Abstract
Butyrivibrio hungatei MB2003 was isolated from the plant-adherent fraction of rumen contents from a pasture-grazed New Zealand dairy cow, and was selected for genome sequencing in order to examine its ability to degrade plant polysaccharides. The genome of MB2003 is 3.39 Mb and consists of four replicons; a chromosome, a secondary chromosome or chromid, a megaplasmid and a small plasmid. The genome has an average G + C content of 39.7%, and encodes 2983 putative protein-coding genes. MB2003 is able to use a variety of monosaccharide substrates for growth, with acetate, butyrate and formate as the principal fermentation end-products, and the genes encoding these metabolic pathways have been identified. MB2003 is predicted to encode an extensive repertoire of CAZymes with 78 GHs, 7 CEs, 1 PL and 78 GTs. MB2003 is unable to grow on xylan or pectin, and its role in the rumen appears to be as a utilizer of monosaccharides, disaccharides and oligosaccharides made available by the degradative activities of other bacterial species.
Electronic supplementary material
The online version of this article (10.1186/s40793-017-0285-8) contains supplementary material, which is available to authorized users.
Keywords: Rumen, Bacteria, Hemicellulose, Pectin, Degradation, Butyrivibrio, Genome
Introduction
10.1601/nm.4129 are important rumen bacteria [1], and are among the small number of rumen genera capable of utilizing the complex plant structural polysaccharides xylan and pectin [2, 3]. They are classified as anaerobic, monotrichous, butyrate-producing, curved rods and have been isolated from the gastrointestinal tracts and feces of various ruminants, monogastric animals and humans [4, 5]. 10.1601/nm.4129 are metabolically versatile and are capable of growing on a range of carbohydrates, from simple mono- or oligosaccharides to complex plant polysaccharides such as pectins, mannans, starch and hemicelluloses [6]. Furthermore, xylans of diverse chemical and physical properties, from a range of forages are degraded by 10.1601/nm.4129 species [7]. Some 10.1601/nm.4129 species show strong proteolytic activity [8], and 10.1601/nm.4129 are thought to be the main butyrate producers in the rumen [9, 10]. The genus 10.1601/nm.4129 is classified within the family 10.1601/nm.4118, order 10.1601/nm.17931, and is phylogenetically diverse. The 10.1601/nm.4129 genus originally consisted of only one species, 10.1601/nm.4130 [2]. In addition to phenotypic characterisations [11, 12], studies have utilized DNA-DNA hybridization [13, 14], 16S rRNA gene sequencing [15, 16] and 16S rRNA-based hybridization probes [17], to differentiate these organisms. To accommodate the observed diversity amongst the newly discovered bacterial strains, a new genus, 10.1601/nm.4145, was described [18]. Four species are currently recognized: 10.1601/nm.4130, 10.1601/nm.4132, 10.1601/nm.13010 and 10.1601/nm.4131 [6], although 10.1601/nm.4131 is more distantly related to the other three. 10.1601/nm.4132 are common anaerobic rumen bacteria found in domestic and wild ruminants and the type strain is JK615T [19]. 10.1601/nm.4132 JK615T is non-proteolytic and non-fibrolytic, but is able to utilize oligo- and monosaccharides as substrates for growth. Gaining an insight into the role of these secondary degrader species in microbial plant polysaccharide breakdown is important for understanding rumen function. Here we present the complete genome sequence of 10.1601/nm.4132 MB2003, a strain isolated from a pasture-grazed dairy cow in New Zealand [20], and describe its comparison with genomes of closely related 10.1601/nm.4132 strains.
Organism information
Classification and features
MB2003 was isolated from the plant-adherent fraction of rumen contents from a New Zealand dairy cow grazing fresh forage [20, 21]. MB2003 cells are Gram positive, short rods, occurring singly or in pairs (Fig. 1). The morphological features of MB2003 cells were determined by electron microscopy of cells grown on RM02 medium [22], negatively stained with 1% phosphotungstic acid, mounted on Formvar-coated copper grids, and examined using a Philips model 201C electron microscope (Eindhoven, The Netherlands). MB2003 cells were observed to have a single polar flagellum (Fig. 2), although cells in growing cultures were non-motile. A phylogenetic analysis of the full-length 16S rRNA gene sequence placed MB2003 within the 10.1601/nm.4132 species, being 98% similar to the 10.1601/nm.4132 type strain JK615T [19] (Fig. 3). Additional characteristics of 10.1601/nm.4132 MB2003 are shown in Table 1.
Fig. 1.
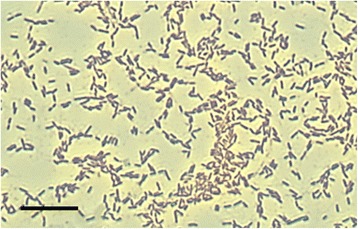
Morphology of B. hungatei MB2003. Micrograph of Gram stained B. hungatei MB2003 cells at 100 × magnification. Bar represents 10 μm
Fig. 2.
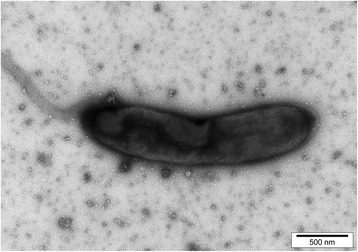
Transmission electron micrograph of B. hungatei MB2003. Micrograph of negatively stained B. hungatei MB2003 cells at 10,000 × magnification
Fig. 3.
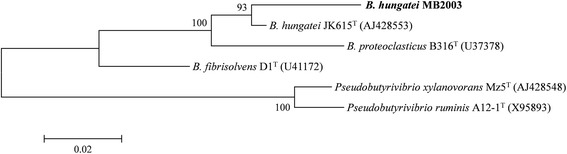
Phylogenetic tree highlighting the relationship of B. hungatei MB2003 relative to the type strains of the other species within the genus Butyrivibrio. The evolutionary history was inferred using the Maximum Likelihood method based on the General Time Reversible model [55]. The tree with the highest log likelihood (−3712.3329) is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (10,000 replicates) is shown next to the branches [56]. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach, and then selecting the topology with superior log likelihood value. A discrete Gamma distribution was used to model evolutionary rate differences among sites (5 categories (+G, parameter = 0.3950)). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved six nucleotide sequences. All positions with less than 95% site coverage were eliminated. There were a total of 1509 positions in the final dataset. Evolutionary analyses were conducted in MEGA6 [55]. GenBank accession numbers of the 16S rRNA gene sequences are shown in parentheses. Bar, 0.02 nucleotide substitutions per site. T, indicates type strain. All the type strains have their genome sequencing projects registered in the Genomes Online Database (GOLD) [57]
Table 1.
Classification and general features of the rumen bacterium B. hungatei MB2003 in accordance with the MIGS recommendations [58]
| MIGS ID | Property | Term | Evidence codea |
|---|---|---|---|
| Current classification | Domain: Bacteria | TAS [59] | |
| Phylum: Firmicutes | TAS [60, 61] | ||
| Class: Clostridia | TAS [62] | ||
| Order: Eubacteriales | TAS [63] | ||
| Family: Lachnospiraceae | TAS [64] | ||
| Genus: Butyrivibrio | TAS [4] | ||
| Species: hungatei | TAS [19] | ||
| Type strain: No | |||
| Strain: MB2003 | TAS [20, 21] | ||
| Gram stain | Positive | TAS [21, 31] | |
| Cell shape | Rod | TAS [21, 31] | |
| Motility | Non-motile | IDA | |
| Sporulation | Not reported | NAS | |
| Temperature range | 37–39 °C | IDA | |
| Optimum temperature | 39 °C | IDA | |
| pH range; Optimum | 6.0–7.0; 6.4 | IDA | |
| Carbon source | Variety of carbohydrates | IDA | |
| Energy metabolism | Fermentative metabolism | IDA | |
| MIGS-6 | Habitat | Bovine rumen | TAS [20] |
| MIGS-6.3 | Salinity | Not reported | |
| MIGS-22 | Oxygen requirement | Anaerobic | IDA |
| MIGS-15 | Biotic relationship | Symbiont of ruminants | TAS [20] |
| MIGS-14 | Pathogenicity | Non-pathogen | NAS |
| MIGS-4 | Geographic location | Ruakura, Hamilton, New Zealand | TAS [20] |
| MIGS-5 | Sample collection time | May 2009 | TAS [20] |
| MIGS-4.1 | Latitude | −37.77 (37°46′28″S) | IDA |
| MIGS-4.2 | Longitude | +175.31 (175°18′31″E) | IDA |
| MIGS-4.4 | Altitude | 40 m | IDA |
aEvidence codes - IDA, Inferred from Direct Assay, NAS, Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). Evidence codes are from the Gene Ontology project [65]
Strain MB2003 grew to highest optical density (OD) at pH values of 6.1 to 6.5 and at a temperature of 39 °C, conditions which are typical of its rumen environment. VFA production was determined from triplicate broth cultures grown overnight in RM02 medium with cellobiose as substrate and analysed for formate, acetate, propionate, n-butyrate, iso-valerate and lactate on a HP 6890 series GC (Hewlett Packard) with 2-ethylbutyric acid (Sigma-Aldrich, St. Louis, MO, USA) as the internal standard. To derivatize formic, lactic and succinic acids, samples were mixed with HCl ACS reagent (Sigma-Aldrich, St. Louis, MO, USA) and diethyl ether, with the addition of N-methyl-N-t-butyldimethylsilyltri-fluoroacetamide (MTBSTFA) (Sigma-Aldrich, St. Louis, MO, USA) [23]. Under these conditions MB2003 produced 16.4 mM formate, 3.6 mM acetate and 4.7 mM butyrate. MB2003 was able to grow in CO2-containing media with various soluble carbon sources and the semi-soluble inulin (all tested at 0.5% w/v final concentration). Growth on soluble substrates was assessed as an increase in culture density OD600nm compared to cultures without carbon source added, whereas total VFA production was used as an indicator of substrate utilization and growth for insoluble polymers (Table 2). All strains tested were net producers of formate, acetate and n-butyrate, which is characteristic of 10.1601/nm.4129 . Cellobiose and glucose supported the growth of MB2003, JK615T and B316T to high cell densities. Therefore, cellobiose was used to examine the growth of MB2003 over a 24 h period. The exponential phase of growth was between 4 and 8 h, with the maximum cell density reached at 8 to 10 h, and stationary phase between 10 to 24 h (Fig. 4).
Table 2.
Carbon source utilization of the Butyrivibrio strains
| Substrate | MB2003 | JK615T | B316T | |
|---|---|---|---|---|
| Monosaccharides | Arabinose | ++ | ++ | ++ |
| Fructose | – | – | ++ | |
| Galactose | ++ | – | ++ | |
| Glucose | ++ | ++ | ++ | |
| Mannose | – | ++ | ++ | |
| Rhamnose | – | – | ++ | |
| Ribose | – | – | – | |
| Xylose | ++ | ++ | ++ | |
| Disaccharides | Cellobiose | ++ | ++ | ++ |
| Lactose | ++ | ++ | ++ | |
| Maltose | ++ | ++ | ++ | |
| Melibiose | – | – | + | |
| Sucrose | ++ | ++ | ++ | |
| Trisaccharides | Melezitose | – | – | ++ |
| Raffinose | – | ++ | ++ | |
| Trehalose | – | – | ++ | |
| Sugar Alcohols | myo-Inositol | – | – | – |
| Mannitol | – | – | + | |
| Sorbitol | – | – | – | |
| Glycosides | Amygdalin | + | – | ++ |
| Esculin | – | ++ | ++ | |
| Rutin | – | ++ | ++ | |
| Salicin | ++ | ++ | ++ | |
| Insoluble substrates | Cellulose | – | – | – |
| Dextrin | – | – | ++ | |
| Inulin | + | – | ++ | |
| Starch | – | – | ++ | |
| Pectin | – | – | ++ | |
| Xylan | – | – | ++ | |
Fig. 4.
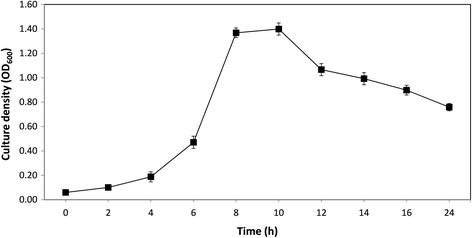
Culture density achieved in 24 h by MB2003 growing in media with cellobiose as the sole substrate. Points indicate means of three replicates, and the error bars represent +/−one standard error
Genome sequencing information
Genome project history
10.1601/nm.4132 MB2003 was selected for genome sequencing as a NZ strain of 10.1601/nm.4132. A summary of the genome project information is shown in Table 3 and in Additional file 1: Table S1.
Table 3.
MB2003 genome project information
| MIGS ID | Property | Term |
|---|---|---|
| MIGS-31 | Finishing quality | High-quality, closed genome |
| MIGS-28 | Libraries used | 454 3 kb mate paired-end library |
| MIGS-29 | Sequencing platforms | 454 GS FLX Titanium chemistry |
| MIGS-31.2 | Fold coverage | 234× |
| MIGS-30 | Assemblers | Newbler version 2.3 |
| MIGS-32 | Gene calling method | Glimmer and BLASTX |
| Locus Tag | bhn and bhn_RS | |
| Genbank ID | CP017830, CP017831, CP017832, CP017833 | |
| Genbank Date of Release | 31 October 2016 | |
| GOLD ID | Ga0074201 | |
| BIOPROJECT ID | PRJNA349214 and PRJNA224116 | |
| BIOSAMPLE ID | SAMN05928573 | |
| MIGS-13 | Source Material Identifier | Butyrivibrio hungatei MB2003 |
| Project relevance | Ruminant plant-fibre degradation |
Growth conditions and genomic DNA preparation
MB2003 was grown in RM02 medium [22] with 10 mM glucose and 0.1% yeast extract but without rumen fluid. Culture purity was confirmed by Gram stain and sequencing of the 16S rRNA gene. Genomic DNA was extracted from freshly grown cells by a modification of the standard cell lysis method of Saito and Miura [24], using lysozyme, proteinase K and sodium dodecyl sulphate, followed by phenol-chloroform extraction, and purification using the Qiagen Genomic-Tip 500 Maxi kit (Qiagen, Hilden, Germany). Genomic DNA was precipitated by the addition of a 0.7 volume of isopropanol, and collected by centrifugation at 12,000×g for 10 min at room temperature. The supernatant was removed, and the DNA pellet was washed in 70% ethanol, re-dissolved in TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 7.5) and stored at −20 °C until required.
Genome sequencing and assembly
The complete genome sequence of MB2003 was determined by pyrosequencing 3 kb mate paired-end sequence libraries using the 454 GS FLX platform with Titanium chemistry (Macrogen, Korea). Pyrosequencing reads provided 234× coverage of the genome and were assembled using the Newbler assembler (version 2.7, Roche 454 Life Sciences, USA) which resulted in 31 contigs across 7 scaffolds. Gap closure was managed using the Staden package [25] and gaps were closed using additional Sanger sequencing by standard and inverse PCR techniques. In addition, MB2003 genomic DNA was sequenced using shotgun sequencing of 2 kb paired-end sequence libraries using the Illumina MiSeq platform (Macrogen, Korea) which provided 800-fold sequencing coverage. Illumina reads were analysed using the Galaxy web-based platform [26] and de novo assembly was performed using the Velvet assembler, version 3.0 [27]. The Velvet assembled MB2003 genome MiSeq sequences were combined with the Newbler assembly using the Staden package and Geneious, version 8.1 [28]. Genome assembly was confirmed by pulsed-field gel electrophoresis.
Genome annotation
Annotation of the MB2003 genome was performed as described previously [29]. The MB2003 genome sequence was prepared for NCBI submission using Sequin [30], and the adenine residue of the start codon of the chromosomal replication initiator protein DnaA1 (bhn_I0001, bhn_RS00450) gene was chosen as the first base for the MB2003 genome.
Genome properties
The genome of 10.1601/nm.4132 MB2003 consists of four replicons [21, 31]; a single chromosome (3,143,784 bp, %G + C 39.91), a chromid or secondary chromosome (BhuII, 91,776 bp, %G + C 37.71), a megaplasmid (pNP144, 144,470 bp, %G + C 36.86) and a plasmid (pNP6, 6284 bp, %G + C 35.71). The total size of the closed genome is 3,386,314 bp with an overall %G + C content of 39.71%. A total of 3064 genes were predicted, of which 2983 (97.4%) were protein-coding genes. A putative function was assigned to 2225 of the protein-coding genes, while 775 protein coding genes were annotated as hypothetical proteins. The MB2003 chromosome encodes 2758 genes, and BhuII, pNP144 and pNP6 encode 89, 147 and 6 genes, respectively. The properties and statistics of the MB2003 genome are summarized in Tables 4, 5 and 6. The nucleotide sequences of the MB2003 chromosome, chromid (BhuII), megaplasmid (pNP144) and plasmid (pNP6) have been deposited in Genbank under accession numbers CP017831, CP017830, CP017832 and CP017833. The genome atlas for 10.1601/nm.4132 MB2003 is shown in Fig. 5.
Table 4.
Summary of MB2003 genome replicon features
| Replicon type | Size (bp) | Topology | INSDC identifier | RefSeq ID |
|---|---|---|---|---|
| Chromosome | 3,143,784 | circular | CP017831 | NZ_CP017831 |
| Chromid_BhuII | 91,776 | circular | CP017830 | NZ_CP017830 |
| Megaplasmid_pNP144 | 144,470 | circular | CP017832 | NZ_CP017832 |
| Plasmid_pNP6 | 6284 | circular | CP017833 | NZ_CP017833 |
Table 5.
MB2003 genome statistics
| Attribute | Value | % of totala |
|---|---|---|
| Genome size (bp) | 3,386,314 | 100 |
| DNA coding (bp) | 3,064,986 | 90.51 |
| DNA G + C (bp) | 1,344,683 | 39.71 |
| DNA scaffolds | 4 | 100 |
| Total genes | 3064 | 100 |
| Protein coding genes | 2983 | 97.36 |
| RNA genes | 60 | 1.96 |
| Pseudogenes | 17 | 0.56 |
| Genes in internal clusters | 160 | 5.22 |
| Genes with function predicted | 2247 | 73.34 |
| Genes assigned to COGs | 1842 | 61.34 |
| Genes with Pfam domains | 2350 | 78.26 |
| Genes with signal peptides | 148 | 4.93 |
| Genes with transmembrane helices | 881 | 29.34 |
| CRISPR repeats | 2 |
aThe total is based on either the size of the genome in base pairs or the total number of genes or protein-coding genes in the annotated genome
Table 6.
Number of genes associated with the general COG functional categories
| Code | Value | % of totala | Description |
|---|---|---|---|
| J | 194 | 9.52 | Translation, ribosomal structure and biogenesis |
| A | 0 | 0 | RNA processing and modification |
| K | 149 | 7.31 | Transcription |
| L | 88 | 4.32 | Replication, recombination and repair |
| B | 0 | 0 | Chromatin structure and dynamics |
| D | 32 | 1.57 | Cell cycle control, Cell division, chromosome partitioning |
| V | 65 | 3.19 | Defense mechanisms |
| T | 139 | 6.82 | Signal transduction mechanisms |
| M | 155 | 7.61 | Cell wall/membrane biogenesis |
| N | 61 | 2.99 | Cell motility |
| U | 22 | 1.08 | Intracellular trafficking and secretion |
| O | 78 | 3.83 | Posttranslational modification, protein turnover, chaperones |
| C | 69 | 3.39 | Energy production and conversion |
| G | 243 | 11.93 | Carbohydrate transport and metabolism |
| E | 177 | 8.69 | Amino acid transport and metabolism |
| F | 80 | 3.93 | Nucleotide transport and metabolism |
| H | 79 | 3.88 | Coenzyme transport and metabolism |
| I | 72 | 3.53 | Lipid transport and metabolism |
| P | 79 | 3.88 | Inorganic ion transport and metabolism |
| Q | 16 | 0.79 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 158 | 7.76 | General function prediction only |
| S | 73 | 3.58 | Function unknown |
| – | 1245 | 40.33 | Not in COGs |
aThe total is based on the total number of protein coding genes in the genome
Fig. 5.
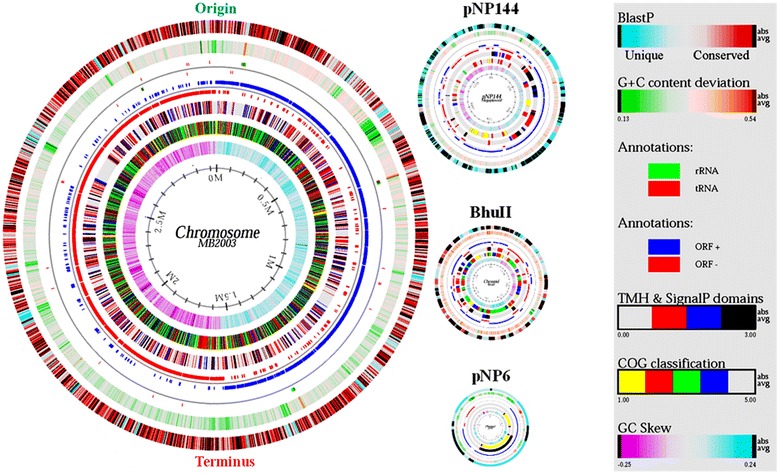
Genome atlas for B. hungatei MB2003. The figure represents a circular view of the four replicons that make up the B. hungatei MB2003 genome. The key at the right describes the concentric circles within each replicon in the outermost to innermost direction. The diagram was created using GENEWIZ [66] and custom-developed software. The innermost circle 1 shows GC-skew; Circle 2 shows COG classification: predicted ORFs were analysed using the COG database and grouped into the five major categories: yellow, information storage and processing; red, cellular processes and signalling; green, metabolism; blue, poorly characterised; and uncoloured, ORFs with uncharacterized COGs or no COG assignment. Circle 3 shows transmembrane helices (TMH) and SignalP domains: the four categories represent, uncoloured, absent; red, TMH; blue, SignalP; and black, both TMH and SignalP present. Circle 4 shows ORF orientation: ORFs in sense orientation (ORF+) are shown in blue; ORFs oriented in antisense direction (ORF-) are shown in red. Circle 5 shows ribosomal machinery: tRNAs and rRNAs are shown as green or red lines, respectively. Clusters are represented as coloured boxes to maintain readability. Circle 6 shows G + C content deviation from the average: GC-content is shown in either green (low GC spike) or orange (high GC spike). A box filter was applied to visualize contiguous regions of low or high GC deviations. Circle 7 shows BLAST similarities: deduced amino acid sequences were compared against the nonredundant (nr) database using gapped BLASTP [67]. Regions in blue represent unique proteins in MB2003, whereas highly conserved features relative to sequences in the nr database are shown in red. The degree of colour saturation corresponds to the level of similarity. The predicted origin and terminus of DNA replication are indicated
Insights from the genome sequence
Comparison of the MB2003, 10.1601/nm.4132 JK615T, and 10.1601/nm.13010 B316T genomes
A comparison of the 10.1601/nm.4132 MB2003 genome with the draft genome of 10.1601/nm.4132 JK615T [32] and the complete 10.1601/nm.13010 B316T genome is shown in Table 7. The MB2003 genome is 8633 bp smaller than JK615T and contains 27 fewer protein-coding genes. Although several plasmid replication genes have been identified in the JK615T draft genome, the presence of extrachromosomal elements requires experimental validation.
Table 7.
Genome statistics of MB2003, JK615T and B316T
| Attribute | B. hungatei MB2003 | B. hungatei JK615Tb | B. proteoclasticus B316T | |||
|---|---|---|---|---|---|---|
| Value | % of totala | Value | % of totala | Value | % of totala | |
| Status | Complete | Draft | Complete | |||
| Isolation source | Bovine rumen | Ovine rumen | Bovine rumen | |||
| Genome size (bp) | 3,386,314 | 100 | 3,394,947 | 100 | 4,404,886 | 100 |
| DNA coding (bp) | 3,064,986 | 90.51 | 3,108,180 | 91.55 | 3,954,077 | 89.77 |
| DNA G + C (bp) | 1,344,683 | 39.71 | 1,353,252 | 39.86 | 1,762,323 | 40.01 |
| Number of replicons | 4 | NA | 4 | |||
| DNA scaffolds | 4 | 100 | 22 | 100 | 4 | 100 |
| Total genes | 3064 | 100 | 3104 | 100 | 3863 | 100 |
| Protein coding genes | 2983 | 97.36 | 2996 | 96.52 | 3739 | 96.79 |
| RNA genes | 60 | 1.96 | 55 | 1.78 | 68 | 1.75 |
| rRNA operons | 4 | 4 | 6 | |||
| tRNA genes | 48 | 1.57 | 46 | 1.49 | 50 | 1.29 |
| Pseudo genes | 17 | 0.56 | 49 | 54 | 1.39 | |
| Genes in internal clusters | 160 | 5.22 | 211 | 6.82 | 327 | 8.43 |
| Genes with function prediction | 2225 | 72.62 | 2314 | 74.55 | 2505 | 64.85 |
| Genes assigned to COGs | 1842 | 61.34 | 1861 | 60.17 | 2075 | 53.49 |
| Genes with Pfam domains | 2350 | 78.26 | 2407 | 77.82 | 2784 | 71.77 |
| Genes with signal peptides | 148 | 4.93 | 137 | 4.43 | 269 | 6.93 |
| Genes with transmembrane helices | 881 | 29.34 | 847 | 27.38 | 1061 | 27.35 |
| CRISPR repeats | 2 | NA | NA | |||
| Reference | This report | [32] | [29] | |||
aThe total is based on either the size of the genome in base pairs or the total number of genes or protein-coding genes in the annotated genome. bIndicates draft genome sequence
A novel feature of both the MB2003 and B316T genomes is the presence of chromids or secondary chromosomes [33]. Chromids are replicons that have %G + C content similar to that of their main chromosome, but have plasmid-type maintenance and replication systems, are smaller than the chromosome, but are usually larger than any other plasmids present. Chromids contain genes essential for growth and maintenance of the organism along with several core genus-specific genes that can be found on the chromosome in other species of bacteria [33]. The Bhu II replicon has most of these characteristics and therefore has been designated as a chromid of MB2003. In B316T, almost 10% of the genes encoding enzymes that have a role in carbohydrate metabolism and transport are found on the chromid [29]. The Bhu II chromid of MB2003 also encodes genes with similar predicted functions (Table 9). Since the Bhu II chromid of MB2003 is smaller than the BPc2 chromid of B316T (186,325 bp), it is now the smallest chromid reported for bacteria. Comparison of MB2003, JK615T and B316T genomes based on COG category (Table 8) and synteny analysis (Fig. 6), show that these 10.1601/nm.4129 species and strains are genetically similar. Although the MB2003 and B316T genome sizes differ, the basic metabolism of these two rumen bacterial species is indicated to be similar.
Table 9.
Genes encoding predicted polysaccharide degrading enzymes in the MB2003 genome
| Locus tag | Name | Annotation | Size (aa) | CAZya | Binding domains |
|---|---|---|---|---|---|
| bhn_I2518 | bga2A | β-galactosidaseb | 1034 | GH2 | |
| bhn_I0827 | bga2C | β-galactosidaseb | 714 | GH2 | |
| bhn_I1587 | bga2B | β-galactosidaseb | 825 | GH2 | |
| bhn_I0200 | gh2B | glycoside hydrolase family 2b | 641 | GH2 | |
| bhn_I1127 | gh2A | glycoside hydrolase family 2b | 912 | GH2 | |
| bhn_I1849 | gh2C | glycoside hydrolase family 2b | 776 | GH2 | |
| bhn_III062 | bgl3A | β-glucosidaseb | 803 | GH3 | |
| bhn_I0707 | bgl3B | β-glucosidaseb | 808 | GH3 | |
| bhn_I0180 | bgl3C | β-glucosidaseb | 671 | GH3 | |
| bhn_I0706 | bgl3D | β-glucosidase b | 982 | GH3 | C-terminal TMH |
| bhn_I0189 | xyl3A | β-xylosidaseb | 707 | GH3 | |
| bhn_I1640 | bhx3A | β-N-acetylhexosaminidase b | 427 | GH3 | |
| bhn_I1693 | cel5C | endo-1,4-β-glucanase b | 543 | GH5 | CBM2a |
| bhn_I0165 | cel5A | endo-1,4-β-glucanase/xylanase b | 417 | GH5 | |
| bhn_I1756 | xyn8A | reducing end xylose-releasing exo-oligoxylanaseb | 383 | GH8 | |
| bhn_I0834 | cel9B | cellodextrinaseb | 552 | GH9 | CelD |
| bhn_I0568 | xyn10B | endo-1,4-β-xylanase b | 425 | GH10 | |
| bhn_I0169 | xyn10A | endo-1,4-β-xylanase b | 451 | GH10 | |
| bhn_I1458 | glgB2 | 1,4-α-glucan branching enzymeb | 824 | GH13 | CBM48 |
| bhn_I0053 | glgB1 | 1,4-α-glucan branching enzymeb | 663 | GH13 | CBM48 |
| bhn_I2702 | amy13A | α-amylaseb | 697 | GH13 | CBM34 |
| bhn_I0634 | amy13B | α-amylase b | 536 | GH13 | |
| bhn_I1680 | amy13C | α-amylaseb | 434 | GH13 | |
| bhn_I0669 | amy13D | α-amylaseb | 511 | GH13 | |
| bhn_I1153 | glgX1 | glycogen debranching enzymeb | 726 | GH13 | CBM48 |
| bhn_I1315 | glgX2 | glycogen debranching enzymeb | 648 | GH13 | |
| bhn_I0652 | suc13P | sucrose phosphorylaseb | 553 | GH13 | |
| bhn_I2526 | chi18A | chitinase b | 567 | GH18 | |
| bhn_I1254 | lyc25A | lysozyme b | 362 | GH25 | |
| bhn_III074 | lyc25B | lysozyme b | 515 | GH25 | |
| bhn_I0191 | lyc25C | lysozyme b | 561 | GH25 | |
| bhn_I1763 | lyc25D | lysozyme b | 242 | GH25 | |
| bhn_I0527 | lyc25E | lysozymeb | 1213 | GH25 | Big2 (×2) |
| bhn_I1287 | aga27A | α-galactosidaseb | 577 | GH27 | |
| bhn_I0082 | gh27A | glycoside hydrolase family 27b | 442 | GH27 | |
| bhn_I1952 | pg128A | polygalacturonaseb | 531 | GH28 | |
| bhn_I2679 | pgl28B | polygalacturonaseb | 519 | GH28 | |
| bhn_I1087 | fuc29A | α-L-fucosidaseb | 475 | GH29 | |
| bhn_I2734 | gh30A | glycoside hydrolase family 30 b | 575 | GH30 | |
| bhn_I1581 | gh31A | glycoside hydrolase family 31b | 756 | GH31 | |
| bhn_I2191 | gh31C | glycoside hydrolase family 31b | 674 | GH31 | |
| bhn_I0283 | gh31B | glycoside hydrolase family 31b | 635 | GH31 | |
| bhn_I0582 | scr32A | sucrose-6-phosphate hydrolaseb | 493 | GH32 | |
| bhn_I0826 | bga35A | β-galactosidaseb | 622 | GH35 | |
| bhn_I1817 | bga35B | β-galactosidaseb | 735 | GH35 | |
| bhn_I0644 | aga36A | α-galactosidaseb | 782 | GH36 | |
| bhn_I1583 | aga36B | α-galactosidaseb | 620 | GH36 | |
| bhn_I1945 | aga36C | α-galactosidaseb | 730 | GH36 | |
| bhn_I0086 | man38A | α-mannosidaseb | 1053 | GH38 | |
| bhn_III010 | bga42A | β-galactosidaseb | 673 | GH42 | |
| bhn_I0167 | xsa43A | xylosidase/arabinofuranosidase b | 543 | GH43 | CBM6 |
| bhn_I0981 | xsa43B | xylosidase/arabinofuranosidaseb | 301 | GH43 | |
| bhn_I2037 | xsa43C | xylosidase/arabinofuranosidaseb | 302 | GH43 | |
| bhn_I2111 | xsa43D | xylosidase/arabinofuranosidaseb | 517 | GH43 | |
| bhn_I2735 | xsa43E | xylosidase/arabinofuranosidaseb | 352 | GH43 | |
| bhn_I0032 | xsa43G | xylosidase/arabinofuranosidaseb | 312 | GH43 | |
| bhn_I0164 | xsa43F | xylosidase/arabinofuranosidase and esteraseb | 925 | GH43 | |
| bhn_I1509 | arf51C | α-L-arabinofuranosidaseb | 630 | GH51 | |
| bhn_I2228 | arf51A | α-L-arabinofuranosidaseb | 502 | GH51 | |
| bhn_I0010 | arf51B | α-L-arabinofuranosidaseb | 504 | GH51 | |
| bhn_I0670 | agn53A | arabinogalactan endo-1,4-β-galactosidase b | 439 | GH53 | |
| bhn_I0183 | agu67A | α-D-glucuronidaseb | 662 | GH67 | |
| bhn_I2177 | mal77A | 4-α-glucanotransferase b | 506 | GH77 | |
| bhn_I0697 | ugl88A | unsaturated glucuronyl hydrolaseb | 385 | GH88 | |
| bhn_I2381 | ugl88B | unsaturated glucuronyl hydrolaseb | 383 | GH88 | |
| bhn_I2196 | cbp94A | cellobiose phosphorylaseb | 814 | GH94 | |
| bhn_I1582 | gh95A | glycoside hydrolase family 95b | 734 | GH95 | |
| bhn_I2548 | gh105A | unsaturated rhamnogalacturonyl hydrolaseb | 349 | GH105 | |
| bhn_I0090 | gh105B | unsaturated rhamnogalacturonyl hydrolaseb | 363 | GH105 | |
| bhn_I2549 | gnpA | D-galactosyl-β-1-4-L-rhamnose phosphorylaseb | 722 | GH112 | |
| bhn_I0185 | gh115A | α-glucuronidaseb | 947 | GH115 | |
| bhn_I1083 | xyl120A | xylosidaseb | 861 | GH120 | |
| bhn_I1738 | xyl120B | xylosidaseb | 664 | GH120 | |
| bhn_III076 | est1A | feruloyl esterase b | 351 | CE1 | |
| bhn_I1244 | est2A | acetyl-xylan esteraseb | 372 | CE2 | |
| bhn_III070 | est4A | polysaccharide deacetylaseb | 207 | CE4 | |
| bhn_I0843 | est4C | polysaccharide deacetylase b | 280 | CE4 | |
| bhn_I0666 | nagA | N-acetylglucosamine-6-phosphate deacetylaseb | 371 | CE9 | |
| bhn_I1609 | est12A | carbohydrate esterase family 12b | 584 | CE12 | |
| bhn_I1927 | est12B | carbohydrate esterase family 12b | 244 | CE12 | |
| bhn_I1926 | pl11A | polysaccharide lyaseb | 746 | PL11 | |
| bhn_I0657 | glgP1 | glycogen phosphorylaseb | 769 | GT35 | |
| bhn_I2673 | glgP2 | glycogen phosphorylaseb | 824 | GT35 | |
| bhn_I1848 | – | carbohydrate binding protein b | 983 | CBM2a (×1), CBM6 (×6) |
aCAZy descriptions and classifications compiled from the CAZy database [68]. bIndicates homologues in the B. hungatei JK615T draft genome. Genes encoding predicted secreted polysaccharide degrading enzymes are in bold
Table 8.
Comparison of MB2003, JK615T and B316T protein coding gene percentages to COG functional categories
| Code | % of totala | Description | ||
|---|---|---|---|---|
| MB2003 | JK615T | B316T | ||
| J | 9.52 | 9.33 | 8.96 | Translation |
| A | RNA processing and modification | |||
| K | 7.31 | 7.59 | 7.30 | Transcription |
| L | 4.32 | 4.59 | 4.63 | Replication, recombination and repair |
| B | Chromatin structure and dynamics | |||
| D | 1.57 | 1.60 | 1.44 | Cell cycle control, mitosis and meiosis |
| V | 3.19 | 2.90 | 3.19 | Defense mechanisms |
| T | 6.82 | 6.72 | 7.47 | Signal transduction mechanisms |
| M | 7.61 | 7.45 | 8.52 | Cell wall/membrane biogenesis |
| N | 2.99 | 3.29 | 2.75 | Cell motility |
| U | 1.08 | 1.26 | 1.14 | Intracellular trafficking and secretion |
| O | 3.83 | 3.63 | 3.89 | Posttranslational modification, protein turnover, chaperones |
| C | 3.39 | 3.63 | 3.72 | Energy production and conversion |
| G | 11.93 | 11.99 | 12.15 | Carbohydrate transport and metabolism |
| E | 8.69 | 8.85 | 7.91 | Amino acid transport and metabolism |
| F | 3.93 | 3.82 | 3.98 | Nucleotide transport and metabolism |
| H | 3.88 | 3.77 | 3.23 | Coenzyme transport and metabolism |
| I | 3.53 | 3.19 | 2.80 | Lipid transport and metabolism |
| P | 3.88 | 4.06 | 2.75 | Inorganic ion transport and metabolism |
| Q | 0.79 | 0.68 | 0.79 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 7.76 | 6.91 | 7.43 | General function prediction only |
| S | 3.58 | 3.53 | 4.11 | Function unknown |
| – | 40.33 | 39.83 | 46.51 | Not in COGs |
aThe percentage is based on the total number of protein coding genes in the genome
Fig. 6.
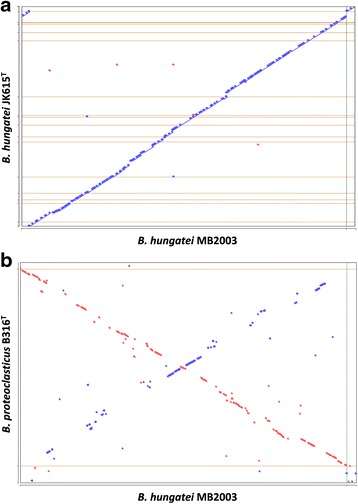
Genome synteny analysis. Alignment of the B. hungatei MB2003 genome against the draft genome of B. hungatei JK615T (a) and the complete genome of B. proteoclasticus B316T (b). Whenever the two sequences agree, a colored line or dot is plotted. If the two sequences were perfectly identical, a single line would go from the bottom left to the top right. Units displayed in base-pairs. Color codes: blue, forward sequence, red, reverse sequence
Butyrate production
For the production of butyrate and H2 from glucose, the MB2003 genome possesses a pyruvate:ferredoxin oxidoreductase gene, nifJ (bhn_I2528) required for pyruvate conversion to acetyl-CoA, as well as a butyryl-CoA dehydrogenase/electron transferring flavoprotein bcd-etfAB (bhn_I2225, bhn_I2221 and bhn_I2222) to generate ATP by classic substrate level phosphorylation. In addition, MB2003 possesses genes that encode all six subunits of the Rnf (rnfA, rnfB, rnfC, rnfD, rnfE, rnfG) and Ech (echA, echB, echC, echD, echE, echF) hydrogenases. These pathways involve the transmembrane ion pumps Ech [34] or Rnf [35–38], that generate a transmembrane proton and/or sodium electrochemical potential from redox cofactors for ATP synthesis by ETP [34, 36]. The MB2003 genome does not possess genes for PorABDG, a pyruvate ferredoxin oxidoreductase similar in function to NifJ or genes for EhaA-R, EhbA-P, HydA-C, MbhLKJ, or MvhADG/HdrABC similar in function to the Fd-dependent Ech hydrogenase. In addition, an alternative pathway exists where formate is predicted to be the end product and involves the decarboxylation of acetyl-CoA by a pyruvate formate lyase pflB (bhn_I0124) instead of NifJ. It has been proposed that Ech and Rnf work in concert with NifJ and Bcd-Etf complex to drive ATP synthesis by ETP during glucose fermentation to butyrate [34, 36, 39]. Interestingly, the vast majority of anaerobic prokaryotes appear to possess either an Ech or Rnf but not both [40, 41]. However, a recent analysis of rumen prokaryotic genomes identified 10.1601/nm.4129 and 10.1601/nm.4145 as a rare group of bacteria that possess genes for both Ech and Rnf [42]. These findings warrant further biochemical investigation to determine the activity of Ech and Rnf in 10.1601/nm.4129.
The MB2003 pathways for butyrate production presume the possession of a complete Embden-Meyerhof-Parnas glycolytic pathway. Enolase (eno, EC4.2.1.11), converts 2-phospho-D-glycerate to phosphoenolpyruvate in the second to last step of the EMP pathway. Previous work has shown that B316T lacks a detectable enolase [29], and the Methylglyoxal Shunt was proposed as a possible alternative to the EMP pathway. In this pathway the dihydroxyacetone phosphate is transformed to pyruvate via methylglyoxal and d-lactate dehydrogenase, encoded by ldhA [43]. The MB2003 genome possesses two methylglyoxal synthase genes, mgsA (bhn_I1328 and bhn_I1996), glyoxalase gene gloA (bhn_I1783) and an alternative l-lactate dehydrogenase, encoded by ldh (bhn_I0363). MB2003 has the same set of genes as B316T for the production of butyrate, formate, acetate and lactate, but also is the only 10.1601/nm.4132 reported to date that lacks a detectable enolase gene. Genome sequences from a wider range of 10.1601/nm.4132 and 10.1601/nm.13010 strains are required to determine if these are common features in these organisms.
Polysaccharide degradation
The Carbohydrate-Active enZYmes database was used to identify glycoside hydrolases, glycosyl transferases, polysaccharide lyases, carbohydrate esterases and carbohydrate-binding protein module families within the MB2003 genome. MB2003 has a similar CAZyme profile to B316T [21, 31], and analysis of the functional domains of enzymes involved in the breakdown or synthesis of complex carbohydrates, has revealed the polysaccharide-degrading potential of this rumen bacterium.
Approximately 3% of the MB2003 genome (90 CDSs) is predicted to encode either secreted or intracellular proteins dedicated to polysaccharide degradation, similar to that found in B316T. The MB2003 genome is predicted to encode 19 secreted (16 GHs, two CEs and one CBP) and 65 intracellular (59 GHs, 5 CEs and one PL) proteins involved in polysaccharide breakdown (Table 9). The enzymatic profiles of MB2003 and JK615T are almost identical, as both possess the same genes encoding predicted secreted and intracellular CAZymes in their genomes (Table 9). Out of the 19 genes predicted to encode secreted polysaccharide degrading enzymes, only two, lysozyme lyc25B (bhn_III074) and feruloyl esterase est1A (bhn_III076), are encoded by the MB2003 chromid (Bhu II). MB2003 has no secreted enzyme larger than 1000 aa in size, with the average size secreted enzymes being 510 aa. The majority (59) of MB2003 genes involved in polysaccharide breakdown (excluding GTs), had corresponding homologues in B316T and JK615T. Three of the genes encoding intracellular proteins were found in the Bhu II chromid: a β-glucosidase bgl3A (bhn_III062), a β-galactosidase bga42A (bhn_III010) and a polysaccharide deacetylase est4A (bhn_III070). The analysis of the Pfam domains from the most abundant GH families (GH2, GH31, GH3, GH13 and GH43) showed they did not contain signal sequences and hence were predicted to be located intracellularly. Similarly, CAZymes with predicted roles in xylan and pectin degradation, the GH8, GH28, GH51, GH67, GH88, GH105, GH115, CE2 and CE10 families were also predicted to be intracellular. Of these, MB2003 contains CAZymes with homologues in B316T except for the α-L-arabinofuranosidase arf51C (bhn_I1509). These findings suggest that a variety of complex oligosaccharides resulting from extracellular hydrolysis are metabolized within the cell.
Growth experiments showed MB2003 to be a metabolically versatile bacterium able to grow on a wide variety of monosaccharides, disaccharides and glycosides (Table 2). However, unlike B316T, MB2003 and JK615T were unable to utilize the insoluble substrates pectin and xylan for growth (Table 2). In addition, MB2003, JK615T and B316T are unable to degrade cellulose, however among these organisms, only B316T is able to utilize a range of other insoluble plant polysaccharides. The ability of B316T to breakdown pectin, starch and xylan is predicted to be based on nine large (>1000 aa) cell-associated proteins shown to be significantly up-regulated in B316T cells grown on xylan [44]. These are: α-amylase amy13A (bpr_I1087), arabinogalactan endo-1,4-β-galactosidase agn53A (bpr_I2041), carbohydrate esterase family 12 est12B (bpr_I1204), endo-1,3(4)-β-glucanase lic16A (bpr_I2326), pectate lyase pel1A (bpr_I2372), pectin methylesterase pme8B (bpr_I2473), xylosidase/arabinofuranosidase xsa43J (bpr_I2935), endo-1,4-β-xylanase xyn10B (bpr_I0026), and the cell wall binding domain-containing protein (bpr_I0264). These proteins contain multiple cell wall binding repeat domains (CW-binding domain, Pfam01473) at their C-termini that are predicted to anchor the protein to the peptidoglycan cell membrane. Among these secreted polysaccharidases, some contain single or combinations of catalytic activities: GH10 (endo-1, 4-β-xylanase, xyn10B), GH43 (xylosidase/arabinofuranosidase, xsa43J), PL1 (pectate lyase, pel1A), CE8 and PL9 (pectin methylesterase, pme8B) [45, 46]. Neither MB2003 nor JK615T contain any genes encoding CW-binding domains and are thus are markedly different from B316T.
A curious feature of MB2003 was the presence of a single large (983 aa) carbohydrate binding protein (CBP, bhn_I1848), also present in JK615T (EJ23DRAFT_00192). The domain structures of bhn_I1848 and EJ23DRAFT_00192 are unusual, containing six CBM6 (Pfam03422) domains towards the N-terminus and a single C-terminal CBM2a (Pfam00553) domain. In contrast, B316T encodes two CBPs (bpr_I0736 and bpr_I1599) where both contain two CBM2a domains, and bpr_I1599 also contains two CBM6 domains [29]. CBM6 non-catalytic modules characteristically bind xylose and are associated with xylanase activity with ligand specificity for xylan [47, 48]. CBM2 domains, are divided into two sub-families: 2a, that bind to crystalline cellulose even when associated with xylanases [49], and 2b, that bind to xylan [50]. Recent studies have shown that in discrete regions of plant cell walls, initial enzymatic attack of pectin increases the access of CBMs to cellulose [51], effectively loosening the polysaccharide interactions to expose the xylan and xyloglucan substrates [52, 53]. This initial stage in enzymatic saccharification of plant cell walls termed amorphogenesis [54], and is a possible role for such CBPs containing multiple non-catalytic domains. In the rumen, MB2003, B316T and JK615T may secrete these non-catalytic CBPs synergistically with polysaccharide-active enzymes as a mechanism to disrupt the interface between polysaccharides to enhance the rate and extent of plant cell wall degradation.
Conclusion
The 10.1601/nm.4132 MB2003 genome sequence adds valuable information regarding the polysaccharide-degrading potential present in the genus 10.1601/nm.4129. Genomic comparisons revealed that 10.1601/nm.4132 MB2003 shows a high level of similarity with 10.1601/nm.4132 JK615T and 10.1601/nm.13010 B316T type strains, including genes involved in production of butyrate, formate, acetate and lactate. While MB2003 and JK615T encode a large repertoire of enzymes predicted to metabolize insoluble polysaccharides such as xylan and pectin, they are unable to grow on these substrates and instead appear to be equipped to utilize mainly oligo- and monosaccharides as substrates for growth. Although MB2003 has similar phenotypic characteristics and occupies the same habitat as other 10.1601/nm.4129 species, its genome encodes fewer extracellular polysaccharide degrading enzymes, in particular, those that contain multiple cell wall binding repeat domains. The overall genome similarities, metabolic versatility and differences in the abundance of CAZymes observed in 10.1601/nm.13010 and 10.1601/nm.4132 offers a new view of the genes required for polysaccharide degradation in the rumen. MB2003 appears to occupy a ruminal niche as a secondary degrader of oligosaccharides, in order to coexist with fibre-degrading organisms in this dynamic and competitive environment.
Acknowledgements
The MB2003 genome sequencing project was funded by the New Zealand Ministry of Business, Innovation and Employment New Economy Research Fund programme: Accessing the uncultured rumen microbiome, contract number C10X0803. Electron microscopy was conducted with the assistance of the Manawatū Microscopy and Imaging Centre at Massey University, Palmerston North, New Zealand.
Abbreviations
- Bp
Base pair(s)
- CAZymes
Carbohydrate-Active enZYmes
- CBMs
Carbohydrate-Binding Module(s)
- CEs
Carbohydrate Esterase(s)
- Ech
Escherichia coli hydrogenase-3-type hydrogenase
- Eha
Energy-converting hydrogenase A
- Ehb
Energy-converting hydrogenase B
- EMP
Embden-Meyerhof-Parnas
- ETP
Electron transport phosphorylation
- GHs
Glycoside Hydrolase(s)
- GTs
Glycosyl Transferase(s)
- Hyd
Ferredoxin hydrogenase
- Mbh
Membrane-bound hydrogenase
- Mvh/Hdr
Methyl viologen hydrogenase/heterodisulfide reductase
- Pfl
Pyruvate formate lyase
- PLs
Polysaccharide Lyase(s)
- Por
Pyruvate ferredoxin oxidoreductase
- Rnf
Rhodobacter nitrogen fixation
Additional file
Associated MIGS record for B. hungatei MB2003, which links to the SIGS supplementary content website. (DOCX 17 kb)
Authors’ contributions
NP, WJK, GTA conceived and designed the experiments. NP performed the sequencing and assembly experiments. NP, WJK, SCL, EA performed the genome annotation and comparative studies. NP performed the bacterial growth studies and polysaccharide utilization profiling. NP, WJK, GTA wrote the manuscript. All authors commented on the manuscript before submission. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s40793-017-0285-8) contains supplementary material, which is available to authorized users.
References
- 1.Henderson G, Cox F, Ganesh S, Jonker A, Young W, Collaborators GRC, Janssen PH. Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range. Sci Rep. 2015;5(14567):1–13. doi: 10.1038/srep14567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bryant MP, Small N. The anaerobic monotrichous butyric acid-producing curved rod-shaped bacteria of the rumen. J Bacteriol. 1956;72(1):16–21. doi: 10.1128/jb.72.1.16-21.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hungate RE. The rumen and its microbes. New York: Academic Press; 1966. [Google Scholar]
- 4.Willems A, Collins MD. Butyrivibrio. Bryant and Small 1956b, 18, emend. Moore, Johnson and Holdeman 1976, 241AL. In: De Vos P, Garrity G, Jones D, Krieg N, Ludwig W, Rainey F, Schleifer K-H, Whitman W, editors. Bergey's manual of systematic bacteriology. Volume 3. 2. New York: Springer; 2009. pp. 927–937. [Google Scholar]
- 5.Hespell RB. The genera Butyrivibrio, Lachnospira, and Roseburia. In: Balows A, Trüper HG, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes: a handbook on the biology of bacteria: ecophysiology, isolation, identification, applications. New York: Springer; 1992. pp. 2023–2033. [Google Scholar]
- 6.Moon CD, Pacheco DM, Kelly WJ, Leahy SC, Li D, Kopecny J, Attwood GT. Reclassification of Clostridium proteoclasticum as Butyrivibrio proteoclasticus comb. nov., a butyrate-producing ruminal bacterium. Int J Syst Evol Microbiol. 2008;58(9):2041–2045. doi: 10.1099/ijs.0.65845-0. [DOI] [PubMed] [Google Scholar]
- 7.Hespell RB, Cotta MA. Degradation and utilization by Butyrivibrio fibrisolvens H17c of xylans with different chemical and physical properties. Appl Environ Microbiol. 1995;61(8):3042–3050. doi: 10.1128/aem.61.8.3042-3050.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Attwood GT, Reilly K. Characterization of proteolytic activities of rumen bacterial isolates from forage-fed cattle. J Appl Bacteriol. 1996;81(5):545–552. doi: 10.1111/j.1365-2672.1996.tb03545.x. [DOI] [PubMed] [Google Scholar]
- 9.Diez-Gonzalez F, Bond DR, Jennings E, Russell JB. Alternative schemes of butyrate production in Butyrivibrio fibrisolvens and their relationship to acetate utilization, lactate production, and phylogeny. Arch Microbiol. 1999;171(5):324–330. doi: 10.1007/s002030050717. [DOI] [PubMed] [Google Scholar]
- 10.Paillard D, McKain N, Rincon MT, Shingfield KJ, Givens DI, Wallace RJ. Quantification of ruminal Clostridium proteoclasticum by real-time PCR using a molecular beacon approach. J Appl Microbiol. 2007;103(4):1251–1261. doi: 10.1111/j.1365-2672.2007.03349.x. [DOI] [PubMed] [Google Scholar]
- 11.Shane BS, Gouws L, Kistner A. Cellulolytic bacteria occurring in the rumen of sheep conditioned to low-protein teff hay. J Gen Microbiol. 1969;55(3):445–457. doi: 10.1099/00221287-55-3-445. [DOI] [PubMed] [Google Scholar]
- 12.Hazlewood GP, Theodorou MK, Hutchings A, Jordan DJ, Galfre G. Preparation and characterization of monoclonal antibodies to a Butyrivibrio sp. and their potential use in the identification of rumen Butyrivibrios, using an enzyme-linked immunosorbent assay. J Gen Microbiol. 1986;132(1):43–52. [Google Scholar]
- 13.Mannarelli BM. Deoxyribonucleic acid relatedness among strains of the species Butyrivibrio fibrisolvens. Int J Syst Bacteriol. 1988;38(4):340–347. doi: 10.1099/00207713-38-4-340. [DOI] [Google Scholar]
- 14.Mannarelli BM, Stack RJ, Lee D, Ericsson L. Taxonomic relatedness of Butyrivibrio, Lachnospira, Roseburia, and Eubacterium species as determined by DNA hybridization and extracellular-polysaccharide analysis. Int J Syst Bacteriol. 1990;40(4):370–378. doi: 10.1099/00207713-40-4-370. [DOI] [Google Scholar]
- 15.Forster R, Teather R, Gong J, Deng S-J. 16S rDNA analysis of Butyrivibrio fibrisolvens: phylogenetic position and relation to butyrate-producing anaerobic bacteria from the rumen of white-tailed deer. Lett Appl Microbiol. 1996;23(4):218–222. doi: 10.1111/j.1472-765X.1996.tb00069.x. [DOI] [PubMed] [Google Scholar]
- 16.Willems A, Amat-Marco M, Collins MD. Phylogenetic analysis of Butyrivibrio strains reveals three distinct groups of species within the Clostridium subphylum of the gram-positive bacteria. Int J Syst Bacteriol. 1996;46(1):195–199. doi: 10.1099/00207713-46-1-195. [DOI] [PubMed] [Google Scholar]
- 17.Forster RJ, Gong J, Teather RM. Group-specific 16S rRNA hybridization probes for determinative and community structure studies of Butyrivibrio fibrisolvens in the rumen. Appl Environ Microbiol. 1997;63(4):1256–1260. doi: 10.1128/aem.63.4.1256-1260.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Gylswyk NO, Hippe H, Rainey FA. Pseudobutyrivibrio ruminis gen. Nov., sp. nov., a butyrate-producing bacterium from the rumen that closely resembles Butyrivibrio fibrisolvens in phenotype. Int J Syst Bacteriol. 1996;46(2):559–563. doi: 10.1099/00207713-46-2-559. [DOI] [Google Scholar]
- 19.Kopecny J, Zorec M, Mrazek J, Kobayashi Y, Marinsek-Logar R. Butyrivibrio hungatei sp. nov. and Pseudobutyrivibrio xylanivorans sp. nov., butyrate-producing bacteria from the rumen. Int J Syst Evol Microbiol. 2003;53(1):201–209. doi: 10.1099/ijs.0.02345-0. [DOI] [PubMed] [Google Scholar]
- 20.Noel S. Cultivation and community composition analysis of plant-adherent rumen bacteria. PhD thesis. Massey University, Institute of Molecular Biosciences; 2013.
- 21.Palevich N. Genome sequencing of rumen bacteria involved in lignocellulose digestion. MSc thesis: Massey University, Institute of Molecular Biosciences; 2011. [Google Scholar]
- 22.Kenters N, Henderson G, Jeyanathan J, Kittelmann S, Janssen PH. Isolation of previously uncultured rumen bacteria by dilution to extinction using a new liquid culture medium. J Microbiol Methods. 2011;84(1):52–60. doi: 10.1016/j.mimet.2010.10.011. [DOI] [PubMed] [Google Scholar]
- 23.Richardson A, Calder A, Stewart C, Smith A. Simultaneous determination of volatile and non-volatile acidic fermentation products of anaerobes by capillary gas chromatography. Lett Appl Microbiol. 1989;9(1):5–8. doi: 10.1111/j.1472-765X.1989.tb00278.x. [DOI] [Google Scholar]
- 24.Saito H, Miura K-I. Preparation of transforming deoxyribonucleic acid by phenol treatment. Biochim Biophys Acta. 1963;72:619–629. doi: 10.1016/0926-6550(63)90386-4. [DOI] [PubMed] [Google Scholar]
- 25.Staden R, Beal KF, Bonfield JK. The staden package. In: Misener S, Krawetz SA, editors. Bioinformatics methods and protocols. New York: Humana Press; 1999. pp. 115–130. [Google Scholar]
- 26.Goecks J, Nekrutenko A, Taylor J. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11(8):R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using De Bruijn graphs. Genome Res. 2008;18(5):821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28(12):1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kelly WJ, Leahy SC, Altermann E, Yeoman CJ, Dunne JC, Kong Z, Pacheco DM, Li D, Noel SJ, Moon CD, Cookson AL, Attwood GT. The glycobiome of the rumen bacterium Butyrivibrio proteoclasticus B316T highlights adaptation to a polysaccharide-rich environment. PLoS One. 2010;5(8):e11942. doi: 10.1371/journal.pone.0011942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benson DA, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2015;43(D1):D30–D35. doi: 10.1093/nar/gku1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palevich N. Comparative genomics of Butyrivibrio and Pseudobutyrivibrio from the rumen. PhD thesis. Massey University, Institute of Fundamental Sciences; 2016.
- 32.Kyrpides NC, Hugenholtz P, Eisen JA, Woyke T, Göker M, Parker CT, Amann R, Beck BJ, Chain PS, Chun J. Genomic encyclopedia of bacteria and archaea: sequencing a myriad of type strains. PLoS Biol. 2014;12(8):e1001920. doi: 10.1371/journal.pbio.1001920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harrison PW, Lower RP, Kim NK, Young JP. Introducing the bacterial 'chromid': not a chromosome, not a plasmid. Trends Microbiol. 2010;18(4):141–148. doi: 10.1016/j.tim.2009.12.010. [DOI] [PubMed] [Google Scholar]
- 34.Welte C, Krätzer C, Deppenmeier U. Involvement of Ech hydrogenase in energy conservation of Methanosarcina mazei. FEBS J. 2010;277(16):3396–3403. doi: 10.1111/j.1742-4658.2010.07744.x. [DOI] [PubMed] [Google Scholar]
- 35.Biegel E, Müller V. Bacterial Na+-translocating ferredoxin: NAD+ oxidoreductase. Proc Natl Acad Sci. 2010;107(42):18138–18142. doi: 10.1073/pnas.1010318107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buckel W, Thauer RK. Energy conservation via electron bifurcating ferredoxin reduction and proton/Na+ translocating ferredoxin oxidation. Biochim Biophys Acta Bioenerg. 2013;1827(2):94–113. doi: 10.1016/j.bbabio.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 37.Schlegel K, Welte C, Deppenmeier U, Müller V. Electron transport during aceticlastic methanogenesis by Methanosarcina acetivorans involves a sodium-translocating Rnf complex. FEBS J. 2012;279(24):4444–4452. doi: 10.1111/febs.12031. [DOI] [PubMed] [Google Scholar]
- 38.Tremblay P-L, Zhang T, Dar SA, Leang C, Lovley DR. The Rnf complex of Clostridium ljungdahlii is a proton-translocating ferredoxin: NAD+ oxidoreductase essential for autotrophic growth. MBio. 2013;4(1):e00406–e00412. doi: 10.1128/mBio.00406-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gutekunst K, Chen X, Schreiber K, Kaspar U, Makam S, Appel J. The bidirectional NiFe-hydrogenase in Synechocystis sp. PCC 6803 is reduced by flavodoxin and ferredoxin and is essential under mixotrophic, nitrate-limiting conditions. J Biol Chem. 2014;289(4):1930–1937. doi: 10.1074/jbc.M113.526376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pereira IAC, Ramos A, Grein F, Marques M, Da Silva S, Venceslau S. A comparative genomic analysis of energy metabolism in sulfate reducing bacteria and archaea. Front Microbiol. 2011;2(69). [DOI] [PMC free article] [PubMed]
- 41.Weghoff MC, Bertsch J, Müller V. A novel mode of lactate metabolism in strictly anaerobic bacteria. Environ Microbiol. 2015;17(3):670–677. doi: 10.1111/1462-2920.12493. [DOI] [PubMed] [Google Scholar]
- 42.Hackmann TJ, Firkins JL. Electron transport phosphorylation in rumen butyrivibrios: unprecedented ATP yield for glucose fermentation to butyrate. Front Microbiol. 2015;6(622). [DOI] [PMC free article] [PubMed]
- 43.Cooper R. Metabolism of methylglyoxal in microorganisms. Annu Rev Microbiol. 1984;38(1):49–68. doi: 10.1146/annurev.mi.38.100184.000405. [DOI] [PubMed] [Google Scholar]
- 44.Leahy SC, Kelly WJ, Altermann E, Ronimus RS, Yeoman CJ, Pacheco DM, Li D, Kong Z, McTavish S, Sang C. The genome sequence of the rumen methanogen Methanobrevibacter ruminantium reveals new possibilities for controlling ruminant methane emissions. PLoS One. 2010;5(1):e8926. doi: 10.1371/journal.pone.0008926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dunne JC, Li D, Kelly WJ, Leahy SC, Bond JJ, Attwood GT, Jordan TW. Extracellular polysaccharide-degrading proteome of Butyrivibrio proteoclasticus. J Proteome Res. 2011;11(1):131–142. doi: 10.1021/pr200864j. [DOI] [PubMed] [Google Scholar]
- 46.Bond JJ, Dunne JC, Kwan FY, Li D, Zhang K, Leahy SC, Kelly WJ, Attwood GT, Jordan TW. Carbohydrate transporting membrane proteins of the rumen bacterium, Butyrivibrio proteoclasticus. J Proteome. 2012;75(11):3138–3144. doi: 10.1016/j.jprot.2011.12.013. [DOI] [PubMed] [Google Scholar]
- 47.McCartney L, Gilbert HJ, Bolam DN, Boraston AB, Knox JP. Glycoside hydrolase carbohydrate-binding modules as molecular probes for the analysis of plant cell wall polymers. Anal Biochem. 2004;326(1):49–54. doi: 10.1016/j.ab.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 48.Gilbert HJ, Knox JP, Boraston AB. Advances in understanding the molecular basis of plant cell wall polysaccharide recognition by carbohydrate-binding modules. Curr Opin Struct Biol. 2013;23(5):669–677. doi: 10.1016/j.sbi.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 49.Gilkes NR, Jervis E, Henrissat B, Tekant B, Miller R, Warren R, Kilburn D. The adsorption of a bacterial cellulase and its two isolated domains to crystalline cellulose. J Biol Chem. 1992;267(10):6743–6749. [PubMed] [Google Scholar]
- 50.Simpson PJ, Bolam DN, Cooper A, Ciruela A, Hazlewood GP, Gilbert HJ, Williamson MP. A family IIb xylan-binding domain has a similar secondary structure to a homologous family IIa cellulose-binding domain but different ligand specificity. Structure. 1999;7(7):853–864. doi: 10.1016/S0969-2126(99)80108-7. [DOI] [PubMed] [Google Scholar]
- 51.Blake AW, McCartney L, Flint JE, Bolam DN, Boraston AB, Gilbert HJ, Knox JP. Understanding the biological rationale for the diversity of cellulose-directed carbohydrate-binding modules in prokaryotic enzymes. J Biol Chem. 2006;281(39):29321–29329. doi: 10.1074/jbc.M605903200. [DOI] [PubMed] [Google Scholar]
- 52.Marcus SE, Verhertbruggen Y, Hervé C, Ordaz-Ortiz JJ, Farkas V, Pedersen HL, Willats WG, Knox JP. Pectic homogalacturonan masks abundant sets of xyloglucan epitopes in plant cell walls. BMC Plant Biol. 2008;8(60):1–12. doi: 10.1186/1471-2229-8-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hervé C, Rogowski A, Gilbert HJ, Paul Knox J. Enzymatic treatments reveal differential capacities for xylan recognition and degradation in primary and secondary plant cell walls. Plant J. 2009;58(3):413–422. doi: 10.1111/j.1365-313X.2009.03785.x. [DOI] [PubMed] [Google Scholar]
- 54.Coughlan MP. The properties of fungal and bacterial cellulases with comment on their production and application. Biotechnol Genet Eng Rev. 1985;3(1):39–110. doi: 10.1080/02648725.1985.10647809. [DOI] [Google Scholar]
- 55.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985:783–91. [DOI] [PubMed]
- 57.Reddy T, Thomas AD, Stamatis D, Bertsch J, Isbandi M, Jansson J, Mallajosyula J, Pagani I, Lobos EA, Kyrpides NC. The genomes OnLine database (GOLD) v. 5: a metadata management system based on a four level (meta) genome project classification. Nucleic Acids Res. 2015;43(D1):D1099–D1106. doi: 10.1093/nar/gku950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol. 2008;26(5):541–547. doi: 10.1038/nbt1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, bacteria, and Eucarya. Proc Natl Acad Sci. 1990;87(12):4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rainey F. Phylum SK-H, XIII. Firmicutes gibbons and Murray 1978, 5 (Fimacutes [sic] gibbons and Murray 1978, 5) In: De Vos P, Garrity G, Jones D, Krieg N, Ludwig W, Rainey F, Schleifer K-H, Whitman W, editors. Bergey’s manual of systematic bacteriology. 2. New York: Springer; 2009. pp. 19–1317. [Google Scholar]
- 61.Ludwig W, Schleifer K-H, Whitman W. Revised road map to the phylum Firmicutes. In: De Vos P, Garrity G, Jones D, Krieg N, Ludwig W, Rainey F, Schleifer K-H, Whitman W, editors. Bergey’s manual of systematic bacteriology. Volume 3. 2. New York: Springer; 2009. pp. 1–13. [Google Scholar]
- 62.Rainey F, Class II. Clostridia class. nov. In: De Vos P, Garrity G, Jones D, Krieg N, Ludwig W, Rainey F, Schleifer K-H, Whitman W, editors. Bergey’s manual of systematic bacteriology. 2. New York: Springer; 2009. p. 736. [Google Scholar]
- 63.Rainey F, Order I. Clostridiales Prévot 1953. In: De Vos P, Garrity G, Jones D, Krieg N, Ludwig W, Rainey F, Schleifer K-H, Whitman W, editors. Bergey’s manual of systematic bacteriology. 2. New York: Springer; 2009. p. 736. [Google Scholar]
- 64.Rainey F, Family V. Lachnospiraceae fam. nov. In: De Vos P, Garrity G, Jones D, Krieg N, Ludwig W, Rainey F, Schleifer K-H, Whitman W, editors. Bergey’s manual of systematic bacteriology. 2. New York: Springer; 2009. pp. 921–968. [Google Scholar]
- 65.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT. Gene ontology: tool for the unification of biology. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jensen LJ, Friis C, Ussery DW. Three views of microbial genomes. Res Microbiol. 1999;150(9):773–777. doi: 10.1016/S0923-2508(99)00116-3. [DOI] [PubMed] [Google Scholar]
- 67.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ, Gapped BLAST. PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The carbohydrate-active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 2009;37(D1):D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]


