Amino acid depletion turns off Ragulator/mTORC1 signaling and causes juxtanuclear clustering of lysosomes, but the mechanisms involved are unclear. Pu et al. show that amino acid depletion enhances a negative regulatory interaction of the Ragulator complex with BORC, inhibiting lysosome transport and causing their juxtanuclear clustering.
Abstract
Lysosomes play key roles in the cellular response to amino acid availability. Depletion of amino acids from the medium turns off a signaling pathway involving the Ragulator complex and the Rag guanosine triphosphatases (GTPases), causing release of the inactive mammalian target of rapamycin complex 1 (mTORC1) serine/threonine kinase from the lysosomal membrane. Decreased phosphorylation of mTORC1 substrates inhibits protein synthesis while activating autophagy. Amino acid depletion also causes clustering of lysosomes in the juxtanuclear area of the cell, but the mechanisms responsible for this phenomenon are poorly understood. Herein we show that Ragulator directly interacts with BLOC-1–related complex (BORC), a multi-subunit complex previously found to promote lysosome dispersal through coupling to the small GTPase Arl8 and the kinesins KIF1B and KIF5B. Interaction with Ragulator exerts a negative regulatory effect on BORC that is independent of mTORC1 activity. Amino acid depletion strengthens this interaction, explaining the redistribution of lysosomes to the juxtanuclear area. These findings thus demonstrate that amino acid availability controls lysosome positioning through Ragulator-dependent, but mTORC1-independent, modulation of BORC.
Introduction
Eukaryotic cells are exquisitely sensitive to the availability of nutrients in their environment. Cells respond to the abundance or scarcity of specific nutrients by modulating signaling pathways that control transcription, translation, degradation, growth, proliferation, metabolism, and other fundamental biological processes (Efeyan et al., 2015). The sensitivity to amino acid availability is particularly strong, owing to the essential roles of amino acids as precursors for proteins, lipids, and carbohydrates. When amino acids are abundant, cells turn on anabolic pathways such as protein synthesis, while simultaneously turning off catabolic pathways such as autophagy. Conversely, when amino acids are scarce, cells shut down protein synthesis and activate autophagy, thereby freeing up amino acids for maintenance of vital cellular processes. Lysosomes play a prominent role in the response to amino acid availability (Lim and Zoncu, 2016). Amino acids such as arginine and glutamine are sensed by the multispanning lysosomal amino acid transporter/receptor (“transceptor”) SLC38A9 (Jung et al., 2015; Rebsamen et al., 2015; Wang et al., 2015). This protein interacts with Ragulator, a complex of five subunits named LAMTOR1-5 that is anchored to the cytosolic face of lysosomes via myristoyl and palmitoyl groups on LAMTOR1 (also known as p18; Teis et al., 2002; Sancak et al., 2010). Ragulator acts as a guanine nucleotide exchange factor for the small GTPases RagA and RagB, which are part of heterodimeric complexes with RagC and RagD (Bar-Peled et al., 2012). Nucleotide exchange activates RagA/B, enabling recruitment of the mammalian target of rapamycin complex 1 (mTORC1) to the lysosomal membrane (Sancak et al., 2010; Bar-Peled et al., 2012). mTORC1 comprises five subunits: the serine/threonine kinase mTOR and the accessory proteins RAPTOR, PRAS40, DEPTOR, and mLST8. Another small GTPase named Rheb functions downstream of growth factor signaling to activate the kinase activity of mTORC1 at the lysosomal membrane (Inoki et al., 2003). Lysosome-associated, activated mTORC1 then promotes protein synthesis by phosphorylation of the translation regulators S6K and 4E-BP1 (Burnett et al., 1998), while simultaneously inhibiting autophagy by phosphorylation of ULK1 (Chan et al., 2007). Amino acid starvation blocks this pathway, leading to the dissociation of mTORC1 from the lysosomal membrane, with consequent inhibition of protein synthesis and activation of autophagy.
In addition to the effects on protein synthesis and autophagy, amino acid starvation induces clustering of lysosomes in the perinuclear area of the cells (Korolchuk et al., 2011; Jung et al., 2015; Starling et al., 2016). This phenomenon is thought to enable sequestration of mTORC1 away from growth factor signaling in the vicinity of the plasma membrane, contributing to mTORC1 inactivation and autophagy enhancement (Korolchuk et al., 2011). In addition, clustering of lysosomes and autophagosomes in the perinuclear area may facilitate encounter of lysosomes with autophagosomes, promoting fusion of these organelles and maintenance of autophagic flux (Korolchuk et al., 2011). The mechanism by which amino acid starvation causes perinuclear clustering of lysosomes, however, is poorly understood.
Lysosomes are capable of moving bidirectionally between the perinuclear and peripheral areas of the cytoplasm along microtubule tracks (Matteoni and Kreis, 1987; Pu et al., 2016; Bonifacino and Neefjes, 2017). Microtubule motors of the dynein and kinesin families are responsible for driving retrograde and anterograde transport of lysosomes, respectively (Hollenbeck and Swanson, 1990; Harada et al., 1998). Coupling of lysosomes to dynein depends on the small GTPase Rab7, the Rab7 effector RILP, and the dynein activator dynactin (Cantalupo et al., 2001; Jordens et al., 2001). On the other hand, the multi-subunit BLOC-1–related complex (BORC) and another small GTPase, Arl8, link lysosomes to the kinesin-1 KIF5 and kinesin-3 KIF1 proteins (Dumont et al., 2010; Rosa-Ferreira and Munro, 2011; Pu et al., 2015; Guardia et al., 2016; Farías et al., 2017). BORC comprises eight subunits named BLOS1 (also known as BORCS1 and BLOC1S1), BLOS2 (BORCS2 and BLOC1S2), snapin (BORCS3), KXD1 (BORCS4), myrlysin (LOH12CR1 and BORCS5), lyspersin (C17orf59 and BORCS6), diaskedin (C10orf32 and BORCS7), and MEF2BNB (BORCS8; Pu et al., 2015). BORC is attached to the cytosolic side of the lysosomal membrane in part through an N-terminal myristoyl group in myrlysin and functions to recruit Arl8 from the cytosol to the lysosomal surface (Pu et al., 2015; Guardia et al., 2016). Interference with these opposing mechanisms results in lysosome clustering at the peripheral or perinuclear areas of the cell, respectively.
Juxtanuclear clustering of lysosomes in response to amino acid and serum withdrawal has been shown to depend on tethering of lysosomes to the Golgi complex by formation of a folliculin-RILP-Rab4 complex (Starling et al., 2016). A similar clustering upon serum removal was also shown to depend on activation of a PI(3,5)P2-TRPML1-ALG-2-dynein complex (Li et al., 2016). We hypothesized that inhibition of kinesin-dependent transport toward the cell periphery could additionally or alternatively contribute to lysosome clustering in nutrient-starved cells. A clue to such a mechanism was provided by our original affinity purification and mass spectrometry (MS) analyses that led to the discovery of BORC (Pu et al., 2015). Using BLOS2 as bait, we observed coisolation of not only the other seven subunits of BORC, but also the five subunits of Ragulator (Pu et al., 2015). Conversely, in studies by other groups, affinity purification with Ragulator subunits resulted in the coisolation of one or more BORC subunits (Jung et al., 2015; Schweitzer et al., 2015). The significance of the BORC–Ragulator copurification, however, was not examined in those studies.
Here we demonstrate that BORC interacts with Ragulator through direct binding of lyspersin to LAMTOR2. This binding involves the predicted domain of unknown function 2365 (DUF2365) in lyspersin and a hydrophobic site on LAMTOR2. We show that DUF2365 comprises two conserved elements (CEs): an N-terminal element (CE1) that mediates interaction with LAMTOR2 and a C-terminal element (CE2) that mediates assembly into BORC. Genetic knockout (KO) and knockdown (KD) experiments reveal an epistatic relationship in which Ragulator opposes the function of BORC to promote lysosome dispersal. These interactions are key to changes in lysosome positioning that occur during amino acid starvation. Indeed, silencing of BORC or Ragulator shifts the distribution of lysosomes toward the perinuclear or peripheral regions of the cell, respectively, in both cases making lysosome positioning unresponsive to amino acid starvation. Moreover, amino acid starvation enhances the interaction of BORC with Ragulator. Remarkably, the negative regulation of BORC by Ragulator is independent of mTORC1 activity. These findings thus demonstrate that amino acid availability controls lysosome positioning through Ragulator-dependent, but mTORC1-independent, modulation of BORC.
Results
BORC interacts with Ragulator through binding of lyspersin to LAMTOR2
BORC and BLOC-1 are structurally related multi-subunit complexes composed of three shared subunits (BLOS1, BLOS2, and snapin) and five complex-specific subunits (Starcevic and Dell’Angelica, 2004; Pu et al., 2015). Despite their structural similarities, BORC regulates lysosome positioning and motility (Pu et al., 2015; Guardia et al., 2016), whereas BLOC-1 participates in the biogenesis of lysosome-related organelles (Falcón-Pérez et al., 2002; Moriyama and Bonifacino, 2002). Previous tandem affinity purification (TAP) and MS analyses in HeLa cells aimed at identifying proteins that interact with human BLOS2 showed that this protein coisolated with all the other subunits of BORC and BLOC-1 (Pu et al., 2015; Fig. 1 A). Additionally, we noticed the coisolation of the five subunits of the Ragulator complex, LAMTOR1-5, albeit with lower total peptide count (Pu et al., 2015; Fig. 1 A). To assess whether the copurification of Ragulator was caused by interaction with BORC, BLOC-1, or both, we performed further TAP-MS experiments using the BORC-specific subunit lyspersin in H4 cells and BLOC1-specific subunit pallidin in HeLa cells as baits. We found that the five Ragulator subunits copurified with lyspersin but not pallidin (Fig. 1 A and Tables S1, S2, and S3). These differential interactions were confirmed by immunoprecipitation-immunoblotting experiments in which tagged forms of BLOS2, lyspersin, and myrlysin, but not pallidin, coprecipitated with endogenous LAMTOR4 (Fig. 1 B). Reciprocally, tagged LAMTOR1 coprecipitated with endogenous myrlysin (Fig. 1 B). Yeast two-hybrid (Y2H) analyses for pairwise interactions between all BORC and Ragulator subunits revealed strong binding of lyspersin to LAMTOR2 and weaker binding of BLOS1 to LAMTOR1 (Fig. 1 C). From these experiments, we concluded that BORC, but not BLOC-1, interacts with Ragulator, mainly through binding of lyspersin to LAMTOR2. Immunofluorescence microscopy of WT HeLa cells transiently expressing GFP-lyspersin showed colocalization of this protein with LAMTOR4 and the lysosomal membrane protein LAMP1 (Fig. 1 D), consistent with BORC–Ragulator interactions occurring on lysosomes.
Figure 1.
BORC interacts with Ragulator on lysosomes. (A) Proteins that interact with BLOS2 (subunit of both BORC and BLOC-1), lyspersin (subunit of BORC), and pallidin (subunit of BLOC-1) tagged with One-STrEP-FLAG (OSF) or FLAG-One-STrEP (FOS) were identified by TAP-MS. Two independent analyses were performed for OSF-BLOS2 and OSF-lyspersin. Total peptide numbers of the coisolated proteins are shown. (B) OSF- or FOS-tagged BLOS2, lyspersin, pallidin, or LAMTOR1 were expressed by stable transfection in WT HeLa cells, and FOS-tagged myrlysin was expressed by stable transfection in myrlysin-KO HeLa cells. Cells were extracted with detergent-containing buffer and subjected to pull-down with Strep-Tactin beads followed by SDS-PAGE. Endogenous myrlysin and LAMTOR4, and the FLAG epitope, were detected by immunoblotting (IB). The positions of molecular mass markers (in kilodaltons) are indicated on the left. (C) Y2H analysis was performed by cotransforming yeast with plasmids encoding BORC subunits fused to the Gal4 binding domain (BD; top) and Ragulator subunits fused to the Gal4 activation domain (AD; left). In this and subsequent Y2H experiments, yeast transformants were grown on −His (top) or +His (bottom) plates. SV40 T antigen and p53 were used as controls. (D) GFP-lyspersin was transiently expressed by transfection in WT HeLa cells, and lysosomes were visualized by immunostaining with antibodies to endogenous LAMP1 and LAMTOR4. Bar, 5 µm. Magnifications of the boxed area are shown in the bottom row. Bar, 11 µm.
Distinct structural elements in the DUF2365 domain of lyspersin mediate interaction with LAMTOR2 and BORC
The 357-aa human lyspersin is the largest of the BORC subunits (Pu et al., 2015). It has an N-terminal unstructured region and a C-terminal structured DUF2365 (Fig. 2, A and B). Secondary structure and phylogenetic conservation analyses indicate that DUF2365 comprises two elements: a short CE1 having β-sheet and α-helix structure and a longer CE2 structured as one or two α-helices (Fig. 2, A and B). The lengths of the N-terminal unstructured region and the intervening segment between CE1 and CE2 vary among lyspersin orthologues from different animal species (Fig. 2 B).
Figure 2.
BORC–Ragulator interactions are mediated by the lyspersin DUF2365 domain and LAMTOR2. (A) Predicted coiled coils (Coils server, embnet.vital-it.ch/software/COILS_form.html) and consensus secondary structure (NPS@, https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_seccons.html) of human lyspersin. Blue, α-helix; red, β-sheet; magenta, random coil structure. Amino acid numbers are indicated. (B) Comparison of the domain organization of lyspersin in different species. DUF2365 comprises two predicted folded segments designated CE1 and CE2. Key residues in CE1 and CE2 and amino acid numbers in the human protein are indicated. H. sapiens, Homo sapiens; X. tropicalis, Xenopus tropicalis; D. rerio, Danio rerio; C. elegans, Caenorhabditis elegans; D. melanogaster, Drosophila melanogaster. (C) GFP or GFP-tagged full-length, truncated, or mutated lyspersin (Lysp) were expressed by transfection into lyspersin-KO HeLa cells. Cells were extracted in detergent and subjected to immunoprecipitation with GFP-Trap beads. Endogenous myrlysin, LAMTOR1, LAMTOR4, and transgenic GFP were detected by immunoblotting (IB). The positions of molecular mass markers (in kilodaltons) are indicated. Arrows indicate the undegraded GFP-fusion proteins. (D and E) Y2H analysis of the interaction of lyspersin constructs fused to Gal4-BD (left) and BORC or Ragulator subunits fused to Gal4-AD (top). “, lyspersin. (F) Structural models generated with UCSF chimera (Pettersen et al., 2004) showing a potential lyspersin-binding site on the reported structure of LAMTOR2-LAMTOR3 (PDB code: 1VET [Kurzbauer et al., 2004]). (Left) Ribbon diagram showing amino acid residues at the potential binding site. (Right) Hydrophobicity surface representation highlights the hydrophobicity of the potential binding site (red patch at the top). Blue for the hydrophilic, to white, and to red for the hydrophobic residues. (G) Y2H analysis of the interaction of lyspersin or LAMTOR3 fused to Gal4-BD (left) and LAMTOR2 or LAMTOR2 mutants fused to Gal4-AD (top).
To identify the regions of human lyspersin that interact with BORC and Ragulator, we expressed different truncation mutants of GFP-lyspersin in lyspersin-KO HeLa cells and examined their ability to coprecipitate the endogenous myrlysin subunit of BORC and the LAMTOR1 and LAMTOR4 subunits of Ragulator. We observed that both BORC and Ragulator subunits coprecipitated with the C-terminal segment comprising DUF2365 (residues 190–357), but not the N-terminal unstructured segment (1–189; Fig. 2 C). Interestingly, mutation of the phylogenetically conserved L221 residue in CE1 (Fig. 2 B; Fig. S1) did not affect coprecipitation of myrlysin but completely abrogated coprecipitation of LAMTOR1 and LAMTOR4 (Fig. 2 C). In contrast, combined mutation of the conserved L349 and L352 residues in CE2 (Fig. 2 B; Fig. S1) abolished coprecipitation of myrlysin, but not LAMTOR1 and LAMTOR4 (Fig. 2 C).
To further dissect the determinants of lyspersin interaction with BORC and Ragulator subunits, we performed Y2H analyses. We observed that lyspersin interacted with itself, as well as with snapin, BLOS1, BLOS2, and KXD1, via the 256–357 segment corresponding to CE2 (Fig. 2 D). In contrast, interaction with LAMTOR2 involved the 190–255 segment encompassing CE1 (Fig. 2 E). In line with these results, combined mutation of the conserved L349 and L352 in CE2 specifically reduced interaction of lyspersin with itself and KXD1, whereas single mutations of F216 or L221 in CE1 specifically prevented interaction with LAMTOR2 (Fig. 2 E). Collectively, the results of the coprecipitation and Y2H analyses demonstrated that lyspersin assembles into the BORC complex via interaction of the DUF2365 CE2 region with lyspersin itself and with KXD1, whereas the DUF2365 CE1 region mediates interaction with LAMTOR2.
Next, we sought to identify structural determinants in LAMTOR2 that are involved in the interaction with lyspersin. The crystal structure of a LAMTOR2-LAMTOR3 (also known as p14/MP1) subcomplex shows that each subunit consists of a single roadblock/longin domain (Kurzbauer et al., 2004; Lunin et al., 2004; Fig. 2 F). Analysis of surface hydrophobicity potential predicted the presence of a hydrophobic patch on a face of LAMTOR2 distal to LAMTOR3 (Fig. 2 F). Y2H analyses showed that single or double substitutions of several hydrophobic residues at this site (L7, L11, V19, L24, L32, V86, L89, V112, and L115) by alanine abolished interaction with lyspersin but not LAMTOR3 (Fig. 2 G). Conservative substitution of some of these residues by other hydrophobic residues decreased but did not abolish binding to lyspersin, and substitution of hydrophilic residues in this region by alanine either had no effect (Q9, Q109, and Q113) or partially decreased binding (N88; Fig. 2 G). These results indicated that the hydrophobic site on LAMTOR2 likely mediates interaction with the DUF2365 CE1 region of lyspersin.
Arl8b recruitment to lysosomes and lysosome dispersal depend on interaction of lyspersin with BORC
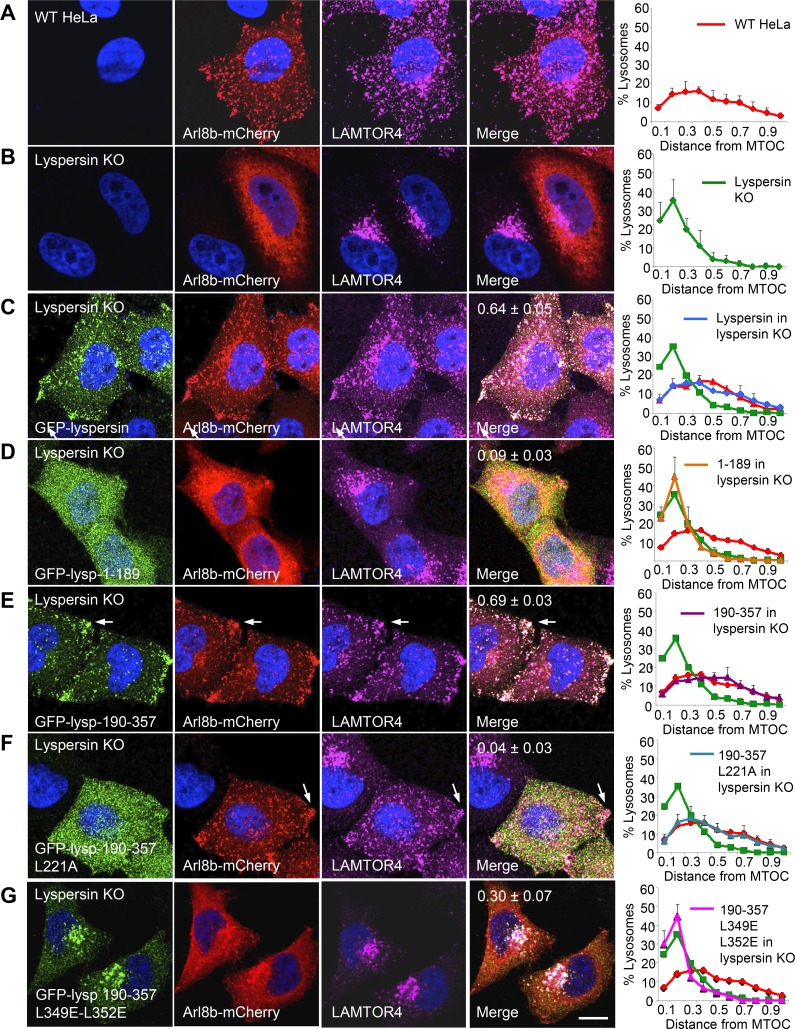
In WT HeLa cells, lysosomes are distributed throughout the cytoplasm, as observed by labeling for transgenic Arl8b-mCherry and endogenous LAMTOR4 (Fig. 3 A). In contrast, in lyspersin-KO cells, Arl8b-mCherry exhibited a more diffuse, cytosolic distribution, and lysosomes stained for LAMTOR4 were clustered in the perinuclear area (Fig. 3 B). This phenotype was similar to that previously described for myrlysin-KO (Pu et al., 2015) and diaskedin-KO cells (Guardia et al., 2016) and reflected the role of BORC in recruitment of Arl8 to lysosomes and lysosome movement toward the cell periphery. This experiment also demonstrated that Ragulator does not require BORC for association with lysosomes (Fig. 3 B). To assess the functional importance of the interactions of lyspersin with Ragulator and BORC, we rescued lyspersin-KO cells with WT and mutant GFP-lyspersin constructs. Full-length GFP-lyspersin expressed in lyspersin-KO cells localized to lysosomes and promoted Arl8b-mCherry recruitment and lysosome dispersal (Fig. 3 C). In contrast, GFP-lyspersin-1–189, corresponding to the N-terminal unstructured region that cannot interact with Ragulator and BORC (Fig. 2), did not associate with lysosomes or rescue Arl8b-mCherry recruitment and lysosome dispersal (Fig. 3 D). The effects of GFP-lyspersin-190–357, comprising the entire DUF2365 domain (Fig. 2), were indistinguishable from those of full-length lyspersin (Fig. 3 E), indicating that this is the functional part of the protein. Mutation of the CE1 L221 residue in GFP-lyspersin-190–357, involved in interaction with LAMTOR2 (Fig. 2), prevented the ability of this construct to be recruited to lysosomes but still rescued Arl8b-mCherry recruitment and lysosome dispersal (Fig. 3 F). Finally, combined mutation of CE2 residues L349 and L352 in GFP-lyspersin-190–357, involved in assembly into BORC (Fig. 2), resulted in a construct that partially rescued Arl8b-mCherry recruitment to lysosomes but did not rescue lysosome dispersal (Fig. 3 G). We concluded that interaction of lyspersin with Ragulator is required for recruitment of transgenic lyspersin to lysosomes, but not for Arl8b-mCherry recruitment and lysosome dispersal. In contrast, assembly of lyspersin into BORC is required for Arl8b-mCherry recruitment and lysosome dispersal, but not for recruitment of transgenic lyspersin to lysosomes.
Figure 3.
The DUF2365 domain mediates lyspersin recruitment to lysosomes via CE1 and Arl8b recruitment to lysosomes and lysosome dispersal via CE2. (A–G) The full-length or mutant GFP-lyspersin constructs indicated in the figure (see Fig. 2 B for scheme) were transiently expressed together with Arl8b-mCherry by transfection into lyspersin-KO HeLa cells. Fixed cells were immunostained with antibodies to GFP, mCherry, and LAMTOR4. WT (A) or lyspersin-KO (B) HeLa cells expressing Arl8b-mCherry were analyzed similarly as controls. Bar, 10 µm. Arrows point to lysosomes accumulated at peripheral sites. Pearson’s coefficients for the colocalization of GFP-lyspersin variants with Arl8b-mCherry are indicated in the merge images. Graphs on the right represent the distribution of lysosomes relative to the MTOC, normalized to the longest distance between the MTOC and the cell periphery. Values are the mean ± SD from 20 cells per condition. Note that overexpression of Arl8b-mCherry causes lysosomes to be more dispersed than in untransfected cells (Rosa-Ferreira and Munro, 2011; Pu et al., 2015).
Ragulator inhibits the function of BORC in lysosome dispersal
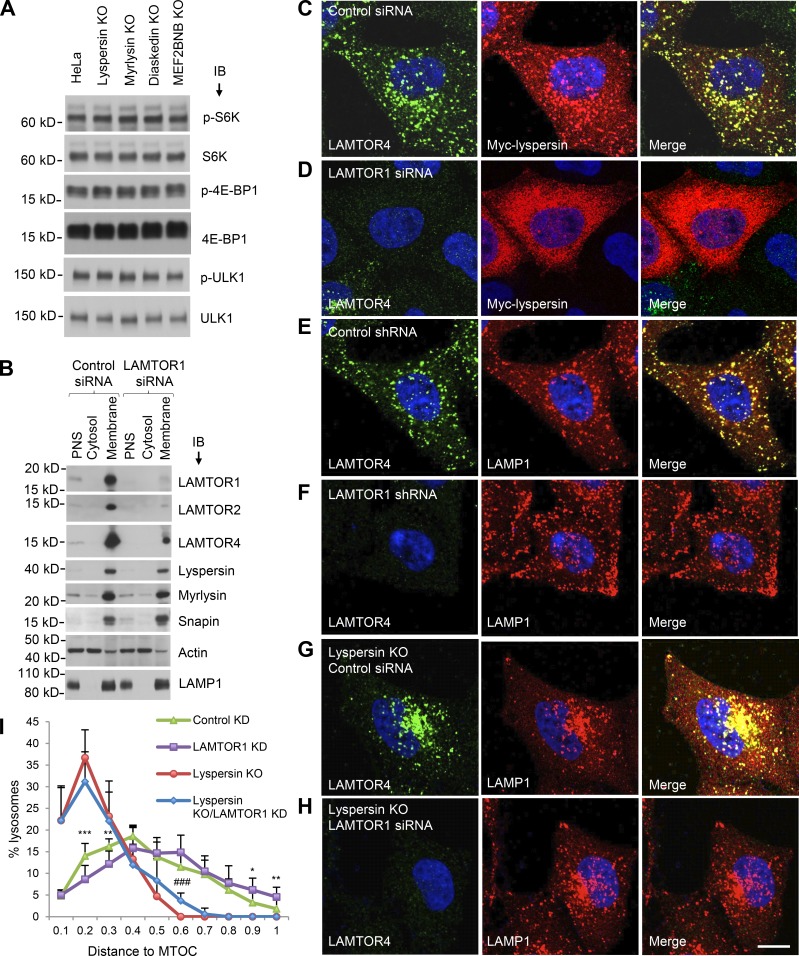
If the interaction of lyspersin with Ragulator is not required for Arl8b recruitment and lysosome dispersal, what is the function of this interaction? Because Ragulator is involved in activation of mTORC1 at the lysosomal membrane, we examined the effect of knocking out BORC subunits on mTORC1 activity. We observed that KO of the lyspersin, myrlysin, diaskedin, or MEF2BNB subunits of BORC had no effect on mTORC1 activity, as determined by immunoblotting for the phosphorylated forms of the S6K, 4EBP, and ULK1 substrates (Fig. 4 A). This experiment demonstrated that BORC, and therefore lysosome positioning, did not affect basal mTORC1 activity under the conditions of our experiments. We next addressed the converse possibility that Ragulator modulated the function of BORC in lysosome dispersal. We found that LAMTOR1 KD did not affect the levels of endogenous BORC subunits or their association with membranes (Fig. 4 B). However, immunofluorescence microscopy showed that LAMTOR1 KD did prevent association of transgenic myc-lyspersin with lysosomes (Fig. 4, C and D). The latter result agreed with the cytosolic localization of the GFP-lyspersin-190–357-L221A mutant (Fig. 3 F), which is unable to interact with Ragulator (Fig. 2). The apparent discrepancy in the requirement of Ragulator for lysosome recruitment of transgenic lyspersin (Fig. 3 F and Fig. 4 D) but not endogenous BORC (Fig. 4 B) can be explained by the fact that Ragulator is more abundant than BORC (∼600,000 vs. ∼100,000 copies per cell, respectively; Itzhak et al., 2016), and overexpressed lyspersin can therefore bind to Ragulator independently of the other BORC subunits. Importantly, LAMTOR1 KD shifted the distribution of lysosomes toward the cell periphery (Fig. 4, compare E and F), the opposite of lyspersin KO (Fig. 4 G). LAMTOR2 KO had a similar effect (Fig. S2). In contrast, LAMTOR1 KD did not cause lysosome dispersal in lyspersin-KO cells (Fig. 4 H), with the cells exhibiting the same perinuclear clustering of lysosomes as lyspersin-KO cells. Quantification of the cytoplasmic distribution of lysosomes confirmed these results (Fig. 4 I). These observations are consistent with Ragulator functioning upstream of BORC to inhibit lysosome movement toward the cell periphery.
Figure 4.
Ragulator inhibits BORC-mediated lysosome dispersal. (A) WT, lyspersin-KO, myrlysin-KO, diaskedin-KO, or MEF2BNB-KO HeLa cells cultured in regular medium were analyzed by SDS-PAGE and immunoblotting (IB) for phosphorylation (p) of the mTORC1 substrates S6K, 4E-BP1, and ULK1. (B) WT HeLa cells were transfected with nontargeting (control) and LAMTOR1 siRNAs and disrupted without detergent. Cytosolic and membrane fractions were separated by centrifugation of PNS for 1 h at 100,000 g and analyzed by SDS-PAGE and IB for the proteins indicated in the figure. In A and B, the positions of molecular mass markers (in kilodaltons) are indicated on the left. (C–H) shRNA- or siRNA-mediated KD was performed in WT HeLa or lyspersin-KO cells, as indicated in the figure. Nontargeting shRNA or siRNA were used as controls. The distribution of lysosomes and the expression and localization of Ragulator were visualized by immunostaining with antibodies to LAMP1 and LAMTOR4, respectively. Myc-lyspersin was detected by immunostaining for the myc epitope. Bar, 10 µm. (I) Lysosome distribution was quantified in the cells indicated in the figure (n = 13 cells from three independent experiments) using Imaris software. The distance between lysosomes and the MTOC was normalized to the longest distance between the cell periphery and the MTOC. Values are the mean ± SD. LAMTOR1 KD versus control KD, *, P < 0.05; **, P < 0.01; ***, P < 0.001; lyspersin KO or LAMTOR1 KD versus lyspersin KO, ###, P < 0.001 (Student’s t test).
Ragulator reduces the population of motile lysosomes
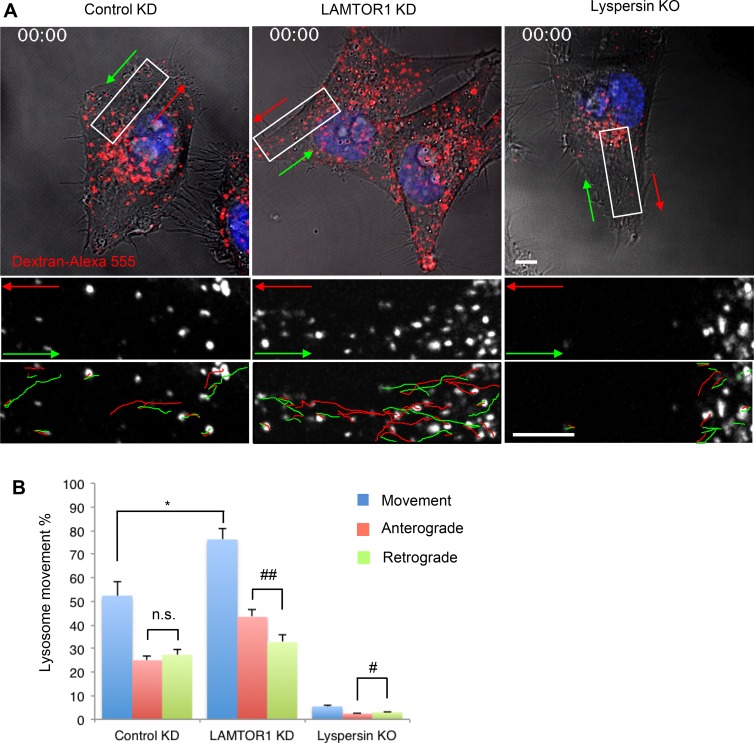
In general, only a fraction of the lysosomal population is motile, and the rest is stationary (Jongsma et al., 2016). Live-cell imaging of lysosomes loaded for 6 h with dextran–Alexa Fluor 555 and chased overnight (Fig. 5 A and Videos 1, 2, and 3) showed that in WT HeLa cells, the percentage of moving lysosomes was 52.4 ± 6.1% (Fig. 5 B). Of these, 47.7 ± 3.9% and 52.3 ± 3.9% moved in anterograde and retrograde directions, respectively (Fig. 5 B). LAMTOR1 KD increased the overall percentage of moving lysosomes to 76.2 ± 4.4% (Fig. 5 B), with anterograde lysosomes increasing to 57.0 ± 4.1% and retrograde lysosomes decreasing to 43.0 ± 4.1% (Fig. 5 B). In contrast, lyspersin KO decreased the percentage of lysosomes that underwent long-range movement to 5.5 ± 0.4%, with 44.2 ± 5.5% and 55.8 ± 5.5% of these moving in anterograde and retrograde directions, respectively (Fig. 5 B). The velocities of movement of anterograde and retrograde lysosomes were not significantly altered by LAMTOR1 KD or lyspersin KO (all in the range of 0.39–0.43 µm/s). These experiments are consistent with Ragulator acting to decrease the number, but not the velocity, of moving lysosomes. Together with the findings described in the previous sections, these experiments indicated that Ragulator inhibits the function of BORC in promoting lysosome movement toward the cell periphery.
Figure 5.
Ragulator KD increases anterograde lysosome transport. (A) Control-KD, LAMTOR1-KD, or lyspersin-KO cells were allowed to internalize dextran–Alexa Fluor 555 for 6 h at 37°C, chased overnight, and analyzed by live-cell imaging. Images are the first frames from Videos 1, 2, and 3. Bottom panels are magnified views of the boxed areas. Anterograde and retrograde trajectories are represented by red and green lines, respectively. Bars, 5 µm. (B) Long-range lysosome movement was tracked and quantified with ImageJ from control-KD, LAMTOR1-KD, or lyspersin-KO cells (five cells from five independent experiments). Values are the mean ± SD. *, P < 0.05; n.s., not significant (Student’s t test); #, P < 0.05; ##, P < 0.01 (ANOVA). In addition to changes in lysosome positioning and motility, we noticed that LAMTOR1 KD increased the number of lysosomes, probably because of enhanced lysosome biogenesis induced by mTORC1 inactivation and consequent TFEB activation and nuclear translocation (Settembre et al., 2012).
The ability of Ragulator to control lysosome positioning is independent of mTORC1
Next, we tested whether the ability of Ragulator to control lysosome positioning was dependent on its role as an mTORC1 regulator. We observed that KD of either LAMTOR1 or RAPTOR (the key scaffold of the mTORC1 complex) inhibited mTORC1 activity (Fig. 6 A) and dissociated mTOR from lysosomes (Fig. 6 B). However, whereas LAMTOR1 KD shifted lysosomes toward the cell periphery (Fig. 4 F and Fig. 6, B and C), RAPTOR KD had no effect on lysosome distribution (Fig. 6, B and C). We also tested the effects of several mTORC1 inhibitors, including PP242, rapamycin, KU-0063794, and Torin1. We found that these compounds were effective at inhibiting mTORC1 (Fig. 6 D), but did not affect lysosome positioning (Fig. 6, E and F). From these experiments, we concluded that lysosome positioning is dependent on Ragulator but not mTORC1. Ragulator is therefore likely to exert its negative regulatory effect on lysosome dispersal positioning directly through its interaction with BORC.
Figure 6.
Ragulator regulates lysosome positioning independently of mTORC1. (A) HeLa cells stably transfected with nontargeting (control), LAMTOR1, or RAPTOR shRNAs and cultured in regular medium were analyzed by SDS-PAGE and immunoblotting for phosphorylation (p) of the mTORC1 substrate S6K and levels of LAMTOR1, LAMTOR2, and RAPTOR. The positions of molecular mass markers (in kilodaltons) are indicated on the left. (B) mTOR localization and lysosome distribution in the cells in A was visualized by immunostaining with antibodies to mTOR and LAMP1. Bar, 10 µm. (C) Lysosome distribution in the cells in B (15 cells from three independent experiments for each cell line) was quantified by measuring lysosome-to-MTOC distance using Imaris software and shown as box-and-whisker plots. The ends of whiskers represent the minima and maxima of the data. ****, P < 0.0001; n.s., not significant (ANOVA). (D) HeLa cells were treated with the mTOR inhibitors PP242 (200 nM), rapamycin (2 µM), KU-0063794 (1 µM), or Torin1 (200 nM) or with DMSO (control), for different times at 37°C, and phosphorylation of the indicated mTORC1 substrates was analyzed by SDS-PAGE and immunoblotting. The positions of molecular mass markers (in kilodaltons) are indicated on the left. (E) HeLa cells were treated with mTOR inhibitors for 2 h at 37°C as in D and immunostained for LAMP1. Bar, 10 µm. (F) Quantification of lysosome distribution from 20 cells in three independent experiments such as that in E using Imaris. Data are shown as box-and-whisker plots with minimum and maximum range. n.s., not significant (ANOVA).
BORC–Ragulator interaction controls changes in lysosome positioning during nutrient starvation
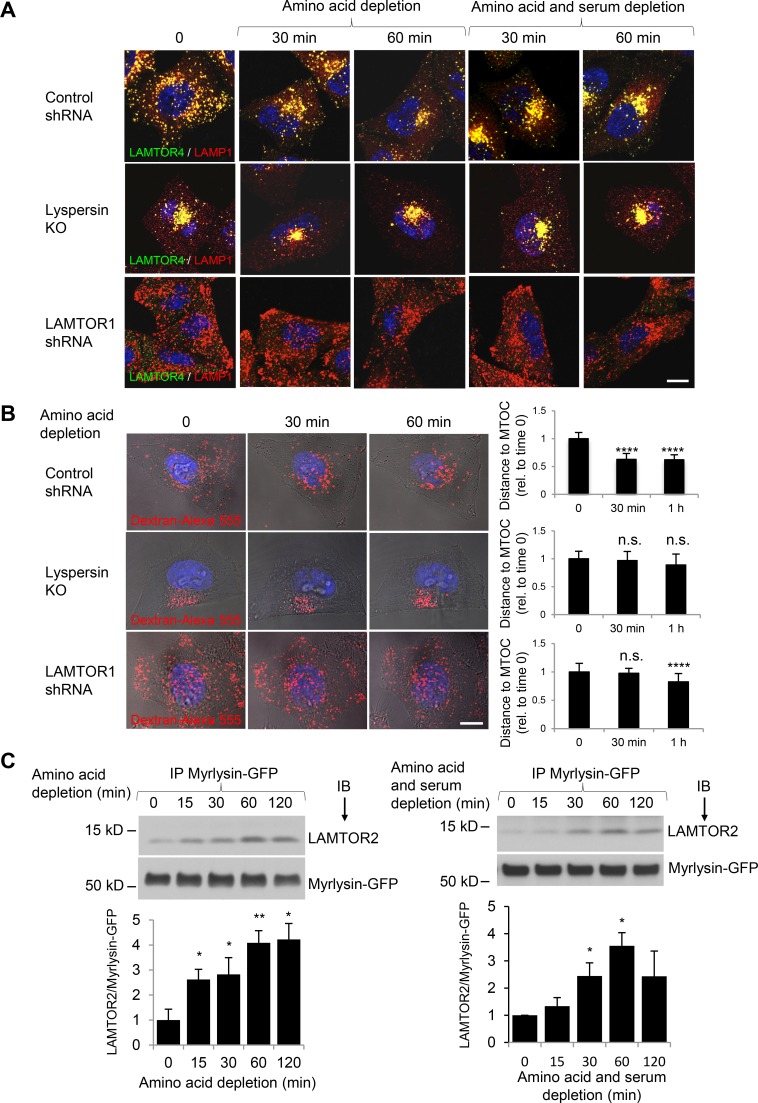
Next, we addressed the question of whether the physical and functional interactions of BORC with Ragulator were involved in the regulation of lysosome positioning during nutrient starvation. In agreement with previous studies (Korolchuk et al., 2011; Jung et al., 2015; Starling et al., 2016), we observed that incubation of WT HeLa cells in amino acid–free medium or amino acid– and serum-free medium (HBSS), caused juxtanuclear clustering of lysosomes, as detected by staining of fixed cells for endogenous LAMP1 and LAMTOR4 (Fig. 7 A). In contrast, in lyspersin-KO cells, lysosomes were already clustered in the juxtanuclear region before nutrient starvation and remained clustered after removal of the nutrients (Fig. 7 A). In LAMTOR1 KD cells, lysosomes were more disperse than in WT cells in nutrient-replete conditions, and this dispersal was only modestly reversed after 60 min of nutrient starvation (Fig. 7 A). Similar observations were made by live-cell imaging of WT, lyspersin-KO, and LAMTOR1-KD cells, in which changes in the distribution of lysosomes loaded with dextran–Alexa Fluor 555 during amino acid depletion could be visualized in the same cell (Fig. 7 B). Therefore, both BORC and Ragulator are required for changes in lysosome positioning observed during nutrient starvation.
Figure 7.
BORC–Ragulator interactions control lysosome positioning during nutrient starvation. (A) WT HeLa cells transfected with control shRNA or LAMTOR1 shRNA and lyspersin-KO cells were incubated in medium without amino acids, or without amino acids and serum (HBSS), for the indicated times. Cells were immunostained with antibodies to LAMTOR4 and LAMP1. Bar, 10 µm. (B) Cells as in A were allowed to internalize dextran–Alexa Fluor 555 for 6 h at 37°C and chased overnight. Cells were placed in amino acid–depleted medium and imaged live. Each row shows images of the same cell at different times of amino acid starvation. Bar, 10 µm. The distance between lysosomes and the MTOC in control-KD (15 cells from four independent experiments), LAMTOR1-KD (18 cells from four independent experiments), and lyspersin-KO cells (15 cells from three independent experiments) was quantified using Imaris, and the mean ± SD at each time point was normalized to the mean distance at time 0. ****, P < 0.0001; n.s., not significant (ANOVA). (C) Myrlysin-KO HeLa cells stably expressing myrlysin-GFP were incubated in medium lacking amino acids or amino acids and serum. Cells were then subjected to immunoprecipitation (IP) with GFP-Trap beads, followed by immunoblotting (IB) with antibodies to LAMTOR2 and GFP. The positions of molecular mass markers (in kilodaltons) are indicated on the left. Bar graphs show the ratios of LAMTOR2 to myrlysin-GFP quantified by densitometry. Values are the mean ± SD from three independent experiments. *, P < 0.05; **, P < 0.01 (ANOVA).
To determine whether the BORC–Ragulator interaction was affected by nutrient starvation, we expressed myrlysin-GFP in myrlysin-KO HeLa cells and analyzed the coprecipitation of this protein with endogenous LAMTOR2 at different times after withdrawal of amino acids or amino acids and serum (Fig. 7 C). We found that both starvation regimens increased the coprecipitation of myrlysin-GFP with LAMTOR2 (Fig. 7 C). Additional experiments showed that amino acid starvation did not significantly alter Arl8b-GFP coprecipitation with myrlysin-FOS or Arl8b association with lysosomes (Fig. S3). Collectively, these observations demonstrated that nutrient starvation enhances the interaction of Ragulator with BORC without affecting the recruitment of Arl8b to lysosomes.
Finally, we examined whether the SLC38A9 amino acid transceptor, previously shown to regulate the ability of Ragulator and the Rag GTPases to recruit mTORC1 to the lysosomal membrane (Jung et al., 2015; Rebsamen et al., 2015; Wang et al., 2015), was also required for Ragulator-BORC–dependent regulation of lysosome positioning. Indeed, we observed that coprecipitation of LAMTOR2 with myrlysin-GFP was already elevated in amino acid–replete SLC38A9-KD cells to levels similar to those in amino acid–depleted control cells, and these levels did not change upon removal of amino acids from the medium (Fig. 8 A). Moreover, lysosomes exhibited more juxtanuclear clustering in SLC38A9-KD cells relative to control cells in amino acid–replete medium (Fig. 8 B), albeit to a lesser extent than in amino acid–depleted medium. These observations are consistent with SLC38A9 functioning upstream of Ragulator and BORC in the regulation of lysosome positioning.
Figure 8.
SLC38A9 is required for amino acid regulation of the Ragulator–BORC interaction and lysosome positioning. (A) Control, nontargeting siRNA (C) or siRNA targeting SLC38A9 (SLC) was electroporated into myrlysin-KO HeLa cells stably rescued with myrlysin-GFP. Cells were incubated in medium with or without amino acids (AA) for 60 min and then subjected to immunoprecipitation (IP) with GFP-Trap beads, followed by immunoblotting (IB) with antibodies to LAMTOR2 or GFP. The positions of molecular mass markers (in kilodaltons) are indicated on the left. Bar graphs show the ratios of LAMTOR2 to myrlysin-GFP quantified by densitometry (in arbitrary units). Values are the mean ± SD from three independent experiments. *, P < 0.05 (Student’s t test). (B) Cells were transfected and treated as in A and subjected to immunostaining with antibodies to GFP and LAMP1. Bar, 10 µm. The distribution of lysosomes relative to the MTOC was quantified using Imaris and normalized to the longest distance between the MTOC and the cell periphery. Values are the mean ± SD from 20 cells per condition. (C) Hypothetical model for the regulation of lysosome positioning by Ragulator and BORC. In amino acid–replete conditions, SLC38A9 tightly binds to Ragulator and the Rag GTPases, leading to RagA/B activation and mTORC1 recruitment to the lysosomal membrane. At the same time, SLC38A9 binding weakens the Ragulator–BORC interaction, allowing BORC and Arl8 to recruit kinesins and thus promote anterograde transport of lysosomes. In amino acid–depleted conditions, weakening of the interaction with SLC38A9 causes a conformational change in Ragulator that prevents activation of the Rags and recruitment of mTORC1 to lysosomes, while simultaneously enhancing an inhibitory effect on BORC. This enhancement does not interfere with Arl8 association with lysosomes, but does prevent the recruitment of kinesins to lysosomes, reducing their anterograde transport and leading to their clustering in the juxtanuclear area. The generic kinesin shown in the scheme represents either KIF1B or KIF5B in complex with the corresponding adaptors (Guardia et al., 2016).
Discussion
The results presented here indicate that Ragulator directly interacts with BORC, through their respective LAMTOR2 and lyspersin subunits, to negatively regulate the function of BORC in movement of lysosomes toward the peripheral cytoplasm. Amino acid starvation strengthens this interaction, resulting in an enhanced negative effect of Ragulator on BORC. This mechanism provides an explanation for the juxtanuclear clustering of lysosomes induced by nutrient depletion. Also importantly, this effect of Ragulator on BORC is independent of mTORC1 signaling, supporting the notion that Ragulator plays additional roles in nutrient signaling that are distinct from its well-established role in mTORC1 regulation.
Structural determinants of the BORC–Ragulator interaction
The interaction of BORC with Ragulator was initially detected by affinity purification/MS (Fig. 1 A) and coimmunoprecipitation (Fig. 1 B). Subsequent Y2H analyses revealed a strong interaction of lyspersin with LAMTOR2 and weak interaction of BLOS1 with LAMTOR1 (Fig. 1 C). The interaction with LAMTOR2 was mediated by DUF2365 at the C terminus of lyspersin, which previously had been only predicted by bioinformatic analyses. We found that lyspersin DUF2365 binds LAMTOR2 via its CE1 subdomain and BORC via its CE2 subdomain (Fig. 2). The latter interaction is essential for the function of BORC in Arl8 recruitment to lysosomes and movement of lysosomes toward the cell periphery (Fig. 3). Previous structural analyses predicted a potential site of protein–protein interactions at the side of LAMTOR2 opposite to that involved in LAMTOR3 binding (Kurzbauer et al., 2004; Lunin et al., 2004). Our mutational and Y2H analyses now show that hydrophobic residues on this site are required for interaction with lyspersin (Fig. 2, F and G). From these findings, we infer that CE1 is a ligand for the hydrophobic patch on LAMTOR2, although definitive demonstration of this interaction will require additional biochemical and structural analyses.
Ragulator is a negative regulator of BORC
The direct interaction of BORC with Ragulator raised the question of who regulates whom. KO or KD of either complex did not affect the association of the other complex with lysosomes (Fig. 2 B and Fig. 4, B and G). Moreover, KO of several BORC subunits did not alter the basal levels of mTORC1 (Fig. 4 A). Schweitzer et al. (2015) showed that overexpression of C17orf59 (lyspersin) inhibited mTORC1 activity by disruption of the interaction of Ragulator with the Rag GTPases. However, BORC is approximately sixfold less abundant than Ragulator (Itzhak et al., 2016), so it is unlikely that the effects of lyspersin overexpression occur under physiological conditions. Thus, at endogenous levels, BORC does not appear to influence Ragulator function. Instead, our findings show that Ragulator regulates BORC. Indeed, Ragulator-KD cells (Figs. 4 F, 5 A, and 6 B) or Ragulator-KO cells (Fig. S2; Teis et al., 2002) exhibit greater dispersal of lysosomes and a higher percentage of motile lysosomes, particularly in the anterograde direction (Fig. 5 B). Importantly, the ability of Ragulator KD to scatter lysosomes is largely dependent on the presence of BORC (Fig. 4 H). These observations indicate that Ragulator functions upstream of BORC to inhibit lysosome dispersal. This function of Ragulator likely contributes to changes in lysosome positioning that occur in response to nutrient availability. Indeed, both Ragulator and BORC are required for these changes (Fig. 7, A and B), and the Ragulator–BORC interaction is enhanced by depletion of amino acids or amino acids and serum combined (Fig. 7 C). These findings are consistent with a mechanism in which nutrient depletion causes a change in Ragulator that enhances its negative regulatory interaction with BORC.
The effects of Ragulator on BORC and lysosome positioning are independent of mTORC1
Despite the direct interaction of Ragulator with BORC, it was still possible that Ragulator could exert additional effects through its role as an mTORC1 activator. However, we found that inhibition of mTORC1 by either KD of its RAPTOR subunit (Fig. 6, A–C) or incubation with pharmacologic inhibitors (Fig. 6, D–F) had no effect on lysosome positioning, in agreement with previous observations by Korolchuk et al. (2011). Therefore, the regulation of BORC by Ragulator is independent of mTORC1 activity. Sensing of amino acid levels through Ragulator thus causes separate effects on lysosome positioning via direct interaction with BORC (this study) and on mTORC1 signaling via activation of RagA/B (Bar-Peled et al., 2012). These findings add to the notion that Ragulator has functions besides regulating mTORC1. In fact, the LAMTOR2 and LAMTOR3 subunits of Ragulator were first recognized as a scaffold complex for the signal transducing kinases MEK1 and ERK1 (Teis et al., 2002). In addition, Ragulator and the Rags were shown to regulate lysosomal activity in zebrafish microglia independently of mTORC1 (Shen et al., 2016).
A hypothetical model for the regulation of lysosome positioning by a direct inhibitory effect of Ragulator on BORC
The scheme in Fig. 8 C depicts a hypothetical model for the role of Ragulator and BORC in the regulation of lysosome positioning. We propose that amino acid levels sensed by SLC38A9 control a switch of Ragulator between two functional states. Under amino acid–replete conditions, Ragulator frees BORC to promote coupling of lysosomes to kinesins 1 and 3 for anterograde movement toward the cell periphery, while at the same time activating RagA/B for recruitment and activation of mTORC1 at the lysosomal membrane. Under amino acid–depleted conditions, on the other hand, Ragulator adopts an alternative state that directly inhibits BORC, uncoupling lysosomes from the kinesins while concomitantly ceasing its activation of RagA/B and mTORC1. This “yin-yang” mechanism of Ragulator function thus coordinates lysosome dispersal with mTORC1 activation and juxtanuclear clustering of lysosomes with mTORC1 inactivation.
The mechanistic events that follow the direct inhibition of BORC by Ragulator remain to be elucidated. Inhibition of BORC does not seem to prevent its interaction with Arl8 or the recruitment of Arl8 to lysosomes (Fig. S3). It is thus possible that Ragulator interferes with the interaction of both BORC and Arl8 with downstream components of the lysosome dispersal machinery. It is also unclear how the effects of nutrient starvation on anterograde transport of lysosomes mediated by the Ragulator-BORC-Arl8-kinesin axis relate to effects on retrograde transport mediated by dynein-dynactin and their regulators (Li et al., 2016; Starling et al., 2016). Lysosome positioning is also controlled by tethering to the perinuclear endoplasmic reticulum via the ubiquitin ligase RNF26 (Jongsma et al., 2016). Future studies will have to address how all of these processes are coordinated to control spatial-temporal changes in lysosome positioning in response to nutrient availability.
Materials and methods
Plasmids and antibodies
Plasmids and antibodies used in this study are listed in Tables 1 and 2, respectively.
Table 1. Plasmids used in this study.
| Vector and insert | Tag | Remarks |
|---|---|---|
| pEGFP-C2 | ||
| Full-length, truncated, and mutated human lyspersin (C17orf59) | N-EGFP | This study |
| pcDNA3.1 | ||
| Human myrlysin | N-OSF; C-FOS | Pu et al., 2015 |
| Human BLOS2 | N-OSF | Pu et al., 2015 |
| Human lyspersin | N-OSF; C-FOS | Pu et al., 2015 |
| Human pallidin | N-OSF | This study |
| Human LAMTOR1 | C-FOS | This study |
| pCI-neo | ||
| Human lyspersin | N-3myc | This study |
| p-mCherry-N1 | ||
| Human Arl8b | C-mCherry | This study |
| pLKO.5-puro | ||
| Nontargeting shRNA | Sigma-Aldrich (SHC216) | |
| Human LAMTOR1 shRNA | Sigma-Aldrich (SHCLNG-NM_017907, TRCN0000282725) | |
| pLKO.1 | ||
| Human RAPTOR shRNA | Addgene (1858) | |
| pSpCas9(BB)-2A-GFP (PX458) | ||
| Sequence from human LAMTOR2 | EGFP, FLAG | Addgene (48138) |
| pQCXIP | ||
| Human SLC38A9 | N-EGFP | This study |
| pGBT9 | ||
| Human lyspersin | This study | |
| Human snapin | This study | |
| Human MEF2BNB | This study | |
| Human myrlysin | This study | |
| Human BLOS1 | This study | |
| Human BLOS2 | This study | |
| Human KXD1 | This study | |
| Human diaskedin | This study | |
| pGADT7 | ||
| Human lyspersin | This study | |
| Human snapin | This study | |
| Human MEF2BNB | This study | |
| Human myrlysin | This study | |
| Human BLOS1 | This study | |
| Human BLOS2 | This study | |
| Human KXD1 | This study | |
| Human diaskedin | This study | |
| Human LAMTOR1 | This study | |
| Human LAMTOR2 and mutants | This study | |
| Human LAMTOR3 | This study | |
| Human LAMTOR4 | This study | |
| Human LAMTOR5 | This study | |
| pGBT9 | ||
| Human lyspersin 190-255 | This study | |
| Human lyspersin 256-357 | This study | |
| Human lyspersin 190-357 | This study | |
| Human lyspersin 190-end F216A | This study | |
| Human lyspersin 190-end L221A | This study | |
| Human lyspersin 190-end L349E, L352E | This study | |
| Human lyspersin 256-357 | This study | |
| Human lyspersin 209-357 | This study |
Table 2. Antibodies used in this study.
| Antigen | Source | Catalog number |
|---|---|---|
| Actin | BD Biosciences | 612656 |
| mCherry | Thermo Fisher Scientific | M11217 |
| p-4E-BP1 (Thr37/46) | Cell Signaling Technology | 2855 |
| 4E-BP1 | Cell Signaling Technology | 9452 |
| FLAG epitope | Sigma-Aldrich | F1804 |
| GFP | Thermo Fisher Scientific | A11122 |
| GFP-HRP | Miltenyi Biotec | 130-091-833 |
| LAMP1 | Developmental Studies Hybridoma Bank | H4A3 |
| LAMTOR1 | Cell Signaling Technology | 8975 |
| LAMTOR2 | Cell Signaling Technology | 8145 |
| LAMTOR4 | Cell Signaling Technology | 13140 |
| Myc epitope | Santa Cruz Biotechnology | sc-40 |
| Myc-HRP | Santa Cruz Biotechnology | sc-40 HRP |
| Myrlysin (LOH12CR1) | Abgent | AP5806b |
| RAPTOR | Cell Signaling Technology | 2280 |
| p-S6K (Thr389) | Cell Signaling Technology | 9234 |
| S6K | Cell Signaling Technology | 2708 |
| Snapin | Synaptic Systems | 148 002 |
| ULK1 | Cell Signaling Technology | 8054 |
| p-ULK1 | Cell Signaling Technology | 14202 |
| mTOR | Cell Signaling Technology | 2983 |
| p-mTOR | Cell Signaling Technology | 5536 |
Cell culture, transfection, and RNAi
HeLa, H4, and HEK293T cells were cultured in DMEM supplemented with 10% FBS and 25 mM Hepes. MycoZap Plus-CL (Lonza) was used to prevent mycoplasma contamination. Plasmids were transfected using Lipofectamine 2000 (Thermo Fisher Scientific). Cells were analyzed 48–72 h after transfection. siRNA was transfected twice with Oligofectamine (Thermo Fisher Scientific) at 24 and 72 h and analyzed 5–6 d after seeding. Alternatively, siRNA was delivered into cells by electroporation using Amaxa Nucleofector (Lonza) twice at 24 and 72 h and analyzed by immunoprecipitation 5 d after seeding. Lentivirus-based shRNA plasmids (Sigma-Aldrich) were cotransfected with lentivirus-packaging plasmids into HEK293T cells. Medium was collected 48 and 72 h after transfection, centrifuged for 3 min at 1,000 g to remove debris, aliquoted, and kept at −80°C for further use. HeLa cells were infected with virus, and stably transduced cells were selected with 2 µg/ml puromycin. Selected cells were reseeded and allowed to form single clones. After 12 d, KD was tested by immunostaining or immunoblotting using antibodies to the target proteins. A similar protocol was used to generate stably transduced cells expressing GFP-SLC38A9 using retrovirus. Nontargeting siRNA was from Eurofins. Smart-pool siRNA targeting LAMTOR1 (L-020916-02) and SLC38A9 (L-007337-02) was purchased from GE Healthcare.
TAP-MS
TAP-MS was performed as previously described (Pu et al., 2015). In brief, HeLa or H4 cells stably expressing proteins tagged with N-terminal OSF or C-terminal FOS were extracted with 1% Triton X-100, 50 mM Tris-HCl, pH 7.4, 300 mM NaCl, and 5 mM EDTA, supplemented with proteinase inhibitor cocktail (Roche). After clearing by centrifugation for 15 min at 17,000 g, proteins were sequentially purified on Strep-Tactin (IBA) and FLAG antibody–coated beads (Sigma-Aldrich). Beads were washed with 0.2% Triton X-100, 50 mM Tris-HCl, pH 7.4, 300 mM NaCl, and 5 mM EDTA, and bound proteins were eluted with 2.5 mM desthiobiotin and 150 ng/ml of 3× FLAG peptide (Sigma-Aldrich), respectively. Proteins were precipitated with 10% TCA, washed with acetone, air-dried, and analyzed by liquid chromatography (LC)/MS at the Taplin MS facility (Harvard Medical School, Boston, MA). The results of MS analysis for OSF-BLOS2 are shown in Table S1 from Pu et al. (2015), and those for OSF-lyspersin, lyspersin-FOS, and OSF-Pallidin are shown in in Tables S1, S2, and S3 of the current study.
Y2H analysis
Complementary DNAs encoding full-length, truncated, or mutated BORC subunits or Ragulator subunits were cloned in-frame into the Gal4-binding domain (BD) plasmid pGBT9 or the Gal4-activation domain (AD) plasmid pGADT7. EZ kit (MP Biomedicals) was used to transform AH109 yeast reporter strain with pGBT9 and pGADT7 constructs. Medium lacking leucine and tryptophan (+His) was used to select double transformants, and medium lacking leucine, tryptophan, and histidine (−His) was used for monitoring the interaction of the fusion proteins.
Immunoprecipitation
Cells were washed twice with ice-cold PBS and extracted with lysis buffer containing proteinase inhibitor cocktail. After clearing by centrifugation at 20,000 g at 4°C for 15 min, cell lysates were incubated with GFP-Trap beads (ChromoTek) for 1 h at 4°C. Bound proteins were washed twice with lysis buffer and once with PBS, eluted with 2× SDS sample buffer, heated at 95°C for 2 min, and subjected to SDS-PAGE and immunoblotting.
Cell fractionation
Cells were washed twice with ice-cold PBS, scraped into 0.25 mM sucrose and 20 mM Tris, pH 7.4, supplemented with proteinase inhibitor cocktail and disrupted by passage through a 25G needle 30 times. Postnuclear supernatant (PNS) was obtained by centrifugation at 1,000 g at 4°C for 2 min and further fractionated by centrifugation at 100,000 g at 4°C for 1 h to separate cytosol and membranes. PBS containing 2% SDS was used to dissolve the membrane fraction. After adding SDS sample buffer and heating at 95°C for 2 min, proteins in PNS, cytosol, and total membranes were analyzed by SDS-PAGE and immunoblotting.
CRISPR/Cas9 KO
We ablated the LAMTOR2 gene using the CRISPR/Cas9 system (Cong et al., 2013). In brief, two 20-bp targeting sequences (5′-GGGCGGCGCGCGGCTCCCTG-3′ and 5′-GCTGCTTCCCGTAGTCCCGT-3′) were synthesized (Eurofins) and introduced separately into the px458 plasmid (Addgene). HeLa cells were cotransfected with both plasmids together with pQCXIP plasmid. Transformants were selected with 2 µg/ml puromycin for 48 h and kept in regular culture medium for another 12 d to allow single colony formation. Genomic DNA was extracted from individual colonies, and cleavage of the target sequence was tested by PCR using a pair of primers (5′-CCTCATCGCACAGAATTGTG-3′ and 5′-ATCTTCCGGAGATCCTGTCC-3′), which produced a smaller band in KO cells relative to WT cells. The KO was confirmed by Sanger sequencing and immunoblotting. A similar protocol was used to knock out the gene encoding lyspersin using the targeting sequence 5′-GGAGGAGGAAGACAACGACG-3′ and SLC38A9 using 5′-GGCTCAAACTGGATATTCATAGG-3′, as reported before (Wang et al., 2015). Other HeLa-KO cells were generated similarly as described previously (Pu et al., 2015; Guardia et al., 2016; Jia et al., 2017). The targeting sequences are as follows: myrlysin, 5′-GCTCAACAGCATGCTGCCCG-3′ and 5′-AGCAGATCCAGAAAGTGAAC-3′; diaskedin, 5′-CCAGAGTCTCAAGCGCGGTT-3′ and 5′-TCTCCCGGCCTGATAGTCCG-3′; and MEF2BNB, 5′-TTTCCCGGTTCGCTCGGCCG-3′ and 5′-CTTTAATTACCGGTCCCCCC-3′.
Nutrient starvation
Complete nutrient starvation was performed by washing the cells once and then incubating for 15 min to 2 h at 37°C in HBSS containing 3 g/liter NaHCO3 and 25 mM Hepes. Amino acid starvation was performed in HBSS containing 3 g/liter NaHCO3 and 25 mM Hepes supplemented with 10% dialyzed FBS (Thermo Fisher Scientific) and 4.5 g/liter glucose.
Live-cell imaging
Cells were incubated in complete medium containing 100 mg/ml dextran–Alexa Fluor 555 (Thermo Fisher Scientific) for 6 h, reseeded onto fibronectin-coated Lab-Tek chambers (Thermo Fisher Scientific), and chased overnight. Live-cell imaging was performed in complete or amino acid–depleted medium using a Zeiss LSM780 confocal microscope equipped with a Plan-Apochromat 63×/1.4 oil immersion objective, a spectral detector system (two PMTs, GaAsp 32× array, and a transmission PMT), and an environmental chamber set at 37°C and 5% CO2 and using Definite Focus. Images were acquired by using software ZEN 2012 (Zeiss) and processed by ImageJ (National Institutes of Health) including brightness adjustment, channel merging, manual tracking, and conversion of images to movies. The software Imaris (Bitplane) was used to measure the distance from the lysosome to the microtubule-organizing center (MTOC).
Statistical methods
Statistical significance was determined by comparing multiple datasets with different numbers of trials using one-way ANOVA or two datasets using Student’s t test with unequal variance. Probability values and number of trials are given in the figure captions and legends where appropriate. All graphs show the mean ± SD.
Online supplemental material
Fig. S1 shows conserved structural elements of the lyspersin DUF2365; Fig. S2 shows how LAMTOR2 KO redistributes lysosomes toward the cell periphery; and Fig. S3 shows the association of Arl8b with lysosomes during amino acid depletion. Tables S1, S2, and S3 show MS data from affinity purifications using OSF-lyspersin, lyspersin-FOS, and OSF-pallidin as bait. Videos show lysosome movement in control (Video 1), LAMTOR1-KD (Video 2), and lyspersin-KO (Video 3) cells.
Supplementary Material
Acknowledgments
We thank X. Zhu for expert technical assistance and all members of the Bonifacino group for helpful discussions.
This work was funded by the Intramural Program of the National Institute of Child Health and Human Development (project ZIA HD001607).
The authors declare no competing financial interests.
Author contributions: J. Pu and J.S. Bonifacino conceived the project. J. Pu performed most of the experiments. T. Keren-Kaplan performed Y2H assays and modeled the structure of LAMTOR2-3. J. Pu and J.S. Bonifacino wrote the manuscript and all three authors edited it.
Footnotes
Abbreviations used:
- BORC
- BLOC-1–related complex
- CE
- conserved element
- KD
- knockdown
- KO
- knockout
- MS
- mass spectrometry
- mTORC
- mammalian target of rapamycin complex
- TAP
- tandem affinity purification
- Y2H
- yeast two-hybrid
References
- Bar-Peled L., Schweitzer L.D., Zoncu R., and Sabatini D.M.. 2012. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell. 150:1196–1208. 10.1016/j.cell.2012.07.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino J.S., and Neefjes J.. 2017. Moving and positioning the endolysosomal system. Curr. Opin. Cell Biol. 47:1–8. 10.1016/j.ceb.2017.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett P.E., Barrow R.K., Cohen N.A., Snyder S.H., and Sabatini D.M.. 1998. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc. Natl. Acad. Sci. USA. 95:1432–1437. 10.1073/pnas.95.4.1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantalupo G., Alifano P., Roberti V., Bruni C.B., and Bucci C.. 2001. Rab-interacting lysosomal protein (RILP): the Rab7 effector required for transport to lysosomes. EMBO J. 20:683–693. 10.1093/emboj/20.4.683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan E.Y., Kir S., and Tooze S.A.. 2007. siRNA screening of the kinome identifies ULK1 as a multidomain modulator of autophagy. J. Biol. Chem. 282:25464–25474. 10.1074/jbc.M703663200 [DOI] [PubMed] [Google Scholar]
- Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., and Zhang F.. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science. 339:819–823. 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont A., Boucrot E., Drevensek S., Daire V., Gorvel J.P., Poüs C., Holden D.W., and Méresse S.. 2010. SKIP, the host target of the Salmonella virulence factor SifA, promotes kinesin-1-dependent vacuolar membrane exchanges. Traffic. 11:899–911. 10.1111/j.1600-0854.2010.01069.x [DOI] [PubMed] [Google Scholar]
- Efeyan A., Comb W.C., and Sabatini D.M.. 2015. Nutrient-sensing mechanisms and pathways. Nature. 517:302–310. 10.1038/nature14190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcón-Pérez J.M., Starcevic M., Gautam R., and Dell’Angelica E.C.. 2002. BLOC-1, a novel complex containing the pallidin and muted proteins involved in the biogenesis of melanosomes and platelet-dense granules. J. Biol. Chem. 277:28191–28199. 10.1074/jbc.M204011200 [DOI] [PubMed] [Google Scholar]
- Farías G.G., Guardia C.M., De Pace R., Britt D.J., and Bonifacino J.S.. 2017. BORC/kinesin-1 ensemble drives polarized transport of lysosomes into the axon. Proc. Natl. Acad. Sci. USA. 114:E2955–E2964. 10.1073/pnas.1616363114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guardia C.M., Farías G.G., Jia R., Pu J., and Bonifacino J.S.. 2016. BORC functions upstream of kinesins 1 and 3 to coordinate regional movement of lysosomes along different microtubule tracks. Cell Reports. 17:1950–1961. 10.1016/j.celrep.2016.10.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada A., Takei Y., Kanai Y., Tanaka Y., Nonaka S., and Hirokawa N.. 1998. Golgi vesiculation and lysosome dispersion in cells lacking cytoplasmic dynein. J. Cell Biol. 141:51–59. 10.1083/jcb.141.1.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenbeck P.J., and Swanson J.A.. 1990. Radial extension of macrophage tubular lysosomes supported by kinesin. Nature. 346:864–866. 10.1038/346864a0 [DOI] [PubMed] [Google Scholar]
- Inoki K., Li Y., Xu T., and Guan K.L.. 2003. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 17:1829–1834. 10.1101/gad.1110003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzhak D.N., Tyanova S., Cox J., and Borner G.H.. 2016. Global, quantitative and dynamic mapping of protein subcellular localization. eLife. 5:e16950 10.7554/eLife.16950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia R., Guardia C.M., Pu J., Chen Y., and Bonifacino J.S.. 2017. BORC coordinates encounter and fusion of lysosomes with autophagosomes. Autophagy. 1–16. 10.1080/15548627.2017.1343768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongsma M.L., Berlin I., Wijdeven R.H., Janssen L., Janssen G.M., Garstka M.A., Janssen H., Mensink M., van Veelen P.A., Spaapen R.M., and Neefjes J.. 2016. An ER-associated pathway defines endosomal architecture for controlled cargo transport. Cell. 166:152–166. 10.1016/j.cell.2016.05.078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordens I., Fernandez-Borja M., Marsman M., Dusseljee S., Janssen L., Calafat J., Janssen H., Wubbolts R., and Neefjes J.. 2001. The Rab7 effector protein RILP controls lysosomal transport by inducing the recruitment of dynein-dynactin motors. Curr. Biol. 11:1680–1685. 10.1016/S0960-9822(01)00531-0 [DOI] [PubMed] [Google Scholar]
- Jung J., Genau H.M., and Behrends C.. 2015. Amino acid-dependent mTORC1 regulation by the lysosomal membrane protein SLC38A9. Mol. Cell. Biol. 35:2479–2494. 10.1128/MCB.00125-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolchuk V.I., Saiki S., Lichtenberg M., Siddiqi F.H., Roberts E.A., Imarisio S., Jahreiss L., Sarkar S., Futter M., Menzies F.M., et al. . 2011. Lysosomal positioning coordinates cellular nutrient responses. Nat. Cell Biol. 13:453–460. 10.1038/ncb2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurzbauer R., Teis D., de Araujo M.E., Maurer-Stroh S., Eisenhaber F., Bourenkov G.P., Bartunik H.D., Hekman M., Rapp U.R., Huber L.A., and Clausen T.. 2004. Crystal structure of the p14/MP1 scaffolding complex: How a twin couple attaches mitogen-activated protein kinase signaling to late endosomes. Proc. Natl. Acad. Sci. USA. 101:10984–10989. 10.1073/pnas.0403435101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R.J., Xu J., Fu C., Zhang J., Zheng Y.G., Jia H., and Liu J.O.. 2016. Regulation of mTORC1 by lysosomal calcium and calmodulin. eLife. 5:e19360 10.7554/eLife.19360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim C.Y., and Zoncu R.. 2016. The lysosome as a command-and-control center for cellular metabolism. J. Cell Biol. 214:653–664. 10.1083/jcb.201607005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunin V.V., Munger C., Wagner J., Ye Z., Cygler M., and Sacher M.. 2004. The structure of the MAPK scaffold, MP1, bound to its partner, p14. A complex with a critical role in endosomal map kinase signaling. J. Biol. Chem. 279:23422–23430. 10.1074/jbc.M401648200 [DOI] [PubMed] [Google Scholar]
- Matteoni R., and Kreis T.E.. 1987. Translocation and clustering of endosomes and lysosomes depends on microtubules. J. Cell Biol. 105:1253–1265. 10.1083/jcb.105.3.1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama K., and Bonifacino J.S.. 2002. Pallidin is a component of a multi-protein complex involved in the biogenesis of lysosome-related organelles. Traffic. 3:666–677. 10.1034/j.1600-0854.2002.30908.x [DOI] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., and Ferrin T.E.. 2004. UCSF Chimera: A visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Pu J., Schindler C., Jia R., Jarnik M., Backlund P., and Bonifacino J.S.. 2015. BORC, a multisubunit complex that regulates lysosome positioning. Dev. Cell. 33:176–188. 10.1016/j.devcel.2015.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu J., Guardia C.M., Keren-Kaplan T., and Bonifacino J.S.. 2016. Mechanisms and functions of lysosome positioning. J. Cell Sci. 129:4329–4339. 10.1242/jcs.196287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebsamen M., Pochini L., Stasyk T., de Araújo M.E., Galluccio M., Kandasamy R.K., Snijder B., Fauster A., Rudashevskaya E.L., Bruckner M., et al. . 2015. SLC38A9 is a component of the lysosomal amino acid sensing machinery that controls mTORC1. Nature. 519:477–481. 10.1038/nature14107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa-Ferreira C., and Munro S.. 2011. Arl8 and SKIP act together to link lysosomes to kinesin-1. Dev. Cell. 21:1171–1178. 10.1016/j.devcel.2011.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y., Bar-Peled L., Zoncu R., Markhard A.L., Nada S., and Sabatini D.M.. 2010. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 141:290–303. 10.1016/j.cell.2010.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweitzer L.D., Comb W.C., Bar-Peled L., and Sabatini D.M.. 2015. Disruption of the Rag-Ragulator complex by c17orf59 inhibits mTORC1. Cell Reports. 12:1445–1455. 10.1016/j.celrep.2015.07.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C., Zoncu R., Medina D.L., Vetrini F., Erdin S., Erdin S., Huynh T., Ferron M., Karsenty G., Vellard M.C., et al. . 2012. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 31:1095–1108. 10.1038/emboj.2012.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen K., Sidik H., and Talbot W.S.. 2016. The Rag-Ragulator complex regulates lysosome function and phagocytic flux in microglia. Cell Reports. 14:547–559. 10.1016/j.celrep.2015.12.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starcevic M., and Dell’Angelica E.C.. 2004. Identification of snapin and three novel proteins (BLOS1, BLOS2, and BLOS3/reduced pigmentation) as subunits of biogenesis of lysosome-related organelles complex-1 (BLOC-1). J. Biol. Chem. 279:28393–28401. 10.1074/jbc.M402513200 [DOI] [PubMed] [Google Scholar]
- Starling G.P., Yip Y.Y., Sanger A., Morton P.E., Eden E.R., and Dodding M.P.. 2016. Folliculin directs the formation of a Rab34-RILP complex to control the nutrient-dependent dynamic distribution of lysosomes. EMBO Rep. 17:823–841. 10.15252/embr.201541382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teis D., Wunderlich W., and Huber L.A.. 2002. Localization of the MP1-MAPK scaffold complex to endosomes is mediated by p14 and required for signal transduction. Dev. Cell. 3:803–814. 10.1016/S1534-5807(02)00364-7 [DOI] [PubMed] [Google Scholar]
- Wang S., Tsun Z.Y., Wolfson R.L., Shen K., Wyant G.A., Plovanich M.E., Yuan E.D., Jones T.D., Chantranupong L., Comb W., et al. . 2015. Lysosomal amino acid transporter SLC38A9 signals arginine sufficiency to mTORC1. Science. 347:188–194. 10.1126/science.1257132 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.