Neutrophils can release their genomic DNA as extracellular traps (NETs), which ensnare bacteria and limit their replication. Stojkov et al. find that modulation of cytoskeletal dynamics by reactive oxygen species and glutathionylation controls the degranulation and release of mitochondrial DNA required for NET formation.
Abstract
The antimicrobial defense activity of neutrophils partly depends on their ability to form neutrophil extracellular traps (NETs), but the underlying mechanism controlling NET formation remains unclear. We demonstrate that inhibiting cytoskeletal dynamics with pharmacological agents or by genetic manipulation prevents the degranulation of neutrophils and mitochondrial DNA release required for NET formation. Wiskott-Aldrich syndrome protein–deficient neutrophils are unable to polymerize actin and exhibit a block in both degranulation and DNA release. Similarly, neutrophils with a genetic defect in NADPH oxidase fail to induce either actin and tubulin polymerization or NET formation on activation. Moreover, neutrophils deficient in glutaredoxin 1 (Grx1), an enzyme required for deglutathionylation of actin and tubulin, are unable to polymerize either cytoskeletal network and fail to degranulate or release DNA. Collectively, cytoskeletal dynamics are achieved as a balance between reactive oxygen species–regulated effects on polymerization and glutathionylation on the one hand and the Grx1-mediated deglutathionylation that is required for NET formation on the other.
Introduction
Degranulation is defined as the secretion of granule-derived substances after granulocyte activation (Borregaard et al., 2007; Lacy and Eitzen, 2008). Granule proteins can be found in the extracellular space, often in association with a DNA scaffold. Together they form net-like structures, the so-called neutrophil extracellular traps (NETs), which are capable of trapping and killing bacteria (Brinkmann et al., 2004; Yousefi et al., 2009). Inflammatory mediators, such as interleukin 8, complement factor 5a (C5a), N-formyl-methionyl-leucyl-phenylalanin (fMLP), lipopolysaccharide (LPS), and TNF, can trigger NET formation (Brinkmann et al., 2004; Yousefi et al., 2009; Lim et al., 2011; Stoiber et al., 2015), causing the release of DNA and an accompanying neutrophil degranulation (Abdel-Latif et al., 2004; Lacy, 2005, 2006; Uriarte et al., 2011; Amini et al., 2016).
Degranulation of neutrophils has been shown to be dependent on actin microfilament (MF) remodeling (Mitchell et al., 2008) mediated by the small GTPase, Rac2; hence, Rac2 knockout neutrophils are unable to generate polymerized F-actin (Gu et al., 2003; Filippi et al., 2004) or to form NETs (Lim et al., 2011). However, whether actin polymerization plays a role also in the release of DNA required for NET formation has remained unclear.
Like almost all cells, neutrophils continuously cycle actin protein subunits between monomeric (G-actin) and polymeric (F-actin) pools, reversibly cross-linking polymeric actin into three-dimensional networks of actin MFs, and assemble and disassemble microtubules (MTs) needed for the transport of proteins and organelles. For instance, it has been suggested that F-actin depolymerization at the cell cortex, coupled with a Rac-dependent F-actin polymerization in the cytoplasm, elicits the degranulation of neutrophils (Mitchell et al., 2008). The assembly of MT and MF networks provides the mechanical propulsion that drives a range of dynamic cellular processes, including cell motility (Kindzelskii et al., 2004; Xu et al., 2005).
In recent years, several proteins that activate the filament nucleation activity of actin-related proteins 2 and 3 have been identified, most notably the Wiskott-Aldrich syndrome protein (WASP) family. WASp is involved in the control of actin dynamics. Products of the WAS gene family thus regulate a variety of actin-dependent processes that range from cell migration to phagocytosis, endocytosis, and membrane trafficking (Millard et al., 2004).
Actin polymerization plays an important role in the initiation of reactive oxygen species (ROS) production in neutrophils by potentiating NADPH oxidase assembly and activity (Suzuki et al., 2003; Lacy, 2005; Shao et al., 2010). On the other hand, ROS levels are in large part responsible for regulating the dynamics of F-actin formation, for example, in neuronal growth cones (Munnamalai and Suter, 2009).
H2O2 is one of the few ROS molecules that can diffuse freely through cellular membranes, oxidizing the -SH group of exposed cysteines to sulfenic acid on target proteins, which can then be reduced back to cysteine by various cellular reducing agents, such as glutathione (GSH). In a process known as S-glutathionylation, under oxidizing conditions, free thiol groups of proteins can be modified to form protein-GSH mixed disulfides (Giustarini et al., 2004). The fact that only a few intracellular proteins carry an oxidizable cysteine at a critical position is the reason why a small molecule such as H2O2 can act as specific second messenger (Reth, 2002). Actin and tubulin are both among the few proteins that can be glutathionylated; the oxidation of a cysteinyl residue in these proteins to a sulfenic acid is followed by glutathionylation, thereby inhibiting their polymerization (Johansson and Lundberg, 2007), and consequently overall cytoskeletal dynamics (Landino et al., 2004). However, glutathionylation is a reversible posttranslational modification, because a reverse reaction, deglutathionylation, is catalyzed specifically and efficiently by glutaredoxin 1 (Grx1). Easy reversibility is critical for the physiological potential of glutathionylation as a means of functional regulation (Shelton and Mieyal, 2008; Wilson and González-Billault, 2015).
Emerging evidence suggests that ROS affects cytoskeletal proteins in multiple ways. For instance, the cellular redox status seems to be tightly coupled with MT formation because ROS signals regulate the organization of the tubulin cytoskeleton and induce tubulin modifications, including glutathionylation (Livanos et al., 2014). Recently, it has been shown that ROS generated in neutrophils by the NADPH oxidase regulates actin polymerization through reversible actin glutathionylation. Grx1 enzyme activity is required to recycle the modified glutathionylated G-actin to free G-actin for F-actin formation. Grx1-deficient mouse neutrophils showed impaired in vivo recruitment to sites of inflammation and reduced bactericidal capability (Oliver et al., 1978; Sakai et al., 2012).
In this article, we confirm that ROS is required for mitochondrial DNA (mtDNA) release in a process of NET formation and newly report that ROS production by the NADPH oxidase is essential for the degranulation of neutrophils. Patients who have chronic granulomatous disease (CGD) have a defect in NADPH oxidase, lack sufficient ROS production, and exhibit defects in both actin and tubulin polymerization, hence also in degranulation, DNA release, and NET formation. We further demonstrate that ROS, in tandem with Grx1, regulates actin and tubulin polymerization through glutathionylation. Moreover, we provide data indicating that actin and tubulin are glutathionylated proteins in neutrophils undergoing physiological activation leading to NET formation. We demonstrate that the cysteine located at the C terminus of actin (Cys374) is required for ROS-induced actin glutathionylation, actin polymerization, and NET formation. Similarly, a lack of Grx1, the key enzyme required for deglutathionylation of actin and tubulin, leads to impaired cytoskeletal dynamics, resulting in defective degranulation and failure of DNA release, the two important requirements for NET formation. Finally, we provide evidence that MT formation is also regulated by a GSH modification, which is essential for neutrophil degranulation, DNA release, and NET formation.
Results
Actin polymerization requires ROS production and is essential for NET formation
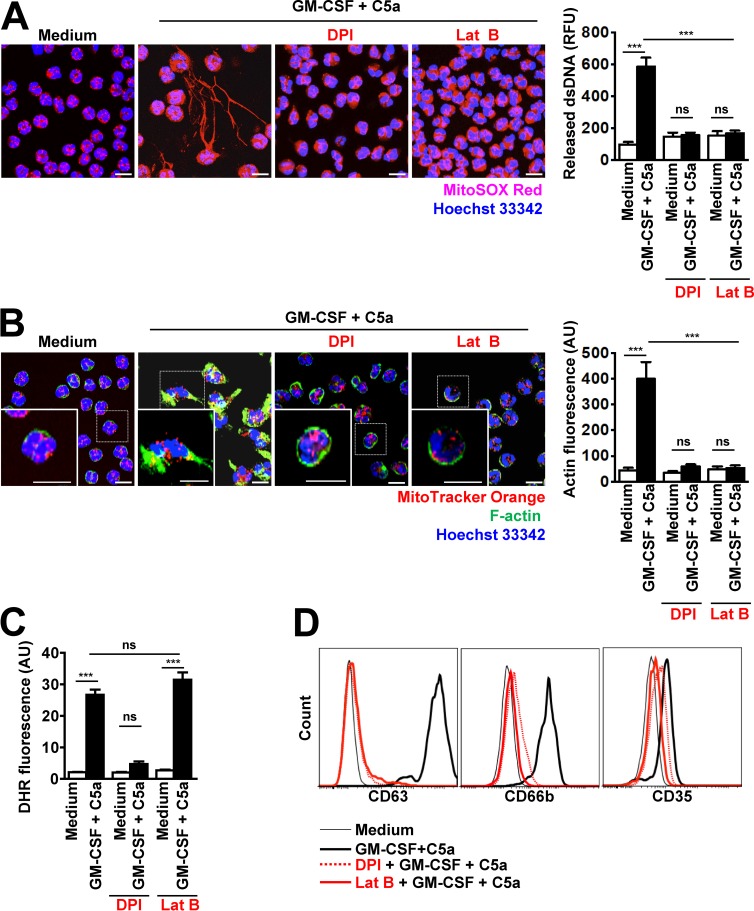
Because primary granule exocytosis in human neutrophils is regulated by Rac2-dependent actin remodeling (Abdel-Latif et al., 2004; Lacy, 2006; Mitchell et al., 2008) and granule proteins bind to the DNA scaffold of NETs (Brinkmann et al., 2004; Yousefi et al., 2009), we studied the role of actin dynamics for mtDNA release and NET formation by activated neutrophils. We first examined actin polymerization in resting and stimulated human neutrophils during NET formation using confocal microscopy. As expected, resting neutrophils exhibited no NET formation (Fig. 1 A), and only cortical F-actin was seen in a ring-like manner close to the cell membrane (Fig. 1 B), confirming previously published work (Mitchell et al., 2008). Granulocyte/macrophage colony-stimulating factor (GM-CSF)–primed and C5a-activated neutrophils demonstrated the release of mtDNA (Fig. 1 A) and F-actin accumulation at one cell pole (Fig. 1 B). To confirm these data, we used mouse neutrophils isolated from Lifeact-EGFP transgenic mice, which we analyzed by live confocal microscopy under resting and activated conditions (Fig. S1 A and Video 1). These mice express 17-amino-acid EGFP-Lifeact peptide, in all cell types including hematopoietic cells, without interference with any cellular processes (Riedl et al., 2010). Ring-like actin was easily detectable in resting neutrophils by live imaging and on activation; rapid actin remodeling was apparent, similar to that seen in images obtained with FITC-phalloidin staining (Fig. 1 B, designated F-actin). In addition, actin polymerization was also evaluated and quantified by using a flow cytometric assay in which the F-actin had been stained with FITC-phalloidin (Fig. S1 B, upper panel). A shift in phalloidin staining was observed only when human blood neutrophils were primed with GM-CSF and activated with C5a, confirming the microscopic data shown in Fig. 1 B. It should be noted that other inflammatory triggers, such as 10−7 M fMLP, 25 nM PMA, as well as the combined 25 ng/ml GM-CSF and 100 ng/ml LPS stimulation, can also lead to NET formation in mouse neutrophils (Fig. S1 C). We also analyzed the F-actin morphology using Lifeact-EGFP mouse neutrophils and consistently observed an accumulation of F-actin at one cell pole with each of these stimuli, but being most prominent in GM-CSF primed and C5a activated and in fMLP-treated cells (Fig. S1 D).
Figure 1.
Actin polymerization requires ROS production and is essential for DNA release. (A) Confocal microscopy. DNA release was analyzed after pretreatment and short-term stimulation (total 35 min) of human neutrophils with the indicated triggers in the presence or absence of 1 µM DPI and 1.25 µM Lat B. Right: Quantification of released dsDNA in supernatants of activated neutrophils. n = 5. (B) Confocal microscopy. F-actin distribution and morphological changes were analyzed after pretreatment and short-term stimulation (total 35 min) of human neutrophils with the indicated inhibitors and triggers. Right: Quantification of F-actin was performed by automated analysis of microscopic images by using Imaris software. n = 4. (C) Flow cytometry. Total ROS activity of human neutrophils after pretreatment and short-term stimulation (total 35 min) with the indicated inhibitors and triggers was assessed by DHR123 fluorescence. n = 5. (D) Flow cytometry. Expression of CD63, CD66b and CD35 was assessed after pretreatment and short-term stimulation (total 35 min) of human neutrophils with the indicated inhibitors, DPI (5 µM), and Lat B (10 µM), and triggers. Representative histograms are presented. n = 5. Data are means ± SEM. ***, P < 0.001. Bars, 10 µm.
The process of NET formation is dependent on the enzymatic activity of the NADPH oxidase (Yousefi et al., 2009; Parker et al., 2012). Diphenylene iodonium chloride (DPI), an inhibitor of NADPH oxidase, completely blocked the release of DNA as previously reported (Yousefi et al., 2008, 2009; Parker et al., 2012; Fig. 1 A). Moreover, blocking ROS production with DPI also greatly reduced the accumulation of F-actin at one cell pole (Fig. 1 B). To demonstrate that the block in DNA release elicited by inhibiting ROS production is mediated by an impaired F-actin polarization, neutrophils were briefly pretreated with latrunculin B (Lat B) that sequesters G-actin monomers and prevents F-actin assembly (Cingolani and Goda, 2008). Lat B blocked the accumulation of F-actin at one cell pole as expected (Fig. 1 B). Interestingly, Lat B also completely prevented DNA release after neutrophil activation (Fig. 1 A), although ROS production was unaffected (Fig. 1 C).
As previously reported (Mitchell et al., 2008), DPI and Lat B also prevented degranulation of azurophilic granules (surrogate marker CD63), specific granules (surrogate marker CD66b), and secretory vesicles (surrogate marker CD35) of GM-CSF–primed and C5a-activated neutrophils, as assessed by flow cytometer (Fig. 1 D). Preliminary experiments were performed to determine the optimal antibody concentrations (Fig. S2 A). Release of N-acetyl-β-glucosaminidase (Sato et al., 2013) was also inhibited under these conditions, confirming that azurophilic granules had not released their contents (Fig. S2 B). In addition, both DPI and Lat B inhibited the release of matrix metalloprotease-9 (MMP-9; Uriarte et al., 2011), suggesting that degranulation of tertiary granules was also blocked (Fig. S2 C).
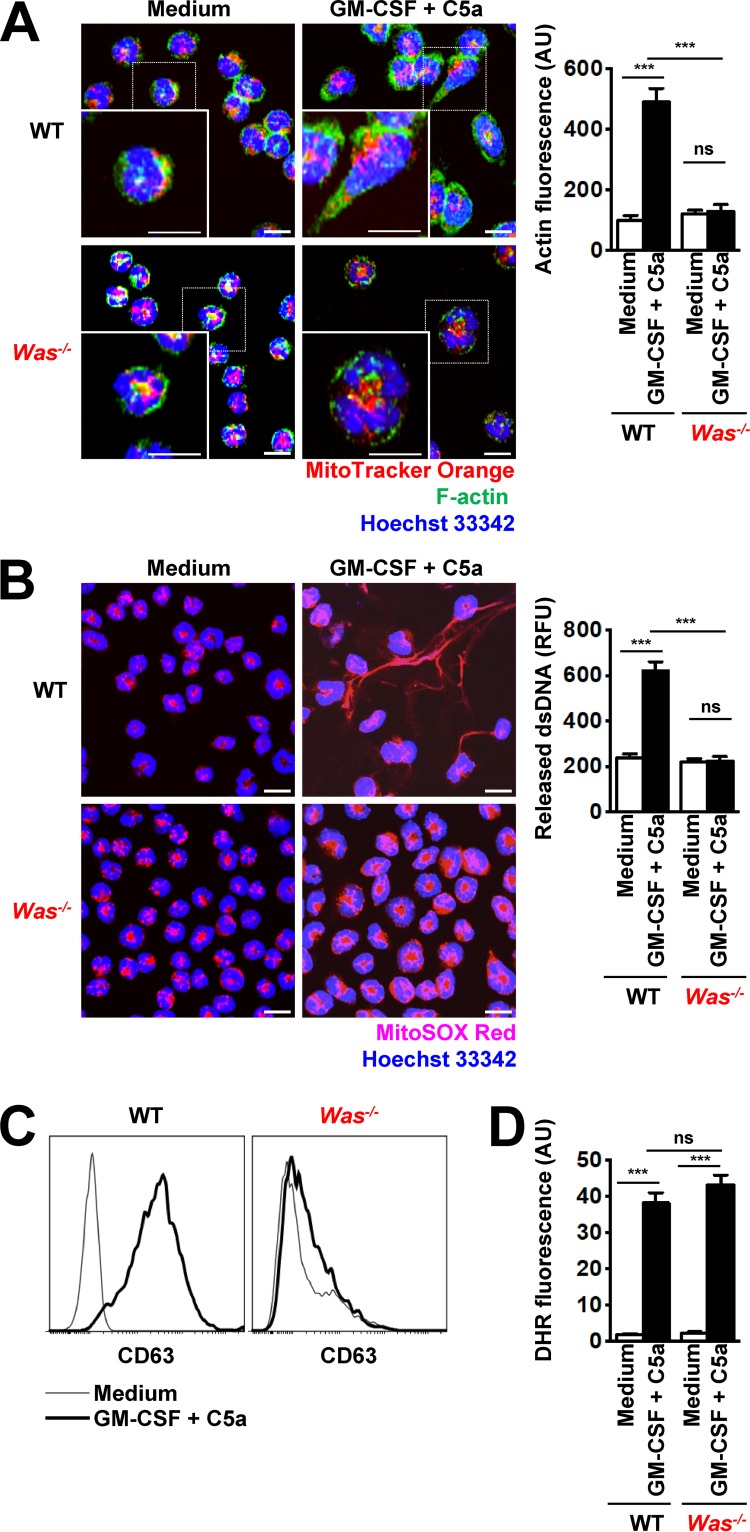
WAS is a severe X-linked immunodeficiency disorder caused by mutations in the WAS gene encoding the WASP, which is implicated in the regulation of the actin cytoskeleton (Snapper et al., 1998). WAS patients exhibit an altered cytoskeleton and develop a variety of bacterial, viral, and fungal respiratory infections (Zhang et al., 2006). To further examine the requirement for cytoskeletal rearrangement in NET formation, we used mouse neutrophils derived from WASP-deficient mice (Was−/− mice). Equal neutrophil differentiation levels and maturity in WT and Was−/− neutrophils were assessed by Ly6G cell surface expression, a myeloid differentiation marker expressed by mature neutrophils (Fig. S3 A) and nuclear morphology (Fig. S3 B). As expected, no actin polarization on physiological activation was seen in Was−/− neutrophils as compared with WT mouse neutrophils (Fig. 2 A). Similar to the pharmacological approach used with human neutrophils, we observed that neutrophils unable to accumulate F-actin exhibited neither DNA release (Fig. 2 B) nor degranulation (Fig. 2 C and Fig. S4, A and B), despite high levels of ROS production (Fig. 2 D). Collectively, these data suggest that F-actin polarization in a ROS-dependent manner is required for the release of both DNA and granule proteins, and, therefore, for NET formation.
Figure 2.
Was−/− mouse neutrophils exhibit defects in actin polymerization, degranulation, and DNA release. (A) Confocal microscopy. F-actin distribution and morphological changes were analyzed after short-term stimulation (total 35 min) of WT and Was−/− mouse neutrophils with the indicated triggers. Right: Quantification of F-actin was performed by automated analysis of microscopic images by using Imaris software. (B) Confocal microscopy. DNA release was analyzed after short-term stimulation (total 35 min) of WT and Was−/− mouse neutrophils, with the indicated triggers. Right: Quantification of released dsDNA in supernatants of activated neutrophils. (C) Flow cytometry. Expression of CD63 was assessed and short-term stimulation (total 35 min) of WT and Was−/− mouse neutrophils with the indicated triggers. Representative histograms are presented. (D) Flow cytometry. Total ROS activity of WT and Was−/− mouse neutrophils after short-term stimulation (total 35 min) with the indicated triggers was assessed by DHR123 fluorescence. Data are means ± SEM. ***, P < 0.001. n = 5. Bars, 10 µm.
Exogenous H2O2 allows actin polymerization and NET formation in the absence of a functional NADPH oxidase
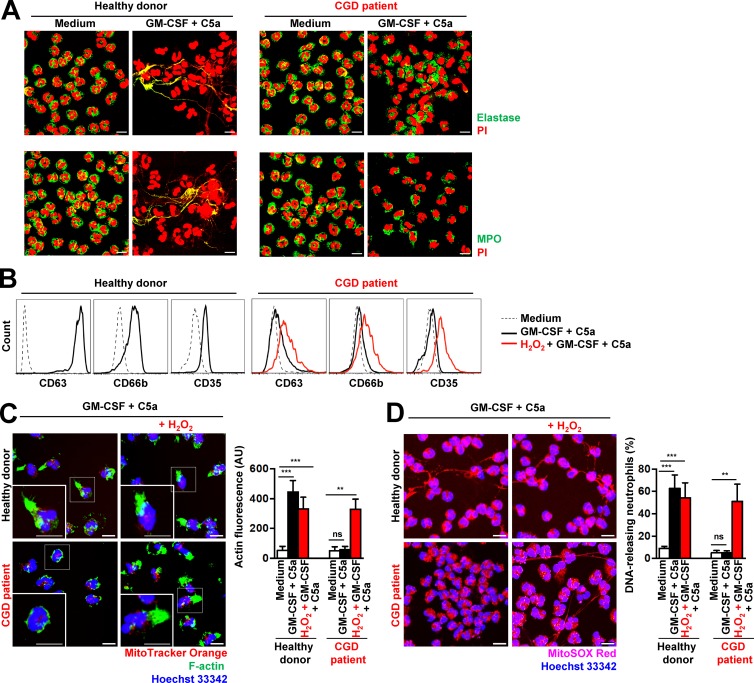
CGD is caused by mutations in genes encoding NADPH oxidase subunits, resulting in poor antimicrobial activity by neutrophils, at least partially because of their inability to generate NETs (Bianchi et al., 2009). Indeed, CGD neutrophils produce very low levels of ROS (Fig. S5 A) and are unable to release DNA (Fig. 3 A), confirming previously published work (Bianchi et al., 2009; Yousefi et al., 2009). Interestingly, we could not also demonstrate degranulation in CGD neutrophils after GM-CSF priming and subsequent C5a activation (Fig. 3 B), despite their normal expression of primary granule proteins (elastase and myeloperoxidase [MPO]; Fig. 3 A). Moreover, CGD neutrophils are unable to polymerize F-actin (Fig. 3 C), confirming a ROS dependency for this event (Fig. 1 B; Bianchi et al., 2009; Yousefi et al., 2009). Addition of H2O2 to CGD neutrophils 30 min before in vitro activation with GM-CSF and C5a was able to correct F-actin polymerization (Fig. 3 C), DNA release (Fig. 3 D), and degranulation (Fig. 3 B). The concentration of 50 µM H2O2 used caused no toxicity to human or mouse neutrophils within a period of 24 h (Fig. S6, A and B).
Figure 3.
Human CGD neutrophils demonstrate defects in actin polymerization, degranulation, and NET formation: restoration by H2O2. (A) Confocal microscopy. NET formation was analyzed after short-term stimulation (total 35 min) of control and CGD human neutrophils with the indicated triggers. Colocalization of elastase (green) or MPO (green) with released DNA (PI, red) is indicated by the yellow color depicting NET formation. (B) Flow cytometry. Expression of CD63, CD66b, and CD35 was assessed after pretreatment and short-term stimulation (total 35 min) of control and CGD human neutrophils with the indicated triggers in the presence and absence of 50 µM H2O2. Representative histograms are presented. (C) Confocal microscopy. F-actin distribution and morphological changes were analyzed after short-term stimulation (total 35 min) of control and CGD human neutrophils with the indicated triggers in the presence and absence of 50 µM H2O2. Right: Quantification of F-actin was performed by automatic analysis of microscopy images by using Imaris software. (D) Confocal microscopy. DNA release was analyzed after short-term stimulation (total 35 min) of control and CGD human neutrophils with the indicated triggers in the presence and absence of 50 µM H2O2. Right: Quantification of DNA-releasing neutrophils in ten high power fields. Data are means ± SEM. ***, P < 0.001; **, P < 0.01. n = 3. Bars, 10 µm.
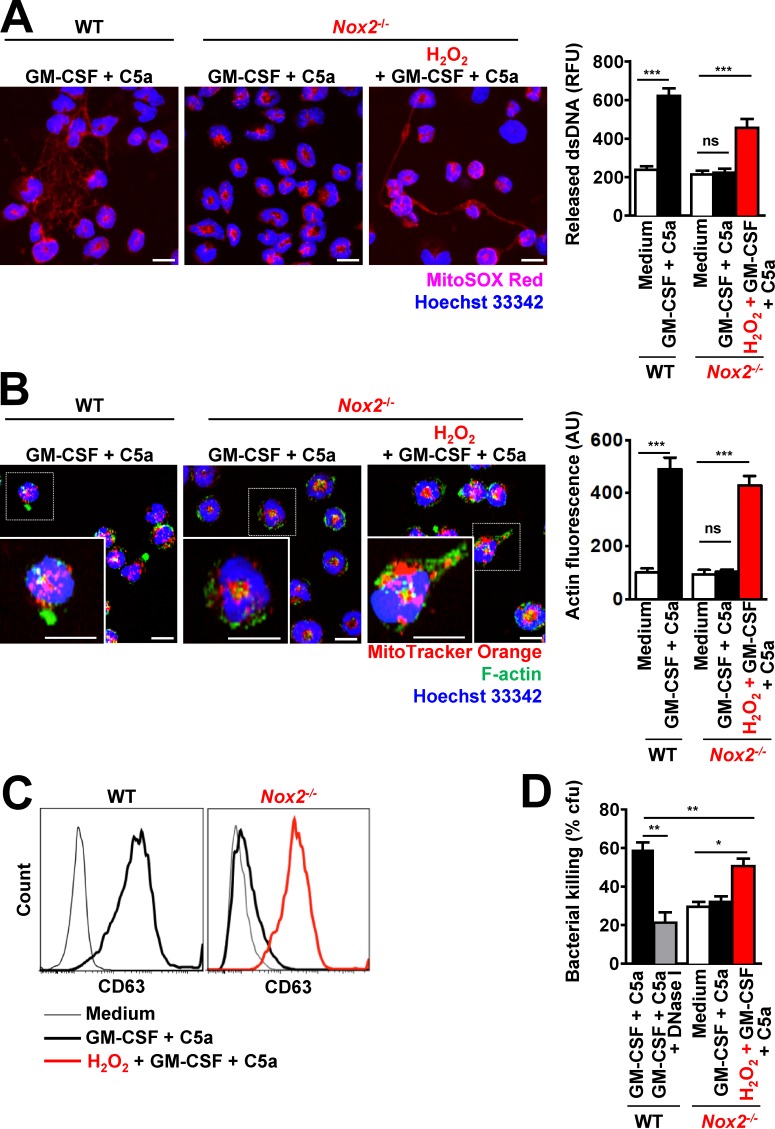
We also used conditional mouse Hoxb8-immortalized progenitor cells to generate large numbers of mouse neutrophils from WT and NADPH oxidase 2 (Nox2; gp91phox)–deficient mice, the latter showing no respiratory burst activity on activation (Fig. S5 B). Equal neutrophil differentiation levels and maturity in WT and Nox2−/− neutrophils were assessed by Ly6G cell surface expression (Fig. S3 A) and nuclear morphology (Fig. S3 B). As expected, Nox2−/− mouse neutrophils were unable to release DNA on activation (Fig. 4 A), accompanied by the absence of degranulation as demonstrated by reduced expression of the surrogate marker CD63 after GM-CSF priming and C5a stimulation, or β-glucosaminidase activity and reduced MMP-9 release (Fig. 4 C as well as Fig. S4, C and D). Exogenous H2O2 not only restored DNA release (Fig. 4 A), it also corrected defective F-actin polymerization (Fig. 4 B) and degranulation (Fig. 4 C). Addition of exogenous H2O2 also restored the bacterial killing capacity of Nox2−/− mouse neutrophils (Fig. 4 D). Collectively, these results indicate that ROS is required for F-actin polymerization, which then is a prerequisite for functional NET formation.
Figure 4.
Exogenous addition of H2O2 restored the ability of Nox2−/− mouse neutrophils to perform actin polymerization, to degranulate, and to release DNA. (A) Confocal microscopy. DNA release was analyzed after short-term stimulation (total 35 min) of WT and Nox2−/− mouse neutrophils with the indicated triggers in the presence and absence of 50 µM H2O2. Right: Quantification of released dsDNA in supernatants of activated neutrophils. RFU, relative fluorescence units. (B) Confocal microscopy. F-actin distribution and morphological changes were analyzed after short-term stimulation (total 35 min) of WT and Nox2−/− mouse neutrophils with the indicated triggers in the presence and absence of 50 µM H2O2. Right: Quantification of F-actin was performed by automated analysis of microscopic images by using Imaris software. AU, arbitrary units. (C) Flow cytometry. Expression of CD63 was assessed after pretreatment and short-term stimulation (total 35 min) of WT and Nox2−/− mouse neutrophils with the indicated triggers in the presence and absence of 50 µM H2O2. Representative histograms are presented. (D) Bacterial killing by colony formation unit (cfu) assay. E. coli killing was analyzed after short-term stimulation (total 35 min) of WT and Nox2−/− mouse neutrophils with the indicated triggers in the presence and absence of 50 µM H2O2. Data are means ± SEM. ***, P < 0.001; **, P < 0.01; *, P < 0.05. n = 5.
F-actin polymerization is regulated by glutathionylation
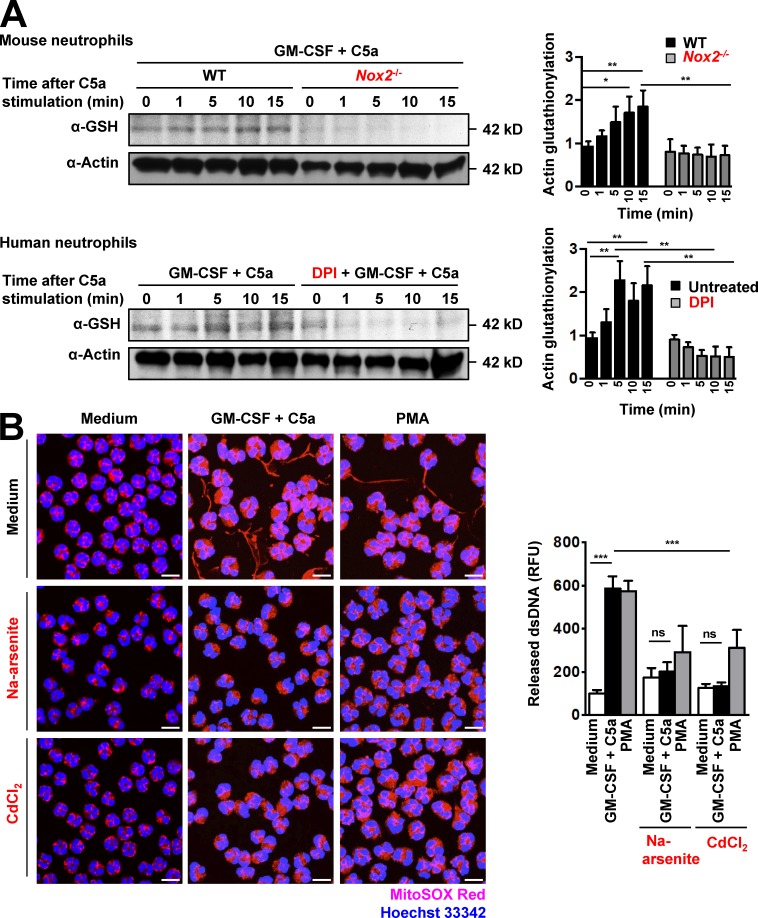
It has been previously reported that H2O2 regulates F-actin polymerization in stimulated neutrophils through reversible actin glutathionylation (Sakai et al., 2012). We asked whether NET formation, which depends on ROS production and F-actin polymerization, is also regulated by actin glutathionylation. We first examined protein glutathionylation with a GSH-specific antibody. A major band corresponding to the size of actin (42 kD) was detected in lysates of activated mouse neutrophils (Fig. 5 A, top). Glutathionylation was time-dependent and reached maximal levels within 10 min after C5a activation in GM-CSF–primed WT mouse neutrophils. In contrast, we could not detect any glutathionylated 42-kD protein in activated Nox2−/− mouse neutrophils. We then examined the expression of glutathionylated actin in human neutrophils in the presence or absence of DPI. GM-CSF–primed human neutrophils displayed maximal glutathionylation levels at 5 min after C5a activation. In contrast, DPI-treated human neutrophils showed only background actin glutathionylation (Fig. 5 A, lbottom). These results confirm that NADPH oxidase–dependent ROS production determines actin glutathionylation in stimulated mouse and human neutrophils.
Figure 5.
Pharmacological inhibition of actin glutathionylation prevents DNA release. (A) Immunoblotting. Actin glutathionylation in activated mouse and human neutrophils is dependent on NADPH oxidase activation. Human and mouse neutrophils were analyzed after short-term stimulation (total 35 min) with the indicated triggers. C5a activation was performed in a time-dependent manner. No glutathionylated actin at the expected size (42 kD) was detected in Nox2−/− mouse neutrophils or in 50-µM-DPI–pretreated human neutrophils. The ratio of glutathionylated actin to total actin is shown as actin glutathionylation in the bar graph (right). Data are representative of three independent experiments. (B) Confocal microscopy. DNA release was analyzed after short-term stimulation (total 35 min) of control human neutrophils with the indicated triggers in the presence and absence of the inhibitors as designated: 50 µM Na-arsenite and 2 µM CdCl2. Right: Quantification of released dsDNA in supernatants of activated neutrophils. Data are means ± SEM. *, P < 0.0242; **, P < 0.0074; ***, P < 0.001. n = 3. Bars, 10 µm.
A previous study suggested that actin glutathionylation occurs via a sulfenic acid intermediary (Johansson and Lundberg, 2007). Accordingly, we tested whether sodium arsenite, a reducing agent that specifically converts sulfenic acids to thiols, would block actin glutathionylation and impair DNA release from neutrophils. As with DPI treatment, a 30-min preincubation of human neutrophils with sodium arsenite before activation significantly decreased actin glutathionylation in neutrophils (not depicted) and completely blocked DNA release (Fig. 5 B). Similarly, cadmium chloride, a nonspecific inhibitor of Grx1 (Liao et al., 2010; Choong et al., 2013), also blocked DNA release in human neutrophils (Fig. 5 B), despite high levels of ROS production (Fig. S5 C). The concentration used for both 50 µM sodium arsenite and 2 µM cadmium chloride was as recommended (Sakai et al., 2012) and was not toxic to human neutrophils within a period of 24 h (Fig. S6 C).
Cys374 is required for actin glutathionylation, actin polymerization, and NET formation
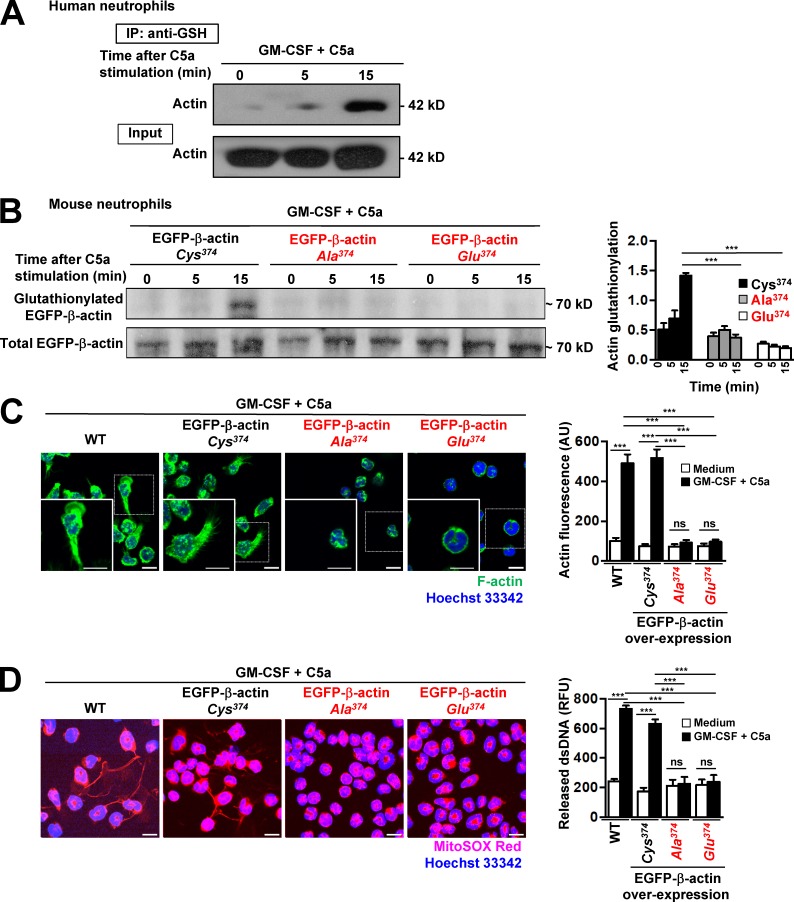
To confirm that one of the glutathionylated proteins was actin, lysates of unactivated and activated human neutrophils were subjected to pulled-down assays by using anti–GSH antibody immobilized on agarose by covalently coupling the antibody to an amine-reactive resin. The materials pulled-down were probed with antibody against pan-actin. The results clearly demonstrated that actin was indeed glutathionylated in a time-dependent manner and that levels of glutathionylation increased within 15 min after C5a treatment of GM-CSF–primed human neutrophils. The total lysates showed equal amounts of actin in the different cell lysates, indicated as “input” (Fig. 6 A).
Figure 6.
Cys374 is required for actin glutathionylation, actin polymerization, and NET formation. (A) Glutathionylated proteins in activated human neutrophil lysates were immunoprecipitated with anti–GSH antibody (time-dependent after C5a stimulation) and immunoblotted by using anti–pan-actin antibody. A total cell lysates (Input) was loaded as control. Data are representative of five independent experiments. (B) Immunoblotting. Actin glutathionylation in activated Hoxb8 mouse neutrophils overexpressing EGFP-β-actin-Cys374 (WT), point-mutated Cys374 to Ala374, or Cys374 to Glu374 was detected by anti–GSH antibody. The overexpression of WT and mutant forms of EGFP-β-actin was confirmed by immunoblotting with anti–GFP antibody. The ratio of glutathionylated EGFP-β-actin to total EGFP-β-actin was calculated as actin-glutathionylation in the bar graph (right panel). (C) Confocal microscopy. F-actin distribution and morphological changes were analyzed after short-term stimulation (total 35 min) of WT and overexpressed EGFP-β-actin-Cys374 (WT), point-mutated Cys374 to Ala374, and Cys374 to Glu374 mouse neutrophils with the indicated triggers. Right: Quantification of F-actin polymerization. (D) Confocal microscopy. DNA release was analyzed after short-term stimulation (total 35 min) of WT and overexpressed EGFP-β-actin-Cys374 (WT), point-mutated Cys374 to Ala374, and Cys374 to Glu374 mouse neutrophils with the indicated triggers. Right: Quantification of released dsDNA in supernatants of activated neutrophils. Data are means ± SEM. ***, P < 0.001. n = 3. Bars, 10 µm.
Actin monomers contain five cysteine (Cys) residues prone to glutathionylation. Previous studies revealed that only Cys374 located at the C terminus of the protein is fully exposed to the cytoplasm and elicits ROS-induced actin glutathionylation (Dalle-Donne et al., 2003; Sakai et al., 2012). To further explore the role of actin glutathionylation in NET formation, we prepared point mutants of EGFP-β-actin and transfected Hoxb8-immortalized myeloid progenitor cells with constructs expressing either WT EGFP-β-actin or the mutant forms that cannot be glutathionylated (Fig. 6 B). On induction of differentiation to mature mouse neutrophils, only WT EGFP-β-actin (Cys374) displayed increased glutathionylation after stimulation. Consistent with previously published work (Sakai et al., 2012), point mutations of the Cys374 to either alanine (Ala374) or glutamic acid (Glu374) abolished actin glutathionylation after neutrophil activation (Fig. 6 B). Moreover, in contrast to control neutrophils expressing normal or increased levels of WT EGFP-β-actin, mouse neutrophils overexpressing mutant forms of actin (Ala374 or Glu374) displayed impaired actin polymerization (Fig. 6 C) and reduced NET formation (Fig. 6 D). It should be noted that neutrophils expressing the actin mutant forms exhibited a normal ability to produce ROS on activation, which was similar to WT mouse neutrophils (Fig. S5 D), suggesting that defective NET formation was most likely a consequence of impaired ROS-induced actin glutathionylation. Collectively, these results strongly suggest that actin glutathionylation plays a critical role in neutrophil NET formation.
ROS and Grx1 in tandem regulate NET formation
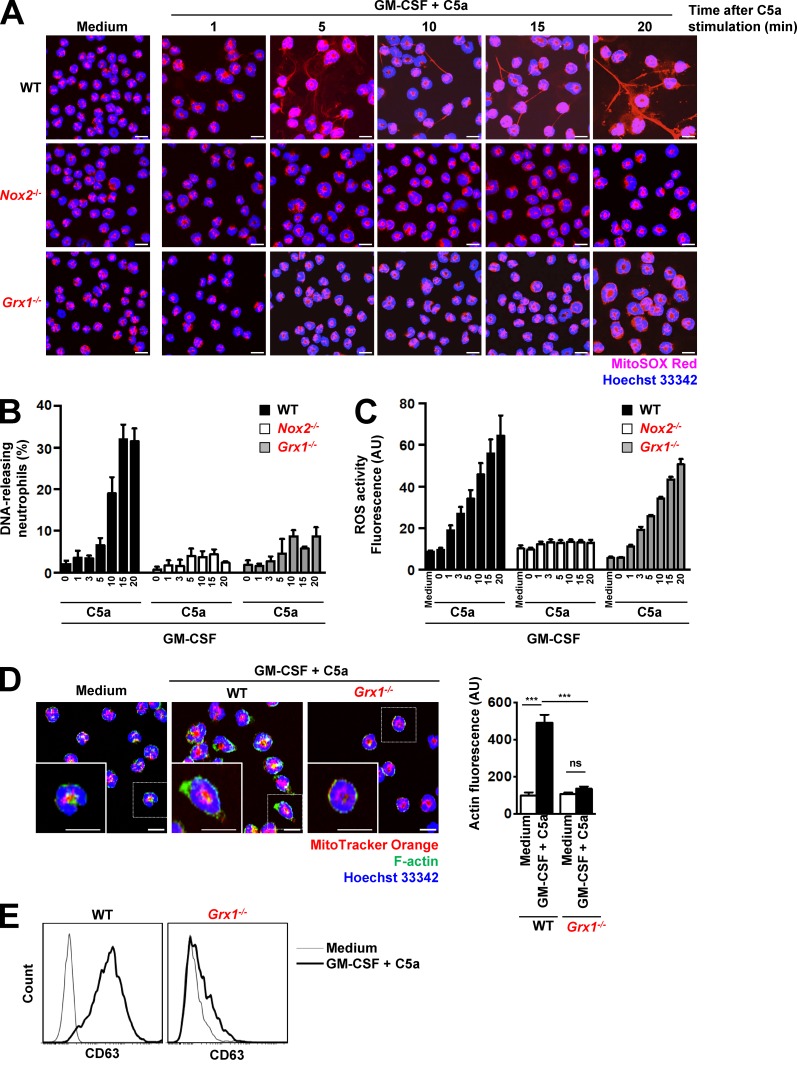
Grx1, a cytosolic thiol disulfide oxidoreductase (thioltransferase) that mediates protein deglutathionylation, is a positive regulator of actin polymerization (Ho et al., 2007). Disruption of Grx1 resulted in elevated actin glutathionylation and attenuated actin polymerization, leading to impaired neutrophil polarization, chemotaxis, adhesion, and phagocytosis (Sakai et al., 2012). We tested the ability of Grx1−/− mouse neutrophils to release DNA and to produce ROS. Once again, equal neutrophil differentiation levels and maturity in WT and Grx1−/− neutrophils were controlled by Ly6G cell surface expression (Fig. S3 A) and nuclear morphology (Fig. S3 B). We set up experiments to quantify DNA release and ROS production in a time-dependent manner and compared the response of Grx1−/− neutrophils with those of WT and Nox2−/− neutrophils. Neutrophils from both knockout mouse lines were completely devoid of any DNA release after GM-CSF and C5a (Fig. 7, A and B; and Fig. S6 D) or PMA stimulation (Fig. S6 D). In contrast to Nox2−/− neutrophils, however, Grx1−/− neutrophils produced the same amounts of ROS in response to combined GM-CSF and C5a stimulation as WT neutrophils (Fig. 7 C). Moreover, GM-CSF-primed Grx1−/− neutrophils showed significantly lower actin polymerization after a 15-min C5a stimulation than WT, similar to Nox2−/− neutrophils (Fig. 7 D). The inability of Grx1−/− neutrophils to release DNA was also accompanied by an absence of degranulation as indicated by the reduced expression of the surrogate marker CD63 after GM-CSF priming and C5a stimulation or reduced β-glucosaminidase activity and reduced MMP-9 release (Fig. 7 E; and Fig. S4, E and F). Collectively, these data further support the notion that actin polymerization is indispensable for NET formation and that it is regulated by the balance between ROS and the Grx1 enzyme.
Figure 7.
Grx1 is a master regulator of actin cytoskeleton rearrangement and DNA release. (A) Confocal microscopy. DNA release was analyzed after short-term stimulation of WT, Nox2−/−, and Grx1−/− mouse neutrophils with the indicated triggers in a time-dependent manner. n = 5. (B) Confocal microscopy. DNA-releasing neutrophils were quantified in ten high-power fields in a time-dependent manner (numbers indicated C5a stimulation time in minutes). In contrast to WT mouse neutrophils, both Nox2−/− and Grx1−/− neutrophils were completely devoid of any extracellular DNA. n = 5. (C) Flow cytometry. Total ROS activity of WT, Nox2−/−, and Grx1−/− mouse neutrophils after short-term stimulation with the indicated triggers was assessed in a time-dependent manner (numbers indicated C5a stimulation time in minutes) by using DHR123 fluorescence. n = 5. (D) Confocal microscopy. F-actin distribution and morphological changes were analyzed after short-term stimulation (total 35 min) of WT and Grx1−/− mouse neutrophils with the indicated triggers. Right: Quantification of F-actin was performed by automated analysis of microscopic images by using Imaris software. n = 3. (E) Flow cytometry. Expression of CD63 was assessed after pretreatment and short-term stimulation (total 35 min) of WT and Grx1−/− mouse neutrophils with the indicated triggers. Representative histograms are presented. n = 5. Data are means ± SEM. ***, P < 0.001. Bars, 10 µm.
Requirement for the MT network in NET formation and its potential regulation by glutathionylation
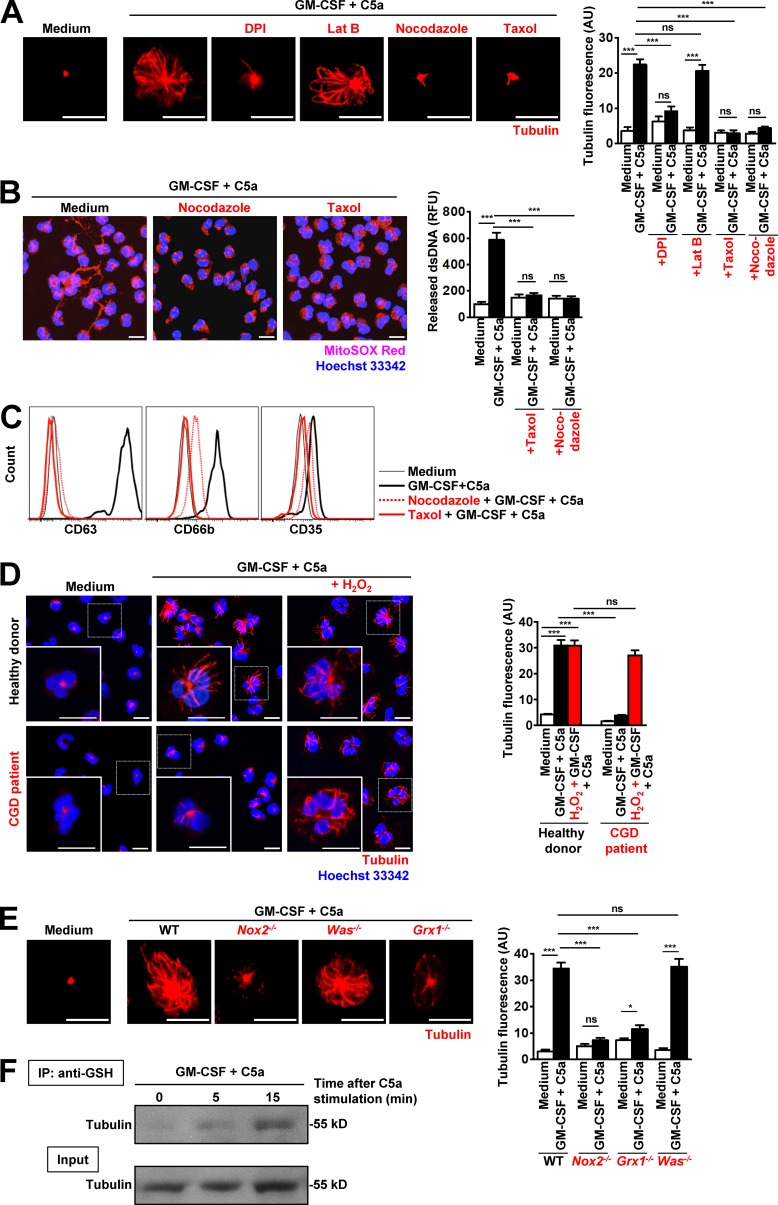
The MT network is important for neutrophil polarization and migration (Eddy et al., 2002). MTs are considered to be of pivotal importance in stabilizing cellular shape and for controlling cellular motility and the intracellular flow of granules, including the process of neutrophil degranulation (Mollinedo et al., 1989). Resting neutrophils exhibit an MT organization center. After priming with GM-CSF and a subsequent 15-min C5a activation, an MT network is established (Fig. S1 A and Video 1). To visualize the MT rearrangement in live cells on activation, we stained neutrophils with SiR-tubulin fluorescent dye before activation (Lukinavičius et al., 2014). To investigate the role of the MT network in NET formation, we used nocodazole and taxol, both known as highly efficient inhibitors of MT formation (Xu et al., 2005). Both cytoskeleton-poisoning drugs completely prevented the formation of the MT network (Fig. 8 A) and blocked DNA release (Fig. 8 B) and degranulation (Fig. 8 C; and Fig. S2, D and E). In contrast, actin polymerization (Fig. S6 E) and ROS production (Fig. S5 E) were unaffected by either nocodazole or taxol.
Figure 8.
Disruption of MT network formation inhibits degranulation and DNA release and is regulated by ROS-dependent glutathionylation. Human CGD neutrophils demonstrate defects in tubulin network formation that can be restored by addition of exogenous H2O2. (A) Confocal microscopy. MT assembly was analyzed after pretreatment and short-term stimulation (total 35 min) of human neutrophils with the indicated inhibitors: 1 µM DPI, 1.25 µM Lat B, 1 µM taxol, 5 µM nocodazole, and triggers. Right: Quantification of MT network formation was performed by automated analysis of microscopic images by using Imaris software. n = 5. (B) Confocal microscopy. DNA release was analyzed after pretreatment and short-term stimulation (total 35 min) of human neutrophils with the indicated inhibitors: 1 µM taxol, 5 µM nocodazole, and triggers. Right: Quantification of released dsDNA in supernatants of activated neutrophils. n = 5. (C) Flow cytometry. Expression of CD63, CD66b, and CD35 was assessed after pretreatment and short-term stimulation (total 35 min) of human neutrophils with the indicated inhibitors: 1 µM taxol, 5 µM nocodazole, and triggers. Representative histograms are presented. n = 5. (D) Confocal microscopy. MT assembly was analyzed after short-term stimulation (total 35 min) of control and CGD human neutrophils in the presence or absence of 50 µM H2O2 with the indicated triggers. Quantification of MT network formation was performed by automated analysis of microscopic images by using Imaris software. n = 3. (E) Confocal microscopy. MT assembly was analyzed after short-term stimulation (total 35 min) of WT, Nox2−/−, Grx1−/−, and Was−/− mouse neutrophils with the indicated triggers. Right: Quantification of MT network formation was performed by automated analysis of microscopic images by using Imaris software. n = 5. (F) Glutathionylated proteins in activated human neutrophil lysates were immunoprecipitated with anti–GSH antibody (time-dependent after C5a stimulation) and immunoblotted for tubulin. A total cell lysate (Input) was used as control. Representative data of three independent experiments are shown. Data are means ± SEM. *, P < 0.0231; ***, P < 0.001. Bars, 10 µm.
Moreover, CGD neutrophils exhibited an impaired MT network (Fig. 8 D), confirming the importance of ROS (Oliver et al., 1976; Landino et al., 2004; Wilson and González-Billault, 2015). Addition of H2O2 to CGD neutrophils, 30 min before in vitro activation, was able to correct MT network formation (Fig. 8 D). The ROS deficiency in CGD patients’ neutrophils with concentrations well below the physiological range was reported to alter the dynamic properties of the MTs in vitro and in vivo (Wilson and González-Billault, 2015).
ROS signals have already been described to regulate the organization of MT cytoskeleton (Livanos et al., 2014), and a lack of Grx1 enzyme activity leads to the accumulation of high levels of glutathionylated tubulin; consequently, GSH levels cannot be maintained, and MT assembly is inhibited (Oliver et al., 1976). We examined MT network formation after stimulation in the presence of DPI. Blocking ROS production reduced MT network formation in human neutrophils (Fig. 8 A). Similarly, Nox2−/− and Grx1−/−, but not Was−/−, mouse neutrophils also demonstrated defects in MT network formation (Fig. 8 E), indicating the crucial role of redox balance in controlling MT network formation.
The protein tubulin is composed of multiple individual α- and β-tubulin forms, and the αβ-tubulin dimer contains multiple cysteines for which cysteine oxidation of tubulin could be shown to be associated with loss of MT polymerization activity (Landino et al., 2011). To test whether tubulin is also glutathionylated in activated neutrophils, lysates of unactivated and activated human neutrophils were subjected to pulled-down assays by using anti–GSH antibody and probed with anti–tubulin antibody. The results clearly demonstrate that tubulin is a glutathionylated protein in GM-CSF–primed and C5a-stimulated human neutrophils (Fig. 8 F). Collectively, one can conclude that the MT network is required for DNA release and degranulation and is at least partially regulated by ROS. The functional role of tubulin glutathionylation will require further experimentation.
Discussion
In this article, we demonstrate that ROS acts as a key signaling molecule to orchestrate essential functions of neutrophils, such as degranulation and NET formation. Mutations resulting in a nonfunctional NADPH oxidase in mouse or human neutrophils result in attenuated actin and tubulin polymerization, degranulation, and NET formation. The same functional consequences were observed when we inactivated the NADPH oxidase pharmacologically (Fig. 9).
Figure 9.
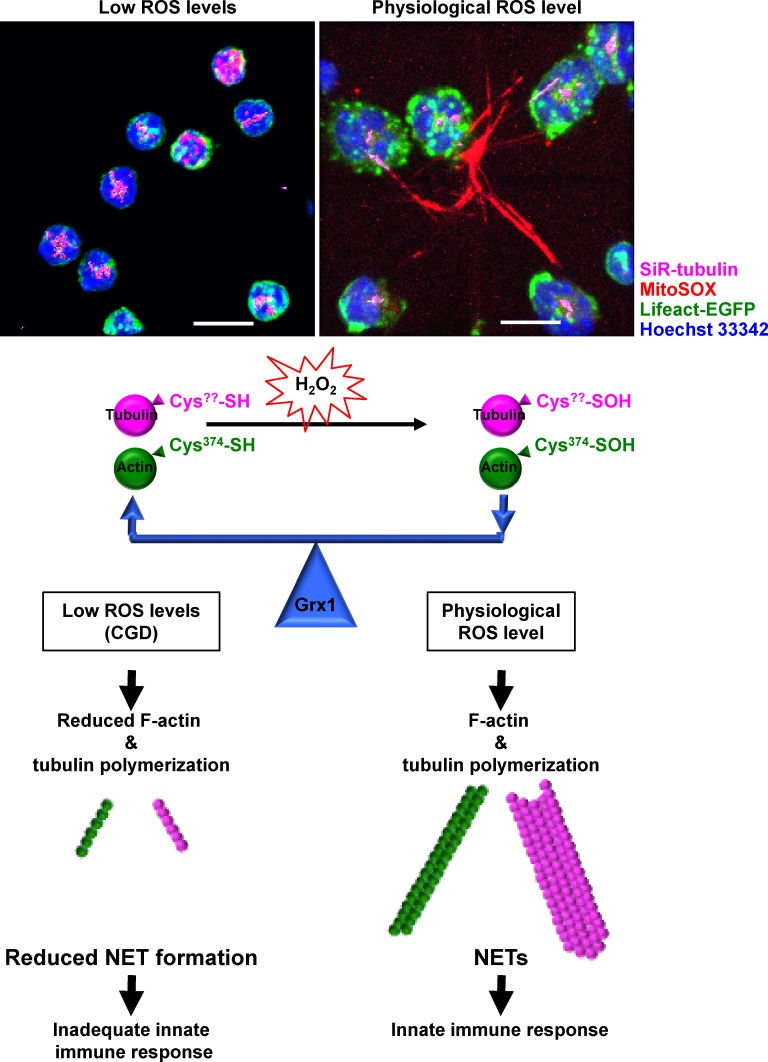
ROS-induced actin and tubulin glutathionylation controls cell cytoskeleton dynamics and NET formation. Physiological activation of neutrophils leads to moderate ROS production and cell cytoskeleton glutathionylation. ROS-induced actin and tubulin glutathionylation is tightly regulated by Grx1. Disruption of Grx1 or deficient ROS production (in CGD patients) unbalances cell cytoskeleton rearrangements and leads to a dysfunctional innate immune response in neutrophils. Confocal microscopy images are back projections of 30 z-stacks, and a visualization of the tubulin (purple) isosurface rendering was applied for better visualization by using Imaris software. Bars, 10 µm.
Interestingly, the literature is controversial regarding the ability of CGD neutrophils to degranulate (Gold et al., 1974; Voetman et al., 1981; Tintinger et al., 2001). Degranulation of neutrophils from CGD patients was variably reported to be normal, decreased, or increased (Baehner et al., 1969). So far, these conclusions have been largely based on older publications that analyzed degranulation by measuring granule enzyme activity during serum-opsonized, zymosan-induced phagocytosis (Voetman et al., 1981). We have analyzed the degranulation ability of mouse and human neutrophils by measuring released granule proteins in the supernatants of activated cells using ELISA and enzymatic β-glucosaminidase activity. Alternatively, we measured surrogate markers of degranulation using the flow cytometer. Our findings clearly demonstrate that both human CGD neutrophils and mouse Nox2-deficient neutrophils exhibit a defect in degranulation.
We newly report that the addition of exogenous H2O2 restored the formation of the MF and MT networks, as well as the ability of CGD neutrophils to degranulate. WASP colocalizes with polymerized actin in hematopoietic cells (Westerberg et al., 2001). Was−/− mouse neutrophils exhibited defects in MF cytoskeletal function, resulting in impaired clustering of β2 integrin after adhesion, leading to reduced outside-in signaling of effector functions, including degranulation (Zhang et al., 2006). Granule movement is mediated by active transport on actin filaments (Burgoyne and Morgan, 2003). The MF cytoskeleton regulates degranulation of all neutrophil granule subsets and controls the access of neutrophil granules to the plasma membrane (Jog et al., 2007). In this study, we confirm that Was−/− mouse neutrophils exhibit a defect in actin polymerization and degranulation. The failure to release DNA and to form NETs by Was−/− mouse neutrophils is a new finding.
Notably, two previous studies suggested a role for the cytoskeleton in the process of NET formation. It was reported that extracellular chromatin release requires functional tubulin and actin filaments. Pretreatment with nocodazole or cytochalasin D blocked NET formation, and cells appeared to have an increased diameter; its interior was filled with homogeneously dispersed chromatin (Neeli et al., 2009). Moreover, neutrophil elastase has been reported to promote F-actin degradation, liberating the protease to enter the nucleus (Metzler et al., 2014). However, it should be noted that this possible role of elastase was observed in long-term (2–4 h), 100 nM PMA-stimulated neutrophils. In contrast to these earlier studies, we show here that the MF network is not just acting as a passive cell structural component to maintain the shape and integrity of the neutrophils but rather directly participates in degranulation and NET formation.
NADPH oxidase–mediated ROS production occurs after the initial actin polymerization (∼0.5–1 min after stimulation) and induces a dysfunctional glutathionylated actin (Sakai et al., 2012). Grx1 is believed to play a pivotal role in the repair of oxidized protein thiols during oxidative stress and in the redox regulation within cells (Shelton and Mieyal, 2008). Targeted disruption of the Grx1 gene did not cause any obvious behavioral phenotype in mice (Ho et al., 2007), but Grx1-deficient mice exhibit defects in chemotaxis and bacterial killing both in vitro and in vivo (Sakai et al., 2012). Our results confirm that physiological levels of ROS drive reversible actin and tubulin glutathionylation. As a new finding, we demonstrate that this mechanism is physiologically involved in cellular processes mediated by MT and MF, such as degranulation and NET formation (Fig. 9).
Collectively, we provide new information regarding the mechanisms of NET formation distal to ROS production in viable neutrophils. ROS stimulates WASp-dependent actin polymerization, which is additionally controlled by the activity of Grx1 enzyme. Both WASP and Grx1 are absolutely required for both degranulation and NET formation. Directly intervening to activate actin polymerization might be a new strategy for improving neutrophil function in cases of NADPH deficiency.
Materials and methods
Reagents
GM-CSF was supplied by Novartis Pharma. Human and mouse C5a were purchased from Hycult Biotech. PMA, DPI, H2O2, and Triton X-100 were purchased from Calbiochem. Nocodazole, paclitaxel (taxol), NaASO2, CdCl2, β-mercaptoethanol, 4-hydroxytamoxifen (4-OHT), BSA, glutaraldehyde, monoclonal anti–α-tubulin (clone DM1A) for immunofluorescence staining, saponinNaBH4, LPS (055:B5), fMLP, and dihydrorhodamine 123 (DHR 123) were purchased from Sigma-Aldrich. Lat B was purchased from Enzo Life Sciences. MitoSOX Red, a derivative of ethidium bromide (a DNA-binding dye) is live-cell permeable and is oxidized by superoxide, exhibiting red fluorescence only when bound to DNA. Alexa Fluor 488 phalloidin (green) stains only polymerized actin (F-actin). MitoTracker Orange CM-H2 TMRos is taken up by mitochondria and stains in proportion to their mass. Prolong Gold mounting media, Hoechst 33342, the Quant-iT PicoGreen dsDNA assay kit, propidium iodide (PI), HBSS, RPMI-1640/GlutaMAX medium, penicillin/streptomycin, as well as Alexa Fluor 488–conjugated anti-mouse and anti-rabbit secondary antibodies were all obtained from Thermo Fisher Scientific (distributed by LuBioScience GmbH). X-VIVO 15 medium without phenol red and antibiotics was purchased from Lonza. DNase I was purchased from Worthington Biochemical Corporation. Mouse anti–human neutrophil elastase monoclonal (clone NP57) and rabbit anti–human neutrophil MPO polyclonal antibodies were obtained from Dako. Polyvalent human IgG was a gift from CSL Behring. Normal goat serum was obtained from Santa Cruz Biotechnology, Inc. ChromPure human IgG was obtained from MILAN ANALYTICA AG. IgG2a mouse anti–GSH monoclonal antibody (clone D8) obtained was from Virogen. Rabbit polyclonal anti–pan-actin and sheep anti-tubulin for immunoblotting were purchased from Cytoskeleton, Inc. The protease inhibitor cocktail was purchased from Roche Diagnostics. Mouse stem cell factor (SCF)–secreting Chinese hamster ovary cells (CHO/SCF) were provided by G. Häcker (University of Freiburg, Freiburg im Breisgau, Germany). German glass coverslips (#1 thickness, 12-mm diameter) were purchased from Hecht-Assistent. Black glass-bottom 96-well and white 96-well plates were purchased from Greiner Bio-One GmbH.
Neutrophils
Human blood neutrophils from healthy individuals and CGD patients were isolated as described previously (Amini et al., 2016). In brief, cell separation was performed by Ficoll-Hypaque centrifugation (Pancoll human from PAN BioTech). The granulocyte fraction was depleted of erythrocytes by lysis with a buffer containing NH4Cl, KHCO3, and EDTA. The resulting cell populations contained >95% neutrophils as assessed by Diff-Quik staining and light microscopy. Written, informed consent was obtained from all blood donors, and the Ethics Committee of Canton Bern had previously approved the study.
Purification of mouse neutrophils
Primary mouse bone marrow neutrophils were isolated from WT, Nox2−/−, and Lifeact-EGFP mice with a negative selection technique. In brief, bone marrow cells were collected by flushing the femur with 5 ml isolation medium (PBS plus 2% FCS, no EDTA added) by using a 26-gauge needle and filtering through a sterile 70-µM nylon cell strainer. Primary bone marrow neutrophils were then isolated with a negative selection technique by using the EasySep Mouse Neutrophil Enrichment kit (StemCell Technologies, Inc). Mouse neutrophil purity was >93% as assessed by staining with the Hematocolor Set (Merck Millipore) followed by light microscopic analysis (Rožman et al., 2015).
Hoxb8 neutrophils
Mouse neutrophils were generated from SCF-dependent, conditional Hoxb8-immortalized myeloid progenitors derived from WT and Nox2−/−, Grx1−/−, and Was−/− mice, as previously described (Morshed et al., 2014). A total of 3 × 105 cells/ml were passaged in RPMI-1640/GlutaMAX with 10% FCS, 100 U/ml penicillin per 100 µg/ml streptomycin, 50 µM 2-β-mercaptoethanol, mouse SCF (5% of CHO/SCF cell–conditioned medium), and 100 nM 4-hydroxytamoxifen for 3–4 d. To initiate differentiation into neutrophils, 2.5 × 104 cells/ml was washed and then cultured in the same medium in the absence of 4-hydroxytamoxifen. On 3 d of differentiation, 5 µg/ml of granulocyte colony-stimulating factor (G-CSF; PeproTech EC Ltd) was added to cell cultures, and after further incubation for 2 d, cells were washed twice with PBS and once with X-VIVO 15 medium before being used for subsequent experiments. Immunophenotypic characterization of Hoxb8 mouse neutrophils was performed by determining the cell surface expression of Ly6G by using flow cytometry. Single-cell suspensions of Hoxb8 neutrophils (0.5–1.0 × 106) were blocked with blocking solution (10% FCS, with 1% normal rat serum and 1% mouse serum in PBS) and stained with FITC-conjugated rat monoclonal anti-mouse (clone 1A8; BioLegend) or appropriate isotype-matched control antibodies for 45 min on ice before flow cytometric measurements (BD FACSCalibur). Data were subsequently analyzed by using FlowJo software (Tree Star). Diff-Quik staining for nuclear morphology was performed by staining the cells with hematoxylin eosin dyes followed by light microscopy. Nox2-deficient mice were purchased from the Jackson Laboratory and provided by K.-H. Krause (University of Geneva, Geneva, Switzerland). Was−/− and Grx1−/− mouse bone marrow samples were supplied by T. Stradal (Westfälische Wilhelms-Universität Münster, Münster, Germany) and K.R. Bracke (University of Ghent, Ghent, Belgium), respectively.
Activation of neutrophils
Human and mouse neutrophils were seeded on 12-mm glass coverslips in X-VIVO 15 medium (Lonza), primed with 25 ng/ml GM-CSF for 20 min, and subsequently stimulated with 10−8 M C5a for 15 min. In certain experiments, differentiated Hoxb8 neutrophils were used.
ROS measurements
ROS measurements of human and mouse neutrophils were performed by using fluorescent detection of ROS activity by flow cytometer as described previously (Amini et al., 2016). Neutrophils (2 × 106/ml) were resuspended in X-VIVO 15 medium; 100 µl cell suspension was preincubated in the presence and absence of different inhibitors for 30 min before priming with 25 ng/ml GM-CSF for 20 min. Cells were then stimulated with 10−8 M C5a. As a control, we stimulated neutrophils with 25 nM PMA for 15 min without prior GM-CSF priming. DHR 123 was added to the cells at the final concentration of 1 µM. The reaction was stopped by adding 200 µl ice-cold PBS, and the ROS activity of the samples was immediately measured by flow cytometry (BD FACSCalibur) and quantified by using FlowJo software (Tree Star).
Determination of cell death
Neutrophils were cultured at 106 cells/ml for the indicated times in the presence or absence of 50 µM H2O2, 25 nM PMA, 2 µM CdCl2, or 50 µM Na-arsenite. Cell viability was determined after 1, 8, and 24 h by using 25-µM ethidium bromide fluorescent dye exclusion assay and assessed by flow cytometry (BD FACSCalibur).
Confocal laser scanning microscopy and image analysis
Neutrophils (2.5 × 106/ml X-VIVO 15 medium) were fixed with 4% paraformaldehyde for 5 min, washed with PBS, pH 7.4, and permeabilized by using 0.05% saponin in PBS, pH 7.4, for 5 min at RT. Subsequently, the staining was performed in the presence of 0.01% saponin. Nonspecific binding was prevented by preincubation of the adherent cells with a blocking buffer (containing human immunoglobulins, normal goat serum, and 7.5% BSA) at RT for 1 h. Indirect immunofluorescence staining was performed as previously described (Amini et al., 2016) by using monoclonal mouse anti–human neutrophil elastase (1:1,000) and polyclonal rabbit anti–human MPO (1:3,000) primary antibodies, diluted in blocking solution, and incubated overnight at 4°C. Staining controls were either mouse or rabbit control Abs at the corresponding concentrations. Appropriate Alexa Fluor 488–conjugated secondary antibody (1:400) was incubated at RT for 1 h. DNA was stained with 10 µg/ml PI. Staining with cell permeable fluorescent dyes, such as 5 µM MitoSox Red or 1 µM MitoTracker Orange, was performed in live cells before fixation according to the corresponding manufacturer’s instructions. Samples were mounted in Prolong Gold mounting medium, and image acquisition was performed by using confocal laser scanning microscopy LSM 700 (Carl Zeiss). Images were sampled at a resolution of 512 × 512 by using a Plan-Apochromat 63x/1.40 Oil DIC M27 objective. Some images were obtained with software zoom (2.5×) and a z-step size of 0.1 µm (30×), and 3D reconstruction was visualized by isosurface rendering by using Imaris software (Bitplane AG).
For live cell imaging and analysis, Lifeact-EGFP mouse neutrophils were incubated with SiR-tubulin dye (100 ng/ml; SC002; Spirochrome AG) for 30 min, washed, and subsequently incubated with 25 ng/ml GM-CSF together with 0.1 µg/ml Hoechst 33342 and 0.5 µM MitoSOX for 20 min in a glass-bottom chamber. 10−8 M C5a was added and further incubated for 15 min. Live images were acquired by using a confocal microscope (inverted) equipped with a motorized stage, an autofocus device (LSM 700; Carl Zeiss) in a full stage-top incubator with humidifier, 5% CO2, and 37°C temperature-control devices. Samples were focused with a 63×/1.4 oil DIC objective, and live acquisition of at least 5 different locations was obtained for a duration of 1 h. Low laser power and fast acquisition were applied to avoid photo toxicity during live cell imaging. Data were analyzed by Imaris software (Bitplane AG).
Quantification of released dsDNA in culture supernatants
Released dsDNA was quantified as previously described (Amini et al., 2016). In brief, neutrophils (2 × 106 neutrophils/500 µl X-VIVO 15 medium) were stimulated as described in the Activation of neutrophils section. At the end of the incubation time, a low concentration of 2.5 U/ml DNase I (Worthington Biochemical Corporation) was added for an additional 10 min. Reactions were stopped by addition of 2.5 mM EDTA, pH 8.0. Cells were centrifuged at 200 g for 5 min. One hundred microliters of supernatant were transferred to black, glass-bottom, 96-well plates (Greiner Bio-One GmbH), and the fluorescent activity of PicoGreen dye bound to dsDNA was excited at 502 nm and the fluorescence emission intensity measured at 523 nm by using a spectrofluorimeter (SpectraMax M2; Molecular Devices), according to the instructions described in the Quant-iT PicoGreen assay kit.
F-actin staining and quantification
Neutrophils (2.5 × 106/ml X-VIVO 15 medium) were seeded on 12-mm glass coverslips and treated as mentioned in the Activation of neutrophils section. Cells were fixed with 4% paraformaldehyde for 5 min, subsequently washed three times in PBS, pH 7.4, and permeabilized with 0.5% saponin in buffer A (1% BSA in PBS) for 5 min at RT. F-Actin was stained using a 1.25% solution of Alexa Fluor 488 phalloidin (green), in PBS, pH 7.4, plus 0.05% BSA and 0.5% of saponin for 15 min at RT protected from light. Cells were washed two times in PBS, pH 7.4, and counterstained with 1 µg/ml Hoechst 33342 to visualize the nuclei. After washing three times in PBS, pH 7.4, samples were mounted in Prolong Gold mounting media. Slides were examined and images acquired by confocal laser scanning microscopy (LSM 700). 10 representative pictures of single-plane images from each condition, each containing more than 20 cells, were subjected to automated analysis to measure fluorescence mean intensity of F-actin before and after the activation period by using automated isosurface module of Imaris software.
Quantification of F-actin by flow cytometer
Neutrophils were cultured at 10 × 106/ml in X-VIVO 15 medium and stimulated as indicated. Cells were fixed 10 min with 4% paraformaldehyde at RT. After centrifugation at 400 g, cells were stained with a 1.25% solution of Alexa Fluor 488 phalloidin (green) in PBS, pH 7.4, with 0.05% BSA (HSA) and 0.5% of saponin for 15 min at RT, protected from light. After two washings with PBS plus 0.05% BSA, F-actin levels were determined by flow cytometer analysis (FACSCalibur) and quantified by using FlowJo software (Tree Star).
Tubulin reorganization and immunofluorescence
Neutrophils were seeded on glass coverslips and stimulated as described in the Activation of neutrophils section. Tubulin staining was performed as previously described (Feltrin and Pertz, 2012). Samples were fixed in mixture of 4% formaldehyde and 0.1% glutaraldehyde in PBS, pH 7.4, and permeabilized first with 0.5% Triton X-100 for 15 min, followed by a 15-min incubation with 0.5% SDS at RT. Autofluorescence was then quenched with 0.5 mg/ml NaBH4 in PBS, pH 7.4, for 10 min on ice. Nonspecific binding to the Fc-receptor was blocked in blocking buffer (7.5% BSA, 30% human IgG polyvalent [IVIG], 30% normal goat sera, and 1% ChromPure human IgG in PBS) for 1 h at RT. Anti–α-tubulin mAb (clone DM1A) was diluted 1:100,000 in blocking buffer and incubated overnight at 4°C and subsequently washed three times in PBS, pH 7.4, with 0.025% Triton X-100 and then further incubated with the secondary antibody Alexa-555 goat anti–mouse IgG (1:1,000 in 7.5% BSA plus 0.025% Triton X-100; Life Technologies Ltd) for 1 h at RT. Samples were extensively washed with PBS, pH 7.4, plus 0.025% Triton X-100 and counterstained with 1 µg/ml Hoechst 33342 for 5 min at RT in the dark. After several washes, the samples were mounted on glass slides by using Prolong Gold mounting medium. Images were acquired by using LSM 700. Ten representative pictures from each condition, each containing more than 20 cells, were subjected to automated analysis to measure fluorescence mean intensity of anti–α-tubulin before and after the activation period by using automated isosurface module of Imaris software.
Glutathionylation immunoblotting
Human neutrophils were incubated in the presence or absence of 50 µM DPI for 30 min at 37°C or mouse neutrophils (WT and Nox2−/−) at a density of 7 × 106/ml in 1× HBSS (with Ca2+ and Mg2+; Invitrogen) and subsequently primed with 25 ng/ml GM-CSF for 20 min. Cells were then stimulated with 10−8 M C5a at the indicated time points. The actin glutathionylation immunoblot was performed as previously described (Sakai et al., 2012). Ice-cold HBSS containing 5 mM N-ethyl maleimide (NEM) was added to samples for 1 min to stop the reaction and to protect the free thiols. Samples were centrifuged and cell pellets lysed in lysis buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 10% glycerol, 1% Triton X-100, 2 mM EDTA, 10 mM sodium pyrophosphate, 50 mM sodium fluoride, 200 µM Na3VO4, and 1 mM PMSF in H2O) plus protease inhibitor cocktail for 25 min on ice with frequent vortexing, and supernatants were collected after high-speed centrifugation (10 min, 13,000 rpm, 4°C). Fifty micrograms of protein was loaded on NuPAGE 12% Bis-Tris Gel (NW00122; Thermo Fisher Scientific) in the absence of denaturing reagents. The lysates were separated by electrophoresis under nonreducing conditions and transferred onto nitrocellulose membranes (RPN2020D; GE Healthcare Life Sciences) by using the NuPAGE blotting system (Thermo Fisher Scientific). Membranes were blocked with 5% nonfat dry milk in Tris-buffered saline solution containing 0.1% Tween 20 (TBST) for 1 h and incubated with primary antibodies α-GSH antibody (clone 8D, 1:1,000) in TBST at 4°C overnight. Membranes were washed by using TBST and further incubated with the corresponding HRP-conjugated secondary antibodies and visualized by chemiluminescence (ECL PLUS Western Blotting Analysis System; GE Healthcare). Total protein loading was evaluated by using rabbit polyclonal anti-actin (Cytoskeleton Inc., 1:1,000) in TBST.
Immunoprecipitation (IP) and immunoblotting of glutathionylated actin and tubulin
Human neutrophils were isolated as described in the Neutrophils section and washed twice with PBS plus 2% FCS and once with X-VIVO 15 medium after which the cell number was adjusted to 15 × 107 cells/ml in X-VIVO 15 medium with 0.1 mM diisopropylfluorophosphate. Cells were primed with 25 ng/ml GM-CSF for 20 min in Eppendorf tubes and stimulated with 10−8 M C5a in a time-dependent manner. On termination of the activation period, 500 ml ice-cold medium (X-VIVO 15) containing 5 mM NEM (04259-5G; Sigma-Aldrich) was added, and samples were placed on ice to stop the activation. Samples were centrifuged at 2,000 g for 3 min at 4°C, and cell pellets were lysed in lysis solution (0.025 M Tris, 0.15 M NaCl, 0.001 M EDTA, 1% NP-40, and 5% glycerol, pH 7.4) provided by the Pierce IP kit (26149; Thermo Fisher Scientific), with addition of the following inhibitors: 5 mM NEM, 1 mM PMSF, and the protease inhibitor cocktail. Cells were lysed for 15 min on ice with frequent vortexing and centrifuged at 13,000 g for 10 min at 4°C. Cell lysates were subjected to preclearing by using Pierce control agarose resin provided by the IP kit for 1 h at 4°C, as described by the IP kit instruction, and the precleared cell lysate protein concentration was measured. Specifically, 1 µg protein from each sample was used to isolate glutathionylated proteins. 50 µg of protein from each sample was stored to be used as “Input.” The IP procedure was performed exactly according to the IP kit instructions, and 5 µg anti–GSH antibody (101-A-100, clone D8; ViroGen Corporation) was cross-linked to aminolink plus coupling resin provided by the IP kit by using sodium cyanoborohydride for 2 h at RT, as recommended by the IP kit instructions. Immobilized antibody on the resin was washed with coupling buffer (0.01 M sodium phosphate and 0.15 M NaCl, pH 7.2) and quenched in quenching buffer (1 M Tris-HCl). After one further treatment with sodium cyanoborohydride, the antibody resin complex was thoroughly washed six times with lysis solution plus 5 mM NEM. After removal of all residual wash solution, immobilized antibody-resin was added to 500 µl (1 µg) precleared cell lysates and incubated overnight at 4°C by rotating end-over-end. On the next day, the immune complexes were washed with lysis solution plus 5 mM NEM and protease cocktail five times, and the captured antigens were eluted using a total of 60 µl elution buffer, and 5× loading buffer in the absence of any reducing agent was added (both provided by the IP kit). Samples were heated at 85°C for 7 min, briefly vortexed, and immediately loaded on NuPAGE 12% Bis-Tris Gel (NW00122; Thermo Fisher Scientific) at 80 V for first 30 min and 140 V for 3.5 h. Blotting was performed on a Hybond nitrocellulose membrane (RPN2020D; GE Healthcare Life Sciences) by using the NuPAGE blotting system (Thermo Fisher Scientific) and transferred at 4°C for 1 h at 30 V. The membrane was washed once with Tris-buffered saline solution containing 0.1% TBST, incubated at RT in blocking buffer (TBST plus 5% nonfat milk, containing 2.5 mM NEM) for 1 h, and further incubated with primary antibodies: rabbit anti-actin (1:1,000; AAN01; Cytoskeleton, Inc) or sheep anti-tubulin (1:1,000; ATN02; Cytoskeleton, Inc) in TBST plus 1% nonfat milk ON at 4°C. Membranes were washed four times, each time for 8 min, with TBST and incubated with secondary antibodies: Amersham ECL donkey anti–rabbit IgG, HRP-linked whole antibody (1:15,000; NA934V; GE Healthcare Life Sciences), or donkey anti–sheep IgG, HRP-linked whole antibody (1:30,000; #A130-100; Bethyl laboratories, Inc). Bands were visualized by chemiluminescence with the ECL PLUS Western blotting system (GE Healthcare Life Sciences).
Overexpression of WT and mutant forms of β-actin in Hoxb8 cells
Human β-actin, ACTB cDNA (NM_001101; encoding identical amino acid sequence to mouse β-actin), was cloned in pcDNA3 expression vector, and the cytomegalovirus promoter was replaced with a long β-actin promoter. N-terminal EGFP was separated from β-actin by a flexible, hydrophilic 5 amino acid spacer (provided by B. Wehrle-Haller, University of Geneva, Geneva, Switzerland; Ballestrem et al., 1998).
To generate point mutations in EGFP-β-actin (Cys374 to Ala374 or Glu374), site-directed mutagenesis, the QuikChange II XL Site-Directed Mutagenesis kit (200521; Agilent Technologies) was used. Mutation primers were as follows: EGFP-β-actin_Ala374 forward primer: 5′-CGTCCACCGCAAAGCCTTCTAGGAATTCTGCAGATATCCA-3′; EGFP-β-actin_Ala374 reverse primer: 5′-TGGATATCTGCAGAATTCCTAGAAGGCTTTGCGGTGGACG-3′; EGFP-β-actin_Glu374 forward: 5′-CGTCCACCGCAAAGAGTTCTAGGAATTCTGCAGATATCCATCA-3′; and EGFP-β-actin_Glu374 reverse: 5′-TGATGGATATCTGCAGAATTCCTAGAACTCTTTGCGGTGGACG-3′, where bold letters indicate the mutated codons. All mutations were introduced by PCR and verified by DNA sequencing. WT and mutant EGFP-β-actin constructs were overexpressed in undifferentiated Hoxb8 cells by using the Neon Transfection System (Thermo Fisher Scientific) according to the manufacturer’s instructions, following specific conditions—voltage: 1,350 V, 10 ms, 4 pulses, Tip, 10 µl (5 µg) plasmid DNA, with a total cell density of 5 × 106. G418-resistant cells (25 µg/ml) were expanded, and cell lysates were tested for expression of EGFP-β-actin and their ability to differentiate to mature mouse neutrophils and then tested for actin polymerization and NET formation.
Degranulation assay
Neutrophils (4 × 106 cells/ml) were resuspended in X-VIVO 15 medium and pretreated for 30 min at 37°C with various inhibitors if needed. Neutrophils were primed with 25 ng/ml GM-CSF and activated by 10−8 M C5a for 15 min. Degranulation of gelatinase granules was determined by ELISA for human MMP-9 (R&D Systems) and mouse MMP-9 (USCN life Science Inc) as previously described (Uriarte et al., 2011). For β-glucosaminidase activity determinations, the reaction was stopped by adding ice-cold HBSS-0.1% BSA, and the supernatants were collected after a centrifugation step (20 min, 2,000 rpm, 4°C). Cell pellets were lysed with 0.12% Triton X-100 for 10 min at RT. The substrate solution (5 mM p-nitrophenyl-2-acetamido-2-deoxy-α-D-glucopyranoside in 25 mM sodium citrate, pH 4.5) was added to both cell lysate and supernatant, and then incubated at 37°C for 1 h. The reaction was stopped by adding 52 mM glycine-NaOH, pH 10.5, and absorbance was measured at 410 nm by using a SpectraMax M2 plate reader (Bucher Biotech). The amount of β-glucosaminidase released in supernatants of neutrophils was expressed as the percentage of the total β-glucosaminidase (Sato et al., 2013).
Degranulation analysis was also performed by flow cytometry after neutrophils had been stimulated as described in the Activation of neutrophils section. For azurophil granule protein, in the final 5 min of GM-CSF priming, 5 µM cytochalasin B was added to the cell suspension as previously described (Mitchell et al., 2008; Uriarte et al., 2011). Degranulation of secretory vesicles, specific granules, and azurophil granules was determined by measuring the increase in plasma membrane expression of surrogate markers by using the following mAbs: APC-conjugated anti–human CD63 (clone H5C6; BioLegend), FITC-conjugated anti–human CD66b (clone G10F5; BioLegend), phycoerythrin-conjugated anti–human CD35 (clone E11; BioLegend), and phycoerythrin-conjugated rat anti–mouse CD63 (clone NVG-2; BD Biosciences). Cells were analyzed by flow cytometry (FACSCalibur) and quantified by using FlowJo software (Tree Star) as previously described (Uriarte et al., 2011).
Bacterial killing assay
The bacterial killing assay was performed as previously described (Amini et al., 2016). A single colony of Escherichia coli GFP M655 was cultured in Luria broth base (LB) medium (Sigma-Aldrich) at 37°C, shaken at 220 rpm, overnight. The bacterial culture was diluted 1:100 in LB medium, grown to midlogarithmic growth phase (OD600 = 0.7), and centrifuged at 1,000 g for 5 min. The bacterial pellets were washed twice with 1× HBSS (LuBioScience GmbH) and gently centrifuged at 100 g for 5 min to remove any clumped bacteria. Bacteria were opsonized with 10% mouse sera (heat-inactivated) in 1× HBSS, rotated end-over-end (6 rpm) for 20 min at 37°C, and then used immediately. Activated neutrophils (107/ml in RPMI-1640 plus 2% FCS in the absence of antibiotics) were mixed with an equal volume of opsonized bacteria (5 × 107/ml) at 1:5 ratios, in the presence and absence of 100 U/ml DNase I (Worthington Biochemical Corporation). The co-cultures of cells and bacteria were rotated end-over-end (6 rpm) for 30 min at 37°C. At the end of the incubation period, an equal volume of ice-cold 1× HBSS was added to each tube to stop the reaction, and cells were pelleted by gentle centrifugation (5 min, 100 g, 4°C) by using a swing-out rotor. Supernatants containing bacteria were collected, diluted 1:300, plated on agar, and grown overnight before the colonies were counted. The tubes containing bacteria alone were treated the same way and used as controls.
Ethics statement
Written, informed consent was obtained from all patients before blood collection. The study was approved by the ethics committee of the Canton of Bern, Switzerland. Animal experiments were reviewed and approved by the animal experimentation review board of the Canton Bern, Switzerland.
Statistical analysis
Analysis of all data was performed by the Prism software (GraphPad Software Inc). All data were expressed as means ± SEM. To compare groups, one-way ANOVA followed by Tukey’s multiple comparisons test or the Kruskal-Wallis test was applied. P-values of <0.05 were considered statistically significant.
Online supplemental material
Fig. S1 shows snapshots of live mouse neutrophils exhibiting actin and tubulin rearrangements on activation and the effects of different stimuli on actin polymerization and NET formation. Fig. S2 shows the inhibition of degranulation by pharmacological inhibitors of the NADPH oxidase, actin polymerization, and MT formation. Fig. S3 confirms the maturation of Hoxb8 neutrophils. Fig. S4 shows defects in degranulation of Was−/−, Nox2−/−, and Grx1−/− neutrophils. Fig. S5 shows that actin glutathionylation and MT formation do not regulate NADPH oxidase activity and that mouse neutrophils overexpressing β-actin with Ala374 or Glu374 mutations are capable of ROS production. Fig. S6 shows that 50 μM H2O2, 50 μM sodium arsenite, and 2 μM cadmium chloride are not toxic for neutrophils. Fig. S6 D shows that Grx1−/− neutrophils are unable to release extracellular DNA. Fig. S6 E shows that pharmacological inhibition of MT formation does not affect actin polymerization.
Supplementary Material
Acknowledgments
Images were acquired on equipment supported by the Microscopy Imaging Centre of the University of Bern.
This work was supported by Swiss National Science Foundation grants 31003A_173215 to S. Yousefi and 310030_166473 to H.-U. Simon. D. Stojkov and P. Amini are PhD students of the Graduate School of Cellular and Biomedical Sciences of the University of Bern.
The authors declare no competing financial interests.
Author contributions: S. Yousefi and H.-U. Simon designed the experiments. D. Stojkov, P. Amini, and K. Oberson performed the experiments. A. Duppenthaler and C. Sokollik organized the patient materials. S. Yousefi, D. Stojkov, P. Amini, and H.-U. Simon analyzed the data. D. Stojkov, H.-U. Simon, and S. Yousefi wrote the paper.
References
- Abdel-Latif D., Steward M., Macdonald D.L., Francis G.A., Dinauer M.C., and Lacy P.. 2004. Rac2 is critical for neutrophil primary granule exocytosis. Blood. 104:832–839. 10.1182/blood-2003-07-2624 [DOI] [PubMed] [Google Scholar]
- Amini P., Stojkov D., Wang X., Wicki S., Kaufmann T., Wong W.W., Simon H.U., and Yousefi S.. 2016. NET formation can occur independently of RIPK3 and MLKL signaling. Eur. J. Immunol. 46:178–184. 10.1002/eji.201545615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baehner R.L., Karnovsky M.J., and Karnovsky M.L.. 1969. Degranulation of leukocytes in chronic granulomatous disease. J. Clin. Invest. 48:187–192. 10.1172/JCI105967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballestrem C., Wehrle-Haller B., and Imhof B.A.. 1998. Actin dynamics in living mammalian cells. J. Cell Sci. 111:1649–1658. [DOI] [PubMed] [Google Scholar]
- Bianchi M., Hakkim A., Brinkmann V., Siler U., Seger R.A., Zychlinsky A., and Reichenbach J.. 2009. Restoration of NET formation by gene therapy in CGD controls aspergillosis. Blood. 114:2619–2622. 10.1182/blood-2009-05-221606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borregaard N., Sørensen O.E., and Theilgaard-Mönch K.. 2007. Neutrophil granules: a library of innate immunity proteins. Trends Immunol. 28:340–345. 10.1016/j.it.2007.06.002 [DOI] [PubMed] [Google Scholar]
- Brinkmann V., Reichard U., Goosmann C., Fauler B., Uhlemann Y., Weiss D.S., Weinrauch Y., and Zychlinsky A.. 2004. Neutrophil extracellular traps kill bacteria. Science. 303:1532–1535. 10.1126/science.1092385 [DOI] [PubMed] [Google Scholar]
- Burgoyne R.D., and Morgan A.. 2003. Secretory granule exocytosis. Physiol. Rev. 83:581–632. 10.1152/physrev.00031.2002 [DOI] [PubMed] [Google Scholar]
- Choong G., Liu Y., Xiao W., and Templeton D.M.. 2013. Cadmium-induced glutathionylation of actin occurs through a ROS-independent mechanism: implications for cytoskeletal integrity. Toxicol. Appl. Pharmacol. 272:423–430. 10.1016/j.taap.2013.07.002 [DOI] [PubMed] [Google Scholar]
- Cingolani L.A., and Goda Y.. 2008. Actin in action: the interplay between the actin cytoskeleton and synaptic efficacy. Nat. Rev. Neurosci. 9:344–356. 10.1038/nrn2373 [DOI] [PubMed] [Google Scholar]
- Dalle-Donne I., Giustarini D., Rossi R., Colombo R., and Milzani A.. 2003. Reversible S-glutathionylation of Cys 374 regulates actin filament formation by inducing structural changes in the actin molecule. Free Radic. Biol. Med. 34:23–32. 10.1016/S0891-5849(02)01182-6 [DOI] [PubMed] [Google Scholar]
- Eddy R.J., Pierini L.M., and Maxfield F.R.. 2002. Microtubule asymmetry during neutrophil polarization and migration. Mol. Biol. Cell. 13:4470–4483. 10.1091/mbc.E02-04-0241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feltrin D., and Pertz O.. 2012. Assessment of Rho GTPase signaling during neurite outgrowth. Methods Mol. Biol. 827:181–194. 10.1007/978-1-61779-442-1_13 [DOI] [PubMed] [Google Scholar]
- Filippi M.D., Harris C.E., Meller J., Gu Y., Zheng Y., and Williams D.A.. 2004. Localization of Rac2 via the C terminus and aspartic acid 150 specifies superoxide generation, actin polarity and chemotaxis in neutrophils. Nat. Immunol. 5:744–751. 10.1038/ni1081 [DOI] [PubMed] [Google Scholar]
- Giustarini D., Rossi R., Milzani A., Colombo R., and Dalle-Donne I.. 2004. S-glutathionylation: from redox regulation of protein functions to human diseases. J. Cell. Mol. Med. 8:201–212. 10.1111/j.1582-4934.2004.tb00275.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold S.B., Hanes D.M., Stites D.P., and Fudenberg H.H.. 1974. Abnormal kinetics of degranulation in chronic granulomatous disease. N. Engl. J. Med. 291:332–337. 10.1056/NEJM197408152910704 [DOI] [PubMed] [Google Scholar]
- Gu Y., Filippi M.D., Cancelas J.A., Siefring J.E., Williams E.P., Jasti A.C., Harris C.E., Lee A.W., Prabhakar R., Atkinson S.J., et al. . 2003. Hematopoietic cell regulation by Rac1 and Rac2 guanosine triphosphatases. Science. 302:445–449. 10.1126/science.1088485 [DOI] [PubMed] [Google Scholar]
- Ho Y.S., Xiong Y., Ho D.S., Gao J., Chua B.H.L., Pai H., and Mieyal J.J.. 2007. Targeted disruption of the glutaredoxin 1 gene does not sensitize adult mice to tissue injury induced by ischemia/reperfusion and hyperoxia. Free Radic. Biol. Med. 43:1299–1312. 10.1016/j.freeradbiomed.2007.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jog N.R., Rane M.J., Lominadze G., Luerman G.C., Ward R.A., and McLeish K.R.. 2007. The actin cytoskeleton regulates exocytosis of all neutrophil granule subsets. Am. J. Physiol. Cell Physiol. 292:C1690–C1700. 10.1152/ajpcell.00384.2006 [DOI] [PubMed] [Google Scholar]
- Johansson M., and Lundberg M.. 2007. Glutathionylation of beta-actin via a cysteinyl sulfenic acid intermediary. BMC Biochem. 8:26–33. 10.1186/1471-2091-8-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindzelskii A.L., Ueki T., Michibata H., Chaiworapongsa T., Romero R., and Petty H.R.. 2004. 6-phosphogluconate dehydrogenase and glucose-6-phosphate dehydrogenase form a supramolecular complex in human neutrophils that undergoes retrograde trafficking during pregnancy. J. Immunol. 172:6373–6381. 10.4049/jimmunol.172.10.6373 [DOI] [PubMed] [Google Scholar]
- Lacy P. 2005. The role of Rho GTPases and SNAREs in mediator release from granulocytes. Pharmacol. Ther. 107:358–376. 10.1016/j.pharmthera.2005.03.008 [DOI] [PubMed] [Google Scholar]
- Lacy P. 2006. Mechanisms of degranulation in neutrophils. Allergy Asthma Clin. Immunol. 2:98–108. 10.1186/1710-1492-2-3-98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacy P., and Eitzen G.. 2008. Control of granule exocytosis in neutrophils. Front. Biosci. 13:5559–5570. 10.2741/3099 [DOI] [PubMed] [Google Scholar]
- Landino L.M., Moynihan K.L., Todd J.V., and Kennett K.L.. 2004. Modulation of the redox state of tubulin by the glutathione/glutaredoxin reductase system. Biochem. Biophys. Res. Commun. 314:555–560. 10.1016/j.bbrc.2003.12.126 [DOI] [PubMed] [Google Scholar]
- Landino L.M., Hagedorn T.D., Kim S.B., and Hogan K.M.. 2011. Inhibition of tubulin polymerization by hypochlorous acid and chloramines. Free Radic. Biol. Med. 50:1000–1008. 10.1016/j.freeradbiomed.2011.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao B.C., Hsieh C.W., Lin Y.C., and Wung B.S.. 2010. The glutaredoxin/glutathione system modulates NF-kappaB activity by glutathionylation of p65 in cinnamaldehyde-treated endothelial cells. Toxicol. Sci. 116:151–163. 10.1093/toxsci/kfq098 [DOI] [PubMed] [Google Scholar]
- Lim M.B.H., Kuiper J.W., Katchky A., Goldberg H., and Glogauer M.. 2011. Rac2 is required for the formation of neutrophil extracellular traps. J. Leukoc. Biol. 90:771–776. 10.1189/jlb.1010549 [DOI] [PubMed] [Google Scholar]
- Livanos P., Galatis B., and Apostolakos P.. 2014. The interplay between ROS and tubulin cytoskeleton in plants. Plant Signal. Behav. 9:e28069 10.4161/psb.28069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukinavičius G., Reymond L., D’Este E., Masharina A., Göttfert F., Ta H., Güther A., Fournier M., Rizzo S., Waldmann H., et al. . 2014. Fluorogenic probes for live-cell imaging of the cytoskeleton. Nat. Methods. 11:731–733. 10.1038/nmeth.2972 [DOI] [PubMed] [Google Scholar]
- Metzler K.D., Goosmann C., Lubojemska A., Zychlinsky A., and Papayannopoulos V.. 2014. A myeloperoxidase-containing complex regulates neutrophil elastase release and actin dynamics during NETosis. Cell Reports. 8:883–896. 10.1016/j.celrep.2014.06.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millard T.H., Sharp S.J., and Machesky L.M.. 2004. Signalling to actin assembly via the WASP (Wiskott-Aldrich syndrome protein)-family proteins and the Arp2/3 complex. Biochem. J. 380:1–17. 10.1042/bj20040176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell T., Lo A., Logan M.R., Lacy P., and Eitzen G.. 2008. Primary granule exocytosis in human neutrophils is regulated by Rac-dependent actin remodeling. Am. J. Physiol. Cell Physiol. 295:C1354–C1365. 10.1152/ajpcell.00239.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollinedo F., Nieto J.M., and Andreu J.M.. 1989. Cytoplasmic microtubules in human neutrophil degranulation: reversible inhibition by the colchicine analogue 2-methoxy-5-(2′,3′,4′-trimethoxyphenyl)-2,4,6-cycloheptatrien-1- one. Mol. Pharmacol. 36:547–555. [PubMed] [Google Scholar]
- Morshed M., Hlushchuk R., Simon D., Walls A.F., Obata-Ninomiya K., Karasuyama H., Djonov V., Eggel A., Kaufmann T., Simon H.U., and Yousefi S.. 2014. NADPH oxidase-independent formation of extracellular DNA traps by basophils. J. Immunol. 192:5314–5323. 10.4049/jimmunol.1303418 [DOI] [PubMed] [Google Scholar]
- Munnamalai V., and Suter D.M.. 2009. Reactive oxygen species regulate F-actin dynamics in neuronal growth cones and neurite outgrowth. J. Neurochem. 108:644–661. 10.1111/j.1471-4159.2008.05787.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neeli I., Dwivedi N., Khan S., and Radic M.. 2009. Regulation of extracellular chromatin release from neutrophils. J. Innate Immun. 1:194–201. 10.1159/000206974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver J.M., Albertini D.F., and Berlin R.D.. 1976. Effects of glutathione-oxidizing agents on microtubule assembly and microtubule-dependent surface properties of human neutrophils. J. Cell Biol. 71:921–932. 10.1083/jcb.71.3.921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver J.M., Spielberg S.P., Pearson C.B., and Schulman J.D.. 1978. Microtubule assembly and function in normal and glutathione synthetase-deficient polymorphonuclear leukocytes. J. Immunol. 120:1181–1186. [PubMed] [Google Scholar]
- Parker H., Dragunow M., Hampton M.B., Kettle A.J., and Winterbourn C.C.. 2012. Requirements for NADPH oxidase and myeloperoxidase in neutrophil extracellular trap formation differ depending on the stimulus. J. Leukoc. Biol. 92:841–849. 10.1189/jlb.1211601 [DOI] [PubMed] [Google Scholar]
- Reth M. 2002. Hydrogen peroxide as second messenger in lymphocyte activation. Nat. Immunol. 3:1129–1134. 10.1038/ni1202-1129 [DOI] [PubMed] [Google Scholar]
- Riedl J., Flynn K.C., Raducanu A., Gärtner F., Beck G., Bösl M., Bradke F., Massberg S., Aszodi A., Sixt M., and Wedlich-Söldner R.. 2010. Lifeact mice for studying F-actin dynamics. Nat. Methods. 7:168–169. 10.1038/nmeth0310-168 [DOI] [PubMed] [Google Scholar]
- Rožman S., Yousefi S., Oberson K., Kaufmann T., Benarafa C., and Simon H.U.. 2015. The generation of neutrophils in the bone marrow is controlled by autophagy. Cell Death Differ. 22:445–456. 10.1038/cdd.2014.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai J., Li J., Subramanian K.K., Mondal S., Bajrami B., Hattori H., Jia Y., Dickinson B.C., Zhong J., Ye K., et al. . 2012. Reactive oxygen species-induced actin glutathionylation controls actin dynamics in neutrophils. Immunity. 37:1037–1049. 10.1016/j.immuni.2012.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Hongu T., Sakamoto M., Funakoshi Y., and Kanaho Y.. 2013. Molecular mechanisms of N-formyl-methionyl-leucyl-phenylalanine-induced superoxide generation and degranulation in mouse neutrophils: phospholipase D is dispensable. Mol. Cell. Biol. 33:136–145. 10.1128/MCB.00869-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao D., Segal A.W., and Dekker L.V.. 2010. Subcellular localisation of the p40phox component of NADPH oxidase involves direct interactions between the Phox homology domain and F-actin. Int. J. Biochem. Cell Biol. 42:1736–1743. 10.1016/j.biocel.2010.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelton M.D., and Mieyal J.J.. 2008. Regulation by reversible S-glutathionylation: molecular targets implicated in inflammatory diseases. Mol. Cells. 25:332–346. [PMC free article] [PubMed] [Google Scholar]
- Snapper S.B., Rosen F.S., Mizoguchi E., Cohen P., Khan W., Liu C.H., Hagemann T.L., Kwan S.P., Ferrini R., Davidson L., et al. . 1998. Wiskott-Aldrich syndrome protein-deficient mice reveal a role for WASP in T but not B cell activation. Immunity. 9:81–91. 10.1016/S1074-7613(00)80590-7 [DOI] [PubMed] [Google Scholar]
- Stoiber W., Obermayer A., Steinbacher P., and Krautgartner W.D.. 2015. The role of reactive oxygen species (ROS) in the formation of extracellular traps (ETs) in humans. Biomolecules. 5:702–723. 10.3390/biom5020702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M., Kato M., Hanaka H., Izumi T., and Morikawa A.. 2003. Actin assembly is a crucial factor for superoxide anion generation from adherent human eosinophils. J. Allergy Clin. Immunol. 112:126–133. 10.1067/mai.2003.1515 [DOI] [PubMed] [Google Scholar]
- Tintinger G.R., Theron A.J., Steel H.C., and Anderson R.. 2001. Accelerated calcium influx and hyperactivation of neutrophils in chronic granulomatous disease. Clin. Exp. Immunol. 123:254–263. 10.1046/j.1365-2249.2001.01447.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uriarte S.M., Rane M.J., Luerman G.C., Barati M.T., Ward R.A., Nauseef W.M., and McLeish K.R.. 2011. Granule exocytosis contributes to priming and activation of the human neutrophil respiratory burst. J. Immunol. 187:391–400. 10.4049/jimmunol.1003112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voetman A.A., Weening R.S., Hamers M.N., Meerhof L.J., Bot A.A.A.M., and Roos D.. 1981. Phagocytosing human neutrophils inactivate their own granular enzymes. J. Clin. Invest. 67:1541–1549. 10.1172/JCI110185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerberg L., Greicius G., Snapper S.B., Aspenström P., and Severinson E.. 2001. Cdc42, Rac1, and the Wiskott-Aldrich syndrome protein are involved in the cytoskeletal regulation of B lymphocytes. Blood. 98:1086–1094. 10.1182/blood.V98.4.1086 [DOI] [PubMed] [Google Scholar]
- Wilson C., and González-Billault C.. 2015. Regulation of cytoskeletal dynamics by redox signaling and oxidative stress: implications for neuronal development and trafficking. Front. Cell. Neurosci. 9:381 10.3389/fncel.2015.00381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Wang F., Van Keymeulen A., Rentel M., and Bourne H.R.. 2005. Neutrophil microtubules suppress polarity and enhance directional migration. Proc. Natl. Acad. Sci. USA. 102:6884–6889. 10.1073/pnas.0502106102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousefi S., Gold J.A., Andina N., Lee J.J., Kelly A.M., Kozlowski E., Schmid I., Straumann A., Reichenbach J., Gleich G.J., and Simon H.U.. 2008. Catapult-like release of mitochondrial DNA by eosinophils contributes to antibacterial defense. Nat. Med. 14:949–953. 10.1038/nm.1855 [DOI] [PubMed] [Google Scholar]
- Yousefi S., Mihalache C., Kozlowski E., Schmid I., and Simon H.U.. 2009. Viable neutrophils release mitochondrial DNA to form neutrophil extracellular traps. Cell Death Differ. 16:1438–1444. 10.1038/cdd.2009.96 [DOI] [PubMed] [Google Scholar]
- Zhang H., Schaff U.Y., Green C.E., Chen H., Sarantos M.R., Hu Y., Wara D., Simon S.I., and Lowell C.A.. 2006. Impaired integrin-dependent function in Wiskott-Aldrich syndrome protein-deficient murine and human neutrophils. Immunity. 25:285–295. 10.1016/j.immuni.2006.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.