Some prenylated proteins such as RAS require carboxyl methylation by ICMT for proper association with cellular membranes. Court et al. show that RAB7 and RAB8 are among these proteins and that by affecting vesicular transport, these GTPases regulate NOTCH signaling in an ICMT-dependent fashion.
Abstract
Isoprenylcysteine carboxyl methyltransferase (ICMT) methylesterifies C-terminal prenylcysteine residues of CaaX proteins and some RAB GTPases. Deficiency of either ICMT or NOTCH1 accelerates pancreatic neoplasia in Pdx1-Cre;LSL-KrasG12D mice, suggesting that ICMT is required for NOTCH signaling. We used Drosophila melanogaster wing vein and scutellar bristle development to screen Rab proteins predicted to be substrates for ICMT (ste14 in flies). We identified Rab7 and Rab8 as ICMT substrates that when silenced phenocopy ste14 deficiency. ICMT, RAB7, and RAB8 were all required for efficient NOTCH1 signaling in mammalian cells. Overexpression of RAB8 rescued NOTCH activation after ICMT knockdown both in U2OS cells expressing NOTCH1 and in fly wing vein development. ICMT deficiency induced mislocalization of GFP-RAB7 and GFP-RAB8 from endomembrane to cytosol, enhanced binding to RABGDI, and decreased GTP loading of RAB7 and RAB8. Deficiency of ICMT, RAB7, or RAB8 led to mislocalization and diminished processing of NOTCH1-GFP. Thus, NOTCH signaling requires ICMT in part because it requires methylated RAB7 and RAB8.
Introduction
Isoprenylcysteine carboxyl methyltransferase (ICMT), a polytopic membrane protein restricted to the ER (Wright et al., 2009), is the third of three enzymes that modify the C terminus of proteins that end with a CaaX motif, such as the products of RAS oncogenes (Wright and Philips, 2006). In addition, ICMT modifies a subset of RAB GTPases that end with a CXC motif (Smeland et al., 1994). The CaaX motif is first prenylated by either farnesyltransferase or geranylgeranyltransferase I and then is acted on by RAS-converting enzyme 1, an ER-restricted endoprotease that removes the aaX sequence, leaving the prenylcysteine as the new C terminus (Wright and Philips, 2006). In the case of CXC RAB proteins, both cysteines are modified by geranylgeranyltransferase II. For both classes of prenylproteins, ICMT acts to methylesterify the α-carboxyl group of the C-terminal prenylcysteine, thereby eliminating a negative charge at physiological pH, adding to the hydrophobicity of the C-terminal domain and promoting membrane association of the modified protein.
Because the RAS oncoproteins are substrates of ICMT, it has long been considered a target for anti-RAS therapy (Cox et al., 2015). To test this idea, we previously crossed mice with a conditional, floxed Icmt allele to mice with a conditional oncogenic KrasG12D allele (Hingorani et al., 2003) and then activated Kras and deleted Icmt simultaneously in the embryonic pancreas by expressing PDX1-Cre (genotype Icmtflx/flx;LSL-KrasG12D;Pdx1-Cre; Court et al., 2013). Surprisingly, ICMT deficiency in the setting of oncogenic KRASG12D exacerbated the neoplastic disease in the pancreas. In addition, cutaneous papillomatous neoplasms of the face were also evident as a result of PDX1-Cre expression in keratinocytes (Mazur et al., 2010). Because the requirement for CaaX processing for KRAS function is well established, we sought to determine whether the accelerated progression of KRAS-driven neoplasms evident in the setting of ICMT deficiency might be attributable not to increased KRAS activity but rather to a tumor suppressor that requires ICMT for full activity. Whereas mammalian NOTCH genes have been shown to be oncogenes or tumor suppressors depending on the cellular context (Radtke and Raj, 2003), in the LSL-KrasG12D;Pdx1-Cre mouse, Notch1 acts as a tumor suppressor (Hanlon et al., 2010; Mazur et al., 2010). Indeed, Notch1flx/flx;LSL-KrasG12D;Pdx1-Cre mice phenocopied Icmtflx/flx;LSL-KrasG12D;Pdx1-Cre both in terms of enhanced pancreatic neoplasia and facial papillomas (Court et al., 2013). This result suggested that NOTCH1 signaling requires ICMT, a hypothesis we confirmed in both mammalian cells and Drosophila melanogaster wing development (Court et al., 2013).
The NOTCH signaling pathway is evolutionarily conserved and is required at many stages of development (Tien et al., 2009; Andersson et al., 2011). NOTCH signaling requires cell–cell contact because the ligands for NOTCH, like the receptor, are transmembrane proteins. In flies, there is one Notch protein and two ligands, Delta and Serrate. Mammalian genomes encode four NOTCH proteins that interact with three Delta-like (DLL) or two Jagged (orthologue of Serrate) proteins (Tien et al., 2009). NOTCH signaling requires three proteolytic events. The first, at site 1 (S1), occurs during biosynthesis of the NOTCH receptor and is catalyzed by a furin-like convertase in the Golgi that cleaves the extracellular domain (ECD) of the receptor, allowing it to form a noncovalently linked transmembrane heterodimer. During canonical NOTCH signaling, a NOTCH receptor interacts with a ligand on an adjacent cell, triggering proteolytic cleavage catalyzed by the A disintegrin and metalloproteinase (ADAM) at site 2 (S2) proximal to the transmembrane segment (Tien et al., 2009). After S2 cleavage, the truncated receptor is endocytosed. The final cleavage at site 3 (S3) is catalyzed by a γ-secretase complex in the endosomal membrane (Andersson et al., 2011). This cleavage liberates the NOTCH intercellular domain (NICD) that upon release from membranes enters the nucleus, where it binds to a CBF1/suppressor of hairless/LAG-1 (CSL) family DNA-binding protein that initiates transcription from NOTCH response elements (NREs).
Based on the well-described elements of canonical NOTCH signaling described above, it is not clear why ICMT activity is required. It is clear that vesicular trafficking is required for NOTCH signaling, both in the biosynthesis of NOTCH and its ligands and in the endocytosis required for NOTCH proteolytic processing. Vesicular trafficking is regulated by the RAB family of small GTPases (Hutagalung and Novick, 2011). Mammalian genomes encode 70 RAB proteins (Colicelli, 2004), many of which are paralogs, that can be organized into at least 14 functional groups (Schwartz et al., 2007). Most RAB proteins end with a CC motif that is geranylgeranylated on both cysteines. These are not substrates for ICMT. A relatively small subset of RAB proteins, those that terminate with a CaaX or CXC sequence, are substrates for ICMTs (Leung et al., 2007). RAB1 and RAB11 have been previously implicated in NOTCH signaling as being required for the trafficking of NOTCH ligands (Emery et al., 2005; Charng et al., 2014). However, neither RAB1 nor RAB11 are ICMT substrates (Leung et al., 2007). In this study, we tested the idea that ICMT may be required for NOTCH signaling because of its ability to modify a subset of RAB proteins. We identified RAB7 and RAB8 as substrates for ICMT that, when silenced, inhibited NOTCH signaling both in D. melanogaster wing development and in mammalian cells. Silencing or disruption of ICMT using CRISPR in mammalian cells caused mislocalization of GFP-RAB7 and GFP-RAB8, and silencing of these RAB transcripts or ICMT resulted in mislocalization of NOTCH1-GFP. The effects of ICMT deficiency could be rescued by overexpression of RAB8. We conclude that ICMT is required for NOTCH signaling in part because it is required for the function of RAB7 and RAB8.
Results
Silencing Rab7 or Rab8 phenocopies loss of ste14 (ICMT) in D. melanogaster
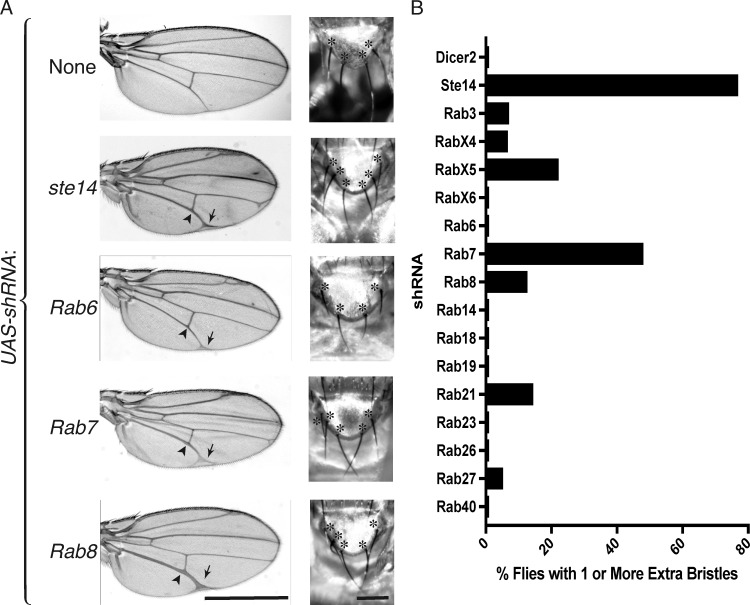
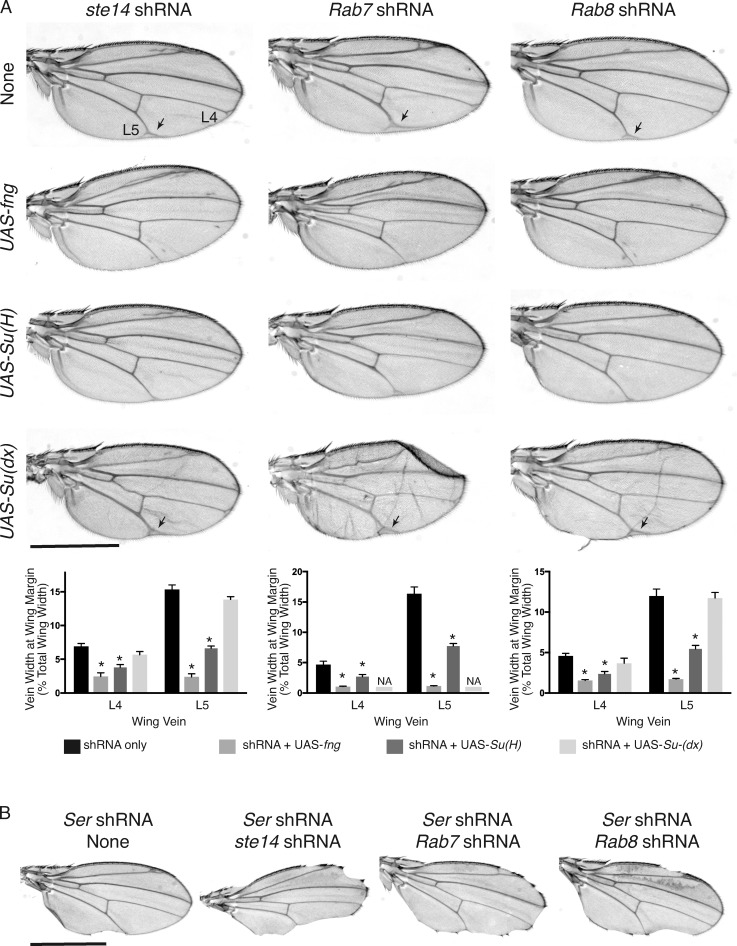
To determine whether the effects of ICMT deficiency on NOTCH signaling might be a consequence of inhibiting RAB protein function, we conducted an shRNA screen of Rab genes in D. melanogaster that are predicted to be substrates of Ste14 (those ending in CaaX or CXC sequences; Figs. S1 and S2 and Table 1; Zhang et al., 2007). We used an Ap-Gal4 driver to express UAS-shRNAs for each of the relevant Rab genes in the developing wing. To enhance the efficacy of shRNA, we crossed Ap-Gal4 flies with those overexpressing UAS-Dicer2. As previously demonstrated (Court et al., 2013), knockdown of ste14 resulted in a broadening of the wing veins, especially L4 and L5, consistent with a Notch loss-of-function phenotype (Parody and Muskavitch, 1993). Silencing Rab 3, 4, 6, 7, 8, and 23 resulted in some degree of wing vein broadening (Figs. S1 and S2), although the effect of silencing Rab 3, 4, 6, and 23 was relatively mild. Silencing Rab 7 or 8 gave a phenotype at least as strong as ste14 (Fig. 1 A).
Table 1. C-terminal amino acid sequence, prenyltransferase, and wing vein phenotype for D. melanogaster Rab proteins predicted to be substrates for ICMT (Ste14).
| Rab protein | C-terminal aa sequence | Predicted prenyltransferase | Wing vein widening |
|---|---|---|---|
| Rab3 | NCNC | GGT II | + |
| Rab4 | CTCRV | ? | + |
| RabX4 | RCTC | GGT II | − |
| RabX5 | GCTC | GGT II | − |
| Rab6 | GCAC | GGT II | ++ |
| RabX6 | SCGC | GGT II | − |
| Rab7 | NCQC | GGT II | +++ |
| Rab8 | CSLL | GGT I | ++ |
| Rab14 | QCSC | GGT II | − |
| Rab18 | TCYC | GGT II | − |
| Rab19 | CNLT | FT | − |
| Rab21 | CCGI | FT | − |
| Rab23 | CGIL | GGT I | + |
| Rab26 | CRMN | FT | − |
| Rab27 | CRNC | FT | − |
| Rab32 | KCSC | GGT II | NA |
| Rab40 | CAIA | FT | − |
FT, farnesyltransferase (CAAX); GGT I, geranylgeranyltransferase type 1 (CAAL); GGT II, geranylgeranyltransferase type 2 (CXC); NA, not assessed.
Figure 1.
Knockdown of Rab7 or Rab8 phenocopies ste14 (ICMT) knockdown in D. melanogaster wing and notum. (A) Adult wing and scutellum of Ap-Gal4; UAS-Dcr2 flies expressing the indicated GAL4-responsive UAS-ShRNAs at 29°C and 25°C, respectively. In left panels, arrowheads indicate widened veins, and arrows mark terminal bifurcations. In right panels, scutellar mechanosensory bristles are marked with asterisks. The normal complement of bristles is four. Bars: (left) 1 mm; (right) 500 µm. (B) Quantification of the percentage of Ap-Gal4; UAS-Dcr2 flies transgenic for the indicated UAS-shRNA that have supernumerary scutellar bristles. Data shown are the means of two independent crosses at 25°C. n ≥ 30.
In addition to the adult wing, the dorsal compartment of the wing imaginal disc gives rise to the adult notum, from which grow mechanosensory scutellar bristles. Notch signaling is critical for patterning these bristles, and loss-of-function Notch alleles lead to supernumerary bristles (Brennan et al., 1999; Yamamoto et al., 2010; Zhang et al., 2012). We observed duplication of one or more scutellar bristles in 77% of Ap-Gal4;UAS-shRNA ste14 flies at 25°C (Fig. 1, A and B), confirming a requirement for Ste14 in Notch signaling. Silencing several of the Rab genes in our screen also resulted in supernumerary bristles, although the phenotype was not as penetrant as that of loss of ste14 (Fig. 1 B). Among the Rab genes required to suppress supernumerary bristles, Rab7, RabX5, and Rab21 had the strongest effects, suggesting that silencing these Rabs causes Notch inactivation. Rab8 but not Rab6 also scored positive in this analysis. These data indicate that silencing of Rab7 or Rab8 phenocopies loss of ste14 and suggest that Rab7 or Rab8 or both may require carboxyl methylation to function efficiently in the Notch pathway.
RAB7 and RAB8 are methylated by ICMT
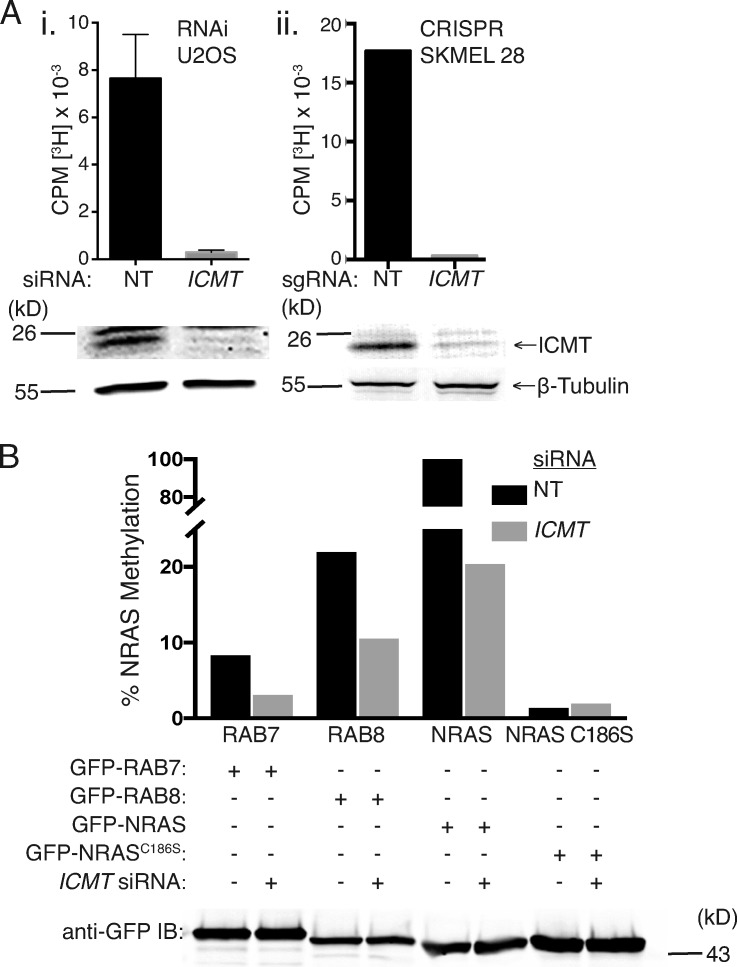
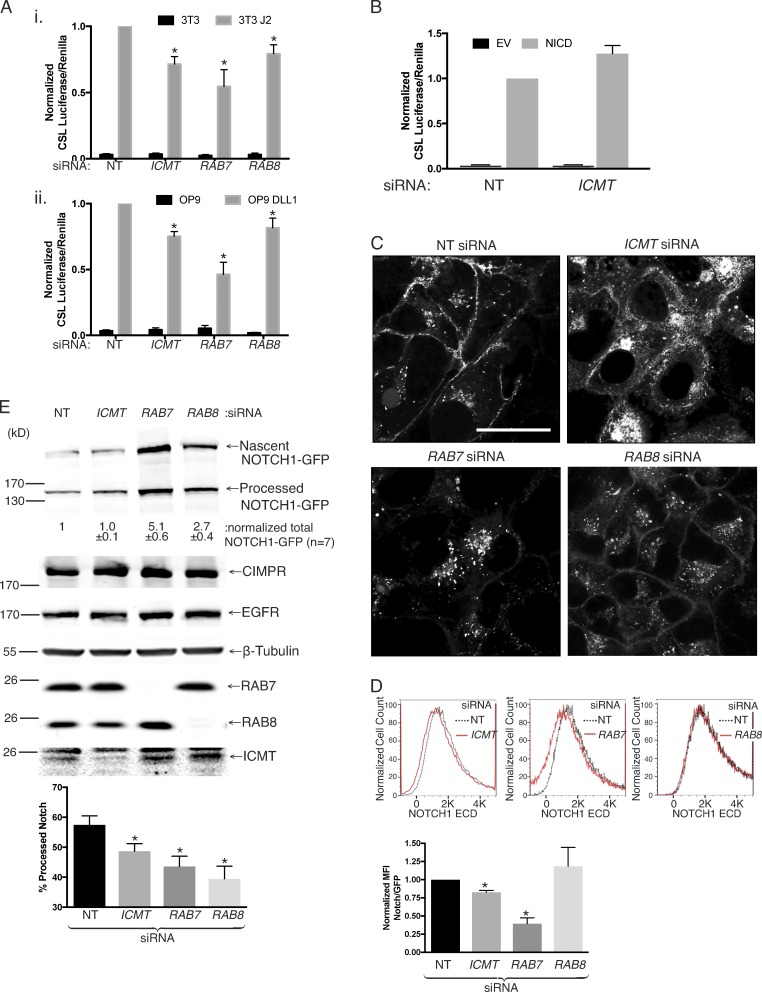
We could silence ICMT expression in mammalian cells as determined by immunoblot either by RNAi or CRISPR/Cas9 genome editing (Fig. 2 A). To confirm that this level of protein depletion resulted in significant decreases in enzyme activity, we homogenized these cells in the absence of detergent with nitrogen cavitation, harvested membranes, and performed a radiometric methylation assay using N-acetyl-S-farnesylcysteine as a methyl acceptor (Choy and Philips, 2000). Both siRNA and CRISPR/Cas9 editing resulted in membranes with <5% of control ICMT activity (Fig. 2 A). RAB proteins ending with CaaX or CXC motifs have been shown to be carboxyl methylated by ICMT (Farnsworth et al., 1991; Smeland et al., 1994; Svensson et al., 2006; Leung et al., 2007; Do et al., 2017). To confirm that RAB7 and RAB8 are substrates for ICMT, we expressed GFP-tagged fusions of each of these GTPases as well as either GFP-NRAS or GFP-NRASC186S, a mutant that cannot be prenylated or carboxyl methylated, in U2OS cells transfected 3 d prior with either nontargeting (NT) or ICMT siRNA (Fig. 2 B). After silencing ICMT, we incubated the cells with l-[methyl-3H]methionine, the precursor of both l-methionyl–tRNA and S-adenosylmethionine, the methyl donor for ICMT, and assayed for carboxyl methylation of immunoprecipitated GFP-GTPases as we have described previously (Choy and Philips, 2000). As expected, GFP-NRAS incorporated alkaline-labile [3H]methyl groups in an ICMT-dependent manner, and GFP-NRASC186S remained unlabeled (Fig. 2 B). Both RAB7 and RAB8 also incorporated [3H]methyl groups in an ICMT-dependent manner, albeit at a level 5–10-fold less than GFP-NRAS. The difference could be either decreased efficiency of carboxyl methylation or decreased stability of the α-carboxyl group. The difference in magnitude of [3H]methyl incorporation notwithstanding, these data demonstrate that both RAB7 and RAB8 are substrates of ICMT.
Figure 2.
RAB7 and RAB8 are ICMT substrates. (A) ICMT activity of membrane fractions isolated from U2OS cells treated with either NT or ICMT siRNA (i) and SKMEL-28 cells with and without genomic disruption of ICMT (ii) using CRIPSR/Cas9 ([3H methyl-]adenosyl-l-methionine and N-acetyl-S-farnesyl-l-cysteine as a methyl donor and acceptor, respectively). The data shown in i are means ± SEM (n = 3), and those in ii are means of triplicates from a single experiment. Shown below the graphs are representative immunoblots (IBs) of lysates from the same cells using ICMT and β-tubulin antibodies. CPM, counts per minute. (B) Incorporation of l-[methyl-3H]methionine into alkaline-labile [3H]methyl esters on the indicated GFP-tagged proteins expressed in U2OS cells with prior transfection with either NT or ICMT siRNA. Data shown are normalized to the level of protein expression as measured by Western blotting using a GFP antibody and to the amount of incorporation in GFP-NRAS without ICMT knockdown (shown as 100%). Data are representative of two experiments.
Unmethylated RAB7 and RAB8 are mislocalized from cellular membranes to the cytosol where they bind RAB–GDP dissociation inhibitor (GDI; RABGDI)
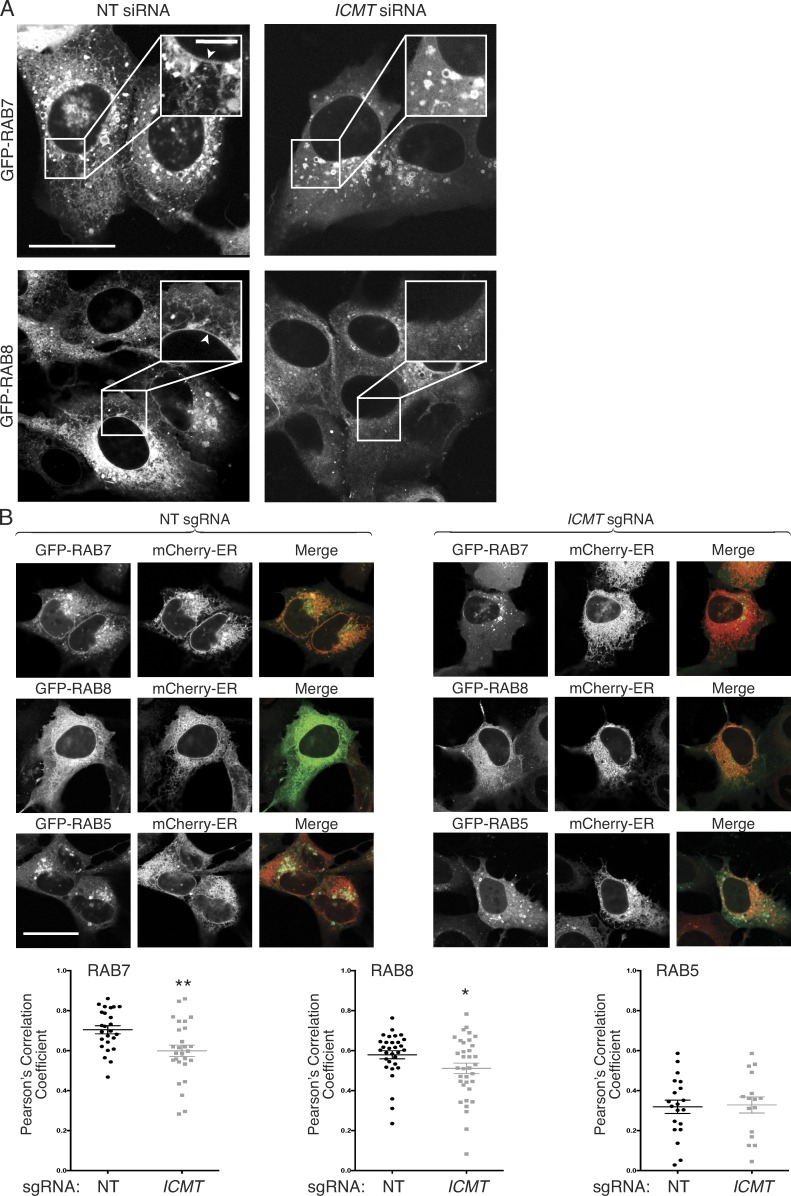
Carboxyl methylation of prenylated proteins serves to neutralize the negative charge of the α-carboxyl group at physiological pH, and this negates an otherwise repulsive force toward negatively charged phospholipids present in cellular membranes (Court et al., 2011). We predicted, therefore, that genomic disruption of ICMT could impact RAB protein activity by diminishing their affinity for membranes and mislocalizing RABs from membranes to cytosol. To test this idea, we performed live-cell confocal imaging of GFP-RAB7 and GFP-RAB8 with and without silencing ICMT. GFP-RAB7 expressed in U2OS cells decorated the nuclear envelope and contiguous ER and was enriched on cytoplasmic vesicles that ringed the nucleus (Fig. 3 A). When ICMT was silenced, the cytoplasmic vesicles persisted, but the ER and nuclear envelope decoration was diminished, and a homogeneous fluorescence, characteristic of a cytosolic distribution, emerged. A similar change in subcellular localization was observed for GFP-RAB8 in U2OS cells when ICMT was silenced (Fig. 3 A). To confirm the decrease in ER decoration of GFP-RAB7 and GFP-RAB8 after silencing of ICMT, we measured colocalization with the ER marker mCherry-ER3 by confocal microscopy using Pearson’s correlation coefficient and Costes method of background correction (Fig. 3 B; Manders et al., 1992; Costes et al., 2004; Dunn et al., 2011). We observed a significant decrease in the colocalization of GFP-RAB7 and GFP-RAB8 but not GFP-RAB5 (not a substrate for ICMT) with mCherry-ER3 after genomic disruption of ICMT in U2OS cells. Identical results were obtained when ICMT was silenced with siRNA rather than disrupted by CRISPR/Cas9 (Fig. S3).
Figure 3.
Unmethylated GFP-RAB7 and GFP-RAB8 are mislocalized from endomembranes to the cytosol. (A) U2OS cells were treated for 3 d with either NT or ICMT siRNA before transfection with either GFP-RAB7 or GFP-RAB8. The cells were imaged live by confocal microscope the following day. Bars: (main images) 10 µm; (insets) 2.5 µm. Arrowheads show prominent nuclear envelope/ER decoration. (B) U2OS cells with or without genomic disruption of ICMT by CRISPR/Cas9 were transfected with constructs for the indicated fluorescent proteins and then imaged 1 d later by confocal microscopy. Bar, 10 µm. Below the representative images are graphs showing the means ± SEM. Pearson’s correlation coefficient for the colocalization of the GFP-tagged protein with mCherry-ER3. Data are from two independent experiments. n ≥ 16. *, P < 0.05; **, P < 0.01 (two-sided t test).
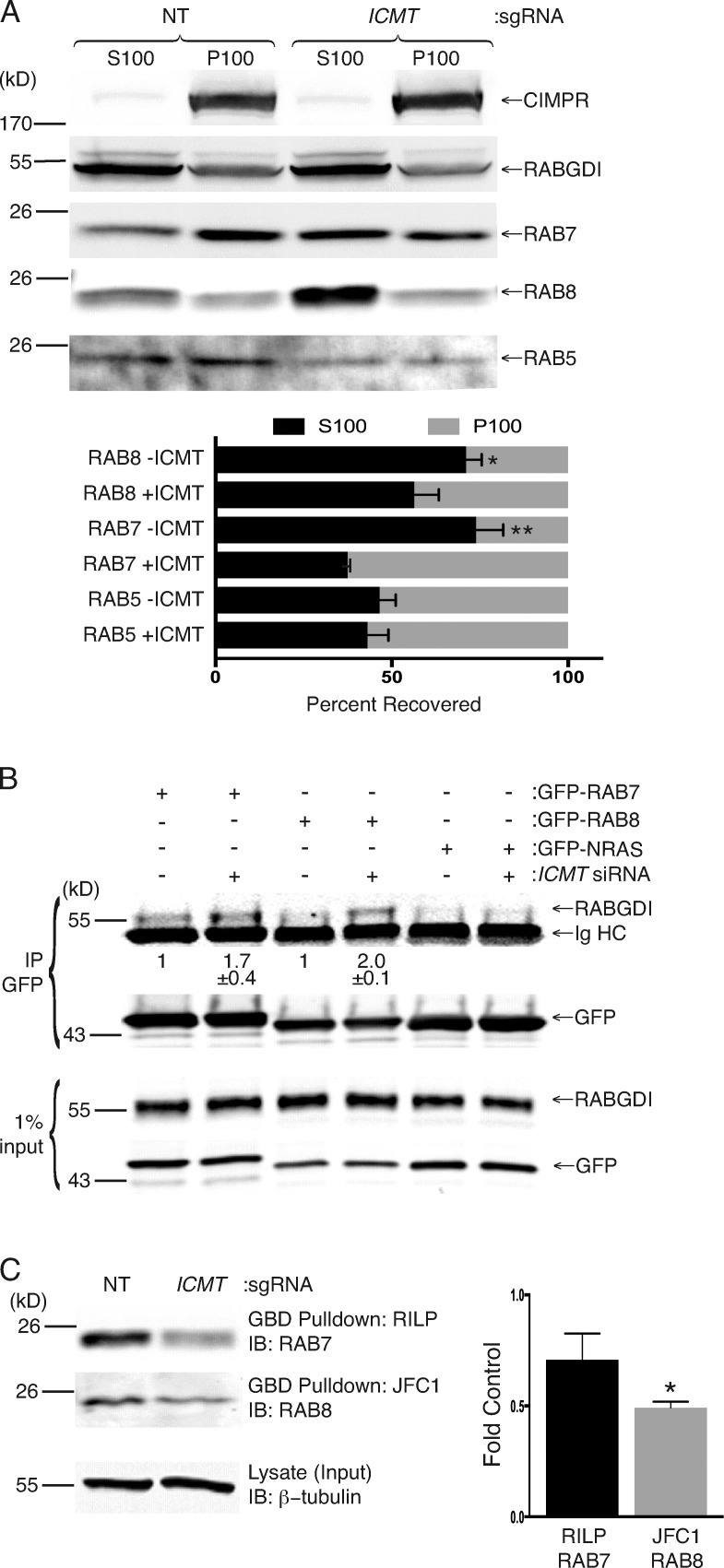
To confirm mislocalization of RAB7 and RAB8, we performed subcellular fractionation. We isolated membrane (P100) and cytosolic (S100) fractions by ultracentrifugation of detergent-free homogenates of SKMEL-28 cells disrupted by nitrogen cavitation after infection with either lentivirus expressing Cas9 alone or Cas9 and a single-guide RNA (sgRNA) targeting ICMT. We observed a reduction in the amounts of both RAB7 and RAB8 in the P100 fraction with a corresponding increase in S100 fractions in cells treated with sgRNA targeting ICMT (Fig. 4 A). In contrast, no difference in the distribution between the S100 or P100 fraction was observed for RAB5. Similar results were obtained in U2OS cells disrupted by nitrogen cavitation 4 d after treatment with either NT or ICMT siRNA (Fig. S4 A).
Figure 4.
Unmethylated RAB7 and RAB8 are enriched in the cytosol, where they interact with the cytosolic chaperone RABGDI, and GTP loading is decreased. (A) Cell equivalents of cytosolic (S100) and membrane (P100) fractions generated from SKMEL-28 cells disrupted by nitrogen cavitation with or without genomic disruption of ICMT by CRISPR/Cas9 were analyzed by SDS-PAGE and immunoblotting with the indicated antibodies. The graph under the representative immunoblots (IBs) shows means ± SEM of the percentage of endogenous RAB5, RAB7, and RAB8 present in each fraction with or without ICMT CRISPR (n = 3). (B) HEK293 cells were treated for 3 d with or without NT or ICMT siRNA before transfection with the indicated GFP constructs. The following day, the cells were lysed, the GFP-tagged proteins were immunoprecipitated (IP), and the precipitates and input were immunoblotted for RABGDI and GFP. Values shown under the representative blot are means ± SEM of the amount of RABGDI coimmunoprecipitated with GFP-RAB7 and GFP-RAB8 normalized to the input as well as the amount immunoprecipitated without ICMT siRNA (n = 3). (C) GTP loading of RAB7 and RAB8 in SKMEL-28 cells with or without genomic disruption of ICMT. GTP-bound RAB7 was quantified by GST-RILP pulldown, and GTP-RAB8 was quantified by GST-JFC1 pulldown. Graphs show the amount of GTP-RAB7 and GTP-RAB8 normalized to the loading control. n = 3. *, P < 0.05; **, P < 0.01 (two-sided t test).
The localization of doubly geranylgeranylated RAB proteins in the aqueous environment of the cytosol presents a conundrum. RAB proteins interact with a protein called RABGDI that was originally identified as a factor that stabilizes inactive GDP-bound RABs by preventing release of GDP (Matsui et al., 1990). By providing a hydrophilic pocket in which the prenyl groups of RAB proteins are sequestered, RABGDI also serves as a carrier protein for geranylgeranylated RAB GTPases, allowing them to dwell in or transit the cytosol. The RAB/RABGDI interaction mediates both the delivery of RABs to membranes and their recycling back to the cytosol (Soldati et al., 1993, 1994; Ullrich et al., 1993; Dirac-Svejstrup et al., 1994). To test the idea that mislocalization of RAB7 and/or RAB8 might be a manifestation of altered binding to RABGDI, we performed coimmunoprecipitation experiments. We coimmunoprecipitated endogenous RABGDI with GFP-RAB7 and RAB8 with or without silencing ICMT. The amount of RABGDI affinity purified in this way in cells deficient in ICMT increased 1.7- and 2.0-fold for GFP-RAB7 and GFP-RAB8, respectively (Fig. 4 B). GFP-NRAS served as a negative control and affinity purified no RABGDI. In contrast with RAB7 and RAB8, RAB5 and RAB23 bound little RABGDI and, although RAB6 bound RABGDI most efficiently, the binding was not affected by silencing ICMT (Fig. S4 B). We conclude that RAB7 and RAB8 are among a subset of RAB proteins that bind RABGDI in a manner inhibited by prenylcysteine carboxyl methylation and that enhanced binding to RABGDI upon silencing ICMT might account, in part, for the observed steady-state localization in the cytosol.
RAB7 and RAB8 require ICMT for efficient GTP loading
RAB proteins generally encounter their cognate exchange factors on membranes such that carboxyl methylation is predicted to promote exchange. To determine whether carboxyl methylation is required for efficient GTP loading of RAB7 and/or RAB8, we performed GST–GTPase binding domain (GBD) pulldown assays for each GTPase in SKMEL-28 melanoma cells with or without CRISPR/Cas9 genomic disruption of ICMT. For RAB7, we used the GBD of RAB-interacting lysosomal protein (GST-RILP) that interacts only with GTP-bound RAB7 (Romero Rosales et al., 2009). For RAB8, we used the GBD of the effector JFC1 (GST-JFC1; Hattula et al., 2006). We detected a reduction in the amount of GTP-RAB7 (Fig. S4 C) but not GTP-RAB8 (Fig. S4 D) pulled down after silencing of ICMT with siRNA in U2OS cells. However, upon genomic disruption of ICMT in SKMEL-28 cells, the GTP-bound fractions of both RAB7 and RAB8 were diminished (Fig. 4 C). We conclude that GTP loading of RAB7 and RAB8 is reduced in the absence of ICMT.
Silencing of Rab7 or Rab8 reduces Notch signaling in D. melanogaster wings
Wing vein patterning in D. melanogaster depends on signaling through several different pathways (Blair, 2007). To confirm that the wing vein phenotypes observed upon silencing of ste14, Rab7, or Rab8 were the result of a reduction in Notch signaling, we sought to determine whether overexpression of elements of the Notch signaling pathway could rescue the Ste14 and Rab7/8 loss-of-function phenotypes. Overexpression of many components of the Notch signaling pathway, including Notch and Delta, in the wing imaginal disc resulted in either a lethal phenotype or crumpled wings that could not be examined. However, we were able to validate Suppressor of Hairless (Su(H)) and Fringe (fng) as genes required for Notch signaling by examining adult wings. Su(H), the orthologue of mammalian CSL, is the nuclear effector of the Notch signaling pathway that binds to the intracellular domain of activated Notch in the nucleus, where it acts as a transcription factor and regulates the expression of Notch target genes (Klein et al., 2000; Kopan and Ilagan, 2009; Auer et al., 2015). Fng is a glycosyltransferase that modifies Notch on its EGF-like repeat sequences and promotes Delta–Notch signaling (Panin et al., 1997; Moloney et al., 2000; Okajima and Irvine, 2002; Haines and Irvine, 2003). Suppressor of deltex (Su(dx)) encodes a HECT domain E3 ubiquitin ligase that directs Notch for lysosomal degradation and is therefore a negative regulator of Notch signaling (Fostier et al., 1998; Cornell et al., 1999; Mazaleyrat et al., 2003; Chastagner et al., 2008).
Expression of UAS-Su(H) or UAS-fng was able to partially rescue the wing vein–broadening phenotype observed in Ap-Gal4; UAS-ShRNA ste14, Ap-Gal4; UAS-ShRNA Rab7, and Ap-Gal4; UAS-ShRNA Rab8 flies at 29°C (Fig. 5 A). In contrast, the wing vein phenotype of Ap-Gal4; UAS-ShRNA ste14, Ap-Gal4; UAS-ShRNA Rab7, and Ap-Gal4; UAS-ShRNA Rab8 flies was not rescued by expression of UAS-Su(dx). We also tested whether concomitant knockdown of ste14, Rab7, or Rab8 could worsen the Notch loss-of-function wing phenotype seen in Ap-Gal4; UAS-ShRNA Serrate flies. Indeed, we found that silencing of ste14, Rab7, or Rab8 enhanced the wing phenotypes (Fig. 5 B), suggesting that diminished Notch signaling upon loss of Serrate (Speicher et al., 1994; Kim et al., 1995) is exacerbated by concomitant loss of ste14, Rab7, or Rab8. Collectively, these data demonstrate that the wing vein phenotypes observed upon silencing ste14, Rab7, or Rab8 in D. melanogaster reflect a loss of Notch signaling.
Figure 5.
Silencing of ste14, Rab7, and Rab8 reduces Notch signaling in D. melanogaster wings. (A) Adult wing of Ap-Gal4; UAS-Dcr2 flies expressing the indicated GAL4-responsive UAS-shRNAs and UAS transgenes at 29°C. Expression of UAS-fng or UAS-Su(H) but not UAS-Su(dx) rescued the wing vein Delta phenotype (arrows) observed in the L4 and L5 veins of the wings expressing UAS-ste14 shRNA, UAS-Rab7 shRNA, or UAS-Rab8 shRNA. Below the representative wings, the widths of the bifurcation of L4 and L5 are plotted as percentages of the wing width. Data shown are means ± SEM. n = 10. *, P < 0.05 (two-sided t test). NA, not assessable because the majority of wings were crumpled and vein width could not be measured accurately. (B) Adult wing of Ap-Gal4; UAS-Dcr2 flies expressing the indicated GAL4-responsive UAS-shRNAs at 25°C. Bar, 1 mm. Knockdown of ste14, Rab7, or Rab8 enhanced the notched wing phenotype observed with knockdown of Serrate (Ser).
Silencing of RAB7 and RAB8 reduces NOTCH trafficking and signaling in mammalian cells
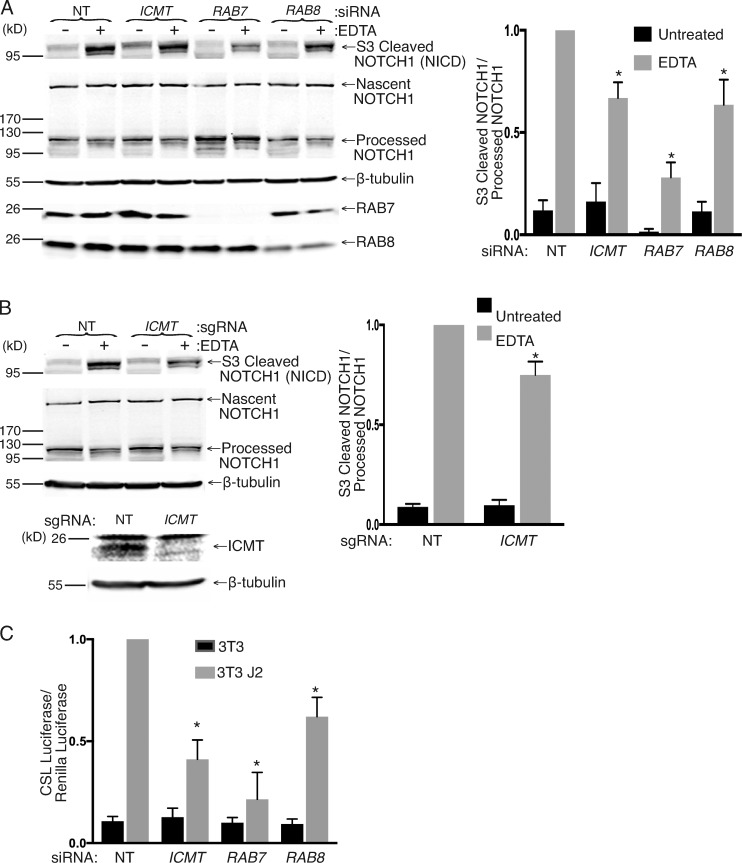
To further confirm that RAB7 and RAB8 play a role in NOTCH signaling, we used a mammalian coculture system that incorporates a luciferase-based assay. Using this assay, we have previously shown that ICMT is required for NOTCH signaling (Court et al., 2013). We used siRNAs to knock down ICMT, RAB7, or RAB8 in U2OS cells expressing NOTCH1-GFP and a NOTCH-responsive CSL-luciferase reporter. To activate the NOTCH pathway, we cocultured these cells with either OP9 cells expressing the NOTCH ligand DLL1 or 3T3 cells expressing the NOTCH ligand Jagged 2 (J2). Upon silencing of ICMT, RAB7, or RAB8, we observed a significant reduction in CSL-luciferase expression when normalized to Renilla luciferase expression (Fig. 6 A). In contrast, silencing ICMT did not affect CSL-luciferase expression driven by ectopic expression of NICD, the product of S3 cleavage of NOTCH that enters the nucleus and binds to CSL (Fig. 6 B), demonstrating that the activity of ICMT is upstream or at the level of S3 cleavage.
Figure 6.
Silencing of ICMT, RAB7, or RAB8 alters the processing and subcellular localization of NOTCH1 and inhibits NOTCH signaling. (A) NOTCH signaling in U2OS cells quantified with a NOTCH-responsive CSL firefly luciferase reporter. Shown are the ratios of CSL firefly/CMV renilla in U2OS cells expressing NOTCH1-GFP cocultured with 3T3 (i) or OP9 (ii) fibroblasts expressing the NOTCH ligands Jagged 2 (J2) or DLL1, respectively. The cells were transfected with the indicated siRNAs 4 d before the luciferase reading. Values were normalized to the maximum firefly/renilla ratio in NT siRNA–treated cells and are given as means ± SEM. n = 7. (B) NOTCH signaling measured as in A in U2OS cells transfected with either empty vector or NICD. The values are normalized to the firefly/renilla measured in NT siRNA–treated cells expressing NICD. Data shown are means ± SEM. n = 3. (C) NOTCH-GFP–expressing U2OS cells were transfected with the indicated siRNAs for 4 d before imaging live by confocal microscopy. Representative images are shown. Bar, 10 µm. (D) Cytofluorometric analysis of surface NOTCH1-GFP by staining intact cells with or without the indicated 4 d siRNA knockdown with an antibody to the ectodomain of NOTCH1. Shown are representative histograms (X axis, linear fluorescence intensity) in the top panels and quantification of normalized mean fluorescence intensity values shown as means ± SEM (n = 3) in the bottom panel. (E) NOTCH1-GFP–expressing U2OS cells were transfected with the indicated siRNAs for 4 d before lysis and analysis by SDS-PAGE and immunoblot with the indicated antibodies. The values shown under the representative blot are means ± SEM of the total amount of NOTCH1-GFP (processed + nascent unprocessed) expressed in each condition normalized to β-tubulin (n = 7). Below is the amount of processed NOTCH1-GFP relative to the total expressed NOTCH1-GFP plotted as means ± SEM. n = 7. *, P < 0.05 (two-sided t test).
We hypothesized that reduced NOTCH signaling in the absence of ICMT, RAB7, or RAB8 was caused by defects in NOTCH trafficking to the plasma membrane (PM). To test this idea, we treated U2OS cells stably expressing NOTCH1-GFP with siRNAs directed toward ICMT, RAB7, or RAB8 and imaged these cells live by confocal microscopy (Fig. 6 C). As a type 1 transmembrane glycoprotein, NOTCH1 is cotranslationally inserted into the ER membrane and trafficked to the PM via the Golgi, where the precursor protein is cleaved by a furin-like convertase at S1 to generate the active processed NOTCH1 that is a noncovalently linked heterodimer (Blaumueller et al., 1997; Logeat et al., 1998). In the control condition, NOTCH1-GFP was detected as expected (van Tetering and Vooijs, 2011) on the PM, on sparse cytoplasmic vesicles, and on a paranuclear compartment consistent with the Golgi apparatus. Upon knockdown of ICMT, RAB7, or RAB8, we observed mislocalization of NOTCH1-GFP, albeit with different steady-state localizations. Silencing of ICMT resulted in loss of NOTCH1-GFP from the PM and accumulation on the endomembrane, including the ER, as confirmed by colocalization with calreticulin (mCherry-ER3; Fig. S5). Silencing RAB7 resulted in a more complete loss of NOTCH1-GFP from the PM, with marked accumulation on cytoplasmic vesicles that did not colocalize with LysoTracker or LAMP1 (Fig. S6, A and B). This compartment is consistent with the late endosomal compartment that is regulated by RAB7 (Vitelli et al., 1997; Bucci et al., 2000; Vanlandingham and Ceresa, 2009). However, the absence of LAMP1 staining suggests that endosomal maturation is altered upon knockdown of RAB7. Silencing RAB8 resulted in an incomplete loss of NOTCH1-GFP from the PM and accumulation on cytoplasmic vesicles. We confirmed loss of NOTCH1-GFP from the PM by staining live cells for the ECD of NOTCH1 and analyzing by flow cytometry (Fig. 6 D). Whereas loss of ICMT and RAB7 reduced surface staining of NOTCH1, silencing RAB8 did not. Thus, although differences were evident among the three knockdowns, all three genes were required for normal NOTCH1-GFP trafficking.
Proteolytic processing of nascent NOTCH is dependent on trafficking through the trans-Golgi network, where S1 cleavage occurs (Blaumueller et al., 1997). Consistent with the trafficking defects detected by NOTCH1-GFP imaging (Fig. 6 C), NOTCH1 S1 cleavage was also affected by silencing ICMT, RAB7, or RAB8 (Fig. 6 E, bar graph). Interestingly, upon knockdown of RAB7 or RAB8, we also observed an increase of total NOTCH1-GFP levels (processed + nascent unprocessed). This suggests a defect in delivery of NOTCH1 to lysosomes or in endosome–lysosome maturation, processes regulated by RAB7. This observation was confirmed in D. melanogaster, where we observed a dramatic accumulation of NOTCH protein in cytoplasmic vesicles of wing imaginal discs in which Rab7 was silenced with shRNA (Fig. S6 C). Thus, the perturbation of NOTCH1 trafficking observed in cells deficient in RAB7 or RAB8 affects not only the steady-state localization and proteolytic maturation of the receptor but also its degradation.
To ascertain that the effects observed on NOTCH1-GFP trafficking and signaling are not artifacts caused by overexpression of GFP-tagged NOTCH1, we examined endogenous NOTCH1 signaling after silencing of ICMT, RAB7, and RAB8 as well as genomic disruption of ICMT in MCF10A cells (Kobia et al., 2014; Vermezovic et al., 2015). We activated NOTCH signaling in these cells by culturing them in the presence of EDTA for 30 min. EDTA treatment dissociates the two domains of NOTCH1, promoting cleavage at the S2 and S3 sites and allowing translocation of the NICD to the nucleus (Rand et al., 2000). We detected formation of the NICD using an antibody specific to the S3-cleaved valine 1,754 residue of NOTCH1 in MCF10A cells. We used siRNAs to knock down ICMT, RAB7, or RAB8 in MCF10A, and after 4 d, we incubated the cells in fresh media for 30 min with and without 10 mM EDTA before lysing the cells and subjecting them to SDS-PAGE and Western blotting. Upon silencing of ICMT, RAB7, and RAB8, we observed a decrease in the ratio of NICD to inactive S1 cleaved NOTCH1 after EDTA treatment (Fig. 7 A). Similar results were observed upon genomic disruption of ICMT in MCF10A cells (Fig. 7 B). Consistent with this finding, we also detected a decrease in CSL-luciferase expression in MCF10A cells transfected with a CSL-luciferase reporter 2 d after treatment with either ICMT, RAB7, or RAB8 siRNA as compared with NT siRNA–treated cells. To activate NOTCH signaling, the cells were cocultured with either 3T3 or 3T3-J2 cells 3 d after siRNA treatment, and NOTCH activation was measured by luciferase assay 1 d later (Fig. 7 C). Thus, loss of ICMT, RAB7, or RAB8 expression affects activation of endogenous NOTCH1 in addition to ectopically expressed NOTCH1-GFP both in response to a physiological ligand in a coculture system and using nonphysiological EDTA to promote NOTCH1 cleavage.
Figure 7.
Loss of ICMT, RAB7, or RAB8 inhibits endogenous NOTCH1 signaling measured as decreased formation of S3 cleaved NICD. (A) Immunoblot for the indicated proteins of MCF10A cell lysates 4 d after treatment with the indicated siRNAs and with or without treatment with media containing 10 mM EDTA for 30 min. Graph to the right shows the normalized ratio of the amount of S3-cleaved NOTCH1 (NICD) to the full-length, uncleaved, S1-processed NOTCH1 (means ± SEM; n = 3). (B) As in A, except using MCF10A with or without genomic disruption of ICMT by CRISPR/Cas9. (C) NOTCH signaling in MCF10A cells quantified with a NOTCH-responsive CSL firefly luciferase reporter. Shown are the ratios of CSL firefly/CMV renilla in MCF10A cells cocultured with 3T3 or 3T3-J2 fibroblasts. The MCF10A cells were transfected with the indicated siRNAs 4 d before the luciferase reading. Values were normalized to the maximum firefly/renilla ratio in NT siRNA–treated cells and are given as means ± SEM. n = 3. *, P < 0.05 (two-sided t test).
Rab8 overexpression rescues the NOTCH1 signaling defect observed in ICMT deficiency
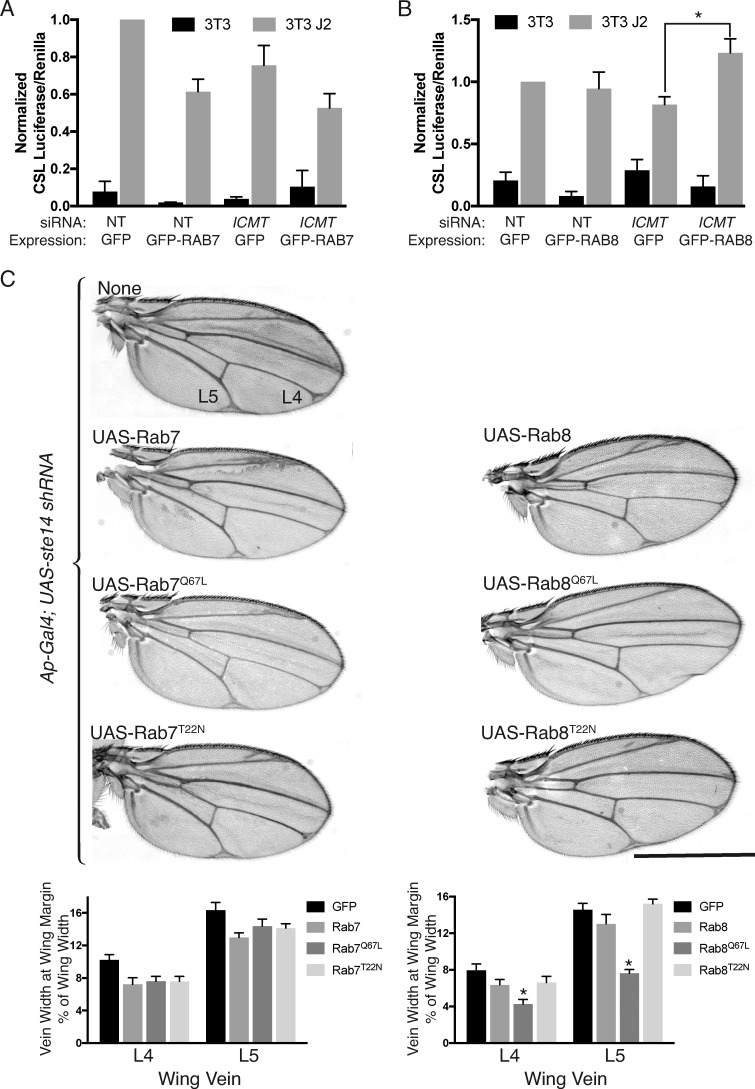
Our results suggest a model in which the effects of ICMT deficiency on NOTCH signaling are caused by diminished function of RAB protein substrates including RAB7 and RAB8. To validate this model, we sought to rescue the effect of ICMT knockdown by overexpressing RAB7 or RAB8. Surprisingly, in U2OS cells, overexpression of GFP-RAB7 inhibited NOTCH1 signaling, obfuscating the ability to detect rescue (Fig. 8 A). This result suggests that, whereas physiological levels of carboxyl methylated RAB7 are required for efficient NOTCH signaling (Fig. 6 A), overexpression of RAB7 somehow dysregulates NOTCH processing and trafficking and also leads to diminished signaling. In contrast, overexpression of GFP-RAB8 had no effect on NOTCH1 signaling but was able to rescue the effect of silencing ICMT (Fig. 8 B).
Figure 8.
NOTCH activity is restored in cells lacking ICMT by overexpression of RAB8 and in fly wings by constitutively active Rab8Q67L. (A) Ratios of CSL firefly/CMV renilla in U2OS cells expressing NOTCH1-GFP and cocultured with 3T3 cells expressing J2 for 24 h. 4 d before coculture, the U2OS cells were transfected with either NT or ICMT siRNA and 48 h later, they were transfected with either GFP or GFP-RAB7 as indicated. Values were normalized to the maximum firefly/renilla ratio in NT siRNA–treated GFP-expressing cells. Data plotted are means ± SEM. n = 3. (B) As in A, except the cells were transfected with either GFP or GFP-RAB8. (C) Virgin female Ap-Gal4; UAS-ste14 shRNA flies were crossed to males carrying the indicated Gal4-responsive UAS-Rab transgenes at 29°C. Bar, 1 mm. Below the representative wings, the widths of the bifurcation of L4 and L5 are plotted as percentages of the wing width. Data shown are means ± SEM. n = 10. *, P < 0.05 (two-sided t test).
We next tested the effects of overexpression of WT, constitutively active (Q67L), and dominant negative (T22N) versions of both UAS-Rab7 and UAS-Rab8 on the wing vein phenotype observed in Ap-Gal4; UAS-shRNA ste14 flies (Fig. 8 C). Overexpression of no form of Rab7 affected the vein width of L4 or L5 in D. melanogaster wings in which ste14 was silenced. In contrast, expression of constitutively active Rab8Q67L reduced the abnormal wing vein width observed with ste14 knockdown. Thus, an increase in the amount of active Rab8 protein, even in the absence of C-terminal methylation, is able to rescue the NOTCH signaling defects seen upon ICMT knockdown. These results establish an epistatic relationship between ICMT and RAB8. However, they do not rule out a functional interaction between ICMT and RAB7 because RAB7 function may depend to a greater degree on carboxyl methylation than does RAB8 function.
Rab7 and Rab8 control different nodes in the Notch pathway and are differentially sensitive to Ste14 deficiency
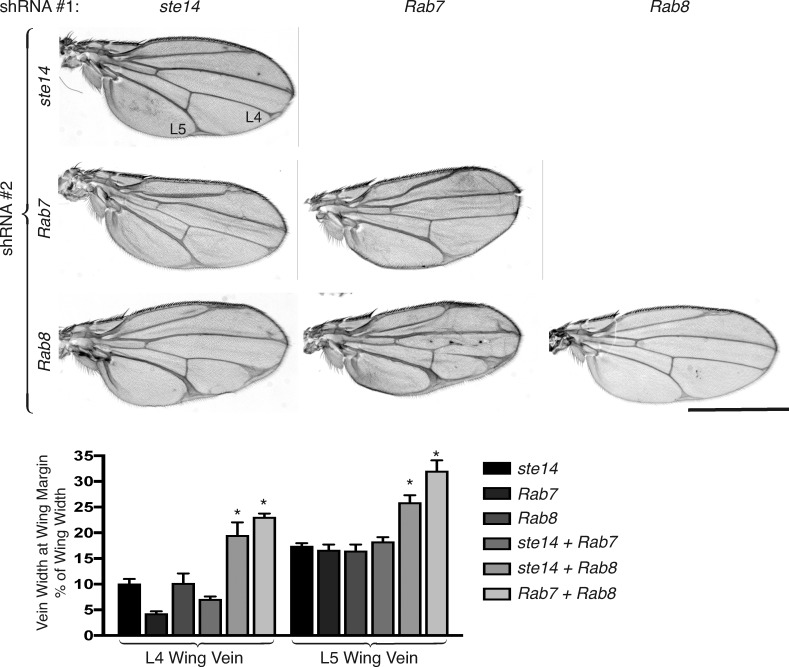
We next examined the effects of simultaneously silencing two of the three genes of interest in the D. melanogaster wing disc. Silencing both Rab7 and Rab8 resulted in a wing vein widening more severe than that of either knockdown alone (Fig. 9). This suggests that Rab7 and Rab8 control different nodes of the Notch trafficking and processing pathway, a conclusion expected because these small GTPases regulate different aspects of vesicular trafficking (Stenmark, 2009). Whereas silencing Rab7 and ste14 in the D. melanogaster wing imaginal disc resulted in a wing vein phenotype no different from silencing either gene alone, silencing Rab8 and ste14 resulted in an exaggerated phenotype (Fig. 9). This result suggests that Rab7 function with regard to Notch signaling is lost in the absence of carboxyl methylation, whereas Rab8 can function without carboxyl methylation, a result consistent with the ability of Rab8 but not Rab7 to rescue the ste14 knockdown phenotype (Fig. 8).
Figure 9.
Rab7 and Rab8 control different nodes in the Notch pathway and are differentially sensitive to Ste14 deficiency. Adult D. melanogaster wings from Ap-Gal4 flies expressing the indicated combinations of UAS-shRNAs at 29°C. Bar, 1 mm. Plotted below the representative wings are the widths of the L4 and L5 vein at the widest part adjacent to the wing margin as a percentage of the total width of the wing for Ap-Gal4 flies expressing the indicated combination of UAS-shRNAs. Data shown are means ± SEM. n = 10. *, P < 0.05 (two-sided t test).
Discussion
Deficiency of NOTCH1 or ICMT affected the Pdx1-Cre;LSL-Kras12D model of KRAS-driven pancreatic neoplasia in the same way; absence of either gene product exacerbated the pancreatic neoplasms and caused cutaneous facial papillomas. Because NOTCH1 acts as a tumor suppressor in this model (Hanlon et al., 2010), one way to interpret these observations is to hypothesize that NOTCH1 signaling requires ICMT. In the NOTCH pathway, the signal-receiving cell requires but one signaling molecule, the NOTCH receptor, which is activated by a series of proteolytic events to generate a fragment, NICD, which interacts with the CSL transcription factor (Tien et al., 2009).
Substrates for ICMT are encoded by ∼200 genes, terminate in a CaaX (Reid et al., 2004) or CXC (Smeland et al., 1994) motif, and are posttranslationally modified to terminate with a prenylcysteine. Neither NOTCH, the NOTCH pathway proteases, nor CSL are substrates for ICMT. However, the NOTCH signaling pathway is dependent on vesicular transport. Specifically, NOTCH and its ligands are all type 1 transmembrane proteins that are transported through the classical secretory pathway. The NOTCH receptor requires O-fucosylation in the Golgi (Okajima and Irvine, 2002), elongation of the polysaccharide by the Fringe β1,3 N-acetylglucosaminyltransferase (Brückner et al., 2000), and proteolytic cleavage (S1) by a furin-like convertase (Logeat et al., 1998), all of which occurs in the Golgi apparatus. Moreover, once engaged by Delta or Serrate/Jagged ligands, NOTCH receptors are cleaved by an ADAM protease (S2), and the truncated receptors are internalized into endosomes, where they undergo proteolytic cleavage by γ-secretase (S3) in the endosomal membrane (Andersson et al., 2011). Among the key regulators of vesicular trafficking are the RAB GTPases, a subset of which are substrates for ICMT. Whereas RAB1 and RAB11 have been implicated in NOTCH signaling, albeit in the biosynthesis of NOTCH ligands on the signal-generating cell (Charng et al., 2014), neither is a substrate for ICMT (Leung et al., 2007). Our screen in flies of RAB genes that encode putative ICMT substrates identified Rab7 and Rab8 as genes that, when silenced, led to NOTCH loss-of-function phenotypes similar to those of ste14 deficiency both in wing vein widening and supernumerary scutellar bristles.
RAB7 is localized on and regulates the biogenesis of late endosomes and lysosomes. It interacts with the homotypic fusion and protein sorting (HOPS) complex of class C vacuolar protein sorting proteins to regulate vesicular fusion events (Zhang et al., 2009). As such, it is not surprising that NOTCH signaling requires RAB7. The function of RAB8 is more obscure (Stenmark, 2009). RAB8 is a homologue of yeast Sec4, which functions in the final stages of the secretory pathway. In mammalian cells, RAB8 has been implicated in Golgi-to-PM transport and exocytosis (Hutagalung and Novick, 2011). RAB8 has also been reported to regulate cell protrusions including filopodia and primary cilia (Peränen, 2011). Besides the trans-Golgi network and secretory vesicles, RAB8 has been reported to associate with recycling endosomes (Ang et al., 2003). Thus, although it is not surprising that RAB8 is required for NOTCH signaling, the precise stage of NOTCH processing that requires this GTPase is not clear.
Our results not only implicate RAB7 and RAB8 in NOTCH signaling but also suggest that carboxyl methylation of RAB7 is required for this function, whereas RAB8 may be able to function without carboxyl methylation. This conclusion is supported by our observations that mammalian cells contain decreased amounts of RAB7-GTP but not RAB8-GTP when ICMT is silenced and by genetic studies of D. melanogaster wing development, particularly the results of our epistasis experiments, where overexpression of RAB8 but not RAB7 was found to rescue ICMT deficiency in both systems.
RAB protein function depends on membrane localization. Like most RAS family GTPases, RAB association with cellular membranes requires posttranslational modification with polyisoprene lipids. For RAB proteins that end in a XXCC or CXC motif, two 20-carbon geranylgeranyl lipids are introduced. For the few RAB proteins that end with a CaaX motif, a single geranylgeranyl or farnesyl lipid modifies the protein. Geranylgeranylated proteins have a higher affinity for membranes than do those proteins modified with the 15-carbon polyisoprene farnesyl, such as RAS (Silvius and l’Heureux, 1994), and doubly geranylgeranylated proteins are predicted to have the highest intrinsic affinity. Using unilamellar liposomes, Silvius and l’Heureux (1994) found that, whereas carboxyl methylation increased membrane affinity for farnesylated proteins ≤10–40-fold, depending on the composition of the bilayer, the boost for geranylgeranylated proteins was <10-fold. The increase in affinity is the result of a decrease in the free energy of lipoprotein partitioning that is caused by the elimination of the negative charge of the α-carboxyl group that is otherwise repelled by anionic phospholipids of the inner leaflet of the PM. The increase in affinity imparted by carboxyl methylation for doubly geranylgeranylated proteins has not been determined but is likely less than that for singly modified proteins. Whereas RAB8 is a CaaX protein that can be modified by no more than one geranylgeranyl lipid, RAB7 ends with a CXC motif that can be doubly geranylgeranylated, suggesting that the localization of RAB7 may be less dependent on carboxyl methylation. Nevertheless, we found that both proteins are substrates for ICMT and both are mislocalized in cells deficient in ICMT. Thus, our data suggest that carboxyl methylation of both proteins is required for efficient partitioning of these proteins into membranes.
Efficiency of partitioning into membranes is but one of two determinants of prenylprotein subcellular distribution. The other is interaction with prenylprotein binding proteins such as RABGDI that can extract RAB proteins from donor membranes, allow them to dwell in the aqueous environment of the cytosol, and later deliver their cargo to acceptor membranes (Ullrich et al., 1993). The effect of carboxyl methylation on the interaction with prenylprotein binding proteins is variable. In the case of PDE6δ, a prenylprotein binding protein that is structurally related to RHOGDI (Ismail et al., 2011), carboxyl methylation promotes the protein–protein interaction (Cook et al., 2000; Mondal et al., 2000; Dharmaiah et al., 2016). In contrast, RAC1 bound more efficiently to RHOGDI (Michaelson et al., 2005), and RAB7 and RAB8 bound more efficiently to RABGDI (Leung et al., 2007) in MEFs null for Icmt. Our results are consistent with the latter studies; silencing ICMT resulted in a greater association of RAB7 and RAB8 with RABGDI. Interestingly, this was not a universal property of carboxyl-methylated RAB proteins because the binding of RAB6 to RABGDI was unaffected by ICMT deficiency and RAB23 did not bind RABGDI even when ICMT was silenced. The increase in RAB7 and RAB8 recovered in the S100 and the redistribution of RAB7 and RAB8 from ER to cytosol that we observed under conditions of ICMT deficiency is consistent with either more protein partitioning alone into the cytosol or more protein complexed with RABGDI in the cytosol or both. Because prenylation is irreversible and the redistributed proteins would be expected to retain either one or two geranylgeranyl modifications, that latter possibility is much more likely. Thus, our results suggest a model in which the function of RAB7 and RAB8 is partially inhibited by redistribution to the cytosol as a consequence of the lack of carboxyl methylation and consequent increase in binding to RABGDI.
The effect of RAB7 deficiency was more dramatic than that of RAB8 deficiency with regard to mislocalization of NOTCH1 in both mammalian cells and D. melanogaster tissues. Indeed, the accumulation of NOTCH1 on cytoplasmic vesicles in the absence of RAB7 was striking in both systems. This is consistent with the idea that RAB7 and RAB8 operate at different stages of NOTCH trafficking, which is not surprising given the well-documented differences in the functions of these GTPases and the fact that NOTCH trafficking and processing requires multiple stages of vesicular transport. The accumulation of NOTCH1 on cytoplasmic vesicles that are negative for markers of lysosomes and the concomitant increase in total NOTCH1 protein suggests that under conditions of RAB7 deficiency, NOTCH1 accumulates on an endosomal compartment but fails to be delivered to lysosomes and thereby avoids degradation, a phenotype consistent with the known function of RAB7. Because RAB7 deficiency is also associated with loss of function of NOTCH, our results suggest that the protein that accumulates cannot be cleaved to generate NICD. Because of the observation that ICMT deficiency precisely phenocopied neither RAB7 nor RAB8 deficiency with regard to the pattern of mislocalization of NOTCH1-GFP, we speculate that other as yet uncharacterized ICMT substrates could also be contributing to the regulation of NOTCH processing and trafficking.
Although our genetic analysis of NOTCH signaling in fly wing vein and scutellar bristle development do not distinguish between a requirement for Rab7, Rab8, or ste14 in signal-generating (Delta- or Ser-expressing) versus signal-receiving (NOTCH-expressing) cells, our results in mammalian cells implicate the signal-receiving cell because these are the only components of the coculture system in which RAB7, RAB8, or ICMT were silenced. Moreover, the accumulation of NOTCH in cytoplasmic vesicles in the portions of wing imaginal discs in which Rab7 was silenced also implicates the signal-receiving cell. Thus, although we cannot rule out effects on both signal-generating and signal-receiving cells, we have established a requirement for RAB7, RAB8, and ICMT in signal-receiving cells expressing NOTCH.
ICMT is considered a target for anti-RAS therapy (Cox et al., 2015), and nanomolar inhibitors have been developed (Judd et al., 2011). Recent evidence has supported the idea that RAS-dependent tumor growth requires ICMT (Lau et al., 2014, 2017). However, because ICMT has >200 substrates, many of them signaling molecules, one should expect off-target effects that are independent of RAS. Our current work suggests that among those effects may be alterations of vesicular trafficking with associated changes in signaling pathways that rely on vesicular transport such as the NOTCH pathway. With regard to inhibition of oncogenic RAS, we have already seen that the effects of ICMT inhibition are highly context dependent. For example, whereas KRAS-driven myeloproliferative disease is ameliorated by ICMT deficiency (Wahlstrom et al., 2008), KRAS-driven pancreatic neoplasia is exacerbated (Court et al., 2013). Thus, although the pursuit of clinically useful ICMT inhibitors is well justified, it is likely that their utility will be disease specific and that a therapeutic window will need to be found.
Materials and methods
D. melanogaster stocks and genetics
All D. melanogaster lines were obtained from The Bloomington Drosophila Stock Center except for Ap-GAL4 (obtained from R. DasGupta, Genome Institute of Singapore, Singapore) and UAS-Dcr2; en-GAL4 UAS-myrRFP NRE-eGFP (obtained from A. Saj; Saj et al., 2010). For the Rab-knockdown adult wing morphology screen, UAS-shRNA males were crossed with Ap-GAL4; UAS-Dcr2 virgin females at 29°C. The notum bristle screen was performed at 25°C. For the wing imaginal disc imaging experiments, UAS-Dcr2; en-GAL4 UAS-myrRFP NRE-eGFP virgins were crossed to UAS-shRNA males at 29°C.
Cell lines
The NOTCH1-Flag-GFP–expressing U2OS cells, DLL1-expressing OP9 cells, and J2-expressing 3T3 cells were gifts from S. Blacklow (Harvard Medical School, Boston, MA). U2OS cells and HEK293 cells were obtained from ATCC. MCF10A cells were a gift from R. Schnieder (New York University Langone Medical Center, New York, NY). SKMEL-28 cells were a gift from E. Hernando (New York University Langone Medical Center, New York, NY). All cells except MCF10A cells were maintained in DMEM containing 10% FBS and 1% penicillin/streptomyces at 37°C and 5% CO2. MCF10A cells were maintained in 50/50 DMEM/Ham’s F12 medium supplemented with 5% horse serum, 20 ng/ml EGF, 100 ng/ml cholera toxin, 0.01 mg/ml bovine insulin, and 500 ng/ml hydrocortisone.
Plasmids
pGFP-RAB8a was a gift from M. Nachury (24898; Addgene; Nachury et al., 2007). EGFP-RAB7a was a gift from Q. Zhong (28047; Addgene; Sun et al., 2010). mCherry-ER3 was a gift from M. Davidson (Florida State University, Tallahassee, FL; 55041; Addgene). LentiCRISPR v2 was a gift from F. Zhang (Massachusetts Institute of Technology, Cambridge, MA; 52961; Addgene). pEGFP-C3 vector was obtained from Takara Bio Inc. CSL-luciferase (8x CBF1; Zhou et al., 2000) and cytomegalovirus (CMV)–renilla luciferase were gifts from R. DasGupta. pGEX 4T3 RILP was a gift from A. Edinger (University of California, Irvine, Irvine, CA). pGEX-JFC-D1 was a gift from J. Peränen (University of Helsinki, Helsinki, Finland).
CRISPR/Cas9
Genomic disruption of ICMT was performed on U2OS cells and SKMEL-28 cells by infecting the cells in six-well plates with lentivirus generated by transfecting HEK293 with LentiCRISPR v2 expressing sgRNA targeting ICMT (5′-CACCGCACCGGGCTGGCGCTCTACG-3′ and 5′-AAACCGTAGAGCGCCAGCCCGGTGC-3′) and Cas9 using Lipofectamine 3000 (Invitrogen). Control cells were generated by infecting cells with lentivirus expressing Cas9 alone. 2 d after infection, cells were selected with 2 µg/ml puromycin and used immediately for experiments.
Cell fractionation
Adherent U2OS and SKMEL-28 cells were washed with cold PBS, scraped off tissue culture plates, and resuspended in ice-cold hypotonic buffer (100 mM KC1, 3 mM NaCl, 3.5 mM MgCl2, 10 mM Hepes, pH 7.3, and protease inhibitors). Cell suspensions were spun in a bomb (Parr Instruments) pressurized with nitrogen to 450 psi for 20 min on ice and then released dropwise to room atmospheric pressure and collected in tubes. Cavitates were centrifuged at 14,000 rpm for 10 min at 4°C to remove nuclei and unbroken cells. The postnuclear supernatant was further centrifuged with a TLA100.1 rotor (Beckman Coulter) at 100,000 rpm (350,000 g) for 30 min. The supernatant (S100 cytosolic fraction) was collected without disturbing the pellet (P100 membrane fraction). The pellet was washed twice with cold hypotonic buffer and resuspended in 25 mM Tris, pH 7.4, plus protease inhibitors to one tenth the volume of the S100 fraction. For SDS-PAGE, the S100 and P100 fractions were mixed with 4× Laemmli sample buffer (Bio-Rad Laboratories) and 2.5% 2-mercaptoethanol and then were loaded in a 10:1 ratio (S100/P100) to maintain cell-equivalent quantities. For ICMT activity assays, the membrane protein concentration was quantified using a BCA protein assay (Thermo Fisher Scientific).
ICMT activity assays
An in vitro assay for ICMT activity was performed using 10 µg of protein in isolated membrane fractions from U2OS and SKMEL-28 cells as described previously (Choy and Philips, 2000). In brief, 10 µg of membrane protein was incubated for 30 min at 37°C with 5 µl of 1 mM N-acetyl-S-farnesyl-l-cysteine (AFC; assay substrate), 3 µl adenosyl-l-S-[3H]methionine ([3H]AdoMet; 60 Ci/mmol, 0.55 mCi/ml; methyl donor), and 12.5 µl 4× TE buffer (200 mM Tris-HCl, pH 8.0, and 4 mM Na-EDTA) in a volume of 50 µl. The reaction was terminated with an equal volume of 20% TCA, 400 µl n-heptane was added, and the tubes were centrifuged to separate the unreacted [3H]AdoMet from the [3H]AFC–methyl ester product in the top organic layer. The top n-heptane layer was removed, placed in a fresh tube, and evaporated overnight. 1 N NaOH was added to promote alkaline hydrolysis of the [3H]AFC-methyl ester to produce volatile [3H]methanol, which was measured using a scintillation counter. An in vivo assay for ICMT activity in cells was performed as described previously (Choy and Philips, 2000). In brief, U2OS cells were transfected with either an ICMT or NT siRNA SMARTpool (GE Healthcare) using DharmaFECT1 transfection reagent in six-well tissue culture plates. 3 d later, cells were transfected with plasmid constructs containing GFP-RAB7a, GFP-RAB8a, GFP-NRAS, or GFP-NRASC186S using Lipofectamine 3000 transfection reagent (Thermo Fisher Scientific). The next day, the cells were washed once in PBS and then incubated in DMEM without methionine/cysteine with 10% dialyzed FBS for 3 h. The cells were treated with 200 µCi of [3H methyl]–methionine for 3 h before being further washed in PBS, and then they were lysed with RIPA buffer (20 mM Tris-HC1, pH 7.5, 150 mM NaCl, 1% NP-40, 0.1% SDS, 0.1% Na-deoxycholate, 0.5 mM EDTA, 1 mM DTT, and protease inhibitors). The GFP-tagged proteins were then immunoprecipitated using anti-GFP agarose beads (MBL International). The beads were washed once in RIPA buffer, divided in half, and then subjected to SDS-PAGE. One gel was used to perform Western blotting using an anti-GFP antibody. The other polyacrylamide gel was dried (DryEase Mini Gel Drying System; Novex), and the regions of the gel containing the proteins of interest were cut out with a razor and treated with 1 N NaOH to cause alkaline hydrolysis of the [3H]methyl ester producing volatile [3H]methanol. The amount of [3H]methanol released was measured using a scintillation counter.
Detection of GTP-bound RAB in cell lysates
GST-JFC1 and GST-RILP were purified from BL21 Escherichia coli using Glutathione agarose beads (Thermo Fisher Scientific) as respectively described previously (Taylor et al., 2001; Romero Rosales et al., 2009). In brief, a 250-ml culture of BL21 was grown in Luria–Bertani media and 100 µg/ml ampicillin to OD600 of 0.6–0.8. Recombinant GST-fusion protein expression was induced with 0.5 mM IPTG at 30°C for 4 h. Bacteria were then harvested by centrifugation and lysed in 1% Triton X-100 with sonication. Lysates were centrifuged at 10,000 g for 10 min. Recombinant GST-fusion proteins were captured using Glutathione agarose beads (Thermo Fisher Scientific). After production of the GST-fusion beads, U2OS cells were treated with either ICMT or NT siRNA SMARTpool for 3 d and then were washed and incubated overnight with DMEM containing either 0.5% or 10% serum. For GTP-RAB8 detection, cells were lysed in lysis buffer (20 mM Tris-HCl, pH 7.4, 100 mM KCl, 5 mM MgCl2, 0.5% Triton X-100, 1 mM DTT, 4 mM Pefabloc SC serine protease inhibitor [Roche], and protease inhibitor cocktail [Roche]). The cell lysates were incubated with GST or GST-JFC1 beads at 4°C for 2 h followed by washing twice with lysis buffer. Bound GTP-RAB8 was detected using SDS-PAGE and immunoblotting using an anti-RAB8 antibody (Cell Signaling Technology). The lysed cellular material (1%) was saved before pulldown as a loading control for the total amount of RAB8 present. For GTP-RAB7 detection, the experiment was repeated, except the cells were lysed in lysis buffer (20 mM Hepes, 100 mM NaCl, 5 mM MgCl2, 1% Triton X-100, and protease inhibitors) and incubated with GST or GST-RILP, and then the GTP-RAB7 was detected with a RAB7 antibody (Abcam).
Coimmunoprecipitation experiments
HEK293 cells were transfected with either an ICMT or NT siRNA SMARTpool using DharmaFECT1 transfection reagent in six-well tissue culture plates. 3 d later, cells were transfected with plasmid constructs containing GFP-RAB7a, GFP-RAB8a, GFP-NRAS GFP-RAB6, CFP-RAB5, or GFP-RAB23 using Lipofectamine 3000 transfection reagent. 24 h later, cells were washed in cold PBS and lysed in lysis buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 2 mM EDTA, 1% NP-40, 10% glycerol, protease inhibitor cocktail, and 4 mM Pefabloc SC serine protease inhibitor). The lysates were centrifuged at 14,000 rpm for 10 min at 4°C to pellet nuclei. GFP-tagged proteins were immunoprecipitated from the postnuclear supernatant by incubating with anti-GFP mAb-agarose beads (RQ2; MBL International) for 2 h at 4°C. The beads were washed twice with lysis buffer and subjected to SDS-PAGE and Western blotting to detect coimmunoprecipitated proteins. The lysed cellular material (1%) was saved before immunoprecipitation as a loading control.
Luciferase assays
Luciferase assays for NOTCH1 activity were performed as described previously (Aste-Amézaga et al., 2010). In brief, U2OS cells stably expressing NOTCH1-GFP or MCF10A cells expressing endogenous NOTCH1 were transfected with CSL-luciferase and CMV-renilla plasmids using Lipofectamine 3000 reagent. The next days, the cells were cocultured 1:1 with “stromal” 3T3, 3T3-J2, OP9, or OP9-DLL1 cells. Luciferase readings were measured 24 h later. All assays were performed in 96-well plates, and luciferase expression was measured using a Dual-Glo Luciferase kit (Promega) and detected in a plate reader. All luciferase readings were normalized to renilla expressed off a CMV promoter. Knockdown of ICMT, RAB7, and RAB8 was performed by transfection of siRNA SMARTpools using the DharmaFECT1 transfection reagent 4 d before the luciferase reading. An NT siRNA pool was used as a negative control. Knockdown was validated by immunoblot using the following antibodies: ICMT antibody (Proteintech), RAB7 antibody (Abcam), and RAB8 antibody (Cell Signaling Technology). For rescue experiments, GFP-RAB7a, GFP-RAB8a, or GFP plasmid constructs were transfected into the cells using Superfect transfection reagent (QIAGEN) 2 d before the luciferase reading.
Flow cytometry
Staining was performed on 106 unfixed, unpermeabilized U2OS cells expressing GFP-NOTCH1 in PBS containing 1% BSA for 30 min on ice. An antibody specific to the ECD of NOTCH1 was used (APC–anti–human NOTCH1; MHN1-519; 352108; BioLegend). 4 d before staining, cells were transfected with siRNA for ICMT, RAB7, RAB8 or NT using DharmaFECT1 transfection reagent. 2 d prior, cells were treated with 1 µg/ml doxycycline to induce NOTCH1-GFP expression. After staining, the cells were washed 2× in 1% BSA and fixed with 1% paraformaldehyde. Flow cytometry was performed using an LSR II (BD) and analyzed using FlowJo software (FlowJo).
Immunoblot
Cells were lysed in Tris-glycine SDS sample buffer (Thermo Fisher Scientific) with 2.5% 2-mercaptoethanol, and the lysates were subjected to SDS-PAGE and Western blotting on nitrocellulose membranes (Bio-Rad Laboratories). After blocking (Odyssey blocking buffer; LI-COR Biosciences), the membranes were incubated with the following primary antibodies: mouse anti-RABGDI α/β (E-5; sc-374649; Santa Cruz Biotechnology, Inc.), mouse anti–β-tubulin E7 antibody (deposited to the Developmental Studies Hybridoma Bank by M. Klymkowsky; DSHB Hybridoma Product E7), rabbit anti-ICMT (51001-2-AP; Proteintech), rabbit anti-RAB8 (D22D8; 6975; Cell Signaling Technology), rabbit anti-RAB7 (EPR7589; ab137029; Abcam), rabbit anti-EGFR (D38B1; 4267; Cell Signaling Technology), rabbit anti-CIMPR (EPR6599 ab124767; Abcam), rabbit anti-GFP (A6455; Thermo Fisher Scientific), rabbit anti–cleaved NOTCH1 (D3B8; 4147; Cell Signaling Technology), and rabbit anti-NOTCH1 (D1E11; 3608; Cell Signaling Technology). Secondary antibodies were IRDye 800–conjugated goat anti–rabbit or IRDye 680–conjugated goat anti–mouse (926-32211 and 926-68070; LI-COR Biosciences). Blots were visualized with the Odyssey infrared imaging system (LI-COR Biosciences) and quantified using Odyssey software.
Immunofluorescence staining and live-cell imaging
U2OS cells grown on glass coverslips were fixed in 2% paraformaldehyde for 10 min. The cells were washed with PBS and then permeabilized with 0.1% Triton X-100 for 5 min. After blocking for 1 h in blocking buffer (5% goat serum [Dako] and 1% IgG-free BSA [Jackson ImmunoResearch Laboratories, Inc.]), the cells were incubated overnight in primary antibody at a 1:500 dilution in blocking buffer (mouse anti-LAMP1 antibody H4A3 was deposited to the Developmental Studies Hybridoma Bank by J.T. August and J.E.K. Hildreth; DSHB Hybridoma Product H4A3). After washing three times in PBS, the cells were incubated for 1 h in the dark with Alexa Fluor 647–conjugated goat anti–mouse secondary antibody (A21236; Thermo Fisher Scientific) and then washed and counter-stained with Hoechst 33342 (Thermo Fisher Scientific). The coverslips were then mounted using ProLong Diamond (Molecular Probes) and imaged with confocal microscopy. LysoTracker Red staining was performed on live cells by incubating cells with prewarmed media (DMEM and 10% FBS) containing 50 nM LysoTracker Red for 30 min before imaging alive by confocal microscopy. An LSM 800 inverted confocal microscope running Zen Blue software (ZEISS) was used for all imaging. All cells were imaged with a 63× 1.4 NA objective. Live-cell imaging was performed at 37°C in 5% CO2 in DMEM/10% FBS media. D. melanogaster wing imaginal discs were dissected from third instar larvae and fixed in 4% paraformaldehyde. Immunofluorescent staining was performed as for the U2OS cells before mounting on glass slides with Vectashield (Vector Laboratories) and imaging by confocal microscopy. The following antibodies were used: Rab7 (deposited to the Developmental Studies Hybridoma Bank by S. Munro; DSHB Hybridoma Product Rab7), C458.2H (deposited to the Developmental Studies Hybridoma Bank by S. Artavanis-Tsakonas; DSHB Hybridoma Product C458.2H), and mouse anti-Rab8 (4/Rab8; 610844; BD). An LSM 800 inverted confocal microscope running Zen Blue software was used for all imaging. All wing discs were imaged with a 40× 1.3 NA objective.
Statistical methods
All p-values were calculated using a Student’s two-sided t test. Data distribution was assumed to be normal, but this was not formally tested. A p-value ≤0.05 was considered statistically significant.
Online supplemental material
We show the full results of the D. melanogaster screen for the Rab substrates of ste14 that phenocopy Ste14 knockdown in the adult wing (Fig. S1 and S2). We also show confirmation of the results obtained using CRISPR/Cas9 disruption of ICMT in Figs. 3 B and 4 (A and C). To control for off-target effects, we repeated these experiments using siRNA knockdown of ICMT and obtained similar results (Figs. S3 and S4 A, C, and D). In Figs. S5 and S6, we provide evidence of the mislocalization of NOTCH1-GFP from the PM to the ER with knockdown of ICMT (Fig. S5) and to cytoplasmic vesicles with knockdown of RAB7 (Fig. S6).
Supplementary Material
Acknowledgments
We thank Stephen Blacklow for cell lines and reagents for the NOTCH1 luciferase assay. We thank Ramanuj DasGupta for fly stocks and discussions. We thank Robert J. Schneider and Eva Hernando (New York University Perlmutter Cancer Center) for MCF10A and SKMEL-28 cells.
This work was supported by the National Institutes of Health (GM55279, CA116034, and CA163489 to M.R. Philips and T32 AR064184 to I.M. Ahearn).
The authors declare no competing financial interests.
Author contributions: H. Court and M.R. Philips designed the study, interpreted the results, prepared the figures, and wrote the manuscript. I.M. Ahearn performed the CRISPR/Cas9 genome editing and assisted with the execution and interpretation of all experiments performed with mammalian cells. M. Amoyel and E.A. Bach provided expertise in D. melanogaster and helped design and interpret the genetic experiments.
Footnotes
Abbreviations used:
- CMV
- cytomegalovirus
- CSL
- CBF1/suppressor of hairless/LAG-1
- DLL
- Delta-like
- ECD
- extracellular domain
- GBD
- GTPase binding domain
- GDI
- GDP dissociation inhibitor
- ICMT
- isoprenylcysteine carboxyl methyltransferase
- NICD
- NOTCH intercellular domain
- NRE
- NOTCH response element
- NT
- nontargeting
- PM
- plasma membrane
- sgRNA
- single-guide RNA
References
- Andersson E.R., Sandberg R., and Lendahl U.. 2011. Notch signaling: simplicity in design, versatility in function. Development. 138:3593–3612. 10.1242/dev.063610 [DOI] [PubMed] [Google Scholar]
- Ang A.L., Fölsch H., Koivisto U.M., Pypaert M., and Mellman I.. 2003. The Rab8 GTPase selectively regulates AP-1B-dependent basolateral transport in polarized Madin-Darby canine kidney cells. J. Cell Biol. 163:339–350. 10.1083/jcb.200307046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aste-Amézaga M., Zhang N., Lineberger J.E., Arnold B.A., Toner T.J., Gu M., Huang L., Vitelli S., Vo K.T., Haytko P., et al. . 2010. Characterization of Notch1 antibodies that inhibit signaling of both normal and mutated Notch1 receptors. PLoS One. 5:e9094 10.1371/journal.pone.0009094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auer J.S., Nagel A.C., Schulz A., Wahl V., and Preiss A.. 2015. Local overexpression of Su(H)-MAPK variants affects Notch target gene expression and adult phenotypes in Drosophila. Data Brief. 5:852–863. 10.1016/j.dib.2015.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair S.S. 2007. Wing vein patterning in Drosophila and the analysis of intercellular signaling. Annu. Rev. Cell Dev. Biol. 23:293–319. 10.1146/annurev.cellbio.23.090506.123606 [DOI] [PubMed] [Google Scholar]
- Blaumueller C.M., Qi H., Zagouras P., and Artavanis-Tsakonas S.. 1997. Intracellular cleavage of Notch leads to a heterodimeric receptor on the plasma membrane. Cell. 90:281–291. 10.1016/S0092-8674(00)80336-0 [DOI] [PubMed] [Google Scholar]
- Brennan K., Tateson R., Lieber T., Couso J.P., Zecchini V., and Arias A.M.. 1999. The abruptex mutations of notch disrupt the establishment of proneural clusters in Drosophila. Dev. Biol. 216:230–242. 10.1006/dbio.1999.9501 [DOI] [PubMed] [Google Scholar]
- Brückner K., Perez L., Clausen H., and Cohen S.. 2000. Glycosyltransferase activity of Fringe modulates Notch-Delta interactions. Nature. 406:411–415. 10.1038/35019075 [DOI] [PubMed] [Google Scholar]
- Bucci C., Thomsen P., Nicoziani P., McCarthy J., and van Deurs B.. 2000. Rab7: a key to lysosome biogenesis. Mol. Biol. Cell. 11:467–480. 10.1091/mbc.11.2.467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charng W.L., Yamamoto S., Jaiswal M., Bayat V., Xiong B., Zhang K., Sandoval H., David G., Gibbs S., Lu H.C., et al. . 2014. Drosophila Tempura, a novel protein prenyltransferase α subunit, regulates notch signaling via Rab1 and Rab11. PLoS Biol. 12:e1001777 10.1371/journal.pbio.1001777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chastagner P., Israël A., and Brou C.. 2008. AIP4/Itch regulates Notch receptor degradation in the absence of ligand. PLoS One. 3:e2735 10.1371/journal.pone.0002735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy E., and Philips M.. 2000. Expression and activity of human prenylcysteine-directed carboxyl methyltransferase. Methods Enzymol. 325:101–114. 10.1016/S0076-6879(00)25435-9 [DOI] [PubMed] [Google Scholar]
- Colicelli J. 2004. Human RAS superfamily proteins and related GTPases. Sci. STKE. 2004:RE13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook T.A., Ghomashchi F., Gelb M.H., Florio S.K., and Beavo J.A.. 2000. Binding of the delta subunit to rod phosphodiesterase catalytic subunits requires methylated, prenylated C-termini of the catalytic subunits. Biochemistry. 39:13516–13523. 10.1021/bi001070l [DOI] [PubMed] [Google Scholar]
- Cornell M., Evans D.A., Mann R., Fostier M., Flasza M., Monthatong M., Artavanis-Tsakonas S., and Baron M.. 1999. The Drosophila melanogaster Suppressor of deltex gene, a regulator of the Notch receptor signaling pathway, is an E3 class ubiquitin ligase. Genetics. 152:567–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costes S.V., Daelemans D., Cho E.H., Dobbin Z., Pavlakis G., and Lockett S.. 2004. Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys. J. 86:3993–4003. 10.1529/biophysj.103.038422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Court H., Hahne K., Philips M.R., and Hrycyna C.A.. 2011. Biochemical and Biological Functions of Isoprenylcysteine Carboxyl Methyltransferase. Enzymes. 30:71–90. 10.1016/B978-0-12-415922-8.00004-5 [DOI] [Google Scholar]
- Court H., Amoyel M., Hackman M., Lee K.E., Xu R., Miller G., Bar-Sagi D., Bach E.A., Bergö M.O., and Philips M.R.. 2013. Isoprenylcysteine carboxylmethyltransferase deficiency exacerbates KRAS-driven pancreatic neoplasia via Notch suppression. J. Clin. Invest. 123:4681–4694. 10.1172/JCI65764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox A.D., Der C.J., and Philips M.R.. 2015. Targeting RAS Membrane Association: Back to the Future for Anti-RAS Drug Discovery? Clin. Cancer Res. 21:1819–1827. 10.1158/1078-0432.CCR-14-3214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmaiah S., Bindu L., Tran T.H., Gillette W.K., Frank P.H., Ghirlando R., Nissley D.V., Esposito D., McCormick F., Stephen A.G., and Simanshu D.K.. 2016. Structural basis of recognition of farnesylated and methylated KRAS4b by PDEδ. Proc. Natl. Acad. Sci. USA. 113:E6766–E6775. 10.1073/pnas.1615316113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirac-Svejstrup A.B., Soldati T., Shapiro A.D., and Pfeffer S.R.. 1994. Rab-GDI presents functional Rab9 to the intracellular transport machinery and contributes selectivity to Rab9 membrane recruitment. J. Biol. Chem. 269:15427–15430. [PubMed] [Google Scholar]
- Do M.T., Chai T.F., Casey P.J., and Wang M.. 2017. Isoprenylcysteine carboxylmethyltransferase function is essential for RAB4A-mediated integrin β3 recycling, cell migration and cancer metastasis. Oncogene. 36 10.1038/onc.2017.183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn K.W., Kamocka M.M., and McDonald J.H.. 2011. A practical guide to evaluating colocalization in biological microscopy. Am. J. Physiol. Cell Physiol. 300:C723–C742. 10.1152/ajpcell.00462.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery G., Hutterer A., Berdnik D., Mayer B., Wirtz-Peitz F., Gaitan M.G., and Knoblich J.A.. 2005. Asymmetric Rab 11 endosomes regulate delta recycling and specify cell fate in the Drosophila nervous system. Cell. 122:763–773. 10.1016/j.cell.2005.08.017 [DOI] [PubMed] [Google Scholar]
- Farnsworth C.C., Kawata M., Yoshida Y., Takai Y., Gelb M.H., and Glomset J.A.. 1991. C terminus of the small GTP-binding protein smg p25A contains two geranylgeranylated cysteine residues and a methyl ester. Proc. Natl. Acad. Sci. USA. 88:6196–6200. 10.1073/pnas.88.14.6196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fostier M., Evans D.A., Artavanis-Tsakonas S., and Baron M.. 1998. Genetic characterization of the Drosophila melanogaster Suppressor of deltex gene: A regulator of notch signaling. Genetics. 150:1477–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haines N., and Irvine K.D.. 2003. Glycosylation regulates Notch signalling. Nat. Rev. Mol. Cell Biol. 4:786–797. 10.1038/nrm1228 [DOI] [PubMed] [Google Scholar]
- Hanlon L., Avila J.L., Demarest R.M., Troutman S., Allen M., Ratti F., Rustgi A.K., Stanger B.Z., Radtke F., Adsay V., et al. . 2010. Notch1 functions as a tumor suppressor in a model of K-ras-induced pancreatic ductal adenocarcinoma. Cancer Res. 70:4280–4286. 10.1158/0008-5472.CAN-09-4645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattula K., Furuhjelm J., Tikkanen J., Tanhuanpää K., Laakkonen P., and Peränen J.. 2006. Characterization of the Rab8-specific membrane traffic route linked to protrusion formation. J. Cell Sci. 119:4866–4877. 10.1242/jcs.03275 [DOI] [PubMed] [Google Scholar]
- Hingorani S.R., Petricoin E.F., Maitra A., Rajapakse V., King C., Jacobetz M.A., Ross S., Conrads T.P., Veenstra T.D., Hitt B.A., et al. . 2003. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 4:437–450. 10.1016/S1535-6108(03)00309-X [DOI] [PubMed] [Google Scholar]
- Hutagalung A.H., and Novick P.J.. 2011. Role of Rab GTPases in membrane traffic and cell physiology. Physiol. Rev. 91:119–149. 10.1152/physrev.00059.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail S.A., Chen Y.-X., Rusinova A., Chandra A., Bierbaum M., Gremer L., Triola G., Waldmann H., Bastiaens P.I.H., and Wittinghofer A.. 2011. Arl2-GTP and Arl3-GTP regulate a GDI-like transport system for farnesylated cargo. Nat. Chem. Biol. 7:942–949. 10.1038/nchembio.686 [DOI] [PubMed] [Google Scholar]
- Judd W.R., Slattum P.M., Hoang K.C., Bhoite L., Valppu L., Alberts G., Brown B., Roth B., Ostanin K., Huang L., et al. . 2011. Discovery and SAR of methylated tetrahydropyranyl derivatives as inhibitors of isoprenylcysteine carboxyl methyltransferase (ICMT). J. Med. Chem. 54:5031–5047. 10.1021/jm200249a [DOI] [PubMed] [Google Scholar]
- Kim J., Irvine K.D., and Carroll S.B.. 1995. Cell recognition, signal induction, and symmetrical gene activation at the dorsal-ventral boundary of the developing Drosophila wing. Cell. 82:795–802. 10.1016/0092-8674(95)90476-X [DOI] [PubMed] [Google Scholar]
- Klein T., Seugnet L., Haenlin M., and Martinez Arias A.. 2000. Two different activities of Suppressor of Hairless during wing development in Drosophila. Development. 127:3553–3566. [DOI] [PubMed] [Google Scholar]
- Kobia F., Duchi S., Deflorian G., and Vaccari T.. 2014. Pharmacologic inhibition of vacuolar H+ ATPase reduces physiologic and oncogenic Notch signaling. Mol. Oncol. 8:207–220. 10.1016/j.molonc.2013.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R., and Ilagan M.X.. 2009. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 137:216–233. 10.1016/j.cell.2009.03.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau H.Y., Ramanujulu P.M., Guo D., Yang T., Wirawan M., Casey P.J., Go M.L., and Wang M.. 2014. An improved isoprenylcysteine carboxylmethyltransferase inhibitor induces cancer cell death and attenuates tumor growth in vivo. Cancer Biol. Ther. 15:1280–1291. 10.4161/cbt.29692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau H.Y., Tang J., Casey P.J., and Wang M.. 2017. Isoprenylcysteine carboxylmethyltransferase is critical for malignant transformation and tumor maintenance by all RAS isoforms. Oncogene. 36:3934–3942. 10.1038/onc.2016.508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung K.F., Baron R., Ali B.R., Magee A.I., and Seabra M.C.. 2007. Rab GTPases containing a CAAX motif are processed post-geranylgeranylation by proteolysis and methylation. J. Biol. Chem. 282:1487–1497. 10.1074/jbc.M605557200 [DOI] [PubMed] [Google Scholar]
- Logeat F., Bessia C., Brou C., LeBail O., Jarriault S., Seidah N.G., and Israël A.. 1998. The Notch1 receptor is cleaved constitutively by a furin-like convertase. Proc. Natl. Acad. Sci. USA. 95:8108–8112. 10.1073/pnas.95.14.8108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manders E.M., Stap J., Brakenhoff G.J., van Driel R., and Aten J.A.. 1992. Dynamics of three-dimensional replication patterns during the S-phase, analysed by double labelling of DNA and confocal microscopy. J. Cell Sci. 103:857–862. [DOI] [PubMed] [Google Scholar]
- Matsui Y., Kikuchi A., Araki S., Hata Y., Kondo J., Teranishi Y., and Takai Y.. 1990. Molecular cloning and characterization of a novel type of regulatory protein (GDI) for smg p25A, a ras p21-like GTP-binding protein. Mol. Cell. Biol. 10:4116–4122. 10.1128/MCB.10.8.4116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazaleyrat S.L., Fostier M., Wilkin M.B., Aslam H., Evans D.A., Cornell M., and Baron M.. 2003. Down-regulation of Notch target gene expression by Suppressor of deltex. Dev. Biol. 255:363–372. 10.1016/S0012-1606(02)00086-6 [DOI] [PubMed] [Google Scholar]
- Mazur P.K., Grüner B.M., Nakhai H., Sipos B., Zimber-Strobl U., Strobl L.J., Radtke F., Schmid R.M., and Siveke J.T.. 2010. Identification of epidermal Pdx1 expression discloses different roles of Notch1 and Notch2 in murine Kras(G12D)-induced skin carcinogenesis in vivo. PLoS One. 5:e13578 10.1371/journal.pone.0013578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelson D., Ali W., Chiu V.K., Bergo M., Silletti J., Wright L., Young S.G., and Philips M.. 2005. Postprenylation CAAX processing is required for proper localization of Ras but not Rho GTPases. Mol. Biol. Cell. 16:1606–1616. 10.1091/mbc.E04-11-0960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moloney D.J., Panin V.M., Johnston S.H., Chen J., Shao L., Wilson R., Wang Y., Stanley P., Irvine K.D., Haltiwanger R.S., and Vogt T.F.. 2000. Fringe is a glycosyltransferase that modifies Notch. Nature. 406:369–375. 10.1038/35019000 [DOI] [PubMed] [Google Scholar]
- Mondal M.S., Wang Z., Seeds A.M., and Rando R.R.. 2000. The specific binding of small molecule isoprenoids to rhoGDP dissociation inhibitor (rhoGDI). Biochemistry. 39:406–412. 10.1021/bi991856n [DOI] [PubMed] [Google Scholar]
- Nachury M.V., Loktev A.V., Zhang Q., Westlake C.J., Peränen J., Merdes A., Slusarski D.C., Scheller R.H., Bazan J.F., Sheffield V.C., and Jackson P.K.. 2007. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 129:1201–1213. 10.1016/j.cell.2007.03.053 [DOI] [PubMed] [Google Scholar]
- Okajima T., and Irvine K.D.. 2002. Regulation of notch signaling by o-linked fucose. Cell. 111:893–904. 10.1016/S0092-8674(02)01114-5 [DOI] [PubMed] [Google Scholar]
- Panin V.M., Papayannopoulos V., Wilson R., and Irvine K.D.. 1997. Fringe modulates Notch-ligand interactions. Nature. 387:908–912. 10.1038/43191 [DOI] [PubMed] [Google Scholar]
- Parody T.R., and Muskavitch M.A.. 1993. The pleiotropic function of Delta during postembryonic development of Drosophila melanogaster. Genetics. 135:527–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peränen J. 2011. Rab8 GTPase as a regulator of cell shape. Cytoskeleton (Hoboken). 68:527–539. 10.1002/cm.20529 [DOI] [PubMed] [Google Scholar]
- Radtke F., and Raj K.. 2003. The role of Notch in tumorigenesis: oncogene or tumour suppressor? Nat. Rev. Cancer. 3:756–767. 10.1038/nrc1186 [DOI] [PubMed] [Google Scholar]
- Rand M.D., Grimm L.M., Artavanis-Tsakonas S., Patriub V., Blacklow S.C., Sklar J., and Aster J.C.. 2000. Calcium depletion dissociates and activates heterodimeric notch receptors. Mol. Cell. Biol. 20:1825–1835. 10.1128/MCB.20.5.1825-1835.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid T.S., Terry K.L., Casey P.J., and Beese L.S.. 2004. Crystallographic analysis of CaaX prenyltransferases complexed with substrates defines rules of protein substrate selectivity. J. Mol. Biol. 343:417–433. 10.1016/j.jmb.2004.08.056 [DOI] [PubMed] [Google Scholar]
- Romero Rosales K., Peralta E.R., Guenther G.G., Wong S.Y., and Edinger A.L.. 2009. Rab7 activation by growth factor withdrawal contributes to the induction of apoptosis. Mol. Biol. Cell. 20:2831–2840. 10.1091/mbc.E08-09-0911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saj A., Arziman Z., Stempfle D., van Belle W., Sauder U., Horn T., Dürrenberger M., Paro R., Boutros M., and Merdes G.. 2010. A combined ex vivo and in vivo RNAi screen for notch regulators in Drosophila reveals an extensive notch interaction network. Dev. Cell. 18:862–876. 10.1016/j.devcel.2010.03.013 [DOI] [PubMed] [Google Scholar]
- Schwartz S.L., Cao C., Pylypenko O., Rak A., and Wandinger-Ness A.. 2007. Rab GTPases at a glance. J. Cell Sci. 120:3905–3910. 10.1242/jcs.015909 [DOI] [PubMed] [Google Scholar]
- Silvius J.R., and l’Heureux F.. 1994. Fluorimetric evaluation of the affinities of isoprenylated peptides for lipid bilayers. Biochemistry. 33:3014–3022. 10.1021/bi00176a034 [DOI] [PubMed] [Google Scholar]
- Smeland T.E., Seabra M.C., Goldstein J.L., and Brown M.S.. 1994. Geranylgeranylated Rab proteins terminating in Cys-Ala-Cys, but not Cys-Cys, are carboxyl-methylated by bovine brain membranes in vitro. Proc. Natl. Acad. Sci. USA. 91:10712–10716. 10.1073/pnas.91.22.10712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldati T., Riederer M.A., and Pfeffer S.R.. 1993. Rab GDI: a solubilizing and recycling factor for rab9 protein. Mol. Biol. Cell. 4:425–434. 10.1091/mbc.4.4.425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldati T., Shapiro A.D., Svejstrup A.B., and Pfeffer S.R.. 1994. Membrane targeting of the small GTPase Rab9 is accompanied by nucleotide exchange. Nature. 369:76–78. 10.1038/369076a0 [DOI] [PubMed] [Google Scholar]
- Speicher S.A., Thomas U., Hinz U., and Knust E.. 1994. The Serrate locus of Drosophila and its role in morphogenesis of the wing imaginal discs: control of cell proliferation. Development. 120:535–544. [DOI] [PubMed] [Google Scholar]
- Stenmark H. 2009. Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 10:513–525. 10.1038/nrm2728 [DOI] [PubMed] [Google Scholar]
- Sun Q., Westphal W., Wong K.N., Tan I., and Zhong Q.. 2010. Rubicon controls endosome maturation as a Rab7 effector. Proc. Natl. Acad. Sci. USA. 107:19338–19343. 10.1073/pnas.1010554107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson A.W., Casey P.J., Young S.G., and Bergo M.O.. 2006. Genetic and pharmacologic analyses of the role of Icmt in Ras membrane association and function. Methods Enzymol. 407:144–159. 10.1016/S0076-6879(05)07013-8 [DOI] [PubMed] [Google Scholar]
- Taylor S.J., Resnick R.J., and Shalloway D.. 2001. Nonradioactive determination of Ras-GTP levels using activated ras interaction assay. Methods Enzymol. 333:333–342. 10.1016/S0076-6879(01)33067-7 [DOI] [PubMed] [Google Scholar]
- Tien A.C., Rajan A., and Bellen H.J.. 2009. A Notch updated. J. Cell Biol. 184:621–629. 10.1083/jcb.200811141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich O., Stenmark H., Alexandrov K., Huber L.A., Kaibuchi K., Sasaki T., Takai Y., and Zerial M.. 1993. Rab GDP dissociation inhibitor as a general regulator for the membrane association of rab proteins. J. Biol. Chem. 268:18143–18150. [PubMed] [Google Scholar]
- Vanlandingham P.A., and Ceresa B.P.. 2009. Rab7 regulates late endocytic trafficking downstream of multivesicular body biogenesis and cargo sequestration. J. Biol. Chem. 284:12110–12124. 10.1074/jbc.M809277200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Tetering G., and Vooijs M.. 2011. Proteolytic cleavage of Notch: “HIT and RUN”. Curr. Mol. Med. 11:255–269. 10.2174/156652411795677972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermezovic J., Adamowicz M., Santarpia L., Rustighi A., Forcato M., Lucano C., Massimiliano L., Costanzo V., Bicciato S., Del Sal G., and d’Adda di Fagagna F.. 2015. Notch is a direct negative regulator of the DNA-damage response. Nat. Struct. Mol. Biol. 22:417–424. 10.1038/nsmb.3013 [DOI] [PubMed] [Google Scholar]
- Vitelli R., Santillo M., Lattero D., Chiariello M., Bifulco M., Bruni C.B., and Bucci C.. 1997. Role of the small GTPase Rab7 in the late endocytic pathway. J. Biol. Chem. 272:4391–4397. 10.1074/jbc.272.7.4391 [DOI] [PubMed] [Google Scholar]
- Wahlstrom A.M., Cutts B.A., Liu M., Lindskog A., Karlsson C., Sjogren A.K., Andersson K.M., Young S.G., and Bergo M.O.. 2008. Inactivating Icmt ameliorates K-RAS-induced myeloproliferative disease. Blood. 112:1357–1365. 10.1182/blood-2007-06-094060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright L.P., and Philips M.R.. 2006. Thematic review series: lipid posttranslational modifications. CAAX modification and membrane targeting of Ras. J. Lipid Res. 47:883–891. 10.1194/jlr.R600004-JLR200 [DOI] [PubMed] [Google Scholar]
- Wright L.P., Court H., Mor A., Ahearn I.M., Casey P.J., and Philips M.R.. 2009. Topology of mammalian isoprenylcysteine carboxyl methyltransferase determined in live cells with a fluorescent probe. Mol. Cell. Biol. 29:1826–1833. 10.1128/MCB.01719-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S., Charng W.L., and Bellen H.J.. 2010. Endocytosis and intracellular trafficking of Notch and its ligands. Curr. Top. Dev. Biol. 92:165–200. 10.1016/S0070-2153(10)92005-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Schulze K.L., Hiesinger P.R., Suyama K., Wang S., Fish M., Acar M., Hoskins R.A., Bellen H.J., and Scott M.P.. 2007. Thirty-one flavors of Drosophila rab proteins. Genetics. 176:1307–1322. 10.1534/genetics.106.066761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Liu M., Su Y., Du J., and Zhu A.J.. 2012. A targeted in vivo RNAi screen reveals deubiquitinases as new regulators of Notch signaling. G3 (Bethesda). 2:1563–1575. 10.1534/g3.112.003780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Chen L., Wang S., and Wang T.. 2009. Rab7: roles in membrane trafficking and disease. Biosci. Rep. 29:193–209. 10.1042/BSR20090032 [DOI] [PubMed] [Google Scholar]
- Zhou S., Fujimuro M., Hsieh J.J., Chen L., Miyamoto A., Weinmaster G., and Hayward S.D.. 2000. SKIP, a CBF1-associated protein, interacts with the ankyrin repeat domain of NotchIC To facilitate NotchIC function. Mol. Cell. Biol. 20:2400–2410. 10.1128/MCB.20.7.2400-2410.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.