Abstract
Background
Calotropis procera is a wild plant species in the family Apocynaceae that is able to grow in harsh, arid and heat stressed conditions. Understanding how this highly adapted plant persists in harsh environments should inform future efforts to improve the hardiness of crop and forage plant species. To study the plant response to droμght and osmotic stress, we treated plants with polyethylene glycol and NaCl and carried out transcriptomic and metabolomics measurements across a time-course of five days.
Results
We identified a highly dynamic transcriptional response across the time-course including dramatic changes in inositol signaling, stress response genes and cytokinins. The resulting metabolome changes also involved sharp increases of myo-inositol, a key signaling molecule and elevated amino acid metabolites at later times.
Conclusions
The data generated here provide a first glimpse at the expressed genome of C. procera, a plant that is exceptionally well adapted to arid environments. We demonstrate, through transcriptome and metabolome analysis that myo-inositol signaling is strongly induced in response to drought and salt stress and that there is elevation of amino acid concentrations after prolonged osmotic stress. This work should lay the foundations of future studies in adaptation to arid environments.
Electronic supplementary material
The online version of this article (10.1186/s12870-017-1155-7) contains supplementary material, which is available to authorized users.
Keywords: Salt stress, Drought stress, Transcriptomics, Metabolomics, Myo-inositol
Background
Calotropis procera is a flowering plant in the family Apocynaceae (Geneianales). It is native to North Africa, Tropical Africa, Western Asia, South Asia, and Indochina. The plant contains a milky sap consisting of a complex mix of chemicals including “cardiac aglycones” belonging to the same chemical family as therapeutic chemicals found in foxgloves (Digitalis purpurea), a related family in the asteroid I clade. It is a desert plant that is commonly known as Ushar or Madar. The medicinal use of this species was known to ancient Egyptians [1]. Its traditional medicinal uses include the treatment of leprosy [2], muscular spasms, dysentery, fever, rheumatism, asthma and as an expectorant and purgative [3–6]. In addition, scientific evidence has shown that C. procera possesses anticancer activity [7] antibacterial [8], nematocidal [9] and larvicidal activities [10–12]. Ecophysiological studies of C. procera have indicated that the photosynthetic capacity of the leaves can change with water availability due to changes in both stomatal and non-stomatal factors. However, the photosynthetic capacity is still relatively high during dry periods compared to ‘wet’ periods and no chronic photoinhibition occurs during dry periods [13] demonstrating that C. procera has a high level of tolerance to drought stress and high light.
Globally, the greatest uncontrolled factor in crop production is the unpredictability of water availability. To achieve sustainable crop production for the future, new crop varieties that have improved water use efficiency will have to be developed. Water use efficiency is given by the ratio of net carbon assimilation in photosynthesis and the water lost by transpiration. Consequently, the ability to regulate water lost by transpiration and maximize photosynthetic carbon assimilation are both potentially important factors when attempting to improve a crop’s water use efficiency. Clearly, C. procera has been shown to withstand arid conditions and maintain high rates of photosynthesis. Photosynthesis is a major determinant of crop growth and productivity, as well as being a crucial parameter in determining the distribution of species and, at an ecosystem level, being the major route by which the biosphere affects the composition of the atmosphere, with all the implications that has for global climate change. Optimizing the rate of photosynthesis will be crucial in maximizing crop productivity and bio-fuel production in arid regions. Understanding the molecular mechanisms by which C. procera tolerates drought successfully will offer a unique insight into how we may modify crops for improved drought tolerance and productivity.
Recent studies have utilized next generation sequencing for molecular discovery in C. procera. RNA-seq approaches have been used to identify a Usp-like gene that is putatively involved in response to heat and/or drought stress [14]. While Pandey et al. [15] have used both transcriptomic and metabolomic approaches to study the glycoside biosynthetic pathway, which is valuable for the production of potential pharmacological compounds. Ramadan et al. [16] have used metabolomics and lipidomics to study the response to water stress and concluded that the plant has a very rapid response to water availability and water loss, which is a likely adaptation to its harsh natural environment.
In all plant systems studied thus far the phytohormone abscisic acid (ABA) has been shown to function as a key regulator of the response to drought and salinity and has a pivotal function as a growth inhibitor [17, 18]. ABA activates reprogramming of cellular mechanisms of abiotic stress adaptation and also carbohydrate and lipid metabolism [19]. Lipids are also thought to be intimately involved in signaling during plant drought and salt stress. Levels of phosphatidylinositol bisphosphate, phosphatidic acid, and DGPP diacylglycerolpyrophosphate are upregulated upon salt stress [20]. The inositol phosphate myo-inositol hexakisphosphate also has a role as a signaling molecule regulating stomatal closure by triggering the release of calcium from endomembrane stores [21].
In this study, we compare the transcriptome and metabolome responses of the C. procera plants exposed to polyethylene glycol (PEG), as a non-absorbed osmoticum often used to assess drought stress, and to osmotic stress induced by salt (NaCl). We demonstrate there are distinct genome-wide responses, which are highly dynamic over the observed time course of treatment. We also integrate this with metabolomics data to compare the differences in accumulated metabolites through the time course. Our data identified large changes in regulation of cytokinins and suggests a key role of inositol signaling in both the transcriptomic and metabolomic profiles. Physiological changes such as the accumulation of amino acid metabolites also appear to be associated with osmotic stress. We discuss how these findings could be further studied to understand adaptation mechanisms to extreme aridity.
Results and discussion
Sequence generation and transcript assembly
The experimental design for the study is shown in Fig. 1. Each sequence library generated a minimum of 16 million reads with the majority of samples producing in excess of 20 million reads. The number of reads generated for each sample is shown in Additional file 1: Table S1. These were combined and assembled into transcripts to give 187,923 transcripts in total with an average length of 1049 nt. Approximately 85% of all reads were included in the final assembly. We assume the remaining reads were either low quality reads or very weakly expressed transcripts which did not form contigs as no major contaminants were identified. The assemblies were annotated using BLAST2GO [22] resulting in 52,108 annotated transcripts of which 28,265 received informative GO terms. Clustering of the all samples based on 20,000 most highly expressed genes using a multidimensional scaling plot based on leading log2 fold changes demonstrates that there is weak clustering replicates however the control samples, and the different treatments form distinct but overlapping clusters (Fig. 2). The fact that discrete clusters are not formed may not be unexpected given the that these are field samples and not cultivated plants which may have substantial genetic diversity, also other processes like circadian regulation of genes will affect sample clustering. From the clustering of the samples demonstrates that circadian effects don’t have a major impact on the data 24 h, 3 days and 5 days do not lie closer to 0 h than 6 and 12 h. There is also good separation between the controls and treated plants at equivalent time points. We also observe that dim 1 (x-axis) separates control from treated samples fairly well, while dim2 (y-axis) separates the treated time points up to 24 h from the 3 and 5 day treatments. There is an unusual pattern of expression at day 3 as the controls are perturbed from the other control samples more than salt and PEG treatments at day 3. This might also explain why day 3 shows fewer apparently differentially expressed genes than 24 h and 5 days.
Fig. 1.
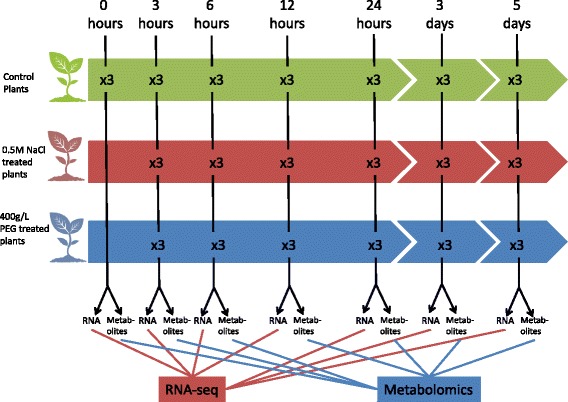
Experimental design for this study. Three groups of plants were used. A control group that were administered water, plants that were administered 0.5 M NaCl and a group administered 400 g/L of PEG at time 0. They were followed for 5 days and three plants samples at each time point. Metabolites and RNA were extracted from 3 replicates from each group and the data compared
Fig. 2.
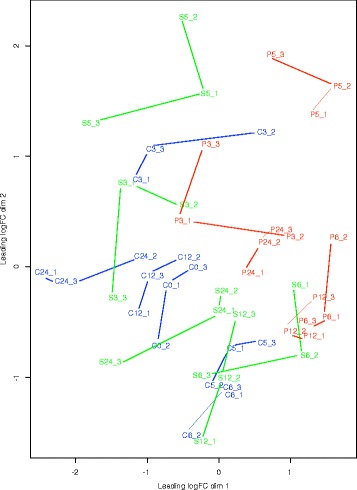
Multi-Dimensional Scaling plot for count data. MDS plot based on raw read count data of 20,000 genes for each sample. C6, C12, C24, C3 and C5 correspond to control plants at 6 h, 12 h, 24 h, 3 days, and 5 days, respectively. S6, S12, S24, S3 and S5 correspond to salt treated plants at 6 h, 12 h, 24 h, 3 days, and 5 days, respectively. P6, P12, P24, P3 and P5 correspond to PEG treated plants at 6 h, 12 h, 24 h, 3 days, and 5 days, respectively. Each replicate is labelled _1, _2, or _3. Lines connect samples from the same treatment and time point
To control for any circadian regulated genes each the gene expression in each group of plants was compared to control plants sampled at the same time point. Therefore, genes that are changing in expression due to circadian regulation alone should have significantly different sequence read counts between the control and treated samples. Additional file 2 shows the annotation, relative fold-changes and P-values for each gene assembly which was significantly regulated between conditions.
Differential response to PEG and NaCl stress
The number of genes regulated in response to NaCl treatment was much higher than that regulated in response to PEG treatment in the first 24 h (Fig. 3). After three days, the number of genes is reduced for NaCl treated plants compared to that of PEG treated plants although as mentioned above, this time-point appears to have an unusual profile (Fig. 2) in the control plants compared to the other time-points. And at 5 days the number of regulated genes are largely similar (Fig. 3).
Fig. 3.
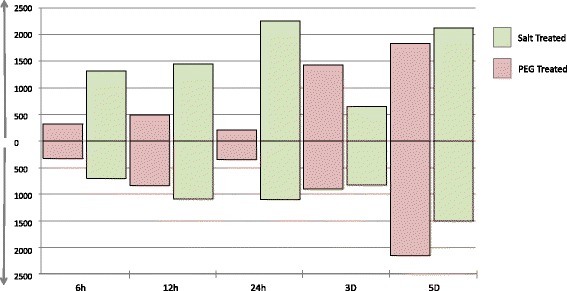
Number of up and downregulated genes in PEG and NaCl treated C. procera plants. Bars above the x-axis represent the number of up regulated genes relative to the control plants sampled at the same time point and bars below the x-axis represent the number of down regulated genes relative to the control plants sampled at the same time point
Expression was also compared between time-points and between treatments. Comparisons are shown in Fig. 4 and Additional file 3: Figure S1. There were very few regulated genes across all time points in either the NaCl or PEG treated plants suggesting that the response in the plant was very dynamic and rapidly changing. Comparing the number of shared regulated genes between the two treatments, it appears that the initial response due to the two different types of stresses is divergent as the overlap of shared genes is relatively small. However, the proportion of regulated genes shared between the treatments increases over time.
Fig. 4.
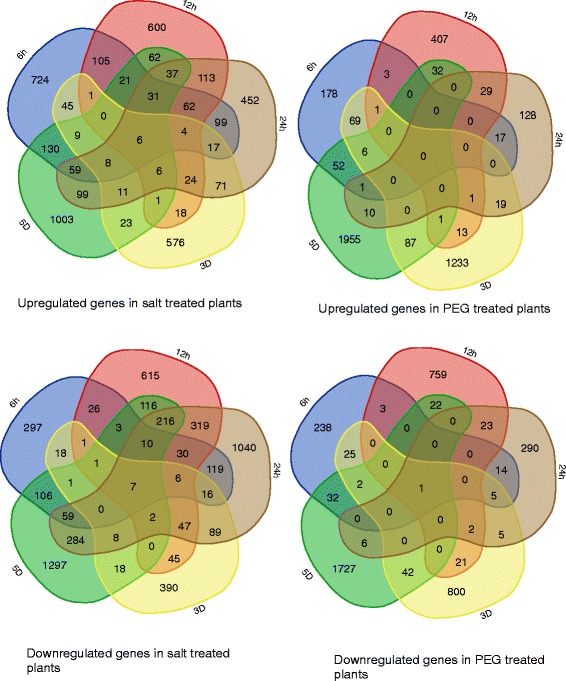
Venn diagrams of shared up and downregulated genes in C. procera across time. Each group is labelled 6 h, 12 h, 24 h, 3D, 5D for 6 h, 12 h, 24 h, 3 days, and 5 days, respectively
Gene ontology enrichment
Figure 5 shows the results of the GO enrichment analysis “Biological Process” GO terms that are enriched for up and downregulated genes for each time point (a full list of enriched GO terms is shown in Additional file 4: Tables S3 and S4). At 6 h of salt treatment, the “Molecular Function” GO terms that are most upregulated are involved in signalling such as DNA binding, kinase and calcium binding (Additional file 4: Table S4).
Fig. 5.
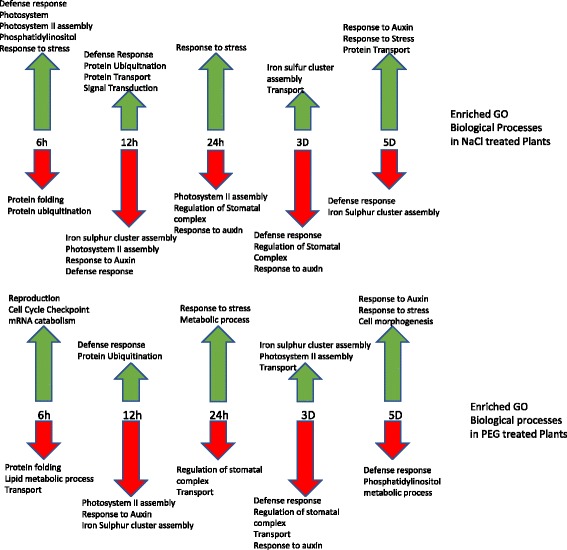
Selected Gene Ontology “Biological Process” terms in C. procera that are enriched in up and downregulated genes in PEG and NaCl treated plants. Enriched GO “Biological Processes” terms in NaCl treated plants, B. Enriched GO “Biological processes” terms in PEG treated plants. Up-arrows correspond to terms enriched in up regulated genes. Down-arrows correspond to terms enriched in down regulated genes
Metabolite analysis
Out of 135 selected signals, 22 matched to authentic standards (out of 148 standard signals). shown in Additional file 1: Table S2. While the transcriptional response to both salt and PEG treatment appears in the first 12 h, the metabolic response was, as expected, more delayed. A key stress related compound that is identified as upregulated in our treatment samples is myo-Inositol. Figure 6 shows myo-inositol is accumulated in the salt treated samples and to a lesser extent in the PEG treated samples relative to the controls. Also, Amino acids and amino acid-related metabolites are observed at greater concentrations in salt treated samples (Fig. 7).
Fig. 6.
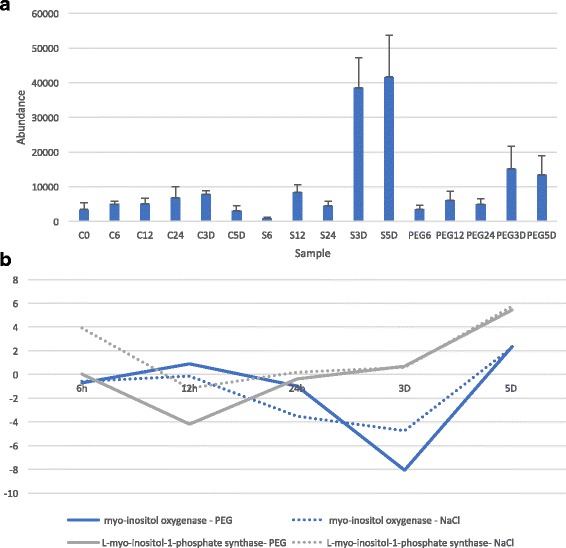
Myo-inositol abundance and myo-inositol metabolism gene expression in C. procera. a: Myo-inositol abundance as measured by metabolome analysis across all sample groups. b: Gene expression of myo-inositol metabolism genes myo-inositol oxygenase and L-myoinositol-1-phosphate synthase in PEG and NaCl treatments
Fig. 7.
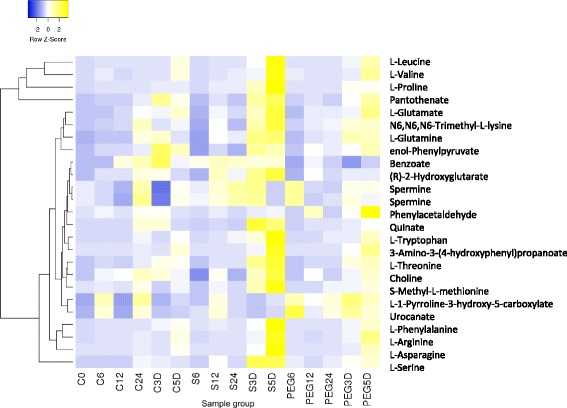
Amino acid pathway metabolite relative abundance across samples in C. procera. Heatmap of Z-scores for amino acid abundance as measured by metabolomics analysis across all sample groups. The dendrogram shows Average linkage clustering of metabolite abundance.
Discussion
In the NaCl treated plants, there are only six upregulated and seven downregulated genes that were shared across time points and in the PEG treated plants, there was just one down-regulated gene (Fig. 4), an O-methyltransferase similar to AT5G53810 in Arabidopsis. Some methyltransferases have been demonstrated to be involved in the response to drought by reducing gibberellin levels [23]. While, similar genes in Brassica have been shown to be downregulated [24]. Since methyltransferases are regulated via a wide range of processes from flowering to lignin accumulation, the significance of this finding is difficult to interpret.
The annotated upregulated genes under salt stress include a xyloglucan specific endoglucanase, and putative calmodulin, peroxidase and lacase genes. Xyloglucan genes are believed to be regulated by abscisic acid [25] and can enable loosening of the cell walls by modifying xyloglucan. Cell wall loosening can become stiffened due to the action reactive oxygen species (ROS) generated by drought or salt stress [26].
Zhou et al. [27] have recently demonstrated that calmodulin genes AtCaM1 and AtCaM4 expression regulate NO2 a key signaling molecule in abiotic stress. The calmodulin gene identified here is most similar to AtCaM3, which is normally associated with response to heat stress [28] and suggests that these signaling pathways have diverged in C. procera.
The peroxidase gene, which was regulated across treatments, is similar to ATPXG2, a lipid body localized calcium dependent peroxidase, that can regulate guard cell membranes and thereby stomatal aperture during the drought response [29]. Therefore, it is likely that this protein is involved in closing the stomata aperture to prevent water loss during drought and salt stress.
The role of laccase in salt response is mysterious. This enzyme is a multi-copper glycoprotein oxidase able to oxidise a wide range of substrates including phenols and amines. Their precise physiological and biochemical role in higher plants remains largely unclear [30]. The identification of this gene as upregulated across the time course should be studied further to elucidate its role.
The downregulated genes across salt stress treatment include a cation antiporter, a peptide transporter, a phosphoribosyl transferase, protochlorophyllide reductase and fructose-1,6-bisphosphatase. Fructose-1,6-bisphosphatase has been demonstrated to be downregulated in tobacco during salt stress as a result of decreased chloroplast activity, which also affects protochlorophyllide reductase. This is consistent with observations in rice where it has been postulated that reduction in photosynthetic activity prevents the accumulation of photosensitive tetrapyrroles that can produce singlet oxygen molecules under stress conditions [31].
The downregulation of the cation antiporter is a surprising finding as it would be expected to be upregulated to help maintaining cytoplasmic homeostasis in response to increased extracellular NaCl levels, as has been reported in many other plants including wheat [32], grape [33], and Arabidopsis [34] and heterologous overexpression has also resulted in greater salt tolerance in many plant systems [35]. However, at six hours of salt treatment, this gene has eight-fold higher activity in the control plant. This unusual response may be associated with Calotropis’ extreme hardiness. It is possible that the activity of this antiporter is replaced by an alternate protein, which is adapted to the high salt conditions. The gene is not significantly regulated in PEG treatments and, therefore, it is specific to osmotic stress associated with high NaCl levels.
Gene ontology enrichment analysis
The pattern of enriched processes from the GO enrichment analysis is consistent with the plant rapidly adapting to stress as it senses an increase of extracellular ions. The PEG treated plants appear to respond much more slowly in the absence of osmotic pressure. At 12 h, the enriched biological processes are not associated with a stress response (Additional file 4: Table S3 and Figure 5). However, both treatment groups at 12 h have upregulation of defence response and ubiquitination processes suggesting protein turnover is occurring in response to stress (Additional file 4: Table S3 and Fig. 5).
The most downregulated gene measured (11 fold change in salt treated plants) is adenine phosphoribosyl transferase, a key metabolic enzyme participating in the cytokinin inactivation by phosphoribosylation. Cytokinin metabolic processes, including cytokinin biosynthesis, degradation, interconversion and inactivation have important regulatory roles in plant development [36, 37]. Analyses of Arabidopsis cytokinin biosynthesis and catabolism genes following NaCl treatment showed upregulation of biosynthesis for several hours before returning to pretreatment levels. Conversely, catabolic genes were initially repressed and followed by a gradual increase. Downregulation of phosphoribosyl transferase will increase cytokinin accumulation and potentially lead to a number of downstream effects including the upregulation xyloglucan endotransglucosylases/hydrolases [38, 39], which we also see upregulated throughout the salt treatment.
At 12 h (Additional file 4: Table S4), carbohydrate binding was identified as upregulated and enriched. Plants are known to synthesize carbohydrate-binding proteins upon exposure to stresses like drought, high salt, hormone treatment, pathogen attack [40]. This class of lectins is located in the cytoplasm and the nucleus. Different lectins are also seen to be downregulated at 24 h in the PEG treated plants (Additional file 4: Table S4). The role of these lectins is still unclear, however recent data from Arabidopsis suggests that some may act as transcriptional regulators in response to abiotic stress [41].
Early in the time course, the biological function ‘response to auxin’ was enriched in downregulated genes which reflects that the plants have slowed growth in response to stress (Additional file 4: Table S3). By day five, the plants have recovered and these genes were enriched in upregulated transcripts relative to the control plants in both treatments and auxin was promoting cell division and growth.
Enriched “Biological Process” GO terms in upregulated genes at six hours include phosphotidylinositol metabolic process (Additional file 4: Table S3 and Fig. 5). Phosphotidylinositol is involved in the generation of the second messengers, inositol 1,4,5-trisphosphate (IP3) and diacylglycerol (DAG). IP3 releases Ca2+ from internal stores, whereas DAG activates protein kinase C. It has been shown in Arabidopsis that IP3 levels rapidly increase in response to hyperosmotic stress [42, 43].
Iron-sulphur (Fe-S) cluster assembly appears to have complex regulatory pattern, being initially downregulated and then upregulated at 24 h and 3 days in both treatments, Iron-sulphur cluster assembly occurs in chloroplasts, mitochondria and cytosol and has been shown to be inhibited by heavy metal stress in rice [44]. Glutaredoxins, which reduce disulphide bridges and involved in iron-sulphur assembly and known to be involved in this process, are sensitive to oxidative stress and are downregulated in our experiment [45].
Myo-inositol metabolism gene regulation is consistent with Myo-inositol abundance
Myo-inositol is a key intermediate in the inositol signaling pathway being a precursor of phosphotidylinositol and a potential entry-point into ascorbate generation [46]. It has been demonstrated in Arabidopsis that constitutive expression of myo-inositol 1-phosphate synthase 1, which is involved in myo-Inositol synthesis, protects against salt stress.
Our observation of myo-inositol from the metabolite data is consistent with the transcriptome data. As previously explained, we saw upregulation of phosphotidylinositol processes in the transcriptome of stressed plants. Also, we observed an increase in the expression of myo- inositol synthase expression peaking at nearly a 6-fold increase by day 5 (Fig. 6). Concurrently, we see a decrease in expression of myo-inositol oxygenase, an enzyme involved in the procession of myo-inositol into the ascorbic pathway and, therefore, acts to reduce myo-inositol [47]. The combined effect of the upregulation of myo-inositol synthesis and reduction in processing, will result in an accumulation of this important signaling molecule.
Amino acids accumulation occurs in response to salt stress
It has been shown that an increase of amino acid concentration in C. procera was closely related to the rapid adjustment of water availability [16]. In other plants, it has also been demonstrated that amino acids such as proline, and amines such as glycine betaine and polyamines accumulate under drought stress act as osmolytes to maintain cell turgor and to stabilize cell proteins and structures during drought stress [48]. Proline acts as a systemic signal to promote stomatal closure in Arabidopsis, it is interesting to observe in C. procera that this is not triggered in the PEG treated plants but only in response to salt treatment (Fig. 7). As amino acids were highly represented in the identified metabolites their role, relative to other osmoprotectants can’t be quantified here and further studies are needed to asses this.
Polyphenols are found to be increasingly abundant in stressed samples, both PEG- and salt stress-induced show the response. Petunidin 3-O-glucoside, p-Coumaroylquinic acid and quinate levels are upregulated across time. It has been shown that phenolics are produced by plants mainly for protection against abiotic stress [49]. The role of these metabolites in abiotic stress is not well understood.
Conclusions
The results of our analyses provide a first glimpse into the response of C. procera to salt and drought stress. Our results show that the plant undergoes complex changes in gene regulation across our five-day time course. The response to salt stress is initiated immediately as evidenced by the fact that at six hours key plant hormones such as cytokinin are being induced through the regulation of adenine phosphoribosyl transferase. These changes induce multiple downstream effects including effects including altering guard cell morphology to reduce water loss, reducing cell wall rigidity and accumulation of metabolites to balance osmotic pressure. It also appears from our study that inositol signaling also plays an important role in regulating stress response. Our approach of integrating metabolome and transcriptome analysis has enabled us to observe transcriptional changes giving rise to physiological changes in the plant, as with the accumulation of myo-inositol.
Understanding the plant’s adaptation to aridity is a vital step in producing crops that are able to cope with current rapid climate change. We argue that C. procera is a useful model for this study due to its remarkable ability to flourish in extremely harsh conditions. We speculate that this study will initiate future analysis of this system.
Methods
Stress treatment and leaf sampling
Seeds were collected from a field site near Jeddah, permission to collect plant material and preform fieldwork at the site was granted by the Governor of Makkah Province, Prince Khalid Al Faisal. All seeds in the experiment were collected from same fruit to reduce genetic heterogeneity. Seedlings were grown at a controlled temperature (25-28 °C) at 8000 lx light intensity for 16 h and an 8 h dark period. Seedlings were grown for one month after sowing. Three conditions were, then, applied across five time points (6 h, 12 h, 24 h, 3D and 5D) with three replicates in each case (Fig. 1). Careful leaf sampling was carried out in order to ensure for environmental conditions and variation between plants. Throughout the time course, samples were collected at regular intervals and sampling was taken from third leaf from top from each plant then with subsequent samples taken from the next lowest node at each time-point. All samples were flash frozen after collection in liquid Nitrogen (N2).
RNA extraction
100 mg of frozen plant leaf material was ground with a pestle and mortar in liquid N2. This was used as input material for the RNeasy Plant Mini kit (B-Mecaptoethanol was added to the RLT lysis buffer) following the manufacturers protocol. RNA was eluted in 30 uL of nuclease-free water.
5 μg of RNA was DNase-treated using Ambion TURBO DNA-Free kit (cat no. AM1907). RNA was cleaned using 1.8× RNA Ampure beads and eluted in 20 uL of nuclease-free water. RNA was then run on the Agilent Bioanalyzer using an RNA Pico chip to check RNA integrity, and the concentration was measured by Qubit of which RIN values were all >7.
2 μg of RNA was RIbozero-depleted using the Illumina RIbozero Plant Leaf kit, and eluted in 12 uL. Ribozero-depleted RNA was, then, ran on the Agilent Bioanalyzer using an RNA pico chip. RNA was, then, used as input material for the Illumina ScriptSeq library preparation. After 13 cycles of PCR, the library was eluted in 20 uL of nuclease-free water. Finally, libraries were run on the Fragment Analyzer using the NGS High Sensitivity kit.
Metabolite extraction
100 mg of the frozen plant tissue was homogenized in 2-mL Eppendorf tubes in a Retschmill. The metabolites were extracted from each aliquot in 1 mL of a homogenous mixture of −20 °C methanol: methyl-tert-butyl-ether: water (1:3:1), with shaking for 30 min at 4 °C, followed by another 10 min of incubation in an ice-cooled ultrasonication bath. Then, 650 μL of UPLC-grade methanol: water (1:3) was added, and the homogenate was vortexed and spun for 5 min at 4 °C in a table-top centrifuge. The addition of methanol: water leads to a phase separation, providing the upper organic phase, containing the lipids, a lower aqueous phase of which we focused on the aqueous metabolites. Metabolites were analysed by the University of Glasgow Polyomics facility using Orbitrap™ Q Exactive™ mass spectrometer with UltiMate™ 3000 RSLCnano separation system at the University of Glasgow Polyomics Facility and the resulting spectra analysed using the IDEOM software package.
RNA sequencing
A number of 48 RNA samples were sequenced across three lanes of a HISEQ 2500 using 2 × 150 bp chemistry. The raw Fastq files were trimmed for the presence of Illumina adapter sequences using Cutadapt version 1.2. [50]. The option -O 3 was used, so the 3′ end of any reads, which match the adapter sequence for ≥ 3 bp, were trimmed. The reads were further trimmed using Sickle version 1.200 with a minimum window quality score of 20. Reads shorter than 10 bp after trimming were removed.
Bioinformatic analysis
De novo assembly of the sequence reads was carried out using Trinity [51], which was also used to generate per-gene read counts. Further analysis was carried out in EdgeR [52] and DEseq. Tagwise dispersions were used to calculate differentially expressed genes with a false discovery rate of ≤ 0.05. Clustering was carried out using Cluster software package (version 3.0) and analysed using Heatmapper [53]. GO enrichment was performed using TopGO (https://bioconductor.org/packages/release/bioc/html/topGO.html) enriched terms were assigned weighted P-values using Fishers exact test.
Additional files
Table S1. Number of sequence reads generated for each sample. Table S2. List of metabolite signals matched to known standards. (DOCX 100 kb)
Annotated regulated genes inferred from the RNAseq data. Description of the data: Genes identified as significantly regulated in PEG treated plants relative to control plants and genes identified as significantly regulated in Salt treated plants relative to control plants. For each gene (rows) normalized read counts and the resulting statistical analysis in EdgeR is presented. Also BLAST2GO annotations are given. (XLSX 4617 kb)
Venn Diagrams showing the number shared and unique up and down regulated genes in each treatment at each time point. Up-regulated genes are on the left and down-regulated genes on the right. Blue circles denote the genes in the NaCl treated plants and red circles denote the genes in the PEG treated plants. (PDF 29 kb)
Enriched Biological GO terms in different treatments and time points. Table S4 Enriched Molecular Function GO terms in different treatments and time points. (DOCX 130 kb)
Acknowledgements
The research team express its gratitude and appreciation to King Abdulaziz City for Science and Technology (KACST) for providing the research grant no. AT-35-9. The research team also expresses its thanks to Deanship of Scientific Research (DSR) and Department of Biological Sciences, Faculty of Science, King Abdulaziz University, Jeddah, for providing services and assistance during the course of the project. NH is supported by the BBSRC, Core Capability Grant BB/CCG1720/1 to the Earlham Institute.”
Funding
The research team express its gratitude and appreciation to King Abdulaziz City for Science and Technology (KACST) for providing the research grant no. T-35-9.
The research team also expresses its thanks to Deanship of Scientific Research (DSR) and Department of Biological Sciences, Faculty of Science, King Abdulaziz University, Jeddah, for providing services and assistance during the course of the project.
Availability of data and materials
All sequence data generated is available in EBI under study accession PRJEB21949.
Abbreviations
- ABA
Abscisic acid
- BLAST
Basic Local Alignment Search Tool
- bp
Base pair
- D
Days
- DAG
Diacylglycerol
- DNA
Deoxyribonucleic acid
- GO
Gene Ontology
- h
Hours
- IP3
Inositol 1,4,5-trisphosphate
- Iron-Sulphur
Fe-S
- mg
milligram
- NaCl
Sodium Chloride
- PCR
Polymerase Chain Reaction
- PEG
Polyethylene glycol
- RNA
Ribonucleic acid
- ug
Microgram
- uL
Microliter
Authors’ contributions
The project was designed and conceived by MZM, JSMS, AB, RKJ and NH. The experimental work was carried out by NHH, AA, MJSMS, CN, RMM, HMA, FME, YG. ASMA. NH, JSMS, AA, YG, HSA, MJS, AKA and AB analysed the data. JSMS, MZM AB, wrote the paper. All authors have read and approved the final version of this manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12870-017-1155-7) contains supplementary material, which is available to authorized users.
Contributor Information
Mohammed Z. Mutwakil, Email: mmutwakil@kau.edu.sa
Nahid H. Hajrah, Email: nhajrah260@gmail.com
Ahmed Atef, Email: ahmed_atefaig2@yahoo.com.
Sherif Edris, Email: sedris@aucegypt.edu.
Mernan J. Sabir, Email: mernan.j.s@hotmail.com
Areej K. Al-Ghamdi, Email: Kalghamdy@kau.edu.sa
Meshaal J. S. M. Sabir, Email: msabir999@gmail.com
Charlotte Nelson, Email: Charlotte.Nelson@liverpool.ac.uk.
Rania M. Makki, Email: rmakki@kau.edu.sa
Hani M. Ali, Email: hmohammedali@kau.edu.sa
Fotouh M. El-Domyati, Email: fm_domyati@hotmail.com
Abdulrahman S. M. Al-Hajar, Email: ahajarr@gmail.com
Yoann Gloaguen, Email: Yoann.Gloaguen@glasgow.ac.uk.
Hassan S. Al-Zahrani, Email: hsazahrani@kau.edu.sa
Jamal S. M. Sabir, Email: jsabir2622@gmail.com
Robert K. Jansen, Email: jansen@austin.utexas.edu
Ahmed Bahieldin, Email: bahieldin55@gmail.com.
Neil Hall, Email: Neil.Hall@earlham.ac.uk.
References
- 1.Greiss E. Anatomical identification of plant remains and other materials from el-Omari excavation at Helwan from the first dynasty. Bull Inst Egypt. 1955;36:227–235. [Google Scholar]
- 2.Ebbell B. A contribution to the earliest history of leprosy. Int J Leprosy. 1935;3:257–263. [Google Scholar]
- 3.Shivkar YM, Kumar VL. Anthelmintic activity of latex of Calotropis Procera. Pharm Biol. 2008;41(4):263–265. doi: 10.1076/phbi.41.4.263.15666. [DOI] [Google Scholar]
- 4.Parhira S, Yang ZF, Zhu GY, Chen QL, Zhou BX, Wang YT, Liu L, Bai LP, Jiang ZH. In Vitro anti-influenza virus activities of a new Lignan glycoside from the latex of Calotropis Gigantea. PLoS One. 2014;9(8) [DOI] [PMC free article] [PubMed]
- 5.Aziz MA, Khan AH, Adnan M, Izatullah I. Traditional uses of medicinal plants reported by the indigenous communities and local herbal practitioners of Bajaur agency, federally administrated tribal areas, Pakistan. J Ethnopharmacol. 2017;198:268–281. doi: 10.1016/j.jep.2017.01.024. [DOI] [PubMed] [Google Scholar]
- 6.Kumar VL, Padhy BM. Protective effect of aqueous suspension of dried latex of Calotropis Procera against oxidative stress and renal damage in diabetic rats. Biocell. 2011;35(3):63–69. [PubMed] [Google Scholar]
- 7.Oliveira JS, Costa-Lotufo LV, Bezerra DP, Alencar NM, Marinho-Filho JD, Figueiredo IS, Moraes MO, Pessoa C, Alves AP, Ramos MV. In vivo growth inhibition of sarcoma 180 by latex proteins from Calotropis Procera. Naunyn Schmiedeberg's Arch Pharmacol. 2010;382(2):139–149. doi: 10.1007/s00210-010-0525-6. [DOI] [PubMed] [Google Scholar]
- 8.Jain SC, Sharma R, Kain R, SHarma RA. Antimicrobial activity of Calotropis Procera. Fitoterapia. 1996;67:275–277. [Google Scholar]
- 9.Iqbal Z, Lateef M, Jabbar A, Muhammad G, Khan MN. Anthelmintic activity of Calotropis Procera (Ait.) Ait. F. Flowers in sheep. J Ethnopharmacol. 2005;102(2):256–261. doi: 10.1016/j.jep.2005.06.022. [DOI] [PubMed] [Google Scholar]
- 10.Ali NO, El-Rabaa FM. Larvicidal activity of some plant extracts to larvae of the mosquito Culex Quinquefasciatus (say 1823) Eur Rev Med Pharmacol Sci. 2010;14(11):925–933. [PubMed] [Google Scholar]
- 11.Bansal SK, Singh KV, Sharma S, Sherwani MR. Laboratory observations on the larvicidal efficacy of three plant species against mosquito vectors of malaria, dengue/dengue hemorrhagic fever (DF/DHF) and lymphatic filariasis in the semi-arid desert. J Environ Biol. 2012;33(3):617–621. [PubMed] [Google Scholar]
- 12.Elimam AM, Elmalik KH, Ali FS. Efficacy of leaves extract of Calotropis Procera Ait. (Asclepiadaceae) in controlling anopheles arabiensis and Culex Quinquefasciatus mosquitoes. Saudi J Biol Sci. 2009;16(2):95–100. doi: 10.1016/j.sjbs.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tezara W, Colombo R, Coronel I, Marin O. Water relations and photosynthetic capacity of two species of Calotropis in a tropical semi-arid ecosystem. Ann Bot-London. 2011;107(3):397–405. doi: 10.1093/aob/mcq245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shokry AM, Al-Karim S, Ramadan A, Gadallah N, Al Attas SG, Sabir JSM, Hassan SM, Madkour MA, Bressan R, Mahfouz M, et al. Detection of a Usp-like.Gene in Calotropis Procera plant from the de novo assembled genome contigs of the high-throughput sequencing dataset. Cr Biol. 2014;337(2):86–94. doi: 10.1016/j.crvi.2013.12.008. [DOI] [PubMed] [Google Scholar]
- 15.Pandey A, Swarnkar V, Pandey T, Srivastava P, Kanojiya S, Mishra DK, Tripathi V. Transcriptome and metabolite analysis reveal candidate genes of the cardiac glycoside biosynthetic pathway from Calotropis Procera. Sci Rep-Uk. 2016;6 [DOI] [PMC free article] [PubMed]
- 16.Ramadan A, Sabir JSM, Alakilli SYM, Shokry AM, Gadalla NO, Edris S, Al-Kordy MA, Al-Zahrani HS, El-Domyati FM, Bahieldin A, et al. Metabolomic response of Calotropis Procera growing in the desert to changes in water availability. PLoS One. 2014;9(2) [DOI] [PMC free article] [PubMed]
- 17.Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol. 2010;61:651–679. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- 18.Raghavendra AS, Gonugunta VK, Christmann A, Grill E. ABA perception and signalling. Trends Plant Sci. 2010;15(7):395–401. doi: 10.1016/j.tplants.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 19.Hey SJ, Byrne E, Halford NG. The interface between metabolic and stress signalling. Ann Bot. 2010;105:197–203. doi: 10.1093/aob/mcp285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Darwish E, Testerink C, Khalil M, El-Shihy O, Munnik T. Phospholipid signaling responses in salt-stressed rice leaves. Plant Cell Physiol. 2009;50(5):986–997. doi: 10.1093/pcp/pcp051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lemtiri-Chlieh F, MacRobbie EA, Webb AA, Manison NF, Brownlee C, Skepper JN, Chen J, Prestwich GD, Brearley CA. Inositol hexakisphosphate mobilizes an endomembrane store of calcium in guard cells. Proc Natl Acad Sci U S A. 2003;100(17):10091–10095. doi: 10.1073/pnas.1133289100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21(18):3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 23.Nir I, Moshelion M, Weiss D. The Arabidopsis gibberellin methyl transferase 1 suppresses gibberellin activity, reduces whole-plant transpiration and promotes drought tolerance in transgenic tomato. Plant Cell Environ. 2014;37(1):113–123. doi: 10.1111/pce.12135. [DOI] [PubMed] [Google Scholar]
- 24.Li W, Lu J, Lu K, Yuan J, Huang J, Du H, Li J. Cloning and phylogenetic analysis of Brassica Napus L. Caffeic acid O-Methyltransferase 1 gene family and its expression pattern under drought stress. PLoS One. 2016;11(11):e0165975. doi: 10.1371/journal.pone.0165975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuluev B, Mikhaylova E, Berezhneva Z, Nikonorov Y, Postrigan B, Kudoyarova G, Chemeris A. Expression profiles and hormonal regulation of tobacco NtEXGT gene and its involvement in abiotic stress response. Plant Physiol Biochem. 2017;111:203–215. doi: 10.1016/j.plaphy.2016.12.005. [DOI] [PubMed] [Google Scholar]
- 26.Tenhaken R. Cell wall remodeling under abiotic stress. Front Plant Sci. 2014;5:771. doi: 10.3389/fpls.2014.00771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou S, Jia L, Chu H, Wu D, Peng X, Liu X, Zhang J, Zhao J, Chen K, Zhao L. Arabidopsis CaM1 and CaM4 promote nitric oxide production and salt resistance by inhibiting S-Nitrosoglutathione Reductase via direct binding. PLoS Genet. 2016;12(9):e1006255. doi: 10.1371/journal.pgen.1006255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xuan Y, Zhou S, Wang L, Cheng Y, Zhao L. Nitric oxide functions as a signal and acts upstream of AtCaM3 in thermotolerance in Arabidopsis seedlings. Plant Physiol. 2010;153(4):1895–1906. doi: 10.1104/pp.110.160424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim YY, Jung KW, Yoo KS, Jeung JU, Shin JS. A stress-responsive caleosin-like protein, AtCLO4, acts as a negative regulator of ABA responses in Arabidopsis. Plant Cell Physiol. 2011;52(5):874–884. doi: 10.1093/pcp/pcr039. [DOI] [PubMed] [Google Scholar]
- 30.Turlapati PV, Kim KW, Davin LB, Lewis NG. The laccase multigene family in Arabidopsis Thaliana: towards addressing the mystery of their gene function(s) Planta. 2011;233(3):439–470. doi: 10.1007/s00425-010-1298-3. [DOI] [PubMed] [Google Scholar]
- 31.Turan S, Tripathy BC. Salt-stress induced modulation of chlorophyll biosynthesis during de-etiolation of rice seedlings. Physiol Plant. 2015;153(3):477–491. doi: 10.1111/ppl.12250. [DOI] [PubMed] [Google Scholar]
- 32.Taneja M, Tyagi S, Sharma S, Upadhyay SK. Ca2+/Cation Antiporters (CaCA): identification, characterization and expression profiling in bread wheat (Triticum Aestivum L) Front Plant Sci. 2016;7:1775. doi: 10.3389/fpls.2016.01775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma Y, Wang J, Zhong Y, Geng F, Cramer GR, Cheng ZM. Subfunctionalization of cation/proton antiporter 1 genes in grapevine in response to salt stress in different organs. Hortic Res. 2015;2:15031. doi: 10.1038/hortres.2015.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barragan V, Leidi EO, Andres Z, Rubio L, De Luca A, Fernandez JA, Cubero B, Pardo JM. Ion exchangers NHX1 and NHX2 mediate active potassium uptake into vacuoles to regulate cell turgor and stomatal function in Arabidopsis. Plant Cell. 2012;24(3):1127–1142. doi: 10.1105/tpc.111.095273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma YC, Auge RM, Dong C, Cheng ZM. Increased salt tolerance with overexpression of cation/proton antiporter 1 genes: a meta-analysis. Plant Biotechnol J. 2017;15(2):162–173. doi: 10.1111/pbi.12599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zwack PJ, Rashotte AM. Interactions between cytokinin signalling and abiotic stress responses. J Exp Bot. 2015;66(16):4863–4871. doi: 10.1093/jxb/erv172. [DOI] [PubMed] [Google Scholar]
- 37.Zhang X, Chen Y, Lin X, Hong X, Zhu Y, Li W, He W, An F, Guo H. Adenine phosphoribosyl transferase 1 is a key enzyme catalyzing cytokinin conversion from nucleobases to nucleotides in Arabidopsis. Mol Plant. 2013;6(5):1661–1672. doi: 10.1093/mp/sst071. [DOI] [PubMed] [Google Scholar]
- 38.Nishiyama R, Watanabe Y, Fujita Y, Le DT, Kojima M, Werner T, Vankova R, Yamaguchi-Shinozaki K, Shinozaki K, Kakimoto T, et al. Analysis of cytokinin mutants and regulation of cytokinin metabolic genes reveals important regulatory roles of cytokinins in drought, salt and abscisic acid responses, and abscisic acid biosynthesis. Plant Cell. 2011;23(6):2169–2183. doi: 10.1105/tpc.111.087395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nishiyama R, Le DT, Watanabe Y, Matsui A, Tanaka M, Seki M, Yamaguchi-Shinozaki K, Shinozaki K, Tran LS. Transcriptome analyses of a salt-tolerant cytokinin-deficient mutant reveal differential regulation of salt stress response by cytokinin deficiency. PLoS One. 2012;7(2):e32124. doi: 10.1371/journal.pone.0032124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lannoo N, Van Damme EJ: Nucleocytoplasmic plant lectins. Biochim Biophys Acta 2010, 1800(2):190–201. [DOI] [PubMed]
- 41.Van Hove J, Stefanowicz K, De Schutter K, Eggermont L, Lannoo N, Al Atalah B, Van Damme EJ. Transcriptional profiling of the lectin ArathEULS3 from Arabidopsis Thaliana toward abiotic stresses. J Plant Physiol. 2014;171(18):1763–1773. doi: 10.1016/j.jplph.2014.08.009. [DOI] [PubMed] [Google Scholar]
- 42.DeWald DB, Torabinejad J, Jones CA, Shope JC, Cangelosi AR, Thompson JE, Prestwich GD, Hama H. Rapid accumulation of phosphatidylinositol 4,5-bisphosphate and inositol 1,4,5-trisphosphate correlates with calcium mobilization in salt-stressed arabidopsis. Plant Physiol. 2001;126(2):759–769. doi: 10.1104/pp.126.2.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu JK. Salt and drought stress signal transduction in plants. Annu Rev Plant Biol. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liang X, Qin L, Liu P, Wang M, Ye H. Genes for iron-sulphur cluster assembly are targets of abiotic stress in rice, Oryza Sativa. Plant Cell Environ. 2014;37(3):780–794. doi: 10.1111/pce.12198. [DOI] [PubMed] [Google Scholar]
- 45.Rouhier N. Plant glutaredoxins: pivotal players in redox biology and iron-sulphur centre assembly. New Phytol. 2010;186(2):365–372. doi: 10.1111/j.1469-8137.2009.03146.x. [DOI] [PubMed] [Google Scholar]
- 46.Lorence A, Chevone BI, Mendes P, Nessler CL. Myo-inositol oxygenase offers a possible entry point into plant ascorbate biosynthesis. Plant Physiol. 2004;134(3):1200–1205. doi: 10.1104/pp.103.033936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Endres S, Tenhaken R. Myoinositol oxygenase controls the level of myoinositol in Arabidopsis, but does not increase ascorbic acid. Plant Physiol. 2009;149(2):1042–1049. doi: 10.1104/pp.108.130948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bartels D, Ramanjulu S. Drought and salt tolerance in plants. Crit Rev Plant Sci. 2005;24(1):23–50. doi: 10.1080/07352680590910410. [DOI] [Google Scholar]
- 49.Watson R: Polyphenols in plants: isolation, Purification and Extract Preparation; 2014.
- 50.Chen C, Khaleel SS, Huang H, CH W. Software for pre-processing Illumina next-generation sequencing short read sequences. Source Code Biol Med. 2014;9:8. doi: 10.1186/1751-0473-9-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, et al. De novo transcript sequence reconstruction from RNA-seq using the trinity platform for reference generation and analysis. Nat Protoc. 2013;8(8):1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Babicki S, Arndt D, Marcu A, Liang Y, Grant JR, Maciejewski A, Wishart DS. Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res. 2016;44(W1):W147–W153. doi: 10.1093/nar/gkw419. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Number of sequence reads generated for each sample. Table S2. List of metabolite signals matched to known standards. (DOCX 100 kb)
Annotated regulated genes inferred from the RNAseq data. Description of the data: Genes identified as significantly regulated in PEG treated plants relative to control plants and genes identified as significantly regulated in Salt treated plants relative to control plants. For each gene (rows) normalized read counts and the resulting statistical analysis in EdgeR is presented. Also BLAST2GO annotations are given. (XLSX 4617 kb)
Venn Diagrams showing the number shared and unique up and down regulated genes in each treatment at each time point. Up-regulated genes are on the left and down-regulated genes on the right. Blue circles denote the genes in the NaCl treated plants and red circles denote the genes in the PEG treated plants. (PDF 29 kb)
Enriched Biological GO terms in different treatments and time points. Table S4 Enriched Molecular Function GO terms in different treatments and time points. (DOCX 130 kb)
Data Availability Statement
All sequence data generated is available in EBI under study accession PRJEB21949.


