Abstract
Spinal muscular atrophy (SMA) is caused by deletions or mutations of the Survival Motor Neuron 1 (SMN1) gene coupled with predominant skipping of SMN2 exon 7. The only approved SMA treatment is an antisense oligonucleotide that targets the intronic splicing silencer N1 (ISS-N1), located downstream of the 5′ splice site (5′ss) of exon 7. Here, we describe a novel approach to exon 7 splicing modulation through activation of a cryptic 5′ss (Cr1). We discovered the activation of Cr1 in transcripts derived from SMN1 that carries a pathogenic G-to-C mutation at the first position (G1C) of intron 7. We show that Cr1-activating engineered U1 snRNAs (eU1s) have the unique ability to reprogram pre-mRNA splicing and restore exon 7 inclusion in SMN1 carrying a broad spectrum of pathogenic mutations at both the 3′ss and 5′ss of the exon 7. Employing a splicing-coupled translation reporter, we demonstrate that mRNAs generated by an eU1-induced activation of Cr1 produce full-length SMN. Our findings underscore a wider role for U1 snRNP in splicing regulation and reveal a novel approach for the restoration of SMN exon 7 inclusion for a potential therapy of SMA.
INTRODUCTION
Alternative pre-mRNA splicing enhances the coding potential of the eukaryotic genome by enabling single genes to produce multiple transcripts/proteins through removal of intronic sequences in different combinations (1). A vast majority of human introns belong to the GUAG type in which the 5′ splice site (5′ss) and the 3′ss are defined by GU and AG dinucleotides at the first and last two positions, respectively (2). In addition, pre-mRNAs harbor a significantly higher number of cryptic splice sites that resemble canonical GUAG type splice sites but are usually suppressed under normal conditions (3). Exon and intron definition models, which are mutually exclusive, are the two primary mechanisms proposed for the selection of splice sites (4,5). These simplistic models do not fully encompass the complexity of splicing regulation that is influenced by both transcription and the overall context of the splice site (6,7). Even more puzzling is the mechanism of suppression of cryptic splice sites that generally overlap with silencer elements (8,9). Diverse factors, including hnRNP C, TDP-43, PTBP1 and PTBP2, have been implicated in the genome-wide suppression of cryptic splice sites (10–12). Point mutations leading to the activation of cryptic splice sites have been associated with various pathological conditions (13). Activation of cryptic splice sites often generates transcripts harboring premature termination codons (PTCs) leading to their degradation by nonsense-mediated decay (NMD) (13). Therefore, the level of transcripts generated by the activation of cryptic splice sites cannot be accurately determined. However, usage of a cryptic splice site downstream of a stop codon or at a site that retains the open-reading frame (ORF) is likely to generate stable transcripts. A recent study attempted to uncover rules why certain point mutations favor activation of a cryptic 5′ss instead of an exon skipping (14). However, there is no systematic study on targeted activation of a cryptic 5′ss that prevents exon skipping caused by a pathogenic mutation at the splice site.
Splicing is catalyzed by the spliceosome, a macromolecular machine in which five small ribonucleoproteins (U1, U2, U4, U5 and U6 snRNPs) play an indispensable role (15,16). Spliceosomal assembly begins with the recruitment of U1 snRNP, which is expressed at much higher levels than other snRNPs in human cells (17). During pre-mRNA splicing, U1 snRNPs are recruited at more sites than are actually utilized (18). Recruitment of U1 snRNP at multiple sites on pre-mRNA offers several benefits, such as suppression of cryptic exons, maintenance of mRNA length and provision of directionality to transcription (19–22). In specific cases when U1 snRNP activates the usage of a 5′ss away from its annealing position, it is referred to as a shift-U1 snRNP (23). Consistently, it has been shown that engineered U1 snRNAs (eU1s) that anneal to different intronic sequences could promote the usage of the upstream 5′ss (24–26). However, proof of the principle that an eU1 could activate a ‘desired’ cryptic 5′ss and neutralize the consequences of a pathogenic mutation at the native 5′ss of an exon remains to be demonstrated.
Humans possess two almost identical copies of the Survival Motor Neuron gene, SMN1 and SMN2 (27). SMN1 predominantly produces full-length SMN, an essential protein involved in snRNP biogenesis, transcription, translation, cell signaling, macromolecular transport and stress granule formation (28). Due to overwhelming skipping of exon 7, SMN2 mostly produces the truncated protein SMNΔ7 (29,30). The 54 nucleotide (nt)-long exon 7 codes for the last 16 amino acids that play a critical role in SMN stability (31). Low levels of SMN due to deletions or mutations of SMN1 leads to spinal muscular atrophy (SMA), one of the leading genetic causes of infant mortality (32). Due to the notion that re-direction of SMN2 exon 7 during pre-mRNA splicing holds the answer for the treatment of most SMA patients, the mechanism of exon 7 splicing regulation has been intensively investigated (33,34). Early studies revealed that skipping of SMN2 exon 7 is triggered by a C-to-T substitution at the 6th position (C6U substitution in RNA) of exon 7 (29,35). Subsequent studies showed that the poor recruitment of U1 snRNP at the 5′ss of exon 7 is also a limiting factor for SMN2 exon 7 inclusion (36–38). In particular, we demonstrated that the intronic splicing silencer N1 (ISS-N1) located downstream of the 5′ss of exon 7 plays a critical role in inducing the skipping of SMN2 exon 7 [Figure 1A, (39)]. Various independent pre-clinical and clinical studies confirmed unparalleled therapeutic benefits of ISS-N1-targeting antisense oligonucleotides (ASOs) (40). The FDA has recently approved nusinersen, an ISS-N1-targeting ASO, as the first medical therapy for SMA (41). Nusinersen is also the first antisense drug that restores the complete ORF of a gene via splicing modulation (42).
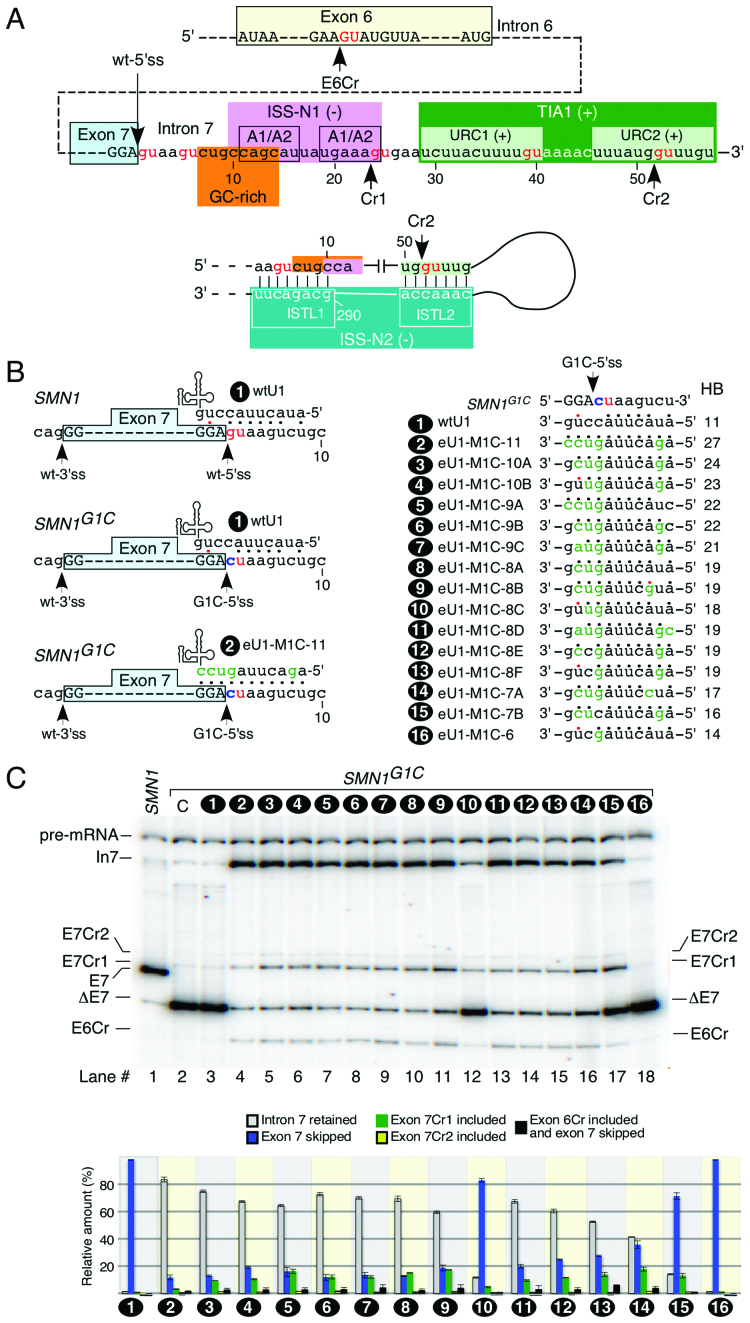
Figure 1.
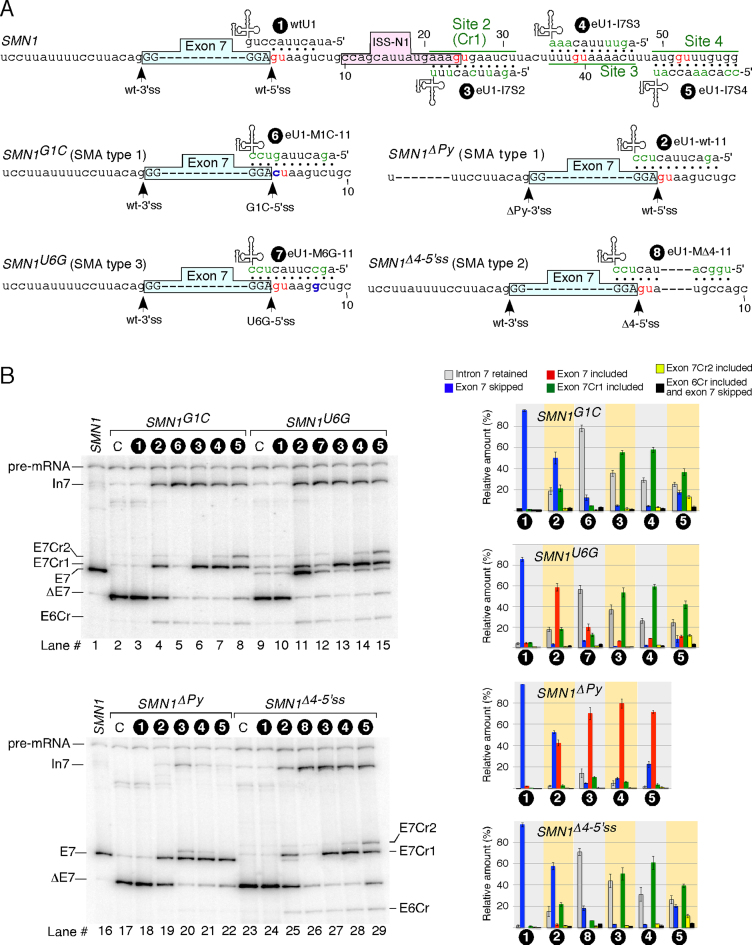
Effect of eU1 base pairing with G1C-5′ss on splicing of exon 7 in SMN1G1C transcripts. (A) Diagrammatic representation of splicing regulatory elements. Exonic and intronic sequences are shown in capital and lower-case letters, respectively. Numbering starts from the first position of intron 7. The GU dinucleotides are indicated in red. Positive (+) and negative (–) regulatory elements of exon 7 splicing are highlighted in colored boxes (also see refs. 39,43,44,46,48). Cryptic 5′ss within intron 7 and exon 6 are indicated by arrows. Abbreviations: Cr1, cryptic site 1; Cr2, cryptic site 2; E6Cr, cryptic site within exon 6. (B) Diagrammatic representation of base pairing formed between eU1s and G1C-5′ss of exon 7. Left panel depicts eU1-M1C-11 that forms the maximum possible 11 base pairs with the G1C-5′ss. The remaining eU1s form progressively decreasing numbers of continuous base pairs with G1C-5′ss (right panel). Number of hydrogen bonds for each U1 construct is shown. Mutated nucleotides within U1 snRNAs are indicated in green. Base pairing is marked by black dots; red dots represent a wobble base pair. G1C mutation at the 5′ss of exon 7 is shown in blue. The 3′ss and 5′ss are indicated by arrows. Abbreviations: HB, hydrogen bonds; wt, wild type. (C) In vivo splicing pattern of transcripts generated by SMN1G1C minigene in the presence of the overexpressed eU1s shown in (B). The identity of eU1 constructs is marked at the top of the gel. The splice products are indicated on the left and the right of the gel. Quantification of the relative amount of the indicated splice products is given in the bottom panel as a bar diagram. Error bars represent standard error. Abbreviations: C, control when SMN1G1C minigene was transfected without a U1 construct. Other abbreviations are described in Supplementary Table S1.
In parallel with the advancements made on the therapeutic front, there has been continued progress towards our understanding of the mechanism by which ISS-N1 and other regulatory elements modulate SMN2 exon 7 splicing. ISS-N1 harbors two adjacent motifs associated with the negative regulator(s) hnRNP A1/A2 and is followed by two uridine-rich clusters (URC1 and URC2) associated with the positive regulator(s) TIA1/TIAR [Figure 1A, (43,44)]. We recently showed that TIA1 is a gender-specific disease modifier in a mouse model of SMA (45). The 5′ end of ISS-N1 partially overlaps with an 8-nt long GC-rich sequence, sequestration of which by an 8-mer ASO fully restores SMN2 exon 7 inclusion and provides therapeutic benefits in mouse models of SMA [Figure 1A, (46,47)]. In addition, the first residue of ISS-N1 strengthens a unique structure formed by long-distance interactions involving ISS-N2, a deep intronic inhibitory element [Figure 1A, (48)]. ASO-mediated sequestration of ISS-N2 has been recently shown to produce a gender-specific amelioration of the phenotype in a mild SMA mouse model (49). There have also been studies on eU1s that promote SMN2 exon 7 inclusion by targeting ISS-N1 or a nearby region (25,26). Sequences downstream of the 5′ss of exon 7 harbor several GU dinucleotides; however, it is not known if any of these dinucleotides is part of a putative cryptic 5′ss. Considering that the stop codon is located within exon 7, transcripts generated by the activation of a cryptic 5′ss downstream of exon 7 may escape NMD. Hence, activation of such a cryptic 5′ss would offer a novel therapeutic avenue for patients carrying deadly mutations at the wild type 5′ss (wt-5′ss). Currently, there is no evidence that a ‘functional’ cryptic 5′ss exists downstream of exon 7, let alone that it could be activated under normal or pathological condition even by the reported eU1s that target ISS-N1 or nearby sequences.
Here, we describe two novel cryptic 5′ss, Cr1 and Cr2, located 23 and 51 nts downstream of the wt-5′ss of SMN exon 7, respectively. The surprising discovery of Cr1 and Cr2 was possible due to the employment of the SMN1G1C minigene that harbors a recently reported dead-end G1C mutation at the first position of intron 7 of SMN1. The G1C mutation was first discovered in a patient that displayed a severe SMA phenotype and died at ∼4 months of age (50). This patient had only a single copy of SMN2 in addition to carrying the G1C mutation in SMN1 (50). Reproducing the lethal effect of G1C, the SMN1G1C minigene showed complete skipping of exon 7. In order to suppress skipping of SMN1G1C exon 7, we screened a library of eU1s that targeted the G1C-5′ss. While most eU1s suppressed exon 7 skipping, they also activated Cr1 and Cr2. Interestingly, Cr1, which partially overlaps with ISS-N1, emerged as the most favorable cryptic 5′ss induced by eU1 overexpression. Our results reveal that ISS-N1 is a suppressor of the Cr1 site. We show strong activation of Cr1 by eU1s targeting both upstream and downstream sequences. Further, we demonstrate that the U1 snRNP-based approach could potentially be employed to restore exon 7 inclusion from SMN1 carrying a variety of pathogenic mutations at both the 3′ and 5′ss of exon 7. Finally, we show that mRNAs generated by an eU1-induced activation of Cr1 can be translated into full-length SMN. Our findings suggest an expanded role of U1 snRNA in splicing regulation and uncover a novel therapeutic strategy applicable to a broad spectrum of SMA patients.
MATERIALS AND METHODS
Cell cultures, siRNAs and ASOs
All tissue culture media and supplements were purchased from Life Technologies. HeLa cells and human neuroblastoma SH-SY5Y cells were obtained from American Type Culture Collection. Mouse motor-neuron-like NSC-34 cells were generously provided by Dr. N. Cashman (51). HeLa and NSC-34 cells were cultured in Dulbecco's modified Eagle's medium (DMEM). SH-SY5Y cells were maintained in 1:1 mixture of Minimum Essential Medium (MEM) and F12 Medium. Both media were supplemented with 10% fetal bovine serum.
Small interfering RNAs (siRNAs, ON-TARGETplus SMARTpool) against hnRNPA1 and A2B1 and antisense oligonucleotides (ASOs) used in this study were purchased from Dharmacon Inc. An ON-TARGETplus nontargeting pool was used as a negative siRNA control. The ASOs incorporated a phosphorothioate backbone and 2′-O-methyl modifications at each base.
Generation of SMN minigenes
The minigenes pSMN1ΔI6 and pSMN2ΔI6 were described previously (52). Here we refer to pSMN1ΔI6 and pSMN2ΔI6 as SMN1 and SMN2, respectively. Minigenes with site-specific mutations, SMA-patient-associated mutations and the insertion of the neutral Sequence 1 and Sequence 2 were generated similarly as described in (52) using a two-step high-fidelity PCR with Phusion DNA polymerase and SMN1/SMN2 as a template. The identities of the newly constructed minigenes were confirmed by sequencing. All primers used were purchased from Integrated DNA Technologies (IDT). Their sequence information may be obtained upon request. Reagents for PCR and cloning were purchased from New England Biolabs.
Generation of U1 snRNA constructs
Plasmids pUCBU1 and pUCBΔU1 were generous gifts from Dr. H. Schaal (53). Site-specific mutations at the 5′ end of U1 RNA were introduced by high-fidelity PCR as described in (38). The identity of all newly generated U1 constructs was confirmed by sequencing. All primers used were purchased from IDT. Their sequence information could be obtained upon request.
Cell transfections
Transient transfections of cells were performed using either X-tremeGene HP (Roche Life Science) or Lipofectamine 2000 (Life Technologies), following the manufacturer's recommendations. Briefly, to study the effect of eU1s on splicing of minigene exon 7, pre-plated cells were co-transfected with 0.05 μg of the SMN minigene of interest and a given U1 snRNA construct (0.05 μg) using 1 μl of X-tremeGene HP reagent. The total amount of plasmid DNA was maintained constant (0.5 μg) using an empty vector (pCI or pCI neo). In the case of Lipofectamine 2000, the total amount of plasmid DNA was maintained at 0.8 μg and 2 μl of the transfection reagent were used. To study the effect of eU1s on splicing of endogenous exon 7, HeLa cells were transfected with 0.5 μg of a given U1 snRNA construct using 1 μl of X-tremeGene HP reagent. Transfections were performed in 24-well plates. Twenty-four hours after transfections, cells were washed with ice-cold phosphate-buffered saline (PBS, Life Technologies) and lysed in TRIzol reagent (Life Technologies) directly in each well. To test the ability of eU1 snRNAs to promote the full-length SMN protein production from the expression vector FLAG-SMN1G1C, HeLa cells were reverse transfected with FLAG-SMN1G1C (0.5 μg) and a U1 snRNA construct of interest (1.0 μg), using Lipofectamine 2000. The FLAG-SMN1 plasmid was used as a control. The total amount of DNA in each transfection reaction was maintained constant (2.0 μg) by adding pCI neo vector. For each transfection, DNA–Lipofectamine 2000 complex was prepared following the manufacturer's recommendations, and combined with HeLa cell suspensions in a total volume of 2 ml. These cells were then plated in six-well plates, 2 ml per one well. Cells were collected for making cell lysates and total RNA preparation 24 h later. Experimental results were confirmed by at least two independent experiments.
Endogenous U1 snRNA inactivation
To test the effect of inactivation of endogenous U1 snRNA on the ability of eU1-I7R7 to promote exon 7 inclusion, pre-plated HeLa cells were co-transfected with 0.05 μg of a minigene of interest, 0.05 μg of an empty vector (pCI) or eU1-I7R7 and 200 nM of either wtU1 ASO or the control oligonucleotide (5′-UUGCCUUUCU-3′; 54) using 2 μl of Lipofectamine 2000. To test the effect of endogenous U1 snRNA inactivation on the ability of F14 ASO (48,54) to promote exon 7 inclusion, pre-plated HeLa cells were co-transfected with 0.05 μg of a minigene of interest, 50 nM of F14 alone or with 200 nM of wtU1 ASO using 2 μl of Lipofectamine 2000. In all transfection reactions, total amount of nucleic acids was maintained constant (0.8 μg) using the control oligonucleotide. HeLa cells were seeded in 24-well plates (∼0.6 × 105 cells per well) 38 h prior to co-transfections and collected 22–24 h after co-transfections took place using TRIzol reagent. Results were confirmed by at least two independent experiments.
hnRNP A1/A2 depletion
hnRNP A1 and hnRNP A2B1 were knocked down simultaneously using siRNAs (ON-TARGETplus SMART pool). To do so, HeLa cells were reverse transfected with siRNAs twice with an interval of ∼48 h. For each siRNA transfection, an siRNA-Lipofectamine 2000 complex was prepared following the manufacturer's recommendations, and combined with HeLa cell suspensions in a total volume of 2 ml. These cells were then plated in six-well plates, 2 ml per one well. For the first transfection, the final concentration for the control siRNA was 50 nM and for the siRNAs against hnRNP A1 and A2B1 it was 25 nM each. The next day, the transfected HeLa cells were trypsinized and moved to 60 mm dishes. The second reverse transfection of HeLa cells with siRNAs was performed ∼48 h after the first one essentially as described above, except that the siRNA concentration was decreased to 12 nM each for the siRNAs against hnRNP A1 and A2B1. Twenty-six hours after the second siRNA transfection, HeLa cells were trypsinized and seeded in 6-well plates to be transfected with the minigenes of interest the next day. The density of plating per well was ∼1.1 × 106 cells. HeLa cells were transfected ∼24 h later with 0.2 μg of a given minigene and the corresponding siRNAs (10 nM each for the siRNAs against hnRNP A1 and A2B1 and 25 nM for the control siRNA) using Lipofectamine 2000. Cells were collected ∼24 h later; one third of the cells was used for total RNA preparation and two-thirds for making whole-cell extracts as previously described (55). Results were confirmed by at least three independent experiments.
In vivo splicing
Total RNA was isolated from cells using TRIzol reagent (Life Technologies), following the manufacturer's recommendations. RNA was treated with RQ1 RNase-free DNase (Promega) followed by phenol:chloroform (OmniPur) extraction and ethanol precipitation. Unless otherwise stated, cDNA was generated using 0.5 or 0.8 μg of total RNA in a 5 μl reaction, employing SuperScript III Reverse Transcriptase (RTase, Life Technologies) and oligo(dT)12–18 primer (Life Technologies). 0.8 μl of RTase reaction was then used as a template for PCR amplification in a 10 μl reaction. Amplification was carried out with Taq DNA Polymerase (New England Biolabs) and the primer pair P1 and P2 (36). Primer P2 was labeled at the 5′-end with [γ-32P] ATP (6000 Ci/mmol, Perkin-Elmer) using T4 Polynucleotide Kinase (New England Biolabs). For amplification of endogenous SMN, PCR reactions were performed using primers P31 and P25 or N-24 and P26 (44) in the presence of a trace amount of [α-32P] dATP (3000 Ci/mmol, Perkin Elmer), followed by phenol:chloroform extraction and ethanol precipitation. To distinguish splice isoforms that originated from SMN2 splicing, ethanol precipitated PCR products were subjected to overnight DdeI digestion as previously described (44). Multi-exon-skipping detection assay (MESDA) was carried out as described previously (55–58), using primers located in exon 1 and exon 8 of SMN (56), where the reverse primer was labeled at the 5′-end with [γ-32P] ATP (6000 Ci/mmol, Perkin-Elmer). Splicing products were resolved on a 6% native polyacrylamide gel. Analysis and quantifications of splice products were performed using a FPL-5000 Image Reader and Multi Gauge software (Fuji Photo Film Inc). For amplification of splice products generated from the FLAG-SMN1G1C and FLAG-SMN1 expression vectors, cDNA was generated using 1 μg of total RNA in a 5 μl reaction and splice products amplified by PCR using primers located in 3XFLAG-tag region and the beginning of exon 8. PCR products were separated on a 5% native acrylamide gel and visualized by ethidium bromide staining. Gel images were obtained using a UVP BioSpectrum AC Imaging System (UVP).
Identification of novel splice isoforms
Bands of interest corresponding to different splice isoforms were excised from a 5% or a 6% native gel, and the DNA was recovered using the ‘crush and soak’ method (44) followed by ethanol precipitation. The recovered products were then cloned in a pGEM-T easy vector (Promega) following the manufacturer's recommendations. Recombinant clones were identified by white/blue colony screening on indicator plates. Several clones were randomly selected for each splice variant. Clones were purified using the QIAprep Spin Miniprep Kit (Qiagen) and sequenced.
Generation of SMN protein expression vectors
Human FLAG-SMN1 and FLAG-SMN1G1C expression vectors were generated as follows. 3XFLAG-tagged SMN cDNA sequence from exon 1 to exon 5 was ligated to the sequence containing exon 6, exon 7, exon 8 and the intervening intronic sequences using a multistep PCR approach in which the 3XFLAG-SMN expression vector (58) and the SMN1 /SMN1G1C minigenes served as templates. The final PCR product was comprised of 3XFLAG tag followed by an in-frame SMN cDNA sequence from exon 1 to exon 6 followed by a shortened version of intron 6 (52), the entire exon 7, the entire intron 7 and 464 nucleotides of exon 8. This PCR fragment was cloned into pCI expression vector digested with NotI and XhoI. The identity of FLAG-SMN1 and FLAG-SMN1G1C constructs was confirmed by sequencing.
Western blot analysis
In hnRNP A1/A2 depletion experiments whole-cell lysates from HeLa cells were prepared using ice-cold radioimmunoprecipitation assay (RIPA) buffer (Boston BioProducts) supplemented with Halt Protease Inhibitor Single-Use cocktail (Thermo Scientific). Protein concentrations were determined using the Bio-Rad protein assay (Bio-Rad). Protein samples (15 μg per lane) were resolved on 10% SDS-polyacrylamide gels, transferred to membranes (Immun-Blot PVDF Membrane for Protein Blotting, Bio-Rad) using the Transfer-Blot Turbo Transfer System (Bio-Rad), and blocked with 5% nonfat milk prepared in Tris-buffered saline containing 0.05% Tween-20 (TBST). After incubation with primary antibodies, membranes were rinsed in TBST at least three times (10 min each) and incubated with the appropriate secondary antibody. The following primary and secondary antibodies were used for Western blot analysis: mouse monoclonal anti-hnRNP A1 (1:5000), clone 9H10 (Abcam; ab5832), mouse monoclonal anti-hnRNP A2B1 (1:1000), clone DP3B3 (Abcam; ab6102), mouse monoclonal anti-GAPDH (1:4000), clone 6C5 (Abcam; ab8245), rabbit polyclonal anti-β-actin (1:2000, Sigma-Aldrich; A2103), mouse monoclonal anti-α-tubulin (1:4000, Sigma-Aldrich; T6199), goat anti-mouse horseradish-peroxidase-conjugated antibody (1:4000, Jackson Immunoresearch; 115-035-003) and donkey anti-rabbit horseradish-peroxidase-conjugated antibody (1:2000, GE Healthcare; NA934V). Membranes were re-probed for proteins of interest following stripping with Restore Western Blot Stripping Buffer (Thermo Scientific). To analyze expression of the FLAG-tagged SMN proteins, HeLa cells were harvested, and cell lysates prepared similarly as previously described (39,44). One-eleventh of each lysate was used for one blot. Protein samples were resolved on 14% SDS-polyacrylamide gels and transferred on PVDF membranes (Immun-Blot PVDF Membrane for Protein Blotting, Bio-Rad) using the Transfer-Blot Turbo Transfer System (Bio-Rad). Membranes were blocked with 5% nonfat milk prepared in TBST followed by incubation with horseradish-peroxidase-conjugated anti-FLAG antibody (1:4000), clone M2 (Sigma-Aldrich). After visualizing the results, membranes were stripped and re-probed for β-actin using the primary and secondary antibodies listed above. Proteins were visualized using Clarity Western ECL Blotting substrate (Bio-Rad), or SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific). Membranes were scanned using a UVP BioSpectrum AC Imaging System (UVP). Results were confirmed by at least two independent experiments.
Computational analysis
The strength of the 5′ and 3′ splice sites was determined using MaxEntScan scoring algorithm (http://genes.mit.edu/cgi-bin/Xmaxentscan_scoreseq.pl) (59). The 5′ splice sites were also scored using the HBond score web interface, version 3.4 (http://www2.hhu.de/rna/html/hbond_score.php). CRYP-SKIP prediction algorithm was used to determine whether a splicing mutation of interest would result in exon skipping or activation of cryptic/di novo splice sites (60).
RESULTS
Engineered U1 snRNAs (eU1s) suppress exon 7 skipping from SMN1 carrying a lethal splice site mutation
To examine the effect of the lethal G1C mutation, we generated the SMN1G1C minigene, in which the G residue at the first position of intron 7 of SMN1 is replaced with a C. Reproducing the splicing pattern in patient tissues (50), the SMN1G1C minigene showed complete skipping of exon 7 in different cell types including HeLa, neuronal SH-SY5Y and mouse motor neuron-like NSC34 cells (Supplementary Figure S1). We conducted all subsequent studies in HeLa cells. To test whether splicing of SMN1G1C exon 7 could be modulated, we generated a library of eU1s predicted to restore base pairing with the +1C position of intron 7 in addition to forming varying degree of complementarity with the G1C-5′ss (Figure 1B). We then co-transfected cells with the SMN1G1C minigene and each eU1 and determined the splicing pattern of SMN1G1C exon 7 ∼24 h post transfection. Overexpression of all eU1s that formed a strong RNA:RNA duplex comprised of 19 or more hydrogen bonds with the G1C-5′ss substantially decreased exon 7 skipping and promoted intron 7 retention (Figure 1C). As expected, wtU1 had no effect on splicing of SMN1G1C exon 7 (Figure 1C). Surprisingly, in the presence of eU1s we observed three novel bands. Cloning and sequencing of these bands confirmed the activation of two cryptic 5′ss (Cr1 and Cr2) within intron 7 and one cryptic 5′ss (E6Cr) within exon 6 (Figures 1A, C and Supplementary Figure S2). Of note, most eU1s we employed had poor complementarity with the cryptic sites (Supplementary Figure S3). Hence, RNA:RNA duplexes formed by the eU1s at the G1C-5′ss appeared to be the sole driving force behind the activation of these cryptic splice sites. While usage of the E6Cr has been previously reported in hybrid SMN1/SMN2 minigenes (18), Cr1 and Cr2 are novel sites, activation of which will increase the size of exon 7 by 23 and 51 nucleotides (nts), respectively (Figure 1A). Since the stop codon is located within exon 7, usage of Cr1 or Cr2 will result in generation of splice products that retain the ORF for the synthesis of full-length SMN.
The Cr2 sequence motif is closest to the consensus 5′ss sequence, since this site contains -1G, +5G and +6U residues. Yet, Cr1 was used the most in SMN1G1Ctranscripts. To better understand the mechanism of cryptic splice site selection, we employed various algorithms to analyze the strengths of all putative cryptic splice sites within the last 105 nts of intron 6, the entire exon 7, and the first 99 nts of intron 7. We began our analysis with the CRYP-SKIP prediction algorithm, which determines whether a splicing mutation will result in exon skipping or activation of a cryptic/di novo splice site (60). This algorithm uses a scale of 0 to 1, with higher values indicating favorability for usage of the cryptic site. The analysis revealed the probability of a cryptic splice site activation (PCR-E) versus exon 7 skipping for SMN1 and SMN2 as 0.39 and 0.12, respectively. CRYP-SKIP algorithm failed to predict activation of either Cr1 or Cr2, while it predicted a cryptic 5′ss within exon 7 (GU dinucleotides at positions 30 and 31). We next analyzed cryptic splice site strengths using MaxEntScan scoring algorithm developed by Yeo and Burge (59). The algorithm simultaneously analyses 9-mers, taking into account both non-adjacent and adjacent dependencies between positions within RNA regulatory sequences, such as the 5′ss. The wt-5′ss of exon 7 was scored as the strongest followed by E6Cr, then Cr2 and finally Cr1 (Supplementary Table S2). Since MaxEntScan also failed to predict Cr1 as a strong site, we infer several factors in addition to base pairing with the U1 snRNP contribute towards the favored selection of Cr1.
Intron 7 retention was the major splicing event in the presence of the G1C-5′ss-targeting eU1s. We observed an inverse correlation between intron 7 retention and exon 7 skipping, suggesting that binding of eU1 to the G1C-5′ss helps define the 3′ss of exon 7 and leads to intron 6 splicing. The relative amount of intron 7-retained product varied with different eU1s, with the highest observed in the presence of eU1-M1C-11 that formed the longest 11-base pair (11-bp) duplex with the G1C-5′ss (Figures 1B and C). Overall, the extent of continuous base pairing between eU1 and G1C-5′ss appeared to correlate with the observed effect on splicing. Consistently, eU1-M1C-6 that formed a 6-bp duplex with the G1C-5′ss completely lost the ability to prevent exon 7 skipping (Figure 1C). However, we observed no correlation between eU1:G1C-5′ss duplex size and activation of the cryptic splice sites. For example, eU1-M1C-10A, eU1-M1C-9B, eU1-M1C-9C and eU1-M1C-8A showed very similar levels of cryptic site usage despite variations in the size of eU1:G1C-5′ss duplexes they formed (Figures 1B and C). The effect on SMN1G1C exon 7 splicing was noticeably different between six eU1s that formed an 8-bp duplex with the G1C-5′ss. While eU1-M1C-8A and eU1-M1C-8B substantially prevented exon 7 skipping, eU1-M1C-8C had only a weak effect on SMN1G1C exon 7 splicing (Figures 1B and C). Of note, eU1-M1C-8A and eU1-M1C-8C target the same sequence; however, eU1-M1C-8C forms one less hydrogen bond compared to eU1-M1C-8A (Figure 1B). This is due to the formation of the wobble and Watson-Crick base pair by eU1-M1C-8C and eU1-M1C-8A with the G residue at the -2 position of the G1C-5′ss, respectively (Figure 1B). Hence, our findings represent a drastic change in the outcome of splicing due to the mere difference in a single hydrogen bond formed between a U1 snRNA and its 5′ss target.
Effect of eU1s targeting intronic GU residues downstream of the G1C-5′ss
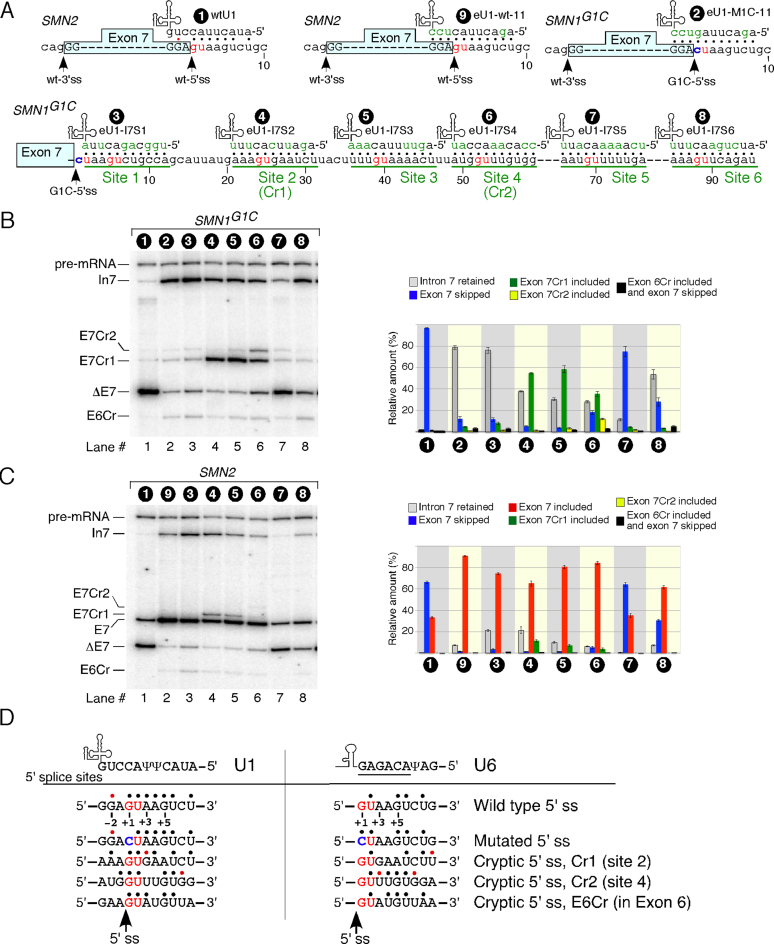
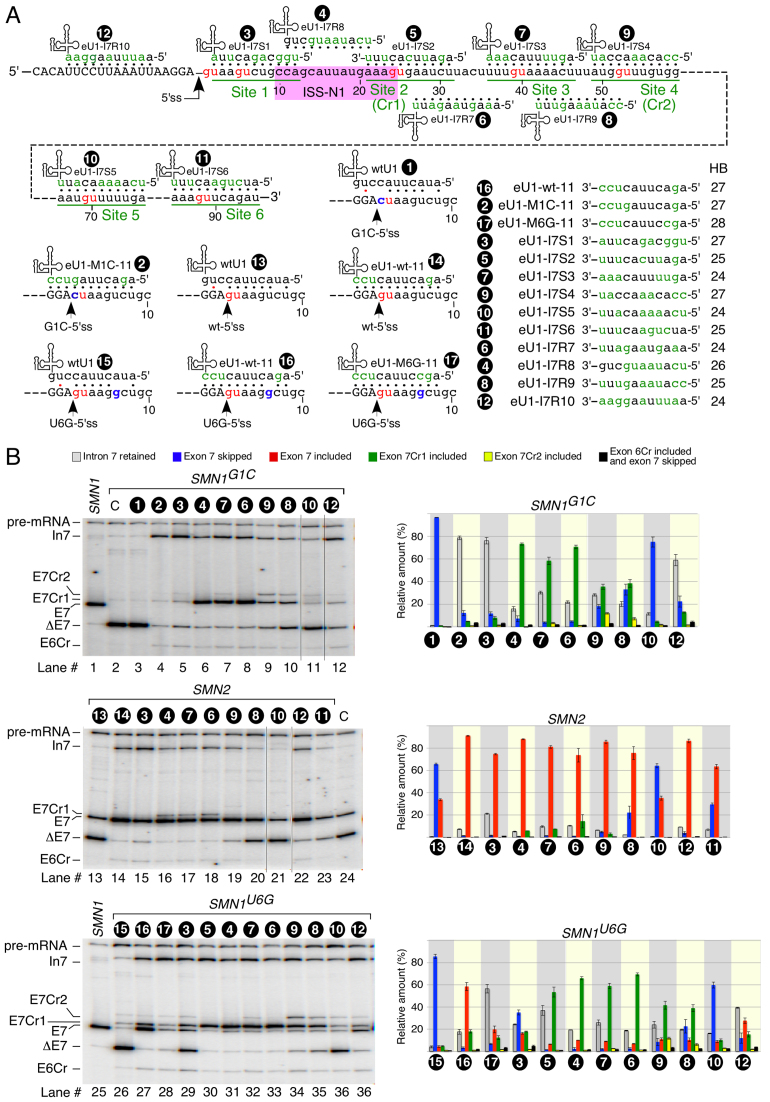
In search of an additional cryptic 5′ss that could be activated downstream of the G1C-5′ss, we employed eU1s that targeted six 5′ss-like sequences harboring a GU dinucleotide (sites 1 through 6) located within the first 90 nucleotides of intron 7 (Figure 2A). We refer to these eU1s as S series (splice-site series) eU1s. They were designed to form an 11-bp duplex with their respective targets, which encompass three nucleotides upstream and six nucleotides downstream of a GU dinucleotide. We used wtU1 and eU1-M1C-11 as controls. Note that sites 2 and 4 correspond to Cr1 and Cr2, respectively (Figures 1A and 2A). As per MaxEntScan algorithm, the strength of Cr2 was greater than Cr1 followed by the rest of the sites (Supplementary Table S2). The HBond algorithm, which scores 5′ss based on the hydrogen bonds of the RNA:RNA duplex (formed between U1 snRNA and the 5′ss) also ranked Cr2 the highest, followed by site 3 and then Cr1 (Supplementary Table S3). The majority of the S series eU1s we employed prevented exon 7 skipping and Cr1 was the most commonly used 5′ss when the six downstream sites were targeted by eU1s (Figure 2B). Both eU1-I7S2 (engineered U1 against Intron 7 Site 2) and eU1-I7S3 that annealed to sites 2 and 3, respectively, produced the highest levels of exon 7 inclusion generated by the activation of Cr1 (Figure 2B). Interestingly, eU1-I7S4 that directly targeted Cr2 activated Cr2 ∼3 times less efficiently than Cr1 (Figure 2B). Also, eU1-I7S4 was less efficient in preventing exon 7 skipping compared to eU1-I7S3. Note that the annealing regions of these two eU1s differ by only 5 nts. In addition, eU1-I7S4 is predicted to form 27 hydrogen bonds with its target versus 24 predicted for eU1-I7S3. Among six intron 7-targeting eU1s, eU1-I7S5 was the least effective in preventing skipping of exon 7 (Figure 2B). Interestingly, eU1-I7S6 that annealed to the most downstream site 6 was more effective in preventing skipping of exon 7 compared to eU1-I7S5, although it was due to increased intron 7 retention rather than the activation of Cr1 or Cr2 (Figure 2B). We also examined whether eU1s could activate Cr1 and/or Cr2 in transcripts derived from endogenous SMN1 and SMN2. Indeed, we captured small but detectable levels of Cr1 activation in both SMN1 and SMN2 (Supplementary Figure S4). These results suggested that the activation of Cr1 is not an artifact of minigenes.
Figure 2.
Effect of eU1s that target ‘GU’ sites within intron 7 on splicing of exon 7. (A) Diagrammatic representation of eU1s and their annealing sites within intron 7. The GU dinucleotides are indicated by red letters and eU1 annealing sites (3 nts upstream and six nts downstream of each GU) are underlined. (B) In vivo splicing pattern of SMN1G1C minigene in the presence of overexpressed eU1s shown in (A). Quantification of the relative amount of the indicated splice products is given in the right panel as a bar diagram. Error bars represent standard error. (C) In vivo splicing pattern of SMN2 minigene in the presence of overexpressed eU1s shown in (A). Quantification of the relative amount of the indicated splice products is given in the right panel as a bar diagram. Error bars represent standard error. (D) Diagrammatic representation of potential base pairing between wild type U1 and U6 snRNAs with the indicated 5′ss. The conserved U6 hexanucleotide is underlined. GU dinucleotides are shown in red letters. Pathogenic G1C mutation is shown in blue. Base pairing is marked by black dots; red dots represent a wobble base pair. Labeling and abbreviations are the same as in Figure 1.
We next examined whether eU1s targeting intronic sequences could also activate Cr1 in the context of the wt-5′ss of exon 7 in the SMN2 minigene. As a control, we used a previously described mutant U1, eU1-wt-11, which possess perfect complementarity to the wt-5′ss exon 7 and prevents SMN2 exon 7 skipping [Figure 2C, (38)]. Except for eU1-I7S5, all eU1s annealing downstream of the wt-5′ss of exon 7 promoted inclusion of exon 7 albeit with varying degrees. While all S series eU1s promoted SMN2 exon 7 inclusion through the usage of the wt-5′ss, we observed slight but noticeable activation of Cr1 when sites 2 and 3 were directly targeted by eU1-I7S2 and eU1-I7S3, respectively. However, we did not observe activation of Cr2 by any of the eU1s employed in the context of SMN2. We noted differential effects of eU1s on exon 7 splicing between the SMN2 and SMN1G1C minigenes. For example, in SMN2 eU1-MI7S1 promoted exon 7 inclusion from the wt-5′ss more effectively than eU1-MI7S2 (Figure 2C). However, the overall exon 7 inclusion using both the wt-5′ss and Cr1 sites was comparable for these two eU1s (Figure 2C). In the context of SMN1G1C, eU1-I7S1 utilized Cr1 very inefficiently as compared to eU1-I7S2 and caused predominant intron 7 retention (Figure 2B). Furthermore, eU1-I7S4 had a markedly better effect on preventing exon 7 skipping in the context of SMN2 than in SMN1G1C (Figures 2B and C). In fact, eU1-I7S4 was among the top two eU1s that caused the highest percentage of exon 7 inclusion when co-expressed with SMN2 (Figure 2C). Levels of exon 7 inclusion promoted by eU1-I7S4 and eU1-I7S3 were comparable to that of eU1-wt-11. Of note, in the context of endogenous SMN2, the effect of overexpressed eU1-I7S3 and eU1-I7S2 was comparable to that produced by eU1-wt-11 (Supplementary Figure S4). Overall, when the wt-5′ss of exon 7 was not mutated, eU1s targeting intron 7 performed much better in promoting exon 7 inclusion predominantly through activation of the wt-5′ss. Similar to its effect on exon 7 in the context of SMN1G1C, eU1-I7S5 was unable to alter the splicing of SMN2 exon 7 (Figures 2B and C).
It has been shown that the base pair interactions between U6 snRNA and the 5′ss determine the selection of the 5′ cleavage site (61). Therefore, we compared potential base pairing of U6 snRNA with the wild, mutated and cryptic 5′ss of exon 7. We observed no clear correlation between the 5′ss usage and the strength of the base pair interactions with U6 snRNA (Figure 2D). We also examined the effect of eU1s targeting intronic sequences on splicing of exon 7 in the SMN1G1A and SMN1G1U minigenes in which the 5′ss of exon 7 was rendered inactive by G1A and G1U mutations, respectively. As with the SMN1G1Cminigene, eU1s did not activate the mutated 5′ss and eU1-I7S3 was most effective in promoting exon 7 inclusion mainly through the usage of Cr1 in both SMN1G1A and SMN1G1U (Supplementary Figure S5).
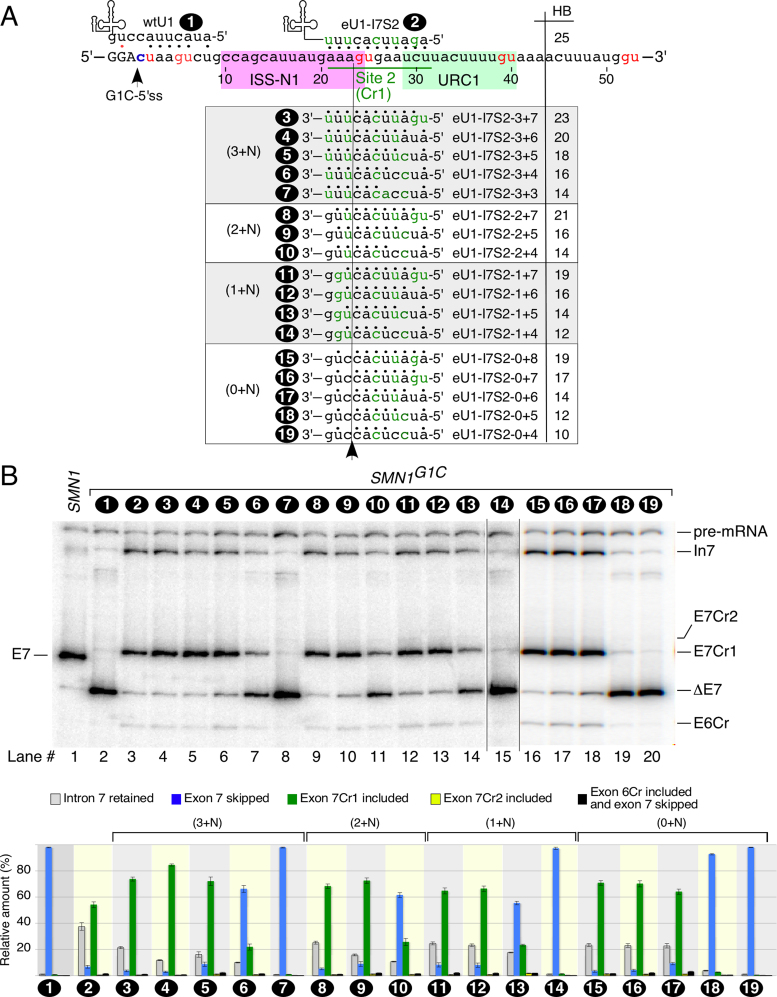
Effect of eU1:Cr1 duplex size on activation of Cr1
Having shown that Cr1 is activated by three S series eU1s that target intronic sequences downstream of the G1C-5′ss, we next asked if the size of the RNA:RNA duplex formed between an eU1 and Cr1 has an effect on the activation of Cr1. To address this question, we generated a library of eU1s that directly targeted Cr1 and for which the number of base pairs with this site ranged from six to eleven (Figure 3A). The exact annealing positions within Cr1 varied for each eU1 (Figure 3A). We co-transfected each eU1 with the SMN1G1Cminigene and monitored exon 7 splicing 24 h post transfection. Irrespective of the size of the eU1:Cr1 duplex, most eU1s rescued exon 7 skipping via Cr1 activation rather than through intron 7 retention (Figure 3B). eU1-I7S2 that formed the largest 11-bp eU1:Cr1 duplex caused the highest intron 7 retention and least Cr1 activation compared to all other ‘effective’ eU1s targeting Cr1 (Figure 3B). eU1-I7S2-3+6 that formed a 9-bp eU1:Cr1 duplex caused the highest eU1-induced exon 7 inclusion through Cr1 activation. This increase in inclusion of exon 7 by eU1-I7S2-3+6 was achieved at the expense of a reduction in intron 7 retention (Figure 3B). Overall, there appeared to be no straightforward correlation between Cr1 usage and the size/strength of eU1:Cr1 duplex. For example, both eU1-I7S2-3+6 and eU1-I7S2-2+7 formed a 9-bp duplex with Cr1, yet eU1-I7S2-3+6 performed noticeably better. Of note, eU1-I7S2-3+6 is calculated to form 20 hydrogen bonds, while eU1-I7S2-2+7 forms 21 hydrogen bonds. While a lesser number of hydrogen bonds may seem to be preferable, eU1-I7S2-3+7 (23 hydrogen bonds) activated Cr1 better than eU1-I7S2-2+7 (Figure 3B). Consistent with these results, eU1s that formed different sizes of eU1:Cr1 duplexes activated Cr1 with similar efficiency (Figure 3B), so long as they formed at least 8 continuous base pairs, with the exception of eU1-I7S2-0+6, whose ability to activate Cr1 was comparable to that of the eU1s that formed much longer duplexes with their corresponding targets within Cr1. This indicates that not only the number of continuous nucleotides involved in base pairing with U1 snRNA is important for the efficiency of the 5′ss activation/usage but also their position relative to a GU dinucleotide within this site.
Figure 3.
Effect of decreased base pairing on the ability of a Cr1-targeting eU1 to promote inclusion of exon 7 in SMN1G1C transcripts. (A) Diagrammatic representation of eU1-I7S2 variants that form different numbers of continuous base pairs with site 2 (Cr1). The eU1-I7S2 variants are grouped in four sets according to their ability to form base pairs with three nts upstream of the GU in Cr1. The names of the U1 variants as well as the number of hydrogen bonds they form with Cr1 are given. (B) In vivo splicing pattern of the SMN1G1C minigene in the presence of overexpressed eU1s shown in (A). Quantification of the relative amount of the indicated splice products is given in the bottom panel as a bar diagram. Error bars represent standard error. Labeling and abbreviations are the same as in Figure 1.
Effect of eU1s on splicing of exon 7 of SMN1 carrying mutations at the 3′ss of SMN1 exon 7
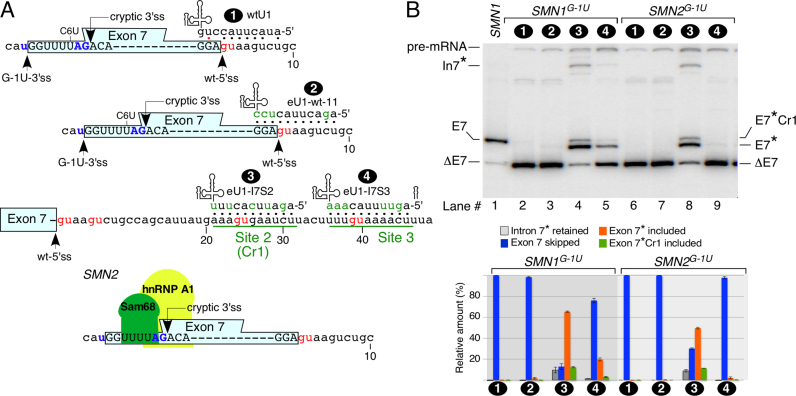
To determine if eU1s that activated Cr1 would also have a stimulatory effect on exon 7 inclusion in the context of an abrogated 3′ss, we generated the SMN1G-1Uand SMN2G-1Uminigenes that carried a G-to-T mutation (G-1U) at the last position of intron 6 (Figure 4A). We co-transfected these minigenes with Cr1-activating eU1s (eU1-I7S2 and eU1-I7S3) and determined the splicing of exon 7 ∼24 h post transfection. As controls, we used wtU1 and eU1-wt-11 that formed 6-bp and 11-bp duplexes at the wt-5′ss of exon 7, respectively. As expected, both SMN1G-1U and SMN2G-1U showed complete skipping of exon 7 (Figure 4B). However, eU1-I7S2 that directly targeted Cr1 rescued exon 7 inclusion predominantly through the usage of the wt-5′ss and the activation of a cryptic 3′ss (E7-3′Cr) located within exon 7 in both SMN1G-1U and SMN2G-1U transcripts (Figure 4B). These results confirmed that an eU1 annealing to a downstream intronic sequence away from a functional 5′ss can define both the 3′ss and 5′ss of an upstream exon. However, there appeared to be a distance limitation for this type of mechanism of action, since eU1-I7S3 that targeted sequences downstream of Cr1 turned out to be less effective in promoting exon 7 inclusion in the context of SMN1G-1U and failed to do so in SMN2G-1U transcripts (Figure 4B). The predicted strengths of E7-3′Cr were very similar for both SMN1 and SMN2 (Supplementary Table S2). Hence, the inability of eU1-I7S3 to promote exon 7 inclusion in the context of SMN2G-1U could be due to several reasons. For example, E7-3′Cr is partially sequestered by terminal stem loop 1 (TSL1) that is strengthened by the SMN2-specific C6U mutation within exon 7 (52). Further, C6U has been proposed to create binding sites for Sam68 and hnRNP A1, which are negative regulators of SMN2 exon 7 splicing [Figure 4A, (62,63)]. Hence, it is likely that the strengthened TSL1 and/or interactions of Sam68 and hnRNP A1 within SMN2 exon 7 renders the E7-3′Cr inaccessible to the spliceosomal machinery.
Figure 4.
Effect of eU1s on splicing of exon 7 with a mutated 3′ ss of SMN exon 7. (A) Diagrammatic representation of eU1s and their target sites within SMN transcripts carrying the mutated 3′ss of exon 7. Labeling is the same as in Figure 2A. Splice sites are indicated by arrows. The binding sites for Sam68 and hnRNP A1, known to promote skipping of exon 7 (62,63), are highlighted (bottom diagram). (B) In vivo splicing pattern of the SMN1G-1U and SMN2G-1U transcripts in the presence of overexpressed eU1s shown in (A). Labeling is the same as in Figure 1C. Quantification of the relative amount of the indicated splice products is given in the bottom panel as a bar diagram. Error bars represent standard error. Asterisks indicate splice products in which the cryptic 3′ss in exon 7 was used for splicing. Labeling and abbreviations are the same as in Figures 1 and 2.
eU1s rescue aberrant splicing caused by a variety of mutations associated with SMA
We expanded our study to examine the effect of eU1s on splicing of exon 7 in the SMN1U6G, SMN1ΔPy and SMN1Δ4-5′ssminigenes encompassing three different pathogenic mutations associated with SMA. SMN1U6Grepresented a SMA type 3 patient with a T-to-G mutation at the sixth position (U6G in RNA) of intron 7 of SMN1 [Figure 5A, (64)]. Of note, while U6G-5′ss is predicted to be weaker than the wt-5′ss of exon 7, it scored better than Cr1 (Supplementary Tables S2 and S3). SMN1ΔPyrepresented a SMA type 1 patient with a 7-nt deletion in the polypyrimidine tract (PPT) upstream of the 3′ss of SMN1 exon 7 [Figure 5A, (27)]. SMN1Δ4-5′ssrepresented a SMA type 2 patient with a 4-nt deletion from the fourth to seventh positions of intron 7 of SMN1 [Figure 5A, (27)]. Similar to U6G-5′ss, Δ4-5′ss is weaker than wt-5′ss but is predicted to be stronger than Cr1 (Supplementary Tables S2 and S3). As expected, all the SMN1 minigenes harboring pathogenic mutations showed massive skipping of exon 7 (Figure 5B). For SMN1U6G and SMN1Δ4-5′ssthat carried mutations within the 5′ss, we also observed usage of Cr1 and to a lesser degree Cr2 and E6Cr (Figure 5B, lanes 9 and 23). While usage of the U6G-5′ss was significantly decreased, we did not detect usage of Δ4-5′ss (Figure 5B). We next compared the effect of the most effective S series eU1s on splicing of exon 7 of the SMN1 minigenes carrying the above-mentioned pathogenic mutations. In the case of SMN1U6G, the annealing position of eU1 dictated which 5′ss would be used. For example, while eU1-wt-11 promoted usage of the U6G-5′ss, eU1-I7S2 and eU1-I7S3 predominantly activated Cr1 (Figure 5B, compare lane 11 to lanes 13 and 14). In the case of SMN1ΔPy, both eU1-I7S2 and eU1-I7S3 prevented SMN1ΔPy exon 7 skipping as well as promoted exon 7 inclusion predominantly through the usage of the wt-5′ss of SMN1ΔPyexon 7 (Figure 5B, lanes 20 and 21). However, eU1-I7S2 also activated Cr1 to a detectable level (Figure 5B, lane 20). Interestingly, despite its perfect complementarity to the wt-5′ss, eU1-wt-11 was less effective in promoting inclusion of SMN1ΔPy exon 7 (Figure 5B, lane 19). This could be due to close proximity of its annealing site to the 3′ss of exon 7. In the context of SMN1Δ4-5′ss, most U1s predominantly activated Cr1 (Figure 5B). eU1-ΔM4-11 that annealed to the Δ4-5′ss was an exception as its overexpression mostly produced intron 7-retained transcripts (Figure 5B, lane 26).
Figure 5.
Effect of eU1s on splicing of transcripts generated by SMN1 minigenes carrying pathogenic mutations. (A) Diagrammatic representation of eU1s and their targets within SMN1 carrying SMA-associated splicing mutations. Labeling is the same as in Figures 1B and 2A. (B) In vivo splicing pattern of the SMN1 mutant minigenes in the presence of overexpressed eU1s shown in (A). Labeling is the same as in Figure 1C. Quantification of the relative amount of the indicated splice products is given in the right panel as a bar diagram. Error bars represent standard error. Labeling and abbreviations are the same as in Figures 1 and 2.
Effect of eU1s that target random sequences on SMN exon 7 splicing
We next examined the effect of what we called R series (random series) eU1s that targeted random sequences downstream and upstream of the 5′ss of exon 7 on splicing of this exon in the context of the SMN2, SMN1G1C and SMN1U6G minigenes (Figure 6A). The targets of R series eU1s lacked GU residues that are essential for the usage of the 5′ss in human U2 type introns. Similar to the S series eU1s, all R series eU1s were designed to form an 11 bp duplex with their respective targets. In the case of SMN1G1C, eU1-I7R7 (engineered U1 against Intron 7 Random sequence 7) and eU1-I7R8 that annealed to sequences immediately downstream and upstream of Cr1, respectively, produced the best effect on activation of Cr1 among all eU1s we used, including S series eU1s (Figure 6B, lanes 6 and 8). There appeared to be no direct correlation between the number of hydrogen bonds and the efficacy of a given eU1, since eU1-I7R7 and eU1-I7R8 showed similar efficacy in Cr1 activation despite forming 24 and 26 hydrogen bonds with their respective targets (Figure 6). In the context of SMN1G1C, eU1-I7R10 that annealed to exon 7 showed a high propensity for intron 7 retention rather than Cr1 activation (Figure 6B). This effect was similar to the one produced by U1s that targeted the mutated 5′ss or site 1. However, eU1s that annealed downstream of site 1 effectively activated Cr1. At the same time, while eU1s targeting sequences downstream of site 3 prevented SMN1G1Cexon 7 skipping to varying degrees, they were not as effective as those targeting site 3 or the sequences upstream of it (Figure 6B). To determine the effect of duplex size formed between a R series eU1 and its target on Cr1 activation, we employed a library of eU1-I7R7 variants with different degree of complementarity to their target site in the context of the SMN1G1C minigene. All eU1-I7R7 variants that formed duplexes of 8 bp or greater activated Cr1 with comparable efficiency (Supplementary Figure S6). eU1-I7R7-D-8 was an exception. In fact, our results suggested that in addition to the duplex size, positions of nucleotides at the 5′ end of eU1 involved in target recognition played an important role in Cr1 activation. For example, eU1-I7R7-D-8 and eU1-I7R7-E-8 are designed to anneal to the same sequence within intron 7, and yet the latter eU1 performed significantly better in Cr1 activation (Supplementary Figure S6, lanes 17 and 19).
Figure 6.
Ability of different eU1s to promote inclusion of exon 7 in transcripts generated by various SMN minigenes. (A) Diagrammatic representation of eU1s and their targets. All eU1s formed 11 base pairs with their corresponding targets. (B) In vivo splicing pattern of the indicated minigenes in the presence of overexpressed eU1s shown in (A). Quantification of the relative amount of the indicated splice products is given as bar diagrams (right panels). Error bars represent standard error. Labeling and abbreviations are the same as in Figures 1 and 2.
In the context of SMN2, all eU1s that we employed effectively promoted exon 7 inclusion utilizing the wt-5′ss; however, we also observed some activation of Cr1 by eU1-I7R7, eU1-I7R8 and eU1-I7S3 that annealed in the vicinity of Cr1 (Figure 6B, lanes 16–18). The efficiency of eU1-I7R7, eU1-I7R8, eU1-I7R9, eU1-I7S3, eU1-I7S4 and eU1-I7R10 in activation of the wt-5′ss was comparable to that of the eU1-wt-11. Of note, employing SMN2 minigene, we also tested the efficacy of eU1s that targeted the predicted cryptic splice site within exon 7 or sequences more than 100 nts downstream of the wt-5′ss of exon 7. None of these eU1s showed a stimulatory effect on SMN2 exon 7 splicing (not shown). We observed somewhat different effects of eU1s on splicing of exon 7 of SMN1U6G in which most eU1s promoted exon 7 inclusion through activation of Cr1 (Figure 6B). The exceptions were eU1-M6G-11 and eU1-I7R10 that annealed to the U6G-5′ss and the upstream sequence within exon 7, respectively. These two eU1s mostly favored intron 7 retention followed by usage of U6G-5′ss. Another engineered U1, eU1-I7S1, activated both U6G-5′ss and Cr1 sites to a comparable degree (Figure 6B, lane 29). Note that eU1-I7S1 annealed to the region that overlapped the U6G-5′ss; however, the complementarity of eU1-I7S1 with its target site is interrupted due to the U6G mutation. eU1s that annealed in the immediate vicinity of Cr1, such as eU1-I7R7 and eU1-I7R8, activated Cr1. The intensity of the band corresponding to the usage of Cr2 was significantly decreased in the presence of these eU1s (Figure 6B, lanes 31 and 33). Similarly, overexpression of eU1s that annealed downstream of Cr1, such as eU1-I7S3, eU1-I7S4 and eU1-I7R9 also promoted the usage of Cr1. In addition, eU1-I7R9 and eU1-I7S4 that targeted site 4 (Cr2) activated Cr2 to some degree (Figure 6B, lanes 34 and 35). Overall, our results show that in the context of several competing 5′ss (as in the case of SMN1U6G exon 7), the proximity of eU1 annealing determines which site will be used.
Point mutations activate Cr1 and Cr2
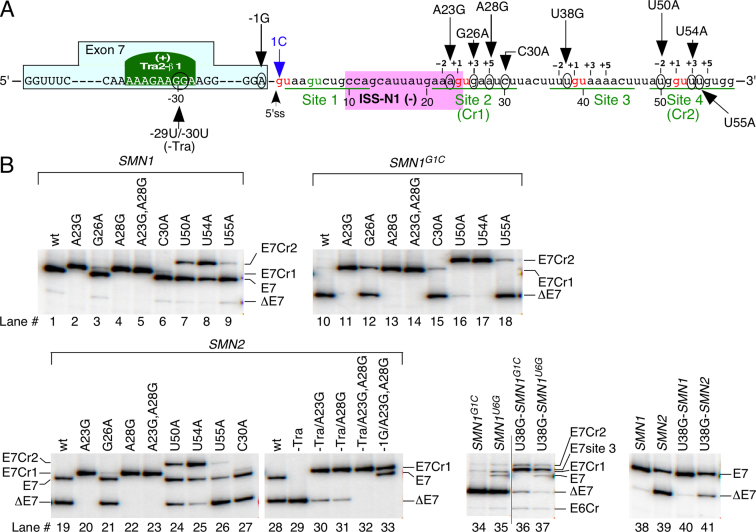
We next tested if strengthening of the cryptic splice sites will enhance their usage in the context of SMN1, SMN2 and SMN1G1C minigenes. We made four point mutations (A23G, G26A, A28G and C30A) and one double mutation (A23G/A28G) in Cr1 and three point mutations (U50A, U54A and U55A) in Cr2 (Figure 7A). All mutations increased complementarity with wtU1. Among all mutations within Cr1 and Cr2, A28G and U54A strengthened their corresponding sites the most (Supplementary Tables S2 and S3). Depending on the minigene used, we observed different effects of mutations on splicing of exon 7 (Figure 7B). While G26A, C30A and U55A did not significantly change the splicing of any minigenes, A23G, A28G and U54A all produced a strong splicing switch. For example, A23G and A28G mutations resulted in complete inclusion of exon 7 through activation of Cr1 exclusively, in both SMN1 and SMN2 transcripts (Figure 7B, lanes 2, 4, 20 and 22). Of note, A23G and A28G extended the size of Cr1:endogenous U1 duplex from 4 to 6 continuous base pairs, which is the same as the size of the duplex formed between endogenous U1 and the wt-5′ss of exon 7. Yet, usage of the latter 5′ss was completely abolished in the presence of A23G or A28G substitutions. Compared to point mutations within the Cr1 site, the U50A and U54A substitutions had a lesser effect on activation of Cr2 in the context of the SMN1 and SMN2 minigenes (Figure 7B). This could be due to the fact that the duplex formed between endogenous U1 and Cr2 carrying either U50A or U54A remained relatively short. However, in the context of the SMN1G1C minigene, both substitutions resulted in almost complete inclusion of exon 7 though activation of Cr2 (Figure 7B, lanes 16 and 17).
Figure 7.
Effect of site-specific mutations on usage of the wt and cryptic 5′ss of SMN. (A) Diagrammatic representation of site-specific mutations within exon 7 and/or intron 7 of SMN. Exon 7 and intron 7 nucleotides are shown in capital and lower-case letters, respectively. Positive and negative numbering starts from the first position of intron 7 and the last position of exon 7, respectively. The location and identity of each mutation is indicated. The GU dinucleotides are shown in red letters. GU-containing sites within intron 7, including site 2 (Cr1) and site 4 (Cr2), are underlined. The 5′ss is indicated by an arrow. ISS-N1 and Tra2-β1 binding site are highlighted with a pink and green boxes, respectively (37,37). (+) and (–) indicate that an element promotes inclusion and skipping of exon 7, respectively. (B) In vivo splicing pattern of the indicated minigenes carrying site-specific mutations as compared to the wild type constructs. Mutations are marked at the top of the gel. Abbreviations: E7 site 3, exon 7-included from the activated site 3. The rest of the labeling and abbreviations are the same as in Figure 1.
It has been previously shown that mutations within the Tra2 exonic splicing enhancer (ESE) cause massive skipping of exon 7 (38). Interestingly, A23G and A28G mutations within intron 7 rescued the inclusion of exon 7 in SMN2 minigene carrying a mutation that abrogated Tra2 ESE (Figure 7B, lanes 29–32). Earlier we reported that A to G substitution at the last position of exon 7 (-1G substitution) confers one of the strongest stimulatory effects on SMN2 exon 7 inclusion through the enhanced usage of the 5′ss (37). When -1G substitution was combined with the intronic A23G/A28G mutations in the SMN2 minigene, we observed an increase in exon 7 inclusion through the usage of both 5′ss and Cr1 (Figure 7B, lane 33). These results confirm that the strength of Cr1 carrying A23G/A28G mutations is comparable to that of the native 5′ss carrying -1G substitution. Of note, –1G produces the strongest stimulatory effect caused by a single nucleotide change within exon 7. While site 3 is not used in the context of any of the minigenes or in the presence of any eU1 we employed, a U-to-G mutation at the 38th intronic position (U38G) did activate this site resulting in predominant inclusion of exon 7, but only in the context of the SMN1G1Cand SMN1U6G transcripts (Figure 7B). In SMN2 transcripts, U38G had some stimulatory effect on exon 7 inclusion as well but through the usage of the wt-5′ss (Figure 7B, lanes 39 and 41).
Effect of mutations within ISS-N1 and URC1 on usage of Cr1
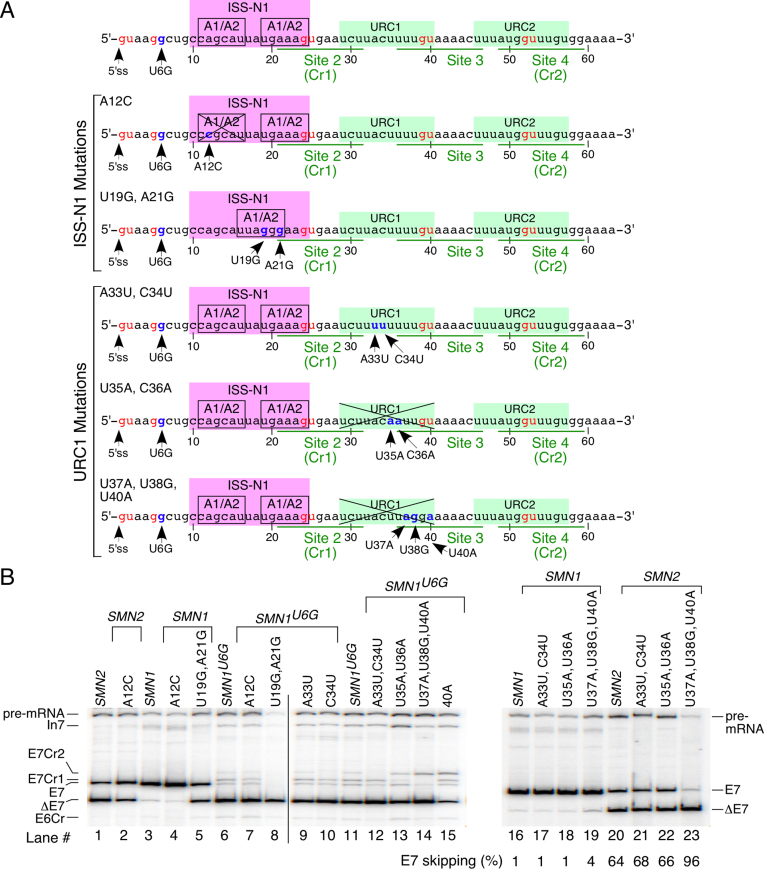
We next attempted to identify splicing regulatory elements that specifically control usage of Cr1. As mentioned above, Cr1 partially overlaps with both the inhibitory element ISS-N1 and the stimulatory element URC1 (Figures 1A and 8A). Consistent with a previous report, an A-to-C mutation at the 12th intronic position (A12C mutation) promoted SMN2 exon 7 inclusion by abrogating one of the hnRNP A1/A2 motives of ISS-N1 [Figure 8B, (43)]. However, A12C mutation did not activate Cr1 in SMN2 (Figure 8B, lane 2). Similarly, we observed no usage of Cr1 in SMN1 and SMN1U6G transcripts in the presence of A12C (Figure 8B, lanes 4 and 7), suggesting that this hnRNP A1/A2 motif is not critical for Cr1 usage in the context of either wt-5′ss or U6G-5′ss. We next introduced a U-to-G and an A-to-G substitution at the 19th and 21st positions (U19G/A21G mutation) of SMN intron 7, respectively. This double mutation created a classical UUAGGG motif that tightly interacts with hnRNP A1 (65). As expected, U19G/A21G mutation increased skipping of SMN1 exon 7 (Figure 8B, lane 5). In SMN1U6G transcripts, U19G/A21G mutation led to complete skipping of exon 7 (Figure 8B, lane 8). We did not observe specific suppression of Cr1 in the presence of U19G/A21G, instead usage of all three competing 5′ss (wt-5′ss, Cr1 and Cr2) was abrogated (Figure 8B, lane 8). These results suggest that although ISS-N1 might contribute to Cr1 suppression, it is not a specific regulator of Cr1.
Figure 8.
Effect of mutations within ISS-N1 and URC1 on Cr1 usage. (A) Diagrammatic representation of mutations within ISS-N1 and URC1. Numbering starts from the first position of intron 7. The location and identity of each mutation is indicated. GU dinucleotides are marked by red letters. GU-containing sites within intron 7 are underlined in green. Mutated bases are marked in blue and indicated with arrows. ISS-N1 with its hnRNPA1/A2 motifs and URCs 1 and 2 are highlighted with pink and green boxes, respectively. (B) In vivo splicing pattern of the indicated minigenes. The percentage of SMN exon 7 skipping was calculated as described previously (52). Labeling and abbreviations are the same as in Figures 1 and 7B.
We also investigated the extent to which natural variation in the sequence of SMN intron 7 may impact usage of Cr1 and/or Cr2. First, we aligned the sequences of SMN exon 7 and the first 100 bases of intron 7 derived from 11 mammalian species (Supplementary Figure S7). As expected, exon 7 was relatively well conserved. Surprisingly, many of the intronic sequences near the 5′ss were also conserved, especially URCs 1 and 2 (Supplementary Figure S7). Cr1 and Cr2 were not universally conserved, but were still present in the majority (7 out of 11 for each site) of species examined. Although the results of U19G/A21G mutant indicate that a strong hnRNP A1/A2 site in ISS-N1 might play a role in Cr1 suppression, this region of ISS-N1 does not appear to be well conserved in mammals (Supplementary Figure S7). Consistently, a common single nucleotide polymorphism (SNP) in humans changes the G residue at position 20 into a C, likely weakening or abrogating the hnRNP A1/A2 motif altogether (Supplementary Figure S7). Indeed, in the context of the SMN2 minigene G20C mutation causes noticeable increase in inclusion of exon 7 (Supplementary Figure S8, lane 3). However, G20C did not increase usage of Cr1, even when the SMN2 minigene carrying this mutation was co-transfected with eU1-I7S3 shown to activate Cr1 (Supplementary Figure S8, lane 6 versus lane 7). While URC1 is well conserved, there is a SNP in humans that alters this motif, changing the U at position 32 to a C nucleotide (Supplementary Figure S7). Consistent with weakening of URC1, SMN2U32C display increased skipping of exon 7 compared to the wild type SMN2 minigene (Supplementary Figure S8). Co-transfection with eU1 is able to efficiently rescue exon 7 skipping in SMN2U32C, but primarily through activation of wt-5′ss rather than Cr1 (Supplementary Figure S8).
To further dissect the potential role of URC1 in Cr1 usage, we introduced A33U and C34U mutations in SMN1, SMN2 and SMN1U6G minigenes. These mutations increased the length of the continuous U-rich tract within URC1. A33U and C34U together or separately had no effect on splicing of exon 7 in SMN1 and SMN2 transcripts (Figure 8B). However, in SMN1U6G transcripts, A33U/C34U double mutation completely suppressed usage of Cr2, and increased usage of Cr1 over the wt-5′ss, granted Cr1 was still used inefficiently (Figure 8B, lane 12 versus lane 11). In our next set of mutations, we changed pyrimidine residues to purines within URC1 based on HEXplorer score predictions. HEXplorer score profiles reflect splicing enhancing and silencing properties of sequences neighboring splice sites and is particularly helpful in analyzing the effect of mutations on splice site usage (66). In SMN1U6G transcripts, U35A/U36A mutation noticeably decreased usage of Cr1 (Figure 8B, lane 13), while U37A/U38G/U40A substitutions completely abrogated Cr1 and simultaneously increased usage of Cr2 (Figure 8B, lane 14). Activation of Cr2 was also stimulated by U40A mutation, which in addition to weakening URC1 abrogated site 3 defined by the presence of a GU dinucleotide (Figure 8B, lane 15). In the context of the SMN1 and SMN2 minigenes, none of the URC1 mutations activated a cryptic splice site. The U37A/U38G/U40A mutant displayed substantial skipping of SMN2 exon 7 (Figure 8B, lane 23). Overall, our results support a point of view that URC1 might control usage of Cr1 in the context of the U6G-mutated 5′ss but not in the context of the wt-5′ss.
Effect of moving Cr1 away from ISS-N1
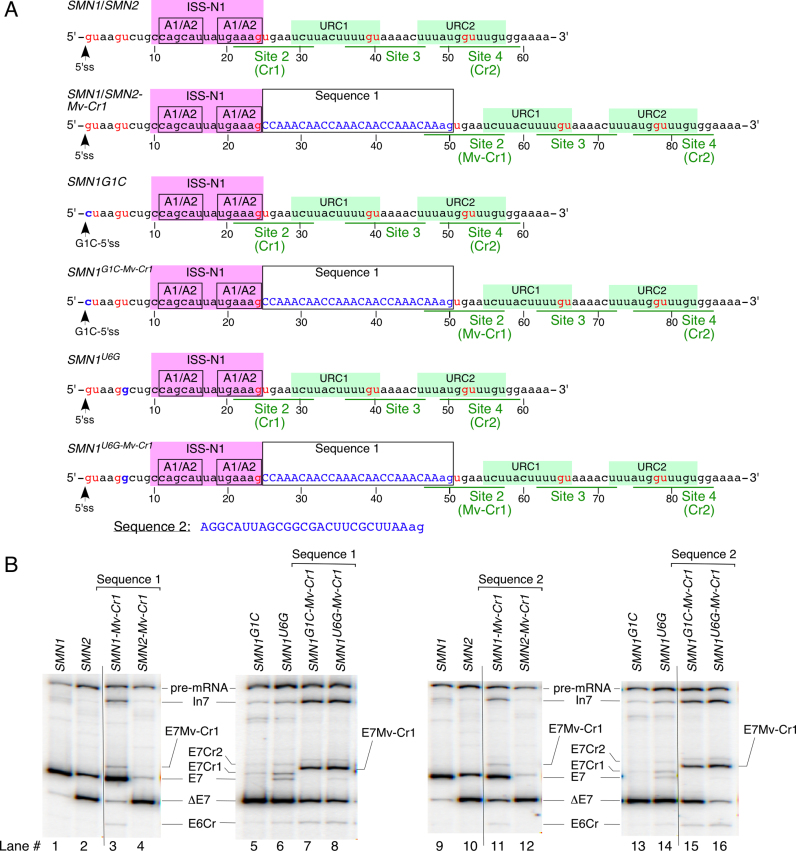
In an effort to determine if the suppression of Cr1 is linked to the presence of ISS-N1, we constructed minigenes in which we moved Cr1 away from ISS-N1 by inserting a 26-nt sequence (Sequence 1) harboring three CCAAACAA motifs (Figure 9A). Our rationale to use these particular motifs was based on its neutral effect on splicing as reported earlier (14,67,68). This insertion will also increase the distance between the 5′ss of exon 7 and URC1/URC2 that are associated with TIA1/TIAR binding (44). Insertion of Sequence 1 in SMN1 caused a small but detectable level of Cr1 activation. We also observed small increase in SMN1 exon 7 skipping and intron 7 retention (Figure 9B, lane 3). In the case of SMN2, insertion of Sequence 1 did not activate Cr1, but caused increased skipping of exon 7 (Figure 9B, lane 4). These results support that a strong 3′ss in SMN1 coupled with moving Cr1 away from ISS-N1 is conducive for Cr1 activation. Enhanced skipping of SMN2 exon 7 upon insertion of Sequence 1 could be due to a weak 3′ss of exon 7 coupled with moving of TIA1/TIAR binding sites (URC1/URC2) away from the weak 5′ss (45). Interestingly, insertion of Sequence 1 in the context of SMN1G1Cand SMN1U6G resulted in robust activation of Cr1 accompanied by a decrease in exon 7 skipping and an increase in intron 7 retention (Figure 9B, lanes 5–8). These results confirm that when Cr1 is presented in the right context it is inherently functional/capable of being activated by the endogenous level of U1 snRNP. Our results support that ISS-N1 contributes towards the suppression of Cr1 usage. As expected, we observed no effect of insertion of CA-rich Sequence 1 on activation of Cr2 (Figure 9B). To rule out the possibility that insertion of Sequence 1 created a binding site(s) for protein factors that affected splicing of exon 7, we tested an additional sequence, Sequences 2, previously employed by others as splicing neutral insert (68,69). Sequence 2 has a length comparable to Sequence 1, but its sequence composition is entirely different: it contains uridines and guanidines that are absent in Sequence 1 (Figure 9A). Insertion of Sequence 2 between ISS-N1 and Cr1 produced splicing patterns nearly identical to the ones observed for four SMN minigenes that contained Sequence 1 (Figure 9). This outcome independently confirmed that the usage of Cr1 is suppressed by ISS-N1.
Figure 9.
Effect of the distance between ISS-N1 and Cr1 on usage of Cr1. (A) Location of Cr1 within intron 7 of the indicated minigenes with respect to ISS-N1. ISS-N1 with its two hnRNP A1/A2 binding sites is highlighted by a pink box. URCs 1 and 2 are marked with green boxes. The 5′ss (wt and mutated) are indicated by arrows. Names of the minigenes are given on the right above each corresponding sequence. Sequence 1 is a 26-nt-long insertion comprised of three repeats of splicing neutral motif CCAAACAA followed by an AG dinucleotide (shown in lower case letters). Sequence 2 is a 25-nt-long insert followed by an AG (shown in lower-case letters). Each sequence was inserted immediately down stream of ISS-N1. The AG dinucleotide was added to restore Cr1. Mutations within intron 7, including the insertion, are shown in blue. The GU dinucleotides are marked by red letters. GU-containing sites within intron 7, including cryptic sites Cr1 and Cr2, are underlined. (B) In vivo splicing pattern of the indicated minigenes carrying the insertions as compared to the wt constructs. The identity of the minigenes is indicated at the top of the gels. The identity of splicing products is marked on the left and the right of the gels. Abbreviations: E7Mv-Cr1, exon 7-included where Mv-Cr1 site was used; the rest of abbreviations are described in Supplementary Table S1.
We expanded our study to examine if F14, an ISS-N1 blocking ASO, was able to activate Cr1 and rescue exon 7 inclusion in four minigenes that carry SMA-associated splicing mutations. We deliberately chose F14 because it sequesters the first 14 residues of ISS-N1 but does not block the GU dinucleotides of Cr1 (Supplementary Figure S9A). We have previously shown that F14 is very effective in restoration of SMN2 exon 7 inclusion (54). Therefore, the SMN2 minigene was used as a positive control. Increase in endogenous SMN2 exon 7 inclusion in samples transfected with the minigenes served as an internal control that F14 was indeed delivered within cells. As expected, F14 restored exon 7 inclusion in SMN2; it also decreased skipping of exon 7 in SMN1ΔPy through activation of the wt-5′ss. (Supplementary Figure S9). However, in the case of SMN1 minigenes with pathogenic mutations at the 5′ss of exon 7, F14 did not produced the expected improvement in exon 7 splicing and further decreased already inefficient usage of Cr1, Cr2 and the U6G-5′ss (Supplementary Figure S9, lanes 7, 8 and 10). These results suggest that the presence of an ISS-N1 targeting ASO interferes with one or more steps of the spliceosomal assembly at the mutated 5′ss as well as at Cr1 and Cr2.
Activation of Cr1 by eU1 is independent of the endogenous U1 snRNP
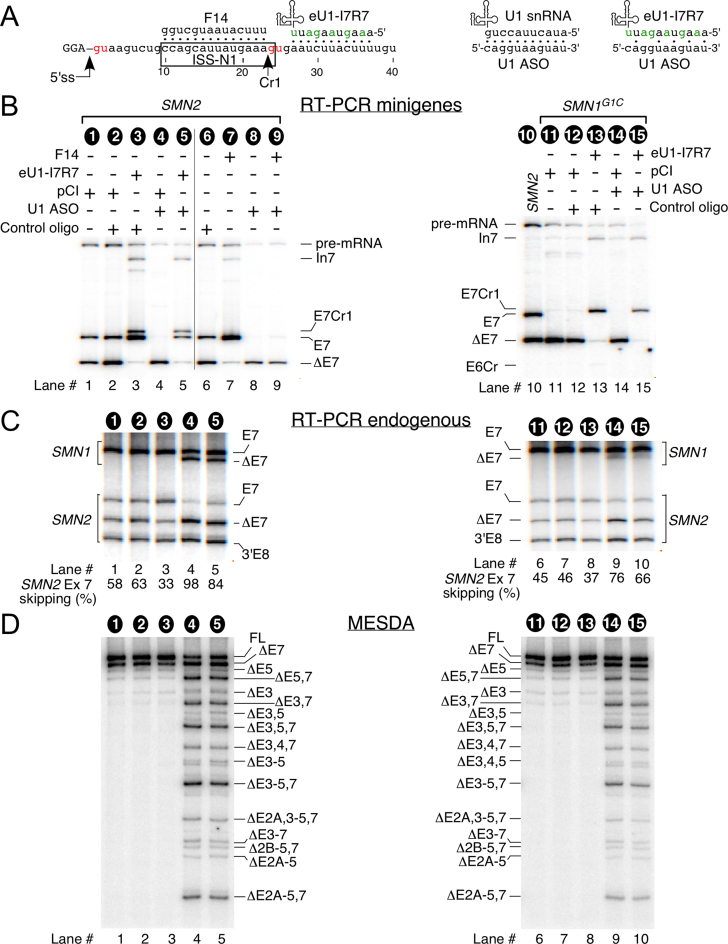
It has been recently shown that endogenous U1 snRNP is not required for an eU1-mediated activation of an authentic 5′ss (26). However, it is not known if this is also the case for the activation of a cryptic splice site. To address this question, we co-transfected SMN2 and SMN1G1C minigenes with eU1-I7R7 with or without an 11-mer U1 ASO complementary to the 5′-tail of the wtU1 (Figure 10A). Note that the annealing site of eU1-I7R7 only partially overlaps with Cr1. The U1 ASO we used has little complementarity with eU1-I7R7; hence it is unlikely to have an effect on the activity of this eU1 (Figure 10A). To monitor that the U1 ASO has inactivated endogenous U1 snRNP, in addition to splicing of exon 7 in SMN2 minigene, we also analyzed splicing of endogenous SMN1 and SMN2 exon 7 as well as splicing of all SMN exons in a single reaction by using MESDA (55). As an additional control for an efficient inactivation of endogenous U1, we assessed the ability of F14 to promote exon 7 inclusion, which was previously shown to rely on the presence of endogenous U1 (26), in the absence and presence of the U1 ASO. Similar to the previous study (26), our treatment of HeLa cells with 200 nM of the U1 ASO abrogated the stimulatory effect of F14 (Figure 10B, lane 7 versus line 9) confirming that endogenous U1 snRNA was sufficiently depleted. Furthermore, in agreement with the prevailing hypothesis that the poor recruitment of U1 snRNP at the wt-5′ss is the limiting factor for exon 7 splicing (33,34), ASO-mediated inactivation of endogenous U1 snRNP caused massive skipping of exon 7 in transcripts derived from SMN2 minigene as well as from endogenous SMN1 and SMN2 (Figure 10B, lane 4 and Figure 10C, lane 4 and 9). In addition, effective inactivation of endogenous U1 was confirmed by the results of MESDA that showed that U1 ASO treatment caused massive skipping of multiple SMN exons (Figure 10D). As expected, when SMN2 and SMN1G1C were co-transfected with eU1-I7R7, we observed Cr1 activation in transcripts derived from both minigenes (Figure 10B, lanes 3 and 13). In the case of SMN1G1C, the activation of Cr1 produced the major spliced product (Figure 10B, lane 13). When overexpression of eU1-I7R7 was combined with the U1 ASO treatment, the usage of Cr1 appears to be mostly unaffected both in SMN2 and SMN1G1C transcripts (Figure 10B, lane 5 and 15). However, eU1-I7R7-induced usage of the wt-5′ss in SMN2 transcripts was significantly decreased in the presence of the U1 ASO (Figure 10B, lane 5). Similarly, eU1-I7R7 was unable to fully prevent skipping of exon 7 from endogenous SMN transcripts when endogenous U1 was depleted (Figure 10C, lane 5 and 10). These results confirmed that eU1 could activate a cryptic splice site without the help of endogenous U1. However, contrary to the previous report (26), our results suggest that the assistance of endogenous U1 is required for an eU1-induced improvement in the usage of the wt-5′ ss of exon 7.
Figure 10.
Ability of an eU1 to activate Cr1 under conditions of the inactivation of the endogenous U1 by an ASO. (A) Diagrammatic representation of eU1 and ASOs used to affect splicing of SMN exon 7. Left panel indicates location of eU1 and F14 ASO within SMN intron 7. Right two panels indicate complementarity of U1 ASO to wtU1 snRNA and eU1-I7R7 snRNA. (B) In vivo splicing pattern of the indicated minigenes in the presence of various ASOs and eU1. Co-transfected minigenes, eU1, and ASO are indicated at the top of each gel. Splice products are indicated on the left and right of each gel. (C) In vivo splicing pattern of endogenous SMN1 and SMN2 in the presence of various ASOs and eU1. Sample identities are indicated at the top of each gel and refer to sample numbers in (B). Spliced products amplified by RT-PCR were digested with DdeI to distinguish between the transcripts originated from SMN1 and SMN2 (29). The percentage of SMN2 exon skipping was calculated as described previously (48). 3′Ex8 represents the cleavage product of DdeI digestion of SMN2 exon 8. The rest of the abbreviations are the same as in Figure 1C. (D) Multi-exon skipping detection assay (MESDA; 55–58) showing splicing pattern of all internal exons of endogenous SMN. Sample identities are indicated at the top of each gel and refer to sample numbers in (B). Identities of splicing products are indicated to the left or right of each gel. Abbreviations: FL, full-length SMN; Δ, skipped exon(s). Labeling is the same as in Figure 1. Abbreviations are described in Supplementary Table S1.
Effect of hnRNP A1/A2 depletion on activation of cryptic splice sites
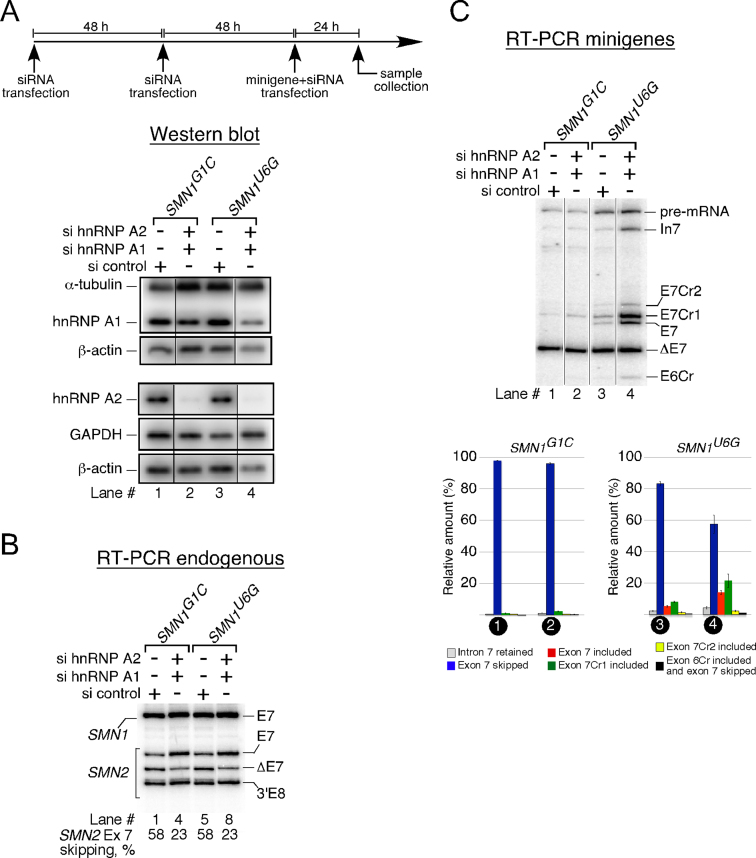
Given the presence of two hnRNP A1/A2 motifs within ISS-N1 that partially overlaps with Cr1 and, as we have shown, suppresses its usage, we asked if the siRNA-mediated depletion of these proteins would activate Cr1. In these experiments we used SMN1G1C and SMN1U6G minigenes to avoid any likely competition between Cr1 and the wt-5′ss. Of note, SMN2 contains several hnRNP A1/A2 binding sites and the depletion of hnRNP A1/A2 has been shown to reduce skipping of exon 7 from endogenous SMN2 (48). Hence, splicing of endogenous exon 7 served as a control for checking the efficiency of the siRNA-mediated depletion of hnRNP A1/A2. To ensure that robust knockdown of hnRNP A1/A2 was achieved, we treated HeLa cells with siRNAs twice before transfecting them with either SMN1G1C or SMN1U6G (Figure 11A). In the case of SMN1G1C, we did not observe a change in the splicing/usage of any of the splice sites, including Cr1, upon depletion of hnRNP A1/A2 (Figure 11C). The absence of Cr1 activation was not due to high levels of residual hnRNP A1/A2, since western blot confirmed substantial depletion of these proteins (Figure 11A). In addition, the levels of hnRNP A1/A2 knockdown were sufficient enough to result in reduced skipping of exon 7 from endogenous SMN2 in cells transfected with SMN1G1Cor SMN1U6G minigene (Figure 11B). Unlike SMN1G1C, SMN1U6G showed significant activation of Cr1 when hnRNP A1/A2 were depleted. The usage of the U6G-5′ss was increased as well (Figure 11C). We also observed small but noticeable increase in the usage of Cr2 and E6Cr (Figure 11C, lane 4 versus lane 3). These results support some role of hnRNP A1/A2 in suppression of Cr1 (at least in the context of SMN1U6G). However, this role is not specific to Cr1, since other 5′ss were activated as well. Our results also suggest that Cr1 is a stronger 5′ss as compared to the U6G-5′ss. The disparity in the splicing pattern between SMN1G1Cand SMN1U6G with respect to the activation of Cr1 under the conditions of hnRNP A1/A2 depletion was somewhat unexpected but not totally surprising. We attribute these differences to the partially functional U6G-5′ss that may recruit stimulatory factors conducive to the activation of Cr1.
Figure 11.
Effect of simultaneous depletion of hnRNP A1 and A2 on usage of Cr1. (A) Top panel shows a flow chart of the depletion experiment. Bottom panel shows western blots after transfection with the indicated siRNAs. Minigenes and siRNAs used for transfection are indicated at the top and antibodies used to probe membranes are indicated on the left of each image. (B) Splicing of endogenous SMN exon 7 in HeLa cells treated with the indicated siRNAs. Spliced products amplified by RT-PCR were digested with DdeI to distinguish between the transcripts originating from SMN1 and SMN2 (29). The percentage of SMN2 exon 7 skipping was calculated as in (48). 3′Ex8 represents the cleavage product of DdeI digestion of SMN2 exon 8. (C) Splicing pattern of transcripts generated by SMN1G1C and SMN1U6G minigenes in HeLa cells treated with the indicated siRNAs. The identity of splice products is marked on the right of the gel. Quantification of the relative amount of the indicated splicing products is given in the bottom panel as a bar diagram. Error bars represent standard error. Abbreviations are described in Supplementary Table S1.
mRNAs generated by the eU1-induced activation of Cr1 produce full-length SMN
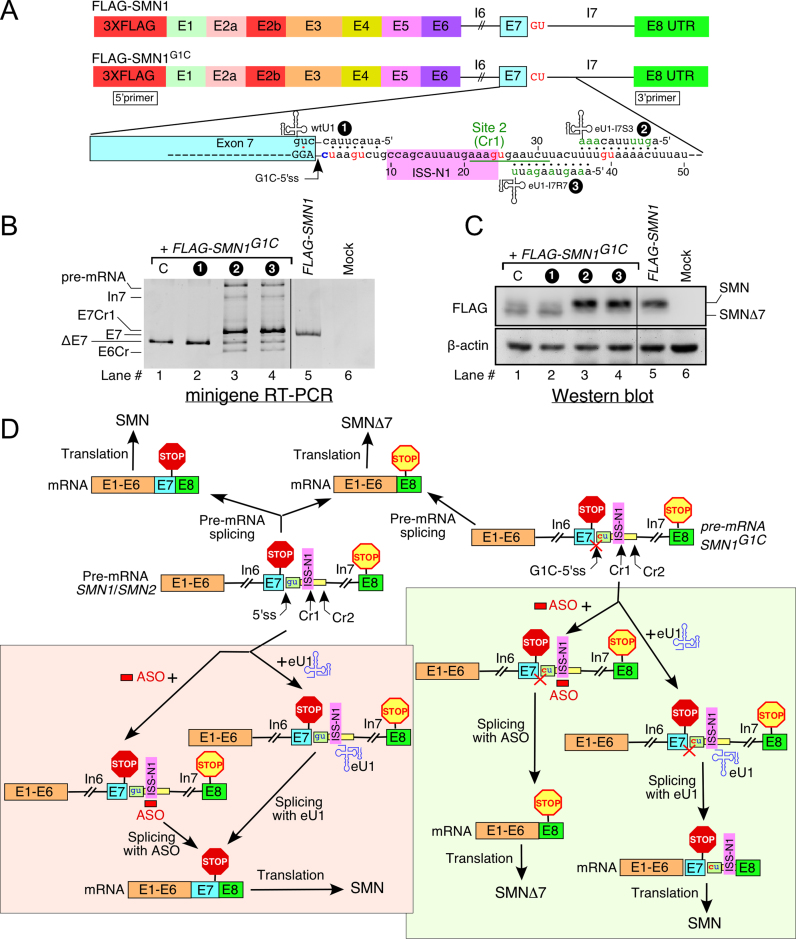
To determine if mRNAs generated by the eU1-induced activation of Cr1 can produce full-length SMN, we employed a splicing-coupled translation reporter. In particular, we constructed FLAG-SMN1 and FLAG-SMN1G1C expression vectors containing a 3XFLAG tag in-frame with the residual SMN coding sequence (cDNA corresponding to exon 1 through exon 5) placed upstream of the SMN1 and SMN1G1C minigenes, respectively (Figure 12A). Inclusion and skipping of exon 7 from these reporter constructs are expected to generate full-length SMN and SMNΔ7, respectively. We co-transfected HeLa cells with FLAG-SMN1G1Cand a Cr1-activating eU1 (eU1-I7R7 or eU1-I7S3) that annealed to sequences downstream of Cr1 (Figure 12A). We specifically chose these eU1s since they will not interfere with nuclear export and translation due to annealing to the processed mRNA. As a negative control, we used wtU1 that has no effect on splicing of SMN1G1C exon 7 due to its poor complementarity/annealing to the G1C-5′ss. In vivo splicing and protein expression from FLAG-SMN1 and FLAG-SMN1G1C constructs were tested ∼24 h post-transfection. As expected, FLAG-SMN1 produced exon 7-included transcripts (Figure 12B, lane 5). In comparison, only exon 7-skipped transcripts were generated from FLAG-SMN1G1C(Figure 12B, lanes 1 and 2). Overexpression of eU1-I7S3 or eU1-I7R7 resulted in predominant inclusion of FLAG-SMN1G1C exon 7 due to activation of Cr1 (Figure 12B, lanes 3 and 4). In the absence of Cr1 activation, low levels of SMNΔ7 were expressed from FLAG-SMN1G1C(Figure 12C, lanes 1 and 2). Of note, a low level of SMNΔ7 can be attributed to its poor stability (31). Consistent with exon 7 inclusion through Cr1 activation, we observed full-length SMN protein as the predominant product generated from FLAG-SMN1G1Cin the presence of overexpressed eU1-I7S3 or eU1-I7R7 (Figure 12C, lanes 3 and 4). FLAG-SMN1, used here as a control, also produced full-length SMN (Figure 12C, lane 5). Our results confirmed that mRNAs generated by the eU1-induced activation of Cr1 from FLAG-SMN1G1C are actively exported from the nucleus and are translationally competent.
Figure 12.
Translation of mRNAs generated by an eU1-induced activation of Cr1. (A) Diagrammatic representation of the human FLAG-SMN1 and FLAG-SMN1G1C expression vectors. Colored boxes represent 3XFLAG tag and exons, lines indicate intronic sequences. eU1s and their annealing sites within intron 7 are indicated. The G1C-5′ss is indicated by an arrow. Site 2 (Cr1) is underlined. Location of primers used for RT-PCR is indicated. Abbreviations: E, exon; I, intron; Cr1, cryptic site 1. (B) In vivo splicing pattern of FLAG-SMN1G1Cand FLAG-SMN1 expression vectors in the presence of eU1s shown in (A). PCR products were resolved on a 5% native gel and visualized by ethidium bromide staining. Sample loading was adjusted to have comparable band intensity. The identity of the SMN expression vectors and eU1 constructs used is marked at the top of the gel. The identity of splice products is indicated on the left of the gel. (C) Western blot showing the effect of the indicated eU1 snRNAs on the levels of 3X-FLAG-tagged SMN protein. The identity of the SMN expression vectors and eU1 constructs is marked at the top of the gel. Antibodies used are indicated on the left. The identity of SMN protein isoforms is marked on the right. SMN refers to the full-length SMN protein; SMNΔ7, to the truncated isoform that lacks the sequence encoded by exon 7. Abbreviations: C, control when FLAG-SMN1G1C was transfected without a U1 construct. (D) Model representing the effects of an ISS-N1-targeting ASO and an eU1-induced activation of Cr1 on the SMN protein production. Exons and introns are indicated by colored boxes and lines, respectively. ASO is shown as a red bar, eU1 as a blue structure. Positioning of ISS-N1 and stop codons are marked. Location of the wt-5′ss, G1C-5′ss and cryptic sites (Cr1 and Cr2) are indicated by arrows. C at the first position of intron 7 is marked in red; red crossing signifies that this mutation renders the native 5′ss of exon 7 inactive. Abbreviations are described in Supplementary Table S1.
DISCUSSION
SMA is a devastating cause of infant mortality that results from low SMN. The only approved therapy for SMA relies upon the availability of two or more SMN2 alleles that could be targeted to elevate the SMN level through splicing correction (70). Here, we report a novel eU1-based approach aimed at benefiting a broad spectrum of SMA patients, including those affected by lethal splice site mutations. We began this study in an effort to find a potential rescue strategy for a severe SMA patient harboring a pathogenic G-to-C mutation at the first position (G1C) of SMN1 intron 7. Reproducing the devastating consequences of the G1C mutation, transcripts derived from the SMN1G1C minigene we generated showed complete skipping of exon 7 (Figure 1). Hence, we used this minigene as a valuable tool to explore potential therapeutic avenues for correction of exon 7 splicing. We first asked if skipping of SMN1G1Cexon 7 could be prevented through promotion of intron 6 splicing employing engineered U1s (eU1s) targeting the G1C-5′ss. We tested a library of eU1s predicted to form duplexes of varying lengths with the G1C-5′ss. Most of these eU1s prevented SMN1G1Cexon 7 skipping; however, the major splice product was intron 7-retained transcript. Supporting a direct correlation between the eU1:G1C-5′ss duplex size and intron 7 retention, eU1s that formed 11 and 6 bp duplexes produced the highest and the lowest levels of SMN1G1C intron 7-retained transcripts, respectively (Figure 1). The translation stop codon of SMN is located within exon 7. Hence, the intron 7-retained transcripts will code for full-length SMN, but will acquire a significantly longer 3′ untranslated region (3′UTR) that might adversely affect nuclear export and translation. Therefore, we turned our attention towards an alternative strategy of cryptic-splice-site activation that could generate mRNAs shorter than intron 7-retained transcripts while maintaining the ability to code for full-length SMN.
An important clue that a cryptic 5′ss immediately downstream of the G1C-5′ss could be activated came from the identification of two novel splice products generated in the presence of eU1s targeting the G1C-5′ss. These products included exon 7 extended by 23 and 51 nts due to the activation of two novel cryptic 5′ss located in intron 7. We named them Cr1 and Cr2, respectively (Figure 1). The relatively strong activation of Cr1 was somewhat surprising, since several existing algorithms scored it as a very weak 5′ss. In search for the strongest cryptic 5′ss that could be activated, we employed eU1s that targeted six sites with GU residues immediately downstream of the G1C-5′ss (Figure 2). While site 1 partially overlapped with the G1C-5′ss, sites 2 and 4 corresponded to Cr1 and Cr2, respectively. All designed eU1s were predicted to form an 11bp duplex with their respective targets to ensure their efficient recruitment. Three eU1s that targeted sites 2 (Cr1), 3 and 4 (Cr2) substantially restored exon 7 inclusion through Cr1 usage. The remaining three eU1s that targeted intronic sequences upstream of site 2 or downstream of site 4 prevented SMN1G1C exon 7 skipping by stimulating intron 7 retention. These results suggested that the redefinition of exon 7 by activation of Cr1 depends upon annealing of an eU1 within a narrow sequence stretch spanning from site 2 through site 4. We hypothesize that the recruitment of eU1s within this narrow sequence stretch is conducive for the spliceosomal assembly leading to the removal of intron 7. An eU1 that targeted site 5 had the smallest effect on SMN1G1C exon 7 splicing. Site 5 corresponds to a uridine-rich region, deletion of which has been shown to promote SMN exon 7 skipping (44). Considering that the site 5-targeting eU1 did not stimulate Cr1 activation nor did it promote intron 7 retention, we hypothesize that a factor(s) interacting with site 5 is critical for the definition of exon 7 and the removal of the upstream intron 6.
Similar to SMN1G1C transcripts, SMN1G1Uand SMN1G1Atranscripts also showed the increase in exon 7 inclusion from Cr1 in the presence of eU1 targeting sites 2, 3 and 4 (Supplementary Figure S5). These results confirmed that Cr1 activation is not specific for the G1C context of the mutated 5′ ss of exon 7. In agreement with a recent report (25), eU1s that targeted sites 1 through 4 promoted robust inclusion of exon 7 through usage of the wt-5′ss in transcripts derived from the SMN2 minigene (Figure 2). We also observed small but noticeable activation of Cr1 in transcripts derived from the SMN2 minigene as well as from endogenous SMN1 and SMN2 genes in the presence of eU1s targeting sites 2 (Cr1) and 3 (Figure 2, Supplementary Figure S4). These results confirmed that Cr1 usage is regulated by the local context rather than by the promoter structure or by sequences upstream of exon 6. Interestingly, similar to SMN1G1C the site 5-targeting eU1 did not promote inclusion of exon 7 in SMN2 transcripts. These results support a hypothesis that a factor(s) interacting with site 5 is critical for the usage of both the wt-5′ss and Cr1.
Our results also captured the effect of eU1:Cr1 duplex size on the usage of Cr1. For instance, shortening of eU1:Cr1 duplex size from 11 to 9 bp improved activation of Cr1 and reduced the levels of intron 7-retained transcripts of SMN1G1C (Figure 3). While the shortest eU1:Cr1 duplex to activate Cr1 was 6 bp-long, not all duplexes of this length were equally potent in doing so. For the most part, eU1:Cr1 duplexes of 7- and 8bp-long activated Cr1 with high efficiency. These results confirmed that SMN1G1C exon 7 inclusion could be rescued by a broad range of eU1:Cr1 duplex sizes. However, no specific rule emerged with respect to the connection between the size of the eU1:Cr1 duplex and the activation of Cr1. Next we asked whether the Cr1-activating eU1 would also rescue inclusion of exon 7 with an abrogated 3′ss. To answer this question, we created the SMNG-1U minigenes carrying a pathogenic G-to-U mutation (G-1U) at the invariant last position of intron 6. As expected, transcripts derived from SMN1G-1U or SMN2G-1U undergo complete skipping of exon 7 (Figure 4). Interestingly, the eU1 that directly targeted Cr1 significantly increased exon 7 inclusion by activating the wt-5′ss and a novel cryptic 3′ss (E7-3′Cr) within exon 7 in both SMN1G-1U and SMN2G-1U. E7-3′Cr usage reduces the size of exon 7 from 54 nts to 46 nts. An early study suggested that ∼50 nts are required between the 3′ss and the 5′ss for an eU1-dependent activation of the upstream 3′ss (71). Consistently, the eU1 that displayed perfect complementarity with the wt-5′ss of exon 7 was unable to activate the E7-3′Cr and did not improve exon 7 inclusion in either SMN1G-1Uor SMN2G-1U transcripts (Figure 4). Interestingly, the site 3-targeting eU1 caused a small increase in exon 7 inclusion and only in SMN1G-1Ubut not in SMN2G-1U. This differential effect could be attributed to a U residue at the sixth position in SMN2 exon 7. This SMN2-associated C6U is known to strengthen the terminal stem–loop 1 (TSL1) structure and/or create a binding site for hnRNP A1/A2 and Sam68 that are the negative regulators of exon 7 splicing [Figure 4, (52,62,63)]. Overall, our results are consistent with the exon definition model, in which the recruitment of eU1 at Cr1 appears to drive cross-exon interactions that define the 3′ss of exon 7. Noticeably, eU1s did not trigger activation of the E7-3′Cr when a pathogenic 7-nt deletion was introduced in the polypyrimidine tract (PPT) of the 3′ss of SMN1 exon 7 (Figure 5, SMN1ΔPy mutant transcript). Instead, in the SMN1ΔPy transcripts eU1s targeting sites 2 through 4 promoted exon 7 inclusion through usage of the wt-3′ss and mostly the wt-5′ss similar to what we observed for the SMN2 transcripts (Figures 2 and 5). It should be noted, however, that despite the difference in what 3′ss was selected, our results confirmed yet again that eU1s we used stimulate the 3′ss recognition, by, for example, assisting in U2AF65 recruitment to PPT as shown in (72).
We observed a clear difference in the effect of eU1s on usage of the 5′ss of SMN1 exon 7 mutated at the first intronic position versus 5′ss mutated at a less conserved +6 position of an intron. In particular, contrary to what we observed in the case of SMN1G1C, eU1 that directly targeted the mutated 5′ss of exon 7 of SMN1U6G activated its usage accompanied by effective exon inclusion (Figure 5). We attribute this difference to the presence of some functionality of U6G-5′ss in carrying out the first step of the splicing reaction, particularly when eU1 is ‘forcefully’ recruited to the U6G-5′ss (Supplementary Tables 2 and 3). However, the rules seemed to change and differences disappear when the eU1 recruitment site was shifted downstream of the mutated 5′ss site, since in both minigenes the usage of Cr1 was activated. In the case of the SMN1 minigene with a 4-nt deletion from fourth to seventh positions of intron 7 (SMN1Δ4-5′ss), the effect of eU1 overexpression was very similar to the one we observed for SMN1G1C. For example, in both minigenes the direct recruitment of the eU1s to the mutated 5′ss (G1C-5′ss and Δ4-5′ss) prevented exon 7 skipping but through retention of intron 7 rather that the usage of the mutated 5′ss or Cr1 (Figure 5). Furthermore, just like in SMN1G1C and SMN1U6G, shifting an eU1 recruitment site downstream of the Δ4-5′ss activated the usage of Cr1 (Figure 5). Overall, our results are consistent with the hypothesis that the recruitment of an eU1 within a narrow sequence stretch spanning from site 2 through site 4 is essential for activation of Cr1.
One of the most surprising findings of our study was the predominant activation of Cr1 by eU1s that targeted random intronic sequences upstream and downstream of Cr1 (Figure 6). None of the eU1 annealing sequences had invariant GU-dinucleotides associated with the 5′ss. Our results refined the sequence range of eU1 annealing for Cr1 activation. In particular, annealing of an eU1 within a ∼50-nt sequence stretch starting from ISS-N1 to site 4 promoted activation of Cr1. However, for reasons not yet understood, the effects of different eU1s targeting this sequence stretch on the activation of Cr1 varied. Our findings are distinct from a previous study in which deletion of four nucleotides (GUAA residues) of the U1 snRNP recruitment site within a cryptic exon of ATM gene promoted usage of this cryptic exon (19). In contrast, Cr1 usage places the annealing site of eU1-I7R8 within the exonic region and yet this eU1 efficiently promoted exon 7 inclusion through activation of Cr1 (Figure 6). At the same time, our findings showed a differential eU1 effect on splicing when the annealing target within the exon is altered. For example, compared to eU1-I7R8, eU1-I7R10 that annealed further upstream within exon 7 predominantly promoted intron 7 retention (Figure 6). This difference could be attributed to the eU1-I7R10 annealing site that falls outside of the Cr1 activation range described above. The effect of eU1s targeting random non-GU sequences was the same in SMN1G1C and SMN1U6G transcripts. In the case of SMN2, all eU1s, including eU1-I7R10, produced a strong stimulatory effect on exon 7 inclusion mainly through the activation of the wt-5′ss. Taken together, our findings confirmed that, while Cr1 was activated by eU1 annealing to a narrow sequence stretch, the usage of the wt-5′ss was promoted by eU1s annealing to a broader region.
We discovered that single nucleotide substitutions within intron 7 strongly activate Cr1 and Cr2. The most interesting observation was that the A23G and A28G substitutions completely abrogated the usage of the wt-5′ss even in the context of SMN1 (Figure 7). The effect of the above mutations on activation of Cr1 appeared to be more robust than the effect caused by targeting Cr1 with an eU1. This is not surprising considering that a mutation aimed at increasing the strength of a 5′ss in question would produce a broader effect, including improved recognition of the site by U1 snRNP, U6 snRNP and possibly other factors involved in pre-mRNA splicing (23). Interestingly, A23G/A28G mutations fully restored SMN2 exon 7 inclusion through Cr1 activation even in the context of the abrogated Tra2-ESE (Figure 7). These results confirmed that the previously described ESEs associated with ASF/SF2 and Tra2 are not required for the activation of Cr1. Further, the first in vivo selection of the entire exon conducted on SMN exon 7 revealed the stimulatory effect of the non-wt G residue at the last position of exon 7 (A-1G) for its inclusion (36). Consistently, A-1G substitution was found to obviate the requirement of all known positive elements within exon 7 (36). Combining A-1G with A23G/A28G mutations resulted in the activation of both the wt-5′ss and Cr1 with nearly equal efficiency (Figure 7). It is likely that the same factors are involved in activation of both the wt-5′ss and Cr.
Consistent with the critical role of ISS-N1 and Cr1 in the regulation of exon 7 splicing, this region was found to be relatively conserved in mammals (Supplementary Figure S7). Since ISS-N1 and Cr1 partially overlap, we wanted to assess the effect of ISS-N1 on Cr1 usage. In an effort to do so, we inserted splicing neutral sequences between these two. Interestingly, insertion of splicing neutral sequences activated Cr1 in the context of SMN1G1C and SMN1U6G (Figure 9). A small but detectable effect of the insertion of splicing neutral sequences on the activation of Cr1 was also observed in SMN1 but not SMN2. This is not surprising, since SMN1, SMN1G1C and SMN1U6G all share identical and relatively strong 3′ss of exon 7. These results suggest that factors interacting with the 3′ss of exon 7 might be important for the activation of Cr1. In addition, we have previously shown that the 5′ss recognition of SMN2 exon 7 is enhanced by TIA1, a splicing factor that binds to the downstream U-rich clusters (URCs) (44). Insertion of the splicing neutral sequences moves the URCs away from the 5′ss of SMN2 exon 7 resulting in its skipping. The results of site-specific mutagenesis suggest that URC1 is also important for Cr1 activation (at least in the context of the SMN1U6G minigene), since abrogation of URC1 decreased/abolished the usage of this site, while strengthening of URC1 improved it (Figure 8).
We observed mixed results with mutagenesis of ISS-N1, which harbors two putative hnRNP A1/A2 motifs. A12C mutation that abrogated the first hnRNP A1/A2 motif did not activate Cr1 (Figure 8), suggesting that this motif is not involved in the suppression of Cr1. However, it is still possible that the second hnRNP A1/A2 motif contributes to Cr1 usage. Our results of depletion experiments only partially supported the role of hnRNP A1/A2 in suppression of Cr1. For instance, depletion of hnRNP A1/A2 activated Cr1 in the case of SMN1U6G but not in the case of SMN1G1C transcripts (Figure 11). Depletion of hnRNP A1/A2 also activated Cr2 and E6Cr in the context of SMN1U6G transcripts (Figure 11). These results are significant because they confirm that the levels of endogenous U1 snRNP are fully capable of activating cryptic splice sites, usage of which we initially captured employing eU1s that formed very strong RNA:RNA duplexes with their respective targets. Our results also confirmed that eU1s that activate Cr1 do not require the assistance of the endogenous U1 snRNP. This is based on the observation that an ASO-mediated suppression of endogenous U1 snRNP barely had any effect on the eU1-induced activation of Cr1, while eU1-induced usage of wt-5′ss of SMN2 exon 7 was severely impacted (Figure 10). These findings underscore that mechanisms of eU1-induced usage of the wt-5′ss and Cr1 could be different. For instance, while Cr1 was substantially activated by eU1-MI7-R7 that does not sequester the invariant GU dinucleotides at the site of catalysis, whereas, sequestration of the GU dinucleotides by endogenous U1 snRNP was critical for the usage of the wt-5′ss (Figure 10). Future studies will determine the mechanism by which the usage of Cr1 is regulated, including participating cis-elements and transacting factors.
Finally, we examined the physiological significance of our findings employing a splicing-coupled translation reporter assay that detects the FLAG-tagged SMN protein produced from transcripts that undergo pre-mRNA splicing, including Cr1 activation. Our results confirmed that transcripts generated by Cr1 activation produce full-length SMN (Figures 12). To the best of our knowledge, this is the first report to show the generation of the full-length SMN protein from SMN1 carrying a pathogenic mutation at the 5′ss of exon 7. Future studies will determine if the addition of 23 nts of intron 7 to the 3′UTR will have an effect on the stability, trafficking and translation of mRNAs generated by Cr1 activation. The mechanism of the full-length SMN restoration via eU1-induced activation of Cr1 is distinct from the action of an ISS-N1-targeting ASO. The latter is incapable of restoring inclusion of SMN1 exon 7 with pathogenic mutations at its 5′ss (Figure 12D; Supplementary Figure S9). Hence, the recently approved antisense drug, nusinersen, based on the ISS-N1 target may not be a viable option for SMA patients with these types of mutations. Similar to an ASO-based approach, an eU1-based approach takes advantage of the endogenous transcript for splicing correction. Employment of transcripts derived from endogenous gene is likely to maintain the physiological levels of the protein. The small size of an eU1 offers a tremendous advantage for its packaging in an adeno-associated virus type 9 (AAV9) vector, which has already shown success in eU1 delivery for the treatment of a genetic disease (73). However, a mouse model carrying a mutated 5′ss of SMN1 exon 7 will need to be generated to test the feasibility of the approach in which eU1s are used.
As per one estimate, about 10% of all human inherited diseases are caused by mutations located within splice sites (74). The G1C mutation is the deadliest mutation among them, since it inevitably leads to the complete skipping of an exon. Consistent with this argument, a recent transcriptome-wide analysis in various tissues did not capture a single incidence of a functional 5′ss carrying a C residue at the first position of an intron, although examples of all other residues at the invariant first and second positions of several functional introns were recorded (75). Hence, until this point, rescuing splicing of an exon carrying a G1C mutation has been an impossible task. In this study, we have shown an alternative strategy using eU1-mediated activation of a cryptic splice site to completely neutralize the consequences of the G1C mutation. This eU1-based strategy is applicable for rescuing splicing defects for a broad range of mutations at both the 3′ and the 5′ss of exon 7 of SMN genes. A vast number of studies have demonstrated the critical role of residues of U1 snRNA in the selection of the 5′ss of an exon (14,76,78). However, the rules by which U1 snRNP affects 5′ss selection from a distance have attracted very little attention. Our findings clearly demonstrate that the U1-snRNP-interacting motifs away from the 5′ss could be utilized for therapeutic purposes. Unlike the general belief that mutations leading to the generation of cryptic splice sites cause genetic diseases, this report demonstrates that the activation of a cryptic splice site employing an eU1 could lead to the rescue of the complete ORF in a pathological condition. As the number of genetic diseases caused by aberrant splicing increases, this work illustrates the promise of eU1-mediated reprogramming of the boundary of an exon for therapeutic solutions.
Supplementary Material
ACKNOWLEDGEMENTS
Authors acknowledge technical support of Dr. Joonbae Seo for the construction of a splicing-coupled translation reporter used in this study. Authors thank Vinita Raj Singh for help with the editing of this manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [R01 NS055925, R21 NS101312]; Iowa Center for Advanced Neurotoxicology (ICAN) and Salsbury Endowment (Iowa State University, Ames, IA, USA) (to R.N.S.). Funding for open access charge: NIH [R01 NS055925, R21 NS101312].
Conflict of interest statement. The ISS-N1 target (US7838657) was discovered in the Singh laboratory at UMass Medical School (MA, USA). Inventors, including R.N. Singh, N.N. Singh and UMASS Medical School, are currently benefiting from licensing of the ISS-N1 target to Ionis Pharmaceuticals. Iowa State University holds intellectual property rights on GC-rich and ISS-N2 targets. Therefore, inventors including R.N. Singh, N.N. Singh and Iowa State University could potentially benefit from any future commercial exploitation of GC-rich and ISS-N2 targets.
REFERENCES
- 1. Raj B., Blencowe B.J.. Alternative splicing in the mammalian nervous system: recent insights into mechanisms and functional roles. Neuron. 2015; 87:14–27. [DOI] [PubMed] [Google Scholar]
- 2. Burge C.B., Tuschl T., Sharp P.A.. Gesteland RF, Cech TR, Atkins JF. Splicing of precursors to mRNAs by the spliceosomes. The RNA World—The Nature of Modern RNA Suggests a Prebiotic RNA. 1999; NY: Cold Spring Harbor Laboratory Press; 525–560. [Google Scholar]
- 3. Kapustin Y., Chan E., Sarkar R., Wong F., Vorechovsky I., Winston R.M., Tatusova T., Dibb N.J.. Cryptic splice sites and split genes. Nucleic Acids Res. 2011; 39:5837–5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Robberson B.L., Cote G.J., Berget S.M.. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol. Cell. Biol. 1990; 10:84–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. De Conti L., Baralle M., Buratti E.. Exon and intron definition in pre-mRNA splicing. Wiley Interdiscip. Rev. RNA. 2013; 4:49–60. [DOI] [PubMed] [Google Scholar]
- 6. Saldi T., Cortazar M.A., Sheridan R.M., Bentley D.L.. Coupling of RNA polymerase II transcription elongation with pre-mRNA splicing. J. Mol. Biol. 2016; 428:2623–2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fu X.-D., Ares M.. Context-dependent control of alternative splicing by RNA-binding proteins. Nat. Rev. Genet. 2014; 15:689–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang Z., Rolish M.E., Yeo G., Tung V., Mawson M., Burge C.B.. Systematic identification and analysis of exonic splicing silencers. Cell. 2004; 119:831–845. [DOI] [PubMed] [Google Scholar]
- 9. Wang Y., Xiao X., Zhang J., Choudhury R., Robertson A., Li K., Ma M., Burge C.B., Wang Z.. A complex network of factors with overlapping affinities represses splicing through intronic elements. Nat. Struct. Mol. Biol. 2013; 20:36–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zarnack K., König J., Tajnik M., Martincorena I., Eustermann S., Stévant I., Reyes A., Anders S., Luscombe N.M., Ule J.. Direct competition between hnRNP C and U2AF65 protects the transcriptome from the exonization of Alu elements. Cell. 2013; 152:453–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ling J.P., Pletnikova O., Troncoso J.C., Wong P.C.. TDP-43 repression of nonconserved cryptic exons is compromised in ALS-FTD. Science. 2015; 349:650–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ling J.P., Chhabra R., Merran J.D., Schaughency P.M., Wheelan S.J., Corden J.L., Wong P.C.. PTBP1 and PTBP2 repress nonconserved cryptic exons. Cell Rep. 2016; 17:104–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sibley C.R., Blazquez L., Ule J.. Lessons from non-canonical splicing. Nat. Rev. Genet. 2016; 17:407–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brillen A.-L., Schöneweis K., Walotka L., Hartmann L., Müller L., Ptok J., Kaisers W., Poschmann G., Stühler K., Buratti E. et al. Succession of splicing regulatory elements determines cryptic 5′ss functionality. Nucleic Acids Res. 2017; 45:4202–4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Matlin A.J., Moore M.J.. Spliceosome assembly and composition. Adv. Exp. Med. Biol. 2007; 623:14–35. [DOI] [PubMed] [Google Scholar]
- 16. Wahl M.C., Will C.L., Lührmann R.. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009; 136:701–718. [DOI] [PubMed] [Google Scholar]
- 17. Baserga S.J., Steitz J.A.. Gesteland RF, Atkins JF. The diverse world of small ribonucleoproteins. The RNA World. 1993; NY: Cold Spring Harbor Laboratory Press; 359–381. [Google Scholar]
- 18. Roca X., Akerman M., Gaus H., Berdeja A., Bennett C.F., Krainer A.R.. Widespread recognition of 5′ splice sites by noncanonical base-pairing to U1 snRNA involving bulged nucleotides. Genes Dev. 2012; 26:1098–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pagani F., Buratti E., Stuani C., Bendix R., Dörk T., Baralle F.E.. A new type of mutation causes a splicing defect in ATM. Nat. Genet. 2002; 30:426–429. [DOI] [PubMed] [Google Scholar]
- 20. Almada A.E., Wu X., Kriz A.J., Burge C.B., Sharp P.A.. Promoter directionality is controlled by U1 snRNP and polyadenylation signals. Nature. 2013; 499:360–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kaida D., Berg M.G., Younis I., Kasim M., Singh L.N., Wan L., Dreyfuss G.. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature. 2010; 468:664–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Berg M.G., Singh L.N., Younis I., Liu Q., Pinto A.M., Kaida D., Zhang Z., Cho S., Sherrill-Mix S., Wan L. et al. U1 snRNP determines mRNA length and regulates isoform expression. Cell. 2012; 150:53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cohen J.B., Snow J.E., Spencer S.D., Levinson A.D.. Suppression of mammalian 5′ splice-site defects by U1 small nuclear RNAs from a distance. Proc. Natl. Acad. Sci. U.S.A. 1994; 91:10470–10474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dal Mas A., Fortugno P., Donadon I., Levati L., Castiglia D., Pagani F.. Exon-specific U1s correct SPINK5 exon 11 skipping caused by a synonymous substitution that affects a bifunctional splicing regulatory element. Hum. Mutat. 2015; 36:504–512. [DOI] [PubMed] [Google Scholar]
- 25. Dal Mas A., Rogalska M.E., Bussani E., Pagani F.. Improvement of SMN2 pre-mRNA processing mediated by exon-specific U1 small nuclear RNA. Am. J. Hum. Genet. 2015; 96:93–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rogalska M.E., Tajnik M., Licastro D., Bussani E., Camparini L., Mattioli C., Pagani F.. Therapeutic activity of modified U1 core spliceosomal particles. Nat. Commun. 2016; 7:11168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lefebvre S., Bürglen L., Reboullet S., Clermont O., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M.. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995; 80:155–165. [DOI] [PubMed] [Google Scholar]
- 28. Singh R.N., Howell M.D., Ottesen E.W., Singh N.N.. Diverse role of survival motor neuron protein. Biochim. Biophys. Acta. 2017; 1860:299–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lorson C.L., Hahnen E., Androphy E.J., Wirth B.. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:6307–6311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vitte J., Fassier C., Tiziano F.D., Dalard C., Soave S., Roblot N., Brahe C., Saugier-Veber P., Bonnefont J.P., Melki J.. Refined characterization of the expression and stability of the SMN gene products. Am. J. Pathol. 2007; 171:1269–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cho S., Dreyfuss G.. A degron created by SMN2 exon 7 skipping is a principal contributor to spinal muscular atrophy severity. Genes Dev. 2010; 24:438–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ahmad S., Bhatia K., Kannan A., Gangwani L.. Molecular mechanisms of neurodegeneration in spinal muscular atrophy. J. Exp. Neurosci. 2016; 10:39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Singh N.N., Singh R.N.. Alternative splicing in spinal muscular atrophy underscores the role of an intron definition model. RNA Biol. 2011; 8:600–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Singh N.N., Lee B.M., Singh R.N.. Splicing regulation in spinal muscular atrophy by an RNA structure formed by long-distance interactions. Ann. N. Y. Acad. Sci. 2015; 1341:176–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Monani U.R., Lorson C.L., Parsons D.W., Prior T.W., Androphy E.J., Burghes A.H., McPherson J.D.. A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum. Mol. Genet. 1999; 8:1177–1183. [DOI] [PubMed] [Google Scholar]
- 36. Singh N.N., Androphy E.J., Singh R.N.. In vivo selection reveals combinatorial controls that define a critical exon in the spinal muscular atrophy genes. RNA. 2004; 10:1291–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Singh N.N., Androphy E.J., Singh R.N.. The regulation and regulatory activities of alternative splicing of the SMN gene. Crit. Rev. Eukaryot. Gene Expr. 2004; 14:271–285. [DOI] [PubMed] [Google Scholar]
- 38. Singh N.N., Singh R.N., Androphy E.J.. Modulating role of RNA structure in alternative splicing of a critical exon in the spinal muscular atrophy genes. Nucleic Acids Res. 2007; 35:371–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Singh N.K., Singh N.N., Androphy E.J., Singh R.N.. Splicing of a critical exon of human Survival Motor Neuron is regulated by a unique silencer element located in the last intron. Mol. Cell. Biol. 2006; 26:1333–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Howell M.D., Singh N.N., Singh R.N.. Advances in therapeutic development for spinal muscular atrophy. Future Med. Chem. 2014; 6:1081–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ottesen E.W. ISS-N1 makes the first FDA-approved drug for spinal muscular atrophy. Translat. Neurosci. 2017; 8:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Singh N.N., Howell M.D., Androphy E.J., Singh R.N.. How the discovery of ISS-N1 led to the first medical therapy for spinal muscular atrophy. Gene Ther. 2017; doi:10.1038/gt.2017.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hua Y., Vickers T.A., Okunola H.L., Bennett C.F., Krainer A.R.. Antisense masking of an hnRNP A1/A2 intronic splicing silencer corrects SMN2 splicing in transgenic mice. Am. J. Hum. Genet. 2008; 82:834–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Singh N.N., Seo J., Ottesen E.W., Shishimorova M., Bhattacharya D., Singh R.N.. TIA1 prevents skipping of a critical exon associated with spinal muscular atrophy. Mol. Cell. Biol. 2011; 31:935–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Howell M.D., Ottesen E.W., Singh N.N., Anderson R.L., Seo J., Sivanesan S., Whitley E.M., Singh R.N.. TIA1 is a gender-specific disease modifier of a mild mouse model of spinal muscular atrophy. Sci. Rep. 2017; 7:7183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Singh N.N., Shishimorova M., Cao L.C., Gangwani L., Singh R.N.. A short antisense oligonucleotide masking a unique intronic motif prevents skipping of a critical exon in spinal muscular atrophy. RNA Biol. 2009; 6:341–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Keil J.M., Seo J., Howell M.D., Hsu W.H., Singh R.N., DiDonato C.J.. A short antisense oligonucleotide ameliorates symptoms of severe mouse models of spinal muscular atrophy. Mol. Ther. Nucleic Acids. 2014; 3:e174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Singh N.N., Lawler M.N., Ottesen E.W., Upreti D., Kaczynski J.R., Singh R.N.. An intronic structure enabled by a long-distance interaction serves as a novel target for splicing correction in spinal muscular atrophy. Nucleic Acids Res. 2013; 41:8144–8165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Howell M.D., Ottesen E.W., Singh N.N., Anderson R.L., Singh R.N.. Gender-specific amelioration of SMA phenotype upon disruption of a deep intronic structure by an oligonucleotide. Mol. Ther. 2017; 25:1328–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ronchi D., Previtali S.C., Sora M.G.N., Barera G., Del Menico B., Corti S., Bresolin N., Comi G.P.. Novel splice-site mutation in SMN1 associated with a very severe SMA-I phenotype. J. Mol. Neurosci. 2015; 56:212–215. [DOI] [PubMed] [Google Scholar]
- 51. Cashman N.R., Durham H.D., Blusztajn J.K., Oda K., Tabira T., Shaw I.T., Dahrouge S., Antel J.P.. Neuroblastoma x spinal cord (NSC) hybrid cell lines resemble developing motor neurons. Dev. Dyn. 1992; 194:209–221. [DOI] [PubMed] [Google Scholar]
- 52. Singh N.N., Androphy E.J., Singh R.N.. An extended inhibitory context causes skipping of exon 7 of SMN2 in spinal muscular atrophy. Biochem. Biophys. Res. Commun. 2004; 315:381–388. [DOI] [PubMed] [Google Scholar]
- 53. Kammler S., Leurs C., Freund M., Krummheuer J., Seidel K., Tange T.O., Lund M.K., Kjems J., Scheid A., Schaal H.. The sequence complementarity between HIV-1 5′ splice site SD4 and U1 snRNA determines the steady-state level of an unstable env pre-mRNA. RNA. 2001; 7:421–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Singh N.N., Hollinger K., Bhattacharya D., Singh R.N.. An antisense microwalk reveals critical role of an intronic position linked to a unique long-distance interaction in pre-mRNA splicing. RNA. 2010; 16:1167–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Singh N.N., Seo J., Rahn S.J., Singh R.N.. A multi-exon-skipping detection assay reveals surprising diversity of splice isoforms of spinal muscular atrophy genes. PLoS ONE. 2012; 7:e49595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ottesen E.W., Howell M.D., Singh N.N., Seo J., Whitley E.M., Singh R.N.. Severe impairment of male reproductive organ development in a low SMN expressing mouse model of spinal muscular atrophy. Sci. Rep. 2016; 6:20193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Seo J., Singh N.N., Ottesen E.W., Sivanesan S., Shishimorova M., Singh R.N.. Oxidative stress triggers body-wide skipping of multiple exons of the spinal muscular atrophy gene. PLoS ONE. 2016; 11:e0154390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Seo J., Singh N.N., Ottesen E.W., Lee B.M., Singh R.N.. A novel human-specific splice isoform alters the critical C-terminus of Survival Motor Neuron protein. Sci. Rep. 2016; 6:30778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yeo G., Burge C.B.. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. 2004; 11:377–394. [DOI] [PubMed] [Google Scholar]
- 60. Divina P., Kvitkovicova A., Buratti E., Vorechovsky I.. Ab initio prediction of mutation-induced cryptic splice-site activation and exon skipping. Eur. J. Hum. Genet. 2009; 17:759–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hwang D.Y., Cohen J.B.. U1 snRNA promotes the selection of nearby 5′ splice sites by U6 snRNA in mammalian cells. Genes Dev. 1996; 10:338–350. [DOI] [PubMed] [Google Scholar]
- 62. Pedrotti S., Bielli P., Paronetto M.P., Ciccosanti F., Fimia G.M., Stamm S., Manley J.L., Sette C.. The splicing regulator Sam68 binds to a novel exonic splicing silencer and functions in SMN2 alternative splicing in spinal muscular atrophy. EMBO J. 2010; 29:1235–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kashima T., Rao N., David C.J., Manley J.L.. hnRNP A1 functions with specificity in repression of SMN2 exon 7 splicing. Hum. Mol. Genet. 2007; 16:3149–3159. [DOI] [PubMed] [Google Scholar]
- 64. Wirth B., Herz M., Wetter A., Moskau S., Hahnen E., Rudnik-Schöneborn S., Wienker T., Zerres K.. Quantitative analysis of survival motor neuron copies: identification of subtle SMN1 mutations in patients with spinal muscular atrophy, genotype-phenotype correlation, and implications for genetic counseling. Am. J. Hum. Genet. 1999; 64:1340–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Han K., Yeo G., An P., Burge C.B., Grabowski P.J.. A combinatorial code for splicing silencing: UAGG and GGGG motifs. PLoS Biol. 2005; 3:e158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Erkelenz S., Theiss S., Otte M., Widera M., Peter J.O., Schaal H.. Genomic HEXploring allows landscaping of novel potential splicing regulatory elements. Nucleic Acids Res. 2014; 42:10681–10697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zhang X.H.-F., Arias M.A., Ke S., Chasin L.A.. Splicing of designer exons reveals unexpected complexity in pre-mRNA splicing. RNA. 2009; 15:367–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Erkelenz S., Mueller W.F., Evans M.S., Busch A., Schöneweis K., Hertel K.J., Schaal H.. Position-dependent splicing activation and repression by SR and hnRNP proteins rely on common mechanisms. RNA. 2013; 19:96–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Caputi M., Freund M., Kammler S., Asang C., Schaal H.. A bidirectional SF2/ASF- and SRp40-dependent splicing enhancer regulates human immunodeficiency virus type 1 rev, env, vpu, and nef gene expression. J. Virol. 2004; 78:6517–6526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Finkel R.S., Chiriboga C.A., Vajsar J., Day J.W., Montes J., De Vivo D.C., Yamashita M., Rigo F., Hung G., Schneider E. et al. Treatment of infantile-onset spinal muscular atrophy with nusinersen: a phase 2, open-label, dose-escalation study. Lancet. 2016; 388:3017–3026. [DOI] [PubMed] [Google Scholar]
- 71. Hwang D.Y., Cohen J.B.. U1 small nuclear RNA-promoted exon selection requires a minimal distance between the position of U1 binding and the 3′ splice site across the exon. Mol. Cell. Biol. 1997; 17:7099–7107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Hoffman B.E., Grabowski P.J.. U1 snRNP targets an essential splicing factor, U2AF65, to the 3′ splice site by a network of interactions spanning the exon. Genes Dev. 1992; 6:2554–2568. [DOI] [PubMed] [Google Scholar]
- 73. Balestra D., Barbon E., Scalet D., Cavallari N., Perrone D., Zanibellato S., Bernardi F., Pinotti M.. Regulation of a strong F9 cryptic 5′ss by intrinsic elements and by combination of tailored U1snRNAs with antisense oligonucleotides. Hum. Mol. Genet. 2015; 24:4809–4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Krawczak M., Thomas N.S.T., Hundrieser B., Mort M., Wittig M., Hampe J., Cooper D.N.. Single base-pair substitutions in exon-intron junctions of human genes: nature, distribution, and consequences for mRNA splicing. Hum. Mutat. 2007; 28:150–158. [DOI] [PubMed] [Google Scholar]
- 75. Parada G.E., Munita R., Cerda C.A., Gysling K.. A comprehensive survey of non-canonical splice sites in the human transcriptome. Nucleic Acids Res. 2014; 42:10564–10578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Carmel I., Tal S., Vig I., Ast G.. Comparative analysis detects dependencies among the 5′ splice-site positions. RNA. 2004; 10:828–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Lund M., Kjems J.. Defining a 5′ splice site by functional selection in the presence and absence of U1 snRNA 5′ end. RNA. 2002; 8:166–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Tan J., Ho J.X.J., Zhong Z., Luo S., Chen G., Roca X.. Noncanonical registers and base pairs in human 5′ splice-site selection. Nucleic Acids Res. 2016; 44:3908–3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.