Abstract
Divalent metal ions, usually Mg2+, are required for both DNA synthesis and proofreading functions by DNA polymerases (DNA Pol). Although used as a non-reactive cofactor substitute for binding and crystallographic studies, Ca2+ supports DNA polymerization by only one DNA Pol, Dpo4. Here, we explore whether Ca2+-driven catalysis might apply to high-fidelity (HiFi) family B DNA Pols. The consequences of replacing Mg2+ by Ca2+ on base pairing at the polymerase active site as well as the editing of terminal nucleotides at the exonuclease active site of the archaeal Pyrococcus abyssi DNA Pol (PabPolB) are characterized and compared to other (families B, A, Y, X, D) DNA Pols. Based on primer extension assays, steady-state kinetics and ion-chased experiments, we demonstrate that Ca2+ (and other metal ions) activates DNA synthesis by PabPolB. While showing a slower rate of phosphodiester bond formation, nucleotide selectivity is improved over that of Mg2+. Further mechanistic studies show that the affinities for primer/template are higher in the presence of Ca2+ and reinforced by a correct incoming nucleotide. Conversely, no exonuclease degradation of the terminal nucleotides occurs with Ca2+. Evolutionary and mechanistic insights among DNA Pols are thus discussed.
INTRODUCTION
Accurate replication and repair of cellular DNA is necessary to preserve the integrity of the genome during cell division (1). DNA synthesis is carried out by HiFi DNA Pols (families A, B, C and D) in the three domains of life (2,3) that selectively incorporate the correct nucleotide opposite the template base. Several factors, such as the structural constraints of the active site, base-pair geometry, base-pair hydrogen bonding and flexibility of the template contribute to base selection (4–6). Additionally, there is a 3′-5′ exonuclease activity or proofreading function which can excise incorrect nucleotides from extending primers, thereby improving fidelity (7). Thus, shuttling the 3′-OH group with the mismatched nucleotide from the polymerase to the exonuclease active site for removal and, thereafter from the exonuclease to the polymerase active site and delays the rate of DNA polymerization (8). Mechanistic details of the interplay between DNA polymerization and proofreading functions which can account for accurate DNA synthesis have mainly been studied in bacterial DNA Pols (9–13). Catalytic centers in charge of DNA polymerase and exonuclease activities are located in separate domains and DNA Pol structures with primer/template (p/t) DNA bound in either the polymerase or the exonuclease mode are distinct (10,14–23).
The polymerase domain is generally composed of three subdomains (the palm, thumb and fingers), each of which plays a crucial role in the polymerase reaction (6,17). Modeled on Escherichia coli DNA PolI, the established dogma for DNA polymerization catalysis utilizes a ‘two-metal-ion’ mechanism for nucleotidyl transfer which involves three highly conserved carboxylate residues and two metal ions (24–26). The catalytic metal ion (metal A) is thought to lower the pKa of the 3′-OH of the terminal primer for attack of the nucleotide α-phosphate. The nucleotide binding metal (metal B) coordinates the triphosphate moiety and assists the departure of the pyrophosphate (PPi). Both metal ions stabilize the expected pentacovalent transition state of the nucleotidyl-transfer reaction (27). Collectively, on the basis of crystal structures for various non-reactive DNA Pol ternary complexes, the two-metal ion mechanisms has been proposed (20,28–33), in which the deprotonation of the 3′-OH of the DNA primer is representing the primary activation event in the chemical step for nucleotide incorporation (34). Recently, high-resolution low fidelity DNA Pol (Families X and Y) structures demonstrated that an additional metal ion is transiently involved in catalysis (35–37), thus reconsidering the metal-ion mechanism for nucleotide incorporation (38). Although not captured by time-resolved X-ray crystallography, a third metal ion has also been observed in the catalytic site of yeast DNA Polδ (19). Whether the three-metal ion mechanism applies to this HiFi family B DNA Pol has to be demonstrated. Overall, these findings indicate that metal ions dynamics plays a crucial role in promoting the catalytic steps during the reaction and account for the flexibility of the active site.
When present, exonuclease activity can be seen in either a separate domain (Eukaryotic Pol ε, δ, γ, bacterial PolII and PolI, bacteriophages T4, T7 and RB69, and archaeal PolB) (19,22,23,39–45) or subunit (Bacterial PolIII and archaeal PolD)(46–50) of the DNA Pol, acting to remove mismatched nucleotides from the 3′-end of the nascent strand. When two metal ions are found in the exonuclease active centers of proofreading DNA Pols, the DNA hydrolysis reaction is thought to proceed by a two-metal-ion mechanism analogous to the one responsible for DNA polymerization (27). However, despite several crystal structures for non-reactive DNA Pol binary complexes and uncountable biochemical studies, the detailed catalytic mechanism and the precise roles of these metal ions in the exonuclease active site still remain open questions (24,33,41,51–53).
Magnesium is by far the most frequently utilized metal ion cofactor by DNA Pols for both DNA synthesis and editing functions, reflected by its natural abundance in the cells (54–56). This metal ion is characterized by a small atomic radius (0.86 Å) and elevated hardness, making it suitable for coordination to oxygen atoms (57). DNA Pols are able to use other divalent cations such as Mn2+, Ni2+, Co2+ and Be2+, but the accuracy and efficiency of DNA synthesis is strongly altered compared to Mg2+ (58–62). Commonly used as non-reactive cofactor of DNA polymerization or for slowing reaction rates, Ca2+ ions are consistently bound to a specific region in the active site in crystals of ternary complexes (Pol/DNA/nucleotide) (19,35,43) and enable probing of local DNA conformational changes at the primer-template junction (11,63). Little is known about the mechanism of inhibitory effects of Ca2+ on exonuclease activity of HiFi DNA Pols and activity-modulating metal ions is usually restricted to Mg2+ and Mn2+ (24,26,41,64). Moreover, metal ion dynamics have not been experimentally observed in exonuclease active sites of HiFi DNA Pols.
To date only the family-Y DNA Pol, namely Dpo4 or Sulfolobus solfataricus P2 (S. solfataricus) DNA Pol IV, has been described to utilize Ca2+ as a cofactor for DNA polymerization. Compared to Mg2+, the DNA synthesis rate is considerably reduced and the formation of full-length products is slower (65). Clearly, Dpo4 seems to have evolved variable metal ion usage for catalysis (65,66), which might serve to regulate polymerase activity in such hyperthermophilic aerobe microorganism.
Here, we sought to analyze whether Dpo4 is the only example evolutionary drifting of the metal ion catalytic mechanism in DNA Pols to satisfy particular environmental requirements. The family-B DNA Pol from the hyperthermophilic archaeal strain Pyrococcus abyssi GE5 (PabPolB) has been used to explore the evolutionary possibilities of metal ion usage in DNA polymerizing and editing functions. PabPolB like most family-B of the Thermococcales order of the Euryarchaeota phylum shows hyperthermostability, accurate and processive DNA synthesis, making the enzyme favored for PCR (67). PabPolB is able to synthesize long DNA fragments (7–8 kb) but cannot extend and displace RNA primers which differs from family-D DNA Pol (PabPolD)(68). Although demonstrated in only a few Archaea (Halobacterium sp. NRC-1, Methanococcus maripaludis and Thermococcus kodakarensis), genetic inactivation of the two encoding subunits of PolD resulted in lethality assigning an essential role of the enzyme to DNA replication (69–71). Interestingly, PolD has a unique architecture sharing molecular core similarities with RNA Pol and Mre11, respectively, within the polymerase and exonuclease domains, which clearly differs from eukaryotic and bacterial replicative DNA Pols (50). On the other hand, the physiological importance of PolB is less clear. While PolB knockout M. maripaludis and T. kodakarensis were viable, Halobacterium sp. NRC-1 PolB-null cell lines were defective for cell proliferation (69–71). Whether PolB and/or PolD act selectively on opposite DNA strands at the replication fork in these euryarchaeal strains is not yet conclusive and remains to be precisely defined. Like many HiFi family-B DNA Pols, PabPolB has been functionally characterized in the presence of Mg2+ neglecting the possibility of using other divalent metal ions. In this study we demonstrate that PabPolB can perform DNA synthesis in the presence of Ca2+ while exonuclease degradation is inactivated. The results also indicate that the rate of DNA synthesis is reduced with Ca2+ compared to Mg2+. Moreover, PabPolB shows a higher degree of selectivity for dNTP and a more pronounced affinity for the DNA template in the presence of Ca2+. Finally, we ask whether Ca2+-induced DNA synthesis is widespread among different DNA Pol families in various organisms from Archaea to eukaryotes and found that it is restricted to thermophilic archaeal families B (B/B1 and B3 only), families Y and D to a lower extent, and bacterial family A DNA Pols. The catalytic promiscuity related to metal ion usage by DNA Pols is thus discussed.
MATERIALS AND METHODS
Enzymes and DNA substrates
Wild-type (PabPolB exo+) and exonuclease-deficient (PabPolB exo−) (D215A) were produced and purified as previously described (22,72). PabPolD was produced and purified as already reported (73). Bacterial recombinant clone for production of family B DNA Pol from the mesophilic crenarchaeon Methanosarcina acetivorans (MacPolB) was kindly gifted by Isaac K. O. Cann, and purification was achieved as previously described (74). Human family X mesophilic DNA Polβ (hPolβ) was kindly provided by Ulrich Hübscher. Family Y DNA Pol Dpo4 from the hyperthermophilic crenarcheaon S. solfataricus (SsoDpo4) was from Enzymax, family B DNA Pol from the hyperthermophilic euryarchaeon Pyrococcus furiosus (PfuPolB) was from Promega, family B DNA Pol from the hyperthermophilic archaeon Thermococcus kodakaraensis (TkoPolB) was from Novagen and family B DNA Pol from the mesophilic bacteriophage T4 (T4Pol) was from NEB. Family A DNA Pol from the thermophilic bacteria Thermus aquaticus (TaqPol) and the mesophilic bacteria E. coli (Klenow) were respectively from MP biomedicals and Promega. Families B DNA Pols from the hyperthermophilic crenarchaeon S. solfataricus (SsoPolB1, SsoPolB2, and SsoPolB3) and Aeropyrum pernix (ApePolB1, ApePolB2, and ApePolB3) were purified as reported previously (75,76) using plasmids kindly provided by Yoshizumi and Sonoko Ishino as well as by Francesca Pisani for SsoPolB1 clone (77). DNA Pols were initially titrated in our reaction conditions and controlled to be defective for polymerase and exonuclease activities in the absence of metal ions.
All oligonucleotides were purchased from Eurogentec (Seraing, Belgium). The unlabeled and labeled (Cy5 and HEX) oligonucleotides were respectively purified by RP-cartridge and RP-HPLC. Complementary primers (P) were annealed respectively to the oligonucleotides and sscM13 (single-stranded circular M13mp18 DNA) templates (T) at a 1:1 and 1:3 molar ratio in 10 mM Tris–HCl, pH 8.0, and 50 mM NaCl, by heating at 75°C for 15 min followed by cooling to room temperature.
To avoid activity because of trace amounts of metal ion contaminations, ultra pure deionized water (Milli-Q system, Millipore) and high purity chemicals were used.
Primer-template extension and exonuclease assays
Extension reactions of the fluorescent-labeled 17-mer primer 5′-Cy5-TGCCAAGCTTGCATGCC-3′ annealed to the 87-mer template 5′-CAGGAAACAGCTATGACCATGATTACGAATTCGAGCTCGG TACCCGGGGATCCTCTAGAGTCGACCTGCAGGCATGCAAGCTTGGC-3′ (25 nM) were carried out in 15 μl of 50 mM Tris (pH 8.8), 10 mM KCl, 1 mM Dithiothreitol (DTT), metal ions at the indicated concentrations and 200 μM each of the dNTPs. Polymerization was initiated by addition of 75 nM of Parabola exo+/exo−, MacPolB, ApePolBI, SsoPolBI, SsoDpo4 and hPolβ; 400 nM of SsoPolB2, SsoPolB3, ApePolB2 and ApePolB3; 148 nM of PabPolD; 0.75 units of T4Pol, TaqPol, Klenow and PfuPolB; 0.06 units of Topology and were conducted at 37°C for mesophilic and 55°C for thermophilic DNA Pols for 30 min or at the indicated times. For comparing the enzymatic activation by metal ions, the concentrations of DNA Pols were chosen such that the completion of full polymerization was readily detectable in the presence of Mg2+, with roughly a minimal extension percentage of ∼15. Reactions were quenched on ice by addition of 15 μl of stop buffer [95% formamide, 10 mM ethylenediaminetetraacetic acid (EDTA), 10 mM NaOH and 1 μM of competitor oligonucleotide (an exact complement of the template strand under study)], before heating at 95°C for 5 min. Products were resolved on 17% acrylamide (19:1 acrylamide:bis acrylamide), 8 M urea denaturing gel, visualized with a Mode Imager Typhoon 9400 and quantified with the ImageQuant TL8.1 software (GE Healthcare) as it follows: Extension (%) is defined as the sum of intermediate and full-length products/total amount of DNA; full-length products (%) represents the band intensity measurement of 87-nt bands/total amount of DNA.
Degradation of the fluorescent-labeled 17-mer primer annealed to the 87-mer template was carried out as described above except that dNTPs were omitted from the reactions. The concentrations of DNA Pols were identical to the ones chosen for extension reactions. Degradation (%) is calculated as band intensity of cleavage products as a percentage of total lane intensity.
Extension reactions of the fluorescent-labeled 32-mer primer 5′-Cy5-TGCCAAGCTTGCATGCCTGCAGGTCGACTCTA-3′ annealed to the M13mp18 template (7 nM) were performed in 20 μl of 50 mM Tris (pH 8.8), 1 mM DTT, 10 mM KCl, MgCl2 or CaCl2 at the indicated concentrations and 200 μM each of dNTPs. DNA polymerization was initiated by addition of 200 nM of Parabola exo+/exo– and was conducted at 60°C for the indicated times. Reactions were quenched on ice by addition of 40% formamide and a 2-fold excess of EDTA over divalent metal ions, before heating at 100°C for 5 min. Products were resolved on 1% (w/v) alkaline agarose gel and visualized with a Mode Imager Typhoon 9400. DNA ladders (Raoul markers, MP Biomedicals) were run into the same gel and revealed separately as described (73).
Single nucleotide incorporation and steady-state kinetic analysis
Single nucleotide incorporation opposite G template base was measured as described above using the fluorescent-labeled 26-mer primer 5′-Cy5-TGCCAAGCTATGCCTGCAGGTCG-3′ annealed to the 34-mer template 5′-GGATCCTGCGACCTGCAGTGCAAGCTTGGCA-3′. For steady-state kinetic analysis, either dCTP or dTTP at the indicated concentrations (Supplementary Figure S3) were used. Time points and Parabola amounts were set so that maximal product formation was ≤20% of the substrate concentration. The evaluation of the kinetic parameters Km, Vm and kcat have been described previously (73).
Primer-template affinity measurements by steady-state fluorescence anisotropy
The binding of 3′-hexachorofluorescein (HEX)-labeled primed synthetic oligodeoxynucleotides by PabPolB exo− was determined by measuring the steady-state fluorescence anisotropy parameter using a spectrofluorometer equipped with polarizers (FL920, Edinburgh Instruments, UK) in a cell thermostatically held at 25°C. The excitation and emission wavelengths were adjusted to 525 nm (2 nm bandpass) and 555 nm (20 nm bandpass), respectively. Primer-templates were prepared by mixing 5′-CGCCGGGCCGAGCCGTGC-3′ (primer) with 5′-HEX-AGGTCGTGCACGGCTCGGCCCGGCG-3′ (template) in 20 mM Tris–HCl (pH 8), 300 mM NaCl and 1mM EDTA a 1:1 molar ratio and by heating at 95°C for 5 min, followed by cooling to room temperature. Titrations were performed in 20 mM sodium-succinate (pH 6), 100 mM NaCl by increasing concentrations of PabPolB exo− (up to 600 nM) to 5 nM of Hex-labeled primer/template complemented with either 5 mM Ca2+ or 5 mM Mg2+ and with saturating amounts of non-hydrolyzable nucleotide dApNHpp (100 μM) or non-hydrolyzable nucleotide dGpNHpp (1000 μM) (Jena Bioscience GmbH). Using a vertical direction for the polarized excitation source, the steady-state fluorescence anisotropy (r) was calculated according to the equation, r= (Iv-Ih)/Iv+2*IH) where IV and IH correspond to the parallel (vertical) and perpendicular (horizontal) fluorescence emission intensity components, respectively. The equilibrium dissociation constant (KD) characterizing the p/t-PabPolB complex was calculated by fitting the plot of r versus PabPolB concentrations with a Hill model using the IgorPro Software (Wavemetrics).
RESULTS
Catalytic activities of PabPolB mediated by Mg2+ or Ca2+
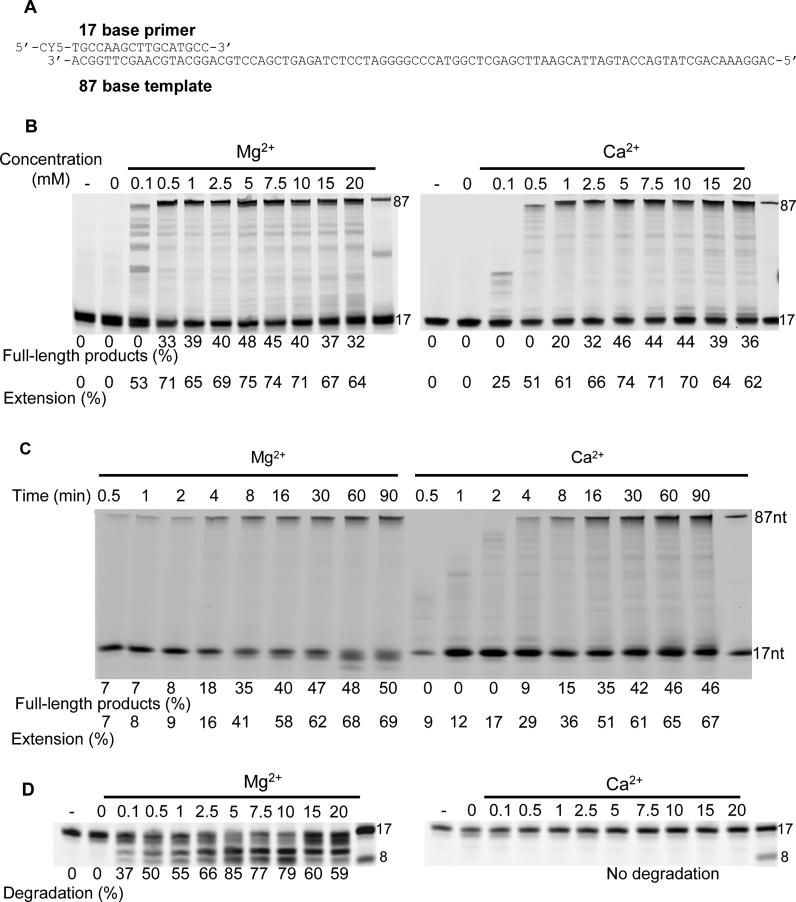
PabPolB, like other HiFi DNA Pols (families A, B, C and D) requires divalent metal ion to support both DNA polymerization and exonuclease degradation. The ability of Ca2+ ions to substitute for Mg2+ during DNA polymerization and primer degradation is shown in Figure 1. When extension of the primer (Cy5-labeled primer, 17 bases long, annealed to a template 87 bases in length) was measured in the presence of Ca2+, DNA synthesis was almost comparable to Mg2+ (Figure 1B). Similar profiles of extended products were observed and the optimum of DNA polymerization was obtained at 5 mM (∼48 and ∼46% full-length products with Mg2+ and Ca2+, respectively). Primer extension was clearly activated by addition of either Ca2+ or Mg2+ and not provided by the enzyme or other contaminants as conferred by the absence of extended products when metal ions were omitted. Interestingly, time course experiments carried out at constant metal ion concentrations (5 mM) highlighted distinct rates of primer-extension (Figure 1C). Full-length extension products began to accumulate at 4 min and only 30 s with Ca2+ and Mg2+, respectively. In general, low molecular weight products accumulated in the shortest time points (≤4 min) in the presence of Ca2+ compared with Mg2+, suggesting that PabPolB likely acts distributively or/and is slow in elongation. At 30 min, similar rates of extended primers were observed. Figure 1D shows the capability of Ca2+ or Mg2+ to serve as metal activator for primer degradation by the exonuclease activity of PabPolB. In the tested conditions, Mg2+ activated hydrolysis and the optimum was 5 mM, with most of the full-length 17-mer primer degraded to ∼8-mer. With Ca2+ or in the absence of an added metal ion, no excision of the 3′-terminal base was detected. These results highlight that Mg2+ triggers both exonuclease and DNA polymerization activities with similar requirement but indicate that Ca2+ can only serve as a metal activator for DNA polymerization. Since the reaction conditions produced no detectable Ca2+-activated hydrolysis, increased amounts of PabPolB, longer incubation time, pH and counterion variations have been applied (Supplementary Figure S1B–D). In any case, no primer degradation was observed compared to Mg2+.
Figure 1.
Catalytic activities of PabPolB mediated by Mg2+ or Ca2+. (A) Primer-template used for primer extension and 3′-exonuclease primer degradation experiments, in panels B–D, respectively. It consists of a Cy5-labeled 17-mer primer annealed to a DNA template of 87-nt in length. (B) Extension of the 17/87 primer-template by PabPolB at the indicated Mg2+ or Ca2+ concentrations. The numbers under the gel lanes represent the total percentage of full-length products and extension. Reference oligodeoxynucleotides of 17 and 87 bases are indicated on the right. (C) Extension of the 17/87 primed-template (25 nM) by PabPolB (75 nM) at fixed Mg2+ or Ca2+ concentrations (5 mM) and 200 μM dNTPs in a time course experiment at 55°C. The numbers under the gel lanes represent the total percentage of full-length products. Reference oligodeoxynucleotides of 17 and 87 bases are indicated on the right. (D) Proofreading exonucleolysis of 17/87 primer-template at the indicated Mg2+ or Ca2+ concentrations. The numbers under the gel lanes represent the percentage of degraded primers. Reference oligodeoxynucleotides of 17 and 8 bases are indicated on the right.
Modulation of DNA polymerization by PabPolB with Mg2+ or Ca2+ on primed-M13mp18 DNA template
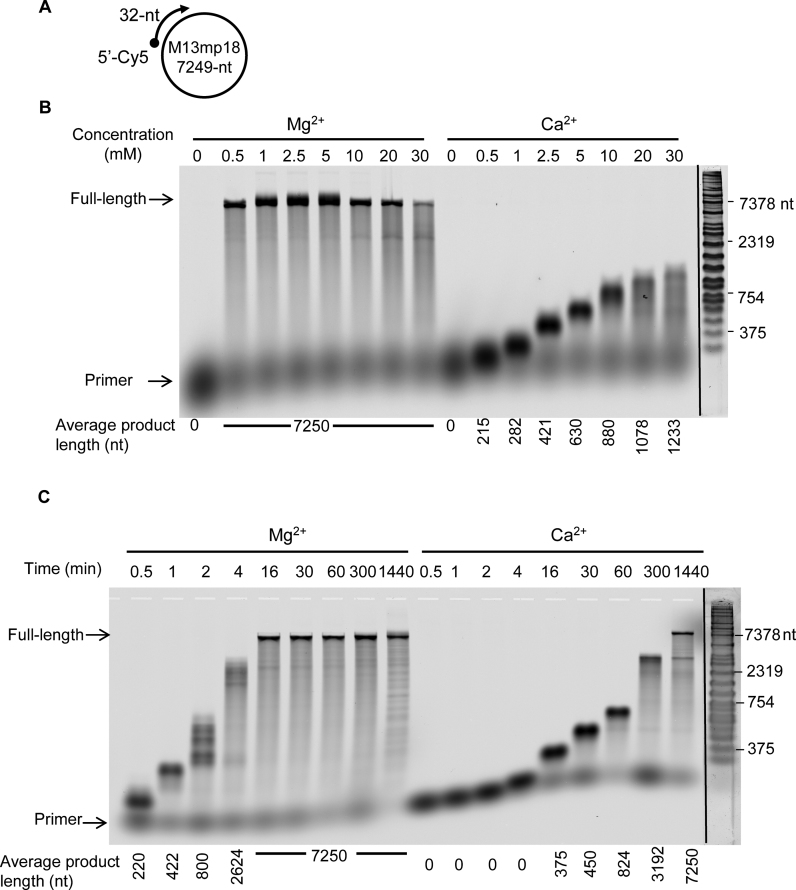
Since Mg2+ and Ca2+-mediated DNA polymerization were almost comparable on short p/t oligonucleotides, this raises the question of DNA polymerization efficiency on p/t with larger size and topology. Using primed-M13mp18 DNA template, PabPolB carried out full-length DNA synthesis (7249-nt) over a broad range of Mg2+ concentrations (Figure 2B). At 5 mM, maximum amount of full-length DNA products were synthesized but higher Mg2+ concentrations (10–30 mM) became inhibitory. Interestingly, full-length DNA synthesis was never obtained in the presence of Ca2+ even at the 5 mM optimum described on short p/t. The longest DNA fragments were ∼1230-nt in length and accumulated at the highest Ca2+ concentration (30 mM). Upon prolonged reaction incubation, full-length DNA products (7249-nt) were detectable with Ca2+ but 24 h-incubation time were required compared to only 16 min with Mg2+ (Figure 2C). Since PabPolB is a thermostable DNA Pol (67) that is known to interact with a variety of replisome components (68,78), elevated temperatures and the inclusion of accessory factor may stimulate full-length DNA synthesis with Ca2+. During a 30-min extension increased temperature (Supplementary Figure S2B) and the addition of the processivity factor PabPCNA (Supplementary Figure S2C) did little to enhance the extension rate to levels comparable to Mg2+.
Figure 2.
Modulation of DNA polymerization PabPolB with Mg2+ or Ca2+ on primed-M13mp18 DNA template. (A) Structure of the primer-template mimic, consisting of a Cy5-labeled 32-mer primer annealed to the circular M13mp18 DNA template of 7249-nt in length. (B) Extension of the primed-M13mp18 DNA template at the indicated Mg2+ or Ca2+ concentrations. The starting primer (32-nt) and full-length product (7249-nt) are shown arrowed on the left. Product length is indicated under the gel lanes. (C) Extension of the primed-M13mp18 DNA template for the times shown above the gels (min) at fixed Mg2+ or Ca2+ concentrations (5 mM). The starting primer (32-nt) and full-length product (7249-nt) are shown arrowed on the left. Product length is indicated under the gel lanes.
Single nucleotide incorporation by PabPolB in the presence of Mg2+ or Ca2+
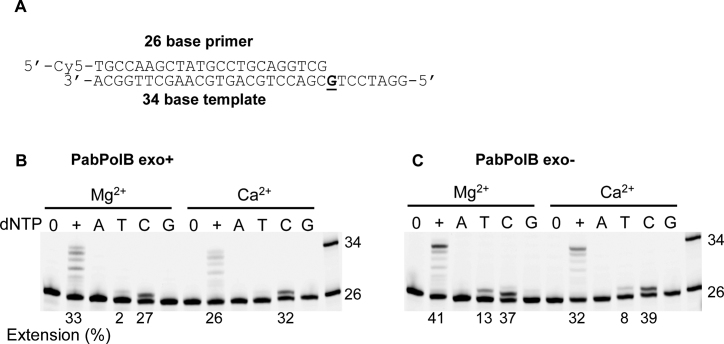
To determine if catalytic metal ion plays any role in DNA polymerase fidelity single nucleotide incorporation assays were carried out with each of the four dNTPs and the defined primer-template (Cy5-labeled primer, 26 bases long, annealed to a template 34 bases in length) (Figure 3A). As can be seen in Figure 3B, PabPolB exo+ incorporated cytosine (∼27%) opposite guanine in the presence of Mg2+ on this primer-template, although a very weak thymidine insertion (∼2%) was also detectable. When Mg2+ was substituted by Ca2+, cytosine was exclusively inserted opposite guanine (∼32%). Similar results were obtained with the exo− variant of PabPolB on the same primer-template from which single incorporation was slightly enhanced due to its inability to proofread mistakes (Figure 3C). While cytosine was mostly inserted opposite guanine with Mg2+ or Ca2+, thymidine incorporation also occurred. Independently of the PabPolB used in the presence of Ca2+ cytosine incorporation appeared slightly higher than with Mg2+, while misincorporation was reduced.
Figure 3.
Single nucleotide incorporation by PabPolB in the presence of Mg2+ or Ca2+. (A) Primer-template used for these experiments consists of a Cy5-labeled 26-mer primer annealed to a DNA template of 34-nt in length. Incorporation using PabPolB exo+ (B) or PabPolB exo− (C) at 5 mM Mg2+ or Ca2+ concentrations. 0 = no dNTPs added; + = all four dNTPs added; A, T, C and G = only dATP or dTTP or dCTP or dGTP added, respectively. The extension (%) for selected lanes is shown under the gels.
Effect of Mg2+ or Ca2+on the steady-state kinetics of wild-type and exonuclease-deficient PabPolB
Steady-state kinetics is addressed by standing start reactions as described in ‘Materials and Methods’ section, and previously published (73). In these conditions, nucleotide incorporation reactions obey Michaelis–Menten kinetics (Supplementary Figure S3) and both PabPolB (exo+ and exo−) incorporated the correct dC opposite template G with slight increased incorporation efficiency (kcat/Km) in the presence of Ca2+ (Table 1). The apparent Km values for the incorporation of cytosine opposite template G were always lower with Ca2+ compared with Mg2+. Respectively, an ∼2-fold and ∼5-fold reduction factor is conferred by the exo− and exo+ variants. While having nearly identical turnover rates (kcat) in cytosine incorporation using Ca2+, these values increased about two–three times with Mg2+. Incorrect incorporation of dT by PabPolB exo− resulted in higher apparent Km values and ∼6-fold reduction of the kcat value with Ca2+ compared with Mg2+. Catalytic efficiencies for incorrect dT is therefore strongly impaired compared to the complementary dC, having much higher dramatically reduced efficiency in the presence of Ca2+ as judged by the ∼15-fold dropped kcat/Km value. Overall these results suggest that Ca2+ seems to play a role in nucleotide selectivity and affects the rate of steady-state turnover of PabPolB.
Table 1. Steady-state kinetic analysis of nucleotide incorporation by exonuclease proficient and deficient PabPolB in the presence of Mg2+ or Ca2+.
| n | PabPolB | Metal ion | Template : incoming dNTP | V max (nM/min) | K m (μM) | K cat (min−1)x10−3 | Catalytic Efficiency Kcat/Km (μM−1min−1)10−3 | Efficiency relative to Mg2+ | f(misinsertion frequency) |
|---|---|---|---|---|---|---|---|---|---|
| 7 | Exo+ | Mg2+ | G : dC | 1.305 ± 0.055 | 0.196 ± 0.038 | 261.00 | 1329.6 | ||
| 8 | Exo+ | Ca2+ | G : dC | 0.348 ± 0.019 | 0.042 ± 0.007 | 69.58 | 1640.7 | 0.81 (↑1.2 x) | |
| 17 | Exo− | Mg2+ | G : dC | 0.736 ± 0.025 | 0.075 ± 0.015 | 147.12 | 1946.5 | 1 | |
| 9 | Exo− | Ca2+ | G : dC | 0.331 ± 0.017 | 0.0327 ± 0.006 | 66.12 | 2024.5 | 0.96 (↑ 1 x) | 1 |
| 5 | Exo− | Mg2+ | G : dT | 0.743 ± 0.082 | 180.90 ± 40.43 | 148.56 | 0.8212 | 0.0004 | |
| 5 | Exo− | Ca2+ | G : dT | 0.111± 0.015 | 396.60± 133.33 | 22.26 | 0.0561 | 0.07 (↓ 15x) | 0.00003 |
Single nucleotide insertion assays were performed as described in ‘Materials and Methods’ section. The observed rates of deoxynucleotide incorporation as a function of dNTP concentration were firstly determined from Lineweaver–Burk plots. The data were fit by non-linear regression using the Marquardt–Levenberg algorithm (EnzFitter 2.0, BioSoft) to the Michaelis–Menten equation describing a hyperbola, v = (Vmax x [dNTP]/Km +[dNTP]) as already described (73) and in Supplementary Figure S3. Apparent Km and Vmax kinetic parameters were obtained from the fit and were used to calculate the efficiency of deoxynucleotide incorporation (kcat/Km). The kinetics values are the average of 5–17 (n) determinations and are shown with standard deviation (SD). The f(misinsertion frequency) is the ratio kcat/Km for the incorrect nucleotide to kcat/Km for the correct nucleotide.
Ionic accessibility of polymerase and exonuclease active sites
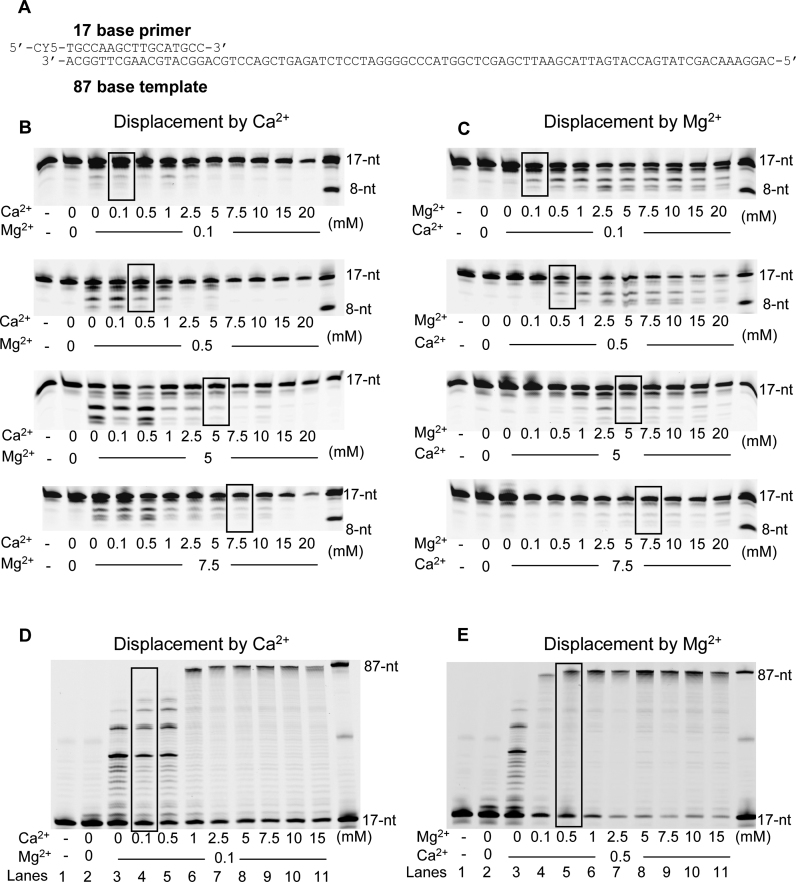
In order to determine whether Ca2+ can enter the exonuclease active site, despite its failure to support DNA degradation, ion displacement assays were performed with either fixed concentrations of Mg2+ and increased concentrations of Ca2+ (displacement by Ca2+), and the reverse (displacement by Mg2+) in the presence of p/t (Figure 4B and C) without dNTPs. When pre-incubated with different concentrations of Mg2+, PabPolB exonuclease activity progressively became repressed by Ca2+ titration (Figure 4B). The resulting loss of activity particularly occurred at equimolar Mg2+ and Ca2+ concentrations (Figure 4B, to see the bands area framed in black). In the reverse experiment using PabPolB pre-incubated with Ca2+ (displacement by Mg2+), the exonuclease activity was gradually restored by titrating Mg2+ and the enhancement of activity was more striking at concentrations ranging from 0.1 to 5 mM Ca2+ (Figure 4C). At 7.5 mM Ca2+ the activity level of primer degradation was poorly enhanced by increasing concentrations of Mg2+, indicating that the catalytic-competent enzyme-metal ion complex was not recovered. From these results, it appears that Ca2+ does not support cleavage activity.
Figure 4.
Ionic accessibility of polymerase and exonuclease active sites of PabPolB. (A) Primer-template used for 3′-exonuclease primer degradation and primer extension experiments, in panels B and C and D and E, respectively. It consists of a Cy5-labeled 17-mer primer annealed to a DNA template of 87-nt in length. Ion displacement experiment is carried out at fixed Mg2+ and Ca2+ concentrations and by increasing metal ion competitor concentrations, respectively, in panels B–D and C–E. PabPolB is pre-incubated at the indicated fixed metal concentrations. Chased experiment is initiated by co-addition of p/t (17/87) and increased ion competitor concentrations. Equimolar concentrations are shown framed. Reference oligodeoxynucleotides of 87, 17 and 8 bases are indicated on the right of each panel.
Since we observed that Ca2+ can be the polymerase catalytic metal ion, we sought to explore the replacement of metal ions in the polymerase active site. To address this question, ion displacement experiments were performed with p/t, dNTPs and increased competitor ion concentrations. PabPolB was pre-incubated with either Mg2+ or Ca2+ at a fixed concentration in order to obtain similar primer extension efficiency, respectively, 0.1 and 0.5 mM (Figure 4D and E, lanes 3). As can been seen from Figure 4D, DNA polymerase activity was progressively stimulated upon addition of Ca2+ to the Mg2+ pre-incubated PabPolB and full-length synthesis was detectable at 2.5 mM Ca2+ (lane 7). In the reverse experiments, only 0.5 mM Mg2+ to the Ca2+ pre-incubated enzyme enhanced full-length DNA synthesis (Figure 4E, lane 5). These results indicate that Ca2+ can replace Mg2+ in magnesium-mediated DNA synthesis by PabPolB and vice versa. Although restoration of full activity occurred at different metal ion concentrations, Ca2+ and Mg2+ can clearly substitute for each other.
Roles of metal ions in primer/template binding by PabPolB
The above results suggest that Ca2+ and Mg2+ are exchangeable in the polymerase and exonuclease active sites. We were thus interested in determining whether the active site bound metal ion are necessary for the binding of PabPolB to the p/t, an essential step in the initiation of DNA polymerization or degradation. Fluorescence anisotropy assays were conducted to evaluate the binding to a 3′-OH primer/template with or without the correct (dApNHpp) or incorrect (dGpNHpp) non-hydrolyzable nucleotide and the exonuclease deficient PabPolB. The results show that p/t binding occurred in the absence of ions (Supplementary Figure S4). It is likely that the apparent KD binding constants represent the sum of any specific and non-specific DNA binding modes (Table 2). On the other hand, a small but significant preference for p/t binding was observed in the presence of metal ions, with a binding activity further enhanced by Ca2+ ions in all three cases (with or without incorrect or correct nucleotides) (Table 2). Interestingly, the highest affinities for p/t were detected with the correct non-hydrolyzable nucleotide in which a polymerase binding mode (ternary complex) is suspected to prevail. In this case, apparent KD values to this primer-template were ∼7 and ∼17 nM, respectively, for Ca2+ and Mg2+. Table 2 indicates that PabPolB was bound less tightly to the p/t in the absence or presence of the incorrect non-hydrolyzable nucleotide. Clearly, the binding affinity of PabPolB decreased ∼5.5-fold and ∼2.5-fold, respectively, with Ca2+ and Mg2+ (binary complex) in the absence of nucleotide. In this particular case, exonuclease DNA binding modes might be preferentially adopted. Upon the addition of the incorrect non-hydrolyzable nucleotide to the p/t DNA, PabPolB bound with a ∼16-fold and ∼8-fold lower affinity than with the correct incoming nucleotide (ternary complex), respectively with Ca2+ and Mg2+. Overall, these binding studies show that PabPolB seems to adopt a polymerase binding mode in the presence of nucleotides and that Ca2+ ions slightly enhance the binding to p/t. Absence of the nucleotide likely triggers exonuclease DNA binding modes by PabPolB with p/t affinities further enhanced by Ca2+ compared to Mg2+.
Table 2. Roles of metal ions in primer/template binding by PabPolB.
| DNA | dNTP | K D (nM) Ca2+ | K D (nM) Mg2+ | K D (nM) no ion |
|---|---|---|---|---|
| 5′-CGCCGGGCCGAGCCGTGC-3′ | no | 38 ± 6 | 43 ± 6 | 20 ± 6 |
| 3′-GCGGCCCGGCTCGGCACGTGCTGGA-HEX 5′ | dApNHpp | 7 ± 1 | 17 ± 2 | 25 ± 3 |
| dGpNHpp | 114 ± 18 | 133 ± 21 | 236 ± 50 |
The KD values for the binding of PabPolB (average ± standard deviation from at least three determinations) to a HEX-labeled primer-template (sequences given in Materials and Methods) are obtained using fluorescence anisotropy titration as described in Supplementary Figure S4. The buffer was complemented (or not = no ion) with metallic cofactor (Ca2+ or Mg2+) and with (or without = no) saturating amounts of non-hydrolyzable nucleotides (dApNHpp for correct or dGpNHpp for incorrect incoming nucleotides).
Contribution of diverse metal ions to exonuclease and DNA polymerase activities by PabPolB
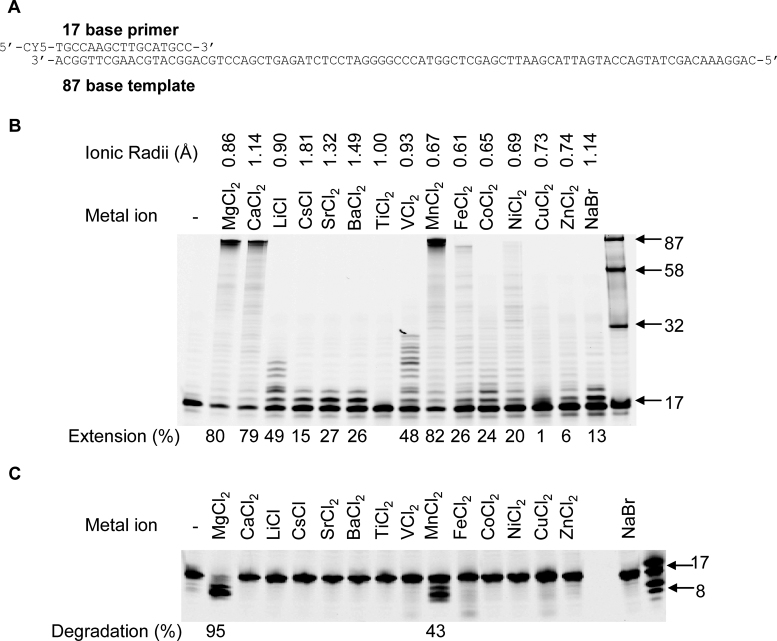
The demonstration that Ca2+ ions can enter the active sites of the exonuclease and polymerase domains (Figure 4) conferring, or not, functionalities prompt us to analyze whether other metal ions could substitute for catalysis. Alkali (Cs1+, Li1+), alkaline earth (Ca2+, Mg2+, Sr2+, Ba2+), transition metals (Ti2+, V2+, Mn2+, Fe2+ Co2+, Ni2+ Cu2+, Zn2+) and the metal halide (NaBr) were thus tested for their ability to support DNA polymerization and primer degradation (Figure 5). In the absence of metal ions, PabPolB did not exhibit DNA synthesis while addition of either Ca2+ or Mg2+ triggered catalysis as expected (Figure 5B). Among alkaline earth metals, primer extension efficiency was the highest with Ca2+ and Mg2+ (∼80% extension). Moreover, increased ionic radii ≥ 1.32 Å (Sr2+and Ba2+) resulted in reduced primer extension, thus indicating size restrictions at the polymerase active site. In the presence of transition metals which have ionic radii (0.61–1.00 Å), Mn2+ was found to be a functional and robust catalytic substitute (∼80% extension) followed to a lesser extent by V2+ (∼48% extension). In this case, most of the metals shared similar ionic radii. For alkali metals, Li1+ supported the highest extension efficiency (∼49% extension), while Cs1+ having a large ionic radii (1.81 Å) was a poor activator. Finally, NaBr slightly promoted primer extension. Overall, efficiency of extension with different metal ions was Mn2+ > Mg2+ > Ca2+ >> Li1+ > V2+ >> Sr2+> Ba2+ = Fe2+ > Co2+ > Ni2+ >> Cs1+ > Zn2+. However, Ti2+ and Cu2+ did not activate DNA synthesis. These results highlight that a broad range of metal ions can occupy and functionally substitute at the polymerase active site.
Figure 5.
Contribution of diverse metal ions to exonuclease and DNA polymerase activities by PabPolB. (A) Primer-template used for primer extension experiments and 3′-exonuclease primer degradation, in panels B and C, respectively. It consists of a Cy5-labeled 17-mer primer annealed to a DNA template of 87-nt in length. (B) Extension of the 17/87 primer-template by PabPolB at 5 mM metal ion concentrations. The numbers under the gel lanes represent the total percentage of extended products. Reference oligodeoxynucleotides of 17, 32, 58 and 87 bases are indicated by the arrows. Ionic radii (Å) are displayed above metal ions. (C) Proofreading exonucleolysis of 17/87 primer-template at 5 mM metal ion concentrations. The numbers under the gel lanes represent the percentage of degraded primers. Reference oligodeoxynucleotides of 17 and 8 bases are indicated by the arrows.
Given the diversity of metal ions that facilitate DNA polymerization by PabPolB, we sought to explore whether they might also participate in exonuclease reactions. Interestingly, only Mg2+ and Mn2+ promoted primer degradation while other metal ions rendered inactive the enzyme (Figure 5C). From these results it appears that the substitutional flexibility of metal ions at exonuclease active site of PabPolB is rather limited.
Effect of calcium on exonuclease and DNA polymerase activities by various families of DNA Pols
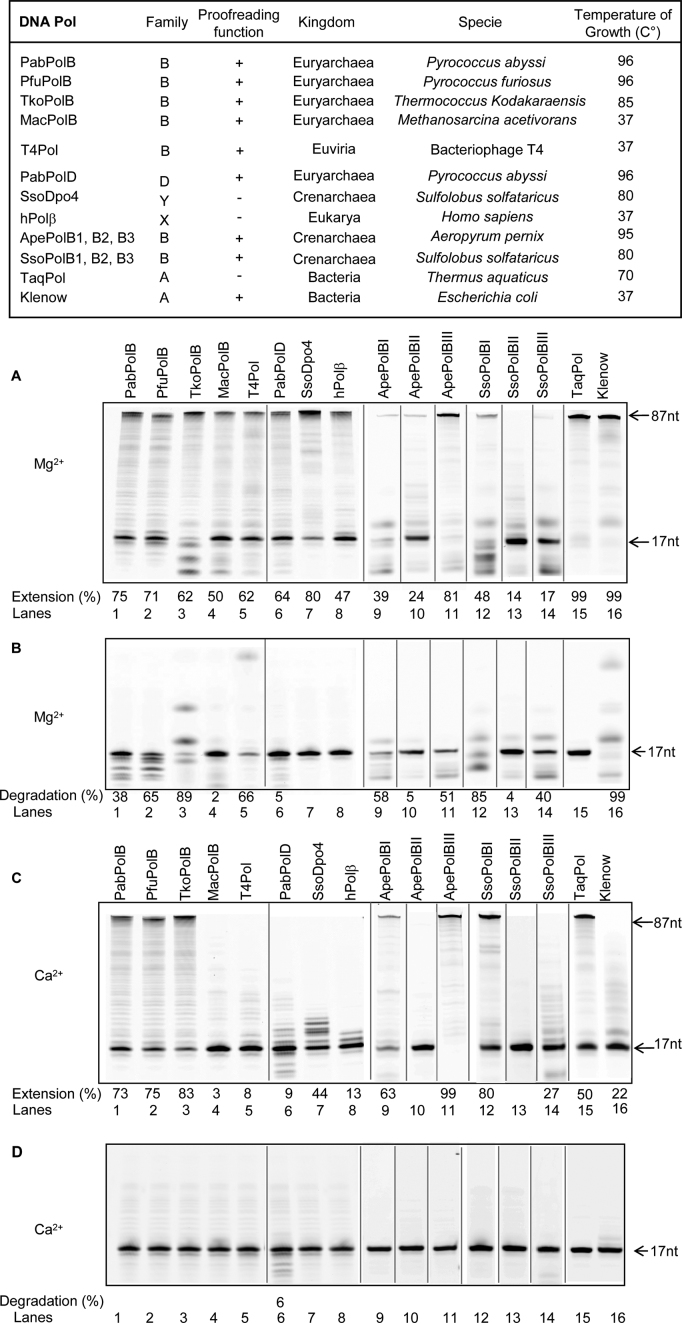
Although the physiological metal ion for different families of DNA Pols appears to be magnesium, they can utilize a variety of divalent cations for in vitro DNA polymerizing and cleavage reactions, this includes Mn2+, Co2+, Ni2+, Zn2+, Cu2+or Cd2+, depending on the enzyme as reviewed recently (62). Along with our findings it has been found that DNA synthesis can be activated by Ca2+ for the archaeal family Y Dpo4 DNA Pol (65). Thus, we sought to analyze whether Ca2+ usage might be expanded to other DNA Pols. To address this question, various families DNA Pols (A, B, D, Y and X) containing or not the proofreading function were tested with either Ca2+ or Mg2+ in primer extension and degradation assays.
As expected, Mg2+ could support both DNA polymerase and exonuclease activities by all families of DNA Pols (Figure 6A and B). Different levels of DNA synthesis was observed (from ∼14 to ∼99% extension) and associated to primer degradation (products shorter than the primer length) to varying degrees for all proficient proofreading enzymes (Figure 6A). Moreover, Mg2+ was required to activate the exonuclease activity to different efficiencies depending on the HiFi DNA pols (Figure 6B). In this case, effective cleavage accounted for short products migrating at a position below the 17-nt oligonucleotide marker. Besides, higher migration products resulting from subsequent exonucleolytic degradation were also detectable (Figure 6B, lanes 3, 5, 9, 12, 14, 16). Although having an altered electrophoretic mobility running at a higher position than the 17-nt oligonucleotide marker these cleavage fragments mostly ranged from 1 to 8-nt in length as previously demonstrated (79).
Figure 6.
Effect of calcium on exonuclease and DNA polymerase activities by various families of DNA Pols. The characteristics of families A, B, D, X and Y DNA Pols distributed across the three kingdom of life are summarized. The primer-template used for primer extension experiments and 3′-exonuclease primer degradation, in panels A–C and B–D, consists of a Cy5-labeled 17-mer primer annealed to a DNA template of 87-nt in length. (A–C) Extension of the 17/87 primer-template by DNA Pols at fixed Mg2+ or Ca2+ concentrations (5 mM). The extension (%) for selected lanes is shown under the gel. Reference oligodeoxynucleotides of 17 and 87 bases are indicated by the arrows. (B–D) Proofreading exonucleolysis of 17/87 primer-template at 5 mM Mg2+ or Ca2+ concentrations. The degradation (%) for selected lanes is shown under the gel. Reference oligodeoxynucleotides of 17 and 8 bases are indicated by the arrows. Black lines separate lanes which were not adjacent in the original gel.
When Mg2+ was replaced by Ca2+ thermophilic DNA Pols retained the highest DNA polymerase activities (Figure 6C, lanes 1–3, 7, 9, 11, 12, 14 and 15). These results are also in agreement with the previous study showing Ca2+-mediated DNA synthesis by Dpo4 (65). However, the level of primer extension was not very effective with mesophilic DNA Pols (Figure 6C, lanes 4, 5, 8 and 16) as well as for the thermostable archaeal HiFi PabPolD (Figure 6C, lane 6). No DNA synthesis was observed for the two thermostable archaeal families B2 of DNA Pols (Figure 6C, lanes 10 and 13). On the other hand, Ca2+ never supported exonuclease degradation of the primer for all DNA Pol families excepted for the thermophilic HiFi PabPolD (Figure 6D, lane 6). In this case, degradation fragments migrating at a position below the 17-nt oligonucleotide marker appeared as clear bands. Although the percentage of cleavage by PabPolD is very low, this result overlaps that seen in Figure 6C wherein DNA synthesis was associated with primer degradation in the presence of Ca2+ (lane 6).
DISCUSSION
The data presented in this publication provide compelling evidence that Ca2+ is used for catalysis of nucleic acid synthesis but not for the excision of the 3′-terminal base by hyperthermophilic euryarchaeal HiFi family-B DNA Pols. Compared to Mg2+, Ca2+-activation of bulk DNA synthesis is relatively slower. Consistently, a lower enzymatic rate constant for single nucleotide incorporation is observed which when extrapolated to multiple rounds of nucleotide incorporation delays full DNA synthesis. Kinetic parameters also indicate that Ca2+-mediated incorporation is more accurate compared to Mg2+ when the incoming nucleotide is correct and that misinsertion events are unfavorable. These findings suggest that PabPolB is endowed with a higher fidelity of DNA synthesis in the presence of Ca2+ metal ions, likely to counteract its Ca2+-inactivated exonuclease proofreading function. Binding studies show that PabPolB likely adopts a polymerase DNA binding mode in the presence of correct or incorrect incoming nucleotides and that Ca2+ ions slightly enhance the binding to p/t. Absence of the nucleotide possibly triggers exonuclease DNA binding modes by PabPolB with p/t affinities further enhanced by Ca2+ compared to Mg2+. Moreover, Ca2+ is not an activator of primer degradation and metal ions substitution is rather limited. Only Mn2+ which is most similar to Mg2+ in chemical nature allows cleavage reactions. On the contrary, the substitutional flexibility of metal ions at the polymerase active site of PabPolB is expansive with Mg2+, Ca2+ and Mn2+ conferring the most efficient DNA synthesis. This study also demonstrates that Ca2+-driven DNA polymerization more generally applies to thermophilic DNA Pols in which family-B DNA Pols (except for B2) retain the highest efficiency.
Although our steady-state kinetic and binding analyses have provided preliminary information on metal activation with Ca2+ by PabPolB, they were not sufficient for understanding the exact detailed reaction mechanism. It was surprising to observe that the higher catalytic efficiency for correct nucleotide insertion as well as the higher affinity for the primer-template did not confer processive DNA synthesis. Indeed, the Ca2+-mediated catalysis by PabPolB resulted in extension products of diminished length in the shortest time points on both synthetic oligonucleotide primer-templates and primed-M13mp18. It is therefore likely that the kinetic mechanism involves the modulation of additional intrinsic parameters during the course of DNA polymerization. Since steady-state measurements are averages of the rates for DNA polymerization and the dissociation of enzyme–DNA complex, pre-steady-state analysis of single turnovers catalyzed by PabPolB would represent an ideal method to provide kinetic schemes of the reaction with Ca2+ compared to Mg2+.
Thanks to the recent development of time-resolved crystallization method allowing the capture of ternary structural intermediates (pre-catalytic, reaction-state and post-catalytic structures), the DNA polymerase catalytic cycle has been revised. A third metal ion (‘C’ site) is likely to be essential in activating the chemical reaction in mesophilic families X and Y DNA Pols (35–37,80). In these time-lapse experiments, pre-catalytic ternary complexes are always obtained with Ca2+ ions because of its inability to trigger DNA synthesis, and structural overlays reveal subtle difference between Ca2+ and Mg2+ coordination geometries at the active site aspartate residues. The longer metal-atom target distances, variable metal-atom numbers and larger atomic radius might explain why Ca2+ ions are not ideally suited for catalysis compared to Mg2+ (81). In addition the dynamics of Ca2+ ion positioned at a varied environment surrounding the C-site might disfavor the three-metal ion catalysis (38). This assumption agrees with the impaired DNA polymerization by most of the mesophilic DNA Pols in the presence of Ca2+ in our study (Figure 6C). In contrast, Ca2+-activated DNA synthesis is observed with thermophilic DNA Pols (archaeal families B and Y, bacterial family A Taq Pol) in which, presumably, a three metal ion mechanism takes place. In Dpo4, these findings are corroborated by subtle changes in the overall architecture and active-site conformations from the structures of ternary Mg2+- and Ca2+-complexes (66). The active site geometry, metal position and composition occupying both the canonical A and B sites are consistent with a pre-formed Pol/DNA/nucleotide catalytic complex. Regards to the fully alignment of the 3′-OH of DNA primer and incoming dNTP (also observed in mesophilic families X and Y DNA Pols), Ca2+ activation awaits the transitory metal ion to occupy the C-site. Interestingly, thermal motion of the fully aligned reactants is proposed to support the transient entrance of metal C in the revised three-metal ion catalysis. As such the third metal ion neutralizes the negative charge in the transition state and facilitates phosphodiester bond formation (36,38). Consistently, the higher temperature reaction required for both Mg2+ and Ca2+ activation might confer an increased flexibility of the polymerase active site of PabPolB to optimize the DNA polymerizing activity at physiological temperature. Interestingly, the regulation of optimal active site flexibility has been recently described in the literature for other thermophilic enzymes (82,83). For PabPolB, Ca2+ or Mg2+ concentrations and the general reaction process are almost indistinguishable with only ∼3.5-fold reduction in enzymatic rate constant for Ca2+. In this case, calcium dynamics within the active site might not be as ideal as Mg2+, possibly delaying enzyme-catalyzed critical steps (adopting the exquisite coordination geometries and metal-ligand distances, positioning of the third metal, releasing of PPi). Thus, it would be interesting to experiment time-lapse X-ray crystallography to more clearly understand the chemical kinetic mechanism of Ca2+-activated DNA synthesis by PabPolB, to which Ti2+ ion represents a good non-catalytic metal surrogate (Figure 5B). Combined to pre-steady-state analysis, PabPolB could therefore provide a good model for Ca2+-catalyzed nucleotidyl transfer reactions in terms of metal ion specificity, nucleotide specificity, fidelity and processivity.
In our study, the bacterial mesophilic family A Klenow Pol displays DNA polymerization in the presence of Ca2+ which is lower than thermophilic enzymes (Figure 6C). This result is consistent with a previous work accounting for nucleotide incorporation. In these experiments, Mg2+ is replaced by Ca2+ in order to slow the reaction rate sufficiently to allow real-time monitoring of primer extension (11). Like the equivalent thermophilic family A Taq Pol, DNA synthesis is activated by Ca2+ with the Klenow Pol but drastically reduced. Again, like for the thermophilic archaeal family B DNA Pol, higher thermal vibrations occurring in the active site of the thermophilic bacterial enzyme might facilitate the chemical reaction.
Surprisingly, only the crenarchaeal hyperthermophilic families B1 and B3 DNA Pols were activated by Ca2+ in our primer extension assays. To date few B3 and B2 DNA Pols compared to B1 have been functionally characterized with Mg2+ (75,84). For instance, it has been shown that SsoPolB1 exhibits the strongest DNA polymerase and 3′-5′ exonuclease activities, while B2 and B3 DNA Pols were less efficient (76). Besides, detailed phylogenetic analyses and amino acid alignments highlighted no striking differences among families B DNA Pols, especially within DNA Pol domains. B1 and B3 DNA Pols are well-represented in Archaea and share almost conserved polymerase and exonuclease motifs, while B2 DNA Pols show a patch distribution in most archaeal lineages and have missing exonuclease motifs (85–87). The fact that B2 DNA Pols are not active with Ca2+ might be explained structurally by their truncated exonuclease domain, thereby influencing the overall architecture of the enzyme and impeding the optimal configuration of the polymerase active site.
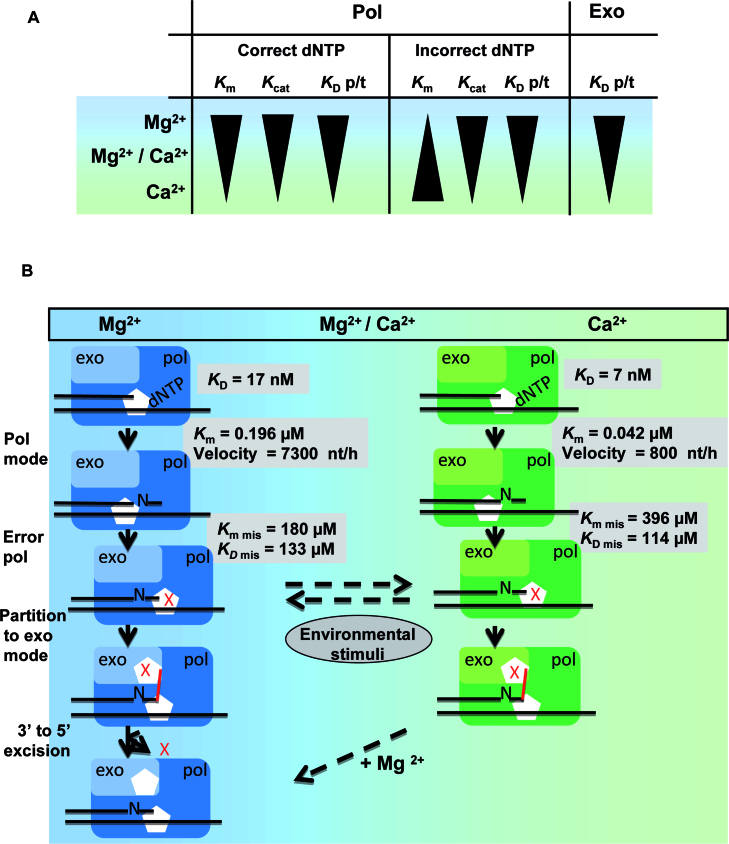
Our findings are illustrated in the diagram in Figure 7A, which describes the altered in vitro kinetic parameters when Mg2+ is replaced by Ca2+ (or vice versa). Compared to Mg2+, Ca2+ confers a higher accuracy of nucleotide incorporation, slows the catalytic rates and prevents misinsertion events by PabPolB. Coupled with this, Ca2+ confers a higher stabilization of PabPolB on p/t, a change that is observed in both polymerase and exonuclease DNA binding mode. On the other hand, Ca2+ is not an activator of primer degradation by PabPolB and represents a potent inhibitor of Mg2+-mediated exonuclease activity. A hybrid exonuclease active site containing both metals is either much less active, or more susceptible to inhibition by Ca2+. The presence of two Mg2+/Ca2+ ions or two Ca2+ ions possibly disrupts the optimal active site configuration. Thus, our data suggest that the polymerase and exonuclease active sites of the euryarchaeal hyperthermophilic family B DNA Pol evolved different chemical and physical constraints on catalysis. Upon variation of metal ions levels, it provides the possibility to tune catalytic functions by PolB without disturbing its biological activity in archaeal cells. An in vivo scenario is presented in Figure 7B. Under conditions where Mg2+ intracellular concentration is imbalanced, Ca2+ can substitute for nucleotide incorporation, thus allowing cells to synthesize DNA. While exonuclease correction is inactivated, Ca2+ slows but renders DNA synthesis more accurate that decreases the probability of mutagenicity. Recovery of physiological Mg2+ concentration then ensures that both the corrective 3′→5′ exonucleolytic and DNA polymerases activities are activated, conferring high fidelity and processive DNA synthesis. Although our working model awaits the full determination of intracellular metal ion concentrations in such a microorganism, there are indications that Archaea evolved elaborate mechanisms to maintain adequate metal ions homeostasis (88,89). Like for eukaryotes (90,91), it is possible that elevation of Ca2+ concentration takes place during replication stress or damage in Archaea. Thus, the effect we have observed in vitro with Ca2+ for PabPolB might occur in vivo. Depending on the stage in the cell cycle in which Ca2+ spikes occur, the recruitment of accessory proteins or enzymes for rendering PabPolB more processive might be specifically regulated and adapted to the DNA synthesis processes (e.g. specialized DNA repair mechanisms, post-replicative repair pathways, translesion DNA synthesis, etc.). In support of this hypothesis, the activation of appropriate DNA repair mechanisms in human cells exposed to ionizing radiation have been shown to be dependent on the cell cycle and measurements of the DNA repair patch sizes clearly reflected the operation of particular pathways (92).
Figure 7.
Schematic illustration of a model for the regulation of exonuclease and DNA polymerase catalyses by metal ions. (A) Diagram of the effects of Mg2+ or/and Ca2+ on PabPolB. The first two columns point out the kinetic parameters (Km and Kcat) and the primer/template binding affinity (KD p/t) when PabPolB selects the correct versus incorrect dNTP. The third column shows the primer/template binding affinity (KD p/t) of PabPolB in exonuclease mode. The black triangles denote the drift constants from low to high. The two-color gradient in the background indicates the presence of Mg2+ (blue) or Ca2+ (green) alone, or a mixture of Mg2+ and Ca2+ (between blue and green). (B) Model of the modulation of DNA polymerase and exonuclease activities by fluctuating metal ion concentration in archaeal cells. PabPolB exonuclease and polymerase domains are light and dark blue with Mg2+ or light and dark green with Ca2+. ‘mis’ denotes misinserted nucleotide which is represented by a red cross. The schematic illustration on the left describes the activation of DNA polymerase and exonuclease activities by Mg2+. The nucleotide incorporation cycle is accurate, fast and balanced by the excision of mismatches. At any time, PabPolB remains susceptible to changes in metal ion content in response to external stress (dashed rightward arrow over leftward arrow). The schematic illustration on the right shows the tuning effects (DNA polymerase activation and exonuclease inactivation) in the presence of Ca2+. PabPolB binds the p/t with higher affinity and DNA synthesis is slower but more accurate. When PabPolB is stalled by a mismatch in an exonuclease mode, displacement of Ca2+ ions by local Mg2+ spikes reactivates its proofreading function (dashed arrow).
On the other hand, our hypothesis that metal-ion catalysis for DNA polymerization involves a combination of metal ions remains to be addressed. To our knowledge, there is no related literature for HiFi DNA Pol which, when available, remains restricted to mesophilic bacterial topoisomerase IV (93) and archaeal topoisomerase III (94). Our study also describes the exonuclease catalytic incompetency for primer degradation by PabPolB which expands to most of the HiFi DNA Pols tested in that study, except for the family D DNA Pol. Interestingly, the exonuclease domain of PabPolD which shares striking structural similarities to the Mre11 exo−/endonuclease likely accounts for the unusual Ca2+-activated DNA degradation (50). So far, the use of Ca2+ ions in the DNA cleavage reaction has been documented for diverse nucleases and topoisomerases from Archaea to Bacteria (93–97).
Supplementary Material
ACKNOWLEDGEMENTS
The technical assistance of Audrey Bossé throughout this project is greatly appreciated. We thank Prof. I.K.O Cann for providing the MacPolB construct, Dr F. Pisani for the SsoPolB1, Prof. U. Hübscher for providing us with purified human Polβ, Prof. Y. Ishino and S. Ishino for the generous gift of the SsoPolB2, SsoPolB3, ApePolB1, ApePolB2 and ApePolB3 plasmids.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: French National Research Agency (ANR) [ANR-10-JCJC-1501–01 to G.H.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Bebenek K., Kunkel T.A.. Functions of DNA polymerases. Adv. Protein Chem. 2004; 69:137–165. [DOI] [PubMed] [Google Scholar]
- 2. Ishino Y., Ishino S.. DNA replication in Archaea, the third domain of life. Tech, Book series: The Mechanisms of DNA Replication. 2013; Croatia: In Tech Open (E-Books) http://www.intechopen.com/articles/show/title/dna-replication-in-archaea-the-third-domain-of-life. [Google Scholar]
- 3. Johansson E., Dixon N.. Replicative DNA polymerases. Cold Spring Harb. Perspect. Biol. 2013; 5:a012799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kool E.T. Active site tightness and substrate fit in DNA replication. Annu. Rev. Biochem. 2002; 71:191–219. [DOI] [PubMed] [Google Scholar]
- 5. Rothwell P.J., Waksman G.. Structure and mechanism of DNA polymerases. Adv. Protein Chem. 2005; 71:401–440. [DOI] [PubMed] [Google Scholar]
- 6. Delagoutte E. DNA polymerases: mechanistic insight from biochemical and biophysical studies. Front. Biosci. (Landmark Ed). 2012; 17:509–544. [DOI] [PubMed] [Google Scholar]
- 7. Kunkel T.A. DNA replication fidelity. J. Biol. Chem. 2004; 279:16895–16898. [DOI] [PubMed] [Google Scholar]
- 8. Goodman M.F., Creighton S., Bloom L.B., Petruska J.. Biochemical basis of DNA replication fidelity. Crit. Rev. Biochem. Mol. Biol. 1993; 28:83–126. [DOI] [PubMed] [Google Scholar]
- 9. Maki H., Kornberg A.. Proofreading by DNA polymerase III of Escherichia coli depends on cooperative interaction of the polymerase and exonuclease subunits. Proc. Natl. Acad. Sci. U.S.A. 1987; 84:4389–4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Joyce C.M. How DNA travels between the separate polymerase and 3′-5′-exonuclease sites of DNA polymerase I (Klenow fragment). J. Biol. Chem. 1989; 264:10858–10866. [PubMed] [Google Scholar]
- 11. Datta K., Johnson N.P., von Hippel P.H.. DNA conformational changes at the primer-template junction regulate the fidelity of replication by DNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:17980–17985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Darmawan H., Harrison M., Reha-Krantz L.J.. DNA polymerase 3′→5′ exonuclease activity: Different roles of the beta hairpin structure in family-B DNA polymerases. DNA Repair (Amst). 2015; 29:36–46. [DOI] [PubMed] [Google Scholar]
- 13. Reha-Krantz L.J. DNA polymerase proofreading: Multiple roles maintain genome stability. Biochim. Biophys. Acta. 2010; 1804:1049–1063. [DOI] [PubMed] [Google Scholar]
- 14. Beese L.S., Derbyshire V., Steitz T.A.. Structure of DNA polymerase I Klenow fragment bound to duplex DNA. Science. 1993; 260:352–355. [DOI] [PubMed] [Google Scholar]
- 15. Li Y., Korolev S., Waksman G.. Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation. EMBO J. 1998; 17:7514–7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Johnson K.A. Conformational coupling in DNA polymerase fidelity. Annu. Rev. Biochem. 1993; 62:685–713. [DOI] [PubMed] [Google Scholar]
- 17. Joyce C.M., Steitz T.A.. Function and structure relationships in DNA polymerases. Annu. Rev. Biochem. 1994; 63:777–822. [DOI] [PubMed] [Google Scholar]
- 18. Johnson S.J., Taylor J.S., Beese L.S.. Processive DNA synthesis observed in a polymerase crystal suggests a mechanism for the prevention of frameshift mutations. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:3895–3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Swan M.K., Johnson R.E., Prakash L., Prakash S., Aggarwal A.K.. Structural basis of high-fidelity DNA synthesis by yeast DNA polymerase delta. Nat. Struct. Mol. Biol. 2009; 16:979–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Xia S., Wang M., Blaha G., Konigsberg W.H., Wang J.. Structural insights into complete metal ion coordination from ternary complexes of B family RB69 DNA polymerase. Biochemistry. 2011; 50:9114–9124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mayanagi K., Kiyonari S., Nishida H., Saito M., Kohda D., Ishino Y., Shirai T., Morikawa K.. Architecture of the DNA polymerase B-proliferating cell nuclear antigen (PCNA)-DNA ternary complex. Proc. Natl. Acad. Sci. U.S.A. 2015; 108:1845–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gouge J., Ralec C., Henneke G., Delarue M.. Molecular recognition of canonical and deaminated bases by P. abyssi family B DNA polymerase. J. Mol. Biol. 2012; 423:315–336. [DOI] [PubMed] [Google Scholar]
- 23. Hopfner K.P., Eichinger A., Engh R.A., Laue F., Ankenbauer W., Huber R., Angerer B.. Crystal structure of a thermostable type B DNA polymerase from Thermococcus gorgonarius. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:3600–3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Beese L.S., Steitz T.A.. Structural basis for the 3′-5′ exonuclease activity of Escherichia coli DNA polymerase I: a two metal ion mechanism. EMBO J. 1991; 10:25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Steitz T.A., Smerdon S.J., Jager J., Joyce C.M.. A unified polymerase mechanism for nonhomologous DNA and RNA polymerases. Science. 1994; 266:2022–2025. [DOI] [PubMed] [Google Scholar]
- 26. Han H., Rifkind J.M., Mildvan A.S.. Role of divalent cations in the 3′,5′-exonuclease reaction of DNA polymerase I. Biochemistry. 1991; 30:11104–11108. [DOI] [PubMed] [Google Scholar]
- 27. Steitz T.A., Steitz J.A.. A general two-metal-ion mechanism for catalytic RNA. Proc. Natl. Acad. Sci. U.S.A. 1993; 90:6498–6502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Doublie S., Ellenberger T.. The mechanism of action of T7 DNA polymerase. Curr. Opin. Struct. Biol. 1998; 8:704–712. [DOI] [PubMed] [Google Scholar]
- 29. Huang H., Chopra R., Verdine G.L., Harrison S.C.. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science. 1998; 282:1669–1675. [DOI] [PubMed] [Google Scholar]
- 30. Ling H., Boudsocq F., Woodgate R., Yang W.. Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell. 2001; 107:91–102. [DOI] [PubMed] [Google Scholar]
- 31. Pelletier H., Sawaya M.R., Kumar A., Wilson S.H., Kraut J.. Structures of ternary complexes of rat DNA polymerase beta, a DNA template-primer, and ddCTP. Science. 1994; 264:1891–1903. [PubMed] [Google Scholar]
- 32. Biertumpfel C., Zhao Y., Kondo Y., Ramon-Maiques S., Gregory M., Lee J.Y., Masutani C., Lehmann A.R., Hanaoka F., Yang W.. Structure and mechanism of human DNA polymerase eta. Nature. 2010; 465:1044–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jain R., Rajashankar K.R., Buku A., Johnson R.E., Prakash L., Prakash S., Aggarwal A.K.. Crystal structure of yeast DNA polymerase epsilon catalytic domain. PLoS One. 2014; 9:e94835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Castro C., Smidansky E., Maksimchuk K.R., Arnold J.J., Korneeva V.S., Gotte M., Konigsberg W., Cameron C.E.. Two proton transfers in the transition state for nucleotidyl transfer catalyzed by RNA- and DNA-dependent RNA and DNA polymerases. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:4267–4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Freudenthal B.D., Beard W.A., Shock D.D., Wilson S.H.. Observing a DNA polymerase choose right from wrong. Cell. 2013; 154:157–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gao Y., Yang W.. Capture of a third Mg(2)(+) is essential for catalyzing DNA synthesis. Science. 2016; 352:1334–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vyas R., Reed A.J., Tokarsky E.J., Suo Z.. Viewing Human DNA Polymerase beta Faithfully and Unfaithfully Bypass an Oxidative Lesion by Time-Dependent Crystallography. J. Am. Chem. Soc. 2015; 137:5225–5230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yang W., Weng P.J., Gao Y.. A new paradigm of DNA synthesis: three-metal-ion catalysis. Cell Biosci. 2016; 6:51–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Szymanski M.R., Kuznetsov V.B., Shumate C., Meng Q., Lee Y.S., Patel G., Patel S., Yin Y.W.. Structural basis for processivity and antiviral drug toxicity in human mitochondrial DNA replicase. EMBO J. 2015; 34:1959–1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Doublie S., Tabor S., Long A.M., Richardson C.C., Ellenberger T.. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution. Nature. 1998; 391:251–258. [DOI] [PubMed] [Google Scholar]
- 41. Freemont P.S., Friedman J.M., Beese L.S., Sanderson M.R., Steitz T.A.. Cocrystal structure of an editing complex of Klenow fragment with DNA. Proc. Natl. Acad. Sci. U.S.A. 1988; 85:8924–8928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang J., Sattar A.K., Wang C.C., Karam J.D., Konigsberg W.H., Steitz T.A.. Crystal structure of a pol alpha family replication DNA polymerase from bacteriophage RB69. Cell. 1997; 89:1087–1099. [DOI] [PubMed] [Google Scholar]
- 43. Franklin M.C., Wang J., Steitz T.A.. Structure of the replicating complex of a pol alpha family DNA polymerase. Cell. 2001; 105:657–667. [DOI] [PubMed] [Google Scholar]
- 44. Lee Y.S., Kennedy W.D., Yin Y.W.. Structural insight into processive human mitochondrial DNA synthesis and disease-related polymerase mutations. Cell. 2009; 139:312–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hogg M., Osterman P., Bylund G.O., Ganai R.A., Lundstrom E.B., Sauer-Eriksson A.E., Johansson E.. Structural basis for processive DNA synthesis by yeast DNA polymerase epsilon. Nat. Struct. Mol. Biol. 2014; 21:49–55. [DOI] [PubMed] [Google Scholar]
- 46. Fernandez-Leiro R., Conrad J., Scheres S.H., Lamers M.H.. Cryo-EM structures of the E. coli replicative DNA polymerase reveal its dynamic interactions with the DNA sliding clamp, exonuclease and tau. Elife. 2015; 4:e11134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bailey S., Wing R.A., Steitz T.A.. The structure of T. aquaticus DNA polymerase III is distinct from eukaryotic replicative DNA polymerases. Cell. 2006; 126:893–904. [DOI] [PubMed] [Google Scholar]
- 48. Lamers M.H., Georgescu R.E., Lee S.G., O’Donnell M., Kuriyan J.. Crystal structure of the catalytic alpha subunit of E. coli replicative DNA polymerase III. Cell. 2006; 126:881–892. [DOI] [PubMed] [Google Scholar]
- 49. Evans R.J., Davies D.R., Bullard J.M., Christensen J., Green L.S., Guiles J.W., Pata J.D., Ribble W.K., Janjic N., Jarvis T.C.. Structure of PolC reveals unique DNA binding and fidelity determinants. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:20695–20700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sauguet L., Raia P., Henneke G., Delarue M.. Shared active site architecture between archaeal PolD and multi-subunit RNA polymerases revealed by X-ray crystallography. Nat. Commun. 2016; 7:12227–12238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hamdan S., Carr P.D., Brown S.E., Ollis D.L., Dixon N.E.. Structural basis for proofreading during replication of the Escherichia coli chromosome. Structure. 2002; 10:535–546. [DOI] [PubMed] [Google Scholar]
- 52. Shamoo Y., Steitz T.A.. Building a replisome from interacting pieces: sliding clamp complexed to a peptide from DNA polymerase and a polymerase editing complex. Cell. 1999; 99:155–166. [DOI] [PubMed] [Google Scholar]
- 53. Elisseeva E., Mandal S.S., Reha-Krantz L.J.. Mutational and pH studies of the 3′ → 5′ exonuclease activity of bacteriophage T4 DNA polymerase. J. Biol. Chem. 1999; 274:25151–25158. [DOI] [PubMed] [Google Scholar]
- 54. Kung F.C., Raymond J., Glaser D.A.. Metal ion content of Escherichia coli versus cell age. J. Bacteriol. 1976; 126:1089–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Maguire M.E., Cowan J.A.. Magnesium chemistry and biochemistry. Biometals. 2002; 15:203–210. [DOI] [PubMed] [Google Scholar]
- 56. Laires M.J., Monteiro C.P., Bicho M.. Role of cellular magnesium in health and human disease. Front. Biosci. 2004; 9:262–276. [DOI] [PubMed] [Google Scholar]
- 57. Cowan J.A. Metal activation of enzymes in nucleic acid biochemistry. Chem. Rev. 1998; 98:1067–1088. [DOI] [PubMed] [Google Scholar]
- 58. Beckman R.A., Mildvan A.S., Loeb L.A.. On the fidelity of DNA replication: manganese mutagenesis in vitro. Biochemistry. 1985; 24:5810–5817. [DOI] [PubMed] [Google Scholar]
- 59. Mendieta J., Cases-Gonzalez C.E., Matamoros T., Ramirez G., Menendez-Arias L.. A Mg2+-induced conformational switch rendering a competent DNA polymerase catalytic complex. Proteins. 2008; 71:565–574. [DOI] [PubMed] [Google Scholar]
- 60. Sirover M.A., Loeb L.A.. Infidelity of DNA synthesis in vitro: screening for potential metal mutagens or carcinogens. Science. 1976; 194:1434–1436. [DOI] [PubMed] [Google Scholar]
- 61. Snow E.T., Xu L.S., Kinney P.L.. Effects of nickel ions on polymerase activity and fidelity during DNA replication in vitro. Chem. Biol. Interact. 1993; 88:155–173. [DOI] [PubMed] [Google Scholar]
- 62. Vashishtha A.K., Wang J., Konigsberg W.H.. Different divalent cations alter the kinetics and fidelity of DNA polymerases. J. Biol. Chem. 2016; 291:20869–20875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Joyce C.M., Potapova O., Delucia A.M., Huang X., Basu V.P., Grindley N.D.. Fingers-closing and other rapid conformational changes in DNA polymerase I (Klenow fragment) and their role in nucleotide selectivity. Biochemistry. 2008; 47:6103–6116. [DOI] [PubMed] [Google Scholar]
- 64. Stano N.M., Chen J., McHenry C.S.. A coproofreading Zn(2+)-dependent exonuclease within a bacterial replicase. Nat. Struct. Mol. Biol. 2006; 13:458–459. [DOI] [PubMed] [Google Scholar]
- 65. Irimia A., Zang H., Loukachevitch L.V., Eoff R.L., Guengerich F.P., Egli M.. Calcium is a cofactor of polymerization but inhibits pyrophosphorolysis by the Sulfolobus solfataricus DNA polymerase Dpo4. Biochemistry. 2006; 45:5949–5956. [DOI] [PubMed] [Google Scholar]
- 66. Irimia A., Loukachevitch L.V., Eoff R.L., Guengerich F.P., Egli M.. Metal-ion dependence of the active-site conformation of the translesion DNA polymerase Dpo4 from Sulfolobus solfataricus. Acta. Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2010; 66:1013–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Dietrich J., Schmitt P., Zieger M., Preve B., Rolland J.L., Chaabihi H., Gueguen Y.. PCR performance of the highly thermostable proof-reading B-type DNA polymerase from Pyrococcus abyssi. FEMS Microbiol. Lett. 2002; 217:89–94. [DOI] [PubMed] [Google Scholar]
- 68. Henneke G., Flament D., Hubscher U., Querellou J., Raffin J.P.. The hyperthermophilic euryarchaeota Pyrococcus abyssi likely requires the two DNA polymerases D and B for DNA replication. J. Mol. Biol. 2005; 350:53–64. [DOI] [PubMed] [Google Scholar]
- 69. Berquist B.R., DasSarma P., DasSarma S.. Essential and non-essential DNA replication genes in the model halophilic Archaeon, Halobacterium sp. NRC-1. BMC Genet. 2007; 8:31–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Sarmiento F., Mrazek J., Whitman W.B.. Genome-scale analysis of gene function in the hydrogenotrophic methanogenic archaeon Methanococcus maripaludis. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:4726–4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Cubonova L., Richardson T., Burkhart B.W., Kelman Z., Connolly B.A., Reeve J.N., Santangelo T.J.. Archaeal DNA polymerase D but not DNA polymerase B is required for genome replication in Thermococcus kodakarensis. J. Bacteriol. 2013; 195:2322–2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Gueguen Y., Rolland J.L., Lecompte O., Azam P., Le Romancer G., Flament D., Raffin J.P., Dietrich J.. Characterization of two DNA polymerases from the hyperthermophilic euryarchaeon Pyrococcus abyssi. Eur. J. Biochem. 2001; 268:5961–5969. [DOI] [PubMed] [Google Scholar]
- 73. Palud A., Villani G., L’Haridon S., Querellou J., Raffin J.P., Henneke G.. Intrinsic properties of the two replicative DNA polymerases of Pyrococcus abyssi in replicating abasic sites: possible role in DNA damage tolerance?. Mol. Microbiol. 2008; 70:746–761. [DOI] [PubMed] [Google Scholar]
- 74. Robbins J.B., Murphy M.C., White B.A., Mackie R.I., Ha T., Cann I.K.. Functional analysis of multiple single-stranded DNA-binding proteins from Methanosarcina acetivorans and their effects on DNA synthesis by DNA polymerase BI. J. Biol. Chem. 2004; 279:6315–6326. [DOI] [PubMed] [Google Scholar]
- 75. Cann I.K., Ishino S., Nomura N., Sako Y., Ishino Y.. Two family B DNA polymerases from Aeropyrum pernix, an aerobic hyperthermophilic crenarchaeote. J. Bacteriol. 1999; 181:5984–5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Choi J.Y., Eoff R.L., Pence M.G., Wang J., Martin M.V., Kim E.J., Folkmann L.M., Guengerich F.P.. Roles of the four DNA polymerases of the crenarchaeon Sulfolobus solfataricus and accessory proteins in DNA replication. J. Biol. Chem. 2011; 286:31180–31193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Pisani F.M., De Felice M., Rossi M.. Amino acid residues involved in determining the processivity of the 3′-5′ exonuclease activity in a family B DNA polymerase from the thermoacidophilic archaeon Sulfolobus solfataricus. Biochemistry. 1998; 37:15005–15012. [DOI] [PubMed] [Google Scholar]
- 78. Pluchon P.F., Fouqueau T., Creze C., Laurent S., Briffotaux J., Hogrel G., Palud A., Henneke G., Godfroy A., Hausner W. et al. An extended network of genomic maintenance in the archaeon Pyrococcus abyssi highlights unexpected associations between eucaryotic homologs. PLoS One. 2013; 8:e79707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Killelea T., Saint-Pierre C., Ralec C., Gasparutto D., Henneke G.. Anomalous electrophoretic migration of short oligodeoxynucleotides labelled with 5′-terminal Cy5 dyes. Electrophoresis. 2014; 35:1938–1946. [DOI] [PubMed] [Google Scholar]
- 80. Nakamura T., Zhao Y., Yamagata Y., Hua Y.J., Yang W.. Watching DNA polymerase eta make a phosphodiester bond. Nature. 2012; 487:196–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Shannon R. Revised effective ionic radii and systematic studies of interatomic distances in halides and chalcogenides. Acta Crystallogr. A. 1976; 32:751–767. [Google Scholar]
- 82. Colombo M., Girard E., Franzetti B.. Tuned by metals: the TET peptidase activity is controlled by 3 metal binding sites. Sci. Rep. 2016; 6:20876–20888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Nagel Z.D., Cun S., Klinman J.P.. Identification of a long-range protein network that modulates active site dynamics in extremophilic alcohol dehydrogenases. J. Biol. Chem. 2013; 288:14087–14097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Sartori A.A., Jiricny J.. Enzymology of base excision repair in the hyperthermophilic archaeon Pyrobaculum aerophilum. J. Biol. Chem. 2003; 278:24563–24576. [DOI] [PubMed] [Google Scholar]
- 85. Iwai T., Kurosawa N., Itoh Y.H., Kimura N., Horiuchi T.. Sequence analysis of three family B DNA polymerases from the thermoacidophilic crenarchaeon Sulfurisphaera ohwakuensis. DNA Res. 2000; 7:243–251. [DOI] [PubMed] [Google Scholar]
- 86. Makarova K.S., Krupovic M., Koonin E.V.. Evolution of replicative DNA polymerases in Archaea and their contributions to the eukaryotic replication machinery. Front. Microbiol. 2014; 5:354–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Rogozin I.B., Makarova K.S., Pavlov Y.I., Koonin E.V.. A highly conserved family of inactivated archaeal B family DNA polymerases. Biol. Direct. 2008; 3:32–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Cameron V., House C.H., Brantley S.L.. A first analysis of metallome biosignatures of hyperthermophilic Archaea. Archaea. 2012; 2012:789278–789289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Srivastava P., Kowshik M.. Mechanisms of metal resistance and homeostasis in haloarchaea. Archaea. 2013; 2013:732864–732879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Popescu R., Heiss E.H., Ferk F., Peschel A., Knasmueller S., Dirsch V.M., Krupitza G., Kopp B.. Ikarugamycin induces DNA damage, intracellular calcium increase, p38 MAP kinase activation and apoptosis in HL-60 human promyelocytic leukemia cells. Mutat. Res. 2011; 709–710:60–66. [DOI] [PubMed] [Google Scholar]
- 91. Schieven G.L., Kirihara J.M., Burg D.L., Geahlen R.L., Ledbetter J.A.. p72syk tyrosine kinase is activated by oxidizing conditions that induce lymphocyte tyrosine phosphorylation and Ca2+ signals. J. Biol. Chem. 1993; 268:16688–16692. [PubMed] [Google Scholar]
- 92. Leadon S.A., Dunn A.B., Ross C.E.. A novel DNA repair response is induced in human cells exposed to ionizing radiation at the G1/S-phase border. Radiat. Res. 1996; 146:123–130. [PubMed] [Google Scholar]
- 93. Pitts S.L., Liou G.F., Mitchenall L.A., Burgin A.B., Maxwell A., Neuman K.C., Osheroff N.. Use of divalent metal ions in the DNA cleavage reaction of topoisomerase IV. Nucleic Acids Res. 2011; 39:4808–4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Morales R., Sriratana P., Zhang J., Cann I.K.. Methanosarcina acetivorans C2A topoisomerase IIIalpha, an archaeal enzyme with promiscuity in divalent cation dependence. PLoS One. 2011; 6:e26903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Leontiou C., Lakey J.H., Lightowlers R., Turnbull R.M., Austin C.A.. Mutation P732L in human DNA topoisomerase IIbeta abolishes DNA cleavage in the presence of calcium and confers drug resistance. Mol. Pharmacol. 2006; 69:130–139. [DOI] [PubMed] [Google Scholar]
- 96. Saravanan M., Vasu K., Kanakaraj R., Rao D.N., Nagaraja V.. R.KpnI, an HNH superfamily REase, exhibits differential discrimination at non-canonical sequences in the presence of Ca2+ and Mg2+. Nucleic Acids Res. 2007; 35:2777–2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Dai P., Wang Y., Ye R., Chen L., Huang L.. DNA topoisomerase III from the hyperthermophilic archaeon Sulfolobus solfataricus with specific DNA cleavage activity. J. Bacteriol. 2003; 185:5500–5507. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.