Wood et al. have developed inducible chemical and optogenetic systems for triggering clathrin-mediated endocytosis using minimal clathrin-binding domains. Bypassing regulatory steps in vesicle creation will enable a better understanding of the factors required for vesicle initiation and processing, which the authors use to define how clathrin functionally interacts with its adaptor AP2.
Abstract
Clathrin-mediated endocytosis (CME) is the major route of receptor internalization at the plasma membrane. Analysis of constitutive CME is difficult because the initiation of endocytic events is unpredictable. When and where a clathrin-coated pit will form and what cargo it will contain are difficult to foresee. Here we describe a series of genetically encoded reporters that allow the initiation of CME on demand. A clathrin-binding protein fragment (“hook”) is inducibly attached to an “anchor” protein at the plasma membrane, which triggers the formation of new clathrin-coated vesicles. Our design incorporates temporal and spatial control by the use of chemical and optogenetic methods for inducing hook–anchor attachment. Moreover, the cargo is defined. Because several steps in vesicle creation are bypassed, we term it “hot-wiring.” We use hot-wired endocytosis to describe the functional interactions between clathrin and AP2. Two distinct sites on the β2 subunit, one on the hinge and the other on the appendage, are necessary and sufficient for functional clathrin engagement.
Introduction
Clathrin-mediated endocytosis (CME) is the major uptake pathway in eukaryotic cells, which influences numerous processes, from nutrition and signaling to organelle biogenesis and cell excitability (Kirchhausen et al., 2014; Robinson, 2015). In clathrin-mediated synaptic vesicle retrieval, the endocytosis of defined cargo is coupled temporally and spatially to an exocytic event (Granseth et al., 2006; Rizzoli, 2014). In contrast, in constitutive CME, the location of clathrin-coated pit formation in space and time is unpredictable (Ehrlich et al., 2004). Moreover, the cargo contained in each vesicle and the proteins contributing to the inner layer of the clathrin coat are variable (Taylor et al., 2011; Borner et al., 2012). This means that we do not know for certain when or where a vesicle will form or what cargo it will contain. Our goal therefore was to design a synthetic system that can be used to trigger endocytosis “on demand.” The aim was to provide temporal and spatial control over the initiation of endocytosis using defined cargo.
A straightforward method to trigger endocytosis is the activation of a receptor at the cell surface, for example, G-protein–coupled receptors activated by their cognate ligands (Puthenveedu et al., 2007). This would provide temporal control and could be adapted for spatial control; however, (a) activation of intracellular signaling would complicate analysis, (b) the precise molecular details for activation-dependent internalization of many receptors are not completely understood, (c) this would not report on constitutive CME, and (d) activated G-protein–coupled receptors may not generate clathrin-coated pits de novo (Lampe et al., 2014). For these reasons, we sought a synthetic system to initiate CME.
The major clathrin adaptor at the plasma membrane is the AP2 complex. AP2 performs the essential function of recognizing cargo and membrane and also contacts clathrin via its β2 subunit, specifically the hinge and appendage domains (Murphy and Keen, 1992; Keyel et al., 2008). The AP2 complex undergoes several large-scale conformational changes to bind membrane, recognize cargo, and become ready for clathrin engagement (Kelly et al., 2008, 2014; Jackson et al., 2010). In designing a synthetic system to trigger endocytosis on demand, these regulatory steps would need to be bypassed so that the process can be “hot-wired.”
Elegant in vitro studies have shown that clathrin-coated pits can be formed by anchoring a clathrin-binding protein (a clathrin “hook”) at a membrane (Dannhauser and Ungewickell, 2012). We reasoned that a similar approach, if it could be made to be inducible, would trigger endocytosis inside living human cells. This paper describes our design and optimization of synthetic reporters to trigger endocytosis on demand in human cells. We show that this system can be applied to answer specific cell biological questions, such as defining the molecular requirements for clathrin-AP2 interaction.
Results
Development of chemically inducible endocytosis
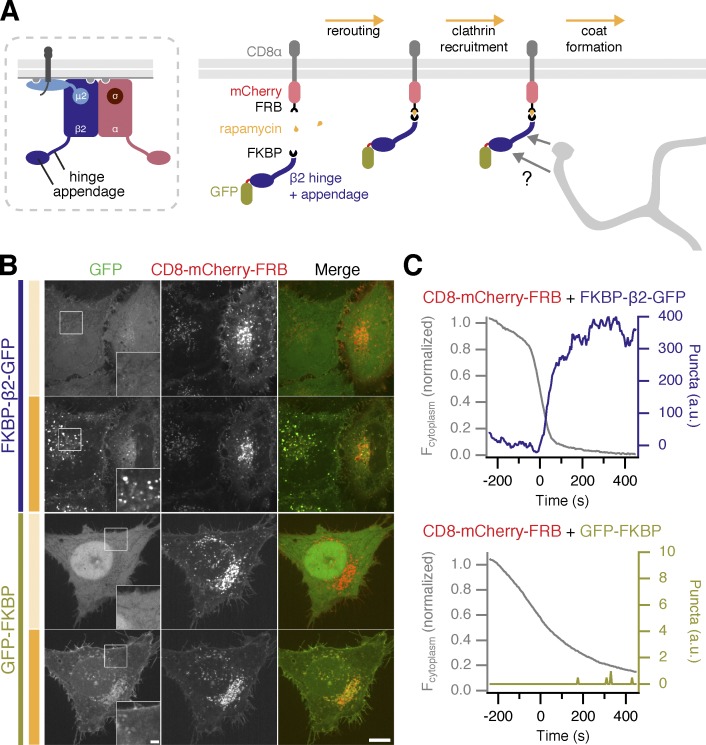
We designed a series of constructs that would allow us to induce endocytosis chemically (Fig. 1). The FK506-binding protein (FKBP)–rapamycin–FK506-binding and rapamycin-binding protein (FRB) system was exploited to induce the dimerization of a clathrin hook with a plasma membrane “anchor” and thereby control the initiation of endocytosis (Fig. 1 A). Live-cell imaging demonstrated that the clathrin hook was rapidly recruited to the plasma membrane in response to rapamycin (200 nM). Immediately afterward, bright green puncta began to form. These bright puncta occurred only when a clathrin hook (FKBP–β2–GFP) was rerouted to the plasma membrane, but not when a construct lacking the hook (GFP–FKBP) was used (Fig. 1, B and C; and Videos 1 and 2). Similar responses were detected using a plasma membrane anchor based on the monomeric transmembrane protein CD4 and even with palmitoylated peripheral membrane proteins acting as the anchor (GAP43–FRB–mRFP or SH4–FRB–mRFP; Fig. S1 A). Interestingly, no bright green puncta were formed when a clathrin hook was sent to the mitochondria, indicating that puncta formation depends on the plasma membrane and on anchors that are correctly addressed there (Fig. S1 B). We suspected that the bright green puncta were clathrin-coated vesicles (CCVs) formed by a hot-wired form of CME.
Figure 1.
Chemically inducible endocytosis. (A) Illustration of chemically inducible endocytosis. Normally, clathrin recognizes the appendage and hinge of the β2 subunit of the AP2 complex after it has engaged cargo and membrane. In chemically induced endocytosis, cells coexpress a plasma membrane anchor (CD8–mCherry–FRB) and a clathrin hook (FKBP–β2–GFP). Rapamycin (200 nM) is added, which causes heterodimerization of FKBP and FRB domains. The clathrin hook is rerouted to the plasma membrane. Clathrin recognizes the clathrin hook, and the plasma membrane anchor is internalized. (B) Chemically inducible endocytosis in live cells. Cells expressing CD8–mCherry–FRB with either FKBP–β2–GFP or GFP–FKBP. Stills from live-cell confocal imaging experiments are shown: the frame before rerouting occurs (top) and 133 frames (665 s) later (bottom). See Videos 1 and 2. Insets show a 2× zoom of the boxed region; intense orange bar indicates rapamycin application. Bars: (main) 10 µm; (insets) 2 µm. (C) Example plots to show chemically inducible endocytosis. Colored traces indicate the total area of bright GFP puncta that form in cells after rerouting. Gray traces show the rerouting of the clathrin hook from the cytoplasm to the plasma membrane (see Materials and methods).
Before application of rapamycin, plasma membrane anchors were also found in intracellular structures to some extent, in addition to the population at the plasma membrane. The clathrin hook is also initially recruited to these structures, forming larger, dimmer spots (Fig. 1 B and Video 1). Using antibody feeding, we found that the dimmer structures do not contain extracellularly applied antibody and that only the bright green puncta do (Fig. S2 A). Using image segmentation, it was possible to distinguish the bright green puncta from the large dimmer accumulations at stationary spots present at the start of the experiment (see Materials and methods and Fig. S2 B). If chemically induced internalization is indeed a hot-wired form of CME, then these observations indicate that the formation of bright green puncta is a readout of this process.
Chemically induced internalization is a hot-wired form of CME
We used four different approaches to test the hypothesis that the bright green puncta generated from the plasma membrane upon heterodimerization of clathrin hook and membrane anchor were CCVs.
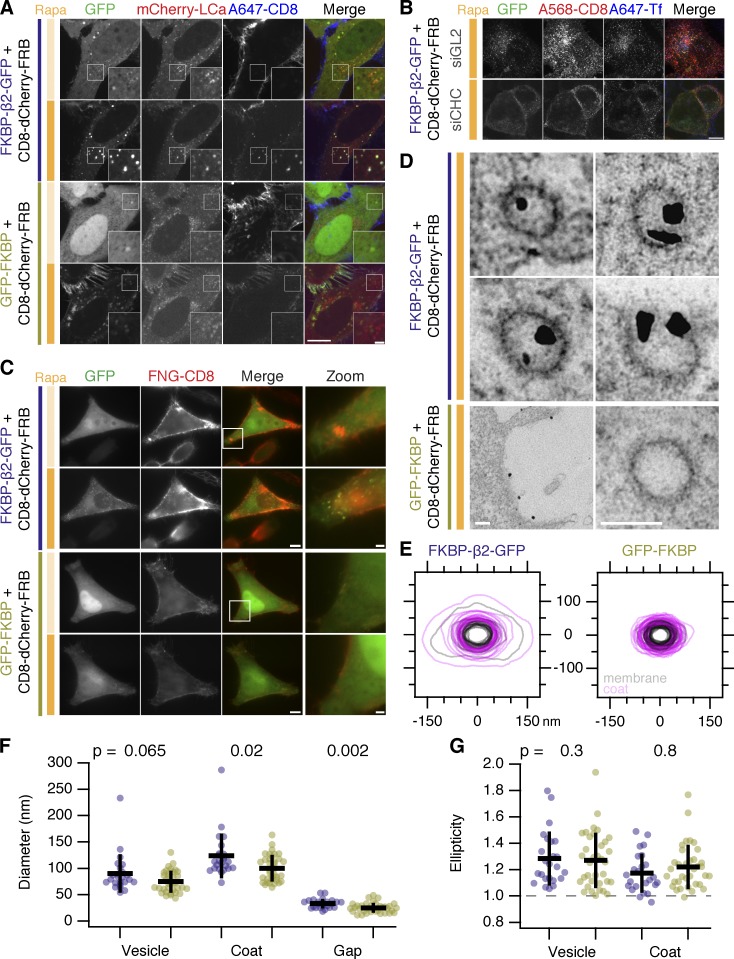
First, we used antibody feeding and simultaneous clathrin imaging in live cells in which chemically induced internalization was triggered. The plasma membrane anchor was labeled extracellularly with anti-CD8/Alexa 647, and the heterodimerization of FKBP and FRB was then triggered with rapamycin (Fig. 2 A). The bright green puncta that formed in cells expressing the clathrin hook contained anti-CD8, which confirmed that these puncta are formed by internalization from the plasma membrane. No bright green puncta or antibody uptake was seen in cells coexpressing GFP–FKBP. Moreover, the bright green puncta also colocalized with clathrin (mCherry–clathrin light chain a [LCa]), suggesting that these vesicles, originating from the plasma membrane, are clathrin coated (Fig. 2 A and Video 3).
Figure 2.
Chemically induced internalization is via CCVs. (A) Bright green spots formed by chemically induced endocytosis colocalize with clathrin and contain anti-CD8–Alexa 647. Cells coexpressing mCherry–LCa, CD8–dCherry–FRB, and either FKBP–β2–GFP or GFP–FKBP were incubated with anti-CD8–Alexa 647 to label the membrane anchor extracellularly, and rapamycin (200 nM) was applied. Insets show a 2× zoom of the boxed region; intense orange bar indicates rapamycin application. Bars: (main) 10 µm; (insets) 2 µm. (B) Live-cell confocal immunolabeling experiments of cells expressing CD8–dCherry–FRB (dark mCherry variant) with FKBP–β2–GFP. Cells were also transfected with siRNAs against GL2 (control) or CHC. Inhibition of uptake of transferrin–Alexa 647 (A647-Tf; blue) was used as a functional test of knockdown efficacy. Bar, 10 µm. (C–G) CLEM to study the uptake of immunolabeled CD8–dCherry–FRB in cells coexpressing either FKBP–β2–GFP or GFP–FKBP. Results from a single experiment are shown, although CLEM experiments were performed three times. (C) Stills from live-cell wide-field imaging showing cells sequentially labeled with anti-CD8 and Alexa 546 FluoroNanoGold-conjugated secondary antibody (FNG; red), before and after the addition of rapamycin (200 nM). The same cell was processed for electron microscopy and imaged. Bars: (main) 10 µm; (zoom) 2 µm. (D) Electron micrographs of the cells shown in A. Clear uptake of NanoGold into CCVs was seen in cells coexpressing FKBP–β2–GFP but not GFP–FKBP. Bars, 100 nm. (E) Segmentation of CCV profiles in rapamycin-treated FKBP–β2–GFP versus GFP–FKBP samples. Membrane (gray) and coat (purple) for multiple vesicles are shown overlaid. Note that CCVs in FKBP–β2–GFP contained NanoGold, whereas GFP–FKBP CCVs did not. (F) Scatterplot of CCV diameters in FKBP–β2–GFP versus GFP–FKBP samples. Diameter refers to the mean of the major and minor axes. (G) Scatterplot of ellipticity of CCV profiles in FKBP–β2–GFP versus GFP–FKBP samples. Ellipticity is the ratio of semimajor and semiminor axis. The p-values from Student’s t test with Welch’s correction are shown. Dots indicate CCVs and bars indicate mean ± SD.
Second, in cells depleted of clathrin heavy chain (CHC) using RNAi, rapamycin-induced heterodimerization of anchor and hook did not result in the formation of bright green puncta (Fig. 2 B). This result confirms that chemically induced internalization is clathrin mediated.
Third, we used total internal reflection fluorescence microscopy (TIRFM) to image chemically induced internalization at the plasma membrane. Internalization was triggered in RPE1 cells coexpressing CD8–dCherry–FRB, FKBP–β2–mRuby2, and either dynamin2–GFP or GFP–LCa. We observed that the FKBP–β2–mRuby2 puncta that form after rapamycin application terminate with an accumulation of dynamin consistent with scission (Fig. S3). As expected, FKBP–β2–mRuby2 puncta colocalize with clathrin throughout their lifetime from appearance to disappearance (Fig. S3). The time course of these events was similar to that reported elsewhere (Aguet et al., 2013).
Fourth, we directly tested whether or not the bright puncta were CCVs using correlative light-electron microscopy (CLEM). To do this, we again took advantage of the ability of our system to be tracked using antibodies against the extracellular domain of CD8. HeLa cells coexpressing CD8–dCherry–FRB and either FKBP–β2–GFP or GFP–FKBP as a control were labeled by anti-CD8/Alexa 546–FluoroNanoGold, and endocytosis was initiated by addition of rapamycin (200 nM). By fluorescence microscopy, the formation of bright puncta was confirmed in cells coexpressing FKBP–β2–GFP but not GFP–FKBP (Fig. 2 C). The same cell observed by light microscopy was then fixed, processed, and imaged by electron microscopy. In cells that formed bright puncta, gold particles were found in CCVs (Fig. 2 D). In controls, gold particles remained at the cell surface and were not found in CCVs (Fig. 2 D). Ultrastructurally, the CCVs triggered by hot-wiring were similar in size and ellipticity to normal CCVs in control cells (Fig. 2, E–G). Minor differences were observed, the membrane vesicle in the CCV was similar in size but the coat, and therefore the gap between vesicle and coat was enlarged in hot-wired CCVs compared with control (Fig. 2 F).
Together, these experiments show that chemically induced internalization is a hot-wired form of CME because (a) internalization occurs at the plasma membrane, (b) it is clathrin dependent and dynamin dependent, and (c) the vesicles are clathrin coated. Therefore, the bright green puncta that form upon chemically induced endocytosis and/or the uptake of anti-CD8 are readouts of hot-wired CME.
Hot-wired endocytic cargo takes a similar intracellular trafficking route to canonical cargo
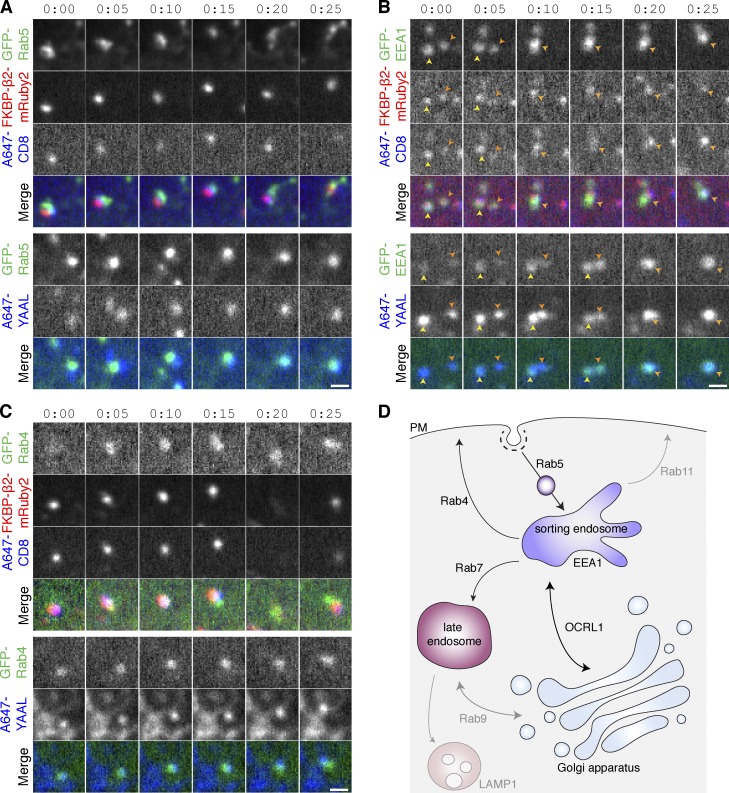
After endocytosis, cargo can be processed in different ways by the cell, and we next wanted to define the fate of the hot-wired endocytic cargo. We used the antibody feeding approach to track the cargo internalized by hot-wiring and compare it with a canonical cargo, CD8–YAAL, which is internalized by constitutive CME (Kozik et al., 2010; Fielding et al., 2012). Colocalization of anti-CD8/Alexa 647 was seen with early or late endosome markers and fast recycling vesicles for both hot-wired cargo and for CD8–YAAL (Rab5, EEA-1, Rab4 [Fig. 3, A–C]; VAMP2, OCRL1, Rab7 [Fig. S4, B–D]). We saw no colocalization of hot-wired cargo with late endosomal or lysosomal markers (Rab9 and LAMP-1) or for slow recycling vesicles marked by Rab11, even at later time points (Fig. S4 E). Limited colocalization was detected between these markers and CD8–YAAL (Fig. S4 F), which suggests that the pathways are similar but not identical. The presence of the R-SNARE VAMP2 on hot-wired vesicles (Fig. S4 D) and the direct observation of vesicle fusion (Fig. 3 B and Fig. S4 B) indicates that vesicles generated by hot-wired CME are competent for intracellular processing. These findings are summarized in Fig. 3 D and Fig. S4 A. Hot-wired cargo is internalized via CCVs, trafficked to the sorting endosome, where some returns to the cell surface via Rab4-positive vesicles. Some cargo continues in maturing vesicles, but there was no evidence of a lysosomal destination of hot-wired cargo.
Figure 3.
CCVs generated by hot-wired endocytosis are trafficked and recycled similarly to normal endocytic cargo. (A–C) Stills from typical live-cell confocal imaging experiments. Vesicles in HeLa cells expressing CD8–dCherry–FRB, FKBP–β2–mRuby2 (top), or CD8–YAAL (bottom) together with the indicated GFP-tagged construct (A, Rab5; B, EEA1; C, Rab4). CD8 was labeled using extracellularly applied anti-CD8/Alexa 647. Endocytosis was triggered by rapamycin addition (200 nM) in the case of hot-wiring, and cells were imaged at 0.2 Hz. Bars, 1 µm. (D) Summary of imaging experiments to delineate the paths taken by hot-wired cargo or by CD8–YAAL. See also Fig. S4.
Hot-wired endocytosis operates independently of endogenous CME
Our working model for hot-wired endocytosis is that vesicles are formed de novo at the plasma membrane. However, an alternative view is that the clathrin hook is bound to preexisting clathrin-coated structures (CCSs) and, upon rapamycin addition, drags the plasma membrane anchor into the preexisting pit and internalization proceeds via endogenous CME.
Our results are compatible with both models. Indeed, before rapamycin addition, a small fraction of FKBP–β2–GFP is localized to preexisting CCSs (Fig. 2 A), which argues for the alternative model. To look at this in detail, we followed GFP spots over time and found that FKBP–β2–GFP leaves CCSs for the general plasma membrane upon rapamycin addition (Fig. S5 A and Video 4). Colocalization between FKBP–β2–GFP spots and mCherry–LCa therefore sharply decreases but then increases, consistent with the formation of CCVs de novo. Furthermore, if we used mCherry–FRB rather than CD8–mCherry–FRB, then upon rapamycin addition, FKBP–β2–GFP actually leaves the CCSs and becomes diffusely localized (Fig. S5 B). This suggests that clathrin hooks cannot drag even freely diffusing proteins into existing CCSs, let alone an anchor in the plasma membrane, making the alternative model implausible.
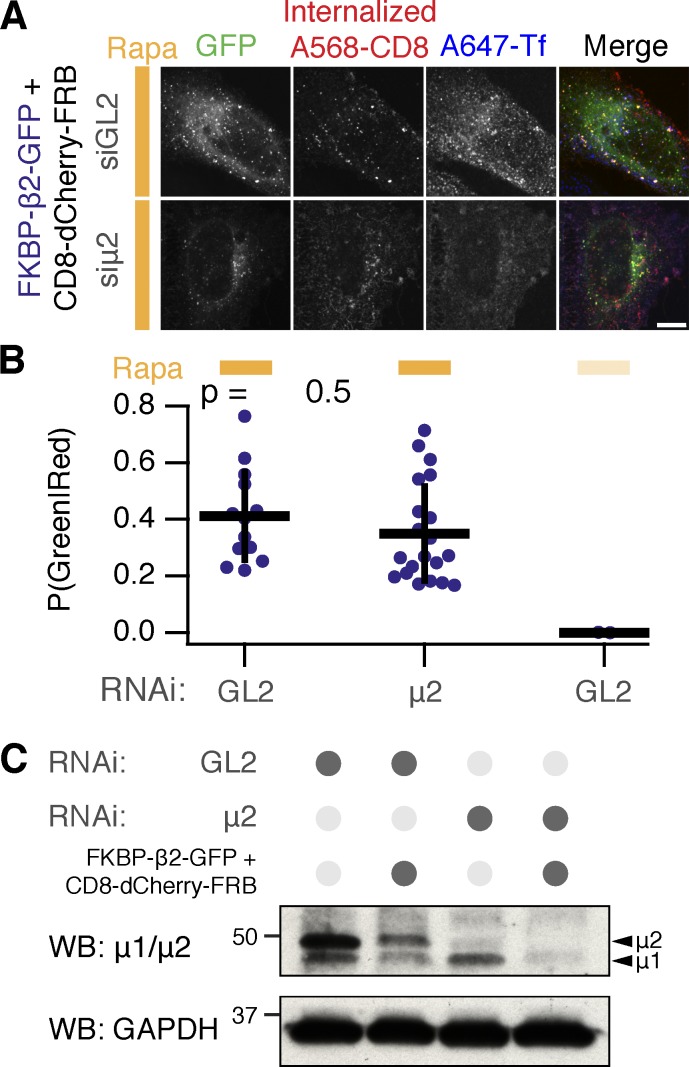
We next tested if hot-wired endocytosis can occur in the absence of endogenous CME. To do this, we triggered endocytosis in cells that were depleted of the μ2 subunit of AP2. We found that uptake of anti–CD8 antibody was similar in depleted cells compared with control RNAi cells (Fig. 4, A and B). The depletion of μ2 by RNAi was good and was sufficient to block endogenous transferrin uptake (Fig. 4, A and C). These results indicate that hot-wired endocytosis can occur via vesicles formed de novo and further argue against the idea that hot-wired endocytosis is simply occurring via endogenous CME.
Figure 4.
Hot-wired endocytosis can function independently of AP2. (A) Chemically induced endocytosis can operate independently of endogenous AP2. Live-cell confocal immunolabeling experiments of cells expressing CD8–dCherry–FRB (dark mCherry variant) with FKBP–β2–GFP. Cells were also transfected with siRNAs against GL2 (control) or μ2 subunit of AP2 complex. Antibody feeding (internalized anti-CD8/Alexa 568) is shown to assess internalization. Inhibition of uptake of transferrin–Alexa 647 was used as a functional test of knockdown efficacy. Bar, 10 µm. (B) Quantification of chemically induced endocytosis. Fraction of above-threshold puncta that were green (FKBP–β2–GFP) and red (A568-CD8) is shown for each cell (spots); bars indicate mean ± SD. Number of experiments = 3. ANOVA, P = 1.48 × 10−8. Tukey test for siGL2 versus siμ2, P = 0.5. (C) Western blot (WB) to assess depletion of μ2 by RNAi. Cells were prepared in parallel with the experiment shown in C. Blotting for μ2 or for GAPDH (as a loading control) is shown.
Hot-wired endocytosis does not inhibit constitutive CME
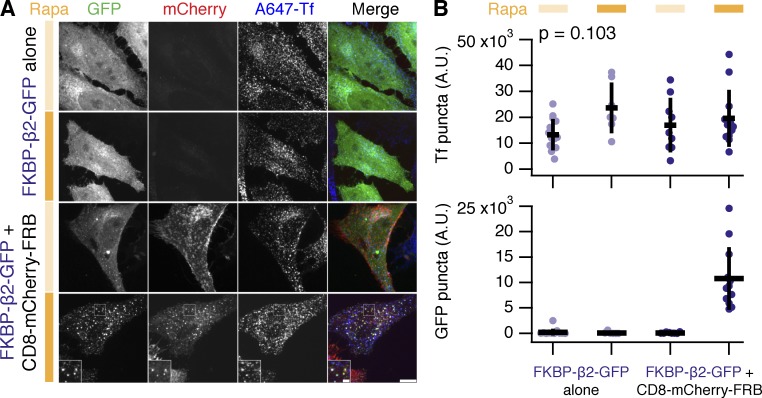
A concern with using our system is that endogenous CME may be affected by triggered endocytosis. To address this, we monitored transferrin uptake in cells expressing FKBP–β2–GFP alone or with CD8–mCherry–FRB. Transferrin uptake appeared normal in the presence or in the absence of rapamycin (Fig. 5). Moreover, there was no obvious detrimental effect on CME by merely expressing our system (Fig. 5). In cells in which hot-wired endocytosis was triggered, we saw three classes of vesicle. The majority were puncta containing only transferrin, others were yellow puncta that were positive for both CD8–mCherry–FRB and FKBP–β2–GFP, of these puncta, some also contained transferrin. This latter class of vesicle suggests interplay of synthetic endocytosis and constitutive CME, which may occur downstream of internalization. This result is to be expected given the similarity in intracellular processing described in the previous section (Fig. 3). These data indicate that triggering endocytosis by hot-wiring is not sufficient to block regular CME.
Figure 5.
Hot-wired endocytosis does not inhibit normal CME. (A) Confocal images of HeLa cells expressing FKBP–β2–GFP alone or together with CD8–mCherry–FRB were tested for ability to internalize transferrin–Alexa 647 (A647-Tf) in the presence or absence of rapamycin (200 nM; filled orange bar). Bars: (main) 10 µm; (inset) 2 µm. (B) Quantification of transferrin uptake and number of GFP-positive puncta in cells. Scatterplot shows values for each cell, bars indicate mean ± SD. Number of experiments = 2. Tf, ANOVA, P = 0.1; GFP, ANOVA, P = 5.89 × 10−6.
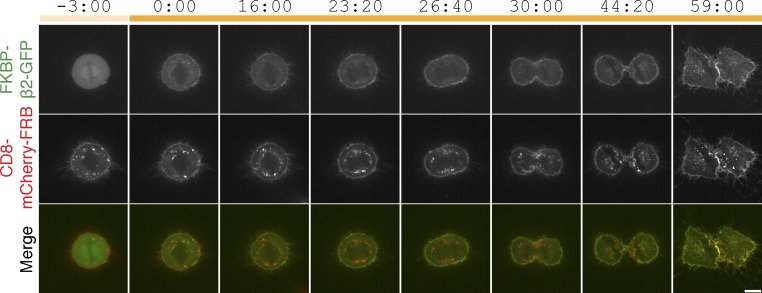
Hot-wired endocytosis cannot override endocytic shutdown mechanisms
One characteristic of CME is that it is inhibited during the early stages of mitosis (Fielding et al., 2012; Kaur et al., 2014). We therefore tested if hot-wired endocytosis could override this inhibition. HeLa cells expressing our system were treated with rapamycin at metaphase. FKBP–β2–GFP relocated to the plasma membrane as usual, but bright green puncta did not form (Fig. 6 and Video 5). About 30 min later, after the cell had undergone cytokinesis, we began to see puncta formation, showing that induction of endocytosis was ultimately possible, just not during mitosis. We imaged 17 cells in four independent experiments. Inhibition of vesicle formation at metaphase and anaphase was observed in all 17 cells. Of these, seven cells could be tracked through to cytokinesis, when all seven began puncta formation. These observations suggest that hot-wiring cannot overcome the mitotic inhibition of CME.
Figure 6.
Chemically induced endocytosis is inhibited during mitosis. Live-cell confocal imaging experiment, mitotic cell expressing CD8–mCherry–FRB with FKBP–β2–GFP. Rapamycin (200 nM) was applied during metaphase. Clear rerouting occurs, but no puncta form in the cytoplasm until after cytokinesis. This cell is shown in Video 5. Bar, 10 µm.
Comparison of different clathrin hooks for chemically inducible endocytosis
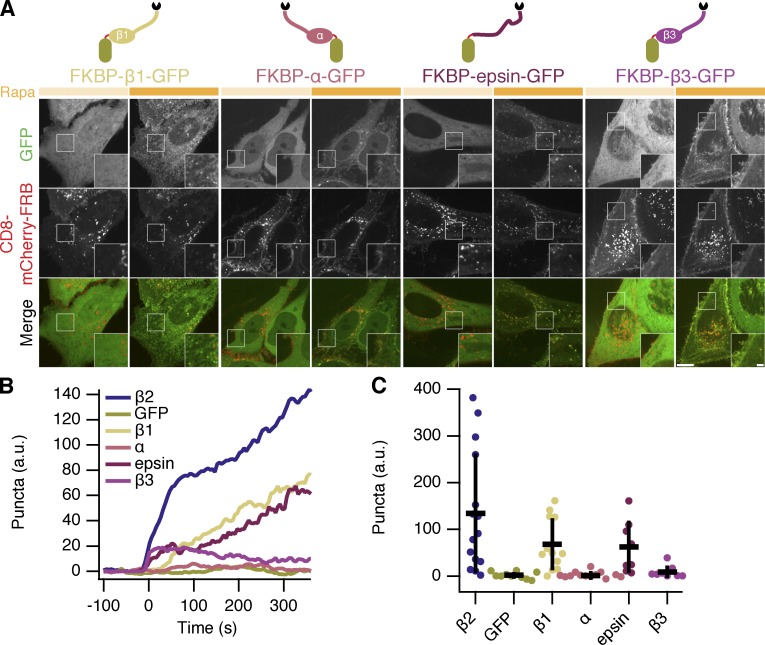
The hinge and appendage of the β2 subunit of AP2 was initially chosen as our clathrin hook because it is AP2 that coordinates endocytosis at the plasma membrane, but we hypothesized that the clathrin-binding region from any adaptor would be sufficient to initiate endocytosis. We next compared various clathrin hooks for their hot-wiring ability. Using the same chemically inducible system, we observed differences in bright green puncta formation despite all proteins being efficiently recruited to CD8–mCherry–FRB. Efficient puncta formation was seen with the hinge and appendage from β1 subunit of the AP1 complex and with a fragment from epsin, which contains clathrin-binding motifs (Fig. 7). No puncta formation was observed with the hinge and appendage from the α subunit of AP2, which was expected because it does not directly bind clathrin. Surprisingly, the hinge and appendage from the β3 subunit of AP3, which can bind clathrin in vitro (Dell’Angelica et al., 1998), was unable to generate bright green puncta. Quantification of this behavior showed that β2 was the most efficient and potent initiator, above β1 and epsin, whereas β3 initiates only marginally more than α adaptin and GFP negative controls (Fig. 7, B and C). These experiments indicate that one use for this system is to test proteins for functional clathrin-binding.
Figure 7.
Chemically inducible internalization: comparison of clathrin hooks. (A) Comparison of four potential clathrin hooks for chemically inducible endocytosis. Cells expressed CD8–mCherry–FRB with FKBP–β1–GFP, FKBP–α–GFP, FKBP–epsin–GFP, or FKBP–β3–GFP. Stills from live-cell confocal imaging experiments are shown: (left) image shows the frame before rerouting occurs and (right) 133 frames (665 s) later. Inset shows a 2× zoom of the boxed region. Bars: (main) 10 µm; (insets) 2 µm. (B) Mean plots to show chemically inducible endocytosis. Colored traces indicate the total area of bright GFP puncta that form in cells after rerouting. Traces were time aligned and averaged. (C) Scatterplot to indicate variability in chemically induced endocytosis. The total normalized area above threshold at 5 min after rerouting for each cell analyzed is shown (spots). ANOVA, P = 1.142 × 10−5. Bars indicate mean ± SD. Number of cells = 8–15 and number of experiments = 3.
Triggering endocytosis using a nonendocytic clathrin hook
The successful clathrin hooks that we tested can all bind clathrin but can also bind other membrane-trafficking proteins. It is possible that these other proteins contribute to the efficacy of triggered endocytosis. We therefore tested a clathrin-binding protein that is not involved in membrane traffic. G2 S-phase expressed protein 1 (GTSE1) is a microtubule-binding protein that can bind clathrin as part of the TACC3–clathrin–ch-TOG complex at the mitotic spindle (Hubner et al., 2010; Cheeseman et al., 2013). We used a C-terminal fragment of GTSE1 as a clathrin hook. First, the FKBP–GTSE1–GFP construct, unlike FKBP–β2–GFP, was not localized to CCSs before rapamycin addition (Fig. 8 A). Second, rapamycin addition to cells coexpressing CD8–mCherry–FRB and FKBP–GTSE1–GFP caused the appearance of bright green puncta (Fig. 8, A and B). We confirmed that these puncta were hot-wired CCVs by visualizing clathrin and the internalization of anti-CD8 from the plasma membrane (Fig. 8, A and C). These experiments therefore indicate that all that is needed to hot-wire endocytosis is a clathrin-binding protein that is correctly addressed to the plasma membrane. Binding of other coated pit components appears to be dispensable, although we note that the rate of accumulation of bright FKBP–GTSE1–GFP puncta was fourfold slower than for the other clathrin hooks that we have tested (Fig. 8 D).
Figure 8.
Chemically inducible internalization: using a clathrin hook of nonendocytic origin. (A) Colocalization of clathrin with bright green puncta formed by chemically induced internalization with a GTSE1 clathrin hook. Cells were coexpressing CD8–dCherry–FRB, mCherry–LCa, and FKBP–GTSE1–GFP. (B) Scatterplot to indicate variability in chemically induced endocytosis. The total normalized area above threshold at 10 min after rerouting for each cell analyzed is shown (spots). Bars: (main) 10 µm; (insets) 2 µm. Bars indicate mean ± SD. Number of cells = 35 and number of experiments = 10. (C) Antibody uptake after hot-wiring using a GTSE1 clathrin hook. Cells coexpressing CD8–mCherry–FRB and FKBP–GTSE1–GFP were incubated with anti-CD8–Alexa 647 to label the membrane anchor extracellularly, and rapamycin (200 nM) was applied. In A and C, stills from live-cell confocal imaging experiments are shown: (top) the frame before rerouting occurs and (bottom) 180 frames (15 min) later; empty and filled orange bars indicate before and after rapamycin addition. Bars: (main) 10 µm; (insets) 2 µm. (D) Table to show the rate of puncta appearance for GTSE1 compared with other clathrin hooks. Slope is the coefficient of a line fit to the averaged puncta data, postrapamycin; units are puncta per second.
Clathrin functionally interacts with AP2 via two binding sites on β2
We next applied our synthetic system to answer a cell biological question: how does clathrin functionally engage with AP2? Two sites on β2 have been implicated in binding clathrin, a clathrin box motif (CBM; LLNLD) in the hinge region (Shih et al., 1995) and a tyrosine residue (Y815) in the appendage (Edeling et al., 2006; Fig. 9 A). However, it is not clear which sites are important in a functional context. We first confirmed that these two sites interact with clathrin biochemically (Fig. 9 B). Deletion of the CBM interfered with binding to a CHC fragment containing the terminal domain and ankle region (residues 1–1,074). Mutation of Y815A (Y-A) also inhibited binding but to a lesser extent.
Figure 9.
Clathrin functionally engages AP2 at the plasma membrane using two distinct interactions. (A) Schematic illustration of the putative interactions between β2 and clathrin. Inset shows mutations to delete the CBM (ΔCBM) or mutation of tyrosine residue Y815 in the appendage (Y-A). Note that Y815 in earlier papers refers to Y829 in our longer isoform. (B) Analysis of clathrin–β2 interaction in vitro. Binding experiments using GST–β2(616–951) WT or mutants to pull down MBP–CHC(1–1074) as indicated. Interaction was assessed by Western blotting (i), and a Coomassie-stained gel was run in parallel to check for equivalent capture on beads. Three independent experiments were performed and analyzed by densitometry (ii). Bar indicates mean. (C) Quantification of live-cell confocal imaging experiments to show extent of endocytosis triggered by FKBP–β2–GFP or related mutants (see inset). Colored traces indicate the total area of bright GFP puncta that form in cells after rerouting. Traces were time aligned and averaged. Mean traces are shown (i), and a scatterplot (ii) of values at 5 min after rerouting for each cell analyzed is shown (spots). Bars indicate mean ± SD. Number of cells = 11–15 and number of experiments = 3. ANOVA, P = 7.92 × 10−5. Note that the values for FKBP–β2–GFP (WT) and GFP–FKBP (GFP) are also shown in Fig. 8. Example live-cell movies are shown in Videos 6, 7, and 8. (D) Representative confocal images (i) of an immunolabeling experiment to assess endocytosis of CD8 by triggered endocytosis. Bars: (main) 10 µm; (insets) 2 µm. Scatterplot (ii) to indicate variability in chemically induced endocytosis. The fraction of green puncta that were also red for each cell analyzed is shown (spots). Bars indicate the mean ± SD. Number of cells = 8–14 and number of experiments = 3. ANOVA, P = 1.66 × 10−6.
Live-cell imaging revealed that deletion of the CBM (ΔCBM) in the hinge of FKBP–β2–GFP and/or mutation of Y815 in the appendage (Y-A) were sufficient to block puncta formation by hot-wiring (Fig. 9 C and Videos 6, 7, and 8). This suggested that both sites were important for internalization. To confirm this result, we used live-cell antibody labeling to further quantify the inhibition (Fig. 9 D). Internalization of antibody upon rapamycin application was clearly observed in cells expressing WT FKBP–β2–GFP with CD8–dCherry–FRB. Partial inhibition of endocytosis was recorded in cells expressing Y-A or ΔCBM forms of FKBP–β2–GFP. Further inhibition was seen in cells expressing the double mutant, such that it was indistinguishable from GFP–FKBP (Fig. 9 D). These data indicate that both clathrin interaction sites on β2 are required for engagement in a functional context.
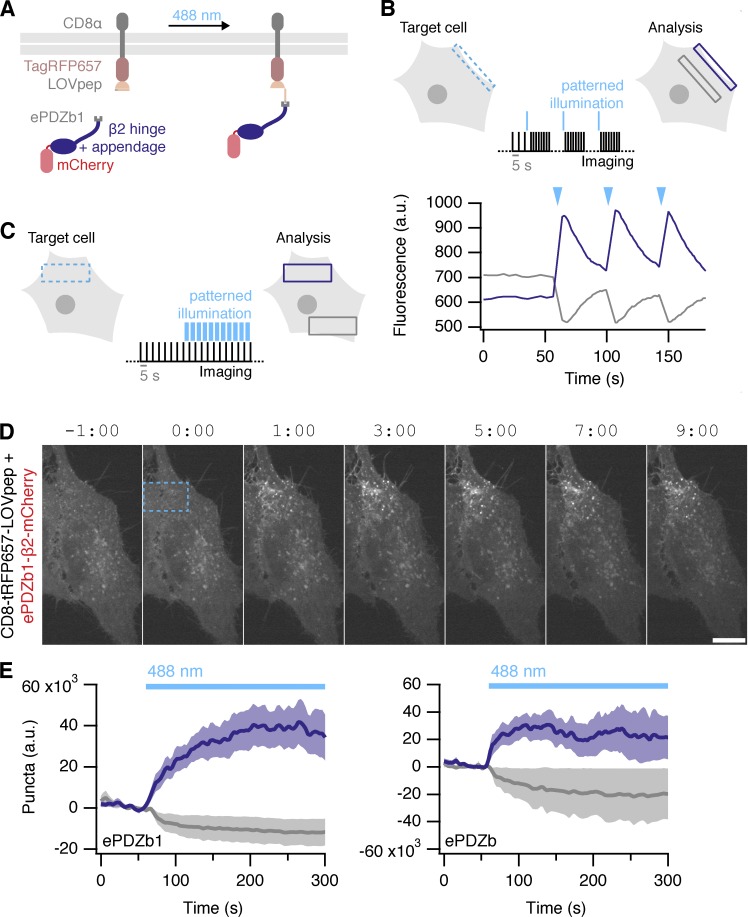
Optogenetic activation of endocytosis
Having successfully implemented temporal control of endocytic initiation using a chemogenetic approach, we next wanted to add spatial control. To do this, we used an optogenetic strategy: using the ability of photosensitive LOV2 domain from Avena sativa phototropin 1 to cage a small peptide that can bind the engineered PDZ domain (ePDZb1) after exposure to <500-nm light (Strickland et al., 2012; van Bergeijk et al., 2015; Wagner and Glotzer, 2016). We designed light-activated versions of our preferred plasma membrane anchor, CD8–TagRFP657–LOVpep(T406A,T407A,I532A), and clathrin hook, ePDZb1–β2–mCherry, which would allow discrete dimerization by blue light (Fig. 10 A). Recruitment of ePDZb1 to the plasma membrane anchor is rapid and reversible (τ = 40 s; Fig. 10 B). Transient recruitment was not sufficient to make vesicles. However sustained, patterned illumination of 488 nm light was sufficient to reroute ePDZb1–β2–mCherry to the plasma membrane and to subsequently initiate vesicle formation (Fig. 10, C and D; and Video 9). Analysis of optogenetic hot-wiring showed that onset of vesicle formation was fast and was less variable than with the chemical method (Fig. 10 E). We found ePDZb1 and ePDZb variants of the clathrin hook to be equivalent and that the LOVpep(T406A,T407A,I532A) version of the plasma membrane anchor was superior to the WT. These data show that spatiotemporal control over initiation of endocytosis is possible and is an efficient method to probe questions related to “on-demand” traffic from the membrane.
Figure 10.
Optically inducible endocytosis. (A) Illustration of optically induced endocytosis. Cells coexpress a plasma membrane anchored LOVpep (CD8–TagRFP657–LOVpep) and a clathrin hook with a PDZb1 tag (PDZb1–β2–mCherry). Upon illumination with blue light, an epitope is exposed to which the PDZ domain can bind, and the clathrin hook is rerouted to the plasma membrane so that endocytosis can occur at that site. (B) Rapid and reversible ePDZb1–mCherry recruitment to CD8–TagRFP657–LOVpep(T406A,T407A,I532A) at the plasma membrane. Brief (<1 s) patterned illumination of plasma membrane areas followed by rapid imaging. Mean pixel density of an ROI at the membrane (purple) compared with one in the cytoplasm (gray) is shown. (C) Endocytosis is activated in a spatiotemporal manner by sustained, patterned light activation within a defined ROI (blue dashed box). For analysis, the number of vesicles formed within the ROI (purple box) is compared with a similar ROI outside the activated region (gray box). (D) Stills from a typical optogenetic hot-wiring experiment. HeLa cell expressing CD8–TagRFP657–LOVpep(T406A,T407A,I532A) and ePDZb1–β2–mCherry. Only the mCherry confocal channel is shown. Time 0 indicates when patterned illumination began. Bar, 10 µm. A movie of this panel is available as Video 9. (E) Plots of mean ± SEM vesicle formation in cells expressing CD8–TagRFP657–LOVpep(T406A,T407A,I532A) and either ePDZb1–β2–mCherry (number of cells = 13 and number of experiments = 4) or ePDZb–β2–mCherry (number of cells = 8 and number of experiments = 3) as indicated. Purple and gray indicate vesicles formed inside and outside the photo-activation zone, respectively.
Discussion
In this paper, we describe systems for triggering endocytosis on demand. These tools are useful because they allow the internalization of defined cargo with temporal or spatiotemporal control via chemical or optical methodology, respectively. Bypassing several regulatory steps in constitutive CME to hot-wire endocytic events has many potential uses, and here we use it to demonstrate how clathrin and AP2 interact functionally to form a CCV and also to test the mitotic inhibition of CME.
Clathrin-coated pits are initiated when AP2 that is bound to PI(4,5)P2 at the plasma membrane simultaneously binds cargo bearing AP2-interacting motifs (Traub, 2009). This exposes the β2 hinge and appendage to bind clathrin and begin polymerization and stabilization of the nascent CCP (Kelly et al., 2014; Kirchhausen et al., 2014). Vesicle formation then proceeds in a regulated manner (Loerke et al., 2009; Mettlen et al., 2009; Taylor et al., 2011; Aguet et al., 2013). Our synthetic systems hot-wire this process by directly recruiting a clathrin hook to the plasma membrane. This mimicking of the PI(4,5)P2-bound, cargo-stabilized form of AP2 eliminates all the preceding steps and immediately starts clathrin polymerization.
A key question is whether the vesicles we generate via hot-wiring are distinct from endogenous CME. Do the hot-wired vesicles exclusively contain the plasma membrane anchor, the hook and endogenous clathrin? If this were the case, the vesicles would probably represent a dead end. For example, without any SNAREs for fusion and onward traffic, the cargo would stall in the first carrier. Instead, we saw that vesicles generated by hot-wiring are processed similarly to cargo bearing a tyrosine-based endocytic motif, exhibiting normal internalization, processing, and recycling in Rab4-containing vesicles. So, although the design of hot-wiring suggests an isolated mechanism of vesicle generation, the reality is an interplay of endogenous resources with our synthetic system. In support of this, we could see transferrin and hot-wired cargo in the same endocytic puncta, as well as in separate vesicles; we found colocalization of hot-wired cargo with VAMP2 and also saw that dynamin was recruited to pinch off hot-wired vesicles. Note that an alternative model, in which the plasma membrane anchor is dragged into preexisting CCSs and internalized via endogenous CME, was not supported by any data.
Any accessory proteins that are recruited to the nascent hot-wired vesicle must be attracted by curvature or by clathrin itself. Two observations support this statement. First, hot-wired endocytosis can occur after depletion of AP2, so AP2 and the plethora of accessory proteins that bind to it are not essential for vesicle formation. Second, clathrin hooks designed from the nonendocytic protein GTSE1 could also generate vesicles. Because this fragment is not known to bind any endocytic proteins, it cannot therefore recruit them. Clathrin and curvature are the only common ingredients between these experiments and our standard hot-wiring procedure. Although it is clear that many proteins modulate vesicle formation, reductionist systems such as hot-wiring are ideal for demonstrating the minimal factors for vesicle creation.
Given that hot-wired events likely share resources with endogenous CME, does hot-wiring affect it? We found no profound impact of hot-wiring on endogenous CME. Our conclusion is that there are sufficient resources in the cell to support endogenous CME as well as the additional, triggered form of CME. This is perhaps to be expected, because cells often have to increase the rate of CCP formation when certain receptors are activated (see Reeves et al., 2016, for a recent example), and this is analogous to hot-wiring. There may be more subtle changes to endogenous CME as a result of hot-wiring. Precisely how, and to what extent, hot-wiring interfaces with endogenous CME needs further examination. We note, however, that potential interference with endogenous CME is less of a concern with optogenetic hot-wiring, because this system is rapidly reversible.
One application of hot-wired endocytosis is to test for functional interactions between clathrin and clathrin-binding proteins. It is clear that binding between protein fragments in a test tube does not necessarily translate to functional interactions in cells. Even in situ, we have previously seen that colocalization of adaptors and clathrin mutants does not reliably indicate functional CME (Willox and Royle, 2012). The inducibility of hot-wiring allows the assessment of functional interactions in ways that expression of mutants do not. We used this technique to demonstrate that two necessary interactions between AP2 and clathrin are both required for CME. A CBM in the β2 hinge binds to the terminal domain, and a site on the β2 appendage interacts with a distinct site, probably on the ankle of CHC. Biochemical evidence for these interactions was in the literature (Owen et al., 2000; ter Haar et al., 2000; Edeling et al., 2006; Hood et al., 2013), but the relative importance of each site for function was previously unknown. A further surprising result was that β3, which is known to bind clathrin in a test tube, was not interchangeable with β1 and β2 as a functional initiator of CME. The β3–clathrin interaction is robust, as CBMs were actually discovered using β3 (Dell’Angelica et al., 1998). Subsequent work questioned whether AP3 was a genuine clathrin adaptor (Peden et al., 2002; Zlatic et al., 2013). Our results suggest that functional clathrin engagement by β3 is not equivalent to β1 and β2. One possible explanation is that the clathrin-binding site on the appendage is not present in β3. Another possibility is that β3 cannot bind the same set of endocytic proteins as β2, epsin, and β1, but this is unlikely given the hot-wiring activity of the nonendocytic protein GTSE1.
Although our systems are ready to use, we think further optimization of the plasma membrane anchor may be possible. First, some internal vesicles, which contained membrane anchor, were present with CD8-, CD4-, GAP43-, and SH4-based anchors. These internal vesicles complicate image analysis and might prevent efficient rerouting exclusively to the plasma membrane. Second, anchors are prone to diffusion in the plasma membrane. For the optogenetic application, this means that we constantly lose photoactivated anchors. Although the anchors at the plasma membrane quickly inactivate once they diffuse out of the illumination zone, which ensures local activation, this behavior reduces the efficiency of vesicle creation. Engineering the anchors to address these issues is key to further improvements in the technology. Moreover, diversifying the cargoes that are programmed for internalization will further extend the capabilities of hot-wiring.
There are numerous uses for our hot-wiring systems. For example, the possible functionalization of the plasma membrane anchor on the extracellular face so that targeted uptake of nanoparticles or other interesting materials is possible. Another goal will be to change cell behaviors by local optogenetic activation of endocytosis. In addition, we anticipate that these systems will be useful for understanding which factors are essential for the initiation of endocytic events as well as for downstream processing of hot-wired vesicles and the machineries that control these aspects of vesicle trafficking.
Materials and methods
Cell culture
HeLa cells (Health Protection Agency/European Collection of Authenticated Cell Cultures 93021013; Research Resource Identifier [RRID]: CVCL_0030) were maintained in DMEM plus 10% FBS and 100 U/ml penicillin/streptomycin. Retinal pigment epithelium (hTERT-RPE1; RRID: CVCL_4388) cells were maintained in DMEM/F12 supplemented with 10% FBS, 100 U/ml penicillin/streptomycin, 0.26% NaHCO3, and 2 mM glutamine. All cells were kept at 37°C and 5% CO2. Cells were confirmed to be mycoplasma free by routine PCR-based testing. DNA transfections were performed with GeneJuice (MerckMillipore) and siRNA transfections with Lipofectamine 2000 (Life Technologies) according to the manufacturer’s instructions. Cells were imaged or fixed 2 d after DNA transfection, and siRNA was used in a “two hit” protocol with transfection at 2 and 4 d before use (Willox and Royle, 2012). Sequences targeting μ2 (μ2-2; 5′-AAGUGGAUGCCUUUCGGGUCA-3′) or CHC (chc-2; 5′-UCCAAUUCGAAGACCAAUU-3′) were as described previously (Motley et al., 2003).
Molecular biology
CD8–FRB was made by PCR amplification of CD8-8A (Fielding et al., 2012) and subcloning into pMito–FRB at AgeI and EcoRI; CD8–mCherry–FRB was made from CD8–FRB by replacing FRB with mCherry–FRB from pMito–mCherry–FRB at the same sites. CD4–mCherry–FRB was made by overlap extension PCR of CD8–mCherry–FRB and CD4–FRB (synthesized by IDT); GAP43–FRB–RFP was available from previous work; SH4–FRB–mRFP was made by replacing the GAP43 sequence in GAP43–FRB–mRFP with the DNA encoding the first 16 residues of Fyn, at NheI–EcoRI sites. Plasmid encoding mCherry–FRB was made by cutting and religating pMito–mCherry–FRB at BglII–BamHI. Dark versions of mCherry constructs (dCherry) were made by site-directed mutagenesis to introduce the K70N mutation.
FKBP–β2–mCherry was made by inserting FKBP PCR product into β2–mCherry at Acc65I and XhoI sites; this was then modified by subcloning of GFP to make FKBP–β2–GFP. β2–mCherry was previously prepared by PCR of GST–β2 adaptin(616–951) (Hood et al., 2013) and insertion into pmCherry–N1. Mutations in FKBP–β2–mCherry were added by site-directed mutagenesis, Y-A by mutation of β2 adaptin residue Y815, and ΔCBM by deletion of regions 627–635 (LLGDLLNLD). The double mutant (ΔCBM/Y-A) was created by switching the β2 adaptin fragment containing the Y815A mutation for the same region in ΔCBM construct using PspOMI and AgeI. Additional FKBP constructs were made by inserting a PCR product in the place of β2 in FKBP–β2–GFP; mouse α adaptin 1 (regions 740–977) and mouse epsins (region 144–575) were inserted at Acc65I and AgeI, and human β1 adaptin (regions 617–949) was added at AgeI and EcoRI. FKBP–β3–GFP was made by insertion of human β3 fragment (regions 702–1,094) into FKBP–β1–GFP. FKBP–GTSE1–GFP was made by amplification of human GTSE1 (638–720) and insertion into FKBP–β2–GFP at BamHI and BspEI and AgeI. GFP–FKBP, pMito–PAGFP–FRB, mCherry–LCa, and GFP–VAMP2 were available from previous work (Granseth et al., 2006; Hood and Royle, 2009; Cheeseman et al., 2013). Endocytic compartment markers were sourced as follows: GFP–EEA1 (42307; Addgene), mCherry–OCRL1 (27675; Addgene), LAMP1–mGFP (34831; Addgene), and mNeonGreen–Rab4a-7 (Allele Biotech). Dynamin2–GFP was a gift from R. Vallee (Columbia University, New York, NY), and GFP–Rab5a, GFP–Rab7, GFP–Rab9a, and GFP–Rab11a were gifts from F. Barr (Oxford University, Oxford, England, UK).
For optogenetic experiments, pTagRFP657–LOVpep(T406A,T407A,I532A) was made by ligation of LOVpep(T406A,T407A,I532A), ordered as a custom gene from IDT, and pTagRFP657 (31959; Addgene). This product was then inserted into CD8–mCherry at EcoRI and AgeI sites. PDZb–β2–GFP and PDZb1–β2–GFP were made by PCR of PDZb (34980; Addgene) or PDZb1 (34981; Addgene), inserted into FKBP–β2–GFP at XhoI and BamHI, replacing FKBP. PDZb–GFP and PDZb1–GFP were made by subcloning of PDZb or PDZb1 (different PCR products to preserve reading frame) into pEGFP–N1. PDZb–β2–mCherry and PDZb1–β2–mCherry were made by subcloning of PDZb/PDZb1 from PDZb–β2–GFP or PDZb1–β2–GFP and inserting into FKBP–β2–mCherry at BamHI and XhoI sites. PDZb–mCherry and PDZb1–mCherry were made by PCR of PDZb/PDZb1–GFP inserted into pmCherry–N1 at BamHI and AgeI. Plasmids to express MBP–CHC(1–1,074)–His6 and GST–β2(616–951) in bacteria were available from previous work (Hood et al., 2013). The GST–β2(616–951) Y-A, ΔCBM, and double mutants were made by inserting into pGEX-6P-1 PCR fragments amplified from CD8–β2–mCherry constructs containing these mutations at EcoRI–NotI. All plasmids described in this paper are available from Addgene (100726–100748).
Immunofluorescence
For live immunolabeling of CD8–β2–mCherry in HeLa cells, 1:100 Alexa 488–conjugated mouse anti-CD8 (MCA1226A488 [AbD Serotec], 0.05 mg/ml; RRID: AB_324877) was added at 4°C or 37°C for 40 min, diluted in DMEM with 10% FBS. The cells were then placed on ice, washed with PBS with 1% BSA, and surface fluorescence quenched with rabbit anti–Alexa 488 (A-11094 [Thermo Fisher Scientific], 1 mg/ml; RRID: AB_221544) before relabeling with 1:500 anti–mouse Alexa 633 (A21052 [Life Technologies], 2 mg/ml; RRID: AB_141459) conjugated secondary antibody.
Live labeling of rapamycin-inducible system was performed in HeLa cells transfected with CD8–mCherry–FRB and FKBP–β2–GFP (or mutant variants) or GFP–FKBP. Live cells were incubated with untagged anti-CD8 (A-11094 [AbD Serotec], 1 mg/ml; RRID: AB_321400) at 1:1,000 at 37°C for 40 min; 200 nM rapamycin was then added for 30 min before cells were put on ice and washed with PBS with 1% BSA. Alexa 647–conjugated anti-mouse secondary (A21235 [Life Technologies], 2 mg/ml; RRID: AB_2535804) was added in DMEM at 1:500 for 1 h, and cells were then fixed and permeabilized with 0.1% Triton X-100 in PBS. Finally, 1:500 Alexa 568–conjugated secondary antibody (A11031 [Life Technologies], 2 mg/ml; RRID: AB_144696) was added in DMEM for 1 h before washing in PBS and mounting.
Live-cell imaging of antibody uptake was performed in cells transfected with FKBP–β2–GFP or GFP–FKBP with CD8–mCherry–FRB or CD8–dCherry–FRB + mCherry LCa. Alexa 647–conjugated anti-CD8 (MCA1226A647 [AbD Serotec]; RRID: AB_324876) was added to HeLa cells at 37°C for 30 min, and cells washed twice in warm medium before imaging using the standard live-cell imaging protocol described in the following section. For colocalization analysis with antibody uptake, cells were imaged and Alexa 647–conjugated anti-CD8 was added at the start of the movie, and endocytosis was triggered by addition of rapamycin (200 nM) after 30 s. For CD8–YAAL, no rapamycin was added. For imaging late trafficking steps, movies were acquired 10–30 min after triggering.
For transferrin uptake analysis, HeLa cells were serum-starved for 20 min in serum-free DMEM and then exposed to 200 nM rapamycin or ethanol vehicle for 20 min, with 0.05 mg/ml Alexa 647–conjugated transferrin (Invitrogen) added for the final 10 min before fixing. All dilutions were performed in serum-free media.
In experiments in which both transferrin uptake and live antibody labeling were performed, cells were first starved for 30 min in serum-free DMEM. Mouse anti-CD8 was applied for 40 min in serum-free DMEM, and cells were then treated with rapamycin (200 nM) or vehicle for 30 min. Alexa 647–conjugated transferrin was applied for the last 10 min of treatment before cells were moved to ice. Unconjugated goat anti–mouse antibody (31164 [Thermo Fisher Scientific]; RRID: AB_228299) was added to block surface anti-CD8. Cells were then fixed with PFA, permeabilized, and then incubated with Alexa 568–conjugated secondary antibody in DMEM for 1 h before washing in PBS and mounting.
Light microscopy
For live-cell imaging of rerouting experiments, HeLa cells were transfected with CD8–mCherry–FRB, CD8–dCherry–FRB, CD4–mCherry–FRB, pMito–PAGFP–FRB, GAP43–FRB–mRFP, mCherry–FRB, or SH4–RFP–FRB with FKBP–β2–GFP (or mutants), FKBP–β2–mRuby2, FKBP–β1–GFP, FKBP–α–GFP, FKBP–β3–GFP, FKBP–epsin–GFP, FKBP–GTSE1–GFP, or GFP–FKBP. For experiments in which the red channel was needed for imaging something other than CD8, dark variants of mCherry constructs were used in which mCherry had been mutated to give K70N (Subach et al., 2009); we refer to this variant as dCherry (dark mCherry). Cells were imaged in glass-bottom fluorodishes (WPI) with Leibovitz L-15 CO2-independent medium (Sigma-Aldrich) supplemented with 10% FBS and kept at 37°C. Imaging was performed using a spinning disc confocal system (Ultraview Vox; PerkinElmer) with a 100× 1.4 NA oil-immersion objective. Images were captured every 5, 20, or 30 s in Volocity using a dual-camera system (ORCA-R2; Hamamatsu) after excitation with lasers of wavelength 488, 561, and 640 nm (if required); 200 nM rapamycin (Alfa Aesar) was added after 1 or 2 min.
Light-induced dimerization experiments were performed similarly. Cells transfected with CD8–TagRFP657–LOVpep(T406A,T407A,I532A) and PDZb1–β2–mCherry or PDZb–β2m–Cherry were imaged with 561- and 640-nm lasers and captured with a single camera at 5-s intervals. Photoexcitation was done using a 488-nm laser in a defined region of interest (ROI). 12 frames of baseline were captured before interleaving photoactivation and imaging, as indicated in Fig. 10 C. Use of the constitutively active form of LOVpep generated many vesicles similar to our CD8–β2–mCherry fusion protein.
All fixed-cell and immunostaining experiments were performed in transiently transfected HeLa cells fixed in PBS with 3% paraformaldehyde and 4% sucrose and mounted in Mowiol with DAPI. Imaging was performed using the same Ultraview spinning disc confocal and 100× objective, and z slices were taken at 0.5-µm intervals.
TIRFM was performed on RPE1 cells expressing CD8–dCherry–FRB and FKBP–β2–mRuby2 with GFP–LCa or dynamin2–GFP. Images were captured at 1 frame/s using an Olympus IX81 equipped with a 100× 1.49 NA objective and Hamamatsu ImagEM-1K electron-multiplying charge-coupled device after excitation with 488- and 561-nm lasers. 200 nM rapamycin was added after 1 min.
CLEM
HeLa cells transfected with CD8–dCherry–FRB and FKBP–β2–GFP or GFP–FKBP were imaged in gridded glass-bottom dishes (P35G-2-14-CGRD; MatTek) after incubation with anti-CD8 (1:1,000) for 30 min and then Alexa 546 FluoroNanoGold–anti-mouse Fab′ (1:200; 7402 [Nanoprobes], 0.08 mg/ml; RRID: AB_2631183) for 10 min. Using the photo-etched coordinates on each grid, the cell location was recorded using bright-field illumination at 20×. After rapamycin addition, fluorescent live-cell imaging determined vesicle formation using a 100× 1.4 NA objective on a Nikon Ti epifluorescence microscope and a Coolsnap Myo camera (Photometrics) using NIS Elements AR software. Once a sufficient number of vesicles had been observed, the sample was immediately fixed with 3% glutaraldehyde and 0.5% paraformaldehyde in 0.05 M phosphate buffer, pH 7.4, for 2 h. Remaining aldehydes were quenched in 50 mM glycine, and cells were subsequently washed in dH2O. Gold enhancement was then performed for 3 min as per the manufacturer’s instructions (GoldEnhance-EM; Nanoprobes). Cells were washed twice in 0.3% Na2S2O3 and then thoroughly in dH2O. Gold-enhanced samples were postfixed in 1% OsO4 for 60 min, rinsed with dH2O, and then stained en bloc with 0.5% uranyl acetate in 30% ethanol for 60 min. Cells were dehydrated in increasing concentrations of ethanol for 10 min each (50%, 70%, 80%, 90%, 100%, and 100%) before being embedded in epoxy resin (TAAB) and left to polymerize for 48 h at 60°C. Each cell of interest was identified by correlating the grid and cell pattern on the surface of the polymerized block with previously acquired bright-field images. Ultrathin (70-nm) sections were collected on formvar-coated copper grids using an EM-UC6 ultra-microtome (Leica Microsystems) and contrasted with saturated aqueous uranyl acetate and Reynolds lead citrate. Sections were imaged on a Jeol 1400 TEM at 100 kV.
Biochemistry
Binding experiments to test the interaction between β2 and clathrin were performed as previously described (Hood et al., 2013). In brief, GST–β2 adaptin(616–951)–His6 and MBP–CHC(1–1,074)–His6 proteins were purified using glutathione Sepharose 4B and amylose resin, respectively. For in vitro interaction studies, equal amounts (50 µg) of GST- and MBP-fused proteins were mixed in reaction buffer I (50 mM Tris-Cl, pH 7.5, 150 mM NaCl, and 0.1 mM EGTA). The mixture was incubated with a 50% slurry of glutathione Sepharose 4B beads (preequilibrated in NET-2 buffer [50 mM Tris-Cl, pH 7.5, 150 mM NaCl, and 0.5% NP-40 substitute]) and left overnight at 4°C with rotation. Next day, beads were collected by spinning down at 1,000 g for 2 min at 4°C and washed four times with NET-2 buffer. Beads were then resuspended in 30 µl of 1× Laemmli buffer, denatured at 95°C, and analyzed by Western blotting after running on 8% SDS-PAGE. Protein samples were also analyzed by staining SDS-PAGE gels with Coomassie brilliant blue. Antibodies used for Western blotting were anti-μ2 (mouse anti-AP50 Clone 31 [BD Transduction Labs]; RRID: AB_398872), anti-GAPDH (rabbit G9545 [Sigma-Aldrich]; RRID: AB_796208), and anti-MBP (mouse 8G1 2396 [Cell Signaling Technology]; RRID: AB_2140060).
Image analysis
All code for automated and semiautomated image analysis is available at https://github.com/quantixed/PaperCode/.
For analysis of triggered endocytosis, a simple thresholding method proved the most straightforward. The green channel was first corrected for photobleaching using the simple ratio method. To examine rerouting kinetics, the mean pixel density within a cytoplasmic ROI was measured for all frames in the movie. Next, a binary threshold was applied to the green channel, which isolated any newly formed vesicles and excluded background signal from other structures such as the plasma membrane. The raw integrated density (number of pixels over threshold × 255) within an ROI containing the cell were measured for all frames in the movie. These values for all cells were fed into Igor Pro 7, and a series of custom-written functions processed the data. The raw integrated densities were aligned to time 0, which was defined as the point at which the mean pixel density of the cytoplasmic ROI fell by 50%. These time-aligned traces were normalized by the size of the whole-cell ROI (in square micrometers) and baseline subtracted. This gave a good approximation of the number of internalized vesicles as defined by bright puncta of FKBP–clathrin hook–GFP fluorescence. For light-activated endocytosis, the same method was used with no time alignment. The raw integrated density was extracted in the same way except that measurements were taken within the illuminated ROI and an equivalent, nonexposed area of the same cell. Because of variability in image capture, time-stamps were extracted from the mvd2 library using IgorPro. These were used for interpolation to calculate precise averages in IgorPro. To examine accumulation of puncta over time, a line was fit to the averaged puncta data; the slope of the line is reported along with the coefficient of determination.
Analysis of CCV morphology from CLEM experiments was done by manual segmentation of membrane and coats in micrographs. The segmented pixels were read into IgorPro and processed automatically. In brief, pixel coordinates were used to find the eigenvectors and eigenvalues via singular value decomposition and rotate the matrix so that the semimajor axis is at y = 0 and the semiminor axis is at x = 0. Maximum values for x and for y were taken as the radii. The diameter was taken as the sum of these two radii. Ellipticity is the ratio of these radii (1 would be a perfect circle).
To examine “colocalization,” bleach-corrected images were searched in each channel using the ComDet v.0.3.5 plugin for ImageJ (https://github.com/ekatrukha/ComDet/). The locations of all spots were loaded into IgorPro and converted to 3 × 3 segments, and the intersection between these segments was calculated and presented in units of 9 pixels. The routine for this analysis (colocAnalysis) randomizes spot locations in the time dimension to test for chance colocalization. For colocalization with endocytic markers, more than three spots per frame (in a single confocal plane) above randomization was used as a benchmark for colocalization.
For analysis of antibody feeding experiments, four z slices from the center of the stack were thresholded in the red and green channels to isolate vesicular structures. Using Fiji and the “analyze particles” plugin, a mask showing only particles of 0.03–0.8 µm and circularity of 0.3–1.0 was created. Particles present in both channels were determined using the image calculator, and a third mask was created. The pixels over threshold within each processed cell mask were measured for red + green and for red alone, and the values combined from each of the z slices. The ratio of (red + green)/red pixels was the proportion of red puncta that were GFP positive, discarding structures containing only CD8. Transferrin uptake was measured using the same method and parameters, using results from the transferrin channel alone.
Analysis of TIRFM images was performed in MATLAB using the cmeAnalysis package (Aguet et al., 2013). Recruitment of GFP–dynamin2 or LCa–GFP to FKBP–β2–mRuby2 positive puncta was observed using the red channel as the master and green as the slave. Additionally, kymographs were created in Fiji.
Figures were made in Fiji, Igor Pro, and Photoshop and assembled in Illustrator. Null hypothesis statistical tests between two groups were done using two-sided Student’s t test with Welch’s correction; among three or more groups, one-way ANOVA was used with Tukey’s post hoc test. The Shapiro-Wilk test was used to assess normality.
Online supplemental material
Fig. S1 shows that plasma membrane anchors are interchangeable for chemically induced internalization, although mitochondrial anchors do not support vesicle generation. Fig. S2 shows that bright green puncta originate from the plasma membrane and can be distinguished from dimmer, nonendocytic spots by a simple threshold. Fig. S3 depicts imaging of chemically inducible endocytosis using TIRFM. Fig. S4 shows that hot-wired endocytic cargo follows a similar route to cargo with a tyrosine-based sorting motif. Fig. S5 shows that hot-wired endocytosis does not result from internalization via preexisting clathrin-coated pits. Video 1 depicts chemical induction of endocytosis: CD8–mCherry–FRB and FKBP–β2–GFP. Video 2 depicts no chemical induction of endocytosis: CD8–mCherry–FRB and GFP–FKBP. Video 3 shows chemical induction of endocytosis: CD8–dCherry–FRB, FKBP–β2–GFP, and mCherry–LCa with anti-CD8/Alexa 647 labeling. Video 4 shows monitoring colocalization of FKBP–β2–GFP with mCherry–LCa during hot-wiring. Video 5 depicts inhibition of hot-wired endocytosis in mitotic cells. Video 6 shows impairment of chemically induced endocytosis using CD8–mCherry–FRB and FKBP–β2–GFP(Y-A). Video 7 shows impairment of chemically induced endocytosis using CD8–mCherry–FRB and FKBP–β2–GFP(ΔCBM). Video 8 depicts impairment of chemically induced endocytosis using CD8–mCherry–FRB and FKBP–β2–GFP(double mutant). Video 9 shows optical induction of endocytosis: CD8–TagRFP657–LOVpep(T406A,T407A,I532A) and PDZb1–β2–mCherry.
Supplementary Material
Acknowledgments
We would like to thank Zuzana Kadlecova for extensive discussions and experimental work not described here and Sandra Schmid for constructive comments on our manuscript. We thank Alex Bird for GTSE1 discussions; Anne Straube for access to TIRFM; Rebecca Milton, Rachel Jones, and Aditi Kibe for technical assistance; and Cristina Gutiérrez Caballero for reagents. Finally, all Royle lab members and Centre for Mechanochemical Cell Biology colleagues gave valuable comments throughout the project.
L.A. Wood was supported by the Medical Research Council Doctoral Training Partnership grant (MR/J003964/1). G. Larocque is supported by Fonds de Recherche du Québec - Nature et Technologies.
The authors declare no competing financial interests.
Author contributions: L.A. Wood did most of the experimental work and analysis, made reagents, and helped write the original draft. G. Larocque carried out additional imaging and analysis during the revision process. N.I. Clarke performed the CLEM experiments. S. Sarkar did binding experiments. S.J. Royle was involved in conceptualization, data analysis, writing software, and writing and revising the manuscript.
Footnotes
Abbreviations used:
- CBM
- clathrin box motif
- CCS
- clathrin-coated structure
- CCV
- clathrin-coated vesicle
- CHC
- clathrin heavy chain
- CLEM
- correlative light-electron microscopy
- CME
- clathrin-mediated endocytosis
- ePDZ
- engineered PDZ
- FKBP
- FK506-binding protein
- LCa
- clathrin light chain a
- ROI
- region of interest
- TIRFM
- total internal reflection fluorescence microscopy
References
- Aguet F., Antonescu C.N., Mettlen M., Schmid S.L., and Danuser G.. 2013. Advances in analysis of low signal-to-noise images link dynamin and AP2 to the functions of an endocytic checkpoint. Dev. Cell. 26:279–291. 10.1016/j.devcel.2013.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner G.H., Antrobus R., Hirst J., Bhumbra G.S., Kozik P., Jackson L.P., Sahlender D.A., and Robinson M.S.. 2012. Multivariate proteomic profiling identifies novel accessory proteins of coated vesicles. J. Cell Biol. 197:141–160. 10.1083/jcb.201111049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman L.P., Harry E.F., McAinsh A.D., Prior I.A., and Royle S.J.. 2013. Specific removal of TACC3-ch-TOG-clathrin at metaphase deregulates kinetochore fiber tension. J. Cell Sci. 126:2102–2113. 10.1242/jcs.124834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dannhauser P.N., and Ungewickell E.J.. 2012. Reconstitution of clathrin-coated bud and vesicle formation with minimal components. Nat. Cell Biol. 14:634–639. 10.1038/ncb2478 [DOI] [PubMed] [Google Scholar]
- Dell’Angelica E.C., Klumperman J., Stoorvogel W., and Bonifacino J.S.. 1998. Association of the AP-3 adaptor complex with clathrin. Science. 280:431–434. 10.1126/science.280.5362.431 [DOI] [PubMed] [Google Scholar]
- Edeling M.A., Mishra S.K., Keyel P.A., Steinhauser A.L., Collins B.M., Roth R., Heuser J.E., Owen D.J., and Traub L.M.. 2006. Molecular switches involving the AP-2 beta2 appendage regulate endocytic cargo selection and clathrin coat assembly. Dev. Cell. 10:329–342. 10.1016/j.devcel.2006.01.016 [DOI] [PubMed] [Google Scholar]
- Ehrlich M., Boll W., Van Oijen A., Hariharan R., Chandran K., Nibert M.L., and Kirchhausen T.. 2004. Endocytosis by random initiation and stabilization of clathrin-coated pits. Cell. 118:591–605. 10.1016/j.cell.2004.08.017 [DOI] [PubMed] [Google Scholar]
- Fielding A.B., Willox A.K., Okeke E., and Royle S.J.. 2012. Clathrin-mediated endocytosis is inhibited during mitosis. Proc. Natl. Acad. Sci. USA. 109:6572–6577. 10.1073/pnas.1117401109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granseth B., Odermatt B., Royle S.J., and Lagnado L.. 2006. Clathrin-mediated endocytosis is the dominant mechanism of vesicle retrieval at hippocampal synapses. Neuron. 51:773–786. 10.1016/j.neuron.2006.08.029 [DOI] [PubMed] [Google Scholar]
- Hood F.E., and Royle S.J.. 2009. Functional equivalence of the clathrin heavy chains CHC17 and CHC22 in endocytosis and mitosis. J. Cell Sci. 122:2185–2190. 10.1242/jcs.046177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood F.E., Williams S.J., Burgess S.G., Richards M.W., Roth D., Straube A., Pfuhl M., Bayliss R., and Royle S.J.. 2013. Coordination of adjacent domains mediates TACC3-ch-TOG-clathrin assembly and mitotic spindle binding. J. Cell Biol. 202:463–478. 10.1083/jcb.201211127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubner N.C., Bird A.W., Cox J., Splettstoesser B., Bandilla P., Poser I., Hyman A., and Mann M.. 2010. Quantitative proteomics combined with BAC TransgeneOmics reveals in vivo protein interactions. J. Cell Biol. 189:739–754. 10.1083/jcb.200911091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson L.P., Kelly B.T., McCoy A.J., Gaffry T., James L.C., Collins B.M., Höning S., Evans P.R., and Owen D.J.. 2010. A large-scale conformational change couples membrane recruitment to cargo binding in the AP2 clathrin adaptor complex. Cell. 141:1220–1229. 10.1016/j.cell.2010.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur S., Fielding A.B., Gassner G., Carter N.J., and Royle S.J.. 2014. An unmet actin requirement explains the mitotic inhibition of clathrin-mediated endocytosis. eLife. 3:e00829 10.7554/eLife.00829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly B.T., McCoy A.J., Späte K., Miller S.E., Evans P.R., Höning S., and Owen D.J.. 2008. A structural explanation for the binding of endocytic dileucine motifs by the AP2 complex. Nature. 456:976–979. 10.1038/nature07422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly B.T., Graham S.C., Liska N., Dannhauser P.N., Höning S., Ungewickell E.J., and Owen D.J.. 2014. Clathrin adaptors. AP2 controls clathrin polymerization with a membrane-activated switch. Science. 345:459–463. 10.1126/science.1254836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyel P.A., Thieman J.R., Roth R., Erkan E., Everett E.T., Watkins S.C., Heuser J.E., and Traub L.M.. 2008. The AP-2 adaptor beta2 appendage scaffolds alternate cargo endocytosis. Mol. Biol. Cell. 19:5309–5326. 10.1091/mbc.E08-07-0712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhausen T., Owen D., and Harrison S.C.. 2014. Molecular structure, function, and dynamics of clathrin-mediated membrane traffic. Cold Spring Harb. Perspect. Biol. 6:a016725 10.1101/cshperspect.a016725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozik P., Francis R.W., Seaman M.N., and Robinson M.S.. 2010. A screen for endocytic motifs. Traffic. 11:843–855. 10.1111/j.1600-0854.2010.01056.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampe M., Pierre F., Al-Sabah S., Krasel C., and Merrifield C.J.. 2014. Dual single-scission event analysis of constitutive transferrin receptor (TfR) endocytosis and ligand-triggered β2-adrenergic receptor (β2AR) or Mu-opioid receptor (MOR) endocytosis. Mol. Biol. Cell. 25:3070–3080. 10.1091/mbc.E14-06-1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loerke D., Mettlen M., Yarar D., Jaqaman K., Jaqaman H., Danuser G., and Schmid S.L.. 2009. Cargo and dynamin regulate clathrin-coated pit maturation. PLoS Biol. 7:e57 10.1371/journal.pbio.1000057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettlen M., Stoeber M., Loerke D., Antonescu C.N., Danuser G., and Schmid S.L.. 2009. Endocytic accessory proteins are functionally distinguished by their differential effects on the maturation of clathrin-coated pits. Mol. Biol. Cell. 20:3251–3260. 10.1091/mbc.E09-03-0256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motley A., Bright N.A., Seaman M.N., and Robinson M.S.. 2003. Clathrin-mediated endocytosis in AP-2-depleted cells. J. Cell Biol. 162:909–918. 10.1083/jcb.200305145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy J.E., and Keen J.H.. 1992. Recognition sites for clathrin-associated proteins AP-2 and AP-3 on clathrin triskelia. J. Biol. Chem. 267:10850–10855. [PubMed] [Google Scholar]
- Owen D.J., Vallis Y., Pearse B.M., McMahon H.T., and Evans P.R.. 2000. The structure and function of the beta 2-adaptin appendage domain. EMBO J. 19:4216–4227. 10.1093/emboj/19.16.4216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peden A.A., Rudge R.E., Lui W.W., and Robinson M.S.. 2002. Assembly and function of AP-3 complexes in cells expressing mutant subunits. J. Cell Biol. 156:327–336. 10.1083/jcb.200107140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthenveedu M.A., Yudowski G.A., and von Zastrow M.. 2007. Endocytosis of neurotransmitter receptors: location matters. Cell. 130:988–989. 10.1016/j.cell.2007.09.006 [DOI] [PubMed] [Google Scholar]
- Reeves P.M., Kang Y.L., and Kirchhausen T.. 2016. Endocytosis of ligand-activated sphingosine 1-phosphate receptor 1 mediated by the clathrin-pathway. Traffic. 17:40–52. 10.1111/tra.12343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzoli S.O. 2014. Synaptic vesicle recycling: steps and principles. EMBO J. 33:788–822. 10.1002/embj.201386357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.S. 2015. Forty years of clathrin-coated vesicles. Traffic. 16:1210–1238. 10.1111/tra.12335 [DOI] [PubMed] [Google Scholar]
- Shih W., Gallusser A., and Kirchhausen T.. 1995. A clathrin-binding site in the hinge of the beta 2 chain of mammalian AP-2 complexes. J. Biol. Chem. 270:31083–31090. 10.1074/jbc.270.52.31083 [DOI] [PubMed] [Google Scholar]
- Strickland D., Lin Y., Wagner E., Hope C.M., Zayner J., Antoniou C., Sosnick T.R., Weiss E.L., and Glotzer M.. 2012. TULIPs: tunable, light-controlled interacting protein tags for cell biology. Nat. Methods. 9:379–384. 10.1038/nmeth.1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subach F.V., Malashkevich V.N., Zencheck W.D., Xiao H., Filonov G.S., Almo S.C., and Verkhusha V.V.. 2009. Photoactivation mechanism of PAmCherry based on crystal structures of the protein in the dark and fluorescent states. Proc. Natl. Acad. Sci. USA. 106:21097–21102. 10.1073/pnas.0909204106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M.J., Perrais D., and Merrifield C.J.. 2011. A high precision survey of the molecular dynamics of mammalian clathrin-mediated endocytosis. PLoS Biol. 9:e1000604 10.1371/journal.pbio.1000604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Haar E., Harrison S.C., and Kirchhausen T.. 2000. Peptide-in-groove interactions link target proteins to the beta-propeller of clathrin. Proc. Natl. Acad. Sci. USA. 97:1096–1100. 10.1073/pnas.97.3.1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traub L.M. 2009. Tickets to ride: selecting cargo for clathrin-regulated internalization. Nat. Rev. Mol. Cell Biol. 10:583–596. 10.1038/nrm2751 [DOI] [PubMed] [Google Scholar]
- van Bergeijk P., Adrian M., Hoogenraad C.C., and Kapitein L.C.. 2015. Optogenetic control of organelle transport and positioning. Nature. 518:111–114. 10.1038/nature14128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner E., and Glotzer M.. 2016. Local RhoA activation induces cytokinetic furrows independent of spindle position and cell cycle stage. J. Cell Biol. 213:641–649. 10.1083/jcb.201603025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willox A.K., and Royle S.J.. 2012. Functional analysis of interaction sites on the N-terminal domain of clathrin heavy chain. Traffic. 13:70–81. 10.1111/j.1600-0854.2011.01289.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlatic S.A., Grossniklaus E.J., Ryder P.V., Salazar G., Mattheyses A.L., Peden A.A., and Faundez V.. 2013. Chemical-genetic disruption of clathrin function spares adaptor complex 3-dependent endosome vesicle biogenesis. Mol. Biol. Cell. 24:2378–2388. 10.1091/mbc.E12-12-0860 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.