Establishment of apicobasal polarity is critical for the lumenal epiblast-like morphogenesis of human pluripotent stem cells (hPSC). Taniguchi et al. show that apical polarization begins on the interior of single hPSC, through dynamic formation of a highly organized, apically charged perinuclear complex called the apicosome.
Abstract
Human pluripotent stem cells (hPSCs) self-organize into apicobasally polarized cysts, reminiscent of the lumenal epiblast stage, providing a model to explore key morphogenic processes in early human embryos. Here, we show that apical polarization begins on the interior of single hPSCs through the dynamic formation of a highly organized perinuclear apicosome structure. The membrane surrounding the apicosome is enriched in apical markers and displays microvilli and a primary cilium; its lumenal space is rich in Ca2+. Time-lapse imaging of isolated hPSCs reveals that the apicosome forms de novo in interphase, retains its structure during mitosis, is asymmetrically inherited after mitosis, and relocates to the recently formed cytokinetic plane, where it establishes a fully polarized lumen. In a multicellular aggregate of hPSCs, intracellular apicosomes from multiple cells are trafficked to generate a common lumenal cavity. Thus, the apicosome is a unique preassembled apical structure that can be rapidly used in single or clustered hPSCs to initiate self-organized apical polarization and lumenogenesis.
Introduction
Epiblast cavity formation is essential for the morphogenesis and survival of the embryo as it implants in the uterine wall. However, because of ethical concerns, this event remains difficult to study in the human embryo. Recently, two groups demonstrated that cultured human blastocysts, generated by in vitro fertilization, can self-organize and polarize to form lumenal proamniotic cavities in the absence of cues from maternal tissues (Deglincerti et al., 2016; Shahbazi et al., 2016). However, limited availability of such in vitro fertilization samples and the inability to perform mechanistic analyses in this type of model continue to hinder studies of peri-implantation human development.
Studies from our laboratory and others reveal that the property of self-organization extends to singly plated human pluripotent stem cells (hPSCs), which readily polarize to form cysts (hPSC cyst) with a central apically marked lumen upon the first cell division (Taniguchi et al., 2015; Shahbazi et al., 2016). During further mitotic expansion of such cysts, all cells retain expression of pluripotency markers. Thus, this process resembles expansion of the lumenal epiblast (proamniotic) cavity of cultured human embryos, and it has been suggested that the molecular pathways driving the polarization of hPSCs in culture may also be involved in epiblast cavity formation in vivo (Taniguchi et al., 2015; Shahbazi et al., 2016; Simunovic and Brivanlou, 2017). Indeed, we recently demonstrated that such hPSC cysts can give rise to squamous amnion-like cells as well as postimplantation amniotic sac–like structures when grown in a specifically engineered environment (Shao et al., 2017a,b), further demonstrating that hPSC cysts self-organize to recapitulate developmental processes associated with the epiblast in vivo.
Although hPSCs indeed display an intrinsic ability to efficiently form lumenal cysts, it is not clear how apical polarization initiates in this model. Here, we show that lumen formation begins on the interior of single cells, with the formation of an apicosome: a highly organized intracellular membrane-bound apical lumenal compartment studded with microvilli and a primary cilium. Time-lapse imaging reveals that the apicosome forms de novo during interphase. In single cells, the apicosome survives through mitosis, is asymmetrically inherited upon cytokinesis, and relocates to the cytokinetic plane following cytokinesis. When hPSCs are plated as aggregates rather than single cells, apicosomes generated in multiple individual cells fuse to generate a single central lumen. We conclude that the apicosome is a major driver of epiblast-like lumen formation in hPSC.
Results and discussion
Formation of an intracellular apically enriched perinuclear complex in dissociated hPSCs
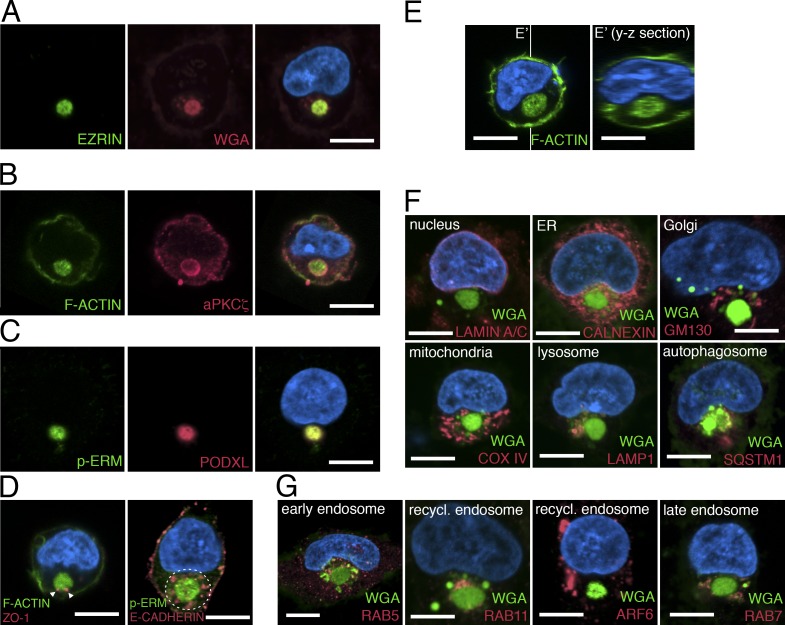
To identify the earliest sign of apical polarization during hPSC lumen formation, we first examined the initial stages of this process in dissociated single hPSCs. 20 h after plating singly isolated H9 (WA09) cells in the presence of the rho-associated kinase inhibitor (ROCK-i), Y-27632 (essential for preventing apoptosis associated with cellular dissociation), 45.33 ± 2.4% of the cells had divided, and the majority (>60%) of two-cell clones showed a single central lumen, centered within the recently formed cytokinetic plane. The lumen was enriched in apical proteins, including F-ACTIN (phalloidin+) and EZRIN (an actin-binding protein), as previously reported (Taniguchi et al., 2015). Of the remaining undivided single cells, 40% contained a perinuclear accumulation of multiple apical markers: F-ACTIN, EZRIN, phosphorylated EZRIN/RADIXIN/MOESIN (p-ERM), atypical PKCζ (aPKCζ), and PODOCALYXIN (PODXL; Fig. 1, A–C; Bryant et al., 2010; Rodríguez-Fraticelli and Martín-Belmonte, 2013; Rodriguez-Boulan and Macara, 2014). This apically enriched structure was membranous, as revealed by WGA staining, a marker for outer cell membrane (Fig. 1 A and Fig. S1 A). The tight junction marker ZO1 was also associated with this perinuclear complex, and the entire structure was surrounded by the basolateral protein E-CADHERIN (Fig. 1 D). Time-course analysis revealed that the number of single cells containing an EZRIN+ perinuclear complex increased from <5% at 30 min after plating single cells to >40% at 20 h (Fig. S1 B). The EZRIN+ structure was entirely intracellular, as optical sectioning (y-z plane) of phalloidin-stained cells demonstrated no obvious connections to the extracellular milieu (Fig. 1 E). Importantly, nuclear expression of the pluripotency markers NANOG and POU5F1 was maintained in cells containing this apical structure (Fig. S1 C). A similar structure also forms in H7 (WA07) and the 1196a human induced pluripotent stem cell (iPSC) line (Chen et al., 2014) as well as in H9 cells grown in several different media formulations and substrates that are used to culture hPSCs, including a recently described laminin-521/E-cadherin matrix substrate that sustains pluripotency and the viability of singly isolated hPSCs in the absence of ROCK-i (Rodin et al., 2014b; Fig. S1, D–H).
Figure 1.
The apicosome is a unique cellular structure. (A–D) Fluorescent confocal images of single H9 cells stained with indicated markers. (E) Fluorescent confocal images of single H9 cells stained for phalloidin (green, F-ACTIN). Image in E′ is the optical y-z section (indicated by white lines) of the 3D rendered image of the cell in E. (F and G) Singly isolated H9 cells are stained for membrane (green, WGA) as well as antibodies specific to distinct organelles (red) as indicated. Bar, 10 µm. For all images, blue indicates DNA staining (HOECHST).
hPSCs share similar developmental and epigenetic characteristics with epiblast cells of peri-implantation mouse embryos (Tesar et al., 2007; De Los Angeles et al., 2015). Interestingly, a recent analysis of polarization in mouse blastocysts revealed apicosome-like intracellular accumulations of Podxl (see Fig. 4 A in Bedzhov and Zernicka-Goetz, 2014). We confirmed those observations in E4.5 mouse blastocysts (Fig. S1 I and Video 1; 7 of 16 cells). However, no such intracellular apical protein accumulations were seen in blastocysts at E3.5 and E4.0 (Fig. S1 J). To further examine this stage-specific formation of apicosome-like structure, we examined whether such structures form in isolated mouse epiblast stem cells (mEpiSCs; epiblast-like) as well as in mouse embryonic stem cells (mESC; inner cell mass-like). Accumulations of the apical marker aPKCζ, surrounded by recycling endosome (Rab11) and Golgi (GM130), were readily seen in sparsely plated, dissociated mEpiSCs but not in mESCs (Fig. S1 K). Thus, formation of these apically enriched intracellular structures is a fundamental property of singly plated hPSC as well as mouse epiblast cells.
The apically enriched complex is a distinct intracellular compartment
To determine the relationship between the apicosome and other cellular organelles, we stained sparsely plated H9 hESCs with markers of the nuclear membrane (LAMIN A/C), ER (CALNEXIN), Golgi (GM130), mitochondria (COX IV), lysosome (lysosomal-associated membrane protein [LAMP] 1), autophagosome (sequestosome [SQSTM] 1), and microtubule (α-TUBULIN; Fig. 1 F and Fig. S2 A; see Fig. S2, B and C, for individual channels and additional cells). The apicosome (marked by WGA or p-ERM) was surrounded by many of these markers but did not costain substantially with any of them (Fig. 1 F and Fig. S2, B and C). Additionally, the apicosome was surrounded by but not colocalized with early endosomes (RAB5), recycling endosomes (RAB11 and ADP-ribosylation factor [ARF] 6), and late endosomes (RAB7; Fig. 1 G; also see Fig. S2, D and E). Together, these results demonstrate that single hPSCs readily form a previously unrecognized highly organized perinuclear complex, hereafter called the apicosome.
The apicosomal membrane surrounds a lumen with extracellular features
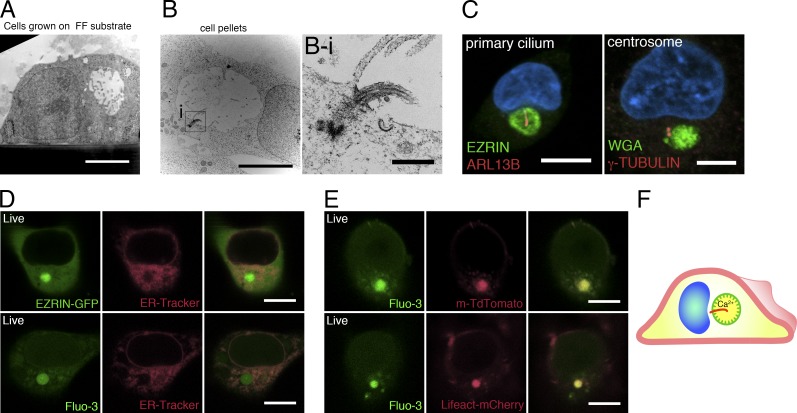
Ultrastructural analyses of isolated H9 cells revealed that the apicosome consists of a membrane-bound lumen with a diameter of 3–5 µm (Fig. 2 A). Strikingly, the demarcating membrane is studded with microvillus-like protrusions (Fig. 2 A) that are highly dynamic, as evident from live imaging of cells expressing EZRIN–GFP fusion protein (Video 2). In some cells, a primary cilium was observed (Fig. 2, B and B-i): staining single H9 cells with ADP-ribosylation factor-like protein [ARL] 13B (Sun et al., 2004), a primary cilium marker, showed that a single primary cilium existed within the apicosome in 86.6 ± 5.8% (n = 3) of the cells containing large (>3 µm) apicosomes (Fig. 2 C) but not in cells containing small apicosomes (Fig. S2 F). Consistent with this observation, centrosomes (γ-TUBULIN) were closely associated with the apicosome (Fig. 2 C). Thus, the membrane demarcating the apicosome has physical characteristics of the exterior surface of the cell (microvilli, cilia). Because the physiological level of Ca2+ is markedly higher in the extracellular environment (1–3 mM) than within cells (100 nm to 1 µM; Hao et al., 2016), we examined the level of Ca2+ in the apicosome using Fluo-3-AM, a cell-permeable fluorescent Ca2+ indicator. The Fluo-3 signal was visible as expected in the ER throughout the cells (Fig. 2, D and E). However, the Fluo-3 signal was particularly robust inside the apicosome (Fig. 2 E), suggesting active accumulation of Ca2+ in this compartment. Collectively, these results suggest that the apicosome provides a fully polarized, membrane-bound lumenal compartment with extracellular characteristics inside of single hPSCs (Fig. 2 F).
Figure 2.
The apicosome has features of the cellular exterior. (A and B) TEM images of single H9 cells grown under feeder-free conditions on a Geltrex-coated Thermanox substrate (A) or on a tissue culture plate (B) for 20 h. (B-i) Magnified image of the boxed region in B, showing a primary cilium within the apicosome. (C) Fluorescent confocal image of a single H9 cell stained with ciliary (ARL13B) and centrosomal (γ-TUBULIN) markers as well as HOECHST (blue). (D) H9 cell expressing EZRIN–EGFP (top), stained with ER-Tracker (red); isolated H9 cell stained with Fluo-3-AM (bottom, green) and ER-Tracker (red). (E) H9 cells expressing m-TdTomato (top) or Lifeact–mCherry (bottom), stained with Fluo-3-AM. (F) Schematic of an isolated hPSC with an internal apicosome, containing microvilli (green protrusions), a primary cilium (red), and accumulated Ca2+. Bars: (A and B) 5 µm; (B-i) 500 nm; (C and E) 10 µm.
Straight and branched actin assembly pathways are required for apicosome formation
We previously demonstrated that the formation of lumens in two-cell hESC clones is dependent on actin assembly pathways (Taniguchi et al., 2015). Indeed, interruption of either Arp2/3-dependent branched actin assembly or mDIA1/formin-dependent straight actin filament assembly reduces the number and size of two cell lumens. Because, similar to the two-cell lumen, the apicosome structure is highly actin enriched (Fig. 1), we investigated whether its formation also requires these actin assembly pathways. Indeed, treatment with either the Arp2/3 inhibitor CK666 (Nolen et al., 2009) or the mDIA/formin inhibitor SMIFH2 (Rizvi et al., 2009) reduced the number of apicosomes in singly plated hESCs (Fig. S2, G and H). Conversely, overexpression of full-length MDIA1 (MDIA1-FL) or a constitutively active truncated form of MDIA1 (MDIA1-δN3; Sakamoto et al., 2012) increased apicosome generation compared with controls (Fig. S2 I). Thus, actin assembly programs play a critical role in both apicosome formation and two-cell lumen generation.
Dynamics of apicosome formation and trafficking in single cells
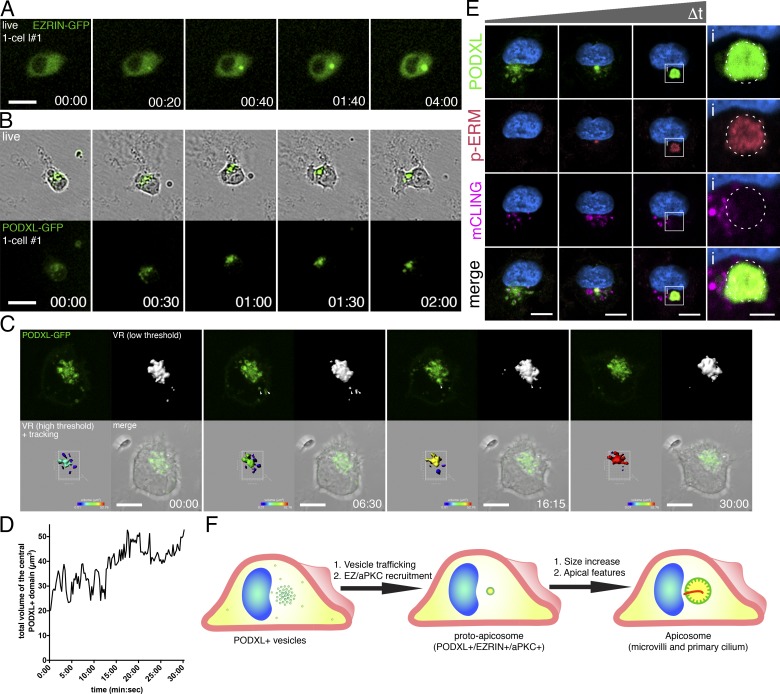
To examine how the apicosome is generated in single cells and to follow the fate of this structure during cell division, live cell imaging was performed using cells expressing EZRIN– or PODXL–GFP. Time-lapse imaging resulted in occasional cell death, likely due to phototoxicity. Thus, only cells that successfully completed mitosis during imaging were considered: 120 cells labeled with EZRIN–GFP as well as 53 cells labeled with PODXL–GFP were followed (in at least three separate imaging experiments for each marker). As early as 30 min after plating, a small concentrated perinuclear EZRIN–GFP signal (Fig. 3 A and Video 3) appeared de novo. Over the next 4–6 h, this signal increased in size and intensity, revealing dynamic growth of the apicosome, until the cell enters mitosis (Fig. 3 A and Fig. S3 A). For cells expressing PODXL–GFP, scattered small PODXL–GFP foci can be seen throughout cytoplasm immediately after plating. Over the course of the next 1–2 h, these foci organize into a single perinuclear structure (Fig. 3 B, Fig. S3 B, and Video 3; see Fig. 3, C and D, and Video 4 for short-term high-resolution imaging and quantitation).
Figure 3.
Formation of the apicosome in singly isolated cells. (A–C) Live imaging of H9 cells expressing EZRIN–GFP (A) or PODXL–GFP (B and C) during apicosome formation (see the detailed legend of Video 4 for C). (D) Quantitation of the total volume of the central perinuclear complex in C during imaging, revealing increase in volume over time (a representative of six independent live-imaged samples). Values were obtained from the high-threshold VR image (C, bottom left) acquired every 15 s. (E) H9 cells dissociated from monolayers incubated for 30 min with mCLING–ATTO647N to label apical membrane. Singly plated cells were examined at several time points (30 min to 20 h after plating), after immunostaining for PODXL and p-ERM. (F) Schematic of an isolated hPSC undergoing apicosome formation. Blue indicates DNA staining (HOECHST). Bars: (A and B) 25 µm; (C and E) 10 µm; (E-i) 3 µm. Time stamp: (A and B) h:min; (C) min:s.
To directly test whether these PODXL+ foci are derived from the preexisting apical domain, hESC monolayers were incubated with mTeSR containing ATTO 657N–labeled mCLING (membrane-binding fluorophore-cysteine-lysine-palmitoyl group) membrane dye for 30 min before dissociation (Revelo et al., 2014). Immediately after individual cell replating and attachment, mCLING signal was indeed colocalized with some (but not all) of the PODXL+ vesicles (Fig. 3 E); p-ERM signal was absent from these early vesicular collections. With time, apicosome structures became PODXL+ and p-ERM+; diffuse mCLING signal was detectable in these assembled apicosome structures (Fig. 3, E and E-i). Thus, at least part of the membrane pool within the apicosome is derived from the preexisting apical surface.
The diffuse mCLING signal within the apicosome led us to additionally examine membrane dynamics within the apicosome using FRAP. Apicosomes labeled with EZRIN– or PODXL–GFP showed rapid fluorescence recovery after laser photobleaching (Fig. S3, C–E; and Videos 5 and 6), indicative of active membrane trafficking at the apicosome. Collectively, these results show that the apicosome forms de novo on the interior of single cells, growing and maturing with time via dynamic membrane trafficking (Fig. 3 F).
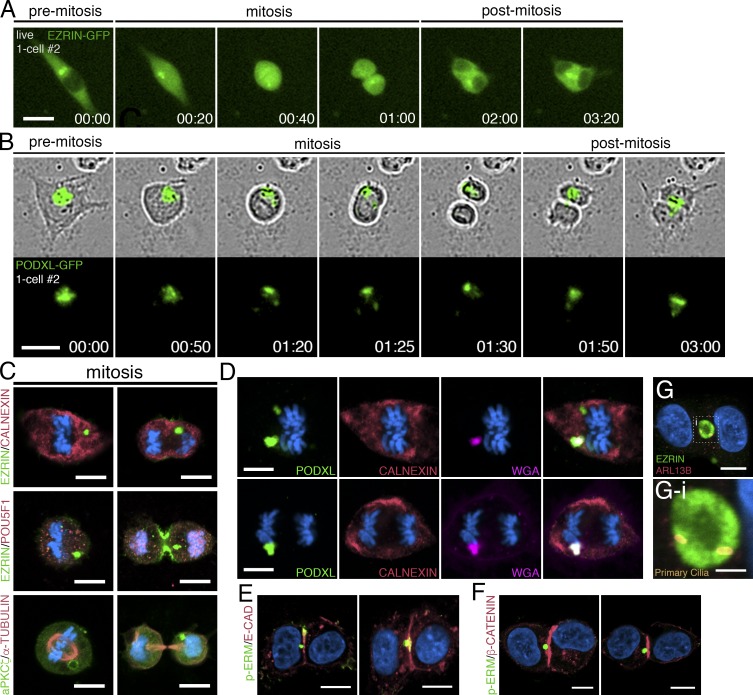
The high background in EZRIN–GFP–labeled cells made it difficult to track the apicosome during mitotic cell rounding using this label. Of 120 individual cells tracked through the entire process of cell division, the EZRIN–GFP-labeled apicosome could be followed throughout the entire mitotic process in only 25 (20.8%; Fig. 4 A and Video 7). In contrast, PODXL–GFP–labeled apicosomes could be tracked in 53 of 56 cells (94%; Fig. 4 B and Video 8). Strikingly, after cytokinesis, the single apicosome was asymmetrically inherited by one daughter cell in all 25 of the tracked EZRIN–GFP cells (Fig. 4 A, cytokinesis at 01:00) and in 41 of 53 (77%) of the PODXL–GFP cells (Fig. 4 B). In the remaining 10 of 53 PODXL–GFP cells, the label appeared to distribute to both daughters as cytokinesis ensued. Immunostained sections confirmed that apicosome-like EZRIN+, aPKCζ+, and PODXL+ domains were present throughout the mitotic cycle in cells expressing either label (Fig. 4, C and D; and Fig. S3 F). Despite the asymmetric inheritance of the apicosome, both daughters remained positive for POU5F1 (Fig. 4 C, see cytokinetic cells in EZRIN/POU5F1 staining).
Figure 4.
Dynamics of the apicosome in mitotic and postmitotic isolated cells. (A and B) Live imaging of mitotic and postmitotic H9 cells expressing EZRIN–GFP (A) or PODXL–GFP (B). Time stamp, h:min. Although most PODXL–GFP is segregated to one daughter cell (top), a fraction of the PODXL–GFP material is occasionally seen in the other daughter cell (bottom), revealing an unequal segregation of PODXL. (C–G) Confocal images of mitotic H9 cells (C and D) and two-cell H9 clones (E–G) stained with indicated markers. (G-i) Magnified image of the boxed region in G, revealing the presence of two primary cilia within a mature lumen shared by two cells. Bars: (A and B) 25 µm; (C–G) 10 µm; (G-i) 3 µm. For all images, blue indicates DNA staining (HOECHST).
Extended tracking showed that for all cells in which the EZRIN– or PODXL–GFP+ structure was still visible, this structure moved to a position near the cytokinetic plane between the two daughter cells within 1–2 h after the completion of cell division (Fig. 4, A [01:00–3:20] and B [01:30–03:00]; see additional cells in Fig. S3 G). This structure was surrounded by RAB11+ recycling endosomes in postmitotic cells, though RAB11 did not show prominent association with the apicosome during mitosis (Fig. S3 H). Further analysis revealed intermediates in which the apicosome was directly adjacent to, or in contact with, the cytokinetic plane in two-cell clones (Fig. 4, E and F; and see Fig. S3 I). Thus, after mitosis, asymmetrically segregated apicosomes relocate to the cytokinetic plane, where they give rise to fully demarcated lumens (Fig. 4 G).
The fate of all individual tracked cells is shown in Table S1. Lumens formed after mitosis in 84.78% or 90% of cells exhibiting apicosome structures labeled with PODXL– or EZRIN–GFP, respectively. Occasionally, cells completed cytokinesis without first forming an apicosome, and no lumen was initially visible (Fig. S3 J). Then, an apicosome appeared in one daughter and was subsequently relocated to the cytokinetic plane to form a lumen (Video 9). Thus, in all tracked cases, apicosome formation preceded lumenogenesis in individually plated hPSCs.
Apicosomes drive lumen formation in hPSC aggregates
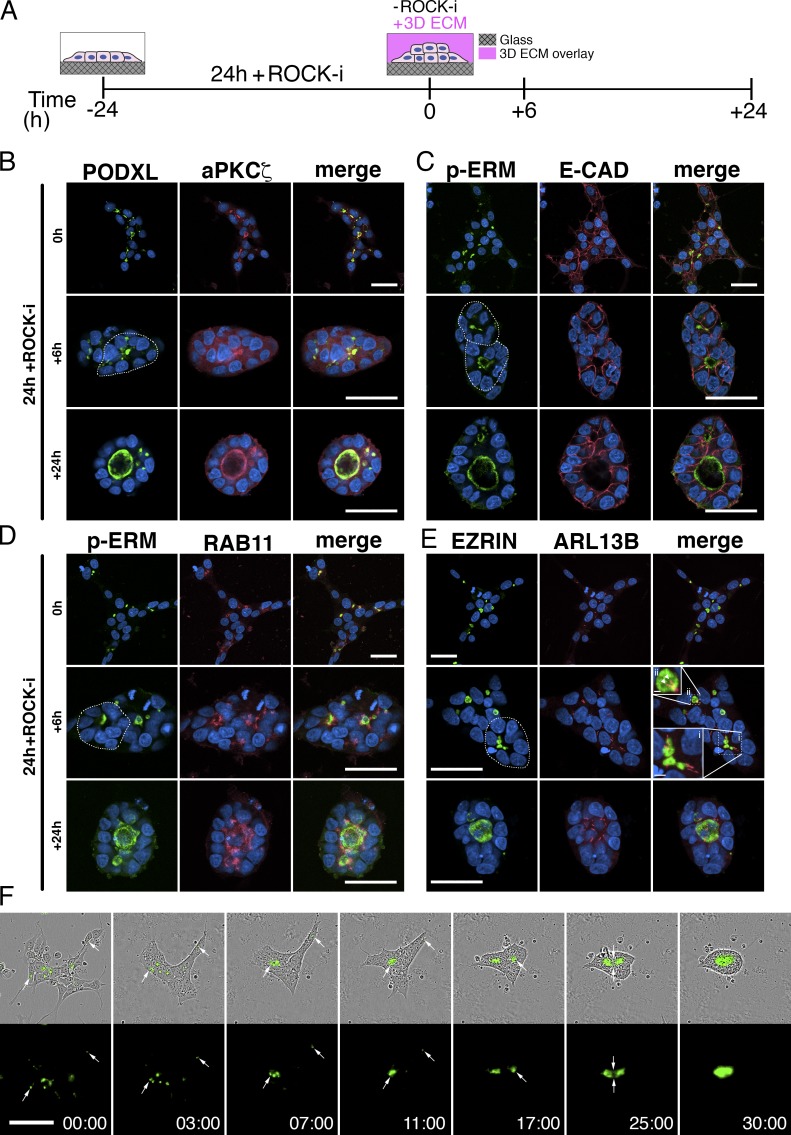
In the blastocyst, pluripotent embryonic cells exist initially as an unpolarized aggregate of cells. Their first clear morphogenic activity is to undergo apicobasal polarization that results in radial organization of epiblast cells and formation of a lumen in the center of the resulting rosette to generate an epiblast cavity (mouse) or proamniotic cavity (human). We therefore explored whether aggregates of hESCs would recapitulate this type of organization and whether apicosomes are involved (Fig. 5). Cells were plated at a density that yielded small cell aggregates in mTeSR1 medium containing Y-27632 (ROCK-i) for the first 24 h; the medium was then changed to mTeSR1 supplemented with 2% Geltrex without ROCK-i for another 24 h (Shao et al., 2017a; Fig. 5 A).
Figure 5.
Dynamics of the apicosome in cell aggregates. (A) Schematic of the hPSC-aggregate lumen formation assay (see the legend to Fig. S3 K for a detailed description). (B–E) hESC aggregates at 0 h, stained with apical markers (PODXL [B], aPKCζ [B], p-ERM [C and D], and EZRIN [E]) in addition to markers of adherence junction (E-CADHERIN [C]), recycling endosome (RAB11 [D]), and primary cilia (ARL13B [E]). Dotted circles in green channel outline radially patterned cells. In E, the boxed regions indicate clustered EZRIN+ apicosomes in the center of radially patterned cells (i) and an EZRIN+ lumen studded with two primary cilia (ii). (F) Time-lapse imaging of H9 PODXL–GFP aggregates. Imaging began immediately after ROCK-i withdrawal (0 h), and the first image is time stamped (00:00). Time stamp, h:min. Bars: (B–F) 50 µm; (E-i) 5 µm; (E-ii) 2 µm. For all images, blue indicates DNA staining (HOECHST).
After culture for 24 h with ROCK-i, 44.9 ± 7.6% (n = 3) of cells in each aggregate exhibited an intracellular concentration of apical markers, including PODXL, aPKCζ, p-ERM, and EZRIN that did not show obvious colocalization with the E-CADHERIN+ cell membrane (Fig. 5, B–E, top row, 0 h groups; higher magnification images in Fig. S3 L). This apically polarized complex was surrounded by RAB11+ early endosomes (Fig. 5 D and Fig. S3 L) and was studded with a single primary cilium in more than half (54.2 ± 2.5%, n = 3) of cases (Fig. 5 E and Fig. S3 L; quantitation in Fig. S3 M). Thus, apicosomes form inside individual hPSCs in the context of cell aggregates.
By 6 h after ROCK-i withdrawal, many cells within the aggregates were oriented radially, and apicosomes were gathered near the center of the radially oriented cells (Fig. 5, B–E, center rows, +6 h, see dotted circles in green channels). Costaining for E-CADHERIN and ARL13B confirmed their intracellular nature and revealed that >40% (46.9 ± 5.9%, n = 3) of these apicosomes continued to exhibit a single primary cilium (Fig. 5, C, E-i, and E-ii; and quantitation in Fig. S3 M). Occasionally, apically charged structures with two or more cilia could be detected (17.3 ± 5.7%, n = 3, Fig. S3 M, see red bars in double and triple+ categories). These structures appear to represent true lumens, as the presence of at least two cilia indicates that they are shared by two or more cells (Fig. 5, E and E-ii, arrowheads). At 24 h after ROCK-i withdrawal, most aggregates consisted of a single cyst with one or multiple dominant lumens that were decorated by multiple cilia (32.8 ± 0.7%, n = 3). Additionally, the total number of intracellular, single-cilium apicosome structures per independent aggregate was significantly reduced over time (Fig. S3 N). Thus, when plated as aggregates, hESCs self-organize to form cysts with a central lumen, and this is concomitant with apicosome fusion and cell rearrangement.
The foregoing analysis suggests a dynamic process of apicosome formation and fusion. To track this process in live cells, PODXL–GFP hESC aggregates were followed using time-lapse imaging. Consistent with the immunostaining results, multiple cells within each aggregate contained a PODXL+ apicosome (75.8 ± 6.1%, n = 3) at 24 h after plating (Fig. 5 F and Video 10). The higher apicosome frequency seen with PODXL (75.8%) versus EZRIN (44.8%) tracking likely reflects the fact that the PODXL label detects preapicosomes that are not yet decorated with EZRIN. Shortly after ROCK-i withdrawal, many cells within the aggregate retracted their protruding processes and compacted with neighboring cells, condensing the colony within 2–3 h (Fig. 5 F, 03:00). Over the next 3–4 h, cells began to orient radially, and PODXL+ apicosomes actively accumulated at the center of these rosette-like structures (Fig. 5 F, 07:00). By 9–11 h after ROCK-i withdrawal, several apicosomes fused to give rise to a common lumen (Fig. 5 F, 11:00). Often, after formation of a dominant lumen, apicosomes in cells on the outskirts of an aggregate began to merge to form a second lumen; multiple fusion events could occur sequentially to give rise to the final shared lumen (Fig. 5 F and Video 10). Quantitation showed that >40% (41.9 ± 6.09%, n = 3) of the independent aggregates formed multiple lumens that fused and gave rise to the final shared lumen by 48 h after ROCK-i withdrawal. Together, these results demonstrate that both in the context of single isolated hPSC and in the context of cell aggregates, apicosomes function as lumenal precursors.
Together, the data presented here establish evidence for a novel type of lumen formation in human and mouse PSC that involves the actin cytoskeleton–dependent formation of an apicosome, a highly organized intracellular structure with lumenal attributes. In both single cells and in cell aggregates, delivery of the apicosome to the cell surface results in a fully formed lumen that is shared by two or more cells, establishing the apicosome as a critical driver of lumenogenesis. Importantly, structures that resemble apicosomes have been previously observed in vivo in developing tissues, including the early mouse epiblast (Bedzhov and Zernicka-Goetz, 2014), as well as in other developing organs, such as the embryonic thyroid (Hick et al., 2013) and pancreas (Kesavan et al., 2009). Moreover, a variety of tumor cells exhibit intracellular lumen-like structures that are studded with microvilli and located in the vicinity of the nucleus (Battifora, 1975; Tsuchiya, 1981; Remy, 1986; Quincey et al., 1991; Gilloteaux et al., 2016), though the molecular characteristics of these structures and their potential relationship to apicosomes remains to be carefully examined. It will be important to explore whether the process of apicosome-dependent lumenogenesis is limited to less differentiated cell types such as those seen in development and cancer. Overall, we demonstrate that the self-organization of hPSC aggregates into lumenal cysts via apicosome trafficking is a biologically relevant process and that this in vitro system provides a valuable model for its further analysis.
Materials and methods
Cell lines used in this study
hPSC lines used in this work included H9 (WA09, P50, and P54; WiCell), H7 (WA07, P52; WiCell), and 1196a (a distinct iPSC line, P42; UM Pluripotent Stem Cell Core). All work with all of these hPSC lines was preapproved by the Human Pluripotent Stem Cell Research Oversight Committee at the University of Michigan. All cell lines have been maintained in a feeder-free system for at least 10 passages and were karyotypically normal at the indicated passage. Karyotype analysis was performed at Cell Line Genetics. These cells are frequently tested for pluripotency markers and differentiation ability. All cell lines tested negative for mycoplasma contamination (LookOut Mycoplasma PCR Detection kit; Sigma-Aldrich). The mESC line E14Tg2a.4 was obtained from the UM Transgenic Core (originally generated by A. Smith; Hooper et al., 1987), and mEpiSCs (22M) were generated in the laboratory of SK as previously described (Tesar et al., 2007) by culturing pre- and peri-implantation embryos on inactivated mouse embryonic fibroblast (MEF) feeder cells in K15F5 medium containing Knockout DMEM (10829-018; GIBCO) supplemented with 15% Knockout Serum Replacement (KSR; A1099201; GIBCO), 5% ESC-qualified FBS (104390924; GIBCO), 2mM l-glutamine (25030; GIBCO), 1× nonessential amino acids (11140-50; GIBCO), and 0.1 mM 2-mercaptoethanol (M7522; Sigma-Aldrich). After 5–6 d, blastocyst outgrowths were dissociated partially with 0.05% trypsin (25300-054; Invitrogen). The partial dissociates were plated individually into a 1.9-cm2 well containing MEF feeder layer and cultured for additional 4–6 d in K15F5 medium. Cells were then dissociated gently by brief exposure (2–3 min) to 0.05% trypsin/EDTA with gentle pipetting to prevent complete single-cell dissociation of pluripotent clusters and plated into a 9.6-cm2 well containing MEF feeders in K15F5 medium. Morphologically distinct mEpiSC colonies appeared over the next 4–8 d. mEpiSC colonies were manually dissociated into small clusters using a glass needle and plated into 1.9-cm2 wells containing MEF feeders in mEpiSC cell medium consisting of knockout DMEM supplemented with 20% KSR, 2 mM Glutamax (35050061; GIBCO), 1× nonessential amino acids, 0.1 mM 2-mercaptoethanol, and 10 ng/ml FGF2 (R&D Systems).
hPSC apicosome formation assays
The hESC and human iPSC lines were cultured and maintained as previously described (Ludwig et al., 2006), using lactate dehydrogenase–elevating virus-free hESC-qualified Geltrex (Life Technologies; derived from reduced growth factor ECM, extracted from murine Engelbreth-Holm-Swarm sarcoma cells similar to Matrigel) to prepare ECM-coated coverslips. Glass coverslips were coated with 1% Geltrex for 1 h at room temperature. To prepare singly dissociated hPSCs, the hPSC monolayer was dissociated using Accutase (Sigma-Aldrich) for 10 min and triturated using a P1000 pipetman to ensure complete dissociation into single cells. Cells were then resuspended in DMEM/F12, centrifuged, resuspended in mTeSR1 (Stem Cell Technologies) containing 10 µM Y-27632 (EMD Millipore) and 2% Geltrex, and plated at 5,000 cells/cm2 on 1% Geltrex–coated coverslips. To test alternative substrates, some experiments were performed using dissociated cells, plated sparsely in mTeSR1 on coverslips thinly coated with 1% Geltrex, Matrigel (diluted as suggested by the manufacturer; Corning), or 10 μg/ml vitronectin (Trevigen). In some cases (Fig. S1 G), cells were plated on irradiated MEFs at 20,000 cells/cm2. H9 cells used for analysis on MEFs had been historically maintained on MEFs. Laminin-521/E-cadherin matrix substrate was generated as previously described using 15 µg/ml laminin-521 (Stem Cell Technologies) and 1.7 µg/ml E-cadherin (R&D Systems; Rodin et al., 2014a). H9 cells used for analysis on laminin-521/E-cadherin matrix were transferred from Geltrex feeder-free system and were maintained on laminin-521 substrate for at least three passages before performing the apicosome formation assay.
Endocytosis assay
5-d hESC monolayer was treated with 0.4 µM mCLING-ATTO647N (710006AT1; Synaptic Systems) for 30 min at 37°C. Cells were then washed with DMEM/F12 before starting the apicosome formation assay. Cells were fixed in 4% paraformaldehyde/0.2% glutaraldehyde solution (Revelo et al., 2014) before immunofluorescent staining.
hPSC aggregate lumen formation assays
Upon single-cell dissociation (as described for the apicosome formation assays), cells were resuspended in mTeSR1 containing 10 µM Y-27632 (ROCK-i) and plated at 30,000 cells/cm2 on 1% Geltrex–coated coverslips. After 24 h, cells were then incubated in mTeSR1 medium containing 2% Geltrex ECM overlay without ROCK-i.
Confocal microscopy
An A-1 confocal microscope (Nikon; equipped with Plan Fluor 40× Oil DIC H N2 [NA 1.3], Plan Apo VC 60× Oil DIC N2 [NA 1.4], Plan Apo VC 60×A WI DIC N2 [NA 1.2] objective lenses, and NIS Elements Confocal software) or a Fluoview 1000 confocal microscope (Olympus; equipped with 60× supercorrected PLAPON 60XOSC [NA 1.4] objective and FV10-ASW software) was used.
Live cell imaging and FRAP assay
For long-term live cell imaging, EZRIN– or PODXL–GFP–expressing hESCs (see cloning in the Constructs and cell lines section) prepared for the apicosome formation assay or the aggregate lumen formation assay were plated on six-well plates (Nunc) in mTeSR1 medium (Stem Cell Technologies), and time-lapse images were taken at 37°C using the IncuCyte Zoom live cell imaging system (Essen Bioscience). Alternatively, cells were plated on a glass bottom culture dish (MatTek) in mTeSR1 medium (Stem Cell Technologies) and were imaged in a LiveCell chamber (Pathology Devices) configured for an A-1 or Fluoview 1000 confocal microscope at 37°C. Live ER-Tracker (E34250; Invitrogen) staining was performed as described in the manufacturer’s manual. FRAP was performed using a Fluoview 1000 confocal microscope with a SIM scanner using PODXL–GFP cells growing in mTeSR1 medium (Stem Cell Technologies) at 37°C. Fig. S3 E is plotted using normalized fluorescent recovery values (Higashi et al., 2016) obtained from 10 quantitated cells. 3D reconstruction, volume rendering, and tracking analyses were performed using Imaris 7.6 (Bitplane), and movies were generated using Imaris 7.6 and Photoshop (Adobe).
Live intracellular Ca2+ imaging
Single cells were plated as for the apicosome formation assay. After 16–20 h, cells were rinsed once with DMEM/F12 before incubation in mTeSR containing 1.5 μM Fluo-3-AM (F1242; Invitrogen) for 30 min. Cells were then washed with DMEM/F12 to remove any residual membrane bound dyes and incubated for an additional 30 min in mTeSR alone to allow de-esterification. Stained cells were imaged in mTeSR1 medium (Stem Cell Technologies) at 37°C in a LiveCell chamber (Pathology Devices) configured for an A-1 confocal microscope.
Small-molecule inhibitor assays
Cells were cultured with or without SMIFH2 and CK-666 (EMD Millipore; concentration given in legends) in standard culture conditions, containing ROCK-i. After 20 h, cells were fixed for downstream analysis.
Immunostaining
Antibodies for immunofluorescence staining were anti-α-tubulin (mouse; T6199; Sigma-Aldrich), Hoechst 33258 (Life Technologies), phalloidin (Life Technologies), anti-GM130 (mouse; 610822; BD), anti-Rab11 (mouse; 610656; BD), anti-Rab7 (rabbit; ab137029; Abcam), anti-ezrin (mouse; E8897; Sigma-Aldrich), anti-p-ERM (rabbit; 3141; Cell Signaling Technology), anti-PKCζ (rabbit; sc-216; Santa-Cruz), anti-PODXL (mouse; MAB1658; R&D Systems), anti-ARF6 (mouse; sc-7971; Santa-Cruz), anti-Arl13b (rabbit; 17711-1-AP; Protein Tech), anti-Lamin A/C (mouse; 4777; Cell Signaling Technology), anti-Calnexin (rabbit, ab22595; Abcam), anti–COX IV (rabbit, 4850; Cell Signaling Technology), anti-LAMP1 (rabbit; ab24170; Abcam), anti-SQSTM1 (rabbit; ab109012; Abcam), WGA (Life Technologies), anti-ZO-1 (mouse; 339100; Life Technologies), anti–γ-tubulin (mouse; T5326; Sigma-Aldrich), anti–E-cadherin (mouse; 610182; BD), anti-Nanog (rabbit; 4903; Cell Signaling), anti–Oct-4 (rabbit; 2750; Cell Signaling), anti-Rab5 (rabbit; 3547; Cell Signaling Technologies), anti-Rab7 (rabbit; ab137029; Abcam), and goat-raised secondary antibodies labeled with various fluorophores (Alexa Fluor 488, 555, and 647; Life Technologies). Imaging was done using an A-1 confocal microscope, and images were analyzed and generated using Imaris (Bitplane), Photoshop CS6 (Adobe), or ImageJ (National Institutes of Health). All stained samples were mounted in 90% glycerol (in 1× PBS) solution.
Constructs and cell lines
The piggyBac transposon system was used to prepare cell lines expressing EZRIN–EGFP and PODXL–EGFP, using a procedure previously described (Taniguchi et al., 2015). EGFP–MDIA1 piggyBac constructs have been described previously (Sakamoto et al., 2012; Taniguchi et al., 2015). pEGFP-C1-MDIA1 full-length or pEGFP-MDIA1-DN3 constructs were obtained from S. Narumiya (Kyoto University, Kyoto, Japan). The coding sequence of mouse EZRIN (IMAGE: 6826190) was PCR-amplified (forward: 5′-GCGAATTCGCCACCATGCCCAAGCCAATC-3′; reverse: 5′-CGGCGGCCGCTTACTTGTACAGCTCGTC-3′). Lifeact–mCherry (46357; Addgene) was PCR-amplified (forward: 5′-GCGAATTCGCCACCATGGGTGTCGCAG-3′; reverse: 5′-CGGCGGCCGCTTACTTGTACAGCTCGTC-3′). Membrane TdTomato (37351; Addgene; Beier et al., 2011) was PCR amplified (forward: 5′-GCGAATTCGCCACCATGCTGTGCTGTATGAG-3′; reverse: 5′-CGGCGGCCGCTTACTTGTACAGCTCGTCC-3′). pCMV6-XL6-human PODXL transcript variant 1 (NM_001018111.1; Origene) was PCR-amplified (forward: 5′-GCGAATTCGCCACCATGCGCTGCGCGCTGGC-3′; reverse: 5′-CGACCGGTGGATCCAAGAGGTGTGTGTCTTCC-3′). Amplified products were then subcloned into the pPBCAG-GFP piggyBac transposon vector (Chen and LoTurco, 2012; Taniguchi et al., 2015) digested with EcoRI and NotI (mouse EZRIN–GFP, Lifeact–mCherry, and membrane TdTomato) and with EcoRI and AgeI (human PODXL–GFP). PiggyBac transposon vectors were obtained from J. LoTurco (University of Connecticut, Storrs, CT). Transfection was done as previously described (Taniguchi et al., 2015). Fluorescence-activated cell sorting was performed to collect cells expressing specific fluorophores. Pooled transiently transfected cells were used in single cell assays using mDIA1 constructs (Fig. S2 I).
Transmission electron microscopy
Cells in Fig. 2 A were grown on Thermanox coverslips. Cells in Fig. 2 B were plated on a tissue culture plate and were scraped and pelleted before processing. These samples were fixed with 2.5% glutaraldehyde in Sorenson’s phosphate buffer for 1 h and postfixed in 1% osmium tetraoxide solution for 1 h. Samples were dehydrated in a series of EtOH solutions (30%, 50%, 70%, 90%, 95%, and 100%) for 5 min each and infiltrated with epoxy resin. These samples were then sectioned at 70 nm (Ultracut E; Reichert-Jung), placed on carbon slotted grids, stained using uranyl acetate, and imaged using the Philips CM100. Samples in Fig. 3 B were grown in a Geltrex feeder-free system, scraped off after the postfixation using osmium tetraoxide, centrifuged briefly, and embedded in HistoGel (Thermo Fisher Scientific) before dehydration. These samples were imaged using a JEM 1400-TEM (JEOL).
Quantitative analyses
Graphs were generated using Prism 6 (GraphPad Software), and Student’s t test was performed to assess significance. P > 0.05 was considered nonsignificant; P ≤ 0.05 was considered significant and marked with an asterisk. For Figs. S1 B and S2 (G–I) 50 cells were counted from three independent samples (total 150) per condition or cell line. For all quantitation of fixed hPSC aggregate samples (in Fig. 5 and Fig. S3, M and N), 20 aggregates were analyzed per independent sample (total 60 aggregates). For time-lapse imaging quantitation of PODXL–GFP aggregates, a total of 53 aggregates (20 in experiment 1, 18 in experiment 2, and 15 in experiment 3) were considered. For quantitation of apicosome formation in cells grown on laminin-521/E-cadherin substrate (Fig. S1 H), 50 cells were counted from three independent samples (total 150). The values to plot the time-course graph in Fig. 3 D was obtained on the basis of the total volume of the central perinuclear PODXL+ domain (calculated using Imaris 7.6; Bitplane) in Fig. 3 C (volume rendered [VR], high threshold + tracking). All experiments were repeated two or three times.
Blastocyst isolation and staining
3.5- to 4.75-dpc mouse blastocysts were recovered by flushing uteri with 1× PBS containing 6 mg/ml BSA and fixed with 1% paraformaldehyde with 0.05% Tergitol (Sigma-Aldrich) for 10 min at room temperature. Blastocysts were washed three times with 1× PBS containing 0.2% Tween 20 (Sigma-Aldrich) and then blocked using 1× PBS containing 5% goat serum, 0.2% Tween 20, and 0.2% Fish Skin Gelatin (Sigma-Aldrich) for 30 min at 37°C. Blastocyts were incubated with primary antibody for 60 min at 37°C, washed with 1× PBS-Tween three times, incubated in secondary antibodies for 30 min at 37°C, and then stained for DNA using Hoechst 33258. At least 10 blastocysts were analyzed.
Online supplemental material
Fig. S1 shows analyses of apicosome formation in different culture conditions as well as in distinct human and mouse pluripotent cell types. Fig. S2 shows the localization of the apicosome with respect to known organelles (individual fluorescent channels of Fig. 1, F and G) and quantitation of CK666 and SMIFH2 treatment as well as cells expressing full-length and constitutively active MDIA1 constructs. Fig. S3 shows additional analyses of the dynamics of apicosome formation in single cells and also shows further apicosome characterization in hPSC aggregates. Table S1 shows quantitation of the fate of each EZRIN– and PODXL–GFP cell during time-lapse imaging in Fig. 3 (A and B) and Fig. 4 (A and B). Video 1 shows serial confocal scans (11 total scans, 1 µm per scan) of late E4.5- to E4.75-dpc mouse blastocysts from Fig. S1 I, stained with indicated markers. Video 2 shows time-lapse imaging of a singly plated EZRIN–GFP cell containing an apicosome. Video 3 shows time-lapse imaging of the cell shown in Fig. 3 A (EZRIN–GFP) and Fig. 3 B (PODXL–GFP). Video 4 shows time-lapse imaging of the PODXL–GFP cell shown in Fig. 3 C (imaging started 30 min after plating), revealing the highly dynamic nature of PODXL vesicles during apicosome formation. Video 5 shows time-lapse imaging of the EZRIN–GFP cell shown in Fig. S3 C during FRAP. Video 6 shows time-lapse imaging of the PODXL–GFP cell shown in Fig. S3 D during FRAP. Video 7 shows time-lapse imaging of the EZRIN–GFP cell shown in Fig. 4 A, showing the dynamics of the apicosome during mitosis. Video 8 shows time-lapse imaging of the PODXL–GFP cells shown in Fig. 4 B, showing the dynamics of the apicosome during mitosis. Video 9 shows time-lapse imaging of the cell shown in Fig. S3 J top (cell 1) and bottom (cell 2), showing delayed apicosome formation. Video 10 shows time-lapse imaging of a cell aggregate (PODXL–GFP) shown in Fig. 5 F.
Supplementary Material
Acknowledgments
We thank the University of Michigan Pluripotent Stem Cell Core. We also thank Torey Arnold for his assistance with FRAP. We thank Drs. Yukiko Yamashita, Ann Miller, and William Shawlot for comments on the manuscript and Drs. Kristen Verhey, Leslie Satin, Benjamin Margolis, Sivaraj Sivaramakrishnan, Ajit Joglekar, Allen Liu, Philip Andrews, and Tristan Frum for insightful discussions. The piggyBac transposon system and human MDIA1-GFP constructs were generously provided by Drs. Joseph LoTurco and Shuh Narumiya, respectively. pQC membrane TdTomato was a gift from Connie Cepko (plasmid 37351; Addgene). pTK_93-Lifeact–mCherry was a gift from Iain Cheeseman (plasmid 46357; Addgene).
This work was supported by National Institutes of Health grants R01-DK089933 (D.L. Gumucio), T32-HD007505 (K. Taniguchi), R21-EB017078 (J. Fu), R21-HL114011 (J. Fu), and DP2-OD-008646 (S. Kalantry); the National Science Foundation (CBET1149401 to J. Fu); the March of Dimes Basil O’Connor Starter Scholar Research Award (5-FY12-119 to S. Kalantry); the University of Michigan Rackham Predoctoral Fellowship (Y. Shao); the University of Michigan Phi-Kappa-Phi Honor Society Student Grant (R.F. Townshend); and the Steven Schwartzberg Memorial Fund and the Prechter Fund (K.S. O’Shea).
The authors declare no competing financial interests.
Author contributions: K. Taniguchi, R.F. Townshend, and C.L. Cortez performed apicosome formation assays. Y. Shao and K. Taniguchi developed and performed the apicosome organization assays in hPSC aggregates. K. Taniguchi, C.E. Harris, and S. Kalantry performed experiments using mouse blastocysts. K. Taniguchi and S. Meshinchi designed and performed apicosome electron microscopy analyses. K. Taniguchi and D.L. Gumucio analyzed data, designed experiments, and wrote the manuscript; all other authors edited the manuscript.
Footnotes
Abbreviations used:
- aPKCζ
- atypical PKCζ
- ARF
- ADP-ribosylation factor
- ARL
- ADP-ribosylation factor-like protein
- hESC
- human embryonic stem cell
- hPSC
- human pluripotent stem cell
- iPSC
- induced pluripotent stem cell
- mCLING
- membrane-binding fluorophore-cysteine-lysine-palmitoyl group
- MEF
- mouse embryonic fibroblast
- mEpiSC
- mouse epiblast stem cell
- mESC
- mouse embryonic stem cell
- p-ERM
- phosphorylated EZRIN/RADIXIN/MOESIN
- ROCK-i
- rho-associated kinase inhibitor
References
- Battifora H. 1975. Intracytoplasmic lumina in breast carcinoma: a helpful histopathologic feature. Arch. Pathol. 99:614–617. [PubMed] [Google Scholar]
- Bedzhov I., and Zernicka-Goetz M.. 2014. Self-organizing properties of mouse pluripotent cells initiate morphogenesis upon implantation. Cell. 156:1032–1044. 10.1016/j.cell.2014.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier K.T., Samson M.E., Matsuda T., and Cepko C.L.. 2011. Conditional expression of the TVA receptor allows clonal analysis of descendents from Cre-expressing progenitor cells. Dev. Biol. 353:309–320. 10.1016/j.ydbio.2011.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant D.M., Datta A., Rodríguez-Fraticelli A.E., Peränen J., Martín-Belmonte F., and Mostov K.E.. 2010. A molecular network for de novo generation of the apical surface and lumen. Nat. Cell Biol. 12:1035–1045. 10.1038/ncb2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., and LoTurco J.. 2012. A method for stable transgenesis of radial glia lineage in rat neocortex by piggyBac mediated transposition. J. Neurosci. Methods. 207:172–180. 10.1016/j.jneumeth.2012.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.M., DeLong C.J., Bame M., Rajapakse I., Herron T.J., McInnis M.G., and O’Shea K.S.. 2014. Transcripts involved in calcium signaling and telencephalic neuronal fate are altered in induced pluripotent stem cells from bipolar disorder patients. Transl. Psychiatry. 4:e375 10.1038/tp.2014.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deglincerti A., Croft G.F., Pietila L.N., Zernicka-Goetz M., Siggia E.D., and Brivanlou A.H.. 2016. Self-organization of the in vitro attached human embryo. Nature. 533:251–254. 10.1038/nature17948 [DOI] [PubMed] [Google Scholar]
- De Los Angeles A., Ferrari F., Xi R., Fujiwara Y., Benvenisty N., Deng H., Hochedlinger K., Jaenisch R., Lee S., Leitch H.G., et al. 2015. Hallmarks of pluripotency. Nature. 525:469–478. 10.1038/nature15515 [DOI] [PubMed] [Google Scholar]
- Gilloteaux J., Bhalla A., Faour O., and Jamison J.M.. 2016. Formation of intracellular lumina in human prostate carcinoma (DU145) cells, maturation into signet cells, and the cribriform morphology of tumors. Ultrastruct. Pathol. 40:189–199. 10.3109/01913123.2016.1155684 [DOI] [PubMed] [Google Scholar]
- Hao B., Webb S.E., Miller A.L., and Yue J.. 2016. The role of Ca(2+) signaling on the self-renewal and neural differentiation of embryonic stem cells (ESCs). Cell Calcium. 59:67–74. 10.1016/j.ceca.2016.01.004 [DOI] [PubMed] [Google Scholar]
- Hick A.C., Delmarcelle A.S., Bouquet M., Klotz S., Copetti T., Forez C., Van Der Smissen P., Sonveaux P., Collet J.F., Feron O., et al. 2013. Reciprocal epithelial:endothelial paracrine interactions during thyroid development govern follicular organization and C-cells differentiation. Dev. Biol. 381:227–240. 10.1016/j.ydbio.2013.04.022 [DOI] [PubMed] [Google Scholar]
- Higashi T., Arnold T.R., Stephenson R.E., Dinshaw K.M., and Miller A.L.. 2016. Maintenance of the epithelial barrier and remodeling of cell-cell junctions during cytokinesis. Curr. Biol. 26:1829–1842. 10.1016/j.cub.2016.05.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper M., Hardy K., Handyside A., Hunter S., and Monk M.. 1987. HPRT-deficient (Lesch-Nyhan) mouse embryos derived from germline colonization by cultured cells. Nature. 326:292–295. 10.1038/326292a0 [DOI] [PubMed] [Google Scholar]
- Kesavan G., Sand F.W., Greiner T.U., Johansson J.K., Kobberup S., Wu X., Brakebusch C., and Semb H.. 2009. Cdc42-mediated tubulogenesis controls cell specification. Cell. 139:791–801. 10.1016/j.cell.2009.08.049 [DOI] [PubMed] [Google Scholar]
- Ludwig T.E., Bergendahl V., Levenstein M.E., Yu J., Probasco M.D., and Thomson J.A.. 2006. Feeder-independent culture of human embryonic stem cells. Nat. Methods. 3:637–646. 10.1038/nmeth902 [DOI] [PubMed] [Google Scholar]
- Nolen B.J., Tomasevic N., Russell A., Pierce D.W., Jia Z., McCormick C.D., Hartman J., Sakowicz R., and Pollard T.D.. 2009. Characterization of two classes of small molecule inhibitors of Arp2/3 complex. Nature. 460:1031–1034. 10.1038/nature08231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quincey C., Raitt N., Bell J., and Ellis I.O.. 1991. Intracytoplasmic lumina—a useful diagnostic feature of adenocarcinomas. Histopathology. 19:83–87. 10.1111/j.1365-2559.1991.tb00898.x [DOI] [PubMed] [Google Scholar]
- Remy L. 1986. The intracellular lumen: origin, role and implications of a cytoplasmic neostructure. Biol. Cell. 56:97–105. 10.1111/j.1768-322X.1986.tb00446.x [DOI] [PubMed] [Google Scholar]
- Revelo N.H., Kamin D., Truckenbrodt S., Wong A.B., Reuter-Jessen K., Reisinger E., Moser T., and Rizzoli S.O.. 2014. A new probe for super-resolution imaging of membranes elucidates trafficking pathways. J. Cell Biol. 205:591–606. 10.1083/jcb.201402066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizvi S.A., Neidt E.M., Cui J., Feiger Z., Skau C.T., Gardel M.L., Kozmin S.A., and Kovar D.R.. 2009. Identification and characterization of a small molecule inhibitor of formin-mediated actin assembly. Chem. Biol. 16:1158–1168. 10.1016/j.chembiol.2009.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodin S., Antonsson L., Hovatta O., and Tryggvason K.. 2014a Monolayer culturing and cloning of human pluripotent stem cells on laminin-521-based matrices under xeno-free and chemically defined conditions. Nat. Protoc. 9:2354–2368. 10.1038/nprot.2014.159 [DOI] [PubMed] [Google Scholar]
- Rodin S., Antonsson L., Niaudet C., Simonson O.E., Salmela E., Hansson E.M., Domogatskaya A., Xiao Z., Damdimopoulou P., Sheikhi M., et al. 2014b Clonal culturing of human embryonic stem cells on laminin-521/E-cadherin matrix in defined and xeno-free environment. Nat. Commun. 5:3195 10.1038/ncomms4195 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Boulan E., and Macara I.G.. 2014. Organization and execution of the epithelial polarity programme. Nat. Rev. Mol. Cell Biol. 15:225–242. 10.1038/nrm3775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Fraticelli A.E., and Martín-Belmonte F.. 2013. Mechanical control of epithelial lumen formation. Small GTPases. 4:136–140. 10.4161/sgtp.24303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto S., Ishizaki T., Okawa K., Watanabe S., Arakawa T., Watanabe N., and Narumiya S.. 2012. Liprin-α controls stress fiber formation by binding to mDia and regulating its membrane localization. J. Cell Sci. 125:108–120. 10.1242/jcs.087411 [DOI] [PubMed] [Google Scholar]
- Shahbazi M.N., Jedrusik A., Vuoristo S., Recher G., Hupalowska A., Bolton V., Fogarty N.N.M., Campbell A., Devito L., Ilic D., et al. 2016. Self-organization of the human embryo in the absence of maternal tissues. Nat. Cell Biol. 18:700–708. 10.1038/ncb3347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y., Taniguchi K., Gurdziel K., Townshend R.F., Xue X., Yong K.M.A., Sang J., Spence J.R., Gumucio D.L., and Fu J.. 2017a Self-organized amniogenesis by human pluripotent stem cells in a biomimetic implantation-like niche. Nat. Mater. 16:419–425. 10.1038/nmat4829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y., Taniguchi K., Townshend R.F., Miki T., Gumucio D.L., and Fu J.. 2017b A pluripotent stem cell-based model for post-implantation human amniotic sac development. Nat. Commun. 8:208 10.1038/s41467-017-00236-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simunovic M., and Brivanlou A.H.. 2017. Embryoids, organoids and gastruloids: new approaches to understanding embryogenesis. Development. 144:976–985. 10.1242/dev.143529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z., Amsterdam A., Pazour G.J., Cole D.G., Miller M.S., and Hopkins N.. 2004. A genetic screen in zebrafish identifies cilia genes as a principal cause of cystic kidney. Development. 131:4085–4093. 10.1242/dev.01240 [DOI] [PubMed] [Google Scholar]
- Taniguchi K., Shao Y., Townshend R.F., Tsai Y.H., DeLong C.J., Lopez S.A., Gayen S., Freddo A.M., Chue D.J., Thomas D.J., et al. 2015. Lumen formation is an intrinsic property of isolated human pluripotent stem cells. Stem Cell Rep. 5:954–962. 10.1016/j.stemcr.2015.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesar P.J., Chenoweth J.G., Brook F.A., Davies T.J., Evans E.P., Mack D.L., Gardner R.L., and McKay R.D.. 2007. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 448:196–199. 10.1038/nature05972 [DOI] [PubMed] [Google Scholar]
- Tsuchiya S. 1981. Intracytoplasmic lumina of human breast cancer--a microscopic study and practical application in cytological diagnosis. Acta Pathol. Jpn. 31:45–54. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.