Abstract
Kallikrein related peptidase 8 (KLK8; also called neuropsin) is a serine protease that plays distinct roles in the skin and hippocampus. In the skin, KLK8 influences keratinocyte proliferation and desquamation, and activates antimicrobial peptides in sweat. In the hippocampus, KLK8 affects memory acquisition. Here, we examined the evolution of KLK8 in mammals and discovered that, out of 70 placental mammals, KLK8 is exclusively lost in three independent fully-aquatic lineages, comprising dolphin, killer whale, minke whale, and manatee. In addition, while the sperm whale has an intact KLK8 reading frame, the gene evolves neutrally in this species. We suggest that the distinct functions of KLK8 likely became obsolete in the aquatic environment, leading to the subsequent loss of KLK8 in several fully-aquatic mammalian lineages. First, the cetacean and manatee skin lacks sweat glands as an adaptation to the aquatic environment, which likely made the epidermal function of KLK8 obsolete. Second, cetaceans and manatees exhibit a proportionally small hippocampus, which may have rendered the hippocampal functions of KLK8 obsolete. Together, our results shed light on the genomic changes that correlate with skin and neuroanatomical differences of aquatic mammals, and show that even pleiotropic genes can be lost during evolution if an environmental change nullifies the need for the different functions of such genes.
Keywords: aquatic mammals, pleiotropy, hippocampus, gene loss
Introduction
Kallikrein related peptidases (KLKs) are serine proteases with diverse physiological functions in different tissues (Lawrence et al. 2010; Prassas et al. 2015). KLKs are secreted as inactive pro-enzymes and are activated by cleavage of the propeptide. KLKs can autoactivate themselves and activate other KLKs, resulting in regulatory cascades and feedback loops, which is also referred to as the KLK activome (Lundwall and Brattsand 2008; Sotiropoulou et al. 2009). In addition to cleaving proteases of the KLK and other families, the substrates of the different KLKs include a wide range of extracellular matrix proteins, receptors, cytokines, hormones, and enzymes (Lawrence et al. 2010). The substrates determine the diverse physiological roles of KLKs that range from semen liquefaction and sperm motility, skin desquamation, hypertension and innate immunity to neuronal function (Lawrence et al. 2010; Lundwall and Brattsand 2008; Prassas et al. 2015). Impaired regulation of KLK genes has been linked to a variety of diseases, including Netherton syndrome, amelogenesis imperfecta, multiple sclerosis, neurodegenerative disorders, and Prostate cancer, making KLKs promising therapeutic targets (Lundwall and Brattsand 2008; Prassas et al. 2015).
KLK8 (kallikrein related peptidase 8), one member of the KLK family, is mainly expressed in the skin and in the brain (Yoshida 2010) and is involved in different physiological functions in both organs. This dual functionality of KLK8 is also reflected by observations that SNPs in human KLK8 are associated with bipolar disorder (Izumi et al. 2008) and that KLK8 is upregulated in psoriasis (Kuwae et al. 2002). In the epidermis, KLK8 is highly expressed in the suprabasal layers and KLK8 secretion increases 14-fold after Ca2+-induced corneocyte formation (Eissa et al. 2011; Komatsu et al. 2005, 2006; Kuwae et al. 2002; Yoshida 2010). KLK8 is an activator of the inactive proenzyme form of KLK1 and KLK11 (Eissa et al. 2011) and influences keratinocyte proliferation and desquamation likely by cleaving corneodesmosomes in conjunction with other kallikreins (Kishibe et al. 2007; Ovaere et al. 2009). In addition, KLK8 is also contained in sweat where it activates antimicrobial peptides (Eissa et al. 2011).
In the brain, KLK8 is expressed in the cornu ammonis areas of the hippocampus and the amygdala (Attwood et al. 2011; Chen et al. 1995), regions of the brain that are involved in stress, learning and memory. KLK8 cleaves a number of synaptic proteins that play a role in learning and stress response, such as ephrin type-B receptor 2 (EPHB2), neural L1 cell adhesion molecule (L1CAM), and neuregulin 1 (NRG1; Attwood et al. 2011; Matsumoto-Miyai et al. 2003; Nakamura et al. 2006; Tamura et al. 2012). The cleavage of L1CAM is an important step in long-term potentiation (the persistent increase in synaptic strength after neuronal stimulation) in the Schaffer axon collateral pathway. Consistent with the L1CAM-cleaving activity of KLK8, mice in which the KLK8 gene has been knocked out have an increased number of immature synapses and exhibit an impaired early-phase long-term potentiation, which affects memory acquisition in comparison to wildtype mice (Attwood et al. 2011; Hirata et al. 2001; Nakamura et al. 2006; Tamura et al. 2006, 2012). KLK8 knockout in mice also has a protective effect against depressive-like behavior and anxiety induced by chronic stress (Attwood et al. 2011; Chang et al. 2016). Thus, KLK8 is a pleiotropic protease with distinct functions in the hippocampus and in the skin.
Here, we investigated the evolution of KLK8 in 70 placental mammals. Our genomic and transcriptomic analysis shows that the pleiotropic KLK8 gene is repeatedly lost in three aquatic mammalian lineages. We suggest that special circumstances in the aquatic environment rendered the two key functions of KLK8 obsolete, which led to the subsequent repeated loss of this gene.
Materials and Methods
Detection and Validation of KLK8 Loss in Aquatic Mammals
We used an alignment between the human hg38 genome assembly and 69 nonhuman placental mammals (Sharma and Hiller 2017) and inspected the coding region of KLK8. Supplementary table 1, Supplementary Material online lists the assemblies of these placental mammals.
Given the fact that genome alignment methods are not aware of the reading frame and splice sites of coding exons, it is possible that an alternative sequence alignment, where the position of a splice site or the position of insertions/deletions is shifted, results in an intact reading frame (Sharma et al. 2016). To rule out that alternative sequence alignments reveal intact exons in the aquatic mammals, we re-aligned every exon with CESAR, a Hidden Markov Model based method that incorporates reading frame and splice sites and is trained to find an intact exon alignment whenever possible (Sharma et al. 2016), using default parameters (https://github.com/hillerlab/CESAR; last accessed November 20, 2017). All inactivating mutations that are shown in figures 1 and 3 were not only observed in the genome alignment but consistently in the CESAR alignment, excluding the possibility that alternative alignments show an intact exon. We only observed two exceptions: The mutations in dolphin exon 2 and manatee exon 5 were only observed in the genome alignment but not in the CESAR alignment (supplementary fig. 1, Supplementary Material online). These two mutations were not considered further, which results in a conservative estimation of the total number of inactivating mutations.
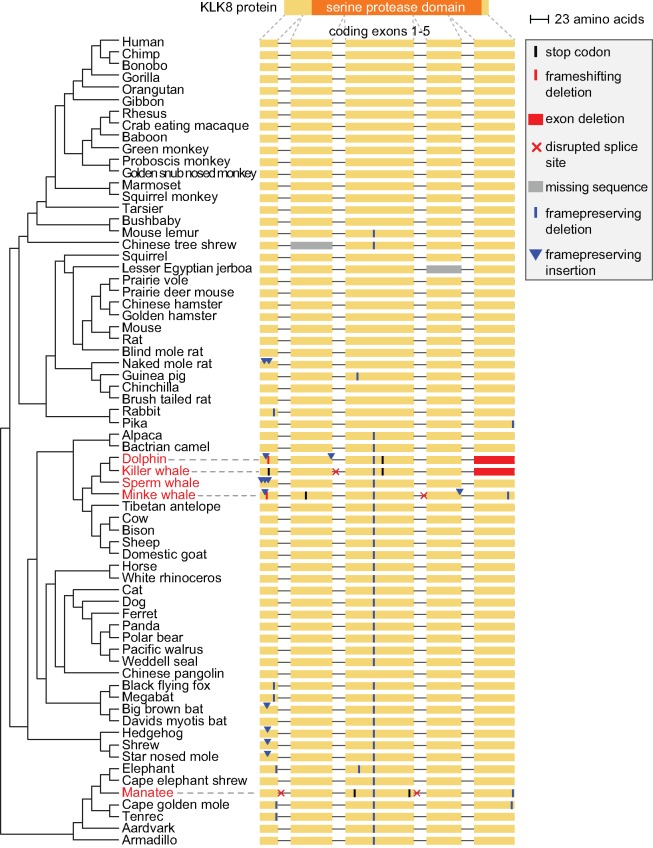
Fig. 1.
—KLK8 is exclusively lost in several aquatic lineages. The phylogenetic tree of sequenced placental mammals is shown on the left. The five coding exons of KLK8 are shown on the right, together with gene-inactivating mutations and reading-frame-preserving insertions and deletions. Inactivating mutations are only observed in the dolphin, killer whale, minke whale, and manatee. These mutations comprise different types (frameshifting deletions, stop codon mutations, splice site disrupting mutations, and exon deletions), are scattered across the entire gene and disrupt the conserved serine protease domain (Pfam PF00089) that is encoded by exons 2–5. Manual inspection shows that the reading frame of KLK8 in all other species is 100% intact; however, KLK8 evolves neutrally in the sperm whale. Exons but not introns are drawn to scale.
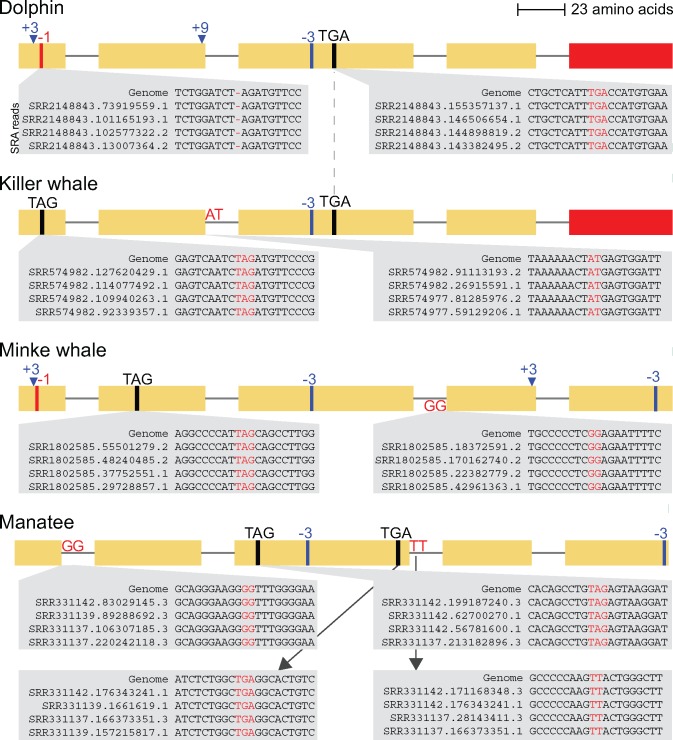
Fig. 3.
—Validation of gene-inactivating mutations in three independent KLK8 loss lineages. The exon–intron structure of KLK8 of the four species that lost this gene is recapitulated from figure 1, showing the mutations in detail. All gene-inactivating mutations were validated by unassembled genome sequencing reads stored in the Short Read Archive, which is shown by insets for 10 of the 12 mutations. The stop codon mutation in exon 3 and the exon 5 deletion are shared between dolphin and killer whale, suggesting that KLK8 loss happened in the common ancestor of both species. However, based on the presence of an intact reading frame in the sperm whale and no shared mutation between dolphin/killer whale and minke whale, KLK8 is independently lost in the minke whale lineage. The loss in the manatee lineage comprises the third independent loss of this gene.
To validate the remaining inactivating mutations in the corresponding aquatic species (dolphin, killer whale, minke whale, and manatee), we searched the SRA for DNA sequences that match to the genomic context comprising at least 50 bp around the mutation. Supplementary table 2, Supplementary Material online lists the SRA identifiers that we used. For validating the loss of exon 5 in dolphin and killer whale, we extracted a ∼100 nt sequence that spans the exon 5 deletion (supplementary table 3, Supplementary Material online). The dolphin sequence was then queried against the dolphin TRACE Archive (Trace\Tursiops_truncatus_WGS) and the killer whale sequence against the SRA (SRX188933 and SRX188930). This search retrieved several reads that were 100% identical to the query sequence and spanned at least 20 nucleotides on either side of the breakpoint, thus confirming the deletion.
To assess whether gene order in the kallikrein gene cluster is conserved, we visualized colinear alignment chains in the UCSC genome browser (Tyner et al. 2016). These chains were generated in a previous study (Sharma and Hiller 2017) using lastz (Harris 2007) to obtain local alignments between two genomes and axtChain (Kent et al. 2003) to obtain colinear alignment chains. We then inspected the locus of other genomes that aligned to the human KLK8 gene, which showed that the entire KLK8 locus is located on a single contig in the genome assembly of all cetaceans and the manatee.
Detection of Inactivating Mutations in Killer Whale Resequencing Data
To test the occurrence of inactivating mutations in different killer whale populations, we downloaded genomic read sequences (FASTQ files) of 48 killer whale individuals which belong to five distinct ecotypes (Foote et al. 2016) from the European Nucleotide Archive (Cochrane et al. 2013). From the killer whale genome assembly, we extracted sequences that contain the inactivating mutations including a 50 nt context, except for exon 1 where we used the entire exon sequence instead. In addition to the sequences that contain the inactivating mutation, we generated artificial query sequences by reversing the inactivating mutation to the ancestral state inferred from the aligned cow sequence (supplementary table 4, Supplementary Material online). After constructing BLAST databases from the killer whale FASTQ files, BlastN in “megablast” mode with default parameters (Zhang et al. 2000) was used to find reads that overlap query sequences for each killer whale individual separately. We then counted the number of reads that either supported the inactivating mutation or the intact ancestral state of each sequence (supplementary table 8, Supplementary Material online).
Investigation of KLK8 Mutations in Other Mammals
To test if KLK8 loss is specific to fully-aquatic mammals, we investigated the coding region of all other 65 mammals that are contained in the genome alignment (Sharma and Hiller 2017). For each putative mutation, we applied the same scrutiny, including CESAR to investigate if a different alignment with an intact reading frame and consensus splice sites is possible. In addition, we examined the correctness of the remaining mutations in armadillo and marmoset with unprocessed DNA sequencing reads from the SRA. For exons that did not align, we used the alignment chains to examine whether the corresponding locus in the other mammal contains assembly gaps. As shown in supplementary figures 3–5, Supplementary Material online, this analysis revealed several sequencing errors, cases of incomplete genome assemblies and alignment ambiguities in species that do have an intact KLK8 gene. For two species (pig and microbat), the kallikrein gene cluster could not be examined because their genome assembly is incomplete at this locus (large assembly gap in pig, locus is not covered by a microbat scaffold). Therefore, these two species were omitted from figure 1.
Analysis of Dolphin Skin Transcriptomes
We downloaded Illumina HiSeq 2500 RNA-seq data of skin biopsies from bottlenose dolphins (GEO-accession: GSE90941) from the SRA (SRA-accession: SRP094638; Kodama et al. 2012; Neely et al. 2017). This data set contained 65 samples that were sequenced in four runs each with an average read length of 100 bp and 51 samples that were sequenced in a single run with an average read length of 125 bp resulting in 311 runs in total. We processed all SRA read files with fastq-dump using parameters for removing technical reads (skip-technical), filtering (read-filter = pass), and removing tags (clip). Next, we extracted the two scaffolds (JH479757 and JH481762) that contain the kallikrein gene cluster from the dolphin turTru2 genome assembly and mapped the reads of all 311 runs separately to the two scaffolds using STAR (version 2.4.2a; Dobin et al. 2013). Given the small total size of the two scaffolds, we set the “genomeSAindexNbases” parameter to eight for building the index file. For mapping reads with STAR, we specified parameters to only consider uniquely mapped reads (outFilterMultimapNmax = 1) and required that reads align with a high sequence identity of at least 99% (outFilterMismatchNoverLmax = 0.01). Afterwards, we randomly picked five skin samples (NCBI BioSample-IDs: SAMN06113305, SAMN06113312, SAMN06113314, SAMN06113320, and SAMN06113334) and visualized the read coverage using bedtools (Quinlan and Hall 2010) and samtools (Li et al. 2009) in the UCSC genome browser (Tyner et al. 2016). These five samples include three samples with 125 bp reads and two samples with 100 bp reads. For samples with more than one run (SAMN06113305, SAMN06113334), we combined the mapped reads of all four runs into a single read profile. Since the turTru2 assembly lacks an Ensembl gene annotation, we combined comparative gene annotations by CESAR (Sharma et al. 2016) and UCSC's TransMap (Zhu et al. 2007) that mapped human genes to the dolphin genome to visualize the exon–intron structure genes in figure 4.
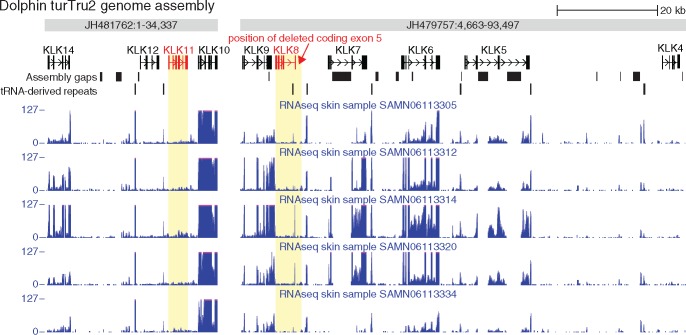
Fig. 4.
—RNA-seq data from the dolphin skin show that KLK8 is not expressed. The coverage of RNA-seq reads obtained from dolphin skin tissue (GEO-accession: GSE90941) is shown for five representative dolphin individuals. The kallikrein gene cluster in the dolphin turTru2 assembly is located on the two scaffolds that are shown here. The exon–intron structure annotation of the genes where obtained by mapping human genes to the dolphin genome using CESAR (Sharma et al. 2016) and TransMap (Zhu et al. 2007). In addition to KLK8 and KLK11, two lost genes (see text), KLK4 that has a role in tooth enamel formation (Lu et al. 2008) and the less well characterized KLK12, do not exhibit a clear expression in the skin, consistent with a previous study (Komatsu et al. 2006). Several tRNA-glu derived SINE repeats (Shimamura et al. 1999) occur in the kallikrein cluster, including the KLK8 locus, and are highly expressed.
Selection Rate Analysis
To assess whether KLK8 in sperm whale and fully-aquatic species in general is under relaxed selection, we analyzed the coding sequences with RELAX to test for significantly relaxed rates of selection compared with the nonfully-aquatic species (Wertheim et al. 2015). For this purpose, we created a multiple-sequence alignment of the coding KLK8 sequence of 68 species. We replaced inactivating stop codons and codons with frameshifts with gaps. Using this alignment as the input for RELAX, we specified 1) the sperm whale branch as test set and all branches of nonfully-aquatic mammals as background, and 2) the branches leading to all fully-aquatic mammals as test set and all branches of all nonfully-aquatic mammals as background.
Results
We used an alignment that comprises 70 placental mammals (Sharma and Hiller 2017) and inspected the coding region of KLK8 in these species (supplementary table 1, Supplementary Material online). Our inspection revealed 12 mutations that would inactivate KLK8 in the bottlenose dolphin, killer whale, minke whale, and the manatee (fig. 1). These mutations include frameshifting deletions, mutations that disrupt the conserved splice site dinucleotides, mutations that terminate translation by creating in-frame stop codons, and the entire deletion of the coding exon 5. In all these aquatic mammals, these mutations are spread across several coding exons and destroy the serine protease domain that is encoded collectively by exons 2–5. This strongly suggests that KLK8 is lost in these species.
To validate the loss of KLK8 in the aquatic mammals, we first sought to rule out that alternative sequence alignments, where the position of frameshifting deletions is shifted into the intron, show an intact reading frame. Likewise, since the position of splice sites can shift in evolution, we wanted to exclude the possibility that other in-frame consensus splice sites exist near the disrupted splice sites. To this end, we applied CESAR, an alignment method trained to find a sequence alignment with an intact reading frame and consensus splice sites whenever possible (Sharma et al. 2016, 2017), to every exon with an inactivating mutation. The alternative CESAR alignment revealed an intact exon for exon 2 in dolphin and exon 5 in manatee (see supplementary fig. 1, Supplementary Material online), therefore, we did not consider these two mutations further. For all other exons, the CESAR alignment consistently showed the inactivating mutation, which excludes the possibility that the exons are intact. These confirmed inactivating mutations are shown in figure 1.
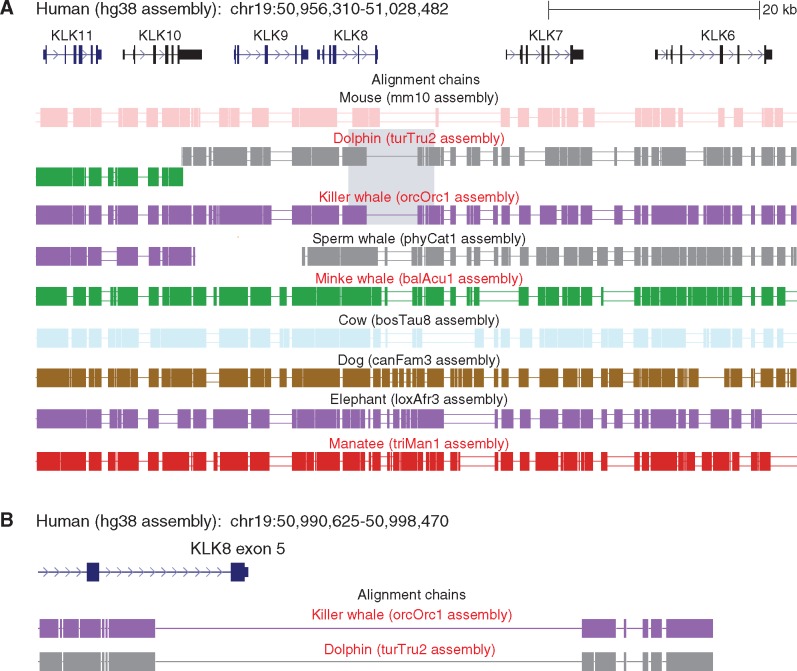
Second, we sought to rule out that misalignments between the KLK8 genomic locus and other genes belonging to the kallikrein family lead to incorrectly inferring KLK8 loss. To this end, we first examined the genome assemblies and found that the entire KLK8 locus is located on a single contig (not interrupted by assembly gaps) in all four cetaceans and the manatee. Next, we assessed if the gene order in the kallikrein gene cluster is conserved in mammals. Pairwise alignments between human and mammals with an intact KLK8 gene show that KLK8 is located in the same context, surrounded by KLK7 and KLK9 (fig. 2A). The remnants of KLK8 in the three cetacean and manatee genome assemblies are also found in this conserved gene context. It should be noted that whereas kallikreins comprise a gene family, the protein and nucleotide sequence of the different members are sufficiently diverged from each other. In particular, KLK8 has at most 51% nucleotide identity to other KLK genes (Prassas et al. 2015). Together with the conserved order of the KLK genes in this cluster, this excludes the possibility of misalignments between the KLK8 genomic locus and other kallikrein genes. Our inspection of the genome alignments also revealed that the exon 5 deletion in dolphin and killer whale has the same breakpoint (fig. 2B). This suggests that the exon 5 deletion occurred in the common ancestor of both species, and rules out the possibility of an assembly error in two independently assembled genomes. In addition, the presence of a shared in-frame stop codon in the third KLK8 exon further corroborates that KLK8 was lost in the common ancestor of dolphin and killer whale.
Fig. 2.
—The remnants of KLK8 in aquatic species are located in a conserved gene context. (A) A UCSC genome browser screenshot of the human genome shows the KLK8 gene and other conserved kallikrein genes up- and downstream. The colinear alignment chains (boxes represent aligning regions, single and double lines represent deletions and regions that do not align between human and the other species) show that the genomic loci that align to KLK8 are in a conserved context, both in the aquatic species that lost KLK8 (red font) and in other mammals that have an intact KLK8. Grey background highlights the exon 5 deletion that is magnified in (B). (B) The deletion of KLK8 coding exon 5 in dolphin and killer whale has the same breakpoints, which indicates that this exon deletion occurred in the common ancestor of both species. For visualization clarity, only the top-scoring chain that aligns to the orthologous locus is shown.
Third, we sought to validate the gene-inactivating mutations by investigating whether unassembled genome sequencing reads from the Short Read Archive (SRA; Kodama et al. 2012) and NCBI Trace Archive clearly support the mutation that is present in the genome assembly (supplementary table 2, Supplementary Material online). While we found conclusive evidence (at least ten reads) for the presence of all 12 inactivating mutations that occur in the four KLK8 loss species, unassembled reads do not support the presence of sequences where the inactivating mutation was reversed to its ancestral state (fig. 3, supplementary tables 4–7, Supplementary Material online). Furthermore, the deletion of exon 5 in dolphin and killer whale could also be confirmed by sequencing reads that span the breakpoint of the exon 5 deletion (supplementary table 3, Supplementary Material online). Together, our validation shows that the inactivating mutations are not polymorphisms or sequencing errors and rules out that an intact KLK8 gene copy exists in the genome of the analyzed species. To test whether any killer whale individual or population has an intact KLK8 gene, we investigated resequencing data of 48 killer whale individuals belonging to five distinct ecotypes (Foote et al. 2016), which confirmed the presence of the inactivating mutations in different killer whale individuals. In total, we found over a hundred reads confirming the inactivating mutations whereas not a single read matched the ancestral intact sequences (supplementary tables 4 and 8, Supplementary Material online). These findings show that the loss of KLK8 is likely fixed in the killer whale species, which corroborates our observation that KLK8 was lost in the common ancestor of dolphin and killer whale.
Finally, to further validate the loss of KLK8, we analyzed available RNA-seq data (GEO-accession: GSE90941) of dolphin skin, a tissue where an intact KLK8 gene is expected to be expressed. This data set contains sequenced RNA of skin biopsies from different bottlenose dolphin individuals. We used this data set to assess if KLK8 is still expressed in dolphin by mapping RNA-seq reads to the bottlenose dolphin genome. We observed no relevant expression of the exons of the inactivated KLK8 gene, compared with other KLK genes that are known to function as active proteins in the skin such as KLK5, KLK7, and KLK14 (Rawlings and Voegeli 2013; fig. 4, supplementary table 9, Supplementary Material online). Interestingly, we also observed that KLK11 is not expressed in the dolphin skin. KLK11 has previously been found to be expressed in the epidermis and is cleaved by KLK8 in vitro (Eissa et al. 2011; Komatsu et al. 2005). Manual inspection shows that KLK11 is lost in all cetaceans (supplementary fig. 2, Supplementary Material online). The loss of KLK11 is likely related to the loss of KLK8 and indicates that parts of a kallikrein activation cascade are lost.
Together, our genome and transcriptome analysis clearly establishes that KLK8 is lost in four fully aquatic species (dolphin, killer whale, minke whale, and manatee), where it does not encode a functional protein anymore.
The loss of KLK8 in three independent fully-aquatic lineages, suggests an association between the loss of KLK8 and a fully-aquatic lifestyle. If this is the case, we expect that KLK8 is not lost in any semi-aquatic (Weddell seal and Pacific walrus) or terrestrial mammal. Therefore, we examined the KLK8 coding sequence in the 65 nonfully-aquatic species in our genome alignment (Sharma and Hiller 2017). Whereas this genome alignment indicates the presence of gene-inactivating mutations in several other species, careful manual inspection showed that all other species have an intact reading frame and that all these putative mutations are sequencing errors or artifacts caused by incomplete assemblies or alignment ambiguities. For example, several mutations in exon 5 in the armadillo genome assembly are sequencing errors as unassembled sequencing reads differ from the genome assembly and have an intact reading frame (supplementary fig. 3A, Supplementary Material online). Similarly, two putative frameshifting mutations in KLK8 exons 1 and 4 in the genome assembly of the marmoset were found to be sequencing errors when the marmoset sequence was queried against raw reads from SRA (supplementary fig. 3B, Supplementary Material online). The apparent deletion of exon 4 in the jerboa assembly overlaps a 1,759 bp assembly gap, suggesting that the sequence of this exon was not added to the genome assembly (supplementary fig. 4, Supplementary Material online). Finally, putative inactivating mutations that the genome alignment showed for naked mole rat exon 1 and for pangolin exon 3 are alignment ambiguities, as an alternative alignment reveals an intact reading frame (supplementary fig. 5, Supplementary Material online). Together, this shows that KLK8 is only lost in the three fully-aquatic lineages.
Although we could not identify inactivating mutations in the sperm whale, RNA-seq data from sperm whale skin (Warren et al. 2017) showed that KLK8 is not expressed anymore in this tissue (supplementary table 10, Supplementary Material online). Furthermore, we found the sperm whale KLK8 gene to be under relaxed selection (P-value < 10−4) using RELAX analysis (Wertheim et al. 2015). In fact, RELAX estimates that all codons evolve neutrally (dN/dS ratio ∼1) on the branch leading to the sperm whale (supplementary table 11, Supplementary Material online) and a recent study found mutations in its catalytic site (Warren et al. 2017), suggesting that KLK8 may accumulate inactivating mutations in future. In contrast, for branches leading to all other semi-aquatic or terrestrial mammals, RELAX estimates that 88% of codons evolve under strong purifying selection (dN/dS 0.13). Consistent with the loss of KLK8, all branches associated with fully-aquatic mammals (four cetaceans and manatee) evolve neutrally (dN/dS ratio ∼1, P-value < 10−5, supplementary table 11, Supplementary Material online).
Discussion
The specific and repeated KLK8 loss pattern together with neutral evolution in the sperm whale suggests that subsequent to the evolution of a fully-aquatic lifestyle, KLK8 became obsolete in these species and evolved neutrally. This in turn resulted in its loss in the manatee lineage and in two cetacean lineages after speciation of the cetacean ancestor. We suggest that neutral evolution and loss of KLK8, a pleiotropic gene with functions in the skin and hippocampus, is likely explained by unique characteristics of the skin and neuroanatomical differences of the aquatic species.
First, in the skin of terrestrial mammals, KLK8 activates antimicrobial proteolytic cascades in sweat (Eissa et al. 2011). As neither cetaceans nor manatees have sweat glands (Berta et al. 2015; Mouton and Botha 2012; Rodrigues et al. 2014), in contrast to the semi-aquatic pinnipeds and terrestrial mammals that have an intact KLK8 gene, the antimicrobial of functions of KLK8 are likely no longer useful for aquatic mammals. Furthermore, the epidermis of KLK8 knockout mouse exhibits delayed recovery from the UVB-induced inflammation (Kirihara et al. 2003). UVB-induced inflammation might be less important for fully-aquatic mammals that live in an environment where UVB light penetration substantially decreases after a few meters of depths (Tedetti and Sempere 2006). Finally, KLK8 also plays a role in keratinocyte proliferation and desquamation (Kishibe et al. 2007). In dolphins, it is known that the desquamation rate of the outermost epidermal cell layers is 8.5 times faster than in humans, which is likely an adaptation that maintains a smooth surface and limits microbe colonization in the aquatic environment (Hicks et al. 1985), and may have further contributed to KLK8 loss in dolphin.
Second, in the hippocampus, KLK8 is involved in neuronal plasticity and long-term potentiation, which plays an important role in memory formation (Attwood et al. 2011; Tamura et al. 2006, 2012). The most plausible explanation for the repeated loss of KLK8, a gene with hippocampus related functions, is the presence of distinct neuroanatomical characteristics of the fully-aquatic mammals compared with other mammals. The size of the hippocampus in relation to the entire brain is much smaller in cetaceans compared with the relative size of the hippocampus in pinnipeds and terrestrial mammals (Marino et al. 2004; Patzke et al. 2015). Similarly, the brain of manatees has a proportionally small hippocampus (Hakeem et al. 2005). These size differences led to suggestions that other brain areas may be utilized for memory-related and other functions that are typically associated with the hippocampus in mammals (Marino et al. 2004). These neuroanatomical differences that evolved repeatedly in the aquatic mammals may have also rendered the hippocampus-related functions of KLK8 obsolete.
Pleiotropic genes are often well conserved in evolution as many mutations in these genes have deleterious consequences for an organism. It is therefore deemed unlikely that pleiotropic genes can be lost during evolution. Here, we present a clear case of a loss of a pleiotropic gene and describe the special circumstances that permitted this loss. Given the distinct skin and neuroanatomical features that characterize fully-aquatic mammals, the repeated loss of KLK8 is likely a consequence of relaxed selection or no selection at all on the dual functions of KLK8, subsequent to the evolution of a fully-aquatic lifestyle. Thus, KLK8 loss is a repeated genomic change that is associated with differences in skin and neuroanatomy of aquatic mammals. While the loss of a pleiotropic gene is typically deleterious as all its functions are lost, the loss of the pleiotropic KLK8 shows that under special circumstances, the loss of genes with a limited pleiotropy may not result in deleterious consequences and thus may be permitted during evolution.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank the Computer Service Facilities of the MPI-CBG and MPI-PKS for their support. This work was supported by the Max Planck Society, the German Research Foundation (HI 1423/3-1), and the Leibniz Association (SAW-2016-SGN-2).
Literature Cited
- Attwood BK, et al. 2011. Neuropsin cleaves EphB2 in the amygdala to control anxiety. Nature 4737347:372–375.http://dx.doi.org/10.1038/nature09938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berta A, Sumich JL, Kovacs KM.. 2015. Chapter 7 – integumentary and sensory systems In: Marine mammals, 3rd ed. San Diego (CA): Academic Press; p. 169–210. [Google Scholar]
- Chang S, Bok P, Sun CP, Edwards A, Huang GJ.. 2016. Neuropsin inactivation has protective effects against depressive-like behaviours and memory impairment induced by chronic stress. PLoS Genet. 1210:e1006356.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZL, et al. 1995. Expression and activity-dependent changes of a novel limbic-serine protease gene in the hippocampus. J Neurosci. 15(7 Pt 2):5088–5097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochrane G, et al. 2013. Facing growth in the European Nucleotide Archive. Nucleic Acids Res. 41(Database issue):D30–D35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, et al. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 291:15–21.http://dx.doi.org/10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissa A, Amodeo V, Smith CR, Diamandis EP.. 2011. Kallikrein-related peptidase-8 (KLK8) is an active serine protease in human epidermis and sweat and is involved in a skin barrier proteolytic cascade. J Biol Chem. 2861:687–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foote AD, et al. 2016. Genome-culture coevolution promotes rapid divergence of killer whale ecotypes. Nat Commun. 7:11693.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakeem AY, et al. 2005. Brain of the African elephant (Loxodonta africana): neuroanatomy from magnetic resonance images. Anat Rec A Discov Mol Cell Evol Biol. 2871:1117–1127. [DOI] [PubMed] [Google Scholar]
- Harris RS. 2007. Improved pairwise alignment of genomic DNA. [PhD thesis]. Pennsylvania State University, University Park, PA. [Google Scholar]
- Hicks BD, St Aubin DJ, Geraci JR, Brown WR.. 1985. Epidermal growth in the bottlenose dolphin, Tursiops truncatus. J Invest Dermatol. 851:60–63.http://dx.doi.org/10.1111/1523-1747.ep12275348 [DOI] [PubMed] [Google Scholar]
- Hirata A, et al. 2001. Abnormalities of synapses and neurons in the hippocampus of neuropsin-deficient mice. Mol Cell Neurosci. 173:600–610.http://dx.doi.org/10.1006/mcne.2000.0945 [DOI] [PubMed] [Google Scholar]
- Izumi A, et al. 2008. Genetic variations of human neuropsin gene and psychiatric disorders: polymorphism screening and possible association with bipolar disorder and cognitive functions. Neuropsychopharmacology 3313:3237–3245.http://dx.doi.org/10.1038/npp.2008.29 [DOI] [PubMed] [Google Scholar]
- Kent WJ, Baertsch R, Hinrichs A, Miller W, Haussler D.. 2003. Evolution's cauldron: duplication, deletion, and rearrangement in the mouse and human genomes. Proc Natl Acad Sci U S A. 10020:11484–11489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirihara T, et al. 2003. Prolonged recovery of ultraviolet B-irradiated skin in neuropsin (KLK8)-deficient mice. Br J Dermatol. 1494:700–706. [DOI] [PubMed] [Google Scholar]
- Kishibe M, et al. 2007. Kallikrein 8 is involved in skin desquamation in cooperation with other kallikreins. J Biol Chem. 2828:5834–5841. [DOI] [PubMed] [Google Scholar]
- Kodama Y, Shumway M, Leinonen R.. International Nucleotide Sequence Database C 2012. The Sequence Read Archive: explosive growth of sequencing data. Nucleic Acids Res. 40(D1):D54–D56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu N, et al. 2005. Multiple tissue kallikrein mRNA and protein expression in normal skin and skin diseases. Br J Dermatol. 1532:274–281. [DOI] [PubMed] [Google Scholar]
- Komatsu N, et al. 2006. Quantification of eight tissue kallikreins in the stratum corneum and sweat. J Invest Dermatol. 1264:925–929. [DOI] [PubMed] [Google Scholar]
- Kuwae K, et al. 2002. Epidermal expression of serine protease, neuropsin (KLK8) in normal and pathological skin samples. Mol Pathol. 554:235–241.http://dx.doi.org/10.1136/mp.55.4.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MG, Lai J, Clements JA.. 2010. Kallikreins on steroids: structure, function, and hormonal regulation of prostate-specific antigen and the extended kallikrein locus. Endocr Rev. 314:407–446.http://dx.doi.org/10.1210/er.2009-0034 [DOI] [PubMed] [Google Scholar]
- Li H, et al. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 2516:2078–2079.http://dx.doi.org/10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, et al. 2008. Functions of KLK4 and MMP-20 in dental enamel formation. Biol Chem. 3896:695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundwall A, Brattsand M.. 2008. Kallikrein-related peptidases. Cell Mol Life Sci. 6513:2019–2038.http://dx.doi.org/10.1007/s00018-008-8024-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino L, et al. 2004. Neuroanatomy of the killer whale (Orcinus orca) from magnetic resonance images. Anat Rec A Discov Mol Cell Evol Biol. 2812:1256–1263. [DOI] [PubMed] [Google Scholar]
- Matsumoto-Miyai K, et al. 2003. NMDA-dependent proteolysis of presynaptic adhesion molecule L1 in the hippocampus by neuropsin. J Neurosci. 2321:7727–7736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouton M, Botha A.. 2012. Cutaneous lesions in cetaceans: an indicator of ecosystem status? In: Romero A, Keith EO, editors. New approaches to the study of marine mammals. InTech. doi: 10.5772/54432. Available from: https://www.intechopen.com/books/new-approaches-to-the-study-of-marine-mammals/cutaneous-lesions-in-cetaceans-an-indicator-of-ecosystem-status-, last accessed November 20, 2017. [Google Scholar]
- Nakamura Y, Tamura H, Horinouchi K, Shiosaka S.. 2006. Role of neuropsin in formation and maturation of Schaffer-collateral L1cam-immunoreactive synaptic boutons. J Cell Sci. 119(Pt 7):1341–1349. [DOI] [PubMed] [Google Scholar]
- Neely MG, et al. 2017. Skin Transcriptomes of common bottlenose dolphins (Tursiops truncatus) from the northern Gulf of Mexico and southeastern U.S. Atlantic coasts. Mar. Genomics. doi:10.1016/j.margen.2017.08.002. [DOI] [PubMed] [Google Scholar]
- Ovaere P, Lippens S, Vandenabeele P, Declercq W.. 2009. The emerging roles of serine protease cascades in the epidermis. Trends Biochem Sci. 349:453–463. [DOI] [PubMed] [Google Scholar]
- Patzke N, et al. 2015. In contrast to many other mammals, cetaceans have relatively small hippocampi that appear to lack adult neurogenesis. Brain Struct Funct. 2201:361–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prassas I, Eissa A, Poda G, Diamandis EP.. 2015. Unleashing the therapeutic potential of human kallikrein-related serine proteases. Nat Rev Drug Discov. 143:183–202.http://dx.doi.org/10.1038/nrd4534 [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM.. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 266:841–842.http://dx.doi.org/10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings AV, Voegeli R.. 2013. Stratum corneum proteases and dry skin conditions. Cell Tissue Res. 3512:217–235. [DOI] [PubMed] [Google Scholar]
- Rodrigues FR, da Silva VM, Barcellos JF.. 2014. The mammary glands of the Amazonian manatee, Trichechus inunguis (Mammalia: Sirenia): morphological characteristics and microscopic anatomy. Anat Rec (Hoboken) 2978:1532–1535.http://dx.doi.org/10.1002/ar.22956 [DOI] [PubMed] [Google Scholar]
- Sharma V, Elghafari A, Hiller M.. 2016. Coding exon-structure aware realigner (CESAR) utilizes genome alignments for accurate comparative gene annotation. Nucleic Acids Res. 4411:e103.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma V, Hiller M.. 2017. Increased alignment sensitivity improves the usage of genome alignments for comparative gene annotation. Nucleic Acids Res. 4514:8369–8377.http://dx.doi.org/10.1093/nar/gkx554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma V, Schwede P, Hiller M.. 2017. CESAR 2.0 substantially improves speed and accuracy of comparative gene annotation. Bioinformatics. doi:10.1093/bioinformatics/btx527 [epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Shimamura M, Abe H, Nikaido M, Ohshima K, Okada N.. 1999. Genealogy of families of SINEs in cetaceans and artiodactyls: the presence of a huge superfamily of tRNA(Glu)-derived families of SINEs. Mol Biol Evol. 168:1046–1060. [DOI] [PubMed] [Google Scholar]
- Sotiropoulou G, Pampalakis G, Diamandis EP.. 2009. Functional roles of human kallikrein-related peptidases. J Biol Chem. 28448:32989–32994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura H, et al. 2006. Neuropsin is essential for early processes of memory acquisition and Schaffer collateral long-term potentiation in adult mouse hippocampus in vivo. J Physiol. 570(Pt 3):541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura H, Kawata M, Hamaguchi S, Ishikawa Y, Shiosaka S.. 2012. Processing of neuregulin-1 by neuropsin regulates GABAergic neuron to control neural plasticity of the mouse hippocampus. J Neurosci. 3237:12657–12672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedetti M, Sempere R.. 2006. Penetration of ultraviolet radiation in the marine environment. A review. Photochem Photobiol. 822:389–397.http://dx.doi.org/10.1562/2005-11-09-IR-733 [DOI] [PubMed] [Google Scholar]
- Tyner C, et al. 2016. The UCSC Genome Browser database: 2017 update. Nucleic Acids Res. 45(D1): D626–D634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren WC, et al. 2017. The novel evolution of the sperm whale genome. Genome Biol Evol. doi:10.1093/gbe/evx187 [epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertheim JO, Murrell B, Smith MD, Kosakovsky Pond SL, Scheffler K.. 2015. RELAX: detecting relaxed selection in a phylogenetic framework. Mol Biol Evol. 323:820–832.http://dx.doi.org/10.1093/molbev/msu400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S. 2010. Klk8, a multifunctional protease in the brain and skin: analysis of knockout mice. Biol Chem. 3914:375–380.http://dx.doi.org/10.1515/bc.2010.034 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Schwartz S, Wagner L, Miller W.. 2000. A greedy algorithm for aligning DNA sequences. J Comput Biol. 7(1–2):203–214. [DOI] [PubMed] [Google Scholar]
- Zhu J, et al. 2007. Comparative genomics search for losses of long-established genes on the human lineage. PLoS Comput Biol. 312:e247. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.