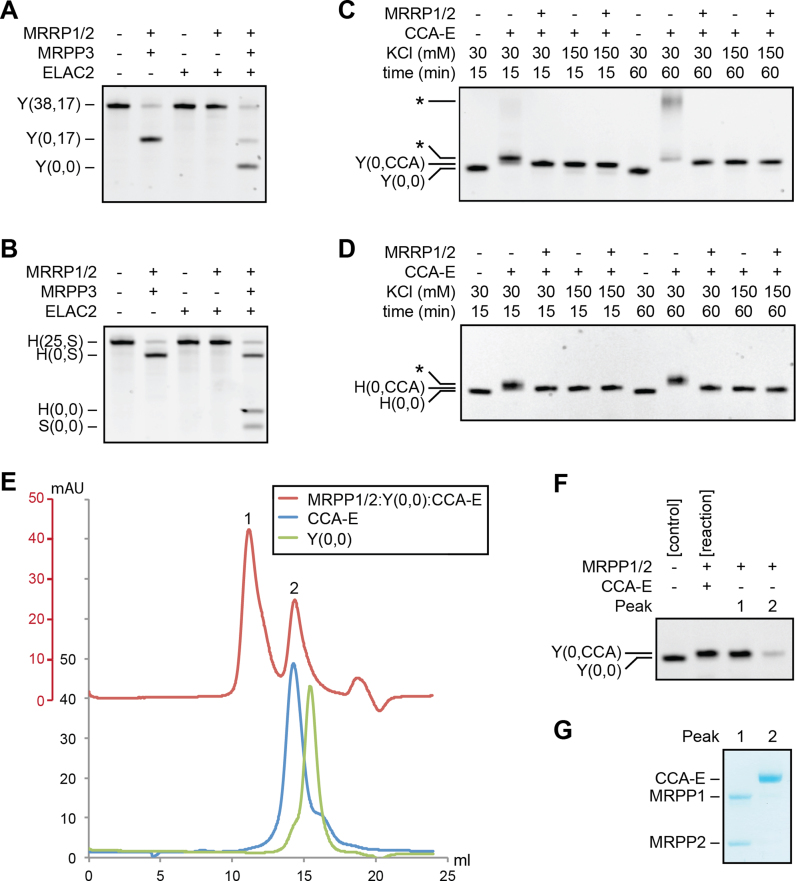
Mitochondrial tRNA processing order and CCA-adding enzyme modify MRPP1/2 bound tRNA. (A and B) Processing order of RNase P and RNase Z on pre-tRNATyr (38,17) (Y (38,17)) (A) and pre-tRNAHis(25,S) (H(25,S)) (B). For activity measurements, 800 nM MRPP1/2, 50 nM MRPP3 and 50 nM ELAC2, or combinations, were used. (C and D) Activity of CCA-adding enzyme (CCA-E, 50 nM) on pre-tRNATyr(0,0) (Y(0,0)) (C) and pre-tRNAHis(0,0) (H(0,0)) (D) in the absence and presence of 800 nM MRPP1/2. Hyper-modified tRNA is indicated by asterisks. (E–G) Gel filtration chromatograms of MRPP1/2:pre-tRNATyr(0,0) after incubation with CCA-adding enzyme and rNTPs (E). Peaks of interested are numbered, and tRNA and protein content of the peaks is visualized by urea PAGE (F) and SDS-PAGE (G), respectively.
MRPP1/2 retains tRNA during and after 3′-CCA addition
After establishing that MRPP1/2 retained the 5′- and 3′-processed pre-tRNA resulting from the cleavage reactions by MRPP3 and ELAC2, we continued by analyzing the tRNA-binding capacity of MRPP1/2. To this end, we reconstituted MRPP1/2:pre-tRNA complexes using pre-tRNATyr samples of different processing status (pre-tRNATyr(5,6), pre-tRNATyr(0,6), pre-tRNATyr(0,0) and pre-tRNATyr(0,CCA)). Using gel filtration, we found that MRPP1/2 was able to form stable complexes with all four tRNA maturation intermediates (Supplementary Figure S5). Since the MRPP1/2-complex was able to bind pre-tRNATyr(0,CCA), we asked whether MRPP1/2 also plays a role in the ensuing CCA-addition reaction catalyzed by the CCA-adding enzyme. To assess this, we analyzed the activity of the CCA-adding enzyme on pre-tRNATyr(0,0) and pre-tRNAHis(0,0) in the absence and presence of MRPP1/2 at low (30 mM) or physiological (150 mM) salt concentrations (Figure 5C and D). We found that at non-physiological, low salt concentrations, the CCA-adding enzyme alone was error-prone; specifically, after 15 min of incubation, more nucleotides than the expected CCA sequence were added to the pre-tRNA substrate. Furthermore, after 60 min incubation, severe hyperactivation with long 3′-tails for pre-tRNATyr was observed. In the presence of MRPP1/2, this hyperactivity was completely abolished. At physiological salt concentrations, there was no difference in the efficiency and fidelity of tRNA 3′-CCA addition regardless of presence or absence of the MRPP1/2 complex.
To test if the 3′-CCA addition was performed on MRPP1/2 bound pre-tRNA substrate, we treated a reconstituted MRPP1/2:pre-tRNATyr(0,0) complex with CCA-adding enzyme in the presence of ribonucleotides, and applied the reaction mixture to a gel filtration column. A stable complex between pre-tRNATyr(0,CCA) and the MRPP1/2 complex could be unambiguously confirmed (Peak 1 in Figure 5E–G). The CCA-adding enzyme in Peak 2 eluted in the same volume as free CCA-adding enzyme. There was a small amount of pre-tRNATyr(0,CCA) visible also in Peak 2 (Peak 2 in Figure 5E–G), which is presumably a tailing effect from Peak 1. In summary, these results showed that the CCA-adding enzyme can perform multiple turnover reactions on mitochondrial tRNA substrates bound to MRPP1/2, leaving behind a stable reaction product of 5′- and 3′-mature pre-tRNA still bound to the MRPP1/2 complex. Further analysis of the three core processing reactions on both pre-tRNATyr (38,17) and pre-tRNAHis(25,S)—including RNase P cleavage, RNase Z cleavage and 3′-CCA addition—confirmed that the presence of the CCA-adding enzyme did not enable MRPP1/2 to perform multiple turnover reactions (Supplementary Figure S6).
DISCUSSION
Here, we have shown that the MRPP1/2 complex is not only an essential component of the mitochondrial RNase P complex (12,19), but that it is also important for the ensuing tRNA 3′-tail removal by ELAC2, at least in vitro and for the majority of human mitochondrial tRNAs. Furthermore, for two representative tRNAs, 3′-CCA addition catalyzed by the CCA-adding enzyme can be performed while the substrate tRNA is bound to the MRPP1/2 complex. The MRPP1/2 complex thereby plays an unexpectedly versatile role as a processing platform in human mitochondrial tRNA maturation.
To investigate the role of MRPP1/2 in the core processing steps of human mitochondrial tRNA maturation, we selected two representative tRNA samples: tRNATyr from the light-strand and tRNAHis from the heavy-strand. For these two tRNAs, the MRPP1/2 complex of RNase P binds stoichiometrically to substrate tRNA, enables cleavage by sub-stoichiometric amounts of MRPP3 and then retains the 5′-mature tRNA product for 3′-tRNA processing (Figures 1 and 2). In the subsequent 3′-processing step, the MRPP1/2 complex ensures efficient and specific cleavage by ELAC2 under all conditions tested, whereas ELAC2 by itself lacked both efficiency and specificity (Figure 3). Similar to our observations regarding the RNase P reaction, MRPP1/2 binds the tRNA substrate stoichiometrically, and the tRNA product remains bound to MRPP1/2 as ELAC2 performs multiple turnover reactions (Figure 4). Even though the presence of MRPP1/2 drastically enhances the activity of ELAC2, the tRNA maturation still follows the canonical order in which the 5′-processing precedes the 3′-processing (Figure 5A and B).
We then expanded our analysis to all mitochondrial tRNAs, showing that under physiological salt conditions, 17 of the 22 tRNAs required MRPP1/2 for efficient and specific 3′-cleavage (Table 1 and Supplementary Figure S1). Hence, for the majority of human mitochondrial tRNAs, the MRPP1/2 complex is a general factor required for both 5′- and 3′-tRNA processing. To our knowledge, all previous studies have used artificially low-ionic strength buffers to investigate ELAC2-based 3′-cleavage (22,32–34). We note that conditions of low ionic strength promote partial unfolding of tRNA (35), which may explain the unspecific cleavage that we observe for tRNATyr (Figure 3A). Moreover, under low ionic-strength conditions, the specificity and overall efficiency of ELAC2 is greatly reduced relative to its activity under physiological conditions when supplemented with MRPP1/2.
Interestingly, there are four mitochondrial tRNAs for which there was no difference in the 3′-processing efficiency in the absence and presence of MRPP1/2. These include the heavy-strand substrate tRNALeu(UUR) as well as the light-strand substrates tRNASer(UCN), tRNAAsn and tRNAGln. Thus, either the MRPP1/2 complex is not required to support the ELAC2-based 3′-processing in these few cases, or our experimental setup is not sensitive enough to allow visualization of this difference (e.g. a time issue). For tRNASer(UCN) this might be explained by the short D-loop and the slightly extended anti-codon stem but most probably not dependent on the lack of N1-methylation of A/G at position 9 since ELAC2-processing of tRNAMet, which contains a C at position 9, is strongly enhanced by MRPP1/2. There is a trend that the unsupported light-strand tRNA substrates tRNASer(UCN), tRNAAsn and tRNAGln contain G as the discriminator base. This trend is, however, contradicted by the observation that tRNATrp, which also contains a G as the discriminator base, is strongly supported by MRPP1/2 during 3′-processing. The experiments in this study were performed in the presence of SAM to ensure that the tRNA substrates were not bound to MRPP1/2 because of a lack of methylation at position 9. Moreover, the tRNA processing efficiency of ELAC2 does not correlate with the location of the tRNA in the genome.
We characterized the processing mechanism of the tightly packed tRNA cluster His-Ser(AGY)-Leu(CUN) from the heavy-strand, in which the three tRNAs directly follow each other. Our data are consistent with previously reported observations that pre-tRNASer(AGY) is released indirectly following ELAC2-processing of pre-tRNAHis and cleavage of tRNALeu(CUN) by RNase P (30) (Figures 2C and 3B). The observation that MRPP1/2 does not bind tRNASer(AGY) during gel filtration experiments leads us to postulate that MRPP1/2 controls the processing mechanism of the His-Ser(AGY)-Leu(CUN) cluster as it recognizes tRNALeu(CUN) and tRNAHis, and thereby supports the cleavage reactions catalyzed by mitochondrial RNase P and RNase Z, respectively, the consequence of which enables the release of tRNASer(AGY). It is notable in this context that tRNASer(AGY) exhibits a truncated cloverleaf structure with absence of the D-arm and an extended anticodon stem (36). This unusual arrangement might be responsible for the failure of MRPP1/2 to recognize tRNASer(AGY), providing a potential hint to the importance of the D-arm for tRNA interaction with the MRPP1/2 complex. Interestingly, processing of the Ser(AGY)-Leu(CUN) junction by ELAC2 is possible, but is severely inhibited in the presence of MRPP1/2 (Supplementary Figure S1), potentially through the binding of MRPP1/2 to tRNALeu(CUN). We further observed that in the presence of MRPP1/2, ELAC2 was capable of removing the 3′-tail from tRNALeu(CUN) even in the presence of tRNASer(AGY) at its 5′-end, yet with a significantly reduced cleavage rate (Supplementary Figures S1C and D). This peculiar observation contradicts the canonical processing order in which RNase P precedes RNase Z processing (24,30).
The last core processing step, 3′-CCA addition by the CCA-adding enzyme, is required for all human mitochondrial tRNAs. In this study, we have shown that the CCA-adding enzyme can perform the 3′-CCA addition while the tRNA substrate remains bound to MRPP1/2 (Figure 5). In contrast to MRPP3 and ELAC2, the presence of MRPP1/2 is not essential for the activity of the CCA-adding enzyme. Importantly, it has been reported previously that the mammalian mitochondrial CCA-adding enzyme does not strictly recognize the conserved T-loop sequence, which is one of the major recognition element for most CCA-adding enzymes from other organisms (28). This altered recognition mechanism may explain why the human mitochondrial CCA-adding enzyme can process MRPP1/2-bound tRNA substrates. Still, we were surprised to see that 3′-CCA addition did not release the tRNA from MRPP1/2 (Figure 5E–G) even if the affinity is slightly lower for tRNATyr with CCA addition than without (Supplementary Figure S5D). The question remains as to how and when does the tRNA leave the MRPP1/2 complex. To answer this question, the roles of aminoacyltransferases, or tRNA-modifying enzymes, will need to be evaluated in future experiments.
Our results allow us to propose an interaction mode between MRPP1/2 and its pre-tRNA substrates. Firstly, the recognition of the pre-tRNA substrate does not seem to involve recognition of the 5′- and 3′-ends. This appears reasonable from a steric point of view, because only then would the termini be accessible for other processing enzymes. Further, we speculated above that the D-arm may be crucial for tRNA-recognition by MRPP1/2. Possibly, MRPP1/2 directs the temporal order of processing events while stabilizing the structurally diverse mitochondrial tRNAs until fully processed.
For mitochondrial RNase P-based removal of the 5′-leader, we have previously suggested that MRPP1/2 induces, and stabilizes, structural rearrangements in MRPP3 that lead to its activation (19). In addition to protein–RNA interactions, this is expected to involve specific protein–protein interactions. Moreover, human mitochondrial ELAC2 is not the only RNase Z whose activity is influenced by other proteins. Yeast La homologous protein 1 (Lhp1p) is required for tRNA 3′-processing by acting as an RNA chaperone that facilitates correct folding of certain pre-tRNAs to allow subsequent endonucleolytic processing (37,38). Considering the structural complexity of human mitochondrial tRNAs, including intricate modification patterns (39,40), the need for a stable processing platform, such as that offered by MRPP1/2, would seem advantageous and reasonable. It remains to be investigated whether other maturation enzymes, such as methyltransferases and aminoacyltransferases, are also able to use MRPP1/2-bound tRNA as substrate. Noteworthy, the interaction of MRPP1/2 with ELAC2 and CCA-adding enzyme is supported by the complex formation of MRPP1 with ELAC2 and CCA-adding enzyme using affinity-capture mass spectrometry (41) as well as of MRPP2 with ELAC2 by co-fractionation (42). Further, ELAC2 and RNase P components co-localizes in vivo in the mitochondrial DNA nucleoids (10).
Mutations in MRPP1/2 have been linked to a range of diseases (43–46). Until now, the effects of mutations have been placed in relation to either the function of MRPP2 (which also performs other metabolic functions), or to an influence on mitochondrial RNase P function (24,43–45,47,48). Additionally, in previous in vivo work, it has been found that patterns of mitochondrial RNA processing intermediates observed when MRPP1 or MRPP2 were knocked down or out clearly differed from patterns observed when MRPP3 was knocked down or out (45,47,49). Though this was clearly observed and noted as putative distinct functions for MRPP1/2 in mitochondrial RNA processing outside their role in the RNase P complex, there was, until now, no molecular basis for this aberrant RNA processing.
The results presented here show that the MRPP1/2 complex plays an important role in downstream mitochondrial tRNA processing and this should be carefully considered in the future when evaluating the effects of MRPP1/2 mutations on 5′-cleavage, 3′-cleavage, 3′-CCA addition and most probably other steps along the tRNA maturation pathway.
Supplementary Material
Footnotes
Present address: Linda Reinhard, III. Department of Medicine, University Medical Center Hamburg-Eppendorf, 20246 Hamburg, Germany.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Swedish Research Council [2011–6510, 2013–5882 to B.M.H.]. Funding for open access charge: Vetenskapsrådet [2011-6510, 2013-5882].
Conflict of interest statement. None declared.
REFERENCES
- 1. Hällberg B.M., Larsson N.G.. Making proteins in the powerhouse. Cell Metab. 2014; 20:226–240. [DOI] [PubMed] [Google Scholar]
- 2. Kauppila T.E., Kauppila J.H., Larsson N.G.. Mammalian mitochondria and aging: an update. Cell Metab. 2017; 25:57–71. [DOI] [PubMed] [Google Scholar]
- 3. Brown T.A., Tkachuk A.N., Shtengel G., Kopek B.G., Bogenhagen D.F., Hess H.F., Clayton D.A.. Superresolution fluorescence imaging of mitochondrial nucleoids reveals their spatial range, limits, and membrane interaction. Mol. Cell. Biol. 2011; 31:4994–5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kukat C., Wurm C.A., Spåhr H., Falkenberg M., Larsson N.G., Jakobs S.. Super-resolution microscopy reveals that mammalian mitochondrial nucleoids have a uniform size and frequently contain a single copy of mtDNA. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:13534–13539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Anderson S., Bankier A.T., Barrell B.G., de Bruijn M.H., Coulson A.R., Drouin J., Eperon I.C., Nierlich D.P., Roe B.A., Sanger F. et al. . Sequence and organization of the human mitochondrial genome. Nature. 1981; 290:457–465. [DOI] [PubMed] [Google Scholar]
- 6. Ojala D., Montoya J., Attardi G.. tRNA punctuation model of RNA processing in human mitochondria. Nature. 1981; 290:470–474. [DOI] [PubMed] [Google Scholar]
- 7. Smeitink J., van den Heuvel L., DiMauro S.. The genetics and pathology of oxidative phosphorylation. Nat. Rev. Genet. 2001; 2:342–352. [DOI] [PubMed] [Google Scholar]
- 8. Antonicka H., Sasarman F., Nishimura T., Paupe V., Shoubridge E.A.. The mitochondrial RNA-binding protein GRSF1 localizes to RNA granules and is required for posttranscriptional mitochondrial gene expression. Cell Metab. 2013; 17:386–398. [DOI] [PubMed] [Google Scholar]
- 9. Jourdain A.A., Koppen M., Wydro M., Rodley C.D., Lightowlers R.N., Chrzanowska-Lightowlers Z.M., Martinou J.C.. GRSF1 regulates RNA processing in mitochondrial RNA granules. Cell Metab. 2013; 17:399–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bogenhagen D.F., Martin D.W., Koller A.. Initial steps in RNA processing and ribosome assembly occur at mitochondrial DNA nucleoids. Cell Metab. 2014; 19:618–629. [DOI] [PubMed] [Google Scholar]
- 11. Bruni F., Lightowlers R.N., Chrzanowska-Lightowlers Z.M.. Human mitochondrial nucleases. FEBS J. 2017; 284:1767–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Holzmann J., Frank P., Loffler E., Bennett K.L., Gerner C., Rossmanith W.. RNase P without RNA: identification and functional reconstitution of the human mitochondrial tRNA processing enzyme. Cell. 2008; 135:462–474. [DOI] [PubMed] [Google Scholar]
- 13. Vilardo E., Nachbagauer C., Buzet A., Taschner A., Holzmann J., Rossmanith W.. A subcomplex of human mitochondrial RNase P is a bifunctional methyltransferase–extensive moonlighting in mitochondrial tRNA biogenesis. Nucleic Acids Res. 2012; 40:11583–11593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Helm M., Brule H., Degoul F., Cepanec C., Leroux J.P., Giege R., Florentz C.. The presence of modified nucleotides is required for cloverleaf folding of a human mitochondrial tRNA. Nucleic Acids Res. 1998; 26:1636–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Helm M., Giege R., Florentz C.. A Watson-Crick base-pair-disrupting methyl group (m1A9) is sufficient for cloverleaf folding of human mitochondrial tRNALys. Biochemistry. 1999; 38:13338–13346. [DOI] [PubMed] [Google Scholar]
- 16. Shafqat N., Marschall H.U., Filling C., Nordling E., Wu X.Q., Bjork L., Thyberg J., Martensson E., Salim S., Jornvall H. et al. . Expanded substrate screenings of human and Drosophila type 10 17beta-hydroxysteroid dehydrogenases (HSDs) reveal multiple specificities in bile acid and steroid hormone metabolism: characterization of multifunctional 3alpha/7alpha/7beta/17beta/20beta/21-HSD. Biochem. J. 2003; 376:49–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yang S.Y., He X.Y., Isaacs C., Dobkin C., Miller D., Philipp M.. Roles of 17beta-hydroxysteroid dehydrogenase type 10 in neurodegenerative disorders. J. Steroid Biochem. Mol. Biol. 2014; 143:460–472. [DOI] [PubMed] [Google Scholar]
- 18. Rossmanith W. Of P and Z: mitochondrial tRNA processing enzymes. Biochim. Biophys. Acta. 2012; 1819:1017–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Reinhard L., Sridhara S., Hällberg B.M.. Structure of the nuclease subunit of human mitochondrial RNase P. Nucleic Acids Res. 2015; 43:5664–5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Melino S., Capo C., Dragani B., Aceto A., Petruzzelli R.. A zinc-binding motif conserved in glyoxalase II, beta-lactamase and arylsulfatases. Trends Biochem. Sci. 1998; 23:381–382. [DOI] [PubMed] [Google Scholar]
- 21. Tavtigian S.V., Simard J., Teng D.H., Abtin V., Baumgard M., Beck A., Camp N.J., Carillo A.R., Chen Y., Dayananth P. et al. . A candidate prostate cancer susceptibility gene at chromosome 17p. Nat. Genet. 2001; 27:172–180. [DOI] [PubMed] [Google Scholar]
- 22. Takaku H., Minagawa A., Takagi M., Nashimoto M.. A candidate prostate cancer susceptibility gene encodes tRNA 3′ processing endoribonuclease. Nucleic Acids Res. 2003; 31:2272–2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Takahashi M., Takaku H., Nashimoto M.. Regulation of the human tRNase ZS gene expression. FEBS Lett. 2008; 582:2532–2536. [DOI] [PubMed] [Google Scholar]
- 24. Brzezniak L.K., Bijata M., Szczesny R.J., Stepien P.P.. Involvement of human ELAC2 gene product in 3′ end processing of mitochondrial tRNAs. RNA Biol. 2011; 8:616–626. [DOI] [PubMed] [Google Scholar]
- 25. Rossmanith W. Localization of human RNase Z isoforms: dual nuclear/mitochondrial targeting of the ELAC2 gene product by alternative translation initiation. PLoS One. 2011; 6:e19152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mineri R., Pavelka N., Fernandez-Vizarra E., Ricciardi-Castagnoli P., Zeviani M., Tiranti V.. How do human cells react to the absence of mitochondrial DNA. PLoS One. 2009; 4:e5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Augustin M.A., Reichert A.S., Betat H., Huber R., Mörl M., Steegborn C.. Crystal structure of the human CCA-adding enzyme: insights into template-independent polymerization. J. Mol. Biol. 2003; 328:985–994. [DOI] [PubMed] [Google Scholar]
- 28. Nagaike T., Suzuki T., Tomari Y., Takemoto-Hori C., Negayama F., Watanabe K., Ueda T.. Identification and characterization of mammalian mitochondrial tRNA nucleotidyltransferases. J. Biol. Chem. 2001; 276:40041–40049. [DOI] [PubMed] [Google Scholar]
- 29. Rossmanith W., Tullo A., Potuschak T., Karwan R., Sbisa E.. Human mitochondrial tRNA processing. J. Biol. Chem. 1995; 270:12885–12891. [DOI] [PubMed] [Google Scholar]
- 30. Rossmanith W. Processing of human mitochondrial tRNA(Ser(AGY))GCU: a novel pathway in tRNA biosynthesis. J. Mol. Biol. 1997; 265:365–371. [DOI] [PubMed] [Google Scholar]
- 31. Rackham O., Busch J.D., Matic S., Siira S.J., Kuznetsova I., Atanassov I., Ermer J.A., Shearwood A.M., Richman T.R., Stewart J.B. et al. . Hierarchical RNA processing is required for mitochondrial ribosome assembly. Cell Rep. 2016; 16:1874–1890. [DOI] [PubMed] [Google Scholar]
- 32. Levinger L., Oestreich I., Florentz C., Mörl M.. A pathogenesis-associated mutation in human mitochondrial tRNA(Leu(UUR)) leads to reduced 3′-end processing and CCA addition. J. Mol. Biol. 2004; 337:535–544. [DOI] [PubMed] [Google Scholar]
- 33. Yan H., Zareen N., Levinger L.. Naturally occurring mutations in human mitochondrial pre-tRNASer(UCN) can affect the transfer ribonuclease Z cleavage site, processing kinetics, and substrate secondary structure. J. Biol. Chem. 2006; 281:3926–3935. [DOI] [PubMed] [Google Scholar]
- 34. Fiedler M., Rossmanith W., Wahle E., Rammelt C.. Mitochondrial poly(A) polymerase is involved in tRNA repair. Nucleic Acids Res. 2015; 43:9937–9949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Urbanke C., Romer R., Maass G.. Tertiary structure of tRNAPhe (yeast): kinetics and electrostatic repulsion. Eur. J. Biochem. 1975; 55:439–444. [DOI] [PubMed] [Google Scholar]
- 36. de Bruijn M.H., Klug A.. A model for the tertiary structure of mammalian mitochondrial transfer RNAs lacking the entire ‘dihydrouridine’ loop and stem. EMBO J. 1983; 2:1309–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yoo C.J., Wolin S.L.. The yeast La protein is required for the 3′ endonucleolytic cleavage that matures tRNA precursors. Cell. 1997; 89:393–402. [DOI] [PubMed] [Google Scholar]
- 38. Chakshusmathi G., Kim S.D., Rubinson D.A., Wolin S.L.. A La protein requirement for efficient pre-tRNA folding. EMBO J. 2003; 22:6562–6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Suzuki T., Nagao A., Suzuki T.. Human mitochondrial tRNAs: biogenesis, function, structural aspects, and diseases. Annu. Rev. Genet. 2011; 45:299–329. [DOI] [PubMed] [Google Scholar]
- 40. Suzuki T., Suzuki T.. A complete landscape of post-transcriptional modifications in mammalian mitochondrial tRNAs. Nucleic Acids Res. 2014; 42:7346–7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Huttlin E.L., Bruckner R.J., Paulo J.A., Cannon J.R., Ting L., Baltier K., Colby G., Gebreab F., Gygi M.P., Parzen H. et al. . Architecture of the human interactome defines protein communities and disease networks. Nature. 2017; 545:505–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wan C., Borgeson B., Phanse S., Tu F., Drew K., Clark G., Xiong X., Kagan O., Kwan J., Bezginov A. et al. . Panorama of ancient metazoan macromolecular complexes. Nature. 2015; 525:339–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vilardo E., Rossmanith W.. Molecular insights into HSD10 disease: impact of SDR5C1 mutations on the human mitochondrial RNase P complex. Nucleic Acids Res. 2015; 43:5112–5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Falk M.J., Gai X., Shigematsu M., Vilardo E., Takase R., McCormick E., Christian T., Place E., Pierce E.A., Consugar M. et al. . A novel HSD17B10 mutation impairing the activities of the mitochondrial RNase P complex causes X-linked intractable epilepsy and neurodevelopmental regression. RNA Biol. 2016; 13:477–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Metodiev M.D., Thompson K., Alston C.L., Morris A.A., He L., Assouline Z., Rio M., Bahi-Buisson N., Pyle A., Griffin H. et al. . Recessive mutations in TRMT10C cause defects in mitochondrial RNA processing and multiple respiratory chain deficiencies. Am J. Hum. Genet. 2016; 98:993–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kopajtich R., Mayr J.A., Prokisch H.. Analysis of mitochondrial RNA-processing defects in patient-derived tissues by qRT-PCR and RNAseq. Methods Mol. Biol. 2017; 1567:379–390. [DOI] [PubMed] [Google Scholar]
- 47. Sanchez M.I., Mercer T.R., Davies S.M., Shearwood A.M., Nygard K.K., Richman T.R., Mattick J.S., Rackham O., Filipovska A.. RNA processing in human mitochondria. Cell Cycle. 2011; 10:2904–2916. [DOI] [PubMed] [Google Scholar]
- 48. Deutschmann A.J., Amberger A., Zavadil C., Steinbeisser H., Mayr J.A., Feichtinger R.G., Oerum S., Yue W.W., Zschocke J.. Mutation or knock-down of 17beta-hydroxysteroid dehydrogenase type 10 cause loss of MRPP1 and impaired processing of mitochondrial heavy strand transcripts. Hum. Mol. Genet. 2014; 23:3618–3628. [DOI] [PubMed] [Google Scholar]
- 49. Wolf A.R., Mootha V.K.. Functional genomic analysis of human mitochondrial RNA processing. Cell Rep. 2014; 7:918–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.