Abstract
Transposable elements (TEs) compose about 40% of the murine genome. Retrotransposition of active TEs such as LINE-1 (L1) tremendously impacts genetic diversification and genome stability. Therefore, transcription and transposition activities of retrotransposons are tightly controlled. Here, we show that the Krüppel-like zinc finger protein Zfp281 directly binds and suppresses a subset of retrotransposons, including the active young L1 repeat elements, in mouse embryonic stem (ES) cells. In addition, we find that Zfp281-regulated L1s are highly enriched for 5-hydroxymethylcytosine (5hmC) and H3K4me3. The COMPASS-like H3K4 methyltransferase Mll2 is the major H3K4me3 methylase at the Zfp281-regulated L1s and required for their proper expression. Our studies also reveal that Zfp281 functions partially through recruiting the L1 regulators DNA hydroxymethylase Tet1 and Sin3A, and restricting Mll2 at these active L1s, leading to their balanced expression. In summary, our data indicate an instrumental role of Zfp281 in suppressing the young active L1s in mouse ES cells.
INTRODUCTION
Transposable elements, also known as jumping genes, comprise ∼40% of mammalian genome (1,2). Transposable elements pervading mammalian genomes mostly originate from retrotransposons, including long interspersed nuclear elements (LINEs), short interspersed nuclear elements (SINEs), and endogenous retroviruses (ERVs) (2,3). Retrotransposons shape host genome landscape and evolution through introducing, deleting or modifying cis-regulatory elements or genes (4,5). However, they can also induce detrimental genome instability and numerous human diseases (3,6).
Host cells have marshaled a variety of defense mechanisms, including nucleic acid editing, RNA-induced silencing, and epigenetic transcriptional repression, to constrain specific subtypes of retrotransposons (7–11). DNA methylation and H3K9 methylation have been considered as the primary host defense against transposable elements (12). DNMT1 and DNMT3A/B are required for the methylation of intracisternal A particle (IAP) retrotransposons in mouse embryos and germ cells (13,14). It has also been reported that the epigenetic regulator KAP1/TRIM28 and its associated histone modifiers ESET, G9a and SUV39H are essential in suppressing distinct subsets of retrotransposons in an H3K9 methylation-dependent manner in mouse embryonic stem (ES) cells (15–18). Members in the fast growing Krüppel associated box (KRAB)-containing zinc finger (KZNF) family can specifically recruit H3K9 methylation machineries to silence subclasses of retrotransposons through interacting with KAP1 (19,20). Nevertheless, the mutations in retrotransposon can lead to evasion from the KZNF mediated repression, providing an interesting genetics model in maintaining an evolutionary balance between retrotransposon and host genome (3,19).
Long interspersed nuclear element-1s (LINE-1s or L1s) are the autonomous retrotransposons in mammals. Hyperactivation of L1s has been linked to tumorigenesis (21,22). There are >100 000 L1 fragments and ∼3000 intact L1s in the mouse genome belonging to different families according to their retrotranposition activities (23). A type (L1Md_A) and F type (L1Md_T and L1Md_F) promoters are carrying retrotranposition activities, at present (24,25). Suv39H-mediated H3K9 methylation has been shown to suppress A type of L1 in mouse ES cells (17). Recent work indicated that 5-hydroxymethylcytosine (5hmC) is highly enriched in F types of young L1 repeat elements (26). Further studies revealed that the co-repressor complex Sin3A could function together with the DNA hydroxymethylases Tet in suppressing the expression of the young L1s (27). However, a full understanding about the regulation of these young L1s is still lacking.
The Krüppel-like zinc finger transcription factor Zfp281 was first identified as a transcriptional repressor, with a preference for binding to the GC-rich DNA sequence (28). Subsequent studies implicated that Zfp281 can either function as a transcriptional repressor or an activator in a context-dependent manner (29–31). Here we show an important role of the Zfp281 in restricting the expression of F type of young L1s in mouse ES cells. The Zfp281 regulated young L1s hijack the H3K4 methyltransferase Mll2 for their proper expression in an H3K4me3-dependent manner. Our further analyses suggest that a functional interplay between Zfp281-Tet1-Sin3A and Mll2 plays critical roles in the regulation of the subsets of L1 repeat elements.
MATERIALS AND METHODS
Expression plasmids and cell lines
Mouse V6.5 embryonic stem cells were cultured on irradiated mouse embryonic fibroblast (iMEF) feeder layers in 0.1% gelatin-coated tissue culture flask. Cells were grown in DMEM (D6546, Sigma) supplemented with 15% ES-certified fetal bovine serum (Hyclone), 2 mM l-glutamine, 0.1 mM nonessential amino acids, 0.1 mM β-mercaptoethanol, and recombinant LIF (Millipore). For ChIP and RNA analyses, cells were grown for one passage off feeders on tissue culture plates for 30 min.
Lentivirus based RNAi
Mouse Zfp281 constructs were discribed previously (32). The Non-targeting shRNA construct (SHC002) was purchased from Sigma. Lentiviral particle preparation and infection were performed as previously described (33). Briefly, ∼70% confluent 293T cells in 150 mm tissue culture plate were co-transfected with 8μg of the shRNA construct or Non-targeting control shRNA, 6 μg of PsPAX2 packaging plasmids and 2 μg of pMD2.G envelope plasmids using X-tremeGENE 9 (Roche). The media was replaced with fresh DMEM supplemented with 10% FBS after 16 h of transfection. The lentiviral supernatants were collected 48 and 72 hours after the transfection, filtered through 0.45 μm filters and concentrated at 18 000 rpm for 2 h. The V6.5 ES cells were infected with concentrated lentiviral particles with polybrene (Sigma) at the concentration of 8 μg/ml. Twenty four hours after infection, the ES cells were subjected to selection with 2 ug/ml of puromycin for an additional 48 hours.
Antibodies
Antibody against Tet1 (GT1462) was purchased from GeteTex and Sin3A (ab3479) from Abcam. H3K4me3 antibody and Mll2 antibody were provided by Shilatifard laboratory. Antibodies against Zfp281 were generated in our laboratory and described previously (32).
ChIP assay
A total of 5 × 107 cells were used per ChIP assay according to the previously described protocol (34). Briefly, cells were cross-linked with 1% paraformaldehyde for 10 min at room temperature; cross-linking was quenched by glycine. Fixed chromatin was sonicated and immunoprecipitated with specific antibodies.
RNA-seq alignment inclusive of none-unique mapped reads
To include as many non-uniquely mapped reads as possible, RNA-seq raw reads were firstly aligned to mouse genome (mm10) by STAR with parameters ‘–alignEndsType EndToEnd –winAnchorMultimapmax 2000 –outFilterMultimapNmax 2000 –outSAMprimaryFlag AllBestScore –outSAMmultNmax 1’. This setting was used to ensure all alignments having the best reported scores. For reads with multiple alignments, only one was randomly picked as the final alignment by customized script.
ChIP-seq alignment inclusive of none-unique mapped reads
Raw reads from ChIP-seq experiment were aligned to mouse genome (mm10) by STAR (2.5.1) with parameter ‘–alignEndsType EndToEnd –winAnchorMultimapmax 2000 –outFilterMultimapNmax 2000 –outSAMprimaryFlag AllBestScore –outSAMmultNmax 1 –alignEndsType EndToEnd –alignIntronMax 1’. With this setting, for each read, if one more equivalent best alignment is found, only one of the hits will be reported randomly. Consequently, none-uniquely mapped reads will be evenly distributed over highly similar repeat elements across genome.
Align RNA-seq and ChIP-seq reads to consensus repeat sequence
The annotation and fasta sequences for consensus repeat sequences were downloaded from Repbase (version 20.01) (35). Reads in fastq format were aligned to consensus using BOWTIE2 with parameter ‘–fast -k 1’.
Peak calling
For ChIP-seq, MACS2 (36) was used to identify peaks with default parameters but including multiple hit reads. The RepeatMasker track in UCSC genome browser was used to identify peaks overlapped with repeat elements. To categorize peaks into weak and strong peaks, we ranked the peaks based on Q-value (minimum FDR) from MACS2. By definition of FDR, for given Q-value 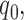 the number of false positive peaks N can be estimated as:
the number of false positive peaks N can be estimated as: 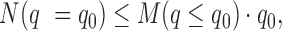 where
where 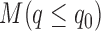 is total number of peaks called with Q-value less than
is total number of peaks called with Q-value less than 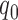 . The lower bound of Qvalue
. The lower bound of Qvalue 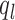 for weak peaks was chosen such as:
for weak peaks was chosen such as: 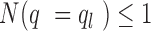 . We next determined the upper or lower bound of Q-value for weak or strong peaks. As illustrated in Supplementary Figure S3B, we first normalized the –log10(Q-value) by the maximum –log10(Q-value) for all of peaks. Then the percentage of peaks given a Q-value was calculated as complementary cumulative distribution. The upper bound of Q-value for weak peaks was chosen as the point where the slope is –1.
. We next determined the upper or lower bound of Q-value for weak or strong peaks. As illustrated in Supplementary Figure S3B, we first normalized the –log10(Q-value) by the maximum –log10(Q-value) for all of peaks. Then the percentage of peaks given a Q-value was calculated as complementary cumulative distribution. The upper bound of Q-value for weak peaks was chosen as the point where the slope is –1.
Phylogenetic analysis of LINE-1 family
According to the annotation of UCSC RepeatMasker Viz tracks (mm10), 21,567 5′ end sequences (including 5′UTR and first 532bp of ORF1) of L1 elements were extracted from genome. The pairwise distance matrix of L1 elements were calculated by CALC_DISTMX in USEARCH package (37). The Neighbour Join tree was then constructed using QUICKTREE (38). To identify the meaningful clusters in large L1 tree, we used an algorithm based on median pairwise patristic distance (39) to cluster L1 tree into 22 clusters, of which the median pairwise patristic distance between the members of its clusters is below a threshold.
ChIP-seq and RNA-seq data sets used in this study are listed below:
| Dataset | GEO | Reference |
|---|---|---|
| Zfp281 KD RNA-seq | GSE77115 | Wang Y et al. Nucleic Acids Res 2017 |
| Zfp281 ChIP-seq | GSE77115 | Wang Y et al. Nucleic Acids Res 2017 |
| Pol II ChIP-seq in Zfp281 KD mESC | GSE77115 | Wang Y et al. Nucleic Acids Res 2017 |
| H3K9me3 ChIP-seq | GSE57092 | Bulut-Karslioglu A et al. Mol Cell 2014 (17) |
| RNA-seq in Mll2 KD and control mESC | GSE48172 | Hu D et al. Nat Struct Mol Biol 2013 (46) |
| Suv39h ChIP-seq | GSE57092 | Bulut-Karslioglu A et al. Mol. Cell. 2014 |
| Tet1 and Sin3A ChIP-seq | GSE24841 | Williams K et al. Nature 2011 |
| H3K9me2 ChIP-seq | GSE54412 | Liu N et al. Genes Dev 2015 (53) |
| Mll2 and H3K4me3 ChIP-seq in Mll2 KD and control mESC | GSE48172,GSE78708 | Hu D et al. Nat Struct Mol Biol 2013 (46); Hu D et al. Mol. Cell. 2017 (44) |
RESULTS
Zfp281 suppresses a subset of retrotransposons in mouse ES cells
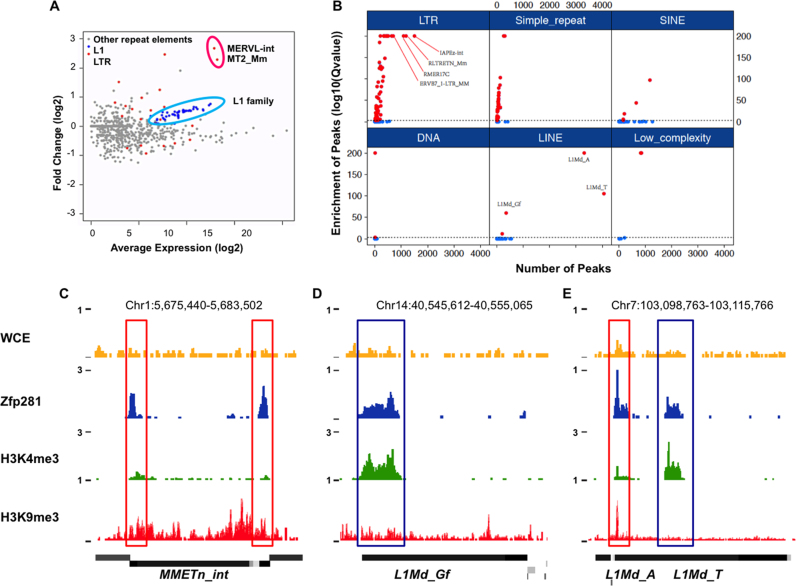
We have previously shown that Zfp281 can recruit the scaffold protein of Super Elongation Complex-Like 3 (SEC-L3), Aff3, to enhancer regions and regulate target gene expression (32). From RNA-seq analyses in Zfp281-depleted mouse ES cells, we noticed that a significant number of genes are upregulated after Zfp281 depletion. Among them, the Zscan4 family genes are ranked on the top of the list (Supplementary Figure S1). It was reported that the Zscan4 cluster genes are flanked and controlled by the endogenous retrovirus MERVL (40). To further explore whether Zfp281 also functions in silencing repeat elements, including MERVL, in mouse ES cells, we analyzed the expression levels of repeat elements following the Zfp281 depletion by mapping RNA-seq reads to the consensus of different repeat elements. MA-plot analysis indicated that the expression of MERVL, MERVL-derived LTR MT2_Mm, and a subset of the active L1 repeat families are significantly upregulated following Zfp281 knockdown (Figure 1A).
Figure 1.
Zfp281 suppresses a subset of retrotransposons in mouse ES cells. (A) Fold change (log2) of retrotransposon expression (RPKM) after Zfp281 depletion in mouse ES cells. The consensus sequences of retrotransposons (extracted from RepBase) (54) were used to calculate their expression change. Other repeat elements, L1 and LTR sequences are color-coded. (B) Zfp281 occupies L1 and LTR repeat elements. For each repeat family, numbers of Zfp281 peaks overlapped with genomic instances of the given family were shown in x-axis. Fisher Exact Test was used to evaluate the significance of overlaps. P-values of Fisher Exact Test were calculated by FISHER in BEDTOOLS, and further adjusted for multiple testing and shown in y-axis. (C–E) Genome browser profiles showing the occupancy of Zfp281, H3K9me3 and H3K4me3 at repeat elements. Red box marks the co-occupied regions by Zfp281 and H3K9me3. Blue box marks the co-occupied regions by Zfp281 and H3K4me3.
Zfp281 is enriched at repeat elements in mouse ES cells
To investigate whether Zfp281 is directly involved in retrotransposon silencing, we examined the enrichment of Zfp281 over repeat elements. We first overlapped the uniquely and non-uniquely mapped Zfp281 ChIP-seq reads with repeat elements from the RepeatMasker Database. The results indicated that Zfp281 occupies a subset of LTR and L1 repeat elements, including L1Md_T, L1Md_A, L1Md_Gf, MMETn and also IAPEY (Figure 1B; Supplementary Figure S2A). However, we did not observe statistically significant enrichment of Zfp281 at MERVL and MERVL MT2_Mm repeats by genome-wide analysis. Since the full-length MERVLs show very high sequence identity, we further mapped the Zfp281 ChIP-seq reads to the MERVL consensus sequence. Only a slight enrichment of Zfp281 over background at MERVL was observed (Data not shown). Thus, we focused on L1s in this study. Further analysis indicated Zfp281 binds to 5′ end of L1 families marked by either H3K9me3 (MMETn and L1Md_A) or H3K4me3 (L1Md_Gf and L1Md_T) (Figure 1C–E). We knocked down Zfp281 by lentivirus-mediated shRNA and observed reduced Zfp281 signals at L1 and LTR elements by ChIP-qPCR analyses, confirming the enrichment of Zfp281 at these regions (Supplementary Figure S2B).
Zfp281 suppresses a subset of active L1 expression
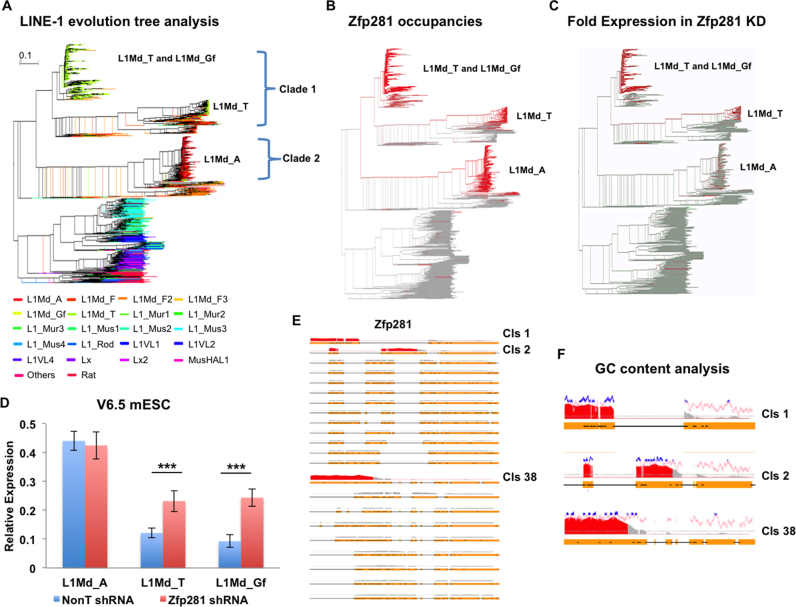
To further measure the occupancy of Zfp281 on 5′ end sequences for all annotated L1 families in the mouse genome, we first performed the phylogenetic analysis of mouse L1 repeat families by using the annotated L1 5′ end sequences (Figure 2A). The enrichment of Zfp281 on L1 elements was then plotted on the L1 phylogenetic tree. The results indicated that Zfp281 is specifically enriched at young L1 subfamilies, including L1Md_A, L1Md_T and L1Md_Gf (Figure 2B). RNA-seq analyses in Zfp281-depleted mouse ES cells further indicated that the depletion of Zfp281 specifically leads to the upregulation of L1Md_T and L1Md_Gf, with little effect on the L1Md_A subfamily (Figure 2C). The roles of Zfp281 in suppressing subsets of young L1 subfamilies expression were validated by RT-qPCR analyses in four different ES cell lines from different mouse strains (Figure 2D; Supplementary Figure S3).
Figure 2.
Zfp281 suppresses subsets of L1s in mouse ES cells. (A) Phylogenetic analysis of the L1 repeat family in the mouse genome. The 5′ end sequences of annotated L1 elements (n = 21 567) were used for constructing the Neighbour Join tree (55). The annotation of L1 subfamily was from UCSC Genome Database. (B) Zfp281 is enriched at subsets of L1 families. The enrichment of Zfp281 on L1 elements was superimposed on the L1 phylogenetic tree. (C) Fold change analysis of the expression of L1 subfamilies after Zfp281 knockdown. The fold expression of L1 elements in Zfp281 knockdown versus control ES cells was superimposed on the L1 phylogenetic tree (A). (D) RT-qPCR analyses of L1 subfamilies L1Md_A, L1Md_T, and L1Md_Gf in Zfp281-depleted ES cells. Expression levels were normalized to Actin mRNA levels. The experiments were biologically repeated more than three times. Error bars represent standard deviations from biological replicates. ***P < 0.001. (E) Occupancy analyses of Zfp281 over the L1 consensus sequences. A consensus sequence was generated from each subfamily in clades 1 and 2 (see Supplementary Figure S5). ChIP-seq signals of Zfp281 were superimposed on these consensus sequences. Gaps are labeled by black lines. Fold enrichments >3 are highlighted in red. (F) GC content analysis at the 5′ end of the L1 families. The percentage of GC within a window of 20 nucleotides was calculated and plotted over the Zfp281 binding signal along the 5′ end of the L1 consensus sequence. Windows with GC content >70% are highlighted in blue.
During evolutionary expansion and suppression cycles, L1 family members have accumulated substantial sequence variation at their 5′ UTRs (41). Accordingly, loss or gain of transcriptional factor binding sites regulates the expansion and suppression of retrotransposons (5). Because Zfp281 functions specifically in repressing the young L1 subfamilies, we hypothesized that Zfp281 might bind to specific sequences on their 5′ UTR. To investigate whether Zfp281 occupies phylogenetic clusters composed of closely related sequences, we clustered Zfp281-enriched Clade 1 and Clade 2 L1 elements from the phylogenetic tree analysis shown in Figure 2A. In total, 22 clusters were identified and the consensus sequence for each cluster was generated (Supplementary Figure S4A). We then performed multiple alignments on these consensus sequences and found that the 5′ UTRs of certain L1 subfamilies, including L1Md_T, L1Md_A and L1Md_Gf, contain GC-rich sequences (Figure 2E). Therefore, Zfp281 is specifically enriched at the unique GC-rich 5′ UTRs of L1 young subfamilies (Figure 2F). This finding is consistent with the previously reported binding preference of Zfp281 over GC-rich sequences (32).
H3K4me3 is enriched at a subset of L1s
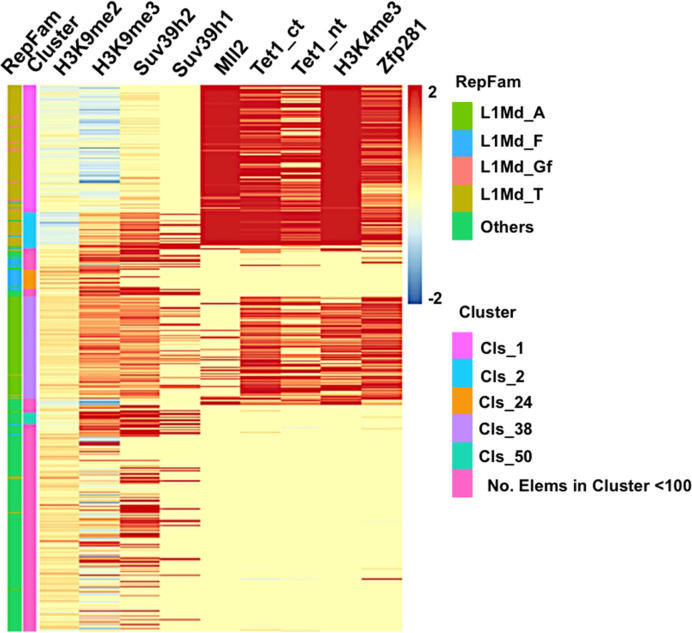
DNA methylation and H3K9 methylation-related machineries are major factors involved in the transcriptional silencing of endogenous retrotransposons (8,12,42). It has been reported that different repeat elements are suppressed via distinct mechanisms (15,17,18,43). H3K9me2 is highly enriched at MERVL repeat regions, while other repeat elements are highly occupied by H3K9me3 (15,17). In order to understand the transcriptional regulation of L1 subfamilies, we performed histone modification analyses on L1 subfamilies. H3K9me3 and its methylase Suv39h are mainly enriched at the L1Md_A subfamily and Suv39h suppresses the expression of L1Md_A (Figure 3) (17). However, the Zfp281-regulated young L1 subfamilies, L1Md_Gf and L1Md_T, are highly occupied by H3K4me3, but largely depleted of H3K9me3 (Figure 3; Supplementary Table S1). These results suggested a potentially distinct mechanism employed by host to suppress the Zfp281-regulated L1s.
Figure 3.
H3K4me3 is enriched at the Zfp281-regulated L1s. Heat maps of binding profiles for Zfp281, Tet1, Sin3A, Suv39h and histone modification are shown over the 5′-UTR of L1s. H3K9me3 is enriched at L1Md_A subfamily, while H3K4me3 is mainly enriched at L1Md_Gf and L1Md_T subfamilies.
The H3K4 methylase Mll2 regulates the expression of subsets of L1s
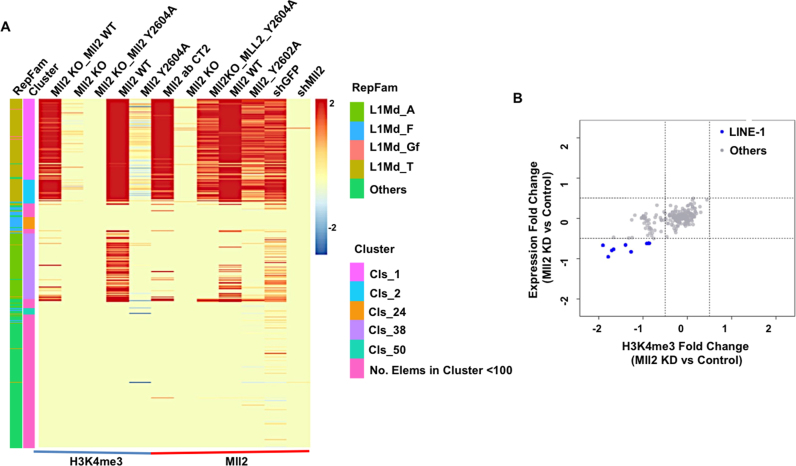
As H3K4me3 is highly enriched at the Zfp281-regulated young L1 subfamilies, we then asked which H3K4 methylase is required for the deposition of H3K4me3 at these regions. We mined the published H3K4me3 ChIP-seq datasets in the H3K4 methylase-depleted mouse ES cells (44,45). The depletion of Cfp1, a shared subunit of Set1A/B COMPASS complexes, does not have dramatic effects on H3K4me3 levels at L1s, except the L1Md_A subfamily (Supplementary Figure S4B-C). In contrast, the depletion of Mll2 results in complete loss of H3K4me3 at the Zfp281-regulated young L1 subfamilies (Figure 4A). It has been previously shown that Mll2 is required for H3K4me3 on bivalent promoters in mouse ES cells (46). However, the expression of bivalent genes remains unchanged after Mll2 depletion. Different from bivalent genes, the expression of these young L1 subfamilies is regulated by Mll2 (Figure 4B; Supplementary Figure S4D-E). The down regulation of these young L1 expression in cells bearing the Mll2 catalytically deficient mutant Y2604A further indicated the requirement of catalytic activity of Mll2 as a methyltransferase in regulating L1 expression (Figure 4A).
Figure 4.
Mll2 is required for the proper expression of subsets of L1s. (A) H3K4me3 and Mll2 occupancy analyses in Mll2 wild type (WT), knockout (KO) and catalytic dead mutant (Y2604A) mouse ES cells at L1s. (B) The MA plot shows that the depletion of Mll2 leads to reduced H3K4me3 and expression levels of subsets of L1s.
Mll2 and Zfp281 antagonize each other in regulating L1 expression
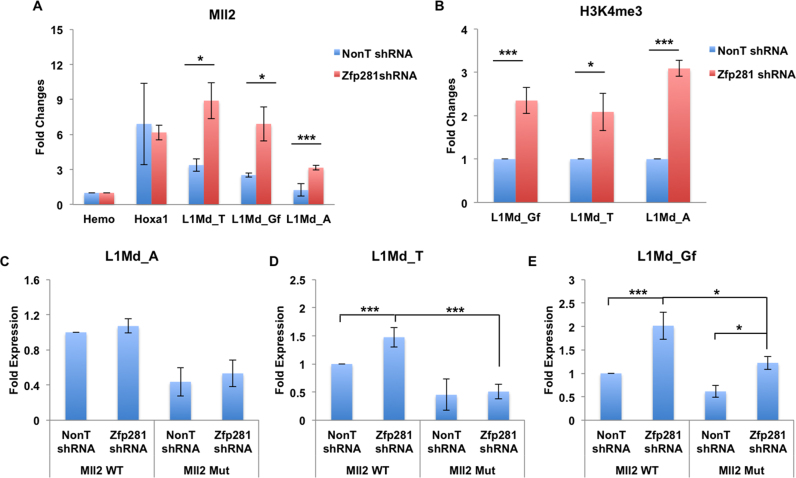
Since Mll2 and Zfp281 play opposite roles in regulating the same subsets of L1s, we further examined the potential functional link between Mll2 and Zfp281 in controlling L1 expression. Our results indicated that the depletion of Zfp281 leads to the increased occupancies of H3K4me3 and Mll2 at L1s (Figure 5A and B). Furthermore, the depletion of Mll2 compromises, despite not abolishing, the derepression of these L1s in Zfp281-depleted cells (Figure 5C–E), indicating that besides Mll2, other epigenetic modifiers could also be involved in the activation of L1s. Thus, these data further support the critical roles of Mll2 in activating the Zfp281-regulated young L1s.
Figure 5.
Mll2 antagonizes Zfp281 in regulating L1 expression. (A) Mll2 enrichment analyses at L1s in the presence and absence of Zfp281. Hemo serves as a non-transcribed control gene. Mll2 enrichment at Hoxa1 and L1s were normalized to its enrichment at the non-transcribed beta-globin gene (Hemo) and fold changes were shown. (B) H3K4me3 enrichment analyses at L1s in the presence and absence of Zfp281. H3K4me3 ChIP signals in Zfp281 depleted cells were calculated to its respective NonT shRNA samples and fold changes were shown. (C–E) Mll2 is required for the increased expression of L1 in the Zfp281-depleted ES cells. The expression levels were normalized to Actin. The relative L1 fold expressions in each sample were normalized to their expression in wild type cells (Mll2 WT) treated with NonT shRNA. The experiments were biologically repeated three times. Error bars represent standard deviations from biological replicates. *P < 0.05, **P < 0.01 and ***P < 0.001.
Zfp281 recruits Tet1 and Sin3A in restricting L1 subfamilies
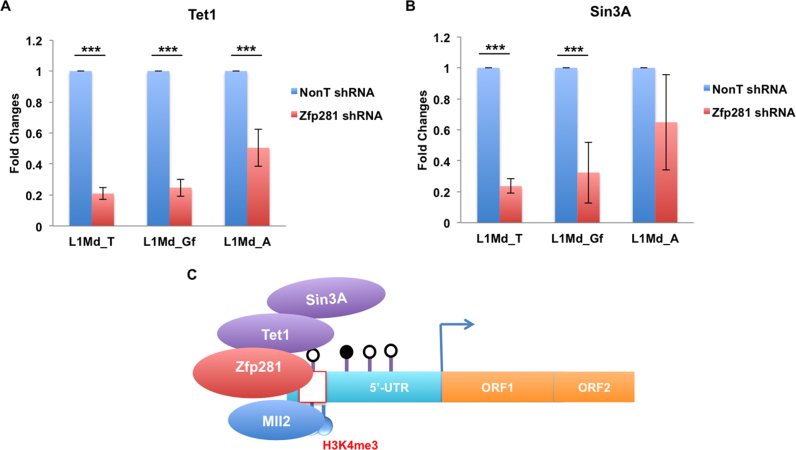
It has been reported that Zfp281 associates with a diverse range of transcriptional regulators, including Tet1 and Sin3A, functioning in different biological processes (47,48). Recently, it has also been shown that Tet1 and Sin3A are both involved in L1 suppression in mouse ES cells (27). However, how the Tet1-Sin3A complex is recruited to L1s remains unclear. The Genome-wide occupancy analyses indicated that Zfp281 and Tet1 co-occupy the 5′end of the L1Md-Gf and L1Md-T subfamilies (Figure 3; Supplementary Table S1). Since Zfp281 bears zinc-finger domains and can directly bind GC-rich DNA sequences in vitro (28), we next tested whether Zfp281 functions through recruiting Tet1 and Sin3A to regulate the L1 repeat elements. Indeed, our results indicated that the depletion of Zfp281 significantly compromises the recruitment of Tet1 and Sin3A to 5′-UTR of the L1 repeat elements (Figure 6A and B), while the expression of Tet1 and Sin3A was not affected by Zfp281 knockdown (Supplementary Figure S5A and B). Furthermore, the double knockdown of both Zfp281 and Sin3A did not lead to further derepression of L1s as compared to the single knockdown of Zfp281 (Supplementary Figure S5C–E). These data indicated that Zfp281 could function partially through Tet1/Sin3A in restricting subsets of L1 expression.
Figure 6.
Zfp281 recruits Tet1 and Sin3A in restricting L1 subfamilies. (A and B) Tet1 and Sin3A enrichment analyses at L1s in the presence and absence of Zfp281. Tet1 and Sin3A ChIP signals in Zfp281 depleted cells were calculated to their respective NonT shRNA samples and fold changes were shown. The experiments were biologically repeated three times. Error bars represent standard deviations from biological replicates. *P < 0.05, **P < 0.01 and ***P < 0.001. (C) A cartoon model for Zfp281-dependent regulation of L1s. Zfp281 binds to the GC-rich DNA region at L1 5′-UTR, and then recruits Tet1 and Sin3A in suppressing their expression. Zfp281-regulated L1s are significantly enriched for H3K4me3, which is mainly deposited by Mll2.
DISCUSSION
Competition between transposable elements and their hosts has led to a co-evolutionary ‘arms race’ scenario in order to maintain the fine balance between genetic diversity and genome instability. Diverse mechanisms have been evolved to regulate the expression of repeat elements during transposable element evolution. Here, we report that the depletion of Zfp281 leads to the up-regulation of a subset of young active L1 subfamilies. Zfp281 is enriched specifically at the unique GC-rich regions of young L1 subfamilies and recruits Tet1 and Sin3A to suppress their expression. Furthermore, Zfp281-controlled L1 subfamilies are highly occupied by H3K4me3 and require the H3K4me3 methylase Mll2 for their proper expression. In summary, our results indicated a coordinated role of the activating H3K4 methyltransferases Mll2 and the suppressing Zfp281-Tet1-Sin3A pathway in the regulation of subsets of young L1 expression (Figure 6C).
A role of Mll2 in L1 expression in mouse ES cells
There are at least 6 different COMPASS and COMPASS-like H3K4 methylases in mammals with functional diversity (49). Set1/COMPASS is responsible for the bulk H3K4me3 in mammals. It has been proposed that Tet1-Sin3A could function through Ogt-Set1/COMPASS in regulating L1 expression (50). However, our analysis indicated that the depletion of Cfp1, a unique subunit of the Set1/COMPASS complex, does not seem to have global effect on H3K4me3 at L1s. Here, we found that Mll2 is specifically enriched at the Zfp281 regulated young active L1s and is the major H3K4 methylase in regulating their proper expression in mouse ES cells. The results from the catalytic dead mutant of Mll2 further suggested the requirement of H3K4me3 in controlling L1 expression. In the past, extensive research has been focused on exploring how repeat elements accumulate mutations to escape host surveillance. Our study suggests that the repeat elements could also hijack host epigenetic modifiers, such as Mll2, to activate their expression during expansion.
Diverse mechanisms in controlling repeat elements
L1 repeat elements are newly expanded active retrotransposons that still carry retrotransposition activity in mammals (3,51). The active young L1Md_Gf and L1Md_T subfamilies are largely depleted of DNA methylation and H3K9me3, but highly enriched for H3K4me3 and 5hmC. The depletion of Tet1 leads to the reduction of 5hmC and further compromises the repression function of Sin3A at L1s (27). However, it is unclear how Tet1 and Sin3A are specifically recruited to these L1 elements. Our results indicated that these young L1 subfamilies contain an extra GC-rich region at their 5′-UTR, to which Zfp281 binds. The depletion of Zfp281 affects the recruitment of Tet1 and Sin3A to L1s. It is likely that the interaction among Zfp281, Tet1, and Sin3A could promote the assembly of the Sin3A repressor complex at the L1 5′-UTR regions to partially restrict their expression. Although both Tet1 and Zfp281 seem also enriched at the L1Md_A subfamily and other ERVs, which are marked by H3K9me3, their depletions do not affect the expression, indicating a context-dependent role of Zfp281 in suppressing L1 expression. In addition, it has been reported that the depletion of Sin3A will lead to the global increases of H3K4me3 (52). Here we observed the increased enrichment of Mll2 and H3K4me3 at L1s upon Zfp281 knockdown. Also, the depletion of Mll2 compromises the repressive function of Zfp281 at L1s, further suggesting that Zfp281 could function partially through Tet1-Sin3A in suppressing Mll2 and thus restricting subsets of young L1 expression in mouse ES cells. Furthermore, since both Zfp281 and Mll2 play critical roles in stem cell pluripotency, it needs to be further explored whether their roles in pluripotency are mediated, at least partially, by L1s.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Prof. Ali Shilatifard for providing the Mll2 antibody, the H3K4me3 antibody and Mll2 KO ES cells for this study. We also thank Zhiqun Tang for technical assistance.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Thousand Young Talents Plan of China [5631006003 to C.L.]; National Natural Science Foundation of China [31671343 to C.L.]; Natural Science Foundation of Jiangsu Province of China [BK20160026 to C.L., BK20160666 to Z.L.]; Fundamental Research Funds for the Central Universities [3231007201 to C.L]. Funding for open access charge: National Natural Science Foundation of ChinaThousand Young Talents Plan of China [5631006003 to C.L.].
Conflict of interest statement. None declared.
REFERENCES
- 1. McClintock B. Controlling elements and the gene. Cold Spring Harbor Symp. Quant. Biol. 1956; 21:197–216. [DOI] [PubMed] [Google Scholar]
- 2. Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W. et al. . Initial sequencing and analysis of the human genome. Nature. 2001; 409:860–921. [DOI] [PubMed] [Google Scholar]
- 3. Burns K.H., Boeke J.D.. Human transposon tectonics. Cell. 2012; 149:740–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cordaux R., Batzer M.A.. The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 2009; 10:691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Goodier J.L., Kazazian H.H. Jr. Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell. 2008; 135:23–35. [DOI] [PubMed] [Google Scholar]
- 6. Hancks D.C., Kazazian H.H. Jr. Active human retrotransposons: variation and disease. Curr. Opin. Genet. Dev. 2012; 22:191–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Walsh C.P., Chaillet J.R., Bestor T.H.. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat. Genet. 1998; 20:116–117. [DOI] [PubMed] [Google Scholar]
- 8. Martens J.H., O'Sullivan R.J., Braunschweig U., Opravil S., Radolf M., Steinlein P., Jenuwein T.. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J. 2005; 24:800–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Esnault C., Heidmann O., Delebecque F., Dewannieux M., Ribet D., Hance A.J., Heidmann T., Schwartz O.. APOBEC3G cytidine deaminase inhibits retrotransposition of endogenous retroviruses. Nature. 2005; 433:430–433. [DOI] [PubMed] [Google Scholar]
- 10. Aravin A., Gaidatzis D., Pfeffer S., Lagos-Quintana M., Landgraf P., Iovino N., Morris P., Brownstein M.J., Kuramochi-Miyagawa S., Nakano T. et al. . A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006; 442:203–207. [DOI] [PubMed] [Google Scholar]
- 11. Aravin A.A., Sachidanandam R., Girard A., Fejes-Toth K., Hannon G.J.. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007; 316:744–747. [DOI] [PubMed] [Google Scholar]
- 12. Yoder J.A., Walsh C.P., Bestor T.H.. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet.: TIG. 1997; 13:335–340. [DOI] [PubMed] [Google Scholar]
- 13. Liang G., Chan M.F., Tomigahara Y., Tsai Y.C., Gonzales F.A., Li E., Laird P.W., Jones P.A.. Cooperativity between DNA methyltransferases in the maintenance methylation of repetitive elements. Mol. Cell. Biol. 2002; 22:480–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Damelin M., Bestor T.H.. Biological functions of DNA methyltransferase 1 require its methyltransferase activity. Mol. Cell. Biol. 2007; 27:3891–3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Maksakova I.A., Thompson P.J., Goyal P., Jones S.J., Singh P.B., Karimi M.M., Lorincz M.C.. Distinct roles of KAP1, HP1 and G9a/GLP in silencing of the two-cell-specific retrotransposon MERVL in mouse ES cells. Epigenet. Chromatin. 2013; 6:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu S., Brind’Amour J., Karimi M.M., Shirane K., Bogutz A., Lefebvre L., Sasaki H., Shinkai Y., Lorincz M.C.. Setdb1 is required for germline development and silencing of H3K9me3-marked endogenous retroviruses in primordial germ cells. Genes Dev. 2014; 28:2041–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bulut-Karslioglu A., De La Rosa-Velazquez I.A., Ramirez F., Barenboim M., Onishi-Seebacher M., Arand J., Galan C., Winter G.E., Engist B., Gerle B. et al. . Suv39h-dependent H3K9me3 marks intact retrotransposons and silences LINE elements in mouse embryonic stem cells. Mol. Cell. 2014; 55:277–290. [DOI] [PubMed] [Google Scholar]
- 18. Rowe H.M., Jakobsson J., Mesnard D., Rougemont J., Reynard S., Aktas T., Maillard P.V., Layard-Liesching H., Verp S., Marquis J. et al. . KAP1 controls endogenous retroviruses in embryonic stem cells. Nature. 2010; 463:237–240. [DOI] [PubMed] [Google Scholar]
- 19. Jacobs F.M., Greenberg D., Nguyen N., Haeussler M., Ewing A.D., Katzman S., Paten B., Salama S.R., Haussler D.. An evolutionary arms race between KRAB zinc-finger genes ZNF91/93 and SVA/L1 retrotransposons. Nature. 2014; 516:242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wolf D., Goff S.P.. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature. 2009; 458:1201–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee E., Iskow R., Yang L., Gokcumen O., Haseley P., Luquette L.J. 3rd, Lohr J.G., Harris C.C., Ding L., Wilson R.K. et al. . Landscape of somatic retrotransposition in human cancers. Science. 2012; 337:967–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shukla R., Upton K.R., Munoz-Lopez M., Gerhardt D.J., Fisher M.E., Nguyen T., Brennan P.M., Baillie J.K., Collino A., Ghisletti S. et al. . Endogenous retrotransposition activates oncogenic pathways in hepatocellular carcinoma. Cell. 2013; 153:101–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Goodier J.L., Ostertag E.M., Du K., Kazazian H.H. Jr. A novel active L1 retrotransposon subfamily in the mouse. Genome Res. 2001; 11:1677–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Padgett R.W., Hutchison C.A. 3rd, Edgell M.H.. The F-type 5′ motif of mouse L1 elements: a major class of L1 termini similar to the A-type in organization but unrelated in sequence. Nucleic Acids Res. 1988; 16:739–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sookdeo A., Hepp C.M., McClure M.A., Boissinot S.. Revisiting the evolution of mouse LINE-1 in the genomic era. Mobile DNA. 2013; 4:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ficz G., Branco M.R., Seisenberger S., Santos F., Krueger F., Hore T.A., Marques C.J., Andrews S., Reik W.. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011; 473:398–402. [DOI] [PubMed] [Google Scholar]
- 27. de la Rica L., Deniz O., Cheng K.C., Todd C.D., Cruz C., Houseley J., Branco M.R.. TET-dependent regulation of retrotransposable elements in mouse embryonic stem cells. Genome Biol. 2016; 17:234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Law D.J., Du M., Law G.L., Merchant J.L.. ZBP-99 defines a conserved family of transcription factors and regulates ornithine decarboxylase gene expression. Biochem. Biophys. Res. Commun. 1999; 262:113–120. [DOI] [PubMed] [Google Scholar]
- 29. Wang Z.X., Teh C.H., Chan C.M., Chu C., Rossbach M., Kunarso G., Allapitchay T.B., Wong K.Y., Stanton L.W.. The transcription factor Zfp281 controls embryonic stem cell pluripotency by direct activation and repression of target genes. Stem Cells. 2008; 26:2791–2799. [DOI] [PubMed] [Google Scholar]
- 30. Fidalgo M., Shekar P.C., Ang Y.S., Fujiwara Y., Orkin S.H., Wang J.. Zfp281 functions as a transcriptional repressor for pluripotency of mouse embryonic stem cells. Stem Cells. 2011; 29:1705–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hahn S., Jackstadt R., Siemens H., Hunten S., Hermeking H.. SNAIL and miR-34a feed-forward regulation of ZNF281/ZBP99 promotes epithelial-mesenchymal transition. EMBO J. 2013; 32:3079–3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang Y., Shen Y., Dai Q., Yang Q., Zhang Y., Wang X., Xie W., Luo Z., Lin C.. A permissive chromatin state regulated by ZFP281-AFF3 in controlling the imprinted Meg3 polycistron. Nucleic Acids Res. 2017; 45:1177–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lin C., Smith E.R., Takahashi H., Lai K.C., Martin-Brown S., Florens L., Washburn M.P., Conaway J.W., Conaway R.C., Shilatifard A.. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol. Cell. 2010; 37:429–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lin C., Garrett A.S., De Kumar B., Smith E.R., Gogol M., Seidel C., Krumlauf R., Shilatifard A.. Dynamic transcriptional events in embryonic stem cells mediated by the super elongation complex (SEC). Genes Dev. 2011; 25:1486–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bao W., Kojima K.K., Kohany O.. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mobile DNA. 2015; 6:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W. et al. . Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Edgar R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010; 26:2460–2461. [DOI] [PubMed] [Google Scholar]
- 38. Howe K., Bateman A., Durbin R.. QuickTree: building huge Neighbour-Joining trees of protein sequences. Bioinformatics. 2002; 18:1546–1547. [DOI] [PubMed] [Google Scholar]
- 39. Prosperi M.C., Ciccozzi M., Fanti I., Saladini F., Pecorari M., Borghi V., Di Giambenedetto S., Bruzzone B., Capetti A., Vivarelli A. et al. . A novel methodology for large-scale phylogeny partition. Nat. Commun. 2011; 2:321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Eckersley-Maslin M.A., Svensson V., Krueger C., Stubbs T.M., Giehr P., Krueger F., Miragaia R.J., Kyriakopoulos C., Berrens R.V., Milagre I. et al. . MERVL/Zscan4 network activation results in transient genome-wide DNA demethylation of mESCs. Cell Rep. 2016; 17:179–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Boissinot S., Chevret P., Furano A.V.. L1 (LINE-1) retrotransposon evolution and amplification in recent human history. Mol. Biol. Evol. 2000; 17:915–928. [DOI] [PubMed] [Google Scholar]
- 42. Gifford W.D., Pfaff S.L., Macfarlan T.S.. Transposable elements as genetic regulatory substrates in early development. Trends Cell Biol. 2013; 23:218–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rowe H.M., Friedli M., Offner S., Verp S., Mesnard D., Marquis J., Aktas T., Trono D.. De novo DNA methylation of endogenous retroviruses is shaped by KRAB-ZFPs/KAP1 and ESET. Development. 2013; 140:519–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hu D., Gao X., Cao K., Morgan M.A., Mas G., Smith E.R., Volk A.G., Bartom E.T., Crispino J.D., Di Croce L. et al. . Not all H3K4 methylations are created equal: Mll2/COMPASS dependency in primordial germ cell specification. Mol. Cell. 2017; 65:460–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Clouaire T., Webb S., Bird A.. Cfp1 is required for gene expression-dependent H3K4 trimethylation and H3K9 acetylation in embryonic stem cells. Genome Biol. 2014; 15:451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hu D., Garruss A.S., Gao X., Morgan M.A., Cook M., Smith E.R., Shilatifard A.. The Mll2 branch of the COMPASS family regulates bivalent promoters in mouse embryonic stem cells. Nat. Struct. Mol. Biol. 2013; 20:1093–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fidalgo M., Huang X., Guallar D., Sanchez-Priego C., Valdes V.J., Saunders A., Ding J., Wu W.S., Clavel C., Wang J.. Zfp281 coordinates opposing functions of Tet1 and Tet2 in pluripotent states. Cell Stem Cell. 2016; 19:355–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fidalgo M., Faiola F., Pereira C.F., Ding J., Saunders A., Gingold J., Schaniel C., Lemischka I.R., Silva J.C., Wang J.. Zfp281 mediates Nanog autorepression through recruitment of the NuRD complex and inhibits somatic cell reprogramming. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:16202–16207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shilatifard A. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu. Rev. Biochem. 2012; 81:65–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. de la Rica L., Deniz Ö., Cheng K.C., Todd C.D., Cruz C., Houseley J., Branco M.R.. TET-dependent regulation of retrotransposable elements in mouse embryonic stem cells. Genome Biol. 2016; 17:234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jachowicz J.W., Torres-Padilla M.E.. LINEs in mice: features, families, and potential roles in early development. Chromosoma. 2015; 125:29–39. [DOI] [PubMed] [Google Scholar]
- 52. Liu M., Pile L.A.. The transcriptional corepressor SIN3 directly regulates genes involved in methionine catabolism and affects histone methylation, linking epigenetics and metabolism. J. Biol. Chem. 2017; 292:1970–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Liu N., Zhang Z., Wu H., Jiang Y., Meng L., Xiong J., Zhao Z., Zhou X., Li J., Li H. et al. . Recognition of H3K9 methylation by GLP is required for efficient establishment of H3K9 methylation, rapid target gene repression, and mouse viability. Genes Dev. 2015; 29:379–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jurka J., Kapitonov V.V., Pavlicek A., Klonowski P., Kohany O., Walichiewicz J.. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005; 110:462–467. [DOI] [PubMed] [Google Scholar]
- 55. Saitou N., Nei M.. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987; 4:406–425. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.