Yeast septins were among the first proteins reported to be SUMOylated, but the impact of this modification on septin function is unclear. Ribet et al. show that septins are SUMOylated in humans and that SUMOylation is critical for septin bundle formation and septin function in cell division.
Abstract
Septins are cytoskeletal proteins that assemble into nonpolar filaments. They are critical in diverse cellular functions, acting as scaffolds for protein recruitment and as diffusion barriers for subcellular compartmentalization. Human septins are encoded by 13 different genes and are classified into four groups based on sequence homology (SEPT2, SEPT3, SEPT6, and SEPT7 groups). In yeast, septins were among the first proteins reported to be modified by SUMOylation, a ubiquitin-like posttranslational modification. However, whether human septins could be modified by small ubiquitin-like modifiers (SUMOs) and what roles this modification may have in septin function remains unknown. In this study, we first show that septins from all four human septin groups can be covalently modified by SUMOs. We show in particular that endogenous SEPT7 is constitutively SUMOylated during the cell cycle. We then map SUMOylation sites to the C-terminal domain of septins belonging to the SEPT6 and SEPT7 groups and to the N-terminal domain of septins from the SEPT3 group. We finally demonstrate that expression of non-SUMOylatable septin variants from the SEPT6 and SEPT7 groups leads to aberrant septin bundle formation and defects in cytokinesis after furrow ingression. Altogether, our results demonstrate a pivotal role for SUMOylation in septin filament bundling and cell division.
Introduction
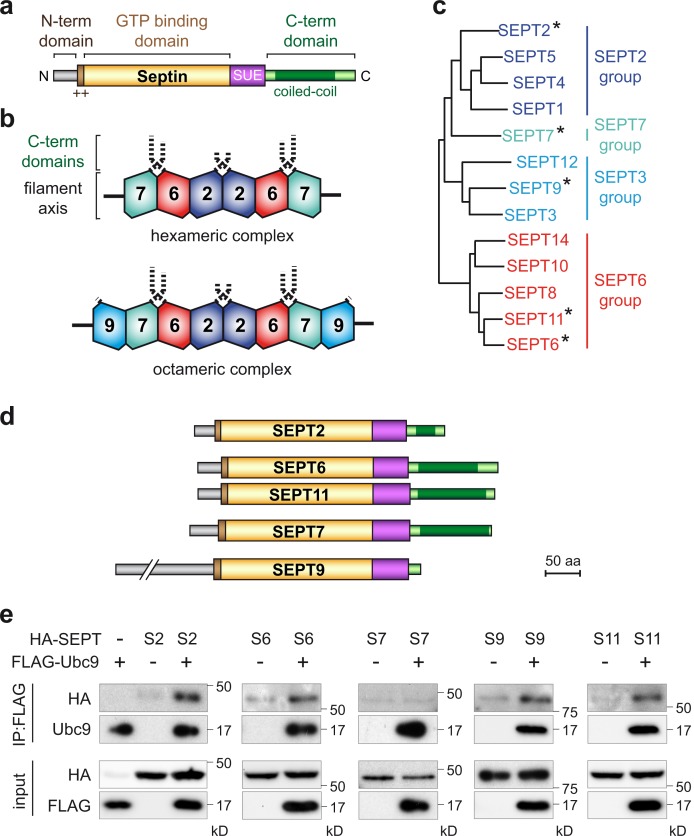
Septins are proteins that assemble into heterooligomeric complexes and form nonpolar filaments that associate with cellular membranes, actin filaments, and microtubules (Hall et al., 2008; Saarikangas and Barral, 2011; Mostowy and Cossart, 2012). Septins act as scaffolds for protein recruitment and as diffusion barriers for subcellular compartmentalization (Caudron and Barral, 2009; Trimble and Grinstein, 2015). They are implicated in numerous biological processes, and their mutation or aberrant expression has been linked to various human pathologies (Mostowy and Cossart, 2012; Dolat et al., 2014; Fung et al., 2014; Angelis and Spiliotis, 2016; Pagliuso et al., 2016). 13 different septin genes have been identified in humans (SEPT1–SEPT12 and SEPT14), which can be classified into four groups based on the encoded protein sequence homology (i.e., the SEPT2 group, the SEPT3 group, the SEPT6 group, and the SEPT7 group; Fig. 1; Kinoshita, 2003; Weirich et al., 2008; Mostowy and Cossart, 2012; Fung et al., 2014). These human genes encode for >30 protein isoforms with tissue-specific expressions. As a result, only a subset of septins may be expressed in a given cell type and the composition of septin filaments may differ in different cells or tissues. All septins contain a central GTP-binding domain flanked by N- and C-terminal domains that vary in length and sequence between different septin groups (Fig. 1 a; Pan et al., 2007; Weirich et al., 2008). Structural studies revealed that the basic unit of septin filaments is composed of the combinatorial association of different septins into hexameric or octameric complexes (Fig. 1 b; Sirajuddin et al., 2007; Kim et al., 2011; Sellin et al., 2011). In humans, these complexes are made up of SEPT7–SEPT6–SEPT2–SEPT2–SEPT6–SEPT7 and/or SEPT9–SEPT7–SEPT6–SEPT2–SEPT2–SEPT6–SEPT7–SEPT9 alternations, with each member of a group possibly able to substitute for another member of the same group. Filaments formed by the end-on-end assembly of these complexes can pair with one another and form higher-order filamentous structures, such as linear bundles, circular rings, or gauzes. Although the GTP-binding domain and part of the N- and C-terminal regions constitute crucial interfaces for septin–septin interactions, the C-terminal domain of septins, which protrudes orthogonally from the filament axis, has been proposed to play a key role in filament stabilization, bundling, and bending and/or in interactions with nonseptin molecules (Fig. 1 b; Sirajuddin et al., 2007; Bertin et al., 2008; de Almeida Marques et al., 2012).
Figure 1.
Interaction between septins and the human SUMOylation machinery. (a) Schematic representation of a prototypical human septin protein (++, phosphoinositide-binding polybasic region; SUE, septin unique element). (b) Schematic organization of typical hexameric and octameric septin complexes. Dashed lines represent extensions formed by septin C-terminal domains. (c) Phylogenetic tree of human septins clustering into four different groups (asterisks denote septins analyzed in this study). (d) Schematic representation of the five human septins analyzed in this study. (e) HeLa cells were cotransfected with HA-tagged septins and FLAG-tagged Ubc9. Cell lysates were subjected to immunoprecipitation (IP) using anti-FLAG antibodies, and coimmunoprecipitation of septins was assayed by immunoblot analysis using anti-HA, anti-FLAG, and anti-Ubc9 antibodies (S2, SEPT2; S6, SEPT6; S7, SEPT7; S9, SEPT9; S11, SEPT11).
Septins play essential roles during mammalian cell division and more particularly during cytokinesis, i.e., the physical separation of the two daughter cells (Cauvin and Echard, 2015; Menon and Gaestel, 2015). Indeed, SEPT2, SEPT6, SEPT7, SEPT9, and SEPT11 are localized along the cleavage furrow or at the cytokinetic bridge, and septin depletion leads to cytokinesis failures, such as delayed or unresolved abscission, generation of binucleated cells, or cell apoptosis (Oegema et al., 2000; Kouranti et al., 2006; Estey et al., 2010; Kim et al., 2011; Founounou et al., 2013; Fung et al., 2014). In mammalian cells, different septins exhibit distinct roles during cytokinesis. Depletion of SEPT2, SEPT7, or SEPT11 leads to abnormal furrow constriction, revealing a role of these septins in cleavage furrow ingression, whereas depletion of SEPT9 strongly delays cell abscission (Joo et al., 2007; Estey et al., 2010).
SUMOylation is a posttranslational modification consisting of the covalent addition of a small ubiquitin-like modifier (SUMO) polypeptide to target proteins. In humans, three functional SUMO isoforms (SUMO1, SUMO2, and SUMO3) are encoded in the genome. They can be conjugated to distinct and overlapping sets of proteins and play essential roles in various cellular processes (Flotho and Melchior, 2013). Pioneering studies in Saccharomyces cerevisiae have shown that yeast septins can be modified by SUMO and that this modification plays a role in the disassembly of bud neck–associated septin rings during yeast cell division (Johnson and Blobel, 1999; Takahashi et al., 1999). Yet, the ability of septins from other species, in particular humans, to be SUMOylated, as well as the role of this modification in septin function, is unknown. Interestingly, two studies have identified components of the SUMOylation machinery as putative interactors of specific human septins, including Ubc9/UBE2I (the E2 SUMO enzyme), PIAS3 (an E3 SUMO enzyme), or SUMO1 itself (Nakahira et al., 2010; Havugimana et al., 2012). Here, by conducting a systematic analysis of all four human septin groups, we unveil that human septins can be modified by SUMO and highlight a critical role for SUMO modification of specific septins (i.e., SEPT6, SEPT7, and SEPT11) in septin filament bundling and cell division.
Results
Human septins from the four different septin groups can be modified by SUMO
To assess whether human septins can be modified by SUMO, we studied members of each of the four human septin groups. We selected SEPT2, SEPT6, and SEPT7, which together form a basic hexameric complex (Sirajuddin et al., 2007). We also selected SEPT9, which occupies a terminal position in septin octameric complexes (Kim et al., 2011; Sellin et al., 2011), and SEPT11, a septin closely related to SEPT6 with unique cellular functions (Fig. 1, c and d; Röseler et al., 2011).
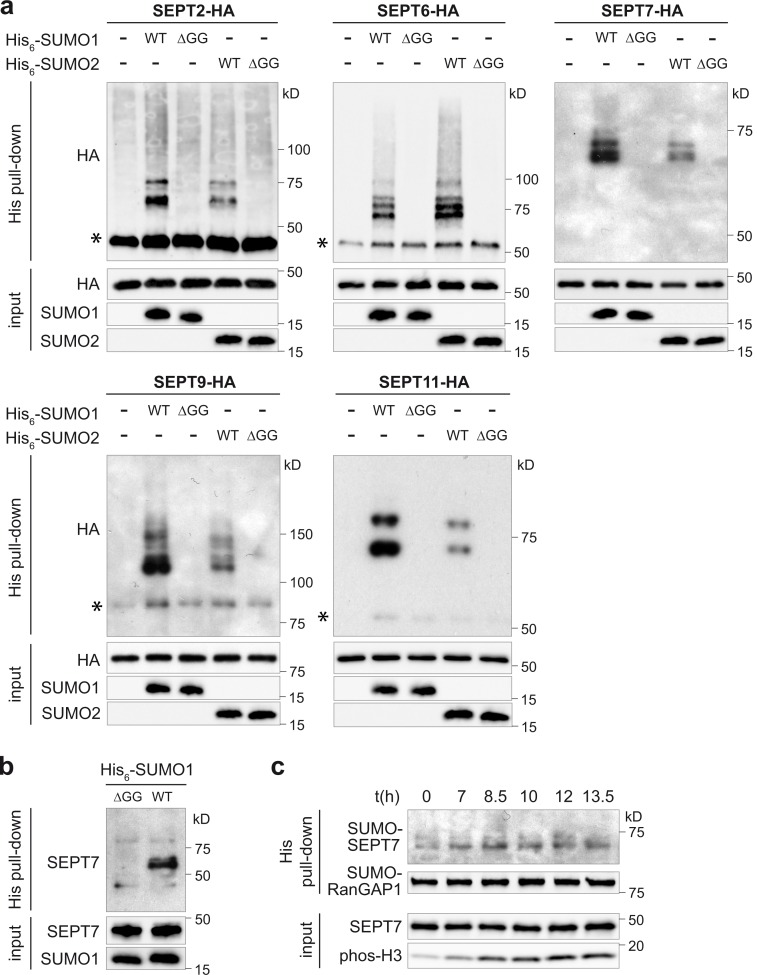
We first addressed whether septins were able to interact with Ubc9, the unique E2 enzyme of the SUMOylation machinery. HeLa cells were cotransfected with expression vectors for FLAG-tagged Ubc9 and different HA-tagged septins belonging to each human septin group. SEPT2, SEPT6, SEPT9, and SEPT11 coimmunoprecipitated with Ubc9, demonstrating that human septins from three different septin groups can interact with the SUMOylation machinery (Fig. 1 e). For SEPT7, we did not observe coimmunoprecipitation with Ubc9 in our experimental conditions. To further demonstrate that mammalian septins could be modified by SUMO, HeLa cells were transfected with expression vectors for HA-tagged septins and plasmids encoding either His6-tagged SUMO1, SUMO2, or nonconjugatable SUMO1ΔGG or SUMO2ΔGG mutants (i.e., SUMO mutants deleted for the C-terminal diglycine motif required for SUMO conjugation; Fig. S1; Flotho and Melchior, 2013). After cell lysis in denaturing conditions, SUMOylated proteins (i.e., proteins covalently attached to His-tagged SUMO) were pulled down, and the presence of SUMOylated septins was assayed by immunoblotting experiments using anti-HA antibodies. We detected one or several bands of higher molecular weight in the His pull-down fraction for each of the five septins tested, corresponding to SUMOylated forms of septins (Fig. 2 a). These SUMO-conjugated forms were observed both with SUMO1 and 2 and were absent with the nonconjugatable SUMO1ΔGG or SUMO2ΔGG mutants, indicating that septins from all four human septin groups can be SUMOylated by both SUMO1 and 2 (Fig. 2 a).
Figure 2.
SUMOylation of human septins. (a) HeLa cells were cotransfected with WT His6-SUMO1, 2, or nonconjugatable (ΔGG) mutants and HA-tagged septins. Cell lysates were then subjected to denaturing His pull-down, and the presence of SUMOylated septins was assayed by immunoblot analysis using anti-HA antibodies (asterisks represent nonspecific binding of un-SUMOylated septins to nickel–nitrilotriacetic acid beads). Input fractions are shown as controls. (b) Immunoblot analysis, using anti-SEPT7 antibodies, of His pull-down proteins from HeLa cells transfected with WT or nonconjugatable (ΔGG) His6-SUMO1. Input fractions are shown as control. (c) Immunoblot analysis of His pull-down proteins from synchronized HeLa cells transfected with WT His6-SUMO1. Anti-RanGAP1 antibodies were used to monitor pull-down of SUMOylated proteins. Antiphosphorylated histone H3 antibodies (phos-H3) were used to control cell cycle synchronization.
To confirm that endogenous septins are SUMO targets, we transfected HeLa cells with a His6-tagged SUMO1 expression vector alone and pulled down SUMOylated proteins. Using immunoblot analysis with antiseptin antibodies, we detected high–molecular weight bands corresponding to SUMOylated forms of endogenous SEPT7 (Fig. 2 b). These bands were not observed when cells were transfected with a nonconjugatable SUMO1ΔGG mutant (Fig. 2 b). This result indicates that endogenous SEPT7 are SUMO modified in human cells. Although we could not detect SUMOylated forms of other septins in our experimental setup, recent proteomic data identified endogenous SEPT2, SEPT5, SEPT6, SEPT7, SEPT8, SEPT9, and SEPT10 as SUMO targets (Becker et al., 2013; Hendriks et al., 2015, 2017; Hendriks and Vertegaal, 2016). Together, these results demonstrate that septins can be conjugated to SUMO in mammals.
SEPT7 is constitutively SUMOylated during the cell cycle
In yeast, the SUMOylation level of septins is regulated during the cell cycle with an increase in SUMOylation appearing during mitosis before anaphase onset and disappearing at cytokinesis (Johnson and Blobel, 1999). To assess whether SUMOylation of septins is cell cycle dependent in human cells, we transfected HeLa cells with His6-tagged SUMO1 expression vector and synchronized them using a double thymidine block procedure. Cells were lysed at different time points after the release from the second thymidine block to cover different stages of the cell cycle (i.e., S phase, G1 phase, mitosis, early cytokinesis, and late cytokinesis). SUMOylated proteins were then pulled down, and the SUMOylation level of endogenous SEPT7 was analyzed by immunoblotting experiments. In contrast to the situation described in yeast, we did not observe abrupt changes in the SUMOylation level of SEPT7 in human cells, indicating that SEPT7 is likely constitutively SUMOylated throughout the cell cycle (Fig. 2 c).
Identification of SUMO sites from human septins in their N- or C-terminal domains
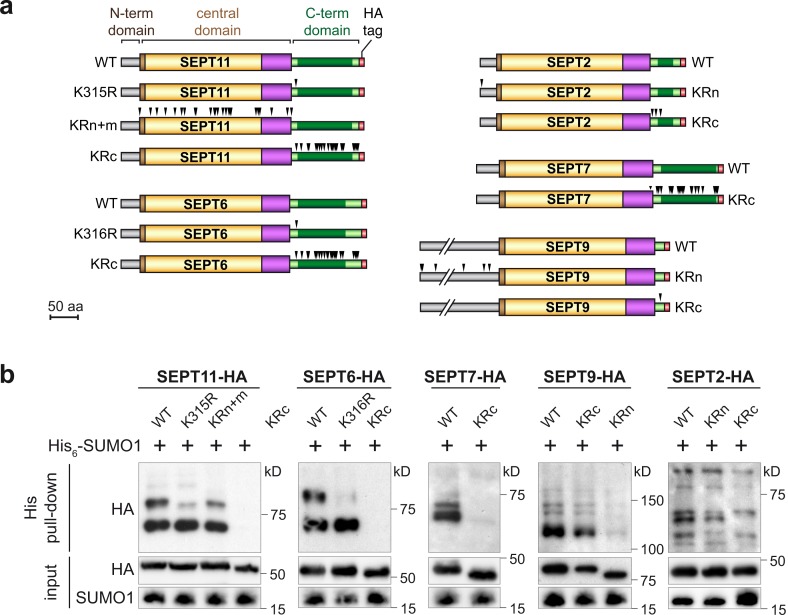
To assess the role of septin SUMOylation, it was important to first map the sites of SUMO conjugation in septins. We thus generated 10 different K to R mutants in SEPT2, SEPT6, SEPT7, SEPT9, and SEPT11 (Fig. 3 a). HeLa cells were transfected with expression vectors for these septin mutants and His6-SUMO1, and the presence of SUMOylated forms of septins was assessed. In the case of SEPT11, mutation of the K315 lysine of the C-terminal domain led to the loss of one of the two SEPT11 SUMOylated forms, strongly suggesting that K315 constitutes a SUMOylation site for SEPT11 (Fig. 3 b). Interestingly, we found that the homologous lysine in SEPT6, belonging to the same septin group, is also a SUMOylation site (K316; Fig. 3 b). Individual mutations of other SEPT11 lysine residues into arginine did not allow us to identify the other SEPT11 SUMO sites, probably because of redundancy between different potential SUMO sites, as classically observed for other SUMO targets. To obtain a non-SUMOylatable mutant of SEPT11, we derived two SEPT11 mutants in which either all the lysines of the N-terminal and GTP-binding domain (SEPT11 KRn+m) or all the lysines of the C-terminal domain (SEPT11 KRc mutant) were changed to arginines (Fig. 3 a). Strikingly, no SUMOylated forms could be observed with the SEPT11 KRc mutant, whereas the SEPT11 KRn+m mutant has a SUMOylation pattern similar to WT SEPT11. Together, these results indicate that the two major SUMO sites of SEPT11 are localized in the C-terminal domain (Fig. 3 b).
Figure 3.
Mapping of human septin SUMOylation sites. (a) Schematic representation of HA-tagged WT and mutant SEPT2, SEPT6, SEPT7, SEPT9, and SEPT11. Black arrowheads indicate K to R mutations (KRn, K to R mutant in all N-terminal lysines; KRn+m, K to R mutant in all N-terminal and central domain lysines; KRc, K to R mutant in all C-terminal lysines). (b) HeLa cells were cotransfected with WT His6-SUMO1 and WT or mutant HA-tagged septins. Cell lysates were then subjected to denaturing His pull-down, and the presence of SUMOylated septins was assayed using anti-HA antibodies. Input fractions are shown as controls.
For SEPT6 and SEPT7, we similarly derived mutants in which all the C-terminal lysines were replaced by arginines (SEPT6 and SEPT7 KRc mutants; Fig. 3 a). As for the SEPT11 KRc mutant, SUMOylation of these septin mutants was abolished (Fig. 3 b), indicating that the major SUMO sites for SEPT6 and SEPT7 are all located in their C-terminal domains. These observations fit well with the structural data showing that the C-terminal domains of these septins protrude from the septin filament core (Sirajuddin et al., 2007) and are thus accessible to the SUMOylation machinery. For SEPT9 (i.e., SEPT9_i1 isoform), which, in contrast to the previously tested septins, possessed a small C-terminal domain and a long N-terminal domain (Fig. 1), a mutation of the unique lysine of the SEPT9 C-terminal domain K579 did not affect its SUMOylation profile (Fig. 3 b). In contrast, mutation of all the lysines from the SEPT9 N-terminal domain impaired its SUMOylation, suggesting that SEPT9 SUMO sites lie in the N-terminal domain of this septin (Fig. 3). Finally, in the case of SEPT2, mutations of the three lysines of its C-terminal domain only partially reduced the number of SUMOylated bands, whereas mutation in the unique lysine of its N-terminal domain had no effect on SEPT2 SUMOylation pattern (Fig. 3). This suggests that at least one SUMO site lies in the SEPT2 C-terminal domain but that other SUMO sites lie in the central domain, although we were unable to generate a non-SUMOylatable variant for this septin. The role of SEPT2 SUMOylation could thus not be assessed.
SUMOylation of SEPT7 and SEPT11 is critical for high-order septin filament organization and dynamics
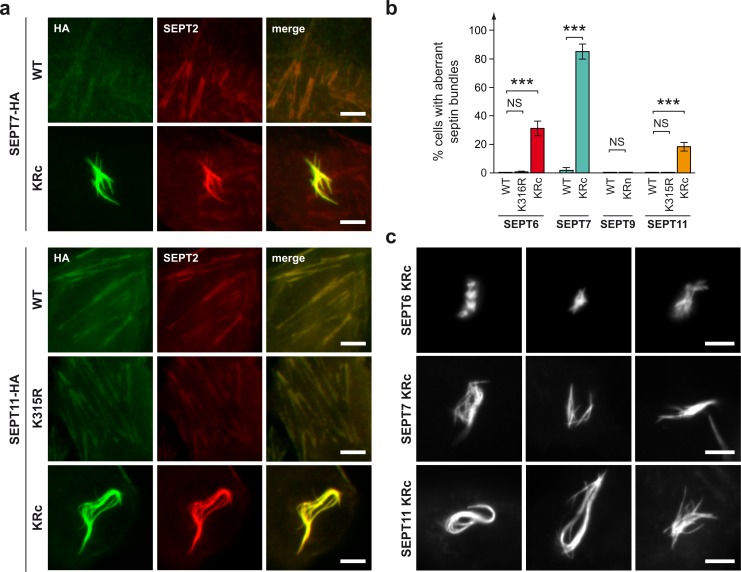
We next used non-SUMOylatable variants of septins to characterize the putative role of SUMO in septin functions. We first investigated whether SUMO was involved in septin filament assembly. We analyzed by immunofluorescence HeLa cells transfected with expression vectors for different versions of HA-tagged septins (Fig. 4 a). For SEPT9, no morphological differences could be observed between filaments formed by WT or non-SUMOylatable septin variants. Strikingly, for SEPT6, SEPT7, and SEPT11, the expression of non-SUMOylatable variants led to the formation of aberrant septin filament assemblies (Fig. 4, a and b). These assemblies, hereafter called aberrant bundles, appeared as thick and intense and clearly differ from septin assemblies formed by WT septins (Fig. 4, a and c). Colabeling with endogenous septins indicates that these aberrant septin bundles incorporate other cellular septins, such as SEPT2 or SEPT9 (Fig. 4 a, Fig. S2, and Fig. S3).
Figure 4.
Non-SUMOylatable septin variants form aberrant bundles. (a) Fluorescent light microscopy images of HeLa cells transfected with HA-tagged WT or non-SUMOylatable SEPT7 and SEPT11 variants. Cells were stained for HA-tagged septins (anti-HA antibodies, green) and endogenous septins (anti-SEPT2 antibodies, red). (b) Percentage of HeLa cells displaying aberrant septin bundles after transfection with WT or mutant SEPT6, SEPT7, SEPT9, or SEPT11 (mean from three independent experiments; error bars, SD; ***, P < 0.001). (c) Examples of aberrant septin bundles observed in cells expressing non-SUMOylatable SEPT6, SEPT7, and SEPT11 variants. Cells were stained for HA-tagged septins (anti-HA antibodies). Bars, 5 µm.
To assess whether SUMOylation of septins modify septin–septin interactions, we transfected HeLa cells with expression vectors for HA-tagged septins. Using immunoprecipitation experiments, we observed that endogenous SEPT2 and SEPT11 coimmunoprecipitate with both WT and non-SUMOylatable HA-tagged SEPT7 (Fig. S4 a). This indicates that a lack of SUMOylation does not inhibit SEPT7’s ability to form complexes with septins from other septin groups. Of note, we observed that the level of coimmunoprecipitated endogenous septins was higher with non-SUMOylatable SEPT7 than with WT, which may indicate stronger interactions (Fig. S4 a). We similarly observed that endogenous SEPT2 and SEPT7 coimmunoprecipitated with both WT and non-SUMOylatable SEPT11. Again, the level of coimmunoprecipitated endogenous septins was higher with non-SUMOylatable SEPT11 than with WT (Fig. S4 a).
Intimate relationships between septin and actin dynamics have been described for a range of cellular processes (Mostowy and Cossart, 2012). By performing the colabeling of septins with actin, we observed a complete lack of colocalization between aberrant bundles formed by non-SUMOylatable SEPT6, SEPT7, or SEPT11 and actin, whereas WT septin assemblies partially colocalized with actin filaments (Fig. S2 and Fig. S3).
Altogether, and given that expression of non-SUMOylatable SEPT6, SEPT7, or SEPT11 variants does not alter endogenous septin protein levels (Fig. S4 b), our results suggest that SUMOylation of these septins is strictly required for the correct organization of septin filaments in HeLa cells. We confirmed these observations in another epithelial cell line, i.e., U-2 OS cells. Expression of non-SUMOylatable septin mutants in U-2 OS cells also led to the formation of aberrant septin bundles that colocalized with endogenous septins, but not with actin (Fig. S5, a and b). This indicates that SUMOylation plays a role in septin filament organization in several human cell types.
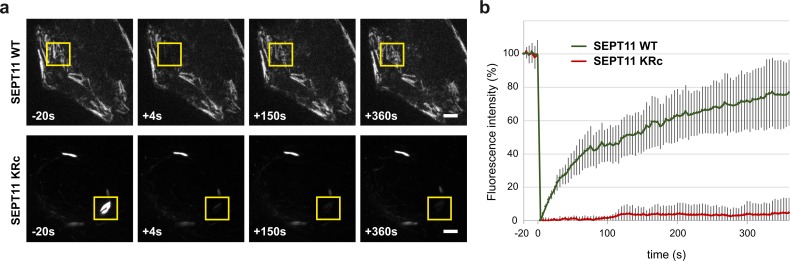
To monitor whether SUMOylation of septins was involved in septin filament dynamics, we performed FRAP experiments on HeLa cells expressing GFP-tagged septins. HeLa cells were transfected with WT or non-SUMOylatable GFP-tagged SEPT11. GFP-labeled filaments were irradiated with a laser beam to irreversibly photobleach GFP. Fluorescence recovery in the bleached area was then monitored to evaluate the exchange of nonbleached septin monomers in preformed filaments (Fig. 5 a). Whereas septin assemblies formed by WT SEPT11 displayed rapid recovery of fluorescence (50% of fluorescence intensity was recovered in <2.5 min), only very little recovery was observed in the case of aberrant bundles formed by non-SUMOylatable septins (<5% of the initial fluorescence intensity was recovered 6 min after bleaching; Fig. 5 b). This indicates that the rate of septin exchange within these aberrant bundles is strongly reduced and that the corresponding septin structures are poorly dynamic compared with WT ones.
Figure 5.
Role of SUMOylation in septin dynamics. HeLa cells were transfected with GFP-tagged WT or non-SUMOylatable (KRc) SEPT11, and GFP-labeled septin filaments were subjected to FRAP. (a) Representative images of fluorescence recovery for WT and KRc SEPT11 filaments. Boxes indicate bleached regions. Bars, 5 µm. (b) Kinetics of fluorescence recovery (mean from six to seven independent fields of one representative experiment; error bars, SD).
Together, these results show that the absence of SUMOylation leads to the formation of septin bundles with aberrant morphology and dynamics and thus imply that SUMOylation is essential for normal septin filament organization.
Absence of SEPT6, SEPT7, and SEPT11 SUMOylation leads to cytokinesis defects
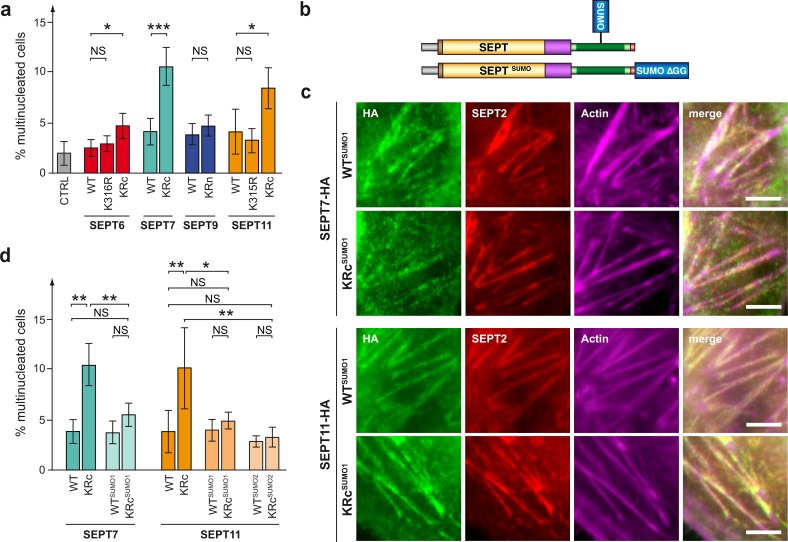
To get further insights into the role of SUMOylation in septin functions, we assessed whether expression of non-SUMOylatable septin variants leads to cell division defects. Indeed, septins were originally characterized for their essential role in cell division in yeast and then in metazoans, and septin depletion has been reported to induce cytokinesis defects classically resulting in the generation of multinucleated cells (Hartwell et al., 1970; Neufeld and Rubin, 1994; Kinoshita et al., 1997; Nagata et al., 2003; Kremer et al., 2005; Spiliotis et al., 2005; Estey et al., 2010; Chesneau et al., 2012; El Amine et al., 2013). Here, we transfected HeLa cells with expression vectors for HA-tagged septins and quantified by immunofluorescence analysis, 72 h after transfection, the frequency of multinucleated cells as a reporter of cytokinesis defects. We observed that the frequency of multinucleated cells was not changed for cells expressing the SEPT9 non-SUMOylatable variant compared with WT SEPT9 (Fig. 6 a). In contrast, expression of SEPT6, SEPT7, and SEPT11 KRc mutants increased the level of multinucleated cells compared with cells expressing WT septins, suggesting that SUMOylation of these septins is required for the correct completion of cell division (Fig. 6 a). Of note, SEPT6 K316R and SEPT11 K315R mutants, which still exhibit partial SUMOylation (Fig. 3), did not show any defect in cytokinesis compared with SEPT6 KRc and SEPT11 KRc mutants, respectively.
Figure 6.
Role of septin SUMOylation in cell division. (a) Percentage of HeLa cells exhibiting multinucleation after transfection with a control plasmid (pCDNA.3; CTRL) or expression vectors for WT or mutant septins (mean from three to five independent experiments). (b) Schematic representation of SUMOylated HA-tagged septin (top) or septin C-terminally fused to SUMO (bottom). (c) Fluorescent light microscopy images of septin filaments formed by WT or non-SUMOylatable SEPT7 and SEPT11 mutants C-terminally fused to SUMO in HeLa cells. Cells were stained for HA-tagged septins (anti-HA antibodies, green), endogenous SEPT2 (anti-SEPT2 antibodies, red), and actin (phalloidin, magenta). Bars, 5 µm. (d) Percentage of HeLa cells exhibiting multinucleation after transfection with WT, non-SUMOylatable, or constitutively SUMOylated SEPT7 and SEPT11 (mean from three to six independent experiments). Error bars, SD; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To confirm that a lack of SUMOylation of SEPT7 or SEPT11 was responsible for the observed defects in septin bundle formation and in cell division, we generated expression vectors for constitutively SUMOylated septins by fusing SUMO to the C-terminal domain of WT and non-SUMOylatable septin variants (SEPT7 WTSUMO, SEPT7 KRcSUMO, SEPT11 WTSUMO, and SEPT11 KRcSUMO; Fig. 6 b). This strategy, which mimics an irreversible covalent linkage of SUMO to a given target, has classically been used to analyze the function of ubiquitin-like modifications (Carter and Vousden, 2008; Choi et al., 2011; Ferreira et al., 2011). By transfecting HeLa cells with these expression vectors, we observed that constitutively SUMOylated SEPT7 and SEPT11 assembled into filaments that colocalize with endogenous SEPT2 and actin (Fig. 6 c). Remarkably, SEPT7 KRcSUMO and SEPT11 KRcSUMO variants, in contrast to SEPT7 KRc and SEPT11 KRc mutants, did not form aberrant septin bundles (Fig. S5 d). This result indicates that the C-terminal fusion of SUMO allows a functional complementation of the corresponding KRc mutant and confirms that the observed defects in the formation of septin bundles observed with SEPT7 and SEPT11 KRc variants are caused by a lack of SUMOylation. In parallel, we quantified the frequency of multinucleated cells in HeLa cells expressing these septins fused to SUMO. Expression of SEPT7 KRcSUMO and SEPT11 KRcSUMO variants, in contrast to SEPT7 KRc and SEPT11 KRc, did not show cytokinesis defects, highlighting that the C-terminal fusion of SUMO restores the phenotype of non-SUMOylatable septin variants (Fig. 6 d). Interestingly, this suggests that the exact location of SUMO is not critical for SEPT7 and SEPT11. We finally assayed whether septin KRcSUMO1 variants could complement cell division defects induced by KRc mutants using cotransfection experiments. For this, HeLa cells were cotransfected with expression vectors for SEPT KRc variants together with expression vectors for SEPT WT, KRc, WTSUMO1, or KRcSUMO1 variants. For SEPT7, we observed that cotransfection of SEPT7 WT with SEPT7 KRc can partially restore the cell division defects induced by SEPT7 KRc alone (Fig. S5 e). We observed a similar complementation when SEPT7 KRcSUMO1 was cotransfected with SEPT7 KRc, indicating that constitutively SUMOylated SEPT7 can functionally restore defects induced by SEPT7 KRc (Fig. S5 e). Similar results were obtained for SEPT11 (Fig. S5 e). To extend our findings, we performed these experiments in U-2 OS cells. We observed similar cytokinesis defects for SEPT7 and SEPT11 non-SUMOylatable mutants that were not observed with the corresponding constitutively SUMOylated variants (Fig. S5 c). This indicates that SUMOylation of these septins plays a role in cell division in several human cell types.
Lack of septin SUMOylation interferes with late steps of cytokinesis
Cytokinesis is a multistep process allowing the physical separation of the daughter cells. It starts with the formation of an actin/myosin-based contractile ring that drives the ingression of the cleavage furrow. Once the ring has contracted, the formed intercellular bridge between the two daughter cells matures and is finally cut through a process called abscission.
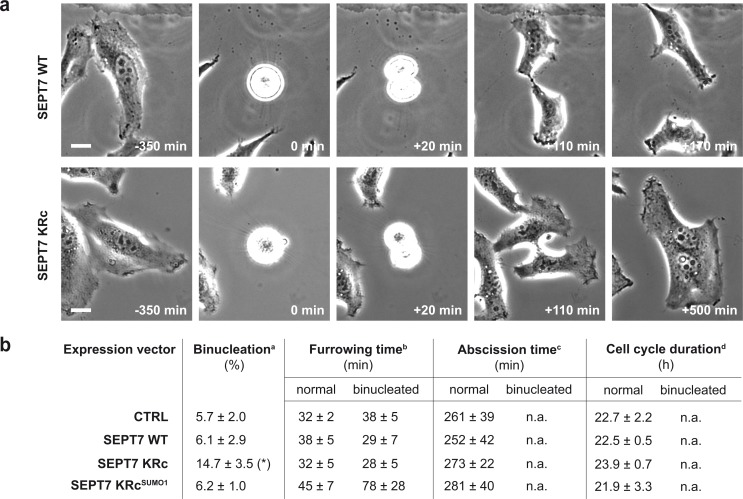
To identify at which step of the cytokinesis process septin SUMOylation is involved, HeLa cells were cotransfected with expression vectors for SEPT7 and for GFP. Cell division of transfected cells (i.e., GFP-positive cells) was followed by time-lapse microscopy for 48 h (Fig. 7 a). We did not observe significant differences in times required for cleavage furrow formation between cells expressing WT or non-SUMOylatable SEPT7 variants (Fig. 7 b). Interestingly, for cells expressing SEPT7 KRc, we always observed cleavage furrow ingression, but in cells failing in cell division, this furrow loosened, leading to a refusion of the two daughter cells and to the generation of binucleated cells (Fig. 7 a). These refusion events occurred on average 249 ± 90 min after cell rounding and were detected significantly more frequently in cells expressing non-SUMOylatable SEPT7 variants than in cells transfected with control plasmids or expressing WT SEPT7 or SEPT7 fused to SUMO (Fig. 7 b). For the remaining cells, no differences were observed in times required for intercellular bridge abscission or between two cell division events (Fig. 7 b). Thus, a lack of septin SUMOylation is unlikely to affect the cell cycle, nor does it induce a delay in cleavage furrow formation or cell–cell separation. Our results show that the presence of non-SUMOylatable septins leads to late cytokinetic defects, characterized by intercellular bridge instability after furrow ingression. Of note, the functional inactivation of most septins leads to a similar postfurrowing regression of the intercellular bridge and the formation of binucleated cells (Kinoshita et al., 1997; Kouranti et al., 2006; Estey et al., 2010).
Figure 7.
Lack of septin SUMOylation interferes with late steps of cytokinesis. (a) Representative images from time-lapse microscopy analysis of HeLa cells expressing WT or non-SUMOylatable SEPT7. Images correspond to different cell division events including cell rounding, cleavage furrow ingression, intercellular bridge formation, and either bridge abscission (top) or bridge regression and formation of a binucleated cell (bottom). Bars, 10 µm. (b) Time-lapse microscopy analysis of HeLa cells transfected with control plasmid (pCDNA.3; CTRL) or expression vectors for SEPT7 WT, KRc, or KRcSUMO1 variants. 24 h after transfection, time-lapse sequences of cells were recorded every 20 min for 48 h. At least 50 transfected cells were analyzed for each condition in each experiment. Means ± standard errors from three independent experiments are indicated. *, P < 0.05 compared with control condition (two-tailed two-sample equal-variance Student’s t test). aPercentage of cells for which cell division results in the formation of binucleated cells. bTime between cell rounding and furrow ingression. cTime between cell rounding and intercellular bridge abscission. dTime between two cell-rounding events. n.a., not applicable.
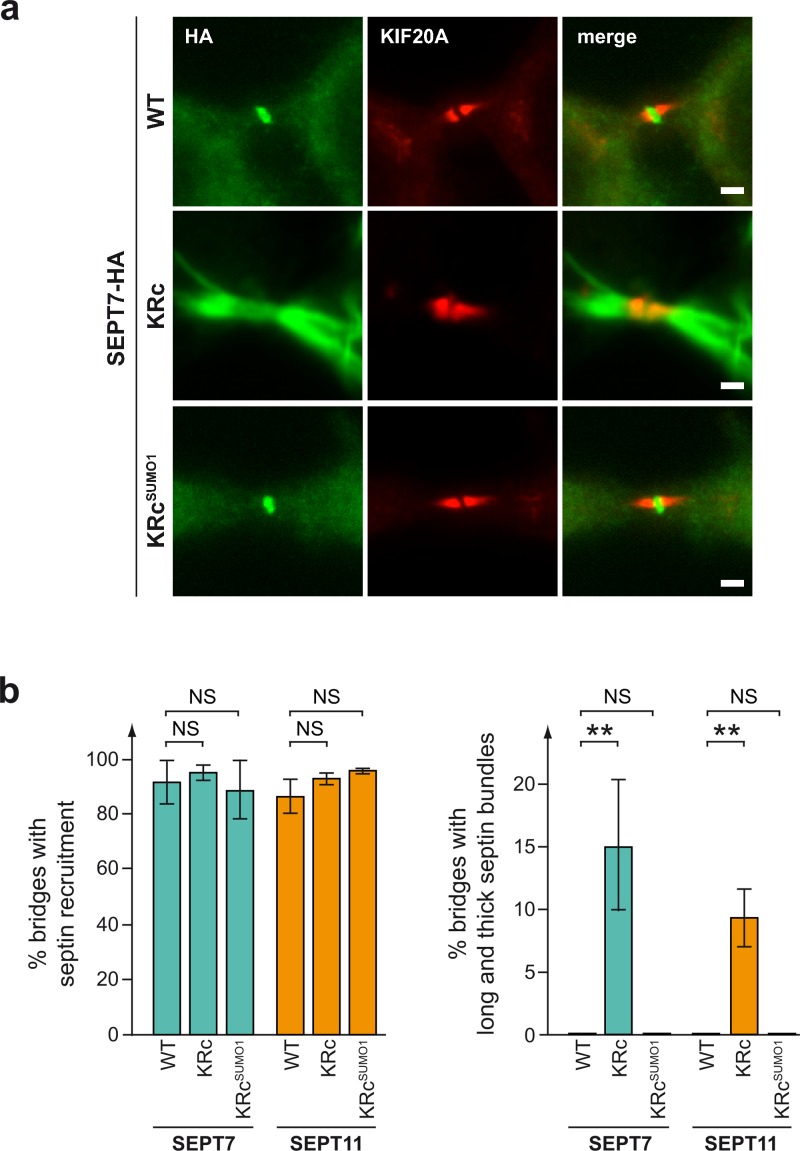
As SEPT7 KRc and SEPT11 KRc variants lead to the formation of aberrant septin bundles, we hypothesized that these structures could be responsible for the observed cytokinesis defects. The presence of such bundles in intercellular bridges during cytokinesis may indeed mechanically hinder the abscission of the two daughter cells and destabilize the bridge. To test this hypothesis, we used KIF20A/Rabkinesin-6 protein as a marker of midbodies (Echard et al., 1998; Hill et al., 2000; Fontijn et al., 2001) and analyzed by immunofluorescence whether aberrant filaments formed by non-SUMOylatable septin variants localize at cell bridges during cytokinesis (Fig. 8 a). We observed that all SEPT7 and SEPT11 variants localized to the intercellular bridge, indicating that cell division defects are not caused by mislocalization of septins out of cell–cell bridges (Fig. 8 b). Interestingly, 15.0 ± 5.3% of the cells in cytokinesis that expressed non-SUMOylatable SEPT7 displayed long (>5 µm) and thick (>1 µm) bundles of septins within the intercellular bridge (as shown in Fig. 8 a), which matches with the 14.7 ± 3.5% of cells that became binucleated as measured by time-lapse microscopy (Fig. 7 b). In contrast, long and thick bundles of septin filaments were not observed in cells expressing WT or KRcSUMO SEPT7 variants (Fig. 8 b), which displayed low levels of binucleated cells. In the case of SEPT11, quantification further revealed that 9.3 ± 2.3% of the cells in cytokinesis that expressed non-SUMOylatable SEPT11 displayed long and thick bundles of septins within the intercellular bridge, which again matches with the 8.4 ± 1.9% of cells that became binucleated (Fig. 6 a). Altogether, these results are consistent with the idea that cells with long and thick septin bundles will not be able to maintain stable bridges and eventually become binucleated.
Figure 8.
SUMO-deficient septin bundles localize at intercellular bridges during cell division. (a) Fluorescent light microscopy images of HeLa cells transfected with HA-tagged WT, non-SUMOylatable (KRc), or constitutively SUMOylated (KRcSUMO1) SEPT7. Cells were stained for HA-tagged septins (anti-HA antibodies, green) and KIF20A to label intercellular midbodies (red). Bars, 2 µm. (b) Percentage of transfected cells undergoing cell division and showing septin recruitment or septin bundles longer than 5 µm and thicker than 1 μm at intercellular midbodies (mean from three independent experiments; error bars, SD; **, P < 0.01).
We conclude that SUMOylation of SEPT7 and SEPT11 plays a pivotal role in the organization of septin filaments and in cell division in mammalian cells.
Discussion
Posttranslational modifications are critical modulators of components of the cell cytoskeleton. In particular, SUMOylation of cytoskeletal proteins, such as actin, tubulin, or intermediate filaments, or of cytoskeleton architecture regulators has been previously reported (Impens et al., 2014; Hendriks and Vertegaal, 2016). Here, we show that septins from all four human septin groups can be covalently modified by either SUMO1 or SUMO2. We provide evidence for interactions of human septins with the cell SUMOylation machinery, identified that at least SEPT7 can be SUMO modified at the endogenous level, and mapped septin SUMO sites in the N-terminal domain for SEPT9 or the C-terminal domain for SEPT6, SEPT7, and SEPT11. Interestingly, the SUMO sites of yeast septins have also been identified in either the C-terminal domains (for Cdc11 and Shs1) or N-terminal domains (for Cdc3), although these domains do not share a sequence homology with human septins (Johnson and Blobel, 1999). Our data extend the first observations of septin SUMOylation initially made in yeast and corroborate previous high-throughput analyses of SUMOylated proteins that identified human septins as genuine SUMO targets (Johnson and Blobel, 1999; Takahashi et al., 1999; Becker et al., 2013; Hendriks and Vertegaal, 2016). Of note, a very recent human SUMO proteome identified some of the SUMO-modified lysines in SEPT2, SEPT6, SEPT7, SEPT8, SEPT9, and SEPT10 (Hendriks et al., 2017). This analysis corroborates our results obtained by the analysis of SUMOylation of septin mutants: two SUMO sites were identified in the C-terminal domain of SEPT6 (including K316), one SUMO site was identified in the C-terminal domain of SEPT7, and five SUMO sites were identified in the N-terminal domain of SEPT9. Additional SUMO sites were identified in SEPT6, SEPT7, and SEPT9 central domains, but our mutagenesis analysis suggests that these do not constitute major SUMO sites.
By analyzing the phenotype of different cell lines expressing non-SUMOylatable septin variants, we demonstrate that septin SUMOylation plays a critical role in the organization of septin filaments. Specifically, SUMOylation of a set of septins prevents their bundling into aberrant structures. We show that a lack of SUMOylation in SEPT7 and SEPT11 does not inhibit canonical septin complex formation. In addition, assembly of septin filaments can be observed with C-terminally truncated septins (in particular with SEPT6 and SEPT7; Sirajuddin et al., 2007). Finally, we could not identify SUMO-interacting motifs (SIMs), i.e., domains mediating noncovalent interactions with SUMO moieties in human septins in silico (Table S1). Collectively, these results suggest that SUMOylation is not required for septin–septin interactions per se. Thus, we propose that SUMOylation of septins instead regulates interactions with nonseptin molecules critical for the correct assembly of septin higher-order structures. This fits well with the mapping of SUMO sites in the C-terminal domain of SEPT6, SEPT7, and SEPT11, which are domains known to be involved in bundling and bending and/or in interactions with nonseptin molecules (Sirajuddin et al., 2007; Bertin et al., 2008; de Almeida Marques et al., 2012). A lack of septin SUMOylation, and thus a lack of recruitment of these septin-interacting factors, would consequently lead to assembly defects and accumulation of bundles with aberrant thickness and dynamics. Of note, as for the vast majority of other SUMO targets, the cellular level of septin SUMOylation is low in steady state. This may indicate that septins are only transiently SUMOylated (for example, during the bundling of different preassembled septin filaments) and/or that only a small fraction of septin monomers is decorated by SUMO in an assembled filament. Our observation that SUMOylation of SEPT6, SEPT7, and SEPT11, but not SEPT9, is critical for septin filament organization furthermore highlights the specific role of SUMOylation of different septins. This result is in agreement with previous studies suggesting that octamers versus hexamers might have different roles, even within the same cell type (Kim et al., 2011; Sellin et al., 2011). Interestingly, other posttranslational modifications have been reported for septins that modulate septin assembly, including phosphorylation, acetylation, and ubiquitylation (Hernández-Rodríguez and Momany, 2012). In particular, phosphorylation of the C-terminal domain of the yeast Shs1 septin was recently shown to regulate the capacity of Shs1-containing septin complexes to form rings (Garcia et al., 2011). Collectively, these data suggest that a complex set of coordinated posttranslational modifications tightly regulates the three-dimensional architecture of the septin cytoskeleton. Of note, the role of septin posttranslational modifications has mainly been characterized in yeast, whereas their function in mammals remains poorly understood. In mammalian cells, defects in septin SUMOylation, and thus in septin cytoskeleton architecture, may have severe outcomes in cell physiology, such as defects in late cytokinetic stages, after furrow ingression. Interestingly, mutations of lysine residues in the C-terminal domain of SEPT6 and SEPT7 have been reported in carcinoma samples (Catalogue of Somatic Mutations in Cancer; Angelis and Spiliotis, 2016). Whether these mutations or other naturally occurring mutations affect septin SUMOylation is unknown. Our results thus open new avenues for better understanding the molecular mechanisms underlying human diseases in which septins have been implicated.
Materials and methods
Plasmids
References and information on plasmids are described in Table S2. pCDNA3-FLAG-Ubc9 plasmid encoding N-terminal FLAG-tagged Ubc9 was obtained by changing the tag from the pCDNA3-HA-Ubc9 plasmid (gift from F. Melchior, Zentrum für Molekulare Biologie der Universität Heidelberg, Heidelberg, Germany; Bossis and Melchior, 2006). pSG5-His6-SUMO1 and SUMO2 plasmids encoding N-terminal His6-tagged mature SUMO isoforms were previously described in Impens et al. (2014). pSG5-His6-SUMO1ΔGG and SUMO2ΔGG plasmids encode SUMO isoforms in which the C-terminal diglycine motif was deleted. Plasmids encoding human septins with a C-terminal HA tag were obtained by inserting into pCDNA3 vector (Invitrogen) HA-tagged versions of human SEPT2, SEPT6, SEPT7, SEPT9, or SEPT11. These plasmids were then used to generate expression vectors for KRn septin mutants (in which all lysines from the N-terminal domain were replaced by arginines), KRn+m septin mutants (in which all lysines from the N-terminal and central domains were replaced by arginines), or KRc septin mutants (in which all lysines from the C-terminal domain were replaced by arginines). Plasmids encoding septins fused to SUMO1 or SUMO2 were obtained by inserting at the C terminus of the corresponding HA-tagged septin the sequence of human mature SUMO1 or SUMO2, deleted for their C-terminal diglycine motifs to avoid aberrant covalent linkage to other cellular proteins. All generated constructs were verified by sequencing.
Antibodies
Primary antibodies used for immunoblot analysis and immunofluorescence are described in Table S3. Rabbit polyclonal antibodies against Ubc9 (R202) was obtained in-house by immunizing rabbits with recombinant proteins produced in Escherichia coli. Antibodies against SEPT6 (R136) and SEPT11 (R174) were obtained by immunizing rabbits with synthetic peptides (CLLQSQGSQAGGSQTLKRDK and CGVQIYQFPTDEETVAEI for SEPT6 and SEPT11, respectively). All rabbit immune sera were further purified by affinity with the corresponding recombinant protein or synthetic peptides.
Cell culture and transfections
HeLa cells (CCL-2; ATCC) were cultivated in MEM-glutamax medium (Life Technologies) supplemented with 10% fetal calf serum, MEM nonessential amino acids (Life Technologies), and 1 mM sodium pyruvate (Life Technologies). U-2 OS cells (HTB-96; ATCC) were cultivated in McCoy’s 5A modified medium (Life Technologies) supplemented with 10% fetal calf serum. For immunofluorescence analyses, HeLa and U-2 OS cells were seeded at a density of 0.5 × 105 and 1.0 × 105 cells on 4-cm2 glass coverslips, respectively. HeLa and U-2 OS cells were then transfected the day after with 1.5 µg and 0.75 µg DNA, respectively, using Lipofectamine LTX reagents (Invitrogen) and processed for immunofluorescence analysis 72 h after transfection. For cotransfection in HeLa cells, plasmids were added at a 1:1 ratio. For video microscopy, HeLa cells were seeded at a density of 0.4 × 105 cells in 35-mm Petri dishes with coverglass (Mattek Corporation). The day after, cells were transfected with 500 ng GFP-SEPT11 expression vectors or cotransfected with 80 ng pEGFP-N2 plasmid (Clontech) and 420 ng SEPT7 expression vectors using Lipofectamine LTX reagents. For analysis of septin expression levels, 1.25 × 105 HeLa cells were seeded in 4-cm2 wells, transfected the day after as previously described, and then directly lysed 48 h after transfection in Laemmli buffer (0.125 M Tris, pH 6.8, 4% SDS, 20% glycerol, and 100 mM DTT). For His pull-down and immunoprecipitation assays, 2.3 × 106 HeLa cells were seeded in 75-cm2 flasks and transfected the day after with 27 µg of plasmids using Lipofectamine LTX reagents. Cells were then lysed and used for His pull-down or immunoprecipitation assays.
Cell synchronization
HeLa cells were grown in 75-cm2 flasks to 40% confluency and blocked for 16 h in culture medium supplemented with 2.5 mM thymidine (Sigma-Aldrich). Cells were then washed four times in PBS, grown in medium supplemented with 24 µM deoxycytidine (Sigma-Aldrich) for 1 h, and transfected for 4 h with His6-SUMO1 expression vector. After transfection, cells were incubated for an additional 3 h in culture medium supplemented with 24 µM deoxycytidine-containing medium and then blocked again in medium supplemented with 2.5 mM thymidine for 16 h. Cells were finally released from the second thymidine block by extensive washes in PBS and incubated in cell culture medium supplemented with 24 µM deoxycytidine.
His pull-down assays
SUMOylated proteins from cell lysates were isolated as described in Tatham et al. (2009). HeLa cells transfected with His6-SUMO expression vectors were washed in PBS and lysed in lysis buffer (6 M guanidium-HCl, 10 mM Tris, 100 mM sodium phosphate buffer, pH 8.0, 5 mM β-mercaptoethanol, and 5 mM imidazole). Nickel–nitrilotriacetic acid agarose resin (Qiagen) prewashed in lysis buffer was incubated with cell lysates overnight at 4°C. Resin was then washed once in lysis buffer, once in wash buffer, pH 8.0 (8 M urea, 10 mM Tris, 100 mM sodium phosphate buffer, pH 8.0, 0.1% Triton X-100, and 5 mM β-mercaptoethanol), and three times in wash buffer, pH 6.3 (8 M urea, 10 mM Tris, 100 mM sodium phosphate buffer, pH 6.3, 0.1% Triton X-100, 5 mM β-mercaptoethanol, and 10 mM imidazole). SUMOylated proteins were then eluted from the resin using elution buffer (200 mM imidazole, 5% SDS, 150 mM Tris-HCl, pH 6.8, 30% glycerol, 720 mM β-mercaptoethanol, and 0.0025% bromophenol blue).
Immunoprecipitations
Transfected HeLa cells were rinsed three times in cold PBS. For immunoprecipitation of FLAG-Ubc9, cells were lysed for 20 min in modified RIPA buffer (50 mM Tris-HCl, pH 7.4, 1% NP-40, 0.25% sodium-deoxycholate, 150 mM NaCl, 1 mM EDTA, 20 mM N-ethylmaleimide, and complete protease inhibitor cocktail [Roche]). For immunoprecipitation of HA-tagged septins, cells were lysed for 20 min in septin lysis buffer (20 mM Tris, pH 8.0, 1% NP-40, 150 mM NaCl, 10% glycerol, 20 mM N-ethylmaleimide, complete protease inhibitor cocktail [Roche], and PhosSTOP phosphatase inhibitors [Sigma-Aldrich]). Lysates were centrifuged for 15 min at 4°C at 16,000 g, and supernatants were incubated for 15 min with protein G–Sepharose beads (GE Healthcare) to eliminate unspecific binding of proteins to beads. After the removal of the beads, cleared lysates were then incubated overnight at 4°C with anti-FLAG (M2; Sigma-Aldrich) or anti-HA (6E2; Cell Signaling Technologies) antibodies. The immunocomplexes were then captured by incubating samples for 3 h with protein G–Sepharose beads. Beads were finally collected and washed four times in the corresponding lysis buffer. For FLAG immunoprecipitation, captured proteins were eluted using 3× FLAG peptide (100 µg/ml of 3× FLAG peptide [Sigma-Aldrich] in 50 mM Tris-HCl, pH 7.4, and 150 mM NaCl). For HA immunoprecipitation, captured proteins were eluted using Laemmli buffer.
Immunoblotting
Cells lysed in Laemmli buffer and proteins eluted from His pull-down or immunoprecipitation assays were separated on SDS-polyacrylamide gels. Proteins were transferred to polyvinylidene fluoride membranes and incubated with primary antibodies (see Table S3 for primary antibody information). Membranes were then incubated with anti–mouse and anti–rabbit HRP-conjugated antibodies (AbCys). Proteins were revealed using Pierce ECL 2 Western blotting substrate (Fisher Scientific). All displayed immunoblots are representative of at least two independent experiments.
Immunofluorescence
HeLa and U-2 OS cells on coverslips were fixed for either 20 min in PBS + 4% paraformaldehyde, followed by a 15-min step of permeabilization in PBS + 0.3% Triton X-100, or fixed for 15 min in ice-cold methanol at −20°C. Cells were then incubated for 3 h with primary antibodies (see Table S3 for primary antibody information) in PBS + 1% BSA, washed, and incubated for 1 h with Alexa Fluor 488– or 546–conjugated anti–mouse and anti–rabbit secondary antibodies (Molecular Probes) in PBS + 1% BSA. F-actin was stained with Alexa Fluor 647–phalloidin (Molecular Probes) and DNA with DAPI. Glass coverslips were finally washed and mounted using Fluoromount (Interchim). Cells were imaged at room temperature using a 200 M inverted microscope (Axiovert; Zeiss) equipped with 100× 1.3 NA NeoFLUAR, 63× 1.4 NA Plan-Apochromat, or 40× 1.3 NA Plan-Apochromat Plan-Neofluar objectives and a camera (Neo sCMOS; Andor) and was controlled by Metamorph software (Universal Imaging). Quantification of aberrant septin bundles and multinucleated cells was performed by analyzing at least 100 transfected cells from 15 independent fields per condition and per experiment (three to five independent experiments). For characterization of septin filaments in cell bridges during cytokinesis, at least 25 transfected cells undergoing cell division were analyzed per condition and per experiment (three independent experiments). P-values were calculated using a two-tailed two-sample equal-variance Student’s t test.
Videomicroscopy and FRAP experiments
For time-lapse phase-contrast microscopy, HeLa cells cotransfected with expression vectors for GFP and SEPT7 were imaged in HeLa cells culture medium in an open chamber (Life Imaging) equilibrated in 5% CO2 and maintained at 37°C. Time-lapse sequences were recorded every 10 min for 48 h using an inverted microscope (Eclipse Ti; Nikon) equipped with a 20× 0.45 NA Plan FluorELWD objective and a camera (CoolSNAP HQ2; Photometrics) and was controlled by Metamorph software. Analyses were performed on at least 50 transfected cells (i.e., GFP-positive cells) per condition and per experiment (three independent experiments). For FRAP experiments, HeLa cells transfected with expression vector for WT or non-SUMOylatable GFP-SEPT11 were imaged in phenol red–free FluoroBrite DMEM (Life Technologies) in an open chamber equilibrated in 5% CO2 and maintained at 37°C. Images were acquired using an inverted microscope (Axio Observer.Z1; Zeiss) equipped with a 100× 1.4 NA Plan-Apochromat objective and a camera (Evolve; Photometrics) and was controlled by Metamorph software. Square areas of 60 µm2 containing GFP-tagged septin filaments were bleached using an iLas2 system (Roper Scientific) with a sequence of 25 irradiations at 100% laser intensity. Equivalent laser intensity, repetition, and exposure time were used for each measurement. Images were taken 20 s before photobleaching and 6 min after photobleaching at a rate of 4 s per frame. Fluorescence intensities were quantified after background subtraction and normalization against unbleached areas using Metamorph software.
Phylogenetic analyses
Sequences of septins were aligned using CLUSTALW2 with default parameters. The corresponding phylogenetic tree was obtained using the neighbor-joining method and rooted using yeast Cdc3 septin as an outgroup. For each septin, one isoform from the UniProt databank was selected: SEPT1, isoform 1 (Q8WYJ6-1); SEPT2, isoform 1 (Q15019-1); SEPT3, isoform 1 (Q9UH03-1); SEPT4, isoform 1 (O43236-1); SEPT5, isoform 1 (Q99719-1); SEPT6, isoform II (Q14141-1); SEPT7, isoform 1 (Q16181-1); SEPT8, isoform 1 (Q92599-1); SEPT9, isoform 1 (Q9UHD8-1); SEPT10, isoform 1 (Q9P0V9-1); SEPT11, isoform 1 (Q9NVA2-1); SEPT12, isoform 1 (Q8IYM1-1); SEPT14 (Q6ZU15); and yeast septin Cdc3 (P32457).
Online supplemental material
Fig. S1 describes the assay used to detect protein SUMOylation. Fig. S2 shows that non-SUMOylatable septins colocalize with endogenous SEPT2, but not with actin filaments in HeLa cells. Fig. S3 shows that non-SUMOylatable septins colocalize with endogenous SEPT9, but not with actin filaments in HeLa cells. Fig. S4 shows that non-SUMOylatable septin variants coimmunoprecipitate with endogenous septins and that expression of these non-SUMOylatable septin variants does not alter endogenous septin protein levels. Fig. S5 shows that non-SUMOylatable SEPT7 and SEPT11 induce defects in septin bundle formation and cell division both in HeLa and U-2 OS cells. Table S1 shows that human septins lack canonical SIMs. Table S2 provides information on the plasmids used in this study. Table S3 provides information on the antibodies used in this study.
Supplementary Material
Acknowledgments
We thank Edith Gouin and Véronique Villiers for their help in antibody production and Frauke Melchior for reagents.
Work in P. Cossart’s laboratory received financial support from Institut Pasteur, Institut National de la Santé et de la Recherche Médicale, Institut National de la Recherche Agronomique, the French National Research Agency (ANR; grant ERANET Infect-ERA PROANTILIS ANR-13-IFEC-0004-02), the French government’s Investissement d’Avenir program, Laboratoire d’Excellence Integrative Biology of Emerging Infectious Diseases (grant ANR-10-LABX-62-IBEID), the European Research Council (grant H2020-ERC-2014-ADG 670823-BacCellEpi), the Fondation le Roch les Mousquetaires, and the International Balzan Prize Foundation. Work from A. Echard’s laboratory received financial support from Institut Pasteur, Centre National de la Recherche Scientifique, Institut National du Cancer (grant 2014-1-PLBIO-04), and ANR (grant AbsyStem 15-CE13-0001-02 and grant CYTOSIGN 16-CE13-004-02). D. Ribet is a Research Associate from INSERM. S. Mostowy is supported by a Wellcome Trust Research Career Development Fellowship (grant WT097411MA) and the Lister Institute of Preventive Medicine. P. Cossart is a Senior International Research Scholar of the Howard Hughes Medical Institute.
The authors declare no competing financial interests.
Author contributions: Conceptualization, D. Ribet, S. Mostowy, A. Echard, and P. Cossart. Investigation, D. Ribet, S. Boscaini, C. Cauvin, and M. Siguier. Original draft, D. Ribet, S. Mostowy, A. Echard, and P. Cossart. Review and editing, D. Ribet, A. Echard, and P. Cossart. Funding acquisition, A. Echard and P. Cossart. Supervision, A. Echard and P. Cossart.
Footnotes
Abbreviations used:
- SIM
- SUMO-interacting motif
- SUMO
- small ubiquitin-like modifier
References
- Angelis D., and Spiliotis E.T.. 2016. Septin Mutations in Human Cancers. Front. Cell Dev. Biol. 4:122 10.3389/fcell.2016.00122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker J., Barysch S.V., Karaca S., Dittner C., Hsiao H.H., Berriel Diaz M., Herzig S., Urlaub H., and Melchior F.. 2013. Detecting endogenous SUMO targets in mammalian cells and tissues. Nat. Struct. Mol. Biol. 20:525–531. 10.1038/nsmb.2526 [DOI] [PubMed] [Google Scholar]
- Bertin A., McMurray M.A., Grob P., Park S.S., Garcia G. III, Patanwala I., Ng H.L., Alber T., Thorner J., and Nogales E.. 2008. Saccharomyces cerevisiae septins: supramolecular organization of heterooligomers and the mechanism of filament assembly. Proc. Natl. Acad. Sci. USA. 105:8274–8279. 10.1073/pnas.0803330105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossis G., and Melchior F.. 2006. Regulation of SUMOylation by reversible oxidation of SUMO conjugating enzymes. Mol. Cell. 21:349–357. 10.1016/j.molcel.2005.12.019 [DOI] [PubMed] [Google Scholar]
- Carter S., and Vousden K.H.. 2008. p53-Ubl fusions as models of ubiquitination, sumoylation and neddylation of p53. Cell Cycle. 7:2519–2528. 10.4161/cc.7.16.6422 [DOI] [PubMed] [Google Scholar]
- Caudron F., and Barral Y.. 2009. Septins and the lateral compartmentalization of eukaryotic membranes. Dev. Cell. 16:493–506. 10.1016/j.devcel.2009.04.003 [DOI] [PubMed] [Google Scholar]
- Cauvin C., and Echard A.. 2015. Phosphoinositides: Lipids with informative heads and mastermind functions in cell division. Biochim. Biophys. Acta. 1851:832–843. 10.1016/j.bbalip.2014.10.013 [DOI] [PubMed] [Google Scholar]
- Chesneau L., Dambournet D., Machicoane M., Kouranti I., Fukuda M., Goud B., and Echard A.. 2012. An ARF6/Rab35 GTPase cascade for endocytic recycling and successful cytokinesis. Curr. Biol. 22:147–153. 10.1016/j.cub.2011.11.058 [DOI] [PubMed] [Google Scholar]
- Choi H.K., Choi K.C., Yoo J.Y., Song M., Ko S.J., Kim C.H., Ahn J.H., Chun K.H., Yook J.I., and Yoon H.G.. 2011. Reversible SUMOylation of TBL1-TBLR1 regulates β-catenin-mediated Wnt signaling. Mol. Cell. 43:203–216. 10.1016/j.molcel.2011.05.027 [DOI] [PubMed] [Google Scholar]
- de Almeida Marques I., Valadares N.F., Garcia W., Damalio J.C., Macedo J.N., de Araújo A.P., Botello C.A., Andreu J.M., and Garratt R.C.. 2012. Septin C-terminal domain interactions: implications for filament stability and assembly. Cell Biochem. Biophys. 62:317–328. 10.1007/s12013-011-9307-0 [DOI] [PubMed] [Google Scholar]
- Dolat L., Hu Q., and Spiliotis E.T.. 2014. Septin functions in organ system physiology and pathology. Biol. Chem. 395:123–141. 10.1515/hsz-2013-0233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echard A., Jollivet F., Martinez O., Lacapère J.J., Rousselet A., Janoueix-Lerosey I., and Goud B.. 1998. Interaction of a Golgi-associated kinesin-like protein with Rab6. Science. 279:580–585. 10.1126/science.279.5350.580 [DOI] [PubMed] [Google Scholar]
- El Amine N., Kechad A., Jananji S., and Hickson G.R.. 2013. Opposing actions of septins and Sticky on Anillin promote the transition from contractile to midbody ring. J. Cell Biol. 203:487–504. 10.1083/jcb.201305053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estey M.P., Di Ciano-Oliveira C., Froese C.D., Bejide M.T., and Trimble W.S.. 2010. Distinct roles of septins in cytokinesis: SEPT9 mediates midbody abscission. J. Cell Biol. 191:741–749. 10.1083/jcb.201006031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira H.C., Luke B., Schober H., Kalck V., Lingner J., and Gasser S.M.. 2011. The PIAS homologue Siz2 regulates perinuclear telomere position and telomerase activity in budding yeast. Nat. Cell Biol. 13:867–874. 10.1038/ncb2263 [DOI] [PubMed] [Google Scholar]
- Flotho A., and Melchior F.. 2013. Sumoylation: a regulatory protein modification in health and disease. Annu. Rev. Biochem. 82:357–385. 10.1146/annurev-biochem-061909-093311 [DOI] [PubMed] [Google Scholar]
- Fontijn R.D., Goud B., Echard A., Jollivet F., van Marle J., Pannekoek H., and Horrevoets A.J.. 2001. The human kinesin-like protein RB6K is under tight cell cycle control and is essential for cytokinesis. Mol. Cell. Biol. 21:2944–2955. 10.1128/MCB.21.8.2944-2955.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Founounou N., Loyer N., and Le Borgne R.. 2013. Septins regulate the contractility of the actomyosin ring to enable adherens junction remodeling during cytokinesis of epithelial cells. Dev. Cell. 24:242–255. 10.1016/j.devcel.2013.01.008 [DOI] [PubMed] [Google Scholar]
- Fung K.Y., Dai L., and Trimble W.S.. 2014. Cell and molecular biology of septins. Int. Rev. Cell Mol. Biol. 310:289–339. 10.1016/B978-0-12-800180-6.00007-4 [DOI] [PubMed] [Google Scholar]
- Garcia G. III, Bertin A., Li Z., Song Y., McMurray M.A., Thorner J., and Nogales E.. 2011. Subunit-dependent modulation of septin assembly: budding yeast septin Shs1 promotes ring and gauze formation. J. Cell Biol. 195:993–1004. 10.1083/jcb.201107123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall P.A., Russel S.E., and Pringle J.R., editors. 2008. The Septins. Wiley, Hoboken, NJ: 380 pp. 10.1002/9780470779705 [DOI] [Google Scholar]
- Hartwell L.H., Culotti J., and Reid B.. 1970. Genetic control of the cell-division cycle in yeast. I. Detection of mutants. Proc. Natl. Acad. Sci. USA. 66:352–359. 10.1073/pnas.66.2.352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havugimana P.C., Hart G.T., Nepusz T., Yang H., Turinsky A.L., Li Z., Wang P.I., Boutz D.R., Fong V., Phanse S., et al. . 2012. A census of human soluble protein complexes. Cell. 150:1068–1081. 10.1016/j.cell.2012.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks I.A., and Vertegaal A.C.. 2016. A comprehensive compilation of SUMO proteomics. Nat. Rev. Mol. Cell Biol. 17:581–595. 10.1038/nrm.2016.81 [DOI] [PubMed] [Google Scholar]
- Hendriks I.A., D’Souza R.C., Chang J.G., Mann M., and Vertegaal A.C.. 2015. System-wide identification of wild-type SUMO-2 conjugation sites. Nat. Commun. 6:7289 10.1038/ncomms8289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks I.A., Lyon D., Young C., Jensen L.J., Vertegaal A.C., and Nielsen M.L.. 2017. Site-specific mapping of the human SUMO proteome reveals co-modification with phosphorylation. Nat. Struct. Mol. Biol. 24:325–336. 10.1038/nsmb.3366 [DOI] [PubMed] [Google Scholar]
- Hernández-Rodríguez Y., and Momany M.. 2012. Posttranslational modifications and assembly of septin heteropolymers and higher-order structures. Curr. Opin. Microbiol. 15:660–668. 10.1016/j.mib.2012.09.007 [DOI] [PubMed] [Google Scholar]
- Hill E., Clarke M., and Barr F.A.. 2000. The Rab6-binding kinesin, Rab6-KIFL, is required for cytokinesis. EMBO J. 19:5711–5719. 10.1093/emboj/19.21.5711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Impens F., Radoshevich L., Cossart P., and Ribet D.. 2014. Mapping of SUMO sites and analysis of SUMOylation changes induced by external stimuli. Proc. Natl. Acad. Sci. USA. 111:12432–12437. 10.1073/pnas.1413825111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson E.S., and Blobel G.. 1999. Cell cycle-regulated attachment of the ubiquitin-related protein SUMO to the yeast septins. J. Cell Biol. 147:981–994. 10.1083/jcb.147.5.981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo E., Surka M.C., and Trimble W.S.. 2007. Mammalian SEPT2 is required for scaffolding nonmuscle myosin II and its kinases. Dev. Cell. 13:677–690. 10.1016/j.devcel.2007.09.001 [DOI] [PubMed] [Google Scholar]
- Kim M.S., Froese C.D., Estey M.P., and Trimble W.S.. 2011. SEPT9 occupies the terminal positions in septin octamers and mediates polymerization-dependent functions in abscission. J. Cell Biol. 195:815–826. 10.1083/jcb.201106131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita M. 2003. The septins. Genome Biol. 4:236 10.1186/gb-2003-4-11-236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita M., Kumar S., Mizoguchi A., Ide C., Kinoshita A., Haraguchi T., Hiraoka Y., and Noda M.. 1997. Nedd5, a mammalian septin, is a novel cytoskeletal component interacting with actin-based structures. Genes Dev. 11:1535–1547. 10.1101/gad.11.12.1535 [DOI] [PubMed] [Google Scholar]
- Kouranti I., Sachse M., Arouche N., Goud B., and Echard A.. 2006. Rab35 regulates an endocytic recycling pathway essential for the terminal steps of cytokinesis. Curr. Biol. 16:1719–1725. 10.1016/j.cub.2006.07.020 [DOI] [PubMed] [Google Scholar]
- Kremer B.E., Haystead T., and Macara I.G.. 2005. Mammalian septins regulate microtubule stability through interaction with the microtubule-binding protein MAP4. Mol. Biol. Cell. 16:4648–4659. 10.1091/mbc.E05-03-0267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon M.B., and Gaestel M.. 2015. Sep(t)arate or not – how some cells take septin-independent routes through cytokinesis. J. Cell Sci. 128:1877–1886. 10.1242/jcs.164830 [DOI] [PubMed] [Google Scholar]
- Mostowy S., and Cossart P.. 2012. Septins: the fourth component of the cytoskeleton. Nat. Rev. Mol. Cell Biol. 13:183–194. [DOI] [PubMed] [Google Scholar]
- Nagata K., Kawajiri A., Matsui S., Takagishi M., Shiromizu T., Saitoh N., Izawa I., Kiyono T., Itoh T.J., Hotani H., and Inagaki M.. 2003. Filament formation of MSF-A, a mammalian septin, in human mammary epithelial cells depends on interactions with microtubules. J. Biol. Chem. 278:18538–18543. 10.1074/jbc.M205246200 [DOI] [PubMed] [Google Scholar]
- Nakahira M., Macedo J.N., Seraphim T.V., Cavalcante N., Souza T.A., Damalio J.C., Reyes L.F., Assmann E.M., Alborghetti M.R., Garratt R.C., et al. . 2010. A draft of the human septin interactome. PLoS One. 5:e13799 10.1371/journal.pone.0013799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld T.P., and Rubin G.M.. 1994. The Drosophila peanut gene is required for cytokinesis and encodes a protein similar to yeast putative bud neck filament proteins. Cell. 77:371–379. 10.1016/0092-8674(94)90152-X [DOI] [PubMed] [Google Scholar]
- Oegema K., Savoian M.S., Mitchison T.J., and Field C.M.. 2000. Functional analysis of a human homologue of the Drosophila actin binding protein anillin suggests a role in cytokinesis. J. Cell Biol. 150:539–552. 10.1083/jcb.150.3.539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliuso A., Tham T.N., Stevens J.K., Lagache T., Persson R., Salles A., Olivo-Marin J.-C., Oddos S., Spang A., Cossart P., and Stavru F.. 2016. A role for septin 2 in Drp1-mediated mitochondrial fission. EMBO Rep. 17:858–873. 10.15252/embr.201541612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan F., Malmberg R.L., and Momany M.. 2007. Analysis of septins across kingdoms reveals orthology and new motifs. BMC Evol. Biol. 7:103 10.1186/1471-2148-7-103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röseler S., Sandrock K., Bartsch I., Busse A., Omran H., Loges N.T., and Zieger B.. 2011. Lethal phenotype of mice carrying a Sept11 null mutation. Biol. Chem. 392:779–781. 10.1515/BC.2011.093 [DOI] [PubMed] [Google Scholar]
- Saarikangas J., and Barral Y.. 2011. The emerging functions of septins in metazoans. EMBO Rep. 12:1118–1126. 10.1038/embor.2011.193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellin M.E., Sandblad L., Stenmark S., and Gullberg M.. 2011. Deciphering the rules governing assembly order of mammalian septin complexes. Mol. Biol. Cell. 22:3152–3164. 10.1091/mbc.E11-03-0253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirajuddin M., Farkasovsky M., Hauer F., Kühlmann D., Macara I.G., Weyand M., Stark H., and Wittinghofer A.. 2007. Structural insight into filament formation by mammalian septins. Nature. 449:311–315. 10.1038/nature06052 [DOI] [PubMed] [Google Scholar]
- Spiliotis E.T., Kinoshita M., and Nelson W.J.. 2005. A mitotic septin scaffold required for Mammalian chromosome congression and segregation. Science. 307:1781–1785. 10.1126/science.1106823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y., Iwase M., Konishi M., Tanaka M., Toh-e A., and Kikuchi Y.. 1999. Smt3, a SUMO-1 homolog, is conjugated to Cdc3, a component of septin rings at the mother-bud neck in budding yeast. Biochem. Biophys. Res. Commun. 259:582–587. 10.1006/bbrc.1999.0821 [DOI] [PubMed] [Google Scholar]
- Tatham M.H., Rodriguez M.S., Xirodimas D.P., and Hay R.T.. 2009. Detection of protein SUMOylation in vivo. Nat. Protoc. 4:1363–1371. 10.1038/nprot.2009.128 [DOI] [PubMed] [Google Scholar]
- Trimble W.S., and Grinstein S.. 2015. Barriers to the free diffusion of proteins and lipids in the plasma membrane. J. Cell Biol. 208:259–271. 10.1083/jcb.201410071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirich C.S., Erzberger J.P., and Barral Y.. 2008. The septin family of GTPases: architecture and dynamics. Nat. Rev. Mol. Cell Biol. 9:478–489. 10.1038/nrm2407 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.