mTORC1 is activated by lysosome positioning and by amino acid–induced phosphatidylinositol 3-phosphate (PtdIns3P). Hong et al. show that amino acids stimulate recruitment of the PtdIns3P-binding protein FYCO1 to lysosomes and promote contacts between FYCO1 lysosomes and ER that contains the PtdIns3P effector Protrudin, mediating lysosome translocation and facilitating mTORC1 activation.
Abstract
The mechanistic target of rapamycin complex 1 (mTORC1) is a protein kinase complex that localizes to lysosomes to up-regulate anabolic processes and down-regulate autophagy. Although mTORC1 is known to be activated by lysosome positioning and by amino acid–stimulated production of phosphatidylinositol 3-phosphate (PtdIns3P) by the lipid kinase VPS34/PIK3C3, the mechanisms have been elusive. Here we present results that connect these seemingly unrelated pathways for mTORC1 activation. Amino acids stimulate recruitment of the PtdIns3P-binding protein FYCO1 to lysosomes and promote contacts between FYCO1 lysosomes and endoplasmic reticulum that contain the PtdIns3P effector Protrudin. Upon overexpression of Protrudin and FYCO1, mTORC1–positive lysosomes translocate to the cell periphery, thereby facilitating mTORC1 activation. This requires the ability of Protrudin to bind PtdIns3P. Conversely, upon VPS34 inhibition, or depletion of Protrudin or FYCO1, mTORC1-positive lysosomes cluster perinuclearly, accompanied by reduced mTORC1 activity under nutrient-rich conditions. Consequently, the transcription factor EB enters the nucleus, and autophagy is up-regulated. We conclude that PtdIns3P-dependent lysosome translocation to the cell periphery promotes mTORC1 activation.
Introduction
The mechanistic target of rapamycin (mTOR) is an evolutionarily conserved serine/threonine kinase that regulates cellular metabolism by sensing growth signals, energy levels, and nutrients such as amino acids. mTOR signaling is critical to organismal homeostasis, and its dysfunction can lead to neurodegeneration, cancer, and metabolic disease (Laplante and Sabatini, 2012; Albert and Hall, 2015; Saxton and Sabatini, 2017).
Lysosomes and late endosomes (LyLEs) play a key role in the signaling from mTOR complex (mTORC) 1, and there is an intimate relationship between mTORC1 activity and lysosomal activity (Betz and Hall, 2013; Puertollano, 2014). When nutrients and growth factors are abundant, mTORC1 facilitates cell growth and suppresses autophagic activity by direct phosphorylation and inhibition of the autophagy initiating kinase complex ULK1/ULK2 (Kim et al., 2011). At the same time, it phosphorylates and inhibits a master regulator of lysosomal activity, the transcription factor EB (TFEB; Settembre et al., 2012). When nutrients are low, the inhibitory phosphorylations are released, and lysosomal activity, such as autophagy, is induced.
LyLEs play an important role in the activation of mTORC1 by amino acids and growth factors, and they serve as signaling platforms for mTORC1 (Betz and Hall, 2013; Dibble and Cantley, 2015). Growth factors can stimulate mTORC1 on the LyLEs via the PIK3C1/AKT pathway at the plasma membrane. Amino acids are crucial to LyLE-mediated mTORC1 activation in several ways (Groenewoud and Zwartkruis, 2013; Jewell et al., 2013; Bar-Peled and Sabatini, 2014). First, they can be rapidly internalized by macropinocytosis and transported to LyLEs where they induce the activation of Rag GTPases at the LyLE membrane. This facilitates the recruitment of mTOR from the cytosol to the LyLE membrane (Sancak et al., 2008; Bar-Peled and Sabatini, 2014; Yoshida et al., 2015). Second, amino acids can act on the microspherule protein 1 to maintain the mTORC1 activator Rheb at the LyLE surface and connect Rheb to mTORC1 (Fawal et al., 2015). Third, amino acids can stimulate mTORC1 by activating the catalytic subunit of the endolysosomal class III phosphatidylinositol 3-kinase complex VPS34/PIK3C3 to produce phosphatidylinositol 3-phosphate (PtdIns3P; Byfield et al., 2005; Nobukuni et al., 2005). However, the mechanism by which PtdIns3P facilitates mTORC1 activity has remained elusive.
Given the close relationship between mTORC1 signaling and LyLEs, it is not surprising that also their intracellular position contributes to the regulation of mTORC1 activity (Korolchuk et al., 2011; Marat et al., 2017). In the presence of nutrients, LyLEs are found to localize close to the plasma membrane, keeping mTORC1 in close proximity to signaling receptors at the cell surface. In nutrient-deprived cells, LyLEs cluster perinuclearly, and this localization is known to suppress mTORC1 activity, facilitate LyLE fusion, and induce lysosomal activity such as autophagy (Korolchuk et al., 2011; Li et al., 2016; Wijdeven et al., 2016). Intracellular pH (pHi) has been implicated in nutrient-dependent LyLE translocation (Korolchuk et al., 2011), but this phenomenon is not yet fully understood.
Here we identify an unexpected connection between LyLE positioning–dependent mTORC1 activation and PtdIns3P-dependent mTORC1 activation. We show that the PtdIns3P-binding FYVE-domain proteins Protrudin and FYCO1 stimulate mTORC1 activity and down-regulate autophagy, presumably by bringing mTOR-positive LyLEs close to the plasma membrane. This process is dependent on amino acid–stimulated VPS34 activity, which implicates VPS34 in nutrient regulated LyLE positioning. Thus, our study reveals a molecular mechanism for how amino acids and VPS34 activate mTORC1, namely, through the regulation of LyLE positioning via Protrudin and FYCO1.
Results
Protrudin makes contact with mTOR and FYCO1-positive lysosomes and mediates their translocation to the cell periphery in a PtdIns3P-dependent manner
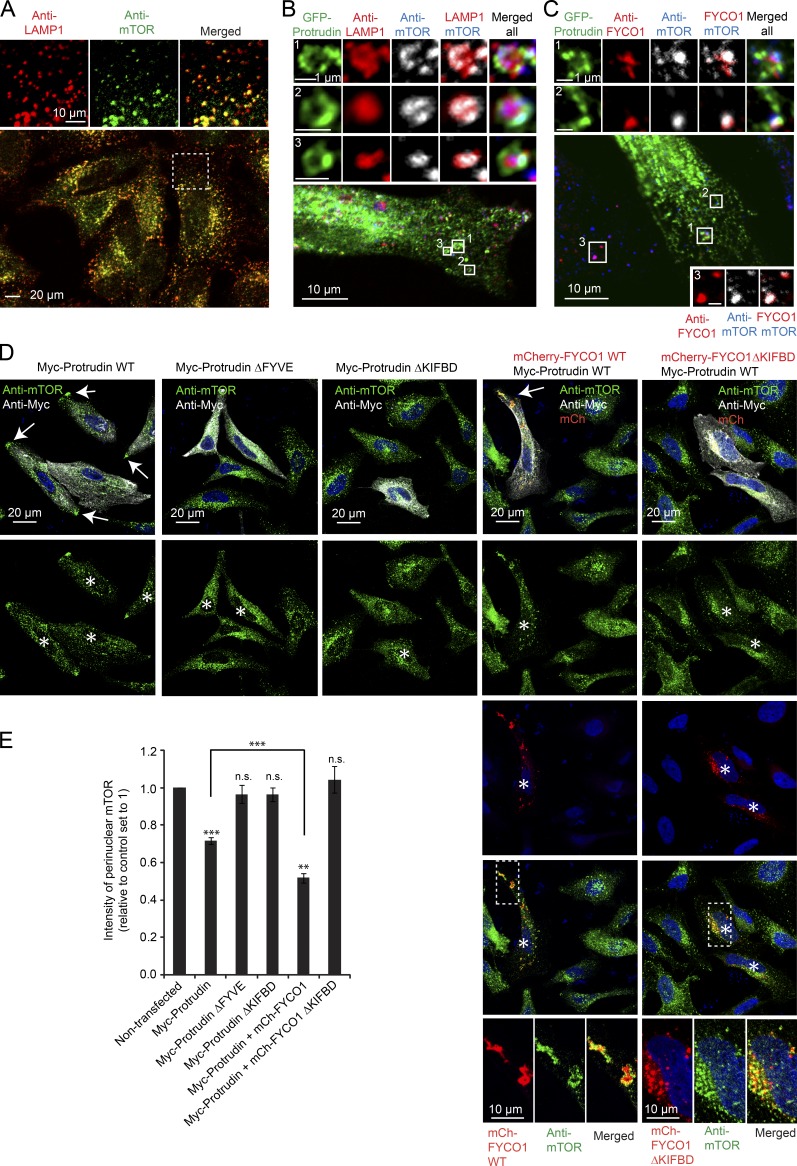
We have recently identified a PtdIns3P-dependent pathway for anterograde translocation of LyLEs (Raiborg et al., 2015). The PtdIns3P-binding FYVE-domain protein Protrudin, which is an integral protein of the ER (Chang et al., 2013), makes contact with PtdIns3P and Rab7-containing LyLEs. In such contact sites, the microtubule motor protein Kinesin-1 is transferred from Protrudin to the LyLE Kinesin-1 adaptor FYCO1, which is also a PtdIns3P-binding protein. This causes translocation of LyLEs to the cell periphery, whereas early and recycling endosomes are unaffected. Given the existence of LyLE subpopulations, we initially investigated if Protrudin could make contact with mTOR-positive LyLEs. In HeLa cells stained with antibodies against mTOR and the LyLE marker LAMP1, we observed a high degree of colocalization, indicating that a large pool of LyLEs is indeed mTOR positive (Fig. 1 A). When the cells were transiently transfected with GFP-tagged Protrudin, the mTOR-positive LyLEs were found in close proximity to, and often surrounded by, the GFP–Protrudin–positive ER (Fig. 1 B), indicating contact between the two cellular compartments. The mTOR-positive LyLEs were also positive for the Kinesin1-adaptor FYCO1 (Fig. 1 C), which cooperates with Protrudin in LyLE translocation (Raiborg et al., 2015). This colocalization was evident both in nontransfected cells and in cells expressing GFP–Protrudin, where the LyLEs were found in close proximity to the Protrudin-positive ER (Fig. 1 C). These findings suggest a possible role for Protrudin and FYCO1 in the translocation of mTOR-positive LyLEs to the cell periphery.
Figure 1.
Protrudin and FYCO1 mediate translocation of mTOR lysosomes to the cell periphery. (A) Confocal micrograph showing colocalization between endogenous LAMP1 and mTOR in HeLa cells grown in complete medium. (B and C) HeLa cells were transfected with GFP Protrudin for 20 h, stained with antibodies against LAMP1 and mTOR (B) or FYCO1 and mTOR (C), and analyzed by confocal microscopy. Insets show magnification of lysosomes in contact with Protrudin-positive ER. (D) HeLa cells were transfected (40 h) with the constructs indicated, labeled for endogenous mTOR, and analyzed by confocal microscopy. Asterisks indicate transfected cells. Arrows point at mTOR in the cell periphery. Insets show magnification of mTOR- and mCherry–FYCO1–positive lysosomes. Note that whereas mCherry–FYCO1WT colocalizes with mTOR in the cell periphery, mCherry–FYCO1ΔKIFBD colocalizes with mTOR in the perinuclear area. (E) Automated quantification of cells treated as in D, using the Olympus ScanR system. Graphs represent the relative total intensities of perinuclear localization of mTOR-positive lysosomes. The transiently transfected cells were compared directly with nontransfected neighboring cells (set to 1) in each sample. Error bars denote ± SEM from independent experiments: GFP–ProtrudinWT (n = 5), GFP–ProtrudinΔFYVE and ΔKIFBD (n = 4), and GFP–ProtrudinWT+mCh-FYCO1WT or ΔKIFBD (n = 3). **, P < 0.01; ***, P < 0.001. Myc–ProtrudinWT and Myc–ProtrudinWT+mChFYCO1WT, unpaired t test. Transfected cells versus control set to 1, one-sample t test. In total 200–800 transfected cells were analyzed per condition.
To induce Protrudin-mediated endosome translocation, HeLa cells were transfected with Myc–Protrudin alone or in combination with mCherry–FYCO1 (Fig. 1 D; Raiborg et al., 2015). In both cases, and especially the latter, the cells displayed reduced intensity of perinuclear mTOR, and mTOR-positive lysosomes were more dispersed and accumulated in the cell periphery compared with nontransfected neighboring cells (Fig. 1, D and E). The peripheral localization of mTOR–lysosomes was prevented by a FYCO1 mutant unable to bind Kinesin-1, as was also the case with a FYVE-deleted version of Protrudin, unable to bind PtdIns3P (Fig. 1, D and E). This indicates that PtdIns3P-dependent ER–endolysosome contact facilitates the translocation of mTOR–lysosomes along microtubules toward the cell periphery.
Protrudin and FYCO1 overexpression increase mTORC1 activity in a PtdIns3P-dependent manner
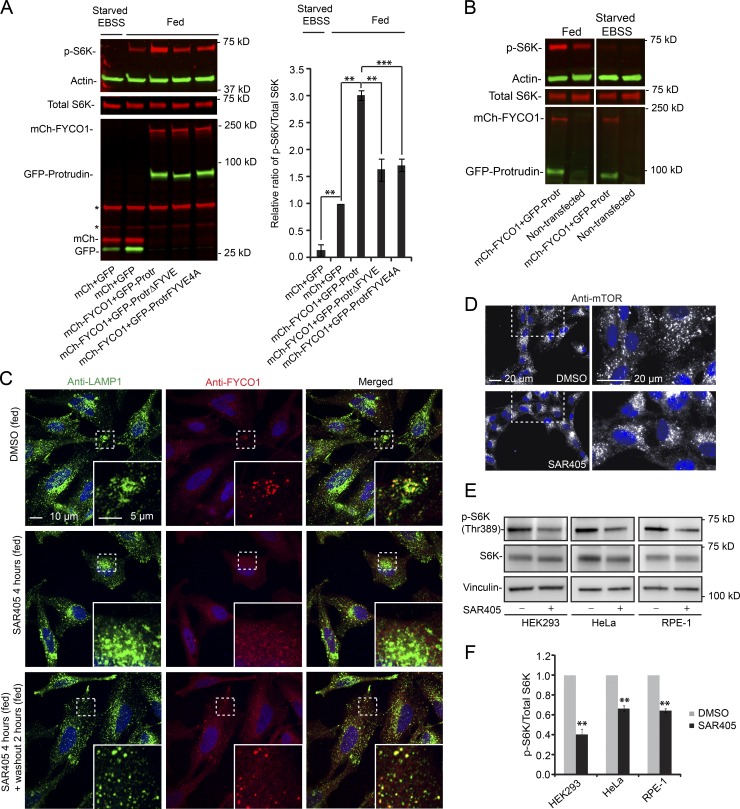
Because mTORC1 activity has been reported to be modulated by LyLE positioning (Korolchuk et al., 2011), we next investigated whether Protrudin and FYCO1-mediated translocation of mTOR-positive LyLEs could influence mTORC1 activity. To induce a strong peripheral localization of mTOR-positive LyLEs, we cotransfected HeLa cells with Protrudin and FYCO1 and monitored the level of phosphorylated S6-kinase (S6K) at the mTORC1-specific phosphosite Thr389 (Pearson et al., 1995) by immunoblotting (Fig. 2 A). As expected, when mock-transfected cells were deprived of growth factors and amino acid by using Earle’s balanced salt solution (EBSS), the level of phosphorylated S6K was strongly reduced compared with cells grown in complete medium. Interestingly, there was an increase in S6K phosphorylation in the GFP–Protrudin– and mCherry–FYCO1–cotransfected cells compared with mock-transfected cells under fed conditions (Fig. 2 A). Importantly, a functional Protrudin FYVE domain was required for the increased S6K phosphorylation, showing that the Protrudin–FYCO1–induced mTORC1 activation (similar to mTOR localization; see, for example, Fig. 1, D and E) is PtdIns3P dependent. Protrudin and FYCO1 coexpression did not facilitate S6K phosphorylation in EBSS-starved cells (Fig. 2 B), indicating that Protrudin and FYCO1 modulate the intensity of mTORC1 activity upon external signaling cues.
Figure 2.
Protrudin and FYCO1 increase S6K phosphorylation in a PtdIns3P-dependent manner. (A) HeLa cells were transfected (40 h) with GFP–Protrudin WT, ΔFYVE, or FYVE4A in combination with mCherry–FYCO1 and analyzed by immunoblotting using antibodies specific to the proteins indicated. GFP and mCherry were used as controls. Where indicated, the cells were starved for 2 h with EBSS. Asterisks indicate unspecific bands from anti-mCherry. Graph shows quantification of immunoblots, representing the relative intensity of phospho-S6K normalized to the total amount of S6K in each sample. Error bars denote ± SEM from independent experiments (n = 3). **, P < 0.01; ***, P < 0.001 (Protrudin WT vs. mutants; unpaired t test; fed control set to 1 vs. starved control or fed Protrudin WT; one-sample t test). (B) HeLa cells were cotransfected (40 h) with GFP–Protrudin WT and mCherry–FYCO1 and analyzed by immunoblotting using antibodies specific to the proteins indicated. Nontransfected cells were used as controls. Where indicated, the cells were starved for 2 h with EBSS. The immunoblot is representative of three independent experiments. (C) HeLa cells were treated with 0.1% DMSO or 3 µM SAR405 for 4 h, before washout of SAR405 for 2 h (all treatments in complete medium), stained with antibodies against LAMP1 and FYCO1, and analyzed by confocal microscopy. (D) RPE-1 cells were treated with 0.1% DMSO or 3 µM SAR405 for 4 h (all treatments in complete medium), stained with an antibody against mTOR, and analyzed by wide-field microscopy. (E) HeLa, HEK293, and RPE-1 cells were treated with 0.1% DMSO or 3 µM SAR405 in complete medium for 4 h and analyzed by immunoblotting. (F) Quantification of immunoblots (as in E), representing the relative intensities of phospho-S6K normalized to the total amount of S6K in each sample. Error bars denote ± SEM from independent experiments (n = 3). **, P < 0.01 (one-sample t test).
Because Protrudin–FYCO1–mediated anterograde LyLE translocation and S6K activation are dependent on PtdIns3P, we next addressed the effect of the PtdIns3P-producing kinase VPS34/PIK3C3 on LyLE positioning and mTORC1 activity, using the specific VSP34 inhibitor SAR405 (Ronan et al., 2014). LAMP1-positive LyLEs clustered perinuclearly in SAR405-treated HeLa cells and were redistributed after washout of SAR405, indicating that the level of PtdIns3P can influence LyLE positioning (Fig. 2 C). Importantly, the PtdIns3P-binding protein FYCO1, which localized to LAMP1-positive LyLEs in DMSO-treated cells, dissociated from the LyLEs in SAR405-treated cells and was relocalized to disperse LyLEs after washout of SAR405, consistent with a role of FYCO1 in VPS34-dependent LyLE positioning (Fig. 2 C). mTOR, on the other hand, did not dissociate from the perinuclearly localizing LyLEs in SAR405-treated cells (Fig. 2 D). As expected, VPS34 inhibition reduced the level of p-S6K in fed HEK293, RPE-1, and HeLa cells (Fig. 2, E and F), in line with the established role of VPS34 in mTORC1 activation (Byfield et al., 2005; Nobukuni et al., 2005).
siRNA-mediated depletion of Protrudin or FYCO1 reduces mTORC1 activity
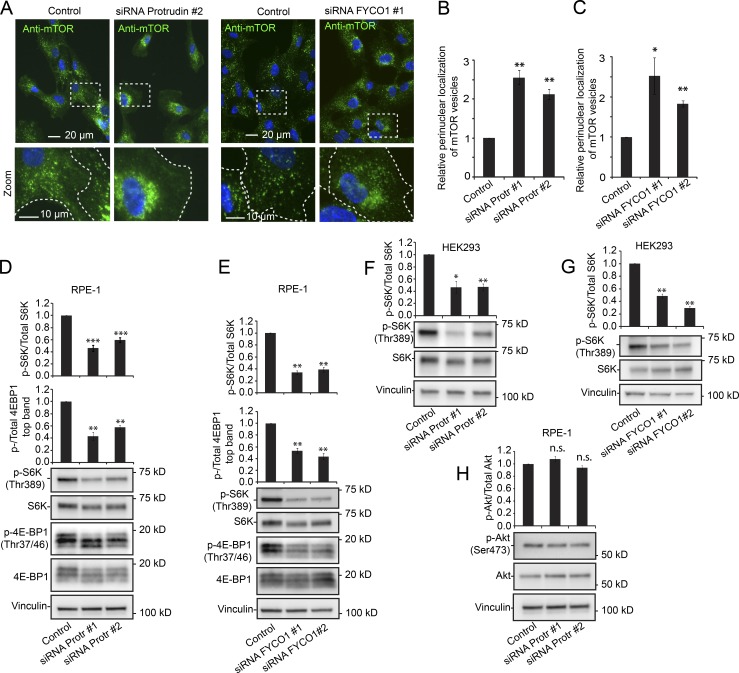
To further test the impact of Protrudin and FYCO1 on mTOR activity, we assayed the localization and activity of mTOR in Protrudin- and FYCO1-depleted RPE-1 cells (knockdown efficiency shown in Fig. S1). Protrudin or FYCO1 depletion by siRNA led, as expected, to perinuclear clustering of LAMP1- and Rab7-positive LyLEs (Fig. S1, C and F; and Fig. S2; Raiborg et al., 2015), whereas the positions of Rab5-positive early endosomes and Rab11-positive recycling endosomes remained unaffected (Fig. S2). Under such conditions, mTOR-positive LyLEs were also found to cluster perinuclearly (Fig. 3, A–C). Similar to mTOR, the mTORC1-specific subcomponent Raptor localized to LAMP1-positive LyLEs and likewise showed perinuclear clustering upon Protrudin depletion (Fig. S3 A). In contrast, the mTORC2-specific subcomponent Rictor predominantly localized to the plasma membrane (and in a few dots not colocalizing with LAMP1), and its localization was not affected by Protrudin depletion (Fig. S3 B). Overall, this indicates that Protrudin specifically regulates the positioning of mTORC1-positive LyLEs.
Figure 3.
Protrudin and FYCO1 depletion leads to perinuclear clustering of mTOR-positive lysosomes and decreases mTORC1 activity. (A) RPE-1 cells grown in complete medium were transfected with control RNA or siRNA targeting Protrudin or FYCO1, stained with an antibody to mTOR, and analyzed by high-content wide-field microscopy. (B and C) Automated quantification of images as in A using the Olympus ScanR system, representing the relative ratios of perinuclear/peripheral intensity of mTOR-positive lysosomes. Error bars denote ± SEM from independent experiments (B); n = 3; **, P ≤ 0.01 (one-sample t test); >1,400 cells per condition. (C) siRNA FYCO1 oligo 1 (n = 6), >5,000 cells. siRNA FYCO1 oligo 2 (n = 3), >900 cells. *, P < 0.05; **, P < 0.01 (one-sample t test). (D) Representative immunoblots showing the reduced levels of phosphorylated S6K and 4E-BP1 upon Protrudin depletion in RPE-1 cells grown in complete medium. Graphs represent quantifications from immunoblots. Error bars denote ± SEM from independent experiments. p-S6K (n = 6) or p-4E-BP1 (n = 4). **, P < 0.01; ***, P < 0.001 (one-sample t test). (E) Representative immunoblots showing the reduced levels of phosphorylated S6K and 4E-BP1 upon FYCO1 depletion in RPE-1 cells grown in complete medium. Graphs represent quantifications from immunoblots. Error bars denote ± SEM from independent experiments. p-S6K (n = 4) or p-4E-BP1 (n = 3). **, P < 0.01 (one-sample t test). (F) Representative immunoblots showing the reduced levels of phosphorylated S6K upon Protrudin depletion in HEK293 cells grown in complete medium. Graph represents quantifications from immunoblots. Error bars denote ± SEM from independent experiments (n = 4). *, P < 0.05; **, P < 0.01 (one-sample t test). (G) Representative immunoblots showing the reduced levels of phosphorylated S6K upon FYCO1 depletion in HEK293 cells grown in complete medium. Graph represents quantifications from immunoblots. Error bars denote ± SEM from independent experiments (n = 3). **, P < 0.01 (one-sample t test). (H) Representative immunoblots showing unaffected levels of phosphorylated AKT (mTORC2 site Ser473) upon Protrudin depletion in RPE-1 cells grown in complete medium. Graph represents quantifications from immunoblots. Error bars denote ± SEM from independent experiments (n = 4).
In line with this, in Protrudin- or FYCO1-depleted RPE-1 or HEK293 cells (knockdown efficiency shown in Fig. S1), mTORC1 activity was significantly reduced compared with control cells, as measured by decreased phosphorylation of S6K(Thr389) and 4E-BP1(Thr37/46) (Fig. 3, D–G), whereas the mTORC2-specific phosphorylation of AKT on Ser 473 was unaffected (Fig. 3 H).
Because the cells were grown in complete medium, the observed decrease in S6K activity upon Protrudin or FYCO1 depletion (Fig. 3, D–G) was less dramatic than in EBSS-starved cells (Fig. 2, A and B), and mTORC1 was still localized to LyLEs (Fig. 3 A and Fig. S3 A). Although mTORC1 deactivation often correlates with its dissociation from LyLEs (Sancak et al., 2008), deactivated mTORC1 can retain on LyLEs in the presence of mTOR inhibitors or upon mild starvation (Ohsaki et al., 2010; Korolchuk et al., 2011; Settembre et al., 2012; Marat et al., 2017), in line with our results. This suggests that Protrudin- and FYCO1-mediated LyLE positioning can regulate the level of mTORC1 activity on the LyLEs in the presence of external signaling.
Protrudin is required for nutrient-dependent lysosomal positioning and full mTORC1 activation
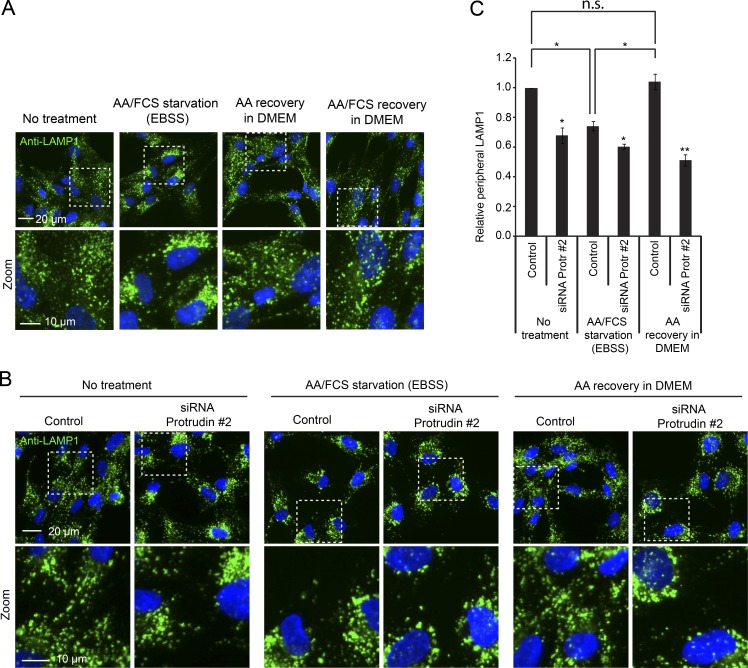
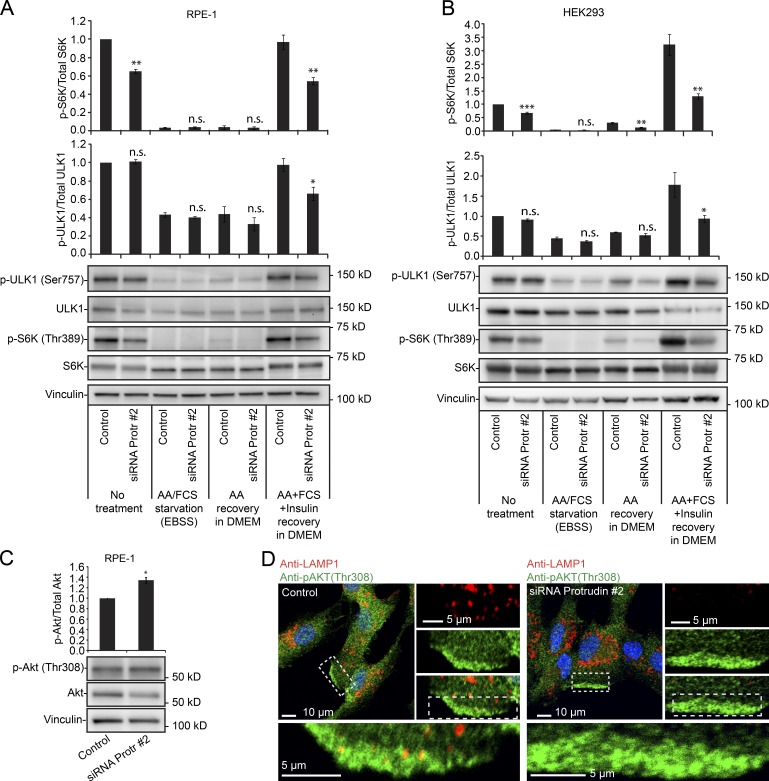
To understand this regulation in more detail, we next addressed whether Protrudin is involved in the previously reported nutrient-dependent LyLE positioning and mTORC1 regulation (Korolchuk et al., 2011). In line with the previous data, complete starvation in EBSS led to perinuclear clustering of LAMP1-positive LyLEs, and after recovery in DMEM containing amino acids alone or in combination with serum, the LyLEs were redispersed (Fig. 4 A; Korolchuk et al., 2011). Interestingly, siRNA-mediated depletion of Protrudin prevented the amino acid–induced dispersion of LyLEs after starvation (Fig. 4, B and C). This shows that Protrudin is important for LyLE redispersion after amino acid depletion–recovery, and we next asked whether Protrudin depletion would also influence the reactivation of mTORC1 after starvation. Complete starvation of RPE-1 or HEK293 cells led to a complete dephosphorylation of S6K, which recovered partially by addition of amino acids, and recovered fully upon addition of amino acids and growth factors, as expected (Fig. 5, A and B). In Protrudin-depleted cells, however, the phosphorylation of S6K did not fully recover after addition of amino acids and growth factors, which was also the case for another mTORC1 substrate, ULK1 (Fig. 5, A and B). The effect of Protrudin depletion was more evident after recovery than before starvation, especially in HEK293 cells.
Figure 4.
Protrudin is required for amino acid–dependent lysosome redispersion after starvation. (A) RPE-1 cells were grown in complete medium, starved with EBSS for 4 h, and stimulated or not with 2× amino acids (AA) in DMEM with or without FCS for 2 h, stained with an antibody to LAMP1, and imaged by wide-field microscopy. (B) RPE-1 cells grown in complete medium were transfected with control RNA or siRNA targeting Protrudin, starved with EBSS for 4 h, and stimulated or not with 1× amino acids in DMEM for 1 h, stained with an antibody to LAMP1, and imaged by high-content wide-field microscopy. (C) Automated quantification of images as in B using the Olympus ScanR system, representing the relative ratios of peripheral/perinuclear intensity of LAMP1-positive lysosomes. Error bars denote ± SEM from independent experiments (n = 3). *, P < 0.05; **, P < 0.01. Treatments versus fed control set to 1 (one-sample t test), samples with equal variance (unpaired t test). Greater than 1,900 cells were analyzed per condition.
Figure 5.
Protrudin is required for full mTORC1 activation after nutrient starvation–recovery. (A and B) RPE-1 (A) or HEK293 (B) cells grown in complete medium were transfected with control RNA or siRNA targeting Protrudin, starved in EBSS for 4 h, and stimulated or not with 1× amino acids (AA) in DMEM with or without growth factors for 2 h, before immunoblotting with the antibodies indicated. Graphs represent the relative levels of p-ULK1 or p-S6K. Error bars denote ± SEM from independent experiments. RPE-1: p-S6K (n = 3), p-ULK1 (n = 4). HEK293: p-S6K (n = 5), p-ULK1 (n = 4). *, P < 0.05; **, P < 0.01; ***, P < 0.001. No treatment (one-sample t test), samples with equal variance (unpaired t test). (C) RPE-1 cells were transfected with control RNA or siRNA targeting Protrudin. The cells were serum starved for 4 h and stimulated with FCS, insulin, and 1× amino acids in DMEM for 15 min and analyzed by immunoblotting. Error bars denote ± SEM from independent experiments (n = 3). *, P < 0.05 (one-sample t test). (D) RPE-1 cells were treated as in C, stained with antibodies against LAMP1 and p-AKT(Thr308), and analyzed by confocal microscopy.
The growth factor–dependent phosphorylation of AKT(Thr308) was not reduced by Protrudin depletion (Fig. 5 C). Because also the mTORC2 site of AKT(Ser473) was not reduced (Fig. 3 H), this shows that AKT can be fully activated in Protrudin-depleted cells, supporting the hypothesis that Protrudin may influence mTORC1 signaling by controlling LyLE positioning. Because mTORC1 can still localize to LyLEs in Protrudin-depleted cells, a plausible explanation for the reduced mTORC1 activation is that the mTORC1-positive LyLEs are kept away from the growth factor–induced signaling at the plasma membrane. In agreement with this, LAMP1-positive LyLEs were found in close proximity to phospho-AKT(Thr308)–positive regions of the plasma membrane in growth factor–stimulated control cells but failed to do so in stimulated Protrudin-depleted cells (Fig. 5 D).
Amino acids stimulate FYCO1 recruitment and ER–lysosome contacts in a VPS34-dependent manner
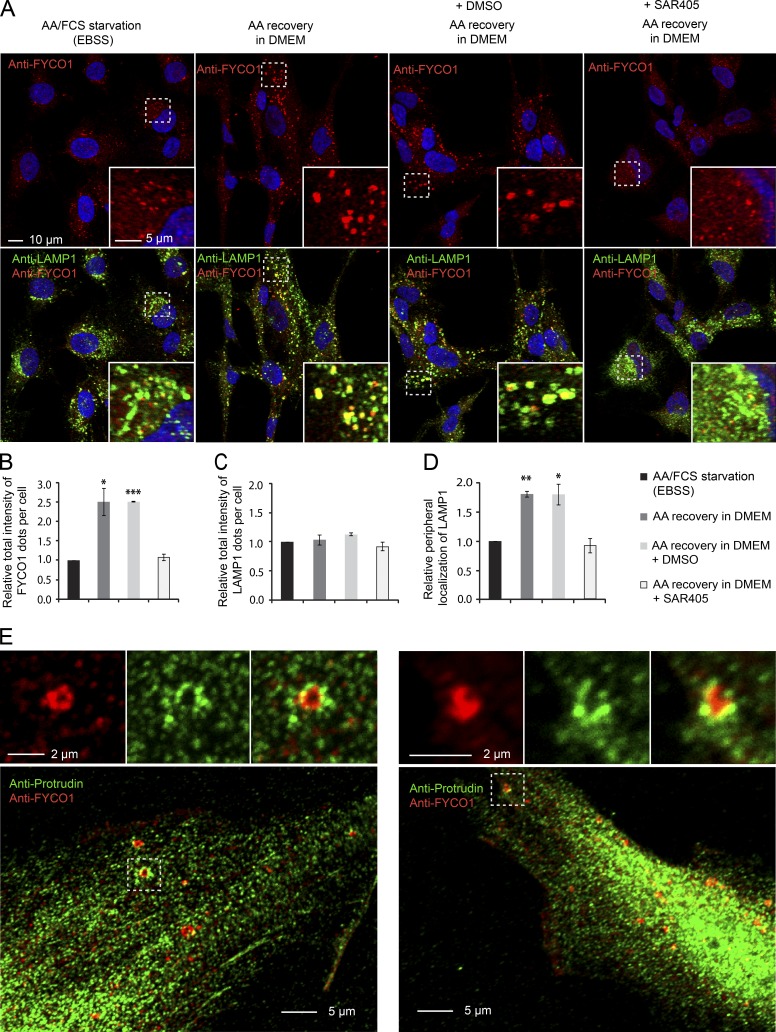
Having established a role for Protrudin in the recovery of lysosomal positioning and mTORC1 activity after full starvation and nutrient/growth factor recovery, we next asked if the PtdIns3P-dependent Protrudin–FYCO1–mediated lysosome translocation could itself be regulated by nutrients. An important clue came from previously published work showing that VPS34 is activated by the addition of amino acids and that this facilitates mTORC1 activation (Byfield et al., 2005; Nobukuni et al., 2005). Because Protrudin depletion prevented the amino acid–induced dispersion of LyLEs (Fig. 4, B and C), one possibility is that Protrudin and FYCO1 are regulated by amino acid–induced VPS34 activity on the LyLEs. If this is the case, we reasoned that the PtdIns3P-binding Kinesin-1 adaptor FYCO1 should be recruited to LyLEs after amino acid stimulation, thereby facilitating their dispersion. To test this hypothesis, we compared the amount of FYCO1 localizing on LAMP1-positive LyLEs in starved versus amino acid–stimulated cells. Indeed, we observed a strong recruitment of FYCO1 to dispersed LyLEs upon amino acid stimulation compared with starved cells, where the LyLEs clustered perinuclearly and had less FYCO1 (Fig. 6, A–D). Moreover, we could easily observe contact between endogenous Protrudin and FYCO1 in amino acid–stimulated cells by confocal microscopy (Fig. 6 E). Contacts between Protrudin in the ER and lysosomal FYCO1 are normally observed in cells overexpressing Protrudin but difficult to visualize endogenously at steady state. This suggests that Protrudin-mediated ER–LyLE contacts and FYCO1 recruitment are stimulated by amino acids, thereby facilitating anterograde LyLE translocation. Importantly, the addition of SAR405 prevented the recruitment of FYCO1 and the dispersion of LyLEs (Fig. 6, A–D), showing that FYCO1 is recruited to LyLEs in a VPS34-dependent manner upon amino acid stimulation. These results implicate VPS34 in amino acid–regulated LyLE positioning through Protrudin and FYCO1.
Figure 6.
Amino acid–stimulated lysosomal FYCO1 recruitment and dispersion. (A) RPE-1 cells were starved with EBSS for 4 h and stimulated or not for 2 h with 2× amino acids (AA) in DMEM with or without 0.1% DMSO or 3 µM SAR405, stained with antibodies against FYCO1 and LAMP1, and analyzed by confocal microscopy. Note that upon amino acid stimulation, FYCO1 is recruited to LAMP1-positive lysosomes that spread, and this is prevented by SAR405. (B–D) High-content imaging and automated analysis of cells treated as in A, using the Olympus ScanR system, representing the relative total intensity of FYCO1 (B) or LAMP1 (C) per cell or the relative ratios of peripheral/perinuclear intensity of LAMP1-positive lysosomes (D). Error bars denote ± SEM from independent experiments (n = 3). *, P < 0.05; **, P < 0.01; ***, P < 0.001 (one-sample t test). More than 1,500 cells were analyzed per condition. (E) RPE-1 cells were starved in EBSS for 4 h and stimulated for 2 h with 2× amino acids in DMEM, stained with antibodies against FYCO1 and Protrudin, and analyzed by confocal microscopy.
Protrudin and FYCO1 depletion induces nuclear translocation of TFEB
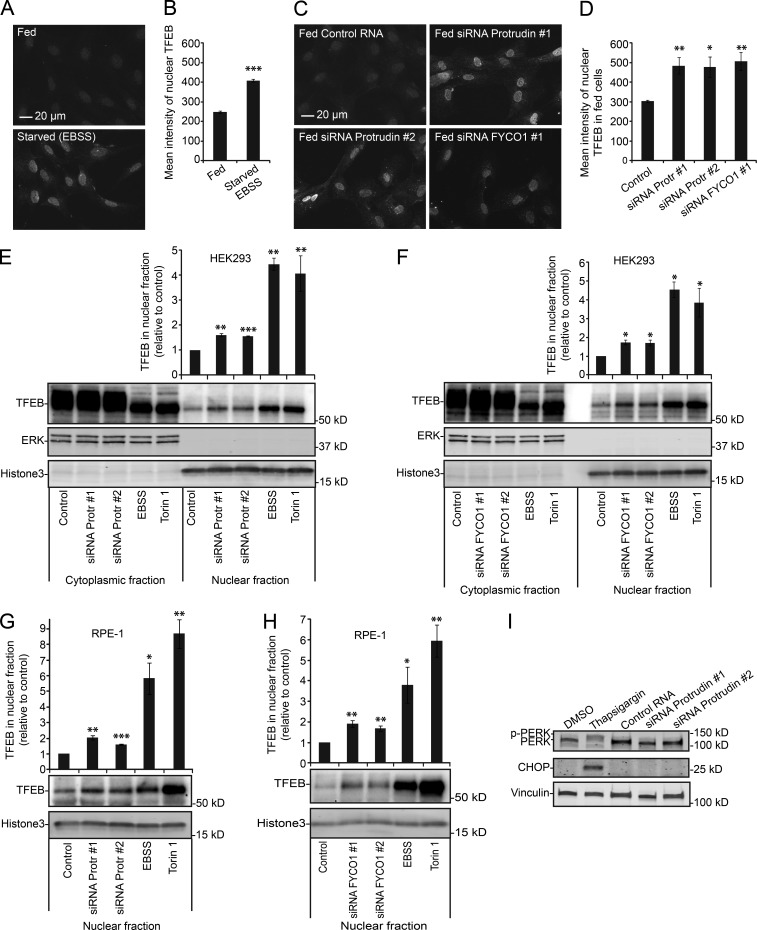
It is well established that mTORC1 deactivation induces dephosphorylation and nuclear translocation of TFEB (Roczniak-Ferguson et al., 2012; Settembre et al., 2012; Napolitano and Ballabio, 2016), and we therefore examined whether Protrudin and FYCO1 depletion could influence such translocation of TFEB. We first measured the mean intensity of endogenous TFEB in nuclei by high-content immunofluorescence microscopy of RPE-1 cells, and as expected, there was an increase in nuclear TFEB intensity in starved cells compared with cells grown in complete medium (Fig. 7, A and B). In fed cells depleted of Protrudin or FYCO1 by siRNA, there was also an increase in the intensity of nuclear TFEB (Fig. 7, C and D), consistent with the observed reduction in mTORC1 activity. We verified the data obtained by microscopy by subcellular fractionation and measurements of the amounts of TFEB in cytoplasmic and nuclear fractions by immunoblotting in HEK293 and RPE-1 cells. As expected, mTOR inhibition by Torin1 (Thoreen et al., 2009) or starvation caused a nuclear translocation of the lower molecular weight band of TFEB in the nuclear fraction compared with control-treated cells, representing the dephosphorylated form of TFEB (Settembre et al., 2012; Fig. 7, E–H). In Protrudin- or FYCO1-depleted cells, we observed a small increase in the amount of nuclear TFEB in both cell lines, consistent with a partial reduction in mTORC1 activity. Collectively, the data indicate that a fraction of the mTORC1 substrate TFEB is dephosphorylated in Protrudin- or FYCO1-depleted cells and can thus enter the nucleus in the presence of nutrients.
Figure 7.
TFEB enters the nucleus in Protrudin- and FYCO1-depleted cells. (A) RPE-1 cells were grown in complete medium or starved in EBSS for 2 h, stained with an antibody to TFEB, and analyzed by high-content wide-field microscopy. (B) Automated quantification of images as in A of the mean intensity of nuclear TFEB using the Olympus ScanR system. Error bars denote ± SEM from independent experiments (n = 3). ***, P < 0.001 (unpaired t test). More than 1,700 cells were analyzed per condition. (C) RPE-1 cells grown in complete medium were transfected with control RNA or siRNA targeting Protrudin or FYCO1, stained with an antibody to TFEB, and analyzed by high-content wide-field microscopy. (D) Automated quantification of images as in C, of the mean intensity of nuclear TFEB using the Olympus ScanR system. Error bars denote ± SEM from independent experiments (n = 3). *, P < 0.05; **, P < 0.01 (unpaired t test). More than 1,200 cells were analyzed per condition. (E) HEK293 cells were transfected with control or Protrudin siRNA or treated with 500 nM Torin1 (1 h) or EBSS (2 h). Cytoplasmic and nuclear fractions were then prepared followed by immunoblotting analysis using indicated antibodies. ERK1/2 and histone H3 were used as the loading control for cytoplasmic and nuclear fractions, respectively. Graphs represent quantifications of TFEB levels in nuclear fractions from immunoblots. TFEB levels were normalized to histone H3 and presented relative to control. Error bars denote ± SEM from independent experiments (n = 3). **, P < 0.01; ***, P = 0.001 (one-sample t test). (F) HEK293 cells were transfected with control or FYCO1 siRNA or treated with Torin1 (1 h) or EBSS (2 h). Separation of cytoplasmic and nuclear fractions and subsequent immunoblotting were performed as described in E. Graphs represent quantifications of TFEB levels in nuclear fractions from immunoblots. TFEB levels were normalized to histone H3 and presented relative to control. Error bars denote ± SEM from independent experiments (n = 3). *, P < 0.05 (one-sample t test). (G) RPE-1 cells were treated as in E. Nuclear fractions were subjected to immunoblotting analysis with indicated antibodies. Graphs represent quantifications of TFEB levels in nuclear fractions from immunoblots. TFEB levels were normalized to histone H3 and presented relative to control. Error bars denote ± SEM from independent experiments (n = 5). *, P < 0.05; **, P < 0.01 (one-sample t test). (H) RPE-1 cells were treated as in F. Nuclear fractions were analyzed using immunoblotting with the indicated antibodies. Graphs represent quantifications of TFEB levels in nuclear fractions from immunoblots. TFEB levels were normalized to histone H3 and presented relative to control. Error bars denote ± SEM from independent experiments (n = 4). *, P < 0.05; **, P < 0.01; ***, P < 0.001 (one-sample t test). (I) Protrudin depletion does not induce ER stress. RPE-1 cells were transfected with control RNA or siRNA targeting Protrudin and analyzed with immunoblotting using antibodies against the ER stress–induced proteins indicated. Thapsigargin was used to induce ER stress in nontransfected cells.
The unfolded protein response is a cellular program that senses various types of cellular stress, including ER stress (Hetz, 2012). Protrudin is a transmembrane ER protein and has a role in ER shaping (Chang et al., 2013; Hashimoto et al., 2014). A Protrudin point mutant, found in a subset of patients with hereditary spastic paraplegia, has been reported to cause increased susceptibility to ER stress (Hashimoto et al., 2014). Because ER stress can influence mTORC1 activity (Di Nardo et al., 2009; Appenzeller-Herzog and Hall, 2012) and TFEB has been shown to enter the nuclei upon ER stress (Martina et al., 2016), we tested whether Protrudin depletion by siRNA could induce ER stress. Thapsigargin, which inhibits the Ca2+ATPase in the ER membrane (Thastrup et al., 1990), was used as a positive control. In cells treated with Thapsigargin, there was a significant mobility shift of the ER resident kinase PERK by immunoblotting, characteristic of its hyperphosphorylation upon ER stress (Harding et al., 1999; Fig. 7 I). Accordingly, a strong elevation of the ER stress-induced protein CHOP (Wang et al., 1996) was observed. Importantly, neither phospho-PERK nor CHOP could be detected in Protrudin-depleted cells, confirming that ER stress was not induced. This indicates that the role of Protrudin in the regulation of mTORC1 and TFEB activity is likely to be mediated by its function in LyLE translocation.
Protrudin depletion triggers autophagy
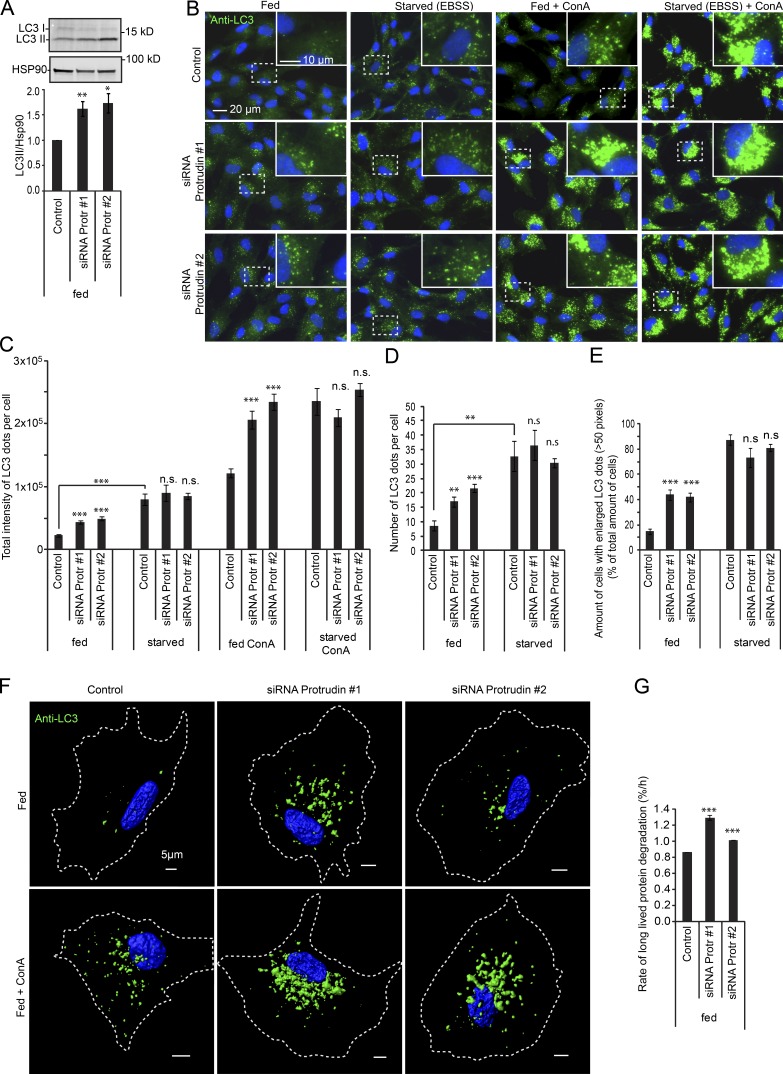
One of the lysosomal activities initiated by loss of mTORC1 activity is autophagy, which is triggered by dephosphorylation of ULK1 and nuclear translocation of TFEB (Napolitano and Ballabio, 2016). We therefore investigated whether Protrudin depletion would cause an up-regulation of autophagy. As a first indication, the level of lipidated LC3 (LC3-II) was measured in fed cells using immunoblotting (Klionsky et al., 2016), and we could indeed observe an increased amount of LC3-II in cells depleted of Protrudin by siRNA compared with control cells (Fig. 8 A). The increase in LC3-II might suggest an increased formation of autophagosomes, consistent with the observed reduction in mTORC1 activity. Alternatively, it could reflect a reduced clearance of LC3 due to impaired autophagosome maturation. To distinguish between these mechanisms, we stained cells with anti–LC3 antibodies and measured the total fluorescence intensity of LC3 dots per cell using high content microscopy in the presence and absence of Concanamycin A (ConA), which blocks acidification of autolysosomes and thereby halts LC3 degradation (Klionsky et al., 2016). Upon starvation of control cells, there was, as expected, a significant increase in LC3 dot intensity compared with cells grown in complete medium, which was further increased in the presence of ConA, consistent with increased autophagosome formation (Fig. 8, B and C). An increase in LC3 intensity was also observed in fed cells depleted of Protrudin by siRNA, in line with the increased level of LC3-II observed by immunoblotting. Importantly, in Protrudin-depleted fed cells, the intensity of LC3 was strongly increased in the presence of ConA, consistent with a role in autophagosome formation (Fig. 8, B and C). Furthermore, Protrudin knockdown did not increase LC3 intensity relative to the control in starved cells (Fig. 8, B and C), when the mTORC1 activity was already fully suppressed. This indicates that Protrudin depletion leads to the initiation of autophagy in cells grown in complete medium, consistent with the observed reduction in mTORC1 activity in fed cells. In line with the increase in LC3 dot intensity, the number and size of the LC3 dots were also increased upon Protrudin or FYCO1 knockdown in fed cells (Fig. 8, D–F; and Fig. S4).
Figure 8.
Protrudin depletion promotes initiation of autophagy. (A) Immunoblot showing an increased level of LC3-II in fed RPE-1 cells transfected with siRNA targeting Protrudin compared with control. The graph represents the relative level of LC3-II normalized to the loading control HSP90 quantified from immunoblots as in A. Error bars denote ± SEM from independent experiments (n = 5). *, P < 0.02; **, P < 0.01 (one-sample t test). (B) RPE-1 cells were siRNA transfected and either grown in complete medium or starved for 2 h in EBSS. ConA was given for 2 h where indicated. The cells were stained with an antibody to LC3 and analyzed by high-content wide-field microscopy. (C) Automated quantification of images as in B of the total intensity of LC3 dots per cell using the Olympus ScanR system. Error bars denote ± SEM from independent experiments (n = 4). ***, P < 0.001 (unpaired t test). More than 1,400 cells were analyzed per condition. (D) Automated quantification of images as in B of the number of LC3 dots per cell using the Olympus ScanR system. Error bars denote ± SEM from independent experiments (n = 4). **, P < 0.01; ***, P < 0.001 (unpaired t test). More than 1,400 cells were analyzed per condition. (E) Automated quantification of images as in B of the amount of cells having LC3 dots with size >50 pixels, using the Olympus ScanR system. Error bars denote ± SEM from independent experiments (n = 4). **, P < 0.01; ***, P < 0.001 (unpaired t test). More than 1,400 cells were analyzed per condition. (F) Imaris representations of confocal z-stacks visualizing the number and size of LC3 positive dots in control- or siRNA-treated RPE-1 cells grown in complete medium with or without ConA. (G) Quantification of the rate of LLPD in control and siRNA-treated RPE-1 cells. Error bars denote ± SEM from independent experiments (n = 3). ***, P < 0.001 (unpaired t test).
Measuring the rate of long-lived protein degradation (LLPD) is another way to quantitatively monitor autophagy, and it has the advantage of measuring the actual autophagic degradation activity (Klionsky et al., 2016). To confirm the effect of Protrudin in basal autophagy, the rate of LLPD was measured in fed RPE-1 cells. Consistent with the observed increase in autophagosome formation, the rate of basal LLPD increased significantly in cells depleted of Protrudin by siRNA compared with control cells (Fig. 8 G). In summary, our data show that Protrudin-depleted cells have increased autophagic activity in cells grown in complete medium, consistent with the observed reduction in mTORC1 activity. Thus, Protrudin levels can influence the rate of autophagy in the presence of nutrients.
Discussion
It has remained a paradox that the PIK3C3 VPS34 has both a positive and negative role in autophagy induction. On one hand, VPS34 produces PtdIns3P, which is crucial for autophagosome biogenesis (Petiot et al., 2000; Ktistakis et al., 2012). On the other hand, VPS34 also produces PtdIns3P, which activates mTORC1 and thereby suppresses autophagy (Byfield et al., 2005; Nobukuni et al., 2005; Ktistakis et al., 2012). Our identification of two LyLE-binding PtIns3P effectors in mTORC1 activation offers a solution to this paradox. VPS34 exists in different subcomplexes depending on its cellular function (Kihara et al., 2001). Whereas autophagy is regulated by VPS34/VPS15/Beclin1 in combination with ATG14 (Itakura et al., 2008), endosomal activities use UVRAG and BIF1 (Thoresen et al., 2010). Our finding that PtdIns3P-dependent LyLE translocation facilitates mTORC1 activation suggests that the UVRAG-containing endosomal VPS34 complex is responsible for mTORC1 activation. Interestingly, two mTOR-specific phosphorylation sites in UVRAG were recently discovered and shown to be important for VPS34 activation, suggesting a positive feedback loop for mTORC1 activation (Munson et al., 2015).
LyLE positioning influences mTORC1 activity, and it has been suggested that activation of mTORC1 is enhanced because of close contact with nutrient signaling complexes in the plasma membrane (Wullschleger et al., 2006; Korolchuk et al., 2011). This is reinforced by our data, showing that Protrudin and FYCO1, which are regulators of LyLE positioning, modulate mTORC1 activity and regulate the position of mTORC1-positive LyLEs relative to phospho-AKT-positive regions near the plasma membrane.
Because LyLE translocation depends on microtubule transport, mTORC1 activity is likely to be influenced by microtubule stability. Indeed, there are reports showing that mTORC1 cannot be fully activated in the presence of Nocodazole or Taxol, which disrupt microtubule dynamics, and that depletion of microtubule motors by siRNA reduces mTORC1 activity (Korolchuk et al., 2011; Li and Guan, 2013). This is in line with our data, in which Kinesin-1 binding–deficient mutants of Protrudin or FYCO1 fail to translocate mTOR-positive LyLEs to the cell periphery. There are, however, other reports showing that mTORC1 activity does not change or increases upon Nocodazole or Taxol treatment (Calastretti et al., 2001; Fawal et al., 2015). The discrepancy between these results could indicate cell type– and context-dependent regulation, which would require future investigation.
Several alternative molecular pathways for LyLE translocation exist that use microtubule-dependent transport (Rocha et al., 2009; Rosa-Ferreira and Munro, 2011; Uusi-Rauva et al., 2012; Pu et al., 2015; Raiborg et al., 2015; Li et al., 2016). Interestingly, only the Protrudin–FYCO1 pathway is dependent on PtdIns3P (Raiborg et al., 2015). We show here that if VPS34 activity or PtdIns3P binding is perturbed in fed cells, translocation of LyLEs to the cell periphery cannot occur, and indeed we observed impaired mTORC1 activation under such conditions. The dependency on PtdIns3P is of special interest in the case of mTORC1 activation, because amino acids and insulin are known to activate mTORC1 through the activation of VPS34 (Byfield et al., 2005; Nobukuni et al., 2005; Gulati et al., 2008; Hirsch et al., 2014; Mohan et al., 2016). However, the molecular mechanism for how VPS34 activity can influence mTORC1 has been elusive. Here we have identified the two PtdIns3P-binding proteins Protrudin and FYCO1 in the activation of mTORC1, providing new molecular insight into this long-standing question of how VPS34 can stimulate mTORC1 activity. Of note, Vps34 is not required for mTORC1 activation in Drosophila melanogaster (Juhász et al., 2008), which lacks a Protrudin counterpart. Protrudin is exclusively expressed in vertebrates, suggesting that this level of mTORC1 activation has evolved in highly complex organisms.
In cells grown under nutrient-rich conditions, LyLEs are distributed more peripherally than in nutrient-deprived cells, in which LyLEs cluster perinuclearly (Korolchuk et al., 2011; Uusi-Rauva et al., 2012; Li et al., 2016), a phenotype also observed by us. The reason for the nutrient-dependent lysosome translocation is not clear, but nutritional status has been shown to affect pHi (Korolchuk et al., 2011). It has been long known that pHi regulates lysosome positioning (Heuser, 1989; Parton et al., 1991), and changes in pHi have been shown to affect the association of lysosomes with the microtubule motor Kinesin-2 as well as with the GTPase ARL8B, which is involved in Kinesin-1–dependent LyLE translocation (Korolchuk et al., 2011). Thus the Protrudin–FYCO1 pathway, which translocates LyLEs using Kinesin-1 (Raiborg et al., 2015), could be susceptible to regulation by nutrient-sensitive changes in pHi. Even though nutrient-sensitive LyLE positioning can be partially dependent on pHi-sensitive microtubule transport, we have shown here that amino acid–induced VPS34 activation stimulates the recruitment of FYCO1 to LyLEs, facilitating Protrudin–FYCO1–mediated ER–LyLE contact and anterograde LyLE translocation. Thus, Protrudin and FYCO1 serve as key regulators of nutrient-dependent LyLE positioning, providing an explanation for the role of amino acids in VPS34-induced mTOR activation and at the same time amino acid–dependent LyLE positioning.
The LyLE-localizing mTORC1 complex phosphorylates a variety of proteins involved in cellular metabolism (Laplante and Sabatini, 2012; Albert and Hall, 2015). We show here that Protrudin and FYCO1 affect the ability of mTORC1 to phosphorylate S6K, 4E-BP1, and ULK1 in several cell lines. Moreover, we find that Protrudin–FYCO1 depletion in fed cells allows a fraction of the transcription factor TFEB to enter the nucleus, presumably by releasing its mTORC1-dependent inhibitory phosphorylation. Consistent with this, we observe an increased onset of autophagy in Protrudin-depleted cells under high-nutrient conditions. FYCO1 has been assigned a role in autophagy (Pankiv et al., 2010; Olsvik et al., 2015), and we could observe increased levels of LC3 in FYCO1-depleted cells. mTORC1 exerts an inhibitory phosphorylation of the autophagy-inducing kinase complex ULK1/ULK2 (Kim et al., 2011). It is likely that the reduced phosphorylation of ULK1 observed in Protrudin-depleted cells contributes to the increased autophagic activity detected. Under high-nutrient conditions, the autophagic pathway plays a crucial role in selective removal of damaged organelles and renewal of macromolecules (Stolz et al., 2014). Because we find that the expression level of Protrudin can influence autophagic activity in fed cells, Protrudin might fine-tune the rate of autophagy under basal conditions.
In the future it will be important to study other physiological roles of LyLE positioning in the fine-tuning of mTORC1 signaling. The regulation of mTORC1 activity mediated by Protrudin and FYCO1 can occur only in the presence of nutrients and growth factors. Consequently, Protrudin and FYCO1 are not able to activate mTORC1 by themselves but instead facilitate full activation in the presence of nutrient and growth factor signaling, which activates mTORC1 directly. Although LyLE positioning as such does not trigger mTORC1 activation, it is likely to modulate context dependent signaling output in complex organisms.
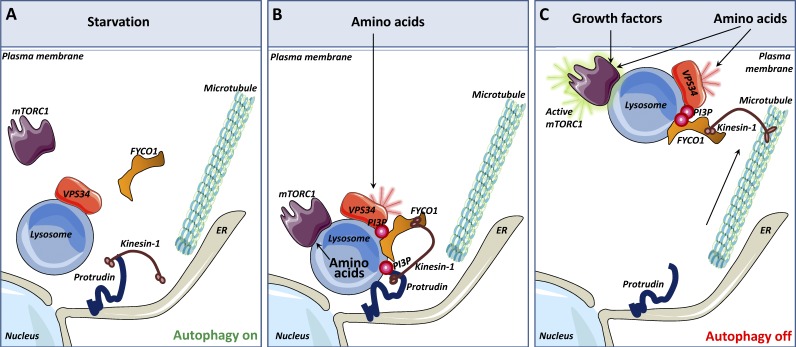
The presence of nutrients is likely to influence Protrudin- and FYCO1-dependent mTORC1 activation in the following way (Fig. 9): Amino acids stimulate PtdIns3P production through the activation of VPS34 (Byfield et al., 2005; Nobukuni et al., 2005). This facilitates the formation of Protrudin-containing ER–LyLE contact sites and promotes FYCO1-mediated LyLE translocation along microtubules in a Kinesin-1-dependent manner toward the plasma membrane. There, the activity of the LyLE-localizing mTORC1 is enhanced because of the close proximity to nutrients and growth factor signaling complexes at the plasma membrane.
Figure 9.
Model for Protrudin- and FYCO1-mediated mTOR activation. (A) Under nutrient starvation conditions, lysosomes localize perinuclearly, mTORC1 is predominantly cytosolic, and VPS34 activity on LyLEs is low. The lack of PtdIns3P prevents Protrudin-mediated ER–endosome contacts. (B) The presence of amino acids stimulates the recruitment of the mTORC1 complex to lysosomes. In addition, amino acids activate VPS34 to produce PtdIns3P at the lysosomal membrane. The PtdIns3P-binding protein FYCO1 is recruited to lysosomes, and Protrudin can mediate PtdIns3P-dependent ER–lysosome contact. In such contact sites, the microtubule motor Kinesin-1 is transferred from Protrudin to FYCO1. (C) Lysosomes loaded with Kinesin-1 are translocated along microtubules to the cell periphery. This brings the lysosomal mTORC1 complex in close apposition to nutrient signaling complexes at the plasma membrane and increases mTORC1 activity.
Materials and methods
Antibodies
Antibodies were obtained from the following resources: mouse anti-LAMP1 (H4A3) was from the Developmental Studies Hybridoma Bank. Rabbit anti-Protrudin (12680-1-AP) was from PTG. Mouse anti-FYCO1 (A01; H00079443-A01) was from Abnova (immunofluorescence and Western blotting). Rabbit anti-LC3 (PM036) was from MBL. Anti-Rab7 (D95F2; 9367), anti-ULK1 (D8H5; 8054), anti-phospho-ULK1 (Ser757; 6888), anti-S6K (9202), anti-phospho-p70 S6K (Thr389; 9205), anti-4E-BP1 (53H11; 9644), anti-phospho-4E-BP1 (Thr37/46; 236B4; 2855), anti-TFEB (4240), anti-Akt (9272), anti-phospho-Akt (Thr308; 9275), anti-phospho-Akt (Ser473; 9271), anti-PERK (D11A8; 5683; gift from N. Engedal, University of Oslo, Oslo, Norway), and anti-Erk1/2 antibody (9102) were rabbit antibodies from Cell Signaling Technology. Mouse anti-CHOP (L63F7; 2895; gift from N. Engedal) was from Cell Signaling Technology. Rabbit anti-Raptor (09-217) was from Merck Millipore. Mouse anti-Rictor (A500-002A) was from Bethyl Laboratories. Rabbit anti–histone H3 (ab1791), goat anti-TFEB (ab122910), mouse anti-GAPDH (ab9484), and rabbit anti-FYCO1 (ab126603; Western blotting) were from Abcam. Mouse anti-Hsp90 (610419) was from BD Biosciences. Rabbit anti-LAMP1 (L1418), mouse anti–β-actin (A5316), and mouse anti-vinculin (V9131) were from Sigma-Aldrich. Mouse anti-Rab5 (4F11) was a gift from C. Bucci (University of Salento, Lecce, Italy). Rabbit anti-Rab11 (715300) was from Zymed Lab. Secondary antibodies (donkey anti–mouse or rabbit Alexa Fluor 488, 555, 568, or 647) were from Jackson Immunoresearch or Molecular Probes, and IRDye680 and IRDye800 were from Odyssey.
Cell culture
Cell lines were grown according to American Type Culture Collection. HeLa and HEK293 cells were maintained in DMEM (D0819; Sigma-Aldrich) supplemented with 10% FCS (F7524; Sigma-Aldrich), 2 mM glutamine (25030-024; Thermo Fisher Scientific), 100 U/ml penicillin, and 100 µg/ml streptomycin at 37°C with 5% CO2. hTERT-RPE-1 cells were grown in DMEM/F12 (31331-028; GIBCO BRL), 10% FCS, and 0.01 mg/ml hygromycin B. For starvation experiments, the growth medium was removed and replaced with EBSS (24010-043; GIBCO BRL). For amino acid stimulation, 1× or 2× amino acids (from 50× amino acid solution; M5550; Sigma-Aldrich) were added to DMEM/F12 or DMEM with or without FCS. The working concentration was for ConA (C9705; Sigma-Aldrich), 100 nM; for Thapsigargin (CAS 67526-95-8; Calbiochem), 100 nM (3 h); for SAR405 (A8883; APEXBIO), 3 µM; for DMSO (D2650; Sigma-Aldrich), 0.1%; and for insulin (I9278; Sigma-Aldrich), 200 nM.
siRNA transfections
Cells were seeded and transfected with siRNA duplexes using Lipofectamine RNAiMax transfection reagent (13778; Thermo Fisher Scientific) following the manufacturer’s instructions. The final concentration of siRNA duplexes was 50 nM. Cells were analyzed 48 h after transfection. The following siRNA targeting sequences for human Protrudin were used. Oligo 1: 5′-CUUCUUGAUCCAGCUGGAGG-3′ (Invitrogen; Raiborg et al., 2015); oligo 2: 5′-AGAAUGAGGUGCUGCGCAG-3′ (J-016349-12-0020; GE Dharmacon). siRNA targeting human FYCO1 oligo 1 was 5′-GGGCCGGCAAGAACCUAUUTT-3′ (Invitrogen; Pankiv et al., 2010); oligo #2: 5′-CCAGUGACUGGUACUAUGC-3′ (AM16708; Thermo Fisher Scientific). Nontargeting control siRNA (D-001810-01-20) was purchased from Dharmacon.
Plasmid transfection
All plasmids were generated by standard molecular biological methods. Human mCherry–FYCO1 was a gift from T. Johansen’s group (University of Tromsø, Tromsø, Norway; Pankiv et al., 2010). Protrudin plasmids were generated by PCR using the cDNA clone BC030621 from ImaGene encoding the canonical human isoform 1 of Protrudin (411 amino acids; Raiborg et al., 2015). The FYVE4A mutant of Protrudin was a gift from W.D. Heo (Korea Advanced Institute of Science and Technology, Daejeon, South Korea). FuGENE6 (E2692; Promega) was used for transient transfection in HeLa cells.
Quantitative real-time PCR of mRNA expression
mRNA expression analysis was done as previously described (Pedersen et al., 2012). In brief, quantitative real-time PCR was performed using QuantiTect SYBR Green PCR kit (QIAGEN), cDNA template, and QuantiTect primers. Reactions were done and analyzed using the Lightcycler 480 (Roche). Cycling conditions were 15 min at 95°C followed by 45 cycles of 15 s at 94°C, 30 s at 55°C, and 30 s at 72°C. Gene amplification was normalized to the expression of TATA-box-binding protein (TBP). The relative quantification of the gene expression was calculated using the ΔΔCt method. QuantiTect primer against ZFYVE27 (QT00088697) and TBP (QT00000721) were purchased from QIAGEN.
Immunoblotting
Cells were washed with ice-cold PBS and lysed with 25 mM Hepes, pH 7.2 (H4034; Sigma-Aldrich), 125 mM potassium acetate (104820; Merck Millipore), 2.5 mM magnesium acetate (105819; Merck Millipore), 5 mM EGTA (E3889; Sigma-Aldrich), 0.5% NP-40 (I18896; Sigma-Aldrich), and 1 mM DTT (D0632; Sigma-Aldrich) supplemented with protease inhibitor cocktail (P9340; Sigma-Aldrich). Cell lysates were then subjected to SDS-PAGE on 10% (567-1034; Bio-Rad) or 4%–20% (567-1094; Bio-Rad) gradient gels and blotted onto Immobilon-P membranes (IPVH00010; Merck Millipore). Membranes incubated with fluorescently labeled secondary antibodies (IRDye680 and IRDye800; LI-COR) were developed by Odyssey infrared scanner (LI-COR). Membranes detected with HRP-labeled secondary antibodies were developed using Clarity Western ECL substrate solutions (Bio-Rad) with a ChemiDoc XRS+ imaging system (Bio-Rad). Quantification of immunoblots was performed using the Odyssey, Volume tools of Image Lab (Bio-Rad), or ImageJ software (National Institutes of Health).
Immunostaining
Cells were seeded on glass coverslips, fixed with 3% formaldehyde (18814; Polysciences) for 15 min on ice, and permeabilized with 0.05% saponin (S7900; Sigma-Aldrich) in PBS. Fixed cells were then stained with primary antibody at room temperature for 1 h, washed in PBS/saponin, stained with fluorescently labeled secondary antibody for 1 h, washed in PBS, and mounted with Mowiol containing 2 µg/ml Hoechst 33342 (H3570; Thermo Fisher Scientific). For the detection of lysosomal mTOR or LC3 and nuclear TFEB, the cells were permeabilized for 5 min on ice with 0.05% saponin in PEM (0.1 M Pipes, pH 6.95 [P7643; Sigma-Aldrich], 2 mM EGTA, and 1 mM MgSO4; 105886; Merck Millipore) buffer before fixation, to decrease the fluorescent signal from the cytosolic pool of the proteins. Before immunostaining with anti-TFEB, the cells were treated with 0.5% Triton X-100 in PBS for 10 min to permeabilize the nuclear membrane.
Confocal fluorescence microscopy
Confocal images were obtained using LSM710 or LSM780 confocal microscope (Carl Zeiss) equipped with an Ar-laser multiline (458/488/514 nm), a DPSS-561 10 (561 nm), a laser diode 405-30 CW (405 nm), and an HeNe laser (633 nm). The objective used was a Plan-Apochromat 63×/1.40 oil DIC III (Carl Zeiss). ZEN 2010 (Carl Zeiss), Photoshop CS4 (Adobe), and ImageJ software were used for brightness and contrast adjustments and for making figures and insets. Imaris 7.1.1 (Bitplane AG) was used for 3D rendering of confocal z-stacks. Illustrator CS4 (Adobe) was used for making figures and insets.
High-content microscopy
Identical imaging and analysis settings were applied for all treatments within one experiment. The Olympus ScanR illumination system with an UPLSAPO 40× objective was used for quantitation of a large number of cells from formaldehyde fixed samples for the following assays. ScanR analysis software was used for background correction and automatic image analysis.
Total intensity of perinuclear mTOR dots in transfected and untransfected cells
Wide-field images of cells transiently transfected with different GFP–Protrudin/mCherry–FYCO1 constructs and labeled for endogenous mTOR were analyzed automatically by the ScanR software. GFP–Protrudin–positive cells were detected by the software (by intensity). Dots of mTOR were segmented automatically in the perinuclear area (determined by distance to Hoechst-positive nuclei) by the ScanR software, and the total fluorescence intensity of mTOR was measured in the transfected and untransfected cells. The total number of cells was quantified automatically by detection of Hoechst nuclear stain.
Relative perinuclear or peripheral localization of mTOR or LAMP1
Wide-field images of siRNA-transfected cells labeled for endogenous mTOR or LAMP1 were analyzed automatically by the ScanR software. The ratios between perinuclear/peripheral or peripheral/perinuclear localization of mTOR- or LAMP1-positive dots (regions defined by the distance to Hoechst-positive nuclei) were calculated after automatic detection of the total intensity of segmented mTOR or LAMP1 dots in the defined regions.
Total intensity of endogenous Protrudin, FYCO1, or LAMP1 in siRNA-treated or amino acid–stimulated cells
Wide-field images of cells labeled for endogenous Protrudin, FYCO1, or LAMP1 were analyzed automatically by the ScanR software. The total fluorescence intensity per cell of Protrudin structures or FYCO1 or LAMP1 dots was measured by intensity-based segmentation. The total number of cells was quantified automatically by detection of Hoechst nuclear stain.
Total intensity of LC3 dots per cell
Wide-field images of control or siRNA-transfected cells labeled for endogenous LC3 were analyzed automatically by the ScanR software. Dots and clusters (appearing in ConA-treated cells) of LC3 were segmented automatically by the ScanR software, and the total fluorescent intensity of LC3 in the dots and clusters was measured. The total number of cells was quantified automatically by detection of Hoechst nuclear stain.
Number and size of LC3 dots per cell
Wide-field images of control or siRNA-transfected cells labeled for endogenous LC3 were analyzed automatically by the ScanR software. Dots of LC3 were segmented automatically by the ScanR software, and the number of LC3 dots per cell was measured. By inducing a threshold of 50 pixels for dot detection, the amount of cells containing >50-pixel-sized LC3 dots was identified. The total number of cells was quantified automatically by detection of Hoechst nuclear stain.
Mean intensity of TFEB in nuclei
Wide-field images of control or siRNA-transfected cells labeled for endogenous TFEB were analyzed automatically by the ScanR software. The mean intensity of TFEB fluorescence was measured in every nucleus defined by the detection of Hoechst nuclear stain.
Detection of TFEB in cytoplasmic and nuclear fractions
RPE-1 or HEK cells were transfected with siRNAs, and nuclear fractionation was performed after 48 h. Starvation treatment: cells were washed once with EBSS and then incubated in EBSS for 2 h. Torin 1 (4247; Tocris Bioscience) treatment: 500 nM for 1 h. After indicated treatments, cells were rinsed twice with ice-cold PBS and scraped into a cold lysis buffer containing 20 mM Tris-Hcl, pH 7.6, 150 mM NaCl, 2 mM EDTA, 0.1% IGEPAL CA-630 (Sigma-Aldrich), and protease and phosphatase inhibitors (Roche). The samples were spun at 13,400 g for 10 s. The supernatant was collected as the cytosolic fraction. The bottom pellet that contains nuclei was washed twice with cold lysis buffer without protease and phosphatase inhibitors. Both cytosolic and nuclear fractions were lysed in 1× Laemmli sample buffer containing 100 mM DTT and then heated at 95°C for 15 min. The denatured fractions were subjected to SDS-PAGE and analyzed by Western blotting.
Degradation of long-lived proteins in fed cells
The degradation of long-lived proteins in fed RPE-1 cells was done according to a protocol described previously (Engedal et al., 2013). In brief, cells seeded in six-well plates were transfected with siRNA duplexes on the same day and radiolabeled with 0.25 µCi/ml L-[14C] valine (NEC291EU050UC; PerkinElmer) in RPMI 1640 (R2405; Sigma-Aldrich) and 10% FBS for 24 h. Cells were then washed and chased with 1.5 ml complete RPMI 1640 supplemented with 10 mM cold valine (V0500; Sigma-Aldrich) for 3 h to allow degradation of short-lived proteins. Then the cells were incubated with 1 ml complete DMEM (10% FBS; supplemented with 10 mM cold valine) for 20 h to allow degradation of long-lived proteins. Subsequently, proteins were precipitated by 50% TCA (100807; Merck Millipore). Supernatant (degraded proteins, TCA-soluble fraction) and pellet (nondegraded protein, TCA-nonsoluble fraction) were obtained through centrifugation at 5,000 g for 10 min at 4°C. The pellet was solubilized in 500 µl 0.2 M KOH (105033; Merck Millipore). Radioactivity was determined by liquid scintillation counting. The degradation rate for long-lived proteins was calculated as the percentage of radioactivity in the TCA-soluble fraction relative to the total radioactivity in the TCA-soluble and nonsoluble fractions, divided by the incubation time.
Statistical analysis and considerations
Values are expressed as mean ± SEM. The number of individual experiments and the number of cells analyzed are indicated in each figure legend. For the calculation of statistical significance, the unpaired t test was used to test to samples with equal variance, and the one-sample t test was used in cases in which the value of the control sample was set to 1 or 100.
Online supplemental material
Fig. S1 shows knockdown efficiency of siRNA oligos targeting Protrudin or FYCO1. Fig. S2 shows that the distribution of Rab7 and LAMP1, but not Rab5 or Rab11, was affected by Protrudin or FYCO1 depletion. Fig. S3 shows that the mTORC1 component Raptor clusters in perinuclear region upon Protrudin depletion, whereas the mTORC2 component Rictor was not affected. Fig. S4 shows that number and size of LC3 dots increase in FYCO1-depleted cells.
Supplementary Material
Acknowledgments
We thank Anne Engen for expert help with cell cultures. We thank Eva Wenzel for methodological teaching and supervision. The Core Facility for Advanced Light Microscopy at Oslo University Hospital is acknowledged for providing access to relevant microscopes.
Z. Hong is a postdoctoral fellow of the Southern and Eastern Norway Regional Health Authority. C. Raiborg is a senior research fellow and N.M. Pedersen is a postdoctoral fellow of Kreftforeningen. M.L. Torgersen is a researcher funded by Norges Forskningsråd. This work was partly supported by Norges Forskningsråd through its Centres of Excellence funding scheme (project number 179571).
The authors declare no competing financial interests.
Author contributions: Z. Hong conducted formal analysis, conceptualization, investigation, methodology, validation, visualization, review, and editing. N.M. Pedersen and L. Wang conducted formal analysis, investigation, methodology, validation, visualization, review, and editing. M.L. Torgersen conducted methodology, supervision, review, and editing. H. Stenmark conceived the study and conducted funding acquisition, resources, supervision, review, and editing. C. Raiborg conceived the study, conducted formal analysis, investigation, methodology, validation, visualization, original draft, review and editing, funding acquisition, resources, supervision, and project administration.
Footnotes
Abbreviations used:
- ConA
- Concanamycin A
- EBSS
- Earle’s balanced salt solution
- LLPD
- long-lived protein degradation
- LyLE
- lysosome and late endosome
- mTOR
- mechanistic target of rapamycin
- mTORC
- mTOR complex
- pHi
- intracellular pH
- PtdIns3P
- phosphatidylinositol 3-phosphate
- S6K
- S6-kinase
- TFEB
- transcription factor EB
References
- Albert V., and Hall M.N.. 2015. mTOR signaling in cellular and organismal energetics. Curr. Opin. Cell Biol. 33:55–66. 10.1016/j.ceb.2014.12.001 [DOI] [PubMed] [Google Scholar]
- Appenzeller-Herzog C., and Hall M.N.. 2012. Bidirectional crosstalk between endoplasmic reticulum stress and mTOR signaling. Trends Cell Biol. 22:274–282. 10.1016/j.tcb.2012.02.006 [DOI] [PubMed] [Google Scholar]
- Bar-Peled L., and Sabatini D.M.. 2014. Regulation of mTORC1 by amino acids. Trends Cell Biol. 24:400–406. 10.1016/j.tcb.2014.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betz C., and Hall M.N.. 2013. Where is mTOR and what is it doing there? J. Cell Biol. 203:563–574. 10.1083/jcb.201306041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byfield M.P., Murray J.T., and Backer J.M.. 2005. hVps34 is a nutrient-regulated lipid kinase required for activation of p70 S6 kinase. J. Biol. Chem. 280:33076–33082. 10.1074/jbc.M507201200 [DOI] [PubMed] [Google Scholar]
- Calastretti A., Bevilacqua A., Ceriani C., Viganò S., Zancai P., Capaccioli S., and Nicolin A.. 2001. Damaged microtubules can inactivate BCL-2 by means of the mTOR kinase. Oncogene. 20:6172–6180. 10.1038/sj.onc.1204751 [DOI] [PubMed] [Google Scholar]
- Chang J., Lee S., and Blackstone C.. 2013. Protrudin binds atlastins and endoplasmic reticulum-shaping proteins and regulates network formation. Proc. Natl. Acad. Sci. USA. 110:14954–14959. 10.1073/pnas.1307391110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibble C.C., and Cantley L.C.. 2015. Regulation of mTORC1 by PI3K signaling. Trends Cell Biol. 25:545–555. 10.1016/j.tcb.2015.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Nardo A., Kramvis I., Cho N., Sadowski A., Meikle L., Kwiatkowski D.J., and Sahin M.. 2009. Tuberous sclerosis complex activity is required to control neuronal stress responses in an mTOR-dependent manner. J. Neurosci. 29:5926–5937. 10.1523/JNEUROSCI.0778-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engedal N., Torgersen M.L., Guldvik I.J., Barfeld S.J., Bakula D., Sætre F., Hagen L.K., Patterson J.B., Proikas-Cezanne T., Seglen P.O., et al. . 2013. Modulation of intracellular calcium homeostasis blocks autophagosome formation. Autophagy. 9:1475–1490. 10.4161/auto.25900 [DOI] [PubMed] [Google Scholar]
- Fawal M.A., Brandt M., and Djouder N.. 2015. MCRS1 binds and couples Rheb to amino acid-dependent mTORC1 activation. Dev. Cell. 33:67–81. 10.1016/j.devcel.2015.02.010 [DOI] [PubMed] [Google Scholar]
- Groenewoud M.J., and Zwartkruis F.J.. 2013. Rheb and Rags come together at the lysosome to activate mTORC1. Biochem. Soc. Trans. 41:951–955. 10.1042/BST20130037 [DOI] [PubMed] [Google Scholar]
- Gulati P., Gaspers L.D., Dann S.G., Joaquin M., Nobukuni T., Natt F., Kozma S.C., Thomas A.P., and Thomas G.. 2008. Amino acids activate mTOR complex 1 via Ca2+/CaM signaling to hVps34. Cell Metab. 7:456–465. 10.1016/j.cmet.2008.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding H.P., Zhang Y., and Ron D.. 1999. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 397:271–274. 10.1038/16729 [DOI] [PubMed] [Google Scholar]
- Hashimoto Y., Shirane M., Matsuzaki F., Saita S., Ohnishi T., and Nakayama K.I.. 2014. Protrudin regulates endoplasmic reticulum morphology and function associated with the pathogenesis of hereditary spastic paraplegia. J. Biol. Chem. 289:12946–12961. 10.1074/jbc.M113.528687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz C. 2012. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat. Rev. Mol. Cell Biol. 13:89–102. [DOI] [PubMed] [Google Scholar]
- Heuser J. 1989. Changes in lysosome shape and distribution correlated with changes in cytoplasmic pH. J. Cell Biol. 108:855–864. 10.1083/jcb.108.3.855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch D.S., Shen Y., Dokmanovic M., Yu J., Mohan N., Elzarrad M.K., and Wu W.J.. 2014. Insulin activation of vacuolar protein sorting 34 mediates localized phosphatidylinositol 3-phosphate production at lamellipodia and activation of mTOR/S6K1. Cell. Signal. 26:1258–1268. 10.1016/j.cellsig.2014.02.009 [DOI] [PubMed] [Google Scholar]
- Itakura E., Kishi C., Inoue K., and Mizushima N.. 2008. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol. Biol. Cell. 19:5360–5372. 10.1091/mbc.E08-01-0080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewell J.L., Russell R.C., and Guan K.L.. 2013. Amino acid signalling upstream of mTOR. Nat. Rev. Mol. Cell Biol. 14:133–139. 10.1038/nrm3522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhász G., Hill J.H., Yan Y., Sass M., Baehrecke E.H., Backer J.M., and Neufeld T.P.. 2008. The class III PI(3)K Vps34 promotes autophagy and endocytosis but not TOR signaling in Drosophila. J. Cell Biol. 181:655–666. 10.1083/jcb.200712051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara A., Noda T., Ishihara N., and Ohsumi Y.. 2001. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J. Cell Biol. 152:519–530. 10.1083/jcb.152.3.519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Kundu M., Viollet B., and Guan K.L.. 2011. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 13:132–141. 10.1038/ncb2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D.J., Abdelmohsen K.. Abe A., Abedin M.J., Abeliovich H., Acevedo Arozena A., Adachi H., Adams C.M., Adams P.D., Adeli K., et al. . 2016. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12:1–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolchuk V.I., Saiki S., Lichtenberg M., Siddiqi F.H., Roberts E.A., Imarisio S., Jahreiss L., Sarkar S., Futter M., Menzies F.M., et al. . 2011. Lysosomal positioning coordinates cellular nutrient responses. Nat. Cell Biol. 13:453–460. 10.1038/ncb2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ktistakis N.T., Manifava M., Schoenfelder P., and Rotondo S.. 2012. How phosphoinositide 3-phosphate controls growth downstream of amino acids and autophagy downstream of amino acid withdrawal. Biochem. Soc. Trans. 40:37–43. 10.1042/BST20110684 [DOI] [PubMed] [Google Scholar]
- Laplante M., and Sabatini D.M.. 2012. mTOR signaling in growth control and disease. Cell. 149:274–293. 10.1016/j.cell.2012.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., and Guan K.L.. 2013. Microtubule-associated protein/microtubule affinity-regulating kinase 4 (MARK4) is a negative regulator of the mammalian target of rapamycin complex 1 (mTORC1). J. Biol. Chem. 288:703–708. 10.1074/jbc.C112.396903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Rydzewski N., Hider A., Zhang X., Yang J., Wang W., Gao Q., Cheng X., and Xu H.. 2016. A molecular mechanism to regulate lysosome motility for lysosome positioning and tubulation. Nat. Cell Biol. 18:404–417. 10.1038/ncb3324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marat A.L., Wallroth A., Lo W.T., Müller R., Norata G.D., Falasca M., Schultz C., and Haucke V.. 2017. mTORC1 activity repression by late endosomal phosphatidylinositol 3,4-bisphosphate. Science. 356:968–972. 10.1126/science.aaf8310 [DOI] [PubMed] [Google Scholar]
- Martina J.A., Diab H.I., Brady O.A., and Puertollano R.. 2016. TFEB and TFE3 are novel components of the integrated stress response. EMBO J. 35:479–495. 10.15252/embj.201593428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan N., Shen Y., Dokmanovic M., Endo Y., Hirsch D.S., and Wu W.J.. 2016. VPS34 regulates TSC1/TSC2 heterodimer to mediate RheB and mTORC1/S6K1 activation and cellular transformation. Oncotarget. 7:52239–52254. 10.18632/oncotarget.10469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munson M.J., Allen G.F., Toth R., Campbell D.G., Lucocq J.M., and Ganley I.G.. 2015. mTOR activates the VPS34-UVRAG complex to regulate autolysosomal tubulation and cell survival. EMBO J. 34:2272–2290. 10.15252/embj.201590992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napolitano G., and Ballabio A.. 2016. TFEB at a glance. J. Cell Sci. 129:2475–2481. 10.1242/jcs.146365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobukuni T., Joaquin M., Roccio M., Dann S.G., Kim S.Y., Gulati P., Byfield M.P., Backer J.M., Natt F., Bos J.L., et al. . 2005. Amino acids mediate mTOR/raptor signaling through activation of class 3 phosphatidylinositol 3OH-kinase. Proc. Natl. Acad. Sci. USA. 102:14238–14243. 10.1073/pnas.0506925102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsaki Y., Suzuki M., Shinohara Y., and Fujimoto T.. 2010. Lysosomal accumulation of mTOR is enhanced by rapamycin. Histochem. Cell Biol. 134:537–544. 10.1007/s00418-010-0759-x [DOI] [PubMed] [Google Scholar]
- Olsvik H.L., Lamark T., Takagi K., Larsen K.B., Evjen G., Øvervatn A., Mizushima T., and Johansen T.. 2015. FYCO1 contains a C-terminally Extended, LC3A/B-preferring LC3-interacting region (LIR) Motif required for efficient maturation of autophagosomes during basal autophagy. J. Biol. Chem. 290:29361–29374. 10.1074/jbc.M115.686915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S., Alemu E.A., Brech A., Bruun J.A., Lamark T., Overvatn A., Bjørkøy G., and Johansen T.. 2010. FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. J. Cell Biol. 188:253–269. 10.1083/jcb.200907015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parton R.G., Dotti C.G., Bacallao R., Kurtz I., Simons K., and Prydz K.. 1991. pH-induced microtubule-dependent redistribution of late endosomes in neuronal and epithelial cells. J. Cell Biol. 113:261–274. 10.1083/jcb.113.2.261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson R.B., Dennis P.B., Han J.W., Williamson N.A., Kozma S.C., Wettenhall R.E., and Thomas G.. 1995. The principal target of rapamycin-induced p70s6k inactivation is a novel phosphorylation site within a conserved hydrophobic domain. EMBO J. 14:5279–5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen N.M., Raiborg C., Brech A., Skarpen E., Roxrud I., Platta H.W., Liestøl K., and Stenmark H.. 2012. The PtdIns3P-binding protein Phafin 2 mediates epidermal growth factor receptor degradation by promoting endosome fusion. Traffic. 13:1547–1563. 10.1111/j.1600-0854.2012.01400.x [DOI] [PubMed] [Google Scholar]
- Petiot A., Ogier-Denis E., Blommaart E.F.C., Meijer A.J., and Codogno P.. 2000. Distinct classes of phosphatidylinositol 3′-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J. Biol. Chem. 275:992–998. 10.1074/jbc.275.2.992 [DOI] [PubMed] [Google Scholar]
- Pu J., Schindler C., Jia R., Jarnik M., Backlund P., and Bonifacino J.S.. 2015. BORC, a multisubunit complex that regulates lysosome positioning. Dev. Cell. 33:176–188. 10.1016/j.devcel.2015.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puertollano R. 2014. mTOR and lysosome regulation. F1000Prime Rep. 6:52 10.12703/P6-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiborg C., Wenzel E.M., Pedersen N.M., Olsvik H., Schink K.O., Schultz S.W., Vietri M., Nisi V., Bucci C., Brech A., et al. . 2015. Repeated ER-endosome contacts promote endosome translocation and neurite outgrowth. Nature. 520:234–238. 10.1038/nature14359 [DOI] [PubMed] [Google Scholar]
- Rocha N., Kuijl C., van der Kant R., Janssen L., Houben D., Janssen H., Zwart W., and Neefjes J.. 2009. Cholesterol sensor ORP1L contacts the ER protein VAP to control Rab7-RILP-p150 glued and late endosome positioning. J. Cell Biol. 185:1209–1225. 10.1083/jcb.200811005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roczniak-Ferguson A., Petit C.S., Froehlich F., Qian S., Ky J., Angarola B., Walther T.C., and Ferguson S.M.. 2012. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci. Signal. 5:ra42 10.1126/scisignal.2002790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronan B., Flamand O., Vescovi L., Dureuil C., Durand L., Fassy F., Bachelot M.F., Lamberton A., Mathieu M., Bertrand T., et al. . 2014. A highly potent and selective Vps34 inhibitor alters vesicle trafficking and autophagy. Nat. Chem. Biol. 10:1013–1019. 10.1038/nchembio.1681 [DOI] [PubMed] [Google Scholar]
- Rosa-Ferreira C., and Munro S.. 2011. Arl8 and SKIP act together to link lysosomes to kinesin-1. Dev. Cell. 21:1171–1178. 10.1016/j.devcel.2011.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y., Peterson T.R., Shaul Y.D., Lindquist R.A., Thoreen C.C., Bar-Peled L., and Sabatini D.M.. 2008. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 320:1496–1501. 10.1126/science.1157535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton R.A., and Sabatini D.M.. 2017. mTOR signaling in growth, metabolism, and disease. Cell. 169:361–371. 10.1016/j.cell.2017.03.035 [DOI] [PubMed] [Google Scholar]
- Settembre C., Zoncu R., Medina D.L., Vetrini F., Erdin S., Erdin S., Huynh T., Ferron M., Karsenty G., Vellard M.C., et al. . 2012. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 31:1095–1108. 10.1038/emboj.2012.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz A., Ernst A., and Dikic I.. 2014. Cargo recognition and trafficking in selective autophagy. Nat. Cell Biol. 16:495–501. 10.1038/ncb2979 [DOI] [PubMed] [Google Scholar]
- Thastrup O., Cullen P.J., Drøbak B.K., Hanley M.R., and Dawson A.P.. 1990. Thapsigargin, a tumor promoter, discharges intracellular Ca2+ stores by specific inhibition of the endoplasmic reticulum Ca2(+)-ATPase. Proc. Natl. Acad. Sci. USA. 87:2466–2470. 10.1073/pnas.87.7.2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen C.C., Kang S.A., Chang J.W., Liu Q., Zhang J., Gao Y., Reichling L.J., Sim T., Sabatini D.M., and Gray N.S.. 2009. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J. Biol. Chem. 284:8023–8032. 10.1074/jbc.M900301200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoresen S.B., Pedersen N.M., Liestøl K., and Stenmark H.. 2010. A phosphatidylinositol 3-kinase class III sub-complex containing VPS15, VPS34, Beclin 1, UVRAG and BIF-1 regulates cytokinesis and degradative endocytic traffic. Exp. Cell Res. 316:3368–3378. 10.1016/j.yexcr.2010.07.008 [DOI] [PubMed] [Google Scholar]
- Uusi-Rauva K., Kyttälä A., van der Kant R., Vesa J., Tanhuanpää K., Neefjes J., Olkkonen V.M., and Jalanko A.. 2012. Neuronal ceroid lipofuscinosis protein CLN3 interacts with motor proteins and modifies location of late endosomal compartments. Cell. Mol. Life Sci. 69:2075–2089. 10.1007/s00018-011-0913-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.Z., Lawson B., Brewer J.W., Zinszner H., Sanjay A., Mi L.J., Boorstein R., Kreibich G., Hendershot L.M., and Ron D.. 1996. Signals from the stressed endoplasmic reticulum induce C/EBP-homologous protein (CHOP/GADD153). Mol. Cell. Biol. 16:4273–4280. 10.1128/MCB.16.8.4273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijdeven R.H., Janssen H., Nahidiazar L., Janssen L., Jalink K., Berlin I., and Neefjes J.. 2016. Cholesterol and ORP1L-mediated ER contact sites control autophagosome transport and fusion with the endocytic pathway. Nat. Commun. 7:11808 10.1038/ncomms11808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullschleger S., Loewith R., and Hall M.N.. 2006. TOR signaling in growth and metabolism. Cell. 124:471–484. 10.1016/j.cell.2006.01.016 [DOI] [PubMed] [Google Scholar]
- Yoshida S., Pacitto R., Yao Y., Inoki K., and Swanson J.A.. 2015. Growth factor signaling to mTORC1 by amino acid-laden macropinosomes. J. Cell Biol. 211:159–172. 10.1083/jcb.201504097 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.