Chang et al. link the RECQ-like helicase BLM and its yeast orthologue Sgs1 to preventing DNA damage caused by the accumulation of DNA:RNA hybrid structures called R-loops. This adds to a growing family of helicases implicated in R-loop resolution.
Abstract
Sgs1, the orthologue of human Bloom’s syndrome helicase BLM, is a yeast DNA helicase functioning in DNA replication and repair. We show that SGS1 loss increases R-loop accumulation and sensitizes cells to transcription–replication collisions. Yeast lacking SGS1 accumulate R-loops and γ-H2A at sites of Sgs1 binding, replication pausing regions, and long genes. The mutation signature of sgs1Δ reveals copy number changes flanked by repetitive regions with high R-loop–forming potential. Analysis of BLM in Bloom’s syndrome fibroblasts or by depletion of BLM from human cancer cells confirms a role for Sgs1/BLM in suppressing R-loop–associated genome instability across species. In support of a potential direct effect, BLM is found physically proximal to DNA:RNA hybrids in human cells, and can efficiently unwind R-loops in vitro. Together, our data describe a conserved role for Sgs1/BLM in R-loop suppression and support an increasingly broad view of DNA repair and replication fork stabilizing proteins as modulators of R-loop–mediated genome instability.
Introduction
Genome instability is an enabling characteristic of tumor formation because it creates mutational diversity in premalignant cell populations, allowing the necessary mutations in driver genes to occur at a sufficiently high frequency (Stratton et al., 2009). One of the best-understood ways in which genome instability arises in cancer is an acquired defect in a DNA repair pathway. Mutations in homologous recombination (HR), nucleotide excision repair, cross-link repair, and mismatch repair are all clearly linked to an increased risk of cancer (Curtin, 2012). Germline mutations in genes coding for DNA repair proteins dramatically increase cancer risk and can be associated with other symptoms, whereas somatically acquired cancer driver mutations in genes that function in the same repair pathways are found in sporadic cancers.
One such DNA repair protein is the RECQ-like helicase, BLM, which is involved in resolution of concatenated DNA molecules during HR, at replication forks, and in anaphase (Böhm and Bernstein, 2014). BLM can act on a wide array of substrates in vitro including Holliday junctions, D-loops, G-quadruplexes, DNA:RNA hybrids, and single-stranded overhangs (Popuri et al., 2008; Croteau et al., 2014). Germline BLM mutations lead to Bloom’s syndrome, which is characterized by cancer predisposition, short stature, and other symptoms (de Renty and Ellis, 2017). At the cellular level, defects in BLM are characterized by high levels of sister chromatid exchanges, and DNA replication and mitotic defects (Böhm and Bernstein, 2014). BLM is highly conserved across evolution, and much of what is known about its function was first described for its orthologue in Saccharomyces cerevisiae, Sgs1. Research on Sgs1 has linked its activity to HR, both at the stage of end-resection and double-Holliday junction resolution, restart of stalled DNA replication forks, meiosis, and telomere maintenance (Ashton and Hickson, 2010). Both BLM and Sgs1 have multiple interacting partners that regulate their activity and cooperate with them in catalyzing DNA transactions. BLM/Sgs1 forms a complex with Topoisomerase III and RMI1/2, which work together with BLM to decatenate DNA molecules. The BLM-Top3-Rmi1/2 complex further associates with members of the Fanconi anemia (FA) pathway to help process stalled DNA replication forks—for example, during interstrand cross-link repair (Suhasini and Brosh, 2012; Ling et al., 2016).
Recently, defects in DNA repair proteins have been linked to a novel mechanism of genome instability involving the formation of excessive DNA:RNA hybrids on genomic DNA. These hybrids form a structure called an R-loop, in which RNA binds to a complementary DNA strand and exposes the nontemplate strand as an single stranded DNA loop (Chan et al., 2014b). R-loops are thought to cause genome instability primarily by interfering with DNA replication. R-loop collision with replication forks leads to fork stalling and an increase in double-strand breaks or error-prone mechanisms of replication (Chan et al., 2014b). The best understood players in R-loop metabolism are those involved in RNA processing, in which normal transcript elongation, termination, splicing, packaging, nuclear export, and RNA degradation have all been shown to suppress R-loop formation (Li and Manley, 2005; Gómez-González et al., 2011; Mischo et al., 2011; Wahba et al., 2011; Stirling et al., 2012). Among these, RNaseH’s have the most direct effects, with RNaseH1 working only on R-loop substrates, and RNaseH2 targeting R-loops and ribonucleotides incorporated into DNA (Cerritelli and Crouch, 2009). The THO complex, made up of Hpr1, Mft1, Tho2, and Thp2 in yeast, is an mRNA export complex whose disruption has been repeatedly associated with R-loop–mediated genome instability (Chávez et al., 2000; Gómez-González et al., 2011). Finally, Sen1, the yeast homologue of senataxin, is a DNA/RNA helicase with multiple functions, including coordination of replication and transcription, likely exploiting a direct R-loop unwinding activity (Mischo et al., 2011).
Interestingly, defects in canonical DNA repair proteins such as HR factors BRCA1 and BRCA2 (Wahba et al., 2013; Bhatia et al., 2014; Hatchi et al., 2015), nucleotide excision repair proteins XPG and XPF (Sollier et al., 2014), the FA pathway (García-Rubio et al., 2015; Schwab et al., 2015), and the DNA damage response kinase ATM (Tresini et al., 2015) have all been associated with stabilization or signaling involving R-loops. Moreover, R-loops have been shown to contribute to DNA replication stress in these DNA repair mutants, and in some cases, a direct role for the repair protein in R-loop removal has been suggested (e.g., R-loop displacement by the FANCM helicase; Schwab et al., 2015). Indeed, the BLM protein is known to cooperate with the HR pathway and is critical for the activation of the FA pathway. Moreover, Sgs1 has synthetic phenotypes with RNaseH2 deletions in yeast, suggesting a functional cooperation between the two proteins (Kim and Jinks-Robertson, 2011; Chon et al., 2013).
Here we show that in yeast deleted for SGS1, genome instability is partially transcription-dependent, and that both R-loops and DNA damage accumulate at sites of Sgs1 action in the genome. By physically mapping Sgs1 binding sites in the yeast genome, we observe a strong association between Sgs1 binding and sites that gain R-loops and DNA damage in sgs1Δ yeast. Indeed, unbiased mutation accumulation in sgs1Δ cells identifies increases in structural rearrangements at R-loop–prone sites. Finally, we confirm R-loop–associated genome instability in BLM-depleted human cells and in Bloom’s syndrome fibroblasts, further showing that BLM localizes near R-loops in cells and is capable of resolving R-loops as efficiently as D-loops in vitro. Together these data establish Sgs1/BLM as a regulator of R-loop–coupled genome instability, adding to the growing repertoire of DNA repair proteins with functions in R-loop mitigation, and extending the notion that transcription–replication collisions are one of the drivers of mutagenesis in cancer.
Results
R-loop formation and consequences in sgs1Δ
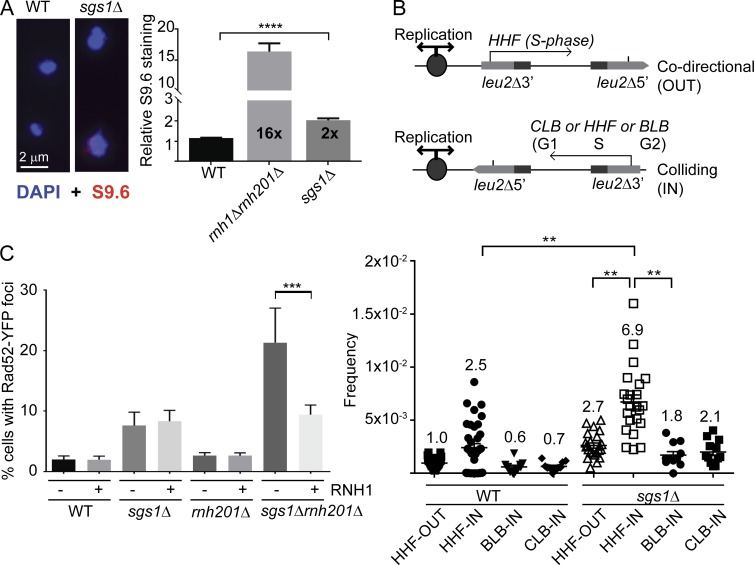
Yeast Sgs1 has been ascribed a downstream or cooperative role with RNaseH2 in suppressing genome instability, presumed to be related to shared roles in DNA replication and potential cooperation at sites of transcription-mediated instability (Kim and Jinks-Robertson, 2011; Chon et al., 2013). However, based on recent studies linking replication fork–associated DNA repair proteins to R-loop suppression, we hypothesized that Sgs1 may be involved in mitigating R-loop–associated genome instability. To directly assess if deletion of SGS1 alters the levels of DNA:RNA hybrids, we used chromosome spreads and quantified staining with the S9.6 mAb, which recognizes DNA:RNA hybrids in a manner largely independent of sequence (Hu et al., 2006). This analysis showed that SGS1 deletion increases S9.6 staining compared with WT (Fig. 1 A). Ectopic R-loops are known to drive transcription–replication conflicts leading to recombination. To determine the effect of Sgs1 on this process, we tested recombination rates in a plasmid system in which transcription is driven by a promoter that is oriented to be colliding (IN) or codirectional (OUT) with the origin of replication (Fig. 1 B). This analysis showed that sgs1Δ–driven hyperrecombination was significantly enhanced when S-phase (Histone H4 promoter [HHF]) transcription is IN with DNA replication but was unaffected in the OUT orientation or by controls in which transcription occurs in G1 (CLB-IN) or G2 (BLB-IN) phase (Fig. 1 B). Transcription–replication conflicts lead to DNA damage, and we found that, whereas deletion of SGS1 leads to a small increase in levels of DNA damage as measured by Rad52-YFP foci, combined loss of RNaseH2A (i.e., sgs1Δrnh201Δ) leads to a synergistic increase in DNA damage (Fig. 1 C; Chon et al., 2013). Importantly, this enhanced damage effect was suppressed by overexpression of RNaseH1. Because RNaseH1 only degrades RNA in DNA:RNA hybrids, as opposed to functioning in replication or ribonucleotide excision repair, the synergistic DNA damage in sgs1Δrnh201Δ cells is a result of R-loops as opposed to the effects of Rnh201 or Sgs1 on replication. Cells lacking SGS1 exhibit hyperrecombination in scenarios independent of R-loops based on Sgs1’s well-established role in HR and replication fork protection. Thus, we propose that a subset of genome instability events in sgs1Δ could be R-loop related.
Figure 1.
Transcription–replication conflicts and R-loops in sgs1Δ cells. (A) S9.6 staining for DNA:RNA hybrids in yeast chromosome spreads (n = 4; for WT, rnh1Δrnh201Δ, and sgs1Δ, 616, 286, and 690 nuclei total were scored). Left, representative images; right, quantification of signal intensity per nucleus. Error bars represent SEM. Fold increase over WT is indicated in each bar. Bar, 2 µm. (B) Hyperrecombination caused by transcription–replication collisions. Top, schematics of transcription direction and cell cycle stage of each promoter are indicated. Bottom, quantification of recombination frequencies for the indicated strain and plasmid. Fold increases over WT +HHF-out are shown above each bar. 11 or more independent frequencies were measured for each sample. (C) DNA damage synergy in sgs1Δrnh201Δ is R-loop–dependent. Quantification of Rad52-YFP foci in the indicated strain with either an empty vector (−) or a Gal-inducible RNH1 (+) construct (n = 3). Error bars are SEM. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Loss of SGS1 exhibits synergistic genome instability with R-loop suppressors
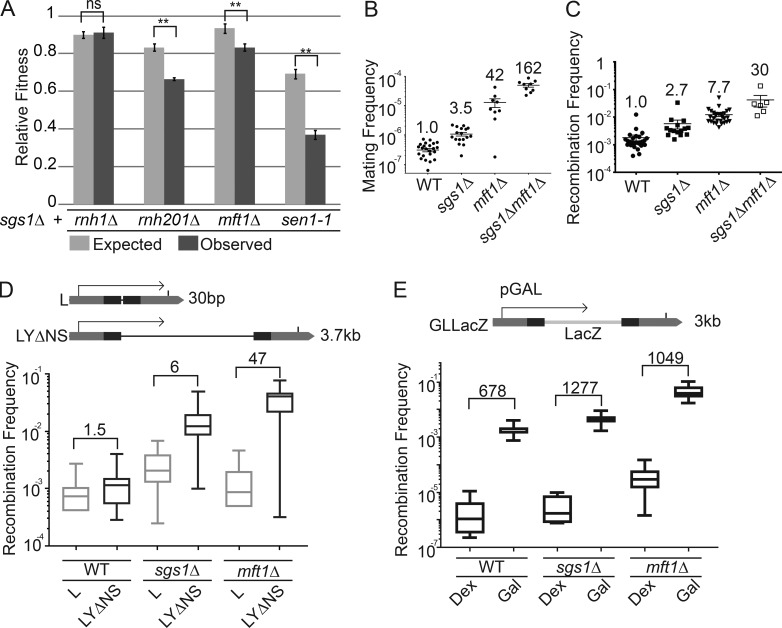
We next sought to further probe the role for R-loop modulators in sgs1Δ phenotypes. We first assessed the fitness of sgs1Δ in combination with deletions of R-loop regulators in the THO complex (MFT1), RNaseH enzymes (RNH1 and RNH201), and senataxin (SEN1) and observed synergistic fitness defects (Huertas and Aguilera, 2003; Gómez-González et al., 2009; Mischo et al., 2011; Skourti-Stathaki et al., 2011). Loss of Sgs1 significantly exacerbated fitness defects in rnh201Δ, sen1-1, and mft1Δ cells (Fig. 2 A). Similar synergy was seen when measuring chromosome instability using the A-like Faker (ALF) assay for disruption of the MAT locus in chromosome III (Fig. 2 B) and a LEU2 plasmid-based direct repeat recombination (Fig. 2 C; Stirling et al., 2011). Double mutants of SGS1 and the THO complex subunit MFT1 caused a dramatic, greater than additive, increase in instability in these assays compared with the single mutants, suggesting a synergistic effect (Fig. 2, B and C). Deletion of known R-loop suppressors with diverse modes of action (i.e., THO complex, Sen1, and RNaseH) all exhibited synergistic chromosome instability phenotypes when combined with sgs1Δ (Fig. S1). These synergies are not surprising because loss of SGS1 is known to promote hyperrecombination through loss of its role in resolution of Holliday junctions (Ashton and Hickson, 2010), although this phenotype would not explain the increases in R-loop staining we see in sgs1Δ (Fig. 1).
Figure 2.
Cooperative genome maintenance by R-loop regulators and Sgs1. (A) Growth defects in strains lacking Sgs1 and the indicated gene. Quantitative growth curve analysis (n = 4 with technical triplicates in each experiment; error bars are SEM) showed observed fitness values were significantly lower than expected values. (B and C) Synergistic genome instability in SGS1, MFT1 double deletions. (B) Mating frequency as a measure of chromosome instability by the ALF assay. (C) Recombination frequency in the direct repeat plasmid system LNA (Prado et al., 1997). Fold increase over WT is shown above each measurement in the indicated strains. Error bars are SEM. (D and E) Plasmid-based LEU2 recombination frequency as a function of transcript length (D) and frequency (E). Above each panel is a schematic of the assay that measures recombination frequency, transcript length dependence, and transcript frequency dependence. For D, the length of the intervening sequence between the leu2 repeats (dark bars) is shown at right. The fold increase over WT is shown above each bar. The direction of transcription is indicated by an arrow. For E, Dex, dextrose (low expression); Gal, galactose (high expression). For D, n ≥ 14 and for E, n ≥ 6 independent frequencies were measured. Boxplots in D and E plot whiskers to the maximum and minimum values, with the box between the 25th and 75th percentile and the line at the median value.
R-loop levels increase with transcript frequency and length in direct repeat recombination assays. Therefore, to implicate Sgs1 further in transcription-associated recombination, we assessed the roles of transcript length and frequency with derivatives of the LEU2 direct repeat systems and compared to mft1Δ as a control. Although sgs1Δ had higher recombination frequencies in both assays, comparing the rates of recombination in a short transcript (L) and a long transcript (LYΔNS) plasmid system revealed only a 1.5-fold increase in recombination in WT, but a sixfold increase in sgs1Δ (Fig. 2 D). Similarly, shifting the galactose-inducible GL-LacZ recombination cassette (González-Aguilera et al., 2008) from dextrose to galactose led to a 678× increase in recombination in WT, but a 1,277× increase in sgs1Δ (Fig. 2 E). These data extend and support previous observations of transcription-associated instability in SGS1 mutants and show how drivers of R-loop stability enhance recombination in sgs1Δ cells (Fig. 1; Kim and Jinks-Robertson, 2011; Chon et al., 2013).
Sgs1 binds genomic sites that show increased R-loops and DNA damage upon deletion of SGS1
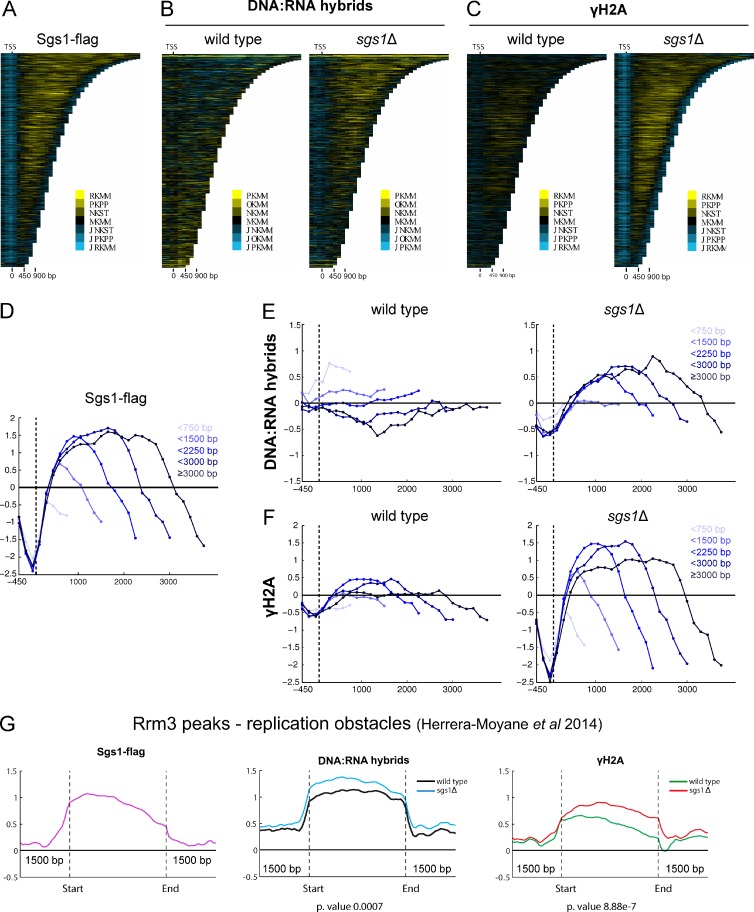
Given the increase in DNA:RNA hybrid levels in the sgs1Δ mutant and the observed relationship between SGS1 and transcription-associated recombination, we hypothesized that Sgs1 might be recruited to sites of transcription and could impact the R-loop and DNA damage landscape at these sites. To test this, we mapped Sgs1 binding by ChIP-chip, and R-loops and γ-H2A levels in sgs1Δ by DRIP-chip and ChIP-chip (Stirling et al., 2012; Chan et al., 2014a). Sgs1 does bind to ORFs, with a preference for longer and more highly transcribed genes (Fig. 3, A and D; and Fig. S2 A; Sgs1-containing genes had a mean length of 1,586 bp, compared with 1,338 bp for all genes; P < 0.0001). Sgs1 also associated at sites bound by the Rrm3 helicase (Fig. 3 G), which functions at stalled replication forks to promote replisome progression, thus demarcating replication obstacles in the yeast genome (Santos-Pereira et al., 2013). Further underscoring a shared role, previous work showed that RRM3 is required for robust growth in sgs1Δ cells (Schmidt and Kolodner, 2004). Finally, we observed some but very sparse binding of Sgs1 along telomeres and the ribosomal DNA (rDNA) loci (Fig. S2, B and C). The relatively low levels of Sgs1 association at the rDNA was surprising given Sgs1’s known role in promoting both replication and transcription of the rDNA loci.
Figure 3.
The genomic binding profile of Sgs1 and the effect of SGS1 deletion on γ-H2A and R-loop occupancy. All profiles were generated in duplicate with quantile normalized and mean data shown here. (A, B, and C) Chromatra plots showing a heat map of Sgs1 (A), DNA:RNA hybrid (B), and γ-H2A (C) occupancy over protein coding genes sorted by length and aligned at the TSS (Hentrich et al., 2012). (D–F) Mean genome-wide Sgs1 (D), DNA:RNA hybrid (E), and γ-H2A (F) occupancy in WT (left) and sgs1Δ (right) as a function of gene length. A total of 4,868 genes were split into the indicated gene length categories (538 genes < 750 bp, 1,861 genes < 1,500 bp, 1,263 genes < 2,250 bp, 636 genes < 3,000 bp, and 570 genes ≤ 3,000 bp) with mean enrichment scores calculated and plotted for each category. We observed Sgs1 binding and increase in DNA:RNA hybrid and γ-H2A levels in the sgs1Δ mutant compared with WT at longer genes. Mean values and statistics are reported in the Results section. (G) Mean Sgs1 (left), DNA:RNA hybrid (center), and γ-H2A (right) occupancy across previously identified Rrm3 peaks (Herrera-Moyano et al., 2014) for the indicated strains. Two-sided Wilcox test p-values comparing mean occupancy scores in sgs1Δ versus WT are noted below.
Given the surprising occupancy of Sgs1 at ORFs, and in particular long genes, we next focused on potential effects of SGS1 deletion on DNA:RNA hybrid and γ-H2A profiles. Consistent with the increase in S9.6 staining and Rad52 foci observed in the sgs1Δ mutant, we also found that loss of SGS1 resulted in increased DNA:RNA hybrids and γ-H2A at a subset of genomic loci. More specifically, we found that loss of SGS1 increased DNA:RNA hybrid and γ-H2A levels at longer genes (Fig. 3, B, C, E, and F), an effect corroborated by our observation of increased recombination at longer versus shorter reporter genes in the sgs1Δ mutant (Fig. 2 D). Analysis of the genes significantly occupied by DNA:RNA hybrids or γ-H2A signal in both replicates of sgs1Δ but not WT confirmed a significant shift toward longer than average genes (i.e., sgs1Δ DNA:RNA hybrid containing genes were 1,745 bp and γ-H2A containing genes were 1,540 bp compared with 1,338 bp for all genes; P < 0.0001 ANOVA with Holm-Sidak correction). The distribution of genes with increased DRIP and γ-H2A signal in sgs1Δ also showed a small bias to subtelomeric regions (Fig. S2 B).
Loss of Sgs1 also increased DNA:RNA hybrid and γ-H2A levels at other sites, namely, regions bound by Rrm3 (Fig. 3 G; DNA replication slow zones), an observation consistent with Sgs1 functioning at replication obstacles (Cobb et al., 2003), and with loss of Sgs1 leading to R-loop stabilization and DNA damage at these sites. Interestingly, our profiles also revealed sites where loss of SGS1 increases DNA:RNA hybrid levels without concomitant effects on γ-H2A: the rDNA loci and a subset of telomeres (Fig. S2). However, we observed higher γ-H2A signal flanking the rDNA, and show that increased rDNA instability in the sgs1Δ mutant could be suppressed to WT levels by ectopic expression of RNaseH1 (Fig. S2 D), suggesting that although these DNA:RNA hybrids were not associated with increased damage at the rDNA loci, as measured by γ-H2A occupancy, they may still contribute to instability.
Normal Sgs1 binding sites define R-loop prone fragile sites in sgs1Δ cells
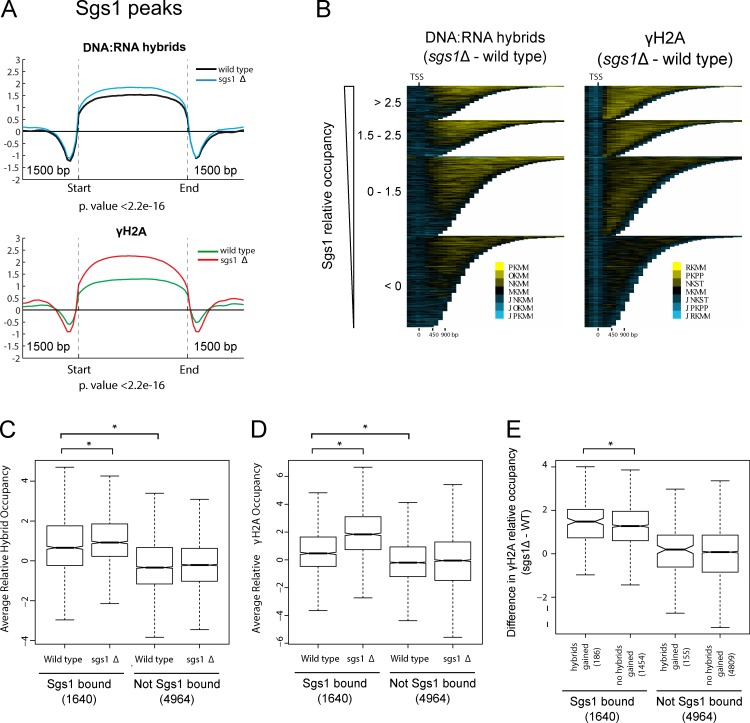
To probe the interdependence of the genome-wide profiles generated, we focused on Sgs1-bound regions and analyzed DNA:RNA hybrid and γ-H2A levels at these sites. Overall, we found that sites of Sgs1 binding were associated with DNA:RNA hybrids and γ-H2A, the levels of which increased when SGS1 was deleted (Fig. 4 A). Focusing on ORFs, we found that increasing levels of Sgs1 binding signal were associated with increased levels of DNA:RNA hybrids and γ-H2A in the sgs1Δ mutant compared with WT (Fig. 4 B). This trend was stronger for γ-H2A than DNA:RNA hybrids, suggesting that Sgs1 functioned directly to prevent DNA damage at a subset of ORFs, although this was not always through a role in preventing DNA:RNA hybrid accumulation at those sites. To probe this relationship further, we divided ORFs into Sgs1-bound and not-bound groups. Consistent with our observation that Sgs1-bound peaks defined sites of R-loop and γ-H2A occupancy in WT cells (Fig. 4 A), we found that under WT conditions, Sgs1-bound ORFs had higher levels of DNA:RNA hybrids and γ-H2A compared with ORFs that were not bound by Sgs1 (Fig. 4, C and D). Importantly, we found that the levels of DNA:RNA hybrids and γ-H2A significantly increased upon deletion of SGS1 only for the group of Sgs1-bound genes, suggesting that the direct association of Sgs1 with these ORFs functioned to mitigate the levels of DNA:RNA hybrids and γ-H2A. To take this analysis a step further, we focused only on ORFs that both were bound by Sgs1 and gained DNA:RNA hybrids in the sgs1Δ mutant compared with WT, and found that upon deletion of SGS1, these genes had a greater increase in γ-H2A levels than genes that were bound by Sgs1 but did not accumulate hybrids upon its loss (Fig. 4 E). Interestingly, this analysis also revealed a set of 155 ORFs that gained DNA:RNA hybrids when SGS1 was deleted but did not pass our binding threshold for Sgs1 binding. Consistent with these sites representing indirect effects of Sgs1 on the DNA:RNA hybrid landscape, they did not show increased γ-H2A levels compared with the rest of ORFs that were not bound by Sgs1 (Fig. 4 E). Together these results suggest that Sgs1 normally binds to fragile, R-loop–prone regions, and that loss of SGS1 activates a subset of these sites to accumulate R-loops and DNA damage.
Figure 4.
Intersection of Sgs1, R-loops and DNA damage. All profiles were generated in duplicate with quantile normalized and averaged data shown here. (A) Mean DNA:RNA hybrid (top) and γ-H2A (bottom) occupancy across Sgs1-binding sites for the indicated strains. Two-sided Wilcox test p-value comparing mean occupancy scores in sgs1Δ versus WT is noted below. (B) Chromatra plots showing a heat map of the difference in DNA:RNA hybrid (left) and γ-H2A (right) occupancy in sgs1Δ compared with WT. Protein coding genes are sorted by length and Sgs1 occupancy and aligned at the TSS (Hentrich et al., 2012). (C and D) Box plots comparing mean occupancy scores of DNA:RNA hybrids (C) and γ-H2A (D) in WT and the sgs1Δ mutant at Sgs1-bound versus not bound ORFs. *, P < 0.05, for two-tailed Wilcox tests. The numbers of genes in each category are shown in brackets. All other comparisons are not significant. (E) Box plot showing increases in γ-H2A levels at genes that are bound by Sgs1 and that gain DNA:RNA hybrids upon loss of SGS1 compared with those that do not gain hybrids. *, P < 0.05, for one-tailed Wilcox test. The numbers of genes in each category are shown in brackets. Changes in γ-H2A were not observed when genes that gained hybrids but were not bound to Sgs1 were compared with those that have neither hybrids nor Sgs1 association. The whiskers on boxplots in C–E represent 1.5× the interquartile range.
Genome instability in sgs1Δ yeast occurs at R-loop–prone regions
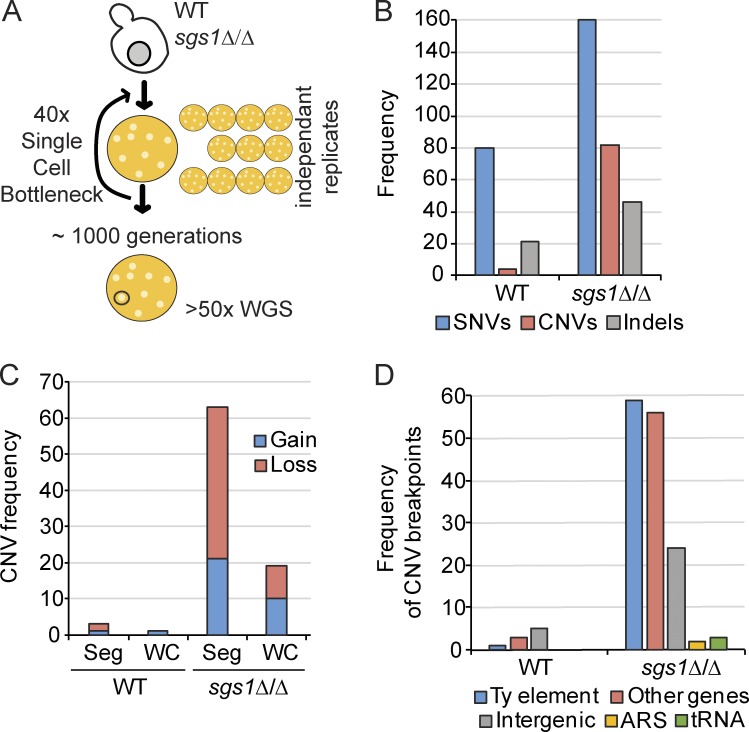
Our data suggest that Sgs1 reduces R-loops and DNA damage at specific genomic loci. To create an unbiased view of ongoing instability in the genome of sgs1Δ cells, we performed a mutation accumulation and whole-genome sequencing experiment (Stirling et al., 2014). Passaging homozygous sgs1Δ/Δ diploids for ∼1,000 generations created a set of 12 mutation accumulation strains that we sequenced at >50× coverage (Fig. 5 A). This analysis revealed a modest approximately twofold increase in single-nucleotide variants (SNVs) for sgs1Δ/Δ compared with WT, which was similar to the rates seen at the CAN1 reporter locus (Segovia et al., 2017). However, sgs1Δ/Δ exhibited a ∼12× increase in copy number variants (CNVs) compared with WT (Fig. 5 B). The majority of these changes were segmental, although aneuploidy also increased in sgs1Δ (Fig. 5 C). Analysis of the predicted breakpoints shows that most originate within or near a TY retrotransposon element (Fig. 5 D and Table S2). Indeed, Sgs1 has been ascribed a role in Ty1 element expansion based on its role in HR (Bryk et al., 2001). Other breakpoints appear to be at telomeres, and at a set of protein coding genes. Breakpoint-associated genes (Table S2) often had paralogs in the yeast genome, and this, along with the TY element enrichment, is consistent with the role for Sgs1 in rejecting HR reactions that could lead to CNVs between repetitive sequences (Myung et al., 2001). Because TY elements and telomeres are known to be hot spots of R-loop formation, we explored the potential correlation of CNV breakpoints within protein coding genes and R-loop occupancy. The R-loop signal was significantly higher than the mean background of WT DRIP peaks (10.2 [n = 19] vs. 4.1 [n = 14,307]; Mann–Whitney test, P < 0.0001; Chan et al., 2014a). Interestingly, as for Sgs1–bound and DRIP- and γ-H2A–associated genes, analysis of genes associated with CNV breakpoints in sgs1Δ were significantly longer than the genome mean (1,745 bp compared with 1,338 bp, Mann–Whitney P = 0.0425). Overall, the mutation signature of sgs1Δ/Δ cells supports its known specific role in promoting noncrossover events and rejecting HR (Myung et al., 2001). In addition, it is consistent with the observed sensitivity of sgs1Δ cells to transcription-associated recombination and the accumulation of DNA:RNA hybrids and DNA damage in sgs1Δ cells.
Figure 5.
The mutation spectrum of SGS1-deficient yeast. (A) Schematic of the 1,000 generation mutation accumulation approach (Segovia et al., 2017). 12 independent lines for each condition were generated and sequenced. WGS, whole genome sequencing. (B) Frequency of SNVs, CNVs, and insertions and deletions (indels) in mutation accumulation lines of the indicated strains. (C) CNV breakpoint classes in mutation accumulation lines. Seg, segmental copy number change; WC, whole chromosome aneuploidy. (D) CNV breakpoint characteristics in mutation accumulation lines. ARS, autonomously replicating sequences (DNA replication origins). For more detailed information, see Table S2.
R-loops accumulate and cause DNA damage in BLM–depleted cell lines
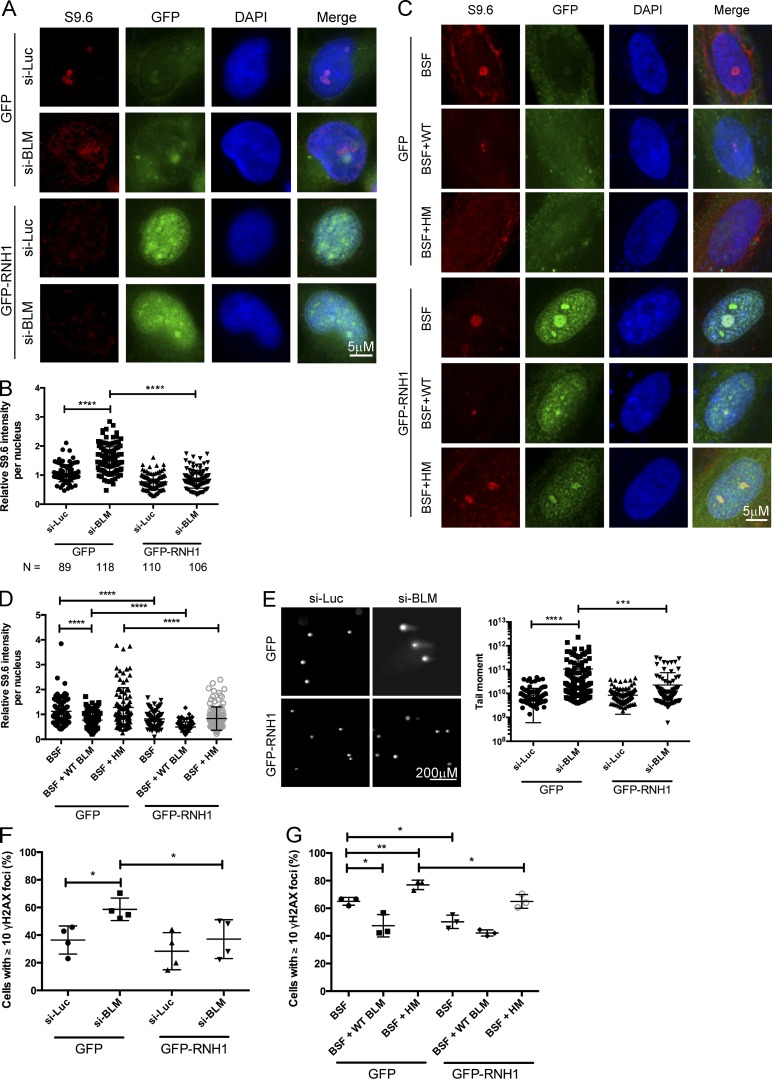
The human orthologue of Sgs1 is the Bloom’s syndrome helicase BLM, one of five RECQ-like helicases in humans and the closest sequence orthologue. To determine whether a role of Sgs1 in R-loop metabolism is conserved in mammalian cells, we used siRNA to target BLM in HeLa cells (Fig. S3 A), and measured R-loop levels by immunofluorescence. Knockdown of BLM leads to a significant increase in S9.6 staining, which could be abolished by overexpression of GFP-RNaseH1 (Fig. 6, A and B). Similar staining results were obtained for BLM−/− knockout derivatives of the near diploid HCT116 cell line compared with an isogenic control line (Fig. S3 C). Importantly, this accumulation was also seen in Bloom’s syndrome fibroblasts relative to an isogenic control complemented with WT BLM (Fig. 6, C and D). Moreover, the reduction of R-loop levels was not seen in fibroblasts complemented with the BLM K695T mutant, which abolishes its helicase activity (Fig. 6, C and D; Neff et al., 1999), suggesting a direct role for the BLM helicase activity in removing R-loops.
Figure 6.
R-loop accumulation and DNA damage in BLM-depleted cells. (A and C) Representative images of S9.6 staining in HeLa cells treated with the indicated siRNA targeted for BLM (si-BLM) or a control luciferase (si-Luc; A) or Bloom’s syndrome fibroblasts complemented with an empty vector control (BSF), WT BLM (BSF + WT), or helicase dead mutant (BSF + HM; C). Cells were transfected with either a control vector (GFP) or one expressing GFP-RNaseH1. (B and D) Quantification of S9.6 signal intensity for nuclear area in the indicated conditions in HeLa cells (B) or Bloom’s syndrome fibroblasts (D). Cell numbers scored across three independent replicates are noted below panel B for HeLa cells. (E) RNaseH-dependent DNA breaks in BLM-deficient HeLa cells. (Left) Representative comet tail images from single-cell electrophoresis. (Right) Quantification of comet tail moment under the indicated conditions. t tests were used for comparisons shown. (F and G) Percentages of cells with ≥10 γ-H2AX foci in HeLa cells treated with indicated siRNA expressing GFP or GFP-RNaseH1 (F) or Bloom’s syndrome fibroblasts expressing GFP or GFP-RNaseH1 (G). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Like Sgs1 in yeast, BLM depletion increases genome instability phenotypes, and we therefore examined the effects of RNaseH1 overexpression in BLM knockdown cells on genome instability. BLM knockdown caused chromosome instability as measured by increased micronucleus formation after 48 h (Fig. S3, D and E), likely as a result of an inability to resolve anaphase bridges at mitosis (Naim and Rosselli, 2009). Importantly, the increase in micronuclei formation was significantly suppressed by overexpression of GFP-RNaseH1 (Fig. S3, D and E). BLM knockdown also induced DNA breaks as measured by the neutral comet assay and γ-H2AX focus accumulation, with both defects significantly reduced by ectopic expression of GFP-RNaseH1 (Fig. 6, E and F). Indeed, Bloom’s syndrome fibroblasts also showed high levels of γ-H2AX foci, which could be reduced by RNaseH1 expression (Fig. 6 G), whereas fibroblasts complemented with WT BLM showed lower levels of γ-H2AX foci that were not reduced by RNaseH1 expression (Fig. 6 G). These data suggest that a considerable proportion of DNA damage and genome instability in BLM-deficient mammalian cells, and potentially in Bloom’s syndrome, may be a result of a reduced ability of these cells to process R-loops.
Assessing potential direct effects of BLM on R-loops
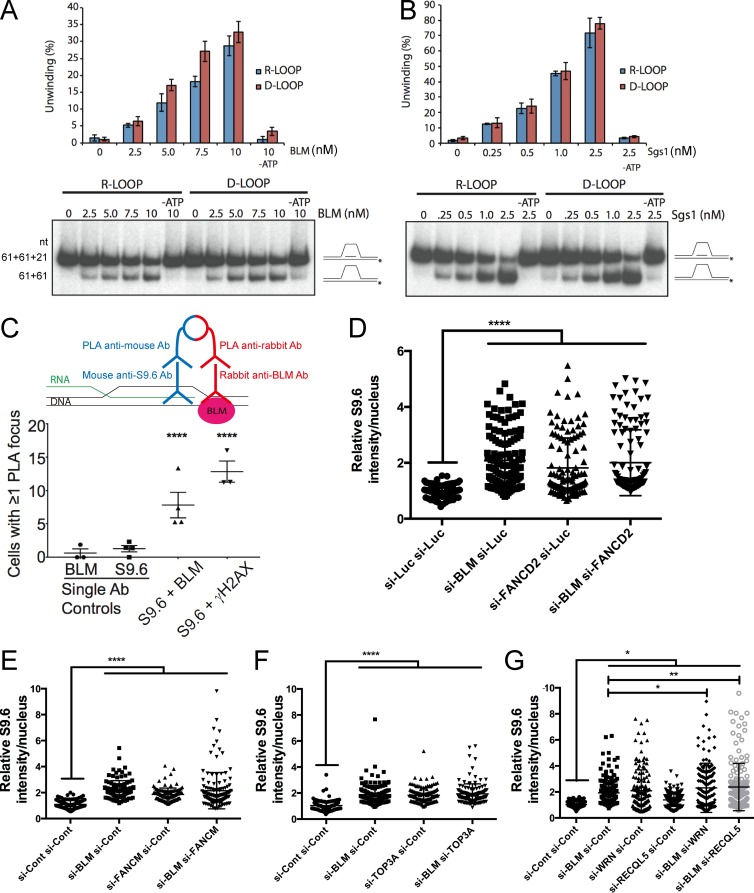
There are several possible mechanisms by which Sgs1 and BLM could impact R-loop–mediated genome instability, including effects on replisome stability at transcription–replication conflicts, or through direct unwinding of R-loops, alone or collaboratively with topoisomerase III or the FA pathway. To support these direct models, and rule out potential indirect effects, for example, through effects on DNA damage signaling pathways or transcriptional effects (Grierson et al., 2012; Tresini et al., 2015), we tested the helicase activity of recombinant BLM and Sgs1 directly on both R- and D-loop substrates. Although BLM has previously been shown to unwind R-loops in vitro (Popuri et al., 2008), how this activity compares to its ability to unwind other structures is unclear, and to our knowledge, whether Sgs1 can unwind R-loops is unknown. We found that either BLM or Sgs1 could unwind R- and D-loop substrates of the same sequence composition with nearly identical efficiencies and that this occurred in an ATP- and concentration-dependent manner (Fig. 7, A and B). Thus, both helicases are highly efficient R-loop resolvases. To determine whether BLM and R-loops are ever proximal in human cells, we performed a proximity ligation assay (PLA) with antibodies targeting BLM and DNA:RNA hybrids. Remarkably, BLM showed a clear and reproducible PLA signal in cells with S9.6 that was significantly higher than in single primary antibody controls, thus showing that BLM comes in close proximity to DNA:RNA hybrids in cells (Fig. 7 C and Fig. S4 A). BLM has many interaction partners and interdependent relationships in DNA damage repair (Suhasini and Brosh, 2012; Chaudhury et al., 2013; Ling et al., 2016). Given the previously reported physical and functional interactions of BLM with the FA pathway, which itself has been implicated in R-loop suppression (García-Rubio et al., 2015; Schwab et al., 2015) we tested potential synergy or epistasis with FA pathway components by siRNA knockdown. Knockdown of FANCD2 or FANCM increased DNA:RNA hybrid staining, but this was epistatic to coincident knockdown of BLM (Fig. 7, D and E). Furthermore, BLM was required for hydroxyurea (HU)-induced increases in FANCD2 foci (Fig. S4, B and C), supporting literature placing these factors in the same pathway during replication stress (Ling et al., 2016; Panneerselvam et al., 2016). Similar epistatic results were found when we performed knockdown of BLM together with topoisomerase III, which forms a complex with BLM to decatenate DNA and to resolve stalled replication forks (Fig. 7 F). Comparison of the effects of knockdown of other RECQ-like helicases, WRN and RECQL5, which have been separately implicated in R-loop biology, again showed S9.6 staining increases. Importantly, double knockdowns of BLM and RECQL5 were not epistatic and led to further enhanced S9.6 staining, whereas knockdown of BLM with WRN showed no additional staining compared with WRN knockdown alone (Fig. 7 G). These data support a model in which local effects of BLM at R-loops occur in the same pathway as those of FANCD2 and FANCM, but not other R-loop suppressors like RECQL5. This provides an important constraint on our model, and shows how R-loop resolution can occur through multiple independent pathways. Together, our data support a conserved mechanism for Sgs1/BLM in suppressing transcription-associated genome instability at R-loop sites (Fig. 8).
Figure 7.
Potential mechanism of BLM-dependent R-loop mitigation. Comparative R-loop and D-loop unwinding by BLM (A) and Sgs1 (representive gel shown of three separate experiments is shown; error bars are SEM; B). Schematics of the slow migrating oligonucleotide loop substrate and faster migrating product are shown at right and nucleotide (nt) sizes are listed to the left of A. Protein concentrations used are listed above. Quantification of unwinding efficiency is shown in the graph. (C) Proximity ligation of BLM and DNA:RNA hybrids in cells. A schematic of PLA (above) shows that signal only emerges when two epitopes (DNA:RNA and BLM) are close enough to ligate oligonucleotides conjugated to secondary antibodies (Ab). S9.6 and γ-H2AX were previously associated in cells using PLA and serve as a positive control (Stork et al., 2016). Cells with any fluorescence signal were scored as positive. Pooled count data for single primary antibody controls and dual antibody PLA reactions were compared using a Fisher’s exact test. (D–G) Nuclear S9.6 staining data for the indicated siRNA treatments. Dot plots show the range of values quantified. Western blots to confirm double knockdown efficiency are shown in Fig. S3. P-values were determined by ANOVA with Sidak’s multiple comparisons test post hoc. *, P < 0.05; **, P < 0.01; ****, P < 0.0001.
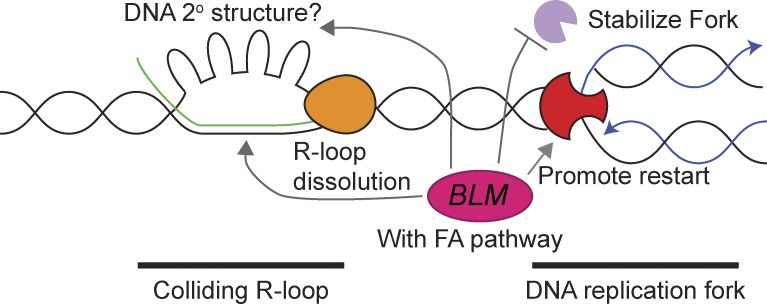
Figure 8.
Model of Sgs1/BLM impact on R-loops. Shown are a replication fork (red) heading toward a stalled RNA polymerase (orange) and associated R-loop. Possible roles for BLM/Sgs1 at these sites are highlighted with arrows and discussed in the Discussion section.
Discussion
Our data show that loss of SGS1 or BLM leads to R-loop accumulation across species and that at least some of the associated genome instability is contingent upon transcription–replication conflicts and/or R-loops. These data match with unbiased mutation accumulation analysis, which shows that loss of SGS1 increased CNVs flanked by homologous repeats and regions of high R-loop occupancy. Published mutation accumulation in rnh1Δrnh201Δ mutants also found increased deletion and duplication events between TY elements, suggesting these could be fragile sites in R-loop–prone mutants (O’Connell et al., 2015). Global profiling of Sgs1 binding sites the in genome in concert with mapping of DNA:RNA hybrids and phospho-H2A-Serine129 in WT and sgs1Δ mutants revealed a remarkably cohesive picture. Sgs1 binding sites define regions that increase in both R-loops and DNA damage when SGS1 is deleted. These sites, many of which are at long genes, overlap with Rrm3 binding sites, indicating that they may be difficult-to-replicate sites of frequent fork stalling. Indeed, long genes have previously been implicated as a binding site for Rrm3 (Santos-Pereira et al., 2013). Moreover, Top2, an interacting partner of Sgs1 that cooperates with Sgs1 and Top3 at rDNA (Mundbjerg et al., 2015), has also been linked to transcription at long protein coding genes in yeast (Joshi et al., 2012). Thus, regions that are sensitive to topological stress may be more likely to form R-loops in sgs1Δ cells. We do not know with high resolution where damage sites occur in BLM-deficient human cells; however, BLM depletion does create ultrafine bridges and DNA damage at common fragile sites (Lukas et al., 2011). Some common fragile sites, in particular those at very long genes, have been linked to transcription–replication conflicts and R-loops (Helmrich et al., 2011), and thus for specific sites of instability in the human genome, we suggest there may be previously unappreciated links between R-loops and BLM.
Recently it was observed that, despite binding them broadly, only a subset of R-loops are degraded by Rnh1 in yeast (Zimmer and Koshland, 2016). This highlights that there may be a poorly understood regulatory distinction between normal and abnormal R-loop formation in cells, and we believe this distinction may be relevant to locus-specific differences seen in DRIP-chip profiles in mutant strains such as sgs1Δ. In the case of the rDNA, Sgs1 is known to have a role in facilitating both replication and transcription (Lee et al., 1999; Versini et al., 2003) and accordingly, we observed that enhanced rDNA instability seen in sgs1Δ cells is completely suppressed by RNaseH1 overexpression. More recently, the chromatin state of histone H3 on DNA-flanking R-loops have been cited as key determinants of whether R-loops will be DNA-damaging (Garcia-Pichardo et al., 2017), and we observed many examples of sites where R-loop occupancy and γ-H2A increase independent of one another. How the replisome and repair proteins like Sgs1/BLM influence transcription–replication conflicts in the context of chromatin is only beginning to be understood.
Possible models of Sgs1/BLM action at R-loops
Overall, we favor a model in which Sgs1/BLM works in proximity to R-loops at sites of transcription–replication conflict, consistent with studies of collaboration between Sgs1 and RNaseH2 (Kim and Jinks-Robertson, 2011; Chon et al., 2013). Within this local model (Fig. 8), there remain several nonmutually exclusive possibilities: the simplest model is that Sgs1/BLM unwinds R-loops directly, as supported by in vitro experiments (Fig. 7; Popuri et al., 2008). Sgs1/BLM could also unfold G-quadruplexes associated with the nontemplate strand opposite a DNA:RNA hybrid in a so-called G-loop (Duquette et al., 2004). The role of Sgs1/BLM in fork stabilization may also allow time for other factors to resolve R-loop blockages. Finally, Sgs1/BLM could direct the activity of another R-loop helicase. For example, our ChIP data link Sgs1 binding sites to those of Rrm3, and both Rrm3 and its paralog Pif1 have been recently implicated in R-loop resolution at specific loci of the yeast genome (Tran et al., 2017). In humans, BLM is known to bind FANCM and FANCJ (Suhasini and Brosh, 2012), and our data suggest that the collaboration between BLM and the FA pathway is likely to be important for mitigating the effects of R-loops in human cells (García-Rubio et al., 2015; Schwab et al., 2015). For example, BLM physically and functionally interacts with FANCM (Ling et al., 2016), potentially coordinating its activity with the activity of the FA pathway, and FANCM can use its strand migration activity to remove R-loops (Schwab et al., 2015). Indeed, FANCD2 also regulates BLM stability and assembly at stalled replication forks, and reciprocally, FANCD2 activation requires BLM (Chaudhury et al., 2013; Panneerselvam et al., 2016). Whether these connections also extend to Sgs1 and the distantly related FA pathway of S. cerevisiae is not known. Thus, there are multiple levels of regulation to be explored across systems in the future. Indeed, the scenario may be more complex in human cells, as BLM paralogs WRN and RECQL5 have both been linked to DNA:RNA hybrid metabolism in the test tube or in cells (Chakraborty and Grosse, 2010; Saponaro et al., 2014).
Defective replisome-associated DNA repair proteins shifting the R-loop landscape
Our data add Sgs1/BLM to a growing list of DNA repair proteins that seem to work toward the error-free resolution of R-loops and the prevention of deleterious transcription–replication conflicts (Bhatia et al., 2014; Sollier et al., 2014; García-Rubio et al., 2015; Hatchi et al., 2015; Schwab et al., 2015; Chang and Stirling, 2017). In addition, there is now evidence that DNA damage alone may induce R-loops in mammalian cells (Britton et al., 2014; Tresini et al., 2015). The idea that normal robust DNA replication itself prevents R-loop accumulation is also gaining support—for example, with recent studies showing effects of the MCM helicase or POLD3 in preventing DNA:RNA hybrid accumulation (Tumini et al., 2016; Vijayraghavan et al., 2016). The abundance of fork-protection factors emerging as R-loop regulators supports a generalized concept that the functional DNA replication machinery is an important way to mitigate deleterious transcription–replication collisions. We speculate that there are common mechanisms at play in cancers experiencing abnormal replication stress and that transcription-mediated genome instability will play a role in tumor mutation accumulation. Recent studies showing that R-loop levels vary among specific cell types in normal primary samples, and that they increase in people carrying cancer risk alleles, raises the hope that the specific role for R-loops in oncogenesis will be elucidated (Zhang et al., 2017).
Materials and methods
Yeast growth and media
Yeast were cultured according to standard conditions in the indicated media at 30°C unless otherwise indicated. Growth curves were conducted in YPD media in 96-well plates using a TECAN M200. The area under the curve was used to compute expected and observed fitness values (Stirling et al., 2011, 2012). A list of yeast strains and plasmids used in this study can be found in Table S1.
Recombination and genome instability assays
Recombination events in L, LYΔNS, LNA, pARSHLB-IN, pARSHLB-OUT, pARSCLB-IN, pARSBLB-IN, L-lacZ, and GL-lacZ systems (gifts of A. Aguilera, CABIMER, Sevilla, Spain) were scored by counting leucine-positive colonies (González-Aguilera et al., 2008; Gómez-González et al., 2009, 2011; Herrera-Moyano et al., 2014). Recombination frequencies and cell viability were obtained from the mean value of three tests performed with 3–9 independent transformants each as described (Stirling et al., 2012). In assays where yeast strains were transformed with a recombination and an overexpression vector, recombination and viability plates maintained both plasmids. To measure rDNA stability, yeast with URA3 inserted into the rDNA locus (a gift of D. Koshland, University of California, Berkeley, Berkeley, CA) was treated as for the recombination assays except that loss of URA3 was measured by the frequency of 5′fluoroorotic acid–resistant colonies (Wahba et al., 2011). Cell viability was measured by growing test strains on SC minus uracil plates. To maintain plasmids, all aspects of the rDNA instability assay were done on media lacking leucine. Finally, frequencies of chromosome III loss were quantified in MATα haploid knockout collection strains using the ALF assay essentially as described (Ang et al., 2016). In brief, overnight cultures of haploid MATα cells were mixed 1:3 with a MATα mating tester strain, pelleted, and spotted in 100 µl of sterile water onto synthetic media (SD) lacking all amino acids, where only prototrophic diploids can grow. Mated ALFs form colonies on the SD plates. Cell viability of the overnight culture was determined by diluting cells 1:100,000, plating 100 µl on YPD, and counting colonies after 48 h. The frequency of mated colonies on the SD plate was expressed as a frequency of the total viable cells plated. Graphing and statistical analyses were done in GraphPad (Prism).
Yeast chromosome spreads and live cell imaging
Chromosome spreads were performed as previously described (Wahba et al., 2011; Chan et al., 2014a). In brief, midlog cells grown at 30°C in YPD were washed and spheroplasted in 1.2 M sorbitol, 0.1 M potassium phosphate, 0.5 M MgCl2, 10 mM dithiothreitol, and 150 µg/ml Zymolyase 20T, pH 7.0, for 20 min at 37°C (Chan et al., 2014a). Ice-cold stop solution (0.1 M 2-[N-morpholino] ethanesulfonic acid, 1 M sorbitol, 1 mM EDTA, and 0.5 mM MgCl2, pH 6.4) was added before lysis with 1% vol/vol Lipsol and fixation and spreading on glass slides in 4% wt/vol paraformaldehyde and 3.4% wt/vol sucrose. Spreads were incubated with a 1:1,000 dilution of S9.6 antibody (mouse; Kerafast) in blocking buffer (5% BSA and 0.2% skim milk powder in 1× PBS) overnight at 4°C before washing three times in blocking buffer and incubating with a 1:1,000 dilution of Cy3-conjugated goat anti–mouse antibody for 1 h (115-165-003; Jackson Laboratories). Slides were washed three times with blocking buffer and mounted in FluorSave mounting media (Calbiochem) before imaging. For each sample, at least 60 nuclei were visualized, and the nuclear fluorescent signal was quantified using ImageJ (Schneider et al., 2012). Each mutant was assayed in quadruplicate. For comparison purposes, the S9.6 median fluorescence intensity of the WT strain of each experiment was used for normalization. Mutants were compared with WT by the unpaired t test.
For live cell imaging, cells expressing Rad52-YFP were grown to logarithmic phase before any indicated treatments. Log-phase cells, treated or untreated, were bound to concanavalin-A–coated slides and imaged on a Leica dmi8 inverted fluorescence microscope using the appropriate filter sets (see Microscope image acquisition; Stirling et al., 2012).
Microscope image acquisition
Both yeast and human cell images were acquired using an Objective HCX PL APO 1.40 NA oil immersion 100× objective (Leica) on an inverted DMi8 microscope (Leica) equipped with a motorized Differential Interference Contrast imaging turret and a filter cube set for FITC/YFP/DAPI/TRITC for multicolor immunofluorescence. All images were captured at room temperature using a scientific complementary metal oxide semiconductor camera (ORCA Flash 4.0 V2; Hamamatsu) and collected using MetaMorph Premier acquisition software and postprocessed (including gamma adjustments, counting of cells with/without foci, and intensity measurements) using ImageJ. For all microscopy experiments, the significance of the differences was determined using Prism5 (GraphPad) or R. For intensity measurements, samples were compared with t tests or ANOVA; GraphPad performs F-tests for variance as part of this analysis. For comparisons of proportions, Fisher’s tests were used and p-values were Holm-Bonferroni–corrected in the event of multiple comparisons. Sample sizes were determined post hoc and are listed in the figure legends.
DRIP-chip and ChIP-chip analysis
DRIP- and ChIP-chip were generated and analyzed as described previously (Stirling et al., 2012; Chan et al., 2014a). For γ-H2A profiles, 5 µl of anti–γ-H2A antibody was used (rabbit; ab15083; Abcam), and profiles were normalized to an h2a-S129A mutant. Sgs1-flag profiles were generated using 4.2 µl of antibody flag (mouse; F3165; Sigma-Aldrich) and profiles were normalized to mock immunoprecipitates. Complete datasets can be found at ArrayExpress: E-MTAB-5582. Data were normalized using the rMAT software (Droit et al., 2010). All profiles were generated in duplicate with averaged and quantile normalized data used for plotting and calculating mean enrichment scores. Mean feature scores were generated by averaging all probes whose start sites fell within the start and end positions of the desired genomic feature. The same was done for Rrm3 peaks (coordinates derived from Herrera-Moyano et al., 2014). CHROMATRA plots were generated as described previously with genes aligned by their transcription start site (TSS) and sorted by length (Hentrich et al., 2012). Mean gene profiles were generated by averaging all probes that mapped to the genes of interest. Here, probes mapping to features of interest were split into 40 bins, and probes matching to the 1,500 bp of flanking sequences were split into 20 bins. Gene length mean gene profiles were generated by splitting all genes into gene-length classes and averaging probes in 150-bp increments. Enriched features were determined as those where at least 50% of the probes had values greater than 1.5. Only genes appearing in both replicates were considered. Comparing the length of enriched genes was done using a Mann–Whitney test (GraphPad).
Mutation accumulation and whole genome sequencing
Mutation accumulation experiments were conducted as described (Segovia et al., 2017). Single colonies from passage 40 were grown overnight in YPD to prepare genomic DNA by two rounds of phenol-chloroform extraction (Stirling et al., 2014). Whole genomes were sequenced using the Illumina HiSeq2500 platform, and sequence files were deposited at the NCBI sequence read archive (accession no. SRP094860 for sgs1Δ/Δ genomes, and no. SRP091984 for WT genomes). Read quality control, alignment to UCSC saccer3, and variant calling were performed exactly as described (Segovia et al., 2017). CNVs were detected using an in-house version of CNAseq (Jones et al., 2010) and Nexus copy number 7.5.2 (Biodiscovery Inc.). Variants were manually checked for read support using the Integrated Genomics Viewer.
Cell culture and transfection
HeLa cells were cultivated in DMEM (Stemcell Technologies), whereas HCT116 was grown in McCoy’s 5A media, both supplemented with 10% FBS (Life Technologies) in 5% CO2 at 37°C. Immortalized Bloom’s syndrome fibroblast lines (gift from J. Campisi, Buck Institute, Novato, CA) were grown as previously described (Davalos and Campisi, 2003). For RNA interference, cells were transfected with either single siRNA sequences targeting BLM (si-BLM, 5′-GCUAGGAGUCUGCGUGCCGA-3′), FANCD2 (si-FANCD2, 5′-GGUCAGAGCUGUAUUAUUC-3′; Blackford et al., 2015; Schwab et al., 2015), Luciferase GL3 Duplex as a control (si-Luc, 5′-GUUACGCUGAGUACUUCGA-3′), or siGENOME-SMARTpool siRNAs from Dharmacon (Non-targeting siRNA Pool 1 as si-Cont, si-BLM, si-FANCM, si-TOP3A, si-RECQL5, and si-WRN). Transfections were done with Dharmafect1 transfection reagent (Dharmacon) according to the manufacturer’s protocol and harvested 48 h after the siRNA administration. For experiments with overexpression of GFP or nuclear-targeting GFP-RNaseH1 (gift from R. Crouch, National Institutes of Health, Bethesda, MD), transfections were performed with Lipofectamine 3000 (Invitrogen) according to manufacturer’s instructions 24 h after the siRNA transfections.
Immunofluorescence
For S9.6 staining, cells were grown on coverslips overnight before siRNA transfection and plasmid overexpression. 48 h after siRNA transfection, cells were washed with PBS, fixed with ice-cold methanol for 10 min, and permeabilized with ice-cold acetone for 1 min. After PBS wash, cells were blocked in 3% BSA and 0.1% Tween 20 in 4× SSC buffer for 1 h at room temperature. Cells were then incubated with primary antibody S9.6 (1:500; mouse, ENH001; Kerafast) overnight at 4°C. For HCT116, nucleolin was costained by coincubating with antinucleolin (rabbit; ab22758; Abcam) at 1:1,000. Cells were then washed three times in PBS and stained with mouse Alexa-Fluoro-568-conjugated secondary antibody (1:1,000; Life Technologies) for 1 h at room temperature, washed three times in PBS, and stained with DAPI for 5 min. Cells were imaged on LeicaDMI8 microscope at 100×, and ImageJ was used for processing and quantification of S9.6 intensity in images. Only GFP-positive cells were quantified, and micronuclei were counted in asynchronous cells from the same slides. For γ-H2AX and FANCD2 foci, the immunostaining was performed the same way with the differences of fixation with 4% paraformaldehyde for 15 min and permeabilization with 0.2% Triton X-100 for 5 min on ice. Primary antibodies for γ-H2AX (rabbit; ab81299; Abcam), FANCD2 (rabbit; NB100-182SS; Novus) and rabbit Alexa-Fluoro-568-conjugated secondary antibody were all diluted 1:1,000. Where indicated, cells were treated with DMSO or 2 mM HU (Sigma-Aldrich) for 2 h before fixing.
Neutral comet assay
The neutral comet assay was performed using the CometAssay Reagent kit for Single Cell Gel Electrophoresis Assay (Trevigen) in accordance with the manufacturer’s instructions. Electrophoresis was performed at 4°C, and slides were stained with PI and imaged on LeicaDMI8 microscope at 20×. Comet tail moments were obtained using an ImageJ plugin as previously described (Mathew et al., 2014). At least 50 cells per sample were analyzed from each independent experiment.
Western blotting
Whole cell lysates were prepared with RIPA buffer containing protease inhibitor (Sigma-Aldrich) and phosphatase inhibitor (Roche Applied Science) cocktail tablets, and the protein concentration were determined by Bio-Rad Protein assay (Bio-Rad). Equivalent amounts of protein were resolved by SDS-PAGE and transferred to polyvinylidene fluoride microporous membrane (Millipore), blocked with 5% skim milk in TBS containing 0.1% Tween 20, and membranes were probed with the following antibodies: BLM (rabbit; ab2179; Abcam), FANCM (rabbit; ab95014; Abcam), GAPDH (mouse; MA5-15738; Thermo Fisher Scientific), FANCD2 (rabbit; NB100-182SS; Novus), RECQL5 (rabbit; A302-520A-T; Bethyl), WRN (rabbit; A300-238A-T; Bethyl), TOP3A (rabbit; 14525–1-AP; Proteintech), and α-tubulin (mouse; 32-2500; Life Technologies). Secondary antibodies were conjugated to HRP, and peroxidase activity was visualized using Chemiluminescent HRP substrate (Thermo Fisher Scientific).
Proximity ligation assay
Cells were grown on coverslips, washed with PBS, and fixed with 4% paraformaldehyde for 15 min. After permeabilization with 0.2% Triton X-100 for 5 min, cells were blocked in 3% BSA, 0.1% Tween 20 in 4× SSC for 1 h at room temperature. Cells were then incubated with primary antibody overnight at 4°C (1:500 rabbit BLM antibody; PLA0029; Sigma-Aldrich,) as negative control; 1:200 mouse S9.6 antibody as negative control; 1:1,000 rabbit BLM with 1:200 mouse S9.6; or 1:1,000 rabbit γ-H2AX (Abcam) with 1:200 mouse S9.6 as positive control. After washing with 1× PBS twice, cells were incubated with premixed PLA probe antimouse minus and PLA probe antirabbit plus (Sigma-Aldrich) for 1 h at 37°C. Binding of PLA probes, ligation, and amplification was performed with the reagents from the Duolink In Situ kit (Sigma-Aldrich) according to the manufacturer’s instructions. Slides were mounted in Duolink In Situ Mounting Medium with DAPI and imaged on LeicaDMI8 microscope at 100×.
In vitro helicase assay
BLM was tagged at the N-terminus with GST and at the C-terminus with His10 and purified as described for PALB2 (Buisson et al., 2010). Sgs1 was purified according to Cejka and Kowalczykowski (2010), with the following modifications. pFB-MBP-Sgs1-His vector (provided by P. Cejka, Institute for Research in Biomedicine, Bellinzona, Switzerland) was used to generate baculoviruses using the Bac-to-Bac system (Invitrogen). Sgs1 was purified from 1 liter of baculovirus-infected Sf9 cells. After PreScission cleavage in P5 buffer (50 mM NaHPO4, pH 7.0, 500 mM NaCl, 10% glycerol, 0.05% Triton X-100; and 5 mM imidazole), proteins were bound to TALON Metal Affinity Resin (Clontech). The resin was washed twice with P30 buffer (50 mM NaHPO4, pH 7.0, 500 mM NaCl, 10% glycerol, 0.05% Triton X-100, and 30 mM imidazole) and eluted with P500 buffer (50 mM NaHPO4, pH 7.0, 500 mM NaCl, 10% glycerol, 0.05% Triton-X-100, and 500 mM imidazole). Sgs1 was dialyzed in storage buffer (20 mM Tris-Cl, pH 7.4, 200 mM NaCl, 10% glycerol, and 1 mM DTT).
After purification, both proteins appeared as a single homogenous band on an SDS-PAGE gel. R-LOOP and D-LOOP substrates were generated by annealing purified oligonucleotides: DNA strand 1, 5′-GGGTGAACCTGCAGGTGGGCGGCTGCTCATCGTAGGTTAGTTGGTAGAATTCGGCAGCGTC-3′; DNA strand 2, 5′-GACGCTGCCGAATTCTACCAGTGCCTTGCTAGGACATCTTTGCCCACCTGCAGGTTCACCC-3′; with either RNA, 5′-AAAGAUGUCCUAGCAAGGCAC-3′, or DNA, 5′-AAAGATGTCCTAGCAAGGCAC-3′. Unwinding assays were performed in MOPS buffer (25 mM MOPS, pH 7.0, 60 mM KCl, 0.2% Tween-20, 2 mM DTT, 2 mM ATP, and 2 mM MgCl2). BLM or Sgs1 and labeled R-LOOP or D-LOOP (100 nM) substrates were incubated in MOPS buffer for 20 min at 37°C, followed by deproteinization in one-fifth volume of stop buffer (20 mM Tris-Cl, pH 7.5, and 2 mg/ml proteinase K) for 20 min at 37°C. Reactions were loaded on an 8% acrylamide gel, run at 150 V for 120 min, dried onto filter paper, and autoradiographed.
Online supplemental material
Fig. S1 shows additional quantification of ALF phenotypes for yeast mutants. Fig. S2 highlights additional analyses of Sgs1 binding to the yeast genome and the position of R-loops and γ-H2A relative to telomeres and rDNA, as well as an rDNA instability assay. Fig. S3 shows BLM levels in different cell types, S9.6 staining data for an additional BLM knockout cell line, and example images and quantification of micronucleus formation in BLM knockdown cells. Fig. S3 also shows Western blots of the various knockdown experiments in Fig. 7. Fig. S4 shows the accumulation of FANCD2 foci in HU-treated cells, and the impact of BLM knockdown on this phenotype. Table S1 is a summary of yeast strains and plasmids used in this study. Table S2 summarizes the positions of mutations, CNVs, and indels detected in the sgs1Δ mutation accumulation experiments.
Acknowledgments
We acknowledge Andres Aguilera, Judith Campisi, Peter Cejka, Frederic Chedin, Karlene Cimprich, Robert Crouch, and Doug Koshland for providing reagents, cell lines and methods. We thank Philip Hieter, in whose laboratory some of the early work was conceived.
This research is funded by the Canadian Cancer Society (grant 703263), the Canadian Institutes of Health Research (grant MOP-136982 to P.C. Stirling and project grant 363317 to J.-Y. Masson), and the Natural Sciences and Engineering Research Council of Canada (grant RGPIN 402095-11 to M.S. Kobor). P.C. Stirling is a Canadian Institutes of Health Research New Investigator and Michael Smith Foundation for Health Research Scholar. J.-Y.M. is a Fond de Recherche du Québec - Santé Chair in genome stability.
The authors declare no competing financial interests.
Author contributions: E.Y.-C. Chang, C.A. Novoa, M.J. Aristizabal, Y. Coulombe, R. Segovia, R. Chaturvedi, C. Keong, and A.S. Tam performed the experiments and analyzed data. Y. Shen provided formal data analysis. S.J.M. Jones, J.-Y. Masson, M.S. Kobor, and P.C. Stirling provided the infrastructure, supervision, and funding. E.Y.-C. Chang, C.A. Novoa, M.J. Aristizabal, J.-Y. Masson, and P.C. Stirling conceived of and designed the experiments. E.Y.-C. Chang, C.A. Novoa, M.J. Aristizabal, and P.C. Stirling wrote the manuscript.
Footnotes
Abbreviations used:
- ALF
- A-like Faker
- CNV
- copy number variant
- FA
- Fanconi anemia
- HR
- homologous recombination
- HU
- hydroxyurea
- PLA
- proximity ligation assay
- rDNA
- ribosomal DNA
- SNV
- single-nucleotide variant
- TSS
- transcription start site
References
- Ang J.S., Duffy S., Segovia R., Stirling P.C., and Hieter P.. 2016. Dosage Mutator Genes in Saccharomyces cerevisiae: A Novel Mutator Mode-of-action of the Mph1 DNA Helicase. Genetics. 204:975–986. 10.1534/genetics.116.192211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashton T.M., and Hickson I.D.. 2010. Yeast as a model system to study RecQ helicase function. DNA Repair (Amst.). 9:303–314. 10.1016/j.dnarep.2009.12.007 [DOI] [PubMed] [Google Scholar]
- Bhatia V., Barroso S.I., García-Rubio M.L., Tumini E., Herrera-Moyano E., and Aguilera A.. 2014. BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature. 511:362–365. 10.1038/nature13374 [DOI] [PubMed] [Google Scholar]
- Blackford A.N., Nieminuszczy J., Schwab R.A., Galanty Y., Jackson S.P., and Niedzwiedz W.. 2015. TopBP1 interacts with BLM to maintain genome stability but is dispensable for preventing BLM degradation. Mol. Cell. 57:1133–1141. 10.1016/j.molcel.2015.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhm S., and Bernstein K.A.. 2014. The role of post-translational modifications in fine-tuning BLM helicase function during DNA repair. DNA Repair (Amst.). 22:123–132. 10.1016/j.dnarep.2014.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton S., Dernoncourt E., Delteil C., Froment C., Schiltz O., Salles B., Frit P., and Calsou P.. 2014. DNA damage triggers SAF-A and RNA biogenesis factors exclusion from chromatin coupled to R-loops removal. Nucleic Acids Res. 42:9047–9062. 10.1093/nar/gku601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryk M., Banerjee M., Conte D. Jr., and Curcio M.J.. 2001. The Sgs1 helicase of Saccharomyces cerevisiae inhibits retrotransposition of Ty1 multimeric arrays. Mol. Cell. Biol. 21:5374–5388. 10.1128/MCB.21.16.5374-5388.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buisson R., Dion-Côté A.M., Coulombe Y., Launay H., Cai H., Stasiak A.Z., Stasiak A., Xia B., and Masson J.Y.. 2010. Cooperation of breast cancer proteins PALB2 and piccolo BRCA2 in stimulating homologous recombination. Nat. Struct. Mol. Biol. 17:1247–1254. 10.1038/nsmb.1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejka P., and Kowalczykowski S.C.. 2010. The full-length Saccharomyces cerevisiae Sgs1 protein is a vigorous DNA helicase that preferentially unwinds holliday junctions. J. Biol. Chem. 285:8290–8301. 10.1074/jbc.M109.083196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerritelli S.M., and Crouch R.J.. 2009. Ribonuclease H: the enzymes in eukaryotes. FEBS J. 276:1494–1505. 10.1111/j.1742-4658.2009.06908.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty P., and Grosse F.. 2010. WRN helicase unwinds Okazaki fragment-like hybrids in a reaction stimulated by the human DHX9 helicase. Nucleic Acids Res. 38:4722–4730. 10.1093/nar/gkq240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Y.A., Aristizabal M.J., Lu P.Y., Luo Z., Hamza A., Kobor M.S., Stirling P.C., and Hieter P.. 2014a Genome-wide profiling of yeast DNA:RNA hybrid prone sites with DRIP-chip. PLoS Genet. 10:e1004288 10.1371/journal.pgen.1004288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Y.A., Hieter P., and Stirling P.C.. 2014b Mechanisms of genome instability induced by RNA-processing defects. Trends Genet. 30:245–253. 10.1016/j.tig.2014.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang E.Y., and Stirling P.C.. 2017. Replication Fork Protection Factors Controlling R-Loop Bypass and Suppression. Genes (Basel). 8:33 10.3390/genes8010033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury I., Sareen A., Raghunandan M., and Sobeck A.. 2013. FANCD2 regulates BLM complex functions independently of FANCI to promote replication fork recovery. Nucleic Acids Res. 41:6444–6459. 10.1093/nar/gkt348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chávez S., Beilharz T., Rondón A.G., Erdjument-Bromage H., Tempst P., Svejstrup J.Q., Lithgow T., and Aguilera A.. 2000. A protein complex containing Tho2, Hpr1, Mft1 and a novel protein, Thp2, connects transcription elongation with mitotic recombination in Saccharomyces cerevisiae. EMBO J. 19:5824–5834. 10.1093/emboj/19.21.5824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chon H., Sparks J.L., Rychlik M., Nowotny M., Burgers P.M., Crouch R.J., and Cerritelli S.M.. 2013. RNase H2 roles in genome integrity revealed by unlinking its activities. Nucleic Acids Res. 41:3130–3143. 10.1093/nar/gkt027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb J.A., Bjergbaek L., Shimada K., Frei C., and Gasser S.M.. 2003. DNA polymerase stabilization at stalled replication forks requires Mec1 and the RecQ helicase Sgs1. EMBO J. 22:4325–4336. 10.1093/emboj/cdg391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croteau D.L., Popuri V., Opresko P.L., and Bohr V.A.. 2014. Human RecQ helicases in DNA repair, recombination, and replication. Annu. Rev. Biochem. 83:519–552. 10.1146/annurev-biochem-060713-035428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtin N.J. 2012. DNA repair dysregulation from cancer driver to therapeutic target. Nat. Rev. Cancer. 12:801–817. 10.1038/nrc3399 [DOI] [PubMed] [Google Scholar]
- Davalos A.R., and Campisi J.. 2003. Bloom syndrome cells undergo p53-dependent apoptosis and delayed assembly of BRCA1 and NBS1 repair complexes at stalled replication forks. J. Cell Biol. 162:1197–1209. 10.1083/jcb.200304016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Renty C., and Ellis N.A.. 2017. Bloom’s syndrome: Why not premature aging?: A comparison of the BLM and WRN helicases. Ageing Res. Rev. 33:36–51. 10.1016/j.arr.2016.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droit A., Cheung C., and Gottardo R.. 2010. rMAT--an R/Bioconductor package for analyzing ChIP-chip experiments. Bioinformatics. 26:678–679. 10.1093/bioinformatics/btq023 [DOI] [PubMed] [Google Scholar]
- Duquette M.L., Handa P., Vincent J.A., Taylor A.F., and Maizels N.. 2004. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev. 18:1618–1629. 10.1101/gad.1200804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Pichardo D., Canas J.C., Garcia-Rubio M.L., Gomez-Gonzalez B., Rondon A.G., and Aguilera A.. 2017. Histone Mutants Separate R Loop Formation from Genome Instability Induction. Mol. Cell. 66:597–609. 10.1016/j.molcel.2017.05.014 [DOI] [PubMed] [Google Scholar]
- García-Rubio M.L., Pérez-Calero C., Barroso S.I., Tumini E., Herrera-Moyano E., Rosado I.V., and Aguilera A.. 2015. The Fanconi Anemia Pathway Protects Genome Integrity from R-loops. PLoS Genet. 11:e1005674 10.1371/journal.pgen.1005674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-González B., Felipe-Abrio I., and Aguilera A.. 2009. The S-phase checkpoint is required to respond to R-loops accumulated in THO mutants. Mol. Cell. Biol. 29:5203–5213. 10.1128/MCB.00402-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-González B., García-Rubio M., Bermejo R., Gaillard H., Shirahige K., Marín A., Foiani M., and Aguilera A.. 2011. Genome-wide function of THO/TREX in active genes prevents R-loop-dependent replication obstacles. EMBO J. 30:3106–3119. 10.1038/emboj.2011.206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Aguilera C., Tous C., Gómez-González B., Huertas P., Luna R., and Aguilera A.. 2008. The THP1-SAC3-SUS1-CDC31 complex works in transcription elongation-mRNA export preventing RNA-mediated genome instability. Mol. Biol. Cell. 19:4310–4318. 10.1091/mbc.E08-04-0355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grierson P.M., Lillard K., Behbehani G.K., Combs K.A., Bhattacharyya S., Acharya S., and Groden J.. 2012. BLM helicase facilitates RNA polymerase I-mediated ribosomal RNA transcription. Hum. Mol. Genet. 21:1172–1183. 10.1093/hmg/ddr545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatchi E., Skourti-Stathaki K., Ventz S., Pinello L., Yen A., Kamieniarz-Gdula K., Dimitrov S., Pathania S., McKinney K.M., Eaton M.L., et al. . 2015. BRCA1 recruitment to transcriptional pause sites is required for R-loop-driven DNA damage repair. Mol. Cell. 57:636–647. 10.1016/j.molcel.2015.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmrich A., Ballarino M., and Tora L.. 2011. Collisions between replication and transcription complexes cause common fragile site instability at the longest human genes. Mol. Cell. 44:966–977. 10.1016/j.molcel.2011.10.013 [DOI] [PubMed] [Google Scholar]
- Hentrich T., Schulze J.M., Emberly E., and Kobor M.S.. 2012. CHROMATRA: a Galaxy tool for visualizing genome-wide chromatin signatures. Bioinformatics. 28:717–718. 10.1093/bioinformatics/bts007 [DOI] [PubMed] [Google Scholar]
- Herrera-Moyano E., Mergui X., García-Rubio M.L., Barroso S., and Aguilera A.. 2014. The yeast and human FACT chromatin-reorganizing complexes solve R-loop-mediated transcription-replication conflicts. Genes Dev. 28:735–748. 10.1101/gad.234070.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z., Zhang A., Storz G., Gottesman S., and Leppla S.H.. 2006. An antibody-based microarray assay for small RNA detection. Nucleic Acids Res. 34:e52 10.1093/nar/gkl142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P., and Aguilera A.. 2003. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol. Cell. 12:711–721. 10.1016/j.molcel.2003.08.010 [DOI] [PubMed] [Google Scholar]
- Jones S.J., Laskin J., Li Y.Y., Griffith O.L., An J., Bilenky M., Butterfield Y.S., Cezard T., Chuah E., Corbett R., et al. . 2010. Evolution of an adenocarcinoma in response to selection by targeted kinase inhibitors. Genome Biol. 11:R82 10.1186/gb-2010-11-8-r82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi R.S., Piña B., and Roca J.. 2012. Topoisomerase II is required for the production of long Pol II gene transcripts in yeast. Nucleic Acids Res. 40:7907–7915. 10.1093/nar/gks626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N., and Jinks-Robertson S.. 2011. Guanine repeat-containing sequences confer transcription-dependent instability in an orientation-specific manner in yeast. DNA Repair (Amst.). 10:953–960. 10.1016/j.dnarep.2011.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.K., Johnson R.E., Yu S.L., Prakash L., and Prakash S.. 1999. Requirement of yeast SGS1 and SRS2 genes for replication and transcription. Science. 286:2339–2342. 10.1126/science.286.5448.2339 [DOI] [PubMed] [Google Scholar]
- Li X., and Manley J.L.. 2005. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 122:365–378. 10.1016/j.cell.2005.06.008 [DOI] [PubMed] [Google Scholar]
- Ling C., Huang J., Yan Z., Li Y., Ohzeki M., Ishiai M., Xu D., Takata M., Seidman M., and Wang W.. 2016. Bloom syndrome complex promotes FANCM recruitment to stalled replication forks and facilitates both repair and traverse of DNA interstrand crosslinks. Cell Discov. 2:16047 10.1038/celldisc.2016.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas C., Savic V., Bekker-Jensen S., Doil C., Neumann B., Pedersen R.S., Grøfte M., Chan K.L., Hickson I.D., Bartek J., and Lukas J.. 2011. 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat. Cell Biol. 13:243–253. 10.1038/ncb2201 [DOI] [PubMed] [Google Scholar]
- Mathew V., Pauleau A.L., Steffen N., Bergner A., Becker P.B., and Erhardt S.. 2014. The histone-fold protein CHRAC14 influences chromatin composition in response to DNA damage. Cell Reports. 7:321–330. 10.1016/j.celrep.2014.03.008 [DOI] [PubMed] [Google Scholar]
- Mischo H.E., Gómez-González B., Grzechnik P., Rondón A.G., Wei W., Steinmetz L., Aguilera A., and Proudfoot N.J.. 2011. Yeast Sen1 helicase protects the genome from transcription-associated instability. Mol. Cell. 41:21–32. 10.1016/j.molcel.2010.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundbjerg K., Jørgensen S.W., Fredsøe J., Nielsen I., Pedersen J.M., Bentsen I.B., Lisby M., Bjergbaek L., and Andersen A.H.. 2015. Top2 and Sgs1-Top3 Act Redundantly to Ensure rDNA Replication Termination. PLoS Genet. 11:e1005697 10.1371/journal.pgen.1005697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung K., Datta A., Chen C., and Kolodner R.D.. 2001. SGS1, the Saccharomyces cerevisiae homologue of BLM and WRN, suppresses genome instability and homeologous recombination. Nat. Genet. 27:113–116. 10.1038/83673 [DOI] [PubMed] [Google Scholar]
- Naim V., and Rosselli F.. 2009. The FANC pathway and BLM collaborate during mitosis to prevent micro-nucleation and chromosome abnormalities. Nat. Cell Biol. 11:761–768. 10.1038/ncb1883 [DOI] [PubMed] [Google Scholar]
- Neff N.F., Ellis N.A., Ye T.Z., Noonan J., Huang K., Sanz M., and Proytcheva M.. 1999. The DNA helicase activity of BLM is necessary for the correction of the genomic instability of bloom syndrome cells. Mol. Biol. Cell. 10:665–676. 10.1091/mbc.10.3.665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell K., Jinks-Robertson S., and Petes T.D.. 2015. Elevated Genome-Wide Instability in Yeast Mutants Lacking RNase H Activity. Genetics. 201:963–975. 10.1534/genetics.115.182725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panneerselvam J., Wang H., Zhang J., Che R., Yu H., and Fei P.. 2016. BLM promotes the activation of Fanconi Anemia signaling pathway. Oncotarget. 7:32351–32361. 10.18632/oncotarget.8707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popuri V., Bachrati C.Z., Muzzolini L., Mosedale G., Costantini S., Giacomini E., Hickson I.D., and Vindigni A.. 2008. The Human RecQ helicases, BLM and RECQ1, display distinct DNA substrate specificities. J. Biol. Chem. 283:17766–17776. 10.1074/jbc.M709749200 [DOI] [PubMed] [Google Scholar]
- Prado F., Piruat J.I., and Aguilera A.. 1997. Recombination between DNA repeats in yeast hpr1delta cells is linked to transcription elongation. EMBO J. 16:2826–2835. 10.1093/emboj/16.10.2826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Pereira J.M., Herrero A.B., García-Rubio M.L., Marín A., Moreno S., and Aguilera A.. 2013. The Npl3 hnRNP prevents R-loop-mediated transcription-replication conflicts and genome instability. Genes Dev. 27:2445–2458. 10.1101/gad.229880.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saponaro M., Kantidakis T., Mitter R., Kelly G.P., Heron M., Williams H., Söding J., Stewart A., and Svejstrup J.Q.. 2014. RECQL5 controls transcript elongation and suppresses genome instability associated with transcription stress. Cell. 157:1037–1049. 10.1016/j.cell.2014.03.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt K.H., and Kolodner R.D.. 2004. Requirement of Rrm3 helicase for repair of spontaneous DNA lesions in cells lacking Srs2 or Sgs1 helicase. Mol. Cell. Biol. 24:3213–3226. 10.1128/MCB.24.8.3213-3226.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., and Eliceiri K.W.. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 9:671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R.A., Nieminuszczy J., Shah F., Langton J., Lopez Martinez D., Liang C.C., Cohn M.A., Gibbons R.J., Deans A.J., and Niedzwiedz W.. 2015. The Fanconi Anemia Pathway Maintains Genome Stability by Coordinating Replication and Transcription. Mol. Cell. 60:351–361. 10.1016/j.molcel.2015.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segovia R., Shen Y., Lujan S.A., Jones S.J., and Stirling P.C.. 2017. Hypermutation signature reveals a slippage and realignment model of translesion synthesis by Rev3 polymerase in cisplatin-treated yeast. Proc. Natl. Acad. Sci. USA. 114:2663–2668. 10.1073/pnas.1618555114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skourti-Stathaki K., Proudfoot N.J., and Gromak N.. 2011. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol. Cell. 42:794–805. 10.1016/j.molcel.2011.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollier J., Stork C.T., García-Rubio M.L., Paulsen R.D., Aguilera A., and Cimprich K.A.. 2014. Transcription-coupled nucleotide excision repair factors promote R-loop-induced genome instability. Mol. Cell. 56:777–785. 10.1016/j.molcel.2014.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling P.C., Bloom M.S., Solanki-Patil T., Smith S., Sipahimalani P., Li Z., Kofoed M., Ben-Aroya S., Myung K., and Hieter P.. 2011. The complete spectrum of yeast chromosome instability genes identifies candidate CIN cancer genes and functional roles for ASTRA complex components. PLoS Genet. 7:e1002057 10.1371/journal.pgen.1002057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling P.C., Chan Y.A., Minaker S.W., Aristizabal M.J., Barrett I., Sipahimalani P., Kobor M.S., and Hieter P.. 2012. R-loop-mediated genome instability in mRNA cleavage and polyadenylation mutants. Genes Dev. 26:163–175. 10.1101/gad.179721.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling P.C., Shen Y., Corbett R., Jones S.J., and Hieter P.. 2014. Genome destabilizing mutator alleles drive specific mutational trajectories in Saccharomyces cerevisiae. Genetics. 196:403–412. 10.1534/genetics.113.159806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stork C.T., Bocek M., Crossley M.P., Sollier J., Sanz L.A., Chédin F., Swigut T., and Cimprich K.A.. 2016. Co-transcriptional R-loops are the main cause of estrogen-induced DNA damage. eLife. 5:e17548 10.7554/eLife.17548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratton M.R., Campbell P.J., and Futreal P.A.. 2009. The cancer genome. Nature. 458:719–724. 10.1038/nature07943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhasini A.N., and Brosh R.M. Jr. 2012. Fanconi anemia and Bloom’s syndrome crosstalk through FANCJ-BLM helicase interaction. Trends Genet. 28:7–13. 10.1016/j.tig.2011.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran P.L.T., Pohl T.J., Chen C.F., Chan A., Pott S., and Zakian V.A.. 2017. PIF1 family DNA helicases suppress R-loop mediated genome instability at tRNA genes. Nat. Commun. 8:15025 10.1038/ncomms15025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tresini M., Warmerdam D.O., Kolovos P., Snijder L., Vrouwe M.G., Demmers J.A., van IJcken W.F., Grosveld F.G., Medema R.H., Hoeijmakers J.H., et al. . 2015. The core spliceosome as target and effector of non-canonical ATM signalling. Nature. 523:53–58. 10.1038/nature14512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumini E., Barroso S., -Calero C.P., and Aguilera A.. 2016. Roles of human POLD1 and POLD3 in genome stability. Sci. Rep. 6:38873 10.1038/srep38873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versini G., Comet I., Wu M., Hoopes L., Schwob E., and Pasero P.. 2003. The yeast Sgs1 helicase is differentially required for genomic and ribosomal DNA replication. EMBO J. 22:1939–1949. 10.1093/emboj/cdg180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayraghavan S., Tsai F.L., and Schwacha A.. 2016. A Checkpoint-Related Function of the MCM Replicative Helicase Is Required to Avert Accumulation of RNA:DNA Hybrids during S-phase and Ensuing DSBs during G2/M. PLoS Genet. 12:e1006277 10.1371/journal.pgen.1006277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahba L., Amon J.D., Koshland D., and Vuica-Ross M.. 2011. RNase H and multiple RNA biogenesis factors cooperate to prevent RNA:DNA hybrids from generating genome instability. Mol. Cell. 44:978–988. 10.1016/j.molcel.2011.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahba L., Gore S.K., and Koshland D.. 2013. The homologous recombination machinery modulates the formation of RNA-DNA hybrids and associated chromosome instability. eLife. 2:e00505 10.7554/eLife.00505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Chiang H.C., Wang Y., Zhang C., Smith S., Zhao X., Nair S.J., Michalek J., Jatoi I., Lautner M., et al. . 2017. Attenuation of RNA polymerase II pausing mitigates BRCA1-associated R-loop accumulation and tumorigenesis. Nat. Commun. 8:15908 10.1038/ncomms15908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer A.D., and Koshland D.. 2016. Differential roles of the RNases H in preventing chromosome instability. Proc. Natl. Acad. Sci. USA. 113:12220–12225. 10.1073/pnas.1613448113 [DOI] [PMC free article] [PubMed] [Google Scholar]