Abstract
Sirtuins, a family of proteins with homology to the yeast silent information regulator 2 (Sir2), are NAD+-dependent histone deacetylases and play crucial roles in energy sensing and regulation in yeast and animal cells. Plants are autotrophic organisms and display distinct features of carbon and energy metabolism. It remains largely unexplored whether and how plant cells sense energy/redox status to control carbon metabolic flux under various growth conditions. In this work, we show that the rice nuclear sirtuin OsSRT1 not only functions as an epigenetic regulator to repress glycolytic genes expression and glycolysis in seedlings, but also inhibits transcriptional activity of glyceraldehyde-3-phosphatedehydrogenase (GAPDH) that is enriched on glycolytic genes promoters and stimulates their expression. We show that OsSRT1 reduces GAPDH lysine acetylation and nuclear accumulation that are enhanced by oxidative stress. Mass spectrometry identified six acetylated lysines regulated by OsSRT1. OsSRT1-dependent lysine deacetylation of OsGAPDH1 represses transcriptional activity of the protein. The results indicate that OsSRT1 represses glycolysis by both regulating epigenetic modification of histone and inhibiting the moonlighting function of GAPDH as a transcriptional activator of glycolytic genes in rice.
INTRODUCTION
Sirtuins are a family of proteins with homology to the silent information regulator 2 (Sir2) in yeast, initially identified as a genetic silencing factor (1). Sir2 is a nicotinamide adenine dinucleotide (NAD+)-dependent deacetylase that targets histones and non-histone proteins. Sir2 has been linked to lifespan extension associated with calorie restriction and regulation of stress response, apoptosis, and DNA repair (2–9). Seven sirtuin homologs (SIRT1–7) have been identified in mammals, of which SIRT1 is the closest homolog in structure and function to the yeast Sir2 (10). The 7 mammalian sirtuins occupy discrete subcellular compartments with SIRT1 mainly found in the nucleus, although its cytoplasmic localization has also been reported (11,12). SIRT2 is mainly in the cytoplasm, while SIRT3, SIRT4, and SIRT5 are in the mitochondrion (13). SIRT6 is predominantly localizes in the nucleus (14), and SIRT7 was reported to reside in the nucleolus (15). Because sirtuins require NAD+ as a substrate, the concentration of which is determined by the nutritional state of the cell, the expression and activity of these enzymes are tightly coupled to changes in cellular energy/redox status in yeast and animal cells (16–18). In animal cells, sirtuins play crucial roles in metabolic regulation to adapt to the changing environmental conditions and are considered to be metabolic and stress-sensor proteins (19). In mammalian cells under calorie restriction (starvation, fasting, and exercise), cells typically shift from growth to survival mode and alter metabolism towards functions important for viability (20). In this situation, acetyl-CoA is oxidized through the full tricarboxylic acid (TCA) cycle in the mitochondria for ATP synthesis, instead of shipping acetyl units out to the cytosol for biosynthesis. The rate of NADH oxidation in the oxidative phosphorylation chain exceeds the reduction of NAD+ thereby increasing the cellular NAD+/NADH ratio. The resulting elevation in NAD+ activates sirtuin deacetylases, which in turn represses glycolysis to favor mitochondrial respiration.
In contrast to in the mammals, only 2 sirtuin genes have been identified in rice, OsSRT1 and OsSRT2. OsSRT1 appeared to be mainly localized in the nucleus (21), while OsSRT2 is in the mitochondria (22,23). Phylogenetic analysis suggests that OsSRT1 is closely related to the mammalian SIRT6, and OsSRT2 to SIRT4 (24). Knock-down of OsSRT1 leads to oxidative burst and cell death, whereas over-expression of the gene enhances tolerance to oxidative stress in rice (21), suggesting that loss- or gain-of-function of OsSRT1 alters the expression of redox-related genes and cellular redox status. Being photosynthetic organisms, plants are autotrophic and there are significant differences in primary carbon and energy metabolisms and in metabolic and signaling pathways. For instance, glycolysis is induced under both biotic and abiotic stresses, and tricarboxylic acid (TCA) cycle is particularly high during light to dark transition (25–27). Although previous data indicate that OsSRT1 is directly involved in epigenetic regulation of metabolic gene expression in both seedlings and developing seeds (28,29), it remains largely unknown whether and how OsSRT1 regulate energy and metabolic flux in plant cells.
Glyceraldehyde-3-phosphatedehydrogenase (GAPDH), an obligatory enzyme in glycolysis, is found in organisms from all kingdoms of life. The enzyme catalyzes the conversion of glyceraldehyde-3-phosphate to 1,3-bisphosphoglyceric acid and reduces NAD+ to NADH in the presence of NAD+ and inorganic phosphate (30). Animal cells contain only one isoform of GAPDH. In addition to the role in glycolysis, animal GAPDH displays many other so-called moonlighting activities in several unrelated non-metabolic processes such as DNA repair, RNA binding, membrane fusion and transport, cytoskeletal dynamics, autophagy and cell death (31–33). This functional versatility is regulated, at least in part, by redox post-translational modifications that alter GAPDH catalytic activity and influence the subcellular localization of the enzyme (34,35).
Plants possess multiple GAPDH isoforms (30,36), including chloroplastic photosynthetic GAPDHs (GAPA, GAPB) that use NADPH to generate NADP+ in the Calvin-Benson reaction, cytosolic glycolytic GAPDHs (GAPC) and plastidic glycolytic GAPDHs (GAPCp), and the NADP-dependent non-phosphorylating cytosolic GAPDH (NP-GAPDH). Therefore, by contributing to the NAD(P)+/NAD(P)H ratio of the cell, plant GAPDHs can influence both cellular redox as well as general metabolism. Similar to mammalian cells, plant glycolytic GAPDH was detected both in the cytoplasm and in the nucleus and the nuclear localization is enhanced under diverse stresses (30,37–39). However, the mechanism by which plant GAPDH trans-localizes to the nucleus remains unknown. In addition, it was shown that GAPDH, as well as many other primary metabolic enzymes, is acetylated at several lysine residues in animal and plant cells (40–43). Although acetylation is shown to affect the enzymatic activity of GAPDH (43,44), it is unclear whether it has any effect on the moonlighting function of the protein.
In this paper, we show that OsSRT1 represses glycolysis by mediating deacetylation of both histone and the glycolytic GAPDH that is found in this work to also function as an activator of glycolytic gene expression. OsGAPDH1 is enriched on the promoter of several glycolytic genes and activates their expression. OsSRT1-mediated lysine deacetylation of GAPDH inhibits its nuclear accumulation and transactivation. The results suggest that OsSRT1 may regulate carbon metabolic flux by coordinating primary metabolism and gene expression for plant to adapt to the changing environmental conditions.
MATERIALS AND METHODS
Plant materials and rice growth conditions
OsSRT1 RNAi (R1, R2 and R3) and over-expression transgenic lines (OxSRT1–1(OX1), OxSRT1–2 (OX2) and OxSRT1–3 (OX3)) used in this study are previously reported by Zhang et al. (45). Both of GAPDH1 RNAi and overexpression transgenic plants were produced in Zhonghua 11 (ZH11) background, genotyped by PCR in the T2 segregating populations.
For germination, seeds were surface-sterilized and germinated in medium containing 0.8% agar supplemented with 3% (w/v) sucrose at 28°C (in light) and 24°C (in dark) with a 14-h-light (full spectrum, 10 000 lX)/10-h-dark cycle. For growth in field, germinated rice seedlings were transplanted in the experimental field in Wuhan (30.4°N, 114.2°E), China.
Characterization of GAPDH1 Insertion mutants
The T-DNA insertion mutants of rice gapdh1 (03Z11EB03) was confirmed by PCR using GAPDH1 specific primers F1 and R1 and a T-DNA right border primer N. For RT-PCR analysis of GAPDH1 transcripts in the mutants, the primer sets OsGAPDH1-F/R were used. Actin1 transcripts were amplified as controls. Nucleotide sequences of the primers are listed in Supplementary Table S4.
Substitution mutagenesis of GAPDH protein
Point mutation was introduced into the GAPDH1 protein by the polymerase chain reaction (PCR) using the mutated oligonucleotide primers. Two fragments, one from the N-terminal half side and the other from the C-terminal half side, were constructed for each mutant GAPDH1 proteins. After amplification of the two fragments in which the mutated sequences were overlapped, they were combined by a polymerization reaction through the mutated sequence using PCR.
qRT-PCR
Total RNA was extracted from tissues using TRIzol reagent (TransGen Biotech) according to the manufacturer's protocol. Four micrograms of total RNA were reverse-transcribed in a reaction of 20 μl by using DNase I and M-MLV Reverse transcriptase (Invitrogen) to obtain cDNA. The qRT-PCR was performed in an optical 96-well plate that included SYBR Premix EX Taq and 0.5 μl of Rox Reference Dye II (Takara), 1 μl of the reverse transcription reaction, and 0.25 μM of each gene-specific primer in a final volume of 25 μl on the ABI PRISM 7500 PCR instrument (Applied Biosystems). Data were collected using the ABI PRISM 7500 sequence detection system following the instruction manual. The relative expression levels were analyzed using the 2–△△CT method (46). The rice Actin1 gene was used as the internal control. All primers were annealed at 60°C. Values represent the means obtained from three biological replicates with independent mRNA isolations, each replicate with 3 technical repeats. The sequences of the primers used are listed in Supplementary Table S4.
Pull-down protein interaction assays
SRT1-His and GAPDH1/2-GST fusion proteins were expressed in Escherichia coli. Equal volumes of His alone or SRT1-His fusion proteins were incubated with GAPDH1/2-GST proteins in total 1.2 ml volume of the pull-down buffer (20 mM Tris–HCl, pH 8.0, 200 mM NaCl, 1 mM EDTA, 0.5% Lgepal CA-630 and protease inhibitor) at 4°C with gentle upside down rotation overnight. His beads (40 μl, Promega REF V8500) were added, followed by six more hours of incubation. After extensive washing, the pulled down proteins were eluted with His elution buffer provided in the kit and denatured for 12% SDS-PAGE. Immunoblots were performed using anti-His antibody (abcam ab9180; lot GR248369–2) and anti-GST antibody (abcam ab19256; lot GR1093331-1).
Chromatin immunoprecipitation (ChIP)-qPCR
The chromatin immunoprecipitation (ChIP) experiments were performed as described previously (47,48). Chromatin was extracted and fragmented via ultrasound to 200–800 bp. Antibodies (5 μl) of anti-H3K9ac (07-352, Millipore), anti-GFP (ab290, Abcam) and anti-OsSRT1 (28) were incubated with 40 μl protein A/G beads (Invitrogen) at 4°C for >4 h, after beads washing, 100 μl fragmented chromatin suspension was added, followed by incubation at 4°C overnight. After extensive washing and de-crosslink, the precipitated and input DNA samples were analyzed by qPCR. Primers for ChIP-qPCR are listed in Supplementary Table S5. Three biological replicates were performed using 10-day-old seedlings harvested from three independent cultures. Each biological replicate was tested with three technical repeats.
Rice protoplast preparation and transformation
Rice protoplasts were prepared from 14-day-old seedlings of rice Zhonghua 11 growing on 1/2 MS-media as described previously (49). The protoplasts were isolated with Digestion Solution (10 mM MES pH 5.7, 0.5 M Mannitol, 1 mM CaCl2, 5 mM beta-mercaptoethanol, 0.1% BSA, 1.5% Cellulase R10 (Yakult Honsha) and 0.75% Macerozyme R10 (Yakult Honsha)) for 5 h. Next, the protoplasts were collected and incubated in W5 solution (2 mM MES, pH 5.7, 154 mM NaCl, 5 mM KCl, 125 mM CaCl2) at 25°C for 30 min. Then, the protoplasts were collected by centrifugation at 100 g for 8 min and re-suspended in MMG solution (4 mM MES, 0.6 M Mannitol, 15 mM MgCl2) to a final concentration of 0.5–1.0 × 107 ml–1. Then, 8 μg plasmid was gently mixed with 100 μl protoplasts and 110 μl PEG-CaCl2 solution (0.6 M Mannitol, 100 mM CaCl2, and 40% PEG4000), and incubated at 25°C for 15 min. Two volumes of W5 solution were added to stop the transformation. Protoplasts were collected by centrifugation and re-suspended in WI solution (4 mM MES, pH 5.7, 0.6 M Mannitol, 4 mM KCl), then cultured in 24-well culture plates for 12 h in the dark. Finally, the transformed protoplasts were collected by centrifugation at 150 g for 8 min.
Transient expression assay in protoplasts
To determine the transcriptional activation activity of OsGAPDH1, full-length OsGAPDH1 was fused to the GAL4 DNA binding domain (GAL4DBD) to generate the GAL4DBD-OsGAPDH1 fusion construct driven by the 35S promoter as effector. Rice GAPDH1 and HXK1 promoters were used to drive the firefly luciferase LUC reporter gene. In order to check the effect of OsSRT1 on the transcriptional activation activity of OsGAPDH1, 35S:OsSRT1 construct as the coeffector of OsGAPDH1. The effector and reporter constructs, together with the construct containing the Renilla luciferase gene driven by the Arabidopsis UBIQUITIN3 promoter (50) as an internal control, were cotransfected into rice protoplasts in a ratio of 6:6:1 (effector:reporter:reference). The cotransfected protoplasts were cultured for 12 h at 25°C in the dark. The luciferase activities were measured using the Dual Luciferase Reporter Assay System (Promega) according to the manufacturer's instructions. The relative reporter gene expression levels were expressed as the ratio of firefly LUC to the Renilla luciferase.
For NAM (Nicotinamide) treatment of protoplasts, NAM was dissolved into 10 and 100 mM with WI solution. Transfected protoplasts were cultured using the WI solution for 12 h at 25°C in the dark, the luciferase activities were measured as described above.
Co-immunoprecipitation (CoIP) assays
For Co-IP assays in tobacco, total proteins were extracted from tobacco leaves that express the two candidate interaction proteins, which were tagged respectively with FLAG and GFP, in Co-IP buffer (50 mM Tris–HCl pH 8.0, 150 mM KCl, 1 mM EDTA, 0.5% Triton X-100, 1 mM DTT, 1 mM PMSF and 1× protease inhibitor cocktail tablets (Roche)). After the immunoprecipitation using GFP beads (GFP-TRAP, Chromo Tec), the samples were detected by the OsSRT1 antibody (28).
For the co-immunoprecipitation (Co-IP) assays in rice protoplasts, total proteins were extracted from OxSRT1–1 leaf protoplasts that were transfected with vectors expressing OsGAPDH1 and OsGAPDH2 tagged by GFP, in Co-IP buffer as described above. The protein extracts were mixed with GFP agarose beads (GFP-TRAP, Chromo Tec) and then incubated at 4°C for 2 h. After being washed at least 5 times, the agarose beads were recovered and mixed with the SDS-PAGE sample buffer. The samples were detected by immunoblot using anti-OsSRT1 antibody (28).
For the co-immunoprecipitation (Co-IP) assays with OsGAPDH1-GFP plants, total proteins were extracted from leaves with Co-IP buffer as described above. The protein extracts were mixed with GFP agarose beads (GFP-TRAP, Chromo Tec) and then incubated at 4°C for 2 h. After being washed at least 5 times, the agarose beads were recovered and mixed with the SDS sample buffer. The samples were detected by immunoblot using anti-SRT1 antibody (28).
Measures of glycolytic enzyme activity
The activities of HXK (51), PFK (52), PK (53) and GAPDH (54) were assayed as described previously, using kits purchased from SuZhou Keming Bioengineer Company, China, following the manufacturer's instructions. Different genotype plants were germinated, and transplanted into 1/2 MS medium. Seedlings (10-day-old) were collected at 9:00 am. The seedlings were grinded in liquid nitrogen and 100 mg grinded powers were incubated in 1 ml related Enzyme Extract Buffers and enzymatic activities were measured according to instructions of the kits purchased from SuZhou Keming Bioengineer Company.
Three biological replicates were performed on plants from 3 independent cultures, each replicate were measured with three technical repeats.
Specifically, HXK activities were based upon the reduction of NADP+ to NADPH through the following reactions:
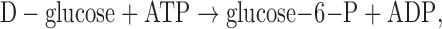 |
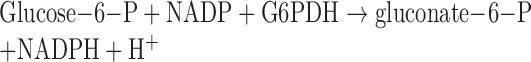 |
HXK activities were determined by NADPH absorbance at the wavelength (λ) 340 nm using the Tecan Microplate Reader.
PFK activities were measured based upon change of NADH concentration, and were determined by absorbance at the wavelength (λ) 340 nm using the Tecan Microplate Reader.
PK activities were based on change of NADH concentration, and measured by absorbance at the wavelength (λ) 340 nm:
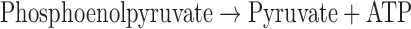 |
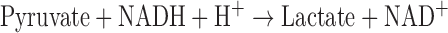 |
For GAPDH activity, chloroplasts was removed by centrifugation and GAPDH activities of cytosolic fractions were measured based on change of NADH concentration through the following reactions and determined by absorbance at the wavelength (λ) 340 nm using the Tecan Microplate Reader:
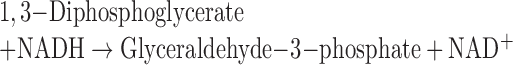 |
For α-KGDH activities, mitochondria were obtained by centrifugation. After sonicate, the α-KGDH activities of mitochondria were based on change of NADH concentration through the following reaction and determined by absorbance at the wavelength (λ) 340 nm using the Tecan Microplate Reader:
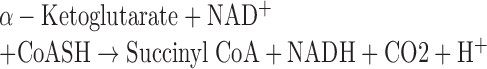 |
For PDH activities, mitochondria were obtained by centrifugation. After sonication, PDH activities of mitochondria were based upon change of 2,6-dDCPIP concentration through the reaction and determined by absorbance at the wavelength (λ) 600 nm using the Tecan Microplate Reader:
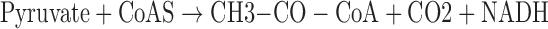 |
LC–MS/MS and data processing
Total proteins were extracted from tobacco leaves that express OsGAPDH1 tagged with GFP, then the protein were immunoprecipitated with anti-GFP antibody-conjugated beads magnetic beads. Next, 0.5 μg of the eluate was performed deacetylation assay with 0.5 μg of the eluate as control. Then the two mixtures were resolved by SDS/PAGE. The bands were excised from SDS/PAGE gel, fully trypsinized, and analyzed by reverse-phase LC-MS/MS. MS was carried out at Thermo Scientific™ Q Exactive™ HF and data were processed using the comparative proteomics analysis software Proteome Discoverer2.1.
Quantitative analysis of sugar content
Different genotype plants were germinated and transplanted into 1/2 MS medium. Seedlings (10-day-old) were collected at 9:00 am. Approximately 100 mg of frozen leaf was used for analysis. Soluble sugar was extracted using a fixation solution (225 μl of methanol, 120 μl of CHCl3, and 240 μl of ddH2O) at 70°C for 15 min. Samples were centrifuged at 12 000 rpm for 10 min; 200 μl of supernatant was transferred to a new tube and then dried at 80°C. For methoximation, 40 μl of methoxyamine hydrochloride in pyridine (20 mg/ml) was added for 90 min at 30°C. Then, 60 μl of N-methyl-N-trimethylsilyltrifluoroacetamide were added, and the mixture was incubated at 37°C for 30 min. The derivatives were analyzed by gas chromatography–mass spectrometry on a Thermo DSQII mass spectrometer using a DB-5 ms column.
In vitro lysine deacetylase enzymes activity assay
The deacetylation reaction was performed in 50 μl of reaction mixture containing Tris–HCl (pH 8.8), 5% glycerol, 50 mM NaCl, 4 mM MgCl2, 1 mM DTT, 500 μM NAD+, 3 μg GST-tagged SRT1 and 0.5 μg GFP-tagged OsGAPDH1 proteins expressed in tobacco leaf cells, than incubated at 25°C for 6 h, the reaction was terminated by boiling. Samples were analyzed by electrophoresis in 12% SDS-PAGE gels followed by immunoblotting.
Immunoblots
Proteins were extracted from wild-type and transgenic plants. After being washed in acetone and dried, the proteins were resuspended in Laemmli sample buffer (62.5 mM Tris–HCl, pH 6.8, 2% SDS, 25% glycerol, 0.01% bromophenol blue and 10% β-mercaptoethanol), then separated by 12% SDS-PAGE and transferred to an Immobilon-P polyvinylidene fluoride (PVDF) transfer membrane (GE Healthcare). The membrane was blocked with 2% BSA in PBS (pH 7.5) and incubated overnight with primary antibodies, such as anti-acetylated lysine (ab21623), anti-GAPDH (ab9485) and anti-histone H3 (ab1791) at room temperature. After three washes (30 min each), the secondary antibody [goat anti-rabbit IgG (Promoter)] at 1:10 000 dilution was used. Visualization was performed using the Super Signal West Pico kit (Thermo) according to the manufacturer's instructions.
Yeast two-hybrid assays
Constructs for yeast two-hybrid analysis were generated using the Matchmaker Gold Yeast Two-Hybrid System (Clontech) vectors pGBKT7 and pGADT7, which express protein fusions to the GAL4DNA-binding domain or transcriptional activation domain, respectively. Full length of cDNA inserts encoding OsSRT1 was introduced in pGADT7 using the restriction enzymes BamHI and EcoRI and the Matchmaker Gold Yeast Two-Hybrid System as described in the user manuals. Full length of cDNA inserts encoding OsGAPDH1 and OsGAPDH2 were introduced in pGBKT7 using the restriction enzymes BamH I and EcoR I and the Matchmaker Gold Yeast Two-Hybrid System as described in the user manuals. The analysis was performed in strain AH109 carrying HIS3 and MEL1 reporters for reconstituted GAL4 activity.
Phylogenetic analysis
The amino acid sequences of Arabidopsis GAPDH proteins were downloaded from The Arabidopsis Information Resource (http://www.arabidopsis.org/). The amino acid sequences of rice GAPDH proteins were downloaded from TIGR Rice Genome Annotation (http://rice.plantbiology.msu.edu/index.shtml). An unrooted neighbor-joining phylogenetic tree was constructed by generating 1000 random bootstrap replicates using MEGA 5.1 software (55).
OsGAPDH Family gene expression profile analysis
Expression profile of OsGAPPDH family genes in 27 tissues was extracted from the Rice Oligonucleotide Array Database (http://www.ricearrary.org/index.shtml). Expression values were obtained by searching the data using Affymetrix probe set ID of each gene for gene cluster analysis by the Cluster 3.0 program and the results were visualized by the Treeview program.
RESULTS
OsSRT1 represses glycolysis in rice seedlings
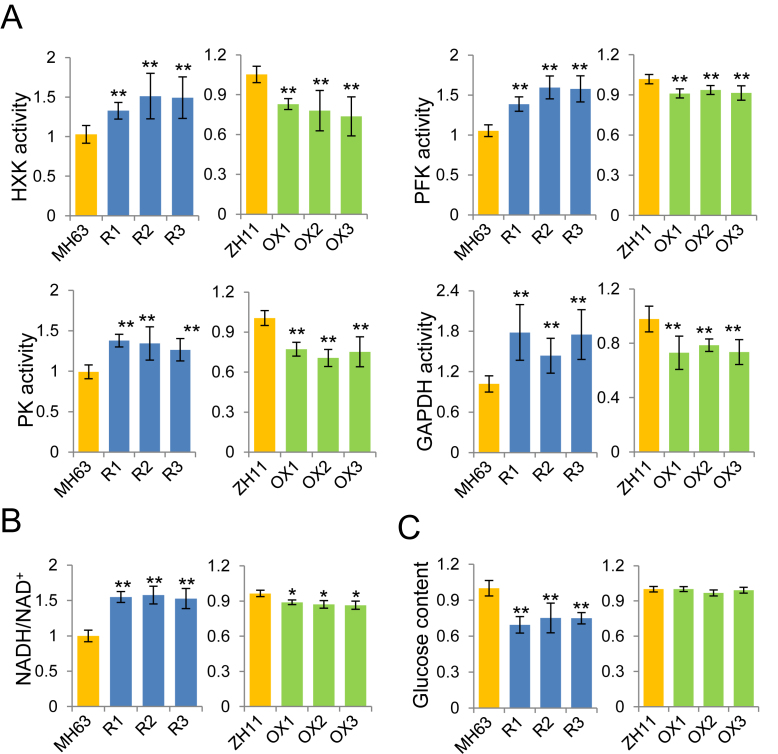
To study sirtuin protein function in plant carbon metabolism, we used RNAi and over-expression lines of OsSRT1 (21,29). Analysis of transcriptome and histone H3 lysine 9 acetylation (H3K9ac) chromatin immunoprecipitation sequencing (ChIP-seq) data of OsSRT1 RNAi seedlings found that genes involved in primary carbon metabolism such as glycolysis were enriched for higher H3K9ac and up-regulation (Supplementary Table S1) (28). qRT-PCR and ChIP-qPCR confirmed the higher H3K9ac and transcript levels of glycolytic genes encoding hexokinase (HXK), 6-phosphofructokinase (PFK), fructose-1,6-bisphosphatase (FBP), aldose 1-epimerase (Ald1-Ep), and GAPDH, (Supplementary Figure S1A and B). ChIP assays with polyclonal antibody of OsSRT1 revealed that the protein was also enriched at the promoter or the 5′ region of the glycolytic genes (Supplementary Figure S1C). Enzymatic activities of HXK, PK, PFK and GAPDH increased in the RNAi lines, but decreased in the over-expression plants (Figure 1A). The NAD+ levels decreased in the RNAi, but increased in the over-expression lines, whereas the NADH levels displayed the opposite accumulations. Consequently, the NADH/NAD+ ratio was clearly augmented in the knockdown plants but reduced in the over-expression lines (Figure 1B). This was consistent with the increases of activity of GAPDH and other glycolytic enzymes in the RNAi plants (Figure 1A). Dosage of sugar contents revealed a clear decrease of glucose accumulation in the RNAi and mutant seedlings (Figure 1C), while no change was observed for fructose, maltose, or sucrose (Supplementary Figure S2A). Collectively, the data indicated that the rice sirtuin protein OsSRT1 has a function to repress glycolysis. By contrast, mitochondrial respiration rate, as measured by oxygen consumption, was much reduced in the RNAi leaves while no clear change was observed in the over-expression plants after light to dark transition (Supplementary Figure S2B), during which period TCA cycle has been shown to be at the highest levels in plant leaves (25,26). Accordingly, key TCA enzymes such as pyruvate dehydrogenase (PDH) and α-ketoglutarate dehydrogenase (α-KGDH) were decreased in the RNAi and increased in the over-expression lines (Supplementary Figure S2C). The data suggest that OsSRT1 may positively regulate TCA cycle in plants.
Figure 1.
Expression levels of OsSRT1 affect glycolytic activity. (A) Relative activities of glycolytic enzymes HXK, PFK, PK, and GAPDH in 12-d-old seedlings of OsSRT1 RNAi (R1, R2 and R3), and over-expression transgenic (OX1, OX2 and OX3) compared to their respective wild type (MH63 and ZH11). (B) NADH/NAD+ ratios in the different genotypes. (C) Glucose contents in the OsSRT1 transgenic plants relative to wild type. Bars are means ± SD from three biological replicates and each replicate with three technical repeats. Significant differences are indicated (t test, n = 3), *P < 0.05; **P < 0.01.
OsSRT1 interacts with GAPDH
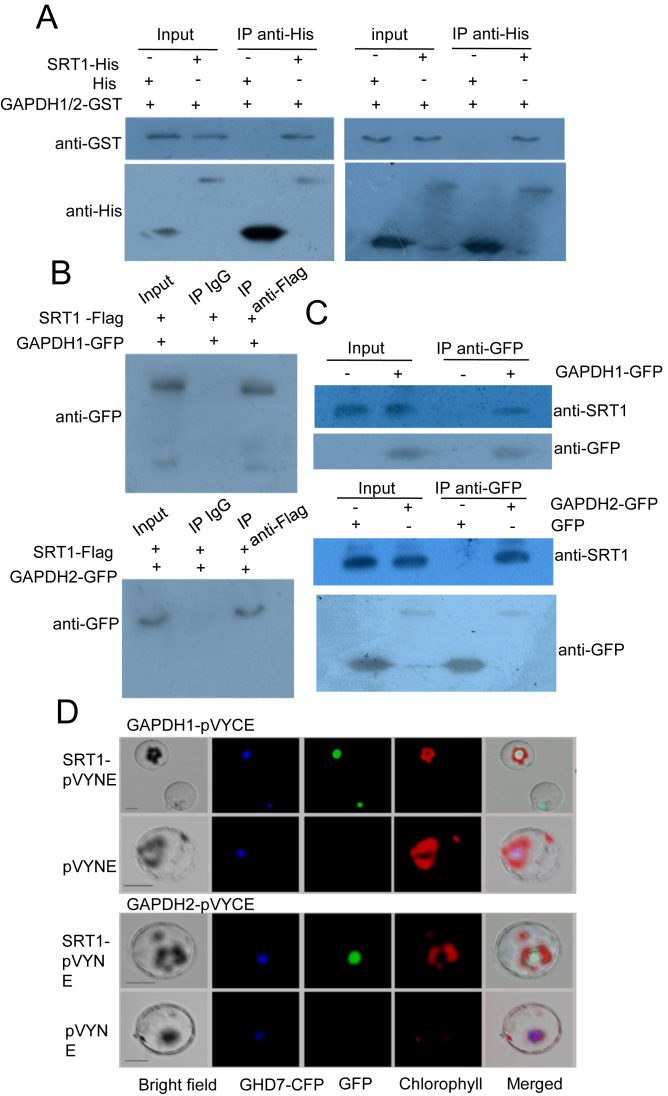
To study the molecular mechanism of OsSRT1-mediated regulation of glycolysis, we undertook yeast two-hybrid screenings with OsSRT1 as bait. GAPDH was found to be among the candidate of OsSRT1-interacting proteins identified. In rice, there are 3 cytosolic glycolytic GAPDH proteins (OsGAPDH1, OsGAPDH2 and OsGAPDH5), closely related to Arabidopsis GAPC1 and GAPC2 (Supplementary Figure S3A). GFP fusions of OsGAPDH1, OsGAPDH2 and OsGAPDH5 were found to be localized in both cytoplasm and nucleus, whereas GFP fusions of the other members were found to be co-localized with chloroplasts (Supplementary Figure S3A). OsGAPDH1, OsGAPDH2 and OsGAPDH5 were highly expressed in different organs and at different developmental stages (Supplementary Figure S3B). OsGAPDH1 and OsGAPDH2 proteins could form dimers in yeast two-hybrid system (Supplementary Figure S3C). To confirm the interaction between OsSRT1 and OsGAPDH1 or OsGAPDH2, we expressed the proteins tagged with 6XHis or GST in E. coli and performed in vitro pull-down assays. The analysis indicated that OsGAPDH1/2-GST fusion proteins could interact with OsSRT1-His in vitro (Figure 2A). The interaction was confirmed in transiently transfected tobacco cells using 35S:OsSRT1-Flag together with 35S:OsGAPDH1-GFP or 35S:OsGAPDH2-GFP. From the transfected tobacco cell extracts, OsGAPDH1-GFP or OsGAPDH2-GFP proteins could be precipitated by anti-Flag but not by IgG (Figure 2B). Furthermore, OsSRT1 protein was shown to interact with OsGAPDH1-GFP or OsGAPDH2-GFP in transgenic rice cells by co-immunoprecipitation with anti-GFP followed by Immunoblots with the antibody of OsSRT1 (Figure 2C). The interaction occurred in the nucleus as demonstrated by BiFC in rice protoplast cells expressing OsGAPDH1/2-pVYCE and OsSRT1-pVYNE (Figure 2D).
Figure 2.
OsSRT1 interacts with OsGAPDH1 and OsGAPDH2. (A) Pull-down assays of interaction between OsSRT1 and OsGAPDH1 (left) and OsGAPDH2 (right). OsSRT1–6XHis fusion protein or 6XHis alone (from PET32a vector) was incubated with OsGAPDH1-GST or OsGAPDH2-GST followed by immunoprecipitation with anti-His antibody and analyzed by immunoblots with anti-GST and anti-HIS. (B) Co-IP assays of in vivo interaction between OsSRT1 and OsGAPDH1 (upper part) and OsGAPDH2 (lower part) in tobacco cells. Nuclear proteins were extracted from tobacco leaves that were injected with Agrobacteria transformed with 35S:OsSRT1-FLAG together with binary vectors 35S:GAPDH1-GFP or 35S:GAPDH2-GFP, and immuno-precipitated with anti-FLAG or with IgG as controls, followed by immunoblots with anti-GFP. (C) Co-IP assays of in vivo interaction between OsSRT1 and OsGAPDH1 (upper part) and OsGAPDH2 (lower part) in rice cells. Nuclear proteins were extracted from wild type or 35S:OsGAPDH1-GFP transgenic rice seedlings (10-day-old), or from rice seedling protoplasts transiently transfected with 35S:OsGAPDH2-GFP or with 35S:GFP, and immuno-precipitated with anti-GFP, followed by immunoblots with anti-OsSRT1 and anti-GFP. (D) Interactions between OsSRT1 and OsGAPDH1/2 occur in the nucleus. Rice protoplasts were transfected by OsGAPDH1-pVYCE (left) or OsGAPDH2-VYCE (right) with OsSRT1-pVYNE or pVYNE alone. Reconstructed fluorescent YFP by the interactions was visualized by excitation at 488 nm. GHD7-CFP, a rice nuclear localized protein, was used as a control. Chlorophyll auto-fluorescence was excited at 543 nm. Bars = 10 μm.
OsGAPDH1 is required for expression of glycolytic genes
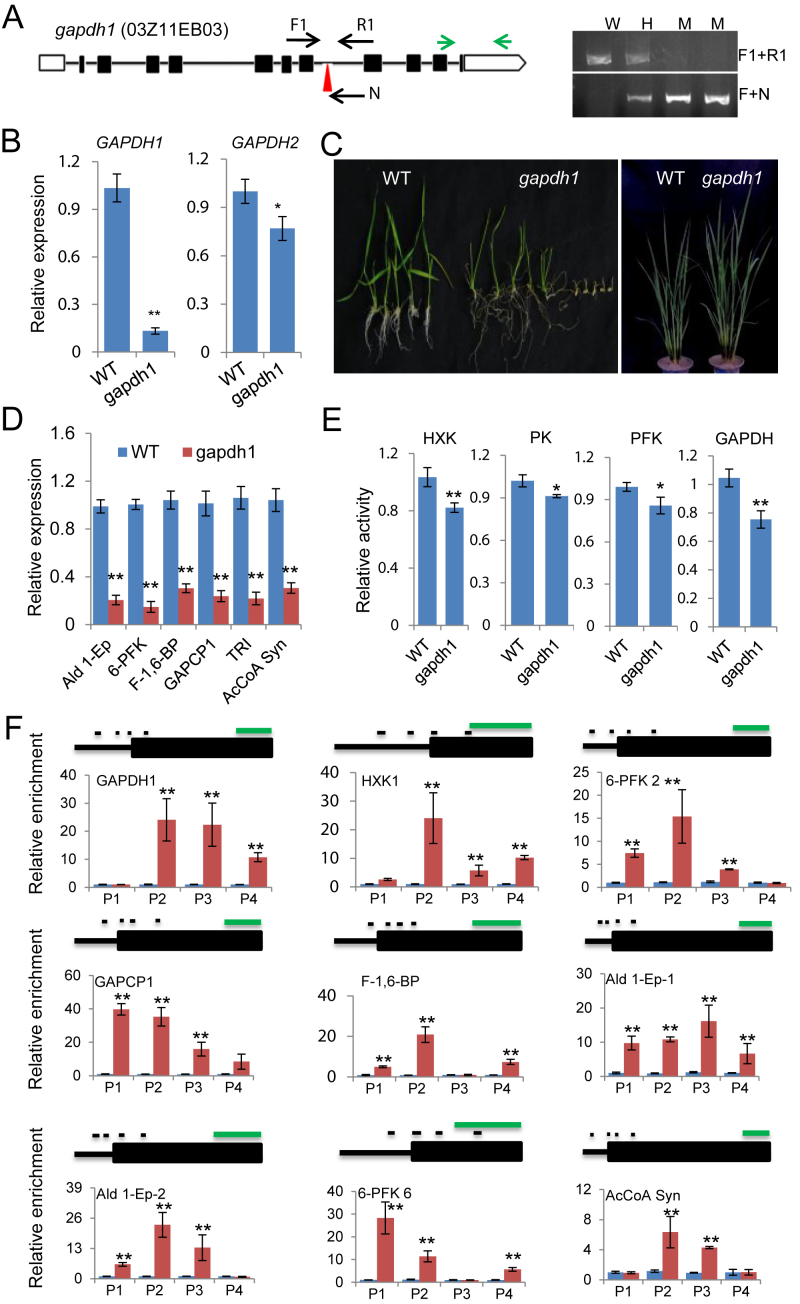
To study functional significance of the interaction between OsSRT1 and OsGAPDH1, we identified a T-DNA mutant line of OsGAPDH1 (Figure 3A), in which the transcript level of the gene was severely reduced (Figure 3B). Interestingly, the expression of OsGAPDH2 was also reduced (Figure 3B), suggesting that OsGAPDH1 loss-of-function may affect the expression of OsGAPDH2. A similar situation was observed in individual knockout lines of Arabidopsis GAPDH genes, in which transcripts and enzymatic activity of most of GAPDH family members are reduced (56). Germination rate of the mutant was highly reduced (Supplementary Figure S3F), and seedling growth was severely retarded, but the delay was made up at mature stages (Figure 3C). In the mutant, transcripts of other glycolytic genes were clearly reduced (Figure 3D). Glycolytic enzymatic activities (i.e. HXK, PK, PFK and GAPDH) were reduced in the mutant seedlings (Figure 3E). The similar effects on gene expression and germination were also produced in several RNAi lines of OsGAPDH1 (Supplementary Figure S3E and F).
Figure 3.
OsGAPDH1 is required for glycolytic enzymes activity and gene expression. (A) Characterization of gapdh1 T-DNA mutant line. Left, Schematic representation of the OsGAPDH1 locus. The T-DNA insertion position in the seventh intron is indicated by red arrow head. The positions of primers F1, R1, and N used for genotyping are indicated. OsGAPDH1 specific primers used for RT-PCR are indicated by green arrows. Right, Genotyping of the gapdh1 T-DNA insertion. W, wild type; H, heterozygosis; M, homozygote. (B) Detection of OsGAPDH1/2 transcripts level in gapdh1. (C) The gapdh1 mutant phenotypes at seedling and maturation stages. (D) Transcript levels of glycolytic genes (Ald 1-Ep, 6-PFK, F-1,6-BP, GAPCP-1, TRI and AcCoA Syn). Actin1 was used as an internal control. (E) Glycolytic enzyme activities in the gapdh1 mutant relative to wild type. (F) OsGAPDH1 associated directly with the glycolytic genes. ChIP-qPCR assays were performed with seedling chromatin fragments of OsGAPDH-GFP plants using anti-GFP and anti-IgG, respectively. Small black lines indicate the relative positions of the primer sets P1 to P4 (left to right). Green line = 1 kb. Anti-IgG was used as controls. Bars in qRT-PCR, enzymatic activities, and ChIP-PCR are means ± SD from three biological replicates. Each replicate was performed using independently cultured plants. Significant enrichments (t test) calculated from three biological replicates are indicated (*P value < 0.05, **P value < 0.01).
We also produced several 35S:OsGAPDH1-GFP over-expression plants in which the OsGAPDH1-GFP fusion protein was detected in both cytoplasm and nucleus (Supplementary Figure S3G). Analysis of the transgenic plants by ChIP-qPCR assays using anti-GFP revealed clear enrichments of OsGAPDH1-GFP mostly in the 5′ regions (near the transcriptional start site, TSS) of 9 glycolytic genes (Figure 3F), suggesting that GAPDH may be associated to chromatin of glycolytic genes promoters.
OsGAPDH1 trans-activates glycolytic gene promoters
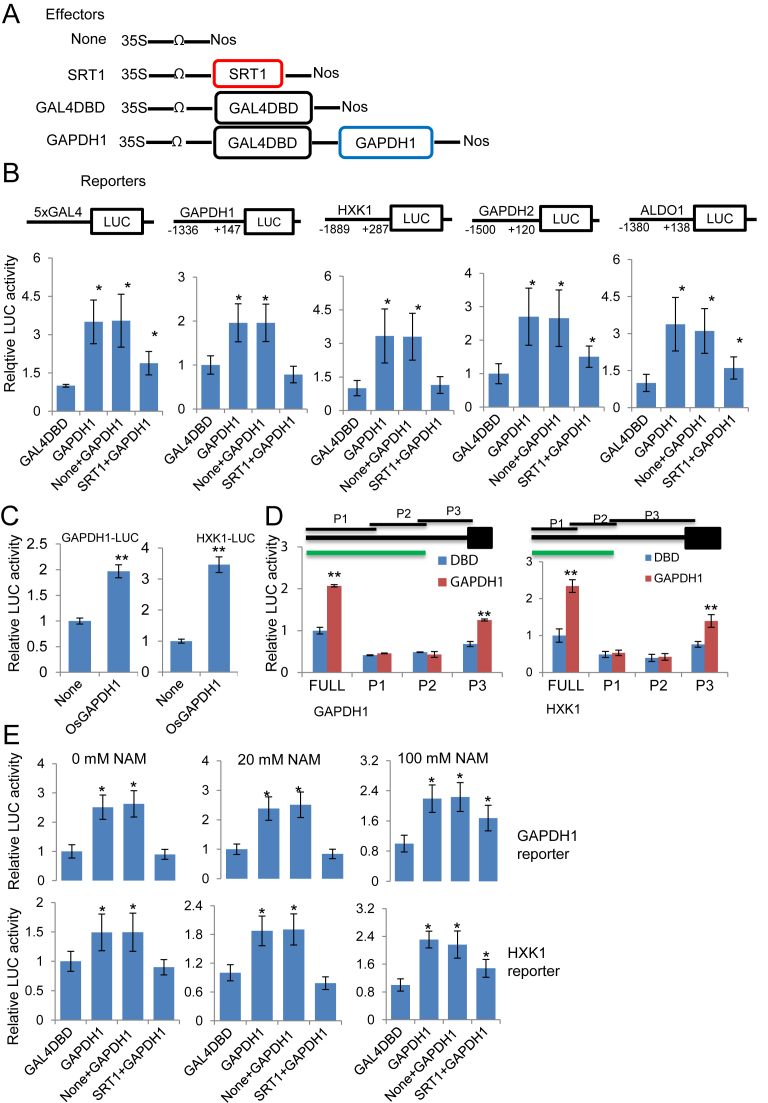
To check whether OsGAPDH1 could trans-activate glycolytic gene promoters, we constructed a vector expressing the fusion between OsGAPDH1 and the GAL4 DNA-binding domain under the control of 35S promoter (35S:GAL4DBD-GAPDH1), and used this effector vector and the control vector (35S:GAL4DBD alone) to co-transfect rice protoplasts together with the GAL4-LUC reporter vector (Figure 4A), as described previously (50). The GAL4DBD-GAPDH1 fusion augmented the LUC activity by ∼5-fold compared to GAL4DBD alone, indicating a transcriptional activation function of OsGAPDH1 (Figure 4B and E). To study whether OsGAPDH1 activates glycolytic gene promoters, we constructed reporter vectors using the promoter region of OsGAPDH1, OsGAPDH2, OsHXK1 and OsALDO1 to drive the expression of luciferase (LUC) gene (Figure 4B), and co-transfected rice protoplasts together with the effector vector 35S:GAL4DBD-GAPDH1 or the control vector 35S:GAL4DBD. The analysis found similar results as with the GAL4-LUC reporter (Figure 4B). Without the DBD domain, OsGAPDH1 could also activate the tested glycolytic promoters (Figure 4C). The data indicated that OsGAPDH1 could bind to and activate glycolytic gene promoters and OsGAPDH1 was auto-regulated. Similarly, OsGAPDH2 and GAPDH5 showed a transactivation function (Supplementary Figure S4A). Deletion analysis of OsGAPDH1 and OsHXK1 promoters revealed that the P3 region (close to TSS) of each promoter was responsive to transactivation by the OsGPADH1 protein (Figure 4D), consistent with the enrichment of the protein on the promoter regions (Figure 3F). However, the transcriptional activity with P3 was lower than with the full promoter, suggesting that sequences in P2 region may be also involved in OsGAPDH1-mediated transactivation. Interestingly, when deleted to the C (catalytic) or the N (NAD binding) domain, OsGAPDH1 was still able to activate the reporters (Supplementary Figure S4B). A possible explanation would be that both the C and N domains served as scaffold for homo-dimerization with endogenous GAPDH proteins that exist in tetramers in rice (57), or interaction with other factors involved in transcription.
Figure 4.
OsGAPDH1 activates glycolytic gene promoters in transient expression assays. (A) Schematic representation of the effectors used in the transient transfection assays of tobacco protoplasts. (B) Luciferase activities from rice protoplasts transfected by the five reporters together with the indicated effectors. (C) 35S:OsGAPDH1 directly activates the OsGAPDH1 and OsHXK promoters. (D) The OsGAPDH1 and OsHXK1 promoter regions near the transcriptional start site are responsive to transactivation by the OsGAPDH1 protein. green line = 1 kb. (E) Luciferase activities from rice protoplasts transfected by OsGAPDH1 and OsHXK1 reporters together with the same effectors as in (B) in the presence of 0, 20 or 100 mM nicotinamide (NAM). The effector and reporter constructs, together with the construct containing the Renilla luciferase gene as an internal control, were cotransfected into rice protoplasts in a ratio of 6:6:1 (effector: reporter: reference). Y-axis, relative reporter gene expression levels expressed as the ratio of firefly LUC to the Renilla LUC activities. Data are values of three independent assays. Significant differences with the corresponding control values are indicated by *P < 0.05 and **P < 0.01, t test, n = 3.
To test whether OsSRT1 had any effect on the OsGAPDH1 function to activate the reporter genes, we included in the transfections assays the 35S:OsSRT1 construct and the empty vector as a control (Figure 4A). The presence of 35S:OsSRT1 abolished the transactivation effect of GAL4DBD-GAPDH1, 2, or 5 (Figure 4B and E, Supplementary Figure S4A), indicating that OsSRT1 inhibited the OsGAPDH1 function of transcriptional activation. Addition of nicotinamide (NAM), an inhibitor of Sir2 family deacetylases, which has been shown to inhibit sirtuins in mammalian/yeast cell culture at 100 mM (58,59), eliminated the inhibitory effect of OsSRT1 on OsGAPDH1 transactivation function (Figure 4E). The results indicated that the deacetylation activity of OsSRT1 was necessary to inhibit OsGAPDH1-mediated gene activation.
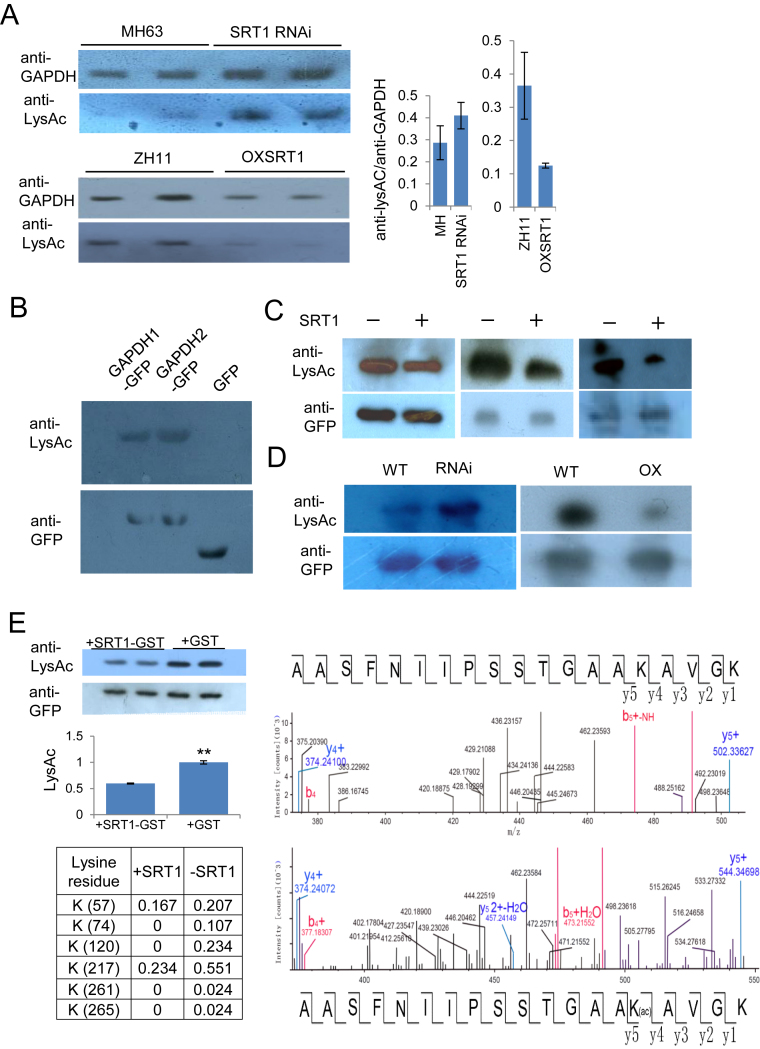
Lysine acetylation of OsGAPDH1 is reversed by OsSRT1
It has been reported that GAPDH proteins are acetylated in animal and Arabidopsis cells (41–43). To check whether rice GAPDHs were acetylated, we precipitated leaf nuclear proteins extracts of wild type and OsSRT1 RNAi and over-expression lines with commercial anti-GAPDH antibodies that recognize plant GAPDHs. The precipitated GAPDH proteins were analyzed by immunoblots using anti-GAPDH and anti-LysAc antibodies. The analysis revealed relatively higher acetylated GAPDH levels in SRT1 RNAi nuclei and lower acetylated GAPDH levels in the over-expression lines than in the respective wild type (Figure 5A). To study whether OsGAPDH1, 2 were acetylated, we isolated OsGAPDH1-GFP and OsGAPDH2-GFP fusion proteins by immune-purification using anti-GFP resins from transiently transfected tobacco leaf cells and analyzed by Immunoblots using antibodies of pan acetylated lysine residues (anti-LysAc). The analysis revealed lysine acetylation of OsGAPDH1-GFP and OsGAPDH2-GFP, but not GFP alone (Figure 5B). To study whether OsSRT1 could deacetylate OsGAPDH1, nuclear proteins of tobacco leaf cells transfected with 35S:OsGAPDH1-GFP together with 35S:SRT1-Flag or the empty vector, were immunoprecipitated with anti-GFP followed by immunoblots with anti-LysAc and anti-GFP. The presence of OsSRT1 reduced the acetylation levels of OsGAPDH1 (Figure 5C). To study whether OsSRT1 deacetylates OsGAPDH1-GFP in rice cells, we introduced the binary vector 35S:OsGAPDH1-GFP into protoplasts of OsSRT1 RNAi, over-expression and wild type leaves by transfection. The OsGAPDH1-GFP fusion proteins were immunopreciptated with anti-GFP antibody. The precipitated proteins were subsequently analyzed by immunoblots using anti-LysAc and anti-GFP. The analysis revealed, for the same amounts of OsGAPDH1-GFP fusion protein, a higher acetylation level of the protein was detected in the RNAi, and a lower acetylation level was detected in the over-expression backgrounds compared to the wild type (Figure 5D). Finally, to test whether OsSRT1 could deacetylate OsGAPDH1 in vitro, we produced and isolated OsGAPDH1, 2 or 5-GFP from tobacco cells and incubated with E coli-produced OsSRT1-GST or GST alone. Lysine acetylation levels of OsGAPDH-GFP fusions were tested by immunoblots with anti-LysAc and anti-GFP (for loading controls). The analysis confirmed that OsSRT1 could deacetylate OsGAPDH1, 2 and 5 (Figure 5E; Supplementary Figure S4D).
Figure 5.
OsSRT1 deacetylates OsGAPDH1. (A) Lysine acetylation of rice endogenous GAPDH is regulated by OsSRT1. Nuclear proteins isolated from wild type (MH63 and ZH11), OsSRT1 RNAi and OXSRT1 transgenic plants leaves were immunoprecipitated by anti-GAPDH and analyzed by immunoblots using anti-GFP and anti-LysAc (acetylated lysine residues). Relative LysAc/GAPDH signals in the different genotypes are measured by ImageJ (right). Bars are means ± SD from three measures of each of the two repeats. (B) In vitro acetylation of OsGAPDH1 and OsGAPDH2. Transiently expressed OsGAPDH1-GFP, OsGAPDH2-GFP or GFP alone were purified from tobacco cell extracts with anti-GFP beads, analyzed by immunoblots with anti-LysAc and anti-GFP as controls. (C) OsSRT1 reduces OsGAPDH1 acetylation levels. Tobacco leaves cells transiently expressed with 35S:GAPDH1-GFP together with (+) or without (–) 35S:OsSRT1-FLAG, immunoprecipitated with anti-GFP, and analyzed by immunoblots with anti-LysAc and anti-GFP. (D) OsSRT1 reduces OsGAPDH1 acetylation levels in rice cells. Protoplasts of wild type (WT), OsSRT1 RNAi or over-expression (OXSRT1) leaves were transiently transfected with 35S:OsGAPDH1-GFP. Nuclear proteins of transfected protoplast were immunoprecipitated by anti-GFP and analyzed by immunoblots with anti-GFP and anti-LysAc, respectively and indicated at left. (E) Left upper part, OsSRT1 deacetylates OsGAPDH1 in vitro. Escherichia coli-produced OsSRT1-GST protein was incubated with tobacco cell-produced OsGAPDH1-GFP. Anti-GFP was used for loading controls and anti-LysAc detected lysine acetylation levels of OsGAPDH1-GFP in the absence or presence of OsSRT1-GST. Left lower part, acetylation ratios of indicated lysine residues of OsGAPDH1 detected by IP-MS analysis of tobacco leaf cell-expressed GAPDH1-GFP protein (0.5 μg) in the presence or absence of OsSRT1-GST (3 μg). Right, molecular mass of an OsGAPDH1 peptide without (upper) or with (lower) K217 acetylation (Ac) detected by MS.
To further confirm the deacetylation of OsGAPDH1 by OsSRT1, we performed mass spectroscopy analysis of tobacco cell-produced OsGAPDH1 and found that 16 lysine residues of OsGAPDH1 were acetylated (Figure 5E, Supplementary Table S2; Supplementary Dataset 1). Incubation with OsSRT1 clearly reduced the acetylation levels of six of them (K57, K74, K120, K217, K261 and K265) (Figure 5E). Previous results have shown that Arabidopsis GAPC2 protein produced in E. coli is acetylated at four lysine residues (K130, K216, K220 and K255) (43), of which K220 corresponded to rice K217 that was the most frequently acetylated among the six lysine residues (Figure 5E).
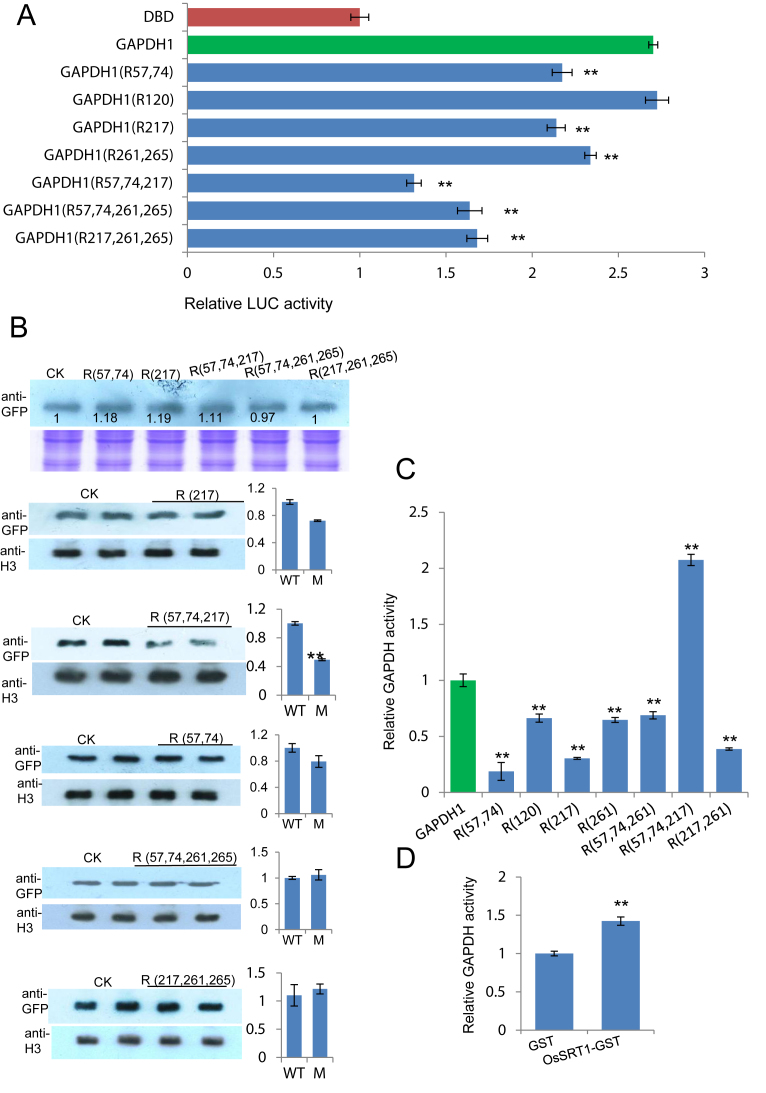
To test whether OsSRT1-regulated lysine acetylation was important for OsGAPDH1, we substituted the six acetylated lysine residues by Arg individually or in combinations of two or three residues and tested transcriptional activity of the mutants in transient transfection assays. The tests revealed that except Lys120, substitution of the other five Lys additively reduced the transactivation activity of the protein, suggesting that lysine acetylation of OsGAPDH1 stimulated its transcriptional activity (Figure 6A). In fact, Arg substitutions of Lys 57, 74 and 217 together showed a clear reduction of relative nuclear accumulation of OsGAPDH1-GFP (Figure 6B). However, the substitutions of one or two lysines reduced the GAPDH enzyme activity while simultaneous substitutions of Lys 57, 74 and 217 resulted in increases of the activity in in vitro tests (Figure 6C). Incubation of with OsSRT1 also increased the enzymatic activity of OsGAPDH1-GFP produced in tobacco cells (Figure 6D).
Figure 6.
Functional analysis of Lys to Arg substitution mutants of OsGAPDH1. (A) Transactivation function of indicated single, double or triple substitution mutants compared to wild type OsGAPDH1 in transient transfection assays. (B) Effect of the substitutions on nuclear accumulation of OsGAPDH1-GFP in transfected rice protoplasts. The first panel shows relative transfection efficiency of each construct tested by immunoblots with anti GFP. Coomassie staining was used for loading controls. The lower panels show nuclear accumulation of the substitution mutants relative to wild type OsGAPDH1 revealed by immunoblots using anti-GFP. Anti-H3 was used to control loadings of nuclear proteins. The ImagJ software was used to quantify the signals indicated on the right. (C) In vitro enzymatic activities of the substitution mutants relative to the wild type OsGAPDH1 protein. The wild type and mutants were produced as GFP fusion proteins in tobacco cells and purified with anti-GFP resins. (D) Enzymatic activity of tobacco cell-produced OsGAPDH1-GFP after incubation with E. coli-produced OsSRT1-GTS or GST alone. Significant differences are indicated by **P value < 0.01.
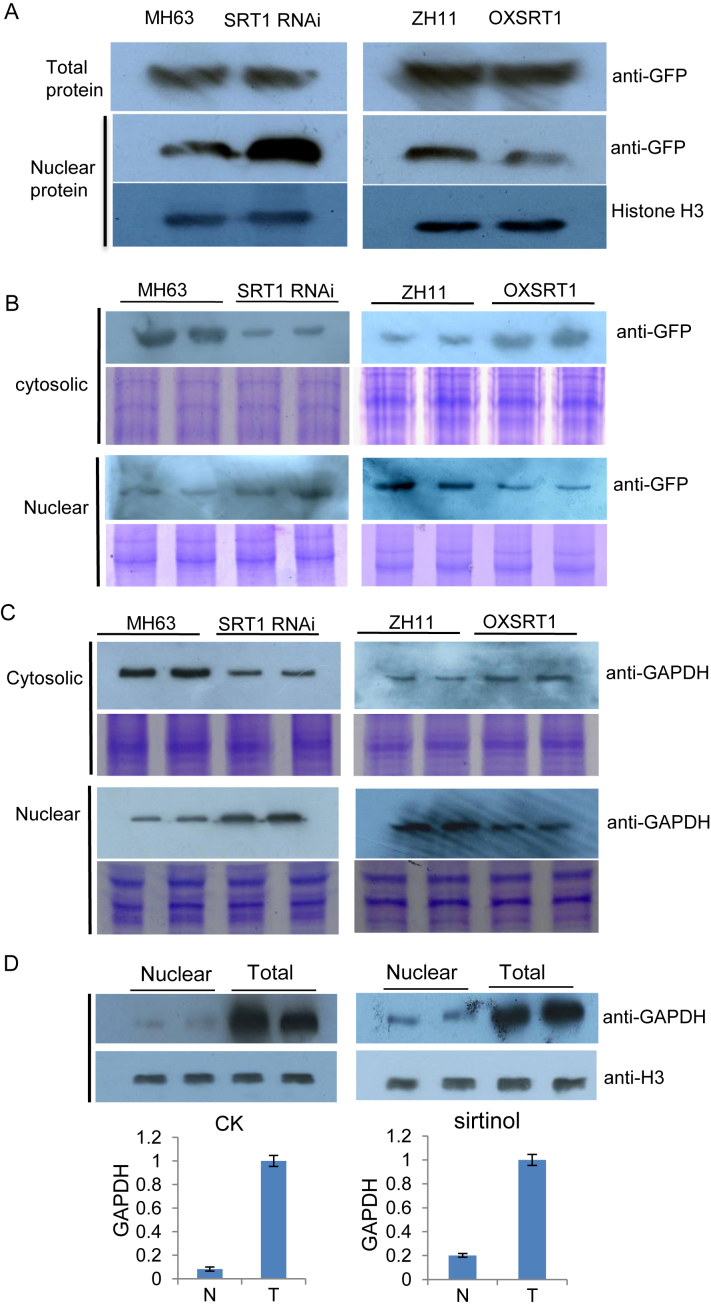
OsSRT1 regulates nuclear accumulation of GAPDH
The OsSRT1 RNAi or over-expression seemed not clearly alter the overall levels of GAPDH proteins, although some increases could be observed in the RNAi plants (Supplementary Figure S5A). This may be also due to the fact that the polyclonal antibody detects all forms of (including chloroplastic) GAPDH which may dilute any increase of OsGAPDH1 levels. Next, we studied whether OsSRT1 was involved in the regulation of rice GAPDH accumulation in the nucleus. Protoplasts were prepared from leaves of OsSRT1 RNAi, over-expression and wild type plants and were transfected with 35S:OsGAPDH1-GFP. Fractions of transfected protoplasts were used for total cellular and nuclear proteins extraction and analyzed by immunoblots using anti-GFP and anti-H3 as controls of nuclear protein loading. The analysis revealed that with similar levels of total OsGAPDH1-GFP protein, the nuclear fraction of the fusion protein was much higher in the OsSRT1 RNAi than in the wild type background. By contrast, in the over-expression plants, the nuclear fraction of the fusion protein was lower (Figure 7A). Analysis of the nuclear and cytosolic fractions by immunoblots using anti-GFP revealed that compared to wild type, much lower and much higher levels of OsGAPDH1-GFP were respectively detected in the RNAi and the over-expression supernatants or cytosolic fractions (Figure 7B). Conversely, compared to wild type, higher levels of the protein were detected in the nuclear fraction of the RNAi and lower levels in the over-expression line (Figure 7B). To study whether endogenous GAPDH protein level in the nucleus was regulated by OsSRT1, nuclear and soluble cytosolic protein fractions of protoplasts isolated from wild type and the transgenic leaves were analyzed by immunoblots using anti-GAPDH. The analysis revealed that there was a relative higher nuclear accumulation of GAPDH in the RNAi and lower accumulation in the over-expression lines compared to their respective wild type (Figure 7C). These data suggested that OsSRT1 may have a function to regulate the nuclear accumulation of OsGAPDH1. OsSRT1-mediated inhibition of GAPDH nuclear accumulation is likely due to its deacetylation function, as treatment of wild type leaf protoplasts with the deacetylase inhibitor sirtinol increased the nuclear proportion of GAPDH (Figure 7D). This hypothesis was supported by reduced accumulation of the substitution mutants of OsGAPDH1 (Figure 6B).
Figure 7.
OsSRT1 reduces nuclear accumulation of OsGAPDH1 in rice protoplasts. (A) Total and nuclear protein extracts of wild type (MH63 or ZH11), OsSRT1 RNAi and over-expression (OXSRT1) leaf protoplasts transfected with 35S:OsGAPDH1-GFP were analyzed by immunoblots using anti-GFP. Anti-H3 was used loading controls. (B) Soluble (supernatant) and insoluble (nuclear pellet) fractions of the lysed transfected protoplasts from the different genotypes (same as A) were analyzed by Western blots using anti-GFP shown at right. (C) Detection of endogenous GAPDH protein levels in the nuclear and supernatant fractions of lysed rice leaf protoplasts detected by anti-GAPDH antibody. Coomassie-staining in B and C are shown as loading controls. (D) Relative nuclear accumulation of GAPDH in rice protoplasts treated with (right) or without (left) the sirtuin inhibitor sirtinol revealed by anti-GAPDH. Anti-H3 was used controls. Nuclear (N) fractions relative to total (T) GAPDH levels were determined by three measures of the two repetitions with ImagJ.
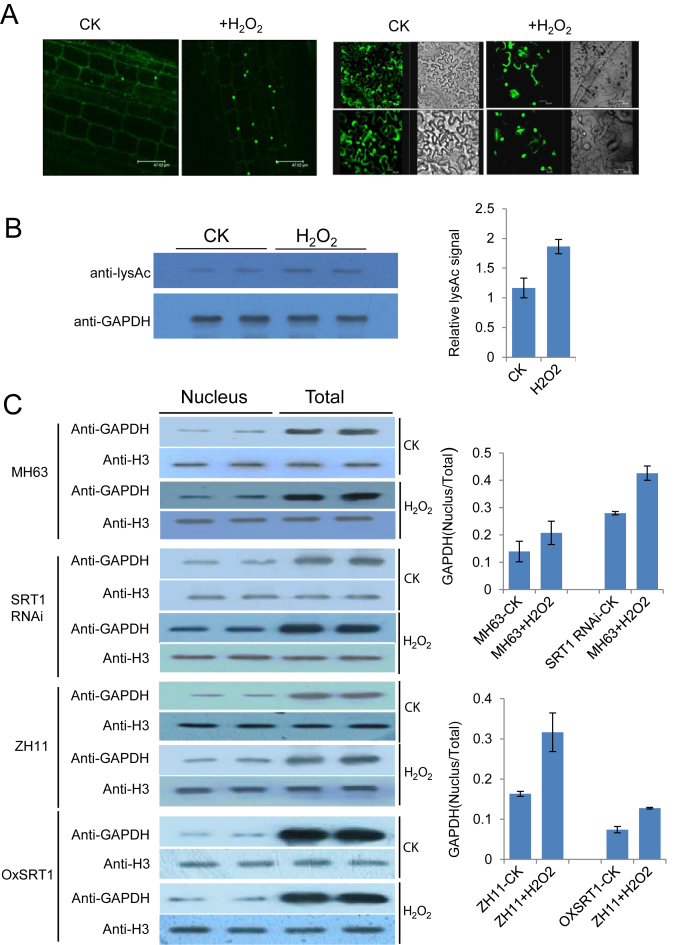
OsSRT1 inhibits stress-induced nuclear accumulation of GAPDH
As mentioned earlier, the nuclear localization of Arabidopsis GAPDH is enhanced by oxidative stresses. Nuclear localization of OsGAPDH1-GFP was also enhanced in rice leaf cells and in transiently transfected tobacco leaf cells when treated with 0.1% H2O2 during 30 min (Figure 8A). Immunoblot analysis with anti-GAPDH in rice leaves indicated that H2O2 treatment did not clearly alter the overall levels of total GADPH proteins in either wild type or the OsSRT1 transgenic lines (Supplementary Figure S5B), but increased acetylated fraction of the protein (Figure 8B). This reinforced the idea that oxidative stress induces GAPDH posttranslational modification to promote nuclear localization in plant cells. However, the GAPDH enzymatic activity was increased (Supplementary Figure S3H), consistent with higher glycolytic activity in stressed plant tissues.
Figure 8.
OsSRT1 inhibits nuclear accumulation of OsGAPDH1 induced by oxidative stress. (A) OsGAPDH1 is enriched in the nucleus under stress conditions. Cellular GFP signal in OsGAPDH1-GFP root cells (left) and tobacco leaf cells (right) transiently expressed 35S:OsGAPDH1-GFP, treated with or without (CK) H2O2 (0.1%) for 30 minutes. Bars = 47 μm (left) or 20 μm (right). (B) H2O2 treatment increased the fraction of acetylated GAPDH in rice leaves. GAPDH proteins were immunoprecipitated from nuclear extracts of leaves treated with or without H2O2 (0.1%) for 30 min and analyzed by immunoblots with anti-LysAc and anti-GAPDH. Relative signals were determined by three measures of the two repetitions with ImagJ (right). (C) Total and nuclear protein extracts of OsSRT1 RNAi, OXSRT1, and their respective wild type (MH63, ZH11) seedling leaves treated with or without H2O2 (0.1%) for 30 min and analyzed by immunoblots using anti-GAPDH antibody. Anti-H3 was used as loading controls. The bands were quantified by ImageJ program. The signals of the nuclear fractions relative to that of total proteins were calculated after normalization with H3 signals. Bars are means ± SD of three measures of each of the two repeats.
Immunoblots with anti-GAPDH were performed to analyze the total and nuclear protein fractions extracted from seedling leaves of wild type and the OsSRT1 transgenic lines treated with or without 0.1% H2O2. Anti-histone H3 antibody was used as loading controls. The analysis revealed that there were relatively higher nuclear accumulations of GAPDH in the RNAi and lower accumulations in the over-expression lines compared to their respective wild type (Figure 8C). The H2O2 treatment increased the nuclear accumulation in the wild type, while the increase was further enhanced in the RNAi but attenuated in the over-expression lines (Figure 8C). The data suggested that OsSRT1 may have a function to prevent stress-induced nuclear accumulation of GAPDH in rice, which is presumably achieved through deacetylation of the protein.
DISCUSSION
OsSRT1 function of glycolysis regulation in rice
Mammalian sirtuin protein SIRT6 is shown to play a role in redirecting carbohydrate flux from glycolysis to mitochondrial respiration through directly suppression of expression of multiple glucose-metabolic genes such as pyruvate dehydrogenase kinase-1 (PDK1), lactate dehydrogenase (LDH), phosphofructokinase-1 (PFK1), and glucose transporter-1 (GLUT1) by interacting with hypoxia inducible factor-1a (HIF1a) and deacetylating H3K9 at the promoter of HIF1a target genes (60,61). HIF1a is known to modulate multiple genes to activate glycolysis and repress simultaneously mitochondrial respiration in a coordinated fashion in mammalian cells. The present work indicates that the rice sirtuin protein OsSRT1, which is most closely related to SIRT6 among the other mammalian sirtuins (24), also represses glycolysis. The OsSRT1-mediated repression of glycolysis would favor TCA cycle to support plant growth and fitness under normal conditions. This hypothesis is supported by the reduced growth of OsSRT1 RNAi plants and more rigorous growth of OsSRT1 overexpression plants (21). However, the present data indicate that in addition to epigenetically repressive glycolytic gene expression, OsSRT1 may repress glycolysis by deacetylating and inactivating the regulatory function of OsGAPDH1 that is shown to activate glycolytic genes. Therefore, substantial mechanistic difference may exist between plant and animal sirtuin proteins for glycolysis repression.
Being influenced by photosynthesis and photorespiration, carbon metabolic flux is complex in plant cells. For instance, in illuminated leaves, glycolysis is low, cytosolic glucose molecules do not enter glycolysis and are committed to sucrose synthesis, and there is only little consumption by glycolysis of triose-phosphates synthesized by photosynthesis (25,26). This indicates that glycolysis is low in photosynthetically active cells. The higher glycolic activity in OsSRT1 RNAi and mutant leaves supports the hypothesis that OsSRT1 may contribute to lower glycolysis in photosynthetic cells. In the dark, enhanced TCA operates in leaf cells producing energy and carbon backbones supporting biosynthesis and growth (26). Intuitively, in the dark OsSRT1 activity in leaf cells would be induced by high levels of NAD+ generated by TCA cycle, which would in turn enhance TCA cycle and reduce glycolysis. This presumption is supported by decreases of mitochondrial respiration rates (consumption of O2) observed in OsSRT1 RNAi leaves after light to dark transition (Supplementary Figure S2B).
Plant glycolysis is induced by diverse biotic and abiotic stresses, such as pathogen attack, oxidative stress, and hypoxia or O2 deficiency (56,62,63). The latter case is particularly important for anaerobic germination and seedling growth in rice (63). Enhanced glycolytic activity has been linked to promotion of cell survival (32,64). In plants undergoing stress responses, enhanced glycolysis may be important for providing additional energy required for resistance or tolerance to stresses and to reactive oxygen species (ROS) scavengers such as pyruvate and NADH. Under hypoxia conditions such as flooding and submergence, oxidative phosphorylation and TCA cycle are inhibited; whereas glycolysis is activated and is supported by fermentation that re-oxidizes NADH to NAD+ to allow the production of ATP required for the survival of plant cells under submergence. This is especially important for rice that has unique capability to germinate and establish seedlings under complete submergence, and to adjust growth to adapt to flooding. The reduced germination rate and seedling growth of OsGAPDH1 knockout/knockdown plants supports this hypothesis (Supplementary Figure S3F, Figure 3C). Under hypoxia conditions, NAD+/NADH ratio is low which may lead to reduced OsSRT1 activity, and consequently enhanced glycolysis enabling plant stress tolerance or resistance, whereas under normal growth conditions, OsSRT1 is likely to favor TCA cycle to support biosynthesis and growth. From this point of view, OsSRT1 may play an important role in the trade-off between stress resistance and growth by coordinating gene expression and carbon metabolic flux, which may be essential for plant adaptation to the changing environmental conditions (65,66).
Nuclear accumulation and transcriptional function of OsGAPDH1
Mounting evidence supports the concept that cytoplasmic GAPDH may fulfill alternative, non-metabolic functions that are triggered by redox post-translational modifications of the protein under stress conditions (35). Recent data indicate that glucose starvation also triggers phosphorylation and nuclear import of GAPDH in animal cells (67). The present and previous data support the notion that nuclear localization of plant cytosolic GAPDHs is stimulated by oxidative modifications which may enhance nuclear functions of the proteins in plant cells (39,56,68). All phosphorylating GAPDHs share a similar structure including a highly reactive catalytic cysteine that can undergo multiple redox-induced post-translational modifications in response to ROS (30). However, recent results indicate that S-nitrosylation of the cysteine residues does not affect stress-induced nuclear accumulation of GAPC1 in tobacco cells (69). The present data suggest that lysine acetylation of GAPDH may be also involved in the accumulation of the protein in the nucleus of rice cells, which is negatively regulated by OsSRT1.
Although the nuclear localization of GAPDH is enhanced under stress conditions in animal and plant cells, a direct transcriptional regulatory function of GAPDH has not been identified so far, while a role has been assigned to yeast GAPDH to promote Sir2-mediated normal silencing at the telomere and rDNA (70). The present data provide clear evidence that a glycolytic GAPDH functions as a transcriptional activator of glycolytic genes in rice. It has been shown that stress conditions, such as hypoxia, induce expression of many genes involved in glycolysis and fermentation in rice coleoptile and root tips of other plant species (63). We hypothesize that the increased expression of glycolytic genes may be partly related to increased nuclear accumulation of GAPDH proteins under stress.
The enrichment of OsGAPDH1 in chromatin of several glycolytic genes (Figure 3F), is consistent with previous results that Arabidopsis cytosolic glycolytic GAPDH (GAPC1 and GAPC2) bind to a gene encoding the NADP-dependent malate dehydrogenase (NADP-MDH) (37). In addition, a recent work using Förster resonance energy transfer experiments shows that tobacco GAPC1 and GAPC2 interact with nucleic acids in the nucleus (69). The enrichment of OsGAPDH1 mostly near the TSS sites of glycolytic genes and the transactivation of proximal promoter regions of OsGAPDH1 and OsHXK1 by the protein suggest that OsGAPDH1 may function in transcriptional activation. However, the enrichment of OsGAPDH1 detected in a relatively large region of glycolytic genes (Figure 3F), is consistent with the binding of Arabidopsis GAPC1 and GAPC2 to several DNA fragments that nearly cover the whole coding region of NADP-MDH (37). In line with the observations that the binding to NADP-MDH by GAPC1 and GAPC2 cannot be narrowed down to a specific motif (37), we failed to detect specific binding of OsGAPDH1 to a promoter region of glycolytic genes by in vitro gel shift assay. Based on these observations, we hypothesize that nuclear localized GAPDH proteins may not function as typical sequence-specific DNA-binding transcription factors.
Regulation of OsGAPDH1 function by OsSRT1-mediated deacetylation
It is well established that histone lysine acetylation plays an essential role in chromatin regulation and gene expression in eukaryotic cells. Recent results have shown that lysine acetylation occurs in a large number of proteins of diverse biological function (41–43). OsSRT1 is a potent histone H3K9 deacetylase in rice (21,28). The finding that OsSRT1 deacetylates OsGAPDH1 indicates that OsSRT1 targets non-histone proteins in the nucleus. Since in the transient expression experiments in protoplasts the transfected DNA does not get a nucleosome-like structure, the repressive effect of OsSRT1 on OsGAPDH1-mediated gene transcription is unlikely due to histone deacetylation, but rather due to deacetylation of the OsGAPDH1 protein. Thus, OsSRT1 also regulates gene expression by deacetylating non-histone proteins. Acetylation of OsSRT1-targetd OsGAPDH1 lysine residues were found to be important for nuclear accumulation and transcriptional activation, indicating that stress-induced higher glycolytic gene expression is counterbalanced by OsSRT1. The increased enzymatic activities in OsGAPDH1 Lys57, 74 and 217 substitution mutants and by OsSRT1 deacetylation of the protein are consistent with previous results in Arabidopsis that deacetylation of GAPC2 increases the enzymatic activity by >3-fold (43). The reason of distinct effects of lysine acetylation on transcriptional function and enzymatic activity of OsGAPDH1 is unclear at this stage. We hypothesize that lysine acetylation may induce higher nuclear accumulation or interaction of the protein with transcriptional apparatus, but may affect substrate binding and/or structure of active center of the enzyme. Although the cytosolic/nucleic ratio of GAPDH is lower under H2O2 stress, the enzymatic activity increased (Supplementary Figure S3H).This suggests that the overall GAPDH activity and/or expression is induced by stress signals, consistent with higher glycolytic activity under stress in plants. We hypothesize that in deacetylated form in cytosol under normal conditions, GAPDH is enzymatically active, whereas in acetylated form in the nucleus, the protein functions as a transcriptional activator to stimulate glycolysis, which is inhibited by OsSRT1 in the nucleus.
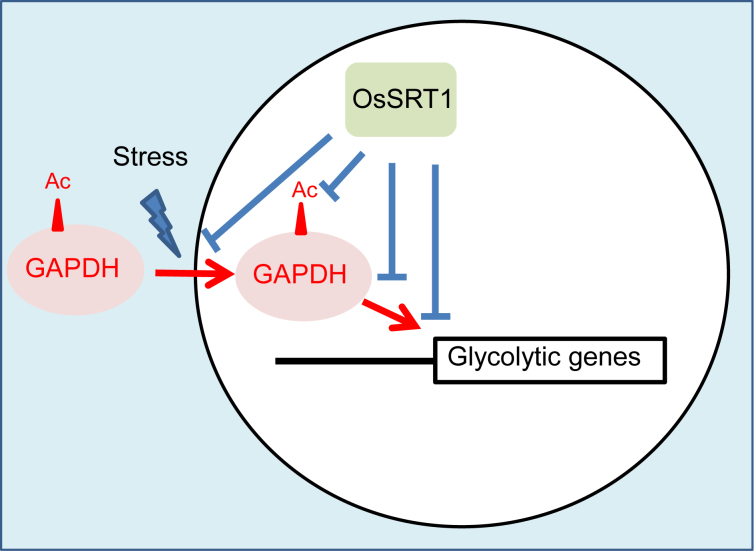
In conclusion, the present results indicate that rice nuclear sirtuin OsSRT1 represses glycolysis by deacetylating both histones and GAPDH, inhibiting its nuclear accumulation and function as a transcriptional activator of glycolytic genes (Figure 9).
Figure 9.
Schematic representation of OsSRT1 actions in repression of glycolysis in rice.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Wei Wei for vector construction and Prof. Xianghua Li and Dr Qinglu Zhang for help in field experiments and management. We also thank Dr. Masaru Ohme-Takagi and Prof. Shouyi Chen for the gift of vectors for the transcriptional regulation activity assay in protoplasts.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [31730049]; National Key Research and Development Program of China [2016YFD0100802]; 111 project [B07041]; Huazhong Agricultural University Scientific & Technological Self-innovation Foundation [2016RC003]; Fundamental Research Funds for the Central Universities [2662015PY228]. Funding for open access charge: National Natural Science Foundation of China [31730049].
Conflict of interest statement. None declared.
REFERENCES
- 1. Imai S., Armstrong C.M., Kaeberlein M., Guarente L.. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000; 403:795–800. [DOI] [PubMed] [Google Scholar]
- 2. Tissenbaum H.A., Guarente L.. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001; 410:227–230. [DOI] [PubMed] [Google Scholar]
- 3. Rogina B., Helfand S.L.. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:15998–16003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lin S.J., Defossez P.A., Guarente L.. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000; 289:2126–2128. [DOI] [PubMed] [Google Scholar]
- 5. Houtkooper R.H., Pirinen E., Auwerx J.. Sirtuins as regulators of metabolism and healthspan. Nat. Rev. Mol. Cell Biol. 2012; 13:225–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kaeberlein M., McVey M., Guarente L.. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999; 13:2570–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Guintini L., Tremblay M., Toussaint M., D’Amours A., Wellinger R.E., Wellinger R.J., Conconi A.. Repair of UV-induced DNA lesions in natural Saccharomyces cerevisiae telomeres is moderated by Sir2 and Sir3, and inhibited by yKu-Sir4 interaction. Nucleic Acids Res. 2017; 45:4577–4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Simoneau A., Ricard E., Weber S., Hammond-Martel I., Wong L.H., Sellam A., Giaever G., Nislow C., Raymond M., Wurtele H.. Chromosome-wide histone deacetylation by sirtuins prevents hyperactivation of DNA damage-induced signaling upon replicative stress. Nucleic Acids Res. 2016; 44:2706–2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hannan A., Abraham N.M., Goyal S., Jamir I., Priyakumar U.D., Mishra K.. Sumoylation of Sir2 differentially regulates transcriptional silencing in yeast. Nucleic Acids Res. 2015; 43:10213–10226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Frye R.A. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem. Biophys. Res. Commun. 2000; 273:793–798. [DOI] [PubMed] [Google Scholar]
- 11. Tanno M., Sakamoto J., Miura T., Shimamoto K., Horio Y.. Nucleocytoplasmic shuttling of the NAD+-dependent histone deacetylase SIRT1. J. Biol. Chem. 2007; 282:6823–6832. [DOI] [PubMed] [Google Scholar]
- 12. Gomes P., Outeiro T.F., Cavadas C.. Emerging role of Sirtuin 2 in the regulation of mammalian metabolism. Trends Pharmacol. Sci. 2015; 36:756–768. [DOI] [PubMed] [Google Scholar]
- 13. Huang J.Y., Hirschey M.D., Shimazu T., Ho L., Verdin E.. Mitochondrial sirtuins. Biochim. Biophys. Acta. 2010; 1804:1645–1651. [DOI] [PubMed] [Google Scholar]
- 14. Mostoslavsky R., Chua K.F., Lombard D.B., Pang W.W., Fischer M.R., Gellon L., Liu P., Mostoslavsky G., Franco S., Murphy M.M. et al. . Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006; 124:315–329. [DOI] [PubMed] [Google Scholar]
- 15. Ford E., Voit R., Liszt G., Magin C., Grummt I., Guarente L.. Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes Dev. 2006; 20:1075–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Guarente L. Sirtuins, aging, and metabolism. Cold Spring Harbor Symp. Quant. Biol. 2011; 76:81–90. [DOI] [PubMed] [Google Scholar]
- 17. Verdin E. NAD(+) in aging, metabolism, and neurodegeneration. Science. 2015; 350:1208–1213. [DOI] [PubMed] [Google Scholar]
- 18. Canto C., Auwerx J.. NAD+ as a signaling molecule modulating metabolism. Cold Spring Harbor Symp. Quant. Biol. 2011; 76:291–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Giblin W., Skinner M.E., Lombard D.B.. Sirtuins: guardians of mammalian healthspan. Trends Genet. 2014; 30:271–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ost A., Pospisilik J.A.. Epigenetic modulation of metabolic decisions. Curr. Opin. Cell Biol. 2015; 33:88–94. [DOI] [PubMed] [Google Scholar]
- 21. Huang L., Sun Q., Qin F., Li C., Zhao Y., Zhou D.X.. Down-regulation of a SILENT INFORMATION REGULATOR2-related histone deacetylase gene, OsSRT1, induces DNA fragmentation and cell death in rice. Plant Physiol. 2007; 144:1508–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Canto C., Gerhart-Hines Z., Feige J.N., Lagouge M., Noriega L., Milne J.C., Elliott P.J., Puigserver P., Auwerx J.. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009; 458:1056–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Konig A.C., Hartl M., Pham P.A., Laxa M., Boersema P.J., Orwat A., Kalitventseva I., Plochinger M., Braun H.P., Leister D. et al. . The Arabidopsis class II sirtuin is a lysine deacetylase and interacts with mitochondrial energy metabolism. Plant Physiol. 2014; 164:1401–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Greiss S., Gartner A.. Sirtuin/Sir2 phylogeny, evolutionary considerations and structural conservation. Mol. Cell. 2009; 28:407–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tcherkez G., Mahe A., Gauthier P., Mauve C., Gout E., Bligny R., Cornic G., Hodges M.. In folio respiratory fluxomics revealed by 13C isotopic labeling and H/D isotope effects highlight the noncyclic nature of the tricarboxylic acid “cycle” in illuminated leaves. Plant Physiol. 2009; 151:620–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tcherkez G., Boex-Fontvieille E., Mahe A., Hodges M.. Respiratory carbon fluxes in leaves. Curr. Opin. Plant Biol. 2012; 15:308–314. [DOI] [PubMed] [Google Scholar]
- 27. Lee C.P., Eubel H., Millar A.H.. Diurnal changes in mitochondrial function reveal daily optimization of light and dark respiratory metabolism in Arabidopsis. Mol. Cell. Proteomics. 2010; 9:2125–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhong X., Zhang H., Zhao Y., Sun Q., Hu Y., Peng H., Zhou D.X.. The rice NAD(+)-dependent histone deacetylase OsSRT1 targets preferentially to stress- and metabolism-related genes and transposable elements. PLoS One. 2013; 8:e66807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang H., Lu Y., Zhao Y., Zhou D.X.. OsSRT1 is involved in rice seed development through regulation of starch metabolism gene expression. Plant Sci. 2016; 248:28–36. [DOI] [PubMed] [Google Scholar]
- 30. Zaffagnini M., Fermani S., Costa A., Lemaire S.D., Trost P.. Plant cytoplasmic GAPDH: redox post-translational modifications and moonlighting properties. Front Plant Sci. 2013; 4:450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tristan C., Shahani N., Sedlak T.W., Sawa A.. The diverse functions of GAPDH: views from different subcellular compartments. Cell. Signal. 2011; 23:317–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Colell A., Ricci J.E., Tait S., Milasta S., Maurer U., Bouchier-Hayes L., Fitzgerald P., Guio-Carrion A., Waterhouse N.J., Li C.W. et al. . GAPDH and autophagy preserve survival after apoptotic cytochrome c release in the absence of caspase activation. Cell. 2007; 129:983–997. [DOI] [PubMed] [Google Scholar]
- 33. Colell A., Green D.R., Ricci J.E.. Novel roles for GAPDH in cell death and carcinogenesis. Cell Death Differ. 2009; 16:1573–1581. [DOI] [PubMed] [Google Scholar]
- 34. Sirover M.A. On the functional diversity of glyceraldehyde-3-phosphate dehydrogenase: biochemical mechanisms and regulatory control. Biochim. Biophys. Acta. 2011; 1810:741–751. [DOI] [PubMed] [Google Scholar]
- 35. Sirover M.A. Subcellular dynamics of multifunctional protein regulation: mechanisms of GAPDH intracellular translocation. J. Cell Biochem. 2012; 113:2193–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Petersen J., Brinkmann H., Cerff R.. Origin, evolution, and metabolic role of a novel glycolytic GAPDH enzyme recruited by land plant plastids. J. Mol. Evol. 2003; 57:16–26. [DOI] [PubMed] [Google Scholar]
- 37. Holtgrefe S., Gohlke J., Starmann J., Druce S., Klocke S., Altmann B., Wojtera J., Lindermayr C., Scheibe R.. Regulation of plant cytosolic glyceraldehyde 3-phosphate dehydrogenase isoforms by thiol modifications. Physiol. Plantarum. 2008; 133:211–228. [DOI] [PubMed] [Google Scholar]
- 38. Wawer I., Bucholc M., Astier J., Anielska-Mazur A., Dahan J., Kulik A., Wyslouch-Cieszynska A., Zareba-Koziol M., Krzywinska E., Dadlez M. et al. . Regulation of Nicotiana tabacum osmotic stress-activated protein kinase and its cellular partner GAPDH by nitric oxide in response to salinity. Biochem. J. 2010; 429:73–83. [DOI] [PubMed] [Google Scholar]
- 39. Vescovi M., Zaffagnini M., Festa M., Trost P., Lo Schiavo F., Costa A.. Nuclear accumulation of cytosolic glyceraldehyde-3-phosphate dehydrogenase in cadmium-stressed Arabidopsis roots. Plant Physiol. 2013; 162:333–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Choudhary C., Kumar C., Gnad F., Nielsen M.L., Rehman M., Walther T.C., Olsen J.V., Mann M.. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009; 325:834–840. [DOI] [PubMed] [Google Scholar]
- 41. Wang Q., Zhang Y., Yang C., Xiong H., Lin Y., Yao J., Li H., Xie L., Zhao W., Yao Y. et al. . Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science. 2010; 327:1004–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhao S., Xu W., Jiang W., Yu W., Lin Y., Zhang T., Yao J., Zhou L., Zeng Y., Li H. et al. . Regulation of cellular metabolism by protein lysine acetylation. Science. 2010; 327:1000–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Finkemeier I., Laxa M., Miguet L., Howden A.J., Sweetlove L.J.. Proteins of diverse function and subcellular location are lysine acetylated in Arabidopsis. Plant Physiol. 2011; 155:1779–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li T., Liu M., Feng X., Wang Z., Das I., Xu Y., Zhou X., Sun Y., Guan K.L., Xiong Y. et al. . Glyceraldehyde-3-phosphate dehydrogenase is activated by lysine 254 acetylation in response to glucose signal. J. Biol. Chem. 2014; 289:3775–3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhang H., Lu Y., Zhao Y., Zhou D.-X.. OsSRT1 is involved in rice seed development through regulation of starch metabolism gene expression. Plant Sci. 2016; 248:28–36. [DOI] [PubMed] [Google Scholar]
- 46. Livak K.J., Schmittgen T.D.. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001; 25:402–408. [DOI] [PubMed] [Google Scholar]
- 47. Huang L., Sun Q., Qin F., Li C., Zhao Y., Zhou D.-X.. Down-regulation of a SILENT INFORMATION REGULATOR2-related histone deacetylase gene, OsSRT1, induces DNA fragmentation and cell death in rice. Plant Physiol. 2007; 144:1508–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhou S., Jiang W., Long F., Cheng S, Yang W., Zhao Y., Zhou D.-X.. Rice homeodomain protein WOX11 recruits a histone acetyltransferase complex to establish programs of cell proliferation of crown root meristem. Plant Cell. 2017; 29:1088–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Xie K., Yang Y.. RNA-guided genome editing in plants using a CRISPR-Cas system. Mol. Plant. 2013; 6:1975–1983. [DOI] [PubMed] [Google Scholar]
- 50. Hao Y.J., Wei W., Song Q.X., Chen H.W., Zhang Y.Q., Wang F., Zou H.F., Lei G., Tian A.G., Zhang W.K. et al. . Soybean NAC transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants. Plant J. 2011; 68:302–313. [DOI] [PubMed] [Google Scholar]
- 51. Pancera S.M., Gliemann H., Schimmel T., Petri D.F.. Adsorption behavior and activity of hexokinase. J. Colloid interface Sci. 2006; 302:417–423. [DOI] [PubMed] [Google Scholar]
- 52. Papagianni M., Avramidis N.. Lactococcus lactis as a cell factory: a twofold increase in phosphofructokinase activity results in a proportional increase in specific rates of glucose uptake and lactate formation. Enzyme Microb. Technol. 2011; 49:197–202. [DOI] [PubMed] [Google Scholar]
- 53. Lepper T.W., Oliveira E., Koch G.D., Berlese D.B., Feksa L.R.. Lead inhibits in vitro creatine kinase and pyruvate kinase activity in brain cortex of rats. Toxicology In Vitro. 2010; 24:1045–1051. [DOI] [PubMed] [Google Scholar]
- 54. Machado A.T., Silva M., Iulek J.. Expression, purification, enzymatic characterization and crystallization of glyceraldehyde-3-phosphate dehydrogenase from Naegleria gruberi, the first one from phylum Percolozoa. Protein Expression Purif. 2016; 127:125–130. [DOI] [PubMed] [Google Scholar]
- 55. Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S.. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011; 28:2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Henry E., Fung N., Liu J., Drakakaki G., Coaker G.. Beyond glycolysis: GAPDHs are multi-functional enzymes involved in regulation of ROS, autophagy, and plant immune responses. PLoS Genet. 2015; 11:e1005199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tien Y.-C., Chuankhayan P., Huang Y.-C., Chen C.-D., Alikhajeh J., Chang S.-L., Chen C.-J.. Crystal structures of rice (Oryza sativa) glyceraldehyde-3-phosphate dehydrogenase complexes with NAD and sulfate suggest involvement of Phe37 in NAD binding for catalysis. Plant Mol. Biol. 2012; 80:389–403. [DOI] [PubMed] [Google Scholar]
- 58. Wurtele H., Tsao S., Lepine G., Mullick A., Tremblay J., Drogaris P., Lee E.H., Thibault P., Verreault A., Raymond M.. Modulation of histone H3 lysine 56 acetylation as an antifungal therapeutic strategy. Nat. Med. 2010; 16:774–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Shakibaei M., Shayan P., Busch F., Aldinger C., Buhrmann C., Lueders C., Mobasheri A.. Resveratrol mediated modulation of Sirt-1/Runx2 promotes osteogenic differentiation of mesenchymal stem cells: potential role of Runx2 deacetylation. PLoS One. 2012; 7:e35712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhong L., D’Urso A., Toiber D., Sebastian C., Henry R.E., Vadysirisack D.D., Guimaraes A., Marinelli B., Wikstrom J.D., Nir T. et al. . The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell. 2010; 140:280–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kugel S., Mostoslavsky R.. Chromatin and beyond: the multitasking roles for SIRT6. Trends Biochem. Sci. 2014; 39:72–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Mutuku J.M., Nose A.. Changes in the contents of metabolites and enzyme activities in rice plants responding to Rhizoctonia solani Kuhn infection: activation of glycolysis and connection to phenylpropanoid pathway. Plant Cell Physiol. 2012; 53:1017–1032. [DOI] [PubMed] [Google Scholar]
- 63. Lee K.W., Chen P.W., Yu S.M.. Metabolic adaptation to sugar/O2 deficiency for anaerobic germination and seedling growth in rice. Plant Cell Environ. 2014; 37:2234–2244. [DOI] [PubMed] [Google Scholar]
- 64. Cheng S.C., Quintin J., Cramer R.A., Shepardson K.M., Saeed S., Kumar V., Giamarellos-Bourboulis E.J., Martens J.H., Rao N.A., Aghajanirefah A. et al. . mTOR- and HIF-1alpha-mediated aerobic glycolysis as metabolic basis for trained immunity. Science. 2014; 345:1250684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Shen Y., Issakidis-Bourguet E., Zhou D.X.. Perspectives on the interactions between metabolism, redox, and epigenetics in plants. J. Exp. Bot. 2016; 67:5291–5300. [DOI] [PubMed] [Google Scholar]
- 66. Shen Y., Wei W., Zhou D.X.. Histone acetylation enzymes coordinate metabolism and gene expression. Trends Plant Sci. 2015; 20:614–621. [DOI] [PubMed] [Google Scholar]
- 67. Chang C., Su H., Zhang D., Wang Y., Shen Q., Liu B., Huang R., Zhou T., Peng C., Wong C.C. et al. . AMPK-dependent phosphorylation of GAPDH triggers Sirt1 activation and is necessary for autophagy upon glucose starvation. Mol. Cell. 2015; 60:930–940. [DOI] [PubMed] [Google Scholar]
- 68. Bae M.S., Cho E.J., Choi E.Y., Park O.K.. Analysis of the Arabidopsis nuclear proteome and its response to cold stress. Plant J. 2003; 36:652–663. [DOI] [PubMed] [Google Scholar]
- 69. Testard A., Da Silva D., Ormancey M., Pichereaux C., Pouzet C., Jauneau A., Grat S., Robe E., Brière C., Cotelle V. et al. . Calcium and nitric oxide-dependent nuclear accumulation of cytosolic glyceraldehyde-3-phosphate dehydrogenase in response to long chain bases in tobacco BY-2 cells. Plant Cell Physiol. 2016; 57:2221–2231. [DOI] [PubMed] [Google Scholar]
- 70. Ringel A.E., Ryznar R., Picariello H., Huang K.L., Lazarus A.G., Holmes S.G.. Yeast Tdh3 (glyceraldehyde 3-phosphate dehydrogenase) is a Sir2-interacting factor that regulates transcriptional silencing and rDNA recombination. PLoS Genet. 2013; 9:e1003871. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.