Alterations of the transcriptome and proteome enable stress recovery, but coordination of these events under stress is only partly understood. Mathew et al. report that under stress, an RNA splicing complex disassembles and the splicing factor Hsh155 moves to protein aggregates, coinciding with a drop in splicing and concomitant repression of ribosome production.
Abstract
Upon genotoxic stress, dynamic relocalization events control DNA repair as well as alterations of the transcriptome and proteome, enabling stress recovery. How these events may influence one another is only partly known. Beginning with a cytological screen of genome stability proteins, we find that the splicing factor Hsh155 disassembles from its partners and localizes to both intranuclear and cytoplasmic protein quality control (PQC) aggregates under alkylation stress. Aggregate sequestration of Hsh155 occurs at nuclear and then cytoplasmic sites in a manner that is regulated by molecular chaperones and requires TORC1 activity signaling through the Sfp1 transcription factor. This dynamic behavior is associated with intron retention in ribosomal protein gene transcripts, a decrease in splicing efficiency, and more rapid recovery from stress. Collectively, our analyses suggest a model in which some proteins evicted from chromatin and undergoing transcriptional remodeling during stress are targeted to PQC sites to influence gene expression changes and facilitate stress recovery.
Introduction
To survive chemical or environmental challenges, organisms have evolved a robust network of stress responses that remodel the transcriptome and proteome to enable recovery and, in some cases, future tolerance (Guan et al., 2012). Damaged proteins and RNAs can be turned over in the context of global changes to the transcriptome and translatome, which help to arrest the cell cycle and promote survival (Gasch et al., 2001; Begley et al., 2007; Tkach et al., 2012). In Saccharomyces cerevisiae (budding yeast), one of the hallmarks of transcriptome remodeling under stress is the suppression of ribosome production, which occurs under various conditions and is regulated by target of rapamycin (TOR)-dependent modulation of transcription factors such as Sfp1 (Gasch et al., 2000; Jorgensen et al., 2004; Marion et al., 2004). The ways in which transcriptome alterations impact proteome dynamics under any given stress condition are only partly understood.
Recent genome-wide cytological screens of yeast strains expressing GFP fusions to most proteins have revealed that genotoxin-induced protein relocalization events affect hundreds of proteins, many of which are not directly involved in DNA repair (Tkach et al., 2012; Mazumder et al., 2013; Chong et al., 2015). For instance, in addition to canonical DNA repair foci, treatment of yeast cells with methyl methanesulfonate (MMS) induced novel perinuclear foci containing the proteins Cmr1, Hos2, Apj1, and Pph21 (Tkach et al., 2012). These foci were subsequently recognized to be sites of molecular chaperone–regulated protein quality control (PQC) and renamed intranuclear quality control (INQ) sites (Gallina et al., 2015; Miller et al., 2015). Dozens of proteins have now been localized to the INQ, and some of which, such as Mrc1, play a role in recovery from genotoxic stress. The localization and composition of PQC sites in yeast depend upon many factors, including the specific type of stress, growth phase, cellular age, and the substrate analyzed (Saarikangas and Barral, 2016). Studies of both endogenous proteins and aggregation-prone model substrates in actively growing cells have identified at least three classes of yeast PQC compartments: INQ/juxtanuclear quality control (JUNQ), cytoplasmic quality control (CytoQ), and the insoluble protein deposit (IPOD). The key players mediating creation and dissolution of these structures are molecular chaperones, in particular small heat shock proteins such as Hsp42, which promote aggregation, and the disaggregation machinery including Hsp104 and Hsp70 family members (Saarikangas and Barral, 2016).
Beginning with a focused screen of >600 yeast chromosome instability (CIN) proteins, we uncover in this study INQ localization of a core-splicing factor, Hsh155, upon MMS treatment. We establish the dynamics of Hsh155 sequestration and define a regulatory network of proteins controlling its relocalization. Furthermore, we link Hsh155 aggregation to transcriptional repression of ribosomal protein (RP) genes (RPGs) in MMS conditions, which dramatically alter the need for splicing in yeast. These observations suggest unappreciated influence of transcriptome changes on the composition of PQC sites under stress. We propose a model wherein repression of RPGs and the concomitant drop in the need for splicing liberates Hsh155 from its spliceosome complex and other factors for sequestration at PQC sites in a TORC1-dependent manner. Collectively, our data provide new links between transcriptome regulators and PQC site composition under stress, where a stress-activated TORC1-regulated transcriptional program is controlling the composition of PQC sites.
Results
A screen for genome stability factors that relocalize after DNA damage identifies Hsh155
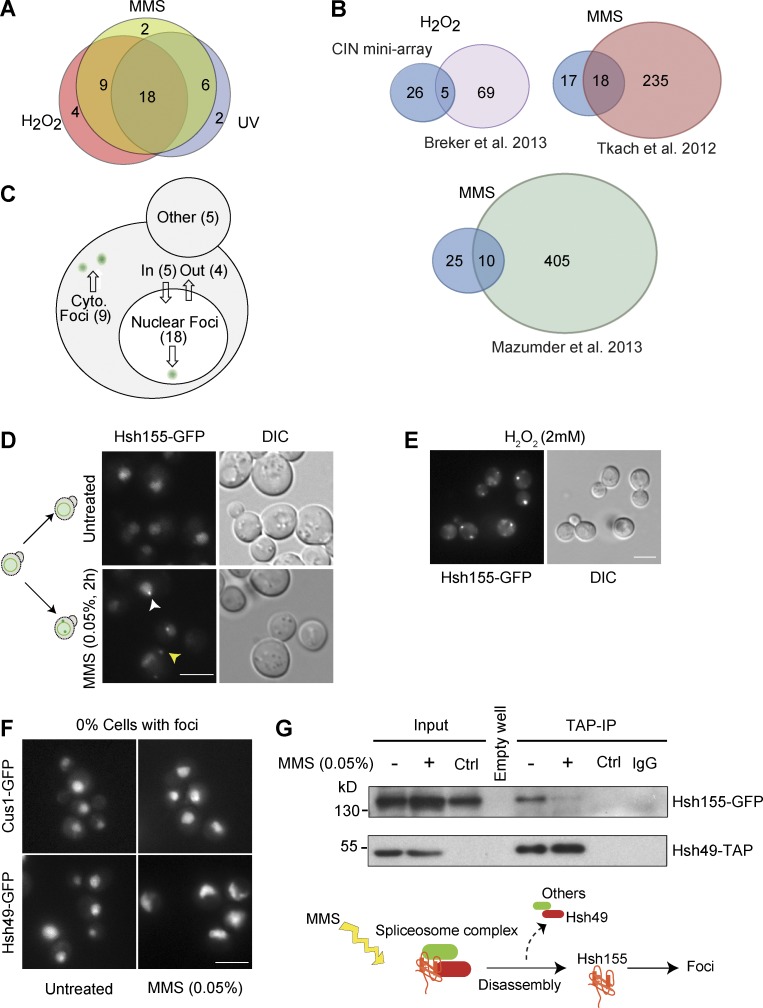
To explore dynamic responses of proteins to genotoxic stresses, we screened a biased miniarray of GFP fusion proteins comprised of proteins whose mutation is linked to an increase in genome instability (Stirling et al., 2011, 2014). The 622 GFP-tagged genome maintenance proteins were imaged at high resolution after no treatment or exposure to the alkylating agent MMS, UV irradiation, or H2O2. Candidate relocalization behaviors from the primary screen were validated in triplicate, leading to a final list of 41 relocalization events after genotoxic stress (Table S1). Most relocalization events occurred in all three stresses, and a large majority occurred in at least two conditions, with only eight appearing under a single stress condition (Fig. 1 A). Comparison of our data to three previously published genome-wide MMS- and H2O2-induced relocalization screens show a degree of overlap, although our screen identified new movements in each case (Fig. 1 B; Tkach et al., 2012; Breker et al., 2013; Mazumder et al., 2013). Most movements occurred into or out of the nucleus or into nuclear or cytoplasmic foci (Figs. 1 C and S1), which, based on their annotation (www.yeastgenome.org), we can ascribe to the formation of aggregates, P-bodies, or DNA repair centers. One unexpected observation was the relocalization of Hsh155-GFP into nuclear and cytoplasmic foci in response to MMS or H2O2 (Fig. 1, D and E). Hsh155 is part of the SF3B subcomplex in the U2 small nuclear ribonucleoprotein of the spliceosome. To assess the specificity of Hsh155 relocalization, we tested the relocalization behavior of Hsh155 binding partners within the spliceosome, Cus1 and Hsh49, after MMS treatment, but we did not observe any change in their localization (Fig. 1 F). In addition, we precipitated a tandem affinity purification (TAP)-tagged Hsh49 allele to isolate Hsh155-GFP with and without MMS treatment (Fig. 1 G). Although MMS treatment did not affect the levels of either protein, it dramatically reduced the association of Hsh155 with Hsh49 by coimmunoprecipitation. Nuclear Hsh155 foci did not colocalize with canonical DNA damage repair proteins, making a direct role in DNA repair unlikely (Fig. S2 A). Our data thus define a previously unrecognized and selective dissociation and relocalization of Hsh155 to foci after alkylating and oxidative genotoxic stress.
Figure 1.
DNA damage relocalization screen of the CIN proteome identifies Hsh155. (A) Overall screen results. Overlap of protein relocalization upon MMS, H2O2, or UV treatment. List of proteins relocalized in each treatment detailed in Table S1. (B) Comparison of screen results with published whole-proteome relocalization screens. The stress is indicated above the diagrams, and the references are below. (C) Yeast cell schematic summarizing relocalizations by destination under stress (see also Fig. S1 and Table S1). (D) MMS-induced relocalization of Hsh155 into nuclear (white arrowhead) and cytoplasmic foci (yellow arrowhead). A schematic (left) summarizes the movements. (E) H2O2-induced relocalization of Hsh155. DIC, differential interference contrast. (F) Spliceosome complex partners of Hsh155, Cus1, and Hsh49 do not form foci after MMS treatment. Bars, 5 µm. (G) Coprecipitation and Western blot of the Hsh155–Hsh49 complex with or without MMS treatment. Control (Ctrl) lanes are mock immunoprecipitations (IPs) from cells expressing Hsh155-GFP only. No nonspecific binding was observed with IgG Sepharose beads or with Hsh155-GFP control. Below is a schematic of proposed SF3b complex disassembly in MMS.
Hsh155 foci are protein aggregates
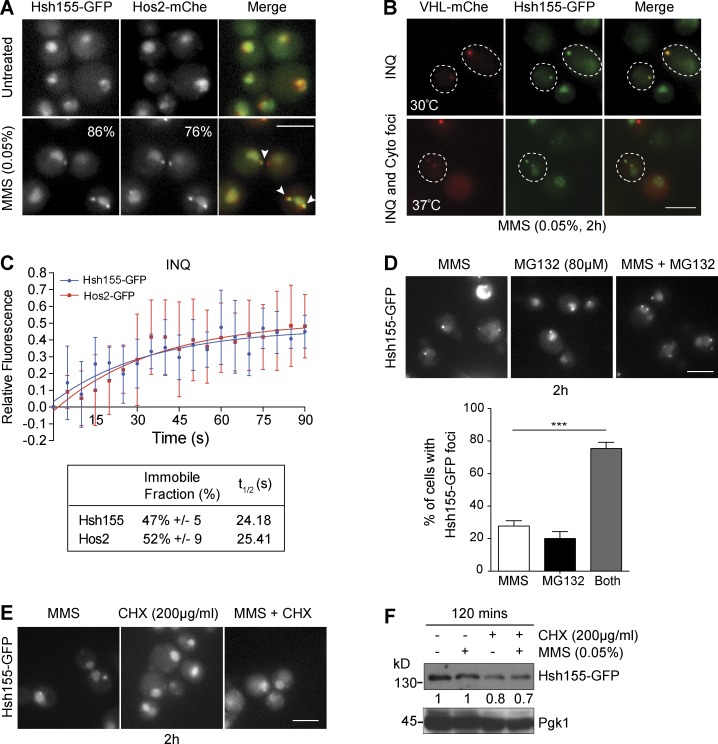
Hsh155 foci resembled the recently described INQ compartment, which contains Cmr1, Hos2, and many other proteins (Tkach et al., 2012; Gallina et al., 2015; Miller et al., 2015). To examine this possibility, we observed Hsh155-GFP and Hos2-mCherry colocalization at both cytoplasmic and nuclear foci after MMS (Fig. 2 A). To confirm that these were indeed sites of protein aggregation, we also colocalized Hsh155-GFP with Von Hippel–Lindau (VHL) tumor suppressor–mCherry, a protein that cannot fold in yeast and is targeted to aggregates under various stresses (McClellan et al., 2005). Hsh155 and VHL were significantly colocalized in MMS (Fig. 2 B), although Hsh155-GFP did not join VHL foci under heat shock (Fig. S2 B). The aggregates of both Hos2 and VHL have been shown clearly to localize in both the nucleus and cytoplasm (Miller et al., 2015). To confirm the compartmental sequestration of Hsh155, we further used Hta2 (histone H2A) and Nic96 (nuclear pore protein) as markers of nuclear area (Fig. S2 C). Consistent with localizing in an aggregated state, FRAP analysis of nuclear Hsh155-GFP or Hos2-GFP foci confirmed a large immobile fraction for each protein (∼50%) similar to known aggregates of PQC (Saarikangas and Barral, 2015) as well as a recovery time (t1/2 ∼25 s) much slower than freely diffusing proteins (Fig. 2 C). These results thus identify the core-splicing factor Hsh155 as a new constituent of the INQ protein aggregate compartment after genotoxic stress.
Figure 2.
Hsh155 relocalizes to nuclear (INQ) and cytoplasmic PQC sites. (A) Colocalization (white arrowheads in merge) of Hsh155-GFP with Hos2-mCherry (mChe) in MMS. The percentage of cells with foci is indicated in the corner of the bottom panels. (B) Colocalization of Hsh155-GFP with VHL-mCherry at both 30°C and 37°C with MMS. Dashed outlines indicate position of the cell body. (C) Quantitative FRAP analysis of GFP-tagged Hsh155 or Hos2 in nuclear foci. The top graph shows the best line of fit curve of relative fluorescence intensities over time for both Hsh155 (blue) and Hos2 (red); the bottom table shows the percentage of Hsh155 and Hos2 in the immobile fraction and diffusion time (t1/2). Error bars are means ± SD of Hsh155 (eight cells) and Hos2 (five cells) analyzed over three independent experiments. (D) Effect of proteasome inhibition by MG132 on Hsh155 aggregation. Representative images (top) and quantification (bottom) are shown. Means ± SEM; n = 3 with >100 cells each. ***, P < 0.001; Fisher’s test. (E) CHX blocks Hsh155 foci formation. Representative image of Hsh155-GFP–tagged cells treated with CHX (200 µg/ml) and MMS (0.05%). Bars, 5 µm. (F) Hsh155 protein levels in MMS by anti-GFP Western blotting relative to Pgk1 levels. Shown is the representative blot from four independent experiments.
To examine the effects of new protein synthesis or degradation on the Hsh155 foci formation, we treated cells with the proteasome inhibitor MG132 or the translation inhibitor cycloheximide (CHX) alone or in combination with MMS. Similar to earlier research on other INQ components (Gallina et al., 2015), MG132 treatment induced Hsh155 sequestration to INQ sites, and this was enhanced significantly in cells treated with MG132 and MMS, suggesting that protein degradation opposes aggregate formation or retention (Fig. 2 D). CHX had no effect on Hsh155 localization but abolished Hsh155 foci in MMS + CHX–treated cells (Fig. 2 E). New protein synthesis is essential for heat-stress aggregates (Zhou et al., 2014), and it is possible that the same is true for INQ. In addition, the stability of Hsh155 in cells did not appear to change significantly after MMS treatment, suggesting that the influence of MG132 on foci formation may be a result of other factors rather than Hsh155 itself being a target of degradation (Figs. 2 F and S2 D).
Dynamic behavior of Hsh155 at PQC sites
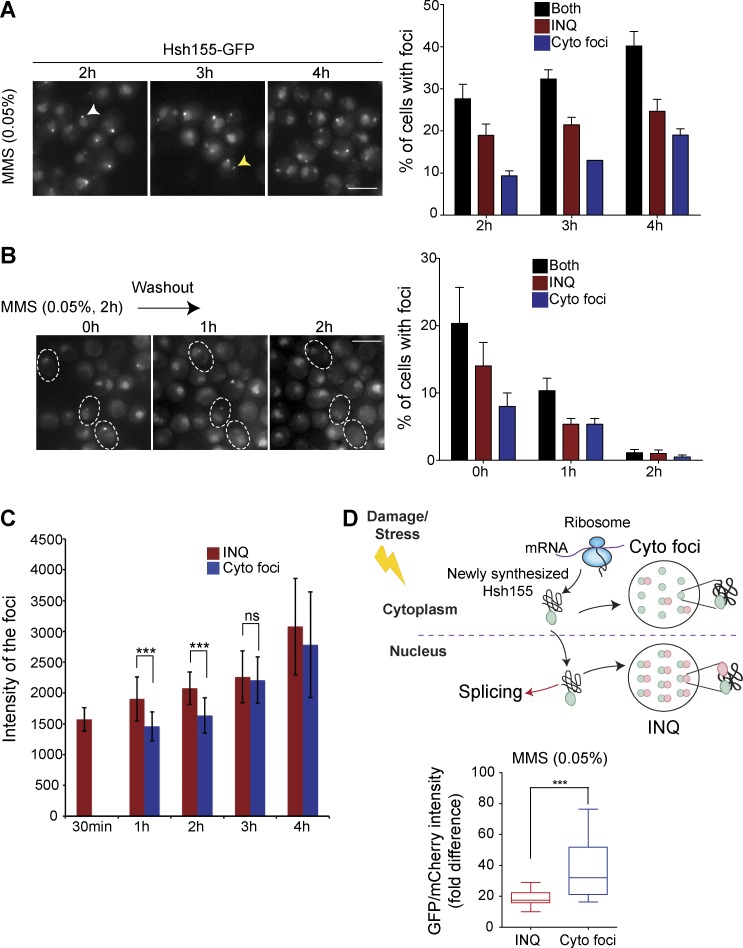
Localization of Hsh155 to protein aggregates was unexpected, and we elected to probe the dynamics of this behavior in greater detail. To explore the nature of Hsh155 localization to PQC sites, we first followed Hsh155 foci formation over time. Hsh155 rapidly accumulated at nuclear foci, followed by gradual increases in the frequency of cytoplasmic foci (Fig. 3 A), whereas washout of MMS led to a gradual decrease of foci and recovery of normal Hsh155 nuclear localization (Fig. 3 B). Cytoplasmic Hsh155-GFP foci were initially dimmer, but their fluorescence intensity gradually increased over time, matching INQ intensities by 3 h (Figs. 3 C and S2 E). These results indicate that the clearance of INQ structures proceeds rapidly after stress removal and that, during prolonged stress, the triage pathway shifts from immediate nuclear deposition to a delayed cytoplasmic aggregate deposition.
Figure 3.
Dynamic behavior of Hsh155 at PQC sites. (A and B) Time course imaging of Hsh155-GFP foci accumulation over time (A; the yellow arrowhead indicates a cytoplasmic focus, and the white arrowhead indicates a nuclear focus) and disappearance after MMS washout (B). Representative images are shown on the left, and quantifications of percentages of cells with Hsh155 foci are on the right. For B, dashed outlines represent time-lapse images of foci disappearance over time in the same set of cells. No claims of significance are made. n = 3 with >200 cells each. Bars, 5 µm. (C) Fluorescence intensity of foci over time show increasing relative GFP in cytoplasmic foci until 2 h of MMS treatment. (D) Tandem fluorescent fusion intensities of Hsh155 in aggregates. Ratios of GFP and mCherry fluorescence in nuclear or cytoplasmic foci are shown (bottom). A schematic of protein lifetime of Hsh155 fusion to a fluorescent timer in PQC (top), showing older (GFP>mCherry) protein in INQ and newer (GFP>>>mCherry) in cytoplasmic foci. For A–C, bars are color-coded to denote INQ (red), cytoplasmic foci (blue), or both (black). For C quantifications: n > 100; Student t test. For D quantifications: n ≥ 28 per replicate; Mann-Whitney test. Both C and D had three replicates; all error bars are means ± SEM. ***, P < 0.001.
To assess how Hsh155 protein pools in nuclear or cytoplasmic foci are related, we used a fluorescent reporter tandem fusion approach in which both a fast-folding GFP and a slowly maturing mCherry are fused in tandem, and the fluorescence ratio of each fluorophore is an indicator of protein turnover rate (Khmelinskii et al., 2012; Gallina et al., 2015). C-terminally tagged Hsh155-GFP/mCherry protein fluorescence ratios in the nuclear or cytoplasmic aggregates were measured after 2 h in MMS. This experiment revealed that a significantly older pool of Hsh155 (i.e., lower GFP/mCherry ratio) appeared in INQ compared with cytoplasmic aggregates (Fig. 3 D), confirming the sequestration of Hsh155 to INQ first and then to cytoplasmic aggregates. The protein turnover rates of the untreated nucleoplasmic signal remained comparable to those after MMS treatment (Fig. S2 F). Collectively, these data suggest that Hsh155 is first sequestered at INQ sites for refolding and reactivation until stress recovery, and only later is it triaged to cytoplasmic aggregates, possibly more so for nascent Hsh155 because a “younger” pool of protein appears to populate cytoplasmic aggregates.
INQ resident proteins regulate Hsh155 sequestration
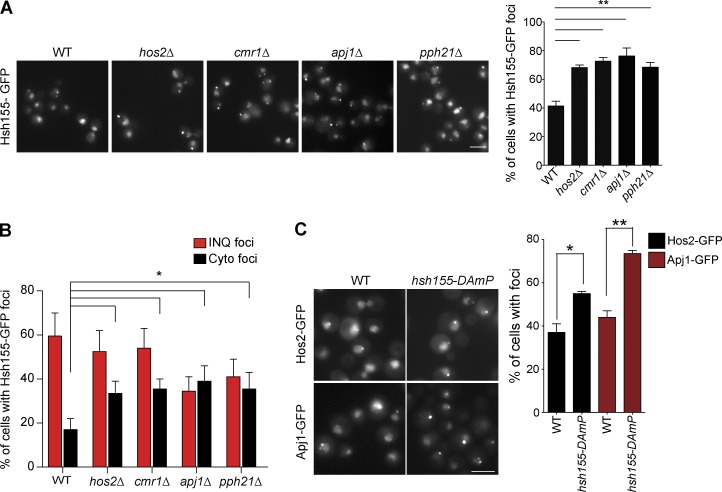
It is possible that known INQ-associated proteins regulate Hsh155 localization. Previous work identified the poorly characterized INQ marker proteins Cmr1, Hos2, Pph21, and Apj1 (Tkach et al., 2012). Cmr1 has been implicated in the recovery from DNA damage stress and has been recently linked, together with Hos2, in global transcriptional regulation (Gallina et al., 2015; Jones et al., 2016). Pph21 is one of two protein phosphatase 2A (PP2A) catalytic subunits with pleiotropic functions in the cell, including opposing TOR functions in nutrient signaling (Jiang and Broach, 1999; Düvel et al., 2003). Apj1 is a poorly characterized Hsp40 molecular chaperone family member. To shed light into their effects on Hsh155, we measured the effects of deleting their encoding genes on Hsh155 foci formation. Deletion of any of these genes significantly increased the number of cells with MMS-induced Hsh155 foci and shifted the distribution to create more cytoplasmic foci (Fig. 4, A and B). How each of these INQ markers regulate Hsh155 sequestration may differ based on their functions in cells (see Discussion). Reciprocally, an HSH155-DAmP (Breslow et al., 2008) allele increased the frequency of Hos2-GFP and Apj1-GFP foci in both INQ and cytoplasmic sites after MMS treatment (Fig. 4 C). Given the core role of Hsh155 in splicing as well as the fact that a DAmP allele would simply reduce the amount of WT protein, this suggests that defective splicing may influence the formation of protein aggregates upon MMS treatment.
Figure 4.
Regulation of Hsh155 foci formation by INQ-resident proteins. (A) Representative images showing foci in MMS-treated cells (left). Quantification of percentage of cells with Hsh155 foci in indicated strains (right). (B) Effect of losing the indicated INQ residents proteins on INQ and cytoplasmic foci distribution. (C) Representative images showing effects of depleting HSH155 on INQ formation (left). Percentage of cells with Hos2 (black) and Apj1 (red) foci in the indicated strain (right). Bars, 5 µm. Error bars are means ± SEM; n = 3 with >50 cells each. *, P < 0.05; **, P < 0.01; Fisher’s test.
Replication stress does not regulate Hsh155 foci formation
Because MMS induces DNA damage, we wondered whether Hsh155 was required to resist chronic genotoxin exposure or whether DNA damage signaling by ATM/Tel1 or ATR/Mec1 was required for Hsh155 relocalization (Gasch et al., 2001; Tkach et al., 2012). However, hsh155–temperature-sensitive (ts) mutants (hsh155-ts) did not show any additional growth defect in MMS after chronic exposure, making a direct role in DNA repair unlikely (Fig. S3 A). Consistent with a recent study (Chong et al., 2015), we saw an increase in Hsh155 signal after hydroxyurea, which induces replication stress without the chemical DNA lesions induced by MMS, but we did not observe any foci, suggesting that stalling DNA replication alone is insufficient for Hsh155 foci formation (Fig. S3 B). To interrogate the function of DNA damage signaling in Hsh155 relocalization, we measured foci formation in a strain lacking both yeast ATM (tel1Δ) and ATR (mec1Δ). Hsh155 foci formed normally in the mec1Δtel1Δ mutant compared with control cells, suggesting that DNA damage signaling is not required for aggregation (Fig. S3, C and D). Interestingly, a significant number of Hos2-GFP aggregates occurred in untreated mec1Δtel1Δ cells, and this was further enhanced in MMS (Fig. S3, E and F). Thus, Hos2 foci formation does not require DNA damage signaling but suggests that stress in the mec1Δtel1Δ mutant is sufficient to promote Hos2 but not Hsh155 aggregation. These data support the idea that although INQ components like Hos2 and Hsh155 can have codependent relationships at PQC sites, they can be governed by independent upstream signals driving sequestration to PQC sites. Collectively, these data show that neither MMS-induced DNA replication stress nor canonical DNA damage signaling is necessary for Hsh155 sequestration at PQC sites.
A network of molecular chaperones regulates Hsh155 PQC site deposition
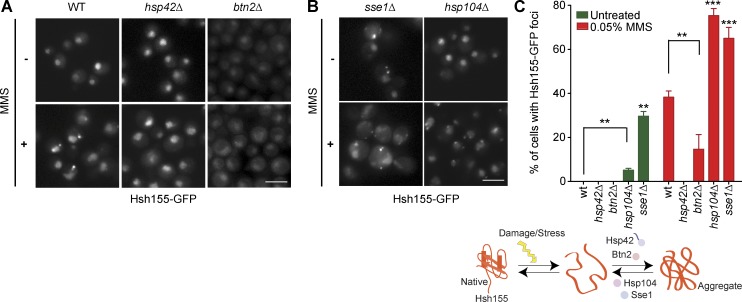
Because Hsh155 localizes to protein aggregates after MMS treatment, it may be regulated by molecular chaperones. Indeed, Apj1 is a molecular chaperone, which our data suggest opposes Hsh155 localization to both nuclear and cytoplasmic foci (Fig. 4 A). Previous work has implicated compartment-specific aggregases Hsp42 and Btn2 in driving substrates to cytoplasmic and nuclear PQC sites, respectively (Miller et al., 2015). Surprisingly, deletion of either HSP42 or BTN2 almost completely abrogated Hsh155 localization to both INQ and cytoplasmic PQC sites (Fig. 5, A and C). Although the distribution of foci between INQ and cytoplasm in btn2Δ was similar to WT, a slight increase in cytoplasmic foci was seen (Fig. S3 G). Although this conflicts with research of compartment-specific functions of Hsp42 and Btn2 when analyzing a model aggregating substrate proteins (Miller et al., 2015), it is consistent with redundant effects of Hsp42 and Btn2 on Cmr1-YFP foci (Gallina et al., 2015). Therefore, at least for some endogenous protein substrates of INQ, both Hsp42 and Btn2 promote aggregate localization in the nucleus and cytoplasm. Our MMS washout results (Fig. 3 B) suggest reactivation of Hsh155 after stress removal, indicating participation of disaggregases such as Hsp104 and Sse1 in the process (O’Driscoll et al., 2015). Deletion of HSP104 and SSE1 dramatically increased the frequency of PQC sites marked by Hsh155 in MMS, increasing the number of cells with predominantly more cytoplasmic aggregates (Figs. 5 B and S3 H). Deletion of SSE1 led to formation of Hsh155 foci even without MMS (Fig. 5 B) and shifted the foci to more cytoplasmic aggregates (Fig. S3 H), indicating that Sse1 may be involved in de novo folding of Hsh155 or that sse1Δ yeast experience ongoing stress that affects spliceosome integrity. Collectively, our data show how a network of chaperones acting as aggregases (Hsp42/Btn2) or disaggregases (Hsp104/Sse1) regulates Hsh155 localization to INQ and cytoplasmic PQC sites after stress (Fig. 5 C).
Figure 5.
Regulation of Hsh155 foci formation by molecular chaperones. (A and B) Effect of aggregases (A) and disaggregases (B) on Hsh155 relocalization in MMS relative to WT. Representative images are shown. Bars, 5 µm. (C) Summary of chaperone regulation of Hsh155 relocalization. Schematic (top) and quantification of cells with foci from A and B (bottom). Error bars are means ± SEM; n = 3 with >50 cells each. Asterisks show p-value thresholds in comparison with WT under the same condition. **, P < 0.01; ***, P < 0.001; Fisher’s test.
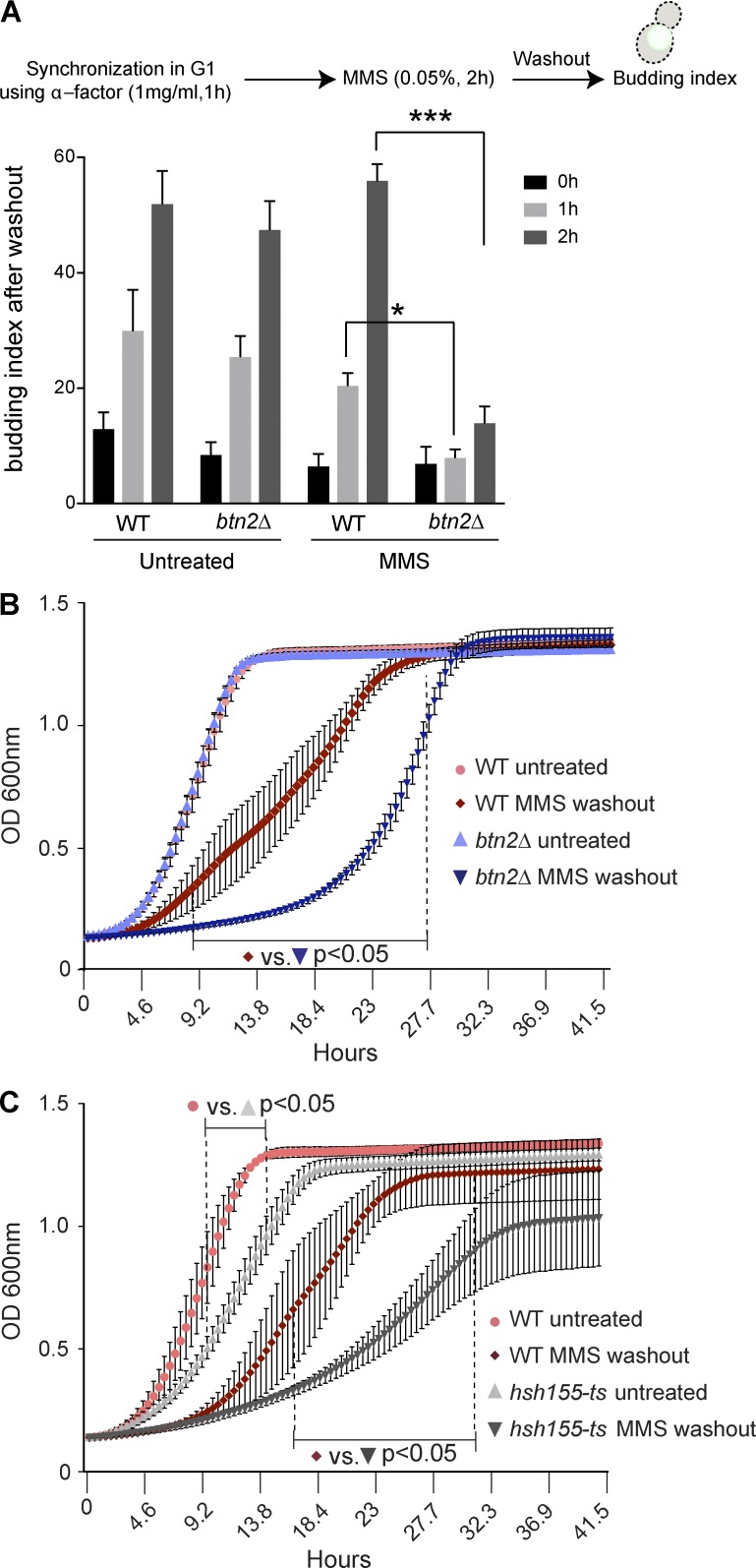
Aggregase-regulated Hsh155 foci formation correlates with efficient stress recovery
Having established the nature and chaperone-regulation of Hsh155 aggregation at INQ, we hypothesized that this must confer an advantage to cells during cellular stress responses, possibly influencing efficient recovery. To test this, we conducted cell cycle reentry analysis by scoring the budding index of cells after MMS washout. These data show that btn2Δ cells are significantly delayed in rebudding after MMS removal compared with WT or hsp104Δ cells (Figs. 6 A and S4 A). Because hsp104Δ cells showed comparable budding index to WT in MMS recovery (Fig. S4 A), the subsequent growth curve analysis was performed on btn2Δ mutants. Growth curve analysis of BTN2 mutants also revealed a significant delay in recovery after MMS treatment compared with WT (Fig. 6 B). Similar delay in recovery can be seen in mutants of HSH155, showing that splicing itself might facilitate efficient recovery (Fig. 6 C). Indeed, previous high-throughput studies have suggested a role for splicing factors in MMS recovery (Svensson et al., 2011; van Pel et al., 2013). Overall, these are strongly supportive of a model linking protein sequestration at INQ sites to splicing suppression and stress recovery.
Figure 6.
Hsh155 foci formation is crucial for efficient stress recovery. (A) Quantification of budded cells in strains synchronized and released from G1 phase ± MMS washout. n = 3 with >30 cells each. *, P < 0.05; ***, P < 0.001; Fisher’s test. (B and C) Growth curve analysis of btn2Δ (B) and hsh155-ts (C) alleles compared with WT strains ± MMS and 2 h after MMS washout at 30°C. The dotted line represents the range wherein all data points significantly differ from WT under the same conditions. n = 3 with triplicates in each. Means ± SEM; two-way ANOVA with Tukey’s post hoc test; p-values range from <0.05–<0.0001.
Hsh155 sequestration influences RPG repression and splicing
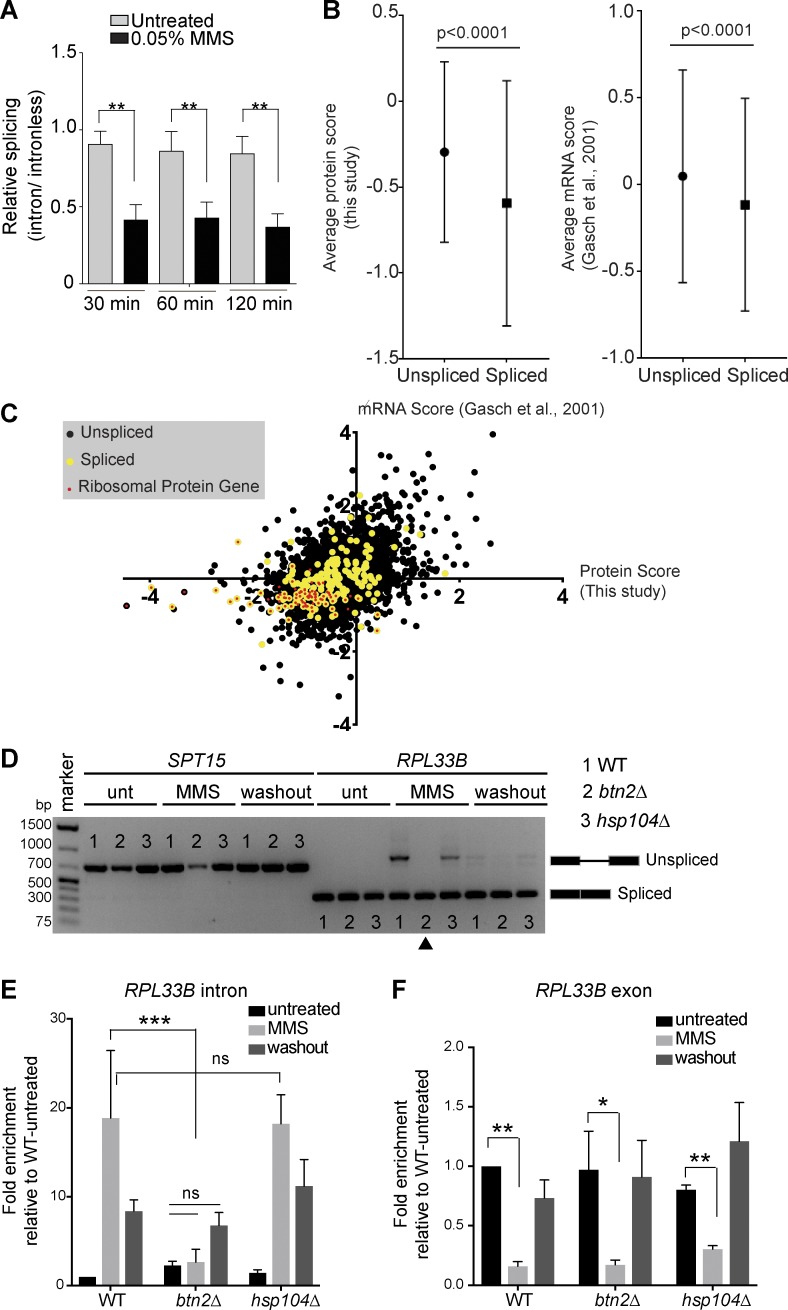
If INQ formation promotes stress recovery, we wondered whether the role of Hsh155 relocalization could be linked to the stress-induced remodeling of the transcriptome known to occur in MMS (Gasch et al., 2001). We first analyzed splicing efficiency using a LacZ reporter construct (Palancade et al., 2005) and found that, whereas MMS treatment reduced LacZ production from both an intronless and intron-containing construct, the intron-containing construct was further repressed (Fig. 7 A). hsh155-ts showed a dramatic splicing defect as expected (Fig. S4 B). Interestingly, the MMS-induced splicing defect was evident by 30 min, well before most cells showed detectable INQ foci (Fig. S4 C), consistent with the idea that spliceosomes disassemble (Fig. 1) in response to MMS treatment and before Hsh155 aggregation.
Figure 7.
Hsh155 sequestration influences RPG repression and splicing. (A) Splicing efficiency of a LacZ reporter in MMS. Quantification of relative LacZ splicing in untreated and MMS-treated normalized to “no intron” control. Student’s t test; n = 3 with individual transformants in triplicates each. (B) Comparison of the spliced gene subset from whole-proteome analysis by mass spectrometry (this study: 4,357 total, 3,864 no change, 75 enriched, and 418 depleted. 39 of the depleted proteins are spliced genes, and 35 are RPs) or microarray data (Gasch et al., 2001) after 2 h in MMS. Mann-Whitney p-value is <0.0001. (C) Scatterplot of proteome and mRNA data for each gene. Spliced genes are highlighted in yellow, and red dots indicate RPG products. Depleted and enriched proteins in our mass spectrometry analysis are listed in Table S2. (D) Hsh155 aggregate formation correlates with RPG intron retention in MMS. Expression and splicing of RPL33B transcripts compared with intronless SPT15 in WT, BTN2-, and HSP104-deleted strains with or without MMS and 2 h after MMS washout. cDNA PCR products representing spliced and unspliced are indicated. The arrowhead indicates the lack of MMS-induced intron retention in btn2Δ cells. Unt, untreated. (E) Quantification of RPL33B intron region in the indicated strains by reverse transcription–quantitative PCR normalized to SPT15 and relative to WT untreated. (F) Quantification of RPL33B mRNA transcript levels from exon region in WT, BTN2-, and HSP104-deleted strains by reverse transcription–quantitative PCR normalized to SPT15 and relative to WT untreated. Three biological replicates; means ± SEM; Asterisks show p-values of ΔΔCt levels. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ANOVA.
Splicing flux in yeast is dominated by the production of RPs, the majority of which encode an intron and whose transcripts account for ∼90% of splicing reactions (Parenteau et al., 2011). It has long been known that RPGs are specifically repressed upon stress as part of a transcriptional program dubbed the environmental stress response (Gasch et al., 2000). Recent studies indicate that RPG expression can also be regulated posttranscriptionally by selective splicing under stresses including MMS (Parenteau et al., 2011; Gabunilas and Chanfreau, 2016). Whole-proteome analysis showed that 418 proteins were repressed, whereas only 75 were more abundant after MMS treatment (Table S2). Gene Ontology analysis of the cellular functions impacted highlights strong repression of transcriptional and translational processes (Table S3). Comparing previous transcriptome data (Gasch et al., 2001) gathered under essentially the same conditions with our proteomics data highlights a significant drop in spliced gene expression and protein production after MMS treatment (Fig. 7, B and C). RPGs are the drivers of this drop in spliced genes as they clustered in the negative quadrant of both datasets (Fig. 7 C and Table S2). Additionally, we conducted intron retention measurements by both qualitative (Figs. 7 D and S4 D) and quantitative (Figs. 7 E and S4 E) RT-PCR for two RP pre-mRNAs in WT, btn2Δ, and hsp104Δ cells. As expected, MMS treatment caused a splicing defect, leading to intron retention in WT cells. Importantly, loss of BTN2 prevented this MMS-induced spike in intron retention, suggesting that the organization of nuclear protein aggregates may be a key step in shutting down splicing in MMS. Cells lacking HSP104 showed the WT spike in intron retention consistent with aggregation competence. These data correlate with the functional data showing that btn2Δ does not recover from MMS-induced stress as quickly as WT and hsp104Δ cells (Fig. 6). Importantly, MMS treatment led to RPL33B gene repression as expected in all the strains (Fig. 7 F). This indicates that although transcriptional repression of RPGs in MMS is normal in btn2Δ, the posttranscriptional effects on splicing are lost.
RPG transcriptional regulation and TORC1 influences Hsh155 sequestration
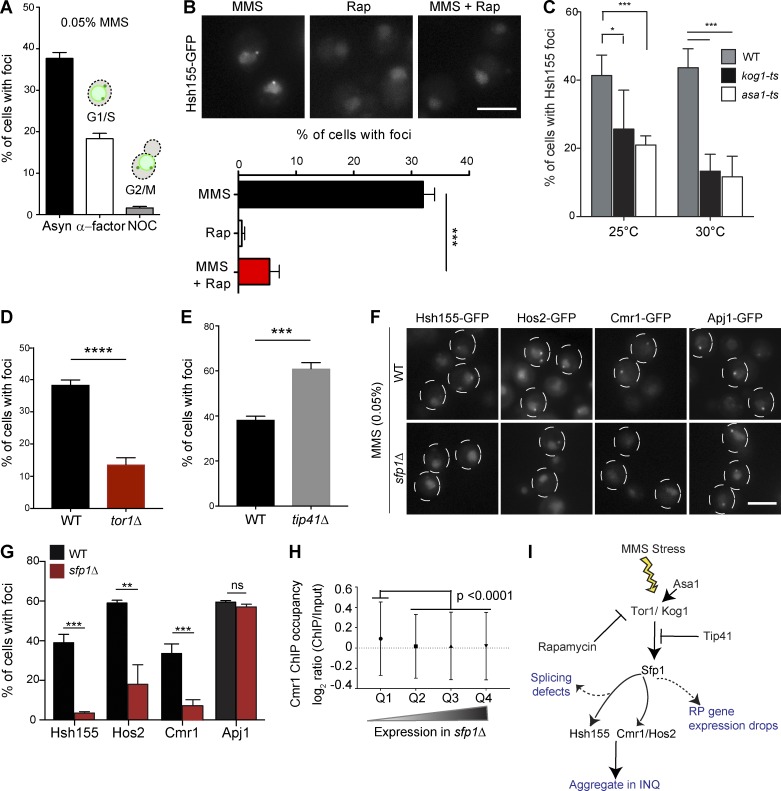
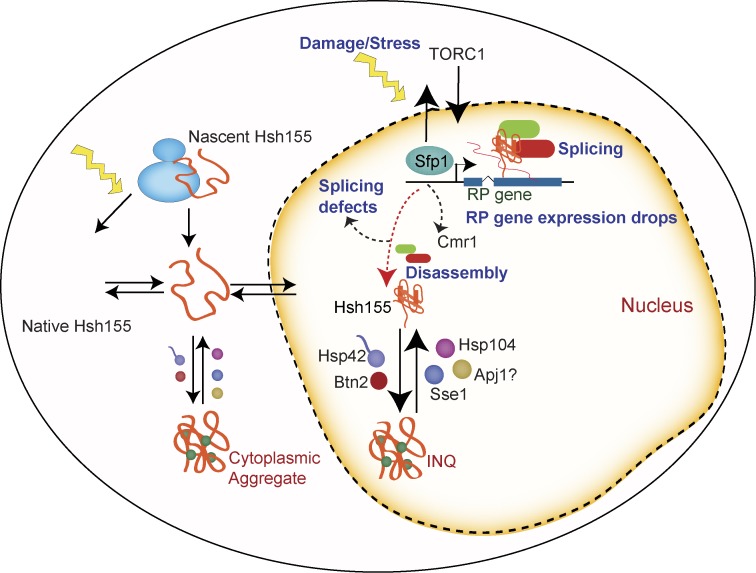
Because MMS is known to arrest cells in S phase (Shirahige et al., 1998), we synchronized cells either in G1 or G2/M before MMS treatment to determine whether passage into S phase was required for foci formation. Although α-factor–arrested G1 cells formed Hsh155 foci readily upon MMS treatment, nocodazole-arrested G2/M cells formed very few foci (Figs. 8 A and S5 A). The presence of aggregates in G1 but not G2/M cells is consistent with a model linked to RPG expression because ribosome production is required to progress through G1 (Bernstein and Baserga, 2004), whereas ribosomal RNA production at least is transiently decreased during mitosis (Clemente-Blanco et al., 2009). TOR signaling regulates ribosome biogenesis normally and coordinates its repression under stress (Martin et al., 2004). Remarkably, co-treatment of cells with the TORC1 inhibitor rapamycin strongly suppressed MMS-induced Hsh155-GFP foci formation, whereas rapamycin treatment alone had no effect on Hsh155 localization (Fig. 8 B). Genetic perturbation of the TORC1 pathway through mutation of a Tor1/2-stabilizing chaperone ASA1 (Stirling et al., 2011) or the TORC1 subunits KOG1 and TOR1 (Loewith et al., 2002) also significantly reduced the frequency of Hsh155-GFP foci (Fig. 8, C and D). Reciprocally, deletion of the TORC1 inhibitor TIP41 significantly increased the number of cells with MMS-induced Hsh155 foci (Fig. 8 E), consistent with data in pph21Δ cells, as Tip41 regulates Pph21 in opposing TORC1 signaling (Jiang and Broach, 1999; Düvel et al., 2003). Overall, this suggests that RPG repression mediated by TORC1 signaling (Fig. 8 I) could be influencing the dynamic behavior of Hsh155 in MMS.
Figure 8.
TORC1 signaling influences sequestration of transcription regulators to PQC through Sfp1. (A) Cell cycle dependence of Hsh155 sequestration. Quantification of cells with Hsh155 foci in asynchronous (Asyn; black), G1 (α-factor; white) and G2/M (nocodazole [NOC]; gray) cells. (B) Rapamycin regulates Hsh155 foci formation in response to MMS. Representative images (top) and quantification (bottom). (C) Effect of ts mutants of TORC1 subunit Kog1 and regulator Asa1 on Hsh155 foci at 25°C and 30°C. (D and E) Opposing effects of TOR1 (D) or TIP41 (E) deletion on Hsh155 foci in MMS. (F) Sfp1 is required for Hsh155, Hos2, and Cmr1 relocalization but not for INQ formation. Representative images of Hsh155-GFP under the indicated conditions. Dashed circles denote two or three cell border outlines each per representative image. Bars, 5 µm. (G) Quantification of Hsh155 foci formation in sfp1Δ cells from F. Three replicates; n > 100; means ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. (H) Cmr1 occupancy at genes affected by sfp1Δ. Quartiles (Q1–Q4) were derived from microarray data in sfp1Δ (Marion et al., 2004) and the Cmr1 ChIP occupancy data in WT (all genes; n = 5,549; listed in Table S4; Jones et al., 2016). Cmr1 occupancy is highest in genes regulated by SFP1 (P < 0.0001; ANOVA with Tukey’s test). (I) Model of TORC1 pathway regulators tested in this study leading to INQ protein aggregation; see also Fig. 9 for integrated model.
TORC1 influences sequestration of transcription regulators to PQC through Sfp1
The effects of TORC1 on RPG expression are mediated through downstream effects on transcriptional activators including Hmo1, Ifh1, and Sfp1 (Schawalder et al., 2004; Xiao et al., 2011; Reja et al., 2015). Mutation of constitutive TORC1-regulated RPG-transcriptional activators such as Hmo1 or Ifh1 had no significant effect on the frequency of Hsh155-GFP foci (Fig. S5 B). Hmo1 and Ifh1 directly regulate RPG expression but do not have an established role in the environmental stress response. On the contrary, Sfp1 is an RPG transcription factor that interacts with and is regulated by TORC1 signaling specifically under stress (Lempiäinen et al., 2009). Normally, Sfp1 is displaced from RPG promoters under stress and is localized to the cytosol to effect rapid adaptation of RPGs to stress (Jorgensen et al., 2004; Marion et al., 2004). In the absence of the Sfp1, RPGs are still transcribed but are not repressed under stress (Marion et al., 2004). Remarkably, although INQ protein aggregates marked by Apj1-GFP still formed normally in MMS-treated sfp1Δ cells, Hsh155-GFP foci were completely abrogated, and Hos2-GFP and Cmr1-GFP foci were significantly reduced (Fig. 8, F and G). Thus, loss of Sfp1 does not preclude the formation of aggregates; rather, it controls the specific endogenous proteins that are recruited to the aggregates under stress.
Interestingly, like the spliceosome, Cmr1 and Hos2 have been linked to RPG expression and bind to RPG promoters (Jones et al., 2016). Together with our data, this suggests that a precipitous drop in RPG expression might be involved in evicting Hsh155, Hos2, and Cmr1 from the chromatin and enabling their sequestration at PQC sites. To assess this possibility, we compared published microarray data on sfp1Δ cells (Marion et al., 2004) with Cmr1 occupancy data in WT cells by chromatin immunoprecipitation (ChIP; Jones et al., 2016). This analysis showed that Cmr1-occupied genes are significantly down-regulated in sfp1Δ cells, supporting the idea that the two factors have common targets (Fig. 8 H and Table S4). Interestingly, analysis of the Cmr1 ChIP occupancy data also indicates that there is significantly more Cmr1 occupancy at spliced genes (mean Cmr1 occupancy for 274 spliced genes = 0.13, and mean occupancy for all genes = 0.046) in WT cells (Fig. S5 C and Table S4). Thus, genes regulated by Cmr1 and Sfp1 overlap and are enriched for spliced genes where Hsh155 will act. Together, these data support a model where stress-induced TORC1-mediated transcriptional changes at RPGs and a precipitous drop in RPG expression regulated by Sfp1 liberate spliceosomes and transcriptional regulators such as Hos2 and Cmr1 (Fig. 8 I). These factors can then be captured in protein aggregates in the nucleus, and eventually the cytoplasm, and then sequestered until stress passes (see Discussion; Fig. 9).
Figure 9.
Model illustrating stress-induced transcriptional changes at RPGs liberating transcriptional regulators such as Sfp1 regulated by TORC1 signaling, Cmr1, and spliceosomes, leading to spliceosome disassembly and splicing defects. Subsequently, Hsh155 is sequestered in INQ and eventually cytoplasmic aggregates, which are regulated by chaperones until stress recovery.
Discussion
Hsh155: a new INQ-localizing protein
The INQ is a relatively poorly characterized PQC site for nuclear proteins. Our data establish several new principles governing INQ formation and substrate protein recruitment. We show that INQ substrates fall into at least two categories, those like Cmr1 that are wholly restricted to nuclear aggregates (Gallina et al., 2015), and those like Hsh155, Hos2, or Apj1 that also accumulate in cytoplasmic foci. This could suggest that the capacity of INQ is limited and that excess proteins are shunted to cytoplasmic aggregates, sequentially making the latter an overflow compartment, or that both the nuclear and cytoplasmic pool of proteins aggregate independently with different kinetics. Our tandem fluorescent fusion data suggest that a younger pool of Hsh155 accumulates in cytoplasmic aggregates compared with INQ, and therefore, we favor a model in which Hsh155 is not actively transported from INQ to cytoplasmic aggregates. In addition, because we show that INQ residents like Apj1 localize to aggregates under conditions where Hsh155 does not (i.e., in MMS-treated sfp1Δ cells), there must be separate upstream signals for the formation of aggregates and the recruitment of specific endogenous proteins. Thus, the formation of the aggregate itself is insufficient to recruit a labile protein; rather, signals which perturb Hsh155 interactions must occur before its recruitment. The observed dissociation of Hsh155 from its binding partner Hsh49 after MMS treatment is consistent with complex disassembly before aggregate recruitment, but the signals controlling this process are unknown.
We also found that INQ markers Cmr1, Hos2, Apj1, and Pph21 affect the frequency of Hsh155 foci. However, based on the annotated functions of these proteins, we propose that there may be different mechanisms by which this occurs. Apj1 is homologous to Hsp40, a molecular chaperone, and we predict that it plays a role in stabilizing soluble Hsh155 in the nucleus and cytoplasm. Cmr1 and Hos2 are now known to affect transcription and RPG expression (Jones et al., 2016), and thus deletions would disrupt the levels of spliced transcripts, potentially sensitizing cells to sequester Hsh155 in PQCs under stress. Finally, Pph21, like Tip41, opposes TORC1-dependent phosphorylation of Tap42 (Jiang and Broach, 1999), which would be partly alleviated in pph21Δ cells and associated with a stronger TORC1 signal. This potentially explains why rapamycin treatment blocked Hsh155 foci formation with MMS, whereas pph21Δ cells were sensitized to accumulate Hsh155 foci. Thus, although each of these INQ proteins fits into a model of stress signaling–induced transcriptional changes leading to protein aggregation, there are many remaining questions about how and why this subset of proteins are sequestered at the INQ and whether they are inactive aggregated substrates or are exerting their enzymatic activities (i.e., lysine deacetylation by Hos2 or S/T dephosphorylation by Pph21) within the INQ. The role of molecular chaperones at protein aggregates is clearer, and we identify well-known disaggregases and aggregases affecting Hsh155 deposition at aggregates. Interestingly, as with Cmr1, Hsh155 localization is regulated by both Btn2 and Hsp42, supporting a common function in aggregation processes as noted in certain contexts previously (Malinovska et al., 2012; Miller et al., 2015).
Linking transcriptome remodeling and functional consequences of Hsh155 aggregation
Coordinating changes in the transcriptome and proteome help to reestablish cellular homeostasis after stress recovery. Temporary protein sequestration or turnover in aggregate structures is one way that such homeostasis is achieved (Wallace et al., 2015; Saarikangas and Barral, 2016). We propose that the transcriptional response to MMS mediated through the TORC1 pathway is the ultimate initiator of Hsh155 aggregation. Spliced transcripts are dramatically affected by MMS treatment because RP production is shut down and RPGs encode the majority of spliced transcripts in yeast. Thus, Hsh155 is no longer engaged in bulk splicing at RPGs under stress. Our data suggest that an additional, posttranscriptional step in RPG repression may occur because we saw a spike in intron retention along with the RPG expression drop consistent with less efficient splicing after MMS treatment. Importantly, loss of BTN2 prevented MMS-induced intron retention, suggesting that Btn2 may play a role in spliceosome inactivation under stress. These data differ from previous work examining RPG repression in meiosis or under rapamycin treatment where splicing of non-RPGs was not inhibited (Munding et al., 2013). However, a direct comparison of our findings to this study is difficult because of the different environmental stimuli used. Indeed, we found that rapamycin treatment reduced MMS-induced foci, suggesting a different overall stress response.
The model in which Hsh155 is evicted from spliced genes after repression could also apply to Cmr1 and Hos2, which bind to RPGs during transcription (Jones et al., 2016). These dynamic changes at RPGs are initiated by TORC1-dependant relocalization of Sfp1 (Jorgensen et al., 2004; Marion et al., 2004), and we show that either TORC1 inhibition or SFP1 deletion strongly represses Hsh155 aggregation in MMS. Additionally, although Hos2 or Cmr1 aggregation in MMS was also blocked by SFP1 deletion, Apj1 foci formation was unaffected. Thus, changing the transcriptional dynamics of the stress response impacts only some INQ-resident proteins. This is important because it may suggest that Hsh155 and Hos2 are not simply aggregation prone in MMS but instead move to INQ or not, based on the transcriptional needs of the cell. Functionally, our data suggest that, although neither defects in splicing nor protein sequestration by Btn2 seem to confer dramatic sensitivity to chronic MMS exposure by spot dilution assays (Gallina et al., 2015), both Btn2 and Hsh155 might aid in normal recovery after MMS removal. Although there was only a correlation between stress recovery and splicing regulation, we hypothesize that any competitive advantage is related to the retention of a pool of near-native proteins for rapid reactivation after stress that would otherwise be lost.
Previous groups have recognized that INQ structures localize adjacent to the nucleolus (Tkach et al., 2012; Miller et al., 2015). Our data link INQ constituents to ribosome production, but the significance of INQ’s nucleolar proximity remains unknown. Another question is why Hsh155 localizes to aggregates when its partners in the spliceosome do not. Hsh155 does not contain known aggregation-prone Q-rich or Q/N domains, suggesting a regulated process that is currently obscure. Our study highlights INQ as an immediate repository for factors perturbed by RPG repression and, by linking dynamic transcriptional changes to PQC, raises important questions about how cells coordinate the assembly and disassembly of chromatin-associated protein complexes during stress and recovery.
Materials and methods
Yeast growth, manipulation, and analysis
Yeast strains were grown in standard rich media YPD or synthetic complete (SC) medium unless otherwise indicated. Serial dilution assays were performed as described previously (Stirling et al., 2011). In brief, an identical OD of cells was serially diluted tenfold and spotted on the indicated plate with a 48-pin replica pinning manifold and incubated at indicated temperatures for 72 h. Yeast growth curves in YPD media at 30°C were performed as previously described by Stirling et al. (2012), and logarithmic phase cultures treated with or without MMS for 2 h were washed twice and then grown for another 2 h after washout. These were diluted to an OD of 0.05 in a 96-well plate in triplicate and grown for 48 h in an M200 plate reader (TECAN) at 30°C. Fig. 6 B shows the growth curves from six biological replicates for each strain. Standard MMS treatments (unless indicated) were at a concentration of 0.05% for 2 h (∼99%; Sigma-Aldrich). All other chemical treatments were MG132 (80 µl), CHX (200 µg/ml), and rapamycin (200 nM) for 2 h. Table S5 contains a list of yeast strains, including database IDs, genotypes, primers, and plasmids used.
Live-cell imaging and CIN-GFP screen
Genes with reported genome instability (Stirling et al., 2011, 2014) were obtained as GFP fusions (Huh et al., 2003). Actively growing cells in SC medium were exposed to H2O2 (2 mM) or MMS (0.05%) for 2 h in batches of 12 strains in well plates before mounting on concanavalin A (ConA)-treated (Stirling et al., 2012) Teflon-masked 12-well slides. For UV exposure, untreated cells were mounted in 12-well slides, and the droplets were irradiated (500 J/m2) in a stratalinker. Irradiated slides were stored in a humid chamber until imaging. The imaging screen was conducted on an Axioscope (ZEISS) at 100× magnification, and candidate relocalizations were retested in triplicate. Imaging of the treated strains in SC medium after the screen was performed live on a DMi8 microscope (Leica Microsystems) at 100× magnification using ConA-treated slides. VHL-mCherry aggregate induction was done as described previously (Miller et al., 2015). In brief, cells were grown 24 h in SC-Ura with 2% raffinose, diluted into SC-Ura with 2% galactose, and grown for 16 h at 30°C to log phase. Before MMS (0.05% for 2 h) or heat-shock treatment (37°C for 20 min), the media were changed to SC with 2% glucose to repress new VHL expression followed by imaging.
Cell cycle and budding index analysis
Budding index
Logarithmic cultures of GFP-tagged Hsh155 cells were grown at 30°C and treated with α-factor (1 mg/ml for 1 h) followed by MMS (0.05%) plus α-factor for a further 2 h. Arrested and/or MMS-treated cells were then washed twice to release the cells from G1/S and to remove MMS. These cells were then imaged over the next 2 h to score rebudding upon stress recovery. Budding was scored using the differential interference contrast channel of the microscope and controlled with Hta2-mCherry signals (nuclear marker).
Cell cycle analysis
Logarithmic cultures of GFP-tagged Hsh155 cells were grown at 30°C and treated with α-factor (1 mg/ml for 1 h) for G1/S arrest and with nocodazole (15 µg/ml for 1 h) for G2/M arrest before MMS treatment (0.05%; 2 h). Arrested and MMS-treated cells were then imaged as described in Live-cell imaging and CIN-GFP screen.
Image acquisition, analysis, and statistical methods
The images were acquired using an Objective HCX Plan Apochromat 1.40 NA oil immersion 100× objective on an inverted DMi8 microscope (Leica Microsystems) equipped with a motorized differential interference contrast turret (for differential interference contrast imaging) and a filter cube set for FITC/TRITC (for GFP and mCherry fluorescence imaging). The images were captured at room temperature using a scientific complementary metal oxide semiconductor camera (ORCA Flash 4.0 V2; Hamamatsu Photonics) and collected using MetaMorph Premier acquisition software (Molecular Devices) and postprocessed (including gamma adjustments, counting of cells with/without foci, budding index quantifications, and foci intensity measurements) using ImageJ (National Institutes of Health). For all microscopy experiments, the significance of the differences was determined using Prism5 (GraphPad Software) or R. For intensity measurements, samples were compared with t tests or ANOVA, and GraphPad performed F tests for variance as part of this analysis. For comparisons of proportions, Fisher’s tests were used, and p-values were Holm-Bonferroni–corrected in the event of multiple comparisons. Sample sizes were determined post hoc and are listed in the figure legends.
FRAP analysis
FRAP experiments were done using an FV1000 confocal imager (Olympus) exactly as described previously (Chao et al., 2014). Hsh155- and Hos2-GFP–tagged cells were grown to log phase and treated with MMS (0.05%) for 2 h. Cells with foci in the nucleus were selected for imaging. FRAP images were collected on an FV1000 microscope with Fluoview (version 3.0; Olympus). Hsh155 and Hos2-GFP INQ foci were bleached, and the recovery of fluorescence in the bleached region of interest (ROI) was monitored every 5 s. The bleaching experiment was performed using a 488-nm laser using 40% bleach laser power and one-frame bleach time. All fluorescence normalization was automated using R and R Studio3. Background fluorescence was monitored in three ROIs, averaged, and removed from the bleached ROI. Three control ROIs in the nuclei of neighboring cells were monitored, averaged, and normalized to the prebleach ROI (T0), and the fluorescence loss over time was calculated and added back to the bleached ROI. Normalized bleached ROI fluorescence data were transformed by setting the prebleached ROI to 100% and the postbleached ROI to 0% to allow for all FRAP curves to be combined. Data were fit using Prism6 (GraphPad Software) by one-phase association nonlinear regression. Mobile fraction and t1/2 values were calculated and obtained from the values output by Prism6.
The R pipeline for the FRAP analysis is available as source code text in Text S1 and on GitHub with the following link: https://github.com/ahofmann4/Open_FRAP_Analysis.
Western blotting and coimmunoprecipitation
Coimmunoprecipitations were performed using yeast strains containing TAP-tagged Hsh49 and/or GFP-tagged Hsh155 treated with or without MMS. TAP-tagged Hsh49 was captured using IgG Sepharose fast-flow beads (Sigma-Aldrich) and proceeded as described previously (Leung et al., 2016). Immunoblotting was performed with mouse anti-GFP (Thermo Fisher Scientific) and rabbit anti-TAP (Thermo Fisher Scientific).
For Western blots, whole-cell extracts were prepared by trichloroacetic acid extraction and blotted with mouse anti-GFP (Thermo Fisher Scientific) or rabbit anti-PGK1 (Abcam) essentially as described previously (Gallina et al., 2015).
Splicing efficiency assay
Splicing assay protocol was adapted and performed as previously described (Galy et al., 2004). All measurements were taken with individual transformants in triplicate. Cells were struck as a patch on SC medium without leucine and then replica plated to glycerol-lactate–containing SC medium without leucine (GGL-leu). Cells from each patch were inoculated in liquid GGL-leu media for 2 h at 30°C and then were induced with final 2% galactose for 1.5 h before treatment with final 0.05% MMS. Time points were taken at 30, 60, and 120 min post-MMS treatment. Cells carrying reporters were lysed and assayed for β-galactosidase assay using a Gal-Screen β-galactosidase reporter gene assay system for yeast or mammalian cells (Applied Biosystems) as per the manufacturer’s instructions and read with a SpectraMax i3 (Molecular Devices). Relative light units were normalized to cell concentration as estimated by measuring OD600.
Whole-proteome analysis by mass spectrometry
Logarithmic cultures of BY4741 WT strain grown at 30°C with or without MMS (0.05% for 2 h) treatment were pelleted and frozen. Frozen pellets were lysed, reduced, alkylated, trypsin digested, and purified using the SP3 method (Hughes et al., 2014) with modifications (Hughes et al., 2016). Samples were analyzed as detailed by Hughes et al. (2016); in brief, prepared peptide samples were labeled with individual tandem mass tags (Thermo Fisher Scientific), combined in sets of 10, and subjected to offline high-pH fractionation/concatenation, and then fractions (12) were analyzed by reverse-phase nanoelectrospray liquid chromatography on an Orbitrap Fusion Tribrid mass spectrometry platform (Thermo Fisher Scientific) using MS3 scanning.
Mass spectrometry data analysis
Data from the Orbitrap fusion were processed using Proteome Discoverer Software (2.1.0.62; Thermo Fisher Scientific). MS2 spectra were searched using Sequest HT against the UniProt S. cerevisiae proteome database appended to a list of common contaminants (6,752 total sequences). Data were filtered at the peptide spectral match level to control for false discoveries using a q-value cutoff of 0.05 as determined by Percolator (MASCOT; Matrix Science). This less-stringent filter was applied to maximize sensitivity, relying on the statistical analyses during peptide quantification to further control for the potential generation of false conclusions within the final dataset. As a result, the final quantitative set of hits that displays significant variance between sample types is enriched in multipeptide-identified high-confidence proteins. A total of 4,357 proteins were reproducibly quantified, and proteins with significant depletion or enrichment are listed in Table S2.
Bioinformatic and statistical analyses
Datasets generated in Proteome Discoverer were exported and analyzed with a combination of scripts built in R designed in-house. Contaminant and decoy proteins were removed from all datasets before analysis. Unless stated otherwise, quantification was performed at the peptide level as discussed previously (Suomi et al., 2015).
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (Vizcaíno et al., 2014, 2016) with the dataset identifier PXD004459.
RNA isolation, cDNA preparation, and reverse transcription–quantitative PCR analysis
Total RNA was isolated from log phase–grown cells treated with or without MMS and 2 h after MMS washout using the yeast RiboPure RNA Purification kit (Ambion). 1 µg of cDNA was reverse transcribed using anchored-oligo(dT)18 primer and Transcriptor Reverse transcription (Roche). Reverse transcription–quantitative PCRs were performed and analyzed using SYBR green PCR Master Mix and a StepOnePlus Real-Time PCR system (Applied Biosystems). cDNA PCR products and reverse transcription–quantitative PCR transcripts were amplified using the primers in Table S5.
Online supplemental material
Supplemental data of this article include five additional figures and five tables showing the results of the CIN miniarray screen (Fig. S1 and Table S1); further characterization of Hsh155 relocalization (Fig. S2); impact of replication stress on Hsh155 foci formation (Fig. S3, A–F); foci distribution in chaperones (Fig. S3, G–H); budding index in hsp104Δ cells (Fig. S4 A); temporal correlation of the drop in splicing and foci formation (Fig. S4, B and C); influence of Hsh155 aggregate formation on RPL27A transcript levels (Fig. S4, D–E); effects of cell cycle arrests and TORC1 signaling on PQC formation (Fig. S5, A and B); additional Cmr1 ChIP occupancy data (Fig. S5 C); whole-proteome abundance data and gene ontology enrichment after MMS treatment (Tables S2 and S3); comparison of the Cmr1 ChIP occupancy data in WT cells and microarray expression data in sfp1Δ strain (Table S4); and yeast strains, primers, and plasmids used in this study (Table S5). Text S1 contains the source code text for the R pipeline used in the FRAP analysis.
Supplementary Material
Acknowledgments
We acknowledge G.W. Brown, S. Ben-Aroya, P. Hieter, B. Palancade, and D. Kaganovich for yeast strains and plasmids, and C. Govind for help accessing the Cmr1 ChIP data.
P.C. Stirling is a Canadian Institutes of Health Research New Investigator and a Michael Smith Foundation for Health Research Scholar. C.S. Hughes and G.B. Morin acknowledge support from the British Columbia Cancer Foundation. This work is supported by operating grants from the Canadian Institutes of Health Research (MOP-136982) and the Natural Sciences and Engineering Research Council of Canada to P.C. Stirling (RGPIN 2014-04490).
The authors declare no competing financial interests.
Author contributions: V. Mathew and P.C. Stirling designed the project and wrote the manuscript. V. Mathew, A.S. Tam, K.L. Milbury, A.K. Hofmann, C.S. Hughes, and P.C. Stirling generated the data. V. Mathew, A.S. Tam, K.L. Milbury, A.K. Hofmann, C.S. Hughes, G.B. Morin, C.J.R. Loewen, and P.C. Stirling analyzed the data.
Footnotes
Abbreviations used:
- ChIP
- chromatin immunoprecipitation
- CHX
- cycloheximide
- CIN
- chromosome instability
- INQ
- intranuclear quality control
- MMS
- methyl methanesulfonate
- PQC
- protein quality control
- ROI
- region of interest
- RP
- ribosomal protein
- RPG
- RP gene
- SC
- synthetic complete
- TAP
- tandem affinity purification
- TOR
- target of rapamycin
- ts
- temperature sensitive
- VHL
- Von Hippel–Lindau
References
- Begley U., Dyavaiah M., Patil A., Rooney J.P., DiRenzo D., Young C.M., Conklin D.S., Zitomer R.S., and Begley T.J.. 2007. Trm9-catalyzed tRNA modifications link translation to the DNA damage response. Mol. Cell. 28:860–870. 10.1016/j.molcel.2007.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein K.A., and Baserga S.J.. 2004. The small subunit processome is required for cell cycle progression at G1. Mol. Biol. Cell. 15:5038–5046. 10.1091/mbc.E04-06-0515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breker M., Gymrek M., and Schuldiner M.. 2013. A novel single-cell screening platform reveals proteome plasticity during yeast stress responses. J. Cell Biol. 200:839–850. (published correction appears in J. Cell Biol. 2013. 201:353) 10.1083/jcb.201301120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslow D.K., Cameron D.M., Collins S.R., Schuldiner M., Stewart-Ornstein J., Newman H.W., Braun S., Madhani H.D., Krogan N.J., and Weissman J.S.. 2008. A comprehensive strategy enabling high-resolution functional analysis of the yeast genome. Nat. Methods. 5:711–718. 10.1038/nmeth.1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao J.T., Wong A.K., Tavassoli S., Young B.P., Chruscicki A., Fang N.N., Howe L.J., Mayor T., Foster L.J., and Loewen C.J.. 2014. Polarization of the endoplasmic reticulum by ER-septin tethering. Cell. 158:620–632. 10.1016/j.cell.2014.06.033 [DOI] [PubMed] [Google Scholar]
- Chong Y.T., Koh J.L., Friesen H., Duffy S.K., Cox M.J., Moses A., Moffat J., Boone C., and Andrews B.J.. 2015. Yeast Proteome Dynamics from Single Cell Imaging and Automated Analysis. Cell. 161:1413–1424. (published erratum appears in Cell 2015. 162:221) 10.1016/j.cell.2015.04.051 [DOI] [PubMed] [Google Scholar]
- Clemente-Blanco A., Mayán-Santos M., Schneider D.A., Machín F., Jarmuz A., Tschochner H., and Aragón L.. 2009. Cdc14 inhibits transcription by RNA polymerase I during anaphase. Nature. 458:219–222. 10.1038/nature07652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Düvel K., Santhanam A., Garrett S., Schneper L., and Broach J.R.. 2003. Multiple roles of Tap42 in mediating rapamycin-induced transcriptional changes in yeast. Mol. Cell. 11:1467–1478. 10.1016/S1097-2765(03)00228-4 [DOI] [PubMed] [Google Scholar]
- Gabunilas J., and Chanfreau G.. 2016. Splicing-Mediated Autoregulation Modulates Rpl22p Expression in Saccharomyces cerevisiae. PLoS Genet. 12:e1005999 10.1371/journal.pgen.1005999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallina I., Colding C., Henriksen P., Beli P., Nakamura K., Offman J., Mathiasen D.P., Silva S., Hoffmann E., Groth A., et al. . 2015. Cmr1/WDR76 defines a nuclear genotoxic stress body linking genome integrity and protein quality control. Nat. Commun. 6:6533 10.1038/ncomms7533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy V., Gadal O., Fromont-Racine M., Romano A., Jacquier A., and Nehrbass U.. 2004. Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell. 116:63–73. 10.1016/S0092-8674(03)01026-2 [DOI] [PubMed] [Google Scholar]
- Gasch A.P., Spellman P.T., Kao C.M., Carmel-Harel O., Eisen M.B., Storz G., Botstein D., and Brown P.O.. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 11:4241–4257. 10.1091/mbc.11.12.4241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch A.P., Huang M., Metzner S., Botstein D., Elledge S.J., and Brown P.O.. 2001. Genomic expression responses to DNA-damaging agents and the regulatory role of the yeast ATR homolog Mec1p. Mol. Biol. Cell. 12:2987–3003. 10.1091/mbc.12.10.2987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Q., Haroon S., Bravo D.G., Will J.L., and Gasch A.P.. 2012. Cellular memory of acquired stress resistance in Saccharomyces cerevisiae. Genetics. 192:495–505. 10.1534/genetics.112.143016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes C.S., Foehr S., Garfield D.A., Furlong E.E., Steinmetz L.M., and Krijgsveld J.. 2014. Ultrasensitive proteome analysis using paramagnetic bead technology. Mol. Syst. Biol. 10:757 10.15252/msb.20145625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes C.S., McConechy M.K., Cochrane D.R., Nazeran T., Karnezis A.N., Huntsman D.G., and Morin G.B.. 2016. Quantitative Profiling of Single Formalin Fixed Tumour Sections: proteomics for translational research. Sci. Rep. 6:34949 10.1038/srep34949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh W.K., Falvo J.V., Gerke L.C., Carroll A.S., Howson R.W., Weissman J.S., and O’Shea E.K.. 2003. Global analysis of protein localization in budding yeast. Nature. 425:686–691. 10.1038/nature02026 [DOI] [PubMed] [Google Scholar]
- Jiang Y., and Broach J.R.. 1999. Tor proteins and protein phosphatase 2A reciprocally regulate Tap42 in controlling cell growth in yeast. EMBO J. 18:2782–2792. 10.1093/emboj/18.10.2782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J.W., Singh P., and Govind C.K.. 2016. Recruitment of Saccharomyces cerevisiae Cmr1/Ydl156w to Coding Regions Promotes Transcription Genome Wide. PLoS One. 11:e0148897 10.1371/journal.pone.0148897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen P., Rupes I., Sharom J.R., Schneper L., Broach J.R., and Tyers M.. 2004. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 18:2491–2505. 10.1101/gad.1228804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khmelinskii A., Keller P.J., Bartosik A., Meurer M., Barry J.D., Mardin B.R., Kaufmann A., Trautmann S., Wachsmuth M., Pereira G., et al. . 2012. Tandem fluorescent protein timers for in vivo analysis of protein dynamics. Nat. Biotechnol. 30:708–714. 10.1038/nbt.2281 [DOI] [PubMed] [Google Scholar]
- Lempiäinen H., Uotila A., Urban J., Dohnal I., Ammerer G., Loewith R., and Shore D.. 2009. Sfp1 interaction with TORC1 and Mrs6 reveals feedback regulation on TOR signaling. Mol. Cell. 33:704–716. 10.1016/j.molcel.2009.01.034 [DOI] [PubMed] [Google Scholar]
- Leung G.P., Brown J.A., Glover J.N., and Kobor M.S.. 2016. Rtt107 BRCT domains act as a targeting module in the DNA damage response. DNA Repair (Amst.). 37:22–32. 10.1016/j.dnarep.2015.10.007 [DOI] [PubMed] [Google Scholar]
- Loewith R., Jacinto E., Wullschleger S., Lorberg A., Crespo J.L., Bonenfant D., Oppliger W., Jenoe P., and Hall M.N.. 2002. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell. 10:457–468. 10.1016/S1097-2765(02)00636-6 [DOI] [PubMed] [Google Scholar]
- Malinovska L., Kroschwald S., Munder M.C., Richter D., and Alberti S.. 2012. Molecular chaperones and stress-inducible protein-sorting factors coordinate the spatiotemporal distribution of protein aggregates. Mol. Biol. Cell. 23:3041–3056. 10.1091/mbc.E12-03-0194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion R.M., Regev A., Segal E., Barash Y., Koller D., Friedman N., and O’Shea E.K.. 2004. Sfp1 is a stress- and nutrient-sensitive regulator of ribosomal protein gene expression. Proc. Natl. Acad. Sci. USA. 101:14315–14322. 10.1073/pnas.0405353101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D.E., Soulard A., and Hall M.N.. 2004. TOR regulates ribosomal protein gene expression via PKA and the Forkhead transcription factor FHL1. Cell. 119:969–979. 10.1016/j.cell.2004.11.047 [DOI] [PubMed] [Google Scholar]
- Mazumder A., Pesudo L.Q., McRee S., Bathe M., and Samson L.D.. 2013. Genome-wide single-cell-level screen for protein abundance and localization changes in response to DNA damage in S. cerevisiae. Nucleic Acids Res. 41:9310–9324. 10.1093/nar/gkt715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClellan A.J., Scott M.D., and Frydman J.. 2005. Folding and quality control of the VHL tumor suppressor proceed through distinct chaperone pathways. Cell. 121:739–748. 10.1016/j.cell.2005.03.024 [DOI] [PubMed] [Google Scholar]
- Miller S.B., Ho C.T., Winkler J., Khokhrina M., Neuner A., Mohamed M.Y., Guilbride D.L., Richter K., Lisby M., Schiebel E., et al. . 2015. Compartment-specific aggregases direct distinct nuclear and cytoplasmic aggregate deposition. EMBO J. 34:778–797. 10.15252/embj.201489524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munding E.M., Shiue L., Katzman S., Donohue J.P., and Ares M. Jr. 2013. Competition between pre-mRNAs for the splicing machinery drives global regulation of splicing. Mol. Cell. 51:338–348. 10.1016/j.molcel.2013.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Driscoll J., Clare D., and Saibil H.. 2015. Prion aggregate structure in yeast cells is determined by the Hsp104-Hsp110 disaggregase machinery. J. Cell Biol. 211:145–158. 10.1083/jcb.201505104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palancade B., Zuccolo M., Loeillet S., Nicolas A., and Doye V.. 2005. Pml39, a novel protein of the nuclear periphery required for nuclear retention of improper messenger ribonucleoparticles. Mol. Biol. Cell. 16:5258–5268. 10.1091/mbc.E05-06-0527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parenteau J., Durand M., Morin G., Gagnon J., Lucier J.F., Wellinger R.J., Chabot B., and Elela S.A.. 2011. Introns within ribosomal protein genes regulate the production and function of yeast ribosomes. Cell. 147:320–331. 10.1016/j.cell.2011.08.044 [DOI] [PubMed] [Google Scholar]
- Reja R., Vinayachandran V., Ghosh S., and Pugh B.F.. 2015. Molecular mechanisms of ribosomal protein gene coregulation. Genes Dev. 29:1942–1954. 10.1101/gad.268896.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saarikangas J., and Barral Y.. 2015. Protein aggregates are associated with replicative aging without compromising protein quality control. eLife. 4:e06197 10.7554/eLife.06197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saarikangas J., and Barral Y.. 2016. Protein aggregation as a mechanism of adaptive cellular responses. Curr. Genet. 62:711–724. 10.1007/s00294-016-0596-0 [DOI] [PubMed] [Google Scholar]
- Schawalder S.B., Kabani M., Howald I., Choudhury U., Werner M., and Shore D.. 2004. Growth-regulated recruitment of the essential yeast ribosomal protein gene activator Ifh1. Nature. 432:1058–1061. 10.1038/nature03200 [DOI] [PubMed] [Google Scholar]
- Shirahige K., Hori Y., Shiraishi K., Yamashita M., Takahashi K., Obuse C., Tsurimoto T., and Yoshikawa H.. 1998. Regulation of DNA-replication origins during cell-cycle progression. Nature. 395:618–621. 10.1038/27007 [DOI] [PubMed] [Google Scholar]
- Stirling P.C., Bloom M.S., Solanki-Patil T., Smith S., Sipahimalani P., Li Z., Kofoed M., Ben-Aroya S., Myung K., and Hieter P.. 2011. The complete spectrum of yeast chromosome instability genes identifies candidate CIN cancer genes and functional roles for ASTRA complex components. PLoS Genet. 7:e1002057 10.1371/journal.pgen.1002057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling P.C., Chan Y.A., Minaker S.W., Aristizabal M.J., Barrett I., Sipahimalani P., Kobor M.S., and Hieter P.. 2012. R-loop-mediated genome instability in mRNA cleavage and polyadenylation mutants. Genes Dev. 26:163–175. 10.1101/gad.179721.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling P.C., Shen Y., Corbett R., Jones S.J., and Hieter P.. 2014. Genome destabilizing mutator alleles drive specific mutational trajectories in Saccharomyces cerevisiae. Genetics. 196:403–412. 10.1534/genetics.113.159806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suomi T., Corthals G.L., Nevalainen O.S., and Elo L.L.. 2015. Using Peptide-Level Proteomics Data for Detecting Differentially Expressed Proteins. J. Proteome Res. 14:4564–4570. 10.1021/acs.jproteome.5b00363 [DOI] [PubMed] [Google Scholar]
- Svensson J.P., Pesudo L.Q., Fry R.C., Adeleye Y.A., Carmichael P., and Samson L.D.. 2011. Genomic phenotyping of the essential and non-essential yeast genome detects novel pathways for alkylation resistance. BMC Syst. Biol. 5:157 10.1186/1752-0509-5-157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkach J.M., Yimit A., Lee A.Y., Riffle M., Costanzo M., Jaschob D., Hendry J.A., Ou J., Moffat J., Boone C., et al. . 2012. Dissecting DNA damage response pathways by analysing protein localization and abundance changes during DNA replication stress. Nat. Cell Biol. 14:966–976. 10.1038/ncb2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Pel D.M., Stirling P.C., Minaker S.W., Sipahimalani P., and Hieter P.. 2013. Saccharomyces cerevisiae genetics predicts candidate therapeutic genetic interactions at the mammalian replication fork. G3 (Bethesda). 3:273–282. 10.1534/g3.112.004754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaíno J.A., Deutsch E.W., Wang R., Csordas A., Reisinger F., Ríos D., Dianes J.A., Sun Z., Farrah T., Bandeira N., et al. . 2014. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32:223–226. 10.1038/nbt.2839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaíno J.A., Csordas A., del-Toro N., Dianes J.A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., et al. . 2016. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44(D1):D447–D456. 10.1093/nar/gkv1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace E.W., Kear-Scott J.L., Pilipenko E.V., Schwartz M.H., Laskowski P.R., Rojek A.E., Katanski C.D., Riback J.A., Dion M.F., Franks A.M., et al. . 2015. Reversible, Specific, Active Aggregates of Endogenous Proteins Assemble upon Heat Stress. Cell. 162:1286–1298. 10.1016/j.cell.2015.08.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L., Kamau E., Donze D., and Grove A.. 2011. Expression of yeast high mobility group protein HMO1 is regulated by TOR signaling. Gene. 489:55–62. 10.1016/j.gene.2011.08.017 [DOI] [PubMed] [Google Scholar]
- Zhou C., Slaughter B.D., Unruh J.R., Guo F., Yu Z., Mickey K., Narkar A., Ross R.T., McClain M., and Li R.. 2014. Organelle-based aggregation and retention of damaged proteins in asymmetrically dividing cells. Cell. 159:530–542. 10.1016/j.cell.2014.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (Vizcaíno et al., 2014, 2016) with the dataset identifier PXD004459.