Functions of lysosomes are tightly associated with their position within the cell. Filipek et al. identify the EGF-dependent LAMTOR/Ragulator-BORC interaction as a negative regulator of Arl8b lysosomal recruitment that triggers plus-end directed lysosome movement.
Abstract
Signaling from lysosomes controls cellular clearance and energy metabolism. Lysosomal malfunction has been implicated in several pathologies, including neurodegeneration, cancer, infection, immunodeficiency, and obesity. Interestingly, many functions are dependent on the organelle position. Lysosomal motility requires the integration of extracellular and intracellular signals that converge on a competition between motor proteins that ultimately control lysosomal movement on microtubules. Here, we identify a novel upstream control mechanism of Arl8b-dependent lysosomal movement toward the periphery of the cell. We show that the C-terminal domain of lyspersin, a subunit of BLOC-1–related complex (BORC), is essential and sufficient for BORC-dependent recruitment of Arl8b to lysosomes. In addition, we establish lyspersin as the linker between BORC and late endosomal/lysosomal adaptor and mitogen activated protein kinase and mechanistic target of rapamycin activator (LAMTOR) complexes and show that epidermal growth factor stimulation decreases LAMTOR/BORC association, thereby promoting BORC- and Arl8b-dependent lysosomal centrifugal transport.
Introduction
Lysosomes can be transported bidirectionally along microtubules (Matteoni and Kreis, 1987). Rab7 regulates centripetal (inward, minus-end directed) movement by interacting with RILP, which mediates the recruitment of the dynein-dynactin motor protein complex (Cantalupo et al., 2001; Jordens et al., 2001). Inward transport can be influenced by many factors including lysosomal cholesterol and Ca2+ content (Ganley and Pfeffer, 2006; Johansson et al., 2007; Li et al., 2016).
Conversely, either Rab7 or Arl8b can mediate centrifugal (outward, plus-end directed) lysosomal movement. In the first case, protrudin, an ER-anchored Rab7-interacting protein, transfers lysosomes to the Rab7 effector FYCO1 and kinesin-1, thereby promoting outward transport (Matsuzaki et al., 2011; Raiborg et al., 2015). In the second case, the small GTPase Arl8b (Bagshaw et al., 2006) interacts with the effector protein SKIP to recruit kinesin-1 or directly binds kinesin-3 to trigger outward movement of lysosomes (Boucrot et al., 2005; Rosa-Ferreira and Munro, 2011; Wu et al., 2013). It remains unclear which stimuli promote Arl8b-dependent lysosomal movement. In general, cells respond to nutrient availability by relocating lysosomes. Starvation triggers perinuclear accumulation of lysosomes, thereby promoting fusion with autophagosomes. In contrast, lysosomes redistribute toward the cell periphery in nutrient-rich conditions (Korolchuk et al., 2011; Li et al., 2016).
It was previously shown that focal adhesion targeting by late endosomal/lysosomal adaptor and MAPK and mechanistic target of rapamycin (mTOR) activator (LAMTOR)–containing late endosomes requires kinesin-1 and Arl8b (Schiefermeier et al., 2014). Interestingly, the nucleotide loading status of Arl8b determines its subcellular localization: GTP-Arl8b associates with lysosomes whereas GDP-Arl8b displays a diffused distribution pattern (Bagshaw et al., 2006). Arl8b also requires the acetylation of its N terminus for correct membrane association (Hofmann and Munro, 2006). Moreover, it has been shown recently that the BLOC-1 (biogenesis of lysosome-related organelles complex 1)–related complex (BORC) is required for the recruitment of Arl8b to lysosomes, a prerequisite for Arl8b-dependent organelle movement (Pu et al., 2015; Guardia et al., 2016). It was suggested that the BORC could function as a guanine nucleotide exchange factor toward Arl8b, but such activity has never been demonstrated. Therefore, it remains largely unclear how BORC performs its function and how the process itself is regulated.
BORC is a multimeric complex consisting of eight subunits (LOH12CR1/myrlysin, C17orf59/lyspersin, C10orf32/diaskedin, KxDL1, MEF2BNB, BLOS1, BLOS2, and snapin; Pu et al., 2015; Guardia et al., 2016). Interestingly, BORC shares three of its subunits with BLOC-1 (Falcón-Pérez et al., 2002; Moriyama and Bonifacino, 2002; Starcevic and Dell’Angelica, 2004; Lee et al., 2012). BLOC-1–dependent cargo-specific sorting regulates maturation of specialized vesicles such as melanosomes and platelet dense granules (Dell’Angelica et al., 2000; Setty et al., 2007). BORC was shown to interact with the LAMTOR complex (Pu et al., 2015), but the function of this interaction remains elusive.
LAMTOR is a pentameric late endosomal/lysosomal scaffold complex that serves as a point of convergence/integration of nutrient status and growth factor signaling. Lipid-modified LAMTOR1 (p18; Nada et al., 2009; Magee and Cygler, 2011) anchors the remaining subunits, LAMTOR2 (p14; Wunderlich et al., 2001), LAMTOR3 (MP1; Schaeffer et al., 1998), LAMTOR4 (C7orf59), and LAMTOR5 (HBXIP; Bar-Peled et al., 2012) to the limiting membrane of the organelle. The LAMTOR2/LAMTOR3 heterodimer was previously shown to scaffold MEK and ERK on late endosomes, thereby providing spatial and temporal specificity in the MAPK pathway (Teis et al., 2002, 2006; Teis and Huber, 2003). In addition, pentameric LAMTOR interacts with the Rag GTPases and SLC38A9 (Jung et al., 2015; Rebsamen et al., 2015; Wang et al., 2015a) and has, therefore, also been named Ragulator. Through these interactions, LAMTOR/Ragulator regulates mTOR signaling in response to amino acids, thus influencing cell growth and proliferation (Sancak et al., 2008, 2010; Bar-Peled et al., 2012; Bar-Peled and Sabatini, 2014).
Genetic ablation of LAMTOR1 or LAMTOR2 in mice causes embryonic lethality (Teis et al., 2006; Nada et al., 2009), thereby identifying LAMTOR genes as essential (Blomen et al., 2015; Wang et al., 2015b). Furthermore, conditional deletion of LAMTOR2 revealed an essential function of the complex in cell/tissue homeostasis and host defense mechanisms against invading pathogens (Teis et al., 2006; Bohn et al., 2007; Taub et al., 2012; Scheffler et al., 2014; Sparber et al., 2014, 2015).
Mouse embryonic fibroblasts (MEFs) devoid of LAMTOR1 or LAMTOR2 were previously established to further elucidate the functions of the complex at the cellular level. LAMTOR2 depletion impairs cell migration by interfering with IQGAP1-dependent focal adhesion turnover (Schiefermeier et al., 2014). In addition, deletion of any one of the LAMTOR subunits leads to depletion of the remaining components (Nada et al., 2009; de Araújo et al., 2013). Finally, knockout (KO) of LAMTOR1 or LAMTOR2 results in displacement of late endosomes/lysosomes toward the cell periphery (Teis et al., 2006; Nada et al., 2009). The impaired organelle positioning (Taub et al., 2007) is associated with cargo trafficking and endosomal maturation defects (Teis et al., 2006; Takahashi et al., 2012). Of note, none of the so far known functions of the LAMTOR complex could explain the altered lysosomal positioning.
Here, we report that the LAMTOR–BORC interaction negatively regulates Arl8b-dependent endosomal positioning in mammalian cells. We show that the association between LAMTOR and the proline-rich region (PRR) of the BORC subunit lyspersin restrains the position of late endosomes/lysosomes to the perinuclear region. Moreover, we find that depletion of the LAMTOR complex increases BORC–Arl8b interaction. Finally, we show that EGF stimulation negatively regulates LAMTOR–BORC interaction, thereby promoting Arl8b-dependent organelle movement. Collectively, our results reveal a previously unknown control mechanism of lysosomal outward transport.
Results
The LAMTOR complex interacts either with the Rag GTPases, SLC38A9 and mTOR complex 1, or with BORC
To understand how LAMTOR controls lysosomal positioning, we characterized LAMTOR interaction partners in HEK293 Flp-In T-REx cells using tandem affinity purification (TAP; Glatter et al., 2009), coupled to liquid chromatography tandem mass spectrometry (LC-MS/MS; Rudashevskaya et al., 2013). LAMTOR subunits 1, 3, 4, and 5 together with their known binding partners RagA and RagC were tagged with HA–Strep II, hereafter HS (HA–Strep II) or SH (Strep II–HA) for N- or C-terminal fusion, respectively, and used as baits. That resulted in the previous identification of SLC38A9 as a component of the amino acid–sensing machinery (Rebsamen et al., 2015).
In addition to those interactions with a described function (Fig. 1 A), we also found BORC components as less prominent binding partners in the LAMTOR interactome (Fig. 1 A) as reported earlier (Jung et al., 2015; Pu et al., 2015), but not in the interactomes of RagA, RagC (Fig. 1 A).
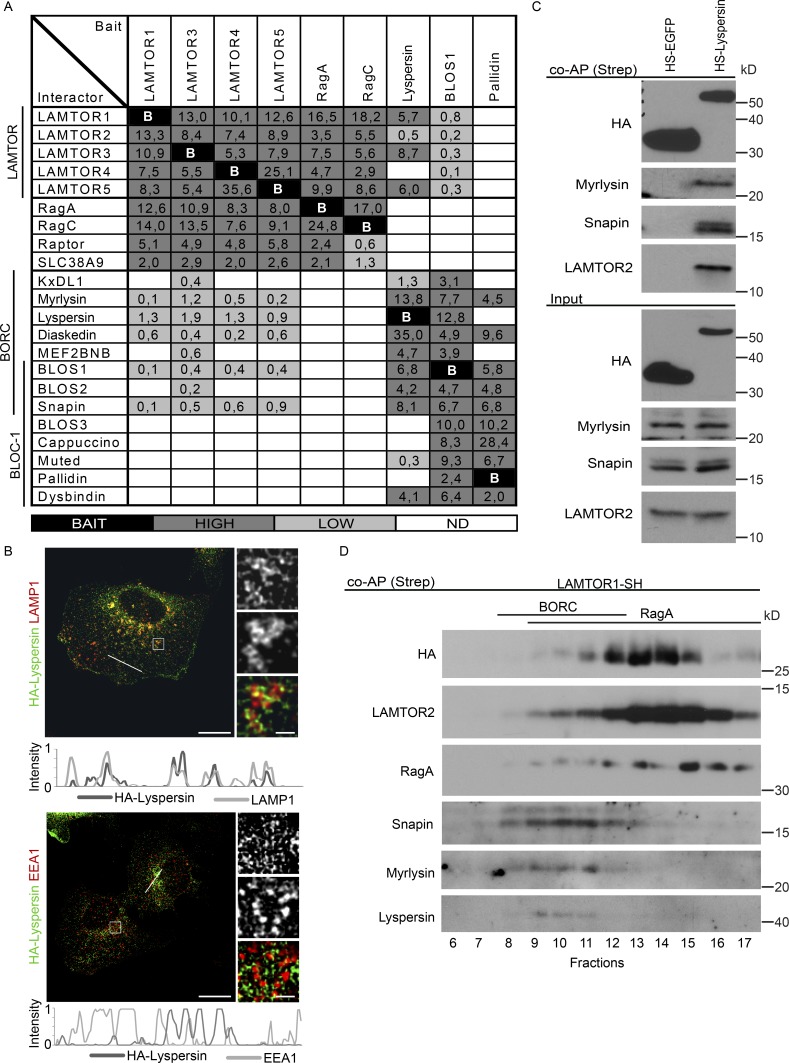
Figure 1.
The LAMTOR pentamer participates in two distinct multimeric complexes on late endosomes/lysosomes. (A) Interactome analysis identifies two LAMTOR populations associating either with RagA/C, SLC38A9, and Raptor or with BORC. HS-tagged baits (LAMTOR1, LAMTOR3, LAMTOR4, LAMTOR5, RagA, RagC, lyspersin, BLOS1, and pallidin) were purified by TAP and analyzed by LC-MS/MS. The normalized abundances (% of total spectral counts [for LAMTOR 1, 3, 4, and 5, RagA, RagC, and BLOS1] or peak areas [for lyspersin and pallidin]) of the interacting partners are shown (Table S1). BAIT (black), bait; HIGH (dark gray), high abundance interactors (≥2%); LOW (bright gray), low abundance interactors (<2%); ND (white), no interaction detected. (B) HA-lyspersin partially colocalizes with late endosomes/lysosomes (LAMP1) but not with early endosomes (EEA1). HeLa cells were transfected with HA-lyspersin. Single plane confocal images of indirect IF of HA-lyspersin (green) and endogenous endosomal markers (red) are shown. Intensity profiles for HA-lyspersin (green) and LAMP1 or EEA1 (red) along the cross section lines are shown. Representative images are shown. Bars: 10 µm; (insets) 1 µm. (C) Immunoblotting analysis of HS-lyspersin Strep-purified complexes confirms the interaction of lyspersin with myrlysin, snapin, and LAMTOR2. (D) The majority of the pentameric LAMTOR complex associates with RagA. Only a minor fraction interacts with BORC components. The eluate from Strep-purified LAMTOR1-SH was subjected to size exclusion chromatography and analyzed by WB with the indicated antibodies.
BORC was previously shown to regulate lysosomal positioning by controlling Arl8b recruitment (Pu et al., 2015), making it a relevant candidate in our search for mediators of LAMTOR-dependent lysosomal distribution. Importantly, subunits unique for BLOC-1 (BLOS3, cappuccino, muted, pallidin, and dysbindin) were not identified as LAMTOR binding partners, indicating a LAMTOR/BORC-specific interaction (Fig. 1 A). To substantiate these findings, we have used three baits belonging to three distinct groups: lyspersin, which is present only in BORC; pallidin, which is specific for BLOC-1; and BLOS1, a subunit that is shared by both complexes. The interactome of pallidin contained all BLOC-1 components together with myrlysin and diaskedin, but not LAMTOR. The BLOS1 interactome contained both BORC and BLOC-1 subunits, confirming BLOS1 as a shared subunit (Fig. 1 A).
Only the lyspersin and the BLOS1 interactomes contained the LAMTOR complex and all remaining BORC proteins, but not the Rag GTPases, Raptor or SLC38A9. These data support the existence of two distinct LAMTOR populations, which can interact either with Rags, SLC38A9 and Raptor, or with the BORC, and complements earlier findings by (Pu et al., 2015), reporting LAMTOR components in BLOS2 interactome. These results also attested to the specificity of the LAMTOR–BORC interaction (Fig. 1 A). Lyspersin was the most abundant BORC interactor of the LAMTOR complex (Fig. 1 A), and immunofluorescence (IF) of HeLa cells showed that it partially associated with late endosomes/lysosomes (Fig. 1 B), as reported previously (Schweitzer et al., 2015), but not with early endosomes (Fig. 1 B).
Using HS-lyspersin as bait, we confirmed the association of lyspersin with both LAMTOR (LAMTOR2) and BORC (snapin, myrlysin) components (Fig. 1 C). Moreover, exogenously expressed lyspersin incorporated into the endogenous BORC complex and interacted with the LAMTOR complex (Fig. S1 A). Finally, to corroborate the presence of LAMTOR in those multimeric complexes and to address the ratio between the two, we have analyzed affinity-purified LAMTOR1-SH–interacting proteins by size exclusion chromatography and Western blot analysis (Fig. 1 D). As a positive control, LAMTOR2, an integral component of the LAMTOR complex, shared the elution profile with the bait. RagA was present in fractions 9–17, whereas BORC subunits (snapin, myrlysin, and lyspersin) were found only in fractions 8–12 (Fig. 1 D). Collectively, fractions 13–14 that contained most of the bait were positive for RagA but negative for BORC. On the other hand, fractions 8–12 that contained the bulk of the interacting BORC components displayed much lower levels of the bait (Fig. 1 D). Collectively, these data indicate that the majority of LAMTOR associates with Rags, SLC38A9 and Raptor, whereas a minor fraction associates with BORC.
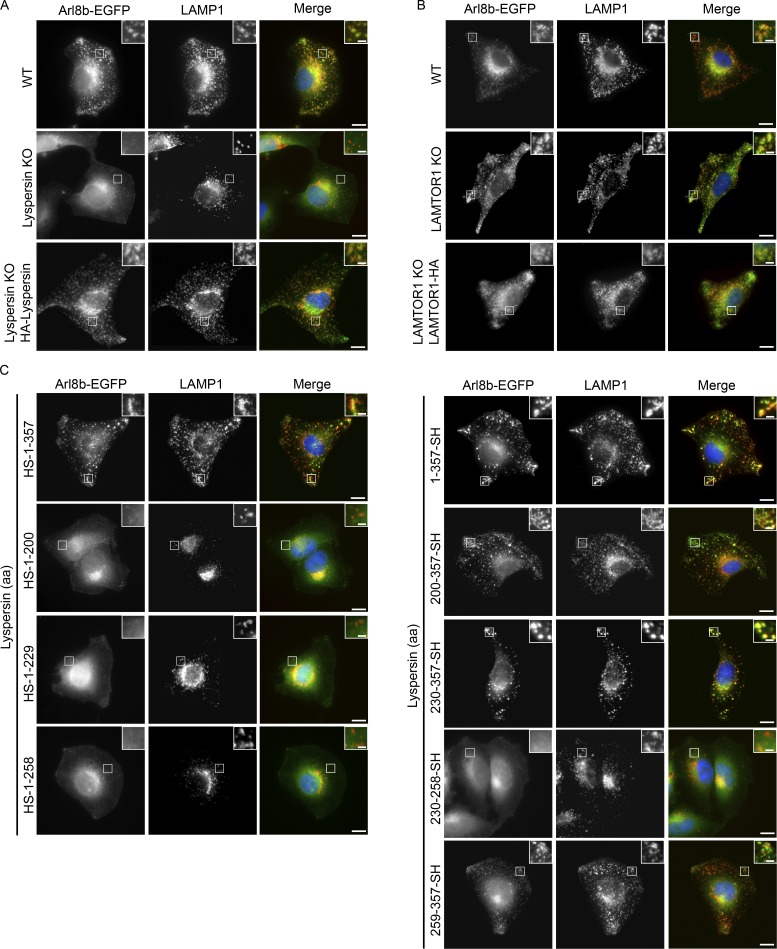
We have shown previously that deletion of LAMTOR2 decreases the protein levels of the remaining LAMTOR subunits (de Araújo et al., 2013), because the pentameric complex can no longer be formed and the individual LAMTOR subunits are degraded. Knowing that LAMTOR and BORC interact with each other, we next addressed if their stability is interdependent. We deleted LAMTOR1 and lyspersin using the CRISPR/Cas9 technology. LAMTOR1 deletion compromised the levels of LAMTOR 2, 3, 4 and 5, and importantly, reconstitution of LAMTOR1 KO cells with LAMTOR1-HA restored the complex stability (Fig. S1 B). The expression levels of BORC subunits (snapin, myrlysin) remained unchanged upon deletion of LAMTOR1 (Fig. S1 B). Interestingly, deletion of lyspersin also had no impact on the protein levels of either BORC or LAMTOR components (Fig. S1 C).
The LAMTOR–BORC interaction is lyspersin-dependent
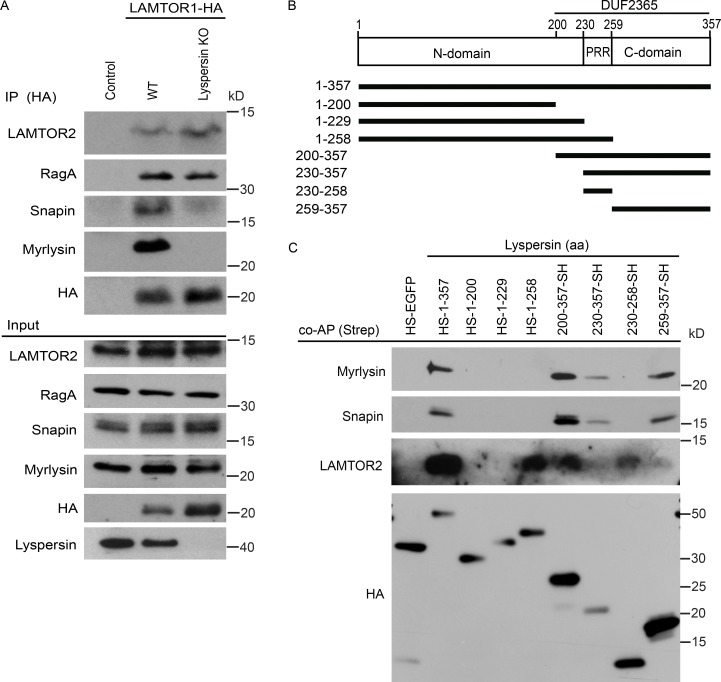
We considered two main possibilities to mechanistically link the two complexes: (1) there are multiple contact sites between LAMTOR and BORC subunits, and deletion of lyspersin will not abolish this interaction; or (2) lyspersin is essential for this interaction, and its depletion will prevent contact between the two complexes. To test these hypotheses, we have expressed LAMTOR1-HA in WT and lyspersin KO cells. In the WT cells, we could detect the interaction of the bait with LAMTOR2, RagA, and BORC (snapin and myrlysin; Fig. 2 A). In the case of lyspersin KO cells, the interaction between LAMTOR1-HA and LAMTOR2 or RagA remained unaffected, but the interaction of LAMTOR1-HA with the remaining BORC subunits (BORCΔlyspersin) was completely abolished (Fig. 2 A).
Figure 2.
The LAMTOR–BORC interaction is lyspersin-dependent. (A) Deletion of lyspersin abolishes the interaction between LAMTOR and the remaining BORC subunits (BORCΔlyspersin). In contrast, the LAMTOR-RagA interaction remains unaffected. HA immunoprecipitates from control, WT, and lyspersin KO HeLa cells stably expressing LAMTOR1-HA were analyzed by immunoblotting. (B) Annotated representation of lyspersin truncation mutants. N-domain, N-terminal region; C-domain, C-terminal region. (C) Lyspersin interacts with the LAMTOR complex through its PRR whereas the C-domain of lyspersin is responsible for the interaction with the remaining BORC subunits. The DUF2365 domain is sufficient to mediate the interaction with both LAMTOR and BORCΔlyspersin. Immunoblotting analysis of Strep coprecipitates from lyspersin truncation mutants inducibly expressed in HEK293 Flp-In T-REx cells.
Lyspersin contains a domain of unknown function (DUF) 2365, which is highly conserved among species. The Pfam protein family’s database search (http://pfam.xfam.org; Finn et al., 2016) identified proteins containing DUF2365 domain in 177 species. Within this domain we found a PRR. The regions flanking the PRR were named N-domain and C-domain, referring to their relative position within the lyspersin sequence (Fig. 2 B). Based on these observations, we have generated truncation mutants of lyspersin, tagged with SH (Fig. 2 B). Coaffinity purification of those mutants allowed us to identify domains required for the interaction with LAMTOR or BORC. The PRR was required and sufficient for the interaction with the LAMTOR complex, whereas lyspersin C-domain was essential for the binding to the BORC (Fig. 2 C). Both regions are part of the DUF2365 domain. Although the relatively low levels of the lyspersin truncation mutant 230–357-SH were sufficient to detect an interaction with myrlysin and snapin, the interaction with LAMTOR2 was below the detection limit. Surprisingly, the N-domain alone did not bind any of the complexes, and its function remains elusive to this date.
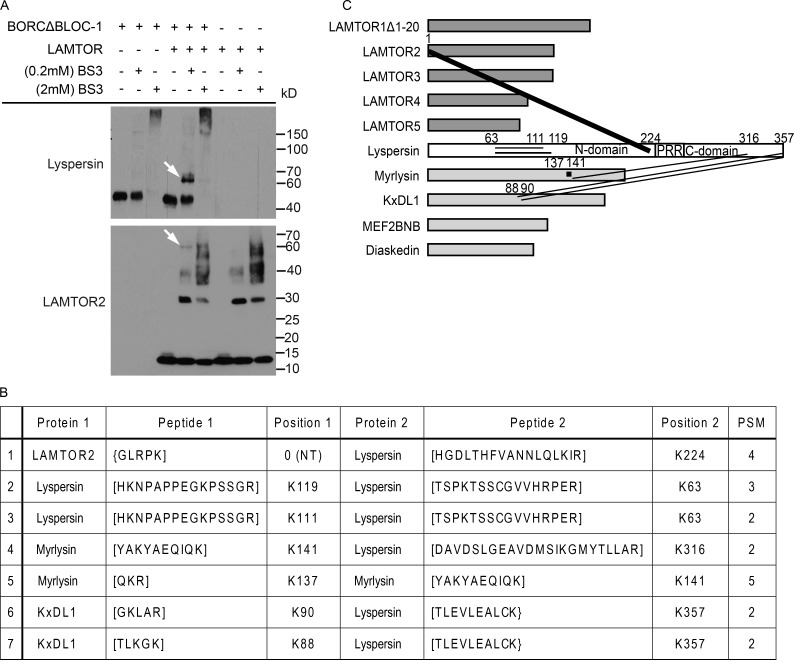
To corroborate this domain mapping, we performed in vitro cross-linking mass-spectrometry experiments (Herzog et al., 2012). The entire LAMTOR complex (LAMTOR1Δ1–20 and LAMTOR2–5) or 5 BORC-specific subunits (lyspersin, myrlysin, KxDL1, MEF2BNB, and diaskedin), hereafter referred to as BORCΔBLOC-1, were purified from Sf21 insect cells. The equimolar mixture of the complexes and individual complexes as controls were cross-linked with isotope-labeled amine-reactive BS3. Western blot analysis revealed an interaction between lyspersin and LAMTOR2 at 0.2 mM cross-linker (Fig. 3 A, arrow). Established cross-linking conditions were applied to further mass spectrometry analysis. After detailed evaluation of the mass spectrometry data (Fig. 3 B), it became clear that the N terminus of LAMTOR2 cross-linked with lysine K224 in lyspersin, the closest to the PRR, whereas BORC subunits (myrlysin and KxDL1) cross-linked with K316 and K357 of the lyspersin C-domain, respectively (Fig. 3 C), consistent with the aforementioned experiments (Fig. 2 C). In addition, we have identified intramolecular cross-links within lyspersin (K63-K111, K63-K119) and myrlysin (K137-K141; Fig. 3, B and C). Collectively, lyspersin binds the LAMTOR complex and BORCΔlyspersin with two distinct domains, PRR and C-domain, respectively, and is crucial for the interaction between the two complexes.
Figure 3.
The direct inter- and intramolecular interactions of LAMTOR and BORCΔBLOC-1 detected by in vitro chemical cross-linking. (A) Cross-linking of in vitro copurified LAMTOR (LAMTOR1Δ1-20, LAMTOR2, LAMTOR3, LAMTOR4, and LAMTOR5) and BORCΔBLOC-1 (lyspersin, myrlysin, KxDL1, MEF2BNB, and diaskedin) complexes with 0.2 mM BS3 results in a higher molecular weight band (arrow) detected by anti-lyspersin and anti-LAMTOR2 antibodies. (B) List of unique cross-links identified by mass spectrometry. Recombinant protein complexes were purified and cross-linked as described in A. The cross-linked proteins were digested and the tryptic peptides analyzed by mass spectrometry. Annotated are the cross-linked proteins, relevant peptide sequences, the lysines being cross-linked and the total number of identified peptide spectra matched (PSM). (C) The N terminus of LAMTOR2 cross-links with K224 of lyspersin (closest lysine to PRR); myrlysin and KxDL1 cross-link with the C-domain of lyspersin. The thickness of the lanes in the graphical representation corresponds to the number of cross-links identified in B.
Lyspersin regulates endosomal positioning
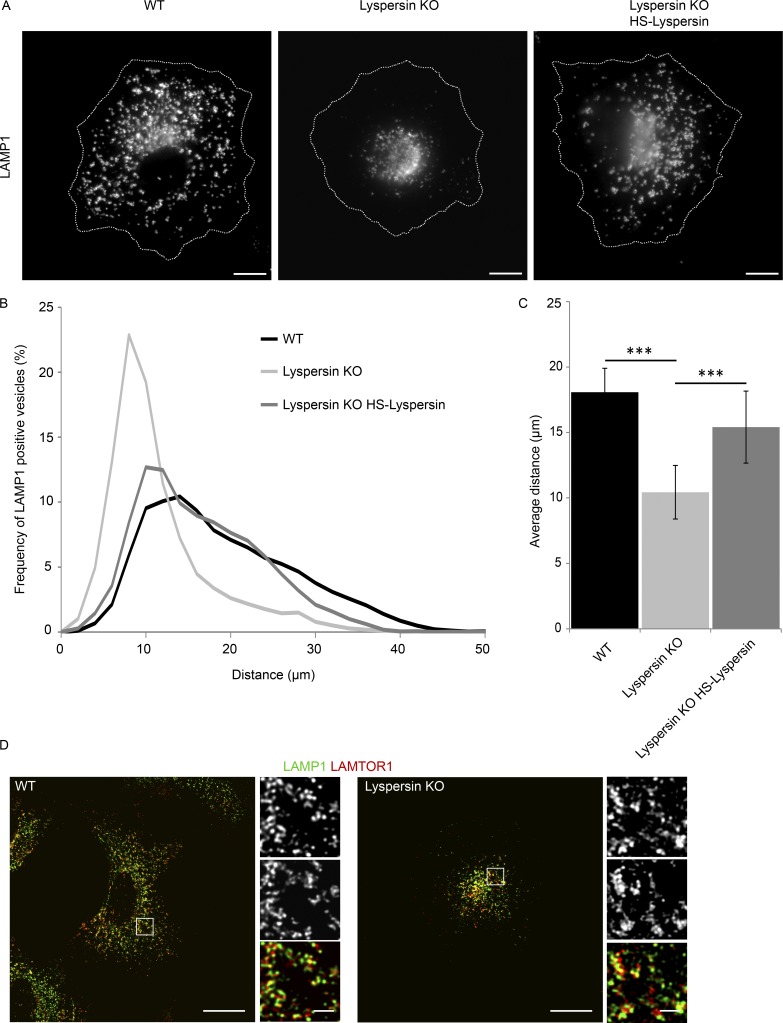
In lyspersin KO cells, the late endosomes/lysosomes clustered in the juxtanuclear region (Fig. 4 A and Video 1), the phenotypical opposite of the peripheral late endosome displacement found in LAMTOR-deficient mutants (Teis et al., 2006; Nada et al., 2009). Reconstitution of lyspersin in KO cells restored the positioning of lysosomes (Fig. 4 A). The same phenotype has been shown for two other BORC subunits, myrlysin and diaskedin (Pu et al., 2015; Guardia et al., 2016). Endosomal distribution analysis confirmed the increased number of perinuclear lysosomes in lyspersin KO cells (Fig. 4 B). As a consequence, the mean distance of the lysosomes to the center of the nucleus was significantly shorter in lyspersin-depleted cells compared with controls (Fig. 4 C). Finally, the clustering of late endosomes/lysosomes observed in lyspersin KO cells had no impact on the association of LAMTOR with lysosomes (Fig. 4 D).
Figure 4.
Lyspersin regulates endosomal positioning. (A) Deletion of lyspersin results in the accumulation of late endosomes/lysosomes in the perinuclear region, and the distribution can be restored by reconstitution of the lyspersin KO cells with a tagged version of the protein. Indirect IF images of HeLa WT, lyspersin KO, and rescue cell line. Representative single-channel images of endogenous lysosomal marker LAMP1 are shown. Bars, 10 µm. (B) Deletion of lyspersin leads to an increase in the number of perinuclear LAMP1-positive endosomes. Images were taken on a confocal microscope and analyzed using Huygens and Imaris software. Number of cells per genotype, n ≥ 15. Depicted is the percentage of late endosomes found at a given distance from the center of nucleus. (C) The mean distance of late endosomes/lysosomes to the center of nucleus is significantly smaller in lyspersin KO cells than in WT or rescued cells. Images were analyzed as in B. Mean ± SD. ***, P ≤ 0.001. n ≥ 15 cells. WT/lyspersin KO, P = 3.0 × 10-12; lyspersin KO HS-lyspersin/lyspersin KO, P = 1.2 × 10-6. (D) LAMTOR1 localizes to late endosomes in a lyspersin independent manner. Control and lyspersin depleted cells were analyzed. Shown are representative, single plane confocal images of indirect IF of endogenous LAMTOR1 (red) and endogenous LAMP1 (green). Bars: 10 µm; (inset) 1 µm.
Arl8b requires lyspersin but not LAMTOR for recruitment to late endosomes/lysosomes
Myrlysin deletion blocks recruitment of Arl8a and Arl8b to lysosomes (Pu et al., 2015; Guardia et al., 2016). Bearing in mind the pivotal function of Arl8b in endosomal transport, we addressed its subcellular localization in lyspersin KO cells. Arl8b-EGFP localized to LAMP1-positive vesicles in WT cells, but deletion of lyspersin blocked this process, leading to a diffused distribution of Arl8b-EGFP and perinuclear accumulation of lysosomes (Figs. 5 A and S2 A). Both phenotypes were restored by lyspersin reconstitution (Figs. 5 A and S2 A). In contrast, Arl8b-EGFP colocalized with LAMP1 in LAMTOR1 KO and controls, indicating that the LAMTOR complex was not required for Arl8b recruitment to late endosomes/lysosomes (Fig. 5 B). A similar analysis was performed in LAMTOR2 −/− MEFs and respective controls, in which we detected lysosomal Arl8b-EGFP under all conditions (Fig. S2 B). In addition, we confirmed that exogenously expressed Arl8b-EGFP did not affect the characteristic peripheral displacement of LAMP1-positive vesicles found in LAMTOR1 KO and LAMTOR2 −/− cells (Fig. S2, C and D; Teis et al., 2006; Nada et al., 2009). Moreover, lyspersin partially localized to lysosomes in LAMTOR2 f/− and −/− MEFs, indicating that the LAMTOR complex was not required for its recruitment to the vesicles (Fig. S2 E). Next, we identified the domain of lyspersin essential for Arl8b recruitment. Arl8b-EGFP was transiently coexpressed with full-length lyspersin and truncation mutants (Fig. 2 B) in lyspersin KO cells. Independent of the tag position, the expression of full-length lyspersin restored lysosomal recruitment of Arl8b-EGFP in lyspersin KO cells (Fig. 5 C, compare top row of images in both panels). Truncation mutants expressing the region complementary to DUF2365 (HS-1-200), the N-domain alone (HS-1-229), or together with the PRR (HS-1-258) were not sufficient to recruit Arl8b-EGFP to LAMP1-positive vesicles in lyspersin KO cells (Fig. 5 C, left). The same diffused localization of Arl8b-EGFP was observed in the KO cells expressing PRR-SH (230-258-SH). In all those conditions, we also observed perinuclear clustering of lysosomes (Fig. S2, F and G), typical for lyspersin (Fig. 4 A), myrlysin, or diaskedin KOs (Pu et al., 2015; Guardia et al., 2016). The truncation mutants expressing DUF2365 (200–357-SH), PRR with C-domain (230-357-SH), and containing the C-domain (259–357-SH) only (Fig. 2 B), were able to restore Arl8b-EGFP recruitment to lysosomes, just like the WT protein. This resulted in centrifugal transport of LAMP1-positive vesicles (Fig. 5 C, right; and Fig. S2 G).
Figure 5.
Arl8b requires lyspersin but not LAMTOR for late endosomal/lysosomal localization. (A) Lyspersin is essential to recruit Arl8b to LAMP1-positive endosomes. Control WT, lyspersin KO, and rescue cell lines were transfected with Arl8b-EGFP. Representative, merged, and single-channel images of endogenous LAMP1 (red) and Arl8b-EGFP (green) are shown. Bars: 10 µm; inset, 2 µm. (B) Arl8b recruitment to late endosomes/lysosomes is LAMTOR-independent. Control WT, LAMTOR1 KO, and rescue cell lines were transfected with Arl8b-EGFP. Experiment was performed as in A. Representative images are shown. Bars: 10 µm; inset, 2 µm. (C) The C-domain of lyspersin or truncation mutants containing the C-domain are sufficient to restore the recruitment of Arl8b to LAMP1-positive endosomes in lyspersin KO cells. Lyspersin KO cells were cotransfected with Arl8b-EGFP and truncation mutants of lyspersin (Fig. 2 B). Experiment was performed as in A. Representative images are shown. Bars: 10 µm; (inset) 2 µm.
We next examined whether the nucleotide load of Arl8b controls its interaction with BORC. Cells expressing control HS-EGFP and HS-lyspersin were transfected with either Arl8b-EGFP QL (Q75L mutant; GTP-locked, active) or Arl8b-EGFP TN (T34N mutant; GDP-locked, inactive; Bagshaw et al., 2006). Coaffinity purification revealed that BORC interacted with Arl8b independent of its GTP/GDP loading status (Fig. S3 A). Taking into consideration the impact of lyspersin deletion on Arl8b distribution (Fig. 5 A), we next investigated whether the localization patterns of the QL and TN mutants were altered upon lyspersin KO. The diffused distribution, characteristic of inactive Arl8b, was not affected by lyspersin depletion (Fig. S3 B). In contrast, the constitutively active QL mutant displayed a dispersed localization on lyspersin KO but remained associated with lysosomes in WT and HS-lyspersin reconstituted cells (Fig. S3 C). Furthermore, transfection of WT Arl8b or Arl8b nucleotide-locked mutants did not alter the perinuclear accumulation of lysosomes found in lyspersin KO cells (Fig. 4 B; Fig. S2 A; and Fig. S3, D and E). Collectively, these data suggested that the C-terminal part of lyspersin mediated the interaction with the remaining BORC components (Figs. 2, B, and C; and Fig. 3 C) and was necessary and sufficient to promote BORC-dependent recruitment of GTP-loaded Arl8b to lysosomes.
LAMTOR controls endosomal positioning by negatively regulating the BORC–Arl8b interaction
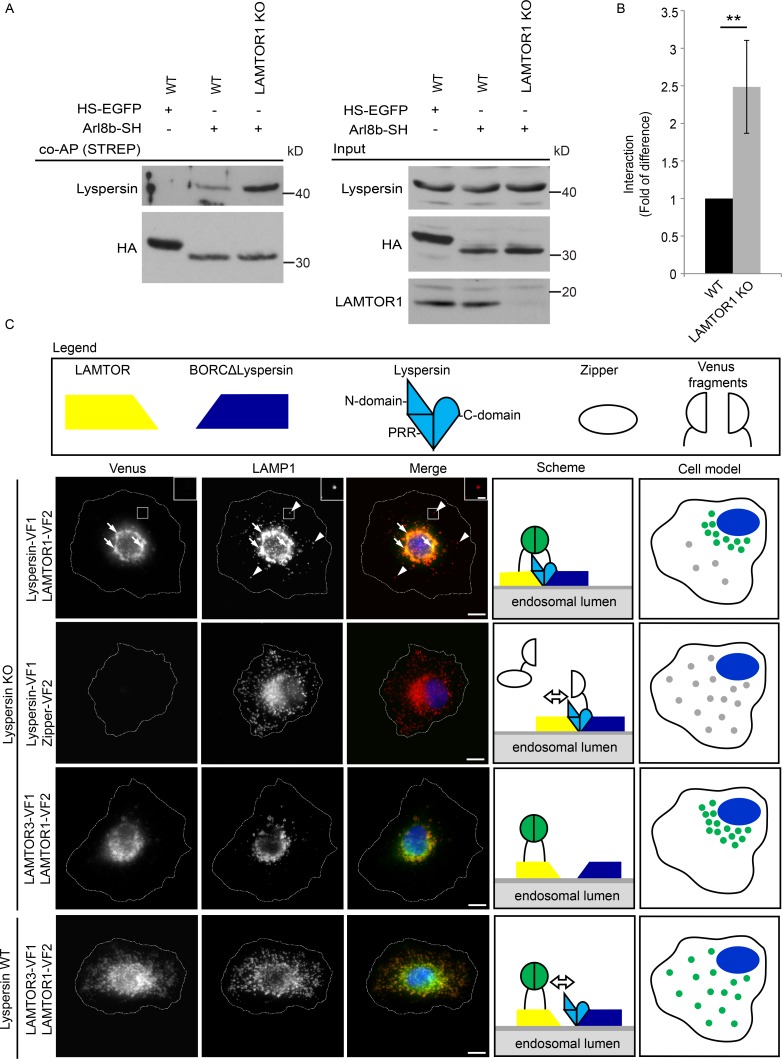
We next asked whether BORC interactions with LAMTOR and Arl8b are mutually exclusive, and therefore affinity-purified HS-Arl8b (Fig. 6 A) from control and LAMTOR1 KO cells was analyzed. Indeed, the BORC–Arl8b (lyspersin) association was considerably increased upon depletion of LAMTOR (Fig. 6, A and B), implicating LAMTOR as a negative regulator of the BORC–Arl8b interaction.
Figure 6.
LAMTOR controls endosomal positioning by negatively regulating the BORC–Arl8b interaction. (A) The interaction of Arl8b-SH with the BORC subunit lyspersin is increased in LAMTOR1 KO cells. Strep-purified Arl8b-SH or HS-EGFP from LAMTOR1 KO and WT cells were analyzed by immunoblotting. (B) Results of four biologically independent experiments were quantified. Mean ± SD. **, P ≤ 0.01; P = 0.0059. (C) In lyspersin KO cells, the irreversible association of BORC with LAMTOR, achieved by Venus fluorophore reconstitution (lyspersin-VF1, LAMTOR1-VF2), immobilizes LAMP1-positive endosomes in the perinuclear region. No interaction (Venus signal) was detected in cells expressing lyspersin-VF1 and the control Zipper-VF2. Lyspersin-VF1 expression in lyspersin KO cells restores transportation of late endosomes toward the cell periphery. The overexpression of two LAMTOR subunits (LAMTOR3-VF1 and LAMTOR1-FV2) results in Venus-fluorophore reconstitution, indicating an interaction between both proteins in lyspersin KO and WT cells. The Venus signal in peripheral LAMP1 vesicles is found exclusively in WT cells. Analysis of Venus-fluorophore reconstitution (green) and indirect IF of endogenous LAMP1 (red). Representative images are shown. Arrows indicate endosomes positive for LAMP1 and BORC–LAMTOR irreversible interaction (Venus). Arrowheads show LAMP1-positive but BORC–LAMTOR interaction–negative late endosomes. Dashed line indicates cell borders. Representative images are shown. Bars: 10 µm; (inset) 2 µm. Legend: yellow trapezoid, LAMTOR complex; dark blue trapezoid, BORCΔlyspersin; light blue shape with discrimination of individual domains (N-domain, PRR, and C-domain), lyspersin; white ellipse, Zipper (negative control); white semicircles, VFs; two green semicircles together, Venus reconstitution and interaction between tagged proteins. The adjacent cell model depicts the presence (green) or absence of interaction (gray) and the overall position of late endosomes in the transfected cells.
To functionally address the LAMTOR–BORC interaction in lysosomal positioning, we used a protein-fragment complementation assay (PCA; Remy and Michnick, 1999; Michnick et al., 2000; Magliery et al., 2005; Remy et al., 2007) to irreversibly lock interactions between LAMTOR and BORC or two LAMTOR subunits on late endosomes/lysosomes. In lyspersin KO cells, cotransfected with lyspersin-Venus fragment (VF) 1 and LAMTOR1-VF2, we observed two populations of LAMP1-positive vesicles by IF: some were localized more toward the cell periphery (Fig. 6 C, arrowheads), whereas others clustered around the nucleus (Fig. 6 C, arrows). The Venus signal, indicating the irreversible interaction of lyspersin with LAMTOR, was exclusively detected on perinuclear LAMP1-positive structures (Fig. 6 C and Fig. S4 A). In contrast, the coexpression of lyspersin-VF1 and the negative control Zipper-VF2 (irrelevant amino acid sequence/nonbinding polypeptide) reconstituted BORC function and restored centrifugal movement of lysosomes (Fig. S4 B). Under these conditions, no Venus signal was detected, indicating no interaction between the cotransfected proteins (Fig. 6 C). Coexpression of LAMTOR3-VF1 with LAMTOR1-VF2 confirmed their interaction on lysosomes in both WT and lyspersin KO cells, but failed to rescue the perinuclear clustering of endosomal vesicles, characteristic of lyspersin KO cells (Figs. 6 C and S4B). Overall, these results explained the observed antagonistic phenotypes of LAMTOR and BORC deletions. Following our rationale, cotransfection of the C-domain lyspersin-VF1, which cannot interact with LAMTOR (Fig. 2, B and C), and LAMTOR1-VF2 in lyspersin KO cells reversed the perinuclear clustering of lysosomes without showing an interaction between the two introduced proteins (no Venus signal; Fig. S5, A and B). We observed the same results when the C-domain of lyspersin was used together with the negative control Zipper-VF2 (Fig. S5, A and B). To confirm the specificity of the assay, we have performed additional controls (Fig. S5 A). When introducing the pairs Zipper-VF1 and LAMTOR1-VF2 or Zipper-VF1 and Zipper-VF2, no Venus signal was detected. In addition, under these conditions, we could not rescue the perinuclear accumulation of lysosomes caused by lyspersin deletion (Fig. 4, A and B; and Fig. S5, A and B). In conclusion, our results indicate that the binding of LAMTOR to BORC (via lyspersin), negatively regulates Arl8b function and thereby controls endosomal positioning.
EGF regulates LAMTOR–BORC interaction, thereby controlling lysosomal movement
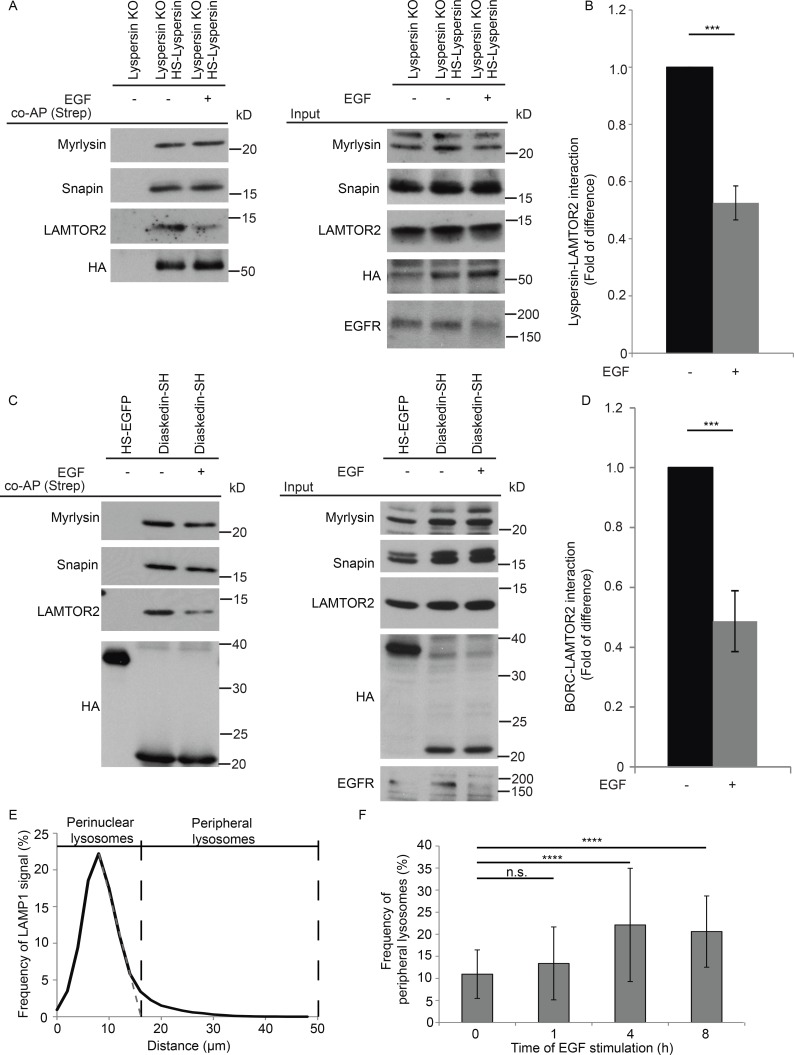
Lysosomal distribution was shown to change in response to availability of nutrients, leading to perinuclear clustering of the organelles in starved conditions (Korolchuk et al., 2011). The same work nicely demonstrates that amino acid–dependent mTOR signaling, which is also controlled by the LAMTOR complex (Sancak et al., 2010; Rebsamen et al., 2015; Wang et al., 2015a), has no impact on the position of lysosomes. In addition, it has been shown that extended EGF stimulation leads to the relocalization of LAMP1-positive vesicles toward the cell periphery (Dykes et al., 2016). Intriguingly, we have previously shown that LAMTOR is crucial for EGF-induced MAPK signaling on late endosomes (Teis et al., 2002). Therefore, we considered that EGF might regulate positioning of lysosomes by affecting the LAMTOR–BORC interaction. To test this hypothesis, we performed coaffinity purification (Strep), using as bait HS-lyspersin stably expressed in lyspersin KO cells. Cells were starved overnight for FBS and stimulated with EGF (100 ng/ml) for 1 h or kept unstimulated (Fig. 7 A). Western blot analysis showed a significant decrease in the interaction between HS-lyspersin and LAMTOR2 upon EGF stimulation (Fig. 7, A and B). Under the same conditions, the interaction between HS-lyspersin and other BORC subunits (myrlysin and snapin) remained unaffected (Fig. 7 A). We corroborated these data in an independent biological system using HEK293 Flp-In T-REx cells expressing diaskedin-SH (Fig. 7, C and D). Collectively, the results indicate that EGF promotes the disassembly of the LAMTOR–BORC interaction.
Figure 7.
EGF regulates LAMTOR–BORC interaction. (A) The interaction between BORC and LAMTOR complexes is decreased upon EGF stimulation. Lyspersin KO cells stably expressing HS-lyspersin were starved for FBS and stimulated with 100 ng/ml EGF for 1 h. Obtained lysates were Strep-purified and analyzed by immunoblotting. EGFR degradation was used as control for the treatment. (B) Quantification of the results of three biologically independent experiments. Mean ± SD. ***, P ≤ 0.001; P = 0.000352. (C) Confirmation that the interaction between BORC and LAMTOR complexes is decreased upon EGF stimulation with HEK293 Flp-In T-REx cells inducibly expressing diaskedin-SH starved and stimulated as in A. Immunoblotting analysis of the Strep-purified complexes. EGFR degradation was used as control for the treatment. (D) Quantification of the results of three biologically independent experiments. Mean ± SD. ***, P ≤ 0.001; P = 0.000938. (E) Arbitrary cutoff for peripheral late endosomes based on LAMP1-positive vesicles distribution in starved WT HeLa cells. Epifluorescence images were analyzed using ImageJ software with our macro RadialIntensityProfile (Materials and methods). Depicted is the signal intensity of LAMP1 found at the indicated distance from the center of nucleus, perinuclear lysosomes ≤16 µm, peripheral lysosomes ≥16 µm. Number of cells per genotype, n ≥ 50 cells. (F) Frequency of peripheral lysosomes increases upon EGF stimulation. WT HeLa cells were starved for FBS overnight and stimulated with EGF for the indicated time points and subsequently treated as in E. Depicted is the signal intensity of LAMP1 found at ≥16 µm from the center of the nucleus. Mean ± SD. Not significant (n.s.), P > 0.05; ****, P ≤ 0.0001 (0 h/1 h P = 0.630, 0 h/4 h P = 1.1 × 10-8, 0 h/8 h P = 3.2 × 10-7). Number of cells per genotype, n ≥ 50 cells.
To confirm the EGF-induced centrifugal movement of lysosomes, we quantified the frequency of peripheral lysosomes over a long stimulation time course. An arbitrary cutoff for peripheral lysosomes was established based on the LAMP1 signal distribution profile in WT HeLa cells starved overnight for FBS (Fig. 7 E). The frequency of peripheral lysosomes remained unchanged after 1 h of EGF stimulation (Fig. 7 F), a period that is sufficient for the disassembly of the LAMTOR–BORC interaction (Fig. 7 A). Later time points significantly increased the frequency of peripheral lysosomes as judged by LAMP1 intensity (Fig. 7 F). This temporal delay in the centrifugal redistribution of the vesicles might be triggered by the fact that EGF was sufficient to regulate LAMTOR–BORC assembly but not the downstream Arl8b-SKIP interaction (Rosa-Ferreira and Munro, 2011). Moreover, it has been recently shown that the majority of late endosomes form a “perinuclear cloud” and that only a minor fraction could move bidirectionally (Jongsma et al., 2016).
Future work will determine if EGF regulates LAMTOR–BORC binding upstream of or directly through MAPK signaling or, for instance, by recruitment of the MEK1-protein to the LAMTOR complex (Teis et al., 2002), independent of its kinase activity toward ERK1.
Discussion
Lysosomes were long considered cellular waste bags, but their function is more complex than previously anticipated (Appelqvist et al., 2013). Lysosomes play a role in killing of intracellular pathogens, antigen presentation, plasma membrane repair, exosome release, cellular adhesion and migration, apoptosis, metabolic signaling and gene regulation, tumor invasion and metastasis, as reviewed recently (Pu et al., 2016). In this review, Pu et al. (2016) beautifully make the case that many of the identified lysosomal functions are dependent on the organelle positioning and motility within the cytoplasm.
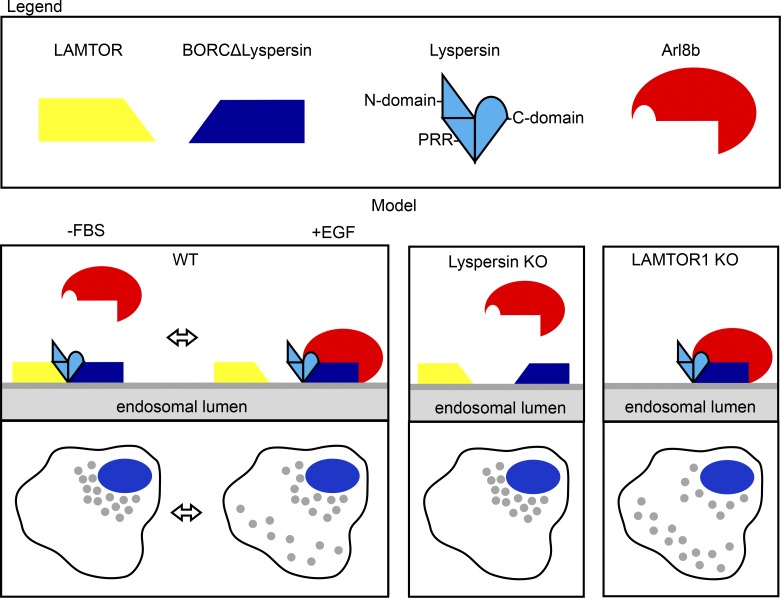
Here we show that LAMTOR is a negative regulator of BORC–Arl8b interaction. BORC binds the LAMTOR complex through the PRR of lyspersin, leading to perinuclear accumulation of lysosomes. The C-domain of lyspersin interacts with the remaining BORC subunits and is essential and sufficient for BORC function toward Arl8b. Moreover, we identified EGF as an upstream trigger regulating the strength of the LAMTOR–BORC interaction. Based on our findings, we propose the following model: at any given moment, different lysosomes coexist in distinct locations within cells. Those in which LAMTOR and BORC interact are unable to recruit Arl8b and remain perinuclear (Fig. 8). This population is increased upon FBS starvation. On the other hand, lysosomes, negative for the LAMTOR–BORC interaction, can be transported toward the cell periphery in an Arl8b-dependent manner. Their number is increased upon EGF stimulation (Fig. 7 F). This model also mechanistically explains the phenotypes previously observed in BORC and LAMTOR KO cells. Arl8b recruitment is compromised by lyspersin deletion (Fig. 8), leading to perinuclear accumulation of lysosomes. In contrast, in LAMTOR KO cells, the absence of the negative regulator enhances BORC–Arl8b interaction, leading to increased centrifugal movement of lysosomes (Fig. 8). In addition, we have shown that BORC interacts with Arl8b independent of its nucleotide-loading status (Fig. S3 A). In contrast, SKIP-kinesin-1 and kinesin-3 were shown to associate with GTP-bound Arl8b (Rosa-Ferreira and Munro, 2011; Wu et al., 2013). This shows that Arl8b-dependent movement of the vesicles is tightly regulated at different levels. Of note, based on overexpression experiments, it has been proposed that lyspersin could be a negative regulator of mTOR complex 1 (mTORC1) activity (Schweitzer et al., 2015). However, the same publication also reports that loss of lyspersin did not result in detectable defects in mTORC1 signaling in response to starvation or restimulation (Schweitzer et al., 2015). In contrast, the results presented here indicate that lyspersin associates with LAMTOR as part of the BORC complex, thereby regulating lysosomal positioning. We would like to emphasize that this function is mediated by a subset of the LAMTOR complexes that does not associate with Rags, SLC38A9, or mTORC1.
Figure 8.
Model. In control cells, two mutually exclusive, synchronous events take place on late endosomes/lysosomes. On the one hand, the BORC–LAMTOR interaction blocks recruitment of Arl8b and leads to the perinuclear position of late endosomes. On the other hand, the BORC–Arl8b interaction promotes the transport of late endosomes, resulting in a peripheral distribution. The steady-state scenario is a combination of both, with some late endosomes being transported toward the cell periphery, whereas others remain perinuclear. In lyspersin KO cells, the remaining subunits of BORC (BORCΔlyspersin) are insufficient to recruit Arl8b to late endosomes, resulting in perinuclear clustering of the organelles. Lyspersin depletion also prevents LAMTOR–BORCΔlyspersin interaction. Depletion of the LAMTOR complex increases BORC–Arl8b interaction, thereby promoting transport and peripheral accumulation of LAMP1-positive endosomes. Yellow trapezoid, LAMTOR complex; dark blue trapezoid, BORCΔlyspersin; light blue shape, lyspersin with discrimination of individual domains (N-domain, PRR, and C-domain); red shape, Arl8b.
Lysosomal movement is a complex mechanism regulated by several upstream stimuli including extracellular pH changes, nutrient conditions, cellular stress, infection by pathogens, and oncogenic transformation. Starvation (Korolchuk et al., 2011) or cytoplasmic alkalization (Heuser, 1989) promotes inward lysosomal movement, whereas acidification of the cytosol leads to outward lysosomal transport (Parton et al., 1991). Interestingly, lysosomes respond to environmental changes by relocalization and their intracellular position also determines their luminal pH, thereby critically influencing lysosomal function (Johnson et al., 2016). A biological process that utterly depends on lysosomal position is autophagy. Perinuclear accumulation of lysosomes increases heterotypic fusion of lysosomes with autophagosomes, thereby promoting content degradation, whereas peripheral lysosomal localization has the opposite effect (Korolchuk et al., 2011). In addition, impaired lysosomal mobility affects antigen presentation by dendritic cells (Chow et al., 2002) and microbial killing by cytotoxic T lymphocytes and natural killer cells, two processes fundamental for an efficient immune response. Finally, outward movement of lysosomes has been implicated in tumor invasiveness and metastasis. The acidification of the tumor microenvironment thereby promotes centrifugal movement of lysosomes (Glunde et al., 2003). Lysosomal exocytosis increases the secretion of hydrolases that may contribute to the digestion of extracellular matrix, facilitating the migration of tumor cells (Mohamed and Sloane, 2006; Dykes et al., 2016).
Collectively, we identified a novel upstream control mechanism of Arl8b-dependent lysosomal positioning. Lysosomes need to move within the cytoplasm to support many cellular functions, and perturbations of lysosome movement thereby contribute to the pathogenesis of various diseases. Because lysosome movement toward the cell periphery seems to be required for cancer invasion and metastasis, one could envisage that pharmacological compounds able to bind lyspersin and mimicking its interaction with LAMTOR might have antimetastatic properties. Therefore, our findings not only contribute to the understanding of lysosomal positioning but also may provide the grounds for novel therapeutic approaches.
Materials and methods
Reagents
Primary antibodies were obtained from the following sources and used according to the manufacturers’ instructions: rabbit anti-C17orf59/lyspersin (Western blotting [WB] 1:1,000; HPA045415; Sigma-Aldrich), rabbit anti-EEA1 (IF 1:100; sc-33585; Santa Cruz), sheep anti–EGF receptor (EGFR; WB 1:3,000; 20-ES04; Fitzgerald), mouse anti-GFP (WB 1:1,000; 11814460001; Roche), mouse anti-HA (WB 1:1,000; 901513; BioLegend), mouse anti-HA (IF 1:1,000; 901501; BioLegend), rabbit anti-HA (IF 1:1,000; 3724; Cell Signaling), mouse anti-LAMP1 (IF 1:100; H4A3; Developmental Studies Hybridoma Bank), rabbit anti-LAMP1 (WB 1:1,000, IF 1:250; HPA002997; Atlas Antibodies), rabbit anti-LAMTOR2 (WB 1:1,000; 8145; Cell Signaling), rabbit anti-LAMTOR3/MP1 (WB 1:1,000; 1525-1; Epitomics), rabbit anti-LAMTOR4 (WB 1:1,000; HPA020998; Atlas Antibodies), rabbit anti-LAMTOR5/HBXIP (WB 1:500; sc-134791; Santa Cruz), rabbit anti-LOH12CR1 (WB 1:1,000; HPA039509; Atlas Antibodies), rabbit anti-RagA (WB 1:1,000; 4357; Cell Signaling), rabbit anti-snapin (WB 1:500; 10055-1-AP; Proteintech), and mouse anti–α-tubulin (WB 1:1,000; 12G10; Developmental Studies Hybridoma Bank). The secondary antibodies HRP-conjugated goat anti–mouse (A4416, 1:5,000; Sigma-Aldrich), HRP-conjugated goat anti–rabbit (1:5,000; A0545; Sigma-Aldrich), and HRP-conjugated donkey anti–sheep (1:5,000; A3415; Sigma-Aldrich) were used for WB. Alexa Fluor 488–conjugated goat anti–mouse (1:1,000; A-11029; Thermo Fisher Scientific), Alexa Fluor 488–conjugated goat anti–rabbit (1:1,000; A-11008; Thermo Fisher Scientific), Alexa Fluor 568–conjugated goat anti–mouse (1:1,000; A-11031; Thermo Fisher Scientific), Alexa Fluor 568–conjugated goat anti–rabbit (1:1,000; A-11036; Thermo Fisher Scientific), and Hoechst 33342 (IF 1:10,000; B2261; Sigma-Aldrich) were used for IF.
Generation of CRISPR/Cas9 KO cell lines
The pLentiCRISPRv2-LoxP plasmid was generated by inserting LoxP sites (5′-ATAACTTCGTATAGCATACATTATACGAAGTTAT-3′) flankingthe elongation factor-1 α short promoter of plentiCRISPRv2 (Addgene 52961; Shalem et al., 2014). gRNA sequences targeting lyspersin (5′-GCCGCAGCTTGACGTCTTCG-3′) and LAMTOR1 (5′-GAGGAGGGACGGCGTCCGTG-3′) were chosen using an online prediction tool (CRISPR Design; Zhang laboratory, MIT; Hsu et al., 2013) and subcloned into pLentiCRISPRv2-LoxP. HEK293LTV were cotransfected with the lentiviral plasmids pVSV-G (631530; Clontech) and psPAX2 (Vogel et al., 2015) and the viral supernatant directly used to infect HeLa cells. Transfected cells were subsequently selected with puromycin (P7255; Sigma-Aldrich). After selection, cells were transiently transfected with pCAG-Cre:GFP (13776; Addgene; Matsuda and Cepko, 2007) to remove the Cas9 and FACS sorted to enrich for the GFP-positive population. Limiting dilution then generated single cell clones. Finally, the KOs of lyspersin and LAMTOR1 (also referred to as LAMTOR1HM; de Araujo et al., 2017) were confirmed by PCR screen (Yu et al., 2014), Western blot analysis, and genotyping of the respective locus.
Plasmids and lentivirus production
The constructs used as cDNA templates were either previously generated in our department or obtained from other researchers. Constructs previously published from our department were LAMTOR2 (Wunderlich et al., 2001) and LAMTOR3 (Wunderlich et al., 2001). We purchased BORCS6 (4672989; Openbiosystems), BORCS7 (4698381; Openbiosystems), LAMP1 (ImaGenes IRAUp969C0275D), LAMTOR4 (5194116; Openbiosystems), and LAMTOR5 (5190406; Openbiosystems). Constructs obtained from other researchers were BLOS1 (Starcevic and Dell’Angelica, 2004), LAMTOR1 (a gift from L. Hengst and K. Ecker, Innsbruck Medical University, Innsbruck, Austria), pallidin (Falcón-Pérez et al., 2002), RagA, and RagC (Rebsamen et al., 2015). As an exception, Arl8b cDNA was obtained by reverse transcription and cDNA synthesis. The truncated versions of lyspersin as well as all additional cDNAs templates were PCR-amplified, adding gateway sites at the 5′ and 3′ ends, and recombined into pENTR207 (12213-013; Invitrogen) using BP clonase (11789-100; Invitrogen). The respective entry constructs were subsequently recombined using LR clonase (11791-100; Invitrogen) into pcDNA5/FRT/TO/SH/GW (a gift from M. Gstaiger, ETH Zurich, Switzerland; Rebsamen et al., 2015) for N-terminal, or into pTO-C FRT/GW/SH (a gift from M. Gstaiger; Rebsamen et al., 2015) for C-terminal tagging. In addition, Arl8b_TN and Arl8b_QL mutants, generated by gene sewing, as well as WT Arl8b, were recombined into pEGFP-N-Dest (a gift from S. Geley, Innsbruck Medical University, Innsbruck, Austria). LAMP1 was recombined into pmCherry-N-Dest (a gift from S. Geley).
The lentiviral constructs were generated as follows: the cDNA of lyspersin was cloned into pENTR4-HA-MCS, pENTR4-HS-MCS (Vogel et al., 2015) using BglII and XbaI sites. LAMTOR1 was C-terminally tagged with HA using gene sewing PCR and BP recombined (11789-100; Invitrogen) into pENTR207 (12213-013; Invitrogen). The generated constructs were subsequently recombined into pQXCIN (631514; Clontech), using LR clonase (11791-100; Invitrogen). The lentiviral plasmids were cotransfected with pVSV-G (631530; Clontech) and psPAX2 (Vogel et al., 2015) into the HEK293LTV producer cell line using Lipofectamine LTX (15338-100; Invitrogen). The viral supernatant was collected 48 and 72 h after transfection and used directly for infection. Finally, cells were selected 72 h after infection with 400 µg/ml G418 (Sigma-Aldrich).
The plasmids required for the PCA assay were generated by cloning lyspersin, C-domain lyspersin, and LAMTOR3 into pcDNA3.1–VF1 (Remy et al., 2004) using NotI and ClaI sites. An identical strategy was used to subclone LAMTOR1 into pcDNA3.1–VF2 (Remy et al., 2004). pcDNA3.1 Zipper-VF1 and pcDNA3.1 Zipper-VF2 have been previously described (Remy et al., 2004). All generated constructs were sequence-verified.
Recombinant protein production and purification
For recombinant expression of 5BORC (BORCΔBLOC-1) and LAMTOR complexes, a synthetic DNA sequence comprising five open reading frames (ORFs) was synthesized by GeneArt AG/Life Technologies and subcloned into pACEBac1 (Geneva Biotech). The first ORF coded for FLAG-tagged diaskedin, followed by Strep lyspersin, and untagged MEF2BNB, myrlysin, and finally KxDL1. A similar design strategy was used for the LAMTOR complex using a N-terminal His6 tag on LAMTOR3. Based on bioinformatical analyses and published data, LAMTOR1 was N-terminally truncated, deleting amino acids 1–20 (LAMTORΔ1–20), to improve its stability and solubility (Nada et al., 2009). Arrangement of the codon-optimized ORFs in the synthetic construct were designed such that each of the genes contained its own transcription initiation and termination site. The synthetic construct was then inserted into the MCS of the acceptor vector pACEBac1, where it is flanked by Tn7 13 transposition elements. Finally, Tn7 transposition in Escherichia coli DH10Multibac allowed the insertion of the expression construct into baculoviral DNA.
Recombinant bacmid DNA was isolated and used to transfect Sf21 insect cells. The initial recombinant baculoviruses were amplified and used for protein expression by infecting cells at a density of 0.5 × 106 cells/ml. Infection was done with 2.5% (vol/vol) of amplified virus stock (MOI > 1). After 78 h, cells were collected by centrifugation, washed in PBS, and temporarly stored as frozen pellets at −20°C. Sf21 cells expressing the pentameric LAMTOR complex were resuspended in lysis buffer (20 mM Hepes, pH 8.0, 150 mM NaCl, and 2 mM DTT) and flash-frozen in liquid nitrogen. After thawing, the cell lysate was supplemented with an EDTA-free protease inhibitors cocktail followed by centrifugation for 30 min at 16.000 g at 4°C. The soluble fraction was added to equilibrated Ni-NTA Sepharose beads (1018244; Qiagen). The beads were washed and the purified complexes eluted in 20 mM Hepes, pH 8.0, 150 mM NaCl, 2 mM DTT, and 250 mM imidazole. After the purification, all tags were cleaved simultaneously by treatment with tobacco etch virus protease (overnight incubation) and removed by preparative size exclusion chromatography on a Superdex 200 prep column. The eluted protein complex was stored in 20 mM Hepes, pH 8.0, supplemented with 150 mM NaCl.
Cell culture
HeLa and HEK293LTV were cultured in DMEM (D6429; Sigma-Aldrich) supplemented with 10% (vol/vol) FBS (10270; Gibco) and antibiotics (100 U/ml penicillin and 100 mg/ml streptomycin; P0781; Sigma-Aldrich) at 37°C in 5% CO2 with 95% humidity. HEK293 Flp-In T-REx were cultured in DMEM (D6429; Sigma-Aldrich) supplemented with 10% (vol/vol) FBS (]10270), 100 U/ml penicillin, 100 mg/ml streptomycin (P0781), 100 µg/ml Zeocin (R25001; Invitrogen), and 10 µg/ml blasticidin (R210-01; Invitrogen). Cells were cotransfected with pOG44 (V600520; Invitrogen) and constructs containing Strep-HA tagged proteins of interest (BLOS1, lyspersin, LAMTOR1, LAMTOR3, LAMTOR4, LAMTOR5, pallidin, RagA, RagC, and truncated versions of lyspersin) using Lipofectamine LTX (15338-100; Invitrogen). 48 h after transfection, cells were selected with 100 µg/ml Hygromycin B (10687010; Invitrogen) for 2 wk.
WT, lyspersin KO, and SH-lyspersin reconstituted cells were cultured in DMEM (D6429; Sigma-Aldrich) overnight and stimulated with EGF (100 ng/ml; E4127; Sigma-Aldrich) for the indicated times.
IF and microscopy
Cells grown on glass coverslips were washed with PBS, fixed with 4% paraformaldehyde in cytoskeleton buffer (CB; 20 mM Pipes, pH 6.8, 300 mM NaCl, 10 mM EGTA, 10 mM glucose, and 10 mM MgCl2), and permeabilized with 0.05% saponin in blocking buffer (CB supplemented with 10% goat serum [G6767; Sigma-Aldrich] and 5% BSA [8076.5; Roth]). Slides were subsequently incubated with primary antibodies in blocking buffer for 1.5 h at RT, washed six times with CB, incubated light-protected with secondary antibody in blocking buffer for 1 h at RT, and washed six times with CB. Coverslips were then mounted in Mowiol. Epifluorescent images were acquired using a 63× oil immersion objective (NA 1.4) and the Axio Imager M1 microscope (Carl Zeiss) equipped with a SPOT Xplorer (Visitron Systems) camera at RT. Acquisition was controlled by the VisiView software (Visitron Systems GmbH). Confocal stacks and single confocal planes were acquired using a Laser Scanning Confocal Microscope (SP5; Leica) equipped with a 63× oil objective (NA 1.4) at RT. Acquisition was controlled by the LAS AF software (Leica). Confocal images were deconvoluted with Huygens Professional Deconvolution and Analysis Software (Scientific Volume Imaging) by applying Classic Maximum Likelihood Estimation, reformatted to TIFF using ImageJ, and adjusted for brightness and contrast using Photoshop CS2.
Live cell imaging
HeLa WT and lyspersin KO cells, transfected with LAMP1-mCherry, were seeded in Leibovitz’s L-15 medium (21083-027; Invitrogen) without phenol red and supplemented with 10% FBS (10270; Gibco Life Technologies). Time-lapse movies were taken with an inverted Axiovert 200M microscope (Carl Zeiss) equipped with a 63× oil objective (NA 1.4) and a CMOS camera (Cool SNAP HQ2; Photonics) at 37°C. Acquisition was controlled by the VisiView software (Visitron Systems GmbH). Images were deconvoluted with Huygens Professional Deconvolution and Analysis Software by applying Classic Maximum Likelihood Estimation and adjusted for brightness and contrast using ImageJ. Images were acquired over a period of 2 min at 1-s intervals.
PCA assay
Lyspersin KO and WT cells were cotransfected with two PCA constructs, one coding for a VF1-tagged protein and the second for a VF2-tagged one. The amount of DNA used was kept at a 2:1 ratio of the VF1-expressing construct over the VF2 construct. Transfection was performed using Lipofectamine LTX (15338-100; Invitrogen) according to the manufacturer’s instructions. The cells were seeded on glass coverslips 24 h after transfection and fixed 24 h later. Finally, cells were stained for late endosomes and nucleus (LAMP1 and Hoechst, respectively) using the IF protocol described in the IF and microscopy section.
Quantification and statistical analysis of endosomal distribution
Confocal images of cells stained for nucleus (Hoechst) and late endosomes (LAMP1) were deconvoluted with Huygens Professional Deconvolution and Analysis Software. The distance of each identified late endosomal structure to the center of the nucleus was measured in 3D-reconstructed cells using the cell module of Imaris 8.2.0 (Bitplane). The mean distance of late endosomes to the center of the nucleus was quantified by applying a Student’s t test (*, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001).
Cells were transfected with Arl8b (WT/QL/TN)-EGFP, cotransfected with the indicated PCA constructs or stimulated with EGF. In all cases, indirect IF analysis of late endosomes (LAMP1) and nucleus (Hoechst) was performed. Epifluorescent images were acquired. To quantify the intracellular distribution of late endosomes, we developed an ImageJ/FIJI (Schindelin et al., 2012, 2015) macro that measures the radial signal intensity profile around the center of the nucleus. In brief, the macro uses the Hoechst staining to determine the center of mass of the nucleus. It subsequently defines concentric annular regions of interest (ROIs) with increasing radii around them. For each ROI, the sum of the pixel intensity values in the target channel (LAMP1 or Venus) are measured. To reduce background intensity, the macro performs ImageJ’s rolling ball background subtraction algorithm on the target channel. Plots show the mean relative intensity of each annular ROI of n ≥ 25 cells. The macro is available at https://github.com/PTschaikner/RadialIntensityProfile. The frequency of peripheral lysosomes after EGF stimulation was quantified by applying ANOVA (NS, P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001; n ≥ 50 cells).
Quantification of changes in protein interaction
The fold of difference of proteins interaction was quantified by applying a Student’s t test (*, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001).
TAP and immunoprecipitation
The generated HEK293 Flip-in T-REx cell lines capable of expressing the HS-tagged proteins of interest were induced with tetracycline for 24 h before harvesting. TAP was performed as described previously (Glatter et al., 2009). In brief, cells were lysed for 20 min in buffer I (50 mM Hepes, pH 8.0, 150 mM NaCl, 5 mM EDTA, 0.5% NP-40, 50 mM NaF, and 1 mM PMSF; P7626; Sigma-Aldrich), 1 µg/ml Pepstatin (P5318; Sigma-Aldrich), 10 µg/ml Leupeptin (L8511; Sigma-Aldrich), and 0.4 mM Pefablock SC (76307; Fluka) supplemented with 1 µg/ml Avidin (2-0204-015; IBA). Lysates were cleared by centrifugation at 16,000 g for 15 min, washed with buffer I, and applied on a column containing Strep-Tactin Sepharose (2-1201-010; IBA). Bound complexes were washed four times with buffer I, subsequently eluted with 2.5 mM D-Biotin (A14207; Alfa Aesar), and applied on columns containing anti–HA-Agarose (A2095; Sigma-Aldrich). The beads were washed three times with buffer I and two times with buffer II (50 mM Hepes, pH 8.0, 150 mM NaCl, and 5 mM EDTA), eluted with 100 mM formic acid (1002640100; Millipore), and neutralized with triethylammonium bicarbonate (17902; Sigma-Aldrich) as described previously (Rudashevskaya et al., 2013). The final eluates were digested with Trypsin (V5111; Promega) and analyzed by LC-MS/MS. Single-step purifications were performed as described for TAP, omitting all the unnecessary steps. Cells expressing HS-EGFP were used as negative control.
Size exclusion chromatography
Protein complexes purified on Strep-Tactin Sepharose were analyzed by size-exclusion chromatography on a HiPrep1660 Superdex200 column (28989335; GE Healthcare) in 50 mM Tris-HCl, pH 8.0, 0.5% NP-40, 150 mM NaCl, and 5 mM EDTA. Proteins in collected fractions were precipitated with 10% TCA and analyzed by WB.
LC-MS/MS
Tryptic peptides of proteins interacting with the LAMTOR subunits 1, 3, 4, and 5 as well as RagA, RagC, and BLOS1 were analyzed by nano–LC-MS/MS on a hybrid linear trap quadrupole-Orbitrap Velos mass spectrometer (Thermo Fisher Scientific) coupled to a 1200 HPLC nanoflow system (Agilent Biotechnologies) via a nanoelectrospray ion source using liquid junction (Proxeon), as previously described (Rudashevskaya et al., 2013). In brief, solvents for HPLC separation were 0.4% formic acid in water (solvent A) and 0.4% formic acid in 70% methanol and 20% isopropanol (solvent B). The gradient profile was as follows: 27 min from 3% to 30% B; 25 min from 30% to 70% B, and 7 min from 70% to 100% B. The analyses were performed in a data-dependent acquisition mode using a top 15 collision-induced dissociation method at 60,000 resolution for peptide identification.
In case of lyspersin and pallidin as baits, digested samples were analyzed using an UltiMate 3000 nano-HPLC system coupled to a Q Exactive Plus mass spectrometer (both Thermo Fisher Scientific) equipped with a Nanospray Flex ionization source. The peptides were separated on a homemade fritless fused-silica microcapillary column (75 µm inner diameter × 280 µm outer diameter × 10 cm length) packed with 3 µm reverse-phase C18 material (Reprosil). Solvents for HPLC were 0.1% formic acid (solvent A) and 0.1% formic acid in 85% acetonitrile (solvent B). The gradient profile was as follows: 0–2 min, 4% B; 2–55 min, 4–50% B; 55–60 min, 50–100% B, and 60–65 min, 100% B. The flow rate was 250 nl/min. The Q Exacitve Plus mass spectrometer was operating in the data-dependent mode selecting the top 12 most abundant isotope patterns with charge >1 from the survey scan with an isolation window of a 1.6 mass-to-charge ratio (m/z). Survey full-scan mass spectrometry spectra were acquired from 300 to 1,750 m/z at a resolution of 70,000 with a maximum injection time of 120 ms and automatic gain control target 106. The selected isotope patterns were fragmented by higher-energy collisional dissociation with normalized collision energy of 28 at a resolution of 35,000 with a maximum injection time of 120 ms and automatic gain control target 5 × 105.
Chemical cross-linking
Sf21 cells expressing BORCΔBLOC-1 were thawed on ice and lysed by sonication five times for 10 s in 50 mM Hepes, pH 8.0, 300 mM NaCl, 50 mM NaF, 5 mM DTT, 5 mM EDTA, 1 µg/ml Pepstatin (P5318; Sigma-Aldrich), 10 µg/ml Leupeptin (L8511; Sigma-Aldrich), and 0.4 mM Pefabloc SC (76307; Fluka) supplemented with 1 µg/ml Avidin (2-0204-015; IBA). After centrifugation at 16,000 g for 15 min, lysates were loaded onto a Strep-Tactin Sepharose (2-1201-010; IBA) spin column (M1003; MoBiTec) and washed five times with lysis buffer without Avidin, followed by seven washes with the same buffer containing 1 M NaCl, and finally equilibrated with the same buffer containing 150 mM NaCl as binding buffer. Purified pentameric LAMTOR complex was added in an approximately equimolar amount and incubated with the beads for 2 h at 4°C. After washing with binding buffer 10 times, protein complexes were eluted with 2.5 mM D-Biotin. Eluted proteins were concentrated by ultracentrifugation using 30MWCO tubes (Vivaspin500; Sartorius), and the buffer was exchanged to cross-linking buffer (50 mM Hepes, pH 8.0, 150 mM NaCl, 2 mM DTT, and 5% glycerol). Protein complexes were cross-linked with 1:1 isotope-labeled 0.2 mM BS3-H12/D12 (001SS; Creative Molecules) for 30 min at 30°C. The cross-linking reaction was quenched by the addition of ammonium bicarbonate to a final concentration of 50 mM. For mass spectrometry, samples were processed applying a filter-aided sample preparation protocol (Wiśniewski et al., 2009) in a 10-kD cutoff filter unit (Vivaspin 500; Z614009; Sigma-Aldrich). Samples were supplemented with 8 M urea, reduced with DTT, alkylated with iodoacetamide, and digested with trypsin (Pierce). Digested samples were analyzed using a Q Exactive HF mass spectrometer (Thermo Fisher Scientific) as described in the LC-MS/MS section with the following adjustments: the top 20 most abundant isotope patterns were selected. Mass spectrometry spectra were acquired at a resolution of 60,000 and higher-energy collisional dissociation fragmentation at a resolution of 15,000. Cross-links were identified by MS/MS spectra searching by the StavroX software (Götze et al., 2012), version 3.6.0. For data analysis, the peptide mass tolerance was set to 4 ppm and the fragment mass tolerance to 10 ppm. Up to three missed cleavages of trypsin were allowed. Carbamidomethylation of cysteine and methionine oxidation as fixed and variable modification, respectively, and reactivity of BS3 with Lys and N terminus were selected. The false discovery rate filter was set to 1%. The data were further evaluated by filtering for pairs of heavy and light peptide precursors with a retention time difference of <30 s. Finally, after manual evaluation of the matching m/z, elution time, charge state, and fragmentation intensities, MS/MS spectra of cross-links identified at least twice (peptide spectra matched ≥ 2) were accepted.
Online supplemental material
Fig. S1 depicts the integration of exogenously expressed HS-lyspersin into the BORC and shows the stability of LAMTOR and BORC complexes upon deletion of LAMTOR1 or lyspersin. Fig. S2 provides additional data on the functional epistatic alignment of Arl8b, lyspersin, and the LAMTOR complex, highlighting the importance of the C-terminal domain of lyspersin in the regulation of lysosomal positioning. Fig. S3 shows HS-lyspersin interacting with Arl8b-EGFP independent of its load status and localization of Arl8b-EGFP mutants (TN and QL) in WT, lyspersin KO, and HS-lyspersin reconstituted cells. Fig. S4 displays the quantifications of the Venus and LAMP1 signals in the PCA assay. Fig. S5 depicts the PCA of control combinations (LAMTOR3-VF1, LAMTOR1-VF2; Zipper-VF1, LAMTOR1-VF2; Zipper-VF1, Zipper-VF2) in lyspersin KO cells and the respective quantification of LAMP1 signal distribution. Table S1 provides the complete mass spectrometry analysis of the LAMTOR, RagA, RagC, and BORC interactomes. Video 1 shows the movement of lysosomes in both lyspersin KO and control cells. Supplemental text contains the custom software, RadialIntensityProfile, which is a FIJI and ImageJ macro for analyzing the radial distribution of a fluorescent signal of interest in relation to a cell nucleus.
Supplementary Material
Acknowledgments
We thank Stephan Geley and Matthias Gstaiger for providing several plasmids, Imre Berger for the MultiBac kit, Sabine Weys and Caroline Herrmann for technical assistance, and David Teis for critically commenting on the manuscript.
This work was supported by the Austrian Science Fund (FWF): P 2668 and the Molecular Cell Biology and Oncology PhD program at Innsbruck Medical University.
The authors declare no competing financial interests.
Author contributions: P.A. Filipek designed and performed experiments and wrote the manuscript. M.E.G. de Araujo and T. Stasyk designed and performed experiments and participated in writing the manuscript. G.F. Vogel, C.H. De Smet, D. Eberharter, M. Rebsamen, E.L. Rudashevskaya, L. Kremser, T. Yordanov, and B.G. Fürnrohr contributed experiments. P. Tschaikner designed the ImageJ/FIJI macro used for the quantification of endosomal distribution. K.L. Bennett, G. Superti-Furga, and H.H. Lindner contributed experiments. K. Scheffzek, S. Lechner, and T. Dunzendorfer-Matt contributed to the design of the LAMTOR and BORC expression vectors and the purification. L.A. Huber initiated and supervised the project, designed experiments, and wrote the manuscript.
Footnotes
Abbreviations used:
- BORC
- BLOC-1–related complex
- DUF
- domain of unknown function
- EGFR
- EGF receptor
- HS
- HA–Strep II
- IF
- immunofluorescence
- KO
- knockout
- LAMTOR
- late endosomal/lysosomal adaptor and MAPK and mTOR activator
- LC-MS/MS
- liquid chromatography tandem mass spectrometry
- MEF
- mouse embryonic fibroblast
- mTOR
- mechanistic target of rapamycin
- PCA
- protein-fragment complementation assay
- PRR
- proline-rich region
- ROI
- region of interest
- SH
- Strep II–HA
- TAP
- tandem affinity purification
- VF
- Venus fragment
- WB
- Western blotting
References
- Appelqvist H., Wäster P., Kågedal K., and Öllinger K.. 2013. The lysosome: from waste bag to potential therapeutic target. J. Mol. Cell Biol. 5:214–226. 10.1093/jmcb/mjt022 [DOI] [PubMed] [Google Scholar]
- Bagshaw R.D., Callahan J.W., and Mahuran D.J.. 2006. The Arf-family protein, Arl8b, is involved in the spatial distribution of lysosomes. Biochem. Biophys. Res. Commun. 344:1186–1191. 10.1016/j.bbrc.2006.03.221 [DOI] [PubMed] [Google Scholar]
- Bar-Peled L., and Sabatini D.M.. 2014. Regulation of mTORC1 by amino acids. Trends Cell Biol. 24:400–406. 10.1016/j.tcb.2014.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Peled L., Schweitzer L.D., Zoncu R., and Sabatini D.M.. 2012. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell. 150:1196–1208. 10.1016/j.cell.2012.07.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomen V.A., Májek P., Jae L.T., Bigenzahn J.W., Nieuwenhuis J., Staring J., Sacco R., van Diemen F.R., Olk N., Stukalov A., et al. . 2015. Gene essentiality and synthetic lethality in haploid human cells. Science. 350:1092–1096. 10.1126/science.aac7557 [DOI] [PubMed] [Google Scholar]
- Bohn G., Allroth A., Brandes G., Thiel J., Glocker E., Schäffer A.A., Rathinam C., Taub N., Teis D., Zeidler C., et al. . 2007. A novel human primary immunodeficiency syndrome caused by deficiency of the endosomal adaptor protein p14. Nat. Med. 13:38–45. 10.1038/nm1528 [DOI] [PubMed] [Google Scholar]
- Boucrot E., Henry T., Borg J.P., Gorvel J.P., and Méresse S.. 2005. The intracellular fate of Salmonella depends on the recruitment of kinesin. Science. 308:1174–1178. 10.1126/science.1110225 [DOI] [PubMed] [Google Scholar]
- Cantalupo G., Alifano P., Roberti V., Bruni C.B., and Bucci C.. 2001. Rab-interacting lysosomal protein (RILP): the Rab7 effector required for transport to lysosomes. EMBO J. 20:683–693. 10.1093/emboj/20.4.683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow A., Toomre D., Garrett W., and Mellman I.. 2002. Dendritic cell maturation triggers retrograde MHC class II transport from lysosomes to the plasma membrane. Nature. 418:988–994. 10.1038/nature01006 [DOI] [PubMed] [Google Scholar]
- de Araujo M.E.G., Naschberger A., Fürnrohr B.G., Stasyk T., Dunzendorfer-Matt T., Lechner S., Welti S., Kremser L., Shivalingaiah G., Offterdinger M., et al. . 2017. Crystal structure of the human lysosomal mTORC1 scaffold complex and its impact on signaling. Science. eaao1583 10.1126/science.aao1583 [DOI] [PubMed] [Google Scholar]
- de Araújo M.E., Stasyk T., Taub N., Ebner H.L., Fürst B., Filipek P., Weys S.R., Hess M.W., Lindner H., Kremser L., and Huber L.A.. 2013. Stability of the endosomal scaffold protein LAMTOR3 depends on heterodimer assembly and proteasomal degradation. J. Biol. Chem. 288:18228–18242. 10.1074/jbc.M112.349480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell’Angelica E.C., Mullins C., Caplan S., and Bonifacino J.S.. 2000. Lysosome-related organelles. FASEB J. 14:1265–1278. 10.1096/fj.14.10.1265 [DOI] [PubMed] [Google Scholar]
- Dykes S.S., Gray A.L., Coleman D.T., Saxena M., Stephens C.A., Carroll J.L., Pruitt K., and Cardelli J.A.. 2016. The Arf-like GTPase Arl8b is essential for three-dimensional invasive growth of prostate cancer in vitro and xenograft formation and growth in vivo. Oncotarget. 7:31037–31052. 10.18632/oncotarget.8832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcón-Pérez J.M., Starcevic M., Gautam R., and Dell’Angelica E.C.. 2002. BLOC-1, a novel complex containing the pallidin and muted proteins involved in the biogenesis of melanosomes and platelet-dense granules. J. Biol. Chem. 277:28191–28199. 10.1074/jbc.M204011200 [DOI] [PubMed] [Google Scholar]
- Finn R.D., Coggill P., Eberhardt R.Y., Eddy S.R., Mistry J., Mitchell A.L., Potter S.C., Punta M., Qureshi M., Sangrador-Vegas A., et al. . 2016. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44(D1):D279–D285. 10.1093/nar/gkv1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganley I.G., and Pfeffer S.R.. 2006. Cholesterol accumulation sequesters Rab9 and disrupts late endosome function in NPC1-deficient cells. J. Biol. Chem. 281:17890–17899. 10.1074/jbc.M601679200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glatter T., Wepf A., Aebersold R., and Gstaiger M.. 2009. An integrated workflow for charting the human interaction proteome: insights into the PP2A system. Mol. Syst. Biol. 5:237 10.1038/msb.2008.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glunde K., Guggino S.E., Solaiyappan M., Pathak A.P., Ichikawa Y., and Bhujwalla Z.M.. 2003. Extracellular acidification alters lysosomal trafficking in human breast cancer cells. Neoplasia. 5:533–545. 10.1016/S1476-5586(03)80037-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götze M., Pettelkau J., Schaks S., Bosse K., Ihling C.H., Krauth F., Fritzsche R., Kühn U., and Sinz A.. 2012. StavroX--a software for analyzing crosslinked products in protein interaction studies. J. Am. Soc. Mass Spectrom. 23:76–87. 10.1007/s13361-011-0261-2 [DOI] [PubMed] [Google Scholar]
- Guardia C.M., Farías G.G., Jia R., Pu J., and Bonifacino J.S.. 2016. BORC Functions Upstream of Kinesins 1 and 3 to Coordinate Regional Movement of Lysosomes along Different Microtubule Tracks. Cell Reports. 17:1950–1961. 10.1016/j.celrep.2016.10.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog F., Kahraman A., Boehringer D., Mak R., Bracher A., Walzthoeni T., Leitner A., Beck M., Hartl F.U., Ban N., et al. . 2012. Structural probing of a protein phosphatase 2A network by chemical cross-linking and mass spectrometry. Science. 337:1348–1352. 10.1126/science.1221483 [DOI] [PubMed] [Google Scholar]
- Heuser J. 1989. Changes in lysosome shape and distribution correlated with changes in cytoplasmic pH. J. Cell Biol. 108:855–864. 10.1083/jcb.108.3.855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann I., and Munro S.. 2006. An N-terminally acetylated Arf-like GTPase is localised to lysosomes and affects their motility. J. Cell Sci. 119:1494–1503. 10.1242/jcs.02958 [DOI] [PubMed] [Google Scholar]
- Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O., et al. . 2013. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31:827–832. 10.1038/nbt.2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M., Rocha N., Zwart W., Jordens I., Janssen L., Kuijl C., Olkkonen V.M., and Neefjes J.. 2007. Activation of endosomal dynein motors by stepwise assembly of Rab7-RILP-p150Glued, ORP1L, and the receptor betalll spectrin. J. Cell Biol. 176:459–471. 10.1083/jcb.200606077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D.E., Ostrowski P., Jaumouillé V., and Grinstein S.. 2016. The position of lysosomes within the cell determines their luminal pH. J. Cell Biol. 212:677–692. 10.1083/jcb.201507112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongsma M.L., Berlin I., Wijdeven R.H., Janssen L., Janssen G.M., Garstka M.A., Janssen H., Mensink M., van Veelen P.A., Spaapen R.M., and Neefjes J.. 2016. An ER-Associated Pathway Defines Endosomal Architecture for Controlled Cargo Transport. Cell. 166:152–166. 10.1016/j.cell.2016.05.078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordens I., Fernandez-Borja M., Marsman M., Dusseljee S., Janssen L., Calafat J., Janssen H., Wubbolts R., and Neefjes J.. 2001. The Rab7 effector protein RILP controls lysosomal transport by inducing the recruitment of dynein-dynactin motors. Curr. Biol. 11:1680–1685. 10.1016/S0960-9822(01)00531-0 [DOI] [PubMed] [Google Scholar]
- Jung J., Genau H.M., and Behrends C.. 2015. Amino Acid-Dependent mTORC1 Regulation by the Lysosomal Membrane Protein SLC38A9. Mol. Cell. Biol. 35:2479–2494. 10.1128/MCB.00125-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolchuk V.I., Saiki S., Lichtenberg M., Siddiqi F.H., Roberts E.A., Imarisio S., Jahreiss L., Sarkar S., Futter M., Menzies F.M., et al. . 2011. Lysosomal positioning coordinates cellular nutrient responses. Nat. Cell Biol. 13:453–460. 10.1038/ncb2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.H., Nemecek D., Schindler C., Smith W.J., Ghirlando R., Steven A.C., Bonifacino J.S., and Hurley J.H.. 2012. Assembly and architecture of biogenesis of lysosome-related organelles complex-1 (BLOC-1). J. Biol. Chem. 287:5882–5890. 10.1074/jbc.M111.325746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Rydzewski N., Hider A., Zhang X., Yang J., Wang W., Gao Q., Cheng X., and Xu H.. 2016. A molecular mechanism to regulate lysosome motility for lysosome positioning and tubulation. Nat. Cell Biol. 18:404–417. 10.1038/ncb3324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee J., and Cygler M.. 2011. Interactions between kinase scaffold MP1/p14 and its endosomal anchoring protein p18. Biochemistry. 50:3696–3705. 10.1021/bi101972y [DOI] [PubMed] [Google Scholar]
- Magliery T.J., Wilson C.G., Pan W., Mishler D., Ghosh I., Hamilton A.D., and Regan L.. 2005. Detecting protein-protein interactions with a green fluorescent protein fragment reassembly trap: scope and mechanism. J. Am. Chem. Soc. 127:146–157. 10.1021/ja046699g [DOI] [PubMed] [Google Scholar]
- Matsuda T., and Cepko C.L.. 2007. Controlled expression of transgenes introduced by in vivo electroporation. Proc. Natl. Acad. Sci. USA. 104:1027–1032. 10.1073/pnas.0610155104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki F., Shirane M., Matsumoto M., and Nakayama K.I.. 2011. Protrudin serves as an adaptor molecule that connects KIF5 and its cargoes in vesicular transport during process formation. Mol. Biol. Cell. 22:4602–4620. 10.1091/mbc.E11-01-0068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matteoni R., and Kreis T.E.. 1987. Translocation and clustering of endosomes and lysosomes depends on microtubules. J. Cell Biol. 105:1253–1265. 10.1083/jcb.105.3.1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michnick S.W., Remy I., Campbell-Valois F.X., Vallée-Bélisle A., and Pelletier J.N.. 2000. Detection of protein-protein interactions by protein fragment complementation strategies. Methods Enzymol. 328:208–230. 10.1016/S0076-6879(00)28399-7 [DOI] [PubMed] [Google Scholar]
- Mohamed M.M., and Sloane B.F.. 2006. Cysteine cathepsins: multifunctional enzymes in cancer. Nat. Rev. Cancer. 6:764–775. 10.1038/nrc1949 [DOI] [PubMed] [Google Scholar]
- Moriyama K., and Bonifacino J.S.. 2002. Pallidin is a component of a multi-protein complex involved in the biogenesis of lysosome-related organelles. Traffic. 3:666–677. 10.1034/j.1600-0854.2002.30908.x [DOI] [PubMed] [Google Scholar]
- Nada S., Hondo A., Kasai A., Koike M., Saito K., Uchiyama Y., and Okada M.. 2009. The novel lipid raft adaptor p18 controls endosome dynamics by anchoring the MEK-ERK pathway to late endosomes. EMBO J. 28:477–489. 10.1038/emboj.2008.308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parton R.G., Dotti C.G., Bacallao R., Kurtz I., Simons K., and Prydz K.. 1991. pH-induced microtubule-dependent redistribution of late endosomes in neuronal and epithelial cells. J. Cell Biol. 113:261–274. 10.1083/jcb.113.2.261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu J., Schindler C., Jia R., Jarnik M., Backlund P., and Bonifacino J.S.. 2015. BORC, a multisubunit complex that regulates lysosome positioning. Dev. Cell. 33:176–188. 10.1016/j.devcel.2015.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu J., Guardia C.M., Keren-Kaplan T., and Bonifacino J.S.. 2016. Mechanisms and functions of lysosome positioning. J. Cell Sci. 129:4329–4339. 10.1242/jcs.196287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiborg C., Wenzel E.M., Pedersen N.M., Olsvik H., Schink K.O., Schultz S.W., Vietri M., Nisi V., Bucci C., Brech A., et al. . 2015. Repeated ER-endosome contacts promote endosome translocation and neurite outgrowth. Nature. 520:234–238. 10.1038/nature14359 [DOI] [PubMed] [Google Scholar]
- Rebsamen M., Pochini L., Stasyk T., de Araújo M.E., Galluccio M., Kandasamy R.K., Snijder B., Fauster A., Rudashevskaya E.L., Bruckner M., et al. . 2015. SLC38A9 is a component of the lysosomal amino acid sensing machinery that controls mTORC1. Nature. 519:477–481. 10.1038/nature14107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remy I., and Michnick S.W.. 1999. Clonal selection and in vivo quantitation of protein interactions with protein-fragment complementation assays. Proc. Natl. Acad. Sci. USA. 96:5394–5399. (published erratum appears in Proc. Natl. Acad. Sci. USA. 1999. 96:7610) 10.1073/pnas.96.10.5394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remy I., Montmarquette A., and Michnick S.W.. 2004. PKB/Akt modulates TGF-beta signalling through a direct interaction with Smad3. Nat. Cell Biol. 6:358–365. 10.1038/ncb1113 [DOI] [PubMed] [Google Scholar]
- Remy I., Campbell-Valois F.X., and Michnick S.W.. 2007. Detection of protein-protein interactions using a simple survival protein-fragment complementation assay based on the enzyme dihydrofolate reductase. Nat. Protoc. 2:2120–2125. 10.1038/nprot.2007.266 [DOI] [PubMed] [Google Scholar]
- Rosa-Ferreira C., and Munro S.. 2011. Arl8 and SKIP act together to link lysosomes to kinesin-1. Dev. Cell. 21:1171–1178. 10.1016/j.devcel.2011.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudashevskaya E.L., Sacco R., Kratochwill K., Huber M.L., Gstaiger M., Superti-Furga G., and Bennett K.L.. 2013. A method to resolve the composition of heterogeneous affinity-purified protein complexes assembled around a common protein by chemical cross-linking, gel electrophoresis and mass spectrometry. Nat. Protoc. 8:75–97. 10.1038/nprot.2012.133 [DOI] [PubMed] [Google Scholar]
- Sancak Y., Peterson T.R., Shaul Y.D., Lindquist R.A., Thoreen C.C., Bar-Peled L., and Sabatini D.M.. 2008. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 320:1496–1501. 10.1126/science.1157535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y., Bar-Peled L., Zoncu R., Markhard A.L., Nada S., and Sabatini D.M.. 2010. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 141:290–303. 10.1016/j.cell.2010.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer H.J., Catling A.D., Eblen S.T., Collier L.S., Krauss A., and Weber M.J.. 1998. MP1: a MEK binding partner that enhances enzymatic activation of the MAP kinase cascade. Science. 281:1668–1671. 10.1126/science.281.5383.1668 [DOI] [PubMed] [Google Scholar]
- Scheffler J.M., Sparber F., Tripp C.H., Herrmann C., Humenberger A., Blitz J., Romani N., Stoitzner P., and Huber L.A.. 2014. LAMTOR2 regulates dendritic cell homeostasis through FLT3-dependent mTOR signalling. Nat. Commun. 5:5138 10.1038/ncomms6138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefermeier N., Scheffler J.M., de Araujo M.E., Stasyk T., Yordanov T., Ebner H.L., Offterdinger M., Munck S., Hess M.W., Wickström S.A., et al. . 2014. The late endosomal p14-MP1 (LAMTOR2/3) complex regulates focal adhesion dynamics during cell migration. J. Cell Biol. 205:525–540. 10.1083/jcb.201310043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. . 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Rueden C.T., Hiner M.C., and Eliceiri K.W.. 2015. The ImageJ ecosystem: An open platform for biomedical image analysis. Mol. Reprod. Dev. 82:518–529. 10.1002/mrd.22489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweitzer L.D., Comb W.C., Bar-Peled L., and Sabatini D.M.. 2015. Disruption of the Rag-Ragulator Complex by c17orf59 Inhibits mTORC1. Cell Reports. 12:1445–1455. 10.1016/j.celrep.2015.07.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setty S.R., Tenza D., Truschel S.T., Chou E., Sviderskaya E.V., Theos A.C., Lamoreux M.L., Di Pietro S.M., Starcevic M., Bennett D.C., et al. . 2007. BLOC-1 is required for cargo-specific sorting from vacuolar early endosomes toward lysosome-related organelles. Mol. Biol. Cell. 18:768–780. 10.1091/mbc.E06-12-1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O., Sanjana N.E., Hartenian E., Shi X., Scott D.A., Mikkelson T., Heckl D., Ebert B.L., Root D.E., Doench J.G., and Zhang F.. 2014. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 343:84–87. 10.1126/science.1247005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparber F., Scheffler J.M., Amberg N., Tripp C.H., Heib V., Hermann M., Zahner S.P., Clausen B.E., Reizis B., Huber L.A., et al. . 2014. The late endosomal adaptor molecule p14 (LAMTOR2) represents a novel regulator of Langerhans cell homeostasis. Blood. 123:217–227. 10.1182/blood-2013-08-518555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparber F., Tripp C.H., Komenda K., Scheffler J.M., Clausen B.E., Huber L.A., Romani N., and Stoitzner P.. 2015. The late endosomal adaptor molecule p14 (LAMTOR2) regulates TGFβ1-mediated homeostasis of Langerhans cells. J. Invest. Dermatol. 135:119–129. 10.1038/jid.2014.324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starcevic M., and Dell’Angelica E.C.. 2004. Identification of snapin and three novel proteins (BLOS1, BLOS2, and BLOS3/reduced pigmentation) as subunits of biogenesis of lysosome-related organelles complex-1 (BLOC-1). J. Biol. Chem. 279:28393–28401. 10.1074/jbc.M402513200 [DOI] [PubMed] [Google Scholar]
- Takahashi Y., Nada S., Mori S., Soma-Nagae T., Oneyama C., and Okada M.. 2012. The late endosome/lysosome-anchored p18-mTORC1 pathway controls terminal maturation of lysosomes. Biochem. Biophys. Res. Commun. 417:1151–1157. 10.1016/j.bbrc.2011.12.082 [DOI] [PubMed] [Google Scholar]
- Taub N., Teis D., Ebner H.L., Hess M.W., and Huber L.A.. 2007. Late endosomal traffic of the epidermal growth factor receptor ensures spatial and temporal fidelity of mitogen-activated protein kinase signaling. Mol. Biol. Cell. 18:4698–4710. 10.1091/mbc.E07-02-0098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taub N., Nairz M., Hilber D., Hess M.W., Weiss G., and Huber L.A.. 2012. The late endosomal adaptor p14 is a macrophage host-defense factor against Salmonella infection. J. Cell Sci. 125:2698–2708. 10.1242/jcs.100073 [DOI] [PubMed] [Google Scholar]
- Teis D., and Huber L.A.. 2003. The odd couple: signal transduction and endocytosis. Cell. Mol. Life Sci. 60:2020–2033. 10.1007/s00018-003-3010-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teis D., Wunderlich W., and Huber L.A.. 2002. Localization of the MP1-MAPK scaffold complex to endosomes is mediated by p14 and required for signal transduction. Dev. Cell. 3:803–814. 10.1016/S1534-5807(02)00364-7 [DOI] [PubMed] [Google Scholar]
- Teis D., Taub N., Kurzbauer R., Hilber D., de Araujo M.E., Erlacher M., Offterdinger M., Villunger A., Geley S., Bohn G., et al. . 2006. p14-MP1-MEK1 signaling regulates endosomal traffic and cellular proliferation during tissue homeostasis. J. Cell Biol. 175:861–868. 10.1083/jcb.200607025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel G.F., Klee K.M.C., Janecke A.R., Müller T., Hess M.W., and Huber L.A.. 2015. Cargo-selective apical exocytosis in epithelial cells is conducted by Myo5B, Slp4a, Vamp7, and Syntaxin 3. J. Cell Biol. 211:587–604. 10.1083/jcb.201506112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Tsun Z.Y., Wolfson R.L., Shen K., Wyant G.A., Plovanich M.E., Yuan E.D., Jones T.D., Chantranupong L., Comb W., et al. . 2015a Metabolism. Lysosomal amino acid transporter SLC38A9 signals arginine sufficiency to mTORC1. Science. 347:188–194. 10.1126/science.1257132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Birsoy K., Hughes N.W., Krupczak K.M., Post Y., Wei J.J., Lander E.S., and Sabatini D.M.. 2015b Identification and characterization of essential genes in the human genome. Science. 350:1096–1101. 10.1126/science.aac7041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiśniewski J.R., Zougman A., Nagaraj N., and Mann M.. 2009. Universal sample preparation method for proteome analysis. Nat. Methods. 6:359–362. 10.1038/nmeth.1322 [DOI] [PubMed] [Google Scholar]
- Wu Y.E., Huo L., Maeder C.I., Feng W., and Shen K.. 2013. The balance between capture and dissociation of presynaptic proteins controls the spatial distribution of synapses. Neuron. 78:994–1011. 10.1016/j.neuron.2013.04.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunderlich W., Fialka I., Teis D., Alpi A., Pfeifer A., Parton R.G., Lottspeich F., and Huber L.A.. 2001. A novel 14-kilodalton protein interacts with the mitogen-activated protein kinase scaffold mp1 on a late endosomal/lysosomal compartment. J. Cell Biol. 152:765–776. 10.1083/jcb.152.4.765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C., Zhang Y., Yao S., and Wei Y.. 2014. A PCR based protocol for detecting indel mutations induced by TALENs and CRISPR/Cas9 in zebrafish. PLoS One. 9:e98282 10.1371/journal.pone.0098282 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.