Assembly of the dynamin GTPase Drp1 into constriction-competent oligomers is a key event in mitochondrial division. Here, Ji et al. show that Drp1 oligomerization can occur on endoplasmic reticulum through an ER-bound population of the tail-anchored protein Mff.
Abstract
Drp1 is a dynamin guanosine triphosphatase important for mitochondrial and peroxisomal division. Drp1 oligomerization and mitochondrial recruitment are regulated by multiple factors, including interaction with mitochondrial receptors such as Mff, MiD49, MiD51, and Fis. In addition, both endoplasmic reticulum (ER) and actin filaments play positive roles in mitochondrial division, but mechanisms for their roles are poorly defined. Here, we find that a population of Drp1 oligomers is associated with ER in mammalian cells and is distinct from mitochondrial or peroxisomal Drp1 populations. Subpopulations of Mff and Fis1, which are tail-anchored proteins, also localize to ER. Drp1 oligomers assemble on ER, from which they can transfer to mitochondria. Suppression of Mff or inhibition of actin polymerization through the formin INF2 significantly reduces all Drp1 oligomer populations (mitochondrial, peroxisomal, and ER bound) and mitochondrial division, whereas Mff targeting to ER has a stimulatory effect on division. Our results suggest that ER can function as a platform for Drp1 oligomerization, and that ER-associated Drp1 contributes to mitochondrial division.
Introduction
Mitochondrial division plays an important role in many cellular processes, facilitating appropriate mitochondrial nucleoid distribution (Lewis et al., 2016), allowing cells to respond to changing metabolic needs (Hatch et al., 2014; Labbé et al., 2014; Mishra and Chan, 2016; Pernas and Scorrano, 2016), and contributing to selective autophagy of damaged mitochondria (Youle and van der Bliek, 2012). Defects in mitochondrial division have been linked to multiple diseases (Nunnari and Suomalainen, 2012; Vafai and Mootha, 2012; DuBoff et al., 2013).
A key component of mitochondrial division is the dynamin family GTPase Drp1. Drp1 is a cytosolic protein that is recruited to the outer mitochondrial membrane (OMM), where it oligomerizes into a spiral around the OMM (Bui and Shaw, 2013). GTP hydrolysis results in Drp1 spiral constriction, providing a driving force for mitochondrial division. Subsequent recruitment of a second dynamin GTPase, dynamin 2, appears necessary for complete membrane division (Lee et al., 2016).
Several features suggest that mitochondrial Drp1 recruitment is a multistep and finely tuned process in mammals. First, mitochondrial division occurs preferentially at contact sites with ER, suggesting that ER contributes components or signaling information to the process (Friedman et al., 2011). Second, Drp1 recruitment to mitochondria is not an all-or-none phenomenon, but rather an equilibrium process in which Drp1 oligomers dynamically assemble on mitochondria independently of signals for mitochondrial division (Ji et al., 2015). A variety of division signals may push Drp1’s ongoing equilibrium toward productive oligomerization on mitochondria, including ER–mitochondrial contact, activated receptors on the OMM, cardiolipin enrichment on the OMM (Bustillo-Zabalbeitia et al., 2014; Macdonald et al., 2014), and modification of Drp1 itself (Chang and Blackstone, 2007, 2010; Cribbs and Strack, 2007; Friedman et al., 2011; Toyama et al., 2016). Another division signal is actin polymerization mediated by the ER-bound formin protein INF2, which stimulates division by shifting the Drp1 oligomerization equilibrium toward productive oligomerization on mitochondria (Korobova et al., 2013, 2014; Ji et al., 2015). Actin’s stimulatory effect may be through direct interaction with Drp1 (Ji et al., 2015; Hatch et al., 2016). Third, there are multiple Drp1 receptors on the OMM in mammals, suggesting two possibilities: (1) there are parallel pathways for Drp1 recruitment, each mediated by one of these receptors, or (2) these receptors act in a common pathway.
Protein receptors for Drp1 are necessary because, unlike other dynamin family members, Drp1 does not contain a specific lipid-binding domain. Four single-pass OMM proteins have been identified as Drp1 receptors in mammals: Mff, Fis1, MiD49, and MiD51 (Richter et al., 2015). Mff and Fis1 are tail-anchored (TA) proteins that are also found on peroxisomes, another organelle that undergoes Drp1-dependent division (Koch and Brocard, 2012; Schrader et al., 2016). In contrast, MiD49 and MiD51 contain N-terminal transmembrane domains and appear to be restricted to mitochondria (Palmer et al., 2013). Our database searches suggest that MiD49 and MiD51 are present only in vertebrates, whereas Mff is found in higher metazoans (coelomates, including arthropods and mollusks but not Caenorhabditis elegans), and Fis1 is expressed in all eukaryotes examined. Mff has consistently been found to be a key Drp1 receptor in mammals, whereas MiD49 and MiD51 are important in specific situations (Losón et al., 2013; Shen et al., 2014; Osellame et al., 2016; Otera et al., 2016). Though Fis1 is the sole known Drp1 receptor in budding yeast, its role in mammals is unclear (Losón et al., 2013; Shen et al., 2014; Richter et al., 2015; Osellame et al., 2016; Otera et al., 2016).
In this study, we examine Drp1 distribution among organelles in mammalian cells. Surprisingly, we find that Drp1 oligomers exist on ER, independent of mitochondrial or peroxisomal association. Populations of both Mff and Fis1 also exist on ER as punctate accumulations. Mff suppression or actin polymerization inhibition eliminates all detectable Drp1 oligomers, including the ER-bound population. We observe Drp1 accumulation at ER-bound Mff punctae, suggesting oligomeric assembly at these sites. Drp1 oligomers can transfer from ER to mitochondria or peroxisomes. Our results suggest a pathway for Drp1 oligomerization on mitochondria involving initial assembly on ER, which is dependent on both Mff and actin.
Results
A subpopulation of oligomeric Drp1 is bound to ER
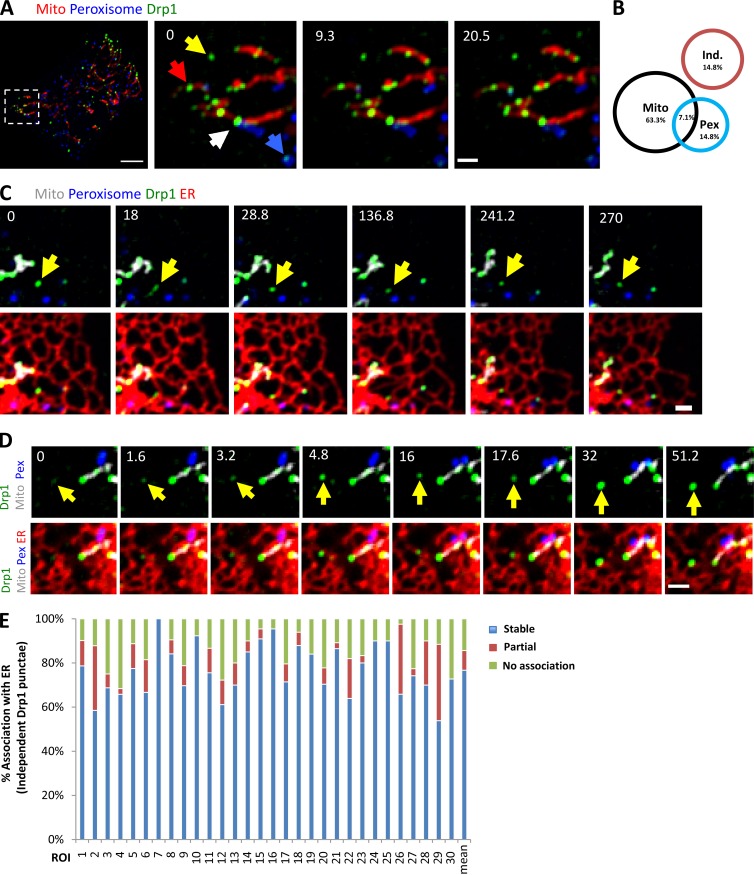
Previously, we used an U2OS cell line stably expressing GFP-Drp1 to show that the majority (∼70%) of large Drp1 “punctae” associate with mitochondria (Ji et al., 2015). These punctae likely represent Drp1 oligomers, which are clearly visible after removing the background GFP signal. To examine Drp1 localization and dynamics in more detail, we developed a GFP-Drp1 CRISPR knock-in U2OS line (called GFP-Drp1-KI) in which ∼50% of the endogenous Drp1 is GFP-tagged and the overall Drp1 level is similar to that of control cells (Fig. S1, A and B). This cell line displays cell growth kinetics similar to WT cells (Fig. S1 C) and a percentage of mitochondrially associated Drp1 punctae (63%) similar to the stably transfected GFP-Drp1 cell line (Fig. 1, A and B).
Figure 1.
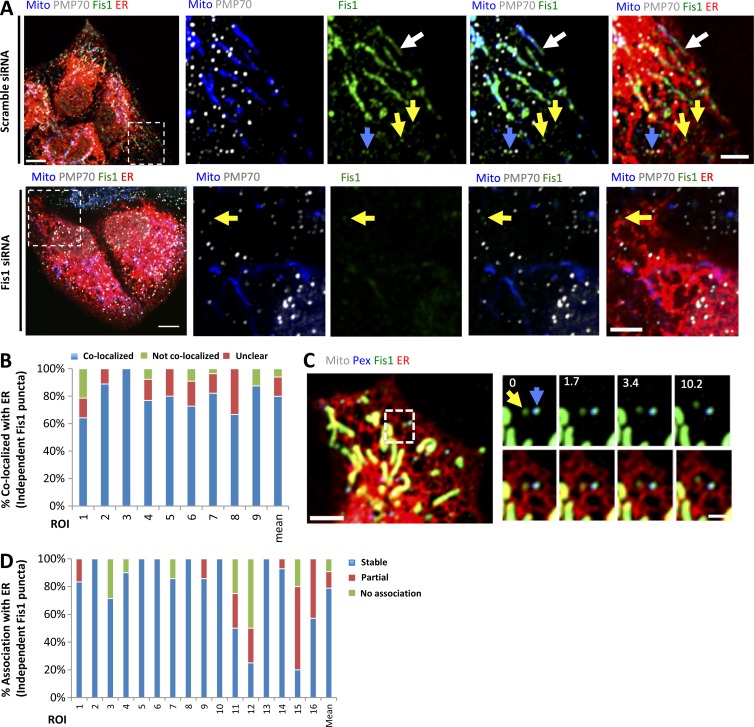
A population of Drp1 associates with ER independently of mitochondria or peroxisomes. (A) Drp1 distribution in GFP-Drp1-KI cells. Left: Merged image of a live Drp1 KI cell transiently expressing mCherry-mito7 (red) and eBFP2-peroxisome (blue). Drp1 in green. Right: Insets from boxed region at three time points. Yellow arrow, independent Drp1 puncta; blue arrow, peroxisome-associated Drp1; red arrow, mitochondrially associated Drp1; white arrowhead, example of Drp1 puncta localizing at the interface of mitochondrion and peroxisome. (B) Venn diagram of Drp1 distribution in GFP-Drp1-KI cells expressing mitochondrial and peroxisomal markers. Black circles, mitochondrially associated Drp1 punctae; blue circles, peroxisomal associated Drp1 punctae (Pex); red circles, independent Drp1 punctae (Ind). The percentage of Drp1 punctae in each category is the mean from 10 consecutive frames with 12-s time intervals from whole-cell videos. Five cells measured (10,761 punctae). (C) Four-color imaging of a live Drp1 KI cell expressing mPlum-mito3 (Mito, gray); eBFP2-peroxisome (Peroxisome, blue); and ER-tagRFP (ER, red); Drp1 in green. Yellow arrows denote independent Drp1 puncta stably associating with ER. See also Video 1. (D) Time-lapse montage showing de novo assembly of an independent Drp1 punctum (yellow arrow) on an ER tubule. Imaging as in C. (E) Graph depicting the degree of association between independent Drp1 punctae and ER during 2.5-min videos imaged every 1.6 s. 30 ROIs from 25 GFP-Drp1-KI cells analyzed (1,003 punctae). Mean values from ROIs: 76.7 ± 11.7%, stable association between Drp1 punctae and ER (no apparent dissociation from ER in any frame); 8.9 ± 9.5%, partial association; 14.4 ± 8.0%, no association. Bars: (A, whole-cell image) 10 µm; (A, inset; C; and D) 2 µm. Time in seconds.
We examined the non–mitochondrially associated Drp1 punctae in more detail, postulating that they would be bound to peroxisomes. Surprisingly, although some of these punctae are peroxisome associated, an equal percentage (14.8%) is not associated with either mitochondria or peroxisomes, which we defined as “independent” Drp1 punctae (Fig. 1, A and B). The remaining punctae (7%) localize to areas of close association between mitochondria and peroxisomes.
We postulated that the independent population might be bound to ER. Indeed, four-color live-cell imaging shows that a population of Drp1 punctae appears to be associated with ER, distinct from mitochondrial or peroxisomal populations (Fig. 1 C and Video 1). Independent Drp1 puncta can arise de novo from ER, maturing within 30 s (Fig. 1 D).
We quantified ER association of independent Drp1 punctae from time-lapse confocal videos, assessing stably associating punctae as those that do not separate from ER during the 2.5-min imaging time (1.6-s frame rate). Although ER occupies a significant portion of the imaging area in these cells (40.9 ± 5.9%, 22 regions of interest [ROIs], and 2,063 individual frames analyzed), there is a significantly higher percentage of independent Drp1 puncta in continual association with ER than would be expected by chance (76.7 ± 11.7%; Fig. 1 E). Other independent Drp1 punctae are associated with ER for a portion of the imaging period (8.9 ± 9.5%), with most only separating for one frame. A third population of independent Drp1 punctae displays no apparent association with ER (14.4 ± 8.0%). Cos7 cells transiently transfected with GFP-Drp1 also display ER-associated Drp1 punctae, independent of either mitochondria or peroxisomes (Fig. S1, D and E). In contrast, independent Drp1 punctae do not display appreciable association with endosomes, as judged by transferrin, Rab4b, and Rab7a markers (Fig. S2).
One possible explanation for independent Drp1 punctae is that they are actually bound to mitochondrially derived vesicles (MDVs) that bud from the OMM (Soubannier et al., 2012). We tested this possibility by imaging GFP-Drp1 and the OMM protein Tom20 in live cells. No overlapping Tom20-only signal is detectable at any time point in videos (4 min, 2-s intervals) for 15 of 16 independent Drp1 punctae analyzed (Fig. S3). These results suggest that the majority of independent Drp1 punctae are not bound to MDVs.
Another explanation for the existence of independent Drp1 punctae could be that they represent unfolded protein aggregates. Indeed, studies in yeast and mammals show that protein aggregates can accumulate on ER, followed by transfer to mitochondria for degradation in the mitochondrial matrix (Zhou et al., 2014; Ruan et al., 2017). Although GFP-Drp1 is not overexpressed in our CRISPR-engineered cell line (Fig. S1, A–C), the GFP tag or other features of this fusion protein could result in unfolding/aggregation. To test this possibility, we examined the distribution of endogenous Drp1 punctae in relation to mitochondria, peroxisomes, and ER by immunofluorescence microscopy. Similar to GFP-Drp1, a subset of endogenous Drp1 punctae is independent of mitochondria or peroxisomes, and 85.5 ± 9.7% of these independent punctae display apparent ER association (Fig. S4, A and B). To confirm specificity of Drp1 immunofluorescence, siRNA suppression significantly reduces staining of all Drp1 populations (Fig. S4 A).
We examined further the effect of the GFP tag by overexpressing oligomerization-deficient mutants of Drp1 that remain monomeric or dimeric at all concentrations tested biochemically (Fröhlich et al., 2013; Hatch et al., 2016). Despite being expressed at significantly higher levels than WT Drp1 in our GFP-Drp1-KI cells, these Drp1 mutants display no apparent punctae (Fig. S4 C). If the GFP tag or overexpression were causing GFP-Drp1 unfolding and aggregation, the mutants might be expected to display similar properties. We conclude that a mechanism exists for Drp1 oligomer assembly on ER.
Transfer of Drp1 from ER to mitochondria
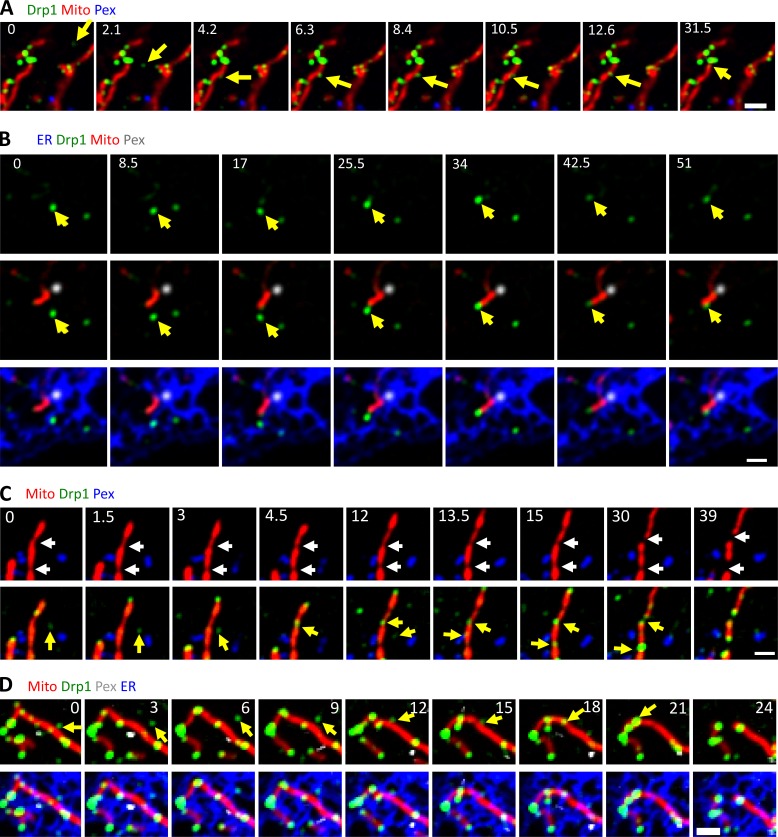
A range of dynamics exists for independent Drp1 punctae, with some puncta displaying little motility over a 5-min period (Fig. 1 C and Video 1) and others displaying periods of rapid directional movement (Fig. 2 A and Video 2). Independent Drp1 punctae can transfer to mitochondria (Fig. 2 A and Video 2), and are ER-associated before transfer (Fig. 2 B and Video 3). We previously reported that most Drp1 oligomerization on mitochondria is nonproductive for mitochondrial division, with only 3% of mitochondrially associated Drp1 punctae resulting in division within a time scale of 10 min (Ji et al., 2015). Similarly, although independent Drp1 punctae can transfer to mitochondria, division rarely occurs after these events. To increase division rate, we treated cells with ionomycin in the presence of serum, which causes a transient fourfold increase in mitochondrial division as well as an increase in Drp1 oligomerization (Ji et al., 2015; Figs. 3 C and S5 B). Upon ionomycin treatment, independent Drp1 puncta transfer to mitochondria followed by division (Fig. 2 C and Video 4), with the puncta maintaining apparent association with ER during the transfer process (Fig. 2 D and Video 5).
Figure 2.
Transfer of Drp1 punctae from ER to mitochondria. (A) Three-color time-lapse images of live GFP-Drp1-KI cell expressing mCherry-mito7 (mitochondria, red), eBFP2-peroxisome (peroxisome, blue), and Drp1 in green. An independent Drp1 puncta (yellow arrow) transfers to a mitochondrion and then translocates along the mitochondrion with no division in the observation time period. See also Video 2. (B) Four-color time-lapse images of live GFP-Drp1-KI cell expressing mito-BFP (mitochondria, gray), mPlum-peroxisome (peroxisome, blue), ER-tagRFP (ER, red) and GFP-Drp1 in green. Yellow arrow denotes an ER-bound Drp1 puncta transferring to mitochondrion. See also Video 3. (C) Three-color time-lapse images of live GFP-Drp1-KI cell expressing mCherry-mito7 (mitochondria, red), eBFP2-peroxisome (peroxisome, blue), and Drp1 in green. Two Drp1 punctae transfer to constriction sites, followed by division. Cells treated with ionomycin (4 µM) to stimulate mitochondrial division. See also Video 4. (D) Four-color time-lapse images of live GFP-Drp1-KI cell expressing mito-BFP (mitochondria, red), mPlum-PMP20 (peroxisome, gray), ER-tagRFP (ER in blue) and GFP-Drp1 in green. Yellow arrow denotes an independent Drp1 puncta transferring to mitochondrion. Cells treated with ionomycin (4 µM) to stimulate mitochondrial division. See also Video 5. Bars, 2 µm. Time in seconds.
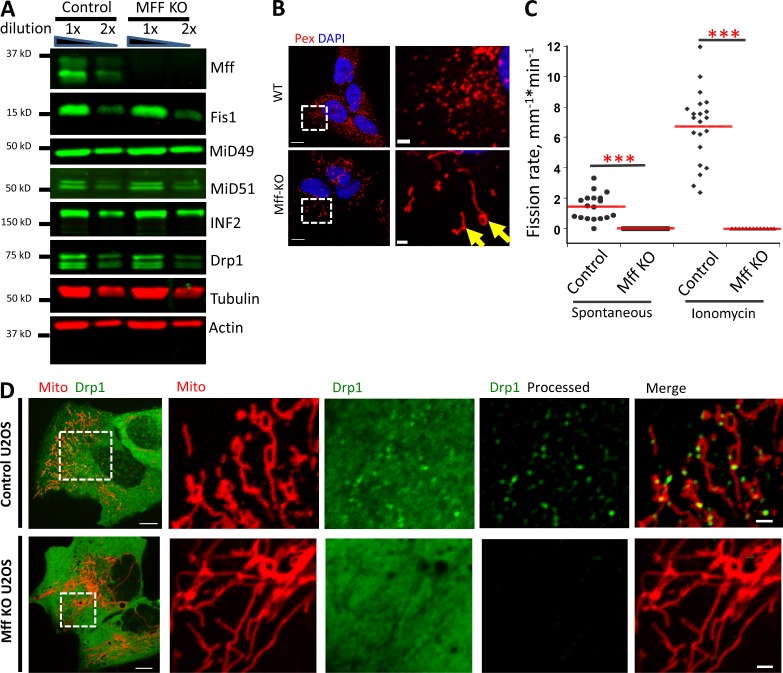
Figure 3.
Mff KO U2OS cells are deficient in mitochondrial and peroxisomal division. (A) Western blotting for Mff and other mitochondrial division proteins in control and Mff KO U2OS cells. (B) Immunofluorescence of fixed cells stained for peroxisomes (red) and DNA (DAPI, blue). Images on right are zoomed regions. Yellow arrows indicate elongated peroxisomes in Mff KO cells. (C) Division rate quantification for both control and Mff KO U2OS cells. For the quantification of spontaneous division rate, 18 ROIs analyzed for either 12 (control) or 14 (Mff KO) cells. For quantification of ionomycin-induced division rate, 21 ROIs (control) and 13 ROIs (Mff KO) were analyzed. ***, P < 0.001 by Student’s t test. (D) Live-cell images of control (top) or Mff KO (bottom) U2OS cells transfected with GFP-Drp1 (green) and mito-RFP (red). Right panels show ROI of selected region (boxed). Raw images shown, except for the rightmost images, which are processed to reveal Drp1 punctae. Bars: (left) 20 µm; (right) 2 µm.
Subpopulations of Mff and Fis1 are ER associated
We postulated that receptors on the ER membrane recruit Drp1 and enhance its oligomerization. Likely candidates for these receptors include proteins involved in mitochondrial Drp1 recruitment: Mff, MiD49, MiD51, and Fis1. There is no published evidence showing ER-bound populations of these proteins.
We first examined Mff, because of its importance for mitochondrial Drp1 recruitment in several studies (Losón et al., 2013; Shen et al., 2014; Osellame et al., 2016; Otera et al., 2016). Mff is a member of the TA family of integral membrane proteins (Gandre-Babbe and van der Bliek, 2008; Otera et al., 2010), with a C-terminal trans-membrane domain that inserts into bilayers posttranslationally. We developed a CRISPR-mediated Mff knockout (KO) cell line that displays no detectable Mff protein but control levels of Drp1, Fis1, MiD49, MiD51, and INF2 (Fig. 3 A). As in past studies, the Mff KO line displays elongated peroxisomes (Fig. 3 B). Mitochondrial division is almost completely eliminated in both unstimulated and ionomycin-stimulated cells (Fig. 3 C). There is also a dramatic reduction in Drp1 punctae (Fig. 3 D). Mff suppression by siRNA causes similar effects, including dramatic inhibition of mitochondrial division in either unstimulated or ionomycin-stimulated cells (Fig. S5, A and B), and near-complete elimination of all Drp1 punctae in GFP-Drp1-KI cells (Figs. S4 C and S5 C). These results show that Mff is a key factor for Drp1 oligomerization in U2OS cells.
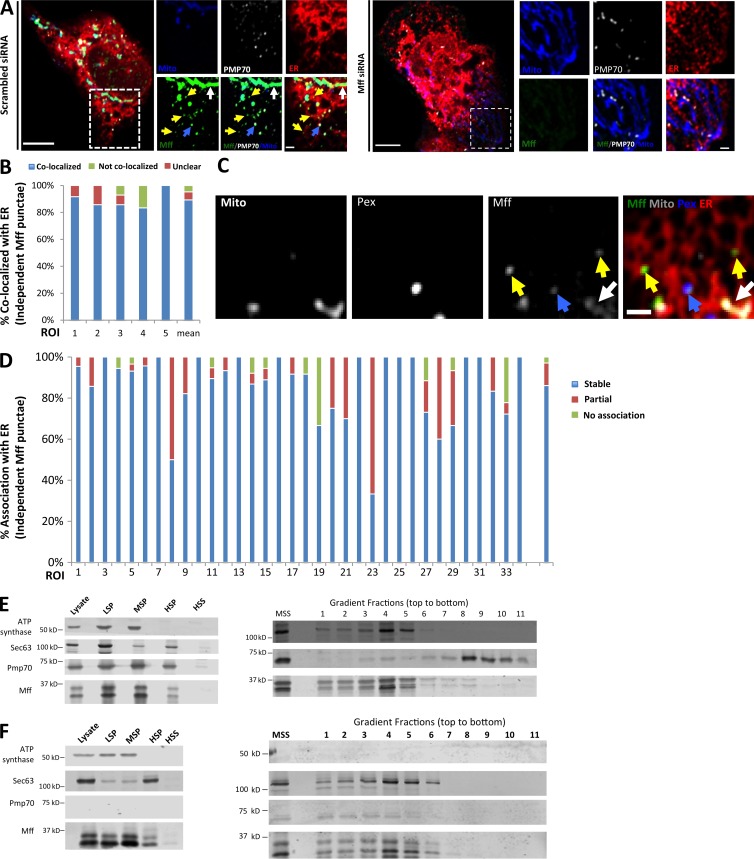
Studies have shown Mff localization on mitochondria and peroxisomes (Gandre-Babbe and van der Bliek, 2008; Otera et al., 2010, 2016; Friedman et al., 2011; Palmer et al., 2013). We asked whether a subpopulation of endogenous Mff was bound to ER. Using immunofluorescence microscopy in U2OS cells, endogenous Mff has a relatively uniform distribution on mitochondria and peroxisomes. In addition, there is a punctate Mff population independent of these organelles, and 89.3 ± 6.7% of these punctae associate with ER (Fig. 4, A and B). This staining is specific for Mff, because Mff knockdown (KD) results in a dramatic reduction in all Mff populations (Fig. 4 A).
Figure 4.
A subpopulation of Mff localizes to ER. (A) Endogenous Mff localization in a fixed U2OS cell by immunofluorescence. Cells labeled with anti-Tom20 (mitochondria, blue), anti-PMP70 (peroxisomes, gray), anti-Mff (green) and transfected with ER-TagRFP (ER, red). Left, scrambled siRNA; right, Mff siRNA. Yellow arrows, independent punctae; blue arrow, peroxisome-associated puncta; white arrow, mitochondrially associated Mff. (B) Graph depicting the percentage of colocalization between independent Mff punctae and ER in U2OS cells (endogenous Mff). 54 independent Mff punctae were counted from five ROIs from four cells. Mean values from ROIs: 89.3 ± 6.7%, colocalized Mff with ER; 6.0 ± 6.1%, not colocalized; 4.8 ± 7.3%, unclear localization. (C) Live-cell time-lapse of GFP-Mff-S (green) in U2OS cell also expressing mCherry-mito3 (gray), eBFP2-peroxisome (blue), and E2-Crimson-ER (red). Yellow arrows, independent Mff punctae associating with ER; blue and gray arrows, peroxisomal and mitochondrial Mff, respectively. See also Video 6. (D) Graph depicting the degree of association between independent GFP-Mff-S punctae and ER from live-cell videos as in C (2.5-min videos imaged every 1.5 s). 34 ROIs from 30 U2OS cells analyzed (441 independent Mff punctae). Mean values from ROIs: 86.1 ± 17.1%, stably associated Mff punctae with ER; 11.0 ± 16.8%, partially associated; 4.6 ± 9.4%, not associated. (E) U2OS fractionation. Left: LSP, MSP, and HSP are low, medium, and high-speed pellets; HSS, high-speed supernatant. Marker proteins are ATP synthase, mitochondria; Sec63, ER; and Pmp70, peroxisomes. Right) Sucrose gradient fractionation of the medium-speed supernatant (MSS). (F) Human PEX3-deficient fibroblast fractionation, similar to U2OS fractionation. Bars: (B, whole cell image) 10 µm; (B, inset; and D) 2 µm. Time in seconds.
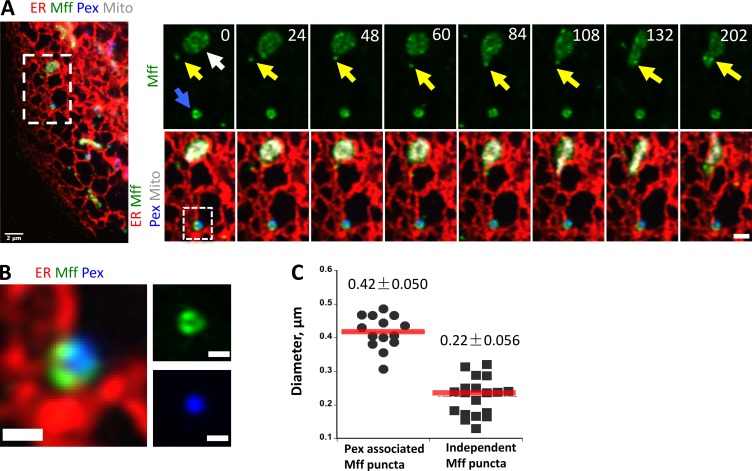
We also examined the localization of exogenously expressed GFP-Mff in live cells. As with endogenous staining, GFP-Mff at low expression levels localizes to both mitochondria and peroxisomes. In addition, a population of independent Mff punctae is present, and 86.1 ± 17.1% of these punctae maintain continuous ER association throughout the imaging period (Fig. 4, C and D; and Video 6). Mff contains four splice insert sites (Gandre-Babbe and van der Bliek, 2008). We used the variant lacking all inserts (termed Mff-S) for most investigations, but found that the variant including all inserts (Mff-L) also displayed this ER-localized subpopulation (Fig. S6, A and B).
As a second approach to examine Mff distribution, we performed cell fractionation studies in U2OS cells. By differential centrifugation, the mitochondrial marker is confined to the low- and medium-speed pellets, whereas ER and peroxisome markers are also present in the high-speed pellet fraction (Fig. 4 E). Similar to past studies (Otera et al., 2016), Mff migrates as a ladder of bands and is present in all membrane fractions. Upon sucrose gradient fractionation of the medium-speed supernatant, ER and peroxisome markers largely separate, with a small fraction of peroxisome marker persisting in the ER fraction. Mff fractionates with the ER (Fig. 4 E). These results suggest that a portion of Mff is bound to ER. To exclude the possibility that peroxisome contamination causes apparent Mff presence in the ER fraction, we used PEX3-deficient human fibroblasts, which lack mature peroxisomes (Sugiura et al., 2017). Similar to U2OS fractionation, PEX3-deficient cells contain an Mff population that fractionates with ER and is devoid of mitochondrial and peroxisomal markers (Fig. 4 F).
We also asked whether a subpopulation of Fis1 is present on ER. Similar to Mff, Fis1 is a TA protein, previously reported on both mitochondria and peroxisomes (Yoon et al., 2003; Stojanovski et al., 2004; Koch et al., 2005; Kobayashi et al., 2007). By immunofluorescence analysis of endogenous protein, we observe three Fis1 populations: mitochondrial, peroxisomal, and independent (Fig. 5 A), with 79.9 ± 11.3% of the independent punctae displaying ER association (Fig. 5 B). Fis1 depletion by siRNA strongly reduces all three of these Fis1 populations (Fig. 5 A). Exogenously expressed GFP-Fis1 displays a similar population of punctae that are independent of the mitochondrial or peroxisomal Fis1 pools (Fig. 5 C). Most of these independent Fis1 punctae are continually ER associated throughout the imaging period (78.8 ± 26.9%, Fig. 5 D).
Figure 5.
A subpopulation of Fis1 localizes to ER. (A) Endogenous Fis1 localization in fixed U2OS cells by immunofluorescence. Cells labeled with anti-Tom20 (mitochondria, blue), anti-PMP70 (peroxisomes, gray), and anti-Fis1 (green) and transfected with ER-TagRFP (ER, red). Left, scrambled siRNA; right, Fis1 siRNA. Yellow arrows, independent punctae; blue arrow, peroxisome-associated puncta; white arrow, mitochondrially associated Fis1. (B) Graph depicting the percentage of colocalization between independent Fis1 punctae and ER in U2OS cells by immunofluorescence (endogenous Fis1). 117 independent Fis1 punctae counted from nine ROIs from four cells. Mean values from ROIs: 79.9 ± 11.3%, colocalized Fis1 punctae with ER; 6.0 ± 7.4%, not colocalized; 14.1 ± 10.2%, unclear localization. (C) Live-cell time-lapse of GFP-Fis1 in U2OS cell also expressing mCherry-mito3 (gray), eBFP2-peroxisome (blue), and E2-Crimson-ER (red). Right: Individual frames from the time course of boxed region, showing independent Fis1 punctae associated with ER (yellow arrow) next to a peroxisome that is positive for Fis1 (blue arrow). (D) Graph depicting the degree of association between independent GFP-Fis1 punctae and ER from live-cell videos as in C (2.5-min videos imaged every 1.7 s). 16 ROIs from 15 U2OS cells (100 independent Fis1 punctae) analyzed. Mean values from ROIs: 78.8% ± 26.9%, stably associated Fis1 punctae with ER; 11.9 ± 18.2%, partially associated; 9.2 ± 14.8%, not associated. Bars: (A and C, whole-cell images) 10 µm; (A, inset) 5 µm; (C, inset) 2 µm inset. Time in seconds.
In contrast to Mff and Fis1, MiD49 and MiD51 contain N-terminal transmembrane domains. We examined the localization of MiD51-GFP expressed at low levels. Similar to past studies (Otera et al., 2016), MiD51 is in punctate accumulations on mitochondria, with no evidence for a peroxisomal population. There is also no evidence for a population of independent MiD51 (Fig. S6 C). We conclude that both Mff and Fis1 display populations that associate with ER independently of mitochondria or peroxisomes, whereas MiD51 is confined to mitochondria.
Dynamic interactions between Drp1 and Mff on ER
GFP-Mff punctae are dynamic on the ER, frequently moving and fluctuating in intensity (Fig. 4 C and Video 6). We examined Mff punctae morphology and dynamics in more detail using Airyscan microscopy. As observed in the confocal images, Mff is generally distributed evenly on the surface of mitochondria and peroxisomes at low expression levels, but has some regions of enrichment on both organelles (Fig. 6 A and Video 7). This enrichment is particularly noticeable on peroxisomes, with one or two highly concentrated regions (Fig. 6 B). The size of ER-bound Mff punctae (220 ± 56 nm, n = 19) is close to the resolution limit of Airyscan and smaller than the enriched Mff regions on peroxisomes (Fig. 6 C). Interestingly, ER-bound Mff punctae periodically appear to transfer to mitochondria (Fig. 6 A and Video 7).
Figure 6.
Dynamics of Mff on ER. (A) Independent Mff punctae dynamics (Airyscan microscopy time-lapse). Left: Merged image of a live U2OS cell expressing ER-tagRFP (ER, red), GFP-Mff-S (green), eBFP2-Peroxisome (blue), and mPlum-mito3 (gray). Right: Time-lapse series of the inset, with independent Mff punctum associating with ER then transferring to mitochondrion (yellow arrow). Blue arrow, peroxisomally associated Mff; white arrow, mitochondrial Mff. Bars: (left) 2 µm; (inset) 1 µm. Time in seconds. See also Video 7. (B) Zoom of A, showing heterogeneous nature of peroxisomally associated Mff. Bars, 0.5 µm. (C) Dot plot showing diameter of peroxisomal Mff and independent Mff punctae from Airyscan images. 14 peroxisomal Mff (0.42 ± 0.050 µm) and 19 independent Mff punctae (0.22 ± 0.056) analyzed.
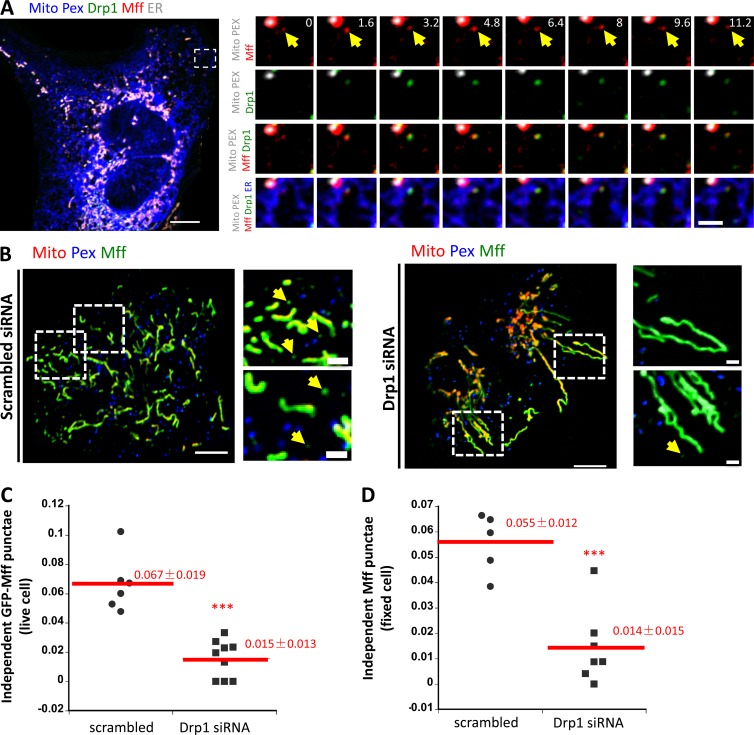
We next examined the relationship between Mff and Drp1 punctae on ER, using our GFP-Drp1-KI cell line transiently expressing mStrawberry-Mff at low levels. Being limited to four-color imaging, we labeled both mitochondria and peroxisomes with BFP and labeled ER with an E2-crimson marker (Fig. 7 A and Video 8). From quantification of live-cell time-lapse images, ∼70% of the ER-bound Drp1 and Mff punctae coassociate for the entirety of the 3-min imaging period (98 of 140 Mff punctae associated with Drp1; 84 of 140 Drp1 punctae associated with Mff). There are also instances of Drp1 appearance from previously existing Mff punctae (Fig. 7 A and Video 8), suggesting that ER-bound Mff punctae are sites of Drp1 oligomerization. Interestingly, the number of independent Mff punctae decreases ∼4-fold upon Drp1 suppression by siRNA, when analyzing either GFP-Mff in live cells (4.5-fold decrease; Fig. 7, B and C) or endogenous Mff by immunofluorescence (3.9-fold decrease; Fig. 7 D).
Figure 7.
Association between Drp1 and Mff on ER. (A) Left: Merged confocal image of a live GFP-Drp1-KI cell expressing mito-BFP (gray), eBFP2-peroxisome (gray), mStrawberry-Mff-S (red), and pLVX-E2-Crimson-ER (blue). Drp1 in green. Right: Time-lapse confocal images of boxed region show example of a Drp1 puncta maturing from an independent Mff puncta (yellow arrows). See also Video 8. (B) Independent Mff punctae in scramble siRNA–treated cells (left) and Drp1 siRNA–treated cells (right). Left: Merged image of live U2OS cells transiently expressing GFP-Mff-S (Mff, green), eBFP2-peroxisome (Pex, blue), and mCherry-mito7 (Mito, red). Right: Insets from boxed regions in whole-cell image. Yellow arrows denote independent Mff punctae. (C) Density of independent Mff punctae in control siRNA– and Drp1 siRNA–treated U2OS cells, quantified from live-cell images of GFP-Mff as in B. Units, number of independent Mff punctae per square micrometer in the ROI. 368 independent punctae from nine control cell ROIs and 106 punctae from nine Drp1 KD cell ROIs. ***, P < 0.0001 by Student’s t test. (D) Density of independent Mff punctae in control siRNA– and Drp1 siRNA–treated U2OS cells, quantified from fixed-cell immunofluorescence of endogenous Mff. Units, number of Mff punctae per square micrometer in ROI. 643 independent punctae from five control cell ROIs and 153 puncta from seven Drp1 KD cell ROIs. ***, P < 0.0005 by Student’s t test. Bars: (whole-cell images) 10 µm; (insets) 2 µm. Time in seconds.
ER-localized Mff enhances mitochondrial division rate
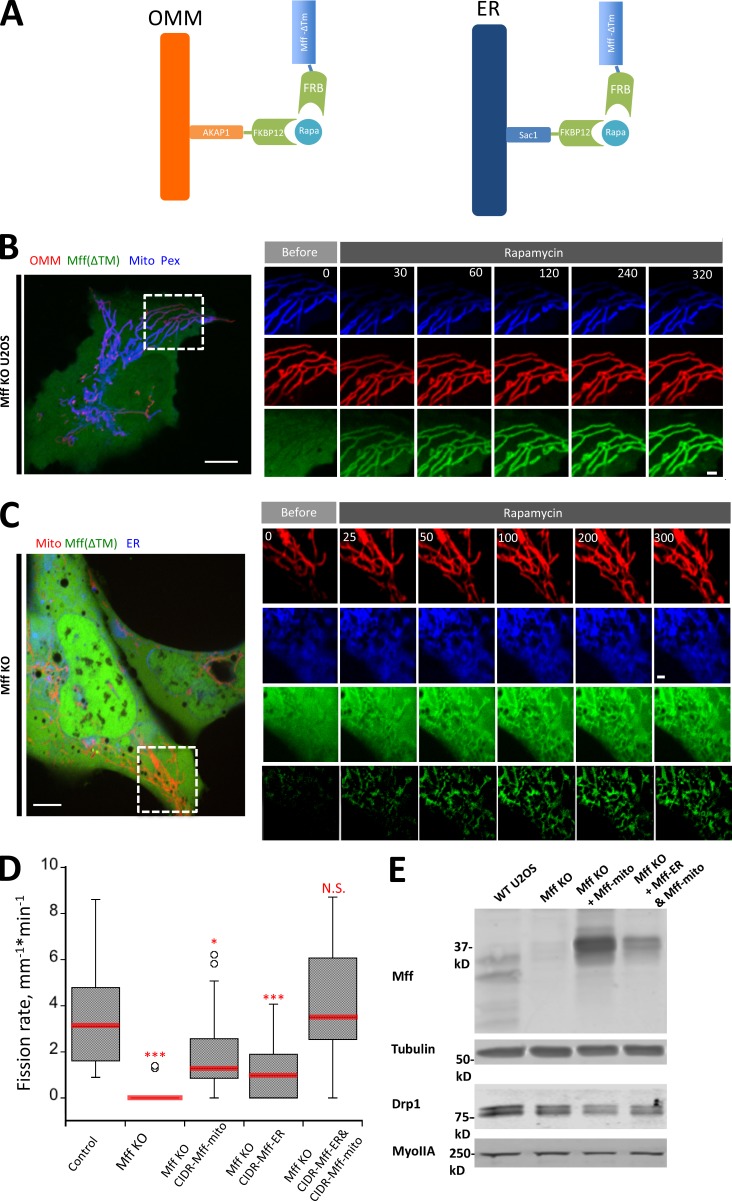
To test the functional significance of ER-targeted Mff, we designed a rapamycin-inducible system in which Mff lacking its transmembrane domain could be targeted to either mitochondria or ER, using the targeting sequences of AKAP1 and Sac1, respectively (Fig. 8 A; Csordás et al., 2010). A similar approach has been used to target Mff to lysosomes (Liu and Chan, 2015). Rapamycin treatment results in rapid Mff translocation from cytosol to mitochondria in Mff KO U2OS cells (Fig. 8 B). Rapamycin-induced translocation to ER is also rapid, but with some Mff still present in cytoplasm (Fig. 8 C). We used this system to test the effect of targeting Mff to specific locations (mitochondria alone, ER alone, or both mitochondria and ER) on mitochondrial division rate in Mff KO cells. Although either mitochondrial or ER targeting causes partial rescue, targeting Mff to both organelles brings the mitochondrial division rate back to the level of control cells (Fig. 8 D). The enhanced effect of expressing both mitochondrial and ER targeting signals is not caused by increased expression of Mff or of Drp1 (Fig. 8 E). These results suggest that ER targeting of Mff has a stimulatory effect on mitochondrial division.
Figure 8.
ER-targeted Mff facilitates mitochondrial division. (A) Schematic cartoon of rapamycin-induced Mff recruitment to either OMM (left) or ER (right). “Mff” refers to the cytoplasmic portion of Mff-S. (B) Dynamics of GFP-Mff-FRB translocation to mitochondria upon rapamycin treatment in Mff KO cells. Live-cell images of cell transfected with AKAP-FKBP12 (red), GFP-Mff-Cyto-FRB (green), eBFP2-peroxisome (peroxisomes, blue), and mitoBFP (mitochondria, blue). Rapamycin (final concentration: 10 µM) added at time 0. (C) Dynamics of GFP-Mff-Cyto translocation to ER upon rapamycin in rapamycin treatment in Mff KO cells. Live-cell images of cells transfected with Sac1-FKBP12 (ER, blue), GFP-Mff-Cyto-FRB (green), and mCherry-mito7 (mitochondria, red). The lower green panel represents GFP-MFF-CytoFRB signal that has been thresholded to remove the cytoplasmic signal. Rapamycin (final concentration: 10 µM) added at time 0. (D) Rapamycin-induced mitochondrial division rates in control U2OS cells (16 ROIs from 15 cells); Mff KO cells (21 ROIs from 21 cells; ***, P = 0.00001); Mff KO cells transfected with mitochondria-targeted Mff (34 ROIs from 30 cells; *, P = 0.0179); Mff KO cells transfected with ER-targeted Mff (20 ROIs from 17 cells; ***, P = 0.0049); or Mff KO cells transfected with both mitochondria- and ER-targeted Mff (34 ROIs from 30 cells; P = 0.4181). Statistical analysis based on comparison to control cells by Student’s t test. N.S., not significant. Error bars represent SD. (E) Western blot showing Mff and Drp1 expression levels in WT cells, Mff KO cells, and Mff KO cells transfected with either the Mff-FRB construct + the mitochondrially targeted FKBP12 construct (Mff KO + Mff-mito) or the Mff-FRB construct + the mitochondrially targeted FKBP12 construct + the ER-targeted FKBP12 construct (Mff KO + Mff-ER & Mff-mito). Tubulin and myosin IIA are loading controls. Endogenous Mff runs as a doublet less than 37 kD, whereas the Mff-FRB construct runs at the 37-kD marker. Bars: (whole-cell images) 10 µm; (insets) 2 µm. Time in seconds.
ER-associated Drp1 oligomers are dependent on INF2-mediated actin polymerization
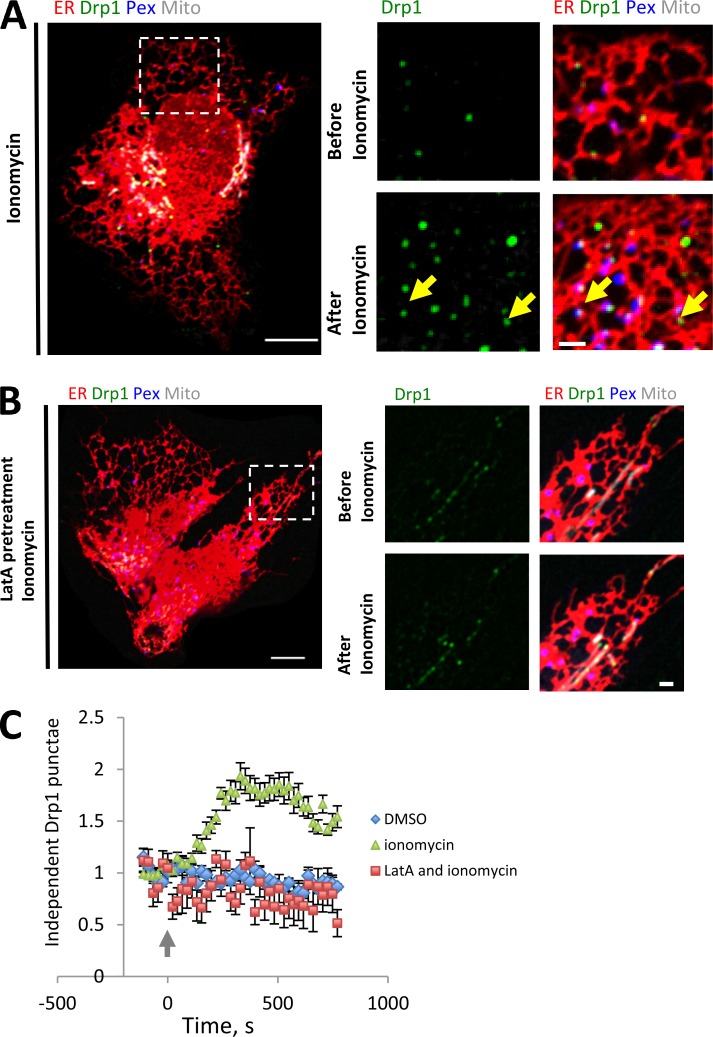
In a previous study (Ji et al., 2015), we found that ionomycin enhances Drp1 maturation on mitochondria. To test whether ionomycin can trigger ER-associated Drp1 maturation as well, we tracked independent Drp1 punctae upon ionomycin treatment. Ionomycin significantly increases both the number (Fig. 9 and Video 9) and size (Fig. 10, A and B) of independent Drp1 punctae.
Figure 9.
Actin-dependent oligomerization of ER-associated Drp1 punctae. (A) Left: Merged image of a live GFP-Drp1-KI cell before ionomycin treatment, transiently expressing mPlum-mito3 (gray), eBFP2-peroxisome (blue), and ER-tagRFP (ER, red). Drp1 in green. Right: Inset from boxed region before (top) and after (bottom) ionomycin treatment (4 µM, 10 min). Yellow arrows denote independent Drp1 maturing upon ionomycin treatment. See also Video 9. (B) Similar experiment as in A, except cells were pretreated for 10 min with 1 µM LatA. See also Video 10. (C) Quantification of independent Drp1 punctae number in response to vehicle treatment (DMSO), ionomycin treatment, and LatA pretreatment followed by ionomycin treatment. Six ROIs from six DMSO-treated cells, 16 ROIs from 14 ionomycin-treated cells, and eight ROIs from six LatA-pretreated/ionomycin-treated cells. Punctae per ROI normalized to 1 at time of ionomycin addition. Error bar, SEM. Arrow indicates time point where ionomycin was added during imaging (time 0). Bars: (left) 10 µm; (right) 2 µm. Time in seconds.
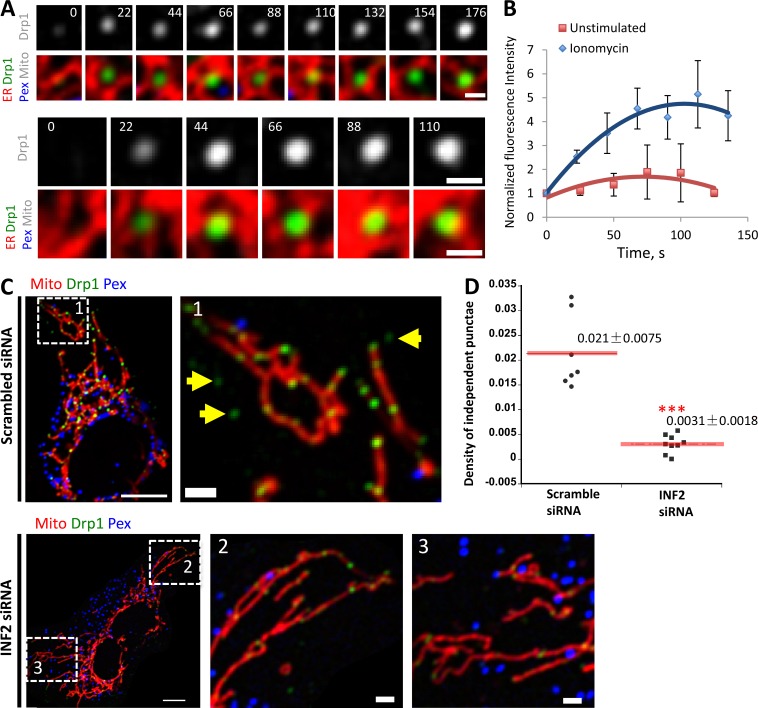
Figure 10.
Maturation of existing independent Drp1 punctae upon ionomycin stimulation. (A) Two examples of independent Drp1 punctae maturation in response to ionomycin. Time-lapse images of live GFP-Drp1-KI cell as in Fig. 9 A. Time indicates seconds after ionomycin treatment. Fluorescence intensity levels modulated uniformly across time course so that final fluorescence is in linear range (resulting in time 0 fluorescence being undetectable as displayed). Bars, 1 µm. Time in seconds. (B) Quantification of mean independent Drp1 punctum intensity in unstimulated or ionomycin-treated conditions. Seven independent Drp1 punctae from unstimulated cells and eight independent Drp1 punctae from ionomycin-treated cells analyzed. Error bars, SD. (C) Effect of INF2 KD on independent Drp1 punctae in GFP-Drp1-KI cells transfected with mCherry-mito7 (mitochondria, red) and eBFP2-peroxisome (blue). Drp1 in green. Top, control siRNA; bottom, INF2 siRNA. Right, zoomed images of boxed regions indicated by numbers. Yellow arrows, independent Drp1 punctae. Bars: (left) 10 µm; (insets, right) 2 µm. (D) Quantification of independent Drp1 punctae density in control (scrambled siRNA) and INF2 siRNA cells. 174 independent punctae from seven control cells; 45 independent punctae from nine INF2 siRNA cells. Density expressed as number of independent Drp1 punctae per area of ROI (in square micrometers). ***, P < 0.001 by Student’s t test.
Our previous studies also showed that mitochondrially bound Drp1 oligomers are significantly decreased by actin polymerization inhibitors (Korobova et al., 2013) and that actin polymerization inhibitors block the ionomycin-induced increase in Drp1 oligomerization (Ji et al., 2015). We tested the effect of Latrunculin A (LatA), an actin polymerization inhibitor, on ER-bound Drp1 oligomers. Pretreatment for 10 min with LatA causes a significant reduction in all Drp1 punctae before ionomycin treatment and a near-complete block of independent Drp1 punctae maturation upon ionomycin treatment (Fig. 9 and Video 10).
We have shown that the formin INF2 is required for actin polymerization leading to efficient mitochondrial division (Korobova et al., 2013), as well as mitochondrial accumulation of oligomeric Drp1 (Ji et al., 2015). The isoform of INF2 responsible for these effects is tightly bound to ER (Chhabra et al., 2009), suggesting that it could also play a role in ER-bound Drp1 oligomerization. We therefore tested whether INF2 played a role in independent Drp1 punctae accumulation. Suppression of INF2 by siRNA causes a 6.8-fold decrease in independent Drp1 punctae (Fig. 10, C and D). These results indicate that INF2-mediated actin polymerization is necessary for ER-associated Drp1 oligomerization.
Discussion
A major finding in this work is the identification of dynamic subpopulations of Drp1, Mff, and Fis1 on ER, distinct from the mitochondrial and peroxisomal populations of these proteins. An earlier study suggested that Drp1 could localize to ER (Yoon et al., 1998), but it did not include mitochondrial or peroxisomal markers, so specific localization to ER is unclear. There has been no previous identification of ER-bound subpopulations of any wild-type Drp1 receptor. We carefully examined previous publications for evidence of such localization for Mff (Gandre-Babbe and van der Bliek, 2008; Friedman et al., 2011; Palmer et al., 2013; Otera et al., 2016) or Fis1 (Yoon et al., 2003; Stojanovski et al., 2004; Koch et al., 2005; Kobayashi et al., 2007). Most of those studies did not stain for both peroxisomes and mitochondria, but in two studies using both markers, we found evidence for Mff (Palmer et al., 2013) and Fis1 (Kobayashi et al., 2007) punctae that are not bound to either organelle. The low abundance of these independent punctae, and their low intensities compared with both the mitochondrial and peroxisomal pools, could explain why this population has not been identified previously. Interestingly, a recent proteomic study identified an apparent ER-linked pool of Mff by proximity ligation (Hung et al., 2017), which could be an ER-bound population but could alternatively represent a population at ER–mitochondrial contact sites.
Mff and Fis1 are TA proteins that are inserted into membranes posttranslationally. TA proteins are found in essentially all cellular membranes, including ER, mitochondria, and peroxisomes. Insertion mechanisms for ER-based TA proteins are best understood, with the GET/TRC40 complex being an important pathway (Stefanovic and Hegde, 2007; Schuldiner et al., 2008; Denic et al., 2013; Mateja et al., 2015), and the recently identified SND pathway being an alternate route (Aviram et al., 2016). At present, the pathways controlling TA protein targeting to mitochondria or peroxisomes are less well understood, with evidence for three routes: (1) protein-free insertion (Krumpe et al., 2012), (2) protein-mediated insertion (Yagita et al., 2013), and (3) delivery from ER (Schuldiner et al., 2005; Lam et al., 2010; van der Zand et al., 2010).
The presence of Mff and Fis1 on all three membranes does not clarify their delivery mechanisms, but their wider distribution suggests mechanisms that would lead to both ER and mitochondrial insertion. Interestingly, one study (Stojanovski et al., 2004) showed that mutagenesis of two C-terminal lysines in mammalian Fis1 caused a shift in its localization from mitochondria to ER, which might suggest mitochondrial localization signals in the C terminus similar to findings for other proteins (Horie et al., 2002). It is also interesting that Mff is undetectable in the peroxisomes present in the “light” membrane fraction of U2OS cells (Fig. 4 E), suggesting that these peroxisomes are different from those in the heavier membrane fractions.
Although we provide evidence that the ER-localized pool of Mff acts in mitochondrial division, there are other possible explanations for Mff’s presence on ER. First, a portion of the ER pool might represent a transient intermediate in Mff’s biosynthetic pathway, in which it is first inserted into the ER membrane then transferred to the OMM. Alternately, a portion ER-bound Mff and Fis1 might represent mislocalized protein that is subsequently sorted to the OMM by a secondary sorting mechanism. These possibilities are not mutually exclusive with the existence of a functional pool of ER-localized Mff. Better understanding of targeting mechanisms for Mff is required, including pulse-chase localization studies to determine whether an ER intermediate exists.
Recent studies in budding yeast and mammals show that aggregates of misfolded protein can bind ER, then move to the mitochondrial matrix for proteolysis (Zhou et al., 2014; Ruan et al., 2017). However, several lines of evidence strongly suggest that the ER-bound punctae of Drp1, Mff, and Fis1 observed here are not protein aggregates. First, in all three cases we observe these punctae using immunofluorescence for endogenous proteins, arguing against overexpression artifact. Second, GFP-fusions of nonoligomerizable Drp1 mutants do not display ER-bound punctae, even when expressed at significantly higher levels than wild-type GFP-Drp1. Third, ER-bound Drp1 punctae are virtually absent in the following conditions: Mff KO, actin polymerization inhibition, and suppression of the actin polymerization factor INF2. All of these conditions inhibit mitochondrial division but are not known to be related to aggregated protein responses. LatA treatment reduces the number of independent Drp1 punctae within 10 min, demonstrating the dynamic nature of this population.
Another possibility is that independent Drp1 or Mff punctae represent MDVs containing OMM but not inner mitochondrial membrane. Our imaging of Drp1 and the OMM protein Tom20 suggest that this is not the case, as we observed no consistent colocalization. Even so, MDVs can have heterogeneous composition (Soubannier et al., 2012), which leaves open the possibility that the independent punctae are bound to a specific MDV subtype. Because the majority of independent Drp1, Mff, and Fis1 punctae track tightly with ER in live-cell imaging, any MDV would likely be associated with ER in this case.
One possible functional role of ER-assembled Drp1 is in mitochondrial division. In support of this function, (a) we observe transfer of Drp1 punctae from ER to mitochondria; (b) we observe mitochondrial division after ER-to-mitochondrial Drp1 transfer; and (c) in Mff KO cells, targeting Mff to both ER and mitochondria is more efficient in rescuing mitochondrial division than is targeting to either ER or mitochondria alone. There are uncertainties in this correlation. Limitations of confocal microscopy in both spatial and temporal resolution make it difficult to be certain of direct ER-to-mitochondrial Drp1 transfer. In addition, there is a significant amount of ER-to-mitochondrial Drp1 transfer that does not result in mitochondrial division. To observe mitochondrial division after ER-to-mitochondrial Drp1 transfer, we stimulate division frequency with the calcium ionophore ionomycin. However, mitochondrial division in general occurs at low frequency, and the vast majority of mitochondrially bound Drp1 punctae are nonproductive for mitochondrial division (Ji et al., 2015), suggesting that Drp1 oligomerization is in dynamic equilibrium independent of mitochondrial division.
From our findings, we propose a working model that includes a role for ER in Drp1 oligomerization and recruitment before interaction with mitochondria or peroxisomes. The combination of ER-bound Mff and INF2-mediated actin polymerization on ER serves as an initiation site for recruitment of Drp1 oligomers. These Drp1 oligomers can be transferred to mitochondria or peroxisomes upon ER contact, where they can serve in the assembly of mitochondrially bound Drp1 oligomers capable of mitochondrial division. This Drp1 transfer can occur without transfer of the receptors themselves, although we have observed movement of Mff punctae between ER and mitochondria. Further study of these dynamics is needed.
Assembly on ER is likely to be only one component of Drp1’s oligomeric equilibrium, in addition to direct assembly on mitochondria or peroxisomes. Another possibility is that the independent Drp1 punctae observed here represent a minor proportion of all ER-bound Drp1 oligomers, with the vast majority being assembled on ER at ER–mitochondrial contact sites. Because of the close proximity of ER–mitochondrial contact sites (Csordás et al., 2010), imaging Drp1 transfer at these sites is challenging by current live-cell techniques.
Our work adds another layer to the understanding of roles for mammalian Drp1 receptors (Mff, Fis1, MiD49, and MiD51) in mitochondrial and peroxisomal division. The current picture is somewhat murky, with recent KD/KO studies in several cell lines providing largely overlapping but at times conflicting results (Losón et al., 2013; Shen et al., 2014; Osellame et al., 2016; Otera et al., 2016). One feature of clear agreement is that neither MiD49 nor MiD51 localizes to peroxisomes, and neither participates in peroxisomal division (Palmer et al., 2013; Otera et al., 2016). Another common theme is that MiD49 and MiD51 are at least partially redundant with each other and have the capability of acting independently of Mff (Losón et al., 2013; Palmer et al., 2013; Osellame et al., 2016; Otera et al., 2016). Most studies find the role of Fis1 in Drp1 recruitment and mitochondrial/peroxisomal division to be minor at best, although one study found more significant effects (Shen et al., 2014). Deletion of Mff typically has the most dramatic effects on both Drp1 recruitment and mitochondrial division, but one study found that MiD49/51 deletion has comparable effects (Osellame et al., 2016). The differing results may be partly explained by cellular context. In mitophagy, for example, Fis1 might play a role in Drp1 recruitment downstream of Mff (Shen et al., 2014). During apoptosis, MiD49 and MiD51 have roles in cristae remodeling (Otera et al., 2016), although other Drp1 receptors clearly function in apoptosis as well (Osellame et al., 2016).
We make three important findings on Drp1 receptors in this work. First, Mff is of fundamental importance in U2OS cells, because either siRNA-mediated suppression or CRISPR-mediated KO strongly reduced both Drp1 oligomerization and mitochondrial division. Second, U2OS cells have ER-bound populations of both Mff and Fis1. Third, the majority of ER-bound Drp1 punctae colocalize with Mff. These populations appear to be codependent, with reduction of either Drp1 or Mff reducing punctae of the other protein on ER. Presumably, Drp1 oligomerization recruits additional Mff from the bulk ER.
One open question concerns why there are so many potential mechanisms for regulating Drp1, including multiple receptors, Drp1 posttranslational modification (Chang and Blackstone, 2007, 2010; Cribbs and Strack, 2007), cardiolipin enrichment on the OMM (Bustillo-Zabalbeitia et al., 2014; Macdonald et al., 2014), actin polymerization (Korobova et al., 2013; Ji et al., 2015; Li et al., 2015; Moore et al., 2016), and ER–mitochondrial contact (Friedman et al., 2011). Do these mechanisms operate in concert or independently? Given that Drp1 oligomer assembly and disassembly are constantly in flux on mitochondria (Ji et al., 2015), the answer could be “both.” A critical threshold of Drp1 oligomerization and mitochondrial recruitment is necessary, regardless of the means by which oligomerization/recruitment are activated. In this model, a variety of combinations of these activators can lead to the final outcome of division-productive Drp1 oligomerization. Other aspects of Drp1-mediated force generation may be similarly nuanced (Ramachandran, 2017). Importantly, the ER-based recruitment of Drp1 oligomers represents only one of these activation mechanisms, and its loss may be compensated by up-regulation of the other mechanisms. An additional step may be recruitment of dynamin 2 late in the process (Lee et al., 2016), which would be subject to its own regulation.
This study extends our findings on the role of actin in mitochondrial division by showing that actin polymerization is necessary for initiation and growth of ER-bound Drp1 oligomers. We have proposed direct binding of Drp1 to actin filaments as a potential mechanism for increasing productive Drp1 oligomerization (Ji et al., 2015; Hatch et al., 2016). The presence of the formin INF2 on ER (Chhabra et al., 2009) and its importance for Drp1 recruitment to mitochondria (Korobova et al., 2013; Ji et al., 2015) suggest that INF2-mediated actin polymerization on ER, in conjunction with Mff on ER, might mediate ER-based Drp1 oligomerization. Actin polymerization has been implicated in mitochondrial division in many contexts (De Vos et al., 2005; DuBoff et al., 2012; Moore et al., 2016) and additional actin-binding proteins on mitochondria (Manor et al., 2015) and in cytosol (Li et al., 2015; Moore et al., 2016) have been implicated. It will be interesting to elucidate if and how these proteins work together in this process.
Given the presence of Drp1 oligomers on ER, and its role in constriction of mitochondria and peroxisomes, it is tempting to speculate that Drp1 might mediate some aspect of ER membrane dynamics. Studies have suggested that dominant-negative Drp1 mutants change ER structure (Pitts et al., 1999). Although we have occasionally observed Drp1 punctae at sites of ER tubule breakage (unpublished data), these instances are rare. Nevertheless, the presence of Drp1, Mff, and Fis1 on ER expands mechanistic possibilities for membrane dynamics in general.
Materials and methods
Plasmids and siRNA oligonucleotides
mCherry-mito-7 was purchased from Addgene (55102), and consists of the mitochondrial targeting sequence from subunit VIII of human cytochrome C oxidase N-terminal to mCherry. mito-BFP constructs were previously described (Friedman et al., 2011) and consist of aa 1–22 of Saccharomyces cerevisiae COX4 N-terminal to BFP. Tom20-mCherry was previously described in Ji et al. (2015). eBFP-Peroxisome was constructed by replacing the CFP sequence of CFP-Peroxisome containing peroxisomal targeting signal 1 (PTS1; 54548; Addgene) with eBFP2, cut from eBFP2-Mito7 (55248; Addgene) with BsrGI/BamHI. mPlum-mito3 and mPlum-peroxisome-2 were purchased from Addgene (55988 and 54509, respectively). ER-tagRFP was a gift from E. Snapp (Albert Einstein College of Medicine, New York, NY), with prolactin signal sequence at 5′ of the fluorescent protein and KDEL sequence at 3′. pEF.myc.ER-E2-Crimson was purchased from Addgene (38770). Mff-S and MiD51 were cloned by reverse transcription-PCR from RNA isolated from HEK293 cells and cloned into eGFP-C1 (Mff) or eGFP-N1 (MiD51) vectors (Clontech). GFP-Mff-L was purchased from Addgene (49153). mStrawberry-Mff-S was constructed by replacing GFP with mStrawberry using SalI–BamHI. MiD51-mStrawberry was constructed by cutting MiD51 from MiD51-GFP and pasting into mStrawberry-N1 vector using Bgl II/BamH1. GFP-Fis1 was a gift from M. Ryan (Monash University, Melbourne, Australia). mStrawberry-Rab4b and mStrawberry-Rab7a were gifts from M. Fukada (Tohoko University, Sendai, Japan; Matsui et al., 2011). In our nomenclature for Mff isoforms, Mff-S corresponds to isoform 8 (no alternately spliced exons), and Mff-S corresponds to isoform 1 (containing all alternately spliced exons) from Gandre-Babbe and van der Bliek (2008). Drp1 mutants that maintain the monomeric (K642E) or dimeric (K401-404A) states were described in Hatch et al. (2016). Rapamycin-inducible constructs include mitochondrial targeting construct, aa 1–31 of mouse AKAP1 fused to FKBP12; ER-targeting construct, C-terminal sequence of human/mouse Sac1 fused to FKBP12; and GFP-Mff inducibly targetable construct, the cytoplasmic region (aa 1–197) of human Mff-S fused to GFP on the N terminus and FRB on the C terminus. Both mitochondrial- and ER-targeted FKBP12 constructs were gifts of G. Hajnoczky (Thomas Jefferson University, Philadelphia, PA; Csordás et al., 2010).
Oligonucleotides for human Mff siRNA were synthesized by Qiagen against target sequence 5′-ACCGATTTCTGCACCGGAGTA-3′. Oligonucleotides for MiD51 were synthesized by Qiagen against sequence 5′-CAGTATGAGCGTGACAAACAT-3′ (siRNA#1) and 5′-CCTGGTCTTTCTCAACGGCAA-3′ (siRNA#2). Oligonucleotides for MiD49 were synthesized by Qiagen against sequence 5′-TTGGGCTATGGTGGCCATAAA-3′ (siRNA#1) and 5′-CTGCTGAGAGAAGGTGACTTA-3′ (siRNA#2). Oligonucleotides for Fis1 were synthesized by IDT against target sequence 5′-GUACAAUGAUGACAUCCUAAAGGC-3′ (siRNA#1) and 5′-ACAAUGAUGACAUCCGUAAAGGCAT-3′ (siRNA#2). Oligonucleotides for human total INF2 siRNA were synthesized by IDT Oligo against target sequence 5′-GGAUCAACCUGGAGAUCAUCCGC-3′. Oligonucleotides for human Drp1siRNA were synthesized by IDT Oligo against target sequence 5′-GCCAGCUAGAUAUUAACAACAAGAA-3′. As a control, Silencer Negative Control, 5′-CGUUAAUCGCGUAUAAUACGCGUAT-3′ (Ambion), was used.
Antibodies
Anti-Mff (17090-1-AP; ProteinTech) was used at 1:1,000 dilution for Western blot and 1:500 dilution for immunofluorescence. Anti-Fis1 (10956-1-AP; ProteinTech) was used at 1:1,000 for Western blot and 1:500 for immunofluorescence. Anti-Tubulin (DM1-α; Sigma-Aldrich) was used at 1:10,000 dilution for Western blot. Drp1 was detected using a rabbit monoclonal antibody (D6C7; Cell Signaling Technologies) at 1:500 dilution for immunofluorescence. Anti-INF2 rabbit polyclonal was described previously (Ramabhadran et al., 2011). Organelle marker antibodies for Western blot included anti-ATP synthase mouse monoclonal (A21351; Molecular Probes), anti-Sec63 (ARP46839; Aviva), and anti-Pmp70 rabbit polyclonal (4200181; Sigma-Aldrich), all used at 1:1,000.
Cell culture and transfection
Human osteosarcoma U2OS cells (HTB96; ATCC) were grown in DMEM (Invitrogen) supplemented with 10% calf serum (Atlanta Biologicals). Human PEX3-deficient fibroblasts (PBD400-T1) were a gift from Heidi McBride (Montreal Neurological Institute, Montreal, Canada) and were grown in DMEM supplemented with 10% FCS (Atlanta Biologicals) and nonessential amino acids (Gibco). To make the GFP-Drp1 KI U2OS cell line by CRISPR-Cas9, we used the GeCKO system (Zhang laboratory, Massachusetts Institute of Technology, Cambridge, MA; http://genome-engineering.org/gecko/). The donor plasmid contained eGFP (A206K mutant) flanked by 445 bases upstream of hDrp1 start codon and 308 bases downstream from start (synthesized by IDT). The target guide sequence (5′-CATTCATTGCCGTGGCCGGC-3′) was predicted using the GeCKO website program and made by IDT. Donor and guide plasmids were transfected into U2OS cells at a 3:1 molar ratio using Lipofectamine 2000 (Invitrogen). Cells were put under puromycin selection, and clones were selected by FACS sorting and single-cell cloning, then verified by immunofluorescence and Western blot.
For transfection of the U2OS or Drp1 KI lines, cells were seeded at 4 × 105 cells per well in a six-well dish ∼16 h before transfection. Plasmid transfections were performed in OPTI-MEM (Invitrogen) with 2 µl Lipofectamine 2000 per well for 6 h, followed by trypsinization and replating onto glass-bottom MatTek dishes (P35G-1.5-14-C) coated with concanavalin A (ConA; C5275; Sigma/Aldrich) at ∼3.5 × 105 cells per well. Cells were imaged in live-cell medium (21063-029; Life Technologies) ∼16–24 h after transfection.
For all experiments, the following amounts of DNA were transfected per well (individually or combined for cotransfection): 500 ng for mito-BFP, eBFP2-peroxisome, and mCherry-mito7; 850 ng for Tom20-mCherry; 1,000 ng for ER-tagRFP, mPlum-mito3, and pEF.myc.ER-E2-Crimson; 100 ng for GFP-Mff-S, mStrawberry-Mff-S, GFP-Mff-S, and GFP-Fis1; 50 ng for MiD51-mStrawberry; 30 ng for mStrawberry-Rab4b and mStrawberry-Rab7a; 1,000 ng for AKAP1-FKBP12; and 500 ng for Sac1-FKBP12 and GFP-Mff-FRB.
For siRNA transfections, cells were plated on six-well plates with 30–40% density, and 2 µl RNAimax (Invitrogen) and 63 ng siRNA were used per well. Cells were analyzed 72–84 h posttransfection for suppression.
Live imaging by confocal and Airyscan microscopy
Cells were grown on glass-bottom matTek dishes coated with ConA (coverslips treated for ∼2 h with 100 µg/ml ConA in water at room temperature). MatTek dishes were loaded to a Wave FX spinning disk confocal microscope (Quorum Technologies) on an Eclipse Ti microscope (Nikon) equipped with Hamamatsu ImageM EM CCD cameras and Bionomic Controller (20/20 Technology) temperature-controlled stage set to 37°C. After equilibrating to temperature for 10 min, cells were imaged with the 60× 1.4-NA Plan Apo objective (Nikon) using the 403-nm laser and 450/50 filter for BFP, 491-nm and 525/20 for GFP, 561-nm and 593/40 for mStrawberry or mCherry, and 640-nm and 700/60 for mPlum and E2-Crimson. For rapamycin induction, cells were treated with freshly prepared rapamycin (Thermo Fisher Scientific; 10 mM Stock in DMSO, 10 µM final concentration on cells) during imaging.
Airyscan images were acquired on LSM 880 equipped with 63×/1.4-NA plan Apochromat oil objective, using the Airyscan detectors (Zeiss). The Airyscan uses a 32-channel array of GaAsP detectors configured as 0.2 Airy units per channel to collect the data that is subsequently processed using Zen2 software. After equilibrating to 37°C for 30 min, cells were imaged with the 405-nm laser and 450/30 filter for BFP, 488-nm and 525/30 for GFP, 561-nm and 595/25 for mStrawberry or mCherry, and 633-nm and LP 625 for mPlum.
Immunofluorescence staining
Cells were fixed with 4% formaldehyde (Electron Microscopy Sciences) in PBS for 10 min at room temperature. After washing with PBS three times, cells were permeabilized with 0.1% Triton X-100 in PBS for 15 min on ice. Cells were then washed three times with PBS, blocked with 0.5% BSA in PBS for 1 h, incubated with primary antibodies in diluted blocking buffer overnight, and washed with PBS three times. Mff or Fis1 polyclonal antibodies (rabbit) were conjugated to Alexa Fluor 488, and PMP70 antibodies (rabbit) were conjugated to Alexa Fluor 647 (Zenon Tricolor Rabbit IgG1 Labeling kit; Invitrogen); Secondary antibodies were applied for 1 h at room temperature. After washing with PBS three times, samples were mounted on Vectashield (H-1000; Vector Laboratories).
Image analysis
ER association of Drp1, Mff, and Fis1
Cells expressing GFP-Drp1, Mff, or Fis1 and markers for ER, mitochondria, and peroxisomes were imaged in a single focal plane for 3 min at 1.5- to 2-s intervals. Regions of cells in which tubular ER could be readily resolved and appeared continuous in a single plane of view were analyzed. Independent Drp1, Mff, or Fis1 punctae were counted as always associated if they remained in contact with the ER during every frame of the video, sometimes ER associated if the independent punctae contacted the ER at least half of total frames where punctae were visible, and not ER associated if no ER contact was visible.
Drp1 punctae quantification
Drp1 KI cells transiently transfected with mitochondrial markers were imaged live by spinning disc confocal fluorescence microscopy for 10 min at 3-s intervals in a single focal plane. ROIs with readily resolvable mitochondria and Drp1 were processed as described previously (Ji et al., 2015). We thresholded mitochondrially associated Drp1 punctae by using the Colocalization ImageJ plugin with the following parameters: ratio, 50% (0–100%); threshold channel 1, 30 (0–255); threshold channel 2, 30 (0–255); and display value, 255 (0–255). Mitochondrially associated Drp1 punctae were further analyzed by Trackmate as described previously (Ji et al., 2015). Numbers of Drp1 punctae were automatically counted frame-by-frame using the Find Stack Maxima ImageJ macro. The density of independent Drp1 punctae was quantified by visual assessment of each Drp1 puncta in an ROI for association with the mitochondria or peroxisome marker. Those punctae associated with neither mitochondria or peroxisomes were classified as independent. The result is expressed as the number of independent Drp1 punctae per area of the ROI in square micrometers.
Mitochondrial division rate
The process is described in detail in Ji et al. (2015). Suitable ROIs were selected for analysis based on whether individual mitochondria were resolvable and did not leave the focal plane. Files of these ROIs were assembled, coded and scrambled by one investigator, and analyzed for division by a second investigator who was blinded as to the treatment condition. The second investigator scanned the ROIs frame-by-frame manually for division events and determined total mitochondrial length within the ROI using the ImageJ macro Mitochondrial Morphology. The results were then given back to the first investigator for decoding. Division rate was analyzed over a 10-min period after DMSO, ionomycin (4 µM), or rapamycin (10 µM) treatment, depending on the experiment.
Cell fractionation
Cell fractionation was a modification of the method in Clayton and Shadel (2014). All protease inhibitors were from EMD Chemicals. For U20S, cells grown to ∼70% confluence in 12 × 75 cm2 flasks were harvested by trypsinization and washed three times with PBS. After trypsinization, all steps were conducted at 4°C or on ice. The cell pellet (∼0.2 ml) was resuspended in 5.4 ml hypotonic buffer (10 mM Tris-HCl, pH 7.5, 10 mM NaCl, 1.5 mM MgCl2, and protease inhibitors [2 µg/ml leupeptin, 10 µg/ml aprotinin, 2 µg/ml pepstatin A, 5 µg/ml calpain inhibitor 1, 5 µg/ml calpeptin, 1 mM benzamidine, and 0.05 µg/ml cathepsin B inhibitor II]), incubated for 10 min, and lysed by Dounce (Wheaton Dura-Grind), followed by addition of 3.6 ml of 2.5× isotonic buffer (525 mM mannitol, 175 mM sucrose, 12.5 mM Tris-HCl, pH 7.5, 2.5 mM EDTA, and protease inhibitors). The lysate was centrifuged at 1,300 g for 5 min (low-speed centrifugation). The low-speed supernatant was centrifuged at 13,000 g for 15 min (medium-speed centrifugation). The medium-speed supernatant was centrifuged at 208,000 g for 1 h (high-speed centrifugation). For PEX3-deficient fibroblasts, conditions were similar except that four centrifugation speeds were used, as follows (Sugiura et al., 2017): 800 g for 10 min (nuclei and unlysed cells, discarded), 2,300 g (low-speed centrifugation), 23,000 g (medium-speed centrifugation), and 208,000 g for 1 h (high-speed centrifugation). All pellets were washed with 1× isotonic buffer then resuspended in SDS-PAGE buffer. For sucrose gradient fractionation, the medium-speed supernatant (3.4 ml) was layered onto a discontinuous gradient containing equal volumes (1.9 ml) of 0.5, 0.75, 1, and 1.3 M sucrose (all in the background of 1× isotonic buffer) and centrifuged for 1 h at 35,000 rpm in an SW41 rotor (Beckman Coulter) with no brake. Fractions (1 ml) were removed from top.
Western blotting
Cells were grown on a six-well plate, trypsinized, washed with PBS, and resuspended 50 µl PBS. This solution was mixed with 34 µl of 10% SDS and 1 µl of 1 M DTT, boiled for 5 min, and cooled to 23°C. 17 µl of 300 mM freshly made NEM in water was added. Just before SDS-PAGE, the protein sample was mixed 1:1 with buffer containing 250 mM Tris-HCl, pH 6.8, 2 mM EDTA, 20% glycerol, 0.8% SDS, 0.02% bromophenol blue, 1,000 mM NaCl, and 4 M urea. Proteins were separated by 7.5% SDS-PAGE and transferred to a PVDF membrane (Millipore). The membrane was blocked with TBS-T (20 mM Tris-HCl, pH 7.6, 136 mM NaCl, and 0.1% Tween-20) containing 3% BSA (Research Organics) for 1 h, then incubated with the primary antibody solution at 4°C overnight. After washing with TBS-T, the membrane was incubated with HRP-conjugated secondary antibody (Bio-Rad) for 1 h at room temperature. Signals were detected by chemiluminescence (Pierce). For Western blot of Mff KO cells, the Odyssey CLx system was used (Li-Cor Biotechnology), as well as IRDye-labeled anti-rabbit and anti-mouse secondary antibodies from the same company.
Online supplemental material
Fig. S1 shows characterization of the GFP-Drp1-KI, as well as GFP-Drp1 dynamics in Cos7 cells. Fig. S2 shows the lack of correlation between independent Drp1 punctae and three endosomal markers. Fig. S3 shows the lack of correlation between GFP-Drp1 independent punctae and the mitochondrial outer membrane marker Tom20. Fig. S4 shows Drp1-independent punctae for endogenous Drp1 in U2OS cells and that GFP-Drp1 punctae are absent in Mff siRNA cells and for oligomerization-deficient GFP-Drp1 mutants. Fig. S5 shows characterization of U2OS cells knocked down for Mff or Fis1, and effects of Mff KD on mitochondrial division and mitochondrial Drp1 oligomer density. Fig. S6 shows that a second splice variant of Drp1 displays independent punctae, but that MiD51 does not. Video 1 shows two independent Drp1 punctae stably associated with ER (one goes out of the focal plane briefly). Videos 2 and 3 show Drp1 punctae translocating to mitochondria (Video 2 does not have an ER marker; Video 3 has an ER marker). Videos 4 and 5 show Drp1 punctae translocating to mitochondria, followed by mitochondrial division, after ionomycin stimulation (Video 4 does not have an ER marker; Video 5 has an ER marker). Video 6 shows an independent GFP-Mff puncta stably on ER. Video 7 shows an independent GFP-Mff puncta transferring from ER to mitochondrion (Airyscan). Video 8 shows colocalization of GFP-Drp1 and mStrawberry-Mff on ER. Videos 9 and 10 show the change in independent Drp1 punctae after ionomycin treatment. In Video 9, the punctae increase, but in Video 10 (pretreated with latrunculin A) the punctae do not increase.
Supplementary Material
Acknowledgments
We thank Mike Ryan for GFP-Fis1, Ayumu Sugiura for advice on organelle fractionation, Heidi McBride for PEX3-deficient cells, Maya Schuldiner for discussions on TA proteins, Anna Hatch for editing, and Peter Rocs for being receptive to anything.
Support was provided by National Institutes of Health GM069818, GM106000, and P20 GM113132 to H.N. Higgs, National Institutes of Health NS056244 and NS 087908 to S. Strack, and National Institutes of Health S10OD010330 to the Norris Cotton Cancer Center.
The authors declare no competing financial interests.
Author contributions: W.-K. Ji: conceptualization, methodology, investigation, formal analysis, writing. R. Chakrabarti: conceptualization, methodology, investigation, formal analysis. X. Fan and L. Schoenfeld: methodology, investigation, formal analysis. S. Strack: methodology, investigation. H.N. Higgs: conceptualization, project administration, writing, funding acquisition.
References
- Aviram N., Ast T., Costa E.A., Arakel E.C., Chuartzman S.G., Jan C.H., Haßdenteufel S., Dudek J., Jung M., Schorr S., et al. . 2016. The SND proteins constitute an alternative targeting route to the endoplasmic reticulum. Nature. 540:134–138. 10.1038/nature20169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui H.T., and Shaw J.M.. 2013. Dynamin assembly strategies and adaptor proteins in mitochondrial fission. Curr. Biol. 23:R891–R899. 10.1016/j.cub.2013.08.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustillo-Zabalbeitia I., Montessuit S., Raemy E., Basañez G., Terrones O., and Martinou J.C.. 2014. Specific interaction with cardiolipin triggers functional activation of dynamin-related protein 1. PLoS One. 9:e102738 10.1371/journal.pone.0102738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.R., and Blackstone C.. 2007. Drp1 phosphorylation and mitochondrial regulation. EMBO Rep. 8:1088–1089. 10.1038/sj.embor.7401118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.R., and Blackstone C.. 2010. Dynamic regulation of mitochondrial fission through modification of the dynamin-related protein Drp1. Ann. N. Y. Acad. Sci. 1201:34–39. 10.1111/j.1749-6632.2010.05629.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhabra E.S., Ramabhadran V., Gerber S.A., and Higgs H.N.. 2009. INF2 is an endoplasmic reticulum-associated formin protein. J. Cell Sci. 122:1430–1440. 10.1242/jcs.040691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton D.A., and Shadel G.S.. 2014. Isolation of mitochondria from tissue culture cells. Cold Spring Harb. Protoc. 10.1101/pdb.prot080002 [DOI] [PubMed] [Google Scholar]
- Cribbs J.T., and Strack S.. 2007. Reversible phosphorylation of Drp1 by cyclic AMP-dependent protein kinase and calcineurin regulates mitochondrial fission and cell death. EMBO Rep. 8:939–944. 10.1038/sj.embor.7401062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csordás G., Várnai P., Golenár T., Roy S., Purkins G., Schneider T.G., Balla T., and Hajnóczky G.. 2010. Imaging interorganelle contacts and local calcium dynamics at the ER-mitochondrial interface. Mol. Cell. 39:121–132. 10.1016/j.molcel.2010.06.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denic V., Dötsch V., and Sinning I.. 2013. Endoplasmic reticulum targeting and insertion of tail-anchored membrane proteins by the GET pathway. Cold Spring Harb. Perspect. Biol. 5:a013334 10.1101/cshperspect.a013334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos K.J., Allan V.J., Grierson A.J., and Sheetz M.P.. 2005. Mitochondrial function and actin regulate dynamin-related protein 1-dependent mitochondrial fission. Curr. Biol. 15:678–683. 10.1016/j.cub.2005.02.064 [DOI] [PubMed] [Google Scholar]
- DuBoff B., Götz J., and Feany M.B.. 2012. Tau promotes neurodegeneration via DRP1 mislocalization in vivo. Neuron. 75:618–632. 10.1016/j.neuron.2012.06.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuBoff B., Feany M., and Götz J.. 2013. Why size matters—Balancing mitochondrial dynamics in Alzheimer’s disease. Trends Neurosci. 36:325–335. 10.1016/j.tins.2013.03.002 [DOI] [PubMed] [Google Scholar]
- Friedman J.R., Lackner L.L., West M., DiBenedetto J.R., Nunnari J., and Voeltz G.K.. 2011. ER tubules mark sites of mitochondrial division. Science. 334:358–362. 10.1126/science.1207385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fröhlich C., Grabiger S., Schwefel D., Faelber K., Rosenbaum E., Mears J., Rocks O., and Daumke O.. 2013. Structural insights into oligomerization and mitochondrial remodelling of dynamin 1-like protein. EMBO J. 32:1280–1292. 10.1038/emboj.2013.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandre-Babbe S., and van der Bliek A.M.. 2008. The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol. Biol. Cell. 19:2402–2412. 10.1091/mbc.E07-12-1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch A.L., Gurel P.S., and Higgs H.N.. 2014. Novel roles for actin in mitochondrial fission. J. Cell Sci. 127:4549–4560. 10.1242/jcs.153791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch A.L., Ji W.K., Merrill R.A., Strack S., and Higgs H.N.. 2016. Actin filaments as dynamic reservoirs for Drp1 recruitment. Mol. Biol. Cell. 27:3109–3121. 10.1091/mbc.E16-03-0193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie C., Suzuki H., Sakaguchi M., and Mihara K.. 2002. Characterization of signal that directs C-tail-anchored proteins to mammalian mitochondrial outer membrane. Mol. Biol. Cell. 13:1615–1625. 10.1091/mbc.01-12-0570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung V., Lam S.S., Udeshi N.D., Svinkina T., Guzman G., Mootha V.K., Carr S.A., and Ting A.Y.. 2017. Proteomic mapping of cytosol-facing outer mitochondrial and ER membranes in living human cells by proximity biotinylation. eLife. 6:e24463 10.7554/eLife.24463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji W.K., Hatch A.L., Merrill R.A., Strack S., and Higgs H.N.. 2015. Actin filaments target the oligomeric maturation of the dynamin GTPase Drp1 to mitochondrial fission sites. eLife. 4:e11553 10.7554/eLife.11553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S., Tanaka A., and Fujiki Y.. 2007. Fis1, DLP1, and Pex11p coordinately regulate peroxisome morphogenesis. Exp. Cell Res. 313:1675–1686. 10.1016/j.yexcr.2007.02.028 [DOI] [PubMed] [Google Scholar]
- Koch J., and Brocard C.. 2012. PEX11 proteins attract Mff and human Fis1 to coordinate peroxisomal fission. J. Cell Sci. 125:3813–3826. 10.1242/jcs.102178 [DOI] [PubMed] [Google Scholar]
- Koch A., Yoon Y., Bonekamp N.A., McNiven M.A., and Schrader M.. 2005. A role for Fis1 in both mitochondrial and peroxisomal fission in mammalian cells. Mol. Biol. Cell. 16:5077–5086. 10.1091/mbc.E05-02-0159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korobova F., Ramabhadran V., and Higgs H.N.. 2013. An actin-dependent step in mitochondrial fission mediated by the ER-associated formin INF2. Science. 339:464–467. 10.1126/science.1228360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korobova F., Gauvin T.J., and Higgs H.N.. 2014. A role for myosin II in mammalian mitochondrial fission. Curr. Biol. 24:409–414. 10.1016/j.cub.2013.12.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumpe K., Frumkin I., Herzig Y., Rimon N., Özbalci C., Brügger B., Rapaport D., and Schuldiner M.. 2012. Ergosterol content specifies targeting of tail-anchored proteins to mitochondrial outer membranes. Mol. Biol. Cell. 23:3927–3935. 10.1091/mbc.E11-12-0994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbé K., Murley A., and Nunnari J.. 2014. Determinants and functions of mitochondrial behavior. Annu. Rev. Cell Dev. Biol. 30:357–391. 10.1146/annurev-cellbio-101011-155756 [DOI] [PubMed] [Google Scholar]
- Lam S.K., Yoda N., and Schekman R.. 2010. A vesicle carrier that mediates peroxisome protein traffic from the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 107:21523–21528. 10.1073/pnas.1013397107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.E., Westrate L.M., Wu H., Page C., and Voeltz G.K.. 2016. Multiple dynamin family members collaborate to drive mitochondrial division. Nature. 540:139–143. 10.1038/nature20555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis S.C., Uchiyama L.F., and Nunnari J.. 2016. ER-mitochondria contacts couple mtDNA synthesis with mitochondrial division in human cells. Science. 353:aaf5549 10.1126/science.aaf5549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Xu S., Roelofs B.A., Boyman L., Lederer W.J., Sesaki H., and Karbowski M.. 2015. Transient assembly of F-actin on the outer mitochondrial membrane contributes to mitochondrial fission. J. Cell Biol. 208:109–123. 10.1083/jcb.201404050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R., and Chan D.C.. 2015. The mitochondrial fission receptor Mff selectively recruits oligomerized Drp1. Mol. Biol. Cell. 26:4466–4477. 10.1091/mbc.E15-08-0591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losón O.C., Song Z., Chen H., and Chan D.C.. 2013. Fis1, Mff, MiD49, and MiD51 mediate Drp1 recruitment in mitochondrial fission. Mol. Biol. Cell. 24:659–667. 10.1091/mbc.E12-10-0721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald P.J., Stepanyants N., Mehrotra N., Mears J.A., Qi X., Sesaki H., and Ramachandran R.. 2014. A dimeric equilibrium intermediate nucleates Drp1 reassembly on mitochondrial membranes for fission. Mol. Biol. Cell. 25:1905–1915. 10.1091/mbc.E14-02-0728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manor U., Bartholomew S., Golani G., Christenson E., Kozlov M., Higgs H., Spudich J., and Lippincott-Schwartz J.. 2015. A mitochondria-anchored isoform of the actin-nucleating spire protein regulates mitochondrial division. eLife. 4:e08828 10.7554/eLife.08828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateja A., Paduch M., Chang H.Y., Szydlowska A., Kossiakoff A.A., Hegde R.S., and Keenan R.J.. 2015. Protein targeting. Structure of the Get3 targeting factor in complex with its membrane protein cargo. Science. 347:1152–1155. 10.1126/science.1261671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T., Itoh T., and Fukuda M.. 2011. Small GTPase Rab12 regulates constitutive degradation of transferrin receptor. Traffic. 12:1432–1443. 10.1111/j.1600-0854.2011.01240.x [DOI] [PubMed] [Google Scholar]
- Mishra P., and Chan D.C.. 2016. Metabolic regulation of mitochondrial dynamics. J. Cell Biol. 212:379–387. 10.1083/jcb.201511036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore A.S., Wong Y.C., Simpson C.L., and Holzbaur E.L.. 2016. Dynamic actin cycling through mitochondrial subpopulations locally regulates the fission-fusion balance within mitochondrial networks. Nat. Commun. 7:12886 10.1038/ncomms12886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunnari J., and Suomalainen A.. 2012. Mitochondria: In sickness and in health. Cell. 148:1145–1159. 10.1016/j.cell.2012.02.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osellame L.D., Singh A.P., Stroud D.A., Palmer C.S., Stojanovski D., Ramachandran R., and Ryan M.T.. 2016. Cooperative and independent roles of the Drp1 adaptors Mff, MiD49 and MiD51 in mitochondrial fission. J. Cell Sci. 129:2170–2181. 10.1242/jcs.185165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otera H., Wang C., Cleland M.M., Setoguchi K., Yokota S., Youle R.J., and Mihara K.. 2010. Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J. Cell Biol. 191:1141–1158. 10.1083/jcb.201007152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otera H., Miyata N., Kuge O., and Mihara K.. 2016. Drp1-dependent mitochondrial fission via MiD49/51 is essential for apoptotic cristae remodeling. J. Cell Biol. 212:531–544. 10.1083/jcb.201508099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer C.S., Elgass K.D., Parton R.G., Osellame L.D., Stojanovski D., and Ryan M.T.. 2013. Adaptor proteins MiD49 and MiD51 can act independently of Mff and Fis1 in Drp1 recruitment and are specific for mitochondrial fission. J. Biol. Chem. 288:27584–27593. 10.1074/jbc.M113.479873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernas L., and Scorrano L.. 2016. Mito-morphosis: Mitochondrial fusion, fission, and cristae remodeling as key mediators of cellular function. Annu. Rev. Physiol. 78:505–531. 10.1146/annurev-physiol-021115-105011 [DOI] [PubMed] [Google Scholar]
- Pitts K.R., Yoon Y., Krueger E.W., and McNiven M.A.. 1999. The dynamin-like protein DLP1 is essential for normal distribution and morphology of the endoplasmic reticulum and mitochondria in mammalian cells. Mol. Biol. Cell. 10:4403–4417. 10.1091/mbc.10.12.4403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramabhadran V., Korobova F., Rahme G.J., and Higgs H.N.. 2011. Splice variant-specific cellular function of the formin INF2 in maintenance of Golgi architecture. Mol. Biol. Cell. 22:4822–4833. 10.1091/mbc.E11-05-0457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran R. 2017. Mitochondrial dynamics: The dynamin superfamily and execution by collusion. Semin. Cell Dev. Biol. 17:30365–30368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter V., Singh A.P., Kvansakul M., Ryan M.T., and Osellame L.D.. 2015. Splitting up the powerhouse: Structural insights into the mechanism of mitochondrial fission. Cell. Mol. Life Sci. 72:3695–3707. 10.1007/s00018-015-1950-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan L., Zhou C., Jin E., Kucharavy A., Zhang Y., Wen Z., Florens L., and Li R.. 2017. Cytosolic proteostasis through importing of misfolded proteins into mitochondria. Nature. 543:443–446. 10.1038/nature21695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader M., Costello J.L., Godinho L.F., Azadi A.S., and Islinger M.. 2016. Proliferation and fission of peroxisomes—An update. Biochim. Biophys. Acta. 1863:971–983. 10.1016/j.bbamcr.2015.09.024 [DOI] [PubMed] [Google Scholar]
- Schuldiner M., Collins S.R., Thompson N.J., Denic V., Bhamidipati A., Punna T., Ihmels J., Andrews B., Boone C., Greenblatt J.F., et al. . 2005. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 123:507–519. 10.1016/j.cell.2005.08.031 [DOI] [PubMed] [Google Scholar]
- Schuldiner M., Metz J., Schmid V., Denic V., Rakwalska M., Schmitt H.D., Schwappach B., and Weissman J.S.. 2008. The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell. 134:634–645. 10.1016/j.cell.2008.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q., Yamano K., Head B.P., Kawajiri S., Cheung J.T., Wang C., Cho J.H., Hattori N., Youle R.J., and van der Bliek A.M.. 2014. Mutations in Fis1 disrupt orderly disposal of defective mitochondria. Mol. Biol. Cell. 25:145–159. 10.1091/mbc.E13-09-0525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soubannier V., Rippstein P., Kaufman B.A., Shoubridge E.A., and McBride H.M.. 2012. Reconstitution of mitochondria derived vesicle formation demonstrates selective enrichment of oxidized cargo. PLoS One. 7:e52830 10.1371/journal.pone.0052830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovic S., and Hegde R.S.. 2007. Identification of a targeting factor for posttranslational membrane protein insertion into the ER. Cell. 128:1147–1159. 10.1016/j.cell.2007.01.036 [DOI] [PubMed] [Google Scholar]
- Stojanovski D., Koutsopoulos O.S., Okamoto K., and Ryan M.T.. 2004. Levels of human Fis1 at the mitochondrial outer membrane regulate mitochondrial morphology. J. Cell Sci. 117:1201–1210. 10.1242/jcs.01058 [DOI] [PubMed] [Google Scholar]
- Sugiura A., Mattie S., Prudent J., and McBride H.M.. 2017. Newly born peroxisomes are a hybrid of mitochondrial and ER-derived pre-peroxisomes. Nature. 542:251–254. 10.1038/nature21375 [DOI] [PubMed] [Google Scholar]
- Toyama E.Q., Herzig S., Courchet J., Lewis T.L. Jr., Losón O.C., Hellberg K., Young N.P., Chen H., Polleux F., Chan D.C., and Shaw R.J.. 2016. Metabolism. AMP-activated protein kinase mediates mitochondrial fission in response to energy stress. Science. 351:275–281. 10.1126/science.aab4138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vafai S.B., and Mootha V.K.. 2012. Mitochondrial disorders as windows into an ancient organelle. Nature. 491:374–383. 10.1038/nature11707 [DOI] [PubMed] [Google Scholar]
- van der Zand A., Braakman I., and Tabak H.F.. 2010. Peroxisomal membrane proteins insert into the endoplasmic reticulum. Mol. Biol. Cell. 21:2057–2065. 10.1091/mbc.E10-02-0082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagita Y., Hiromasa T., and Fujiki Y.. 2013. Tail-anchored PEX26 targets peroxisomes via a PEX19-dependent and TRC40-independent class I pathway. J. Cell Biol. 200:651–666. 10.1083/jcb.201211077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon Y., Pitts K.R., Dahan S., and McNiven M.A.. 1998. A novel dynamin-like protein associates with cytoplasmic vesicles and tubules of the endoplasmic reticulum in mammalian cells. J. Cell Biol. 140:779–793. 10.1083/jcb.140.4.779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon Y., Krueger E.W., Oswald B.J., and McNiven M.A.. 2003. The mitochondrial protein hFis1 regulates mitochondrial fission in mammalian cells through an interaction with the dynamin-like protein DLP1. Mol. Cell. Biol. 23:5409–5420. 10.1128/MCB.23.15.5409-5420.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle R.J., and van der Bliek A.M.. 2012. Mitochondrial fission, fusion, and stress. Science. 337:1062–1065. 10.1126/science.1219855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C., Slaughter B.D., Unruh J.R., Guo F., Yu Z., Mickey K., Narkar A., Ross R.T., McClain M., and Li R.. 2014. Organelle-based aggregation and retention of damaged proteins in asymmetrically dividing cells. Cell. 159:530–542. 10.1016/j.cell.2014.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.