Abstract
A special class of poorly characterized architectural proteins is required for chromatin topology and enhancer–promoter interactions. Here, we identify Opbp as a new Drosophila architectural protein, interacting with CP190 both in vivo and in vitro. Opbp binds to a very restrictive set of genomic regions, through a rare sequence specific motif. These sites are co-bound by CP190 in vivo, and generally located at bidirectional promoters of ribosomal protein genes. We show that Opbp is essential for viability, and loss of opbp function, or destruction of its motif, leads to reduced ribosomal protein gene expression, indicating a functional role in promoter activation. As characteristic of architectural/insulator proteins, the Opbp motif is sufficient for distance-dependent reporter gene activation and enhancer-blocking activity, suggesting an Opbp-mediated enhancer–promoter interaction. Rather than having a constitutive role, Opbp represents a new type of architectural protein with a very restricted, yet essential, function in regulation of housekeeping gene expression.
INTRODUCTION
Insulators, in both Drosophila and vertebrates, have been defined based on their ability to disrupt the communication between an enhancer and a promoter when inserted in between (1–6). Accumulating evidence suggests that some insulator proteins also play a positive role in gene expression, providing an architectural function in mediating inter- and intra-chromosomal interactions (7–9). These insulator proteins have been attributed to the category of architectural proteins (10).
Genomic studies indicate that known architectural proteins most frequently bind near transcription start sites, suggesting a general role in promoter activity and enhancer–promoter communication (11–18). In vertebrates, CTCF is currently the only architectural protein identified to date, and plays a prominent role in establishing chromatin loops (9,19). CTCF is a highly conserved protein, with eleven Zinc finger domains (C2H2-ZF) many of which mediate DNA-binding, targeting CTCF complexes to a wide range of diverse sites throughout the genome (20). In Drosophila in addition to CTCF a number of other architectural proteins are known. All of these proteins (Su(Hw), Pita, ZIPIC, Zw5) contain C2H2-ZF domains for specific DNA-binding. Zw5, Pita and ZIPIC, also contain a characteristic N-terminal ZAD domain that is responsible for protein–protein interactions (17,21–24).
The majority of known Drosophila architectural proteins interact with a multifunctional co-factor named the Centrosomal Protein 190 kDa, CP190 (14,25–33). CP190 (a 1096 amino acid protein) contains an N-terminal BTB/POZ domain, an aspartic-acid-rich D-region, four C2H2 zinc finger motifs and a C-terminal E-rich domain (29,34). The BTB domain of CP190 forms stable homodimers that may be involved in protein–protein interactions (29,35,36). In addition to these motifs, CP190 also contains a centrosomal targeting domain (M) responsible for its localization to centrosomes during mitosis (37). CP190 is recruited to chromatin via its interaction with DNA-binding architectural proteins, mediated through its BTB and M domains (14,38).
The CP190 protein preferentially binds near the transcription start sites of genes suggesting a role in the organization of promoter architecture (13,25,34). CP190 has major effects on chromatin, such as the depletion of nucleosomes, high nucleosomal turnover and the prevention of heterochromatin expansion (9,25,39). There is also some evidence suggesting that CP190 can support long-distance interactions between distantly located binding sites for recruiter proteins (40) and in this way CP190 might also be involved in the organization of enhancer–promoter or promoter-promoter interactions (14,27). Interestingly, CP190 are highly enriched near housekeeping (hk) genes’ promoters (25,41,42) and active hk enhancers (18,43), and CP190 can recruit NURF, dREAM and SAGA complexes to chromatin (39,44–46), all of which are essential for the activation of hk genes expression. How CP190 is specially recruited to hk genes is not known - the architectural proteins CTCF, Pita and ZIPIC do not display specific binding to hk promoters (18), suggesting the existence of an unknown architectural proteins that recruits CP190 preferentially to this class of genes.
Here we report a new DNA-binding transcription factor, previously named Optix binding protein (Opbp), as a sequence-specific recruiter of CP190. This protein was identified as a putative partner of a transcription factor Optix (47). In contrast to all other known DNA-bound insulator proteins, which bind to thousands of sites throughout the genome (11,31), we show that Opbp occupies a very restricted set of specific sites, which are generally located at divergent promoters often involving ribosomal protein genes. Opbp is essential for the expression of ribosomal protein genes in vivo and interacts with CP190 and its sequence-specific DNA motif via distinct protein domains. Using transgenic assays, we show that Opbp has characteristic enhancer blocking activity in vivo and mediates distance-dependent promoter activation, suggesting that it mediates chromatin loops essential for enhancer–promoter interactions. These results identify Opbp as a new architectural protein with insulator like properties, similar to Pita, dCTCF and ZIPIC, but with a more specific role in the regulation of ribosomal protein gene expression.
MATERIALS AND METHODS
Yeast two-hybrid assay
Yeast two-hybrid assay was carried out using yeast strain pJ69-4A, with plasmids and protocols from Clontech. For growth assays, plasmids were transformed into yeast strain pJ69–4A by the lithium acetate method, as described by the manufacturer, and plated on media without tryptophan and leucine. After 2 days of growth at 30°C, the cells were plated on selective media without tryptophan, leucine, histidine and adenine, and their growth was compared after 2–3 days. Each assay was repeated three times.
Pull-down assay
BL21 cells co-transformed with plasmids expressing GST-fused Opbp derivatives and 6xHis-fused CP190[245-606] were grown in LB media to an A600 of 1.0 at 37°C and then induced with 1 mM IPTG at 18°C overnight. ZnCl2 was added to final concentration 100 μM before induction. For GST-pulldown cells were disrupted by sonication in buffer C (20 mM HEPES–KOH, pH 7.7, 150 mM NaCl, 10 mM MgCl2, 0.1 mM ZnCl2, 0.1% NP40, 10% (w/w) glycerol) containing 1 mM PMSF and Calbiochem Complete Protease Inhibitor Cocktail VII (5 μL/ml), centrifuged, applied to Immobilized Glutathione Agarose (Pierce) for 10 min at +4°C, after that resin was washed four times with buffer C containing 500 mM NaCl and elution performed with 50 mM reduced glutathione, 100 mM Tris, pH 8.0, 100 mM NaCl for 15 min. 6xHis-pulldown was performed similarly with Co-IDA resin (Biontex) in buffer A (see Supplementary Methods), washed with buffer A containing 30 mM imidazole and proteins were eluted with buffer B.
Protein chemical crosslinking
Chemical crosslinking with glutaraldehyde was carried out for 10 min at room temperature in PBS buffer containing 1 mM β-mercaptoethanol. Prior to crosslinking protein concentration was adjusted to 5 μM. Crosslinking was quenched with 50 mM glycine and samples were resolved using SDS-PAGE followed by silver-staining.
Antibodies
Antibodies against Opbp [aa 1–331] and CP190 [aa 308–1096] were raised in rats and rabbits and purified from the sera by ammonium sulfate fractionation followed by affinity purification on CNBr-activated Sepharose (GE Healthcare, USA) according to standard protocols. Anti-FLAG M2 antibodies were from Sigma (USA).
Generation and validation of fly lines with deletion of the opbp gene using CRISPR/Cas9
We used the fly CRISPR Optimal Target Finder tool (University of Wisconsin) to design a CRISPR target sequence for the 5′ and 3′ end of the opbp gene (48) (Supplementary Table S1). The 5′ target site was selected in the gene's first intron (Supplementary Figure S1). 5′ and 3′ target sequences were cloned into the pCR vector based on pCFD4-U6:1_U6:3tandemgRNAs plasmid (Addgene # 49411), using BbsI. Correct ligation of the opbp CRISPR target sequences was confirmed by sequencing (49).
The 5′ and 3′ 500 bp flanking regions (Supplementary Table S1) surrounding the target sites were cloned (Supplementary Figure S1) into the plasmid for homological recombination (pHR) (Supplementary Figure S2A). This vector contains φC31 attP and mCherry under actin promoter surrounded by loxP sites. Plasmids mixture (10:1—pHR:pCR, concentration 500 ng/μl) was injected in the embryos of the flies expressing Cas9 under control of the nanos promoter (Bloomington stock center: 54591) (49). Flies with potential opbp deletions were identified by mCherry fluorescence (Supplementary Figure S2B). Flies were returned balanced over the CyO second chromosome balancer. Gene deletion was confirmed using qPCR with primers Opbp RT (Supplementary Figure S2C) and primers targeted the inserted vector - using conventional PCR (Supplementary Figure S2D). 5′ and 3′ junctions were also sequenced. For detection of ΔOpbp lethality, flies were crossed with CyO GFP balancer (Bloomington stock center: 5194).
Rescuing of the opbp gene
The opbp gene was obtained by PCR and cloned to plasmid with the white reporter gene, loxP site, SV40 terminator and attB (Supplementary Figure S3A). A line with ΔOpbp flies was crossed with flies with φC31 under control of the vasa promotor (50). The embryos from this cross were injected with the rescuing plasmid. The mCherry and white reporter genes were removed by crossing with Cre recombinase-expressing line (y1w1; Cyo, P[w+,cre]/Sco; +) (Supplementary Figure S3A). The loxP site remained in the first intron of the gene and does not affected opbp mRNA (Supplementary Figure S3B and C). As a result, the opbp gene was re-inserted. Correct splicing of the opbp gene with loxP in the first intron was confirmed with PCR from cDNA (Supplementary Figure S3B).
Opbp chromatin immunoprecipitation (ChIP-seq)
Chromatin was prepared as described in the Supplementary Methods.
Immunoprecipitation
Debris was removed by centrifugation at 14 000 g, 4°C, for 10 min, and chromatin was pre-cleared with Protein A agarose (Pierce) blocked with BSA and salmon sperm DNA, with 50 μl aliquots of such pre-cleared chromatin being stored as input material. Samples containing 10–20 μg of DNA equivalent in 1 mL of nuclear lysis buffer were incubated overnight, at 4°C, with rabbit antibodies against Opbp (1:500) and CP190 (1:1000) or with nonspecific IgG purified from rat preimmune sera (control). Chromatin–antibody complexes were collected using blocked Protein A or G agarose at 4°C over 5 h. After several rounds of washing with lysis buffer (as such and with 500 mM NaCl), LiCl buffer (20 mM Tris–HCl, pH 8; 250 mM LiCl, 1 mM EDTA, 0.5% NP-40, 0.5% sodium deoxycholate, protease inhibitors) and TE buffer (10 mM Tris-HCl, pH 8; 1 mM EDTA), the DNA was eluted with elution buffer (50 mM Tris-HCl, pH 8.0; 1 mM EDTA, 1% SDS), the cross-links were reversed, and the precipitated DNA was extracted by the phenol–chloroform method. The enrichment of specific DNA fragments was analyzed by real-time PCR, using a StepOne Plus Thermal Cycler (Applied Biosystems). The primers used for PCR in ChIP experiments for genome fragments are shown in Supplementary Table S1.
Illumina libraries were prepared according to manufacturers recommendations with small modifications. In short, 1–10 ng of purified, RNase treated, and reverse cross-linked genomic DNA was end-repaired and terminal adenosine residues were added using the NEBNext reagents. Custom-made indexed adapters were ligated, after which the material was size selected at ∼200–600 bp with Ampure XP beads (Beckman Coulter). PCR amplification was performed using PE1.0 and PE2.0 primers (custom-made) for 12 cycles for Input samples and 14–15 cycles for IP-ed samples using the Q5 Hot Start HiFi PCR Master Mix (NEB). The PCR-amplified library was purified using Ampure XP beads and its quality was assessed on a Bioanalyzer 2100 system (Agilent). The libraries were sequenced on a HiSeq 2000 (Illumina) in single end mode.
Electrophoretic mobility shift assay (EMSA)
Aliquots of purified recombinant proteins (10–15 μg) were incubated with fluorescently labeled DNA fragments of Opbp-binding sites in the presence of nonspecific binding competitor poly(dI-dC). Incubation was performed in PBS (pH 8.0) containing 5 mM MgCl2, 0.1 mM ZnSO4, 1 mM DTT, 0.1% NP-40 and 10% glycerol at room temperature for 30 min. The mixtures were then resolved by nondenaturing 5% PAGE (79 AA:1 BAA) in 0.5 × TBE buffer at 5 V/cm.
RESULTS
Identification of Opbp as a partner of the CP190 protein
To identify new DNA binding factors that are associated with CP190’s regulation of chromatin architecture, we searched for zinc finger proteins that can interact with CP190 using the yeast two-hybrid assay with a custom library of zinc finger proteins cloned in the corresponding vectors. As a result, we identified Opbp as a specific interactor of CP190. Opbp contains eight C2H2-ZF at the carboxy-terminus and one additional C2H2-ZF at the amino-terminus (Figure 1A, Supplementary Figure S4). We confirmed this interaction by co-immunoprecipitation of CP190 and 3 × FLAG-tagged Opbp transfected S2 cells (Figure 1C). To further examine the function of this new protein, we prepared polyclonal affinity purified antibodies against Opbp (amino-acids 1–331), which we show by RNAi knockdown are specific for Opbp (Supplementary Figure S5).
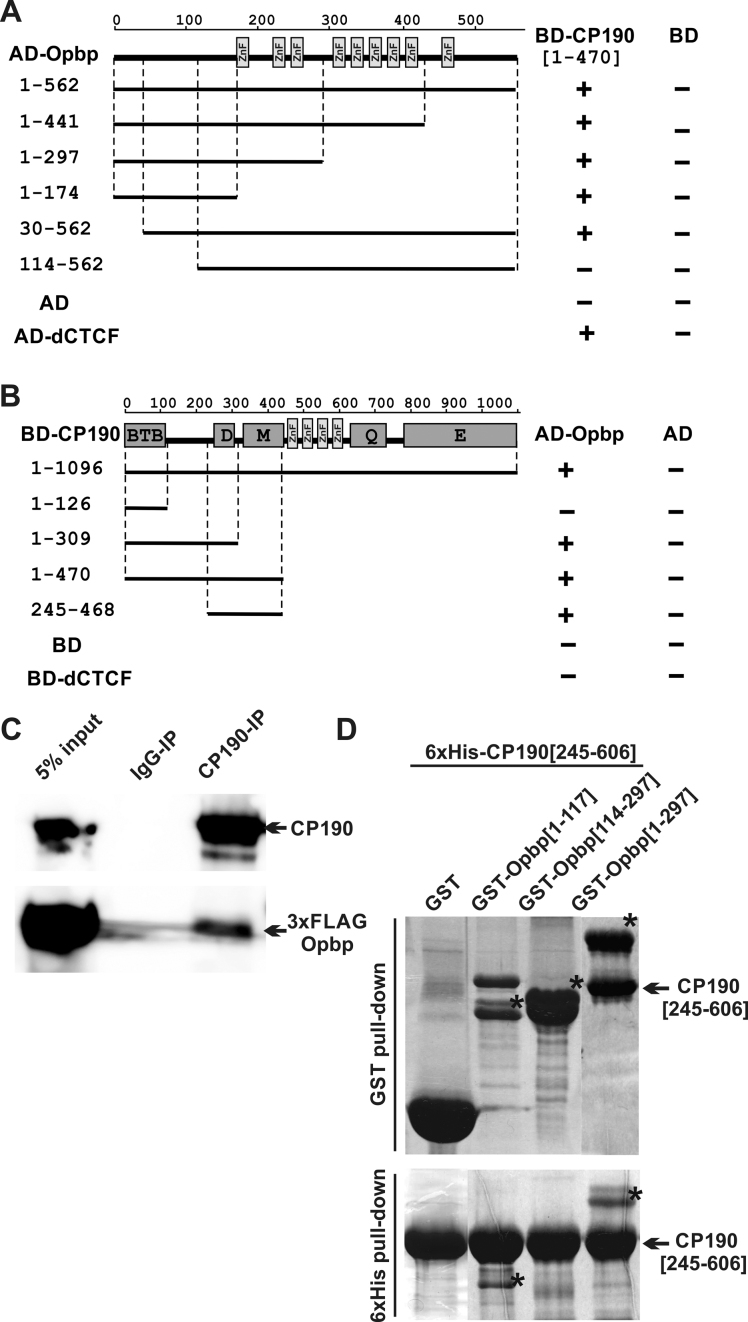
Figure 1.
Opbp is a new C2H2 protein that interacts with CP190. (A) Localization of Opbp domains interacting with CP190 in yeast two-hybrid assay. Different fragments of Opbp were fused to the GAL4 activating domain and tested for interaction with CP190 fused to the GAL4 DNA-binding domain. All Opbp fragments were tested for the absence of interaction with the GAL4 DNA-binding domain alone (BD). (B) Localization of CP190 domains interacting with Opbp in yeast two-hybrid assay. Different fragments of CP190 were fused to the GAL4 DNA-binding domain and tested for interaction with Opbp fused to the GAL4 activating domain. All CP190 fragments were tested for the absence of interaction with the GAL4 activating domain alone (AD). (A, B) Protein domains of full-length CP190 and Opbp are indicated as boxes, and lines represent the different deletion fragments (amino acid residues indicated on the left). Previously described interaction between CP190 and dCTCF was used as a positive control. The results are summarized in columns on the right (BD-CP190 or AD-Opbp), with the ‘+’ and ‘–‘ signs referring to the presence and absence of interaction, respectively. (C) Nuclear extracts from Drosophila S2 cells co-transfected with CP190 and 3 × FLAG-Opbp were immunoprecipitated with antibodies against CP190 (using nonspecific IgG as a negative control), and the immunoprecipitates were analyzed by Western blotting for the presence of FLAG-tagged proteins in immunoprecipitated sample. (D) Interaction of the recombinant 6xHis 245–606 aa region of CP190 with different N-terminal GST-tagged fragments of Opbp (indicated by *) in GST and 6xHis pull-down assays. The precipitated proteins were resolved by SDS-PAGE and stained with Coomassie. The positions of amino acids are indicated by square brackets.
To determine which protein domains are involved in the interaction between CP190 and Opbp we used the yeast two-hybrid assay, which narrowed down the interacting region within Opbp to amino-acids 30–114 (Figure 1A). Conversely, after testing different fragments of the CP190 protein, we identified the region that overlaps the D domain between amino-acid 245–309 of CP190 to be essential for Opbp interaction (Figure 1B). It is worth noting that in some cases a lack of interaction can be attributed to misfolding of truncated proteins. We therefore confirmed the interactions, and the domains, obtained in yeast two-hybrid assays with in vitro pull-down assay using recombinant proteins (Figure 1D). The ability of the 245–606 aa region of CP190, which includes the D, M and ZnF domains fused to 6xHis, to interact with different N-terminal fragments of Opbp fused to GST, was examined under stringent salt conditions. These experiments clearly identify an interaction between CP190[245–606] with Opbp[1–117] and Opbp[1–297], but not with Opbp[114–297] or GST alone (Figure 1D). Unfortunately shortened versions of CP190 (corresponding to 245–309 aa region) demonstrated characteristics of natively unstructured molecules that are not able to interact with any proteins in vitro. Taken together, these results indicate a direct protein–protein interaction between CP190 and Opbp that is mediated via amino-acids 245–606 of CP190 and amino-acids 30–114 of Opbp.
Opbp is essential for Drosophila viability
To determine the function of Opbp in vivo, we used a two-step genome engineering platform that combines CRISPR-mediated HDR (Homology-directed repair) with φC31 recombinase-mediated cassette exchange (RMCE) (51). This enables both the efficient identification of the opbp deletion and by using attP sites to insert any DNA of choice using RMCE (50,52). As a result, we substituted the opbp gene (Supplementary Figures S1 and S2) with a mCherry reporter under the control of the Actin 5C promoter and flanked by attP sites. The obtained deletions were balanced against a CyO,GFP balancer. The homozygotes for the opbp deletion died mostly late in development, often surviving to pupal stages (45% from expected number of homozygous pupae). The lethality is not fully penetrant as several adults (1.2% from expected numbers of homozygous adult flies) eclosed, and displayed thinner bristles than wild-type (WT), changes in wing blade and eyes, reduced viability and complete sterility (Supplementary Figure S6). Such phenotypes are similar to those previously described for the ‘Minute’ phenotype, which is induced by the disruption of ribosomal protein genes. The ‘Minute’ syndrome of dominant, haploinsufficient phenotypes, includes prolonged development, short and thin bristles, poor fertility and viability (53,54).
Larvae with opbp deletion emerge from the food 3–4 days later than WT larvae (9 days versus 5 days), indicating a developmental delay. To confirm that the lethal phenotype is a consequence of the opbp deletion, we re-inserted the deleted opbp gene using RMCE (Supplementary Figure S3A). The opbp deletion was fully rescued as homozygotes, demonstrating that the lethality is due to opbp and that opbp is an essential gene for Drosophila viability (Supplementary Figure S3D).
Given our observed protein–protein interaction between Opbp and CP190, we next assessed if there is a genetic interaction between both proteins. We used the previously characterized cp190 null alleles named cp2 and cp3 (37). The cp2/cp3 transgeterozygote showed some larval mortality, but approximately half of the zygotic mutants survived until late pupal stages of development, dying as pharate adults, most likely due to the presence of maternal protein. The cp2/+ or cp3/+ heterozygous displayed normal viability and a WT phenotype. Placing even one copy of the cp190 loss-of-function allele, cp2/+ or cp3/+ heterozygous, altered the phenotype of the Δopbp homozygotes that all died before pupae stage. Double homozygous were not observed even at larvae stage. These results suggest a functional interaction between cp190 and opbp.
Opbp binds to a specific set of divergent promoters in the vicinity of ribosomal protein genes
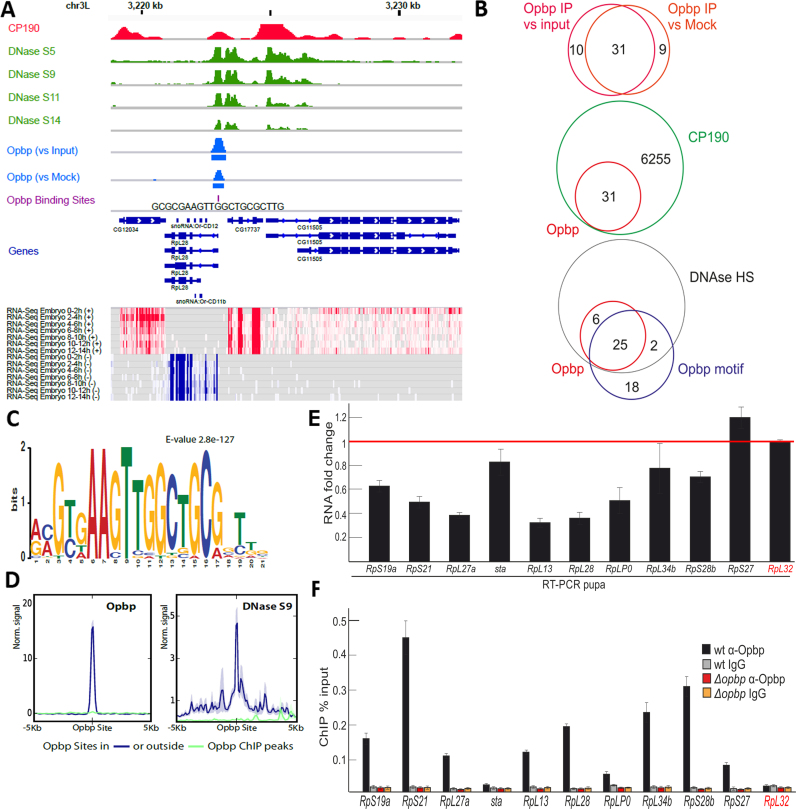
To discern the molecular function of Opbp, we first identified regions bound by Opbp in vivo during Drosophila embryogenesis by performing chromatin immunoprecipitation followed by next generation sequencing (ChIP-seq) on chromatin extracted from 0 to 12 h embryos. A total of four independent biological replicates were performed with antibodies against Opbp (Supplementary Figure S5). We first performed two biological replicates and compared ChIP enrichment to matched input controls, using an Irreproducible Discovery Rate (IDR) threshold of 0.05 (Methods). Based on this stringent cut-off, we identified 41 high confidence Opbp-bound regions, genome-wide. Given this low number, we performed a second two completely independent biological replicates and compared Opbp ChIP enrichment to mock immunoprecipitations, using the pre-immune serum of the rabbit used to generate the Opbp antibody (Figure 2A). This identified 40 high confidence Opbp-bound regions; with 31 peaks common to both data sets (Supplementary Table S2). The similar number of peaks obtained (40,41), as well as the very high overlap between these two sets (Figure 2B) indicates a high reproducibility of the ChIP-seq experiments.
Figure 2.
Opbp binds to a restricted number of genomic loci via a specific motif. (A) Opbp binding at a representative locus showing Opbp IP signal scaled to library size and normalized to input or mock IP (middle blue tracks, rectangles indicate MACS2 peaks at 5% IDR) together with DNase hypersensitivity at embryonic stages 5, 9, 11 and 14 from (57) (green tracks), CP190 binding from (59) (red track), Opbp Binding Sites (violet track, Patser predictions), Transcript models (Flybase 5.57) and heatmaps of RNA-seq embryonic time course (total RNA from modENCODE (76)) for sense (red) and antisense (blue) strands. (B) Overlap between Opbp ChIP peaks, reported by MACS2, when contrasting Opbp IP with Mock versus input (upper Venn) at an IDR cutoff of 5%. Overlap between Opbp and CP190 bound regions (middle Venn) and Opbp bound regions with DNAse hypersensitivity sites and Opbp motifs. (C) Opbp motif discovered by MEME. (D) Opbp binds exclusively to regions containing its motif and in open chromatin. The quantitative ChIP signal for Opbp (mean profiles of normalized signal) centered around the Opbp motif for bound (blue) and unbound (green) sites. DNase hypersensitivity (embryonic stage 9, right) centered at all Opbp binding sites at Opbp-bound (blue) or unbound (green) regions. Semi-transparent ribbons around profiles show standard errors. (E) Expression levels of the ribosomal protein genes with the Opbp-bound promoter regions at pupae 2d stage in the wt and opbp− backgrounds. Individual transcript levels were determined by RT-qPCR with corresponding primers normalized relative to RpL32 (marked by red) for the amount of input cDNA. Histogram shows the changes of mRNAs for tested ribosomal protein genes comparing with expression levels in the wild type pupa (corresponding to the red line at the ‘1’ on the scale). Error bars show standard deviation of triplicate PCR measurements. (F) Comparing binding of Opbp to the promoters of the ribosomal protein genes in the wt and opbp− (Δopbp) background. Histogram shows ChIP enrichments at the promoter regions on chromatin isolated from wild type (wt) and Δopbp pupa with antibodies against Opbp and non-specific IgG. The results are presented as a percentage of input genomic DNA. Error bars show standard deviations of triplicate PCR measurements for three independent experiments. RpL32 (marked by red) is the negative control negative controls for Opbp binding.
The small number of peaks for such a genome-wide experiment is unusual. De novo motif discovery using MEME (55) on the 31 Opbp overlapping peaks identified a new and very significant (E-value 2.8e–127) sequence motif, a 21 base-pair long element (Figure 2C). Such long motifs are common for TFs with many C2H2 zinc fingers. The length and high sequence information content of this motif provides the specificity to search the entire Drosophila genome to identify all Opbp binding sites genome-wide (using patser described in (56)). This identified only 45 Opbp binding sites throughout the entire Drosophila genome. Although surprisingly low, this number of binding sites is in line with the low number of ChIP-peaks we identified, and attests to the sensitivity of our in vivo binding data. Moreover, 81% of the 31 Opbp ChIP-seq common peaks contain this 21bp motif, indicating the specificity by which this transcription factor binds to DNA. Examining the remaining 20 Opbp binding sites located away from Opbp ChIP peaks revealed that they are almost completely devoid of any Opbp ChIP signal and moreover, 80% (18) are located in regions devoid of any DNAse signal, indicating that they are in closed chromatin (Figure 2B, data from (57)). Taken together these results indicate that Opbp binds directly, and very specifically, to DNA using its own sequence specific motif. 92% (25/27) of the Opbp motifs found in open chromatin (as assessed by DNAse accessibility) are occupied by Opbp - a level of occupancy very rarely observed for transcription factors (Figure 2B and D). We note that the Opbp motifs found in closed chromatin may also be real Opbp binding sites targeted at different developmental stage(s).
The small number of regions specifically bound by Opbp (as determined by ChIP-seq) thereby reflects the very low number of its specific motifs present within the genome, and indicates a very specific function, perhaps at specialized regulatory elements. We manually inspected the distribution of all 31 common Opbp bound regions (see Supplementary Table S2): 97% (30/31) are located in promoter regions, while the remaining peak was in an intron. Interestingly, 75% (23/30) of Opbp bound regions are between divergent genes in a head-to-head gene pair (P-value is 0.0001) (as seen in the example in Figure 3A); in this arrangement, the Opbp bound region is always located at one of the two promoters, and not at equal distance between the two, suggesting that Opbp specifically regulates only one the two divergent genes. This systematic identification of Opbp occupancy also revealed a high number of ribosomal protein genes among Opbp targeted loci (e.g. RpL28 shown in Figure 2A). This enrichment for ribosomal gene loci is highly significant compared to expectations from the whole genome (using DAVID (58), Methods, Supplementary Table S3).
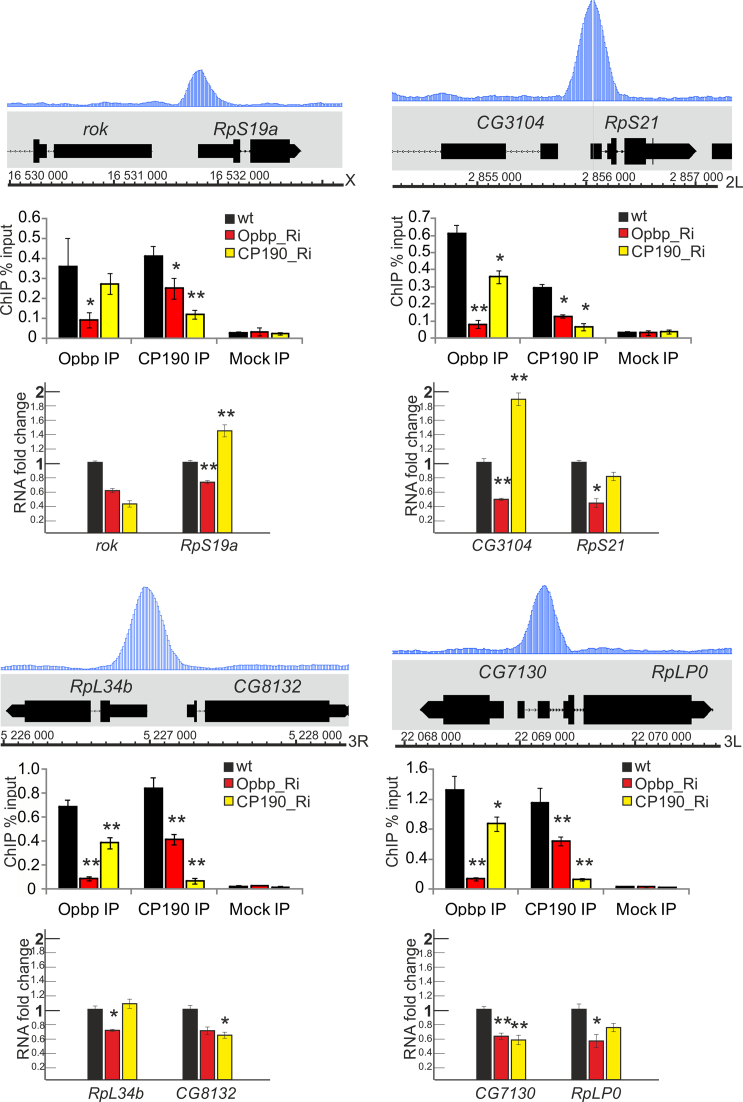
Figure 3.
CP190 binding is dependent on Opbp recruitment to Opbp-binding sites. Tracks for Opbp binding profiles are presented at the selected genome regions. Histograms show ChIP enrichments for Opbp and CP190 in Opbp-binding regions on chromatin isolated from S2 cells treated with specific Opbp (red) or CP190 (yellow) long dsRNAs (Ri) and incubated with antibodies against Opbp and CP190. WT is the mock-treated S2 cells (non-specific eGFP dsRNA). The results are presented as a percentage of input genomic DNA. Error bars show standard deviations of triplicate PCR measurements for three independent experiments. Below: Bar charts indicate changes in gene expression in S2 cells treated with specific dsRNA (Ri) against Opbp or CP190 coding regions according to quantitative real-time PCR with cDNAs synthesized on RNAs extracted from S2 cells after treatment with dsRNAs. Individual transcript levels determined by quantitative PCR with corresponding primers were normalized relative to RpL32 and γTub37C for the amount of input cDNA. Error bars show standard deviations of three replicas. *P-value < 0.05; **P-value < 0.01.
Importantly, Opbp bound regions are also bound by CP190 in vivo, using ChIP-seq data from (59): Opbp and CP190 are co-bound at all Opbp bound regions (Figure 2B) (P-value is 0.0001). This, combined with the direct protein–protein interaction (Figure 1) and genetic interaction we observed, strongly suggests that Opbp is required for CP190 recruitment and function at a specific sub-set of its target genes. To directly assess this, we first examined if Opbp is required for CP190 recruitment. Depletion of Opbp in S2 cells by RNAi (Supplementary Figure S5) lead to a significant reduction in CP190 binding at most of the Opbp bound regions (Figure 3, Supplementary Figure S8). The binding of Opbp itself was also drastically reduced, as expected, demonstrating the effectiveness of the protein depletion, although we note that this was not a complete protein knockdown (Supplementary Figure S5B). Thus, Opbp is essential for recruiting of CP190 to these promoters. CP190 depletion by RNAi also affected Opbp binding at three of the four analyzed sites. It seems likely that interaction with CP190 increases the efficiency of Opbp binding to these sites. These results indicate that CP190 recruitment to a very specific set of genomic regions is largely dependent on Opbp binding.
We next examined the expression of all ribosomal protein genes with Opbp occupancy in opbp− loss-of-function mutants at the pupa stage (Figure 2E and F, Supplementary Figures S7 and S8). In the majority of cases the Opbp bound genes had significantly reduced expression in opbp mutants, indicating that the occupancy is functional (Figure 2E). Importantly, Opbp inactivation in S2 cells leads to approximately a two-fold decrease in the expression of both the upstream (further away from the binding) and downstream targeted genes, suggesting an essential role in promoter activation (Figure 3). It is also interesting to note that cp190 depletion seems to have a mixed effect on gene expression, sometimes decreasing, sometimes increasing transcription. This suggests that CP190 has both Opbp-dependent and independent mechanisms of action in the regulation of these genes’ expression.
Opbp is required for the regulation of ribosomal protein gene expression in vivo
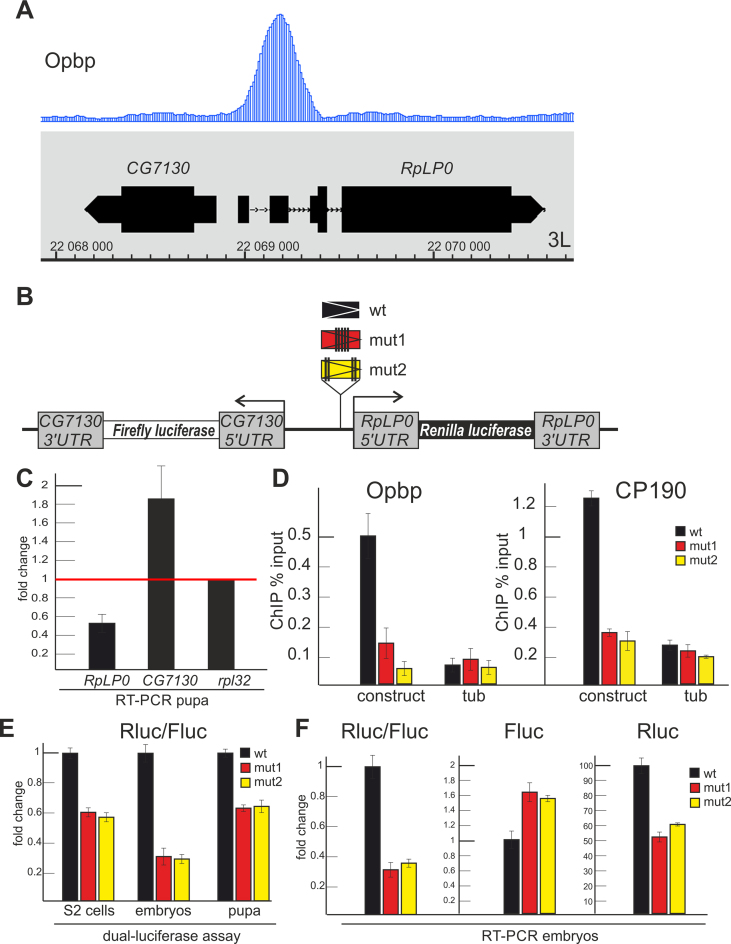
To further confirm the functional requirement of Opbp in the activation of ribosomal protein genes’ promoters in vivo, within a genomic context, we examined a locus containing two divergently orientated promoters of the RplP0 and CG7130 genes (Figure 4A). The occupied Opbp binding sites is located at –12 bp relative to the transcription start of RplP0 (isoform A), a ubiquitously expressed ribosomal protein gene. The transcription start site of the weakly expressed CG7130 gene is located at –335 bp relative to the transcription start site of RplP0. To measure the regulatory effect of Opbp, we generated a transgenic reporter construct where the CG7130 and RplP0 coding regions were substituted by Firefly and Renilla luciferase, respectively (Figure 4B) and the Opbp site was either present or mutated. Importantly, electrophoretic mobility shift assay showed that recombinant Opbp can bind to the ∼200 bp wild-type promoter region, confirming the in vivo occupancy data, but not to the promoter when the Opbp sites were mutated (Supplementary Figure S9). To facilitate a direct comparison of the activation of both promoters in the presence or absence of an Opbp binding site, constructs were inserted in the same genomic region, 86Fb, using the phiC31-based integration system (50). Upon obtaining the homozygous transgenic lines, the binding of both Opbp and CP190 to the wild-type and two mutant promoters (where the Opbp binding site was mutated) in the transgenic locus was determined by ChIP-qPCR using embryonic chromatin (Figure 4D).
Figure 4.
Opbp regulates the RplP0 promoter in transgenic flies. (A) Track for Opbp binding profile is presented at the CG7130/RplP0 genome region. (B) Scheme of transgenic constructs used to examine the role of Opbp (wt and two mutants of Opbp motifs) at the RplP0 promoter. (C) Changes of expression levels of RplP0 and CG7130 measured by RT-qPCR with cDNAs synthesized from RNA extracted from wt and opbp− pupa. Individual transcript levels were determined by RT-qPCR with corresponding primers normalized relative to RpL32 for the amount of input cDNA. Histogram shows the changes of RplP0 and CG7130 relatively expression levels in wild type pupa (red line at the ‘1’ on the scale). Error bars show standard deviation of triplicate PCR measurements. (D) Both Opbp and CP190 are recruited to the transgenic construct containing the Opbp motif. ChIP-real-time-PCR of Opbp and CP190 to the transgenic constructs; percentage of input recovery is shown. Error bars indicate standard deviations of quadruplicate PCR measurements for three independent experiments. (E), (F) Opbp/CP190 recruitment is required for reporter expression driven by the RplP0 promoter. Histogram shows the ratio of Renilla to Firefly luciferases in S2 cells (left) and transgenic fly lines (right) in dual-luciferase assay (E) and relative amount of luciferase mRNAs (F) in transgenic fly lines.
In contrast, to the wild-type locus where both proteins could bind, mutation of the Opbp binding site prevented Opbp, and also importantly, CP190 binding to the RplP0 promoter (Figure 4D), consistent with the possibility that CP190 recruitment is dependent on Opbp binding. To examine the transcriptional response caused by a lack of Opbp-CP190 recruitment, we measured both luciferase activity and transcript levels (Figure 4E and F) directed from both promoters in the transgenic embryos. This identified a two to three fold decrease in gene expression directed from the RplP0 promoter in the absence of Opbp-CP190 recruitment (Figure 4E and F). At the same time, CG7130 expression appears to increase in the absence of Opbp-CP190 recruitment. These results are in agreement with the effect of the opbp gene deletion in pupa (Figure 4C), and strongly suggests that an Opbp directed CP190 complex is required for RplP0 promoter activation in vivo.
Opbp motifs are sufficient to support distance chromatin interactions to activate transcription
The C2H2-ZF protein Opbp has a similar structure to the previously described architectural/insulator proteins dCTCF, Su(Hw), Pita, Zw5 and ZIPIC. These proteins also typically bind to promoter regions, like Opbp, via a single binding site. The characteristic property of architectural proteins is to efficiently bind to their consensus sites and to direct specific long-distant genomic interactions and enhancer blocking (4,7). However, Su(Hw) (60) and dCTCF (O.M., O.K. unpublished) proteins only display enhancer blocking if they bind to multimerised 4–5 sites. To directly determine if the Opbp protein can function in a similar manner as these architectural proteins, we constructed a synthetic DNA fragment containing six equally-oriented consensus sites (Supplementary Figure S10).
All C2H2-ZF architectural proteins characterized to date can support distance-dependent chromatin interactions in the transgenic GAL4/white model system (61,62). The model system is based on the inability of the GAL4 activator to stimulate the white promoter across the yellow gene in the absence of an insulator mediated chromatin loop (Figure 5A, Supplementary Figure S11). Architectural proteins generally function through protein–protein interactions directed from two distally located sites, which come together bringing the flanking DNA closer to each other while the intervening DNA is ‘looped out’ of this conformation. The loop formation brings the GAL4 binding sites and the promoter together to facilitate GAL4-mediated white gene activation.
Figure 5.
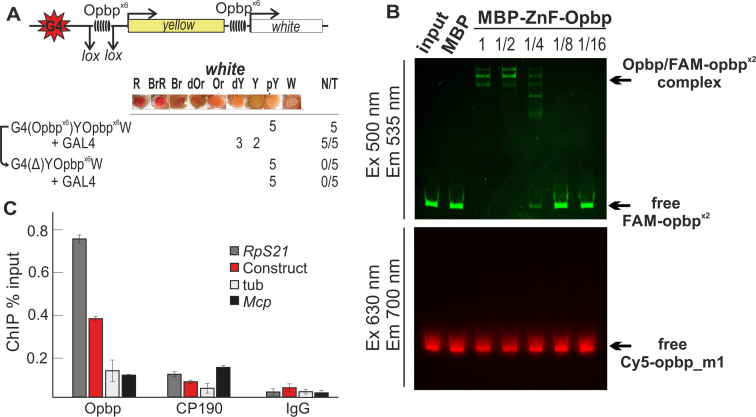
Opbp sites are sufficient to mediate long-distance chromatin interactions. (A) Schematic of GAL4/white transgenic model system used to examine functional interactions between two Opbp binding sites (upper). GAL4 binding sites (indicated as G4) are at a distance of 5 kb from the white promoter. The level of white eye colour was assessed in five independent transgenic lines for each construct, the content of which is indicated as G4(Opbpx6)Y Opbpx6 W, etc ‘+GAL4’ indicates that eye phenotypes were examined after the induction of GAL4 expression (lower). The symbol Δ indicates the deletion of corresponding element from transgenic line. In the N/T ratio, N = number of transgenic lines that acquired a new white phenotype after GAL4 induction or deletion of a DNA fragment flanked by loxP sites, T = total number of transgenic lines examined. Level of eye pigmentation was estimated on an arbitrary nine-grade scale, from wild-type expression (R—red), BrR—brown-red, Br—brown, dOR—dark orange, Or—orange, dY—dark yellow, Y—yellow, pY—pale yellow, W—white. (B) Electrophoretic mobility shift assay of recombinant Zinc finger domains of Opbp with a DNA fragment containing Opbp binding sites used in the transgenic construct. Zinc finger domains of Opbp fused with MBP (7.2 ng) or alone MBP (5.2 ng) was incubated with fluorescently labeled DNA fragments; Opbp binding sites labeled with FAM (wt) and a fragment with mutated Opbp site labeled with Cy5 (used as a negative control). Signals were detected for FAM-labeled fragment at the Ex 500 nm/Em 535 nm and for Cy5-labeled fragment at the Ex 630 nm/Em 700 nm. Specificity of interaction was demonstrated by incubation of DNA fragments with different amount of Opbp protein presented as a series of 2-fold dilutions. The specific Opbp bound region (indicated by arrow) decreased with decreasing Opbp protein, as seen by the decrease in FAM-signal. Note, no binding was observed on the mutated Opbp site (Cy5 signal). (C) Opbp binding sites are sufficient to recruit both Opbp and CP190 to a transgenic loci. Chromatin was isolated from embryos carrying the construct and treated with antibodies to Opbp and CP190. Nonspecific IgG was used as a negative control. ChIP recovery is presented as a percentage of input DNA. The tubulin-γ37C (tub) coding region (devoid of binding sites for the test proteins) was used as a negative control; Mcp and Rps21 as CP190-binding regions, Rps21 as Opbp-binding region were used as positive controls. Error bars indicate standard deviations of quadruplicate PCR measurements for three independent experiments.
To determine if Opbp can mediate such a loop, we placed six Opbp binding sites, flanked by loxP sites, near the GAL4 binding sites and an additional six Opbp sites close to the white promoter (Figure 5A, Supplementary Figure S11). The binding of Opbp to these sites was confirmed in vitro by EMSA using recombinant Opbp protein (Figure 5B), and importantly in vivo by ChIP-qPCR using chromatin isolated from pupae (Figure 5C). In all five transgenic lines tested, GAL4, under the control of the ubiquitous tubulin promoter, could activate white transcription in the presence of both clusters of Opbp motifs (Figure 5A). In contrast, deletion of the distal Opbp binding site cluster, by Cre-mediated recombination, abrogated the ability of GAL4 to activate the white gene (Figure 5A), demonstrating that Opbp binding regions are essential to support distance interaction, presumably by facilitating chromatin loop formation.
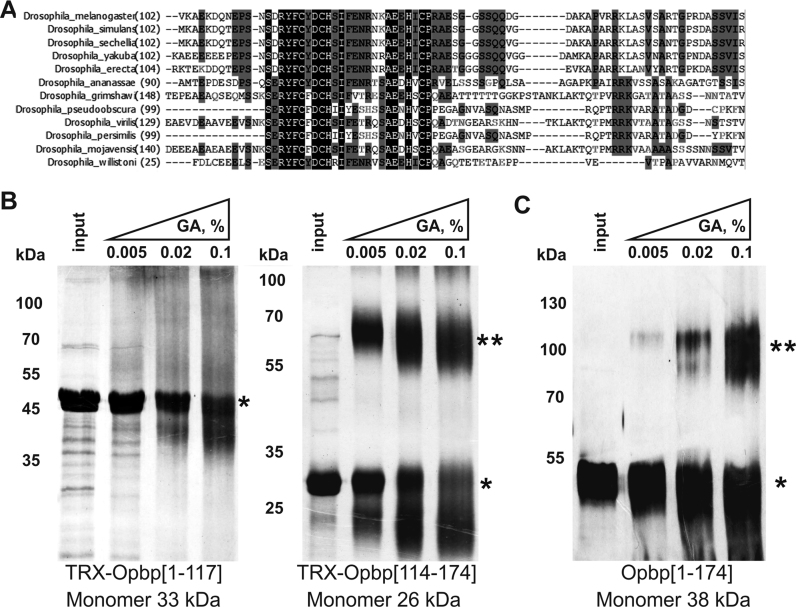
The requirement of both Opbp binding regions to support distant interactions suggests that homotypic Opbp protein–protein interactions may be required, and therefore that Opbp protein could contain a protein homo-dimerization domain. To examine this possibility, we tested parts of the Opbp protein for self-interaction in the yeast two-hybrid assay and found that a region at the N-terminus (within amino-acids 1–297) is able to self-associated (data not shown). The 102–174 aa region is more conserved among Drosophilids than the N-terminal 1–102 aa region (Figure 6A). We therefore split the N-terminus into two regions (amino acids [1–117] and [114–174]), and tested their ability to homodimer in a glutaraldehyde cross-linking assay that can assess specific oligomer formation in vitro (35) (Figure 6B). Only Opbp amino-acids from [114–174] gave a prominent cross-linked band at the approximate size expected for the dimer, while Opbp[1–117] was not able to homo-dimerize. We also confirmed homo-dimerization of the Opbp[1–174] fragment in the absence of Thioredoxin, which can affect the detection of dimerization in glutaraldehyde cross-linking assay (Figure 6C). Taken together these results indicate that Opbp can support distance interaction between its binding sites, presumably with help of dimerization domain.
Figure 6.
Opbp homo-dimerizes using a domain at its N-terminus. (A) Sequence alignment of dimerization domain of Opbp protein from different Drosophila species. (B, C) Cross-linking of Opbp N-terminal Thioredoxin-tagged domains (B) and Opbp N-terminal (1–174 aa) domain after deletion of TRX (C) using increasing concentrations of glutaraldehyde (GA). Protein was separated in 5–12% gradient SDS-PAGE gels and visualized with silver-staining. Monomer is labeled by *, homodimer – by **.
Opbp binding sites have enhancer-blocking activity
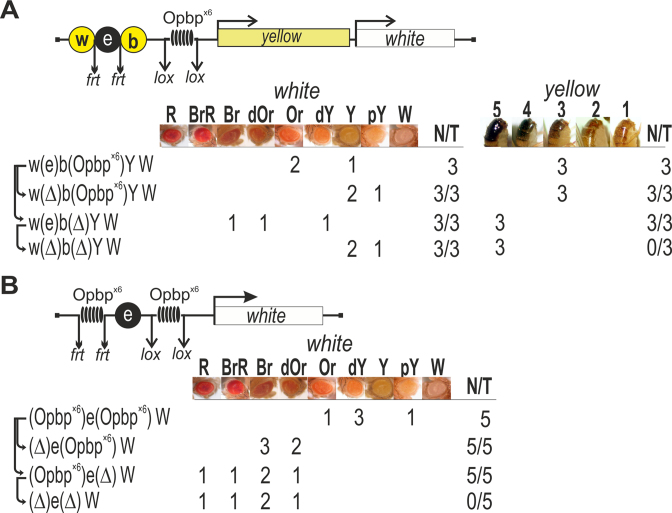
Another key property of reiterated binding sites for architectural proteins is their ability to block enhancer (or silencer) activity, when placed between the regulatory element and its target promoter. To test the ability of Opbp binding sites to block enhancer and silencer activity, we used an established transgenic reporter assay (Figure 7A, Supplementary Figure S11) based on the yellow gene, responsible for the dark pigmentation of the larval and adult cuticle and its derivatives and the white gene, responsible for eye pigmentation. Two upstream enhancers activate yellow expression in the body cuticle and wing blades (indicated by w and b in Figure 7A) (63). We inserted an eye enhancer for the white gene, flanked by frt sites, in between the wing and body enhancers at −1870 bp relative to the yellow transcription start site, while the white gene was downstream of the yellow gene. To test the ability of Opbp to act as an insulator protein, we inserted the same DNA fragment containing six consensus binding sites for Opbp (Opbp × 6), flanked by loxP sites, at −893 bp between the yellow promoter and the enhancers (Figure 7A). In all three independent transgenic lines, flies displayed significantly reduced pigmentation of body and wing (yellow expression) and eyes (white expression). Deletion of the eye enhancer by Flp-mediated recombination further reduced eye pigmentation suggesting that the eye enhancer was only partially blocked. While deletion of the Opbp binding sites by Cre-recombinase restored yellow and white expression. Thus, Opbp binding sites are able to partially block enhancer activity. These results are similar to those obtained with binding sites for the known insulator proteins Su(Hw), Pita, ZIPIC and Zw5 (24,60,61,64–66). Opbp binding sites can therefore function like other insulator elements to block enhancer activity.
Figure 7.
Opbp binding sites mediate enhancer-blocking activity. (A) Schematic of yellow/white transgenic model system used to examine enhancer blocking activity (upper). Downward arrows indicate target sites for Flp recombinase (frt) or Cre recombinase (loxP). The symbol Δ indicates the deletion of corresponding element from transgenic line. The relative locations of the body cuticle (b) and wing blades (w) enhancers of yellow, a white gene enhancer (e) in between and six Opbp consensus binding sites (Opbp×6) are indicated. The level of white eye colour was assessed in three independent transgenic lines for each construct, the content of which is indicated as w(e)b(Opbpx6)YW, etc. In the N/T ratio, N = number of transgenic lines that acquired a new white or yellow phenotype, T = total number of transgenic lines examined. Level of eye pigmentation was estimated on an arbitrary nine-grade scale, from wild-type expression (R—red), BrR—brown-red, Br—brown, dOR—dark orange, Or—orange, dY—dark yellow, Y—yellow, pY—pale yellow, W—white. The yellow pigmentation level in the abdominal cuticle (reflecting the activity of the body enhancer) was estimated on an arbitrary five-grade scale, with wild-type expression and the absence of expression assigned scores 5 and 1, respectively. (B) Functional interactions between Opbp binding sites enhance enhancer blocking enhancer activity. A cluster of Opbp binding sties (Opbp×6) placed upstream (flanked by frt sites) and downstream (flanked by loxP sites) of the eye enhancer (e). The level of eye colour was assessed in five independent transgenic lines for each construct, the content of which is indicated as (Opbpx6)e(Opbpx6)W etc. In the N/T ratio, N = number of transgenic lines that acquired a new white phenotype, T = total number of transgenic lines examined. Level of eye pigmentation was estimated on a nine-grade scale, from wild-type expression (R—red), BrR—brown-red, Br—brown, dOR—dark orange, Or—orange, dY—dark yellow, Y—yellow, pY—pale yellow, W—white.
Pairing between gypsy insulators flanking an enhancer was previously shown to strongly improve enhancer blocking activity (67). Given this, and the homotypic Opbp dimerization interaction observed, we assessed if pairing of Opbp sites improves enhancer blocking activity by engineering constructs where the eye enhancer was flanked by a pair of Opbp × 6 fragments (Figure 7B). The Opbp × 6 fragments were flanked by either frt or loxP sites. In all five independent transgenic lines carrying the construct with two sets of Opbp × 6 sites (Figure 7B), flies had eye pigmentation ranging from pale yellow to orange, indicating that the eye enhancer activity was strongly suppressed. Deletion of the upstream Opbp sites by Flp-recombinase partially restored eye pigmentation in all transgenic lines (Figure 7B). However, deletion of the Opbp sites located between the enhancer and promoter by Cre-recombinase completely restored activity of the white promoter. The subsequent deletion of the upstream Opbp sites by Flp-mediated recombination did not have any further affect on eye pigmentation. These results indicate that Opbp sites located upstream of the eye enhancer do not influence its activity when present alone. However, in the presence of a second bound region, interaction between the Opbp binding sites flanking the eye enhancer improves their insulator activity.
To examine possible barrier activity of Opbp × 6 sites we used the 661-bp PcG-responsive element (PRE) from the regulatory region of homeotic gene Ultrabithorax (Ubx), which is often used in anti-silencing assays (68–70). The PRE flanked by frt sites was inserted between the wing and body enhancers at −1870 bp relative to the yellow transcription start site (Supplementary Figure S12). The tested DNA fragments contained the Opbp sites, flanked by loxP sites, was inserted at −893 bp between the yellow promoter and the regulatory region including PRE and the enhancers (Supplementary Figure S12). Previously, we found that CTCF or Su(Hw) binding sites protected yellow expression from PRE-mediated silencing in the same transgenic assay (71,72). In contrast, the percentage of flies displaying yellow bristle pigmentation did not change after deletion of the Opbp sites in all fifteen transgenic lines (Supplementary Figure S12), while deletion of the PRE did restore yellow pigmentation. Thus, Opbp sites are not sufficient to protect from PRE-mediated silencing, at least in the context of yellow expression in this transgenic model system, while it is sufficient for enhancer blocking activity.
DISCUSSION
Here we identify a new architectural protein, Opbp, with a very restricted, yet essential role in genome regulation. Opbp contains a cluster of five DNA-binding C2H2 zinc-fingers (C2H2-ZF), which allows it to bind with very high specificity to a long consensus sequence. In contrast to other architectural proteins, which bind to thousands of sites throughout the genome, Opbp binds to fewer than 50 sites genome-wide. These sites are generally found close to divergent promoters, often in the vicinity of highly transcribed ribosomal protein genes. Although very unusual, this type of very restricted occupancy is not unprecedented. Chromatin immunoprecipitation using an antibody directed against the histone modification H2AQ105me identified a single site throughout the entire genome (73). In the case of Opbp, its very restrictive binding is essential for ribosomal protein genes expression, both in vivo and in vitro. Moreover, Opbp function is essential for Drosophila viability, speaking to its general importance in this fundamental biological process. Interestingly, the one site targeted by the histone modification H2AQ105me was a ribosomal locus, the 35S rDNA array (73), suggesting that perhaps ribosomal genes require a specific chromatin architecture to facilitate transcription.
Opbp both physically and genetically interacts with CP190, a protein known to interact at promoters with several architectural proteins containing arrays of C2H2 zinc-fingers (14,26,28,30,38). In these contexts, CP190 is thought to act as a cofactor, being recruited to DNA indirectly via its interactions with C2H2 partner proteins. In the case of Opbp, here we show that CP190 co-binds with Opbp to endogenous sites, as well as to Opbp transgenic sites in model constructs, indicating that this Opbp participates in targeting CP190 to chromatin. Moreover, our studies in S2 cells and in transgenic reporter assays demonstrate that CP190 binding depends on the presence of Opbp. Opbp interacts directly with CP190, as demonstrated by co-immunoprecipitation, yeast two-hybrid analysis and by pull-down experiments in vitro. Protein interaction requires the D domain of CP190 to bind the N-terminus of Opbp. The same domain and adjacent M domain of CP190 are involved in interaction with ZIPIC (14). The BTB domain of CP190 is required for interactions with other insulator proteins, like Pita and dCTCF (14,38). Therefore, at least three different domains of CP190 are involved in interactions with DNA-binding proteins. CP190 may therefore function to facilitate distance interactions (34,40) by using its domains to bridge genomic regions bound by different transcription factors located at a distance.
Opbp is recruited to highly specific sites in a restricted set of promoters, where it is required for their expression through the organization of open transcriptionally active chromatin. As mentioned above, in many cases Opbp binds near TSS of ribosomal protein genes. Interestingly, strongly expressed ribosomal promoters are usually located in close vicinity (200–300 bp) to promoters that drive relatively weak transcription in the opposite direction to the ribosomal protein genes. Inactivation of Opbp can affect transcription of the weak promoters in opposite ways. The striking example is the CG7130 gene, whose promoter is divergent with the promoter of the RpLP0 gene. Inactivation of Opbp led to opposite outcomes for CG7130 transcription in flies (two-fold increase) and S2 cells (2-fold decrease). Thus, it seems likely that Opbp only indirectly influences transcription of the weak divergent promoters that are located in the vicinity of promoters of ribosomal protein genes.
With the exception of Su(Hw), most known architectural proteins (dCTCF, ZIPIC, ZW5 and Pita) with C2H2-ZF domains are generally associated with active promoters (11–14). It seems therefore very likely that architectural proteins organize chromatin architecture at promoter regions to facilitate the recruitment of other transcription factors and to organize enhancer–promoter interactions (74). Inactivation of the Opbp binding site in the RplPO promoter only partially reduced transcription suggesting that additional architectural proteins are required for the architecture of this promoter. It seems likely that several architectural proteins can bind to regulatory elements cooperatively and facilitate binding of other transcription factors like CP190. In turn, CP190 participates in recruiting transcriptional complexes like NURF, dREAM and SAGA that are critical for promoter stimulation (9,25,39,44–46).
Our results demonstrate that Opbp displays all of the characteristics of an architectural/insulator protein. The multimerized binding sites for the architectural protein Pita, ZIPIC, Su(Hw) and CTCF form functional insulator-like elements that support distance interactions and display enhancer blocking activity (7,14,60,61). The binding sites for Opbp have enhancer blocking activity in established transgenic model systems that have been used for other well characterized insulator proteins. We also showed that interaction between two Opbp binding regions can improve enhancer blocking activity for the strong eye enhancer located between them. Previously, we found that enhancer blocking activity (using the eye enhancer) by the best studied gypsy insulator consisting of twelve Su(Hw) binding sites is strongly improved by a chromatin loop that physically interferes with the ability of the protein complexes bound to the eye enhancer and promoter to interact with each other (67). Here, we observed a similar effect for the Opbp binding sites: enhancer blocking was improved when two sets of Opbp binding sites flanked the eye enhancer (Figure 7B). Thus, interaction between Opbp proteins can form a chromatin loop domain that blocks an enhancer–promoter interaction. As previously shown for the C2H2-ZF architectural proteins (61), Opbp binding sites can support ‘long’ distance interactions in transgenic model system. The distance interactions might be organized by a dimerization domain located at the N-terminus of Opbp and by CP190, that according to a recent model is a key organizer of distance interactions in Drosophila (34,40).
In contrast to dCTCF, Su(Hw), Pita and ZIPIC (14,71,72,75), the Opbp binding sites did not protect reporter expression from PRE-mediated silencing. The highly conserved ENY2 protein is required for this barrier activity, by interaction with the C2H2-ZF domains of Su(Hw) and dCTCF (71,72). Interestingly, Opbp does not interact with ENY2 (O.M., unpublished results), providing a plausible explanation for why it does not block PRE-mediated silencing activity. In addition, as ribosomal protein genes are ubiquitously and highly expressed, it is also conceivable that Polycomb-mediated repression is never required at loci where Opbp functions.
In vitro genome-wide studies identified enhancer-core-promoter specificity that separates developmental and housekeeping genes (43). The housekeeping (hk) promoters extensively interact with hk enhancers and each other (18). These findings suggest that special architectural proteins are involved in the organization of hk promoters that may facilitate the recruitment of hk-specific transcription factors and the organization of specific enhancer–promoter and promoter-promoter chromatin topologies. We speculate that Opbp belongs to this class of architectural proteins to facilitate specific distance interactions between hk regulatory elements. Further study is required to elucidate the mechanisms of how Opbp fulfills architectural functions in the organization of hk promoters.
DATA AVAILABILITY
All ChIP data are available at ArrayExpress (http://www.ebi.ac.uk/arrayexpress) under accession numbers E-MTAB-4146 (Opbp). The high-confidence transcription factor (TF) binding information is provided in Supplementary Table S2 and also on the Furlong lab web page at http://furlonglab.embl.de/
Supplementary Material
ACKNOWLEDGEMENTS
We thank Farhod Hasanov and Aleksander Parshikov for fly injection. This work was supported by the Russian Science Foundation; part of the work devoted to delete the opbp gene was supported by the RFBR. The work was technically supported by the EMBL Genomics Core facility. This study was performed using the equipment of the IGB RAS facilities supported by the Ministry of Science and Education of the Russian Federation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Russian Science Foundation [14-24-00166 to P.G.]; RFBR [16-04-01531 to P.G.]. Funding for open access charge: Russian Science Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1. Raab J.R., Kamakaka R.T.. Insulators and promoters: closer than we think. Nat. Rev. Genet. 2010; 11:439–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ghirlando R., Giles K., Gowher H., Xiao T., Xu Z., Yao H., Felsenfeld G.. Chromatin domains, insulators, and the regulation of gene expression. Biochim. Biophys. Acta. 2012; 1819:644–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Herold M., Bartkuhn M., Renkawitz R.. CTCF: insights into insulator function during development. Development. 2012; 139:1045–1057. [DOI] [PubMed] [Google Scholar]
- 4. Kyrchanova O., Georgiev P.. Chromatin insulators and long-distance interactions in Drosophila. FEBS Lett. 2014; 588:8–14. [DOI] [PubMed] [Google Scholar]
- 5. Matzat L.H., Lei E.P.. Surviving an identity crisis: A revised view of chromatin insulators in the genomics era. Biochim. Biophys. Acta. 2013; 1839:203–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chetverina D., Aoki T., Erokhin M., Georgiev P., Schedl P.. Making connections: Insulators organize eukaryotic chromosomes into independent cis-regulatory networks. BioEssays. 2014; 36:163–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Maksimenko O., Georgiev P.. Mechanisms and proteins involved in long-distance interactions. Front. Genet. 2014; 5:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Matharu N.K., Ahanger S.H.. Chromatin insulators and topological domains: adding new dimensions to 3D genome architecture. Genes. 2015; 6:790–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ali T., Renkawitz R., Bartkuhn M.. Insulators and domains of gene expression. Curr. Opin. Genet. Dev. 2016; 37:17–26. [DOI] [PubMed] [Google Scholar]
- 10. Fedotova A.A., Bonchuk A.N., Mogila V.A., Georgiev P.G.. C2H2 zinc finger proteins: the largest but poorly explored family of higher eukaryotic transcription factors. Acta Naturae. 2017; 9:47–58. [PMC free article] [PubMed] [Google Scholar]
- 11. Negre N., Brown C.D., Shah P.K., Kheradpour P., Morrison C.A., Henikoff J.G., Feng X., Ahmad K., Russell S., White R.A. et al. . A comprehensive map of insulator elements for the Drosophila genome. PLoS Genet. 2010; 6:e1000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Negre N., Brown C.D., Ma L., Bristow C.A., Miller S.W., Wagner U., Kheradpour P., Eaton M.L., Loriaux P., Sealfon R. et al. . A cis-regulatory map of the Drosophila genome. Nature. 2011; 471:527–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schwartz Y.B., Linder-Basso D., Kharchenko P.V., Tolstorukov M.Y., Kim M., Li H.B., Gorchakov A.A., Minoda A., Shanower G., Alekseyenko A.A. et al. . Nature and function of insulator protein binding sites in the Drosophila genome. Genome Res. 2012; 22:2188–2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Maksimenko O., Bartkuhn M., Stakhov V., Herold M., Zolotarev N., Jox T., Buxa M.K., Kirsch R., Bonchuk A., Fedotova A. et al. . Two new insulator proteins, Pita and ZIPIC, target CP190 to chromatin. Genome Res. 2015; 25:89–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hnisz D., Day D.S., Young R.A.. Insulated neighborhoods: structural and functional units of mammalian gene control. Cell. 2016; 167:1188–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Merkenschlager M., Nora E.P.. CTCF and cohesin in genome folding and transcriptional gene regulation. Annu. Rev. Genomics Hum. Genet. 2016; 17:17–43. [DOI] [PubMed] [Google Scholar]
- 17. Zolotarev N., Fedotova A., Kyrchanova O., Bonchuk A., Penin A.A., Lando A.S., Eliseeva I.A., Kulakovskiy I.V., Maksimenko O., Georgiev P.. Architectural proteins Pita, Zw5,and ZIPIC contain homodimerization domain and support specific long-range interactions in Drosophila. Nucleic Acids Res. 2016; 44:7228–7241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cubenas-Potts C., Rowley M.J., Lyu X., Li G., Lei E.P., Corces V.G.. Different enhancer classes in Drosophila bind distinct architectural proteins and mediate unique chromatin interactions and 3D architecture. Nucleic Acids Res. 2017; 45:1714–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Merkenschlager M., Odom D.T.. CTCF and cohesin: linking gene regulatory elements with their targets. Cell. 2013; 152:1285–1297. [DOI] [PubMed] [Google Scholar]
- 20. Nakahashi H., Kwon K.R., Resch W., Vian L., Dose M., Stavreva D., Hakim O., Pruett N., Nelson S., Yamane A. et al. . A genome-wide map of CTCF multivalency redefines the CTCF code. Cell Rep. 2013; 3:1678–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chung H.R., Schafer U., Jackle H., Bohm S.. Genomic expansion and clustering of ZAD-containing C2H2 zinc-finger genes in Drosophila. EMBO Rep. 2002; 3:1158–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chung H.R., Lohr U., Jackle H.. Lineage-specific expansion of the zinc finger associated domain ZAD. Mol. Biol. Evol. 2007; 24:1934–1943. [DOI] [PubMed] [Google Scholar]
- 23. Jauch R., Bourenkov G.P., Chung H.R., Urlaub H., Reidt U., Jackle H., Wahl M.C.. The zinc finger-associated domain of the Drosophila transcription factor grauzone is a novel zinc-coordinating protein–protein interaction module. Structure. 2003; 11:1393–1402. [DOI] [PubMed] [Google Scholar]
- 24. Gaszner M., Vazquez J., Schedl P.. The Zw5 protein, a component of the scs chromatin domain boundary, is able to block enhancer–promoter interaction. Genes Dev. 1999; 13:2098–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bartkuhn M., Straub T., Herold M., Herrmann M., Rathke C., Saumweber H., Gilfillan G.D., Becker P.B., Renkawitz R.. Active promoters and insulators are marked by the centrosomal protein 190. EMBO J. 2009; 28:877–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gerasimova T.I., Lei E.P., Bushey A.M., Corces V.G.. Coordinated control of dCTCF and gypsy chromatin insulators in Drosophila. Mol. Cell. 2007; 28:761–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liang J., Lacroix L., Gamot A., Cuddapah S., Queille S., Lhoumaud P., Lepetit P., Martin P.G., Vogelmann J., Court F. et al. . Chromatin immunoprecipitation indirect peaks highlight long-range interactions of insulator proteins and Pol II pausing. Mol. Cell. 2014; 53:672–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mohan M., Bartkuhn M., Herold M., Philippen A., Heinl N., Bardenhagen I., Leers J., White R.A., Renkawitz-Pohl R., Saumweber H. et al. . The Drosophila insulator proteins CTCF and CP190 link enhancer blocking to body patterning. EMBO J. 2007; 26:4203–4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Oliver D., Sheehan B., South H., Akbari O., Pai C.Y.. The chromosomal association/dissociation of the chromatin insulator protein Cp190 of Drosophila melanogaster is mediated by the BTB/POZ domain and two acidic regions. BMC Cell Biol. 2010; 11:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pai C.Y., Lei E.P., Ghosh D., Corces V.G.. The centrosomal protein CP190 is a component of the gypsy chromatin insulator. Mol. Cell. 2004; 16:737–748. [DOI] [PubMed] [Google Scholar]
- 31. Bushey A.M., Ramos E., Corces V.G.. Three subclasses of a Drosophila insulator show distinct and cell type-specific genomic distributions. Genes Dev. 2009; 23:1338–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dai Q., Ren A., Westholm J.O., Duan H., Patel D.J., Lai E.C.. Common and distinct DNA-binding and regulatory activities of the BEN-solo transcription factor family. Genes Dev. 2015; 29:48–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cuartero S., Fresan U., Reina O., Planet E., Espinas M.L.. Ibf1 and Ibf2 are novel CP190-interacting proteins required for insulator function. EMBO J. 2014; 33:637–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ahanger S.H., Shouche Y.S., Mishra R.K.. Functional sub-division of the Drosophila genome via chromatin looping: the emerging importance of CP190. Nucleus. 2013; 4:115–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bonchuk A., Denisov S., Georgiev P., Maksimenko O.. Drosophila BTB/POZ domains of “ttk group" can form multimers and selectively interact with each other. J. Mol. Biol. 2011; 412:423–436. [DOI] [PubMed] [Google Scholar]
- 36. Plevock K.M., Galletta B.J., Slep K.C., Rusan N.M.. Newly characterized region of CP190 associates with microtubules and mediates proper spindle morphology in Drosophila stem cells. PLoS One. 2015; 10:e0144174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Butcher R.D., Chodagam S., Basto R., Wakefield J.G., Henderson D.S., Raff J.W., Whitfield W.G.. The Drosophila centrosome-associated protein CP190 is essential for viability but not for cell division. J. Cell Sci. 2004; 117:1191–1199. [DOI] [PubMed] [Google Scholar]
- 38. Bonchuk A., Maksimenko O., Kyrchanova O., Ivlieva T., Mogila V., Deshpande G., Wolle D., Schedl P., Georgiev P.. Functional role of dimerization and CP190 interacting domains of CTCF protein in Drosophila melanogaster. BMC Biol. 2015; 13:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bohla D., Herold M., Panzer I., Buxa M.K., Ali T., Demmers J., Kruger M., Scharfe M., Jarek M., Bartkuhn M. et al. . A functional insulator screen identifies NURF and dREAM components to be required for enhancer-blocking. PLoS One. 2014; 9:e107765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vogelmann J., Le Gall A., Dejardin S., Allemand F., Gamot A., Labesse G., Cuvier O., Negre N., Cohen-Gonsaud M., Margeat E. et al. . Chromatin insulator factors involved in long-range DNA interactions and their role in the folding of the Drosophila genome. PLoS Genet. 2014; 10:e1004544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jiang N., Emberly E., Cuvier O., Hart C.M.. Genome-wide mapping of boundary element-associated factor (BEAF) binding sites in Drosophila melanogaster links BEAF to transcription. Mol. Cell. Biol. 2009; 29:3556–3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Emberly E., Blattes R., Schuettengruber B., Hennion M., Jiang N., Hart C.M., Kas E., Cuvier O.. BEAF regulates cell-cycle genes through the controlled deposition of H3K9 methylation marks into its conserved dual-core binding sites. PLoS Biol. 2008; 6:2896–2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zabidi M.A., Arnold C.D., Schernhuber K., Pagani M., Rath M., Frank O., Stark A.. Enhancer-core-promoter specificity separates developmental and housekeeping gene regulation. Nature. 2015; 518:556–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lhoumaud P., Hennion M., Gamot A., Cuddapah S., Queille S., Liang J., Micas G., Morillon P., Urbach S., Bouchez O. et al. . Insulators recruit histone methyltransferase dMes4 to regulate chromatin of flanking genes. EMBO J. 2014; 33:1599–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kwon S.Y., Grisan V., Jang B., Herbert J., Badenhorst P.. Genome-wide mapping targets of the metazoan chromatin remodeling factor NURF reveals nucleosome remodeling at enhancers, core promoters and gene insulators. PLoS Genet. 2016; 12:e1005969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ali T., Kruger M., Bhuju S., Jarek M., Bartkuhn M., Renkawitz R.. Chromatin binding of Gcn5 in Drosophila is largely mediated by CP190. Nucleic Acids Res. 2017; 45:2384–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kenyon K.L., Li D.J., Clouser C., Tran S., Pignoni F.. Fly SIX-type homeodomain proteins Sine oculis and Optix partner with different cofactors during eye development. Dev. Dyn. 2005; 234:497–504. [DOI] [PubMed] [Google Scholar]
- 48. Gratz S.J., Ukken F.P., Rubinstein C.D., Thiede G., Donohue L.K., Cummings A.M., O’Connor-Giles K.M.. Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics. 2014; 196:961–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Port F., Chen H.M., Lee T., Bullock S.L.. Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:E2967–E2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bischof J., Maeda R.K., Hediger M., Karch F., Basler K.. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:3312–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang X., Koolhaas W.H., Schnorrer F.. A versatile two-step CRISPR- and RMCE-based strategy for efficient genome engineering in Drosophila. G3 (Bethesda). 2014; 4:2409–2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Venken K.J., Schulze K.L., Haelterman N.A., Pan H., He Y., Evans-Holm M., Carlson J.W., Levis R.W., Spradling A.C., Hoskins R.A. et al. . MiMIC: a highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nat. Methods. 2011; 8:737–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Marygold S.J., Roote J., Reuter G., Lambertsson A., Ashburner M., Millburn G.H., Harrison P.M., Yu Z., Kenmochi N., Kaufman T.C. et al. . The ribosomal protein genes and Minute loci of Drosophila melanogaster. Genome Biol. 2007; 8:R216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lambertsson A. The minute genes in Drosophila and their molecular functions. Adv. Genet. 1998; 38:69–134. [DOI] [PubMed] [Google Scholar]
- 55. Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S.. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009; 37:W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hertz G.Z., Stormo G.D.. Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics. 1999; 15:563–577. [DOI] [PubMed] [Google Scholar]
- 57. Thomas S., Li X.Y., Sabo P.J., Sandstrom R., Thurman R.E., Canfield T.K., Giste E., Fisher W., Hammonds A., Celniker S.E. et al. . Dynamic reprogramming of chromatin accessibility during Drosophila embryo development. Genome Biol. 2011; 12:R43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Huang da W., Sherman B.T., Lempicki R.A.. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009; 4:44–57. [DOI] [PubMed] [Google Scholar]
- 59. Bowman S.K., Deaton A.M., Domingues H., Wang P.I., Sadreyev R.I., Kingston R.E., Bender W.. H3K27 modifications define segmental regulatory domains in the Drosophila bithorax complex. eLife. 2014; 3:e02833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Scott K.C., Taubman A.D., Geyer P.K.. Enhancer blocking by the Drosophila gypsy insulator depends upon insulator anatomy and enhancer strength. Genetics. 1999; 153:787–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kyrchanova O., Chetverina D., Maksimenko O., Kullyev A., Georgiev P.. Orientation-dependent interaction between Drosophila insulators is a property of this class of regulatory elements. Nucleic Acids Res. 2008; 36:7019–7028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kyrchanova O., Toshchakov S., Podstreshnaya Y., Parshikov A., Georgiev P.. Functional interaction between the Fab-7 and Fab-8 boundaries and the upstream promoter region in the Drosophila Abd-B gene. Mol. Cell. Biol. 2008; 28:4188–4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Geyer P.K., Corces V.G.. Separate regulatory elements are responsible for the complex pattern of tissue-specific and developmental transcription of the yellow locus in Drosophila melanogaster. Genes Dev. 1987; 1:996–1004. [DOI] [PubMed] [Google Scholar]
- 64. Golovnin A., Biryukova I., Romanova O., Silicheva M., Parshikov A., Savitskaya E., Pirrotta V., Georgiev P.. An endogenous Su(Hw) insulator separates the yellow gene from the Achaete-scute gene complex in Drosophila. Development. 2003; 130:3249–3258. [DOI] [PubMed] [Google Scholar]
- 65. Kyrchanova O., Leman D., Parshikov A., Fedotova A., Studitsky V., Maksimenko O., Georgiev P.. New properties of Drosophila scs and scs' insulators. PLoS One. 2013; 8:e62690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Tikhonov M., Gasanov N.B., Georgiev P., Maksimenko O.. A model system in S2 cells to test the functional activities of Drosophila insulators. Acta Naturae. 2015; 7:97–106. [PMC free article] [PubMed] [Google Scholar]
- 67. Kyrchanova O., Maksimenko O., Stakhov V., Ivlieva T., Parshikov A., Studitsky V.M., Georgiev P.. Effective blocking of the white enhancer requires cooperation between two main mechanisms suggested for the insulator function. PLoS Genet. 2013; 9:e1003606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Comet I., Savitskaya E., Schuettengruber B., Negre N., Lavrov S., Parshikov A., Juge F., Gracheva E., Georgiev P., Cavalli G.. PRE-mediated bypass of two Su(Hw) insulators targets PcG proteins to a downstream promoter. Dev. Cell. 2006; 11:117–124. [DOI] [PubMed] [Google Scholar]
- 69. Mallin D.R., Myung J.S., Patton J.S., Geyer P.K.. Polycomb group repression is blocked by the Drosophila suppressor of Hairy-wing [su(Hw)] insulator. Genetics. 1998; 148:331–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Sigrist C.J., Pirrotta V.. Chromatin insulator elements block the silencing of a target gene by the Drosophila polycomb response element (PRE) but allow trans interactions between PREs on different chromosomes. Genetics. 1997; 147:209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Maksimenko O., Kyrchanova O., Bonchuk A., Stakhov V., Parshikov A., Georgiev P.. Highly conserved ENY2/Sus1 protein binds to Drosophila CTCF and is required for barrier activity. Epigenetics. 2014; 9:1261–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kurshakova M., Maksimenko O., Golovnin A., Pulina M., Georgieva S., Georgiev P., Krasnov A.. Evolutionarily conserved E(y)2/Sus1 protein is essential for the barrier activity of Su(Hw)-dependent insulators in Drosophila. Mol. Cell. 2007; 27:332–338. [DOI] [PubMed] [Google Scholar]
- 73. Tessarz P., Kouzarides T.. Histone core modifications regulating nucleosome structure and dynamics. Nat. Rev. Mol. Cell Biol. 2014; 15:703–708. [DOI] [PubMed] [Google Scholar]
- 74. Ghavi-Helm Y., Klein F.A., Pakozdi T., Ciglar L., Noordermeer D., Huber W., Furlong E.E.. Enhancer loops appear stable during development and are associated with paused polymerase. Nature. 2014; 512:96–100. [DOI] [PubMed] [Google Scholar]
- 75. Kuhn-Parnell E.J., Helou C., Marion D.J., Gilmore B.L., Parnell T.J., Wold M.S., Geyer P.K.. Investigation of the properties of non-gypsy suppressor of hairy-wing-binding sites. Genetics. 2008; 179:1263–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Graveley B.R., Brooks A.N., Carlson J.W., Duff M.O., Landolin J.M., Yang L., Artieri C.G., van Baren M.J., Boley N., Booth B.W. et al. . The developmental transcriptome of Drosophila melanogaster. Nature. 2011; 471:473–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All ChIP data are available at ArrayExpress (http://www.ebi.ac.uk/arrayexpress) under accession numbers E-MTAB-4146 (Opbp). The high-confidence transcription factor (TF) binding information is provided in Supplementary Table S2 and also on the Furlong lab web page at http://furlonglab.embl.de/