A new common mechanism for increasing RhoA-GTP is identified in Tripathi et al. The increased RhoA-GTP results from signaling mechanisms that phosphorylate and attenuate the DLC1 tumor suppressor, which encodes RhoGAP. The potentially reversible nature of this attenuation may have therapeutic relevance in cancer.
Abstract
We report several receptor tyrosine kinase (RTK) ligands increase RhoA–guanosine triphosphate (GTP) in untransformed and transformed cell lines and determine this phenomenon depends on the RTKs activating the AKT serine/threonine kinase. The increased RhoA-GTP results from AKT phosphorylating three serines (S298, S329, and S567) in the DLC1 tumor suppressor, a Rho GTPase-activating protein (RhoGAP) associated with focal adhesions. Phosphorylation of the serines, located N-terminal to the DLC1 RhoGAP domain, induces strong binding of that N-terminal region to the RhoGAP domain, converting DLC1 from an open, active dimer to a closed, inactive monomer. That binding, which interferes with the interaction of RhoA-GTP with the RhoGAP domain, reduces the hydrolysis of RhoA-GTP, the binding of other DLC1 ligands, and the colocalization of DLC1 with focal adhesions and attenuates tumor suppressor activity. DLC1 is a critical AKT target in DLC1-positive cancer because AKT inhibition has potent antitumor activity in the DLC1-positive transgenic cancer model and in a DLC1-positive cancer cell line but not in an isogenic DLC1-negative cell line.
Introduction
The RhoA GTPase (RhoA) is an essential, widely expressed, membrane-associated, guanine nucleotide–binding protein that contributes to various physiologic processes, including cell proliferation, cytoskeletal dynamics, cell migration, cell metabolism, cytokinesis, and vesicle trafficking. It is frequently activated in advanced cancer and has also been implicated in cardiovascular and other diseases (Zhou and Zheng, 2013; Loirand, 2015; Ricker et al., 2016; Shimokawa et al., 2016; Wu and Xu, 2016).
RhoA acts as a molecular switch that is active when bound to GTP and inactive when bound to GDP. Regulation of RhoA by ligands for G protein–coupled receptors, especially those for lysophosphatidic acid (LPA), has been recognized for many years (Xiang et al., 2013; Yu and Brown, 2015). RhoA can also be regulated by adhesion and mechanical factors (Marjoram et al., 2014). In addition, receptor tyrosine kinases (RTKs) may up-regulate RhoA under some conditions, secondary to RTK-dependent activation of Rho guanine nucleotide exchange factors (Rho-GEFs), which catalyze replacement of GDP-bound RhoA with GTP-bound RhoA (Schiller, 2006).
RhoA can also be negatively regulated by Rho guanine nucleotide dissociation inhibitors, which sequester RhoA from the membrane (Garcia-Mata et al., 2011), and Rho GTPase-activating proteins (RhoGAPs), which inactivate RhoA by catalyzing the hydrolysis of GTP-bound RhoA to GDP-bound RhoA. However, their role in ligand-dependent RhoA signaling is not well established. Here, we report that ligand-dependent stimulation of RTKs in epithelial cells and fibroblasts can stimulate the activation of RhoA, and we determined that the activation was attributable to a previously unknown mechanism, down-regulated activity of a specific, widely expressed RhoGAP DLC1 by a process that involves its phosphorylation by the serine/threonine kinase AKT.
Results
EGF, insulin, and insulin-like growth factor-1 (IGF-1) positively regulate RhoA-GTP in a DLC1-dependent manner
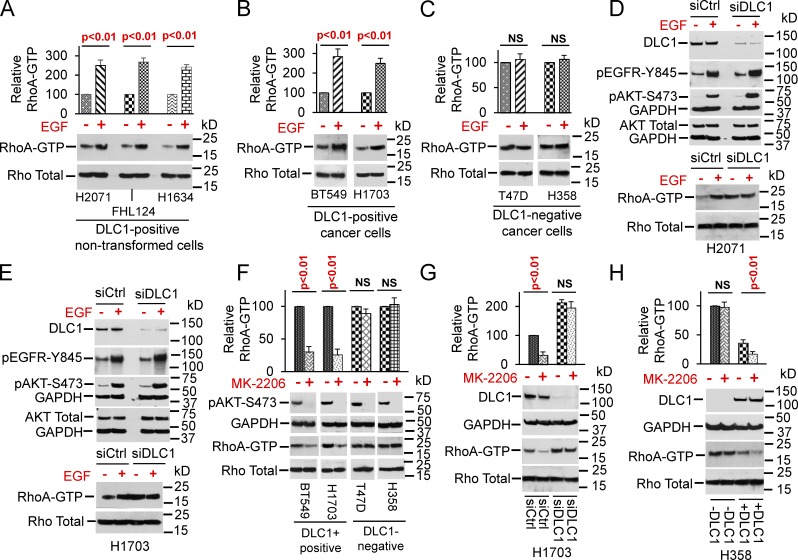
We observed that stimulation of the EGF RTK, with its cognate ligand EGF, could activate RhoA in two nontransformed epithelial cell lines, a fibroblast line, and a subset of cancer cell lines. Analysis of the lines unexpectedly found an excellent correlation between the ability of EGF to increase RhoA-GTP and the expression of endogenous DLC1, which is a tumor-suppressor gene that encodes a 1091 amino acid protein containing a highly conserved RhoGAP domain and is required for its tumor-suppressor function (Durkin et al., 2005; Lukasik et al., 2011). The nontransformed lines H2071 (skin epithelial cells), FHL124 (lens epithelial cells), and H1634 (fibroblasts) all express DLC1, and EGF increased RhoA-GTP in each of them (Fig. 1 A), as did all four DLC1-positive cancer lines tested: two breast cancer lines, BT549 and MCF10Ca1h, and two non–small cell lung cancer (NSCLC) lines, H1703 and H157 (Fig. 1 B and Fig. S1 A). However, EGF did not increase RhoA-GTP in the DLC1-negative lines examined: two breast cancer lines, T47D and MDA-MB-468, and two NSCLC lines, H358 and A549 (Fig. 1 C and Fig S1 B).
Figure 1.
EGF-induced AKT activity increases RhoA-GTP through DLC1. (A and B) EGF increases RhoA-GTP, but not total Rho, in DLC1-positive nontransformed (A) and cancer (B) cells. (C) EGF does not alter RhoA-GTP in DLC1-negative lines. (D and E) DLC1 siRNA renders RhoA-GTP unresponsive to EGF. EGF-induced EGFR activity (phosphorylation of EGFR-Y845) and AKT activity (phosphorylation of AKT-S473) in DLC1-expressing and DLC1-knockdown cells. DLC1 knockdown abrogates the ability of EGF to increase RhoA-GTP in nontransformed (D) and cancer (E) cells. (F) MK-2206 decreases RhoA-GTP in DLC1-positive lines (BT549 and H1703) but not in DLC1-negative lines (T47D and H358), although MK-2206 inhibits AKT activity in all lines. (G) MK-2206 suppresses RhoA-GTP in DLC1-expressing cells but not in DLC1-knockdown cells. (H) Stable DLC1 transfection of DLC1-negative H358 cells decreases basal RhoA-GTP and enables MK-2206 to further reduce RhoA-GTP. MK-2206 does not affect RhoA-GTP in parental H358 cells. Each graph shows relative RhoA-GTP means ± SD from three experiments. Parametric two-tailed t tests were performed for statistical analysis.
To determine whether the EGF-induced increase in RhoA-GTP depended on DLC1, the effect of DLC1 knockdown by siRNA was tested in DLC1-positive lines: two nontransformed, H2071 and H1634, and two transformed lines, H1703 and BT549. The siRNAs efficiently suppressed DLC1 expression in each line, leading, as expected, to an increase in basal RhoA-GTP levels (Fig. 1, D and E; and Fig. S1, C and D). Although EGF activated the EGF receptor (EGFR), as measured by EGFR-Y845 phosphorylation, whether or not the cells had been treated with DLC1 siRNAs, EGF increased RhoA-GTP only under conditions with continued expression of DLC1 (Fig. 1, D and E; Fig. S1, C and D; and Fig. S1 E, which shows the relative DLC1 expression of all lines used in this study). Thus, the EGF-induced increase in RhoA-GTP depended on the presence of DLC1.
In a preliminary exploration of the mechanism by which EGF might be regulating DLC1, the level of serine phosphorylation of DLC1 increased after EGF treatment (Fig. S1 F), whereas tyrosine phosphorylation was unchanged (unpublished data). One serine/threonine kinase known to be activated by EGF is AKT (Fig. S1 G; Garay et al., 2015; Nishimura et al., 2015). A previous study identified one serine in DLC1 (S567) as being phosphorylated by AKT (Ko et al., 2010), which decreased the growth inhibitory activity of DLC1 by a mechanism the authors concluded was not associated with a change in RhoA-GTP. Two additional serines in DLC1 are possible AKT substrates (S298 and S329), but Ko et al. (2010) concluded they were not phosphorylated.
We evaluated whether AKT activation might be associated with the EGF-induced stimulation of RhoA. As expected, EGF treatment led to increased AKT activity, as measured by AKT-S473 phosphorylation, which was not attenuated by siRNA knockdown of DLC1 (Fig. 1, D and E; and Fig. S1, C and D). To explore the temporal relationship between EGF-induced changes in EGFR activation, AKT activation, and RhoA-GTP level, we evaluated two nontransformed DLC1-positive lines (H2071 and H1634) with low AKT activity and three DLC1-positive cancer lines (BT549, H1703, and H157) with high AKT activity. After 15 min of EGF treatment, EGFR was activated in each line; it remained high at 30 min and then decreased at 60 and 120 min (Fig. S1 H). The kinetic profiles of AKT activity, as measured by pAKT-S473 and RhoA-GTP, paralleled that of EGFR activity (Fig. S1 H).
To test whether AKT activity was mechanistically linked to the observed changes in RhoA-GTP, two DLC1-positive lines (BT549 and H1703) and two DLC1-negative lines (T47D and H358) were treated with an AKT inhibitor, MK-2206 (Hirai et al., 2010). Although AKT activity was efficiently inhibited in all lines, MK-2206 substantially reduced RhoA-GTP in the DLC1-positive lines (Fig. 1 F, two left columns), but not in the DLC1-negative lines (Fig. 1 F, two right columns). To verify that the observed effect of AKT activity on RhoA-GTP depended on DLC1, H1703 cells in which DLC1 had been knocked down by siRNAs were treated with MK-2206. Under those conditions of low DLC1, MK-2206 did not affect RhoA-GTP (Fig. 1 G). We also examined MK-2206 on an H358-derived cell line that, although the parental line does not express DLC1, had been stably transfected with DLC1. MK-2206 had no effect on RhoA-GTP in the parental cells, but it did reduce RhoA-GTP in the DLC1 transfectant (Fig. 1 H). Thus, AKT-regulated RhoA-GTP was dependent on DLC1. AKT also negatively regulated the RhoGAP activity of other DLC family members, DLC2 and DLC3 (Lukasik et al., 2011; Braun and Olayioye, 2015), but the effect of AKT inhibition on DLC2 and DLC3 on RhoA-GTP was less than that on DLC1 when cells transfected with the DLC genes were treated with MK-2206 (Fig. S1 I).
Because EGFR is an RTK, we explored whether ligands that activate other RTKs and AKT might also increase RhoA-GTP. Analogous to EGF, insulin (Fig. S2 A) and IGF-1 (Fig. S2 B) activated AKT in all lines tested, but they increased RhoA-GTP only in the DLC1-positive lines.
We also examined LPA, a ligand frequently used to increase RhoA-GTP, to see whether DLC1 was involved in that process. However, LPA, which increases RhoA-GTP through G protein–coupled receptors, rather than through RTKs (Yu and Brown, 2015), did not activate AKT and induced RhoA-GTP in a DLC1-independent manner in all lines examined (Fig. S2, C–E). Thus, LPA and RTK ligands induce RhoA-GTP by distinct mechanisms.
LPA treatment also enabled us to verify that the observed inability of AKT to increase RhoA-GTP in cells that were deficient for DLC1 expression (Figs 1, D and E; and Fig. S1, C and D) was not because they were incapable of further increasing their RhoA-GTP. Indeed, LPA increased RhoA-GTP in cells that were DLC1-negative (Fig. S2 C, right) and in cells whose DLC1 had been subjected to knockdown by siRNAs (Fig. S2 E).
AKT forms an endogenous complex with DLC1 and interacts with two regions of DLC1
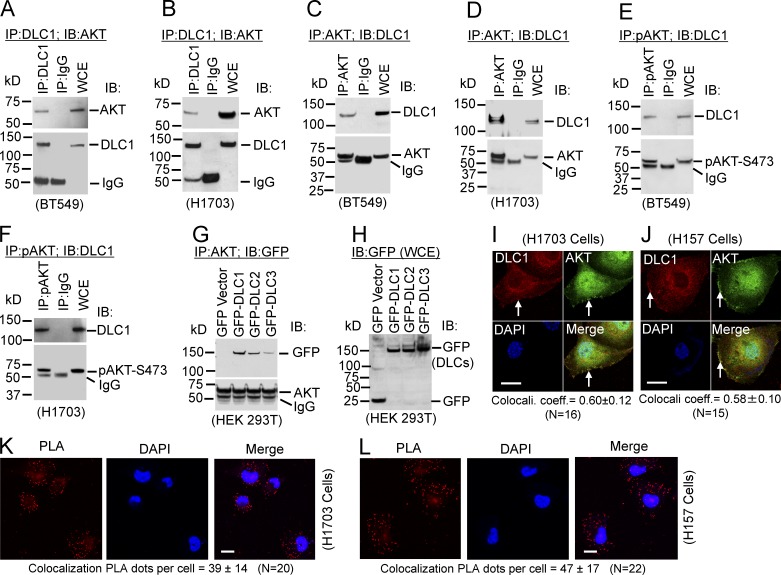
The aforementioned results suggest that AKT might bind DLC1. To determine whether the two proteins form a complex in cells, we performed coimmunoprecipitation (coIP) experiments in cells expressing both proteins: two nontransformed lines (H2071 and H1634) and three cancer lines (BT549, H1703, and H157). DLC1 and AKT formed a complex in each line, whether cell lysates were immunoprecipitated (IP), first with a DLC1 antibody, and then immunoblotted (IB) for AKT (Fig. 2, A and B; and Fig. S3, A, C, and D) or by the reciprocal coIP (Fig. 2, C and D; and Fig S3 B), implying the interaction is physiologically relevant.
Figure 2.
Endogenous protein complex formation between DLC1 and AKT. (A–D) Complex between DLC1 and AKT. (A and B) Lysates from BT549 and H1703 were IP with DLC1 or mock IgG antibodies, followed by IB with AKT (top) or DLC1 (bottom) antibodies. WCE, whole cell extract. (C and D) AKT/DLC1 complex by reciprocal coIP. (E and F) Some AKTs associated with DLC1 were enzymatically active. Protein complex between DLC1 and kinase-active AKT (pAKT-S473). (E) Lysates from BT549 cells were IP with pAKT-S473 or mock IgG antibodies followed by IB with DLC1 (top) or pAKT-S473 (bottom) antibodies. (F) Enzymatically active complex was detected by the reciprocal coIP. (G and H) AKT interacts with DLC2 and DLC3 but less strongly than with DLC1. (G) Lysates from HEK 293T cells transfected with GFP-tagged DLC1, DLC2, or DLC3 were IP with AKT antibody, followed by IB with GFP (top) or AKT (bottom) antibodies. (H) Expression of GFP and various GFP-tagged DLC constructs for G. (I–L) Colocalization of AKT with DLC1. Colacali. coeff., colocalization coefficient. (I) H1703 cells were stained with DLC1 (red) and AKT (green) antibodies. Colocalization of DLC1 and AKT is highlighted in yellow (arrow) in the merged image. Overlapping colocalization coefficient means ± SD (below panel) was calculated from 16 cells randomly selected from several fields. Bar, 20 µm. (J) Colocalization of endogenous DLC1 with AKT in H157 cells. Experimental conditions were similar to those in I. Bar, 20 µm. (K and L) Colocalization of DLC1 and AKT was confirmed by PLA. Bars, 10 µm.
Some AKT associated with DLC1 was inferred to be enzymatically active because positive coIP results were obtained from all three lines when the lysates were IP with a phospho-specific antibody against pAKT-S473, followed by IB for DLC1 (Fig. 2, E and F; and Fig. S3 E). However, MK-2206 did not reduce the amount of the AKT/DLC1 complex (Fig. S3, F–J), and AKT activity stimulation by EGF did not increase the AKT/DLC1 complex (Fig. S3, K–N), implying complex formation does not depend on the enzymatic activity of AKT. AKT also formed a complex with DLC2 and DLC3, but less efficiently than it did with DLC1 (Fig. 2, G and H). Colocalization of AKT and DLC1 was confirmed by confocal microscopy in the two NSCLC lines, with colocalization coefficients of ∼0.60 (Fig. 2, I and J), and by proximity ligation assay (PLA; Fig. 2, K and L).
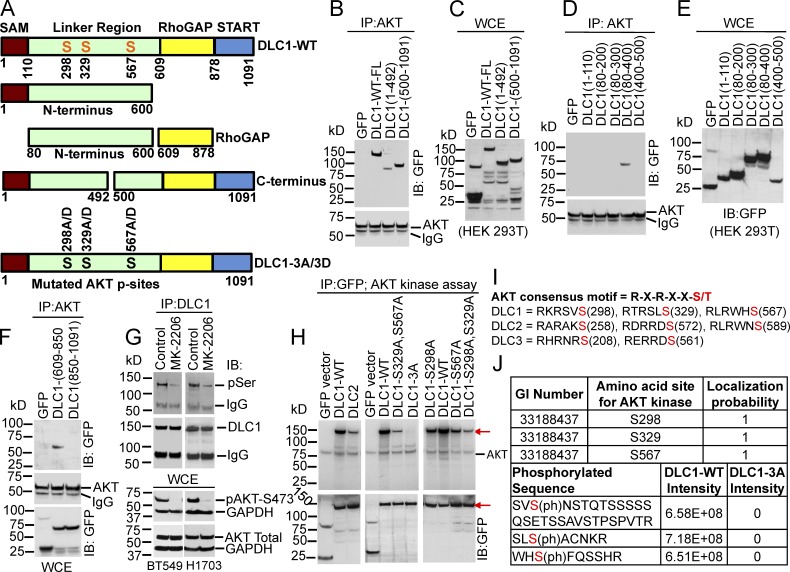
DLC1 has four recognized regions: an N-terminal SAM domain, a linker region, a RhoGAP domain, and a C-terminal START domain (Fig. 3 A; Tripathi et al., 2014). To map the regions of DLC1 required for AKT binding, lysates from human embryonic kidney (HEK) 293T cells expressing various GFP-tagged DLC1 fragments, which were numbered by their respective N-terminal– and C-terminal–encoded amino acids (Fig. 3 A), were IP with AKT antibody, followed by IB with a GFP antibody. Interestingly, AKT bound to some nonoverlapping N-terminal and C-terminal DLC1 fragments, implying that AKT interacts with more than one region of DLC1 (Fig. 3, B and C). To more precisely map the DLC1 regions required for AKT binding, smaller N-terminal and C-terminal DLC1 fragments were used. N-terminal AKT binding required at least some DLC1 amino acids 300–400 because DLC1 (80–400) was positive, whereas DLC1 (80–300) was negative (Fig. 3, D and E). The C-terminal DLC1 fragment that bound AKT was mapped to the RhoGAP domain because AKT interacted with DLC1 (609–850) but not with DLC1 (850–1091; Fig. 3 F).
Figure 3.
AKT binds two nonoverlapping regions of DLC1 and phosphorylates three serines in DLC1. (A) Schematic representation of DLC1 domains. DLC1-3A and DLC1-3D mutants have three AKT phosphorylatable serines mutated to alanine or aspartate, respectively. All constructs were GFP tagged. (B and C) AKT binds to N- and C-terminal sequences of DLC1. WCE, whole cell extract. (B) Lysates from HEK 293T cells transfected with indicated DLC1 constructs were IP with AKT antibody, followed by IB with GFP (top) and AKT (bottom) antibodies. (C) Expression of DLC1 constructs for B. (D and F) Experimental conditions were as in B and C. (D) AKT binds to DLC1 amino acids 80–400 but not to the other N-terminal DLC1 fragments. (E) Expression of DLC1 constructs for D. (F) AKT binds to the RhoGAP domain (amino acids 609–850) of DLC1, but not the C-terminal fragment (amino acids 850–1091; top). IP pellets were IB with AKT antibody (middle). Expression of GFP and DLC1 constructs (bottom). (G) MK-2206 reduces phosphorylation of AKT serines in DLC1 (pSer) without reducing total DLC1. Lysates from BT549 and H1703 cells, treated without or with MK-2206, which reduced pAKT-473 but not its protein level, were IP with DLC1 antibody followed by IB with phospho-AKT substrate-specific (top) or DLC1 antibodies. MK-2206 inhibited AKT activity in both lines (bottom). (H) AKT phosphorylates DLC1 in vitro. (H, top) IP DLC1-WT from transfected HEK 293T cells was strongly phosphorylated by recombinant AKT kinase (left, middle lane), as detected with 32P autoradiography; GFP control was negative (lane 1 in both left and middle). Combined DLC1-S329A,S567A mutant was weakly phosphorylated (middle, lane 3), but the DLC1-3A mutant (combined serine-to-alanine mutation of S298, S329, and S567) gave no phospho-signal (middle, lane 4). DLC2 was also phosphorylated (left, lane 3) but weaker than it was with DLC1. (H, bottom) Expression of DLC1 constructs. (I) Consensus motifs for AKT kinase for indicated serines (red) in DLC1, DLC2, and DLC3. (J) Phosphorylation of DLC1 by AKT in cells. DLC1 phosphopeptides were detected by mass spectrometry. The AKT phosphorylation signals were fully localized to the indicated serines and were absent from the DLC1-3A mutant protein.
AKT phosphorylates three serines in DLC1
Although a previous study (Ko et al., 2010) concluded only S567 in DLC1 was phosphorylated by AKT, we evaluated whether S298 and/or S329 (Fig. 3, A and I) might be phosphorylated under our growth conditions using 32P labeling. First, we confirmed that most of the serine phosphorylation of DLC1 was dependent on AKT by treating the cells with MK-2206, which greatly reduced both AKT activity and serine phosphorylation of DLC1 (Fig. 3 G). Using partially purified, full-length DLC1, we observed DLC1 was strongly phosphorylated by AKT kinase in vitro, unlike the GFP control (Fig. 3 H, left, lanes 1 and 2). Thus, DLC1 is a direct substrate for AKT. DLC2 and DLC3 were also phosphorylated in vitro but less strongly than DLC1 (Fig. 3 H, left; and Fig. S4 A; see Fig. 3 I for consensus motifs).
To determine whether S298, S329, and S567 are the major AKT phosphorylation sites in DLC1, all three were mutated to alanine (DLC1-3A), resulting in a drastically reduced in vitro phospho signal (Fig. 3 H, middle, lane 4). When serine-to-alanine double mutants were analyzed, the phospho signal was reduced but was greater than the DLC1-3A mutant; single serine-to-alanine mutants retained a strong phospho signal (Fig. 3 H, middle and right; S329A mutant not depicted). Liquid chromatography–mass spectrometry confirmed those results, suggesting that all three serines were phosphorylated under these growth conditions because the analysis of the partially purified DLC1 from cells transfected with DLC1-WT or the DLC1-3A mutant detected phosphorylation in S298, S329, and S567 in the relevant DLC1-WT peptides, but not in the DLC1-3A mutant (Fig. 3 J and Fig. S4 B).
Phosphorylation of the three “AKT” serines in DLC1 greatly attenuates its RhoGAP and tumor-suppressor functions
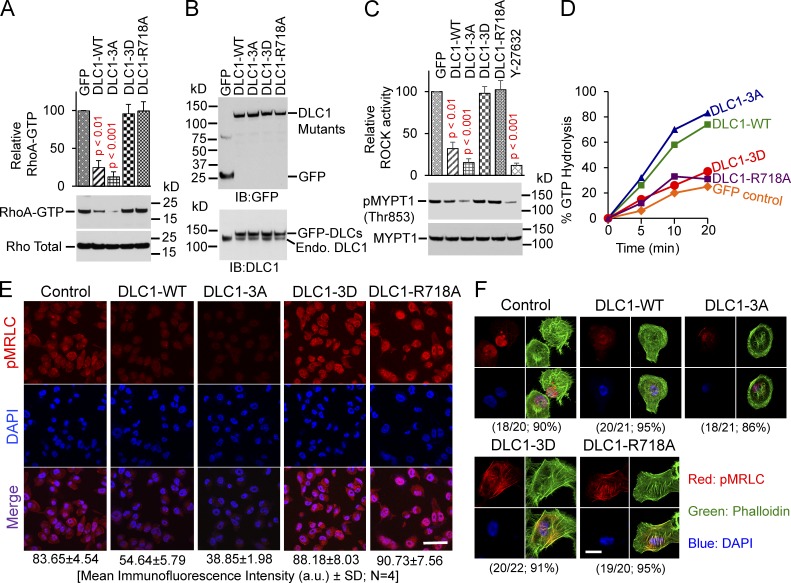
Our observation that AKT could increase RhoA-GTP in a DLC1-dependent manner suggested that AKT phosphorylation of the three DLC1 serines would attenuate its RhoGAP and tumor suppressor functions. To evaluate those possibilities, we compared the activities of DLC1-WT with the nonphosphorylatable, triple alanine mutant (DLC1-3A). We also constructed and analyzed the phosphomimetic, triple serine-to-aspartate mutant (DLC1-3D) and included a “GAP-dead” mutant (DLC1-R718A) as a control.
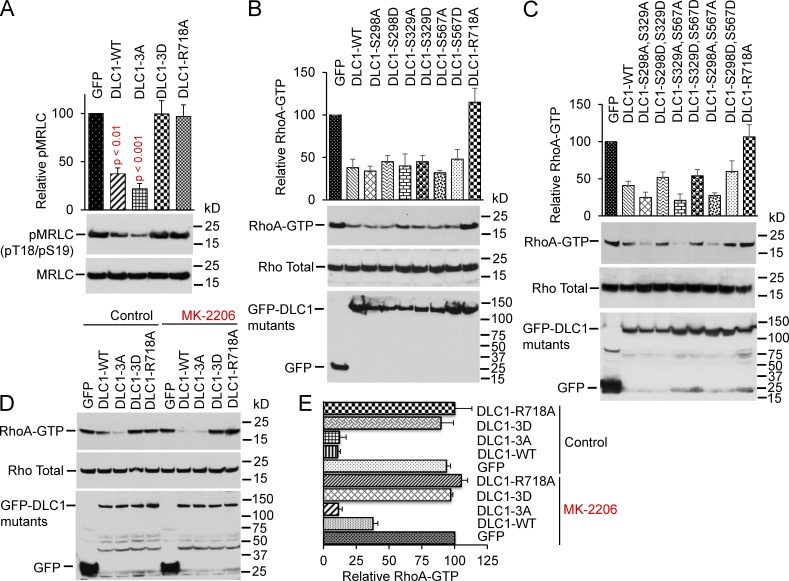
In H1703 cells that stably expressed similar levels of the constructs, the RhoA-GTP level in the DLC1-3D transfectant was similar to the GAP-dead DLC1 (DLC1-R718A; Fig. 4, A and B). However, the RhoA-GTP in the DLC1-3A mutant was at least as low as that induced by DLC1-WT. Analogous results were seen when the DLC1 mutants were analyzed for Rho kinase (ROCK) activity (Fig. 4 C), in vitro RhoGAP activity (Fig. 4 D), and phosphorylation of myosin regulatory light chain (pMRLC; Fig. 5 A), a major downstream effector of the RhoA-ROCK pathway. By immunofluorescence, cells transfected with GFP, DLC1-3D, or DLC1-R718A showed similar strong staining of pMRLC (Fig. 4 E), with well-formed stress fibers (Fig. 4 F). In contrast, cells transfected with DLC1-WT or DLC1-3A showed less pMRLC staining, fewer stress fibers, and few, if any, concave boundaries, consistent with reduced RhoA-ROCK signaling and reduced cell contraction.
Figure 4.
Phosphomimetic DLC1-3D mutant has attenuated RhoGAP activity. (A) Graph shows relative RhoA-GTP means ± SD for indicated DLC1 mutants from three experiments. DLC1-3D mutant is as defective as GAP-dead DLC1-R718A mutant for reducing RhoA-GTP. DLC1-3A mutant is more active than DLC1-WT. Parametric two-tailed t tests were performed for statistical analysis. P < 0.01 for DLC1-WT and P < 0.001 for DLC1-3A compared with other transfectants. (B) Expression of stable DLC1 transfectants in H1703 cells for A. Lysates were IB with GFP (top) or DLC1 (bottom) antibodies. (C) Experimental conditions and data displays were similar to A, but for ROCK activity, analogous to the results in A. (D) Relative RhoGAP activity of DLC1 mutants. In vitro RhoA-GTP hydrolysis by DLC1 mutants. Hydrolysis of DLC1-3D was similar to GFP control or GAP-dead DLC1-R718A. Hydrolysis of DLC1-3A was greater than it was with DLC1-WT. (E) Cells with stably transfected DLC1-WT or DLC1-3A have less phospho-MRLC (red) compared with the other transfectants. DAPI (blue) represents the nuclei. Bar, 100 µm. (F) Cells transfected with DLC1-WT or DLC1-3A have fewer concave boundaries, consistent with reduced contraction, and fewer stress fibers (green), especially in the central region, compared with the other transfectants. The confocal images are representative of most of the cells. Bar, 20 µm.
Figure 5.
RhoGAP activity of DLC1 is inversely related to the number of S-to-D mutations in DLC1. (A) DLC1-WT–positive control has less pMRLC (P < 0.01) than GFP-transfected cells have. DLC1-3D mutant is as defective as a GAP-dead DLC1-R718A mutant for inhibition of pMRLC. Graph shows relative pMRLC/MRLC means ± SD from three experiments. Parametric two-tailed t tests were performed for statistical analysis. (B and C) RhoA-GTP (top) and total Rho (middle) in individual or combined mutants of DLC1 transfected in the DLC1-negative A549 cells. RhoA-GTP levels are inversely related to the number of S-to-D mutations, and the degree of RhoGAP reduction is additive for each mutation. Expression of indicated DLC1 constructs (bottom). Graph shows relative RhoA-GTP means ± SD from two experiments. (D) Effect of MK-2206 on RhoGAP activity of DLC1 mutants. In control panel, RhoA-GTP was reduced by DLC1-WT or DLC1-3A mutant compared with GFP control, DLC1-3D, or DLC1-R718A. MK-2206 further reduces RhoA-GTP in DLC1-WT (compare lanes 2 and 7). MK-2206 does not influence RhoA-GTP in GFP, DLC1-3A, DLC1-3D, or DLC1-R718A. (E) Graph shows relative RhoA-GTP means ± SD from three experiments, as in D. A significant decrease (P < 0.01) in RhoA-GTP in DLC1-WT–transfected cells by MK-2206 (compare DLC1-WT control versus DLC1-WT treated with MK-2206).
When less-drastic individual or combined mutants were analyzed in DLC1-negative A549 cells, RhoA-GTP was inversely related to the number of phosphomimetic serine-to-aspartate (S-to-D) mutations in DLC1 (Fig. 5, B and C). These results suggest the effects of each serine phosphorylation on RhoA-GTP are additive.
We examined MK-2206 treatment of stable DLC1 transfectants in DLC1-negative H358 cells. The findings verified that MK-2206 did not affect RhoA-GTP in DLC1-3A or DLC1-3D mutants, although, consistent with earlier results (Fig. 1 H), it did decrease RhoA-GTP for DLC1-WT (Fig. 5, D and E).
We had previously found that CDK5 phosphorylation of four other serines in the linker region (S120, S205, S422, and S509) increased the RhoGAP activity of DLC1 (Tripathi et al., 2014), which is the opposite of the effects of AKT phosphorylation of DLC1. To determine whether AKT phosphorylation of DLC1 was phenotypically dominant over CDK5 phosphorylation of DLC1, we studied A549 cells, in which both endogenous CDK5 and endogenous AKT are active and stably transfected with mutants of all four “CDK5” serines in DLC1 (DLC1-4A-CDK5 and DLC1-4D-CDK5) and those transfected with the AKT serine mutants DLC1-3A and DLC1-3D mutants described here (for clarity, they are designated DLC1-3A-AKT and DLC1-3D-AKT in this paragraph), as well as with WT DLC1. The effects of AKT inhibition or CDK5 inhibition on the phosphorylation of the AKT-serines and the CDK5-serines and on RhoA-GTP were evaluated. Consistent with AKT activity being phenotypically dominant over CKD5 activity, CKD5 inhibition with roscovitine did not increase RhoA-GTP for the DLC1-3A-AKT mutant, but AKT inhibition with MK-2206 did reduce RhoA-GTP for the DLC1-4A-CDK5 mutant (Fig. S5, A and B).
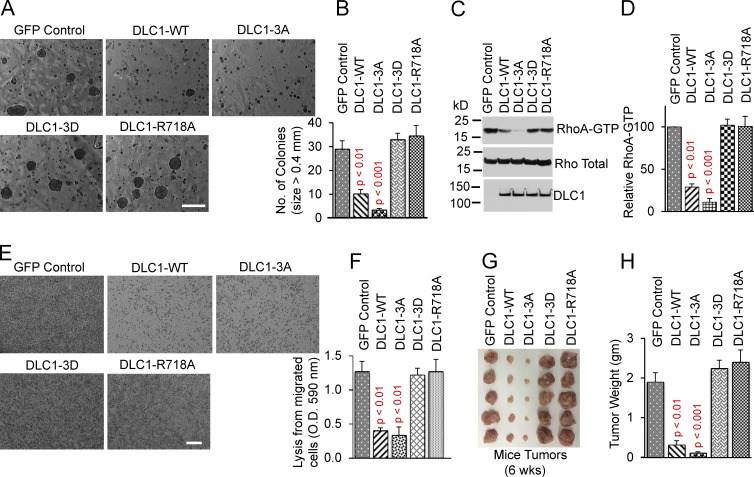
To assess the biological significance of DLC1 phosphorylation by AKT, we evaluated the DLC1-3A and DLC1-3D mutants in several bioassays. Stable DLC1-WT transfectants in the H358 line reduced the following functional bioassays: anchorage-independent cell growth (Fig. 6, A and B), RhoA-GTP level (Fig. 6, C and D), transwell cell migration (Fig. 6, E and F), and xenograft tumors in immunodeficient mice (Fig. 6, G and H). The DLC1-3D mutant was as deficient as the GAP-dead DLC1-R718A mutant in those bioassays, whereas the DLC1-3A mutant was even more active than DLC1-WT.
Figure 6.
DLC1-3D mutant has attenuated tumor-suppressor functions. (A and B) Anchorage-independent growth: DLC1-3D is as defective as GAP-dead DLC1-R718A; DLC1-3A is even more active than DLC1-WT. (A) Photomicrographs of representative agar colonies. Bar, 2 mm. (B) Quantification of agar colonies (>0.4 mm) from three experiments. (C and D) RhoA-GTP of the transfected mutants parallels the results in A and B. (D) Immunoblots for RhoA-GTP and total Rho, as shown in C, were quantified, and the ratio of RhoA-GTP to total Rho was normalized. Parametric two-tailed t tests were performed for statistical analysis. A significant decrease in RhoA-GTP/total Rho in DLC1-WT (P < 0.01) and in DLC1-3A (P < 0.001) compared with GFP control, DLC1-3D, or DLC1-R718A transfected cells. Graph shows relative RhoA-GTP means ± SD from three experiments. (E and F) Cell migration assay: DLC1-3D is as defective as GAP-dead DLC1-R718A; DLC1-3A is similar to DLC1-WT. (E) Photomicrograph of representative migrated cells. Bar, 100 mm. (F) Graph shows transwell cell migration from three experiments. (G and H) Xenograft tumors from mice excised 6 wk after injecting stable transfectants. (G) Photographs of excised tumors. (H) Graph shows tumor weight (g) means ± SD for each group. Parametric two-tailed t tests were performed for statistical analysis. DLC1-3D is as defective for tumor suppression as GAP-dead DLC1-R718A. The tumor suppressor activity of DLC1-3A mutant is even greater than DLC1-WT.
AKT phosphorylation of DLC1 increases binding between the linker region and the RhoGAP domain
Given the location of the three AKT serine phosphorylation sites in the linker region (Fig. 3 A), we speculated their phosphorylation might attenuate the RhoGAP activity of DLC1 by increasing the binding of the linker region to the RhoGAP domain, placing the protein in a closed conformation. Such a mechanism would be the opposite of the one by which CDK5 activates DLC1 (Tripathi et al., 2014).
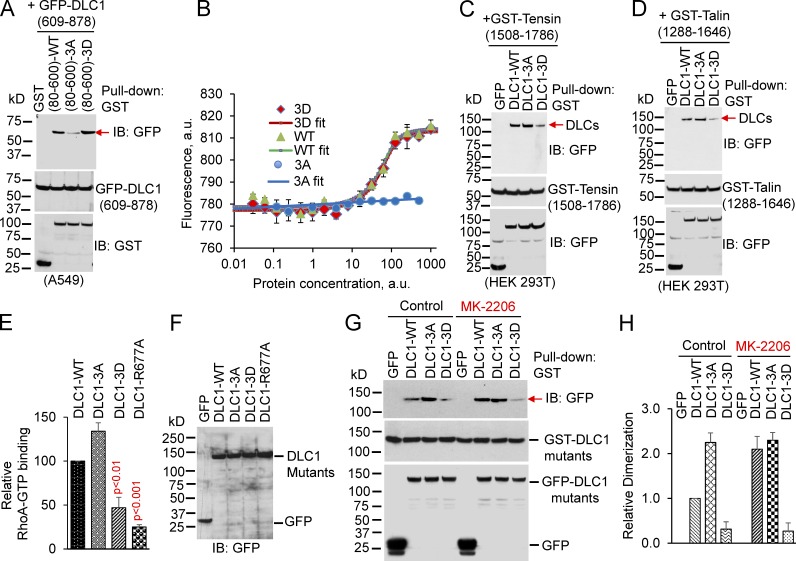
To test that hypothesis, we generated three isogenic versions of a GST-tagged DLC1 linker region encoding amino acids 80–600, which contains the three serines phosphorylatable by AKT: a WT fragment [GST-DLC1 (80–600)–WT], its nonphosphorylatable 3A mutant [GST-DLC1 (80–600)–3A], and its phosphomimetic 3D mutant [GST-DLC1 (80–600)–3D]. A549 cells, which have high AKT activity and are DLC1 negative, were cotransfected with a fragment encoding the GFP-tagged RhoGAP domain [GFP-DLC1 (609–878)] and each of the linker region constructs. Consistent with our hypothesis, GST pull-downs indicated the RhoGAP domain efficiently formed a complex with GST-DLC1 (80–600)–WT and with the GST-DLC1 (80–600)–3D mutant, whereas the GST-DLC1 (80–600)–3A mutant had weaker binding (Fig. 7 A). Binding of the constructs in A549 cells was also analyzed by immobilization-free, microscale thermophoresis (MST), which provides quantitative in vitro measurements of protein interactions in close-to-native conditions. Similar to the GST pull-down assays, binding between the RhoGAP domain and DLC1 (80–600)–WT was similar to that of the phosphomimetic DLC1 (80–600)–3D mutant, whereas the nonphosphorylatable DLC1 (80–600)–3A mutant did not bind the RhoGAP domain even at very high concentrations (Fig. 7 B).
Figure 7.
AKT phosphorylation of the AKT serines in DLC1 reduces several DLC1 functions. (A and B) Phosphorylation increases binding of linker region to RhoGAP domain. (A) GST-tagged N-terminal DLC1 fragment or its indicated mutants were cotransfected with GFP-tagged RhoGAP domain of DLC1 in DLC1-negative A549 cells, which have high AKT activity. The expressed GST-tagged fragments were pulled down with GST antibody, followed by IB with GFP antibody (top). IB with GFP (middle) or GST (bottom) antibodies show expression of DLC1 fragments in whole cell extract (WCE). (B) In vitro binding by MST of DLC1 fragments. The partially purified WT and 3D mutant demonstrate binding to the RhoGAP domain by the s-shape of the binding plot; the 3A mutant shows no binding (flat line). (C and D) Phosphorylation decreases tensin and talin binding to DLC1. In HEK 293T, which have low AKT activity, GST-tensin (1508–1786; C) or GST-talin (1288–1646; D) bind more efficiently to DLC1-WT or DLC1-3A compared with DLC1-3D mutant (top). IB with GST (middle) or GFP (bottom) antibodies show expression of each construct in WCE. (E) DLC1-3D binds RhoA-GTP in vitro less efficiently compared with DLC1-WT or DLC1-3A. DLC1 proteins were partially purified from transfected HEK 293T cells, and bound in vitro to RhoA-GTPγS, a nonhydrolyzable GTP analogue. GAP-dead DLC1-R677A, which has reduced binding in this assay (Jaiswal et al., 2014), was included as a control. Graph shows relative RhoA-GTPγS binding to each DLC1 mutant from three experiments. Parametric two-tailed t tests were performed for statistical analysis. (F) Expression of GFP and GFP-tagged DLC1 mutants for E. (G) In A549 cells, inactive DLC1 is mainly a monomer; AKT inhibition converts DLC1 to an active dimer. Dimerization between DLC1-WT and DLC1-WT, DLC1-3A, or DLC1-3D mutant in the absence (control) or presence of MK-2206. (H) Graph shows relative dimerization of each DLC1 mutant, relative to DLC1-WT, from two experiments. Error bars in all graphs indicate the SD.
AKT phosphorylation of DLC1 decreases the binding of RhoA-GTP and other ligands
The linker region of DLC1 also binds tensin (Liao et al., 2007; Qian et al., 2007) and talin (Li et al., 2011), two ligands implicated in integrin signaling, whose binding contributes to the tumor-suppressor function of DLC1. We speculated the closed conformation induced by AKT phosphorylation might also reduce DLC1 binding of tensin and talin. Indeed, they interacted less efficiently with the phosphomimetic full-length DLC1-3D mutant than they did with DLC1-WT (Fig. 7, C and D) in HEK 293T cells, which have lower AKT activity than A549 cells. The decreased binding was attributable to the closed conformation, rather than to a putative, intrinsic decrease in binding to the linker region because the N-terminal 3D fragment [GST-DLC1 (80–600)–3D] bound tensin and talin as efficiently as the N-terminal WT fragment [GST-DLC1 (80–600)–WT] or the N-terminal 3A fragment [GST-DLC1 (80–600)–3A; Fig. S5, C and D].
The results also suggested the closed conformation of DLC1 would reduce the efficiency of the interaction between the RhoGAP domain and RhoA-GTP. Consistent with that hypothesis, the DLC1-3D mutant, which places the protein in the closed conformation, bound RhoA-GTPγS in vitro almost as weakly as the GAP-dead DLC1-R677A mutant, which is known to have reduced RhoA-GTPγS binding (Fig. 7, E and F; Jaiswal et al., 2014). In contrast, the binding efficiency of RhoA-GTPγS to the DLC1-3A mutant, which is in the open conformation, was at least as strong as that of DLC1-WT.
DLC1 is mainly a monomer in the closed conformation induced by AKT
Because active DLC1 is a dimer (Ko et al., 2013), we asked whether DLC1 might be a monomer when in the closed conformation induced by AKT. To evaluate that possibility, we cotransfected GST-tagged, full-length DLC1 together with GFP-tagged, full-length DLC1 into A549 cells, performed GST pull-downs, and IB the pulled-down proteins for GFP (Fig. 7, G and H). Consistent with the Ko et al. (2013) findings, there was a strong GFP band in cells cotransfected with the DLC1-3A mutant, which has the open conformation. However, the GFP band was much weaker with the DLC1-3D mutant, which is closed, implying most of that mutant protein is monomeric. The intensity of the GFP band with DLC1-WT, which is mainly closed, was reduced compared with DLC1-3A, consistent with the closed conformation being a monomer. When the cells were treated with MK-2206, the intensity of the GFP band from the DLC1-WT transfectants increased, consistent with them becoming active dimers. We conclude that the closed conformation induced by AKT arises via an intramolecular interaction because that form is mainly monomeric.
AKT phosphorylation of DLC1 alters its focal adhesion (FA) localization
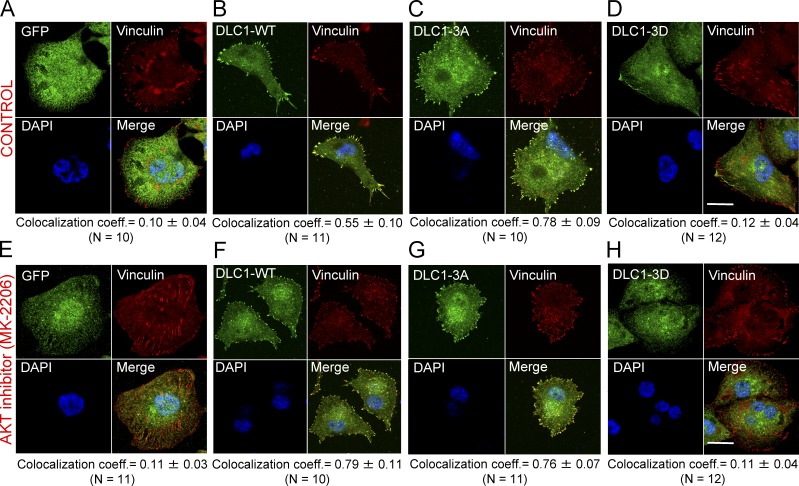
Part of the linker region has been referred to as the FA targeting domain (Liao et al., 2007) because it participates in DLC1 localization to FAs. We used H1703 cells to test the hypothesis that FA localization would be affected by the closed conformation induced by AKT (Fig. 8). Consistent with that prediction, the phosphomimetic DLC1-3D mutant colocalized poorly with the FA protein vinculin (Fig. 8 D; colocalization coefficient = 0.12), whereas the nonphosphorylatable DLC1-3A mutant was strongly colocalized (Fig. 8 C; colocalization coefficient = 0.78). Colocalization was intermediate for the DLC1-WT (Fig. 8 B; colocalization coefficient = 0.55), suggesting that some DLC1-WT is phosphorylated by AKT in H1703, which implies that MK-2206 treatment could further increase its FA colocalization. Consistent with that hypothesis, MK-2206 treatment increased FA colocalization of the DLC1-WT (Fig. 8 F; colocalization coefficient = 0.79) but did not affect colocalization of GFP or the DLC1-3A or DLC1-3D mutants (Fig. 8, E, G, and H).
Figure 8.
The closed conformation of the DLC1-3D mutant affects its subcellular localization. (A–D) Colocalization of GFP-tagged DLC1 constructs (green) with FA protein vinculin (red) in stable H1703 cells. Unlike the GFP control (A), DLC1-WT (B) and DLC1-3A (C) efficiently colocalize to FAs, whereas DLC1-3D (D) and GFP control (A) do not. (E–H) Experimental conditions were similar to A–D, but cells were treated with MK-2206. MK-2206 increases colocalization of DLC1-WT to FAs, but does not influence GFP control, DLC1-3A, or DLC1-3D. Images are representative of most cells. Averaged overlapping colocalization coefficient means ± SD (below each panel) was calculated from ≥10 cells per condition randomly selected from several fields. Bar, 20 µm.
AKT inhibition has antitumor activity in DLC1-positive tumors
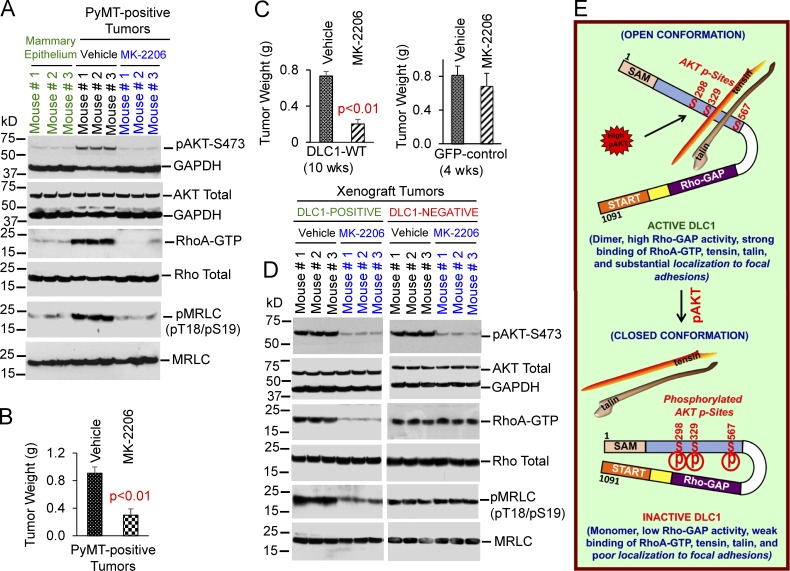
One potential implication of these findings is that inhibition of AKT, a candidate approach for cancer treatment (Nitulescu et al., 2016), could reverse the attenuation of DLC1 activity that depends on AKT. Although AKT has many targets, if DLC1 is a critical one, AKT inhibition might have more potent activity against tumors that express WT DLC1 compared with those that do not. To test that possibility, we evaluated MK-2206 in the MMTV-PyMT transgenic breast cancer model, in which the MMTV promoter drives the polyomavirus middle T antigen, which stimulates AKT (Meili et al., 1998; Summers et al., 1998). First, we confirmed that, compared with mammary epithelial tissue from pregnant mice, MMTV-PyMT tumors have greater AKT activity (Fig. 9 A and Fig. S5 E). Consistent with the cell culture findings that linked AKT, DLC1, and RhoA-GTP, AKT activity in the tumors was associated with higher levels of RhoA-GTP, greater downstream effector phospho-MRLC, and greater phosphorylation of DLC1 serine residues (Fig. 9 A and Fig. S5, E and F). We then treated randomized tumor-bearing mice with oral MK-2206 for 5 d, which induced a threefold reduction in mean tumor weight (Fig. 9 B) and was correlated with decreased AKT activity and reduced RhoA-GTP, phospho-MRLC, and DLC1 serine phosphorylation by AKT (Fig. 9 A and Fig. S6, A and B).
Figure 9.
MK-2206 can reactivate DLC1 and has greater antitumor activity in DLC1-positive tumors than it has in DLC1-negative tumors. (A) Tumors from MMTV-PyMT–positive mice have high AKT activity (lanes 4–6), high RhoA-GTP (lanes 4–6), and high RhoA/ROCK-dependent myosin phosphorylation (pMRLC pT18/pS19; lanes 4–6) compared with mammary epithelium from pregnant mice (lanes 1–3 in the same blot). MK-2206 treatment for 5 d reduced AKT activity, RhoA-GTP, and pMRLC pT18/pS19. Total AKT, Rho, and MRLC were similar in each condition. (B) MK-2206 reduces tumor weight. Graph shows tumor weight means ± SD (C and D) MK-2206 suppresses tumor growth more strongly in DLC1-positive tumors than it does in DLC1-negative tumors. Isogenic H358 stable clones expressing DLC1-WT or GFP control were injected subcutaneously into NOD-SCID mice. When tumors were ∼1.0 cm in diameter (4 wk for GFP- and 10 wk for DLC1-expressing cells), mice were treated with MK-2206 for 5 d. (C) MK-2206 reduced tumor weight by more than 70% in DLC1-positive tumors (left), but <20% in DLC1-negative tumors (right). Parametric two-tailed t tests were performed for statistical analysis. Error bars in the graph indicate the SD. (D) MK-2206 efficiently inhibited AKT activity in DLC1-positive and DLC1-negative tumors. MK-2206 reduced RhoA-GTP– and RhoA-ROCK–dependent myosin phosphorylation in DLC1-positive tumors (left) but not in the DLC1-negative tumors (right). (E) Model for regulation of DLC1 by AKT. Top part shows that, in the absence of serine phosphorylation (S298, S329, and S567) by AKT, DLC1 has an open conformation with high RhoGAP activity; forms dimers; strongly binds RhoA-GTP, tensin, and talin; and colocalizes to FAs. Bottom part shows that, in the presence of serine phosphorylation by AKT, DLC1 has a closed conformation with low RhoGAP activity; is mainly a monomer; weakly binds RhoA-GTP, tensin, and talin; and poorly colocalizes to FAs.
We speculated the DLC1 reactivation from MK-2206 treatment had contributed to the observed antitumor activity. To evaluate that hypothesis more directly, we used two isogenic tumor cell lines, from H358, which differed only in their DLC1 expression (GFP versus GFP-DLC1-WT), and evaluated the ability of MK-2206 to affect their growth in agar and tumor growth in nonobese diabetic–severe combined immunodeficiency (NOD-SCID) mice. In both bioassays, MK-2206 potently reduced the growth of GFP-DLC1-WT cells (Fig. 9 C, for mouse tumors; and Fig. S6, C and D, for agar growth), whereas its effects were less pronounced in GFP control cells. These results were correlated with MK-2206 reducing RhoA-GTP and its RhoA-dependent signaling (pMRLC level) in the GFP-DLC1-WT cells but not in the GFP control cells, although the AKT activity was similarly reduced in both isogenic lines (Fig. 9 D). A separate experiment, which included H358 cells stably expressing DLC1-3A and DLC1-3D in addition to those expressing DLC1-WT, confirmed the specificity of the Fig. 9 C results. MK-2206 treatment for 5 d decreased the size of DLC1-WT tumors by 69%, compared with decreases of 18–20% for tumors expressing DLC1-3A, DLC1-3D, or vector control (Fig. S6, E and F).
Discussion
This study identified increased RhoA-GTP by RTK ligands (EGF, IGF-1, and insulin) as a previously unrecognized, but common, physiological determinant of RhoA activation. We found the kinase activity of AKT, which is stimulated by the cognate RTKs these ligands activate, is required for the increased RhoA-GTP. DLC1, a tumor-suppressor gene that encodes a RhoGAP, is the key downstream target for the AKT kinase in this phenomenon, which also occurs in tumor cell lines that express DLC1 but not in those with down-regulated DLC1.
In DLC1-positive cells, AKT phosphorylated three serines in DLC1 (S298, S329, and S567, the AKT serines), which greatly attenuated its RhoGAP and tumor-suppressor activities in several bioassays. Analysis of single, double, and triple serine-to-alanine (nonphosphorylatable) and serine-to-aspartate (phosphomimetic) mutants of the three AKT serines indicated that each phosphorylation contributed to the reduction in those activities, suggesting an electrostatic mechanism. In cells expressing WT DLC1, AKT inhibition by MK-2206 greatly reduced serine phosphorylation of DLC1 and RhoA-GTP, specific changes that were not seen in cells expressing either of the triple mutants of the AKT serines treated with MK-2206. In contrast to the RTK ligands, increased RhoA-GTP induced by LPA, which is mediated by G protein–coupled receptors, occurs by a distinct mechanism that is not associated with AKT activation or DLC1.
A notable feature of the AKT serines in DLC1 is that they are located in the linker region of DLC1, rather than in the RhoGAP domain, which is necessary and sufficient for the RhoGAP activity of DLC1 (Healy et al., 2008). To explore the mechanism by which those phosphorylations outside the RhoGAP domain can reduce the RhoGAP activity, we studied complex formation between the RhoGAP domain and WT or serine mutants of the linker region after their cotransfection in cells and in vitro by immobilization-free MST to compare relative binding efficiencies of those polypeptides. The data led us to conclude the phosphorylations of the AKT serines induce strong binding between the linker region and the RhoGAP domain, which places DLC1 in a closed, inactive conformation that is mainly monomeric (Fig. 9 E) and reduces RhoA-GTP binding to DLC1, tensin and talin binding to DLC1, and DLC1 colocalization to FAs. That mechanism is essentially the opposite of what we found from phosphorylation of four serines in the DLC1 linker region by CDK5 (Tripathi et al., 2014), which placed DLC1 in an open, active conformation by decreasing an autoinhibitory interaction between the linker region and the RhoGAP domain.
Our results differ from, and extend in important ways, previous studies that DLC1 is an AKT substrate. Hers et al. (2006) found that insulin induces phosphorylation in the rat version of DLC1 at S322 (which corresponds to S329 in human DLC1) and showed that phosphorylation was mediated by AKT, but they did not examine the possible downstream consequences. Ko et al. (2010) made the important observation that AKT reduces the tumor-suppressor activity of DLC1 and concluded that phosphorylation of S567 by AKT was responsible for that reduction because they did not detect phosphorylation of S298 or S329. Under our growth conditions, however, all three AKT serines are phosphorylated in cells, as documented with the DLC1 mutants and mass spectrometry results. Consistent with these findings, our analysis of phosphoproteomic data in the National Cancer Institute’s Clinical Proteomic Tumor Analysis Consortium (CPTAC) database indicates a significant correlation in human breast cancer (Mertins et al., 2016) between AKT activity, as measured by phosphorylation of AKT T308 and S473, and phosphorylation of DLC1 S298 and S329 (P = 0.03; the CPTAC screen did not identify the peptide with the S567 residue).
Ko et al. (2010) concluded the reduced tumor-suppressor activity induced by AKT was not associated with a decrease in the RhoGAP activity of DLC1 because they did not detect an increase in RhoA-GTP. However, we developed strong evidence that AKT-dependent phosphorylation of DLC1 is associated with an increase in RhoA-GTP in nontransformed lines, tumor-derived lines, and the MMTV-PyMT transgenic breast cancer model. One possible way to reconcile the RhoA-GTP conclusions of Ko et al. (2010) with ours is that they focused on the RhoA-GTP levels of the single DLC1-S567D mutant; in agreement with their results, we found the RhoGAP activity in that mutant was close to that of WT DLC1. In our experiments, the serine-to-aspartate mutation of at least two of the three AKT serines was needed to detect a decrease in RhoGAP activity.
EGF treatment of some cell lines has been reported to have somewhat different effects on DLC1. Cao et al. (2012) reported that EGF can activate DLC1 in MCF10A breast epithelial cells via a mechanism that includes replacement of tensin-3 binding to DLC1 with CTEN (C-terminal tensin-like) binding, a process that takes much longer than the one described here. MCF10A is the only nontransformed line we have examined in which endogenous DLC1 is inactive (Tripathi et al., 2014), so it may not be surprising that it behaves differently. Ravi et al. (2015) examined the effects of EGF on a DLC1-negative, HeLa-derived cancer line. In agreement with our data, EGF treatment of the parental line had no effect on RhoA-GTP. However, they found that, although transient transfection with WT DLC1 RhoGAP did not alter RhoA-GTP, EGF treatment of those transfectants resulted in reduced RhoA-GTP, in contrast to the increased RhoA-GTP we observed for endogenous DLC1 and the stable transfectants in several cell lines we tested. The two-step mechanism that they concluded was responsible for their observation is unrelated to the one described here.
Our observations may have therapeutic implications in cancer. The AKT-induced attenuation of the tumor-suppressor function of DLC1 is potentially reversible because AKT inhibition can decrease phosphorylation of the three AKT serines and reactivate DLC1. In the transgenic MMTV-PyMT cancer model, which we determined has constitutive activation of AKT and high RhoA-GTP, pharmacologic AKT inhibition by MK-2206 decreased phosphorylation of the AKT serines in DLC1, decreased RhoA-GTP, and reduced tumor size. We extended those results by testing an isogenic version of a tumor line with constitutively active AKT that either expressed DLC1 or did not express DLC1. Similar to the MMTV-PyMT, MK-2206 reduced RhoA-GTP and the size of xenograft tumors from the DLC1-positive subline. However, in the DLC1-negative subline, AKT inhibition by MK-2206 did not change RhoA-GTP and had only marginal antitumor activity. The results strongly imply the antitumor activity observed in the DLC1-positive line was attributable, at least in part, to reactivation of DLC1 by MK-2206.
These findings highlight the potential utility of monitoring the reactivation of tumor-suppressor genes when inhibiting an oncoprotein, such as AKT, which has a clinical potential that is still being explored (Nitulescu et al., 2016). Such reactivation may also be relevant for inhibition of other oncoproteins that inactivate tumor-suppressor genes indirectly or directly, as occurs with CDK4/6 and pRB via phosphorylation (Hamilton and Infante, 2016) or MDM2 and p53 via the ubiquitin system (Wasylishen and Lozano, 2016).
Materials and methods
Plasmid constructs
GFP-tagged DLC1 WT (GFP-DLC1-WT); DLC1 GAP-dead mutant (GFP-DLC1-R718A); GFP-tagged DLC1 fragments encoding DLC1 residues 1–492, 500–1091, 1–110, 80–200, 80–300, 80–400, 400–500, 609–850, 850–1091, and 609–878; and GFP-DLC3 were constructed by PCR and subcloned into a modified pEGFP-C1 vector (Takara Bio Inc.) through Kpn1-NotI sites, as described (Qian et al., 2007). GST-tagged tensin and talin fragments were described previously (Qian et al., 2007; Li et al., 2011). GFP-DLC2 was a gift from M. Mowat (Manitoba Institute of Cell Biology, Winnipeg, Manitoba, Canada). A series of individual and combined serine-to-Alanine (S-to-A) and serine-to-aspartate (S-to-D) mutations were introduced into full-length DLC1-WT and into DLC1 fragments encoding residues 80–600, using a site-directed mutagenesis kit (Agilent Technologies). Table S1 lists the primers used. Full-length DLC1 and DLC1 (80–600) linker region fragments, with or without 3A or 3D mutations, were engineered into the PEBG vector with BamHI and NotI, resulting in GST-tagged DLC1 constructs. All PCR regions were confirmed by sequencing.
Antibodies and fluorescent probes
The following antibodies were purchased from Cell Signaling Technology: AKT mouse (2920), AKT rabbit (4691), phospho-AKT-pS473 rabbit (4060) and phospho-AKT-pT308 rabbit (13038), phospho-Akt substrate (RXXS*/T*) rabbit (9614), phosphorylated Thr18/Ser19 pMRLC rabbit (3674), and GAPDH (2118) rabbit. Two DLC1 antibodies, which gave similar results, were used: one, generated in our laboratory (DLC1 antibody; clone 428; 24), and the other, DLC1 mouse mAb (612021), purchased from BD. pEGFR-Y845 rabbit (ab5636), MRLC mouse (ab11082), GFP mouse (ab1218), and GFP rabbit (ab290) antibodies were purchased from Abcam. RhoA mouse (ARH04) and pMRLC goat (sc-12896) antibodies were obtained from Cytoskeleton, Inc., and Santa Cruz Biotechnology, Inc., respectively. The phospho-serine mouse (612547) antibody was purchased from BD. Anti–rabbit and anti–mouse IgG HRP-linked secondary antibodies were obtained from GE Healthcare. Alexa Fluor 568 anti–rabbit IgG, Alexa Fluor 488 anti–mouse IgG, Alexa Fluor 488 phalloidin, and DAPI were purchased from Thermo Fisher Scientific.
Cell lines, culture conditions, and DNA transfection
HEK 293T, human skin epithelial H2071, human lens epithelial FHL124, and human fibroblastic H1634 cells were cultured in DMEM supplemented with 10% FBS. Human breast cancer lines (BT549, MCF10A1Ca1h, MDA-MB-468, and T47D) and NSCLC lines (H1703, H157, A549, and H358, provided by C. Harris, National Cancer Institute, Bethesda, MD) were cultured in RPMI-1640 supplemented with 10% FBS. Transient transfections were performed with Lipofectamine 3000 (Thermo Fisher Scientific) and cultured for 48 h. Stable clones expressing GFP or DLC1 mutants were made by transfecting H1703 or H358 cells with Lipofectamine 3000, followed by G418 selection (0.9 µg/ml).
siRNA transfection and treatment of cells with EGF, insulin, IGF-1, LPA, and AKT-inhibitor MK-2206
To suppress DLC1 expression, cells were transfected with 160 nM of DLC1 siRNAs or with scrambled control siRNAs, and harvested 48 h later. Suppression of protein expression, with two different siRNAs, was confirmed by immunoblotting. Validated siRNAs for human DLC1 (Hs_DLC1 siRNA_5, SI03219909, and Hs_DLC1 siRNA_11, SI04952213) were from QIAGEN, as were negative control siRNAs (control siRNA 1, 1027280; and control siRNA 2, 1027310). The sequence for each DLC1 siRNA was as follows: Hs_DLC1 siRNA _5 sense sequence: 5′-CGAUGUCGUAAUUCCUAUATT-3′; Hs_DLC1_5 antisense sequence: 3′-CGGCUACAGCAUUAAGGAUAU-5′; Hs_DLC1_11 sense sequence: 5′-GGAGUGUAGGAAUUGACUATT-3′; Hs_DLC1_11 antisense sequence: 3′-gaCCUCACAUCCUUAACUGAU-5′.
The final concentration of ligands was as follow: EGF (100 ng/ml), insulin (100 nM), IGF-1 (100 ng/ml), and LPA (1.0 µM). All four ligands were procured from Sigma-Aldrich. AKT inhibitor MK-2206 (used at 10 µM) was from Selleck Chemicals. After overnight incubation in serum-free medium, cells were treated with the indicated ligands or inhibitors for 15–120 min.
In vitro AKT kinase assay
Lysates from transfected cells were IP with GFP antibody, and immunopellets were sequentially washed once with high-salt HNTG buffer (20 mM Hepes, 500 mM NaCl, 0.1% Triton X-100, and 10% glycerol), twice with low-salt HNTG buffer (20 mM Hepes, 150 mM NaCl, 0.1% Triton X-100, and 10% glycerol), and once with kinase reaction buffer (35 mM Hepes, pH 7.4, 10 mM MgCl2, 1 mM EGTA, 1% Tween 20, 0.1 mM sodium vanadate, and 1 mM DTT). The kinase reaction was performed in 30 µl of reaction buffer containing 15 µM cold ATP, 2.5 µCi γ-[32P]ATP, and 100 ng of recombinant active AKT (EMD Millipore) at 30°C for 45 min. The reaction was stopped by adding 10 µl of 4× Laemmli sample buffer and heating at 95°C for 5 min. Proteins were separated by gel electrophoresis and autoradiographed to detect 32P incorporation.
RhoA-GTP (Rhotekin-Rho binding domain pull-down) assay
A Rho activation assay kit (EMD Millipore) was used to measure GTP-bound RhoA, as described previously (Tripathi et al., 2014). In brief, equal amounts (1,000 µg) of each cell lysate were incubated with 30 µg GST-Rhotekin Rho-binding domain coupled to glutathione-agarose beads for 45 min. Beads were washed three times with washing buffer, samples were subjected to 4–12% SDS-PAGE, transferred onto nitrocellulose membranes (Thermo Fisher Scientific), and detected by IB, using RhoA antibody (ARH04 from Cytoskeleton, Inc., and 05–778 from EMD Millipore; clone 55).
ROCK (Rho kinase) assay
Cells were fixed and harvested in 10% TCA containing 10 mM DDT. Pellets were dissolved in 10 µl of 1 M Tris base and mixed with 100 µl of extraction buffer (8 M urea, 2% SDS, 5% sucrose, and 5% 2-mercaptoethanol). Equal amounts of protein from each cell extract were subjected to 10% SDS-PAGE, transferred onto nitrocellulose membranes, and incubated with an antibody specific for phospho-myosin binding subunit (phospho-Thr853-MYPT1) or myosin binding subunit (MYPT1), and bands were visualized by enhanced chemiluminescence. ROCK activity was expressed as the ratio of phospho-MYPT1 to total MYPT1.
RhoGAP activity assay
GFP-tagged DLC1 constructs were purified by IP using GFP rabbit polyclonal (ab290) antibody from transfected cells using a high-stringency buffer (20 mM Tris-HCl, pH 8.0, 100 mM NaCl, 5 mM MgCl2, NP-40 [0.5%], 1 mM DTT, and protease and phosphatase inhibitor). Highly purified Rho was bound to γ-labeled [P32]GTP. The GTPase-accelerating activity (RhoGAP activity) of various DLC1 mutants at each time point was determined by incubating with GTP-bound RhoA at 18°C with shaking and removing samples at the indicated time points. The guanidine nucleotides were separated by chromatography on cellulose filter paper dissolved in buffer. The γ-P32 signal was then determined, and the net GTP hydrolysis was calculated.
In vivo pull-down assay, coIP, and immunoblotting
Cells were transiently cotransfected with plasmids expressing GST or the indicated GST fusion constructs together with GFP or the indicated GFP-DLC1 constructs. 48 h after transfection, cells were lysed with golden lysis buffer. The cleared supernatants were collected, and a small portion of supernatants was taken to determine the protein concentration using the DC protein assay (Bio-Rad Laboratories). For the pull-down assay, 1.0 mg of total protein from each cell extract was used, to which 30 µl of glutathione sepharose-4B slurry (GE Healthcare) was added, with continuous rotation for 3 h at 4°C. The pellets were sequentially washed once with golden lysis buffer, once with high-salt HNTG buffer, and twice with low-salt HNTG buffer. The beads were incubated with 30 µl Laemmli sample buffer, subjected to 10% SDS-PAGE, transferred onto nitrocellulose membranes (Thermo Fisher Scientific), and detected by IB using specific antibodies. A portion of the cell extracts was used as a loading control to verify expression of the GFP fusion proteins and the GPF control. For coIP experiments, equal amounts of protein from each cell lysate were precleared with protein G slurry (Thermo Fisher Scientific) and incubated with the indicated antibodies or control IgG for 1 h at RT. After incubation, 30 µl of protein G slurry was added to each immune reaction and rotated at 4°C overnight. The immunopellets were washed three times with IP buffer. Coimmunoprecipitated proteins were eluted by boiling for 5 min in 30 µl Laemmli sample buffer containing 5% (vol/vol) 2-mercaptoethanol. Eluted proteins were resolved on a NuPage 4–12% BisTris gel and detected by immunoblotting using specific antibodies. Immunoreactive bands were detected by enhanced chemiluminescence (ECL-Plus; GE Healthcare) using HRP-linked anti–rabbit or anti–mouse secondary antibodies (1:5,000 dilutions).
MST
Binding efficiency of DLC1 N terminus to its RhoGAP domain was studied by MST (Khavrutskii et al., 2013; Seidel et al., 2013) using a Monolith NT.115 instrument (NanoTemper). Serial 1:1 dilutions of the DLC1 N-terminal fragment (80–600) WT or its combined 3A or 3D mutant versions were mixed with each aliquot and a fixed volume of fluorescent GFP-tagged RhoGAP domain [GFP-DLC1 (609–878)], the resulting probes were placed in glass capillaries (NanoTemper), and assayed with “blue” light-emitting diode excitation (excitation, 460–480 nm, and emission, 515–530 nm), observing the fluorescence signal upon start of illumination with a pin-point infrared laser (1,480 nm). The MST plots for each capillary set were analyzed by NT.Analysis 1.5.41 software (NanoTemper), providing fluorescence intensity versus protein concentration plots for each binding pair. The inflection point of those plots corresponds to the dissociation constant of each interaction.
Immunofluorescent staining
Transiently or stably transfected cells were seeded onto glass chambers, incubated for 24 h, and fixed with 4% PFA for 20 min. Fixed cells were permeabilized with 0.25% Triton X-100 in PBS and then blocked with 3% BSA in PBS for 2 h. The cells were incubated with a 1:200 dilution of the indicated primary antibodies at 4°C overnight. After being thoroughly washed in PBS, the cells were incubated with the appropriate 1:250 Alexa Fluor–conjugated secondary antibodies for 1 h. To visualize actin or nuclei, cells were incubated with phalloidin (1:50) or DAPI (1:2,500) for 1 h. After staining, the cells were thoroughly washed with PBS and mounted with gel-mounting solution (BIOMEDA).
Fluorescent confocal microscopy
Confocal microscopy of fluorescent-labeled cells was performed with a microscope (LSM 780; ZEISS) with an excitation wavelength of 488 nm to detect transfected GFP fusion proteins. Alexa Fluor probes were viewed with excitation wavelengths of 488 (Alexa Fluor 488) and 568 (Alexa Fluor 568) nm. Images were made at RT using photomultiplier tubes with a Plan-Apochromat 63×/1.4 NA oil-differential, interference-contrast, objective lens with a 2× magnifier to produce a 125× magnification. The colocalization of two proteins was analyzed by confocal software (ZEN 2012; ZEISS). For quantification of representative morphology in each group, ∼15 cells per condition, randomly selected from several fields, were analyzed. The Mander’s overlapping colocalization coefficient means ± SD were calculated and are shown below each figure panel. The overlapping colocalization coefficients can range from 0 to 1, where 0 means no colocalization and 1 means full colocalization of the two proteins. The images were minimally processed for levels/contrast adjustment in DAPI panels, and the adjustment was performed on entire images using Photoshop CC software (Adobe). The adjustments do not enhance, erase, or misrepresent any information present in the original images.
PLA
PLA was used to visualize proximity colocalization (<40 nm) of DLC1 and AKT in NSCLC lines using the Duolink Detection kit (Olink Proteomics). The cells were fixed with 4% PFA for 20 min at RT and then incubated with 0.25% Triton-X-100 for 5 min. After blocking with 3% BSA, cells were incubated overnight at 4°C with rabbit anti–DLC1 (1:100) plus mouse anti–AKT (1:500) antibodies. After washing, cells were incubated with secondary antibodies with PLA probes (MINUS probe–conjugated anti–rabbit IgGplus PLUS probe–conjugated anti–mouse IgG (Olink Proteomics). Circularization and ligation of the oligonucleotides in the probes were followed by an amplification step. A complementary fluorescent-labeled probe was used to detect the product of the rolling-circle amplification. Slides were mounted with Duolink II mounting medium containing DAPI. Images were obtained with an LSM 780 (ZEISS) using ZEN software (ZEISS). The colocalization PLA dots in cells were counted using ImageJ software (National Institutes of Health). Quantifications are given as means ± SD.
Mass spectrometry analysis
Lysates from HEK 293T cells expressing GFP-tagged DLC1 constructs, either untreated or treated with the MK-2206 AKT inhibitor, were IP with GFP antibody. The immunopellets were resolved on a NuPage 4–12% BisTris gel. The Coomassie-stained DLC1 gel band was destained, and proteins were reduced, alkylated, and digested with trypsin or LysC as described (Shevchenko et al., 2006). Digested peptides were further desalted using StageTip C18 columns (Rappsilber et al., 2007) and were analyzed on a Q-Exactive instrument (Thermo Fisher Scientific) equipped with a Proxeon EASY-nLC 1,000 UHPLC System. Liquid chromatography–tandem mass spectrometry data were searched against a human Refseq database using MaxQuant v1.3.0.5 (Cox and Mann, 2008; Cox et al., 2011) with carbamidomethylation as a fixed modification and the following variable modifications: oxidation of methionine, acetylation of protein N termini, deamidation of asparagine, and phosphorylation of serine, threonine, and tyrosine residues. For peptide identification, we applied a 1% false-discovery rate using a target–decoy search strategy (Elias and Gygi, 2010).
Cell migration assay
Cell migration was measured by 6.5-mm-diam Falcon cell culture inserts (8 µm pore size; BD). Transiently transfected cells or stable clones were trypsinized, resuspended in serum-free RPMI-1640 medium, and transferred to the upper chamber (7.5 × 104 cells in 300 µl). 600 µl of 10% FBS in RPMI-1640 was placed in the lower chamber. After 18 h incubation, the cells remaining on the upper surface of the insert were removed five times with a cotton swab moistened in PBS. Migrated cells on the lower surface were fixed in methanol for 20 min at RT, followed by staining with 2% crystal violet (Sigma-Aldrich) in methanol for 30 min, destained, examined, and photographed by microscopy. For quantification, migrated cells were solubilized with 1% Triton X-100 and counted in a spectrophotometer at an OD of 590 nm.
Soft agar and anchorage-independent growth assay
For soft agar assays, a 0.6% agar (BD) base in RPMI-1640 medium was placed in 60-mm dishes for 1 h at RT. 1.0 × 105 cells were mixed with complete medium containing 0.4% agar and placed over 0.6% basal agar in 60-mm dishes. Cells were grown for 3 wk, and colonies were photographed microscopically and quantified with a colony counter. For clonogenic assays, 0.6 × 105 cells were seeded in six-well plates and cultured in 0.9 µg/ml G418 RPMI-1640 medium with 10% FBS for 3 wk. Colonies were fixed, stained with 4% crystal violet, and counted.
In vivo tumorigenesis and treatment of mice with AKT inhibitor MK-2206
The mouse studies were approved by the National Cancer Institute Animal Care and Use Committee and were conducted in compliance with the approved protocols. For the tumor xenograft, H358 stable clones expressing GFP, GFP-DLC1-WT, GFP-DLC1-3A, GFP-DLC1-3D, and GFP-DLC1-R718A were trypsinized, washed with cold PBS, diluted to 108 cells/ml with serum-free medium/Matrigel basement membrane matrix (BD) at a ratio of 3:1, and injected subcutaneously into NOD-SCID mice (1.0 × 107 cells/injection). The animals were monitored for tumor growth, and tumor masses were weighed (in grams) 6 wk after injection.
For mice with xenograft tumors, H358 stable clones expressing various GFP-tagged DLC1 constructs were trypsinized, washed with cold PBS, diluted to 108/ml with serum-free medium/Matrigel basement membrane matrix (BD) at a ratio of 3:1, and injected subcutaneously into NOD-SCID mice (1.0 × 107 cells/injection). When tumors were 0.5–1.0 cm, mice were treated with MK-2206 or vehicle control for 5 d, and the remaining tumor tissues were then excised, weighed, and processed for biochemical assays. For MMTV-PyMT mice, tumors were randomly divided into two groups. Mice were treated orally with 50 mg/kg MK-2206 AKT inhibitor for five consecutive days or the vehicle control. The remaining tumor tissues were then excised, weighed, and processed for biochemical assays.
Data analysis
At least two independent experiments were performed for all in vitro analysis. Immunoblots were quantified by densitometric scanning using ImageQuant software. Results are expressed as density means ± SD from two or three experiments. All experiments were designed with matched control conditions within each experiment. Data distribution was assumed normal, but that was not tested formally. For statistical analysis, parametric two-tailed t tests were performed using PRISM software (version 7.0a; GraphPad Software), and P < 0.05 was considered statistically significant.
Online supplemental material
Fig. S1 shows that EGF-induced AKT activity, which increases RhoA-GTP through DLC1 and AKT, regulates all three DLC family members. Fig. S2 shows that insulin and ILGF-1 also induce AKT activity and increase RhoA-GTP through DLC1. However, LPA induces RhoA-GTP in a DLC1-independent manner and does not activate AKT. Fig. S3 shows DLC1 and AKT form a protein complex in human cell lines, and the complex formation does not require enzymatic activity of AKT. Fig. S4 shows all three members of the DLC gene family, DLC1, DLC2, and DLC3, are AKT substrates. The phosphorylation of the three AKT serines in DLC1 (S298, S329, and S567) was confirmed by mass spectrometry. Fig. S5 shows that the AKT phenotype was dominant over the CDK5 phenotype. PyMT-positive tumors have high AKT activity, high RhoA-GTP, and high MRLC phosphorylation. Fig. S6 shows that the MMTV-PyMT–positive tumors have high AKT activity, high RhoA-GTP, and high DLC1 phosphorylation. Treatment with MK-2206 inhibits colony formation, growth in soft agar, and tumor formation in mice of DLC1-WT transfected cells only, but does not alter anchorage-independent growth or tumor formation by cells transfected with the GFP control, DLC1-3A, DLC1-3D, or DLC1-R718A.
Supplementary Material
Acknowledgments
We thank Marian Durkin for critically reading the manuscript, the National Cancer Institute Center for Cancer Research Imaging Core Facility for confocal microscopy, Michael Mowat for the GFP-DLC2 construct, and Curt Harris for H1703 and H358 cell lines.
This research was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, and Center for Cancer Research.
The authors declare no competing financial interests.
Author contributions: B.K. Tripathi and D.R. Lowy conceived the project, designed the experiments, and analyzed the data. B.K. Tripathi performed most of the experiments with the help of T. Grant; X. Qian engineered the DLC1 plasmid constructs. M. Zhou, P. Mertins, and S.A. Carr performed mass spectrometric analyses. D. Wang performed bioinformatics analyses of proteomics data. S.G. Tarasov helped with the MST assay, and A.G. Papageorge helped with in vitro RhoA-GTP binding assay. K.W. Hunter provided the MMTV-PyMT transgenic mice. B.K. Tripathi and D.R. Lowy wrote the manuscript with input from all authors.
Footnotes
Abbreviations used:
- coIP
- coimmunoprecipitation
- EGFR
- EGF receptor
- FA
- focal adhesion
- HEK
- human embryonic kidney
- IB
- immunoblotted
- IGF
- insulin-like growth factor
- IP
- immunoprecipitated
- LPA
- lysophosphatidic acid
- MST
- microscale thermophoresis
- NOD-SCID
- nonobese diabetic–severe combined immunodeficiency
- NSCLC
- non–small cell lung cancer
- PLA
- proximity ligation assay
- pMRLC
- phosphorylation of myosin regulatory light chain
- Rho-GEF
- Rho guanine nucleotide exchange factor
- RhoGAP
- Rho GTPase-activating protein
- RTK
- receptor tyrosine kinase
References
- Braun A.C., and Olayioye M.A.. 2015. Rho regulation: DLC proteins in space and time. Cell. Signal. 27:1643–1651. 10.1016/j.cellsig.2015.04.003 [DOI] [PubMed] [Google Scholar]
- Cao X., Voss C., Zhao B., Kaneko T., and Li S.S.. 2012. Differential regulation of the activity of deleted in liver cancer 1 (DLC1) by tensins controls cell migration and transformation. Proc. Natl. Acad. Sci. USA. 109:1455–1460. (published erratum appears in Proc. Natl. Acad. Sci. USA. 2012. 109:4708) 10.1073/pnas.1114368109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J., and Mann M.. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26:1367–1372. 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- Cox J., Neuhauser N., Michalski A., Scheltema R.A., Olsen J.V., and Mann M.. 2011. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10:1794–1805. 10.1021/pr101065j [DOI] [PubMed] [Google Scholar]
- Durkin M.E., Avner M.R., Huh C.G., Yuan B.Z., Thorgeirsson S.S., and Popescu N.C.. 2005. DLC-1, a Rho GTPase-activating protein with tumor suppressor function, is essential for embryonic development. FEBS Lett. 579:1191–1196. 10.1016/j.febslet.2004.12.090 [DOI] [PubMed] [Google Scholar]
- Elias J.E., and Gygi S.P.. 2010. Target-decoy search strategy for mass spectrometry-based proteomics. Methods Mol. Biol. 604:55–71. 10.1007/978-1-60761-444-9_5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garay C., Judge G., Lucarelli S., Bautista S., Pandey R., Singh T., and Antonescu C.N.. 2015. Epidermal growth factor-stimulated Akt phosphorylation requires clathrin or ErbB2 but not receptor endocytosis. Mol. Biol. Cell. 26:3504–3519. 10.1091/mbc.E14-09-1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Mata R., Boulter E., and Burridge K.. 2011. The ‘invisible hand’: regulation of RHO GTPases by RHOGDIs. Nat. Rev. Mol. Cell Biol. 12:493–504. 10.1038/nrm3153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton E., and Infante J.R.. 2016. Targeting CDK4/6 in patients with cancer. Cancer Treat. Rev. 45:129–138. 10.1016/j.ctrv.2016.03.002 [DOI] [PubMed] [Google Scholar]
- Healy K.D., Hodgson L., Kim T.Y., Shutes A., Maddileti S., Juliano R.L., Hahn K.M., Harden T.K., Bang Y.J., and Der C.J.. 2008. DLC-1 suppresses non-small cell lung cancer growth and invasion by RhoGAP-dependent and independent mechanisms. Mol. Carcinog. 47:326–337. 10.1002/mc.20389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hers I., Wherlock M., Homma Y., Yagisawa H., and Tavaré J.M.. 2006. Identification of p122RhoGAP (deleted in liver cancer-1) Serine 322 as a substrate for protein kinase B and ribosomal S6 kinase in insulin-stimulated cells. J. Biol. Chem. 281:4762–4770. 10.1074/jbc.M511008200 [DOI] [PubMed] [Google Scholar]
- Hirai H., Sootome H., Nakatsuru Y., Miyama K., Taguchi S., Tsujioka K., Ueno Y., Hatch H., Majumder P.K., Pan B.S., and Kotani H.. 2010. MK-2206, an allosteric Akt inhibitor, enhances antitumor efficacy by standard chemotherapeutic agents or molecular targeted drugs in vitro and in vivo. Mol. Cancer Ther. 9:1956–1967. 10.1158/1535-7163.MCT-09-1012 [DOI] [PubMed] [Google Scholar]
- Jaiswal M., Dvorsky R., Amin E., Risse S.L., Fansa E.K., Zhang S.C., Taha M.S., Gauhar A.R., Nakhaei-Rad S., Kordes C., et al. 2014. Functional cross-talk between ras and rho pathways: a Ras-specific GTPase-activating protein (p120RasGAP) competitively inhibits the RhoGAP activity of deleted in liver cancer (DLC) tumor suppressor by masking the catalytic arginine finger. J. Biol. Chem. 289:6839–6849. 10.1074/jbc.M113.527655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khavrutskii L., Yeh J., Timofeeva O., Tarasov S.G., Pritt S., Stefanisko K., and Tarasova N.. 2013. Protein purification-free method of binding affinity determination by microscale thermophoresis. J. Vis. Exp. 78:50541 10.3791/50541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko F.C., Chan L.K., Tung E.K., Lowe S.W., Ng I.O., and Yam J.W.. 2010. Akt phosphorylation of deleted in liver cancer 1 abrogates its suppression of liver cancer tumorigenesis and metastasis. Gastroenterology. 139:1397–1407. 10.1053/j.gastro.2010.06.051 [DOI] [PubMed] [Google Scholar]
- Ko F.C., Chan L.K., Sze K.M., Yeung Y.S., Tse E.Y., Lu P., Yu M.H., Ng I.O., and Yam J.W.. 2013. PKA-induced dimerization of the RhoGAP DLC1 promotes its inhibition of tumorigenesis and metastasis. Nat. Commun. 4:1618 10.1038/ncomms2604 [DOI] [PubMed] [Google Scholar]
- Li G., Du X., Vass W.C., Papageorge A.G., Lowy D.R., and Qian X.. 2011. Full activity of the deleted in liver cancer 1 (DLC1) tumor suppressor depends on an LD-like motif that binds talin and focal adhesion kinase (FAK). Proc. Natl. Acad. Sci. USA. 108:17129–17134. 10.1073/pnas.1112122108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y.C., Si L., deVere White R.W., and Lo S.H.. 2007. The phosphotyrosine-independent interaction of DLC-1 and the SH2 domain of cten regulates focal adhesion localization and growth suppression activity of DLC-1. J. Cell Biol. 176:43–49. 10.1083/jcb.200608015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loirand G. 2015. Rho Kinases in Health and Disease: From Basic Science to Translational Research. Pharmacol. Rev. 67:1074–1095. 10.1124/pr.115.010595 [DOI] [PubMed] [Google Scholar]
- Lukasik D., Wilczek E., Wasiutynski A., and Gornicka B.. 2011. Deleted in liver cancer protein family in human malignancies (Review). Oncol. Lett. 2:763–768. 10.3892/ol.2011.345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marjoram R.J., Lessey E.C., and Burridge K.. 2014. Regulation of RhoA activity by adhesion molecules and mechanotransduction. Curr. Mol. Med. 14:199–208. 10.2174/1566524014666140128104541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meili R., Cron P., Hemmings B.A., and Ballmer-Hofer K.. 1998. Protein kinase B/Akt is activated by polyomavirus middle-T antigen via a phosphatidylinositol 3-kinase-dependent mechanism. Oncogene. 16:903–907. 10.1038/sj.onc.1201605 [DOI] [PubMed] [Google Scholar]
- Mertins P., Mani D.R., Ruggles K.V., Gillette M.A., Clauser K.R., Wang P., Wang X., Qiao J.W., Cao S., Petralia F., et al. NCI CPTAC . 2016. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature. 534:55–62. 10.1038/nature18003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura Y., Takiguchi S., Ito S., and Itoh K.. 2015. EGF‑stimulated AKT activation is mediated by EGFR recycling via an early endocytic pathway in a gefitinib‑resistant human lung cancer cell line. Int. J. Oncol. 46:1721–1729. 10.3892/ijo.2015.2871 [DOI] [PubMed] [Google Scholar]
- Nitulescu G.M., Margina D., Juzenas P., Peng Q., Olaru O.T., Saloustros E., Fenga C., Spandidos D.Α., Libra M., and Tsatsakis A.M.. 2016. Akt inhibitors in cancer treatment: The long journey from drug discovery to clinical use (Review). Int. J. Oncol. 48:869–885. 10.3892/ijo.2015.3306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X., Li G., Asmussen H.K., Asnaghi L., Vass W.C., Braverman R., Yamada K.M., Popescu N.C., Papageorge A.G., and Lowy D.R.. 2007. Oncogenic inhibition by a deleted in liver cancer gene requires cooperation between tensin binding and Rho-specific GTPase-activating protein activities. Proc. Natl. Acad. Sci. USA. 104:9012–9017. 10.1073/pnas.0703033104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J., Mann M., and Ishihama Y.. 2007. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2:1896–1906. 10.1038/nprot.2007.261 [DOI] [PubMed] [Google Scholar]
- Ravi A., Kaushik S., Ravichandran A., Pan C.Q., and Low B.C.. 2015. Epidermal growth factor activates the Rho GTPase-activating protein (GAP) Deleted in Liver Cancer 1 via focal adhesion kinase and protein phosphatase 2A. J. Biol. Chem. 290:4149–4162. 10.1074/jbc.M114.616839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricker E., Chowdhury L., Yi W., and Pernis A.B.. 2016. The RhoA-ROCK pathway in the regulation of T and B cell responses. F1000 Res. Faculty Rev-2295:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller M.R. 2006. Coupling receptor tyrosine kinases to Rho GTPases--GEFs what’s the link. Cell. Signal. 18:1834–1843. 10.1016/j.cellsig.2006.01.022 [DOI] [PubMed] [Google Scholar]
- Seidel S.A., Dijkman P.M., Lea W.A., van den Bogaart G., Jerabek-Willemsen M., Lazic A., Joseph J.S., Srinivasan P., Baaske P., Simeonov A., et al. 2013. Microscale thermophoresis quantifies biomolecular interactions under previously challenging conditions. Methods. 59:301–315. 10.1016/j.ymeth.2012.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A., Tomas H., Havlis J., Olsen J.V., and Mann M.. 2006. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1:2856–2860. 10.1038/nprot.2006.468 [DOI] [PubMed] [Google Scholar]
- Shimokawa H., Sunamura S., and Satoh K.. 2016. RhoA/Rho-Kinase in the Cardiovascular System. Circ. Res. 118:352–366. 10.1161/CIRCRESAHA.115.306532 [DOI] [PubMed] [Google Scholar]
- Summers S.A., Lipfert L., and Birnbaum M.J.. 1998. Polyoma middle T antigen activates the Ser/Thr kinase Akt in a PI3-kinase-dependent manner. Biochem. Biophys. Res. Commun. 246:76–81. 10.1006/bbrc.1998.8575 [DOI] [PubMed] [Google Scholar]
- Tripathi B.K., Qian X., Mertins P., Wang D., Papageorge A.G., Carr S.A., and Lowy D.R.. 2014. CDK5 is a major regulator of the tumor suppressor DLC1. J. Cell Biol. 207:627–642. 10.1083/jcb.201405105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasylishen A.R., and Lozano G.. 2016. Attenuating the p53 Pathway in Human Cancers: Many Means to the Same End. Cold Spring Harb. Perspect. Med. 6:a026211 10.1101/cshperspect.a026211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., and Xu X.M.. 2016. RhoA/Rho kinase in spinal cord injury. Neural Regen. Res. 11:23–27. 10.4103/1673-5374.169601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang S.Y., Dusaban S.S., and Brown J.H.. 2013. Lysophospholipid receptor activation of RhoA and lipid signaling pathways. Biochim. Biophys. Acta. 1831:213–222. 10.1016/j.bbalip.2012.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu O.M., and Brown J.H.. 2015. G Protein-Coupled Receptor and RhoA-Stimulated Transcriptional Responses: Links to Inflammation, Differentiation, and Cell Proliferation. Mol. Pharmacol. 88:171–180. 10.1124/mol.115.097857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., and Zheng Y.. 2013. Cell type-specific signaling function of RhoA GTPase: lessons from mouse gene targeting. J. Biol. Chem. 288:36179–36188. 10.1074/jbc.R113.515486 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.