Abstract
Background
Cerebellum is an area of the brain particularly sensitive to the effects of acute and chronic alcohol consumption. Alcohol exposure decreases cerebellar Purkinje cell output by increasing GABA release from Golgi cells onto extrasynaptic α6/δ-containing GABAA receptors located on glutamatergic granule cells. Here, we studied whether chronic alcohol consumption induces changes in GABAA receptor subunit expression and whether these changes are associated with alterations in epigenetic mechanisms via DNA methylation.
Methods
We used a cohort of postmortem cerebellum from control and chronic alcoholics, here defined as alcohol use disorders subjects (n=25/group). S-adenosyl-methionine/S-adenosyl-homocysteine were measured by high-performance liquid chromatography. mRNA levels of various genes were assessed by reverse transcriptase-quantitative polymerase chain reaction. Promoter methylation enrichment was assessed using methylated DNA immunoprecipitation and hydroxy-methylated DNA immunoprecipitation assays.
Results
mRNAs encoding key enzymes of 1-carbon metabolism that determine the S-adenosyl-methionine/S-adenosyl-homocysteine ratio were increased, indicating higher “methylation index” in alcohol use disorder subjects. We found that increased methylation of the promoter of the δ subunit GABAA receptor was associated with reduced mRNA and protein levels in the cerebellum of alcohol use disorder subjects. No changes were observed in α1- or α6-containing GABAA receptor subunits. The expression of DNA-methyltransferases (1, 3A, and 3B) was unaltered, whereas the mRNA level of TET1, which participates in the DNA demethylation pathway, was decreased. Hence, increased methylation of the δ subunit GABAA receptor promoter may result from alcohol-induced reduction of DNA demethylation.
Conclusion
Together, these results support the hypothesis that aberrant DNA methylation pathways may be involved in cerebellar pathophysiology of alcoholism. Furthermore, this work provides novel evidence for a central role of DNA methylation mechanisms in the alcohol-induced neuroadaptive changes of human cerebellar GABAA receptor function.
Keywords: alcohol, cerebellum, GABAA receptor, one-carbon metabolism, methylation
Significance Statement
The cerebellum, an area of the brain exerting a strong control on posture, motor, and cognitive function, is particularly sensitive to the effects of both acute and chronic alcohol exposure. Alcohol-induced facilitation of GABAergic neurotransmission impairs cerebellar function by increasing GABA release from Golgi cells onto the extrasynaptic GABAA receptors. Our study demonstrates an enhancement of the 1-carbon metabolism leading to an increased DNA “methylation index” in cerebellum of alcohol use disorders (AUD) subjects. These changes were associated with a selective hypermethylation of the promoter region and a reduced expression of the extrasynaptic δ-containing GABAA receptor located on cerebellar granule cells. This work provides evidence for a central role of DNA methylation in the chronic alcohol-induced neuroadaptive changes of extrasynaptic (tonic) GABAA receptor structure and function in the human cerebellum.
Introduction
Excessive alcohol consumption is a major public health concern that leads to nearly 80,000 deaths and 2.3 million years of potential life lost in the United States each year, making it the third-leading preventable cause of death in the country (Bouchery et al., 2011). Excessive alcohol drinking for long periods of time (more than 80 g/d/20–30 years), leading to alcohol use disorders (AUD), has been linked to alterations of brain epigenetic mechanisms producing deficits in inhibitory and excitatory neurotransmission (Ponomarev et al., 2012; Guidotti et al., 2013; Barbier et al., 2015). These deficits include the downregulation of GABAergic (e.g., glutamic acid decarboxylase-67 [GAD1], and GABAA receptor subtypes) and glutamatergic (e.g., glutamate ionotropic receptor NMDA type subunit 1 [GRIN1], GIPC PDZ domain containing family member 1 [GIPC1]) neurotransmitter associated genes (Zhou et al., 2011; Enoch et al., 2012; Warden and Mayfield, 2017) as well as decreased expression of genes encoding synaptic plasticity proteins, such as brain derived nerve growth factor (BDNF) (McGough et al., 2004; Davis, 2008; Kyzar and Pandey, 2015), reelin (Skorput and Yeh, 2015), synaptotagmin 2, serine-threonine protein kinase 2, and calcium channel alpha1 subunits (Ponomarev et al., 2012; Barbier et al., 2015). These studies are performed mainly in cortical and limbic structures and suggest the important role of epigenetic processes in molecular mechanisms of alcoholism.
The cerebellum, an area of the brain responsible for coordinating movements and motor learning, appears to be particularly sensitive to the effects of acute and chronic alcohol consumption (Dar, 2015). The neuronal activity of cerebellar Purkinje cells is controlled by the phasic release of GABA from basket and stellate cells and the phasic release of glutamate from the axon terminals (parallel fibers) of granule cells, whose function is in turn regulated by GABAergic input from Golgi interneurons (Ito, 2008). Mounting evidence suggests that chronic alcohol-induced cerebellar dysfunction is partly mediated by alterations in GABAergic neurotransmission (Valenzuela and Jotty, 2015). For example, the sedative-anesthetic action of alcohol is reversed by the administration of GABAA receptor antagonists such as bicuculline and pentamethylenetetrazole. Furthermore, alcohol effects are potentiated by benzodiazepine ligands that act as positive allosteric modulators of GABAA receptors (i.e., diazepam and congeners) but are inhibited by benzodiazepine ligands that act as antagonists or negative allosteric modulators of GABAA receptors (flumazenil, Ro15–4513, FG7142, DMCM) (Korpi, 1994). Studies on GABAA receptor subunit expression point to a possible decrease in α1 (GABRA1) and α2 mRNA expression in cortex and hippocampus during chronic, heavy ethanol administration in rodents (Mhatre and Ticku, 1989; Buck et al., 1991; Montpied et al., 1991). Further electrophysiological investigation of cerebellar slices obtained from laboratory rodents shows that alcohol exposure alters cerebellar function by increasing tonic release of GABA from Golgi cells onto the extrasynaptic α6/δ-containing GABAA receptors located on cerebellar granule cells (Valenzuela and Jotty, 2015). Excessive and protracted alcohol intake in rats may damage cerebellar granular neuron function by downregulating the δ subunit of the GABAA receptor (GABRD) expression (Marutha Ravindran et al., 2007).
To the best of our knowledge, the mechanisms whereby chronic alcohol intake induces GABAA receptor expression downregulation have been poorly investigated in the brain. However, recently we showed that chronic alcohol exposure alters transmethylation reactions and induces DNA hypermethylation status in the cerebellum of rats (Auta et al., 2017).
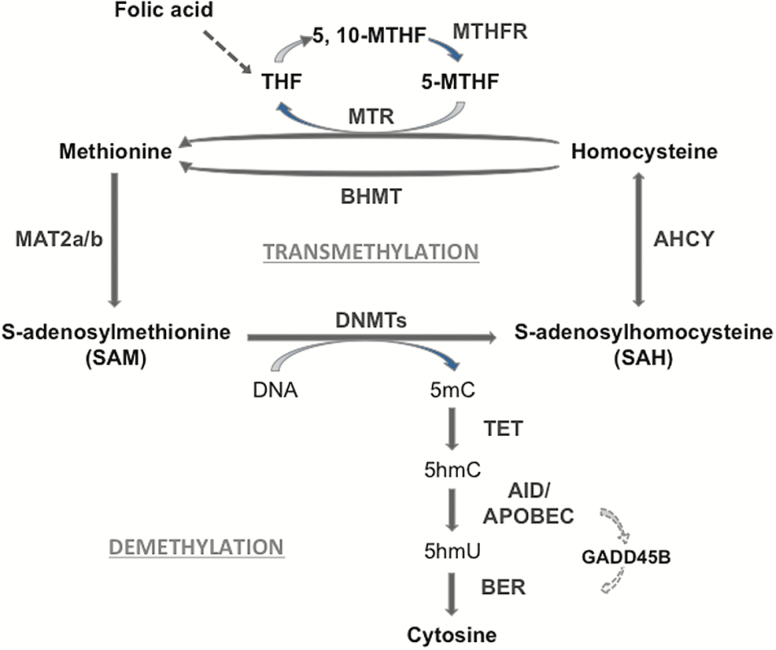
DNA methylation in the brain, the most significant of epigenetic gene regulation, is uniquely able to account for the gene expression changes that may underlie chronic alcohol-induced behavior and neuronal plasticity changes (Ponomarev et al., 2012; Guidotti et al., 2013; Barbier et al., 2015). Steady-state DNA methylation levels in mammalian brain are the result of a dynamic equilibrium between the actions of enzymes (DNA-methyltransferases [DNMTs]) that add methyl groups at the fifth position of cytosine (5mC) (Goll and Bestor, 2005; Ma et al., 2009; Guo et al., 2011; Grayson and Guidotti, 2013) and the action of an active DNA-demethylation pathway including the Ten-Eleven Translocation Methylcytosine Dioxygenase enzyme family (TET1, 2, 3) that oxidize 5mC to 5-hydroxymethylcytosine (5hmC), and 5hmC to 5-formylcytosine and 5-carboxylcytosine (Fu and He, 2012). The Activation-Induced Deaminase/Apolipoprotein B mRNA editing enzyme catalytic (APOBEC) family of cytidine deaminases promotes demethylation processes leading to 5-hydroxymethyluridine (5hmU) (Guo et al., 2011; Bochtler et al., 2017). The latter is then excised by base excision repair pathway leading to cytosine (Figure 1).
Figure 1.
One-carbon metabolism, transmethylation reactions and DNA demethylation. DNA methylation on specific cytosine moieties is catalyzed by DNA-methyltransferases (DNMTs). This reaction is dependent on the levels of S-adenosyl-methionine (SAM), which is synthesized in the brain by the methionine-adenosyl-transferase (MAT) 2. After the DNA methyl transfer reaction, S-adenosyl-homocysteine (SAH) is formed as a by-product. SAH exerts a feedback inhibitory activity on DNMT and is used as a substrate for the adenosyl homocysteine hydrolase (AHCY) leading to the synthesis of homocysteine. The remethylation of homocysteine to form methionine is catalyzed via the folate cycle, that is, the conversion of folic acid into tetrahydrofolate (THF) and 5,10-methylenete-trahydrofolate (5,10-MTHF). The methylene tetrahydrofolate reductase (MTHFR) will produce 5-methyltetrahydrofolate (5-MTHF) that is used as a substrate by methionine synthase (MTR) to form methionine. The synthesis of methionine may also involve the activation of the betaine homocysteine methyltransferase (BHMT) pathway. DNA demethylation is initiated by ten-eleven-translocase (TET) enzymes that hydroxylate 5-methylcytosine (5mC) forming 5-hydroxymethylcytosine (5hmC). In the end, through a GADD45B coordinated process, 5hmC is deaminated to 5-hydroxyuracil (5hmU) by cytidine deaminases (i.e., AID/APOBECs). The base excision repair (BER) pathway removes 5hmU and substitutes it with cytosine.
The addition of a methyl group to cytosine in CpG-rich DNA regulatory regions is dependent on the availability of S-adenosyl-methionine (SAM), which is synthesized in the brain by the enzyme methionine-adenosyl-transferase 2 (Figure 1). Following the methyl transfer reaction, S-adenosyl-homocysteine (SAH) is formed as a by-product with strong feedback inhibitory activity on transmethylation reactions (Yi et al., 2000). SAH is in turn rapidly hydrolyzed into homocysteine in a reversible reaction catalyzed by the adenosyl homocysteine hydrolase (AHCY). The remethylation of homocysteine to form methionine is catalyzed via methylene tetrahydrofolate reductase (MTHFR). These enzymes are part of the so-called 1-carbon metabolism pathway and are crucial for the biosynthesis of the methyl donor SAM and the efficiency of transmethylation reactions (Figure 1; Auta et al., 2017).
Chronic alcohol-induced aberrant DNA methylation has been extensively studied in the liver where global DNA hypomethylation has been associated with reductions in DNMT (i.e., DNMT3A and DNMT3B) activities and a decreased SAM/SAH ratio (decreased methylation index) (Tsukamoto and Lu, 2001; Lu and Mato 2005; Bönsch et al., 2006; Varela-Rey et al., 2013; Auta et al., 2017). However, the relationship between the expression of DNMTs, the levels of SAM/SAH ratio, and the expression of DNA demethylating enzymes in the brain of AUD subjects has not been extensively studied. To the best of our knowledge, there are no detailed studies on 1-carbon metabolism and DNA methylation mechanisms of GABAergic neurotransmitter-related genes in the cerebellum of subjects with AUD.
The goal of this work is to study whether a history of long-term alcohol drinking results in aberrant 1-carbon metabolism and DNA methylation reactions and whether such alterations underlie the altered promoter methylation of GABAergic (GAD1 and GABAA receptor subunits) genes in postmortem cerebellum of control and AUD subjects.
Materials and Methods
Subjects
We received brain tissue from a cohort of 25 control and 25 AUD subjects from the New South Wales Brain Tissue Resource Center (Australia). Demographic characteristics are provided in Table 1. Both AUD and control subjects met strict inclusion criteria, and an attempt was made to match control cases for sex, age, body mass index (BMI), race, and postmortem interval (PMI).
Table 1.
Demographic Characteristics of Brain Samples from Control and Alcohol Use Disorders (AUD) Subjects
| Subjects | Control | AUD | |
|---|---|---|---|
| n | 25 | 25 | |
| Age | 58 ± 1.7 | 58 ± 1.8 | |
| Sex | 5F 20M | 5F 20M | |
| PMI (h) | 33 ± 2.7 | 39 ± 3.3 | |
| pH | 6.6 ± 0.06 | 6.6 ± 0.05 | |
| RIN | 5.9 ± 0.3 | 6.2 ± 0.3 | |
| BMI | 30 ± 1.2 | 26 ± 1.2* | |
| Total Drinking Years | 33 ± 3.3 | 35 ± 1.8 | |
| EtOH daily use (g) | 15 ± 3.7 | 205 ± 32.8* | |
| Drinks per weeks | 9.1 ± 2.6 | 105 ± 20.1* | |
| Pack of cigarettes per year | 25.5 ± 7 | 37.9 ± 4.8 | |
| Cause of death | R | 2 | 3 |
| T | 1 | 5 | |
| C | 19 | 12 | |
| H | 0 | 1 | |
| V | 1 | 0 | |
| U | 0 | 1 | |
| I | 0 | 2 | |
| Cv | 1 | 1 | |
| Cc | 1 | 0 | |
Abbreviations: BMI, body mass index; C, cardiac; Cc, cancer; Cv, cardiovascular; F,female; H, hepatic; I, infection; M, male; PMI, postmortem interval (hours); R, respiratory; RIN, RNA integrity number; T, toxicity; U, unknown; V, vascular.
Indicated values are mean ± SEM. *P < .05, Student’s t test.
Inclusion criteria for AUD cases were: age > 18 years, no developmental disorders, no history of other major psychiatric disorders (such as schizophrenia or bipolar disorders) or history of toxicology of other substance abuse disorders (according to the criteria in the DSM-IV, American Psychiatric Association, 1994) with the exception of nicotine (cigarette smoking) that was used by most of the AUD and control subjects. The AUD cohort included 8 AUD subjects that had alcohol toxicology at death (0.03–0.430 g/100 mL) and 17 AUD subjects that had unknown (n=4) or not detected blood alcohol levels (n=13). Hence, this cohort offers the opportunity of establishing whether the presence of alcohol or its withdrawal is related to changes in the expression of DNA-methylation or demethylation genes in brain of AUD subjects.
Postmortem Brain Collection
The cerebellum was divided in the mid-sagittal plane and hemispheres were cut into approximately 8- to 10-mm coronal slices. A small tissue segment, usually the lateral cerebellar hemisphere, was taken for pH and RNA integrity number (RIN) measurements, which provides an estimation of tissue quality for molecular studies. Although PMI may be a factor that contributes to changes in RIN, PMI values do not differ in control and AUD subjects (Table 1) and no significant correlation was found between RIN and PMI (r=0.018, P=.905). Moreover, ANCOVA analysis shows that changes in the variables (i.e., MTHFR, GADD45B, TET1, and TET2) that show significant correlation with PMI remain statistically significant when adjusting for PMI.
Reverse Transcriptase-Quantitative Polymerase Chain Reaction (qRT-PCR)
mRNA expression levels were measured by qRT-PCR using the Applied Biosystems/Life Technologies SYBR green master mix (Thermo Fisher Scientific). qRT-PCR was run on a Stratagene Mx3005P QPCR System. Samples were homogenized with BeadBug prefilled tubes (Sigma) and total RNA was isolated using the Qiagen miRNeasy Mini Kit. RIN was measured with the Agilent 2100 Bioanalyzer (Agilent Technologies). Primer sequences used for mRNA expression studies are listed in Table 2. Primers were designed by selecting non-intron sequences to amplify most transcript variants of the target gene. Samples with a RIN number<3 were excluded from the analyses (Gavin et al., 2012). Samples were run in duplicate and were repeated. Three reference genes (i.e., Beta-2-Microglobulin, Glyceraldehyde-3-Phosphate Dehydrogenase [GAPDH], and ß-actin) were chosen as internal controls for normalizing mRNA levels.
Table 2.
Primer Sequences
| Gene symbol | 5’-Primer | 3’-Primer |
|---|---|---|
| mRNA Expression | ||
| MAT2A | 5’-GATGCCAAAGTAGCTTGTGAAACT | 5’-CGTTACAAGTCTTGTAGTCAAAACCT |
| MAT2B | 5’-GAGAACAATCTAGGAGCTGCTGT | 5’-CCTTAATTGATGGATCCAGCATTCTC |
| MTHFR | 5’-CTTGAAGGAGAAGGTGTCTGCG | 5’-ATCTCCTGTGGCACCTCCA |
| AHCY | 5’-GACCGGTATCGGTTGAAGAATGG | 5’-GGTACTTGTCTGGATGGGTCCA |
| MTR | 5’-CGCAACCCGAAGGTCTGAA | 5’-TGGTAAATGACATCAGGCTGAGTTATA |
| DNMT1 | 5’-AGAAGCTGTCCATCTTTGAT | 5’-CATAGATTGGTTTTGCTGAA |
| DNTM3A | 5’-TCTTTGATGGAATCGCTAC | 5’-GCGGTAGAACTCAAAGAAGA |
| DNTM3B | 5’- GCTCTTACCTTACCATCGAC | 5’-GAGACGAGCTTATTGAAGGT |
| TET1 | 5’-CCCGGGCTCCAAAGTTGTG | 5’-GCAGGAAACAGAGTCATT |
| TET2 | 5’-TGTGTGGCACTAGATTTCAT | 5’-AGTCTCTGAAGCCTGTTGAT |
| TET3 | 5’-CAGTGGCTTCTTGGAGTCACCTC | 5’-GGATGGCTTTCCCCTTCTCTCC |
| GADD45B | 5’-ATTGCAACATGACGCTGGAA | 5’-CTGTCTGGGTCCACATTC |
| APOBEC3C | 5’-TGTATCCAGGCACATTCTACTTC | 5’-AACTGAGCGGCGCTTTAT |
| GABRD | 5’-CGACATGGACCTGGCCAAATA | 5’-CCTTCTGCTTCTTCCTGTAGTCG |
| GABRA6 | 5’-ACCTGGACTGATGAGAGGTTGAAG | 5’-CAGAAAGACACCTGGGAAAGAATG |
| GABRA1 | 5’-ACAACACTTACGCTCCAACA | 5’-TTTCGGGCTTGACCTCTTTAG |
| GAD1 | 5’-AGGCAATCCTCCAAGAACCT | 5’-GGTGGAGCGATCAAATGTCT |
| SRD5A1 | 5’-GGCGCTTCTCTATGGACTTT | 5’-CAACCTCCATTTCAGCGTATTTAG |
| AIF1 | 5’-CTGAAACGAATGCTGGAGAAAC | 5’-GAGAAAGTCAGGGTAGCTGAAC |
| ENO2 | 5’-CTGATCCTTCCCGATACATCAC | 5’-CTGGTCAAATGGGTCCTCAA |
| B2M | 5’-CTATCCAGCGTACTCCAAA | 5’-GCTCCACTTTTTCAATTCTC |
| ACTB | 5’-CTCCCTGGAGAAGAGCTAC | 5’-GATCCACACGGAGTACTTG |
| GAPDH | 5’-CGAGATCCCTCCAAAATCAA | 5’-TTCACACCCATGACGAACAT |
| Methylated DNA Immunoprecipitation | ||
| TSH2B | Promoter region, Diagenode (Denville, NJ), Catalog-#C17011041 | |
| GAPDH | TSS region, Diagenode (Denville, NJ), Catalog-#pp-1047–050 | |
| GABRD | ||
| -633-414bp | 5’-AAGGCAGGAGAGTGTGAGGCC | 5’-TGCTTCCAAAACCCTCCTGCCTT |
| -346-153b | 5’-CCAGAAGCAGCCACGCAGA | 5’-GGCAGCTCTCCCTGGCC |
| GABRA6 | ||
| -215-111bp | 5’-GGAATCCAAGAGGGTCACAATAG | 5’-TTCCAATAGGCTCAGCGTTTAT |
| -37 + 63bp | 5’-GGCAGTGGATTTCTTCCTTCTA | 5’-GTTGGGAAAGGAGAGTCTGAAG |
| +215 + 325bp | 5’-AGGCAAACAAGGAACAGAGATA | 5’-AAATGCAGAATTCACCCTCCTA |
High-Performance Liquid Chromatography
Samples were homogenized in 2 mL/mg wet tissue weight of 0.4 M HClO4, then centrifuged at 10000 g for 5 minutes. Supernatants were filtered through 0.22-µm Millipore membranes, and 100-µL aliquots were injected into a reverse-phase high-performance liquid chromatography column, Symmetry C18 4.6 x 250 nm (Waters). The mobile phase consisted of 3 solvents: solvent A, 40 mM NaH2PO4 and 8 mM 1-octanesulfonic acid sodium salt adjusted to pH 3.0 with H3PO4; solvent B, 40% methanol in solvent A; and solvent C, 100% methanol. The column was equilibrated with solvent A at a flow rate of 1 mL/min and developed using a multistep gradient: 45 minutes at the equilibration conditions, up to 50% solvent B in 1 minute and maintained for 8 minutes, up to 100% solvent B in 30 seconds and maintained for 16.5 minutes, then down to 50% solvent B and 50% solvent C in 1 minute and maintained for 10 minutes to wash and return to equilibration conditions for a minimum of 30 minutes before subsequent injections. Five point calibration curves for SAM and SAH (Sigma-Aldrich) were linear in the range of 25 to 800 ng of the respective standards.
One limitation of the measurements of SAM and SAH in postmortem tissues is their chemical stability. To address this, we measured SAM and SAH content in mouse brain that was left at room temperature for 6 hours. The levels of SAM and SAH in these brains were comparable with the levels of SAM and SAH measured in mouse brains immediately frozen after decapitation.
DNA Isolation and Global Methylation/Hydroxymethylation Analysis
DNA was extracted using QIAamp DNA Mini Kit (Qiagen). Global DNA methylation and hydroxymethylation were measured using Methylamp Global DNA Methylation Quantification Ultra Kit and MethylFlash Hydroxymethyl DNA Quantification Kit, respectively (Epigentek) following the manufacturer’s instructions. Signals obtained were quantified by measuring optical density, and total methylation level was estimated by generating a standard curve from a methylated DNA standard (Epigentek). Values are presented as methylation percentage relative to controls.
Methylated DNA Immunoprecipitation
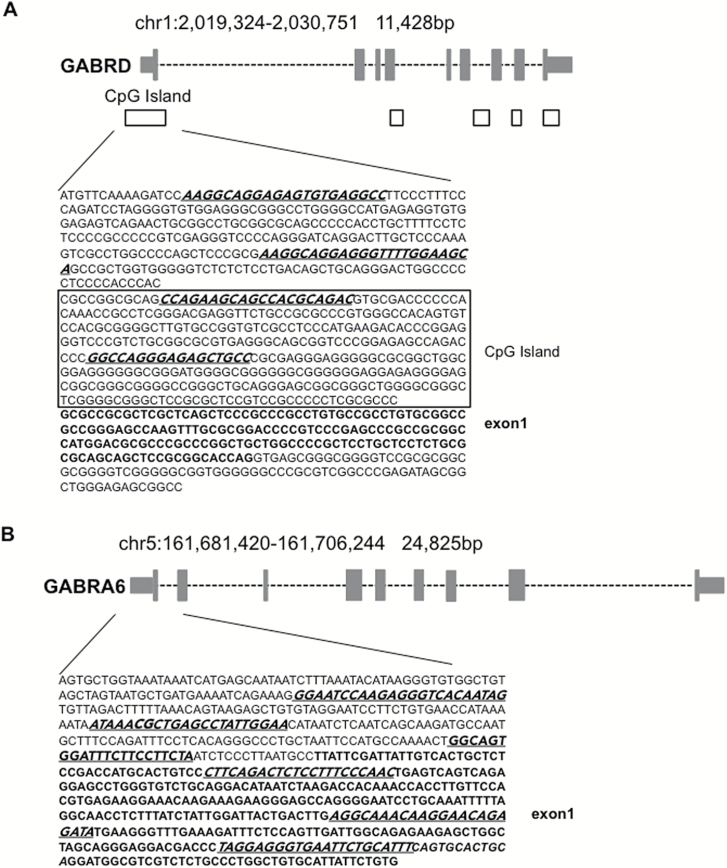
Methylated DNA immunoprecipitation experiments were conducted as previously described (Dong et al., 2008; Gavin et al., 2012) using MagMeDIP kit and hMeDIP kit from Diagenode. DNA was precipitated according to the manufacturer’s instructions with anti-5mC and anti-5hmC monoclonal mouse antibodies, respectively. The percentage of (hydroxy)methylated vs unmethylated promoter was calculated using the following equation: %(meDNA−IP/total input)=2^[(Ct(10% input) −3.32) −Ct(meDNA−IP)] × 100. For the GABRD gene, primers were chosen to target the CpG island, as well as the upstream region of the promoter, while for the α6-containing GABAA receptor (GABRA6) gene, primers were selected to amplify 2 regions, that is, the upstream region of the promoter and exon 1 (Figure 2; Table 2). MeDIP efficiency was validated by qRT-PCR using internal positive and negative DNA controls (methylated/hydroxymethylated and unmethylated DNA) as well as control primers for testis-specific H2B histone gene (which is methylated in all somatic cells but not in testis), and GAPDH promoter, which is poorly methylated (Table 2). Data obtained with internal positive and negative DNA controls indicate a specificity of 96% (specificity %=1−(enrichment unmeth/enrichment meth) x 100). Values obtained for % (meDNA − IP/total input) are in the same range as those obtained in other individual genes studied by us (Dong et al., 2016; Zhubi et al., 2017) and other groups (Sunahori et al., 2013; Hong et al., 2015).
Figure 2.
Location of human GABAA receptor (A) δ (GABRD) and (B) α6 (GABRA6) subunit promoter regions. CpG islands are illustrated as black squares and exons (grey rectangles) are in bold letters. Methylated DNA immunoprecipitation and hydroxymethylated DNA immunoprecipitation were performed using the primers underlined and italicized in the sequence.
Western Blot
Samples were homogenized in HEPES-buffered sucrose (320 mM sucrose, 4 mM HEPES, pH 7.4, 1% SDS), separated by electrophoresis on Novex 4–12% Tris-Glycine gels (Invitrogen) and transferred to PVDF membrane (Millipore). Transfer was performed at 4°C in a buffer containing 35 mM Tris, 192 mM glycine, and 20% methanol. Membranes were blocked using 5% milk and later incubated with the primary antibody, anti-GABAA receptor delta subunit (1:1000, Novus). Protein levels were normalized with GAPDH (1:5000 Millipore). HRP-conjugated secondary anti-mouse or anti-rabbit antibodies (1:10000, GE Healthcare) were used. Membranes were developed with Immobilon Western Chemiluminescent HRP Substrate (Millipore) and densitometric analysis was performed with Image StudioTM (LI-COR Bioscience).
Statistical Analysis
Student’s t tests were performed and comparisons were considered statistically significant if P<.05. For many comparisons, Bonferroni’s correction is too conservative; hence, it is likely that no significance will be observed (Benjamini and Hochberg, 1995). In such situations (for many multiple comparisons), to control for the false discovery rate (FDR), we used the Benjamini-Hochberg approach (1995) with FDR of q=0.1 (the concept of q is different from the significant level α, the value of q=0.1 is well established in the literature; Benjamini and Hochberg, 1995). In addition, 1-way ANOVA followed by Tukey’s posthoc comparisons was performed for the comparison of the subgroup 1 and 2 (with and without blood alcohol level) with control group (i.e., a comparison for a total of 3 groups). ANCOVA was performed for adjusting covariants, that is, smoking, pH, and PMI on the results. All statistical tests were run using PASW v.18 software (SPSS).
Results
DNA Methylation Reaction and 1-Carbon Metabolism
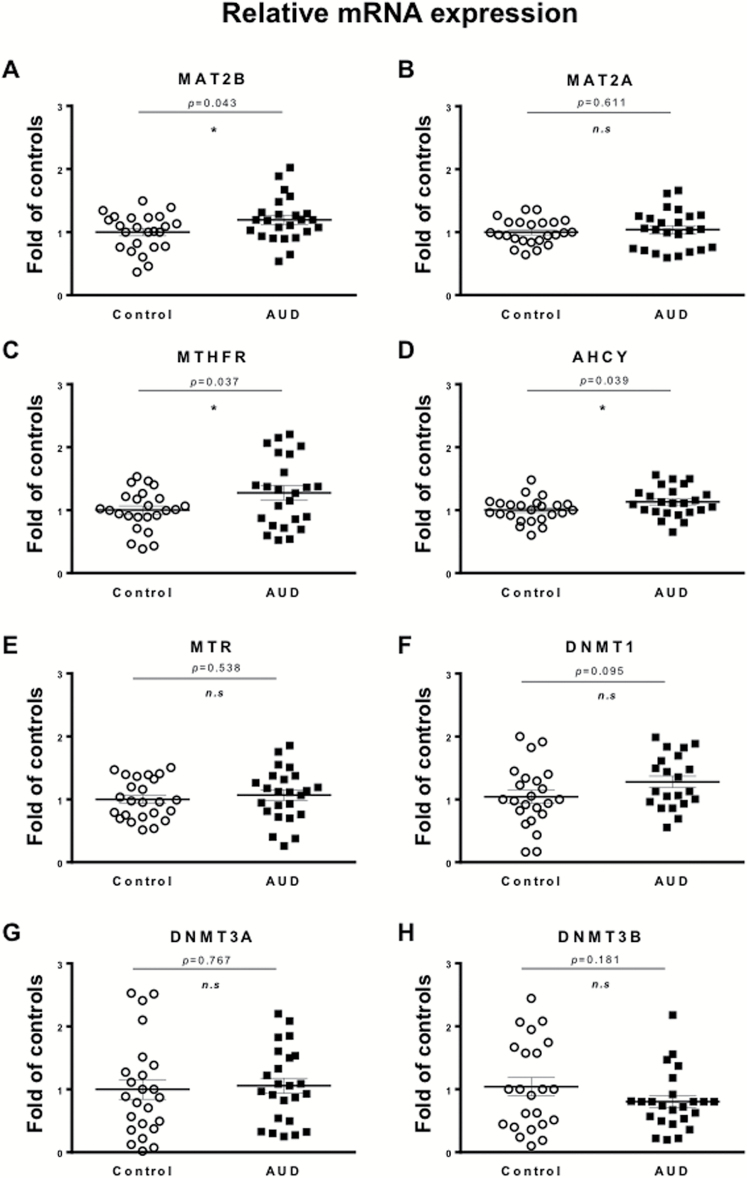
To study whether AUD affect DNA methylation dynamics by altering the availability of SAM, we first measured the mRNA expression of key enzymes of 1-carbon metabolism and transmethylation reactions in the cerebellar cortex (Figure 3). While comparing the AUD group with the control group, we found a 15% to 30% increase of MAT2B (Figure 3A; t(1,46)=2.085, P=.043) while no changes were observed for MAT2A mRNA levels (Figure 3B). MTHFR (Figure 3C; t(1,46)=2.146, P=.037) and AHCY mRNA (Figure 3D; t(1,46)=2.115, P=.039) were also increased in AUD subjects. However, methionine synthase (MTR; Figure 3E), betaine-homocysteine S-methyltransferase (data not shown), as well as DNMT1, DNMT3A, and DNMT3B mRNA transcripts remained unchanged (Figures 3F, G, and H, respectively).
Figure 3.
Chronic ethanol intake increases expression of key enzymes of the 1-carbon metabolism in cerebellum. mRNA levels of (A) methionine adenosyltransferase (MAT) 2B, (B) MAT2A, (C) methylenetetrahydrofolate reductase (MTHFR), (D) adenosyl homocysteine hydrolase (AHCY), (E) methionine synthase (MTR), and (F–H) DNA methyltransferase (DNMT) 1, 3A, and 3B, respectively, are expressed as fold change of controls. Values are mean ± SEM of 24 samples per group with exception of DNMT1, where only 21 samples per group were measured. *P < .05, Student’s t test vs controls.
When we performed Pearson’s correlations between the demographic variables and the mRNA expression of the 1-carbon metabolism enzymes, we found positive correlations between various drinking-related measures (i.e., ethanol daily use (g), number of drinks per week) and AHCY, MTR, and MTHFR mRNA levels (Table 3). Since most of the AUD subjects are smokers (n=23, n=2 not reported), we also assessed the correlation between mRNA levels and smoking habit (pack of cigarettes per year), and no significant correlation was observed (Table 3). We did not find any significant correlation between PMI, pH, sex, smoking habit, age, and mRNA levels with the exception of MAT2B, which shows a significant correlation with age. Hence, using ANCOVA, we tested the changes in MAT2B after adjusting for the effect of age. The mRNA expression of MAT2B remained significantly different between control and AUD groups (F(1,47)=4.961, P=.031).
Table 3.
Pearson’s Correlation Analysis of 1-Carbon Metabolism Enzymes with Total Drinking Years, Ethanol Daily Use (g), Number of Drinks Per Week, and Cigarette Smoking Behavior
| Gene expression | Total drinking years | Ethanol daily use (g) | Drinks per week |
Smoking
behavior |
|---|---|---|---|---|
| MAT2A | r=0.206 P=.241 |
r =0.210 P=.234 |
r =0.221 P=.208 |
r =-0.101 P= .566 |
| MAT2B | r =-0.105 P=.560 |
r =0.211 P=.230 |
r =0.184 P=.297 |
r =0.089 P=.089 |
| AHCY | r =0.180 P=.308 |
r =369
P=.032 |
r =0.350
P=.042 |
r =-0.050 P=.776 |
| MTR | r =0.037 P=.837 |
r =0.340
P=.049 |
r =0.317 P=.068 |
r =-0.112 P=.522 |
| MTHFR | r =0.028 P=.874 |
r =0.382
P=.026 |
r =0.313 P=.071 |
r =-0.147 P=.399 |
Significant correlations are highlighted in bold.
To control for the FDR, we performed the Benjamini-Hochberg approach for the 1-carbon metabolism enzymes (MAT2A/B, MTR, MTHFR, and AHCY). With an FDR level of q=0.1, MAT2B, MTHFR, and AHCY remained statistically significant (adjusted P =.032). DNMTs levels were not significantly different using this FDR approach (Figure 3).
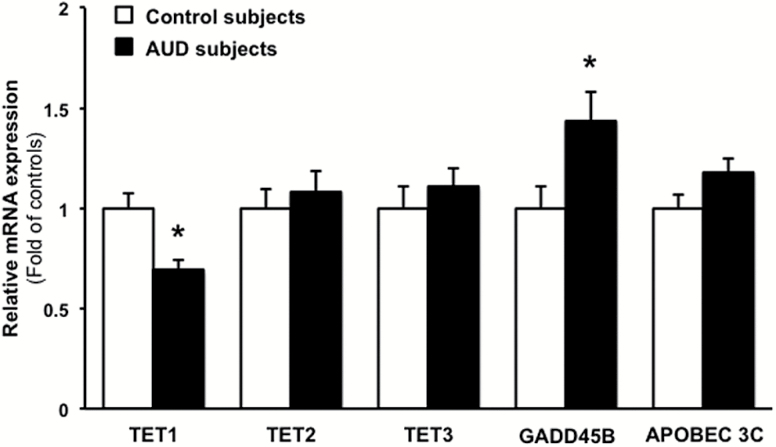
DNA Demethylation Pathways
The mRNA expression of TETs enzymes (TET1, TET2, and TET3) is differentially altered in the cerebellum of AUD subjects. For example, TET1 mRNA is decreased in the whole AUD group (Figure 4; t(1,46)=3.526, P=.001), whereas TET2 and TET3 mRNA expression fail to change in the AUD group (Figure 4). Growth Arrest and DNA Damage Inducible Beta (GADD45B), a protein that facilitates deamination of 5hmC to 5hmU, is also increased in AUD subjects (Figure 4; t(1,46)=2.371, P=.022), while APOBEC 3C does not significantly change (Figure 4; t(1,46)=1.918, P=.0614) compared with controls. The Benjamini-Hochberg approach, using FDR with q=0.1, shows that the differences between groups in TET1, GADD45B, and APOBEC 3C are statistically different (adjusted P value=.003, .036, .0675, respectively). Although a significant correlation was found between PMI and TET1 (r=-0.416, P=.003), TET3 (r=0.348, P=.015), and GADD45B (r=0.343, P=.017), the difference between control and AUD subjects remained statistically significant following ANCOVA analysis adjusting for PMI (TET1: F(1,48)=10.24, P=.003; TET3: F(1,48)=0.113, P=.738; GADD45B: F(1,48)=4.108, P=.049).
Figure 4.
Chronic ethanol intake impairs demethylation pathways. mRNA levels of Ten-Eleven Translocase (TET) 1, 2, and 3, as well as Growth Arrest and DNA Damage Inducible Beta (GADD45B) and Apolipoprotein B mRNA editing enzyme catalytic subunit 3C (APOBEC 3C) are expressed as fold change of controls. Values are mean ± SEM of 24 samples per group. *P < .05, Student’s t test vs controls.
Measurements of blood alcohol levels carried out at the time of death suggest that the cohort of chronic alcoholics can be divided in 2 subgroups: subgroup 1 including 7 alcoholic subjects with detectable alcohol levels and subgroup 2 including 13 alcoholic subjects whose blood alcohol levels were undetectable at the time of death (most likely these subjects were either in withdrawal or abstinent). The demethylating enzyme TET1 is decreased (~43%) in subgroup 2 compared with controls (1-way ANOVA approach F(2,43)=5.058; P=.011; Tukey’s posthoc test P=.01), whereas GADD45B is increased (~64.5%, 1-way ANOVA approach F(2,43)=4.01; P=.028; Tukey’s posthoc test P= 0.021). MAT2B, AHCY, MTHFR, MTR, DNTM1, 3A, and 3B were not significantly different in subgroups 1 and 2.
SAM/SAH Ratio
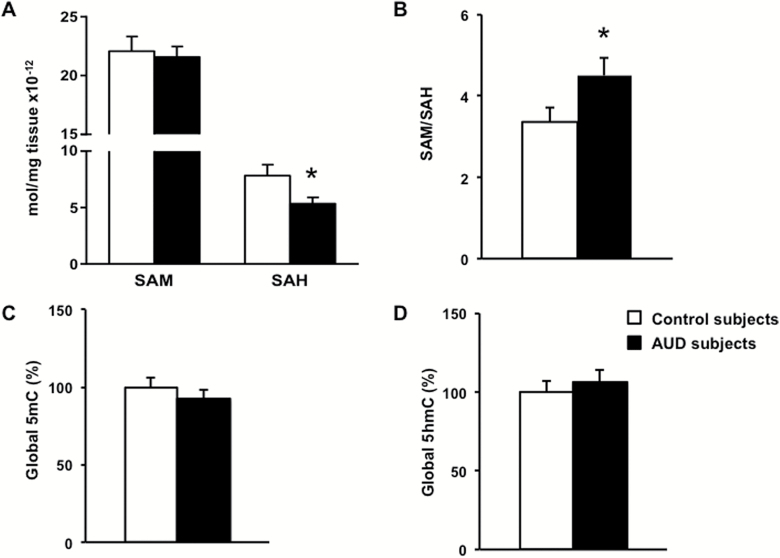
Figure 5A shows the average values of SAM and SAH in the cerebellum of control and AUD subjects. The levels of SAM are 2- to 3-fold higher than those of SAH and are almost identical in both groups. In contrast, SAH is significantly reduced in cerebellum of AUD subjects (Figure 5A; t(1,32)=2.249, P=.031). This reduction in SAH shifts the SAM/SAH ratio to values higher in AUD than in control subjects (Figure 5B; t(1,32)=2.273, P=.029). Although a significant correlation was found between SAM/SAH ratio and PMI (r=0.323, P=.048), the difference between control and AUD subjects remained statistically significant following ANCOVA analysis adjusting for PMI (F(1,34)=5.306, P=.027).
Figure 5.
Chronic ethanol intake changes the “methylation index” in cerebellum. (A) Amounts of S-adenosyl-methionine (SAM) and S-adenosyl-homocysteine (SAH) were measured (mol/mg tissue x10-12) and (B) expressed as a ratio, indicating the methylation index. Global (C) methylation (5mC) and (D) hydroxymethylation (5hmC) were determined. Values are mean ± SEM of 18 samples per group. *P < .05, Student’s t test vs controls.
Since most of the subjects were heavy smokers, we introduced smoking as a covariant, but the outcome of this analysis did not influence the results (SAH: ANCOVA diagnosis effect F(1,24)=6.178, P=.021; SAM/SAH: diagnosis effect F(1,24)=5.829, P=.025). There is no apparent influence of sex on SAM/SAH ratio, but the significance of this observation remains inconclusive due to the small number of females (n=5).
Global DNA Methylation/Hydroxymethylation
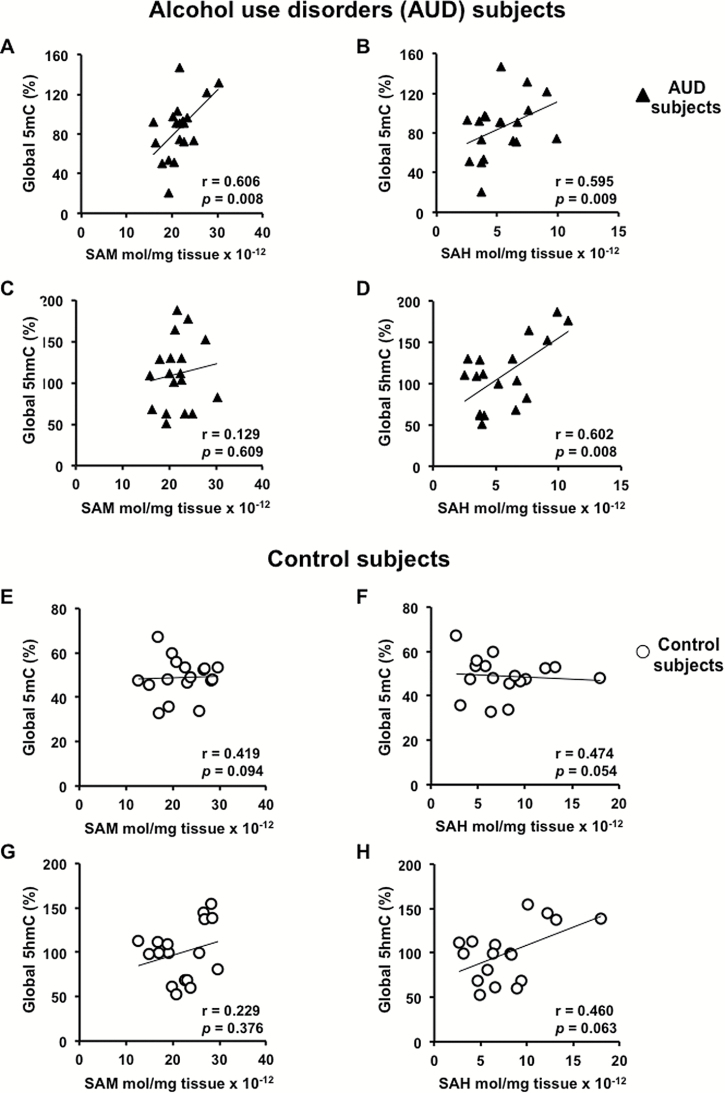
Global 5mC and 5hmC DNA levels measured in the cerebellum of control and AUD subjects are similar (Figure 5C–D). However, global 5mC DNA enrichment positively correlates with the level of SAM (Figure 6A; r=0.606, P=.008) and SAH (Figure 6B; r=0.595, P=.009) in AUD subjects only. Interestingly, SAM does not correlate with 5hmC (Figure 6C) but SAH does (Figure 6D; r=0.602, P=.008). No significant correlation is found with SAM/SAH in AUD subjects (5mC and SAM/SAH: r=-0.337, P=.171; 5hmC and SAM/SAH: r=-0.382, P=.117). Correlations between 5mC, 5hmC and SAM, SAH (Figure 6E–H) or SAM/SAH are not significant in control subjects.
Figure 6.
Pearson’s correlation analysis of global methylation (5mC) and hydroxymethylation (5hmC) with S-adenosyl-methionine (SAM) and S-adenosyl-homocysteine (SAH). Correlations were performed in alcohol use disorders (AUD) subjects (A–D) and control subjects (E–H), n =18 samples per group.
Promoter Methylation and Transcriptional Expression of Alcohol-Related GABAergic Genes
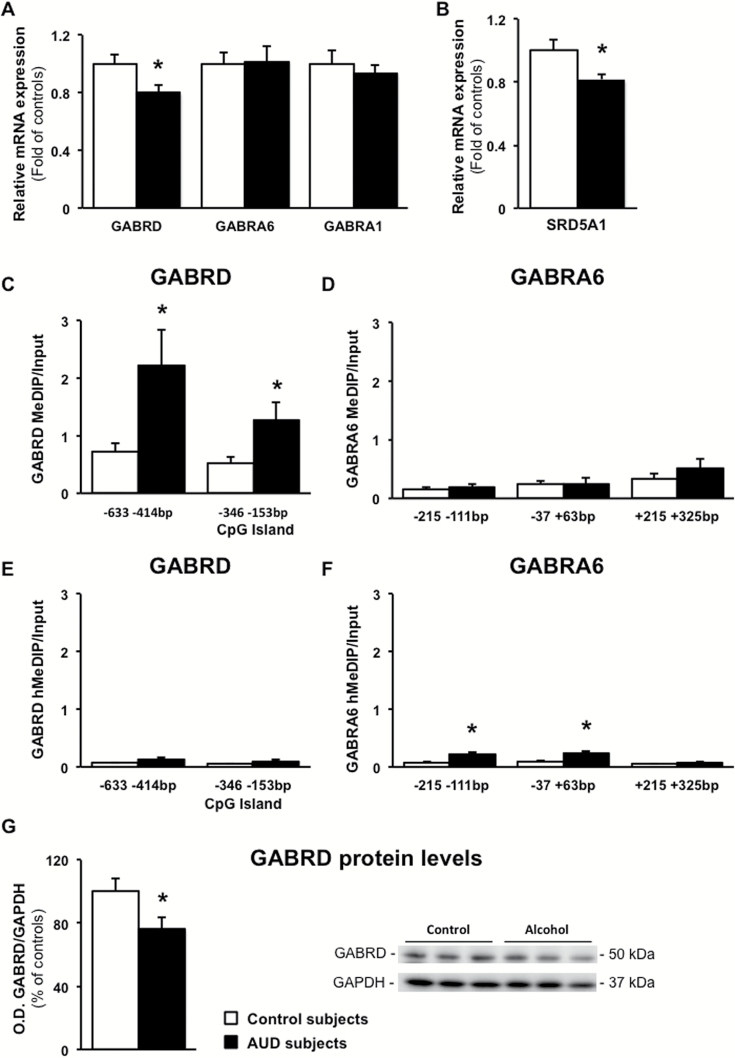
We next measured promoter methylation enrichment and mRNA expression levels of GABAergic genes in the cerebellum of control and AUD subjects. As shown in Figure 7, GABRD mRNA (Figure 7A, t(1,46)=2.376, P=.022) and protein (Figure 7G, t(1,48)=2.164, P=.035) levels were significantly decreased in AUD subjects. The reduced GABRD mRNA expression in AUD subjects correlated with the number of total drinking years (r=0.408, P=.048). A higher promoter methylation (Figure 7C) but not hydroxymethylation (Figure 7E) at base pairs -633 to -414 (t(1,42)=2.294, P=.027) and -346 to -153 (CpG island, t(1,42)=2.189, P=.034; see Figure 2 for primer locations) was also observed. However, no significant correlation was found between methylation levels and transcriptional changes. As proof of specificity for the effect of chronic alcohol on GABRD expression, we show that GABRA6 and GABRA1 subunit mRNA levels were not altered (Figure 7A) and that the promoter methylation at regulatory regions of the GABRA6 subunit, which is poor in CpG nucleotide repeats (Figure 2), was barely at the level of detection and similar in control and AUD subjects (Figure 7D). Interestingly, hydroxymethylation of GABRD (Figure 7E) and GABRA6 (Figure 7F) promoter regions was present only in a small proportion (~10%) of the total methylated promoters, and the significance of this nuclear marking remains unclear.
Figure 7.
GABAA receptor delta subunit expression is reduced in the cerebellum of alcohol use disorders subjects. (A) mRNA levels of GABAA receptor δ (GABRD), α6 (GABRA6), α1 (GABRA1) subunits, and (B) 5-α-reductase 1 (SRD5A1) are expressed as fold change of controls. Methylated DNA immunoprecipitation (MeDIP) of the regulatory promoter regions of (C) GABRD and (D) GABRA6 are measured as percentage of input. Hydroxymethylated DNA immunoprecipitation (hMeDIP) of the regulatory promoter regions of (E) GABRD and (F) GABRA6. (G) Protein levels of GABRD measured as ratio of GABRD/GAPDH O.D. values are expressed as percentage of controls. Representative western blot is shown in the lower right-hand corner. Values are mean ± SEM of 24 samples per group for mRNA assays, 11 to 13 samples per group for the immunoprecipitation analysis; 25 samples per group were used for the western blot. *P < .05, Student’s t test vs controls.
To establish whether the altered expression of GABRD and 1-carbon metabolism enzymes could be the consequence of neuronal loss, we measured the levels of the neuronal marker Neuronal Specific Enolase (ENO2) (Guidotti et al., 2000). The levels of ENO2 mRNA were similar in control and AUD subjects (relative mRNA expression in AUD subjects [fold of controls] 1.10±0.06, t(1,46)=1.277, P=.208), suggesting that altered expression of GABRD and 1-carbon metabolism genes is independent of neuronal loss. We also studied whether the expression of 1-carbon metabolism enzymes and GABRD are related with changes in mRNA expression of the microglia marker, ionized calcium-binding adapter molecule 1 (AIF1). This marker was decreased by approximately 30% in the cerebellum of AUD subjects (relative mRNA expression 0.72±0.04, t(1,46)=2.297, P=.026), but its decrease was not accompanied by a parallel decrease of 1-carbon metabolism enzymes or GABRD, suggesting that these genes are largely expressed in neurons. Importantly, when AIF1 mRNA expression was tested for interaction with the expression of GABRD, no correlation was detected (r=-0.039, P=.793).
To study whether the effect of alcohol on GABAergic neurotransmission is limited to the GABAA receptor or also implicates a coordinated involvement of other GABAergic neurotransmitter components, we measured the expression of type-1 5α-reductase (SRD5A1) in the cerebellum of AUD subjects. This enzyme initiates the conversion of pregnenolone to allopregnenolone, which is a potent GABAA receptor modulator (Lambert et al., 2003). We found reduced levels of SRD5A1 in AUD subjects (Figure 6B, t(1,46) =2.448, P=.018). The GABAergic synthesizing enzyme GAD1 (relative mRNA expression in AUD subjects 1.15± 0.13, t(1,46)=1.528, P=.133) failed to change in AUD subjects compared with control. The difference between control and AUD groups remained statistically significant for GABRD and SRD5A1 with Benjamini-Hochberg analysis (FDR with q=0.1) for the GABAergic neurotransmitter genes studied (adjusted Pvalue=.036). To study whether SRD5A1 promoter is differentially methylated would be of interest, and we will focus on this issue in other brain regions of the same cohort, where changes in this enzyme activity have important roles in the regulation of the GABAergic neurotransmission.
Discussion
Our study focuses on the epigenetic DNA methylation marking of GABAergic neurotransmitter genes in cerebellum of AUD subjects. To the best of our knowledge, this is the first study that reports the increased mRNA expression of 3 key enzymes of 1-carbon metabolism (MAT2B, AHCY, MTHFR) and changes in the bioavailability of SAM, SAH, and SAM/SAH ratio in human cerebellum. These changes are associated with higher DNA methylation of regulatory regions and transcriptional repression of GABRD in the cerebellum of AUD subjects.
All 3 one-carbon metabolism enzymes that increase in the cerebellum of AUD subjects (i.e., MAT2B, AHCY, MTHFR) may participate in the control of the levels of SAM, making it available for DNMT-mediated methylation reactions and consequently to modulate gene promoter methylation, as suggested in the scheme of Figure 1. MAT2 utilizes methionine for the biosynthesis of SAM and is highly expressed in cerebellar Purkinje neurons. MAT2 consists of a catalytic (MAT2A) and a regulatory (MAT2B) subunit. The MAT2B subunit regulates MAT2A activity by reducing its KM for L-methionine and by rendering the enzyme more susceptible to feedback inhibition by SAH (Halim et al., 1999). Our results suggest that the increased expression of MAT2B might be an important mechanism to modulate the activity of the catalytic subunit of MAT2A in the cerebellum of AUD subjects. AHCY may modulate transmethylation reactions by facilitating the conversion of SAH into homocysteine, thus reducing the feedback inhibition of SAH on transmethylation reactions (Yi et al., 2000). Our data show that AHCY expression is increased in the cerebellum of AUD subjects (Figure 3), and this increase is associated with a reduction of SAH and an increased SAM/SAH ratio (Figure 5), that is, an increase in the “methylation index”. MTHFR, which increases by ~30% in the cerebellum of AUD subjects, may indirectly contribute to the biosynthesis of SAM by catalyzing the reduction of 5,10-methylene-trahydrofolate to 5-methylene-trahydrofolate, which is then utilized as a methyl donor by methionine synthase (MTR) to remethylate homocysteine and produce methionine.
In transmethylation reactions, DNMTs and other methylating enzymes transfer methyl groups from SAM to a number of molecules, including DNA, RNA, histones, biogenic amines, and other acceptor molecules, and release SAH as by-product. SAH is a potent competitive inhibitor of most methyltransferases (Clarke and Banfield, 2001; Kharbanda, 2007). Hence, either an increase of SAM or a decrease in SAH levels results in an increased SAM/SAH ratio (methylation index), which is associated with an increase in transmethylation reactions. Our findings of increased expression of key enzymes of 1-carbon metabolism, SAM/SAH ratio, as well as DNA methylation in cerebellum of AUD subjects contrast with results reported in liver (Tsukamoto and Lu, 2001; Bönsch et al., 2006; Varela-Rey et al., 2013; Auta et al., 2017), but are similar to previous studies in rats’ cerebellum (Auta et al., 2017). AUD subjects have reduced hepatic MAT (MAT1 and MAT3) and decreased 1-carbon metabolism activity, which leads to decreased SAM biosynthesis and consequently decreased DNMT-mediated reactions or DNA hypomethylation (Lee et al., 2004; Varela-Rey et al., 2013).
The changes observed in the expression of DNA methylation or demethylation genes in the brain of AUD subjects might be related to the presence of alcohol or its withdrawal. Indeed, we found that in AUD subjects with no blood alcohol level detected at the time of death, the expression of TET1 was significantly decreased and that of GADD45B was significantly increased. The neuroadaptive changes in the expression of TET1 and GADD45B in AUD subjects during withdrawal may be the direct result of a transient epigenetic effect of alcohol on transcriptional regulation.
We next studied whether the alcohol-facilitated transmethylation reaction could influence DNA methylation patterns of GABAergic neurotransmitter components. Distinct GABAA receptors participate in 2 types of inhibitory control in cerebellum: (1) the α1-containing GABAA receptor located on the dendritic shafts of Purkinje cells, responsible for conventional synaptic (phasic) inhibition and, (2) the GABRD receptors located on the cell body of cerebellar granule cells that are activated tonically by micromolar GABA levels present in the extracellular milieu and are responsible for inhibiting granule cell firing rates (Mody, 2005). We found that the expression of GABRD (mRNA and protein) but not α1-containing GABAA receptor subunit is significantly reduced in cerebellum of AUD subjects, suggesting that alcohol-induced alterations of GABAA receptor subtype are specific to GABRD. Accordingly, 5mC levels were higher at the GABRD promoter in AUD subjects (Figure 7B). Although the percentage of GABRD promoter pulled-down with the anti-5mC antibody is low, we have established in previous studies using MeDIP that the level of methylation at GAD1, Reelin, and BDNFix promoters (~0.5% to 3%) is comparable and proportional to the percentage of MeCP2 binding in the same samples (Dong et al., 2016; Zhubi et al., 2017). Interestingly, the chronic alcohol-related increased GABRD promoter methylation we measured in the cerebellum of AUD subjects has also been reported in a downstream intronic CpG island of the GABRD isolated from whole-blood samples (Liu et al., 2016), suggesting that GABRD may be considered a putative candidate biomarker for alcohol abuse. Further studies will establish whether regulatory regions of GABRD human gene are characterized by enrichment of 5mC near regions with lower levels of transcription.
One of the more striking physiologically relevant differences between the modulation of GABRA and GABRD inhibitory activity is their sensitivity to neurosteroids (Lambert et al., 2003). Concentrations of tetrahydrodeoxycorticosterone or allopregnanolone as low as 10 nM significantly potentiate GABA-mediated tonic conductance in cerebellar granule cells (Mihalek et al., 1999; Carver and Reddy, 2013). Here, we show that SRD5A1, the enzyme that initiates the transformation of pregnenolone into allopregnenolone, is reduced in the cerebellum of AUD subjects. We previously reported that SRD5A1 is highly expressed in the cerebellar granule cell layer (Agis-Balboa et al., 2006). Taken together, these data suggest that chronic alcohol exposure, in addition to changes in DNA methylation at promoters of GABAA receptor subunit genes, may also affect GABAergic neurotransmission by decreasing allopregnenolone biosynthesis, reducing the sensitivity of extrasynaptic GABRD to ambient GABA and thus contributing to the reduced sensitivity of the GABRD subunit to GABA in the cerebellum of AUD subjects.
Measurements of DNA demethylation components show that the cerebellum of AUD subjects expresses reduced levels of TET1 mRNA, the key enzyme that catalyzes the hydroxymethylation of 5mC (Guo et al., 2011). The reduced expression of TET1 may facilitate the presence of DNA hypermethylation in cerebellum of AUD subjects. Our data indicate that the increased methylation of GABRD promoter in the absence of significant changes in DNMTs expression in the cerebellum of AUD subjects (Figure 3) may be the result of reduced active demethylation mechanisms (i.e., decreased TET1) (Figure 4). Alternatively, gain of methylation could be due to a positive (allosteric?) effect of alcohol on DNMT binding (via the CXXC binding domain; Jurkowska and Jeltsch, 2016). Future studies are needed to explore these possibilities.
We cannot predict whether the effect of alcohol on 1-carbon and DNA methylation reactions observed in the cerebellum of AUD subjects can be extrapolated to other brain areas. In fact, the location of the 1-carbon metabolism enzymes may be characteristic for each brain region depending on their localization in specific cells. Therefore, the regulation of their expression may be characteristic of the function of Purkinje cells in this brain region. Interestingly, the 1-carbon metabolism enrichment observed in the cerebellum of AUD subjects has also been found in the cerebellum of rats chronically treated with ethanol (Auta et al., 2017), further supporting our hypothesis that DNA hypermethylation may be involved in alcohol-induced cerebellar pathophysiology.
There are reports that suggest a sexually dimorphic effect of chronic alcohol exposure (Becker and Koob, 2016; Priddy et al., 2017). In our cohort, we had 5 AUD females. One female had a low RIN value (2.5) and was not included in the analysis. In the 4 remaining females, the levels of MAT2A, MAT2B, MTHFR, AHCY, DNMT1, DNMT3A, and DNMT3B mRNA as well as SAM and SAH were not different from the average values found in male AUD subjects. Obviously, a larger number of female subjects is needed for more meaningful comparisons.
Conclusion
Accumulating evidence indicates that alcohol abuse can alter the methylation status of specific gene promoters in brain and liver (Berkel and Pandey, 2017). In liver, alcohol abuse elicits DNA hypomethylation (Valera-Rey et al., 2013), whereas our data show that in cerebellum it elicits DNA hypermethylation of specific promoters.
GABRD is an inhibitory extrasynaptic receptor subunit preferentially expressed on glutamatergic cerebellar granule cells. The alteration of its expression may profoundly impact the function of granule and Purkinje cell output from the cerebellum. Hence, the decreased expression of GABRD may also alter the function of the cerebellum in AUD subjects. In future studies, we will investigate whether the correlation between 1-carbon metabolism, active DNA demethylation pathway, and local or global DNA methylation found in cerebellum also occurs in other brain areas of AUD subjects. Nonetheless, our study could establish a central role for the activation of 1-carbon metabolism and DNA methylation reactions as an emerging epigenetic mechanism in the neuroadaptive changes that might occur in the cerebellum of AUD subjects. Furthermore, understanding the role of transmethylation reactions in the brain of AUD subjects has potential for the use of novel effective drugs in the treatment of alcoholism.
Statement of Interest
None.
Acknowledgments
We thank New South Wales Brain Tissue Resource Centre (NSW BTRC), the University of Sydney, Australia for providing postmortem brain tissues for this study, which is supported by NIH-NIAAA R28AA012725.
This work was supported by the National Institute on Alcohol Abuse and Alcoholism (NIAAA) grant P50AA022538 (Center for Alcohol Research in Epigenetics) to S.C.P. and A.G. as well as a senior VA research career scientist award to S.C.P.
References
- Agís-Balboa RC, Pinna G, Zhubi A, Maloku E, Veldic M, Costa E, Guidotti A (2006) Characterization of brain neurons that express enzymes mediating neurosteroid biosynthesis. Proc Natl Acad Sci USA 103:14602–14607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Psychiatric Association (1994) Diagnostic and Statistical Manual of Mental Disorders, 4th ed Washington, DC: American Psychiatric Association. [Google Scholar]
- Auta J, Zhang H, Pandey SC, Guidotti A (2017) Chronic alcohol exposure differentially alters one-carbon metabolism in rat liver and brain. Alcohol Clin Exp Res 41:1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbier E, Tapocik JD, Juergens N, Pitcairn C, Borich A, Schank JR, Sun H, Schuebel K, Zhou Z, Yuan Q, Vendruscolo LF, Goldman D, Heilig M (2015) DNA methylation in the medial prefrontal cortex regulates alcohol-induced behavior and plasticity. J Neurosci 35:6153–6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker JB, Koob GF (2016) Sex differences in animal models: focus on addiction. Pharmacol Rev 68:242–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery fate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol 57:289–300. [Google Scholar]
- Berkel TDM, Pandey SC (2017) Emerging role of epigenetic mechanisms in alcohol addiction. Alcohol Clin Exp Res 41:666–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochtler M, Kolano A, Xu GL (2017) DNA demethylation pathways: additional players and regulators. Bioessays 39:1–13. [DOI] [PubMed] [Google Scholar]
- Bönsch D, Lenz B, Fiszer R, Frieling H, Kornhuber J, Bleich S (2006) Lowered DNA methyltransferase (DNMT-3b) mRNA expression is associated with genomic DNA hypermethylation in patients with chronic alcoholism. J Neural Transm (Vienna) 113:1299–1304. [DOI] [PubMed] [Google Scholar]
- Bouchery EE, Harwood HJ, Sacks JJ, Simon CJ, Brewer RD (2011) Economic costs of excessive alcohol consumption in the U.S., 2006. Am J Prev Med 41:516–524. [DOI] [PubMed] [Google Scholar]
- Buck KJ, Hahner L, Sikela J, Harris RA (1991) Chronic ethanol treatment alters brain levels of gamma-aminobutyric acidA receptor subunit mRNAs: relationship to genetic differences in ethanol withdrawal seizure severity. J Neurochem 57:1452–1455. [DOI] [PubMed] [Google Scholar]
- Carver CM, Reddy DS (2013) Neurosteroid interactions with synaptic and extrasynaptic GABA(A) receptors: regulation of subunit plasticity, phasic and tonic inhibition, and neuronal network excitability. Psychopharmacology (Berl) 230:151–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke S, Banfield K (2001) Homocysteine in health and disease. Cambridge: Cambridge University Press. [Google Scholar]
- Dar MS. (2015) Ethanol-induced cerebellar ataxia: cellular and molecular mechanisms. Cerebellum 14:447–465. [DOI] [PubMed] [Google Scholar]
- Davis MI. (2008) Ethanol-BDNF interactions: still more questions than answers. Pharmacol Ther 118:36–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong E, Nelson M, Grayson DR, Costa E, Guidotti A (2008) Clozapine and sulpiride but not haloperidol or olanzapine activate brain DNA demethylation. Proc Natl Acad Sci USA 105:13614–13619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong E, Tueting P, Matrisciano F, Grayson DR, Guidotti A (2016) Behavioral and molecular neuroepigenetic alterations in prenatally stressed mice: relevance for the study of chromatin remodeling properties of antipsychotic drugs. Transl Psychiatry 6:e711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoch MA, Zhou Z, Kimura M, Mash DC, Yuan Q, Goldman D (2012) GABAergic gene expression in postmortem hippocampus from alcoholics and cocaine addicts; corresponding findings in alcohol-naïve P and NP rats. PLoS ONE 7:e29369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, He C (2012) Nucleic acid modifications with epigenetic significance. Curr Opin Chem Biol 16:516–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin DP, Sharma RP, Chase KA, Matrisciano F, Dong E, Guidotti A (2012) Growth arrest and DNA-damage-inducible, beta (GADD45b)-mediated DNA demethylation in major psychosis. Neuropsychopharmacology 37:531–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goll MG, Bestor TH (2005) Eukaryotic cytosine methyltransferases. Annu Rev Biochem 74:481–514. [DOI] [PubMed] [Google Scholar]
- Grayson DR, Guidotti A (2013) The dynamics of DNA methylation in schizophrenia and related psychiatric disorders. Neuropsychopharmacology 38:138–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidotti A, Auta J, Davis JM, Di-Giorgi-Gerevini V, Dwivedi Y, Grayson DR, Impagnatiello F, Pandey G, Pesold C, Sharma R, Uzunov D, Costa E, DiGiorgi Gerevini V (2000) Decrease in reelin and glutamic acid decarboxylase67 (GAD67) expression in schizophrenia and bipolar disorder: a postmortem brain study. Arch Gen Psychiatry 57:1061–1069. [DOI] [PubMed] [Google Scholar]
- Guidotti A, Dong E, Gavin DP, Veldic M, Zhao W, Bhaumik DK, Pandey SC, Grayson DR (2013) DNA methylation/demethylation network expression in psychotic patients with a history of alcohol abuse. Alcohol Clin Exp Res 37:417–424. [DOI] [PubMed] [Google Scholar]
- Guo JU, Su Y, Zhong C, Ming G, Song H (2011) Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell 145:423–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halim AB, LeGros L, Geller A, Kotb M (1999) Expression and functional interaction of the catalytic and regulatory subunits of human methionine adenosyltransferase in mammalian cells. J Biol Chem 274:29720–29725. [DOI] [PubMed] [Google Scholar]
- Hong S, Zheng G, Wiley JW (2015) Epigenetic regulation of genes that modulate chronic stress-induced visceral pain in the peripheral nervous system. Gastroenterology 148:148–157.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M. (2008) Control of mental activities by internal models in the cerebellum. Nat Rev Neurosci 9:304–313. [DOI] [PubMed] [Google Scholar]
- Jurkowska RZ, Jeltsch A (2016) Enzymology of mammalian DNA methyltransferases. Adv Exp Med Biol 945:87–122. [DOI] [PubMed] [Google Scholar]
- Kharbanda KK. (2007) Role of transmethylation reactions in alcoholic liver disease. World J Gastroenterol 13:4947–4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korpi ER. (1994) Role of GABAA receptors in the actions of alcohol and in alcoholism: recent advances. Alcohol Alcohol 29:115–129. [PubMed] [Google Scholar]
- Kyzar EJ, Pandey SC (2015) Molecular mechanisms of synaptic remodeling in alcoholism. Neurosci Lett 601:11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert JJ, Belelli D, Peden DR, Vardy AW, Peters JA (2003) Neurosteroid modulation of GABAA receptors. Prog Neurobiol 71:67–80. [DOI] [PubMed] [Google Scholar]
- Lee TD, Sadda MR, Mendler MH, Bottiglieri T, Kanel G, Mato JM, Lu SC (2004) Abnormal hepatic methionine and glutathione metabolism in patients with alcoholic hepatitis. Alcohol Clin Exp Res 28:173–181. [DOI] [PubMed] [Google Scholar]
- Liu C, et al. (2016) A DNA methylation biomarker of alcohol consumption. Mol Psychiatry [Epub ahead of print]. doi: 10.1038/mp.2016.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu SC, Mato JM (2005) Role of methionine adenosyltransferase and S-adenosylmethionine in alcohol-associated liver cancer. Alcohol 35:227–234. [DOI] [PubMed] [Google Scholar]
- Ma DK, Jang M-H, Guo JU, Kitabatake Y, Chang M-L, Pow-Anpongkul N, Flavell RA, Lu B, Ming GL, Song H (2009) Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science 323:1074–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marutha Ravindran CR, Mehta AK, Ticku MK (2007) Effect of chronic administration of ethanol on the regulation of the delta-subunit of GABA(A) receptors in the rat brain. Brain Res 1174:47–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGough NNH, He DY, Logrip ML, Jeanblanc J, Phamluong K, Luong K, Kharazia V, Janak PH, Ron D (2004) RACK1 and brain-derived neurotrophic factor: a homeostatic pathway that regulates alcohol addiction. J Neurosci 24:10542–10552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhatre M, Ticku MK (1989) Chronic ethanol treatment selectively increases the binding of inverse agonists for benzodiazepine binding sites in cultured spinal cord neurons. J Pharmacol Exp Ther 251:164–168. [PubMed] [Google Scholar]
- Mihalek RM, Banerjee PK, Korpi ER, Quinlan JJ, Firestone LL, Mi ZP, Lagenaur C, Tretter V, Sieghart W, Anagnostaras SG, Sage JR, Fanselow MS, Guidotti A, Spigelman I, Li Z, DeLorey TM, Olsen RW, Homanics GE (1999) Attenuated sensitivity to neuroactive steroids in gamma-aminobutyrate type A receptor delta subunit knockout mice. Proc Natl Acad Sci USA 96:12905–12910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mody I. (2005) Aspects of the homeostatic plasticity of GABAA receptor-mediated inhibition. J Physiol (Lond) 562:37–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montpied P, Morrow AL, Karanian JW, Ginns EI, Martin BM, Paul SM (1991) Prolonged ethanol inhalation decreases gamma-aminobutyric acidA receptor alpha subunit mRNAs in the rat cerebral cortex. Mol Pharmacol 39:157–163. [PubMed] [Google Scholar]
- Ponomarev I, Wang S, Zhang L, Harris RA, Mayfield RD (2012) Gene coexpression networks in human brain identify epigenetic modifications in alcohol dependence. J Neurosci 32:1884–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priddy BM, Carmack SA, Thomas LC, Vendruscolo JCM, Koob GF, Vendruscolo LF (2017) Sex, strain, and estrous cycle influences on alcohol drinking in rats. Pharmacol Biochem Behav 152:61–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skorput AGJ, Yeh HH (2015) Effects of ethanol exposure in utero on Cajal-Retzius cells in the developing cortex. Alcohol Clin Exp Res 39:853–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunahori K, Nagpal K, Hedrich CM, Mizui M, Fitzgerald LM, Tsokos GC (2013) The catalytic subunit of protein phosphatase 2A (PP2Ac) promotes DNA hypomethylation by suppressing the phosphorylated mitogen-activated protein kinase/extracellular signal-regulated kinase (ERK) kinase (MEK)/phosphorylated ERK/DNMT1 protein pathway in T-cells from controls and systemic lupus erythematosus patients. J Biol Chem 288:21936–21944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto H, Lu SC (2001) Current concepts in the pathogenesis of alcoholic liver injury. FASEB J 15:1335–1349. [DOI] [PubMed] [Google Scholar]
- Valenzuela CF, Jotty K (2015) Mini-Review: effects of ethanol on GABAA receptor-mediated neurotransmission in the cerebellar cortex-recent advances. Cerebellum 14:438–446. [DOI] [PubMed] [Google Scholar]
- Varela-Rey M, Woodhoo A, Martinez-Chantar ML, Mato JM, Lu SC (2013) Alcohol, DNA methylation, and cancer. Alcohol Res 35:25–35. [PMC free article] [PubMed] [Google Scholar]
- Warden AS, Mayfield RD (2017) Gene expression profiling in the human alcoholic brain. Neuropharmacology [Epub ahead of print]. doi: 10.1016/j.neuropharm.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi P, Melnyk S, Pogribna M, Pogribny IP, Hine RJ, James SJ (2000) Increase in plasma homocysteine associated with parallel increases in plasma S-adenosylhomocysteine and lymphocyte DNA hypomethylation. J Biol Chem 275:29318–29323. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Yuan Q, Mash DC, Goldman D (2011) Substance-specific and shared transcription and epigenetic changes in the human hippocampus chronically exposed to cocaine and alcohol. Proc Natl Acad Sci USA 108:6626–6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhubi A, Chen Y, Guidotti A, Grayson DR (2017) Epigenetic regulation of RELN and GAD1 in the frontal cortex (FC) of autism spectrum disorder (ASD) subjects. Int J Dev Neurosci [Epub ahead of print]. doi: 10.1016/j.ijdevneu.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]